
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 24 May 2022
Sec. Hematologic Malignancies
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.878098
This article is part of the Research Topic Acute Leukemias: Molecular Characterization, Leukemia-Initiating Cells, and Influence of the Microenvironment View all 18 articles
To further emphasize the clinical–genetic features and prognosis of CDKN2A/B deletions in childhood acute lymphoblastic leukemia (ALL), we retrospectively analyzed 819 consecutive B-ALL patients treated with the Chinese Children’s Cancer Group ALL-2015 (CCCG-ALL-2015) protocol, and fluorescence in situ hybridization (FISH) analysis on CDKN2A/B deletion was available for 599 patients. The prevalence of CDKN2A/B gene deletions was 20.2% (121/599) of B-ALL. CDKN2A/B deletions were significantly associated with older age, higher leukocyte counts, a higher percentage of hepatosplenomegaly, and a higher frequency of BCR-ABL (p < 0.05). Those patients achieved similar minimal residual disease (MRD) clearance and complete remission compared to patients without CDKN2A/B deletion. The CDKN2A/B deletions were correlated with inferior outcomes, including a 3-year event-free survival (EFS) rate (69.8 ± 4.6 vs. 89.2 ± 1.6%, p = 0.000) and a 3-year overall survival (OS) rate (89.4% ± 2.9% vs. 94.7% ± 1.1%, p = 0.037). In multivariable analysis, CDKN2A/B deletion was still an independent prognostic factor for EFS in total cohorts (p < 0.05). We also detected a multiplicative interaction between CDKN2A/B deletions and TP53 deletion on dismal prognosis (p-interaction < 0.05). In conclusion, CDKN2A/B deletion is associated with distinct characteristics and serves as a poor prognostic factor in pediatric ALL, especially in TP53 deletion carriers.
Pediatric acute lymphoblastic leukemia (ALL) is one of the most curable malignancies, with 5-year event-free survival rates exceeding 80% in many developed countries and even exceeding 90% in high-income countries (1). With the increased understanding of genetic alterations in ALL, many molecular markers have been identified and applied to risk stratification and treatment protocols in leukemia. CDKN2A/B is one of the most frequent abnormal genes in ALL and can be detected by fluorescence in situ hybridization (FISH), multiplex ligation-dependent probe amplification (MLPA), array-based comparative genomic hybridization (aCGH) analysis, and single-nucleotide polymorphism array (SNPA) (2–6). As a secondary genetic event in the development of leukemia, the CDKN2A/B deletions were found in approximately 20%–25% of B-cell precursor acute lymphoblastic leukemia (BCP-ALL) and 38.5%–50% of T-ALL patients (2, 5, 7, 8). Despite the high frequency of CDKN2A/B deletions in pediatric ALL, the prognostic importance of the deletions is still inconclusive. Most investigators have concluded that the deletions were associated with the recurrence of pediatric ALL (7, 9, 10), and some researchers found that the inactivation of CDKN2A/B did not influence the outcome of childhood B-lineage ALL (4, 11). As previous inconsistent conclusions were drawn from small cohorts, large sample-sized studies are needed to clarify the prognostic impact of CDKN2A/B deletion in pediatric ALL. In this study, we assessed the clinical and biological characteristics and prognostic factors of CDKN2A/B deletions in 662 pediatric ALL patients.
The cohort included 902 patients with newly diagnosed pediatric ALL who were treated according to the CCCG-ALL 2015 protocol (ClinicalTrials.gov identifier: ChiCTRIPR-14005706) (12) registered at the Blood Disease Hospital of CAMS & PUMC between May 2015 and December 2019. As the FISH test has been performed since September 2016, FISH data from 662 patients were collected in this cohort finally (Figure 1). The protocol described in this study was approved by the Ethics Committee, Institute of Hematology and Blood Disease Hospital, Diseases Hospital, Chinese Academy of Medical Sciences (CAMS) and Peking Union Medical College (PUMC) (No. IIT2015010-EC-1). All patients or their legal guardians signed written informed consent before treatment.
Patients enrolled in the CCCG-ALL-2015 study were assigned to different risk groups based on morphology and immunophenotypic and genetic features of leukemia cells. Patients were assigned to the low-risk group if they had B-cell ALL and were aged between 1 year and 10 years; had a leukocyte count of less than 50 × 109/L, a chromosome number of more than 50, or the ETV6–RUNX1 fusion gene; and did not have CNS 3 status or testicular leukemia, and had a minimal residual disease of less than 1% on day 19 of induction and less than 0.01% on day 46 of induction. Patients with a minimal residual disease of 1% or more (or ≥5% blasts morphologically without suitable markers for minimal residual disease) in bone marrow on day 46 of induction and infants younger than 6 months with KMT2A rearrangement and a leukocyte count of 300 × 109/L or more were classified as high-risk ALL. The remaining participants were assigned to the intermediate-risk group (12). All eligible patients received minimal residual disease-directed, risk-stratified treatment modified from the St Jude Children’s Research Hospital Total Therapy 15 and 16 studies (13, 14) and the Shanghai Children’s Medical Center ALL-2005 trial (15), with IR/HR patients receiving more intensive treatment.
