- 1Clinical Research Development Unit of Tabriz Valiasr Hospital, Tabriz University of Medical Sciences, Tabriz, Iran
- 2Department of Medical Genetics, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
- 3Woman’s Reproductive Health Research Center, Tabriz University of medical sciences, Tabriz, Iran
- 4Department of Pharmacognosy, College of Pharmacy, Hawler Medical University, Erbil, Iraq
- 5Center of Research and Strategic Studies, Lebanese French University, Erbil, Iraq
- 6Urology and Nephrology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 7Institute of Human Genetics, Jena University Hospital, Jena, Germany
- 8Cancer Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Retinoblastoma (RB) is one of the most common childhood cancers caused by RB gene mutations (tumor suppressor gene in various patients). A better understanding of molecular pathways and the development of new diagnostic approaches may lead to better treatment for RB patients. The number of studies on ceRNA axes is increasing, emphasizing the significance of these axes in RB. Circular RNAs (circRNAs) play a vital role in competing endogenous RNA (ceRNA) regulatory axes by sponging microRNAs and regulating gene expression. Because of the broadness of ceRNA interaction networks, they may assist in investigating treatment targets in RB. This study conducted a systematic scoping review to evaluate verified loops of ceRNA in RB, focusing on the ceRNA axis and its relationship to circRNAs. This scoping review was carried out using a six-step strategy and the Prisma guideline, and it involved systematically searching the publications of seven databases. Out of 363 records, sixteen articles were entirely consistent with the defined inclusion criteria and were summarized in the relevant table. The majority of the studies focused on the circRNAs circ_0000527, circ_0000034, and circTET1, with approximately two-fifths of the studies focusing on a single circRNA. Understanding the many features of this regulatory structure may help elucidate RB’s unknown causative factors and provide novel molecular potential therapeutic targets and medical fields.
1 Introduction
Retinoblastoma (RB) is the most frequent type of childhood eye tumor. RB is caused by a mutation in the tumor suppressor gene Rb1. It is well-known that RB is a typical illustration of Knudson’s two-hit theory (1). It is a rapidly developing tumor, with a doubling period of 15 days in highly young patients (2). The global prevalence of RB ranges between 1/15,000 and 1/20,000 live births (3). The International Classification of RB categorizes intraocular tumors based on tumorigenesis and pattern. They are divided into five categories: A through E. The odds of saving the eye diminish as the group progresses from A to E. Enucleation is favored in unilateral patients (tumor in a single eye) when the malignancy reaches Groups D and E. In bilateral patients (tumors in both eyes), the eye with the most advanced tumor is enucleated (4). External beam radiation, episcleral plaque radiotherapy, enucleation, cryotherapy, and photocoagulation were formerly the standard therapies for children with RB (5). RB therapy has evolved significantly during the last decade. In this regard, with shifting views on radiotherapy concerns, intravenous and intra-arterial chemotherapies have become the cornerstone of RB treatment since they have been proven to efficiently reduce tumor size, metastasis, and preserve eyesight (6). However, their clinical applicability is restricted owing to the possibility of systemic toxicity, drug resistance, and fast blood clearance (7). Remarkably, attention to the molecular mechanisms like competing endogenous RNA (ceRNA) axes involved in all RB processes, from formation to even resistance to chemotherapy, provides the basis for this disease to be recognized as more fundamental.
CeRNA are transcripts that compete for shared microRNAs to cross-regulate each other (miRNAs) (8). The latter are short RNAs of 21–23 nucleotides in length that may lead Argonaute proteins to target transcripts by base pairing, causing them to degrade or repress translation (9, 10). Franco-Zorrilla et al. described a phenomenon known as ‘target mimicry’ in which a non-coding RNA in plants may sequester miR-399 and de-repress its target (11). Not long after, Ebert et al. discovered a similar phenomenon in animal cells. Ectopic production of a miRNA that has a large number of binding sites (also referred to as miRNA response elements (MREs)) results in miRNA sequestration that is hardly visible but leads to a 1.5- to 2.5-fold overexpression of the targets of the miRNA in this research (12). As a result, the term “RNA sponge” was created to characterize the phenomenon of miRNAs being absorbed by overexpressed MRE-containing transcripts. Following that, the RNA sponge phenomenon was found in various malignancies (13, 14). The term ‘ceRNA’ was created in 2011 to characterize this additional layer of posttranscriptional control (15).
Circular RNA (circRNA) is a non-coding RNA that primarily regulates biological processes via gene control. CircRNAs contain many binding sites for miRNA and hence operate like sponge, absorbing miRNAs (16). CircRNA may also interact with other RNAs through base pairing (17). Furthermore, by interacting with proteins, circRNA may limit their function (18). Although circRNA is a form of non-coding RNA, under specific circumstances, it may serve as a translation template for protein synthesis and yield functional proteins (19). CircRNAs have recently been discovered to affect host gene expression, interact with RNA-binding proteins that govern transcription, function in cis transcriptional regulation, and even regulate and control alternative splicing (20). The fact that ceRNA interaction networks are multifactorial means that they may be valuable in investigating complicated diseases such as RB in terms of therapeutic targets since by targeting just one of them, the levels of several disease-related RNAs change simultaneously (21). We performed a systematic scoping review in this study to look into validated ceRNA loops in RB. The focus of our study was the circRNA-ceRNA axes, which have been associated with RB etiology and might be utilized as therapeutic targets.
2 Methods
The approach for this study was based on Arksey and O’Malley’s (22) scoping review framework, which was subsequently revised by Levac et al. (23). This process consists of five unique steps: (1) identifying the research question, (2) identifying relevant studies, (3) study selection, (4) data charting, and (5) collating, summarizing, and reporting results. We did not use consultation in our research, an optional sixth stage in the scoping review. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews (PRISMA-ScR) Checklist also effectively guided this review (24).
2.1 Identifying the Research Question
The purpose of this study was to compile and organize the existing information on circRNA-associated ceRNA loops in RB. In order to achieve this goal, we aimed to respond to the following question: What is known about circRNA-associated ceRNA regulatory axes in RB based on the extant literature?
2.2 Identifying Relevant Studies
An initial restricted search of PubMed and Embase was carried out, and then the keywords in the title and abstract, as well as the index terms that were used in the articles, were evaluated for significance. A second search was conducted throughout the PubMed, Embase, Scopus, Web of Science, and Cochrane databases without any restrictions, using keywords, MeSH, or Emtree terms identified in the original search. Additionally, searches were conducted in two gray-listed (i.e., difficult to find or unpublished) literature databases: Google Scholar and ProQuest. The most recent search was conducted on March 23, 2022. We also looked through the reference lists of relevant literature and reviewed papers for additional sources.
2.3 Study Selection
The included papers met the following criteria: (1) explicitly discussing the circRNA-associated ceRNA axis in RB, (2) published in English, and (3) be original research. The exclusion criteria were: (1) non-RB or any ocular tumor studies, (2) studies that did not utilize human specimens or cell lines, and (3) studies that did not use a molecular approach to verify the ceRNA loop components. The title and abstract of each article were separately reviewed for eligibility according to the criteria mentioned earlier by two reviewers (MRA, HS). The remaining publications’ entire texts were assessed, and those that met the eligibility requirements were included in the final data analysis. Any differences were resolved via conversation or, if necessary, with a third reviewer (MR).
2.4 Data Charting
Two reviewers (MRA and HS) extracted data separately into a predesigned charting template in Microsoft Excel. It listed the first author, the year of publication, the country, the type of study, the cell line(s), human samples, animal models, methods, ceRNAs, shared miRNAs (s), and the key findings.
2.5 Collating, Summarizing, and Reporting the Results
We conducted both quantitative and qualitative analyses. We produced a descriptive numerical overview of the features of the included articles for the quantitative section. As part of the qualitative analysis, we wrote a narrative review of the current information that answered our research question, focusing on how important the results were in a larger context, as suggested by Levac et al. (23).
3 Results
3.1 Search Results
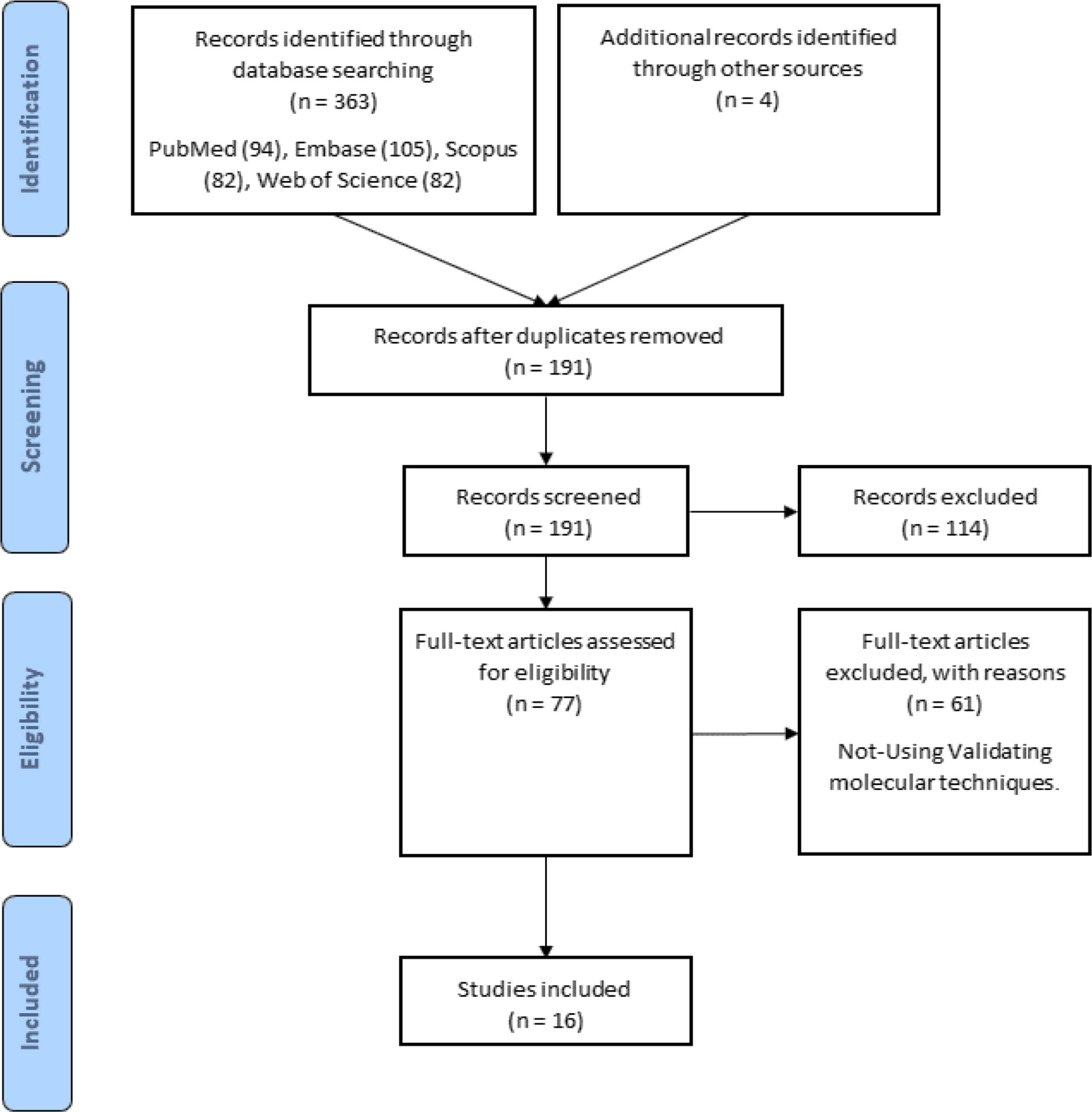
The flow chart in Figure 1 depicts the various steps involved in locating eligible studies. A total of 363 articles were identified from multiple sources, of which 176 were duplicates. One hundred seventy-one articles were omitted due to their insignificance. After reading the full texts of the remaining 16 articles for the purpose of evaluation, the results are presented in Table 1.
