
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 28 February 2022
Sec. Genitourinary Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.801842
This article is part of the Research Topic Advances in Molecular Targeted Therapies of Urologic Cancers View all 10 articles
 Soudeh Ghafouri-Fard1
Soudeh Ghafouri-Fard1 Sajad Najafi2
Sajad Najafi2 Bashdar Mahmud Hussen3
Bashdar Mahmud Hussen3 Abbas Basiri4
Abbas Basiri4 Hazha Jamal Hidayat5
Hazha Jamal Hidayat5 Mohammad Taheri6*
Mohammad Taheri6* Fariborz Rashnoo7*
Fariborz Rashnoo7*Circular RNAs (circRNAs) are a group of transcripts with enclosed configurations which can regulate gene expression. These transcripts have important roles in normal development and in the pathogenesis of disorders. Recent evidence has supported involvement of circRNAs in the development of bladder cancer. Several circRNAs such as circ_0058063, hsa-circRNA-403658, circPDSS1, circCASC15, circRNA-MYLK, and circRNA_103809 have been upregulated in bladder cancer samples. On the other hand, hsa_circ_0137606, BCRC-3, circFUT8, hsa_circ_001598, circSLC8A1, hsa_circ_0077837, hsa_circ_0004826, and circACVR2A are among downregulated circRNAs in bladder cancer. Numerous circRNAs have diagnostic or prognostic value in bladder cancer. In this review, we aim to outline the latest findings about the role of circRNAs in bladder cancer and introduce circRNAs for further investigations as therapeutic targets.
Non-coding RNAs (ncRNAs) comprise several groups of RNA transcripts whose no protein is known to be encoded and thus considered as junk; however, they constitute a majority of expressed RNAs compared to protein-coding transcripts (1). Circular RNAs (circRNAs) are a distinct class of ncRNAs in eukaryotic cells which have been identified via electron microscopy in 1979 for the first time (2). Unlike coding-RNAs, circRNAs lack the 5′ cap and 3′ polyadenylated tail and do not mainly encode any protein; therefore, no primary function has been described for them (3). However, peptide-coding circRNAs have also been recognized. Some findings have revealed developmental, pathogenic, and especially regulatory roles for circRNAs. As their name suggests, a close circular loop in circRNAs is formed by covalent linkages between the 5′ and 3′ ends of their transcripts. CircRNAs compared to their linear counterparts show higher stability against degrading agents like RNase R due to closed ends (4) but are found in lower quantities within the animal cells (5), although higher abundance is reported for some circRNAs (6). Their sequence is evolutionarily conserved, indicating selective pressure for them (6). CircRNAs also show specific cell and tissue tendencies (7). Their elevated levels in several diseases demonstrate their potentials as diagnostic biomarkers and also therapeutic potentials especially in cancers (8). Today, due to development of advanced technologies like high-throughput RNA sequencing (RNA seq) and in situ experiments, a huge number of circRNAs have been recognized in animal cells (2). Moreover, their regulatory roles in gene expression and pathogenesis of disorders have been recognized. Similar to other regulatory non-coding RNAs, regulatory functions of circRNAs are suggested to be exerted through modulating gene expression at different levels. Based on the gene region, circRNAs can be divided into three types: those originating from exons (exonic circRNAs), introns (intronic circRNAs), or exon–intron junctions (exon–intron circRNAs) (9). Exonic circRNAs have been found in higher concentrations in cytoplasm compared to the nucleus showing capability of sponging microRNAs (miRNAs) and so can positively affect the expression of target genes leading to their overexpression. Unlike the first type, the other types are more concentrated in the cell nucleus and thus regulate gene expression at the primary steps of transcriptional and posttranscriptional levels (10, 11). CircRNAs have been widely detected in different cells, tissues, and organisms and also during various stages of organism development, playing a role in controlling cell growth and stress (12). Through their regulatory mechanisms in the cell cycle, circRNAs have been found to apply surveillance on eukaryotic cell proliferation and homeostasis. Consistent with these findings, dysregulation of circRNAs has been reported in a vast number of proliferative disorders like different tumors. Bladder cancer (BCa) is an example in which the role of aberrantly expressed circRNAs in tumor development and progression has been studied. Similar to other malignancies, response to treatment in BCa requires early diagnosis which also guarantees better prognosis for the patients. CircRNAs not only have acted as potential biomarkers for BCa with promising characteristics in diagnosis and prediction of prognosis in BCa patients but also have been suggested as therapeutical targets in fighting against malignancy. In this review, we aim to outline the latest findings about the role of circRNAs in BCa.
In expression analyses via high-throughput technologies like microarray and sequencing and also in quantitative PCR studies, a number of circRNAs have been found to be upregulated in samples taken from patients with BCa compared to healthy controls. These kinds of circRNAs are suggested as oncogenes with carcinogenic roles. Accordingly, their overexpressed levels have been shown to promote tumor cell proliferation and invasion in cell studies and also in vivo experiments, while their downregulation or knockdown reverses these effects.
Microarray analysis provides a possibility to screen a large number of aberrantly expressed circRNA candidates in a single platform. CircRNA_0058063 is an example which was reported recently by Liang et al. (13) as an upregulated circRNA in cancerous tissues of BCa patients compared to adjacent normal tissues. Microarray results revealed 312 aberrantly expressed circRNAs including 195 upregulated and 117 downregulated ones. CircRNA _0058063 showed a significantly increased expression in both BCa cell lines and tissues. A reverse correlation was seen between circRNA _0058063 expression level and overall survival (OS) in patients. Consistent with expectations, circRNA _0058063 knockdown suppressed tumor cell proliferation and metastasis in BCa BIU-87 cell lines. CircRNA_0058063 was found to act as a miRNA sponge to decrease the expression level of miR-486-3p by making interaction via a complementary sequence and induction of silencing. miR-486-3p inhibits the expression of the FOXP4 transcription factor which promotes tumorigenicity in various cancers.
Li et al. (14) using high-throughput sequencing found a number of upregulated RNA transcripts including long non-coding RNAs (lncRNAs), protein-coding mRNAs, and 34 circRNAs in 20 BCa tissues compared with a matched number of adjacent normal bladder tissues in addition to another set of transcripts which were downregulated. In GO and KEGG pathway enrichment analyses, dysregulated RNAs were associated with several signaling pathways controlling different critical cellular processes particularly DNA replication and cell cycle which play a role in the pathogenesis of BCa. In addition, 3 circRNAs including circPGM5 and circKIAA1462 were validated by qPCR. CircRNA PGM5 was demonstrated in the competing endogenous RNA (ceRNA) network to possess recognition sites for miRNAs associated with BCa along with lncRNA MIR194-2HG and AATBC.
