
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 08 June 2022
Sec. Gynecological Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.797453
This article is part of the Research Topic Global Excellence in Oncology: Europe 2021 View all 6 articles
 Dieuwke L. Marvin1†
Dieuwke L. Marvin1† Vivian M. Spaans2,3†
Vivian M. Spaans2,3† Cor D. de Kroon2
Cor D. de Kroon2 Roderick C. Slieker4,5
Roderick C. Slieker4,5 Maryam Khelil6
Maryam Khelil6 Peter ten Dijke1
Peter ten Dijke1 Laila Ritsma1†
Laila Ritsma1† Ekaterina S. Jordanova3,6,7*
Ekaterina S. Jordanova3,6,7*Cervical cancer is the fourth most common cancer in women worldwide. Squamous cell carcinoma (SCC) and adenocarcinoma (AC) are the most common histological types, with AC patients having worse prognosis. Over the last two decades, incidence rates of AC have increased, highlighting the importance of further understanding AC tumorigenesis, and the need to investigate new treatment options. The cytokine TGF-β functions as a tumour suppressor in healthy tissue. However, in tumour cells this suppressive function can be overcome. Therefore there is an increasing interest in using TGF-β inhibitors in the treatment of cancer. Here, we hypothesize that TGF-β plays a different role in SCC and AC. Analysis of RNA-seq data from the TCGA, using a TGF-β response signature, resulted in separate clustering of the two subtypes. We further investigated the expression of TGF-β-signalling related proteins (TβR1/2, SMAD4, pSMAD2, PAI-1, αvβ6 and MMP2/9) in a cohort of 62 AC patients. Low TβR2 and SMAD4 expression was associated with worse survival in AC patients and interestingly, high PAI-1 and αvβ6 expression was also correlated with worse survival. Similar correlations of TβR2, PAI-1 and αvβ6 with clinical parameters were found in previously reported SCC analyses. However, when comparing expression levels between SCC and AC patient samples, pSMAD2, SMAD4, PAI-1 and αvβ6 showed lower expression in AC compared to SCC. Because of the low expression of core TβR1/2, (p-)SMAD2 and SMAD4 proteins and the correlation with worse prognosis, TGF-β pathway most likely leads to tumour inhibitory effects in AC and therefore the use of TGF-β inhibitors would not be recommended. However, given the correlation of PAI-1 and αvβ6 with poor prognosis, the use of TGF- β inhibitors might be of interest in SCC and in the subsets of AC patients with high expression of these TGF-β associated proteins.
Cervical cancer is the fourth most common cancer in women worldwide (1). It is caused by a persistent infection with high risk human papillomavirus (HPV) (2). Tumour progression is facilitated by the failing control of the host’s immune response, combined with several immune escape mechanisms of the virally infected cells (3).
Histologically, cervical cancer can be classified as squamous cell carcinoma (SCC), adenocarcinoma (AC), adenosquamous carcinoma (ASC), and rare variants (4), each accounting for approximately 70-75%, 20-25%, 3-5%, and <1% of the cases, respectively (5–8). In contrast to SCC, the absolute and relative incidence rates of cervical AC have increased over the last two decades, predominantly in young women, and in developed countries (6, 8–12). When compared to SCC, AC histology was associated with higher recurrence rates, worse prognosis and survival, showed different dissemination patterns, and a different response to similar treatment regimens (11, 13). Furthermore, we and others have shown that the AC subtype represents a different spectrum of oncogenic mutations and has a different immunological and genomic pathway activation profile, compared to SCC (14–21).
Transforming growth factor-β (TGF-β) is a multifunctional cytokine that functions as a tumour suppressor in healthy cells, by being a potent inducer of growth arrest and apoptosis, maintaining tissue (microenvironment) homeostasis (22). In malignancy, tumour cells can overcome the cytostatic effects of TGF‐β signalling, mostly by acquiring mutations in the canonical signalling components (eliminating the tumour suppressor function) or by adaptation of TGF‐β signalling through non‐canonical signalling (leading to tumour promoting effects) (22–24). Accordingly, interest in TGF-β counteracting immunotherapies is gaining momentum (25), (26).
