- Department of Neurosurgery, The affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University, Changzhou, China
Although patients with lower-grade gliomas (LGGs; grades II and III) have a relatively favorable prognosis, patients frequently relapse and tend to progress to higher-grade gliomas, leading to treatment resistance, poor survival, and ultimately treatment failure. However, until now, thorough research has not yet been reported on the relationship between PD-L2 and immune infiltration and therapeutic sensitivity to immunotherapy and TMZ-based chemotherapy of LGGs. In this study, we found that the expression of PD-L2 is upregulated in glioma, with high PD-L2 expression predicting a worse prognosis. Univariate and multivariate Cox regression analysis both indicated that PD-L2 represented an independent prognostic factor with high accuracy in survival prediction for LGGs. A nomogram comprising of age, grade, IDH mutation, and PD-L2 was established for predicting OS. Additionally, PD-L2 was found to be remarkably correlated with immune infiltration and some anti-tumor immune functions. The degree of PD-L2 expression was also found to be strongly related to the prediction of therapeutic sensitivity to immunotherapy and TMZ-based chemotherapy. Furthermore, immunohistochemistry demonstrated that PD-L2 and the macrophage biomarker CD68 were both increased in glioma, with PD-L2 expression having a strong positive connection with CD68 expression. Taken together, PD-L2 is a prognostic biomarker for LGGs patients that may provide novel insights into glioma individualized therapeutic strategies and guide effective immunotherapy and chemotherapy.
Introduction
Gliomas are the most prevalent and fatal primary malignant intracranial tumors of the central nervous system (CNS), with a poor prognosis and fast progression (1, 2). According to the World Health Organization (WHO) classification criterion, gliomas can be classified as grade I-IV based on histopathological characteristics and biological behaviors (3). Lower-grade gliomas (LGGs) are classified as WHO grades II and III, while glioblastoma multiforme is classified as WHO grade IV (GBM) (4, 5). Despite the recent multimodal therapeutic strategies that have yielded some recent advances, including surgical resection, radio-chemotherapy, and immunotherapy, the survival and prognosis of glioma patients are still dismal with a 5-year survival rate of 20-30% (6–8). Studies have shown that the median survival time of LGGs is highly variable, and its biological behavior shows great intrinsic heterogeneity, which can rapidly develop into high-grade glioblastoma (GBM) (9, 10). Novel and effective prognostic biomarkers for early diagnosis, prognosis evaluation, and treatment response prediction of LGGs patients are urgently needed due to the lack of accurate and effective approaches to predict the prognosis of LGGs patients.
PD-L2 is the second ligand for PD-1, with a two- to six-fold affinity for PD-1 compared to PD-L1, and it can be produced by stromal, immune, or tumor cells (11).PD-L2 is a less studied PD-1 ligand, which plays an important role in cancer progression and immune regulation. Recent studies have linked PD-L2 expression to a poor prognosis in a variety of malignancies, such as lung adenocarcinoma (12), esophageal cancer (13), renal cell carcinoma (14), gastric cancer (15), colorectal cancer (16). Multiple recent studies have revealed that the tumor immune microenvironment (TIME) works as an immunosuppressive therapeutic barrier and is increasingly recognized as a crucial regulator of tumorigenesis, progression, maintenance, and therapy resistance (17–20). Tumor-infiltrating immune cells (TIICs), such as tumor-associated macrophages (TAMs), may be able to predict cancer prognosis and the efficacy of chemotherapy and immunotherapy in the TIME, which might be a viable target for anti-cancer therapy (21, 22). Quantifying tumor-infiltrating immune cells and revealing the significance of tumor immune microenvironment components and signatures may thus aid in better understanding their role in tumor immune escape, predicting patient prognosis, and guiding the development of innovative therapeutic strategies (23). A growing body of research suggests that transcriptional signatures could well evaluate the tumor immune microenvironment and predict clinical prognosis in glioma (24, 25). Nevertheless, until now, the literature lacks a comprehensive analysis of the association between PD-L2 and infiltrating immune cells in the LGGs TME, as well as the relationship between PD-L2 and therapeutic sensitivity to immunotherapy and chemotherapy.
In this study, we comprehensively analyzed the expression pattern and prognostic value of PD-L2 in glioma with RNA-seq data and corresponding clinical data from The Cancer Genome Atlas (TCGA) (n = 529) and the Chinese Glioma Genome Atlas (CGGA) (n = 443) datasets. Furthermore, an accurate nomogram integrating PD-L2, age, grade, and IDH mutation status was constructed to predict the 1-, 3- and 5-year OS of patients with LGGs, which revealed a high efficacy for prognosis prediction. The relationship between PD-L2 and tumor-infiltrating immune cells (TIICs) infiltration in the tumor immune microenvironment (TIME) was then investigated. Subsequently, we further explored the potential biological processes and pathways of PD-L2 involvement in the pathogenesis of LGGs. Furthermore, the predictive value of PD-L2 in the efficacy of therapeutic sensitivity to immunotherapy and chemotherapy was also investigated. Our findings could shed light on the critical role of PD-L2 as a novel biomarker and potential therapeutic target in LGGs, as well as provide an underlying mechanism between PD-L2 and tumor-immune interactions.
Materials and Methods
Ethics Statement
The experiments were undertaken with the understanding and written consent of each subject. The study was approved by the Ethical Review Boards of the Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University and conformed to the standards set by the Declaration of Helsinki.
Gene Expression Data Acquisition and Analysis
Glioma tissues (WHO grade II and grade III, n=20) and normal brain tissue (n=2) embedded in paraffin were collected from patients undergoing surgery in the Department of Neurosurgery, The Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University. The RNA-seq data and corresponding clinical data of LGG patients were downloaded from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) and the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn). The collected clinical data included age, gender, grade, IDH mutation status, 1p/19q codeletion status, and histological types.
Evaluation of the Independent Prognostic Factor and Survival Analysis
Correlations between PD-L2 expression and the clinicopathological and molecular features were analyzed by the “ComplexHeatmap”, “ggalluvial”, and “ggpubr” R package. Univariate and multivariate Cox regression analyses were used to determine whether PD-L2 could be used as an independent prognostic factor in patients with LGGs, regardless of clinical and molecular characteristics such as age, gender, grade, IDH mutation status, 1p/19q codeletion status, and histological types. According to the median expression level of PD-L2, glioma patients were divided into high-expression and low-expression groups. The Kaplan–Meier survival analysis was performed to evaluate overall survival (OS) using the ‘survival’ R package.
Development and Validation of the Nomogram Model
The nomogram including age, grade, IDH mutation status, and PD-L2 for prediction of 1-, 3-, and 5-years survival probability was conducted and verified in the TCGA cohort as well as CGGA cohort. The concordance index (C-index) and a calibration curve plot were then used to evaluate the nomogram’s predictive accuracy and discriminative ability.
Infiltration Patterns in the Tumor Microenvironment
The ESTIMATE algorithm (Estimation of Stromal and Immune cells in Malignant Tumors using Expression data) was applied to calculate the immune score, stromal score, estimate score, and tumor purity based on the transcriptome profile of TCGA and CGGA LGG cohorts (26). Single-sample gene set enrichment analysis (ssGSEA) is an algorithm performed to quantify immune cell infiltration in a single sample according to the expression levels of immune cell-specific markers in the tumor microenvironment using the “limma”, “GSEABase”, and “GSVA” packages (27–29). To identify TME infiltrating immune cells, the list of immune cell-specific markers gene sets was collected from a previous study (30). The correlation between PD-L2 expression and tumor-infiltrating immune cells (TIICs) infiltration in the tumor immune microenvironment was analyzed by the ssGSEA analysis (31). To validate the accuracy of the ssGSEA, we analyzed the correlation of PD-L2 expression with the abundances of six types of immune cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) from the online TIMER (https://cistrome.shinyapps.io/timer/) database (32). Currently acknowledged methods including TIMER, QUANTISEQ, CIBERSORT−ABS, CIBERSORT were performed to calculate the relationship between the expression of PD-L2 and the abundance of infiltrating immune cells among the LGGs samples from the TCGA dataset. The “GSVA” R package was used to compare the ssGSEA scores of immune cell infiltration and immune-related functions or pathways between high-PD-L2 and low-PD-L2 expression groups. Subsequently, the KEGG, GOBP, and hallmark pathways were used to identify the potential biological functions and pathways associated with PD-L2 expression level using the “GSVA” R package. Among them, Hallmark (h.all.v7.4.symbols.gmt), KEGG (c2.cp.kegg.v7.4.symbols.gmt), and GOBP (c5.go.bp.v7.4.symbols.gmt) gene sets were downloaded from Molecular Signatures Database (http://www.gsea-msigdb.org/). Statistical significance was determined for functional categories with an adjusted P-value < 0.05 or a false discovery rate (FDR) < 0.05.
Therapeutic Sensitivity Prediction
Next, we further predict the therapeutic sensitivity of LGGs patients to anti-PD-1 and anti-CTLA-4 immunotherapy and chemosensitivity of temozolomide (TMZ). To better predict the therapeutic sensitivity of immune checkpoint inhibitors (ICIs), we downloaded the immune cell and immunophenotype data from The Cancer Immunome Atlas (TCIA) (https://tcia.at/home). The immunophenogram was used to predict anti-PD1/PD-L1 therapy responses in LGGs. The immunophenogram was used to calculate the immunophenoscore (IPS) among four types (CTLA4 positive + PD-1 positive, CTLA4 negative + PD-1 negative, CTLA4 positive + PD-1 negative, CTLA4 negative + PD-1 positive, CTLA4 negative + PD-1 positive) from the TCGA-GBM database. The IPS scale ranged from 0 to 10.A high PD-1 positive IPS predicts a good response to anti-PD-1/PD-L1 therapy. Furthermore, we used the R package “pRRophetic” to predict the chemosensitivity of temozolomide (TMZ) based on half-maximal inhibitory concentration (IC50) in different groups (33, 34).
