- 1Department II, University of Medicine and Pharmacy “Carol Davila” Bucharest, Bucharest, Romania
- 2Department of Surgery, “Sf. Ioan” Clinical Emergency Hospital, Bucharest, Romania
- 3Department of Biochemistry and Molecular Biology, University of Bucharest, Bucharest, Romania
- 4OncoTeam Diagnostic S.A., Bucharest, Romania
- 5Department of Toxicology, University of Medicine and Pharmacy “Carol Davila” Bucharest, Bucharest, Romania
- 6Department of Oncology, Elias University Emergency Hospital, Bucharest, Romania
Colorectal cancer (CRC) is the second most frequently diagnosed type of cancer and a major worldwide public health concern. Despite the global efforts in the development of modern therapeutic strategies, CRC prognosis is strongly correlated with the stage of the disease at diagnosis. Early detection of CRC has a huge impact in decreasing mortality while pre-lesion detection significantly reduces the incidence of the pathology. Even though the management of CRC patients is based on robust diagnostic methods such as serum tumor markers analysis, colonoscopy, histopathological analysis of tumor tissue, and imaging methods (computer tomography or magnetic resonance), these strategies still have many limitations and do not fully satisfy clinical needs due to their lack of sensitivity and/or specificity. Therefore, improvements of the current practice would substantially impact the management of CRC patients. In this view, liquid biopsy is a promising approach that could help clinicians screen for disease, stratify patients to the best treatment, and monitor treatment response and resistance mechanisms in the tumor in a regular and minimally invasive manner. Liquid biopsies allow the detection and analysis of different tumor-derived circulating markers such as cell-free nucleic acids (cfNA), circulating tumor cells (CTCs), and extracellular vesicles (EVs) in the bloodstream. The major advantage of this approach is its ability to trace and monitor the molecular profile of the patient’s tumor and to predict personalized treatment in real-time. On the other hand, the prospective use of artificial intelligence (AI) in medicine holds great promise in oncology, for the diagnosis, treatment, and prognosis prediction of disease. AI has two main branches in the medical field: (i) a virtual branch that includes medical imaging, clinical assisted diagnosis, and treatment, as well as drug research, and (ii) a physical branch that includes surgical robots. This review summarizes findings relevant to liquid biopsy and AI in CRC for better management and stratification of CRC patients.
1 Introduction
Colorectal cancer (CRC) is the second most frequently diagnosed type of cancer and a major worldwide public health concern (1, 2). Despite the efforts in the development of modern therapeutic strategies, the prognosis of CRC mostly depends on the disease stage, and specific biological features. Despite that the management of CRC patients has been based for decades on robust diagnostic methods such as colonoscopy, histopathological analysis of tumor tissue, molecular biology assays for molecular profiling, imaging methods (computer tomography or magnetic resonance), and serum tumor markers analysis (Carcinoembryonic antigen (CEA), Carbohydrate antigen 19-9 (CA 19-9), etc.), these strategies still have many limitations and do not fully satisfy clinical needs due to their lack of sensitivity and/or specificity to early disease detection even before clinical onset. Clinical investigations that leverage knowledge on tumor classifications enabled the development of new treatments by providing output data from effective screening and surveillance strategies. Therefore, improvements of the current practice would substantially impact the management of CRC patients and this review provides an in-depth particular discussion on two major approaches that hold great promise in this respect. On one hand, liquid biopsy, as a minimally-invasive procedure that involves the real-time analysis of tumor-derived material isolated from different body fluids (3) is a promising tool that could improve the current practice by helping clinicians screen for disease, stratify patients to the best treatment and monitor treatment response and resistance mechanisms. On the other hand, the prospective use of artificial intelligence (AI) in medicine holds great promise in oncology, not only for early and precise diagnosis but also treatment, therapy, and prognosis prediction of disease.
2 Standardization in Colorectal Cancer Molecular Profiling
Significant advances have been made in the past 20 years with respect to diagnostic by refining tumor classification, enabling this way pathologists and molecular biologists to better define both pathologic and molecular colorectal cancer features (4). In the current clinical landscape, the most common tools used for CRC classification are the histological subtype, tumor localization, and the molecular pathway underlying carcinogenesis (5). Regarding the histopathology of CRC, adenocarcinoma is the most common type of CRC, accounting for more than 90% of the CRC cases diagnosed worldwide, including mucinous adenocarcinoma (10%) and signet ring cell CRC (2%) (6, 7). Other rare types of CRC are medullary, squamous cell, adenosquamous, micropapillary, serrated, cribriform comedo−type, spindle cell, neuroendocrine, and undifferentiated carcinomas (8–10), that are more aggressive and present a significantly worse prognosis than classic adenocarcinomas (11). More, due to the embryonic origin, local vascular and nervous anatomy as well as the distribution of the gut microbiota, the tumor localization along the colon can be divided into: right sided tumors arising from cecum, ascending colon, hepatic flexure and/or transverse colon up to splenic flexure and left sided tumors arising from descending, sigmoid and/or rectosigmoid parts of the colon, potentially influences progression, prognosis and therapy. As a result of the different physiological functions of the colon segments, the patient’s clinical symptoms also significantly differ: right sided colon cancer frequently determine occult blood loss and consequently, iron deficiency anemia, while left sided colon cancer indices changes in bowel habits (12, 13). Finally, a complex picture of the CRC can be obtained by identification of the molecular pathways underlying CRC carcinogenesis, at least three major molecular pathways being related to CRC development and progression: chromosomal instability pathway (CIN), microsatellite instability (MSI), and the CpG island methylation pathway (CIMP), pathways that are not mutually exclusive (14–16). Most frequently identified (>85% cases) is CIN, which presents as tumor genetic signature aberrations in oncogenes and/or tumor suppressor genes involved in key signaling pathways for cancer initiation and progression such as APC, KRAS, NRAS, PIK3CA, BRAF, and TP53 (17–20).
Considering the heterogeneity of tumors sharing the same histology, starting with 1963, the World Health Organization (WHO) implemented a worldwide standardization by classifying the tumor diagnosis and providing internationally acknowledged standards. Currently, in its fifth edition (21), this classification is organized considering the tumor site, category, family, and type and, in addition to histopathology, it includes clinical appearances, epidemiology, etiology and pathogenesis, imaging studies, genetics, epigenetics, and other molecular investigations knowledge, highlighting the expanding concern of the multidisciplinary approach. The WHO new classification of tumors includes for the first time a volume exclusively dedicated to the Digestive System Tumors that now contains separate chapters for soft tissue and hematolymphoid tumors, inherited genetic tumor susceptibility syndromes and a broad classification of neuroendocrine tumors and carcinomas, malignancies that share common histology but occur in different anatomic sites (22). This classification was possible due to the accelerated growth in understanding cancer biology that includes the unrevealing of the genomic makeup of all known cancer types. Currently, sequencing of tumor-derived DNA is used as an investigational tool, aiming to match patients with their most appropriate treatments based on their particular target mutations. US Food and Drug Administration (FDA) labeling, National Comprehensive Cancer Network (NCCN) guidelines, conference proceedings, disease-focused expert group recommendations, and the scientific literature are relevant sources of information regarding the clinical significance in a specific gene’s alteration. To ease the interpretation of the genomic alterations detected in patient tumors, several support tools such as My Cancer Genome (23), CIViC (24, 25), the Precision Medicine Knowledge Base (26, 27), The Jackson Laboratory Clinical Knowledgebase (28, 29), Cancer Genome Interpreter (30), Cancer Driver Log (31, 32), Tumor Portal (33, 34), Targeted Cancer Care (35), Personalized Cancer Therapy (36, 37), and OncoKB (38) were developed. For example, OncoKB is the first racking that assigned each mutation to one of four levels defined based on the data availability supporting the use of the mutation as a predictive biomarker. Off-label use of cancer drugs in oncology has been practiced for many years (39–41) especially in the case of rare cancers where a randomized clinical trial may not be done. Notably, there are particular situations where FDA-approved drugs are not warranted as off-label use due to clear evidence of low efficiency. Such an example is vemurafenib, a BRAF inhibitor and a standard treatment option for patients with BRAFV600E mutant melanoma or NSCLC, which is not efficient at least as monotherapy in patients with BRAFV600E mutant CRC (42). More, aiming to support oncologists to assign potential targets when a broad gene sequencing panel is available, the European Society for Medical Oncology (ESMO) proposed a six levels of evidence framework classifying targets for precision cancer medicine (43). In this sense, the ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT) has been developed (44). It measures how targetable colorectal tumors’ gene alterations are, based on data that support the effectiveness of their corresponding drugs. By far, the most important in terms of treatment decision are hot-spot RAS mutations (NRAS and KRAS), which predict resistance to epidermal growth factor receptor (EGFR) therapy. Decisions in metastatic colorectal cancer in the first line are made depending on this and the location of the tumor (right colon versus left colon) (45–47). From a molecular point of view, right sided colon cancer and left sided colon cancer are very different as the first one is frequently associated with defective mismatch repair genes, KRAS and BRAF mutations and miRNA-31, while the latter is commonly associated with CIN, p53, NRAS, miRNA-146a, microRNA-147b, and microRNA-1288 (48). As a consequence, this different molecular makeover of the tumors impacts on the therapeutically approach (49–51). The BRAFV600E mutation is next as importance for treatment decision. The prevalence of genetic alterations in CRC was reported in the literature as follows: KRAS mutation 44%, NRAS mutation 4%, BRAF mutation 8.5%, NTRK1 fusion 0.5%, ERBB2 amplification 2%, PIK3CA hotspot mtation 17%, ATM mutation 5%, MET amplification 1.7%, RET fusion 0.3%, ALK fusion 0.2% (44). The combination of encorafenib and cetuximab has been shown to be effective in terms of overall survival in patients with this mutation according to a phase III study (52). Another very useful information in the therapeutic decision is about microsatellite instability. Patients with altered mismatch repair proteins (MLH1, MSH2, MSH6 and PMS2) had a very good oncological response to pembrolizumab and nivolumab in phase 2 and 3 trials (53, 54). Other gene alterations that may influence the therapeutic decision are NTRK fusions (which may correspond to an NTRK inhibitor, like entrectinib in the metastatic stage) or the amplification of ERBB2 (which can be associated with dual anti-HER-2 blockade) (36, 55, 56).
3 Liquid Biopsy in Colorectal Malignancies
For a long time, solid biopsy, a procedure based on analyzing tissue samples harvested during endoscopic tests or surgical specimens, was the only available option for diagnosis and tumor profiling in CRC patients’ management and it still represent the standard approach, widely available in clinical practice (57, 58). Tissue biopsy represents a static snapshot of the tumor that lacks to capture the CRC intratumor heterogeneity and dynamic evolution of CRC disease, also determined by clonal pressure induced by the administered treatment (59). More, it is an invasive procedure, which cannot be commonly repeated on-demand, making this approach unfeasible to be performed as a routine procedure for CRC patients’ long-term monitoring and treatment readjustment. As solid biopsy still represents a critical tool for CRC diagnosis, staging, and tumor molecular characterization at the sampling time, the disadvantages associated with this classical approach opened the opportunity for exploring novel analysis methods to improve the current clinical practice in CRC. In this context, the appearance of liquid biopsy represented a game-changer for the current clinical landscape and holds great promise for improving the ongoing CRC patients’ approach in terms of diagnosis, prognosis, and treatment personalization. Liquid biopsy is considered a surrogate of the traditional biopsy, being a minimally-invasive procedure that involves the real-time analysis of tumor-derived material isolated from different body fluids, such as peripheral blood, urine, pleural liquid, saliva, or ascites (3). Based on the anatomical localization of the CRC primary tumors and metastases, peripheral blood remains the main sample employed for liquid biopsy analysis, although urine can be also a viable alternative (60, 61). Unlike traditional biopsy, this modern approach is not limited spatially and temporally and provides clinical decision-worthy information associated with both primary and metastatic cancerous lesions. Among tumor-derived components, circulating tumor cells (CTCs), cell-free circulating nucleic acids (cfNAs), and extracellular vesicles (cEVs) are the most extensively studied and well-characterized markers in connection with CRC and are used for different purposes such as early cancer detection, staging, prognosis, or drug resistance and minimal residual disease (MRD) monitoring (Figure 1).
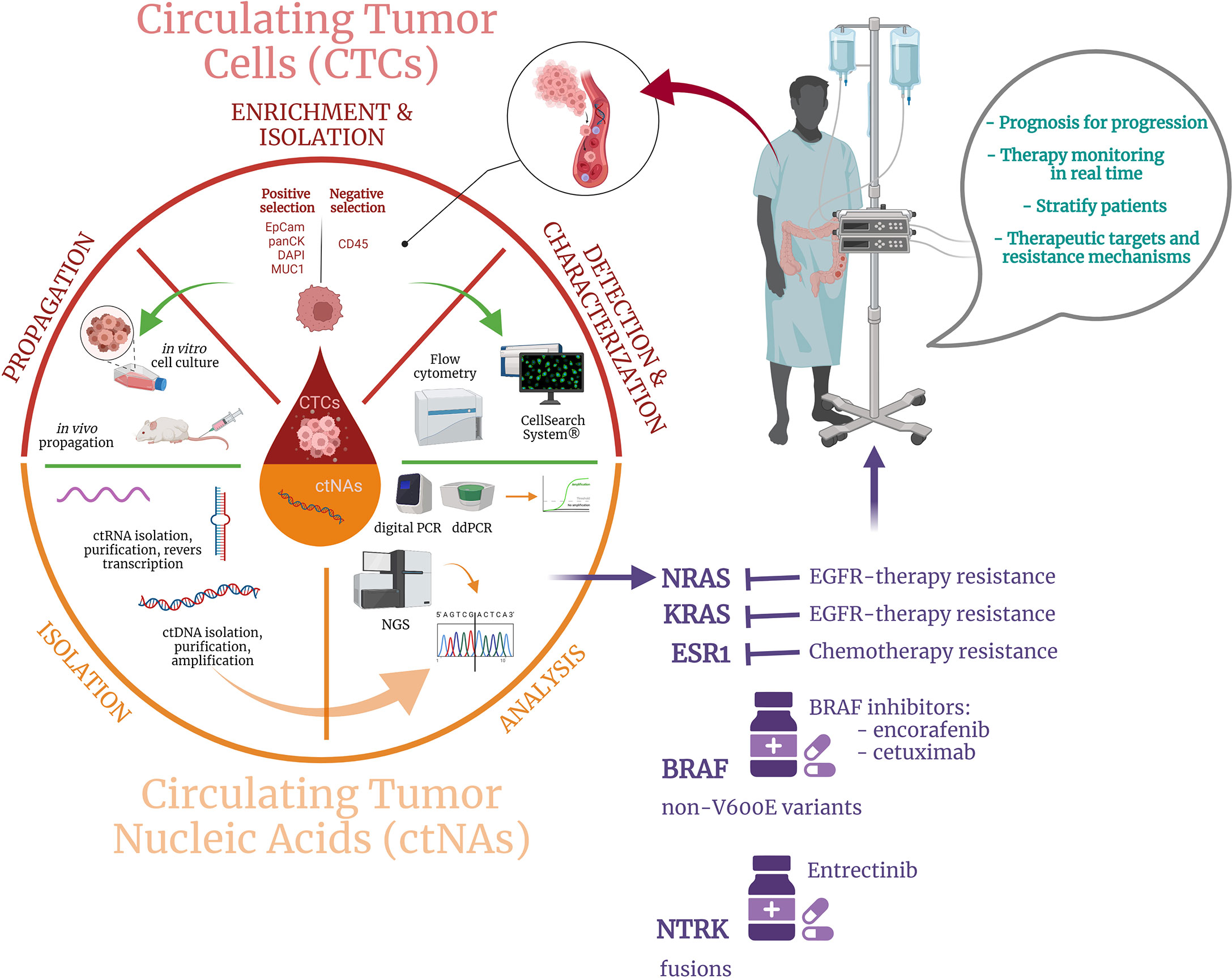
Figure 1 Schematic depiction of the liquid biopsy approach including: (i) CTCs and ctNAs enrichment, isolation, and characterization by specific techniques, (ii) output data analysis, and (iii) potential benefits for CRC patients.
3.1 Circulating Tumor Cells (CTCs)
The first biomarkers associated with the liquid biopsy concept were the CTCs, but their discovery is not state of the art. Since 1869, when Thomas Ashworth first reported the presence of a subpopulation of cells in the bloodstream of metastatic cancer patients, cells that shared similar features to the primary tumor where they detached from (62), numerous successive studies reported the existence of CTCs and their role in the metastatic cascade (63–65). However, since the number of CTCs is extremely low, the promotion of these circulating biomarkers as promising tools in oncological clinical practice was dependent on the development of novel analysis methods with superior sensitivity that emerged from the recent progress made in the biomedical field (66). The frequency of CTCs found in circulation is directly related to the tumor burden, but even in metastatic CRC (mCRC) patients, only 1-10 CTCs/mL of blood can be detected in peripheral blood samples (67). Considered initiators of the metastatic cascade, CTCs are cells of epithelial origin that shed into the blood and lymphatic systems, exit routes that allow them to colonize distant tissues and generate secondary cancer lesions. These cells depart from solid tumors as single intact cells or cell clusters, a feature that impacts their metastatic potential and half-life in circulation (68). CTCs can agglomerate and form homotypic cell clusters or can interact with macrophages, platelets, and/or stromal cells to form heterotypic clusters (68). Of note, in mCRC, CTCs originate both from primary and metastatic lesions, showing the effectiveness of these biomarkers to capture the global landscape of CRC tumor heterogeneity (69, 70). However, independent of the departure point, the capacity of CTCs to cause metastatic lesions is dependent on their stability in circulation, as most of CTCs die as a result of their inability to surpass physical and oxidative stress, anoikis, and absence of the tumor-specific microenvironment (71–73). Based on studies performed on preclinical models, it is estimated that of all detached CTCs, 0.1% remain viable in circulation, from which only 0.01 – 0.2% ultimately will grow into a metastatic lesion (74).
CTCs are characterized by the expression of the epithelial cell adhesion molecule (EpCAM) (75) and the cytokeratins (panCK) (76). However, there are serious concerns on using EpCAM only to capture and define CTCs as these cells undergo epithelial to mesenchymal transition (EMT) and down-regulate the expression of this marker (77, 78). Additional markers MUC-1 (79, 80) could be also relevant for targeting CRC cells. More, circulating cells of tumor origin undergo epithelial to mesenchymal transition (EMT) a process that was first identified in embryogenesis and that refers to epithelial cells reprogramming by acquiring a mesenchymal phenotype. EMT is involved in crucial physiological processes such as development and wound healing, but it is also involved in malignant progression. Upon activation of this reprogramming process, CTCs undergo a series of physical changes that enable their long-distance dissemination, invasion, and survival in the bloodstream (81).
