
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 26 July 2022
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.922430
This article is part of the Research TopicAdvances in Molecular Biology Knowledge of Rectal Cancer and Forthcoming Role of Liquid BiopsyView all 10 articles
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers and among the leading causes of death in both men and women. Rectal cancer (RC) is particularly challenging compared with colon cancer as the treatment after diagnosis of RC is more complex on account of its narrow anatomical location in the pelvis adjacent to the urogenital organs. More and more existing studies have begun to refine the research on RC and colon cancer separately. Early diagnosis and multiple treatment strategies optimize outcomes for individual patients. However, the need for more accurate and precise models to facilitate RC research is underscored due to the heterogeneity of clinical response and morbidity interrelated with radical surgery. Organoids generated from biopsies of patients have developed as powerful models to recapitulate many aspects of their primary tissue, consisting of 3-D self-organizing structures, which shed great light on the applications in both biomedical and clinical research. As the preclinical research models for RC are usually confused with colon cancer, research on patient-derived RC organoid models enable personalized analysis of cancer pathobiology, organizational function, and tumor initiation and progression. In this review, we discuss the various applications of patient-derived RC organoids over the past two years in basic cancer biology and clinical translation, including sequencing analysis, drug screening, precision therapy practice, tumor microenvironment studies, and genetic engineering opportunities.
Cancer is a leading cause of noncommunicable disease deaths (9.9 million people annually) and has sparked a health crisis worldwide (1). Colorectal cancer (CRC) is the third most common type of cancer and second in terminology of cancer mortality. CRC includes colon cancer (CC) and rectal cancer (RC); 40% of CRC is represented by RC (2). Although early diagnosis and multiple treatment strategies optimize outcomes for individual patients, we still need a more accurate and precise model to facilitate cancer research, including predicting response to standard therapies, as large numbers of newly developed strategies fail in clinical trials but go well on cancer models (3).
Traditional immortalized two-dimensional cancer cell lines and patient-derived xenografts (PDX) have long been applied in CRC research (4–11). Several studies from Bardelli’s laboratory are focused on 151 established CRC lines (6) along with xenopatient-derived cell lines or xenolines (XLs) (7, 8). The authors later improve this platform to compare drug responses and better carry on genomic analysis. As a result, this system is able to detect CRC dependencies on kinases for which clinically approved drugs are available and to discern CRC cross-sensitivity to Olaparib and Oxaliplatin. However, the translation of the obtained results from laboratory to bedside is often hampered as cell lines fail to recapitulate primary tumor characteristics due to lack of heterogeneity, fewer cancer cells expand from the microenvironment, and experience loss of their primary polarity (12). Furthermore, different types of PDX models, such as murine (9) and zebrafish (10, 11) are developed to study CRC tumor initiation, progression, immune response, and response of novel strategies. The histology and genomic mutational patterns of their primary tissue are maximally retained in PDX (13). Yet, due to high costs and low success rates, their use in cancer research is limited. In addition, animal models have conquered some of these drawbacks, but they do not mirror human physiology, resulting in the high failure rate of new cancer strategies in the clinic (14).
Organoid technology bridges the gap between cell culture and animal models, providing in vivo-like conditions. In general, organoids involve most types of cells from primary organs and can recapitulate the key features, main structure, and tissue functions along with gene expression profiles of the organs from which they were derived. To date, organoid technology nowadays has shown great capacity in breaking down mechanisms associated with gastrointestinal cancer and improving patient outcomes as recently reviewed by Lau et al (15). The novel technology based on generating patients’ tumors from biopsies as patient-derived organoids (PDOs) constitutes a major breakthrough for the study of translational cancer research, such as studying tumor biology, discovering new biomarkers, drug screenings, and testing targeted personalized therapies. Furthermore, huge efforts have been made in CRC research using the technology of tumor-derived organoids. The success establishment rate could reach around 63% by using small amounts of starting material from tissue biopsy samples (16, 17). These cancer organoids can be accomplished in a few weeks with high efficiency, and most of them have inheritable stability after several generations.
More than one third of CRC is represented by RC (2). Although CC and RC are synonymously called CRC, as they share the same histological classification and characteristics (18), RC is particularly challenging compared with CC since treatment of the RC patient is usually more complex than that of the CC patient due to their differences in anatomical position (19). Some RC patients can avoid surgery if they respond completely to neoadjuvant chemoradiotherapy alone (20, 21), and others responding poorly to CRT require radical surgery (22). Therefore, it is promising to use a correct and specific research model for identifying patients’ response to treatment in order to minimize potential harm from overtreatment and enable personalized therapy of RC.
Despite the fact that RC treatment is more complicated by utilizing trimodal therapy consisting of neoadjuvant chemoradiation, surgical resection, and 5-fluorouracil-based chemotherapy (19), few efforts have been made to develop RC-specific research models—not to mention that one preclinical practice for treatment of RC has unexpectedly depended on CC cell lines (23). Cell lines are also established from RCs (SW837, Caco-2, etc) (24–26), but whether they were derived from the correct side of the rectum or from patients in the context of multimodal therapy is hard to prove. As for the PDX platform, the first study to establish a PDX model from RC patients’ biopsies before treatment is reported recently, supporting the translational suitability of the PDX platform that predicted the response of corresponding patients, and to identify cetuximab as a strategy to enhance the efficacy of 5-fluorouracil/radiotherapy (27).
As PDOs can be easily converted into PDX models and represent one of the models that comes closest to the primary patient’s tumor, it currently shows potential in high-throughput drug screening and personalized therapy testing. Whereas extensive research is available for CRC organoids, most studies have not distinguished CC organoids from RC. For instance, we can find 214 full text results from the past two years through searching “(Patient-derived organoid) AND (colorectal cancer)” in PubMed and 132 full text results by applying the MESH “Patient derived organoid AND Colorectal Cancer” with the same year range, whereas only 26 full text results were found with the same year range by using “(Patient-derived organoid) AND (rectal cancer)” and 14 full text results were found by applying the MESH “Patient derived organoid AND rectal Cancer.” Furthermore, we find that some CRC-associated research did not mention whether it used samples from CC or RC patients (Table 1), and some clinically derived CRC organoids or biobanks exclude tissue from RC patients and mainly focus on CC specimens (30, 38) with RC PDOs remaining an underexplored field. This may be because patients with RC are often irradiated prior to surgery or treatment, which can affect the efficacy of derived organoids to assess the effectiveness of subsequent treatment strategies. However, with the continuous improvement of technology, we are looking forward to seeing that research in the field of CRC can be more refined in the future, and research models specifically used for RC will continue to be applied. Recently, specific research or protocols on the organoid model of RC is gradually garnering attention from scientists in the last two years, and translational applications are springing up (16, 39–41)(Table 2). The translational application of the RC PDO model is also mentioned in some recent literature and systematic reviews (44, 46, 50, 52, 53). There is a need for standard procedures and methods to improve the reproducibility and stability during laboratory practice.