Also, robust next-generation sequencing (NGS) technique and confirmatory qRT-PCR have been used to screen dysregulated circRNAs in BCa samples (15). A significant differential expression of a single circRNA is assessed in BCa tissues relative to adjacent normal tissues via quantitative reverse transcription polymerase chain reaction (qRT-PCR; also known as real-time RT-PCR). An increasing number of circRNAs have been reported in separate studies to be aberrantly expressed in BCa tissues or in serum or urine samples of patients; thus, these circRNAs have been suggested as potential diagnostic and prognostic biomarkers for BCa patients (16). Hsa-circRNA-403658 is a good instance for a series of circRNAs found to be upregulated in BCa tissues via the latter method. Wei et al. (17) demonstrated a differential expression of a number of circRNAs in BCa SW780, 5637, T24, J82, and RT4 cell lines cultured in hypoxic conditions in comparison with CCC-HB-2 healthy bladder epithelial cells using circRNA microarray. Hsa-circRNA-403658 was one of these circRNAs which showed the highest level of increased expression in further evaluation by qRT-PCR assay. Clinical samples of patients with BCa in which higher levels of hsa-circRNA-403658 were seen showed poorer prognosis, larger tumor size, increased metastasis, and higher clinicopathological stage (TNM staging) compared to patients with lower hsa-circRNA-403658 expression levels. As expected, hsa-circRNA-403658 knockdown using specific silencing RNA (siRNA) inhibited circRNA tumorigenic effects. Furthermore, in vitro and in vivo studies showed that hsa-circRNA-403658 controls the anaerobic glycolysis in hypoxic culture via enhancing the promotor activity and consequently positive regulation of lactate dehydrogenase A (LDHA) expression.
Computational studies and network analyses using bioinformatics methods have also facilitated the detection of differentially expressed circRNAs and their pathological roles and potential application as novel biomarkers for BCa (18–20), suggesting their application in early diagnosis and prediction of prognosis as well as their therapeutic potentials. Figure 1 demonstrates the role of circRNAs in modulating bladder cancer development via promoting glycolysis.
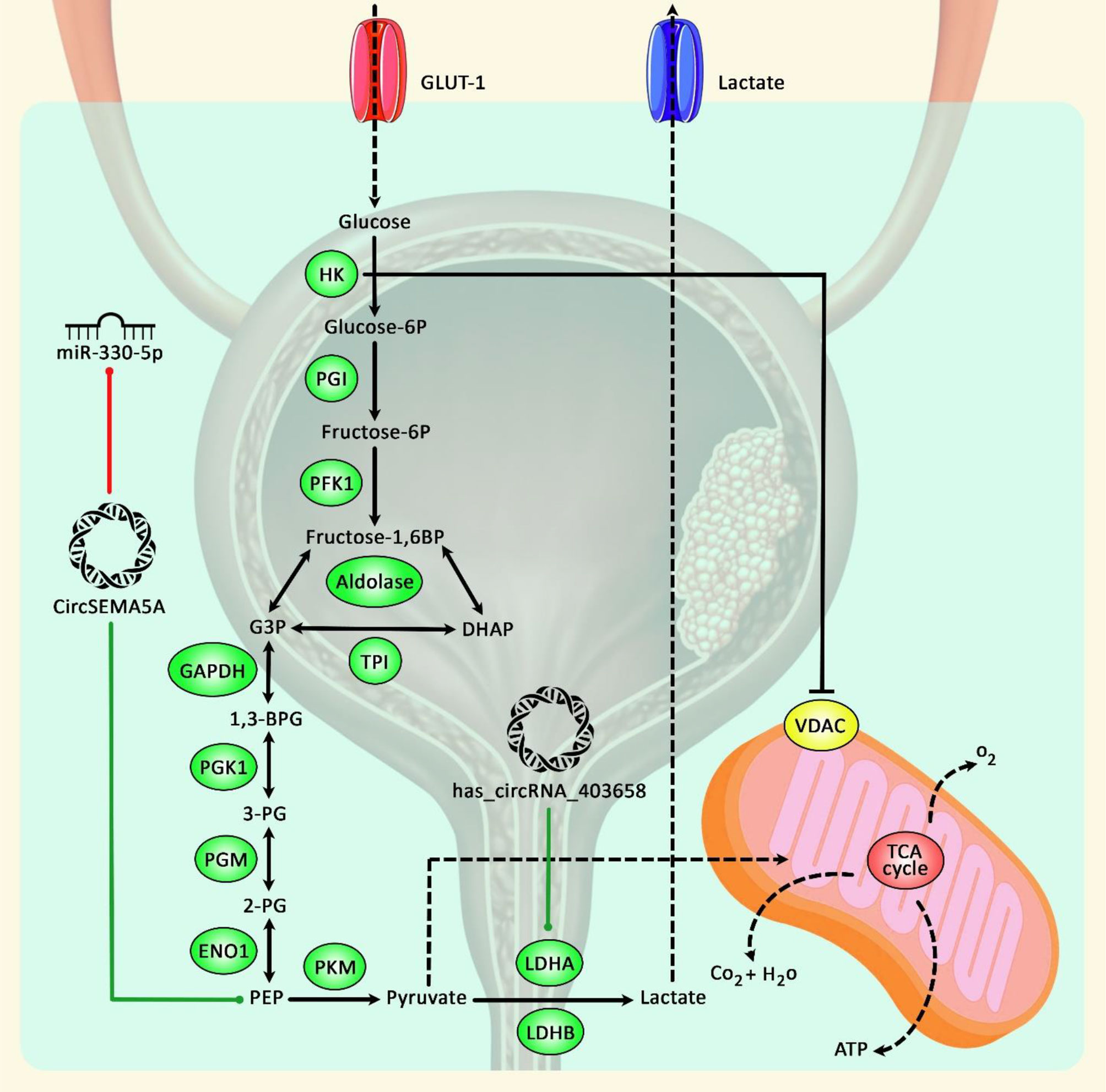
Figure 1 A schematic diagram of the role of circRNAs in promoting bladder cancer progression via enhancing glycolysis. Accumulating findings have suggested that upregulation of key glycolysis proteins could play a crucial role in cancer development. As an illustration, a previous study has authenticated that has_circRNA_403658 through upregulation of LDHA-mediated aerobic glycolysis could have a significant part in enhancing bladder cancer cell growth (17). In addition, another research has detected that circSEMA5A via sponging miR-330-5p could upregulate the expression level of ENO1, thereby elevating proliferation, invasion, angiogenesis, and glycolysis of bladder cancer cells by facilitating the activation of Akt and β-catenin signaling cascades (21). Green arrows indicate the upregulation of target genes by circRNAs, and red arrows depict inhibitory effects.
Table 1 outlines the most important circRNAs with an elevated expression level in the BCa cell line and also in patients’ samples.
These kinds of circRNAs have been found to exhibit lower expression levels in BCa samples compared to normal adjacent tissues. They are suggested to play a role as tumor suppressors with biological functions controlling the critical cellular processes cell proliferation and extracellular matrix stability and so their downregulation in a set of studies has been shown to facilitate tumor cell proliferation, migration, and invasion. In vivo experiments have also demonstrated accelerated tumor progression in the presence of decreased expression of these circRNAs. Similar to the former circRNAs, these kinds have been linked with cell cycle regulation through different miRNA–protein axes, among which are some oncogenes or tumor-suppressor genes which are dysregulated. Bioinformatic analyses, RNA pull-down assays, and luciferase reporter assays have shown single or several miRNAs being sponged in close interaction with circRNAs. The expression of these miRNAs is mainly suppressed via upstream circRNAs, and they themselves regulate some actions through affecting downstream molecules. Yet, some miRNAs act upstream of circRNAs exerting a regulatory role on them. Downregulated circRNAs are mainly located in the cytoplasm, so it is suggested that they play their regulatory roles at posttranscriptional or translational steps.