Immunohistochemistry
In formalin-fixed, paraffin-embedded surgical specimens of normal brains and different grades (WHO grade II-III) of human gliomas, immunohistochemical (IHC) staining was used to detect PD-L2 and CD68 expression. First, all paraffin specimens were cut into 4 μm thick slices, dewaxed in xylene (10 min x 2) and rehydrated with graded ethanol (100%, 95%, and 75% for 2 min, respectively), and then washed with phosphate-buffered saline (PBS; 3 times for 3 min). After antigen retrieval in 10 mM sodium citrate buffer (pH 6.0) and blocking endogenous peroxidase activity with 3% H2O2 (five minutes), the slides were incubated with 10% normal goat serum for 30 minutes at 37°C. Then the sections were incubated overnight with rabbit polyclonal anti-PD-L2 (Invitrogen, PA5-82484, 1:300), anti-CD68 (ZSGB-BIO, ZM-0464, ready-to-use) at 4°C, followed by incubation with biotinylated goat anti-rabbit immunoglobulin G (IgG) secondary antibody (ZSGB-BIO, ZDR5306, ready-to-use). The sections were then stained with DAB (3,3-diaminobenzidine), counterstained with hematoxylin, dehydrated, dried, and mounted. Three experienced pathologists were invited to evaluate all immunostained slides for IHC scores. The numbers of positively stained PD-L2 cells and CD68 cells were counted under the high-magnification lens (400×) in six randomly selected visual fields (35). The immunoreactive score (IRS) was calculated by multiplying the intensity of the staining by the percentage of positive cells. The staining intensity was scored as follows: 0 (negative), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). The percentage of cells that were positive was scored as follows: 0 score (0% positive cells), 1 score (1%–25% positive cells), 2 score (26%–50% positive cells), 3 score (51%–75% positive cells), and 4 score (76%–100% positive cells) (36).
Statistical Analysis
R (version 4.1.0) and publicly available packages were used for all statistical studies. P value<0.05 was considered statistically significant. (*P<0.05, **P<0.01, and ***P<0.001).
Results
PD-L2 Expression Was Upregulated in Glioma and Associated With Clinical and Molecular Characteristics
The PD-L2 expression level was significantly increased with an increasing grade of glioma in the TCGA cohort (Figures 1A, B). We then performed to investigate the relationship between the expression of PD-L2 and clinicopathological characteristics of LGGs patients in the TCGA and CGGA databases (Table 1). The results showed that PD-L2 was significantly higher expressed in IDH wildtype than that in the IDH mutation (p< 0.001), and in 1p19q non-codeletion than 1p19q codeletion (p<0.001). Furthermore, PD-L2 expression was significantly higher in LGGs patients with a dead status than in those with an alive status(p<0.05). The expression level of PD-L2 was also found to be highly correlated with histological types (p < 0.05, Table 1 and Figure 1). Consistent with the above findings, we found substantial differences between the high- and low-PD-L2 expression groups in terms of survival, grade, IDH mutation status, 1p/19q codeletion status, and histological types in the CGGA cohort (P < 0.05, Supplementary Figure 1 and Supplementary Figure 2).
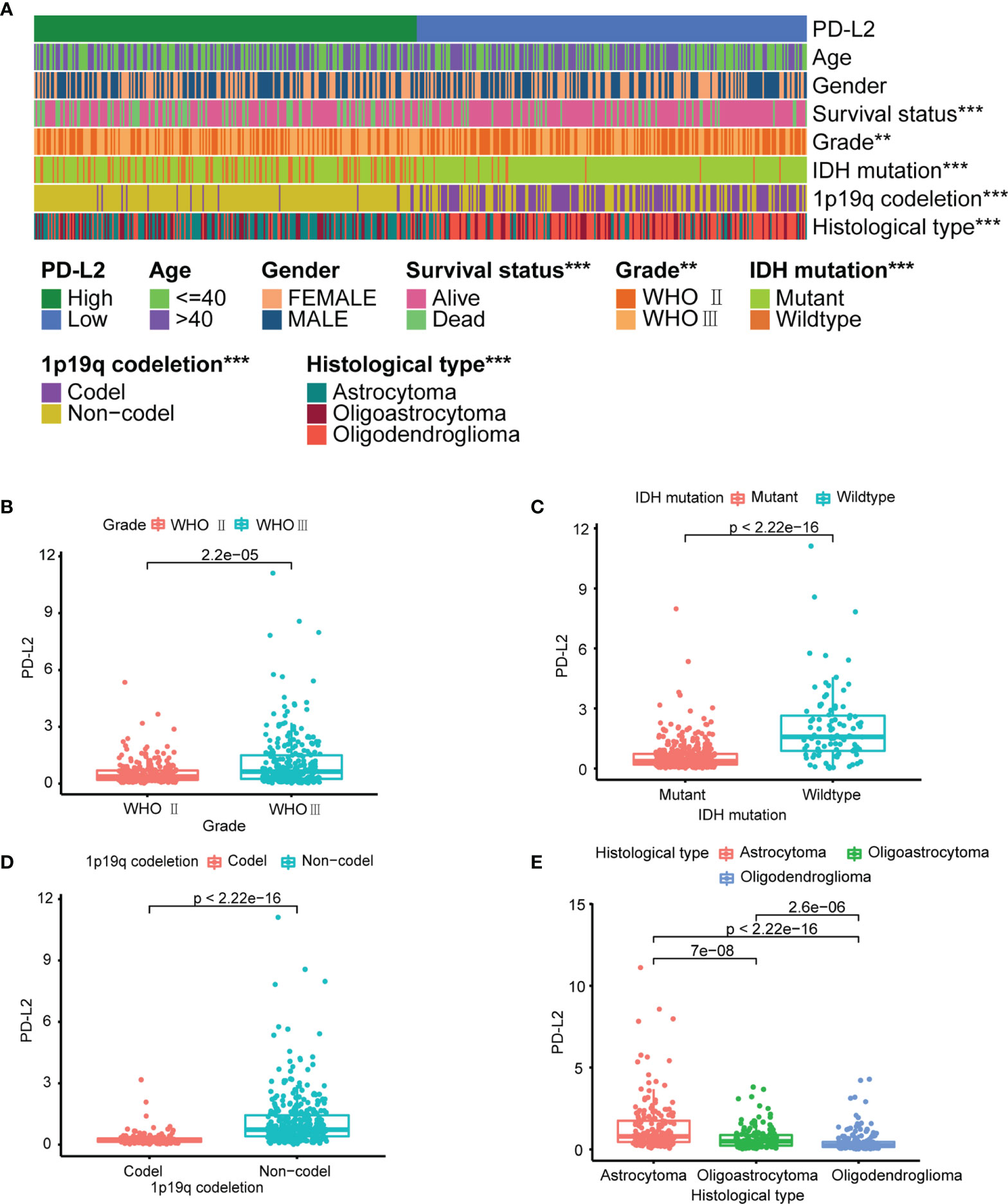
Figure 1 PD-L2 expression is associated with clinicopathological features in the TCGA dataset. (A) The heatmap showing the correlation between the expression levels of PD-L2 and the clinicopathological features including age, gender, survival status, grade, IDH mutation status, 1p/19q codeletion status, and histological types. (B–E) The scatter diagram showed that grade, IDH mutation status, 1p/19q codeletion status and histological types were significantly associated with PD-L2 expression (***P<0.001; **P<0.01).
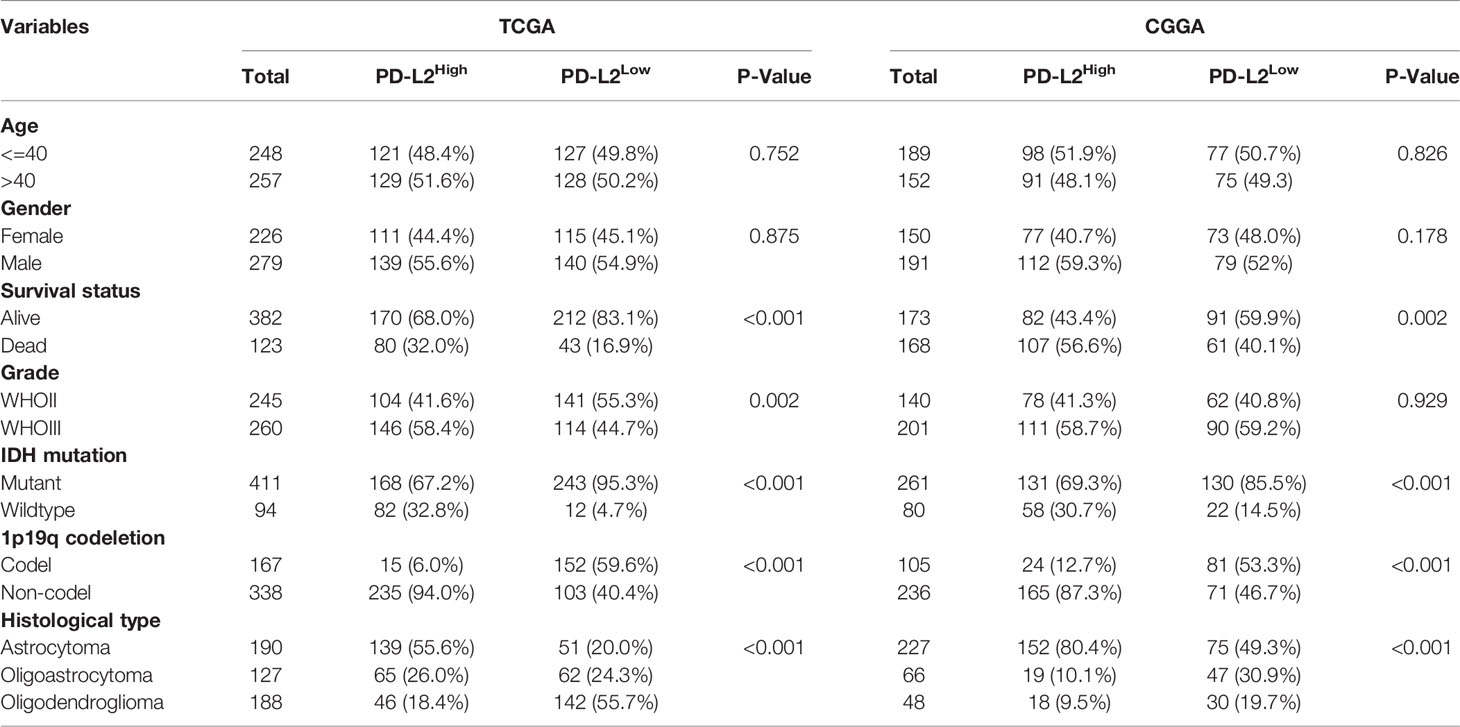
Table 1 Correlation between PD-L2 expression and clinicopathologic characteristics of patients with LGGs in TCGA and CGGA cohorts.
