- 1Department of Biochemistry and Molecular Biology, School of Life Sciences, Pondicherry University, Pondicherry, India
- 2Centre of Molecular Medicine and Diagnostics, Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical Sciences, Saveetha University, Chennai, India
- 3Department of Biological Sciences, Aliah University, Kolkata, India
- 4Department of Basic Medical Sciences, College of Medicine, University of Sharjah, Sharjah, United Arab Emirates
- 5Department of Basic Medical Sciences, College of Medicine, AlMaarefa University, Riyadh, Saudi Arabia
- 6Department of Clinical Medical Sciences, College of Medicine, AlMaarefa University, Riyadh, Saudi Arabia
- 7Departments of Biochemistry, Molecular Virology, Research, Clinical Skills, and Simulation, Panimalar Medical College Hospital and Research Institute, Chennai, India
Replicating the naturalistic biomechanical milieu of cells is a primary requisite to uncover the fundamental life processes. The native milieu is significantly not replicated in the two-dimensional (2D) cell cultures. Alternatively, the current three-dimensional (3D) culture techniques can replicate the properties of extracellular matrix (ECM), though the recreation of the original microenvironment is challenging. The organization of cells in a 3D manner contributes to better insight about the tumorigenesis mechanism of the in vitro cancer models. Gene expression studies are susceptible to alterations in their microenvironment. Physiological interactions among neighboring cells also contribute to gene expression, which is highly replicable with minor modifications in 3D cultures. 3D cell culture provides a useful platform for identifying the biological characteristics of tumor cells, particularly in the drug sensitivity area of translational medicine. It promises to be a bridge between traditional 2D culture and animal experiments and is of great importance for further research in tumor biology. The new imaging technology and the implementation of standard protocols can address the barriers interfering with the live cell observation in a natural 3D physiological environment.
Introduction
Since the 1940s, cells are consistently maintained by proliferation in specially designed culture media, adhered to glass or plastic surfaces (1). Hela cells are among the first cancer cells to be maintained and cultured consistently since 1951 (2). Such adherent-type cells proliferate in a specific culture medium and grow in a two-dimensional (2D) pattern. Such cell types may play a significant role in various cancer-based experiments; however, they have their share of limitations. The major hurdle in the 2D cell culture techniques is that they fail to replicate the three-dimensional (3D) microenvironment of biological tissues (3). The microenvironment includes the extracellular matrix (ECM) and neighboring cells, which are elementary for the formation and functioning of tissues and play a pivotal role in regulating tissue growth and development (4). The microenvironment is a connecting structure that explains cell behavior, identity, and function. It is not simply an arena to hold the cells in. The present 3D culture techniques can imitate the ECM elements, though the recreation of the exact microenvironment is still challenging (5).
Standard 2D cell cultures do not perform a satisfactory role in recreating a natural cellular environment (6). Hence, there is a demand for creating a reliable, controllable, and realistic culture systems that assist cell growth, differentiation, and organization resembling natural tissues and organs (7). Biomaterials like hydrogels have been developed to grow cells in three dimensions and to investigate countless cellular processes towards understanding morphogenesis, aging, and disease (8). 3D culture systems help in covering the gap between 2D monolayer cell cultures and the complexity of the organism (9). Cells growing in 3D cultures behave differently, and physiologically significant aspects can be studied more realistically. Those aspects include cell morphology, behavior, differentiation, proliferation rate, gene expression, and genomic profiles; malignant tumor formation, drug development, and testing; and in vitro culture of multi-cellular tissue for later implantation (10).
The applications of 3D culture include tissue engineering or cell biology studies that emphasize the presence of cells in an artificial environment. Currently, there are no such models that can imitate the intricacy of the natural environment in vivo. In 2D cell culture, cells were grown only on the surface; on the other hand, in 3D culture, cells can migrate into the matrix and develop contacts in all dimensions (10, 11). The generation of 3D cell cultures may be broadly categorized depending on the presence of scaffolds. Techniques such as hanging drop, magnetic levitation, and ultralow attachments are typically classified as scaffold-independent 3D cell culture techniques (Figure 1), whereas 3D techniques including hydrogel, sponge, and microcarriers are categorized as scaffold-dependent 3D cell culture techniques (Figure 2). In many 3D cell cultures, the migration of cells in a confined 3D environment depends on microtubule dynamics, whereas in 2D, the migration depends on cycles of myosin-mediated contraction, integrin-mediated adhesion, and actin protrusion (12, 13).
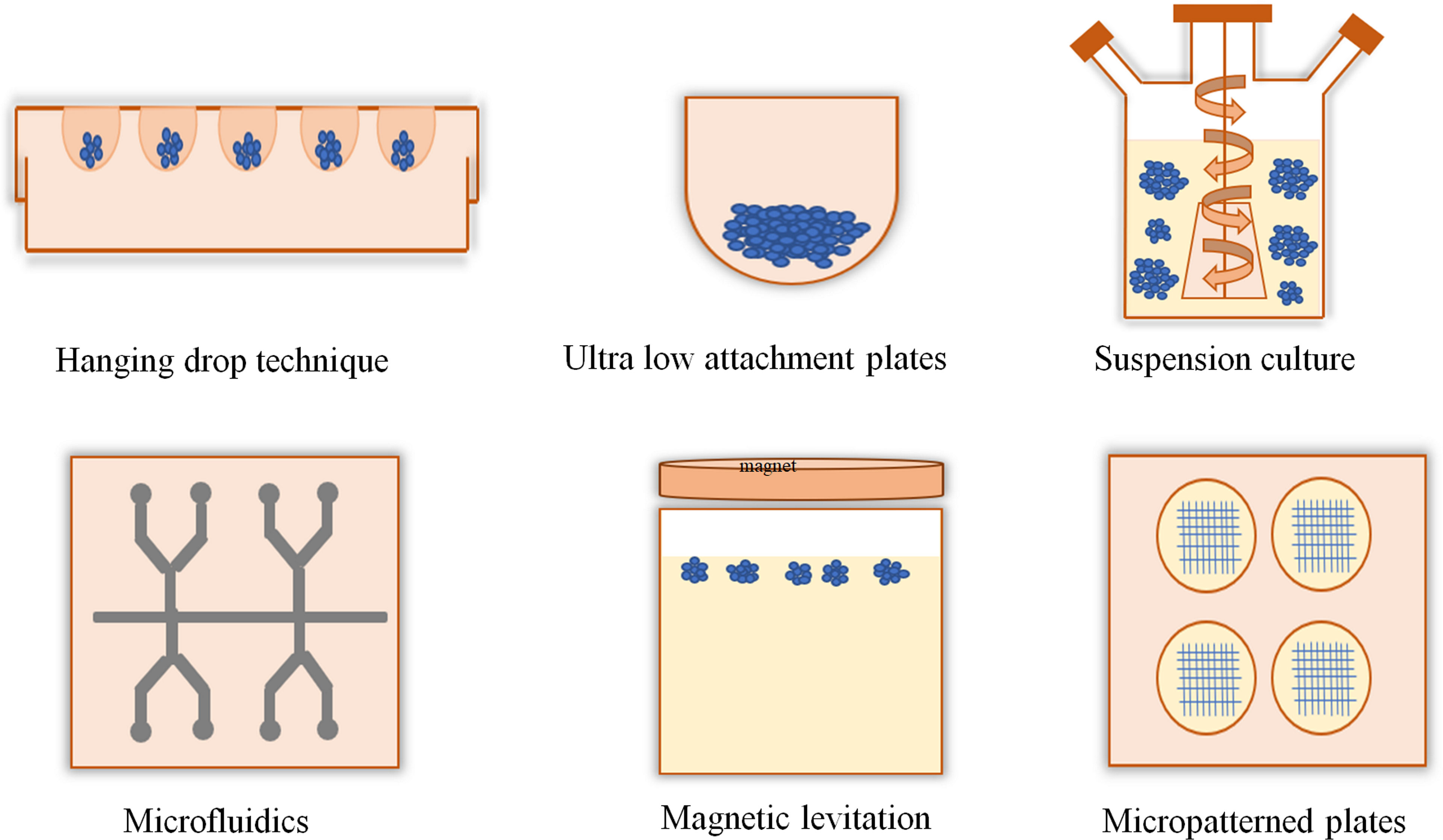
Figure 1 Scaffold-free and anchor-independent three-dimensional cell cultures. The hanging drop technique uses the surface tension of a droplet of cell suspension and gravity to suspend the droplet of cells onto the base of a lid which could promote cell aggregation into a spheroid. In the ultra-low attachment plate technique, the plate surface is coated with an inert substance that minimizes cell attachment and promotes cell aggregation. Suspension culture methods produce tumor spheroids using bioreactors, such as spinner flask and rotating flasks. Microfluidics can create uniform-sized spheroids for high-throughput screening applications. In the magnetic levitation method, cells with magnetic iron oxide were held at the air–medium interface using magnetic force. Micropatterning techniques control the position of cells and their shape.
