- 1Center for Tumor Diagnosis & Therapy, Jinshan Hospital, Fudan University, Shanghai, China
- 2Liver Cancer Institute, Zhongshan Hospital, and Key Laboratory of Carcinogenesis and Cancer Invasion (Ministry of Education), Fudan University, Shanghai, China
Liver cancer is the fourth leading cause of cancer-related death worldwide. Hepatocellular carcinoma (HCC) accounts for about 85%-90% of all primary liver malignancies. However, only 20-30% of HCC patients are eligible for curative therapy mainly due to the lack of early-detection strategies, highlighting the significance of reliable and accurate biomarkers. The integration of multi-omics became an important tool for biomarker screening and unique alterations in tumor-associated genes, transcripts, proteins, post-translational modifications and metabolites have been observed. We here summarized the novel biomarkers for HCC diagnosis based on multi-omics technology as well as the clinical significance of these potential biomarkers in the early detection of HCC.
Introduction
Liver cancer is one of the leading causes of cancer-related death worldwide (1). Hepatocellular carcinoma (HCC) accounts for > 80% of liver cancer and is usually developed from advanced chronic liver diseases (CLD) with hepatitis virus (mainly HBV and HCV) infection and alcoholic/nonalcoholic liver diseases (2–4). α-fetoprotein (AFP) (5) and Lens culinaris agglutinin-reactive fraction of AFP (AFP-L3) (6, 7), des-gamma-carboxy prothrombin (DCP) (6) and glypican-3 (GPC3) (8, 9) have been used for the clinical diagnosis of HCC. However, the complex pathology and individual heterogeneity of HCC pose great challenges for its early detection (10). Most HCC patients were found at late-stage and had a 5-year survival rate as low as 10.0% (11, 12). It was reported that the 5-year survival rate would be over 86.2% if the patients were given intervention at an early phase (13).
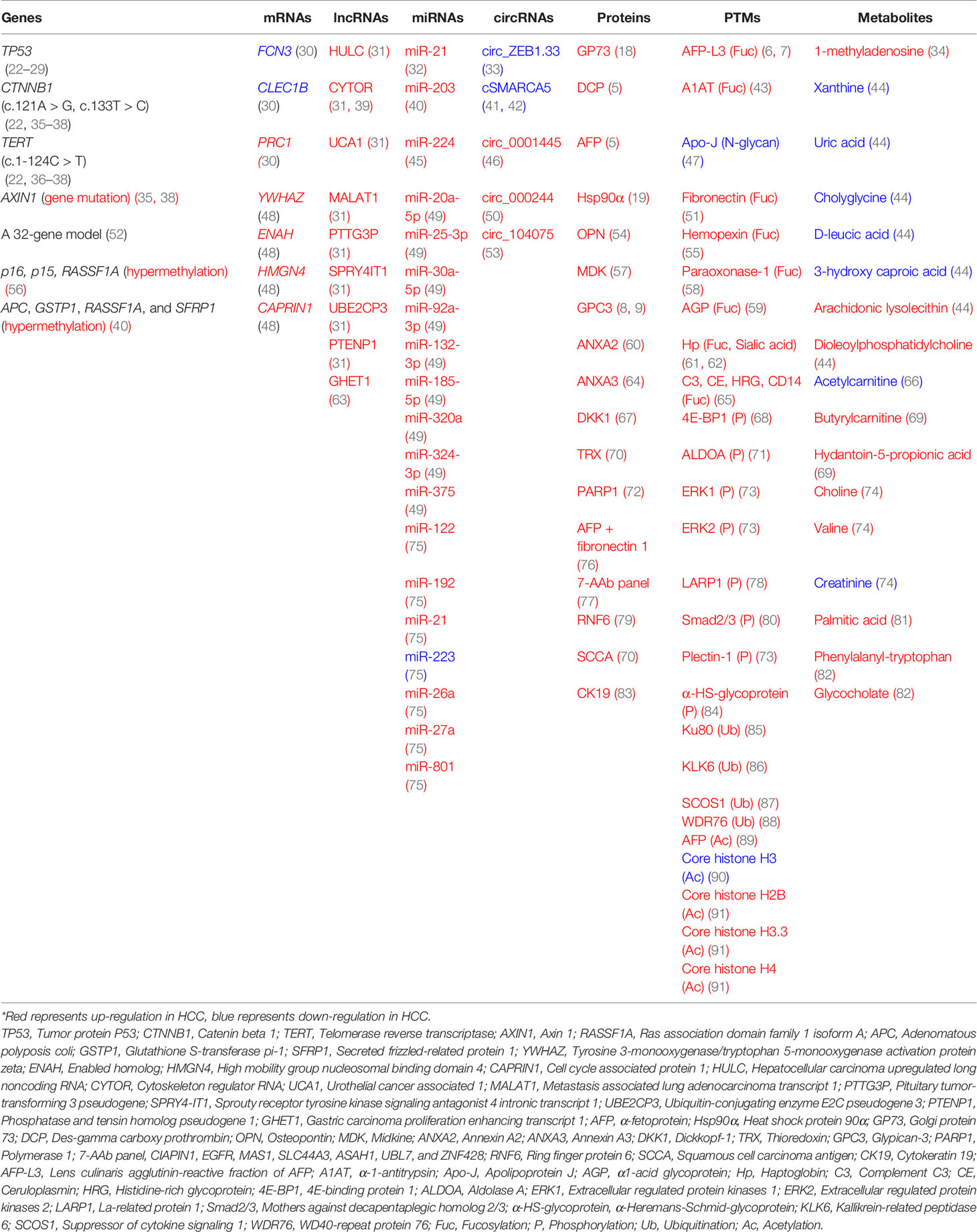
Multi-omics including genomics, epigenomics, transcriptomics, proteomics, glycomics/glycoproteomics and metabolomics can provide novel insights for HCC detection. For genomics and epigenomics, more evidence has shown that circulating tumor DNAs (ctDNAs) and their epigenetic changes could be used as reliable biomarkers (14–16). For transcriptomics, significant changes were observed in mRNAs and noncoding RNAs (miRNAs, lncRNAs, circRNAs) (17). For proteomics, potential protein biomarkers such as Golgi protein-73 (GP73) (18) and heat shock protein 90α (Hsp90α) (19) were identified for HCC detection. For post-translational modifications (PTMs), glycosylation, phosphorylation, acetylation and ubiquitination can be considered for discovering novel biomarkers. Mass spectrometry (MS)-based glycomics/glycoproteomics technology enabled the researchers to characterize aberrant glycoforms and site-specific glycans (20). In addition, metabolomics has contributed to the diagnosis of HCC (21). This review focused on potential biomarkers for liver cancer diagnosis based on multi-omics strategies. Potential HCC biomarkers including genetic mutations, epigenetic changes, mRNAs, noncoding RNAs, proteins, PTMs and metabolites have been summarized in Table 1.
1 Genetic Alteration of HCC
Circulating cell-free DNAs (cfDNAs) are DNA fragments released into the peripheral circulation after the degradation of cell components. The increased proliferation and metabolism of tumor cells would release abundant cfDNAs, which could be used as biomarkers (92–96). The conventional genetic changes of HCC, including point mutations, microsatellite changes and chromosomal rearrangements are reflected in cfDNAs. It was reported that about 50 somatic alterations had been detected and then they could cause changes of their corresponding proteins in HCC (97, 98). Among 48 patients, 56.3% of patients had at least one mutation of the four sites in the following three genes (c.747 for TP53, c.121, c.133 for CTNNB1 and c.1-124 for TRET), which could be further found in 22.2% of HCC patients’ tissues (22, 23). Moreover, the R249S mutation in TP53 was proved to have a potential diagnostic value in the test of 895 HCC patients (24). Other mutation sites of TP53, such as 157 (25), 175 (26), 245 (27), 248 (28) and 273 (29) have been considered for HCC detection. In addition, amino acid changes, such as S37 and S33, could be used as available monitoring indicators for HCC (35–37). The combination of cfDNA mutations and protein changes can increase the diagnosis accuracy of HCC. For example, HCC diagnosis using TP53, TERT, CTNNB1, AFP and DCP has achieved satisfactory results (38).
Epigenetic alterations of ctDNAs were associated with HCC. Methylation changes of ctDNAs often occur in the early stage of tumorigenesis, particularly those alterations in the CpG islands of anti-oncogenes, which may play critical roles in the initiation and progression of HCC (99). The 5-hydroxymethylcytosines (5hmC) are abundantly expressed epigenetic markers (100). A 32-gene diagnostic model was developed using the 5hmC-Seal technique, which accurately distinguished early-stage HCC from non-HCC (52). Accumulating studies have also reported the aberrant methylation of glutathione S-transferase pi-1 (GSTP1) promoter region and cyclin-dependent kinase inhibitor p15 and p16 in HCC patients (101–103). Moreover, the combination of several hot methylated genes was utilized for HCC diagnosis. For example, p16, p15 and ras association domain family 1A (RASSF1A) were assessed in 50 HCC patients and they provided an overall predictive accuracy of 89% with a sensitivity of 84% and a specificity of 94% (56). A panel of four genes (APC, GSTP1, RASSF1A and SFRP1) could make a distinction between HCC and normal controls with a sensitivity of 92.7% and a specificity of 81.9% (104). A predictive model that consisted of three abnormally methylated genes (APC, COX2 and RASSF1A) and one miRNA (miR-203) could be considered to diagnose HCC (40, 105).