In normal tissue homeostasis, TGF-β is secreted into the extracellular matrix as part of a latent complex. TGF-β activators, such as integrins and proteases, release TGF-β from its latent state. Once activated, TGF-β initiates signalling by binding TGF-β receptor 2 (TβR2), which then forms a heteromeric complex with TGFβ receptor 1 (TβR1). TβR2 phosphorylates TβR1, which subsequently phosphorylates Sma and Mad related protein 2 (SMAD2) or SMAD3. These SMADs form heteromeric complexes with common mediator SMAD4, allowing for translocation into the nucleus. There, they interact with various transcription factors to regulate transcription of genes such as plasminogen activator inhibitor 1 (PAI-1), SMAD7, matrix metalloproteinases (MMP’s), collagen type I, and fibronectin (27, 28).
The TGF-β signalling pathway has been investigated predominantly in cervical cancer cell lines and in SCC histology (29–35). Previous studies of our group found that TGF-β1 expression in SCC patients was strongly associated with tumour infiltration within surrounding stroma and thus poor outcome, and TGF-β1 expression positively correlated with PAI-1 expression (31, 33). PAI-1 expression levels correlated with poor overall and disease free survival, while high SMAD4 expression was related to low infiltration depth (33, 34). A potential difference in TGF-β signalling between SCC and AC subtypes was suggested in these studies. However, too few AC patients were included to allow for a meaningful comparison.
Research concerning TGF-β signalling in cervical AC is scarce. With the rising incidence rates of cervical AC, it is important to further distinguish the biological and immunological processes that drive the tumorigenesis of this histological subtype, as this knowledge will contribute to the development of more precise, tumour-specific treatment approaches. Here, we hypothesised that the TGF-β pathway plays a different role in cervical AC compared to SCC (19, 33).
The aim of this study was to investigate the TGF-β pathway in cervical AC. We first analysed RNA-seq data from the TCGA, using a TGF-β response signature. Subsequently, we systematically investigated the protein expression of core canonical pathway members (SMAD4, pSMAD2, TβR1 and TβR2) and TGF-β pathway regulators and transcriptional targets (PAI-1, alpha-v beta-6 integrin (αvβ6 integrin), MMP2 and MMP9), and determined associations between protein expression and clinicopathological parameters in a well-defined, consecutive cohort of cervical AC patients.
This is a retrospective, single-centre- cohort study. All human tissues were used according to the Code of Conduct for responsible use of human tissues in the context of health research 2011 (https://www.bbmri.nl/sites/bbmri/files/styles/Federa_code_of_conduct_english.pdf). (All women included, were diagnosed with cervical AC, International Federation of Gynaecology and Obstetrics (FIGO) stage 1b-2a, and underwent a radical hysterectomy with lymphadenectomy as primary treatment at the Leiden University Medical Center (LUMC) between January 1990 and December 2005. Patients of whom sufficient representative tumour material, preserved in formalin-fixed, paraffin-embedded tissue (FFPE) blocks, was available in the archives of the department of Pathology of the LUMC were included. Clinical files were reviewed to collect data including age, FIGO stage, tumour diameter, invasion depth, lymph-vascular space infiltration (LVSI), parametrial involvement, tumour positivity of the surgical resection margins, tumour positivity of the pelvic lymph nodes, adjuvant treatment, disease recurrence and survival.
Conventional histology sections were stained with haematoxylin and eosin, and reviewed by an experienced pathologist to select only the ‘usual type’ endocervical ACs, according to the World Health Organization (WHO) classification of tumours (4). All tumours were typed for HPV, previously (18).
Immunohistochemistry was performed to investigate expression of TβR1 and TβR2, pSMAD2, SMAD4, PAI-1, αvβ6, MMP2 (gelatinase-A) and MMP9 (gelatinase-B). The primary antibodies used are listed in Supplementary Table 1.
Immunohistochemistry was performed and evaluated essentially as described before (29, 31, 32, 36). See Supplementary Methods for further details. To compare AC and SCC, immunohistochemistry data from SCC performed in previous studies was used, in which immunohistochemistry and evaluation was essentially as described in this study (29, 32, 33, 37). As a correction for comparing immunohistochemistry stainings, 3-5 SCC cases were taken along in the current AC immunohistochemistry staining.
RNA-seq and clinical data of AC and SCC were obtained from TCGA. The core set described in (19), was used for bioinformatical analysis. The most optimal number of groups within the TCGA data, was based on the silhouette value where a higher value indicates a better underlying number of groups. Overall similarities between AC and SCC, were visualized based on multidimensional scaling and the heatmap was based on scaled expression data and coloured by AC or SCC status.
