
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 30 June 2022
Sec. Breast Cancer
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.899622
This article is part of the Research Topic Cancer Stem Cells as Attractive Targets for Breast Cancer Therapy View all 5 articles
 Hossam Taha Mohamed1,2*†
Hossam Taha Mohamed1,2*† Aya Ali El-Sharkawy1†
Aya Ali El-Sharkawy1† Mohamed El-Shinawi3,4†
Mohamed El-Shinawi3,4† Robert J. Schneider5†
Robert J. Schneider5† Mona Mostafa Mohamed1,6†
Mona Mostafa Mohamed1,6†Inflammatory breast cancer (IBC) is a highly aggressive phenotype of breast cancer that is characterized by a high incidence early metastasis. We previously reported a significant association of human cytomegalovirus (HCMV) DNA in the carcinoma tissues of IBC patients but not in the adjacent normal tissues. HCMV-infected macrophages serve as “mobile vectors” for spreading and disseminating virus to different organs, and IBC cancer tissues are highly infiltrated by tumor-associated macrophages (TAMs) that enhance IBC progression and promote breast cancer stem cell (BCSC)-like properties. Therefore, there is a need to understand the role of HCMV-infected TAMs in IBC progression. The present study aimed to test the effect of the secretome (cytokines and secreted factors) of TAMs derived from HCMV+ monocytes isolated from IBC specimens on the proliferation, invasion, and BCSC abundance when tested on the IBC cell line SUM149. HCMV+ monocytes were isolated from IBC patients during modified radical mastectomy surgery and tested in vitro for polarization into TAMs using the secretome of SUM149 cells. MTT, clonogenic, invasion, real-time PCR arrays, PathScan Intracellular Signaling array, and cytokine arrays were used to characterize the secretome of HCMV+ TAMs for their effect on the progression of SUM149 cells. The results showed that the secretome of HCMV+ TAMs expressed high levels of IL-6, IL-8, and MCP-1 cytokines compared to HCMV- TAMs. In addition, the secretome of HCMV+ TAMs induced the proliferation, invasion, colony formation, and expression of BCSC-related genes in SUM149 cells compared to mock untreated cells. In addition, the secretome of HCMV+ TAMs activated the phosphorylation of intracellular signaling molecules p-STAT3, p-AMPKα, p-PRAS40, and p-SAPK/JNK in SUM149 cells. In conclusion, this study shows that the secretome of HCMV+ TAMs enhances the proliferation, invasion, colony formation, and BCSC properties by activating the phosphorylation of p-STAT3, p-AMPKα, p-PRAS40, and p-SAPK/JNK intracellular signaling molecules in IBC cells.
IBC is a highly metastatic and lethal subtype of breast cancer presenting with high incidence among young women in the Middle East and North Africa (1). TAMs of IBC patients were found to contribute to high levels of growth factors and cytokines in the TME and enhance metastasis via different mechanisms (2, 3).
In breast cancer, studies showed that the incidence of BCSCs plays a key role in recurrence and metastasis, which is associated to a certain extent with the presence of macrophages in the TME (4). For instance, β-Catenin activation attracts monocytes and induces the expression of CCL2 which changes monocytes to macrophages (needs a reference). BCSCs are associated in-vivo with increased metastasis activity promoted by β-Catenin-mediated CCL2 and TAMs (CD163) (5). TAMs in breast cancer TME induce stem cell properties via different mechanisms (6). Our previous studies identified different molecular, cellular, viral, immunological, and stem cell drivers involved in the poor prognosis of IBC (3, 7–10). For instance, we have been studying the role of TAMs in IBC progression. We showed that monocytes/macrophages identified as CD14+ cells highly infiltrate IBC cancer tissues and are localized with high density around IBC tumor emboli, clusters of highly metastatic IBC cells (3). The detected cytokines secreted by TAMs suggest a role for viral infection in the etiology of IBC (3).
Even though several studies showed links between viral infection and poor breast cancer prognosis (11), a causative role for viral infection in breast cancer development and progression is still uncertain, including in IBC. Previous studies by the authors assessed the incidence of different viral DNAs including HPV-16 and -18, HCMV, HBV, HSV-1, HSV-2, and HHV-8 in the cancer tissues of IBC versus non-IBC patients (9). Results showed that the incidence of multiple viral DNAs in IBC tissues was higher compared to non-IBC tissues, with the most prevalent detected viruses identified as HCMV and HPV-16. Moreover, detection of the DNA of different viruses positively correlated with the upregulation of the Ki-67 proliferation marker in IBC cells. Our results suggested a significant association between viral infection and IBC disease etiology (9). Herpes virus association with IBC may not be unique (7). HCMV is detected in more aggressive forms of malignancies including colorectal cancer (12), malignant glioblastoma (13), and breast cancer (14, 15). Regarding IBC, we found that HCMV-DNA is more prevalent in the cancer tissues of IBC but not in the adjacent non-cancer tissues (8). HCMV-infected IBC cancer tissues also exhibit higher expression and activation (phosphorylation) of NF-κB/p65 in cancer tissues of IBC patients than those of non-IBC patients (8), although a causal relationship to increased HCMV presence has not been established. In addition, we showed that certain polymorphisms in HCMV genotypes may be associated with increased involvement of HCMV in IBC disease etiology (9).
Monocytes/macrophages are a major site of persistence of HCMV in the peripheral blood (16). Whereas HCMV does not replicate in monocytes, primary HCMV infection to monocytes induces differentiation and biological turnover of monocytes to tissue macrophages. In addition, infected macrophages serve as “mobile vectors” for virus spreading and dissemination to different organs mainly by trans-endothelial migration (17). CD14+ monocytes were found to be a reservoir for HCMV latency (18). Furthermore, monocyte HCMV infection induces the differentiation of monocytes into macrophages characterized by the expression of Toll-like receptor (TLR) and inflammatory cytokines TNF-α, IL-6, and IL-8 (19). Notably, and possibly related, gene expression profile studies showed that TLR and IL-6 are more highly expressed in IBC patients compared to non-IBC patients (20, 21). Among subpopulations of macrophages associated with viral infection are macrophages expressing the marker MAC387 (22). MAC387 is associated with poor prognosis and increased distant metastasis of cholangiocancers (CCA) (23). HCMV+ macrophages are associated with HCMV viral replication and are characterized by significantly high expression of TNF-α, IL-6, and. IL-8 at gene and protein levels in response to stimulation by TLR4 ligand (LPS) compared with mock uninfected macrophages (19). Using in-vitro co-culture models seeding MRC-5 cells infected with HCMV-encoded UL128 protein (HCMV-UL128+) with peripheral blood mononuclear cells (PBMCs), significantly induced secretion of IL-6 and TNF-α in the culture media was found compared to those co-cultured with HCMV-UL128- (24). IL-6 binds to tumor cell transmembrane IL-6 receptors (IL-6R) and subsequently dimerizes with glycoprotein 130 (gp130), a transmembrane protein that behaves like a signal transducer following cytokine engagement. This binding activates the Janus Kinases/Signal Transducers and Activators of Transcription (JAK/STAT) pathway, which ultimately induces invasion and motility of cancer cells (25). This links back to BCSCs, because the Jak-STAT pathway modulates BCSC activity (26).
Several in vitro studies on different cancer cell lines revealed that HCMV infection in the TME promotes stemness and an epithelial-to-mesenchymal transition (EMT), leading to increased cancer aggressiveness (19). Moreover, HCMV infection in breast cancer cells inhibits EMT and promotes the mesenchymal-epithelial transition (MET), which is necessary for colonization of metastases (27). Supporting this hypothesis, it was found that 96% of brain metastases derived from primary breast cancers were HCMV+, while adjacent normal tissues are virus-free (28). Moreover, high expression of GM-CSF is associated with the prevalence of CCL18-secreting macrophages and the realization of EMT by breast cancer cells (29).
In the present study, we showed that HCMV+ IBC tissues are characterized by higher infiltration of CD163+ and MAC387+ TAMs compared to HCMV- IBC tissues. SUM149 cells stimulated by the secretome of HCMV+ TAMs showed an increase in proliferation, invasion, colony formation, and expression of BCSC-related genes compared to mock infected cells. A probable mechanism of action involves activation of p-STAT3, p-AMPKα, p-PRAS40, and p-SAPK/JNK intracellular signaling molecules. These results highlight a potential critical onco-modulatory role of HCMV infection in IBC.
The study protocol was approved by the Institutional Review Board (IRB#00006379), Faculty of Medicine, Ain Shams University, Egypt. A total of 28 IBC patients were enrolled in the present study. IBC patients were clinically and pathologically diagnosed as previously described (3, 30). Patients suffering from Hepatitis or AIDS or COVID-19 or auto-immune diseases were excluded from this study. Before participation, every patient signed a consent form, including approval for publication of the study results.
Five ml of peripheral blood was collected from each IBC patient in red-top tubes, which contained no anticoagulant or preservative (Greiner Bio-one, Kremsmünster, Austria) for serological diagnosis of HCMV IgG antibodies. Blood was collected and centrifuged at 1500 rpm for 10 min to isolate serum for serological tests. IgG antibodies against HCMV were measured in serum samples using HCMV IgG Chemiluminescence detection kit (Diasorin, Liaison, Italy) and a Liaison chemiluminescence device. The kit measures HCMV IgG antibody concentrations which are expressed as IU/ml. Samples with HCMV IgG concentrations equal to or more than 0.6 IU/ml were considered positive (31).
During axillary dissection in modified radical mastectomy (MRM) operations, 10 ml of blood was withdrawn from the identified axillary tributaries in a heparinized syringe with an angular needle as described before (32). Plasma was collected from the whole blood sample via centrifugation at 1500 rpm for 20 min. Mononuclear cells were separated from the precipitated blood content by Ficoll-Hypaque density gradient centrifugation (Lonza, ME, USA) at 1500 rpm for 30 min. The buffy coat layer containing mononuclear cells was separated and washed twice with PBS. The TME CD14+ monocytes were purified from the mononuclear cells using “EasySep™ Human Monocyte Enrichment Kit without CD16 Depletion” (StemCell Technologies, Vancouver, Canada). Purified TME CD14+ monocytes were grown overnight in Petri dishes (35 X 10 mm) at a concentration of (1 x 106 cells/ml) in RPMI media with 1% of penicillin/streptomycin antibiotic mixture in 3% fetal bovine serum (FBS) and incubated at 37°C in 5% CO2 for 24 h. Media conditioned by TME CD14+ TAMs secretions were collected as TAMs secretome and concentrated 1:100 using Vivaspin™ protein concentrator column (Sartorius, Goettingen, Germany) with 10,000 molecular weight cutoff value (MWCO value), and its protein content was determined via Bradford assay (Biorad Laboratories, CA, USA) and measured by Multiskan SkyHigh microplate spectrophotometer (ThermoFisher Scientific, MA, USA). Cells were aliquoted and stored at −80°C for DNA, RNA, and protein extraction.
DNA was extracted from purified IBC cancer tissues and isolated TME CD14+ monocytes/macrophages (1 x 106 cells/ml) of HCMV seropositive and seronegative IBC patients using GeneJETTM Genomic DNA purification Kit (Thermo Scientific, MA, USA). The extracted DNA was used in nested PCR for detecting the fourth exon of the HCMV Immediate Early (IE) gene was performed as described before (8, 33). Amplified PCR products (293 bp) were visualized on 1.5% agarose gels (Bio Basic INC, ON, Canada) stained with ethidium bromide, and photographed by the GBOX-F3 gel documentation system (Syngene, MD, USA).
Immunohistochemical (IHC) staining was performed after chemical dewaxing of 4 μm thick formalin-fixed paraffin-embedded (FFPE) tissue sections as described before (30, 34) using antibodies for CD14 (diluted at 1:50 in PSA) (Chemicon, CA, USA), CD68 (diluted at 1:50 in PSA) (M0814) from Dako (Agilent, CA, USA), CD163 (1:500) (Abcam, Cambridge, UK), and MAC387 (1µg/ml) (Abcam, Cambridge, UK). IHC Staining was carried out by adding 100 µl of DAB+ chromogen diluted at 1:50 in substrate buffer [(EnVision+ Dual Link System-HRP (DAB+)] for 10 min. Finally, tissue specimens were washed in phosphate buffer saline (PBS), the nuclei were counterstained with hematoxylin and mounted using Permount® (Fisher Scientific, PA, USA) for microscopic examination. Negative control slides were run in parallel with each marker where primary antibody was replaced by PBS. The stained area fractions were calculated using ImageJ software (National Institutes of Health, Bethesda, MD, USA) (7, 8).
For cellular morphology of in vitro polarized HCMV+ TAMs, microscopic images were taken at 40x magnification. Three distinct morphologic subtypes were identified, (M0) were small/roundish cells characterized by a cellular diameter ≤ 10 μm with the absence of cytoplasmic projections on the cell surface, (M1) were enlarged amoeboid cells characterized by a cellular diameter ≥ 10 μm and presence of many delicate cytoplasmic extensions on the cell surface with visible intracellular vacuoles, and (M2) demonstrated a large and elongated “spindeloid” cells with a cellular diameter between 10 to 30 μm and cytoplasmic extensions on the apical ends of the cell bodies (35, 36).
Studies confirmed the effect of the secretome of SUM149 on the polarization status of in vitro polarized HCMV- and HCMV+ TAMs. Based on previous studies (37–44), we carefully designed and customized a PCR array that included 43 macrophages polarization-related genes (Vivantis, Selangor, Malaysia) to analyze the gene expression signature of in vitro polarized HCMV- and HCMV+ TAMs. Total RNA was purified from mock and polarized HCMV- and HCMV+ TAMs using QIAzol lysis reagent (Qiagen, Hilden, Germany). The total RNA concentration was measured by Multiskan SkyHigh microplate spectrophotometer (ThermoFisher Scientific, MA, USA), and RNA integrity was tested by separating the RNA on a 1% standard agarose gel and examining the ribosomal RNA bands. One µg of RNA was transcribed into complementary DNA (cDNA) using a High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific, MA, USA). A list of all primers included in this array is described in Supplementary Table S1. The quantitative real-time PCR was conducted using SYBR Green dye (Qiagen, Hilden, Germany) and gene expression levels were measured using AriaMX System (Agilent, CA, USA).). PCR thermal profile was 95°C initial denaturation for 10 minutes, followed by 40 cycles at 94°C for 15 seconds and 60°C for 1 minute. The obtained CTs were normalized to the CTs of 18S, ACTB, B2M, GAPDH, and HPRT housekeeping genes and analyzed using Pfaffl’s method (45).
