- 1Department of Orthodontics II, Affiliated Stomatological Hospital of Zunyi Medical University, Zunyi, China
- 2Oral Disease Research Key Laboratory of Guizhou Tertiary Institution, School of Stomatology, Zunyi Medical University, Zunyi, China
- 3Microbial Resources and Drug Development Key Laboratory of Guizhou Tertiary Institution, Life Sciences Institute, Zunyi Medical University, Zunyi, China
- 4Department of Gastroenterology, Affiliated Hospital of Zunyi Medical University, Zunyi, China
- 5Department of Urology, The Third Affiliated Hospital of Zunyi Medical University, Zunyi, China
The interaction of non-kinase transmembrane glycoprotein CD44 with ligands including hyaluronic acid (HA) is closely related to the occurrence and development of tumors. Changes in CD44 glycosylation can regulate its binding to HA, Siglec-15, fibronectin, TM4SF5, PRG4, FGF2, collagen and podoplanin and activate or inhibit c-Src/STAT3/Twist1/Bmi1, PI3K/AKT/mTOR, ERK/NF-κB/NANOG and other signaling pathways, thereby having a profound impact on the tumor microenvironment and tumor cell fate. However, the glycosylation of CD44 is complex and largely unknown, and the current understanding of how CD44 glycosylation affects tumors is limited. These issues must be addressed before targeted CD44 glycosylation can be applied to treat human cancers.
Introduction
CD44 is a nonkinase family and single-transmembrane glycoprotein that is expressed at different levels on the cell membranes of embryonic stem cells, bone marrow cells, tumor cells, etc. (1). In humans, CD44 is encoded by 19 exons, 10 of which are constant across all subtypes. The canonical forms of CD44 (CD44s) are encoded by 10 constant exons. CD44 variant isoforms (CD44v1-10) are produced by alternative splicing with any combination of 10 constant exons and the remaining 9 variant exons (2, 3). CD44s and various CD44v isoforms have overlapping and distinct functional roles. CD44v isoforms have additional binding motifs that facilitate CD44 interactions with molecules in the microenvironment (4). CD44v isoforms can act as coreceptors by binding/sequestering growth factors on the cell surface and presenting these growth factors to their specific receptors (5). CD44 promotes the stemness of cancer stem cells through interactions with HA, extracellular matrix components, growth factors, and cytokines (1). CD44 has been identified as a surface marker of cancer stem cell (CSC), especially the CD44v subtype is widely used to isolate and enrich CSC in different types of cancers (6). The CD44 transmembrane glycoprotein family not only establishes specific transmembrane complexes but also organizes signaling cascades through association with the actin cytoskeleton (7). Thus, CD44 is a signaling platform that integrates cellular microenvironmental signals, growth factor and cytokine signals and transduces signals to membrane-associated cytoskeletal proteins or the nucleus to regulate cell-matrix adhesion, cell migration, proliferation, differentiation and survival (6). Therefore, targeting different CD44 variants may be a promising therapeutic target for malignancies.
As a common feature of cancers, aberrant glycosylation is involved in fundamental molecular and cellular biological processes in cancer such as cell signaling and communication, tumor cell division and invasion, cell-matrix interactions, tumor angiogenesis, immune regulation, and metastasis formation (8). A growing body of biochemical, molecular and genetic studies suggests that alterations in protein glycosylation may be a major contributor to the tumorigenic transformation process, with significant effects on tumor disease progression (9). In this review, we describe the glycosylation modification characteristics and the potential of glycosylation alterations as a tumor therapy involving the glycoprotein CD44.
The Structure and Glycosylation Domain of CD44
The CD44 protein has four main structures: an extracellular region, a stem region (standard stem region and/or variable stem region), a transmembrane region (TM) and a short C-terminal intracellular/cytoplasmic region (CP) (10). The extracellular part consists of 7 extracellular domains (constant exons 1, 5, 6, 7), including the N-terminal domain (ligand binding region). The stem region (alternatively spliced region) inserts one or more variant exons between exon 5 and exon 6. The transmembrane region is encoded by exon 8, whereas the cytoplasmic region is encoded by exons 9 or 10. However, exon 9 is spliced out in almost all CD44 cDNA isoforms (11). CD44v has different structures in the stem region (Figure 1A), resulting in different functions (10). Due to the attachment of side chains, the conserved form of CD44 (37 kDa) expands to 80-100 kDa, with some isoforms exceeding 200 kDa due to the high degree of glycosylation (11). The extracellular domain of the CD44 protein is also known as the extracellular HA-binding domain (HABD) (13). CD44-HABD mainly binds to hyaluronic acid (HA), collagen, laminin and fibronectin (14–16).
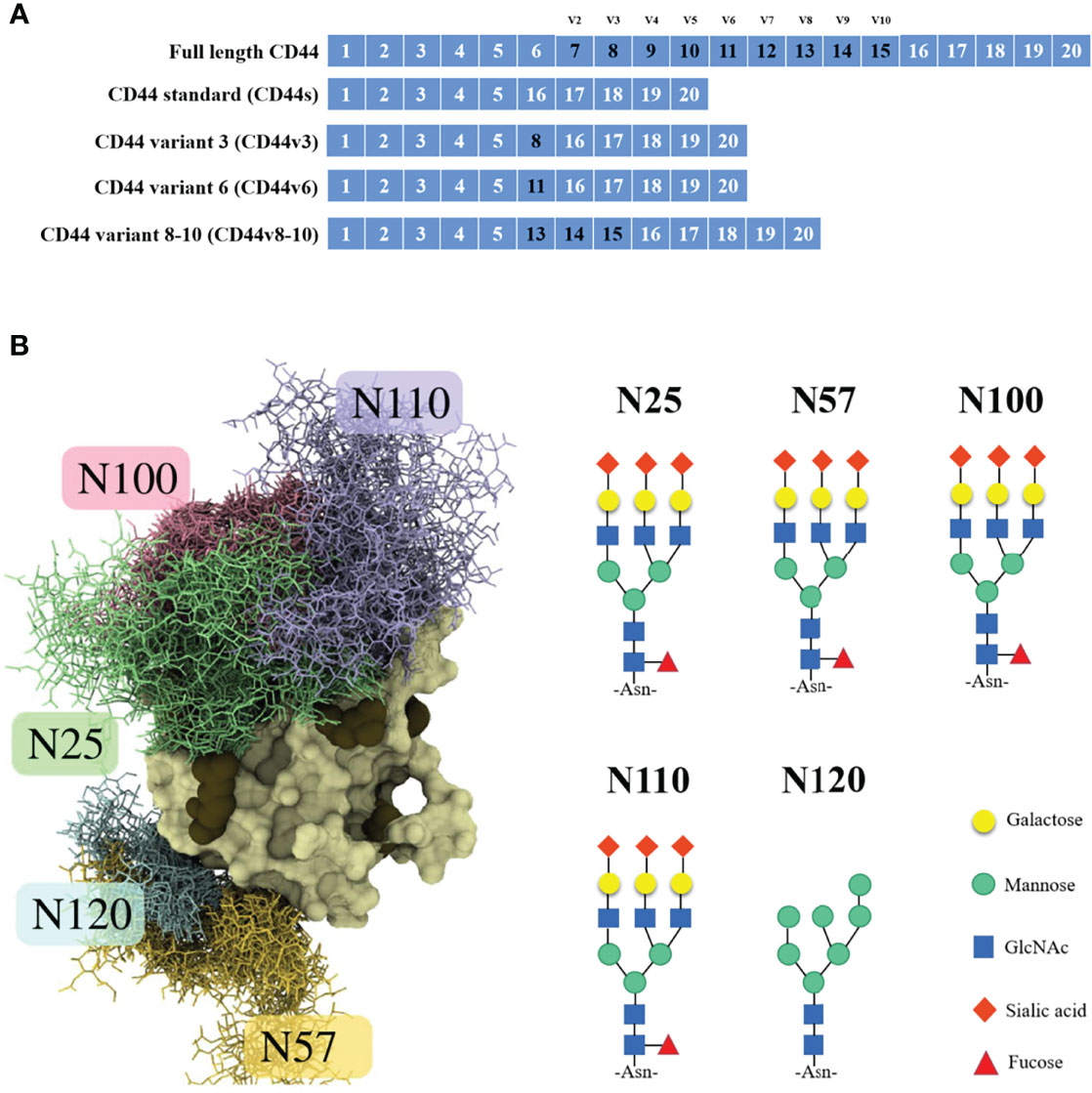
Figure 1 CD44 gene and CD44s protein structure. (A) Schematic representation of full-length CD44, CD44s, CD44v3, CD44v6 and CD44v8-10. (B) Example structure and N-glycan pattern of myeloma CD44-HABD monosialo (12).
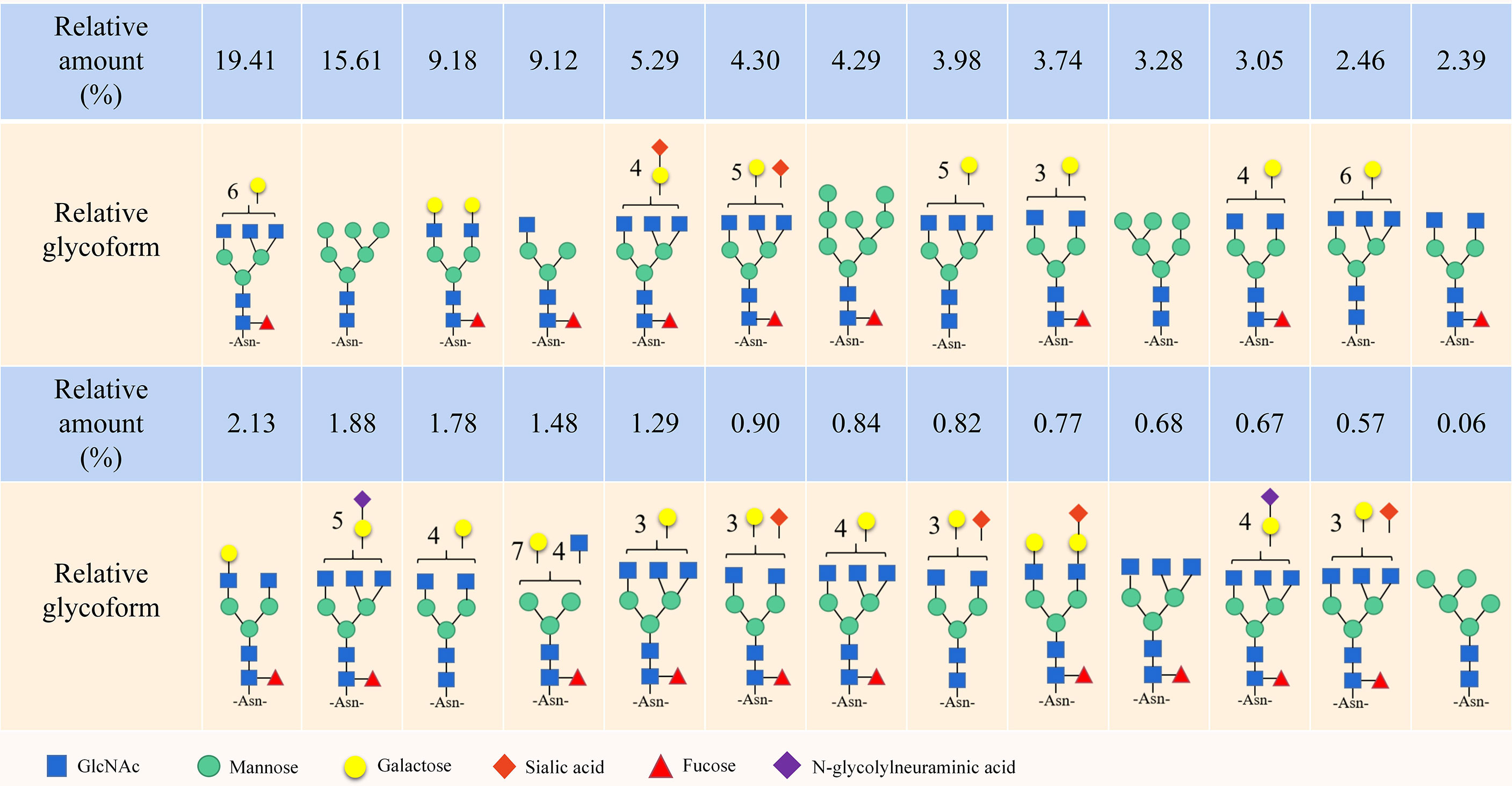
Notably, the CD44 extracellular structure is posttranslationally modified by N-glycans, O-glycans and glycosaminoglycans (heparan sulfate [HS], chondroitin sulfate [CS] and keratan sulfate) (17–19). Five conserved N-glycosylation consensus sites are located at amino-terminal 120 aa of CD44 (Figure 1B) (20). Mutation of any one of the five N-glycosylation sites of human CD44 results in the loss of wild-type CD44-mediated adhesion in active human cell lines (21). Nonsialylated and nonsucucosylated complex glycans dominate the N-glycans of CD44s (Figure 2) . In addition, the site-specific N-glycan profiles analyzed using LC-ESI-MS (E) showed that the vast majority of glycosyl groups, except for glycosyl N100, contained complex-type sugars, and high-mannose-type N-glycans occupied glycosyl N100 (22). The level of N-glycosylation may play a key role in CD44 activation-specific ligand binding (23–25). Most of the potential sites for O-glycosylation are located at the membrane proximal end of the CD44 ectodomain. A total of 146 O-glycan sites were predicted (26). Colon cancer cells modify CD44 with O-linked glycosyl groups, blocking CD44-mediated adhesion to HA (27). However, some studies suggest that the binding of HA to CD44 is not related to the level of CD44 O-glycosylation (25, 28).
