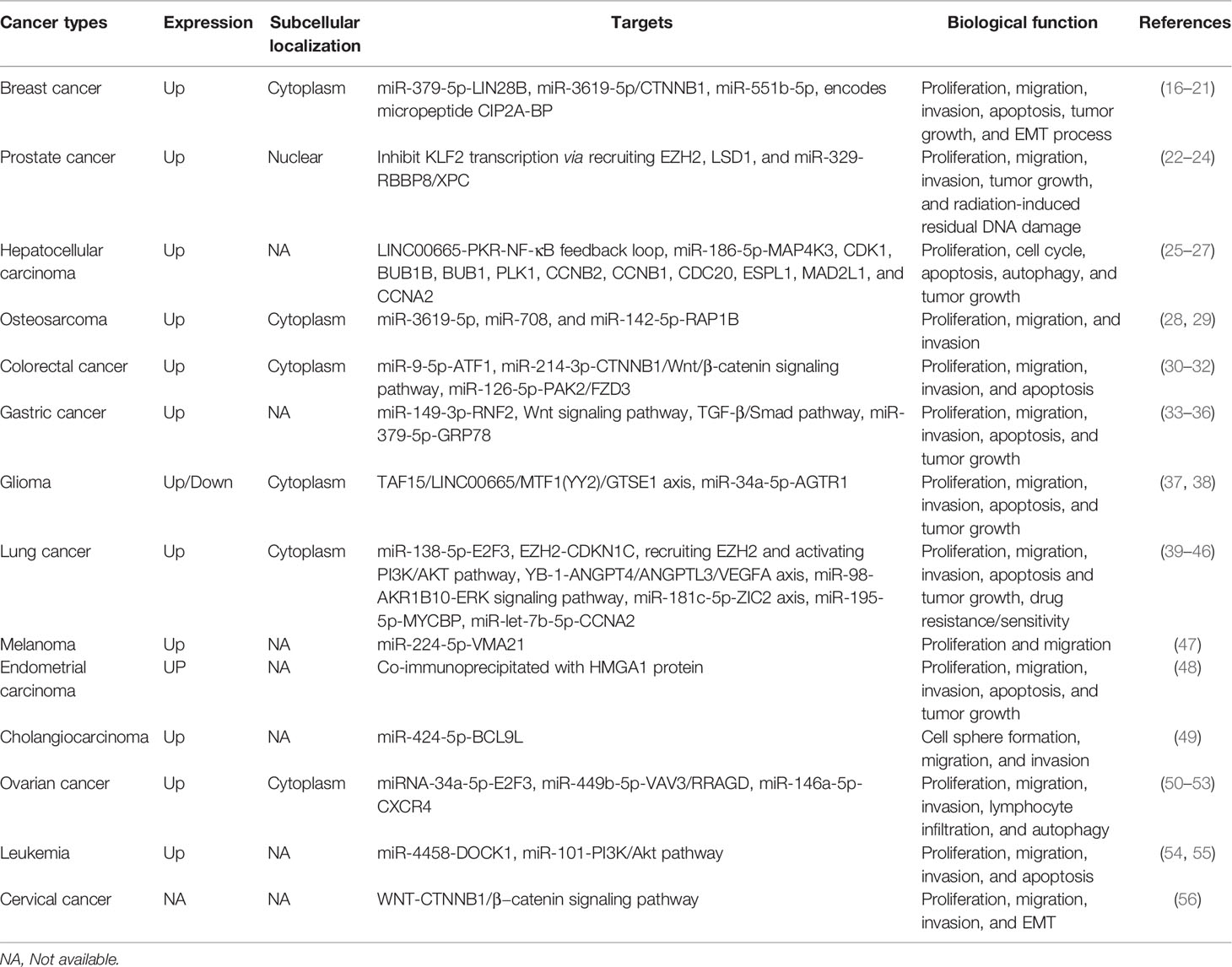
- Department of Oncology, The Affiliated Cancer Hospital of Zhengzhou University and Henan Cancer Hospital, Zhengzhou, China
Long non-coding RNAs (lncRNAs) are more than 200 nucleotides in length and are implicated in the development of human cancers, without protein-coding function. Mounting evidence indicates that cancer initiation and progression are triggered by lncRNA dysregulation. Recently, a growing number of studies have found that LINC00665, a long intergenic non-protein coding RNA, may be associated with various cancers, including gastrointestinal tumors, gynecological tumors, and respiratory neoplasms. LINC00665 was reported to be significantly dysregulated in cancers and has an important clinical association. It participates in cell proliferation, migration, invasion, and apoptosis through different biological pathways. In this review, we summarize the current findings on LINC00665, including its biological roles and molecular mechanisms in various cancers. LINC00665 may be a potential prognostic biomarker and novel therapeutic target for cancers.
Introduction
With an increase in aging and population growth, the incidence and mortality of human cancers are growing rapidly worldwide (1). According to statistical estimations from 185 countries, there were more than 18 million new cancer cases and almost 10 million cancer-related deaths in 2018 (1). Although numerous treatment strategies for cancers have been developed and improved, including classical therapies (surgery, radiotherapy, and chemotherapy) and immune and molecular targeted therapies, the therapeutic effects remain unsatisfactory (2, 3). Thus, there is an urgent need to explore valuable targets and novel biomarkers for cancer therapy, diagnosis, and prognostic evaluation.
Long non-coding RNAs (lncRNAs), a class of linear RNA molecules with a length greater than 200 nt, have no protein-coding ability, and their average expression and abundance are lower than that of mRNA (4, 5). Mounting studies have found that lncRNAs were dysregulated in human diseases, especially in cancers, and could act as mediators in tumorigenesis and metastasis. LncRNAs have a wide subcellular distribution in cells, which determines the diversity of their functional mechanisms (6, 7). For example, lncRNAs located in the cytoplasm can regulate mRNA stability as competing endogenous RNAs (ceRNAs) through sponging specific miRNAs (8–10). In the nucleus, the lncRNA PVT1 could increase MYC stability and its expression level in cancers by interfering with the phosphorylation of MYC at Thr58 (11). Furthermore, emerging evidence has shown that lncRNAs might be valuable diagnostic and prognostic biomarkers or therapeutic targets for cancers (12–15). Although lots of lncRNAs have been identified in cancers, the biological function of several remains unclear.
Long intergenic non-protein coding RNA 665 (LINC00665), also known as CIP2A-BP, is a novel lncRNA that is dysregulated in different human cancers. Recent studies have found that LINC00665 expression was significantly upregulated in most cancers and could be used as a valuable diagnostic, prognostic, and therapeutic target. LINC00665 plays an oncogenic role in cancer cell proliferation, migration, and invasion through various molecular mechanisms. All these findings indicate that LINC00665 has a key function in cancer. The present review summarizes current findings of LINC00665 in tumorigenesis and progression, including aberrant expression, clinical value, and molecular mechanism (Tables 1, 2).