Proteomic composition of the secretome of HCMV- and HCMV+ TAMs were characterized quantitatively using RayBio™ human cytokine antibody array 3 (RayBiotech Life, GA, USA). Briefly, antibody array membranes were incubated in blocking buffer for 1 h and then incubated overnight at 4°C with 1 mL of the secretome of HCMV- and HCMV+ TAMs (2000 µg/ml). After washing, membranes were incubated with the biotinylated antibody cocktail for 2 h at room temperature with shaking, and then incubated with HRP-streptavidin at room temperature for 2 h. Finally, membranes were developed using the chemiluminescence detection reagent provided with the kit. The quantification of each cytokine was achieved by densitometry analysis using ImageJ software (National Institutes of Health, Bethesda, MD, USA) as we described before (46).
The SUM149 breast cancer cell line was provided by Prof. Schneider’s lab in the Department of Microbiology, New York University School of Medicine, USA, to be used in this study representing the IBC. SUM149 cells were cultured in HAM’s F12 medium supplemented with 5% fetal bovine serum (FBS), 5mM HEPES, 1µg/ml Hydrocortisone, 5µg/ml insulin, and 1% of penicillin/streptomycin antibiotic mixture. SUM149 cells were incubated in a humidified atmosphere at 37°C in 5% CO2. SUM149 cells were authenticated by STR profiling and routinely checked for mycoplasma contamination.
SUM149 cells were cultured in 6 well plates (3x105 cell/well) in 2 ml Ham/s F12 culture medium supplemented with 5% FBS and incubated for 48h in a humidified atmosphere at 37°C in 5% CO2. When cells reached 75-80% confluency, they were seeded in culture media containing 1% FBS and conditioned with concentrated secretome of HCMV- and HCMV+ TAMs (2000 ng/mL). Control SUM149 cells (Mock) were seeded in a complete culture media with 1% FBS. Cells were incubated for 72 h in a 5% CO2 humidified incubator at 37 °C and then culture media was discarded. Cells were washed twice with room temperature PBS and aliquoted to be stored at −80°C for RNA and protein extraction.
MTT assay was used to test the effect of the secretome of HCMV- and HCMV+ TAMs on the proliferation of SUM149 cells. Mock and stimulated SUM-149 cells were placed in a 96-well plate at a density of 4×103/well and were incubated for 48 hours under optimum conditions. After that, 20 μL (5 g/L) of MTT (3-(4,5-dimethyl-2-thiazolyl)- 2,5-diphenyl-2-H-tetrazolium bromide) reagent was added to the designated wells. After a 4 h incubation, the MTT formazan precipitate was dissolved in dimethylsulphoxide (DMSO) (150 μL/well, Sigma-Aldrich, St. Louis, MO, USA) in a shaker for 5 min before reading the absorbance at 570 nm using a 96-well plate reader (Bio-Rad, Winooski, VT USA) (47–49).
To test the effect of the secretome of HCMV- and HCMV+ TAMs (2000 ng/mL) on the invasive properties of SUM149 cells, we used Corning® BioCoat™ Matrigel® Invasion Chambers (Corning, MA, USA). According to the kit guidelines, cells that invade should be able to secrete proteases and degrade the Matrigel matrix-coat insert of the chamber. Thus, the invasion assay was conducted as described in the instruction guidelines of the kit. SUM149 cells were grown in the upper chamber with Ham’s F12 culture media 1% FBS in the lower chamber (control). The secretome of HCMV- and HCMV+ TAMs (2000 ng/ml) were added to the lower chamber. Invasion chambers were incubated in humidified CO2 incubator at 37°C for 24 h. The percentage of invasion was calculated by counting the number of invaded cells in response to the chemotactic factor and dividing them by the control and multiplying by 100 as described by the equation in the kit manual.
To test the effect of the TAM secretions prepared from HCMV- and HCMV+ on the clonogenic ability of SUM149 cells, 1000 mock and stimulated SUM-149 cells were seeded in six-well plates and maintained in Ham-F12 with 5% FBS for 10–14 days as previously performed (50). Cells were washed with PBS, fixed in methanol for 20 min, and stained with 0.05% crystal violet for 20 min and the excess stain was removed by water. The stained colonies were counted as described before (10, 51).
RT2 extracellular matrix and adhesion molecule PCR array (Qiagen, Hilden, Germany) was used to study the gene expression profiles of 84 extracellular matrix and cell adhesion molecules in SUM149 cells seeded in growth media conditioned by the secretome of HCMV- and HCMV+ TAMs. Briefly, PCR array was performed using AriaMX System (Agilent, CA, USA) in a 25 µL total volume using RT2 SYBR Green Master Mix (Qiagen, Hilden, Germany). PCR thermal profile was 95°C initial denaturation for 10 minutes, followed by 40 cycles at 94°C for 15 seconds and 60°C for 1 minute. Data were analyzed using the Qiagen Gene globe web tool (https://geneglobe.qiagen.com/analyze/) after normalization to ACTB, B2M, GAPDH, HPRT1, and RPLP0 housekeeping genes.
Based on previous studies (52–56), we carefully designed and customized BCSC-related gene PCR array including 43 genes (Vivantis, Selangor, Malaysia) to test the effect of the secretome of HCMV- and HCMV+ TAMs on the expression of BCSC-related genes in SUM149 cells. The quantitative real-time PCR was conducted using SYBR Green dye (Qiagen, Hilden, Germany) and gene expression levels were measured using AriaMX System (Agilent, CA, USA). A list of all primers included in this array is described in Supplementary Table S2. PCR thermal profile was 95°C initial denaturation for 10 minutes, followed by 40 cycles at 94°C for 15 seconds and 60°C for 1 minute. The obtained CTs were normalized to the CTs of 18S, ACTB, B2M, GAPDH, and HPRT housekeeping genes and analyzed using Pfaffl’s method (45).
GeneMANIA (http://genemania.org/) is an online tool that provides large information about protein-DNA, protein-protein, and genetic interactions, reactions, pathways, and protein domains of related genes. Herein, we used GeneMANIA to compare the correlation between DEGs in SUM149 cells stimulated by the secretome of HCMV+ TAMs and neighboring related genes. In addition, Metascape (http://metascape.org/gp/index.html#/main/step1) is a public online analysis database that was used for functional annotation and enrichment analysis based on Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Reactome, canonical pathways, and hallmark gene terms for differentially expressed genes (DEGs) in SUM149 cells stimulated by the secretome of HCMV+ TAMs. The analysis was based on the relevant parameters as follows: minimum overlap, 3; P-value cutoff, 0.01 and minimum enrichment 1.5. All protein-protein interactions among input genes were extracted from the PPI data source and formed a PPI network. GO enrichment analysis was applied to the network to extract “biological meanings”. The MCODE algorithm was then applied to this network to identify neighborhoods where proteins are densely connected (57–60).
PathScan® Intracellular Signaling Array Kit (Cell signaling technology, MA, USA) was used to study the effect of the secretome of HCMV- and HCMV+ TAMs on activating different signaling molecules in SUM149 cells. Mock and stimulated SUM149 cells’ lysates were diluted to 0.2 – 1.0 mg/mL using array diluent buffer before performing the Pathscan array. Pathscan glass slides were incubated in blocking buffer for 1h at room temperature with shaking. After washing, 50 - 75 μL of each cell lysate was added to each well and covered with sealing tape and incubated overnight at 4°C on an orbital shaker. After that, cell lysate was removed and the Pathscan glass slides were washed 3 successive times using the washing buffer provided with the kit. Detection Antibody Cocktail was added to each well and covered with sealing tape and incubated for 1 h at room temperature with shaking. After washing 3 times, 75 μL (1X) HRP-linked Streptavidin was added to each well and covered with sealing tape, and incubated for 30 minutes at room temperature with shaking. Subsequently, Pathscan glass slides were washed 3 successive times using washing buffer and covered with LumiGLO®/Peroxide reagent. Finally, Pathscan slides were transferred to the chemiluminescent development folder and images taken using ChemiDoc XRS+ Gel Imaging System (Bio-Rad, CA, USA). Relative density values were calculated via densitometric methods using ImageJ software (National Institutes of Health, MD, USA).
The Statistical Package of the Social Sciences software (SPSS, Chicago, IL, USA), version 22.0 was used for data analysis. The data were presented as the mean ± standard deviation (SD). In addition, differences among two groups of variables were evaluated using Student’s t-test and Chi-square test. The statistical difference between more than two groups was evaluated using one-way ANOVA followed by Tukey’s HSD Post Hoc tests. The level of significance was set at P < 0.05 (61–63).
Results of HCMV IgG serological assay of peripheral blood showed that 19 out of 28 IBC patients were HCMV IgG positive (Figure 1A). HCMV-DNA of the fourth exon of the HCMV IE gene was detected in all cancer tissues of HCMV seropositive IBC patients (Figure 1B). Moreover, HCMV-DNA was detected in the isolated TME monocytes of HCMV seropositive IBC patients (Figure 1B). In contrast, HCMV-DNA was not detected in the cancer tissues or the isolated TME derived monocytes of all HCMV seronegative IBC patients. However, it should be noted that we did not detect HCMV-DNA in 4 out of 19 (21%) TME derived monocytes isolated from HCMV seropositive IBC patients.
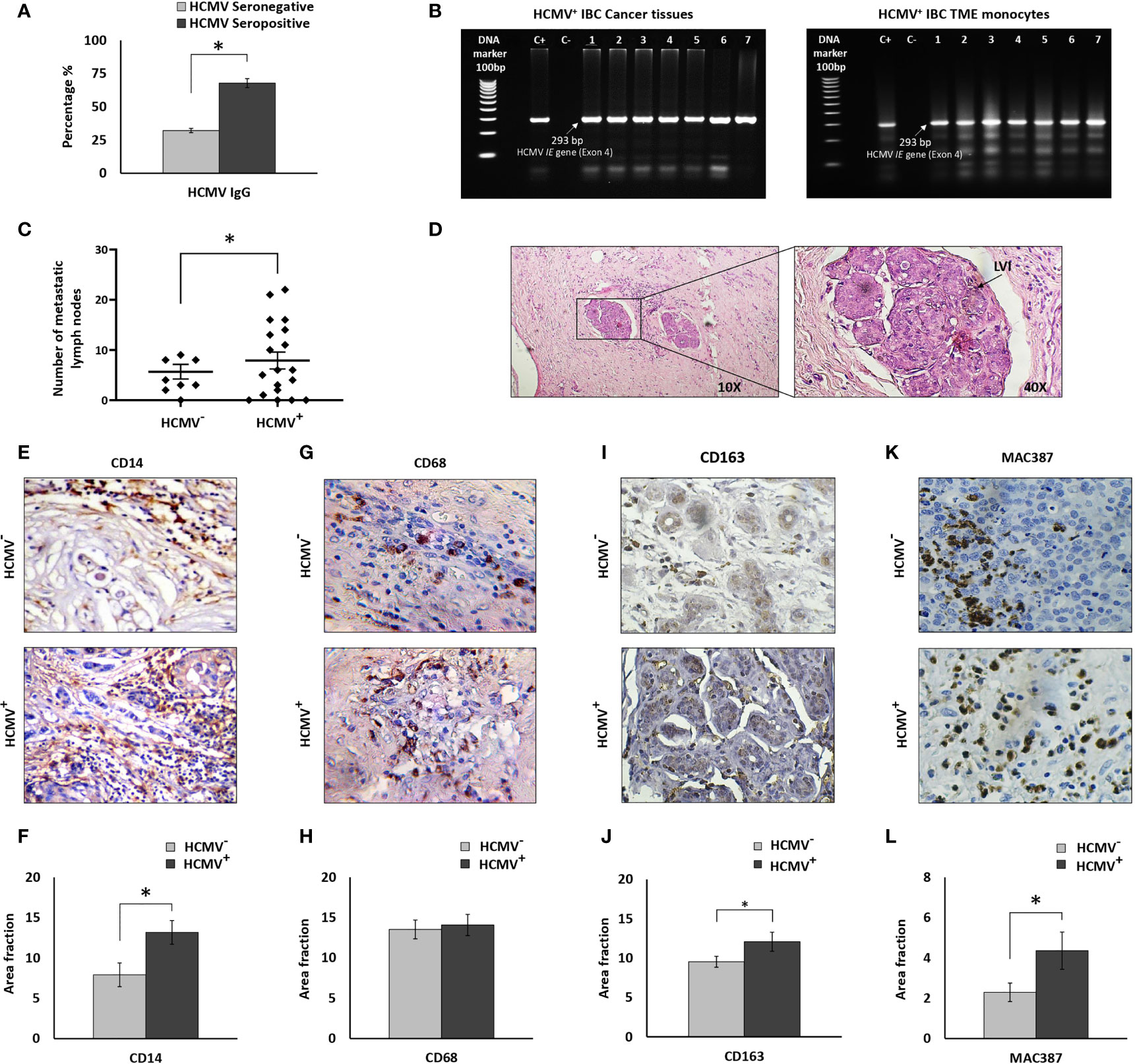
Figure 1 Prevalence of HCMV-DNA among IBC patients and IHC staining of IBC cancer tissues against CD14, CD68, CD163, and MAC387. (A) Bars represent prevalence of HCMV IgG among IBC patients. (B) Representative agarose gel electrophoresis showing amplicons of HCMV Immediate Early (IE) gene nested PCR (296 bp) among HCMV+ IBC cancer tissues and isolated TME CD14+ monocytes. (C) Scatter plot showing the number of metastatic lymph nodes among HCMV- versus HCMV+ IBC patients. (D) Microscopic images of H and E stained paraffin embedded tissue section of HCMV+ IBC patients showing invasion of cancer cells to lymphatic vessels. (E, F) Microscopic images of CD14 stained paraffin-embedded tissue sections in HCMV- versus HCMV+ IBC cancer tissues and bars showing a significantly high prevalence of CD14+ monocytes among HCMV+ compared to HCMV- IBC cancer tissues. (G, H) Microscopic images of CD14 stained paraffin-embedded tissue sections in HCMV- versus HCMV+ IBC cancer tissues. Bars show no significant difference in the prevalence of CD68+ TAMs among HCMV+ compared to HCMV- IBC cancer tissues. (I, J) Microscopic images of CD163 stained paraffin-embedded tissue sections in HCMV- versus HCMV+ IBC cancer tissues. Bars show a significantly high prevalence of CD163+ TAMs among HCMV+ compared to HCMV- IBC cancer tissues. (K, L) Microscopic images of MAC387 stained paraffin-embedded tissue sections in HCMV- versus HCMV+ IBC cancer tissues and bars showed a significantly high prevalence of MAC387+ TAMs among HCMV+ compared to HCMV- IBC cancer tissues. Data represented the mean of ± SD. P values were calculated using Student t-test, where * represented (P < 0.05).
