- 1Department of Breast Surgery, The Second Hospital, Cheeloo College of Medicine, Shandong University, Jinan, China
- 2School of Medicine, Cheeloo College of Medicine, Shandong University, Jinan, China
- 3Institute of Translational Medicine of Breast Disease Prevention and Treatment, Shandong University, Jinan, China
- 4Shandong Provincial Engineering Laboratory of Translational Research on Prevention and Treatment of Breast Disease, Jinan, China
- 5Department of Breast Surgery, Affiliated Tumor Hospital of Zhengzhou University, Zhengzhou, China
- 6Department of Breast Surgery, Shanxi Cancer Hospital, Taiyuan, China
- 7Department of Breast Surgery, The First Hospital of Jilin University, Changchun, China
- 8Department of Breast Center, The Fourth Hospital of Hebei Medical University, Shijiazhuang, China
- 9Department of Breast Surgery, Tianjin Medical University Cancer Institute and Hospital, Tianjin, China
- 10Department of Thyroid and Breast Surgery, The First Affiliated Hospital of Binzhou Medical University, Binzhou, China
- 11Department of Breast Surgery, Cancer Hospital, Chinese Academy of Medical Sciences, Beijing, China
- 12Department of General Surgery, Linyi People’s Hospital, Linyi, China
- 13Department of Breast Disease Center, Peking University People’s Hospital, Beijing, China
- 14Department of General Surgery, Beijing Chaoyang Hospital, Beijing, China
- 15Department of Breast Disease Center, Peking University First Hospital, Beijing, China
- 16Department of Breast Center, Qingdao University Affiliated Hospital, Qingdao, China
- 17Department of Breast and Thyroid Surgery, Weifang Traditional Chinese Hospital, Weifang, China
- 18Department of Breast Surgery, The Second Affiliated Hospital of Qingdao Medical College, Qingdao Central Hospital, Qingdao, China
- 19Department of General Surgery, The Second Affiliated Hospital of Harbin Medical University, Harbin, China
- 20Department of Breast Surgery, The First Affiliated Hospital of China Medical University, Shenyang, China
- 21Department of General Surgery, Nanjing Medical University Affiliated Cancer Hospital, Cancer Institute of Jiangsu Province, Nanjing, China
- 22Department of Breast and Thyroid Surgery, Zibo Central Hospital, Zibo, China
- 23Department of Breast Surgery, Yantai Yuhuangding Hospital, Yantai, China
- 24Department of Breast Cancer Center, Shandong Cancer Hospital, Jinan, China
Background: Leptin (LEP) plays a physiological role through its specific receptor (LEPR) and is involved in the occurrence and development of breast cancer. Our current study aimed at determining the influence of single-nucleotide polymorphisms (SNPs) in the genes coding for LEP and LEPR on breast cancer risk.
Methods: In the present study, 963 breast cancer cases and 953 controls were enrolled. Five SNPs of LEP and two of LEPR were chosen to evaluate the correlation of selected SNPs with breast cancer susceptibility among women in northern and eastern China. Analyses were further stratified by body mass index (BMI), waist–hip rate (WHR), estrogen receptor, and progesterone receptor status. The expression patterns of risk variant-associated genes were detected by expression quantitative trait locus (eQTL) analysis with eQTLGen and The Cancer Genome Atlas database.
Results: There were significant differences between breast cancer cases and control groups in the menopausal status and family history of breast cancer. Two SNPs (rs1137101 and rs4655555) of the LEPR gene decreased overall breast cancer risk, and other five SNPs showed no significant association with breast cancer risk. rs1137101 (GA vs. GG; adjusted OR = 0.719, 95% CI = 0.578–0.894, p = 0.003) and rs4655555 (TT vs. AA; adjusted OR = 0.574, 95% CI = 0.377–0.873, p = 0.009) significantly decreased breast cancer risk after Bonferroni correction for multiple testing. In subgroup analyses, the GA and GA + AA genotypes of LEPR rs1137101 associated with decreased breast cancer risk in the subgroup of BMI ≤ 24 kg/m2 or WHR ≥ 0.85 after Bonferroni correction. Furthermore, we found that the expressions of rs4655555-associated gene LEPR and leptin receptor overlapping transcript (LEPROT) were upregulated in breast cancer tumor tissues compared with adjacent normal tissues, and a higher expression of LEPR in tumor tissues was correlated with poor prognosis of breast cancer patients using The Cancer Genome Atlas Breast Invasive Carcinoma (TCGA-BRCA) data.
Conclusion: Our study demonstrated that the polymorphisms rs1137101 and rs4655555 located in the LEPR gene decreased breast cancer risk in Chinese females, which might be a research-worthy bio-diagnostic marker and applied for early prediction and risk assessment of breast cancer.
Introduction
According to Global Cancer Statistics 2021, breast cancer is the most commonly diagnosed cancer worldwide, which accounts for 30% of all new cancers in women. Furthermore, breast cancer is the second leading cause of cancer death among women (1). China is undergoing cancer transition with an increasing burden of breast cancer, and female breast cancer patients took up 18.41% of breast cancer deaths across the world (2). Breast cancer incidence rates continue to increase by about 0.5% per year, which is attributed at least in part to continued declines in fertility rate and increased body weight (3). There are varieties of factors that can increase the risk of breast cancer, including family history of breast cancer, breastfeeding, secondhand smoke, eating habits, obesity, and diabetes mellitus (4). Based on larger observational studies, obesity is associated with a higher risk of developing breast cancer, particularly in postmenopausal women (5, 6). Obesity induces the dysfunction of adipocyte and changes the expression levels of adipokines and hormones, which will promote the progress of obesity related-tumors (7). A high expression of leptin (LEP) and low levels of adiponectin in obese patients were identified associated with the occurrence of breast cancer (8).
Leptin (LEP), the circulating product of the obesity gene, is a 16-kDa glycoprotein expressed and secreted primarily by the adipocyte. LEP plays an important role in body weight homeostasis by influencing food intake and energy expenditure and maintaining constant energy stores (9). In addition to the regulation of body weight, LEP was also identified to be involved in insulin resistance, cancer cell inflammation, oxidative stress, cell proliferation, apoptosis, angiogenesis, and antitumor immune regulation (10, 11). Leptin exerts its biological action majorly through binding to and activating the leptin receptors (LEPR) (12) and the gene encoding LEPR overlapping transcript (LEPROT).
The expressions of LEP and LEPR were associated with enhanced cell proliferation and angiogenesis in both benign and malignant breast epithelial cells (13–17). Higher circulating leptin concentrations were significantly associated with an increased risk of breast cancer (18). Previous studies indicated that LEPROT could negatively regulate the cell surface expression of LEPR and the silencing LEPROT expression in the mouse hypothalamic arcuate nucleus prevented the development of high-fat-diet-induced obesity (19). In contrast, LEPROT was found to activate the JAK/STAT pathway and may facilitate cancer development (20). A recent study identified an aberrant expression of LEPROT in 78.9% cancers compared with corresponding normal tissues (21).
