- 1Department of Hepatobiliary Surgery, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China
- 2The Key Laboratory of Tumor Molecular Diagnosis and Individualized Medicine of Zhejiang Province, Zhejiang Provincial People’s Hospital, Affiliated People’s Hospital, Hangzhou Medical College, Hangzhou, China
- 3Department of Hepatobiliary and Pancreatic Surgery, Zhejiang Provincial People’s Hospital, Affiliated People’s Hospital, Hangzhou Medical College, Hangzhou, China
- 4Institute of Basic Medicine and Forensic Medicine, Hangzhou Medical College, Hangzhou, China
- 5Qingdao Medical College, Qingdao University, Qingdao, China
- 6The Second Clinical Medical College, Zhejiang Chinese Medical University, Hangzhou, China
- 7Department of Hepatobiliary and Pancreatic Surgery, The First Affiliated Hospital, Zhejiang University School of Medicine; Zhejiang Provincial Key Laboratory of Pancreatic Disease; Innovation Center for the Study of Pancreatic Diseases, Hangzhou, China
Pancreatic cancer (PC) is one of the most common malignant cancers, ranking the seventh highest causes of cancer-related deaths globally. Recently, RNA N6-methyladenosine (m6A) is emerging as one of the most abundant RNA modifications in eukaryote cells, involved in multiple RNA processes including RNA translocation, alternative splicing, maturation, stability, and degradation. As reported, m6A was dynamically and reversibly regulated by its “writers”, “erasers”, and “readers”, Increasing evidence has revealed the vital role of m6A modification in the development of multiple types of cancers including PC. Currently, aberrant m6A modification level has been found in both PC tissues and cell lines. Moreover, abnormal expressions of m6A regulators and m6A-modified genes have been reported to contribute to the malignant development of PC. Here in this review, we will focus on the function and molecular mechanism of m6A-modulated RNAs including coding RNAs as well as non-coding RNAs. Then the m6A regulators will be summarized to reveal their potential applications in the clinical diagnosis, prognosis, and therapeutics of PC.
Introduction
Pancreatic cancer (PC) is the seventh highest leading cause of cancer mortality worldwide accompanied by poor prognosis as well as a 5-year survival rate of about 10% (1, 2). With the development of clinical diagnosis and treatment for PC in the past two decades, the survival rate of PC patients has increased yearly, while the mortality rate of PC patients remains high. According to the statistics, the death cases (466,000) of PC are close to its new cases (496,000) globally (1). Lacking typical clinical symptoms and early diagnosis biomarkers, numerous PC patients are diagnosed at an advanced stage and miss the chance to get a surgical resection, resulting in the worse clinical outcome of PC patients. Thus, there is great urgency to clarify the initiation and progression of PC. Currently, aberrant genetic mutations (KRAS, p53, CDKN2A, SMAD4), dysregulation of the key signaling pathway (TGF-β, Wnt/β-catenin, Notch, Hippo, YAP), and epigenetic alterations (DNA methylation, RNA methylation, posttranslational modifications) have been reported to participate in PC development (3, 4). However, the molecular mechanism of PC progression remains largely unknown. Therefore, a comprehensive understanding of the pathogenesis and molecular regulatory mechanism of PC will greatly contribute to the early diagnosis, prognosis, and targeted therapeutics development of PC.
In recent years, RNA modifications, such as N6-methyladenosine (m6A), 5-methylcytosine (m5C), N1-methylguanosine(m1A), have been extensively reported in many cancers including PC (5). As one of the most abundant RNA modifications in eukaryotes, m6A has attracted more and more attention, which existed in RNAs including protein-coding RNAs as well as non-coding RNAs (ncRNAs). As reported, m6A modification is catalyzed by the methyltransferase (also called “m6A writers”) and meanwhile can also be removed by the demethylases (also called “m6A eraser”) (5, 6). Additionally, m6A-binding proteins (also called “m6A readers”) recognize and bind to the m6A-modified RNAs to regulate RNA fate (5). In PC, a significantly increased m6A level has been observed in both PC tissues and cell lines, and an elevated m6A level was associated with poor prognosis of PC patients (7–11). So far, dysregulation of m6A-associated modulators and m6A-modified RNAs has been associated with PC cell growth, iron metabolism, glycolysis metabolism, stemness-like property, and metastasis.
In this review, we will systemically summarize the molecular mechanisms and biological functions of m6A modifications in both mRNA and ncRNAs as well as the m6A regulators in the initiation and progression of PC and then discuss the potential applications of m6A modifications in the clinical diagnosis, prognosis, and targeted therapy of PC.
m6A Modification
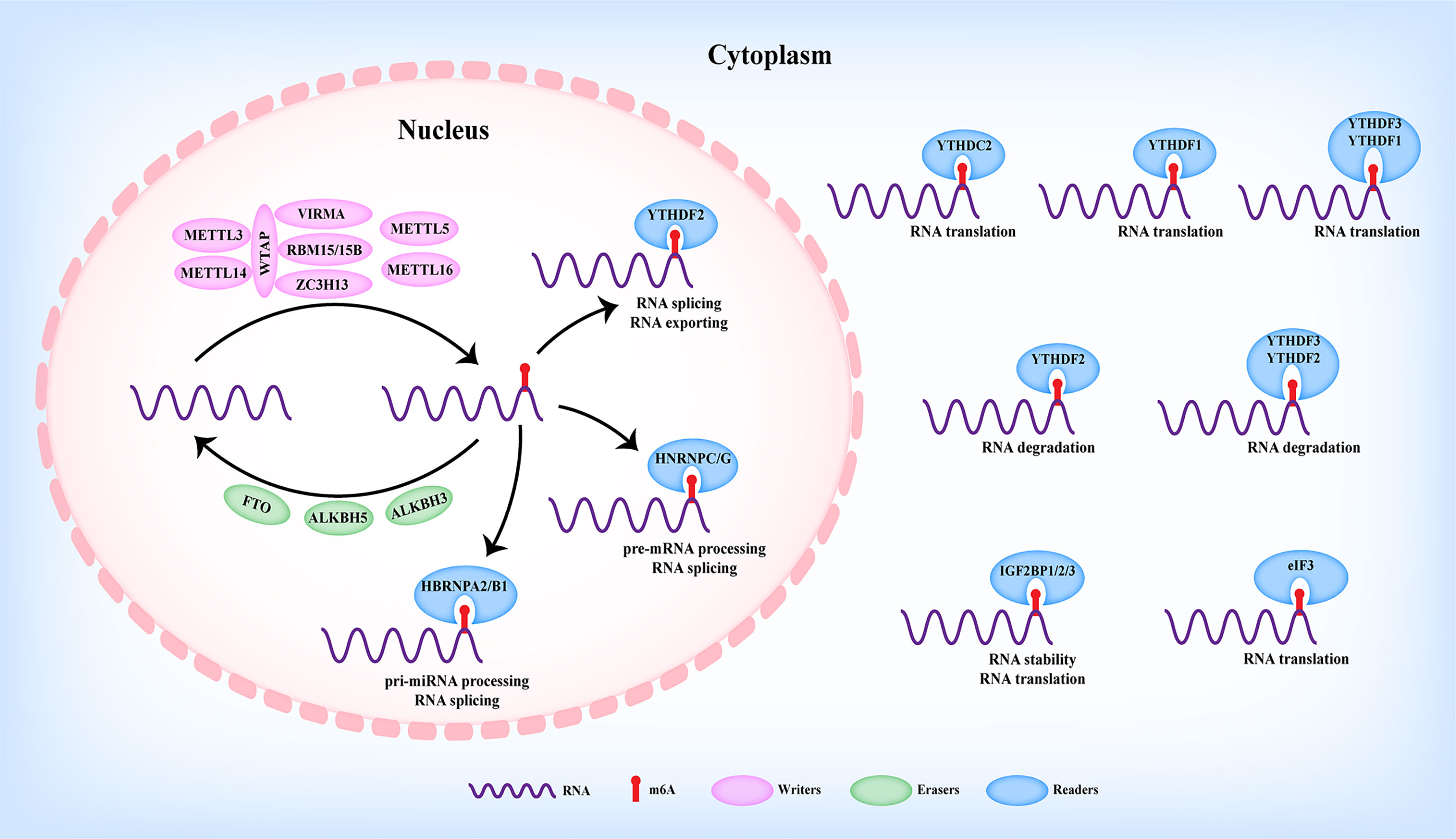
The RNA N6-methyladenosine (m6A) modification was defined to methylate the N6 position of adenosine, which was firstly reported in eukaryotic cells by Desrosiers et al. in 1974 (12). Up to now, m6A modification was considered to be the most abundant modification in RNAs (13, 14) and prefers to occur at the consensus motif RRACH (R=A or G, H=A, C, or U) of 3′-untranslated regions (3′UTRs), long internal exons (CDS), and near stop codons rather than randomly happens (15, 16). Nowadays, with the development of m6A detection-associated technologies, m6A modifications have been revealed in various types of RNAs including protein-coding RNA and ncRNAs, such as long non-coding RNAs (LncRNAs), microRNAs (miRNAs), circular RNAs (circRNAs), ribosomal RNA (rRNA), and transfer RNA (tRNA) (5, 17, 18). As shown in Figure 1, m6A modifications have been shown to be involved in RNA processes including nuclear export, miRNA maturation, alternative splicing, stability, translation, and degradation, thus participating in the initiation and progression of various diseases (19). As reported, m6A modification was dynamically and reversibly regulated by m6A methyltransferase (“writers”), m6A demethylases (“erasers”), and m6A-binding proteins (“readers”) (Figure 1) (20).
Writers
m6A modification was installed by the methyltransferase complex (MTC), in which Wilms’ tumor-1-associated protein (WTAP), methyltransferase-like 3 (METTL3), and methyltransferase-like 14 (METTL14) formed the core component (Figure 1). METTL3 was the firstly identified m6A writer with catalytic activity to trigger m6A modification via the S-adenosylmethionine (SAM)-binding motif. Serving as a supporting structure without catalytic activity, METTL14 formed a METTL3/METTL14 complex with METTL3 to recognize the conserved RRACH motif (21, 22). WTAP was further revealed to interact with the METTL3/METTL14 complex to mediate their nuclear speckle localization, thus modulating target RNA m6A modification (23). Other m6A readers including Vir-like m6A RNA methyltransferase-associated protein (VIRMA/KIAA1429), RNA-binding motif protein 15/15B (RBM15/15B), and zinc finger CCCH domain-containing protein 13 (ZC3H13) have also been identified to participate in the m6A modification of MTC (Figure 1). For example, VIRMA recruited the catalytic METTL3/METTL14/WTAP complex to mediate m6A modification in the 3′UTR and near the stop codon (24). RBM15/15B could interact with and recruit the MTC to a specific site to enhance the m6A modification of the LncRNA X-inactive specific transcript (XIST), thereby facilitating XIST-mediated gene silencing (25). Moreover, ZC3H13 enhanced the nuclear translocation of the ZC3H13–WTAP–Virilizer–Hakai complex to facilitate m6A modification (26). Apart from the above writers, methyltransferase-like protein 5 (METTL5) and methyltransferase-like protein 16 (METTL16) were identified as m6A writers (Figure 1) (27–30).
Erasers
Contrary to the function of writers, m6A erasers exerted the demethylation of m6A modification of RNAs. Currently, erasers mainly contain fat mass and obesity-associated protein (FTO), alkB homolog 5 (ALKBH5) (Figure 1) (31, 32), both of which are primarily located in the nucleus and belong to the alkB family (32). As reported, FTO was the first identified eraser, participating in the m6A modification of nuclear RNAs (31). To date, FTO and ALKBH5 have been widely reported in modulating RNA m6A modification in various human cancers (19). Recently, alkB homolog 3 (ALKBH3) was revealed as a novel m6A eraser in mediating tRNA demethylation and protein translation (Figure 1) (33).
Readers
Unlike m6A writers or m6A erasers to add or remove the m6A modification of RNAs, readers could recognize and interact with m6A-motified RNAs, thereby modulating various RNA processes, such as alternative splicing, nuclear export, miRNA maturation, stability, degradation, and translation (Figure 1) (34). Currently, insulin-like growth factor 2-binding proteins (IGF2BPs), YTH family proteins (YTHDC1/2 and YTHDF1/2/3), heterogeneous nuclear ribonucleoprotein family (HNRNPC, HNRNPG, HNRNPA2B1), and eIF3 have been identified as m6A readers (Figure 1). Based on their cellular localization, m6A readers exerted different functions. On the one hand, m6A readers with nuclear localization including YTHDC1 (35–38), HNRNPA2B1 (39, 40), and HNRNPC/G (41, 42) were reported to be involved in pri-miRNA processing, splicing, and nuclear exporting of m6A-modified RNAs (Figure 1). On the other hand, m6A readers with cytoplasmic localization, such as YTHDF1 (43), YTHDF2 (44, 45), YTHDF3 (46, 47), YTHDC2 (48, 49), IGF2BPs (50), and eIF3 (51), were demonstrated to participate in the stability, translation, and degradation of m6A-modified RNAs (Figure 1). In addition, CCHC-type zinc finger nucleic-acid binding protein (CNBP) (52) and NF-κB associated protein (NKAP) (53) were recently uncovered as novel m6A readers involved in modulating stability and pri-miRNA processing.
