- 1College of Biotechnology and Bioengineering, Zhejiang University of Technology, Hangzhou, China
- 2Laboratory of Tumor Molecular Diagnosis and Individualized Medicine of Zhejiang Province, Zhejiang Provincial People’s Hospital, Affiliated People’s Hospital, Hangzhou Medical College, Hangzhou, China
- 3Qingdao Medical College, Qingdao University, Qingdao, China
- 4Department of Hepatobiliary and Pancreatic Surgery, Zhejiang Provincial People’s Hospital, Affiliated People’s Hospital, Hangzhou Medical CollegeHangzhou, China
- 5The Key Laboratory of Tumor Molecular Diagnosis and Individualized Medicine of Zhejiang Province, Zhejiang Provincial People’s Hospital, Affiliated People’s Hospital, Hangzhou Medical College, Hangzhou, China
According to GLOBOCAN 2021 cancer incidence and mortality statistics compiled by the International Agency for Research on Cancer, hepatocellular carcinoma (HCC) is the most common malignancy in the human liver and one of the leading causes of cancer death worldwide. Although there have been great advances in the treatment of HCC, such as regofenib, sorafenib, and lomvatinib, which have been developed and approved for the clinical treatment of advanced or metastatic HCC. However, they only prolong survival by a few months, and patients with advanced liver cancer are susceptible to tumor invasion metastasis and drug resistance. Ubiquitination modification is a type of post-translational modification of proteins. It can affect the physiological activity of cells by regulating the localization, stability and activity of proteins, such as: gene transcription, DNA damage signaling and other pathways. The reversible process of ubiquitination is called de-ubiquitination: it is the process of re-releasing ubiquitinated substrates with the participation of de-ubiquitinases (DUBs) and other active substances. There is growing evidence that many dysregulations of DUBs are associated with tumorigenesis. Although dysregulation of deuquitinase function is often found in HCC and other cancers, The mechanisms of action of many DUBs in HCC have not been elucidated. In this review, we focused on several deubiquitinases (DUBs) associated with hepatocellular carcinoma, including their structure, function, and relationship to hepatocellular carcinoma. hepatocellular carcinoma was highlighted, as well as the latest research reports. Among them, we focus on the USP family and OTU family which are more studied in the HCC. In addition, we discussed the prospects and significance of targeting DUBs as a new strategy for the treatment of hepatocellular carcinoma. It also briefly summarizes the research progress of some DUB-related small molecule inhibitors and their clinical application significance as a treatment for HCC in the future.
Introduction
Liver cancer is a common cause of cancer death worldwide and is one of the ten cancers with a high incidence (1). Due to the asymptomatic nature of early hepatocellular carcinoma (HCC), HCC can only be evaluated by some early biomarkers in the patient’s body, such as serum α-fetoprotein (AFP) (2), Glypican-3 (GPC3) (3), and tumor-associated antigens (TAAs) (4). As a result, most patients are unable to detect and treat HCC at an early stage; moreover, HCC has a poor prognosis and a high mortality rate (5). For patients with early and intermediate HCC, surgical therapies such as hepatic resection and liver transplantation have good results (6). However, surgical therapy needs to consider factors such as the patient’s tumor stage and physical condition, so surgical therapy is not suitable for some patients. At present, systemic therapy and some adjuvant therapies of clinical surgery have become new research strategies for the treatment of HCC (6), such as transarterial chemoembolization (TACE), transarterial radioembolization (TARE), external beam radiation therapy, and oncolytic virus (7), but the effect of these treatments is not ideal. Systemic drug therapy has also become an important means of current liver cancer treatment (8). At present, many targeted drugs have been approved for the clinical treatment of HCV patients, for example, Nexavar (sorafenib), an oral drug first approved to target multiple kinases (9); regorafenib (Stivarga) was approved in June 2017 (10); and lenvatinib (11). These drugs all provide new treatment directions for HCC patients.
As we all know, the pathogenesis of human HCC is more complex, and an in-depth understanding of the molecular mechanism of HCC pathogenesis can provide an effective treatment strategy for improving the survival rate of HCC patients. At present, the development of targeted drugs provides new therapeutic prospects for the current treatment of HCC. Signaling pathways and potential targets related to the pathogenesis of HCC have become important methods for the development of drugs targeted for the treatment of advanced HCC (12). Studies have reported many key targets associated with HCC, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) (13), programmed cell death-1 and its ligands (PD-1/PD-L1) (14), hypoxia-inducible factor (HIF) (15), and deubiquitinases (DUBs) (16).
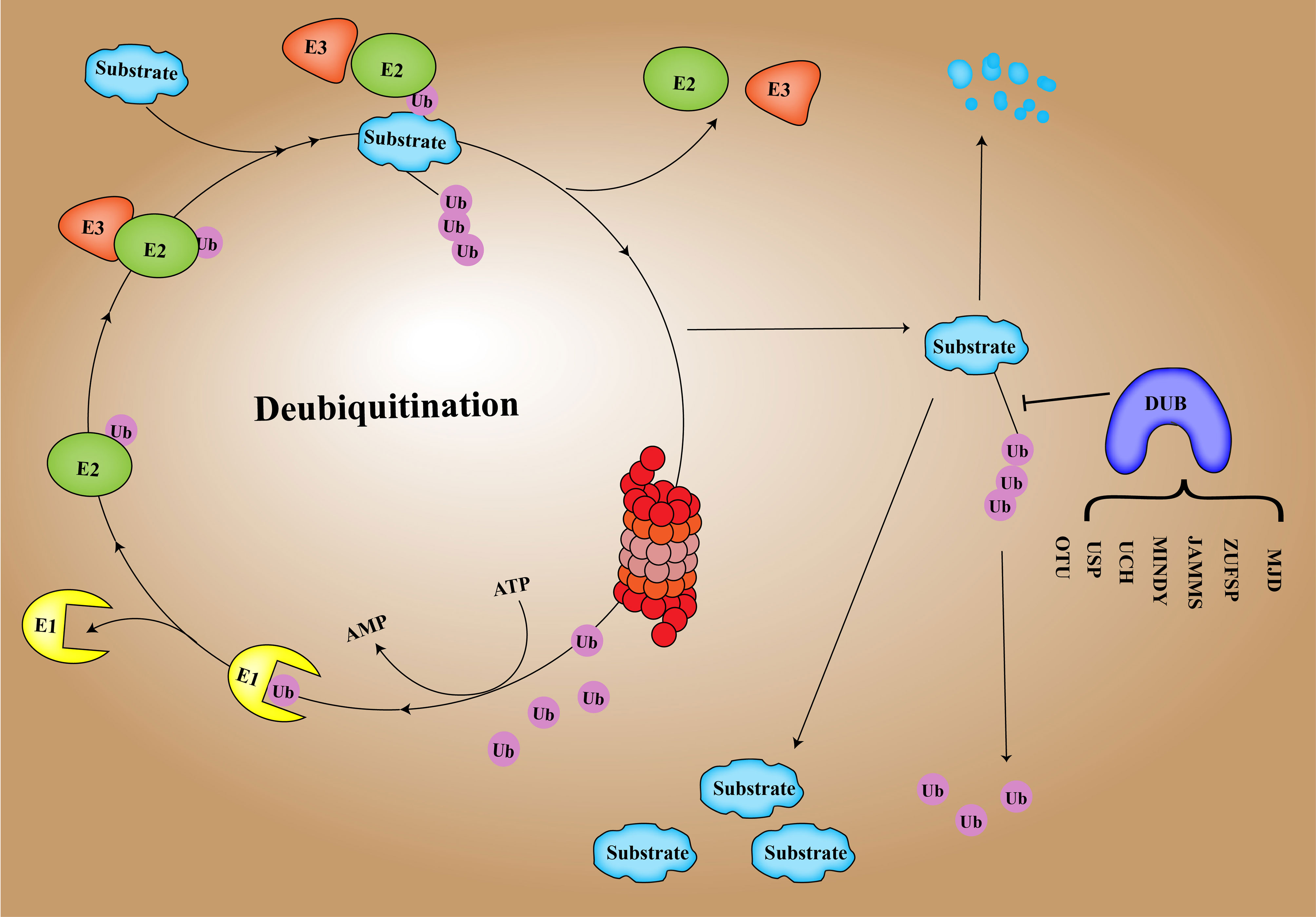
DUB is an important regulator of the process of deubiquitination and ubiquitination balance in human cells (17). Ubiquitination of proteins is a process in which multiple ubiquitin molecules are covalently attached to the protein substrate and then degraded by the 26S proteasome complex under the combined action of three types of enzymes: ubiquitin-activated enzyme (E1), ubiquitin-coupled enzyme (E2), and ubiquitin ligase (E3) (18, 19). Ubiquitination involves seven lysine residues: K6, K11, K27, K29, K33, K48, and K63 and N-Teline (Met1) (20). These residues can be ubiquitinated to form isopeptide-linked ubiquitin chains (21). DUBs include cysteine proteases as well as metalloproteinases that specifically cleave ubiquitin molecules in protein substrates (22). Regulating the homeostasis of ubiquitination and deubiquitination is conducive to the normal progress of human cell activities and maintains homeostasis in the human body (23). There are approximately 100 DUBs in humans, and DUB enzymes can be divided into 7 families based on structure and function (24), including ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), proteases containing the Machado–Joseph domain (MINDYs), ovarian tumor proteases (OTUs), newly discovered zinc finger protease (ZUPs/ZUFSPs), JAM/MPN domain-related Zn-dependent metalloproteinases (JAMMs), and Machado–Josephin domain-containing proteases (MJDs) (25) (Figure 1).
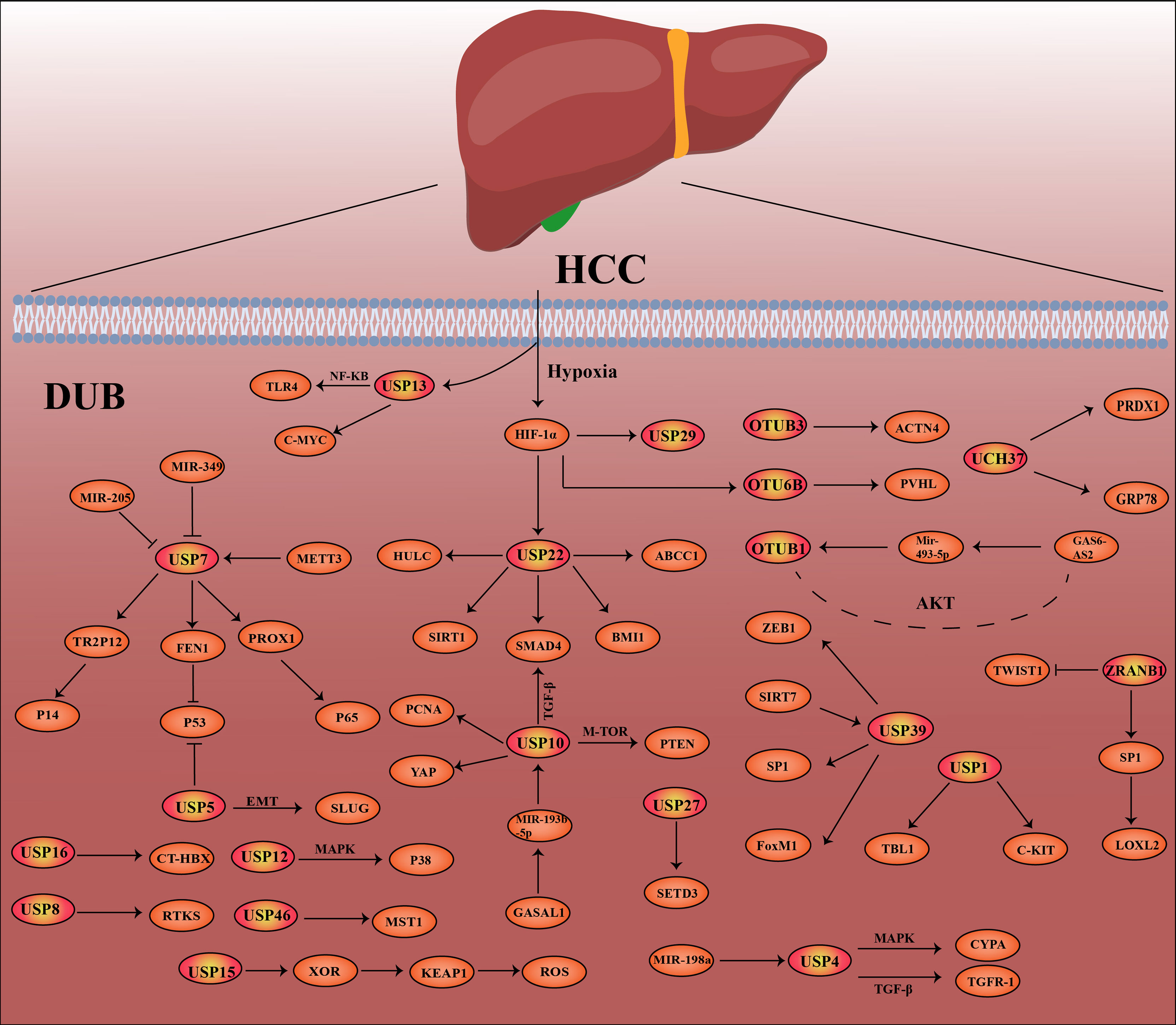
At present, a number of studies have shown that the deubiquitination effect of proteins is closely related to the occurrence and development of cancer, such as breast cancer, lung cancer, stomach cancer and hepatocellular carcinoma (26–30). In this review, we highlight hepatocellular carcinoma -related DUBs, including their structure, mechanisms of action in hepatocellular carcinoma, and recent research advances. In Figure 2, the related pathways and target proteins of DUBs in HCC are shown (Figure 2). Last but not least, we discussed the prospects and implications of DUBs and DUB-related small molecule inhibitors as potential protein targets for hepatocellular carcinoma treatment.