Studies have identified genetic variants of LEP and LEPR correlated with susceptibility of various malignant tumors, including breast cancer (22, 23). Several single-nucleotide polymorphisms (SNPs) of LEP and LEPR were found correlated with breast cancer risk, including LEP-2548G/A (rs7799039), LEPR K109R (rs1137100), and LEPR Q223R (rs1137101) (24–27), but the results are not exactly consistent. Based on these results, in the current study, we selected 7 polymorphisms located in the LEP and LEPR genes and identified the association with breast cancer risk in northern and eastern Chinese Han females by conducting a multicenter case–control study. This exploration could further provide a research basis for discovering pathogenic targets of breast cancer prevention.
Materials and Methods
Participants
Characteristics of the study participants have been reported previously (4). Briefly, participants in the case group were Han ethnic female patients aged 25 to 70 years, who had newly diagnosed, histologically confirmed breast cancer and were recruited at 21 hospitals located in 11 provinces of northern and eastern China between April 2012 and April 2013. The control group comprised age-matched (± 3 years) volunteers recruited at the same hospital who were examined within 2 months of the case group and were confirmed as being breast cancer free by negative physical and imaging findings. Participants with other malignant tumors were excluded from the study. The ethics committee of the Second Hospital, Cheeloo College of Medicine, Shandong University, had approved this study, and all participants signed informed consent.
Personal information and samples from each participant were obtained after signing the informed consent. The demographic information and lifestyle habits of the participants, clinical data including age, body mass index (BMI), waist–hip rate (WHR), personal medical history, family backgrounds, the clinical examination results of visual examination, palpation, and related diagnostic tests such as breast ultrasound and mammography were documented. The statuses of the estrogen receptor (ER) and progesterone receptor (PR) were determined by immunohistochemical staining and obtained from the patients’ medical records. According to the American Society of Clinical Oncology/College of American Pathologists (2020) guideline recommendations, samples with 1% to 100% of tumor nuclei positive for ER or PR are interpreted as positive (28). For each participant, a 4-ml non-fasting blood sample was collected using an EDTA vacutainer (Becton Dickinson, New York). Each blood sample was stored vertically in a freezer at -80°C after sedimentation.
Genotyping and Laboratory Methods
Blood DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, Madison, USA). According to previous studies, 7 SNPs were reported in relation with obesity, weight, or cancer risk including rs10244329 (29), rs10954173 (30), rs2167270 (31), rs3828942 (29), and rs4731426 (32) of LEP and rs1137101 (33) and rs4655555 (34) of LEPR (Supplementary Table 1) with a minor allelic frequency (MAF) > 5% according to the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/), which were selected for further analysis. All participants were genotyped using the Sequenom MassARRAY SNP system (CapitalBio Technology, Beijing, China), as previously described (35).
Statistical Analysis
SPSS 26.0 statistical software (IBM, New York) was used to analyze the data. Among them, χ2 tests were used to compare the differences in demographic and lifestyle data between the case and control groups. A population representative was detected using Hardy–Weinberg equilibrium (HWE) in the control group. Unconditional logistics regression was used to assess the co-dominant (heterozygous or mutant homozygous vs. wild-type homozygous), dominant (heterozygous and mutant homozygous vs. wild-type homozygous), and recessive (mutant homozygous vs. wild-type homozygous and heterozygous) models of genetic variants and breast cancer risk. Odds ratios (OR) with 95% confidence interval (95% CI) were estimated after adjustment for menstrual status and family history of breast cancer. Subjects were further stratified into subgroups according to BMI, WHR, ER, and PR statuses. Bonferroni correction was used to adjust for multiple testing, and the level of significance was set at α < 0.01 (0.05/5) for testing the five loci of LEP and α < 0.025 (0.05/2) for testing the two loci of LEPR.
The eQTLGen database was used to identify affected genes related to the risk SNPs (36). The expressions of related genes in paired breast tumor tissue and normal tissue (n = 112) of The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) were further analyzed by using the two-tailed paired Wilcoxon rank-sum test. The association between related genes and the survival rate of breast cancer patients were analyzed by the Kaplan–Meier survival analysis based on TCGA Breast Invasive Carcinoma (TCGA-BRCA) data. The curves were generated with an optimum cutoff value for LEPR or LEPROT expression. A p-value < 0.05 was considered as statistically significant unless otherwise specified.
Results
General Demographic Characteristics of Participants
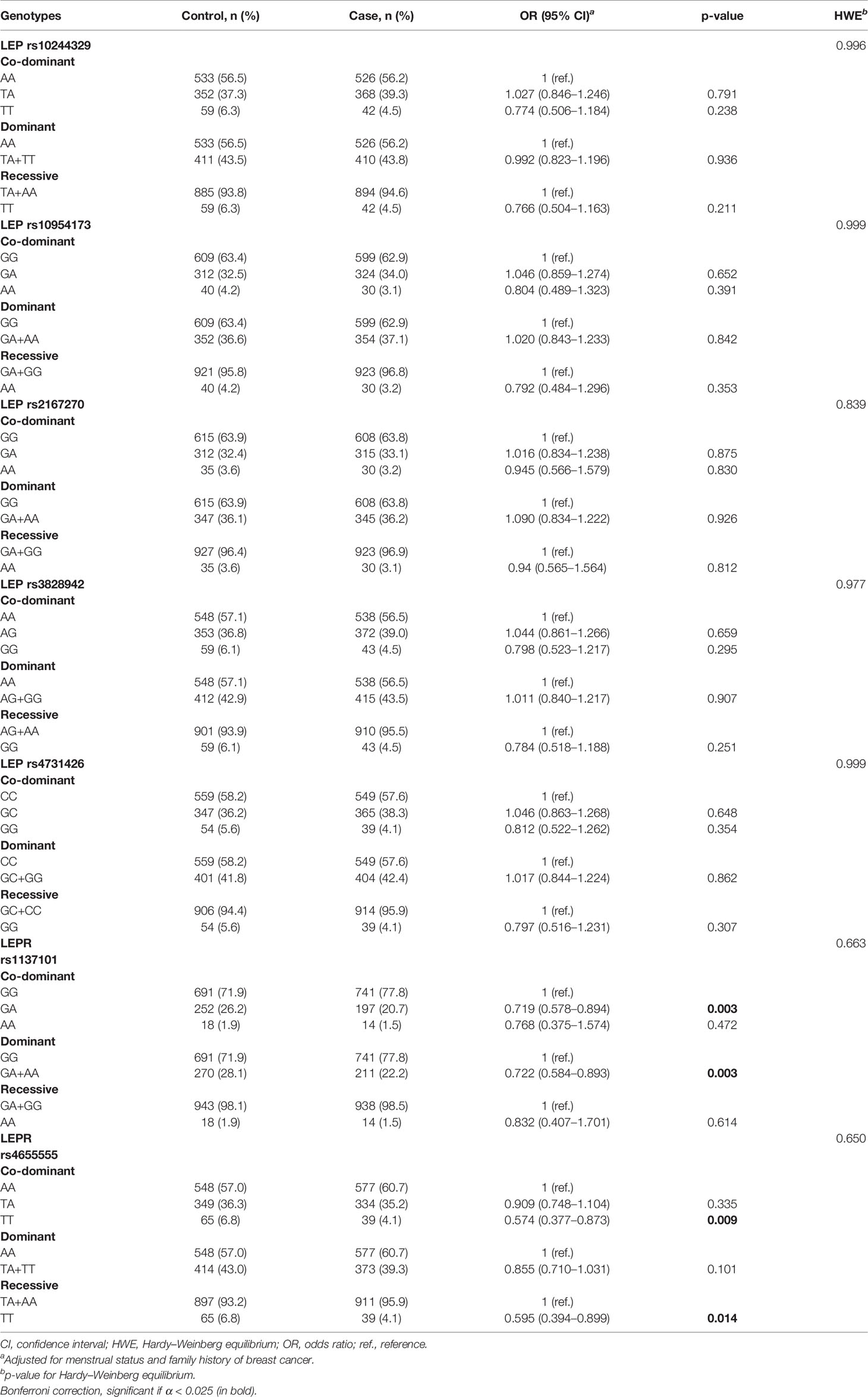
In this study, we enrolled 963 controls and 953 cases. The general demographic characteristics of the participants are presented in Table 1. The menstrual status and family history of breast cancer showed significant differences between the case and control groups (p < 0.05). There was no significant difference for other factors between the two groups.