The major clinicopathological characteristics of the 20 patients with LGGs are summarized in Supplementary Table 1. The average age of patients was 50 (range, 13–73 years), 15 males and 5 females. As for grade, 45% patients were in grade II. Astrocytoma was the main histological type (n = 11), followed by oligodendroglioma (n = 7) and oligoastrocytoma (n = 2). Other clinical and molecular characteristics of these patients are presented in Supplementary Table 1. Subsequently, the protein levels of PD-L2 were further validated via IHC staining in the normal brain tissue and gliomas samples from the Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University (n=20). IHC staining of PD-L2 in tissue samples from primary gliomas WHO grade II-III patients (n = 20) and normal brain (n = 2) demonstrated that PD-L2 protein levels were more often increased in LGGs (WHO grade II-III) samples, while expression was low/undetectable in normal brain tissues (Figure 2). Taken together, these results indicated that PD-L2 levels were considerably higher in glioma tissue than in normal brain tissue.
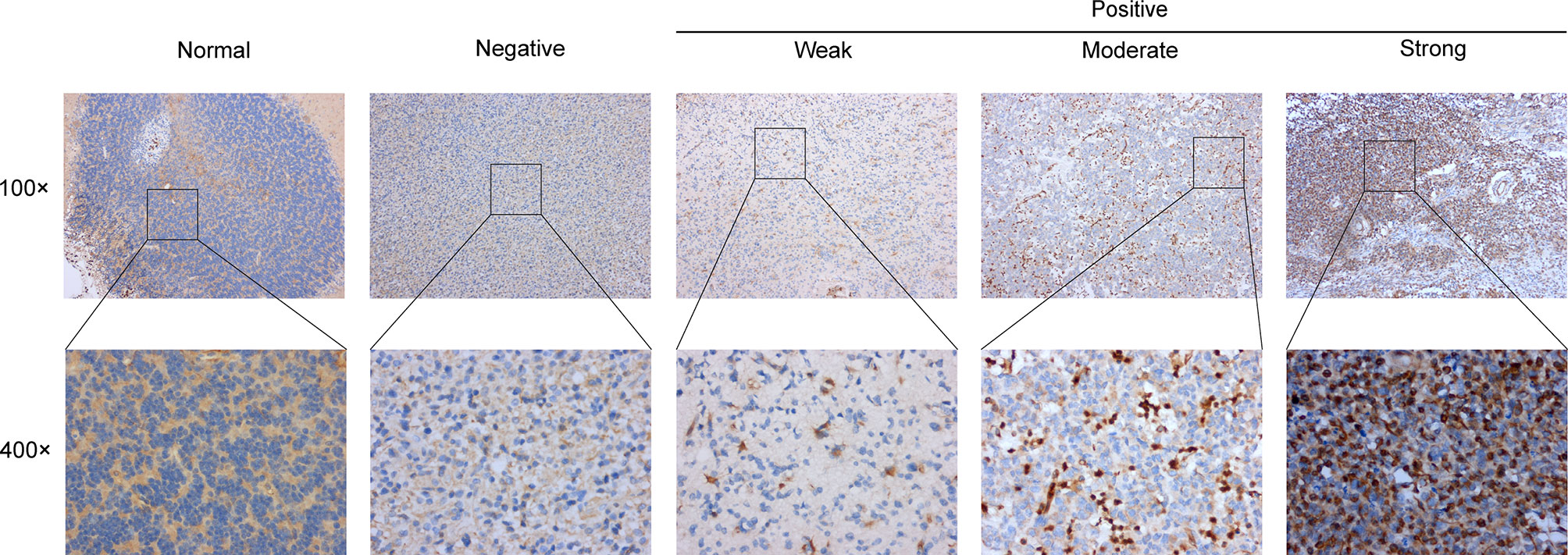
Figure 2 Representative images of immunohistochemical (IHC) staining of PD-L2 protein in glioma tissues and normal brain tissue (×100 and ×400).
PD-L2 Was an Independent Prognostic Factor in LGGs
We investigated the impact of PD-L2 expression on overall survival (OS). Patients were divided into a high expression group and a low expression group based on the median PD-L2 expression level. The Kaplan-Meier survival analysis revealed that patients with higher levels of PD-L2 expression had a worse prognosis than those with lower levels of PD-L2 expression in the TCGA and CGGA cohorts (P < 0.001, Figures 3A, B).
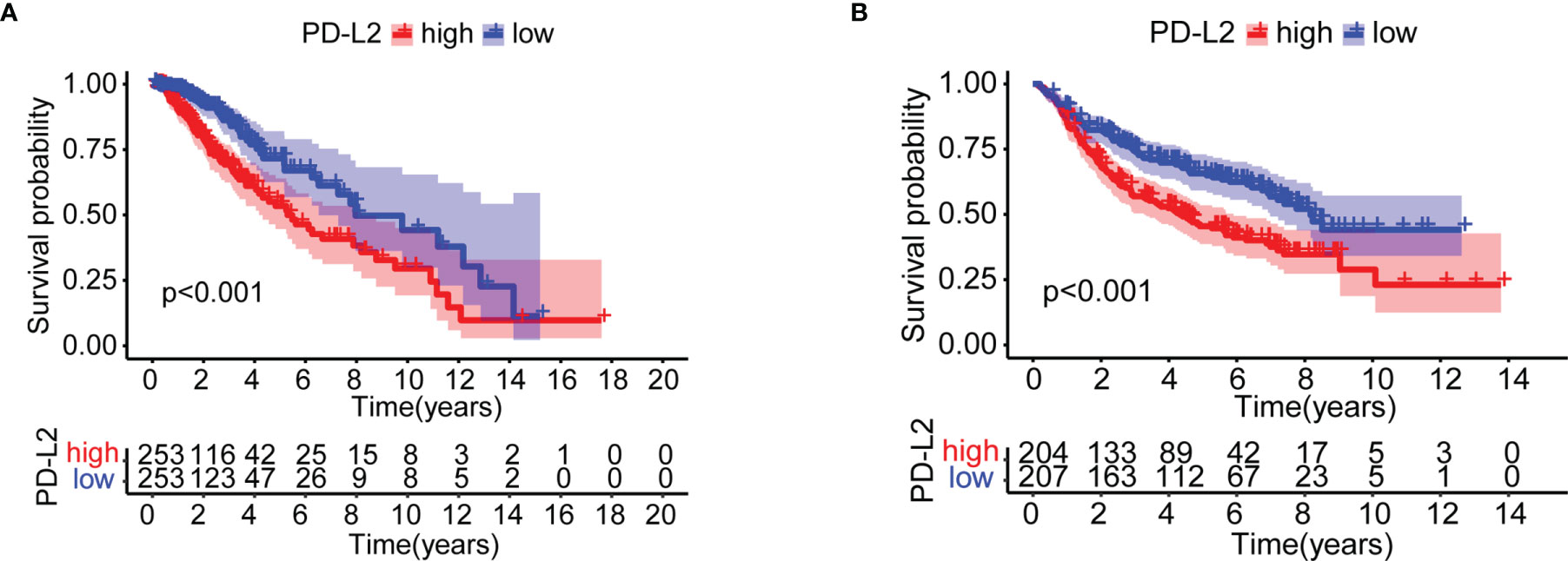
Figure 3 Kaplan–Meier overall survival (OS) curves for patients in the TCGA (A) and CGGA (B) datasets assigned to high- and low-expression groups.
Next, to assess the independent prognostic value of PD-L2 and other clinical-pathological factors in LGGs, Univariate and multivariate Cox regression analysis indicated that PD-L2 expression remained an independent prognostic factor of OS for patients with LGGs (HR: 1.939; 95% confidence interval [CI]: 1.355-2.775) after adjusting for the six clinicopathological factors (age, gender, grade, IDH mutation status, 1p/19q codeletion status, and histological types) (Table 2).We developed a nomogram model based on independent prognostic factors (age, grade, IDH mutation status, and PD-L2 expression) to quantitatively evaluate the 1-, 3-, and 5-year OS of patients with LGGs (Figure 4A). The C-index of the nomogram was calculated to be 0.826, and the calibration curves for the 1-, 3- and 5-year OS had an excellent predictive effect (Figures 4B–D). Additionally, external validation was done by the CGGA samples. The C-index was 0.701 and the calibration curves for probabilities for 1-, 3- and 5-year OS revealed good agreement between the predicted nomogram and actual survival (Figures 4E-G). Conclusively, this nomogram model based on PD-L2 expression may be a promising prognostic model for evaluating the clinical prognosis of LGGs.
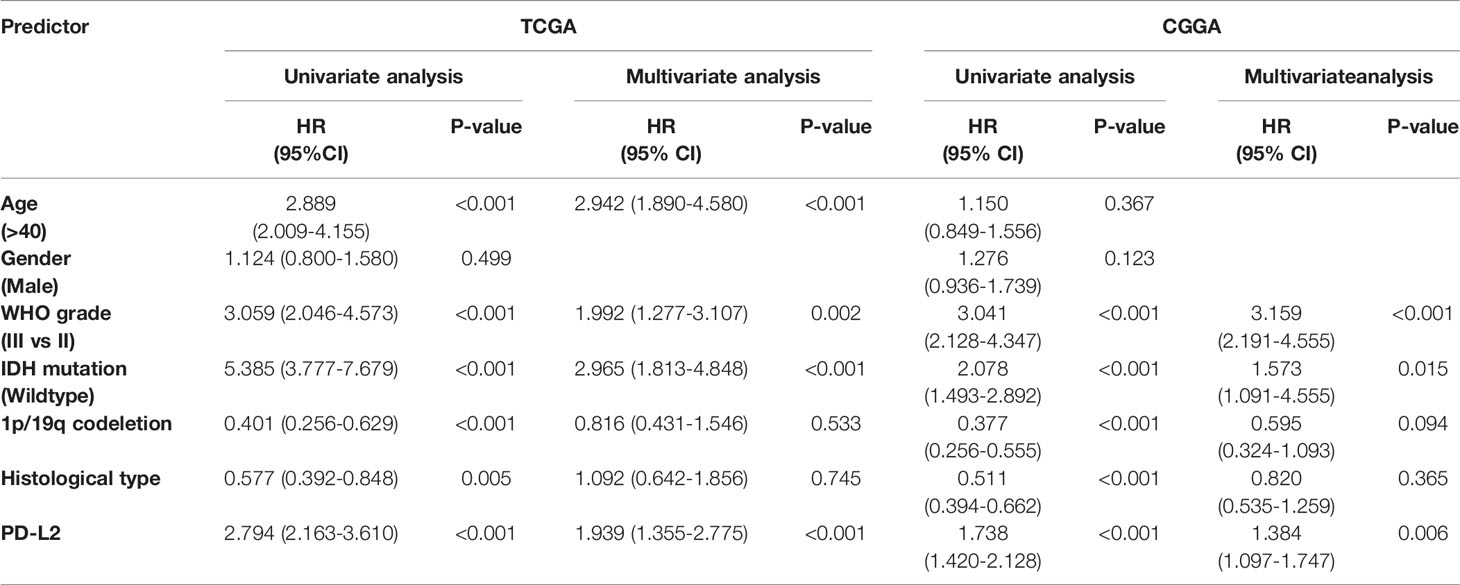
Table 2 Univariate and multivariate analysis of association of PD-L2 and prognostic factors with overall survival in LGGs based on the TCGA and CGGA databases.