Characterization of LINC00665
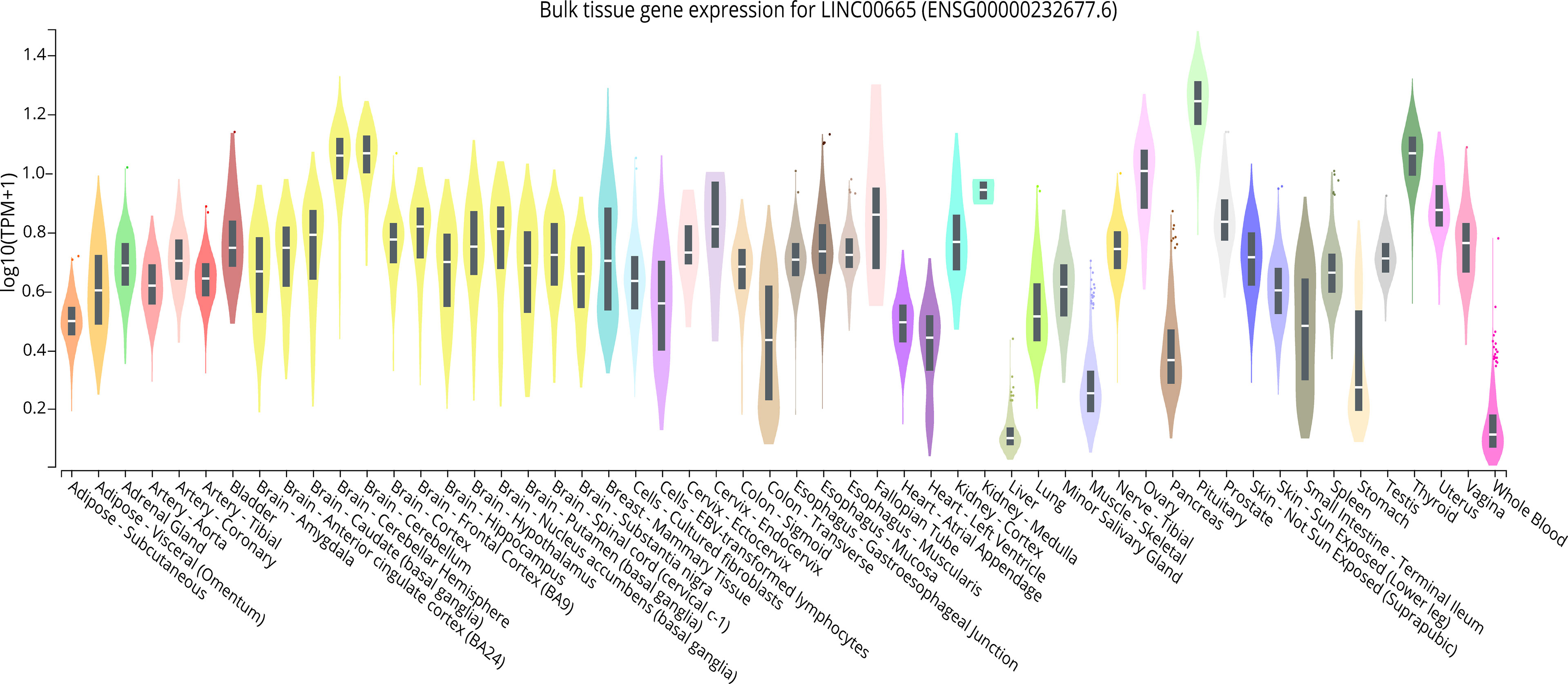
LINC00665,on chromosome19q13.12, is a RNA gene with a length of 1,749 bp containing eight exons (Figure 1A). First, we explored the subcellular localization of LINC00665 using the COMPARTMENTS tools (57) and it showed that LINC00665 was expressed in the nucleus, cytosol, and cytoskeleton (Figure 1B). The lncLocator (58) predicted that the score of LINC00665 located in the cytosol was the highest (Figure 1C). CPC algorithm V2.0 (59) was used to calculate the coding potential of LINC00665. As shown in Figure 1D, LINC00665 was predicted as a non-coding transcript. The expression levels of LINC00665 in human cancers and normal tissues were explored using the Genotype-Tissue Expression (GTEx) Project (60) and Gene Expression Profiling Interactive Analysis (GEPIA) (61). It was found that LINC00665 was broadly expressed in different human tissues (Figure 2). The data from TCGA and GTEx revealed that LINC00665 was significantly differentially expressed in breast invasive carcinoma, cholangiocarcinoma, colon adenocarcinoma, lymphoid neoplasm diffuse large B-cell lymphoma, head and neck squamous cell carcinoma, acute myeloid leukemia, liver hepatocellular carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, ovarian serous cystadenocarcinoma, rectum adenocarcinoma, stomach adenocarcinoma, testicular germ cell tumors, thymoma, and uterine carcinosarcoma (Figure 3). The expression of LINC00665 in cancer and normal tissues analyzed using TCGA data is shown in Figure 4.
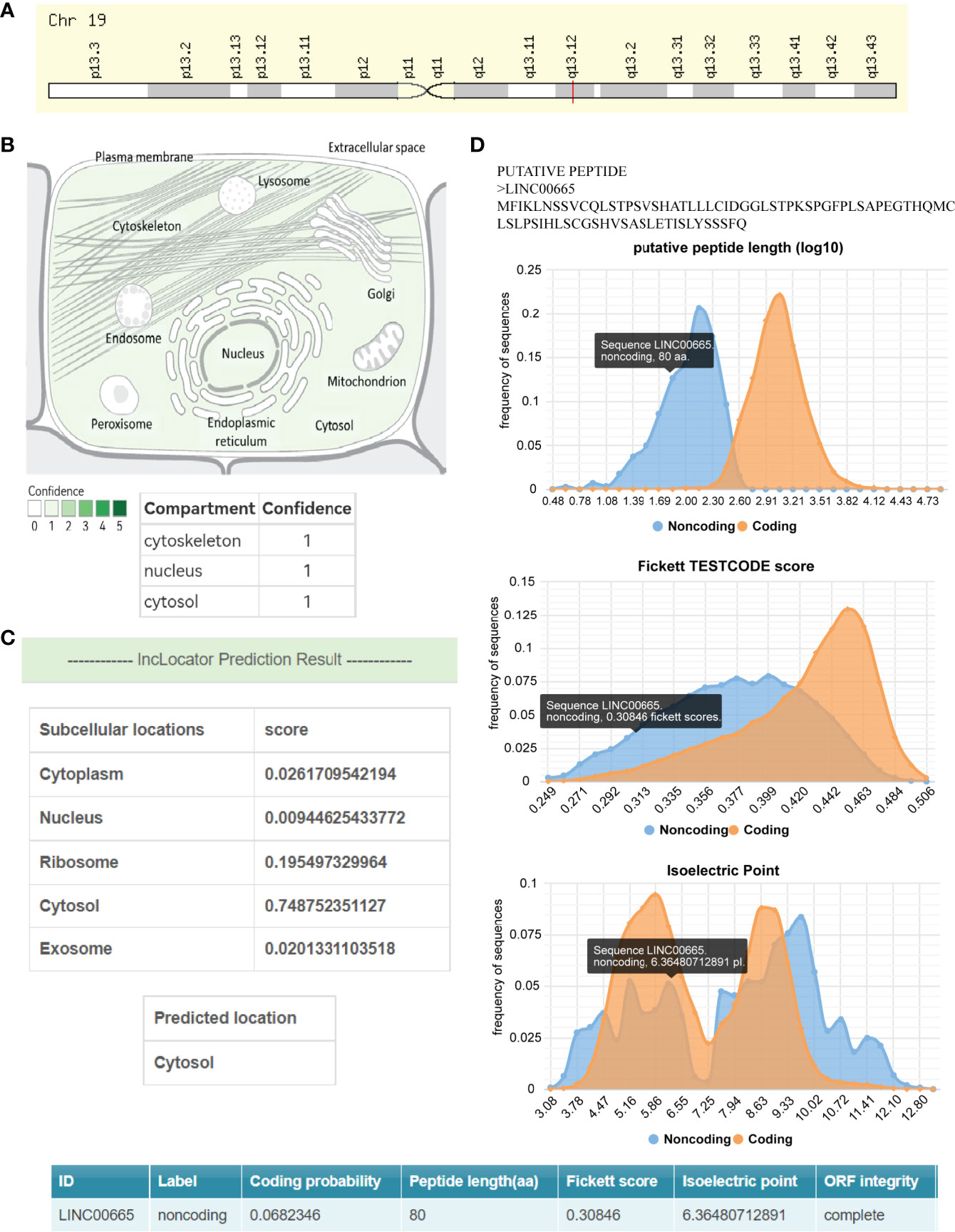
Figure 1 The subcellular localization and coding potential of LINC00665. (A) Chromosome 19q13.12 has a cytogenetic band for LINC00665. (B) LINC00665 expression concentrates in the nucleus, cytosol, and cytoskeleton. (C) The subcellular localization of LINC00665 predicted by lncLocator was the cytoplasm. (D) The protein-coding probability features of LINC00665, including Fickett score, peptide length (synonymous with ORF length), and isoelectric point.
