
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 07 June 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.908162
This article is part of the Research TopicEpigenetic regulation of lncRNAs and role in human cancerView all 7 articles
 Shengnan Jiang1,2†
Shengnan Jiang1,2† Qian Zhang1,2†
Qian Zhang1,2† Jiaqi Li1,2
Jiaqi Li1,2 Khadija Raziq1,2
Khadija Raziq1,2 Xinyu Kang1,2
Xinyu Kang1,2 Shiyin Liang1,2
Shiyin Liang1,2 Chaoyue Sun1,2
Chaoyue Sun1,2 Xiao Liang1,2
Xiao Liang1,2 Di Zhao3
Di Zhao3 Songbin Fu1,2
Songbin Fu1,2 Mengdi Cai1,2*
Mengdi Cai1,2*LINC01133 is a long intergenic non-coding RNA that regulates malignancy in several cancers, including those of the digestive, female reproductive, respiratory, and urinary system. LINC01133 is an extensively studied lncRNA that is highly conserved, and its relatively stable expression is essential for its robust biological function. Its expression is highly tissue-specific with a distinct subcellular localization. It functions as an oncogene or a tumor suppressor gene in different cancers via multiple mechanisms, such as those that involve competing with endogenous RNA and binding to RNA-binding proteins or DNA. Moreover, the secretion and transportation of LINC01133 by extracellular vesicles in the tumor micro-environment is regulated by other cells in the tumor micro-environment. To date, two mechanisms, an increase in copy number and regulation of transcription elements, have been found to regulate LINC01133 expression. Clinically, LINC01133 is an ideal marker for cancer prognosis and a potential therapeutic target in cancer treatment regimes. In this review, we aimed to summarize the aforementioned information as well as posit future directions for LINC01133 research.
Long noncoding RNAs (lncRNAs) are vital regulators of cancer progression. Several studies have reported various mechanisms of regulation of cancers by lncRNA. However, not all lncRNAs are functionally significant in cancer; and their function is highly conserved and tissue specific (1, 2). Recently, a set of star lncRNAs with significant functions had been identified. Of these, LINC01133 has been found to be a regulator of cancers of the digestive, female reproductive, urinary, respiratory, and skeletal systems. The carcinogenic nature of high LINC01133 expression levels has been reported in renal cancer (3), cervical cancer (4),Lung adenocarcinoma (5) and sarcoma (6). However, LINC01133 also acts as a tumor suppressor that inhibits the invasion and metastasis of cancer cells in bladder cancer (7), nasopharyngeal carcinoma (8), oral epithelial cancer (9)and melanoma cancer (10).
As per the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov), LINC01133 is located on the long arm of chromosome 1, zone 2 (1q32.2; hg38 159,961,224-159,979,086 bp) and has four transcripts: ENST00000635112.1, ENST00000657602.1, ENST00000423943.2, and ENST00000443364.6. The transcript ENST00000635112.1 has 1996 bases and 3 exons, the transcript ENST00000657602.1 has 1418 bases and 4 exons, the transcript ENST00000423943.2 has 1405 bases and 3 exons, and the transcript ENST00000443364.6 has 1266 bases and 4 exons. LINC01133 is a long intergenic non-coding RNA (lincRNA) that does not share sequences with the coding regions of the genes present on chromosome 1. LincRNAs bind to DNA (11), RNA (6), and proteins (6) and mediate various functions; such as chromatin and genome architecture remodeling, RNA stabilization, and enhancer-associated transcription (12–14).
The subcellular localization of lncRNAs is closely correlated to their function. LINC01133 is located in both the nucleus and cytoplasm, and this subcellular localization depends on the type of cancer. For example, LINC01133 tends to localize in both the nucleus and cytoplasm in ovarian cancer (15), and it tends to localize in the cytoplasm rather than in the nucleus in gastric cancers (16).
Based on its genomic location (intergenic), structural features, and subcellular localization, LINC01133 regulates biological processes in cancers via multiple mechanisms that are activated simultaneously. For example, LINC01133 exerts tumor suppressor effects in hepatocellular carcinoma not only through the competing endogenous RNAs (ceRNA) mechanism that involves binding to miR-199a-5p, but also by binding to the ANXA2 protein (17). In this review, we describe the molecular mechanism underlying the various biological functions of LINC01133, its role in cancer, and future research directions.
The subcellular localization of lncRNA largely determines its biological function. Post-transcriptional regulation is mainly mediated by the ceRNA mechanism, whereas translation and post-translational protein degradation are mediated by binding to the protein in the cytoplasm (18). Linc00483 has been found to act as a potential tumor suppressor in colorectal cancer (CRC) via ceRNA mechanism in the cytoplasm (19). Cytoplasmic lncRNA ALAL-1 regulates lung cancer immune evasion by binding to SART3 and affecting nuclear translocation of USP4 in (20).One of the most important mechanisms which ncRNA regulates cancer progression is interacting with miRNA or protein. CircRNA, another famous ncRNA, binds to miRNA or protein to exert the function in cancer. CircSMARCA5 acts as a tumor suppressor by binding SRSF1 in glioblastoma (21)or binding miR-181b-5p/miR-17-3p-TIMP3 axis in prostate cancer (22). When lncRNAs are localized in the nucleus, they regulate gene transcription or pre-transcription levels by binding to the DNA promoter region or transcription factors, respectively. LncRNA Haunt competitively interacts with enhancer-promoter region of HOXA gene during embryonic stem cell differentiation (23). The interaction between lncRNA CCAT1-Land transcription factor CTCF has a vital role in mediating MYC chromatin looping in CRC (24). The primary and most prevalent way for LINC01133 to the regulate biological progression of cancers is through the ceRNA and protein binding mechanisms in the cytoplasm and the regulation of transcription in the nucleus (Figure 1).

Figure 1 Subcellular localization and function of LINC01133. In the cytoplasm, LINC01133 functions by the mechanisms of ceRNA and binding to RBPs which promotes the phosphorylation of target protein or makes it dysfunction. In the nucleus, LINC01133 functions by the mechanisms of inhibiting transcription and binding to DNA or RNA. The blue rectangle represents the cytoplasm and the yellow oval represents the nucleus. The black dashed line separates the different mechanisms of LINC01133 in the cytoplasm.
ceRNAs are transcripts that competitively bind to miRNAs to regulate their abundance. In a ceRNA network of lncRNA-miRNA-mRNA, the miRNA directs the RNA-induced silencing complex by binding to the miRNA response element to inhibit protein production by inhibiting translation or promoting mRNA degradation. However, lncRNAs compete with mRNA to bind to miRNAs and attenuate the inhibition of mRNA translation to restore expression levels (25) (Figure 2A).