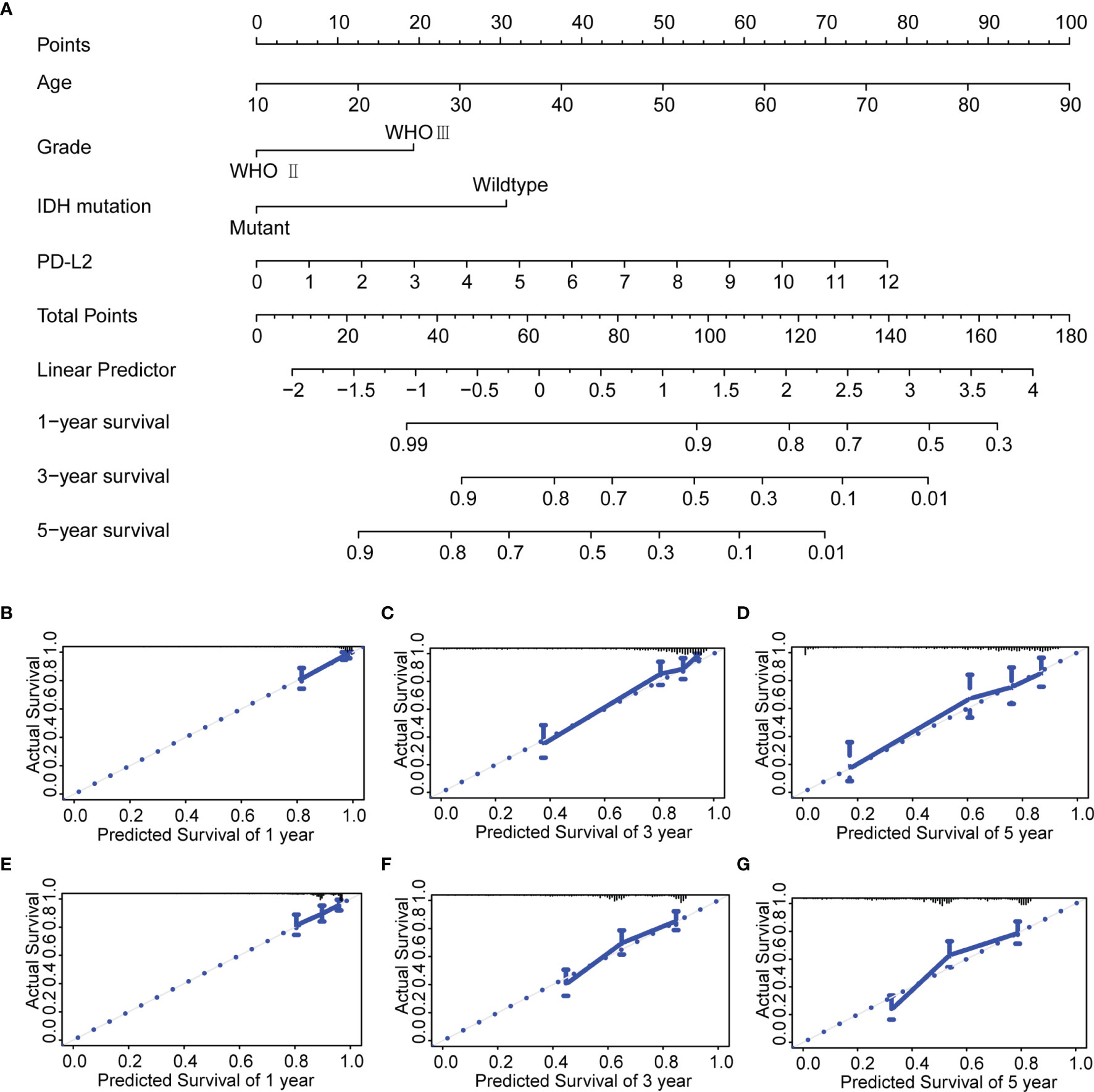
Figure 4 Construction and evaluation of a prognostic nomogram. (A) The prognostic nomogram predicts the probability of the 1-, 3-, and 5-year OS. (B–D) The calibration curve of the nomogram for predicting the 1-, 3-, and 5-year OS in the TCGA database. (E–G) The calibration curve of the nomogram for predicting the 1-, 3-, and 5-year OS in the CGGA database.
PD-L2 Expression Correlated With Immune Cell Infiltration and Tumor Immune Microenvironment
Then, using the ESTIMATE algorithm, we investigated the potential relationship between PD-L2 expression and immune infiltration and discovered that PD-L2 expression was positively correlated with the stroma score, immune score, and ESTIMATE score, as well as negatively correlated with tumor purity in the TCGA cohort (Figure 5A). To further explore the immune infiltration difference of 23 immune cells and the immune status betweenthehigh-PD-L2 and low-PD-L2 expression groups in the TIME, we quantified the infiltrating scores of diverse immune cell subpopulations and the activity of immune-related functions or pathways with ssGSEA. The difference and correlation analyses revealed that the high-PD-L2 expression group was associated with more tumor-infiltrating immune cells, including CD8+ T cells, CD4+ T cells, B cells, macrophages, neutrophils, dendritic cells, regulatory T cells (Tregs), MDSCs, and natural killer T cells (NKT) (Figures 5B, C and Supplementary Figure 3). Additionally, the analysis by the TIMER database showed that PD-L2 expression was negatively correlated with tumor purity and positively associated with the infiltrating levels of B cells, CD8+T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (Supplementary Figure 4). Furthermore, immune-related functions or pathways such as antigen-presenting cell (APC) co-inhibition, APC co-stimulation, chemokine receptor (CCR), Check-point, cytolytic activity, human leukocyte antigen (HLA), inflammation-promoting, MHC class I, para-inflammation, T-cell co-inhibition, T-cell co-stimulation, and type I and II interferon responses were associated with significantly higher ssGSEA scores in the high-PD-L2 expression group (P < 0.05, Figure 5D). Notably, we got similar results in the validation set, the results from the CGGA cohort also indicated that PD-L2 played a major role in immune cell infiltration in the tumor immune microenvironment of gliomas (Supplementary Figures 5, 6).
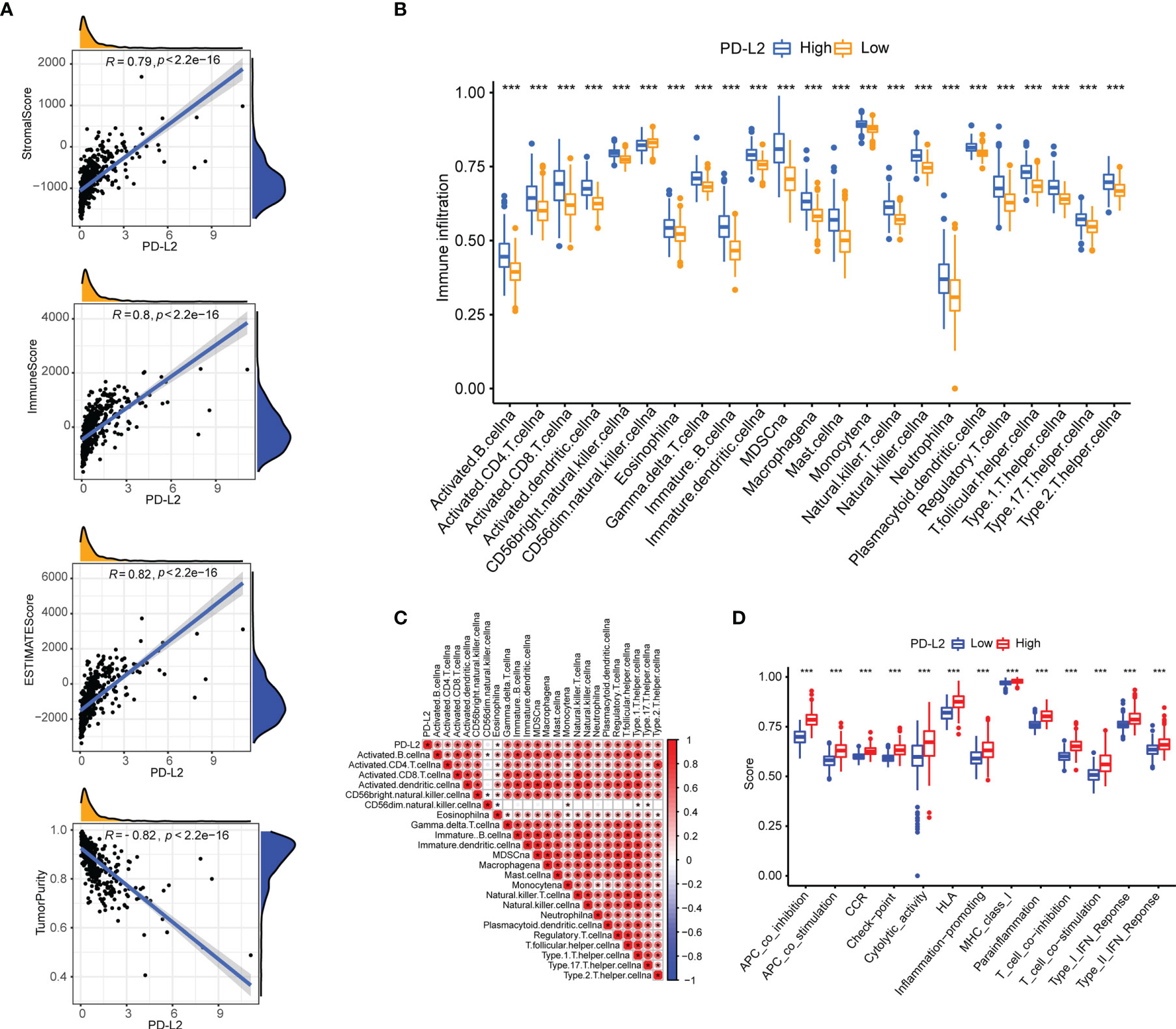
Figure 5 The association of PD-L2 with the tumor immune microenvironment and immune status in the TCGA cohort. (A) PD-L2 was positively correlated with the stroma score, immune score, and ESTIMATE score, as well as negatively correlated with tumor purity in the TCGA cohort. (B) The abundance of tumor-infiltrating immune cells in the high-PD-L2 and low-PD-L2 expression groups. (C) Pearson correlation analysis ofPD-L2 expression and immune cell infiltration. (D) The immune pathway functions between the high- and low- expression patients. ***P<0.001.