Clinical and pathological characterizations of HCMV- and HCMV+ IBC patients are described in Table 1. Statistical analysis revealed that there are no significant differences in age, tumor size, tumor grade, status of lymph node metastasis, hormonal receptors, and receiving neoadjuvant chemotherapy between HCMV- and HCMV+ IBC patients. On the contrary, the number of metastatic lymph nodes and the presence of lymphovascular invasion in HCMV+ IBC patients were statistically significantly higher (P= 0.03 and 0.021, respectively) compared to HCMV- IBC patients (Figures 1C, D). This confirmed our previous results (8).
CD14+ monocytes were found to be a reservoir for HCMV latency (18) and infected macrophages serve as “mobile vectors” for HCMV spreading and dissemination to breast tissue (17). In addition, we previously found high infiltration of CD14+ monocytes in IBC cancer tissues compared to non-IBC (64). There is also an increased incidence of CD68+ and CD163+ TAMs associated with breast cancer progression (65–67), including MAC387+ macrophages that are associated with viral infection (22). Thus, we determined the incidence of CD14+ monocytes, and CD68+, CD163+ and MAC387+ TAMs in the cancer tissues of HCMV+ IBC patients. Statistical analysis of IHC imaging data showed a significant (P= 0.04) infiltration of CD14+ monocytes in HCMV+ IBC cancer tissues compared to the HCMV- tissues (Figures 1E, F). It was demonstrated that a high incidence of migratory TAMs correlates with aggressiveness and worse outcomes in breast cancer (68). We therefore determined the infiltration of CD68+ and MAC387+ TAMs in HCMV- and HCMV+ cancer tissues of IBC patients. Statistical analysis showed no significant differences in the infiltration of CD68+ TAMs (Figures 1G, H) in HCMV+ compared to HCMV- IBC cancer tissues, while HCMV+ IBC cancer tissues were characterized by significant high infiltration of CD163+ (Figures 1I, J) and MAC387+ TAMs (Figures 1K, L) (P= 0.48 and 0.04, respectively) compared to HCMV- IBC cancer tissues.
Microscopic examination showed that the secretome of SUM149 cells polarized isolated HCMV- and HCMV+ TME CD14+ monocytes towards M1/M2 TAMs (Figure 2A). The gene expression signature showed that the secretome of SUM149 cells activates the polarization of both HCMV- and HCMV+ TME CD14+ monocytes towards M1/M2 TAMs. Moreover, the mRNA level of IL-6, IL-8 and CD163 were significantly (P = 0.03, 0.03 and 0.04, respectively) more highly expressed in HCMV+ TAMs compared to HCMV- TAMs (Figure 2B).
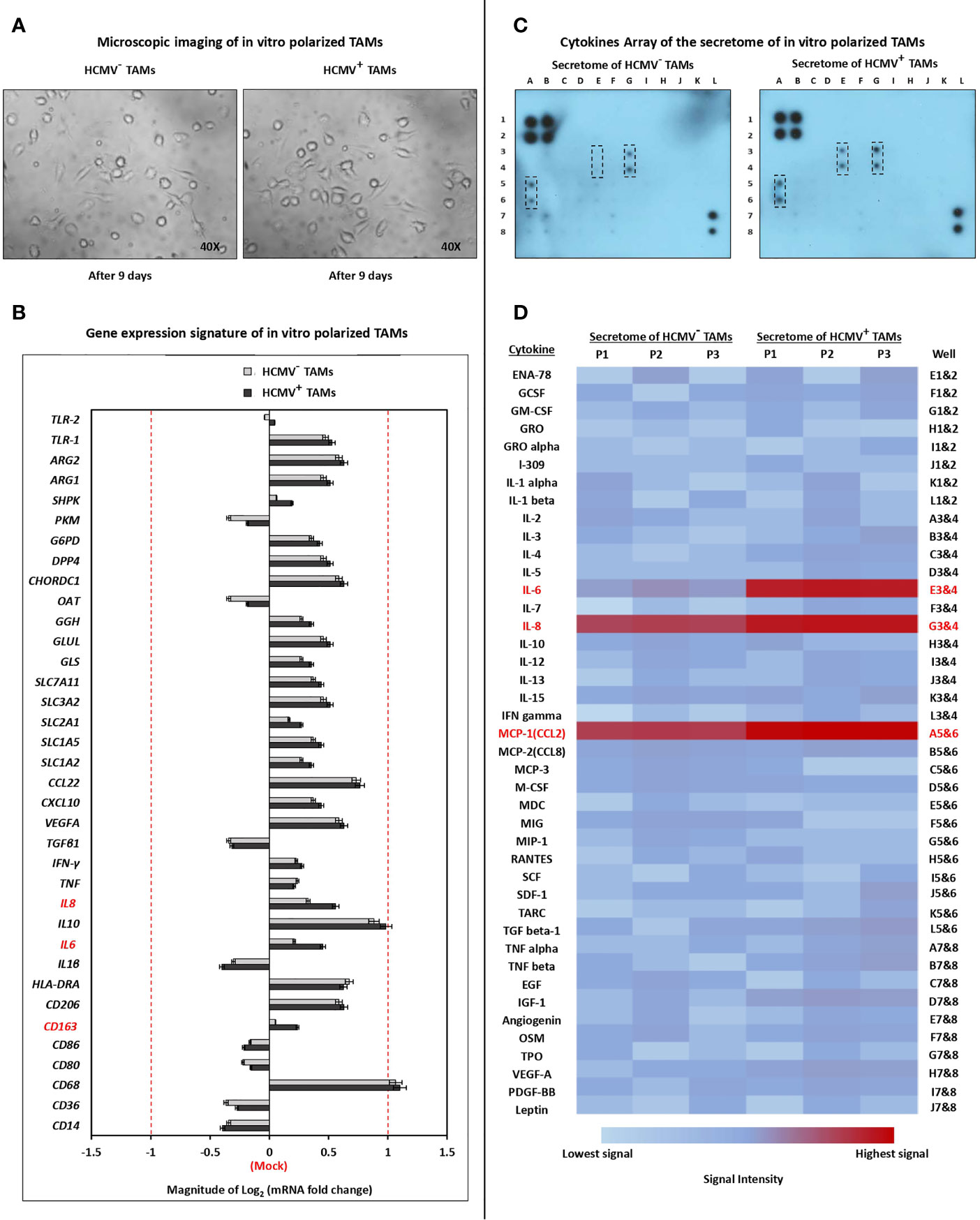
Figure 2 Characterization of polarized TAMs via microscopic examination, quantitative Real-time PCR, and cytokine array. (A) Microscopic images show polarization status of TME CD14+ monocytes isolated from HCMV- and HCMV+ IBC patients. (M0) were identified as small/roundish cells without cytoplasmic projections on the cell surface, (M1) were identified as enlarged amoeboid cells characterized by presence of many delicate cytoplasmic extensions on the cell surface with visible intracellular vacuoles, and (M2) were identified as a large and elongated “spindeloid” cells with cytoplasmic extensions on the apical ends of the cell bodies. (B) Clustered bars represent the mRNA expression of macrophage polarization-related genes in HCMV- and HCMV+ polarized TAMs compared to mock cells. (C) Membrane-based cytokine profiling array. Spots represent the signal intensity value of each cytokine, calculated using ImageJ software (NIH, Bethesda, MA, USA) and normalized according to an algorithm provided in the cytokine antibody array kit instruction manual. (D) Heat map showing expression levels of 42 cytokines in HCMV- versus HCM+ polarized TAMs. Genes and cytokines in red color are significantly highly expressed in HCMV+ TAMs compared to HCMV- TAMs. Data represented the mean of ± SD (number of experimental replicates = 3). P < 0.05 consider significant as calculated using Student t-test.
The human cytokine array analysis showed that the secretome of HCMV+ TAMs is characterized by significantly higher expression level of IL-6 (P= 0.01), IL-8 (P= 0.03), and MCP-1 (P= 0.04) compared to the secretome of HCMV- TAMs (Figures 2C, D).
MTT assay results showed a non-significant increase in the proliferation of SUM149 cells in the presence of the secretome of HCMV- TAMs, while showing a significant increase (P= 0.04) in the proliferation of SUM149 cells in the presence of the secretome of HCMV+ TAMs compared to control cells (Figure 3A). To test whether the secretome of HCMV- and HCMV+ TAMs promote the invasive properties of SUM149 cells via secretion of proteases, we used BD Matrigel invasion chambers. Invading cells are those that can degrade Matrigel via secretion of proteases and pass through the coated filters of Matrigel invasion chambers. Statistical analysis showed that the secretome of HCMV- and HCMV+ TAMs significantly (P= 0.04 and 0.01, respectively) enhanced the invasive properties of SUM149 cells by 155.3% and 208.5%, respectively compared to control SUM149 cells (Figure 3B). In addition, clonogenic assay results revealed a significant increase in the colony formation potential of SUM149 cells seeded in presence of the secretome of both HCMV- and HCMV+ TAMs (P= 0.04 and 0.03, respectively) compared to control cells. In contrast, there are no significant differences in the colony formation potential of SUM149 cells seeded in presence of the secretome of both HCMV- and HCMV+ TAMs (Figure 3C).
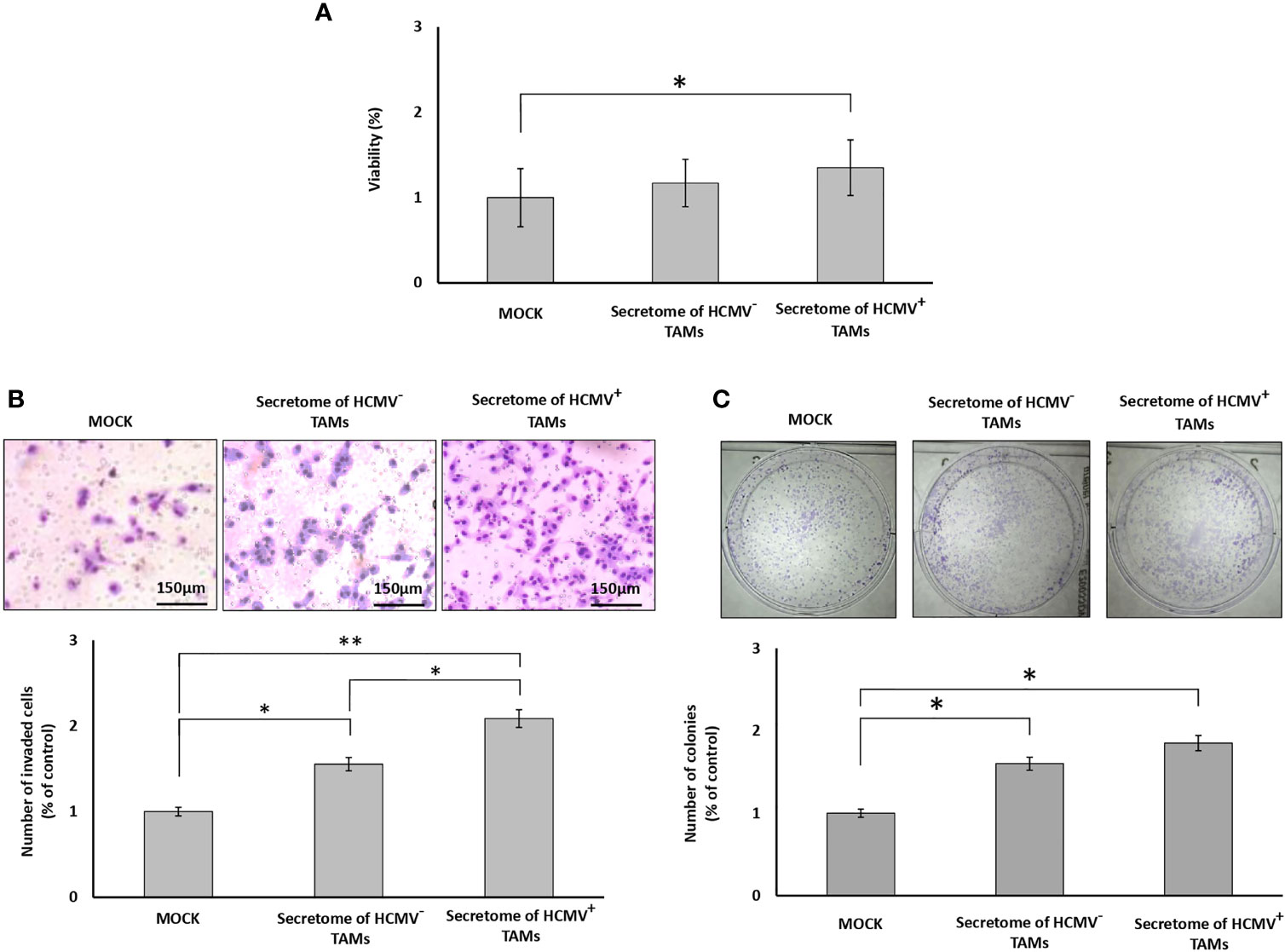
Figure 3 Secretome of HCMV+ TAMs enhance proliferation, invasion, and colony formation in SUM149 cells. (A) Bars show cell proliferation percentages of SUM149 cells stimulated by the secretome of HCMV- and HCMV+ compared to mock cells. (B) Microscopic images of SUM149 cells that invaded the Coat Matrigel-coated lower chamber in response to the secretome of HCMV- and HCMV+ TAMs compared to mock cells. Bars represent the percentage of invasive SUM149 cells in response to the secretome of HCMV- and HCMV+ TAMs compared to mock cells. (C) Wells showed the clonogenic ability of SUM149 cells in response to secretome of HCMV- and HCMV+ TAMs compared to mock cells. Bars represent the number of colonies of SUM149 cells in response to the secretome of HCMV- and HCMV+ TAMs compared to mock cells. Data represented the mean of ± SD (number of experimental replicates = 3). P values were as determined by one-way ANOVA test followed by Tukey’s HSD and Post Hoc tests, where * represented (P < 0.05) and ** represented (P < 0.01).