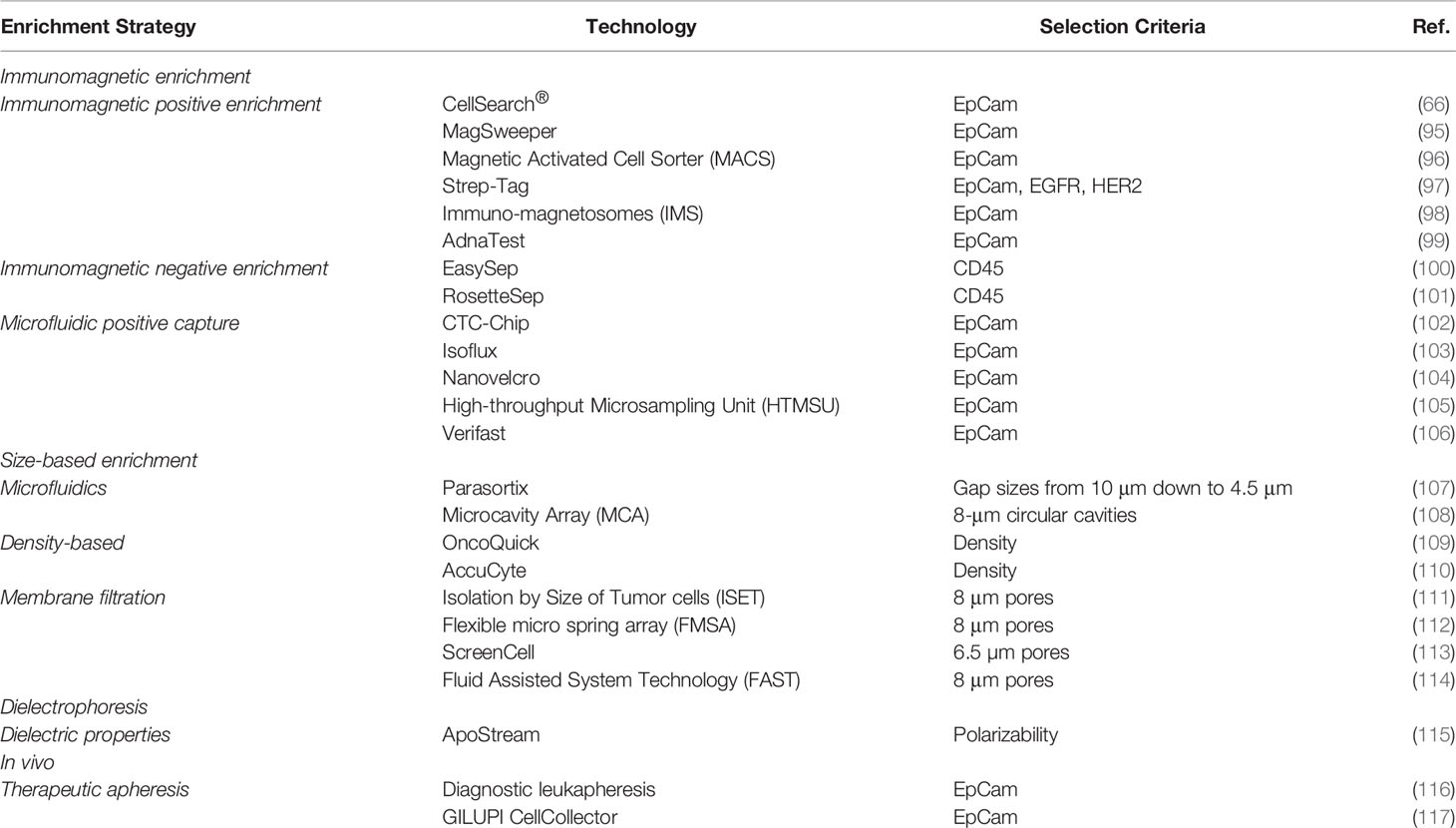
Due to the rarity of CTCs in circulation, the identification, capture, and analysis of these cells are challenging issues. In this view, to improve the success of CTCs detection, the process is preceded by an enrichment technique to unmask these low-frequency cells from the myriad of blood cells: white blood cells (5-10 × 106/mL), red blood cells (5-9 × 109/mL), and platelets (2.5-4 × 108/mL) (67). The enrichment strategies for CTCs detection take advantage of their specific physical and biological characteristics in terms of size, density, deformability, or specific cell surface markers (82). Among all the enrichment strategies explored, the most popular choice remains immunocapturing which can be performed either for CTCs collection (positive enrichment) or removal of blood cell subpopulations (negative enrichment).
The CellSearch® platform is so far the only system that has been approved by the Food and Drug Administration (FDA) for CTCs identification, isolation, and enumeration in whole-blood samples collected from metastatic colorectal (83), breast (84, 85), and prostate cancer (86) patients. The technology uses a positive enrichment strategy based on selection and immunomagnetic capture of Epithelial Cell Adhesion molecule (EpCam) positive cells (87) through ferrofluidic nanoparticles functionalized with the EpCam antigen that select and capture a subpopulation of cells which are subsequently discriminated from residual hematopoietic cells by CD45 negative expression after an immunofluorescence assay. Using the CellSearch® system, EpCam positive cells that are cytokeratin+/DAPI+/CD45- are selected as CTCs and counted for cancer prognosis (88, 89). The platform was successfully employed in various studies that aimed to quantify CTCs in CRC patients and revealed the importance of these as independent prognostic markers (66, 90, 91). However, despite being the only system FDA approved, this platform presents several disadvantages such as low sensitivity and specificity and most important, the failure of detecting CTCs that lose their epithelial features as a result of EMT (92, 93). As a result of undergoing EMT, CTCs acquire a mesenchymal phenotype and thus lack specific epithelial markers such as EpCam and cytokeratins (94), pillar molecules for numerous CTCs enrichment and detection strategies, including the FDA-approved CellSearch® System. From this point of view, Flow Cytometry which is extensively used in hematology could be successfully employed in CTCs detection and count as a versatile approach considering that the instruments work with full customizable protocols. Fluorescence-activated Cell Sorting (FACS) which is also a flow cytometry assay allows also the isolation of the CTCs sorted based on their specific markers expression. Other methods for CTCs isolation, divided by the enrichment strategy, are presented in Table 1.
The low frequency of CTCs in blood severely impacts the clinical utility of these biomarkers for CRC screening and early detection as CTCs detection is almost impossible in early-stage cases (118). In this case, their detection has been a challenging issue that is reflected by the reduced number of studies available regarding the use of CTCs as CRC screening and early diagnosis tools. However, promising results for employing CTCs in screening protocols were shown by Yang et al. (119) that used a negative enrichment strategy for CTCs detection by fluorescence in situ hybridization in blood samples of patients with colorectal polyps and non-metastatic CRC. This study revealed the presence of CTCs in both benign and malign disease, with a significant increase of CTCs in the metastatic setup, where the CTCs count was modulated by the tumor anatomical location and degree of tissue differentiation. More, Tsai et al. employed the CellMax biomimetic platform to detect and enumerate CTCs in subjects with adenoma, polyps, or stage I-IV CRC and revealed an 88% accuracy of this approach for detection of CTCs in all tumor stages, including precancerous lesions (120). The CellMax platform’s unique design enables a superior detection method of CTCs in terms of sensitivity and purity and leads to recovery of viable CTCs that can be employed in downstream applications, making this approach appealing not only for early detection of CRC (121). Another approach to overcome the low frequency of CTCs for early CRC cancer could be repositioning diagnosis leukapheresis (DLA) as a CTCs enrichment tool based on the similar density these cells have with the mononuclear cells from peripheral blood (116, 122, 123), a method that has been successfully employed by Soya and colleagues for CRC patients (124).
Nevertheless, the most common clinical utility of CTCs remains cancer prognosis, a high count of CTCs being associated with a worse prognosis. A prospective study on preoperative patients with stage I-IV CRC showed that CTCs frequency, quantified using the CellSearch® system, was increased in mCRC patients as compared with non-mCRC patients. For non-mCRC, CTCs detection represented a strong and independent prognostic marker and was associated with worse overall survival (OS) and progression-free survival (PFS) compared to CTCs negative patients (90). Using CTC for mCRC patients exclusively revealed that the quantification of CTCs represents an independent predictor of PFS and OS as both indicators were shorter for patients with ≥3 CTCs/7.5mL as compared with those of patients with <3 CTCs (83).
Regarding the clinical implications of CTCs analysis with impact on the post-operative therapy regimen setting in early and mCRC, Guadagni et al. (125) showed that CTCs from 79% of patient exhibited moderate to high sensitivity to mitomycin as compared to only 7% for irinotecan, 30% for oxaliplatin, 2.3% for 5-fuorouracil, 14% for raltitrexed, 4.6% for alkeran, and 4.6% for carboplatin. More, the same group used CTCs to select drug regimens and subsequent target therapy that was subsequent successfully administered by hepatic artery infusion to patients with colorectal cancer liver metastasis (126).
3.2 Circulating Tumor Nucleic Acids (ctNAs)
The limited clinical applicability of CTCs for CRC diagnosis and treatment personalization has led to a shift of liquid biopsy studies in recent years to employ preferentially cell-free circulating nucleic acids (cfNAs) in CRC patient management, cfNAs including cell-free circulating DNA (cfDNA) and cell-free circulating RNA (cfRNA). Indeed, analyzing cfNAs by molecular technologies can lead to real-time genomic profiling of the tumor, an extremely useful approach for the modern trends in CRC management that are directed towards precision medicine and patient-oriented treatment. As the tumor molecular profile dynamically changes in response to the immune system and administered drugs, it is mandatory to implement a novel analysis tool that can be used on demand for ensuring the appropriate treatment regimens for CRC patients (127).
cfDNA is comprised of multiple degraded fragments of DNA that are released in circulation by cell apoptosis, necrosis, or active release mechanisms (128, 129). Apoptosis generates short fragments of cfDNA (< 1000 bp) while following necrosis longer fragments are released (130). The presence of cfDNA in circulation is not exclusively associated with cancer, and this observation was made since its discovery in the blood plasma harvested from healthy subjects (131). However, in cancer patients, the levels of cfDNA are increased due to the supplementary cfDNA released by tumor cells. This small fraction (<1%) of cfDNA, known as cell-free circulating tumor DNA (ctDNA), originates from primary tumors, metastases, or CTCs and harbors specific tumor-related alterations that mirror the genomic status of CRC tumors and epigenetic alterations, thus unraveling the tumor molecular make-up (130, 132). More, cfDNA levels are directly dependent on the cancer stage, mCRC patients showing significantly increased levels of cfDNA in comparison with non-mCRC patients, highlighting the prospective use of cfDNA analysis for early detection of CRC (132–134). More, the ctDNA concentrations are correlated not only with CRC stage, but also with tumor size (135), and could be also employed as a prognostic tool since high levels of cfDNA reveal a shorter OS (136).
In early CRC, ctDNA can be used for investigating DNA methylation, a feature that is closely related to initiating and sustaining the transition of colon polyps to CRC (137). This alteration can be monitored through the FDA-approved Epi proColon® assay, designed for identifying septin-9 (SEPT9) methylation in CRC patients. SEPT9 gene is a tumor suppressor gene that loses its role as a result of hypermethylation of the CpG island in the promoter region, a molecular aberration that promotes the development of CRC (138). The Epi proColon® test aids the early detection of CRC and showed good sensitivity and specificity when compared with the other available non-invasive tests: stool-based guaiac fecal occult blood tests (FOBT) and fecal immunochemical tests (FIT) and blood-based CEA test (139, 140). A study conducted by Tóth and colleagues (141) that included both left- and right-sided CRC cases revealed that this assay is a reliable screening method in both cases, showing superior sensitivity and specificity than FOBT and CEA. Superior performance was observed also comparing SEPT9 assay with FIT in terms of sensitivity (73.3% vs. 68%), while the specificity was significantly better for FIT (97.4% vs. 81.5%). However, the combinatorically approach of using both tests improved these numbers to 88.7% sensitivity and 78.8% specificity (142).
At the moment, for ctDNA analysis, PCR-based and next-generation sequencing (NGS)-based approaches are explored. While PCR remains the backbone for ctDNA analysis, this approach is limited to CRC-specific driver mutations or requires prior knowledge of genetic alterations associated with the investigated CRC tumor. However, PCR-derived techniques are cost-affordable compared with NGS and allow the identification of therapeutically actionable targets or acquired mutations responsible for resistance to therapy. By qPCR-derived techniques, therapeutic vulnerabilities of CRC patients can be easily identified, allowing to discriminate between patients that are suitable for a particular treatment regimen or to readjust treatment according to acquired mutations. Currently, for mCRC patients, fluoropyrimidines remain the backbone of more intensive treatment strategies, including doublet (FOLFOX or FOLFIRI) or triplet combinations (FOLFOXIRI) with leucovorin, oxaliplatin (OXP), irinotecan (IRI), and capecitabine (CAPE) (143–145), added to target agents such as bevacizumab and/or anti-EGFR (146). With this respect, pivotal phase 3 randomized studyAVF2107g proves that the addition of bevacizumab to FOLFIRI significantly improved the overall survival. More, based on promising results obtained in phase 2 studies on the combination of bevacizumab with a FOLFOXIRI, a phase 3 randomized study (TRIBE) showed that this approach displays improved progression-free survival among patients with mCRC as compared with FOLFIRI and bevacizumab (hazard ratio for progression, 0.75; 95% CI, 0.62 to 0.90; P = 0.003) and and reached a median OS of 37.1 months in the RAS wild-type (147). More, the addition of cetuximab (CET) and panitumumab (PAN), which are anti-EGFR drugs, to FOLFOX or FOLFIRI chemotherapy in mCRC patients with KRAS exon 2 wild type and overexpression of EGFR led to improved results with respect to objective response rate, progression-free survival and OS (148). The main concern in the association of anti-EGFR drugs with triplet therapy regimens is the significant toxicity rate in therms of diarrhea, asthenia, mucositis and neutropenia. However, previous analysis of some pharmacogenomics markers such as 5-FUDR for 5-FU degradation rate or and single nucleotide polymorphisms (SNPs) in ABCB1, CYP3A4, DYPD, UGT1A1 which encode for enzymes responsible for 5-FU metabolism, are usefull to predict chemotherapy induced gastrointestinal toxicity (148). Consequently, a shift towards the administration of biological therapies is observed based on the success of these to improve the OS of mCRC patients, among which the best outcomes were observed following treatment with human vascular endothelial growth factor (VEGF) monoclonal antibody bevacizumab and the EGFR monoclonal antibodies cetuximab and panitumumab (149, 150). Independent of the therapeutic choice, the success of the therapy is based on the correct identification of the existing patients’ genetic abnormalities. In contrast to tissue biopsy, identifying genetic alteration by ctDNAs analysis can be employed on-demand establishing, therefore, a patient’s real-time treatment sensitivity pattern that helps avoid the administration of drugs that lack their antitumor effects and exert unnecessary toxicity. More, in absence of tumor tissue for CRC patient molecular characterization, blood can be used for ctDNA mutation screening, therefore providing a correct and efficient targeted therapy scheme (151).
Several genes alterations are screened for aiding CRC treatment, most usual for identifying anti-EGFR-resistant-CRC (152). EGFR is a transmembrane receptor tyrosine kinase with a crucial role in CRC development and progression, being, therefore, an important target for therapy by administration of EGFR monoclonal antibodies or tyrosine kinase inhibitors (153). Following administration of EGFR monoclonal antibodies, these bind to the extracellular domain of EGFR and prevent the activation of the receptor tyrosine kinase and multiple downstream signaling pathways related to cell survival, proliferation, and migration, such as RAS-RAF-MAPK, PI3K-PTEN-AKT, and JAK/STAT pathways (154). Half of the CRC patients harbor RAS mutation, a particularity that renders the anti-EGFR drugs ineffective (155). While KRAS-mutated patients are not exclusively associated with EGFR resistance, mutations in KRAS codons 12 and 13 at exon 2 are clear indicators of EGFR therapy resistance (156, 157), together with mutations in exons 3 and 4 that can indicate a poor response to anti-EGFR (149, 158–160). Another mutation that can overdrive the MAPK pathway, thus promoting cell survival and proliferation, is BRAF identified in 5% - 10% of CRC cases, the commonest being the BRAFV600E mutation (>95%) (161). The presence of BRAF mutation is an indicator of poor prognosis in patients with mCRC and a marker of resistance to anti-EGFR therapy, and it was considered to be a signature of patients not harboring RAS mutations (154, 162, 163). However, recent studies showed the presence of concomitant KRAS and BRAF mutations (164–167). Other EGFR-resistant related mutations are identified with significantly low frequency than KRAS and BRAF mutation, making it difficult to unravel their role in anti-EGFR therapy resistance. The phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)-Phosphatase and Tensin Homolog (PTEN) pathway is an oncogenic signaling network that is deregulated in CRC by aberrant activation of the prooncogenic gene PI3K or through loss of function of the tumor suppressor PTEN (43). PI3K mutations are identified in 10-20% mCRC in exon 9 or exon 20 and lead to constitutive activation of the p110a protein kinase and its downstream pathway, which promotes tumor cell proliferation and survival (168, 169). Both PI3K mutations and loss of PTEN function have been associated with EGFR blockade resistance (170, 171). More, although HER2 amplification was considered relevant for breast and gastric cancer, it appears that it represents also a negative predictor for anti-EGFR therapy (172). With a lower rate of incidence (2% - 4%), MET amplification can determine resistance to anti-EGFR drugs (173).
KIT proto-oncogene receptor tyrosine kinase and platelet-derived growth factor receptor (PDGFR) mutations play a key role in the prognosis of gastrointestinal stromal tumors (4), while ALK receptor tyrosine kinase fusion genes (174) and beta-catenin 1 (CTNNB1) mutations (175, 176) are crucial in inflammatory myofibroblastic tumors and desmoid fibromatosis, respectively. More, the key feature of the CRC occurring neuroendocrine neoplasms (NENs) classification is the discrimination between well-differentiated neuroendocrine tumors (NETs), the carcinoid tumors, and the poorly differentiated neuroendocrine carcinomas (NECs). Despite that NETs and NECs share the expression of neuroendocrine markers, they display significant genetic differences, and therefore, they are not considered to be akin neoplasms (177–180). The main genetic differences between them rely on the Retinoblastoma (RB) and Tumor Protein p53 (TP53) mutations that are commonly displayed by NECs. High-grade colorectal NECs are one of the most lethal types of neuroendocrine neoplasm (181). This poor prognosis pushes the exploration of more effective treatments. Late studies aimed to unreveal colorectal NECs genomic fingerprint. Considering the low incidence of colorectal NECs, only a few studies included highly significant amounts of patients. However, some noteworthy findings show that BRAF gene is frequently altered (182–186). Chen et al. (187) explored the genomic characteristics, and potentially targetable gene alterations in colorectal NEC and compared these characteristics with those of colorectal adenocarcinomas and gastrointestinal NETs using the American Association of Cancer Research (AACR) Project Genomics, Evidence, Neoplasia, Information, Exchange (GENIE) public NGS database (188). They discovered that the genetic characteristics of colorectal NEC are more similar to colorectal adenocarcinomas than gastrointestinal NETs and that the Wnt, MAPK, and PI3K signaling pathways as well as the cell cycle regulation are frequently aberrant in colorectal NECs. More, they confirmed that a significant group of colorectal NEC patients was bearing potentially targetable alterations such as BRAFV600E (187).
There is no doubt that the identification of actionable mutations in mCRC has significantly contributed to the expansion of the therapeutic options available for these patients, but unfortunately very few of the drugs used are at the moment FDA-approved. However, besides the use of monoclonal antibodies to target EGFR, anti-angiogenic and anti-kinase drugs are also used for mCRC management, and studies have shown that these can significantly increase OS and PFS of mCRC patients. Generally, anti-angiogenic and anti-kinase agents are combinatorically administrated with fluoropyrimidine-based chemotherapy plus OXP and/or IRI (189, 190). Among these, the following are clinically approved and used in mCRC treatment regimens: bevacizumab (VEGF inhibitor), aflibercept (blocks activation of VEGF-A, VEGF-B, and PIGF), ramucirumab (anti-VEGFR-2 monoclonal antibody), and regorafenib (191) with approval and reimburse differently applied in the countries. This last one is a multikinase inhibitor that not only acts on angiogenic protein kinases (VEGFR-1, VEGFR-2, VEGFR-3, TIE-2), but as well on proteins involved in oncogenesis (KIT, RET, RAF-1, BRAF, BRAFV600), metastasis (PDGFR, FGFR), and tumor immunity (CSF1R) (192, 193). Therefore, as in CRC cases mutations in genes encoding protein kinases are frequently identified, these drugs can be used to target and block the activity of these overexpressed proteins, improving, therefore, the OS and PFS of mCRC patients (194).