Our results have demonstrated that PD-L2 expression was found to be positively associated with macrophages in patients with LGGs in the TCGA (r = 0.59, P < 0.05) and CGGA cohorts (r = 0.69, P < 0.05) (Supplementary Figures 3, 6) and the TIMER database’s correlation analysis showed that PD-L2 was substantially associated with macrophage (r = 0.753, P < 0.05) (Supplementary Figure 4). In consistence with the findings described above, PD-L2 was more positively correlated with macrophages (Supplementary Figure 7). Therefore, we detected the expression of CD68 in glioma and normal brain tissues using IHC staining to further explore the relationship between PD-L2 and CD68. IHC staining results showed that the CD68 expression was observed in normal brain tissues with weak staining, while moderate or strong CD68 staining was detected in LGGs tissues (WHO II and WHO III) in almost all cases (Figure 6A). Furthermore, correlation analysis revealed a positive connection between PD-L2 and CD68 (r = 0.59, P 0.05) in glioma specimens (Figure 6B), which was comparable with the results of correlation analysis in the TCGA and CGGA cohorts (Figures 6C, D). Taken together, our findings confirmed the positive correlation between PD-L2 levels and the abundance of macrophage infiltration in LGGs.
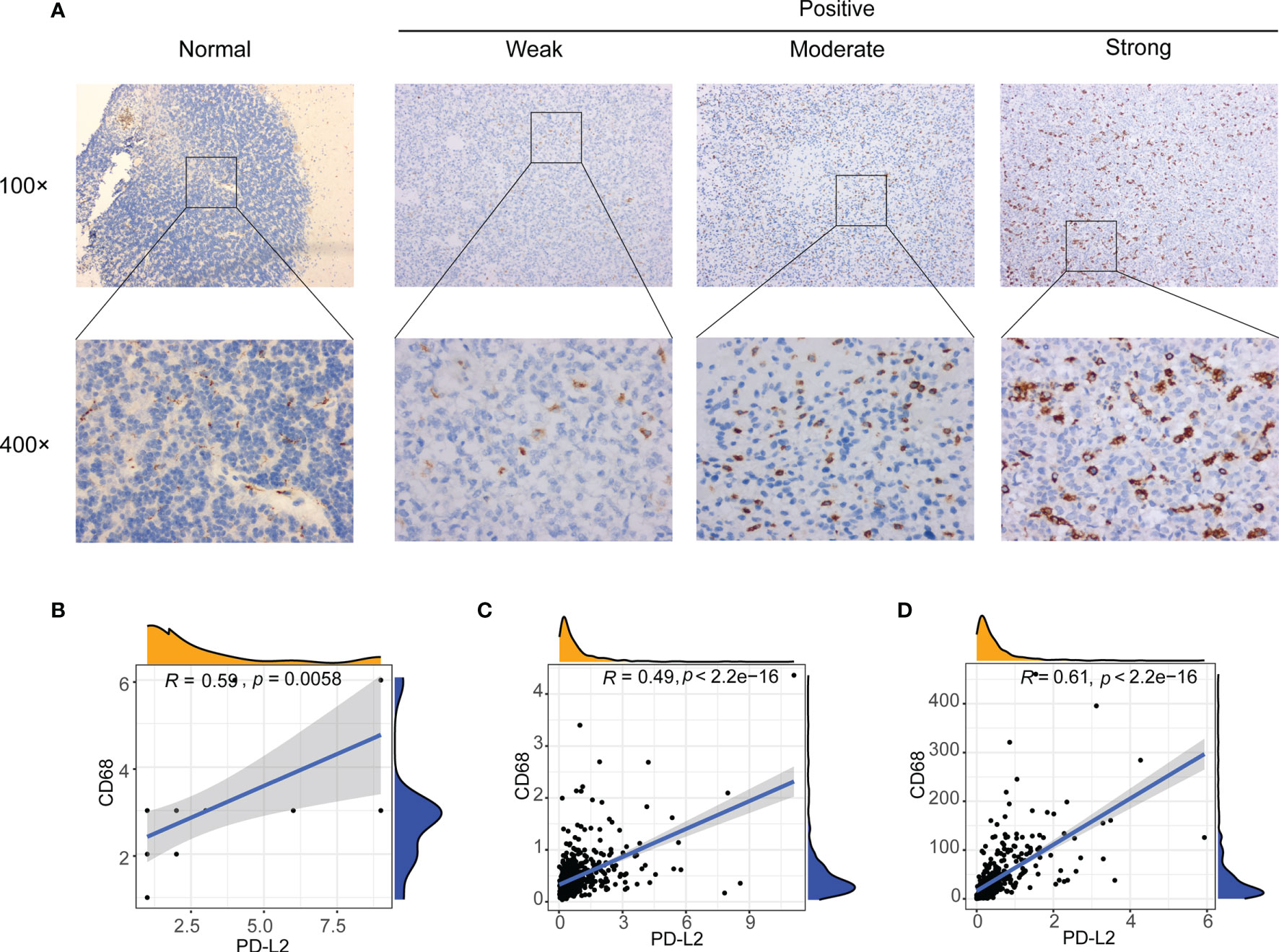
Figure 6 Correlation between PD-L2 expression and macrophages marker (CD68 expression) in gliomas. (A) Representative images of immunohistochemical (IHC) staining of CD68 protein in glioma tissues and normal brain tissue (×100 and ×400). (B–D) PD-L2 expression was significantly associated with macrophages marker (CD68 expression) in glioma specimens (B), TCGA database (C), and CGGA database (D).
Functional Annotation and Pathway Enrichment of PD-L2
To further investigate the underlying immune-related biological processes and tumor-related pathways associated with the expression of PD-L2, we performed the KEGG, GOBP, and hallmark pathways enrichment analysis in the TCGA cohort. The findings revealed that the high-PD-L2 expression group was considerably enriched in several biological processes, including that for cytokine-cytokine receptor interaction, response to interferon−gamma, inflammatory response, angiogenesis, chemokine production, apoptosis, hypoxia, interleukin (IL)-6, and IL-8 production. Moreover, tumor-related pathways such as JAK/STAT signaling pathway, Toll-like receptor signaling pathway, PI3K/AKT/mTOR signaling, TGF-beta signaling, IL-2-STAT5 signaling pathway, IL-6-JAK-STAT3 signaling pathway, P53 pathway signaling, TNFA signaling via NFKB were found to be enriched in the high-PD-L2 expression group (Figures 7A-C).
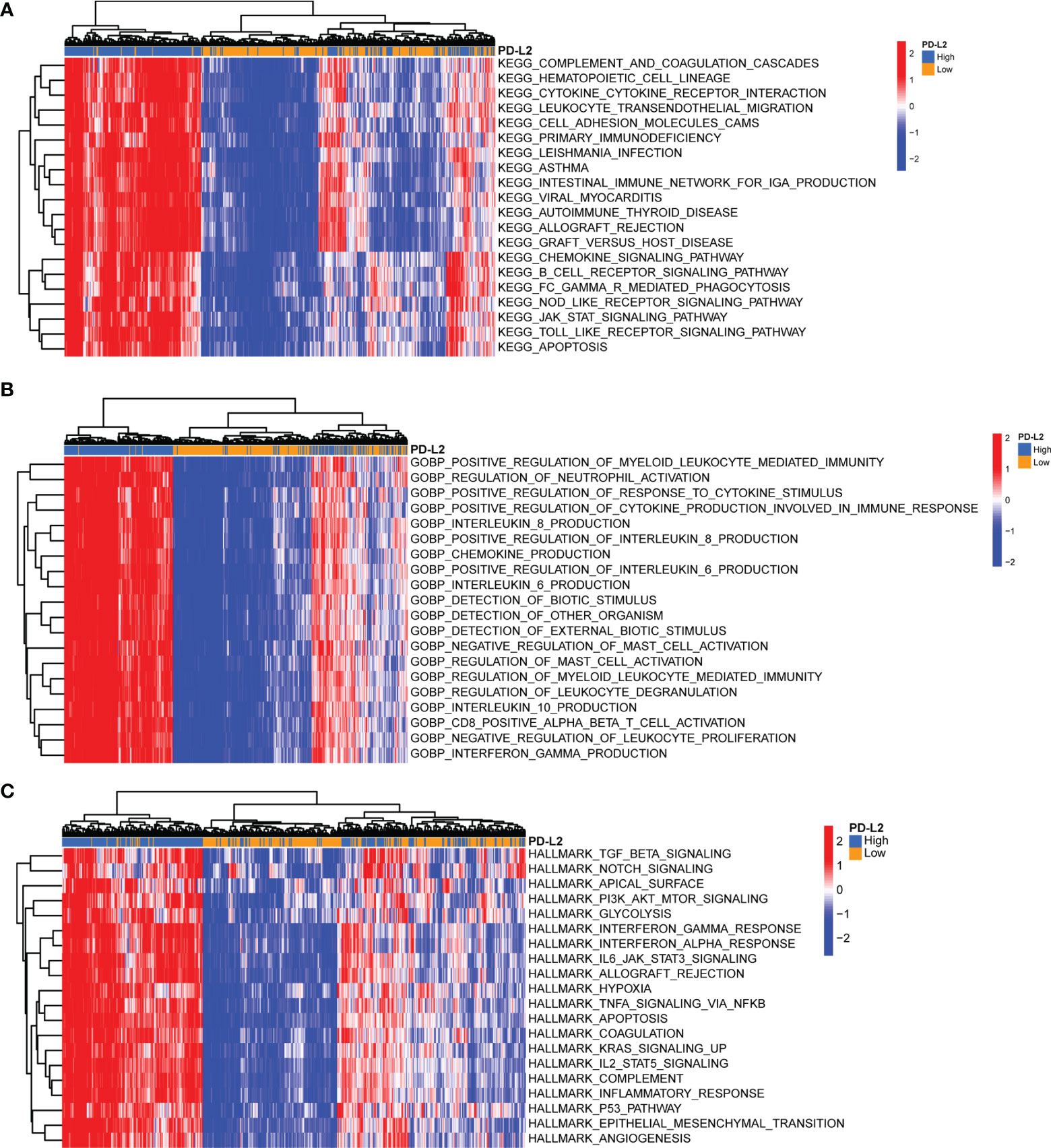
Figure 7 Gene Set Variation Analysis (GSVA) of biological processes and tumor-related pathways associated with the expression of PD-L2 in the TCGA cohort. Top 20 significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (A), Gene Ontology (GO) biological processes (B), and hallmark pathways (C) identified by GSVA analysis. GSVA, Gene Set Variation Analysis; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Prediction of Therapeutic Sensitivity to Immunotherapy and TMZ-Based Chemotherapy
Subsequently, to predict the potential chemosensitivity of temozolomide (TMZ) in LGGs, the relationship between PD-L2 expression and TMZ-based chemotherapy was explored and the results revealed a substantial positive correlation between PD-L2 expression and TMZ-based chemotherapy (r = 0.54, P < 0.05) (Figure 8A). Additionally, patients in the low-expression group showed a more sensitive response to TMZ-based chemotherapy based on the estimated IC50 (P < 0.05, Figure 8B). In consideration of the importance of ICIs in immunotherapy, the immunophenogram was then used to predict the response to anti-PD-1/PD-L1 therapy. We discovered that the high-PD-L2 expression group had significantly higher IPS than the low-PD-L2 expression group in the CTLA4 positive + PD-1 positive, CTLA4 positive + PD-1 negative, and CTLA4 negative + PD-1 positive types (P< 0.05) (Figure 8C). These findings suggested that patients with high levels of PD-L2 expression were more likely to respond well to anti-PD-1/PD-L1 therapy or a combination of anti-PD-1/PD-L1 and anti-CTLA-4 therapy, suggesting that PD-L2 had good potential in predicting ICIs response and may benefit from immunotherapies. Conclusively, PD-L2 could contribute to the selection of optimal immunotherapy and chemotherapy strategy in glioma patients.
