- 1Department of Urology, Affiliated Zhongda Hospital of Southeast University, Nanjing, China
- 2Surgical Research Center, Institute of Urology, Southeast University Medical School, Nanjing, China
- 3Sichuan Cancer Hospital & Institute, Sichuan Cancer Center, Cancer Hospital affiliate to School of Medicine, UESTC, Chengdu, China
- 4Department of Urology, Wuxi No.2 People’s Hospital of Nanjing Medical University, Wuxi, China
- 5Nanjing Lishui District People’s Hospital, Zhongda Hospital Lishui Branch, Southeast University, Nanjing, China
Background: BAP1 is an important tumor suppressor involved in various biological processes and is commonly lost or inactivated in clear-cell renal cell carcinoma (ccRCC). However, the role of the BAP1-deficient tumor competing endogenous RNA (ceRNA) network involved in ccRCC remains unclear. Thus, this study aims to investigate the prognostic BAP1-related ceRNA in ccRCC.
Methods: Raw data was obtained from the TCGA and the differentially expressed genes were screened to establish a BAP1-related ceRNA network. Subsequently, the role of the ceRNA axis was validated using phenotypic experiments. Dual-luciferase reporter assays and fluorescence in situ hybridization (FISH) assays were used to confirm the ceRNA network.
Results: Nuclear enriched abundant transcript 1 (NEAT1) expression was significantly increased in kidney cancer cell lines. NEAT1 knockdown significantly inhibited cell proliferation and migration, which could be reversed by miR-10a-5p inhibitor. Dual-luciferase reporter assay confirmed miR-10a-5p as a common target of NEAT1 and Serine protease inhibitor family E member 1 (SERPINE1). FISH assays revealed the co-localization of NEAT1 and miR-10a-5p in the cytoplasm. Additionally, the methylation level of SERPINE1 in ccRCC was significantly lower than that in normal tissues. Furthermore, SERPINE1 expression was positively correlated with multiple immune cell infiltration levels.
Conclusions: In BAP1-deficient ccRCC, NEAT1 competitively binds to miR-10a-5p, indirectly upregulating SERPINE1 expression to promote kidney cancer cell proliferation. Furthermore, NEAT1/miR-10a-5p/SERPINE1 were found to be independent prognostic factors of ccRCC.
Introduction
Kidney cancer is a common malignancy, with approximately 430,000 new global cases in 2020 and approximately 170,000 kidney cancer-related deaths (1). The pathology of clear-cell renal cell carcinoma (ccRCC) is characterized by a ‘clear cytoplasm’, owing to its ability to accumulate glycogen and lipids in the cytoplasm. It accounts for up to 80% of all renal cell carcinomas (RCC) and is also considered the most aggressive subtype. Loss or inactivation of tumor suppressors is crucial in tumorigenesis. The role of the classic VHL gene and its pathway in ccRCC have been extensively studied (2). Moreover, drugs targeting the VHL–HIF–VEGF pathway, such as sunitinib, sorafenib and axitinib, have been shown to benefit patients with ccRCC, becoming the standard treatment for patients in the advanced stages of ccRCC (3). Unlike other epithelial tumors, mutations in the classic tumor suppressors, such as BRAF, TP53, PTEN, RB1, and EGFR, are rare in ccRCC (4, 5). In addition to VHL inactivation, a recurrent loss of chromosome 3p fragments in ccRCC has been reported (6, 7). Furthermore, BRCA1-Associated Protein 1 (BAP1) on chromosome 3p was identified as a novel tumor-driver gene in ccRCC using extensive parallel sequencing techniques (8, 9).
BAP1 is a novel ubiquitin carboxy-terminal hydrolase and a subfamily member of deubiquitinating enzymes (10, 11). BAP1 is located in the nucleus and binds to BRCA1 to enhance its tumor suppressor activity (12). Additionally, BAP1 is involved in many biological processes, such as DNA damage repair, cell differentiation and cell proliferation, via its deubiquitinating activity (12). Various studies have reported that BAP1 is commonly lost or inactivated in numerous human malignancies, especially in ccRCC, hepatocellular carcinoma and mesothelioma (13). The majority of chromosomes have a 3p deletion, which is an initial marker for nonhereditary ccRCC (14). The mutated frequency of BAP1 was also reported to be as high as 20% in ccRCC (15, 16), with RCC accounting for 9% of BAP1 tumor predisposition syndrome (BAP1-TPDS) (17). Compared with sporadic tumors, BAP1-TPDS RCC has earlier onset, more aggressive tumors and poorer patient survival (17, 18). Due to the poor treatment response, a standard treatment for BAP1 mutated tumors is yet to be identified (19). Recently, several studies have reported the direct targeting of differentially expressed genes in BAP1-deficient tumors (20, 21). For example, histone deacetylase and enhancer of Zeste 2 Polycomb repressive complex 2 are upregulated in BAP1-deficient tumors; therefore, targeting these genes could improve BAP1-deficient tumor sensitivity to treatment (22, 23). Therefore, understanding differentially expressed genes in BAP1-deficient tumors could provide a novel perspective for targeted therapy.
Long noncoding RNAs (lncRNA), longer than 200 nucleotides (nt), are non-protein-coding RNAs that are involved in various tumor developments, including ccRCC, and specific lncRNAs are associated with tumor migration, invasion and poor prognosis (24, 25). MicroRNA (miRNA) is a single-stranded non-coding RNA of approximately 19–25 nt, which can bind to the 3’ untranslated mRNA region and regulate target gene expression (26). Non-coding RNAs (namely, lncRNAs and circular RNAs) could serve as competitive endogenous RNAs (ceRNA) that competitively bind to miRNAs (a post-transcriptional regulator) for cell–cell communication and gene expression co-regulation (27–30).
Methods and Materials
Data Collection and Processing
RNAseq (lncRNA, mRNA and miRNA) and relevant clinical data for kidney renal clear cell carcinoma (KIRC) were obtained from the TCGA database (https://portal.gdc.cancer.gov/).
Identification of Differentially Expressed lncRNAs, miRNAs and mRNAs
Based on the median expression of BAP1, patients were divided into the BAP1high and the BAP1low groups. For differential expression analysis, the cutoff value of DElncRNA was set at |log2 FC| >0.5 and P. adj <0.05; DEmiRNA with cutoff value of |log2 FC| >0.7 and P. adj <0.05; DEmRNA with cutoff value of |log2 FC| >0.5 and P. adj <0.05.
Construction of ceRNA Networks
Highly conserved microRNA family files were downloaded from the miRcode database (http://mircode.org/). Subsequently, the obtained DElncRNAs were used to find potential miRNAs targeting these DElncRNAs. Then, the selected miRNAs were inputted into the miRBD database (http://mirdb.org/) (31) and the Targetscan database (http://www.targetscan.org/vert_72/) (32) to explore target mRNAs. Finally, the screened DElncRNAs, DEmiRNAs, and DEmRNAs were put into Cytoscape (version 3.6.1) for ceRNA network construction, using the plug-in ‘cytoHubba’ for hub genes network construction (33, 34).
Functional Annotation
To investigate the functional annotation implicated with DEmRNAs, the Gene Ontology annotation and the Kyoto Encyclopedia of Genes and Genomes pathway analysis were performed using the Metascape website (https://metascape.org/) (35).
Expression of Hub Genes and Survival Analysis
The expression of the hub genes in tumor and normal tissues based on the ccRCC dataset were compared using the Wilcoxon rank-sum test (P <0.05). Overall survival (OS) analysis for the expression of the hub genes between the high- and low-expression groups was performed, with P <0.05 indicating statistical significance.
Clinical Relevance of the Nuclear Enriched Abundant Transcript 1 (NEAT1)/miR-10a-5p/SERPINE1 Axis in Patients With ccRCC
To explore the clinical relevance of the ceRNA axis, the expression levels of NEAT1, miR-10a-5p, and SERPINE1 with different clinical characteristics were evaluated, and the Bonferroni method was used to correct the results of multiple hypothesis testing (Dunn’s test, P. adj <0.05). Moreover, univariate and multivariate Cox regression analyses were conducted to investigate the prognostic significance of clinical features.
Cell Culture
The ACHN and 786-O cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). Both cell lines were cultured in Dulbecco’s modified Eagle’s medium (Gibco) containing 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin and incubated at 37°C in a humidified 5% CO2 atmosphere.
Cell Transfection
Cells were seeded into six-well plates and cultured until the cell density reached approximately 60%. The cells were then transfected with small-interfering RNA (siRNA-NEAT1#1, #2) or miR-10a-5p mimic/inhibitor, using jetPRIME® transfection reagent (NY, USA) following the protocol of the manufacturer. Additionally, a relevant negative control (NC) was used. These siRNAs, miR-10a-5p mimic/inhibitor and relevant NC were designed by GenePharma (Shanghai, China).
RT-qPCR (Quantitative Reverse Transcription-Polymerase Chain Reaction) and Western Blotting
Total RNA was extracted using RNAeasy™ (Beyotime; Shanghai, China), following the instructions of the manufacturer. HiScript® II reverse transcriptase (Vazyme; Nanjing, China) was used to convert RNA to cDNA. RT-qPCR was performed in a LineGene 9600 Plus system (Bioer Technology, Hangzhou, China), using 2 × SYBR Green qPCR Master Mix (High ROX; Servicebio; Wuhan, China), following the protocol of the manufacturer. The expression of miR-10a-5p was normalized to control U6, while that of the others were normalized to GAPDH, and relative expression was calculated using the 2−ΔΔCt method. Western blotting was performed as previously described (36), and anti-SERPINE1 (Rabbit Polyclonal Antibody, AF7965, Beyotime), anti-GAPDH (Mouse Monoclonal Antibody, AF5009, Beyotime) antibodies were used for further experimentation.
Cell Viability
After 48 h of transfection, 100 µl cell suspension containing 1 × 103 cells were seeded into 96-well plates and cultured. After 24, 48, 72, and 96 h, 10 µl of CCK-8 (Beyotime; Shanghai, China) was added to each well and incubated at 37°C for another 2 h. The absorbance was detected using a microplate reader at 450 nm.
Colony Formation Assay
After transfection for 48 h, the cell suspension was added to six-well plates with approximately 1 × 103 cells per well and incubated at 37°C in a humidified 5% CO2 atmosphere for 2 weeks. After culturing, the cells were washed twice with PBS, fixed with 4% paraformaldehyde for 15 min, stained with 0.1% crystal violet for 15 min, washed thrice with PBS again. The number of cell colonies with a diameter of >0.1 mm was further observed under the microscope.
Wound Healing Assay
The treated cells were seeded into six-well plates. When the cell density was close to 90%, a linear scratch was made using a 200 µl plastic pipette tip. The wound-healing time in different treatment groups were observed under a microscope and photographed at 0 and 12 h. The wound closure rate was measured thrice and averaged.
Luciferase Reporter Assays
The dual-luciferase reporter gene plasmids containing wild-type and mutant sequences were synthesized using Promega (Madison, WI, USA). The wild-type or mutant (NEAT1, SERPINE1) reporter plasmid and miR-10a-5p mimic or mimic-NC were co-transfected into 293T cells. Luciferase activity was measured 48 h after transfection.
