- 1Department of Obstetrics and Gynecology, Taizhou Central Hospital (Taizhou University, Hospital), Taizhou, China
- 2Zhongnan Hospital of Wuhan University, Institute of Hepatobiliary Diseases of Wuhan University, Transplant Center of Wuhan University, National Quality Control Center for Donated Organ Procurement, Hubei Key Laboratory of Medical Technology on Transplantation, Hubei Clinical Research Center for Natural Polymer Biological Liver, Hubei Engineering Center of Natural Polymer-based Medical Materials, Wuhan, China
- 3Department of Anesthesiology, Xiangya Hospital of Central South University, Changsha, China
- 4National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Changsha, China
- 5Department of General Surgery, Taizhou Central Hospital (Taizhou University Hospital), Taizhou, China
Gynecological cancers seriously affect the reproductive system of females; diseases include ovarian tumors, uterine tumors, endometrial cancers, cervical cancers, and vulva and vaginal tumors. At present, the diagnosis methods of gynecological cancer are insufficiently sensitive and specific, leading to failure of early disease detection. N6-methyladenosine (m6A) plays various biological functions in RNA modification and is currently studied extensively. m6A modification controls the fate of transcripts and regulates RNA metabolism and biological processes through the interaction of m6A methyltransferase (“writer”) and demethylase (“erasers”) and the binding protein decoding m6A methylation (“readers”). In the field of epigenetics, m6A modification is a dynamic process of reversible regulation of target RNA through its regulatory factors. It plays an important role in many diseases, especially cancer. However, its role in gynecologic cancers has not been fully investigated. Thus, we review the regulatory mechanism, biological functions, and therapeutic prospects of m6A RNA methylation regulators in gynecological cancers.
1 Introduction
Gynecological cancers are a series of tumors that seriously damage the female reproductive system; diseases include ovarian cancer, uterine cancer, endometrial cancer, cervical cancer, and vulva and vaginal cancer (1). Gynecological cancers become a serious global public health challenge due to their high incidence in women of all ages (2). Among them, ovarian cancer, endometrial cancer, and cervical cancer are the most common gynecological tumors. Considerable studies have shown that the occurrence and development of gynecological cancers are related to the activation of oncogenes, the inactivation of tumor suppressor genes, and the activation of abnormal cell signaling pathways. In addition, epigenetic processes regulate gene expression through DNA methylation, histone modification, and noncoding RNA, thereby affecting the occurrence and development of gynecological cancer (3).
Ovarian cancer is one of the most common malignant tumors in women (3). It has the characteristic of insidious onset and has no specific clinical symptoms in the early stage of the disease. Moreover, sensitive and effective clinical screening methods for ovarian cancer are currently limited, and approximately 70% of patients are advanced at the time of diagnosis (4). According to the American Cancer Society, the United States records approximately 21,000 new cases of ovarian Cancer each year, accounting for 5% of all female malignancies, with a mortality rate of 62% and a five-year survival rate of 20%–30%, seriously affecting women’s health (5). At present, the pathogenesis of ovarian cancer is still unclear. The development of epigenetics provides new means to discover specific biomarkers and treatment methods, which greatly improve the diagnosis and treatment prospects of ovarian cancer.
Cervical cancer is the fourth most common female malignancy in the world and a leading cause of cancer-related death in women (6, 7). Cervical cancer is diagnosed in more than 500,000 patients worldwide each year and leads to more than 300,000 deaths (8, 9). Although HPV vaccination is effective in preventing cervical cancer, it remains the fourth most common cancer among women globally due to inadequate screening programs in many parts of the world (10, 11). Despite the continuous innovation of radiotherapy and/or chemotherapy on the basis of surgery, early lymph node metastasis still occurs in some patients with cervical cancer, resulting in poor prognosis and low survival rate. The five-year survival rate is still approximately 40%, posing a serious threat to women’s health (12–15). Thus, elucidating the molecular mechanism of cervical cancer occurrence and metastasis has great clinical importance.
Endometrial cancer, as one of the most common gynecological cancers, has become the fourth most common malignant tumor and the fifth most common cause of death among women in the United States (16, 17). According to WHO statistics in 2021, the total incidence of endometrial cancer in the United States was 7%, with 66,570 new cases (5). Despite advances in drugs and surgical treatments for endometrial cancer, recent studies have shown that survival rates for endometrial cancer have not improved significantly, but death rates have increased. Thus, improving the ability to identify the prognostic risk factors of endometrial cancer and formulating reasonable new treatment plans are greatly important for improving the survival rate and prognosis of patients with endometrial cancer (18). In recent years, the study of tumor genesis, intracellular signaling pathway changes, and epigenetics in tumor microenvironment has developed rapidly, providing a new means for the discovery of specific biomarkers and therapeutic methods (19).
m6A methylation was first identified in 1974 and subsequently proven to be the most common and abundant RNA modification in eukaryotic cells (20). m6A modification not only exists in mRNA, but also in various noncoding RNAs (21, 22). m6A modification affects cell function by regulating the function and metabolism of RNA; it is involved in various pathophysiological processes, such as cell division, immune regulation, and regulation of the occurrence of various cancers (23, 24). A large number of studies have shown that m6A modification is related to the proliferation, differentiation, tumorigenesis, invasion, and metastasis of gynecological cancers; it can function as an oncogene or anticancer gene (25–28). Here, we comprehensively review the modification of m6A and analyze the potential molecular mechanism of m6A in gynecological cancers. The prospect of m6A modification as a new marker and therapeutic target for gynecological cancer was further clarified.
2 Molecular Mechanisms of m6A Modification
2.1 m6A Writers
Modification of m6A is dynamic and reversible, and methyltransferases (writers) are mainly composed of KIAA1429(VIRMA), METTL3, RBM15, WTAP, ZC3H13, METTL16, METTL14, and CBLL1 (29). KIAA1429, also known as Virlike m6A methyltransferase-associated protein (VIRMA), a newly identified component of the RNA m6A methyltransferase complex, plays a key role in guiding regionally selective m6A deposition (30). Meanwhile, it regulates the expression of sex-lethal genes by selective splicing of premRNA with WTAP (31). Interestingly, METTL14 and METTL3, as core components of m6A methyltransferase, form A stable METTL3-METTL14 heterodimer core complex, which plays A role in cell m6A deposition on mammalian nuclear RNA (32). As a mammalian splicing factor, WTAP has no methylation activity by itself, but can interact with METTL14/METTL3 complex and affect methylation (32). In addition, WTAP is a regulatory subunit required for the formation of the m6A methyltransferase complex (including METTL3 and METTL14), which plays an important role in gene expression regulation and alternative splicing. Moreover, in vivo localization to pre-mRNA rich nuclear spots and catalytic m6A methyl transferase activity. In the absence of WTAP, the RNA binding ability of METTL3 was significantly weakened (33).The main feature of METTL3 and METTL14 is that they contain methyltransferase domains (S-adenosine methylene thiocyanine binding motifs, SAM-binding), which transfer methyl groups to adenosine at N6 (34, 35). RBM15, an RNA-binding protein, is involved in m6A modification and the regulation of alternative splicing (AS) through the regulation of Notch, Wnt, and other signaling pathways; it has inhibitory functions in multiple signaling pathways (36). ZC3H13 is a typical CCCH zinc finger protein, which acts as a tumor suppressor and inhibits tumor development by regulating Ras-ERK signaling pathway (37). METTTL16 is an emerging player in the field of RNA modification in human cells. Originally thought to be a ribosomal RNA methyltransferase, it has currently been shown to bind and methylate MAT2A messenger RNA (mRNA) and U6 small nuclear RNA (snRNA) (38). Casitas B family lymphoma transforming sequence-like protein 1 (CBLL1), also known as Hakai, was originally identified as the E3 ubiquitin ligase of the E-cadherin complex (39). The molecular mechanisms of m6A writers as show in Figure 1.
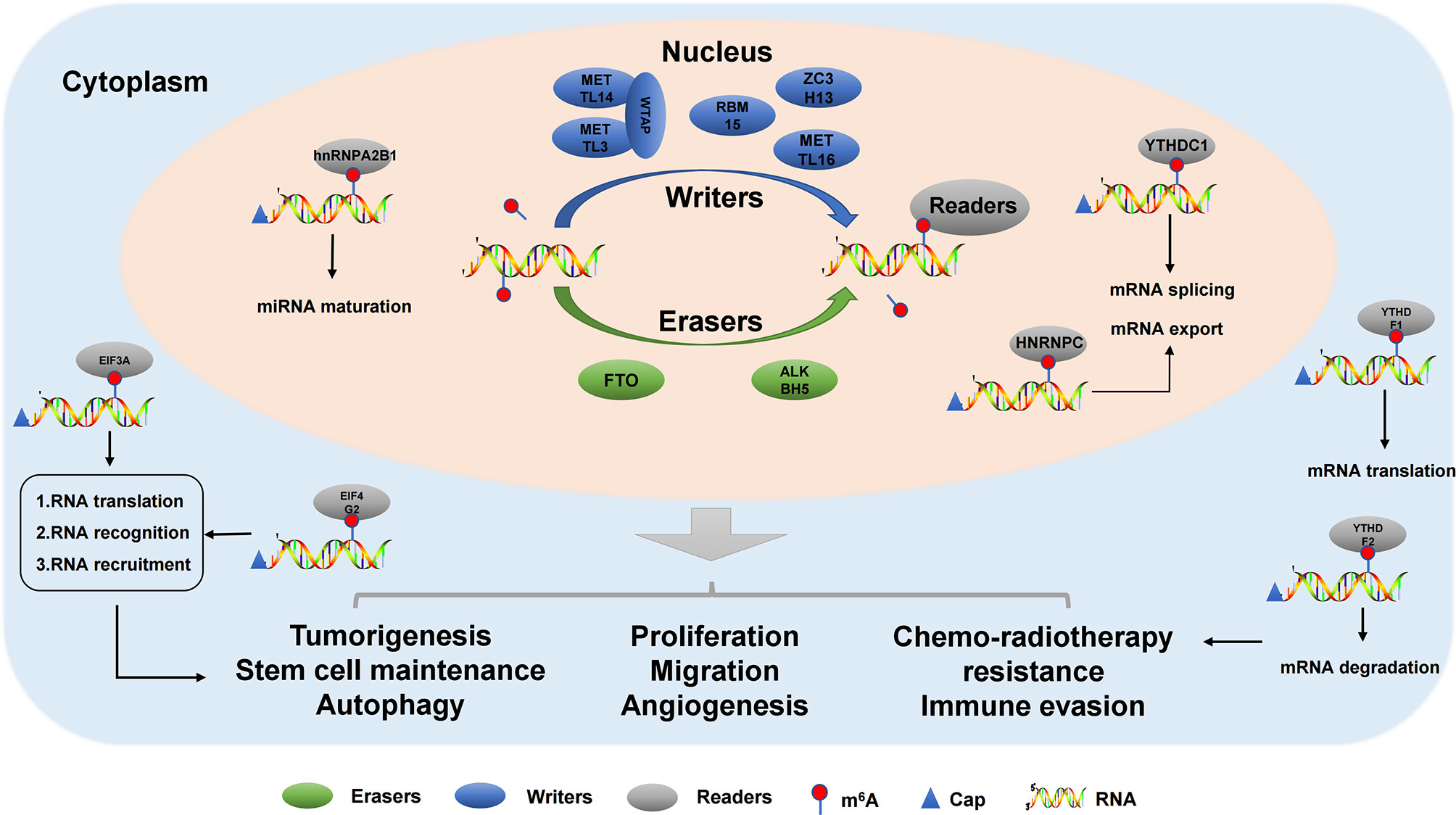
Figure 1 The molecular mechanisms of m6A modification in cancers. m6A modification is a dynamic and reversible process. m6A methylation is catalysed by methyltransferase complex (writers), reversed by demethylases (erasers) and functionally facilitated by m6A-binding proteins (readers). m6A methylation participates in carcinogenesis and tumor progression.