Statistical analysis was performed with IBM Statistical Package for the Social Sciences (SPSS) Statistics (version 23, IBM corp., Chicago, IL, USA). Data were processed using the Chi-square test or Fisher’s exact test for categorical variables, Student’s t-test for parametric continuous variables, or one-way analysis of variance for numerical data when comparing more than two groups. Correlation was tested by the Spearman rho correlation coefficient in non-parametric data and the Pearson correlation in cases of normality. Kaplan Meier survival curves and the Log Rank test were used to determine between-group differences in disease-specific survival (DSS) and recurrence-free survival (RFS). Multivariate analysis was performed using a Cox proportional hazard model with stepwise regression. For all reported tests, P values were two-sided and P values <0.05 were considered to indicate statistical significance.
To determine if the TGF-β pathway plays a different role in AC versus SCC, expression data from TCGA on cervical cancer were analyzed (19). In this dataset, 144 squamous cell carcinomas and 31 adenocarcinomas were present. We determined the expression of a 153-gene TGF-β response signature (38) (Supplementary Table 2) in every tumor, and used these data to analyze the clustering of AC and SCC patients. The optimal amount of clusters in this data was two, based on the silhouette value (Figure 1A). Multidimensional scaling (Figure 1B) showed the clustering into two major subclasses; subclass A, presenting mostly AC samples (yellow), and subclass B, presenting mostly SCC patients (dark blue). Unsupervised hierarchical clustering of both the patient samples and signature genes, resulted in a heatmap confirming clustering of the SCC samples (dark gray) separately from the AC samples (light gray) (Figure 1C). As AC and SCC clustered mostly separately based on the TGF-β response signature, this indeed confirmed our hypothesis that AC is different from SCC in their response to TGF-β. We investigated this further at the protein level.
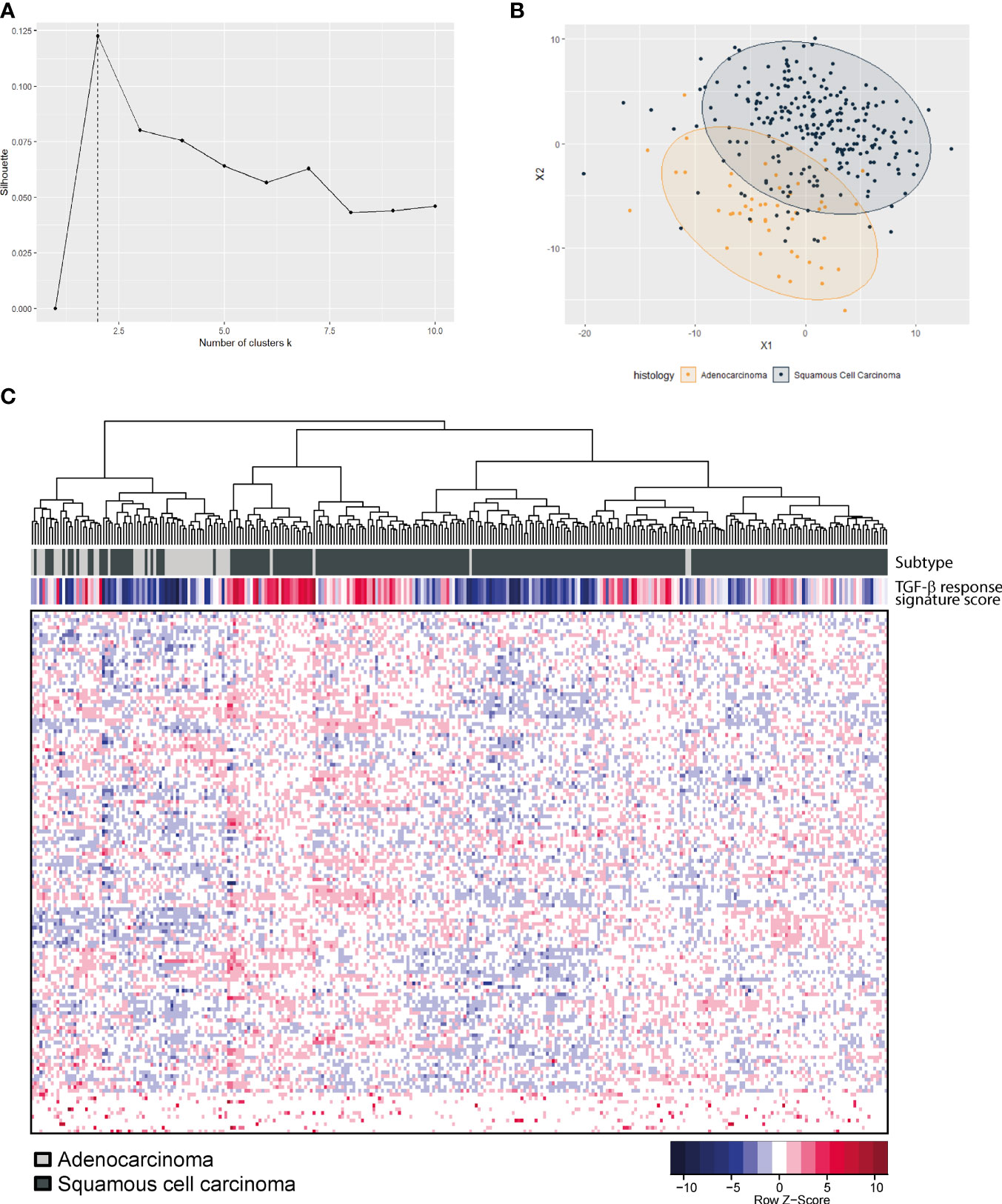
Figure 1 TGF-β pathway gene expression in the TCGA cervical cancer dataset. (A) Optimal amount of clusters was calculated based on the silhouette value and was found to be two. (B) Multidimensional scaling shows a separate clustering of AC (yellow) and SCC (dark blue). (C) Heatmap showing unsupervised hierarchical clustering analysis of 144 SCC (dark grey) and 31 AC (light grey) patient samples on the X-axis and 153-gene TGF-β response signature genes on the Y-axis.
