
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 07 April 2022
Sec. Cancer Genetics
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.845703
This article is part of the Research Topic Reviews in Cancer Genetics View all 20 articles
Circular RNAs (circRNAs) are non-coding single-stranded covalently closed circular RNA, mainly produced by reverse splicing of exons of precursor mRNAs (pre-mRNAs). The characteristics of high abundance, strong specificity, and good stability of circRNAs have been discovered. A large number of studies have reported its various functions and mechanisms in biological events, such as the occurrence and development of cancer. In this review, we focus on the classification, characterization, biogenesis, functions of circRNAs, and the latest advances in cancer research. The development of circRNAs as biomarkers in cancer diagnosis and treatment also provides new ideas for studying circRNAs research.
CircRNAs were first discovered in RNA viruses in 1976 (1). Subsequently, it was discovered in eukaryotic cells and humans (2–4). CircRNAs are covalently closed ring structures with 5 ‘and 3’ ends directly linked together, which makes them more stable than linear RNA. They were originally thought to be the product of splicing errors during low abundance transcription. With the development of high-throughput RNA sequencing technology and bioinformatics algorithms, a new understanding of circRNAs has emerged. The functions and mechanisms of new types of circRNAs during biogenesis have been identified. CircRNAs have been found to act as ceRNA or miRNA sponges and bind to proteins. As well as some newly discovered functions, such as regulating parental gene expression, regulating pre-RNA splicing and potential translation templates for proteins (5). Many circular RNAs have been discovered to be biomarkers that impact the onset and growth of malignancies in recent years, attracting a lot of attention. They have been identified in lung cancer (6), hepatocellular carcinoma (7), gastric cancer (8), colorectal cancer (9), and so on. However, the molecular mechanisms and early diagnosis of cancer are not well understood. And, diagnosis and treatment based on circRNAs are still lacking. Therefore, it is urgent to explore new molecular mechanisms and effective biomarkers for the diagnosis of cancer.
In this review, we focused on the biological characteristics, functions, mechanisms, and detection techniques of circRNAs associated with cancer, and discussed their potential application as biomarkers and therapeutic targets. Thus, provide valuable clinical information for the diagnosis and timely treatment of cancer in the future.
According to the formed sequence, circRNAs can be divided into six categories: exonic circular RNAs (ecircRNAs), circular intronic RNAs (ciRNAs), exon-intron circular RNAs (EIciRNAs), intergenic circRNAs, anti-sense circRNAs, and tRNA intronic circRNAs (tricRNAs) (10).
CircRNAs have no ends so it is highly stable and have specific spatiotemporal expression patterns. Numerous circRNAs usually express in specific tissues and specific developmental stages (11). CircRNAs were found to be evolutionarily conserved in diversity and the conservation is different in different tissues, among which the most conservative in the brain (12). Jeck et al. identified over 25,000 different circRNAs in human fibroblasts (13). In addition, circRNAs are widely distributed and have been reported in thousands of animal cells, such as humans, mice, and nematodes, and expressed in high abundance (11, 14). Rybak-wolf et al. found that circRNAs are abnormally enriched in the mammalian brain and are specifically and dynamically expressed in neuronal differentiation (15). New classification methods such as differences in length, stability, function, and characteristics of circRNAs still need to be continuously explored.
CircRNAs are derived from the reverse splicing mechanism of pre-mRNAs to form a single-stranded closed loop (16). However, the biogenesis mechanism has not been fully elucidated, circRNAs can be derived from exons, introns, 3’UTR, 5’UTR, intergenic regions, or antisense sequences (Figure 1). As early as 2013, Jeck et al. proposed two circRNAs cyclization models: Lariat-driven circularization and intron-pairing-driven circularization (13). Lariat-driven circularization, also known as exon skipping, is connected by non-adjacent exons as donors and acceptors to form a lasso structure. Then the introns in the lasso are removed, resulting in ecircRNAs. The biogenesis of circRNAs is mainly studied in ecircRNAs, other types of circRNAs are rarely studied. EcircRNAs account for 80% of identified circRNAs (17). The biogenesis of eicircRNAs is similar to ecircRNAs. In some cases, the intron portion is completely sheared to form eicircRNAs. Another model is intron-pairing-driven circularization, also known as direct reverse splicing. This model is based on the direct base pairing of the flanking introns, and then the introns are removed to form a ring structure (17). Zhang et al. reported a class of intron-derived circRNAs, namely CiRNAs (5). The formation of ciRNAs depends on the 7 nucleotides GU enrichment element near the 5’ splicing site and the 11 nucleotides C-rich element near the branch site. Zhang et al. believed that the exon cycle depends on the complementary sequences of the flanking introns. Intergenic circRNAs are formed from genes outside known genetic loci (11). TricRNA is formed by intron excision of the pre-tRNA by the tRNA splicing mechanism, followed by intron release and binding into tRNA and TricRNA (18).
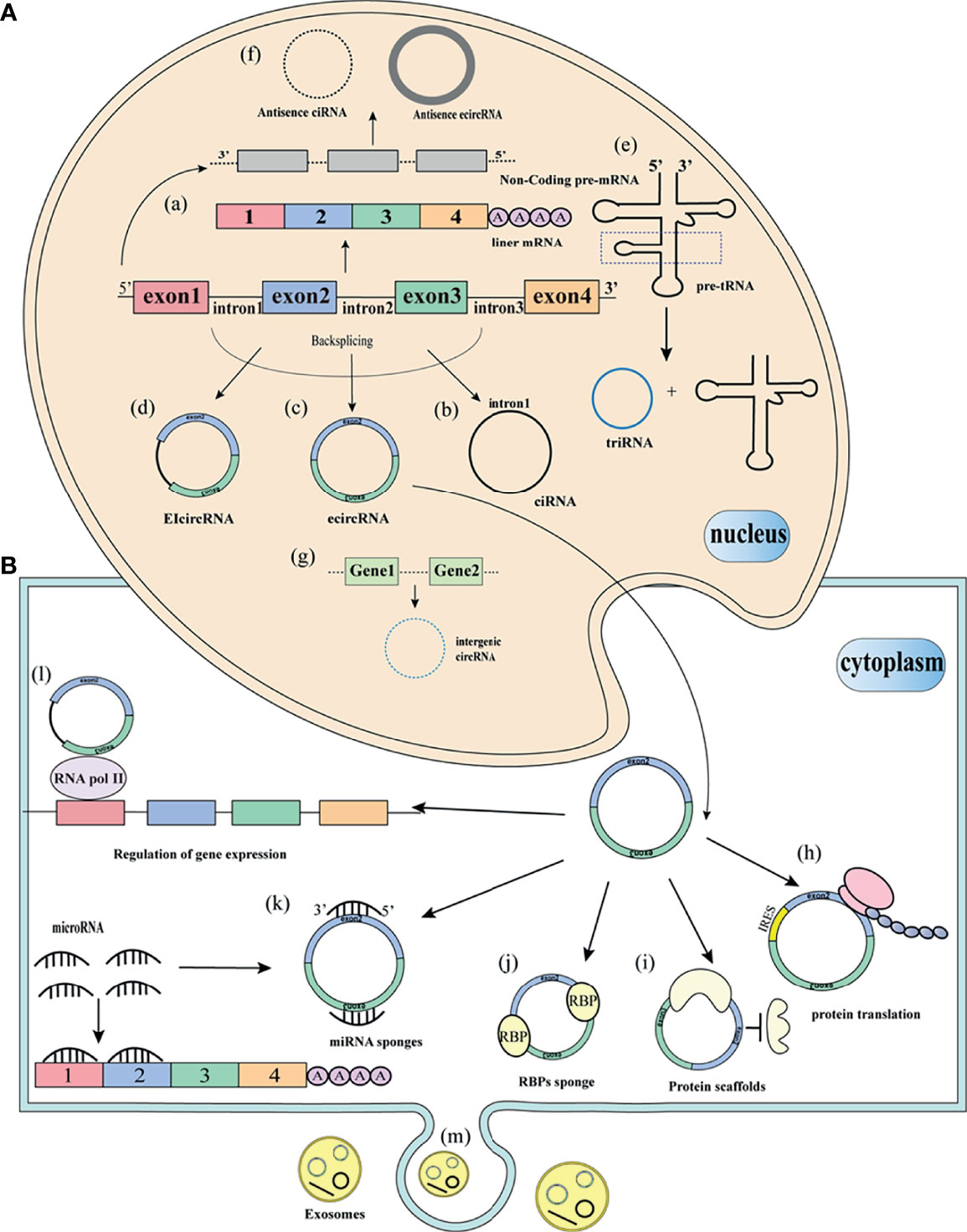
Figure 1 Biogenesis and function of circRNAs (A) Biogenesis of circRNAs. (a) Pre-mRNA splicing removes introns to form mature linear mRNA. (b) The introns removed by pre-mRNA splicing form circularization to form a stable ciRNA. (c) Circularization of pre-mRNA exons into ecircRNA. (d) pre-mRNA exons and introns are circularized into ecircRNAs. (e) removal of pre-tRNA introns, release to form triRNA and tRNA. (f) form antisense circRNAs from non-coding regions of pre-mRNA. (g) from two different intergenic sequences to form intergenic circRNAs. (B) Functions and of circRNAs. (h) circRNAs translation protein with similar IRES sequence. (i) circRNAs as protein scaffolds. (j) circRNAs bind to RBPs. (k) circRNAs as sponges for miRNA. (l) circRNAs interact with RNA polymerase II to regulate gene expression. (m) circRNAs are packaged into vesicles and released outside the cell to perform biological functions.
In addition, multiple factors are involved in the biogenesis of circRNAs. Zhang et al. reported that the exon cycle depends on the complementary sequences of the flanking introns (19). Some RBPs play an important role in reverse splicings, such as MBL(splicing factor muscleblind), QKI(Quaking), and FUS(fused in sarcoma), binding to both sides of the flanking intron sequence enhances exon cycling by tightly linking the 3 ‘and 5’ ends of circRNAs. Thereby promoting exon circulation. Muscleblind is a splicing factor for MBL-derived genes and MBL in Drosophila promotes the production of circRNAs from the second exon of its own pre-mRNA by binding to flanking introns (20). The QKI of the STAR family is a tumor suppressor protein with three isoforms, all of which have the same KH domain but have different 3’UTRs. Among them, QKI-5, the most abundant nuclear isoform, acts on circRNAs during splicing. QKI dimerizes through its N-terminal Qua1 domain and binds to two-part sequence motifs that can be located on the same or separate RNA molecules (21). The investigation of PAR-CLIP cross-linking in human embryonic kidney cells (HEK293T) indicated that the majority of QKI binding occurs within introns and is responsible for circRNA synthesis, which limits proliferation and EMT during human cancer (22). In addition, Conn et al. also introduced consensus binding sequences for QKI in flanking introns to enable circRNAs to be generated from exons that normally only undergo canonical linear splicing (21). A recent study showed that overexpression of circ-SHPRH in cadmium-transformed BEAS-2B cells promoted the expression of QKI and significantly inhibited cell proliferation, EMT, invasion, migration, and non-anchored growth. Conclusions of Conn et al. (23). FUS was first reported to be involved in circRNA generation in the nervous system in 2017 (24). Cao et al. recently discovered that the nematode homologous gene FUST-1 promotes the creation of numerous circRNAs while having no effect on the analogous linear mRNA, regulating exon skipping and reverse splicing, surprisingly, CLIP-seq results suggest that FUS attaches to stem-loop secondary structure rather than particular sequences (25).
Besides, negative regulators destroy the stability of intron interactions, thereby reducing the cyclization efficiency, such as adenosine deaminases acting on RNA 1 (ADAR1) reduces the efficiency of cyclization by disrupting the base pairing between flanking introns through the A to I RNA editing mechanism (15). ADAR is an adenosine deaminase that is widely expressed in humans and can be applied to RNA modification. ADAR systems are used for programmable RNA editing in vitro and in vivo by recruiting ADARs to target RNA sequences using ADAR recruitment guide RNAs (adRNAs). Two recent studies have designed circRNAs that can recruit ADARs to improve RNA editing efficiency. Katrekar et al. engineered a highly stable circular ADAR-recruiting guide RNA (cadRNA) to recruit endogenous ADARs, improving the efficiency and durability of RNA editing (26). The engineered circ-arRNAs designed by Yi et al.’s LEAPER2.0 system have much higher editing efficiency than the corresponding linear arRNAs, which greatly improves the efficiency and robustness of RNA editing (27).
Interestingly, UAP56 and URH49 proteins can assist the transport of circRNAs from the nucleus to the cytoplasm (28). This discovery is novel, however, many of the regulatory factors involved in circRNAs biogenesis remain unclear and require more research. A better understanding of the biogenesis mechanism of circRNAs will lead to a better understanding of their specific roles in cancer development. Future studies can explore the levels of specific key factors that regulate the biogenesis of circRNAs, which will also provide innovative strategies for cancer treatment and prevention.
In addition to its unique way of formation, how circRNAs participate in the process of biogenesis has also deepened our understanding of circRNAs. However, the functions of most circRNAs are still unknown. Recent studies show that the functions of circRNAs are mainly ceRNA or miRNA sponging, binding with proteins, regulation of pre-RNA splicing, regulation of parental gene expression, and potential translation templates for proteins or peptides (Figure 1).
