
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 28 April 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.848341
 Zeping Han1,2†
Zeping Han1,2† Huafang Chen3†
Huafang Chen3† Zhonghui Guo1,2†
Zhonghui Guo1,2† Jian Shen1
Jian Shen1 Wenfeng Luo1
Wenfeng Luo1 Fangmei Xie1
Fangmei Xie1 Yu Wan4
Yu Wan4 Shengbo Wang4
Shengbo Wang4 Jianhao Li5*
Jianhao Li5* Jinhua He1,2*
Jinhua He1,2*As a novel class of endogenous non-coding RNAs discovered in recent years, circular RNAs (circRNAs) are highly conserved and stable covalently closed ring structures with no 5′-end cap or 3′-end poly(A) tail. CircRNAs are formed by reverse splicing, mainly by means of a noose structure or intron complementary pairing. Exosomes are tiny discoid vesicles with a diameter of 40-100 nm that are secreted by cells under physiological and pathological conditions. Exosomes play an important role in cell-cell communication by carrying DNA, microRNAs, mRNAs, proteins and circRNAs. In this review, we summarize the biological functions of circRNAs and exosomes, and further reveal the potential roles of exosomal circRNAs in different diseases, providing a scientific basis for the diagnosis, treatment, and prognosis of a wide variety of diseases.
Non-coding RNAs (ncRNAs) are a class of endogenous RNAs that do not encode proteins, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), which are involved in post-transcriptional regulation (1). miRNAs are 18 -22 nucleotides small ncRNAs that modulate the translation of more than 60% of all protein-coding mRNAs in the cell, providing an intermediate regulatory step between gene transcription and translation (2). In contrast with miRNAs, lncRNA segments are longer in sequences, typically more than 200 nucleotides. LncRNAs have similar functions to miRNAs and can regulate gene expressions, such as protein translation and post-transcriptional silencing. Moreover, lncRNAs inhibit the translation of cis and trans genes through histone modifications or disrupting the miRNA regulation (3). CircRNAs were originally considered to be by-products of aberrant splicing or intermediate products of intron lariats that have escaped degradation (4). With the rapid development of various accurate detection technologies, it was found that circRNAs are a kind of ncRNAs molecule composed of hundreds or even thousands of nucleotides, which can directly connect to the 5′- and 3′-ends of linear RNAs, as an intermediate product of the RNA processing reaction. CircRNAs can also be produced by “reverse splicing.” The downstream 5′ splice site (splice donor) is connected to the upstream 3′ splice site (splice receptor), which are found in prokaryotes, eukaryotes, and viruses (5). According to their sequence, circRNAs are divided into three categories: (1) intronic circRNAs (ciRNAs) are composed only of introns and are located mainly in the nucleus (6); (2) exonic circRNAs are produced by the Epstein–Barr virus from the BART gene region and are distributed in the cytoplasm and nucleus (7); and (3) exon–intron circRNAs (EIciRNAs) retain introns between the exons, are located mainly in the nucleus, like circRNAs, and can enhance the expression of parent genes in cis (8). Recently, a special type of circRNAs has been identified, called tRNA intronic circular RNAs, which is spliced from endogenous tRNAs under the action of RtcB ligase and the tRNA splicing endonuclease complex (9). CircRNAs have various biological functions and play vital roles in the occurrence, development, invasion, diagnosis, and prognosis of tumors (10–14), and are also important contributors to the development of cardiovascular (15), neurological (16), orthopedic (17), and other diseases.
Exosomes are one of the three main subtypes of extracellular vesicles (EVs) (18), which have attracted considerable attention in clinical research. They are nanoscale round or oval capsule vesicles that are released from cells after the contact and fusion of multiple vesicles with the cell membrane. They were first discovered by Wolf (19) in 1967, who observed vesicle structures he called “platelet dust” that were secreted by cells in vitro, with a size of 40–100 nm. In 1983, Pan and Johnstone identified membranous vesicles in the culture medium of reticulocytes and named them exosomes (20). Since then, exosomes have been shown to carry nucleic acids, proteins, lipids, and other biological molecules as important mediators of intercellular communication and its regulation. Their role in the occurrence, development, metastasis, invasion, and drug resistance of tumors has been studied in depth (21), and the content and functions of exosomes are constantly being supplemented and revised.
CircRNAs are competitive endogenous RNA molecules with abundant miRNA binding sites, which are also called miRNA response elements, that can competitively bind to miRNAs to remove their inhibitory effects on downstream target genes, thereby affecting intracellular signal transduction pathways and the expression of target genes (22). The role of circRNAs as miRNA sponges is the classic model of their function. For example, Hansen et al. identified the circular RNA sponge for miRNA (miR)-7 (ciRS-7) (23), also known as miRNA sponge cerebellar degeneration-related protein 1 antisense, which negatively regulates miR-7 expression. In addition, more than 70 miR-7 binding sites have been found on ciRS-7 (24), which is highly expressed in HEK293 cells and can bind up to 20,000 miR-7 molecules in each cell. Other studies have confirmed that the downregulation of circRNA-PVT1 inhibits the expression of sirtuin 7 by upregulating the expression of miR-3666, and finally inhibits the proliferation and metastasis of hepatocellular carcinoma cells (25). Moreover, circRNAs not only act as miRNA sponges in humans but also in parasites such as nematodes (26). Some circRNAs with miRNA sponge function have been found in plants such as Arabidopsis (27), wheat (28), and citrus (29).
CiRNAs and EIciRNAs competitively regulate the transcription of parental genes through linear splicing. CiRNAs can act as active regulators of RNA polymerase (Pol) II to regulate the transcription of parental genes (30). EIciRNAs can interact with U1 small nuclear RNA (snRNP) to form the EIciRNA-U1 snRNP complex, which then interacts with the transcriptional complex of RNA Pol II to affect parental gene expression (8, 31). In addition, non-coding intron transcripts, such as ci-ankrd52, act as positive regulators of RNA Pol II transcription, have cis-regulatory effects on their parental coding genes, and are associated with a mechanism for the extension of RNA Pol II (6). Subsequently, a study in 2020 showed that circ-DAB1 upregulates the expression of recombination signal-binding protein for immunoglobulin kappa J region (RBPJ), which leads to the increased binding of RBPJ to the DAB adaptor protein 1 (DAB1) promoter, thereby activating the transcription of the parental gene, DAB1 (32).
CircRNAs can also act as sponges of proteins and combine with RNA-binding proteins (RBPs) to form RNA-protein complexes that affect protein expression (33). For example, the RNA splicing factor MBL is an RBP that binds to the second exon of its parent gene and promotes its cyclization to form circ-MBL in drosophila (31). At the same time, there are multiple sites on circ-MBL that bind to MBL protein, which reduces the effective concentration of MBL (31). CircRNAs can also interact with specific target proteins and participate in cell proliferation, differentiation, and apoptosis (34). For example, circ-Foxo3, cyclin-dependent kinase inhibitor 1 (p21), and cyclin-dependent kinase 2 (CDK2) form the ternary complex circ-Foxo3-p21-CDK2, which inhibits the function of CDK2 and blocks the cell cycle. In addition, directly silencing endogenous circ-Foxo3 promotes cell proliferation (35). Another report shows that the circ-ANRIL can bind to PES1 protein, thereby preventing pre-rRNA binding and exonuclease-mediated rRNA maturation. Consequently, circANRIL impairs ribosome biogenesis, leading to activation of p53 and a subsequent increase in apoptosis and decrease in proliferative rate (36). Because proteins have many functions, the other effects of circRNAs binding to proteins need to be studied in more depth.
Initially, circRNAs were considered as non-coding RNA molecules, but in recent years, many studies have shown that circRNAs have roles in protein translation (37–40), which disproved a long-believed concept. It was originally thought that eukaryotic ribosomes could initiate translation from circRNAs only if the circRNAs contained an internal ribosome entry site (IRES) (41). For example, circ-PINT can be translated via its IRES into the PINT87aa polypeptide, which contains 87 amino acids. PINT87aa binds to the polymerase-associated factor complex gene and inhibits the development of malignant glioma (42). The IRES domain of circRNAs was subsequently predicted to be a binding site for many RBPs, including HUR and PTB, which modulate the translation of IRES element-driven proteins (43). As research continues to advance, it has been discovered that circRNAs without IRES components can be translated into multiple functional proteins. For example, N6-methyladenosine (m6A) not only affects mRNA translation under heat shock stress conditions (44) but also regulates the translation of circRNAs. The presumed mechanism is m6A modification by the METTL3/METTL14-WTAP protein complex, the regulation of m6A de-modification by FTO, and the site containing the m6A modification ultimately initiates protein translation by recruiting YTHDF3 and thus eIF4G2 (45), which fills a gap in the study of the chemical modifications of circRNAs. In addition, it was discovered that circRNAs can use overlapping codons to translate proteins, which is a unique method of translation (46). For example, circ-AKT3 uses overlapping codons to generate a new functional protein, AKT3-174aa. This protein affects the phosphorylation of AKT2/3 molecules by binding to p-PDK1, and then negatively regulates the PI3K/AKT signaling pathway, inhibiting the occurrence and development of brain tumors (47). This unique translation process has significance for the study of new functions of circRNAs (Figure 1).