For instance, circSLC8A1 is a circRNA which has been reported by Lu et al. (66) to be downregulated in BCa tissues compared to healthy adjacent tissues in a study of 70 patients diagnosed with BCa. They detected a number of aberrantly expressed circRNAs through RNA sequencing. qRT-PCR confirmed a decreased expression of circSLC8A1 in 81% (57/70) of total BCa tissues compared to their matched adjacent tissues. Expression assay in 6 BCa cell lines using qRT-PCR also showed a decreased expression of circSLC8A1 compared to SV-HUC-1 normal bladder cells.
In vitro analyses revealed suppression of tumorigenic impacts following circSLC8A1 overexpression in BCa cell lines. By using different prediction tools, it was demonstrated that circSLC8A1 potentially sponges 7 miRNAs, among which were miR-130b and miR-494 whose interactions with circSLC8A1 were confirmed by RNA pull-down assay and biotin labeling. Overexpression of both miRNAs was associated with oncogenesis in BCa cells. Furthermore, luciferase reporter assay and Western blotting analysis demonstrated that miRNAs can bind to the 3′ end of the phosphatase and tensin homolog (PTEN) tumor suppressor and inhibit its expression. Rescue experiments and immunohistochemistry (IHC) analysis showed that circSLC8A1 acts as a tumor suppressor via the miR-130b and miR-494/PTEN/PI3k/Akt signaling axis. Table 2 summarizes the recent findings of tumorigenicity studies on downregulated circRNAs in BCa. Figure 2 represents the role of several circRNAs in bladder cancer cells via regulating some key signaling cascades.
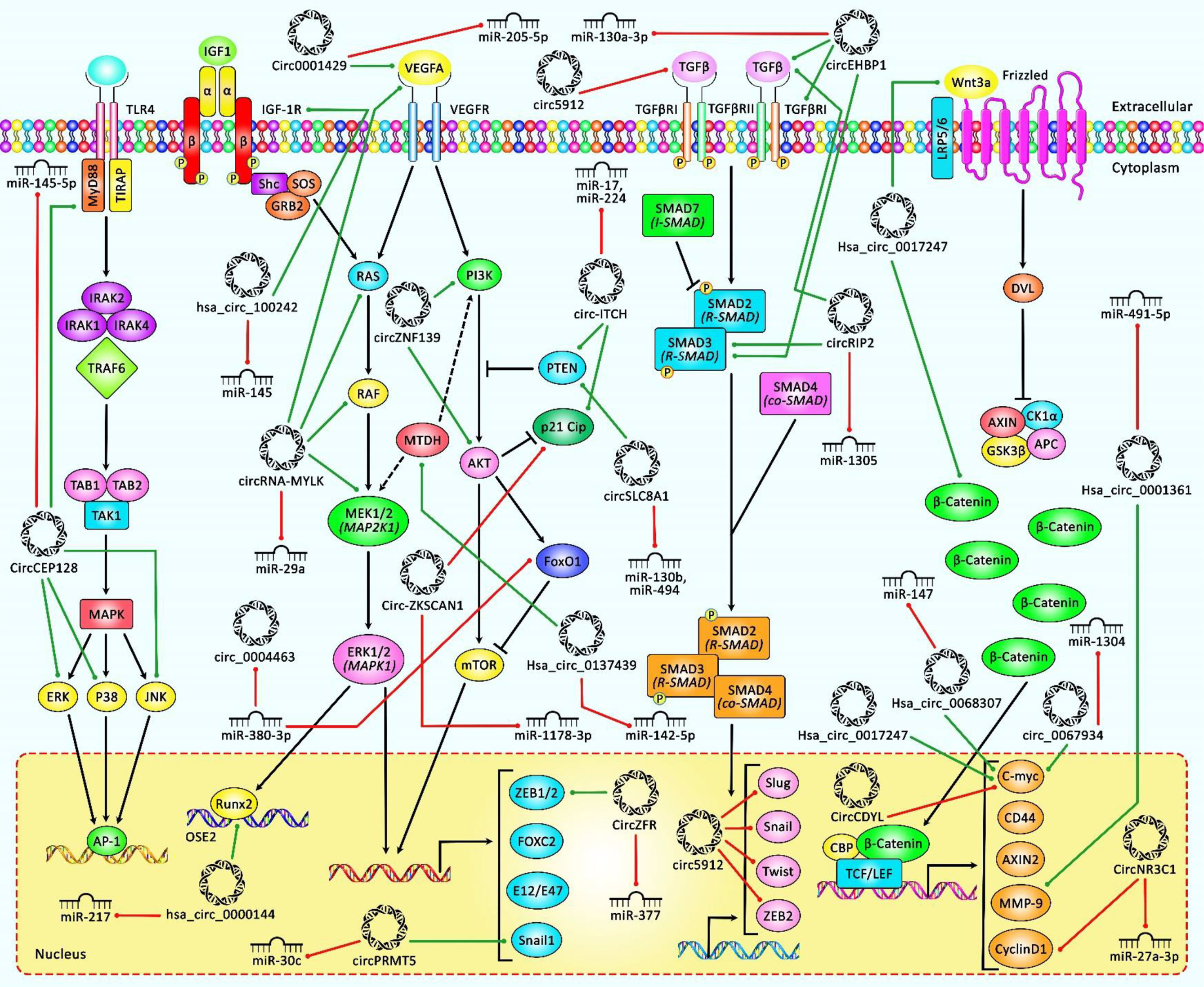
Figure 2 A schematic representation of the role of various circRNAs in human bladder cancer through modulating the PI3K/AKT, MAPK/ERK, TGFβ/SMAD3, and Wnt/β-catenin signaling pathways. According to the diagram, the upregulation or downregulation of several circRNAs could have a considerable role in bladder cancer development through modulation of miRNA levels. Green arrows indicate upregulation of target genes by circRNAs, and red arrows depict inhibition regulated by them. All the information regarding the role of these circRNAs in bladder cancer is shown in Tables 1, 2.
As explained above, dysregulated circRNAs cause disturbance in the cellular proliferation leading to malignancies, particularly BCa. On the other side, in the majority of the studies, dysregulation in circRNA expression has been statistically correlated with unfavorable clinicopathological features including high tumor size, histological grade, pathological stage, and presence of distant or lymph node metastasis in uni- or multivariate analyses in BCa patients. Therefore, as a consequence, it has been found that a dysregulation in the circRNA level can predict poorer survival (in terms of overall survival (OS), recurrence-free survival (RFS), disease-free survival (DFS), or progression-free survival (PFS)) and worse prognosis in Kaplan–Meier analyses.