Role of RNA m6A Modification in PC
m6A Modification of Coding RNAs in PC
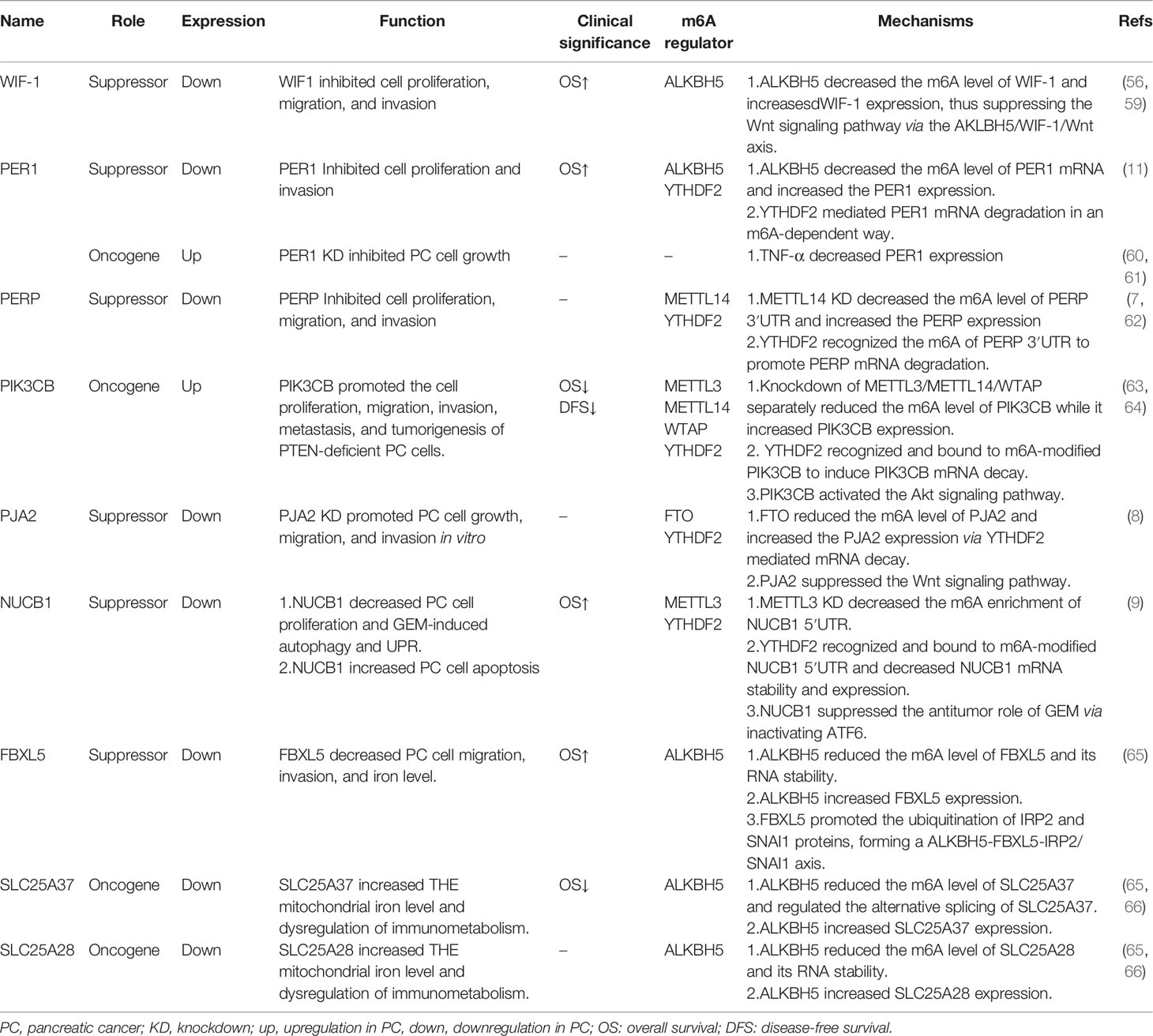
WIF-1
Wnt inhibitory factor 1 (WIF-1) was firstly identified by Hsieh et al. as a secreted Wnt-binding protein to suppress Wnt signaling activity (54). Currently, the tumor-suppressive role of WIF-1 has been clarified in various cancers including PC (55–58).
As shown in Table 1, the WIF-1 protein level was downregulated in PC tissues, which was correlated with poor overall survival (OS) of PC patients (59). WIF-1 was further identified as a downstream target of ALKBH5 via m6A-seq and RNA-seq. The demethylase-ALKBH5 increases WIF-1 expression through reducing the m6A level of WIF-1 mRNA, which inhibits the Wnt signaling pathway via the WIF-1/Wnt axis. Functionally, knockdown of WIF-1 alleviated the ALKBH5-induced suppression of cell growth, migration, and invasion, while overexpression of WIF-1 attenuated the ALKBH5 deficiency-induced promotion of cell growth, migration, and invasion (56) (Table 1; Figure 2). All the above findings suggests the antitumor role of WIF-1 in the malignant progression of PC.
PER1
PERIOD1 (PER1) is a clock gene involved in circadian rhythm regulation, DNA damage, cell cycle, cell proliferation, and metastasis. The abnormal expression of PER1 has been shown in various types of cancers, and both oncogenic and tumor-suppressive roles of PER1 have been revealed (11, 60, 61, 67–76).
As reported, the expression of PER1 was significantly decreased in PC tissues as compared to the benign and adjacent normal tissues (11, 77) (Table 1). Moreover, the reduced expression of PER1 was associated with shorter OS of PC patients (11, 77). Analysis of RNA-seq and m6A-seq further revealed that PER1 was a downstream target of ALKBH5 (11) (Table 1). The expression of PER1 was increased in the presence of ALKBH5, whereas the deficiency of ALKBH5 led to a reduced PER1 expression, which was confirmed in PC through immunohistochemistry (IHC) and TCGA dataset analysis (11). The MeRIP-qPCR analysis revealed that the m6A level of PER1 was negatively regulated by the demethylase ALKBH5 (11) (Table 1; Figure 2). Additionally, YTHDF2 served as an m6A reader to trigger PER1 mRNA degradation. Furthermore, a positive feedback loop between ALKBH5 and PER1 was revealed, since PER1 attenuated the enhanced cell proliferation and invasion induced by ALKBH5 deficiency (11), while PER1 could in turn enhance p53 activation to elevate the ALKBH5 expression through the PER1–P53–ALKBH5 axis (11) (Table 1; Figure 2). Taken together, there is a novel positive feedback loop between ALKBH5 and PER1, and ALKBH5 triggers PER1 expression via in an m6A-YTHDF2-dependent manner to suppress PC progression. On the contrary, studies have also shown that PER1 was upregulated in PC tissues as compared to normal tissues (60). Furthermore, TNF-α treatment suppressed PER1 expression, and loss of PER1 suppressed cell proliferation and increased apoptosis of PC cells acting as an oncogene (60, 61). Therefore, PER1 might be a potential biomarker for PC prognosis and also a promising therapeutics target for PC treatment.
PERP
The P53 effector related to pmp-22 (PERP) was firstly reported as a transcriptional target of p53 involved in cell apoptosis (78). At present, a tumor-suppressive role of PERP has been confirmed in various cancers (62, 79–82).
Zhao et al. revealed that PERP was highly expressed in PC (83) (Table 1). PERP was further identified as a target of METTL14 through RNA sequencing and m6A sequencing. Acting as an m6A writer, METTL14 knockdown significantly decreased the m6A level of PERP-3′UTR, which stabilized the mRNA of PERP and further increased the expression of PERP. In contrast, METTL14 could reduce the stability and expression of PERP mRNA. Mechanistically, METTL14 deficiency promoted PERP expression in an m6A-YTHDF2-dependented manner, in which YTHDF2 mediated PERP mRNA degradation (Table 1; Figure 2). Moreover, PERP inhibition could rescue the decreased cell proliferation, migration, and invasion induced by METTL14 knockdown (7) (Table 1). The above findings suggest a tumor-suppressive role of PERP in the malignant progression of PC.
PIK3CB
p110β/PIK3CB is a p110 catalytic subunit of phosphoinositide 3-kinases (PI3Ks), which together with p110α/PIK3CA, p110δ/PIK3CB, and the p85 regulatory subunit formed class IA PI3Ks (84). Aberrant expression of PIK3CB has been found in multiple cancers and is involved in tumor cell growth, metabolism, angiogenesis, metastasis, and multidrug resistance (63, 64, 85–93).
As reported, PIK3CB expression was remarkably elevated in PC tissues and a high expression of PIK3CB predicted poor prognosis of PC patients (63, 64) (Table 1). PIK3CB promoted cell proliferation, migration, invasion, and tumorigenesis in PTEN-deficient PC cells both in vitro and in vivo via activating the Akt signaling pathway (64) (Table 1; Figure 2). Furthermore, m6A writers including METTL3, METTL14, and WTAP could positively regulate the m6A level of PIK3CB and then reduce its expression in both mRNA and protein levels (Table 1, Figure 2). On the contrary, FTO, as an m6A eraser, significantly reduced the m6A level of PIK3CB and subsequently enhanced the PIK3CB expression (Table 1; Figure 2). Moreover, YTHDF2 could interact with m6A-modified PIK3CB to decrease PIK3CB mRNA stability as well as its protein expression (64) (Table 1; Figure 2). In summary, m6A regulators (METTL3, METTL14, WTAP, FTO) inhibited PIK3CB expression via an m6A-YTHDF2-dependent way. The oncogenic effect of PIK3CB in PTEN-deficient PC, indicating that PIK3CB is an emerging therapeutic target for PC.
PJA2
Praja ring finger ubiquitin ligase 2 (PJA2) is a RING-H2-type E3 ubiquitin ligase, which was firstly identified as an axotomy-suppressed gene in nerve cells (94) and played key roles in regulating the cAMP-dependent activation of PKA (95). Emerging evidence has shown that PJA2 was aberrantly expressed across cancers and acts as an oncogene or a tumor suppressor in thyroid cancer (96), glioblastoma (97), and gastric cancer (98, 99).
As shown, PJA2 was significantly upregulated in PC cells (8). Furthermore, PJA2 underwent m6A regulation of FTO, since FTO increased while loss of FTO decreased the expression and m6A level of PJA2 (8) (Table 1). Subsequently, Zeng et al. demonstrated that YTHDF2 but not YTHDF1 acted as an m6A reader to mediate PJA2 mRNA degradation and its downregulation (Table 1; Figure 2). Moreover, PJA2 deficiency could rescue the FTO-induced suppression of cell growth and metastasis via inhibiting the Wnt signaling pathway (8) (Table 1). Taken together, PJA2 acted as a tumor suppressor to regulate PC malignant behaviors via the FTO–PJA2–Wnt axis in an m6A-YTHDF2-dependent way, indicating that PJA2 is a new promising molecular target for PC therapeutic treatment.
NUCB1
Nucleobindins including NUCB1 and NUCB2 are DNA- and calcium-binding proteins (100, 101), involved in the development of various cancers (9, 102–107). Currently, an oncogenic role of NUCB1 has been found in colon cancer (105) and breast cancer (106).
As regards PC, NUCB1 was downregulated in both mRNA and protein levels of PC tissues, and an obviously poor prognosis was observed in PC patients with a lower expression of NUCB1 (9) (Table 1). In contrast to the effect of NUCB1 in colon cancer and breast cancer, NUCB1 suppressed the cell growth and promoted the apoptosis of PC cells both in vitro and in vivo, while loss of NUCB1 in turn promoted PC cell growth and inhibited cell apoptosis (9) (Table 1). Hua et al. further clarified that METTL3 induced the m6A modification of NUCB1 5′UTR, in which YTHDF2 mediated m6A-modified NUCB1 mRNA degradation (9) (Table 1; Figure 2). Functionally, NUCB1 inhibited the cell proliferation and promoted the antitumor effect of gemcitabine (GEM) on PC cells in vitro and in vivo (9). Moreover, NUCB1 also decreased autophagy and unfolded protein response (UPR)-induced by GEM via suppressing ATF6 activity (9) (Table 1; Figure 2). All the above findings demonstrate the m6A modulation of NUCB1 in an m6A–METTL3–YTHDF2-dependent way and a tumor-suppressive role of NUCB1 in PC progression.
FBXL5
F-box and leucine-rich repeat protein 5 (FBXL5) was firstly reported as a subunit of E3 ubiquitin ligase to promote the ubiquitination of p150Glued (108). IRP2 and Snail1 have been identified as substrates of FBXL5 (109, 110). FBXL5 has been reported to contain a hemerythrin-like domain that binds to iron and oxygen, thereby being involved in iron homeostasis (109, 111). Likewise, iron and oxygen conditions could in turn regulate FBXL5 (109). Currently, both the oncogenic and tumor-suppressive roles of FBXL5 have been found across cancers including colon cancer and HCC (112–114).
Downregulation of FBXL5 has been detected in PC tissues, which was associated with poor prognosis of PC patients (65) (Table 1). FBXL5 was subsequently identified as a target of ALKBH5, which reduced the FBXL5 m6A level and enhanced mRNA stability to increase the FBXL5 expression (Table 1). Moreover, FBXL5 depletion successfully rescued the ALKBH5-mediated inhibition of intracellular iron accumulation, cell migration, and invasion through downregulating IRP2 and Snail (65). In a summary, FBXL5 served as a tumor suppressor in PC carcinogenesis through the ALKBH5–FBXL5–IRP2/SNAI1 axis in an m6A-dependent manner (Figure 2), indicating a potential role of FBXL5 in the diagnosis, prognosis, and target therapy of PC.
SLC25A28 and SLC25A37
Mitoferrin (MFRN) belongs to the mitochondrial solute carrier family (SLC25), located in the inner membrane (115). Mitoferrin consists of two isoforms: mitoferrin-1 (SLC25A37) and mitoferrin-2 (SLC25A28), which transport iron to the mitochondria (115). So far, both SLC25A37 and SLC25A28 have been involved in dysregulation of mitochondrial iron (116, 117). The tumor-suppressor roles of SLC25A37 and SLC25A28 have been confirmed since they were involved in tumor cell growth, ROS production, mitochondrial iron uptake, and ferroptosis (117–121).