Signals for glycosaminoglycan (GAG) assembly are encoded by the proteoglycan backbone. GAG synthesis occurs at serine followed by glycine, one or more proximal acidic amino acids. In addition, some acceptor sites are modified with CS only, and some acceptor sites are modified with both CS and HS (29). This suggests that CS synthesis occurs by default wherever GAGs can be attached, whereas HS assembly requires an additional signal. Duplication of the SG motif and signals found in the proteoglycan backbone also include proximal hydrophobic residues, when HS is assembled (30–32). The SGSG motif in the exon of CD44v3 is the only assembly site for HS, and the HS and CS linking at this site is abolished by serine to alanine (AGAG) mutation in the V3 motif. Eight amino acids downstream of the SGSG site in the V3 region are responsible for the specific addition of HS to this site (29). The E5 exon is the only exon in CD44 that supports GAG assembly and is modified by CS. If the 8 amino acids downstream of the first SG site of the CD44 E5 exon are exchanged with the 8 amino acids downstream of the SGSG site of the V3 exon, the SG site of E5 is modified by HS and CS. The 8 amino acids downstream of the first SG found in E5 are located downstream of SGSG in V3, and this site is modified by CS but not HS (29).
N-Glycosylation and Sialylation Levels Regulate CD44 Binding to HA
HA is a group of polysaccharides, originally designated acid mucopolysaccharides, which are now labeled as glycosaminoglycans and are usually found in the connective tissue of vertebrates (33). Although HA is similar to other glycosaminoglycans, it consists of a single polysaccharide chain due to its molecular weight, which is in the millions of daltons (34). HA is composed of repeating disaccharide units of N-acetyl-D-glucosamine and D-glucuronic acid and is abundantly present in the extracellular matrix (35). HA is a polyanion that can self-associate or bind to water molecules, creating a viscous, gel-like environment when it is not bound to other molecules (35). The size of the HA polymer ranges from 50 to 20 million Da, and the function of HA is largely determined by its size (36). High molecular weight HA is present in normal, intact, healthy tissues and helps maintain normal homeostasis by inhibiting cell proliferation, migration, angiogenesis, inflammation and immunogenicity (37). Low-molecular-weight HA regulates tumor cell motility by binding to the CD44 receptor (38, 39). The interaction between HA and the CD44 receptor affects cell proliferation, survival, motility, invasiveness and chemoresistance (40). Posttranslational modifications, including glycosylation, sulfation, phosphorylation and clustering can all regulate the binding capacity of CD44 to HA (38, 41).
CD44 N-glycosylation regulates the "on" or "off" status of CD44 binding to HA (12, 24). Of the five N-glycosylation sites in CD44-HABD, Asn25 and Asn120 are required for HA binding and subsequent biological functions (20, 42). The negatively charged sugar chain attached to Asn25 directly hinders the binding of CD44 to HA (12), and N25 glycans can interact closely with nearby N100 and N110 glycans to form a sugar shield covering the typical binding sites of HA (43). Asn120 is located on the backside of the HABD close to Arg29 and does not appear to directly hinder the binding of CD44 to HA (12). A possible explanation is that charged glycans attached to Asn120 interfere with CD44 self-association and optimal binding of cross-linked CD44 molecules by directing them to adjacent sites on the same HA molecule (44). Furthermore, two mutations (IRAWB14 and IRAWB26) immediately adjacent to Asn120 in the CD44 structure, centered on Lys68, affect CD44 binding to HA (45).
The CD44 antigen is modified with sialic acids at the terminus of its glycans (46). The sialidase inhibitor 2-deoxy-2,3-dehydro-N-acetylneuraminic acid blocks HA recognition (47). When CD44 N-glycans are modified with sialic acid, sialic acid forms a competing intramolecular contact with the arginine side chain, thereby hindering the binding of HA (48). When CD44 N-glycans are modified with sialic acid, sialic acid forms a competing intramolecular contact with the arginine side chain, thereby hindering the binding of HA (48). Sialidase catalyzes the removal of sialic acid (49), including NEU1, NEU2, NEU3 and NEU4 and is associated with apoptosis, neuronal differentiation, and tumorigenesis (50). After NEU4 overexpression or activation, the modification of α-(2,3)-sialylation on CD44 decreases, and the binding affinity of CD44 for HA increases in HCC (51). Similarly, in airway inflammation, CD44 HA binding activity is also dependent on Neu1 enzymatic activity (52). The sialylation status of CD44 may be more important than its degree of expression for binding to HA (53).
N-glycosylation at specific sites of CD44 and the level of sialylation modification on N-glycans together affect HA binding. The extent of primary CD44 glycosylation and the size of the attached oligosaccharides determine the coverage of the binding site to HA. It is worth noting that the addition of sialic acid had little effect on binding site coverage compared with similarly sized nonsialylated N-glycans (43). The inhibition of HA binding by sialylation may be due to the increased degree of negative charge. Smaller N-glycan types, such as simple GlcNAc residues, lack the range and charge necessary to negatively affect HA binding. Conversely, the presence of GlcNAc residues provides HA with an additional binding surface, potentially facilitating HA recognition by providing additional polar interaction sites and minimal binding barriers (43, 54). In addition, N-glycosylation of CD44-HABD promotes a secondary, less shielded but weaker (>10 μM) HA binding site (12, 55). In the case of CD44 N-glycosylation, the flanking arginines (R150, R154 and R162) are relatively more likely to interact with HA (43). Modulation of glycosylation alone cannot completely block CD44 binding to HA; however, this binding is weak enough that modulation of the CD44 N-glycan/HA axis remains a potential therapeutic target.
CD44 O-Glycosylation Regulates The Aggressiveness of Cancer
The O-glycosylation process refers to the addition of N-acetylgalactosamine (GalNAc) to serine or threonine residues in proteins and the addition of other sugar branches to create more complex structures (56, 57). The biosynthesis of O-glycans is controlled by T-synthase (C1GalT1) (58, 59). The endoplasmic reticulum-localized chaperone Cosmc is required for C1GalT1 activity and expression (60, 61). Cosmc dysfunction results in inactive C1GalT1 and subsequent expression of the Tn antigen (62). Cosmc deficiency disrupts the CD44 O-glycosylation structure and subsequent inhibition of MAPK signaling leads to the inhibition of breast cancer cell growth in vivo (63). Deficiency or overexpression of Tn antigens occurs in many types of cancer, including gastric, colon, breast, lung, esophageal, prostate and endometrial cancers (64), which suggests that aberrant O-glycosylation occurs frequently during tumor development. In the complex O-glycosylation process, the formation of the Core 1 O-glycosyl is considered to be one of the main modes of glycosylation. Core 1-mediated disruption of CD44 O-glycosylation allows truncated CD44 in human colon cancer cell lines to be secreted into the extracellular environment via microvesicles; thus, exosome CD44 may be a potential vehicle for aberrant O-glycosylation (65). In addition, higher levels of serum CD44 protein containing the STn structure have been shown to distinguish gastric cancer patients from healthy subjects (66, 67).
C1GalT1 is overexpressed in many cancers of epithelial origin, including colon, breast, gastric, HNSCC, esophagus, prostate, and hepatocellular carcinoma. C1GalT1 overexpression is also frequently associated with poorer prognosis and poorer patient survival (68). The oncogenic effects of C1GalT1 may be achieved by altering the glycosylation and function of receptor tyrosine kinases, cell surface integrins, and cell surface death receptors (68). However, human pancreatic cancer cells with knockout of C1GALT1 had an increased tendency for tumorigenesis and metastasis (69). By localizing Tn-containing glycoproteins in C1GalT1 KO cells, Leon et al. (70) found that Tn was significantly enriched on the CSC glycoprotein CD44. C1GalT1-mediated truncation of O-glycans on CD44 inactivates the ERK/NF-κB signaling pathway, resulting in NANOG expression in pancreatic cancer (PC) cells and changes in tumor stem cell characteristics. However, high C1GALT1 expression is associated with poor survival in pancreatic duct adenocarcinoma patients (71). Why does truncation of CD44 O-glycosylation contribute to downstream signaling? We speculate that excessive O-glycosylation, similar to incomplete O-glycosylation of CD44, affects the efficiency of CD44 function (68–71). Furthermore, the increased tumorigenicity of pancreatic cancer caused by knockdown of C1GalT1 may be also related to the truncation of o-glycosylation on MUC16 (69).
Proteoglycan 4 (PRG4) is a mucin-like glycoprotein originally found in synovial fluid, a secreted product of the intimal cells of joint tissue and present on the surface of articular cartilage (72). Extensive O-glycosylated mucin-like domains are required for the boundary lubrication and lytic properties of PRG4 at various biological interfaces in vivo, including articular cartilage, tendon, pericardium, and the ocular surface (72). PRG4 is a ligand for CD44 (73–75). Removal of CD44 O-glycosylation significantly increases rhPRG4 binding to CD44 (76). PRG4 has been observed to inhibit cancer progression through the CD44/TGF-β pathway (77, 78). However, it is unknown whether PRG4 inhibits tumor progression by binding to CD44, thereby competing with HA or other ligands.
Fucosyltransferase-Mediated CD44 Fucosylation Promotes Tumor Progression
Fucosylation comprises the attachment of a fucose residue to N-glycans, O-glycans and glycolipids, and is one of the most common modifications (79). All fucosylation reactions in cells are catalyzed by focusyltransferases (FUTs). To date, 13 FUTs—FUT1 to 11, protein O-focusyltransferase 1 (POFUT1) and POFUT—have been identified (80). After CD44 N-acetylglucosamine is modified by α-(1,3)-focusylation, it can further form a sialyl-Lewis X (sLeX) structure, which can effectively bind to E-selectin (81, 82). Cell adhesion mediated by selectin and its carbohydrate ligand sLeX plays an important role in cancer metastasis (83). CD44 fucosylation improves the stemness of mouse bone marrow mesenchymal stem cells (84–87). During chemotherapy in canine lymphosarcoma, Xiong et al. (88) found that the concentration of CD44 fucosylated protein decreased by more than 2-fold, which may be related to tumor cell adhesion and migration. Increasing some specific forms of CD44 fucosylation in tumors is also thought to promote tumor progression.