Pretreatment bone marrow aspirates were taken at diagnosis and at least 1–2 ml of bone marrow aspirates was analyzed by FISH for cytogenetic abnormalities (including CDKN2A/B, KMT2A rearrangement, TP53 deletion, and BCR-ABL1), polymerase chain reaction (PCR) for fusion gene (including ETV6-RUNX1, BCR-ABL, and TCF3-PBX1) (16), and karyotyping. We analyzed interphase cells according to the instructions of the probe manufacturer (America Abbott). The CDKN2A/B probe spanned approximately 222 kilobases (kb) and contained many genes, including methylthioadenosine phosphorylase, CDKN2A (INK4A and ARF), and CDKN2B (INK4B) in the 9p21 chromosome region. The cutoff level for positive results was calculated to be 5%, and at least five hundred cells were analyzed (5, 17). Some cases with two different deleted populations (one biallelic and one monoallelic) were classified as having a biallelic deletion.
To identify early prognostic factors, we evaluated leukemic blast counts in bone marrow by morphology on day 46 and minimal residual disease (MRD) on days 19 and 46 during the first induction. A total of 1–2×106 leukocytes from bone marrow were incubated with marker panel [B-lineage cells (CD10, CD19, TdT, cyμ, sIgM, CD20, cyCD22, CD22, and cyCD79a), T-lineage cells (CD1a, CD2, CD3, CD4, CD5, CD7, CD8, TCRαβ, TCRγδ, and cyCD3), or myeloid cells (CD11b, CD13, CD14, CD15, CD33, CD41, CD61, CD64, CD65, CD71, GPA, and cyMPO)] for 30 min, washed twice, and then resuspended in 100 μl of PBS. The cells were acquired on a Navios (Beckman Coulter, UK) and analyzed using Kaluza software. Samples were defined as MRD-negative if no baseline and/or different-from-normal leukemia-associated immunophenotypes (LAIP) cells could be quantitated above the limit of detection (approximately 0.01%) (18).
Diagnosis of complete remission (CR) was based solely on bone marrow morphology with a cutoff value of 5% of leukemic blasts on treatment day 46. Bone marrow MRD that is not lower than 10-5 was defined as positive. A poor response to prednisone was considered if the peripheral blast count on treatment day 5 was not lower than 1×109/L.
The primary endpoint event-free survival was calculated from the date of diagnosis until the following events: induction treatment failure, any relapse after CR, second malignancy, and death due to any cause. Overall survival was considered as the time from diagnosis to death due to any cause.
Chi-squared test and Mann–Whitney U tests were used to compare categorical and continuous variables, respectively. Event-free and overall survival were calculated and compared using Kaplan–Meier analysis and log-rank tests. Cox proportional hazard regression analyses were performed using univariable and multivariable regression approaches. Variables that were found statistically significant and a p-value of approximately 0.1 were included in multivariable Cox regression analysis. We performed subgroup analyses for prespecified baseline factors with rates of inferior events by factor interaction with the use of Cox models. Statistical significance was defined as a p-value of less than 0.05. All analyses were performed using SPSS v. 24. and SAS v. 9.4 software.
In total, 599 patients were included in the analysis. The final follow-up was in December 2020 and the median follow-up time was 34 months (range: 0 to 58 months). The median age was 5 years (range, 0 to 14), and the male/female ratio was 1.4 (350/249). The prevalence of CDKN2A/B gene deletions in B-ALL was 20.2% (121/599). Among patients with CDKN2A/B deletions, 57 (45.3%) harbored CDKN2A/B biallelic deletions, whereas 82 (54.7%) harbored monoallelic deletions, and 11 (7.3%) patients harbored both biallelic and monoallelic deletions.
Compared to patients with wild-type CDKN2A/B (Table 1), patients with CDKN2A/B deletions were significantly associated with older age (age >10 years; 30.6% vs. 15.2%, p < 0.001), a higher leukocyte count (median: 24.7 vs. 8.9×109/L, p < 0.001), a lower platelet count (median: 51 vs. 64 g/L, p = 0.049), and a higher percentage of hepatosplenomegaly (64.5% vs. 43.9%, p < 0.001). A higher rate of central nervous system status 2(CNS 2)/traumatic lumbar puncture was found among patients with CDKN2A/B deletion, but no statistical significance was found in the two groups. Patients with CDKN2A/B deletions belonged more to the intermediate-risk groups (58.7% vs. 32.6%, p < 0.01) than patients without CDKN2A/B deletions based on CCCG-ALL 2015 risk stratification. There was no significant difference between patients with biallelic deletions and monoallelic deletions. The comparison of characteristics for the two groups is shown in Table S1.
