- 1Department of Neurosurgery, Xiangya Hospital, Central South University, Changsha, China
- 2Department of Pathology, Xiangya Hospital, Central South University, Changsha, China
- 3Department of Pathology, Xiangya Changde Hospital, Changde, China
- 4National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China
The AlkB family of Fe (II) and α-ketoglutarate-dependent dioxygenases works by removing alkyl substituents from alkylation-damaged nucleic acid bases through oxidative dealkylation, subsequently affecting tumor progression and patient prognosis. However, the specific roles of the AlkB family in Glioblastoma remain to be elucidated. By taking advantage of the abundant bioinformatics databases, such as GEPIA2, cBioPortal and TIMER, we performed a comprehensive analysis of the AlkB family in GBM, and managed to identify the significant prognostic hallmarks and therapeutic targets within this family. We found that the expression levels of ALKBH2 and ALKBH8 were significantly up-regulated in GBM compared with normal tissues. Meanwhile, the patients with high levels of ALKBH2 and ALKBH8 possessed significant poor overall survival (OS). In addition, the results suggested that the biological function of the AlkB family was closely related to DNA damage repair, cell metabolism, cell proliferation and tumor immune infiltration in GBM. Furthermore, the high expression of ALKBH8 in GBM was verified by immunohistochemistry. Taken together, this study could provide meaningful information about the aberrant AlkB family associated with GBM initiation and progression, and help clinicians precisely predict patient survival and select alternative therapeutic drugs.
Introduction
Glioblastoma (GBM) is the most common malignant primary tumor of the central nervous system, accounting for about 14.5% of all primary brain tumors and 48% of all malignant primary brain tumors (1). It has the characteristics of easy recurrence, high malignancy and poor prognosis (2, 3). In the United States, the average age-adjusted incidence of GBM is 3.21 per 100,000 population (1). GBM patients have a poor prognosis with a 5-year survival rate of 7.2% and a median survival of 8 months (1). The standard approach for GBM is maximal safe surgical resection followed by radiotherapy with concomitant temozolomide (TMZ, an alkylating agent) and a further 6 cycles of temozolomide (4). Even after the most effective treatment, the average survival time of GBM patients is only 14.5-16.6 months (5). The illustration of the molecular profile of GBM confers great significance to understand the occurrence and development of the disease. It was reported that patients with as isocitrate dehydrogenase 1 (IDH-1), IDH-2 mutation or MGMT methylation have relatively longer survival time (6, 7). However, we need to continue to depict the molecular features to combat this devastating disease.
The AlkB family of Fe (II) and α-ketoglutarate-dependent dioxygenases is a group of direct reversal DNA repair enzymes (8). The family of proteins works by removing alkyl substituents from alkylation-damaged nucleic acid bases through oxidative dealkylation (9, 10). So far, nine homologues (ALKBH1–8 and FTO) have been identified in human cells (11). The class of proteins possess a variety of biological functions, such as DNA repair, participating in the RNA metabolism, acting as demethylase or participating in fatty acid metabolism (12–14). And this family has been reported as prognostic biomarkers and therapeutic targets of different kinds of tumor, including head and neck cancer, ovarian serous carcinoma, lung adenocarcinoma, and colorectal cancer (15–18). In particular, ALKBH2 was discovered to play a significant role in TMZ and photodynamic therapy resistance in GBM (19, 20). However, the specific role of the AlkB family in GBM remains to be furtherly elucidated.
By taking advantage of the abundant bioinformatics databases (Table 1), we performed a comprehensive analysis of the AlkB family in GBM, and managed to identify the significant prognostic hallmarks and therapeutic targets within this family, which we believe could help clinicians precisely predict patient survival and select alternative therapeutic drugs.
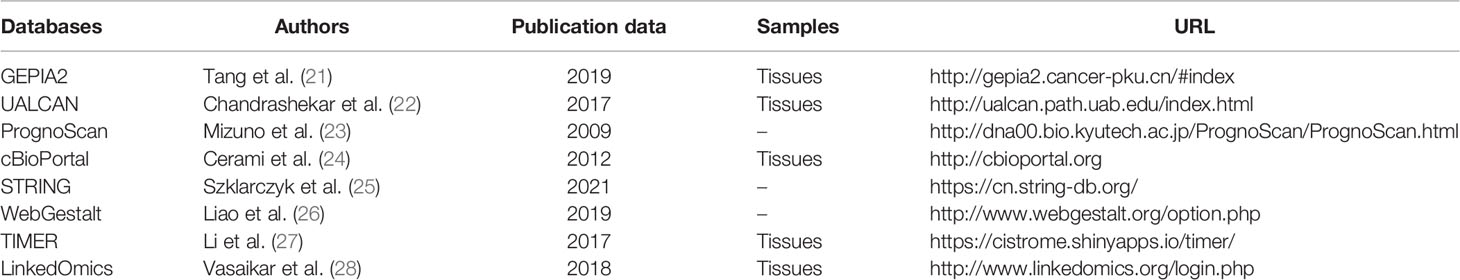
Table 1 The online databases applied to evaluate the expression and biological functions of AlkB family in GBM.
Materials and Methods
GEPIA2
GEPIA2 (Gene Expression Profiling Interactive Analysis, http://gepia2.cancer-pku.cn/#index) is a web resource based om tumor and normal samples from the TCGA and the GTEx databases, which provides differential gene expression profiling, correlation analysis, and patient survival analysis (21, 29). In this study, we utilized the “single gene analysis” module of GEPIA2 to compare mRNA expression differences between GBM and normal brain tissue. Student’s t test was used to generate p values for mRNA expression, and p < 0.05 was considered statistically significant.
UALCAN
UALCAN (http://ualcan.path.uab.edu/index.html) is a convenient, interactive web-portal to perform analyses of TCGA gene expression data, which provides analyses of relative expression of genes across tumor and normal samples, estimation of the effect of gene expression level and clinicopathologic features on patient survival, and identification of the top up and down regulated genes in individual cancer types (22). In this research, the expression levels of the AlkB family were obtained through the “TCGA Gene analysis” module of the website. Differences in mRNA expression were calculated by Student’s t test, and p < 0.05 was considered statistically significant.
The Human Protein Atlas
The Human Protein Atlas website (http://www.proteinatlas.org/) is an information database of protein expression patterns in normal human tissues and in cancer, which can identify clinically useful biomarkers by applying antibodies and protein expression data as tools (30). In this study, we obtained the protein expression profiles of the AlkB family members in normal brain and GBM tissues through this database.
PrognoScan
PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/PrognoScan.html) is a database offering a tool for assessing the relationship between gene expression and tumor patient prognosis, which employs the minimum P-value approach for grouping patients so as to find the optimal cut point in continuous gene expression measurement (23). In our study, we utilized this useful database to evaluate the relationship between the AlkB family gene expression and GBM patient overall survival.
cBioPortal
The cBioPortal (cBio Cancer Genomics Portal, http://cbioportal.org) is a free resource for interactive exploration of multidimensional cancer genomics, through which we can get access to molecular profiles and clinical attributes from large-scale cancer genomics projects (24). In this study, we acquired the genetic alteration map of the AlkB family members in GBM tissues by searching this database.
STRING
The STRING (https://cn.string-db.org/) is an open-access database providing integration of all known and predicted protein-protein interactions (PPIs), including physical interactions and functional associations, by automated text mining of the scientific literature, databases of interaction experiments, computational interaction predictions and systematic transfers of interaction evidence from one organism to another (25, 31). In our study, we utilized this database to analyze the interaction of altered proteins in tumor samples with mutation of at least one gene of AlkB family.
Cytoscape
Cytoscape is an open software which can visually integrate biomolecular interaction networks with high-throughput expression data and other molecular states into a unified conceptual framework (32). In the study, we performed functional integration of 38 altered proteins in tumor samples with mutation of at least one gene of AlkB family.
WebGestalt
WebGestalt (WEB-based Gene SeT AnaLysis Toolkit, http://www.webgestalt.org/option.php) is a functional enrichment analysis web tool, which supports three methods for enrichment analysis, including Over-Representation Analysis (ORA), Gene Set Enrichment Analysis (GSEA), and Network Topology-based Analysis (NTA) (26). In our study, we performed Gene ontology (GO) enrichment analysis by applying the WebGestalt website.
TIMER
TIMER web server (https://cistrome.shinyapps.io/timer/) is a comprehensive resource for systematical analysis of immune infiltrates across diverse cancer types (27, 33). In this study, we utilized “gene module” to explore the correlation between AlkB family gene expression and abundance of immune infiltrates. And the “survival module” was applied to assess the association of clinical outcome with immune cell infiltration and the AlkB family gene expression. The multivariable cox proportional hazard model was used as the statistical method.