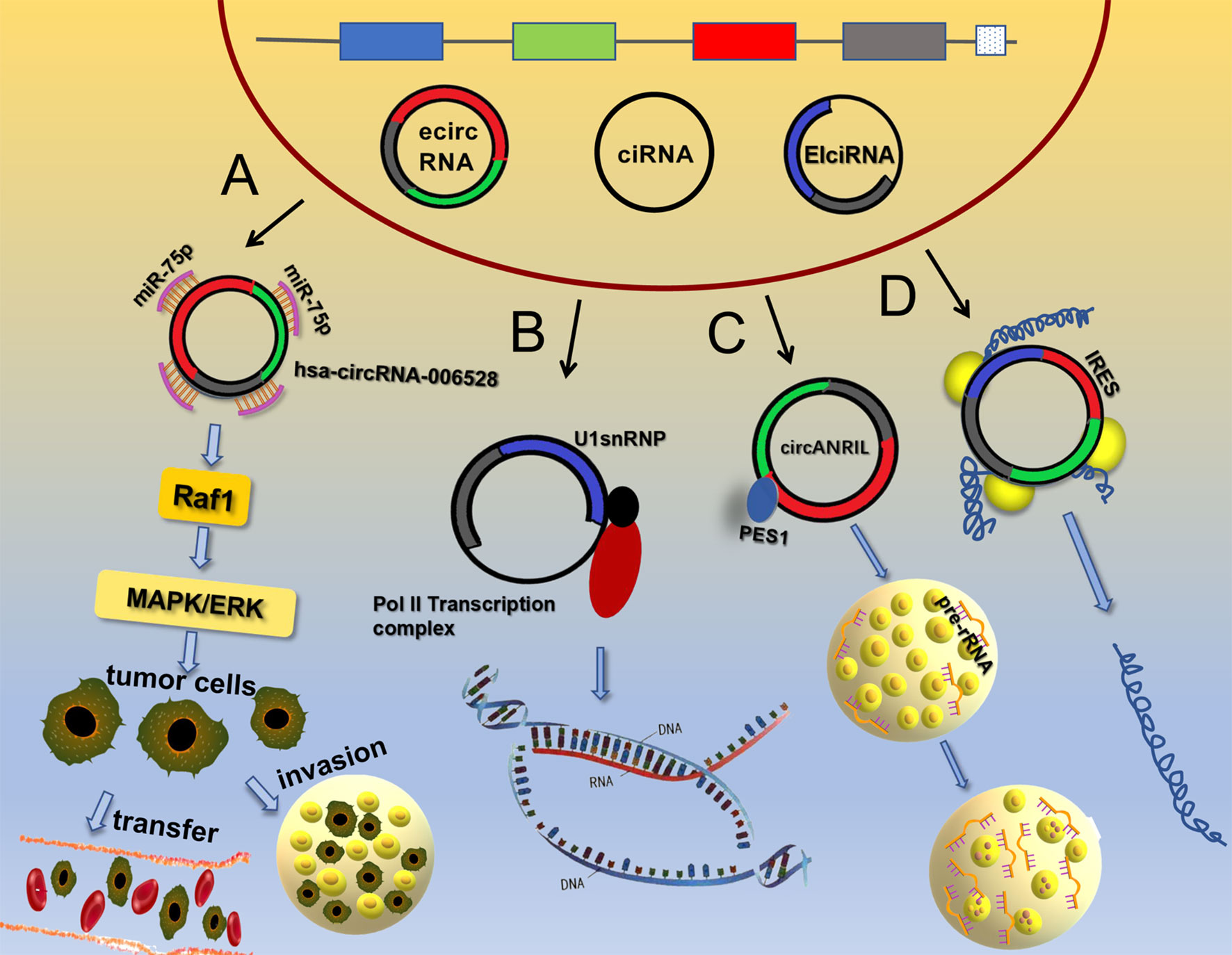
Figure 1 Main functions of circRNAs. (A) CircRNAs can adsorb miRNAs to activate signaling pathways. (B) EIciRNA and U1 snRNP form a complex that interacts with the RNA Pol II complex to regulate gene transcription. (C) Circ-ANRIL binds to PES1 protein to increase the number of cell nucleoli and pre-rRNA accumulation, thereby promoting apoptosis. (D) CircRNAs can be translated into protein through an IRES.
In 2015, it was first reported that circRNAs are enriched in exosomes and have good stability (48). The association of circRNAs with exosomes, followed by genome-wide analysis of RNA-sequencing data, showed that the abundance of circRNAs in exosomes and the circular–linear splicing rate is increased by at least 2- to 6-fold compared with their levels in producer cells, suggesting that circRNAs are actively incorporated into exosomes. Furthermore, there are more than 1,000 different circRNA candidates in human serum exosomes (49). Dou et al. used three colon cancer cell lines to show that circRNAs are more abundant in exosomes than in cells. In addition, different mutations of KRAS (proto-oncogene) were shown to have different effects on the exosomal content of circRNAs. These findings indicate that a large number of circRNAs are present in exosomes and that KRAS mutations affect their abundance (50).
Although the presence of circRNAs in exosomes has been confirmed, the mechanism by which circRNAs enter exosomes remains unclear. It has been found that circRNAs are selectively packaged into EVs, such as exosomes and microvesicles, but it is more pronounced in exosomes. circRNAs sorting into exosomes may be regulated by the following mechanisms: (1) lncRNAs competitively regulate circRNAs sorting into exosomes. Barbagallo C et al. found that knockdown of lncRNA UCA1 in serum exosomes could inhibit mitogen activated protein kinase (MAPK) signaling pathway, resulting in up regulation of circHIPK3 expression, suggesting that the competitive mechanism of lncRNA UCA1 may regulate the sorting of circHIPK3 (51). (2) circRNAs act as miRNA sponges. It’s reported that exosomal circCDR1as acted as a sponge for miR-7. When miR-7 was ectopically expressed in liver cancer cells, the expression level of circCDR1as in exosomes was significantly down-regulated, while the expression of circCDR1as in cells was increased (48). (3) RBPs recognize RNA with specific binding sequences and regulate the sorting of exosomal circRNAs. DKs-8 cells secreted exosomes enrich RBPs, which were involved in regulating the sorting process of circRNAs by binding with circFAT1 (50). (4) Exosomes preferentially release smaller circRNAs. Cells secrete exosomes circRNAs and release them to the extracellular environment may be related to the size of circRNAs. Preußer C et al. found that the average size of circRNAs that were not secreted from cells was 459 nts (52), while the average size of circRNAs released by exosomes was 435 nts, suggesting that size appears to be an important determinant for selective vesicle export of circRNAs (Figure 2). Nevertheless, the exact mechanism regarding the sorting and release of circRNAs is still largely unknown and awaits further study.
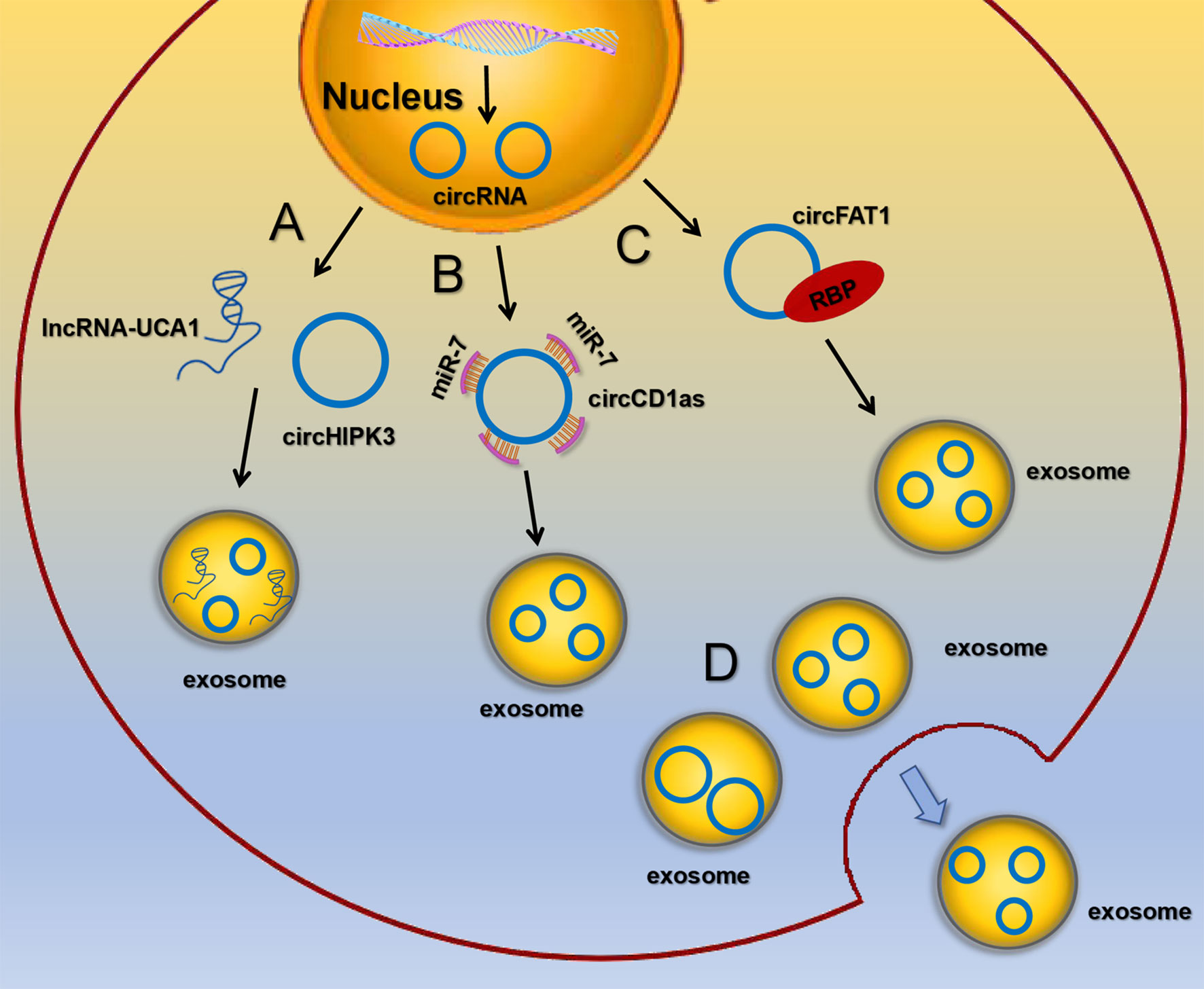
Figure 2 The sorting and secretion mechanisms of circRNA in exosomes (A) lncRNAs competitively regulate circRNAs sorting into exosomes. (B) circRNAs act as miRNA sponges. (C) RBPs recognize RNA with specific binding sequences and regulate the sorting of exosomal circRNAs. (D) Exosomes preferentially release smaller circRNAs.