SLC25A37 and SLC25A28 are lowly expressed in PC tissues (65). Li et al. found that a high expression of SLC25A37 but not SLC25A28 indicated shorter OS of PC patients (66) (Table 1). Huang et al. demonstrated that either SLC25A37 or SLC25A28 underwent m6A modification modulated by ALKBH5. Detailly, ALKBH5 demethylated SLC25A37 mRNA to modulate its alternative splicing (65) (Table 1). As for SLC25A28, ALKBH5 removed the m6A modification of SLC25A28 and thus promoted its mRNA stability and expression (65) (Table 1; Figure 2). More importantly, SLC25A37 or SLC25A28 was elevated by depletion of PINK1 or PARK2, enhanced mitochondrial iron level, and dysregulation of immunometabolism via the PINK1/PARK2–SLC25A37/SLC25A28–HIF1A–AIM2–HNGB1–CD274 axis and thereby triggered PC carcinogenesis (66). Therefore, SLC25A37 and SLC25A28 were regulated through both m6A-dependent and m6A-independent regulations in the modulation of PC progression.
m6A Modification of Non-Coding RNAs in PC
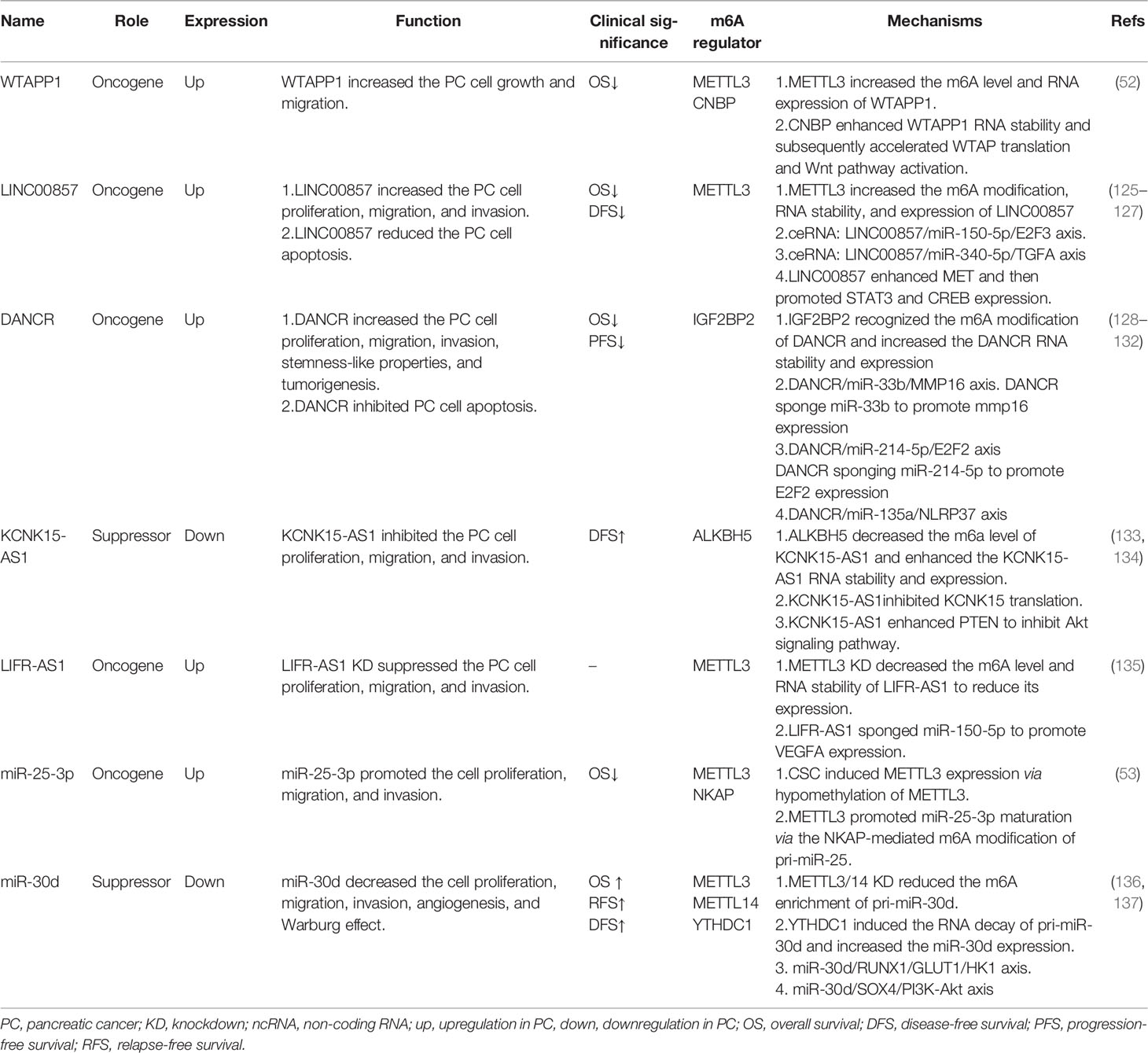
WTAPP1
LncRNA WTAPP1, short for Wilms tumor 1-associated protein pseudogene 1, has been shown to play key roles in tumor cell proliferation, migration, invasion, and angiogenesis (122–124).
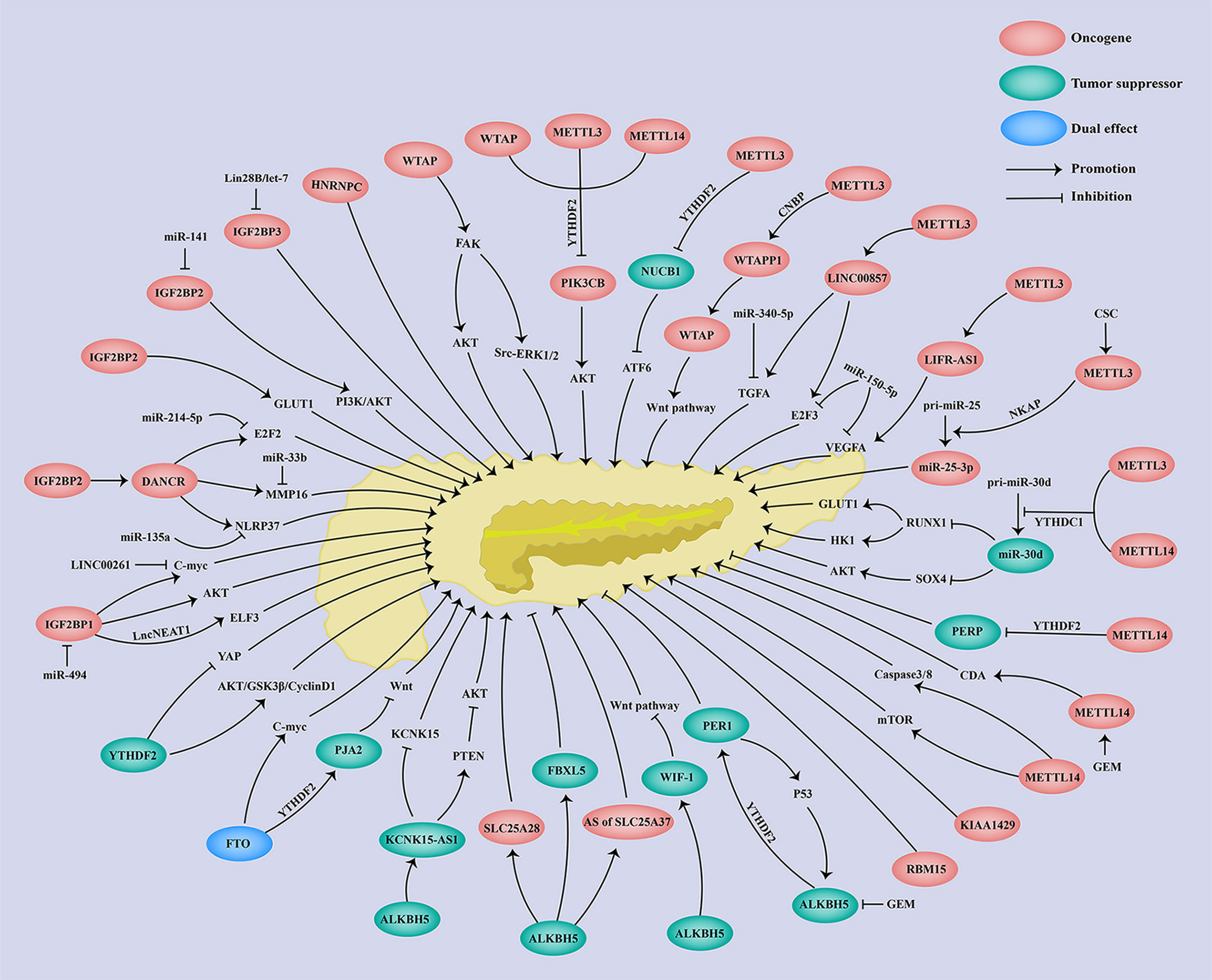
Here in PC, WTAPP1 was markedly overexpressed in tumor tissues, which was correlated with poor prognosis of PC patients (52) (Table 2). An oncogenic role of WTAPP1 was found in PC, showing that WTAPP1 enhanced PC cell proliferation and invasion both in vitro and in vivo (Table 2). Deng et al. further revealed that METTL3 mediated the m6A modification of WTAPP1, in which CCHC-type zinc finger nucleic-acid binding protein (CNBP) served as an m6A reader and stabilized the RNA of WTAPP1 (52) (Table 2). Moreover, the pseudogene WTAPP1 could enhance the translation of WTAP and activate the Wnt signaling, contributing to the malignant progression of PDAC (52) (Table 2, Figure 2). In summary, WTAPP1 may be a potential diagnostic and prognostic biomarker as well as a promising therapeutic target for PC.
LINC00857
Long non-coding RNA LINC00857 was firstly revealed to be highly expressed in lung cancer and indicated poor survival of lung cancer patients (138). Up to now, the upregulation of LINC00857 has been found in multiple types of cancers and is involved in tumor cell growth, migration, invasion, glycolysis, autophagy, and radiosensitivity (139–143).
As reported, LINC00857 was overexpressed in both PC cells and tissues, and its upregulation was associated with shorter OS and disease-free survival of PC patients (125–127) (Table 2). Studies have shown that LINC00857 could increase PC cell proliferation, migration, and invasion and decrease cell apoptosis (125–127) (Table 2). In regard to the molecular regulation of LINC00857, Meng et al. demonstrated that m6A writer METTL3 elevated both the m6A level and RNA stability of LINC00857 to promote LINC00857 expression, and then LINC00857 functioned as a ceRNA to regulate E2F3 expression through sponging miR-150-5p, contributing to the malignant development of PC cells (125) (Table 2; Figure 2). Moreover, Li et al. further demonstrated that LINC00857 could also sponge miR-340-5p to enhance the TGFA expression in PC cells and then accelerate PC cell migration and invasion (126) (Table 2, Figure 2). Additionally, LINC00857 could also upregulate MET, STAT3, and CREB expression to enhance PC cell proliferation (127). Above all, LINC00857 exerts an oncogenic role in PC and may provide a possible therapeutic target for PC treatment.
DANCR
Long non-coding RNA differentiation antagonizing non-protein-coding RNA (DANCR) was firstly identified as a progenitor differentiation suppressor in 2012 (144). Later, both the oncogenic and tumor-suppressive roles of DANCR have been identified across cancers (128, 145), participating in modulating tumor cell growth, stemness-like properties, EMT, and chemoresistance (145).
Consistent with previous reports in various cancers, DANCR was also highly expressed in both PC cells and tissues (129, 130) (Table 2). Moreover, a high expression of DANCR predicted poor clinical outcomes in PC (130) (Table 2). Luo et al. demonstrated that DANCR deficiency significantly decreased the cell proliferation, migration, and invasion of PC through the DANCR/miR-33b/MMP16 axis, in which DANCR served as a miR-33b sponge (129) (Table 2; Figure 2). Consistent with Yao et al.’s reports, Tang et al. also revealed that DANCR enhanced cell proliferation and invasion via the DANCR/miR-214-5p/E2F2 axis or DANCR/miR-135a/NLRP37 (130, 131) (Table 2; Figure 2). In addition to ceRNA regulation, DANCR also underwent m6A regulation. In detail, IGF2BP2 acted as an m6A reader and could recognize and bind to DANCR to enhance its RNA stability and expression, thereby facilitating PC cell proliferation and stemness-like properties (128) (Table 2; Figure 2). The above findings indicate the oncogenic role of DANCR and its potential clinical application in PC prognosis and treatment.
KCNK15-AS1
LncRNA KCNK15 and WISP2 antisense RNA 1 (KCNK15-AS1 or RP11-445H22.4) were firstly found to be upregulated in osteoarthritis (146). Subsequently, an abnormal expression of KCNK15-AS1 was also observed among cancers.
As reported, there was a remarkable downregulation of KCNK15-AS1 in both PC tissues and cell line, and patients with a low expression of KCNK-AS1 have shown shorter OS (133, 134) (Table 2). RNA m6A demethylase ALKBH5 could bind to KCNK15-AS1 and thus enhance its RNA stability and expression through eliminating m6A modification (133, 134) (Table 2; Figure 2). More importantly, KCNK15-AS1 suppressed KCNK15 translation through interacting with the 5′UTR of KCNK15, while KCNK15-AS1 also promoted PTEN expression and thus inhibited the Akt pathway, thereby suppressing PC cell proliferation, migration, and invasion (133, 134) (Table 2; Figure 2). In summary, KCNK15-AS1 regulated by ALKBH5-mediated m6A demethylation acted as a tumor suppressor to suppress the malignant progression of PC cells through the KCNK15–AS1/KCNK15 axis and KCNK15–AS1/PTEN/Akt axis, suggesting a promising therapeutic target for PC clinical treatment.
LncRNA LIFR-AS1
Leukemia inhibitory factor receptor antisense RNA 1 (LIFR-AS1) is a long non-coding RNA, which is transcribed from the LIFR gene. An abnormal expression of LIFR-AS1 has been found in various cancers and is involved in cancer development (147). The function of LIFR-AS1 is complex, since an antitumor role of LIFR-AS1 has been shown in glioma, breast cancer, and lung cancer, while an oncogenic role has been found in thyroid cancer, colorectal cancer, and osteosarcoma (147). Furthermore, both the oncogenic role and tumor suppressive role of LIFR-AS1 were revealed in gastric cancer (148, 149).
An obvious upregulation of LIFR-AS1 was observed in PC tissue and cell lines, which was correlated with poor clinical outcomes of PC patients (135) (Table 2). Knockdown of METTL3 reduced the m6A level of LIFR-AS1 and thus suppressed its RNA stability and expression (135) (Table 2, Figure 2). Moreover, a significant decrease in cell proliferation, migration, and invasion was observed following LIFR-AS1 inhibition (135) (Table 2). Additionally, a ceRNA regulation was also revealed showing that LIFR-AS1 could sponge miR-150-5p, thus activating downstream target VEGFA and promoting PC progression (135) (Table 2, Figure 2). These results revealed that LIFR-AS1 is an oncogenic gene in PC via the METTL3/LIFR-AS1/miR-150-5p/VEGFA axis in an m6A-dependent manner.
miR-25-3p
pri-miR-25 is the primary miRNA of miR-25-3p. miR-25 has been widely reported as an oncogenic miRNA. An abnormal expression of miR-25 has been found in multiple cancers (150, 151).
Here in PC, miR-25-3p was found to be highly expressed, which predicted the poor prognosis of PC patients (53) (Table 2). Zhang et al. further demonstrated that METTL3 induced by cigarette smoke condensate (CSC) could increase the m6A modification of pri-miR-25 via in a NKAP-dependent manner, in which NF-κB-associated protein (NKAP) functioned as an m6A reader of pri-miR-25 (53), thereby enhancing miR-25-3p maturation. Additionally, upregulated miR-25-3p could then activate the Akt signaling pathway through inhibiting the expression of its target-PHLPP2, thus promoting the cell migration and invasion of PC (53) (Table 2; Figure 2). In summary, the METTL3/miR-25-3p/PHLPP20/Akt axis exerts an oncogenic role in the carcinogenesis of PC patients who smoke.
miR-30d
miR-30d belongs to the miR-30 family, which is abnormally expressed across cancers. So far, both the oncogenic and antitumor roles of miR-30d have been revealed. miR-30d has been reported to remarkably suppress cell growth and metastasis of breast cancer (152), whereas upregulation of miR-30d enhanced tumor growth and angiogenesis of prostate cancer (153).