In this review, we aim to review the latest in RC organoid development and its cutting-edge applications for basic or translational cancer research (Figure 1), including mutational analysis, drug screening, personalized medicine therapy, tumor heterogeneity, and microenvironment study as well as cancer modeling.
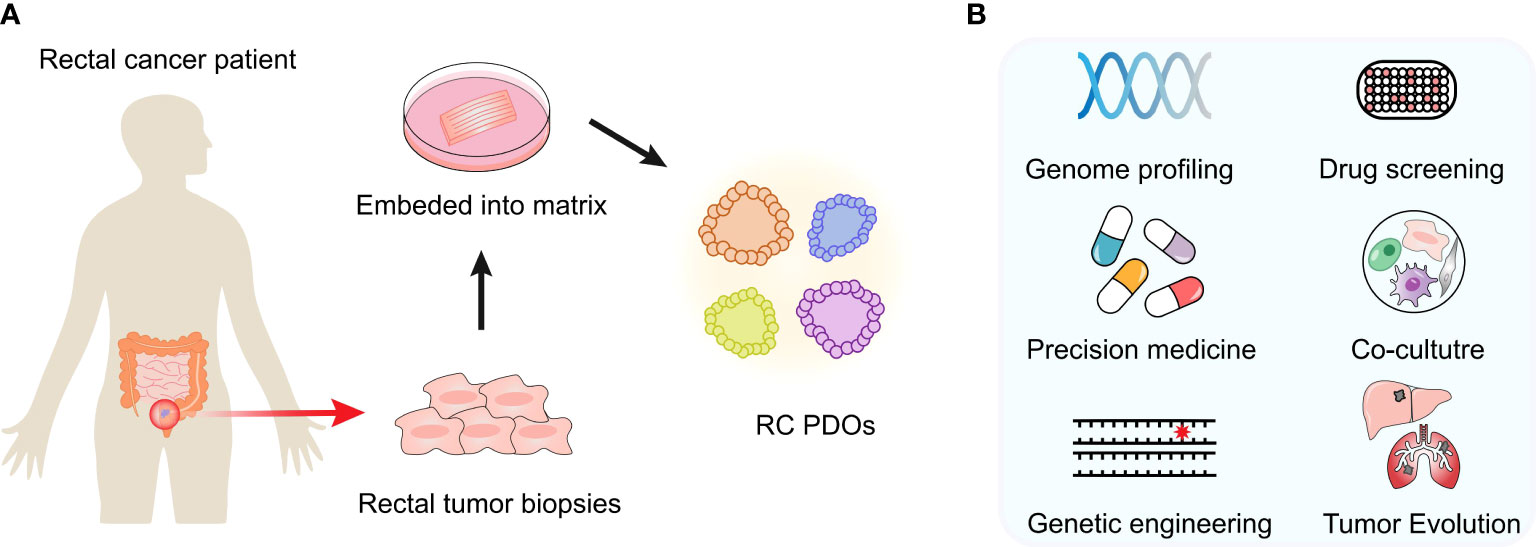
Figure 1 Developments and cutting-edge applications of the PDO model. (A) After an RC patient tumor’s surgery or biopsy, the tumor specimen is collected. PDOs can be generated in the laboratory and expanded to create sufficient material for biobank building and storage. Once established, these models are expanded in order to create sufficient material for storage and biobanking. (B) Multiple applications in vitro or ex vivo can be performed, such as genome profiling, drug screening, coculture experiments, etc.
Organoids over a period of continuous culture represent one of the in vitro models that comes closest to the primary tumor tissue, and they retain the function, histology, and genomic mutational patterns of the primary tumor, thereby preserving the heterogeneity of the origin tissue. By undergoing RC PDO sequencing, even low-frequency subclonal mutations can be detected and identified, and this gives scientists a hand in studying cancer pathobiology.
In the field of RC PDOs, two published articles in Cell Stem Cell and Nature Medicine (16, 39) illustrate the successful practice of RC organoids derived from patients, representing a major breakthrough in the precision medicine field. One group generated a biobank of 80 locally advanced RC PDOs from biopsy samples. Whole-exome sequencing was constructed in 18 culture lines, and the results displayed the mutational profile level of the corresponding tumor tissue. A 94.4% overlay between known cancer driver mutations and the TCGA data set was found in PDO samples. In 12 of 18 cases, they compared the genome-wide gene copy number variations of RC PDO cultures with that of primary tumors. The results displayed that the DNA copy number losses and gains of the original tissue were retained in PDOs. At the same time, WNT signaling was reported to represent the most related mutated genes pathway (88.9%), including APC (72.2%), FBXW7, TCF7L2, ARID1A, LRP5, and SOX9 mutation (16). Another group investigated the mutational fingerprint and larger landscape of molecular alterations in RC PDOs by undergoing the Food and Drug Administration–cleared tumor-profiling panel, MSK Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT). The data displayed that 92% (range: 0.66–1.00) of the oncogenic mutation was retained in the paired PDO cultures compared with an 88% concordance (range: 0.62–1.00) reported of organoids from colon and colon metastases (with RCs excluded), and 77% of the clonal oncogenic mutations in the original tumors were also detected as clonal mutations in PDO cultures. The top mutations in the RC PDOs included the alterations of genes APC, tumor protein P53 (TP53), KRAS, and F-Box and WD Repeat Domain Containing 7 (FBXW7) (39).