MicroRNA 29b is a tumor suppressor with important effects on cancer progression (89). Specificity protein 1 (Sp1) is a well-known member of a family of transcription factors that also includes Sp2, Sp3, and Sp4, which are involved in processes such as cell growth, differentiation, apoptosis, and carcinogenesis (90). Sp1 is one of the targets of miR-29b (91, 92). Liu et al. (93) found that miR-29b/Sp1 mediates the malignancy of leukemia stem cells by regulating FUT4-mediated CD44 fucosylation in acute myeloid leukemia. CD44/E-selectin-binding signaling upregulates intercellular adhesion molecule 1 (ICAM-1) expression on the cell surface via the PKCα/p38/Sp1 pathway, thereby promoting melanoma cell metastasis (94). The E-selectin ligand activity of CD44 is conferred by sialylation and fucosylation modification of CD44 N-glycans (95). The lncRNA HOX transcribed antisense RNA (HOTAIR) is elevated in broad-spectrum tumors and is associated with metastasis and poor prognosis (96). In human tumors, HOTAIR competitively binds to miR-326, thereby regulating miR-326 expression (97–99). The HOTAIR/miR-326 axis was shown to regulate FUT6 expression to promote fucosylation of CD44 in colorectal cancer, and fucosylated CD44 could activate the PI3K/AKT/mTOR pathway to promote tumor progression (100). FUT6 mediates cell surface α-(1,3)-fucosylation, induces sLeX expression, and converts CD44 to hematopoietic cell E-/L-selectin ligand (HCELL) (82). Both FUT4 and FUT6 can modify α-(1,3)fucosyl bond formation. In addition, transfection of α-(1,2)-focusyltransferase (FUT1/2) cDNA into colon cancer cell lines resulted in cell surface expression of CD44 variants carrying the amino acid sequence encoded by exon v6 (CD44v6), thereby promoting tumorigenesis progress (101–104). CD44v6 is thought to be expressed and to have increased tumorigenicity in various human tumors, including colorectal, and head and neck squamous cell carcinoma (HNSCC) (105–107).
The enzyme α-(1,6)-focusyltransferase (FUT8) is the only fucosyltransferase responsible for protein N-glycan core fucosylation (108). In the Golgi apparatus, FUT8 transfers an L-focal site from guanosine diphosphate (GDP-β-L-focus) (GDP-fuc) to the innermost GlcNAc of N-glycans to form an α-1,6 fucosyl bond (109, 110). FUT8 knockout homozygous mice experience early postnatal death, severe growth retardation, and emphysema-like changes in the lungs (111). FUT8 has an important stabilizing effect on N-glycan structure, which allows FUT8 to regulate key glycoproteins. FUT8-mediated core fucosylation plays an important role in regulating the biological functions of EGFR, TGFBR, E-cadherin, PD1/PD-L1 and α3β1 integrin (80). However, there is still no report on the effect of removing core fucosylation modification from the CD44 protein.
CD44 N-Glycosylation Regulates its Binding to TM4SF5
Hepatic transmembrane 4 L six family member 5 (TM4SF5) is a membrane protein and a member of the tetrase protein family, with four transmembrane domains, a cytoplasmic N- and C-terminus, and an intracellular loop. TM4SF5 is N-glycosylated on residues N138 and N155 and palmitoylated on cysteine residues near the cytoplasmic boundary of the transmembrane domain (112). Similar to other tetraterpenoids, TM4SF5 has been shown to interact with a variety of membrane proteins and receptors on the cell membrane, resulting in TM4SF5-enriched microdomains (T5ERMs) (113). TM4SF5 has been shown to form protein–protein complexes with CD44 (114), CD133 (115), CD151 (116), epidermal growth factor receptor (EGFR) (117), insulin-like growth factor 1 receptor (IGF1R) (118) and integrin-α5 (119) on the cell surface and play roles in tumor cell migration and anticancer drug resistance. The TM4SF5/CD44 interaction activates the proto-oncogene tyrosine-protein kinase Src (c-Src)/signal transducer and activator of transcription 3 (STAT3)/Twist-related protein 1 (Twist1)/B-cell-specific Moloney murine leukemia virus integration site 1 (Bmi1) signaling pathway (120), and epithelial-mesenchymal transition (EMT) (111), which makes TM4SF5/CD44 a potential target for tumor-targeted therapy with coexpression of TM4SF5 and CD44. The interaction between TM4SF5 and CD44 and the activation of the c-Src/STAT3/Twist1/Bmi1 pathway occur through the N-glycosylation modification of the extracellular domain of TM4SF5 and CD44 (114). Targeted therapy of TM4SF5 or TM4SF5/CD44 interaction may effectively inhibit tumor progression.
CD44 Terminal Sialic Acids on Tumors Regulate Their Binding to Siglec-15
Siglec-15 was originally identified as a member of the Siglec family with structural features of sialic acid-binding immunoglobulin-type lectins (121). Siglec15 is upregulated in many human cancers, and as an immunosuppressive molecule that plays a role in the tumor microenvironment (TME), it is mutually exclusive with PD-L1 and has potential implications in patients with anti-PD-1/PD-L1 resistance (122, 123). CD44 is a ligand for Siglec-15 (124). In the presence of CD44 N-glycan α-(2,6)-linked sialic acid modifications, Siglec-15 interacts with CD44 and mediates liver cancer progression and metastasis by preventing CD44 lysosome-mediated degradation (125).
The ST3GAL4 gene encodes β-galactosidase α-(2,3) sialyltransferase 4 and is involved in the biosynthesis of tumor antigens sLeX and sulfo sLeX (126). MicroRNA 193b targets ST3GAL4 to regulate CD44 sialylation via the NF-κB pathway, thereby accelerating osteoarthritis progression (127). The α-(2,3)-sialyltransferases ST3GAL1 and ST3GAL4 are the main enzymes for the synthesis of Siglec7 and Siglec-9 in tumor cells. Both ST3GAL4 and Siglec-7/9 have been shown to play important roles in human tumors (128, 129). However, whether ST3GAL4 regulates CD44 sialylation and plays a role in tumors still needs to be confirmed. In conclusion, inhibition of tumor progression by modulating CD44 sialylation appears to be a viable option.
HCELLs Are Major E-/L-Selectin Ligands in Tumors
E-selectin is a cytokine-activated cell adhesion molecule expressed on endothelial cells and plays an important role in the adhesion of inflammatory and metastatic cancer cells to endothelial cells (130). L-selectin is an L-type transmembrane glycoprotein and cell adhesion molecule expressed on most circulating leukocytes and plays a role in regulating monocyte protrusion during monocyte transendothelial migration (131). Hematopoietic cell E/L-selectin ligand (HCELL) is a specific glycated form of sialofucosylated CD44 that is characteristically expressed on human hematopoietic stem cells and is the most potent E-selectin and L-selectin expressed on human cell prime ligands (132). According to the current commonly used CD44 nomenclature, the activities of HCELL on standard and variant CD44 isoforms are designated as HCELLs and HCELLv, respectively (133). The two isoforms of HCELL differ from each other at the protein backbone and glycosylation levels. The two HCELL isoforms have significantly different molecular weights: HCELL migrates as a band of 90-100 kDa on SDS–PAGE gels, whereas the HCELLv of colon cancer cells typically has a molecular weight of approximately 150 kDa (134). Metastatic MDA-MB-231 breast cancer cells express high levels of the ~170 kDa HCELLv4 isoform (135). HCELLs have been shown to be present at high levels in human malignant hematopoietic cells, including neonatal acute myeloid leukemia (AML) cells and the AML-derived primitive human hematopoietic progenitor cell line KG1a (95, 132, 136). In contrast, malignant cells of solid cancers predominantly express the HCELLv subtype (132, 133, 137).
HCELLv is characterized by the presentation of sialofucosylated glycosyl groups on O-glycans of the CD44 subtype. Despite these differences, cleavage-based in vitro assays suggest that HCELLs and HCELLv are equally potent ligands for E- and L-selectin (81). Current evidence suggests that HCELL is a key E-selectin ligand in breast cancer (138) and a major E-/L-selectin ligand in colon cancer (139). Human bone marrow-derived mesenchymal stem cells migrate across endothelial cells without the need for chemokine signaling through a VLA-4/vcam-1-dependent "Step 2-bypass pathway" after forced targeted expression of HCELL (140, 141). The discovery of HCELL and its function as a bone marrow homing receptor may have enormous implications for the realization of inverted glycobiology in clinical medicine.
CD44 Chondroitin Sulfate Modification Mediates CD44 Binding to Fibronectin and Collagen in Tumors
Chondroitin sulfate (CS) is a sulfated glycosaminoglycan (GAG) distributed on the cell surface and in the extracellular space. CS chains are covalently linked to a core protein called CS proteoglycan (CSPG), which mediates protein–protein interactions between cells and the extracellular matrix (ECM) by maintaining the physical structure of the tissue, supporting various biological functions of CSPG (142). CSPG interacts with multiple transmembrane proteins, including integrins and receptor tyrosine kinases, and modulates cell signaling related to tumor cell proliferation, invasion, migration, angiogenesis, and metastasis (143, 144). CS and keratin sulfate are found on CD44 in certain cell types (145, 146). The addition of this glycosaminoglycan has been shown to modify the function of CD44. The keratin sulfate side chain on CD44H in highly metastatic colon cancer cells significantly reduces HA binding (146). Keratin sulfate modification of CD44 regulates hyaluronic acid adhesion through its B-loop domain (146). Tumor necrosis factor alpha or lipopolysaccharide and interferon gamma (LPS/IFNγ) stimulation in mouse bone marrow-derived macrophages induces HA binding by downregulating the sulfation of CD44 by CS (147). Chondroitin sulfate-modified CD44 (110 kDa) in mouse melanoma promotes melanoma cell motility (not adhesion and spread) on type I collagen. Furthermore, the binding of CD44 to type I collagen was mediated by chondroitin sulfate by affinity chromatography and solid-phase binding assays (148). The nonspliced core protein of CD44 is glycosylated by chondroitin sulfate, which promotes the migration of fibroblasts to the fibrin clot and the migration of endothelial cells on the fibrin matrix. The attachment of CD44 to fibronectin requires chondroitin sulfate modification (16, 149). CD44 chondroitin sulfate modification may be a target for inhibiting tumor cell motility and metastasis.
CD44 Heparan Sulfate Modification Mediates CD44 Binding to Growth Factors
HS proteoglycan is ubiquitously expressed on the surface of most animal cells and in the extracellular matrix, and its function mainly depends on the interaction of the HS side chain with various proteins such as cytokines, growth factors and their receptors, and it plays an important role in tumor progression (150). In HS polysaccharides, negatively charged sulfate and carboxylic acid groups are arranged in various domains and generated through tightly regulated biosynthetic reactions, with great potential for structural change (151). The CD44v3 isoform containing the HS attachment site is overexpressed on the tumor epithelium of colorectal adenomas and most carcinomas compared with normal colon (152). CD44v3 is modified by HS and has been shown to bind growth factors (153, 154), promoting the binding of CD44 molecules to the cytoskeleton in colon cancer (155). In colorectal cancer, the heparin-like and chondroitin sulfate B side chains of CD44 bind to the laminin A5G27 peptide (156, 157). The A5G27 peptide has excellent specificity for cancer cells overexpressing CD44v3 and CD44v6 and inhibits the migration and invasion of cancer cells (158). A5G27 shares considerable sequence homology (69%) with a sequence in fibroblast growth factor 2 (FGF2) that binds heparin and the FGF receptor, which is essential for central cavity formation in FGF2 (159). CD44 binding to FGF2 specifically increases FGF2-mediated proliferation, migration and survival of tumor and endothelial cells, thereby increasing tumor growth and metastasis (160, 161). The A5G27 peptide inhibits melanoma metastasis and angiogenesis by reducing the biological activity of FGF2 by blocking the binding of FGF2 to the HS side chain of CD44v3 (162). Modulation of CD44 heparan sulfate modification may be a promising antitumor therapy.