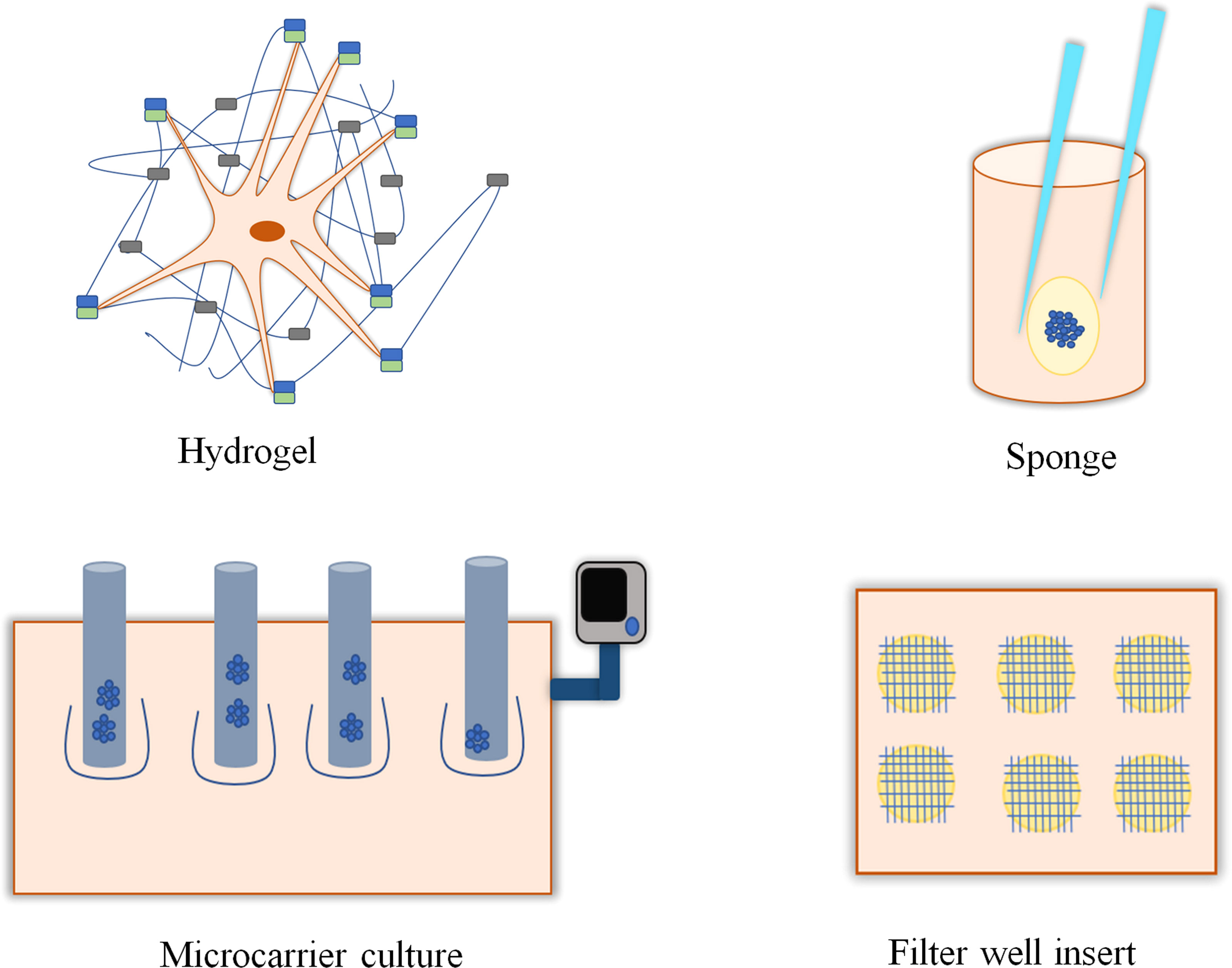
Figure 2 Scaffold-based and anchor-dependent three-dimensional cell cultures. Hydrogels with interconnected pores promote cell growth by providing a 3D environment and supporting the transport of oxygen, nutrients, and metabolites required for cell growth. Sponges, with their porous structure, support cell growth and migration. Microcarrier is a support matrix in bioreactors that supports the development of multi-cellular spheroids. Filter-well insert in culture plates promotes the growth of 3D cell culture.
Major Research Challenges in 2D Cell Culture
The native 3D milieu of the cells is not sufficiently taken into account by 2D culture systems, i.e., cells in 2D culture appear stretched and flat compared to those in in vivo conditions (14). In traditional 2D cultures, cells are grown as a monolayer on flat plastic surfaces, allowing the cells to receive oxygen and nutrients from the medium throughout their growth in a homogenous manner. Proliferating cells form monolayers wherein the necrotic cells are easily removed while changing the culture medium. The abnormal morphology of cells in monolayers influences cellular differentiation, proliferation, gene expression, protein expression, and apoptosis (15). There is an increasing demand for in vitro models that can recapture the relevant complexity of in vivo milieu other than monolayer cultures (16, 17). Appropriate platforms of 3D culture act as outstanding in vitro models that allow the study of cell responses in a setting that mimics the native in vivo environments and curtail the need for animal trials, especially for toxicity assays (10). The physical and spatial aspects of the 3D platform influence the signal transduction between the cells, which, in turn, affects their behavior and gene expression (18). The 3D culture system offers spheroids, which have a higher ability to adhere and shield cells from harsh conditions. In spheroids, there is close mimicking of cell–ECM and cell–cell interactions that imitate the natural milieu found in vivo so that the morphology of the cells mimic the actual shape of cells in the tissues (19). Cells in a spheroid usually contain cells at various stages comprising necrotic, hypoxic, quiescent, apoptotic, and proliferating cells. The outer layer of the spheroid has viable and proliferating cells because it is more exposed to the medium. On moving to the core, cells receive fewer nutrients, growth factors, and oxygen from the medium and are present in a hypoxic or quiescent state (20). This cellular heterogeneity is a good replica of in vivo tissues, mainly in tumors. The cellular processes in 3D cultures are closely imitated in vivo because of the morphology and interaction of the cells (21).
The rate of proliferation of cells in 2D and 3D usually varies and depends on the matrix and cell line used. An array of endometrial cell lines (RL95-2, EN-1078D, KLE, and Ishikawa) display reduced proliferation rates when grown in 3D reconstituted basement membrane (rBM) compared to 2D culture and was identified with a drop in the proliferating cell nuclear antigen (PCNA) protein marker expression. The total number of cells in 3D rBM is decreased after 8 days of growth (22).
The differential expression of genes and proteins in a monolayer and 3D cultures is generally the speculation for the different behavior of mainly the genes which are involved in chemosensitivity, proliferation, migration, invasion, and angiogenesis (23). Loessner et al. (2010) (24) has reported that 3D ovarian cancer lines depict a significant increase in the mRNA expression levels of the receptors on the cell surface like β1, α3, α5 integrins, and protease—matrix metalloproteinase 9 when compared to cells in 2D culture. Monolayer culturing of cells is well adopted because it has helped in understanding the cell behavior; however, it is now established that 2D systems cannot recapture the complete depiction of the cells as present in vivo—for example, few vital cancer cell features like tumoral heterogeneity, genetic profile, and morphology are not accurately recaptured in monolayer cultures. The 3D cell culture models help to preserve the original shape, polarization, heterogeneity, and genetic profile of stromal and cancer cells (25).
Importance of Extracellular Matrix
There is a growing interest in 3D cell culture after recognizing the importance of ECM in various cell functions and behavior. ECM affects the interaction of cells with other cells and their microenvironment, along with the spatial arrangement of cells in their milieu (26). ECM is not an irregular mixture of secreted components; it has instead a defined composition of biochemicals and typical geometrical structures triggering individual cellular responses like interaction and differentiation (27). The elemental composition of ECM includes water, polysaccharides, and proteins. ECM molecules consist of fibrous matrix proteins (e.g., elastin, collagens), glycoproteins (e.g., fibronectin), proteoglycans (e.g., syndecan, perlecan), glycosaminoglycans (e.g., hyaluronan, heparan sulfate), ECM-sequestered growth factors, e.g., hepatocyte growth factor, platelet-derived growth factor, transforming growth factor-β, vascular endothelial growth factor, and other secreted proteins such as protease inhibitors and proteolytic enzymes (28). Proteoglycans form a natural, hydrated, gel-like structure to fill the extracellular interstitial space. Proteoglycans perform various functions, including buffering, hydration, binding, and conferring force resistance properties (29). The composition of the ECM influences cell adhesion and migration (30). The sensitivity of ECM components to proteolytic enzymes decides the capacity of the cell to redesign the matrix to migrate through it. Basement membrane is secreted basically by epithelial and endothelial cells, which interrupts the transport of proteins and enhances mechanical stiffness (31). In the in vivo study model, cells are surrounded by ECM-associated signaling molecules and connective tissues consisting of mesenchymal cells, fibroblasts, and adipocytes. Every tissue has its own ECM with a particular topology and composition that is produced during tissue development (29, 32).