After a previous successful practice, more specific works are springing up. Costales-Carrera A et al. analyze the RNA-seq transcriptomes of six normal rectum or colonoscopy and rectal tumor organoid cultures by establishing a biobank of 50 organoids from therapy-naive RC patients. They confirm that these established RC organoids involve classic genetic alterations of sporadic CRC (APC, TP53, KRAS). Interestingly, some aberrant genes presented expression differences in RC and CC organoids. A cluster of genes regulating protein synthesis (ribosomal biogenesis, translation components, and regulation) was only expressed at a high level in rectal tumor organoids, indicating that CC and RC mainly differ in protein expression rather than the RNA level (55). In a new coclinical trial, Park et al. compares the result of PDTOs after irradiation with the individual patients’ response of radiotherapy. They used targeted next-generation sequencing analysis to check the ability of organoids to recapitulate the paired patients’ tissue and found that the match rate of KRAS, NRAS, and BRAF mutations in PDTOs could reach to 86.6%, 100%, and 100%, respectively. WNT signaling pathway–associated genes (including APC and FBXW7) were mutated in 68.4% of established cultures with identified APC (68.4%) and FBXW7 (31.5%). But, there was a problem in that the sample size was small, and they did not mention whether these PDOs were derived from patients who were therapy-naïve or not (48).
To analyze relevance between gene expression and the radiosensitivity of PDOs, Lee et al. checked 27,685 genes and identified 1741 differentially expressed genes from radio-resistant (RR) and radio-sensitive (RS) organoids by RNA-seq. In RR organoids, up-regulation genes that encode the control protein include calcium-dependent interactions (ANXA2, S100A4), immune cell activation (CD55, IL18, RUNX3), receptor catabolic processes (NPC1, APOE, LGMN), and plasma membrane proteolysis (ADAM9, BACE1, CTSE). They found the enhanced expression of CTSE regulated by DNA methylation status in RR organoids, suggesting that CTSE could be applied as a biomarker for radio-responsiveness (49).
One group combined the results from PDX, RC organoids generated from xenograft mouse models, and RC cell lines and identified ST6GAL-1 protein as a mediator for resistance to clinical chemoradiation therapy through restraining apoptosis. The data might need further studies in a larger sample size system (42).
Another report mentions that the high expression of VSTM2L could be one of the bad factors leading to patient resistance to CRT through diminished γ-H2AX expression, but they all did not mention about the correlation between PDTO response and patient outcomes (41).
Overall, these studies confirm the feasibility that rectal organoids can be generated and cultured from the clinical patients’ specimens and the great potential of PDO libraries as powerful platforms for RC research in vitro since the gene expression profiles of PDOs are very close to the organs from which they were derived.
Some RC patients can avoid surgery if they respond completely to CRT alone (20, 21), whereas others responding poorly to CRT require radical surgery (22). So it is crucial to undergo drug screening to minimize potential harm from overtreatment and enable personalized therapy.
PDOs now represent the most precise and effective models for drug screening and research. To date, Yao et al. generated 80 organoid cultures derived from patients with locally advanced RC and tested their susceptibility to 5-FU, irinotecan, or radiation. These data support that RC PDOs can function as a biomarker to help identify therapy-sensitive patients and promote precision medicine (16). Another group further leveraged an RC PDO platform to test responses to treatment in the front line, including FOLFOX (5-FU, leucovorin, and oxaliplatin) or radiation. Particularly, the responses of RC PDOs were consistent with the progression-free survival of the patients (39). It is also reported that PDOs derived from RC patients are used as a system to access the effect of combined radiotherapy with the short-chain fatty acids (such as butyrate, propionate, and acetate) by regulating HDAC activity. They further identified FOXO3A as a factor with cases nonresponsive to butyrate in PDOs. These data displayed the advantages of PDOs for testing novel drugs and modulating drug response (54). Kryeziu K et al. tested the drug efficacy of a medium-throughput drug library comprising 33 single agents and three 5-FU-based drug combinations containing Leucovorin (FLV), Oxaliplatin (FLOX), and SN-38 (FLIRI) by screening RC PDOs. They applied an interdisciplinary method to analyze spatiotemporal pharmacogenomic heterogeneity in a recurrent patient with KRAS-mutated liver metastases from RC. Coclinical ex vivo analyzes of front-line neoadjuvant combination chemotherapy regimens in three subsequent liver resections simulate actual responses to treatment, including indications of acquired resistance to FLOX. This case sets a great example for clinical application except for the small sample size. It is promising to conduct practice based on more large samples for evaluating the potential of PDOs in precision medicine (51).
These studies provide evidence that the capacity of PDOs to discover novel strategies and undergo drug screening overcome the flaws of 2-D cancer cell lines and upgrade the success of newly developed drugs.
RC is an ideal disease type to conduct precision medicine practice due to the different clinical responses of different patients and the subsequent tailored treatment. If the expected radiosensitivity is poor, patients may need more intensive chemotherapy application, and if the pretreatment worked well with the patients, they could avoid radical surgery. Accordingly, stable and reliable prediction tools play an important role in clinical practice for RC patients. It is reported that PDOs preserve high predictive value and the possibility to validate clinical biomarkers.
Ganesh et al. treated organoid culture from RC patients’ tumors of different stages with clinical chemotherapy (5-FU alone or with oxaliplatin together) or radiation and analyzed the corresponding clinical response of each patient. They revealed that the PDO predictions were correlated with the results from clinical patients after treatment. They also successfully used tumoroids to establish the endoluminal model as a feasible and reproducible in vivo platform of RC and invested its chemosensitivity. They found that the in vivo model could also recapitulate the clinical response to therapy such as 5-FU and FOLFOX (39). In a clinical phase III study, Yao et al. reported an LARC organoid biobank that could foresee the clinical response to 5-FU, irinotecan chemotherapy, irradiation, or combined treatment (16). They compared the radiosensitivity of organoids with clinical patients’ responses and found that 16 out of 17 patients with irradiation-sensitive organoids displayed positive response upon noadjuvant chemoradiation (NACR) treatment. Among the 64 patients whose organoids were resistant to irradiation, 42 of them had a poor response to NACR. They also exposed RCOs to 10 μM 5-Fu or 10 μM CPT-11. Twenty-seven patients with 5-Fu-sensitive organoids achieved a good response except for five poor response cases, and 38 of the 53 patients with 5-Fu–resistant PDOs had a poor response, and another 15 performed well. As for CPT-11, 32 sensitive PDOs of paired patients achieved a good response except for seven cases, and 27 of 34 patients with resistant organoids had a poor response. They, together, reveal that patients can acquire a good clinical response if their paired tumor organoids were sensitive to at least one of the three preceding treatments, which means that those with 5-Fu– or CPT-11–sensitive PDOs would probably benefit from 5-Fu or CPT-11 treatment alone although their PDOs were resistant to irradiation. Although PDOs generated by Yao et al. lack sources from patients of different stages compared with that from Ganesh et al., the data from these two groups could provide the evidence that the PDOs may enable tailored, personalized strategies with chemoradiation or chemotherapy and reduce overtreatment or toxicities for RC patients.