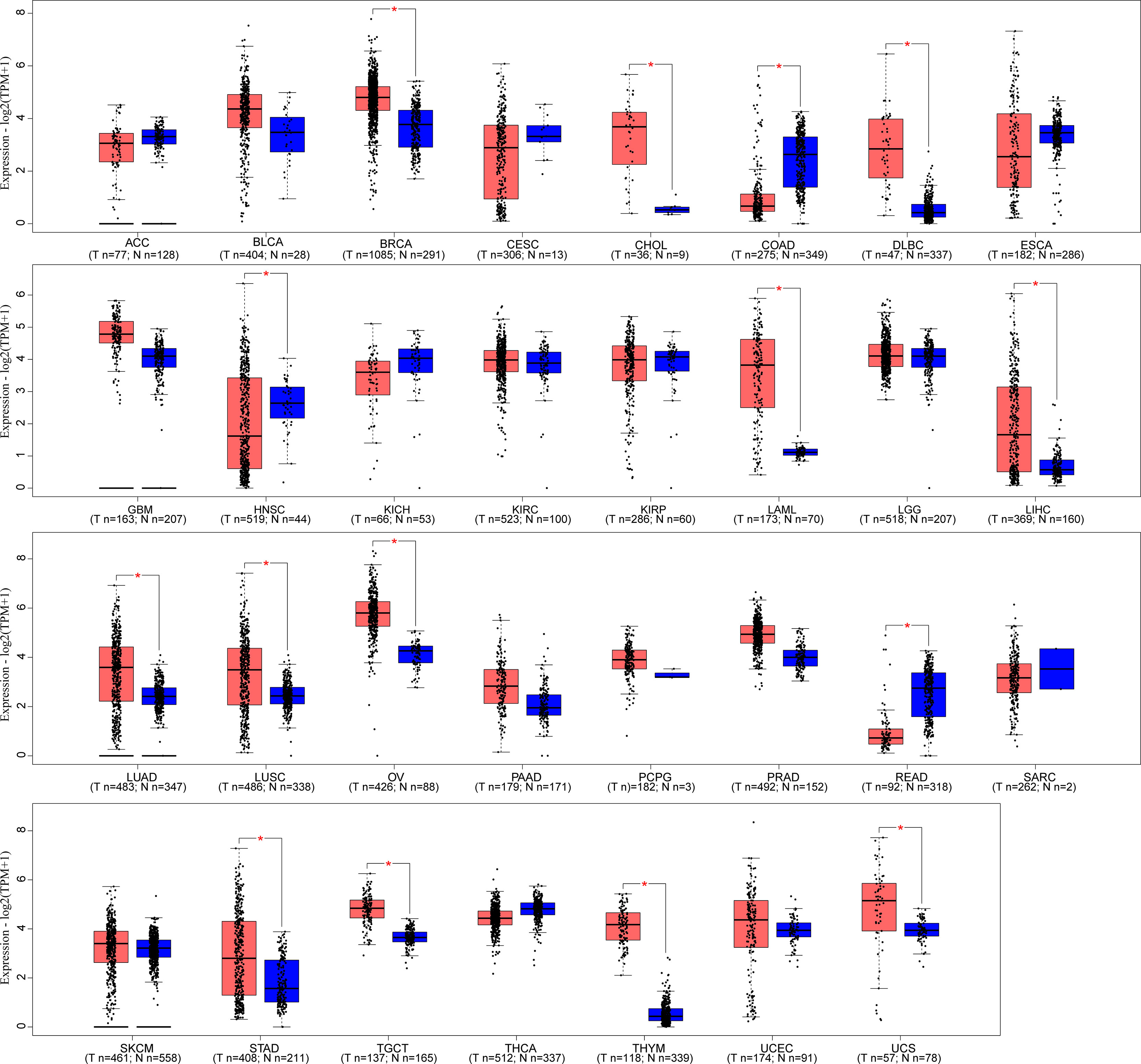
Figure 3 LINC00665 expression in human cancers and corresponding normal tissues (data from TCGA and GTEx). *p < 0.05.