Natural ECM gels like fibrin and type I collagen are readily available and contribute to a number of physical and chemical cues which are required for inducing morphogenesis (33). The diverse cues which are present in natural ECM may become a limitation when trying to isolate an effect of a particular factor. In this case, many new synthetic biomaterials are being developed at a rapid pace and are used as 3D ECM to resemble the regulatory characteristics of natural ECM-bound growth factors for studying the basic biological processes. Synthetic gels are being tailored to imitate particular properties of ECM to provide a reproducible and well-controlled cellular niche. Synthetic ECM is particularly developed to trigger cellular adhesion through short peptide sequences that are recognized as integrin-binding domains within ECM proteins (34). Arginine–glycine–aspartic acid peptides derived from fibronectin are prototypes, and many other sequences which are recognized by several integrins and other adhesion receptors have been identified (35).
Luca et al. (2013) (36) have reported the precise analysis of how ECM can control the genotype and phenotype of routinely used colorectal cancer (CRC) cell lines like HT-29, CACO-2, LOVO, DLD-1, COLO-206F, and SW480 in a laminin-rich ECM 3D model. They studied the expression of EGFR, protein kinase B, and p42/44 mitogen-activated protein kinases (MAPKs) as EGFR stimulates the proliferation of cells via MAPKs. It is a well-established therapeutic target for the treatment of advanced CRC. The results suggest that there are alterations in the gene and protein expression levels of EFGR, phospho-AKT, and phospho-MAPK in 3D cells when compared to 2D culture. Kiss et al. (2013) (37) reported that interactions between cells and ECM in a matrigel culture can regulate cell morphology and upregulate the expression of chemokine receptors—CXCR7 and CXCR4—in prostate cancer cell lines like LNCaP, PC3, and DU145.
Significance of 3D Cell Culture
3D cell cultures facilitate the study of challenges that are posed to an organism (4). Monolayer culturing triggers the cells to adopt to an artificial, rigid, and flat surface which may reshape the cell metabolism and reduce the functionality of cells. The results obtained from monolayer culture methods may not match the in vivo behavior of cells (1). In 3D culture, cells have interacted with ECM and neighboring cells in all dimensions, while in 2D cell culture, only a part of the cell surface is exposed to the adjacent cells in the monolayer. Several studies have demonstrated that 3D cellular morphological organizations have an intense impact on the morphogenesis, development, response to therapy, and gene expression profiles of the cell. Contrary to the monolayer models, 3D culture systems differentiate tumor phenotypes from non-malignant ones, where the tumor phenotype is embodied as multi-cellular organizations (37).
The contemporary 3D cell cultures, however, still lack the complex vascular systems that provide oxygen and nutrients and facilitate waste removal compared to in vivo systems. Cells grown in 3D cultures perform those functions by diffusion. Small spheroids do not face significant problems, but it becomes a challenge for large spheroids due to the lack of vasculature (38). Some reports suggest substantial progress in this direction (39). Expensive and highly complex culturing with complex downstreaming processes and an endpoint assay dependent on the culture method utilized are the various challenges of 3D cell culture (40). The advantages and disadvantages of different types of 3D culture techniques are summarized in Table 1.
Role of 3D Cell Culture in Cancer Research
Cancer is the second major cause of death worldwide (48). Loss of tissue organization and the aberrant behavior of cellular components are characteristically observed in the pre-metastatic niche of cancer tissue (49). Early carcinogenesis stages include loss in cell polarity and detachment from the basement membrane, which allow the discrete accumulation of cells that communicate among them and with the surrounding microenvironment (50). Cancer cells reside in a complex microenvironment referred to as tumor microenvironment (TME), which contains various types of non-cancer cells and their stroma cells like endothelial cells, immune cells (lymphocytes and macrophages), fibroblasts, and mesenchymal cells. All these have a particular role in forming a specific structure, functioning, and physiology of the tumor (51).
The tumor and TME induce bidirectional changes in their functions and phenotypes that sustain the continuing process of tumor progression and metastasis under ephrin receptors and ligand interaction. These interactions evolve along with the progression of the disease (52). Hallmarks of cells in a tumor tissue include sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, and activating invasion and metastasis (53).
In the last few decades, we have arrived at a better understanding of the genetic and molecular underpinnings of tumor etiology. However, concerns regarding the fast adaptation of drug resistance, dormancy, indolent disease, relapse, and metastatic colonization are inadequately answered by the classic monolayer culture models (54). Currently, the majority of cancer biologists rely on monolayer culture techniques to test the efficacy of experimental anti-tumor drug candidates before proceeding to in vivo testing because of their convenience (55). Conventional 2D platforms are well rooted and uncomplicated to use, but the lack of structural architecture and proper environment cause alterations in cell functions. When normal epithelial cells are grown in 2D monolayers, cells frequently lose the ability to differentiate, become highly plastic, and display characteristics exhibited by tumor cells. The malignant cells differ from their benign tumor counterparts (56). Tibbitt and Anseth (57) have illustrated that human breast epithelial cells grow like tumor cells when grown as monolayers, but when cultured in 3D fashion, they return to their natural growth behavior. A lack of a three-dimensional approach can generate uncertain experimental observations and produce inconsistent results. Furthermore, when cells grown in monolayer cultures are screened, they may negate the actions of some important compounds because of the adherence of the cells to plastic surfaces. Cells grown in monolayer culture are exposed to a homogenous environment with adequate oxygen and nutrients, whereas solid tumor cells are exposed to fluctuated critical biological and chemical signals, which can apply both inhibitory and stimulatory effects on tumor progression (58). Drug testing results may be affected due to the lack of 3D ECM network structures in 2D cultures. During anti-tumor drug testing, drugs applied to 2D cell culture can reach the cells without encountering any physical barriers, but when the same drug is delivered in vivo, they face a completely contrasting environment that originally reduces the drug’s partition throughout the complete tumor (59). Some vital cancer cell characteristics like morphology, tumoral heterogeneity, or genetic profile are not correctly imitated in monolayer cultures (11).
Researchers have developed 3D models on realizing the impediments of 2D cultures and the need for a complex native tissue microenvironment. 3D models can certainly recapitulate definite features of solid tumor tissue, like tumor morphology, multi-drug-resistant proteins and pro-angiogenic expression, varying distribution of biological and chemical factors, and reciprocal interactions between tumor and stroma (60).
The relentless development of tumor cell culture systems is crucial for studying tumor cell biology. In the last few decades, 3D cell culture technology has become an essential platform for basic research in cancer biology, utilizing an array of materials and processes to replicate the in vivo TME of cultured tumor cells (55). A number of studies highlight the organization of cells in 3D manner, revealing unique and unexpected visions into the tumorigenesis mechanism, and could show a valuable missing component in the in vitro cancer studies (61). Engineered 3D systems maintain a controllable and replicative microhabitat for incorporating ECM molecules, growth factors, specific cells, and other biochemical signals to better replicate the natural TME, favoring the growth and progression of tumor (59). Table 2 summarizes various 3D cell culture models that have been designed and explored for cancer research.
Role of Microfluidics in Cancer Metastasis Research
Tumor metastasis is the migration of tumor cells from the primary site to colonize distant organs progressively. The progression of tumor metastasis takes place in a stepwise cascade manner, including tumor growth, angiogenesis, stromal invasion, intravasation, extravasation, and colonization at secondary sites within the body (76). The establishment and survival of metastases depend upon the microenvironment of specific receptive organs and the specific metastatic properties of the tumor cells inherited from the parental/primary tumor (77).
Metastasis is the major contributor to most cancer-related deaths. Treating cancer is a challenge because of biological heterogeneity and resistance of metastases to conventional drug therapy. Animal models have been instrumental in studying the cellular and molecular basis of the metastatic process. Due to the physiological difference between animals and humans, it has been problematic to determine the cause-and-effect relationships between specific biological cues and the resulting cancer cell behavior (76). Intermediate steps of the metastatic cascade cannot be satisfactorily studied using in vivo models. Hence, there is an immediate urge to develop technologies that directly control the physical and biochemical factors influencing tumor–vessel interactions (78).