Genotype Distribution of LEP/LEPR and Breast Cancer Risk
The association of LEP and LEPR SNP genotypes with breast cancer risk after adjustments for risk factors, including menstrual status and family history of breast cancer, is shown in Table 2. All SNPs were consistent with HWE in the control group (p > 0.05). Among the 7 SNPs, LEPR rs1137101 showed a significantly decreased breast cancer risk under the dominant genetic model (GA + AA vs. GG, adjusted OR = 0.722, 95% CI = 0.584–0.893, p = 0.003) and co-dominant genetic model (GA vs. GG, adjusted OR = 0.719, 95% CI = 0.578–0.894, p = 0.003). For LEPR rs4655555, a significant association with decreased breast cancer risk was also identified in the co-dominant genetic model (TT vs. AA, adjusted OR = 0.574, 95% CI = 0.377–0.873, p = 0.009) and recessive model (TT vs. TA + AA, adjusted OR = 0.595, 95% CI = 0.394–0.899, p = 0.014). However, the other five SNPs showed no significant association with overall breast cancer risk.
Associations Between LEP/LEPR Polymorphisms and Risk of ER+/PR+ or ER-/PR- Breast Cancer Patients
Among the 953 cases, 848 (89.0%) patients had explicit joint ER and PR statuses. Overall, 572 (60.0%) cases were ER+/PR+, 189 (19.8%) cases were ER-/PR-, 72 (7.6%) cases were ER+/PR−, and 15 (1.6%) cases were ER-/PR+. Due to the limited sample size, we excluded ER+/PR- and ER-/PR+ cases for further analysis. The association between the genotypes of LEP/LEPR and the risk of ER+/PR+ or ER-/PR- cases is shown in Table 3 and Supplementary Table 2. LEPR rs1137101 decreased ER+/PR+ breast cancer risk in the dominant genetic model (GA + AA vs. GG, adjusted OR = 0.727, 95% CI = 0.568–0.930, p = 0.011) and the co-dominant genetic model (GA vs. GG, adjusted OR = 0.739, 95% CI = 0.575–0.951, p = 0.019). For LEPR rs4655555, a significant association with decreased ER+/PR+ breast cancer risk in the co-dominant genetic model (TT vs. AA, adjusted OR = 0.533, 95% CI = 0.321–0.884, p = 0.015) and the recessive model (TT vs. TA + AA, adjusted OR = 0.562, 95% CI = 0.341–0.925, P = 0.023) was identified.
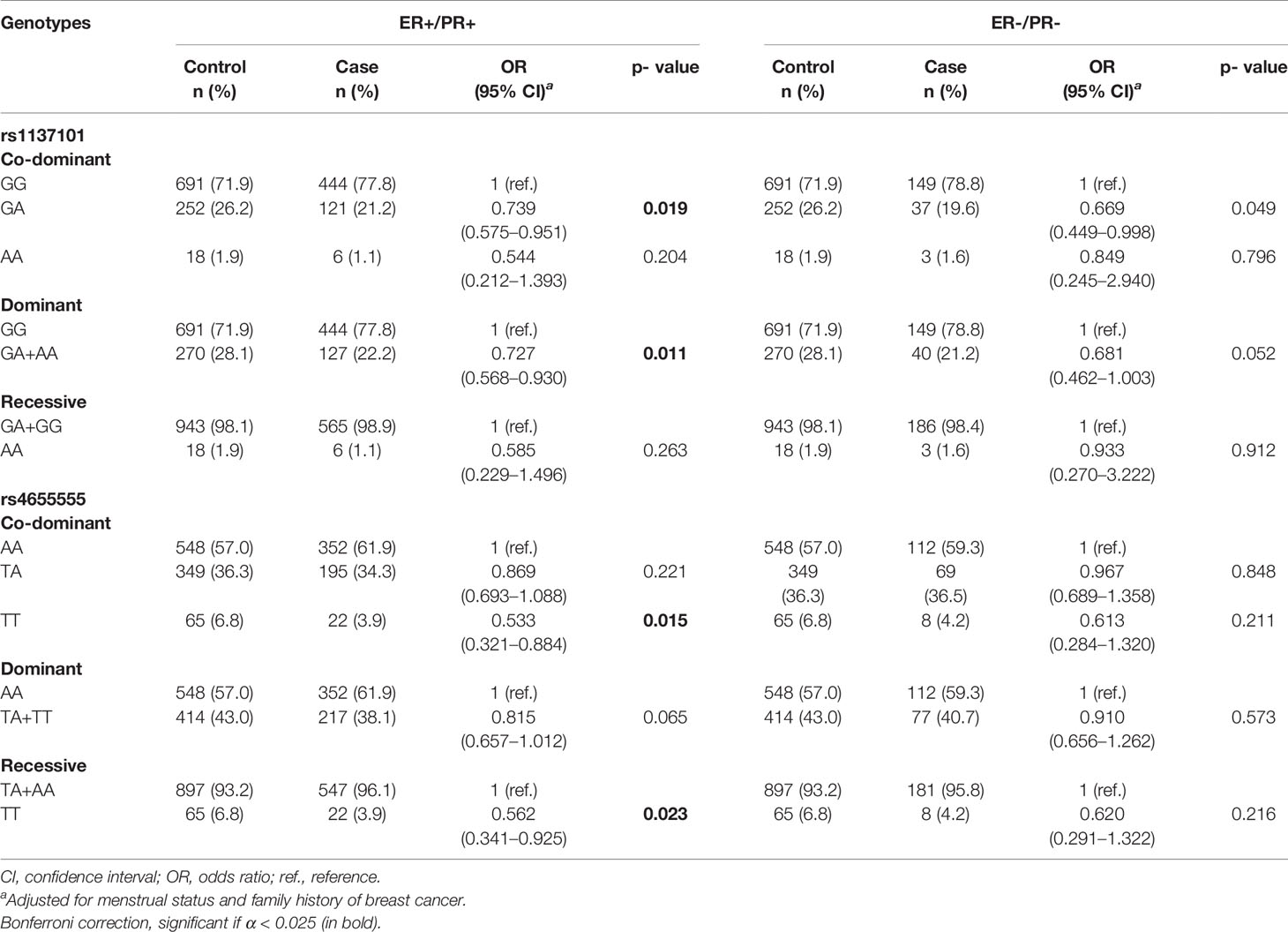
Table 3 The association of LEP/LEPR genetic variations with risk of ER+/PR+ and ER-/PR- breast cancer.