Karyotype analysis, 43 fusion genes, and FISH studies of pretreatment bone marrow were available for 599 patients. Table 1 summarizes the correlations of CDKN2A/B deletions with other cytogenetic abnormalities. The CDKN2A/B deletion group had a higher prevalence of the BCR/ABL fusion gene (12.4% vs. 4.4%, p = 0.001). A higher co-occurrence of the ETV6/RUNX1 fusion gene (19.8% vs. 26.8%) was found in patients with CDKN2A/B deletion; however, no statistical significance was identified. A higher incidence of chromosome < 44 (2.5% vs. 0.6%) and TP53 deletions (8.3% vs. 4.0%) was detected by FISH, but this did not reach statistical significance (p ≥ 0.05). The genetic feature of patients with CDKN2A/B gene deletions was also related to the risk stratification distribution at the first diagnosis.
A chi-squared test showed a high rate of poor responses to prednisone (PPR) (28.1% vs. 20.9%, p = 0.091) in the CDKN2A/B deletion group. No significant difference was observed between the two groups for MRD on day 19 (61.2% vs. 69.4%, p = 0.946) and 46 (18.5% vs. 18.2%, p = 0.946). The CR rate (98.4% vs. 99.2%, p = 0.719) for each group was equivalent.
A high CR rate was achieved in 593 of 599 patients (99.0%) (Figure 1). Based on their MRD levels on days 19 and 46 during remission induction treatment, 286 (48.2%) of 593 patients were classified as having low-risk ALL, 296 (49.9%) as having intermediate-risk ALL, and 11 (1.9%) as having high-risk ALL. All six patients who did not achieve the first CR died of severe pneumonia. After remission induction, 74 patients had adverse events, including 68 relapses (n = 54 hematologic relapses, n = 4 combined hematologic and CNS relapses, n = 2 combined hematologic and testicular relapses, n = 1 combined hematologic, CNS, and testicular