LinkedOmics
LinkedOmics (http://www.linkedomics.org/login.php) is publicly available website which includes multi-omics data from all 32 TCGA Cancer types and 10 Clinical Proteomics Tumor Analysis Consortium (CPTAC) cancer cohorts (28). In our study, we applied the “LinkFinder” module to explore the correlated significant genes of AlkB family genes, and the “LinkInterpreter” module to perform the GO pathway enrichment analysis based on the association results.
Immunohistochemistry
5 cases of normal brain and 20 cases of GBM formalin-fixed, paraffin-embedded (FFPE) tumor tissue blocks were submitted for immunohistochemistry (IHC). IHC was performed following standard protocols. The slides were dewaxed, boiled in 0.01 M citrate buffer (pH 6.0) for 30 minutes, and then incubated with 3% hydrogen peroxide for 15 minutes. After washing with PBS, the slides were incubated with 5% normal bovine serum albumin (BSA) for 1 hour, followed by incubation with rabbit polyclonal antibody recognizing ALKBH8 (A7142, 1: 100, ABclonal Technology, Wuhan, China) overnight. After incubating with secondary antibody, the slides were then incubated with 3,3′ diaminobenzidine (DAB) (PV-6000D, ZSGB-BIO, Beijing, China) for staining. The slides were then counterstained with hematoxylin, dehydrated, cleared, and mounted. The study was approved by the Ethics Committee of Xiangya Hospital (Ethics approval No. 202201015).
Statistical Analyses
ALKBH8 expression profile difference between normal brain and GBM tissues was analyzed using Wilcoxon rank testing. P-value < 0.05 was considered to be statistically significant.
Results
Aberrant Expression of AlkB Family Members in GBM Patients
We firstly searched the GEPIA2 database to explore the mRNA expression levels of the AlkB family (ALKBH1-8 and FTO) in GBM and normal brain tissue. The results revealed that the expression levels of ALKBH1/2/3/4/5/7/8/FTO were elevated, while the expression level of ALKBH6 was reduced compared with normal brain tissues (Figure 1A). We also utilized GEPIA2 to analyze the relative expression level of the AlkB family in GBM, and the result illustrated that ALKBH5/7/FTO had relatively higher expression levels among the family members (Figure 1B). Besides the GEPIA2 database, we applied UALCAN to explore the mRNA expression levels of the AlkB family in GBM and normal brain tissue. The results showed that the expression levels of ALKBH1/2/8 were significantly up-regulated and the expression levels of ALKBH5/6/FTO were significantly down-regulated in GBM compared with normal tissues (Figure 2A). After description of the mRNA expression levels of AlkB family, we applied the Human Protein Atlas to explore the protein expression levels of AlkB family in GBM. The results revealed that ALKBH2/4/7/8/FTO were highly or mediumly expressed, while ALKBH3/6 were lowly or not detected in GBM tissues (Figure 2B). Taken together, these results indicated the potential biological roles of up-regulated ALKBH2/8 in GBM pathogenesis.
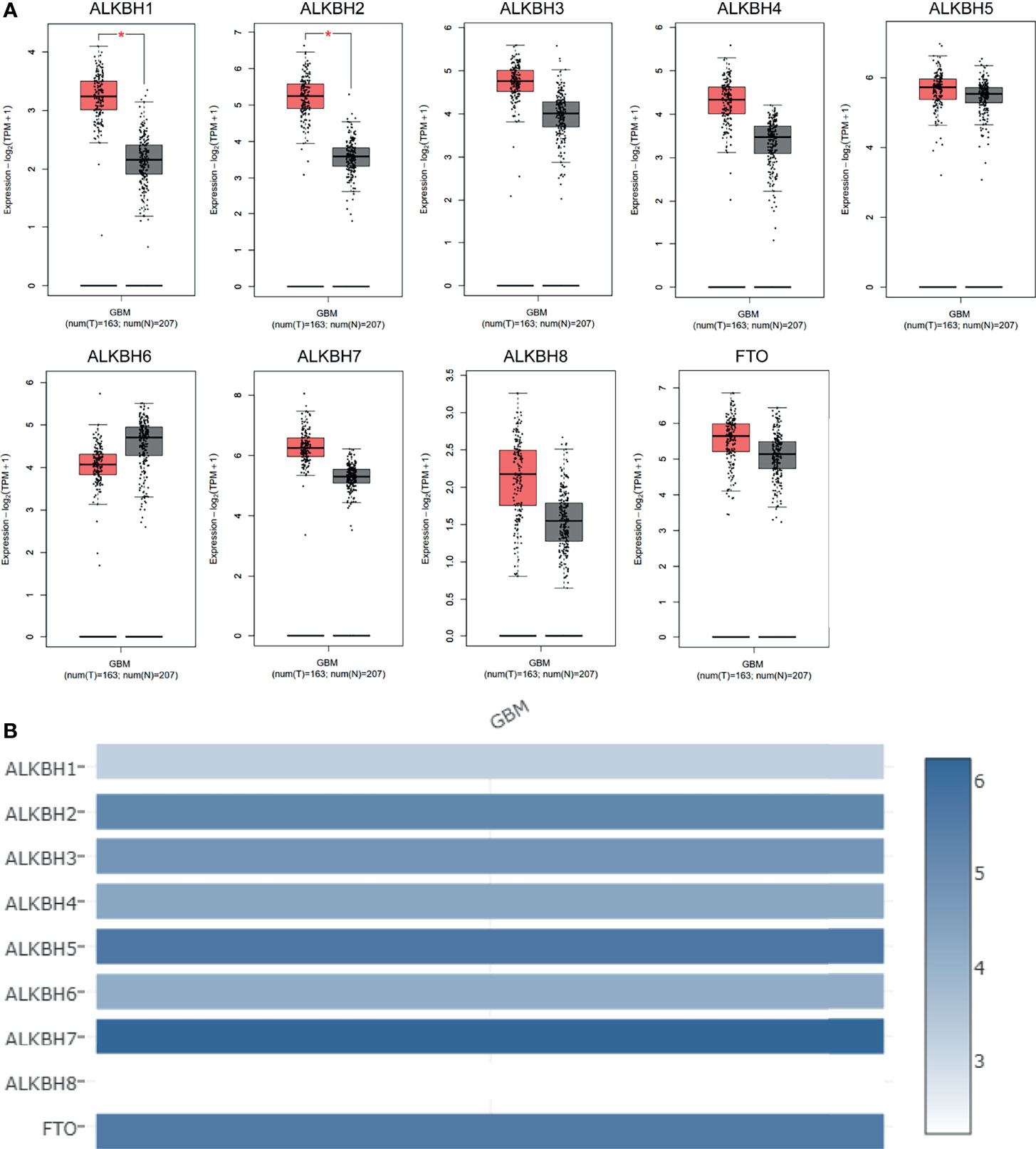
Figure 1 mRNA expression levels of the AlkB family members in GBM. (A) mRNA expression levels of the AlkB family members in GBM and normal brain tissues based on the GEPIA2 database. T and N indicated the tumor and normal tissues, respectively. (B) The relative mRNA expression levels of the AlkB family in GBM based on the GEPIA2 database. *p < 0.05.

Figure 2 mRNA and protein expression levels of the AlkB family members in GBM. (A) mRNA expression levels of the AlkB family members in GBM and normal brain tissues by searching the UALCAN website. (B) Representative immunohistochemical staining of the AlkB family members in GBM and normal brain tissues based on the Human Protein Atlas database.
Prognostic Value of AlkB Family mRNA Expression in GBM Patients
We next managed to investigate whether the mRNA expression levels of AlkB family genes were associated with GBM patients’ survival. After searching the PrognoScan database, we found intriguingly that patients with higher transcription levels of ALKBH2 and ALKBH8 possessed significant shorter overall survival (OS) time based on the GEO GSE4271 dataset (34) (Figure 3). Conversely, patients with higher transcription level of FTO displayed significant longer OS time.