The Association Between LEP/LEPR Genotypes and Breast Cancer Risk According to BMI and/or WHR
We further performed stratified analysis to determine the association between LEP/LEPR polymorphisms and breast cancer risk according to obesity indicators including BMI and WHR (Table 4 and Supplementary Tables 3, 4).
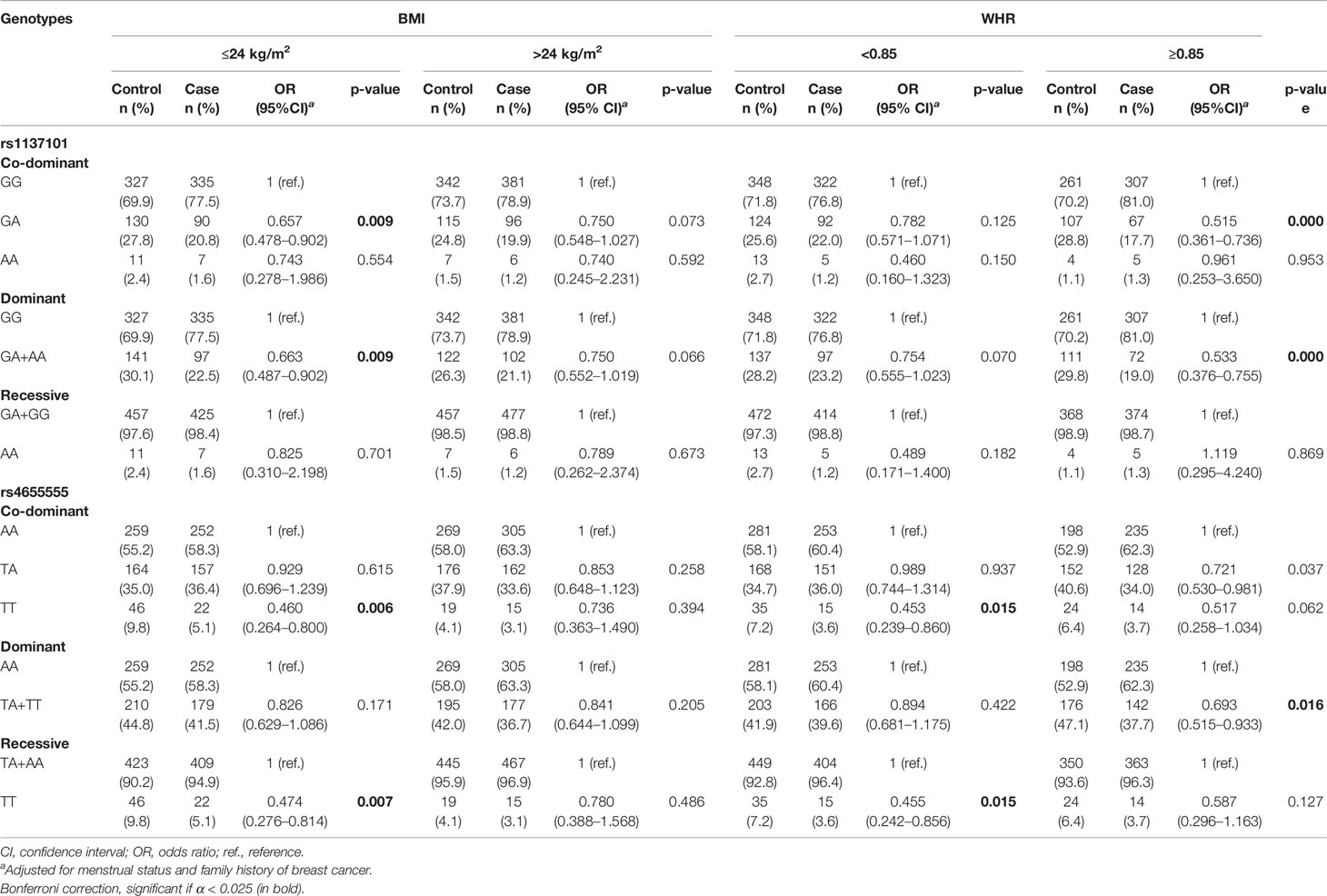
Table 4 The association of LEP/LEPR genetic variations with breast cancer risk according to BMI or WHR category.
According to the BMI category, rs1137101 and rs4655555 only showed a significant association with decreased breast cancer risk in the subgroup of BMI ≤ 24 kg/m2 (Table 4, p < 0.01). In the subgroup of WHR ≥ 0.85, rs1137101 and rs4655555 showed a lower breast cancer risk in the dominant genetic model (Table 4, rs1137101, GA + AA vs. GG, adjusted OR = 0.533, 95% CI = 0.376–0.755, p = 0.000; rs4655555, TA + TT vs. AA, adjusted OR = 0.693, 95% CI = 0.515–0.933, p = 0.016). A similar association between rs4655555 and breast cancer risk was also identified in women with WHR < 0.85 (Table 4, TT vs. AA, adjusted OR = 0.453, 95% CI = 0.239–0.860, p = 0.015; TT vs. TA+ AA, adjusted OR = 0.455, 95% CI = 0.242–0.856, p = 0.015).
We further conducted a stratified analysis by combining BMI and WHR. The results indicated a similar association between the rs1137101 genotype and decreased breast cancer risk in the BMI ≤ 24.0 kg/m2 and WHR ≥ 0.85 subgroup (Supplementary Table 4, GA+AA vs. GG, adjusted OR = 0.482, 95% CI = 0.288–0.807, p = 0.006) and BMI > 24.0 kg/m2 and WHR ≥ 0.85 subgroup (Supplementary Table 4, GA+AA vs. GG, adjusted OR = 0.563, 95% CI = 0.344–0.921, p = 0.022). The TT genotype of rs4655555 showed a decreased breast cancer risk in women of BMI ≤ 24.0 kg/m2 and WHR < 0.85 (Supplementary Table 4, TT vs. AA, adjusted OR = 0.397, 95% CI = 0.179–0.884, p = 0.024; TT vs. TA+ AA, adjusted OR = 0.389, 95% CI = 0.177–0.852, p = 0.018).
Expression of rs4655555-Associated Genes in Breast Cancer Tissues
To further identify the downstream associated genes of breast cancer risk-related variants, we found that 2 cis-eQTL genes (LEPR and LEPROT) were associated with rs4655555 based on the eQTLGen database (36) (Table 5). The expression pattern of LEPR and LEPROT in breast cancer tissues was analyzed using TCGA data. The results indicated that compared to matched adjacent normal tissues, both LEPR and LEPROT showed a higher expression in breast cancer tumor tissues (p < 0.001, Figures 1A, B).