MicroRNAs (miRNAs) are a class of small non-coding RNAs, which play a regulatory role in various cellular activities including cancer by pairing regulatory genes with mRNAs target bases. Hansen et al. first proposed the concept of miRNAs sponges in 2013 (29). There are miRNAs response elements on circRNAs, which can competitively bind to miRNAs, eliminate the inhibitory effect of miRNAs on target genes, and regulate the expression of related genes. They demonstrated for the first time that circCIRS-7 (CDR1as) can be a sponge of miRNAs. CIRS-7 promotes the progression of a variety of tumors. There are more than 70 miR-7 sponge binding sites on CIRS-7. CIRS-7 inhibits miR-7 and participates in various events in tumorigenesis, such as cell proliferation, migration, invasion, and differentiation (30). With the development of scientific research, more and more circRNAs have been found to act as sponges for miRNAs. This mechanism affects cell proliferation, migration, invasion, and blood vessel formation, and has been widely reported in the cancer field. For example, the combination of circSATB2 and miR-326 regulates the expression of FSCN1 and further promotes the proliferation, migration, and invasion of NSCLC cells (31). In addition, some circRNAs have also been found to combine multiple miRNAs to act on different systems. For example, cirMAT2B can be combined with miR-515-5p to increase the expression of HIF-1α and promote the growth of gastric cancer (32). Moreover, circMTO1 can also combine with miR-541-5p to inhibit the progression of liver cancer (33).
Another function of circRNAs is to directly bind proteins to participate in physiological and pathological processes (Figure 1). More than 800 RNA-binding proteins (RBPs) have been identified in the human genome (34). RBPs play a role in circRNAs splicing, processing, folding, stabilization, and positioning. For example, CircFoxo3 is formed by Foxo3 exon 2 and has a wide range of complex biological functions, which are related to cell differentiation, apoptosis, and cycle. It has been reported that in the cytoplasm, the senescence-related proteins ID-1, E2F1, FAK, and HIF1α interact with circFoxo3 and no longer exert their anti-aging and anti-stress effects, leading to the promotion of cell senescence (35). CircFoxo3 can also bind to cyclin cells cyclin-dependent-kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (p21) to form a ternary complex to inhibit the binding of CDK2 and p21, and in the G1 phase inhibits the cell cycle progression (36). In addition, circMBL can bind to mannose-binding lectin (MBL) protein to control excess MBL protein (20). Two circRNAs, KIRKOS-73 and KIRKOS-71, are able to regulate the exosomal metastasis of p53 expression in recipient cells, and p53 plays a key role in metastasis and tumorigenesis (37). CircAgo2 transfers HuR protein from the nucleus to the cytoplasm, stabilizing the binding of mRNA and AU-rich elements in UTR (38). CircPABPN1 competitively binds to HuR, prevents HuR from binding to PABPN1 mRNA, and subsequently inhibits the translation of PABPN1 (39). The above studies have proved that the interaction between circRNAs and proteins plays an important role.
CircRNAs may affect the splicing of pre-RNA and can compete with pre-RNA for splicing sites. For example, circUBR5 may be involved in the RNA splicing regulation process, it can be combined with the splicing regulator QKI in the nucleus, NOVA alternative splicing regulator 1 (NOVA1), and U1 small nuclear RNA (snRNA) (40). CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1 (41).
EIciRNAs and ciRNAs are circRNAs with intron sequences, which are mainly located in the nucleus. Experiments have shown that EIciRNAs and ciRNAs can regulate gene expression. For example, the knockdown of circEIF3J and circPAIP2 can cause the transcription level of EIF3J and PAIP2 to decrease (42). EIciRNAs can promote the transcription of their parental genes in cis by interacting with U1 snRNA, revealing a new regulatory strategy for gene expression in RNA-RNA interactions (43). Li et al. found that ci-ankrd52 shows a different open structure conformation from pre-mRNA with the same sequence, which can replace pre-mRNA to form more stable R-loops (44). It can be seen that ci-ankrd52 plays a potential role in promoting transcription elongation. However, the effect of EIciRNAs and ciRNAs regulation still need to study in-depth.
Although circRNAs have long been considered non-coding RNAs that cannot translate proteins, recent studies have shown that circRNAs do not rely on conventional translation modes and have translation potential. Previous studies have found that some circRNAs have internal ribosome entry (IRE) site sequences or open reading frame (ORF) translatable proteins such as circMAPK1 and circMBL3. CircMAPK1 encodes a new protein with a length of 109 amino acids that competitively binds to MEK1 to inhibit the phosphorylation of MAPK1 (45). CircMBI translates to a small peptide in the head of a fly (46). However, recent studies have found that circRNAs lacking this sequence can also translate proteins. The N6-methyladenosine (M6A) modification allows circRNAs to be translated in a cap-independent manner (47). In addition, bioinformatics tools have been developed to predict translation potential, but the accuracy needs to be verified. Although these new discoveries are exciting, the function and efficiency of these translated proteins or peptides need further research (48, 49).
Early RNA sequencing did not identify circular RNAs without a ployA tail, and non-linear fragments were often considered errors and were ignored. This section introduces some traditional circRNAs detection techniques and emerging technical methods (Table 1). Northern blot, qRT-PCR, RNA-seq, and Microarrays are examples of traditional circRNA detection techniques (63). However, previous RNA analysis methods are also difficult to study circRNAs. RNA-seq detection of RNA detection efficiency is low, so many low abundance circRNAs may be missed. And microarray technology has been used to detect linear RNA for a long time. The detection efficiency of circRNAs microarray is much higher than RNA-seq because it contains probes that target the head-to-tail connection (51). But it produces data that is difficult to compare between studies. Therefore, there is an urgent need to develop simple, effective, and sensitive new methods to study circRNAs.
Recently, a newly reported exome capture RNA sequencing technology can detect and characterize circRNAs in more than 2000 cancer samples (64). In addition, Zhang et al. developed a new method for quantitative detection of circular RNA with high sensitivity and specificity (56). The two cleverly designed DNA probes can be precisely connected by using ligase at the connection site of circular RNA. Distinguish circular RNA from corresponding linear RNA. Liu et al. proposed a reverse transcription-rolling cycle amplification (RT-RCA) process that can selectively amplify target circular RNA (57). Zhang et al. designed a pair of Stem-loop primers (SLPs) based on loop-mediated isothermal amplification (LAMP), an excellent nucleic acid amplification method, that could accurately recognize circRNA junction sequences, thereby establishing an SLP-induced dual amplification system (58).The exponential amplification method makes circRNA detection simple and accurate. Additionally, Li et al. designed an electrochemical method for the recognition and capture of circRNAs with hairpin probes, avoiding additional RNase R treatment, and the method exhibited good sensitivity and selectivity (62).
According to the characteristics of circRNAs, some detection methods for circRNAs have been developed, including traditional detection methods and newly developed methods, which have their own advantages and disadvantages. However, the way forward is clear, and the detection method of circRNAs still needs to be developed or improved, so that its sensitivity, specificity, rapidity, and convenience can be applied to biomedical research and clinical detection. All these make it possible for circRNAs to become diagnostic tools and therapeutic targets.
In recent years, with the intensive study of circRNAs, researchers have developed many high-quality online databases. This section introduces some databases that can be used for circRNAs research in Table 2. These artificially established databases are of great significance to the study of the biological functions of circRNAs. In addition to the online database listed in the table, in 2022, the University of Padova developed a circRNA function prediction software CRAFT, which can predict circRNA sequences and molecular interactions with miRNAs and RBPs, as well as their coding potential (81). Nevertheless, the existing circRNAs collection is largely limited to certain well-studied species, such as humans and mice. In addition, the current annotations and naming are rather incomplete. Most databases only use one or two resources for annotations. Searching for the same circRNAs has different naming methods in different databases, which increases the difficulty of studying circRNAs.
To date, a large number of reports have found that circRNAs are abnormally expressed in tumor tissues, and more and more evidence shows that circRNAs play a critical role in the occurrence and development of tumors (82, 83). Most of the abnormal circRNAs discovered are the sponges or proteins of miRNAs (Table 2). In addition to affecting cancer cell proliferation, migration, invasion, and escape from apoptosis and angiogenesis. CircRNAs can also regulate these cancer markers by regulating signal pathways such as Wnt/β-catenin (7), PIK3/AKT (84), and MAPK/ERK pathways (85). Among them, CIRS-7 is widely studied in cancer and is usually up-regulated in cancer cells. It has been described to be expressed in liver cancer, lung cancer, gastric cancer, colorectal cancer, breast cancer, and other cancers (30). The miRNA regulated by CIRS-7 are miR-7 (86), miR-135a-5p (87, 88), miR-1270 (89), miR-26a-5p (90) and so on.
In addition to the circRNAs described above, some circRNAs expressed in common cancers have recently been discovered, as shown in Table 3 and Figure 2.
Lung cancer is the malignant tumor with the highest mortality rate in the world (199). Circ0003222 sponges miR-527 to down-regulate the expression of PHF21B and its downstream β-catenin. Thereby promoting the proliferation, migration, and invasion of tumor cells. Yu et al. (91) discovered a new type of circHMGA2 (hsa_circ_0027446) molecule through microarray, which is highly expressed in lung adenocarcinoma (LUAD). Mechanically, circHMGA2 promotes LUAD cell metastasis through the miR-1236-3p/ZEB1 axis. Yao et al. (99) found that circ_0018414 was down-regulated in LUAD tissues and cells, and inhibited the progression of LUAD through the Wnt/β-catenin pathway of miR-6807-3p/DKK1 axis inactivation. Some circRNAs have been described as binding to proteins in lung cancer. For example, circNDUFB2 (101), which is down-regulated in non-small cell lung cancer, acts as a scaffold to enhance the interaction between TRIM25 and IGF2BPs. It inhibits the growth and metastasis of NSCLC cells by regulating protein ubiquitination and degradation and cellular immune responses. In addition, Huang et al. (103)found that circXPO1 is highly expressed in LUAD through RNA sequencing. In terms of mechanism, circXPO1 can bind to IGF2BP1 to enhance the stability of CTNNB1 mRNA, thereby promoting the progress of LUAD.
The development of new NSCLC-specific biomarkers to aid in diagnosis and clinical decision-making has always been a pressing concern. Li et al. found that circ0003222 is related to the staging, metastasis, and survival rate of patients with non-small cell lung cancer (NSCLC) (6). Additionally, high expression of circ 0070354 was demonstrated to be substantially linked to advanced TNM staging and poor differentiation in NSCLC and was an independent predictor of poor prognosis. CEA, SCC, and Cyfra21-1 are the acronyms for CEA, SCC, and Cyfra21-1, respectively. The AUC of circ0070354, when combined with the other three mature tumor markers, was 0.730, which was much higher than the solitary diagnosis (200). According to the findings, some circRNAs potentially outperform traditional tumor markers in terms of diagnosis, and the combined diagnosis has higher sensitivity and specificity for lung cancer diagnosis and treatment.
Colorectal cancer (CRC) is the second leading cause of death from cancer worldwide (199). Jian et al. (110) tested the gene expression in 42 pairs of colorectal cancer tissues and normal tissues adjacent to cancer. The results showed that circ001680 was overexpressed in 71.4% of colorectal cancer tissues. In terms of mechanism, circ001680 promotes the proliferation and migration of colorectal cancer cells by targeting miR-340. Yang et al. discovered a new circRNA, circPTK2, and found that circPTK2 binds to the Ser38, Ser55, and Ser82 sites of vimentin protein to promote EMT of CRC cells in vivo and in vitro (112). In addition to interacting with proteins to regulate the expression of target genes, circRNAs encoding proteins or peptides have also been found in colorectal cancer, such as circ0006401 (116), circPLCE1 (117), and circFNDC3B (118). Among them, circ000641 encoding peptide fragment promotes the proliferation and migration of CRC and promotes the stability of the host gene col6a3 mRNA, and thus promotes the proliferation and metastasis of CRC. The circPLCE1-411 protein encoded by circPLCE1 combined with the HSP90α/RPS3 complex plays a key role in the NF-κB activation of CRC and ultimately inhibits tumor proliferation and metastasis in CRC cells (117). The tumor suppressor circFNDC3B is mainly located in the cytoplasm and encodes a new protein circFNDC3B-218aa, thereby inhibiting the proliferation, invasion, and migration of colon cancer cells (118).
Wang et al. (108) found that circSPARC is highly expressed in the tissues and plasma of CRC patients, is associated with advanced TNM staging, lymph node metastasis, and a low survival rate of CRC. Mechanistically, circSPARC can upregulate the expression of JAK2 by sponge miR-485-3p, and ultimately promote the accumulation of phosphorylated p-STAT3, thereby promoting the proliferation and migration of cancer cells. The most commonly used colorectal tumor marker CEA has limited sensitivity in early CRC (201, 202). While circRNAs can be employed as reliable biomarker complements for CEA in CRC early diagnosis and treatment monitoring. According to the ROC curve analysis of Hu et al., the AUC (0.831 vs 0.657), sensitivity (0.677 vs 0.532), and specificity (0.915 vs 0.675) values of circ 001659 in the early diagnosis of CRC were higher than those of CEA as a novel biomarker of successful treatment and response for cancer tracking thing (203). These findings indicate that circRNAs can become potential diagnostic and prognostic biomarkers and therapeutic targets for the treatment of CRC.