2.2 m6A Erasers
Thus far, only two m6A demethylases have been identified, namely, ALKBH5 and FTO (40). ALKHB5 is a member of the AlkB family and plays an important regulatory role in many biological processes, such as mRNA modification and regulation (41, 42). ALKBH5 also plays a regulatory role in the occurrence and development of tumors (41). For example, ALKBH5 inhibits pancreatic cancer by decreasing WIF-1 RNA methylation and mediating Wnt signaling (43). ALKBH5 promotes the invasion and metastasis of gastric cancer by reducing the methylation of lncRNA NEAT1 (44). At the same time, autophagy in epithelial ovarian cancer was inhibited by miR-7 and Bcl-2 (45). In addition, ALKBH5 can regulate the expression of PD-L1 in cholangiocarcinoma, promote the expression of PD-L1 on monocytes/macrophages, and reduce the infiltration of bone marrow-derived inhibitory cells, making tumors with strong ALKBH5 nuclear expression pattern more sensitive to PD1 immunotherapy (46). Fat mass and obesity-related protein (FTO), as the first m6A demethylase responsible for RNA modification in cells, is involved in various physiological processes, and its dysregulation is closely related to various human diseases, especially the occurrence and development of tumors (47). The FTO gene was originally identified as being involved in obesity and type 2 diabetes. This gene encodes the FTO protein, belonging to the AlkB dioxygenase family dependent on Fe2+ and 2-oxoglutarate (2OG) (48). FTO shows complex biological functions in physiological process, and its disorder is related to various human diseases (49). The molecular mechanisms of m6A erasers as show in Figure 1.
2.3 m6A Readers
m6A binding protein, composed of YT521-B homolog (YTH) domain protein, is mainly composed of HNRNP family (HNRNPA2/B1, HNRNPC, and HNRNPG) and YTH domain protein family (YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3). In addition, it includes FXR, IGF2BP, eIF, and G3BP family members (50). Heterogeneous nuclear ribonucleoproteins (hnRNPs) represent a large family of RNA-binding proteins (RBPs) that play roles in nucleic acid metabolism including selective splicing, mRNA stabilization, transcription, and translation regulation (51). The expression levels of hnRNPs are altered in many types of cancer, suggesting that they play an important regulatory role in tumorigenesis. The YTH domain recognizes m6A modifications through a conserved aromatic ring. This RNA binding domain is dependent on m6A modification (52). Reading proteins recognize and read information from m6A RNA in a methylation-dependent manner. YTHDC1-2 and YTHDF1-3 are the main intracellular proteins in the human body. YTHDC2 is located in the cytoplasm of meiosis spermatocytes, and YTHDF1-3 mainly recognizes the information of m6A methylation in the cytoplasm (53, 54). The five proteins not only contain the same domain, but also have special domains that determine their different roles. The molecular mechanisms of m6A readers as show in Figure 1.
3 m6A and Cancers
Previous studies have shown that the role of m6A methylation regulators in carcinogenesis and tumor progression is mainly achieved by regulating oncogene expression and inhibiting gene expression (40). m6A methylation regulator plays a “double-edged sword” role in tumor progression, which can promote the expression of oncogenes and inhibit the expression of oncogenes to promote tumor progression or the expression of oncogenes and inhibit the expression of oncogenes to inhibit tumor progression (55). With the deepening of studies on m6A methylation, the pathophysiological processes of m6A modification and regulation also expand, including mRNA regulation, immune regulation, biorhythm, neural development, and autophagy (21, 56). The dual role of m6A in cancer is increasingly recognized (57, 58). On the one hand, m6A regulates the expression of oncogenes or tumor suppressor genes, thereby affecting tumor progression. On the other hand, the level of m6A and the expression and activity of m6A enzyme can be regulated, affecting the role of m6A in cancer (58). Thus, m6A methylation regulator is expected to be a potential target for cancer therapy. The relationship between m6A modification and various gynecological cancers is reviewed, and the role of m6A regulatory factors in different gynecological cancers is further clarified.
3.1 m6A in Cervical Cancer
3.1.1 Function of m6A on mRNA in Cervical Cancer
Post-expression regulation of genes is mainly carried out in four aspects, namely, transcription, post-transcription, translation, and post-translation. m6A modification is mainly manifested in the RNA transcription process, which regulates gene expression after RNA transcription by modifying the structure of RNA or specific binding in the form of binding protein (59). As an important m6A “reader”, YTHDF1 regulates the fate of m6A-modified mRNA. Studies have found that the up-regulation of YTHDF1 in cervical cancer is closely related to the poor prognosis of cervical cancer patients. YTHDF1 regulates RANBP2 translation in an m6A-dependent manner without affecting its mRNA expression. RANBP2 can promote the growth, migration, and invasion of cervical cancer cells. Thus, YTHDF1 has a carcinogenic effect in cervical cancer by regulating the expression of RANBP2, and YTHDF1 is a potential target for cervical cancer treatment (26). YTHDF2, as another member of the YTH domain protein family, is also up-regulated in cervical cancer, and the higher expression in cervical cancer indicates shorter survival time. After YTHDF2 knockdown, the proliferation of cervical cancer cells was transplanted to promote cell apoptosis, and the tumor cells stagnated in the S phase (60).
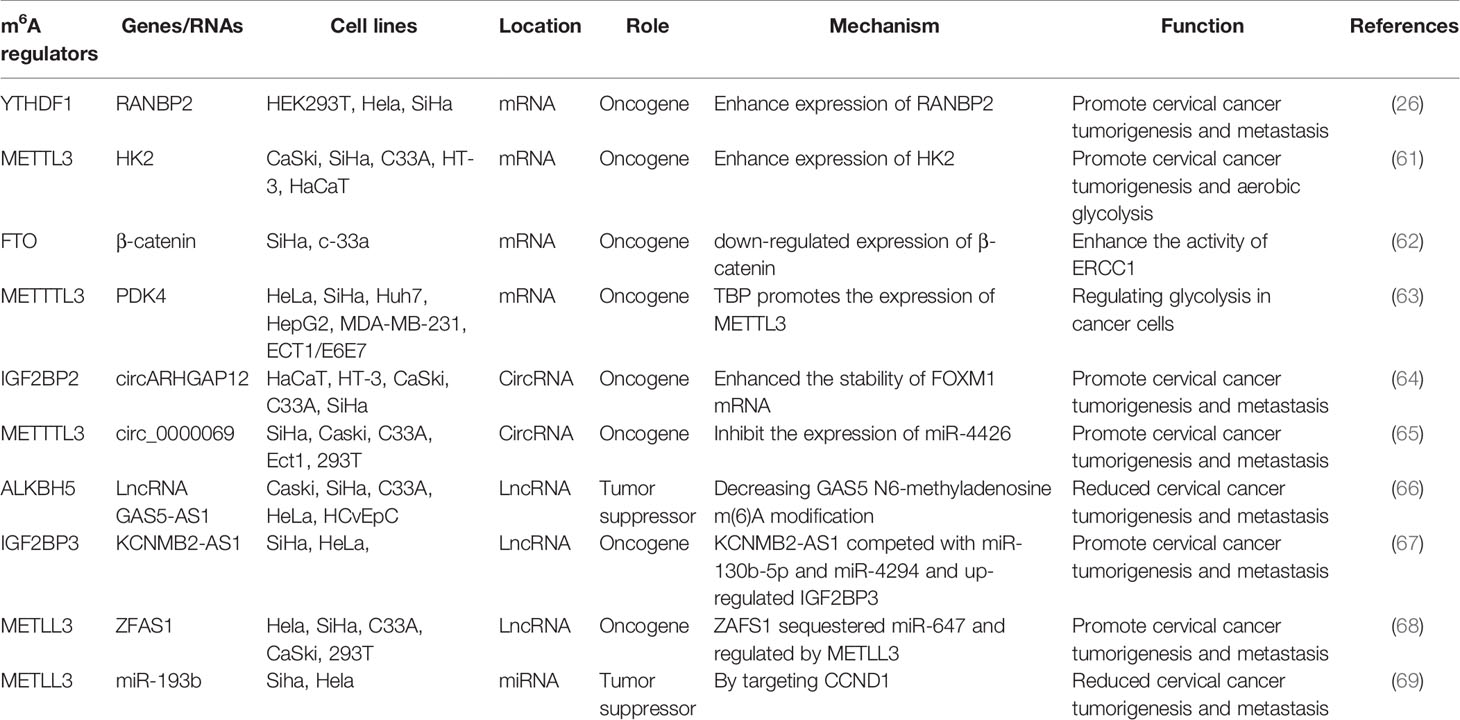
In addition to m6A “readers” that play an important regulatory role in cervical cancer, m6A “writers” play an important regulatory role in the occurrence and progression of cervical cancer. For example, METTL3 is significantly up-regulated in cervical cancer tissues and cells; it is closely associated with lymph node metastasis and poor prognosis in cervical cancer patients. Moreover, METTL3 can promote the proliferation and decrease apoptosis of cervical cancer cells in vitro. METTL3 promotes the proliferation and aerobic glycolysis of cervical cancer cells by targeting the 3’ -untranslated region (3’-UTR) of hexokinase 2 (HK2) mRNA. In addition, METTL3 in combination with YTHDF1 enhances the stability of HK2. These data suggest that METTTL3 may be a carcinogenic factor in the development of cervical tumors. In the existing studies, the role of METTTL3 in aerobic glycolysis of tumors is rarely reported. Thus, METTTL3 regulates the stability of HK2 mRNA by recruiting YTHDF1 and acts as a carcinogen by accelerating glycolysis through the YTHDF1/HK2 axis, which provides a potential prognostic biomarker for cervical cancer treatment (61). FTO mRNA level, as an m6A demethylase, is up-regulated in cervical cancer tissues, and FTO regulates chemotherapy resistance of cervical squamous cell carcinoma (CSCC) through mRNA demethylation targeting β-catenin. FTO regulates β-catenin expression by reducing m6A level in β-catenin mRNA transcripts. Furthermore, the excision repair cross-complementary group 1 (ERCC1) activity was improved to enhance chemotherapy and radiotherapy resistance in vitro and in vivo (62). In addition, Li et al. found that TATA binding protein (TBP) can increase the expression of METTL3 in cervical cancer cells by binding to the promoter of METTL3. In vivo and clinical data confirm that m6A/PDK4 plays an active role in the growth and progression of cervical cancer and liver cancer. Furthermore, m6A regulates glycolysis of cancer cells through PDK4, and the methylation of PDK4 is regulating the stability and translation of its mRNA, thereby regulating glycolysis in cancer cells (63). The roles of different m6A regulators in regulating RNAs in cervical cancer are shown in Figure 2 and Table 1.