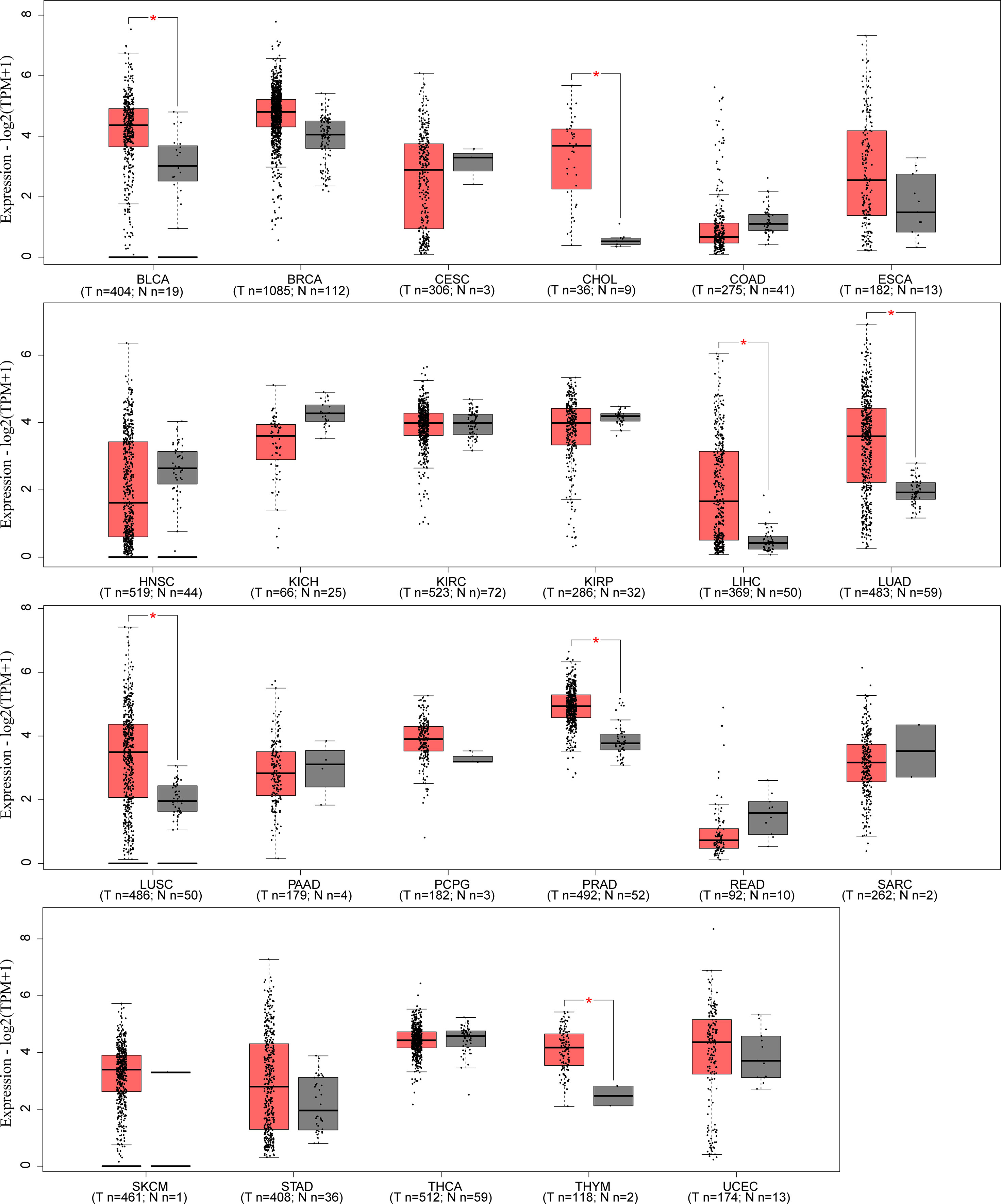
Figure 4 LINC00665 expression in human cancers and corresponding normal tissues (data from TCGA). *p < 0.05.
LINC00665 Dysregulation and Biological Roles in Human Cancers
LINC00665 in Breast Cancer
The subcellular localization of LINC00665 in BCa cells is well studied. Evidence based on fluorescence in situ hybridization (FISH) and subcellular fractionation followed by RT-qPCR showed that LINC00665 was mainly located in the cytoplasm (16, 17). LINC00665 is remarkably elevated in 106 BCa tissues and cell lines (MDA-MB-231 and MCF-7), and its expression is significantly associated with tumor size and TNM stage. LINC00665 knockdown suppresses BCa cell proliferation, migration, and invasion, but promotes cell apoptosis. Mechanically, LINC00665 silencing could inhibit β-catenin expression through competitively binding miR-3619-5p in BCa cells (18). Increasing evidence found that LINC00665 acted as a sponge for miR-379-5p (16) and miR-551b-5p (19) in BCa. Dai et al. (20) explored the correlation between LINC00665 expression and pathological complete response (pCR) in 102 neoadjuvant chemotherapy BCa patients. The results showed that LINC00665 expression was an independent predictor of pCR (OR = 0.351, 95% CI: 0.125–0.936, p = 0.040), especially in patients with hormone receptor-positive/human epidermal growth factor receptor 2-negative subtypes (OR = 0.272, 95% CI: 0.104–0.664, p = 0.005). Although LINC00665 has no protein-coding function, it was found to encode biologically active micropeptide. Guo et al. (17) revealed that LINC00665 was regulated by TGF-β at the translational level and encoded micropeptide CIP2A‐BP, thus participating in BCa progression. LINC00665 may promote BCa cell metastasis by triggering the epithelial–mesenchymal transition (EMT) process (21).
LINC00665 in Prostate Cancer
Xue et al. (22) observed that LINC00665 expression was upregulated in 50 PCa tissues and cell lines (LNCaP, 22RV1, PC-3,and DU-145) compared with corresponding control groups. Upregulated LINC00665 expression was correlated with the advanced T stage, lymph node metastasis, and the poor survival of PCa patients. Moreover, LINC00665 knockdown significantly suppressed PCa cell proliferation and migration. To further explore the biological mechanism of LINC00665, the subcellular fractionation assay found that LINC00665 was mainly located in the nucleus of PCa (22RV1 and DU-145 cells). Mechanically, LINC00665 may epigenetically inhibit KLF2 transcription and expression through recruiting EZH2 and LSD1 to its promoter region, thus playing an oncogenic role in PCa. LINC00665 could also promote PCa progression as a ceRNA through the miR-1224-5p/SND1 axis (23). Eke et al. (24) studied radiation-induced lncRNA dysregulation and found that LINC00665 was upregulated in LNCaP PCa cells at 2 months after a single dose of 10 Gy but not after multifractionated irradiation. Subsequent cell function assays showed that LINC00665 regulated the expression of the DNA repair proteins CtIP (RBBP8) and XPC.
LINC00665 in Hepatocellular Carcinoma
Compared with adjacent normal tissues and a normal liver cell line HL-7702, LINC00665 was highly expressed in 76 HCC tissues and HCC cell lines (Huh-7, HepG2, HCCLM6, MHCC-97H, and Hep3B) (25). High LINC00665 expression was associated with the advanced tumor size, Edmondson grade, and the poor survival of HCC patients. In vitro and in vivo assays indicated that LINC00665 knockdown inhibited HCC cell viability and tumor growth, and induced apoptosis and autophagy. Further molecular regulatory investigation revealed that LINC00665 modulated the expression of MAP4K3 by sponging miR-186-5p. The findings of upregulated LINC00665 in HCC were also confirmed by Wen et al. (26) using TCGA, GEO, and quantitative real-time polymerase chain reaction (qRT-PCR) data. Ding et al. (27) performed an RNA-sequencing analysis in Huh-7 cells that were treated with TNFα, IL-1β, or INF-γ to identify the NF-κB-associated lncRNAs in HCC, and found that LINC00665 was the most highly induced upregulated lncRNA. In line with other studies, LINC00665 was upregulated in (84/122) HCC tissues and its expression was positively correlated with TNM stages, Barcelona Clinic Liver Cancer (BCLC) stages, and the poor prognosis of HCC patients. Furthermore, LINC00665 physically interacted with PKR and played an oncogenic role by promoting PKR activation and stability, thus giving feedback on prosperous NF-κB signaling in HCC.
LINC00665 in Colorectal Cancer
Wu et al. (30) measured the expression level of LINC00665 in 67 pairs of CRC tissues and cell lines by qRT-PCR. The findings showed that LINC00665 was overexpressed in CRC tissues and cell lines, and LINC00665 knockdown suppressed CRC cell proliferation and promoted cell apoptosis. The subcellular distribution of LINC00665 in CRC cells was mainly in the cytoplasm. Mechanistically, LINC00665 could sponge miR-126-5p to regulate PAK2 and FZD3 expression. A study by Han et al. (31) also found that LINC00665 was abnormally upregulated in CRC cells and mainly located in the cytoplasm of HCT-116 and SW480 cells. Molecular investigations found that LINC00665 could increase the expression of CTNNB1 by sponging miR-214-3p or binding to U2AF2 protein, further activating the Wnt/β-catenin signaling pathway to promote the tumorigenesis of CRC. In addition, upregulated LINC00665 stimulated CRC progression by regulating the miR-9-5p/ATF1 axis (32).