Fluorescence In Situ Hybridization (FISH)
FISH analysis on the ACHN cells was performed to determine the subcellular localization of NEAT1 and miR-10a-5p. The RNA probes were designed and synthesized via Servicebio (Wuhan, China). Briefly, the cells were fixed with 4% paraformaldehyde for 20 min and washed with PBS. Then, proteinase K (20 ug/ml) was digested for 8 min, followed by PBS washes. Subsequently, a pre-hybridization solution was added for 1 h at 37°C and then discarded. RNA probes containing a hybridization solution was added to the cells and hybridized overnight at 37°C. Next, DAPI staining solution was added after the washes and incubated for 8 min, followed by rinsing and the addition of an anti-fluorescence quenching blocker dropwise to seal the slice. Finally, the slices were visualized under a Nikon fluorescence microscope.
DNA Methylation Analysis of SERPINE1
To investigate the DNA methylation level of SERPINE1, co-expression analysis of SEPRINE1 was conducted using three DNA methyltransferases (DNMT1, DNMT3A, DNMT3B). Following this, methylation analysis of SERPINE1 was performed between tumor and normal tissues using the online database DiseaseMeth version 2.0 (http://bio-bigdata.hrbmu.edu.cn/diseasemeth/). MEXPRESS (https://mexpress.be/) was used to further determine the relationship between SERPINE1 expression and DNA methylation status (37).
Correlation Between Immune Infiltration and Expression of SERPINE1 in KIRC
TIMER (https://cistrome.shinyapps.io/timer/) was used for the comprehensive analysis of the relationship between SERPINE1 expression and tumor-infiltrating immune cell levels, namely, neutrophils, macrophages, dendritic cells, B cells, CD4+ T cells, and CD8+ T cells (38).
Software and Versions
R software (x64, version 4.0.3) was used for the statistical analyses (https://www.r-project.org/).
Results
BAP1 Acts as a Tumor Suppressor in ccRCC
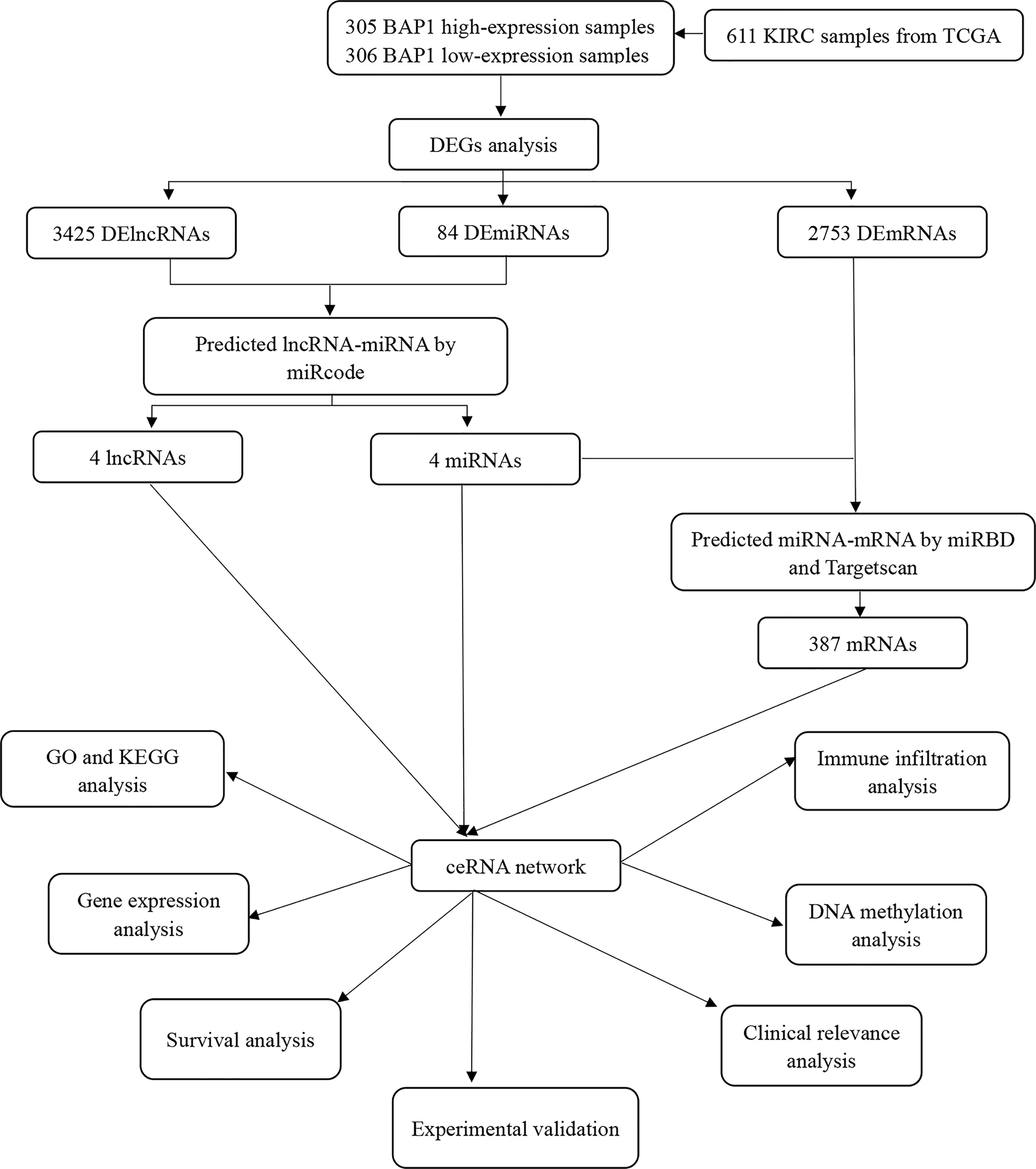
Figure 1 presents a flow diagram of ceRNA construction and analysis. A pan-cancer analysis was performed to evaluate the RNA and protein expression level of BAP1 using data from the UCSC XENA (https://xenabrowser.net/datapages/) and the Human Protein Atlas (HPA) databases (https://www.proteinatlas.org/), respectively. BAP1 RNA expression was found to be significantly downregulated in ccRCC (Figure 2A), while BAP1 protein level was the lowest in renal cancer (Figure S1). Furthermore, immunohistochemistry staining data obtained from the HPA validated BAP1 downregulation in tumor tissue (Figure 2B and Table S1). Additionally, Kaplan–Meier survival curves indicated that the low expression of BAP1 was associated with poor OS in ccRCC (Figure 2C).
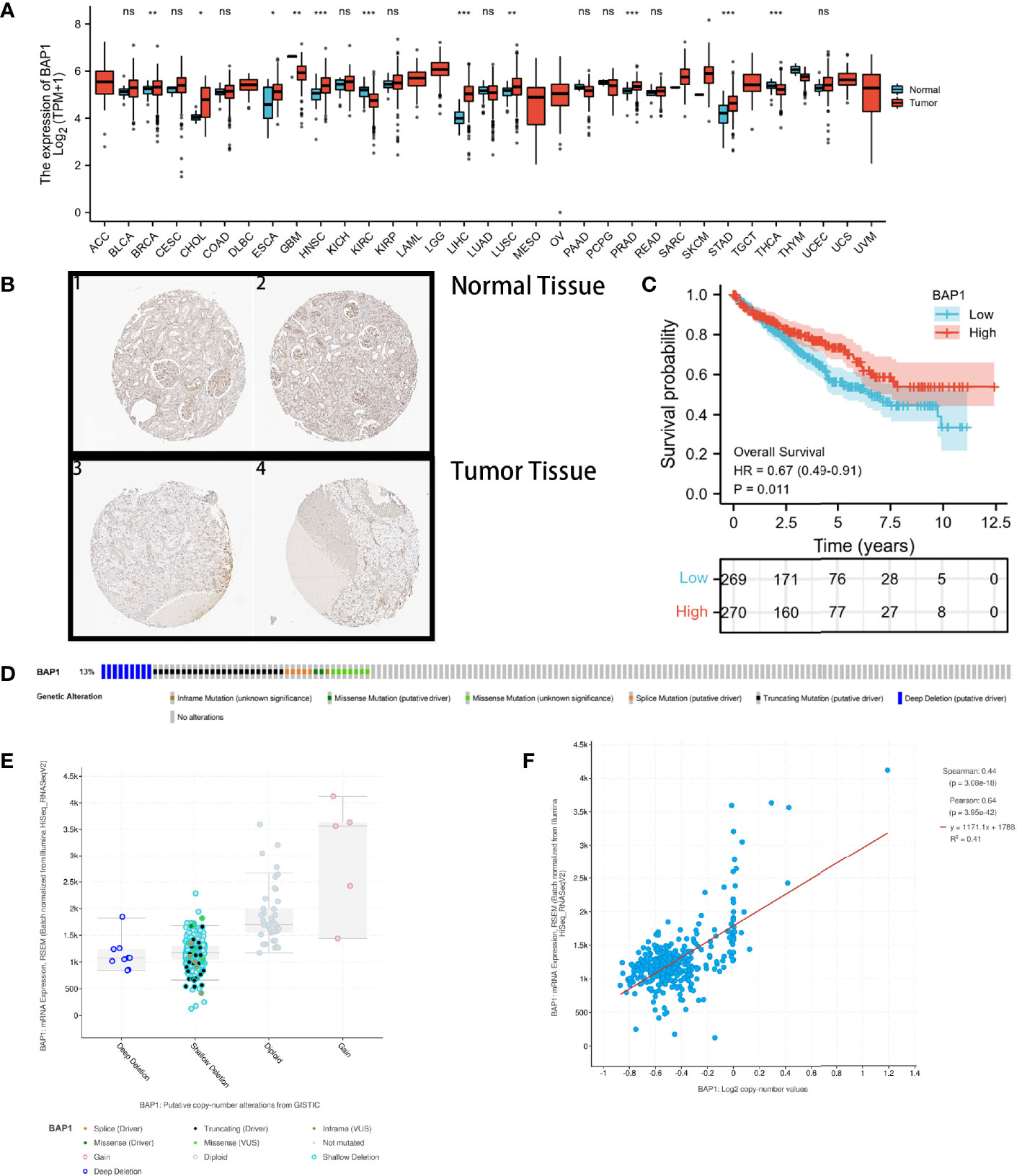
Figure 2 BAP1 acts as a tumor suppressor in kidney renal clear cell carcinoma (KIRC). (A) Pan-cancer analysis of BAP1; (B) Immunohistochemical analysis of BAP1 in renal tumor and normal tissues; (C) Survival analysis comparing high- and low expression of BAP1; (D) Distribution of BAP1 genomic alterations inTCGA KIRC; (E, F) Relationship between copy number alterations and BAP1 expression: scatter plot (E), correlation plot (F). ns, not significant, p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001.
