- 1Department of Radiation Oncology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Department of Surgical Oncology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
Objective: To probe into the role of pyroptosis-related genes in gastric cancer.
Methods: To establish pyroptosis-related genes, observe their expression in gastric cancer, and analyze the prognosis of pyroptosis-related genes in gastric cancer by single-factor COX, which showed that only GSDME had prognostic significance in gastric cancer. The mRNA expression profiles and lncRNA expression profiles of gastric cancer downloaded from the Cancer Genome Atlas were combined for weighted gene regulatory network analysis, after which the lncRNA nodes of the module to which GSDME belongs were extracted to obtain the lncRNAs−GSDME interactions, which were visualized with Cytoscape network plots. Finally, the effects of GSDME on the proliferation, migration, and apoptosis of gastric cancer cells were observed with CCK8, and flow cytometry.
Results: Our results show that only GSDME has prognostic significance in gastric cancer, and show that it has an important role in a variety of tumors. In addition, our results show that 16 lncRNAs have a significant interaction with GSDME. Finally, the experimental analysis showed that knocking down the expression level of GSDME could affect the growth as well as apoptosis of gastric cancer cells.
Conclusion: The significant prognostic significance of GSDME in gastric cancer and the fact that affecting GSDME expression inhibits gastric cancer cell growth suggest that GSDME can be used as a predictive biomarker.
Introduction
Stomach cancer has caused many deaths. Gastric cancer causes more than 720,000 deaths per year (1, 2). More than 90% of gastric cancers are adenocarcinomas, which can be subdivided into cardia and non-cardia tumors, respectively, according to the site of the tumor (3). Cardia cancer occurs in the region adjacent to the esophagogastric junction and therefore has the same epidemiological features as esophageal adenocarcinomas. Non-cardia carcinomas, also known as distal gastric carcinomas, often occur in the lower part of the stomach (4). Due to the lack of typical early signs, 40% of them have metastatic disease at the time of diagnosis (5). When patients are found to have metastases, they are already in the middle to late stages of gastric cancer, and Patients with advanced gastric cancer are bound to die, ranging from approximately 10% to 30% at five years (6).
Pyroptosis amplifies or maintains inflammation by releasing pro-inflammatory cellular contents that tightly control the inflammatory response and orchestrate antimicrobial host defenses. It is an innate immune effector mechanism for fighting intracellular pathogens (7, 8). Heat illnesses are linked to many diseases, with different tissue and genetic backgrounds having different effects on cancer. On the one hand, pyroptosis can inhibit tumor development; on the other hand, as pro-inflammatory death, pyroptosis can form a microenvironment suitable for tumor cell growth, thus promoting tumor growth (9, 10). With the progress of research, the role of pyroptosis in tumors has become more prominent, as it may affect all level of carcinogenesis. Therefore, this study investigated the potential impact of genes related to pyrogenesis in gastric cancer.
Methods
Data Acquisition
There were 375 gastric cancer tissue samples and 32 corresponding normal tissue samples. We also downloaded gastric cancer mRNA expression profiles and lncRNA expression profiles from the TCGA dataset for subsequent analysis.
Pyroptosis-Related Gene Acquisition
The human genes with BP_PYROPTOSIS in their annotation results were NLRC4, NLRP1, AIM2, TREM2, GSDMC, GSDMB, NLRP9, NLRP6, ELANE, NAIP, CASP8, CASP4 GZBP1, APIP, GZMB, GZMA, CASP1, DHX9, GSDMA, GSDME, and GSDMD. Thus, the set of functional genes related to focal death was constructed.
Expression Analysis
Statistical significance was considered achieved when p < 0.05.
Pan-Cancer Analysis
The correlation between MSI and GSDME expression levels in pan-cancer was analyzed using the R language with the fmsb package. It was considered statistically significant when p < 0.05. Table 1 displays 33 tumor abbreviations in the TCGA database.
GSEA Analysis
To understand the impact of various biological functional gene sets of GSDME in gastric cancer, GSEA was used for enrichment analysis. GSDME was classified into high and low expression parts. Subsequently, GSEA V3.0 software was used to analyze the enrichment results of genes. A nominal P < 0.05 and a false discovery rate (FDR) < 25% were selected as cut-off criteria. We selected the top five-ranked analysis results.
Weighted Gene Regulatory Network Analysis
A soft threshold for network construction is first selected. Second, the adjacency matrix is transformed into a topology matrix. The genes are clustered based on the topology matrix using the average chain hierarchy clustering method. We set the minimum base number for each gene network module at 30. After determining the gene modules by the dynamic cut method, the feature vectors of each module are calculated in turn. Next, the modules are clustered, and the closer modules are merged into new modules.
Sample Collection
The gastric cancer tissues and the corresponding adjacent cancer tissues were collected from 5 GC patients, and the liquid nitrogen was preserved. Two cases were male and three cases were female. Sample collection excluded patients with major diseases. The study was informed by the patient and obtained by the Ethics committee of our hospital (ethical batch number: IIT20210716A).
Cell Source and Culture
Human gastric cancer cell lines (HS-746T, HT-X2557, MKN74, HTX2401C) and human gastric mucosal cells (GES-1, HT-X1964) were got from Shenzhen Haodi Huatuo Biotechnology Co. These cells were cultured with Dulbecco’s modified Eagle medium. Cells were supplemented with 10% fetal bovine serum and penicillin-streptomycin solution at 37°C and 5% CO2. After reaching 80–90% confluence with wall growth, cells were digested with 25% trypsin for passaging.
Cell Transfection
Cells were transferred to six-well plates at 2 x 10 five cells/well and incubated overnight at 37°C in an incubator. Si-NC and Si-GSDME were transfected with HS-746T and MKN74 cells using a Lipofectamine 2000 kit according to the kit instructions into DMEM containing 10% FBS and incubated for 6 h after transfection.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Assay
Grind the gastric cancer tissue to prepare a cell suspension. Total RNA was extracted from cells using the TRIzol Kit (Invitrogen, USA).Then, 1.7 ≤ OD260/OD280 ≤ 2.1 RNA samples could be used for analysis. Subsequently, the total RNA was reverse transcribed using a reverse transcription kit. The volume was adjusted by 20 μL of total reaction volume and 10 μL of SYBR Premix Ex Taq II (2X), 2 μL of cDNA, 0.8 μL of upstream and downstream primers, and the addition of sterile purified water. The amplification conditions were: 95°C for 30 s, 95°C for 5 s, 60°C for 30 s, and 40 cycles. GAPDH was used as an internal reference for GSDME. All primers were purchased from Shanghai Gene Chemistry Co. The forward primer for GSDME was 5′-TGCCTACGGTGTCATTGAGTT-3′; its reverse primer was TCTGGCATGTCTATGAATGCAAA-3′. The forward primer for GAPDH was 5′-GAGTCAACGGATTTGGTCGT-3′, and its reverse primer was 5′-GACAAGCTTCCCGTTCTCAG-3′. Now, 2–ΔΔCt was used to analyze the data, and the experiment was repeated three times.