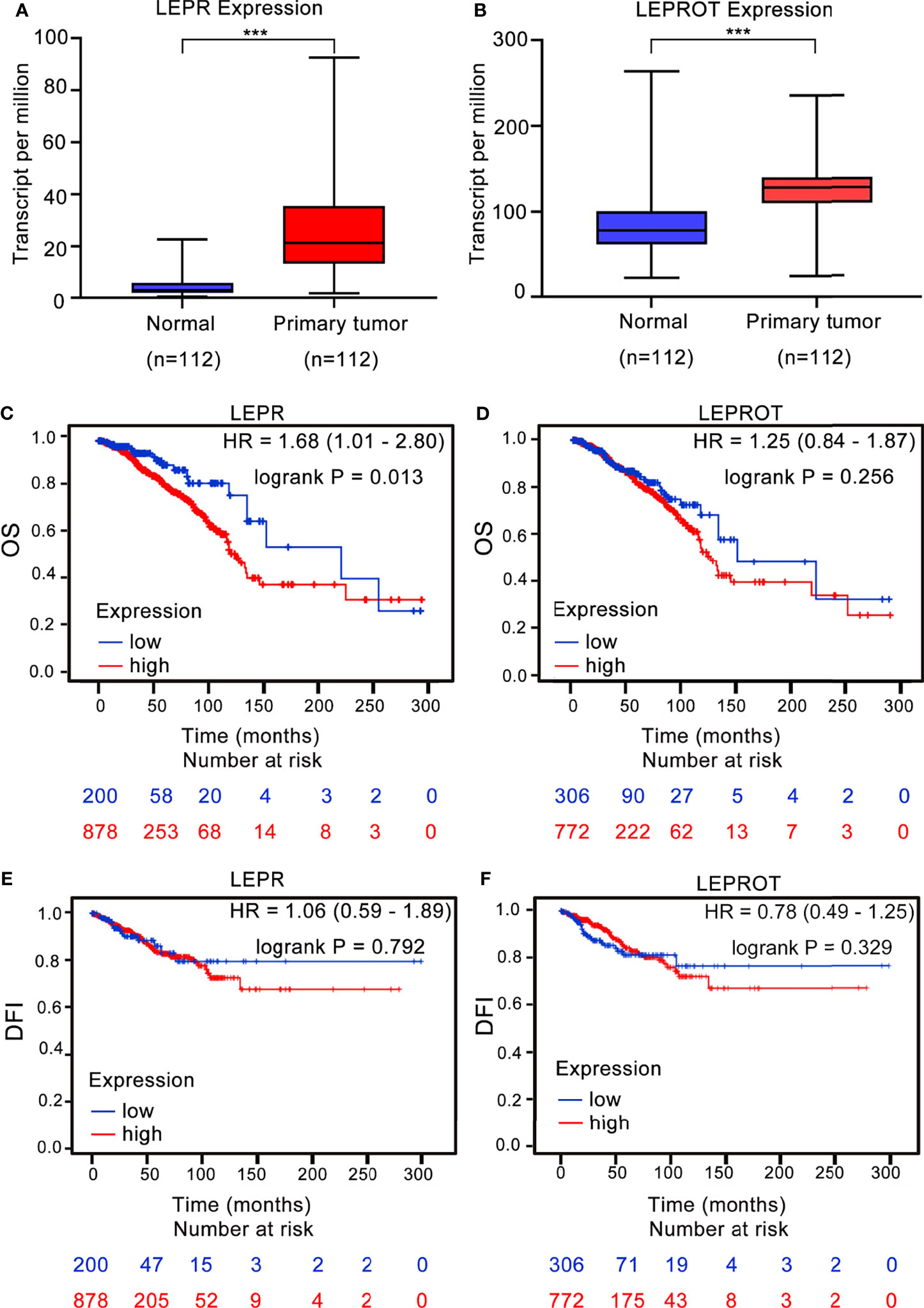
Figure 1 The association between LEPR/LEPROT expression and prognosis of breast cancer patients. The expression of LEPR (A) and LEPROT (B) in adjacent normal tissues and primary tumor issues of paired TCGA-BRCA data as analyzed by two-tailed paired Wilcoxon’s rank-sum test. ***p-value < 0.001. Kaplan–Meier curves for overall survival (OS) and disease-free interval (DFI) by using TCGA-BRCA data according to high and low LEPR (C, E) or LEPROT (D, F) gene expression. HR with 95% CI and log-rank p values were calculated. A log-rank p value < 0.05 was considered as statistically significant.
The association between LEPR or LEPROT level and outcomes of breast cancer patients was further evaluated by Kaplan–Meier survival analysis based on TCGA-BRCA data. A high expression of LEPR is significantly associated with poor OS of breast cancer patients, with HRs (95% CIs) of 1.68 (95% CI: 1.01–2.80) (Figure 1C). However, the association between LEPROT expression and OS of breast cancer patients was not significant (Figure 1D, HR = 1.25, 95% CI = 0.84–1.87). Furthermore, no significant association between LEPR or LEPROT expression and disease-free interval (DFI) of breast cancer patients was identified (Figures 1E, F).
Discussion
Obesity is a common public health problem nowadays. A number of studies have shown that obesity is associated with the occurrence of breast cancer (5, 6). The molecular mechanisms of the relationship between obesity and breast cancer involve estrogens, insulin, leptin, adiponectin, and inflammatory cytokines. Specifically, the activation of leptin signaling leads to simultaneous activation of multiple oncogenic pathways, leading to increased breast cancer cell proliferation, epithelial–mesenchymal transformation, migration, and invasion (37, 38). Previous studies have shown that elevated leptin levels are associated with aggressiveness and poor prognosis of breast cancer patients (18, 39). In addition, studies also showed that variants of LEP and LEPR gene are associated with breast cancer susceptibility (24–26). In the current study, we recruited 1,616 participants including 963 breast cancer cases and 953 cancer-free controls to assess the correlation between LEP/LEPR polymorphisms and susceptibility of breast cancer. Our current study found a significant association between the LEPR rs1137101 and rs4655555 variants and decreased risk of breast cancer in a Chinese Han population.
The LEPR rs1137101 (Arg223Gln), a missense SNP, has been analyzed for the correlation with cancer risk and development; however, the previous results were not consistent. In the current study, we found that the GA and GA+AA genotypes of rs1137101 were associated with decreased breast cancer risk. Some previous case–control studies reported that the A allele of rs1137101 is a protective factor against cancer occurrence (26, 40, 41), which were consistent with our results, but some studies have shown the opposite results (42, 43). The inconsistent results may be due to differences in race, genetic background, environment, or lifestyle.