relapses, n = 4 isolated CNS relapse, n = 2 testicular relapse, and n = 1 ocular relapse). Six patients died [n = 2 died of severe pneumonia and emesis in remission and n = 4 died after chimeric antigen receptor T-cell immunotherapy (CAR-T) therapy or hematopoietic stem cell transplantation (HSCT) because of elevated MRD level]. The 3-year OS and EFS were 93.6 ± 1.1% and 85.2 ± 1.6%, respectively, for the whole series.
The CDKN2A/B deletion group is related to a higher relapse rate (28.1% for the deletions group vs. 7.1% for the wild-type group, p = 0.000) (Table 2). Patients with CDKN2A/B deletions seemed to have a higher CNS relapse rate than patients with CDKN2A/B deletions, although the p-value was not significant (3.3% vs. 1.1%, p = 0.087). Patients with CDKN2A/B deletions had lower 3-year EFS and OS rates than patients without CDKN2A/B deletions (3-year EFS: 69.8 ± 4.6 vs. 89.2 ± 1.6%, p = 0.000; and 3-year OS: 89.4% ± 2.9% vs. 94.7% ± 1.1%, p = 0.037) (Figures 2A, B). Furthermore, CDKN2A/B deletion serves as an independent prognosis factor (HR = 3.4 [95% CI 2.0–5.6]; p = 0.000) for EFS in whole cohorts (Figure 3). MRD on day 46 was a strong predictor (HR = 3.3 [95% CI 2.0–5.3]; p = 0.000) for all cases.
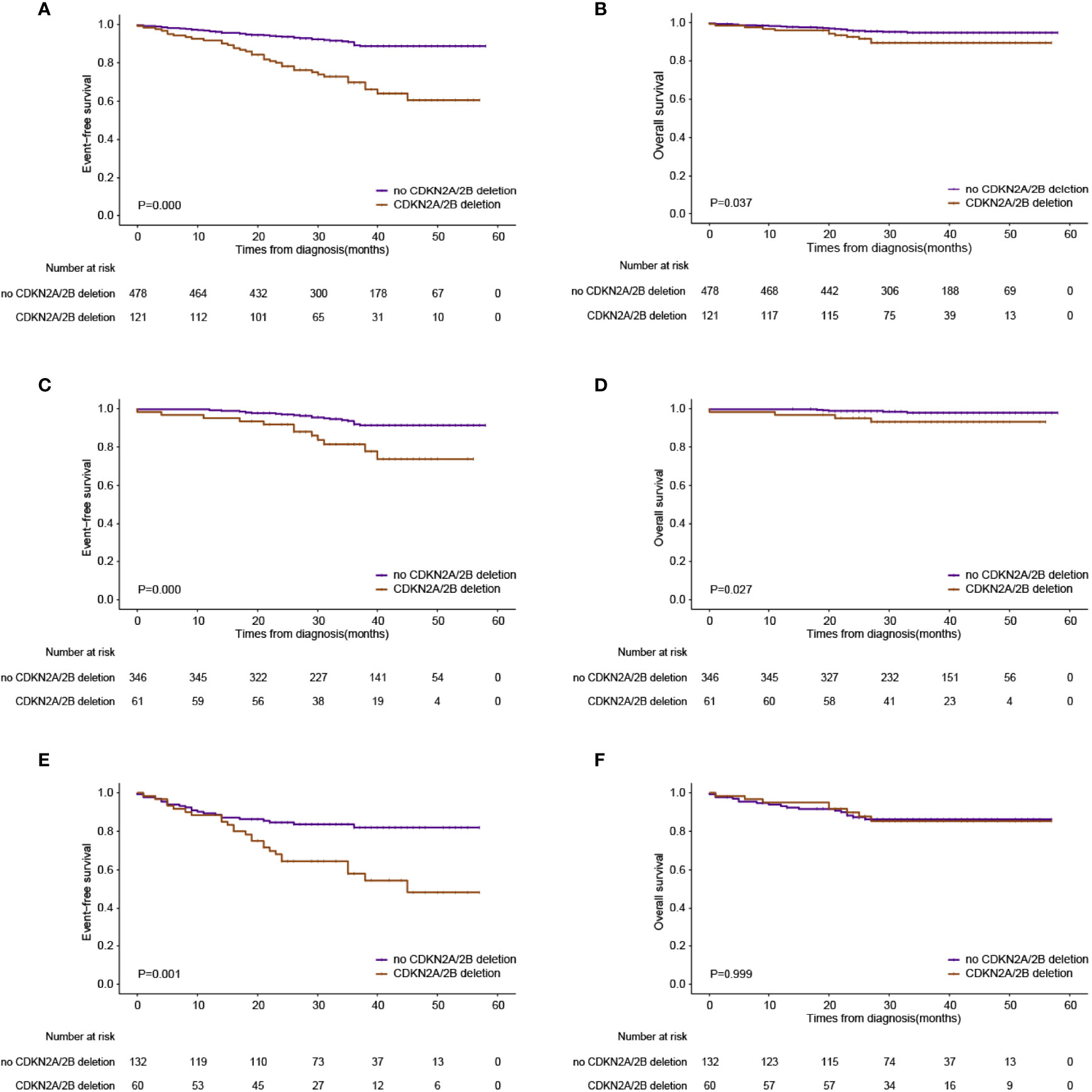
Figure 2 Outcomes based on the presence or absence of CDKN2A deletions. (A) Event-free survival in the whole series. (B) Overall survival in the whole series. (C) Event-free survival in the low-risk group. (D) Overall survival in the low-risk group. (E) Event-free survival in the intermediate-risk group. (F) Overall survival in the intermediate-risk group.
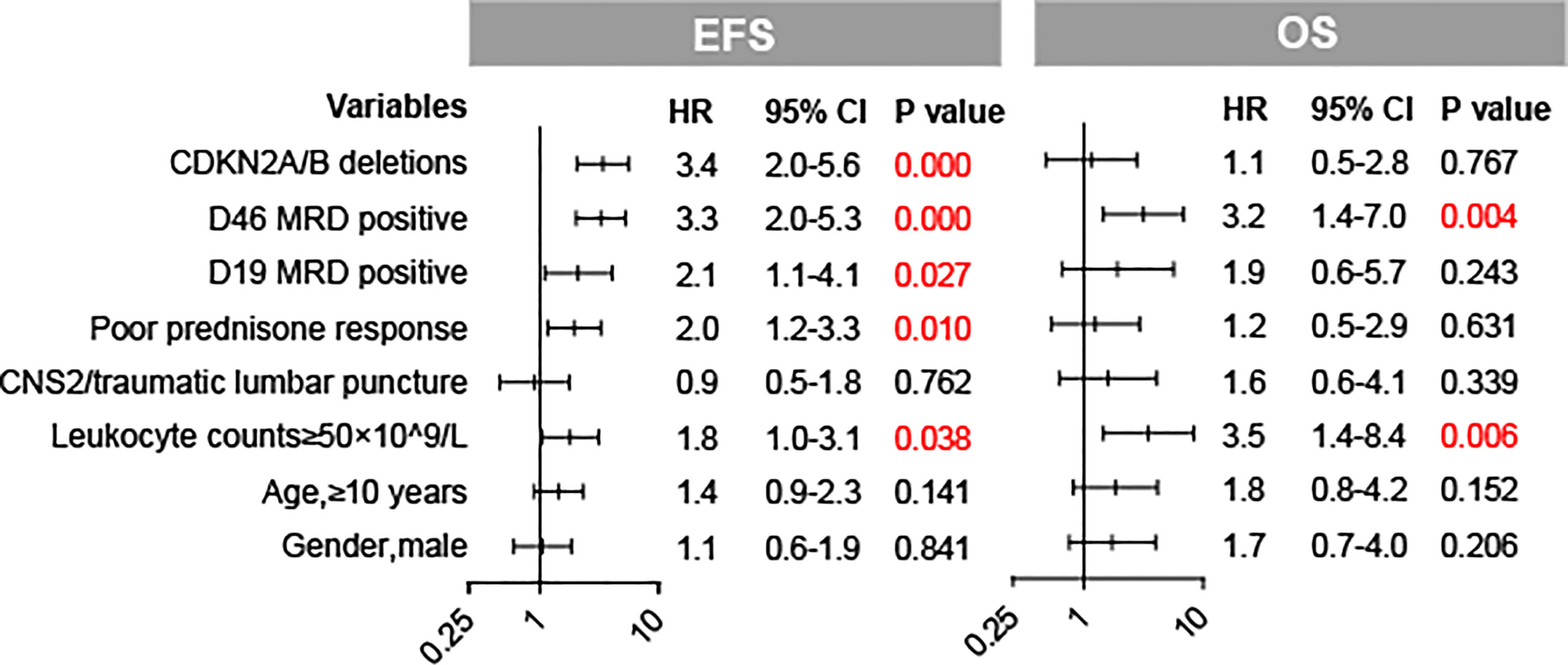
Figure 3 Forest plots of the multivariate analysis of CDKN2A/B deletions and baseline factors for EFS and OS in our cohort.