Cell Counting Kit-8 (CCK8) Assay
Cell proliferation assays were performed using CCK-8 (Beyotime Biotechnology, China) according to the kit’s instructions: Cells were transfected for 24 h and incubated in 96-well plates at 2.5×10 3 cells/well. After 24, 48, and 72 h of incubation, 10 μL of CCK-8 solution was added to each well and incubated at room temperature for 2 h. Subsequently, the optical density of each well at 450 nm was detected using an enzyme marker. The experiment was repeated three times.
Flow Cytometry
The Annexin V-FITC Apoptosis Detection Kit (Beyotime, China) measured the apoptosis in gastric cancer cells treated as indicated. The experiments were repeated three times.
Statistical Analysis
The data obtained were plotted and analyzed using GraphPad 8. Comparisons between groups were made using independent t-tests, and comparisons between multiple parts were made using one-way ANOVA.
Results
Expression Levels of Pyroptosis-Related Genes in Gastric Cancer
We performed bulk expression analysis obtained and observed the expression of pyroptosis-related genes in gastric cancer (n = 375) and paracancerous tissue (n = 32). Most of the pyroptosis-related genes were upregulated in gastric cancer. See Figure 1.
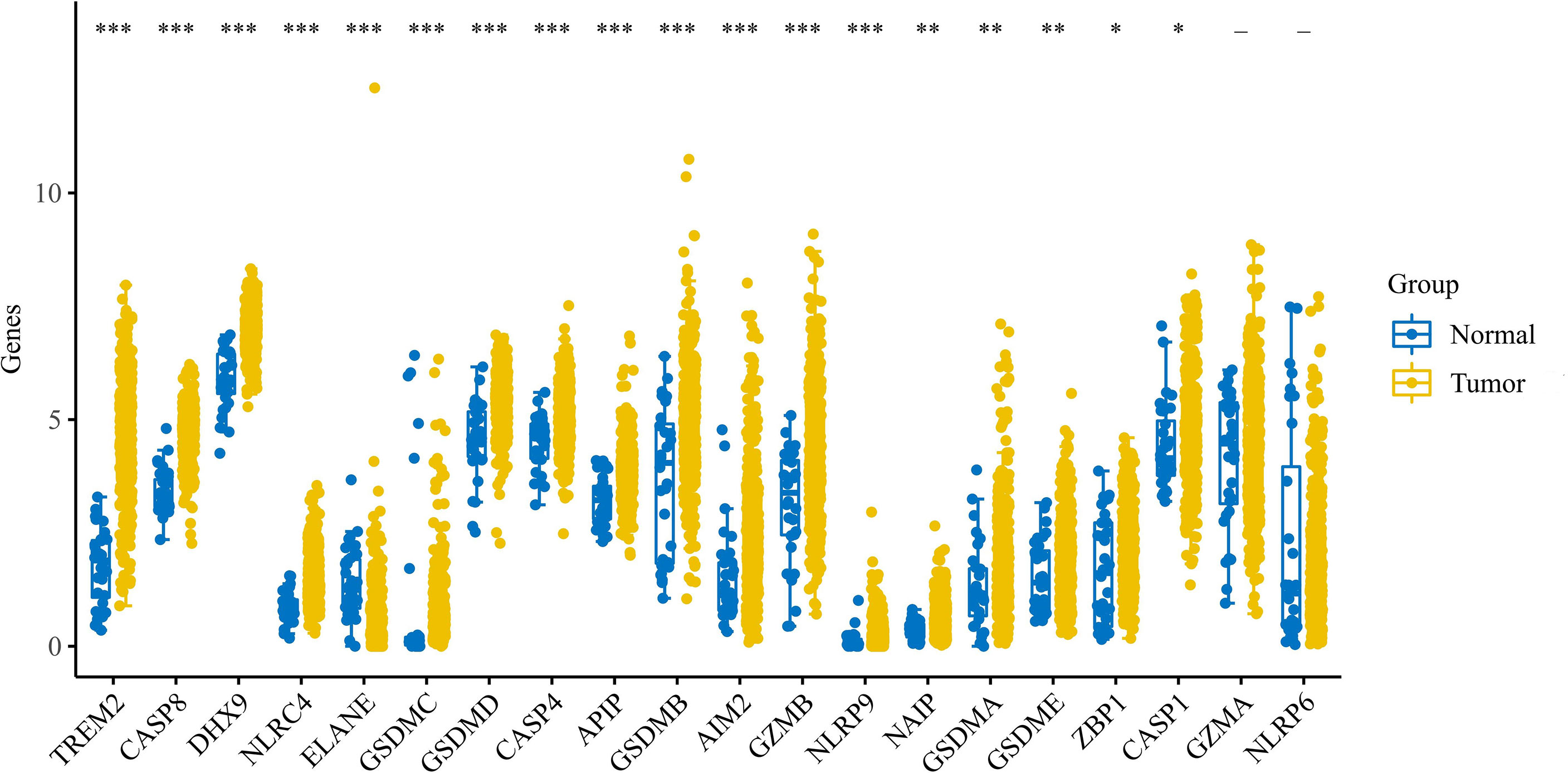
Figure 1 Expression of pyroptosis-related genes in gastric cancer. Expression of pyroptosis-related genes in gastric cancer. * indicates p < 0.05 for comparison between gastric cancer and paraneoplasia; ** indicates p < 0.01 for comparison between gastric cancer and paraneoplasia; *** indicates p < 0.001 for comparison between gastric cancer and paraneoplasia.
Prognostic Analysis of Pyroptosis-Related Genes
The results showed that only GSDME had prognostic significance in gastric cancer. The KM survival curve analysis also showed that the high-level survival of GSDME was lower than the low-level survival of GSDME. See Figure 2.