We further identified that the genotype of LEPR rs1137101 was associated with decreased breast cancer risk under the dominant genetic model in the subgroup of WHR ≥ 0.85 regardless of BMI by stratified analysis (Table 4 and Supplementary Table 4). The risk of breast cancer in rs1137101 GA carriers was most significantly reduced in women with WHR ≥ 0.85 and normal BMI (Supplementary Table 4, BMI ≤ 24.0 kg/m2 and WHR ≥ 0.85, adjusted OR = 0.446, 95% CI: 0.264–0.754, p = 0.003), which was the indicator of central obesity. Our previous study indicated that central obesity was positive with ER-/PR- breast cancer risk (44). A similar trend of the GA genotype of rs1137101 was associated with a decreased risk in ER-/PR- breast cancer (Table 3), indicating that rs1137101 may participate in mediating the correlation between central obesity and ER-/PR- breast cancer risk. We also found that the GA + AA genotype of rs1137101 was significantly associated with decreased breast cancer risk in women of WHR ≥ 0.85 and BMI > 24.0 kg/m2, which may partially explain for the reduced risk of ER+/PR+ cases (Table 3 and Supplementary Table 4). A previous study has shown that serum leptin levels are positively associated with increased expression of ER and PR in breast cancer patients (45). Similarly, LEPR expression was positively correlated with tumor size and ER expression in breast cancer (46). However, the association between rs1137101 genotypes and LEPR expression is still unknown. Therefore, the in-depth mechanism between rs1137101 and breast cancer risk in different hormone receptor statuses needs to be further explored.
To our knowledge, the association between rs465555 and breast cancer susceptibility has not been demonstrated in previous studies. We found that the TT genotype of rs4655555 was associated with decreased overall breast cancer risk. A similar association was identified in the subgroup of BMI ≤ 24.0 kg/m2 and BMI ≤ 24.0 kg/m2 and WHR < 0.85. Based on the eQTLGen database, we identified 2 cis-eQTL genes of rs4655555, including LEPR and LEPROT, the receptors for leptin action (47). A previous genome-wide association study also identified a strong association between rs4655555 and circulating soluble leptin receptor (sOB-R) levels (34). There is no previous correlation study between rs465555 and LEPROT expression. To evaluate the potential causal function of rs4655555-associated genes in breast cancer risk, based on the TCGA database, we found a higher expression of LEPR and LEPROT in breast cancer tumor tissues compared to that of adjacent normal tissues. Previous studies indicated that leptin signaling played a key role in breast cancer incidence and development, and a higher expression of leptin and LEPR was also validated in breast cancer tissues (48, 49). The expression of LEPR is also necessary for maintaining cancer stem cell-like and metastatic properties in triple-negative breast cancer (50). We further identified that a high expression of LEPR significantly correlated with worse prognosis of breast cancer patients, which suggesting that the rs4655555 variant may affect breast cancer risk and development through regulating the expression of LEPR, and subsequently prognosis. Further confirmatory studies are necessary to validate the regulatory mechanism in variant-associated breast carcinogenesis.
The strengths of this study include the multicenter retrospective study design, the large sample size, the availability of fasting blood samples, and the measurement and examination of relevant indicators in participants using standardized procedures. However, some limitations of this study need to be addressed. First, although the number of participants was relatively large, the sample sizes of some subgroups were small. A larger external multicenter prospective cohort study needs to be further conducted to validate our identification. Furthermore, the correlation between rs1137101, rs4655555, and downstream causal genes needs to be further validated by conducting experimental analyses.
In conclusion, our study provides evidence that rs1137101 and rs4655555 of the LEPR gene are associated with breast cancer susceptibility in Chinese women. In addition, rs1137101 may have the potential to inhibit the occurrence and development of breast cancer in centrally obese women, providing new ideas for the prevention of obesity-associated breast cancer.
Data Availability Statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding authors on reasonable request.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of The Second Hospital, Cheeloo College of Medicine, Shandong University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
ZGY conceived and designed the study. XM and SH performed statistical analyses. LYL and FW organized the database. LL (1st author), XM, and SH wrote the first draft of the manuscript. LY, FZ, and CZ contributed to the manuscript revision and statistical analyses. YX, WZZ, CY, GY, and ZM contributed to the DNA extraction. SC, FT, ZF, CG, XC, ZLY, XW, HL, SW, HJ, XD, HW, GL, QW, JZ, FJ, JT, LL (27th author), SZ, and WSZ contributed to the collection of the data and biological samples. All authors contributed to the manuscript revision and read and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (82072914), Natural Science Foundation of Shandong Province (ZR2019PH016), and National Key Research and Development Program of China (2016YFC0901300).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank all the individuals involved in the case–control study for their participation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.809570/full#supplementary-material
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
2. Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing Profiles of Cancer Burden Worldwide and in China: A Secondary Analysis of the Global Cancer Statistics 2020. Chin Med J (Engl) (2021) 134(7):783–91. doi: 10.1097/CM9.0000000000001474
3. Pfeiffer RM, Webb-Vargas Y, Wheeler W, Gail MH. Proportion of U.S. Trends in Breast Cancer Incidence Attributable to Long-Term Changes in Risk Factor Distributions. Cancer Epidemiol Biomarkers Prev (2018) 27(10):1214–22. doi: 10.1158/1055-9965.EPI-18-0098
4. Liu LY, Wang F, Cui SD, Tian FG, Fan ZM, Geng CZ, et al. A Case-Control Study on Risk Factors of Breast Cancer in Han Chinese Women. Oncotarget (2017) 8(57):97217–30. doi: 10.18632/oncotarget.21743
5. Picon-Ruiz M, Morata-Tarifa C, Valle-Goffin JJ, Friedman ER, Slingerland JM. Obesity and Adverse Breast Cancer Risk and Outcome: Mechanistic Insights and Strategies for Intervention. CA Cancer J Clin (2017) 67(5):378–97. doi: 10.3322/caac.21405
6. Gravena AAF, Romeiro Lopes TC, Demitto MO, Borghesan DHP, Dell’ Agnolo CM, Brischiliari SCR, et al. The Obesity and the Risk of Breast Cancer Among Pre and Postmenopausal Women. Asian Pac J Cancer Prev (2018) 19(9):2429–36. doi: 10.22034/APJCP.2018.19.9.2429
7. Atoum MF, Alzoughool F, Al-Hourani H. Linkage Between Obesity Leptin and Breast Cancer. Breast Cancer (Auckl) (2020) 14:1178223419898458. doi: 10.1177/1178223419898458
8. Pu X, Chen D. Targeting Adipokines in Obesity-Related Tumors. Front Oncol (2021) 11:685923. doi: 10.3389/fonc.2021.685923
9. Pan WW, Myers MG Jr. Leptin and the Maintenance of Elevated Body Weight. Nat Rev Neurosci (2018) 19(2):95–105. doi: 10.1038/nrn.2017.168
10. Uddin S, Hussain AR, Siraj AK, Khan OS, Bavi PP, Al-Kuraya KS. Role of Leptin and Its Receptors in the Pathogenesis of Thyroid Cancer. Int J Clin Exp Pathol (2011) 4(7):637–43. doi: 10.1002/ijc.25536
11. Ray A, Cleary MP. The Potential Role of Leptin in Tumor Invasion and Metastasis. Cytokine Growth Factor Rev (2017) 38:80–97. doi: 10.1016/j.cytogfr.2017.11.002
12. de Luis DA, Perez Castrillon JL, Duenas A. Leptin and Obesity. Minerva Med (2009) 100(3):229–36.