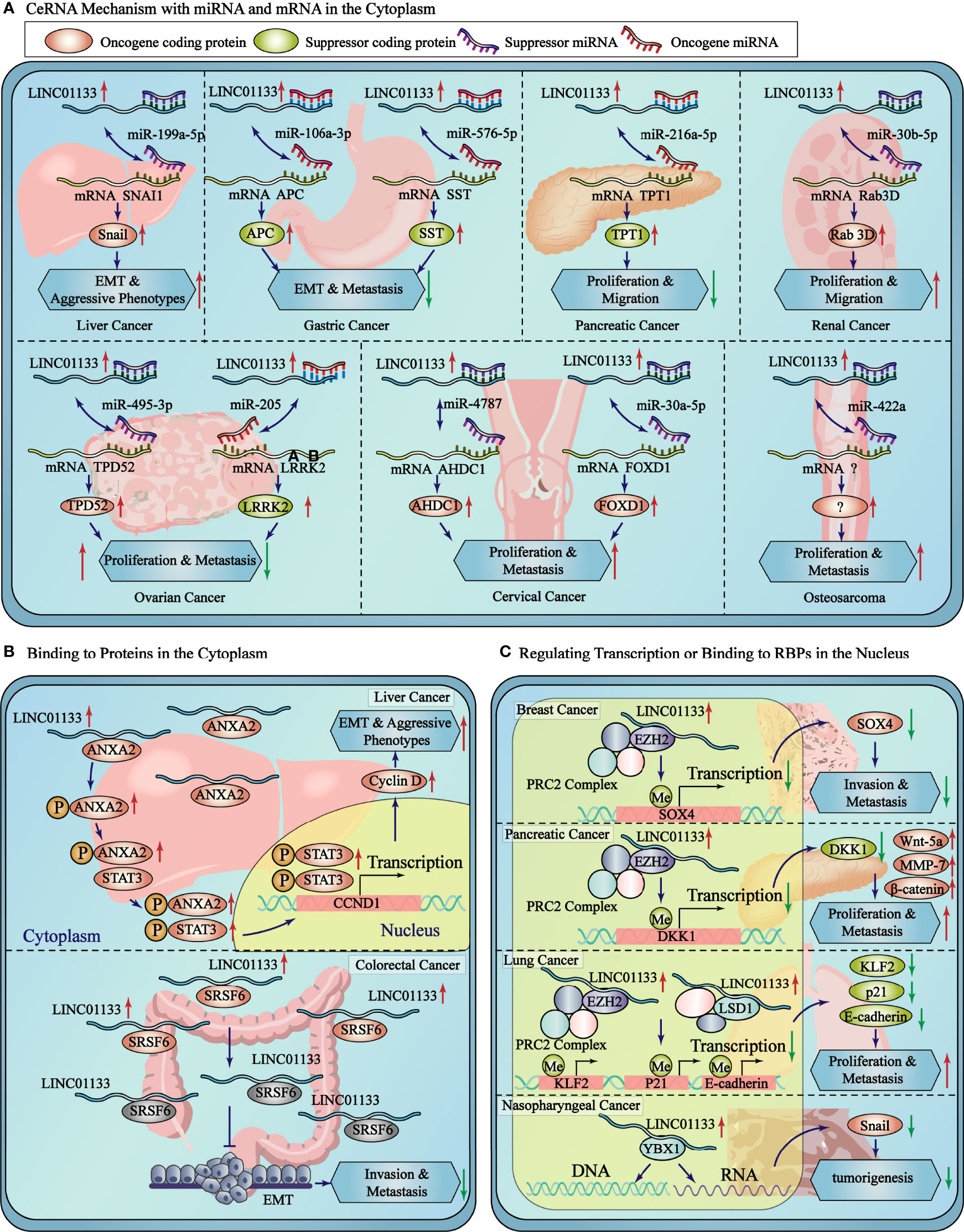
Figure 2 LINC01133 is involved in ceRNA regulation and protein binding in cytoplasm and protein/DNA binding in nucleus. (A) The detailed ceRNA mechanisms of LINC01133 in the cancers of different tissue type. (B) LINC01133 bounds to proteins in cytoplasm to regulate the modification or activity of the target proteins. (C) LINC01133 bounds to proteins or DNA in nucleus to regulate the transcription of target genes. The blue rectangle represents the cytoplasm, and the yellow block represents the nucleus. The black dashed lines separate the cancers of different tissue types.
The ceRNA mechanism of regulation by LINC01133 is commonly observed during the regulation of malignancy of cancers. Studies have reported that LINC01133, which is mainly enriched in the cytoplasm of gastric cancer cells, binds to miR-106a-3p and promotes the expression of proteins of the anaphase promoting complex (APC). Overexpression of the APC blocks the nuclear accumulation of β-catenin, leading to the inactivation of Wnt signaling; in turn inhibiting the proliferation, migration, and epithelial-mesenchymal transition (EMT) of gastric cancer cells (26). LINC01133 was also found to be enriched in the cytoplasm of liver cancer cells. It sponges up miR-199a-5p, leading to an increase in the expression of SNAI1, thereby inducing EMT in liver cancer cells (17). LINC01133 was found to be primarily located in the cytoplasm of cervical cancer cells, wherein it binds to miR-4784 and competitively inhibits the binding of AHDC1 mRNA to miR-4784. This results in the enhanced expression of AHDC1; thereby promoting the proliferation, migration, and EMT of cervical cancer cells (4). The oncogenic and tumor suppressor effect of LINC01133 via the ceRNA mechanism in various cancers have been presented in Table 1.
LINC01133 regulates protein function by directly binding to proteins to activate or inhibit its expression or by blocking its post-translational modification. Using an RNA pull-down assay and mass spectrometry analysis, Yi et al. showed that LINC01133 has nine potential binding proteins (with a peptide count > 5 and a unique peptide count > 5). The interaction between LINC01133 and ANXA2 had the highest score for protein identification analysis and was also identified with the RNA immunoprecipitation chip (RIP) assay in MHCC97H hepatocellular carcinoma (HCC) cells. ANXA2 was found to have higher expression levels in HCC tissues than that in matched peritumoral tissues, and this higher expression level of ANXA2 represented a shorter tumor-free survival period in the Kaplan-Meyer analysis. The phosphorylation levels of ANXA2 and STAT3 and the expression level of cyclin D1 were both increased by LINC01133 overexpression and decreased by its silencing. In conclusion, LINC01133 promotes HCC progression by binding to and promoting ANXA2 and STAT3 signaling (17). Similarly, Kong et al. overexpressed LINC01133 in HCT116 colon cancer cells and found that an increase in LINC01133 expression significantly repressed the migration and invasion of CRC cells. In addition, RNA pull-down and mas spectroscopy screening assays showed the direct binding of SRSF6 and LINC01133, which was confirmed using western blot and RIP assays. However, SRSF6 promoted CRC metastasis by inducing EMT independent of LINC01133, and the malignant phenotype was altered significantly by depleting SRSF6 in HT29 cells with high LINC01133 expression, in HCT116 cells with low LINC01133 expression, and in SW620 cells without LINC01133 expression. However, silencing LINC01133 in both SRSF6-expressing or -knocked down cells led to an interesting observation. The effect of LINC01133 silencing on EMT markers was attenuated in SRSF6-depleted cells, indicating that LINC01133 inhibited EMT and metastasis in CRC cells by binding to SRSF6 and blocking its function (17) (Figure 2B).
LncRNAs localized in the nucleus regulate the transcriptional expression of multiple proximal or distal genes by binding to chromatin, transcription factors, or RBPs. Approximately 20% of lncRNAs have been reported to associate with polycomb repressive complex 2 (PRC2) and induce the trimethylation of H3K27 (30). Song et al. suggested that LINC01133 potentially suppresses SOX4 expression using this mechanism in breast cancer cells. RIP assay using an EZH2 antibody, which is an important subunit of the PRC2 complex, and RNA pull-down assay identified the interaction between EZH2 and LINC01133. Chromatin immunoprecipitation (ChIP) assay in LINC01133-overexpressing cells revealed that LINC01133 increased EZH2 and H3K27me3 at the promoter region of SOX4 to regulate its transcription and inhibit its expression (31). Weng et al. also found that LINC01133 binds to the promoter region of DKK1, resulting in H3K27 trimethylation and decreased DKK1 expression; while the expression of Wnt-5a, MMP-7, and β-catenin increased upon LINC01133 binding to their respective promoting regions in pancreatic cancer (32). Similarly, Zang et al. found that the nucleus-localized LINC01133 could interact with EZH2 and LSD1 and repress the transcription of KLF2, P21, or E-cadherin by recruiting EZH2 and LSD1 to the promoter regions of the inhibited genes in non-small cell lung cancer (33).