Later, Janakiraman et al. found that cetuximab has the potential to selectively sensitize the radiation effect with a KRAS mutational condition existing, and this group also detected cetuximab sensitivity gene impactors by establishing novel PDX and related PDO platforms from RC patients under pre-neoadjuvant therapy (27). Another observational coclinical study checked the standard combination of chemotherapy regimens in a PDO model generated from a patient with recurrent KRAS-mutated liver metastases from RC. They identified the SMAC mimetic as a unique therapeutic option in PDOs from the recurrent tumor tissue (51). Park et al. developed a prediction model to analyze clinical and laboratory patients’ radiotherapy response data by using a machine learning algorithm. The whole coclinical experiment involved 33 patients with diagnosed mid-to-low RC, and the machine prediction accuracy could achieve over 89% (48). The other study from Ting et al. displayed that the sensitivity, specificity, accuracy, and positive and negative predictive value of the PDO model for predicting chemotherapy drug responses were 63.33%, 94.12%, 79.69%, 90.48%, and 74.42%, which indicated that PDO responses consisted of the sensitivity in the respective patient in the clinic, which indicates that PDTOs can serve as a prediction model to guide the individualized selection of chemotherapy regimens for patients with RC (38). The work from another group identifies acid ceramidase (AC) as a target for improving radiotherapy treatment with the finding that PDOs presenting low AC expression were more radiosensitive than other models possessing higher levels of AC. However, the sample size of this study was small, and they did not demonstrate the original sample collection location of the organoid cultures (56). Another group investigated the radiosensitivity of CRC with PDOs of both colon and rectal tumors and focused on how the radiosensitivity of 13 RC PDOs correlated with clinical treatment outcomes. A significant mutual correlation is seen between the D0 (from the single hit multitarget algorithm, which is a single value to evaluate tissue radiosensitivity) of primary tumor PDO and the clinical response to neoadjuvant therapy (45). In a nutshell, the prediction accuracy could reach a level of 60%–90%, demonstrating the potential of RC PDOs to correctly identify clinical therapy responders and nonresponders and guide personalized therapy selection for RC patients.
In a newly published study, PDOs were very helpful in finding a new resistance mechanism of iCAFs and IL1α in RC patients. These PDOs were generated from biopsies collected prior to CRT from (non-pCRs) patients with poor along with fine prognoses and (pCRs) patients with pathological complete responses, and subsequently, these organoids detected different IL-1ra expression levels but comparable IL-1α expression. This group also created a preclinical RC mouse model (APTKA) through orthotopically transplanting organoids into C57BL/6 mice. Through a series of experiments, they identified the therapy-resistant capacity of iCAFs in RC and proposed the IL-1 pathway as a potential target for matrix repolarization and prevention for cancer-associated fibroblast senescence (43). Based on the previous achievement, this group recently further started an ACO/ARO/AIO-21 phase I trial to test the IL-1 receptor antagonist (IL-1 RA) anakinra combining with CRT therapy for RC, which set up a great example for a translational application from bench to bedside (47).
According to the above research, PDOs show high accuracy in predicting therapy responses and guiding personalized strategies. There is a matter of modifying culture methods to achieve a stable growth rate and improve the prediction accuracy in drug tests and targeted therapy research, which enables the application of RC PDOs in clinical practice and trials.
It is currently reported that PDOs facilitate the study of intratumoral heterogeneity and tumor evolution by analyzing the differences between PDOs from primary tissue and from the metastatic sites in a patient. Organoids derived from different sites in the same CRC patient possess different patterns and distinct responses to diverse therapies (26, 57, 58), whereas there are only rare reports about RC PDOs in this field.
One RC-related study from Ganesh et al. mentions the metastatic potential of an in vivo transplanted RC organoid model. They created a patient-derived orthotopic xenograft model with human tumoroids, which could reflect the actual process, including cancer initiation, invasion, and metastasis in the rectum. After 22 and 30 weeks post-transplant, these in vivo models turned into invasive adenocarcinoma, which is consistent with the stage I or II characteristics in human RC. They also found lung and liver metastases with infiltrating normal parenchyma by poorly differentiated carcinoma. It is worth mentioning the observation that metastases beginning from the endoluminal rectum shared the same appearance with the metastases found in the individual patients. Their success in the generation of a RC PDO endoluminal model paves the way forward in the field of tumor evolution research (39). This application of the PDO system again proves the potential and sheds light on the RC metastasis assays field.
RC has a poor prognosis for both metastases and a higher risk of local recurrence than CC in CRC. Four different molecular CRC consensus subtypes (CMS1–S4) have recently been delineated according to their transcriptome profiles and cellular heterogeneity. The CMS4 type was identified as having the worst prognosis with a feature of an abundant mesenchymal signature (59). Therefore, the tumor microenvironment (TME) and, particularly, cancer-associated fibroblasts (CAFs) consisting of heterogeneous populations have great importance in CRC research.
In vivo models are familiar tools to study the interaction between cancer cells in the microenvironment, whereas organoids as a model in vitro make the interactions visible. With the purpose of overcoming the limitation that organoids do not harbor diverse cell types or complex tissue compositions, scientists developed a cocultivation method incubating human organoids with human CAFs in order to activate fibroblasts and promote the growth of desmoplastic stroma (60). Immune cells can also be successfully activated and possess killing capacity when cocultured with PDOs.
For instance, a cocultivation method is reported by a research group in which circulating tumor reactive T lymphocytes could be coincubated together with human CRC organoids and stimulated into active status (61, 62). By establishing these models, it is promising that we could assess the efficacy of immune therapy under a controlled environment and produce individual T cells for adoptive T cell transferring. Another study treated CRC organoids with chemoradiotherapy and examined the capability of tumor-infiltrating T lymphocytes within the culture (63). They checked the cytotoxic effects as well in the co-culture system containing RC organoids and infiltrating T lymphocytes and found that the killing ability of PDOs from patients with a good therapy response was stronger than in PDOs from nonresponders. In newly published research, Nicolas et al. cocultured the supernatants from low or high IL-1ra expression organoids together with matched human intestinal fibroblasts and found that supernatants with insufficient IL-1ra expression activated a series of pro-inflammatory gene expression profiles (43). Patient RC organoid application in the study of the TME is indeed a remarkable achievement. We expect to see more high-quality research using PDOs as a tool in RC microenvironment field.