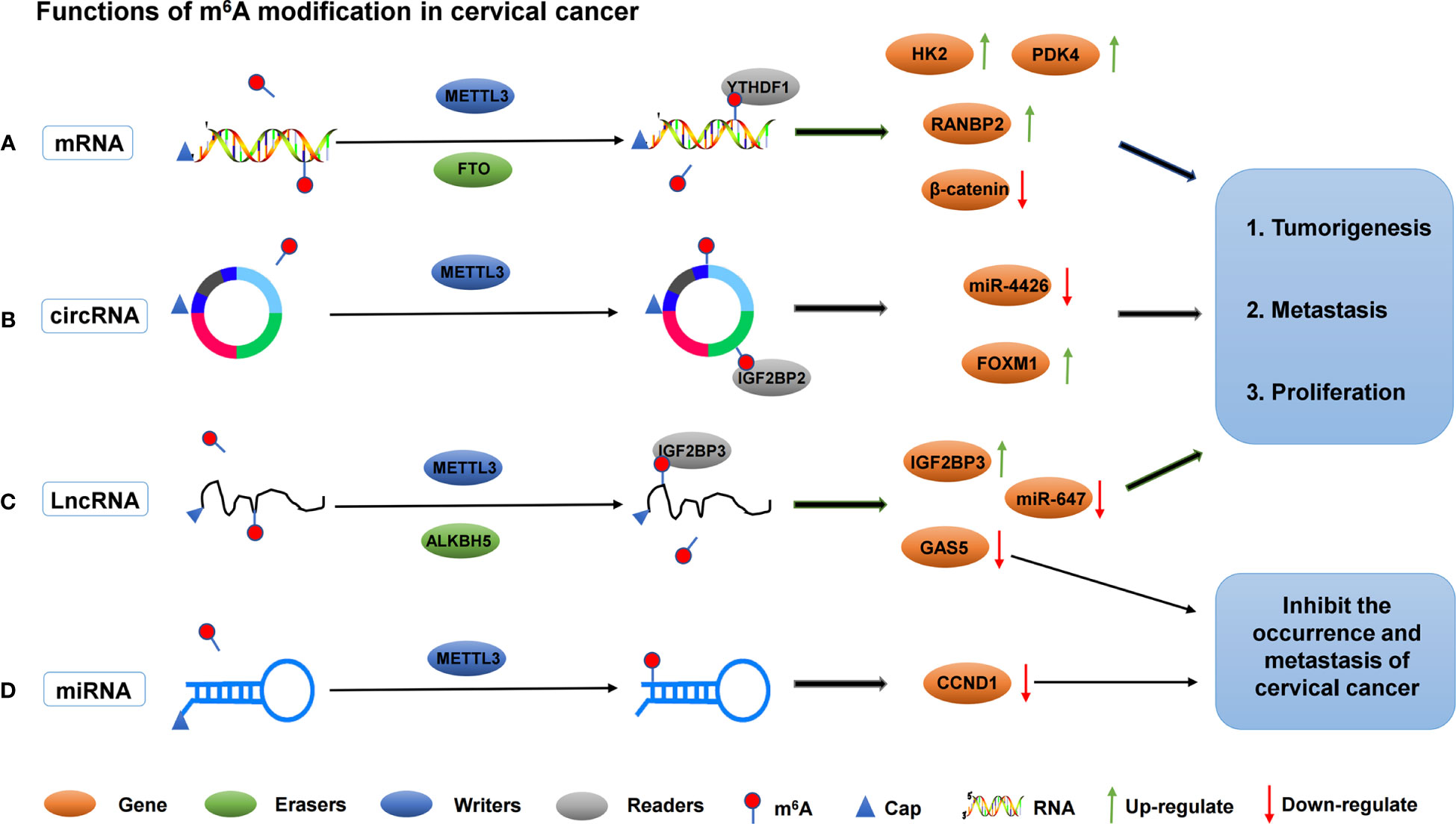
Figure 2 The roles of different m6A regulators in regulating RNAs in cervical cancer. (A) m6A methylation regulators METTL3, FTO and YTHDF1 promote the invasion and metastasis of cervical cancer by binding to mRNA and regulating mRNA expression. (B) m6A methylation regulators METTL3 and IGF2BP2 promote the invasion and metastasis of cervical cancer by binding to circRNA and regulating target genes expression. (C) m6A methylation regulators METTL3, ALKBH5 and IGF2BP3 promote/inhibit the invasion and metastasis of cervical cancer by binding to lncRNA and regulating target genes expression. (D) m6A methylation regulators METTL3 inhibit the invasion and metastasis of cervical cancer by binding to lncRNA and regulating target genes expression.
3.1.2 Function of m6A on ncRNA In Cervical Cancer
Noncoding RNAs (ncRNAs) have been shown to be involved in the development and progression of cervical cancer (65). Circ-RNAs are a class of ncRNAs with covalently closed circular structures, which are generated by reverse splicing of exon precursor mRNA or lasso intron splicing (70, 71). The most studied function of circ-RNA is being a major regulator of gene expression, and its role is to isolate or “sponge” other gene expression regulators, especially miRNAs. They have also been shown to work by directly regulating transcription and interfering with splicing mechanisms (72). At present, with the deepening of people’s understanding of cervical cancer, the pathogenesis of circ-RNA in cervical cancer is widely studied. Fei et al. found, by analyzing cervical cancer RNA sequencing data, that a new m6A modified circ-RNA (circARHGAP12, hsa_circ_0000231) was up-regulated in cervical cancer tissues and cells. Further studies found that circARHGAP12 can promote tumor progression of cervical cancer. In addition, circARHGAP12 interacts with m6A reader IGF2BP2 to bind to FOXM1 mRNA and enhance the stability of FOXM1 mRNA (64). Another circ-RNA (hsa_circ_0000069) was also regulated by m6A modification. Hsa_circ_0000069 expression was up-regulated in cervical cancer, and m6A modification enhanced the stability of circ_0000069. The proliferation and migration of cervical cancer cells were promoted by inhibiting miR-4426 (65). miR-4426 expression in cervical cancer cells was down-regulated due to the up-regulation of circ_0000069. Therefore, m6A modification indirectly inhibits the expression of miR-4426, which in turn inhibits cell proliferation and migration. However, the downstream targets of miR-4426 remain unclear.
Long ncRNAs (LncRNAs), a group of large transcripts (more than 200 nucleotides in length) without protein-coding potential, play an important role in various human diseases, including cancer. LncRNAs are involved in various pathophysiological processes, such as cell proliferation, migration, invasion, apoptosis, and chemotherapy resistance (73). m6A modification is the most abundant internal modification of RNA and exists in various RNAs, such as mRNA and lncRNA. At present, only few lncRNAs have been functionally verified in cervical cancer, especially those regulated by m6A modification. For example, the expression of lncRNA GAS5-AS1 is significantly down-regulated in cervical cancer tissues, and studies have found that the down-regulation of GAS5-AS1 is significantly correlated with late, distant, lymphatic metastasis, and poor prognosis of FIGO in cervical cancer patients. Moreover, GAS5-AS1 interacts with ALKBH5, which reduces the m6A modification of GAS5 and increases its stability, revealing the important mechanism of epigenetic changes in the occurrence and metastasis of cervical cancer (66). Another lncRNA, KCNMB2-AS1, is significantly overexpressed in cervical cancer and is associated with poor prognosis. KCNMB2-AS1 is predominantly located in the cytoplasm, and leads to upregulation of IGF2BP3, which is a proven oncogene in cervical cancer, as endogenous RNA competes with a large number of miR-130b-5p and miR-4294. In addition, IGF2BP3 binds to KCNMB2-AS1 via three m6A modification sites on KCNMB2-AS1. IGF2BP3, as the “reader” of m6A, plays a stabilizing role in KCNMB2-AS1 and promotes the occurrence and development of cervical cancer (67). LncRNA ZFAS1 has been observed to be abnormally expressed in cervical carcinoma (74). Yang et al. found that the expression of ZFAS1 was up-regulated in cervical cancer, and the up-regulation of ZFAS1 was correlated with FIGO stage, lymph node, and distant metastasis. This finding also suggested that the overall survival of cervical cancer patients was poor (68). In addition, ZAFS1 isolated miR-647, an RNA–RNA interaction regulated by METTL3-mediated m6A modification. Another study found that METTL3 enhanced the stability of lncRNA FOXD2-AS1 and maintained its expression, and promoted the development of cervical cancer. FOXD2-AS1 is significantly up-regulated in cervical cancer cells and tissues, which is closely associated with poor prognosis. Moreover, FOXD2-AS1 promotes the migration and proliferation of cervical cancer cells. Mechanistically, METTL3 enhances the stability of FOXD2-AS1 and maintains its expression. Meanwhile, FOXD2-AS1 recruits LSD1 to be silenced on the promoter of P21, thus accelerating the progression of cervical cancer (75).
MiRNAs are small ncRNAs (18-24 nucleotides) that negatively regulate gene expression by binding to target mRNA in the 3 ‘-untranslated region (UTR) during the post-transcriptional stage. Many physiological and pathophysiological processes, including cancer, are affected by miRNA activity (76). Huang et al. found from the cervical cancer specimens that the low expression of miR-193b was closely related to cervical cancer staging and interstitial invasion. miR-193b, as a tumor suppressor, is regulated by the m6A methylation regulator METTL3 in cervical cancer. miR-193b inhibits the occurrence and development of cervical cancer by targeting CCND1 (69). Currently, studies on how m6A regulates miRNA expression in cervical cancer are relatively few, and more studies are still required to further reveal the regulatory mechanism of m6A in miRNA.
3.1.3 Prognostic Effect of m6A RNA Methylation Regulators on Cervical Cancer
Worldwide, cervical cancer remains one of the most common type of cancer with a major treatment challenge facing mankind (77). The carcinogenesis of cervical cancer is a complex multistep process characterized by a wide range of molecular abnormalities, providing many potential therapeutic targets. Understanding the mechanism of these molecules is crucial for their potential therapeutic uses (78, 79). In recent years, RNA modification plays an important role in various biological processes, and its abnormal regulation has become an important factor affecting the occurrence and development of tumors. Several m6A RNA methylation regulators have been found to be prognostic factors for various cancers (80, 81). Wu et al. compared the differential expression of 20 m6A RNA methylation regulators in cervical cancer tissues by using RNA sequence data and clinical information in TCGA database. Among them, five m6A RNA methylation regulators (FTO, HNRNPA2B1, RBM15, IGF2BP1, and IGF2BP3) were significantly correlated with the status of cervical cancer. In addition, six m6A RNA methylation regulators (YTHDC2, YTHDC1, ALKBH5, ZC3H13, RBMX, and YTHDF1) were selected to construct risk markers. The overall survival of cervical cancer patients in the high-risk group was significantly lower than that in the low-risk group, with an area under the curve (AUC) of 0.718. Thus, this risk model can be used as an independent prognostic factor for cervical cancer patients; it can predict the overall survival of cervical cancer patients with different clinical factors (82). METTL3 is a member of the m6A methyltransferase family, which acts as an oncogene in cancer. Ni et al. found that METTL3 and CD33(+) MDSCs were up-regulated in cervical cancer tissues by analyzing paraffin-embedded tumor specimens from 197 patients with cervical cancer. METTTL3 expression was positively correlated with CD33(+) cell density in tumor tissues. Meanwhile, METTL3 level in tumor microenvironment was significantly correlated with tumor-advanced stage. The levels of METTTL3 and CD33(+) MDSCs in tumor tissue were significantly correlated with the reduction of DFS or OS. Thus, Cox model analysis showed that the METTL3 level in cervical cancer cells was an independent factor for patient survival (83). Pan et al. obtained clinical and survival data and RNA sequencing data of 13 m6A RNA methylation regulators from the TCGA database. Consensus cluster analysis was performed to identify different cervical cancer clusters according to the expression differences of regulatory factors. Four regulatory factors (RBM15, METTTL3, FTO, and YTHDF2) were abnormally expressed in cervical cancer tissues. LASSO Cox regression analysis showed that ZC3H13, YTHDC1, and YTHDF1 were independent prognostic indicators of cervical cancer (84).
3.2 m6A in Endometrial Cancer
3.2.1 Function of m6A on mRNA in Endometrial Cancer
m6A dynamic methylation mRNA may affect cell physiology, especially key transcripts, which may lead to significant changes in biological functions (49). In recent years, studies have shown that m6A modified mRNA methylation plays an important role in cell proliferation and tumorigenicity of endometrial cancer, and the decrease or increase in m6A mRNA methylation is likely to be the carcinogenic mechanism of most endometrial cancer, promoting the occurrence and development of endometrial cancer (27). m6A-dependent mRNA regulation affects various biological processes in endometrial cancer and is involved in the regulation of RNA structure, translation, and degradation (85). ALKBH5, an RNA demethylase, is significantly up-regulated in endometrial carcinoma and promotes the proliferation and invasion of endometrial carcinoma. Studies have shown that ALKBH5 mainly regulates the demethylation of target gene IGF1R and enhances the stability of IGF1R mRNA, thereby promoting IGF1R translation and activating IGF1R signaling pathway. It further promotes the proliferation and invasion of endometrial cancer, suggesting a potential therapeutic target for endometrial cancer (27). FTO, another demethylase, can eliminate m6A modification and regulate the metabolism of mRNA. Although many studies have confirmed the relationship between obesity and endometrial cancer, the molecular mechanism of obesity and endometrial cancer progression has not been clarified. Zhang et al. found that the expression of FTO was up-regulated in endometrial cancer, and this effect promoted the metastasis and invasion of endometrial cancer. In addition, FTO catalyzed the demethylation of the 3’UTR region of HOXB13 mRNA, thereby eliminating the recognition of m6A modification from YTHDF2 protein. Decreased mRNA attenuation of HOXB13 leads to increased protein expression, whereas WNT signaling pathway activation and downstream protein expression lead to metastasis and invasion of endometrial carcinoma (86).