2 Transcriptomics of HCC
Analysis of differential gene expression was important for HCC detection. Three genes (FCN3, CLEC1B and PRC1) were explored to be HCC biomarkers based on large-scale transcriptome datasets (30). It was found that YWHAZ, ENAH, HMGN4 and CAPIRN1 changed significantly in HCC (48). Transcriptomics can be integrated into other omics for biomarkers screening. A transcriptome-proteome assay was performed to track the possible biomarkers from HCC-derived gene expression to its protein product released into serum, and a candidate biomarker, Hsp90α, was identified (106).
Long noncoding RNAs (lncRNAs), microRNAs (miRNAs) and circular RNAs (circRNAs) play important roles in the epigenetic regulation of gene expression (107–109). Especially, lncRNAs can affect the expression and stability of miRNAs and messenger RNAs (mRNAs) (110–112). It was reported that lncRNAs HULC and CYTOR were used for joint diagnosis of HCC (39). Moreover, a diagnostic panel including lncRNAs CYTOR, UCA1 and AFP had satisfactory sensitivity and specificity (31).
miRNAs are small noncoding RNAs composed of 20-24 nucleotides. Elevated miR-21 levels in HCC patients have been widely reported (32). The high expression of miR-224 had a good diagnostic value with an AUC of 0.888-0.899 for patients with early HCC (45). It was reported that the overexpression of eight miRNAs (miR-20a-5p, miR-25-3p, miR-30a-5p, miR-92a-3p, miR-132-3p, miR-185-5p, miR-320a and miR-324-3p) had diagnostic significance in HBV positive HCC patients. Besides, the four miRNAs (miR-20a-5p, miR-320a, miR-324-3p and miR-375) panel could contribute to the early screening of HCC (49). Zhou et al. found plasma miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a and miR-801 were potential circulating biomarkers for HCC (75). This seven-miRNA panel showed high accuracy in the diagnosis of HCC, even in patients with early stages. The miRNA panel enabled a more detailed distinction among HCC, healthy, chronic HBV and cirrhosis. It has been evaluated by multi-center clinical detection and authorized by National Medical Products Administration in China.
The changes in circRNA expression were also observed in HCC patients (113). For example, cSMARCA5 (41, 42), circ-ZEB1.33 (33) and circ_0001445 (46) levels were notably decreased in HCC patients. Two circRNAs (circ_000244 and circ_104075) were upregulated in HCC tissues and sera. The circ_104075 exhibited better sensitivity and specificity than some traditional HCC biomarkers (53). The circ_000244 showed better diagnostic performance than AFP (50).
3 Proteomics of HCC
Many classical proteins, which served as reliable biomarkers for other cancers, have been posing new value in HCC diagnosis. Squamous cell carcinoma antigen (SCCA) was previously reported to be associated with cervical cancer (114). Recent studies also indicated that it had a significant contribution to the early diagnosis of HCC (70). Protein expression of cytokeratin 19 (CK19) in HCC was low, however, it would reflect the malignant progression of hepatoma cells (83). GP73 (18), osteopontin (OPN) (54), midkine (MDK) (57), annexin A2 (ANXA2) (60), annexin A3 (ANXA3) (64), dickkopf-1 (DKK1) (67), thioredoxin (TRX) (70) and polymerase 1 (PARP1) (72) have shown diagnostic value for the early diagnosis of liver cancer. Ring finger protein 6 (RNF6) was upregulated and promoted the tumorigenicity of HCC, which might be useful for the detection of HCC at the initial stage (79). A combination of protein markers was also considered for HCC detection, for example, the joint diagnosis with AFP and fibronectin 1 (76). A novel 7-autoantibody (AAb) panel containing CIAPIN1, EGFR, MAS1, SLC44A3, ASAH1, UBL7 and ZNF428 was identified using HCC-focused array (77). The artificial neural network model was established for this panel and it was also able to detect AFP-negative HCC with AUC values of 0.841-0.948. Further, proteogenomics was used to address the complex biological properties of cancer and it could incorporate proteomics into genomic-level studies to obtain more accurate cancer information (115).
4 Post-Translational Modifications (PTMs) of HCC
PTMs such as glycosylation, phosphorylation, acetylation and ubiquitination play vital roles in multiple physiological processes and disease progression, including control of the cell cycle progression, changes of chromatin structures and transduction of cellular signals (116). We have summarized potential biomarkers with different PTMs in Figure 1.
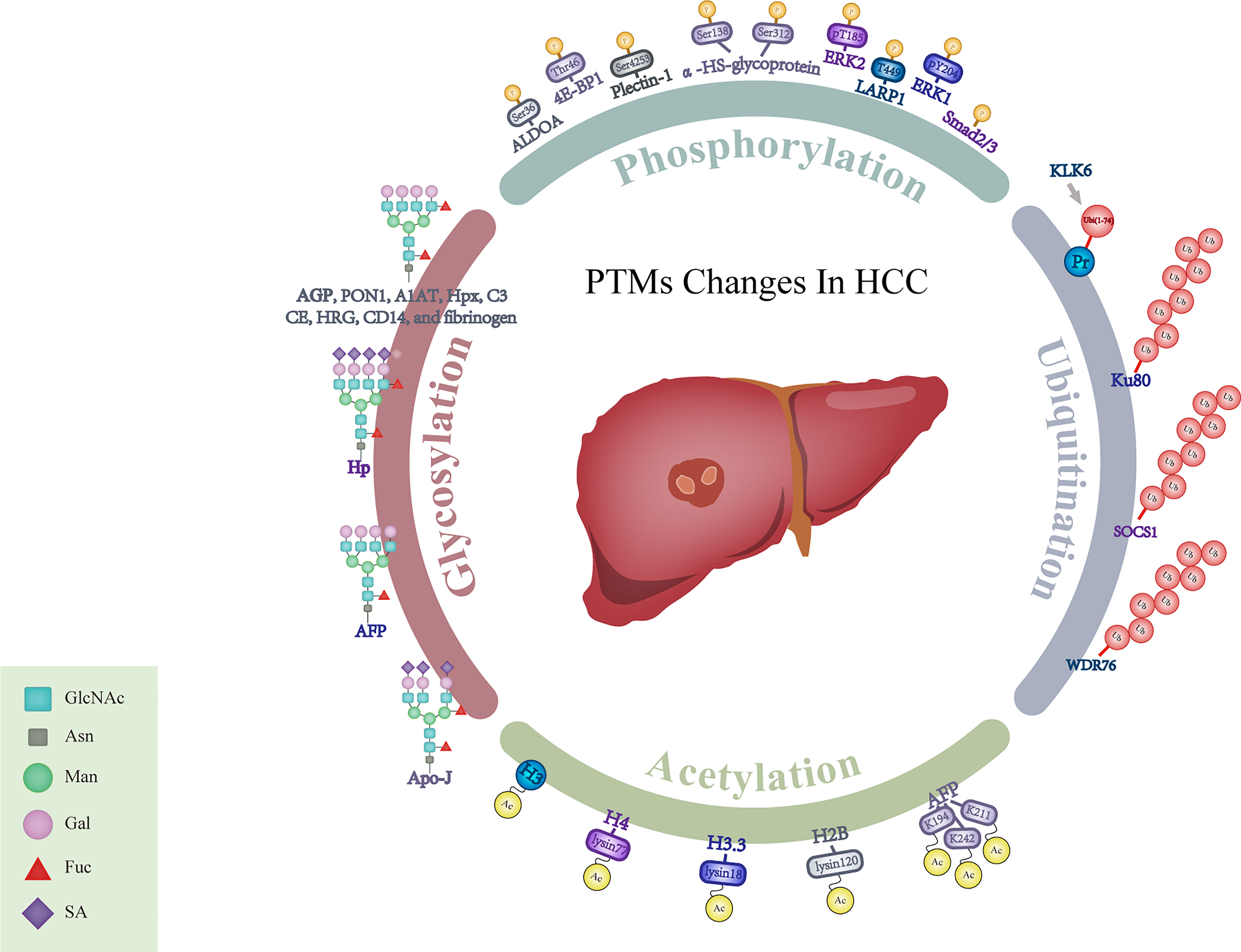
Figure 1 Schematic representation for alterations of PTMs in HCC. GlcNAc, N-acetylglucosamine; Asn, Asparagine; Man, Mannose; Gal, Galactose; Fuc, Fucose; SA, Sialic Acid; AGP, α1-acid glycoprotein; PON1, Paraoxonase 1; A1AT, α-1- antitrypsin; Hpx, Hemopexin; C3, Complement C3; CE, Ceruloplasmin; HRG, Histidine-rich glycoprotein; Hp, Haptoglobin; AFP, α-fetoprotein; Apo-J, Apolipoprotein J; ALDOA, Aldolase A; 4E-BP1, 4E-binding protein 1; ERK1, Extracellular regulated protein kinases 1; ERK2, Extracellular regulated protein kinases 2; LARP1, La-related protein 1; Smad2/3, Mothers against decapentaplegic homolog 2/3; α-HS-glycoprotein, α-Heremans-Schmid-glycoprotein; KLK6, Kallikrein-related peptidase 6; Pr, Protein; SOCS1, Suppressor of cytokine signaling 1; WDR76, WD40-repeat protein 76; H2B, Histone 2B; H3.3, Histone 3.3; H4, Histone 4; H3, Histone 3.