As an example, it was formerly stated that hsa_circRNA_403658 is upregulated in BCa tissues compared to adjacent tissues (17). Kaplan–Meier analysis for the evaluation of survival showed that a high level of hsa_circRNA_403658 expression correlates with shorter survival in BCa patients. Implementation of the χ2 test to assess the association between circRNA expression and clinicopathological features has revealed a positive correlation between high hsa_circRNA_403658 expression and malignant characteristics including higher tumor volume (size ≥3 cm related to <3 cm), metastasis to distant places and advanced TNM stage (III–IV). In the univariate and multivariate Cox regression test for assessment of prognostic factors, it was demonstrated that hsa_circRNA_403658 is an independent factor for prediction of prognosis in BCa patients (17).
In another study (70), downregulated circRNAs hsa_circ_0077837 and hsa_circ_0004826 in BCa were found to be significantly associated with worse OS and RFS in BCa patients in Kaplan–Meier analysis. Univariate and multivariate Cox regression analyses confirmed that both circRNAs can act as independent prognostic factors compared to other factors in BCa patients. The area under the curve (AUC) for assessment of prognostic power of circRNAs revealed 0.775 and 0.790 values for hsa_circ_0077837 and hsa_circ_0004826, respectively, showing acceptable measures and suggesting their potentials as reliable biomarkers for prediction of worse prognosis in BCa patients.
Among other circRNAs, circASXL1 has been reported to have sensitivity and specificity of 0.686 and 0.769, respectively, which suggests its reliable diagnostic power in distinguishing the BCa patients from healthy people (38). A number of circRNAs whose prognostic or diagnostic values have been studied in BCa are shown in Table 3.
Circular RNAs (circRNAs) are covalently closed nucleic acid strands which are classified as non-coding RNAs and mainly do not code any protein. They have been found to play a role in gene regulation in several stages. CircRNAs show cell-, tissue-, or species-specific tropism and are known to be dysregulated in tissues in a number of cancers. They have been found to either act as tumor suppressors or exhibit oncogenic roles on overexpression. The causative mechanisms of circRNAs’ role in tumorigenicity are vastly being studied. Their dysregulation has mainly been associated with disturbances in cell cycle regulation through activation of several signaling pathways.
In this review, we summarized a number of studies conducted on dysregulated circRNAs in BCa. Dysregulation includes any increase or decrease in circRNA expression levels in BCa tissues or cell studies compared to normal adjacent tissues. To assess the circRNA dysregulation, some high-throughput technologies like RNA sequencing and confirmatory qRT-PCR have been employed. Upregulated circRNAs in the first section (Table 1) have been found to accelerate tumor cell proliferation and enhance migration and invasion of cancer cell in vitro. In vivo studies have shown increased tumor progression when these circRNAs are overexpressed. Suppression of upregulated circRNAs via specific siRNAs in BCa has confirmed repression of proliferative and migratory potential following their silencing.
Downregulated circRNAs (Table 2), on the other hand, have been known to exert their anti-tumorigenic roles via suppression of tumor cell proliferation, migration, invasion, and metastasis. Overexpression experiments have confirmed tumor-suppressing roles of the second class of circRNAs via diminishing tumor cell malignant behaviors.
Furthermore, correlation studies have shown a significant association between dysregulated circRNA expression levels and worse clinicopathological features including higher tumor size, distant or lymph node metastasis, higher histological grade, pathological stage, and advanced TNM stage in BCa patients (Table 3). Statistical analyses have also demonstrated that dysregulation of circRNAs can be used as independent prognostic factors for BCa patients. Acceptable AUC, specificity, or sensitivity values in diagnostic analyses have revealed the diagnostic power of circRNAs in distinguishing BCa from other diseases. Taken together, circRNAs have been suggested as markers with reliable prognostic and diagnostic potential which can be used as biomarkers in either diagnosis or prediction of prognosis in BCa patients. Not only circRNAs but also other ncRNAs like lncRNAs have shown high stability in biological fluids, making them good biomarkers with easy detection for a number of human diseases particularly diverse types of cancers including BCa, breast cancer, hepatocellular carcinoma, and colorectal cancer (97, 98). Exosomes as extracellular vesicles involved in cellular communications are particularly shown to contain circRNAs in high concentrations and in a stabile form (99). These membranes can be beneficial in the detection of malignancies when derived from cancer cells spreading to blood and detectable in serum. Newly identified circRNAs have been introduced through high-throughput approaches such as next-generation sequencing (NGS), and microarray analysis, which make the huge identification of ncRNAs possible, while qRT-PCR is the main technique with potential in clinical diagnosis and quantification of circRNAs for both prognostic and diagnostic goals, and it is also used as the confirmatory method upon a novel ncRNA as previously reported (100). Vast application of qRT-PCR in quantification of a circRNA, however, is possible when the junction/fusion site is identified (8). Moreover, other technological drawbacks require to be addressed for clinical applications of circRNAs.
In vitro and xenograft studies have confirmed the suitability of circRNAs as therapeutic targets in cancers. However, several issues such as biosafety ones should be solved before application of circRNA-targeting methods in clinical settings.
Taken together, although circRNAs have shown roles in the development and progression of various human cancers, and their quantification have demonstrated excellent potential in distinguishing patients with BCa from healthy individuals, it seems that application of circRNAs as novel biomarkers needs further investigations, more time, and addressing of technological problems to enter in clinical settings.
SGF and SN wrote the draft and revised it. MT designed and supervised the study. BMH, HJH, FR, and AB collected the data and designed the figures and tables. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Najafi S, Tan SC, Raee P, Rahmati Y, Asemani Y, Lee EHC, et al. Gene Regulation by Antisense Transcription: A Focus on Neurological and Cancer Diseases. Biomed Pharmacother (2022) 145:112265. doi: 10.1016/j.biopha.2021.112265
2. Hsu M-T, Coca-Prados M. Electron Microscopic Evidence for the Circular Form of RNA in the Cytoplasm of Eukaryotic Cells. Nature (1979) 280(5720):339–40. doi: 10.1038/280339a0
3. Rahmati Y, Asemani Y, Aghamiri S, Ezzatifar F, Najafi S. CiRS-7/CDR1as; An Oncogenic Circular RNA as a Potential Cancer Biomarker. Pathol - Res Pract (2021) 227:153639. doi: 10.1016/j.prp.2021.153639
4. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-Digested Cellular RNA Source That Consists of Lariat and Circular RNAs From pre-mRNA Splicing. Nucleic Acids Res (2006) 34(8):e63. doi: 10.1093/nar/gkl151
5. Lasda E, Parker R. Circular RNAs: Diversity of Form and Function. RNA (New York NY) (2014) 20(12):1829–42. doi: 10.1261/rna.047126.114
6. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs Are Abundant, Conserved, and Associated With ALU Repeats. RNA (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
7. Jeck WR, Sharpless NE. Detecting and Characterizing Circular RNAs. Nat Biotechnol (2014) 32(5):453–61. doi: 10.1038/nbt.2890
8. Taheri M, Najafi S, Basiri A, Hussen BM, Baniahmad A, Jamali E, et al. The Role and Clinical Potentials of Circular RNAs in Prostate Cancer. Front Oncol (2021) 11:781414. doi: 10.3389/fonc.2021.781414
9. Liu J, Liu T, Wang X, He A. Circles Reshaping the RNA World: From Waste to Treasure. Mol Cancer (2017) 16(1):58. doi: 10.1186/s12943-017-0630-y
10. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a Large Class of Animal RNAs With Regulatory Potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
11. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
12. Fischer JW, Leung AKL. CircRNAs: A Regulator of Cellular Stress. Crit Rev Biochem Mol Biol (2017) 52(2):220–33. doi: 10.1080/10409238.2016.1276882
13. Liang H, Huang H, Li Y, Lu Y, Ye T. CircRNA_0058063 Functions as a ceRNA in Bladder Cancer Progression via Targeting miR-486-3p/FOXP4 Axis. Biosci Rep (2020) 40(3). doi: 10.1042/BSR20193484
14. Li M, Liu Y, Zhang X, Liu J, Wang P. Transcriptomic Analysis of High-Throughput Sequencing About circRNA, lncRNA and mRNA in Bladder Cancer. Gene (2018) 677:189–97. doi: 10.1016/j.gene.2018.07.041
15. Yang X, Yuan W, Tao J, Li P, Yang C, Deng X, et al. Identification of Circular RNA Signature in Bladder Cancer. J Cancer (2017) 8(17):3456–63. doi: 10.7150/jca.19345
16. Pan J, Xie X, Li H, Li Z, Ren C, Ming L. Detection of Serum Long Non-Coding RNA UCA1 and Circular RNAs for the Diagnosis of Bladder Cancer and Prediction of Recurrence. Int J Clin Exp Pathol (2019) 12(8):2951–8.