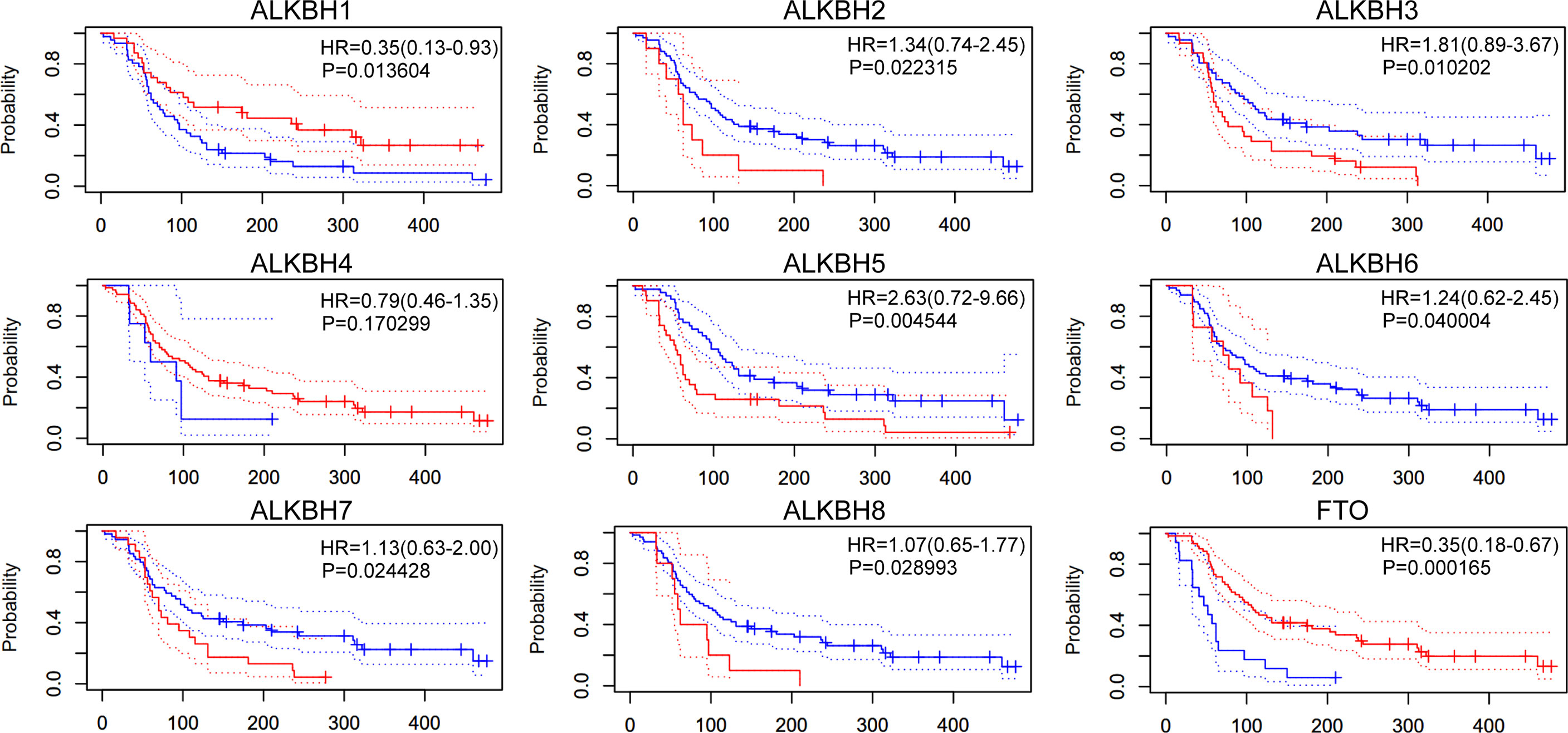
Figure 3 Overall survival of GBM patients. The overall survival of GBM patients based on the AlkB family genes expression in GEO GSE4271 dataset.
Genetic Alterations and Functional Analysis of the AlkB Family in GBM Patients
To furtherly explore the biological function of AlkB Family in GBM, we first evaluated the genetic alterations of the AlkB family genes by searching the cBioPortal database. The results presented that the genetic alteration rates of ALKBH1-8/FTO in GBM tumor samples were 2.9%, 1.9%, 1.7%, 10%, 1.5%, 2.4%, 2.2%, 1% and 3%, respectively (Figure 4A). We also found that “mRNA high” was the most common alteration mode for most of the AlkB family members. After confirming the genetic alteration profiling of AlkB family in GBM, we explored the altered proteins and genes in GBM samples with mutation of at least one gene of AlkB family by utilizing the cBioPortal database, and identified 38 proteins (Supplementary Table 1) and 183 genes (Supplementary Table 2) most related to AlkB family genetic alterations. Then we conducted protein interaction analysis by applying the STRING website and Cytoscape software to depict the PPIs among these 38 altered proteins based on the corresponding nodes and combined scores (Supplementary Table 3). We found that a variety of proteins displayed the critical functions determining cell fate. Specifically, a series of DNA damage repair related proteins, including ATM, CHEK2 and MSH2, were closely associated with the functions of AlkB family in GBM. Moreover, we also identified other proteins playing pivotal roles in controlling metabolism and immunity (Figure 4B). After analyzing the altered proteins, we performed GO annotation and pathway enrichment analysis by applying the WebGestalt website based on the 183 altered genes. The GO annotation results suggested that the altered genes were mainly associated with biological regulation, response to stimulus and metabolic process. Regarding cellular components, the altered genes were mostly enriched in membrane and endomembrane system. In terms of molecular function categories, the altered genes were mostly related to protein binding, ion binding and nucleic acid binding (Figure 5A). The corresponding enriched GO pathways included leukocyte migration, extracellular structure organization and T cell activation (Figure 5B).
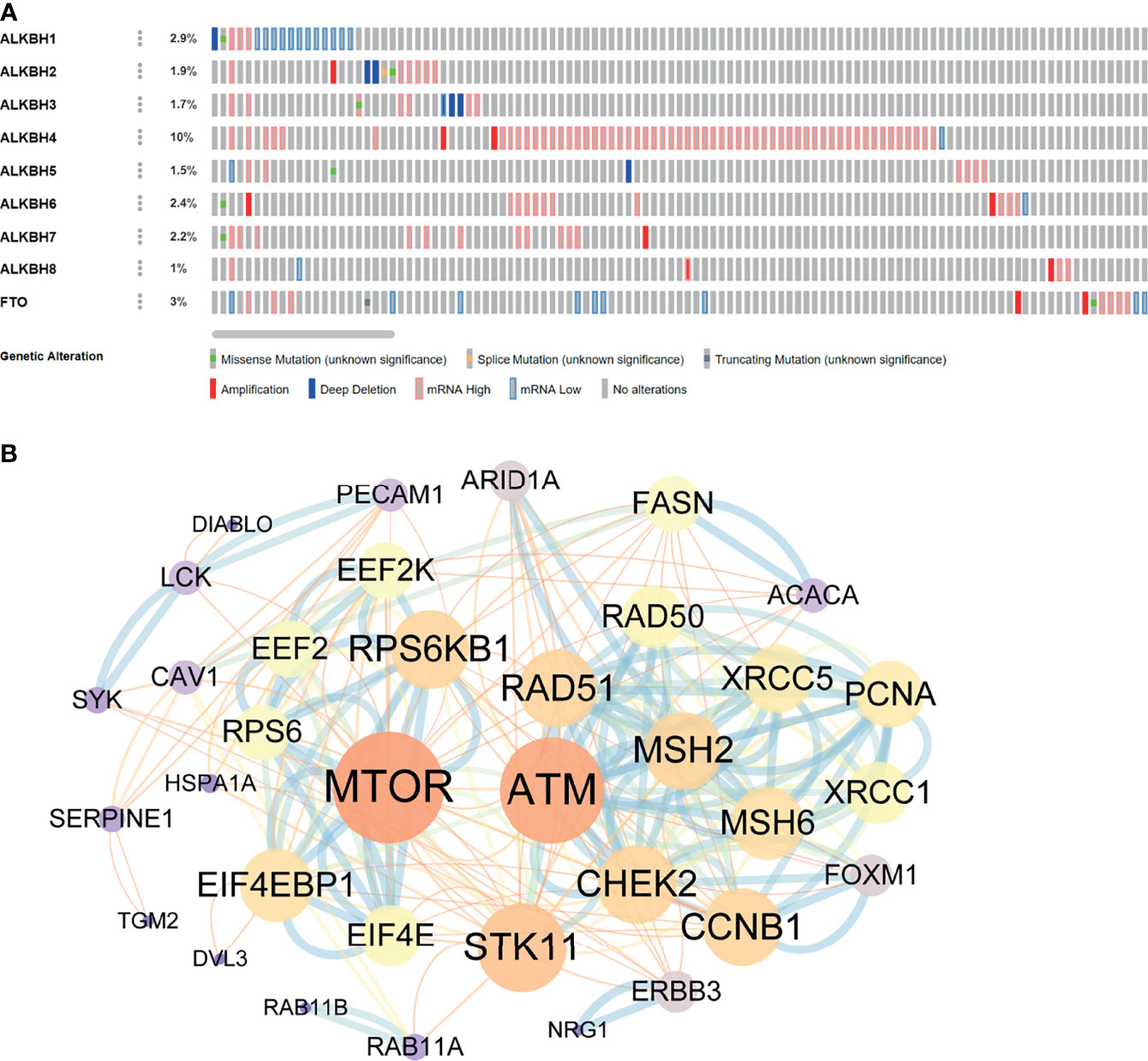
Figure 4 Genetic alterations of the AlkB family genes and related PPIs. (A) The genetic alterations of the AlkB family genes in GBM by searching the cBioPortal database. (B) The PPIs among the altered proteins most related to AlkB family genetic alterations generated by applying the STRING website and Cytoscape software.