On the basis of the CCCG-ALL 2015 risk stratification, more patients were classified as intermediate risk in the CDKN2A/B deletion group at diagnosis. For patients assigned into the low-risk group, patients with CDKN2A/B deletions had adverse clinical outcomes (3-year EFS: 81.4 ± 5.4% vs. 91.9 ± 1.7%, p = 0.000; 3-year OS: 93.2 ± 3.3% vs. 97.9 ± 0.9%, p = 0.027) compared to patients without CDKN2A/B deletions (Figures 2C, D). Intermediate-risk patients with CDKN2A/B deletions had inferior 3-year EFS (58.0 ± 7.1% vs. 82.0 ± 3.6%, p = 0.001), but no inferior 3-year OS (85.3 ± 4.9% vs. 86.3 ± 3.1%, p = 0.999) compared to intermediate-risk patients without CDKN2A/B deletions (Figures 2E, F).
We performed stratified analyses by subgroups defined by major co-variables that might have been related to EFS and further quantified the effect modification of major co-variables on the influence of CDKN2A/B deletions in EFS, accounting for important covariates (Table 3). The hazard rates of CDKN2A/B deletions in EFS were consistent in most subgroups. The hazard rate of CDKN2A/B deletions for the adverse events was quite different for patients with and without TP53 deletions (HR = 24.080 [95% CI 2.978–194.738]; p = 0.0029 and HR = 2.993 [95% CI 1.868–4.796]; p < 0.0001, respectively), while there were no significant interactions between two groups (p-interaction = 0.0631).
In this study, we present a large retrospective study of pediatric ALL patients with CDKN2A/B deletions treated in a single center and demonstrated the adverse effect of CDKN2A/B deletions on clinical outcomes.
The prevalence of CDKN2A/B deletions in our study was 20.2%, which was higher than Agarwal’s research (19.8%) but lower than Sulong’s research (22.0%) (2, 5). Previous investigators have reported the characteristics and clinical impact of CDKN2A/B deletions in pediatric ALL, and the prognostic importance of CDKN2A/B deletions in pediatric ALL is still controversial (2, 4, 5, 10, 19). We conducted a comprehensive analysis of CDKN2A/B deletions in 599 pediatric B-ALL patients. Data from our cohort showed that CDKN2A/B deletions were associated with older age at diagnosis, higher white blood cell counts, and prominent hepatosplenomegaly, which were consistent with the previous studies (5).
Many researchers concluded that CDKN2A/B deletions in childhood ALL were associated with an increased probability of relapse and death (2, 9, 10, 19–21), whereas Kima et al. and Mirebeau et al. concluded that homozygous CDKN2A/B deletion was not a poor prognostic factor in childhood B-ALL (4, 11). In our study, CDKN2A/B deletion carriers had decreased endpoints for 3-year EFS and OS compared to CDKN2A/B wild-type patients. Furthermore, our data showed that patients with the biallelic deletions had a worse survival rate (3-year EFS: 71.3 ± 6.0% vs. 67.7 ± 7.0% vs. 89.2 ± 1.6%, p = 0.000; 3-year OS: 922.2 ± 3.4% vs. 85.2 ± 5.2% vs. 94.7 ± 1.1%, p = 0.025) than patients without biallelic deletions (Figure S1).
The long-term outcome of pediatric ALL in China is optimal, especially for low-risk patients, and the OS rate remains 97.8% (12). Notably, in our cohorts, low-risk patients with CDKN2A/B deletions had inferior outcomes (3-year OS: 93.2 ± 3.3% vs. 97.9 ± 0.9%) compared to low-risk patients without CDKN2A/B deletions (Figures 2C, D). The result indicated that CDKN2A/B deletion patients even stratified in the low-risk group urgently needed intensive chemotherapy. Furthermore, allo-HSCT (n = 14) has improved overall survival than chemotherapy alone did (n = 20) in relapse patients with CDKN2A/B deletions, though no significance was observed between the two groups (3-year OS: 85.7 ± 9.4% vs. 60.5 ± 11.9%, p = 0.192) (Figure S2).