RT2 extracellular matrix (ECM) and cell adhesion (CA) mRNA PCR array results showed that the CA mRNAs CDH1, CNTN1, ITGA1, ITGA4, ITGA7, ITGAL, ITGB1, ITGB3, NCAM1, PECAM1, SELE, SELL and SGCE, and ECM proteins ADAMTS1, ADAMTS8, CLEC3B, COL11A1, COL7A1, CTGF, LAMA2, MMP1, MMP10, MMP13, MMP8, SPARC, TGFBI, and THBS2 were significant more highly expressed than other CA and ECM mRNAs in SUM149 cells stimulated by the secretome of HCMV+ TAMs, compared to SUM149 cells stimulated by the secretome of HCMV- TAMs. On the other hand, the CA mRNAs ITGA5 and those corresponding to ECM COL1A1, SPG7, LAMB3, and TIMP1 were significantly more reduced in expression than other CA and ECM mRNAs in SUM149 cells stimulated by the secretome of HCMV+ TAMs compared to SUM149 cells stimulated by the secretome of HCMV- TAMs. (Figures 4A–C).
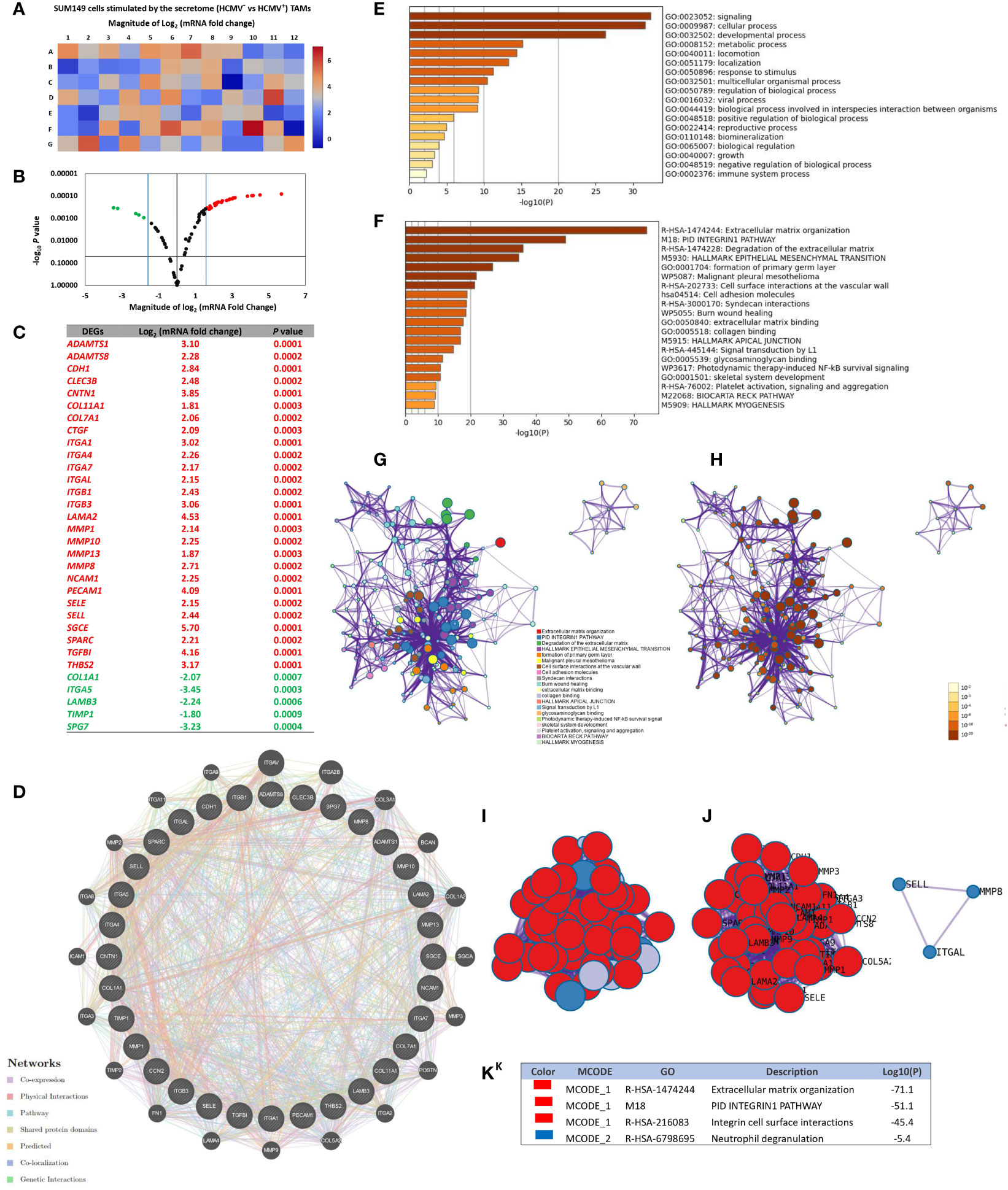
Figure 4 Up- and downregulated extracellular matrix and cell adhesion genes of SUM149 cells stimulated by the secretome of HCMV+ polarized TAMs versus HCMV+ polarized TAMs. (A, B) Heat map and volcano plot showing significant up- and downregulated genes in SUM149 cells stimulated by the secretome of HCMV+ polarized TAMs compared to HCMV+ polarized TAMs. (C) Table showing the Log2 mRNA fold change and significant P values of up- and downregulated genes in SUM149 cells stimulated by the secretome of HCMV+ polarized TAMs versus HCMV+ polarized TAMs. (D) Gene-gene interaction network of different expressed extracellular matrix and cell adhesion molecule genes and their neighboring genes. Each node represents a gene, and the size of which represents the strength of the interaction. (E) Bar graph of the top-level Gene Ontology biological processes, with P-values. (F) Bar graph of enriched terms across input gene lists, with P -values. (G, H) Network of enriched terms: colored by cluster ID, where nodes that share the same cluster ID are typically close to each other; colored by p-value, where terms containing more genes tend to have a more significant P-value. (I, J) Protein-protein interaction network and MCODE components identified in the gene list. (K) Table showed the functional analysis of MCODE1 and MCODE2 components.
Through GeneMANIA analysis, we identified the top 20 neighboring genes with the highest frequency association with differential expressed CA and ECM-related genes in SUM149 cells stimulated by the secretome of HCMV+ TAMs. These data indicate that ITGAV, ITGA2B, COL3A1, BCAN, COL1A2, SGCA, MMP3, POSTN, ITGA2, COL5A2, MMP9, LAMA4, FN1, TIMP2, ITGA3, ICAM1, ITGA8, MMP2, ITGA11, and ITGA9 were associated with the function and pathway of differential expressed CA and ECM -related genes (Figure 4D). The functions of these CA and ECM -related genes and their neighboring genes were predicted using Metascape. The top 18 GO enrichment items of these CA and ECM -related genes and their neighboring genes were described in (Figure 4E), which mainly included signaling, cellular process, locomotion, localization, and viral processes. Pathway enrichment analysis represented pathways associated with IBC progression and metastasis including extracellular matrix organization, PID Integrin-1 pathway, degradation of the extracellular matrix, hallmark epithelial mesenchymal transition and cell adhesion molecules, involved in IBC, the top 20 associated pathways are described in (Figure 4F) and Supplementary Table S3. Moreover, to better understand the relationship between CA and ECM -related genes and their neighboring genes, and IBC, we then performed a Metascape protein-protein interaction (PPI) enrichment analysis. The PPI network and MCODE components are shown in (Figures 4G–K). Data showed that the biological functions of CA and ECM-related genes and their neighboring genes are mainly enriched in formation of Extracellular matrix organization, PID Integrin1 pathway, Integrin cell surface interactions, and Neutrophil degranulation in IBC.
HCMV infection in the TME can induce stemness and an EMT program, leading to a potential increase in the progression of tumors including breast cancer (69–71). In addition, a higher incidence of CD133+/HCMV-IE+ cells is associated with poor patient survival (72). Herein, the BCSC-related gene PCR array results showed that the mRNA expression of 26 genes out of 43 genes was significantly (P < 0.05) differentially expressed (2-fold cut off) in both stimulated SUM149 cells compared to mock cells Supplementary Data (Figure S1).
More precisely, SUM149 cells stimulated by the secretome of HCMV+ TAMs were characterized by significantly (P < 0.05) higher mRNA expression (2-fold cut off) of POU5F1, NANOG, MSI1, SLC2A1, DPP4, CD44, CD133, CD29, CAIX, and ALDH1A1, while significantly lower expression of CD24 was found compared to SUM149 cells stimulated by the secretome of HCMV- TAMs (Figure 5A).
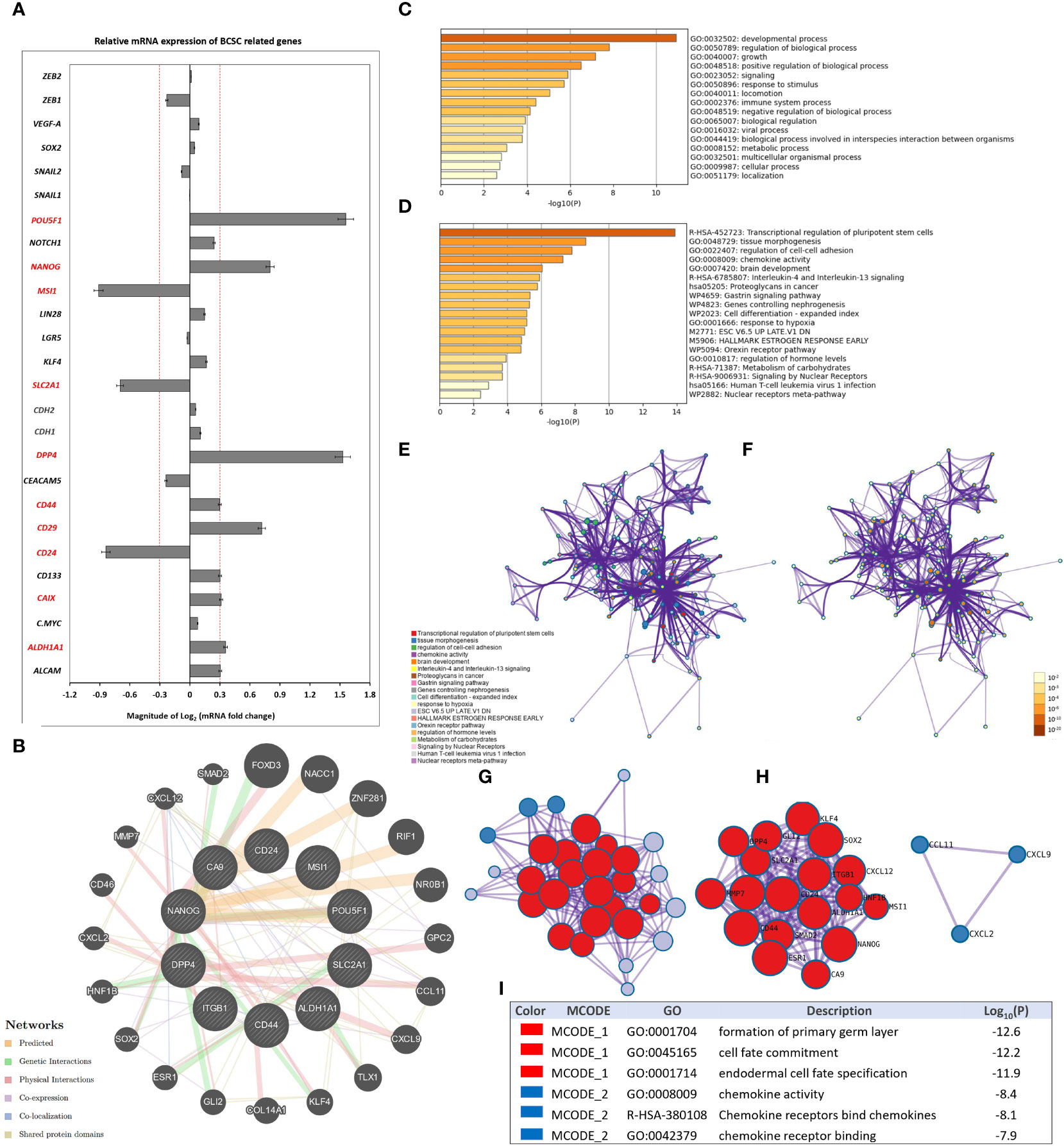
Figure 5 Secretome of HCMV+ TAMs enhance the expression of BCSC-related genes in SUM149 cells. (A) Clustered bars represent the mRNA expression of BCSC-related genes in SUM149 cells stimulated by the secretome of HCMV- and HCMV+ TAMs compared to mock cells. Data represent the mean of ± SD (number of experimental replicates = 3). P < 0.05 consider significant as calculated using Student t-test. (B) Gene-gene interaction network of upregulated BCSC-related genes and their neighboring genes. Each node represents a gene, and the size of which represents the strength of the interaction. (C) Bar graph of the top-level Gene Ontology biological processes, colored by P-values. (D) Bar graph of enriched terms across input gene lists, colored by P-values. (E, F) Network of enriched terms: firstly colored by cluster ID, where nodes that share the same cluster ID are typically close to each other; secondly colored by p-value, where terms containing more genes tend to have a more significant P-value. (G, H) Protein-protein interaction network and MCODE components identified in the gene list. (I) Table showed the functional analysis of MCODE1 and MCODE2 components.
GeneMANIA analysis identified the top 20 neighboring genes with the highest frequency association with differentially expressed BCSC-related genes in SUM149 cells stimulated by the secretome of HCMV+ TAMs. These data indicate that FOXD3, NACC1, ZNF281, RIF1, NR0B1, GPC2, CCL11, CXCL9, TLX1, KLF4, COL14A1, GLI2, ESR1, SOX2, HNF1B, CXCL2, CD46, MMP7, CXCL12, and SMAD2 were associated with the function and pathway of differentially expressed BCSC-related genes (Figure 5B). The functions of these BSCS genes and their neighboring genes were predicted using Metascape. The top 16 GO enrichment items of those upregulated BSCS-related genes and their neighboring genes described in (Figure 5C), which mainly included developmental process, regulation of biological process, signaling, response to stimulus, locomotion, immune system process, and viral process. Pathway enrichment analysis represented pathways associated with IBC progression and stemness including transcriptional regulation of pluripotent stem cells, tissue morphogenesis, chemokine activity, Interleukin-4 and Interleukin-13 signaling, and proteoglycans in cancer, the top 19 associated pathways are described in (Figure 5D) and Supplementary Table S4. Moreover, to better understand the relationship between differentially expressed BSCS-related genes with their neighboring genes, and IBC, we performed a Metascape protein-protein interaction (PPI) enrichment analysis. The PPI network and MCODE components are shown in (Figures 5E–I). Data showed that the biological functions of upregulated BSCS-related genes and their neighboring genes are mainly enriched in formation of primary germ layer, cell fate commitment, endodermal cell fate specification, chemokine activity, chemokine receptors bind chemokines, and chemokine receptor binding in IBC.