CircRNAs are highly stable, evolutionarily conserved, and have no 3′- and 5′-ends (53). They also have built-in resistance to the major enzymes responsible for mRNA degradation, and thus may accumulate in cells. Lasda and Parker (54) analyzed the relative number of circRNAs and linear RNAs in EVs (including exosomes and microvesicles) in HeLa, 239T, and U-2OS cell lines. They found that circRNAs were more abundant compared with linear RNAs. A large number of circRNAs can be packaged into EVs, which bind to EVs and co-precipitate for their removal from cells. In addition, reducing the accumulation of circRNAs induced by reverse splicing may allow them to be exported preferentially from cells. This may be a mechanism for clearing accumulated circRNAs from cells. Furthermore, the authors suspected that there are other mechanisms for the removal or degradation of circRNAs, including transport mechanisms of other vesicles or endonuclease cleavage. Recently, a new process for RNA degradation mediated by m6A has been identified that can be applied to mRNAs and circRNAs. The m6A-recognition protein YTHDF2 binds to target molecules and recruits HRSP12. CircRNAs can be bound to and be degraded through the YTHDF2-HRSP12-mediated RNase P/MRP complex (55).
Exosomes or microvesicles can be taken up by other cells. They stimulate target cells directly through receptor-mediated interactions, and also transfer biologically active molecules such as membrane receptors, proteins, and mRNAs from source cells to target cells. Exosomes or microvesicles participate in communication between cells under physiological and pathological conditions (56). Therefore, Lasda and Parker believed that, in some cases, circRNAs may be packaged into EVs for cell-to-cell communication. Another study also suggested that the mechanism for the selective release of circRNAs is a way to transfer information from donor cells to recipient cells (52). However, the elimination mechanism and the signal communication pathway need to be explored further and confirmed.
Cancer is a global public health problem that poses a serious threat to human life and physical health, resulting in significant social and economic burdens. According to a report by GLOBOCAN, in 2020, the number of new cancer cases worldwide reached 19.3 million, and approximately 10 million people died from cancer. In addition, breast cancer has replaced lung cancer as the most common cancer worldwide, and it is estimated that by 2040, the global cancer burden will be 28.4 million cases, an increase of 47% compared to 2020 (57). Exosomal circRNAs play an important role in a wide range of pathological processes, especially in a variety of tumors, including occurrence, development, invasion, and other processes, and might make a significant contribution to the diagnosis and prognosis of tumors (58). Table 1 summarizes some of the functions and clinical significance of exosomal circRNAs in a range of different tumors.
As dysproliferation is one of the important factors in tumor transformation, the regulation mechanism of the cell cycle has received increasing attention (111). In recent years, an increasing number of exosomal circRNAs have been revealed to regulate cell proliferation in various cancers. Ding C et al. found that the level of circ-MEMO1 in serum of patients with non-small cell lung cancer (NSCLC) was higher than that in healthy people (59). Knockout of circ-MEMO1 inhibited NSCLC cell proliferation and blocked cell cycle in G0/G1 phase, while up-regulated circ-MEMO1 expression promoted cell proliferation, cell cycle progression and glycolysis metabolism and inhibited cell apoptosis. The possible mechanism might be exosomal circ-MEMO1 up-regulated KRAS expression in NSCLC cells by competitively binding miR-101-3p. Another study showed that exosomal circRASSF2 could promote the progression of laryngeal squamous cell carcinoma (LSCC). Compared with the control group, the expression of circRASSF2 in tumor tissues and serum exosomes were significantly up-regulated, and the down-regulation of exosomal circRASSF2 through the miRNA-302b-3p/IGF-1R axis could significantly suppress cell proliferation (60). Yin K et al. found that CircMMP1 was abnormally up-regulated in glioma tissues and serum exosomes compared with corresponding counterparts, and CircMMP1 promoted the proliferation and motility and impeded the apoptosis of glioma cells by enhanced high mobility group box 3 (HMGB3) level through sponging miR-433 (61). Exosomal circDB was up-regulated in hepatocellular carcinoma (HCC) patients with a high body fat rate. Exosomal circDB secreted by adipocytes could regulate deubiquitination of HCC by inhibiting miR-34a and activating USP7/Cyclin A2 signaling pathway, which promoted cell growth and reduced DNA damage (62). Lai Z et al. reported that CircFBLIM1 was highly expressed in HCC serum exosomes and HCC cells (63). Their further findings verified that exosomal circFBLIM1 contributed to the progression and glycolysis of HCC by sponging miR-338 and upregulating LRP6 expression. Chen W et al. show that circ0051443 expression was significantly lower in the plasma exosomes and tissues from patients with HCC than healthy controls (64). Exosomes containing circ-0051443 were secreted from normal cells, with exosomal circ-0051443 transported from normal cells to HCC cells. Exogenous circ-0051443 bound to miR-331-3p competitively and reduced the expression of Bcl2 Antagonist/Killer 1 (BAK1), a crucial cell death regulator, thereby suppressing the malignant biological behaviors by promoting cell apoptosis and arresting the cell cycle.
Exosomes can mediate molecular communication and material transfer between primary tumor sites and distant metastasis sites. By regulating a series of cell activities including epithelial mesenchymal transition (EMT) and angiogenesis, exosomes play crucial roles in tumor cell metastasis and invasion (112, 113). In colorectal cancer (CRC), circIFT80 accelerated the tumor progression by entering exosomes promoting CRC cell growth, migration, and invasion through mir-1236-3p/HOXB7 axis (68). circFMN2 was highly expressed in serum exosomes of patients with CRC and negatively correlated with the level of miRNA-1182. circFMN2 could increase the expression of hTERT by binding with miR-1182, which significantly promoted the proliferation and migration of CRC cells, suggesting that exosomal circFMN2 play an important role in promoting the growth of colorectal cancer (69). Exosomes from CD133+ CRC cells carrying circ-ABCC1 could activate the Wnt/β-catenin pathway to mediate cell stemness and metastasis in CRC (70). Shang A et al. also demonstrated that CRC-derived exosomes promote CRC proliferation, migration, and invasion (71). Their further research confirmed that circPACRGL was significantly up-regulated in CRC cells with tumor-derived exosomes addition. And CRC-derived exosomal circPACRGL regulated differentiation of N1-N2 neutrophils and promoted CRC proliferation, migration, and invasion via miR-142-3p/miR-506-3p-TGF-β1 axis. Zeng W et al. found that the expression of circFNDC3B was significantly decreased in CRC tissues, CRC cell lines and exosomes. circFNDC3B-enriched exosomes inhibited angiogenesis and CRC progression (72). This study also demonstrated that circFNDC3B-enriched exosomes could repress angiogenesis and CRC progression by decreasing VEGFR expression. Interestingly, another research showed that circFNDC3B was highly expressed in extracted serum exosomes derived from papillary thyroid cancer (PTC) patients, and the expression levels of circFNDC3B and Mir-1178 have significantly negatively correlated (73). Their further research demonstrated that circFNDC3B inhibited PTC cell proliferation, migration, and invasion and promoted cell apoptosis through the miR-1178/TLR4 pathway. In addition to making cancer cells more invasive, exosomal circRNAs can also promote the invasion of cancer cells by changing the tumor microenvironment. Huang C et al. found that exosome-mediated the intercellular transmission of circANTXR1 in HCC cells (75). Overexpressed exosomal circANTXR1 could depress miR-532-5p expression and promote XRCC5 mRNA and protein expression, promoting the proliferation, migration and invasion of HCCLM3 cells. Another research suggested that exosomal circRNA Cdr1as from HCC cells enhanced circRNA Cdr1as expression and accelerated proliferative and migratory abilities to surround normal cells (76). It’s reported that exosomal circRNA-100338 was up-regulated in a highly metastatic HCC cell line. The metastatic ability of HCC cells could be enhanced by transferring exosomal circRNA-100338 to recipient human umbilical vein endothelial cells (HUVECs) and also promoted cell proliferation, angiogenesis, permeability, and vasculogenic mimicry (VM) formation ability and tumor metastasis (77).