In total 62 patients with usual type cervical adenocarcinoma were included in this study. Clinicopathological characteristics are summarized in Table 1. Protein expression scores are summarized in Table 2 and Figure 2, examples of stainings are shown in Figure 2. Univariate survival analysis is shown in Table 3 and illustrative Kaplan Meier Curves for RFS and DSS are shown in Figure 3 and 4. A comparison of protein expression scores between this cohort of 62 ACs and a previously analysed and reported cohort of SCCs (29, 32, 33, 37) is summarized in Table 4 and shown in graphs in Figure 2.
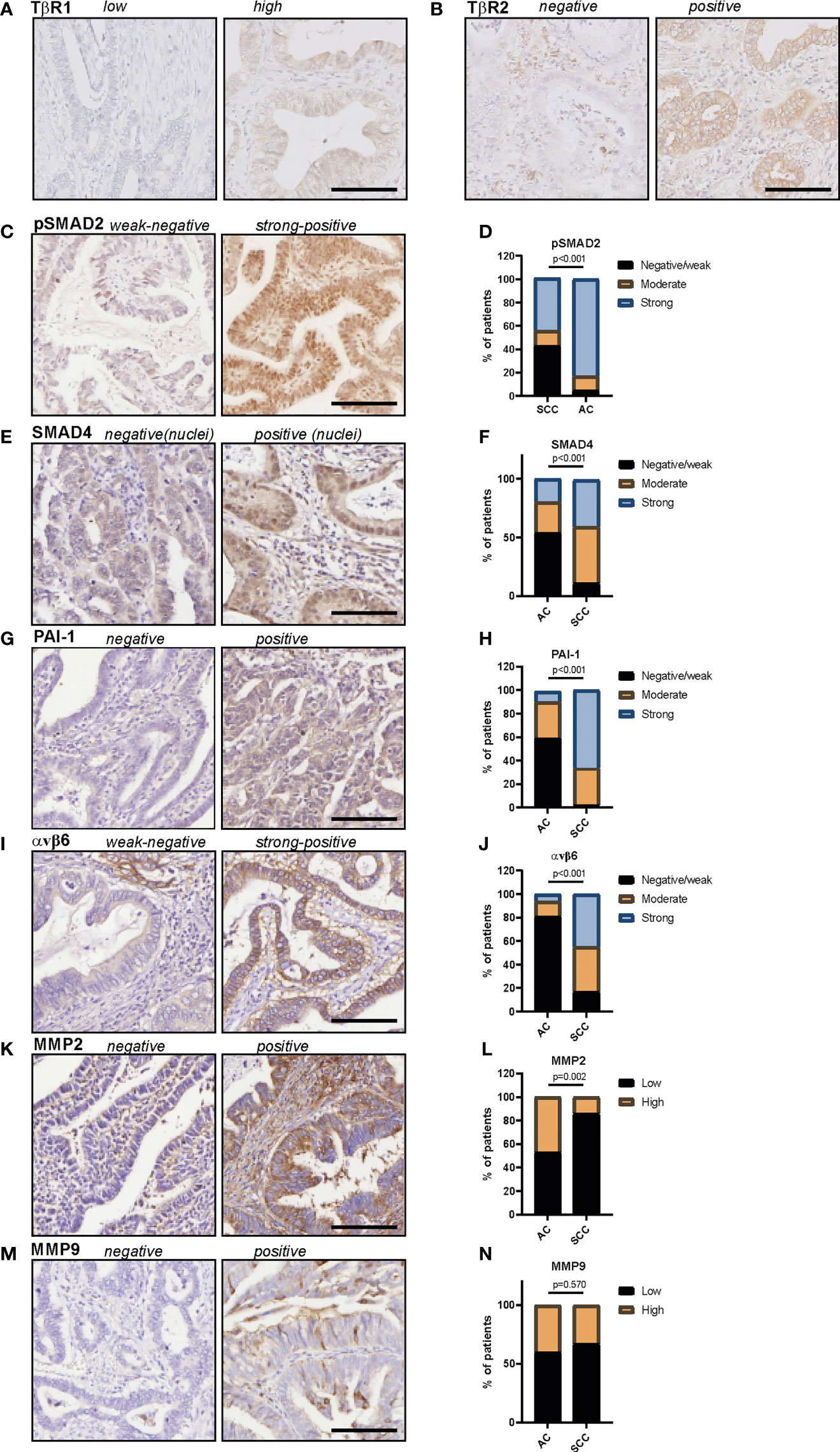
Figure 2 Protein expression of TβR1, TβR2, pSMAD2, SMAD4, PAI-1, αvβ6 and MMP2 and MMP9 in cervical adenocarcinomas. (A) example of TβR1 staining, (B) example of TβR2 staining, (C) example of pSMAD2 staining, (D) pSMAD2 staining in AC and SCC samples. (E) Example of SMAD4 staining, (F) SMAD4 staining in AC and SCC samples. (G) Example of PAI-1 staining, (H) PAI-1 staining in AC and SCC samples. (I) Example of αvβ6 staining, (J) αvβ6 staining in AC and SCC samples. (K) Example of MMP2 staining, (L) MMP2 staining in AC and SCC samples. (M) Example of MMP9 staining, (N) MMP9 staining in AC and SCC samples. All protein expression scores can be found in Table 2 (AC) and Table 4 (SCC and AC). P values are calculated using Chi-squared test. AC, adenocarcinoma; SCC, squamous cell carcinoma. Scale bar represents 100 μm.
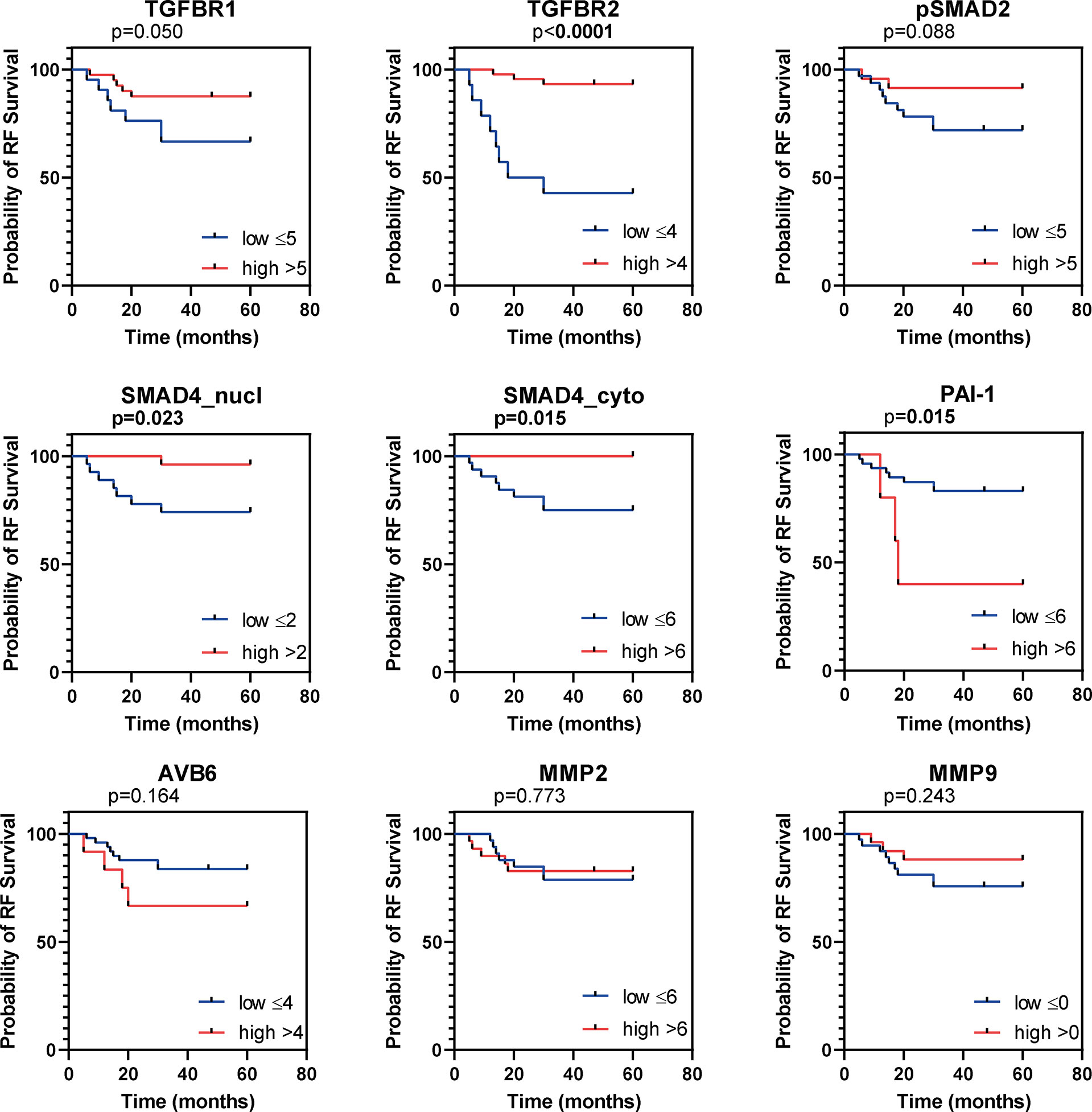