Sulong et al. reported that pediatric ALL with CDKN2A/B deletions had recurrent cytogenetic abnormalities including high frequencies of TCF/PBX1, BCR/ABL, hyperdiploidy, and KMT2A, and low frequencies of ETV6/RUNX1 compared to patients without CDKN2A/B deletions (5, 11). We conclude a similar result in which CDKN2A/B deletion carriers had a higher prevalence of BCR/ABL and a low prevalence of ETV6/RUNX1. However, we did not find ph+ patients had inferior outcomes than ph- patients in CDKN2A/B deletion groups; this is most likely due to the utilization of tyrosine kinase inhibitors (TKIs) in ph+ patients for the entire duration of ALL therapy (22, 23). Pfeifer et al. demonstrated that CDKN2A/B deletions were adverse despite allogeneic stem cell transplantation in adult Philadelphia chromosome-positive (Ph+) ALL (24, 25); further studies in a larger cohort of pediatric Ph+ ALL with CDKN2A/B deletions are needed.
In our cohort, patients with CDKN2A/B deletions were more frequently steroid-resistant, whereas they had a better MRD clearance on day 19 of induction therapy compared with patients without CDKN2A/B deletions. These results differed from a study by Braun who found higher MRD levels on day 15 of induction therapy in patients with CDKN2A/B deletions (26). The differences in MRD clearance may be related to the patients’ race, chemotherapy protocols, and examination methods. An earlier study demonstrated that CDKN2A/B deletions were associated with unfavorable outcomes independent of MRD level in adult patients (27). However, both MRD positive on day 46 of induction therapy and CDKN2A/B deletion were still independent poor indicators for childhood ALL patients in our study.
Our study also indicated that patients with CDKN2A/B deletion had the worst prognosis in the TP53 deletion subgroup. A similar outcome was found by Delfau-Larue for mantle cell lymphoma; i.e., patients with both CDKN2A/B and TP53 deletions had the worst prognosis (28). The CDKN2A/B gene controls the cell cycle through the P53-MDM pathway; thus, co-occurrence of CDKN2A/B deletions and TP53 deletions might enhance the aggressiveness of disease by strongly increasing the self-renewal capacity of leukemia cells (29–31). The hypothesis needs to be proved in future research.
Several limitations exist in this study. First, because of the limited sample size and short follow-up duration in relapse patients with CDKN2A/B deletions, the role of allogeneic transplant in the treatment needs to be interpreted carefully. Second, for the same reason, we did not conclude whether there had been significant interactions between two TP53 deletions and CDKN2A/B deletions in pediatric ALL. CDKN2A gene can control cell cycle through the P53-MDM pathway; it might be attributed to the fact that CDKN2A deletion cooperates with the TP53 deletion to enhance the aggressiveness of the disease by strongly increasing the self-renewal capacity of leukemia cells, but there is no theory to support this hypothesis.
In conclusion, a significant proportion of pediatric ALL patients still experience relapse, particularly patients with both CDKN2A/B and TP53 deletions despite the high survival rate of childhood ALL. MDM2-P53 targeted agents are still in experimental research. In addition, CDK4/6 inhibitors (e.g., palbociclib) combined with chemotherapy are in clinical trials for the management of pediatric patients with relapsed/refractory ALL (32, 33). In the future, a new therapeutic strategy (e.g., target drug) based on genetic events might be applied in some subtypes of pediatric ALL.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The protocols described in this study were approved by the Ethics Committee, Institute of Hematology and Blood Disease Hospital, CAMS & PUMC. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The work is supported by the National Nature Science Foundation of China grants 81770175 (YCZ), 81870131 (XZ), and 81670112 (XZ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the patients for participating in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.878098/full#supplementary-material
1. Inaba H, Mullighan CG. Pediatric Acute Lymphoblastic Leukemia. Haematol (2020) 105(11):2524–39. doi: 10.3324/haematol.2020.247031