Nowadays, the combination of radiotherapy and chemotherapy after surgical resection is the main clinical treatment of malignant tumors. Although the initial efficacy is remarkable, with the emergence of chemoresistance, it has become a huge obstacle to the prognosis. Studies have confirmed that epigenetic variation, oncogene activation, anti-oncogene inactivation, tumor heterogeneity and apoptosis dysregulation are the main causes of tumor drug resistance (114). Current studies have shown that circRNAs can play crucial roles in mediating tumor chemoresistance (115), and exosomes, as essential mediators of communication between tumor cells or between tumor cells and stromal cells, can play a role in the transmission of chemoresistance through the transfer of circRNA (86). At present, the study of circRNA-mediated tumor treatment resistance through exosome delivery is at the forefront of academic research and has important scientific significance.
In serum exosomes of osimertinib-resistance NSCLC patients, Ma J et al. found that the expression of hsa_circ_0002130 was up-regulated, while knockdown of hsa_circ_0002130 significantly inhibited cell proliferation, glycolysis, and promoted cell apoptosis in osimertinib-resistant NSCLC, thereby inhibiting tumor growth (94). The mechanism was to regulate the expression of glucose transporter 1 (GLUT1), hexokinase-2 (HK2) and lactate dehydrogenase A (LDHA) by sponging miR-498, which promoted osimertinib-resistant NSCLC cells to respond to osimertinib-resistance. Hence, hsa_circ_0002130 could act as a new therapeutic target for osimertinib-resistant NSCLC. HCC-derived exosomal circUHRF1 expression was elevated compared with healthy control and was associated with poor clinical prognosis and natural killer (NK) cells dysfunction. Additionally, exosomal circUHRF1 could inhibit IFN-γ and TNF-α secretion in NK cells by upregulating the expression of T cell immunoglobulin and mucin domain 3 (TIM-3) via degradation of miR-449c-5p, thereby promoting HCC immune avoidance and anti-PD1 immunotherapy resistance (97). Wang X et al. demonstrated that ciRS-122 could be transferred from oxaliplatin-resistant CRC cell exosomes to sensitive cells, which increased the expression of the M2 isoform of pyruvate kinase (PKM2) by suppressing miR-122, then promoting glycolysis and drug resistance (100). In addition, si-ciRS-122 could block the ciRS-122/miR-122/PKM2 axis at the post-transcriptional level and reverse the resistance of CRC cells to oxaliplatin. In epithelial ovarian cancer (EOC) patients, the expression of exosomal circFoxp1 in cisplatin (DDP)-resistant patients was significantly higher than that in DDP-sensitive patients. Overexpressed exosomal circFoxp1 could enhance the survival and proliferation of EOC cells and promote the resistance of EOC cells to DDP, which could be an independent predictor of survival outcome and DDP resistance in EOC patients. The mechanism might be that the expression of CCAAT enhancer binding protein gamma (CEBPG) and formin like 3 (FMNL3) are positively regulated by miR-22 and miR-150-3p (102). Furthermore, Zhao M et al. found that circATP8B4 from radioresistant U251 extracellular vesicles might be transferred to normal glioma U251 cells and act as an miR−766 sponge to promote cell radioresistance (108). The above studies suggest that a variety of exosomal circRNAs are closely related to treatment resistance and may become a new target for tumor therapy.
Exosomal circRNAs not only play a role in a variety of biological processes in tumors but also have an impact on the mechanisms of a variety of other diseases (116). For instance, 5,095 circRNAs were identified in cerebrospinal fluid-derived exosomes from patients with immune-mediated demyelinating disease. Among them, the expression of 26 circRNAs was found to be significantly different in cerebrospinal fluid exosomes. Further study showed that hsa_circ_0087862 and hsa_circ_0012077 could act as molecular markers for the diagnosis of immune-mediated demyelinating disease and could also indicate cellular metabolism and disease progression (117). Secondly, circrNA-EP400, significantly increased in M2 macrophage exosomes, was reported to inhibit miR-15b-5p expression and increase the expression of fibroblast growth factor (FGF)-1/7/9, thereby promoting fibrosis, proliferation, and migration of fibroblasts and tenocytes (118). This study provides a new therapy for tendon injury. In polycystic ovary syndrome, the expression level of exosomal circLDLR decreased significantly. As an important mediator, circLDLR could directly bind to miR-129, inhibit the expression of CYP19A1 in KGN cells, and reduce the secretion of estradiol (119). Exosomal circ-Ehmt1 regulates retinal microvascular dysfunction through the NFIA/NLRP3 signaling pathway by inhibiting the formation of NLRP3 inflammasomes in endothelial cells to protect endothelial cells from high glucose-induced damage, which could be a therapeutic target for diabetic retinopathy (120). Besides, some differentially expressed exosomal circRNAs with miRNA-binding sites in umbilical cord blood are essential for the progression of gestational diabetes and fetal growth and development (121). Exosomal circ_0000253 is upregulated in nucleus pulposus cells (NPCs) during intervertebral disc degeneration, and competitively adsorbs miRNA-141-5p and downregulates SIRT1 expression to promote the apoptosis of NPCs and inhibit the proliferation, blocking circRNA_0000253 as a potential treatment for IDD (122). Notably, exosomal circRNAs have also been demonstrated to have a role in the pathogenesis of Corona Virus Disease 2019 (COVID-19). Studies have shown that differentially expressed circRNAs and long non-coding RNAs in exosomes may be involved in regulating host cell immunity and inflammation, substance and energy metabolism, cell cycle, and apoptosis (123). All of these studies provide a new direction for the diagnosis and treatment of various diseases. Table 2 summarizes some of the functions and clinical significance of exosomal circRNAs in other diseases.
Exosomal circRNAs not only participate in pathological processes but also play key roles in some physiological processes. For example, hsa_circ_0075932 has a low basal expression level in dermal keratinocytes, but its overexpression in these cells has no deleterious effect. However, if it is significantly expressed in adipose tissue, it directly combines with PUM2 to activate the AuroraA/NF-κB pathway, thereby promoting cell inflammation and apoptosis (135). In addition, Zhao et al. conducted experiments in mice, showing that exosomal circRNAs may participate in neuron growth and repair, nervous system development, and nerve signal transmission through glutamatergic synapses and cGMP-PKG signaling pathways (136). Therefore, it is necessary to further study exosomal circRNAs, which might help us better understand some important physiological processes.
Exosomes and circRNAs are both hot research topics. In this review, several important functions of circular RNA are summarized in detail. For example, circRNAs can act as miRNA sponges to affect gene expression, interact with Pol II to regulate the transcription of parental genes, act as a protein sponge to affect protein expression, and participate in protein translation through multiple pathways. Moreover, experiments have shown that circRNAs can enter exosomes. Although its regulatory mechanism is not completely clear, exosomal circRNAs play an important role in pathological processes such as tumors, nerves, cardiovascular diseases, as well as physiological processes such as inflammation and cell apoptosis. Exosomes are rich in lipid components such as cholesterol, neuramide, and sphingolipids, which are not easily degraded by proteases and ribonucleases, so circRNAs are stable in exosomes and can perform their functions. In addition, exosomes are found widely in various body fluids, such as blood, tears, saliva, sputum, semen, urine, and breast milk (137, 138). Therefore, liquid biopsies can be performed; it is convenient and simple for clinicians to take specimens and it is not traumatic for patients. Therefore, the study of exosomal circRNAs will provide a biological basis for the identification of new markers for early diagnosis, targeted therapy, and prognostic evaluation of various diseases in the future.
Even though significant breakthroughs have been made in the study of exosomes and circRNAs, there are still many unanswered questions. First, what conditions affect the interactions of circRNAs with other molecules, and what other types of modifications other than m6A in circRNAs are involved in protein translation? Second, are there differences between exosomes in cells and those in plasma, and do these differences affect the expression of circRNAs? Third, what is the specific mechanism by which circRNAs enter exosomes, and how are they cleared or involved in cell-to-cell communications? Fourth, blood, urine, sputum, and other liquids also contain many other impurities, including protein complexes and nucleic acid lysates, which interfere with the extraction of exosomes. Therefore, there are still many difficulties and challenges in the clinical application of exosomal circRNAs, and thus continuous in-depth research and exploration are needed.
JH and JL provided direction and guidance throughout the preparation of this manuscript. ZH, HC, and ZG wrote and edited the manuscript. JS and WL reviewed and made significant revisions to the manuscript. FX, YW, and SW collected and prepared the related papers. All authors contributed to the article and approved the submitted version.