Ovarian Tumor Protease OTU
There are 16 species of cysteine protease OTU family members, which can be divided into four different subfamilies: OTUB subfamilies (OTUB1 and OTUB2), OTUD subfamilies (OTUD1, OTUD2/YOD1, OTUD3, OTUD4, OTUD5/DUBA, OTUD6A, OTUD6B, and ALG13), A20-like subfamilies (A20, Cezanne, Cezanne2, TRABID, and VCPIP), and OTULIN subfamily (OTULIN) (31). Studies have shown that the Cys catalytic residues present in the OTU subfamily protease active site make it susceptible to reverse oxidation (32). Here, we introduce the structure and function of “OTUB1 OTUD3 OTUD6B ZRANB1” in the OTU family and the research progress in HCC.
OTUB1
Structure of OTUB1
OTUB1 is a founding member of the ovarian tumor (OTU) domain family of DUBs and belongs to the OTUB subfamily (33). In addition to the OTU domain containing 130 amino acids, OTUB1’s unique crystal structure has two different ubiquitin-binding sites (34). During the deubiquitination process, OTUB1 preferentially cleaves the polyubiquitin chains connected by Lys (34, 35) while using the active center to catalyze the substrate reaction. The catalytic domain of OTUB1 consists of three parts: Cys(C)91, His(H)265, and Asp(D)268 (36). Studies have shown that in the presence of free ubiquitin molecules, the activity of the OTUB1 enzyme is regulated by the E2 enzyme: the uncharged E2 enzyme can activate the activity of the OTUB1 enzyme by stabilizing the N-terminal structure of OTUB1. In addition, OTUB1 is able not only to remove the ubiquitin molecules linked to the substrate but also to inhibit the ubiquitination process through binding to the E2 enzyme (37).
The Function of OTUB1 and Research Progress in Hepatocellular Carcinoma
OTUB1 is expressed in a variety of tissues in the body, such as the kidneys, colorectum, stomach, brain, and liver (38). In human liver cancer and other tumor tissues, OTUB1 has been shown to have a high expression and is associated with a poor prognosis in patients (38, 39). Inhibiting the expression of OTUB1 by shRNA will weaken the proliferation, migration, and invasion ability of HCC cells (38).
LncRNAs are a class of RNAs that are not protein-coding and can bind to downstream MIR genes through endogenous competition and targeted action (40, 41). It is widely believed to be associated with many diseases in the human body and is also a related causative agent of cancer (42). OTUB1 is also associated with lncRNA in liver cancer. LncRNA GAS6-AS2 was shown to be upregulated in liver cancer cells as well as tissues. GAS6-AS2 regulates the expression of downstream OTUB1 by targeting miR-493-5p with 3′UTR (39). The hyperactivated PI3K/Akt signaling pathway plays a central role in cancer cell metabolism and is also thought to be associated with the occurrence of HCC as well as metastasis (43, 44). LncRNA GAS6-AS2 knockdown can promote HCC cell proliferation, invasion, metastasis, and apoptosis by mediating the miR-493-5p/OTUB1 axis to activate the PI3K/AKT/FoxO3a pathway (39). The above studies show that OTUB1 can be used as a novel marker for targeted therapy for liver cancer.
OTUD3
Structure of OTUD3
OTUD3 belongs to the subfamily OTUD and is structurally similar to OTUD2. Its active domain is approximately 52–209 amino acids, which also includes the DUB family’s classic catalytic triplet residues (31). During the deubiquitination process, OTUD3 is the only DUB that tends to cleave k6-linked double ubiquitin and bind it to the S1 and S1′ sites (31). The Lys6-linked polyUb is a mysterious type of chain whose role in cells has not yet been elucidated (45).
The Function of OTUD3 and Research Progress in Hepatocellular Carcinoma
A growing number of reports suggest the role of OTUD3 in human cancers, such as breast cancer as well as lung cancer (35, 46). Studies have demonstrated that OTUD3 is expressed in high amounts in HCC tissues and is associated with a poor prognosis in HCC patients (47). α-Actin 4 (ACTN4) is called an actin-binding protein and belongs to a family of actin-binding proteins. OTUD3 can affect the expression of α-actin 4 (ACTN4) at the protein level and promote the proliferation, invasion, and metastasis of HCC by stabilizing ACTN4 by deubiquitination (47).
Other Enzymes of the OTU Family
OTUD6B
Studies have shown that OTUD6B can regulate HCC metastasis by regulating the activity of HIF under hypoxic conditions. Mechanistically, OTUD6B directly interacts with pVHL and enhances its stability. In human HCC tissues, the protein level of OTUD6B was positively correlated with pVHL, whereas HIF-1α and vascular endothelial growth factor were negatively correlated. This study demonstrates that OTUD6B is a direct transcriptional target of HIF-1α, providing a new strategy for targeting hypoxic microenvironments for HCC therapy (48).
ZRANB
ZRANB1 overexpression was associated with poorer survival in patients with HCC, and there was a significant positive correlation between the expressions of ZRANB1 and LOXL2 in clinical HCC specimens, which can regulate the expression of LOXL2 through specific protein 1 (SP1). Mechanistically, ZRANB1 stabilizes and binds SP1 through deubiquitination, which promotes liver cancer progression (17). However, another study reported that the deletion or downregulation of ZRANB1 was closely associated with the recurrence, metastasis, tumor volume, and disease stage of liver cancer significantly increased. Knockdown of ZRANB1 promotes HCC growth and metastasis by regulating Twist1 K63 ubiquitination (49).
Ubiquitin-Specific Protease
The USP family is the most frequently studied DUB family and is a large family of more than 60 DUBs. The USP protein is considered an antagonist of the E3 ligase and is a potential target for cancer treatment (50). Here, we introduce the structure and function of “USP14, USP1, USP10, USP39, USP22, USP9X, USP2, USP7, USP4, USP5, USP29, USP15, USP12, USP16, USP27, USP46, and USP8” in the USP family research progress in HCC.
USP14
Structure of USP14
The full length of the protein sequence of USP14 contains 494 amino acids. Its structure can be roughly divided into the N-terminal ubiquitination active center and C-terminal deubiquitination catalytic activity domain. The N-terminus has a 9-kDa ubiquitin-like (Ubl) domain, which is an important regulator of proteasome activity (51, 52); the C-terminus is a 45-kDa catalytic domain responsible for its DUB activity (53). The catalytic domain of USP14 is similar to the structure of the HAUSP catalytic core domain, which is an extended right hand consisting of three domains of fingers, palm, and thumb (51). When the apolipoprotein USP14 binds to the proteasome, the conformation of the two surface rings (BL1 and BL2) changes to bring the ubiquitin C-terminus into the catalytically active site (54, 55).
The Function of USP14 and Research Advances in Hepatocellular Carcinoma
Many studies have shown that USP14 can be involved in modulating a variety of signaling pathways associated with human diseases, such as cancer, autophagy, immune response, and viral infections (56, 57). In HCC, USP14 is highly expressed in liver cancer and is associated with a poor prognosis in patients with HCC. In the hypoxic environment of liver cancer (58), USP14 can enhance the transcriptional activity of HIF-1α and the stability of HIF-1α through deubiquitination, which in turn promotes the migration and invasion of HCC cells in a HIF-1α-dependent manner (59). This suggests that USP14 is a potential diagnostic biomarker for HCC as well as a therapeutic target. IU1, an inhibitor of USP14, can significantly inhibit the proliferation of liver cancer cells and liver cancer tissue tumors. It can be used as a potential HCC treatment agent in vivo and in vitro.
USP1
Structure of USP1
USP1 regulates cellular DNA repair processes (60). USP1 has highly conserved USP domains of His and Cys and also has a catalytic triad consisting of C90, H593, and D751 (61). The protein sequence of USP1 consists of 785 amino acids, and the protein molecular weight is about 88.2 kDa (62). The cofactor UAF1 is a related factor of USP1 (63), which regulates the activity of USP1 isopeptidase by combining with UAF1 into a unique exogenous dimer complex. The enzyme activity of USP1 alone is low, but the activity of the enzyme is increased when combined with UAF1 (64, 65).
The Function and Research Progress of USP1 in Hepatocellular Carcinoma
In addition to being a regulator of cellular DNA repair (60), USP1 is also involved in the occurrence and development of various human diseases, such as USP1, plays a key role in the Fanconi anemia pathway (60), is a potential target for differentiation therapy (66), is upregulated in breast cancer, and is associated with poor patient prognosis (67). USP1 can also affect the development of lung cancer by regulating the PHLPP1-Akt signaling axis (68). In liver cancer, USP1 is thought to play a key role in the immune infiltration process of tumors. Drugs such as pimozide and ML-323 can inhibit the promotion of USP1 on the cell cycle and proliferation of HCC (69).
Ribosomal protein S16 (RPS16) is a highly conserved 40S ribosomal protein, which has been reported to be highly expressed in various cancers, such as colorectal cancer (CRC) (70). Studies have shown that USP1 can promote the stability of RPS16 protein and promote the proliferation and migration of liver cancer cells by binding to the cys90 (C90) site at the N-terminus of UAF1 (a cofactor of USP1) (71).
Protein transduction protein (TBL1) is a key regulator of the Wnt pathway and is proven to be associated with tumors in several studies, such as in cervical (72), prostate (73), and ovarian cancers (74). In liver cancer, USP1 can maintain the survival of hepatic circulating tumor cells by deubiquitinating and stabilizing TBL1 protein (75).
Lenvatinib (Lenvima) is an oral small-molecule inhibitor of multiple receptor tyrosine kinases for the treatment of advanced liver cancer patients (76). However, most patients will develop resistance to lenvatinib (77), so research on the mechanism of drug resistance in patients will help the development of targeted therapy for liver cancer (78). USP1 can promote the proliferation and migration of HCC cells by promoting the expression and stability of c-kit protein, and USP1 also promotes the efficacy of lenvatinib in HCC (79). In conclusion, USP1, as a novel diagnostic and predictive marker in the treatment of liver cancer, can provide new ideas for the development of targeted drugs for liver cancer treatment.
USP10
Structure of USP10
USP10 is a cysteine protease of approximately 798 amino acids in length and is a highly conserved protein in eukaryotes (80). The catalytic domain of USP10 is located at 415 amino acids at the N-terminus of the protein and is about 380 amino acids in size. USP10 can remove Ub from the target protein by undergoing a hydrolysis reaction (80).
The Function of USP10 and Research Progress in Hepatocellular Carcinoma
USP10 is involved in many physiological activities in the human body, such as promoting cell proliferation and differentiation by targeting p53 protein (81); USP10 can activate the downstream protein AMPK through deubiquitination and form a feedforward loop with it (82). In addition, USP10 is also a tumor-related factor in human lung cancer (83), CRC (84), liver cancer, etc. (30). In HCC, multiple studies have shown that the transforming growth factor β (TGF-β) pathway is closely related to the metastasis of HCC (85, 86). USP10 can directly bind to Smad4 and act on the Lys-48-linked polyubiquitin chain on Smad4 to stabilize it; USP10 regulates the abundance and function of Smad4 protein through deubiquitination and activates the TGF-β pathway to further promote the migration of hepatoma cells (87). In addition, the USP10 inhibitor Spautin-1 can inhibit HCC metastasis in a dose-dependent manner, which makes it a targeted drug for effective anti-metastatic agents in the treatment of HCC.
mTOR signaling is highly expressed in liver cancer and other cancers (88). PTEN and AMPKα signaling pathways are regulators upstream of mTOR activation (89). USP10 acts as a tumor suppressor and acts as a tumor suppressor protein in HCC. USP10 stabilizes PTEN and AMPKα in HCC cells through deubiquitination and can inhibit AKT 329 phosphorylation and mTORC1 activation in HCC cells, thereby inhibiting the mTOR pathway (90).
A study showed that USP10 interacts with lncRNA GASAL1 to promote the malignancy of HCC (91). Mechanistic analysis revealed that lncRNA-GASAL1 could upregulate USP10 expression by targeting downstream miR-193b-5p through competitive binding. In addition, USP10 can stabilize proliferating cell nuclear antigen (PCNA) through deubiquitination to enhance the proliferation and migration of hepatoma cells (92).
YAP protein is a regulator found in Drosophila to control organ size (93, 94). Studies have shown that the Hippo-YAP/TAZ pathway is closely related to human metabolism, organ regeneration, and cancer (95–97). In HCC, USP10 was shown to activate YAP/TAZ protein and stabilize its activity through deubiquitination. USP10 can upregulate the abundance of YAP/TAZ protein in HCC and promote the proliferation and migration of HCC in vivo and in vitro (30). These provide new ideas and research proof for the mechanism of USP10 in HCC.
USP39
Structure of USP39
Family member ubiquitin-specific peptidase 39 (USP39) is the homolog of Sad1p in yeast, also known as the human 65-kDa SR-related protein (98, 99). The structure of USP39 includes a central zinc finger ubiquitin domain and a canonical UCH domain (100). Studies have shown that there are no active site residues of cysteine and histidine in the structure of USP39, so there is no DUB enzyme activity (101), and it is also classified as a DUB (99).
The Function and Research Progress of USP39 in Hepatocellular Carcinoma
USP 39 (USP39) is an important regulator of human mRNA splicing and is highly expressed in a variety of cancers (100, 102). New research shows that USP39 plays a key role in the occurrence and development of liver cancer. The Kaplan–Meier analysis found that the high expression of USP39 in liver cancer was closely related to the poor prognosis of patients. USP39 may promote the malignancy of liver cancer by participating in the regulation of the epithelial–mesenchymal transition (EMT) pathway of HCC. ZEB1 is a key factor in the human tumor EMT pathway (103, 104). Mechanistic studies suggest that USP39 stabilizes ZEB1 protein through deubiquitination and activates the development of the EMT pathway and the proliferation and migration of hepatoma cells (105).