Microfluidic platforms provide modeling systems for investigating various complex phenomena by combining an array of controllable biophysical and biochemical microenvironments with high-resolution real-time imaging. Bersini et al. devised a tri-culture microfluidic system with bone marrow-derived human mesenchymal cells lined with endothelium and MDA-MB-231 human breast cancer cell line to provide an osteo-conditioned environment. They have shown how breast cancer receptor CXCR2 and bone-secreted chemokine CXCL5 are involved in the breast cancer cell’s extravasation process. A profound understanding of the metastatic process can help develop therapeutic approaches that can increase the survival rate of cancer patients (79).
Cancer-on-Chip
Cancer-on-Chip (CoC) is a microfluidic platform containing micrometer-sized compartments to millimeter-sized microchannels to facilitate controlled fluid transport. These compartments provide a space for recreating the niche where the mini-tumors can grow, develop, and interact with its specific microenvironment like in vivo. These chips contain chambers for cell culture where fluid flow, tissue mechanics, and composition of the microenvironment can be controlled. The present models of CoC help us to understand the role of the composition and structure of ECM in cancer cell invasion by using different imaging techniques to visualize the matrix (79). The successful development of a CoC model is based on the critical focus that mimics organ-level physiology or pathophysiology observed in vivo. Microfluidic models provide a significant advantage over static models, including transwells, spheroids, and organoid cultures (80). Wong and Searson (2014) (81) developed an artificial microvessel model which positions the tumor cells (dual-labeled MDA-MB-231 adenocarcinoma and HT-1080 fibrosarcoma cells) next to the artificial microvessel lined with endothelial cells [human umbilical vein endothelial cells (HUVEC) and adult human dermal microvascular endothelial cells] embedded in the ECM. They used live-cell fluorescence microscopy to inspect the interplay between the metastatic cancer cells and the endothelial cells in the artificial microvessel. They suggested that optimizing the pore size and stiffness in the ECM can enhance invasion by using low collagen concentration in dense matrices. Xie et al. (2020) (82) developed a unique 3D tumor array chip with a layer cake structure to screen the anti-cancer drugs epirubicin and paclitaxel. The chip contains the droplets of MDA-MB-231 breast cancer lines encapsulated with gelatin methacryloyl hydrogel. They observed that epirubicin could cause apoptosis at the core of the cell clusters at lower concentrations compared to paclitaxel. Based on this study, it was concluded that epirubicin was a more effective anti-cancer drug compared to paclitaxel (82).
A study on the co-culture of T47D human breast carcinoma cells with immortalized human mammary fibroblasts in fibronectin-rich ECM in a microfluidic device increased the growth of breast cancer cell clusters. This platform simplified the investigation of the morphology and growth of T47D cell clusters in response to broad-spectrum inhibitors of matrix metalloproteinases (83). A microfluidic model with a heterotypic co-culture approach using three different cell types (breast cancer cells, stromal cells, and monocytes) coupled with gene expression analysis exposed the interaction between different cell types through paracrine signaling via transforming growth factor-β production by breast cancer cells and corresponding receptor expression by stromal cells (84). Angiogenesis is a vital step in the cancer cascade as neovascularization is an essential control element restricting cancer progression and growth. Hence, researchers developed multiple microfluidic angiogenesis models that recreate capillary sprouting and vessel formation. The microfluidic angiogenesis model using primary human renal carcinoma cells produced a gradient of angiogenic stimuli that induced capillary outspread in co-cultured HUVECs (85, 86). A microfluidic device using ECM gel was developed, and it demonstrated the trans-endothelial cell migration of neutrophils in situ in response to chemical gradients (80, 87). A reconstituted glioblastoma tumor-on-a-chip model replicating the patient-specific resistances developed from concurrent chemoradiation and temozolomide treatment can be used to test different drug combinations with superior tumor killing potential (88).
CoC platforms have the potential to curtail the use of animal models as a complementary research tool. They can revolutionize the way of cancer research in terms of disease modeling, drug screening, and developing new therapeutic approaches specifically to prevent metastasis.
3D Cell Culture-Based Gene Expression Studies of Cancer
Gene expressions in the cells are susceptible to respond to even slight alterations in their microenvironment. Because of the physiological interactions with neighboring cells in 3D models, gene expression of cells is highly reproducible with minor modifications in 3D cultures (89). Spheroids of glioblastoma grown in 3D conditions are used in examining the different expressions of various genes, which play a vital role in tumor biosynthesis, metabolism, cell architecture, cellular transport, and signal transduction. It was observed that over-expression is analogous to in vivo, but not in the monolayer cultures. Kumar et al. (2008) (90) reported that when multiple genes of human neuroblastoma cells were screened through high-throughput screening methods, they revealed that the 3D matrix structural properties influence changes in gene expression. A microarray analysis of 1,766 genes featured the importance of neuroblastoma cells grown in 3D rather than as monolayers for studying diversity in gene expression (90). Similarly, Juillerat-Jeanneret et al. (2008) (91) reported a similarity in the heterogeneous gene expression of enzymes involved in detoxification and DNA repair, like O6-methylguanine-DNA-methyltransferase and glutathione-S-transferase, in 3D models of human glioblastoma cell line and in vivo (91). Such studies reveal that flexible 3D matrices should be optimized according to the application.
The advantage of gene expression measurements is that they provide insight into the biological mechanisms associated with the adaptation to a 3D environment (92). In in vitro studies, the adaptation of cells to a 3D environment is integral for maintaining transcriptional and translational functions. So that gene expression can match the in vivo level, Takahashi et al. (2015) (93) demonstrated that when hepatoma cell lines like HepG2 and Hepa RG were cultured using the hanging drop method, the expression of genes related to glucose, lipid, and drug metabolism was remarkably increased, the secretion of albumin and Apo B (Apolipoprotein B) was enhanced, and the mRNA levels of CYP (cytochrome P450) enzymes, on exposure to specific inducers, increased. While comparing the 2D and 3D culture models as drug-testing platforms in breast cancer, Imamura et al. (2015) (18) reported that the 3D model with the breast cancer cell line BT-474 had a lower level of caspase-3 than the 2D model, suggesting the anti-apoptotic nature of dense multi-cellular spheroid model of BT-474 cell line. Rodriguez et al. (2018) (94) reported that 3D architecture can modulate the breast cancer stem cell population and expression and distribution of HER2 receptors and create the environmental conditions to allow trastuzumab to exert its effect over the cells that are clinically defined as HER2-negative. Melissardou et al. (2019) (95) reported that when head and neck squamous cell carcinoma cell lines were cultured using ultra-low attachment plates, spheroids with varied sizes and densities were formed. All spheroids showed the upregulation of CDH1 (epithelial to mesenchymal transition-associated gene), NANOG, and SOX2 (stem cell markers) as compared to 2D cultures. This 3D model showed decreased sensitivity to cisplatin and cetuximab. When Fontoura et al. (2020) (96) compared the cell culture models of mouse melanoma (in vivo and cell line B16F10 GFP cells) using different scaffold-based techniques like electrospinning, solvent-casting particle-leaching, and Engelbreth–Holm swarm gel with 2D cultures, the microarray analysis of RNA showed the upregulation of lymphoid enhancer-binding factor 1 in 3D models than in 2D.
Role of 3D Cultures in Cancer Drug Discovery and Development
Drug discovery and development is a complex and lengthy process and generally involves the same trend of progression (97, 98). Cell-based assays have become an integral part of the drug discovery process, facilitating a fast, simple, and cost-effective tool and thereby minimizing cost-intensive and large-scale animal testing (28). Lack of efficacy is the main reason for the failure of about 65% of drugs in phase II and phase III clinical trials. About one-third of drugs fail due to safety issues and inadequate therapeutic index (99). Although conventional in vitro human cell culture platforms are economical and easy for screening experimental targets, they cannot imitate the complex and dynamic responses of human organs. Phenotype recreation of the target tissue in cultured cells is crucial for procuring reliable bio-medical data. Drug discovery and development researchers are interested in 3D culture systems because of their advantages in contributing to more anticipating and physiologically consistent data for in vivo tests (11). 3D cultures assist the initial drug discovery process starting from disease modeling for identifying the target and its validation, screening, lead selection, analysis of efficiency, and safety assessment (100). Among the various criteria for drug development, hepatotoxicity is an essential determining parameter for the approval, non-approval, or limitation in drug usage by the Food and Drug Association (101). Utilization of hepatocarcinoma cell line (HepG2) grown as monolayers has been the gold standard in the initial screening of drug candidates for hepatotoxicity activity; however, the spheroid culture of hepatocytes is fast replacing the 2D culture (102). Furthermore, the cells in this setup either wholly lack or express drastically low levels of transporters and many cytochrome P450s (CYPs), which are drug-metabolizing enzymes found in hepatocytes in vivo (103). When hepatocyte cultures are accomplished in bioreactors, they have been shown to imitate in vivo characteristics to such an extent that they can be utilized for the bio-fabrication of a functional artificial organ for transplantation (103). The structure and function of the mammary gland and/or steps involved in breast cancer progression have been studied in the in vitro 3D scaffolds that imitate the same. Silk fibroin hydrogels with good biocompatibility and processability can be engineered further to encourage normal tissue regeneration or to enhance the tumorigenicity of cancer cells.