To further investigate whether the secretome of HCMV+ TAMs may alter intracellular signaling pathways of SUM149 cells, we used a PathScan Intracellular Signaling Array that detects 18 phosphorylated or cleaved signaling molecules per test. We measured changes in the expression of signaling molecules by SUM149 cells before and after stimulation by the secretome of HCMV- and HCMV+ TAMs (Figure 6A). Statistical analysis revealed that secretome of HCMV- and HCMV+ TAMs significantly upregulated p-STAT3 (P= 0.03 and 0.01, respectively), p-AMPKα (P= 0.04 and 0.03, respectively), p-PRAS40 (P= 0.03 and 0.04, respectively) and p-SAPK/JNK (P= 0.04 and 0.03, respectively) signaling molecules compared to control SUM149 cells. Moreover, statistical analysis revealed that secretome of HCMV+ TAMs significantly upregulated p-STAT3 (P= 0.04) in stimulated SUM149 cells compared to secretome of HCMV- TAMs (Figures 6B, C).
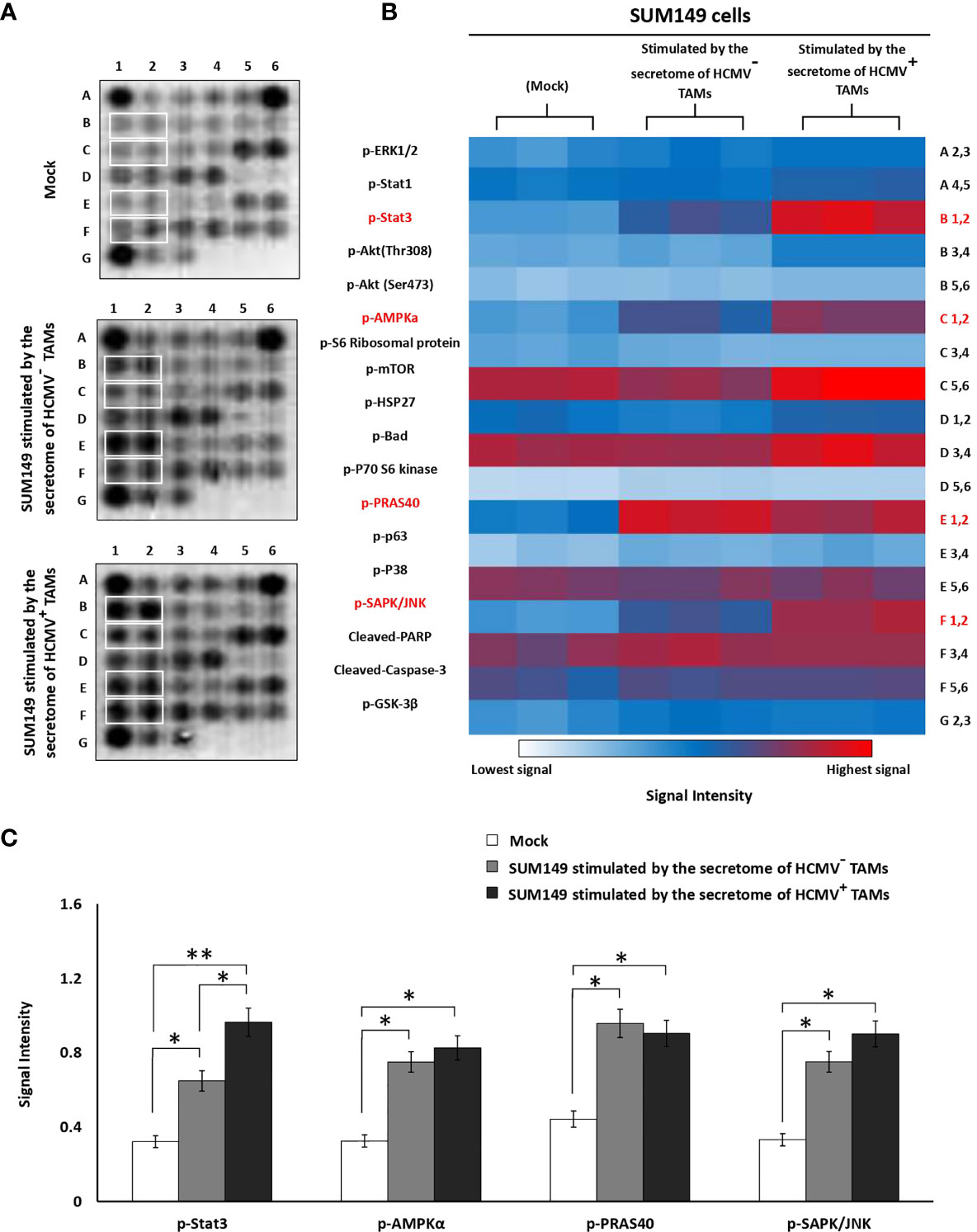
Figure 6 Intracellular pathways array analysis of SUM149 cells stimulated by the secretome of HCMV- and HCMV+ TAMs compared to mock cells. (A) PathScan® intracellular array membranes of SUM149 cells stimulated by the secretome of HCMV- and HCMV+ TAMs compared to mock cells. (B) Heat map showing the different activation levels of eighteen signaling molecules in SUM149 cells stimulated by the secretome of HCMV- and HCMV+ TAMs compared to mock cells. (C) Bars represent a high expression of p-STAT3, p-AMPKa, p-PRAS40, and p-SAPK/JNK intracellular cell signaling molecules in SUM149 cells stimulated by the secretome of HCMV- and HCMV+ TAMs compared to mock cells. * Represented (P < 0.05) and ** represented (P < 0.01) as determined by one-way ANOVA test followed by Tukey’s HSD and Post Hoc tests. Data represented as means ± SD (number of experimental replicates = 3).
The DNA and proteins of several onco-modulatory viruses have been found in breast cancers and associated with the disease initiation process although causality has not been established (73). HCMV is an onco-modulatory virus was found in more than 90% of primary and metastatic breast cancers (71). In addition, increased expression of HCMV proteins in breast cancer tissues was correlated with more aggressive breast cancer phenotypes (14, 71, 74, 75). HCMV is also known to promote a cellular secretome of several immunosuppressive or tumor-promoting mediators that affect the tumor microenvironment (76). These include the expression of transforming growth factor β (TGF-β) and IL-10 by T regulatory (Treg) cells, which inhibit the functions of natural killer (NK) cells and T cells and promote the EMT (77, 78). Furthermore, HCMV produces its immunosuppressive protein cmvIL-10, encoded by the UL111A gene, which induces maturation of pro-tumoral M2 macrophages and expression of the proto-oncogene Bcl-3 (77). cmvIL-10, UL37/vMIA, pUL144 which is a tumor necrosis factor (TNF) receptor homolog, and pUL128, a CC-like chemokine encoded by HCMV, modulate monocyte activity (79). In glioblastoma, HCMV infection confers glioblastoma stem cell (GSC) properties (80). The interplay of cancer cell stemness with commonly detected HCMV infection is poorly understood in medulloblastomas, where HCMV infection induces stem cell properties in the TMA (81). In this regard, Chan and colleagues suggested that HCMV stimulates monocyte reprogramming into M1/M2 macrophage phenotype via activation of NF-κB and P13K signaling pathway (82).
HCMV induces STAT3, TGF-β, IL-10 and cmvIL-10 in polarized M2 macrophages, leading to an immunosuppressive phenotype that is closely associated with TAMs (77, 82). Thus, HCMV infection of breast epithelial cells may contribute to neoplastic transformation and differentiation of M2-macrophages into the same tissue compartment. In vitro studies using different cancer cell lines revealed that HCMV infection in the TME may induce stemness and EMT in cancer cells, leading to increased cancer aggressiveness (72). Of note, EMT has a complex association with increased cancer stem cell properties that facilitates cancer cell motility and invasion (83). Thus, our results agree with Zhu and colleagues indicating that HCMV infection can induce EMT and stem cell properties of cancer cells (glioblastoma), in turn promoting increased cancer cell invasion and dissemination, through a mechanism that involves increased activity of JNK pathway (84). Moreover, latent CMV infection in a mouse xenograft tumor model, showed an increase in cell proliferation in number of lung metastatic nodules (69). In the spontaneous HCMV mouse glioblastoma model, the immediate-early (IE) proteins also augment stem cell properties in cancer cells (85).
There are therefore a number of ways in which HCMV can promote greater malignancy and BCSC activity. In breast cancer, HCMV+ TAMs secrete CCL18, which promotes the EMT of breast cancer cells, leading to increased metastasis (73, 86). Our previous studies showed an association between the incidence of HCMV DNA and IBC poor prognosis (7–9), but the potential onco-modulatory effect of HCMV in IBC is still not clearly understood. Here, we determined the prevalence of CD163+ and MAC387+ TAMs in the cancer tissues of HCMV+ compared to HCMV- IBC patients. In addition, we tested the effect of the secretome of HCMV+ TAMs compared to HCMV- TAMs on the proliferation, invasion, colony formation, and expression of BCSC-related genes in IBC cell line SUM149.
Previously, we found that CD14+ cells highly infiltrate the tumor microenvironment of IBC compared to non-IBC patients (9). In our present study, IHC analysis showed a significantly high infiltration of CD14+ monocytes in HCMV+ IBC cancer tissues compared to the HCMV- tissues. During HCMV primary infection, HCMV+ monocytes migrate into tissues and differentiate into macrophages (87). HCMV-infected macrophages generate an appropriate microenvironment for virus replication, and these macrophages have a high survival rate (87, 88). Infected macrophages serve as “mobile vectors” for virus spreading and dissemination to different organs mainly by transendothelial migration (17). HCMV-infected macrophages show classical macrophage markers but less phagocytic and immunogenic capacity (89). Furthermore, HCMV+ IBC cancer tissues are characterized by significantly high infiltration of CD163+ and MAC387+ TAMs in comparison with HCMV- IBC cancer tissues. High infiltration of CD163+ TAMs are associated with poor survival in breast cancer patients (90). Patients with CD163+ cancer cells are characterized by shorter disease-free survival after radiotherapy (91). CD163+ cancer cells were suggested to be caused via fusion between TAMs and cancer cells (92, 93). It was demonstrated that a high incidence of migratory MAC387+ TAMs correlated with aggressiveness and worse outcomes in breast cancer (68). MAC387+ TAMs were associated with high-grade, HR-negative breast cancers and early recurrence (68, 94, 95). It was demonstrated that MAC387 was expressed by a subset of TAMs as well as by a subset of cancer cells as a result of the fusion between TAMs and cancer cells leading to increase disease aggressiveness (96–98). HCMV infects macrophages and induces M1/M2 phenotype, close to the TAMs, which is associated with breast cancer poor prognosis (15). For instance, HCMV-DB clinical isolates trigger M2 polarization, and upregulates the proto-oncogene Bcl-3 (99). Our gene expression signature analysis showed that the secretome of SUM149 cells activates the polarization of both HCMV- and HCMV+ TME CD14+ monocytes towards TAMs. Moreover, the mRNA levels of IL-6, IL-8, and CD163 were significant highly expressed in HCMV+ TAMs compared to HCMV- TAMs. In addition, our human cytokine array analysis showed that the secretome of HCMV+ TAMs is characterized by the significantly higher expression level of IL-6, IL-8, and MCP-1/CCL2 compared to the secretome of HCMV- TAMs. IL-6 and IL-8 are cytokines associated with BCSCs and treatment resistance in breast cancer patients (100). Benner and colleagues demonstrated that TAMs stimulated by conditioned media of MDA-MB-231 cells displayed an increase in the co-expression of CD163/CD206 compared to in vitro M2- like macrophages (101). Indeed, these generated in vitro TAMs exhibited high transcriptional levels of IL-6, IL-10, CCL2, c-Myc, iNOS, and arginase compared to in vitro M2-like macrophages (101). Reinartz and colleagues showed that TAMs significantly express high M2 marker mRNA levels of CD163, IL10, and VEGFA compared to monocyte-derived macrophages (MDMs). In contrast, TAMs significantly express high M2 marker mRNA levels of IL27RA, CCL18, CCL22 and MMP9 compared to MDMs. In addition, TAMs significantly express high levels of several M1 markers such as CD86, CCR2, and TNF relative to MDMs (102). Our results showed that the secretome of HCMV+ TAMs enhances cell proliferation, invasion, colony formation, and alters the expression of many CA and ECM mRNAs. In agreement with our results, it was demonstrated that TAMs have potential roles in cancer cell proliferation (103), invasion (104), angiogenesis (105), and metastasis (106). EGF, FGFs, and VEGFs secreted by TAMs promote cancer cell proliferation, and angiogenesis (107, 108).
HCMV infection enhances cell proliferation, migration, and upregulation of EMT markers in colorectal cancer-derived stem cell-like cells (109). In line with our findings, HCMV infection in the TME can induce a cancer cell EMT program, resulting in increasing the aggressiveness of cancers (72). Fiallos and colleagues revealed that long-term HCMV infection promotes cell proliferation in glioma stem-like cells (GSC) (110).Moreover, cmvIL-10 facilitates cell migration and invasion via upregulation of both urokinase plasminogen receptor (uPAR) and plasminogen activator inhibitor-1 (PAI-1), which can stimulate MMP-3 activity in MDA-MB-231 cells (111). Moreover, HCMV infection alters the MMP-9/TIMP-1 balance in macrophages through immediate-early (IE) gene and late viral gene expression (112). Herein, SUM149 cells stimulated by the secretome of HCMV+ TAMs are characterized by the significantly high mRNA expression level of 10 BCSC-related genes as described in (Figure 5A) compared to SUM149 cells that are stimulated by the secretome of HCMV¯ TAMs.
We used gene enrichment analysis to identify the neighboring genes and the main functions and pathways of the differential expressed CA and ECM molecules and BCSC-related genes. The pathway enrichment and PPI network analysis of these differential expressed genes and their neighboring genes showed strong association mainly with degradation of the extracellular matrix, hallmark epithelial mesenchymal transition and cell adhesion molecules, transcriptional regulation of pluripotent stem cells, tissue morphogenesis, chemokine activity, Interleukin-4 and Interleukin-13 signaling, and proteoglycans in cancer. In agreement with previous studies (113–119), this bioinformatic analysis support our results which indicate a strong association between incidence of HCMV+ TAMs and IBC stemness properties and progression.