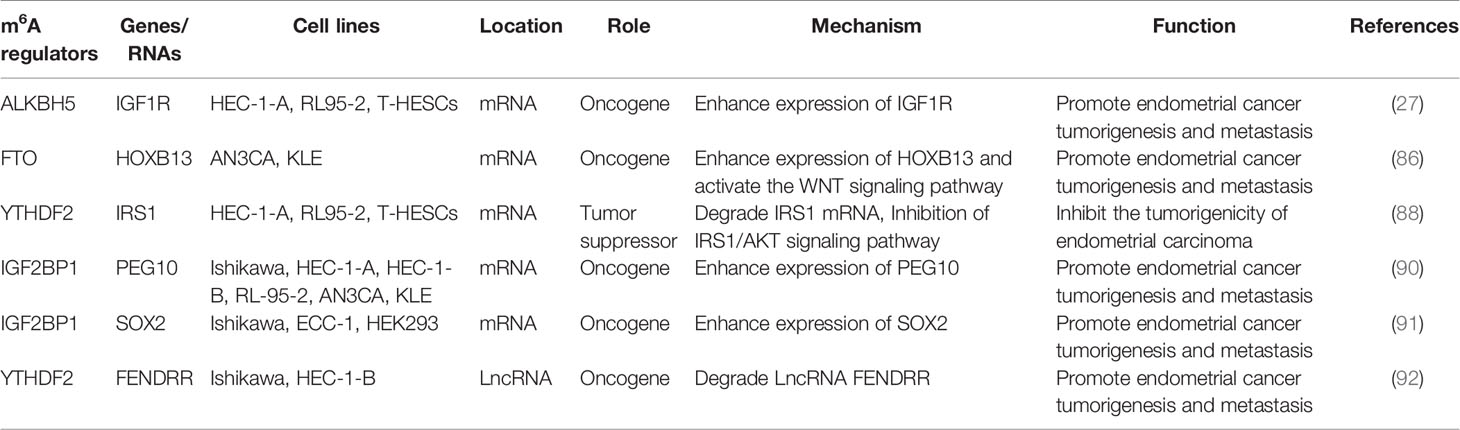
In addition to demethylase, methyltransferase plays an important role in the development of endometrial carcinoma. Liu et al. found that approximately 70% of tumor samples from patients with endometrial cancer showed decreased m6A levels, either due to reduced METTL3 expression or loss of functional mutations in METTL14. METTL14 mutations and METTL3 downregulation down-regulated m6A mRNA methylation levels and enhanced endometrial carcinoma proliferation and tumorigenicity. In addition, downregulation of m6A methylation decreases the expression of the negative AKT regulator PHLPP2 and increases the expression of the positive AKT regulator mTORC2, thereby activating the AKT pathway. Therefore, m6A modification mediated by METTL14 and METTL3 is a regulator of the AKT signaling pathway (87). In addition, the expression of m6A reader protein YTHDF2 was significantly up-regulated in endometrial cancer. YTHDF2 promoted the degradation of IRS1 mRNA by binding to the methylation site of the target transcript of IRS1, thereby inhibiting the expression of IRS1, inhibiting the IRS1/AKT signaling pathway, and ultimately inhibiting the tumorigenicity of endometrial cancer (88). IGF2BP1 belongs to the IGF2BP family. Studies have shown that IGF2BP1 is involved in the regulation of mRNA and affects the function of tumor cells (89). Zhang et al. found that up-regulation of IGF2BP1 expression in endometrial cancer is a factor affecting patient survival. Moreover, IGF2BP1 enhances PEG10 expression and promotes endometrial cancer cell proliferation by recognizing the m6A site of PEG10 mRNA (90). Another study found that PADI2-catalyzed MEK1 citrulline activates ERK1/2 and promotes IGF2BP1-mediated SOX2 mRNA stability. PADI2-catalyzed MEK1 R113/189 citrulline is a key factor in endometrial cancer. These findings suggest that targeting PADI2/MEK1 may be a potential therapy for endometrial cancer patients (91). The roles of different m6A regulators in regulating RNAs in endometrial cancer are shown in Figure 3 and Table 2.
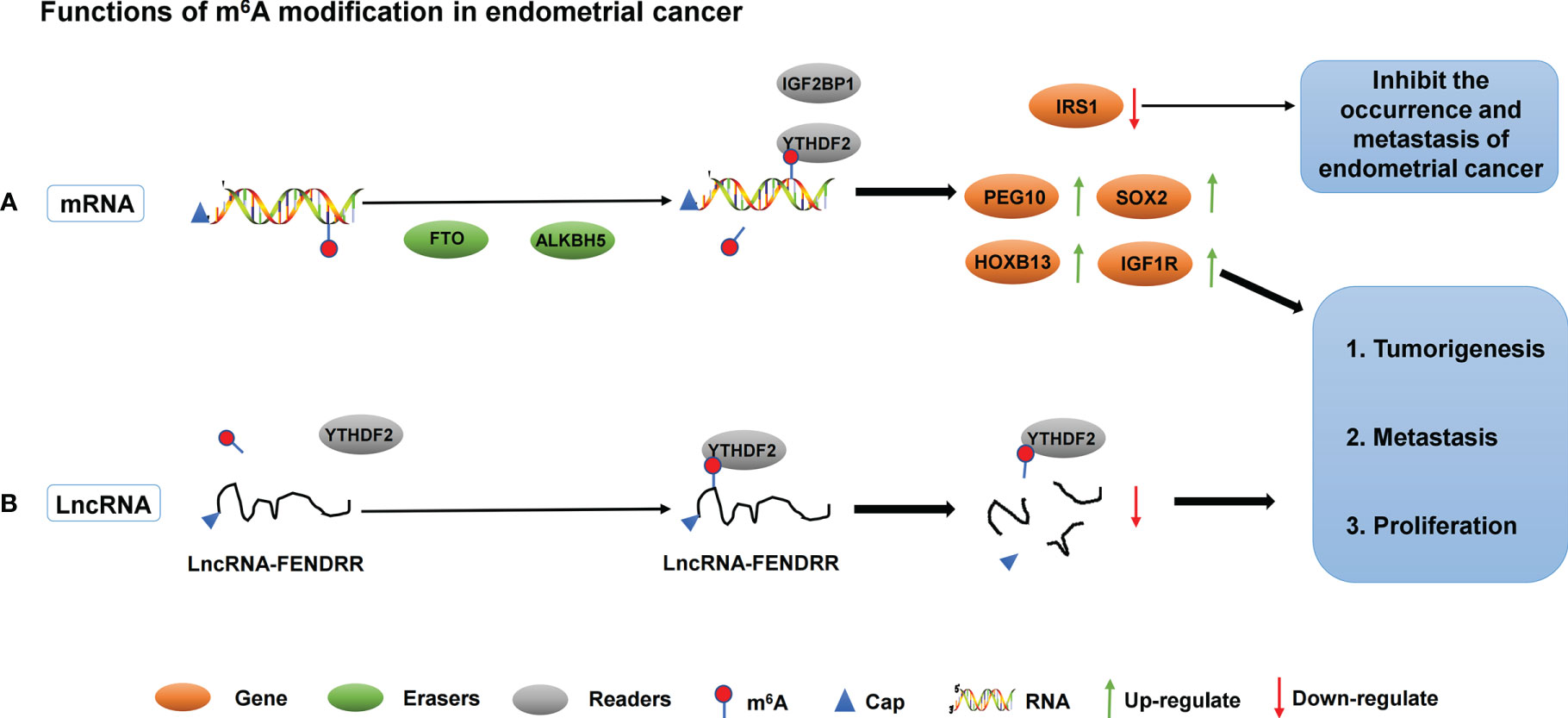
Figure 3 The roles of different m6A regulators in regulating RNAs in endometrial cancer. (A) m6A methylation regulators ALKBH5, FTO, IGF2BP1 and YTHDF2 promote/inhibit the invasion and metastasis of endometrial cancer by binding to mRNA and regulating mRNA expression. (B) m6A methylation regulators YTHDF2 promote the invasion and metastasis of endometrial cancer by binding to lncRNA-FENDRR and down- regulating lncRNA-FENDRR expression.
3.2.2 Function of m6A on ncRNA in Endometrial Cancer
Abnormal regulation of ncRNA has been shown to be closely associated with the progression of endometrial cancer. At present, the study on the relationship between m6A and ncRNA in endometrial cancer is still in the early stage. Shen et al. studied 60 cases of endometrial carcinoma from tumor tissues, cell lines, and xenograft mouse models. He found that the LncRNA FENDRR expression level decreased and the m6A methylation level increased in the cancer tissues of patients with endometrial cancer. In vitro experiments showed that YTHDF2 could recognize the abundance of m6A modified LncRNA FENDRR in endometrial cancer cells and promote its degradation. Overexpression of LncRNA FENDRR inhibited the proliferation and promoted apoptosis of HEC-1B cells by reducing the mRNA level of SRY-related HMG box transcription factor 4 (SOX4) protein. In vivo experiments confirmed that LncRNA FENDRR overexpression inhibited the growth of endometrial cancer cells. Therefore, in endometrial cancer, the m6A modification level of lncRNA FENDRR is increased, and YTHDF2 is recruited to promote the degradation of FENDRR. Subsequently, the downregulation of FENDRR leads to the accumulation of SOX4 protein, thereby promoting the proliferation of EEC cells (92).
3.2.3 Prognostic Effect of m6A RNA Methylation Regulators on Endometrial Cancer
Endometrial cancer is the sixth most common cancer in women worldwide. The expression level of m6A regulatory factor may be used for stratification of cancer prognosis, including endometrial cancer. Wang et al. determined, by analyzing matched clinical information from the TCGA database of endometrial cancer patients, that replication number variations (CNVs) in the m6A regulatory gene had a significant negative impact on patient survival. Univariate Cox regression analysis showed that IGF2BP1, KIAA1429, IGF2BP3, YTHDF3, and IGF2BP2 were closely related to the survival and prognosis of endometrial cancer patients. Among them, IGF2BP3, KIAA1429, and IGF2BP1 can effectively predict the prognosis of patients (93). In addition, Pang et al. found that IGF2BP1 and YTHDF3 had a strong ability to stratify the prognosis of different endometrial cancer patients (94). Song et al. downloaded the human endometrial carcinoma m6A sequencing dataset “GSE93911” from the comprehensive gene expression database. A total of 181 genes with significantly differentially expressed and differentially methylated loci in endometrial carcinoma were screened. Among them, 31 genes were associated with survival, and 11 genes were identified as risk prognosis models, including GDF7, BNC2, SLC8A1, B4GALNT3, DHCR24, ESRP1, HOXB9, IGSF9, KIAA1324, MSnX1, and PHGDH (95). In addition, Ma et al. analyzed the sequences, copy number variation, and clinical data obtained from the TCGA database. The changes in the m6A RNA methylation regulators are closely related to the clinicopathological stage and prognosis of endometrial carcinoma. Among them, ZC3H13, YTHDC1, and METTTL14 have been identified as potential markers for the diagnosis and prognosis of endometrial cancer. TIMER algorithm suggested that immune cell infiltration was related to the expression changes of ZC3H13, YTHDC1, and METTTL14. Meanwhile, ZC3H13 or YTHDC1 knockdown can promote the proliferation and invasion of endometrial cancer cells (96). Zhai et al. analyzed 406 cases of endometrial adenocarcinoma and 19 controls using a TCGA dataset. FTO, RBM15, and YTHDF1 were identified as independent prognostic markers for endometrial cancer, and FTO and RBM15 were differentially expressed between endometrial adenocarcinoma and hyperplasia. These data suggest that FTO, RBM15, and YTHDF1 are critical in the progression and prognosis of endometrial cancer (82, 97). Interestingly, Zhang et al. found that CpG sites located at the m6A regulatory site may be considered a potential prognostic feature of endometrial cancer patients (98). In a more detailed study, 19 m6A RNA methylation regulators were abnormally expressed in endometrial carcinoma. Univariate and multivariate Cox regression analyses showed that age, grade, and risk score were independent risk factors. High FTO expression was associated with poor overall survival (99).