On the other hand, decellularized ECM plays a significant role as a novel tissue-specific ECM material that matches the native tissue matrix (104, 105). Yi et al. (2016) developed a 3D printed polymeric patch loaded with 5-fluorouracil using an extrusion-based printing system made of PCL and poly(lactide-coglycolide). This resulted in sustained drug release and significantly reduced pancreatic tumor in a rabbit model (106).
Cells in 3D culture typically display decreased sensitivity to chemotherapeutic agents when compared to cells in 2D culture (107). The benefits of using 3D cultures in drug discovery include cell–cell contact, communication, and signaling pathway activation due to the ECM components of matrices. In addition, 3D culture helps in the morphological and functional differentiation of cells (108). 3D culture contributes to in vitro models for building multi-cellular systems. It covers the divergence between in vitro and in vivo drug screening, possibly reducing animal trials.
Limitations and Challenges of 3D Cell Culture
In spite of the continued effort and considerable success in making biomimetic 3D tumor models, major challenges and impediments still accompany the existing models. Several 3D models are generated using the longstanding 2D adapted cancer cell lines, which might not exactly replicate the pathology of the disease (109). Ex vivo culturing of patient cells provides access to a high level of accuracy in finding the biological insights of metastasis and recognizing the susceptibility to drugs targeting the pathways in a correct manner (110). The remodeling of the complex vascular system associated with tumor in vitro is another important challenge necessary to be addressed because cancer progression is influenced by the abnormal blood vessels and greatly affect drug transport within the tumor tissues. A quantitative understanding of the movement of tumor cell aggregates in a cancer microenvironment is needed. Cancer cells are generally trapped at a single-cell state, proliferate within the matrix, and simultaneously migrate through mesenchymal or amoeboid movement, which is vital for forming tumor cell aggregates (111). Cancer biologists and tissue engineers are required to team up to create an optimized 3D in vitro model of drug testing and to make progress in this field. Systems biology, computational modeling approaches, detection and analysis modalities, and in situ real-time imaging, are severely required for a better understanding of 3D tumor models. These advancements are expected to lead to better insights into the largely illusory aspects of cancers.
Differences in the biologically derived matrices might generate non-reproducible results. High-throughput assays and large-scale studies are expensive. Within the same flask, spheroids of varying sizes may be produced. 3D models do not contain vasculature, which is important in drug delivery. The three-dimensional cultures must benefit the drug discovery and clinical research as it generates physiologically relevant screening assays (112). The national institutes of health, hospitals, and university laboratories are shifting their emphasis on identifying culture models with clinical potential.
Conclusion
Cell culture has been the elemental tool to unveil the biochemical mechanisms underlying the assembly of cells into tissues, organs and, finally, an organism. Conventional cell culture models have played a significant role in achieving so, but they fail to exactly translate the natural in vivo setting. The process of carcinogenesis is not revealed entirely because implementing the culture techniques could not recreate the milieu of the tumor and its TME. 3D cultures can explore new ways to understand the biological processes of various diseases, mainly cancer. Bioengineered 3D tumor models have the potential to reveal the whole scenery of cancer and can help in developing new therapeutic approaches.
Author Contributions
Conceptualization: APF and RR. Writing—original draft preparation: KP, SS, and MH. Writing—review and editing: MH, APF, and RR. Resources: VPV, UM, and KMS. Funding acquisition: MAE, ME-S, SMA, and ABAH. All authors contributed to the article and approved the submitted version.
Funding
This publication was supported by AlMaarefa University Researchers Supporting Program (grant number: MA-006), AlMaarefa University, Riyadh, Saudi Arabia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Andersen T, Auk-Emblem P, Dornish M. 3d Cell Culture in Alginate Hydrogels. Microarrays (2015) 4:133–61. doi: 10.3390/microarrays4020133
2. Lucey BP, Nelson-Rees WA, Hutchins GM. Henrietta Lacks, HeLa Cells, and Cell Culture Contamination. Arch Pathol Lab Med (2009) 133:1463–7. doi: 10.5858/133.9.1463
3. Khoruzhenko AI. 2D- and 3D-Cell Culture. Biopolymers Cell (2011) 27:17–24. doi: 10.7124/bc.00007D
4. Koledova Z. 3d Cell Culture - Methods and Protocols. New York. U.S.A.: Springer Nature (2017). doi: 10.1007/978-1-4939-7021-6
5. Justice BA, Badr NA, Felder RA. 3D Cell Culture Opens New Dimensions in Cell-Based Assays. Drug Discov Today (2009) 14:102–7. doi: 10.1016/j.drudis.2008.11.006
6. Caliari SR, Burdick JA. A Practical Guide to Hydrogels for Cell Culture. Nat Methods (2016) 13:405–14. doi: 10.1038/nmeth.3839
7. Rebelo SP, Pinto C, Martins TR, Harrer N, Estrada MF, Loza-Alvarez P, et al. 3D-3-Culture: A Tool to Unveil Macrophage Plasticity in the Tumour Microenvironment. Biomaterials (2018) 163:185–97. doi: 10.1016/j.biomaterials.2018.02.030
8. Caló E, Khutoryanskiy VV. Biomedical Applications of Hydrogels: A Review of Patents and Commercial Products. Eur Polymer J (2015) 65:252–67. doi: 10.1016/j.eurpolymj.2014.11.024
9. Speroni L, Sweeney MF, Sonnenschein C, Soto AM. A Hormone-Responsive 3D Culture Model of the Human Mammary Gland Epithelium. J Visualized Experiments (2016) 2016:e53098. doi: 10.3791/53098
10. Hoarau-Véchot J, Rafii A, Touboul C, Pasquier J. Halfway Between 2D and Animal Models: Are 3D Cultures the Ideal Tool to Study Cancer-Microenvironment Interactions? Int J Mol Sci (2018) 19:181. doi: 10.3390/ijms19010181
11. Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay Drug Dev Technol (2014) 12:207–18. doi: 10.1089/adt.2014.573
12. Thiele J, Ma Y, Bruekers SMC, Ma S, Huck WTS. 25th Anniversary Article: Designer Hydrogels for Cell Cultures: A Materials Selection Guide. Advanced Mater (2014) 26:125–48. doi: 10.1002/adma.201302958
13. Yamada KM, Sixt M. Mechanisms of 3D Cell Migration. Nat Rev Mol Cell Biol (2019) 20:738–52. doi: 10.1038/s41580-019-0172-9
14. Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, et al. Effects of Substrate Stiffness on Cell Morphology, Cytoskeletal Structure, and Adhesion. Cell Motil Cytoskeleton (2005) 60:24–34. doi: 10.1002/cm.20041
15. van der Hee B, Loonen LMP, Taverne N, Taverne-Thiele JJ, Smidt H, Wells JM. Optimized Procedures for Generating an Enhanced, Near Physiological 2D Culture System From Porcine Intestinal Organoids. Stem Cell Res (2018) 28:165–71. doi: 10.1016/j.scr.2018.02.013