This work was supported by Science and Technology Project of Panyu District, Guangzhou (2019-Z04-02; 2020-Z04-026), Guangzhou Health and Family Planning Commission Program (No. 20181A011118; 20192A011027; 20191A011119; 20201A010085; 20212A010025), Guangzhou Science and Technology Plan Project (No. 202002030032), Medical Science and Technology Research Foundation of Guangdong Province (No. A2020304; A2022524), Scientific Research Fund project of Guangzhou Panyu Central Hospital (No. 2021Y002; 2021Y004).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Zhu L, Li N, Sun L, Zheng D, Shao G. Non-Coding RNAs: The Key Detectors and Regulators in Cardiovascular Disease. Genomics (2021) 113(Pt 2):1233–46. doi: 10.1016/j.ygeno.2020.10.024
2. Khan S, Ayub H, Khan T, Wahid F. MicroRNA Biogenesis, Gene Silencing Mechanisms and Role in Breast, Ovarian and Prostate Cancer. Biochimie (2019) 167:12–24. doi: 10.1016/j.biochi.2019.09.001
3. Huang Y, Guo Q, Ding XP, Wang X. Mechanism of Long Noncoding RNAs as Transcriptional Regulators in Cancer. RNA Biol (2020) 17(11):1680–92. doi: 10.1080/15476286.2019.1710405
4. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD. Scrambled Exons. Cell (1991) 64(3):607–13. doi: 10.1016/0092-8674(91)90244-S
5. Lasda E, Parker R. Circular RNAs: Diversity of Form and Function. RNA (2014) 20(12):1829–42. doi: 10.1261/rna.047126.114
6. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular Intronic Long Noncoding RNAs. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
7. Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, Lee N, et al. Circular DNA Tumor Viruses Make Circular RNAs. Proc Natl Acad Sci USA (2018) 115(37):E8737–45. doi: 10.1073/pnas.1811728115
8. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
9. Schmidt CA, Giusto JD, Bao A, Hopper AK, Matera AG. Gregory.Molecular Determinants of Metazoan tricRNA Biogenesis. Nucleic Acids Res (2019) 47(12):6452–65. doi: 10.1093/nar/gkz311
10. Chen J, Shi P, Zhang J, Li Y, Ma J, Zhang Y, et al. CircRNA_0044556 Diminishes the Sensitivity of Triple−Negative Breast Cancer Cells to Adriamycin by Sponging Mir−145 and Regulating NRAS. Mol Med Rep (2022) 25(2):51. doi: 10.3892/mmr.2021.12567
11. Xie L, Pan Z. Circular RNA Circ_0000467 Regulates Colorectal Cancer Development via miR-382-5p/EN2 Axis. Bioengineered (2021) 12(1):886–97. doi: 10.1080/21655979.2021.1889130
12. Lai Q, Wang M, Hu C, Tang Y, Li Y, Hao S. Circular RNA Regulates the Onset and Progression of Cancer Through the Mitogen-Activated Protein Kinase Signaling Pathway. Oncol Lett (2021) 22(6):817. doi: 10.3892/ol.2021.13078
13. Cui X, Wang J, Guo Z, Li M, Li M, Liu S, et al. Emerging Function and Potential Diagnostic Value of Circular RNAs in Cancer. Mol Cancer (2018) 17(1):123. doi: 10.1186/s12943-018-0877-y
14. Yuan G, Ding W, Sun B, Zhu L, Gao Y, Chen L. Upregulated circRNA_102231 Promotes Gastric Cancer Progression and Its Clinical Significance. Bioengineered (2021) 12(1):4936–45. doi: 10.1080/21655979.2021.1960769
15. Du N, Li M, Yang D. Hsa_circRNA_102541 Regulates the Development of Atherosclerosis by Targeting miR-296-5p/PLK1 Pathway. Ir J Med Sci (2021) doi: 10.1007/s11845-021-02708-x.
16. Sun Y, Zhou Y, Shi X, Ma X, Peng X, Xie Y, et al. CircTYW1 Serves as a Sponge for microRNA-380 in Accelerating Neurological Recovery Following Spinal Cord Injury via Regulating Fgf9. Cell Cycle (2021) 20(18):1828–44. doi: 10.1080/15384101.2021.1962634
17. Sun N, Liang Y, Hu B, Feng J, Lin G, Chen X, et al. circSKIL Promotes the Ossification of Cervical Posterior Longitudinal Ligament by Activating the JNK/STAT3 Pathway. Exp Ther Med (2021) 22(1):761. doi: 10.3892/etm.2021.10193
18. Raposo G, Stoorvogel W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. Cell Biol (2013) 200(4):373–83. doi: 10.1083/jcb.201211138
19. Wolf P. The Nature and Significance of Platelet Products in Human Plasma. Br J Haematol (1967) 13(3):269–88. doi: 10.1111/j.1365-2141.1967.tb08741.x
20. Pan B, Johnstone R. Fate of the Transferrin Receptor During Maturation of Sheep Reticulocytes In Vitro: Selective Externalization of the Receptor. Cell (1983) 33(3):967–78. doi: 10.1016/0092-8674(83)90040-5
21. Li SP, Lin ZX, Jiang XY, Yu XY. Exosomal Cargo-Loading and Synthetic Exosome-Mimics as Potential Therapeutic Tools. Acta Pharmacol Sin (2018) 39(4):542–51. doi: 10.1038/aps.2017.178
22. Singh D, Kesharwani P, Alhakamy NA, Siddique HR. Accentuating CircRNA-miRNA-Transcription Factors Axis: A Conundrum in Cancer Research. Front Pharmacol (2022) 12:784801. doi: 10.3389/fphar.2021.784801
23. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
24. Hansen T, Wiklund E, Bramsen J. MiRNA-Dependent Gene Silencing Involving A902-Mediated Cleavage of a Circular Antisense RNA. EMBOJ (2011) 30(21):4414–22. doi: 10.1038/emboj.2011.359
25. Li Y, Shi H, Yuan J, Qiao L, Dong L, Wang Y. Downregulation of Circular RNA Circpvt1 Restricts Cell Growth of Hepatocellular Carcinoma Through Downregulation of Sirtuin 7 via microRNA-3666. Clin Exp Pharmacol Physiol (2020) 47(7):1291–300. doi: 10.1111/1440-1681.13273
26. Kim SS, Lee SV. Non-Coding RNAs in Caenorhabditis Elegans Aging. Mol Cells (2019) 42(5):379–85. doi: 10.14348/molcells.2019.0077
27. Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread Noncoding Circular RNAs in Plants. New Phytol (2015) 208(1):88–95. doi: 10.1111/nph.13585
28. Wang Y, Yang M, Wei S, Qin F, Zhao H, Suo B. Identification of Circular RNAs and Their Targets in Leaves of Triticum Aestivum L. Under Dehydration Stress. Front Plant Sci (2017) 7:2024. doi: 10.3389/fpls.2016.02024.
29. Zeng RF, Zhou JJ, Hu CG, Zhang JZ. Transcriptome-Wide Identification and Functional Prediction of Novel and Flowering-Related Circular RNAs From Trifoliate Orange (Poncirus Trifoliata L. Raf.). Planta (2018) 247(5):1191–202. doi: 10.1007/s00425-018-2857-2
30. Bose R, Ain R. Regulation of Transcription by Circular RNAs. Adv Exp Med Biol (2018) 1087:81–94. doi: 10.1007/978-981-13-1426-1_7
31. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA Biogenesis Competes With pre-mRNA Splicing. Mol Cell (2014) 56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
32. Chia W, Liu J, Huang YG, Zhang C. A Circular RNA Derived From DAB1 Promotes Cell Proliferation and Osteogenic Differentiation of BMSCs via RBPJ/DAB1 Axis. Cell Death Dis (2020) 11(5):372. doi: 10.1038/s41419-020-2572-3
33. Okholm TLH, Sathe S, Park SS, Kamstrup AB, Rasmussen AM, Shankar A, et al. Transcriptome-Wide Profiles of Circular RNA and RNA-Binding Protein Interactions Reveal Effects on Circular RNA Biogenesis and Cancer Pathway Expression. Genome Med (2020) 12(1):112. doi: 10.1186/s13073-020-00812-8
34. Zhang L, Wang Y, Zhang Y, Zhao Y, Li P. Pathogenic Mechanisms and the Potential Clinical Value of Circfoxo3 in Cancers. Mol Ther Nucleic Acids (2021) 23:908–17. doi: 10.1016/j.omtn.2021.01.010
35. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 Circular RNA Retards Cell Cycle Progression via Forming Ternary Complexes With P21 and CDK2. Nucleic Acids Res (2016) 44(6):2846–58. doi: 10.1093/nar/gkw027
36. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular Non-Coding RNA ANRIL Modulates Ribosomal RNA Maturation and Atherosclerosis in Humans. Nat Commun (2016) 7:12429. doi: 10.1038/ncomms12429
37. Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A Novel Protein Encoded by a Circular RNA Circppp1r12a Promotes Tumor Pathogenesis and Metastasis of Colon Cancer via Hippo-YAP Signaling. Mol Cancer (2019) 18(1):47. doi: 10.1186/s12943-019-1010-6
38. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 Is a Circular RNA That Can Be Translated and Functions in Myogenesis. Mol Cell (2017) 66(1):22–37. doi: 10.1016/j.molcel.2017.02.017
39. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst (2018) 110(3):304–15. doi: 10.1093/jnci/djx166
40. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66(1):9–21. doi: 10.1016/j.molcel.2017.02.021
41. Chen CY, Sarnow P. Initiation of Protein Synthesis by the Eukaryotic Translational Apparatus on Circular RNAs. Science (1995) 268(5209):415–7. doi: 10.1126/science.7536344
42. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A Peptide Encoded by Circular Form of LINC-PINT Suppresses Oncogenic Transcriptional Elongation in Glioblastoma. Nat Commun (2018) 9(1):4475. doi: 10.1038/s41467-018-06862-2
43. Martínez-Salas E, Lozano G, Fernandez-Chamorro J, Francisco-Velilla R, Galan A, Diaz R. RNA-Binding Proteins Impacting on Internal Initiation of Translation. Int J Mol Sci (2013) 14(11):21705–26. doi: 10.3390/ijms141121705
44. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic M(6)A mRNA Methylation Directs Translational Control of Heat Shock Response. Nature (2015) 526(7574):591–4. doi: 10.1038/nature15377
45. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive Translation of Circular RNAs Driven by N6-Methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
46. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A Novel Protein Encoded by the Circular Form of the SHPRH Gene Suppresses Glioma Tumorigenesis. Oncogene (2018) 37(13):1805–14. doi: 10.1038/s41388-017-0019-9
47. Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, et al. A Novel Tumor Suppressor Protein Encoded by Circular AKT3 RNA Inhibits Glioblastoma Tumorigenicity by Competing With Active Phosphoinositide-Dependent Kinase-1. Mol Cancer (2019) 18(1):131. doi: 10.1186/s12943-019-1056-5
48. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA Is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res (2015) 25(8):981–4. doi: 10.1038/cr.2015.82
49. Bao C, Lyu D, Huang S. Circular RNA Expands Its Territory. Mol Cell Oncol (2016) 3(2):e1084443. doi: 10.1080/23723556.2015.1084443
50. Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, et al. Circular RNAs Are Down-Regulated in KRAS Mutant Colon Cancer Cells and Can be Transferred to Exosomes. Sci Rep (2016) 6:37982. doi: 10.1038/srep37982
51. Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A, et al. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol Ther Nucleic Acids (2018) 12:229–41. doi: 10.1016/j.omtn.2018.05.009
52. Preußer C, Hung LH, Schneider T, Schreiner S, Hardt M, Moebus A, et al. Selective Release of circRNAs in Platelet-Derived Extracellular Vesicles. J Extracellular Vesicles (2018) 7(1):1424473. doi: 10.1080/20013078.2018.1424473
53. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs Are Abundant, Conserved, and Associated With ALU Repeats. RNA (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
54. Lasda E, Parker R. Circular RNAs Co-Precipitate With Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PloS One (2016) 11(2):e0148407. doi: 10.1371/journal.pone.0148407
55. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic Cleavage of M6a-Containing RNAs by RNase P/MRP Complex. Mol Cell (2019) 74(3):494–507. doi: 10.1016/j.molcel.2019.02.034
56. Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a Mechanism of Cell-To-Cell Communication. Kidney Int (2010) 78(9):838–48. doi: 10.1038/ki.2010.278
57. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
58. Bai H, Lei K, Huang F, Jiang Z, Zhou X. Exo-circRNAs: A New Paradigm for Anticancer Therapy. Mol Cancer (2019) 18(1):56. doi: 10.1186/s12943-019-0986-2
59. Ding C, Xi G, Wang G, Cui D, Zhang B, Wang H, et al. Exosomal Circ-MEMO1 Promotes the Progression and Aerobic Glycolysis of Non-Small Cell Lung Cancer Through Targeting MiR-101-3p/KRAS Axis. Front Genet (2020) 11:962. doi: 10.3389/fgene.2020.00962
60. Tian L, Cao J, Jiao H, Zhang J, Ren X, Liu X, et al. CircRASSF2 Promotes Laryngeal Squamous Cell Carcinoma Progression by Regulating the miR-302b-3p/IGF-1r Axis. Clin Sci (Lond) (2019) 133:1053–66. doi: 10.1042/CS20190110
61. Yin K, Liu X. CircMMP1 Promotes the Progression of Glioma Through miR-433/HMGB3 Axis In Vitro and In Vivo. IUBMB Life (2020) 72(11):2508–24. doi: 10.1002/iub.2383
62. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA Secreted From Adipocytes Promotes the Growth of Hepatocellular Carcinoma by Targeting Deubiquitination-Related Usp7. Oncogene (2019) 38(15):2844–59. doi: 10.1038/s41388-018-0619-z
63. Lai Z, Wei T, Li Q, Wang X, Zhang Y, Zhang S. Exosomal Circfblim1 Promotes Hepatocellular Carcinoma Progression and Glycolysis by Regulating the miR-338/LRP6 Axis. Cancer Biother Radiopharm (2020) doi: 10.1089/cbr.2020.3564
64. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-Transmitted Circular RNA Hsa_Circ_0051443 Suppresses Hepatocellular Carcinoma Progression. Cancer Lett (2020) 475:119–28. doi: 10.1016/j.canlet.2020.01.022
65. Luo Y, Liu F, Guo J, Gui R. Upregulation of Circ_0000199 in Circulating Exosomes Is Associated With Survival Outcome in OSCC. Sci Rep (2020) 10(1):13739. doi: 10.1038/s41598-020-70747-y
66. Xiao H, Shi J. Exosomal Circular RNA_400068 Promotes the Development of Renal Cell Carcinoma via the miR-210-5p/SOCS1 Axis. Mol Med Rep (2020) 22(6):4810–20. doi: 10.3892/mmr.2020.11541
67. Sun R, Liu W, Zhao Y, Chen H, Wang Z, Zhang Y, et al. Exosomal circRNA as a Novel Potential Therapeutic Target for Multiple Myeloma-Related Myocardial Damage. Cancer Cell Int (2021) 21(1):311. doi: 10.1186/s12935-021-02011-w
68. Feng W, Gong H, Wang Y, Zhu G, Xue T, Wang Y, et al. Circift80 Functions as a ceRNA of miR-1236-3p to Promote Colorectal Cancer Progression. Mol Ther Nucleic Acids (2019) 18:375–87. doi: 10.1016/j.omtn.2019.08.024
69. Li Y, Li C, Xu R, Wang Y, Li D, Zhang B. A Novel Circfmn2 Promotes Tumor Proliferation in CRC by Regulating the miR-1182/hTERT Signaling Pathways. Clin Sci (London England: 1979) (2019) 133(24):2463–79. doi: 10.1042/CS20190715
70. Hong Z, Chen S, Fu Q. Exosomes From CD133 Cells Carrying Circ-ABCC1 Mediate Cell Stemness and Metastasis in Colorectal Cancer. J Cell Biochem (2020) 121:3286–97. doi: 10.1002/jcb.29600
71. Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL Promotes Progression of Colorectal Cancer via the miR-142-3p/miR-506-3p- TGF-β1 Axis. Mol Cancer (2020) 19(1):117. doi: 10.1186/s12943-020-01235-0
72. Zeng W, Liu Y, Li WT, Li Y, Zhu JF. CircFNDC3B Sequestrates miR-937-5p to Derepress TIMP3 and Inhibit Colorectal Cancer Progression. Mol Oncol (2020) 14(11):2960–84. doi: 10.1002/1878-0261.12796
73. Wu G, Zhou W, Pan X, Sun Z, Sun Y, Xu H, et al. Circular RNA Profiling Reveals Exosomal Circ_0006156 as a Novel Biomarker in Papillary Thyroid Cancer. Mol Ther Nucleic Acids (2020) 19:1134–44. doi: 10.1016/j.omtn.2019.12.025
74. Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, et al. Hypoxia Induced Exosomal circRNA Promotes Metastasis of Colorectal Cancer via Targeting GEF-H1/RhoA Axis. Theranostics (2020) 10:8211–26. doi: 10.7150/thno.44419