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths. There have been multiple reports that a variety of circRNAs inhibit or promote tumor progression in liver cancer. Hu et al. (120) found that circASAP1 promotes the proliferation and invasion of liver cancer cells by regulating the miR-326/miR-532-5p-MAPK1 signaling pathway, and then mediates tumor-associated macrophages by regulating the miR-326/miR-532-5p-CSF-1 pathway Cell infiltration. The circRNAs array analyzes the expression of circRNAs in tumor tissues and normal tissues. In a study by Dong et al. (125), it was found that 28 up-regulated and 18 down-regulated circRNAs were found in liver cancer tissues. circMEMO1 is significantly down-regulated in HCC samples and can act as a sponge of miR-106b-5p to regulate TCF21 promoter methylation and gene expression, thereby regulating HCC progression. Li et al. (128)found that circMRPS35 was highly expressed in 35 pairs of HCC patients compared with normal tissues. It is worth noting that circMRPS35 can not only adsorb miR-148a-3p, regulate the expression of Syntaxin 3 (STX3), thereby regulating the ubiquitination and degradation of phosphatase and tensin homolog (PTEN) but can also encode a peptide (circMRPS35-168aa), this peptide promotes cisplatin resistance in HCC cells. CircLRIG3 is significantly up-regulated in HCC, forming a ternary complex with EZH2 and STAT3, promoting EZH2-induced STAT3 methylation and subsequent phosphorylation, leading to the activation of STAT3 signal, thereby promoting the proliferation, migration, and invasion of liver cancer cells. Reduce cell apoptosis (129).
Wei et al. reported that the expression of circCDYL or the combined expression of HDGF and HIF1AN are independent markers for distinguishing early HCC, providing the possibility for the detection and early treatment of liver cancer (204). Yang et al. found that circFN1 promotes sorafenib resistance by regulating the miR-1205/E2F1 signaling pathway, which is a potentially valuable target for HCC resistance (205).
Gastric cancer (GC) is the fourth leading cause of death in the world (199), especially in Asian countries, the incidence of gastric cancer is increasing year by year. Cao et al. found that circ0008259 (circLMO7) is highly expressed in GC tissues, circLMO7 sponge miR-30a-3p regulates the WNT2/β-Catenin pathway and affects the glutamine metabolism of GC cells, and ultimately promotes the growth and migration of GC (131). Peng et al. found that the level of circCUL2 in GC tissues and cells was significantly reduced, and the sponge engulfed miR-142-3p to regulate ROCK2, thereby inhibiting malignant transformation and inhibiting tumorigenicity in vivo (140). In addition, Yan et al. (206) found that circEVI5 was significantly down-regulated in GC tissues and cells. circEVI5 sponges swallowed miR-4793-3p and increased the expression level of FOXO1 to inhibit the proliferation of GC and delay the cell cycle. Wang et al. (148)analyzed the circRNAs of five pairs of human stomachs and corresponding non-tumor adjacent specimens and found that circURI1 was significantly highly expressed in GC and metastasized in GC. It regulates a small part of genes involved in cell movement by isolating hnRNPM protein to inhibit GC metastasis. Zhang et al. (149) found that circDIDO1 is down-regulated in gastric cancer tissues, and its low level is associated with larger tumors, distant metastasis, and poor prognosis. In mechanism, circDIDO1 encodes a new 529aa protein, which interacts with poly ADP-ribose polymerase 1 (PARP1). Effect and inhibit its activity. Interestingly, circDIDO1 also binds to peroxide reduction protein 2 (PRDX2), which promotes the ubiquitination and degradation of PRDX2 mediated by rbx1, leading to inactivation of its downstream signaling pathways.
Further, numerous research has explored the clinical utility of circRNAs as biomarkers for the early detection and prognosis of gastric cancer. For instance, Song et al. (207) detected the expression profile of circRNAs and found that hsa_circ_000780 was significantly downregulated in GC tissue samples, and its level was correlated with the level of tumor clinicopathological features. Interestingly, circ000780 was also found in gastric juice of patients with early GC. In another report, circERBB2 (208) in plasma can be used as a prognostic biomarker for gastric cancer patients. CircERBB2 levels in preoperative plasma (high group) were significantly correlated with lymph node metastasis (P = 0.035), suggesting that it could be used to predict noninvasively the prognosis of GC.
Breast cancer (BC) is the main cause of cancer in women and the main cause of death in women. Wang et al. (151) used the circRNAs microarray data set and found that four circRNAs were abnormally expressed in TNBC. Among them, circBACH2 is most significantly elevated in BC tissues, and its high expression promotes epithelial-mesenchymal transition and cell proliferation and is positively correlated with the malignant progression of BC patients. Mechanistically, circBACH2 sponges miR-186-5p and miR-548c-3p, thereby releasing the expression of C-X-C chemokine receptor type 4 (CXCR4). Li et al. (160) discovered that circ-EIF6 encodes a new peptide called EIF6-224 amino acid (aa). EIF6-224aa directly interacts with the oncogene MYH9 in BC and inhibits the ubiquitin-proteasome pathway and subsequently activates the Wnt/β-catenin pathway to reduce the degradation of MYH9, thereby playing a carcinogenic effect. In addition, Wang et al. reported that circACTN4 can competitively bind to far upstream element-binding protein 1 (FUBP1) to prevent FUBP1 from binding to FIR, thereby activating MYC transcription and promoting tumor progression in breast cancer (158).
Some potential circRNAs biomarkers for early diagnosis of BC and prediction of recurrence and metastasis have emerged from the detection of clinical samples in the tissues and peripheral blood of BC patients and healthy controls, combined with the correlation analysis of clinicopathological factors and the analysis of prognosis and survival. For example, CircSMARCA5 can form an R-loop with its parental locus, causing a transcriptional pause at SMARCA5 exon 15, and SMARCA5 DNA is involved in chromatin remodeling in damaged regions.circSMARCA5 may serve as a therapeutic target for patients with drug-resistant BC (209). The above studies have provided new insights into the role of circRNAs in BC.
According to recent research findings, the involvement of circRNAs in hematological malignancies is becoming more widely recognized (210). Among them, aberrant circRNAs expression might upset the balance between self-renewal and differentiation of hematopoietic stem cells (HSCs), resulting in bone marrow failure or hematological malignancies.
Acute myeloid leukemia (AML), the most common leukemia in adults, is uncommon in children, but has a poor prognosis and is prone to relapse (211). Liu et al. discovered that circRNF220 is abundantly and precisely expressed in children’s peripheral blood and bone marrow using microarray technology. CircRNF220 knockout can reduce the proliferation of AML cell lines and primary cells while also promoting cell death (162). FLT3-ITD+ AML is a significant subtype of AML, and Zhang et al. observed that the effect of circ0000370 on the development of FLT3-ITD-positive AML may be directly connected to miR-1299 and S100A7A (164). Acute lymphoblastic leukemia (ALL) is the most prevalent malignant tumor in children, and circ0000094 has been demonstrated to be a molecular sponge of miR-223-3p, which can upregulate the expression of FBW7 by limiting the expression of miR-223-3p, hence preventing ALL progression (165). Zhu et al. found that circADD2, as a tumor suppressor gene in ALL, inhibited cell proliferation and promoted cell apoptosis both in vitro and in vivo. Mechanistically, circADD2, which can sponge miR-149-5p, may serve as a potential biomarker for ALL diagnosis or treatment (166). Interestingly, circRNAs expression profiles can also clearly distinguish Acute leukemia (AL). For example, Guo et al. reported circ0001857 and circ0012152 ALL and AML (212). The recently discovered Circ0009910 can regulate ULK1-induces autophagy by targeting miR-34a-5p and accelerating the resistance of CML cells to imatinib (167). High expression of circ-RPL15 was positively correlated with IGHV mutation status, which is crucial for evaluating CLL prognosis. MiR-146b-3p-mediated RAS/RAF1/MEK/ERK pathway inhibition could be alleviated by circRPL15 overexpression in CLL. CircRPL15 may represent a promising novel plasma biomarker for the diagnosis of CLL (168). Mei et al. found that the relative expression of circADARB1 was significantly increased in the plasma of Natural killer/T-cell lymphoma (NKTCL), which binds to miR-214-3p and regulates p-Stat3, promotes the proliferation of NKTCL cells, and inhibits apoptosis (169). Furthermore, Zhao et al. reported that CircEAF2 inhibited Epstein-Barr infection positive diffuse large B cell expansion and advanced apoptosis via the miR-BART19-3p/APC/-catenin axis (170).
Some of the circRNAs reported above play a regulatory role in the occurrence and development of hematological tumors through various molecular mechanisms, suggesting some potentials of circRNAs in the research of hematological malignancies in the future. These findings also aid in the diagnosis and prediction of hematological malignancies.
The abnormal expression of many circRNAs has been verified in many cancers. In renal cell carcinoma (RCC), Cen et al. found that circSDHC competitively binds to miR-127-3p, preventing it from inhibiting the downstream genes CDKN3 and E2F1 pathways, leading to RCC Malignant progress (172). NONO-TFE3 TRCC (Xp11.2 translocation/NONO-TFE3 fusion renal cell carcinoma) is a subgroup of renal cell carcinoma. Yang et al. found that highly expressed circMET accelerates the decay of CDKN2A mRNA by recruiting YTHDF2, while competitively binding miR- 1197, Regulates SMAD3 expression (177). In bladder cancer (BC), Yang et al. (179) used high-throughput sequencing and RT-qPCR to verify the abnormally high expression of circUBE2K BC tissue. As a ceRNA, the expression of ARHGAP5 was regulated by sponge miR-516b-5p to promote tumor development. The down-regulated circZKSCAN1 in BC tissues and cell lines up-regulates the expression of p21 through sponge miR-1178-3p, which inhibits the proliferation, migration, and invasion of bladder cancer (185). In addition, some circRNAs have also been found to play an important role in prostate cancer and cervical cancer, as shown in Table 3.
In short, various studies have shown that circRNAs are involved in the occurrence and development of various cancers. However, the role of circRNAs in the diagnosis and treatment of cancers needs to be further studied.
There are RNase in human body fluids, and circRNAs can resist this enzyme, thus being a stable biomarker for the detection of body fluids such as blood, exosomes, saliva, and urine (213). In addition, the half-life of circRNAs in the blood is longer than that of mRNA. Coupled with the high abundance and specificity of circRNAs, circRNAs are expected to become an excellent non-invasive biomarker for tumor diagnosis and prognosis. Xu et al. reported that compared with breast cancer and adjacent normal tissues, the expression of circRNAs in peripheral blood was significantly higher than that of host genes (209). This discovery helps to explore diagnostic biomarkers for breast cancer.
CircRNAs may be used as biomarkers for cancer diagnosis and prognosis. It is worth noting that exosomes can protect RNA RNases from degradation, so circRNAs are also enriched and stably expressed in exosomes (214). Exosomes derived from cancer cells can target specific organs to promote the formation of pre-metastasis niches (215) and tumor microenvironment (216). Exosomal circRNAs participate in cell proliferation, invasion, EMT, and metastasis through intercellular communication. Shang et al. (217) discovered a new circRNA in colorectal cancer exosomes, circPACRGL, which acts as a sponge for miR-142-3p/miR-506-3p and promotes the expression of transforming growth factor-β1 (TGF-β1). It has been reported that exosomal circSHKBP1 inhibits HSP90 degradation and promotes GC progress through miR-582-3p/HUR/VEGF pathway (134). Recently, Li et al. used circRNA deep sequencing and bioinformatics methods to build a circRNA repertoire, and 3 up-regulated serum exosomal circRNAs (circ0075828, circ0003828, and circ0002976) could be used to screen for high-grade astrocytoma (HGA). Five highly expressed exosomal circRNAs (circ0005019, circ0000880, circ0051680, and circ0006365) were used as HGA prognostic markers. revealed that circular RNAs in HGA exosomes are targets for HGA liquid biopsy and prognostic monitoring (218). At present, progress has been made in the research of exosomal circRNAs, but the mechanism of circRNAs entering exosomes and the role of circRNAs in exosomes are still unclear.
In summary, the prospects of circRNAs as biomarkers for liquid biopsy and therapeutic targets are promising, but there are few studies at present.
CircRNAs are attractive targets for cancer therapy and offer novel cancer treatment techniques. In this section, we will discuss some future perspectives on the usage of circRNAs in cancer therapy.
As mentioned above, more and more studies have demonstrated that dysregulation of circRNAs in cancer can promote or inhibit cancer (Table 3). While the up-or down-regulation of certain circRNAs is linked to clinical aspects such as TNM and other related phases, differentiation, or survival (142, 219). This shows that circRNAs actions are context-dependent, making it difficult to categorize circRNAs as oncogenic or tumor suppressors.