Based on the low content of ctDNA, its small fragment size, and limited high-life, qPCR-based methods were insufficient to detect mutations in ctDNA due to their limited sensitivity. Therefore, new techniques such as droplet digital PCR (ddPCR) or its high-throughput version BEAMing (beads, emulsion, amplification, and magnetics) showed superior efficiency in ctDNA analysis. The main advantage of ddPCR is the use of nanoliter-sized water-in-oil emulsion droplet technology to partition the cfDNA sample into numerous independent PCR sub-reaction (195), the target sequence being therefore concentrated between the aqueous droplets. In this way, after detection by fluorescence of the amplified targeted sequences and comparison of the signals obtained, rare mutations are discriminated from the wild-type sequences. ddPCR principle by which multiple PCR individual microreactors are generated, favors the quantification of low-abundance point mutations in cfDNA from a background of wild-type sequences, with high sensitivity ranging from 0.001% to 0.1% (173, 196).
In contrast, the modern NGS platform opens new opportunities for CRC patients by employing massively parallel sequencing techniques for genome-wide assessment and screen for multiple mutations with greater sensitivity without prior knowledge of the patient`s existing genetic abnormalities and can lead to the discovery of novel therapeutic targets (197). Undoubtedly, the main advantage of NGS analysis compared with the targeted approaches is the possibility of investigating a large range of markers reaching up to several hundred genes in a single panel. Also, a single test provides valuable information on single nucleotide polymorphisms, short indels, copy number variations, gene fusions, TMB (tumor mutational burden), or MSI (198). New NGS panels have been developed to detect the pathological variants associated with cancer at both cfDNA and cfRNA levels (e.g., Oncomine Pan-Cancer Cell-Free Assay - Thermo Fisher, Waltham, MA, USA). NGS approaches are based in fact on massively parallel sequencing technologies that offer high throughput, reproducibility, and speed. High-throughput sequencing, which includes Next-Generation “short-read” and “long-read” Sequencing methods allows scientists to perform a wide variety of applications in terms of genomics such as genome and exome de novo sequencing and resequencing, study of DNA-protein interactions, and epigenome characterization. Nowadays, there are at least 9 different NGS platforms using different template preparation and sequencing chemistry. However, all NGS platforms perform sequencing of millions of small fragments of DNA in parallel producing thousands or millions of sequences concurrently. Bioinformatics analyses are used to piece together these fragments by mapping the individual reads to a reference. NGS platforms allow deeply sequence target regions, analyze epigenetic factors such as genome-wide DNA methylation and DNA-protein interactions, and also sequencing cancer samples to study rare somatic variants or tumor subclones. Many methods are applying NGS to target panels for specific and highly sensitive detection of targeted ctDNA mutations (Tagged-Amplicon deep sequencing - TAm-seq; Safe-Sequencing System - Safe-SeqS or CAncer Personalized Profiling by deep sequencing - CAPP-Seq) (199). However, the use of NGS for detecting rare mutations provides a lower technical sensitivity than ddPCR (200), highlighting that PCR-based methods and NGS could be employed as complementary techniques for clinical applications.
Various commercial platforms are available for identifying actionable mutations in CRC such as Idylla or OncoBEAM, Idylla platform being used also for MSI detection (201–203). Vidal and colleagues (204) revealed a 93% overall agreement between the mutational status of RAS when comparing tissue and plasma samples. From 55 patients presenting RAS mutations in investigated tumor tissue, 53 had RAS mutational status confirmed by ctDNA analysis using OncoBEAM. Poor results were obtained by García-Foncillas et al. that showed just 89% agreement between results obtained by analyzing the mutational status of KRAS from tissue vs. plasma (201). For detecting MRD, Tie et al. (205) successfully used massively parallel sequencing-based assays to predict recurrence in patients with resected stage II CRC by analyzing ctDNA from postoperatively blood samples and can measure in real-time the effectiveness of treatment for patients receiving adjuvant chemotherapy.
Other valuable biomarkers in the liquid biopsy are ctRNAs such as micro-RNA (miRNA), messenger RNA (mRNA), and noncoding RNA (ncRNA) that are usually released in circulation within extracellular vesicles or associated with RNA-binding proteins (206). ncRNAs for example, are involved in all CRC stages of tumorigenesis and progression influencing key signaling pathways such as: WNT/-catenin, phosphoinositide-3-kinase (PI3K)/protein kinase B (Akt), epidermal growth factor receptor (EGFR), NOTCH1, mechanistic target of rapamycin (mTOR) and TP53 (207). Although an incomplete domain, in the last years many gene fusions have been identified in colorectal cancer only due to the possibility of high throughput NGS. The most frequent gene rearrangements detected in CRC with prognostic value and treatment perspectives are: NTRK fusions, ALK and ROS1 rearrangements, RET fusions and BRAF translocations. NTRK fusions were detected especially in women, elder people with RAS and BRAF wild-type genes and were presumably associated with primary resistance to EGFR-targeted treatment. Conventionally gene fusions are investigated by FISH, IHC or RT-PCR. Unfortunately, these methods can test for only one fusion gene at a time, being low-throughput and sometimes costly. The great advantage NGS brings in clinics is the large amount of information obtained. In just one test hundreds of gene rearrangements, known and unknown, frequent or rare can be detected. It can also investigate very small rearrangements that would be impossible to be evaluated using classical methods (208).
4 Biomedical Applications of Artificial Intelligence in Colorectal Cancer Management
4.1 General Background
Nowadays we are all surrounded and widely interacting in our daily lives with AI in the form of consumer technology, whether we think about smartphones, wearable devices, search engines, social media channels, personalized advertising, facial recognition, autonomous vehicles, energy-efficient buildings, smart toys and many more. The concept of AI is not new (209), but only in the past few years, it has raised interest due to its potential as a virtual assistant able to serve various domains of human activity, especially the biomedical field. These AI experiences aim to improve human lives by increasing efficiency and by tailoring solutions for each individual. From this point of view, AI may be defined as a combination of theories, algorithms, and computing frameworks enabling a machine to independently reproduce intellectual processes associated with human cognition to decide on an action in response to its perceived environment to achieve a predetermined goal. Consequently, AI empowers humans to become faster in analyzing big amounts of data and smarter in decision-making by augmenting, however without replacement, human intelligence, and intuition (210). At its core, AI is a branch of computer science that has gradually changed not only our daily lives but also the landscape of healthcare and biomedical research for medical image analysis, intraoperative imaging, and genomics.
Starting with the ‘50 when the concept was first defined, there have been developed rule-based clinical decision support systems (211) that helped physicians to diagnose disease (212, 213) or decide the best treatment (214). Despite all these advantages and clear progress in the current practice, the rule-based systems displayed a series of inconveniences such as high building cost and system limitation by the difficult encoding of complex interactions and requirement of solid medical knowledge (215). Therefore, this first generation of AI systems has been improved by the integration of machine learning (ML) methods, which apply complex interactions to identify data patterns. ML is a subfield of AI that uses both mathematical and statistical approaches to improve the performance of computers. More specifically, ML resides in the development and implementation of dynamic algorithms that are able to take particular actions in response to particular inputs (environment), analyze the data to determine the actions (216, 217). These algorithms are able to self-improve (learn) as more data is available and the optimization process is called “training”. Depending on the tasks to be solved, basic ML algorithms can be divided into supervised, unsupervised (218), and reinforcement learning.
Supervised learning (SL) algorithms such as Naive Bayes classification (217), linear and logistic regression (219), support vector machines (SVMs) (216), or random forests (RF) (220) analyses the training data (input-output pairs) and map an input to output (221). A good example, in this case, is the histopathology grading of digitalized tissue biopsy slides. First, a set of images are labeled by the pathologists as positive or negative for a specific type of cancer and then SVMs for example learn to classify new, unlabeled images and tag them on a “probability map”. Zhi et al. (222) used five data sets containing metastatic and non-metastatic CRC samples to identify potential biomarkers for CRC metastasis. For this, the authors used a meta-analysis method to identify the metastatic and non-metastatic CRC differentially expressed genes and then, used the SVM classifier to select the top candidates having as reference a CRC dataset from The Cancer Genome Atlas database. The authors report a 100% precision of the SVM-classified 40 gene signatures and highlight the CREB1, CUL7, and SSR3 genes as biomarkers for the prognosis of metastatic CRC.
Unsupervised learning (UL) algorithms such as k-means clustering (217), principal component analysis (223), and autoencoders (224) identify patterns from untagged data by separating the items into different classes based on the training data features to find sub-clusters and outliers in the data. Bae et al. (225) proposed a feature selection method able to distinguish CRC patients from normal individuals using K-means clustering and the modified harmony search algorithm. To classify CRC using gene information the authors analyzed 6500 genes in 40 CRC tissue biopsies and 22 normal colonic tissue samples by a 4 step hybrid method consisting of: (i) Z-normalization of gene information values, (ii) Fisher score based reduction of redundant genes, (iii) K-means clustering of representative genes and (iv) Harmony Search (HS) algorithm based selection.
Reinforcement learning (RL), consists of a set of algorithms (agents) that operate sequentially by predicting the features of each step based on the past and present features while assigning a reward or a penalty on the prediction basis. The purpose of RL is for the agent to learn an optimal or nearly-optimal policy that maximizes the “reward function” (226).
Deep learning (DL) is a subfield of ML which involves training an artificial neural network (ANN) with many layers. DL recapitulates the biological neural network of the human brain and uses a layered structure of algorithms to analyze data, identify patterns, draw conclusions, and make decisions. The basic architecture of deep neural networks (DNNs) consists of an input and an output layer together with a variable number of hidden layers in between. In such a network, the input is provided to the input layer, which transfers its computed value towards the hidden layers that are finally linked to an output layer. DL can be further classified into the deep neural network (DNN), recurrent neural network (RNN), and convolutional neural network (CNN). CNNs are particularly important to identify patterns from unprocessed images (227). CNNs apply nonlinear transformations to structured data, for example the raw pixels within an image, to automatically learn relevant features. In this approach, it is mandatory to process accurately the images before analysis in order to reduce the risk of the model to learn from artifacts. CNNs basically uses two main models: the first one uses images from a large collection such as ImageNet to train the initial layers, and the second one is based on an auto encoder where the model learns background features from a subset of representative images (228). DL has been widely applied to empower medical procedures mainly involving analysis of images resulting from a wide range of procedures (229–232) and genomics (233).
In brief, biomedical applications of AI can be divided into two main branches: virtual and physical. The virtual component consists of ML algorithms: supervised learning, unsupervised learning, and reinforcement learning, and DL as an ML subset (Figure 2). CNN is the most prominent DL construction, representing a particular type of multilayer artificial neural network that is highly efficient for image classification (227). While the virtual branch of AI-powered biomedical applications refers to image analysis and genomic (big data) analysis, the physical component includes medical devices, surgery robots, and devices for automated therapy formulation and nanorobots for targeted drug delivery (234).
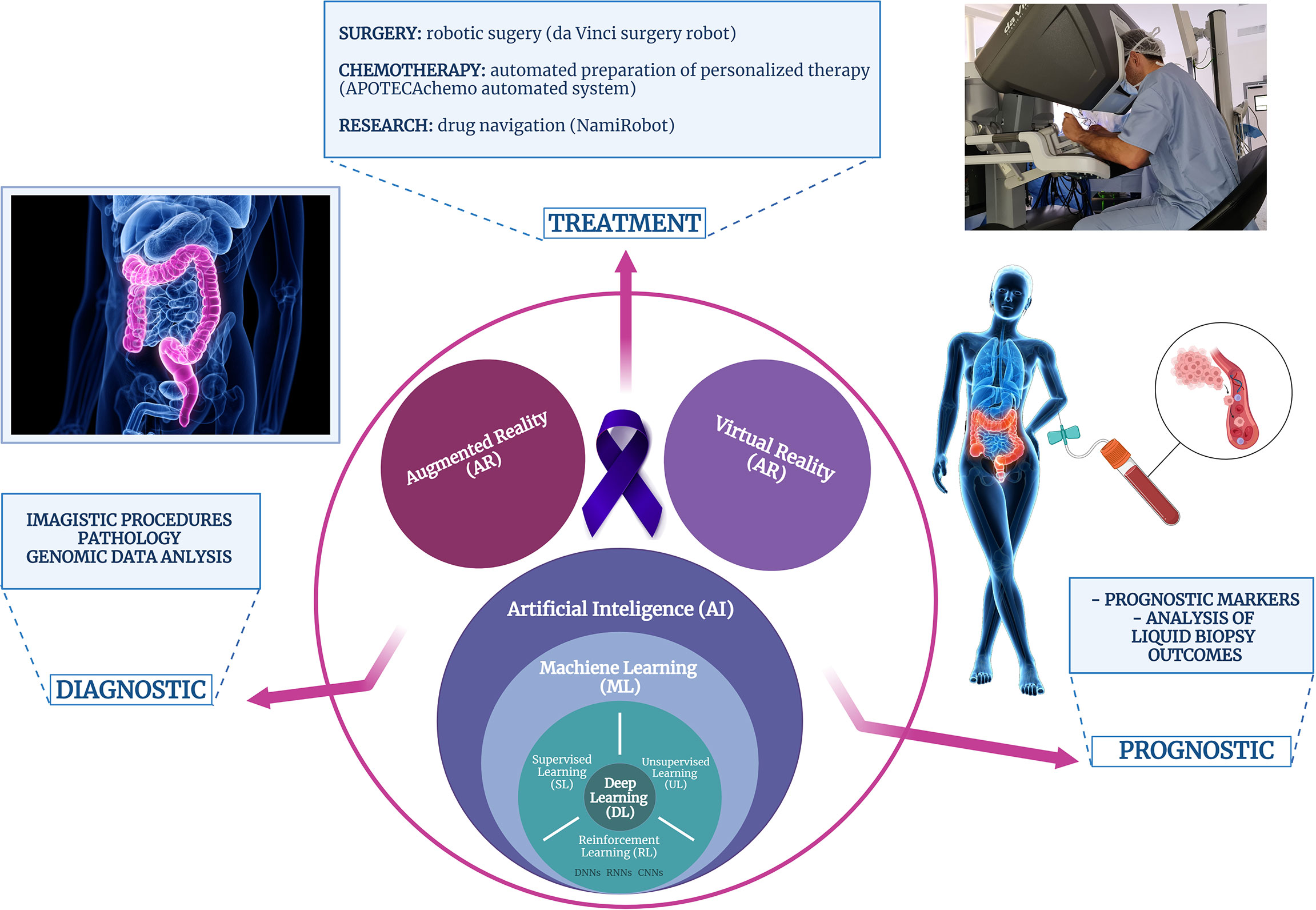
Figure 2 Schematic representation of the computer-assisted technologies applications in the biomedical field, particularly, in patients with colorectal cancer.
Virtual reality (VR) is a simulated experience that generates an immersive, artificial image and/or environment with real-time interaction. Applications of VR include entertainment (video games), education (virtual training in a wide range of fields including medicine), and even business (virtual meetings). In medicine, one of the earliest VR platforms developed was Minimally Invasive Surgical Trainer-Virtual Reality (MIST-VR) (Mentice Medical Simulation, Gothenburg, Sweden) for endoscopic training (235). VR 3D technology is being used for simulation-based training due to its major advantage of creating near true experiences overcoming the disadvantages of the current distance learning and 2D videos (236). Currently, using VR (237), previously segmented organs and structures can be highlighted by streaming a digital full image, often visualized as a 3D render in a virtual environment.
Augmented reality (AR) is an interactive experience consisting of the addition of artificial information to the objects in the real world through computer-generated perceptual information that enables the user to perform tasks easily. This can be achieved across multiple sensory modalities like overlapping images and generating video or computer models. The first headset equipped with an overlapping display was introduced by Ivan Sutherland in 1965 and served for military purposes (238). Some examples with applications in the biomedical field are AccuVein (AccuVein Inc., NY, USA), a projector-like device that displays a map of the vasculature on the skin surface (239), or Google Glass which is a head-mounted display with generated objects superimposed onto real-time images. Unlike the virtual environment provided with VR, AR has been used in real surgery for many years (240) as it can overlay “invisible” information such as pre-or intraoperatively obtained imaging findings in the real environment.
4.2 AI in CRC Screening and Diagnostic
CRC is a highly preventable disease with very good survival rates when diagnosed in the early stages. This is why routine screening is a crucial step in lowering the incidence rates of this pathology (241) while offering patients the hope for disease-free. The premalignant alterations from normal to malignant lesions take almost over 15 years. Efficient screening methods are permanently developed to accurately diagnose abnormal modifications with potential pathologic signification (242, 243). Available CRC screening procedures include colonoscopy, capsule endoscopy, imaging examinations, and/or blood and stool tests (244, 245). ML algorithms may be used as non-invasive and cost-effective methods to screen the CRC risk in large populations using personal health data (246).
4.2.1 AI in CRC Imagistic Screening Procedures
4.2.1.1 Colonoscopy and Virtual Colonoscopy
Colonoscopy is currently the gold-standard method for CRC screening displaying several advantages such as high sensitivity, specificity, and ability to directly visualize the tissue and act on cancerous and precancerous lesions by polyp resection and/or tissue biopsy harvest. Multiple works have shown that colonoscopy significantly reduces CRC incidence and mortality (247–250). However, due to the significant risk of the human eye to missing spotting small or flat polyps, computer-aided detection (CADe) and diagnosis (CADx) systems were developed to assist colonoscopy in real-time automated polyp detection (251). DL techniques such as CNNs algorithms were used to improve colorectal adenoma detection rates and proved to accurately spot the presence of premalignant lesions (252). With this respect, the DL-based CADe model effect on polyp and adenoma detection rates was investigated by Wang et al. (253) in a randomized controlled trial, while the CADx potential assistance in discrimination between neoplastic and non-neoplastic polyps during a colonoscopy was investigated by Mori et al. (254). To improve even more the CRC polyp detection and classification, AI-based tools were employed furthermore in colonoscopy. This approach led to the development of the virtual colonoscopy or computed tomographic colonoscopy described for the first time by Lefere in 2006 (255) and originating from the computed tomographic colonography described in 1994 (256, 257). This AI-powered tool may also contribute to the automatic detection of flat neoplastic lesions, which may represent an aggressive tumorigenesis and a determining factor in increased adenoma miss rate (258). Numerous strategies for small and/or flat polyps detection and their discrimination between neoplastic and non-neoplastic were developed and already reviewed in recent literature (259).
4.2.1.2 Capsule Endoscopy (CE)
Capsule endoscopy (CE) is a minimally invasive diagnostic technique, well tolerated by patients, and a powerful alternative approach in incomplete colonoscopy cases. Traditionally colon CE is a time-consuming procedure requiring human interpretation and analysis of the captured images for the detection of potential CRC lesions. In this view, AI-powered technologies have the potential to increase the detection rates of adenomas by automating the analysis of the results (260, 261) reported two algorithms, one able to match CE with polyps identified by colonoscopy and another one based on deep CNNs for automatic colorectal polyp detection and localization.
4.2.2 AI in CRC Molecular Imaging
Molecular imaging is a modern approach of medical imaging that consists of the in vivo visualization of anatomical structures based on the presence of a radioactive, fluorescent, or magnetic label. This approach can be used for early diagnostics, therapy planning, and resection. Nuclear medicine remains the gold standard for non-invasive molecular imaging, using PET imaging, combined with CT or MRI anatomical imaging (262). Alternatively, an optical or electrical expression of the target molecule in a non-contrasted fashion (263, 264) is applied. More, the concept of interventional molecular imaging refers to the use of tracers and other surgical molecular imaging approaches, to target and help surgeons precisely resect tissues based on molecular features (265) during radio-guided surgery (266, 267) or fluorescence-guided surgery (268).