USP39 can directly bind and interact with the ubiquitinated E3 ligase TRIM26 (105). Studies have shown that the E3 ligase TRIM26 can inhibit the occurrence and development of several tumors in humans (106). USP39 and TRIM26 promote HCC progression through antagonism to balance the expression level of ZEB1 (105).
USP39 can be acetylated by the acetyltransferases HAT and MYST1. Acetylated USP39 can be degraded by E3 ubiquitin ligase (VHL)-mediated proteasome (107). SIRT7 has been reported to be an oncogenic potential factor in HCC and can form a regulatory loop with miRNAs to promote HCC progression (108). In the development of hepatoma cells, SIRT7 can deacetylate USP39, which improves the stability of USP39 and promotes the proliferation of HCC (107).
FoxM1 is widely recognized as a key factor in the transcriptional regulation of human cancers (109). It can promote the occurrence and development of HCC by regulating the expression of KIF4A (110). USP39 has been reported to promote the cleavage of forkhead box protein M1 (FoxM1) in hepatoma cells to promote the occurrence and development of HCC (111); USP39 knockdown can also induce apoptosis by targeting FoxM1 shear force on mRNA and promote the growth of hepatoma cell SMMC-7721 in vitro and in vivo (112).
Specific protein 1 (SP1) belongs to the Sp/KLF transcription factor family (113) and is considered to be the basal transcription factor in humans. Sp1 is also associated with a variety of human diseases, such as Huntington’s disease (113, 114). SP1 is also associated with poor prognosis in a variety of cancers (115). Studies have shown that USP39 can stabilize Sp1 and prolong its half-life through deubiquitination in HCC (116). In addition, USP39 can also promote the SP1-dependent pathway. Therefore, USP39 can target Sp1 to promote liver cancer cell proliferation (116).
USP22
Structure of USP22
USP 22 (USP22) is an important member of the USP family. Its protein structure consists of 525 amino acids, including the structural sequence of a putative ubiquitin hydrolase containing a C-terminal peptidase domain and an N-terminal UBP-type zinc finger motif (117). In addition, USP22, ATXN7L3, ATXN7, and ENY2 are transcriptional cofactors of human Spt-Ada-Gcn5 acetyltransferase (hSAGA) and key subunits of the SAGA complex (118, 119).
The Function and Research Progress of USP22 in Hepatocellular Carcinoma
As an important member of the USP family, USP22 also plays a very important role in the occurrence and development of HCC. Among them, the expression of USP22 and survivin was shown to be closely related to the malignant behavior of HCC cases, including tumor size, stage, and differentiation (120). Several studies have reported that USP22 is closely related to the drug resistance mechanism of HCC. For example, in sorafenib-resistant cell lines, USP22 can regulate and upregulate ABCC1 (121). In addition, USP22 can directly interact with SIRT1 and regulate the protein expression level of SIRT1, which promotes the resistance of hepatoma cells to 5-fluorouracil (5-FU) (122). Previous reports have demonstrated that SIRT1 can deacetylate and activate the AKT pathway (123), and USP22 can promote MDR in HCC cells by activating the SIRT1/AKT/MRP1 pathway (124); USP22 is also able to regulate chemotolerance in HCC through Smad4/Akt-dependent MDR-related gene regulation (117). Relevant drug resistance genes include BMI1 and EZH2. Co-expression of USP22 and BMI1 is associated with poor prognosis and enhanced anticancer drug resistance in HCC (125). Some researchers have proposed a self-activating cascade reaction—the co-delivery system of sorafenib and shUSP22 (Gal-SLP), aiming at the effect of USP22 on the drug resistance of liver cancer cells. This delivery system exhibits potent antitumor efficiency through three synergistic effects (126). This is also a major advance in the use of DUBs for the treatment of HCC. We presume that with the in-depth study of DUB, DUBs can provide new approaches and strategies for the treatment of cancer in humans. In addition to affecting the drug resistance of liver cancer cells, USP22 can also regulate peroxisome proliferator-activated receptor γ (PPARγ) in HCC through deubiquitination to promote fatty acid synthesis and tumorigenesis. These findings provide a new therapeutic strategy for patients with high USP22 expression in HCC (127).
In addition to the effect on drug resistance of liver cancer cells, other studies have also reported that USP22 can significantly affect the glycolysis and stemness characteristics of liver cancer cells under hypoxic conditions: HIF-1α knockdown inhibits USP22-induced and hypoxia-induced effects (128). USP22 can also affect the transcription of the phosphatase DUSP1 by E2F6 protein through deubiquitination, which can activate the AKT pathway in hepatoma cells (129). In addition, USP22 can also be regulated by lncRNA HULC to further affect the drug resistance and tumor growth of liver cancer cells (130, 131).
Other Enzymes of the USP Family
In addition to the abovementioned USP family DUBs, there are other liver cancer-related USP family DUBs, including USP9X, USP2, USP7, USP4, USP5, USP29, USP15, USP12, USP16, USP27, USP46, and USP8.
Their Function and Research Progress in Hepatocellular Carcinoma
USP9X
USP9X has been proved by many studies to affect the occurrence and development of HCC (132). For example, by promoting HCC cell proliferation by regulating the expression of β-catenin (eta-catenin) (133), USP9X is able to affect hepatoma cells with ARID1A mutations through the AMPK pathway (134), miR-26b can regulate USP9X-mediated p53 deubiquitination to enhance the sensitivity of HCC cells to doxorubicin (135), miR-26b is also able to target USP9X expression to suppress EMT in hepatocytes (136), and usp9x can affect the drug sensitivity of hepatoma cells to doxorubicin and WP1130 through p53 (137). The lncRNA LINC00473 is also able to exert its oncogenic function in HCC by interacting with USP9X and may be a therapeutic target for HCC treatment (138).
USP2
USP2a is significantly upregulated in HCC tissues and positively correlated with poor patient prognosis, and USP2a can promote HCC progression by deubiquitinating and stabilizing RAB1A (139). In addition, USP2a is also believed to be involved in the production of nascent adipose to further regulate the progression of HCC, which has pathogenic and prognostic significance for HCC (140). USP2b has been shown to be dysregulated in HCC patients, promoting apoptosis and necrosis of HepG2 and Huh 7 cells. This study demonstrates that USP2 contributes to the pathogenesis of HCC and provides a molecular basis for the development of HCC therapies by modulating USP2b expression or activity (141).
USP7
The expression of USP7 is significantly increased in HCC and has been reported to have clinical significance in the prognosis and functional mechanism of HCC (142). USP7 may be a drug target for chemoresistance in HCC (143). MicroRNA-205 (miR-205) may negatively regulate the UPS7 protein level by targeting the 3′-untranslated region in HCC cells (144). Adipocyte-secreted exosomal circRNAs promote tumor growth and reduce DNA damage by inhibiting miR-34a and activating USP7/Cyclin A2 signaling pathway (145). METTL3 can regulate the expression of USP7 through m6A methylation and promote the invasion, migration, and proliferation of HCC cells (146). Furthermore, the homolog of Usp7, HAUSP, is able to regulate the Hippo pathway and stabilize Yorkie (Yki) and HAUSP as potential therapeutic targets for HCC (147). USP7 can also bind to FEN1, a poor prognostic molecule in HCC through deubiquitination, which can reduce the expression of p53 and promote the progression of HCC (148). In liver cancer, PROX1 can also enhance the stability of p65 by binding USP7 to affect angiogenesis in liver cancer cells (149). USP7 promotes HCC cell growth by forming a complex with thyroid hormone receptor-interacting protein 12 (TRIP12) and stabilizing p14 (ARF) ubiquitination, thereby promoting HCC progression (150).
USP4
Kaplan-Meier survival analysis showed that patients whose tumors overexpressed USP4 had poor overall survival, and it combined with cyclophilin A (CypA) to form a complex to activate the MAPK signaling pathway in HCC (151). In addition, USP4 can directly interact with TGF-β receptor type I (TGFR-1) through deubiquitination and activate the TGF-β signaling pathway, which can induce EMT in hepatoma cells, providing a new therapeutic target for the treatment of HCC (152). USP4 is able to act as a downstream target of miR-148a in hepatoma cells, and overexpression may contribute to the progression of HCC to more aggressive features (153).
Others
USP5 has been reported to be highly expressed in human hepatoma cells and can inhibit the expression of p53 and DNA repair function (154). It also binds to SLUG and regulates the EMT pathway associated with hepatoma cells (155).
The expression of USP13 was significantly upregulated in HCC cells, and studies showed that USP13 knockdown could inhibit the activation of the TLR4/MyD88/NF-κB pathway in hypoxia-induced HCC cells. In addition, studies have shown that USP13 can affect the growth of liver cancer cells by regulating the expression of c-Myc (156, 157).
Studies have shown that USP29 is related to HIF-1α in hepatoma cells. Mechanistically, USP29 promotes sorafenib resistance in HCC cells by upregulating glycolysis, thus opening a new avenue for therapeutic targeting of patients with sorafenib-resistant HCC (158).
USP15 is highly expressed in liver cancer tissues and cell lines, and high expression is significantly positively correlated with HCC recurrence. Studies have shown that downregulation of USP15 expression can inhibit the proliferation and apoptosis of liver cancer cells (159). In addition, xanthine oxidoreductase (XOR) can interact with USP15 to enhance the stability of Kelch-like ECH-associated protein 1 (KEAP1), which ultimately promotes the accumulation of reactive oxygen species (ROS) and liver cancer stem cells (CSCs) (160).
USP12 promotes HCC proliferation and apoptosis by affecting p38 and MAPK pathways (161). USP16 is downregulated in HCC, leading to Ct-HBx promoting the tumorigenicity and malignancy of HCC (162). USP27 promotes its stability by interacting with SETD3 and accelerates the growth of hepatoma tumor cells, and higher expression of USP27 and SETD3 predicts poorer survival in HCC patients (163). USP46 can promote MST1 kinase activity through deubiquitination to inhibit tumor growth and metastasis, suggesting that USP46 may be a potential therapeutic strategy for HCC (164). USP8 can regulate the expression of multiple receptor tyrosine kinases (RTKs) to affect the drug resistance of liver cancer cells (165).
Ubiquitin C-Terminal Hydrolase
The family of UCHs includes UCH-L1, UCH-L3, UCHL5/UCH37, and BRCA1-associated protein-1 (BAP1) (166). The UCH family has a classically conserved catalytic domain of about 230 amino acids in size (167). The domains of UCH-L5, UCH-L1, and UCH-L3 contain an active site crossover loop (116, 166). UCHL1 has been reported to be strongly associated with Parkinson’s disease (PD) (168, 169) and Alzheimer’s disease (AD) in humans (170). In view of the lack of current research reports on the UCH family, here we only introduce the structure and function of UCH37 and the research progress in HCC.
UCH37
Structure of UCH37
UCH37, also known as UCHL5, belongs to the human UCH family and is the only DUB in the family that is associated with the mammalian proteasome (171, 172). The protein structural sequence of the protease UCH37 contains 329 amino acids and is mainly associated with the Ub isopeptidase activity in the 19S proteasome regulatory complex (173). It is also the only UCH family of proteases capable of acting on the 19S proteasome complex and cleaving Lys48-linked polyubiquitin molecules in a unique manner (174). The three-dimensional structure of UCH37 consists of two parts, a globular UCH domain and a fibrillar unique C-terminal extension (175). Studies have shown that NFRKB can inhibit its activity by interacting with the extended structure of the C-terminus of UCH37 (173). During deubiquitination, UCH37 is able to associate with the 26S proteasome via Rpn13.
The Function and Research Progress of UCH37 in Hepatocellular Carcinoma
Studies have shown that UCH37 is highly expressed in liver cancer cells (HCC) and cancer tissues, and the prognosis of patients is poor (176). Peroxiredoxin 1 (Prdx1) belongs to the peroxidase family and plays a dual role in human tumorigenesis (177). Multiple studies have shown that Prdx1 is involved in the progression of human liver cancer, including tumor angiogenesis (178), apoptosis, autophagy (179), and poor patient prognosis in HCC (180). PRDX1 low expression can promote the proliferation, migration, and invasion of HCC cells in vitro. New research shows that the interaction of Prdx1 with UCH37 attenuates the effects of UCH37 on cell migration and invasion; this interaction may be through the formation of a complex rather than the deubiquitination of UCH37 itself, but the mechanism of the two on the development of liver cancer has not yet been elucidated (181). UCH37 can also act on the RNA splicing factor PRP19 through deubiquitination (182), and their interaction can promote HCC migration and invasion (176).
The protein chaperone GRP78 is often highly expressed in human cancers (183), such as lung cancer (184), pancreatic cancer (185), and breast cancer (186). GRP78 is also associated with diseases such as tumor resistance, patient prognosis (187), M2 macrophage polarization (188), and folding of nervous system proteins (189). The latest study shows that UCH37 can interact with the protein chaperone GRP78 by co-immunoprecipitation and confocal laser scanning microscopy, which provides new ideas and directions for the mechanism of UCH37 in HCC (190).
DUB-Related Inhibitors
As we know, proteasome inhibitors have been developed and used successfully in the treatment of some diseases (191, 192), which lays the foundation for the development of DUB as a drug research target. Currently reported inhibitors such as PR-619 and WP1130 can inhibit a variety of DUBs, of which WP1130 inhibits at least five DUBs: USP5, UCH-L1, USP9X, USP14, and UCH37 (193). However, the development of specific inhibitors has been challenging, which is related to the highly conserved structural features of the DUB catalytic site.
