
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Oncol. , 11 May 2022
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.903586
This article is part of the Research Topic Recent Advances in Liquid Biopsy in Colorectal Cancer View all 9 articles
This article is a correction to:
Longitudinal Circulating Tumor DNA Profiling in Metastatic Colorectal Cancer During Anti-EGFR Therapy
 Wentao Yang1,2†
Wentao Yang1,2† Jianling Zou1,2†
Jianling Zou1,2† Ye Li2,3†
Ye Li2,3† Rujiao Liu1,2
Rujiao Liu1,2 Zhengqing Yan4
Zhengqing Yan4 Shiqing Chen4
Shiqing Chen4 Xiaoying Zhao1,2
Xiaoying Zhao1,2 Weijian Guo1,2
Weijian Guo1,2 Mingzhu Huang1,2
Mingzhu Huang1,2 Wenhua Li1,2
Wenhua Li1,2 Xiaodong Zhu1,2
Xiaodong Zhu1,2 Zhiyu Chen1,2*
Zhiyu Chen1,2*A Corrigendum on
Longitudinal Circulating Tumor DNA Profiling in Metastatic Colorectal Cancer During Anti-EGFR Therapy
By Yang W, Zou J, Li Y, Liu R, Yan Z, Chen S, Zhao X, Guo W, Huang M, Li W, Zhu X and Chen Z (2022) Front. Oncol. 12:830816. doi: 10.3389/fonc.2022.830816
In the original article, there was a mistake in Figure 1 as published. The figure illustration is wrong due to a software bug during picture output. The corrected Figure 1 appears below.
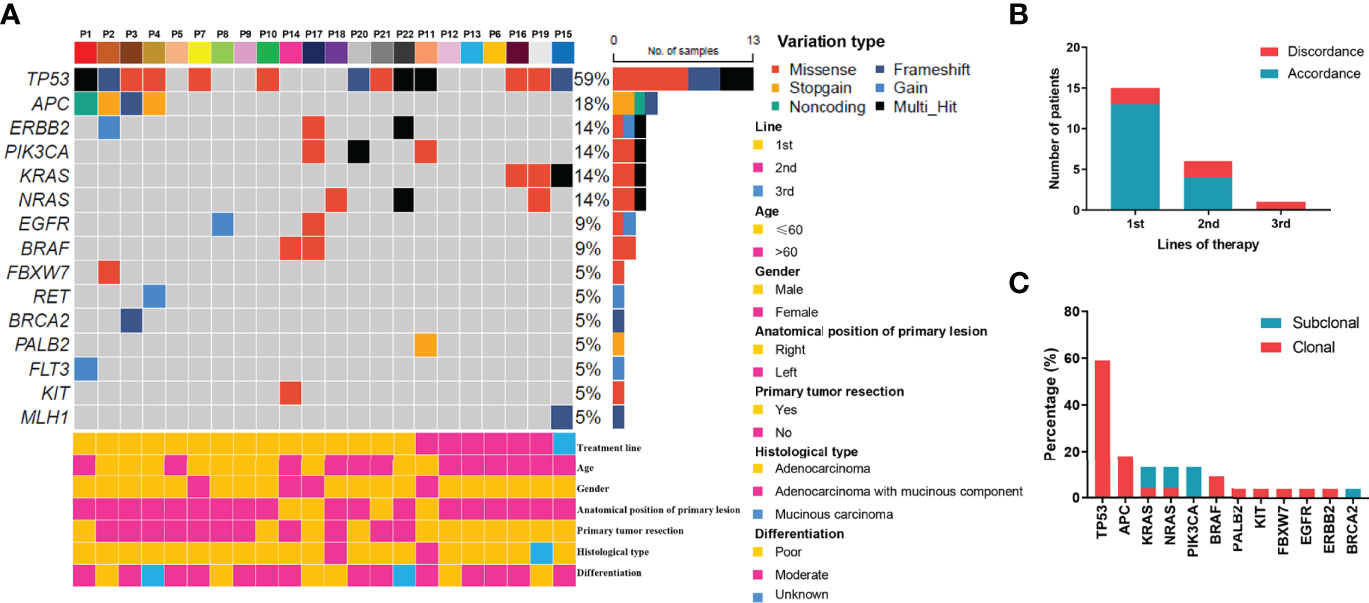
Figure 1 Mutation profiling of pre-treatment ctDNA. (A) Genomic profiles of 22 advanced colorectal cancer patients from pre-treatment ctDNA. (B) The consistency of the RAS mutations detected in paired tissues and plasma. (C) The clonal and subclonal landscapes in 22 mCRC patient at baseline. Gain: segments with log ratio more than 3 times of standard deviation of all segment level were considered as “gain”.
In the original article, references 14 and reference 37 are the same. We have renumbered the references.
In the original article, there was an additional error in Results, Mutation Profiles at Baseline, paragraph two. In the sentence “For patients who received cetuximab as second-line treatment, the RAS mutation discrepancy was 13.3% (2/6)”, the percentage is wrong and needs to be changed from 13.3% to 33.3%. The corrected paragraph appears below.
“For the KRAS, NRAS, and BRAF V600E genes, the concordance rates between the tumor tissue test by PCR and ctDNA test by NGS were 86.4%, 86.4%, and 100%, respectively. The RAS mutation discrepancy was also compared among treatments (Figure 1B). For patients who received cetuximab as first-line treatment, the RAS mutation discrepancy was 13.3% (2/15). Both of these patients also had NRAS mutations. The mutation sites were NRAS p.Q61K (0.31%), NRAS p.G13R (0.07%), and NRAS p.G12R (0.37%). For patients who received cetuximab as second-line treatment, the RAS mutation discrepancy was 33.3% (2/6). One patient had a KRAS p.G12V mutation (2.17%) and the other patient had both KRAS p.Q61H (0.02%) and NRAS p.G13C (0.03%) mutations. The only patient who received cetuximab as third-line treatment had a KRAS mutation. The mutation sites included KRAS p.Q61Hc.183A>T (0.05%), KRAS p.Q61Hc.183A>C (0.91%), and KRAS p.G12A (0.58%). The clonal and subclonal landscapes were detected at baseline (Figure 1C). Subclonal mutations were found in 31.8% (7/22) of the patients. The three most common clonal mutation genes were TP53, APC, and BRAF, while the three most common subclonal mutation genes were PIK3CA, KRAS, and NRAS.”
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.903586/full#supplementary-material
Keywords: colorectal cancer, next-generation sequencing, circulating tumor DNA, dynamic monitoring, prognosis
Citation: Yang W, Zou J, Li Y, Liu R, Yan Z, Chen S, Zhao X, Guo W, Huang M, Li W, Zhu X and Chen Z (2022) Corrigendum: Longitudinal Circulating Tumor DNA Profiling in Metastatic Colorectal Cancer During Anti-EGFR Therapy. Front. Oncol. 12:903586. doi: 10.3389/fonc.2022.903586
Received: 24 March 2022; Accepted: 11 April 2022;
Published: 11 May 2022.
Edited by:
Antonio Avallone, G. Pascale National Cancer Institute Foundation (IRCCS), ItalyReviewed by:
Luca Lazzari, IFOM - The FIRC Institute of Molecular Oncology, ItalyCopyright © 2022 Yang, Zou, Li, Liu, Yan, Chen, Zhao, Guo, Huang, Li, Zhu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyu Chen, Y2hhbmhqNzVAYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.