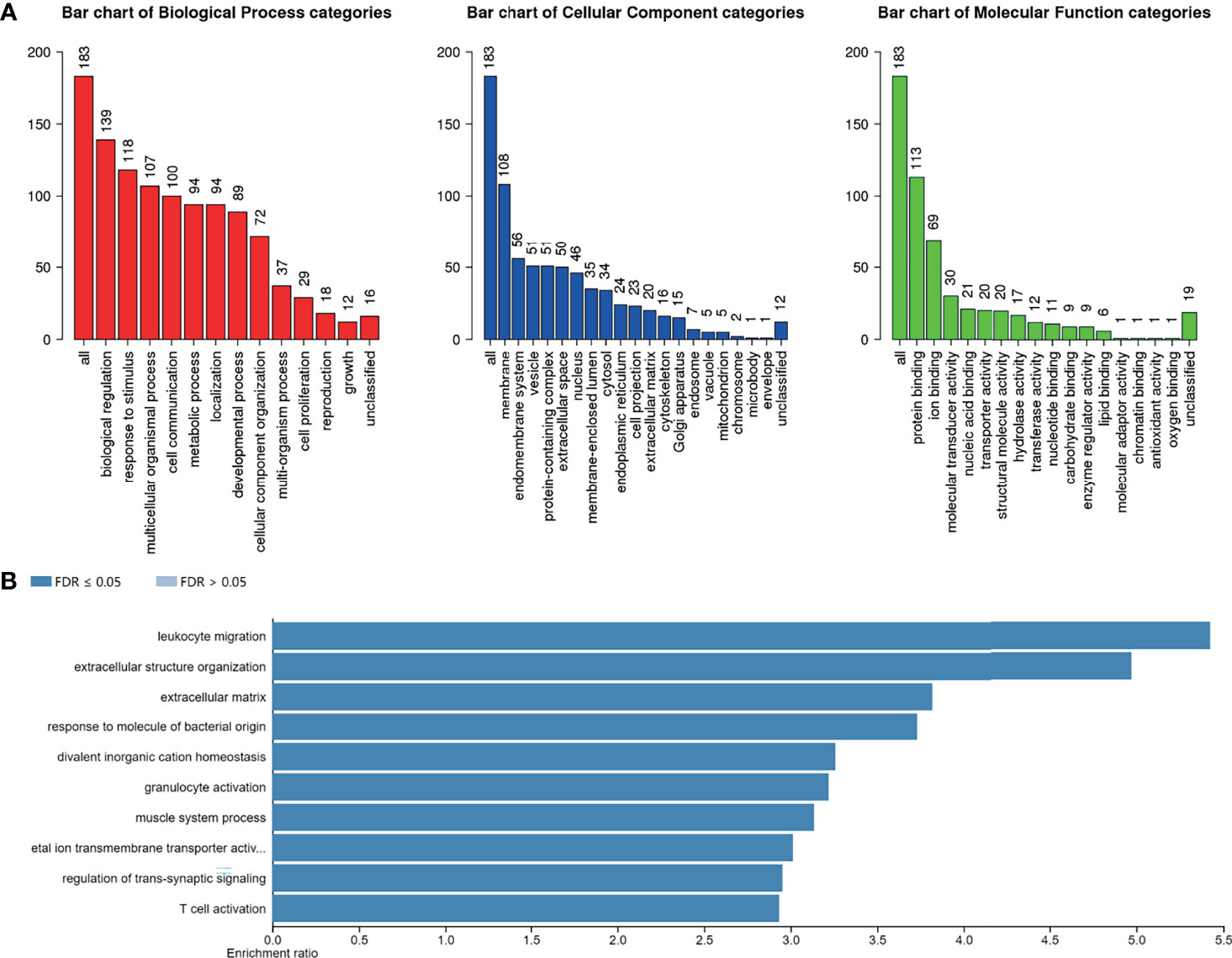
Figure 5 GO annotation and pathway enrichment analysis of the altered genes. (A) GO annotation of altered genes related to the AlkB family genetic alterations including biological process, cellular component and molecular function categories by applying the WebGestalt website. (B) GO pathway enrichment analysis of the altered genes by applying the WebGestalt website.
Immune Cell Infiltration Based on the AlkB Family Gene Expression in GBM Patients
On account of the GO pathway enrichment results, we became interested in the correlation between AlkB family gene expression and abundance of immune infiltrates in GBM tissues. By searching the TIMER web server, we found that ALKBH1 expression level was positively correlated with CD8+ T cells, macrophages and neutrophils infiltration (Figure 6A). Regarding ALKBH2, its expression level was positively correlated with B cells and CD8+ T cells, and negatively correlated with CD4+ T cells, neutrophils and dendritic cells infiltration (Figure 6B). For ALKBH3, the only significant immune cells were macrophages, which was negatively correlated with the gene expression level (Figure 6C). Regarding ALKBH4, its expression level was positively correlated with CD8+ T cells, CD4+ T cells, macrophages and neutrophils (Figure 6D). For ALKBH5, its expression level was positively correlated with dendritic cells, and negatively correlated with B cells and CD8+ T cells infiltration (Figure 6E). Regarding ALKBH6, its expression level was not significantly tied to immune cell infiltration (Figure 6F). For ALKBH7, its expression level was positively correlated with B cells and CD8+ T cells, and negatively correlated with CD4+ T cells and dendritic cells infiltration (Figure 6G). Regarding ALKBH8, its expression level was not significantly related to immune cell infiltration (Figure 6H). For FTO, its expression level was positively correlated with CD8+ T cells, CD4+ T cells, macrophages and neutrophils, which suggested active anti-tumor microenvironment and might explain patients with higher transcription level of FTO possessed significant longer OS time (Figure 6I). After analyzing the correlation between AlkB family gene expression and immune infiltrates in GBM tissues, we furtherly assessed the association of clinical outcome with immune cell infiltration and the AlkB family gene expression by applying the multivariable Cox proportional hazard model. The results revealed that CD4+ T cells, dendritic cells and ALKBH4/6/8 were significantly associated with patient survival (Table 2).
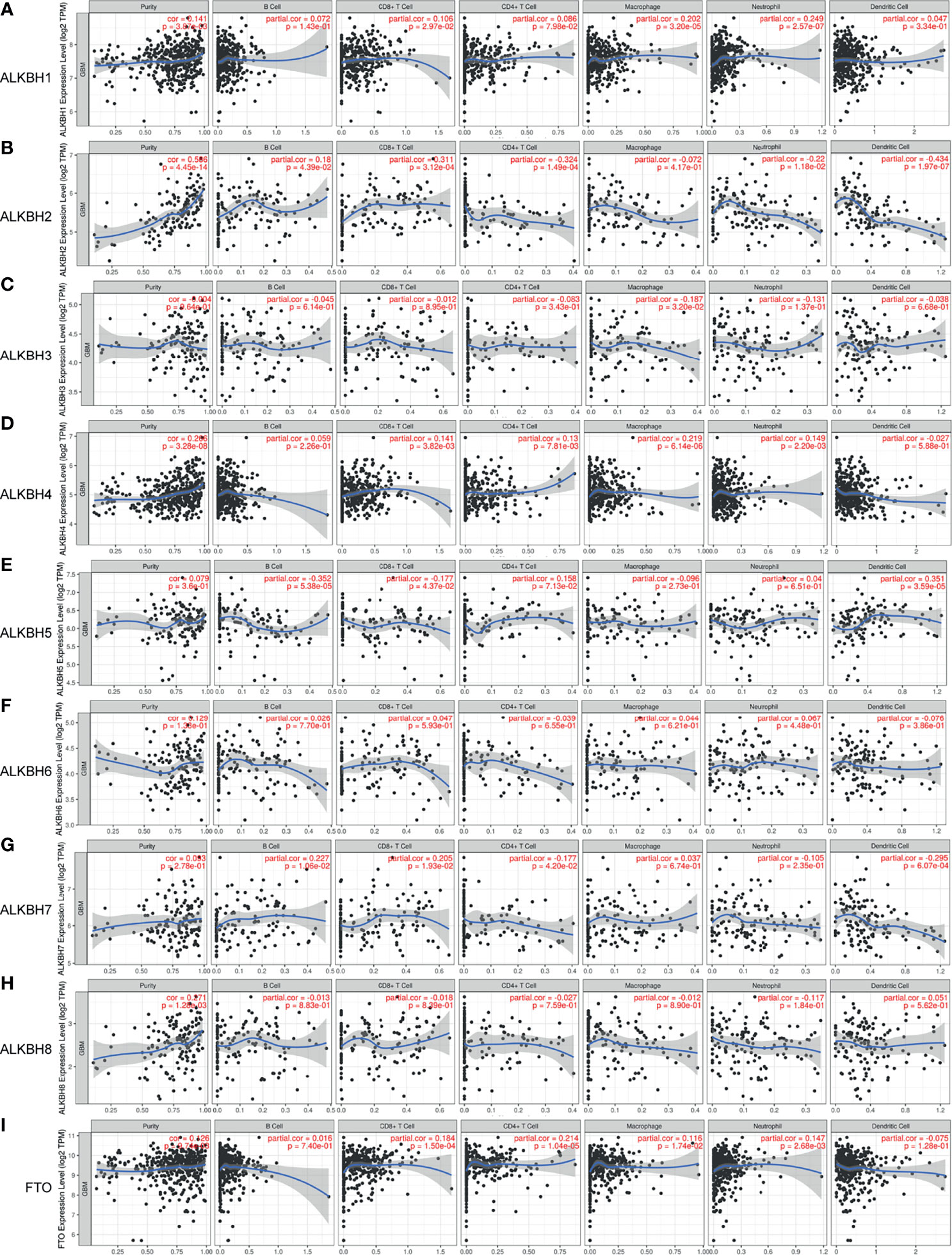
Figure 6 Correlation between the AlkB family gene expression and abundance of immune infiltrates in GBM. (A-I) The effect of ALKBH1 (A), ALKBH2 (B), ALKBH3 (C), ALKBH4 (D), ALKBH5 (E), ALKBH6 (F), ALKBH7 (G), ALKBH8 (H), and FTO (I) on the tumor immune cell infiltration analyzed by the TIMER database.