CD44 Glycosylation Negatively Regulates Podoplanin-CD44 Binding in Squamous Cell Carcinoma
Podoplanin (PDPN) is a type I transmembrane mucin-like sialoglycoprotein (163). PDPN is highly expressed on lymphatic endothelial cells and used as a marker of lymphangiogenesis (164). Mice deficient in PDPN die soon after birth due to abnormalities in the lungs, heart, and lymphatic vasculature (165). PDPN increases tumor cell clonality, EMT, migration, invasion, metastasis and inflammation in tumors including glioma, squamous cell carcinoma, mesothelioma and melanoma and is considered a potential tumor biomarker and therapy target (166). Podoplanin molecules lack obvious enzymatic motifs, so they must exert their biological and pathological functions through protein–protein interactions. The C-type lectin-like receptor 2 (CLEC-2), ERM (ezrin, radixin, moesin) protein family members ezrin and moesin, CD9 tetraspanin, standard isoforms of CD44s and CD44s have been found in different cell types and environments in which PDPN interacts (167). In squamous cell carcinoma (SCC) cells, CD44 and PDPN colocalize on cell surface protrusions, and CD44 is required for PDPN to promote directional and sustained movement of epithelial cells (168). CD44v3-10 is the main variant isoform coexpressed by CD44s and PDPN in human SCC cell lines (169). The interaction of PDPN and CD44 is mediated by the transmembrane and cytoplasmic domains and is negatively regulated by glycosylation of the extracellular domain of CD44 (169). Inhibition of PDPN-mediated tumor progression by regulating CD44 glycosylation deserves further in-depth research.
Conclusion and Prospect
The level of CD44 N-glycosylation and sialylation negatively regulates the binding to HA; however, whether O-glycosylation has a similar effect still needs further study. The functioning of CD44 generally involves combining with some membrane proteins or the extracellular matrix, and N-glycosylation and O-glycosylation on the extracellular domain of CD44 cover or constitute the corresponding binding site, thereby regulating the tumor microenvironment and intracellular signal transduction.
The development of glycosyl-based cancer neoantigens as cancer vaccines and targeted therapies may pave the way for more effective and specific tumor targeting (170). The monoclonal antibody F77 is highly specific for prostate cancer and can recognize the glycosylation structure of CD44v10 (171). Bivalent F77 can induce apoptosis in prostate cancer cells, particularly at 4°C (172), where F77 has a higher binding affinity to its antigen (173). Downregulation of CD44 or FUT1 genes significantly reduced F77-induced apoptosis in prostate cancer cell lines, suggesting that the binding site of F77 may require fucosylation modification (172). KMP1 is an IgG1 antibody that specifically binds EJ, BIU-87, and T24 bladder cancer cell lines and bladder cancer tissue but not Lovo, HeLa, K562, HepG2, Jurkat, 293, or HCV29 cell lines, human erythrocytes, human lymphocytes or normal bladder tissue (174). However, the effectiveness of CD44 as a therapeutic or diagnostic target has not been fully demonstrated in some other studies. In addition, polysaccharide-based biomaterials HA and CS have attracted great interest as tumor drug delivery systems due to their good biocompatibility with and targeting to CD44. The use of such drugs, combined with drugs targeting the CD44 glycosylation site, may be able to achieve better therapeutic effects (175).
The critical biological roles of CD44s and CD44v in tumors have been extensively studied; however, the glycosylation patterns of CD44s and CD44v in different tumors are unknown. How to engineer the glycosylation pattern of the CD44 extracellular domain to achieve antitumor effects still requires much effort. Notably, the CD44 glycosylation pattern as a precise diagnostic marker in different tumors has also not been reported. The identification of glycan epitopes by tumor subtype may have potential applications in patient treatment stratification. Furthermore, the prospects for creating CAR-T cells specific to CD44 glycosylation could be very interesting work. The complexity of the glycosylation process and the lack of specific methods to study it hinder related research progress. However, recent advances in new methods such as cellular glycoengineering and high-throughput screening (HTS) have opened new avenues of discovery (176), which helps to explore the effect of different glycosylation of specific amino acid residues on the binding of different ligands
Author Contributions
XG, LX and JL contributed to the conception and design of the review. CL wrote the manuscript. QW, JA and QL validate the manuscript. XL and JC contributed to the visualization of CD44s glycosylation. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was supported by the National Key R&D Program (2016YFC1102800); Guizhou Province Medical Biomaterials R&D Talent Base (QianRenLingFa [2018] No. 3); the Sixth Talent Foundation in Guizhou province (rcjd2019-9); the Graduate Research Fund of Guizhou Province (Qian-Jiao-He YJSCXJH [2019]087); Zunyi Medical Biomaterials R&D and Innovative Talent Base (ZunWei [2019] No. 69); the Youth Science and Technology Talents Growth Project of Guizhou Education Department (Qian-Jiao-He KY ZI [2018]236); Zunyi City and Zunyi Medical University Unite Fund [QianShiKeHe HZ Zi (2022)393], Outstanding Young Talent Project of Zunyi Medical University (17zy-002) (F-801); The Project to Cultivate Young Scientific; Master Fund of Zunyi Medical University (S-81) and Technological Talents in Colleges and Universities of Guizhou Province (Qian Jiao He KY [2021] 215).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chen C, Zhao S, Karnad A, Freeman JW. The Biology and Role of CD44 in Cancer Progression: Therapeutic Implications. J Hematol Oncol (2018) 11(1):64. doi: 10.1186/s13045-018-0605-5
2. Screaton GR, Bell MV, Bell JI, Jackson DG. The Identification of a New Alternative Exon With Highly Restricted Tissue Expression in Transcripts Encoding the Mouse Pgp-1 (CD44) Homing Receptor. Comparison of All 10 Variable Exons Between Mouse, Human, and Rat. J Biol Chem (1993) 268(17):12235–8. doi: 10.1111/j.1432-1033.1993.tb17998.x
3. Batsché E, Yi J, Mauger O, Kornobis E, Hopkins B, Hanmer-Lloyd C, et al. CD44 Alternative Splicing Senses Intragenic DNA Methylation in Tumors via Direct and Indirect Mechanisms. Nucleic Acids Res (2021) 49(11):6213–37. doi: 10.1093/nar/gkab437
4. Yamano S, Gi M, Tago Y, Doi K, Okada S, Hirayama Y, et al. Role of Deltanp63(Pos)CD44v(pos) Cells in the Development of N-Nitroso-Tris-Chloroethylurea-Induced Peripheral-Type Mouse Lung Squamous Cell Carcinomas. Cancer Sci (2016) 107(2):123–32. doi: 10.1111/cas.12855
5. Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H. CD44 is Required for Two Consecutive Steps in HGF/c-Met Signaling. Genes Dev (2002) 16(23):3074–86. doi: 10.1101/gad.242602
6. Wang L, Zuo X, Xie K, Wei D. The Role of CD44 and Cancer Stem Cells. Methods Mol Biol (2018) 1692:31–42. doi: 10.1007/978-1-4939-7401-6_3
7. Roedig H, Damiescu R, Zeng-Brouwers J, Kutija I, Trebicka J, Wygrecka M, et al. Danger Matrix Molecules Orchestrate CD14/CD44 Signaling in Cancer Development. Semin Cancer Biol (2020) 62:31–47. doi: 10.1016/j.semcancer.2019.07.026
8. Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in Health and Disease. Nat Rev Nephrol (2019) 15(6):346–66. doi: 10.1038/s41581-019-0129-4
9. Johannes L, Billet A. Glycosylation and Raft Endocytosis in Cancer. Cancer Metastasis Rev (2020) 39(2):375–96. doi: 10.1007/s10555-020-09880-z
10. Chen KL, Li D, Lu TX, Chang SW. Structural Characterization of the CD44 Stem Region for Standard and Cancer-Associated Isoforms. Int J Mol Sci (2020) 21(1):336. doi: 10.3390/ijms21010336
11. Mishra MN, Chandavarkar V, Sharma R, Bhargava D. Structure, Function and Role of CD44 in Neoplasia. J Oral Maxillofac Pathol (2019) 23(2):267–72. doi: 10.4103/jomfp.JOMFP_246_18
12. Teriete P, Banerji S, Noble M, Blundell CD, Wright AJ, Pickford AR, et al. Structure of the Regulatory Hyaluronan Binding Domain in the Inflammatory Leukocyte Homing Receptor CD44. Mol Cell (2004) 13(4):483–96. doi: 10.1016/s1097-2765(04)00080-2
13. Perschl A, Lesley J, English N, Trowbridge I, Hyman R. Role of CD44 Cytoplasmic Domain in Hyaluronan Binding. Eur J Immunol (1995) 25(2):495–501. doi: 10.1002/eji.1830250228
14. Banerji S, Day AJ, Kahmann JD, Jackson DG. Characterization of a Functional Hyaluronan-Binding Domain From the Human CD44 Molecule Expressed in Escherichia Coli. Protein Expr Purif (1998) 14(3):371–81. doi: 10.1006/prep.1998.0971
15. Ishii S, Ford R, Thomas P, Nachman A, Steele G Jr, Jessup JM. CD44 Participates in the Adhesion of Human Colorectal Carcinoma Cells to Laminin and Type IV Collagen. Surg Oncol (1993) 2(4):255–64. doi: 10.1016/0960-7404(93)90015-q
16. Jalkanen S, Jalkanen M. Lymphocyte CD44 Binds the COOH-Terminal Heparin-Binding Domain of Fibronectin. J Cell Biol (1992) 116(3):817–25. doi: 10.1083/jcb.116.3.817
17. Lokeshwar VB, Bourguignon LY. Post-Translational Protein Modification and Expression of Ankyrin-Binding Site(s) in GP85 (Pgp-1/CD44) and its Biosynthetic Precursors During T-Lymphoma Membrane Biosynthesis. J Biol Chem (1991) 266(27):17983–9. doi: 10.1007/BF00016160
18. Stephenson EL, Yong VW. Pro-Inflammatory Roles of Chondroitin Sulfate Proteoglycans in Disorders of the Central Nervous System. Matrix Biol (2018) 71-72:432–42. doi: 10.1016/j.matbio.2018.04.010
19. Brown TA, Bouchard T, St John T, Wayner E, Carter WG. Human Keratinocytes Express a New CD44 Core Protein (CD44E) as a Heparan-Sulfate Intrinsic Membrane Proteoglycan With Additional Exons. J Cell Biol (1991) 113(1):207–21. doi: 10.1083/jcb.113.1.207
20. English NM, Lesley JF, Hyman R. Site-Specific De-N-Glycosylation of CD44 can Activate Hyaluronan Binding, and CD44 Activation States Show Distinct Threshold Densities for Hyaluronan Binding. Cancer Res (1998) 58(16):3736–42. doi: 10.1016/0017-9310(63)90063-2
21. Bartolazzi A, Nocks A, Aruffo A, Spring F, Stamenkovic I. Glycosylation of CD44 is Implicated in CD44-Mediated Cell Adhesion to Hyaluronan. J Cell Biol (1996) 132(6):1199–208. doi: 10.1083/jcb.132.6.1199
22. Han H, Stapels M, Ying W, Yu Y, Tang L, Jia W, et al. Comprehensive Characterization of the N-Glycosylation Status of CD44s by Use of Multiple Mass Spectrometry-Based Techniques. Anal Bioanal Chem (2012) 404(2):373–88. doi: 10.1007/s00216-012-6167-4
23. Hathcock KS, Hirano H, Murakami S, Hodes RJ. CD44 Expression on Activated B Cells. Differential Capacity for CD44-Dependent Binding to Hyaluronic Acid. J Immunol (1993) 151(12):6712–22.