3.3 m6A in Ovarian Cancer
3.3.1 Function of m6A on mRNA in Ovarian Cancer
Ovarian cancer is one of the most deadly gynecological malignancies. It often leads to poor prognosis due to the insidious onset and lack of effective early detection indicators (100). YTHDF1 is a member of the YT521-B homologous domain (YTH) protein family. The protein recognizes m6A through a conserved aromatic cage in its YTH domain and mediates gene regulation at the post-transcriptional level (52). Hao et al. found that TRIM29 expression increased at the translational level in cisplatin-resistant ovarian cancer cells and clinical tissues. Increased TRIM29 expression is associated with poor prognosis in patients with ovarian cancer. In addition, YTHDF1’s recruitment of m6A-modified TRIM29 is involved in promoting TRIM29 translation in cisplatin-resistant ovarian cancer cells. Knockout of YTHDF1 inhibits the tumor stem cell-like characteristics of cisplatin-resistant ovarian cancer cells, which can be salvaged by ectopic expression of TRIM29 (101). YTHDF1, as an upstream molecule of TRIM29, can recognize its 3’UTR and promote its translation in ovarian cancer. Thus, TRIM29 is expected to be a potential therapeutic target for ovarian cancer. In addition, YTHDF1 can promote ovarian cancer progression by increasing EIF3C translation. YTHDF1, as a direct target, binds to m6A-modified EIF3C mRNA to increase EIF3C translation in an m6A-dependent manner and simultaneously promotes overall translation output. Thus, the occurrence and metastasis of ovarian cancer are promoted (102). Therefore, the YTHDF1-EIF3C axis is expected to be a target for the development of cancer treatment drugs.
Another study showed that the RNA demethylase ALKBH5 was upregulated in ovarian cancer tissues but lower in ovarian cancer cell lines than in normal ovarian cell lines. Interestingly, the molecular function toll-like receptor (TLR4) in the tumor microenvironment also showed the same expression trend. NANOG is a target of ALKBH5-mediated m6A modification. The expression of NANOG increased after mRNA demethylation, thereby enhancing the aggressiveness of ovarian cancer cells. In addition, the high expression of TLR4 activated the NF-Kappa B pathway, upregulated the expression of ALKBH5, and increased the level of m6A and the expression of NANOG, all of which contributed to the occurrence of ovarian cancer (103). FBW7, a tumor suppressor, is a substrate recognition component of the SCF e3-ubiquitin ligase complex, which mediates protein degradation of various carcinogenic proteins. Xu et al. used MeRIP-Seq and RNA-Seq to evaluate downstream targets of YTHDF2. They found that FBW7 was significantly down-regulated in ovarian cancer tissues, and its high expression was associated with a good prognosis and increased m6A modification. FBW7 counteracts the tumor-promoting effect of YTHDF2 by inducing proteasome degradation of YTHDF2 in ovarian cancer. In addition, YTHDF2 can regulate the turnover of m6A-modified mRNA, including pro-apoptotic gene BMF. Therefore, FBW7 inhibits tumor growth and progression by antagonizing the YTHDF2-mediated attenuation of BMF mRNA in ovarian cancer tissues (28). FTO, an m6A demethylase, plays an important role in the progression of ovarian cancer. One study found that FTO enhanced the second messenger 3’, 5’ -cyclic adenosine phosphate (cAMP) signaling by decreasing the m6A modification level of 3’ UTR and the mRNA stability of two phosphodiesterase genes (PDE1C and PDE4B), inhibiting the dry character of ovarian cancer cells. Therefore, FTO plays a tumor suppressor role in ovarian cancer by inhibiting cAMP signaling (104).
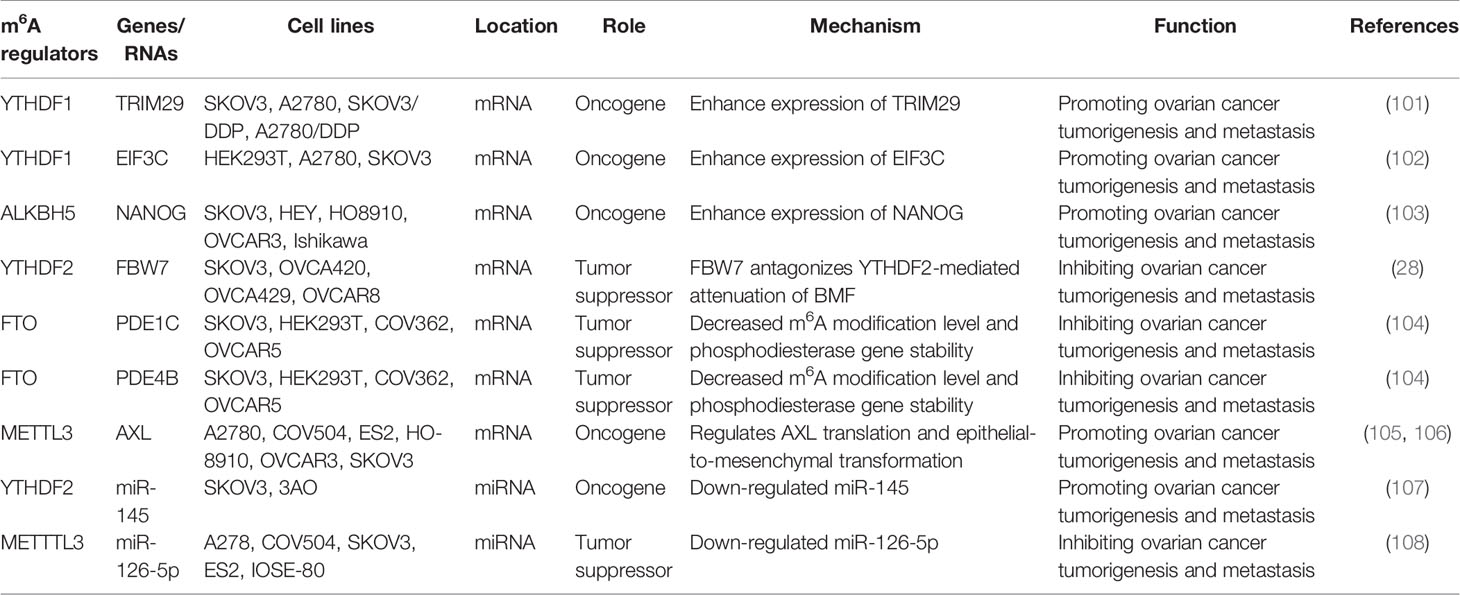
Progress has also been made in the role of methyltransferase in ovarian cancer. For example, methyltransferase METTL3 not only promotes the growth and invasion of ovarian cancer by regulating AXL translation and epithelial-to-mesenchymal transformation (105), but also plays a carcinogenic role in the progression of ovarian cancer cells by regulating the phosphorylation of AKT and the expression of Cyclin D1, a downstream effector (106). Thus, METTL3 may be a new prognostic and/or therapeutic target for ovarian cancer. The roles of different m6A regulators in regulating RNAs in ovarian cancer are shown in Figure 4 and Table 3.
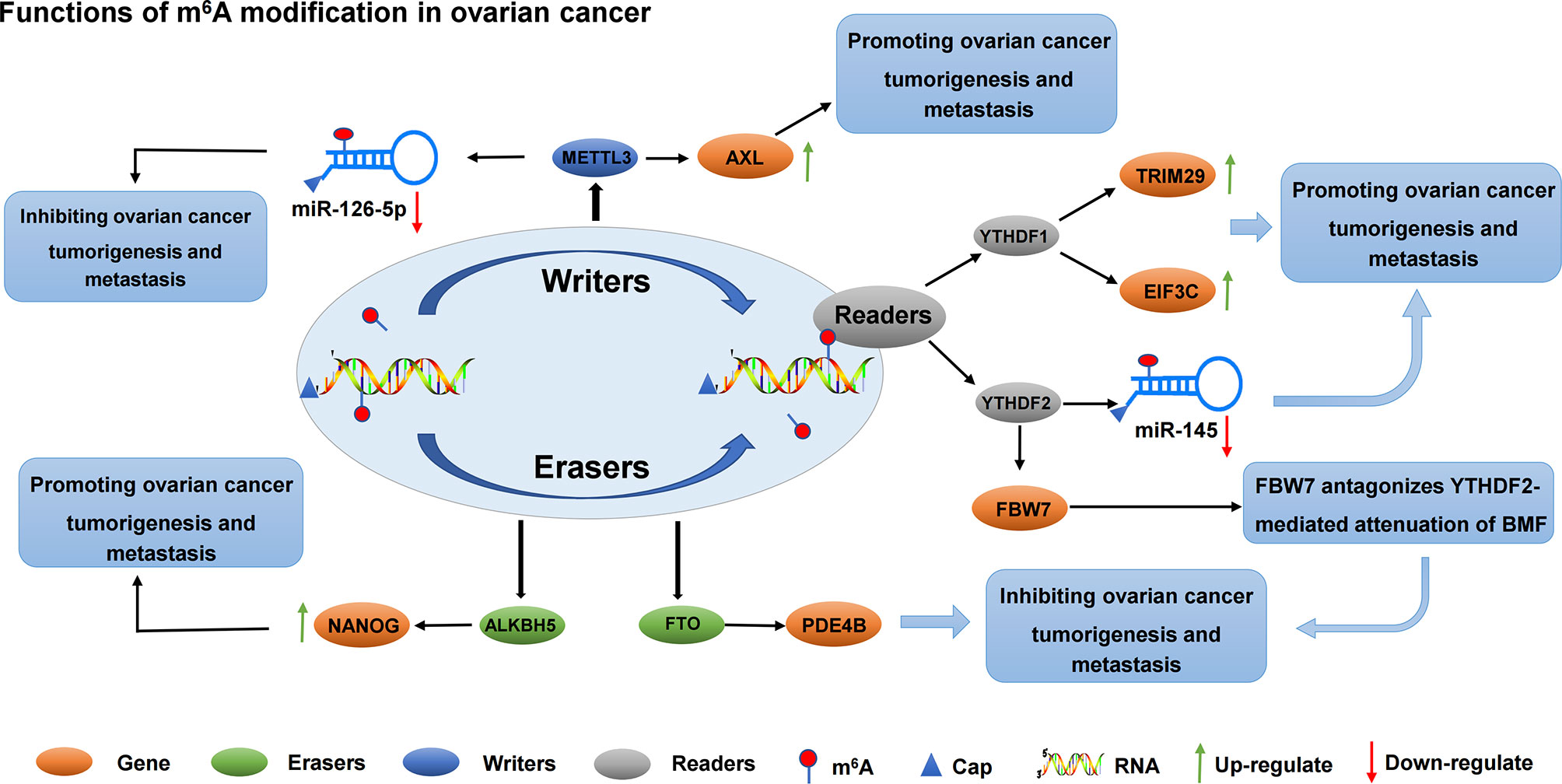
Figure 4 The roles of different m6A regulators in regulating RNAs in ovarian cancer. m6A methylation regulators METTL3, YTHDF1, YTHDF2, FTO and ALKBH5 promote/inhibit the invasion and metastasis of ovarian cancer by binding to mRNA and regulating mRNA expression. In addition, METTL3 regulates the expression of miR-126-5P and inhibits the proliferation and metastasis of ovarian cancer. YTHDF2 regulates the expression of miR-145 and promotes the proliferation and metastasis of ovarian cancer.
3.3.2 Function of m6A on ncRNA in Ovarian Cancer
RNA methylation can be methylated at the RNA level, which is an extremely important epigenetic modification. Methylation of RNA m6A was correlated with miRNA. On the one hand, the target sites of miRNA showed m6A enrichment, and miRNA positively regulated m6A modification activity. On the other hand, miRNA synthesis relies on m6A methylation modification (109). Li et al. found, by studying how YTHDF2 and miR-145 regulate the progression of ovarian cancer through m6A modification, that YTHDF2 was significantly up-regulated in ovarian cancer tissues compared with normal ovarian tissues. Meanwhile, YTHDF2 significantly promoted the proliferation and migration of ovarian cancer cell lines, and reduced the modification level of m6A mRNA. The expression level of miR-145 in ovarian cancer tissues and cells was negatively correlated with that of YTHDF2, which is the direct target gene of miR-145. Key crosstalk occurred between miR-145 and YTHDF2 through a double negative feedback loop. The overexpression of YTHDF2 rescues the decreased proliferation and migration of miR-145-induced ovarian cancer cells, suggesting a new target for the treatment of ovarian cancer (107). Another miRNA, miR-126-5p, is up-regulated in ovarian cancer. The overexpression of miR-126-5P can promote proliferation, migration, and invasion of ovarian cancer cells and inhibit apoptosis. In addition, miR-126-5p activates the PI3K/Akt/mTOR pathway by targeting PTEN. Moreover, RNA methyltransferase METTL3 promoted the maturation of miR-126-5p through m6A modification of pri-miR-126-5p. Finally, in vitro and in vivo experiments confirmed that METTL3 silencing blocks the PI3K/AKT/mTOR pathway by disrupting miR-126-5P targeted inhibition of PTEN, thereby hindering ovarian cancer progression and tumorgenesis. Therefore, the down-regulation of METTL3 can inhibit the up-regulation of PTEN by miR-126-5p, and prevent the activation of PI3K/AKT/mTOR pathway, inhibiting the occurrence and development of ovarian cancer (108). The role and possible mechanisms of circRNAs in autophagy in ovarian cancer have not been systematically studied. Zhang et al. screened circRNA, miRNA, and mRNA expression profiles of Torin 1-induced ovarian cancer cells. They found that circRNA circRAB11FIP1 was up-regulated in ovarian cancer cells, and silencing circRAB11FIP1 could inhibit autophagy of ovarian cancer cells. CIRCRAB11FIP1-induced autophagy accelerated the proliferation and invasion of ovarian cancer cells. In addition, circRAB11FIP1 directly binds to Mir-129 and regulates its targets ATG7 and ATG14. CircRAB11FIP1 mediates ATG5 and ATG7 mRNA expression levels depending on m6A modification (110). Few studies on lncRNAs in ovarian cancer have also been conducted. In recent years, studies have found that lncRNA is an important functional regulator in ovarian cancer. Wang et al. found that lncRNA RHPT1-AS1 was up-regulated in ovarian cancer tissues and was closely associated with poor prognosis. However, m6A modification improved the stability of RHPT1-AS1 methylated transcription by reducing RNA degradation, leading to the up-regulation of RHPT1-AS1 expression in ovarian cancer and promoting the proliferation and metastasis of ovarian cancer (111).