Figure 3 Kaplan Meier curves for recurrence free survival (RFS). Kaplan Meier curves for RFS of the institutional AC cohort, dichotomized as indicated by the numbers in the legend. P values based on log-rank test, bold indicates significance (p ≤ 0.05).
All samples presented positive TβR1 expression (Table 2). A cytoplasmic staining was more prevalent than membranous staining (Figure 2A). TβR1 expression scores were dichotomized based on the 25th percentile of the total score (=5). Low expression was associated with increased tumour size (mean diameter 35 ± 16mm vs. 22 ± 10mm, p=0.001). A trend was observed for association between low expression and lymph node positivity (6/20 (30%) vs. 4/39 (10%), p=0.056). TβR1 expression was not associated with survival (Table 3; Figures 3, 4).
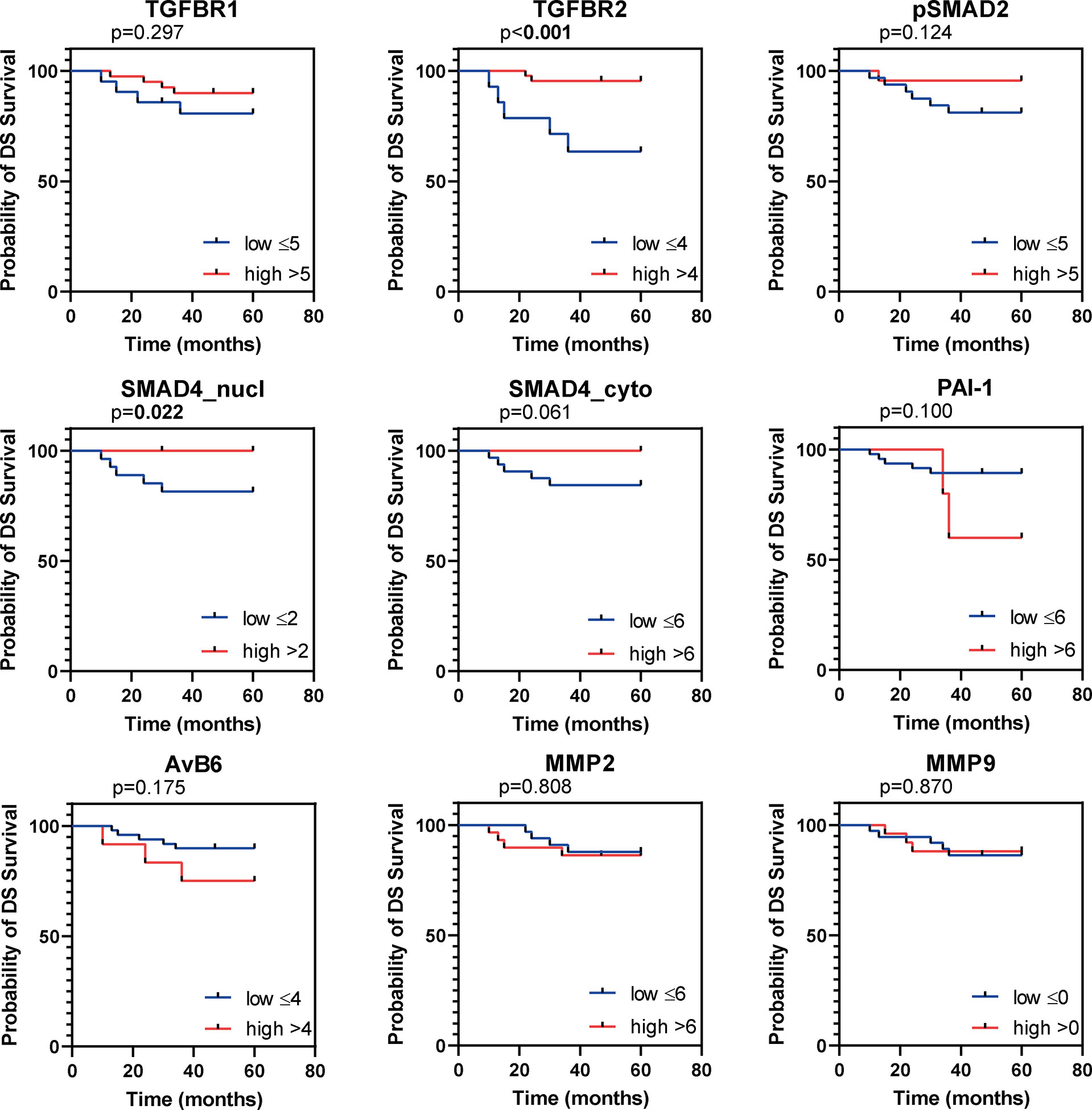
Figure 4 Kaplan Meier figures for disease specific survival (DSS). Kaplan Meier curves for DSS of the institutional AC cohort, dichotomized as indicated by the numbers in the legend. P values based on log-rank test, bold indicates significance (p ≤ 0.05).
Moderate/strong TβR2 expression was seen in the majority (74%) of cervical ACs (Table 2; Figure 2B). Low expression was associated with advanced tumour size (35 ± 17mm vs. 23 ± 11mm, p=0.002) and recurrent disease [8/15 (53%) vs. 4/43 (9%), p=0.001]. Univariate