4.2.2.1 AI in CRC Molecular Imaging for Diagnostic
Molecular imaging powered by AI strategies holds great potential to improve the CRC diagnosis as AI algorithms allow the comparison of the information available for a current patient with huge databases of previously treated patients potentially empowering the selection of the most optimal treatment strategy and the prediction of the treatment outcome (269).
Magnifying chromoendoscopy is an optical diagnosis procedure, that proved to spot colorectal lesions with over 90% of sensitivity, specificity, and accuracy (270). This technique consists in the analysis of the polyp surface with a high-resolution magnifying colonoscope after indigo carmine or crystal violet dye spray. With this respect, Häfner et al. (271) developed a texture feature extraction algorithm, while Takemura et al. (272) used the quantitative analysis of pit patterns to propose a software model for the differential diagnosis of colorectal lesions.
Endocytoscopy refers to an endoscopic imaging procedure, which is an in vivo imaging microscopy technique that allows a real-time diagnosis based on the analysis of the cells patterns observed at high magnification (273). Endocytoscopy relies on the contact light microscopy principle and requires colonic mucosa prestaining with absorptive contrast agents, such as toluidine blue (273, 274). Computer-assisted algorithms were developed for in vivo discrimination of colonic lesions using endocytoscopy and even further improved by the use of SVM as classifiers for benign, adenomatous lesions or invasive carcinoma (275).
Confocal laser endomicroscopy is a microscopic imaging technique powering the in vivo inspection of cellular and subcellular structures (276). K-nearest neighbor classification was used by Andre et al. (277) to build an automated polyp characterization system that proved to distinguish between malignant and benign lesions with an accuracy rate of almost 90%.
Laser-induced fluorescence spectroscopy (LIFS) and autofluorescence imaging (AFI) endoscopy are used for in vivo detection of targeted tissue fluorescence emission. The autofluorescence could emerge from endogenous molecules such as collagen or flavins after excitation with an appropriate light source. Based on color differences these procedures can predict in real-time the lesion pathology and accurately characterize the colorectal polyps. Using computer-aided software for color analysis models these techniques may significantly contribute to the differentiation of non-neoplastic from neoplastic lesions during colonoscopy (278–281).
4.2.2.2 CRC Surgery Procedures Powered by Molecular Imaging and AI
Classically, preoperative imaging techniques are routinely used to create detailed roadmaps that help surgeons to plan the surgery and execute the desired resection. However, the up growing interest in the development of minimally invasive surgical strategies, such as robotic surgery has empowered molecular imaging as a valuable tool for precision surgery. Intuitive AR visualizations and AI integration for an optimized perception into the video console used by surgeons in robotic surgery brought the classical practice to the next level. For example, the TilePro extension of the da Vinci surgical console shows preoperative images as windows in the surgeon’s display (282, 283). Beyond planning and guidance through the preoperative scans, intraoperative molecular imaging approaches (radioactive, fluorescent, magnetic, multispectral optoacoustic tomography (284), fiber-based microscopy (285), Raman spectrometry (286), etc.) play an important role in intraoperative lesion localization, decision making, and subsequent confirmation.
4.2.3 AI in CRC Imagistic Pathology Diagnostic
Despite that other tools have been proposed with great promise for the future, tissue biopsy remains the gold standard for colon cancer diagnosis and staging. Therefore, much effort has been invested to improve this approach. With this respect, some groups have developed support-vector machines (SVMs) to enable the automatic classification and diagnose CRC based on biopsy samples, significantly improving the accuracy of diagnosis while reducing time and costs (287–289). Convolutional neural networks (CNNs) are the most commonly used AI technology in pathology image analysis (290). For example, CNNs were used to detect and classify nuclei in colon cancer biopsies (291). Histopathology images of cancerous tissues stained with standard hematoxylin and eosin (HE) stains were used to develop technologies with significantly enhanced reading accuracy (291). More, the TuPaQ algorithm using immunohistochemistry (IHC) staining was developed to segment CRC tumor epitheliums, providing a basis for automated biomarker quantification (292). Recently, Yu et al. (293) developed a recognition system for CRC which achieved one of the highest diagnosis accuracies in cancer diagnosis with AI using SL. More, AI has made a huge step in the field of intraoperative pathology, providing preliminary evaluations or highlighting suspicious areas (294). For example, one-step nucleic acid amplification (OSNA), a non-imaging approach of the intraoperative biochemical analyses of lymph nodes has been successfully implemented into the clinical routine in several countries showing excellent diagnostic accuracy in CRC (295).
4.2.4 AI in CRC Genomic Data Analysis
AI algorithms have shown several promising results with genetic testing for CRC. For example, Hu et al. (296) classified CRC patients with the Union for International Cancer Control (UICC) II into two groups based on their relapse status and compared the classification accuracy using the following neural networks: S-Kohonen (91%), Back-propagation (BP, 66%), and SVM (70%). Xu et al. (297) used SVM to identify differentially expressed genes and identified a 15-genes with relevance in distinguishing patients with a high CRC recurrence risk. Zhang et al. (298) developed a counter propagation artificial neural network (CP-ANN) able to detect BRAF mutation. AI algorithms such as but not limited to BP were used by Wang et al. (299) to combine gene expression profiling data from The Cancer Genome Atlas (TCGA) database for CRC diagnosis improvement. In the context of the up growing interest of the liquid biopsy approach development, Wan et al. (300) proposed a ML method using tumor-derived cfDNA. Chang et al. (301) compared the expression profiles of 380 miRNAs in stage II CRC versus normal tissue and identified a 3-miRNA signature potentially predicting the tumor status in stage II CRC. However, during this time, many AI-based technologies were developed targeting the using miRNAs (302–305). Ge et al. (306) used CIBERSORT, a deconvolution algorithm, to study the role of 22 immune cells types and 404 immune-related gene expression in the CRC surrounding microenvironment.
Liquid biopsies output data are large and complex and therefore, traditional methods fail their efficient process. In this view, ML represents a promising tool for automated analyses of these data and future prediction (307). Different ML algorithms, such as SVM, random forest (RF) and ANNs, have been widely used in the field of medicine (308–310). SML is usually implemented for liquid biopsy data analysis and involves model evaluation and selection methods. Data preprocessing consists of missing-value solution, normalization, dimension reduction, and feature reconstruction. Regarding the CTCs in particular, the CellMax (CMx®) platform was developed based on AI achieved clinical sensitivity and specificity of 80% (121). In addition, ML algorithms could assist in analyzing data on specific serum protein biomarkers such as LRG1, EGFR, ITIH4, HPX and SOD3 in order to identify CRC with 70% sensitivity at over 89% specificity (311).
4.3 AI in CRC Surgery
Along with chemotherapy administration, surgery remains the main curative procedure for CRC management. With the development of robotic surgery as a minimally invasive procedure, CRC management enters a new era. To date, the da Vinci System developed based on the physical branch of AI technologies, is the most used surgical robot worldwide. The major advantages of its use reside both on the surgeon’s and patient’s sites. Robot-assisted surgery needs smaller incisions that will conduct minimal scarring and a significant decrease in the risk of surgical site infections, faster recovery and decrease of hospitalization period. Postoperative pain and bleeding are significantly lower as compared to traditional open surgery (312, 313). On the other side, using computer-controlled devices surgeons benefit from enhanced visual field, more flexibility, increased precision, and minimal fatigue. Besides the above-mentioned advantages, the da Vinci dual-console also allows integrated teaching and supervising. Several retrospective studies on robotic-assisted CRC resections showed that the procedure significantly reduced the complications rate and resulted in less evident inflammatory response as compared with the open surgery (314, 315). Other robots for surgery such as The Senhance System (TransEnterix Surgical Inc., Morrisville, NC, USA) which is a laparoscopy-based system are developed. Overall, robot-assisted colorectal surgery has better performance in terms of both short- and long-term outcomes (316).
4.4 AI in CRC Therapy
Concerning the actual preparation of the administered therapy for patients, manual handling of cytotoxic drugs is a high-risk activity due to the prolonged exposure to carcinogens (317) and also a major error generating point in the patient’s management (318). Therefore, the introduction of robots such as the APOTECAchemo automated system (319) improved the safety of cytotoxic drug preparation, almost excluding the errors associated with human handling (320). Regarding CRC therapy research, Martel et al. (321) developed the NamiRobot System that can deliver drugs to cancer cells and hypoxic regions based on the reduced oxygen levels caused by the proliferation of cancer cells. More, AI technology can also promote research on new drugs. For example, Cruz et al. (322) used machine learning to detect with a prediction accuracy of over 63% the half-maximal inhibitory concentration (IC50) of a new drug on the HCT116 colon cancer cell line.
5 Conclusions and Perspectives
The emerging field of research in CRC pathology empowered the development of new approaches and technologies for better management of this malignancy in terms of: diagnostic, treatment, and prognostic. Specifically, on one hand, liquid biopsy is a promising tool revealing the residual cancer cells, and patient’s disease progression in real-time. This valuable outcome is enabled by the access to worthy decision-making information like genomic profiling for eligibility to targeted therapy, identification of chemotherapy-induced genomic alterations as compared to the initial tissue biopsy, detection of disease relapse, and/or MDR. Moreover, this innovative approach could be applied in clinical practice to enhance the knowledge of the specific biological feature of CRC disease including temporal and spatial heterogeneity. For example, the analysis of NGS data might reveal RAS mutations, which predict resistance to epidermal growth factor receptor (EGFR) therapy and/or BRAF mutations which indicate that encorafenib and cetuximab therapy might be effective. Moreover, liquid biopsy is a powerful tool also for driving anti-EGFR rechallenge therapy in mCRC as shown within CHRONOS, a phase 2 trial aiming to rechallenge anti-EGFR therapy by monitoring of the mutational status of RAS, BRAF and EGFR in circulating tumor DNA (ctDNA) (323). Lastly, but not least, the same analysis might spot NTRK fusions or the amplification of ERBB2 which can be associated with dual anti-HER-2 blockade. On the other hand, the development of AI-based technologies for biomedical applications opened a new era in the field of automated detection of colorectal polyps during screening procedures such as but not limited to colonoscopy and automated detection of malignant tissues in pathology analysis. More, AI meets AR and VR in robotics, with specific applications in CRC surgery (example: the highly versatile da Vinci robotic surgery system) and also in traceable and personalized proper preparation of anti-blastic drugs in hospitals (example: APOTECAchemo automated system).
Author Contributions
OG reviewed the whole content regarding CRC pathology, screening methods and robotic surgery. He also contributed with content for the images. AH wrote and reviewed the whole section 3. MZ, AS, MM, and IB wrote sections 4.1, 4.2 and 4.3. LB, SG, and MC wrote and reviewed the content regarding ctDNA (Section 3.2.). CNe wrote section 4.4. CNi wrote section 2 and BG wrote sections 1 and 5, reviewed the whole manuscript and coordinated the whole team. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant of the Romanian Ministry of Education and Research, CCCDI - UEFISCDI,project number PN-III-P2-2.1-PTE-2019-0577, within PNCDI III.
Conflict of Interest
Author LB was employed by company OncoTeam Diagnostic S.A.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This work was supported by the project 41PFE/30.12.2021, financed by the Ministry of Research, Innovation and Digitalization through Program 1 – Development of the National R&D System, Subprogram 1.2. Institutional performance – Financing projects for excellence in RDI.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
3. Patel K, Tsui D. The Translational Potential of Circulating Tumour DNA in Oncology. Clin Biochem (2015) 48(15):957–61. doi: 10.1016/j.clinbiochem.2015.04.005
4. Washington MK, Goldberg RM, Chang GJ, Limburg P, Lam AK, Salto-Tellez M, et al. Diagnosis of Digestive System Tumours. Int J Cancer (2021) 148(5):1040–50. doi: 10.1002/ijc.33210
5. Alzahrani SM, Al Doghaither HA, Al-Ghafari AB. General Insight Into Cancer: An Overview of Colorectal Cancer (Review). Mol Clin Oncol (2021) 15(6):271–1. doi: 10.3892/mco.2021.2433
6. Thrumurthy SG, Thrumurthy SSD, Gilbert CE, Ross P, Haji A. Colorectal Adenocarcinoma: Risks, Prevention and Diagnosis. BMJ (2016) 354:i3590. doi: 10.1136/bmj.i3590
7. Fleming M, Ravula S, Tatishchev SF, Wang HL. Colorectal Carcinoma: Pathologic Aspects. J Gastrointest Oncol (2012) 3(3):153–73. doi: 10.3978/j.issn.2078-6891.2012.030
8. Wu X, Lin H, Li S. Prognoses of Different Pathological Subtypes of Colorectal Cancer at Different Stages: A Population-Based Retrospective Cohort Study. BMC Gastroenterol (2019) 19(1):164. doi: 10.1186/s12876-019-1083-0
9. Li C, Zheng H, Jia H, Huang D, Gu W, Cai S, et al. Prognosis of Three Histological Subtypes of Colorectal Adenocarcinoma: A Retrospective Analysis of 8005 Chinese Patients. Cancer Med (2019) 8(7):3411–9. doi: 10.1002/cam4.2234
10. Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. The 2019 WHO Classification of Tumours of the Digestive System. Histopathol (2020) 76(2):182–8. doi: 10.1111/his.13975
11. Hyngstrom JR, Hu C-Y, Xing Y, You YN, Feig BW, Skibber JM, et al. Clinicopathology and Outcomes for Mucinous and Signet Ring Colorectal Adenocarcinoma: Analysis From the National Cancer Data Base. Ann Surg Oncol (2012) 19(9):2814–21. doi: 10.1245/s10434-012-2321-7
12. Mukund K, Syulyukina N, Ramamoorthy S, Subramaniam S. Right and Left-Sided Colon Cancers - Specificity of Molecular Mechanisms in Tumorigenesis and Progression. BMC Cancer (2020) 20(1):317. doi: 10.1186/s12885-020-06784-7
13. Petrelli F, Tomasello G, Borgonovo K, Ghidini M, Turati L, Dallera P, et al. Prognostic Survival Associated With Left-Sided vs Right-Sided Colon Cancer: A Systematic Review and Meta-Analysis. JAMA Oncol (2017) 3(2):211–9. doi: 10.1001/jamaoncol.2016.4227
14. Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci (2017) 18(1):197. doi: 10.3390/ijms18010197
15. Pino MS, Chung DC. The Chromosomal Instability Pathway in Colon Cancer. Gastroenterol (2010) 138(6):2059–72. doi: 10.1053/j.gastro.2009.12.065
16. Nojadeh JN, Behrouz Sharif S, Sakhinia E. Microsatellite Instability in Colorectal Cancer. EXCLI J (2018) 17:159–68. doi: 10.17179/excli2017-948
17. Tijhuis AE, Johnson SC, McClelland SE. The Emerging Links Between Chromosomal Instability (CIN), Metastasis, Inflammation and Tumour Immunity. Mol Cytogenet (2019) 12(1):17. doi: 10.1186/s13039-019-0429-1
18. Hoevenaar WHM, Janssen A, Quirindongo AI, Ma H, Klaasen SJ, Teixeira A, et al. Degree and Site of Chromosomal Instability Define its Oncogenic Potential. Nat Commun (2020) 11(1):1501. doi: 10.1038/s41467-020-15279-9
19. Thompson SL, Bakhoum SF, Compton DA. Mechanisms of Chromosomal Instability. Curr Biol CB (2010) 20(6):R285–95. doi: 10.1016/j.cub.2010.01.034
20. Fodde R. The APC Gene in Colorectal Cancer. Eur J Cancer Oxf Engl 1990 (2002) 38(7):867–71. doi: 10.1016/s0959-8049(02)00040-0
21. WHO Classification of Tumours Online. Available at: https://tumourclassification.iarc.who.int/welcome/.
23. Home - My Cancer Genome. Available at: https://www.mycancergenome.org/.
24. Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM, et al. CIViC is a Community Knowledgebase for Expert Crowdsourcing the Clinical Interpretation of Variants in Cancer. Nat Genet (2017) 49(2):170–4. doi: 10.1038/ng.3774
25. CIViC: Clinical Interpretations of Variants in Cancer. McDonnell Genome Institute (MGI) (2018). Available at: https://www.genome.wustl.edu/items/civic-clinical-interpretations-of-variants-in-cancer/.
26. Huang L, Fernandes H, Zia H, Tavassoli P, Rennert H, Pisapia D, et al. The Cancer Precision Medicine Knowledge Base for Structured Clinical-Grade Mutations and Interpretations. J Am Med Inform Assoc (2017) 24(3):513–9. doi: 10.1093/jamia/ocw148
27. PMKB. Available at: https://pmkb.weill.cornell.edu/.
28. Patterson SE, Liu R, Statz CM, Durkin D, Lakshminarayana A, Mockus SM. The Clinical Trial Landscape in Oncology and Connectivity of Somatic Mutational Profiles to Targeted Therapies. Hum Genomics (2016) 10(1):1–13. doi: 10.1186/s40246-016-0061-7
29. Clinical Knowledgebase (CKB). The Jackson Laboratory. Available at: https://www.jax.org/clinical-genomics/ckb.
30. Cancer Genome Interpreter - Identification of therapeutically actionable genomic alterations in tumors. Available at: https://www.cancergenomeinterpreter.org/home.
31. Damodaran S, Miya J, Kautto E, Zhu E, Samorodnitsky E, Datta J, et al. Cancer Driver Log (CanDL): Catalog of Potentially Actionable Cancer Mutations. J Mol Diagn (2015) 17(5):554–9. doi: 10.1016/j.jmoldx.2015.05.002
32. CanDL: Cancer Data Log Full-Text Search. Available at: http://candl.osu.edu/.
33. Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and Saturation Analysis of Cancer Genes Across 21 Tumour Types. Nature (2014) 505(7484):495–501. doi: 10.1038/nature12912
34. TumorPortal. Available at: http://tumorportal.org/.
35. Clinical Trials and Research. Massachusetts General Hospital. Available at: https://www.massgeneral.org/cancer-center/clinical-trials-and-research.