3.3.3 Prognostic Effect of m6A RNA Methylation Regulators on Ovarian Cancer
m6A RNA methylation is involved in the initiation and progression of various cancers. Therefore, m6A RNA methylation regulators are greatly important in tumor prognosis. Fan et al. analyzed the prognostic value of the transcription levels of 18 m6A RNA methylation regulators in ovarian cancer and found that IGF2BP1, VIRMA, and ZC3H13 predicted the highest prognostic score of ovarian cancer. Therefore, the authors suggest that IGF2BP1, VIRMA, and ZC3H13 mRNA levels are important factors in predicting prognosis and developing treatment strategies (112). In addition, Han et al. downloaded the mutation and copy number variation (CNV) data from 579 ovarian cancer patients from TCGA database and analyzed gene expression and prognostic value using integrated bioinformatics. Bioinformatics and Cox multivariate analysis showed the significant correlation between high expression of WTAP and ovarian cancer prognosis (113). Meanwhile, analysis of other gene sets found that the prognosis of ovarian cancer was associated with HNRNPA2B1, KIAA1429, and WTAP (114). Na et al. identified NEBL, PDGFRA, WDR91, and ZBTB4 genes as potential independent prognostic risk characteristics of ovarian cancer (115).
4 Conclusions and Perspectives
RNA m6A modification, as a hotspot of epigenetics research, plays a crucial role in the post-transcriptional regulation of gene expression; it has attracted increased attention. It is also involved in various biological processes and disease progression. RNA m6A modification plays an important role in promoting or inhibiting the growth, proliferation, migration, invasion, specific metastasis, drug resistance, and prognosis of gynecological cancers through three effector factors, namely, writer, erasers, and readers. From the viewpoint of the epitome, the tissue specificity and uneven distribution of m6A modification provide new directions for understanding the pathogenesis of multiple diseases, especially tumors. m6A modification is a “double-edged sword”, promoting or inhibiting the occurrence and development of tumor mainly by regulating the mRNA level of oncogene or tumor suppressor gene. The role of m6A modification in gynecological cancer is further clarified with the deepening of the research on the network mechanism of m6A modification regulation.
At present, studies on the biological effects of m6A modification on gynecological cancers have made some progress, but some problems should still be further studied and solved. Multiple studies have shown that m6A RNA methylation regulators have the potential for prognostic assessment and as biomarkers for early diagnosis of gynecological cancers. For example, METTL3, ZC3H13, YTHDC1, and YTHDF1 have been found to be prognostic markers of cervical cancer (84). IGF2BP3, KIAA1429, IGF2BP1, YTHDF3, ZC3H13, YTHDC1, and METTL14 can be used as prognostic markers of endometrial cancer (93, 94). HNRNPA2B1, KIAA1429, and WTAP can be used as prognostic markers of ovarian cancer (114). However, these studies are based only on systematic analyses of public databases. The prognostic ability of m6A RNA methylation regulators in gynecological cancers remains limited due to the difficulty of obtaining sufficient detection samples. Therefore, large-scale experimental verification and clinical trials on m6A modification should be conducted to remedy this defect in future studies. In addition, most current studies on m6A modification in gynecological cancer are limited to the mechanism of action in gynecological cancer cells. Thus, more translational studies are required in the future to further clarify the use of m6A alone or in combination with other therapies for gynecological cancers for effective application to clinical treatment. Finally, the immune system is the host’s defense system against infection and disease. Meanwhile, immunotherapy is a new cancer treatment strategy, which has been widely used to treat various solid tumors, including gastrointestinal tumors and gynecological tumors (116, 117). In recent years, m6A regulatory factors have been widely studied in tumor immunotherapy and immune evasion. Moreover, tumor immunotherapy is the most promising therapeutic strategy, and m6A modification-mediated immune invasion becomes a hotspot in the study of the pathogenesis and prognosis of gynecological tumors. However, the study of immune invasion mediated by m6A modification in gynecological tumors is still in its infancy, and m6A modification is expected to make new breakthroughs in this field in future studies.
Author Contributions
WH, FK, and KW contributed to conceive and design the study. RL and XC performed the systematic searching. WH and RL extracted the data. FK and KW wrote the manuscript. XC and WH supervised the manuscript. All of the authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Rahimian N, Razavi ZS, Aslanbeigi F, Mirkhabbaz AM, Piroozmand H, Shahrzad MK, et al. Non-Coding RNAs Related to Angiogenesis in Gynecological Cancer. Gynecol Oncol (2021) 161(3):896–912. doi: 10.1016/j.ygyno.2021.03.020
2. Di Fiore R, Suleiman S, Pentimalli F, O’Toole SA, O’Leary JJ, Ward MP, et al. Could MicroRNAs Be Useful Tools to Improve the Diagnosis and Treatment of Rare Gynecological Cancers? A Brief Overview. Int J Mol Sci (2021) 22(8):3822–47. doi: 10.3390/ijms22083822
3. Xie W, Sun H, Li X, Lin F, Wang Z, Wang X. Ovarian Cancer: Epigenetics, Drug Resistance, and Progression. Cancer Cell Int (2021) 21(1):434. doi: 10.1186/s12935-021-02136-y
4. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
5. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
6. Laios A, Duarte Portela S, Papadopoulou A, Gallos ID, Otify M, Ind T. Ovarian Transposition and Cervical Cancer. Best Pract Res Clin Obstet Gynaecol (2021) 75:37–53. doi: 10.1016/j.bpobgyn.2021.01.013
7. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global Cancer Statistics, 2012. CA Cancer J Clin (2015) 65(2):87–108. doi: 10.3322/caac.21262
8. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical Cancer. Lancet (2019) 393(10167):169–82. doi: 10.1016/s0140-6736(18)32470-x
9. World Health Organization. Cervical Cancer (2021). Available at: https://www.who.int/health-topics/cervical-cancer#tab=tab_2.
10. Koh WJ, Abu-Rustum NR, Bean S, Bradley K, Campos SM, Cho KR, et al. Cervical Cancer, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw (2019) 17(1):64–84. doi: 10.6004/jnccn.2019.0001
11. Wang JY, Chen LJ. The Role of miRNAs in the Invasion and Metastasis of Cervical Cancer. Biosci Rep (2019) 39(3):1–16. doi: 10.1042/bsr20181377
12. Xu F, Li Y, Fan L, Ma J, Yu L, Yi H, et al. Preoperative SCC-Ag and Thrombocytosis as Predictive Markers for Pelvic Lymphatic Metastasis of Squamous Cervical Cancer in Early FIGO Stage. J Cancer (2018) 9(9):1660–6. doi: 10.7150/jca.24049
13. Nanthamongkolkul K, Hanprasertpong J. Predictive Factors of Pelvic Lymph Node Metastasis in Early-Stage Cervical Cancer. Oncol Res Treat (2018) 41(4):194–8. doi: 10.1159/000485840
14. Wright JD, Huang Y, Ananth CV, Tergas AI, Duffy C, Deutsch I, et al. Influence of Treatment Center and Hospital Volume on Survival for Locally Advanced Cervical Cancer. Gynecol Oncol (2015) 139(3):506–12. doi: 10.1016/j.ygyno.2015.07.015
15. Vaccarella S, Lortet-Tieulent J, Plummer M, Franceschi S, Bray F. Worldwide Trends in Cervical Cancer Incidence: Impact of Screening Against Changes in Disease Risk Factors. Eur J Cancer (2013) 49(15):3262–73. doi: 10.1016/j.ejca.2013.04.024
16. Brooks RA, Fleming GF, Lastra RR, Lee NK, Moroney JW, Son CH, et al. Current Recommendations and Recent Progress in Endometrial Cancer. CA Cancer J Clin (2019) 69(4):258–79. doi: 10.3322/caac.21561
17. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated Genomic Characterization of Endometrial Carcinoma. Nature (2013) 497(7447):67–73. doi: 10.1038/nature12113
18. Di Tucci C, Capone C, Galati G, Iacobelli V, Schiavi MC, Di Donato V, et al. Immunotherapy in Endometrial Cancer: New Scenarios on the Horizon. J Gynecol Oncol (2019) 30(3):e46. doi: 10.3802/jgo.2019.30.e46
19. Chen W, Zhang Z, Fang X, Xiong L, Wen Y, Zhou J, et al. Prognostic Value of the ALBI Grade Among Patients With Single Hepatocellular Carcinoma Without Macrovascular Invasion. Med (Baltimore) (2021) 100(24):e26265. doi: 10.1097/md.0000000000026265
20. Desrosiers R, Friderici K, Rottman F. Identification of Methylated Nucleosides in Messenger RNA From Novikoff Hepatoma Cells. Proc Natl Acad Sci USA (1974) 71(10):3971–5. doi: 10.1073/pnas.71.10.3971
21. Huang H, Weng H, Chen J. The Biogenesis and Precise Control of RNA M(6)A Methylation. Trends Genet (2020) 36(1):44–52. doi: 10.1016/j.tig.2019.10.011
22. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The Interplay Between M6a RNA Methylation and Noncoding RNA in Cancer. J Hematol Oncol (2019) 12(1):121. doi: 10.1186/s13045-019-0805-7
23. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic Transcriptomic M(6)A Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cell Res (2018) 28(6):616–24. doi: 10.1038/s41422-018-0040-8
24. Zhao W, Qi X, Liu L, Ma S, Liu J, Wu J. Epigenetic Regulation of M(6)A Modifications in Human Cancer. Mol Ther Nucleic Acids (2020) 19:405–12. doi: 10.1016/j.omtn.2019.11.022
25. Wang T, Kong S, Tao M, Ju S. The Potential Role of RNA N6-Methyladenosine in Cancer Progression. Mol Cancer (2020) 19(1):88. doi: 10.1186/s12943-020-01204-7
26. Wang H, Luo Q, Kang J, Wei Q, Yang Y, Yang D, et al. YTHDF1 Aggravates the Progression of Cervical Cancer Through M(6)A-Mediated Up-Regulation of RANBP2. Front Oncol (2021) 11:650383. doi: 10.3389/fonc.2021.650383
27. Pu X, Gu Z, Gu Z. ALKBH5 Regulates IGF1R Expression to Promote the Proliferation and Tumorigenicity of Endometrial Cancer. J Cancer (2020) 11(19):5612–22. doi: 10.7150/jca.46097
28. Xu F, Li J, Ni M, Cheng J, Zhao H, Wang S, et al. FBW7 Suppresses Ovarian Cancer Development by Targeting the N(6)-Methyladenosine Binding Protein YTHDF2. Mol Cancer (2021) 20(1):45. doi: 10.1186/s12943-021-01340-8
29. Shen H, Lan Y, Zhao Y, Shi Y, Jin J, Xie W. The Emerging Roles of N6-Methyladenosine RNA Methylation in Human Cancers. biomark Res (2020) 8:24. doi: 10.1186/s40364-020-00203-6
30. Hu Y, Ouyang Z, Sui X, Qi M, Li M, He Y, et al. Oocyte Competence is Maintained by M(6)A Methyltransferase KIAA1429-Mediated RNA Metabolism During Mouse Follicular Development. Cell Death Differ (2020) 27(8):2468–83. doi: 10.1038/s41418-020-0516-1
31. Bansal H, Yihua Q, Iyer SP, Ganapathy S, Proia DA, Penalva LO, et al. WTAP is a Novel Oncogenic Protein in Acute Myeloid Leukemia. Leukemia (2014) 28(5):1171–4. doi: 10.1038/leu.2014.16
32. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat Chem Biol (2014) 10(2):93–5. doi: 10.1038/nchembio.1432
33. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3
34. Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell (2016) 63(2):306–17. doi: 10.1016/j.molcel.2016.05.041
35. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA Cloning of the AdoMet-Binding Subunit of the Human mRNA (N6-Adenosine)-Methyltransferase. Rna (1997) 3(11):1233–47.