13. Hu X, Juneja SC, Maihle NJ, Cleary MP. Leptin–a Growth Factor in Normal and Malignant Breast Cells and for Normal Mammary Gland Development. J Natl Cancer Inst (2002) 94(22):1704–11. doi: 10.1093/jnci/94.22.1704
14. Strong AL, Ohlstein JF, Biagas BA, Rhodes LV, Pei DT, Tucker HA, et al. Leptin Produced by Obese Adipose Stromal/Stem Cells Enhances Proliferation and Metastasis of Estrogen Receptor Positive Breast Cancers. Breast Cancer Res (2015) 17:112. doi: 10.1186/s13058-015-0622-z
15. Dieudonne MN, Machinal-Quelin F, Serazin-Leroy V, Leneveu MC, Pecquery R, Giudicelli Y. Leptin Mediates a Proliferative Response in Human MCF7 Breast Cancer Cells. Biochem Biophys Res Commun (2002) 293(1):622–8. doi: 10.1016/S0006-291X(02)00205-X
16. Dubois V, Jarde T, Delort L, Billard H, Bernard-Gallon D, Berger E, et al. Leptin Induces a Proliferative Response in Breast Cancer Cells But Not in Normal Breast Cells. Nutr Cancer (2014) 66(4):645–55. doi: 10.1080/01635581.2014.894104
17. Laud K, Gourdou I, Pessemesse L, Peyrat JP, Djiane J. Identification of Leptin Receptors in Human Breast Cancer: Functional Activity in the T47-D Breast Cancer Cell Line. Mol Cell Endocrinol (2002) 188(1-2):219–26. doi: 10.1016/s0303-7207(01)00678-5
18. Wu MH, Chou YC, Chou WY, Hsu GC, Chu CH, Yu CP, et al. Circulating Levels of Leptin, Adiposity and Breast Cancer Risk. Br J Cancer (2009) 100(4):578–82. doi: 10.1038/sj.bjc.6604913
19. Couturier C, Sarkis C, Seron K, Belouzard S, Chen P, Lenain A, et al. Silencing of OB-RGRP in Mouse Hypothalamic Arcuate Nucleus Increases Leptin Receptor Signaling and Prevents Diet-Induced Obesity. Proc Natl Acad Sci USA (2007) 104(49):19476–81. doi: 10.1073/pnas.0706671104
20. Li X, Shi W, Wu G, Qin X, Wan G, Zeng Q, et al. OB-RGRP Regulates the Phosphorylation of JAK2 and STAT3 in Primary Rat Adipocytes. Artif Cells Nanomed Biotechnol (2019) 47(1):3664–70. doi: 10.1080/21691401.2019.1632322
21. Li B, He Y, Li P, Chen X. Leptin Receptor Overlapping Transcript (LEPROT) Is Associated With the Tumor Microenvironment and a Prognostic Predictor in Pan-Cancer. Front Genet (2021) 12:749435. doi: 10.3389/fgene.2021.749435
22. Lin J, Xie Z, Lan B, Guo Z, Tang WF, Liu C, et al. Investigation of Leptin and Its Receptor (LEPR) for Single Nucleotide Polymorphisms in Colorectal Cancer: A Case-Control Study Involving 2,306 Subjects. Am J Transl Res (2020) 12(7):3613–28. doi: 10.1200/JCO.2020.38.15_suppl.e16100
23. Unsal M, Kara N, Karakus N, Tural S, Elbistan M. Effects of Leptin and Leptin Receptor Gene Polymorphisms on Lung Cancer. Tumour Biol (2014) 35(10):10231–6. doi: 10.1007/s13277-014-2293-2
24. Mohammadzadeh G, Ghaffari MA, Bafandeh A, Hosseini SM, Ahmadi B. The Relationship Between -2548 G/A Leptin Gene Polymorphism and Risk of Breast Cancer and Serum Leptin Levels in Ahvazian Women. Iran J Cancer Prev (2015) 8(2):100–8.
25. El-Hussiny MA, Atwa MA, Rashad WE, Shaheen DA, Elkady NM. Leptin Receptor Q223R Polymorphism in Egyptian Female Patients With Breast Cancer. Contemp Oncol (Pozn) (2017) 21(1):42–7. doi: 10.5114/wo.2017.66655
26. Mohammadzadeh G, Ghaffari MA, Bafandeh A, Hosseini SM. Effect of Leptin Receptor Q223R Polymorphism on Breast Cancer Risk. Iran J Basic Med Sci (2014) 17(8):588–94.
27. Wang LQ, Shen W, Xu L, Chen MB, Gong T, Lu PH, et al. The Association Between Polymorphisms in the Leptin Receptor Gene and Risk of Breast Cancer: A Systematic Review and Pooled Analysis. Breast Cancer Res Treat (2012) 136(1):231–9. doi: 10.1007/s10549-012-2228-9
28. Allison KH, Hammond MEH, Dowsett M, McKernin SE, Carey LA, Fitzgibbons PL, et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Guideline Update. Arch Pathol Lab Med (2020) 144(5):545–63. doi: 10.5858/arpa.2019-0904-SA
29. Labayen I, Ruiz JR, Moreno LA, Ortega FB, Beghin L, DeHenauw S, et al. The Effect of Ponderal Index at Birth on the Relationships Between Common LEP and LEPR Polymorphisms and Adiposity in Adolescents. Obesity (2011) 19(10):2038–45. doi: 10.1038/oby.2011.74
30. Brandl EJ, Frydrychowicz C, Tiwari AK, Lett TA, Kitzrow W, Buttner S, et al. Association Study of Polymorphisms in Leptin and Leptin Receptor Genes With Antipsychotic-Induced Body Weight Gain. Prog Neuropsychopharmacol Biol Psychiatry (2012) 38(2):134–41. doi: 10.1016/j.pnpbp.2012.03.001
31. Zhang A, Wang S, Zhang F, Li W, Li Q, Liu X. The Prognosis of Leptin Rs2167270 G > A (G19A) Polymorphism in the Risk of Cancer: A Meta-Analysis. Front Oncol (2021) 11:754162. doi: 10.3389/fonc.2021.754162
32. Erez G, Tirosh A, Rudich A, Meiner V, Schwarzfuchs D, Sharon N, et al. Phenotypic and Genetic Variation in Leptin as Determinants of Weight Regain. Int J Obes (2011) 35(6):785–92. doi: 10.1038/ijo.2010.217
33. Bienkiewicz J, Romanowicz H, Wilczynski M, Jablonski G, Stepowicz A, Oblekowska A, et al. Association of Single Nucleotide Polymorphism LEP-R C.668A>G (P.Gln223Arg, Rs1137101) of Leptin Receptor Gene With Endometrial Cancer. BMC Cancer (2021) 21(1):925. doi: 10.1186/s12885-021-08620-y
34. Sun Q, Cornelis MC, Kraft P, Qi L, van Dam RM, Girman CJ, et al. Genome-Wide Association Study Identifies Polymorphisms in LEPR as Determinants of Plasma Soluble Leptin Receptor Levels. Hum Mol Genet (2010) 19(9):1846–55. doi: 10.1093/hmg/ddq056