36. Meric-Bernstam F, Johnson A, Holla V, Bailey AM, Brusco L, Chen K, et al. A Decision Support Framework for Genomically Informed Investigational Cancer Therapy. JNCI J Natl Cancer Inst (2015) 107(7):djv098. doi: 10.1093/jnci/djv098
37. Knowledge Base for Precision Oncology. Available at: https://pct.mdanderson.org/#/.
38. Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol (2017) 1:1–16. doi: 10.1200/PO.17.00011
39. Conti RM, Bernstein AC, Villaflor VM, Schilsky RL, Rosenthal MB, Bach PB. Prevalence of Off-Label Use and Spending in 2010 Among Patent-Protected Chemotherapies in a Population-Based Cohort of Medical Oncologists. J Clin Oncol (2013) 31(9):1134. doi: 10.1200/JCO.2012.42.7252
40. Fairman KA, Curtiss FR. Regulatory Actions on the Off-Label Use of Prescription Drugs: Ongoing Controversy and Contradiction in 2009 and 2010. J Manag Care Pharm (2010) 16(8):629–39. doi: 10.18553/jmcp.2010.16.8.629
41. Largent EA, Miller FG, Pearson SD. Going Off-Label Without Venturing Off-Course: Evidence and Ethical Off-Label Prescribing. Arch Intern Med (2009) 169(19):1745–7. doi: 10.1001/archinternmed.2009.314
42. Corcoran RB, Atreya CE, Falchook GS, Kwak EL, Ryan DP, Bendell JC, et al. Combined BRAF and MEK Inhibition With Dabrafenib and Trametinib in BRAF V600–mutant Colorectal Cancer. J Clin Oncol (2015) 33(34):4023. doi: 10.1200/JCO.2015.63.2471
43. Mateo J, Chakravarty D, Dienstmann R, Jezdic S, Gonzalez-Perez A, Lopez-Bigas N, et al. A Framework to Rank Genomic Alterations as Targets for Cancer Precision Medicine: The ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT). Ann Oncol (2018) 29(9):1895–902. doi: 10.1093/annonc/mdy263
44. Mosele F, Remon J, Mateo J, Westphalen C, Barlesi F, Lolkema M, et al. Recommendations for the Use of Next-Generation Sequencing (NGS) for Patients With Metastatic Cancers: A Report From the ESMO Precision Medicine Working Group. Ann Oncol (2020) 31(11):1491–1505. doi: 10.1016/j.annonc.2020.07.014
45. Cutsem EV, Lenz H-J, Kohne C, Heinemann V, Tejpar S, Melezínek I, et al. Fluorouracil, Leucovorin, and Irinotecan Plus Cetuximab Treatment and RAS Mutations in Colorectal Cancer. J Clin Oncol (2015) 33(7):692–700. doi: 10.1200/JCO.2014.59.4812
46. Douillard J-Y, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab–FOLFOX4 Treatment and RAS Mutations in Colorectal Cancer. N Engl J Med (2013) 369(11):1023–34. doi: 10.1056/NEJMoa1305275
47. Sorich M, Wiese M, Rowland A, Kichenadasse G, McKinnon R, Karapetis C. Extended RAS Mutations and Anti-EGFR Monoclonal Antibody Survival Benefit in Metastatic Colorectal Cancer: A Meta-Analysis of Randomized, Controlled Trials. Ann Oncol (2015) 26(1):13–21. doi: 10.1093/annonc/mdu378
48. Shen H, Yang J, Huang Q, Jiang M-J, Tan Y-N, Fu J-F, et al. Different Treatment Strategies and Molecular Features Between Right-Sided and Left-Sided Colon Cancers. World J Gastroenterol WJG (2015) 21(21):6470. doi: 10.3748/wjg.v21.i21.6470
49. Sinicrope FA, Mahoney MR, Smyrk TC, Thibodeau SN, Warren RS, Bertagnolli MM, et al. Prognostic Impact of Deficient DNA Mismatch Repair in Patients With Stage III Colon Cancer From a Randomized Trial of FOLFOX-Based Adjuvant Chemotherapy. J Clin Oncol (2013) 31(29):3664. doi: 10.1200/JCO.2013.48.9591
50. Weiss JM, Pfau PR, O’Connor ES, King J, LoConte N, Kennedy G, et al. Mortality by Stage for Right-Versus Left-Sided Colon Cancer: Analysis of Surveillance, Epidemiology, and End Results–Medicare Data. J Clin Oncol (2011) 29(33):4401. doi: 10.1200/JCO.2011.36.4414
51. Loupakis F, Yang D, Yau L, Feng S, Cremolini C, Zhang W, et al. Primary Tumor Location as a Prognostic Factor in Metastatic Colorectal Cancer. JNCI J Natl Cancer Inst (2015) 107(3):dju427. doi: 10.1093/jnci/dju427
52. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E–mutated Colorectal Cancer. N Engl J Med (2019) 381(17):1632–43. doi: 10.1056/NEJMoa1908075
53. Le DT, Kim TW, Van Cutsem E, Geva R, Jäger D, Hara H, et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability–High/Mismatch Repair–Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J Clin Oncol (2020) 38(1):11. doi: 10.1200/JCO.19.02107
54. Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz H-J, Morse MA, et al. Nivolumab in Patients With Metastatic DNA Mismatch Repair-Deficient or Microsatellite Instability-High Colorectal Cancer (CheckMate 142): An Open-Label, Multicentre, Phase 2 Study. Lancet Oncol (2017) 18(9):1182–91. doi: 10.1016/S1470-2045(17)30422-9
55. Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-Targeted Therapy With Trastuzumab and Lapatinib in Treatment-Refractory, KRAS Codon 12/13 Wild-Type, HER2-Positive Metastatic Colorectal Cancer (HERACLES): A Proof-of-Concept, Multicentre, Open-Label, Phase 2 Trial. Lancet Oncol (2016) 17(6):738–46. doi: 10.1016/S1470-2045(16)00150-9
56. Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, et al. Entrectinib in Patients With Advanced or Metastatic NTRK Fusion-Positive Solid Tumours: Integrated Analysis of Three Phase 1–2 Trials. Lancet Oncol (2020) 21(2):271–82. doi: 10.1016/S1470-2045(19)30691-6
57. Waters GS, Gardner EM. Colorectal Cancer Imaging and Management: A Surgeon’s Perspective. Semin Roentgenol (2021). 56(2):158–63. doi: 10.1053/j.ro.2020.07.004
58. Ginghina O, Nitipir C. Liquid Biopsy for Colorectal Cancer Screening, A Modern Approach for Patients Stratification and Monitoring. In: Colorectal Cancer-Diagnosis, Screening and Management. London: IntechOpen (2017).
59. Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, et al. Liquid Versus Tissue Biopsy for Detecting Acquired Resistance and Tumor Heterogeneity in Gastrointestinal Cancers. Nat Med (2019) 25(9):1415–21. doi: 10.1038/s41591-019-0561-9
60. Lu T, Li J. Clinical Applications of Urinary Cell-Free DNA in Cancer: Current Insights and Promising Future. Am J Cancer Res (2017) 7(11):2318.
61. Heitzer E, Haque IS, Roberts CE, Speicher MR. Current and Future Perspectives of Liquid Biopsies in Genomics-Driven Oncology. Nat Rev Genet (2019) 20(2):71–88. doi: 10.1038/s41576-018-0071-5
62. Ashworth T. A Case of Cancer in Which Cells Similar to Those in the Tumours Were Seen in the Blood After Death. Aust Med J (1869) 14:146.
63. Griffiths J, McKinna J, Rowbotham H, Tsolakidis P, Salsbury A. Carcinoma of the Colon and Rectum: Circulating Malignant Cells and Five-Year Survival. Cancer (1973) 31(1):226–36. doi: 10.1002/1097-0142(197301)31:1<226::AID-CNCR2820310130>3.0.CO;2-M
64. Zeidman I. The Fate of Circulating Tumor Cells: I. Passage of Cells Through Capillaries. Cancer Res (1961) 21(1):38–9.
65. Skolnik G, Alpsten M, Ivarsson L. Studies on Mechanisms Involved in Metastasis Formation From Circulating Tumor Cells. J Cancer Res Clin Oncol (1980) 97(3):249–56. doi: 10.1007/BF00405776
66. Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients With Metastatic Breast Colorectal and Prostate Cancer. J Oncol (2010) 2010:617421. doi: 10.1155/2010/617421
67. Cubero MA, Lorente J, Robles-Fernandez I, Rodriguez-Martinez A, Puche J, Serrano M. Circulating Tumor Cells: Markers and Methodologies for Enrichment and Detection. In: Circulating Tumor Cells. New York: Springer (2017). p. 283–303.
68. Amintas S, Bedel A, Moreau-Gaudry F, Boutin J, Buscail L, Merlio J-P, et al. Circulating Tumor Cell Clusters: United We Stand Divided We Fall. Int J Mol Sci (2020) 21(7):2653. doi: 10.3390/ijms21072653
69. Yanai Y, Hayashi T, Akazawa Y, Yatagai N, Tsuyama S, Yao T, et al. Clinicopathological and Mutational Differences Between Tumors With Multiple Metastases and Single Lung Metastasis in Colorectal Cancer. Oncol Lett (2020) 20(1):541–50. doi: 10.3892/ol.2020.11627
70. Kim M-Y, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH-F, Norton L, et al. Tumor Self-Seeding by Circulating Cancer Cells. Cell (2009) 139(7):1315–26. doi: 10.1016/j.cell.2009.11.025
71. Krog BL, Henry MD. Biomechanics of the Circulating Tumor Cell Microenvironment. Biomech Oncol (2018) 1092:209–33. doi: 10.1007/978-3-319-95294-9_11
72. Marrella A, Fedi A, Varani G, Vaccari I, Fato M, Firpo G, et al. High Blood Flow Shear Stress Values are Associated With Circulating Tumor Cells Cluster Disaggregation in a Multi-Channel Microfluidic Device. PloS One (2021) 16(1):e0245536. doi: 10.1371/journal.pone.0245536
73. Wang W-C, Zhang X-F, Peng J, Li X-F, Wang A-L, Bie Y-Q, et al. Survival Mechanisms and Influence Factors of Circulating Tumor Cells. BioMed Res Int (2018) 2018:6304701. doi: 10.1155/2018/6304701
74. Fidler IJ. Metastasis: Quantitative Analysis of Distribution and Fate of Tumor Emboli Labeled With 125I-5-Iodo-2′-Deoxyuridine. J Natl Cancer Inst (1970) 45(4):773–82.
75. Nicolazzo C, Raimondi C, Francescangeli F, Ceccarelli S, Trenta P, Magri V, et al. EpCAM-Expressing Circulating Tumor Cells in Colorectal Cancer. Int J Biol Markers (2017) 32(4):415–20. doi: 10.5301/ijbm.5000284
76. Lu S-H, Tsai W-S, Chang Y-H, Chou T-Y, Pang S-T, Lin P-H, et al. Identifying Cancer Origin Using Circulating Tumor Cells. Cancer Biol Ther (2016) 17(4):430–8. doi: 10.1080/15384047.2016.1141839
77. Bednarz-Knoll N, Alix-Panabières C, Pantel K. Plasticity of Disseminating Cancer Cells in Patients With Epithelial Malignancies. Cancer Metastasis Rev (2012) 31(3):673–87. doi: 10.1007/s10555-012-9370-z
78. Kang Y, Pantel K. Tumor Cell Dissemination: Emerging Biological Insights From Animal Models and Cancer Patients. Cancer Cell (2013) 23(5):573–81. doi: 10.1016/j.ccr.2013.04.017
79. Zeng Y, Zhang Q, Zhang Y, Lu M, Liu Y, Zheng T, et al. MUC1 Predicts Colorectal Cancer Metastasis: A Systematic Review and Meta-Analysis of Case Controlled Studies. PloS One (2015) 10(9):e0138049. doi: 10.1371/journal.pone.0138049
80. Takahashi H, Jin C, Rajabi H, Pitroda S, Alam M, Ahmad R, et al. MUC1-C Activates the TAK1 Inflammatory Pathway in Colon Cancer. Oncogene (2015) 34(40):5187–97. doi: 10.1038/onc.2014.442
81. Zhang N, Ng AS, Cai S, Li Q, Yang L, Kerr D. Novel Therapeutic Strategies: Targeting Epithelial–Mesenchymal Transition in Colorectal Cancer. Lancet Oncol (2021) 22(8):e358–68. doi: 10.1016/S1470-2045(21)00343-0
82. Rushton AJ, Nteliopoulos G, Shaw JA, Coombes RC. A Review of Circulating Tumour Cell Enrichment Technologies. Cancers (2021) 13(5):970. doi: 10.3390/cancers13050970
83. Cohen SJ, Punt C, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients With Metastatic Colorectal Cancer. Clin Oncol (2008) 26:3213–21. doi: 10.1200/JCO.2007.15.8923
84. Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, et al. Circulating Tumor Cells: A Novel Prognostic Factor for Newly Diagnosed Metastatic Breast Cancer. J Clin Oncol (2005) 23(7):1420–30. doi: 10.1200/JCO.2005.08.140
85. Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N Engl J Med (2004) 351(8):781–91. doi: 10.1056/NEJMoa040766
86. De Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating Tumor Cells Predict Survival Benefit From Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin Cancer Res (2008) 14(19):6302–9. doi: 10.1158/1078-0432.CCR-08-0872
87. Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas But Not in Healthy Subjects or Patients With Nonmalignant Diseases. Clin Cancer Res (2004) 10(20):6897–904. doi: 10.1158/1078-0432.CCR-04-0378
88. Riethdorf S, O’Flaherty L, Hille C, Pantel K. Clinical Applications of the CellSearch Platform in Cancer Patients. Nanotechnol Enabling Use Circ Tumor Cells CTCs Reliab Cancer biomark (2018) 125:102–21. doi: 10.1016/j.addr.2018.01.011
89. Wang L, Balasubramanian P, Chen AP, Kummar S, Evrard YA, Kinders RJ. Promise and Limits of the CellSearch Platform for Evaluating Pharmacodynamics in Circulating Tumor Cells. Pharmacodyn Cancer Drug Dev (2016) 43(4):464–75. doi: 10.1053/j.seminoncol.2016.06.004
90. Bork U, Rahbari N, Schölch S, Reissfelder C, Kahlert C, Büchler M, et al. Circulating Tumour Cells and Outcome in non-Metastatic Colorectal Cancer: A Prospective Study. Br J Cancer (2015) 112(8):1306–13. doi: 10.1038/bjc.2015.88
91. Sastre J, Maestro ML, Gómez-España A, Rivera F, Valladares M, Massuti B, et al. Circulating Tumor Cell Count is a Prognostic Factor in Metastatic Colorectal Cancer Patients Receiving First-Line Chemotherapy Plus Bevacizumab: A Spanish Cooperative Group for the Treatment of Digestive Tumors Study. oncologist (2012) 17(7):947. doi: 10.1634/theoncologist.2012-0048
92. Alix-Panabières C, Schwarzenbach H, Pantel K. Circulating Tumor Cells and Circulating Tumor DNA. Annu Rev Med (2012) 63:199–215. doi: 10.1146/annurev-med-062310-094219
93. Swennenhuis JF, van Dalum G, Zeune LL, Terstappen LWMM. Improving the CellSearch® System. Expert Rev Mol Diagn (2016) 16(12):1291–305. doi: 10.1080/14737159.2016.1255144
94. Vu T, Datta PK. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers (2017) 9(12):171. doi: 10.3390/cancers9120171
95. Sieuwerts AM, Jeffrey SS. Multiplex Molecular Analysis of CTCs. Minimal Residual Dis Circ Tumor Cells Breast Cancer (2012) 195:125–40. doi: 10.1007/978-3-642-28160-0_11
96. Miltenyi S, Müller W, Weichel W, Radbruch A. High Gradient Magnetic Cell Separation With MACS. Cytom J Int Soc Anal Cytol (1990) 11(2):231–8. doi: 10.1002/cyto.990110203
97. Lu N-N, Xie M, Wang J, Lv S-W, Yi J-S, Dong W-G, et al. Biotin-Triggered Decomposable Immunomagnetic Beads for Capture and Release of Circulating Tumor Cells. ACS Appl Mater Interfaces (2015) 7(16):8817–26. doi: 10.1021/acsami.5b01397
98. Xiong K, Wei W, Jin Y, Wang S, Zhao D, Wang S, et al. Biomimetic Immuno-Magnetosomes for High-Performance Enrichment of Circulating Tumor Cells. Adv Mater (2016) 28(36):7929–35. doi: 10.1002/adma.201601643
99. Vojtechova G, Benesova L, Belsanova B, Minarikova P, Levy M, Lipska L, et al. Monitoring of Circulating Tumor Cells by a Combination of Immunomagnetic Enrichment and RT-PCR in Colorectal Cancer Patients Undergoing Surgery. Adv Clin Exp Med (2016) 25(6):1273–9. doi: 10.17219/acem/63824
100. Drucker A, Teh EM, Kostyleva R, Rayson D, Douglas S, Pinto DM. Comparative Performance of Different Methods for Circulating Tumor Cell Enrichment in Metastatic Breast Cancer Patients. PloS One (2020) 15(8):e0237308. doi: 10.1371/journal.pone.0237308
101. Hudiță A, Ioana-Lavric V, Zamfir A, Buburuzan L, Ginghină O, Negrei C, et al. Optimization of a Flow Cytometry Method for the Approach of Liquid Biopsy as a Therapy Modulation Tool in Patients With Colorectal Cancer. Farmacia (2018) 66(5):853–860. doi: 10.31925/farmacia.2018.5.16
102. Kure K, Hosoya M, Ueyama T, Fukaya M, Sugimoto K, Tomiki Y, et al. Using the Polymeric Circulating Tumor Cell Chip to Capture Circulating Tumor Cells in Blood Samples of Patients With Colorectal Cancer. Oncol Lett (2020) 19(3):2286–94. doi: 10.3892/ol.2020.11335
103. Harb W, Fan A, Tran T, Danila DC, Keys D, Schwartz M, et al. Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay. Transl Oncol (2013) 6(5):528–IN1. doi: 10.1593/tlo.13367
104. Jan YJ, Chen J-F, Zhu Y, Lu Y-T, Chen SH, Chung H, et al. NanoVelcro Rare-Cell Assays for Detection and Characterization of Circulating Tumor Cells. Adv Drug Delivery Rev (2018) 125:78–93. doi: 10.1016/j.addr.2018.03.006
105. Dharmasiri U, Njoroge SK, Witek MA, Adebiyi MG, Kamande JW, Hupert ML, et al. High-Throughput Selection, Enumeration, Electrokinetic Manipulation, and Molecular Profiling of Low-Abundance Circulating Tumor Cells Using a Microfluidic System. Anal Chem (2011) 83(6):2301–9. doi: 10.1021/ac103172y
106. Casavant BP, Berry SM, Lang J, Guckenberger DJ, Beebe DJ. VERIFAST: AN INTEGRATED SYSTEM FOR FLEXIBLE CTC ISOLATION AND ANALYSIS. (2012) 1:1393–5.
107. Miller MC, Robinson PS, Wagner C, O’Shannessy DJ. The ParsortixTM Cell Separation System—A Versatile Liquid Biopsy Platform. Cytometry A (2018) 93(12):1234–9. doi: 10.1002/cyto.a.23571
108. Hosokawa M, Hayata T, Fukuda Y, Arakaki A, Yoshino T, Tanaka T, et al. Size-Selective Microcavity Array for Rapid and Efficient Detection of Circulating Tumor Cells. Anal Chem (2010) 82(15):6629–35. doi: 10.1021/ac101222x
109. Clawson GA, Kimchi E, Patrick SD, Xin P, Harouaka R, Zheng S, et al. Circulating Tumor Cells in Melanoma Patients. PloS One (2012) 7(7):e41052. doi: 10.1371/journal.pone.0041052
110. Ramirez AB, U’Ren L, Campton DE, Stewart D, Nordberg JJ, Stilwell JL, et al. RareCyte® CTC Analysis Step 1: AccuCyte® Sample Preparation for the Comprehensive Recovery of Nucleated Cells From Whole Blood. In: Circulating Tumor Cells. New York: Springer (2017). p. 163–72.