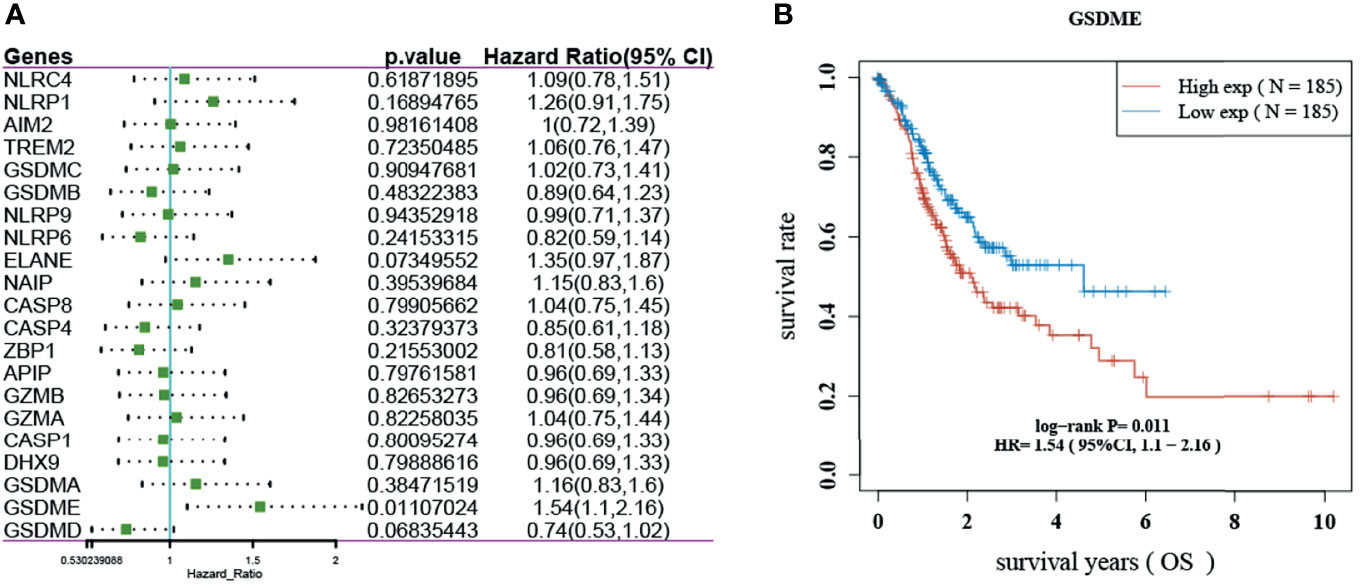
Figure 2 Prognosis of pyroptosis-related genes in gastric cancer. (A) Forest plot showing the OS prognosis of pyroptosis-related genes in gastric cancer. (B) KM survival curve of GSDME in gastric cancer.
Role of GSDME in Multiple Tumors
To continue to probe the role of GSDME in tumors, we also observed the expression of GSDME in multiple tumors. GSDME was highly expressed in CHOL, ESCA, GBM and so on, and low expressed in BLCA, BRCA, KICH, PRAD, THCA, and UCEC. A univariate cox analysis revealed the prognostic significance of GSDME in BLCA, KIRP, LIHC, KIRC, HNSC, LGG, KICH, and ACC. An immune checkpoint correlation analysis demonstrated that GSDME was closely associated with immune checkpoints TNFSF9, TNFSF15, TNFSF18, TNFSF4, TNFRSF25, TNFRSF4, TNFRSF8, LGALS9, NRP1, CD276, CD40, and CD200 in gastric cancer. A neoantigen analysis depicted that GSDME was only correlated in KIRP. An immunomutational load correlation indicated that GSDME was significantly correlated with immunomutational load in STAD, MESO, PAAD, TGCT, BLCA, and LUSC. Microsatellite correlation analysis showed that GSDME was significantly correlated with microsatellite instability in STAD and SARC. See Figure 3.
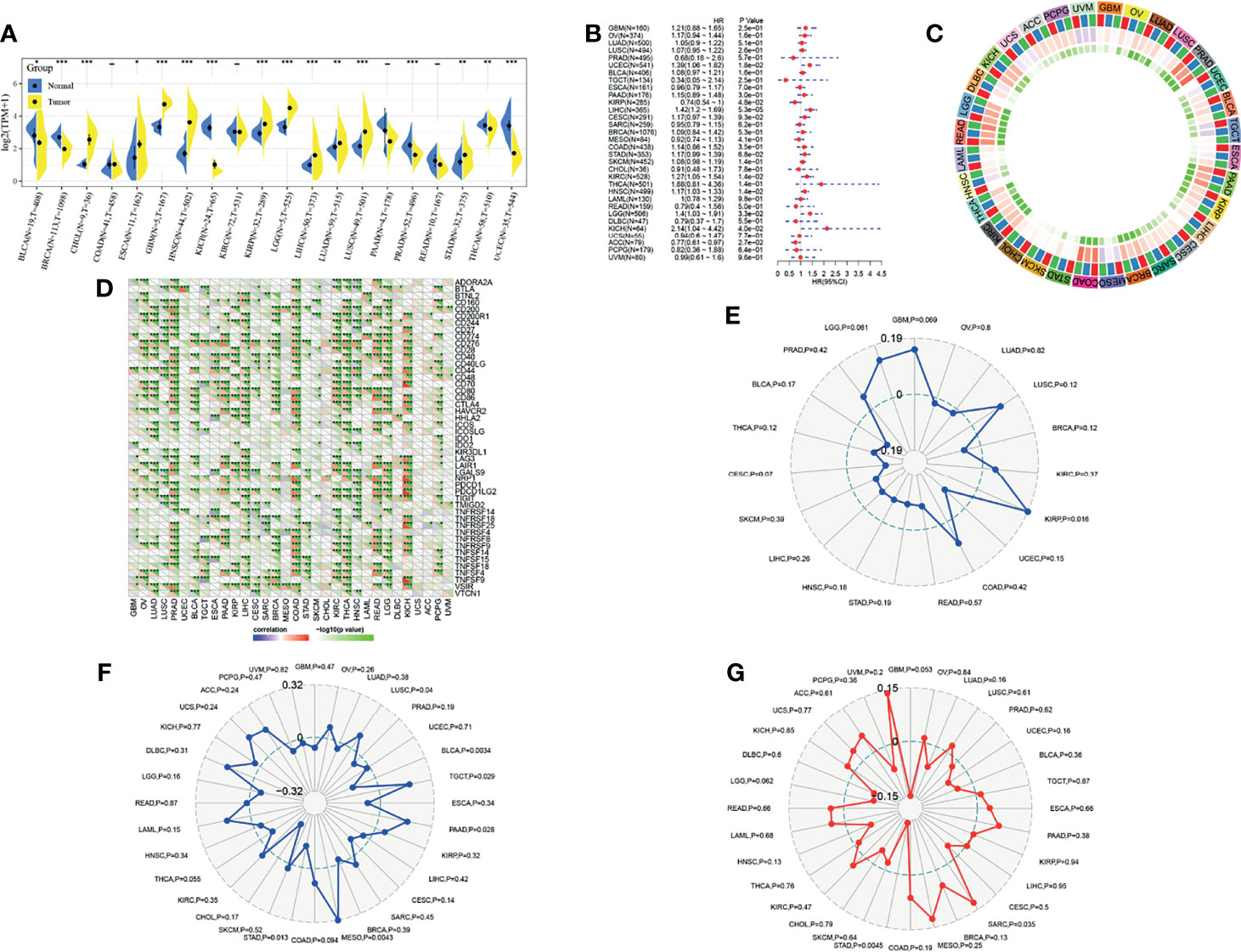
Figure 3 Analysis of GSDME in multiple tumors. (A) Expression of GSDME in multiple tumors (* indicates p < 0.05 for comparison between cancer and paracancer; ** indicates p < 0.01 for comparison between cancer and paracancer; *** indicates p < 0.001 for comparison between cancer and paracancer). (B) Prognostic analysis of GSDME in multiple tumors. (C) Correlation of GSDME with immune microenvironment scores in multiple tumors (blue, minimum color; red, maximum value color; green, minimum p-value color). (D) Correlation of GSDME with immune checkpoints in multiple tumors. (E) Correlation of GSDME with immune neoantigens in multiple tumors. (F) Correlation of GSDME with immune mutational load in multiple tumors. (G) correlation of GSDME with microsatellites in multiple tumors.