The results of PathScan Intracellular Signaling Array revealed that the secretome of HCMV+ TAMs significantly activated p-STAT3, p-AMPKα, p-PRAS40, and p-SAPK/JNK intracellular signaling molecules in SUM149 cells compared to mock cells. Furthermore, p-STAT3 significantly upregulated in SUM149 cells stimulated by the secretome of HCMV+ TAMs compared to SUM149 cells stimulated by the secretome of HCMV- TAMs. Polyak and colleagues showed that in samples from IBC patients 40% of CD44+/CD24- cells were positive for p-STAT3. In addition, they demonstrated that the inhibition of JAK2 decreases the proliferation rate of p-STAT3+ IBC cells in vitro and in vivo (120). AMP-activated protein kinase (AMPK) promotes EMT in breast cancer cells via Twist1 upregulation (121). Phospho-PRAS40 upregulates and activates EMT-related factors to induce metastasis via the phosphorylation of smad (122). JNK activity promotes invasion, and EMT in breast cancer cells via ERK activation (123). IL-6 and IL-8 secreted from adjacent cells in the TME such as TAMs activate the STAT3 signaling pathway in breast cancer cells (124). It is demonstrated that IL-6 is significantly expressed in IBC compared to non-IBC patients (125, 126). It was found that M2 macrophage-educated mesenchymal stem/stromal cells highly expressed IL-6 and increased IBC cell invasion and mammosphere formation (127). Moreover, a decrease in the infiltration of TAMs downregulates p-STAT3 and IL-6 secretion within the IBC TME, leading to reduced skin invasion and local recurrence (127). IBC cells highly expressed IL8 and growth-regulated oncogene (GRO) chemokines that activate the STAT3 signaling pathway, which promotes the EMT program. In addition, IBC cells attract and differentiate monocytes into TAMs, which were found to secrete high levels of IL8 and GRO chemokines that further promote the IBC EMT program (115).
The secretome of HCMV+ TAMs increases the proliferation, invasion, colony formation, and increases expression of CA and ECM mRNAs and BCSC-related genes compared to mock cells by acting on p-STAT3, p-AMPKα, p-PRAS40, and p-SAPK/JNK intracellular signaling molecules. Our results highlight the critical onco-modulatory role of HCMV infection in IBC. Further studies are necessary to begin to establish whether there is causality in these associations, and to identify the clinical significance of our findings. A potential combination therapy would be to use anti-HCMV drugs with chemotherapy to improve the effectiveness of chemotherapy and decrease the resistance of cancer cells.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The study protocol was reviewed and approved by the Institutional Review Board (IRB#00006379), Faculty of Medicine, Ain Shams University, Egypt. Before participation, all patients signed written informed consent forms, including approval for publication of the study results.
RJS and MMM applied for funding and directed the project. HTM and MMM suggested the idea and designed the research strategy and experimental protocols. Surgeon MES was responsible for enrolling patients. HTM collected patients’ clinical and pathological data. HTM and AAE conducted all practical experiments of the study. HTM analyzed the data using Statistical Package of the Social Sciences and biomedical informatics software. HTM, RJS, and MMM drafted and wrote the manuscript with the input of all co-authors.
The study was supported by Avon Foundation Grants # 02-2009-085 a and b (RJS and MMM) and the Cairo University Scientific Research Sector (MMM), and Breast Cancer Research Foundation (BCRF 18-146) (RJS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This work was conducted in the Cancer Biology Research Laboratory, Faculty of Science, Cairo University, Egypt. Special thanks to Egyptian breast cancer patients who participated in the present study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.899622/full#supplementary-material
1. Soliman AS, Banerjee M, Lo AC, Ismail K, Hablas A, Seifeldin IA, et al. High Proportion of Inflammatory Breast Cancer in the Population-Based Cancer Registry of Gharbiah, Egypt. Breast J (2009) 15(4):432–4. doi: 10.1111/j.1524-4741.2009.00755.x
2. Allen SG, Chen YC, Madden JM, Fournier CL, Altemus MA, Hiziroglu AB, et al. Macrophages Enhance Migration in Inflammatory Breast Cancer Cells via RhoC GTPase Signaling. Sci Rep (2016) 6:39190. doi: 10.1038/srep39190
3. Mohamed MM, El-Ghonaimy EA, Nouh MA, Schneider RJ, Sloane BF, El-Shinawi M. Cytokines Secreted by Macrophages Isolated From Tumor Microenvironment of Inflammatory Breast Cancer Patients Possess Chemotactic Properties. Int J Biochem Cell Biol (2014) 46:138–47. doi: 10.1016/j.biocel.2013.11.015
4. Choi J, Gyamfi J, Jang H, Koo JS. The Role of Tumor-Associated Macrophage in Breast Cancer Biology. Histol Histopathol (2018) 33(2):133–45. doi: 10.14670/hh-11-916
5. Aretz P, Maciaczyk D, Yusuf S, Sorg RV, Hänggi D, Liu H, et al. Crosstalk Between β-Catenin and CCL2 Drives Migration of Monocytes Towards Glioblastoma Cells. Int J Mol Sci (2022) 23(9):1–16. doi: 10.3390/ijms23094562
6. Aramini B, Masciale V, Grisendi G, Banchelli F, D'Amico R, Maiorana A, et al. Cancer Stem Cells and Macrophages: Molecular Connections and Future Perspectives Against Cancer. Oncotarget (2021) 12(3):230–50. doi: 10.18632/oncotarget.27870
7. El-Shinawi M, Mohamed HT, Abdel-Fattah HH, Ibrahim SA, El-Halawany MS, Nouh MA, et al. Inflammatory and Non-Inflammatory Breast Cancer: A Potential Role for Detection of Multiple Viral DNAs in Disease Progression. Ann Surg Oncol (2016) 23(2):494–502. doi: 10.1245/s10434-015-4888-2
8. El-Shinawi M, Mohamed HT, El-Ghonaimy EA, Tantawy M, Younis A, Schneider RJ, et al. Human Cytomegalovirus Infection Enhances NF-Kappab/P65 Signaling in Inflammatory Breast Cancer Patients. PLoS One (2013) 8(2):e55755. doi: 10.1371/journal.pone.0055755
9. Mohamed HT, El-Shinawi M, Nouh MA, Bashtar AR, Elsayed ET, Schneider RJ, et al. Inflammatory Breast Cancer: High Incidence of Detection of Mixed Human Cytomegalovirus Genotypes Associated With Disease Pathogenesis. Front Oncol (2014) 4:246. doi: 10.3389/fonc.2014.00246
10. Ibrahim SA, Gadalla R, El-Ghonaimy EA, Samir O, Mohamed HT, Hassan H, et al. Syndecan-1 is a Novel Molecular Marker for Triple Negative Inflammatory Breast Cancer and Modulates the Cancer Stem Cell Phenotype via the IL-6/STAT3, Notch and EGFR Signaling Pathways. Mol Cancer (2017) 16(1):57. doi: 10.1186/s12943-017-0621-z
11. Akhter J, Ali Aziz MA, Al Ajlan A, Tulbah A, Akhtar M. Breast Cancer: Is There a Viral Connection? Adv Anat Pathol (2014) 21(5):373–81. doi: 10.1097/PAP.0000000000000037
12. Harkins L, Volk AL, Samanta M, Mikolaenko I, Britt WJ, Bland KI, et al. Specific Localisation of Human Cytomegalovirus Nucleic Acids and Proteins in Human Colorectal Cancer. Lancet (2002) 360(9345):1557–63. doi: 10.1016/S0140-6736(02)11524-8
13. dos Santos CJ, Stangherlin LM, Figueiredo EG, Correa C, Teixeira MJ, da Silva MC. High Prevalence of HCMV and Viral Load in Tumor Tissues and Peripheral Blood of Glioblastoma Multiforme Patients. J Med Virol (2014) 86(11):1953–61. doi: 10.1002/jmv.23820
14. Taher C, de Boniface J, Mohammad AA, Religa P, Hartman J, Yaiw KC, et al. High Prevalence of Human Cytomegalovirus Proteins and Nucleic Acids in Primary Breast Cancer and Metastatic Sentinel Lymph Nodes. PLoS One (2013) 8(2):e56795. doi: 10.1371/journal.pone.0056795
15. Herbein G, Kumar A. The Oncogenic Potential of Human Cytomegalovirus and Breast Cancer. Front Oncol (2014) 4:230. doi: 10.3389/fonc.2014.00230
16. Taylor-Wiedeman J, Sissons JG, Borysiewicz LK, Sinclair JH. Monocytes are a Major Site of Persistence of Human Cytomegalovirus in Peripheral Blood Mononuclear Cells. J Gen Virol (1991) 72(Pt 9):2059–64. doi: 10.1099/0022-1317-72-9-2059
17. Smith MS, Bentz GL, Alexander JS, Yurochko AD. Human Cytomegalovirus Induces Monocyte Differentiation and Migration as a Strategy for Dissemination and Persistence. J Virol (2004) 78(9):4444–53. doi: 10.1128/JVI.78.9.4444-4453.2004
18. Hargett D, Shenk TE. Experimental Human Cytomegalovirus Latency in CD14+ Monocytes. Proc Natl Acad Sci U S A (2010) 107(46):20039–44. doi: 10.1073/pnas.1014509107
19. Smith PD, Shimamura M, Musgrove LC, Dennis EA, Bimczok D, Novak L, et al. Cytomegalovirus Enhances Macrophage TLR Expression and MyD88-Mediated Signal Transduction to Potentiate Inducible Inflammatory Responses. J Immunol (2014) 193(11):5604–12. doi: 10.4049/jimmunol.1302608
20. Van Laere SJ, Van den Eynden GG, van der Auwera I, Vandenberghe M, van Dam P, Van Marck EA, et al. Identification of Cell-of-Origin Breast Tumor Subtypes in Inflammatory Breast Cancer by Gene Expression Profiling. Breast Cancer Res Treat (2006) 95(3):243–55. doi: 10.1007/s10549-005-9015-9
21. Morrow RJ, Etemadi N, Yeo B, Ernst M. Challenging a Misnomer? The Role of Inflammatory Pathways in Inflammatory Breast Cancer. Mediators Inflammation (2017) 2017:4754827. doi: 10.1155/2017/4754827
22. Soulas C, Conerly C, Kim WK, Burdo TH, Alvarez X, Lackner AA, et al. Recently Infiltrating MAC387(+) Monocytes/Macrophages a Third Macrophage Population Involved in SIV and HIV Encephalitic Lesion Formation. Am J Pathol (2011) 178(5):2121–35. doi: 10.1016/j.ajpath.2011.01.023
23. Subimerb C, Pinlaor S, Khuntikeo N, Leelayuwat C, Morris A, McGrath MS, et al. Tissue Invasive Macrophage Density is Correlated With Prognosis in Cholangiocarcinoma. Mol Med Rep (2010) 3(4):597–605. doi: 10.3892/mmr_00000303
24. Zheng Q, Tao R, Gao H, Xu J, Shang S, Zhao N. HCMV-Encoded UL128 Enhances TNF-Alpha and IL-6 Expression and Promotes PBMC Proliferation Through the MAPK/ERK Pathway In Vitro. Viral Immunol (2012) 25(2):98–105. doi: 10.1089/vim.2011.0064
25. Scheller J, Rose-John S. Interleukin-6 and its Receptor: From Bench to Bedside. Med Microbiol Immunol (2006) 195(4):173–83. doi: 10.1007/s00430-006-0019-9
26. Hernandez-Vargas H, Ouzounova M, Le Calvez-Kelm F, Lambert MP, McKay-Chopin S, Tavtigian SV, et al. Methylome Analysis Reveals Jak-STAT Pathway Deregulation in Putative Breast Cancer Stem Cells. Epigenetics (2011) 6(4):428–39. doi: 10.4161/epi.6.4.14515
27. Andreoni M, Faircloth M, Vugler L, Britt WJ. A Rapid Microneutralization Assay for the Measurement of Neutralizing Antibody Reactive With Human Cytomegalovirus. J Virol Methods (1989) 23(2):157–67. doi: 10.1016/0166-0934(89)90129-8
28. Fearon WF, Potena L, Hirohata A, Sakurai R, Yamasaki M, Luikart H, et al. Changes in Coronary Arterial Dimensions Early After Cardiac Transplantation. Transplantation (2007) 83(6):700–5. doi: 10.1097/01.tp.0000256335.84363.9b
29. Shimamura M, Murphy-Ullrich JE, Britt WJ. Human Cytomegalovirus Induces TGF-β1 Activation in Renal Tubular Epithelial Cells After Epithelial-to-Mesenchymal Transition. PLoS Pathog (2010) 6(11):e1001170. doi: 10.1371/journal.ppat.1001170
30. Nouh MA, Mohamed MM, El-Shinawi M, Shaalan MA, Cavallo-Medved D, Khaled HM, et al. Cathepsin B: A Potential Prognostic Marker for Inflammatory Breast Cancer. J Transl Med (2011) 9:1. doi: 10.1186/1479-5876-9-1
31. El-Shinawi M, Mohamed HT, El-Ghonaimy EA, Tantawy M, Younis A, Schneider RJ, et al. Human Cytomegalovirus Infection Enhances NF-κb/P65 Signaling in Inflammatory Breast Cancer Patients. PLoS One (2013) 8(2):e55755. doi: 10.1371/journal.pone.0055755
32. El-Shinawi M, Abdelwahab SF, Sobhy M, Nouh MA, Sloane BF, Mohamed MM. Capturing and Characterizing Immune Cells From Breast Tumor Microenvironment: An Innovative Surgical Approach. Ann Surg Oncol (2010) 17(10):2677–84. doi: 10.1245/s10434-010-1029-9
33. Zhang S, Zhou YH, Li L, Hu Y. Monitoring Human Cytomegalovirus Infection With Nested PCR: Comparison of Positive Rates in Plasma and Leukocytes and With Quantitative PCR. Virol J (2010) 7:73 p 1–7. doi: 10.1186/1743-422x-7-73