111. Matikas A, Voutsina A, Lagoudaki E, Hatzidaki D, Trypaki M, Stoupis G, et al. Detection of KRAS Exon 2 Mutations in Circulating Tumor Cells Isolated by the ISET System From Patients With RAS Wild Type Metastatic Colorectal Cancer. Transl Oncol (2017) 10(4):693–8. doi: 10.1016/j.tranon.2017.06.005
112. Harouaka RA, Zhou M-D, Yeh Y-T, Khan WJ, Das A, Liu X, et al. Flexible Micro Spring Array Device for High-Throughput Enrichment of Viable Circulating Tumor Cells. Clin Chem (2014) 60(2):323–33. doi: 10.1373/clinchem.2013.206805
113. Dolfus C, Piton N, Toure E, Sabourin J-C. Circulating Tumor Cell Isolation: The Assets of Filtration Methods With Polycarbonate Track-Etched Filters. Chin J Cancer Res (2015) 27(5):479. doi: 10.3978/j.issn.1000-9604.2015.09.01
114. Kang HM, Kim GH, Jeon HK, Kim DH, Jeon TY, Park DY, et al. Circulating Tumor Cells Detected by Lab-on-a-Disc: Role in Early Diagnosis of Gastric Cancer. PloS One (2017) 12(6):e0180251. doi: 10.1371/journal.pone.0180251
115. Gupta V, Jafferji I, Garza M, Melnikova VO, Hasegawa DK, Pethig R, et al. ApoStream(TM), a New Dielectrophoretic Device for Antibody Independent Isolation and Recovery of Viable Cancer Cells From Blood. Biomicrofluidics (2012) 6(2):24133–3. doi: 10.1063/1.4731647
116. Fischer JC, Niederacher D, Topp SA, Honisch E, Schumacher S, Schmitz N, et al. Diagnostic Leukapheresis Enables Reliable Detection of Circulating Tumor Cells of Nonmetastatic Cancer Patients. Proc Natl Acad Sci (2013) 110(41):16580–5. doi: 10.1073/pnas.1313594110
117. Saucedo-Zeni N, Mewes S, Niestroj R, Gasiorowski L, Murawa D, Nowaczyk P, et al. A Novel Method for the In Vivo Isolation of Circulating Tumor Cells From Peripheral Blood of Cancer Patients Using a Functionalized and Structured Medical Wire. Int J Oncol (2012) 41(4):1241–50. doi: 10.3892/ijo.2012.1557
118. Marcuello M, Vymetalkova V, Neves RP, Duran-Sanchon S, Vedeld HM, Tham E, et al. Circulating Biomarkers for Early Detection and Clinical Management of Colorectal Cancer. Mol Aspects Med (2019) 69:107–22. doi: 10.1016/j.mam.2019.06.002
119. Yang C, Zhuang W, Hu Y, Zhu L. Clinical Significance of Peripheral Circulating Tumor Cell Counts in Colorectal Polyps and non-Metastatic Colorectal Cancer. World J Surg Oncol (2018) 16(1):13. doi: 10.1186/s12957-017-1305-2
120. Tsai W-S, Nimgaonkar A, Segurado O, Chang Y, Hsieh B, Shao H-J, et al. Prospective Clinical Study of Circulating Tumor Cells for Colorectal Cancer Screening. J Clin Oncol (2018) 36(4_suppl):556–6. doi: 10.1200/JCO.2018.36.4_suppl.556
121. Gupta P, Gulzar Z, Hsieh B, Lim A, Watson D, Mei R. Analytical Validation of the CellMax Platform for Early Detection of Cancer by Enumeration of Rare Circulating Tumor Cells. J Circ biomark (2019) 8:1849454419899214. doi: 10.1177/1849454419899214
122. Fehm TN, Meier-Stiegen F, Driemel C, Jäger B, Reinhardt F, Naskou J, et al. Diagnostic Leukapheresis for CTC Analysis in Breast Cancer Patients: CTC Frequency, Clinical Experiences and Recommendations for Standardized Reporting. Cytometry A (2018) 93(12):1213–9. doi: 10.1002/cyto.a.23669
123. Tamminga M, Andree KC, van den Bos H, Hiltermann TJN, Mentink A, Spierings DC, et al. Leukapheresis Increases Circulating Tumour Cell Yield in non-Small Cell Lung Cancer, Counts Related to Tumour Response and Survival. Br J Cancer (2021) 126(3):1–10. doi: 10.1038/s41416-021-01634-0
124. Soya R, Taguchi J, Nagakawa Y, Takahashi O, Sandoh N, Hosokawa Y, et al. A Large Number of Circulating Tumor Cells (CTCs) Can Be Isolated From Samples Obtained by Using Leukapheresis Procedures. Gan To Kagaku Ryoho (2015) 42(9):1069–72.
125. Guadagni S, Fiorentini G, De Simone M, Masedu F, Zoras O, Mackay AR, et al. Precision Oncotherapy Based on Liquid Biopsies in Multidisciplinary Treatment of Unresectable Recurrent Rectal Cancer: A Retrospective Cohort Study. J Cancer Res Clin Oncol (2020) 146(1):205–19. doi: 10.1007/s00432-019-03046-3
126. Guadagni S, Clementi M, Mackay AR, Ricevuto E, Fiorentini G, Sarti D, et al. Real-Life Multidisciplinary Treatment for Unresectable Colorectal Cancer Liver Metastases Including Hepatic Artery Infusion With Chemo-Filtration and Liquid Biopsy Precision Oncotherapy: Observational Cohort Study. J Cancer Res Clin Oncol (2020) 146(5):1273–90. doi: 10.1007/s00432-020-03156-3
127. Diaz LA Jr, Bardelli A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J Clin Oncol (2014) 32(6):579. doi: 10.1200/JCO.2012.45.2011
128. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch R-D, et al. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin From Apoptotic and Necrotic Cells. Cancer Res (2001) 61(4):1659–65.
129. Mouliere F, Thierry AR. The Importance of Examining the Proportion of Circulating DNA Originating From Tumor, Microenvironment and Normal Cells in Colorectal Cancer Patients. Expert Opin Biol Ther (2012) 12(sup1):S209–15. doi: 10.1517/14712598.2012.688023
130. Wan JC, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid Biopsies Come of Age: Towards Implementation of Circulating Tumour DNA. Nat Rev Cancer (2017) 17(4):223. doi: 10.1038/nrc.2017.7
131. Mandel P, Metais P. Nuclear Acids in Human Blood Plasma. C R Seances Soc Biol Fil (1948) 142(3–4):241–3.
132. Leon S, Shapiro B, Sklaroff D, Yaros M. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res (1977) 37(3):646–50.
133. Hao T, Shi W, Shen X, Qi J, Wu X, Wu Y, et al. Circulating Cell-Free DNA in Serum as a Biomarker for Diagnosis and Prognostic Prediction of Colorectal Cancer. Br J Cancer (2014) 111(8):1482–9. doi: 10.1038/bjc.2014.470
134. Vymetalkova V, Cervena K, Bartu L, Vodicka P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int J Mol Sci (2018) 19(11):3356. doi: 10.3390/ijms19113356
135. Yang Y-C, Wang D, Jin L, Yao H-W, Zhang J-H, Wang J, et al. Circulating Tumor DNA Detectable in Early-and Late-Stage Colorectal Cancer Patients. Biosci Rep (2018) 38(4):BSR20180322. doi: 10.1042/BSR20180322
136. El Messaoudi S, Mouliere F, Du Manoir S, Bascoul-Mollevi C, Gillet B, Nouaille M, et al. Circulating DNA as a Strong Multimarker Prognostic Tool for Metastatic Colorectal Cancer Patient Management Care. Clin Cancer Res (2016) 22(12):3067–77. doi: 10.1158/1078-0432.CCR-15-0297
137. Zitt M, Zitt M, Müller HM. DNA Methylation in Colorectal Cancer–Impact on Screening and Therapy Monitoring Modalities? Dis Markers (2007) 23(1, 2):51–71. doi: 10.1155/2007/891967
138. Toth K, Sipos F, Kalmar A, Patai AV, Wichmann B, Stoehr R, et al. Detection of Methylated SEPT9 in Plasma is a Reliable Screening Method for Both Left-and Right-Sided Colon Cancers. PLoS One (2012) 7(9):e46000. doi: 10.1371/journal.pone.0046000
139. Molnár B, Tóth K, Barták BK, Tulassay Z. Plasma Methylated Septin 9: A Colorectal Cancer Screening Marker. Expert Rev Mol Diagn (2015) 15(2):171–84. doi: 10.1586/14737159.2015.975212
140. Lamb YN, Dhillon S. Epi Procolon® 2.0 CE: A Blood-Based Screening Test for Colorectal Cancer. Mol Diagn Ther (2017) 21(2):225–32. doi: 10.1007/s40291-017-0259-y
141. Tóth K, Galamb O, Spisák S, Wichmann B, Sipos F, Valcz G, et al. The Influence of Methylated Septin 9 Gene on RNA and Protein Level in Colorectal Cancer. Pathol Oncol Res (2011) 17(3):503–9. doi: 10.1007/s12253-010-9338-7
142. Johnson DA, Barclay RL, Mergener K, Weiss G, König T, Beck J, et al. Plasma Septin9 Versus Fecal Immunochemical Testing for Colorectal Cancer Screening: A Prospective Multicenter Study. PloS One (2014) 9(6):e98238. doi: 10.1371/journal.pone.0098238
143. Gustavsson B, Carlsson G, Machover D, Petrelli N, Roth A, Schmoll H-J, et al. A Review of the Evolution of Systemic Chemotherapy in the Management of Colorectal Cancer. Clin Colorectal Cancer (2015) 14(1):1–10. doi: 10.1016/j.clcc.2014.11.002
144. Meyerhardt JA, Mayer RJ. Systemic Therapy for Colorectal Cancer. N Engl J Med (2005) 352(5):476–87. doi: 10.1056/NEJMra040958
145. Mohelnikova-Duchonova B, Melichar B, Soucek P. FOLFOX/FOLFIRI Pharmacogenetics: The Call for a Personalized Approach in Colorectal Cancer Therapy. World J Gastroenterol WJG (2014) 20(30):10316. doi: 10.3748/wjg.v20.i30.10316
146. Bruera G, Santomaggio A, Cannita K, Baldi PL, Tudini M, De Galitiis F, et al. “ Poker” Association of Weekly Alternating 5-Fluorouracil, Irinotecan, Bevacizumab and Oxaliplatin (FIr-B/FOx) in First Line Treatment of Metastatic Colorectal Cancer: A Phase II Study. BMC Cancer (2010) 10(1):1–10. doi: 10.1186/1471-2407-10-567
147. Loupakis F, Cremolini C, Masi G, Lonardi S, Zagonel V, Salvatore L, et al. Initial Therapy With FOLFOXIRI and Bevacizumab for Metastatic Colorectal Cancer. N Engl J Med (2014) 371(17):1609–18. doi: 10.1056/NEJMoa1403108
148. Bruera G, Massacese S, Pepe F, Malapelle U, Dal Mas A, Ciacco E, et al. Intensive First-Line FIr-C/FOx-C Association of Triplet Chemotherapy Plus Cetuximab in RAS Wild-Type Metastatic Colorectal Cancer Patients: Preliminary Phase II Data and Prediction of Individual Limiting Toxicity Syndromes by Pharmacogenomic Biomarkers. Ther Adv Med Oncol (2019) 11:1758835919846421. doi: 10.1177/1758835919846421
149. Xie Y-H, Chen Y-X, Fang J-Y. Comprehensive Review of Targeted Therapy for Colorectal Cancer. Signal Transduct Target Ther (2020) 5(1):1–30. doi: 10.1038/s41392-020-0116-z
150. Martini G, Troiani T, Cardone C, Vitiello P, Sforza V, Ciardiello D, et al. Present and Future of Metastatic Colorectal Cancer Treatment: A Review of New Candidate Targets. World J Gastroenterol (2017) 23(26):4675. doi: 10.3748/wjg.v23.i26.4675
151. Morelli M, Overman M, Dasari A, Kazmi S, Mazard T, Vilar E, et al. Characterizing the Patterns of Clonal Selection in Circulating Tumor DNA From Patients With Colorectal Cancer Refractory to Anti-EGFR Treatment. Ann Oncol (2015) 26(4):731–6. doi: 10.1093/annonc/mdv005
152. Wee P, Wang Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (2017) 9(5):52. doi: 10.3390/cancers9050052
153. Yarom N, Jonker DJ. The Role of the Epidermal Growth Factor Receptor in the Mechanism and Treatment of Colorectal Cancer. Discovery Med (2011) 11(57):95–105.
154. Zhao B, Wang L, Qiu H, Zhang M, Sun L, Peng P, et al. Mechanisms of Resistance to Anti-EGFR Therapy in Colorectal Cancer. Oncotarget (2017) 8(3):3980. doi: 10.18632/oncotarget.14012
155. Kratz JD, Turk AA, Sievers CK, Clipson L, Matkowskyj KA, Deming DA. Molecular Profiles Guide Colorectal Cancer Treatment. Fed Pract (2016) 33(Suppl 4):50S.
156. Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to Anti-EGFR Therapy in Colorectal Cancer: From Heterogeneity to Convergent Evolution. Cancer Discovery (2014) 4(11):1269–80. doi: 10.1158/2159-8290.CD-14-0462
157. Van Cutsem E, Köhne C-H, Láng I, Folprecht G, Nowacki MP, Cascinu S, et al. Cetuximab Plus Irinotecan, Fluorouracil, and Leucovorin as First-Line Treatment for Metastatic Colorectal Cancer: Updated Analysis of Overall Survival According to Tumor KRAS and BRAF Mutation Status. J Clin Oncol (2011) 29(15):2011–9. doi: 10.1200/JCO.2010.33.5091
158. Bruera G, Cannita K, Di Giacomo D, Lamy A, Giordano AV, Vicentini R, et al. Worse Prognosis of KRAS C. 35 G> A Mutant Metastatic Colorectal Cancer (MCRC) Patients Treated With First-Line Triplet Chemotherapy Plus Bevacizumab (FIr-B/FOx) and Post-Progression. BMC Med (2013) 11:59. doi: 10.1186/1741-7015-11-59
159. Bruera G, Cannita K, Di Giacomo D, Lamy A, Troncone G, Dal Mas A, et al. Prognostic Value of KRAS Genotype in Metastatic Colorectal Cancer (MCRC) Patients Treated With Intensive Triplet Chemotherapy Plus Bevacizumab (FIr-B/FOx) According to Extension of Metastatic Disease. BMC Med (2012) 10(1):1–10. doi: 10.1186/1741-7015-10-135
160. Bruera G, Cannita K, Tessitore A, Russo A, Alesse E, Ficorella C, et al. The Prevalent KRAS Exon 2 C. 35 G> A Mutation in Metastatic Colorectal Cancer Patients: A Biomarker of Worse Prognosis and Potential Benefit of Bevacizumab-Containing Intensive Regimens? Crit Rev Oncol Hematol (2015) 93(3):190–202. doi: 10.1016/j.critrevonc.2014.10.004
161. Grothey A, Fakih M, Tabernero J. Management of BRAF-Mutant Metastatic Colorectal Cancer: A Review of Treatment Options and Evidence-Based Guidelines. Ann Oncol (2021) 32(8):959–67. doi: 10.1016/j.annonc.2021.03.206
162. Rowland A, Dias M, Wiese M, Kichenadasse G, McKinnon R, Karapetis C, et al. Meta-Analysis of BRAF Mutation as a Predictive Biomarker of Benefit From Anti-EGFR Monoclonal Antibody Therapy for RAS Wild-Type Metastatic Colorectal Cancer. Br J Cancer (2015) 112(12):1888–94. doi: 10.1038/bjc.2015.173
163. De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA Mutations on the Efficacy of Cetuximab Plus Chemotherapy in Chemotherapy-Refractory Metastatic Colorectal Cancer: A Retrospective Consortium Analysis. Lancet Oncol (2010) 11(8):753–62. doi: 10.1016/S1470-2045(10)70130-3
164. Garcia-Carbonero N, Martinez-Useros J, Li W, Orta A, Perez N, Carames C, et al. KRAS and BRAF Mutations as Prognostic and Predictive Biomarkers for Standard Chemotherapy Response in Metastatic Colorectal Cancer: A Single Institutional Study. Cells (2020) 9(1):219. doi: 10.3390/cells9010219
165. Cafiero C, Re A, D’Amato G, Surico PL, Surico G, Pirrelli M, et al. KRAS and BRAF Concomitant Mutations in a Patient With Metastatic Colon Adenocarcinoma: An Interesting Case Report. Case Rep Oncol (2020) 13(2):595–600. doi: 10.1159/000507882
166. Midthun L, Shaheen S, Deisch J, Senthil M, Tsai J, Hsueh C-T. Concomitant KRAS and BRAF Mutations in Colorectal Cancer. J Gastrointest Oncol (2019) 10(3):577. doi: 10.21037/jgo.2019.01.10
167. Bruera G, Pepe F, Malapelle U, Pisapia P, Dal Mas A, Di Giacomo D, et al. KRAS, NRAS and BRAF Mutations Detected by Next Generation Sequencing, and Differential Clinical Outcome in Metastatic Colorectal Cancer (MCRC) Patients Treated With First Line FIr-B/FOx Adding Bevacizumab (BEV) to Triplet Chemotherapy. Oncotarget (2018) 9(41):26279. doi: 10.18632/oncotarget.25180
168. Mao C, Yang Z, Hu X, Chen Q, Tang J. PIK3CA Exon 20 Mutations as a Potential Biomarker for Resistance to Anti-EGFR Monoclonal Antibodies in KRAS Wild-Type Metastatic Colorectal Cancer: A Systematic Review and Meta-Analysis. Ann Oncol (2012) 23(6):1518–25. doi: 10.1093/annonc/mdr464
169. Mazouji O, Ouhajjou A, Incitti R, Mansour H. Updates on Clinical Use of Liquid Biopsy in Colorectal Cancer Screening, Diagnosis, Follow-Up, and Treatment Guidance. Front Cell Dev Biol (2021) 9:962. doi: 10.3389/fcell.2021.660924
170. Bronte G, Silvestris N, Castiglia M, Galvano A, Passiglia F, Sortino G, et al. New Findings on Primary and Acquired Resistance to Anti-EGFR Therapy in Metastatic Colorectal Cancer: Do All Roads Lead to RAS? Oncotarget (2015) 6(28):24780. doi: 10.18632/oncotarget.4959
171. Jhawer M, Goel S, Wilson AJ, Montagna C, Ling Y-H, Byun D-S, et al. PIK3CA Mutation/PTEN Expression Status Predicts Response of Colon Cancer Cells to the Epidermal Growth Factor Receptor Inhibitor Cetuximab. Cancer Res (2008) 68(6):1953–61. doi: 10.1158/0008-5472.CAN-07-5659
172. Wang G, He Y, Sun Y, Wang W, Qian X, Yu X, et al. Prevalence, Prognosis and Predictive Status of HER2 Amplification in Anti-EGFR-Resistant Metastatic Colorectal Cancer. Clin Transl Oncol (2020) 22(6):813–22. doi: 10.1007/s12094-019-02213-9
173. Li Q-H, Wang Y-Z, Tu J, Liu C-W, Yuan Y-J, Lin R, et al. Anti-EGFR Therapy in Metastatic Colorectal Cancer: Mechanisms and Potential Regimens of Drug Resistance. Gastroenterol Rep (2020) 8(3):179–91. doi: 10.1093/gastro/goaa026
174. Mariño-Enríquez A, Dal Cin P. ALK as a Paradigm of Oncogenic Promiscuity: Different Mechanisms of Activation and Different Fusion Partners Drive Tumors of Different Lineages. Cancer Genet (2013) 206(11):357–73. doi: 10.1016/j.cancergen.2013.07.001
175. Crago AM, Chmielecki J, Rosenberg M, O’Connor R, Byrne C, Wilder FG, et al. Near Universal Detection of Alterations in CTNNB1 and Wnt Pathway Regulators in Desmoid-Type Fibromatosis by Whole-Exome Sequencing and Genomic Analysis. Genes Chromosomes Cancer (2015) 54(10):606–15. doi: 10.1002/gcc.22272
176. Penel N, Chibon F, Salas S. Adult Desmoid Tumors: Biology, Management and Ongoing Trials. Curr Opin Oncol (2017) 29(4):268. doi: 10.1097/CCO.0000000000000374
177. Jiao Y, Shi C, Edil BH, De Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, MEN1, and mTOR Pathway Genes are Frequently Altered in Pancreatic Neuroendocrine Tumors. Science (2011) 331(6021):1199–203. doi: 10.1126/science.1200609
178. Yachida S, Vakiani E, White CM, Zhong Y, Saunders T, Morgan R, et al. Small Cell and Large Cell Neuroendocrine Carcinomas of the Pancreas are Genetically Similar and Distinct From Well-Differentiated Pancreatic Neuroendocrine Tumors. Am J Surg Pathol (2012) 36(2):173. doi: 10.1097/PAS.0b013e3182417d36