LINC00665 in Gastric Cancer
In the digestive tract, GC remains a common malignant tumor with a high incidence and mortality and a poor prognosis (62). LINC00665 was first identified to be overexpressed in 49 paired GC tissues and cell lines in a study by Qi et al. (33). High LINC00665 expression was also found to be associated with an advanced TNM stage and the histological grade of GC. Moreover, in vitro assays showed that LINC00665 may promote the proliferation and invasion of GC cells, and as a ceRNA for miR-149-3p to regulate the expression of RNF2. Yang et al. (34) demonstrated that LINC00665 silencing could significantly inhibit GC cell proliferation, migration, invasion, and induce apoptosis in vitro, as well as restrain tumor growth in vivo. The Wnt pathway, also named Wnt/β-catenin, was known to play a crucial role in GC tumorigenesis and was found to be activated by LINC00665. In addition, LINC00665 could facilitate the proliferation, invasion, and metastasis of GC cells via activating the TGF-β signal pathway (35). Acquired resistance to cisplatin (DDP)-based chemotherapy is a common clinical issue in the treatment of GC. Yue et al. (36) reported that LINC00665 expression was higher in DDP-resistant GC cell lines than that in normal gastric mucosal epithelial cell lines and DDP-sensitive GC cell lines. LINC00665 knockdown inhibited DDP-resistant GC cell proliferation, induced apoptosis, and improved its DDP sensitivity by suppressing endoplasmic reticulum (ER) stress.
LINC00665 in Lung Cancer
According to the pathological pattern, lung cancer cases are divided into non-small cell lung cancer (NSCLC) and small cell lung cancer. NSCLC accounts for approximately 85% and its primary subtype is lung adenocarcinoma. Yang et al. (39) found that LINC00665 was upregulated in lung cancer tissues by analyzing the TCGA database, and further qRT-PCR assay confirmed that it was highly expressed in 51 of 60 NSCLC tissues. High LINC00665 expression was associated with advanced tumor size, TNM stage, and lymph node metastasis. Kaplan–Meier survival analysis indicated that patients with high LINC00665 expression had poorer overall survival (OS) and progression-free survival than those with low expression. Functionally, LINC00665 knockdown suppressed NSCLC cell proliferation and migration and promoted cell apoptosis. Further assays in vitro and in vivo showed that LINC00665 knockdown improved the sensitivity of NSCLC cells to DDP. Based on the findings that LINC00665 was primarily distributed in the cytoplasm of NSCLC cells, LINC00665 was found to recruit EZH2 to the CDKN1C promoter region to facilitate the demethylation of histone H3K27 and inhibit CDKN1C transcription. As a ceRNA, LINC00665 could regulate the expression of E2F3, AKR1B10, ZIC2, MYCBP, and CCNA2 by sponging miR-138-5p (40), miR-98 (41), miR-181c-5p (42), miR-195-5p (43), and miR-let-7b (44), respectively. LINC00665 also participated in tumor angiogenesis. Cong et al. (45) uncovered that YB-1 protein stability was improved by directly interacting with LINC00665, and thus activated the transcription and expression of ANGPT4, ANGPTL3, and VEGFA by binding to their promoters in the process of tumor angiogenesis. A recent study also found that LINC00665 was overexpressed in lung cancer tissues and cells with acquired gefitinib resistance (46). A series of assays in vitro and in vivo confirmed that LINC00665 promoted the resistance of NSCLC cells to gefitinib by increasing EZH2 and activating the PI3K/AKT pathway.
LINC00665 in Ovarian Cancer
OC, with high mortality and poor prognosis, is one of the most common malignant tumors in the female reproductive system. Gao et al. (50) found that LINC00665 was upregulated in OC tissues by analyzing the GEPIA database and constructing a prognosis correlated with the LINC00665-miR-146a-5p-CXCR4 regulatory network. LINC00665 expression was positively correlated with infiltrating levels of CD4+ T cells and negatively correlated with CD8+ T cells, neutrophils, macrophages, and dendritic cells, which demonstrated the correlation between LINC00665 and lymphocyte infiltration in high-grade serous OC (51). Moreover, a nine-autophagy-related lncRNA signature (including LINC00665) was constructed as independent prognostic factors for the OS of OC patients (52). LINC00665 may regulate E2F3 expression through competitively binding to miRNA-34a-5p in promoting OC progression (53).
LINC00665 in Glioma
Through lncRNA microarray analysis, LINC00665 was identified to be differentially expressed in glioma. Further RT-qPCR assays confirmed the high expression levels of LINC00665 in 48 glioma tissues and cell lines. Based on the evidence that LINC00665 was mainly distributed in the cytoplasm, Dai et al. discovered that the LINC00665/miR-34a-5p/AGTR1 axis contributed to the development and progression of glioma (37). In addition to the ceRNA regulatory pathway, lncRNAs, which were located in the cytoplasm, could regulate mRNA expression through STAU1-mediated mRNA degradation (63). Ruan et al. (38) reported that LINC00665 was downregulated in glioma tissues and cells and acted as a tumor suppressor gene. Further exploration found that LINC00665 facilitated the degradation of MTF1 and YY2 mRNA via interacting with a double-stranded RNA-binding protein STAU1. The Alu elements on LINC00665 and MTF1 or YY2 mRNA 3’UTR constructed a STAU1-binding site through complementary base pairing; thus, STAU1 executed mRNA decay via binding to the constructed site.
LINC00665 in Other Cancers
LINC00665 expression was upregulated in 42 osteosarcoma tissues and four cell lines. Further clinicopathological correlation analysis indicated that higher LINC00665 expression was correlated with larger tumor size, later clinical stages, and poorer OS. Mechanistically, LINC00665 facilitated osteosarcoma progression by increasing RAP1B expression via targeting miR-708 and miR-142-5p (28). LINC00665 could also promote osteosarcoma progression by sponging miR-3619 (29). Compared with normal human epidermal melanocytes, LINC00665 expression was significantly increased in four melanoma cells lines (A375, M21, A2058, and A-875). A series of assays in vitro and in vivo showed that LINC00665 was mainly expressed in the cytoplasm and could promote the malignant behaviors of melanoma cells through the miR-224-5p/VMA21 axis (47). Xia et al. (56) demonstrated that LINC00665 may promote HeLa cell proliferation, metastasis, and EMT via the WNT-CTNNB1/β−catenin signaling pathway. In endometrial carcinoma, LINC00665 was overexpressed in endometrial carcinoma tissues and cell lines. Mechanistically, LINC00665 co-immunoprecipitated with the HMGA1 protein and promoted the tumorigenicity of endometrial carcinoma in vitro and in vivo (48). Lu et al. (49) explored the role of LINC00665 in gemcitabine resistance of cholangiocarcinoma and found that LINC00665 was upregulated in gemcitabine-resistant cells. High LINC00665 expression was positively correlated to advanced TNM stage, lymph node/distant metastasis, and poor prognosis. Assays in vitro and in vivo indicated that LINC00665 increased the gemcitabine tolerance of cholangiocarcinoma cells by regulating EMT, stemless properties, and the miR-424-5p/BCL9L axis. Through extracting the RNAs from acute myeloid leukemia (AML) or normal bone marrow tissues and cells, an RT-qPCR assay displayed that LINC00665 was upregulated in AML tissues and cell lines. LINC00665 could accelerate the progression of AML by regulating the miR-4458/DOCK1 axis (54). LINC00665 was also found to be upregulated in T-cell acute lymphoblastic leukemia (T-ALL) and could promote T-ALL progression through the miR-101/PI3K/Akt pathway (55).