survival analysis showed a worse RFS (HR 0.1, 95% CI 0.04-0.41) and DSS (HR 0.15, 95% CI 0.05-0.52) for low expression (Table 3; Figures 3, 4). However, in multivariate survival analysis, TβR2 expression was not an independent predictor for RFS (HR 0.3, 95% CI 0.1-1.6) or for DSS (HR 0.5, 95% CI 0.74-3.0).
Overall, pSMAD2 and nuclear SMAD4 staining was more weakly expressed in AC compared to SCC (Table 4, Figures 2C–F). Nuclear pSMAD2 staining was observed in almost all AC tumours (only one tumour was completely negative, Table 2 and Figures 2C, D). Nuclear SMAD4 staining was present in 68% of the tumours, and cytoplasmic SMAD4 staining in 87% of the tumours (Table 2; Figures 2E, F). For SMAD4, four cases showed weak nuclear staining but absent cytoplasmic staining, whilst 13 cases showed absent nuclear staining but positive cytoplasmic staining. For SMAD4, as well as for pSMAD2, a typical staining pattern was observed with an intense positive nuclear staining on the tumour edges, and a weak/negative staining pattern towards the centre of the tumour fields.
Low pSMAD2 expression was associated with larger tumour size (mean diameter 32 ± 15mm vs. 20 ± 10mm, p=0.003), higher invasiveness (mean infiltration depth 14 ± 8mm vs. 8 ± 4mm, p=0.001), but pSMAD2 expression was not associated with RFS or DSS (Table 3; Figures 3, 4).