In PC, the expression of miR-30d was significantly decreased in both PC tissues and cell lines, which predicted a shorter OS, RFS, and DFS of PC patients (136, 137) (Table 2). More importantly, miR-30d overexpression inhibited PC cell growth, metastasis, and angiogenesis both in vitro and in vivo (136, 137) (Table 2). miR-30d was further shown to be involved in glycolysis regulation since miR-30d decreased the lactic acid level, glucose uptake, and ATP level while miR-30d inhibition increased the lactic acid level, glucose uptake, and ATP level of PC (Table 2). miR-30d has m6A modification and is regulated by METTL3, METTL14, and YTHDC1. In detail, YTHDC1 significantly promotes the RNA degradation of pri-miR-30d and increases the expression of miR-30d, and knockdown of METTL3/14 significantly reduces the m6A enrichment of pri-miR-30d (136) (Table 2; Figure 2). Moreover, RUNX1 and SOX4 were identified as a downstream target of miR-30d. Hou et al. demonstrated that RUNX1 deficiency could attenuate miR-30d inhibition-induced PC cell growth, metastasis, angiogenesis, and glycolysis via the miR-30d/RUNX1/GLUT1/HK1 axis (136) (Table 2; Figure 2). Xu et al. further revealed that SOX4 overexpression successfully rescued the antitumor effect of miR-30d via the miR-30d/SOX4/PI3K-Akt axis (137) (Table 2; Figure 2). Taking all the above into consideration, miR-30d is shown to be modulated by YTHDC1-mediated m6A modification and there is a tumor-suppressive role of the miR-30d/RUNX1/GLUT1/HK1 axis and miR-30d/SOX4/PI3K-Akt axis in PC progression, providing a possible application of miR-30d as a diagnosis and prognosis biomarker and also a therapeutic target for PC treatment.
Functions of m6A Regulators in Pancreatic Cancers
Writers
WTAP
WTAP has been found to be highly expressed in PC tissue, which was correlated with shorter survival of PC patients (7, 52, 65, 154) (Table 3). As reported, WTAP triggered the cell proliferation, migration, and invasion and GEM resistance of PC cells, while knockdown of WTAP suppressed cell proliferation migration, and invasion and GEM resistance (52, 155) (Table 3).
As an m6A methylase, WTAP increased the m6A level of its target PIK3CB and enhanced the PIK3CB expression via an m6A-YTHDF2-mediated RNA decay of PIK3CB (64) (Table 3; Figure 2). Apart from m6A regulation, WTAP could also stabilize FAK mRNA and increase its expression through an m6A-indenpendent way, thereby activating the FAK-PI3K-AKT and FAK-SRC-GRB2-ERK1/2 signaling pathways (155) (Table 3; Figure 2). Moreover, GSK2256098, a specific FAK inhibitor, could attenuate WTAP-induced cell migration and invasion and GEM resistance of PC cells (155). As a key modulator to affect its downstream target genes, WTAP could also be regulated by its upstream genes. Deng et al. have shown that LncRNA WTAPP1 could recruit EIF3B to enhance WTAP translation, and then WTAP activated the Wnt signaling pathway, forming a functional WTAPP1/WTAP/Wnt axis[75] (Table 3; Figure 2). Additionally, knockdown of WTAP attenuated WTAPP1-induced PC cell proliferation, migration, and invasion (52) (Table 3). All the above findings indicate the oncogenic role of WTAP in PC in an m6A-dependent and m6A-independent way as well as the potential application of WTAP in PC prognosis and targeted therapy.
METTL3
Similar to WTAP, METTL3 was also shown to be upregulated in PC, which was correlated with shorter OS of PC patients (7, 10) (Table 3). According to the studies, METTL3 deficiency decreased PC cell proliferation, migration, invasion, stemness, and radio- and chemoresistance (10, 156) (Table 3). Acting as an m6A methylase, METTL3 knockdown obviously reduced the total RNA m6A level of PC cells (10). Several m6A-regulated targets of METTL3 have been identified, such as PIK3CB, NUCB1, WTAPP1, LINC00857, LIFR-AS1, pir-miR-25, and pri-miR-30d (Table 3).
Firstly, METTL3 inhibition reduced the m6A level of PIK3CB and increased the PIK3CB expression via YTHDF2-mediated mRNA decay (64). Later, Hua et al. found that the m6A enrichment of the NUCB1 5′UTR was notably decreased upon knockdown of METTL3 via in an m6A-YTHDF2-dependent way (9) (Table 3; Figure 2). In addition to coding RNAs, METTL3 also mediated the m6A regulation of non-coding RNAs. For instance, loss of METTL3 significantly reduced the m6A level of WTAPP1 as well as its expression, in which m6A reader CNBP enhanced the mRNA stability of WTAPP1 (52) (Table 3; Figure 2). Furthermore, METTL3 deficiency also decreased the m6A level and RNA stability of LINC00857 and LIFR-AS1, resulting in their downregulation (135) (Table 3; Figure 2). Smoke was a high-risk factor of PC, and smokers were reported to have a two-fold higher risk of PC than non-smokers (164). Interestingly, there was a significant upregulation of METTL3 in smokers as compared with non-smokers (53). Zhang et al. have further shown that cigarette smoke condensate (CSC) promoted METTL3 expression through hypomethylating the promoter of METTL3 (53), which enhanced the m6A level and expression of pri-miR-25 and miR-25-3p maturation via in an m6A-NKAP-dependent manner, in which NF-κB-associated protein (NKAP) functioned as an m6A reader of pri-miR-25 (53) (Table 3; Figure 2). Additionally, METTL3 was also shown to affect the m6A enrichment of pri-miR-30d (136). In summary, METTL3 served as an oncogene in PC progression and provides a possible prognosis biomarker and therapeutic target for PC.
METTL14
An obvious upregulation of METTL14 has been found in both RNA and protein levels in PC tissues (7, 157) (Table 3). Patients with a higher expression of METTL14 have shown shorter OS (7) (Table 2). As mentioned before, METTL14 was involved in various cellular processes of PC cells, since METTL14 remarkably promoted PC cell proliferation, migration, and invasion; metastasis; cisplatin resistance; and GEM resistance while it inhibited the apoptosis and autophagy of PC cells (7, 157–159) (Table 3).
It was shown that loss of METTL14 increased cisplatin-induced cell apoptosis and autophagy by activating caspase3/8 and mTOR pathway, thereby enhancing the antitumor effect of cisplatin (157) (Table 3; Figure 2). Interestingly, GEM treatment specifically upregulated the expression of METTL14, without changes in other m6A regulators (158). However, GEM-induced METTL14 could in turn increase the GEM resistance via promoting CDA expression both in vitro and in vivo (158) (Table 3; Figure 2). Additionally, SRFR5 was shown to regulate the alternative splicing of METTL14, which formed a SRSF5–METTL14 axis to enhance PC cell growth and metastasis in vitro and in vivo (159) (Table 3). For m6A regulation, Tian et al. revealed that loss of METTL14 suppressed the m6A level and promoted the expression of PIK3CB via m6A-YTHDF2-mediated RNA decay of PIK3CB (64) (Table 3; Figure 2). Moreover, Wang et al. demonstrated that METTL14 deficiency also decreased the m6A level and thus increased the expression of PERP through m6A-YTHDF2-mediated degradation of PERP mRNA (7), resulting in cell growth and metastasis of PC (Table 3; Figure 2). In addition, METTL14 knockdown also deceased the m6A enrichment of pri-miR-30d (136). The above results suggest that both the m6A-dependent and m6A-independent regulation of METTL14 are involved in the carcinogenesis of PC, and METTL14 is a promising diagnosis and prognosis biomarker and chemotherapy resistance target for PC treatment.
KIAA1429
Vir-like m6A methyltransferase-associated (VIRMA, also named KIAA1429) was significantly upregulated in PC tissues as compared to normal tissues (160) (Table 3). Moreover, KIAA1429 was revealed as an independent risk factor for PC prognosis (165), and high expression of KIAA1429 was associated with shorter OS of PC patients (161, 162) (Table 3). Depletion of KIAA1429 remarkably reduced the cell proliferation of PC cells (162), indicating an oncogenic role of KIAA1429 in PC (Table 3; Figure 2).
RBM15
RNA-binding motif protein 15 (RBM15) has been identified as a methylase during m6A modification. According to TCGA and GTEx databases, RBM15 was highly upregulated in PC tissues and loss of RBM15 suppressed the cell proliferation of PC cells (163) (Table 3). Moreover, PC patients with a high expression of RBM15 have shown decreased OS, DFI, PFI, and DSS (163) (Table 3). Additionally, a highly correlated relationship between RBM15 expression and immune checkpoint markers was also revealed. The above findings suggest a favorable application of RBM15 in the prognosis and immunotherapy of PC.
Readers
IGF2BP1
It has been shown that an upregulation of IGF2BP1 was observed in PC tissues (166), which was associated with shorter OS of PC patients (166) (Table 4). Knockdown of IGF2BP1 dramatically reduced cell proliferation and induced G1 cell-cycle arrest and apoptosis (166, 167) (Table 4). IGF2BP1, an RNA-binding protein, attenuated the Linc00261-induced suppression of c-myc RNA stability through binding to Linc00261 (168) (Table 4; Figure 2). Moreover, IGF2BP1 cooperated with LncNEAT1 to increase the RNA stability of ELF3 and enhanced PC cell proliferation, migration, and invasion (167) (Table 4; Figure 2). In addition, miR-494 could target IGF2BP1 to suppress IGF2BP1 expression and then IGF2BP1 promoted PC progression via activating the Akt pathway (166) (Table 4; Figure 2). Therefore, IGF2BP1 might serve as a potential therapeutic target and prognostic biomarker for PC.
IGF2BP2
Consistent with IGF2BP1, IGF2BP2 was also significantly upregulated in PC tissues and cell lines (128, 169–172) (Table 4). A high expression of IGF2BP2 predicted a shorter OS of PC patients (128, 160–162, 169–173) (Table 4). Knockdown of IGF2BP2 significantly reduced PC cell growth and invasion (128, 162, 169, 171, 172) (Table 4). It has been reported that IGF2BP2 promotes the aerobic glycolysis of PC cells by binding to and stabilizing GLUT1 mRNA (169) (Table 4; Figure 2). IGF2BP2 was subsequently revealed as a potential target of miR-141 and to be involved in miR-141-induced PC cell growth via activating the PI3K-Akt signaling pathway (171) (Table 4; Figure 2). Acting as an m6A reader, IGF2BP2 could bind to DANCR to increase RNA stability via in an m6A-dependent manner and enhance the cell proliferation and stemness-like properties of PC cells (128) (Table 4; Figure 2). Thus, IGF2BP2 plays an oncogenic role in PC.
IGF2BP3
A high expression of IGF2BP3 in both PC tissues and cell lines was observed (172, 174, 175) (Table 4) and was correlated with shorter OS as well as PFS of PC patients (162, 172, 174–177) (Table 4). Knockdown of IGF2BP3 reduced the cell proliferation, migration, and invasion of PC cells (172, 178, 179). Mechanism-wise, IGF2BP3, located in cytoplasmic stress granules along with its downstream targets ARF6 and ARHGE4, promoted cell protrusion formation and enhanced PC cell invasion and metastasis (179, 180). A genome-wide analysis upon IGF2BP3 knockdown has further shown that IGF2BP3 was strongly correlated with genes regulating cell migration, proliferation, and adhesion (178). Moreover, IGF2BP3 was identified as a target of Lin28B/Let7 and enhanced the cell growth and stemness-like properties of PC cells (177) (Table 4; Figure 2). Overall, IGF2BP3 might be a promising diagnostic and prognostic biomarker as well as therapeutic target for PC.
YTHDFs
YTHDF1, YTHDF2, and YTHDF3 were upregulated in PC tissues (65, 181–183) (Table 4). Among these YTHDF family genes, YTHDF2 has been extensively studied in PC, while few studies have reported the roles of YTHDF1 and YTHDF3 in PC. As previously reported, PC patients with a higher expression of YTHDF2 have shown a shorter OS (181) and advanced stage (183) (Table 4). Chen et al. found that knockdown of YTHDF2 inhibited cell growth through inhibiting the Akt/GSK3β/CyclinD1 signaling pathway (183) (Table 4; Figure 2). However, an enhancement of cell migration, invasion, and EMT was also observed upon YTHDF2 deficiency (183). Furthermore, loss of YTHDF2-mediated YAP signaling activation may participate in PC cell EMT (183) (Table 4; Figure 2). Referring to m6A regulation, YTHDF2 served as an m6A reader which could recognize and bind to m6A-modified PIK3CB, PERP, PER1, PJA2, and NUCB1 RNA, thereby mediating their RNA degradation (7–9, 11, 64) (Table 4; Figure 2). The above findings indicate the critical roles of YTHDF2 in PC malignant progression.
YTHDCs
YTHDC1 and YTHDC2 were downregulated in PC tissues when compared to normal tissues (136) (Table 4). The upregulation of YTHDC1 predicted the longer OS and relapse-free survival (RFS) of PC patients (136) (Table 4). As an m6A reader, YTHDC1 triggered the degradation of pri-miR-30d and enhanced the maturation of miR-30d in an m6A-dependent manner (136) (Table 4; Figure 2). Finally, YTHDC1 was further found to suppress cell growth induced by miR-30d inhibition (136). Therefore, YTHDC1 might be a possible biomarker for PC prognosis and targeted therapy due to its antitumor effect.
HNRNPC
Few studies have reported the function of HNRNPC in PC. Hou et al. have shown that knockdown of HNRNPC significantly reduced the cell proliferation of PC cells (162) (Table 4). A high expression of HNRNPC was associated with a shorter OS of PC patients (162) (Table 4).