Moreover, the cBioPortal (http://www.cbioportal.org/) was used to explore the potential mechanisms underlying the abnormally low expression of BAP1 in ccRCC (39). Figure 2D shows that the genetic alteration rate of BAP1 was found to be 13%, with gene deletions (deep deletion and shallow deletion) accounting for more than half of the copy number alterations in ccRCC samples (Figure 2E). Additionally, the mRNA expression level of BAP1 was found to be positively correlated with the copy number value (Figure 2F).
Identification of DERNAs and lncRNA–miRNA–mRNA Networks
DERNAs were screened according to the cut-off value (Figure 3) and intersected with the predicted RNAs. The shortlisted genes were inputted into Cytoscape for hub genes network construction (Figure 4A). Finally, lncRNAs (NEAT1, HELLPAR, PURPL), miRNAs (miR-10a-5p, miR-508-3p, miR-135a-5p) and mRNAs (IRS1, SERPINE1, KCAN1, TRIM2, RORB, SIX4) were identified (Figure 4B). A functional enrichment analysis of DEmRNAs demonstrated their involvement in mesenchyme development, transmembrane receptor protein tyrosine kinase signaling pathway and cell adhesion regulation (Figure 4C).
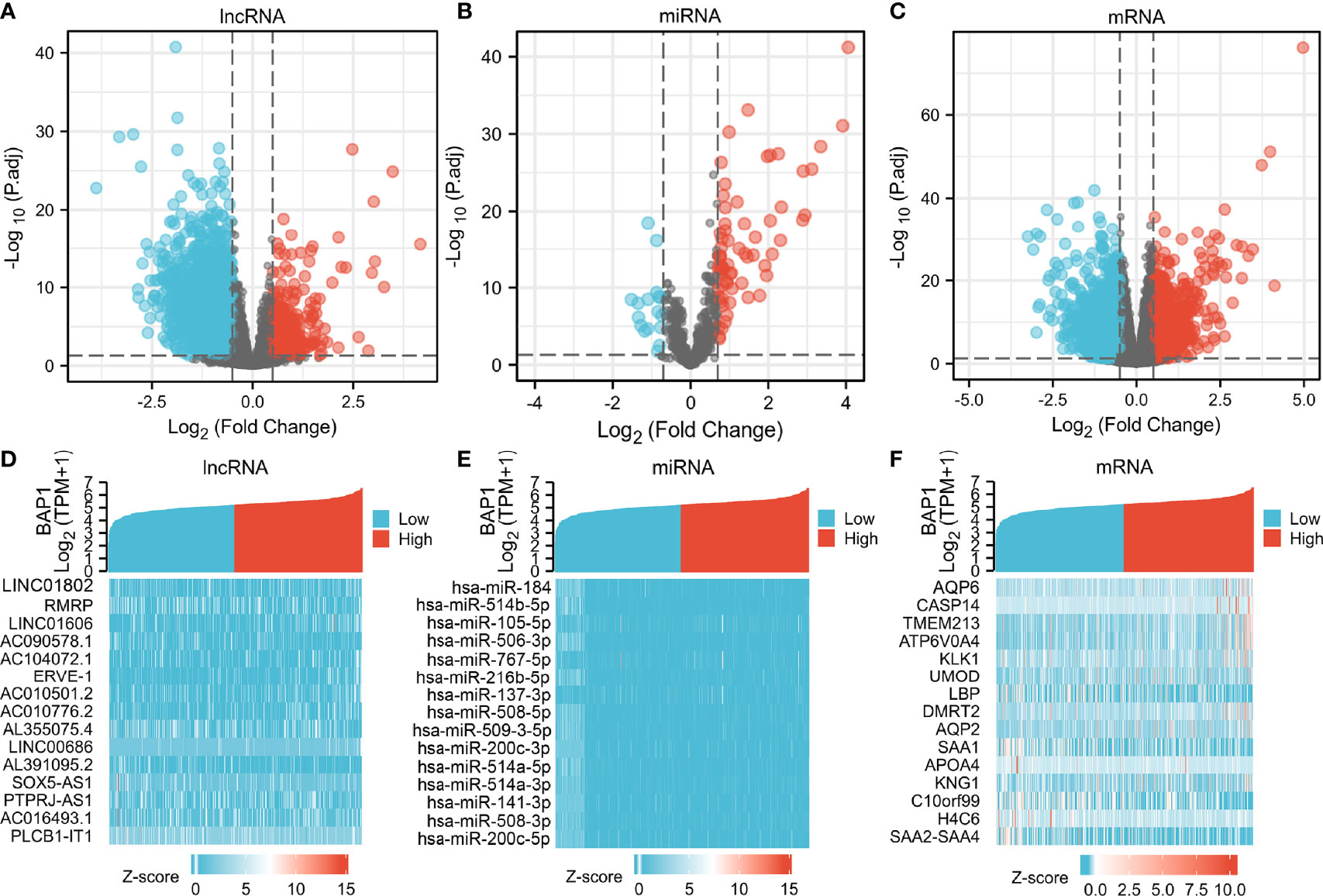
Figure 3 Volcano plots and heatmap plots of DElncRNAs, DEmiRNAs, and DEmRNAs between the expression of BAP1high and BAP1low in KIRC samples. Red color represents up-regulated genes, blue represents down-regulated genes. (A) 3425 DElncRNAs (|log2 FC| > 0.5 and P. adj < 0.05); (B) 84 DEmiRNAs (|log2 FC| > 0.7 and P. adj < 0.05); (C) 2753 DEmRNAs with cutoff value of |log2 FC| > 0.5 and P. adj < 0.05; (D–F) Heatmaps of the top 15 significant DElncRNAs, miRNAs and mRNAs.

Figure 4 Construction of ceRNA networks and functional annotation. (A) A triple regulatory network based on 4 lncRNAs, 4 miRNAs, and 387 mRNAs; (B) Hub genes network was constructed using plug-in “cytoHubba”; (C) Functional enrichment analysis of DEmRNAs.
Construction of Prognostic-Related ceRNA in ccRCC
Expression and survival analyses of hub genes are shown in Figures 5, 6. In total, two DElncRNAs (NEAT1, HELLPAR), one DEmiRNA (miR-10a-5p) and four DEmRNAs (SERPINE1, TRIM2, RORB, SIX4) were found to be prognostic-related genes. Moreover, the lncLocator (www.csbio.sjtu.edu.cn/bioinf/lncLocator/) showed that NEAT1 was mainly distributed in the cytoplasm (Figure S2), indicating its role as a ceRNA in enhancing SERPINE1 expression by sponging miR-10a-5p. Finally, a prognostic-related NEAT1/miR-10a-5p/SERPINE1 ceRNA network was constructed (Figure 7A).
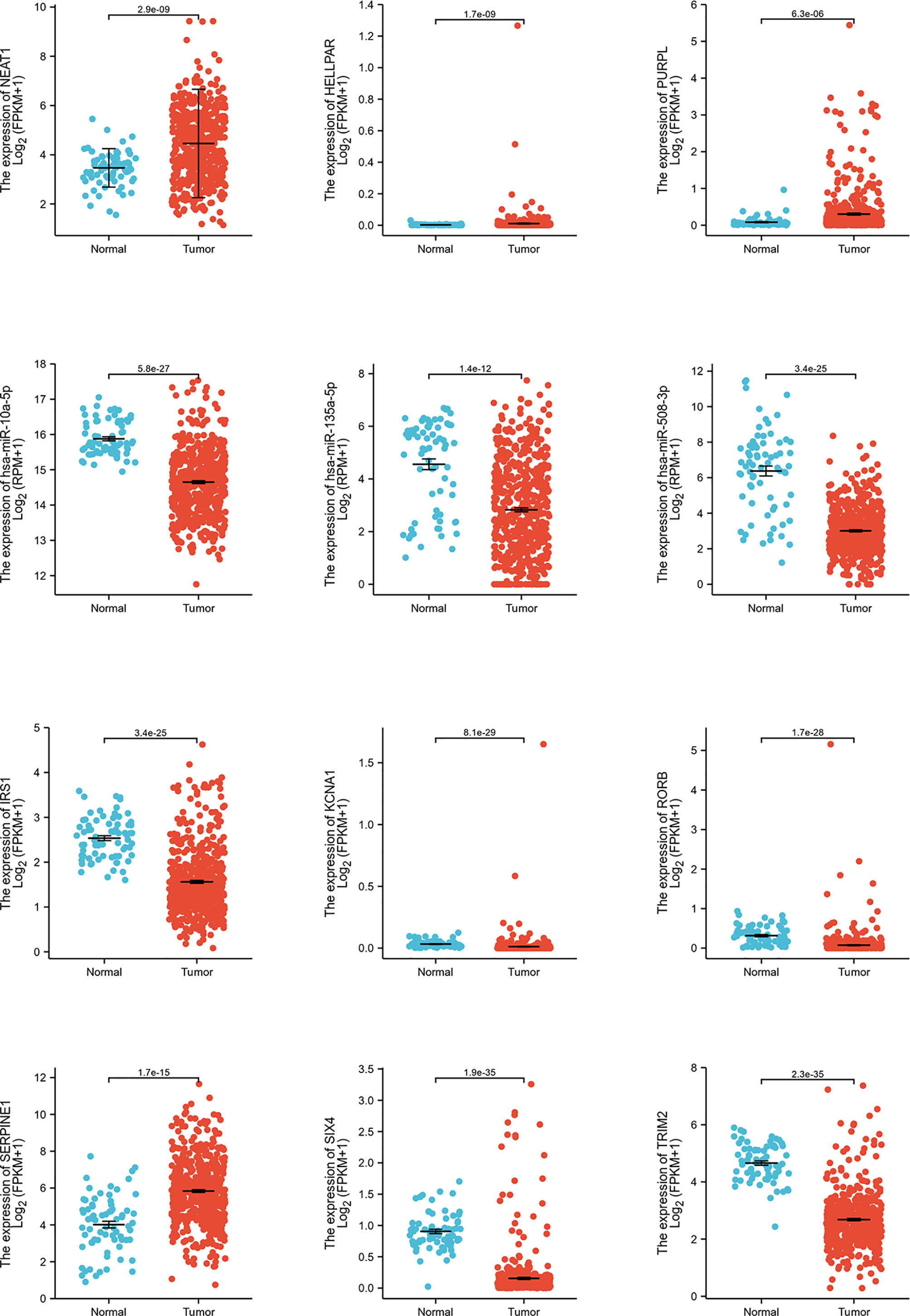
Figure 5 Hub genes expression analysis. Expression analysis of 12 hub genes (3 lncRNAs, 3 miRNAs, 6 mRNAs) comparing tumor and normal tissues.