3.2 Study Characteristics
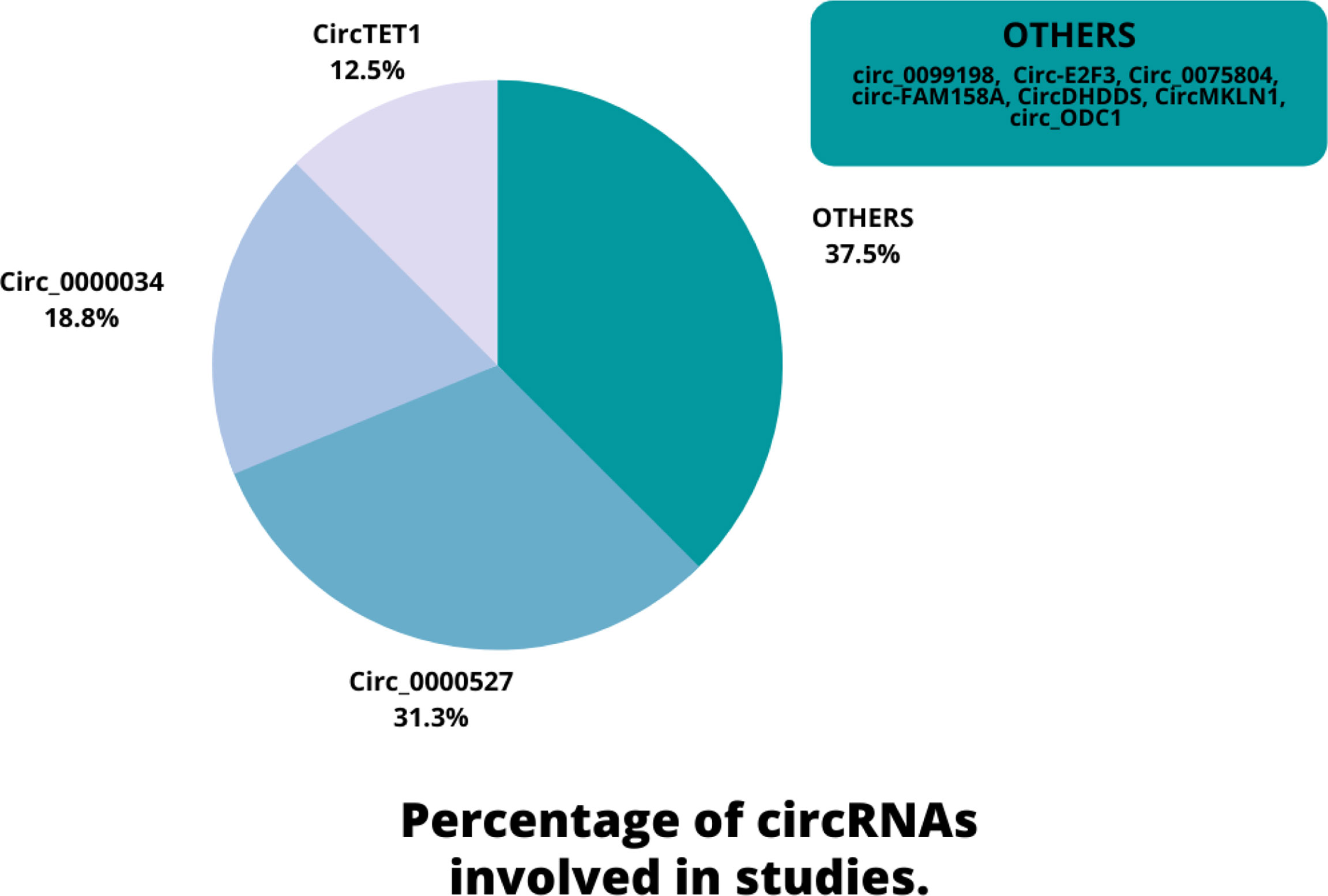
Eligible studies were published between 2019 and 2022. These studies included 465 samples of RB patients and 343 healthy controls, based on the mentioned number. The gender of patients and controls is rarely mentioned. An animal model was used in 13 studies, and mice were the only animals involved. Weri-Rb-1, Y79, SO-RB50, SO-Rb70, RB355, HRA, HXO-RB44, ARPE-19, and hTERT-RPE1 are among the cell lines used in these studies. Figure 2 depicts the frequency of circRNAs. Notably, the distribution of studies is restricted to only one country, namely China.
4 Discussion
4.1 CircRNA Features and Biogenesis
A recent study has shown that circRNAs are commonly formed from one to five exons, with lengths ranging from just a few hundred to thousands of nucleotides (41). CircRNAs produced via back-splicing have numerous fundamental properties: I. Circular RNAs have a closed ring shape, which means they lack either 5′–3′ polarity or a polyadenylated tail, making them resistant to exonuclease degradation and much more stable than linear RNA (42). II. CircRNAs are frequently expressed in eukaryotic cells, and high-throughput sequencing has shown that more than one million circRNAs exist in human tissues (43). III. The majority of circRNAs are found in the cytoplasm, with a minor number of circRNAs found in the nucleus as well (44). IV. The majority of circRNAs contain substantially conserved sequences across various species (45). V. CircRNAs demonstrate tissue-and developmental stage-specific and dynamic expression patterns (46). VI. CircRNAs act as transcriptional or posttranscriptional regulators (44).
It is critical to understand how circRNAs are created in order to classify them. In conjunction with the activation of the spliceosomal machinery, canonical splicing is a process in which the introns of precursor messenger RNAs (pre-mRNAs) are removed, and exons are then joined to generate mature messenger RNAs (47). In contrast to canonical splicing of mRNAs, circular RNAs are produced by nonsequential back-splicing of pre-mRNAs by spliceosomes rather than conventional splicing (5). Back-splicing is the process of reversibly joining the upstream 5′ splice donor site to the downstream 3′ splice acceptor site to form a covalently closed structure (5, 48). According to recent research, back-splicing requires both conventional splicing signals and canonical splicing machinery (49, 50).
Two key factors may aid back-splicing circularization: intron pairing and RNA-binding proteins (RBPs). On one hand, intron pairing causes exons to approach one another through ALU elements (51) or nonrepetitive yet complementary sequences (52). Long flanking introns with complementary ALU repeats are essential for developing circHIPK3 (53). On the other hand, some RBPs, such as MBL (MBNL1), QKI, and FUS (54), have the ability to bind firmly and precisely to conserved regions of flanking introns, thus promoting exon circularization and, ultimately, the production of circular RNAs. Furthermore, the action of adenosine deaminase (ADAR) specific to double-stranded RNA (dsRNA) on the editing of adenosine to inosine (50) and the unwinding of the dsRNA helix structure by ATP-dependent RNA helicase A (also known as DHX9) suppresses the synthesis of circRNAs (55, 56). Furthermore, circRNA production may be aided by SFs such as ESRP1 (57) and the elongation velocity of RNA polymerase II (58).
4.2 CircRNAs’ Functional Mechanisms
4.2.1 MiRNA Sponge
CircRNAs are involved in a wide range of biological activities because of their distinct structures and other characteristics (59). The capacity of circRNAs to act as miRNA sponges is their most notable function. CircRNAs that include miRNA binding sites bind directly to the matching miRNAs, inhibiting miRNA function and regulating target gene expression. The first circRNA to act as a ceRNA was CDR1as (antisense to the cerebellar degeneration-related protein 1 transcript), which has more than 60 conserved binding sites for miR-7 (44, 60). It bonded to miR-7 and inhibited midbrain growth in zebrafish, comparable to miR-7 knockdown. Hansen et al. hypothesized the same year that mouse circRNA circSRY functioned as a miR-138 sponge with more than ten binding sites (61, 62). Despite the recent identification of numerous circRNAs that operate as miRNA sponges (63, 64), it is essential to note that this represents just a fraction of the hundreds of human circRNAs that have several computational binding sites for specific microRNAs. Interestingly, P. falciparum and S. cerevisiae do not have a miRNA pathway (65), but they do have a large number of circRNAs. As a result, the idea of a miRNA sponge may not represent a universal function for circRNAs.
4.2.2 Splicing or Transcriptional Regulation
Alternative splicing is frequently related to a variety of biological activities (66). CircRNAs were discovered to influence alternative splicing through RNA-mediated interaction, therefore influencing gene expression (67). According to Ashwal-Fluss et al., circMBL, produced from the second exon of the splicing factor muscle blind (MBL/MBNL1), might compete with linear splicing pre-mRNA (50). MBL levels considerably influence circMbl biosynthesis depending on the MBL binding sites. CircMbl has the potential to inhibit the synthesis of its parental mRNA. Another noteworthy example was published in 2015, demonstrating that circRNAs might govern the transcription of the genes they regulate (68). Li et al. discovered that a subclass of circRNA called EIciRNA was mainly located in the nucleus and stimulated transcription of their parental genes by interacting with U1 snRNP, revealing a unique regulatory mechanism for transcriptional control via specialized RNA-RNA interactions (68).
4.2.3 Protein Scaffolding and Interaction With RBPs
In addition to MREs, circRNAs include a large number of RBP binding sites. For example, in human cervical cancer HeLa cells, it has been revealed that the antigen R (HuR) interacts with a large number of circRNAs (69). It has been shown that CircPABPN1, which is generated from the PABPN1 gene, may compete with PABPN1 mRNA for binding to HuR, hence limiting the translation of PABPN1. Also discovered were elciRNAs and ciRNAs, which are mainly found in the nucleus and have the ability to bind with small nuclear ribonucleoprotein U1 (snRNP U1) and boost RNA polymerase II (Pol II) transcriptional activity on their parental genes, acting as cis regulators of their respective genes (68, 70). Also of note is that the binding of circRNAs to specific functional proteins may have an impact on several signaling pathways, resulting in alterations in homeostasis (71). In their study, Du et al. discovered that the circular RNA circ-Foxo3 is associated with the anti-senescence protein ID-1, the transcription factor E2F1, and the anti-stress proteins FAK and HIF1α and holds them in the cytoplasm, preventing them from performing their respective duties (72).
4.2.4 Translate Proteins
Although circRNAs are classified as non-coding RNAs, growing evidence suggests that certain circRNAs are translatable (47). Unlike mature mRNA translation, which generally requires the presence of a 5′ end 7-methylguanosine (m7G) cap structure and a 3′ poly(A) tail, circRNA translation is distinct because it does not need the presence of these structures (73). It was proven in 1998 that a circular mRNA containing the whole open reading frame (ORF) of green fluorescent protein (GFP) could effectively encode GFP in Escherichia coli (74). Legnini et al. discovered that circ-ZNF609 includes an ORF and may be translated into a protein in murine myoblasts when activated by IRES (75). Further research found that certain circRNAs containing internal ribosome entry sites (IRESs) may function as the entrance point for ribosomes, therefore triggering translation (76, 77). Surprisingly, the most prevalent RNA modification, N6-methyladenosine (m6A), was discovered to improve circRNA protein-coding capacity (78). According to recent research, FBXW7-185aa, a new 21-kDa protein, was encoded via a spanning junction open reading frame in circ-FBXW7 that was directed by an internal ribosome entry site. In glioblastoma, upregulation of FBXW7-185aa suppressed proliferation and cell cycle progression, as well as reduced the half-life of c-Myc by antagonizing USP28-induced c-Myc stabilization, while knockdown of FBXW7-185aa increased malignant phenotypes both in vitro and in vivo, as previously reported (79). Similarly, Pamudurti N. R. et al. discovered that circMbl could also translate proteins without the need for a cap (80). The protein-coding function sheds new light on the involvement of circRNAs in diseases.