17. Wei Y, Zhang Y, Meng Q, Cui L, Xu C. Hypoxia-Induced Circular RNA has_circRNA_403658 Promotes Bladder Cancer Cell Growth Through Activation of LDHA. Am J Transl Res (2019) 11(11):6838–49.
18. Kouhsar M, Azimzadeh Jamalkandi S, Moeini A, Masoudi-Nejad A. Detection of Novel Biomarkers for Early Detection of Non-Muscle-Invasive Bladder Cancer Using Competing Endogenous RNA Network Analysis. Sci Rep (2019) 9(1):8434. doi: 10.1038/s41598-019-44944-3
19. Yang J, Chen J, Wu S, Fei X, Wang X, Wang K. Microarray Expression Profiles and Bioinformatics Analyses Reveal Aberrant Circular RNAs Expression in Bladder Cancer. Onco Targets Ther (2020) 13:10889–99. doi: 10.2147/OTT.S270747
20. Cai D, Liu Z, Kong G. Molecular and Bioinformatics Analyses Identify 7 Circular RNAs Involved in Regulation of Oncogenic Transformation and Cell Proliferation in Human Bladder Cancer. Med Sci Monitor Int Med J Exp Clin Res (2018) 24:1654–61. doi: 10.12659/MSM.908837
21. Wang L, Li H, Qiao Q, Ge Y, Ma L, Wang Q. Circular RNA Circsema5a Promotes Bladder Cancer Progression by Upregulating ENO1 and SEMA5A Expression. Aging (Albany NY) (2020) 12(21):21674–86. doi: 10.18632/aging.103971
22. Sun M, Zhao W, Chen Z, Li M, Li S, Wu B, et al. Circ_0058063 Regulates CDK6 to Promote Bladder Cancer Progression by Sponging miR-145-5p. J Cell Physiol (2019) 234(4):4812–24. doi: 10.1002/jcp.27280
23. Yu Q, Liu P, Han G, Xue X, Ma D. CircRNA Circpdss1 Promotes Bladder Cancer by Down-Regulating miR-16. Biosci Rep (2020) 40(1):BSR20191961. doi: 10.1042/BSR20191961
24. Zhuang C, Huang X, Yu J, Gui Y. Circular RNA Hsa_Circ_0075828 Promotes Bladder Cancer Cell Proliferation Through Activation of CREB1. BMB Rep (2020) 53(2):82–7. doi: 10.5483/BMBRep.2020.53.2.059
25. Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, et al. Circular RNA MYLK as a Competing Endogenous RNA Promotes Bladder Cancer Progression Through Modulating VEGFA/VEGFR2 Signaling Pathway. Cancer Lett (2017) 403:305–17. doi: 10.1016/j.canlet.2017.06.027
26. Liu F, Zhang H, Xie F, Tao D, Xiao X, Huang C, et al. Hsa_circ_0001361 Promotes Bladder Cancer Invasion and Metastasis Through miR-491-5p/MMP9 Axis. Oncogene (2020) 39(8):1696–709. doi: 10.1038/s41388-019-1092-z
27. Zeng Z, Zhou W, Duan L, Zhang J, Lu X, Jin L, et al. Circular RNA Circ-VANGL1 as a Competing Endogenous RNA Contributes to Bladder Cancer Progression by Regulating miR-605-3p/VANGL1 Pathway. J Cell Physiol (2019) 234(4):3887–96. doi: 10.1002/jcp.27162
28. Zhong Z, Lv M, Chen J. Screening Differential Circular RNA Expression Profiles Reveals the Regulatory Role of Circtcf25-miR-103a-3p/miR-107-CDK6 Pathway in Bladder Carcinoma. Sci Rep (2016) 6(1):30919. doi: 10.1038/srep30919
29. Song Z, Zhang Q, Zhu J, Yin G, Lin L, Liang C. Identification of Urinary Hsa_Circ _0137439 as Potential Biomarker and Tumor Regulator of Bladder Cancer. Neoplasma (2020) 67(1):137–46. doi: 10.4149/neo_2018_181214N970
30. Xu Z-Q, Yang M-G, Liu H-J, Su C-Q. Circular RNA Hsa_Circ_0003221 (Circptk2) Promotes the Proliferation and Migration of Bladder Cancer Cells. J Cell Biochem (2018) 119(4):3317–25. doi: 10.1002/jcb.26492
31. Sun M, Zhao W, Chen Z, Li M, Li S, Wu B, et al. Circular RNA CEP128 Promotes Bladder Cancer Progression by Regulating Mir-145-5p/Myd88 via MAPK Signaling Pathway. Int J Cancer (2019) 145(8):2170–81. doi: 10.1002/ijc.32311
32. Cao W, Zhao Y, Wang L, Huang X. Circ0001429 Regulates Progression of Bladder Cancer Through Binding miR-205-5p and Promoting VEGFA Expression. Cancer Biomarkers (2019) 25:101–13. doi: 10.3233/CBM-182380
33. Yang C, Li Q, Chen X, Zhang Z, Mou Z, Ye F, et al. Circular RNA circRGNEF Promotes Bladder Cancer Progression via miR-548/KIF2C Axis Regulation. Aging (Albany NY) (2020) 12(8):6865–79. doi: 10.18632/aging.103047
34. Gu C, Zhou N, Wang Z, Li G, Kou Y, Yu S, et al. Circgprc5a Promoted Bladder Oncogenesis and Metastasis Through Gprc5a-Targeting Peptide. Mol Ther - Nucleic Acids (2018) 13:633–41. doi: 10.1016/j.omtn.2018.10.008
35. Huang W, Lu Y, Wang F, Huang X, Yu Z. Downregulation of Circular RNA Hsa_Circ_0000144 Inhibits Bladder Cancer Progression via Stimulating miR-217 and Suppressing RUNX2 Expression. Gene (2018) 678:337–42. doi: 10.1016/j.gene.2018.08.036
36. Zhang X, Liu X, Jing Z, Bi J, Li Z, Liu X, et al. The Circints4/miR-146b/CARMA3 Axis Promotes Tumorigenesis in Bladder Cancer. Cancer Gene Ther (2020) 27(3):189–202. doi: 10.1038/s41417-019-0085-y
37. Zhang W-Y, Liu Q-H, Wang T-J, Zhao J, Cheng X-H, Wang J-S. CircZFR Serves as a Prognostic Marker to Promote Bladder Cancer Progression by Regulating miR-377/ZEB2 Signaling. Biosci Rep (2019) 39(12). doi: 10.1042/BSR20192779
38. Tang G, Xie W, Qin C, Zhen Y, Wang Y, Chen F, et al. Expression of Circular RNA Circasxl1 Correlates With TNM Classification and Predicts Overall Survival in Bladder Cancer. Int J Clin Exp Pathol (2017) 10(8):8495–502.