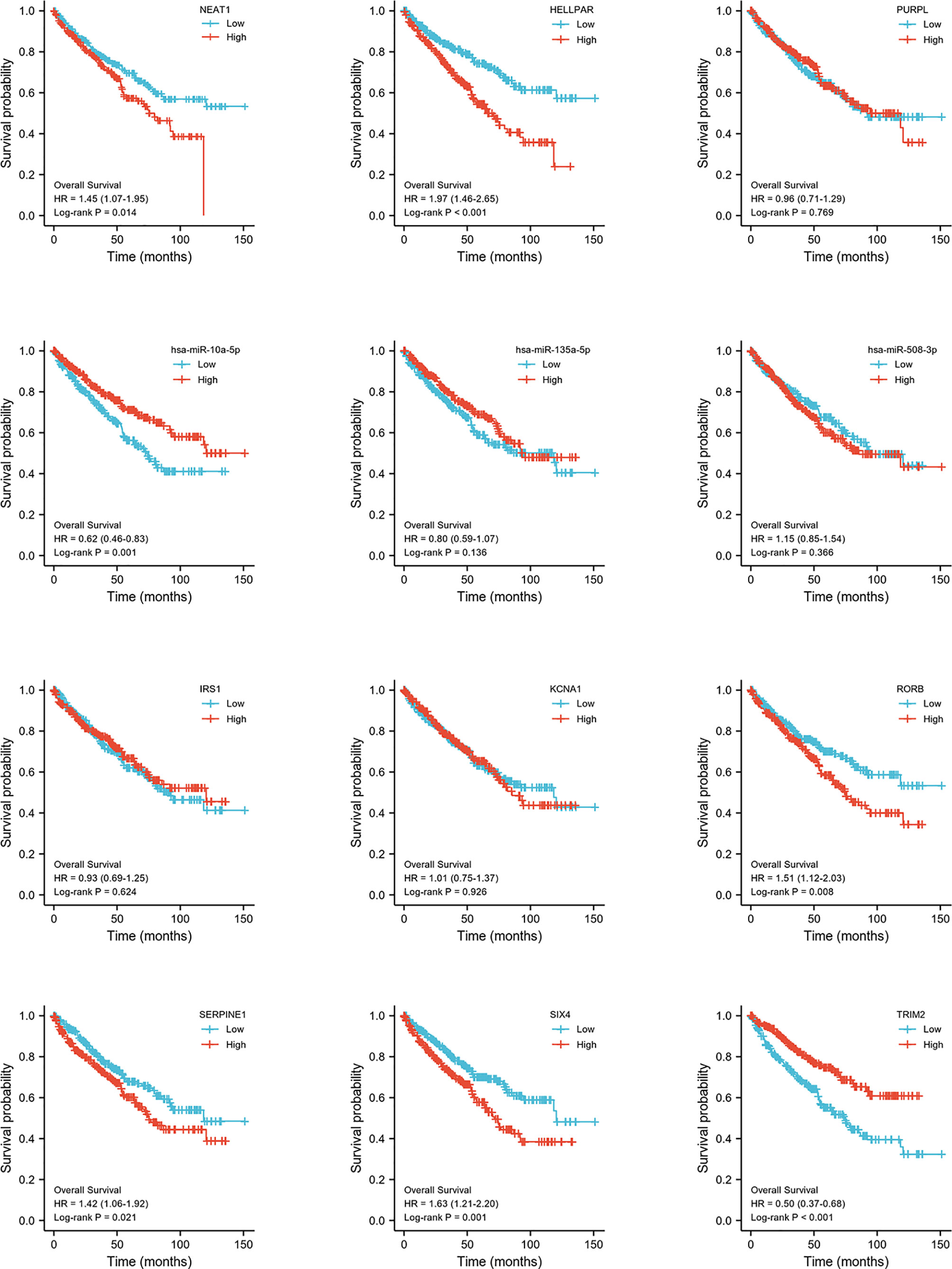
Figure 6 Hub genes survival analysis. Survival analysis of 12 hub genes (3 lncRNAs, 3 miRNAs, 6 mRNAs) comparing high- and low expression group.
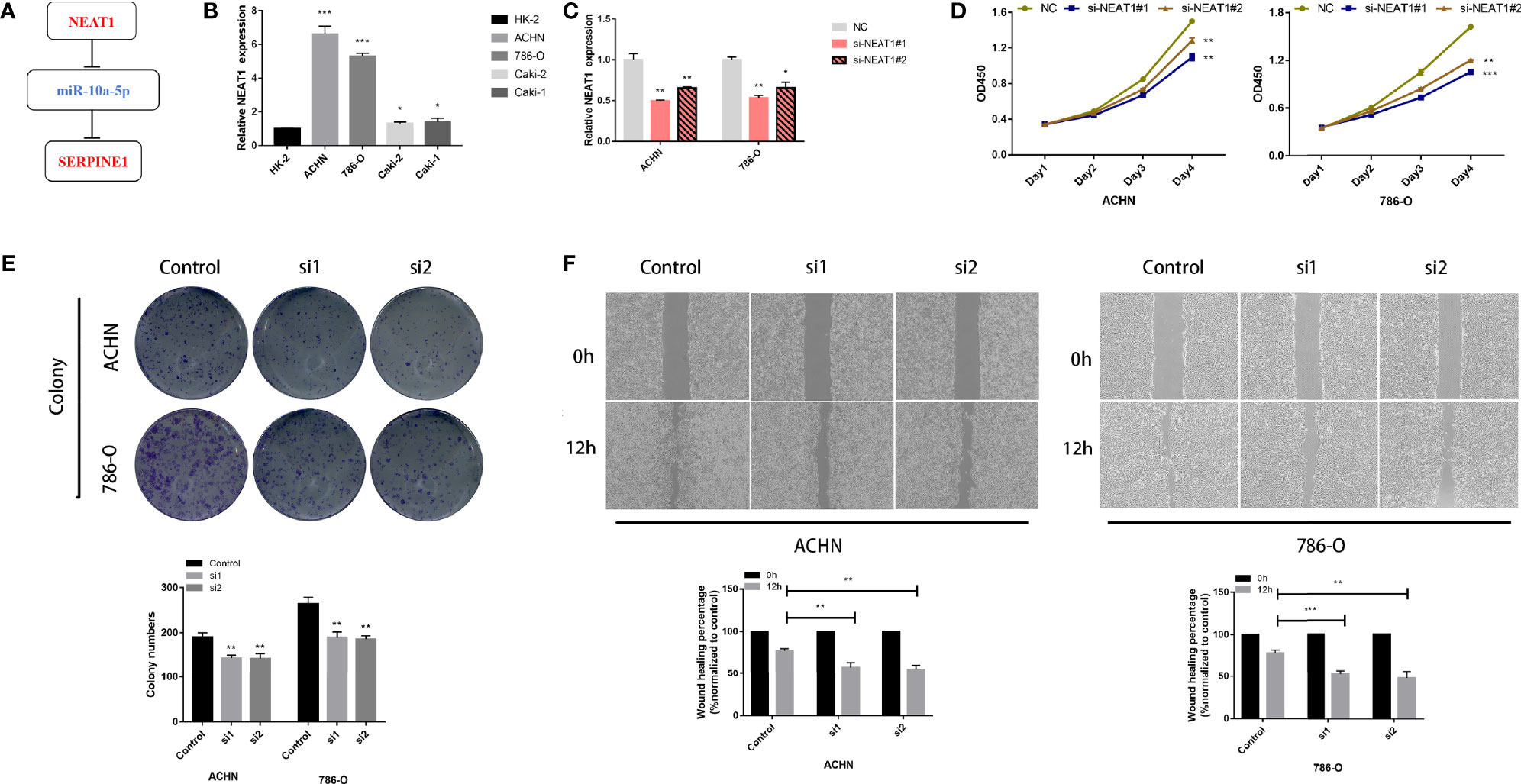
Figure 7 LncRNA NEAT1 promotes proliferation and migration in kidney cancer cell lines. (A) Construction of a ceRNA axis; (B) NEAT1 is upregulated in kidney cancer cell lines; (C) Validation of knockdown efficiency of siNEAT1 by RT-qPCR; Knockdown of NEAT1 inhibits proliferation (D), colony formation (E) and migration (F) of kidney cancer cells. p≥0.05; *p < 0.05; **p < 0.01; ***p < 0.001.
Clinical Relevance of the NEAT1/miR-10a-5p/SERPINE1 Axis in Patients With ccRCC
In this study, NEAT1 was correlated with gender (P <0.05, Figure S3A). The lower expression level of miR-10a-5p was associated with higher T stage, M stage, pTNM stage, tumor grade and gender (P <0.05, Figure S3B). Moreover, SERPINE1 was strongly correlated with T stage, N stage, pTNM stage, tumor grade and gender (P <0.05, Figure S3C). The multivariate Cox regression analysis showed that NEAT1 (hazard ratio (HR) = 1.488, P = 0.011), SERPINE1 (HR = 1.456, P = 0.015) and miR-10a-5p (HR = 0.681, P = 0.014) were independent prognostic factors in ccRCC (Tables S2–4). The AUC (area under the curve) of the receiver operating characteristics (ROC) analysis (Figure S4) indicated a good prognostic performance of SERPINE1 (AUC = 0.789) and miR-10a-5p (AUC = 0.892). Additionally, the pan-cancer analysis showed that SERPINE1 mRNA was highly expressed in kidney cancer (Figure S5A). Furthermore, immunohistochemical analysis revealed that SERPINE1 was located in the cytoplasmic/membranous area (Figure S5B).
lncRNA NEAT1 Regulates Tumor Proliferation and Migration in Kidney Cancer Cell Lines
RT-qPCR analysis showed that NEAT1 was highly expressed in kidney cancer cell lines compared to that in HK-2, with more significant differences in 786-O and ACHN cell lines (Figure 7B). The knockdown efficiency of the two siRNAs was verified using RT-qPCR (Figure 7C). CCK-8 and colony formation assays showed that 786-O and ACHN cell proliferation was significantly suppressed after NEAT1 knockdown (Figures 7D, E). Wound healing assays showed that the migration ability of the cells was inhibited after transfection with siNEAT1 (Figure 7F).
NEAT1 Serves as a Sponge for miR-10a-5p and Upregulates SERPINE1 to Regulate the Proliferation of Kidney Cancer Cells
The target site of NEAT1 to miR-10a-5p was predicted using the starBase (https://starbase.sysu.edu.cn/), and the wild-type and mutant sequences are shown in Figure 8A. A dual fluorescein reporter gene plasmid (NEAT1-WT/NEAT1-MUT) was constructed and co-transfected into 293T cells with miR-10a-5p mimic and miR-NC. Overexpression of miR-10a-5p significantly reduced luciferase activity in the NEAT1-WT group but not in the NEAT1-MUT group (Figure 8B), confirming that miR-10a-5p binds directly to NEAT1. Moreover, FISH assays revealed that miR-10a-5p co-localized with NEAT1 in the cytoplasm of ACHN cells (Figure 8C). Therefore, NEAT1 serves as a sponge for miR-10a-5p and inhibit its function in kidney cancer cells.