CircRNAs rely on the sponge action of miRNAs to promote cancer progression. In recent years, small molecule inhibitors (SMIs) and small molecule degradants (SMDs) of miRNAs have been reported for drug therapy, so whether it is possible to develop blockers targeting miRNAs to reduce the cancer-promoting activity of circRNAs (220). Of course, the specificity of the drug requires other biotechnological validation and the safety of the drug also needs to be assessed. In addition, when cancer develops, some critical circRNAs are greatly up-regulated. Can it decrease cancers by reducing the number of cancer-promoting circRNAs without influencing the expression of their parental genes? It may be able to regulate the occurrence of back-splicing events by focusing on the splicing mechanisms that affect circRNAs. For example, Tassinari et al. demonstrated that downregulation of the RBP splicing factor ADAR1, which controls circular RNA biogenesis, is sufficient to strongly inhibit glioblastoma growth in vivo (221). This inspires the prospect of a technique that modulates RBP to suppress circular RNA expression.
Finally, gene editing techniques such as CRISPR/Cas13 has been applied to RNA editing (222), Whether circRNAs can also be edited to reduce or increase activity. Recently, Ishola et al. found that CRISPR/Cas13a-mediated knockdown of circ0000190 reduced the proliferation and migration of non-small cell lung cancer cells in vitro and inhibited tumor growth in vivo (223). This also confirms the potential of the novel CRISPR/Cas13a system as a cancer therapy tool.
CircRNAs have been considered splicing errors before, but they have attracted widespread attention in recent years. A lot of innovative research has emerged in the field of circRNAs, but there are still many challenges and problems that need to be solved. From the above-mentioned large number of retrospective reports, it can be seen that the importance of circRNAs is beyond doubt. However, the function of most circRNAs is still unclear, whether there are new undiscovered functions. In addition, the coding potential of circRNAs is often overlooked. And whether the proteins encoded by circRNAs have the functions of conventional proteins. Thousands of circRNAs have been detected, some of them are highly abundant in cancer and some are low in abundance. The detection method for low-abundance circRNAs is not yet mature, and their use as non-invasive biomarkers requires a large number of clinical sample collections. Moreover, their sensitivity and specificity are controversial. In addition, packaging circRNAs into cells to regulate cell activities also requires a lot of research and exploration, so that these studies can truly produce clinical application value. Standardization is needed in many aspects, such as the extraction of differences between detection technologies and the standardization of naming.
In summary, circRNAs play an important role in cancer and provide new insights for cancer management, but the mechanism of action is still in its infancy. The research of circRNAs still has a long way to go.
ZL, HQ, and WX contributed to conception and design of this review. YZ wrote the first draft of the manuscript. XZ, YX, and SF wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (no. 81602883), Zhenjiang Key Laboratory of High Technology Research on Exosomes Foundation and Transformation Application (Grant SS2018003), The Foundation for Excellent Young Teachers of Jiangsu University, a project of social development in Zhenjiang (No. SH2021045), and Clinical Medical Science and Technology Development Foundation of Jiangsu University (No. JLY2021013).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids Are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-Like Structures. Proc Natl Acad Sci USA (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
2. Hsu MT, Coca-Prados M. Electron Microscopic Evidence for the Circular Form of RNA in the Cytoplasm of Eukaryotic Cells. Nature (1979) 280:339–40. doi: 10.1038/280339a0
3. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The Hepatitis Delta (Delta) Virus Possesses a Circular RNA. Nature (1986) 323:558–60. doi: 10.1038/323558a0
4. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled Exons. Cell (1991) 64:607–13. doi: 10.1016/0092-8674(91)90244-s
5. Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H, et al. Circular Intronic Long Noncoding RNAs. Mol Cell (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
6. Li C, Zhang J, Yang X, Hu C, Chu T, Zhong R, et al. Hsa_Circ_0003222 Accelerates Stemness and Progression of non-Small Cell Lung Cancer by Sponging miR-527. Cell Death Dis (2021) 12:807. doi: 10.1038/s41419-021-04095-8
7. Huang G, Liang M, Liu H, Huang J, Li P, Wang C, et al. CircRNA Hsa_circRNA_104348 Promotes Hepatocellular Carcinoma Progression Through Modulating miR-187-3p/RTKN2 Axis and Activating Wnt/β-Catenin Pathway. Cell Death Dis (2020) 11:1065. doi: 10.1038/s41419-020-03276-1
8. Yang D, Hu Z, Zhang Y, Zhang X, Xu J, Fu H, et al. CircHIPK3 Promotes the Tumorigenesis and Development of Gastric Cancer Through miR-637/AKT1 Pathway. Front Oncol (2021) 11:637761. doi: 10.3389/fonc.2021.637761
9. Yang B-L, Liu G-Q, Li P, Li X-H. Circular RNA CUL2 Regulates the Development of Colorectal Cancer by Modulating Apoptosis and Autophagy via miR-208a-3p/PPP6C. Aging (Albany NY) (2022) 14:497–508. doi: 10.18632/aging.203827
10. Solé C, Lawrie CH. Circular RNAs and Cancer: Opportunities and Challenges. Adv Clin Chem (2020) 99:87–146. doi: 10.1016/bs.acc.2020.02.008
11. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs Are a Large Class of Animal RNAs With Regulatory Potency. Nature (2013) 495:333–8. doi: 10.1038/nature11928
12. Gokool A, Loy CT, Halliday GM, Voineagu I. Circular RNAs: The Brain Transcriptome Comes Full Circle. Trends Neurosci (2020) 43:752–66. doi: 10.1016/j.tins.2020.07.007
13. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs Are Abundant, Conserved, and Associated With ALU Repeats. RNA (2013) 19:141–57. doi: 10.1261/rna.035667.112
14. Glažar P, Papavasileiou P, Rajewsky N. Circbase: A Database for Circular RNAs. RNA (2014) 20:1666–70. doi: 10.1261/rna.043687.113
15. Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell (2015) 58:870–85. doi: 10.1016/j.molcel.2015.03.027
16. Suzuki H, Tsukahara T. A View of pre-mRNA Splicing From RNase R Resistant RNAs. Int J Mol Sci (2014) 15:9331–42. doi: 10.3390/ijms15069331
17. Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L. Complementary Sequence-Mediated Exon Circularization. Cell (2014) 159:134–47. doi: 10.1016/j.cell.2014.09.001
18. Bach D-H, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids (2019) 16:118–29. doi: 10.1016/j.omtn.2019.02.005
19. Schmidt CA, Giusto JD, Bao A, Hopper AK, Matera AG. Molecular Determinants of Metazoan tricRNA Biogenesis. Nucleic Acids Res (2019) 47:6452–65. doi: 10.1093/nar/gkz311
20. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA Biogenesis Competes With pre-mRNA Splicing. Mol Cell (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
21. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell (2015) 160:1125–34. doi: 10.1016/j.cell.2015.02.014
22. Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-Wide Identification of RNA-Binding Protein and microRNA Target Sites by PAR-CLIP. Cell (2010) 141:129–41. doi: 10.1016/j.cell.2010.03.009
23. Zhou M, Li L, Chen B, Pan S, Tu W, Hou Y, et al. Circ-SHPRH Suppresses Cadmium-Induced Transformation of Human Bronchial Epithelial Cells by Regulating QKI Expression via miR-224-5p. Ecotoxicol Environ Saf (2021) 220:112378. doi: 10.1016/j.ecoenv.2021.112378
24. Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, et al. FUS Affects Circular RNA Expression in Murine Embryonic Stem Cell-Derived Motor Neurons. Nat Commun (2017) 8:14741. doi: 10.1038/ncomms14741
25. Cao D. An Autoregulation Loop in Fust-1 for Circular RNA Regulation in Caenorhabditis Elegans. Genetics (2021) 219:iyab145. doi: 10.1093/genetics/iyab145
26. Katrekar D, Yen J, Xiang Y, Saha A, Meluzzi D, Savva Y, et al. Efficient In Vitro and In Vivo RNA Editing via Recruitment of Endogenous ADARs Using Circular Guide RNAs. Nat Biotechnol (2022). doi: 10.1038/s41587-021-01171-4
27. Yi Z, Qu L, Tang H, Liu Z, Liu Y, Tian F, et al. Engineered Circular ADAR-Recruiting RNAs Increase the Efficiency and Fidelity of RNA Editing In Vitro and In Vivo. Nat Biotechnol (2022). doi: 10.1038/s41587-021-01180-3
28. Wan Y, Hopper AK. Size Matters: Conserved Proteins Function in Length-Dependent Nuclear Export of Circular RNAs. Genes Dev (2018) 32:600–1. doi: 10.1101/gad.316216.118
29. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495:384–8. doi: 10.1038/nature11993
30. Chen J, Yang J, Fei X, Wang X, Wang K. CircRNA ciRS-7: A Novel Oncogene in Multiple Cancers. Int J Biol Sci (2021) 17:379–89. doi: 10.7150/ijbs.54292
31. Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, et al. Circular RNA Circsatb2 Promotes Progression of non-Small Cell Lung Cancer Cells. Mol Cancer (2020) 19:101. doi: 10.1186/s12943-020-01221-6
32. Liu J, Liu H, Zeng Q, Xu P, Liu M, Yang N. Circular RNA Circ-MAT2B Facilitates Glycolysis and Growth of Gastric Cancer Through Regulating the miR-515-5p/HIF-1α Axis. Cancer Cell Int (2020) 20:171. doi: 10.1186/s12935-020-01256-1
33. Li D, Zhang J, Yang J, Wang J, Zhang R, Li J, et al. CircMTO1 Suppresses Hepatocellular Carcinoma Progression via the miR-541-5p/ZIC1 Axis by Regulating Wnt/β-Catenin Signaling Pathway and Epithelial-to-Mesenchymal Transition. Cell Death Dis (2021) 13:12. doi: 10.1038/s41419-021-04464-3
34. Lunde BM, Moore C, Varani G. RNA-Binding Proteins: Modular Design for Efficient Function. Nat Rev Mol Cell Biol (2007) 8:479–90. doi: 10.1038/nrm2178
35. Du WW, Yang W, Chen Y, Wu Z-K, Foster FS, Yang Z, et al. Foxo3 Circular RNA Promotes Cardiac Senescence by Modulating Multiple Factors Associated With Stress and Senescence Responses. Eur Heart J (2017) 38:1402–12. doi: 10.1093/eurheartj/ehw001
36. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 Circular RNA Retards Cell Cycle Progression via Forming Ternary Complexes With P21 and CDK2. Nucleic Acids Res (2016) 44:2846–58. doi: 10.1093/nar/gkw027
37. O’Leary VB, Smida J, Matjanovski M, Brockhaus C, Winkler K, Moertl S, et al. The circRNA Interactome-Innovative Hallmarks of the Intra- and Extracellular Radiation Response. Oncotarget (2017) 8:78397–409. doi: 10.18632/oncotarget.19228
38. Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, et al. Circular RNA Circago2 Drives Cancer Progression Through Facilitating HuR-Repressed Functions of AGO2-miRNA Complexes. Cell Death Differ (2019) 26:1346–64. doi: 10.1038/s41418-018-0220-6
39. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR Target Circular RNAs Uncovers Suppression of PABPN1 Translation by Circpabpn1. RNA Biol (2017) 14:361–9. doi: 10.1080/15476286.2017.1279788
40. Qin M, Wei G, Sun X. Circ-UBR5: An Exonic Circular RNA and Novel Small Nuclear RNA Involved in RNA Splicing. Biochem Biophys Res Commun (2018) 503:1027–34. doi: 10.1016/j.bbrc.2018.06.112
41. Barbagallo D, Caponnetto A, Brex D, Mirabella F, Barbagallo C, Lauretta G, et al. CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1. Cancers (Basel) (2019) 11:E194. doi: 10.3390/cancers11020194
42. Hu Q, Zhou T. EIciRNA-Mediated Gene Expression: Tunability and Bimodality. FEBS Lett (2018) 592:3460–71. doi: 10.1002/1873-3468.13253
43. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat Struct Mol Biol (2015) 22:256–64. doi: 10.1038/nsmb.2959
44. Li X, Zhang J-L, Lei Y-N, Liu X-Q, Xue W, Zhang Y, et al. Linking Circular Intronic RNA Degradation and Function in Transcription by RNase H1. Sci China Life Sci (2021) 64:1795–809. doi: 10.1007/s11427-021-1993-6
45. Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S, et al. A Novel Protein Encoded by Circmapk1 Inhibits Progression of Gastric Cancer by Suppressing Activation of MAPK Signaling. Mol Cancer (2021) 20:66. doi: 10.1186/s12943-021-01358-y
46. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021
47. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive Translation of Circular RNAs Driven by N6-Methyladenosine. Cell Res (2017) 27:626–41. doi: 10.1038/cr.2017.31
48. Liu M, Wang Q, Shen J, Yang BB, Ding X. Circbank: A Comprehensive Database for circRNA With Standard Nomenclature. RNA Biol (2019) 16:899–905. doi: 10.1080/15476286.2019.1600395
49. Wu W, Ji P, Zhao F. CircAtlas: An Integrated Resource of One Million Highly Accurate Circular RNAs From 1070 Vertebrate Transcriptomes. Genome Biol (2020) 21:101. doi: 10.1186/s13059-020-02018-y
50. Tabak HF, van der Horst G, Smit J, Winter AJ, Mul Y, Groot Koerkamp MJ. Discrimination Between RNA Circles, Interlocked RNA Circles and Lariats Using Two-Dimensional Polyacrylamide Gel Electrophoresis. Nucleic Acids Res (1988) 16:6597–605. doi: 10.1093/nar/16.14.6597
51. Li S, Teng S, Xu J, Su G, Zhang Y, Zhao J, et al. Microarray Is an Efficient Tool for circRNA Profiling. Brief Bioinform (2019) 20:1420–33. doi: 10.1093/bib/bby006
52. Zirkel A, Papantonis A. Detecting Circular RNAs by RNA Fluorescence In Situ Hybridization. Methods Mol Biol (2018) 1724:69–75. doi: 10.1007/978-1-4939-7562-4_6
53. Oudejans C, Manders V, Visser A, Keijser R, Min N, Poutsma A, et al. Circular RNA Sequencing of Maternal Platelets: A Novel Tool for the Identification of Pregnancy-Specific Biomarkers. Clin Chem (2021) 67:508–17. doi: 10.1093/clinchem/hvaa249
54. Vromman M, Yigit N, Verniers K, Lefever S, Vandesompele J, Volders P-J. Validation of Circular RNAs Using RT-qPCR After Effective Removal of Linear RNAs by Ribonuclease R. Curr Protoc (2021) 1:e181. doi: 10.1002/cpz1.181
55. Chen D-F, Zhang L-J, Tan K, Jing Q. Application of Droplet Digital PCR in Quantitative Detection of the Cell-Free Circulating circRNAs. Biotechnol Biotechnol Equip (2018) 32:116–23. doi: 10.1080/13102818.2017.1398596
56. Zhang P, Guo N, Gao K, Su F, Wang F, Li Z. Direct Recognition and Sensitive Detection of Circular RNA With Ligation-Based PCR. Org Biomol Chem (2020) 18:3269–73. doi: 10.1039/d0ob00625d
57. Liu Y, Zhang X, Liu M, Xu F, Zhang Q, Zhang Y, et al. Direct Detection of circRNA in Real Samples Using Reverse Transcription-Rolling Circle Amplification. Anal Chim Acta (2020) 1101:169–75. doi: 10.1016/j.aca.2019.12.027
58. Zhang P, Gao K, Liang Y, Su F, Wang F, Li Z. Ultrasensitive Detection of Circular RNA by Accurate Recognition of the Specific Junction Site Using Stem-Loop Primer Induced Double Exponential Amplification. Talanta (2020) 217:121021. doi: 10.1016/j.talanta.2020.121021
59. Liu Z, Tao C, Li S, Du M, Bai Y, Hu X, et al. circFL-Seq Reveals Full-Length Circular RNAs With Rolling Circular Reverse Transcription and Nanopore Sequencing. eLife (2021) 10:e69457. doi: 10.7554/eLife.69457
60. Zhang J, Hou L, Zuo Z, Ji P, Zhang X, Xue Y, et al. Comprehensive Profiling of Circular RNAs With Nanopore Sequencing and CIRI-Long. Nat Biotechnol (2021) 39:836–45. doi: 10.1038/s41587-021-00842-6
61. Xin R, Gao Y, Gao Y, Wang R, Kadash-Edmondson KE, Liu B, et al. Isocirc Catalogs Full-Length Circular RNA Isoforms in Human Transcriptomes. Nat Commun (2021) 12:266. doi: 10.1038/s41467-020-20459-8
62. Jiao J, Li C, Ning L, Shi L, Wang L, Xiang Y, et al. Electrochemical Detection of circRNAs Based on the Combination of Back-Splice Junction and Duplex-Specific Nuclease. Sensors Actuators B: Chem (2020) 302:127166. doi: 10.1016/j.snb.2019.127166
63. Mi Z, Zhongqiang C, Caiyun J, Yanan L, Jianhua W, Liang L. Circular RNA Detection Methods: A Minireview. Talanta (2022) 238:123066. doi: 10.1016/j.talanta.2021.123066
64. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell (2019) 176:869–881.e13. doi: 10.1016/j.cell.2018.12.021
65. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A Web Tool for Exploring Circular RNAs and Their Interacting Proteins and microRNAs. RNA Biol (2016) 13:34–42. doi: 10.1080/15476286.2015.1128065
66. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: A Comprehensive Database for Human Circular RNAs With Protein-Coding Annotations. Sci Rep (2016) 6:34985. doi: 10.1038/srep34985
67. Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. Starbase V2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and Protein-RNA Interaction Networks From Large-Scale CLIP-Seq Data. Nucleic Acids Res (2014) 42:D92–97. doi: 10.1093/nar/gkt1248
68. Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: A Comprehensive Database for Circular RNA Potentially Associated With Disease and Traits. Front Genet (2013) 4:283. doi: 10.3389/fgene.2013.00283
69. Zhao Z, Wang K, Wu F, Wang W, Zhang K, Hu H, et al. circRNA Disease: A Manually Curated Database of Experimentally Supported circRNA-Disease Associations. Cell Death Dis (2018) 9:475. doi: 10.1038/s41419-018-0503-3
70. Wu S-M, Liu H, Huang P-J, Chang IY-F, Lee C-C, Yang C-Y, et al. Circlncrnanet: An Integrated Web-Based Resource for Mapping Functional Networks of Long or Circular Forms of Noncoding RNAs. Gigascience (2018) 7:1–10. doi: 10.1093/gigascience/gix118
71. Feng J, Chen W, Dong X, Wang J, Mei X, Deng J, et al. CSCD2: An Integrated Interactional Database of Cancer-Specific Circular RNAs. Nucleic Acids Res (2022) 50:D1179–83. doi: 10.1093/nar/gkab830
72. Xie F, Liu S, Wang J, Xuan J, Zhang X, Qu L, et al. Deepbase V3.0: Expression Atlas and Interactive Analysis of ncRNAs From Thousands of Deep-Sequencing Data. Nucleic Acids Res (2021) 49:D877–83. doi: 10.1093/nar/gkaa1039
73. Dong R, Ma X-K, Li G-W, Yang L. CIRCpedia V2: An Updated Database for Comprehensive Circular RNA Annotation and Expression Comparison. Genomics Proteomics Bioinf (2018) 16:226–33. doi: 10.1016/j.gpb.2018.08.001
74. Tang Z, Li X, Zhao J, Qian F, Feng C, Li Y, et al. TRCirc: A Resource for Transcriptional Regulation Information of circRNAs. Brief Bioinform (2019) 20:2327–33. doi: 10.1093/bib/bby083
75. Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, et al. Exorbase: A Database of circRNA, lncRNA and mRNA in Human Blood Exosomes. Nucleic Acids Res (2018) 46:D106–12. doi: 10.1093/nar/gkx891
76. Chen Y, Yao L, Tang Y, Jhong J-H, Wan J, Chang J, et al. CircNet 2.0: An Updated Database for Exploring Circular RNA Regulatory Networks in Cancers. Nucleic Acids Res (2022) 50:D93–D101. doi: 10.1093/nar/gkab1036
77. Fan C, Lei X, Fang Z, Jiang Q, Wu F-X. CircR2Disease: A Manually Curated Database for Experimentally Supported Circular RNAs Associated With Various Diseases. Database (Oxford) (2018) 2018:bay044. doi: 10.1093/database/bay044
78. Gaffo E, Buratin A, Dal Molin A, Bortoluzzi S. Sensitive, Reliable and Robust circRNA Detection From RNA-Seq With Circompara2. Brief Bioinform (2022) 23:bbab418. doi: 10.1093/bib/bbab418
79. Zhang W, Liu Y, Min Z, Liang G, Mo J, Ju Z, et al. Circmine: A Comprehensive Database to Integrate, Analyze and Visualize Human Disease-Related circRNA Transcriptome. Nucleic Acids Res (2022) 50:D83–92. doi: 10.1093/nar/gkab809
80. Lee BD, Neri U, Oh CJ, Simmonds P, Koonin EV. ViroidDB: A Database of Viroids and Viroid-Like Circular RNAs. Nucleic Acids Res (2022) 50:D432–8. doi: 10.1093/nar/gkab974
81. Dal Molin A, Gaffo E, Difilippo V, Buratin A, Tretti Parenzan C, Bresolin S, et al. CRAFT: A Bioinformatics Software for Custom Prediction of Circular RNA Functions. Brief Bioinform (2022) 23(2):bbab601. doi: 10.1093/bib/bbab601
82. Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, et al. Roles of circRNAs in the Tumour Microenvironment. Mol Cancer (2020) 19:14. doi: 10.1186/s12943-019-1125-9
83. Lei B, Tian Z, Fan W, Ni B. Circular RNA: A Novel Biomarker and Therapeutic Target for Human Cancers. Int J Med Sci (2019) 16:292–301. doi: 10.7150/ijms.28047
84. Wang Y, Yin L, Sun X. CircRNA Hsa_Circ_0002577 Accelerates Endometrial Cancer Progression Through Activating IGF1R/PI3K/Akt Pathway. J Exp Clin Cancer Res (2020) 39:169. doi: 10.1186/s13046-020-01679-8
85. Gao D, Qi X, Zhang X, Fang K, Guo Z, Li L. Hsa_circRNA_0006528 as a Competing Endogenous RNA Promotes Human Breast Cancer Progression by Sponging miR-7-5p and Activating the MAPK/ERK Signaling Pathway. Mol Carcinog (2019) 58:554–64. doi: 10.1002/mc.22950
86. Zhou X, Li J, Zhou Y, Yang Z, Yang H, Li D, et al. Down-Regulated ciRS-7/Up-Regulated miR-7 Axis Aggravated Cartilage Degradation and Autophagy Defection by PI3K/AKT/mTOR Activation Mediated by IL-17A in Osteoarthritis. Aging (Albany NY) (2020) 12:20163–83. doi: 10.18632/aging.103731
87. Cai W, Zhang Y, Su Z. ciRS-7 Targeting miR-135a-5p Promotes Neuropathic Pain in CCI Rats via Inflammation and Autophagy. Gene (2020) 736:144386. doi: 10.1016/j.gene.2020.144386
88. Zhang Z, Ren L, Zhao Q, Lu G, Ren M, Lu X, et al. TRPC1 Exacerbate Metastasis in Gastric Cancer via ciRS-7/miR-135a-5p/TRPC1 Axis. Biochem Biophys Res Commun (2020) 529:85–90. doi: 10.1016/j.bbrc.2020.05.181