24. Katoh S, Zheng Z, Oritani K, Shimozato T, Kincade PW. Glycosylation of CD44 Negatively Regulates its Recognition of Hyaluronan. J Exp Med (1995) 182(2):419–29. doi: 10.1084/jem.182.2.419
25. Lesley J, English N, Perschl A, Gregoroff J, Hyman R. Variant Cell Lines Selected for Alterations in the Function of the Hyaluronan Receptor CD44 Show Differences in Glycosylation. J Exp Med (1995) 182(2):431–7. doi: 10.1084/jem.182.2.431
26. Mereiter S, Martins ÁM, Gomes C, Balmaña M, Macedo JA, Polom K, et al. O-Glycan Truncation Enhances Cancer-Related Functions of CD44 in Gastric Cancer. FEBS Lett (2019) 593(13):1675–89. doi: 10.1002/1873-3468.13432
27. Dasgupta A, Takahashi K, Cutler M, Tanabe KK. O-Linked Glycosylation Modifies CD44 Adhesion to Hyaluronate in Colon Carcinoma Cells. Biochem Biophys Res Commun (1996) 227(1):110–7. doi: 10.1006/bbrc.1996.1475
28. Zheng Z, Cummings RD, Pummill PE, Kincade PW. Growth as a Solid Tumor or Reduced Glucose Concentrations in Culture Reversibly Induce CD44-Mediated Hyaluronan Recognition by Chinese Hamster Ovary Cells. J Clin Invest (1997) 100(5):1217–29. doi: 10.1172/JCI119635
29. Greenfield B, Wang WC, Marquardt H, Piepkorn M, Wolff EA, Aruffo A, et al. Characterization of the Heparan Sulfate and Chondroitin Sulfate Assembly Sites in CD44. J Biol Chem (1999) 274(4):2511–7. doi: 10.1074/jbc.274.4.2511
30. Zhang L, Esko JD. Amino Acid Determinants That Drive Heparan Sulfate Assembly in a Proteoglycan. J Biol Chem (1994) 269(30):19295–9. doi: 10.1016/S0021-9258(17)32166-X
31. Fritz TA, Lugemwa FN, Sarkar AK, Esko JD. Biosynthesis of Heparan Sulfate on Beta-D-Xylosides Depends on Aglycone Structure. J Biol Chem (1994) 269(1):300–7. doi: 10.1016/S0021-9258(17)42349-0
32. Zhang L, David G, Esko JD. Repetitive Ser-Gly Sequences Enhance Heparan Sulfate Assembly in Proteoglycans. J Biol Chem (1995) 270(45):27127–35. doi: 10.1074/jbc.270.45.27127
33. Balazs EA, Laurent TC, Jeanloz RW. Nomenclature of Hyaluronic Acid. Biochem J (1986) 235(3):903. doi: 10.1042/bj2350903
34. Fraser JR, Laurent TC, Laurent UB. Hyaluronan: Its Nature, Distribution, Functions and Turnover. J Intern Med (1997) 242(1):27–33. doi: 10.1046/j.1365-2796.1997.00170.x
35. Fakhari A, Berkland C. Applications and Emerging Trends of Hyaluronic Acid in Tissue Engineering, as a Dermal Filler and in Osteoarthritis Treatment. Acta Biomater (2013) 9(7):7081–92. doi: 10.1016/j.actbio.2013.03.005
36. Zhu Y, Kruglikov IL, Akgul Y, Scherer PE. Hyaluronan in Adipogenesis, Adipose Tissue Physiology and Systemic Metabolism. Matrix Biol (2019) 78-79:284–91. doi: 10.1016/j.matbio.2018.02.012
37. Stecco A, Gesi M, Stecco C, Stern R. Fascial Components of the Myofascial Pain Syndrome. Curr Pain Headache Rep (2013) 17(8):352. doi: 10.1007/s11916-013-0352-9
38. Cichy J, Puré E. Cytokines Regulate the Affinity of Soluble CD44 for Hyaluronan. FEBS Lett (2004) 556(1-3):69–74. doi: 10.1016/s0014-5793(03)01370-x
39. Koyama H, Hibi T, Isogai Z, Yoneda M, Fujimori M, Amano J, et al. Hyperproduction of Hyaluronan in Neu-Induced Mammary Tumor Accelerates Angiogenesis Through Stromal Cell Recruitment: Possible Involvement of Versican/PG-M. Am J Pathol (2007) 170(3):1086–99. doi: 10.2353/ajpath.2007.060793
40. Pratt RL. Hyaluronan and the Fascial Frontier. Int J Mol Sci (2021) 22(13):6845. doi: 10.3390/ijms22136845
41. Camp RL, Kraus TA, Puré E. Variations in the Cytoskeletal Interaction and Posttranslational Modification of the CD44 Homing Receptor in Macrophages. J Cell Biol (1991) 115(5):1283–92. doi: 10.1083/jcb.115.5.1283
42. Heldin P, Kolliopoulos C, Lin CY, Heldin CH. Involvement of Hyaluronan and CD44 in Cancer and Viral Infections. Cell Signal (2020) 65:109427. doi: 10.1016/j.cellsig.2019.109427
43. Vuorio J, Škerlová J, Fábry M, Veverka V, Vattulainen I, Řezáčová P, et al. N-Glycosylation can Selectively Block or Foster Different Receptor-Ligand Binding Modes. Sci Rep (2021) 11(1):5239. doi: 10.1038/s41598-021-84569-z
44. Lesley J, Hascall VC, Tammi M, Hyman R. Hyaluronan Binding by Cell Surface CD44. J Biol Chem (2000) 275(35):26967–75. doi: 10.1074/jbc.M002527200
45. Bajorath J, Greenfield B, Munro SB, Day AJ, Aruffo A. Identification of CD44 Residues Important for Hyaluronan Binding and Delineation of the Binding Site. J Biol Chem (1998) 273(1):338–43. doi: 10.1074/jbc.273.1.338
46. Guvench O. Revealing the Mechanisms of Protein Disorder and N-Glycosylation in CD44-Hyaluronan Binding Using Molecular Simulation. Front Immunol (2015) 6:305. doi: 10.3389/fimmu.2015.00305
47. Katoh S, Miyagi T, Taniguchi H, Matsubara Y, Kadota J, Tominaga A, et al. Cutting Edge: An Inducible Sialidase Regulates the Hyaluronic Acid Binding Ability of CD44-Bearing Human Monocytes. J Immunol (1999) 162(9):5058–61.
48. Faller CE, Guvench O. Terminal Sialic Acids on CD44 N-Glycans can Block Hyaluronan Binding by Forming Competing Intramolecular Contacts With Arginine Sidechains. Proteins (2014) 82(11):3079–89. doi: 10.1002/prot.24668
49. Schauer R, Kamerling JP. Exploration of the Sialic Acid World. Adv Carbohydr Chem Biochem (2018) 75:1–213. doi: 10.1016/bs.accb.2018.09.001
50. Bigi A, Tringali C, Forcella M, Mozzi A, Venerando B, Monti E, et al. A Proline-Rich Loop Mediates Specific Functions of Human Sialidase NEU4 in SK-N-BE Neuronal Differentiation. Glycobiology (2013) 23(12):1499–509. doi: 10.1093/glycob/cwt078
51. Zhang X, Dou P, Akhtar ML, Liu F, Hu X, Yang L, et al. NEU4 Inhibits Motility of HCC Cells by Cleaving Sialic Acids on CD44. Oncogene (2021) 40(35):5427–40. doi: 10.1038/s41388-021-01955-7
52. Katoh S, Maeda S, Fukuoka H, Wada T, Moriya S, Mori A, et al. A Crucial Role of Sialidase Neu1 in Hyaluronan Receptor Function of CD44 in T Helper Type 2-Mediated Airway Inflammation of Murine Acute Asthmatic Model. Clin Exp Immunol (2010) 161(2):233–41. doi: 10.1111/j.1365-2249.2010.04165.x
53. Matsubara Y, Katoh S, Taniguchii H, Oka M, Kadota J, Kohno S. Expression of CD44 Variants in Lung Cancer and its Relationship to Hyaluronan Binding. J Int Med Res (2000) 28(2):78–90. doi: 10.1177/147323000002800203
54. Skelton TP, Zeng C, Nocks A, Stamenkovic I. Glycosylation Provides Both Stimulatory and Inhibitory Effects on Cell Surface and Soluble CD44 Binding to Hyaluronan. J Cell Biol (1998) 140(2):431–46. doi: 10.1083/jcb.140.2.431
55. Vuorio J, Vattulainen I, Martinez-Seara H. Atomistic Fingerprint of Hyaluronan-CD44 Binding. PloS Comput Biol (2017) 13(7):e1005663. doi: 10.1371/journal.pcbi.1005663
56. Magalhães A, Duarte HO, Reis CA. The Role of O-Glycosylation in Human Disease. Mol Aspects Med (2021) 79:100964. doi: 10.1016/j.mam.2021.100964
57. Springer AD, Dowdy SF. GalNAc-siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic Acid Ther (2018) 28(3):109–18. doi: 10.1089/nat.2018.0736
58. Xia L, McEver RP. Targeted Disruption of the Gene Encoding Core 1 Beta1-3-Galactosyltransferase (T-Synthase) Causes Embryonic Lethality and Defective Angiogenesis in Mice. Methods Enzymol (2006) 416:314–31. doi: 10.1016/S0076-6879(06)16021-8
59. Ju T, Xia B, Aryal RP, Wang W, Wang Y, Ding X, et al. A Novel Fluorescent Assay for T-Synthase Activity. Glycobiology (2011) 21(3):352–62. doi: 10.1093/glycob/cwq168
60. Wang Y, Ju T, Ding X, Xia B, Wang W, Xia L, et al. Cosmc is an Essential Chaperone for Correct Protein O-Glycosylation. Proc Natl Acad Sci U.S.A. (2010) 107(20):9228–33. doi: 10.1073/pnas.0914004107
61. Ju T, Cummings RD. A Unique Molecular Chaperone Cosmc Required for Activity of the Mammalian Core 1 Beta 3-Galactosyltransferase. Proc Natl Acad Sci U.S.A. (2002) 99(26):16613–8. doi: 10.1073/pnas.262438199
62. Ju T, Wang Y, Aryal RP, Lehoux SD, Ding X, Kudelka MR, et al. Tn and Sialyl-Tn Antigens, Aberrant O-Glycomics as Human Disease Markers. Proteomics Clin Appl (2013) 7(9-10):618–31. doi: 10.1002/prca.201300024
63. Du T, Jia X, Dong X, Ru X, Li L, Wang Y, et al. Cosmc Disruption-Mediated Aberrant O-Glycosylation Suppresses Breast Cancer Cell Growth via Impairment of CD44. Cancer Manag Res (2020) 12:511–22. doi: 10.2147/CMAR.S234735