39. Mao W, Huang X, Wang L, Zhang Z, Liu M, Li Y, et al. Circular RNA Hsa_Circ_0068871 Regulates FGFR3 Expression and Activates STAT3 by Targeting miR-181a-5p to Promote Bladder Cancer Progression. J Exp Clin Cancer Res (2019) 38(1):169. doi: 10.1186/s13046-019-1136-9
40. Gong P, Xu R, Zhuang Q, He X. A Novel Circular RNA (Hsa_circRNA_102336), a Plausible Biomarker, Promotes the Tumorigenesis by Sponging miR-515-5p in Human Bladder Cancer. Biomed Pharmacother (2020) 126:110059. doi: 10.1016/j.biopha.2020.110059
41. Chen Q, Yin Q, Mao Y, Zhang Z, Wu S, Cheng Z, et al. Hsa_circ_0068307 Mediates Bladder Cancer Stem Cell-Like Properties via miR-147/C-Myc Axis Regulation. Cancer Cell Int (2020) 20(1):151. doi: 10.1186/s12935-020-01235-6
42. Li M, Liu Y, Liu J, Li W, Li N, Xue D, et al. Circ_0006332 Promotes Growth and Progression of Bladder Cancer by Modulating MYBL2 Expression via miR-143. Aging (Albany NY) (2019) 11(22):10626–43. doi: 10.18632/aging.102481
43. Liu Z, Yang Y, Yang Z, Xia S, Lin D, Xiao B, et al. Novel circRNA_0071196/miRNA−19b−3p/CIT Axis Is Associated With Proliferation and Migration of Bladder Cancer. Int J Oncol (2020) 57(3):767–79. doi: 10.3892/ijo.2020.5093
44. Yao J, Qian K, Chen C, Liu X, Yu D, Yan X, et al. ZNF139/circZNF139 Promotes Cell Proliferation, Migration and Invasion via Activation of PI3K/AKT Pathway in Bladder Cancer. Aging (Albany NY) (2020) 12(10):9915–34. doi: 10.18632/aging.103256
45. Liu P, Li X, Guo X, Chen J, Li C, Chen M, et al. Circular RNA DOCK1 Promotes Bladder Carcinoma Progression via Modulating Circdock1/hsa-miR-132-3p/Sox5 Signalling Pathway. Cell Proliferation (2019) 52(4):e12614. doi: 10.1111/cpr.12614
46. Shi Y-R, Wu Z, Xiong K, Liao Q-J, Ye X, Yang P, et al. Circular RNA Circkif4a Sponges miR-375/1231 to Promote Bladder Cancer Progression by Upregulating NOTCH2 Expression. Front Pharmacol (2020) 11:605–. doi: 10.3389/fphar.2020.00605
47. Jin M, Lu S, Wu Y, Yang C, Shi C, Wang Y, et al. Hsa_circ_0001944 Promotes the Growth and Metastasis in Bladder Cancer Cells by Acting as a Competitive Endogenous RNA for miR-548. J Exp Clin Cancer Res (2020) 39(1):186. doi: 10.1186/s13046-020-01697-6
48. Fang Y, Chen S, Liu Z, Ai W, He X, Wang L, et al. Endothelial Stem Cells Attenuate Cardiac Apoptosis via Downregulating Cardiac microRNA−146a in a Rat Model of Coronary Heart Disease. Exp Ther Med (2018) 16(5):4246–52.
49. Wu S, Yang J, Xu H, Wang X, Zhang R, Lu W, et al. Circular RNA Circglis3 Promotes Bladder Cancer Proliferation via the miR-1273f/SKP1/Cyclin D1 Axis. Cell Biol Toxicol (2022) 38:129–46. doi: 10.1007/s10565-021-09591-3
50. Chen LQ, Yi CL, Liu DC, Wang P, Zhu YF, Yuan LP. Hsa_circ_0041103 Induces Proliferation, Migration and Invasion in Bladder Cancer via the miR-107/FOXK1 Axis. Eur Rev Med Pharmacol Sci (2021) 25(3):1282–90. doi: 10.26355/eurrev_202102_24832
51. Zhu J, Luo Y, Zhao Y, Kong Y, Zheng H, Li Y, et al. Circehbp1 Promotes Lymphangiogenesis and Lymphatic Metastasis of Bladder Cancer via miR-130a-3p/Tgfβr1/VEGF-D Signaling. Mol Ther J Am Soc Gene Ther (2021) 29(5):1838–52. doi: 10.1016/j.ymthe.2021.01.031
52. Zheng H, Yang C, Tang J. Cyclic RNA Circ_0000735 Sponges miR-502-5p to Promote Bladder Cancer Cell Proliferation and Invasion and Inhibit Apoptosis. Int J Clin Exp Pathol (2020) 13(12):2994–3003.
53. Tong L, Yang H, Xiong W, Tang G, Zu X, Qi L. Circ_100984-miR-432-3p Axis Regulated C-Jun/YBX-1/β-Catenin Feedback Loop Promotes Bladder Cancer Progression. Cancer Sci (2020) 112(4):1429–42. doi: 10.1111/cas.14774
54. Wang F, Fan M, Cai Y, Zhou X, Tai S, Yu Y, et al. Circular RNA Circrims1 Acts as a Sponge of miR-433-3p to Promote Bladder Cancer Progression by Regulating CCAR1 Expression. Mol Ther Nucleic Acids (2020) 22:815–31. doi: 10.1016/j.omtn.2020.10.003
55. Du L, Xu Z, Wang X, Liu F. Integrated Bioinformatics Analysis Identifies microRNA-376a-3p as a New microRNA Biomarker in Patient With Coronary Artery Disease. Am J Trans Res (2020) 12(2):633.