GSEA Analysis
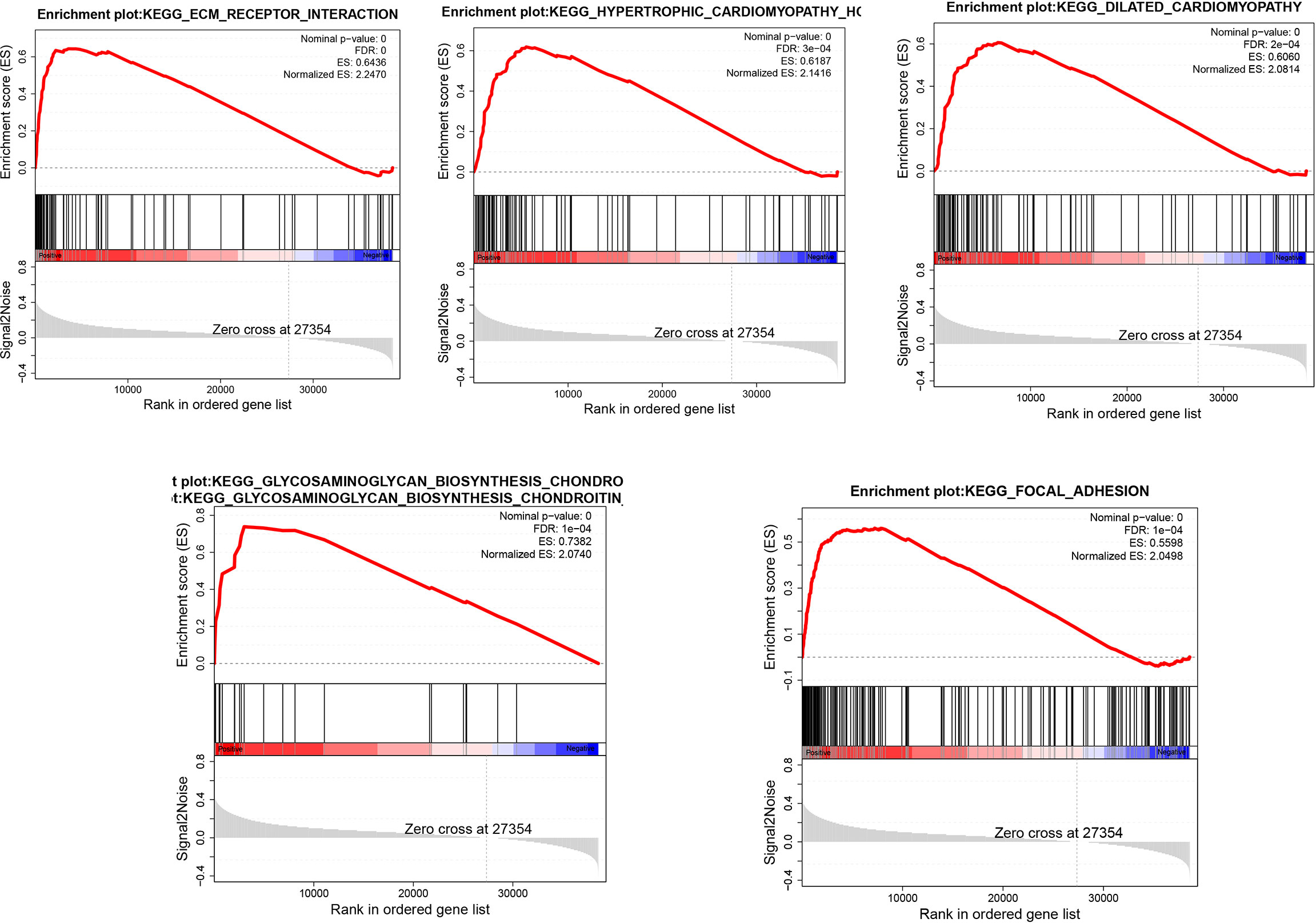
We divided the samples into 2parts according to the median expression levels of GSDME in gastric cancer and then analyzed the biological processes or signaling pathways involved in high GSDME expression in gastric cancer. We only screened the top five-ranked signaling pathways. The analysis revealed that the high expression of GSDME in gastric cancer was found in ECM_RECEPTOR_INTERACTION, HYPERTROPHIC_CARDIOMYOPATHY_ HCM, DILATED_CARDIOMYOPATHY, GLYCOSAMINOGLYCAN_ BIOSYNTHESIS_CHONDROITIN_SULFATE, and FOCAL_ADHESION, which have important roles. See Figure 4.