2. Agarwal M, Bakhshi S, Dwivedi SN, Kabra M, Shukla R, Seth R. Cyclin Dependent Kinase Inhibitor 2A/B Gene Deletions are Markers of Poor Prognosis in Indian Children With Acute Lymphoblastic Leukemia. Pediatr Blood Cancer (2018) 65(6):e27001. doi: 10.1002/pbc.27001
3. Boldrin E, Gaffo E, Niedermayer A, Boer JM, Zimmermann M, Weichenhan D, et al. MicroRNA-497/195 is Tumor-Suppressive and Cooperates With CDKN2A/B in Pediatric Acute Lymphoblastic Leukemia. Blood (2021) 138:1953–65. doi: 10.1182/blood.2020007591
4. Kim M, Yim S-H, Cho N-S, Kang S-H, Ko D-H, Oh B, et al. Homozygous Deletion of CDKN2A (P16, P14) and CDKN2B (P15) Genes is a Poor Prognostic Factor in Adult But Not in Childhood B-Lineage Acute Lymphoblastic Leukemia: A Comparative Deletion and Hypermethylation Study. Cancer Genet Cytogenetics (2009) 195:59–65. doi: 10.1016/j.cancergencyto.2009.06.013
5. Sulong S, Moorman AV, Irving JAE, Strefford JC, Konn ZJ, Case MC, et al. A Comprehensive Analysis of the CDKN2A Gene in Childhood Acute Lymphoblastic Leukemia Reveals Genomic Deletion, Copy Number Neutral Loss of Heterozygosity, and Association With Specific Cytogenetic Subgroups. Blood (2009) 113:100–7. doi: 10.1182/blood-2008-07-166801
6. Usvasalo A, Savola S, Räty R, Vettenranta K, Harila-Saari A, Koistinen P, et al. CDKN2A Deletions in Acute Lymphoblastic Leukemia of Adolescents and Young Adults—An Array CGH Study. Leukemia Res (2008) 32:1228–35. doi: 10.1016/j.leukres.2008.01.014
7. Bertin R, Acquaviva C, Mirebeau D, Guidal-Giroux C, Vilmer E, Cavé H. CDKN2A, CDKN2B, and MTAP Gene Dosage Permits Precise Characterization of Mono- and Bi-Allelic 9p21 Deletions in Childhood Acute Lymphoblastic Leukemia: CDKN2A , CDKN2B , and MTAP Dosage in Leukemia. Genes Chromosom Cancer (2003) 37(1):44–57. doi: 10.1002/gcc.10188
8. Karrman K, Castor A, Behrendtz M, Forestier E, Olsson L, Ehinger M, et al. Deep Sequencing and SNP Array Analyses of Pediatric T-Cell Acute Lymphoblastic Leukemia Reveal NOTCH1 Mutations in Minor Subclones and a High Incidence of Uniparental Isodisomies Affecting CDKN2A. J Hematol Oncol (2015) 8:42. doi: 10.1186/s13045-015-0138-0
9. Graf Einsiedel H, Taube T, Hartmann R, Wellmann S, Seifert G, Henze G, et al. Deletion Analysis of P16inka and P15inkb in Relapsed Childhood Acute Lymphoblastic Leukemia. Blood (2002) 99:4629–31. doi: 10.1182/blood.V99.12.4629
10. Kees UR, Burton PR, Lu C, Baker DL. Homozygous Deletion of the P16/MTS1 Gene in Pediatric Acute Lymphoblastic Leukemia Is Associated With Unfavorable Clinical Outcome. (1997) 89(11):4161–6. doi: 10.1182/blood.V89.11.4161
11. Mirebeau D, Acquaviva C, Suciu S, Bertin R, Dastugue N, Robert A, et al. The Prognostic Significance of CDKN2A, CDKN2B and MTAP Inactivation in B-Lineage Acute Lymphoblastic Leukemia of Childhood. Results of the EORTC Studies 58881 and 5895. haematologica (2006) 91:881–5.
12. Yang W, Cai J, Shen S, Gao J, Yu J, Hu S, et al. Pulse Therapy With Vincristine and Dexamethasone for Childhood Acute Lymphoblastic Leukaemia (CCCG-ALL-2015): An Open-Label, Multicentre, Randomised, Phase 3, Non-Inferiority Trial. Lancet Oncol (2021) 22:1322–32. doi: 10.1016/S1470-2045(21)00328-4
13. Pui C-H, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC, et al. Treating Childhood Acute Lymphoblastic Leukemia Without Cranial Irradiation. N Engl J Med (2009) 360:2730–41. doi: 10.1056/NEJMoa0900386
14. Jeha S, Pei D, Choi J, Cheng C, Sandlund JT, Coustan-Smith E, et al. Improved CNS Control of Childhood Acute Lymphoblastic Leukemia Without Cranial Irradiation: St Jude Total Therapy Study 16. JCO (2019) 37:3377–91. doi: 10.1200/JCO.19.01692
15. Liu Y, Chen J, Tang J, Ni S, Xue H, Pan C. Cost of Childhood Acute Lymphoblastic Leukemia Care in Shanghai, China. Pediatr Blood Cancer (2009) 53:557–62. doi: 10.1002/pbc.22127
16. Viehmann S, Borkhardt A, Lampert F, Harbott J. Multiplex PCR - a Rapid Screening Method for Detection of Gene Rearrangements in Childhood Acute Lymphoblastic Leukemia. Ann Hematol (1999) 78:157–62. doi: 10.1007/s002770050494
17. Wang H, Zhou Y, Huang X, Zhang Y, Qian J, Li J, et al. CDKN2A Deletions are Associated With Poor Outcomes in 101 Adults With T‐Cell Acute Lymphoblastic Leukemia. Am J Hematol (2021) 96:312–9. doi: 10.1002/ajh.26069
18. Wood BL. Principles of Minimal Residual Disease Detection for Hematopoietic Neoplasms by Flow Cytometry: Principles of MRD. Cytometry (2016) 90:47–53. doi: 10.1002/cyto.b.21239