Conclusion and Future Perspectives
Mounting evidence has indicated that lncRNAs were dysregulated in human cancers and act as critical regulators in tumorigenesis and tumor progression. Although lncRNA was known as a kind of non-coding RNA, several studies have reported that it has the capacity to code small proteins or micropeptides. LncRNA LOC90024 was found to encode a splicing regulatory small 130-amino acid protein, which could promote the tumorigenesis and progression of CRC (64). In BCa, LINC00665 could encode a biologically active micropeptide CIP2A-BP. However, whether LINC00665 could encode micropeptides in other kinds of cancers remains unclear and needs further exploration. The roles played by lncRNAs differ depending on the subcellular location. LINC00665 was found to be mainly located in the cytoplasm in BCa, osteosarcoma, CRC, glioma, lung cancer, and OC, thus participating in biological regulation through ceRNA, STAU1-mediated mRNA degradation, interfering with RNA-binding proteins, and so on. In PCa, LINC00665 was mainly expressed in the nucleus and may function at the transcriptional level. The subcellular location of LINC00665 in HCC, GC, cervical cancer, and melanoma is still unclear. The present findings showed that LINC00665 was highly expressed in most cancers and functioned as an oncogene in cell proliferation, migration, invasion, and apoptosis. However, the expression status and specific roles of LINC00665 in esophagus cancer, pancreatic cancer, and so on are unknown, and its expression level in glioma is controversial. Further studies should enroll a larger cohort of clinical samples to improve the reliability of studies and focus more on exploring the precise biological regulatory mechanisms of LINC00665.
Author Contributions
YL and CZ designed the study. CZ, S-NX, KL, and J-HC helped with data processing and reference collection. CZ, J-HC, and QL prepared the figures and tables. All authors participated in revising the final manuscript and approved it for publication.
Funding
This work was supported by Wu Jieping Medical Foundation (No. 320.6750.19028) and PhD Start-up Fund of Henan Cancer Hospital.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Carrieri FA, Smack C, Siddiqui I, Kleinberg LR, Tran PT. Tumor Treating Fields: At the Crossroads Between Physics and Biology for Cancer Treatment. Front Oncol (2020) 10:575992. doi: 10.3389/fonc.2020.575992
3. Mun EJ, Babiker HM, Weinberg U, Kirson ED, Von Hoff DD. Tumor-Treating Fields: A Fourth Modality in Cancer Treatment. Clin Cancer Res (2018) 24(2):266–75. doi: 10.1158/1078-0432.CCR-17-1117
4. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE V7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res (2012) 22(9):1775–89. doi: 10.1101/gr.132159.111
5. Orom UA, Shiekhattar R. Long Non-Coding RNAs and Enhancers. Curr Opin Genet Dev (2011) 21(2):194–8. doi: 10.1016/j.gde.2011.01.020
6. Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci (2016) 41(9):761–72. doi: 10.1016/j.tibs.2016.07.003
7. Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, et al. Localization and Abundance Analysis of Human lncRNAs at Single-Cell and Single-Molecule Resolution. Genome Biol (2015) 16:20. doi: 10.1186/s13059-015-0586-4
8. Liu Z, Wang Y, Wang L, Yao B, Sun L, Liu R, et al. Long Non-Coding RNA AGAP2-AS1, Functioning as a Competitive Endogenous RNA, Upregulates ANXA11 Expression by Sponging miR-16-5p and Promotes Proliferation and Metastasis in Hepatocellular Carcinoma. J Exp Clin Cancer Res (2019) 38(1):194. doi: 10.1186/s13046-019-1188-x
9. Cao C, Li J, Li G, Hu G, Deng Z, Huang B, et al. Long Non-Coding RNA TMEM220-AS1 Suppressed Hepatocellular Carcinoma by Regulating the miR-484/MAGI1 Axis as a Competing Endogenous RNA. Front Cell Dev Biol (2021) 9:681529. doi: 10.3389/fcell.2021.681529
10. Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, et al. A Novel lncRNA MCM3AP-AS1 Promotes the Growth of Hepatocellular Carcinoma by Targeting miR-194-5p/FOXA1 Axis. Mol Cancer (2019) 18(1):28. doi: 10.1186/s12943-019-0957-7
11. Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 Dependence in Cancer With MYC Copy-Number Increase. Nat (2014) 512(7512):82–6. doi: 10.1038/nature13311
12. Xu Z, Peng B, Liang Q, Chen X, Cai Y, Zeng S, et al. Construction of a Ferroptosis-Related Nine-lncRNA Signature for Predicting Prognosis and Immune Response in Hepatocellular Carcinoma. Front Immunol (2021) 12:719175. doi: 10.3389/fimmu.2021.719175
13. Liang Y, Zhang CD, Zhang C, Dai DQ. DLX6-AS1/miR-204-5p/OCT1 Positive Feedback Loop Promotes Tumor Progression and Epithelial-Mesenchymal Transition in Gastric Cancer. Gastric Cancer (2020) 23(2):212–27. doi: 10.1007/s10120-019-01002-1
14. Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M, et al. LincRNAFEZF1-AS1 Represses P21 Expression to Promote Gastric Cancer Proliferation Through LSD1-Mediated H3K4me2 Demethylation. Mol Cancer (2017) 16(1):39. doi: 10.1186/s12943-017-0588-9
15. Zhang C, Liang Y, Zhang CD, Pei JP, Wu KZ, Li YZ, et al. The Novel Role and Function of LINC01235 in Metastasis of Gastric Cancer Cells by Inducing Epithelial-Mesenchymal Transition. Genomics (2021) 113(3):1504–13. doi: 10.1016/j.ygeno.2021.03.027
16. Ji W, Diao YL, Qiu YR, Ge J, Cao XC, Yu Y. LINC00665 Promotes Breast Cancer Progression Through Regulation of the miR-379-5p/LIN28B Axis. Cell Death Dis (2020) 11(1):16. doi: 10.1038/s41419-019-2213-x
17. Guo B, Wu S, Zhu X, Zhang L, Deng J, Li F, et al. Micropeptide CIP2A-BP Encoded by LINC00665 Inhibits Triple-Negative Breast Cancer Progression. EMBO J (2020) 39(1):e102190. doi: 10.15252/embj.2019102190