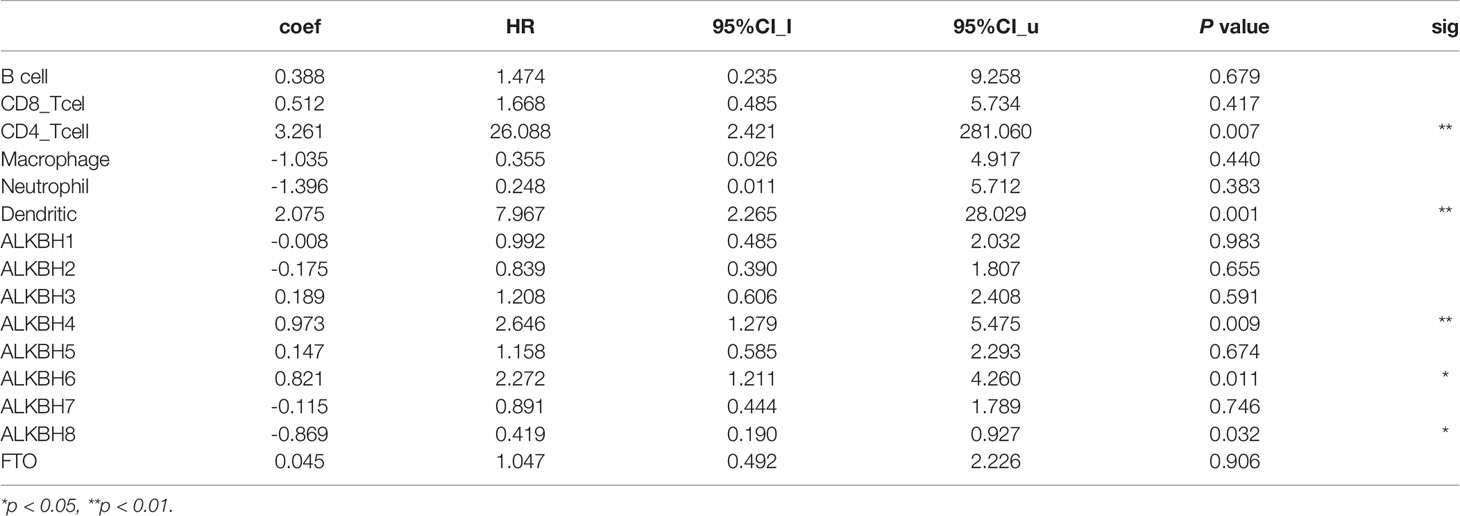
Table 2 The association of clinical outcome of GBM patients with immune cell infiltration and the AlkB family gene expression by applying the multivariable Cox proportional hazard model from the TIMER database.
Correlated Significant Genes and Corresponding Enriched GO Pathways of ALKBH2/8
After identifying ALKBH2/8/FTO as significant prognostic biomarkers based on the mRNA expression levels, we proceeded to explore the correlated significant genes of ALKBH2/8, and to perform the GO pathway enrichment analysis based on the association results by taking advantage of the LinkedOmics website. The positively and negatively correlated significant genes were exhibited in Figures 7A, B. We noticed that the correlated genes consisted of DNA damage repair related genes such as NTHL1, PCNA and ERCC8, and cell metabolism related genes such as MRPS26, EIF5A and EEF1G, which was consistent with the previous results. Concerning the GO pathway enrichment results, we found that mRNA processing was positively correlated with the expression level of both ALKBH2 and ALKBH8, and immune response was negatively correlated with ALKBH2/8 expression level, which might partly explain the worse survival of GBM patients with higher transcription levels of ALKBH2/8 (Figures 8A, B). The high expression of ALKBH8 in GBM was verified by IHC.
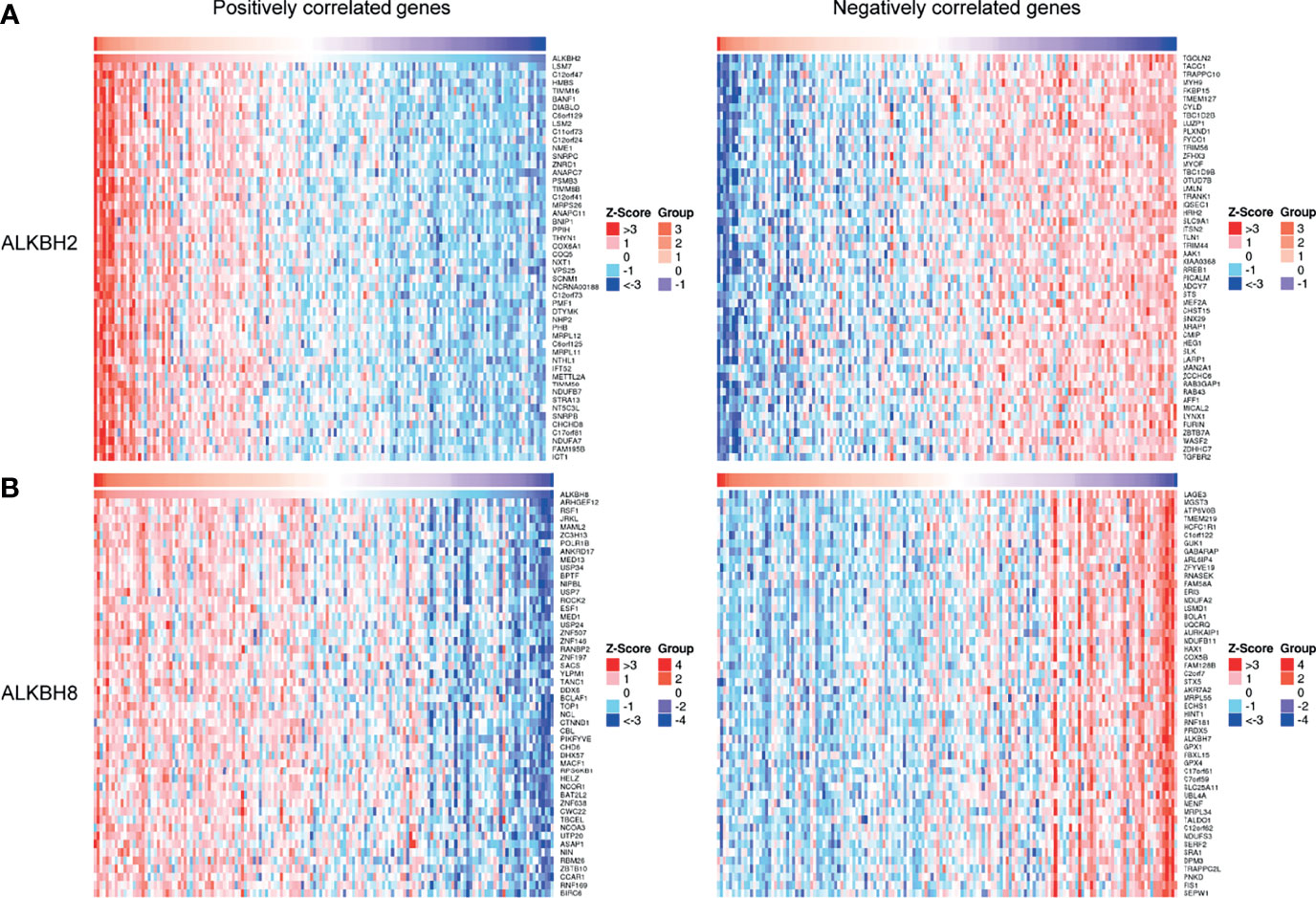
Figure 7 Correlated significant genes of ALKBH2/8 in GBM. (A, B) The positively and negatively correlated significant genes of ALKBH2 (A) and ALKBH8 (B) by taking advantage of the LinkedOmics website.