19. Kathiravan M, Singh M, Bhatia P, Trehan A, Varma N, Sachdeva MS, et al. Deletion of CDKN2A/B is Associated With Inferior Relapse Free Survival in Pediatric B Cell Acute Lymphoblastic Leukemia. Leukemia Lymphoma (2019) 60:433–41. doi: 10.1080/10428194.2018.1482542
20. Carter TL. Hemizygous P16ink4a Deletion in Pediatric Acute Lymphoblastic Leukemia Predicts Independent Risk of Relapse. Blood (2001) 97:572–4. doi: 10.1182/blood.V97.2.572
21. Dalle JH, Fournier M, Nelken B, Mazingue F, Laıü J-L, Bauters F, et al. P16ink4a Immunocytochemical Analysis is an Independent Prognostic Factor in Childhood Acute Lymphoblastic Leukemia. Blood (2002) 99:2620–3. doi: 10.1182/blood.V99.7.2620
22. Schultz KR, Carroll A, Heerema NA, Bowman WP, Aledo A, Slayton WB, et al. Long-Term Follow-Up of Imatinib in Pediatric Philadelphia Chromosome-Positive Acute Lymphoblastic Leukemia: Children’s Oncology Group Study Aall0031. Leukemia (2014) 28(7):1467–71. doi: 10.1038/leu.2014.30
23. Shen S, Chen X, Cai J, Yu J, Gao J, Hu S, et al. Effect of Dasatinib vs Imatinib in the Treatment of Pediatric Philadelphia Chromosome–Positive Acute Lymphoblastic Leukemia: A Randomized Clinical Trial. JAMA Oncol (2020) 6:358. doi: 10.1001/jamaoncol.2019.5868
24. Pfeifer H, Raum K, Markovic S, Nowak V, Fey S, Obländer J, et al. Genomic CDKN2A/2B Deletions in Adult Ph+ ALL are Adverse Despite Allogeneic Stem Cell Transplantation. Blood (2018) 131:1464–75. doi: 10.1182/blood-2017-07-796862
25. Iacobucci I, Ferrari A, Lonetti A, Papayannidis C, Paoloni F, Trino S, et al. CDKN2A/B Alterations Impair Prognosis in Adult BCR-ABL1-Positive Acute Lymphoblastic Leukemia Patients. Clin Cancer Res (2011) 17(23):7413–23. doi: 10.1158/1078-0432.CCR-11-1227
26. Braun M, Pastorczak A, Fendler W, Madzio J, Tomasik B, Taha J, et al. Biallelic Loss of CDKN2A is Associated With Poor Response to Treatment in Pediatric Acute Lymphoblastic Leukemia. Leukemia Lymphoma (2017) 58:1162–71. doi: 10.1080/10428194.2016.1228925
27. Ribera J, Zamora L, Morgades M, Vives S, Granada I, Montesinos P, et al. Molecular Profiling Refines Minimal Residual Disease‐Based Prognostic Assessment in Adults With Philadelphia Chromosome‐Negative B‐Cell Precursor Acute Lymphoblastic Leukemia. Genes Chromosomes Cancer (2019) 58:815–9. doi: 10.1002/gcc.22788
28. Delfau-Larue M-H, Klapper W, Berger F, Jardin F, Briere J, Salles G, et al. High-Dose Cytarabine Does Not Overcome the Adverse Prognostic Value of CDKN2A and TP53 Deletions in Mantle Cell Lymphoma. Blood (2015) 126:604–11. doi: 10.1182/blood-2015-02-628792
29. Zhao R, Choi BY, Lee MH, Bode AM, Dong Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (P16ink4a) in Cancer. EBioMedicine (2016) 8:30–9. doi: 10.1016/j.ebiom.2016.04.017
30. Otsuki T, Clark HM, Wellmann A, Jaffe ES, Raffeld M. Involvement of CDKN2 (P16ink4a/MTS1) and P15ink4b/MTS2 in Human Leukemias and Lymphomas. Cancer Res (1995) 55(7):1436.
31. Johnson M, Dimitrov D, Vojta PJ, Barrett JC, Noda A, Pereira-Smith OM, et al. Evidence for a P53-Independent Pathway for Upregulation Ofsdi1/CIP1/WAF1/P21 RNA in Human Cells. Mol Carcinog (1994) 11:59–64. doi: 10.1002/mc.2940110202
32. Bride KL, Hu H, Tikhonova A, Fuller TJ, Vincent TL, Shraim R, et al. Rational Drug Combinations With CDK4/6 Inhibitors in Acute Lymphoblastic Leukemia. haematol (2021). doi: 10.3324/haematol.2021.279410
Keywords: CDKN2A/B, pediatric acute lymphoblastic leukemia, fluorescence in situ hybridization, prognosis, TP53
Citation: Feng J, Guo Y, Yang W, Zou Y, Zhang L, Chen Y, Zhang Y, Zhu X and Chen X (2022) Childhood Acute B-Lineage Lymphoblastic Leukemia With CDKN2A/B Deletion Is a Distinct Entity With Adverse Genetic Features and Poor Clinical Outcomes. Front. Oncol. 12:878098. doi: 10.3389/fonc.2022.878098
Received: 17 February 2022; Accepted: 14 April 2022;
Published: 24 May 2022.
Edited by:
Lokman Varisli, Dicle University, TurkeyReviewed by:
Luca Lo Nigro, Azienda Ospedaliero Universitaria Policlinico - San Marco, ItalyCopyright © 2022 Feng, Guo, Yang, Zou, Zhang, Chen, Zhang, Zhu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofan Zhu, eGZ6aHVAaWhjYW1zLmFjLmNu; Xiaojuan Chen, Y2hlbnhpYW9qdWFuQGloY2Ftcy5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.