In nasopharyngeal carcinoma (NPC), RNA pull-down and RIP assays was used to validate the binding of YBX1 and LINC01133. YBX1 is a protein that can activate Snail translation and induce EMT. YBX1 knockdown inhibited the upregulation of cell migration and invasion in vitro, and this was affected by the depletion of LINC01133. This suggests that LINC01133 promoted NPC tumorigenesis by inhibiting YBX1 (8) (Figure 2C).
The abnormal expression of LINC01133 is extremely common in cancers. Therefore, elucidating the specific mechanism of regulating LINC01133 expression in tumors is of great significance for cancer therapies targeting the abnormal expression of LINC01133. Recently, studies have shown that the differential regulatory mechanism of LINC01133 primarily includes an increase in copy number and the regulation of transcription elements and entry from the tumor microenvironment (TME) (Figure 3).
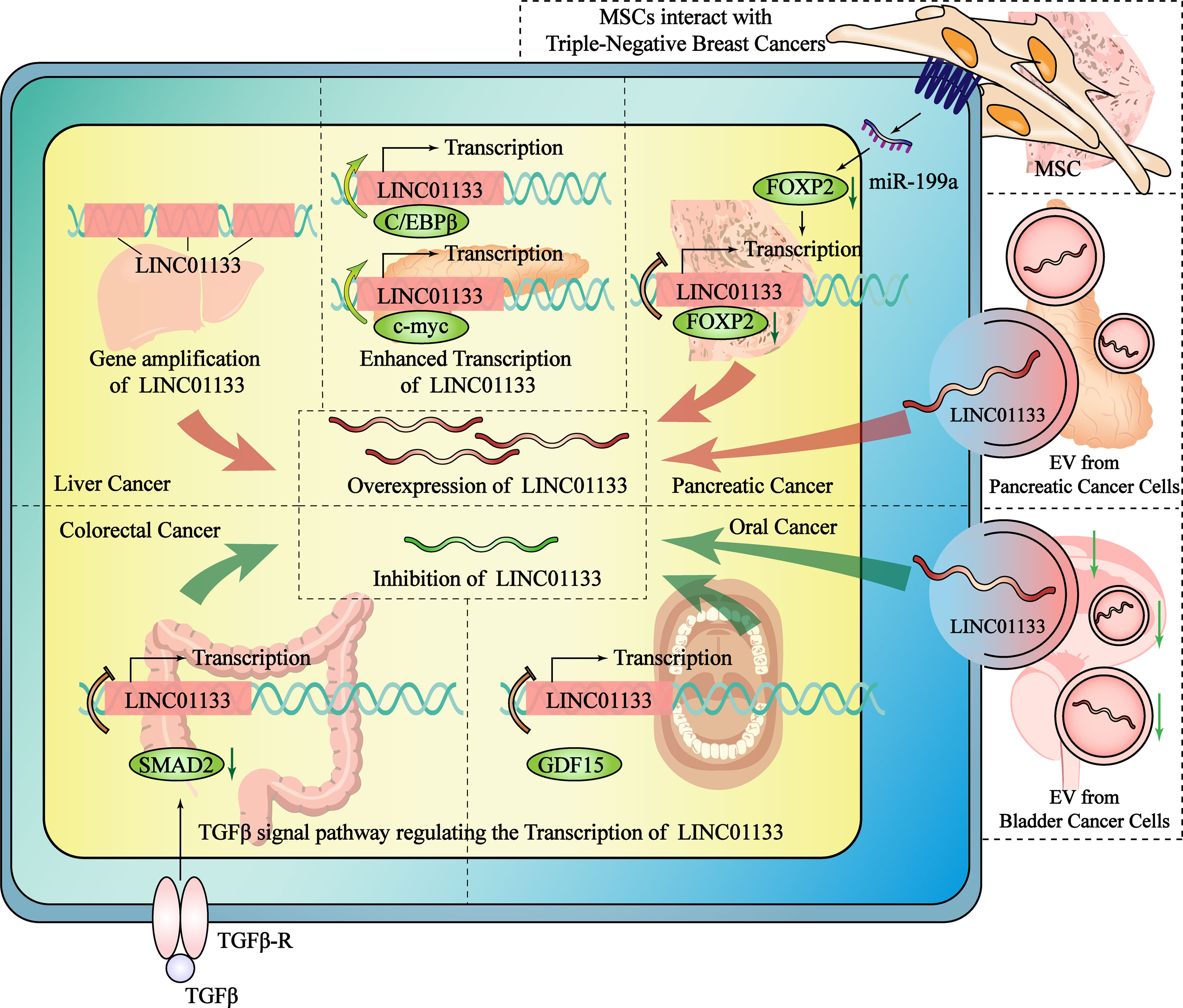
Figure 3 The mechanisms regulate the expression of LINC01133 in the cancers of different tissue type. The overexpression of LINC01133 could be regulated by gene amplification of LINC01133, the enhanced transcription of LINC01133, the increased transport of LINC01133 by EVs and the promoting effect from MSC in TME. The inhibition of LINC01133 could be regulated by the TGF-β pathway. GDF15 belongs to TGF-βsubfamily and also inhibit LINC01133 transcription. Compared with normal tissue cells, the number of LINC01133-containing exosomes secreted and received by cancer cells is reduced, which is one of the reasons why the concentration of LINC01133 in tumor cells decreases. The red arrow indicates the mechanisms that promote the expression of LINC01133, while the green arrow indicates the mechanisms that inhibit it. The black dashed lines separate different studies of the mechanisms that regulate the expression of LINC01133.
Multiple duplications of the LINC01133 gene may directly increase its RNA expression. It has been reported that lncRNA gene amplification occurs frequently in liver cancer. Screening of copy number variations from the whole genome sequencing data of 49 Chinese patients and a validation cohort of another 238 patients with HCC revealed that the copy number and expression level of LINC01133 were significantly higher in HCC tissue than that in the para-cancer tissue. Additionally, there was a positive relationship between these two indices (R2 = 0.535), indicating that a high amplification at the level of the DNA might be the cause of LINC01133 dysregulation (17).
Two studies on pancreatic ductal adenocarcinoma (PDAC) have revealed the underlying mechanism of transcription factor-mediated regulation of LINC01133 expression. Higher expression of LINC01133 and CCAAT/enhancer-binding protein β (C/EBPβ) was observed in PDAC compared to that in control conditions. C/EBPβ was found to positively regulate LINC01133 expression by binding to response elements within the LINC01133 promoter. The high expression of C/EBPβ was an indicator of a poor prognosis of PDAC (11). C-myc also binds to the promoter region of LINC01133 and is a key regulator of expression levels. C-myc overexpression was induced using periostin, which is a 90 kDa protein that is specifically secreted by pancreatic stellate cells, and this resulted in enhanced LINC01133 expression compared to that in control conditions (34). In oral squamous cell carcinoma, a transcriptional regulatory mechanism was found to regulate LINC01133 expression. The TGF-β canonical signaling pathway-mediated regulation of transcription is known to be associated with the occurrence and development of malignant tumors (35). The transcription of LINC01133 is also regulated by the TGF-β superfamily. In CRC, TGF-β inhibits the transcription of LINC01133, leading to EMT (36). In a study by Kong et al., a negative feedback inhibitory pathway was found between the TGFβ superfamily member GDF15 and LINC01133, causing a decrease in LINC01133 expression in oral squamous cell carcinoma and thereby promoting tumor metastasis (9).