It is reported that cancer genetic mutations can be generated in wild-type organoids through technology such as CRISPR/Cas9 to facilitate research in oncology pathway mutants, tumor origin, and invasion. To date, there are fewer reports about RC organoid application in this field, so we use the research of CRC organoids.
As for CRC metastasis research, it has underscored the need for engineered organoids and emphasized the importance of genetic composition (64, 65).
By using CRISPR/Cas9 technology, oncologists could establish both advanced cancer and early tumor organoids with various mutations. The Medema group generated sessile serrated adenoma (SSA) organoids with the BRAFV600E mutation by engineering normal colon organoids to ensure regular growth. They later put forward that the CRC mesenchymal subtype could originate by activating RAS signaling through TGFβ stimulation together with BRAFV600E alternation (66). Another group made use of the SSAs characteristic of Rspondin gene fusions and first established chromosomal rearrangements with CRISPR/Cas9 to dig the impact of Rspondin fusions (67). This study introduced the idea that CRISPR/Cas9 technology can also be applied to delete, insert, or translocate chromosomes in addition to the use for mutating genes, which led the way to discover new therapy through targeting a specific pathway.
It is worth mentioning that Nicolas et al. activated Il1a transcription in irradiation-sensitive APTK organoids (Apc, Trp53, Tgfbr2, and K-rasG12D mutant) by CRISPR/Cas9 (43). Their data supports the conclusion that tumor cell–derived IL-1α polarizes CAFs toward an inflammatory phenotype, which then causes resistance to irradiation. A strong expression of inflammatory genes has been discovered in fibroblasts exposed to the fluid of APTK-sgIl1a organoids. APTK-sgIl1a organoids show an increased postirradiation stromal response with the features of suppressed CD8+ T cell infiltration along with increased Sirius red staining. However, irradiation did not further improve the level of macrophage and neutrophil infiltration with the IL-1α-dependent increase. Nicolas et al. combined CRISPR/Cas9 technology with RC organoids, enriching the field of study known as RC modeling.
In summary, CRISPR/Cas9 technology in combination with organoids suggests a potential strategy for basic cancer research and exploration in clinical application. We expect to see the development of this application in RC field.
In recent decades, organoid cultures of various types of tumors have been widely used in basic and translational cancer research, which is a major boost for the field. PDOs can truly reflect an individual’s condition and possess the capacity to predict individual responses to diverse clinical therapies, allowing clinicians to tailor treatment strategies for each patient and practice precision medicine. The establishment of living biological sample banks enables us to detect a wide range of molecular and different patterns of one cancer entity to further promote new medicine development and fully exploit existing drugs. The presented studies also reveal the possible applications in cocultivation assays with stromal, immune, or CAR cells, which may facilitate research in targeting stromal compositions to lead immune cells to their points.
However, current PDO systems do show some limitations, and there are gaps together with challenges between knowledge and the translational application of PDO models. First, culturing PDOs still costs a great deal, and the materials and culturing conditions used vary between laboratories. We present a summary in Table 3 with methodologic differences and limitations of selected studies (Table 3). An agreement needs to be reached. Second, successful establishment of a PDO platform depends on many factors, such as the size of the biopsy samples, amount of living tumor cells, and cancer cell proliferation rate. Contamination and residue of other cell types could also be problems in the biopsy material. In addition, as for rectal cancer, successful PDOs biobanks are precious resources since most RC patients have received neoadjuvant therapy prior to surgical resection, which may affect its role as prognosis tool in preclinical and clinical practice. At the same time, the frequency of eligible organoids for RC entities, such as sarcomas, is far from 80%–90%, suggesting the existence of relevant subtype conditions that may require specific growth factors. The greatest current challenge in patient-derived RC organoid culture is the absence of a TME. PDOs cannot recapitulate the full structure and function (such as the ability to grow blood vessels) of primary tissue for lacking CAFs, stroma cells, and immune cells in their culture system. Breakthroughs are being made in this field with continuous improvement of existing culture methods (e.g., an air–liquid interface PDO model enabling coculture with endogenous tumor-infiltrating lymphocytes). Finally, in view of inter- and intratumor heterogeneity, PDOs may need more in vivo model practice compared with PDX. Thus, the RC PDO model used in precision medicine requires extensive optimization and more focused research along with prospective study.
New technology, such as genomic, transcriptomic, and proteomic analyses, are used on organoids and help us further understand tumor biology to define targeted treatment. It is promising to see that PDOs allow direct prediction of the individual response to identified RC therapies. In a nutshell, patient-derived cancer organoids show the potential to bridge the gap between the present basic research and clinical practice in the near future, but more detail is needed before we can really identify the areas of most impact in terms of research and clinical application.
This review followed the PRISMA guidelines. Two investigators (IC and YY) independently conducted a literature search using as combined keywords rectal cancer and organoid, patient-derived organoid on Pubmed (https://www.ncbi.nlm.nih.gov/pubmed/) and Web of Science (v. 5.35). The database search was run on published articles in the last two years, all languages, from database inception until May 20, 2022. In both databases, the following search strategy was used: terms were searched as follows: organoid AND rectal cancer; organoid AND rectal carcinoma; patient-derived organoid AND rectal cancer; patient-derived organoid AND rectal carcinoma. It is thought that these terms would identify the majority of manuscripts within a narrow definition of rectal cancer and organoid, patient-derived organoid, though it remains likely that relevant sections might be embedded within the methodology sections of particular projects and, thus, more challenging to identify. In addition, we conducted a search on ClinicalTrials (https://clinicaltrials.gov/) using the keywords colorectal cancer organoid or rectal cancer organoid.
All studies reporting information on RC and organoids, PDOs were included. One hundred twenty-four articles were identified and reviewed independently by two authors (IC and YY), and after all duplicates were removed, 41 articles were considered. After removing articles that were not in English and those that had simply a mention of the words with no further expansion, 21 articles were considered. Sixteen articles (of the 21) devoted a considerable amount of the manuscript to expand on those topics, and five articles (of the 21) are reviews or systematic reviews.
As for the search on ClinicalTrials, 24 studies were identified using the term “colorectal cancer organoid” and “rectal cancer organoid.” Eighteen studies were considered after all duplicates and studies unrelated to CRC or organoid were removed.