179. Konukiewitz B, Jesinghaus M, Steiger K, Schlitter AM, Kasajima A, Sipos B, et al. Pancreatic Neuroendocrine Carcinomas Reveal a Closer Relationship to Ductal Adenocarcinomas Than to Neuroendocrine Tumors G3. Hum Pathol (2018) 77:70–9. doi: 10.1016/j.humpath.2018.03.018
180. Konukiewitz B, Schlitter AM, Jesinghaus M, Pfister D, Steiger K, Segler A, et al. Somatostatin Receptor Expression Related to TP53 and RB1 Alterations in Pancreatic and Extrapancreatic Neuroendocrine Neoplasms With a Ki67-Index Above 20%. Mod Pathol (2017) 30(4):587–98. doi: 10.1038/modpathol.2016.217
181. Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, et al. Trends in the Incidence, Prevalence, and Survival Outcomes in Patients With Neuroendocrine Tumors in the United States. JAMA Oncol (2017) 3(10):1335–42. doi: 10.1001/jamaoncol.2017.0589
182. Klempner SJ, Gershenhorn B, Tran P, Lee TK, Erlander MG, Gowen K, et al. BRAFV600E Mutations in High-Grade Colorectal Neuroendocrine Tumors may Predict Responsiveness to BRAF–MEK Combination Therapy. Cancer Discovery (2016) 6(6):594–600. doi: 10.1158/2159-8290.CD-15-1192
183. Olevian DC, Nikiforova MN, Chiosea S, Sun W, Bahary N, Kuan S-F, et al. Colorectal Poorly Differentiated Neuroendocrine Carcinomas Frequently Exhibit BRAF Mutations and are Associated With Poor Overall Survival. Hum Pathol (2016) 49:124–34. doi: 10.1016/j.humpath.2015.11.004
184. Burkart J, Owen D, Shah MH, Abdel-Misih SR, Roychowdhury S, Wesolowski R, et al. Targeting BRAF Mutations in High-Grade Neuroendocrine Carcinoma of the Colon. J Natl Compr Canc Netw (2018) 16(9):1035–40. doi: 10.6004/jnccn.2018.7043
185. Idrees K, Padmanabhan C, Liu E, Guo Y, Gonzalez RS, Berlin J, et al. Frequent BRAF Mutations Suggest a Novel Oncogenic Driver in Colonic Neuroendocrine Carcinoma. J Surg Oncol (2018) 117(2):284–9. doi: 10.1002/jso.24834
186. Dizdar L, Werner TA, Drusenheimer JC, Möhlendick B, Raba K, Boeck I, et al. BRAFV600E Mutation: A Promising Target in Colorectal Neuroendocrine Carcinoma. Int J Cancer (2019) 144(6):1379–90. doi: 10.1002/ijc.31828
187. Chen L, Liu M, Zhang Y, Guo Y, Chen M, Chen J. Genetic Characteristics of Colorectal Neuroendocrine Carcinoma: More Similar to Colorectal Adenocarcinoma. Clin Colorectal Cancer (2021) 20(2):177–85. doi: 10.1016/j.clcc.2020.09.001
188. Project Genie Consortium AACR. AACR Project GENIE: Powering Precision Medicine Through an International Consortium. Cancer Discovery (2017) 7(8):818–31. doi: 10.1158/2159-8290.CD-17-0151
189. Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken J, Aderka D, et al. ESMO Consensus Guidelines for the Management of Patients With Metastatic Colorectal Cancer. Ann Oncol (2016) 27(8):1386–422. doi: 10.1093/annonc/mdw235
190. Scartozzi M, Vincent L, Chiron M, Cascinu S. Aflibercept, a New Way to Target Angiogenesis in the Second Line Treatment of Metastatic Colorectal Cancer (mCRC). Target Oncol (2016) 11(4):489–500. doi: 10.1007/s11523-016-0447-4
191. Lai E, Cascinu S, Scartozzi M. Are All Anti-Angiogenic Drugs the Same in the Treatment of Second-Line Metastatic Colorectal Cancer? Expert Opin Clin Pract Front Oncol (2021) 11:1626. doi: 10.3389/fonc.2021.637823
192. Loupakis F, Antonuzzo L, Bachet J-B, Kuan F-C, Macarulla T, Pietrantonio F, et al. Practical Considerations in the Use of Regorafenib in Metastatic Colorectal Cancer. Ther Adv Med Oncol (2020) 12:1758835920956862. doi: 10.1177/1758835920956862
193. Novakova-Jiresova A, Kopeckova K, Boublikova L, Chloupkova R, Melichar B, Petruzelka L, et al. Regorafenib for Metastatic Colorectal Cancer: An Analysis of a Registry-Based Cohort of 555 Patients. Cancer Manag Res (2020) 12:5365. doi: 10.2147/CMAR.S255332
194. García-Aranda M, Redondo M. Targeting Receptor Kinases in Colorectal Cancer. Cancers (2019) 11(4):433. doi: 10.3390/cancers11040433
195. Quan P-L, Sauzade M, Brouzes E. dPCR: A Technology Review. Sensors (2018) 18(4):1271. doi: 10.3390/s18041271
196. Huerta M, Roselló S, Sabater L, Ferrer A, Tarazona N, Roda D, et al. Circulating Tumor DNA Detection by Digital-Droplet PCR in Pancreatic Ductal Adenocarcinoma: A Systematic Review. Cancers (2021) 13(5):994. doi: 10.3390/cancers13050994
197. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An Ultrasensitive Method for Quantitating Circulating Tumor DNA With Broad Patient Coverage. Nat Med (2014) 20(5):548–54. doi: 10.1038/nm.3519
198. So M-K, Park J-H, Kim J-W, Jang J-H. Analytical Validation of a Pan-Cancer Panel for Cell-Free Assay for the Detection of EGFR Mutations. Diagnostics (2021) 11(6):1022. doi: 10.3390/diagnostics11061022
199. Chen M, Zhao H. Next-Generation Sequencing in Liquid Biopsy: Cancer Screening and Early Detection. Hum Genomics (2019) 13(1):1–10. doi: 10.1186/s40246-019-0220-8
200. Ding Y, Li W, Wang K, Xu C, Hao M, Ding L. Perspectives of the Application of Liquid Biopsy in Colorectal Cancer. BioMed Res Int (2020) 2020:6843180. doi: 10.1155/2020/6843180
201. García-Foncillas J, Tabernero J, Élez E, Aranda E, Benavides M, Camps C, et al. Prospective Multicenter Real-World RAS Mutation Comparison Between OncoBEAM-Based Liquid Biopsy and Tissue Analysis in Metastatic Colorectal Cancer. Br J Cancer (2018) 119(12):1464–70. doi: 10.1038/s41416-018-0293-5
202. Harle A, Gavoille C, Bouche O, Ben Abdelghani M, Plaza JE, Lambert A, et al. cfDNA for Accurate Determination of RAS and BRAF Mutations Using OncoBEAM Liquid Biopsy in Metastatic Colorectal Cancer Patients: Results of the Real-World Multicentric ColoBEAM Study. J Clin Oncol (2019) 37(15):3542–3542. doi: 10.1200/JCO.2019.37.15_suppl.3542
203. De Craene B, Van de Velde J, Bellon E, Gazin M, Rondelez E, Vandenbroeck L, et al. Detection of Microsatellite Instability (MSI) With a Novel Set of 7 Idylla Biomarkers on Colorectal Cancer Samples in a Multi-Center Study. Ann Oncol (2018) 29:viii51. doi: 10.1093/annonc/mdy269.160
204. Vidal J, Muinelo L, Dalmases A, Jones F, Edelstein D, Iglesias M, et al. Plasma ctDNA RAS Mutation Analysis for the Diagnosis and Treatment Monitoring of Metastatic Colorectal Cancer Patients. Ann Oncol (2017) 28(6):1325–32. doi: 10.1093/annonc/mdx125
205. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating Tumor DNA Analysis Detects Minimal Residual Disease and Predicts Recurrence in Patients With Stage II Colon Cancer. Sci Transl Med (2016) 8(346):346ra92–346ra92. doi: 10.1126/scitranslmed.aaf6219
206. Cheung KWE, Choi SR, Lee LTC, Lee NLE, Tsang HF, Cheng YT, et al. The Potential of Circulating Cell Free RNA as a Biomarker in Cancer. Expert Rev Mol Diagn (2019) 19(7):579–90. doi: 10.1080/14737159.2019.1633307
207. Galamb O, Barták BK, Kalmár A, Nagy ZB, Szigeti KA, Tulassay Z, et al. Diagnostic and Prognostic Potential of Tissue and Circulating Long non-Coding RNAs in Colorectal Tumors. World J Gastroenterol (2019) 25(34):5026. doi: 10.3748/wjg.v25.i34.5026
208. Pagani F, Randon G, Guarini V, Raimondi A, Prisciandaro M, Lobefaro R, et al. The Landscape of Actionable Gene Fusions in Colorectal Cancer. Int J Mol Sci (2019) 20(21):5319. doi: 10.3390/ijms20215319
209. Russell S, Norvig P. Artificial Intelligence: A Modern Approach. Pearson Education Limited, London (2002).
210. Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med (2019) 380(14):1347–58. doi: 10.1056/NEJMra1814259
211. Miller RA. Medical Diagnostic Decision Support Systems—Past, Present, and Future: A Threaded Bibliography and Brief Commentary. J Am Med Inform Assoc (1994) 1(1):8–27. doi: 10.1136/jamia.1994.95236141
212. De Dombal F, Leaper D, Staniland JR, McCann A, Horrocks JC. Computer-Aided Diagnosis of Acute Abdominal Pain. Br Med J (1972) 2(5804):9–13. doi: 10.1136/bmj.2.5804.9
213. Miller RA, McNeil MA, Challinor SM, Masarie FE Jr, Myers JD. The INTERNIST-1/Quick Medical REFERENCE Project—Status Report. West J Med (1986) 145(6):816.
214. Shortliffe EH, Davis R, Axline SG, Buchanan BG, Green CC, Cohen SN. Computer-Based Consultations in Clinical Therapeutics: Explanation and Rule Acquisition Capabilities of the MYCIN System. Comput BioMed Res (1975) 8(4):303–20. doi: 10.1016/0010-4809(75)90009-9
215. Berner ES, Webster GD, Shugerman AA, Jackson JR, Algina J, Baker AL, et al. Performance of Four Computer-Based Diagnostic Systems. N Engl J Med (1994) 330(25):1792–6. doi: 10.1056/NEJM199406233302506
218. Deo RC. Machine Learning in Medicine. Circulation (2015) 132(20):1920–30. doi: 10.1161/CIRCULATIONAHA.115.001593
219. Kotz S, Balakrishnan N, Read CB, Vidakovic B. Encyclopedia of Statistical Sciences, Volume 1. New Jersey: John Wiley & Sons (2005).
221. Yu K-H, Snyder M. Omics Profiling in Precision Oncology. Mol Cell Proteomics (2016) 15(8):2525–36. doi: 10.1074/mcp.O116.059253
222. Zhi J, Sun J, Wang Z, Ding W. Support Vector Machine Classifier for Prediction of the Metastasis of Colorectal Cancer. Int J Mol Med (2018) 41(3):1419–26. doi: 10.3892/ijmm.2018.3359
223. Everitt BS, Howell DC. Encyclopedia of Statistics in Behavioral Science–Volume 2. New Jersey: John Wiley & Sons, Ltd (2021).
225. Bae JH, Kim M, Lim J, Geem ZW. Feature Selection for Colon Cancer Detection Using K-Means Clustering and Modified Harmony Search Algorithm. Mathematics (2021) 9(5):570. doi: 10.3390/math9050570
227. Ruffle JK, Farmer AD, Aziz Q. Artificial Intelligence-Assisted Gastroenterology—Promises and Pitfalls. Off J Am Coll Gastroenterol ACG (2019) 114(3):422–8. doi: 10.1038/s41395-018-0268-4
228. Bhinder B, Gilvary C, Madhukar NS, Elemento O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discovery (2021) 11(4):900–15. doi: 10.1158/2159-8290.CD-21-0090
229. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A Survey on Deep Learning in Medical Image Analysis. Med Image Anal (2017) 42:60–88. doi: 10.1016/j.media.2017.07.005
230. Shen D, Wu G, Suk H-I. Deep Learning in Medical Image Analysis. Annu Rev BioMed Eng (2017) 19:221–48. doi: 10.1146/annurev-bioeng-071516-044442
231. Suzuki K. Overview of Deep Learning in Medical Imaging. Radiol Phys Technol (2017) 10(3):257–73. doi: 10.1007/s12194-017-0406-5
232. Alam IS, Steinberg I, Vermesh O, van den Berg NS, Rosenthal EL, van Dam GM, et al. Emerging Intraoperative Imaging Modalities to Improve Surgical Precision. Mol Imaging Biol (2018) 20(5):705–15. doi: 10.1007/s11307-018-1227-6
233. Angermueller C, Pärnamaa T, Parts L, Stegle O. Deep Learning for Computational Biology. Mol Syst Biol (2016) 12(7):878. doi: 10.15252/msb.20156651
234. Hamet P, Tremblay J. Artificial Intelligence in Medicine. Metabolism (2017) 69:S36–40. doi: 10.1016/j.metabol.2017.01.011
235. Wilson M, Middlebrook A, Sutton C, Stone R, McCloy R. MIST VR: A Virtual Reality Trainer for Laparoscopic Surgery Assesses Performance. Ann R Coll Surg Engl (1997) 79(6):403.
236. Khor WS, Baker B, Amin K, Chan A, Patel K, Wong J. Augmented and Virtual Reality in Surgery—the Digital Surgical Environment: Applications, Limitations and Legal Pitfalls. Ann Transl Med (2016) 4(23):454. doi: 10.21037/atm.2016.12.23
237. Volonté F, Buchs NC, Pugin F, Spaltenstein J, Schiltz B, Jung M, et al. Augmented Reality to the Rescue of the Minimally Invasive Surgeon. The Usefulness of the Interposition of Stereoscopic Images in the Da VinciTM Robotic Console. Int J Med Robot (2013) 9(3):e34–8. doi: 10.1002/rcs.1471
239. Miyake RK, Zeman HD, Duarte FH, Kikuchi R, Ramacciotti E, Lovhoiden G, et al. Vein Imaging: A New Method of Near Infrared Imaging, Where a Processed Image is Projected Onto the Skin for the Enhancement of Vein Treatment. Dermatol Surg (2006) 32(8):1031–8. doi: 10.1111/j.1524-4725.2006.32226.x
240. Alberti O, Dorward N, Kitchen N, Thomas D. Neuronavigation–impact on Operating Time. Stereotact Funct Neurosurg (1997) 68(1–4):44–8. doi: 10.1159/000099901
241. Lauby-Secretan B, Vilahur N, Bianchini F, Guha N, Straif K. The IARC Perspective on Colorectal Cancer Screening. N Engl J Med (2018) 378(18):1734–40. doi: 10.1056/NEJMsr1714643
242. Hewitson P, Glasziou PP, Irwig L, Towler B, Watson E. Screening for Colorectal Cancer Using the Faecal Occult Blood Test, Hemoccult. Cochrane Database Syst Rev (2007) 1:CD001216. doi: 10.1002/14651858.CD001216.pub2
243. Doubeni CA, Corley DA, Quinn VP, Jensen CD, Zauber AG, Goodman M, et al. Effectiveness of Screening Colonoscopy in Reducing the Risk of Death From Right and Left Colon Cancer: A Large Community-Based Study. Gut (2018) 67(2):291–8. doi: 10.1136/gutjnl-2016-312712
244. Smith RA, Andrews KS, Brooks D, Fedewa SA, Manassaram-Baptiste D, Saslow D, et al. Cancer Screening in the United States, 2019: A Review of Current American Cancer Society Guidelines and Current Issues in Cancer Screening. CA Cancer J Clin (2019) 69(3):184–210. doi: 10.3322/caac.21557
245. Issa IA, Noureddine M. Colorectal Cancer Screening: An Updated Review of the Available Options. World J Gastroenterol (2017) 23(28):5086. doi: 10.3748/wjg.v23.i28.5086
246. Nartowt BJ, Hart GR, Muhammad W, Liang Y, Stark GF, Deng J. Robust Machine Learning for Colorectal Cancer Risk Prediction and Stratification. Front Big Data (2020) 3:6. doi: 10.3389/fdata.2020.00006
247. Singh H, Turner D, Xue L, Targownik LE, Bernstein CN. Risk of Developing Colorectal Cancer Following a Negative Colonoscopy Examination: Evidence for a 10-Year Interval Between Colonoscopies. Jama (2006) 295(20):2366–73. doi: 10.1001/jama.295.20.2366
248. Brenner H, Chang-Claude J, Seiler CM, Rickert A, Hoffmeister M. Protection From Colorectal Cancer After Colonoscopy: A Population-Based, Case–Control Study. Ann Intern Med (2011) 154(1):22–30. doi: 10.7326/0003-4819-154-1-201101040-00004
249. Baxter NN, Goldwasser MA, Paszat LF, Saskin R, Urbach DR, Rabeneck L. Association of Colonoscopy and Death From Colorectal Cancer. Ann Intern Med (2009) 150(1):1–8. doi: 10.7326/0003-4819-150-1-200901060-00306
250. Kahi CJ, Imperiale TF, Juliar BE, Rex DK. Effect of Screening Colonoscopy on Colorectal Cancer Incidence and Mortality. Clin Gastroenterol Hepatol (2009) 7(7):770–5. doi: 10.1016/j.cgh.2008.12.030
251. Mori Y, Kudo S, Berzin TM, Misawa M, Takeda K. Computer-Aided Diagnosis for Colonoscopy. Endoscopy (2017) 49(08):813–9. doi: 10.1055/s-0043-109430
252. Nogueira-Rodríguez A, Domínguez-Carbajales R, Campos-Tato F, Herrero J, Puga M, Remedios D, et al. Real-Time Polyp Detection Model Using Convolutional Neural Networks. Neural Comput Appl (2021), 1–22. doi: 10.1007/s00521-021-06496-4
253. Wang P, Berzin TM, Brown JRG, Bharadwaj S, Becq A, Xiao X, et al. Real-Time Automatic Detection System Increases Colonoscopic Polyp and Adenoma Detection Rates: A Prospective Randomised Controlled Study. Gut (2019) 68(10):1813–9. doi: 10.1136/gutjnl-2018-317500
254. Mori Y, Kudo S, Misawa M, Saito Y, Ikematsu H, Hotta K, et al. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med (2018) 169(6):357–66. doi: 10.7326/M18-0249
255. Lefere P, Gryspeerdt S, Schotte K. Virtual Colonoscopy: An Overview. Oncol Res Treat (2006) 29(6):281–6. doi: 10.1159/000093125
256. Vining D. Technical Feasibility of Colon Imaging With Helical CT and Virtual Reality. AJR. Am J Roentgenol, Leesburg, VA (1994).
257. Halligan S, Altman DG, Taylor SA, Mallett S, Deeks JJ, Bartram CI, et al. CT Colonography in the Detection of Colorectal Polyps and Cancer: Systematic Review, Meta-Analysis, and Proposed Minimum Data Set for Study Level Reporting. Radiology (2005) 237(3):893–904. doi: 10.1148/radiol.2373050176
258. O’Brien MJ, Winawer SJ, Zauber AG, Bushey MT, Sternberg SS, Gottlieb LS, et al. Flat Adenomas in the National Polyp Study: Is There Increased Risk for High-Grade Dysplasia Initially or During Surveillance? Clin Gastroenterol Hepatol (2004) 2(10):905–11. doi: 10.1016/S1542-3565(04)00392-1
259. Wang Y, He X, Nie H, Zhou J, Cao P, Ou C. Application of Artificial Intelligence to the Diagnosis and Therapy of Colorectal Cancer. Am J Cancer Res (2020) 10(11):3575. doi: 10.7150/thno.49168
260. Blanes-Vidal V, Baatrup G, Nadimi ES. Addressing Priority Challenges in the Detection and Assessment of Colorectal Polyps From Capsule Endoscopy and Colonoscopy in Colorectal Cancer Screening Using Machine Learning. Acta Oncol (2019) 58(sup1):S29–36. doi: 10.1080/0284186X.2019.1584404
261. Hosoe N, Limpias Kamiya KJ, Hayashi Y, Sujino T, Ogata H, Kanai T. Current Status of Colon Capsule Endoscopy. Dig Endosc (2021) 33(4):529–37. doi: 10.1111/den.13769
262. Wester H-J, Schottelius M. PSMA-Targeted Radiopharmaceuticals for Imaging and Therapy. Amsterdam: Elsevier (2019) p. 302–12.