In addition to endogenous LINC01133 present in cancer cells, exogenous LINC01133 present in exosomes in the TME can also affect the malignancy of cancer cells. Extracellular vesicles (EVs) or exosomes may originate from constituent cells in the TME, even from tumor cells themselves. Liu et al. have reported that the high secretion of exosomes containing LINC01133 in PDAC is correlated with a poor overall survival rate of PDAC patients. In their study, periostin was found to promote exosome secretion and to induce EMT. They demonstrated that exosomal LINC01133 extracted from a LINC01133-overexpressing cell line promoted proliferation, migration, invasion, and EMT and inhibited apoptosis in PDAC cells. In order to identify the mechanism underlying the regulation of LINC01133 expression, the nuclear localization of LINC01133 was first confirmed by a fluorescence in situ hybridization assay. Then, chromatin isolation by RNA purification and CHIP assay was used to identify a binding interaction between EZH2 and LINC01133 that promotes H3K27 trimethylation and inhibits the transcription of AXIN2 by targeting its promoter region. LINC01133-mediated silencing of AXIN2 further suppressed GSK3 activity, ultimately activating β-catenin (34). In bladder cancer (BC), LINC01133 expression was found to be high in exosomes of the SV‐HUC‐1 human uroepithelial cell line compared to the low expression in BC cells. Similar to the function of LINC01133, exosomes containing LINC01133 inhibited cell viability, proliferation, migration, and invasion. Exosomal LINC01133 was found to repress BC progression by regulating the Wnt signaling pathway (7).
The complex and diverse molecular regulatory mechanisms of LINC01133 determine the molecular basis for its functional specificity in different cancers. The abnormal expression and dysfunction of LINC01133 in different cancers are distinct in nature (Figure 4).
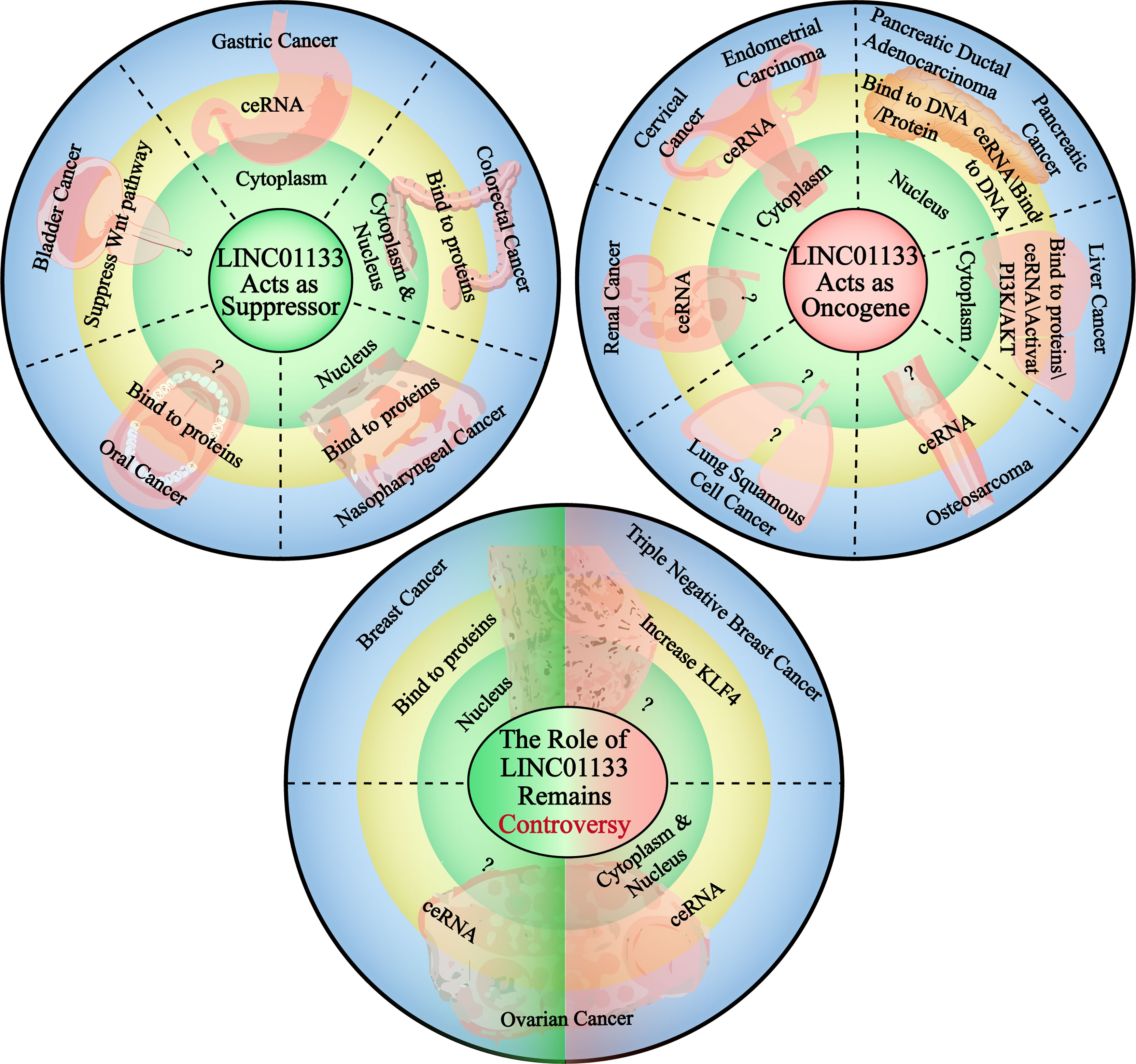
Figure 4 The carcinogenetic or suppressing role of LINC01133 in the cancers of different tissue type. The circle in the upper left corner demonstrates the cancers in which LINC01133 acts as a cancer suppressor, the subcellular localization of LINC01133 in each type of cancer and the main mechanism of it to function. The circle in the upper right corner demonstrates the cancers in which LINC01133 acts as an oncogene, and also the subcellular localization and functional mechanisms. The circle lower in the middle states the cancers in which the role of LINC01133 is still controversy. The green left side represents the suppressor role of LINC01133, the localization and functional mechanisms, and the red right side represents the oncogene role of LINC01133, the localization and functional mechanisms. The black dashed lines separate the cancers of different tissue types.
LINC01133 functions as a tumor suppressor in CRC. Compared with 219 normal tissue samples, LINC01133 was found to be significantly downregulated in CRC tissues. The low expression of LINC01133 was positively correlated with tumor metastasis, TNM staging, and low overall survival rate of CRC patients. Attenuating LINC01133 function by shRNA or miRNA binding promotes metastasis of CRC cells both in vivo and in vitro (36). In gastric cancer, the low expression and anti-tumor effect of LINC01133 was predicted using bioinformatics analysis and verified experimentally as well. Yang et al. quantified the level of LINC01133 in 200 cases of gastric cancer and paired adjacent tissues using qPCR and found a significantly lower expression level of LINC01133 in gastric cancer tissues compared to that in the paired tissues (26). This result was also consistent with that of another study that analyzed a cohort of 254 pairs of gastric cancer and adjacent tissues (16). In both these studies, LINC01133 acted as a tumor suppressor via the ceRNA mechanism. Bioinformatics analysis using the Gene Expression Omnibus and The Cancer Genome Atlas databases and subsequent verification revealed a tissue-specific downregulation of LINC01133 expression in gastrointestinal tissues compared to that in control tissues (37). In Table 2, we summarize the types of cancers in which LINC01133 exerts a tumor suppressor effect and the underlying mechanism of action.