Thematic groupings and analyses were reviewed by an additional author (IC). All outcomes were included due to the wide range of use of the terminologies.
The manuscripts identified in this review (n = 21) followed six loosely defined thematic groups: a) analysis of the mutational landscape using high-throughput sequencing technologies (n = 7), b) drug screening to delineate novel treatment strategies (n = 4), c) personalized medicine based on the testing of individual PDOs (n = 10), d) investigation of intratumoral heterogeneity and tumor evolution (n = 1), e) studying the TME with PDOs (n = 2) and f) cancer modeling by genetic engineering of organoids (n = 2).
In terms of the search on ClinicalTrials (n =18), 13 studies (of the 18) were excluded for the following reasons: a) the study did not mention whether RC patients were involved in the studies (n = 10), b) studies related to CC PDO (n = 2), c) RC study used a malignant colonic organoid model (n = 1). Five studies associated with patient-derived RC organoid application were finally considered.
All publications or clinical studies that met the criteria were included in Table 2, and some of the publications and clinical studies excluded are shown in Table 1 (28–37). They are presented subsequently in this order, reflecting the scientific continuum, moving from the characterization and acquisition of knowledge to the interpretation and finally toward clinical implementation.
YY and IC conducted the review and applied the eligibility selection criteria for the identified manuscripts. IC validated the selected manuscripts and arbitrated any queries. CP participated in literature collecting. YY and IC wrote the manuscript. HW, XL and XW supervised the whole article. All authors contributed to the article and approved the submitted version.
This study was supported by grants from the National Natural Science Foundation of China(82030099), the National Key R&D Program of China (2018YFC2000700), Shanghai Public Health System Construction Three-Year Action Plan(GWV-10.1-XK15), Innovative research team of high-level local universities in Shanghai.
We thank all colleagues in the School of Public Health for their unconditional support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. World Health Organization. Global health estimates: Leading causes of death . Available at: https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates/ghe-leading-causes-of-death.
2. Global Cancer Observatory. . Available at: https://gco.iarc.fr/.
3. Caponigro G, Sellers WR. Advances in the preclinical testing of cancer therapeutic hypotheses. Nat Rev Drug Discovery (2011) 10(3):179–87. doi: 10.1038/nrd3385
4. Auman JT, McLeod HL. Colorectal cancer cell lines lack the molecular heterogeneity of clinical colorectal tumors. Clin Colorectal Cancer (2010) 9(1):40–7. doi: 10.3816/CCC.2010.n.005
5. Kapalczynska M, Kolenda T, Przybyla W, Zajaczkowska M, Teresiak A, Filas V, et al. 2d and 3d cell cultures - a comparison of different types of cancer cell cultures. Arch Med Sci (2018) 14(4):910–9. doi: 10.5114/aoms.2016.63743
6. Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun (2015) 6:7002. doi: 10.1038/ncomms8002
7. Lazzari L, Corti G, Picco G, Isella C, Montone M, Arcella P, et al. Patient-derived xenografts and matched cell lines identify pharmacogenomic vulnerabilities in colorectal cancer. Clin Cancer Res (2019) 25(20):6243–59. doi: 10.1158/1078-0432.CCR-18-3440
8. Arena S, Corti G, Durinikova E, Montone M, Reilly NM, Russo M, et al. A subset of colorectal cancers with cross-sensitivity to olaparib and oxaliplatin. Clin Cancer Res (2020) 26(6):1372–84. doi: 10.1158/1078-0432.CCR-19-2409
9. Burtin F, Mullins CS, Linnebacher M. Mouse models of colorectal cancer: Past, present and future perspectives. World J Gastroenterol (2020) 26(13):1394–426. doi: 10.3748/wjg.v26.i13.1394
10. Fior R, Povoa V, Mendes RV, Carvalho T, Gomes A, Figueiredo N, et al. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc Natl Acad Sci U.S.A. (2017) 114(39):E8234–E43. doi: 10.1073/pnas.1618389114
11. Costa B, Ferreira S, Povoa V, Cardoso MJ, Vieira S, Stroom J, et al. Developments in zebrafish avatars as radiotherapy sensitivity reporters - towards personalized medicine. EBioMedicine (2020) 51:102578. doi: 10.1016/j.ebiom.2019.11.039
12. Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst (2013) 105(7):452–8. doi: 10.1093/jnci/djt007
13. Bhimani J, Ball K, Stebbing J. Patient-derived xenograft models-the future of personalised cancer treatment. Br J Cancer (2020) 122(5):601–2. doi: 10.1038/s41416-019-0678-0
14. Wong CH, Siah KW, Lo AW. Estimation of clinical trial success rates and related parameters. Biostatistics (2019) 20(2):273–86. doi: 10.1093/biostatistics/kxx069
15. Lau HCH, Kranenburg O, Xiao H, Yu J. Organoid models of gastrointestinal cancers in basic and translational research. Nat Rev Gastroenterol Hepatol (2020) 17(4):203–22. doi: 10.1038/s41575-019-0255-2
16. Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell (2020) 26(1):17–26.e6. doi: 10.1016/j.stem.2019.10.010
17. Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, van Werkhoven E, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med (2019) 11(513):eaay2574. doi: 10.1126/scitranslmed.aay2574
18. Nagtegaal ID, Arends M, Odze R. Tumours of the colon and rectum: Who classification of tumours of the colon and rectum, tnm staging of carcinomas of the colon and rectum and the introduction. 5th. France: IARC Press (2019)
19. Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. Nccn guidelines insights: Rectal cancer, version 6.2020. J Natl Compr Canc Netw (2020) 18(7):806–15. doi: 10.6004/jnccn.2020.0032
20. van der Valk MJM, Hilling DE, Bastiaannet E, Meershoek-Klein Kranenbarg E, Beets GL, Figueiredo NL, et al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the international watch & wait database (Iwwd): An international multicentre registry study. Lancet (2018) 391(10139):2537–45. doi: 10.1016/s0140-6736(18)31078-x
21. Smith JJ, Strombom P, Chow OS, Roxburgh CS, Lynn P, Eaton A, et al. Assessment of a watch-and-Wait strategy for rectal cancer in patients with a complete response after neoadjuvant therapy. JAMA Oncol (2019) 5(4):e185896. doi: 10.1001/jamaoncol.2018.5896
22. Park IJ, You YN, Agarwal A, Skibber JM, Rodriguez-Bigas MA, Eng C, et al. Neoadjuvant treatment response as an early response indicator for patients with rectal cancer. J Clin Oncol (2012) 30(15):1770–6. doi: 10.1200/jco.2011.39.7901
23. Midostaurin (Pkc412) for locally advanced rectal Cancer.Nct01282502 . Available at: https://clinicaltrials.gov/ct2/show/NCT01282502.