35. Huang S, Liu L, Xiang Y, Wang F, Yu L, Zhou F, et al. Association of PTPN1 Polymorphisms With Breast Cancer Risk: A Case-Control Study in Chinese Females. J Cell Biochem (2019) 120(7):12039–50. doi: 10.1002/jcb.28490
36. Vosa U, Claringbould A, Westra HJ, Bonder MJ, Deelen P, Zeng B, et al. Large-Scale Cis- and trans-eQTL Analyses Identify Thousands of Genetic Loci and Polygenic Scores That Regulate Blood Gene Expression. Nat Genet (2021) 53(9):1300–10. doi: 10.1038/s41588-021-00913-z
37. Barone I, Giordano C, Bonofiglio D, Ando S, Catalano S. Leptin, Obesity and Breast Cancer: Progress to Understanding the Molecular Connections. Curr Opin Pharmacol (2016) 31:83–9. doi: 10.1016/j.coph.2016.10.003
38. Sanchez-Jimenez F, Perez-Perez A, de la Cruz-Merino L, Sanchez-Margalet V. Obesity and Breast Cancer: Role of Leptin. Front Oncol (2019) 9:596. doi: 10.3389/fonc.2019.00596
39. Guo S, Liu M, Wang G, Torroella-Kouri M, Gonzalez-Perez RR. Oncogenic Role and Therapeutic Target of Leptin Signaling in Breast Cancer and Cancer Stem Cells. Biochim Biophys Acta (2012) 1825(2):207–22. doi: 10.1016/j.bbcan.2012.01.002
40. Gu F, Kraft P, Rice M, Michels KB. Leptin and Leptin Receptor Genes in Relation to Premenopausal Breast Cancer Incidence and Grade in Caucasian Women. Breast Cancer Res Treat (2012) 131(1):17–25. doi: 10.1007/s10549-011-1778-6
41. Abdu Allah AM, El-Hefnway SM, Alhanafy AM, Zahran AM, Kasem HE. Leptin Receptor Gene (A/G) Polymorphism Rs1137101 and Renal Cell Carcinoma. Mol Cell Biochem (2018) 448(1-2):137–44. doi: 10.1007/s11010-018-3320-1
42. Luan H, Zhang H, Li Y, Wang P, Cao L, Ma H, et al. Association of Two Obesity-Related Gene Polymorphisms LEPG2548A Rs7799039 and LEPRQ223R Rs1137101 With the Risk of Breast Cancer. Oncotarget (2017) 8(35):59333–44. doi: 10.18632/oncotarget.19580
43. Li Z, Yuan W, Ning S, Li J, Zhai W, Zhang S. Role of Leptin Receptor (LEPR) Gene Polymorphisms and Haplotypes in Susceptibility to Hepatocellular Carcinoma in Subjects With Chronic Hepatitis B Virus Infection. Mol Diagn Ther (2012) 16(6):383–8. doi: 10.1007/s40291-012-0008-1
44. Wang F, Liu L, Cui S, Tian F, Fan Z, Geng C, et al. Distinct Effects of Body Mass Index and Waist/Hip Ratio on Risk of Breast Cancer by Joint Estrogen and Progestogen Receptor Status: Results From a Case-Control Study in Northern and Eastern China and Implications for Chemoprevention. Oncol (2017) 22(12):1431–43. doi: 10.1634/theoncologist.2017-0148
45. Tessitore L, Vizio B, Pesola D, Cecchini F, Mussa A, Argiles JM, et al. Adipocyte Expression and Circulating Levels of Leptin Increase in Both Gynaecological and Breast Cancer Patients. Int J Oncol (2004) 24(6):1529–35. doi: 10.3892/ijo.24.6.1529
46. Jarde T, Caldefie-Chezet F, Damez M, Mishellany F, Penault-Llorca F, Guillot J, et al. Leptin and Leptin Receptor Involvement in Cancer Development: A Study on Human Primary Breast Carcinoma. Oncol Rep (2008) 19(4):905–11. doi: 10.3892/or.19.4.905
47. Poetsch MS, Strano A, Guan K. Role of Leptin in Cardiovascular Diseases. Front Endocrinol (2020) 11:354. doi: 10.3389/fendo.2020.00354
48. Cleary MP, Juneja SC, Phillips FC, Hu X, Grande JP, Maihle NJ. Leptin Receptor-Deficient MMTV-TGF-Alpha/Lepr(db)Lepr(db) Female Mice do Not Develop Oncogene-Induced Mammary Tumors. Exp Biol Med (2004) 229(2):182–93. doi: 10.1177/153537020422900207
49. Hosney M, Sabet S, El-Shinawi M, Gaafar KM, Mohamed MM. Leptin Is Overexpressed in the Tumor Microenvironment of Obese Patients With Estrogen Receptor Positive Breast Cancer. Exp Ther Med (2017) 13(5):2235–46. doi: 10.3892/etm.2017.4291
Keywords: breast cancer risk, leptin, LEPR, single-nucleotide polymorphisms, case–control study
Citation: Li L, Meng X, Liu L, Xiang Y, Wang F, Yu L, Zhou F, Zheng C, Zhou W, Cui S, Tian F, Fan Z, Geng C, Cao X, Yang Z, Wang X, Liang H, Wang S, Jiang H, Duan X, Wang H, Li G, Wang Q, Zhang J, Jin F, Tang J, Li L, Zhu S, Zuo W, Ye C, Yin G, Ma Z, Huang S and Yu Z (2022) Single-Nucleotide Polymorphisms in LEP and LEPR Associated With Breast Cancer Risk: Results From a Multicenter Case–Control Study in Chinese Females. Front. Oncol. 12:809570. doi: 10.3389/fonc.2022.809570
Received: 05 November 2021; Accepted: 14 January 2022;
Published: 10 February 2022.
Edited by:
Hajo Zeeb, Leibniz Institute for Prevention Research and Epidemiology (LG), GermanyReviewed by:
Ke-Da Yu, Fudan University, ChinaShanchun Guo, Xavier University of Louisiana, United States
Copyright © 2022 Li, Meng, Liu, Xiang, Wang, Yu, Zhou, Zheng, Zhou, Cui, Tian, Fan, Geng, Cao, Yang, Wang, Liang, Wang, Jiang, Duan, Wang, Li, Wang, Zhang, Jin, Tang, Li, Zhu, Zuo, Ye, Yin, Ma, Huang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuya Huang, aHVhbmdzeWFAc2R1LmVkdS5jbg==; Zhigang Yu, eXV6aGlnYW5nQHNkdS5lZHUuY24=
†These authors have contributed equally to this work
 Liang Li1,2,3,4†
Liang Li1,2,3,4† Xingchen Meng
Xingchen Meng Liyuan Liu
Liyuan Liu Chao Zheng
Chao Zheng Wenzhong Zhou
Wenzhong Zhou Zhimin Fan
Zhimin Fan Xuchen Cao
Xuchen Cao Xiang Wang
Xiang Wang Feng Jin
Feng Jin Chunmiao Ye
Chunmiao Ye Shuya Huang
Shuya Huang Zhigang Yu
Zhigang Yu