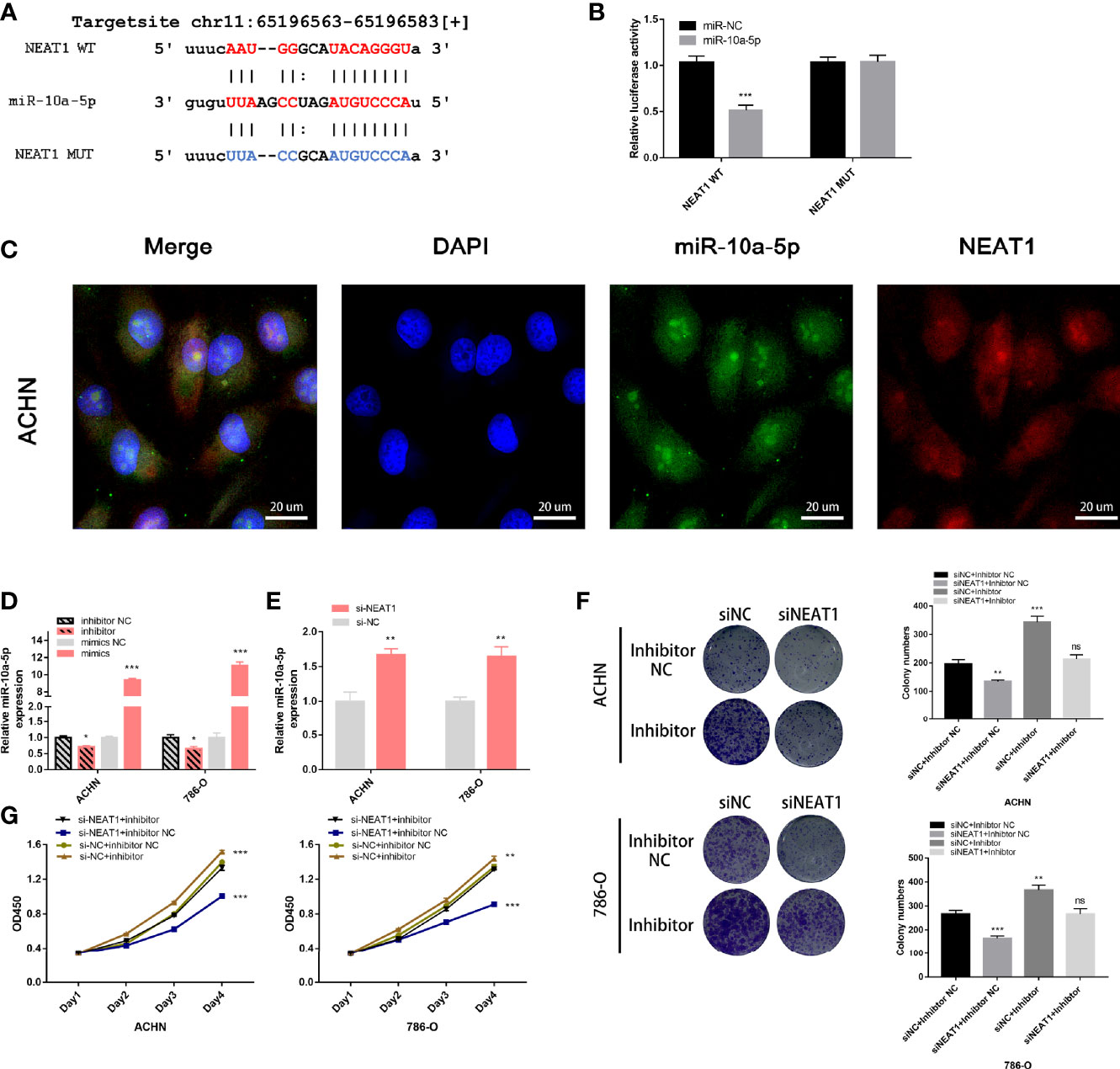
Figure 8 NEAT1 serves as sponge for miR‑10a-5p in kidney cancer cells. (A) Prediction the target sequence of NEAT1 bonding to miR-10a-5p by starBase; (B) Dual luciferase reporter gene assays verify the direct binding of miR-10a-5p to NEAT1 target sequence; (C) FISH assays confirm that NEAT1 co-localizes with miR-10a-5p in the cytoplasm of kidney cancer cells; (D) Validation the efficiency of miR-10a-5p mimic and inhibitor by RT-qPCR; (E) Knockdown of NEAT1 enhances the expression of miR-10a-5p; (F, G) miR-10a-5p inhibitor reverses the inhibition of cell proliferation caused by siNEAT1. ns, not significant, p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001.
Next, the efficiency of the miR-10a-5p mimic and inhibitor on miR-10a-p expression was verified using RT-qPCR (Figure 8D). On transfection with siNEAT1 in ACHN and 786-O cells, miR-10a-5p expression was significantly enhanced (Figure 8E). Therefore, co-transfection siRNA and miR-inhibitor (siNC + inhibitor NC; siNC + inhibitor; siNEAT1 + inhibitor NC; siNEAT1 + inhibitor) in ACHN and 786-O cells, CCK-8 and colony formation assays suggested that the knockdown of NEAT1 on the suppression of cell proliferation and colony formation in ACHN and 786-O cells could be reversed by a miR-10a-5p inhibitor (Figures 8F, G).
To confirm that miR-10a-5p regulates the expression of SERPINE1 by binding directly to the target sequence, a luciferase reporter gene plasmid was constructed for SERPINE1-WT/MUT (Figure 9A). The results showed that miR-10a-5p mimic significantly reduced the luciferase activity of SERPINE1-WT; however, no significant changes were observed for SERPINE1-MUT (Figure 9B). siNEAT1 was co-transfected with miR-10a-5p inhibitor into ACHN and 786-O cells and evaluated using RT-qPCR and western blot to verify the regulation of SERPINE1 expression. The results demonstrated that the knockdown of NEAT1 significantly downregulated the expression of SERPINE1; however, this effect could be reversed by a miR-10a-5p inhibitor (Figures 9C, D).
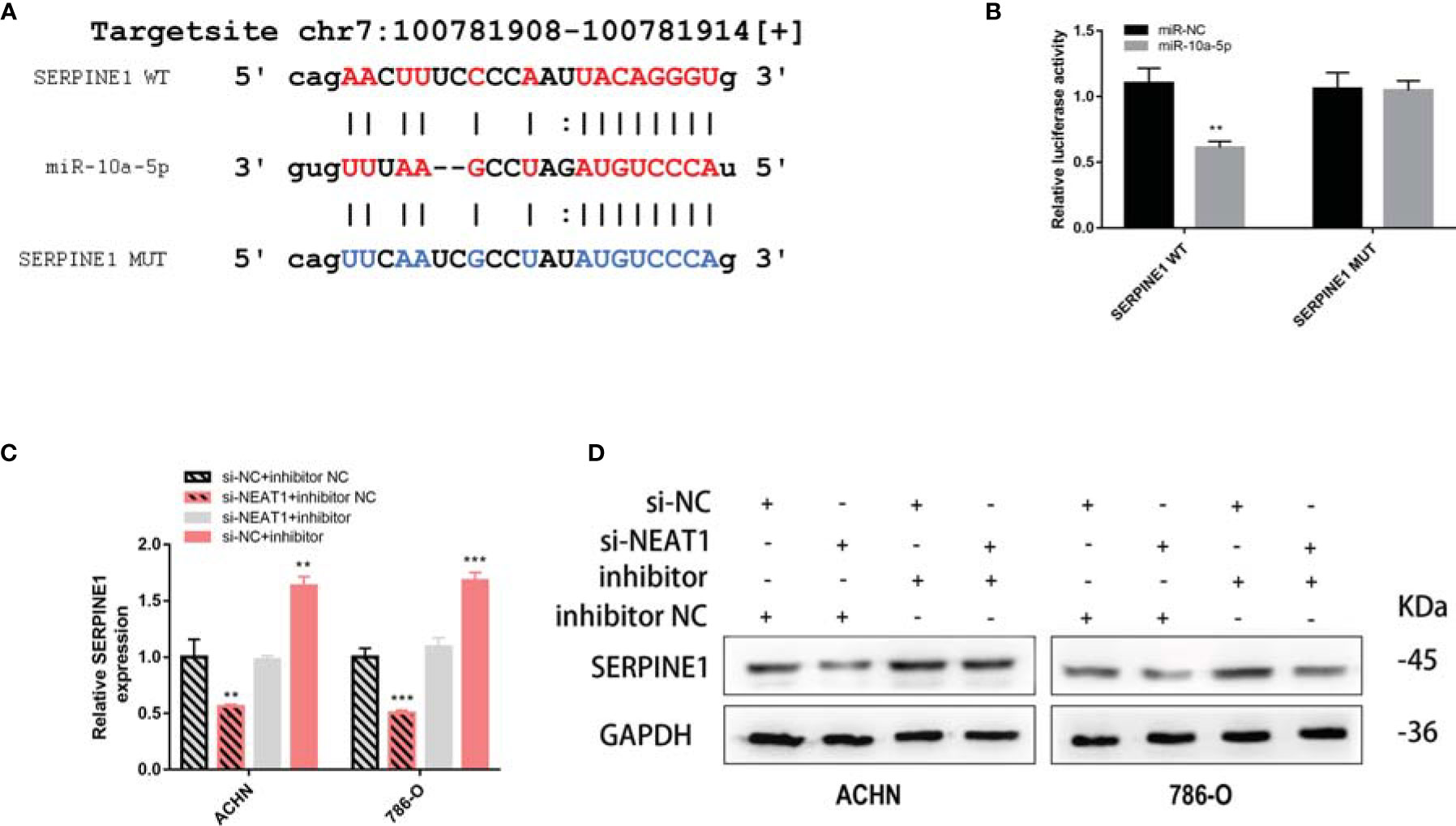
Figure 9 NEAT1 sponging miR-10a-5p to regulate SERPINE1. (A) Prediction the target sequence of SERPINE1 bonding to miR-10a-5p by starBase; (B) Dual luciferase reporter gene assays verify the direct binding of miR-10a-5p to SERPINE1 target sequence; (C, D) Knockdown of NEAT1 can downregulate the expression of SERPINE1 but can be reversed by miR-10a-5p inhibitor. **p < 0.01; ***p < 0.001.
DNA Methylation Analysis of SERPINE1
To elucidate the mechanism of the abnormally high expression of SERPINE1 in ccRCC, a series of methylation analyses of SERPINE1 was performed. Co-expression analysis suggested that SERPINE1 expression was positively correlated with DNMT1, DNMT3A, and DNMT3B expression levels (P <0.05, Figure 10A). Additionally, the methylation level of SERPINE1 in normal tissues was much higher than ccRCC tissue samples (P <0.001, Figure 10B). Moreover, 12 DNA methylation sites that were negatively correlated with SERPINE1 expression were identified (Figure 10C).