56. Jiang P, Zhu Y, Xu Z, Chen X, Xie L. Interference With Circbc048201 Inhibits the Proliferation, Migration, and Invasion of Bladder Cancer Cells Through the miR-1184/ITGA3 Axis. Mol Cell Biochem (2020) 474(1-2):83–94. doi: 10.1007/s11010-020-03835-2
57. Feng F, Chen AP, Wang XL, Wu GL. Circ_0061140 Promotes Metastasis of Bladder Cancer Through Adsorbing microRNA-1236. Eur Rev Med Pharmacol Sci (2020) 24(10):5310–9. doi: 10.26355/eurrev_202103_25199
58. Peng G, Meng H, Pan H, Wang W. CircRNA 001418 Promoted Cell Growth and Metastasis of Bladder Carcinoma via EphA2 by miR-1297. Curr Mol Pharmacol (2021) 14(1):68–78. doi: 10.2174/1874467213666200505093815
59. Liu Q, Zhou Q, Zhong P. Circ_0067934 Increases Bladder Cancer Cell Proliferation, Migration and Invasion Through Suppressing miR-1304 Expression and Increasing Myc Expression Levels. Exp Ther Med (2020) 19(6):3751–9. doi: 10.3892/etm.2020.8648
60. Gao W, Cui H, Li Q, Zhong H, Yu J, Li P, et al. Upregulation of microRNA-218 Reduces Cardiac Microvascular Endothelial Cells Injury Induced by Coronary Artery Disease Through the Inhibition of HMGB1. J Cell Physiol (2020) 235(3):3079–95. doi: 10.1002/jcp.29214
61. Chen J, Sun Y, Ou Z, Yeh S, Huang CP, You B, et al. Androgen Receptor-Regulated circFNTA Activates KRAS Signaling to Promote Bladder Cancer Invasion. EMBO Rep (2020) 21(4):e48467. doi: 10.15252/embr.201948467
62. Su Y, Feng W, Shi J, Chen L, Huang J, Lin T. Circrip2 Accelerates Bladder Cancer Progression via miR-1305/Tgf-β2/Smad3 Pathway. Mol Cancer (2020) 19(1):23–. doi: 10.1186/s12943-019-1129-5
63. Yang C, Wu S, Wu X, Zhou X, Jin S, Jiang H. Silencing Circular RNA UVRAG Inhibits Bladder Cancer Growth and Metastasis by Targeting the microRNA-223/Fibroblast Growth Factor Receptor 2 Axis. Cancer Sci (2019) 110(1):99–106. doi: 10.1111/cas.13857
64. Bi J, Liu H, Cai Z, Dong W, Jiang N, Yang M, et al. Circ-BPTF Promotes Bladder Cancer Progression and Recurrence Through the miR-31-5p/RAB27A Axis. Aging (Albany NY) (2018) 10(8):1964–76. doi: 10.18632/aging.101520
65. Wu L, Zhang M, Qi L, Zu X, Li Y, Liu L, et al. Erα-Mediated Alterations in Circ_0023642 and miR-490-5p Signaling Suppress Bladder Cancer Invasion. Cell Death Dis (2019) 10(9):635. doi: 10.1038/s41419-019-1827-3
66. Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, et al. Circular RNA Circslc8a1 Acts as a Sponge of miR-130b/miR-494 in Suppressing Bladder Cancer Progression via Regulating PTEN. Mol Cancer (2019) 18(1):111. doi: 10.1186/s12943-019-1040-0
67. Xie F, Li Y, Wang M, Huang C, Tao D, Zheng F, et al. Circular RNA BCRC-3 Suppresses Bladder Cancer Proliferation Through miR-182-5p/P27 Axis. Mol Cancer (2018) 17(1):144. doi: 10.1186/s12943-018-0892-z
68. He Q, Yan D, Dong W, Bi J, Huang L, Yang M, et al. circRNA Circfut8 Upregulates Krüpple-Like Factor 10 to Inhibit the Metastasis of Bladder Cancer via Sponging miR-570-3p. Mol Ther - Oncolytics (2020) 16:172–87. doi: 10.1016/j.omto.2019.12.014
69. Faccini J, Ruidavets J-B, Cordelier P, Martins F, Maoret J-J, Bongard V, et al. Circulating miR-155, miR-145 and Let-7c as Diagnostic Biomarkers of the Coronary Artery Disease. Sci Rep (2017) 7:42916. doi: 10.1038/srep42916
70. Shen C, Wu Z, Wang Y, Gao S, Da L, Xie L, et al. Downregulated Hsa_Circ_0077837 and Hsa_Circ_0004826, Facilitate Bladder Cancer Progression and Predict Poor Prognosis for Bladder Cancer Patients. Cancer Med (2020) 9(11):3885–903. doi: 10.1002/cam4.3006
71. Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q, et al. Circular RNA ACVR2A Suppresses Bladder Cancer Cells Proliferation and Metastasis Through miR-626/EYA4 Axis. Mol Cancer (2019) 18(1):95. doi: 10.1186/s12943-019-1025-z
72. Jiang Y, Wei T, Li W, Zhang R, Chen M. Circular RNA Hsa_Circ_0002024 Suppresses Cell Proliferation, Migration, and Invasion in Bladder Cancer by Sponging miR-197-3p. Am J Transl Res (2019) 11(3):1644–52.