16. Griffith LG, Swartz MA. Capturing Complex 3D Tissue Physiology In Vitro. Nat Rev Mol Cell Biol (2006) 7:211–24. doi: 10.1038/nrm1858
17. Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking Cell-Matrix Adhesions to the Third Dimension. Sci (2001) 294:1708–12. doi: 10.1126/science.1064829
18. Imamura Y, Mukohara T, Shimono Y, Funakoshi Y, Chayahara N, Toyoda M, et al. Comparison of 2D- and 3D-Culture Models as Drug-Testing Platforms in Breast Cancer. Oncol Rep (2015) 33:1837–43. doi: 10.3892/or.2015.3767
19. Nath S, Devi GR. Three-Dimensional Culture Systems in Cancer Research: Focus on Tumor Spheroid Model. Pharmacol Ther (2016) 163:94–108. doi: 10.1016/j.pharmthera.2016.03.013
20. Schmitz A, Fischer SC, Mattheyer C, Pampaloni F, Stelzer EHK. Multiscale Image Analysis Reveals Structural Heterogeneity of the Cell Microenvironment in Homotypic Spheroids. Sci Rep (2017) 7:1–13. doi: 10.1038/srep43693
21. Laschke MW, Menger MD. Life is 3D: Boosting Spheroid Function for Tissue Engineering. Trends Biotechnol (2017) 35:133–44. doi: 10.1016/j.tibtech.2016.08.004
22. Qu W, Zhao Y, Wang X, Qi Y, Zhou C, Hua Y, et al. Culture Characters, Genetic Background, Estrogen/Progesterone Receptor Expression, and Tumorigenic Activities of Frequently Used Sixteen Endometrial Cancer Cell Lines. Clin Chim Acta (2019) 489:225–32. doi: 10.1016/j.cca.2018.08.013
23. Duggal S, Frønsdal KB, Szöke K, Shahdadfar A, Melvik JE, Brinchmann JE. Phenotype and Gene Expression of Human Mesenchymal Stem Cells in Alginate Scaffolds. Tissue Eng Part A (2009) 15:1763–73. doi: 10.1089/ten.tea.2008.0306
24. Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA, Rizzi SC. Bioengineered 3D Platform to Explore Cell-ECM Interactions and Drug Resistance of Epithelial Ovarian Cancer Cells. Biomaterials (2010) 31:8494–506. doi: 10.1016/j.biomaterials.2010.07.064
25. Duval K, Grover H, Han LH, Mou Y, Pegoraro AF, Fredberg J, et al. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology (2017) 32:266–77. doi: 10.1152/physiol.00036.2016
26. Gattazzo F, Urciuolo A, Bonaldo P. Extracellular Matrix: A Dynamic Microenvironment for Stem Cell Niche. Biochim Biophys Acta Gen Subj (2014) 1840:2506–19. doi: 10.1016/j.bbagen.2014.01.010
27. Young JL, Holle AW, Spatz JP. Nanoscale and Mechanical Properties of the Physiological Cell-ECM Microenvironment. Exp Cell Res (2016) 343:3–6. doi: 10.1016/j.yexcr.2015.10.037
28. Langhans SA. Three-Dimensional In Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front Pharmacol (2018) 9:6. doi: 10.3389/fphar.2018.00006
29. Frantz C, Stewart KM, Weaver VM. The Extracellular Matrix at a Glance. J Cell Sci (2010) 123:4195–200. doi: 10.1242/jcs.023820
30. Nagano M, Hoshino D, Koshikawa N, Akizawa T, Seiki M. Turnover of Focal Adhesions and Cancer Cell Migration. Int J Cell Biol (2012) 2012:310616. doi: 10.1155/2012/310616
31. Candiello J, Balasubramani M, Schreiber EM, Cole GJ, Mayer U, Halfter W, et al. Biomechanical Properties of Native Basement Membranes. FEBS J (2007) 274:2897–908. doi: 10.1111/j.1742-4658.2007.05823.x
32. Rozario T, DeSimone DW. The Extracellular Matrix in Development and Morphogenesis: A Dynamic View. Dev Biol (2010) 341:126–40. doi: 10.1016/j.ydbio.2009.10.026
33. Clause KC, Barker TH. Extracellular Matrix Signaling in Morphogenesis and Repair. Curr Opin Biotechnol (2013) 24:830–3. doi: 10.1016/j.copbio.2013.04.011
34. Lutolf MP, Hubbell JA. Synthetic Biomaterials as Instructive Extracellular Microenvironments for Morphogenesis in Tissue Engineering. Nat Biotechnol (2005) 23:47–55. doi: 10.1038/nbt1055
35. DeRoock IB, Pennington ME, Sroka TC, Lam KS, Bowden GT, Bair EL, et al. Synthetic Peptides Inhibit Adhesion of Human Tumor Cells to Extracellular Matrix Proteins. Cancer Res (2001) 61:3308–13.
36. Luca AC, Mersch S, Deenen R, Schmidt S, Messner I, Schäfer KL, et al. Impact of the 3D Microenvironment on Phenotype, Gene Expression, and EGFR Inhibition of Colorectal Cancer Cell Lines. PloS One (2013) 8:59689. doi: 10.1371/journal.pone.0059689
37. Kiss DL, Windus LCE, Avery VM. Chemokine Receptor Expression on Integrin-Mediated Stellate Projections of Prostate Cancer Cells in 3D Culture. Cytokine (2013) 64:122–30. doi: 10.1016/j.cyto.2013.07.012
38. Bilgin CC, Kim S, Leung E, Chang H, Parvin B. Integrated Profiling of Three Dimensional Cell Culture Models and 3D Microscopy. Bioinformatics (2013) 29:3087–93. doi: 10.1093/bioinformatics/btt535
39. Pimentel CR, Ko SK, Caviglia C, Wolff A, Emnéus J, Keller SS, et al. Three-Dimensional Fabrication of Thick and Densely Populated Soft Constructs With Complex and Actively Perfused Channel Network. Acta Biomater (2018) 65:174–84. doi: 10.1016/j.actbio.2017.10.047
40. Laib AM, Bartol A, Alajati A, Korff T, Weber H, Augustin HG. Spheroid-Based Human Endothelial Cell Microvessel Formation In Vivo. Nat Protoc (2009) 4:1202–15. doi: 10.1038/nprot.2009.96
41. Ware MJ, Colbert K, Keshishian V, Ho J, Corr SJ, Curley SA, et al. Generation of Homogenous Three-Dimensional Pancreatic Cancer Cell Spheroids Using an Improved Hanging Drop Technique. Tissue Eng Part C: Methods (2016) 22:312–21. doi: 10.1089/ten.tec.2015.0280
42. Santo VE, Estrada MF, Rebelo SP, Abreu S, Silva I, Pinto C, et al. Adaptable Stirred-Tank Culture Strategies for Large Scale Production of Multi-CellularSpheroid-Based Tumor Cell Models. J Biotechnol (2016) 221:118-29. doi: 10.1016/j.jbiotec.2016.01.031
43. Zhong Q, Ding H, Gao B, He Z, Gu Z. Advances of Microfluidics in Biomedical Engineering. Advanced Mater Technol (2019) 4:1800663. doi: 10.1002/admt.201800663
44. Anil-Inevi M, Yaman S, Yildiz AA, Mese G, Yalcin-Ozuysal O, Tekin HC, et al. Biofabrication of in Situ Self Assembled 3d Cell Cultures in a Weightlessness Environment Generated Using Magnetic Levitation. Sci Rep (2018) 8:7239. doi: 10.1038/s41598-018-25718-9
45. Madoux F, Tanner A, Vessels M, Willetts L, Hou S, Scampavia L, et al. A 1536-Well 3d Viability Assay to Assess the Cytotoxic Effect of Drugs on Spheroids. SLAS Discov (2017) 22:516-24. doi: 10.1177/2472555216686308
46. Klotz BJ, Gawlitta D, Rosenberg AJWP, Malda J, Melchels FPW. Gelatin-Methacryloyl Hydrogels: Towards Biofabrication-Based Tissue Repair. Trends Biotechnol (2016) 34:394–407. doi: 10.1016/j.tibtech.2016.01.002
47. Ruedinger F, Lavrentieva A, Blume C, Pepelanova I, Scheper T. Hydrogels for 3D Mammalian Cell Culture: A Starting Guide for Laboratory Practice. Appl Microbiol Biotechnol (2015) 99:623–36. doi: 10.1007/s00253-014-6253-y
48. Ryan SL, Baird AM, Vaz G, Urquhart AJ, Senge M, Richard DJ, et al. Drug Discovery Approaches Utilizing Three-Dimensional Cell Culture. Assay Drug Dev Technol (2016) 14:19–28. doi: 10.1089/adt.2015.670
49. Cancer IA for R on. Latest Global Cancer Data: Cancer Burden Rises to 18.1 Million New Cases and 9.6 Million Cancer Deaths in 2018. Press Release (2018).