GSDME and the Stemness Score of Gastric Cancer Cells
We then divided the group into high and low expression groups based on the median GSDME expression. The mRNAsi scores between the two groups and the normal group were observed. The results showed that the mRNAsi score was significantly higher in the GSDME high expression group compared to the normal group. Surprisingly, the mRNAsi score was again significantly higher in the GSDME low expression group compared to the high expression. This may be a shift in the progression of gastric cancer, but the exact reason for this remains to be explored. See Figure 5.
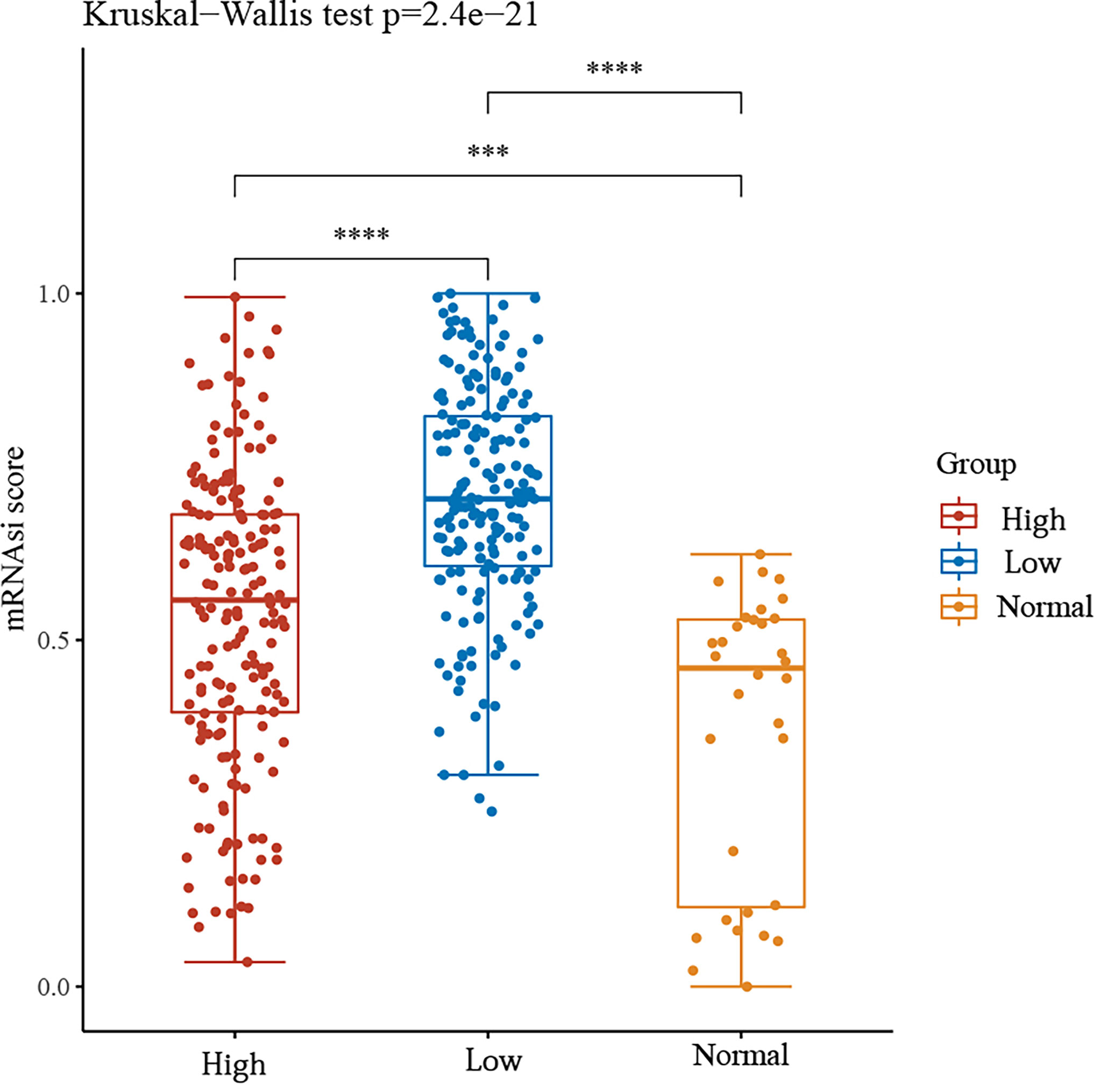
Figure 5 mRNAsi scores between high and low GSDME expression groups and normal groups. ***P < 0.001, ****P < 0.0001.
WGCNA Analysis
We combined the mRNA expression profiles obtained from the TCGA database with lncRNA expression profiles and performed WGCNA analysis. We found the module darkorange2 to which GSDME belongs. Next, we extracted the lncRNA nodes belonging to this module and obtained 16 lncRNAs with interactions with GSDME, namely, LINC01106, LINC01547, LINC00265, LINC00910, LINC01560, LINC01123, LINC00654, LINC01521, LINC00998, LINC00680, LINC00339, LINC00630, LINC00205, LINC01057, LINC00539, and LINC00863. Finally, we used Cytoscape (https://cytoscape.org/) for visualization. See Figure 6.