Erasers
FTO
A contradictory expression of FTO has been reported in PC. On the one hand, Tang et al. found that FTO was highly expressed in both PC tissues and cell lines (185) (Table 4). Loss of FTO inhibited cell proliferation and also enhanced the apoptosis of PC cells (185). Meanwhile, a significantly elevated m6A level of PC cells was detected after FTO knockdown through m6A dot-blot (185). Furthermore, FTO could interact with c-myc and enhance the expression and mRNA stability of c-myc, forming a functional FTO/c-myc axis (185) (Table 4; Figure 2). On the other hand, downregulation of FTO in PC tissues was also observed (8, 65) (Table 3). Furthermore, a low expression of FTO predicted a shorter OS of PC patients (8). FTO suppressed PC cell proliferation, migration, and invasion (8). Acting as an m6A demethylase, FTO reduced while FTO deficiency enhanced the total RNA m6A level of PC cells (8). Moreover, FTO reduced the m6A level of PJA2 and increased PJA2 expression via YTHDF2-mdeidated RNA degradation of PJA2, thereby suppressing the Wnt pathway and forming a functional FTO/YTHDF2/PJA2/Wnt axis to inhibit PC malignant progression (8) (Table 4; Figure 2). Taken together, FTO exerted both oncogenic and antitumor roles in the carcinogenesis of PC.
ALKBH5
Unlike the m6A writer expression in PC, ALKBH5 expression is significantly reduced in PC tissues, and PC patients with a low expression of ALKBH5 have shown a shorter OS (11, 56, 65). In contrast, Cho et al. revealed that a high expression of ALKBH5 was associated with a shorter OS of PC patients (184) (Table 4). So far, ALKBH5 has been reported to negatively regulate PC cell proliferation, migration, and invasion; iron metabolism; and GEM resistance (11, 56, 65, 133, 134) (Table 4). Currently, several m6A targets of ALKBH5 have been identified, such as WIF-1, PER1, FBXL5, SLC25A28, SLC25A37, and KCNK15-AS1. As reported, ALKBH5 reduced the m6A level of PER1 and then increased PER1 expression via in an m6A-YTHDF2-dependent way (11) (Table 4; Figure 2). Moreover, PER1 could in turn increase the ALKBH5 expression through activating p53 (11), suggesting a positive feedback loop between ALKBH5 and PER1 in promoting tumor growth and metastasis of PC (Table 4; Figure 2). Tang et al. found that ALKBH5, downregulated by GEM treatment, could also decrease the m6A level of WIF-1 and promote its expression to suppress the Wnt pathway, leading to PC cell growth, metastasis, and GEM resistance (56) (Table 4; Figure 2). In addition, ALKBH5 modulated the RNA stability of FBXL5 and SLC25A28, as well as the alternative splicing of SLC25A37 in an m6A-dependent manner (65), and reduced the cell migration and invasion and the intracellular iron level, thus preventing PC progression (65) (Table 4; Figure 2). He et al. further demonstrated that ALKBH5 remarkably enhanced the KCNK15-AS1 expression through decreasing the m6A level and stabilizing the KCNK15-AS1 mRNA, thereby suppressing cancer development (133, 134) (Table 4; Figure 2). Therefore, the above results revealed a tumor-suppressive role of ALKBH5 in PC, indicating a possible application of ALKBH5 for PC prognosis and chemoresistance prediction.
Conclusions and Perspectives
In recent years, the molecular mechanisms of genetic and epigenetic regulation have been extensively studied in the occurrence and progression of PC. Notably, increasing attention has been paid to the m6A modifications in PC development. Here in this review, we focused on the function and molecular mechanism of m6A regulators and m6A-regulated genes. We summarized that m6A modifications exerted their functions mainly in two ways. Firstly, m6A modifications modulate mRNA methylation to affect their RNA stability as well as protein expression (Table 1; Figure 2). Secondly, m6A modifications regulate the methylation of ncRNAs including long non-coding RNAs (LncRNAs) and miRNAs and alter the ncRNA expression to participate in PC carcinogenesis (Table 2; Figure 2). In spite of the above findings, the molecular mechanism of m6A regulation in PC remains largely unknown. Increasing comprehensive and in-depth studies are required to elucidate the critical roles of m6A modification in the malignant progression of PC and to further identify novel promising diagnostic and prognostic biomarkers as well as therapeutic targets for PC, and finally explore their possible clinical applications. Moreover, further research is also required to be done to illustrate the m6A modulation in higher-risk factors of PC, such as smoking, obesity, diabetes, and chronic pancreatitis. Therefore, systematic and comprehensive studies are urgently needed to clarify the interplay between m6A regulation and PC malignant progression, paving the way for exploring more approaches for PC treatment.
Author Contributions
XH, WSu, QX, and KT contributed to the idea and the manuscript. XL, WF, WSun, and QL collected the literature. JG contributed to the figure preparation. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (82103295, 81801642), Natural Science Foundation of Zhejiang Province (LQ22H160062), and Medical and Health Science Technology Project of Zhejiang Province (2019RC105, 2022KY516).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic Cancer. Lancet (2020) 395(10242):2008–20. doi: 10.1016/S0140-6736(20)30974-0
3. Khan AA, Liu X, Yan X, Tahir M, Ali S, Huang H. An Overview of Genetic Mutations and Epigenetic Signatures in the Course of Pancreatic Cancer Progression. Cancer Metastasis Rev (2021) 40(1):245–72. doi: 10.1007/s10555-020-09952-0
4. Javadrashid D, Baghbanzadeh A, Derakhshani A, Leone P, Silvestris N, Racanelli V, et al. Pancreatic Cancer Signaling Pathways, Genetic Alterations, and Tumor Microenvironment: The Barriers Affecting the Method of Treatment. Biomedicines (2021) 9(4):373. doi: 10.3390/biomedicines9040373
5. Barbieri I, Kouzarides T. Role of RNA Modifications in Cancer. Nat Rev Cancer (2020) 20(6):303–22. doi: 10.1038/s41568-020-0253-2
6. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The Role of M6a Modification in the Biological Functions and Diseases. Signal Transduct Target Ther (2021) 6(1):74. doi: 10.1038/s41392-020-00450-x
7. Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, et al. Upregulation of METTL14 Mediates the Elevation of PERP mRNA N(6) Adenosine Methylation Promoting the Growth and Metastasis of Pancreatic Cancer. Mol Cancer (2020) 19(1):130. doi: 10.1186/s12943-020-01249-8
8. Zeng J, Zhang H, Tan Y, Wang Z, Li Y, Yang X. M6a Demethylase FTO Suppresses Pancreatic Cancer Tumorigenesis by Demethylating PJA2 and Inhibiting Wnt Signaling. Mol Ther Nucleic Acids (2021) 25:277–92. doi: 10.1016/j.omtn.2021.06.005
9. Hua YQ, Zhang K, Sheng J, Ning ZY, Li Y, Shi WD, et al. NUCB1 Suppresses Growth and Shows Additive Effects With Gemcitabine in Pancreatic Ductal Adenocarcinoma via the Unfolded Protein Response. Front Cell Dev Biol (2021) 9:641836. doi: 10.3389/fcell.2021.641836
10. Xia T, Wu X, Cao M, Zhang P, Shi G, Zhang J, et al. The RNA M6a Methyltransferase METTL3 Promotes Pancreatic Cancer Cell Proliferation and Invasion. Pathol Res Pract (2019) 215(11):152666. doi: 10.1016/j.prp.2019.152666
11. Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, et al. RNA Demethylase ALKBH5 Prevents Pancreatic Cancer Progression by Posttranscriptional Activation of PER1 in an M6a-YTHDF2-Dependent Manner. Mol Cancer (2020) 19(1):91. doi: 10.1186/s12943-020-01158-w
12. Desrosiers R, Friderici K, Rottman F. Identification of Methylated Nucleosides in Messenger RNA From Novikoff Hepatoma Cells. Proc Natl Acad Sci U S A (1974) 71(10):3971–5. doi: 10.1073/pnas.71.10.3971
13. Fu Y, Dominissini D, Rechavi G, He C. Gene Expression Regulation Mediated Through Reversible M(6)A RNA Methylation. Nat Rev Genet (2014) 15(5):293–306. doi: 10.1038/nrg3724
14. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the Human and Mouse M6a RNA Methylomes Revealed by M6a-Seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
15. Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. A Majority of M6a Residues are in the Last Exons, Allowing the Potential for 3’ UTR Regulation. Genes Dev (2015) 29(19):2037–53. doi: 10.1101/gad.269415.115
16. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3’ UTRs and Near Stop Codons. Cell (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
17. Huang H, Weng H, Chen J. M(6)A Modification in Coding and Non-Coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell (2020) 37(3):270–88. doi: 10.1016/j.ccell.2020.02.004
18. Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, et al. The Role of N(6)-Methyladenosine (M(6)A) Modification in the Regulation of circRNAs. Mol Cancer (2020) 19(1):105. doi: 10.1186/s12943-019-1112-1
19. Chen XY, Zhang J, Zhu JS. The Role of M(6)A RNA Methylation in Human Cancer. Mol Cancer (2019) 18(1):103. doi: 10.1186/s12943-019-1033-z
20. Zhao W, Qi X, Liu L, Ma S, Liu J, Wu J. Epigenetic Regulation of M(6)A Modifications in Human Cancer. Mol Ther Nucleic Acids (2020) 19:405–12. doi: 10.1016/j.omtn.2019.11.022
21. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural Basis of N(6)-Adenosine Methylation by the METTL3-METTL14 Complex. Nature (2016) 534(7608):575–8. doi: 10.1038/nature18298
22. Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell (2016) 63(2):306–17. doi: 10.1016/j.molcel.2016.05.041
23. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3
24. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA Mediates Preferential M(6)A mRNA Methylation in 3’UTR and Near Stop Codon and Associates With Alternative Polyadenylation. Cell Discovery (2018) 4:10. doi: 10.1038/s41421-018-0019-0
25. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. M(6)A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature (2016) 537(7620):369–73. doi: 10.1038/nature19342
26. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 Regulates Nuclear RNA M(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell (2018) 69(6):1028–38 e6. doi: 10.1016/j.molcel.2018.02.015
27. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA M(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell (2017) 169(5):824–35.e14. doi: 10.1016/j.cell.2017.05.003
28. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hobartner C, et al. Human METTL16 Is a N(6)-Methyladenosine (M(6)A) Methyltransferase That Targets pre-mRNAs and Various Non-Coding RNAs. EMBO Rep (2017) 18(11):2004–14. doi: 10.15252/embr.201744940
29. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The Human 18S rRNA M6a Methyltransferase METTL5 is Stabilized by TRMT112. Nucleic Acids Res (2019) 47(15):7719–33. doi: 10.1093/nar/gkz619
30. Wang L, Liang Y, Lin R, Xiong Q, Yu P, Ma J, et al. Mettl5 Mediated 18S rRNA N6-Methyladenosine (M(6)A) Modification Controls Stem Cell Fate Determination and Neural Function. Genes Dis (2022) 9(1):268–74. doi: 10.1016/j.gendis.2020.07.004
31. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in Nuclear RNA is a Major Substrate of the Obesity-Associated FTO. Nat Chem Biol (2011) 7(12):885–7. doi: 10.1038/nchembio.687
32. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a Mammalian RNA Demethylase That Impacts RNA Metabolism and Mouse Fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
33. Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, et al. AlkB Homolog 3-Mediated tRNA Demethylation Promotes Protein Synthesis in Cancer Cells. Sci Rep (2017) 7:42271. doi: 10.1038/srep42271
34. Wang T, Kong S, Tao M, Ju S. The Potential Role of RNA N6-Methyladenosine in Cancer Progression. Mol Cancer (2020) 19(1):88. doi: 10.1186/s12943-020-01204-7
35. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear M(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell (2016) 61(4):507–19. doi: 10.1016/j.molcel.2016.01.012
36. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear M6a Reader YTHDC1 Regulates Alternative Polyadenylation and Splicing During Mouse Oocyte Development. PloS Genet (2018) 14(5):e1007412. doi: 10.1371/journal.pgen.1007412
37. Kim GW, Imam H, Siddiqui A. The RNA Binding Proteins YTHDC1 and FMRP Regulate the Nuclear Export of N(6)-Methyladenosine-Modified Hepatitis B Virus Transcripts and Affect the Viral Life Cycle. J Virol (2021) 95(13):e0009721. doi: 10.1128/JVI.00097-21
38. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 Mediates Nuclear Export of N(6)-Methyladenosine Methylated mRNAs. Elife (2017) 6. doi: 10.7554/eLife.31311
39. Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of M(6)A-Dependent Nuclear RNA Processing Events. Cell (2015) 162(6):1299–308. doi: 10.1016/j.cell.2015.08.011
40. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-Methyladenosine Marks Primary microRNAs for Processing. Nature (2015) 519(7544):482–5. doi: 10.1038/nature14281
41. McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. hnRNP C Tetramer Measures RNA Length to Classify RNA Polymerase II Transcripts for Export. Science (2012) 335(6076):1643–6. doi: 10.1126/science.1218469
42. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-Methyladenosine Alters RNA Structure to Regulate Binding of a Low-Complexity Protein. Nucleic Acids Res (2017) 45(10):6051–63. doi: 10.1093/nar/gkx141
43. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The M6a Reader YTHDF1 Promotes Ovarian Cancer Progression via Augmenting EIF3C Translation. Nucleic Acids Res (2020) 48(7):3816–31. doi: 10.1093/nar/gkaa048
44. Li J, Xie H, Ying Y, Chen H, Yan H, He L, et al. YTHDF2 Mediates the mRNA Degradation of the Tumor Suppressors to Induce AKT Phosphorylation in N6-Methyladenosine-Dependent Way in Prostate Cancer. Mol Cancer (2020) 19(1):152. doi: 10.1186/s12943-020-01267-6
45. Liao J, Wei Y, Liang J, Wen J, Chen X, Zhang B, et al. Insight Into the Structure, Physiological Function, and Role in Cancer of M6a Readers-YTH Domain-Containing Proteins. Cell Death Discovery (2022) 8(1):137. doi: 10.1038/s41420-022-00947-0
46. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 Facilitates Translation and Decay of N(6)-Methyladenosine-Modified RNA. Cell Res (2017) 27(3):315–28. doi: 10.1038/cr.2017.15
47. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long Noncoding RNA GAS5 Inhibits Progression of Colorectal Cancer by Interacting With and Triggering YAP Phosphorylation and Degradation and is Negatively Regulated by the M(6)A Reader YTHDF3. Mol Cancer (2019) 18(1):143. doi: 10.1186/s12943-019-1079-y
48. IMao Y, Dong L, Liu XM, Guo J, Ma H, Shen B, et al. M(6)A in mRNA Coding Regions Promotes Translation via the RNA Helicase-Containing YTHDC2. Nat Commun (2019) 10(1):5332. doi: 10.1038/s41467-019-13317-9
49. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)-Methyladenosine Binding Protein That Regulates Mammalian Spermatogenesis. Cell Res (2017) 27(9):1115–27. doi: 10.1038/cr.2017.99
50. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat Cell Biol (2018) 20(3):285–95. doi: 10.1038/s41556-018-0045-z
51. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5’ UTR M(6)A Promotes Cap-Independent Translation. Cell (2015) 163(4):999–1010. doi: 10.1016/j.cell.2015.10.012
52. Deng J, Zhang J, Ye Y, Liu K, Zeng L, Huang J, et al. N(6) -Methyladenosine-Mediated Upregulation of WTAPP1 Promotes WTAP Translation and Wnt Signaling to Facilitate Pancreatic Cancer Progression. Cancer Res (2021) 81(20):5268–83. doi: 10.1158/0008-5472.CAN-21-0494
53. Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25-3p Maturation via N(6)-Methyladenosine Stimulated by Cigarette Smoke Promotes Pancreatic Cancer Progression. Nat Commun (2019) 10(1):1858. doi: 10.1038/s41467-019-09712-x
54. Hsieh JC, Kodjabachian L, Rebbert ML, Rattner A, Smallwood PM, Samos CH, et al. A New Secreted Protein That Binds to Wnt Proteins and Inhibits Their Activities. Nature (1999) 398(6726):431–6. doi: 10.1038/18899
55. Tang Y, Simoneau AR, Liao WX, Yi G, Hope C, Liu F, et al. WIF1, a Wnt Pathway Inhibitor, Regulates SKP2 and C-Myc Expression Leading to G1 Arrest and Growth Inhibition of Human Invasive Urinary Bladder Cancer Cells. Mol Cancer Ther (2009) 8(2):458–68. doi: 10.1158/1535-7163.MCT-08-0885
56. Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, et al. M(6)A Demethylase ALKBH5 Inhibits Pancreatic Cancer Tumorigenesis by Decreasing WIF-1 RNA Methylation and Mediating Wnt Signaling. Mol Cancer (2020) 19(1):3. doi: 10.1186/s12943-019-1128-6
57. Lin YC, You L, Xu Z, He B, Yang CT, Chen JK, et al. Wnt Inhibitory Factor-1 Gene Transfer Inhibits Melanoma Cell Growth. Hum Gene Ther (2007) 18(4):379–86. doi: 10.1089/hum.2006.005
58. Ohigashi T, Mizuno R, Nakashima J, Marumo K, Murai M. Inhibition of Wnt Signaling Downregulates Akt Activity and Induces Chemosensitivity in PTEN-Mutated Prostate Cancer Cells. Prostate (2005) 62(1):61–8. doi: 10.1002/pros.20117
59. Zhu D, Yang Z, Liu Z, Zou Q, Yuan Y, Hu C. Association Between Wnt Inhibitory Factor 1 and Receptor Tyrosine Kinase-Like Orphan Receptor 2 Protein Expression and the Clinical Pathological Significance in Benign and Malignant Pancreatic Lesions. Oncol Lett (2017) 13(4):2244–52. doi: 10.3892/ol.2017.5704
60. Sato F, Nagata C, Liu Y, Suzuki T, Kondo J, Morohashi S, et al. PERIOD1 is an Anti-Apoptotic Factor in Human Pancreatic and Hepatic Cancer Cells. J Biochem (2009) 146(6):833–8. doi: 10.1093/jb/mvp126
61. Suzuki T, Sato F, Kondo J, Liu Y, Kusumi T, Fujimoto K, et al. Period is Involved in the Proliferation of Human Pancreatic MIA-PaCa2 Cancer Cells by TNF-Alpha. BioMed Res (2008) 29(2):99–103. doi: 10.2220/biomedres.29.99
62. Hildebrandt T, Preiherr J, Tarbe N, Klostermann S, Van Muijen GN, Weidle UH. Identification of THW, a Putative New Tumor Suppressor Gene. Anticancer Res (2000) 20(5A):2801–9.