73. Li Y, Qiao L, Zang Y, Ni W, Xu Z. Circular RNA FOXO3 Suppresses Bladder Cancer Progression and Metastasis by Regulating MiR-9-5p/TGFBR2. Cancer Manage Res (2020) 12:5049–56. doi: 10.2147/CMAR.S253412
74. Wang C, Tao W, Ni S, Chen Q. Circular RNA Circ-Foxo3 Induced Cell Apoptosis in Urothelial Carcinoma via Interaction With miR-191-5p. Onco Targets Ther (2019) 12:8085–94. doi: 10.2147/OTT.S215823
75. Zheng F, Wang M, Li Y, Huang C, Tao D, Xie F, et al. CircNR3C1 Inhibits Proliferation of Bladder Cancer Cells by Sponging miR-27a-3p and Downregulating Cyclin D1 Expression. Cancer Lett (2019) 460:139–51. doi: 10.1016/j.canlet.2019.06.018
76. Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA Circmto1 Suppresses Bladder Cancer Metastasis by Sponging miR-221 and Inhibiting Epithelial-to-Mesenchymal Transition. Biochem Biophys Res Commun (2019) 508(4):991–6. doi: 10.1016/j.bbrc.2018.12.046
77. Yang C, Yuan W, Yang X, Li P, Wang J, Han J, et al. Circular RNA Circ-ITCH Inhibits Bladder Cancer Progression by Sponging miR-17/miR-224 and Regulating P21, PTEN Expression. Mol Cancer (2018) 17(1):19. doi: 10.1186/s12943-018-0771-7
78. Tan S, Kang Y, Li H, He H-Q, Zheng L, Wu S-Q, et al. Circst6galnac6 Suppresses Bladder Cancer Metastasis by Sponging miR-200a-3p to Modulate the STMN1/EMT Axis. Cell Death Dis (2021) 12(2):168. doi: 10.1038/s41419-021-03459-4
79. Abulizi R, Li B, Zhang CG. Circ_0071662, a Novel Tumor Biomarker, Suppresses Bladder Cancer Cell Proliferation and Invasion by Sponging miR-146b-3p. Oncol Res (2019). doi: 10.3727/096504019X15740729375088
80. Li M, Wang Y, Liu Y, Zhang X, Liu J, Wang P. Low Expression of Hsa_Circ_0018069 in Human Bladder Cancer and Its Clinical Significance. BioMed Res Int (2019). doi: 10.1155/2019/9681863
81. Yan D, Dong W, He Q, Yang M, Huang L, Kong J, et al. Circular RNA circPICALM Sponges miR-1265 to Inhibit Bladder Cancer Metastasis and Influence FAK Phosphorylation. EBioMedicine (2019) 48:316–31. doi: 10.1016/j.ebiom.2019.08.074
82. Zhang L, Xia HB, Zhao CY, Shi L, Ren XL. Cyclic RNA Hsa_Circ_0091017 Inhibits Proliferation, Migration and Invasiveness of Bladder Cancer Cells by Binding to microRNA-589-5p. Eur Rev Med Pharmacol Sci (2020) 24(1):86–96. doi: 10.26355/eurrev_202001_19898
83. Bi J, Liu H, Dong W, Xie W, He Q, Cai Z, et al. Circular RNA Circ-ZKSCAN1 Inhibits Bladder Cancer Progression Through miR-1178-3p/P21 Axis and Acts as a Prognostic Factor of Recurrence. Mol Cancer (2019) 18(1):133. doi: 10.1186/s12943-019-1060-9
84. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 Sponges miR-558 to Suppress Heparanase Expression in Bladder Cancer Cells. EMBO Rep (2017) 18(9):1646–59. doi: 10.15252/embr.201643581
85. Liu T, Lu Q, Liu J, Xie S, Feng B, Zhu W, et al. Circular RNA FAM114A2 Suppresses Progression of Bladder Cancer via Regulating ∆NP63 by Sponging miR-762. Cell Death Dis (2020) 11(1):47. doi: 10.1038/s41419-020-2226-5
86. He Q, Huang L, Yan D, Bi J, Yang M, Huang J, et al. CircPTPRA Acts as a Tumor Suppressor in Bladder Cancer by Sponging miR-636 and Upregulating KLF9. Aging (Albany NY) (2019) 11(23):11314–28. doi: 10.18632/aging.102530
87. Su Y, Feng W, Zhong G, Ya Y, Du Z, Shi J, et al. ciRs-6 Upregulates March1 to Suppress Bladder Cancer Growth by Sponging miR-653. Aging (Albany NY) (2019) 11(23):11202–23. doi: 10.18632/aging.102525
88. Liu H, Bi J, Dong W, Yang M, Shi J, Jiang N, et al. Invasion-Related Circular RNA Circfndc3b Inhibits Bladder Cancer Progression Through the miR-1178-3p/G3BP2/SRC/FAK Axis. Mol Cancer (2018) 17(1):161–. doi: 10.1186/s12943-018-0908-8
89. Liu H, Chen D, Bi J, Han J, Yang M, Dong W, et al. Circular RNA Circubxn7 Represses Cell Growth and Invasion by Sponging miR-1247-3p to Enhance B4GALT3 Expression in Bladder Cancer. Aging (Albany NY) (2018) 10(10):2606–23. doi: 10.18632/aging.101573
90. Sun J, Zhang H, Tao D, Xie F, Liu F, Gu C, et al. CircCDYL Inhibits the Expression of C-MYC to Suppress Cell Growth and Migration in Bladder Cancer. Artif Cells Nanomed Biotechnol (2019) 47(1):1349–56. doi: 10.1080/21691401.2019.1596941
91. Su Y, Du Z, Zhong G, Ya Y, Bi J, Shi J, et al. Circ5912 Suppresses Cancer Progression via Inducing MET in Bladder Cancer. Aging (Albany NY) (2019) 11(23):10826–38. doi: 10.18632/aging.102464
92. Yang C, Mou Z, Zhang Z, Wu S, Zhou Q, Chen Y, et al. Circular RNA RBPMS Inhibits Bladder Cancer Progression via miR-330-3p/RAI2 Regulation. Mol Ther Nucleic Acids (2021) 23:872–86. doi: 10.1016/j.omtn.2021.01.009
93. Lin G, Sheng H, Xie H, Zheng Q, Shen Y, Shi G, et al. Circlpar1 Is a Novel Biomarker of Prognosis for Muscle-Invasive Bladder Cancer With Invasion and Metastasis by miR-762. Oncol Lett (2019) 17(3):3537–47. doi: 10.3892/ol.2019.9970
94. Chi BJ, Zhao DM, Liu L, Yin XZ, Wang FF, Bi S, et al. Downregulation of Hsa_Circ_0000285 Serves as a Prognostic Biomarker for Bladder Cancer and Is Involved in Cisplatin Resistance. Neoplasma (2019) 66(2):197–202. doi: 10.4149/neo_2018_180318N185
95. Yuan W, Zhou R, Wang J, Han J, Yang X, Yu H, et al. Circular RNA Cdr1as Sensitizes Bladder Cancer to Cisplatin by Upregulating APAF1 Expression Through miR-1270 Inhibition. Mol Oncol (2019) 13(7):1559–76. doi: 10.1002/1878-0261.12523
96. Su H, Tao T, Yang Z, Kang X, Zhang X, Kang D, et al. Circular RNA cTFRC Acts as the Sponge of MicroRNA-107 to Promote Bladder Carcinoma Progression. Mol Cancer (2019) 18(1):27. doi: 10.1186/s12943-019-0951-0
97. Najafi S, Ghafouri-Fard S, Hussen BM, Hidayat H, Taheri M, Hallajnejad M. Oncogenic Roles of Small Nucleolar RNA Host Gene 7 (SNHG7) Long Non-Coding RNA in Human Cancers and Potentials. Front Cell Dev Biol (2022) 9:809345. doi: 10.3389/fcell.2021.809345.
98. Zhang H-D, Jiang L-H, Sun D-W, Hou J-C, Ji Z-L. CircRNA: A Novel Type of Biomarker for Cancer. Breast Cancer (2018) 25(1):1–7. doi: 10.1007/s12282-017-0793-9
99. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res (2015) 25(8):981–4. doi: 10.1038/cr.2015.82
Keywords: bladder cancer, ncRNAs, circRNAs, expression, biomarker
Citation: Ghafouri-Fard S, Najafi S, Hussen BM, Basiri A, Hidayat HJ, Taheri M and Rashnoo F (2022) The Role of Circular RNAs in the Carcinogenesis of Bladder Cancer. Front. Oncol. 12:801842. doi: 10.3389/fonc.2022.801842
Received: 25 October 2021; Accepted: 28 January 2022;
Published: 28 February 2022.
Edited by:
Bianca Nitzsche, Charité Universitätsmedizin Berlin, GermanyReviewed by:
Ning Li, Fourth Affiliated Hospital of China Medical University, ChinaCopyright © 2022 Ghafouri-Fard, Najafi, Hussen, Basiri, Hidayat, Taheri and Rashnoo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, TW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Fariborz Rashnoo, ZmFyaWJvcnoucmFzaG5vb0B5YWhvby5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.