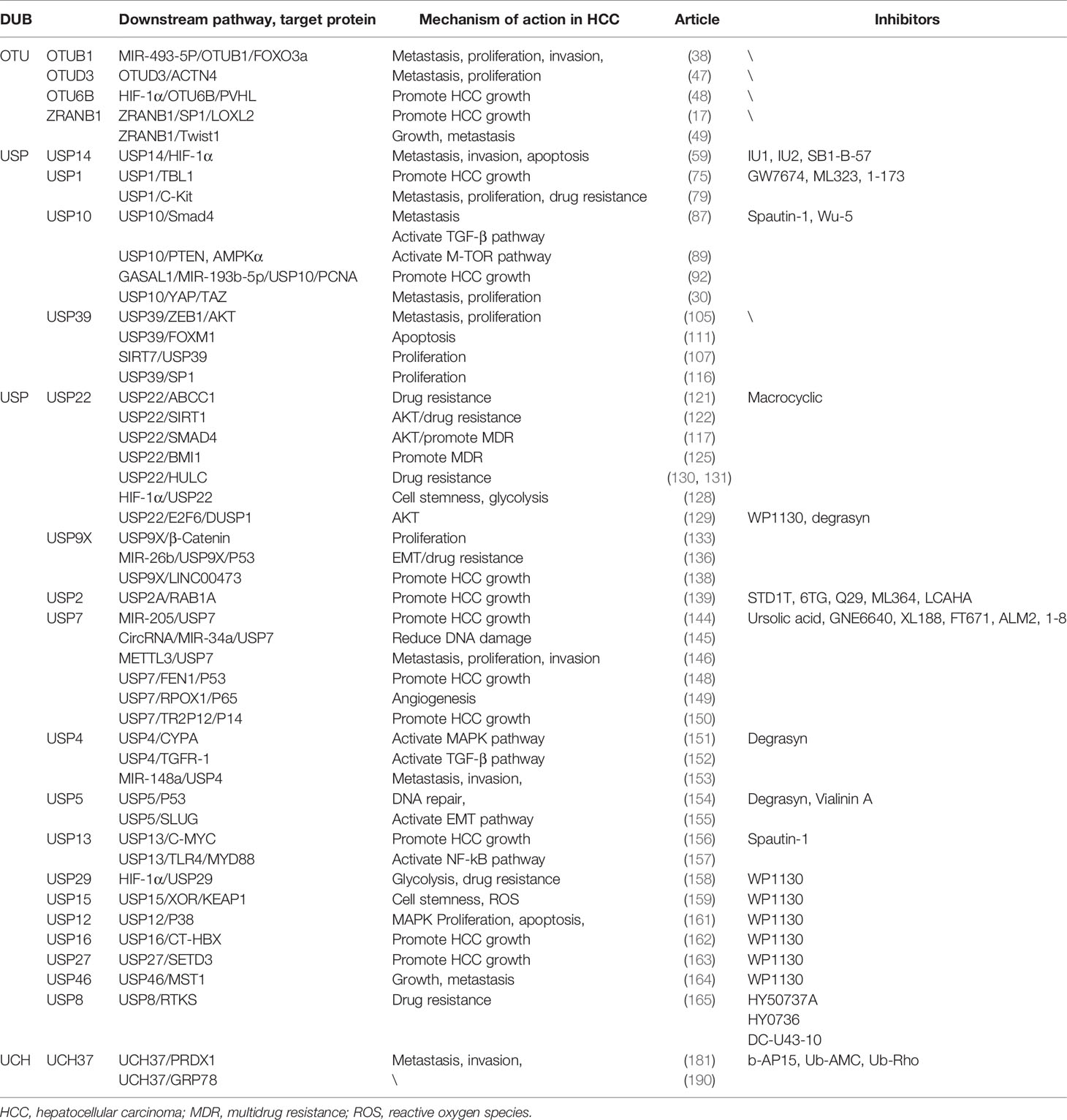
In the DUB family, UPS has been clearly regarded as one of the most important drug targets, and the research and development of inhibitors are more in-depth than those of other families. Among them, USP14 has been researched and developed as a more mature inhibitor. The research group of Finley et al. identified more than two hundred inhibitors of USP14 based on high-throughput screening of Ub-AMC hydrolysis assays; IU1 is the first specific inhibitor targeting USP14, named IU1 (194). In addition, other inhibitors have been developed: IU1 analogs, such as IU1-206 and IU1-248 and IU2 series (195); spautin-1 is a small molecule inhibitor of USP10 and also inhibits USP13 (196). Recent research reports identify Wu-5 as a novel USP10 inhibitor that induces degradation of the FLT3 mutant protein (197). A ubiquitin variant (UbV) phage library has also been used to develop an inhibitor-UbV.7.2 that can target USP7 and USP10, which structurally enhances the affinity for USP7 (198). A leukemia drug, 6-thioguanine, was found to be a potent inhibitor of USP2, exhibiting a non-competitive and slow-binding inhibitory mechanism for USP2 (199). Studies have reported that inhibitors of USP8 include RA-9, DUB-IN-1, DUBs-IN-2, and a novel inhibitor DC-U43 (200–202). Among them, DC-U43-10 is a USP8 inhibitor with a novel scaffold, which can bring new research directions for the development and clinical research of USP8 inhibitors (203). Morgan et al. screened a large number of cyclic peptide combinatorial libraries and identified the first inhibitors of USP22, which have broad prospects for development (204). Furthermore, WP1130 is a general inhibitor. UCH37-specific inhibitors have not yet been developed, but there are some non-specific DUB inhibitors targeting UCH37 activity, such as b-AP15, Ub-AMC, Ub-Rho, and WP1130 (195). Although UCH37 lacks specific inhibitors, the developed multi-target inhibitors can also provide new strategies and ideas for clinical drug development. We summarize some inhibitors of DUB in Table 1.
Summary and Outlook
Based on the introduction and analysis of some liver cancer-related DUBs in this paper, it can be seen that DUBs play a unique regulatory role in the occurrence and development of HCC.
However, the regulatory mechanism of DUBs in liver cancer is relatively complex, involving many pathways and targets, and the development of targeted drugs has become an important treatment method for patients with high DUB expression in HCC. At present, molecularly targeted drugs and small molecule inhibitors for ubiquitination and deubiquitination-related enzymes have been used in the clinical treatment of cancer (205, 206). Drugs such as oprozomib, ixazomib, and bortezomib have achieved remarkable therapeutic results (207). In the article, we also summarize some inhibitors of liver cancer-related DUBs. The current research results show that the USP family-related inhibitors are widely studied. Represented by USP14, the research on IU1 is relatively mature and has great potential for clinical application. USP22, USP14, USP10, USP13, USP7, USP2, and USP8, liver cancer-related DUBs, have also been reported to have related small molecule inhibitors, but the research and development are not mature enough. Using a large library of cyclic peptides in high-throughput screening, researchers recently identified the first inhibitors of USP22—macrocyclic inhibitors. In addition, UCH37 of the UCH family also has small molecule inhibitors, but these are not specific inhibitors of UCH37. Due to the unique active structure of DUB, there are still some difficulties in the development of many small molecule inhibitors. In conclusion, the prospect of DUB inhibitors as drug targets is still very impressive, and it will also have great clinical application significance for the treatment of human diseases in the future.
In this article, we introduce that OTUB1 OTUD3 OTUD6B ZRANB1 USP14, USP1, USP10, USP39, USP22, USP9X, USP2, USP7, USP4, USP5, USP29, USP15, USP12, USP16, USP27, USP46, USP8, and UCH37 can affect the malignant degree of HCC through the corresponding mechanism. Among them, USP22, USP1, and USP9X were all related to drug resistance to HCC; USP14, USP13, USP29, and OTU6B were all related to the hypoxic microenvironment and HIF in HCC. As can be seen, the relationship between the USP family and liver cancer is currently the most frequently studied with the participation of a class of DUBs, and many corresponding small molecule inhibitors have also been studied for such DUBs. Therefore, we presume that the USP family is the most promising biomarker for DUB for the diagnosis and treatment of liver cancer. Among them, USP14 small molecule inhibitors are the most clinically significant drug targets. However, the research progress on OTU and UCH family in liver cancer is less. There are also very few reports on Machado–Joseph domain-containing proteases (MINDY) and zinc-dependent metalloproteinases (JAMMs), so the research on the mechanism of DUBs in liver cancer is far from being in-depth. There is still a lot of room for development in the study of DUBs on the pathogenesis and treatment of liver cancer. The development of related targeted drugs and the clinical application of small molecule inhibitors will also become a research hotspot in the future. These can provide new ideas and research directions for the treatment of liver cancer in the future.
Author Contributions
(I) Conception and design: JieZ, JG. (II) Administrative support: QX, XH. (III) Collection and assembly of data: YW, QM, YS, QL, WF, FC, GO, JiZ. All authors contributed to this article and agree to the submission of the article version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA: A Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
2. Aghoram R, Cai P, Dickinson JA. Alpha-Foetoprotein and/or Liver Ultrasonography for Screening of Hepatocellular Carcinoma in Patients With Chronic Hepatitis B. Cochrane Database Systematic Rev (2012) 2012(9):Cd002799. doi: 10.1002/14651858.CD002799.pub2
3. Nakatsura T, Yoshitake Y, Senju S, Monji M, Komori H, Motomura Y, et al. Glypican-3, Overexpressed Specifically in Human Hepatocellular Carcinoma, is a Novel Tumor Marker. Biochem Biophys Res Commun (2003) 306(1):16–25. doi: 10.1016/S0006-291X(03)00908-2
4. Xing M, Wang X, Kiken RA, He L, Zhang JY. Immunodiagnostic Biomarkers for Hepatocellular Carcinoma (HCC): The First Step in Detection and Treatment. Int J Mol Sci (2021) 22(11):6139. doi: 10.3390/ijms22116139
5. Couri T, Pillai A. Goals and Targets for Personalized Therapy for HCC. Hepatol Int (2019) 13(2):125–37. doi: 10.1007/s12072-018-9919-1
6. Chen Z, Xie H, Hu M, Huang T, Hu Y, Sang N, et al. Recent Progress in Treatment of Hepatocellular Carcinoma. Am J Cancer Res (2020) 10(9):2993–3036.
7. Lawal G, Xiao Y, Rahnemai-Azar AA, Tsilimigras DI, Kuang M, Bakopoulos A, et al. The Immunology of Hepatocellular Carcinoma. Vaccines (2021) 9(10):1184. doi: 10.3390/vaccines9101184
8. Foerster F, Galle PR. The Current Landscape of Clinical Trials for Systemic Treatment of HCC. Cancers (2021) 13(8):1962. doi: 10.3390/cancers13081962
9. Zhai B, Sun XY. Mechanisms of Resistance to Sorafenib and the Corresponding Strategies in Hepatocellular Carcinoma. World J Hepatol (2013) 5(7):345–52. doi: 10.4254/wjh.v5.i7.345
10. El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in Patients With Advanced Hepatocellular Carcinoma (CheckMate 040): An Open-Label, non-Comparative, Phase 1/2 Dose Escalation and Expansion Trial. Lancet (London England) (2017) 389(10088):2492–502. doi: 10.1016/S0140-6736(17)31046-2
11. Tai WT, Chu PY, Shiau CW, Chen YL, Li YS, Hung MH, et al. STAT3 Mediates Regorafenib-Induced Apoptosis in Hepatocellular Carcinoma. Clin Cancer Res (2014) 20(22):5768–76. doi: 10.1158/1078-0432.CCR-14-0725
12. Aitcheson G, Pillai A, Dahman B, John BV. Recent Advances in Systemic Therapies for Advanced Hepatocellular Carcinoma. Curr Hepatol Rep (2021) 20(1):23–33. doi: 10.1007/s11901-021-00560-2
13. Wei L, Wang X, Lv L, Liu J, Xing H, Song Y, et al. The Emerging Role of microRNAs and Long Noncoding RNAs in Drug Resistance of Hepatocellular Carcinoma. Mol Cancer (2019) 18(1):147. doi: 10.1186/s12943-019-1086-z
14. Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and Tumor Responses With Lambrolizumab (Anti-PD-1) in Melanoma. New Engl J Med (2013) 369(2):134–44. doi: 10.1056/NEJMoa1305133
15. Semenza GL. Targeting HIF-1 for Cancer Therapy. Nat Rev Cancer (2003) 3(10):721–32. doi: 10.1038/nrc1187
16. Mennerich D, Kubaichuk K, Kietzmann T. DUBs, Hypoxia, and Cancer. Trends Cancer (2019) 5(10):632–53. doi: 10.1016/j.trecan.2019.08.005
17. Li Q, Chao Q, Liu Y, Fang J, Xie J, Zhen J, et al. Deubiquitinase ZRANB1 Drives Hepatocellular Carcinoma Progression Through SP1-LOXL2 Axis. Am J Cancer Res (2021) 11(10):4807–25.