34. Mohamed MM, Sabet S, Peng DF, Nouh MA, El-Shinawi M, El-Rifai W. Promoter Hypermethylation and Suppression of Glutathione Peroxidase 3 are Associated With Inflammatory Breast Carcinogenesis. Oxid Med Cell Longev (2014) 2014:787195. doi: 10.1155/2014/787195
35. Heinrich F, Lehmbecker A, Raddatz BB, Kegler K, Tipold A, Stein VM, et al. Morphologic, Phenotypic, and Transcriptomic Characterization of Classically and Alternatively Activated Canine Blood-Derived Macrophages In Vitro. PLoS One (2017) 12(8):e0183572–e0183572. doi: 10.1371/journal.pone.0183572
36. Shi J, Li Q, Sheng M, Zheng M, Yu M, Zhang L. The Role of TLR4 in M1 Macrophage-Induced Epithelial-Mesenchymal Transition of Peritoneal Mesothelial Cells. Cell Physiol Biochem (2016) 40(6):1538–48. doi: 10.1159/000453204
37. Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS–) vs. Alternatively Activated Macrophages. Front Immunol. (2019) 10(1):14. doi: 10.3389/fimmu.2019.01084
38. Eissa N, Hussein H, Ghia JE. A Gene Expression Analysis of M1 and M2 Polarized Macrophages. Methods Mol Biol (2020) 2184:131–44. doi: 10.1007/978-1-0716-0802-9_10
39. Unuvar Purcu D, Korkmaz A, Gunalp S, Helvaci DG, Erdal Y, Dogan Y, et al. Effect of Stimulation Time on the Expression of Human Macrophage Polarization Markers. PLoS One (2022) 17(3):e0265196. doi: 10.1371/journal.pone.0265196
40. Murray PJ. Macrophage Polarization. Annual Review of Physiology (2017) 79(1):541–66. doi: 10.1146/annurev-physiol-022516-034339
41. Palma A, Jarrah AS, Tieri P, Cesareni G, Castiglione F. Gene Regulatory Network Modeling of Macrophage Differentiation Corroborates the Continuum Hypothesis of Polarization States. Front Physiol (2018) 9:1659. doi: 10.3389/fphys.2018.01659
42. Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional Profiling of the Human Monocyte-to-Macrophage Differentiation and Polarization: New Molecules and Patterns of Gene Expression. J Immunol (2006) 177(10):7303–11. doi: 10.4049/jimmunol.177.10.7303
43. Zhao Y, Li M, Yang Y, Wu T, Huang Q, Wu Q, et al. Identification of Macrophage Polarization-Related Genes as Biomarkers of Chronic Obstructive Pulmonary Disease Based on Bioinformatics Analyses. BioMed Res Int (2021) 2021:9921012. doi: 10.1155/2021/9921012
44. Geiß C, Alanis-Lobato G, Andrade-Navarro M, Régnier-Vigouroux A. Assessing the Reliability of Gene Expression Measurements in Very-Low-Numbers of Human Monocyte-Derived Macrophages. Sci Rep (2019) 9(1):17908. doi: 10.1038/s41598-019-54500-8
45. Pfaffl MW. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res (2001) 29(9):e45. doi: 10.1093/nar/29.9.e45
46. Ahmed S, Mohamed HT, El-Husseiny N, El Mahdy MM, Safwat G, Diab AA, et al. IL-8 Secreted by Tumor Associated Macrophages Contribute to Lapatinib Resistance in HER2-Positive Locally Advanced Breast Cancer via Activation of Src/STAT3/ERK1/2-Mediated EGFR Signaling. Biochim Biophys Acta Mol Cell Res (2021) 1868(6):118995. doi: 10.1016/j.bbamcr.2021.118995
47. Lafta IJ. E2F6 is Essential for Cell Viability in Breast Cancer Cells During Replication Stress. Turkish J Biol = Turk biyoloji dergisi (2019) 43(5):293–304. doi: 10.3906/biy-1905-6
48. Lothong M, Sakares W, Rojsitthisak P, Tanikawa C, Matsuda K, Yodsurang V. Collagen XVII Inhibits Breast Cancer Cell Proliferation and Growth Through Deactivation of the AKT/mTOR Signaling Pathway. PLoS One (2021) 16(7):e0255179. doi: 10.1371/journal.pone.0255179
49. Normann LS, Aure MR, Leivonen S-K, Haugen MH, Hongisto V, Kristensen VN, et al. MicroRNA in Combination With HER2-Targeting Drugs Reduces Breast Cancer Cell Viability In Vitro. Sci Rep (2021) 11(1):10893. doi: 10.1038/s41598-021-90385-2
50. Debeb BG, Cohen EN, Boley K, Freiter EM, Li L, Robertson FM, et al. Pre-Clinical Studies of Notch Signaling Inhibitor RO4929097 in Inflammatory Breast Cancer Cells. Breast Cancer Res Treat (2012) 134(2):495–510. doi: 10.1007/s10549-012-2075-8
51. Mazurek A, Luo W, Krasnitz A, Hicks J, Powers RS, Stillman B. DDX5 Regulates DNA Replication and is Required for Cell Proliferation in a Subset of Breast Cancer Cells. Cancer Discovery (2012) 2(9):812–25. doi: 10.1158/2159-8290.Cd-12-0116
52. Wang W-J, Wang H, Wang M-s, Huang Y-Q, Ma Y-Y, Qi J, et al. Assessing the Prognostic Value of Stemness-Related Genes in Breast Cancer Patients. Sci Rep (2020) 10(1):18325. doi: 10.1038/s41598-020-73164-3
53. Pei J, Wang Y, Li Y. Identification of Key Genes Controlling Breast Cancer Stem Cell Characteristics via Stemness Indices Analysis. J Trans Med (2020) 18(1):74. doi: 10.1186/s12967-020-02260-9
54. Jonasson E, Ghannoum S, Persson E, Karlsson J, Kroneis T, Larsson E, et al. Identification of Breast Cancer Stem Cell Related Genes Using Functional Cellular Assays Combined With Single-Cell RNA Sequencing in MDA-MB-231 Cells. Front Genet (2019) 10:500. doi: 10.3389/fgene.2019.00500
55. Zhang JY, Luo Q, Xu JR, Bai J, Mu LM, Yan Y, et al. Regulating Stem Cell-Related Genes Induces the Plastic Differentiation of Cancer Stem Cells to Treat Breast Cancer. Mol Ther Oncolytics (2020) 18:396–408. doi: 10.1016/j.omto.2020.07.009
56. Parsana P, Amend SR, Hernandez J, Pienta KJ, Battle A. Identifying Global Expression Patterns and Key Regulators in Epithelial to Mesenchymal Transition Through Multi-Study Integration. BMC Cancer (2017) 17(1):447. doi: 10.1186/s12885-017-3413-3
57. Wang T, Zhang Y, Bai J, Xue Y, Peng Q. MMP1 and MMP9 are Potential Prognostic Biomarkers and Targets for Uveal Melanoma. BMC Cancer (2021) 21(1):1068–8. doi: 10.1186/s12885-021-08788-3
58. Li M, He M, Xu F, Guan Y, Tian J, Wan Z, et al. Abnormal Expression and the Significant Prognostic Value of Aquaporins in Clear Cell Renal Cell Carcinoma. PLoS One (2022) 17(3):e0264553–e0264553. doi: 10.1371/journal.pone.0264553
59. Jin Y, Lin Q, Fei H, Xue L, Li L, Xi Q, et al. Bioinformatics Analysis of Potential Therapeutic Targets and Prognostic Biomarkers Amid CXC Chemokines in Ovarian Carcinoma Microenvironment. J Oncol (2021) 2021:8859554:1–18. doi: 10.1155/2021/8859554
60. Ota A, Hyon S-H, Sumi S, Matsumura K. Gene Expression Analysis of Human Induced Pluripotent Stem Cells Cryopreserved by Vitrification Using StemCell Keep. Biochem biophysics Rep (2021) 28:101172–2. doi: 10.1016/j.bbrep.2021.101172
61. McCrum-Gardner E. Which is the Correct Statistical Test to Use? Br J Oral Maxillofac Surg (2008) 46(1):38–41. doi: 10.1016/j.bjoms.2007.09.002
62. Nayak BK, Hazra A. How to Choose the Right Statistical Test? Indian J Ophthalmol (2011) 59(2):85–6. doi: 10.4103/0301-4738.77005
63. Ranganathan P. An Introduction to Statistics: Choosing the Correct Statistical Test. Indian J Crit Care Med peer-reviewed Off Publ Indian Soc Crit Care Med (2021) 25(Suppl 2):S184–6. doi: 10.5005/jp-journals-10071-23815
64. Mohamed HT, El-Husseiny N, El-Ghonaimy EA, Ibrahim SA, Bazzi ZA, Cavallo-Medved D, et al. IL-10 Correlates With the Expression of Carboxypeptidase B2 and Lymphovascular Invasion in Inflammatory Breast Cancer: The Potential Role of Tumor Infiltrated Macrophages. Curr Probl Cancer (2018) 42(2):215–30. doi: 10.1016/j.currproblcancer.2018.01.009
65. Jamiyan T, Kuroda H, Yamaguchi R, Abe A, Hayashi M. CD68- and CD163-Positive Tumor-Associated Macrophages in Triple Negative Cancer of the Breast. Virchows Archiv (2020) 477(6):767–75. doi: 10.1007/s00428-020-02855-z
66. Pelekanou V, Villarroel-Espindola F, Schalper KA, Pusztai L, Rimm DL. CD68, CD163, and Matrix Metalloproteinase 9 (MMP-9) Co-Localization in Breast Tumor Microenvironment Predicts Survival Differently in ER-Positive and -Negative Cancers. Breast Cancer Res (2018) 20(1):154. doi: 10.1186/s13058-018-1076-x
67. Ni C, Yang L, Xu Q, Yuan H, Wang W, Xia W, et al. CD68- and CD163-Positive Tumor Infiltrating Macrophages in non-Metastatic Breast Cancer: A Retrospective Study and Meta-Analysis. J Cancer (2019) 10(19):4463–72. doi: 10.7150/jca.33914
68. Mukhtar RA, Moore AP, Tandon VJ, Nseyo O, Twomey P, Adisa CA, et al. Elevated Levels of Proliferating and Recently Migrated Tumor-Associated Macrophages Confer Increased Aggressiveness and Worse Outcomes in Breast Cancer. Ann Surg Oncol (2012) 19(12):3979–86. doi: 10.1245/s10434-012-2415-2
69. Yang Z, Tang X, Meng G, Benesch MGK, Mackova M, Belon AP, et al. Latent Cytomegalovirus Infection in Female Mice Increases Breast Cancer Metastasis. Cancers (Basel) (2019) 11(4):1–20. doi: 10.3390/cancers11040447
70. Oberstein A, Shenk T. Cellular Responses to Human Cytomegalovirus Infection: Induction of a Mesenchymal-to-Epithelial Transition (MET) Phenotype. Proc Natl Acad Sci U S A (2017) 114(39):E8244–e8253. doi: 10.1073/pnas.1710799114
71. Taher C, Frisk G, Fuentes S, Religa P, Costa H, Assinger A, et al. High Prevalence of Human Cytomegalovirus in Brain Metastases of Patients With Primary Breast and Colorectal Cancers. Transl Oncol (2014) 7(6):732–40. doi: 10.1016/j.tranon.2014.09.008
72. Fornara O, Bartek J Jr., Rahbar A, Odeberg J, Khan Z, Peredo I, et al. Cytomegalovirus Infection Induces a Stem Cell Phenotype in Human Primary Glioblastoma Cells: Prognostic Significance and Biological Impact. Cell Death Differentiation (2016) 23(2):261–9. doi: 10.1038/cdd.2015.91
73. Geisler J, Touma J, Rahbar A, Söderberg-Nauclér C, Vetvik K. A Review of the Potential Role of Human Cytomegalovirus (HCMV) Infections in Breast Cancer Carcinogenesis and Abnormal Immunity. Cancers (2019) 11(12):1842. doi: 10.3390/cancers11121842
74. Richardson AK, Walker LC, Cox B, Rollag H, Robinson BA, Morrin H, et al. Breast Cancer and Cytomegalovirus. Clin Transl Oncol (2020) 22(4):585–602. doi: 10.1007/s12094-019-02164-1
75. El Shazly DF, Bahnassey AA, Omar OS, Elsayed ET, Al-Hindawi A, El-Desouky E, et al. Detection of Human Cytomegalovirus in Malignant and Benign Breast Tumors in Egyptian Women. Clin Breast Cancer (2018) 18(4):e629–42. doi: 10.1016/j.clbc.2017.10.018
76. Pocock JM, Storisteanu DML, Reeves MB, Juss JK, Wills MR, Cowburn AS, et al. Human Cytomegalovirus Delays Neutrophil Apoptosis and Stimulates the Release of a Prosurvival Secretome. Front Immunol (2017) 8:1185. doi: 10.3389/fimmu.2017.01185
77. Dziurzynski K, Wei J, Qiao W, Hatiboglu MA, Kong LY, Wu A, et al. Glioma-Associated Cytomegalovirus Mediates Subversion of the Monocyte Lineage to a Tumor Propagating Phenotype. Clin Cancer Res (2011) 17(14):4642–9. doi: 10.1158/1078-0432.Ccr-11-0414
78. Tang B, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, et al. TGF-Beta Switches From Tumor Suppressor to Prometastatic Factor in a Model of Breast Cancer Progression. J Clin Invest (2003) 112(7):1116–24. doi: 10.1172/jci18899
79. Avdic S, McSharry BP, Steain M, Poole E, Sinclair J, Abendroth A, et al. Human Cytomegalovirus-Encoded Human Interleukin-10 (IL-10) Homolog Amplifies Its Immunomodulatory Potential by Upregulating Human IL-10 in Monocytes. J Virol (2016) 90(8):3819–27. doi: 10.1128/jvi.03066-15
80. Singh P, Neumann DM. Persistent HCMV Infection of a Glioblastoma Cell Line Contributes to the Development of Resistance to Temozolomide. Virus Res (2020) 276:197829. doi: 10.1016/j.virusres.2019.197829
81. Bartek J Jr., Merchut-Maya JM, Maya-Mendoza A, Fornara O, Rahbar A, Bröchner CB, et al. Cancer Cell Stemness, Responses to Experimental Genotoxic Treatments, Cytomegalovirus Protein Expression and DNA Replication Stress in Pediatric Medulloblastomas. Cell Cycle (2020) 19(7):727–41. doi: 10.1080/15384101.2020.1728025
82. Chan G, Bivins-Smith ER, Smith MS, Yurochko AD. NF-kappaB and Phosphatidylinositol 3-Kinase Activity Mediates the HCMV-Induced Atypical M1/M2 Polarization of Monocytes. Virus Res (2009) 144(1-2):329–33. doi: 10.1016/j.virusres.2009.04.026
83. Ribatti D, Tamma R, Annese T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Trans Oncol (2020) 13(6):100773–3. doi: 10.1016/j.tranon.2020.100773
84. Zhu X, Hu B, Hu M, Qian D, Wang B. Human Cytomegalovirus Infection Enhances Invasiveness and Migration of Glioblastoma Cells by Epithelial-to-Mesenchymal Transition. Int J Clin Exp Pathol (2020) 13(10):2637–47.