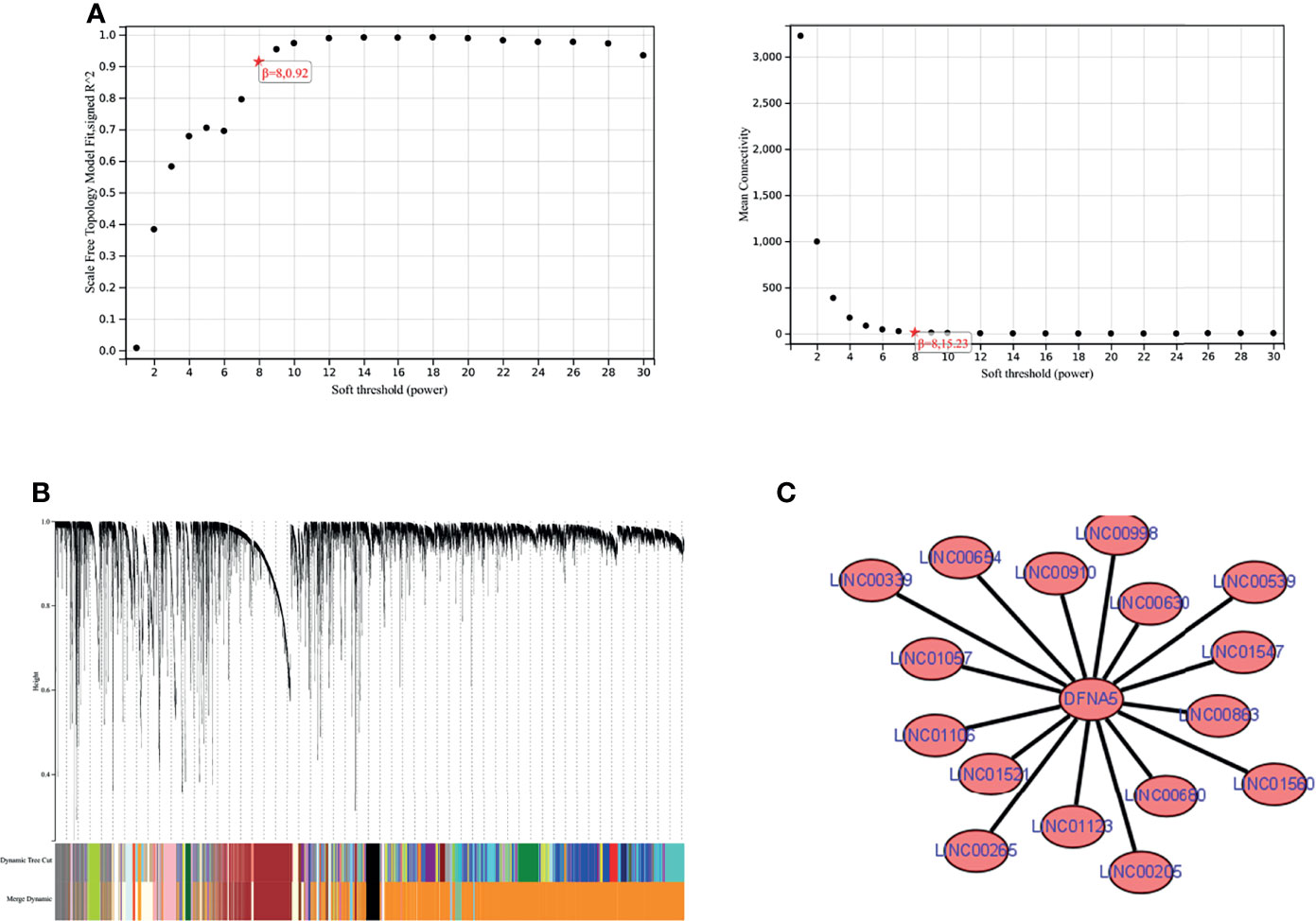
Figure 6 WGCNA analysis. (A) soft threshold determination. (B) gene clustering map. (C) lncRNAs-GSDME interaction network map.
Experimental Observation of the Effect of GSDME on Gastric Cancer Cells
Finally, we performed experiments to observe whether GSDME influences the biological behavior of gastric cancer cells. We first observed the expression of GSDME in gastric cancer tissue, gastric cancer cell lines HS-746T and MKN74 and gastric mucosal cells GES-1. The expression level of GSDME was significantly higher in gastric cancer tissue, HS-746T and MKN74 than in GES-1. We then further observed the effect of GSDME on gastric cancer cells by knocking down the expression level of GSDME to observe its effect on gastric cancer cells. The expression level of GSDME was reduced, the growth ability of gastric cancer cells were significantly slowed, and the apoptosis of cells was markedly increased. See Figure 7.
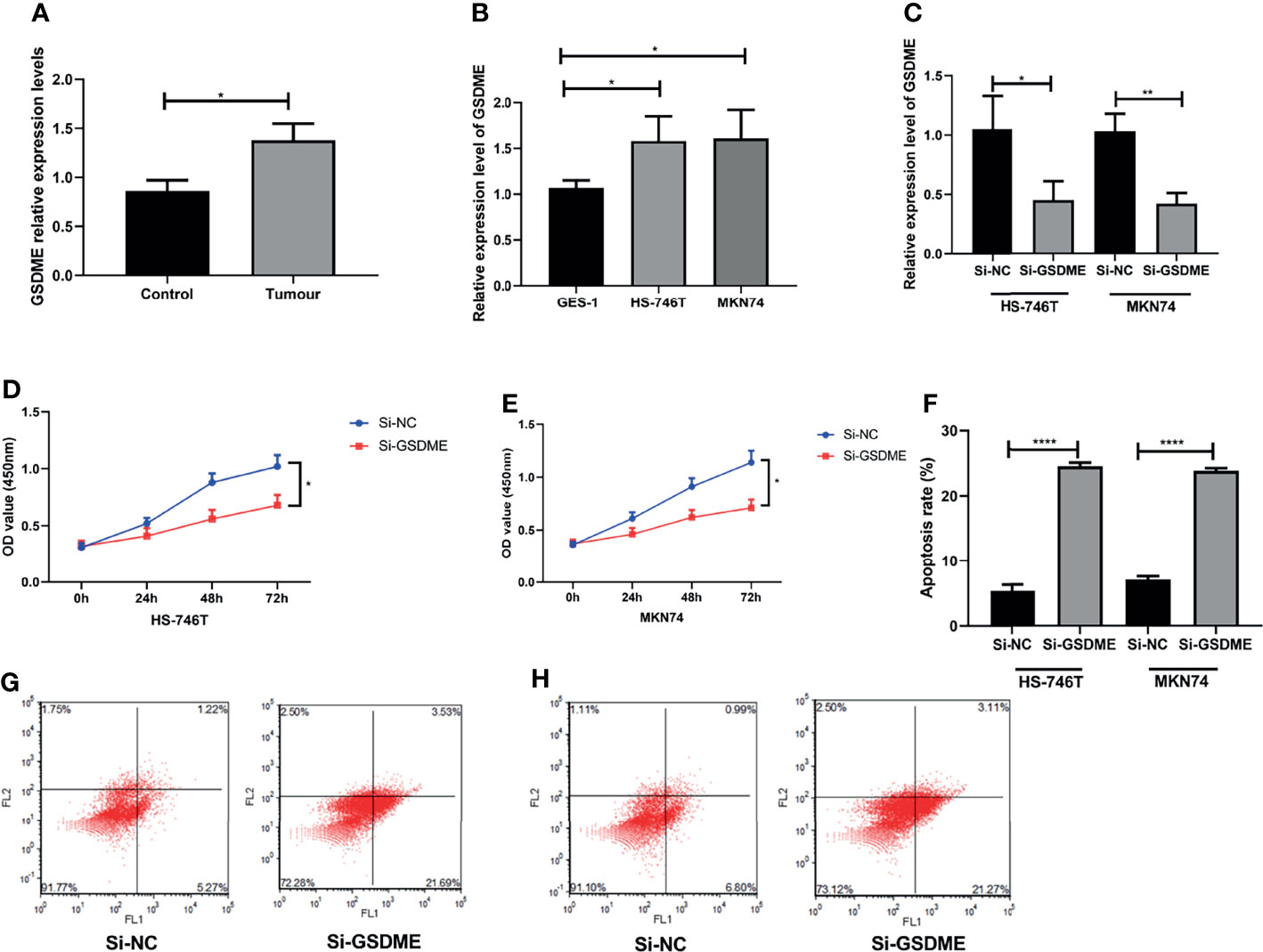
Figure 7 Effect of the knockdown of GSDME expression on gastric cancer cells. (A). Relative expression levels of GSDME in gastric cancer and adjacent to paracancerous tissues. (B) Expression of GSDME in human gastric cancer cell lines HS-746T and MKN74 and gastric mucosal cells GES-1. (C) Transfection efficiency. (D) Effect of reduced GSDME levels on the proliferation of HS-746T cells. (E) Effect of reduced GSDME levels on the proliferation of MKN74 cells. (F) Promotion of apoptosis after GSDME levels were reduced. (G) Flow apoptosis diagram based on HS-746T cells. (H) Flow apoptosis diagram based on MKN74 cells. *P < 0.05, **P < 0.01, ****P < 0.0001.