4.3 CircRNA: Oncogene or Tumor Suppressor Gene
Emerging evidence suggests that dysregulated circRNAs have both tumor-suppressive and oncogenic roles in cancer origin and development, affecting a wide range of cellular processes. The tumor structure is comprised of cells that have mutations in the genes involved in proliferation and differentiation (81). Oncogenes contribute to the stimulation of cell proliferation. Changes in these genes may vary from the development of novel oncogenes to the overexpression of previously proto-oncogenes. In contrast, tumor suppressor genes (TSGs) inhibit cell growth through the opposite mechanism (82). In this regard, the role of several circRNAs as oncogenes has been investigated. Circ-ABCB10 has been identified as a breast cancer oncogene, and circ-ABCB10 knockdown suppresses proliferation and increases apoptosis in breast cancer cells (83). Similarly, circ-ZEB1 was one of the oncogenic circRNAs that was significantly higher in triple-negative breast cancer (TNBC) tumor tissues and tumor cell lines (84). Circ-ZEB1 knockdown has been shown to reduce TNBC cell proliferation and tumor formation by releasing miR-448 and consequently decreasing the expression of the miR-448 target, eEF2K (84). CircCD44 with oncogenic roles is substantially expressed in TNBC and is adversely correlated with TNBC patient prognosis (85). CircCD44 enhances TNBC proliferation, migration, invasion, and tumorigenesis (85). Remarkably, the oncogenic role of circRNAs in other cancers has also been considered, including circRNA_100859 (86), circAGO2 (87), PIP5K1A (88), circPPP1R12A (89), CCDC66 (90), GLIS2 (91), and circ_0084615 (92) in colon cancer, circRNA-C190 (93), circNDUFB2 (94), circ_100146 (95), circRNA_001846 (96), and circ_0087862 (97) in lung cancer, circAF4 (98), and circSPI1 (99) in leukemia.
On the contrary, studies have shown changes in circRNAs and their functions as TSGs. Circ_0006220 expression was lower in TNBC compared to other subtypes of BC tissues and cell lines. Notably, circ_0006220 functions as a TSG, inhibiting TNBC cell proliferation, migration, and invasion (100). CircCDYL was down-regulated in TNBC tissues, and its expression was found to be positively associated with patient survival rates (101). CircCDYL induces apoptosis and suppresses the proliferation of breast cancer cells with a malignant phenotype (101). The function of circRNAs as TSGs has also been proven in many cancers, including circDDX17 (102), circ_PLXNB1 (103), circTADA2A (104), and Circ-SMARCA5 (105) in colorectal cancer, circ_0004872 (106), circDIDO1 (107), circMAPK1 (108), circ_0035445 (109) and circCCDC9 (110) in gastric cancer, circPLEKHM3 (111) in ovarian cancer, circ-MYBL2 (112) in multiple melanoma, and circSLC8A1 (113) in prostate cancer.
4.4 Potentials of CircRNAs in CeRNA Axes
Several circRNAs exhibit tissue and developmental stage-specific expression patterns (44). According to recent research, the global abundance of circRNAs was decreased in colorectal cancer (CRC) tissues compared to normal tissues (114). The abundance of a circRNA known as Has-circ-002059 was likewise lower in malignant vs. non-cancerous tissues in gastric cancer (115). Another recent study detected circRNAs in exosomes (small membrane vesicles with endocytic origins released by cells) and discovered that the profiles of seral exosomal circRNAs may identify CRC patients from healthy controls (116). Figure 3 illustrates a schematic view of the overall function of circRNAs.
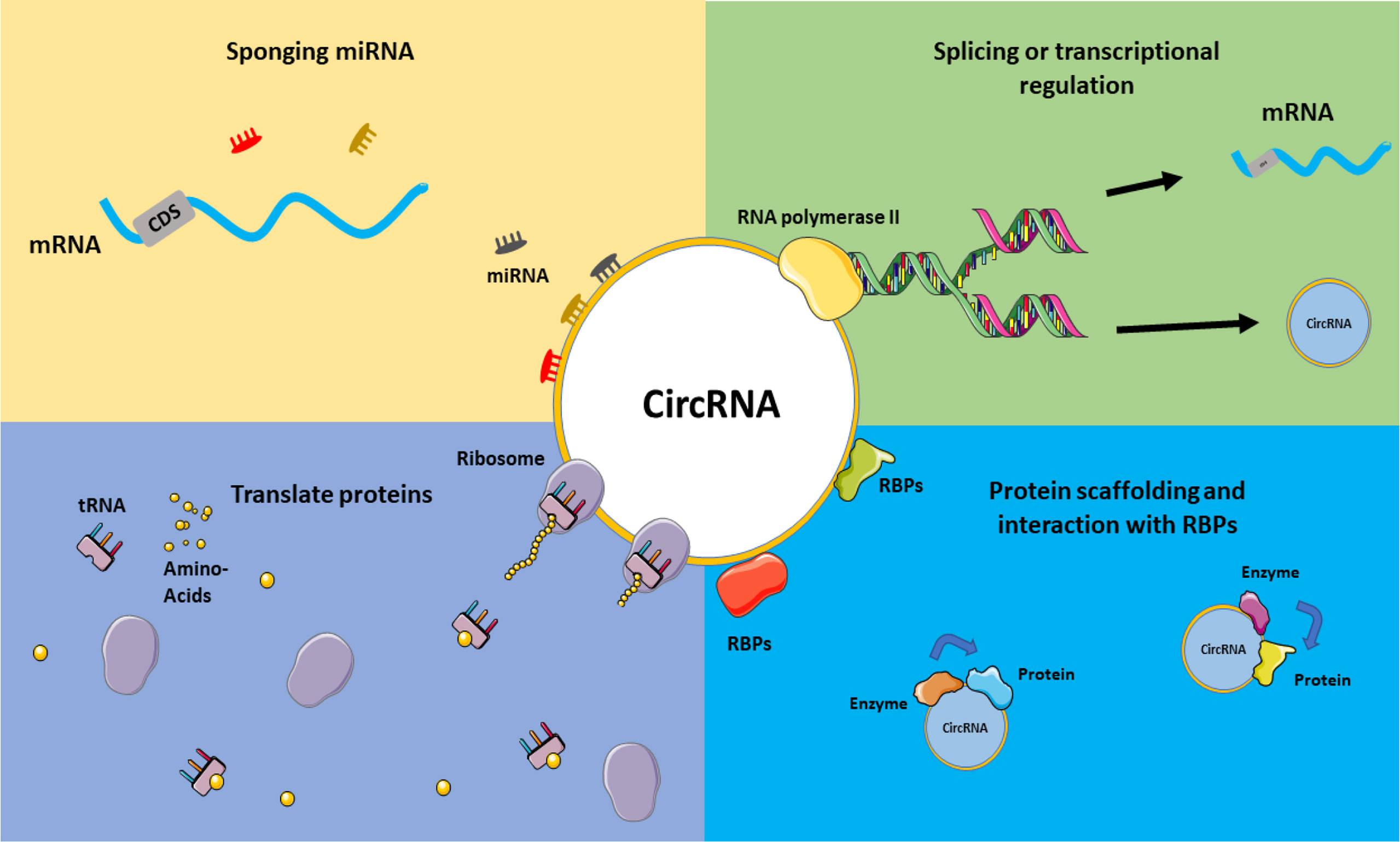
Figure 3 CircRNAs mechanism of actions. Various functions can be considered for circRNAs, including acting in splicing or transcriptional regulations, sponging miRNAs and acting on ceRNA axes, translating proteins and interacting with RBPs, and their role in the protein scaffolding process. This graphical figure was created using the vector image bank of Servier Medical Art (http://smart.servier.com).
The structure of circular RNAs is relatively similar to that of linear RNAs, but they are typically more stable than their linear counterparts because they lack accessible ends and are hence resistant to exonucleases (44). It has been hypothesized that circRNA may create active ceRNA pairs with their linear counterparts due to their unique characteristics like miRNA sponging. CDR1as, a brain-enriched circRNA, has been reported to act as a miRNA sponge for miR-7 in a variety of human disorders (44, 61), including colon cancer (117), gastric cancer (118), esophageal cancer (119), and myocardial infarction (120). Other ceRNA connections discovered in cancer include circPVT1-miR125 in gastric cancer (64), circITCH-miR7/miR214 in lung cancer (121), circHIPK3-miR124 in liver cancer (122), and circTTBK2-miR217 in glioma (123).
In addition, it is interesting that the ceRNA axes in RB have received much attention as a representative of ocular tumors in recent years, and the potential of these regulatory axes is becoming more and more meaningful. On the other hand, studies on axes involving circRNAs in RB are still in their infancy and can play an essential role in the cell by considering the properties of a circRNA. In the following, studies on ceRNA axes in which circRNAs play a crucial role have been obtained in the form of a systematic review study and will be further discussed, and it is expected that this study can be a guide for further studies in this field and cause these studies to receive more attention (Figure 4).
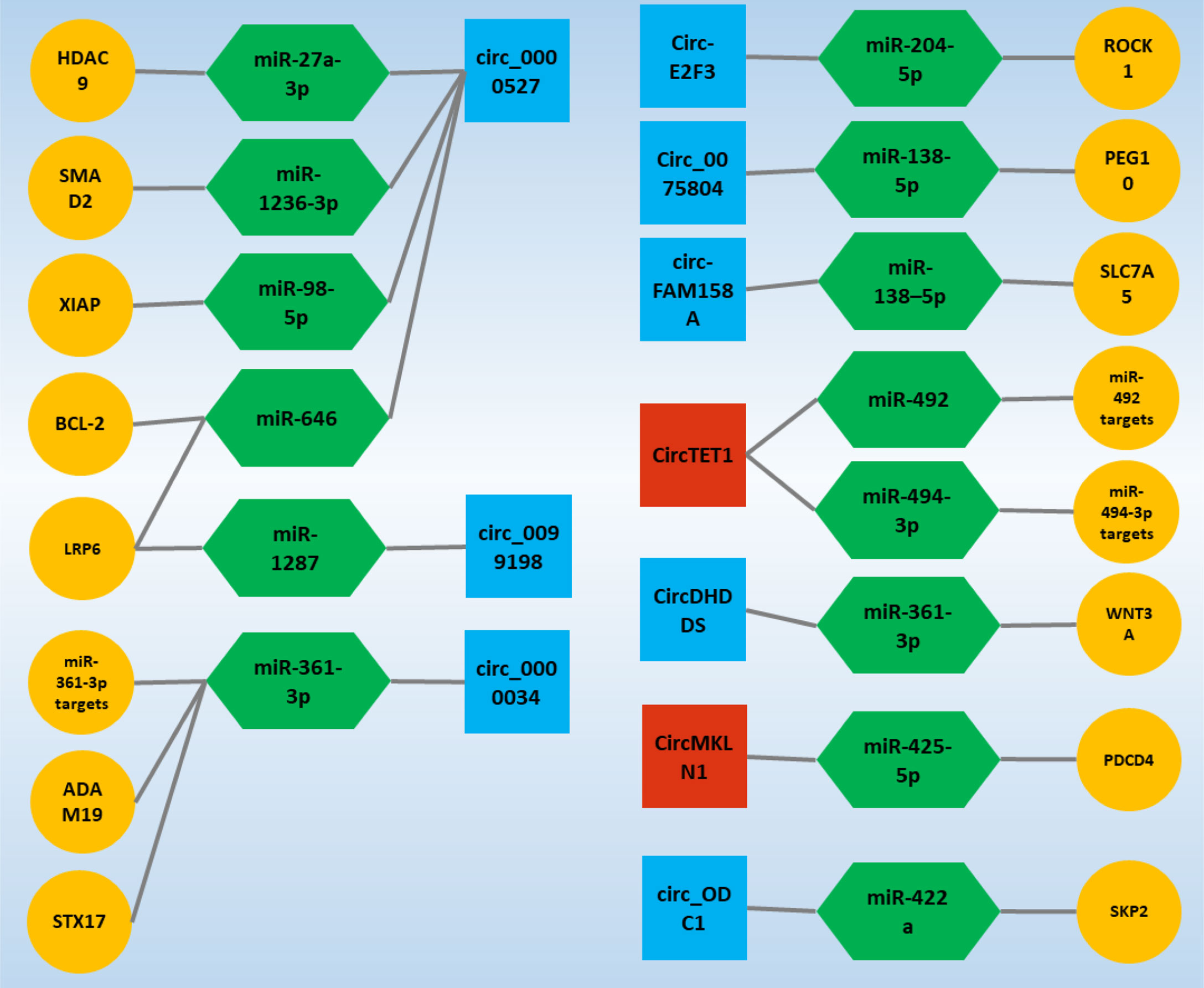
Figure 4 Validated circ-associated ceRNA axes in RB. CircRNAs, miRNAs, and mRNAs are represented by the square, Hexagonal, and circle, respectively. The blue and red colors of the squares indicate an increase and decrease in the expression of circRNAs, respectively. CircRNA, circular RNA; miRNA, microRNA.