24. Ye X, Yin H, Lu Y, Zhang H, Wang H. Evaluation of hydrogel suppositories for delivery of 5-aminolevulinic acid and hematoporphyrin monomethyl ether to rectal tumors. Molecules (2016) 21(10):1347. doi: 10.3390/molecules21101347
25. Emons G, Spitzner M, Reineke S, Möller J, Auslander N, Kramer F, et al. Chemoradiotherapy resistance in colorectal cancer cells is mediated by Wnt/B-catenin signaling. Mol Cancer Res (2017) 15(11):1481–90. doi: 10.1158/1541-7786.Mcr-17-0205
26. Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell (2016) 18(6):827–38. doi: 10.1016/j.stem.2016.04.003
27. Janakiraman H, Zhu Y, Becker SA, Wang C, Cross A, Curl E, et al. Modeling rectal cancer to advance neoadjuvant precision therapy. Int J Cancer (2020) 147(5):1405–18. doi: 10.1002/ijc.32876
28. Petti L, Rizzo G, Rubbino F, Elangovan S, Colombo P, Restelli S, et al. Unveiling role of sphingosine-1-Phosphate receptor 2 as a brake of epithelial stem cell proliferation and a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res (2020) 39(1):253. doi: 10.1186/s13046-020-01740-6
29. Park SY, Kim JH, Choi JH, Lee CJ, Lee WJ, Park S, et al. Lipid raft-disrupting miltefosine preferentially induces the death of colorectal cancer stem-like cells. Clin Transl Med (2021) 11(11):e552. doi: 10.1002/ctm2.552
30. van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell (2015) 161(4):933–45. doi: 10.1016/j.cell.2015.03.053
31. Cho EJ, Kim M, Jo D, Kim J, Oh JH, Chung HC, et al. Immuno-genomic classification of colorectal cancer organoids reveals cancer cells with intrinsic immunogenic properties associated with patient survival. J Exp Clin Cancer Res (2021) 40(1):230. doi: 10.1186/s13046-021-02034-1
32. De Oliveira T, Goldhardt T, Edelmann M, Rogge T, Rauch K, Kyuchukov ND, et al. Effects of the novel Pfkfb3 inhibitor Kan0438757 on colorectal cancer cells and its systemic toxicity evaluation in vivo. Cancers (Basel) (2021) 13(5):1011. doi: 10.3390/cancers13051011
33. Blackmur JP, Vaughan-Shaw PG, Donnelly K, Harris BT, Svinti V, Ochocka-Fox AM, et al. Gene Co-expression network analysis identifies vitamin d-associated gene modules in adult normal rectal epithelium following supplementation. Front Genet (2021) 12:783970. doi: 10.3389/fgene.2021.783970
34. Tirado FR, Bhanja P, Castro-Nallar E, Olea XD, Salamanca C, Saha S. Radiation-induced toxicity in rectal epithelial stem cell contributes to acute radiation injury in rectum. Stem Cell Res Ther (2021) 12(1):63. doi: 10.1186/s13287-020-02111-w
35. Mare M, Colarossi L, Veschi V, Turdo A, Giuffrida D, Memeo L, et al. Cancer stem cell biomarkers predictive of radiotherapy response in rectal cancer: A systematic review. Genes (Basel) (2021) 12(10):1502. doi: 10.3390/genes12101502
36. Rana T, Korolkova OY, Rachakonda G, Williams AD, Hawkins AT, James SD, et al. Linking bacterial enterotoxins and alpha defensin 5 expansion in the crohn's colitis: A new insight into the etiopathogenetic and differentiation triggers driving colonic inflammatory bowel disease. PLoS One (2021) 16(3):e0246393. doi: 10.1371/journal.pone.0246393
37. Rana N, Privitera G, Kondolf HC, Bulek K, Lechuga S, De Salvo C, et al. Gsdmb is increased in ibd and regulates epithelial Restitution/Repair independent of pyroptosis. Cell (2022) 185(2):283–98.e17. doi: 10.1016/j.cell.2021.12.024
38. Wang T, Pan W, Zheng H, Zheng H, Wang Z, Li JJ, et al. Accuracy of using a patient-derived tumor organoid culture model to predict the response to chemotherapy regimens in stage iv colorectal cancer: A blinded study. Dis Colon Rectum (2021) 64(7):833–50. doi: 10.1097/dcr.0000000000001971
39. Ganesh K, Wu C, O'Rourke KP, Szeglin BC, Zheng Y, Sauvé CG, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med (2019) 25(10):1607–14. doi: 10.1038/s41591-019-0584-2
40. Chen Y, Probro Chongqing Biotechnology Co (PROB-Non-standard). Patent No. CN112176021-A; Derwent Master Accession Number: 2021-07659E. (2021)
41. Liu H, Zhang Z, Zhen P, Zhou M. High expression of Vstm2l induced resistance to chemoradiotherapy in rectal cancer through downstream il-4 signaling. J Immunol Res (2021) 2021:6657012. doi: 10.1155/2021/6657012
42. Smithson M, Irwin R, Williams G, Alexander KL, Smythies LE, Nearing M, et al. Sialyltransferase St6gal-1 mediates resistance to chemoradiation in rectal cancer. J Biol Chem (2022) 298(3):101594. doi: 10.1016/j.jbc.2022.101594
43. Nicolas AM, Pesic M, Engel E, Ziegler PK, Diefenhardt M, Kennel KB, et al. Inflammatory fibroblasts mediate resistance to neoadjuvant therapy in rectal cancer. Cancer Cell (2022) 40(2):168–84.e13. doi: 10.1016/j.ccell.2022.01.004
44. Li M, Xiao Q, Venkatachalam N, Hofheinz RD, Veldwijk MR, Herskind C, et al. Predicting response to neoadjuvant chemoradiotherapy in rectal cancer: From biomarkers to tumor models. Ther Adv Med Oncol (2022) 14:17588359221077972. doi: 10.1177/17588359221077972
45. Hsu KS, Adileh M, Martin ML, Makarov V, Chen J, Wu C, et al. Colorectal cancer develops inherent radiosensitivity that can be predicted using patient-derived organoids. Cancer Res (2022) 82(12):2298–312. doi: 10.1158/0008-5472.Can-21-4128