64. Fu C, Zhao H, Wang Y, Cai H, Xiao Y, Zeng Y, et al. Tumor-Associated Antigens: Tn Antigen, sTn Antigen, and T Antigen. HLA (2016) 88(6):275–86. doi: 10.1111/tan.12900
65. Gao T, Wen T, Ge Y, Liu J, Yang L, Jiang Y, et al. Disruption of Core 1-Mediated O-Glycosylation Oppositely Regulates CD44 Expression in Human Colon Cancer Cells and Tumor-Derived Exosomes. Biochem Biophys Res Commun (2020) 521(2):514–20. doi: 10.1016/j.bbrc.2019.10.149
66. Oliveira FMS, Mereiter S, Lönn P, Siart B, Shen Q, Heldin J, et al. Detection of Post-Translational Modifications Using Solid-Phase Proximity Ligation Assay. N Biotechnol (2018) 45:51–9. doi: 10.1016/j.nbt.2017.10.005
67. Campos D, Freitas D, Gomes J, Magalhães A, Steentoft C, Gomes C, et al. Probing the O-Glycoproteome of Gastric Cancer Cell Lines for Biomarker Discovery. Mol Cell Proteomics (2015) 14(6):1616–29. doi: 10.1074/mcp.M114.046862
68. Wan Y, Yu LG. Expression and Impact of C1GalT1 in Cancer Development and Progression. Cancers (Basel) (2021) 13(24):6305. doi: 10.3390/cancers13246305
69. Chugh S, Barkeer S, Rachagani S, Nimmakayala RK, Perumal N, Pothuraju R, et al. Disruption of C1galt1 Gene Promotes Development and Metastasis of Pancreatic Adenocarcinomas in Mice. Gastroenterology (2018) 155(5):1608–24. doi: 10.1053/j.gastro.2018.08.007
70. Leon F, Seshacharyulu P, Nimmakayala RK, Chugh S, Karmakar S, Nallasamy P, et al. Reduction in O-Glycome Induces Differentially Glycosylated CD44 to Promote Stemness and Metastasis in Pancreatic Cancer. Oncogene (2022) 41(1):57–71. doi: 10.1038/s41388-021-02047-2
71. Kuo TC, Wu MH, Yang SH, Chen ST, Hsu TW, Jhuang JY, et al. C1GALT1 High Expression is Associated With Poor Survival of Patients With Pancreatic Ductal Adenocarcinoma and Promotes Cell Invasiveness Through Integrin αv. Oncogene (2021) 40(7):1242–54. doi: 10.1038/s41388-020-01594-4
72. Jay GD, Waller KA. The Biology of Lubricin: Near Frictionless Joint Motion. Matrix Biol (2014) 39:17–24. doi: 10.1016/j.matbio.2014.08.008
73. Qadri M, Jay GD, Zhang LX, Richendrfer H, Schmidt TA, Elsaid KA. Proteoglycan-4 Regulates Fibroblast to Myofibroblast Transition and Expression of Fibrotic Genes in the Synovium. Arthritis Res Ther (2020) 22(1):113. doi: 10.1186/s13075-020-02207-x
74. Bennett M, Chin A, Lee HJ, Morales Cestero E, Strazielle N, Ghersi-Egea JF, et al. Proteoglycan 4 Reduces Neuroinflammation and Protects the Blood-Brain Barrier After Traumatic Brain Injury. J Neurotrauma (2021) 38(4):385–98. doi: 10.1089/neu.2020.7229
75. Iqbal SM, Leonard C, Regmi SC, De Rantere D, Tailor P, Ren G, et al. Lubricin/Proteoglycan 4 Binds to and Regulates the Activity of Toll-Like Receptors In Vitro. Sci Rep (2016) 6:18910. doi: 10.1038/srep18910
76. Al-Sharif A, Jamal M, Zhang LX, Larson K, Schmidt TA, Jay GD, et al. Lubricin/Proteoglycan 4 Binding to CD44 Receptor: A Mechanism of the Suppression of Proinflammatory Cytokine-Induced Synoviocyte Proliferation by Lubricin. Arthritis Rheumatol (2015) 67(6):1503–13. doi: 10.1002/art.39087
77. Sarkar A, Chanda A, Regmi SC, Karve K, Deng L, Jay GD, et al. Recombinant Human PRG4 (Rhprg4) Suppresses Breast Cancer Cell Invasion by Inhibiting Tgfβ-Hyaluronan-CD44 Signalling Pathway. PloS One (2019) 14(7):e0219697. doi: 10.1371/journal.pone.0219697
78. Dituri F, Scialpi R, Schmidt TA, Frusciante M, Mancarella S, Lupo LG, et al. Proteoglycan-4 is Correlated With Longer Survival in HCC Patients and Enhances Sorafenib and Regorafenib Effectiveness via CD44 In Vitro. Cell Death Dis (2020) 11(11):984. doi: 10.1038/s41419-020-03180-8
79. Shan M, Yang D, Dou H, Zhang L. Fucosylation in Cancer Biology and its Clinical Applications. Prog Mol Biol Transl Sci (2019) 162:93–119. doi: 10.1016/bs.pmbts.2019.01.002
80. Liao C, An J, Yi S, Tan Z, Wang H, Li H, et al. FUT8 and Protein Core Fucosylation in Tumours: From Diagnosis to Treatment. J Cancer (2021) 12(13):4109–20. doi: 10.7150/jca.58268
81. Sackstein R. Glycosyltransferase-Programmed Stereosubstitution (GPS) to Create HCELL: Engineering a Roadmap for Cell Migration. Immunol Rev (2009) 230(1):51–74. doi: 10.1111/j.1600-065X.2009.00792.x
82. Pachón-Peña G, Donnelly C, Ruiz-Cañada C, Katz A, Fernández-Veledo S, Vendrell J, et al. A Glycovariant of Human CD44 Is Characteristically Expressed on Human Mesenchymal Stem Cells. Stem Cells (2017) 35(4):1080–92. doi: 10.1002/stem.2549
83. Kannagi R. Molecular Mechanism for Cancer-Associated Induction of Sialyl Lewis X and Sialyl Lewis A Expression-The Warburg Effect Revisited. Glycoconj J (2004) 20(5):353–64. doi: 10.1023/B:GLYC.0000033631.35357.41
84. Shi H, Jiang C, Yao H, Zhang Y, Zhang Q, Hou X, et al. CD44 Fucosylation on Bone Marrow-Derived Mesenchymal Stem Cells Enhances Homing and Promotes Enteric Nervous System Remodeling in Diabetic Mice. Cell Biosci (2021) 11(1):118. doi: 10.1186/s13578-021-00632-2
85. Chou KJ, Lee PT, Chen CL, Hsu CY, Huang WC, Huang CW, et al. CD44 Fucosylation on Mesenchymal Stem Cell Enhances Homing and Macrophage Polarization in Ischemic Kidney Injury. Exp Cell Res (2017) 350(1):91–102. doi: 10.1016/j.yexcr.2016.11.010
86. Xiao F, Chen L. Effects of Extracorporeal Fucosylation of CD44 on the Homing Ability of Rabbit Bone Marrow Mesenchymal Stem Cells. J Orthop Sci (2019) 24(4):725–30. doi: 10.1016/j.jos.2018.11.010
87. García-Bernal D, García-Arranz M, Yáñez RM, Hervás-Salcedo R, Cortés A, Fernández-García M, et al. The Current Status of Mesenchymal Stromal Cells: Controversies, Unresolved Issues and Some Promising Solutions to Improve Their Therapeutic Efficacy. Front Cell Dev Biol (2021) 9:650664. doi: 10.3389/fcell.2021.650664
88. Xiong L, Andrews D, Regnier F. Comparative Proteomics of Glycoproteins Based on Lectin Selection and Isotope Coding. J Proteome Res (2003) 2(6):618–25. doi: 10.1021/pr0340274
89. Yan B, Guo Q, Fu FJ, Wang Z, Yin Z, Wei YB, et al. The Role of miR-29b in Cancer: Regulation, Function, and Signaling. Onco Targets Ther (2015) 8:539–48. doi: 10.2147/OTT.S75899
90. Vizcaíno C, Mansilla S, Portugal J. Sp1 Transcription Factor: A Long-Standing Target in Cancer Chemotherapy. Pharmacol Ther (2015) 152:111–24. doi: 10.1016/j.pharmthera.2015.05.008
91. Jia LF, Huang YP, Zheng YF, Lyu MY, Wei SB, Meng Z, et al. miR-29b Suppresses Proliferation, Migration, and Invasion of Tongue Squamous Cell Carcinoma Through PTEN-AKT Signaling Pathway by Targeting Sp1. Oral Oncol (2014) 50(11):1062–71. doi: 10.1016/j.oraloncology.2014.07.010
92. Amodio N, Di Martino MT, Foresta U, Leone E, Lionetti M, Leotta M, et al. miR-29b Sensitizes Multiple Myeloma Cells to Bortezomib-Induced Apoptosis Through the Activation of a Feedback Loop With the Transcription Factor Sp1. Cell Death Dis (2012) 3(11):e436. doi: 10.1038/cddis.2012.175
93. Liu B, Ma H, Liu Q, Xiao Y, Pan S, Zhou H, et al. MiR-29b/Sp1/FUT4 Axis Modulates the Malignancy of Leukemia Stem Cells by Regulating Fucosylation via Wnt/β-Catenin Pathway in Acute Myeloid Leukemia. J Exp Clin Cancer Res (2019) 38(1):200. doi: 10.1186/s13046-019-1179-y
94. Zhang P, Goodrich C, Fu C, Dong C. Melanoma Upregulates ICAM-1 Expression on Endothelial Cells Through Engagement of Tumor CD44 With Endothelial E-Selectin and Activation of a Pkcα-P38-SP-1 Pathway. FASEB J (2014) 28(11):4591–609. doi: 10.1096/fj.11-202747
95. Dimitroff CJ, Lee JY, Rafii S, Fuhlbrigge RC, Sackstein R. CD44 is a Major E-Selectin Ligand on Human Hematopoietic Progenitor Cells. J Cell Biol (2001) 153(6):1277–86. doi: 10.1083/jcb.153.6.1277
96. Qu X, Alsager S, Zhuo Y, Shan B. HOX Transcript Antisense RNA (HOTAIR) in Cancer. Cancer Lett (2019) 454:90–7. doi: 10.1016/j.canlet.2019.04.016
97. Wang R, Chen X, Xu T, Xia R, Han L, Chen W, et al. MiR-326 Regulates Cell Proliferation and Migration in Lung Cancer by Targeting Phox2a and is Regulated by HOTAIR. Am J Cancer Res (2016) 6(2):173–86.