4.5 Circ_0000527
CircRNA’s function in carcinogenesis and cancer development has received much interest. CircRNA contributes to tumor development by controlling cancer cells’ malignant biological activities such as proliferation, apoptosis, and metastasis (124). Circ_0001649 is claimed to inhibit RB cell proliferation and apoptosis by blocking the AKT/mTOR signaling pathway, and its low expression level implies a bad prognosis in patients (125); circ_ODC1 positively regulates SKP2 and increases RB cell proliferation (25). Surprisingly, it was revealed in another study that the expression level of circ_0000527 in RB tissues and cells was dramatically elevated and that its high expression was highly connected with the degree of differentiation and cTNM staging (30). These findings suggest that circ_0000527 was likely to predict a poor prognosis in RB patients (30). Furthermore, circ_0000527 overexpression significantly increased cell proliferation, motility, and invasion while significantly decreasing cell apoptosis (26). On the other hand, knocking down circ_0000527 had the opposite effect (30, 38). At the same time, miR-646 expression in RB showed a significant decrease, and since miR-646 has several binding sites for circ_0000527, further studies confirmed that circ_0000527 negatively regulates miR-646 expression (26). Interestingly, LRP6, which is regulated by miR-646 and is one of the primary expression genes expressed in malignancies, also experienced a significant increase in expression in the form of a ceRNA axis due to the sponging of miR-646 by circ_0000527 (30). However, studies on increasing the expression of LRP6 in the form of the ceRNA axis in RB were not stopped, and sponge miR-1287 confirmed the increase in the expression of this gene by circ_0099198 in another study (33). Circ_0099198 in RB experiences a significant increase in expression, and this increase in expression shows its effects by decreasing the expression of miR-1287, which is responsible for regulating the expression of LRP6 (33).
Most notably, miR-646 has been identified as a tumor suppressor in a variety of human malignancies. For example, miR-646 inhibits cell growth and migration in lung cancer by negatively regulating the EGFR/Akt pathway (126); in colorectal cancer, miR-646 inhibits cancer development by suppressing NOB1 (127). MiR-646 inhibits the spread of clear cell renal carcinoma cells by downregulating the nin one binding protein (128). MiR-646 suppresses non-small cell lung cancer growth and metastasis through binding to FGF2 and CCND2 (129). In this regard, miR-646 transfection in RB cells was associated with decreased viability, migration, and invasion of these cells (26). Although the oncogenic effects of circ_0000527 have been demonstrated by sponge miR-646, another ceRNA axis was identified by miR-646 targeting BCL-2. Increased circ_0000527 expression by miR-646 sponging increased BCL-2 expression (26), which is one of the significant genes in various malignancies (130).
Circ_0000527 acted as a sponge for miR-1236-3p, which targeted SMAD2 directly, and as a result, miR-1236-3p expression was reduced in RB tissues and cells (37). Circ_0000527 knockdown on RB cell malignancy reduced the effects of MiR-1236-3p inhibition. Furthermore, miR-1236-3p overexpression inhibited RB cell development, whereas SMAD2 elevation reversed the impact (37). Moreover, circ_0000527 knockdown inhibited tumor development in vivo (38). Circ_0000527 also sponges miR-98-5p to form another ceRNA axis, promoting the development of RB by increasing the expression of the miR-98-5p target gene, which is XIAP (38).
Nonetheless, investigations in RB on circ_0000527 have demonstrated an increase of this circRNA in the presence of the ceRNA axis, which enhances the expression of genes that lead to the formation and progression of RB and even respond to role treatment sponging particular target miRs. These findings highlight circ_0000527’s significant potential in the processes behind RB and establish circ_0000527 as one of the primary circRNAs in RB.
4.6 Circ_0000034
Circ_0000034 is one of the most studied circRNAs in RB. All studies around this circRNA have shown an increase in the expression of this transcript (28, 35, 55). Interestingly, in studies of circ_0000034, the only target of this circRNA was miR−361−3p, whose targets had the potential to influence the mechanisms involved in RB and are essential goals in the process of formation and progression this malignancy. STX17 is a target for miR-361-3p that regulates its expression. Increased circ_0000034 expression due to the binding sites on miR-361-3p sponging miR-361-3p, which increases STX17 expression. On the other hand, the miR-361-3p expression has decreased in studies in RB (27). ADAM19 is another miR-361-3p target gene that experiences increased expression in the circ_0000034/miR-361-3p/ADAM19 ceRNA axis due to miR-361-3p sponging (35). Silencing circ_0000034 also showed that RB progress was suppressed due to circ_0000034’s inability to sponge miR-361-3p. In this condition, miR-361-3p inhibits RB progression by regulating ADAM19 expression (35). Similarly, CircDHDDS, a miR-361-3p molecule sponge, regulated WNT3A expression. As a result, the circDHDDS/miR-361-3p/WNT3A axis promoted RB development by regulating RB cell proliferation, cell cycle program, migration, and invasion (29). In general, since the expression of circ_0000034 increases in RB and because silencing this circRNA inhibits the progression and development of RB, this circRNA can be recognized as one of the main circRNAs in RB.
4.7 Other CircRNAs
The ceRNA axes inside the cell are in equilibrium under normal conditions. Increasing or decreasing the expression of each component of an axis can advance the trend in favor of abnormal conditions. The majority of circRNAs studied in the form of ceRNA axes are associated with increased expression, but circTET1 in RB is disturbed by decreased expression of the desired ceRNA axis. It was discovered that CircTET1 expression was downregulated in RB tissues and cells and that its upregulation hampered the growth of RB cells in culture. CircTET1 was shown to be a miR-492/miR-494-3p sponge, and it blocked the Wnt/-catenin pathway via miR-492/miR-494-3p (31). Similarly, circMKLN1 expression was reduced in RB tissues and cells. Elevated levels of circMKLN1 were linked to better results in RB patients. CircMKLN1 overexpression inhibited RB cell proliferation, migration, and invasion in vitro. MiR-425-5p was shown to be a target of circMKLN1, and it was discovered that increasing miR-425-5p levels might counteract the effects of circMKLN1 overexpression on RB cell malignant tendencies. Furthermore, because circMKLN1 is a target gene for miR-425-5p, suppressing PDCD4 may enhance circMKLN1’s inhibitory effects in RB cell development and metastasis (34).
Nonetheless, the rest of the circRNAs, which have only been studied in one study, have been shown to experience increased expression. Among these, miR-138-5p is considered one of the potential miRs in RB, which is located in specific ceRNA axes due to binding sites for circ_0075804 and circ-FAM158A (36, 39). The circ_0075804/miR-138-5p/PEG10 axis was identified as a ceRNA axis in RB. Through its interaction with miR-138-5p, Circ_0075804 led to the development of the malignant phenotypes of RB cells. Circ_0075804 By sponging, miR-138-5p acts as a barrier to the activity of this miR and cannot regulate the expression of its target gene, which is PEG10, and as a result, PEG10 expression is significantly increased, which acts to cause oncogenic effects in RB (36). Similarly, circ-FAM158A and SLC7A5 were up-regulated, and miR-138-5p was down-regulated in RB. Circ-FAM158A silencing inhibited RB cell proliferation, invasion, and migration while promoting G0/G1 cell cycle arrest and cell apoptosis. Circ-FAM158A functioned as an oncogene in RB by sponging miR-138-5p, which regulates SLC7A5 expression (39).
On the other hand, Circ-E2F3 was shown to be increased in RB tissues and cells, and silencing of circ-E2F3 suppressed RB cell proliferation, migration, invasion, and triggered apoptosis in vitro, as well as reduced RB tumor development in vivo. Circ-E2F3 sponged MiR-204-5p, and its inhibitor restored the inhibitory impact of circ-E2F3 silencing on RB progression. Furthermore, ROCK1 is known as the target for miR-204-5p, and it is able to control RB development by regulating ROCK1. Moreover, through sponging miR-204-5p, circ-E2F3 favorably regulated ROCK1 expression (32). Finally, given that studies of ceRNA axes centered on circRNAs are at the beginning of the path, and given what genes with potential are involved in these regulatory axes, it will demonstrate the increasing importance of these regulatory axes in RB. The importance of these genes involved in ceRNA axes will be discussed in the following.
4.8 Genes Involved in CircRNA-Associated CeRNA Axes
It is desirable to further discuss the genes located in the ceRNA axes in these studies, which are among the genes that have the potential for various cancerous processes. The genes in these ceRNA axes include SMAD2, ROCK1, WNT3A, STX17, LRP6, HDAC9, BCL-2, SLC7A5, ADAM19, XIAP, PEG10, PDCD4, and SKP2. The majority of these genes experience increased expression due to up-regulation of their ceRNA axis circRNA, but PDCD4 experiences reduced expression due to decreased circMKLN1 expression and a reduced ability to sponge miR-425-5p (34). PDCD4 is a new tumor suppressor that exhibits a wide range of actions, including the inhibition of cell proliferation, tumor invasion, metastasis, and the induction of apoptosis. In addition to binding to the translation initiation factor eIF4A and several transcription factors, the PDCD4 protein interacts with a variety of other factors, modulating their functions (131). Decreased expression of this tumor suppressor gene in RB acts in favor of the progression of this malignancy and leaves its effects. Among these, Wnt3A, a member of the Wnt family, is a transcription factor that regulates various cellular processes, including self-renewal, proliferation, differentiation, and motility. Increasing evidence suggests that Wnt3A, depending on the cancer type, either stimulates or inhibits tumor growth via the canonical Wnt signaling pathway (132). In RB development, it acts by increasing the expression induced by the circDHDDS/miR-361-3p/WNT3A ceRNA axis by influencing cell proliferation and cell cycle programming, migration, and invasion (29).
Interestingly, SMAD2 is one of the genes that in other cancers, such as prostate cancer (133) and breast cancer (134) are among the genes that appear to play a role in suppressing tumors, and silencing this gene works to promote malignancy (134), it interacts with and promotes RB progression via a ceRNA axis, circ_0000527/miR-1236-3p/SMAD2 (37). On the other hand, it is well established that LRP6 regulates breast cancer. LRP6’s expression is most often up-regulated in breast cancer tissue, and overexpression or knockdown of LRP6 causes or halts breast tumorigenesis (135). In line with the study conducted in RB, the expression of LRP6 in the form of a ceRNA axis also experiences an increase in expression, and in the direction of RB development, this increase in expression demonstrates its oncogenic effects (30, 33). SKP2 (S-phase kinase-associated protein 2) is an SCF E3 ligase-specific factor promoting cell cycle progression by degrading its targets. SKP2 substrates other than p27 have been found, including p57 (136), p130 (137), p21 (138), Tob1 (139), and FOXO1 (140). These substrates have a role in various biological activities, including cell cycle regulation, growth, differentiating, apoptosis, and survival. SKP2 has an undeniably essential role in controlling various cellular processes owing to the degradation of its substrates, the majority of which are tumor suppressor proteins (141). SKP2 is hypothesized to operate as an oncoprotein since it is responsible for the degradation of the tumor, as mentioned earlier, suppressor proteins. SKP2 overexpression has been found in a wide range of human malignancies, including lymphomas (142), prostate cancer (143), colorectal cancer (144), melanoma (145), nasopharyngeal carcinoma (146), pancreatic cancer (147), and breast carcinomas (148). In addition, the expression of SKP2 in RB in the form of a ceRNA axis also experiences an increase in expression due to the effect of circ_ODC1 on miR‐422a and the oncogenic effects of SKP2 in RB appear (25).