In most gynecological cancers, LINC01133 plays the role of an oncogene. Yang et al. found that LINC01133 is highly expressed in endometrial cancer and its high expression was positively correlated with the occurrence of this cancer. Knockdown of LINC01133 reduced the vitality of endometrial cancer cells by arresting the cell cycle and increasing apoptosis of cancer cells, while simultaneously inhibiting their migration and invasion ability in vitro (39). LINC01133 levels were also found to be significantly elevated in the serum of patients with cervical squamous carcinoma (CESC) and cervical intraepithelial neoplasia (CIN) when compared to that in the control group; but there was no difference in LINC01133 expression between the CESC and CIN groups (43). Moreover, high LINC01133 levels were associated with a shorter survival period in CSEC patients (44). LINC01133 is also likely to function as an oncogene in non-digestive tract cancers. An in vitro study by Zheng et al. showed that the expression of LINC01133 was increased in liver cancer cell lines compared to that in control cells, and this increased expression promoted malignancy in cancer cells via the PI3K/AKT pathway (40). This was consistent with the result of Dan Yin’s study. Further, a ceRNA network between miR-199a-5p, LINC01133, and SNAI1 was identified to regulate the growth, invasion, and metastasis of liver cancer cells (17). Several studies have reported the carcinogenicity of LINC01133 in pancreatic cancer, (Table 2). In urinary system cancers, LINC01133 has been found to act as an oncogene in renal cancer cells. Zhai et al. reported that LINC01133 was highly expressed in renal cancer cell lines and in 34 renal cancer tissue specimens when compared with that of normal cell lines and tissues. The depletion of LINC01133 inhibited the proliferation, migration, and invasion of renal cancer cell lines. In vivo knockdown of LINC01133 inhibited subcutaneous tumor formation in nude mice (3). Details of the oncogenic function of LINC01133 in cancer are shown in Table 2.
The role of LINC01133 in some gynecological tumors remains controversial. For example, Song et al. found that LINC01133 expression was significantly downregulated in breast cancer samples compared to that it control samples, and this was associated with the progression and poor prognosis of breast cancer. As mentioned previously, the overexpression of LINC01133 inhibits the invasion and metastasis of breast cancer cells both in vitro and in vivo via a transcription-mediated regulatory mechanism (31). However, a study by Zhenbo Tu showed that mesenchymal stem/stromal cells (MSCs) played a critical role in promoting the initiation and progression of triple-negative breast cancers by inducing LINC01133 expression. In this study, LINC01133 promoted the phenotypic and growth characteristics of cancer stem cell-like cells and was a direct mediator of the MSC-triggered miR-199a-FOXP2 pathway (42). A similar contradiction regarding LINC01133 function also exists in ovarian cancer. Liu et al. (2019) found that LINC01133 expression was reduced significantly in ovarian cancer when compared with that in normal tissue, and this downregulation was an indicator of poor prognosis. The low levels of LINC01133 regulated by miR-205 promoted tumor proliferation and metastasis both in vivo and in vitro (27). However, in a study by Liu et al. (2020), LINC01133 was reported to be upregulated in epithelial ovarian cancer tissues and cell lines compared with that in control tissues and cell lines, and this increase facilitated cancer cell migration and invasion in vitro and tumor metastasis in vivo (15). Detailed information regarding the contradictory functions of LINC01133 had been presented in Table 2.
The abnormal expression of LINC01133 in cancers makes it a potential biomarker of the disease. In some cancers with well-defined and -documented effects, LINC01133 has great potential to be a prognostic marker. For example, the results of bioinformatics analysis showed evidence of a correlation between high LINC01133 expression and poor PDAC prognosis. LINC01133 was identified by weighted gene co-expression network analysis, and a survival analysis was performed to verify its role in advancing cancer progression (45). In pancreatic cancer (45), cervical cancer (44), and lung adenocarcinoma (5), increased LINC01133 levels represent poor patient survival. While in CRC (38), gastric cancer (26), and other cancers, a high level of LINC01133 was an indicator of a good prognosis (Figure 5).
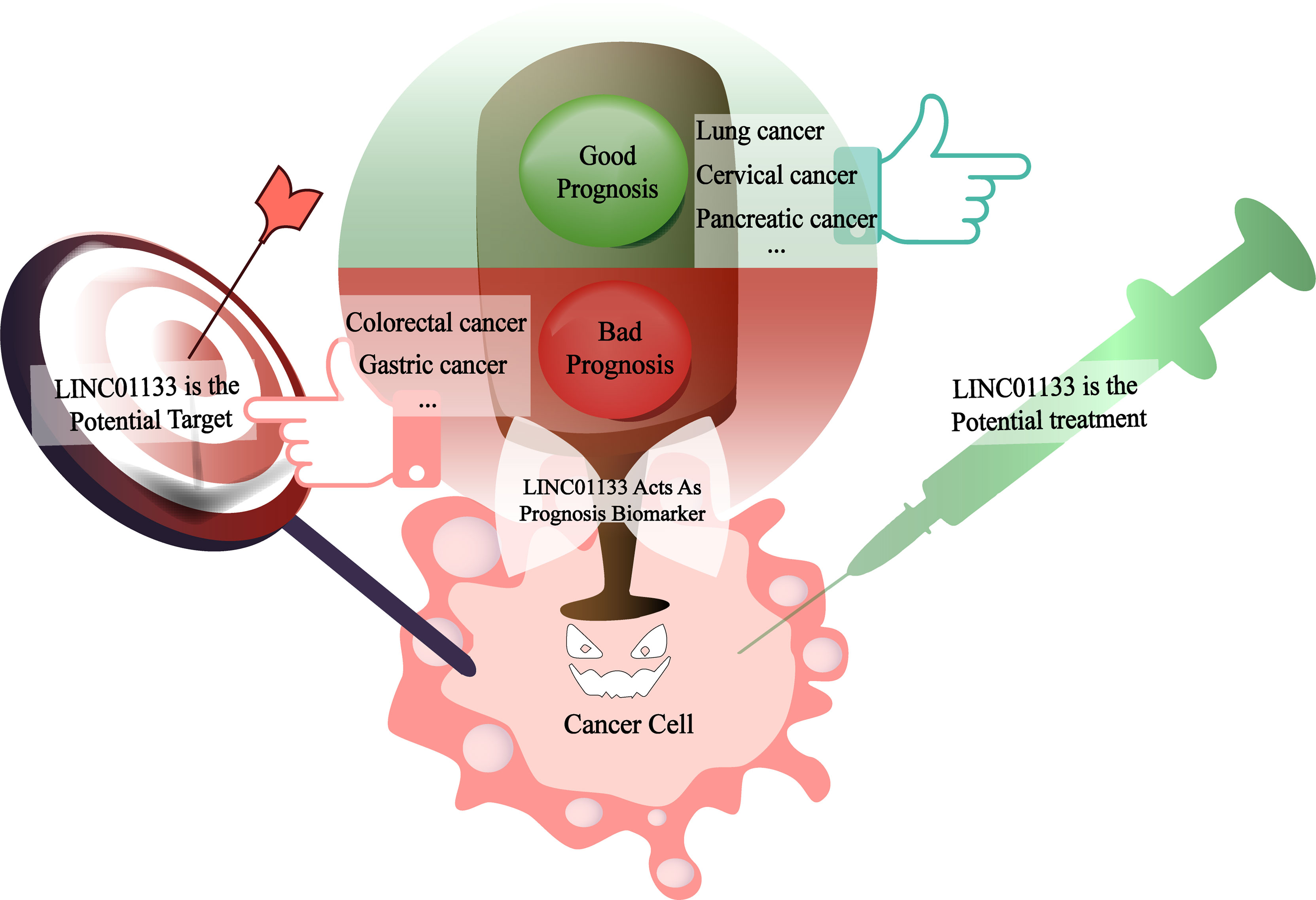
Figure 5 LINC01133 is the biomarker of cancer prognosis as well as the potential therapeutic target or treatment reagent in the future. When LINC01133 acts as an oncogene, its high expression suggests a bad prognosis of patients as shown in the red section and might be a potential therapeutic target molecular for the clinical treatment. In the contrary, when LINC01133 acts as a cancer suppressor, its high expression indicates a good prognosis of patients as shown in the green section and might be a potential treatment reagent in the future.