Glycosylation of HCC
Tremendous evidence illustrated that glycan structures were altered in cancers (117, 118). Cancer-associated glycosylation aberration provides novel biomarkers by utilizing glycomic/glycoproteomic technologies (119–123). For example, the glycan profile has been assessed to predict the development of HCC in cirrhosis (124). Glycomics is to detect glycans attached to macromolecules such as proteins (7, 119, 121, 125) and glycoproteomics is a high-throughput technique that could reveal glycosylation sites and site-specific glycoforms (126).
Different glycosylation patterns mainly occur in fucosylation, glycan branching, sialylation and terminal N-acetylgalactosamine (127–129). It was reported that increased fucosylated N-glycans played crucial roles in cancer development, such as core-α-1,6-fucosylated triantennary glycan (130–132). Fucosylation of α1-acid glycoprotein (AGP) was increased in patients with liver cirrhosis and HCC. Meanwhile, different degrees of fucosylation could further distinguish HCC from liver cirrhosis. Thus, determining the specific changes of AGP glycan structures could be helpful for HCC detection (59). The combination of trifucosylated N-glycan of AGP, AFP and AGP showed superiority in discriminating HCC from liver cirrhosis. Zhu et al. investigated the alterations in fucosylation degree of serum haptoglobin (Hp) in a cohort, which included healthy controls, liver cirrhosis and HCC patients, and also confirmed that the fucosylation abnormalities of Hp were closely related to HCC (62). The monofucosylated triantennary glycan at Asn184 and Asn241 of Hp had the diagnostic potential for HCC patients (133). In addition, enhanced fucosylation of serum paraoxonase 1 (PON1) (58), α-1-antitrypsin (A1AT) (43), hemopexin (Hpx) (55), complement C3 (C3), ceruloplasmin (CE), histidine-rich glycoprotein (HRG), CD14 (65) and fibrinogen (51) have been reported to be potential glycobiomarkers for early-stage HCC detection.
High-mannose levels were reported to be associated with HCC (134, 135). Previous studies have shown that N-glycosylation changes occurred in the progression of HCC (136). A total of 83 N-glycans was identified in HCC, and among them, 57 had alterations (137). Two glycopeptides of IgA2 might be unique glycan signatures and provided diagnostic clues in HBV-related liver cancer (138). Apolipoprotein J (Apo-J) in HCC had decreased levels of triantennary glycan and the level of glycosylation of Apo-J could differentiate HCC from cirrhosis with an AUC of 0.852 (47). Besides, sialylation played an important role in cell recognition, adhesion and signal transduction. The high content of sialic acid has been observed in HCC (139). Three glycans containing sialic acid have been regarded as candidate markers for the detection of HCC and they could differentiate HCC from CLD with the AUC of 0.89-0.93 (140). The sialylated glycans of serum Hp were also elevated in HCC (61, 141).
Phosphorylation of HCC
Aberrant protein phosphorylation is associated with HCC (142) and more phosphorylation alterations have been elucidated with the development of omics methods. Elevating phosphorylation levels of 4E-binding protein 1 (4E-BP1) on Thr46 could be used to predict the early recurrence and metastasis of HCC (68). The level of Ser36 phosphorylation of aldolase A (ALDOA) was increased and could be used as a potential biomarker for HCC (71). Changes of some phosphorylation sites, such as the remarkable downregulation of pT185 on extracellular regulated protein kinases 2 (ERK2) and pY204 on extracellular regulated protein kinases 1 (ERK1), have contributed to the progression of HCC (73). Phosphorylation of plectin-1 (phospho-Ser-4253) and α-HS-glycoprotein (phospho-Ser 138 and 312) were also found to be potential HCC biomarkers (84). Furthermore, phosphorylation of la-related protein 1 (LARP1)-T449 and mothers against decapentaplegic homolog 2/3 (Smad2/3)-Thr8 could be useful for HCC detection (78, 80).
Ubiquitination of HCC
Ubiquitination modification, mediating protein enzymatic degradation by labeling proteins, plays a critical role in tumorigenesis (143). The change of Lys48-linked ubiquitination in heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) inhibited HNRNPA1-dependent pyruvate kinase isozyme splicing, and subsequently promoted glucose metabolism reprogramming and malignant behavior of cells in HCC (144). Kallikrein-related peptidase 6 (KLK6) and its catalytic products Ubi1-74 could identify cirrhotic patients at risk of developing HCC (86). The ubiquitination levels of several proteins could serve as HCC indicators, such as lupus Ku autoantigen protein p80 (Ku80) (85), suppressor of cytokine signaling 1 (SCOX1) (87) and WD40-repeat protein 76 (WDR76) (88).
Acetylation of HCC
Acetylation modification, as a dynamic and particular component of PTMs, has attracted more attention in recent years. Lysine acetylation is regulated by the interaction between acetylase and deacetylase (145). Increasing evidence has shown that lysine acetylation played a pivotal role in metabolic function and cellular signaling transduction in the occurrence and development of HCC (63, 146). The sites of lysine acetylation in non-histone proteins and histone proteins have been studied in liver tissues (147, 148). Using MS detection, the acetylation at K194, K211 and K242 of AFP provided novel markers and therapeutic targets for HCC (89). Additionally, the acetylation levels of lysine 120 in histone H2B, lysine 18 in histone H3.3 and lysine 77 in histone H4 were found to be increased in HCC (91). Core histone H3 is another highly conserved protein in cell nucleus and its acetylation has indicated diagnostic significance in HCC (90).
5 Metabolomics of HCC
Metabolites with low molecular weight such as < 1.5 kDa can be defined as “metabolome”, and these small molecular metabolites can dynamically change in liver diseases (149). Metabolomics is a high-throughput method to identify and measure metabolites and offers an opportunity to discover biomarkers (150). Many metabolites were identified including xanthine, uric acid, cholyglycine, D-leucic acid, 3-hydroxy caproic acid, arachidonic acid lysolecithin and dioleoylphosphatidylcholine. They could be effective for the discrimination of HCC from HCV (44). Serum acetylcarnitine enabled clinicians to detect HCC from liver cirrhosis (66). In addition, palmitic acid made a distinction between liver cirrhosis and HBV. The 5-methoxytryptamine, malic acid and phenylalanine were used to discriminate HBV and normal controls. The β-glutamate and asparagine were potential liver disease-specific biomarkers to distinguish HCC from liver cirrhosis (74). Serum 1-methyladenosine was identified as a characteristic metabolite for HCC (34). Two metabolites, butyrylcarnitine and hydantoin-5-propionic acid could be combined together to detect HCC (69). A total of 169 genes and 28 metabolites was reported to be associated with HCC (81). The product of stearoyl CoA desaturase, monounsaturated palmitic acid, increased the invasiveness of HCC, enhanced the migration ability of HCC cells in vitro and might be helpful for HCC diagnosis.
Discussion
Omics technologies and analytical software have been improved. For glycosylation, pGlyco (151), StrucGP (152), GPQuest and GlycoPAT (153) helped to obtain detailed and accurate data. Multi-omics proposed more biomarkers and the specificity and sensitivity of these biomarkers still need to be comprehensively evaluated. Previous studies showed that the profile of DNA methylation had high tissue specificity and helped to determine the tissue origin of cfDNAs (154–157). The combination of different omics biomarkers and the application of computational models can increase diagnostic accuracy. For example, monitoring the change of HCC-specific CpG island methylator phenotype in company with AFP was proved to have better diagnostic performance (158). Measuring both Mac-2 binding protein glycosylation isomer (M2BPGi) and AFP improved the detection sensitivity (159). Metabolites such as phenylalanyl-tryptophan and glycocholate could be added to the traditional HCC diagnostic process to achieve early detection (82).
Different types of biomarkers and detection methods have their advantages and applicable fields. The changes of some proteins and ctDNAs can be detected in the early stages of cancer and they may have high sensitivity for detecting high-risk patients. Considering the affinity of lectin and glycan-specific antibodies to their corresponding glycosylated structures may be low, so its detection usually needs more complex methods (160, 161). Milliliters of plasma were often used for cfDNA extraction (162); micrograms of proteins or microliters of serum seem to be enough for PTM determination (163); for metabolites, microliters of serum were often considered (164). Thus, different methods need to be considered and improved to promote clinic application of multi-omics biomarkers.
Conclusion
The combination of multi-omics, including genomics, transcriptomics, proteomics, glycomics, glycoproteomics and metabolomics would provide more sensitive and accurate detection for HCC, especially in the early stage. Multi-omics approaches also enable the researchers to gain deeper insight into the molecular mechanism of HCC development. With optimized technologies and clinical validation, multi-omics biomarkers would become practical in clinic for HCC diagnosis.