Discussion
GSDME related to the induction of secondary necrosis/scarring death. GSDME expression may also control the transition, such that in response to apoptotic stimuli, cells lacking GSDME expression undergo apoptosis without progressing. GSDME also plays distinct roles in a variety of tumors. In the present study, the expression of GSDME also differed in assorted tumors. Moreover, GSDME was closely associated with immune checkpoints in most tumors, including gastric cancer. GSDME expression enhanced the phagocytosis of tumor cells by tumor-associated macrophages and enhanced the number and function of tumor-infiltrating natural killer cells and CD8+ T cells, It can be cleaved by activated caspase-3 to produce its N-terminal fragment (GSDME-NT) (11, 12). It has also been indicated that targeted drugs induce GSDME-mediated cellular scorching in melanoma, linking the tumor immune microenvironment to T cell-mediated anti-tumor immunity (13). It is suggested that GSDME may related to cancer therapy and antitumor immunity, but the potential effect of GSDME on immune cells in gastric cancer needs to be investigated in more depth.
In rectal and esophageal squamous cell carcinoma also significantly higher than normal tissue and promote tumor progression (14, 15). In the present study, only GSDME among several pyroptosis genes had prognostic significance in gastric cancer. The KM curve revealed that the high expression of GSDME was associated with a poor prognosis for patients. The stemness score also showed a significantly higher mRNAsi score in the GSDME high expression group compared to the normal group. Interestingly, the mRNAsi score was again significantly higher in the GSDME low expression group compared to the high expression. However, the exact mechanism of development remains to be investigated.
Tumor development is inseparable from the proliferation and migration of cancer cells. Conversely, the inhibition of cancer cell proliferation may stunt tumor growth. In breast cancer, cell survival is strongly correlated with GSDMB expression. In addition, reports have demonstrated that GSDMB overexpression reduces cell viability (16). Gene deletion of GSDME promoted drug resistance (17). Another study reported that the knockdown of GSDME shifted loplatin-induced cell death from cell scorching to apoptosis, but did not affect the growth and tumor formation of colon cancer cells treated with loplatin (18). Although GSDME has been less studied in gastric cancer, one study has shown that GSDME converts chemotherapy-induced caspase-3-dependent apoptosis to cellular scorching in gastric cancer cells (19).Our in vitro experiments also show that changes in GSDME expression may affect the biological behavior of gastric cancer cells, which may provide new ideas for the treatment of gastric cancer.
LncRNA is a transcript with more than 200 nucleotides and no protein-coding potential. However, it is frequently dysregulated in cancer (20, 21). In therapeutic agents, LncRNAs can exert clinical therapeutic effects on tumors by inhibiting the transcription of mRNAs and blocking their function in combination with proteins (22, 23). In the present study, we found that 16 lncRNAs had reciprocal relationships with GSDME by WGCNA analysis, namely, LINC01106, LINC01547, LINC00265, LINC00910, LINC01560, LINC01123, LINC00654, LINC01521 LINC00998, LINC00680, LINC00339, LINC00630, LINC00205, LINC01057, LINC00539, and LINC00863. However, no clinical studies indicate that these lncRNAs have a regulatory relationship with GSDME. Correspondingly, more in-depth studies are needed.
In conclusion, GSDME is highly expressed in gastric cancer. The knockdown of GSDME expression can inhibit the growth of gastric cancer cells. GSDME, then, has prognostic significance in gastric cancer and can be used as a predictive biomarker.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author Contributions
These authors contributed equally: JY and GC. All authors contributed to the article and approved the submitted version.