36. Wang X, Tian L, Li Y, Wang J, Yan B, Yang L, et al. RBM15 Facilitates Laryngeal Squamous Cell Carcinoma Progression by Regulating TMBIM6 Stability Through IGF2BP3 Dependent. J Exp Clin Cancer Res (2021) 40(1):80. doi: 10.1186/s13046-021-01871-4
37. Zhu D, Zhou J, Zhao J, Jiang G, Zhang X, Zhang Y, et al. ZC3H13 Suppresses Colorectal Cancer Proliferation and Invasion via Inactivating Ras-ERK Signaling. J Cell Physiol (2019) 234(6):8899–907. doi: 10.1002/jcp.27551
38. Satterwhite ER, Mansfield KD. RNA Methyltransferase METTL16: Targets and Function. Wiley Interdiscip Rev RNA (2021):e1681. doi: 10.1002/wrna.1681
39. Cao M, Zheng H, Tan X, Xu W, Rui Y, Li L, et al. Upregulation of CBLL1 in Rat Brain Cortex After Lipopolysaccharide Treated. J Mol Histol (2013) 44(2):135–45. doi: 10.1007/s10735-012-9467-2
40. Huo FC, Zhu ZM, Pei DS. N(6) -Methyladenosine (M(6) A) RNA Modification in Human Cancer. Cell Prolif (2020) 53(11):e12921. doi: 10.1111/cpr.12921
41. Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, et al. M(6)A Demethylase ALKBH5 Inhibits Tumor Growth and Metastasis by Reducing YTHDFs-Mediated YAP Expression and Inhibiting miR-107/LATS2-Mediated YAP Activity in NSCLC. Mol Cancer (2020) 19(1):40. doi: 10.1186/s12943-020-01161-1
42. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. M(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-Like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell (2017) 31(4):591–606.e6. doi: 10.1016/j.ccell.2017.02.013
43. Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, et al. M(6)A Demethylase ALKBH5 Inhibits Pancreatic Cancer Tumorigenesis by Decreasing WIF-1 RNA Methylation and Mediating Wnt Signaling. Mol Cancer (2020) 19(1):3. doi: 10.1186/s12943-019-1128-6
44. Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, et al. ALKBH5 Promotes Invasion and Metastasis of Gastric Cancer by Decreasing Methylation of the lncRNA Neat1. J Physiol Biochem (2019) 75(3):379–89. doi: 10.1007/s13105-019-00690-8
45. Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 Inhibited Autophagy of Epithelial Ovarian Cancer Through miR-7 and BCL-2. J Exp Clin Cancer Res (2019) 38(1):163. doi: 10.1186/s13046-019-1159-2
46. Qiu X, Yang S, Wang S, Wu J, Zheng B, Wang K, et al. M6A Demethylase ALKBH5 Regulates PD-L1 Expression and Tumor Immunoenvironment in Intrahepatic Cholangiocarcinoma. Cancer Res (2021) 81(18):4778–93. doi: 10.1158/0008-5472.can-21-0468
47. Chen J, Du B. Novel Positioning From Obesity to Cancer: FTO, an M(6)A RNA Demethylase, Regulates Tumour Progression. J Cancer Res Clin Oncol (2019) 145(1):19–29. doi: 10.1007/s00432-018-2796-0
48. Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, et al. The Obesity-Associated FTO Gene Encodes a 2-Oxoglutarate-Dependent Nucleic Acid Demethylase. Science (2007) 318(5855):1469–72. doi: 10.1126/science.1151710
49. Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, et al. FTO-Dependent Demethylation of N6-Methyladenosine Regulates mRNA Splicing and is Required for Adipogenesis. Cell Res (2014) 24(12):1403–19. doi: 10.1038/cr.2014.151
50. Zheng F, Du F, Zhao J, Wang X, Si Y, Jin P, et al. The Emerging Role of RNA N6-Methyladenosine Methylation in Breast Cancer. biomark Res (2021) 9(1):39. doi: 10.1186/s40364-021-00295-8
51. Geuens T, Bouhy D, Timmerman V. The hnRNP Family: Insights Into Their Role in Health and Disease. Hum Genet (2016) 135(8):851–67. doi: 10.1007/s00439-016-1683-5
52. Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J. Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins. J Biol Chem (2015) 290(41):24902–13. doi: 10.1074/jbc.M115.680389
53. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-Methyladenosine Modulates Messenger RNA Translation Efficiency. Cell (2015) 161(6):1388–99. doi: 10.1016/j.cell.2015.05.014
54. Bailey AS, Batista PJ, Gold RS, Chen YG, de Rooij DG, Chang HY, et al. The Conserved RNA Helicase YTHDC2 Regulates the Transition From Proliferation to Differentiation in the Germline. Elife (2017) 6:e26116–44. doi: 10.7554/eLife.26116
55. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-Methyladenosine and Its Role in Cancer. Mol Cancer (2019) 18(1):176. doi: 10.1186/s12943-019-1109-9
56. Huang J, Yin P. Structural Insights Into N(6)-Methyladenosine (M(6)A) Modification in the Transcriptome. Genomics Proteomics Bioinf (2018) 16(2):85–98. doi: 10.1016/j.gpb.2018.03.001
57. Wang S, Chai P, Jia R, Jia R. Novel Insights on M(6)A RNA Methylation in Tumorigenesis: A Double-Edged Sword. Mol Cancer (2018) 17(1):101. doi: 10.1186/s12943-018-0847-4
58. He L, Li J, Wang X, Ying Y, Xie H, Yan H, et al. The Dual Role of N6-Methyladenosine Modification of RNAs is Involved in Human Cancers. J Cell Mol Med (2018) 22(10):4630–9. doi: 10.1111/jcmm.13804
59. Huang H, Weng H, Chen J. M(6)A Modification in Coding and Non-Coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell (2020) 37(3):270–88. doi: 10.1016/j.ccell.2020.02.004
60. Li Z, Luo Q, Wang H, Liu Y, Feng X, Li Z, et al. Knockdown of YTH N(6)-Methyladenosine RNA Binding Protein 2 (YTHDF2) Inhibits Cell Proliferation and Promotes Apoptosis in Cervical Cancer Cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi (2020) 36(3):255–63.
61. Wang Q, Guo X, Li L, Gao Z, Su X, Ji M, et al. N(6)-Methyladenosine METTL3 Promotes Cervical Cancer Tumorigenesis and Warburg Effect Through YTHDF1/HK2 Modification. Cell Death Dis (2020) 11(10):911. doi: 10.1038/s41419-020-03071-y
62. Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, et al. FTO Regulates the Chemo-Radiotherapy Resistance of Cervical Squamous Cell Carcinoma (CSCC) by Targeting β-Catenin Through mRNA Demethylation. Mol Carcinog (2018) 57(5):590–7. doi: 10.1002/mc.22782
63. Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J, et al. N(6)-Methyladenosine Regulates Glycolysis of Cancer Cells Through PDK4. Nat Commun (2020) 11(1):2578. doi: 10.1038/s41467-020-16306-5
64. Ji F, Lu Y, Chen S, Yu Y, Lin X, Zhu Y, et al. IGF2BP2-Modified Circular RNA Circarhgap12 Promotes Cervical Cancer Progression by Interacting M(6)A/FOXM1 Manner. Cell Death Discovery (2021) 7(1):215. doi: 10.1038/s41420-021-00595-w
65. Chen Z, Ling K, Zhu Y, Deng L, Li Y, Liang Z. Circ0000069 Promotes Cervical Cancer Cell Proliferation and Migration by Inhibiting miR-4426. Biochem Biophys Res Commun (2021) 551:114–20. doi: 10.1016/j.bbrc.2021.03.020
66. Wang X, Zhang J, Wang Y. Long Noncoding RNA GAS5-AS1 Suppresses Growth and Metastasis of Cervical Cancer by Increasing GAS5 Stability. Am J Transl Res (2019) 11(8):4909–21.