50. Liu Y, Cao X. Characteristics and Significance of the Pre-Metastatic Niche. Cancer Cell (2016) 30:668–81. doi: 10.1016/j.ccell.2016.09.011
51. Quail DF, Joyce JA. Microenvironmental Regulation of Tumor Progression and Metastasis. Nat Med (2013) 19:1423–37. doi: 10.1038/nm.3394
52. Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell (2015) 162:1229–41. doi: 10.1016/j.cell.2015.08.016
53. Pasquale EB. Eph Receptors and Ephrins in Cancer: Bidirectional Signalling and Beyond. Nat Rev Cancer (2010) 10:165–80. doi: 10.1038/nrc2806
54. Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular Principles of Metastasis: A Hallmark of Cancer Revisited. Signal Transduction Targeted Ther (2020) 5:1–17. doi: 10.1038/s41392-020-0134-x
55. Tanner K, Gottesman MM. Beyond 3D Culture Models of Cancer. Sci Trans Med (2015) 7:283ps9–9. doi: 10.1126/scitranslmed.3009367
56. Lv D, Hu Z, Lu L, Lu H, Xu X. Three-Dimensional Cell Culture: A Powerful Tool in Tumor Research and Drug Discovery. Oncol Lett (2017) 14:6999–7010. doi: 10.3892/ol.2017.7134
57. Tibbitt MW, Anseth KS. Hydrogels as Extracellular Matrix Mimics for 3D Cell Culture. Biotechnol Bioeng (2009) 103:655–63. doi: 10.1002/bit.22361
58. Christofori G. Changing Neighbours, Changing Behaviour: Cell Adhesion Molecule-Mediated Signalling During Tumour Progression. EMBO J (2003) 22:2318–23. doi: 10.1093/emboj/cdg228
59. Mitra A, Mishra L, Li S. Technologies for Deriving Primary Tumor Cells for Use in Personalized Cancer Therapy. Trends Biotechnol (2013) 31:347–54. doi: 10.1016/j.tibtech.2013.03.006
60. Xu X, Farach-Carson MC, Jia X. Three-Dimensional In Vitro Tumor Models for Cancer Research and Drug Evaluation. Biotechnol Adv (2014) 32:1256–68. doi: 10.1016/j.biotechadv.2014.07.009
61. Breslin S, O’Driscoll L. The Relevance of Using 3D Cell Cultures, in Additionto 2D Monolayer Cultures, When Evaluating Breast Cancer Drug Sensitivityand Resistance. Oncotarget (2016) 7:45745–56. doi: 10.18632/oncotarget.9935
62. Deweerd ES, Hoffman-kim D, Ph D. Genomic and Morphological Changes of Neuroblastoma Cells in Response to Three-Dimensional Matrices. Tissue Eng (2007) 13:1035-47. doi: 10.1089/ten.2006.0251
63. Mcgrath J, Panzica L, Ransom R, Withers HG, Gelman IH. Identification of Genes Regulating Breast Cancer Dormancy in 3D Bone Endosteal Niche Cultures. Mol Cancer Res (2019) 17:860–70. doi: 10.1158/1541-7786.MCR-18-0956
64. Rogers M, Sobolik T, Schaffer DK, Samson PC, Johnson AC, Owens P, et al. Engineered Microfluidic Bioreactor for Examining the Three-Dimensional Breast Tumor Microenvironment. Biomicrofluidics (2018) 12:034102 doi: 10.1063/1.5016433
65. Tsai S, Mcolash L, Palen K, Johnson B, Duris C, Yang Q, et al. Development of Primary Human Pancreatic Cancer Organoids, Matched Stromal and Immune Cells and 3D Tumor Microenvironment Models. BMC Cancer (2018) 18:335. doi: 10.1186/s12885-018-4238-4
66. Lazzari G, Nicolas V, Matsusaki M, Akashi M, Couvreur P, Mura S. Multicellular Spheroid Based on a Triple Co-Culture: A Novel 3D Model to Mimic Pancreatic Tumor Complexity. Acta Biomater (2018) 78:296–307. doi: 10.1016/j.actbio.2018.08.008
67. Fontana F, Raimondi M, Marzagalli M, Sommariva M, Limonta P, Gagliano N. Epithelial-To-Mesenchymal Transition Markers and CD44 Isoforms Are Differently Expressed in 2D and 3D Cell Cultures of Prostate Cancer Cells. Cells (2019) 8:143. doi: 10.3390/cells8020143
68. Jakubikova J, Cholujova D, Hideshima T, Gronesova P, Szalat E, Richardson PG, et al. A Novel 3D Mesenchymal Stem Cell Model of the Multiple Myeloma Bone Marrow Niche : Biologic and Clinical Applications. Oncotarget (2016) 7:77326–41. doi: 10.18632/oncotarget.12643
69. Petrikaite V. Differences of Statin Activity in 2D and 3D Pancreatic Cancer Cell Cultures. Drug Des Devel Ther (2017), 3273–80. doi: 10.2147/DDDT.S149411
70. Zhu W, Holmes B, Glazer RI, Zhang LG. 3D Printed Nanocomposite Matrix for the Study of Breast Cancer Bone Metastasis. Nanomed: Nanotechnol Biol Med (2016) 12:69–79. doi: 10.1016/j.nano.2015.09.010
71. Hübner J, Raschke M, Rütschle I, Gräßle S, Hasenberg T, Schirr K, et al. Simultaneous Evaluation of Anti- EGFR-Induced Tumour and Adverse Skin Effects in a Microfluidic Human 3D Co-Culture Model. Sci Rep (2018) 8:15010. doi: 10.1038/s41598-018-33462-3
72. Mishra DK, Sakamoto JH, Thrall MJ, Baird BN, Blackmon SH, Ferrari M, et al. Human Lung Cancer Cells Grown in an Ex Vivo 3d Lung Model Produce Matrix Metalloproteinases Not Produced in 2D Culture. PLoS One (2012) 7:e45308.. doi: 10.1371/journal.pone.0045308
73. Mazzocchi A, Devarasetty M, Herberg S, Petty WJ, Miller L, Kucera G, et al. Pleural Effusion Aspirate for Use in 3D Lung Cancer Modeling and Chemotherapy Screening. ACS Biomater Sci Eng (2019) 5:1937–43. doi: 10.1021/acsbiomaterials.8b01356
74. Deepa K, Dorayappan P, Gardner ML, Hisey CL, Roman A, Smith BQ, et al. A Microfluidic Chip Enables Isolation of Exosomes and Establishment of Their Protein Profiles and Associated Signaling Pathways in Ovarian Cancer. Cancer Res (2020) 79:3503–13. doi: 10.1158/0008-5472.CAN-18-3538
75. Rashidi MRW, Mehta P, Bregenzer M, Raghavan S, Ravikumar V, Brady S. Engineered 3d Model of Cancer Stem Cell Enrichment and Chemoresistance. Neoplasia (2019) 21:822–36. doi: 10.1016/j.neo.2019.06.005
77. Peela N, Truong D, Saini H, Chu H, Mashaghi S, Ham SL, et al. Advanced Biomaterials and Microengineering Technologies to Recapitulate the Stepwise Process of Cancer Metastasis. Biomaterials (2017) 133:176–207. doi: 10.1016/j.biomaterials.2017.04.017
78. Fidler IJ, Kripke ML. The Challenge of Targeting Metastasis. Cancer Metastasis Rev (2015) 34:635–41. doi: 10.1007/s10555-015-9586-9
79. Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, et al. A Microfluidic 3D Invitro Model for Specificity of Breast Cancer Metastasis to Bone. Biomaterials (2014) 35:2454–61. doi: 10.1016/j.biomaterials.2013.11.050
80. Sontheimer-Phelps A, Hassell BA, Ingber DE. Modelling Cancer in Microfluidic Human Organs-on-Chips. Nat Rev Cancer (2019) 19:65–81. doi: 10.1038/s41568-018-0104-6
81. Wong AD, Searson PC. Live-Cell Imaging of Invasion and Intravasation in an Artificial Microvessel Platform. Cancer Res (2014) 74:4937–45. doi: 10.1158/0008-5472.CAN-14-1042
82. Xie M, Gao Q, Fu J, Chen Z, He Y. Bioprinting of Novel 3D Tumor Array Chip for Drug Screening. Bio-Design Manufacturing (2020) 3:175–88. doi: 10.1007/s42242-020-00078-4
83. Montanez-Sauri SI, Sung KE, Berthier E, Beebe DJ. Enabling Screening in 3D Microenvironments: Probing Matrix and Stromal Effects on the Morphology and Proliferation of T47D Breast Carcinoma Cells. Integr Biol (2013) 5:631–40. doi: 10.1039/C3IB20225A
84. Regier MC, Maccoux LJ, Weinberger EM, Regehr KJ, Berry SM, Beebe DJ, et al. Transitions From Mono- to Co- to Tri-Culture Uniquely Affect Gene Expression in Breast Cancer, Stromal, and Immune Compartments. BioMed Microdevices (2016) 18:70. doi: 10.1007/S10544-016-0083-X