18. Lv M, Mao Q, Li J, Qiao J, Chen X, Luo S. Knockdown of LINC00665 Inhibits Proliferation and Invasion of Breast Cancer via Competitive Binding of miR-3619-5p and Inhibition of Catenin Beta 1. Cell Mol Biol Lett (2020) 25:43. doi: 10.1186/s11658-020-00235-8
19. Qi L, Sun B, Yang B, Lu S. LINC00665 Stimulates Breast Cancer Progression via Regulating miR-551b-5p. Cancer Manag Res (2021) 13:1113–21. doi: 10.2147/CMAR.S275096
20. Dai H, Sheng X, Sha R, Peng J, Yang F, Zhou L, et al. Linc00665 Can Predict the Response to Cisplatin-Paclitaxel Neoadjuvant Chemotherapy for Breast Cancer Patients. Front Oncol (2021) 11:604319. doi: 10.3389/fonc.2021.604319
21. Zhou JL, Zou L, Zhu T. Long Non-Coding RNA LINC00665 Promotes Metastasis of Breast Cancer Cells by Triggering EMT. Eur Rev Med Pharmacol Sci (2020) 24(6):3097–104. doi: 10.26355/eurrev_202003_20674
22. Xue P, Yan M, Wang K, Gu J, Zhong B, Tu C. Up-Regulation of LINC00665 Facilitates the Malignant Progression of Prostate Cancer by Epigenetically Silencing KLF2 Through EZH2 and LSD1. Front Oncol (2021) 11:639060. doi: 10.3389/fonc.2021.639060
23. Chen W, Yu Z, Huang W, Yang Y, Wang F, Huang H. LncRNA LINC00665 Promotes Prostate Cancer Progression via miR-1224-5p/SND1 Axis. Onco Targets Ther (2020) 13:2527–35. doi: 10.2147/OTT.S241578
24. Eke I, Bylicky MA, Sandfort V, Chopra S, Martello S, Graves EE, et al. The lncRNAs LINC00261 and LINC00665 Are Upregulated in Long-Term Prostate Cancer Adaptation After Radiotherapy. Mol Ther Nucleic Acids (2021) 24:175–87. doi: 10.1016/j.omtn.2021.02.024
25. Shan Y, Li P. Long Intergenic Non-Protein Coding RNA 665 Regulates Viability, Apoptosis, and Autophagy via the MiR-186-5p/MAP4K3 Axis in Hepatocellular Carcinoma. Yonsei Med J (2019) 60(9):842–53. doi: 10.3349/ymj.2019.60.9.842
26. Wen DY, Lin P, Pang YY, Chen G, He Y, Dang YW, et al. Expression of the Long Intergenic Non-Protein Coding RNA 665 (LINC00665) Gene and the Cell Cycle in Hepatocellular Carcinoma Using The Cancer Genome Atlas, the Gene Expression Omnibus, and Quantitative Real-Time Polymerase Chain Reaction. Med Sci Monit (2018) 24:2786–808. doi: 10.12659/MSM.907389
27. Ding J, Zhao J, Huan L, Liu Y, Qiao Y, Wang Z, et al. Inflammation-Induced Long Intergenic Noncoding RNA (LINC00665) Increases Malignancy Through Activating the Double-Stranded RNA-Activated Protein Kinase/Nuclear Factor Kappa B Pathway in Hepatocellular Carcinoma. Hepatol (2020) 72(5):1666–81. doi: 10.1002/hep.31195
28. Wang L, Song X, Yu L, Liu B, Ma J, Yang W. LINC00665 Facilitates the Malignant Processes of Osteosarcoma by Increasing the RAP1B Expression via Sponging miR-708 and miR-142-5p. Anal Cell Pathol (Amst) (2021) 2021:5525711. doi: 10.1155/2021/5525711
29. Zhang DW, Gu GQ, Chen XY, Zha GC, Yuan Z, Wu Y. LINC00665 Facilitates the Progression of Osteosarcoma via Sponging miR-3619-5p. Eur Rev Med Pharmacol Sci (2020) 24(19):9852–9. doi: 10.26355/eurrev_202010_23195
30. Wu CL, Shan TD, Han Y, Kong Y, Li YB, Peng XG, et al. Long Intergenic Noncoding RNA 00665 Promotes Proliferation and Inhibits Apoptosis in Colorectal Cancer by Regulating miR-126-5p. Aging (Albany NY) (2021) 13(10):13571–84. doi: 10.18632/aging.202874
31. Han T, Gao M, Wang X, Li W, Zhuo J, Qu Z, et al. LINC00665 Activates Wnt/Beta-Catenin Signaling Pathway to Facilitate Tumor Progression of Colorectal Cancer via Upregulating CTNNB1. Exp Mol Pathol (2021) 120:104639. doi: 10.1016/j.yexmp.2021.104639
32. Zhao X, Weng W, Long Y, Pan W, Li Z, Sun F. LINC00665/miR-9-5p/ATF1 Is a Novel Axis Involved in the Progression of Colorectal Cancer. Hum Cell (2020) 33(4):1142–54. doi: 10.1007/s13577-020-00393-z
33. Qi H, Xiao Z, Wang Y. Long Non-Coding RNA LINC00665 Gastric Cancer Tumorigenesis by Regulation miR-149-3p/RNF2 Axis. Onco Targets Ther (2019) 12:6981–90. doi: 10.2147/OTT.S214588
34. Yang B, Bai Q, Chen H, Su K, Gao C. LINC00665 Induces Gastric Cancer Progression Through Activating Wnt Signaling Pathway. J Cell Biochem (2020) 121(3):2268–76. doi: 10.1002/jcb.29449
35. Zhang X, Wu J. LINC00665 Promotes Cell Proliferation, Invasion, and Metastasis by Activating the TGF-Beta Pathway in Gastric Cancer. Pathol Res Pract (2021) 224:153492. doi: 10.1016/j.prp.2021.153492
36. Yue C, Yu C, Peng R, Wang J, Li G, Xu L. LINC00665/miR-379-5p/GRP78 Regulates Cisplatin Sensitivity in Gastric Cancer by Modulating Endoplasmic Reticulum Stress. Cytotechnol (2021) 73(3):413–22. doi: 10.1007/s10616-021-00466-3
37. Dai Y, Zhang Y, Hao M, Zhu R. LINC00665 Functions as a Competitive Endogenous RNA to Regulate AGTR1 Expression by Sponging Mir34a5p in Glioma. Oncol Rep (2021) 45(3):1202–12. doi: 10.3892/or.2021.7949
38. Ruan X, Zheng J, Liu X, Liu Y, Liu L, Ma J, et al. lncRNA LINC00665 Stabilized by TAF15 Impeded the Malignant Biological Behaviors of Glioma Cells via STAU1-Mediated mRNA Degradation. Mol Ther Nucleic Acids (2020) 20:823–40. doi: 10.1016/j.omtn.2020.05.003
39. Yang D, Feng W, Zhuang Y, Liu J, Feng Z, Xu T, et al. Long Non-Coding RNA Linc00665 Inhibits CDKN1C Expression by Binding to EZH2 and Affects Cisplatin Sensitivity of NSCLC Cells. Mol Ther Nucleic Acids (2021) 23:1053–65. doi: 10.1016/j.omtn.2021.01.013
40. Wang H, Wang L, Zhang S, Xu Z, Zhang G. Downregulation of LINC00665 Confers Decreased Cell Proliferation and Invasion via the miR-138-5p/E2F3 Signaling Pathway in NSCLC. BioMed Pharmacother (2020) 127:110214. doi: 10.1016/j.biopha.2020.110214