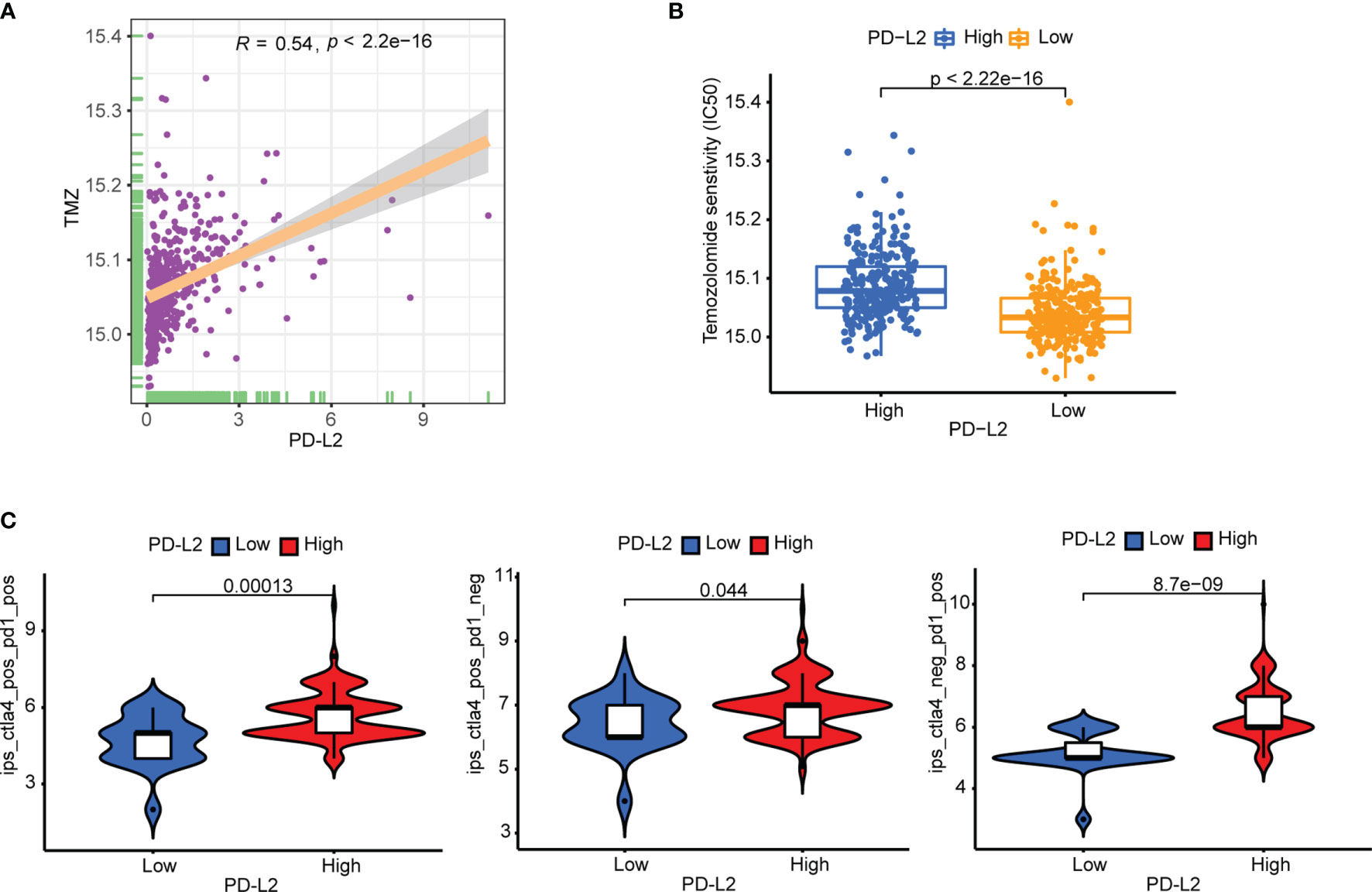
Figure 8 Therapeutic sensitivity prediction to immunotherapy and TMZ-based chemotherapy in the TCGA cohort. (A) PD-L2 was significantly positively correlated with the TMZ-based chemotherapy. (B) The low-PD-L2 expression patients showed a more sensitive response to TMZ-based chemotherapy based on the estimated IC50. (C) Patients with high PD-L2 expression had higher immunophenoscores (IPS) in three subgroups: CTLA-4 positive PD-1 positive, CTLA-4 negative PD-1 positive, and CTLA-4 positive PD-1 negative. TMZ, temozolomide; IC50, half-maximal inhibitory concentration; IPS, immunophenoscore.
Discussion
Patients with LGGs have a more favorable prognosis than those with GBM, but many advances to higher-grade gliomas, resulting in recurrence and poor survival, posing a therapeutic challenge to physicians (37–39).Despite standard treatment of maximally safe surgical resection followed by radiotherapy and adjuvant chemotherapy with temozolomide, the prognosis of glioma, particularly glioblastoma, remains poor and almost all patients relapse inevitably (40, 41). Identifying new prognostic markers of LGGs at an early stage of the tumor, thus, can successfully predict and enhance the clinical prognosis of glioma patients. A previous study showed that PD-L2 was found to be able to predict the poor prognosis of glioma patients (42, 43). However, the specific role of PD-L2 in LGGs and their therapeutic sensitivity to immunotherapy and TMZ-based chemotherapy have not been reported, and few studies have been performed to explore the relationship between PD-L2 and TIICs in the LGGs tumor immune microenvironment (TIME). In this study, we discovered that PD-L2 is elevated in LGGs samples and is substantially associated with a poor prognosis in glioma patients in the CGGA and TCGA cohorts. Univariate and multivariate Cox regression analysis confirmed that high PD-L2 expression was found to be an independent prognostic factor of patients with LGGs. Additionally, we established a nomogram that integrates PD-L2 with clinicopathological factors (age, grade, and IDH mutation status) and discovered that it performs exceptionally well in predicting 1-, 3-, and 5-year OS in glioma patients.
The role of tumor immune microenvironment in the occurrence, development, invasion, and drug-resistance of gliomas is becoming increasingly recognized (44, 45), with numerous published studies demonstrating that tumor-infiltrating immune cells (TIICs) play a critical role in the tumorigenesis, invasion, metastasis, diagnosis, treatment, and prognosis of gliomas (46–48). In our study, PD-L2 expression was found to positively correlate with the immune score, stromal score, and ESTIMATE score, as well as negatively correlate with tumor purity, indicating that high PD-L2 expression was positively correlated with immune infiltration in LGGs. Numerous studies have previously described that the role of tumor-infiltrating immune cells in various cancers is increasingly being viewed as a critical factor driving or mediating tumor progression and influencing therapeutic outcomes and patient prognosis (49, 50).Unlike a previous study (42), we further investigate the correlation between PD-L2 and TIICs via GSVA analysis and the online website TIMER. Notably, our results revealed that PD-L2 is closely associated with TIICs infiltration in LGGs, especially macrophages, which can be validated by the analysis of the online website TIMER.
Tumor-associated macrophages (TAMs), which play a significant role in carcinogenesis, progression, metastasis, invasion, immunosuppression, angiogenesis, and the immune response to cancer, have been linked to treatment failure and poor prognosis in LGGs patients in several recent studies. Multiple recent studies have shown that TAMs, which play an important role in tumorigenesis, progression, metastasis, invasion, immunosuppression, angiogenesis, and immune response to cancer, are linked to treatment failure and poor prognosis in LGGs patients (51, 52). Our immune cell correlation analysis revealed that PD-L2 was positively associated with macrophages in the TCGA and CGGA cohorts (r = 0.59, r = 0.69, respectively, P < 0.05). Our immunohistochemistry results are in accordance with a previous study that CD68 expression in lower-grade glioma tissues (LGGs, WHO II and WHO III) was significantly higher than the expression in normal brain tissues. Further analysis demonstrated PD-L2 expression was significantly positively correlated with macrophage infiltration in our glioma samples (r = 0.59, P < 0.05). Additionally, several lines of evidence indicate that a high TAMs infiltration, especially M2 macrophages, are closely associated with poor clinical outcomes in a wide variety of tumors and are now being recognized as potential biomarkers for diagnosis and prognosis of malignant tumors, as well as an attractive therapeutic target in cancer therapy (53–56). Chanmee et al. have reported that achieving TAMs-targeting cancer therapy by changing its polarization from M2 macrophages to M1 macrophages, inhibiting macrophage recruitment, and inhibiting the survival of TAMs, can enhance the response to treatment (57, 58).Therefore, targeting the tumor immune checkpoints and macrophages targeting may be a novel strategy for effective cancer treatments.