85. Wang X, Phan DTT, Sobrino A, George SC, Hughes CCW, Lee AP. Engineering Anastomosis Between Living Capillary Networks and Endothelial Cell-Lined Microfluidic Channels. Lab Chip (2016) 16:282–90. doi: 10.1039/C5LC01050K
86. Miller CP, Tsuchida C, Zheng Y, Himmelfarb J, Akilesh S. A 3d Human Renal Cell Carcinoma-On-a-Chip for the Study of Tumor Angiogenesis. Neoplasia (2018) 20:610–20. doi: 10.1016/J.NEO.2018.02.011
87. Wu X, Newbold MA, Haynes CL. Recapitulation of In Vivo-Like Neutrophil Transendothelial Migration Using a Microfluidic Platform. Analyst (2015) 140:5055–64. doi: 10.1039/C5AN00967G
88. Yi HG, Jeong YH, Kim Y, Choi YJ, Moon HE, Park SH, et al. A Bioprinted Human-Glioblastoma-on-a-Chip for the Identification of Patient-Specific Responses to Chemoradiotherapy. Nat Biomed Eng (2019) 3:509–19. doi: 10.1038/s41551-019-0363-x
89. Amirghasemi F, Adjei-Sowah E, Pockaj BA, Nikkhah M. Microengineered 3d Tumor Models for Anti-Cancer Drug Discovery in Female-Related Cancers. Ann Biomed Eng (2021) 49:1943-72. doi: 10.1007/s10439-020-02704-9
90. Kumar HR, Zhong X, Hoelz DJ, Rescorla FJ, Hickey RJ, Malkas LH, et al. Three-Dimensional Neuroblastoma Cell Culture: Proteomic Analysis Between Monolayer and Multi-Cellular Tumor Spheroids. Pediatr Surg Int (2008) 24:1229–34. doi: 10.1007/s00383-008-2245-2
91. Juillerat-Jeanneret L, Bernasconi CC, Bricod C, Gros S, Trepey S, Benhattar J, et al. Heterogeneity of Human Glioblastoma: Glutathione-S-Transferase and Methylguanine-Methyltransferase. Cancer Invest (2008) 26:597–609. doi: 10.1080/07357900802072913
92. Smolina M, Goormaghtigh E. Gene Expression Data and FTIR Spectra Provide a Similar Phenotypic Description of Breast Cancer Cell Lines in 2D and 3D Cultures. Analyst (2018) 143:2520–30. doi: 10.1039/c8an00145f
93. Takahashi Y, Hori Y, Yamamoto T, Urashima T, Ohara Y, Tanaka H. 3D Spheroid Cultures Improve the Metabolic Gene Expression Profiles of HepaRG Cells. Biosci Rep (2015) 35:1–7. doi: 10.1042/BSR20150034
94. Rodríguez CE, Berardi DE, Abrigo M, Todaro LB, Bal de Kier Joffé ED, Fiszman GL. Breast Cancer Stem Cells are Involved in Trastuzumab Resistance Through the HER2 Modulation in 3D Culture. J Cell Biochem (2018) 119:1381–91. doi: 10.1002/jcb.26298
95. Melissaridou S, Wiechec E, Magan M, Jain MV, Chung MK, Farnebo L, et al. The Effect of 2D and 3D Cell Cultures on Treatment Response, EMT Profile and Stem Cell Features in Head and Neck Cancer. Cancer Cell Int (2019) 19:16. doi: 10.1186/s12935-019-0733-1
96. Fontoura JC, Viezzer C, dos Santos FG, Ligabue RA, Weinlich R, Puga RD, et al. Comparison of 2D and 3D Cell Culture Models for Cell Growth, Gene Expression and Drug Resistance. Mater Sci Eng C (2020) 107:110264. doi: 10.1016/j.msec.2019.110264
97. Breslin S, O’Driscoll L. Three-Dimensional Cell Culture: The Missing Link in Drug Discovery. Drug Discov Today (2013) 18:240–9. doi: 10.1016/j.drudis.2012.10.003
98. Fang Y, Eglen RM. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov (2017) 22:456–72. doi: 10.1177/1087057117696795
99. Thiers FA, Sinskey AJ, Berndt ER. Trends in the Globalization of Clinical Trials. Nat Rev Drug Discov (2008) 7:13–4. doi: 10.1038/nrd2441
100. Pampaloni F, Reynaud EG, Stelzer EHK. The Third Dimension Bridges the Gap Between Cell Culture and Live Tissue. Nat Rev Mol Cell Biol (2007) 8:839–45. doi: 10.1038/nrm2236
101. Thoma CR, Zimmermann M, Agarkova I, Kelm JM, Krek W. 3D Cell Culture Systems Modeling Tumor Growth Determinants in Cancer Target Discovery. Advanced Drug Deliv Rev (2014) 69–70:29–41. doi: 10.1016/j.addr.2014.03.001
102. Dash A, Inman W, Hoffmaster K, Sevidal S, Kelly J, Obach RS, et al. Liver Tissue Engineering in the Evaluation of Drug Safety. Expert Opin Drug Metab Toxicol (2009) 5:1159–74. doi: 10.1517/17425250903160664
103. Shin DS, Seo H, Yang JY, Joo J, Im SH, Kim SS, et al. Quantitative Evaluation of Cytochrome P450 3a4 Inhibition and Hepatotoxicity in HepaRG 3-D Spheroids. Int J Toxicol (2018) 37:393–403. doi: 10.1177/1091581818780149
104. Maghdouri-White Y, Bowlin GL, Lemmon CA, Dréau D. Bioengineered Silk Scaffolds in 3D Tissue Modeling With Focus on Mammary Tissues. Mater Sci Engineering: C (2016) 59:1168–80. doi: 10.1016/J.MSEC.2015.10.007
105. Yi H-G, Kim H, Kwon J, Choi Y-J, Jang J, Cho D-W. Application of 3D Bioprinting in the Prevention and the Therapy for Human Diseases. Signal Transduction Targeted Ther (2021) 6:77. doi: 10.1038/s41392-021-00566-8
106. Yi HG, Choi YJ, Kang KS, Hong JM, Pati RG, Park MN, et al. A 3D-Printed Local Drug Delivery Patch for Pancreatic Cancer Growth Suppression. J Controlled Release (2016) 238:231–41. doi: 10.1016/J.JCONREL.2016.06.015
107. Lai Y, Asthana A, Kisaalita WS. Biomarkers for Simplifying HTS 3D Cell Culture Platforms for Drug Discovery: The Case for Cytokines. Drug Discovery Today (2011) 16:293–7. doi: 10.1016/j.drudis.2011.01.009
108. Benton G, Arnaoutova I, George J, Kleinman HK, Koblinski J. Matrigel: From Discovery and ECM Mimicry to Assays and Models for Cancer Research. Advanced Drug Deliv Rev (2014) 79:3–18. doi: 10.1016/j.addr.2014.06.005
109. Horvath P, Aulner N, Bickle M, Davies AM, Del NE, Ebner D, et al. Screening Out Irrelevant Cell-Based Models of Disease. Nat Rev Drug Discov (2016) 15:751–69. doi: 10.1038/nrd.2016.175
110. Clark AM, Ma B, Taylor DL, Griffith L, Wells A. Liver Metastases: Microenvironments and Ex-Vivo Models. Exp Biol Med (2016) 241:1639–52. doi: 10.1177/1535370216658144
111. van Duinen V, Zhu D, Ramakers C, van Zonneveld AJ, Vulto P, Hankemeier T. Perfused 3D Angiogenic Sprouting in a High-Throughput In Vitro Platform. Angiogenesis (2019) 22:157–65. doi: 10.1007/s10456-018-9647-0
Keywords: 3D culture, biomimetic, microenvironment, cancer, gene expression, drug discovery
Citation: Poornima K, Francis AP, Hoda M, Eladl MA, Subramanian S, Veeraraghavan VP, El-Sherbiny M, Asseri SM, Hussamuldin ABA, Surapaneni KM, Mony U and Rajagopalan R (2022) Implications of Three-Dimensional Cell Culture in Cancer Therapeutic Research. Front. Oncol. 12:891673. doi: 10.3389/fonc.2022.891673
Received: 08 March 2022; Accepted: 11 April 2022;
Published: 12 May 2022.
Edited by:
Shanmugasundaram Ganapathy-Kanniappan, Johns Hopkins Medicine, United StatesReviewed by:
Hilary Ann Kenny, The University of Chicago, United StatesHee-Gyeong Yi, Chonnam National University, South Korea
Copyright © 2022 Poornima, Francis, Hoda, Eladl, Subramanian, Veeraraghavan, El-Sherbiny, Asseri, Hussamuldin, Surapaneni, Mony and Rajagopalan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rukkumani Rajagopalan, cnVrczJrMkBnbWFpbC5jb20=; cnVra3VtYW5pLmJibUBwb25kaXVuaS5lZHUuaW4=
 Kolluri Poornima1
Kolluri Poornima1 Muddasarul Hoda
Muddasarul Hoda Mohamed Ahmed Eladl
Mohamed Ahmed Eladl Vishnu Priya Veeraraghavan
Vishnu Priya Veeraraghavan Mohamed El-Sherbiny
Mohamed El-Sherbiny Krishna Mohan Surapaneni
Krishna Mohan Surapaneni Rukkumani Rajagopalan
Rukkumani Rajagopalan