It was recently discovered that ROCK1 plays a critical role in regulating cell motility, angiogenesis, and migration (149). Furthermore, ROCK1 has been found to be overexpressed in a variety of solid tumors, including glioblastoma (150), melanoma (151), osteosarcoma (152), and hepatocellular carcinoma (153). According to published reports, ROCK1 overexpression is strongly associated with more highly metastatic and invasive phenotypes in human cancers (149). In this regard, ROCK1 is overexpressed in RB as part of a ceRNA axis, which promotes RB proliferation, metastasis, and invasion while decreasing apoptosis (32). Similarly, according to several research findings, HDAC9 has been implicated in the development of malignant tumors. HDAC9 overexpression has been linked to cell growth in medulloblastoma (154), lymphoma (155), oral squamous cell carcinoma (156), and breast cancer (157). It should be noted that the overexpression of this gene in RB was also proven in a study, but this increase in expression was not detected in the form of a ceRNA axis (158). HDAC9 experiences increased expression in the form of the circ_0000527/miR-27a-3p/HDAC9 ceRNA axis and exerts its oncogenic properties by activating the PI3K/AKT pathway (40). STX17 (Syntaxin 17) is an autophagy-related ER-resident SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein. Various roles for STX17 in terms of its mechanisms of action have been proposed (159).
XIAP overexpression in tumor cells has been linked to tumor aggressiveness and has been described as a mediator of resistance to chemotherapy and targeted therapy in various malignancies (160). In RB, the increase in XIAP expression also occurs due to the sponging of miR-98-5p by circ_0000527, which acts in the direction of the oncogenic properties of XIAP (38). Likewise, PEG10 appears to play a critical role in tumor growth in a variety of cancers, including hepatocellular carcinoma (161), lung cancer (162), and prostate cancer (163). In RB, miR-138-5p is sponged by circ_0075804 and forms a ceRNA axis with one of the targets of miR-138-5p, PEG10, which increases its expression and leads to a malignant phenotype (36). Bcl-2-family proteins play critical roles in cell death regulation, regulating various cell death mechanisms such as apoptosis, necrosis, and autophagy (164, 165). Increased viability, metastasis, and invasion in RB occur due to increased expression levels of BCL-2 in the form of the ceRNA axis circ_0000527/miR-646/BCL-2 (26). SLC7A5 imports essential amino acids into cancer cells, and research has shown that amino acids, particularly leucine, activate mTORC1, which regulates protein translation and cell growth, and inhibits apoptosis in malignant cells by activating mTORC1 (166, 167). Circ-FAM158A and SLC7A5 are overexpressed in RB. SLC7A5 acts as a lever to demonstrate the oncogenic properties of circ-FAM158A (39).
As mentioned, the genes considered so far in the studies, including genes with potential and influence in the processes involved in cancer, play an essential role in the development of RB. The localization of these essential genes in the ceRNA axes indicates the potential of these axes in cancer causation and emphasizes the principle that molecular mechanisms, especially ceRNA axes, should be further studied.
5 Limitations
The present study had several limitations. On the one hand, ceRNA studies centered on circRNAs are just beginning, and the number of such studies is limited. This study tried to be a roadmap for further studies in this field and increase attention and enthusiasm for future studies in RB. On the other hand, we attempted to cover all of the details pertaining to these regulatory axes and circRNAs in Table 1 in the section on key findings. All care was taken not to miss a study during the screening process, and three persons collaborated on this part so that this study would include all of the studies conducted in the field of RB, but this is conceivable due to an individual error that may have left a study off the table.
6 Conclusion
Numerous circRNAs have been discovered and studied in recent years. CircRNAs act as miRNA sponges, influencing the expression of related genes and forming a regulatory axis called ceRNA. CeRNA axes are potential axes that function from disease onset to diagnosis and treatment. This study gathered evidence that circRNAs had a fascinating impact on essential genes in RB, each of which has the potency to be a defining characteristic of this cancer. Thus, our efforts to understand different aspects of ceRNA regulation processes in RB pathogenesis provide new insights into potential molecular targets, identify ceRNA-based diagnostics, and develop ceRNA-based therapeutic potential.
Author Contributions
MT and MA wrote the draft and revised it. MR and AR designed and supervised the study. BH, HS, MM, MS-B and PH collected the data and designed the figures and tables. All the authors read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Clinical Research Development Unit of Tabriz Valiasr Hospital, Tabriz University of Medical Sciences, Tabriz, Iran for their assistance in this research.
Abbreviations
RB, Retinoblastoma; ceRNA, Competing endogenous RNA; CircRNA, Circular RNA; MRE, MicroRNA Response Element; PRISMA-ScR, Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews; RBP, RNA-binding protein; dsRNA, double-stranded RNA; snRNP, small nuclear ribonucleoprotein; ORF, open reading frame; GFP, green fluorescent protein; CRC, Colorectal Cancer; TSGs, tumor suppressor genes; TNBC, triple negative breast cancer.
References
1. Knudson AG Jr. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc Natl Acad Sci U S A (1971) 68(4):820–3. doi: 10.1073/pnas.68.4.820
2. Abramson DH, Schefler AC, Beaverson KL, Rollins IS, Ruddat MS, Kelly CJ. Rapid Growth of Retinoblastoma in a Premature Twin. Arch Ophthalmol (2002) 120(9):1232–3. doi: 10.1001/archopht.120.9.1232
3. Fabian ID, Onadim Z, Karaa E, Duncan C, Chowdhury T, Scheimberg I, et al. The Management of Retinoblastoma. Oncogene (2018) 37(12):1551–60. doi: 10.1038/s41388-017-0050-x
4. Kiss S, Leiderman YI, Mukai S. Diagnosis, Classification, and Treatment of Retinoblastoma. Int Ophthalmol Clin (2008) 48(2):135–47. doi: 10.1097/IIO.0b013e3181693670
5. Shields CL, Shields JA, De Potter P, Minelli S, Hernandez C, Brady LW, et al. Plaque Radiotherapy in the Management of Retinoblastoma. Use as a Primary and Secondary Treatment. Ophthalmology (1993) 100(2):216–24. doi: 10.1016/S0161-6420(93)31667-2
6. Murphree AL, Villablanca JG, Deegan WF3, Sato JK, Malogolowkin M, Fisher A, et al. Chemotherapy Plus Local Treatment in the Management of Intraocular Retinoblastoma. Arch Ophthalmol (1996) 114(11):1348–56. doi: 10.1001/archopht.1996.01100140548005
7. Yanık Ö, Gündüz K, Yavuz K, Taçyıldız N, Ünal E. Chemotherapy in Retinoblastoma: Current Approaches. Turk J Ophthalmol (2015) 45(6):259–67. doi: 10.4274/tjo.06888
8. Sabaie H, Amirinejad N, Asadi MR, Jalaiei A, Daneshmandpour Y, Rezaei O, et al. Molecular Insight Into the Therapeutic Potential of Long Non-Coding RNA-Associated Competing Endogenous RNA Axes in Alzheimer’s Disease: A Systematic Scoping Review. Front Aging Neurosci (2021) 13:742242. doi: 10.3389/fnagi.2021.742242
9. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) (2018) 9:402. doi: 10.3389/fendo.2018.00402
10. Singh G, Storey KB. MicroRNA Cues From Nature: A Roadmap to Decipher and Combat Challenges in Human Health and Disease? Cells (2021) 10(12). doi: 10.3390/cells10123374
11. Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target Mimicry Provides a New Mechanism for Regulation of microRNA Activity. Nat Genet (2007) 39(8):1033–7. doi: 10.1038/ng2079
12. Ebert MS, Neilson JR, Sharp PA. MicroRNA Sponges: Competitive Inhibitors of Small RNAs in Mammalian Cells. Nat Methods (2007) 4(9):721–6. doi: 10.1038/nmeth1079
13. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A Coding-Independent Function of Gene and Pseudogene mRNAs Regulates Tumour Biology. Nature (2010) 465(7301):1033–8. doi: 10.1038/nature09144
14. Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread Changes in Protein Synthesis Induced by microRNAs. Nature (2008) 455(7209):58–63. doi: 10.1038/nature07228
15. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell (2011) 146(3):353–8. doi: 10.1016/j.cell.2011.07.014
16. Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR Recruits Let-7/RISC to Repress C-Myc Expression. Genes Dev (2009) 23(15):1743–8. doi: 10.1101/gad.1812509
17. Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of Epidermal Growth Factor Receptor Signaling in Human Cancer Cells by microRNA-7. J Biol Chem (2009) 284(9):5731–41. doi: 10.1074/jbc.M804280200
18. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 Circular RNA Retards Cell Cycle Progression via Forming Ternary Complexes With P21 and CDK2. Nucleic Acids Res (2016) 44(6):2846–58. doi: 10.1093/nar/gkw027
19. Sinha T, Panigrahi C, Das D, Chandra Panda A. Circular RNA Translation, a Path to Hidden Proteome. Wiley Interdiscip Rev RNA (2022) 13(1):e1685. doi: 10.1002/wrna.1685
20. Verduci L, Tarcitano E, Strano S, Yarden Y, Blandino G. CircRNAs: Role in Human Diseases and Potential Use as Biomarkers. Cell Death Dis (2021) 12(5):468. doi: 10.1038/s41419-021-03743-3
21. Qi X, Lin Y, Chen J, Shen B. Decoding Competing Endogenous RNA Networks for Cancer Biomarker Discovery. Brief Bioinform (2020) 21(2):441–57. doi: 10.1093/bib/bbz006
22. Arksey H, O’Malley L. Scoping Studies: Towards a Methodological Framework. Int J Soc Res Methodol (2005) 8(1):19–32. doi: 10.1080/1364557032000119616
23. Levac D, Colquhoun H, O’Brien KK. Scoping Studies: Advancing the Methodology. Implement Sci (2010) 5:69. doi: 10.1186/1748-5908-5-69
24. Tricco AC, Lillie E, Zarin W, O’Brien KK, Colquhoun H, Levac D, et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann Intern Med (2018) 169(7):467–73. doi: 10.7326/M18-0850
25. Du S, Wang S, Zhang F, Lv Y. SKP2, Positively Regulated by Circ_ODC1/miR-422a Axis, Promotes the Proliferation of Retinoblastoma. J Cell Biochem (2020) 121(1):322–31. doi: 10.1002/jcb.29177
26. Chen NN, Chao DL, Li XG. Circular RNA has_Circ_0000527 Participates in Proliferation, Invasion and Migration of Retinoblastoma Cells via miR-646/BCL-2 Axis. Cell Biochem Funct (2020) 38(8):1036–46. doi: 10.1002/cbf.3535
27. Liu H, Yuan HF, Xu D, Chen KJ, Tan N, Zheng QJ. Circular RNA Circ_0000034 Upregulates STX17 Level to Promote Human Retinoblastoma Development via Inhibiting miR-361-3p. Eur Rev Med Pharmacol Sci (2020) 24(23):12080–92.