As mentioned, the abnormal expression of LINC01133 directly affects the prognosis of cancer patients. The molecular mechanisms underlying its biological functions are gradually being elucidated. Therefore, in cancers where LINC01133 exerts a carcinogenic effect, LINC01133 is a potential target for cancer therapies. However, considering that LINC01133 acts as a tumor suppressor gene in some cancers, importing functional LINC01133 is also a viable treatment option. Recent studies have shown that LINC01133 can be encapsulated into EVs (7, 34) and transported into tumor cells to regulate the traits of cancer cells. With further advances in research that aims to develop EVs as targeted drug carriers, it will become possible to deliver LINC01133 as a targeted therapeutic drug to cancer foci via EVs in the future (Figure 5).
Although several diverse studies have examined the regulatory mechanism of LINC01133 on cancer; as a non-coding RNA with obvious tissue-specific expression, further research in this area is still needed. Various mechanisms that can affect the function and expression of lncRNA; such as mutation (46), amplification or deletion of lncRNA at the genomic level (20), DNA methylation (47), histone modification (48), RNA modification (for example, m6A) (49), binding with RBPs [for example, transcription factor (50) or transcription enhancer (51)], and the interaction with bound RNAs (for example, miRNA) at the epigenetic level are all potential future directions in understanding the regulation of LINC01133 expression.
Further, additional mechanisms underlying the biological function of LINC01133 still need to be explored. Considering that lncRNAs require proteins to execute their regulatory roles, proteins that directly or indirectly interact with LINC01133 need to be identified. RBPs are well known for their roles in regulating RNA fate (from synthesis to decay) and protein translation (by assisting and/or directing RNAs) (52). However, the only RBPs reported to be associated to LINC01133 are the translation-promoting protein YBX1 (8) and the transcription repressor EZH2 (31). Therefore, cross-linking immunoprecipitation and RIP assays can be used to identify RBPs that bind to LINC01133.
More notably, the impact of the TME on cancer cells is receiving increasing attention (53–55), and the transport and protection of lncRNAs by exosomes when secreted in the TME to regulate malignancy of cancers is also a new mechanism that has been recently discovered (56–58). Numerous studies have found that lncRNAs can be secreted from different types of TME stromal cells, including cancer-associated fibroblasts (57), cancer-associated macrophages (59), and cancer cells packaged in EVs. EVs act as carriers of lncRNAs and are swallowed by cancer cells to allow functional lncRNAs to enter tumor cells and execute their biological functions. LINC01133 has been shown to be secreted into EVs from pancreatic ductal adenocarcinoma cells and is transferred to cancer cells to silence AXIN2/GSK3 and activate the Wnt/β-catenin pathway, which promotes EMT (34). In bladder cancer, LINC01133 is packaged into EVs from both cancer and normal tissue cells. Interestingly, the concentration of LINC01133 in EVs from normal tissue cells was higher than that in EVs from cancer cells. EVs are taken up by cancer cells, wherein they regulate the Wnt signaling pathway and inhibit the progression of bladder cancer (7). However, the types of stromal cells that secrete EVs containing LINC01133, the function of LINC01133 in the stromal cell itself, and the distinct roles of LINC01133 in the TME of different tissues are yet to be elucidated.
Similar to other star lncRNA molecules, the functions and underlying mechanisms of LINC01133 in cancers of different tissue types needs to be extensively studied. In future studies, additional mechanisms affecting the expression and regulatory mechanisms of LINC01133 should be explored. In addition, the role of LINC01133 in the interaction between cells in the TME should be studied in different cell types and in EVs. Due to the current contradiction with respect to its carcinogenic and tumor suppressor status, LINC01133 is a potential target for cancer therapies and a candidate molecule for the development of tumor treatment reagents.
SJ: Conceptualization, Investigation, Writing – original draft, Writing – review &editing. QZ: Writing-original draft, Writing – review &editing. JL: Writing – review &editing. KR: Language editing. XK, SL, and CS: Visualization, review & editing. XL: Writing – original draft. DZ: Conceptualization, review & editing. SF: Conceptualization, editing& Funding acquisition. MC: Conceptualization, Investigation, Writing – original draft, Writing-review, editing & Funding acquisition. All authors contributed to the article and approved the submitted version.
The present study was supported by HMU Marshal Initiative Funding (No. HMUMIF-21007) and the Open Project Program of Key Laboratory of Preservation of Human Genetic Resources and Disease Control in China (Harbin Medical University), Ministry of Education (No. LPHGRDC2020-001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Thanks to Pro. Wenjing Sun and Pro. Jie Wu for their help with writing skills in the review writing.
CeRNA, competing endogenous RNA; CRC colorectal cancer,EMT, epithelial-mesenchymal transition; EVs, extracellular vesicles; HCC, hepatocellular carcinoma; LncRNA, long non-coding RNA; LincRNA, long intergenic non-coding RNA; LINC01133, long intergenic non-coding RNA01133; PDAC, pancreatic ductal adenocarcinoma; RBPs, RNA binding proteins; RIP, RNA immunoprecipitation chip assay; TME, Tumor micro-environment.
1. Palazzo AF, Lee ES. Non-Coding Rna: What Is Functional and What Is Junk? Front Genet (2015) 6:2. doi: 10.3389/fgene.2015.00002
2. Zhou B, Yang Y, Zhan J, Dou X, Wang J, Zhou Y. Predicting Functional Long Non-Coding Rnas Validated by Low Throughput Experiments. RNA Biol (2019) 16(11):1555–64. doi: 10.1080/15476286.2019.1644590
3. Zhai X, Wu Y, Wang Z, Zhao D, Li H, Chong T, et al. Long Noncoding Rna Linc01133 Promotes the Malignant Behaviors of Renal Cell Carcinoma by Regulating the Mir-30b-5p/Rab3d Axis. Cell Transplant (2020) 29:963689720964413. doi: 10.1177/0963689720964413
4. Feng Y, Qu L, Wang X, Liu C. Linc01133 Promotes the Progression of Cervical Cancer by Sponging Mir-4784 to Up-Regulate Ahdc1. Cancer Biol Ther (2019) 20(12):1453–61. doi: 10.1080/15384047.2019.1647058
5. Geng W, Lv Z, Fan J, Xu J, Mao K, Yin Z, et al. Identification of the Prognostic Significance of Somatic Mutation-Derived Lncrna Signatures of Genomic Instability in Lung Adenocarcinoma. Front Cell Dev Biol (2021) 9:657667. doi: 10.3389/fcell.2021.657667
6. Zeng HF, Qiu HY, Feng FB. Long Noncoding Rna Linc01133 Functions as an Mir-422a Sponge to Aggravate the Tumorigenesis of Human Osteosarcoma. Oncol Res (2018) 26(3):335–43. doi: 10.3727/096504017X14907375885605
7. Yang H, Qu H, Huang H, Mu Z, Mao M, Xie Q, et al. Exosomes-Mediated Transfer of Long Noncoding Rna Linc01133 Represses Bladder Cancer Progression Via Regulating the Wnt Signaling Pathway. Cell Biol Int (2021) 45(7):1510–22. doi: 10.1002/cbin.11590
8. Zhang W, Du M, Wang T, Chen W, Wu J, Li Q, et al. Long Non-Coding Rna Linc01133 Mediates Nasopharyngeal Carcinoma Tumorigenesis by Binding to Ybx1. Am J Cancer Res (2019) 9(4):779–90.