Author Contributions
FC and SZ collected information and wrote the manuscript. JW made the table and drew the figure. YW made modifications to the manuscript. QG and SZ formulated the writing frame and made important modifications to the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the Science and Technology Commission of Shanghai Municipality (20JC1418900) and Shanghai Pujiang Program (2020PJD012).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Villanueva A. Hepatocellular Carcinoma. N Engl J Med (2019) 380(15):1450–62. doi: 10.1056/NEJMra1713263
2. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A Global View of Hepatocellular Carcinoma: Trends, Risk, Prevention and Management. Nat Rev Gastroenterol Hepatol (2019) 16(10):589–604. doi: 10.1038/s41575-019-0186-y
3. Rinella ME. Nonalcoholic Fatty Liver Disease: A Systematic Review. JAMA (2015) 313(22):2263–73. doi: 10.1001/jama.2015.5370
4. Vandenbulcke H, Moreno C, Colle I, Knebel JF, Francque S, Serste T, et al. Alcohol Intake Increases the Risk of HCC in Hepatitis C Virus-Related Compensated Cirrhosis: A Prospective Study. J Hepatol (2016) 65(3):543–51. doi: 10.1016/j.jhep.2016.04.031
5. Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, Roberts LR, et al. Alpha-Fetoprotein, Des-Gamma Carboxyprothrombin, and Lectin-Bound Alpha-Fetoprotein in Early Hepatocellular Carcinoma. Gastroenterol (2009) 137(1):110–8. doi: 10.1053/j.gastro.2009.04.005
6. Choi J, Kim GA, Han S, Lee W, Chun S, Lim YS. Longitudinal Assessment of Three Serum Biomarkers to Detect Very Early-Stage Hepatocellular Carcinoma. Hepatol (2019) 69(5):1983–94. doi: 10.1002/hep.30233
7. Nakagawa T, Miyoshi E, Yakushijin T, Hiramatsu N, Igura T, Hayashi N, et al. Glycomic Analysis of Alpha-Fetoprotein L3 in Hepatoma Cell Lines and Hepatocellular Carcinoma Patients. J Proteome Res (2008) 7(6):2222–33. doi: 10.1021/pr700841q
8. Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, et al. Glypican-3: A Novel Serum and Histochemical Marker for Hepatocellular Carcinoma. Gastroenterol (2003) 125(1):89–97. doi: 10.1016/s0016-5085(03)00689-9
9. Zhou F, Shang W, Yu X, Tian J. Glypican-3: A Promising Biomarker for Hepatocellular Carcinoma Diagnosis and Treatment. Med Res Rev (2018) 38(2):741–67. doi: 10.1002/med.21455
10. El-Serag HB, Rudolph KL. Hepatocellular Carcinoma: Epidemiology and Molecular Carcinogenesis. Gastroenterol (2007) 132(7):2557–76. doi: 10.1053/j.gastro.2007.04.061
11. Liu XN, Cui DN, Li YF, Liu YH, Liu G, Liu L. Multiple "Omics" Data-Based Biomarker Screening for Hepatocellular Carcinoma Diagnosis. World J Gastroenterol (2019) 25(30):4199–212. doi: 10.3748/wjg.v25.i30.4199
12. Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, et al. Cancer Survival in China, 2003-2005: A Population-Based Study. Int J Cancer (2015) 136(8):1921–30. doi: 10.1002/ijc.29227
13. Tsilimigras DI, Bagante F, Sahara K, Moris D, Hyer JM, Wu L, et al. Prognosis After Resection of Barcelona Clinic Liver Cancer (BCLC) Stage 0, A, and B Hepatocellular Carcinoma: A Comprehensive Assessment of the Current BCLC Classification. Ann Surg Oncol (2019) 26(11):3693–700. doi: 10.1245/s10434-019-07580-9
14. Campos-Carrillo A, Weitzel JN, Sahoo P, Rockne R, Mokhnatkin JV, Murtaza M, et al. Circulating Tumor DNA as an Early Cancer Detection Tool. Pharmacol Ther (2020) 207:107458. doi: 10.1016/j.pharmthera.2019.107458
15. Zhou J, Shi YH, Fan J. Circulating Cell-Free Nucleic Acids: Promising Biomarkers of Hepatocellular Carcinoma. Semin Oncol (2012) 39(4):440–8. doi: 10.1053/j.seminoncol.2012.05.013
16. Tang JC, Feng YL, Guo T, Xie AY, Cai XJ. Circulating Tumor DNA in Hepatocellular Carcinoma: Trends and Challenges. Cell Biosci (2016) 6:32. doi: 10.1186/s13578-016-0100-z
17. Supplitt S, Karpinski P, Sasiadek M, Laczmanska I. Current Achievements and Applications of Transcriptomics in Personalized Cancer Medicine. Int J Mol Sci (2021) 22(3):1422. doi: 10.3390/ijms22031422
18. Block TM, Comunale MA, Lowman M, Steel LF, Romano PR, Fimmel C, et al. Use of Targeted Glycoproteomics to Identify Serum Glycoproteins That Correlate With Liver Cancer in Woodchucks and Humans. Proc Natl Acad Sci USA (2005) 102(3):779–84. doi: 10.1073/pnas.0408928102
19. Liu W, Li J, Zhang P, Hou Q, Feng S, Liu L, et al. A Novel Pan-Cancer Biomarker Plasma Heat Shock Protein 90alpha and its Diagnosis Determinants in Clinic. Cancer Sci (2019) 110(9):2941–59. doi: 10.1111/cas.14143
20. Peng W, Gutierrez Reyes CD, Gautam S, Yu A, Cho BG, Goli M, et al. MS-Based Glycomics and Glycoproteomics Methods Enabling Isomeric Characterization. Mass Spectrom Rev (2021). doi: 10.1002/mas.21713
21. Banales JM, Inarrairaegui M, Arbelaiz A, Milkiewicz P, Muntane J, Munoz-Bellvis L, et al. Serum Metabolites as Diagnostic Biomarkers for Cholangiocarcinoma, Hepatocellular Carcinoma, and Primary Sclerosing Cholangitis. Hepatol (2019) 70(2):547–62. doi: 10.1002/hep.30319
22. Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, et al. Detecting Circulating Tumor DNA in Hepatocellular Carcinoma Patients Using Droplet Digital PCR Is Feasible and Reflects Intratumoral Heterogeneity. J Cancer (2016) 7(13):1907–14. doi: 10.7150/jca.15823
23. Pelizzaro F, Cardin R, Penzo B, Pinto E, Vitale A, Cillo U, et al. Liquid Biopsy in Hepatocellular Carcinoma: Where Are We Now? Cancers (Basel) (2021) 13(9):2274. doi: 10.3390/cancers13092274
24. Shen T, Li SF, Wang JL, Zhang T, Zhang S, Chen HT, et al. TP53 R249S Mutation Detected in Circulating Tumour DNA Is Associated With Prognosis of Hepatocellular Carcinoma Patients With or Without Hepatectomy. Liver Int (2020) 40(11):2834–47. doi: 10.1111/liv.14581
25. Villanueva A, Hoshida Y. Depicting the Role of TP53 in Hepatocellular Carcinoma Progression. J Hepatol (2011) 55(3):724–5. doi: 10.1016/j.jhep.2011.03.018
26. Ikeda S, Lim JS, Kurzrock R. Analysis of Tissue and Circulating Tumor DNA by Next-Generation Sequencing of Hepatocellular Carcinoma: Implications for Targeted Therapeutics. Mol Cancer Ther (2018) 17(5):1114–22. doi: 10.1158/1535-7163.MCT-17-0604
27. Paget V, Sichel F, Garon D, Lechevrel M. Aflatoxin B1-Induced TP53 Mutational Pattern in Normal Human Cells Using the FASAY (Functional Analysis of Separated Alleles in Yeast). Mutat Res (2008) 656(1-2):55–61. doi: 10.1016/j.mrgentox.2008.07.009
28. Joseph NM, Tsokos CG, Umetsu SE, Shain AH, Kelley RK, Onodera C, et al. Genomic Profiling of Combined Hepatocellular-Cholangiocarcinoma Reveals Similar Genetics to Hepatocellular Carcinoma. J Pathol (2019) 248(2):164–78. doi: 10.1002/path.5243
29. Li VD, Li KH, Li JT. TP53 Mutations as Potential Prognostic Markers for Specific Cancers: Analysis of Data From The Cancer Genome Atlas and the International Agency for Research on Cancer TP53 Database. J Cancer Res Clin Oncol (2019) 145(3):625–36. doi: 10.1007/s00432-018-2817-z
30. Kaur H, Dhall A, Kumar R, Raghava GPS. Identification of Platform-Independent Diagnostic Biomarker Panel for Hepatocellular Carcinoma Using Large-Scale Transcriptomics Data. Front Genet (2019) 10:1306. doi: 10.3389/fgene.2019.01306
31. Huang J, Zheng Y, Xiao X, Liu C, Lin J, Zheng S, et al. A Circulating Long Noncoding RNA Panel Serves as a Diagnostic Marker for Hepatocellular Carcinoma. Dis Markers (2020) 2020:5417598. doi: 10.1155/2020/5417598
32. Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, et al. Circulating microRNA-21 as a Novel Biomarker for Hepatocellular Carcinoma. J Hepatol (2012) 56(1):167–75. doi: 10.1016/j.jhep.2011.04.026
33. Gong Y, Mao J, Wu D, Wang X, Li L, Zhu L, et al. Circ-ZEB1.33 Promotes the Proliferation of Human HCC by Sponging miR-200a-3p and Upregulating CDK6. Cancer Cell Int (2018) 18:116. doi: 10.1186/s12935-018-0602-3
34. Chen F, Xue JH, Zhou LF, Wu SS, Chen Z. Identification of Serum Biomarkers of Hepatocarcinoma Through Liquid Chromatography/Mass Spectrometry-Based Metabonomic Method. Anal Bioanal Chem (2011) 401(6):1899–904. doi: 10.1007/s00216-011-5245-3
35. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated Analysis of Somatic Mutations and Focal Copy-Number Changes Identifies Key Genes and Pathways in Hepatocellular Carcinoma. Nat Genet (2012) 44(6):694–8. doi: 10.1038/ng.2256
36. Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, et al. Whole-Genome Mutational Landscape and Characterization of Noncoding and Structural Mutations in Liver Cancer. Nat Genet (2016) 48(5):500–9. doi: 10.1038/ng.3547
37. Cancer Genome Atlas Research Network. Electronic Address Wbe, Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell (2017) 169(7):1327–41 e23. doi: 10.1016/j.cell.2017.05.046
38. Qu C, Wang Y, Wang P, Chen K, Wang M, Zeng H, et al. Detection of Early-Stage Hepatocellular Carcinoma in Asymptomatic HBsAg-Seropositive Individuals by Liquid Biopsy. Proc Natl Acad Sci USA (2019) 116(13):6308–12. doi: 10.1073/pnas.1819799116
39. Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J, et al. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem (2015) 37(2):687–96. doi: 10.1159/000430387
40. Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant Methylation of Multiple Tumor Suppressor Genes in Aging Liver, Chronic Hepatitis, and Hepatocellular Carcinoma. Hepatol (2008) 47(3):908–18. doi: 10.1002/hep.22110
41. Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang L, et al. Using Circular RNA SMARCA5 as a Potential Novel Biomarker for Hepatocellular Carcinoma. Clin Chim Acta (2019) 492:37–44. doi: 10.1016/j.cca.2019.02.001
42. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA Csmarca5 Inhibits Growth and Metastasis in Hepatocellular Carcinoma. J Hepatol (2018) 68(6):1214–27. doi: 10.1016/j.jhep.2018.01.012
43. Comunale MA, Rodemich-Betesh L, Hafner J, Wang M, Norton P, Di Bisceglie AM, et al. Linkage Specific Fucosylation of Alpha-1-Antitrypsin in Liver Cirrhosis and Cancer Patients: Implications for a Biomarker of Hepatocellular Carcinoma. PloS One (2010) 5(8):e12419. doi: 10.1371/journal.pone.0012419
44. Bowers J, Hughes E, Skill N, Maluccio M, Raftery D. Detection of Hepatocellular Carcinoma in Hepatitis C Patients: Biomarker Discovery by LC-MS. J Chromatogr B Analyt Technol BioMed Life Sci (2014) 966:154–62. doi: 10.1016/j.jchromb.2014.02.043
45. Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, et al. Serum Exosomal microRNAs as Novel Biomarkers for Hepatocellular Carcinoma. Exp Mol Med (2015) 47:e184. doi: 10.1038/emm.2015.68
46. Zhang X, Zhou H, Jing W, Luo P, Qiu S, Liu X, et al. The Circular RNA Hsa_Circ_0001445 Regulates the Proliferation and Migration of Hepatocellular Carcinoma and may Serve as a Diagnostic Biomarker. Dis Markers (2018) 2018:3073467. doi: 10.1155/2018/3073467
47. Comunale MA, Wang M, Rodemich-Betesh L, Hafner J, Lamontagne A, Klein A, et al. Novel Changes in Glycosylation of Serum Apo-J in Patients With Hepatocellular Carcinoma. Cancer Epidemiol Biomarkers Prev (2011) 20(6):1222–9. doi: 10.1158/1055-9965.EPI-10-1047
48. Xia QL, Li ZH, Zheng JH, Zhang X, Di Y, Ding J, et al. Identification of Novel Biomarkers for Hepatocellular Carcinoma Using Transcriptome Analysis. J Cell Physiol (2019) 234(4):4851–63. doi: 10.1002/jcp.27283
49. Wen Y, Han J, Chen JG, Dong J, Xia YX, Liu JB, et al. Plasma miRNAs as Early Biomarkers for Detecting Hepatocellular Carcinoma. Int J Cancer (2015) 137(7):1679–90. doi: 10.1002/ijc.29544
50. Matboli M, Shafei AE, Ali MA, Ashry AM, Kamal KM, Agag MA, et al. circRNAs (Hsa_Circ_00156, Hsa_Circ _000224, and Hsa_Circ _000520) are Novel Potential Biomarkers in Hepatocellular Carcinoma. J Cell Biochem (2018) 120(5):7711–24. doi: 10.1002/jcb.28045
51. Yin H, Tan Z, Wu J, Zhu J, Shedden KA, Marrero J, et al. Mass-Selected Site-Specific Core-Fucosylation of Serum Proteins in Hepatocellular Carcinoma. J Proteome Res (2015) 14(11):4876–84. doi: 10.1021/acs.jproteome.5b00718
52. Cai J, Chen L, Zhang Z, Zhang X, Lu X, Liu W, et al. Genome-Wide Mapping of 5-Hydroxymethylcytosines in Circulating Cell-Free DNA as a non-Invasive Approach for Early Detection of Hepatocellular Carcinoma. Gut (2019) 68(12):2195–205. doi: 10.1136/gutjnl-2019-318882
53. Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, et al. circRNA_104075 Stimulates YAP-Dependent Tumorigenesis Through the Regulation of HNF4a and may Serve as a Diagnostic Marker in Hepatocellular Carcinoma. Cell Death Dis (2018) 9(11):1091. doi: 10.1038/s41419-018-1132-6
54. Shang S, Plymoth A, Ge S, Feng Z, Rosen HR, Sangrajrang S, et al. Identification of Osteopontin as a Novel Marker for Early Hepatocellular Carcinoma. Hepatol (2012) 55(2):483–90. doi: 10.1002/hep.24703
55. Benicky J, Sanda M, Pompach P, Wu J, Goldman R. Quantification of Fucosylated Hemopexin and Complement Factor H in Plasma of Patients With Liver Disease. Anal Chem (2014) 86(21):10716–23. doi: 10.1021/ac502727s
56. Zhang YJ, Wu HC, Shen J, Ahsan H, Tsai WY, Yang HL, et al. Predicting Hepatocellular Carcinoma by Detection of Aberrant Promoter Methylation in Serum DNA. Clin Cancer Res (2007) 13(8):2378–84. doi: 10.1158/1078-0432.CCR-06-1900
57. Zhu WW, Guo JJ, Guo L, Jia HL, Zhu M, Zhang JB, et al. Evaluation of Midkine as a Diagnostic Serum Biomarker in Hepatocellular Carcinoma. Clin Cancer Res (2013) 19(14):3944–54. doi: 10.1158/1078-0432.CCR-12-3363
58. Zhang S, Jiang K, Zhang Q, Guo K, Liu Y. Serum Fucosylated Paraoxonase 1 as a Potential Glycobiomarker for Clinical Diagnosis of Early Hepatocellular Carcinoma Using ELISA Index. Glycoconj J (2015) 32(3-4):119–25. doi: 10.1007/s10719-015-9576-8
59. Astrom E, Stal P, Zenlander R, Edenvik P, Alexandersson C, Haglund M, et al. Reverse Lectin ELISA for Detecting Fucosylated Forms of Alpha1-Acid Glycoprotein Associated With Hepatocellular Carcinoma. PloS One (2017) 12(3):e0173897. doi: 10.1371/journal.pone.0173897
60. Sun Y, Gao G, Cai J, Wang Y, Qu X, He L, et al. Annexin A2 Is a Discriminative Serological Candidate in Early Hepatocellular Carcinoma. Carcinogenesis (2013) 34(3):595–604. doi: 10.1093/carcin/bgs372
61. Ang IL, Poon TC, Lai PB, Chan AT, Ngai SM, Hui AY, et al. Study of Serum Haptoglobin and its Glycoforms in the Diagnosis of Hepatocellular Carcinoma: A Glycoproteomic Approach. J Proteome Res (2006) 5(10):2691–700. doi: 10.1021/pr060109r
62. Zhu J, Lin Z, Wu J, Yin H, Dai J, Feng Z, et al. Analysis of Serum Haptoglobin Fucosylation in Hepatocellular Carcinoma and Liver Cirrhosis of Different Etiologies. J Proteome Res (2014) 13(6):2986–97. doi: 10.1021/pr500128t
63. Ding GQ, Li W, Liu JP, Zeng YL, Mao CS, Kang Y, et al. LncRNA GHET1 Activated by H3K27 Acetylation Promotes Cell Tumorigenesis Through Regulating ATF1 in Hepatocellular Carcinoma. Biomed Pharmacother (2017) 94:326–31. doi: 10.1016/j.biopha.2017.07.046
64. Ma XL, Jiang M, Zhao Y, Wang BL, Shen MN, Zhou Y, et al. Application of Serum Annexin A3 in Diagnosis, Outcome Prediction and Therapeutic Response Evaluation for Patients With Hepatocellular Carcinoma. Ann Surg Oncol (2018) 25(6):1686–94. doi: 10.1245/s10434-018-6402-0
65. Liu Y, He J, Li C, Benitez R, Fu S, Marrero J, et al. Identification and Confirmation of Biomarkers Using an Integrated Platform for Quantitative Analysis of Glycoproteins and Their Glycosylations. J Proteome Res (2010) 9(2):798–805. doi: 10.1021/pr900715p
66. Lu Y, Li N, Gao L, Xu YJ, Huang C, Yu K, et al. Acetylcarnitine Is a Candidate Diagnostic and Prognostic Biomarker of Hepatocellular Carcinoma. Cancer Res (2016) 76(10):2912–20. doi: 10.1158/0008-5472.CAN-15-3199
67. Shen QJ, Fan J, Yang XR, Tan YX, Zhao WF, Xu Y, et al. Serum DKK1 as a Protein Biomarker for the Diagnosis of Hepatocellular Carcinoma: A Large-Scale, Multicentre Study. Lancet Oncol (2012) 13(8):817–26. doi: 10.1016/S1470-2045(12)70233-4
68. Lin X, Huang Y, Sun Y, Tan X, Ouyang J, Zhao B, et al. 4e-BP1(Thr46) Phosphorylation Association With Poor Prognosis in Quantitative Phosphoproteomics of Portal Vein Tumor Thrombus Revealed That 4E-BP1Thr46 Phosphorylation Is Associated With Poor Prognosis in HCC. Cancer Manag Res (2020) 12:103–15. doi: 10.2147/CMAR.S230849