98. Yu GJ, Sun Y, Zhang DW, Zhang P. Long non-Coding RNA HOTAIR Functions as a Competitive Endogenous RNA to Regulate PRAF2 Expression by Sponging miR-326 in Cutaneous Squamous Cell Carcinoma. Cancer Cell Int (2019) 19:270. doi: 10.1186/s12935-019-0992-x
99. Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, et al. Knockdown of Long non-Coding RNA HOTAIR Inhibits Malignant Biological Behaviors of Human Glioma Cells via Modulation of miR-326. Oncotarget (2015) 6(26):21934–49. doi: 10.18632/oncotarget.4290
100. Pan S, Liu Y, Liu Q, Xiao Y, Liu B, Ren X, et al. HOTAIR/miR-326/FUT6 Axis Facilitates Colorectal Cancer Progression Through Regulating Fucosylation of CD44 via PI3K/AKT/mTOR Pathway. Biochim Biophys Acta Mol Cell Res (2019) 1866(5):750–60. doi: 10.1016/j.bbamcr.2019.02.004
101. Labarrière N, Piau JP, Otry C, Denis M, Lustenberger P, Meflah K, et al. H Blood Group Antigen Carried by CD44V Modulates Tumorigenicity of Rat Colon Carcinoma Cells. Cancer Res (1994) 54(23):6275–81. doi: 10.1007/BF01534430
102. Goupille C, Hallouin F, Meflah K, Le Pendu J. Increase of Rat Colon Carcinoma Cells Tumorigenicity by Alpha(1-2) Fucosyltransferase Gene Transfection. Glycobiology (1997) 7(2):221–9. doi: 10.1093/glycob/7.2.221
103. Hallouin F, Goupille C, Bureau V, Meflah K, Le Pendu J. Increased Tumorigenicity of Rat Colon Carcinoma Cells After Alpha1,2-Fucosyltransferase FTA Anti-Sense cDNA Transfection. Int J Cancer (1999) 80(4):606–11. doi: 10.1002/(sici)1097-0215(19990209)80:4<606::aid-ijc20>3.0.co;2-m
104. Montesino B, Steenackers A, Lozano JM, Young GD, Hu N, Sackstein R, et al. Identification of α1,2-Fucosylated Signaling and Adhesion Molecules in Head and Neck Squamous Cell Carcinoma. Glycobiology (2022) 32(5):441–55. doi: 10.1093/glycob/cwab131
105. Ma L, Dong L, Chang P. CD44v6 Engages in Colorectal Cancer Progression. Cell Death Dis (2019) 10(1):30. doi: 10.1038/s41419-018-1265-7
106. Liao C, An J, Tan Z, Xu F, Liu J, Wang Q. Changes in Protein Glycosylation in Head and Neck Squamous Cell Carcinoma. J Cancer (2021) 12(5):1455–66. doi: 10.7150/jca.51604
107. Heider KH, Kuthan H, Stehle G, Munzert G. CD44v6: A Target for Antibody-Based Cancer Therapy. Cancer Immunol Immunother (2004) 53(7):567–9. doi: 10.1007/s00262-003-0494-4
108. Bastian K, Scott E, Elliott DJ, Munkley J. FUT8 Alpha-(1,6)-Fucosyltransferase in Cancer. Int J Mol Sci (2021) 22(1):455. doi: 10.3390/ijms22010455
109. Becker DJ, Lowe JB. Fucose: Biosynthesis and Biological Function in Mammals. Glycobiology (2003) 13(7):41R–53R. doi: 10.1093/glycob/cwg054
110. Miyoshi E, Moriwaki K, Nakagawa T. Biological Function of Fucosylation in Cancer Biology. J Biochem (2008) 143(6):725–9. doi: 10.1093/jb/mvn011
111. Wang X, Gu J, Miyoshi E, Honke K, Taniguchi N. Phenotype Changes of Fut8 Knockout Mouse: Core Fucosylation is Crucial for the Function of Growth Factor Receptor(s). Methods Enzymol (2006) 417:11–22. doi: 10.1016/S0076-6879(06)17002-0
112. Lee JW. Transmembrane 4 L Six Family Member 5 (TM4SF5)-Mediated Epithelial-Mesenchymal Transition in Liver Diseases. Int Rev Cell Mol Biol (2015) 319:141–63. doi: 10.1016/bs.ircmb.2015.06.004
113. Jung JW, Kim JE, Kim E, Lee JW. Amino Acid Transporters as Tetraspanin TM4SF5 Binding Partners. Exp Mol Med (2020) 52(1):7–14. doi: 10.1038/s12276-019-0363-7
114. Lee D, Na J, Ryu J, Kim HJ, Nam SH, Kang M, et al. Interaction of Tetraspan(in) TM4SF5 With CD44 Promotes Self-Renewal and Circulating Capacities of Hepatocarcinoma Cells. Hepatology (2015) 61(6):1978–97. doi: 10.1002/hep.27721
115. Kim S, Cho CY, Lee D, Song DG, Kim HJ, Jung JW, et al. CD133-Induced TM4SF5 Expression Promotes Sphere Growth via Recruitment and Blocking of Protein Tyrosine Phosphatase Receptor Type F (PTPRF). Cancer Lett (2018) 438:219–31. doi: 10.1016/j.canlet.2018.09.009
116. Kang M, Ryu J, Lee D, Lee MS, Kim HJ, Nam SH, et al. Correlations Between Transmembrane 4 L6 Family Member 5 (TM4SF5), CD151, and CD63 in Liver Fibrotic Phenotypes and Hepatic Migration and Invasive Capacities. PloS One (2014) 9(7):e102817. doi: 10.1371/journal.pone.0102817
117. Lee MS, Kim HP, Kim TY, Lee JW. Gefitinib Resistance of Cancer Cells Correlated With TM4SF5-Mediated Epithelial-Mesenchymal Transition. Biochim Biophys Acta (2012) 1823(2):514–23. doi: 10.1016/j.bbamcr.2011.11.017
118. Choi J, Kang M, Nam SH, Lee GH, Kim HJ, Ryu J, et al. Bidirectional Signaling Between TM4SF5 and IGF1R Promotes Resistance to EGFR Kinase Inhibitors. Lung Cancer (2015) 90(1):22–31. doi: 10.1016/j.lungcan.2015.06.023
119. Kim HJ, Kwon S, Nam SH, Jung JW, Kang M, Ryu J, et al. Dynamic and Coordinated Single-Molecular Interactions at TM4SF5-Enriched Microdomains Guide Invasive Behaviors in 2- and 3-Dimensional Environments. FASEB J (2017) 31(4):1461–81. doi: 10.1096/fj.201600944RR
120. Lee D, Lee JW. Self-Renewal and Circulating Capacities of Metastatic Hepatocarcinoma Cells Required for Collaboration Between TM4SF5 and CD44. BMB Rep (2015) 48(3):127–8. doi: 10.5483/bmbrep.2015.48.3.047
121. Angata T, Tabuchi Y, Nakamura K, Nakamura M. Siglec-15: An Immune System Siglec Conserved Throughout Vertebrate Evolution. Glycobiology (2007) 17(8):838–46. doi: 10.1093/glycob/cwm049
122. Sun J, Lu Q, Sanmamed MF, Wang J. Siglec-15 as an Emerging Target for Next-Generation Cancer Immunotherapy. Clin Cancer Res (2021) 27(3):680–8. doi: 10.1158/1078-0432.CCR-19-2925
123. Wang J, Sun J, Liu LN, Flies DB, Nie X, Toki M, et al. Siglec-15 as an Immune Suppressor and Potential Target for Normalization Cancer Immunotherapy. Nat Med (2019) 25(4):656–66. doi: 10.1038/s41591-019-0374-x
124. Chang L, Chen YJ, Fan CY, Tang CJ, Chen YH, Low PY, et al. Identification of Siglec Ligands Using a Proximity Labeling Method. J Proteome Res (2017) 16(10):3929–41. doi: 10.1021/acs.jproteome.7b00625
125. Liu W, Ji Z, Wu B, Huang S, Chen Q, Chen X, et al. Siglec-15 Promotes the Migration of Liver Cancer Cells by Repressing Lysosomal Degradation of CD44. FEBS Lett (2021) 595(17):2290–302. doi: 10.1002/1873-3468.14169
126. Roa-de La Cruz L, Martínez-Morales P, Morán-Cruz I, Milflores-Flores L, Rosas-Murrieta N, González-Ramírez C, et al. Expression Analysis of ST3GAL4 Transcripts in Cervical Cancer Cells. Mol Med Rep (2018) 18(1):617–21. doi: 10.3892/mmr.2018.8938
127. Wang T, Hao Z, Liu C, Yuan L, Li L, Yin M, et al. MiR-193b Modulates Osteoarthritis Progression Through Targeting ST3GAL4 via Sialylation of CD44 and NF-Кb Pathway. Cell Signal (2020) 76:109814. doi: 10.1016/j.cellsig.2020.109814
128. Rodriguez E, Boelaars K, Brown K, Eveline Li RJ, Kruijssen L, Bruijns SCM, et al. Sialic Acids in Pancreatic Cancer Cells Drive Tumour-Associated Macrophage Differentiation via the Siglec Receptors Siglec-7 and Siglec-9. Nat Commun (2021) 12(1):1270. doi: 10.1038/s41467-021-21550-4
129. Zheng Y, Ma X, Su D, Zhang Y, Yu L, Jiang F, et al. The Roles of Siglec7 and Siglec9 on Natural Killer Cells in Virus Infection and Tumour Progression. J Immunol Res (2020) 2020:6243819. doi: 10.1155/2020/6243819
130. Jubeli E, Moine L, Vergnaud-Gauduchon J, Barratt G. E-Selectin as a Target for Drug Delivery and Molecular Imaging. J Control Release (2012) 158(2):194–206. doi: 10.1016/j.jconrel.2011.09.084
131. Ivetic A, Hoskins Green HL, Hart SJ. L-selectin: A Major Regulator of Leukocyte Adhesion, Migration and Signaling. Front Immunol (2019) 10:1068. doi: 10.3389/fimmu.2019.01068
132. Dimitroff CJ, Lee JY, Fuhlbrigge RC, Sackstein R. A Distinct Glycoform of CD44 is an L-Selectin Ligand on Human Hematopoietic Cells. Proc Natl Acad Sci USA (2000) 97(25):13841–6. doi: 10.1073/pnas.250484797
133. Jacobs PP, Sackstein R. CD44 and HCELL: Preventing Hematogenous Metastasis at Step 1. FEBS Lett (2011) 585(20):3148–58. doi: 10.1016/j.febslet.2011.07.039
134. Hanley WD, Napier SL, Burdick MM, Schnaar RL, Sackstein R, Konstantopoulos K. Variant Isoforms of CD44 are P- and L-Selectin Ligands on Colon Carcinoma Cells. FASEB J (2006) 20(2):337–9. doi: 10.1096/fj.05-4574fje
135. Zen K, Liu DQ, Guo YL, Wang C, Shan J, Fang M, et al. CD44v4 is a Major E-Selectin Ligand That Mediates Breast Cancer Cell Transendothelial Migration. PloS One (2008) 3(3):e1826. doi: 10.1371/journal.pone.0001826
136. Sackstein R, Dimitroff CJ. A Hematopoietic Cell L-Selectin Ligand That is Distinct From PSGL-1 and Displays N-Glycan-Dependent Binding Activity. Blood (2000) 96(8):2765–74. doi: 10.1046/j.1537-2995.2000.40111421.x
137. Hanley WD, Burdick MM, Konstantopoulos K, Sackstein R. CD44 on LS174T Colon Carcinoma Cells Possesses E-Selectin Ligand Activity. Cancer Res (2005) 65(13):5812–7. doi: 10.1158/0008-5472.CAN-04-4557
138. Carrascal MA, Silva M, Ferreira JA, Azevedo R, Ferreira D, Silva AMN, et al. A Functional Glycoproteomics Approach Identifies CD13 as a Novel E-Selectin Ligand in Breast Cancer. Biochim Biophys Acta Gen Subj (2018) 1862(9):2069–80. doi: 10.1016/j.bbagen.2018.05.013
139. Burdick MM, Chu JT, Godar S, Sackstein R. HCELL is the Major E- and L-Selectin Ligand Expressed on LS174T Colon Carcinoma Cells. J Biol Chem (2006) 281(20):13899–905. doi: 10.1074/jbc.M513617200
140. Sackstein R. Fulfilling Koch's Postulates in Glycoscience: HCELL, GPS and Translational Glycobiology. Glycobiology (2016) 26(6):560–70. doi: 10.1093/glycob/cww026