75. Huang C, Yu W, Wang Q, Huang T, Ding Y. CircANTXR1 Contributes to the Malignant Progression of Hepatocellular Carcinoma by Promoting Proliferation and Metastasis. J Hepatocell Carcinoma (2021) 8:1339–53. doi: 10.2147/JHC.S317256
76. Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, et al. CircRNA Cdr1as Functions as a Competitive Endogenous RNA to Promote Hepatocellular Carcinoma Progression. Aging (2019) 11(19):8183–203. doi: 10.18632/aging.102312
77. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal Circrna-100338 Promotes Hepatocellular Carcinoma Metastasis via Enhancing Invasiveness and Angiogenesis. J Exp Clin Cancer Res (2020) 39(1):20. doi: 10.1186/s13046-020-1529-9
78. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three Isoforms of Exosomal Circptgr1 Promote Hepatocellular Carcinoma Metastasis via the Mir449a-MET Pathway. EBioMedicine (2019) 40:432–45. doi: 10.1016/j.ebiom.2018.12.062
79. Lin Y, Zheng ZH, Wang JX, Zhao Z, Peng TY. Tumor Cell-Derived Exosomal Circ-0072088 Suppresses Migration and Invasion of Hepatic Carcinoma Cells Through Regulating MMP-16. Front Cell Dev Biol (2021) 9:726323. doi: 10.3389/fcell.2021.726323
80. Zhu C, Su Y, Liu L, Wang S, Liu Y, Wu J. Circular RNA Hsa_Circ_0004277 Stimulates Malignant Phenotype of Hepatocellular Carcinoma and Epithelial-Mesenchymal Transition of Peripheral Cells. Front Cell Dev Biol (2021) 8:585565. doi: 10.3389/fcell.2020.585565
81. Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, et al. Circular RNA IARS (Circ-IARS) Secreted by Pancreatic Cancer Cells and Located Within Exosomes Regulates Endothelial Monolayer Permeability to Promote Tumor Metastasis. J Exp Clin Cancer Res: CR (2018) 37(1):177. doi: 10.1186/s13046-018-0822-3
82. Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, et al. Tumor-Released Exosomal Circular RNA PDE8A Promotes Invasive Growth via the miR-338/MACC1/MET Pathway in Pancreatic Cancer. Cancer Lett (2018) 432:237–50. doi: 10.1016/j.canlet.2018.04.035
83. Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder Through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin Cancer Res (2018) 24(24):6319–30. doi: 10.1158/1078-0432.CCR-18-1270
84. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA Circnrip1 Acts as a microRNA-149-5p Sponge to Promote Gastric Cancer Progression via the AKT1/mTOR Pathway. Mol Cancer (2019) 18(1):20. doi: 10.1186/s12943-018-0935-5
85. Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, et al. Circular RNA Circ-RanGAP1 Regulates VEGFA Expression by Targeting miR-877-3p to Facilitate Gastric Cancer Invasion and Metastasis. Cancer Lett (2020) 471:38–48. doi: 10.1016/j.canlet.2019.11.038
86. Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, et al. Exosomal Circshkbp1 Promotes Gastric Cancer Progression via Regulating the miR-582-3p/HUR/VEGF Axis and Suppressing HSP90 Degradation. Mol Cancer (2020) 19(1):112. doi: 10.1186/s12943-020-01208-3
87. Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, et al. Circular RNA Circsatb2 Promotes Progression of Non-Small Cell Lung Cancer Cells. Mol Cancer (2020) 19:101. doi: 10.1186/s12943-020-01221-6
88. Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y, et al. FLI1 Exonic Circular RNAs as a Novel Oncogenic Driver to Promote Tumor Metastasis in Small Cell Lung Cancer. Clin Cancer Res (2019) 25(4):1302–17. doi: 10.1158/1078-0432.CCR-18-1447
89. Xu L, Liao WL, Lu QJ, Zhang P, Zhu J, Jiang GN, et al. Hypoxic Tumor-Derived Exosomal Circular RNA SETDB1 Promotes Invasive Growth and EMT via the miR-7–Sp1 Axis in Lung Adenocarcinoma. Mol Ther Nucleic Acids (2021) 23:1078–92. doi: 10.1016/j.omtn.2021.01.019
90. Guan X, Zong ZH, Liu Y, Chen S, Wang LL, Zhao Y. Circpum1 Promotes Tumorigenesis and Progression of Ovarian Cancer by Sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids (2019) 18:882–92. doi: 10.1016/j.omtn.2019.09.032
91. Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 Promotes Ovarian Cancer Progression by Regulating MUC1 and hTERT Through Sponging miR-145 and miR-1182. J Exp Clin Cancer Res (2019) 38:437. doi: 10.1186/s13046-019-1437-z
92. Wang S, Hu Y, Lv X, Li B, Gu D, Li Y, et al. Circ-0000284 Arouses Malignant Phenotype of Cholangiocarcinoma Cells and Regulates the Biological Functions of Peripheral Cells Through Cellular Communication. Clin Sci (Lond) (2019) 133(18):1935–53. doi: 10.1042/CS20190589
93. Li T, Sun X, Chen L. Exosome Circ_0044516 Promotes Prostate Cancer Cell Proliferation and Metastasis as a Potential Biomarker. J Cell Biochem (2020) 121:2118–266. doi: 10.1002/jcb.28239
94. Ma J, Qi G, Li L. A Novel Serum Exosomes-Based Biomarker Hsa_Circ_0002130 Facilitates Osimertinib-Resistance in Non-Small Cell Lung Cancer by Sponging miR-498. OncoTargets Ther (2020) 13:5293–307. doi: 10.2147/OTT.S243214
95. Xu X, Tao R, Sun L, Ji X. Exosome-Transferred Hsa_Circ_0014235 Promotes DDP Chemoresistance and Deteriorates the Development of Non-Small Cell Lung Cancer by Mediating the miR-520a-5p/CDK4 Pathway. Cancer Cell Int (2020) 20(1):552. doi: 10.1186/s12935-020-01642-9
96. Yang B, Teng F, Chang L, Wang J, Liu DL, Cui YS, et al. Tumor-Derived Exosomal circRNA_102481 Contributes to EGFR-TKIs Resistance via the miR-30a-5p/ROR1 Axis in Non-Small Cell Lung Cancer. Aging (2021) 13(9):13264–86. doi: 10.18632/aging.203011
97. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer Cell-Derived Exosomal Circuhrf1 Induces Natural Killer Cell Exhaustion and May Cause Resistance to Anti-PD1 Therapy in Hepatocellular Carcinoma. Mol Cancer (2020) 19(1):110. doi: 10.1186/s12943-020-01222-5
98. Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, et al. CircRNA-SORE Mediates Sorafenib Resistance in Hepatocellular Carcinoma by Stabilizing Ybx1. Signal Transduct Target Ther (2020) 5(1):298. doi: 10.1038/s41392-020-00375-5
99. Lu JC, Zhang PF, Huang XY, Guo XJ, Gao C, Zeng HY, et al. Amplification of Spatially Isolated Adenosine Pathway by Tumor-Macrophage Interaction Induces Anti-PD1 Resistance in Hepatocellular Carcinoma. J Hematol Oncol (2021) 14(1):200. doi: 10.1186/s13045-021-01207-x
100. Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, et al. Exosome-Delivered circRNA Promotes Glycolysis to Induce Chemoresistance Through the miR-122-PKM2 Axis in Colorectal Cancer. Mol Oncol (2020) 14:539–55. doi: 10.1002/1878-0261.12629
101. Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal R, Abu N. Extracellular Vesicle-Derived Circular RNAs Confers Chemoresistance in Colorectal Cancer. Sci Rep (2019) 9(1):16497. doi: 10.1038/s41598-019-53063-y
102. Luo Y, Gui R. Circulating Exosomal Circfoxp1 Confers Cisplatin Resistance in Epithelial Ovarian Cancer Cells. J Gynecol Oncol (2020) 31(5):e75. doi: 10.3802/jgo.2020.31.e75
103. Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as Upregulates SCAI to Suppress Cisplatin Resistance in Ovarian Cancer via miR-1270 Suppression. Mol Ther Nucleic Acids (2019) 18:24–33. doi: 10.1016/j.omtn.2019.07.012
104. Yao W, Guo P, Mu Q, Wang Y. Exosome-Derived Circ-PVT1 Contributes to Cisplatin Resistance by Regulating Autophagy, Invasion, and Apoptosis Via miR-30a-5p/YAP1 Axis in Gastric Cancer Cells. Cancer Biother Radiopharm (2020) 36(4):347-59. doi: 10.1089/cbr.2020.3578
105. Zhong Y, Wang D, Ding Y, Tian G, Jiang B. Circular RNA Circ_0032821 Contributes to Oxaliplatin (OXA) Resistance of Gastric Cancer Cells by Regulating SOX9 via miR-515-5p. Biotechnol Lett (2021) 43(2):339–51. doi: 10.1007/s10529-020-03036-3.