28. Sun Z, Zhang A, Hou M, Jiang T. Circular RNA Hsa_Circ_0000034 Promotes the Progression of Retinoblastoma via Sponging microRNA-361-3p. Bioengineered (2020) 11(1):949–57. doi: 10.1080/21655979.2020.1814670
29. Wang H, Li M, Cui H, Song X, Sha Q. CircDHDDS/miR-361-3p/WNT3A Axis Promotes the Development of Retinoblastoma by Regulating Proliferation, Cell Cycle, Migration, and Invasion of Retinoblastoma Cells. Neurochem Res (2020) 45(11):2691–702. doi: 10.1007/s11064-020-03112-0
30. Zhang L, Wu J, Li Y, Jiang Y, Wang L, Chen Y, et al. Circ_0000527 Promotes the Progression of Retinoblastoma by Regulating miR-646/LRP6 Axis. Cancer Cell Int (2020) 20(1):301. doi: 10.1186/s12935-020-01396-4
31. Fu C, Wang S, Jin L, Zhang M, Li M. CircTET1 Inhibits Retinoblastoma Progression via Targeting miR-492 and miR-494-3p Through Wnt/β-Catenin Signaling Pathway. Curr Eye Res (2021) 46(7):978–87. doi: 10.1080/02713683.2020.1843685
32. Huang Y, Xue B, Pan J, Shen N. Circ-E2F3 Acts as a ceRNA for miR-204-5p to Promote Proliferation, Metastasis and Apoptosis Inhibition in Retinoblastoma by Regulating ROCK1 Expression. Exp Mol Pathol (2021) 120:104637. doi: 10.1016/j.yexmp.2021.104637
33. Jiang Y, Xiao F, Wang L, Wang T, Chen L. Hsa_circ_0099198 Facilitates the Progression of Retinoblastoma by Regulating miR-1287/LRP6 Axis. Exp Eye Res (2021) 206:108529. doi: 10.1016/j.exer.2021.108529
34. Xu L, Long H, Zhou B, Jiang H, Cai M. CircMKLN1 Suppresses the Progression of Human Retinoblastoma by Modulation of miR-425-5p/PDCD4 Axis. Curr Eye Res (2021) 46(11):1751–61. doi: 10.1080/02713683.2021.1927110
35. Jiang Y, Xiao F, Wang L, Wang T, Chen L. Circular RNA has_Circ_0000034 Accelerates Retinoblastoma Advancement Through the miR-361-3p/ADAM19 Axis. Mol Cell Biochem (2021) 476(1):69–80. doi: 10.1007/s11010-020-03886-5
36. Zhang Y, Dou X, Kong Q, Li Y, Zhou X. Circ_0075804 Promotes the Malignant Behaviors of Retinoblastoma Cells by Binding to miR-138-5p to Induce PEG10 Expression. Int Ophthalmol (2022) 42(2):509–23. doi: 10.1007/s10792-021-02067-7
37. Liang T, Fan M, Meng Z, Sun B, Mi S, Gao X. Circ_0000527 Drives Retinoblastoma Progression by Regulating miR-1236-3p/SMAD2 Pathway. Curr Eye Res (2021), 1–10.
38. Yu B, Zhao J, Dong Y. Circ_0000527 Promotes Retinoblastoma Progression Through Modulating miR-98-5p/XIAP Pathway. Curr Eye Res (2021) 46(9):1414–23. doi: 10.1080/02713683.2021.1891255
39. Zheng T, Chen W, Wang X, Cai W, Wu F, Lin C. Circular RNA Circ-FAM158A Promotes Retinoblastoma Progression by Regulating miR-138-5p/SLC7A5 Axis. Exp Eye Res (2021) 211:108650. doi: 10.1016/j.exer.2021.108650
40. Zuo X, Fu C, Xie J, Wang X, Yan Z. Hsa_circ_0000527 Downregulation Suppresses the Development of Retinoblastoma by Modulating the miR-27a-3p/HDAC9 Pathway. Curr Eye Res (2022) 47(1):115–26. doi: 10.1080/02713683.2021.1925697
41. Iparraguirre L, Prada-Luengo I, Regenberg B, Otaegui D. To Be or Not to Be: Circular RNAs or mRNAs From Circular DNAs? Front Genet (2019) 10. doi: 10.3389/fgene.2019.00940
42. Suzuki H, Tsukahara T. A View of pre-mRNA Splicing From RNase R Resistant RNAs. Int J Mol Sci (2014) 15(6):9331–42. doi: 10.3390/ijms15069331
43. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive Characterization of Tissue-Specific Circular RNAs in the Human and Mouse Genomes. Brief Bioinform (2017) 18(6):984–92.
44. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs Are a Large Class of Animal RNAs With Regulatory Potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
45. Chen X, Fan S, Song E. Noncoding RNAs: New Players in Cancers. Adv Exp Med Biol (2016) 927:1–47. doi: 10.1007/978-981-10-1498-7_1
46. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-Type Specific Features of Circular RNA Expression. PLoS Genet (2013) 9(9):e1003777. doi: 10.1371/journal.pgen.1003777
47. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-Like Structures. Proc Natl Acad Sci U S A (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852
48. Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse Alternative Back-Splicing and Alternative Splicing Landscape of Circular RNAs. Genome Res (2016) 26(9):1277–87. doi: 10.1101/gr.202895.115
49. Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, et al. Exon Circularization Requires Canonical Splice Signals. Cell Rep (2015) 10(1):103–11. doi: 10.1016/j.celrep.2014.12.002
50. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA Biogenesis Competes With pre-mRNA Splicing. Mol Cell (2014) 56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
51. Chen L-L, Carmichael G. Gene Regulation by SINES and Inosines: Biological Consequences of A-To-I Editing of Alu Element Inverted Repeats. Cell Cycle (Georgetown Tex) (2008) 7:3294–301. doi: 10.4161/cc.7.21.6927
52. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary Sequence-Mediated Exon Circularization. Cell (2014) 159(1):134–47. doi: 10.1016/j.cell.2014.09.001
53. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA Profiling Reveals an Abundant Circhipk3 That Regulates Cell Growth by Sponging Multiple miRNAs. Nat Commun (2016) 7:11215. doi: 10.1038/ncomms11215
54. Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, et al. FUS Affects Circular RNA Expression in Murine Embryonic Stem Cell-Derived Motor Neurons. Nat Commun (2017) 8:14741. doi: 10.1038/ncomms14741
55. Lin YC, Yu YS, Lin HH, Hsiao KY. Oxaliplatin-Induced DHX9 Phosphorylation Promotes Oncogenic Circular RNA CCDC66 Expression and Development of Chemoresistance. Cancers (Basel) (2020) 12(3). doi: 10.3390/cancers12030697
56. Aktaş T, Avşar Ilık İ, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, et al. DHX9 Suppresses RNA Processing Defects Originating From the Alu Invasion of the Human Genome. Nature (2017) 544(7648):115–9. doi: 10.1038/nature21715
57. Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY, et al. The Circular RNA Circbirc6 Participates in the Molecular Circuitry Controlling Human Pluripotency. Nat Commun (2017) 8(1):1149. doi: 10.1038/s41467-017-01216-w
58. Ebbesen KK, Hansen TB, Kjems J. Insights Into Circular RNA Biology. RNA Biol (2017) 14(8):1035–45. doi: 10.1080/15476286.2016.1271524
59. Zhao X, Cai Y, Xu J. Circular RNAs: Biogenesis, Mechanism, and Function in Human Cancers. Int J Mol Sci (2019) 20(16). doi: 10.3390/ijms20163926
60. Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, et al. The Emerging Landscape of Circular RNA in Life Processes. RNA Biol (2017) 14(8):992–9. doi: 10.1080/15476286.2016.1220473
61. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
62. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded Identification and Characterization of Mammalian Circular RNAs. Genome Biol (2014) 15(7):409. doi: 10.1186/s13059-014-0409-z
63. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in Cancer: Opportunities and Challenges in the Field. Oncogene (2018) 37(5):555–65. doi: 10.1038/onc.2017.361
64. Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA Profile Identifies Circpvt1 as a Proliferative Factor and Prognostic Marker in Gastric Cancer. Cancer Lett (2017) 388:208–19. doi: 10.1016/j.canlet.2016.12.006
65. Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is Expressed Across the Eukaryotic Tree of Life. PLoS One (2014) 9(6):e90859. doi: 10.1371/journal.pone.0090859
66. Wang Y, Liu J, Huang BO, Xu YM, Li J, Huang LF, et al. Mechanism of Alternative Splicing and its Regulation. BioMed Rep (2015) 3(2):152–8. doi: 10.3892/br.2014.407
67. Li X, Zhang B, Li F, Yu K, Bai Y. The Mechanism and Detection of Alternative Splicing Events in Circular RNAs. PeerJ (2020) 8:e10032. doi: 10.7717/peerj.10032
68. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
69. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR Target Circular RNAs Uncovers Suppression of PABPN1 Translation by Circpabpn1. RNA Biol (2017) 14(3):361–9. doi: 10.1080/15476286.2017.1279788
70. Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H, et al. Circular Intronic Long Noncoding RNAs. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
71. Fischer JW, Leung AK. CircRNAs: A Regulator of Cellular Stress. Crit Rev Biochem Mol Biol (2017) 52(2):220–33. doi: 10.1080/10409238.2016.1276882
72. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, et al. Foxo3 Circular RNA Promotes Cardiac Senescence by Modulating Multiple Factors Associated With Stress and Senescence Responses. Eur Heart J (2017) 38(18):1402–12.
73. Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell (2018) 71(3):428–42. doi: 10.1016/j.molcel.2018.06.034
74. Perriman R, Ares M Jr. Circular mRNA can Direct Translation of Extremely Long Repeating-Sequence Proteins In Vivo. Rna (1998) 4(9):1047–54. doi: 10.1017/S135583829898061X
75. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 Is a Circular RNA That Can Be Translated and Functions in Myogenesis. Mol Cell (2017) 66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017
76. Wang Y, Wang Z. Efficient Backsplicing Produces Translatable Circular mRNAs. Rna (2015) 21(2):172–9. doi: 10.1261/rna.048272.114
77. Xin Z, Ma Q, Ren S, Wang G, Li F. The Understanding of Circular RNAs as Special Triggers in Carcinogenesis. Brief Funct Genomics (2017) 16(2):80–6.
78. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive Translation of Circular RNAs Driven by N6-Methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
79. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst (2018) 110(3):304–15. doi: 10.1093/jnci/djx166
80. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021
82. Zhu K, Liu Q, Zhou Y, Tao C, Zhao Z, Sun J, et al. Oncogenes and Tumor Suppressor Genes: Comparative Genomics and Network Perspectives. BMC Genomics (2015) 16(7):1–11. doi: 10.1186/1471-2164-16-S7-S8
83. Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA Circ-ABCB10 Promotes Breast Cancer Proliferation and Progression Through Sponging miR-1271. Am J Cancer Res (2017) 7(7):1566–76.
84. Pei X, Zhang Y, Wang X, Xue B, Sun M, Li H. Circular RNA Circ-ZEB1 Acts as an Oncogene in Triple Negative Breast Cancer via Sponging miR-448. Int J Biochem Cell Biol (2020) 126:105798. doi: 10.1016/j.biocel.2020.105798
85. Li J, Gao X, Zhang Z, Lai Y, Lin X, Lin B, et al. CircCD44 Plays Oncogenic Roles in Triple-Negative Breast Cancer by Modulating the miR-502-5p/KRAS and IGF2BP2/Myc Axes. Mol Cancer (2021) 20(1):138. doi: 10.1186/s12943-021-01444-1
86. Zhou P, Xie W, Huang HL, Huang RQ, Tian C, Zhu HB, et al. circRNA_100859 Functions as an Oncogene in Colon Cancer by Sponging the miR-217-HIF-1α Pathway. Aging (Albany NY) (2020) 12(13):13338–53.
87. Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, et al. Circular RNA Circago2 Drives Cancer Progression Through Facilitating HuR-Repressed Functions of AGO2-miRNA Complexes. Cell Death Differ (2019) 26(7):1346–64. doi: 10.1038/s41418-018-0220-6
88. Zhang Q, Zhang C, Ma JX, Ren H, Sun Y, Xu JZ. Circular RNA PIP5K1A Promotes Colon Cancer Development Through Inhibiting miR-1273a. World J Gastroenterol (2019) 25(35):5300–9. doi: 10.3748/wjg.v25.i35.5300
89. Wei Z, Tian Z, Zhang L. CircPPP1R12A Promotes the Progression of Colon Cancer Through Regulating CTNNB1 via Sponging miR-375. Anticancer Drugs (2021) 32(6):635–46. doi: 10.1097/CAD.0000000000001037
90. Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, et al. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res (2017) 77(9):2339–50. doi: 10.1158/0008-5472.CAN-16-1883
91. Chen J, Yang X, Liu R, Wen C, Wang H, Huang L, et al. Circular RNA GLIS2 Promotes Colorectal Cancer Cell Motility via Activation of the NF-κb Pathway. Cell Death Dis (2020) 11(9):788.
92. Zhang B, Yang S, Wang J. Circ_0084615 is an Oncogenic Circular RNA in Colorectal Cancer and Promotes DNMT3A Expression via Repressing miR-599. Pathol Res Pract (2021) 224:153494. doi: 10.1016/j.prp.2021.153494
93. Ishola AA, Chien CS, Yang YP, Chien Y, Yarmishyn AA, Tsai PH, et al. Oncogenic circRNA C190 Promotes Non-Small Cell Lung Cancer via Modulation of the EGFR/ERK Pathway. Cancer Res (2022) 82(1):75–89. doi: 10.1158/0008-5472.CAN-21-1473
94. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. Circndufb2 Inhibits non-Small Cell Lung Cancer Progression via Destabilizing IGF2BPs and Activating Anti-Tumor Immunity. Nat Commun (2021) 12(1):295. doi: 10.1038/s41467-020-20527-z
95. Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, et al. Circular RNA 100146 Functions as an Oncogene Through Direct Binding to miR-361-3p and miR-615-5p in Non-Small Cell Lung Cancer. Mol Cancer (2019) 18(1):13. doi: 10.1186/s12943-019-0943-0
96. Yang F, Ma C, Qiu J, Feng X, Yang K. Identification of circRNA_001846 as Putative Non-Small Cell Lung Cancer Biomarker. Bioengineered (2021) 12(1):8690–7. doi: 10.1080/21655979.2021.1991161
97. Li L, Wan K, Xiong L, Liang S, Tou F, Guo S. CircRNA Hsa_Circ_0087862 Acts as an Oncogene in Non-Small Cell Lung Cancer by Targeting miR-1253/RAB3D Axis. Onco Targets Ther (2020) 13:2873–86. doi: 10.2147/OTT.S243533
98. Huang W, Fang K, Chen TQ, Zeng ZC, Sun YM, Han C, et al. circRNA Circaf4 Functions as an Oncogene to Regulate MLL-AF4 Fusion Protein Expression and Inhibit MLL Leukemia Progression. J Hematol Oncol (2019) 12(1):103. doi: 10.1186/s13045-019-0800-z
99. Wang X, Jin P, Zhang Y, Wang K. CircSPI1 Acts as an Oncogene in Acute Myeloid Leukemia Through Antagonizing SPI1 and Interacting With microRNAs. Cell Death Dis (2021) 12(4):297. doi: 10.1038/s41419-021-03566-2
100. Shi Y, Han T, Liu C. CircRNA Hsa_Circ_0006220 Acts as a Tumor Suppressor Gene by Regulating miR-197-5p/CDH19 in Triple-Negative Breast Cancer. Ann Transl Med (2021) 9(15):1236–. doi: 10.21037/atm-21-2934
101. Wang S, Liu F, Ma H, Cui X, Yang S, Qin R. circCDYL Acts as a Tumor Suppressor in Triple Negative Breast Cancer by Sponging miR-190a-3p and Upregulating Tp53inp1. Clin Breast Cancer (2020) 20(5):422–30. doi: 10.1016/j.clbc.2020.04.006
102. Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA Sequencing Reveals the Expression Profiles of circRNA and Indicates That Circddx17 Acts as a Tumor Suppressor in Colorectal Cancer. J Exp Clin Cancer Res (2018) 37(1):325. doi: 10.1186/s13046-018-1006-x
103. Yan D, Liu W, Liu Y, Zhu X. Circular RNA Circ_0065378 Upregulates Tumor Suppressor Candidate 1 by Competitively Binding With miR-4701-5p to Alleviate Colorectal Cancer Progression. J Gastroenterol Hepatol (2022):1107–1118. doi: 10.1111/jgh.15862
104. Li Z, Yao H, Wang S, Li G, Gu X. CircTADA2A Suppresses the Progression of Colorectal Cancer via miR-374a-3p/KLF14 Axis. J Exp Clin Cancer Res (2020) 39(1):160. doi: 10.1186/s13046-020-01642-7
105. Miao X, Xi Z, Zhang Y, Li Z, Huang L, Xin T, et al. Circ-SMARCA5 Suppresses Colorectal Cancer Progression via Downregulating miR-39-3p and Upregulating ARID4B. Dig Liver Dis (2020) 52(12):1494–502. doi: 10.1016/j.dld.2020.07.019
106. Ma C, Wang X, Yang F, Zang Y, Liu J, Wang X, et al. Circular RNA Hsa_Circ_0004872 Inhibits Gastric Cancer Progression via the miR-224/Smad4/ADAR1 Successive Regulatory Circuit. Mol Cancer (2020) 19(1):157. doi: 10.1186/s12943-020-01268-5
107. Zhang Y, Jiang J, Zhang J, Shen H, Wang M, Guo Z, et al. CircDIDO1 Inhibits Gastric Cancer Progression by Encoding a Novel DIDO1-529aa Protein and Regulating PRDX2 Protein Stability. Mol Cancer (2021) 20(1):101. doi: 10.1186/s12943-021-01390-y
108. Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S, et al. A Novel Protein Encoded by Circmapk1 Inhibits Progression of Gastric Cancer by Suppressing Activation of MAPK Signaling. Mol Cancer (2021) 20(1):66. doi: 10.1186/s12943-021-01358-y
109. Sun W, Zhang H, Duan W, Zhu H, Gu C. Tumor Suppressor Role of Hsa_Circ_0035445 in Gastric Cancer. J Clin Lab Anal (2021) 35(6):e23727. doi: 10.1002/jcla.23727
110. Luo Z, Rong Z, Zhang J, Zhu Z, Yu Z, Li T, et al. Circular RNA Circccdc9 Acts as a miR-6792-3p Sponge to Suppress the Progression of Gastric Cancer Through Regulating CAV1 Expression. Mol Cancer (2020) 19(1):86. doi: 10.1186/s12943-020-01203-8
111. Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J, et al. CircPLEKHM3 Acts as a Tumor Suppressor Through Regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 Axis in Ovarian Cancer. Mol Cancer (2019) 18(1):144. doi: 10.1186/s12943-019-1080-5
112. Yu S, Ai L, Wei W, Pan J. circRNA Circ-MYBL2 is a Novel Tumor Suppressor and Potential Biomarker in Multiple Myeloma. Hum Cell (2021) 34(1):219–28. doi: 10.1007/s13577-020-00441-8
113. Wang D, Yan S, Wang L, Li Y, Qiao B. Circslc8a1 Acts as a Tumor Suppressor in Prostate Cancer via Sponging miR-21. BioMed Res Int (2021) 2021:6614591. doi: 10.1155/2021/6614591
114. Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of Circular RNA Abundance With Proliferation–Exemplified With Colorectal and Ovarian Cancer, Idiopathic Lung Fibrosis, and Normal Human Tissues. Sci Rep (2015) 5:8057. doi: 10.1038/srep08057
115. Li P, Chen S, Chen H, Mo X, Li T, Shao Y, et al. Using Circular RNA as a Novel Type of Biomarker in the Screening of Gastric Cancer. Clin Chim Acta (2015) 444:132–6. doi: 10.1016/j.cca.2015.02.018
116. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA Is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res (2015) 25(8):981–4. doi: 10.1038/cr.2015.82
117. Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res (2017) 23(14):3918–28. doi: 10.1158/1078-0432.CCR-16-2541
118. Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, et al. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J Cell Biochem (2018) 119(1):440–6. doi: 10.1002/jcb.26201
119. Li RC, Ke S, Meng FK, Lu J, Zou XJ, He ZG, et al. CiRS-7 Promotes Growth and Metastasis of Esophageal Squamous Cell Carcinoma via Regulation of miR-7/Hoxb13. Cell Death Dis (2018) 9(8):838. doi: 10.1038/s41419-018-0852-y
120. Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS One (2016) 11(3):e0151753. doi: 10.1371/journal.pone.0151753
121. Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/β-Catenin Pathway. BioMed Res Int (2016) 2016:1579490. doi: 10.1155/2016/1579490
122. Chen G, Shi Y, Liu M, Sun J. Circhipk3 Regulates Cell Proliferation and Migration by Sponging miR-124 and Regulating AQP3 Expression in Hepatocellular Carcinoma. Cell Death Dis (2018) 9(2):175. doi: 10.1038/s41419-017-0204-3
123. Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z, et al. TTBK2 Circular RNA Promotes Glioma Malignancy by Regulating miR-217/Hnf1β/Derlin-1 Pathway. J Hematol Oncol (2017) 10(1):1–19.