9. Kong J, Sun W, Zhu W, Liu C, Zhang H, Wang H. Long Noncoding Rna Linc01133 Inhibits Oral Squamous Cell Carcinoma Metastasis Through a Feedback Regulation Loop With Gdf15. J Surg Oncol (2018) 118(8):1326–34. doi: 10.1002/jso.25278
10. Yan H, Tan D, Xie P, Liu Z, Li X. [Multiple Lncrnas Affect the Incidence and Development of Melanoma]. Zhong Nan Da Xue Xue Bao Yi Xue Ban (2017) 42(2):134–8. doi: 10.11817/j.issn.1672-7347.2017.02.003
11. Huang CS, Chu J, Zhu XX, Li JH, Huang XT, Cai JP, et al. The C/Ebpbeta-Linc01133 Axis Promotes Cell Proliferation in Pancreatic Ductal Adenocarcinoma Through Upregulation of Ccng1. Cancer Lett (2018) 421:63–72. doi: 10.1016/j.canlet.2018.02.020
12. Ransohoff JD, Wei Y, Khavari PA. The Functions and Unique Features of Long Intergenic Non-Coding Rna. Nat Rev Mol Cell Biol (2018) 19(3):143–57. doi: 10.1038/nrm.2017.104
13. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding Rna Gas5 Is a Growth Arrest- and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci Signal (2010) 3(107):ra8. doi: 10.1126/scisignal.2000568
14. Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, et al. A Long Noncoding Rna Lincrna-Eps Acts as a Transcriptional Brake to Restrain Inflammation. Cell (2016) 165(7):1672–85. doi: 10.1016/j.cell.2016.05.075
15. Liu S, Xi X. Linc01133 Contribute to Epithelial Ovarian Cancer Metastasis by Regulating Mir-495-3p/Tpd52 Axis. Biochem Biophys Res Commun (2020) 533(4):1088–94. doi: 10.1016/j.bbrc.2020.09.074
16. Zhang L, Pan K, Zuo Z, Ye F, Cao D, Peng Y, et al. Linc01133 Hampers the Development of Gastric Cancer Through Increasing Somatostatin Via Binding to Microrna-576-5p. Epigenomics (2021) 13(15):1205–19. doi: 10.2217/epi-2020-0377
17. Yin D, Hu ZQ, Luo CB, Wang XY, Xin HY, Sun RQ, et al. Linc01133 Promotes Hepatocellular Carcinoma Progression by Sponging Mir-199a-5p and Activating Annexin A2. Clin Transl Med (2021) 11(5):e409. doi: 10.1002/ctm2.409
18. Ferre F, Colantoni A, Helmer-Citterich M. Revealing Protein-Lncrna Interaction. Brief Bioinform (2016) 17(1):106–16. doi: 10.1093/bib/bbv031
19. Brex D, Barbagallo C, Mirabella F, Caponnetto A, Battaglia R, Barbagallo D, et al. Linc00483 Has a Potential Tumor-Suppressor Role in Colorectal Cancer Through Multiple Molecular Axes. Front Oncol (2020) 10:614455. doi: 10.3389/fonc.2020.614455
20. Athie A, Marchese FP, Gonzalez J, Lozano T, Raimondi I, Juvvuna PK, et al. Analysis of Copy Number Alterations Reveals the Lncrna Alal-1 as a Regulator of Lung Cancer Immune Evasion. J Cell Biol (2020) 219(9):e201908078. doi: 10.1083/jcb.201908078
21. Barbagallo D, Caponnetto A, Barbagallo C, Battaglia R, Mirabella F, Brex D, et al. The Gaugaa Motif Is Responsible for the Binding Between Circsmarca5 and Srsf1 and Related Downstream Effects on Glioblastoma Multiforme Cell Migration and Angiogenic Potential. Int J Mol Sci (2021) 22(4):1678. doi: 10.3390/ijms22041678
22. Xie X, Sun FK, Huang X, Wang CH, Dai J, Zhao JP, et al. A Circular Rna, Circsmarca5, Inhibits Prostate Cancer Proliferative, Migrative, and Invasive Capabilities Via the Mir-181b-5p/Mir-17-3p-Timp3 Axis. Aging (Albany NY) (2021) 13(15):19908–19. doi: 10.18632/aging.203408
23. Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z, et al. Opposing Roles for the Lncrna Haunt and Its Genomic Locus in Regulating Hoxa Gene Activation During Embryonic Stem Cell Differentiation. Cell Stem Cell (2015) 16(5):504–16. doi: 10.1016/j.stem.2015.03.007
24. Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, et al. Human Colorectal Cancer-Specific Ccat1-L Lncrna Regulates Long-Range Chromatin Interactions at the Myc Locus. Cell Res (2014) 24(5):513–31. doi: 10.1038/cr.2014.35
25. Raziq K, Cai M, Dong K, Wang P, Afrifa J, Fu S. Competitive Endogenous Network of Lncrna, Mirna, and Mrna in the Chemoresistance of Gastrointestinal Tract Adenocarcinomas. BioMed Pharmacother (2020) 130:110570. doi: 10.1016/j.biopha.2020.110570
26. Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, et al. Linc01133 as Cerna Inhibits Gastric Cancer Progression by Sponging Mir-106a-3p to Regulate Apc Expression and the Wnt/Beta-Catenin Pathway. Mol Cancer (2018) 17(1):126. doi: 10.1186/s12943-018-0874-1
27. Liu M, Shen C, Wang C. Long Noncoding Rna Linc01133 Confers Tumor-Suppressive Functions in Ovarian Cancer by Regulating Leucine-Rich Repeat Kinase 2 as an Mir-205 Sponge. Am J Pathol (2019) 189(11):2323–39. doi: 10.1016/j.ajpath.2019.07.020
28. Zhang D, Zhang Y, Sun X. Linc01133 Promotes the Progression of Cervical Cancer Via Regulating Mir-30a-5p/Foxd1. Asia Pac J Clin Oncol (2021) 17(3):253–63. doi: 10.1111/ajco.13451
29. Zhang J, Gao S, Zhang Y, Yi H, Xu M, Xu J, et al. Mir-216a-5p Inhibits Tumorigenesis in Pancreatic Cancer by Targeting Tpt1/Mtorc1 and Is Mediated by Linc01133. Int J Biol Sci (2020) 16(14):2612–27. doi: 10.7150/ijbs.46822
30. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many Human Large Intergenic Noncoding Rnas Associate With Chromatin-Modifying Complexes and Affect Gene Expression. Proc Natl Acad Sci USA (2009) 106(28):11667–72. doi: 10.1073/pnas.0904715106
31. Song Z, Zhang X, Lin Y, Wei Y, Liang S, Dong C. Linc01133 Inhibits Breast Cancer Invasion and Metastasis by Negatively Regulating Sox4 Expression Through Ezh2. J Cell Mol Med (2019) 23(11):7554–65. doi: 10.1111/jcmm.14625
32. Weng YC, Ma J, Zhang J, Wang JC. Long Non-Coding Rna Linc01133 Silencing Exerts Antioncogenic Effect in Pancreatic Cancer Through the Methylation of Dkk1 Promoter and the Activation of Wnt Signaling Pathway. Cancer Biol Ther (2019) 20(3):368–80. doi: 10.1080/15384047.2018.1529110
33. Zang C, Nie FQ, Wang Q, Sun M, Li W, He J, et al. Long Non-Coding Rna Linc01133 Represses Klf2, P21 and E-Cadherin Transcription Through Binding With Ezh2, Lsd1 in Non Small Cell Lung Cancer. Oncotarget (2016) 7(10):11696–707. doi: 10.18632/oncotarget.7077
34. Liu Y, Tang T, Yang X, Qin P, Wang P, Zhang H, et al. Tumor-Derived Exosomal Long Noncoding Rna Linc01133, Regulated by Periostin, Contributes to Pancreatic Ductal Adenocarcinoma Epithelial-Mesenchymal Transition Through the Wnt/Beta-Catenin Pathway by Silencing Axin2. Oncogene (2021) 40(17):3164–79. doi: 10.1038/s41388-021-01762-0
35. Derynck R, Zhang YE. Smad-Dependent and Smad-Independent Pathways in Tgf-Beta Family Signalling. Nature (2003) 425(6958):577–84. doi: 10.1038/nature02006
36. Kong J, Sun W, Li C, Wan L, Wang S, Wu Y, et al. Long Non-Coding Rna Linc01133 Inhibits Epithelial-Mesenchymal Transition and Metastasis in Colorectal Cancer by Interacting With Srsf6. Cancer Lett (2016) 380(2):476–84. doi: 10.1016/j.canlet.2016.07.015
37. Foroughi K, Amini M, Atashi A, Mahmoodzadeh H, Hamann U, Manoochehri M. Tissue-Specific Down-Regulation of the Long Non-Coding Rnas Pcat18 and Linc01133 in Gastric Cancer Development. Int J Mol Sci (2018) 19(12):3881. doi: 10.3390/ijms19123881
38. Zhang JH, Li AY, Wei N. Downregulation of Long Non-Coding Rna Linc01133 Is Predictive of Poor Prognosis in Colorectal Cancer Patients. Eur Rev Med Pharmacol Sci (2017) 21(9):2103–7.