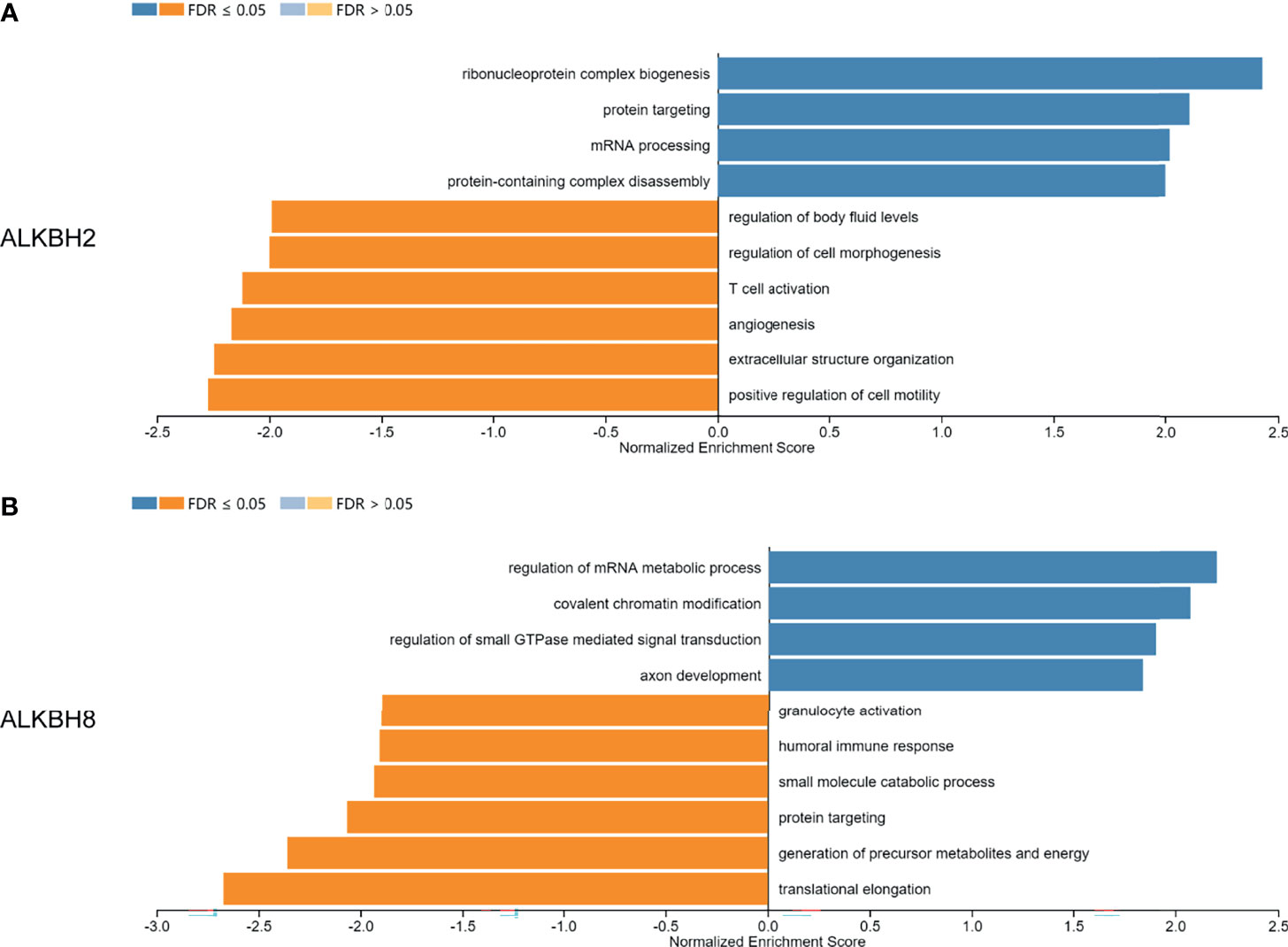
Figure 8 GO pathway enrichment analysis based on the correlated genes of ALKBH2/8 in GBM. (A, B) The GO pathway enrichment results based on the correlated genes of ALKBH2 (A) and ALKBH8 (B) by taking advantage of the LinkedOmics website.
After analyzing the online databases, we performed the ALKBH8 staining in 5 normal brain tissues and 20 GBM tissues. The IHC results illustrated that ALKBH8 was highly expressed in GBM tissue compared with normal brain, which is consistent with the results obtained from the databases (Figures 9A, B).
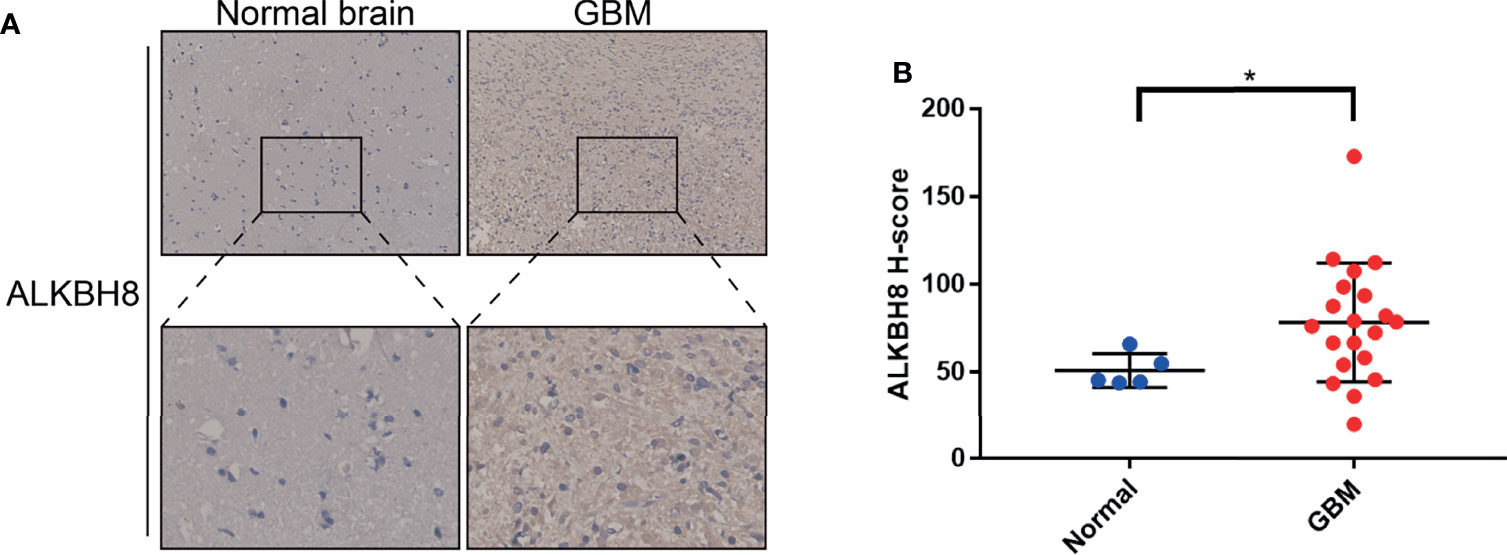
Figure 9 Representative IHC images and H-score of ALKBH8 protein expression in GBM and normal brain tissues. (A) Immunohistochemical staining for ALKBH8 in human GBM and normal brain tissues. The specific ALKBH8 signal is shown in brown (DAB staining). (B) The difference of ALKBH8 protein expression between GBM and normal brain was assessed by Wilcoxon rank testing. *p < 0.05.