89. Yin Y-L, Liu T-L, Yao Q, Wang Y-X, Wu X-M, Wang X-T, et al. Circular RNA ciRS-7 Affects the Propagation of Cryptosporidium Parvum in HCT-8 Cells by Sponging miR-1270 to Activate the NF-κb Signaling Pathway. Parasit Vectors (2021) 14:238. doi: 10.1186/s13071-021-04739-w
90. Cui M, Shen W, Qin W, Wang X, Li Y, Xu F, et al. Circular RNA ciRS-7 Promotes Tube Formation in Microvascular Endothelial Cells Through Downregulation of miR-26a-5p. J Biochem Mol Toxicol (2020) 34:e22468. doi: 10.1002/jbt.22468
91. Yu Z, Zhu X, Li Y, Liang M, Liu M, Liu Z, et al. Circ-HMGA2 (Hsa_Circ_0027446) Promotes the Metastasis and Epithelial-Mesenchymal Transition of Lung Adenocarcinoma Cells Through the miR-1236-3p/ZEB1 Axis. Cell Death Dis (2021) 12:313. doi: 10.1038/s41419-021-03601-2
92. Shi J, Lv X, Zeng L, Li W, Zhong Y, Yuan J, et al. CircPVT1 Promotes Proliferation of Lung Squamous Cell Carcinoma by Binding to miR-30d/E. J Exp Clin Cancer Res (2021) 40:193. doi: 10.1186/s13046-021-01976-w
93. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA Circ-CPA4/let-7 miRNA/PD-L1 Axis Regulates Cell Growth, Stemness, Drug Resistance and Immune Evasion in Non-Small Cell Lung Cancer (NSCLC). J Exp Clin Cancer Res (2020) 39:149. doi: 10.1186/s13046-020-01648-1
94. Xu Y, Yu J, Huang Z, Fu B, Tao Y, Qi X, et al. Circular RNA Hsa_Circ_0000326 Acts as a miR-338-3p Sponge to Facilitate Lung Adenocarcinoma Progression. J Exp Clin Cancer Res (2020) 39:57. doi: 10.1186/s13046-020-01556-4
95. Yu C, Cheng Z, Cui S, Mao X, Li B, Fu Y, et al. Circfoxm1 Promotes Proliferation of non-Small Cell Lung Carcinoma Cells by Acting as a ceRNA to Upregulate FAM83D. J Exp Clin Cancer Res (2020) 39:55. doi: 10.1186/s13046-020-01555-5
96. Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, et al. Circular RNA 100146 Functions as an Oncogene Through Direct Binding to miR-361-3p and miR-615-5p in Non-Small Cell Lung Cancer. Mol Cancer (2019) 18:13. doi: 10.1186/s12943-019-0943-0
97. Ishola AA, Chien C-S, Yang Y-P, Chien Y, Yarmishyn AA, Tsai P-H, et al. Oncogenic circRNA C190 Promotes Non-Small Cell Lung Cancer via Modulation of the EGFR/ERK Pathway. Cancer Res (2022) 82:75–89. doi: 10.1158/0008-5472.CAN-21-1473
98. Chi Y, Zheng W, Bao G, Wu L, He X, Gan R, et al. Circular RNA Circ_103820 Suppresses Lung Cancer Tumorigenesis by Sponging miR-200b-3p to Release LATS2 and SOCS6. Cell Death Dis (2021) 12:185. doi: 10.1038/s41419-021-03472-7
99. Yao Y, Zhou Y, Hua Q. circRNA Hsa_Circ_0018414 Inhibits the Progression of LUAD by Sponging miR-6807-3p and Upregulating DKK1. Mol Ther Nucleic Acids (2021) 23:783–96. doi: 10.1016/j.omtn.2020.12.031
100. Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, et al. Circular RNA Circhipk3 Modulates Autophagy via MIR124-3p-STAT3-PRKAA/Ampkα Signaling in STK11 Mutant Lung Cancer. Autophagy (2020) 16:659–71. doi: 10.1080/15548627.2019.1634945
101. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. Circndufb2 Inhibits non-Small Cell Lung Cancer Progression via Destabilizing IGF2BPs and Activating Anti-Tumor Immunity. Nat Commun (2021) 12:295. doi: 10.1038/s41467-020-20527-z
102. Liang Y, Wang H, Chen B, Mao Q, Xia W, Zhang T, et al. Circdcun1d4 Suppresses Tumor Metastasis and Glycolysis in Lung Adenocarcinoma by Stabilizing TXNIP Expression. Mol Ther Nucleic Acids (2021) 23:355–68. doi: 10.1016/j.omtn.2020.11.012
103. Huang Q, Guo H, Wang S, Ma Y, Chen H, Li H, et al. A Novel Circular RNA, Circxpo1, Promotes Lung Adenocarcinoma Progression by Interacting With IGF2BP1. Cell Death Dis (2020) 11:1031. doi: 10.1038/s41419-020-03237-8
104. Lv X, Huang H, Feng H, Wei Z. Circ-MMP2 (Circ-0039411) Induced by FOXM1 Promotes the Proliferation and Migration of Lung Adenocarcinoma Cells In Vitro and In Vivo. Cell Death Dis (2020) 11:426. doi: 10.1038/s41419-020-2628-4
105. Wang J, Zhou L, Chen B, Yu Z, Zhang J, Zhang Z, et al. Circular RNA Circcspp1 Promotes the Occurrence and Development of Colon Cancer by Sponging miR-431 and Regulating ROCK1 and ZEB1. J Transl Med (2022) 20:58. doi: 10.1186/s12967-022-03240-x
106. Chen C, Huang Z, Mo X, Song Y, Li X, Li X, et al. The Circular RNA 001971/miR-29c-3p Axis Modulates Colorectal Cancer Growth, Metastasis, and Angiogenesis Through VEGFA. J Exp Clin Cancer Res (2020) 39:91. doi: 10.1186/s13046-020-01594-y
107. Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, et al. Circ3823 Contributes to Growth, Metastasis and Angiogenesis of Colorectal Cancer: Involvement of miR-30c-5p/TCF7 Axis. Mol Cancer (2021) 20:93. doi: 10.1186/s12943-021-01372-0
108. Wang J, Zhang Y, Song H, Yin H, Jiang T, Xu Y, et al. The Circular RNA circSPARC Enhances the Migration and Proliferation of Colorectal Cancer by Regulating the JAK/STAT Pathway. Mol Cancer (2021) 20:81. doi: 10.1186/s12943-021-01375-x
109. Zhou C, Liu H-S, Wang F-W, Hu T, Liang Z-X, Lan N, et al. Circcamsap1 Promotes Tumor Growth in Colorectal Cancer via the miR-328-5p/E2F1 Axis. Mol Ther (2020) 28:914–28. doi: 10.1016/j.ymthe.2019.12.008
110. Jian X, He H, Zhu J, Zhang Q, Zheng Z, Liang X, et al. Hsa_circ_001680 Affects the Proliferation and Migration of CRC and Mediates its Chemoresistance by Regulating BMI1 Through miR-340. Mol Cancer (2020) 19:20. doi: 10.1186/s12943-020-1134-8
111. Zheng L, Liang H, Zhang Q, Shen Z, Sun Y, Zhao X, et al. Circpten1, a Circular RNA Generated From PTEN, Suppresses Cancer Progression Through Inhibition of TGF-β/Smad Signaling. Mol Cancer (2022) 21:41. doi: 10.1186/s12943-022-01495-y
112. Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, et al. CircPTK2 (Hsa_Circ_0005273) as a Novel Therapeutic Target for Metastatic Colorectal Cancer. Mol Cancer (2020) 19:13. doi: 10.1186/s12943-020-1139-3
113. Liu X, Liu Y, Liu Z, Lin C, Meng F, Xu L, et al. CircMYH9 Drives Colorectal Cancer Growth by Regulating Serine Metabolism and Redox Homeostasis in a P53-Dependent Manner. Mol Cancer (2021) 20:114. doi: 10.1186/s12943-021-01412-9
114. Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A Novel Protein Encoded by a Circular RNA Circppp1r12a Promotes Tumor Pathogenesis and Metastasis of Colon Cancer via Hippo-YAP Signaling. Mol Cancer (2019) 18:47. doi: 10.1186/s12943-019-1010-6
115. Chen J, Wu Y, Luo X, Jin D, Zhou W, Ju Z, et al. Circular RNA Circrhobtb3 Represses Metastasis by Regulating the HuR-Mediated mRNA Stability of PTBP1 in Colorectal Cancer. Theranostics (2021) 11:7507–26. doi: 10.7150/thno.59546
116. Zhang C, Zhou X, Geng X, Zhang Y, Wang J, Wang Y, et al. Circular RNA Hsa_Circ_0006401 Promotes Proliferation and Metastasis in Colorectal Carcinoma. Cell Death Dis (2021) 12:443. doi: 10.1038/s41419-021-03714-8
117. Liang Z-X, Liu H-S, Xiong L, Yang X, Wang F-W, Zeng Z-W, et al. A Novel NF-κb Regulator Encoded by Circplce1 Inhibits Colorectal Carcinoma Progression by Promoting RPS3 Ubiquitin-Dependent Degradation. Mol Cancer (2021) 20:103. doi: 10.1186/s12943-021-01404-9
118. Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang J, et al. A Novel Protein Encoded by Circfndc3b Inhibits Tumor Progression and EMT Through Regulating Snail in Colon Cancer. Mol Cancer (2020) 19:71. doi: 10.1186/s12943-020-01179-5
119. Han K, Wang F-W, Cao C-H, Ling H, Chen J-W, Chen R-X, et al. CircLONP2 Enhances Colorectal Carcinoma Invasion and Metastasis Through Modulating the Maturation and Exosomal Dissemination of microRNA-17. Mol Cancer (2020) 19:60. doi: 10.1186/s12943-020-01184-8
120. Hu Z-Q, Zhou S-L, Li J, Zhou Z-J, Wang P-C, Xin H-Y, et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology (2020) 72:906–22. doi: 10.1002/hep.31068
121. Huang X-Y, Zhang P-F, Wei C-Y, Peng R, Lu J-C, Gao C, et al. Circular RNA circMET Drives Immunosuppression and Anti-PD1 Therapy Resistance in Hepatocellular Carcinoma via the miR-30-5p/Snail/DPP4 Axis. Mol Cancer (2020) 19:92. doi: 10.1186/s12943-020-01213-6
122. Zhao Z, Song J, Tang B, Fang S, Zhang D, Zheng L, et al. CircSOD2 Induced Epigenetic Alteration Drives Hepatocellular Carcinoma Progression Through Activating JAK2/STAT3 Signaling Pathway. J Exp Clin Cancer Res (2020) 39:259. doi: 10.1186/s13046-020-01769-7
123. Wu D, Xia A, Fan T, Li G. Circrasgrf2 Functions as an Oncogenic Gene in Hepatocellular Carcinoma by Acting as a miR-1224 Sponge. Mol Ther Nucleic Acids (2021) 23:13–26. doi: 10.1016/j.omtn.2020.10.035
124. Song L-N, Qiao G-L, Yu J, Yang C-M, Chen Y, Deng Z-F, et al. Hsa_circ_0003998 Promotes Epithelial to Mesenchymal Transition of Hepatocellular Carcinoma by Sponging miR-143-3p and PCBP1. J Exp Clin Cancer Res (2020) 39:114. doi: 10.1186/s13046-020-01576-0
125. Dong Z-R, Ke A-W, Li T, Cai J-B, Yang Y-F, Zhou W, et al. CircMEMO1 Modulates the Promoter Methylation and Expression of TCF21 to Regulate Hepatocellular Carcinoma Progression and Sorafenib Treatment Sensitivity. Mol Cancer (2021) 20:75. doi: 10.1186/s12943-021-01361-3
126. Xu L, Feng X, Hao X, Wang P, Zhang Y, Zheng X, et al. CircSETD3 (Hsa_circ_0000567) Acts as a Sponge for microRNA-421 Inhibiting Hepatocellular Carcinoma Growth. J Exp Clin Cancer Res (2019) 38:98. doi: 10.1186/s13046-019-1041-2
127. Cao P, Ma B, Sun D, Zhang W, Qiu J, Qin L, et al. Hsa_Circ_0003410 Promotes Hepatocellular Carcinoma Progression by Increasing the Ratio of M2/M1 Macrophages Through the miR-139-3p/CCL5 Axis. Cancer Sci (2022) 113:634–47. doi: 10.1111/cas.15238
128. Li P, Song R, Yin F, Liu M, Liu H, Ma S, et al. Circmrps35 Promotes Malignant Progression and Cisplatin Resistance in Hepatocellular Carcinoma. Mol Ther (2022) 30(1):431–47. doi: 10.1016/j.ymthe.2021.08.027
129. Sun S, Gao J, Zhou S, Li Y, Wang Y, Jin L, et al. A Novel Circular RNA Circ-LRIG3 Facilitates the Malignant Progression of Hepatocellular Carcinoma by Modulating the EZH2/STAT3 Signaling. J Exp Clin Cancer Res (2020) 39:252. doi: 10.1186/s13046-020-01779-5
130. Liu H, Lan T, Li H, Xu L, Chen X, Liao H, et al. Circular RNA Circdlc1 Inhibits MMP1-Mediated Liver Cancer Progression via Interaction With HuR. Theranostics (2021) 11:1396–411. doi: 10.7150/thno.53227
131. Cao J, Zhang X, Xu P, Wang H, Wang S, Zhang L, et al. Circular RNA Circlmo7 Acts as a microRNA-30a-3p Sponge to Promote Gastric Cancer Progression via the WNT2/β-Catenin Pathway. J Exp Clin Cancer Res (2021) 40:6. doi: 10.1186/s13046-020-01791-9
132. Xia Y, Lv J, Jiang T, Li B, Li Y, He Z, et al. CircFAM73A Promotes the Cancer Stem Cell-Like Properties of Gastric Cancer Through the miR-490-3p/HMGA2 Positive Feedback Loop and HNRNPK-Mediated β-Catenin Stabilization. J Exp Clin Cancer Res (2021) 40:103. doi: 10.1186/s13046-021-01896-9