18. Schwartz AL, Ciechanover A. The Ubiquitin-Proteasome Pathway and Pathogenesis of Human Diseases. Annu Rev Med (1999) 50:57–74. doi: 10.1146/annurev.med.50.1.57
19. Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of Ubiquitin Conjugates by an Isopeptidase in the 26S Proteasome. Nature (1997) 385(6618):737–40. doi: 10.1038/385737a0
20. Zhu Q, Fu Y, Li L, Liu CH, Zhang L. The Functions and Regulation of Otubains in Protein Homeostasis and Diseases. Ageing Res Rev (2021) 67:101303. doi: 10.1016/j.arr.2021.101303
21. Kwon YT, Ciechanover A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem Sci (2017) 42(11):873–86. doi: 10.1016/j.tibs.2017.09.002
22. D'Andrea A, Pellman D. Deubiquitinating Enzymes: A New Class of Biological Regulators. Crit Rev Biochem Mol Biol (1998) 33(5):337–52. doi: 10.1080/10409239891204251
23. Gallo LH, Ko J, Donoghue DJ. The Importance of Regulatory Ubiquitination in Cancer and Metastasis. Cell Cycle (Georgetown Tex) (2017) 16(7):634–48. doi: 10.1080/15384101.2017.1288326
24. Mevissen TET, Komander D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu Rev Biochem (2017) 86:159–92. doi: 10.1146/annurev-biochem-061516-044916
25. Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating Enzymes and Drug Discovery: Emerging Opportunities. Nat Rev Drug Discov (2018) 17(1):57–78. doi: 10.1038/nrd.2017.152
26. Sacco JJ, Coulson JM, Clague MJ, Urbé S. Emerging Roles of Deubiquitinases in Cancer-Associated Pathways. IUBMB Life (2010) 62(2):140–57. doi: 10.1002/iub.300
27. Li W, Shen M, Jiang YZ, Zhang R, Zheng H, Wei Y, et al. Deubiquitinase USP20 Promotes Breast Cancer Metastasis by Stabilizing SNAI2. Genes Dev (2020) 34(19-20):1310–5. doi: 10.1101/gad.339804.120
28. Dai X, Lu L, Deng S, Meng J, Wan C, Huang J, et al. USP7 Targeting Modulates Anti-Tumor Immune Response by Reprogramming Tumor-Associated Macrophages in Lung Cancer. Theranostics (2020) 10(20):9332–47. doi: 10.7150/thno.47137
29. Wang Z, Kang W, Li O, Qi F, Wang J, You Y, et al. Abrogation of USP7 is an Alternative Strategy to Downregulate PD-L1 and Sensitize Gastric Cancer Cells to T Cells Killing. Acta Pharm Sin B (2021) 11(3):694–707. doi: 10.1016/j.apsb.2020.11.005
30. Zhu H, Yan F, Yuan T, Qian M, Zhou T, Dai X, et al. USP10 Promotes Proliferation of Hepatocellular Carcinoma by Deubiquitinating and Stabilizing YAP/TAZ. Cancer Res (2020) 80(11):2204–16. doi: 10.1158/0008-5472.CAN-19-2388
31. Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, et al. OTU Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis. Cell (2013) 154(1):169–84. doi: 10.1016/j.cell.2013.05.046
32. Tyagi A, Haq S, Ramakrishna S. Redox Regulation of DUBs and its Therapeutic Implications in Cancer. Redox Biol (2021) 48:102194. doi: 10.1016/j.redox.2021.102194
33. Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, et al. Chemistry-Based Functional Proteomics Reveals Novel Members of the Deubiquitinating Enzyme Family. Chem Biol (2002) 9(10):1149–59. doi: 10.1016/S1074-5521(02)00248-X
34. Edelmann MJ, Iphöfer A, Akutsu M, Altun M, di Gleria K, Kramer HB, et al. Structural Basis and Specificity of Human Otubain 1-Mediated Deubiquitination. Biochem J (2009) 418(2):379–90. doi: 10.1042/BJ20081318
35. Du T, Li H, Fan Y, Yuan L, Guo X, Zhu Q, et al. The Deubiquitylase OTUD3 Stabilizes GRP78 and Promotes Lung Tumorigenesis. Nat Commun (2019) 10(1):2914. doi: 10.1038/s41467-019-10824-7
36. Sun XX, Dai MS. Deubiquitinating Enzyme Regulation of the P53 Pathway: A Lesson From Otub1. World J Biol Chem (2014) 5(2):75–84. doi: 10.4331/wjbc.v5.i2.75
37. Saldana M, VanderVorst K, Berg AL, Lee H, Carraway KL. Otubain 1: A non-Canonical Deubiquitinase With an Emerging Role in Cancer. Endocrine-related Cancer (2019) 26(1):R1–r14. doi: 10.1530/ERC-18-0264
38. Ni Q, Chen J, Li X, Xu X, Zhang N, Zhou A, et al. Expression of OTUB1 in Hepatocellular Carcinoma and its Effects on HCC Cell Migration and Invasion. Acta Biochim Biophys Sinica (2017) 49(8):680–8. doi: 10.1093/abbs/gmx056
39. Liang C, Pang L, Ke Y, Ji W, Xiong J, Ding R, et al. LncRNA GAS6-AS2 Facilitates Tumor Growth and Metastasis of Hepatocellular Carcinoma by Activating the PI3K/AKT/FoxO3a Signaling Pathway. Int J Clin Exp Pathol (2019) 12(11):4011–23.
40. Wilusz JE, Sunwoo H, Spector DL. Long Noncoding RNAs: Functional Surprises From the RNA World. Genes Dev (2009) 23(13):1494–504. doi: 10.1101/gad.1800909
41. Peng WX, Koirala P, Mo YY. LncRNA-Mediated Regulation of Cell Signaling in Cancer. Oncogene (2017) 36(41):5661–7. doi: 10.1038/onc.2017.184
42. Wapinski O, Chang HY. Long Noncoding RNAs and Human Disease. Trends Cell Biol (2011) 21(6):354–61. doi: 10.1016/j.tcb.2011.04.001
43. Dimri M, Humphries A, Laknaur A, Elattar S, Lee TJ, Sharma A, et al. NAD(P)H Quinone Dehydrogenase 1 Ablation Inhibits Activation of the Phosphoinositide 3-Kinase/Akt Serine/Threonine Kinase and Mitogen-Activated Protein Kinase/Extracellular Signal-Regulated Kinase Pathways and Blocks Metabolic Adaptation in Hepatocellular Carcinoma. Hepatol (Baltimore Md) (2020) 71(2):549–68. doi: 10.1002/hep.30818
44. Tang Z, Tang Y, Li L, Liu T, Yang J. Limonin Provokes Hepatocellular Carcinoma Cells With Stemness Entry Into Cycle via Activating PI3K/Akt Signaling. Biomed Pharmacother Biomed Pharmacother (2019) 117:109051. doi: 10.1016/j.biopha.2019.109051
45. Kulathu Y, Komander D. Atypical Ubiquitylation - the Unexplored World of Polyubiquitin Beyond Lys48 and Lys63 Linkages. Nat Rev Mol Cell Biol (2012) 13(8):508–23. doi: 10.1038/nrm3394
46. Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y, et al. Deubiquitylase OTUD3 Regulates PTEN Stability and Suppresses Tumorigenesis. Nat Cell Biol (2015) 17(9):1169–81. doi: 10.1038/ncb3218
47. Xie P, Chen Y, Zhang H, Zhou G, Chao Q, Wang J, et al. The Deubiquitinase OTUD3 Stabilizes ACTN4 to Drive Growth and Metastasis of Hepatocellular Carcinoma. Aging (2021) 13(15):19317–38. doi: 10.18632/aging.203293
48. Liu X, Zhang X, Peng Z, Li C, Wang Z, Wang C, et al. Deubiquitylase OTUD6B Governs pVHL Stability in an Enzyme-Independent Manner and Suppresses Hepatocellular Carcinoma Metastasis. Adv Sci (Weinheim Baden-Wurttemberg Germany) (2020) 7(8):1902040. doi: 10.1002/advs.201902040
49. Zhu Y, Qu C, Hong X, Jia Y, Lin M, Luo Y, et al. Trabid Inhibits Hepatocellular Carcinoma Growth and Metastasis by Cleaving RNF8-Induced K63 Ubiquitination of Twist1. Cell Death Diff (2019) 26(2):306–20. doi: 10.1038/s41418-018-0119-2
50. Hoeller D, Dikic I. Targeting the Ubiquitin System in Cancer Therapy. Nature (2009) 458(7237):438–44. doi: 10.1038/nature07960
51. Hu M, Li P, Li M, Li W, Yao T, Wu JW, et al. Crystal Structure of a UBP-Family Deubiquitinating Enzyme in Isolation and in Complex With Ubiquitin Aldehyde. Cell (2002) 111(7):1041–54. doi: 10.1016/S0092-8674(02)01199-6
52. Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, et al. Structure and Mechanisms of the Proteasome-Associated Deubiquitinating Enzyme USP14. EMBO J (2005) 24(21):3747–56. doi: 10.1038/sj.emboj.7600832
53. Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, et al. Rpn1 Provides Adjacent Receptor Sites for Substrate Binding and Deubiquitination by the Proteasome. Sci (New York NY) (2016) 351(6275). doi: 10.1126/science.aad9421
54. Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, et al. Structural Characterization of the Interaction of Ubp6 With the 26S Proteasome. Proc Natl Acad Sci United States America (2015) 112(28):8626–31. doi: 10.1073/pnas.1510449112
55. Lee JH, Shin SK, Jiang Y, Choi WH, Hong C, Kim DE, et al. Facilitated Tau Degradation by USP14 Aptamers via Enhanced Proteasome Activity. Sci Rep (2015) 5:10757. doi: 10.1038/srep10757
56. Wang F, Ning S, Yu B, Wang Y. USP14: Structure, Function, and Target Inhibition. Front Pharmacol (2021) 12:801328. doi: 10.3389/fphar.2021.801328
57. Crimmins S, Jin Y, Wheeler C, Huffman AK, Chapman C, Dobrunz LE, et al. Transgenic Rescue of Ataxia Mice With Neuronal-Specific Expression of Ubiquitin-Specific Protease 14. J Neurosci (2006) 26(44):11423–31. doi: 10.1523/JNEUROSCI.3600-06.2006
58. Feng J, Li J, Wu L, Yu Q, Ji J, Wu J, et al. Emerging Roles and the Regulation of Aerobic Glycolysis in Hepatocellular Carcinoma. J Exp Clin Cancer Res CR (2020) 39(1):126. doi: 10.1186/s13046-020-01629-4
59. Lv C, Wang S, Lin L, Wang C, Zeng K, Meng Y, et al. USP14 Maintains HIF1-α Stabilization via its Deubiquitination Activity in Hepatocellular Carcinoma. Cell Death Dis (2021) 12(9):803. doi: 10.1038/s41419-021-04089-6
60. Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, et al. The Deubiquitinating Enzyme USP1 Regulates the Fanconi Anemia Pathway. Mol Cell (2005) 17(3):331–9. doi: 10.1016/j.molcel.2005.01.008
61. Villamil MA, Chen J, Liang Q, Zhuang Z. A Noncanonical Cysteine Protease USP1 is Activated Through Active Site Modulation by USP1-Associated Factor 1. Biochemistry (2012) 51(13):2829–39. doi: 10.1021/bi3000512
62. Fujiwara T, Saito A, Suzuki M, Shinomiya H, Suzuki T, Takahashi E, et al. Identification and Chromosomal Assignment of USP1, a Novel Gene Encoding a Human Ubiquitin-Specific Protease. Genomics (1998) 54(1):155–8. doi: 10.1006/geno.1998.5554
63. Li H, Lim KS, Kim H, Hinds TR, Jo U, Mao H, et al. Allosteric Activation of Ubiquitin-Specific Proteases by β-Propeller Proteins UAF1 and WDR20. Mol Cell (2016) 63(2):249–60. doi: 10.1016/j.molcel.2016.05.031
64. Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, et al. A UAF1-Containing Multisubunit Protein Complex Regulates the Fanconi Anemia Pathway. Mol Cell (2007) 28(5):786–97. doi: 10.1016/j.molcel.2007.09.031
65. García-Santisteban I, Peters GJ, Giovannetti E, Rodríguez JA. USP1 Deubiquitinase: Cellular Functions, Regulatory Mechanisms and Emerging Potential as Target in Cancer Therapy. Mol Cancer (2013) 12:91. doi: 10.1186/1476-4598-12-91
66. Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, et al. USP1 Deubiquitinates ID Proteins to Preserve a Mesenchymal Stem Cell Program in Osteosarcoma. Cell (2011) 146(6):918–30. doi: 10.1016/j.cell.2011.07.040
67. Ma A, Tang M, Zhang L, Wang B, Yang Z, Liu Y, et al. USP1 Inhibition Destabilizes KPNA2 and Suppresses Breast Cancer Metastasis. Oncogene (2019) 38(13):2405–19. doi: 10.1038/s41388-018-0590-8
68. Zhiqiang Z, Qinghui Y, Yongqiang Z, Jian Z, Xin Z, Haiying M, et al. USP1 Regulates AKT Phosphorylation by Modulating the Stability of PHLPP1 in Lung Cancer Cells. J Cancer Res Clin Oncol (2012) 138(7):1231–8. doi: 10.1007/s00432-012-1193-3
69. Zhao Y, Xue C, Xie Z, Ouyang X, Li L. Comprehensive Analysis of Ubiquitin-Specific Protease 1 Reveals its Importance in Hepatocellular Carcinoma. Cell Proliferation (2020) 53(10):e12908. doi: 10.1111/cpr.12908
70. Chen J, Wei Y, Feng Q, Ren L, He G, Chang W, et al. Ribosomal Protein S15A Promotes Malignant Transformation and Predicts Poor Outcome in Colorectal Cancer Through Misregulation of P53 Signaling Pathway. Int J Oncol (2016) 48(4):1628–38. doi: 10.3892/ijo.2016.3366
71. Liao Y, Shao Z, Liu Y, Xia X, Deng Y, Yu C, et al. USP1-Dependent RPS16 Protein Stability Drives Growth and Metastasis of Human Hepatocellular Carcinoma Cells. J Exp Clin Cancer Res CR (2021) 40(1):201. doi: 10.1186/s13046-021-02008-3
72. Wang J, Ou J, Guo Y, Dai T, Li X, Liu J, et al. TBLR1 is a Novel Prognostic Marker and Promotes Epithelial-Mesenchymal Transition in Cervical Cancer. Br J Cancer (2014) 111(1):112–24. doi: 10.1038/bjc.2014.278
73. Daniels G, Li Y, Gellert LL, Zhou A, Melamed J, Wu X, et al. TBLR1 as an Androgen Receptor (AR) Coactivator Selectively Activates AR Target Genes to Inhibit Prostate Cancer Growth. Endocrine-Related Cancer (2014) 21(1):127–42. doi: 10.1530/ERC-13-0293
74. Wu X, Zhan Y, Li X, Wei J, Santiago L, Daniels G, et al. Nuclear TBLR1 as an ER Corepressor Promotes Cell Proliferation, Migration and Invasion in Breast and Ovarian Cancer. Am J Cancer Res (2016) 6(10):2351–60.