106. Ding C, Yi X, Wu X, Bu X, Wang D, Wu Z, et al. Exosome-Mediated Transfer of circRNA CircNFIX Enhances Temozolomide Resistance in Glioma. Cancer Lett (2020) 479:1–12. doi: 10.1016/j.canlet.2020.03.002
107. Han C, Wang S, Wang H, Zhang J. Exosomal Circ-HIPK3 Facilitates Tumor Progression and Temozolomide Resistance by Regulating miR-421/ZIC5 Axis in Glioma. Cancer Biother Radiopharm (2021) 36(7):537–48. doi: 10.1089/cbr.2019.3492
108. Zhao M, Xu J, Zhong S, Liu Y, Xiao H, Geng L, et al. Expression Profiles and Potential Functions of Circular RNAs in Extracellular Vesicles Isolated From Radioresistant Glioma Cells. Oncol Rep (2019) 41(3):1893–900. doi: 10.3892/or.2019.6972
109. Pan Y, Lin Y, Mi C. Cisplatin-Resistant Osteosarcoma Cell-Derived Exosomes Confer Cisplatin Resistance to Recipient Cells in an Exosomal Circ_103801-Dependent Manner. Cell Biol Int (2021) 45(4):858–68. doi: 10.1002/cbin.11532
110. Hu K, Liu X, Li Y, Li Q, Xu Y, Zeng W, et al. Exosomes Mediated Transfer of Circ_UBE2D2 Enhances the Resistance of Breast Cancer to Tamoxifen by Binding to MiR-200a-3p. Med Sci Monit: Int Med J Exp Clin Res (2020) 26:e922253. doi: 10.12659/MSM.922253
111. Hsieh Y, Tseng JJ. Azidothymidine (AZT) Inhibits Proliferation of Human Ovarian Cancer Cells by Regulating Cell Cycle Progression. Anticancer Res (2020) 40(10):5517–27. doi: 10.21873/anticanres
112. Qin W, Tsukasaki Y, Dasgupta S, Mukhopadhyay N, Ikebe M, Sauter ER. Exosomes in Human Breast Milk Promote EMT. Clin Cancer Res (2016) 22(17):4517–24. doi: 10.1158/1078-0432.CCR-16-0135
113. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: Composition, Biogenesis, and Mechanisms in Cancer Metastasis and Drug Resistance. Mol Cancer (2019) 18(1):75. doi: 10.1186/s12943-019-0991-5
114. Zhang Z, Yin J, Lu C, Wei Y, Zeng A, You Y, et al. Exosomal Transfer of Long Non-Coding RNA SBF2-AS1 Enhances Chemoresistance to Temozolomide in Glioblastoma. J Exp Clin Cancer Res (2019) 38(1):166. doi: 10.1186/s13046-019-1139-6
115. Wang M, Yu F, Zhang Y, Zhang L, Chang W, Wang K. The Emerging Roles of Circular RNAs in the Chemoresistance of Gastrointestinal Cancer. Front Cell Dev Biol (2022) 10:821609. doi: 10.3389/fcell.2022.821609
116. Fanale D, Taverna S, Russo A, Bazan V. Circular RNA in Exosomes. Adv Exp Med Biol (2018) 1087:109–17. doi: 10.1007/978-981-13-1426-1_9
117. He J, Ren M, Li H, Yang L, Wang X, Yang Q. Exosomal Circular RNA as a Biomarker Platform for the Early Diagnosis of Immune-Mediated Demyelinating Disease. Front Genet (2019) 10:860. doi: 10.3389/fgene.2019.00860
118. Yu Y, Sun B, Wang Z, Yang M, Cui Z, Lin S, et al. Exosomes From M2 Macrophage Promote Peritendinous Fibrosis Posterior Tendon Injury via the MiR-15b-5p/FGF-1/7/9 Pathway by Delivery of circRNA-Ep400. Front Cell Dev Biol (2021) 9:595911. doi: 10.3389/fcell.2021.595911
119. Huang X, Wu B, Chen M, Hong L, Kong P, Wei Z, et al. Depletion of Exosomal circLDLR in Follicle Fluid Derepresses miR-1294 Function and Inhibits Estradiol Production via CYP19A1 in Polycystic Ovary Syndrome. Aging (2020) 12(15):15414–35. doi: 10.18632/aging.103602
120. Ye L, Guo H, Wang Y, Peng Y, Zhang Y, Li S, et al. Exosomal Circehmt1 Released From Hypoxia-Pretreated Pericytes Regulates High Glucose-Induced Microvascular Dysfunction via the NFIA/NLRP3 Pathway. Oxid Med Cell Longevity (2021) 2021:8833098. doi: 10.1155/2021/8833098
121. Cao M, Zhang L, Lin Y, Li Z, Xu J, Shi Z, et al. Circular RNA Expression Profiles in Umbilical Cord Blood Exosomes From Normal and Gestational Diabetes Mellitus Patients. Biosci Rep (2020) 40(11):BSR20201946. doi: 10.1042/BSR20201946
122. Song J, Chen ZH, Zheng CJ, Song KH, Xu GY, Xu S, et al. Exosome-Transported circRNA_0000253 Competitively Adsorbs microRNA-141-5p and Downregulates SIRT1 to Increase Intervertebral Disc Degeneration. Mol Ther Nucleic Acids (2020) 21:1087–99. doi: 10.1016/j.omtn.2020.07.039
123. Wu Y, Zhao T, Deng R, Xia X, Li B, Wang X. A Study of Differential circRNA and lncRNA Expressions in COVID-19-Infected Peripheral Blood. Sci Rep (2021) 11(1):7991. doi: 10.1038/s41598-021-86134-0
124. Zhang J, Zhang Y, Ma Y, Luo L, Chu M, Zhang Z, et al. Therapeutic Potential of Exosomal circRNA Derived From Synovial Mesenchymal Cells via Targeting Circedil3/miR-485-3p/PIAS3/STAT3/VEGF Functional Module in Rheumatoid Arthritis. Int J Nanomed (2021) 16:7977–94. doi: 10.2147/IJN.S333465
125. Tian C, Liu J, Di X, Cong S, Zhao M, Wang K. Exosomal Hsa_circRNA_104484 and Hsa_circRNA_104670 May Serve as Potential Novel Biomarkers and Therapeutic Targets for Sepsis. Sci Rep (2021) 11(1):14141. doi: 10.1038/s41598-021-93246-0
126. Tian T, Li F, Chen R, Wang Z, Su X, Yang C. Therapeutic Potential of Exosomes Derived From circRNA_0002113 Lacking Mesenchymal Stem Cells in Myocardial Infarction. Front Cell Dev Biol (2021) 9:779524. doi: 10.3389/fcell.2021.779524
127. Cao G, Meng X, Han X, Li J. Exosomes Derived From circRNA Rtn4-Modified BMSCs Attenuate TNF-α-Induced Cytotoxicity and Apoptosis in Murine MC3T3-E1 Cells by Sponging miR-146a. Biosci Rep (2020) 40(5):BSR20193436. doi: 10.1042/BSR20193436
128. Guo Z, Wang H, Zhao F, Liu M, Wang F, Kang M, et al. Exosomal Circ-BRWD1 Contributes to Osteoarthritis Development Through the Modulation of miR-1277/TRAF6 Axis. Arthritis Res Ther (2021) 23(1):159. doi: 10.1186/s13075-021-02541-8
129. Mao G, Xu Y, Long D, Sun H, Li H, Xin R, et al. Exosome-Transported circRNA_0001236 Enhances Chondrogenesis and Suppress Cartilage Degradation via the miR-3677-3p/Sox9 Axis. Stem Cell Res Ther (2021) 12(1):389. doi: 10.1186/s13287-021-02431-5
130. Wen Y, Chun Y, Lian ZQ, Yong ZW, Lan YM, Huan L, et al. CircRNA−0006896−miR1264−DNMT1 Axis Plays an Important Role in Carotid Plaque Destabilization by Regulating the Behavior of Endothelial Cells in Atherosclerosis. Mol Med Rep (2021) 23(5):311. doi: 10.3892/mmr.2021.11950
131. Li B, Sun G, Yu H, Meng J, Wei F. Exosomal Circtaok1 Contributes to Diabetic Kidney Disease Progression Through Regulating SMAD3 Expression by Sponging miR-520h. Int Urol Nephrol (2022) doi: 10.1007/s11255-022-03139-y
132. Bai S, Xiong X, Tang B, Ji T, Li X, Qu X, et al. Exosomal Circ_DLGAP4 Promotes Diabetic Kidney Disease Progression by Sponging miR-143 and Targeting ERBB3/NF-κb/MMP-2 Axis. Cell Death Dis (2020) 11(11):1008. doi: 10.1038/s41419-020-03169-3
133. Zhu M, Liu X, Li W, Wang L. Exosomes Derived From Mmu_Circ_0000623-Modified ADSCs Prevent Liver Fibrosis via Activating Autophagy. Hum Exp Toxicol (2020) 39(12):1619–27. doi: 10.1177/0960327120931152
134. Li F, Liu Z, Zhang B, Jiang S, Wang Q, Du L, et al. Circular RNA Sequencing Indicates Circ-IQGAP2 and Circ-ZC3H6 as Noninvasive Biomarkers of Primary Sjgren’s Syndrome. Rheumatol (Oxford) (2020) 59(9):2603–15. doi: 10.1093/rheumatology/keaa163
135. Zhang X, Chen L, Xiao B, Liu H, Su Y. Circ_0075932 in Adipocyte-Derived Exosomes Induces Inflammation and Apoptosis in Human Dermal Keratinocytes by Directly Binding With PUM2 and Promoting PUM2-Mediated Activation of AuroraA/NF-κb Pathway. Biochem Biophys Res Commun (2019) 511(3):551–8. doi: 10.1016/j.bbrc.2019.02.082
136. Zhao RT, Zhou J, Dong XL, Bi CW, Jiang RC, Dong JF, et al. Circular Ribonucleic Acid Expression Alteration in Exosomes From the Brain Extracellular Space After Traumatic Brain Injury in Mice. J Neurotrauma (2018) 35(17):2056–66. doi: 10.1089/neu.2017.5502
137. Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of Extracellular Vesicles (EV): Exosomes, Microvesicles, Retrovirus-Like Vesicles, and Apoptotic Bodies. J Neuro-Oncol (2013) 113(1):1–11. doi: 10.1007/s11060-013-1084-8
Keywords: non-coding RNA (ncRNA), circRNA, exosome, exosomal circRNAs, disease
Citation: Han Z, Chen H, Guo Z, Shen J, Luo W, Xie F, Wan Y, Wang S, Li J and He J (2022) Circular RNAs and Their Role in Exosomes. Front. Oncol. 12:848341. doi: 10.3389/fonc.2022.848341
Received: 04 January 2022; Accepted: 01 April 2022;
Published: 28 April 2022.
Edited by:
César López-Camarillo, Universidad Autónoma de la Ciudad de México, MexicoReviewed by:
Yongsoo Park, Qatar Biomedical Research Institute, QatarCopyright © 2022 Han, Chen, Guo, Shen, Luo, Xie, Wan, Wang, Li and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhao Li, MTg5MjIyMzgwMzJAMTYzLmNvbQ==; Jinhua He, MzMyNTE4NTc5QHFxLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.