133. Liang M, Yao W, Shi B, Zhu X, Cai R, Yu Z, et al. Circular RNA Hsa_Circ_0110389 Promotes Gastric Cancer Progression Through Upregulating SORT1 via Sponging miR-127-5p and miR-136-5p. Cell Death Dis (2021) 12:639. doi: 10.1038/s41419-021-03903-5
134. Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, et al. Exosomal Circshkbp1 Promotes Gastric Cancer Progression via Regulating the miR-582-3p/HUR/VEGF Axis and Suppressing HSP90 Degradation. Mol Cancer (2020) 19:112. doi: 10.1186/s12943-020-01208-3
135. Lu J, Wang Y-H, Yoon C, Huang X-Y, Xu Y, Xie J-W, et al. Circular RNA Circ-RanGAP1 Regulates VEGFA Expression by Targeting miR-877-3p to Facilitate Gastric Cancer Invasion and Metastasis. Cancer Lett (2020) 471:38–48. doi: 10.1016/j.canlet.2019.11.038
136. Zhang Z, Wang C, Zhang Y, Yu S, Zhao G, Xu J. CircDUSP16 Promotes the Tumorigenesis and Invasion of Gastric Cancer by Sponging miR-145-5p. Gastric Cancer (2020) 23:437–48. doi: 10.1007/s10120-019-01018-7
137. Niu Q, Dong Z, Liang M, Luo Y, Lin H, Lin M, et al. Circular RNA Hsa_Circ_0001829 Promotes Gastric Cancer Progression Through miR-155-5p/SMAD2 Axis. J Exp Clin Cancer Res (2020) 39:280. doi: 10.1186/s13046-020-01790-w
138. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA Circnrip1 Acts as a microRNA-149-5p Sponge to Promote Gastric Cancer Progression via the AKT1/mTOR Pathway. Mol Cancer (2019) 18:20. doi: 10.1186/s12943-018-0935-5
139. Sang H, Zhang W, Peng L, Wei S, Zhu X, Huang K, et al. Exosomal Circrell1 Serves as a miR-637 Sponge to Modulate Gastric Cancer Progression via Regulating Autophagy Activation. Cell Death Dis (2022) 13:56. doi: 10.1038/s41419-021-04364-6
140. Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, et al. Circcul2 Regulates Gastric Cancer Malignant Transformation and Cisplatin Resistance by Modulating Autophagy Activation via miR-142-3p/ROCK2. Mol Cancer (2020) 19:156. doi: 10.1186/s12943-020-01270-x
141. Luo Z, Rong Z, Zhang J, Zhu Z, Yu Z, Li T, et al. Circular RNA Circccdc9 Acts as a miR-6792-3p Sponge to Suppress the Progression of Gastric Cancer Through Regulating CAV1 Expression. Mol Cancer (2020) 19:86. doi: 10.1186/s12943-020-01203-8
142. Sun G, Li Z, He Z, Wang W, Wang S, Zhang X, et al. Circular RNA MCTP2 Inhibits Cisplatin Resistance in Gastric Cancer by miR-99a-5p-Mediated Induction of MTMR3 Expression. J Exp Clin Cancer Res (2020) 39:246. doi: 10.1186/s13046-020-01758-w
143. Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang W, et al. CircPSMC3 Suppresses the Proliferation and Metastasis of Gastric Cancer by Acting as a Competitive Endogenous RNA Through Sponging miR-296-5p. Mol Cancer (2019) 18:25. doi: 10.1186/s12943-019-0958-6
144. Liu Y, Xu J, Jiang M, Ni L, Ling Y. CircRNA DONSON Contributes to Cisplatin Resistance in Gastric Cancer Cells by Regulating miR-802/BMI1 Axis. Cancer Cell Int (2020) 20:261. doi: 10.1186/s12935-020-01358-w
145. Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu X, et al. Circular RNA Circ-DONSON Facilitates Gastric Cancer Growth and Invasion via NURF Complex Dependent Activation of Transcription Factor SOX4. Mol Cancer (2019) 18:45. doi: 10.1186/s12943-019-1006-2
146. Jie M, Wu Y, Gao M, Li X, Liu C, Ouyang Q, et al. CircMRPS35 Suppresses Gastric Cancer Progression via Recruiting KAT7 to Govern Histone Modification. Mol Cancer (2020) 19:56. doi: 10.1186/s12943-020-01160-2
147. Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y, et al. Circ-HuR Suppresses HuR Expression and Gastric Cancer Progression by Inhibiting CNBP Transactivation. Mol Cancer (2019) 18:158. doi: 10.1186/s12943-019-1094-z
148. Wang X, Li J, Bian X, Wu C, Hua J, Chang S, et al. CircURI1 Interacts With hnRNPM to Inhibit Metastasis by Modulating Alternative Splicing in Gastric Cancer. Proc Natl Acad Sci USA (2021) 118:e2012881118. doi: 10.1073/pnas.2012881118
149. Zhang Y, Jiang J, Zhang J, Shen H, Wang M, Guo Z, et al. CircDIDO1 Inhibits Gastric Cancer Progression by Encoding a Novel DIDO1-529aa Protein and Regulating PRDX2 Protein Stability. Mol Cancer (2021) 20:101. doi: 10.1186/s12943-021-01390-y
150. Wang Z, Yang L, Wu P, Li X, Tang Y, Ou X, et al. The Circrobo1/KLF5/FUS Feedback Loop Regulates the Liver Metastasis of Breast Cancer by Inhibiting the Selective Autophagy of Afadin. Mol Cancer (2022) 21:29. doi: 10.1186/s12943-022-01498-9
151. Wang X, Xue B, Zhang Y, Guo G, Duan X, Dou D. Up-Regulated Circbach2 Contributes to Cell Proliferation, Invasion, and Migration of Triple-Negative Breast Cancer. Cell Death Dis (2021) 12:412. doi: 10.1038/s41419-021-03684-x
152. Wang X, Ji C, Hu J, Deng X, Zheng W, Yu Y, et al. Hsa_circ_0005273 Facilitates Breast Cancer Tumorigenesis by Regulating YAP1-Hippo Signaling Pathway. J Exp Clin Cancer Res (2021) 40:29. doi: 10.1186/s13046-021-01830-z
153. Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, et al. Autophagy-Associated circRNA circCDYL Augments Autophagy and Promotes Breast Cancer Progression. Mol Cancer (2020) 19:65. doi: 10.1186/s12943-020-01152-2
154. Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R, et al. The circRNA Circsept9 Mediated by E2F1 and EIF4A3 Facilitates the Carcinogenesis and Development of Triple-Negative Breast Cancer. Mol Cancer (2020) 19:73. doi: 10.1186/s12943-020-01183-9
155. Guo X-Y, Liu T-T, Zhu W-J, Liu H-T, Zhang G-H, Song L, et al. CircKDM4B Suppresses Breast Cancer Progression via the miR-675/NEDD4L Axis. Oncogene (2022). doi: 10.1038/s41388-022-02232-x
156. Fan Y, Wang J, Jin W, Sun Y, Xu Y, Wang Y, et al. CircNR3C2 Promotes HRD1-Mediated Tumor-Suppressive Effect via Sponging miR-513a-3p in Triple-Negative Breast Cancer. Mol Cancer (2021) 20(1):25. doi: 10.1186/s12943-021-01321-x
157. Wang F, Wang X, Li J, Lv P, Han M, Li L, et al. CircNOL10 Suppresses Breast Cancer Progression by Sponging miR-767-5p to Regulate SOCS2/JAK/STAT Signaling. J BioMed Sci (2021) 28:4. doi: 10.1186/s12929-020-00697-0
158. Wang X, Xing L, Yang R, Chen H, Wang M, Jiang R, et al. The Circactn4 Interacts With FUBP1 to Promote Tumorigenesis and Progression of Breast Cancer by Regulating the Expression of Proto-Oncogene MYC. Mol Cancer (2021) 20:91. doi: 10.1186/s12943-021-01383-x
159. Du WW, Yang W, Li X, Fang L, Wu N, Li F, et al. The Circular RNA Circska3 Binds Integrin β1 to Induce Invadopodium Formation Enhancing Breast Cancer Invasion. Mol Ther (2020) 28:1287–98. doi: 10.1016/j.ymthe.2020.03.002
160. Li Y, Wang Z, Su P, Liang Y, Li Z, Zhang H, et al. Circ-EIF6 Encodes EIF6-224aa to Promote TNBC Progression via Stabilizing MYH9 and Activating the Wnt/beta-Catenin Pathway. Mol Ther (2022) 30:415–30. doi: 10.1016/j.ymthe.2021.08.026
161. Li J, Ma M, Yang X, Zhang M, Luo J, Zhou H, et al. Circular HER2 RNA Positive Triple Negative Breast Cancer Is Sensitive to Pertuzumab. Mol Cancer (2020) 19:142. doi: 10.1186/s12943-020-01259-6
162. Liu X, Liu X, Cai M, Luo A, He Y, Liu S, et al. CircRNF220, Not its Linear Cognate Gene RNF220, Regulates Cell Growth and Is Associated With Relapse in Pediatric Acute Myeloid Leukemia. Mol Cancer (2021) 20:139. doi: 10.1186/s12943-021-01395-7
163. Wang X, Jin P, Zhang Y, Wang K. CircSPI1 Acts as an Oncogene in Acute Myeloid Leukemia Through Antagonizing SPI1 and Interacting With microRNAs. Cell Death Dis (2021) 12:297. doi: 10.1038/s41419-021-03566-2
164. Zhang L, Bu Z, Shen J, Shang L, Chen Y, Wang Y. A Novel Circular RNA (Hsa_Circ_0000370) Increases Cell Viability and Inhibits Apoptosis of FLT3-ITD-Positive Acute Myeloid Leukemia Cells by Regulating miR-1299 and S100A7A. BioMed Pharmacother (2020) 122:109619. doi: 10.1016/j.biopha.2019.109619
165. Hou Y, Sun J, Huang J, Yao F, Chen X, Zhu B, et al. Circular RNA circRNA_0000094 Sponges microRNA-223-3p and Up-Regulate F-Box and WD Repeat Domain Containing 7 to Restrain T Cell Acute Lymphoblastic Leukemia Progression. Hum Cell (2021) 34:977–89. doi: 10.1007/s13577-021-00504-4
166. Zhu Y, Ma X, Zhang H, Wu Y, Kang M, Fang Y, et al. Mechanism of Circadd2 as ceRNA in Childhood Acute Lymphoblastic Leukemia. Front Cell Dev Biol (2021) 9:639910. doi: 10.3389/fcell.2021.639910
167. Cao H-X, Miao C-F, Sang L-N, Huang Y-M, Zhang R, Sun L, et al. Circ_0009910 Promotes Imatinib Resistance Through ULK1-Induced Autophagy by Sponging miR-34a-5p in Chronic Myeloid Leukemia. Life Sci (2020) 243:117255. doi: 10.1016/j.lfs.2020.117255
168. Wu Z, Sun H, Liu W, Zhu H, Fu J, Yang C, et al. Circ-RPL15: A Plasma Circular RNA as Novel Oncogenic Driver to Promote Progression of Chronic Lymphocytic Leukemia. Leukemia (2020) 34:919–23. doi: 10.1038/s41375-019-0594-6
169. Mei M, Wang Y, Song W, Li Z, Wang Q, Li J, et al. CircADARB1 Serves as a New Biomarker in Natural Killer T-Cell Lymphoma and a Potential Regulator of P-Stat3. Cancer Cell Int (2021) 21:594. doi: 10.1186/s12935-021-02296-x
170. Zhao C-X, Yan Z-X, Wen J-J, Fu D, Xu P-P, Wang L, et al. CircEAF2 Counteracts Epstein-Barr Virus-Positive Diffuse Large B-Cell Lymphoma Progression via miR-BART19-3p/APC/β-Catenin Axis. Mol Cancer (2021) 20:153. doi: 10.1186/s12943-021-01458-9
171. Li W, Yang F-Q, Sun C-M, Huang J-H, Zhang H-M, Li X, et al. Circprrc2a Promotes Angiogenesis and Metastasis Through Epithelial-Mesenchymal Transition and Upregulates TRPM3 in Renal Cell Carcinoma. Theranostics (2020) 10:4395–409. doi: 10.7150/thno.43239
172. Cen J, Liang Y, Huang Y, Pan Y, Shu G, Zheng Z, et al. Circular RNA circSDHC Serves as a Sponge for miR-127-3p to Promote the Proliferation and Metastasis of Renal Cell Carcinoma via the CDKN3/E2F1 Axis. Mol Cancer (2021) 20:19. doi: 10.1186/s12943-021-01314-w
173. Li J, Huang C, Zou Y, Ye J, Yu J, Gui Y. CircTLK1 Promotes the Proliferation and Metastasis of Renal Cell Carcinoma by Sponging miR-136-5p. Mol Cancer (2020) 19:103. doi: 10.1186/s12943-020-01225-2
174. Lv Q, Wang G, Zhang Y, Shen A, Tang J, Sun Y, et al. CircAGAP1 Promotes Tumor Progression by Sponging miR-15-5p in Clear Cell Renal Cell Carcinoma. J Exp Clin Cancer Res (2021) 40:76. doi: 10.1186/s13046-021-01864-3
175. Liu H, Hu G, Wang Z, Liu Q, Zhang J, Chen Y, et al. Circptch1 Promotes Invasion and Metastasis in Renal Cell Carcinoma via Regulating miR-485-5p/MMP14 Axis. Theranostics (2020) 10:10791–807. doi: 10.7150/thno.47239
176. Wen G, Zhou T, Gu W. The Potential of Using Blood Circular RNA as Liquid Biopsy Biomarker for Human Diseases. Protein Cell (2021) 12:911946. doi: 10.1007/s13238-020-00799-3
177. Yang L, Chen Y, Liu N, Lu Y, Ma W, Yang Z, et al. CircMET Promotes Tumor Proliferation by Enhancing CDKN2A mRNA Decay and Upregulating SMAD3. Mol Cancer (2022) 21:23. doi: 10.1186/s12943-022-01497-w