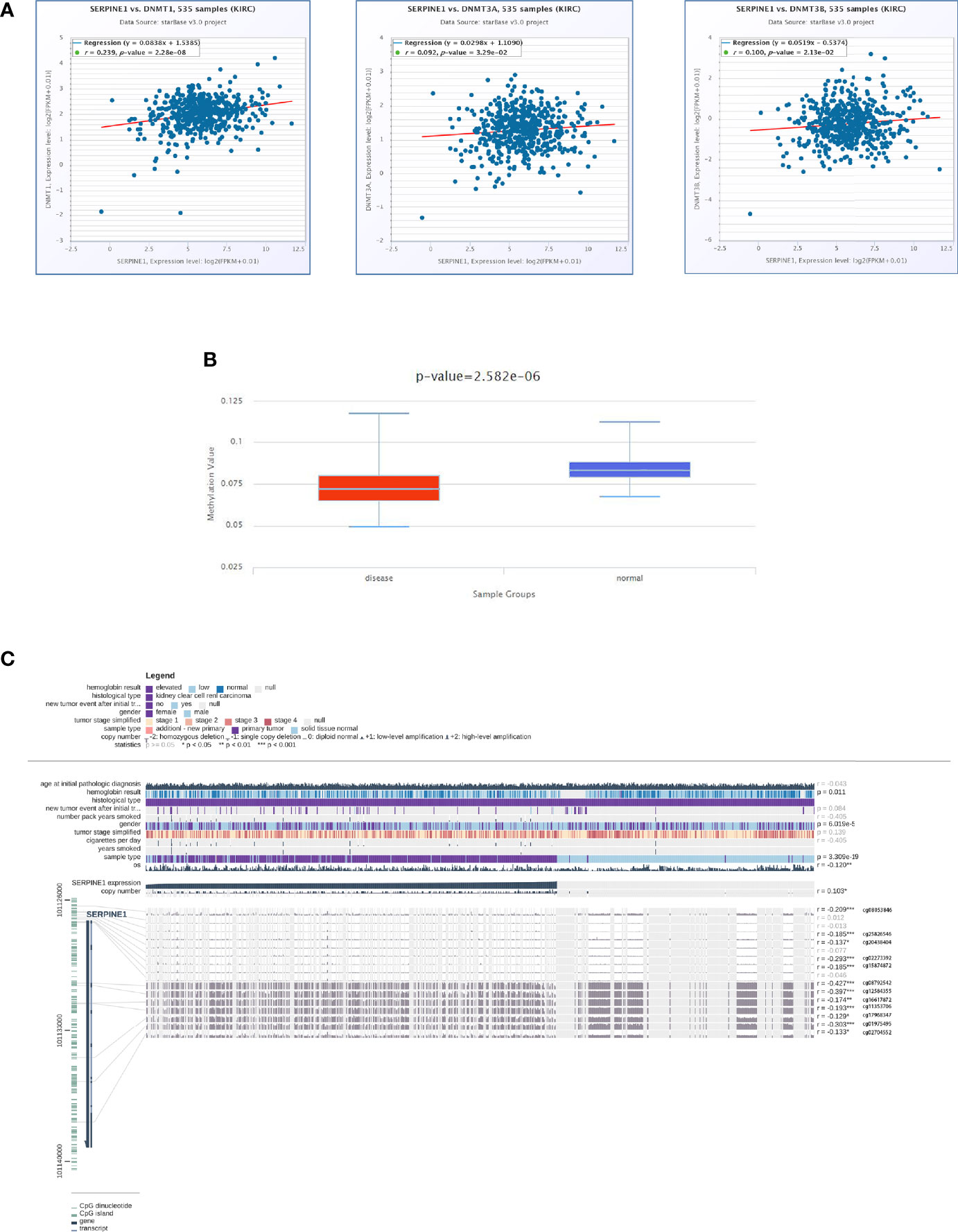
Figure 10 DNA methylation analysis of SERPINE1. (A) Co-expression analysis of SERPINE1 with three DNMTS (DNMT1, DNMT3A and DNMT3B); (B) Methylation level of SERPINE1 comparing KIRC and normal samples; (C) Relationship between SERPINE1 expression and genome-wide methylation by MEXPRESS.
Immune Infiltration Analysis of SERPINE1 in KIRC
To further investigate the relationship between SERPINE1 expression and the immune microenvironment in ccRCC, an immune infiltration analysis was performed using TIMER. ‘SCNA’ module analysis indicated that the immune infiltration level of CD 4+ T cell was associated with the altered copy numbers of SERPINE1 (Figure 11A). Moreover, ‘Gene’ module analysis showed that the expression of SERPINE1 was positively related to the immune infiltration level of CD4+ T cell, CD8+ T cell, macrophages, dendritic cells and neutrophils (Figure 11B).
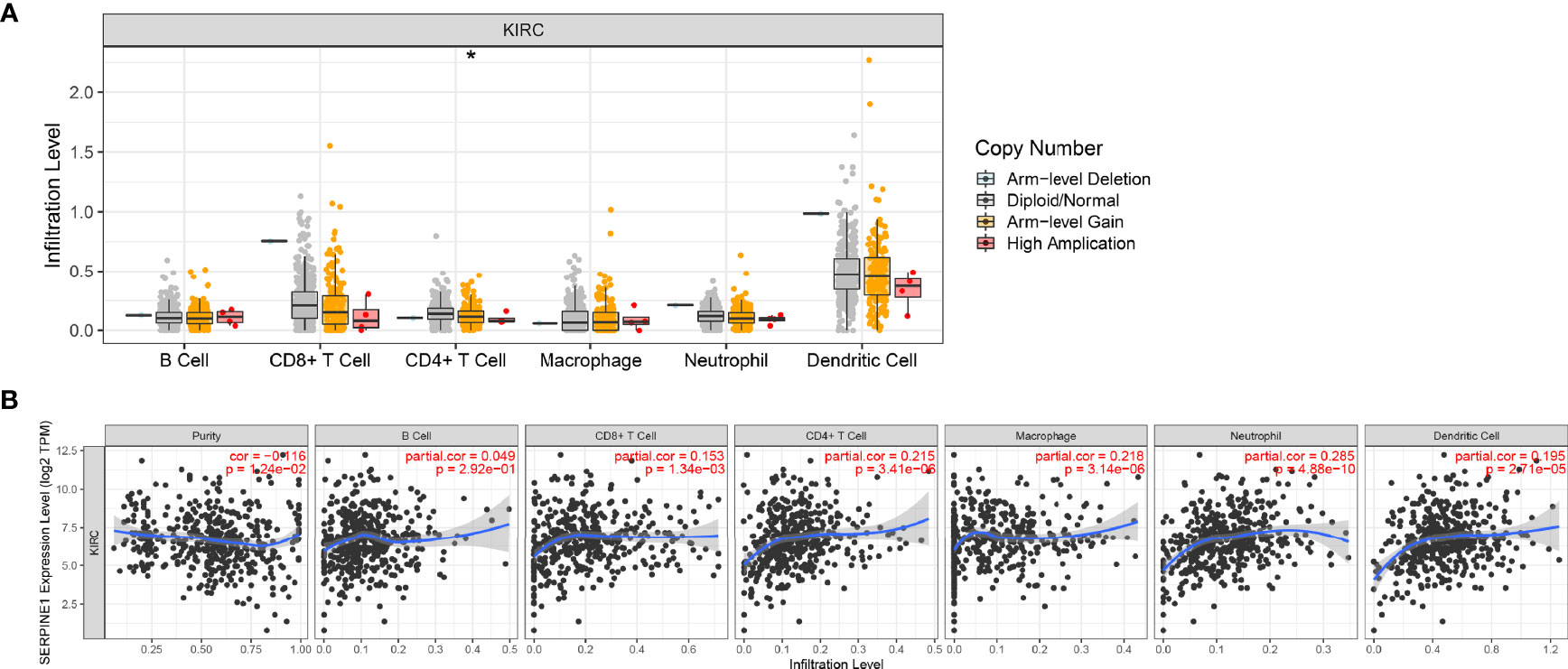
Figure 11 Immune infiltration analysis of SERPINE1 in KIRC. (A) Relationship between the level of immune cell infiltration and the copy number of SERPINE1 in KIRC. (B) Correlation of SERPINE1 expression and immune cell infiltration in KIRC. *p < 0.05.
Discussion
Numerous studies have shown that transcription occurs in approximately 80% of the human genome, with protein-coding genes accounting for only 2% of the human genome. This suggests that the majority of RNAs are non-coding genes (40). Cancer is often associated with abnormal transcriptomes; moreover, increasing evidence indicates that the non-coding transcriptome is often dysregulated in cancer and plays an important role in determining its pathogenesis (41, 42). Therefore, on analyzing the differentially expressed non-coding RNAs in BAP1-deficient ccRCC, this study identified NEAT1/miR-10a-5p/SERPINE1 as a BAP1-related prognostic ceRNA.
NEAT1, which is located in the nuclear paraspeckles, has been reported to be involved in various biological processes, such as tumorigenesis, infection, neuropathy, and immunity (43–46). Previous studies have shown that NEAT1 plays a key role in carcinogenesis by mediating gene expression. For example, NEAT1 induces transcription factors to relocate from the promoters of downstream genes to the paraspeckles, altering gene transcription (44). NEAT1 also acts as a scaffold to bind multiple proteins located in the paraspeckles together or as a bridge for protein complexes (47, 48). Recently, NEAT1 has been reported to act as a ceRNA to regulate the expression of downstream genes by sponging miRNAs in malignant tumors (49–51). Previous studies have demonstrated that BRCA1 as a transcription factor can directly bind 1.4 kb upstream of the TSS region of NEAT1 and negatively regulate its expression (52). In the nucleus, BAP1 binds to BRCA1 and enhances its tumor suppressive activity. Thereby, the loss of BAP1 was hypothesized to indirectly affect the tumor suppressor activity of BRCA1, resulting in an abnormally high expression of NEAT1.
SERPINE1, encoding plasminogen activator inhibitor 1 (PAI-1), is an essential inhibitor of tissue plasminogen activator and urokinase (uPA). Previous studies have focused on the effect of SERPINE1 on human thrombosis, cardiac fibrosis, inflammatory injury, ageing and metabolism (53–57). However, current studies report on the importance of SERPINE1 in promoting tumor malignant progression, distant metastasis and chemotherapy resistance through multiple pathways. Moreover, high SERPINE1 expression is significantly associated with poor prognosis (58, 59). The underlying mechanism has been speculated to be the migratory effect of uPA–uPAR–PAI-1 systems on endothelial cells, with fibrin deposition playing an important role in tumor angiogenesis (60); SERPINE1 also functions as an extracellular matrix (ECM) component to stabilize tumor cell adhesion in migration (61). ECM is composed of proteins and proteoglycans that are secreted by keratinocytes, fibroblasts and immune cells (62). In the complex tumor microenvironment (TME), dynamic cell–cell and cell–ECM interactions play a crucial role in tumor initiation and immune cell regulation (63). The tumor immune microenvironment determines the biological behaviour of tumour cells, with immune cell infiltration levels correlating with tumor prognosis (64–66). In this study, the copy number of SERPINE1 was found to be associated with the immune infiltration of CD4+ T cells, and the expression of SERPINE1 was related to the level of immune infiltration of CD4+ T cells, CD8+ T cells, macrophages, dendritic cells and neutrophils. Roelofs et al. demonstrated that SERPINE1 regulated neutrophil influx during renal fibrosis, suggesting that SERPINE1 acts as a chemokine to mediate immune cell infiltration (67). Moreover, in oral squamous cell carcinoma, PAI-1 has been shown to induce CD14+ monocytes to differentiate into CD206+ tumor-associated macrophages (TAMs), producing epidermal growth factors to mediate tumor cell migration (68). In the TME, cancer-associated fibroblasts induce the M2-polarization of macrophages and produce chemokine ligand 12 to promote the secretion of PAI-1 in TAMs, leading to the malignant process of hepatocellular carcinoma (69).