63. Qu J, Zheng B, Ohuchida K, Feng H, Chong SJF, Zhang X, et al. PIK3CB is Involved in Metastasis Through the Regulation of Cell Adhesion to Collagen I in Pancreatic Cancer. J Adv Res (2021) 33:127–40. doi: 10.1016/j.jare.2021.02.002
64. Tian J, Zhu Y, Rao M, Cai Y, Lu Z, Zou D, et al. N(6)-Methyladenosine mRNA Methylation of PIK3CB Regulates AKT Signalling to Promote PTEN-Deficient Pancreatic Cancer Progression. Gut (2020) 69(12):2180–92. doi: 10.1136/gutjnl-2019-320179
65. Huang R, Yang L, Zhang Z, Liu X, Fei Y, Tong WM, et al. RNA M(6)A Demethylase ALKBH5 Protects Against Pancreatic Ductal Adenocarcinoma via Targeting Regulators of Iron Metabolism. Front Cell Dev Biol (2021) 9:724282. doi: 10.3389/fcell.2021.724282
66. Li C, Zhang Y, Cheng X, Yuan H, Zhu S, Liu J, et al. PINK1 and PARK2 Suppress Pancreatic Tumorigenesis Through Control of Mitochondrial Iron-Mediated Immunometabolism. Dev Cell (2018) 46(4):441–55.e8. doi: 10.1016/j.devcel.2018.07.012
67. Cadenas C, van de Sandt L, Edlund K, Lohr M, Hellwig B, Marchan R, et al. Loss of Circadian Clock Gene Expression is Associated With Tumor Progression in Breast Cancer. Cell Cycle (2014) 13(20):3282–91. doi: 10.4161/15384101.2014.954454
68. Chen M, Zhang L, Liu X, Ma Z, Lv L. PER1 Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Ovarian Cancer. Front Genet (2021) 12:697471. doi: 10.3389/fgene.2021.697471
69. Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu J, et al. A Role for the Clock Gene Per1 in Prostate Cancer. Cancer Res (2009) 69(19):7619–25. doi: 10.1158/0008-5472.CAN-08-4199
70. Mostafaie N, Kallay E, Sauerzapf E, Bonner E, Kriwanek S, Cross HS, et al. Correlated Downregulation of Estrogen Receptor Beta and the Circadian Clock Gene Per1 in Human Colorectal Cancer. Mol Carcinog (2009) 48(7):642–7. doi: 10.1002/mc.20510
71. Gery S, Komatsu N, Kawamata N, Miller CW, Desmond J, Virk RK, et al. Epigenetic Silencing of the Candidate Tumor Suppressor Gene Per1 in non-Small Cell Lung Cancer. Clin Cancer Res (2007) 13(5):1399–404. doi: 10.1158/1078-0432.CCR-06-1730
72. Wang J, Huang Q, Hu X, Zhang S, Jiang Y, Yao G, et al. Disrupting Circadian Rhythm via the PER1-HK2 Axis Reverses Trastuzumab Resistance in Gastric Cancer. Cancer Res (2022) 82(8):1503–17. doi: 10.1158/0008-5472.CAN-21-1820
73. Yang Y, Tang H, Zheng J, Yang K. The PER1/HIF-1alpha Negative Feedback Loop Promotes Ferroptosis and Inhibits Tumor Progression in Oral Squamous Cell Carcinoma. Transl Oncol (2022) 18:101360. doi: 10.1016/j.tranon.2022.101360
74. Yang G, Yang Y, Tang H, Yang K. Loss of the Clock Gene Per1 Promotes Oral Squamous Cell Carcinoma Progression via the AKT/mTOR Pathway. Cancer Sci (2020) 111(5):1542–54. doi: 10.1111/cas.14362
75. Liu Y, Hao J, Yuan G, Wei M, Bu Y, Jin T, et al. PER1 as a Tumor Suppressor Attenuated in the Malignant Phenotypes of Breast Cancer Cells. Int J Gen Med (2021) 14:7077–87. doi: 10.2147/IJGM.S328184
76. Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The Circadian Gene Per1 Plays an Important Role in Cell Growth and DNA Damage Control in Human Cancer Cells. Mol Cell (2006) 22(3):375–82. doi: 10.1016/j.molcel.2006.03.038
77. Relles D, Sendecki J, Chipitsyna G, Hyslop T, Yeo CJ, Arafat HA. Circadian Gene Expression and Clinicopathologic Correlates in Pancreatic Cancer. J Gastrointest Surg (2013) 17(3):443–50. doi: 10.1007/s11605-012-2112-2
78. Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, et al. PERP, an Apoptosis-Associated Target of P53, is a Novel Member of the PMP-22/Gas3 Family. Genes Dev (2000) 14(6):704–18. doi: 10.1101/gad.14.6.704
79. Awais R, Spiller DG, White MR, Paraoan L. P63 is Required Beside P53 for PERP-Mediated Apoptosis in Uveal Melanoma. Br J Cancer (2016) 115(8):983–92. doi: 10.1038/bjc.2016.269
80. Paraoan L, Gray D, Hiscott P, Ebrahimi B, Damato B, Grierson I. Expression of P53-Induced Apoptosis Effector PERP in Primary Uveal Melanomas: Downregulation is Associated With Aggressive Type. Exp Eye Res (2006) 83(4):911–9. doi: 10.1016/j.exer.2006.04.016
81. Beaudry VG, Jiang D, Dusek RL, Park EJ, Knezevich S, Ridd K, et al. Loss of the P53/P63 Regulated Desmosomal Protein Perp Promotes Tumorigenesis. PloS Genet (2010) 6(10):e1001168. doi: 10.1371/journal.pgen.1001168
82. Kong CS, Cao H, Kwok S, Nguyen CM, Jordan RC, Beaudry VG, et al. Loss of the P53/P63 Target PERP is an Early Event in Oral Carcinogenesis and Correlates With Higher Rate of Local Relapse. Oral Surg Oral Med Oral Pathol Oral Radiol (2013) 115(1):95–103. doi: 10.1016/j.oooo.2012.10.017
83. Zhao X, Li H, Lyu S, Zhai J, Ji Z, Zhang Z, et al. Single-Cell Transcriptomics Reveals Heterogeneous Progression and EGFR Activation in Pancreatic Adenosquamous Carcinoma. Int J Biol Sci (2021) 17(10):2590–605. doi: 10.7150/ijbs.58886
84. Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The Emerging Mechanisms of Isoform-Specific PI3K Signalling. Nat Rev Mol Cell Biol (2010) 11(5):329–41. doi: 10.1038/nrm2882
85. Azad AK, Zhabyeyev P, Vanhaesebroeck B, Eitzen G, Oudit GY, Moore RB, et al. Inactivation of Endothelial Cell Phosphoinositide 3-Kinase Beta Inhibits Tumor Angiogenesis and Tumor Growth. Oncogene (2020) 39(41):6480–92. doi: 10.1038/s41388-020-01444-3
86. Zhang Z, Zhang L, Wang B, Wei R, Wang Y, Wan J, et al. MiR-337-3p Suppresses Proliferation of Epithelial Ovarian Cancer by Targeting PIK3CA and PIK3CB. Cancer Lett (2020) 469:54–67. doi: 10.1016/j.canlet.2019.10.021
87. Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, et al. Essential Roles of PI(3)K-P110beta in Cell Growth, Metabolism and Tumorigenesis. Nature (2008) 454(7205):776–9. doi: 10.1038/nature07091
88. Zhang L, Li Y, Wang Q, Chen Z, Li X, Wu Z, et al. The PI3K Subunits, P110alpha and P110beta are Potential Targets for Overcoming P-Gp and BCRP-Mediated MDR in Cancer. Mol Cancer (2020) 19(1):10. doi: 10.1186/s12943-019-1112-1
89. Hong BS, Ryu HS, Kim N, Kim J, Lee E, Moon H, et al. Tumor Suppressor miRNA-204-5p Regulates Growth, Metastasis, and Immune Microenvironment Remodeling in Breast Cancer. Cancer Res (2019) 79(7):1520–34. doi: 10.1158/0008-5472.CAN-18-0891
90. Pu P, Kang C, Zhang Z, Liu X, Jiang H. Downregulation of PIK3CB by siRNA Suppresses Malignant Glioma Cell Growth In Vitro and In Vivo. Technol Cancer Res Treat (2006) 5(3):271–80. doi: 10.1177/153303460600500308
91. Hill KM, Kalifa S, Das JR, Bhatti T, Gay M, Williams D, et al. The Role of PI 3-Kinase P110beta in AKT Signally, Cell Survival, and Proliferation in Human Prostate Cancer Cells. Prostate (2010) 70(7):755–64. doi: 10.1002/pros.21108
92. Chen H, Mei L, Zhou L, Shen X, Guo C, Zheng Y, et al. PTEN Restoration and PIK3CB Knockdown Synergistically Suppress Glioblastoma Growth In Vitro and in Xenografts. J Neurooncol (2011) 104(1):155–67. doi: 10.1007/s11060-010-0492-2
93. Pridham KJ, Le L, Guo S, Varghese RT, Algino S, Liang Y, et al. PIK3CB/p110beta is a Selective Survival Factor for Glioblastoma. Neuro Oncol (2018) 20(4):494–505. doi: 10.1093/neuonc/nox181
94. Nakayama M, Miyake T, Gahara Y, Ohara O, Kitamura T. A Novel RING-H2 Motif Protein Downregulated by Axotomy: Its Characteristic Localization at the Postsynaptic Density of Axosomatic Synapse. J Neurosci (1995) 15(7 Pt 2):5238–48. doi: 10.1523/JNEUROSCI.15-07-05238.1995
95. Hedrick ED, Agarwal E, Leiphrakpam PD, Haferbier KL, Brattain MG, Chowdhury S. Differential PKA Activation and AKAP Association Determines Cell Fate in Cancer Cells. J Mol Signal (2013) 8(1):10. doi: 10.1186/1750-2187-8-10
96. Cantara S, D’Angeli F, Toti P, Lignitto L, Castagna MG, Capuano S, et al. Expression of the Ring Ligase PRAJA2 in Thyroid Cancer. J Clin Endocrinol Metab (2012) 97(11):4253–9. doi: 10.1210/jc.2012-2360
97. Lignitto L, Arcella A, Sepe M, Rinaldi L, Delle Donne R, Gallo A, et al. Proteolysis of MOB1 by the Ubiquitin Ligase Praja2 Attenuates Hippo Signalling and Supports Glioblastoma Growth. Nat Commun (2013) 4:1822. doi: 10.1038/ncomms2791
98. Liu J, Li Z, Teng W, Ye X. Identification of Downregulated circRNAs From Tissue and Plasma of Patients With Gastric Cancer and Construction of a circRNA-miRNA-mRNA Network. J Cell Biochem (2020) 121(11):4590–600. doi: 10.1002/jcb.29673
99. Song Y, Lee S, Kim JR, Jho EH. Pja2 Inhibits Wnt/beta-Catenin Signaling by Reducing the Level of TCF/Lef1. Int J Stem Cells (2018) 11(2):242–7. doi: 10.15283/ijsc18032
100. Miura K, Titani K, Kurosawa Y, Kanai Y. Molecular Cloning of Nucleobindin, a Novel DNA-Binding Protein That Contains Both a Signal Peptide and a Leucine Zipper Structure. Biochem Biophys Res Commun (1992) 187(1):375–80. doi: 10.1016/S0006-291X(05)81503-7
101. Lin P, Le-Niculescu H, Hofmeister R, McCaffery JM, Jin M, Hennemann H, et al. The Mammalian Calcium-Binding Protein, Nucleobindin (CALNUC), is a Golgi Resident Protein. J Cell Biol (1998) 141(7):1515–27. doi: 10.1083/jcb.141.7.1515
102. Liu GM, Xu ZQ, Ma HS. Nesfatin-1/Nucleobindin-2 Is a Potent Prognostic Marker and Enhances Cell Proliferation, Migration, and Invasion in Bladder Cancer. Dis Markers (2018) 2018:4272064. doi: 10.1155/2018/4272064
103. Zhang D, Lin J, Chao Y, Zhang L, Jin L, Li N, et al. Regulation of the Adaptation to ER Stress by KLF4 Facilitates Melanoma Cell Metastasis via Upregulating NUCB2 Expression. J Exp Clin Cancer Res (2018) 37(1):176. doi: 10.1186/s13046-018-0842-z
104. Kan JY, Yen MC, Wang JY, Wu DC, Chiu YJ, Ho YW, et al. Nesfatin-1/Nucleobindin-2 Enhances Cell Migration, Invasion, and Epithelial-Mesenchymal Transition via LKB1/AMPK/TORC1/ZEB1 Pathways in Colon Cancer. Oncotarget (2016) 7(21):31336–49. doi: 10.18632/oncotarget.9140
105. Sinha S, Pattnaik S, Aradhyam GK. Molecular Evolution Guided Functional Analyses Reveals Nucleobindin-1 as a Canonical E-Box Binding Protein Promoting Epithelial-To-Mesenchymal Transition (EMT). Biochim Biophys Acta Proteins Proteom (2019) 1867(9):765–75. doi: 10.1016/j.bbapap.2019.05.009