85. Soroceanu L, Matlaf L, Khan S, Akhavan A, Singer E, Bezrookove V, et al. Cytomegalovirus Immediate-Early Proteins Promote Stemness Properties in Glioblastoma. Cancer Res (2015) 75(15):3065–76. doi: 10.1158/0008-5472.Can-14-3307
86. Pasquereau S, Al Moussawi F, Karam W, Diab Assaf M, Kumar A, Herbein G. Cytomegalovirus, Macrophages and Breast Cancer. Open Virol J (2017) 11:15–27. doi: 10.2174/1874357901711010015
87. Min C-K, Shakya AK, Lee B-J, Streblow DN, Caposio P, Yurochko AD. The Differentiation of Human Cytomegalovirus Infected-Monocytes Is Required for Viral Replication. Front Cell Infect Microbiology. (2020) 10(1):11. doi: 10.3389/fcimb.2020.00368
88. Stevenson EV, Collins-McMillen D, Kim JH, Cieply SJ, Bentz GL, Yurochko AD. HCMV Reprogramming of Infected Monocyte Survival and Differentiation: A Goldilocks Phenomenon. Viruses (2014) 6(2):782–807. doi: 10.3390/v6020782
89. Gredmark S, Tilburgs T, Soderberg-Naucler C. Human Cytomegalovirus Inhibits Cytokine-Induced Macrophage Differentiation. J Virol (2004) 78(19):10378–89. doi: 10.1128/JVI.78.19.10378-10389.2004
90. Ramos RN, Rodriguez C, Hubert M, Ardin M, Treilleux I, Ries CH, et al. CD163+ Tumor-Associated Macrophage Accumulation in Breast Cancer Patients Reflects Both Local Differentiation Signals and Systemic Skewing of Monocytes. Clinical & Translat Immunol (2020) 9(2). doi: 10.1002/cti2.1108
91. Garvin S, Oda H, Arnesson L-G, Lindström A, Shabo I. Tumor Cell Expression of CD163 is Associated to Postoperative Radiotherapy and Poor Prognosis in Patients With Breast Cancer Treated With Breast-Conserving Surgery. J Cancer Res Clin Oncol (2018) 144(7):1253–63. doi: 10.1007/s00432-018-2646-0
92. Powell AE, Anderson EC, Davies PS, Silk AD, Pelz C, Impey S, et al. Fusion Between Intestinal Epithelial Cells and Macrophages in a Cancer Context Results in Nuclear Reprogramming. Cancer Res (2011) 71(4):1497–505. doi: 10.1158/0008-5472.Can-10-3223
93. Shabo I, Midtbö K, Andersson H, Åkerlund E, Olsson H, Wegman P, et al. Macrophage Traits in Cancer Cells are Induced by Macrophage-Cancer Cell Fusion and Cannot be Explained by Cellular Interaction. BMC Cancer (2015) 15:922. doi: 10.1186/s12885-015-1935-0
94. Shabo I, Svanvik J. Expression of Macrophage Antigens by Tumor Cells. Adv Exp Med Biol (2011) 714:141–50. doi: 10.1007/978-94-007-0782-5_7
95. Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The Inflammatory Micro-Environment in Tumor Progression: The Role of Tumor-Associated Macrophages. Crit Rev Oncol Hematol (2008) 66(1):1–9. doi: 10.1016/j.critrevonc.2007.07.004
96. Pawelek JM. Cancer-Cell Fusion With Migratory Bone-Marrow-Derived Cells as an Explanation for Metastasis: New Therapeutic Paradigms. Future Oncol (2008) 4(4):449–52. doi: 10.2217/14796694.4.4.449
97. Pawelek JM. Tumour-Cell Fusion as a Source of Myeloid Traits in Cancer. Lancet Oncol (2005) 6(12):988–93. doi: 10.1016/s1470-2045(05)70466-6
98. Pawelek JM, Chakraborty AK. Fusion of Tumour Cells With Bone Marrow-Derived Cells: A Unifying Explanation for Metastasis. Nat Rev Cancer (2008) 8(5):377–86. doi: 10.1038/nrc2371
99. Khan KA, Coaquette A, Davrinche C, Herbein G. Bcl-3-Regulated Transcription From Major Immediate-Early Promoter of Human Cytomegalovirus in Monocyte-Derived Macrophages. J Immunol (2009) 182(12):7784. doi: 10.4049/jimmunol.0803800
100. Chen W, Qin Y, Liu S. Cytokines, Breast Cancer Stem Cells (BCSCs) and Chemoresistance. Clin Trans Med (2018) 7(1):27–7. doi: 10.1186/s40169-018-0205-6
101. Benner B, Scarberry L, Suarez-Kelly LP, Duggan MC, Campbell AR, Smith E, et al. Generation of Monocyte-Derived Tumor-Associated Macrophages Using Tumor-Conditioned Media Provides a Novel Method to Study Tumor-Associated Macrophages In Vitro. J immunotherapy Cancer (2019) 7(1):140–0. doi: 10.1186/s40425-019-0622-0
102. Reinartz S, Schumann T, Finkernagel F, Wortmann A, Jansen JM, Meissner W, et al. Mixed-Polarization Phenotype of Ascites-Associated Macrophages in Human Ovarian Carcinoma: Correlation of CD163 Expression, Cytokine Levels and Early Relapse. Int J Cancer (2014) 134(1):32–42. doi: 10.1002/ijc.28335
103. Bohrer LR, Schwertfeger KL. Macrophages Promote Fibroblast Growth Factor Receptor-Driven Tumor Cell Migration and Invasion in a CXCR2-Dependent Manner. Mol Cancer Res (2012) 10(10):1294–305. doi: 10.1158/1541-7786.Mcr-12-0275
104. Baghel KS, Tewari BN, Shrivastava R, Malik SA, Lone MU, Jain NK, et al. Macrophages Promote Matrix Protrusive and Invasive Function of Breast Cancer Cells via MIP-1β Dependent Upregulation of MYO3A Gene in Breast Cancer Cells. Oncoimmunology (2016) 5(7):e1196299. doi: 10.1080/2162402x.2016.1196299
105. Lin EY, Pollard JW. Tumor-Associated Macrophages Press the Angiogenic Switch in Breast Cancer. Cancer Res (2007) 67(11):5064–6. doi: 10.1158/0008-5472.Can-07-0912
106. Qian BZ, Pollard JW. Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell (2010) 141(1):39–51. doi: 10.1016/j.cell.2010.03.014
107. Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, et al. Macrophages Promote the Invasion of Breast Carcinoma Cells via a Colony-Stimulating Factor-1/Epidermal Growth Factor Paracrine Loop. Cancer Res (2005) 65(12):5278–83. doi: 10.1158/0008-5472.Can-04-1853
108. Linde N, Lederle W, Depner S, van Rooijen N, Gutschalk CM, Mueller MM. Vascular Endothelial Growth Factor-Induced Skin Carcinogenesis Depends on Recruitment and Alternative Activation of Macrophages. J Pathol (2012) 227(1):17–28. doi: 10.1002/path.3989
109. Teo WH, Chen HP, Huang JC, Chan YJ. Human Cytomegalovirus Infection Enhances Cell Proliferation, Migration and Upregulation of EMT Markers in Colorectal Cancer-Derived Stem Cell-Like Cells. Int J Oncol (2017) 51(5):1415–26. doi: 10.3892/ijo.2017.4135
110. Fiallos E, Judkins J, Matlaf L, Prichard M, Dittmer D, Cobbs C, et al. Human Cytomegalovirus Gene Expression in Long-Term Infected Glioma Stem Cells. PLoS One (2014) 9(12):e116178. doi: 10.1371/journal.pone.0116178
111. Valle Oseguera CA, Spencer JV. Human Cytomegalovirus Interleukin-10 Enhances Matrigel Invasion of MDA-MB-231 Breast Cancer Cells. Cancer Cell Int (2017) 17(1):24. doi: 10.1186/s12935-017-0399-5
112. Strååt K, de Klark R, Gredmark-Russ S, Eriksson P, Söderberg-Nauclér C. Infection With Human Cytomegalovirus Alters the MMP-9/TIMP-1 Balance in Human Macrophages. J Virol (2009) 83(2):830–5. doi: 10.1128/JVI.01363-08
113. Deligne C, Midwood KS. Macrophages and Extracellular Matrix in Breast Cancer: Partners in Crime or Protective Allies? Frontiers in Oncology.(2021) 11(1):12. doi: 10.3389/fonc.2021.620773
114. Bednarczyk RB, Tuli NY, Hanly EK, Rahoma GB, Maniyar R, Mittelman A, et al. Macrophage Inflammatory Factors Promote Epithelial-Mesenchymal Transition in Breast Cancer. Oncotarget (2018) 9(36):24272–82. doi: 10.18632/oncotarget.24917
115. Valeta-Magara A, Gadi A, Volta V, Walters B, Arju R, Giashuddin S, et al. Inflammatory Breast Cancer Promotes Development of M2 Tumor-Associated Macrophages and Cancer Mesenchymal Cells Through a Complex Chemokine Network. Cancer Res (2019) 79(13):3360–71. doi: 10.1158/0008-5472.Can-17-2158
116. Ingthorsson S, Briem E, Bergthorsson JT, Gudjonsson T. Epithelial Plasticity During Human Breast Morphogenesis and Cancer Progression. J mammary gland Biol neoplasia (2016) 21(3-4):139–48. doi: 10.1007/s10911-016-9366-3
117. Little AC, Pathanjeli P, Wu Z, Bao L, Goo LE, Yates JA, et al. IL-4/IL-13 Stimulated Macrophages Enhance Breast Cancer Invasion Via Rho-GTPase Regulation of Synergistic VEGF/CCL-18 Signaling. Frontiers in Oncology. (2019) 9(1):13. doi: 10.3389/fonc.2019.00456
118. Tzanakakis G, Giatagana E-M, Kuskov A, Berdiaki A, Tsatsakis A, Neagu M, et al. Proteoglycans in the Pathogenesis of Hormone-Dependent Cancers: Mediators and Effectors. Cancers (2020) 12(9):2401. doi: 10.3390/cancers12092401
119. Rahal OM, Wolfe AR, Mandal PK, Larson R, Tin S, Jimenez C, et al. Blocking Interleukin (IL)4- and IL13-Mediated Phosphorylation of STAT6 (Tyr641) Decreases M2 Polarization of Macrophages and Protects Against Macrophage-Mediated Radioresistance of Inflammatory Breast Cancer. Int J Radiat Oncol Biol Phys (2018) 100(4):1034–43. doi: 10.1016/j.ijrobp.2017.11.043
120. Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 Signaling Pathway is Required for Growth of CD44+CD24- Stem Cell-Like Breast Cancer Cells in Human Tumors. J Clin Invest (2011) 121(7):2723–35. doi: 10.1172/jci44745
121. Saxena M, Balaji SA, Deshpande N, Ranganathan S, Pillai DM, Hindupur SK, et al. AMP-Activated Protein Kinase Promotes Epithelial-Mesenchymal Transition in Cancer Cells Through Twist1 Upregulation. J Cell Sci (2018) 131(14):jcs208314. doi: 10.1242/jcs.208314
122. Lv D, Guo L, Zhang T, Huang L. PRAS40 Signaling in Tumor. Oncotarget (2017) 8(40):69076–85. doi: 10.18632/oncotarget.17299
123. Wang J, Kuiatse I, Lee AV, Pan J, Giuliano A, Cui X. Sustained C-Jun-NH2-Kinase Activity Promotes Epithelial-Mesenchymal Transition, Invasion, and Survival of Breast Cancer Cells by Regulating Extracellular Signal-Regulated Kinase Activation. Mol Cancer Res MCR (2010) 8(2):266–77. doi: 10.1158/1541-7786.MCR-09-0221
124. Banerjee K, Resat H. Constitutive Activation of STAT3 in Breast Cancer Cells: A Review. International Journal of Cancer (2016) 138(11):2570–8. doi: 10.1002/ijc.29923
125. Ortiz-Montero P, Londoño-Vallejo A, Vernot JP. Senescence-Associated IL-6 and IL-8 Cytokines Induce a Self- and Cross-Reinforced Senescence/Inflammatory Milieu Strengthening Tumorigenic Capabilities in the MCF-7 Breast Cancer Cell Line. Cell Commun Signal (2017) 15(1):17. doi: 10.1186/s12964-017-0172-3
126. Iliopoulos D, Hirsch HA, Struhl K. An Epigenetic Switch Involving NF-Kappab, Lin28, Let-7 MicroRNA, and IL6 Links Inflammation to Cell Transformation. Cell (2009) 139(4):693–706. doi: 10.1016/j.cell.2009.10.014
Keywords: inflammatory breast cancer, breast cancer stem cell, human cytomegalovirus, tumor-associated macrophages, secretome
Citation: Mohamed HT, El-Sharkawy AA, El-Shinawi M, Schneider RJ and Mohamed MM (2022) Inflammatory Breast Cancer: The Secretome of HCMV+ Tumor-Associated Macrophages Enhances Proliferation, Invasion, Colony Formation, and Expression of Cancer Stem Cell Markers. Front. Oncol. 12:899622. doi: 10.3389/fonc.2022.899622
Received: 18 March 2022; Accepted: 25 May 2022;
Published: 30 June 2022.
Edited by:
Martin Götte, University of Münster, GermanyReviewed by:
Marco A. Velasco-Velazquez, National Autonomous University of Mexico, MexicoCopyright © 2022 Mohamed, El-Sharkawy, El-Shinawi, Schneider and Mohamed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hossam Taha Mohamed, aG90YWhhQG1zYS5lZHUuZWc=
†ORCID ID: Hossam Taha Mohamed, orcid.org/0000-0003-1989-5783
Aya Ali El-Sharkawy, orcid.org/0000-0002-5915-6953
Mohamed El-Shinawi, orcid.org/0000-0003-3645-2343
Robert J. Schneider, orcid.org/0000-0001-5807-5564
Mona Mostafa Mohamed, orcid.org/0000-0003-0640-8894
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.