263. Weissleder R. Molecular Imaging: Exploring the Next Frontier. Radiology (1999) 212(3):609–14. doi: 10.1148/radiology.212.3.r99se18609
264. van Leeuwen FW, Schottelius M, Brouwer OR, Vidal-Sicart S, Achilefu S, Klode J, et al. Trending: Radioactive and Fluorescent Bimodal/Hybrid Tracers as Multiplexing Solutions for Surgical Guidance. J Nucl Med (2020) 61(1):13–9. doi: 10.2967/jnumed.119.228684
265. Mondal SB, O’Brien CM, Bishop K, Fields RC, Margenthaler JA, Achilefu S. Repurposing Molecular Imaging and Sensing for Cancer Image–Guided Surgery. J Nucl Med (2020) 61(8):1113–22. doi: 10.2967/jnumed.118.220426
266. Povoski SP, Neff RL, Mojzisik CM, O’Malley DM, Hinkle GH, Hall NC, et al. A Comprehensive Overview of Radioguided Surgery Using Gamma Detection Probe Technology. World J Surg Oncol (2009) 7(1):1–63. doi: 10.1186/1477-7819-7-11
267. Van Oosterom MN, Rietbergen DD, Welling MM, van der Poel HG, Maurer T, Van Leeuwen FW. Recent Advances in Nuclear and Hybrid Detection Modalities for Image-Guided Surgery. Expert Rev Med Devices (2019) 16(8):711–34. doi: 10.1080/17434440.2019.1642104
268. van Leeuwen FW, Hardwick JC, van Erkel AR. Luminescence-Based Imaging Approaches in the Field of Interventional Molecular Imaging. Radiology (2015) 276(1):12–29. doi: 10.1148/radiol.2015132698
269. Wang C, Liu C, Chang Y, Lafata K, Cui Y, Zhang J, et al. Dose-Distribution-Driven PET Image-Based Outcome Prediction (DDD-PIOP): A Deep Learning Study for Oropharyngeal Cancer IMRT Application. Front Oncol (2020) 10:1592. doi: 10.3389/fonc.2020.01592
270. Kudo S, Mori Y, Wakamura K, Ikehara N, Ichimasa K, Wada Y, et al. Endocytoscopy can Provide Additional Diagnostic Ability to Magnifying Chromoendoscopy for Colorectal Neoplasms. J Gastroenterol Hepatol (2014) 29(1):83–90. doi: 10.1111/jgh.12374
271. Häfner M, Liedlgruber M, Uhl A, Vécsei A, Wrba F. Delaunay Triangulation-Based Pit Density Estimation for the Classification of Polyps in High-Magnification Chromo-Colonoscopy. Comput Methods Programs BioMed (2012) 107(3):565–81. doi: 10.1016/j.cmpb.2011.12.012
272. Takemura Y, Yoshida S, Tanaka S, Onji K, Oka S, Tamaki T, et al. Quantitative Analysis and Development of a Computer-Aided System for Identification of Regular Pit Patterns of Colorectal Lesions. Gastrointest Endosc (2010) 72(5):1047–51. doi: 10.1016/j.gie.2010.07.037
273. Neumann H, Fuchs FS, Vieth M, Atreya R, Siebler J, Kiesslich R, et al. In Vivo Imaging by Endocytoscopy. Aliment Pharmacol Ther (2011) 33(11):1183–93. doi: 10.1111/j.1365-2036.2011.04647.x
274. Mori Y, Kudo S, Wakamura K, Misawa M, Ogawa Y, Kutsukawa M, et al. Novel Computer-Aided Diagnostic System for Colorectal Lesions by Using Endocytoscopy (With Videos). Gastrointest Endosc (2015) 81(3):621–9. doi: 10.1016/j.gie.2014.09.008
275. Mori Y, Kudo S, Chiu PWY, Singh R, Misawa M, Wakamura K, et al. Impact of an Automated System for Endocytoscopic Diagnosis of Small Colorectal Lesions: An International Web-Based Study. Endoscopy (2016) 48(12):1110–8. doi: 10.1055/s-0042-113609
276. Neumann H, Kiesslich R, Wallace MB, Neurath MF. Confocal Laser Endomicroscopy: Technical Advances and Clinical Applications. Gastroenterology (2010) 139(2):388–92. doi: 10.1053/j.gastro.2010.06.029
277. André B, Vercauteren T, Buchner AM, Krishna M, Ayache N, Wallace MB. Software for Automated Classification of Probe-Based Confocal Laser Endomicroscopy Videos of Colorectal Polyps. World J Gastroenterol WJG (2012) 18(39):5560. doi: 10.3748/wjg.v18.i39.5560
278. Kuiper T, Alderlieste YA, Tytgat KM, Vlug MS, Nabuurs JA, Bastiaansen BA, et al. Automatic Optical Diagnosis of Small Colorectal Lesions by Laser-Induced Autofluorescence. Endoscopy (2015) 47(01):56–62. doi: 10.1055/s-0034-1378112
279. Rath T, Tontini GE, Vieth M, Nägel A, Neurath MF, Neumann H. In Vivo Real-Time Assessment of Colorectal Polyp Histology Using an Optical Biopsy Forceps System Based on Laser-Induced Fluorescence Spectroscopy. Endoscopy (2016) 48(06):557–62. doi: 10.1055/s-0042-102251
280. Aihara H, Saito S, Inomata H, Ide D, Tamai N, Ohya TR, et al. Computer-Aided Diagnosis of Neoplastic Colorectal Lesions Using ‘Real-Time’numerical Color Analysis During Autofluorescence Endoscopy. Eur J Gastroenterol Hepatol (2013) 25(4):488–94. doi: 10.1097/MEG.0b013e32835c6d9a
281. Inomata H, Tamai N, Aihara H, Sumiyama K, Saito S, Kato T, et al. Efficacy of a Novel Auto-Fluorescence Imaging System With Computer-Assisted Color Analysis for Assessment of Colorectal Lesions. World J Gastroenterol WJG (2013) 19(41):7146. doi: 10.3748/wjg.v19.i41.7146
282. Bhayani SB, Das N. Robotic Assisted Laparoscopic Partial Nephrectomy for Suspected Renal Cell Carcinoma: Retrospective Review of Surgical Outcomes of 35 Cases. BMC Surg (2008) 8(1):1–4. doi: 10.1186/1471-2482-8-16
283. Porpiglia F, Fiori C, Checcucci E, Amparore D, Bertolo R. Hyperaccuracy Three-Dimensional Reconstruction is Able to Maximize the Efficacy of Selective Clamping During Robot-Assisted Partial Nephrectomy for Complex Renal Masses. Eur Urol (2018) 74(5):651–60. doi: 10.1016/j.eururo.2017.12.027
284. MacCuaig WM, Jones MA, Abeyakoon O, McNally LR. Development of Multispectral Optoacoustic Tomography as a Clinically Translatable Modality for Cancer Imaging. Radiol Imaging Cancer (2020) 2(6):e200066. doi: 10.1148/rycan.2020200066
285. Ferrer-Roca O. Telepathology and Optical Biopsy. Int J Telemed Appl (2009) 2009:740712. doi: 10.1155/2009/740712
286. Pinto M, Zorn KC, Tremblay J-P, Desroches J, Dallaire F, Aubertin K, et al. Integration of a Raman Spectroscopy System to a Robotic-Assisted Surgical System for Real-Time Tissue Characterization During Radical Prostatectomy Procedures. J BioMed Opt (2019) 24(2):025001. doi: 10.1117/1.JBO.24.2.025001
287. Acs B, Rantalainen M, Hartman J. Artificial Intelligence as the Next Step Towards Precision Pathology. J Intern Med (2020) 288(1):62–81. doi: 10.1111/joim.13030
288. Rathore S, Hussain M, Iftikhar MA, Jalil A. Novel Structural Descriptors for Automated Colon Cancer Detection and Grading. Comput Methods Programs BioMed (2015) 121(2):92–108. doi: 10.1016/j.cmpb.2015.05.008
289. Rathore S, Iftikhar MA, Chaddad A, Niazi T, Karasic T, Bilello M. Segmentation and Grade Prediction of Colon Cancer Digital Pathology Images Across Multiple Institutions. Cancers (2019) 11(11):1700. doi: 10.3390/cancers11111700
290. Chang HY, Jung CK, Woo JI, Lee S, Cho J, Kim SW, et al. Artificial Intelligence in Pathology. J Pathol Transl Med (2019) 53(1):1. doi: 10.4132/jptm.2018.12.16
291. Sirinukunwattana K, Raza SEA, Tsang Y-W, Snead DR, Cree IA, Rajpoot NM. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans Med Imaging (2016) 35(5):1196–206. doi: 10.1109/TMI.2016.2525803
292. Abdelsamea MM, Grineviciute RB, Besusparis J, Cham S, Pitiot A, Laurinavicius A, et al. Tumour Parcellation and Quantification (TuPaQ): A Tool for Refining Biomarker Analysis Through Rapid and Automated Segmentation of Tumour Epithelium. Histopathology (2019) 74(7):1045–54. doi: 10.1111/his.13838
293. Yu G, Sun K, Xu C, Shi X-H, Wu C, Xie T, et al. Accurate Recognition of Colorectal Cancer With Semi-Supervised Deep Learning on Pathological Images. Nat Commun (2021) 12(1):1–13. doi: 10.1038/s41467-021-26643-8
294. Kim Y-G, Kim S, Cho CE, Song IH, Lee HJ, Ahn S, et al. Effectiveness of Transfer Learning for Enhancing Tumor Classification With a Convolutional Neural Network on Frozen Sections. Sci Rep (2020) 10(1):1–9. doi: 10.1038/s41598-020-78129-0
295. Zhou M, Wang X, Jiang L, Chen X, Bao X, Chen X. The Diagnostic Value of One Step Nucleic Acid Amplification (OSNA) in Differentiating Lymph Node Metastasis of Tumors: A Systematic Review and Meta-Analysis. Int J Surg (2018) 56:49–56. doi: 10.1016/j.ijsu.2018.05.010
296. Hu H, Niu Z, Bai Y, Tan X. Cancer Classification Based on Gene Expression Using Neural Networks. Genet Mol Res (2015) 14(4):17605–11. doi: 10.4238/2015.December.21.33
297. Xu G, Zhang M, Zhu H, Xu J. A 15-Gene Signature for Prediction of Colon Cancer Recurrence and Prognosis Based on SVM. Gene (2017) 604:33–40. doi: 10.1016/j.gene.2016.12.016
298. Zhang X, Yang Y, Wang Y, Fan Q. Detection of the BRAF V600E Mutation in Colorectal Cancer by NIR Spectroscopy in Conjunction With Counter Propagation Artificial Neural Network. Molecules (2019) 24(12):2238. doi: 10.3390/molecules24122238
299. Wang Q, Wei J, Chen Z, Zhang T, Zhong J, Zhong B, et al. Establishment of Multiple Diagnosis Models for Colorectal Cancer With Artificial Neural Networks. Oncol Lett (2019) 17(3):3314–22. doi: 10.3892/ol.2019.10010
300. Wan N, Weinberg D, Liu T-Y, Niehaus K, Ariazi EA, Delubac D, et al. Machine Learning Enables Detection of Early-Stage Colorectal Cancer by Whole-Genome Sequencing of Plasma Cell-Free DNA. BMC Cancer (2019) 19(1):1–10. doi: 10.1186/s12885-019-6003-8
301. Chang KH, Miller N, Kheirelseid EA, Lemetre C, Ball GR, Smith MJ, et al. MicroRNA Signature Analysis in Colorectal Cancer: Identification of Expression Profiles in Stage II Tumors Associated With Aggressive Disease. Int J Colorectal Dis (2011) 26(11):1415–22. doi: 10.1007/s00384-011-1279-4
302. Amirkhah R, Farazmand A, Gupta SK, Ahmadi H, Wolkenhauer O, Schmitz U. Naive Bayes Classifier Predicts Functional microRNA Target Interactions in Colorectal Cancer. Mol Biosyst (2015) 11(8):2126–34. doi: 10.1039/C5MB00245A
303. Herreros-Villanueva M, Duran-Sanchon S, Martín AC, Pérez-Palacios R, Vila-Navarro E, Marcuello M, et al. Plasma microRNA Signature Validation for Early Detection of Colorectal Cancer. Clin Transl Gastroenterol (2019) 10(1):e00003. doi: 10.14309/ctg.0000000000000003
304. Afshar S, Afshar S, Warden E, Manochehri H, Saidijam M. Application of Artificial Neural Network in miRNA Biomarker Selection and Precise Diagnosis of Colorectal Cancer. Iran BioMed J (2019) 23(3):175. doi: 10.29252/ibj.23.3.175
305. Xuan P, Dong Y, Guo Y, Zhang T, Liu Y. Dual Convolutional Neural Network Based Method for Predicting Disease-Related miRNAs. Int J Mol Sci (2018) 19(12):3732. doi: 10.3390/ijms19123732
306. Ge P, Wang W, Li L, Zhang G, Gao Z, Tang Z, et al. Profiles of Immune Cell Infiltration and Immune-Related Genes in the Tumor Microenvironment of Colorectal Cancer. BioMed Pharmacother (2019) 118:109228. doi: 10.1016/j.biopha.2019.109228
307. Liu L, Chen X, Petinrin OO, Zhang W, Rahaman S, Tang Z-R, et al. Machine Learning Protocols in Early Cancer Detection Based on Liquid Biopsy: A Survey. Life (2021) 11(7):638. doi: 10.3390/life11070638
308. Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine Learning Applications in Cancer Prognosis and Prediction. Comput Struct Biotechnol J (2015) 13:8–17. doi: 10.1016/j.csbj.2014.11.005
309. Pérez MJ, Grande RG. Application of Artificial Intelligence in the Diagnosis and Treatment of Hepatocellular Carcinoma: A Review. World J Gastroenterol (2020) 26(37):5617. doi: 10.3748/wjg.v26.i37.5617
310. Jin P, Ji X, Kang W, Li Y, Liu H, Ma F, et al. Artificial Intelligence in Gastric Cancer: A Systematic Review. J Cancer Res Clin Oncol (2020) 146(9):2339–50. doi: 10.1007/s00432-020-03304-9
311. Ivancic MM, Megna BW, Sverchkov Y, Craven M, Reichelderfer M, Pickhardt PJ, et al. Noninvasive Detection of Colorectal Carcinomas Using Serum Protein Biomarkers. J Surg Res (2020) 246:160–9. doi: 10.1016/j.jss.2019.08.004
312. Hussain A, Malik A, Halim MU, Ali AM. The Use of Robotics in Surgery: A Review. Int J Clin Pract (2014) 68(11):1376–82. doi: 10.1111/ijcp.12492
314. Spanheimer PM, Armstrong JG, Fu S, Liao J, Regenbogen SE, Byrn JC. Robotic Proctectomy for Rectal Cancer: Analysis of 71 Patients From a Single Institution. Int J Med Robot (2017) 13(4):e1841. doi: 10.1002/rcs.1841
315. Zawadzki M, Krzystek-Korpacka M, Gamian A, Witkiewicz W. Comparison of Inflammatory Responses Following Robotic and Open Colorectal Surgery: A Prospective Study. Int J Colorectal Dis (2017) 32(3):399–407. doi: 10.1007/s00384-016-2697-0
316. Yoon SN, Kim KY, Kim JW, Lee SC, Kwon YJ, Cho JW, et al. Comparison of Short-and Long-Term Outcomes of an Early Experience With Robotic and Laparoscopic-Assisted Resection for Rectal Cancer. Hepatogastroenterology (2015) 62(137):34–9.
317. Connor TH, DeBord DG, Pretty JR, Oliver MS, Roth TS, Lees PS, et al. Evaluation of Antineoplastic Drug Exposure of Health Care Workers at Three University-Based US Cancer Centers. J Occup Environ Med (2010) 52(10):1019–27. doi: 10.1097/JOM.0b013e3181f72b63
318. Martini M, Pelati C, Ghirardini A. La Gestione Del Rischio Clinico. New York: McGraw-Hill (2011).
319. Masini C, Nanni O, Antaridi S, Gallegati D, Marri M, Paolucci D, et al. Automated Preparation of Chemotherapy: Quality Improvement and Economic Sustainability. Am J Health Syst Pharm (2014) 71(7):579–85. doi: 10.2146/ajhp130489
320. Kaushal R, Bates D. Information Technology and Medication Safety: What is the Benefit? BMJ Qual Saf (2002) 11(3):261–5. doi: 10.1136/qhc.11.3.261
321. Felfoul O, Mohammadi M, Taherkhani S, De Lanauze D, Xu YZ, Loghin D, et al. Magneto-Aerotactic Bacteria Deliver Drug-Containing Nanoliposomes to Tumour Hypoxic Regions. Nat Nanotechnol (2016) 11(11):941–7. doi: 10.1038/nnano.2016.137
322. Cruz S, Gomes SE, Borralho PM, Rodrigues CM, Gaudêncio SP, Pereira F. In Silico HCT116 Human Colon Cancer Cell-Based Models En Route to the Discovery of Lead-Like Anticancer Drugs. Biomolecules (2018) 8(3):56. doi: 10.3390/biom8030056
323. Sartore-Bianchi A, Pietrantonio F, Lonardi S, Mussolin B, Rua F, Fenocchio E, et al. Phase II Study of Anti-EGFR Rechallenge Therapy With Panitumumab Driven by Circulating Tumor DNA Molecular Selection in Metastatic Colorectal Cancer: The CHRONOS Trial. J Clin Oncol (2021) 39(15):3506–3506. doi: 10.1200/JCO.2021.39.15_suppl.3506
Keywords: liquid biopsy, colorectal cancer, patients stratification, artificial intelligence, robotic surgery
Citation: Ginghina O, Hudita A, Zamfir M, Spanu A, Mardare M, Bondoc I, Buburuzan L, Georgescu SE, Costache M, Negrei C, Nitipir C and Galateanu B (2022) Liquid Biopsy and Artificial Intelligence as Tools to Detect Signatures of Colorectal Malignancies: A Modern Approach in Patient’s Stratification. Front. Oncol. 12:856575. doi: 10.3389/fonc.2022.856575
Received: 17 January 2022; Accepted: 16 February 2022;
Published: 08 March 2022.
Edited by:
Stefano Guadagni, University of L’Aquila, ItalyReviewed by:
Ioannis Papasotiriou, RGCC international GmbH, SwitzerlandGane Ka-Shu Wong, University of Alberta, Canada
Gemma Bruera, University of L’Aquila, Italy
Copyright © 2022 Ginghina, Hudita, Zamfir, Spanu, Mardare, Bondoc, Buburuzan, Georgescu, Costache, Negrei, Nitipir and Galateanu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ariana Hudita, YXJpYW5hLmh1ZGl0YUBiaW8udW5pYnVjLnJv
 Octav Ginghina
Octav Ginghina Ariana Hudita
Ariana Hudita Marius Zamfir2
Marius Zamfir2 Sergiu Emil Georgescu
Sergiu Emil Georgescu Carolina Negrei
Carolina Negrei Bianca Galateanu
Bianca Galateanu