124. Rajappa A, Banerjee S, Sharma V, Khandelia P. Circular RNAs: Emerging Role in Cancer Diagnostics and Therapeutics. Front Mol Biosci (2020) 7. doi: 10.3389/fmolb.2020.577938
125. Xing L, Zhang L, Feng Y, Cui Z, Ding L. Downregulation of Circular RNA Hsa_Circ_0001649 Indicates Poor Prognosis for Retinoblastoma and Regulates Cell Proliferation and Apoptosis via AKT/mTOR Signaling Pathway. BioMed Pharmacother (2018) 105:326–33. doi: 10.1016/j.biopha.2018.05.141
126. Pan Y, Chen Y, Ma D, Ji Z, Cao F, Chen Z, et al. miR-646 Is a Key Negative Regulator of EGFR Pathway in Lung Cancer. Exp Lung Res (2016) 42(6):286–95. doi: 10.1080/01902148.2016.1207726
127. Dai H, Hou K, Cai Z, Zhou Q, Zhu S. Low-Level miR-646 in Colorectal Cancer Inhibits Cell Proliferation and Migration by Targeting NOB1 Expression. Oncol Lett (2017) 14(6):6708–14. doi: 10.3892/ol.2017.7032
128. Li W, Liu M, Feng Y, Xu YF, Huang YF, Che JP, et al. Downregulated miR-646 in Clear Cell Renal Carcinoma Correlated With Tumour Metastasis by Targeting the Nin One Binding Protein (NOB1). Br J Cancer (2014) 111(6):1188–200. doi: 10.1038/bjc.2014.382
129. Wang J, Shu H, Guo S. MiR-646 Suppresses Proliferation and Metastasis of Non-Small Cell Lung Cancer by Repressing FGF2 and CCND2. Cancer Med (2020) 9(12):4360–70. doi: 10.1002/cam4.3062
130. de Ridder I, Kerkhofs M, Veettil SP, Dehaen W, Bultynck G. Cancer Cell Death Strategies by Targeting Bcl-2's BH4 Domain. Biochim Biophys Acta Mol Cell Res (2021) 1868(5):118983. doi: 10.1016/j.bbamcr.2021.118983
131. Matsuhashi S, Manirujjaman M, Hamajima H, Ozaki I. Control Mechanisms of the Tumor Suppressor PDCD4: Expression and Functions. Int J Mol Sci (2019) 20(9). doi: 10.3390/ijms20092304
132. He S, Lu Y, Liu X, Huang X, Keller ET, Qian CN, et al. Wnt3a: Functions and Implications in Cancer. Chin J Cancer (2015) 34(12):554–62. doi: 10.1186/s40880-015-0052-4
133. Yang J, Wahdan-Alaswad R, Danielpour D. Critical Role of Smad2 in Tumor Suppression and Transforming Growth Factor-Beta-Induced Apoptosis of Prostate Epithelial Cells. Cancer Res (2009) 69(6):2185–90. doi: 10.1158/0008-5472.CAN-08-3961
134. Petersen M, Pardali E, van der Horst G, Cheung H, van den Hoogen C, van der Pluijm G, et al. Smad2 and Smad3 Have Opposing Roles in Breast Cancer Bone Metastasis by Differentially Affecting Tumor Angiogenesis. Oncogene (2010) 29(9):1351–61. doi: 10.1038/onc.2009.426
135. Jeong W, Jho EH. Regulation of the Low-Density Lipoprotein Receptor-Related Protein LRP6 and Its Association With Disease: Wnt/β-Catenin Signaling and Beyond. Front Cell Dev Biol (2021) 9:714330. doi: 10.3389/fcell.2021.714330
136. Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, et al. Degradation of p57Kip2 Mediated by SCFSkp2-Dependent Ubiquitylation. Proc Natl Acad Sci U S A (2003) 100(18):10231–6. doi: 10.1073/pnas.1831009100
137. Bhattacharya S, Garriga J, Calbó J, Yong T, Haines DS, Graña X. SKP2 Associates With P130 and Accelerates P130 Ubiquitylation and Degradation in Human Cells. Oncogene (2003) 22(16):2443–51. doi: 10.1038/sj.onc.1206339
138. Yu ZK, Gervais JL, Zhang H. Human CUL-1 Associates With the SKP1/SKP2 Complex and Regulates P21(CIP1/WAF1) and Cyclin D Proteins. Proc Natl Acad Sci U S A (1998) 95(19):11324–9. doi: 10.1073/pnas.95.19.11324
139. Hiramatsu Y, Kitagawa K, Suzuki T, Uchida C, Hattori T, Kikuchi H, et al. Degradation of Tob1 Mediated by SCFSkp2-Dependent Ubiquitination. Cancer Res (2006) 66(17):8477–83. doi: 10.1158/0008-5472.CAN-06-1603
140. Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, et al. Skp2 Inhibits FOXO1 in Tumor Suppression Through Ubiquitin-Mediated Degradation. Proc Natl Acad Sci U S A (2005) 102(5):1649–54. doi: 10.1073/pnas.0406789102
141. Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, et al. Skp2 is Oncogenic and Overexpressed in Human Cancers. Proc Natl Acad Sci U S A (2001) 98(9):5043–8. doi: 10.1073/pnas.081474898
142. Seki R, Okamura T, Koga H, Yakushiji K, Hashiguchi M, Yoshimoto K, et al. Prognostic Significance of the F-Box Protein Skp2 Expression in Diffuse Large B-Cell Lymphoma. Am J Hematol (2003) 73(4):230–5. doi: 10.1002/ajh.10379
143. Wang Z, Gao D, Fukushima H, Inuzuka H, Liu P, Wan L, et al. Skp2: A Novel Potential Therapeutic Target for Prostate Cancer. Biochim Biophys Acta (2012) 1825(1):11–7. doi: 10.1016/j.bbcan.2011.09.002
144. Li JQ, Wu F, Masaki T, Kubo A, Fujita J, Dixon DA, et al. Correlation of Skp2 With Carcinogenesis, Invasion, Metastasis, and Prognosis in Colorectal Tumors. Int J Oncol (2004) 25(1):87–95. doi: 10.3892/ijo.25.1.87
145. Rose AE, Wang G, Hanniford D, Monni S, Tu T, Shapiro RL, et al. Clinical Relevance of SKP2 Alterations in Metastatic Melanoma. Pigment Cell Melanoma Res (2011) 24(1):197–206. doi: 10.1111/j.1755-148X.2010.00784.x
146. Xu HM, Liang Y, Chen Q, Wu QN, Guo YM, Shen GP, et al. Correlation of Skp2 Overexpression to Prognosis of Patients With Nasopharyngeal Carcinoma From South China. Chin J Cancer (2011) 30(3):204–12. doi: 10.5732/cjc.010.10403
147. Schüler S, Diersch S, Hamacher R, Schmid RM, Saur D, Schneider G. SKP2 Confers Resistance of Pancreatic Cancer Cells Towards TRAIL-Induced Apoptosis. Int J Oncol (2011) 38(1):219–25.
148. Zheng WQ, Zheng JM, Ma R, Meng FF, Ni CR. Relationship Between Levels of Skp2 and P27 in Breast Carcinomas and Possible Role of Skp2 as Targeted Therapy. Steroids (2005) 70(11):770–4. doi: 10.1016/j.steroids.2005.04.012
149. Rath N, Olson MF. Rho-Associated Kinases in Tumorigenesis: Re-Considering ROCK Inhibition for Cancer Therapy. EMBO Rep (2012) 13(10):900–8. doi: 10.1038/embor.2012.127
150. Xu S, Guo X, Gao X, Xue H, Zhang J, Guo X, et al. Macrophage Migration Inhibitory Factor Enhances Autophagy by Regulating ROCK1 Activity and Contributes to the Escape of Dendritic Cell Surveillance in Glioblastoma. Int J Oncol (2016) 49(5):2105–15. doi: 10.3892/ijo.2016.3704
151. Huang GX, Wang Y, Su J, Zhou P, Li B, Yin LJ, et al. Up-Regulation of Rho-Associated Kinase 1/2 by Glucocorticoids Promotes Migration, Invasion and Metastasis of Melanoma. Cancer Lett (2017) 410:1–11. doi: 10.1016/j.canlet.2017.09.005
152. Wang Z, Wang Z, Liu J, Yang H. Long non-Coding RNA SNHG5 Sponges miR-26a to Promote the Tumorigenesis of Osteosarcoma by Targeting ROCK1. BioMed Pharmacother (2018) 107:598–605. doi: 10.1016/j.biopha.2018.08.025
153. Zhan Y, Zheng N, Teng F, Bao L, Liu F, Zhang M, et al. MiR-199a/B-5p Inhibits Hepatocellular Carcinoma Progression by Post-Transcriptionally Suppressing ROCK1. Oncotarget (2017) 8(40):67169–80. doi: 10.18632/oncotarget.18052
154. Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P, et al. HDAC5 and HDAC9 in Medulloblastoma: Novel Markers for Risk Stratification and Role in Tumor Cell Growth. Clin Cancer Res (2010) 16(12):3240–52. doi: 10.1158/1078-0432.CCR-10-0395
155. Gil VS, Bhagat G, Howell L, Zhang J, Kim CH, Stengel S, et al. Deregulated Expression of HDAC9 in B Cells Promotes Development of Lymphoproliferative Disease and Lymphoma in Mice. Dis Model Mech (2016) 9(12):1483–95.
156. Rastogi B, Raut SK, Panda NK, Rattan V, Radotra BD, Khullar M. Overexpression of HDAC9 Promotes Oral Squamous Cell Carcinoma Growth, Regulates Cell Cycle Progression, and Inhibits Apoptosis. Mol Cell Biochem (2016) 415(1-2):183–96. doi: 10.1007/s11010-016-2690-5
157. Lapierre M, Linares A, Dalvai M, Duraffourd C, Bonnet S, Boulahtouf A, et al. Histone Deacetylase 9 Regulates Breast Cancer Cell Proliferation and the Response to Histone Deacetylase Inhibitors. Oncotarget (2016) 7(15):19693–708. doi: 10.18632/oncotarget.7564
158. Zhang Y, Wu D, Xia F, Xian H, Zhu X, Cui H, et al. Downregulation of HDAC9 Inhibits Cell Proliferation and Tumor Formation by Inducing Cell Cycle Arrest in Retinoblastoma. Biochem Biophys Res Commun (2016) 473(2):600–6. doi: 10.1016/j.bbrc.2016.03.129
159. Xian H, Yang Q, Xiao L, Shen HM, Liou YC. STX17 Dynamically Regulated by Fis1 Induces Mitophagy via Hierarchical Macroautophagic Mechanism. Nat Commun (2019) 10(1):2059.
160. Evans MK, Sauer SJ, Nath S, Robinson TJ, Morse MA, Devi GR. X-Linked Inhibitor of Apoptosis Protein Mediates Tumor Cell Resistance to Antibody-Dependent Cellular Cytotoxicity. Cell Death Dis (2016) 7(1):e2073. doi: 10.1038/cddis.2015.412
161. Bang H, Ha SY, Hwang SH, Park CK. Expression of PEG10 Is Associated With Poor Survival and Tumor Recurrence in Hepatocellular Carcinoma. Cancer Res Treat (2015) 47(4):844–52. doi: 10.4143/crt.2014.124
162. Deng X, Hu Y, Ding Q, Han R, Guo Q, Qin J, et al. PEG10 Plays a Crucial Role in Human Lung Cancer Proliferation, Progression, Prognosis and Metastasis. Oncol Rep (2014) 32(5):2159–67. doi: 10.3892/or.2014.3469
163. Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, et al. The Placental Gene PEG10 Promotes Progression of Neuroendocrine Prostate Cancer. Cell Rep (2015) 12(6):922–36. doi: 10.1016/j.celrep.2015.07.012
164. Reed J. Bcl-2 - Family Proteins and Hematologic Malignancies: History and Future Prospects. Blood (2008) 111:3322–30. doi: 10.1182/blood-2007-09-078162
165. Cory S, Huang DC, Adams JM. The Bcl-2 Family: Roles in Cell Survival and Oncogenesis. Oncogene (2003) 22(53):8590–607. doi: 10.1038/sj.onc.1207102
166. Bond P. Regulation of Mtorc1 by Growth Factors, Energy Status, Amino Acids and Mechanical Stimuli at a Glance. J Int Soc Sports Nutr (2016) 13:8. doi: 10.1186/s12970-016-0118-y
Keywords: retinoblastoma, circular RNA, CeRNA, sponge, circ_0000527, circ_0000034, circTET1
Citation: Asadi MR, Moslehian MS, Sabaie H, Sharifi-Bonab M, Hakimi P, Hussen BM, Taheri M, Rakhshan A and Rezazadeh M (2022) CircRNA-Associated CeRNAs Regulatory Axes in Retinoblastoma: A Systematic Scoping Review. Front. Oncol. 12:910470. doi: 10.3389/fonc.2022.910470
Received: 01 April 2022; Accepted: 10 June 2022;
Published: 05 July 2022.
Edited by:
FangXiang Wu, University of Saskatchewan, CanadaReviewed by:
Brian D Adams, Brain Institute of America, United StatesBekesho Geleta, Children’s Cancer Institute Australia, Australia
Copyright © 2022 Asadi, Moslehian, Sabaie, Sharifi-Bonab, Hakimi, Hussen, Taheri, Rakhshan and Rezazadeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, TW9oYW1tYWQudGFoZXJpQHVuaS1qZW5hLmRl; Azadeh Rakhshan, YXphZGVocmFraHNoYW5AeWFob28uY29t; Maryam Rezazadeh, UmV6YXphZGVobWFAdGJ6bWVkLmFjLmly
 Mohammad Reza Asadi
Mohammad Reza Asadi Marziyeh Sadat Moslehian1
Marziyeh Sadat Moslehian1 Hani Sabaie
Hani Sabaie Mirmohsen Sharifi-Bonab
Mirmohsen Sharifi-Bonab Bashdar Mahmud Hussen
Bashdar Mahmud Hussen Mohammad Taheri
Mohammad Taheri