69. Shao Y, Zhu B, Zheng R, Zhao X, Yin P, Lu X, et al. Development of Urinary Pseudotargeted LC-MS-Based Metabolomics Method and its Application in Hepatocellular Carcinoma Biomarker Discovery. J Proteome Res (2015) 14(2):906–16. doi: 10.1021/pr500973d
70. Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T. Biomarkers for the Early Diagnosis of Hepatocellular Carcinoma. World J Gastroenterol (2015) 21(37):10573–83. doi: 10.3748/wjg.v21.i37.10573
71. Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell (2019) 179(2):561–77.e22. doi: 10.1016/j.cell.2019.08.052
72. Goh WW, Lee YH, Zubaidah RM, Jin J, Dong D, Lin Q, et al. Network-Based Pipeline for Analyzing MS Data: An Application Toward Liver Cancer. J Proteome Res (2011) 10(5):2261–72. doi: 10.1021/pr1010845
73. Xu B, Wang F, Song C, Sun Z, Cheng K, Tan Y, et al. Large-Scale Proteome Quantification of Hepatocellular Carcinoma Tissues by a Three-Dimensional Liquid Chromatography Strategy Integrated With Sample Preparation. J Proteome Res (2014) 13(8):3645–54. doi: 10.1021/pr500200s
74. Wei S, Suryani Y, Gowda GA, Skill N, Maluccio M, Raftery D. Differentiating Hepatocellular Carcinoma From Hepatitis C Using Metabolite Profiling. Metabolites (2012) 2(4):701–16. doi: 10.3390/metabo2040701
75. Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA Panel to Diagnose Hepatitis B Virus-Related Hepatocellular Carcinoma. J Clin Oncol (2011) 29(36):4781–8. doi: 10.1200/JCO.2011.38.2697
76. Krishnan MS, Rajan Kd A, Park J, Arjunan V, Garcia Marques FJ, Bermudez A, et al. Genomic Analysis of Vascular Invasion in HCC Reveals Molecular Drivers and Predictive Biomarkers. Hepatol (2021) 73(6):2342–60. doi: 10.1002/hep.31614
77. Zhang S, Liu Y, Chen J, Shu H, Shen S, Li Y, et al. Autoantibody Signature in Hepatocellular Carcinoma Using Seromics. J Hematol Oncol (2020) 13(1):85. doi: 10.1186/s13045-020-00918-x
78. Ramani K, Robinson AE, Berlind J, Fan W, Abeynayake A, Binek A, et al. S-Adenosylmethionine Inhibits La Ribonucleoprotein Domain Family Member 1 in Murine Liver and Human Liver Cancer Cells. Hepatology (2022) 75(2):280–96. doi: 10.1002/hep.32130
79. Jin ZD, Chang L, Zhu LL. RNF6 Serves as a Diagnostic Hallmark of non-Alcoholic Fatty Liver Disease With Hepatocellular Carcinoma: A Clinical Research. Eur Rev Med Pharmacol Sci (2020) 24(21):11052–7. doi: 10.26355/eurrev_202011_23590
80. Zhang Y, Tao R, Wu SS, Xu CC, Wang JL, Chen J, et al. TRIM52 Up-Regulation in Hepatocellular Carcinoma Cells Promotes Proliferation, Migration and Invasion Through the Ubiquitination of PPM1A. J Exp Clin Cancer Res (2018) 37(1):116. doi: 10.1186/s13046-018-0780-9
81. Budhu A, Roessler S, Zhao X, Yu Z, Forgues M, Ji J, et al. Integrated Metabolite and Gene Expression Profiles Identify Lipid Biomarkers Associated With Progression of Hepatocellular Carcinoma and Patient Outcomes. Gastroenterol (2013) 144(5):1066–75.e1. doi: 10.1053/j.gastro.2013.01.054
82. Luo P, Yin P, Hua R, Tan Y, Li Z, Qiu G, et al. A Large-Scale, Multicenter Serum Metabolite Biomarker Identification Study for the Early Detection of Hepatocellular Carcinoma. Hepatol (2018) 67(2):662–75. doi: 10.1002/hep.29561
83. Ding SJ, Li Y, Tan YX, Jiang MR, Tian B, Liu YK, et al. From Proteomic Analysis to Clinical Significance: Overexpression of Cytokeratin 19 Correlates With Hepatocellular Carcinoma Metastasis. Mol Cell Proteomics (2004) 3(1):73–81. doi: 10.1074/mcp.M300094-MCP200
84. Lee HJ, Na K, Kwon MS, Kim H, Kim KS, Paik YK. Quantitative Analysis of Phosphopeptides in Search of the Disease Biomarker From the Hepatocellular Carcinoma Specimen. Proteomics (2009) 9(12):3395–408. doi: 10.1002/pmic.200800943
85. Sun Y, Zheng X, Yuan H, Chen G, Ouyang J, Liu J, et al. Proteomic Analyses Reveal Divergent Ubiquitylation Patterns in Hepatocellula Carcinoma Cell Lines With Different Metastasis Potential. J Proteomics (2020) 225:103834. doi: 10.1016/j.jprot.2020.103834
86. Laouirem S, Le Faouder J, Alexandrov T, Mestivier D, Leger T, Baudin X, et al. Progression From Cirrhosis to Cancer Is Associated With Early Ubiquitin Post-Translational Modifications: Identification of New Biomarkers of Cirrhosis at Risk of Malignancy. J Pathol (2014) 234(4):452–63. doi: 10.1002/path.4398
87. Santharam MA, Shukla A, Ihsan AU, Cloutier M, Levesque D, Ramanathan S, et al. SILAC Proteomics Implicates SOCS1 in Modulating Cellular Macromolecular Complexes and the Ubiquitin Conjugating Enzyme UBE2D Involved in MET Receptor Tyrosine Kinase Downregulation. Biochimie (2021) 182:185–96. doi: 10.1016/j.biochi.2021.01.012
88. Jeong WJ, Park JC, Kim WS, Ro EJ, Jeon SH, Lee SK, et al. WDR76 Is a RAS Binding Protein That Functions as a Tumor Suppressor via RAS Degradation. Nat Commun (2019) 10(1):295. doi: 10.1038/s41467-018-08230-6
89. Xue J, Cao Z, Cheng Y, Wang J, Liu Y, Yang R, et al. Acetylation of Alpha-Fetoprotein Promotes Hepatocellular Carcinoma Progression. Cancer Lett (2020) 471:12–26. doi: 10.1016/j.canlet.2019.11.043
90. Wang W, Xu L, Kong J, Fan H, Yang P. Quantitative Research of Histone H3 Acetylation Levels of Human Hepatocellular Carcinoma Cells. Bioanalysis (2013) 5(3):327–39. doi: 10.4155/bio.12.324
91. Chai XQ, Guo JF, Dong RZ, Yang X, Deng C, Wei CY, et al. Quantitative Acetylome Analysis Reveals Histone Modifications That may Predict Prognosis in Hepatitis B-Related Hepatocellular Carcinoma. Clin Trans Med (2021) 11(3):e313. doi: 10.1002/ctm2.313
92. Campos-Carrillo A, Weitzel JN, Sahoo P, Rockne R, Mokhnatkin JV, Murtaza M, et al. Circulating Tumor DNA as an Early Cancer Detection Tool. Pharmacol Therapeut (2020) 207:107458. doi: 10.1016/j.pharmthera.2019.107458
93. Ye Q, Ling S, Zheng S, Xu X. Liquid Biopsy in Hepatocellular Carcinoma: Circulating Tumor Cells and Circulating Tumor DNA. Mol Cancer (2019) 18(1):114. doi: 10.1186/s12943-019-1043-x
94. Kaseb AO, Sanchez NS, Sen S, Kelley RK, Tan B, Bocobo AG, et al. Molecular Profiling of Hepatocellular Carcinoma Using Circulating Cell-Free DNA. Clin Cancer Res (2019) 25(20):6107–18. doi: 10.1158/1078-0432.CCR-18-3341
95. Piciocchi M, Cardin R, Vitale A, Vanin V, Giacomin A, Pozzan C, et al. Circulating Free DNA in the Progression of Liver Damage to Hepatocellular Carcinoma. Hepatol Int (2013) 7(4):1050–7. doi: 10.1007/s12072-013-9481-9
96. Yan LL, Chen YH, Zhou JY, Zhao H, Zhang HH, Wang GQ. Diagnostic Value of Circulating Cell-Free DNA Levels for Hepatocellular Carcinoma. Int J Infect Dis (2018) 67:92–7. doi: 10.1016/j.ijid.2017.12.002
97. Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular Carcinoma. Nat Rev Dis Primers (2016) 2:16018. doi: 10.1038/nrdp.2016.18
98. Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome Sequencing of Hepatocellular Carcinomas Identifies New Mutational Signatures and Potential Therapeutic Targets. Nat Genet (2015) 47(5):505–11. doi: 10.1038/ng.3252
99. Mann J, Reeves HL, Feldstein AE. Liquid Biopsy for Liver Diseases. Gut (2018) 67(12):2204–12. doi: 10.1136/gutjnl-2017-315846
100. Branco MR, Ficz G, Reik W. Uncovering the Role of 5-Hydroxymethylcytosine in the Epigenome. Nat Rev Genet (2011) 13(1):7–13. doi: 10.1038/nrg3080
101. Matsuda Y, Ichida T, Matsuzawa J, Sugimura K, Asakura H. P16(INK4) is Inactivated by Extensive CpG Methylation in Human Hepatocellular Carcinoma. Gastroenterol (1999) 116(2):394–400. doi: 10.1016/S0016-5085(99)70137-X
102. Roncalli M, Bianchi P, Bruni B, Laghi L, Destro A, Di Gioia S, et al. Methylation Framework of Cell Cycle Gene Inhibitors in Cirrhosis and Associated Hepatocellular Carcinoma. Hepatol (2002) 36(2):427–32. doi: 10.1053/jhep.2002.34852
103. Zhong S, Tang MW, Yeo W, Liu CL, Li D, Johnson PJ. Silencing of GSTP1 Gene by CpG Island DNA Hypermethylation in HBV-Associated Hepatocellular Carcinomas. Clin Cancer Res (2002) 8(4):1087–92.