Immunotherapy is thought to be one of the most promising glioma treatments (59). Emerging evidence has indicated that immune checkpoint blockade (ICB) therapies such as anti-programmed death 1 (PD-1), ant-programmed cell death-ligand 1 (PD-L1) and anti-cytotoxic T lymphocyte-associated protein 4 (CTLA-4) have emerged as a promising approach as anti-cancer immunotherapy, dramatically changing treatment in solid tumors (60–62). However, the majority of patients still showed adaptive resistance with a low response rate in most cancers, especially for tumors with a low mutational burden (63–65). As a result, to improve the clinical prognosis of cancer patients, we must investigate new targets and predict the therapeutic sensitivity of immune checkpoint inhibitors (ICIs). Here, our study indicated that patients with high expression of PD-L2 are associated with a high response rate to treatment by ICIs, suggesting that PD-L2 had good potential in predicting ICIs response and high expression of PD-L2 may more likely to yield considerable clinical benefit from immunotherapy. So far, temozolomide (TMZ), a DNA alkylating agent, has become the first-line chemotherapy for the treatment of glioma, and resistance to it posed a major challenge (66–68). Therefore, it is crucial to investigate promising and reliable biomarkers for glioma therapy that can predict potential the chemosensitivity of TMZ-based chemotherapy. We discovered that patients with a low-PD-L2 expression group exhibited a more sensitive response to TMZ-based chemotherapy based on the estimated IC50, suggesting that patients with low PD-L2 were more likely to benefit from TMZ-based chemotherapy in LGGs. Combining the therapeutic sensitivity to immunotherapy and TMZ-based chemotherapy, patients with high PD-L2 may benefit from immunotherapy, whereas patients with low PD-L2 may benefit from TMZ-based chemotherapy, which implies the possible values of PD-L2 for selecting an optimal therapeutic strategy. These findings, however, will need to be further verified in the future.
Nevertheless, there are still some limitations to the present study. Firstly, we should consider the effect of tissue heterogeneity on prognostic assessments. Secondly, although we found that PD-L2 was associated with macrophage infiltration in the IHC analysis, the potential biological mechanism and pathway of PD-L2 promoting macrophage infiltration in the glioma immune microenvironment needed to be further validated using in-vivo or in-vitro experiments. Thus, we will expand the sample size and collect more complete clinical samples for further verification in our future study. Thirdly, the present findings may be the basis for further studies to validate the outcomes, such as we can further explore the development of single-cell sequencing in glioma to provide a stronger predictive value to a large extent.
Conclusion
In conclusion, our study demonstrates that PD-L2 is overexpressed in glioma tissues and is associated with a poor prognosis. Moreover, PD-L2 overexpression correlates with immune cell infiltration, especially macrophages. The relationship between PD-L2 and the tumor immune microenvironment, as well as therapeutic sensitivity to immunotherapy and TMZ-based chemotherapy in LGGs, has never been investigated before. More importantly, PD-L2 may be a promising predictive biomarker that contributes to the selection of optimal individualized therapeutic strategy by predicting therapeutic sensitivity to immunotherapy and TMZ-based chemotherapy for future studies of patients with LGGs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by The affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
QX contributed to the study conceptualization, methodology, software, investigation, writing - original draft. XH contributed to the study writing - review & editing, data curation. WH contributed to the study supervision, material preparation, data collection, formal analysis, visualization. FL writing - review & editing, supervision, conceptualization writing - review & editing, language revision, funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
Sponsored by Key Research and Development Program of Jiangsu Province (No.BE2019652), Changzhou international cooperation program (CZ20200039) and Development Program of Changzhou City (CE20205024).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.860640/full#supplementary-material
Abbreviations
CGGA, Chinese Glioma Genome Atlas; GBM, Glioblastoma; GO, Gene Ontology; GSVA, Gene Set Variation Analysis; ICIs, Immune checkpoint inhibitors; IC50, half-maximal inhibitory concentration; IHC, Immunohistochemical; IPS, Immunophenoscore; KEGG, Kyoto Encyclopedia of Genes and Genomes; LGGs, lower-grade gliomas; OS, Overall survival; ssGSEA, Single Sample Gene Set Enrichment Analysis; TCGA, The Cancer Genome Atlas; TCIA, The Cancer Immunome Atlas; TIME, Tumor immune microenvironment; TMZ, Temozolomide; TIICs, Tumor-infiltrating immune cells; TAMs, tumor-associated macrophages.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
2. Lapointe S, Perry A, Butowski NA. Primary Brain Tumours in Adults. Lancet (2018) 392(10145):432–46. doi: 10.1016/s0140-6736(18)30990-5
3. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol (2016) 131(6):803–20. doi: 10.1007/s00401-016-1545-1
4. Gusyatiner O, Hegi ME. Glioma Epigenetics: From Subclassification to Novel Treatment Options. Semin Cancer Biol (2018) 51:50–8. doi: 10.1016/j.semcancer.2017.11.010
5. Gittleman H, Sloan AE, Barnholtz-Sloan JS. An Independently Validated Survival Nomogram for Lower-Grade Glioma. Neuro Oncol (2020) 22(5):665–74. doi: 10.1093/neuonc/noz191
6. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy Plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med (2005) 352(10):987–96. doi: 10.1056/NEJMoa043330
7. Omuro A, DeAngelis LM. Glioblastoma and Other Malignant Gliomas: A Clinical Review. Jama (2013) 310(17):1842–50. doi: 10.1001/jama.2013.280319
8. Lim M, Xia Y, Bettegowda C, Weller M. Current State of Immunotherapy for Glioblastoma. Nat Rev Clin Oncol (2018) 15(7):422–42. doi: 10.1038/s41571-018-0003-5
9. Zhang M, Wang X, Chen X, Zhang Q, Hong J. Novel Immune-Related Gene Signature for Risk Stratification and Prognosis of Survival in Lower-Grade Glioma. Front Genet (2020) 11:363. doi: 10.3389/fgene.2020.00363
10. Yu H, Zhang D, Lian M. Identification of an Epigenetic Prognostic Signature for Patients With Lower-Grade Gliomas. CNS Neurosci Ther (2021) 27(4):470–83. doi: 10.1111/cns.13587
11. Solinas C, Aiello M, Rozali E, Lambertini M, Willard-Gallo K, Migliori E. Programmed Cell Death-Ligand 2: A Neglected But Important Target in the Immune Response to Cancer? Transl Oncol (2020) 13(10):100811. doi: 10.1016/j.tranon.2020.100811
12. Takamori S, Takada K, Azuma K, Jogo T, Shimokawa M, Toyokawa G, et al. Prognostic Impact of Programmed Death-Ligand 2 Expression in Primary Lung Adenocarcinoma Patients. Ann Surg Oncol (2019) 26(6):1916–24. doi: 10.1245/s10434-019-07231-z
13. Okadome K, Baba Y, Nomoto D, Yagi T, Kalikawe R, Harada K, et al. Prognostic and Clinical Impact of PD-L2 and PD-L1 Expression in a Cohort of 437 Oesophageal Cancers. Br J Cancer (2020) 122(10):1535–43. doi: 10.1038/s41416-020-0811-0
14. Erlmeier F, Weichert W, Autenrieth M, Wiedemann M, Schrader AJ, Hartmann A, et al. PD-L2: A Prognostic Marker in Chromophobe Renal Cell Carcinoma? Med Oncol (2017) 34(5):71. doi: 10.1007/s12032-017-0926-1
15. Nakayama Y, Mimura K, Kua LF, Okayama H, Min AKT, Saito K, et al. Immune Suppression Caused by PD-L2 Expression on Tumor Cells in Gastric Cancer. Gastric Cancer (2020) 23(6):961–73. doi: 10.1007/s10120-020-01079-z
16. Wang H, Yao H, Li C, Liang L, Zhang Y, Shi H, et al. PD-L2 Expression in Colorectal Cancer: Independent Prognostic Effect and Targetability by Deglycosylation. Oncoimmunology (2017) 6(7):e1327494. doi: 10.1080/2162402x.2017.1327494
17. Bornhorst M. New Insights Into Low Grade Glioma Tumor Microenvironment for Improved Patient Management. Neuro Oncol (2021) 24(1):27–8. doi: 10.1093/neuonc/noab224
18. Deep G, Agarwal R. Targeting Tumor Microenvironment With Silibinin: Promise and Potential for a Translational Cancer Chemopreventive Strategy. Curr Cancer Drug Targets (2013) 13(5):486–99. doi: 10.2174/15680096113139990041
19. Relation T, Dominici M, Horwitz EM. Concise Review: An (Im)Penetrable Shield: How the Tumor Microenvironment Protects Cancer Stem Cells. Stem Cells (2017) 35(5):1123–30. doi: 10.1002/stem.2596
20. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
21. Sica A, Schioppa T, Mantovani A, Allavena P. Tumour-Associated Macrophages are a Distinct M2 Polarised Population Promoting Tumour Progression: Potential Targets of Anti-Cancer Therapy. Eur J Cancer (2006) 42(6):717–27. doi: 10.1016/j.ejca.2006.01.003
22. Waniczek D, Lorenc Z, Śnietura M, Wesecki M, Kopec A, Muc-Wierzgoń M. Tumor-Associated Macrophages and Regulatory T Cells Infiltration and the Clinical Outcome in Colorectal Cancer. Arch Immunol Ther Exp (Warsz) (2017) 65(5):445–54. doi: 10.1007/s00005-017-0463-9
23. Finotello F, Trajanoski Z. Quantifying Tumor-Infiltrating Immune Cells From Transcriptomics Data. Cancer Immunol Immunother (2018) 67(7):1031–40. doi: 10.1007/s00262-018-2150-z
24. Lin W, Wu S, Chen X, Ye Y, Weng Y, Pan Y, et al. Characterization of Hypoxia Signature to Evaluate the Tumor Immune Microenvironment and Predict Prognosis in Glioma Groups. Front Oncol (2020) 10:796. doi: 10.3389/fonc.2020.00796