178. Wu S, Yang J, Xu H, Wang X, Zhang R, Lu W, et al. Circular RNA Circglis3 Promotes Bladder Cancer Proliferation via the miR-1273f/SKP1/Cyclin D1 Axis. Cell Biol Toxicol (2022) 38:129–46. doi: 10.1007/s10565-021-09591-3
179. Yang C, Mou Z, Wu S, Ou Y, Zhang Z, Chen X, et al. High-Throughput Sequencing Identified Circular RNA Circube2k Mediating RhoA Associated Bladder Cancer Phenotype via Regulation of miR-516b-5p/ARHGAP5 Axis. Cell Death Dis (2021) 12:719. doi: 10.1038/s41419-021-03977-1
180. Qiu F, Liu Q, Xia Y, Jin H, Lin Y, Zhao X. Circ_0000658 Knockdown Inhibits Epithelial-Mesenchymal Transition in Bladder Cancer via miR-498-Induced HMGA2 Downregulation. J Exp Clin Cancer Res (2022) 41:22. doi: 10.1186/s13046-021-02175-3
181. Jin M, Lu S, Wu Y, Yang C, Shi C, Wang Y, et al. Hsa_circ_0001944 Promotes the Growth and Metastasis in Bladder Cancer Cells by Acting as a Competitive Endogenous RNA for miR-548. J Exp Clin Cancer Res (2020) 39:186. doi: 10.1186/s13046-020-01697-6
182. Tan S, Kang Y, Li H, He H-Q, Zheng L, Wu S-Q, et al. Circst6galnac6 Suppresses Bladder Cancer Metastasis by Sponging miR-200a-3p to Modulate the STMN1/EMT Axis. Cell Death Dis (2021) 12:168. doi: 10.1038/s41419-021-03459-4
183. Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q, et al. Circular RNA ACVR2A Suppresses Bladder Cancer Cells Proliferation and Metastasis Through miR-626/EYA4 Axis. Mol Cancer (2019) 18:95. doi: 10.1186/s12943-019-1025-z
184. Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, et al. Circular RNA Circslc8a1 Acts as a Sponge of miR-130b/miR-494 in Suppressing Bladder Cancer Progression via Regulating PTEN. Mol Cancer (2019) 18:111. doi: 10.1186/s12943-019-1040-0
185. Bi J, Liu H, Dong W, Xie W, He Q, Cai Z, et al. Circular RNA Circ-ZKSCAN1 Inhibits Bladder Cancer Progression Through miR-1178-3p/P21 Axis and Acts as a Prognostic Factor of Recurrence. Mol Cancer (2019) 18:133. doi: 10.1186/s12943-019-1060-9
186. Xie F, Xiao X, Tao D, Huang C, Wang L, Liu F, et al. Circnr3c1 Suppresses Bladder Cancer Progression Through Acting as an Endogenous Blocker of BRD4/C-Myc Complex. Mol Ther Nucleic Acids (2020) 22:510–9. doi: 10.1016/j.omtn.2020.09.016
187. Chen W, Cen S, Zhou X, Yang T, Wu K, Zou L, et al. Circular RNA CircNOLC1, Upregulated by NF-KappaB, Promotes the Progression of Prostate Cancer via miR-647/PAQR4 Axis. Front Cell Dev Biol (2020) 8:624764. doi: 10.3389/fcell.2020.624764
188. Shan G, Shao B, Liu Q, Zeng Y, Fu C, Chen A, et al. Circfmn2 Sponges miR-1238 to Promote the Expression of LIM-Homeobox Gene 2 in Prostate Cancer Cells. Mol Ther Nucleic Acids (2020) 21:133–46. doi: 10.1016/j.omtn.2020.05.008
189. Mao Y, Li W, Hua B, Gu X, Pan W, Chen Q, et al. Circular RNA_PDHX Promotes the Proliferation and Invasion of Prostate Cancer by Sponging MiR-378a-3p. Front Cell Dev Biol (2020) 8:602707. doi: 10.3389/fcell.2020.602707
190. Chao F, Song Z, Wang S, Ma Z, Zhuo Z, Meng T, et al. Novel Circular RNA circSOBP Governs Amoeboid Migration Through the Regulation of the miR-141-3p/MYPT1/p-MLC2 Axis in Prostate Cancer. Clin Transl Med (2021) 11:e360. doi: 10.1002/ctm2.360
191. Yu Y-Z, Lv D-J, Wang C, Song X-L, Xie T, Wang T, et al. Hsa_circ_0003258 Promotes Prostate Cancer Metastasis by Complexing With IGF2BP3 and Sponging miR-653-5p. Mol Cancer (2022) 21:12. doi: 10.1186/s12943-021-01480-x
192. Hong H, Zhu H, Zhao S, Wang K, Zhang N, Tian Y, et al. The Novel Circclk3/miR-320a/FoxM1 Axis Promotes Cervical Cancer Progression. Cell Death Dis (2019) 10:950. doi: 10.1038/s41419-019-2183-z
193. Ou R, Lv J, Zhang Q, Lin F, Zhu L, Huang F, et al. Circamotl1 Motivates AMOTL1 Expression to Facilitate Cervical Cancer Growth. Mol Ther Nucleic Acids (2020) 19:50–60. doi: 10.1016/j.omtn.2019.09.022
194. Li X, Ma N, Zhang Y, Wei H, Zhang H, Pang X, et al. Circular RNA Circnrip1 Promotes Migration and Invasion in Cervical Cancer by Sponging miR-629-3p and Regulating the PTP4A1/ERK1/2 Pathway. Cell Death Dis (2020) 11:399. doi: 10.1038/s41419-020-2607-9
195. Ji F, Du R, Chen T, Zhang M, Zhu Y, Luo X, et al. Circular RNA Circslc26a4 Accelerates Cervical Cancer Progression via miR-1287-5p/HOXA7 Axis. Mol Ther Nucleic Acids (2020) 19:413–20. doi: 10.1016/j.omtn.2019.11.032
196. Ou R, Mo L, Tang H, Leng S, Zhu H, Zhao L, et al. circRNA-AKT1 Sequesters miR-942-5p to Upregulate AKT1 and Promote Cervical Cancer Progression. Mol Ther Nucleic Acids (2020) 20:308–22. doi: 10.1016/j.omtn.2020.01.003
197. Xu J, Zhang Y, Huang Y, Dong X, Xiang Z, Zou J, et al. Circeya1 Functions as a Sponge of miR-582-3p to Suppress Cervical Adenocarcinoma Tumorigenesis via Upregulating Cxcl14. Mol Ther Nucleic Acids (2020) 22:1176–90. doi: 10.1016/j.omtn.2020.10.026
198. Zhou M, Yang Z, Wang D, Chen P, Zhang Y. The Circular RNA circZFR Phosphorylates Rb Promoting Cervical Cancer Progression by Regulating the SSBP1/CDK2/cyclin E1 Complex. J Exp Clin Cancer Res (2021) 40:48. doi: 10.1186/s13046-021-01849-2
199. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
200. Huang Y, Qin S, Gu X, Zheng M, Zhang Q, Liu Y, et al. Comprehensive Assessment of Serum Hsa_Circ_0070354 as a Novel Diagnostic and Predictive Biomarker in Non-Small Cell Lung Cancer. Front Genet (2021) 12:796776. doi: 10.3389/fgene.2021.796776
201. Kim SS, Donahue TR, Girgis MD. Carcinoembryonic Antigen for Diagnosis of Colorectal Cancer Recurrence. JAMA (2018) 320:298–9. doi: 10.1001/jama.2018.8424
202. Tsai I-J, Su EC-Y, Tsai I-L, Lin C-Y. Clinical Assay for the Early Detection of Colorectal Cancer Using Mass Spectrometric Wheat Germ Agglutinin Multiple Reaction Monitoring. Cancers (Basel) (2021) 13:2190. doi: 10.3390/cancers13092190
203. He B, Chao W, Huang Z, Zeng J, Yang J, Luo D, et al. Hsa_circ_001659 Serves as a Novel Diagnostic and Prognostic Biomarker for Colorectal Cancer. Biochem Biophys Res Commun (2021) 551:100–6. doi: 10.1016/j.bbrc.2021.02.121
204. Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, et al. A Noncoding Regulatory RNAs Network Driven by Circ-CDYL Acts Specifically in the Early Stages Hepatocellular Carcinoma. Hepatology (2020) 71:130–47. doi: 10.1002/hep.30795
205. Yang C, Dong Z, Hong H, Dai B, Song F, Geng L, et al. Circfn1 Mediates Sorafenib Resistance of Hepatocellular Carcinoma Cells by Sponging miR-1205 and Regulating E2F1 Expression. Mol Ther Nucleic Acids (2020) 22:421–33. doi: 10.1016/j.omtn.2020.08.039
206. Yan M, Niu L, Liu J, Yao Y, Li H. Circevi5 Acts as a miR-4793-3p Sponge to Suppress the Proliferation of Gastric Cancer. Cell Death Dis (2021) 12:774. doi: 10.1038/s41419-021-04061-4
207. Song J, Yu S, Zhong D, Yang W, Jia Z, Yuan G, et al. The Circular RNA Hsa_Circ_000780 as a Potential Molecular Diagnostic Target for Gastric Cancer. BMC Med Genomics (2021) 14:282. doi: 10.1186/s12920-021-01096-6
208. Nanishi K, Konishi H, Shoda K, Arita T, Kosuga T, Komatsu S, et al. Circulating Circerbb2 as a Potential Prognostic Biomarker for Gastric Cancer: An Investigative Study. Cancer Sci (2020) 111:4177–86. doi: 10.1111/cas.14645
209. Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, et al. CircRNA Inhibits DNA Damage Repair by Interacting With Host Gene. Mol Cancer (2020) 19:128. doi: 10.1186/s12943-020-01246-x
210. Lin Z, Long F, Zhao M, Zhang X, Yang M. The Role of Circular RNAs in Hematological Malignancies. Genomics (2020) 112:4000–8. doi: 10.1016/j.ygeno.2020.06.051
211. Cammarata-Scalisi F, Girardi K, Strocchio L, Merli P, Garret-Bernardin A, Galeotti A, et al. Oral Manifestations and Complications in Childhood Acute Myeloid Leukemia. Cancers (Basel) (2020) 12:E1634. doi: 10.3390/cancers12061634
212. Guo S, Li B, Chen Y, Zou D, Yang S, Zhang Y, et al. Hsa_circ_0012152 and Hsa_circ_0001857 Accurately Discriminate Acute Lymphoblastic Leukemia From Acute Myeloid Leukemia. Front Oncol (2020) 10:1655. doi: 10.3389/fonc.2020.01655
213. Wen G, Zhou T, Gu W. The Potential of Using Blood Circular RNA as Liquid Biopsy Biomarker for Human Diseases. Protein Cell (2021) 12:911–46. doi: 10.1007/s13238-020-00799-3
214. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA Is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res (2015) 25:981–4. doi: 10.1038/cr.2015.82
215. Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-Mediated Metastasis: Communication From a Distance. Dev Cell (2019) 49:347–60. doi: 10.1016/j.devcel.2019.04.011
216. Tian X, Shen H, Li Z, Wang T, Wang S. Tumor-Derived Exosomes, Myeloid-Derived Suppressor Cells, and Tumor Microenvironment. J Hematol Oncol (2019) 12:84. doi: 10.1186/s13045-019-0772-z
217. Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL Promotes Progression of Colorectal Cancer via the miR-142-3p/miR-506-3p- TGF-β1 Axis. Mol Cancer (2020) 19:117. doi: 10.1186/s12943-020-01235-0
218. Li P, Xu Z, Liu T, Liu Q, Zhou H, Meng S, et al. Circular RNA Sequencing Reveals Serum Exosome Circular RNA Panel for High-Grade Astrocytoma Diagnosis. Clin Chem (2022) 68:332–43. doi: 10.1093/clinchem/hvab254
219. Xu H, Liu Y, Cheng P, Wang C, Liu Y, Zhou W, et al. CircRNA_0000392 Promotes Colorectal Cancer Progression Through the miR-193a-5p/PIK3R3/AKT Axis. J Exp Clin Cancer Res (2020) 39:283. doi: 10.1186/s13046-020-01799-1
220. Liu D, Wan X, Shan X, Fan R, Zha W. Drugging the “Undruggable” microRNAs. Cell Mol Life Sci (2021) 78:1861–71. doi: 10.1007/s00018-020-03676-8
221. Tassinari V, Cesarini V, Tomaselli S, Ianniello Z, Silvestris DA, Ginistrelli LC, et al. ADAR1 Is a New Target of METTL3 and Plays a Pro-Oncogenic Role in Glioblastoma by an Editing-Independent Mechanism. Genome Biol (2021) 22:51. doi: 10.1186/s13059-021-02271-9
222. Zeballos C MA, Gaj T. Next-Generation CRISPR Technologies and Their Applications in Gene and Cell Therapy. Trends Biotechnol (2021) 39:692–705. doi: 10.1016/j.tibtech.2020.10.010
Keywords: circRNAs, biogenesis, cancer, biomarkers, mechanisms
Citation: Zhang Y, Zhang X, Xu Y, Fang S, Ji Y, Lu L, Xu W, Qian H and Liang ZF (2022) Circular RNA and Its Roles in the Occurrence, Development, Diagnosis of Cancer. Front. Oncol. 12:845703. doi: 10.3389/fonc.2022.845703
Received: 20 January 2022; Accepted: 14 March 2022;
Published: 07 April 2022.
Edited by:
Xiao Zhu, Guangdong Medical University, ChinaReviewed by:
Chongming Jiang, Baylor College of Medicine, United StatesCopyright © 2022 Zhang, Zhang, Xu, Fang, Ji, Lu, Xu, Qian and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhao Feng Liang, bGlhbmd6aGFvZmVuZ0B1anMuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.