39. Yang W, Yue Y, Yin F, Qi Z, Guo R, Xu Y. Linc01133 and Linc01243 Are Positively Correlated With Endometrial Carcinoma Pathogenesis. Arch Gynecol Obstet (2021) 303(1):207–15. doi: 10.1007/s00404-020-05791-0
40. Zheng YF, Zhang XY, Bu YZ. Linc01133 Aggravates the Progression of Hepatocellular Carcinoma by Activating the Pi3k/Akt Pathway. J Cell Biochem (2019) 120(3):4172–9. doi: 10.1002/jcb.27704
41. Zhang J, Zhu N, Chen X. A Novel Long Noncoding Rna Linc01133 Is Upregulated in Lung Squamous Cell Cancer and Predicts Survival. Tumour Biol (2015) 36(10):7465–71. doi: 10.1007/s13277-015-3460-9
42. Tu Z, Schmollerl J, Cuiffo BG, Karnoub AE. Microenvironmental Regulation of Long Noncoding Rna Linc01133 Promotes Cancer Stem Cell-Like Phenotypic Traits in Triple-Negative Breast Cancers. Stem Cells (2019) 37(10):1281–92. doi: 10.1002/stem.3055
43. Wang WJ, Wang D, Zhao M, Sun XJ, Li Y, Lin H, et al. Serum Lncrnas (Ccat2, Linc01133, Linc00511) With Squamous Cell Carcinoma Antigen Panel as Novel Non-Invasive Biomarkers for Detection of Cervical Squamous Carcinoma. Cancer Manag Res (2020) 12:9495–502. doi: 10.2147/CMAR.S259586
44. Mao X, Qin X, Li L, Zhou J, Zhou M, Li X, et al. A 15-Long Non-Coding Rna Signature to Improve Prognosis Prediction of Cervical Squamous Cell Carcinoma. Gynecol Oncol (2018) 149(1):181–7. doi: 10.1016/j.ygyno.2017.12.011
45. Giulietti M, Righetti A, Principato G, Piva F. Lncrna Co-Expression Network Analysis Reveals Novel Biomarkers for Pancreatic Cancer. Carcinogenesis (2018) 39(8):1016–25. doi: 10.1093/carcin/bgy069
46. Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, et al. The Norad Lncrna Assembles a Topoisomerase Complex Critical for Genome Stability. Nature (2018) 561(7721):132–6. doi: 10.1038/s41586-018-0453-z
47. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long Noncoding Rna as Modular Scaffold of Histone Modification Complexes. Science (2010) 329(5992):689–93. doi: 10.1126/science.1192002
48. Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang K, et al. Lncrna Casc9 Promotes Esophageal Squamous Cell Carcinoma Metastasis Through Upregulating Lamc2 Expression by Interacting With the Creb-Binding Protein. Cell Death Differ (2018) 25(11):1980–95. doi: 10.1038/s41418-018-0084-9
49. Wen S, Wei Y, Zen C, Xiong W, Niu Y, Zhao Y. Long Non-Coding Rna Neat1 Promotes Bone Metastasis of Prostate Cancer Through N6-Methyladenosine. Mol Cancer (2020) 19(1):171. doi: 10.1186/s12943-020-01293-4
50. Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A Strategy for Probing the Function of Noncoding Rnas Finds a Repressor of Nfat. Science (2005) 309(5740):1570–3. doi: 10.1126/science.1115901
51. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A Long Noncoding Rna Maintains Active Chromatin to Coordinate Homeotic Gene Expression. Nature (2011) 472(7341):120–4. doi: 10.1038/nature09819
52. Gerstberger S, Hafner M, Tuschl T. A Census of Human Rna-Binding Proteins. Nat Rev Genet (2014) 15(12):829–45. doi: 10.1038/nrg3813
53. Fane M, Weeraratna AT. How the Ageing Microenvironment Influences Tumour Progression. Nat Rev Cancer (2020) 20(2):89–106. doi: 10.1038/s41568-019-0222-9
54. Ho WJ, Jaffee EM, Zheng L. The Tumour Microenvironment in Pancreatic Cancer - Clinical Challenges and Opportunities. Nat Rev Clin Oncol (2020) 17(9):527–40. doi: 10.1038/s41571-020-0363-5
55. Pitt JM, Marabelle A, Eggermont A, Soria JC, Kroemer G, Zitvogel L. Targeting the Tumor Microenvironment: Removing Obstruction to Anticancer Immune Responses and Immunotherapy. Ann Oncol (2016) 27(8):1482–92. doi: 10.1093/annonc/mdw168
56. Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, et al. Emerging Role of Exosome-Derived Long Non-Coding Rnas in Tumor Microenvironment. Mol Cancer (2018) 17(1):82. doi: 10.1186/s12943-018-0831-z
57. Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, et al. Carcinoma-Associated Fibroblasts Promote the Stemness and Chemoresistance of Colorectal Cancer by Transferring Exosomal Lncrna H19. Theranostics (2018) 8(14):3932–48. doi: 10.7150/thno.25541
58. Fan JT, Zhou ZY, Luo YL, Luo Q, Chen SB, Zhao JC, et al. Exosomal Lncrna Neat1 From Cancer-Associated Fibroblasts Facilitates Endometrial Cancer Progression Via Mir-26a/B-5p-Mediated Stat3/Ykl-40 Signaling Pathway. Neoplasia (2021) 23(7):692–703. doi: 10.1016/j.neo.2021.05.004
Keywords: LINC01133, cancer, CeRNA, RBP, TME, transcription, LncRNA
Citation: Jiang S, Zhang Q, Li J, Raziq K, Kang X, Liang S, Sun C, Liang X, Zhao D, Fu S and Cai M (2022) New Sights Into Long Non-Coding RNA LINC01133 in Cancer. Front. Oncol. 12:908162. doi: 10.3389/fonc.2022.908162
Received: 30 March 2022; Accepted: 13 May 2022;
Published: 07 June 2022.
Edited by:
Subash Gupta, Banaras Hindu University, IndiaReviewed by:
Vivek Sharma, Birla Institute of Technology and Science, IndiaCopyright © 2022 Jiang, Zhang, Li, Raziq, Kang, Liang, Sun, Liang, Zhao, Fu and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mengdi Cai, Y2FpbWVuZ2RpQGVtcy5ocmJtdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.