25. Zheng J, Zhou Z, Qiu Y, Wang M, Yu H, Wu Z, et al. A Pyroptosis-Related Gene Prognostic Index Correlated With Survival and Immune Microenvironment in Glioma. J Inflamm Res (2022) 15:17–32. doi: 10.2147/jir.S341774
26. Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring Tumour Purity and Stromal and Immune Cell Admixture From Expression Data. Nat Commun (2013) 4:2612. doi: 10.1038/ncomms3612
27. Li H, Wang G, Wang W, Pan J, Zhou H, Han X, et al. A Focal Adhesion-Related Gene Signature Predicts Prognosis in Glioma and Correlates With Radiation Response and Immune Microenvironment. Front Oncol (2021) 11:698278. doi: 10.3389/fonc.2021.698278
28. Zhao B, Wang Y, Wang Y, Dai C, Wang Y, Ma W, et al. Investigation of Genetic Determinants of Glioma Immune Phenotype by Integrative Immunogenomic Scale Analysis. Front Immunol (2021) 12:557994. doi: 10.3389/fimmu.2021.557994
29. Hänzelmann S, Castelo R, Guinney J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinform (2013) 14:7. doi: 10.1186/1471-2105-14-7
30. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-Cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep (2017) 18(1):248–62. doi: 10.1016/j.celrep.2016.12.019
31. Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The Prognostic Landscape of Genes and Infiltrating Immune Cells Across Human Cancers. Nat Med (2015) 21(8):938–45. doi: 10.1038/nm.3909
32. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77(21):e108–10. doi: 10.1158/0008-5472.Can-17-0307
33. Geeleher P, Cox NJ, Huang RS. Clinical Drug Response can be Predicted Using Baseline Gene Expression Levels and In Vitro Drug Sensitivity in Cell Lines. Genome Biol (2014) 15(3):R47. doi: 10.1186/gb-2014-15-3-r47
34. Geeleher P, Cox N, Huang RS. Prrophetic: An R Package for Prediction of Clinical Chemotherapeutic Response From Tumor Gene Expression Levels. PLoS One (2014) 9(9):e107468. doi: 10.1371/journal.pone.0107468
35. Roongrotwattanasiri K, Pawankar R, Kimura S, Mori S, Nonaka M, Yagi T. Decreased Expression of FOXP3 in Nasal Polyposis. Allergy Asthma Immunol Res (2012) 4(1):24–30. doi: 10.4168/aair.2012.4.1.24
36. Zhang GC, Yu XN, Sun JL, Xiong J, Yang YJ, Jiang XM, et al. UBE2M Promotes Cell Proliferation via the β-Catenin/Cyclin D1 Signaling in Hepatocellular Carcinoma. Aging (Albany NY) (2020) 12(3):2373–92. doi: 10.18632/aging.102749
37. Chaichana KL, McGirt MJ, Laterra J, Olivi A, Quiñones-Hinojosa A. Recurrence and Malignant Degeneration After Resection of Adult Hemispheric Low-Grade Gliomas. J Neurosurg (2010) 112(1):10–7. doi: 10.3171/2008.10.Jns08608
38. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med (2015) 372(26):2481–98. doi: 10.1056/NEJMoa1402121
39. Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, et al. Mutational Landscape and Clonal Architecture in Grade II and III Gliomas. Nat Genet (2015) 47(5):458–68. doi: 10.1038/ng.3273
40. Khasraw M, Lassman AB. Advances in the Treatment of Malignant Gliomas. Curr Oncol Rep (2010) 12(1):26–33. doi: 10.1007/s11912-009-0077-4
41. Li T, Yang W, Li M, Zhang S, Zhou X, Zuo M, et al. Engrailed 2 (EN2) Acts as a Glioma Suppressor by Inhibiting Tumor Proliferation/Invasion and Enhancing Sensitivity to Temozolomide. Cancer Cell Int (2020) 20:65. doi: 10.1186/s12935-020-1145-y
42. Wang ZL, Li GZ, Wang QW, Bao ZS, Wang Z, Zhang CB, et al. PD-L2 Expression is Correlated With the Molecular and Clinical Features of Glioma, and Acts as an Unfavorable Prognostic Factor. Oncoimmunology (2019) 8(2):e1541535. doi: 10.1080/2162402x.2018.1541535
43. Fu Y, Liu CJ, Kobayashi DK, Johanns TM, Bowman-Kirigin JA, Schaettler MO, et al. GATA2 Regulates Constitutive PD-L1 and PD-L2 Expression in Brain Tumors. Sci Rep (2020) 10(1):9027. doi: 10.1038/s41598-020-65915-z
44. Barthel L, Hadamitzky M, Dammann P, Schedlowski M, Sure U, Thakur BK, et al. Glioma: Molecular Signature and Crossroads With Tumor Microenvironment. Cancer Metastasis Rev (2021) 41(1):53–75. doi: 10.1007/s10555-021-09997-9
45. Li G, Qin Z, Chen Z, Xie L, Wang R, Zhao H. Tumor Microenvironment in Treatment of Glioma. Open Med (Wars) (2017) 12:247–51. doi: 10.1515/med-2017-0035
46. Kim AR, Choi KS, Kim MS, Kim KM, Kang H, Kim S, et al. Absolute Quantification of Tumor-Infiltrating Immune Cells in High-Grade Glioma Identifies Prognostic and Radiomics Values. Cancer Immunol Immunother (2021) 70(7):1995–2008. doi: 10.1007/s00262-020-02836-w
47. Gieryng A, Pszczolkowska D, Walentynowicz KA, Rajan WD, Kaminska B. Immune Microenvironment of Gliomas. Lab Invest (2017) 97(5):498–518. doi: 10.1038/labinvest.2017.19
48. Gajewski TF, Schreiber H, Fu YX. Innate and Adaptive Immune Cells in the Tumor Microenvironment. Nat Immunol (2013) 14(10):1014–22. doi: 10.1038/ni.2703
49. Roxburgh CS, McMillan DC. The Role of the in Situ Local Inflammatory Response in Predicting Recurrence and Survival in Patients With Primary Operable Colorectal Cancer. Cancer Treat Rev (2012) 38(5):451–66. doi: 10.1016/j.ctrv.2011.09.001
50. Bykov Y, Kim SH, Zamarin D. Preparation of Single Cells From Tumors for Single-Cell RNA Sequencing. Methods Enzymol (2020) 632:295–308. doi: 10.1016/bs.mie.2019.05.057
51. Tong N, He Z, Ma Y, Wang Z, Huang Z, Cao H, et al. Tumor Associated Macrophages, as the Dominant Immune Cells, Are an Indispensable Target for Immunologically Cold Tumor-Glioma Therapy? Front Cell Dev Biol (2021) 9:706286. doi: 10.3389/fcell.2021.706286
52. Zhu C, Kros JM, Cheng C, Mustafa D. The Contribution of Tumor-Associated Macrophages in Glioma Neo-Angiogenesis and Implications for Anti-Angiogenic Strategies. Neuro Oncol (2017) 19(11):1435–46. doi: 10.1093/neuonc/nox081
53. Xiang X, Wang J, Lu D, Xu X. Targeting Tumor-Associated Macrophages to Synergize Tumor Immunotherapy. Signal Transduct Target Ther (2021) 6(1):75. doi: 10.1038/s41392-021-00484-9
54. Ngambenjawong C, Gustafson HH, Pun SH. Progress in Tumor-Associated Macrophage (TAM)-Targeted Therapeutics. Adv Drug Deliv Rev (2017) 114:206–21. doi: 10.1016/j.addr.2017.04.010
55. Yang L, Zhang Y. Tumor-Associated Macrophages: From Basic Research to Clinical Application. J Hematol Oncol (2017) 10(1):58. doi: 10.1186/s13045-017-0430-2
56. Komohara Y, Fujiwara Y, Ohnishi K, Takeya M. Tumor-Associated Macrophages: Potential Therapeutic Targets for Anti-Cancer Therapy. Adv Drug Deliv Rev (2016) 99(Pt B):180–5. doi: 10.1016/j.addr.2015.11.009
57. Brown JM, Recht L, Strober S. The Promise of Targeting Macrophages in Cancer Therapy. Clin Cancer Res (2017) 23(13):3241–50. doi: 10.1158/1078-0432.Ccr-16-3122
58. Chanmee T, Ontong P, Konno K, Itano N. Tumor-Associated Macrophages as Major Players in the Tumor Microenvironment. Cancers (Basel) (2014) 6(3):1670–90. doi: 10.3390/cancers6031670
59. Chen Z, Wu H, Yang H, Fan Y, Zhao S, Zhang M. Identification and Validation of RNA-Binding Protein-Related Gene Signature Revealed Potential Associations With Immunosuppression and Drug Sensitivity in Glioma. Cancer Med (2021) 10(20):7418–39. doi: 10.1002/cam4.4248
60. Pardoll DM. The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat Rev Cancer (2012) 12(4):252–64. doi: 10.1038/nrc3239
61. He X, Xu C. Immune Checkpoint Signaling and Cancer Immunotherapy. Cell Res (2020) 30(8):660–9. doi: 10.1038/s41422-020-0343-4
62. Ribas A, Wolchok J. Cancer Immunotherapy Using Checkpoint Blockade. Science (2018) 359(6382):1350–5. doi: 10.1126/science.aar4060
63. Qin S, Xu L, Yi M, Yu S, Wu K, Luo S. Novel Immune Checkpoint Targets: Moving Beyond PD-1 and CTLA-4. Mol Cancer (2019) 18(1):155. doi: 10.1186/s12943-019-1091-2
64. Marin-Acevedo J, Kimbrough E, Lou Y. Next Generation of Immune Checkpoint Inhibitors and Beyond. J Hematol Oncol (2021) 14(1):45. doi: 10.1186/s13045-021-01056-8
65. Pérez-Ruiz E, Melero I, Kopecka J, Sarmento-Ribeiro A, García-Aranda M, De Las Rivas J. Cancer Immunotherapy Resistance Based on Immune Checkpoints Inhibitors: Targets, Biomarkers, and Remedies. Drug Resist Updat (2020) 53:100718. doi: 10.1016/j.drup.2020.100718
66. Choi S, Yu Y, Grimmer MR, Wahl M, Chang SM, Costello JF. Temozolomide-Associated Hypermutation in Gliomas. Neuro Oncol (2018) 20(10):1300–9. doi: 10.1093/neuonc/noy016
67. Chang L, Su J, Jia X, Ren H. Treating Malignant Glioma in Chinese Patients: Update on Temozolomide. Onco Targets Ther (2014) 7:235–44. doi: 10.2147/ott.S41336
Keywords: lower-grade gliomas (LGGs), PD-L2, tumor immune microenvironment, immune infiltration, prognosis, therapeutic sensitivity
Citation: Xie Q, Huang X, Huang W and Liu F (2022) PD-L2 Serves as a Potential Prognostic Biomarker That Correlates With Immune Infiltration and May Predict Therapeutic Sensitivity in Lower-Grade Gliomas. Front. Oncol. 12:860640. doi: 10.3389/fonc.2022.860640
Received: 23 January 2022; Accepted: 13 May 2022;
Published: 08 June 2022.
Edited by:
Wen Cheng, The First Affiliated Hospital of China Medical University, ChinaReviewed by:
Alexander David Barrow, The University of Melbourne, AustraliaEdjah K Nduom, National Institutes of Health (NIH), United States
Zhe Wang, City of Hope National Medical Center, United States
Yumeng Huang, Shanghai Jiao Tong University, China
Copyright © 2022 Xie, Huang, Huang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Liu, Y3pkb2N0b3JsaXVAaG90bWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
 Qijun Xie
Qijun Xie Xianlong Huang†
Xianlong Huang† Fang Liu
Fang Liu