75. Li Y, Xu Y, Gao C, Sun Y, Zhou K, Wang P, et al. USP1 Maintains the Survival of Liver Circulating Tumor Cells by Deubiquitinating and Stabilizing Tblr1. Front Oncol (2020) 10:554809. doi: 10.3389/fonc.2020.554809
76. Al-Salama ZT, Syed YY, Scott LJ. Lenvatinib: A Review in Hepatocellular Carcinoma. Drugs (2019) 79(6):665–74. doi: 10.1007/s40265-019-01116-x
77. Wei L, Lee D, Law CT, Zhang MS, Shen J, Chin DW, et al. Genome-Wide CRISPR/Cas9 Library Screening Identified PHGDH as a Critical Driver for Sorafenib Resistance in HCC. Nat Commun (2019) 10(1):4681. doi: 10.1038/s41467-019-12606-7
78. Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, et al. Immunotherapies for Hepatocellular Carcinoma. Nat Rev Clin Oncol (2022) 19(3):151–72. doi: 10.1038/s41571-021-00573-2
79. Chen Z, Ma Y, Guo Z, Song D, Chen Z, Sun M. Ubiquitin-Specific Protease 1 Acts as an Oncogene and Promotes Lenvatinib Efficacy in Hepatocellular Carcinoma by Stabilizing C-Kit. Ann Hepatol (2022) 27(2):100669. doi: 10.1016/j.aohep.2022.100669
80. Bhattacharya U, Neizer-Ashun F, Mukherjee P, Bhattacharya R. When the Chains do Not Break: The Role of USP10 in Physiology and Pathology. Cell Death Dis (2020) 11(12):1033. doi: 10.1038/s41419-020-03246-7
81. Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 Regulates P53 Localization and Stability by Deubiquitinating P53. Cell (2010) 140(3):384–96. doi: 10.1016/j.cell.2009.12.032
82. Deng M, Yang X, Qin B, Liu T, Zhang H, Guo W, et al. Deubiquitination and Activation of AMPK by USP10. Mol Cell (2016) 61(4):614–24. doi: 10.1016/j.molcel.2016.01.010
83. Wang X, Xia S, Li H, Wang X, Li C, Chao Y, et al. The Deubiquitinase USP10 Regulates KLF4 Stability and Suppresses Lung Tumorigenesis. Cell Death Diff (2020) 27(6):1747–64. doi: 10.1038/s41418-019-0458-7
84. Kim K, Huh T, Park Y, Koo DH, Kim H, Hwang I, et al. Prognostic Significance of USP10 and P14arf Expression in Patients With Colorectal Cancer. Pathol Res Practice (2020) 216(6):152988. doi: 10.1016/j.prp.2020.152988
85. Eichhorn PJ, Rodón L, Gonzàlez-Juncà A, Dirac A, Gili M, Martínez-Sáez E, et al. USP15 Stabilizes TGF-β Receptor I and Promotes Oncogenesis Through the Activation of TGF-β Signaling in Glioblastoma. Nat Med (2012) 18(3):429–35. doi: 10.1038/nm.2619
86. Guturi KKN, Bohgaki M, Bohgaki T, Srikumar T, Ng D, Kumareswaran R, et al. RNF168 and USP10 Regulate Topoisomerase Iiα Function via Opposing Effects on its Ubiquitylation. Nat Commun (2016) 7:12638. doi: 10.1038/ncomms12638
87. Yuan T, Chen Z, Yan F, Qian M, Luo H, Ye S, et al. Deubiquitinating Enzyme USP10 Promotes Hepatocellular Carcinoma Metastasis Through Deubiquitinating and Stabilizing Smad4 Protein. Mol Oncol (2020) 14(1):197–210. doi: 10.1002/1878-0261.12596
88. Matter MS, Decaens T, Andersen JB, Thorgeirsson SS. Targeting the mTOR Pathway in Hepatocellular Carcinoma: Current State and Future Trends. J Hepatol (2014) 60(4):855–65. doi: 10.1016/j.jhep.2013.11.031
89. Garcia-Cao I, Song MS, Hobbs RM, Laurent G, Giorgi C, de Boer VC, et al. Systemic Elevation of PTEN Induces a Tumor-Suppressive Metabolic State. Cell (2012) 149(1):49–62. doi: 10.1016/j.cell.2012.02.030
90. Lu C, Ning Z, Wang A, Chen D, Liu X, Xia T, et al. USP10 Suppresses Tumor Progression by Inhibiting mTOR Activation in Hepatocellular Carcinoma. Cancer Lett (2018) 436:139–48. doi: 10.1016/j.canlet.2018.07.032
91. González-Magaña A, Blanco FJ. Human PCNA Structure, Function and Interactions. Biomolecules (2020) 10(4):570. doi: 10.3390/biom10040570
92. Shen C, Li J, Zhang Q, Tao Y, Li R, Ma Z, et al. LncRNA GASAL1 Promotes Hepatocellular Carcinoma Progression by Up-Regulating USP10-Stabilized PCNA. Exp Cell Res (2022) 415(1):112973. doi: 10.1016/j.yexcr.2021.112973
93. Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo Signaling Pathway Coordinately Regulates Cell Proliferation and Apoptosis by Inactivating Yorkie, the Drosophila Homolog of YAP. Cell (2005) 122(3):421–34. doi: 10.1016/j.cell.2005.06.007
94. Wu S, Huang J, Dong J, Pan D. Hippo Encodes a Ste-20 Family Protein Kinase That Restricts Cell Proliferation and Promotes Apoptosis in Conjunction With Salvador and Warts. Cell (2003) 114(4):445–56. doi: 10.1016/S0092-8674(03)00549-X
95. Koo JH, Guan KL. Interplay Between YAP/TAZ and Metabolism. Cell Metab (2018) 28(2):196–206. doi: 10.1016/j.cmet.2018.07.010
96. Moya IM, Halder G. Hippo-YAP/TAZ Signalling in Organ Regeneration and Regenerative Medicine. Nat Rev Mol Cell Biol (2019) 20(4):211–26. doi: 10.1038/s41580-018-0086-y
97. Nguyen CDK, Yi C. YAP/TAZ Signaling and Resistance to Cancer Therapy. Trends Cancer (2019) 5(5):283–96. doi: 10.1016/j.trecan.2019.02.010
98. Ding K, Ji J, Zhang X, Huang B, Chen A, Zhang D, et al. RNA Splicing Factor USP39 Promotes Glioma Progression by Inducing TAZ mRNA Maturation. Oncogene (2019) 38(37):6414–28. doi: 10.1038/s41388-019-0888-1
99. Makarova OV, Makarov EM, Lührmann R. The 65 and 110 kDa SR-Related Proteins of the U4/U6. U5 tri-snRNP are Essential for the Assembly of Mature Spliceosomes. EMBO J (2001) 20(10):2553–63. doi: 10.1093/emboj/20.10.2553
100. Zhao Y, Zhang B, Lei Y, Sun J, Zhang Y, Yang S, et al. Knockdown of USP39 Induces Cell Cycle Arrest and Apoptosis in Melanoma. Tumour Biol (2016) 37(10):13167–76. doi: 10.1007/s13277-016-5212-x
101. van Leuken RJ, Luna-Vargas MP, Sixma TK, Wolthuis RM, Medema RH. Usp39 is Essential for Mitotic Spindle Checkpoint Integrity and Controls mRNA-Levels of Aurora B. Cell Cycle (Georgetown Tex) (2008) 7(17):2710–9. doi: 10.4161/cc.7.17.6553
102. Wang L, Chen T, Li X, Yan W, Lou Y, Liu Z, et al. USP39 Promotes Ovarian Cancer Malignant Phenotypes and Carboplatin Chemoresistance. Int J Oncol (2019) 55(1):277–88. doi: 10.3892/ijo.2019.4818
103. Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, et al. The EMT-Activator Zeb1 is a Key Factor for Cell Plasticity and Promotes Metastasis in Pancreatic Cancer. Nat Cell Biol (2017) 19(5):518–29. doi: 10.1038/ncb3513
104. Sheng W, Shi X, Lin Y, Tang J, Jia C, Cao R, et al. Musashi2 Promotes EGF-Induced EMT in Pancreatic Cancer via ZEB1-ERK/MAPK Signaling. J Exp Clin Cancer Res CR (2020) 39(1):16. doi: 10.1186/s13046-020-1521-4
105. Li X, Yuan J, Song C, Lei Y, Xu J, Zhang G, et al. Deubiquitinase USP39 and E3 Ligase TRIM26 Balance the Level of ZEB1 Ubiquitination and Thereby Determine the Progression of Hepatocellular Carcinoma. Cell Death Diff (2021) 28(8):2315–32. doi: 10.1038/s41418-021-00754-7
106. Wang K, Chai L, Qiu Z, Zhang Y, Gao H, Zhang X. Overexpression of TRIM26 Suppresses the Proliferation, Metastasis, and Glycolysis in Papillary Thyroid Carcinoma Cells. J Cell Physiol (2019) 234(10):19019–27. doi: 10.1002/jcp.28541
107. Dong L, Yu L, Li H, Shi L, Luo Z, Zhao H, et al. An NAD(+)-Dependent Deacetylase SIRT7 Promotes HCC Development Through Deacetylation of USP39. iScience (2020) 23(8):101351. doi: 10.1016/j.isci.2020.101351
108. Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, et al. Sirtuin7 Oncogenic Potential in Human Hepatocellular Carcinoma and its Regulation by the Tumor Suppressors MiR-125a-5p and MiR-125b. Hepatol (Baltimore Md) (2013) 57(3):1055–67. doi: 10.1002/hep.26101
109. Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, et al. Regulation of the Master Regulator FOXM1 in Cancer. Cell Commun Signaling CCS (2018) 16(1):57. doi: 10.1186/s12964-018-0266-6
110. Hu G, Yan Z, Zhang C, Cheng M, Yan Y, Wang Y, et al. FOXM1 Promotes Hepatocellular Carcinoma Progression by Regulating KIF4A Expression. J Exp Clin Cancer Res CR (2019) 38(1):188. doi: 10.1186/s13046-019-1202-3
111. Yuan X, Sun X, Shi X, Jiang C, Yu D, Zhang W, et al. USP39 Promotes the Growth of Human Hepatocellular Carcinoma In Vitro and In Vivo. Oncol Rep (2015) 34(2):823–32. doi: 10.3892/or.2015.4065
112. Yuan X, Sun X, Shi X, Jiang C, Yu D, Zhang W, et al. USP39 Regulates the Growth of SMMC-7721 Cells via Foxm1. Exp Ther Med (2017) 13(4):1506–13. doi: 10.3892/etm.2017.4115
113. O'Connor L, Gilmour J, Bonifer C. The Role of the Ubiquitously Expressed Transcription Factor Sp1 in Tissue-Specific Transcriptional Regulation and in Disease. Yale J Biol Med (2016) 89(4):513–25.
114. Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, et al. Sp1 and TAFII130 Transcriptional Activity Disrupted in Early Huntington's Disease. Sci (New York NY) (2002) 296(5576):2238–43. doi: 10.1126/science.1072613
115. Vizcaíno C, Mansilla S, Portugal J. Sp1 Transcription Factor: A Long-Standing Target in Cancer Chemotherapy. Pharmacol Ther (2015) 152:111–24. doi: 10.1016/j.pharmthera.2015.05.008
116. Dong X, Liu Z, Zhang E, Zhang P, Wang Y, Hang J, et al. USP39 Promotes Tumorigenesis by Stabilizing and Deubiquitinating SP1 Protein in Hepatocellular Carcinoma. Cell Signalling (2021) 85:110068. doi: 10.1016/j.cellsig.2021.110068
117. Zhang J, Luo N, Tian Y, Li J, Yang X, Yin H, et al. USP22 Knockdown Enhanced Chemosensitivity of Hepatocellular Carcinoma Cells to 5-Fu by Up-Regulation of Smad4 and Suppression of Akt. Oncotarget (2017) 8(15):24728–40. doi: 10.18632/oncotarget.15798
118. Roedig J, Kowald L, Juretschke T, Karlowitz R, Ahangarian Abhari B, Roedig H, et al. USP22 Controls Necroptosis by Regulating Receptor-Interacting Protein Kinase 3 Ubiquitination. EMBO Rep (2021) 22(2):e50163. doi: 10.15252/embr.202050163
119. Cai Z, Zhang MX, Tang Z, Zhang Q, Ye J, Xiong TC, et al. USP22 Promotes IRF3 Nuclear Translocation and Antiviral Responses by Deubiquitinating the Importin Protein KPNA2. J Exp Med (2020) 217(5):e20191174. doi: 10.1084/jem.20191174
120. Tang B, Liang X, Tang F, Zhang J, Zeng S, Jin S, et al. Expression of USP22 and Survivin is an Indicator of Malignant Behavior in Hepatocellular Carcinoma. Int J Oncol (2015) 47(6):2208–16. doi: 10.3892/ijo.2015.3214
121. Chang YS, Su CW, Chen SC, Chen YY, Liang YJ, Wu JC. Upregulation of USP22 and ABCC1 During Sorafenib Treatment of Hepatocellular Carcinoma Contribute to Development of Resistance. Cells (2022) 11(4):634. doi: 10.3390/cells11040634
122. Wen X, Ling S, Wu W, Shan Q, Liu P, Wang C, et al. Ubiquitin-Specific Protease 22/Silent Information Regulator 1 Axis Plays a Pivotal Role in the Prognosis and 5-Fluorouracil Resistance in Hepatocellular Carcinoma. Digestive Dis Sci (2020) 65(4):1064–73. doi: 10.1007/s10620-019-05844-8
123. Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, et al. The Deacetylase SIRT1 Promotes Membrane Localization and Activation of Akt and PDK1 During Tumorigenesis and Cardiac Hypertrophy. Sci Signaling (2011) 4(182):ra46. doi: 10.1126/scisignal.2001465
124. Ling S, Li J, Shan Q, Dai H, Lu D, Wen X, et al. USP22 Mediates the Multidrug Resistance of Hepatocellular Carcinoma via the SIRT1/AKT/MRP1 Signaling Pathway. Mol Oncol (2017) 11(6):682–95. doi: 10.1002/1878-0261.12067
125. Zhai R, Tang F, Gong J, Zhang J, Lei B, Li B, et al. The Relationship Between the Expression of USP22, BMI1, and EZH2 in Hepatocellular Carcinoma and Their Impacts on Prognosis. OncoTarg Ther (2016) 9:6987–98. doi: 10.2147/OTT.S110985
126. Xu S, Ling S, Shan Q, Ye Q, Zhan Q, Jiang G, et al. Self-Activated Cascade-Responsive Sorafenib and USP22 shRNA Co-Delivery System for Synergetic Hepatocellular Carcinoma Therapy. Adv Sci (Weinheim Baden-Wurttemberg Germany) (2021) 8(5):2003042. doi: 10.1002/advs.202003042
127. Ning Z, Guo X, Liu X, Lu C, Wang A, Wang X, et al. USP22 Regulates Lipidome Accumulation by Stabilizing Pparγ in Hepatocellular Carcinoma. Nat Commun (2022) 13(1):2187. doi: 10.1038/s41467-022-29846-9
128. Ling S, Shan Q, Zhan Q, Ye Q, Liu P, Xu S, et al. USP22 Promotes Hypoxia-Induced Hepatocellular Carcinoma Stemness by a HIF1α/USP22 Positive Feedback Loop Upon TP53 Inactivation. Gut (2020) 69(7):1322–34. doi: 10.1136/gutjnl-2019-319616
129. Jing T, Wang B, Yang Z, Liu Y, Xu G, Xu X, et al. Deubiquitination of the Repressor E2F6 by USP22 Facilitates AKT Activation and Tumor Growth in Hepatocellular Carcinoma. Cancer Lett (2021) 518:266–77. doi: 10.1016/j.canlet.2021.07.044
130. Xiong H, Ni Z, He J, Jiang S, Li X, He J, et al. LncRNA HULC Triggers Autophagy via Stabilizing Sirt1 and Attenuates the Chemosensitivity of HCC Cells. Oncogene (2017) 36(25):3528–40. doi: 10.1038/onc.2016.521
131. Xiong H, Li B, He J, Zeng Y, Zhang Y, He F. lncRNA HULC Promotes the Growth of Hepatocellular Carcinoma Cells via Stabilizing COX-2 Protein. Biochem Biophys Res Commun (2017) 490(3):693–9. doi: 10.1016/j.bbrc.2017.06.103
132. Hu H, Tang C, Jiang Q, Luo W, Liu J, Wei X, et al. Reduced Ubiquitin-Specific Protease 9X Expression Induced by RNA Interference Inhibits the Bioactivity of Hepatocellular Carcinoma Cells. Oncol Lett (2015) 10(1):268–72. doi: 10.3892/ol.2015.3152
133. Chen MY, Li ZP, Sun ZN, Ma M. USP9X Promotes the Progression of Hepatocellular Carcinoma by Regulating Beta-Catenin. Irish J Med Sci (2020) 189(3):865–71. doi: 10.1007/s11845-020-02199-2
134. Zhang FK, Ni QZ, Wang K, Cao HJ, Guan DX, Zhang EB, et al. Targeting USP9X-AMPK Axis in ARID1A-Deficient Hepatocellular Carcinoma. Cell Mol Gastroenterol Hepatol (2022) 14(1):101–27. doi: 10.1016/j.jcmgh.2022.03.009
135. Chen E, Li E, Liu H, Zhou Y, Wen L, Wang J, et al. miR-26b Enhances the Sensitivity of Hepatocellular Carcinoma to Doxorubicin via USP9X-Dependent Degradation of P53 and Regulation of Autophagy. Int J Biol Sci (2021) 17(3):781–95. doi: 10.7150/ijbs.52517
136. Shen G, Lin Y, Yang X, Zhang J, Xu Z, Jia H. MicroRNA-26b Inhibits Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma by Targeting USP9X. BMC Cancer (2014) 14:393. doi: 10.1186/1471-2407-14-393
137. Liu H, Chen W, Liang C, Chen BW, Zhi X, Zhang S, et al. WP1130 Increases Doxorubicin Sensitivity in Hepatocellular Carcinoma Cells Through Usp9x-Dependent P53 Degradation. Cancer Lett (2015) 361(2):218–25. doi: 10.1016/j.canlet.2015.03.001
138. Chen H, Yang F, Li X, Gong ZJ, Wang LW. Long Noncoding RNA LNC473 Inhibits the Ubiquitination of Survivin via Association With USP9X and Enhances Cell Proliferation and Invasion in Hepatocellular Carcinoma Cells. Biochem Biophys Res Commun (2018) 499(3):702–10. doi: 10.1016/j.bbrc.2018.03.215
139. Xiong B, Huang J, Liu Y, Zou M, Zhao Z, Gong J, et al. Ubiquitin-Specific Protease 2a Promotes Hepatocellular Carcinoma Progression via Deubiquitination and Stabilization of RAB1A. Cell Oncol (Dordrecht) (2021) 44(2):329–43. doi: 10.1007/s13402-020-00568-8
140. Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S, et al. Increased Lipogenesis, Induced by AKT-Mtorc1-RPS6 Signaling, Promotes Development of Human Hepatocellular Carcinoma. Gastroenterology (2011) 140(3):1071–83. doi: 10.1053/j.gastro.2010.12.006
141. Nadolny C, Zhang X, Chen Q, Hashmi SF, Ali W, Hemme C, et al. Dysregulation and Activities of Ubiquitin Specific Peptidase 2b in the Pathogenesis of Hepatocellular Carcinoma. Am J Cancer Res (2021) 11(10):4746–67.
142. Wang X, Zhang Q, Wang Y, Zhuang H, Chen B. Clinical Significance of Ubiquitin Specific Protease 7 (USP7) in Predicting Prognosis of Hepatocellular Carcinoma and its Functional Mechanisms. Med Sci Monitor (2018) 24:1742–50. doi: 10.12659/MSM.909368
143. Zhang W, Zhang J, Xu C, Zhang S, Bian S, Jiang F, et al. Ubiquitin-Specific Protease 7 is a Drug-Able Target That Promotes Hepatocellular Carcinoma and Chemoresistance. Cancer Cell Int (2020) 20:28. doi: 10.1186/s12935-020-1109-2
144. Zhu L, Liu R, Zhang W, Qian S, Wang JH. MicroRNA-205 Regulates Ubiquitin Specific Peptidase 7 Protein Expression in Hepatocellular Carcinoma Cells. Mol Med Rep (2015) 12(3):4652–6. doi: 10.3892/mmr.2015.3998
145. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA Secreted From Adipocytes Promotes the Growth of Hepatocellular Carcinoma by Targeting Deubiquitination-Related USP7. Oncogene (2019) 38(15):2844–59. doi: 10.1038/s41388-018-0619-z
146. Li Y, Cheng X, Chen Y, Zhou T, Li D, Zheng WV. METTL3 Facilitates the Progression of Hepatocellular Carcinoma by Modulating the M6a Level of USP7. Am J Trans Res (2021) 13(12):13423–37.
147. Sun X, Ding Y, Zhan M, Li Y, Gao D, Wang G, et al. Usp7 Regulates Hippo Pathway Through Deubiquitinating the Transcriptional Coactivator Yorkie. Nat Commun (2019) 10(1):411. doi: 10.1038/s41467-019-08334-7
148. Bian S, Ni W, Zhu M, Zhang X, Qiang Y, Zhang J, et al. Flap Endonuclease 1 Facilitated Hepatocellular Carcinoma Progression by Enhancing USP7/MDM2-Mediated P53 Inactivation. Int J Biol Sci (2022) 18(3):1022–38. doi: 10.7150/ijbs.68179
149. Liu Y, Zhang Y, Wang S, Dong QZ, Shen Z, Wang W, et al. Prospero-Related Homeobox 1 Drives Angiogenesis of Hepatocellular Carcinoma Through Selectively Activating Interleukin-8 Expression. Hepatol (Baltimore Md) (2017) 66(6):1894–909. doi: 10.1002/hep.29337
150. Cai JB, Shi GM, Dong ZR, Ke AW, Ma HH, Gao Q, et al. Ubiquitin-Specific Protease 7 Accelerates P14(ARF) Degradation by Deubiquitinating Thyroid Hormone Receptor-Interacting Protein 12 and Promotes Hepatocellular Carcinoma Progression. Hepatol (Baltimore Md) (2015) 61(5):1603–14. doi: 10.1002/hep.27682
151. Li T, Yan B, Ma Y, Weng J, Yang S, Zhao N, et al. Ubiquitin-Specific Protease 4 Promotes Hepatocellular Carcinoma Progression via Cyclophilin A Stabilization and Deubiquitination. Cell Death Dis (2018) 9(2):148. doi: 10.1038/s41419-017-0030-7
152. Qiu C, Liu Y, Mei Y, Zou M, Zhao Z, Ye M, et al. Ubiquitin-Specific Protease 4 Promotes Metastasis of Hepatocellular Carcinoma by Increasing TGF-β Signaling-Induced Epithelial-Mesenchymal Transition. Aging (2018) 10(10):2783–99. doi: 10.18632/aging.101587
153. Heo MJ, Kim YM, Koo JH, Yang YM, An J, Lee SK, et al. microRNA-148a Dysregulation Discriminates Poor Prognosis of Hepatocellular Carcinoma in Association With USP4 Overexpression. Oncotarget (2014) 5(9):2792–806. doi: 10.18632/oncotarget.1920
154. Liu Y, Wang WM, Lu YF, Feng L, Li L, Pan MZ, et al. Usp5 Functions as an Oncogene for Stimulating Tumorigenesis in Hepatocellular Carcinoma. Oncotarget (2017) 8(31):50655–64. doi: 10.18632/oncotarget.16901
155. Meng J, Ai X, Lei Y, Zhong W, Qian B, Qiao K, et al. USP5 Promotes Epithelial-Mesenchymal Transition by Stabilizing SLUG in Hepatocellular Carcinoma. Theranostics (2019) 9(2):573–87. doi: 10.7150/thno.27654
156. Gao S, Chen T, Li L, Liu X, Liu Y, Zhao J, et al. Hypoxia-Inducible Ubiquitin Specific Peptidase 13 Contributes to Tumor Growth and Metastasis via Enhancing the Toll-Like Receptor 4/Myeloid Differentiation Primary Response Gene 88/Nuclear Factor-κb Pathway in Hepatocellular Carcinoma. Front Cell Dev Biol (2020) 8:587389. doi: 10.3389/fcell.2020.587389
157. Huang J, Gu ZL, Chen W, Xu YY, Chen M. Knockdown of Ubiquitin-Specific Peptidase 13 Inhibits Cell Growth of Hepatocellular Carcinoma by Reducing C-Myc Expression. Kaohsiung J Med Sci (2020) 36(8):615–21. doi: 10.1002/kjm2.12209
158. Gao R, Buechel D, Kalathur RKR, Morini MF, Coto-Llerena M, Ercan C, et al. USP29-Mediated HIF1α Stabilization is Associated With Sorafenib Resistance of Hepatocellular Carcinoma Cells by Upregulating Glycolysis. Oncogenesis (2021) 10(7):52. doi: 10.1038/s41389-021-00338-7
159. Yao XQ, Li L, Piao LZ, Zhang GJ, Huang XZ, Wang Y, et al. Overexpression of Ubiquitin-Specific Protease15 (USP15) Promotes Tumor Growth and Inhibits Apoptosis and Correlated With Poor Disease-Free Survival in Hepatocellular Carcinoma. Technol Cancer Res Treat (2020) 19:1533033820967455. doi: 10.1177/1533033820967455
160. Sun Q, Zhang Z, Lu Y, Liu Q, Xu X, Xu J, et al. Loss of Xanthine Oxidoreductase Potentiates Propagation of Hepatocellular Carcinoma Stem Cells. Hepatol (Baltimore Md) (2020) 71(6):2033–49. doi: 10.1002/hep.30978
161. Liu C, Li X, Feng G, Cao M, Liu F, Zhang G, et al. Downregulation of USP12 Inhibits Tumor Growth via the P38/MAPK Pathway in Hepatocellular Carcinoma. Mol Med Rep (2020) 22(6):4899–908. doi: 10.3892/mmr.2020.11557
162. Qian Y, Wang B, Ma A, Zhang L, Xu G, Ding Q, et al. USP16 Downregulation by Carboxyl-Terminal Truncated HBx Promotes the Growth of Hepatocellular Carcinoma Cells. Sci Rep (2016) 6:33039. doi: 10.1038/srep33039
163. Zou T, Wang Y, Dong L, Che T, Zhao H, Yan X, et al. Stabilization of SETD3 by Deubiquitinase USP27 Enhances Cell Proliferation and Hepatocellular Carcinoma Progression. Cell Mol Life Sci CMLS (2022) 79(1):70. doi: 10.1007/s00018-021-04118-9
164. Qiu Y, Huang D, Sheng Y, Huang J, Li N, Zhang S, et al. Deubiquitinating Enzyme USP46 Suppresses the Progression of Hepatocellular Carcinoma by Stabilizing MST1. Exp Cell Res (2021) 405(1):112646. doi: 10.1016/j.yexcr.2021.112646
165. Zhu Y, Xu J, Hu W, Wang F, Zhou Y, Gong W, et al. Inhibiting USP8 Overcomes Hepatocellular Carcinoma Resistance via Suppressing Receptor Tyrosine Kinases. Aging (2021) 13(11):14999–5012. doi: 10.18632/aging.203061
166. Fang Y, Shen X. Ubiquitin Carboxyl-Terminal Hydrolases: Involvement in Cancer Progression and Clinical Implications. Cancer Metastasis Rev (2017) 36(4):669–82. doi: 10.1007/s10555-017-9702-0
167. Todi SV, Paulson HL. Balancing Act: Deubiquitinating Enzymes in the Nervous System. Trends Neurosci (2011) 34(7):370–82. doi: 10.1016/j.tins.2011.05.004
168. Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, et al. The Ubiquitin Pathway in Parkinson's Disease. Nature (1998) 395(6701):451–2. doi: 10.1038/26652
169. Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT Jr. The UCH-L1 Gene Encodes Two Opposing Enzymatic Activities That Affect Alpha-Synuclein Degradation and Parkinson's Disease Susceptibility. Cell (2002) 111(2):209–18. doi: 10.1016/S0092-8674(02)01012-7
170. Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, et al. Oxidative Modifications and Down-Regulation of Ubiquitin Carboxyl-Terminal Hydrolase L1 Associated With Idiopathic Parkinson's and Alzheimer's Diseases. J Biol Chem (2004) 279(13):13256–64. doi: 10.1074/jbc.M314124200
171. Vander Linden RT, Hemmis CW, Schmitt B, Ndoja A, Whitby FG, Robinson H, et al. Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase. Mol Cell (2015) 57(5):901–11.
172. Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, et al. Proteasome Recruitment and Activation of the Uch37 Deubiquitinating Enzyme by Adrm1. Nat Cell Biol (2006) 8(9):994–1002. doi: 10.1038/ncb1460
173. Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, et al. Distinct Modes of Regulation of the Uch37 Deubiquitinating Enzyme in the Proteasome and in the Ino80 Chromatin-Remodeling Complex. Mol Cell (2008) 31(6):909–17. doi: 10.1016/j.molcel.2008.08.027
174. Nishio K, Kim SW, Kawai K, Mizushima T, Yamane T, Hamazaki J, et al. Crystal Structure of the De-Ubiquitinating Enzyme UCH37 (Human UCH-L5) Catalytic Domain. Biochem Biophys Res Commun (2009) 390(3):855–60. doi: 10.1016/j.bbrc.2009.10.062
175. Burgie SE, Bingman CA, Soni AB, Phillips GN Jr. Structural Characterization of Human Uch37. Proteins (2012) 80(2):649–54. doi: 10.1002/prot.23147
176. Fang Y, Fu D, Tang W, Cai Y, Ma D, Wang H, et al. Ubiquitin C-Terminal Hydrolase 37, a Novel Predictor for Hepatocellular Carcinoma Recurrence, Promotes Cell Migration and Invasion via Interacting and Deubiquitinating PRP19. Biochim Biophys Acta (2013) 1833(3):559–72. doi: 10.1016/j.bbamcr.2012.11.020
177. Sun YL, Cai JQ, Liu F, Bi XY, Zhou LP, Zhao XH. Aberrant Expression of Peroxiredoxin 1 and its Clinical Implications in Liver Cancer. World J Gastroenterol (2015) 21(38):10840–52. doi: 10.3748/wjg.v21.i38.10840
178. Sun QK, Zhu JY, Wang W, Lv Y, Zhou HC, Yu JH, et al. Diagnostic and Prognostic Significance of Peroxiredoxin 1 Expression in Human Hepatocellular Carcinoma. Med Oncol (Northwood London England) (2014) 31(1):786. doi: 10.1007/s12032-013-0786-2
179. An Y, Jiang J, Zhou L, Shi J, Jin P, Li L, et al. Peroxiredoxin 1 is Essential for Natamycin-Triggered Apoptosis and Protective Autophagy in Hepatocellular Carcinoma. Cancer Lett (2021) 521:210–23. doi: 10.1016/j.canlet.2021.08.023
180. Xu M, Xu J, Zhu D, Su R, Zhuang B, Xu R, et al. Expression and Prognostic Roles of PRDXs Gene Family in Hepatocellular Carcinoma. J Trans Med (2021) 19(1):126. doi: 10.1186/s12967-021-02792-8
181. Fang Y, He J, Janssen HLA, Wu J, Dong L, Shen XZ. Peroxiredoxin 1, Restraining Cell Migration and Invasion, is Involved in Hepatocellular Carcinoma Recurrence. J Digestive Dis (2018) 19(3):155–69. doi: 10.1111/1751-2980.12580
182. Srivastava A, Ambrósio DL, Tasak M, Gosavi U, Günzl A. A Distinct Complex of PRP19-Related and Trypanosomatid-Specific Proteins is Required for pre-mRNA Splicing in Trypanosomes. Nucleic Acids Res (2021) 49(22):12929–42. doi: 10.1093/nar/gkab1152
183. Bailly C, Waring MJ. Pharmacological Effectors of GRP78 Chaperone in Cancers. Biochem Pharmacol (2019) 163:269–78. doi: 10.1016/j.bcp.2019.02.038
184. Xia S, Duan W, Liu W, Zhang X, Wang Q. GRP78 in Lung Cancer. J Trans Med (2021) 19(1):118. doi: 10.1186/s12967-021-02786-6
185. Samanta S, Yang S, Debnath B, Xue D, Kuang Y, Ramkumar K, et al. The Hydroxyquinoline Analogue YUM70 Inhibits GRP78 to Induce ER Stress-Mediated Apoptosis in Pancreatic Cancer. Cancer Res (2021) 81(7):1883–95. doi: 10.1158/0008-5472.CAN-20-1540
186. Zheng Y, Liu P, Wang N, Wang S, Yang B, Li M, et al. Betulinic Acid Suppresses Breast Cancer Metastasis by Targeting GRP78-Mediated Glycolysis and ER Stress Apoptotic Pathway. Oxid Med Cell Longev (2019) 2019:8781690. doi: 10.1155/2019/8781690
187. Gifford JB, Hill R. GRP78 Influences Chemoresistance and Prognosis in Cancer. Curr Drug Targ (2018) 19(6):701–8. doi: 10.2174/1389450118666170615100918
188. Zhang H, Wang SQ, Hang L, Zhang CF, Wang L, Duan CJ, et al. GRP78 Facilitates M2 Macrophage Polarization and Tumour Progression. Cell Mol Life Sci CMLS (2021) 78(23):7709–32. doi: 10.1007/s00018-021-03997-2
189. Moreno JA, Tiffany-Castiglioni E. The Chaperone Grp78 in Protein Folding Disorders of the Nervous System. Neurochem Res (2015) 40(2):329–35. doi: 10.1007/s11064-014-1405-0
190. Fang Y, Mu J, Ma Y, Ma D, Fu D, Shen X. The Interaction Between Ubiquitin C-Terminal Hydrolase 37 and Glucose-Regulated Protein 78 in Hepatocellular Carcinoma. Mol Cell Biochem (2012) 359(1-2):59–66. doi: 10.1007/s11010-011-0999-7
191. Richardson PG, Hideshima T, Anderson KC. Bortezomib (PS-341): A Novel, First-in-Class Proteasome Inhibitor for the Treatment of Multiple Myeloma and Other Cancers. Cancer Control (2003) 10(5):361–9. doi: 10.1177/107327480301000502
192. Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the First Proteasome Inhibitor Anticancer Drug: Current Status and Future Perspectives. Curr Cancer Drug Targ (2011) 11(3):239–53. doi: 10.2174/156800911794519752
193. Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase Inhibition by Small-Molecule WP1130 Triggers Aggresome Formation and Tumor Cell Apoptosis. Cancer Res (2010) 70(22):9265–76. doi: 10.1158/0008-5472.CAN-10-1530
194. Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of Proteasome Activity by a Small-Molecule Inhibitor of USP14. Nature (2010) 467(7312):179–84. doi: 10.1038/nature09299
195. Moon S, Muniyappan S, Lee SB, Lee BH. Small-Molecule Inhibitors Targeting Proteasome-Associated Deubiquitinases. Int J Mol Sci (2021) 22(12):6213. doi: 10.3390/ijms22126213
196. Pesce E, Sondo E, Ferrera L, Tomati V, Caci E, Scudieri P, et al. The Autophagy Inhibitor Spautin-1 Antagonizes Rescue of Mutant CFTR Through an Autophagy-Independent and USP13-Mediated Mechanism. Front Pharmacol (2018) 9:1464. doi: 10.3389/fphar.2018.01464
197. Yu M, Fang ZX, Wang WW, Zhang Y, Bu ZL, Liu M, et al. Wu-5, a Novel USP10 Inhibitor, Enhances Crenolanib-Induced FLT3-ITD-Positive AML Cell Death via Inhibiting FLT3 and AMPK Pathways. Acta Pharmacol Sinica (2021) 42(4):604–12. doi: 10.1038/s41401-020-0455-x
198. Zhang W, Sartori MA, Makhnevych T, Federowicz KE, Dong X, Liu L, et al. Generation and Validation of Intracellular Ubiquitin Variant Inhibitors for USP7 and USP10. J Mol Biol (2017) 429(22):3546–60. doi: 10.1016/j.jmb.2017.05.025
199. Chuang SJ, Cheng SC, Tang HC, Sun CY, Chou CY. 6-Thioguanine is a Noncompetitive and Slow Binding Inhibitor of Human Deubiquitinating Protease USP2. Sci Rep (2018) 8(1):3102. doi: 10.1038/s41598-018-21476-w
200. Treppiedi D, Di Muro G, Marra G, Barbieri AM, Mangili F, Catalano R, et al. USP8 Inhibitor RA-9 Reduces ACTH Release and Cell Growth in Tumor Corticotrophs. Endocrine-Related Cancer (2021) 28(8):573–82. doi: 10.1530/ERC-21-0093
201. Sha B, Sun Y, Zhao S, Li M, Huang W, Li Z, et al. USP8 Inhibitor-Induced DNA Damage Activates Cell Cycle Arrest, Apoptosis, and Autophagy in Esophageal Squamous Cell Carcinoma. Cell Biol Toxicol (2022). doi: 10.1007/s10565-021-09686-x
202. Kageyama K, Asari Y, Sugimoto Y, Niioka K, Daimon M. Ubiquitin-Specific Protease 8 Inhibitor Suppresses Adrenocorticotropic Hormone Production and Corticotroph Tumor Cell Proliferation. Endocrine J (2020) 67(2):177–84. doi: 10.1507/endocrj.EJ19-0239
203. Han J, Tian Y, Yu L, Zhang Q, Xu X, Zhang Y, et al. Discovery of Novel USP8 Inhibitors via Ubiquitin-Rho-110 Fluorometric Assay Based High Throughput Screening. Bioorg Chem (2020) 101:103962. doi: 10.1016/j.bioorg.2020.103962
204. Morgan M, Ikenoue T, Suga H, Wolberger C. Potent Macrocycle Inhibitors of the Human SAGA Deubiquitinating Module. Cell Chem Biol (2022) 29(4):544–54.e4. doi: 10.1016/j.chembiol.2021.12.004
205. Wang D, Ma L, Wang B, Liu J, Wei W. E3 Ubiquitin Ligases in Cancer and Implications for Therapies. Cancer Metastasis Rev (2017) 36(4):683–702. doi: 10.1007/s10555-017-9703-z
206. Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-Like Protein Conjugation and the Ubiquitin-Proteasome System as Drug Targets. Nat Rev Drug Discov (2011) 10(1):29–46. doi: 10.1038/nrd3321
Keywords: ubiquitin, deubiquitinating enzymes, hepatocellular carcinoma, targeted, structure, inhibitors
Citation: Zhao J, Guo J, Wang Y, Ma Q, Shi Y, Cheng F, Lu Q, Fu W, Ouyang G, Zhang J, Xu Q and Hu X (2022) Research Progress of DUB Enzyme in Hepatocellular Carcinoma. Front. Oncol. 12:920287. doi: 10.3389/fonc.2022.920287
Received: 14 April 2022; Accepted: 24 May 2022;
Published: 27 June 2022.
Edited by:
Fan Feng, The 302nd Hospital of PLA, ChinaReviewed by:
Nurulisa Zulkifle, Universiti Sains Malaysia (USM), MalaysiaPengyun Li, Beijing Institute of Pharmacology & Toxicology, China
Copyright © 2022 Zhao, Guo, Wang, Ma, Shi, Cheng, Lu, Fu, Ouyang, Zhang, Xu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoge Hu, aHV4aWFvZ2UyMDA4eG11QDE2My5jb20=; Qiuran Xu, d2luZHdheTYyNkBzaW5hLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Jie Zhao
Jie Zhao Jinhui Guo
Jinhui Guo Yanan Wang1
Yanan Wang1 Qiuran Xu
Qiuran Xu