Discussion
The AlkB family members possess a variety of important biological functions and play significant roles in the development of tumor including GBM. By generating the genetic alteration analysis and associated altered proteins interaction interpretation, we identified a series of DNA damage repair related proteins such as ATM and CHEK2, suggesting the critical role of this family played in GBM. As a DNA repair enzyme, ALKBH2 functions through removing alkyl substituents from alkylation-damaged nucleic acid bases. It was reported that ALKBH2 was abundantly expressed in GBM cell lines and human GBM tissues, and enhanced resistance to the alkylating agents TMZ (20). Another study suggested ALKBH2 expression was implicated in resistance to photodynamic therapy in U87 GBM cells (19). Though the potential roles of this family members in cancer are rapidly uncovered, the comprehensive function of this particular family in GBM has not been elucidated yet.
Besides the DNA damage repair related proteins, we also found mTOR and its downstream effector molecules such as EIF4E and RPS6 were closely related to the AlkB family genetic alterations. In eukaryotes, mTOR signaling pathway integrates both intracellular and extracellular signals and serves as a central regulator of cellular metabolism and survival (35, 36). Moreover, major DNA damage sensors such as ATM, ATR and DNA-PK communicate with the mTORC1 pathway to ensure an efficient response to the metabolic and genotoxic stresses (37). Beuvink et al. reported that mTOR inhibitor everolimus sensitizes tumor cells to DNA-damaged induced apoptosis (38). Another study suggested that m6A RNA modification catalyzed by enzymes including ALKBH5 and FTO had a fundamental role in the regulation of PI3K/Akt/mTOR signaling pathway in cancer (39). HT Zhu et al. revealed that ALKBH5 was upregulated and activated EGFR-PIK3CA-AKT-mTOR signaling pathway in epithelial ovarian cancer tissue (40). Yet, the crosstalk between AlkB family members and the metabolic pathways in GBM need to be furtherly elucidated.
Aberrant epigenetic landscapes largely affected tumor initiation and progression, and The AlkB family was continuingly studied within this aspect. DNA methylation is a critical epigenetic mark which plays a fundamental role in cancer. Xie et al. demonstrated that ALKBH1 was a critical demethylase for DNA N6-mA and regulated specific genetic pathways in GBM. Depletion of ALKBH1 could cause transcriptional silencing of oncogenic pathways through decreasing chromatin accessibility and inhibit tumor cell proliferation (41). Besides DNA modifications, RNA processing also plays major roles in cells and tumors. N6-Methyladenosine (m6A) is the most prevalent internal chemical modification of mRNAs in eukaryotes, which can affect the stability and translation efficiency of mRNA (42–44). Our results suggested that mRNA processing was positively correlated with the expression level of both ALKBH2 and ALKBH8 based on the GO pathway enrichment analysis. Zhang et al. revealed that ALKBH5, a m6A demethylase, was highly expressed in glioblastoma stem-like cells (GSCs) and demethylated FOXM1 nascent transcripts leading to enhanced FOXM1 expression. Their work suggested ALKBH5-FOXM1 pathway was critical for GSC proliferation and tumorigenesis, and uncovered the critical function of ALKBH5 in GBM (45). FTO is another frequently studied RNA m6A demethylase. Rui Su et al. reported that R-2HG displayed anti-tumor activity in leukemia and glioma through targeting FTO, leading to MYC mRNA decay and downregulation of MYC/CEBPA-associated oncogenic pathways (46).
Accumulating evidence suggests that the tumor immune microenvironment plays a critical role in anti-cancer immunity, which may affect patient prognosis and the effect of immunotherapy (47, 48). Our results suggested significant correlation between AlkB family gene expression and abundance of immune infiltrates in GBM tissues based on the TIMER website, and the corresponding enriched GO pathways included leukocyte migration and T cell activation associated with the family genes genetic alterations. In addition to the significant roles of RNA m6A modifications in tumor-intrinsic oncogenic pathways, increasing evidence supports that RNA m6A metabolism also functions in tumor immune regulation (49, 50). Li and his colleague reported that ALKBH5 modulated Mct4/Slc16a3 expression and lactate content of the tumor microenvironment and promoted the composition of tumor-infiltrating Treg and myeloid-derived suppressor cells in melanoma and colorectal cancer, thus leading to resistance to immunotherapy. And deletion of ALKBH5 sensitized tumors to cancer immunotherapy (51). Another study revealed that FTO expression level was increased in human melanoma and contributed to promoting melanoma tumorigenesis and anti-PD-1 resistance. Knockdown of FTO could sensitize melanoma cells to interferon gamma and anti-PD-1 treatment in mice (52). However, the relevance between AlkB family members and tumor immunity in GBM need to be furtherly explored.
We are aware of the limitations of this study. Firstly, the study mainly contains results from the public database, but is short of experimental verification. Though the expression of ALKBH8 in GBM was verified by IHC, more in vitro and in vivo experiments are needed to explore the biological function of this family in GBM. Secondly, the data sets used in different databases are inconsistent, and some contradictory data need to be further confirmed. Thirdly, we need further research to verify the therapeutic significance of AlkB family members in GBM patients.
In conclusion, by taking advantage of the abundant bioinformatics databases, we conducted this comprehensive analysis of the AlkB family in GBM for the first time, and managed to identify the value of evaluating patients’ prognosis based on the mRNA expression levels of ALKBH2/8/FTO. In addition, we revealed that the biological function of the AlkB family was closely related to DNA damage repair, cell metabolism, cell proliferation and tumor immune infiltration in GBM, which needed to be furtherly explored in the future. We believe that the existing results could help researchers discover novel mechanisms associated with GBM initiation and progression, and help clinicians precisely predict patient survival and select alternative therapeutic drugs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Xiangya Hospital. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
Acquisition of Data: SF and ZX. Analysis and Interpretation of Data: ZX and SF. Conception and Design: ZX and JP. Data Curation: SF. Development of Methodology: ZX. Performing the experiment: MZ. Writing the Manuscript: SF, JP, and MZ. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82102848) and the Hunan Provincial Natural Science Foundation of China (No. 2020JJ8111).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
All authors thank TCGA and GEO databases for providing the worthy data and all the online-tools which are the free platforms for data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.847821/full#supplementary-material
References
1. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011-2015. Neuro Oncol (2018) 20:iv1–iv86. doi: 10.1093/neuonc/noy131
2. Wen PY, Reardon DA. Neuro-Oncology in 2015: Progress in Glioma Diagnosis, Classification and Treatment. Nat Rev Neurol (2016) 12:69–70. doi: 10.1038/nrneurol.2015.242
3. Tan AC, Ashley DM, Lopez GY, Malinzak M, Friedman HS, Khasraw M. Management of Glioblastoma: State of the Art and Future Directions. CA Cancer J Clin (2020) 70:299–312. doi: 10.3322/caac.21613
4. Stupp R, Mason WP, Van Den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy Plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med (2005) 352:987–96. doi: 10.1056/NEJMoa043330
5. Stupp R, Hegi ME, Mason WP, Van Den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of Radiotherapy With Concomitant and Adjuvant Temozolomide Versus Radiotherapy Alone on Survival in Glioblastoma in a Randomised Phase III Study: 5-Year Analysis of the EORTC-NCIC Trial. Lancet Oncol (2009) 10:459–66. doi: 10.1016/S1470-2045(09)70025-7
6. Hegi ME, Diserens AC, Gorlia T, Hamou MF, De Tribolet N, Weller M, et al. MGMT Gene Silencing and Benefit From Temozolomide in Glioblastoma. N Engl J Med (2005) 352:997–1003. doi: 10.1056/NEJMoa043331
7. Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med (2015) 372:2499–508. doi: 10.1056/NEJMoa1407279
8. Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. The AlkB Family of Fe(II)/alpha-Ketoglutarate-Dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J Biol Chem (2015) 290:20734–42. doi: 10.1074/jbc.R115.656462
9. Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of Alkylated DNA: Recent Advances. DNA Repair (Amst) (2007) 6:429–42. doi: 10.1016/j.dnarep.2006.10.005
10. Xie LJ, Liu L, Cheng L. Selective Inhibitors of AlkB Family of Nucleic Acid Demethylases. Biochemistry (2020) 59:230–9. doi: 10.1021/acs.biochem.9b00774
11. Aravind L, Koonin EV. The DNA-Repair Protein AlkB, EGL-9, and Leprecan Define New Families of 2-Oxoglutarate- and Iron-Dependent Dioxygenases. Genome Biol (2001) 2:RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007
12. Rajecka V, Skalicky T, Vanacova S. The Role of RNA Adenosine Demethylases in the Control of Gene Expression. Biochim Biophys Acta Gene Regul Mech (2019) 1862:343–55. doi: 10.1016/j.bbagrm.2018.12.001
13. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat Chem Biol (2011) 7:885–7. doi: 10.1038/nchembio.687
14. Delaney JC, Essigmann JM. Mutagenesis, Genotoxicity, and Repair of 1-Methyladenine, 3-Alkylcytosines, 1-Methylguanine, and 3-Methylthymine in alkB Escherichia Coli. Proc Natl Acad Sci USA (2004) 101:14051–6. doi: 10.1073/pnas.0403489101
15. Pilzys T, Marcinkowski M, Kukwa W, Garbicz D, Dylewska M, Ferenc K, et al. ALKBH Overexpression in Head and Neck Cancer: Potential Target for Novel Anticancer Therapy. Sci Rep (2019) 9:13249. doi: 10.1038/s41598-019-49550-x
16. Cai Y, Wu G, Peng B, Li J, Zeng S, Yan Y, et al. Expression and Molecular Profiles of the AlkB Family in Ovarian Serous Carcinoma. Aging (Albany NY) (2021) 13:9679–92. doi: 10.18632/aging.202716
17. Wu G, Yan Y, Cai Y, Peng B, Li J, Huang J, et al. ALKBH1-8 and FTO: Potential Therapeutic Targets and Prognostic Biomarkers in Lung Adenocarcinoma Pathogenesis. Front Cell Dev Biol (2021) 9:633927. doi: 10.3389/fcell.2021.633927
18. Ke B, Ye K, Cheng S. ALKBH2 Inhibition Alleviates Malignancy in Colorectal Cancer by Regulating BMI1-Mediated Activation of NF-KappaB Pathway. World J Surg Oncol (2020) 18:328. doi: 10.1186/s12957-020-02106-0
19. Lee SY, Luk SK, Chuang CP, Yip SP, To SS, Yung YM. TP53 Regulates Human AlkB Homologue 2 Expression in Glioma Resistance to Photofrin-Mediated Photodynamic Therapy. Br J Cancer (2010) 103:362–9. doi: 10.1038/sj.bjc.6605797
20. Johannessen TC, Prestegarden L, Grudic A, Hegi ME, Tysnes BB, Bjerkvig R. The DNA Repair Protein ALKBH2 Mediates Temozolomide Resistance in Human Glioblastoma Cells. Neuro Oncol (2013) 15:269–78. doi: 10.1093/neuonc/nos301
21. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res (2019) 47:W556–60. doi: 10.1093/nar/gkz430
22. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (2017) 19:649–58. doi: 10.1016/j.neo.2017.05.002
23. Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: A New Database for Meta-Analysis of the Prognostic Value of Genes. BMC Med Genomics (2009) 2:18. doi: 10.1186/1755-8794-2-18
24. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov (2012) 2:401–4. doi: 10.1158/2159-8290.CD-12-0095
25. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res (2021) 49:D605–12. doi: 10.1093/nar/gkaa1074
26. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: Gene Set Analysis Toolkit With Revamped UIs and APIs. Nucleic Acids Res (2019) 47:W199–205. doi: 10.1093/nar/gkz401
27. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77:e108–10. doi: 10.1158/0008-5472.CAN-17-0307
28. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing Multi-Omics Data Within and Across 32 Cancer Types. Nucleic Acids Res (2018) 46:D956–63. doi: 10.1093/nar/gkx1090
29. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45:W98–102. doi: 10.1093/nar/gkx247
30. Asplund A, Edqvist PH, Schwenk JM, Ponten F. Antibodies for Profiling the Human Proteome-The Human Protein Atlas as a Resource for Cancer Research. Proteomics (2012) 12:2067–77. doi: 10.1002/pmic.201100504
31. Snel B, Lehmann G, Bork P, Huynen MA. STRING: A Web-Server To Retrieve And Display The Repeatedly Occurring Neighbourhood Of A Gene. Nucleic Acids Res (2000) 28:3442–4. doi: 10.1093/nar/28.18.3442
32. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment For Integrated Models Of Biomolecular Interaction Networks. Genome Res (2003) 13:2498–504. doi: 10.1101/gr.1239303
33. Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive Analyses of Tumor Immunity: Implications for Cancer Immunotherapy. Genome Biol (2016) 17:174. doi: 10.1186/s13059-016-1028-7
34. Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular Subclasses of High-Grade Glioma Predict Prognosis, Delineate a Pattern of Disease Progression, and Resemble Stages in Neurogenesis. Cancer Cell (2006) 9:157–73. doi: 10.1016/j.ccr.2006.02.019
35. Sabatini DM. Twenty-Five Years of mTOR: Uncovering the Link From Nutrients to Growth. Proc Natl Acad Sci USA (2017) 114:11818–25. doi: 10.1073/pnas.1716173114
36. Xie X, Hu H, Tong X, Li L, Liu X, Chen M, et al. The mTOR-S6K Pathway Links Growth Signalling to DNA Damage Response by Targeting RNF168. Nat Cell Biol (2018) 20:320–31. doi: 10.1038/s41556-017-0033-8
37. Ma Y, Vassetzky Y, Dokudovskaya S. Mtorc1 Pathway in DNA Damage Response. Biochim Biophys Acta Mol Cell Res (2018) 1865:1293–311. doi: 10.1016/j.bbamcr.2018.06.011
38. Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O’reilly T, et al. The mTOR Inhibitor RAD001 Sensitizes Tumor Cells to DNA-Damaged Induced Apoptosis Through Inhibition of P21 Translation. Cell (2005) 120:747–59. doi: 10.1016/j.cell.2004.12.040
39. Zhao Q, Zhao Y, Hu W, Zhang Y, Wu X, Lu J, et al. M(6)A RNA Modification Modulates PI3K/Akt/mTOR Signal Pathway in Gastrointestinal Cancer. Theranostics (2020) 10:9528–43. doi: 10.7150/thno.42971
40. Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 Inhibited Autophagy of Epithelial Ovarian Cancer Through miR-7 and BCL-2. J Exp Clin Cancer Res (2019) 38:163. doi: 10.1186/s13046-019-1159-2
41. Xie Q, Wu TP, Gimple RC, Li Z, Prager BC, Wu Q, et al. N(6)-Methyladenine DNA Modification in Glioblastoma. Cell (2018) 175:1228–43.e20. doi: 10.1016/j.cell.2018.10.006
42. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-Methyladenosine-Dependent Regulation of Messenger RNA Stability. Nature (2014) 505:117–20. doi: 10.1038/nature12730
43. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the Human and Mouse M6a RNA Methylomes Revealed by M6a-Seq. Nature (2012) 485:201–6. doi: 10.1038/nature11112
44. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-Methyladenosine Modulates Messenger RNA Translation Efficiency. Cell (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
45. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. M(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-Like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell (2017) 31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013
46. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2hg Exhibits Anti-Tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell (2018) 172:90–105.e23. doi: 10.1016/j.cell.2017.11.031
47. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the Tumor Immune Microenvironment (TIME) for Effective Therapy. Nat Med (2018) 24:541–50. doi: 10.1038/s41591-018-0014-x
48. Tang T, Huang X, Zhang G, Hong Z, Bai X, Liang T. Advantages of Targeting the Tumor Immune Microenvironment Over Blocking Immune Checkpoint in Cancer Immunotherapy. Signal Transduct Target Ther (2021) 6:72. doi: 10.1038/s41392-020-00449-4
49. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-Tumour Immunity Controlled Through mRNA M(6)A Methylation and YTHDF1 in Dendritic Cells. Nature (2019) 566:270–4. doi: 10.1038/s41586-019-0916-x
50. Yan G, An Y, Xu B, Wang N, Sun X, Sun M. Potential Impact of ALKBH5 and YTHDF1 on Tumor Immunity in Colon Adenocarcinoma. Front Oncol (2021) 11:670490. doi: 10.3389/fonc.2021.670490
51. Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, et al. ALKBH5 Regulates Anti-PD-1 Therapy Response by Modulating Lactate and Suppressive Immune Cell Accumulation in Tumor Microenvironment. Proc Natl Acad Sci USA (2020) 117:20159–70. doi: 10.1073/pnas.1918986117
Keywords: glioblastoma, AlkB family, patient prognosis, DNA damage repair, immune infiltration
Citation: Feng S, Xu Z, Peng J and Zhang M (2022) The AlkB Family: Potential Prognostic Biomarkers and Therapeutic Targets in Glioblastoma. Front. Oncol. 12:847821. doi: 10.3389/fonc.2022.847821
Received: 03 January 2022; Accepted: 21 February 2022;
Published: 10 March 2022.
Edited by:
Wen Cheng, The First Affiliated Hospital of China Medical University, ChinaCopyright © 2022 Feng, Xu, Peng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijie Xu, eHpqMTMyMjAwN0Bjc3UuZWR1LmNu; Mingyu Zhang, aG5jc3pteUAxNjMuY29t
 Songshan Feng
Songshan Feng Zhijie Xu
Zhijie Xu Jinwu Peng
Jinwu Peng Mingyu Zhang1*
Mingyu Zhang1*