Current anti-cancer drug research uses a two-dimensional model of cytotoxicity in vitro (63), which does not accurately represent the three-dimensional TME. Moreover, the individual differences and intra-tumor cell heterogeneity are not sufficiently considered, resulting in many clinically ineffective anti-cancer drugs (70). Therefore, targeting the abnormally elevated functional proteins of ECMs in the TME could be a new direction for the development of anti-tumor drugs. In this study, the loss of BAP1 resulted in an abnormal upregulation of SERPINE1 (PAI-1) and hence, SERPINE1 could be a new target for the treatment of BAP1-deficient ccRCC.
Conclusions
A ceRNA (NEAT1/miR-10a-5p/SERPINE1) network was constructed that could be used as a prognostic biomarker of BAP1-deficient ccRCC. Furthermore, miR-10a-5p/SERPINE1 was significantly associated with clinical features, indicating their role as independent prognostic factors of ccRCC.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
RJL designed the study and conducted data extraction, analysis and experimentation. RJL, ZPX, SYL, JJY, and BX wrote the manuscript. RJL, ZPX, NHF, BX, and MC reviewed and revised the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This study was funded by The National Natural Science Foundation of China (Nos. 81872089, 81370849, 81672551, 81300472, 81070592, 81202268, 81202034); the Six Talent Peaks Project in Jiangsu Province, Jiangsu Provincial Medical Innovation Team (CXTDA2017025); the Natural Science Foundation of Jiangsu Province (BK20161434, BL2013032, BK20150642, and BK2012336); the Major Project of Jiangsu Commission of Health: (No: ZD2021002); the Wuxi ‘Taihu Talents Program’ Medical Expert Team Project (Nos. THRCJH20200901, THRCJH20200902).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all authors for their support in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.852515/full#supplementary-material
References
1. Sung HA-O, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Gossage L, Eisen T, Maher ER. VHL, the Story of a Tumour Suppressor Gene. Nat Rev Cancer (2015) 15(1):55–64. doi: 10.1038/nrc3844
3. Escudier B, Szczylik C, Porta C, Gore M. Treatment Selection in Metastatic Renal Cell Carcinoma: Expert Consensus. Nat Rev Clin Oncol (2012) 9(6):327–37. doi: 10.1038/nrclinonc.2012.59
4. Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, et al. Systematic Sequencing of Renal Carcinoma Reveals Inactivation of Histone Modifying Genes. Nature (2010) 463(7279):360–3. doi: 10.1038/nature08672
5. Tate JG, Bamford S, Jubb H, Sondka Z, Beare D, Bindal N, et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res (2019) 47(D1):D941–7. doi: 10.1093/nar/gky1015
6. van den Berg A, Dijkhuizen T, Draaijers T, Hulsbeek M, Maher E, Berg E, et al. Analysis of Multiple Renal Cell Adenomas and Carcinomas Suggests Allelic Loss at 3p21 to be a Prerequisite for Malignant Development. Genes Chromosomes Cancer (1997) 19(4):228–32. doi: 10.1002/(SICI)1098-2264(199708)19:4<228::AID-GCC4>3.0.CO;2-Z
7. Clifford SC, Prowse AH, Affara NA, Buys CH, Maher ER. Inactivation of the Von Hippel-Lindau (VHL) Tumour Suppressor Gene and Allelic Losses at Chromosome Arm 3p in Primary Renal Cell Carcinoma: Evidence for a VHL-Independent Pathway in Clear Cell Renal Tumourigenesis. Genes Chromosomes Cancer (1998) 22(3):200–9. doi: 10.1002/(SICI)1098-2264(199807)22:3<200::AID-GCC5>3.0.CO;2-#
8. Guo G, Gui YT, Gao SJ, Tang AF, Hu XD, Huang Y, et al. Frequent Mutations of Genes Encoding Ubiquitin-Mediated Proteolysis Pathway Components in Clear Cell Renal Cell Carcinoma. Nat Genet (2011) 44(1):17–9. doi: 10.1038/ng.1014
9. Creighton CJ. Comprehensive Molecular Characterization of Clear Cell Renal Cell Carcinoma. Nature (2013) 499:43–9. doi: 10.1038/nature12222
10. Carbone M, Harbour JW, Brugarolas J, Bononi A, Pagano I, Dey A, et al. Biological Mechanisms and Clinical Significance of BAP1 Mutations in Human Cancer. Cancer Discovery (2020) 10(8):1103–20. doi: 10.1158/2159-8290.CD-19-1220
11. Carbone M, Yang HN, Pass H, Krausz T, Testa JR, Gaudino M. BAP1 and Cancer. Nat Rev Cancer (2013) 13(3):153–9. doi: 10.1038/nrc3459
12. Wang A, Papneja A, Hyrcza M, Al-Habeeb A, Ghazarian D. Gene of the Month: BAP1. J Clin Pathol (2016) 69(9):750–3. doi: 10.1136/jclinpath-2016-203866
13. Masclef L, Ahmed O, Estavoyer B, Larrivée B, Labrecque N, Nijnik A, et al. Roles and Mechanisms of BAP1 Deubiquitinase in Tumor Suppression. Cell Death Differ (2021) 28(2):606–25. doi: 10.1038/s41418-020-00709-4
14. Jonasch EA-O, Walker CL, Rathmell WA-O. Clear Cell Renal Cell Carcinoma Ontogeny and Mechanisms of Lethality. Nat Rev Nephrol (2021) 17(4):245–61. doi: 10.1038/s41581-020-00359-2
15. Kapur P, Peña-Llopis S, Christie A, Zhrebker L, Pavía-Jiménez A, Rathmell WK, et al. Effects on Survival of BAP1 and PBRM1 Mutations in Sporadic Clear-Cell Renal-Cell Carcinoma: A Retrospective Analysis With Independent Validation. Lancet Oncol (2013) 14(2):159–67. doi: 10.1016/S1470-2045(12)70584-3
16. Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, Leng N, Pavía-Jiménez A, Wang S, et al. BAP1 Loss Defines a New Class of Renal Cell Carcinoma. Nat Genet (2012) 44(7):751–9. doi: 10.1038/ng.2323
17. Haugh AM, Njauw CN, Bubley JA, Verzì AE, Zhang B, Kudalkar E, et al. Genotypic and Phenotypic Features of BAP1 Cancer Syndrome: A Report of 8 New Families and Review of Cases in the Literature. JAMA Dermatol (2017) 153(10):999–1006. doi: 10.1001/jamadermatol.2017.2330
18. Rai K, Pilarski R, Cebulla CM, Abdel-Rahman MH. Comprehensive Review of BAP1 Tumor Predisposition Syndrome With Report of Two New Cases. Clin Genet (2016) 89(3):285–94. doi: 10.1111/cge.12630
19. Louie BH, Kurzrock R. BAP1: Not Just a BRCA1-Associated Protein. Cancer Treat Rev (2020) 90:102091. doi: 10.1016/j.ctrv.2020.102091
20. Landreville S, Agapova OA, Matatall KA, Kneass ZT, Onken MD, Lee RS, et al. Histone Deacetylase Inhibitors Induce Growth Arrest and Differentiation in Uveal Melanoma. Clin Cancer Res (2012) 18(2):408–16. doi: 10.1158/1078-0432.CCR-11-0946
21. LaFave LM, Béguelin W, Koche R, Teater M, Spitzer B, Chramiec A, et al. Loss of BAP1 Function Leads to EZH2-Dependent Transformation. Nat Med (2015) 21(11):1344–9. doi: 10.1038/nm.3947
22. Sacco JJ, Kenyani J, Butt Z, Carter R, Chew HY, Cheesemanet LP, et al. Loss of the Deubiquitylase BAP1 Alters Class I Histone Deacetylase Expression and Sensitivity of Mesothelioma Cells to HDAC Inhibitors. Oncotarget (2015) 6(15):13757–71. doi: 10.18632/oncotarget.3765
23. Sun C, Zhao CC, Li SG, Wang JQ, Zhou QD, Sun JL, et al. EZH2 Expression is Increased in BAP1-Mutant Renal Clear Cell Carcinoma and is Related to Poor Prognosis. J Cancer (2018) 9(20):3787–96. doi: 10.7150/jca.26275
24. Hu G, Ma J, Zhang J, Chen Y, Liu H, Huang Y, et al. LncRNA HOTAIR Regulates HIF-1a/AXL Signaling Through Inhibition of miR-217 in Renal Cell Carcinoma. Mol Ther (2021) S1525-0016(21):00302–6. doi: 10.1016/j.ymthe.2021.05.020.