104. Huang ZH, Hu Y, Hua D, Wu YY, Song MX, Cheng ZH. Quantitative Analysis of Multiple Methylated Genes in Plasma for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Exp Mol Pathol (2011) 91(3):702–7. doi: 10.1016/j.yexmp.2011.08.004
105. Um TH, Kim H, Oh BK, Kim MS, Kim KS, Jung G, et al. Aberrant CpG Island Hypermethylation in Dysplastic Nodules and Early HCC of Hepatitis B Virus-Related Human Multistep Hepatocarcinogenesis. J Hepatol (2011) 54(5):939–47. doi: 10.1016/j.jhep.2010.08.021
106. Zhou Y, Deng X, Zang N, Li H, Li G, Li C, et al. Transcriptomic and Proteomic Investigation of HSP90A as a Potential Biomarker for HCC. Med Sci Monit (2015) 21:4039–49. doi: 10.12659/msm.896712
107. Chen X, Tang FR, Arfuso F, Cai WQ, Ma Z, Yang J, et al. The Emerging Role of Long Non-Coding RNAs in the Metastasis of Hepatocellular Carcinoma. Biomolecules (2019) 10(1):66. doi: 10.3390/biom10010066
108. Lin YH, Wu MH, Yeh CT, Lin KH. Long Non-Coding RNAs as Mediators of Tumor Microenvironment and Liver Cancer Cell Communication. Int J Mol Sci (2018) 19(12):3742. doi: 10.3390/ijms19123742
109. Dragomir MP, Kopetz S, Ajani JA, Calin GA. Non-Coding RNAs in GI Cancers: From Cancer Hallmarks to Clinical Utility. Gut (2020) 69(4):748–63. doi: 10.1136/gutjnl-2019-318279
110. El Khodiry A, Afify M, El Tayebi HM. Behind the Curtain of non-Coding RNAs; Long non-Coding RNAs Regulating Hepatocarcinogenesis. World J Gastroenterol (2018) 24(5):549–72. doi: 10.3748/wjg.v24.i5.549
111. Zhang H, Chen X, Zhang J, Wang X, Chen H, Liu L, et al. Long Noncoding RNAs in HBV-Related Hepatocellular Carcinoma (Review). Int J Oncol (2020) 56(1):18–32. doi: 10.3892/ijo.2019.4909
112. Lim LJ, Wong SYS, Huang F, Lim S, Chong SS, Ooi LL, et al. Roles and Regulation of Long Noncoding RNAs in Hepatocellular Carcinoma. Cancer Res (2019) 79(20):5131–9. doi: 10.1158/0008-5472.CAN-19-0255
113. Chen B, Butte AJ. Leveraging Big Data to Transform Target Selection and Drug Discovery. Clin Pharmacol Ther (2016) 99(3):285–97. doi: 10.1002/cpt.318
114. Lee WK, Chong GO, Jeong SY, Lee HJ, Park SH, Ryu JM, et al. Prognosis-Predicting Model Based on [18F]Fluorodeoxyglucose PET Metabolic Parameters in Locally Advanced Cervical Cancer Patients Treated With Concurrent Chemoradiotherapy: Multi-Center Retrospective Study. J Clin Med (2020) 9(2):427. doi: 10.3390/jcm9020427
115. Zhang B, Whiteaker JR, Hoofnagle AN, Baird GS, Rodland KD, Paulovich AG. Clinical Potential of Mass Spectrometry-Based Proteogenomics. Nat Rev Clin Oncol (2019) 16(4):256–68. doi: 10.1038/s41571-018-0135-7
116. Chen L, Liu S, Tao Y. Regulating Tumor Suppressor Genes: Post-Translational Modifications. Signal Transduct Target Ther (2020) 5(1):90. doi: 10.1038/s41392-020-0196-9
117. Zhang S, Cao X, Gao Q, Liu Y. Protein Glycosylation in Viral Hepatitis-Related HCC: Characterization of Heterogeneity, Biological Roles, and Clinical Implications. Cancer Lett (2017) 406:64–70. doi: 10.1016/j.canlet.2017.07.026
118. Zhu J, Warner E, Parikh ND, Lubman DM. Glycoproteomic Markers of Hepatocellular Carcinoma-Mass Spectrometry Based Approaches. Mass Spectrom Rev (2019) 38(3):265–90. doi: 10.1002/mas.21583
119. An HJ, Kronewitter SR, de Leoz ML, Lebrilla CB. Glycomics and Disease Markers. Curr Opin Chem Biol (2009) 13(5-6):601–7. doi: 10.1016/j.cbpa.2009.08.015
120. Callewaert N, Van Vlierberghe H, Van Hecke A, Laroy W, Delanghe J, Contreras R. Noninvasive Diagnosis of Liver Cirrhosis Using DNA Sequencer-Based Total Serum Protein Glycomics. Nat Med (2004) 10(4):429–34. doi: 10.1038/nm1006
121. Tian Y, Zhang H. Glycoproteomics and Clinical Applications. Proteomics Clin Appl (2010) 4(2):124–32. doi: 10.1002/prca.200900161
122. Wang M, Zhu J, Lubman DM, Gao C. Aberrant Glycosylation and Cancer Biomarker Discovery: A Promising and Thorny Journey. Clin Chem Lab Med (2019) 57(4):407–16. doi: 10.1515/cclm-2018-0379
123. Xiao H, Sun F, Suttapitugsakul S, Wu R. Global and Site-Specific Analysis of Protein Glycosylation in Complex Biological Systems With Mass Spectrometry. Mass Spectrom Rev (2019) 38(4-5):356–79. doi: 10.1002/mas.21586
124. Verhelst X, Vanderschaeghe D, Castera L, Raes T, Geerts A, Francoz C, et al. A Glycomics-Based Test Predicts the Development of Hepatocellular Carcinoma in Cirrhosis. Clin Cancer Res (2017) 23(11):2750–8. doi: 10.1158/1078-0432.CCR-16-1500
125. An HJ, Lebrilla CB. A Glycomics Approach to the Discovery of Potential Cancer Biomarkers. Methods Mol Biol (2010) 600:199–213. doi: 10.1007/978-1-60761-454-8_14
126. Fry SA, Afrough B, Lomax-Browne HJ, Timms JF, Velentzis LS, Leathem AJ. Lectin Microarray Profiling of Metastatic Breast Cancers. Glycobiol (2011) 21(8):1060–70. doi: 10.1093/glycob/cwr045
127. Adamczyk B, Tharmalingam T, Rudd PM. Glycans as Cancer Biomarkers. Bba-Gen Subjects (2012) 1820(9):1347–53. doi: 10.1016/j.bbagen.2011.12.001
128. Kailemia MJ, Park D, Lebrilla CB. Glycans and Glycoproteins as Specific Biomarkers for Cancer. Anal Bioanal Chem (2017) 409(2):395–410. doi: 10.1007/s00216-016-9880-6
129. Mehta A, Herrera H, Block T. Glycosylation and Liver Cancer. Adv Cancer Res (2015) 126:257–79. doi: 10.1016/bs.acr.2014.11.005
130. Miyoshi E, Moriwaki K, Terao N, Tan CC, Terao M, Nakagawa T, et al. Fucosylation Is a Promising Target for Cancer Diagnosis and Therapy. Biomolecules (2012) 2(1):34–45. doi: 10.3390/biom2010034
131. Liu XE, Desmyter L, Gao CF, Laroy W, Dewaele S, Vanhooren V, et al. N-Glycomic Changes in Hepatocellular Carcinoma Patients With Liver Cirrhosis Induced by Hepatitis B Virus. Hepatol (2007) 46(5):1426–35. doi: 10.1002/hep.21855
132. Agrawal P, Fontanals-Cirera B, Sokolova E, Jacob S, Vaiana CA, Argibay D, et al. A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis. Cancer Cell (2017) 31(6):804–19.e7. doi: 10.1016/j.ccell.2017.05.007
133. Zhu J, Huang J, Zhang J, Chen Z, Lin Y, Grigorean G, et al. Glycopeptide Biomarkers in Serum Haptoglobin for Hepatocellular Carcinoma Detection in Patients With Nonalcoholic Steatohepatitis. J Proteome Res (2020) 19(8):3452–66. doi: 10.1021/acs.jproteome.0c00270
134. Li T, Dong ZR, Guo ZY, Wang CH, Zhi XT, Zhou JW, et al. Mannose-Mediated Inhibitory Effects of PA-MSHA on Invasion and Metastasis of Hepatocellular Carcinoma via EGFR/Akt/IkappaBbeta/NF-kappaB Pathway. Liver Int (2015) 35(4):1416–29. doi: 10.1111/liv.12644
135. Yuen MF, Lau CS, Lau YL, Wong WM, Cheng CC, Lai CL. Mannose Binding Lectin Gene Mutations are Associated With Progression of Liver Disease in Chronic Hepatitis B Infection. Hepatol (1999) 29(4):1248–51. doi: 10.1002/hep.510290417