41. Cong Z, Diao Y, Xu Y, Li X, Jiang Z, Shao C, et al. Long Non-Coding RNA Linc00665 Promotes Lung Adenocarcinoma Progression and Functions as ceRNA to Regulate AKR1B10-ERK Signaling by Sponging miR-98. Cell Death Dis (2019) 10(2):84. doi: 10.1038/s41419-019-1361-3
42. Wei W, Zhao X, Liu J, Zhang Z. Downregulation of LINC00665 Suppresses the Progression of Lung Adenocarcinoma via Regulating miR-181c-5p/ZIC2 Axis. Aging (Albany NY) (2021) 13(13):17499–515. doi: 10.18632/aging.203240
43. Wang A, Zhang T, Wei W, Wang H, Zhang Z, Yang W, et al. The Long Noncoding RNA LINC00665 Facilitates C-Myc Transcriptional Activity via the miR-195-5p MYCBP Axis to Promote Progression of Lung Adenocarcinoma. Front Oncol (2021) 11:666551. doi: 10.3389/fonc.2021.666551
44. Huang Y, Zhong L, Nie K, Li L, Song S, Liu F, et al. Identification of LINC00665-miR-Let-7b-CCNA2 Competing Endogenous RNA Network Associated With Prognosis of Lung Adenocarcinoma. Sci Rep (2021) 11(1):4434. doi: 10.1038/s41598-020-80662-x
45. Cong Z, Diao Y, Li X, Jiang Z, Xu Y, Zhou H, et al. Long Non-Coding RNA Linc00665 Interacts With YB-1 and Promotes Angiogenesis in Lung Adenocarcinoma. Biochem Biophys Res Commun (2020) 527(2):545–52. doi: 10.1016/j.bbrc.2020.04.108
46. Liu X, Lu X, Zhen F, Jin S, Yu T, Zhu Q, et al. LINC00665 Induces Acquired Resistance to Gefitinib Through Recruiting EZH2 and Activating PI3K/AKT Pathway in NSCLC. Mol Ther Nucleic Acids (2019) 16:155–61. doi: 10.1016/j.omtn.2019.02.010
47. Wang X, Wang Y, Lin F, Xu M, Zhao X. Long Non-Coding RNA LINC00665 Promotes Melanoma Cell Growth and Migration via Regulating the miR-224-5p/VMA21 Axis. Exp Dermatol (2022) 31(1):64–73. doi: 10.1111/exd.14246
48. Cai Y, Hao M, Chang Y, Liu Y. LINC00665 Enhances Tumorigenicity of Endometrial Carcinoma by Interacting With High Mobility Group AT-Hook 1. Cancer Cell Int (2021) 21(1):8. doi: 10.1186/s12935-020-01657-2
49. Lu M, Qin X, Zhou Y, Li G, Liu Z, Geng X, et al. Long Non-Coding RNA LINC00665 Promotes Gemcitabine Resistance of Cholangiocarcinoma Cells via Regulating EMT and Stemness Properties Through miR-424-5p/BCL9L Axis. Cell Death Dis (2021) 12(1):72. doi: 10.1038/s41419-020-03346-4
50. Gao L, Li X, Nie X, Guo Q, Liu Q, Qi Y, et al. Construction of Novel mRNA-miRNA-lncRNA Regulatory Networks Associated With Prognosis of Ovarian Cancer. J Cancer (2020) 11(23):7057–72. doi: 10.7150/jca.49557
51. Wu M, Shang X, Sun Y, Wu J, Liu G. Integrated Analysis of Lymphocyte Infiltration-Associated lncRNA for Ovarian Cancer via TCGA, GTEx and GEO Datasets. PeerJ (2020) 8:e8961. doi: 10.7717/peerj.8961
52. Meng C, Zhou JQ, Liao YS. Autophagy-Related Long Non-Coding RNA Signature for Ovarian Cancer. J Int Med Res (2020) 48(11):300060520970761. doi: 10.1177/0300060520970761
53. Xu D, Song Q, Liu Y, Chen W, Lu L, Xu M, et al. LINC00665 Promotes Ovarian Cancer Progression Through Regulating the miRNA-34a-5p/E2F3 Axis. J Cancer (2021) 12(6):1755–63. doi: 10.7150/jca.51457
54. Yang X, Wang Y, Pang S, Li X, Wang P, Ma R, et al. LINC00665 Promotes the Progression of Acute Myeloid Leukemia by Regulating the miR-4458/DOCK1 Pathway. Sci Rep (2021) 11(1):5009. doi: 10.1038/s41598-021-82834-9
55. Abuduer M, EZG A. LINC00665 Promotes the Viability, Migration and Invasion of T Cell Acute Lymphoblastic Leukemia Cells by Targeting miR-101 via Modulating PI3K/Akt Pathway. Tissue Cell (2021) 71:101579. doi: 10.1016/j.tice.2021.101579
56. Xia L, Chen YX, Lian JB. LINC00665 Promotes HeLa Cell Proliferation, Migration, Invasion and Epithelial-Mesenchymal Transition by Activating the WNT-CTNNB1/betacatenin Signaling Pathway. Sheng Li Xue Bao (2021) 73(2):233–43. doi: 10.13294/j.aps.2021.0025
57. Binder JX, Pletscher-Frankild S, Tsafou K, Stolte C, O'Donoghue SI, Schneider R, et al. COMPARTMENTS: Unification and Visualization of Protein Subcellular Localization Evidence. Database (Oxf) (2014) 2014:bau012. doi: 10.1093/database/bau012
58. Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The Lnclocator: A Subcellular Localization Predictor for Long Non-Coding RNAs Based on a Stacked Ensemble Classifier. Bioinformatics (2018) 34(13):2185–94. doi: 10.1093/bioinformatics/bty085
59. Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, et al. CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res (2017) 45(W1):W12–W6. doi: 10.1093/nar/gkx428
60. Consortium GT. The Genotype-Tissue Expression (GTEx) Project. Nat Genet (2013) 45(6):580–5. doi: 10.1038/ng.2653
61. Li C, Tang Z, Zhang W, Ye Z, Liu F. GEPIA2021: Integrating Multiple Deconvolution-Based Analysis Into GEPIA. Nucleic Acids Res (2021) 49(W1):W242–W6. doi: 10.1093/nar/gkab418
62. Thrift AP, El-Serag HB. Burden of Gastric Cancer. Clin Gastroenterol Hepatol (2020) 18(3):534–42. doi: 10.1016/j.cgh.2019.07.045
63. Gong C, Maquat LE. lncRNAs Transactivate STAU1-Mediated mRNA Decay by Duplexing With 3' UTRs via Alu Elements. Nat (2011) 470(7333):284–8. doi: 10.1038/nature09701
Keywords: cancer, lncRNA, LINC00665, biological roles, biomarker
Citation: Zhang C, Xu S-N, Li K, Chen J-H, Li Q and Liu Y (2022) The Biological and Molecular Function of LINC00665 in Human Cancers. Front. Oncol. 12:886034. doi: 10.3389/fonc.2022.886034
Received: 01 March 2022; Accepted: 25 April 2022;
Published: 19 May 2022.
Edited by:
Peixin Dong, Hokkaido University, JapanReviewed by:
Wei He, University of Texas MD Anderson Cancer Center, United StatesDeepak Kumar Singh, Albert Einstein College of Medicine, United States
Copyright © 2022 Zhang, Xu, Li, Chen, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Liu, emx5eWxpdXlpbmcxNjY0QHp6dS5lZHUuY24=
 Cheng Zhang
Cheng Zhang Shu-Ning Xu
Shu-Ning Xu Qun Li
Qun Li Ying Liu
Ying Liu