106. Pacheco-Fernandez N, Pakdel M, Blank B, Sanchez-Gonzalez I, Weber K, Tran ML, et al. Nucleobindin-1 Regulates ECM Degradation by Promoting Intra-Golgi Trafficking of MMPs. J Cell Biol (2020) 219(8). doi: 10.1083/jcb.201907058
107. Barbazan J, Dunkel Y, Li H, Nitsche U, Janssen KP, Messer K, et al. Prognostic Impact of Modulators of G Proteins in Circulating Tumor Cells From Patients With Metastatic Colorectal Cancer. Sci Rep (2016) 6:22112. doi: 10.1038/srep22112
108. Zhang N, Liu J, Ding X, Aikhionbare F, Jin C, Yao X. FBXL5 Interacts With p150Glued and Regulates its Ubiquitination. Biochem Biophys Res Commun (2007) 359(1):34–9. doi: 10.1016/j.bbrc.2007.05.068
109. Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, et al. An E3 Ligase Possessing an Iron-Responsive Hemerythrin Domain is a Regulator of Iron Homeostasis. Science (2009) 326(5953):722–6. doi: 10.1126/science.1176326
110. Vinas-Castells R, Frias A, Robles-Lanuza E, Zhang K, Longmore GD, Garcia de Herreros A, et al. Nuclear Ubiquitination by FBXL5 Modulates Snail1 DNA Binding and Stability. Nucleic Acids Res (2014) 42(2):1079–94. doi: 10.1093/nar/gkt935
111. Wang H, Shi H, Rajan M, Canarie ER, Hong S, Simoneschi D, et al. FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2fe2s] Cluster. Mol Cell (2020) 78(1):31–41.e5. doi: 10.1016/j.molcel.2020.02.011
112. Yao H, Su S, Xia D, Wang M, Li Z, Chen W, et al. F-Box and Leucine-Rich Repeat Protein 5 Promotes Colon Cancer Progression by Modulating PTEN/PI3K/AKT Signaling Pathway. BioMed Pharmacother (2018) 107:1712–9. doi: 10.1016/j.biopha.2018.08.119
113. He ZJ, Li W, Chen H, Wen J, Gao YF, Liu YJ. miR-1306-3p Targets FBXL5 to Promote Metastasis of Hepatocellular Carcinoma Through Suppressing Snail Degradation. Biochem Biophys Res Commun (2018) 504(4):820–6. doi: 10.1016/j.bbrc.2018.09.059
114. Muto Y, Moroishi T, Ichihara K, Nishiyama M, Shimizu H, Eguchi H, et al. Disruption of FBXL5-Mediated Cellular Iron Homeostasis Promotes Liver Carcinogenesis. J Exp Med (2019) 216(4):950–65. doi: 10.1084/jem.20180900
115. Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, et al. Mitoferrin is Essential for Erythroid Iron Assimilation. Nature (2006) 440(7080):96–100. doi: 10.1038/nature04512
116. Paradkar PN, Zumbrennen KB, Paw BH, Ward DM, Kaplan J. Regulation of Mitochondrial Iron Import Through Differential Turnover of Mitoferrin 1 and Mitoferrin 2. Mol Cell Biol (2009) 29(4):1007–16. doi: 10.1128/MCB.01685-08
117. Hung HI, Schwartz JM, Maldonado EN, Lemasters JJ, Nieminen AL. Mitoferrin-2-Dependent Mitochondrial Iron Uptake Sensitizes Human Head and Neck Squamous Carcinoma Cells to Photodynamic Therapy. J Biol Chem (2013) 288(1):677–86. doi: 10.1074/jbc.M112.422667
118. Wang C, Chen X, Zou H, Chen X, Liu Y, Zhao S. The Roles of Mitoferrin-2 in the Process of Arsenic Trioxide-Induced Cell Damage in Human Gliomas. Eur J Med Res (2014) 19:49. doi: 10.1186/s40001-014-0049-5
119. Zhang T, Sun L, Hao Y, Suo C, Shen S, Wei H, et al. ENO1 Suppresses Cancer Cell Ferroptosis by Degrading the mRNA of Iron Regulatory Protein 1. Nat Cancer (2022) 3(1):75–89. doi: 10.1038/s43018-021-00299-1
120. Ni S, Kuang Y, Yuan Y, Yu B. Mitochondrion-Mediated Iron Accumulation Promotes Carcinogenesis and Warburg Effect Through Reactive Oxygen Species in Osteosarcoma. Cancer Cell Int (2020) 20:399. doi: 10.1186/s12935-020-01494-3
121. Zhang Z, Guo M, Shen M, Kong D, Zhang F, Shao J, et al. The BRD7-P53-SLC25A28 Axis Regulates Ferroptosis in Hepatic Stellate Cells. Redox Biol (2020) 36:101619. doi: 10.1016/j.redox.2020.101619
122. Shi XY, Lin JJ, Ge XJ, Shi Y. LncRNA WTAPP1 Promotes Proliferation of Laryngeal Carcinoma Cells Through Regulating microRNA-592. Eur Rev Med Pharmacol Sci (2020) 24(18):9532–40. doi: 10.26355/eurrev_202009_23038
123. Li WD, Zhou DM, Sun LL, Xiao L, Liu Z, Zhou M, et al. LncRNA WTAPP1 Promotes Migration and Angiogenesis of Endothelial Progenitor Cells via MMP1 Through MicroRNA 3120 and Akt/PI3K/Autophagy Pathways. Stem Cells (2018) 36(12):1863–74. doi: 10.1002/stem.2904
124. Zhang L, Jin C, Yang G, Wang B, Hua P, Zhang Y. LncRNA WTAPP1 Promotes Cancer Cell Invasion and Migration in NSCLC by Downregulating lncRNA HAND2-As1. BMC Pulm Med (2020) 20(1):153. doi: 10.1186/s12890-020-01180-0
125. Meng X, Deng Y, He S, Niu L, Zhu H. M(6)A-Mediated Upregulation of LINC00857 Promotes Pancreatic Cancer Tumorigenesis by Regulating the miR-150-5p/E2F3 Axis. Front Oncol (2021) 11:629947. doi: 10.3389/fonc.2021.629947
126. Li T, Zhao H, Zhou H, Geng T. LncRNA LINC00857 Strengthens the Malignancy Behaviors of Pancreatic Adenocarcinoma Cells by Serving as a Competing Endogenous RNA for miR-340-5p to Upregulate TGFA Expression. PloS One (2021) 16(3):e0247817. doi: 10.1371/journal.pone.0247817
127. Song Y, Liang Y, Zou Q, Zeng S, Lin H, Liu M, et al. LINC00857 Promotes the Proliferation of Pancreatic Cancer via MET, STAT3, and CREB. J Gastrointest Oncol (2021) 12(6):2622–30. doi: 10.21037/jgo-21-723
128. Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 Regulates DANCR by Serving as an N6-Methyladenosine Reader. Cell Death Differ (2020) 27(6):1782–94. doi: 10.1038/s41418-019-0461-z
129. Luo Y, Wang Q, Teng L, Zhang J, Song J, Bo W, et al. LncRNA DANCR Promotes Proliferation and Metastasis in Pancreatic Cancer by Regulating miRNA-33b. FEBS Open Bio (2020) 10(1):18–27. doi: 10.1002/2211-5463.12732
130. Yao Z, Chen Q, Ni Z, Zhou L, Wang Y, Yang Y, et al. Long Non-Coding RNA Differentiation Antagonizing Nonprotein Coding RNA (DANCR) Promotes Proliferation and Invasion of Pancreatic Cancer by Sponging miR-214-5p to Regulate E2F2 Expression. Med Sci Monit (2019) 25:4544–52. doi: 10.12659/MSM.916960
131. Tang Y, Cao G, Zhao G, Wang C, Qin Q. LncRNA Differentiation Antagonizing non-Protein Coding RNA Promotes Proliferation and Invasion Through Regulating miR-135a/NLRP37 Axis in Pancreatic Cancer. Invest New Drugs (2020) 38(3):714–21. doi: 10.1007/s10637-019-00798-0
132. Chen L, Liu J, Tang T, Zhang YC, Liu MZ, Xu LY, et al. lncRNA Differentiation Antagonizing Nonprotein Coding RNA Overexpression Accelerates Progression and Indicates Poor Prognosis in Pancreatic Ductal Adenocarcinoma. Onco Targets Ther (2018) 11:7955–65. doi: 10.2147/OTT.S167065
133. He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cell Physiol Biochem (2018) 48(2):838–46. doi: 10.1159/000491915
134. He Y, Yue H, Cheng Y, Ding Z, Xu Z, Lv C, et al. ALKBH5-Mediated M(6)A Demethylation of KCNK15-AS1 Inhibits Pancreatic Cancer Progression via Regulating KCNK15 and PTEN/AKT Signaling. Cell Death Dis (2021) 12(12):1121. doi: 10.1038/s41419-021-04401-4
135. Chen JQ, Tao YP, Hong YG, Li HF, Huang ZP, Xu XF, et al. M(6)A-Mediated Up-Regulation of LncRNA LIFR-AS1 Enhances the Progression of Pancreatic Cancer via miRNA-150-5p/VEGFA/Akt Signaling. Cell Cycle (2021) 20(23):2507–18. doi: 10.1080/15384101.2021.1991122
136. Hou Y, Zhang Q, Pang W, Hou L, Liang Y, Han X, et al. YTHDC1-Mediated Augmentation of miR-30d in Repressing Pancreatic Tumorigenesis via Attenuation of RUNX1-Induced Transcriptional Activation of Warburg Effect. Cell Death Differ (2021) 28(11):3105–24. doi: 10.1038/s41418-021-00804-0
137. Xu X, Zong K, Wang X, Dou D, Lv P, Zhang Z, et al. miR-30d Suppresses Proliferation and Invasiveness of Pancreatic Cancer by Targeting the SOX4/PI3K-AKT Axis and Predicts Poor Outcome. Cell Death Dis (2021) 12(4):350. doi: 10.1038/s41419-021-03576-0
138. Wang L, He Y, Liu W, Bai S, Xiao L, Zhang J, et al. Non-Coding RNA LINC00857 is Predictive of Poor Patient Survival and Promotes Tumor Progression via Cell Cycle Regulation in Lung Cancer. Oncotarget (2016) 7(10):11487–99. doi: 10.18632/oncotarget.7203
139. Zhou D, He S, Zhang D, Lv Z, Yu J, Li Q, et al. LINC00857 Promotes Colorectal Cancer Progression by Sponging miR-150-5p and Upregulating HMGB3 (High Mobility Group Box 3) Expression. Bioengineered (2021) 12(2):12107–22. doi: 10.1080/21655979.2021.2003941
140. Su W, Wang L, Zhao H, Hu S, Zhou Y, Guo C, et al. LINC00857 Interacting With YBX1 to Regulate Apoptosis and Autophagy via MET and Phosphor-AMPKa Signaling. Mol Ther Nucleic Acids (2020) 22:1164–75. doi: 10.1016/j.omtn.2020.10.025
141. Han F, Yang S, Wang W, Huang X, Huang D, Chen S. Silencing of lncRNA LINC00857 Enhances BIRC5-Dependent Radio-Sensitivity of Lung Adenocarcinoma Cells by Recruiting NF-Kappab1. Mol Ther Nucleic Acids (2020) 22:981–93. doi: 10.1016/j.omtn.2020.09.020
142. Lin X, Feng D, Li P, Lv Y. LncRNA LINC00857 Regulates the Progression and Glycolysis in Ovarian Cancer by Modulating the Hippo Signaling Pathway. Cancer Med (2020) 9(21):8122–32. doi: 10.1002/cam4.3322
143. Wang L, Cao L, Wen C, Li J, Yu G, Liu C. LncRNA LINC00857 Regulates Lung Adenocarcinoma Progression, Apoptosis and Glycolysis by Targeting miR-1179/SPAG5 Axis. Hum Cell (2020) 33(1):195–204. doi: 10.1007/s13577-019-00296-8
144. Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, et al. Suppression of Progenitor Differentiation Requires the Long Noncoding RNA ANCR. Genes Dev (2012) 26(4):338–43. doi: 10.1101/gad.182121.111
145. Wang M, Gu J, Zhang X, Yang J, Zhang X, Fang X. Long Non-Coding RNA DANCR in Cancer: Roles, Mechanisms, and Implications. Front Cell Dev Biol (2021) 9:753706. doi: 10.3389/fcell.2021.753706
146. Xing D, Liang JQ, Li Y, Lu J, Jia HB, Xu LY, et al. Identification of Long Noncoding RNA Associated With Osteoarthritis in Humans. Orthop Surg (2014) 6(4):288–93. doi: 10.1111/os.12147
147. Bai Z, Wang X, Zhang Z. Long Noncoding RNA LIFR-AS1: A New Player in Human Cancers. BioMed Res Int (2022) 2022:1590815. doi: 10.1155/2022/1590815
148. Wang Q, Wu J, Huang H, Jiang Y, Huang Y, Fang H, et al. lncRNA LIFR-AS1 Suppresses Invasion and Metastasis of non-Small Cell Lung Cancer via the miR-942-5p/ZNF471 Axis. Cancer Cell Int (2020) 20:180. doi: 10.1186/s12935-020-01228-5