136. Pinho SS, Reis CA. Glycosylation in Cancer: Mechanisms and Clinical Implications. Nat Rev Cancer (2015) 15(9):540–55. doi: 10.1038/nrc3982
137. Goldman R, Ressom HW, Varghese RS, Goldman L, Bascug G, Loffredo CA, et al. Detection of Hepatocellular Carcinoma Using Glycomic Analysis. Clin Cancer Res (2009) 15(5):1808–13. doi: 10.1158/1078-0432.CCR-07-5261
138. Zhang S, Cao X, Liu C, Li W, Zeng W, Li B, et al. N-Glycopeptide Signatures of IgA2 in Serum From Patients With Hepatitis B Virus-Related Liver Diseases. Mol Cell Proteomics (2019) 18(11):2262–72. doi: 10.1074/mcp.RA119.001722
139. Yu H, Zhu M, Qin Y, Zhong Y, Yan H, Wang Q, et al. Analysis of Glycan-Related Genes Expression and Glycan Profiles in Mice With Liver Fibrosis. J Proteome Res (2012) 11(11):5277–85. doi: 10.1021/pr300484j
140. Kamiyama T, Yokoo H, Furukawa J, Kurogochi M, Togashi T, Miura N, et al. Identification of Novel Serum Biomarkers of Hepatocellular Carcinoma Using Glycomic Analysis. Hepatol (2013) 57(6):2314–25. doi: 10.1002/hep.26262
141. Zhang S, Shu H, Luo K, Kang X, Zhang Y, Lu H, et al. N-Linked Glycan Changes of Serum Haptoglobin Beta Chain in Liver Disease Patients. Mol Biosyst (2011) 7(5):1621–8. doi: 10.1039/c1mb05020f
142. Tomasi ML, Ramani K. SUMOylation and Phosphorylation Cross-Talk in Hepatocellular Carcinoma. Transl Gastroenterol Hepatol (2018) 3:20. doi: 10.21037/tgh.2018.04.04
143. Deng L, Meng T, Chen L, Wei W, Wang P. The Role of Ubiquitination in Tumorigenesis and Targeted Drug Discovery. Signal Transduct Target Ther (2020) 5(1):11. doi: 10.1038/s41392-020-0107-0
144. Chen D, Wang YJ, Lu RX, Jiang XF, Chen XH, Meng N, et al. E3 Ligase ZFP91 Inhibits Hepatocellular Carcinoma Metabolism Reprogramming by Regulating PKM Splicing. Theranostics (2020) 10(19):8558–72. doi: 10.7150/thno.44873
145. Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and Functional Diversity of Lysine Acetylation Revealed by a Proteomics Survey. Mol Cell (2006) 23(4):607–18. doi: 10.1016/j.molcel.2006.06.026
146. Huang Z, Zhao J, Deng W, Chen Y, Shang J, Song K, et al. Identification of a Cellularly Active SIRT6 Allosteric Activator. Nat Chem Biol (2018) 14(12):1118–26. doi: 10.1038/s41589-018-0150-0
147. Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, et al. Proteomic Analysis of Lysine Acetylation Sites in Rat Tissues Reveals Organ Specificity and Subcellular Patterns. Cell Rep (2012) 2(2):419–31. doi: 10.1016/j.celrep.2012.07.006
148. Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of Cellular Metabolism by Protein Lysine Acetylation. Sci (2010) 327(5968):1000–4. doi: 10.1126/science.1179689
149. Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, et al. HMDB 4.0: The Human Metabolome Database for 2018. Nucleic Acids Res (2018) 46(D1):D608–D17. doi: 10.1093/nar/gkx1089
150. Ganti S, Weiss RH. Urine Metabolomics for Kidney Cancer Detection and Biomarker Discovery. Urol Oncol (2011) 29(5):551–7. doi: 10.1016/j.urolonc.2011.05.013
151. Zeng WF, Cao WQ, Liu MQ, He SM, Yang PY. Precise, Fast and Comprehensive Analysis of Intact Glycopeptides and Modified Glycans With Pglyco3. Nat Methods (2021) 18(12):1515–23. doi: 10.1038/s41592-021-01306-0
152. Shen J, Jia L, Dang L, Su Y, Zhang J, Xu Y, et al. StrucGP: De Novo Structural Sequencing of Site-Specific N-Glycan on Glycoproteins Using a Modularization Strategy. Nat Methods (2021) 18(8):921–9. doi: 10.1038/s41592-021-01209-0
153. Kawahara R, Chernykh A, Alagesan K, Bern M, Cao W, Chalkley RJ, et al. Community Evaluation of Glycoproteomics Informatics Solutions Reveals High-Performance Search Strategies for Serum Glycopeptide Analysis. Nat Methods (2021) 18(11):1304–16. doi: 10.1038/s41592-021-01309-x
154. Guo S, Diep D, Plongthongkum N, Fung HL, Zhang K, Zhang K. Identification of Methylation Haplotype Blocks Aids in Deconvolution of Heterogeneous Tissue Samples and Tumor Tissue-of-Origin Mapping From Plasma DNA. Nat Genet (2017) 49(4):635–42. doi: 10.1038/ng.3805
155. Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, et al. Identification of Tissue-Specific Cell Death Using Methylation Patterns of Circulating DNA. Proc Natl Acad Sci USA (2016) 113(13):E1826–34. doi: 10.1073/pnas.1519286113
156. Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, et al. Comprehensive Human Cell-Type Methylation Atlas Reveals Origins of Circulating Cell-Free DNA in Health and Disease. Nat Commun (2018) 9(1):5068. doi: 10.1038/s41467-018-07466-6
157. Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, et al. Plasma DNA Tissue Mapping by Genome-Wide Methylation Sequencing for Noninvasive Prenatal, Cancer, and Transplantation Assessments. Proc Natl Acad Sci USA (2015) 112(40):E5503–12. doi: 10.1073/pnas.1508736112
158. Cheng J, Wei D, Ji Y, Chen L, Yang L, Li G, et al. Integrative Analysis of DNA Methylation and Gene Expression Reveals Hepatocellular Carcinoma-Specific Diagnostic Biomarkers. Genome Med (2018) 10(1):42. doi: 10.1186/s13073-018-0548-z
159. Inoue T, Tanaka Y. Novel Biomarkers for the Management of Chronic Hepatitis B. Clin Mol Hepatol (2020) 26(3):261–79. doi: 10.3350/cmh.2020.0032
160. Zhang S, Li W, Lu H, Liu Y. Quantification of N-Glycosylation Site Occupancy Status Based on Labeling/Label-Free Strategies With LC-Ms/MS. Talanta (2017) 170:509–13. doi: 10.1016/j.talanta.2017.04.053
161. Du X, Lu J, Yang L, Bao H, Zhang L, Yan G, et al. Rapid and Easy Enrichment Strategy for Naturally Acetylated N Termini Based on LysN Digestion and Amine-Reactive Resin Capture. Anal Chem (2020) 92(12):8315–22. doi: 10.1021/acs.analchem.0c00695
162. Del Giacco L, Cattaneo C. Introduction to Genomics. Methods Mol Biol (2012) 823:79–88. doi: 10.1007/978-1-60327-216-2_6
163. He Y, Mohamedali A, Huang C, Baker MS, Nice EC. Oncoproteomics: Current Status and Future Opportunities. Clin Chim Acta (2019) 495:611–24. doi: 10.1016/j.cca.2019.06.006
Keywords: hepatocellular carcinoma, multi-omics, biomarker, clinical diagnosis, early detection
Citation: Chen F, Wang J, Wu Y, Gao Q and Zhang S (2022) Potential Biomarkers for Liver Cancer Diagnosis Based on Multi-Omics Strategy. Front. Oncol. 12:822449. doi: 10.3389/fonc.2022.822449
Received: 25 November 2021; Accepted: 17 January 2022;
Published: 03 February 2022.
Edited by:
David Mitchell Lubman, University of Michigan, United StatesReviewed by:
Yu Lin, University of Michigan, United StatesZhijing Tan, Reidge Inc., United States
Haidi Yin, Shenzhen Bay Laboratory, China
Copyright © 2022 Chen, Wang, Wu, Gao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu Zhang, emhhbmcuc2h1QHpzLWhvc3BpdGFsLnNoLmNu
 Fanghua Chen
Fanghua Chen Junming Wang
Junming Wang Yingcheng Wu
Yingcheng Wu Qiang Gao
Qiang Gao Shu Zhang
Shu Zhang