25. Hong Q, Li O, Zheng W, Xiao WZ, Zhang L, Wu D, et al. LncRNA HOTAIR Regulates HIF-1α/AXL Signaling Through Inhibition of miR-217 in Renal Cell Carcinoma. Cell Death Dis (2017) 8(5):e2772. doi: 10.1038/cddis.2017.181
26. Fabian MR, Sonenberg N. The Mechanics of miRNA-Mediated Gene Silencing: A Look Under the Hood of miRISC. Nat Struct Mol Biol (2012) 9(6):1586–93. doi: 10.1038/nsmb.2296
27. Cazalla D, Yario T Fau - Steitz JA, Steitz JA. Down-Regulation of a Host microRNA by a Herpesvirus Saimiri Noncoding RNA. Science (2010) 328(5985):1563–6. doi: 10.1126/science.1187197
28. Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target Mimicry Provides a New Mechanism for Regulation of microRNA Activity. Nat Genet (2007) 39(8):1033–7. doi: 10.1038/ng2079
29. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A Coding-Independent Function of Gene and Pseudogene mRNAs Regulates Tumour Biology. Nature (2010) 465(7301):1033–8. doi: 10.1038/nature09144
30. Mao W, Wang K, Xu B, Zhang H, Sun S, Hu Q, et al. ciRS-7 is a Prognostic Biomarker and Potential Gene Therapy Target for Renal Cell Carcinoma. Mol Cancer (2021) 20(1):142. doi: 10.1186/s12943-021-01443-2
31. Chen Y, Wang X. miRDB: An Online Database for Prediction of Functional microRNA Targets. Nucleic Acids Res (2020) 48(D1):127–31. doi: 10.1093/nar/gkz757
32. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting Effective microRNA Target Sites in Mammalian mRNAs. Elife (2015) 4:e05005. doi: 10.7554/eLife.05005.028
33. Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko MT, Lin C-Y. Cytohubba: Identifying Hub Objects and Sub-Networks From Complex Interactome. BMC Syst Biol (2014) Suppl 4:S11. doi: 10.1186/1752-0509-8-S4-S11
34. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res (2003) 13(11):2498–504. doi: 10.1101/gr.1239303
35. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat Commun (2019) 10(1):1523. doi: 10.1038/s41467-019-09234-6
36. Mao W, Wang K, Sun S, Wu J, Chen M, Geng J. ID2 Inhibits Bladder Cancer Progression and Metastasis via PI3K/AKT Signaling Pathway. Front Cell Dev Biol (2021) 9:738364. doi: 10.3389/fcell.2021.738364
37. Koch A, Jeschke J, Van Criekinge W, van Engeland M, De Meyer T. MEXPRESS Update 2019. Nucleic Acids Res (2019) 47(W1):W561–5. doi: 10.1093/nar/gkz445
38. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77(21):e108-e110. doi: 10.1158/0008-5472.CAN-17-0307
39. Cerami E, Gao JJ, Dogrusoz U, Gross EB, Sumer SO, Aksoy BA, et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery (2012) 2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095
40. Tay Y, Rinn J, Pandolfi PP. The Multilayered Complexity of ceRNA Crosstalk and Competition. Nature (2014) 505(7483):344–52. doi: 10.1038/nature12986
41. Mattick JS. Non-Coding RNAs: The Architects of Eukaryotic Complexity. EMBO Rep (2001) 2(11):986–91. doi: 10.1093/embo-reports/kve230
42. Mattick JS, Gagen MJ. The Evolution of Controlled Multitasked Gene Networks: The Role of Introns and Other Noncoding RNAs in the Development of Complex Organisms. Mol Biol Evol (2001) 18(9):1611–30. doi: 10.1093/oxfordjournals.molbev.a003951
43. Zhang P, Cao L, Zhou R, Yang X, Wu M. The lncRNA Neat1 Promotes Activation of Inflammasomes in Macrophages. Nat Commun (2019) 10(1):1495. doi: 10.1038/s41467-019-09482-6
44. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long Noncoding RNA NEAT1-Dependent SFPQ Relocation From Promoter Region to Paraspeckle Mediates IL8 Expression Upon Immune Stimuli. Mol Cell (2014) 53(3):393–406. doi: 10.1016/j.molcel.2014.01.009
45. Li Y, Cheng C. Long Noncoding RNA NEAT1 Promotes the Metastasis of Osteosarcoma via Interaction With the G9a-DNMT1-Snail Complex. Am J Cancer Res (2018) 8(1):81–90.
46. Boros FA, Vécsei L, Klivényi P. NEAT1 on the Field of Parkinson's Disease: Offense, Defense, or a Player on the Bench? J Parkinsons Dis (2021) 11(1):123–38. doi: 10.3233/JPD-202374
47. Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan C, et al. The lncRNA NEAT1 Activates Wnt/β-Catenin Signaling and Promotes Colorectal Cancer Progression via Interacting With DDX5. J Hematol Oncol (2018) 11(1):113. doi: 10.1186/s13045-018-0656-7
48. Guttman M, Rinn JL. Modular Regulatory Principles of Large non-Coding RNAs. Nature (2012) 482(7385):339–46. doi: 10.1038/nature10887
49. Wang C, Chen Y, Wang Y, Liu X, Liu Y, Li Y, et al. Inhibition of COX-2, mPGES-1 and CYP4A by Isoliquiritigenin Blocks the Angiogenic Akt Signaling in Glioma Through ceRNA Effect of miR-194-5p and lncRNA Neat1. J Exp Clin Cancer Res (2019) 22:371. doi: 10.1186/s13046-019-1361-2
50. Yu H, Xu A, Wu B, Wang M, Chen Z. Long Noncoding RNA NEAT1 Promotes Progression of Glioma as a ceRNA by Sponging miR-185-5p to Stimulate DNMT1/mTOR Signaling. J Cell Physiol (2021) 236(1):121–30. doi: 10.1002/jcp.29644
51. Luo Y, Chen J, Lv Q, Qin J, Huang Y, Yu M, et al. Long non-Coding RNA NEAT1 Promotes Colorectal Cancer Progression by Competitively Binding miR-34a With SIRT1 and Enhancing the Wnt/β-Catenin Signaling Pathway. Cancer Lett (2019) 440-441:11–22. doi: 10.1016/j.canlet.2018.10.002
52. Lo PK, Zhang YS, Wolfson B, Gernapudi R, Yao Y, Duru N, et al. Dysregulation of the BRCA1/long non-Coding RNA NEAT1 Signaling Axis Contributes to Breast Tumorigenesis. Oncotarget (2016) 7(40):65067–89. doi: 10.18632/oncotarget.11364
53. Khan SS, Shah SJ, Strande JL, Baldridge AS, Flevaris P, Puckelwartz MJ, et al. Identification of Cardiac Fibrosis in Young Adults With a Homozygous Frameshift Variant in SERPINE1. JAMA Cardiol (2021) 6(7):841–6. doi: 10.1001/jamacardio.2020.6909
54. Li S, Wei X, He J, Tian X, Yuan S, Sun L. Plasminogen Activator Inhibitor-1 in Cancer Research. BioMed Pharmacother (2018) 105:83–94. doi: 10.1016/j.biopha.2018.05.119
55. Wang T, Lu H, Li D, Huang TWG. TGF-β1-Mediated Activation of SERPINE1 is Involved in Hemin-Induced Apoptotic and Inflammatory Injury in HT22 Cells. Neuropsychiatr Dis Treat (2021) 17:423–33. doi: 10.2147/NDT.S293772
56. Khan SA-O, Shah SJ, Klyachko E, Baldridge AS, Eren M, Place AT, et al. A Null Mutation in SERPINE1 Protects Against Biological Aging in Humans. Sci Adv (2017) 3(11):1617. doi: 10.1126/sciadv.aao1617
57. Xu Y, Chen W, Liang J, Zeng X, Ji K, Zhou J, et al. The miR-1185-2-3p-GOLPH3L Pathway Promotes Glucose Metabolism in Breast Cancer by Stabilizing P53-Induced SERPINE1. J Exp Clin Cancer Res (2021) 40(1):47. doi: 10.1186/s13046-020-01767-9
58. Chen S, Li Y, Zhu Y, Fei J, Song L, Sun G, et al. SERPINE1 Overexpression Promotes Malignant Progression and Poor Prognosis of Gastric Cancer. J Oncol (2022) 2022:2647825. doi: 10.1155/2022/2647825
59. Zhang Q, Lei L, Jing D. Knockdown of SERPINE1 Reverses Resistance of Triple−Negative Breast Cancer to Paclitaxel via Suppression of VEGFA. Oncol Rep (2020) 44(5):1875–84. doi: 10.3892/or.2020.7770
60. Binder BR, Mihaly J Fau - Prager GW, Prager GW. uPAR-uPA-PAI-1 Interactions and Signaling: A Vascular Biologist's View. Thromb Haemost (2007) 97(3):336–42. doi: 10.1160/TH06-11-0669
61. Boccaccio C, Comoglio PM. A Functional Role for Hemostasis in Early Cancer Development. Cancer Res (2005) 65(19):8579–82. doi: 10.1158/0008-5472.CAN-05-2277
62. Bhattacharjee O, Ayyangar U, Kurbet A, Ashok D, Raghavan S. Unraveling the ECM-Immune Cell Crosstalk in Skin Diseases. Front Cell Dev Biol (2019) 7:68. doi: 10.3389/fcell.2019.00068
63. Bahcecioglu G, Basara G, Ellis BW, Ren X, Zorlutuna P. Breast Cancer Models: Engineering the Tumor Microenvironment. Acta Biomater (2020) 106:1–21. doi: 10.1016/j.actbio.2020.02.006
64. Pagès F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector Memory T Cells, Early Metastasis, and Survival in Colorectal Cancer. N Engl J Med (2005) 353(25):2654–66. doi: 10.1056/NEJMoa051424
65. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, et al. Type, Density, and Location of Immune Cells Within Human Colorectal Tumors Predict Clinical Outcome. Science (2006) 313(5795):960–4. doi: 10.1126/science.1129139
66. Mlecnik B, Tosolini M, Kirilovsky A, Berger A, Bindea G, Meatchi T, et al. Histopathologic-Based Prognostic Factors of Colorectal Cancers are Associated With the State of the Local Immune Reaction. J Clin Oncol (2011) 29(6):610–8. doi: 10.1200/JCO.2010.30.5425
67. Roelofs JJ, Teske GJ, Bonta PI, Vries CJ, Meijers JC, Weening JJ, et al. Plasminogen Activator Inhibitor-1 Regulates Neutrophil Influx During Acute Pyelonephritis. Kidney Int (2009) 75(1):52–9. doi: 10.1038/ki.2008.454
68. Kai K, Moriyama M, Haque A, Hattori T, Chinju A, Hu C, et al. Oral Squamous Cell Carcinoma Contributes to Differentiation of Monocyte-Derived Tumor-Associated Macrophages via PAI-1 and IL-8 Production. Int J Mol Sci (2021) 22(17):9475. doi: 10.3390/ijms22179475
69. Chen S, Morine Y, Tokuda K, Yamada S, Saito Y, Nishi M, et al. Cancer−associated Fibroblast−Induced M2−polarized Macrophages Promote Hepatocellular Carcinoma Progression via the Plasminogen Activator Inhibitor−1 Pathway. Int J Onco (2021) 59(2):59. doi: 10.3892/ijo.2021.5239
Keywords: ceRNA, DNA methylation, immune microenvironment, clear-cell renal cell carcinoma, prognosis
Citation: Liu R-j, Xu Z-P, Li S-Y, Yu J-J, Feng N-h, Xu B and Chen M (2022) BAP1-Related ceRNA (NEAT1/miR-10a-5p/SERPINE1) Promotes Proliferation and Migration of Kidney Cancer Cells. Front. Oncol. 12:852515. doi: 10.3389/fonc.2022.852515
Received: 11 January 2022; Accepted: 25 February 2022;
Published: 29 March 2022.
Edited by:
Hernandes F. Carvalho, State University of Campinas, BrazilReviewed by:
Shan Gao, Suzhou Institute of Biomedical Engineering and Technology (CAS), ChinaTing Li, University of Pennsylvania, United States
Copyright © 2022 Liu, Xu, Li, Yu, Feng, Xu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ning-han Feng, bi5mZW5nQG5qbXUuZWR1LmNu; Bin Xu, bmp4YjE5ODJAMTI2LmNvbQ==; Ming Chen, bWluZ2NoZW5zZXVAMTI2LmNvbQ==
†ORCID: Ming Chen, orcid.org/0000-0002-3572-6886
‡These authors have contributed equally to this work
 Rui-ji Liu
Rui-ji Liu Zhi-Peng Xu1,2‡
Zhi-Peng Xu1,2‡ Ning-han Feng
Ning-han Feng Ming Chen
Ming Chen