149. Pan H, Ding Y, Jiang Y, Wang X, Rao J, Zhang X, et al. LncRNA LIFR-AS1 Promotes Proliferation and Invasion of Gastric Cancer Cell via miR-29a-3p/COL1A2 Axis. Cancer Cell Int (2021) 21(1):7. doi: 10.1186/s12935-020-01644-7
150. Sarkozy M, Kahan Z, Csont T. A Myriad of Roles of miR-25 in Health and Disease. Oncotarget (2018) 9(30):21580–612. doi: 10.18632/oncotarget.24662
151. Caiazza C, Mallardo M. The Roles of miR-25 and its Targeted Genes in Development of Human Cancer. Microrna (2016) 5(2):113–9. doi: 10.2174/2211536605666160905093429
152. Yin H, Wang Y, Wu Y, Zhang X, Zhang X, Liu J, et al. EZH2-Mediated Epigenetic Silencing of miR-29/miR-30 Targets LOXL4 and Contributes to Tumorigenesis, Metastasis, and Immune Microenvironment Remodeling in Breast Cancer. Theranostics (2020) 10(19):8494–512. doi: 10.7150/thno.44849
153. Lin ZY, Chen G, Zhang YQ, He HC, Liang YX, Ye JH, et al. MicroRNA-30d Promotes Angiogenesis and Tumor Growth via MYPT1/c-JUN/VEGFA Pathway and Predicts Aggressive Outcome in Prostate Cancer. Mol Cancer (2017) 16(1):48. doi: 10.1186/s12943-017-0615-x
154. Li BQ, Huang S, Shao QQ, Sun J, Zhou L, You L, et al. WT1-Associated Protein is a Novel Prognostic Factor in Pancreatic Ductal Adenocarcinoma. Oncol Lett (2017) 13(4):2531–8. doi: 10.3892/ol.2017.5784
155. Li BQ, Liang ZY, Seery S, Liu QF, You L, Zhang TP, et al. WT1 Associated Protein Promotes Metastasis and Chemo-Resistance to Gemcitabine by Stabilizing Fak mRNA in Pancreatic Cancer. Cancer Lett (2019) 451:48–57. doi: 10.1016/j.canlet.2019.02.043
156. Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, et al. The Epitranscriptome M6a Writer METTL3 Promotes Chemo- and Radioresistance in Pancreatic Cancer Cells. Int J Oncol (2018) 52(2):621–9. doi: 10.3892/ijo.2017.4219
157. Kong F, Liu X, Zhou Y, Hou X, He J, Li Q, et al. Downregulation of METTL14 Increases Apoptosis and Autophagy Induced by Cisplatin in Pancreatic Cancer Cells. Int J Biochem Cell Biol (2020) 122:105731. doi: 10.1016/j.biocel.2020.105731
158. Zhang C, Ou S, Zhou Y, Liu P, Zhang P, Li Z, et al. M(6)A Methyltransferase METTL14-Mediated Upregulation of Cytidine Deaminase Promoting Gemcitabine Resistance in Pancreatic Cancer. Front Oncol (2021) 11:696371. doi: 10.3389/fonc.2021.696371
159. Chen S, Yang C, Wang ZW, Hu JF, Pan JJ, Liao CY, et al. CLK1/SRSF5 Pathway Induces Aberrant Exon Skipping of METTL14 and Cyclin L2 and Promotes Growth and Metastasis of Pancreatic Cancer. J Hematol Oncol (2021) 14(1):60. doi: 10.1186/s13045-021-01072-8
160. Yao Y, Luo L, Xiang G, Xiong J, Ke N, Tan C, et al. The Expression of M(6)A Regulators Correlated With the Immune Microenvironment Plays an Important Role in the Prognosis of Pancreatic Ductal Adenocarcinoma. Gland Surg (2022) 11(1):147–65. doi: 10.21037/gs-21-859
161. Wang L, Zhang S, Li H, Xu Y, Wu Q, Shen J, et al. Quantification of M6a RNA Methylation Modulators Pattern was a Potential Biomarker for Prognosis and Associated With Tumor Immune Microenvironment of Pancreatic Adenocarcinoma. BMC Cancer (2021) 21(1):876. doi: 10.1186/s12885-021-08550-9
162. Hou J, Wang Z, Li H, Zhang H, Luo L. Gene Signature and Identification of Clinical Trait-Related M(6) A Regulators in Pancreatic Cancer. Front Genet (2020) 11:522. doi: 10.3389/fgene.2020.00522
163. Zhao Z, Ju Q, Ji J, Li Y, Zhao Y. N6-Methyladenosine Methylation Regulator RBM15 is a Potential Prognostic Biomarker and Promotes Cell Proliferation in Pancreatic Adenocarcinoma. Front Mol Biosci (2022) 9:842833. doi: 10.3389/fmolb.2022.842833
164. Bosetti C, Lucenteforte E, Silverman DT, Petersen G, Bracci PM, Ji BT, et al. Cigarette Smoking and Pancreatic Cancer: An Analysis From the International Pancreatic Cancer Case-Control Consortium (Panc4). Ann Oncol (2012) 23(7):1880–8. doi: 10.1093/annonc/mdr541
165. Fang K, Qu H, Wang J, Tang D, Yan C, Ma J, et al. Characterization of Modification Patterns, Biological Function, Clinical Implication, and Immune Microenvironment Association of M(6)A Regulators in Pancreatic Cancer. Front Genet (2021) 12:702072. doi: 10.3389/fgene.2021.702072
166. Wan BS, Cheng M, Zhang L. Insulin-Like Growth Factor 2 mRNA-Binding Protein 1 Promotes Cell Proliferation via Activation of AKT and is Directly Targeted by microRNA-494 in Pancreatic Cancer. World J Gastroenterol (2019) 25(40):6063–76. doi: 10.3748/wjg.v25.i40.6063
167. Feng Y, Gao L, Cui G, Cao Y. LncRNA NEAT1 Facilitates Pancreatic Cancer Growth and Metastasis Through Stabilizing ELF3 mRNA. Am J Cancer Res (2020) 10(1):237–48.
168. Zhai S, Xu Z, Xie J, Zhang J, Wang X, Peng C, et al. Epigenetic Silencing of LncRNA LINC00261 Promotes C-Myc-Mediated Aerobic Glycolysis by Regulating miR-222-3p/HIPK2/ERK Axis and Sequestering IGF2BP1. Oncogene (2021) 40(2):277–91. doi: 10.1038/s41388-020-01525-3
169. Huang S, Wu Z, Cheng Y, Wei W, Hao L. Insulin-Like Growth Factor 2 mRNA Binding Protein 2 Promotes Aerobic Glycolysis and Cell Proliferation in Pancreatic Ductal Adenocarcinoma via Stabilizing GLUT1 mRNA. Acta Biochim Biophys Sin (Shanghai) (2019) 51(7):743–52. doi: 10.1093/abbs/gmz048
170. Dahlem C, Barghash A, Puchas P, Haybaeck J, Kessler SM. The Insulin-Like Growth Factor 2 mRNA Binding Protein IMP2/IGF2BP2 is Overexpressed and Correlates With Poor Survival in Pancreatic Cancer. Int J Mol Sci (2019) 20(13):3204. doi: 10.3390/ijms20133204
171. Xu X, Yu Y, Zong K, Lv P, Gu Y. Up-Regulation of IGF2BP2 by Multiple Mechanisms in Pancreatic Cancer Promotes Cancer Proliferation by Activating the PI3K/Akt Signaling Pathway. J Exp Clin Cancer Res (2019) 38(1):497. doi: 10.1186/s13046-019-1470-y
172. Cui XH, Hu SY, Zhu CF, Qin XH. Expression and Prognostic Analyses of the Insulin-Like Growth Factor 2 mRNA Binding Protein Family in Human Pancreatic Cancer. BMC Cancer (2020) 20(1):1160. doi: 10.1186/s12885-020-07590-x
173. Yan X, Wan H, Hao X, Lan T, Li W, Xu L, et al. Importance of Gene Expression Signatures in Pancreatic Cancer Prognosis and the Establishment of a Prediction Model. Cancer Manag Res (2019) 11:273–83. doi: 10.2147/CMAR.S185205
174. Schaeffer DF, Owen DR, Lim HJ, Buczkowski AK, Chung SW, Scudamore CH, et al. Insulin-Like Growth Factor 2 mRNA Binding Protein 3 (IGF2BP3) Overexpression in Pancreatic Ductal Adenocarcinoma Correlates With Poor Survival. BMC Cancer (2010) 10:59. doi: 10.1186/1471-2407-10-59
175. Wang BJ, Wang L, Yang SY, Liu ZJ. Expression and Clinical Significance of IMP3 in Microdissected Premalignant and Malignant Pancreatic Lesions. Clin Transl Oncol (2015) 17(3):215–22. doi: 10.1007/s12094-014-1216-4
176. Johnson B, Khalil M, Blansfield J, Lin F, Zhu S, Kirchner HL, et al. Investigating the Prognostic Value of KOC (K Homology Domain Containing Protein Overexpressed in Cancer) Overexpression After Curative Intent Resection of Pancreatic Ductal Adenocarcinoma. J Gastrointest Oncol (2016) 7(6):E113–E7. doi: 10.21037/jgo.2016.11.05
177. Kugel S, Sebastian C, Fitamant J, Ross KN, Saha SK, Jain E, et al. SIRT6 Suppresses Pancreatic Cancer Through Control of Lin28b. Cell (2016) 165(6):1401–15. doi: 10.1016/j.cell.2016.04.033
178. Ennajdaoui H, Howard JM, Sterne-Weiler T, Jahanbani F, Coyne DJ, Uren PJ, et al. IGF2BP3 Modulates the Interaction of Invasion-Associated Transcripts With RISC. Cell Rep (2016) 15(9):1876–83. doi: 10.1016/j.celrep.2016.04.083
179. Taniuchi K, Furihata M, Hanazaki K, Saito M, Saibara T. IGF2BP3-Mediated Translation in Cell Protrusions Promotes Cell Invasiveness and Metastasis of Pancreatic Cancer. Oncotarget (2014) 5(16):6832–45. doi: 10.18632/oncotarget.2257
180. Taniuchi K, Furihata M, Saibara T. KIF20A-Mediated RNA Granule Transport System Promotes the Invasiveness of Pancreatic Cancer Cells. Neoplasia (2014) 16(12):1082–93. doi: 10.1016/j.neo.2014.10.007
181. Tang R, Zhang Y, Liang C, Xu J, Meng Q, Hua J, et al. The Role of M6a-Related Genes in the Prognosis and Immune Microenvironment of Pancreatic Adenocarcinoma. PeerJ (2020) 8:e9602. doi: 10.7717/peerj.9602
182. Xu F, Zhang Z, Yuan M, Zhao Y, Zhou Y, Pei H, et al. M6A Regulatory Genes Play an Important Role in the Prognosis, Progression and Immune Microenvironment of Pancreatic Adenocarcinoma. Cancer Invest (2021) 39(1):39–54. doi: 10.1080/07357907.2020.1834576
183. Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, et al. YTH Domain Family 2 Orchestrates Epithelial-Mesenchymal Transition/Proliferation Dichotomy in Pancreatic Cancer Cells. Cell Cycle (2017) 16(23):2259–71. doi: 10.1080/15384101.2017.1380125
184. Cho SH, Ha M, Cho YH, Ryu JH, Yang K, Lee KH, et al. ALKBH5 Gene is a Novel Biomarker That Predicts the Prognosis of Pancreatic Cancer: A Retrospective Multicohort Study. Ann Hepatobil Pancreat Surg (2018) 22(4):305–9. doi: 10.14701/ahbps.2018.22.4.305
Keywords: pancreatic cancer, non-coding RNAs, coding RNAs, RNA methylation, N6-methyladenosine (m6A)
Citation: Hu X, Lei X, Guo J, Fu W, Sun W, Lu Q, Su W, Xu Q and Tu K (2022) The Emerging Role of RNA N6-Methyladenosine Modification in Pancreatic Cancer. Front. Oncol. 12:927640. doi: 10.3389/fonc.2022.927640
Received: 25 April 2022; Accepted: 15 June 2022;
Published: 22 July 2022.
Edited by:
Xiangsong Wu, Shanghai Jiao Tong University School of Medicine, ChinaReviewed by:
Yiyin Zhang, Sir Run Run Shaw Hospital, ChinaYinu Wang, Northwestern University, United States
Copyright © 2022 Hu, Lei, Guo, Fu, Sun, Lu, Su, Xu and Tu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Su, c2Fnb3dlaUBvdXRsb29rLmNvbQ==; Qiuran Xu, d2luZHdheTYyNkBzaW5hLmNvbQ==; Kangsheng Tu, dGtzMDkxMkBmb3htYWlsLmNvbQ==
 Xiaoge Hu
Xiaoge Hu Xiangxiang Lei
Xiangxiang Lei Jinhui Guo
Jinhui Guo Wen Fu
Wen Fu Wen Sun
Wen Sun Qiliang Lu
Qiliang Lu Wei Su
Wei Su Qiuran Xu
Qiuran Xu Kangsheng Tu
Kangsheng Tu