46. Flood M, Narasimhan V, Wilson K, Lim WM, Ramsay R, Michael M, et al. Organoids as a robust preclinical model for precision medicine in colorectal cancer: A systematic review. Ann Surg Oncol (2022) 29(1):47–59. doi: 10.1245/s10434-021-10829-x
47. Fleischmann M, Diefenhardt M, Nicolas AM, Rödel F, Ghadimi M, Hofheinz RD, et al. Aco/Aro/Aio-21 - capecitabine-based chemoradiotherapy in combination with the il-1 receptor antagonist anakinra for rectal cancer patients: A phase I trial of the German rectal cancer study group. Clin Transl Radiat Oncol (2022) 34:99–106. doi: 10.1016/j.ctro.2022.04.003
48. Park M, Kwon J, Kong J, Moon SM, Cho S, Yang KY, et al. A patient-derived organoid-based radiosensitivity model for the prediction of radiation responses in patients with rectal cancer. Cancers (Basel) (2021) 13(15):3760. doi: 10.3390/cancers13153760
49. Lee J, Kwon J, Kim D, Park M, Kim K, Bae I, et al. Gene expression profiles associated with radio-responsiveness in locally advanced rectal cancer. Biol (Basel) (2021) 10(6):500. doi: 10.3390/biology10060500
50. Le Compte M, Komen N, Joye I, Peeters M, Prenen H, Smits E, et al. Patient-derived organoids as individual patient models for chemoradiation response prediction in gastrointestinal malignancies. Crit Rev Oncol Hematol (2021) 157:103190. doi: 10.1016/j.critrevonc.2020.103190
51. Kryeziu K, Moosavi SH, Bergsland CH, Guren MG, Eide PW, Totland MZ, et al. Increased sensitivity to smac mimetic Lcl161 identified by longitudinal ex vivo pharmacogenomics of recurrent, kras mutated rectal cancer liver metastases. J Transl Med (2021) 19(1):384. doi: 10.1186/s12967-021-03062-3
52. Gillespie MA, Steele CW, Lannagan TRM, Sansom OJ, Roxburgh CSD. Pre-clinical modelling of rectal cancer to develop novel radiotherapy-based treatment strategies. Oncol Rev (2021) 15(1):511. doi: 10.4081/oncol.2021.511
53. Alkan A, Hofving T, Angenete E, Yrlid U. Biomarkers and cell-based models to predict the outcome of neoadjuvant therapy for rectal cancer patients. biomark Res (2021) 9(1):60. doi: 10.1186/s40364-021-00313-9
54. Park M, Kwon J, Shin H-J, Moon SM, Kim SB, Shin US, et al. Butyrate enhances the efficacy of radiotherapy Via Foxo3a in colorectal cancer patient-derived organoids. Int J Oncol (2020) 57(6):1307–18. doi: 10.3892/ijo.2020.5132
55. Costales-Carrera A, Fernández-Barral A, Bustamante-Madrid P, Domínguez O, Guerra-Pastrián L, Cantero R, et al. Comparative study of organoids from patient-derived normal and tumor colon and rectal tissue. Cancers (Basel) (2020) 12(8):2302. doi: 10.3390/cancers12082302
56. Clifford RE, Govindarajah N, Bowden D, Sutton P, Glenn M, Darvish-Damavandi M, et al. Targeting acid ceramidase to improve the radiosensitivity of rectal cancer. Cells (2020) 9(12):2693. doi: 10.3390/cells9122693
57. Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature (2018) 556(7702):457–62. doi: 10.1038/s41586-018-0024-3
58. Schumacher D, Andrieux G, Boehnke K, Keil M, Silvestri A, Silvestrov M, et al. Heterogeneous pathway activation and drug response modelled in colorectal-Tumor-Derived 3d cultures. PLoS Genet (2019) 15(3):e1008076. doi: 10.1371/journal.pgen.1008076
59. Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med (2015) 21(11):1350–6. doi: 10.1038/nm.3967
60. Luo X, Fong ELS, Zhu C, Lin QXX, Xiong M, Li A, et al. Hydrogel-based colorectal cancer organoid Co-culture models. Acta Biomater (2021) 132:461–72. doi: 10.1016/j.actbio.2020.12.037
61. Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of tumor-reactive T cells by Co-culture of peripheral blood lymphocytes and tumor organoids. Cell (2018) 174(6):1586–98.e12. doi: 10.1016/j.cell.2018.07.009
62. Cattaneo CM, Dijkstra KK, Fanchi LF, Kelderman S, Kaing S, van Rooij N, et al. Tumor organoid-T-Cell coculture systems. Nat Protoc (2020) 15(1):15–39. doi: 10.1038/s41596-019-0232-9
63. Kong JCH, Guerra GR, Millen RM, Roth S, Xu H, Neeson PJ, et al. Tumor-infiltrating lymphocyte function predicts response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. JCO Precis Oncol (2018) 2:1–15. doi: 10.1200/po.18.00075
64. Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol (2017) 35(6):569–76. doi: 10.1038/nbt.3836
65. O'Rourke KP, Loizou E, Livshits G, Schatoff EM, Baslan T, Manchado E, et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat Biotechnol (2017) 35(6):577–82. doi: 10.1038/nbt.3837
66. Fessler E, Drost J, van Hooff SR, Linnekamp JF, Wang X, Jansen M, et al. Tgfβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol Med (2016) 8(7):745–60. doi: 10.15252/emmm.201606184
Keywords: rectal cancer, organoids, patient-derived, precision medicine, treatment prediction, tumor microenvironment, cancer modeling
Citation: Yan Y, Cheong IH, Chen P, Li X, Wang X and Wang H (2022) Patient-derived rectal cancer organoids—applications in basic and translational cancer research. Front. Oncol. 12:922430. doi: 10.3389/fonc.2022.922430
Received: 18 April 2022; Accepted: 28 June 2022;
Published: 26 July 2022.
Edited by:
Francesca Negri, University Hospital of Parma, ItalyReviewed by:
J. Joshua Smith, Memorial Sloan Kettering Cancer Center, United StatesCopyright © 2022 Yan, Cheong, Chen, Li, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Wang, aHVpd2FuZ0BzaHNtdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.