67. Zhang Y, Wang D, Wu D, Zhang D, Sun M. Long Noncoding RNA KCNMB2-AS1 Stabilized by N(6)-Methyladenosine Modification Promotes Cervical Cancer Growth Through Acting as a Competing Endogenous RNA. Cell Transplant (2020) 29:963689720964382. doi: 10.1177/0963689720964382
68. Yang Z, Ma J, Han S, Li X, Guo H, Liu D. ZFAS1 Exerts an Oncogenic Role via Suppressing miR-647 in an M(6)A-Dependent Manner in Cervical Cancer. Onco Targets Ther (2020) 13:11795–806. doi: 10.2147/ott.s274492
69. Huang C, Liang J, Lin S, Wang D, Xie Q, Lin Z, et al. N(6)-Methyladenosine Associated Silencing of miR-193b Promotes Cervical Cancer Aggressiveness by Targeting Ccnd1. Front Oncol (2021) 11:666597. doi: 10.3389/fonc.2021.666597
70. Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell (2018) 71(3):428–42. doi: 10.1016/j.molcel.2018.06.034
71. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a Large Class of Animal RNAs With Regulatory Potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
72. Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie CH. CircRNAs and Cancer: Biomarkers and Master Regulators. Semin Cancer Biol (2019) 58:90–9. doi: 10.1016/j.semcancer.2018.12.002
73. D’Angelo E, Agostini M. Long non-Coding RNA and Extracellular Matrix: The Hidden Players in Cancer-Stroma Cross-Talk. Noncoding RNA Res (2018) 3(4):174–7. doi: 10.1016/j.ncrna.2018.08.002
74. Meng Q, Zhang R, Ding W, Mao B. Long Noncoding RNA ZFAS1 Promotes Cell Proliferation and Tumor Growth by Upregulating LIN28 in Cervical Carcinoma. Minerva Med (2020) 111(5):511–4. doi: 10.23736/s0026-4806.19.06123-8
75. Ji F, Lu Y, Chen S, Lin X, Yu Y, Zhu Y, et al. M(6)A Methyltransferase METTL3-Mediated lncRNA FOXD2-AS1 Promotes the Tumorigenesis of Cervical Cancer. Mol Ther Oncolytics (2021) 22:574–81. doi: 10.1016/j.omto.2021.07.004
76. Kong F, Zou H, Liu X, He J, Zheng Y, Xiong L, et al. miR-7112-3p Targets PERK to Regulate the Endoplasmic Reticulum Stress Pathway and Apoptosis Induced by Photodynamic Therapy in Colorectal Cancer CX-1 Cells. Photodiagn Photodyn Ther (2020) 29:101663. doi: 10.1016/j.pdpdt.2020.101663
77. Mayadev JS, Enserro D, Lin YG, Da Silva DM, Lankes HA, Aghajanian C, et al. Sequential Ipilimumab After Chemoradiotherapy in Curative-Intent Treatment of Patients With Node-Positive Cervical Cancer. JAMA Oncol (2020) 6(1):92–9. doi: 10.1001/jamaoncol.2019.3857
78. Lin C, Slama J, Gonzalez P, Goodman MT, Xia N, Kreimer AR, et al. Cervical Determinants of Anal HPV Infection and High-Grade Anal Lesions in Women: A Collaborative Pooled Analysis. Lancet Infect Dis (2019) 19(8):880–91. doi: 10.1016/s1473-3099(19)30164-1
79. Kong FH, Miao XY, Zou H, Xiong L, Wen Y, Chen B, et al. End-Stage Liver Disease Score and Future Liver Remnant Volume Predict Post-Hepatectomy Liver Failure in Hepatocellular Carcinoma. World J Clin cases (2019) 7(22):3734–41. doi: 10.12998/wjcc.v7.i22.3734
80. Kandimalla R, Gao F, Li Y, Huang H, Ke J, Deng X, et al. RNAMethyPro: A Biologically Conserved Signature of N6-Methyladenosine Regulators for Predicting Survival at Pan-Cancer Level. NPJ Precis Oncol (2019) 3:13. doi: 10.1038/s41698-019-0085-2
81. Kong F, Liu X, Zhou Y, Hou X, He J, Li Q, et al. Downregulation of METTL14 Increases Apoptosis and Autophagy Induced by Cisplatin in Pancreatic Cancer Cells. Int J Biochem Cell Biol (2020) 122:105731. doi: 10.1016/j.biocel.2020.105731
82. Wu H, Dong H, Fu Y, Tang Y, Dai M, Chen Y, et al. Expressions of M6a RNA Methylation Regulators and Their Clinical Predictive Value in Cervical Squamous Cell Carcinoma and Endometrial Adenocarcinoma. Clin Exp Pharmacol Physiol (2021) 48(2):270–8. doi: 10.1111/1440-1681.13412
83. Ni HH, Zhang L, Huang H, Dai SQ, Li J. Connecting METTL3 and Intratumoural CD33(+) MDSCs in Predicting Clinical Outcome in Cervical Cancer. J Transl Med (2020) 18(1):393. doi: 10.1186/s12967-020-02553-z
84. Pan J, Xu L, Pan H. Development and Validation of an M6a RNA Methylation Regulator-Based Signature for Prognostic Prediction in Cervical Squamous Cell Carcinoma. Front Oncol (2020) 10:1444. doi: 10.3389/fonc.2020.01444
85. Zhao BS, Roundtree IA, He C. Post-Transcriptional Gene Regulation by mRNA Modifications. Nat Rev Mol Cell Biol (2017) 18(1):31–42. doi: 10.1038/nrm.2016.132
86. Zhang L, Wan Y, Zhang Z, Jiang Y, Lang J, Cheng W, et al. FTO Demethylates M6a Modifications in HOXB13 mRNA and Promotes Endometrial Cancer Metastasis by Activating the WNT Signalling Pathway. RNA Biol (2021) 18(9):1265–78. doi: 10.1080/15476286.2020.1841458
87. Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. M(6)A mRNA Methylation Regulates AKT Activity to Promote the Proliferation and Tumorigenicity of Endometrial Cancer. Nat Cell Biol (2018) 20(9):1074–83. doi: 10.1038/s41556-018-0174-4
88. Hong L, Pu X, Gan H, Weng L, Zheng Q. YTHDF2 Inhibit the Tumorigenicity of Endometrial Cancer via Downregulating the Expression of IRS1 Methylated With M(6)a. J Cancer (2021) 12(13):3809–18. doi: 10.7150/jca.54527
89. Ghoshal A, Rodrigues LC, Gowda CP, Elcheva IA, Liu Z, Abraham T, et al. Extracellular Vesicle-Dependent Effect of RNA-Binding Protein IGF2BP1 on Melanoma Metastasis. Oncogene (2019) 38(21):4182–96. doi: 10.1038/s41388-019-0797-3
90. Zhang L, Wan Y, Zhang Z, Jiang Y, Gu Z, Ma X, et al. IGF2BP1 Overexpression Stabilizes PEG10 mRNA in an M6a-Dependent Manner and Promotes Endometrial Cancer Progression. Theranostics (2021) 11(3):1100–14. doi: 10.7150/thno.49345
91. Xue T, Liu X, Zhang M, E Q, Liu S, Zou M, et al. PADI2-Catalyzed MEK1 Citrullination Activates ERK1/2 and Promotes IGF2BP1-Mediated SOX2 mRNA Stability in Endometrial Cancer. Adv Sci (Weinh) (2021) 8(6):2002831. doi: 10.1002/advs.202002831
92. Shen J, Feng XP, Hu RB, Wang H, Wang YL, Qian JH, et al. N-Methyladenosine Reader YTHDF2-Mediated Long Noncoding RNA FENDRR Degradation Promotes Cell Proliferation in Endometrioid Endometrial Carcinoma. Lab Invest (2021) 101(6):775–84. doi: 10.1038/s41374-021-00543-3
93. Wang Y, Ren F, Song Z, Wang X, Ma X. Multiomics Profile and Prognostic Gene Signature of M6a Regulators in Uterine Corpus Endometrial Carcinoma. J Cancer (2020) 11(21):6390–401. doi: 10.7150/jca.46386
94. Pang X, Zhang X, Huang Y, Qian S. Development and Validation of M6a Regulators’ Prognostic Significance for Endometrial Cancer. Med (Baltimore) (2021) 100(26):e26551. doi: 10.1097/md.0000000000026551
95. Song K, Xu H, Wang C. The Role of N6-Methyladenosine Methylation in the Progression of Endometrial Cancer. Cancer Biother Radiopharm (2020) 10(14):1–13. doi: 10.1089/cbr.2020.3912
96. Ma J, Yang D, Ma XX. Immune Infiltration-Related N6-Methyladenosine RNA Methylation Regulators Influence the Malignancy and Prognosis of Endometrial Cancer. Aging (Albany NY) (2021) 13(12):16287–315. doi: 10.18632/aging.203157
97. Zhai J, Li S, Li Y, Du Y. Data Mining Analysis of the Prognostic Impact of N(6)-Methyladenosine Regulators in Patients With Endometrial Adenocarcinoma. J Cancer (2021) 12(15):4729–38. doi: 10.7150/jca.50868
98. Zhang X, Pang X, Huang Y, Qian S. A Seven-M6a Regulator-Related CpG Site-Based Prognostic Signature for Endometrial Carcinoma. Med (Baltimore) (2021) 100(29):e26648. doi: 10.1097/md.0000000000026648
99. Zhang Y, Yang Y. Effects of M6a RNA Methylation Regulators on Endometrial Cancer. J Clin Lab Anal (2021) 35(9):e23942. doi: 10.1002/jcla.23942
100. Yang D, Li H, Sun X, Yang S, Wang K, Liu Z. Clinical Usefulness of High Levels of C-Reactive Protein for Diagnosing Epithelial Ovarian Cancer. Sci Rep (2020) 10(1):20056. doi: 10.1038/s41598-020-77167-y
101. Hao L, Wang JM, Liu BQ, Yan J, Li C, Jiang JY, et al. M6a-YTHDF1-Mediated TRIM29 Upregulation Facilitates the Stem Cell-Like Phenotype of Cisplatin-Resistant Ovarian Cancer Cells. Biochim Biophys Acta Mol Cell Res (2021) 1868(1):118878. doi: 10.1016/j.bbamcr.2020.118878
102. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The M6a Reader YTHDF1 Promotes Ovarian Cancer Progression via Augmenting EIF3C Translation. Nucleic Acids Res (2020) 48(7):3816–31. doi: 10.1093/nar/gkaa048
103. Jiang Y, Wan Y, Gong M, Zhou S, Qiu J, Cheng W. RNA Demethylase ALKBH5 Promotes Ovarian Carcinogenesis in a Simulated Tumour Microenvironment Through Stimulating NF-κb Pathway. J Cell Mol Med (2020) 24(11):6137–48. doi: 10.1111/jcmm.15228
104. Huang H, Wang Y, Kandpal M, Zhao G, Cardenas H, Ji Y, et al. FTO-Dependent N (6)-Methyladenosine Modifications Inhibit Ovarian Cancer Stem Cell Self-Renewal by Blocking cAMP Signaling. Cancer Res (2020) 80(16):3200–14. doi: 10.1158/0008-5472.can-19-4044
105. Hua W, Zhao Y, Jin X, Yu D, He J, Xie D, et al. METTL3 Promotes Ovarian Carcinoma Growth and Invasion Through the Regulation of AXL Translation and Epithelial to Mesenchymal Transition. Gynecol Oncol (2018) 151(2):356–65. doi: 10.1016/j.ygyno.2018.09.015
106. Liang S, Guan H, Lin X, Li N, Geng F, Li J. METTL3 Serves an Oncogenic Role in Human Ovarian Cancer Cells Partially via the AKT Signaling Pathway. Oncol Lett (2020) 19(4):3197–204. doi: 10.3892/ol.2020.11425
107. Li J, Wu L, Pei M, Zhang Y. YTHDF2, a Protein Repressed by miR-145, Regulates Proliferation, Apoptosis, and Migration in Ovarian Cancer Cells. J Ovarian Res (2020) 13(1):111. doi: 10.1186/s13048-020-00717-5
108. Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, et al. METTL3-Mediated Maturation of miR-126-5p Promotes Ovarian Cancer Progression via PTEN-Mediated PI3K/Akt/mTOR Pathway. Cancer Gene Ther (2021) 28(3-4):335–49. doi: 10.1038/s41417-020-00222-3
109. Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-Methyladenosine Marks Primary microRNAs for Processing. Nature (2015) 519(7544):482–5. doi: 10.1038/nature14281
110. Zhang Z, Zhu H, Hu J. CircRAB11FIP1 Promoted Autophagy Flux of Ovarian Cancer Through DSC1 and miR-129. Cell Death Dis (2021) 12(2):219. doi: 10.1038/s41419-021-03486-1
111. Wang J, Ding W, Xu Y, Tao E, Mo M, Xu W, et al. Long non-Coding RNA RHPN1-AS1 Promotes Tumorigenesis and Metastasis of Ovarian Cancer by Acting as a ceRNA Against miR-596 and Upregulating LETM1. Aging (Albany NY) (2020) 12(5):4558–72. doi: 10.18632/aging.102911
112. Fan L, Lin Y, Lei H, Shu G, He L, Yan Z, et al. A Newly Defined Risk Signature, Consisting of Three M(6)A RNA Methylation Regulators, Predicts the Prognosis of Ovarian Cancer. Aging (Albany NY) (2020) 12(18):18453–75. doi: 10.18632/aging.103811
113. Han X, Liu J, Cheng G, Cui S. Gene Signatures and Prognostic Values of M6a RNA Methylation Regulators in Ovarian Cancer. Cancer Control (2020) 27(1):1073274820960460. doi: 10.1177/1073274820960460
114. Zhang C, Liu J, Guo H, Hong D, Ji J, Zhang Q, et al. M6a RNA Methylation Regulators Were Associated With the Malignancy and Prognosis of Ovarian Cancer. Bioengineered (2021) 12(1):3159–76. doi: 10.1080/21655979.2021.1946305
115. Na Z, Fan L, Wang X. Gene Signatures and Prognostic Values of N6-Methyladenosine Related Genes in Ovarian Cancer. Front Genet (2021) 12:542457. doi: 10.3389/fgene.2021.542457
116. Lee JC, Green MD, Huppert LA, Chow C, Pierce RH, Daud AI. The Liver-Immunity Nexus and Cancer Immunotherapy. Clin Cancer Res (2021) 28(1):5–12. doi: 10.1158/1078-0432.ccr-21-1193
Keywords: gynecological cancer, N6-methyladenosine (m6A), epigenetics, cervical cancer, endometrial cancer, ovarian cancer
Citation: Huang W, Kong F, Li R, Chen X and Wang K (2022) Emerging Roles of m6A RNA Methylation Regulators in Gynecological Cancer. Front. Oncol. 12:827956. doi: 10.3389/fonc.2022.827956
Received: 02 December 2021; Accepted: 13 January 2022;
Published: 28 January 2022.
Edited by:
Florentia Fostira, National Centre of Scientific Research Demokritos, GreeceReviewed by:
Yanhong Cui, University of Chicago, United StatesParaskevi Apostolou, National Centre of Scientific Research Demokritos, Greece
Copyright © 2022 Huang, Kong, Li, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruolan Li, OTY1ODUyMjc0QHFxLmNvbQ==; Xiang Chen, Y2hlbnhpYW5neHRAY3N1LmVkdS5jbg==; Kunpeng Wang, d2FuZ2twOTY0NEB0enp4eXkuY29t
†These authors have contributed equally to this work and share first authorship
 Wanjun Huang1†
Wanjun Huang1† Fanhua Kong
Fanhua Kong Xiang Chen
Xiang Chen Kunpeng Wang
Kunpeng Wang