Funding
The Key Research and Development Project of Zhejiang Province (2019C03071).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Noto JM, Peek RM Jr. The Gastric Microbiome, its Interaction With Helicobacter Pylori, and Its Potential Role in the Progression to Stomach Cancer. PLoS Pathog (2017) 13. doi: 10.1371/journal.ppat.1006573
2. Poorolajal J, Moradi L, Mohammadi Y, Cheraghi Z, Gohari-Ensaf F. Risk Factors for Stomach Cancer: A Systematic Review and Meta-Analysis. Epidemiol Health (2020) 42. doi: 10.4178/epih.e2020004
3. Etemadi A, Safiri S, Sepanlou SG, Ikuta K, Bisignano C, Shakeri R, et al. The Global, Regional, and National Burden of Stomach Cancer in 195 Countries, 1990–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol (2020) 5:42–54. doi: 10.1016/S2468-1253(19)30328-0
4. Rawla P, Barsouk A. Epidemiology of Gastric Cancer: Global Trends, Risk Factors and Prevention. Prz Gastroenterol (2019) 14:26. doi: 10.5114/pg.2018.80001
5. Koemans W, van der Kaaij R, Boot H, Buffart T, Veenhof A, Hartemink K, et al. Cytoreductive Surgery and Hyperthermic Intraperitoneal Chemotherapy Versus Palliative Systemic Chemotherapy in Stomach Cancer Patients With Peritoneal Dissemination, the Study Protocol of a Multicentre Randomised Controlled Trial (PERISCOPE Ii). BMC Cancer (2019) 19:1–8. doi: 10.1186/s12885-019-5640-2
6. Cao F, Hu Y, Chen Z, Han W, Lu W, Xu J, et al. Circulating Long Noncoding RNAs as Potential Biomarkers for Stomach Cancer: A Systematic Review and Meta-Analysis. World J Surg Oncol (2021) 19:1–13. doi: 10.1186/s12957-021-02194-6
7. Wang Y-Y, Liu X-L, Zhao R. Induction of Pyroptosis and its Implications in Cancer Management. Front Oncol (2019) 9:971. doi: 10.3389/fonc.2019.00971
8. Zeng C, Wang R, Tan H. Role of Pyroptosis in Cardiovascular Diseases and its Therapeutic Implications. Int J Biol Sci (2019) 15:1345. doi: 10.7150/ijbs.33568
9. Fang Y, Tian S, Pan Y, Li W, Wang Q, Tang Y, et al. Pyroptosis: A New Frontier in Cancer. BioMed Pharmacother (2020) 121:109595. doi: 10.1016/j.biopha.2019.109595
10. Xia X, Wang X, Cheng Z, Qin W, Lei L, Jiang J, et al. The Role of Pyroptosis in Cancer: Pro-Cancer or Pro-“Host”? Cell Death Dis (2019) 10:1–13. doi: 10.1038/s41419-019-1883-8
11. Zhang C-C, Li C-G, Wang Y-F, Xu L-H, He X-H, Zeng Q-Z, et al. Chemotherapeutic Paclitaxel and Cisplatin Differentially Induce Pyroptosis in A549 Lung Cancer Cells via Caspase-3/GSDME Activation. Apoptosis (2019) 24:312–25. doi: 10.1007/s10495-019-01515-1
12. Zhang Z, Zhang Y, Xia S, Kong Q, Li S, Liu X, et al. Gasdermin E Suppresses Tumour Growth by Activating Anti-Tumour Immunity. Nature (2020) 579:415–20. doi: 10.1038/s41586-020-2071-9
13. Erkes DA, Cai W, Sanchez IM, Purwin TJ, Rogers C, Field CO, et al. Mutant BRAF and MEK Inhibitors Regulate the Tumor Immune Microenvironment via Pyroptosis. Cancer Discov (2020) 10:254–69. doi: 10.1158/2159-8290.CD-19-0672
14. Tan G, Huang C, Chen J, Zhi FJ. HMGB1 Released From GSDME-Mediated Pyroptotic Epithelial Cells Participates in the Tumorigenesis of Colitis-Associated Colorectal Cancer Through the ERK1/2 Pathway. J Hematol Oncol Cancer Discov (2020) 13:1–11. doi: 10.1186/s13045-020-00985-0
15. Wu M, Wang Y, Yang D, Gong Y, Rao F, Liu R, et al. A PLK1 Kinase Inhibitor Enhances the Chemosensitivity of Cisplatin by Inducing Pyroptosis in Oesophageal Squamous Cell Carcinoma. EBioMedicine (2019) 41:244–55. doi: 10.1016/j.ebiom.2019.02.012
16. Zhang Z, Zhang H, Li D, Zhou X, Qin Q, Zhang Q. Caspase-3-Mediated GSDME Induced Pyroptosis in Breast Cancer Cells Through the ROS/JNK Signalling Pathway. J Cell Mol Med (2021) 25:8159–68. doi: 10.1111/jcmm.16574
17. Lu H, Zhang S, Wu J, Chen M, Cai M-C, Fu Y, et al. Molecular Targeted Therapies Elicit Concurrent Apoptotic and GSDME-Dependent Pyroptotic Tumor Cell Death. Clin Cancer Res (2018) 24:6066–77. doi: 10.1158/1078-0432.CCR-18-1478
18. Yu J, Li S, Qi J, Chen Z, Wu Y, Guo J, et al. Cleavage GSDME by caspase-3 Determines Lobaplatin-induced Pyroptosis Colon Cancer Cells. Cell Death Dis (2019) 10:1–20. doi: 10.1038/s41419-019-1441-4
19. Wang Y, Yin B, Li D, Wang G, Han X, Sun XJB. GSDME Mediates Caspase-3-Dependent Pyroptosis in Gastric Cancer. Clin Cancer Res (2018) 495:1418–25. doi: 10.1016/j.bbrc.2017.11.156
20. Liang Y, Song X, Li Y, Chen B, Zhao W, Wang L, et al. LncRNA BCRT1 Promotes Breast Cancer Progression by Targeting miR-1303/PTBP3 Axis. Mol Cancer (2020) 19(1):85. doi: 10.1186/s12943-020-01206-5
21. Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging Role of Tumor-Related Functional Peptides Encoded by lncRNA and circRNA. Mol Cancer (2020) 19:1–14. doi: 10.1186/s12943-020-1147-3
22. Jiang M-C, Ni J-J, Cui W-Y, Wang B-Y, Zhuo W. Emerging Roles of lncRNA in Cancer and Therapeutic Opportunities. Am J Cancer Res (2019) 9:1354.
23. He S, Yang S, Zhang Y, Li X, Gao D, Zhong Y, et al. LncRNA ODIR1 Inhibits Osteogenic Differentiation of hUC-MSCs Through the FBXO25/H2BK120ub/H3K4me3/OSX Axis. Cell Death Dis (2019) 10:1–16. doi: 10.1038/s41419-019-2148-2
Keywords: gastric cancer, pyroptosis, GSDME, cell proliferation, pyroptosis-related genes
Citation: Yin J, Che G, Wang W, Chen S and Liu J (2022) Investigating the Prognostic Significance of Pyroptosis-Related Genes in Gastric Cancer and Their Impact on Cells’ Biological Functions. Front. Oncol. 12:861284. doi: 10.3389/fonc.2022.861284
Received: 24 January 2022; Accepted: 01 March 2022;
Published: 28 March 2022.
Edited by:
Ye Wang, The Second Affiliated Hospital of Medical College of Qingdao University, ChinaReviewed by:
Xuefeng Xia, Nanjing Drum Tower Hospital, ChinaFei Liu, The Fifth Affiliated Hospital of Sun Yat-Sen University, China
Copyright © 2022 Yin, Che, Wang, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Liu, bGl1amlhbnp5QHpqdS5lZHUuY24=; Shitu Chen, MTE2MTgzMzlAemp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Jie Yin1†
Jie Yin1† Jian Liu
Jian Liu