141. Sackstein R. The Biology of CD44 and HCELL in Hematopoiesis: The 'Step 2-Bypass Pathway' and Other Emerging Perspectives. Curr Opin Hematol (2011) 18(4):239–48. doi: 10.1097/MOH.0b013e3283476140
142. Chajra H, Auriol D, Joly F, Pagnon A, Rodrigues M, Allart S, et al. Reactivating the Extracellular Matrix Synthesis of Sulfated Glycosaminoglycans and Proteoglycans to Improve the Human Skin Aspect and its Mechanical Properties. Clin Cosmet Investig Dermatol (2016) 9:461–72. doi: 10.2147/CCID.S116548
143. Kim Y, Kang H, Powathil G, Kim H, Trucu D, Lee W, et al. Role of Extracellular Matrix and Microenvironment in Regulation of Tumor Growth and LAR-Mediated Invasion in Glioblastoma. PloS One (2018) 13(10):e0204865. doi: 10.1371/journal.pone.0204865
144. Mellai M, Casalone C, Corona C, Crociara P, Favole A, Cassoni P, et al. Chondroitin Sulphate Proteoglycans in the Tumour Microenvironment. Adv Exp Med Biol (2020) 1272:73–92. doi: 10.1007/978-3-030-48457-6_5
145. Henke CA, Roongta U, Mickelson DJ, Knutson JR, McCarthy JB. CD44-Related Chondroitin Sulfate Proteoglycan, a Cell Surface Receptor Implicated With Tumor Cell Invasion, Mediates Endothelial Cell Migration on Fibrinogen and Invasion Into a Fibrin Matrix. J Clin Invest (1996) 97(11):2541–52. doi: 10.1172/JCI118702
146. Takahashi K, Stamenkovic I, Cutler M, Dasgupta A, Tanabe KK. Keratan Sulfate Modification of CD44 Modulates Adhesion to Hyaluronate. J Biol Chem (1996) 271(16):9490–6. doi: 10.1074/jbc.271.16.9490
147. Ruffell B, Poon GF, Lee SS, Brown KL, Tjew SL, Cooper J, et al. Differential Use of Chondroitin Sulfate to Regulate Hyaluronan Binding by Receptor CD44 in Inflammatory and Interleukin 4-Activated Macrophages. J Biol Chem (2011) 286(22):19179–90. doi: 10.1074/jbc.M110.200790
148. Faassen AE, Schrager JA, Klein DJ, Oegema TR, Couchman JR, McCarthy JB. A Cell Surface Chondroitin Sulfate Proteoglycan, Immunologically Related to CD44, is Involved in Type I Collagen-Mediated Melanoma Cell Motility and Invasion. J Cell Biol (1992) 116(2):521–31. doi: 10.1083/jcb.116.2.521
149. Clark RA, Lin F, Greiling D, An J, Couchman JR. Fibroblast Invasive Migration Into Fibronectin/Fibrin Gels Requires a Previously Uncharacterized Dermatan Sulfate-CD44 Proteoglycan. J Invest Dermatol (2004) 122(2):266–77. doi: 10.1046/j.0022-202X.2004.22205.x
150. Bartolini B, Caravà E, Caon I, Parnigoni A, Moretto P, Passi A, et al. Heparan Sulfate in the Tumor Microenvironment. Adv Exp Med Biol (2020) 1245:147–61. doi: 10.1007/978-3-030-40146-7_7
151. Kaczor-Kamińska M, Kamiński K, Wróbel M. Heparan Sulfate, Mucopolysaccharidosis IIIB and Sulfur Metabolism Disorders. Antioxid (Basel) (2022) 11(4):678. doi: 10.3390/antiox11040678
152. Wielenga VJ, van der Voort R, Taher TE, Smit L, Beuling EA, van Krimpen C, et al. Expression of C-Met and Heparan-Sulfate Proteoglycan Forms of CD44 in Colorectal Cancer. Am J Pathol (2000) 157(5):1563–73. doi: 10.1016/S0002-9440(10)64793-1
153. Bennett KL, Jackson DG, Simon JC, Tanczos E, Peach R, Modrell B, et al. CD44 Isoforms Containing Exon V3 Are Responsible for the Presentation of Heparin-Binding Growth Factor. J Cell Biol (1995) 128(4):687–98. doi: 10.1083/jcb.128.4.687
154. Jackson DG, Bell JI, Dickinson R, Timans J, Shields J, Whittle N. Proteoglycan Forms of the Lymphocyte Homing Receptor CD44 are Alternatively Spliced Variants Containing the V3 Exon. J Cell Biol (1995) 128(4):673–85. doi: 10.1083/jcb.128.4.673
155. Kuniyasu H, Oue N, Tsutsumi M, Tahara E, Yasui W. Heparan Sulfate Enhances Invasion by Human Colon Carcinoma Cell Lines Through Expression of CD44 Variant Exon 3. Clin Cancer Res (2001) 7(12):4067–72. doi: 10.1159/000048556
156. Barbour AP, Reeder JA, Walsh MD, Fawcett J, Antalis TM, Gotley DC. Expression of the CD44v2-10 Isoform Confers a Metastatic Phenotype: Importance of the Heparan Sulfate Attachment Site CD44v3. Cancer Res (2003) 63(4):887–92. doi: 10.1002/cncr.11257
157. Jones M, Tussey L, Athanasou N, Jackson DG. Heparan Sulfate Proteoglycan Isoforms of the CD44 Hyaluronan Receptor Induced in Human Inflammatory Macrophages can Function as Paracrine Regulators of Fibroblast Growth Factor Action. J Biol Chem (2000) 275(11):7964–74. doi: 10.1074/jbc.275.11.7964
158. Zaiden M, Rütter M, Shpirt L, Ventura Y, Feinshtein V, David A. CD44-Targeted Polymer-Paclitaxel Conjugates to Control the Spread and Growth of Metastatic Tumors. Mol Pharm (2018) 15(9):3690–9. doi: 10.1021/acs.molpharmaceut.8b00269
159. Ogura K, Nagata K, Hatanaka H, Habuchi H, Kimata K, Tate S, et al. Solution Structure of Human Acidic Fibroblast Growth Factor and Interaction With Heparin-Derived Hexasaccharide. J Biomol NMR (1999) 13(1):11–24. doi: 10.1023/a:1008330622467
160. Bourguignon LY. CD44-Mediated Oncogenic Signaling and Cytoskeleton Activation During Mammary Tumor Progression. J Mammary Gland Biol Neoplasia (2001) 6(3):287–97. doi: 10.1023/a:1011371523994
161. Mylona E, Jones KA, Mills ST, Pavlath GK. CD44 Regulates Myoblast Migration and Differentiation. J Cell Physiol (2006) 209(2):314–21. doi: 10.1002/jcp.20724
162. Hibino S, Shibuya M, Hoffman MP, Engbring JA, Hossain R, Mochizuki M, et al. Laminin Alpha5 Chain Metastasis- and Angiogenesis-Inhibiting Peptide Blocks Fibroblast Growth Factor 2 Activity by Binding to the Heparan Sulfate Chains of CD44. Cancer Res (2005) 65(22):10494–501. doi: 10.1158/0008-5472.CAN-05-0314
163. Suzuki-Inoue K. Platelets and Cancer-Associated Thrombosis: Focusing on the Platelet Activation Receptor CLEC-2 and Podoplanin. Blood (2019) 134(22):1912–8. doi: 10.1182/blood.2019001388
164. Cobec IM, Sas I, Pirtea L, Cimpean AM, Moatar AE, Ceauşu RA, et al. Podoplanin as Key Player of Tumor Progression and Lymph Vessel Proliferation in Ovarian Cancer. Anticancer Res (2016) 36(10):5265–72. doi: 10.21873/anticanres.11097
165. Mir Seyed Nazari P, Riedl J, Pabinger I, Ay C. The Role of Podoplanin in Cancer-Associated Thrombosis. Thromb Res (2018) 1:S34–9. doi: 10.1016/j.thromres.2018.01.020
166. Krishnan H, Rayes J, Miyashita T, Ishii G, Retzbach EP, Sheehan SA, et al. Podoplanin: An Emerging Cancer Biomarker and Therapeutic Target. Cancer Sci (2018) 109(5):1292–9. doi: 10.1111/cas.13580
167. Quintanilla M, Montero-Montero L, Renart J, Martín-Villar E. Podoplanin in Inflammation and Cancer. Int J Mol Sci (2019) 20(3):707. doi: 10.3390/ijms20030707
168. Martín-Villar E, Fernández-Muñoz B, Parsons M, Yurrita MM, Megías D, Pérez-Gómez E, et al. Podoplanin Associates With CD44 to Promote Directional Cell Migration. Mol Biol Cell (2010) 21(24):4387–99. doi: 10.1091/mbc.E10-06-0489
169. Montero-Montero L, Renart J, Ramírez A, Ramos C, Shamhood M, Jarcovsky R, et al. Interplay Between Podoplanin, CD44s and CD44v in Squamous Carcinoma Cells. Cells (2020) 9(10):2200. doi: 10.3390/cells9102200
170. Mereiter S, Balmaña M, Campos D, Gomes J, Reis CA. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell (2019) 36(1):6–16. doi: 10.1016/j.ccell.2019.06.006
171. Zhang G, Zhang H, Wang Q, Lal P, Carroll AM, de la Llera-Moya M, et al. Suppression of Human Prostate Tumor Growth by a Unique Prostate-Specific Monoclonal Antibody F77 Targeting a Glycolipid Marker. Proc Natl Acad Sci U.S.A. (2010) 107(2):732–7. doi: 10.1073/pnas.0911397107
172. Chen X, Nagai Y, Zhu Z, Ruan H, Peehl DM, Greene MI, et al. A Spliced Form of CD44 Expresses the Unique Glycan That is Recognized by the Prostate Cancer Specific Antibody F77. Oncotarget (2017) 9(3):3631–40. doi: 10.18632/oncotarget.23341
173. Gao C, Liu Y, Zhang H, Zhang Y, Fukuda MN, Palma AS, et al. Carbohydrate Sequence of the Prostate Cancer-Associated Antigen F77 Assigned by a Mucin O-Glycome Designer Array. J Biol Chem (2014) 289(23):16462–77. doi: 10.1074/jbc.M114.558932
174. Ding M, Chong LI, Zuo Y, Hui Z, Zhan Z. Aberrant Glycosylation of CD44 in Oncogenesis of EJ Cells and Biological Effects of Anti-CD44 Monoclonal Antibody Kmp1. Chin J Clin Oncol (2011) 38(13):759–62. doi: 10.3969/j.issn.1000-8179.2011.13.003
175. Li M, Sun J, Zhang W, Zhao Y, Zhang S, Zhang S. Drug Delivery Systems Based on CD44-Targeted Glycosaminoglycans for Cancer Therapy. Carbohydr Polym (2021) 251:117103. doi: 10.1016/j.carbpol.2020.117103
Keywords: CD44, glycosylation, hyaluronic acid, fucosylation, sialylation, HCELL
Citation: Liao C, Wang Q, An J, Chen J, Li X, Long Q, Xiao L, Guan X and Liu J (2022) CD44 Glycosylation as a Therapeutic Target in Oncology. Front. Oncol. 12:883831. doi: 10.3389/fonc.2022.883831
Received: 25 February 2022; Accepted: 10 June 2022;
Published: 21 July 2022.
Edited by:
Deilson Elgui De Oliveira, São Paulo State University, BrazilReviewed by:
Alexander Filatov, Federal Medical & Biological Agency of Russia, RussiaWenjun Guo, Albert Einstein College of Medicine, United States
Copyright © 2022 Liao, Wang, An, Chen, Li, Long, Xiao, Guan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linlin Xiao, OTQ1NTI3NDYwQHFxLmNvbQ==; Xiaoyan Guan, MTI3ODI3OTEyNUBxcS5jb20=; Jianguo Liu, MTMwODc4OTEwMDFAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Chengcheng Liao
Chengcheng Liao Qian Wang2,3
Qian Wang2,3 Jie Chen
Jie Chen Xiaolan Li
Xiaolan Li Linlin Xiao
Linlin Xiao