Nuclear- nor cytoplasmic SMAD4 expression was associated with any of the clinicopathological parameters. However, univariate survival analysis revealed a trend for worse RFS (HR 0.3, 95% CI 0.05-1.19), and worse DSS (HR 0.1, 95%CI 0.02-1.01) for low nuclear SMAD4 expression and a worse RFS (HR 0.2, 95% CI 0.02-1.32) and DSS (HR 0.2, 95% CI 0.02-1.5) for low cytoplasmic SMAD4 expression (Table 3; Figures 3, 4). Multivariate analysis revealed that nuclear SMAD4 expression was an independent predictor for RFS (HR 0.2, 95% CI 0.04-0.99) besides tumour size (HR 1.1, 95% CI 1.01-1.11), but not for DSS (HR 0.1, 95% CI 0.01-1.64). Cytoplasmic SMAD4 expression was not an independent predictor for RFS (HR 0.2, 95% CI 0.02-1.59) or DSS (HR 0.1, 95%CI 0.01-1.11).
Weak PAI-1 expression was observed in the majority of cases, and was more weakly expressed in AC compared to SCC (Table 3; Table 4 and Figures 2G, H). PAI-1 expression was not associated with the clinicopathological parameters, although a trend association was observed between moderate/strong PAI-1 expression and lymph node metastasis (5/20 (25%) vs. 2/32 (6%), p=0.054).
Univariate survival analysis revealed the worst RFS and DSS for patients with high PAI-1 expression (Table 3; Figures 3, 4). In multivariate analyses for RFS and DSS, PAI-1 expression was not an independent predictor for survival (HR 3.2, 95%CI 0.6-16.3, and HR 3.1, 95%CI 0.6-15.6, respectively).
Overall, αvβ6 was weakly expressed in AC compared to SCC (Table 4; Figures 2I, J). The characteristic αvβ6 staining of intense staining at peripheral tumour borders and weaker staining centrally within the tumour nest was less obvious in AC compared to SCC, and was only found in larger, undifferentiated AC samples (32).
High αvβ6 expression was associated with tumour size (mean tumour size 34 ± 18mm for high expression vs. 25 ± 13mm for low expression, p=0.049). Univariate survival analysis revealed a worse RFS and DSS for high αvβ6 expression (Table 3; Figures 3, 4). In multivariate analysis αvβ6 expression was not an independent predictor for RFS (HR 2.1, 95% CI 0.6-7.3) or DSS (HR 2.1, 95% CI 0.6-7.2).
Moderate to strong MMP2 expression was shown in 76% of the cases (Table 2; Figures 2K, L). The typical tumour border staining [as previously reported in SCC (37)] was less obvious in AC. Although a stronger staining pattern on the edges of the tumour outline was observed in 20 tumours (32%) versus homogenous staining in all other tumours.
Low MMP2 expression was associated with deeper tumour infiltration (mean infiltration depth 13 ± 8mm vs. 9 ± 4mm, p=0.025), but not with any of the other clinicopathological characteristics. MMP2 expression was not associated with RFS or DSS (Table 3; Figure 3, 4). High MMP2 expression occurred more frequently in AC compared to SCC (Table 4; Figure 2L).
Weak MMP9 expression was observed in 40% of the tumours, whilst 60% stained negative (Table 2; Figures 2M, N). MMP9 expression was also seen in different amounts at the tumour stroma near the tumour border and scored as abundant (n=15, 24%), sporadic (n=34, 55%), or negative (n=13, 21%). Neither MMP9 expression in tumour cells, nor MMP9 stromal expression was associated with any of the clinicopathological characteristics, and there were no associations with survival (Table 3; Figures 3 and 4). No difference in MMP9 expression intensity was found between AC and SCC (Table 4; Figure 2N).
Previously, the TCGA research network identified a high number of aberrations in tumour suppressor genes related to the TGF-β pathway in both SCC and AC (19). In SCC, both SMAD4 and TGFBR2 were mutated in 4% and 8% of the cases, respectively, whereas in AC only SMAD4 and not TGFBR2 mutations were observed in 12% of cases. Mutations in these genes are expected to result in absence of or aberrant protein expression. Hence, we assessed the absence of TβR2 and SMAD4 expression in our AC dataset. Indeed, SMAD4 was completely absent (SMAD4null, for both nuclear and cytoplasmic staining) in 4/52 AC cases (7.54%) (Figures 5A, B). Unexpectedly, also TβR2 expression was absent (TβR2null) in 5/58 (8.62%) AC cases (Figure 5C). PAI-1 expression and MMP2 expression were both significantly reduced in TβR2null patients (Figure 5D). Kaplan Meier survival analysis indicated that the TβR2null group and the TβR2null or SMAD4 null group had a significantly worse RFS compared to the wild type group (Figures 5C, D and E).
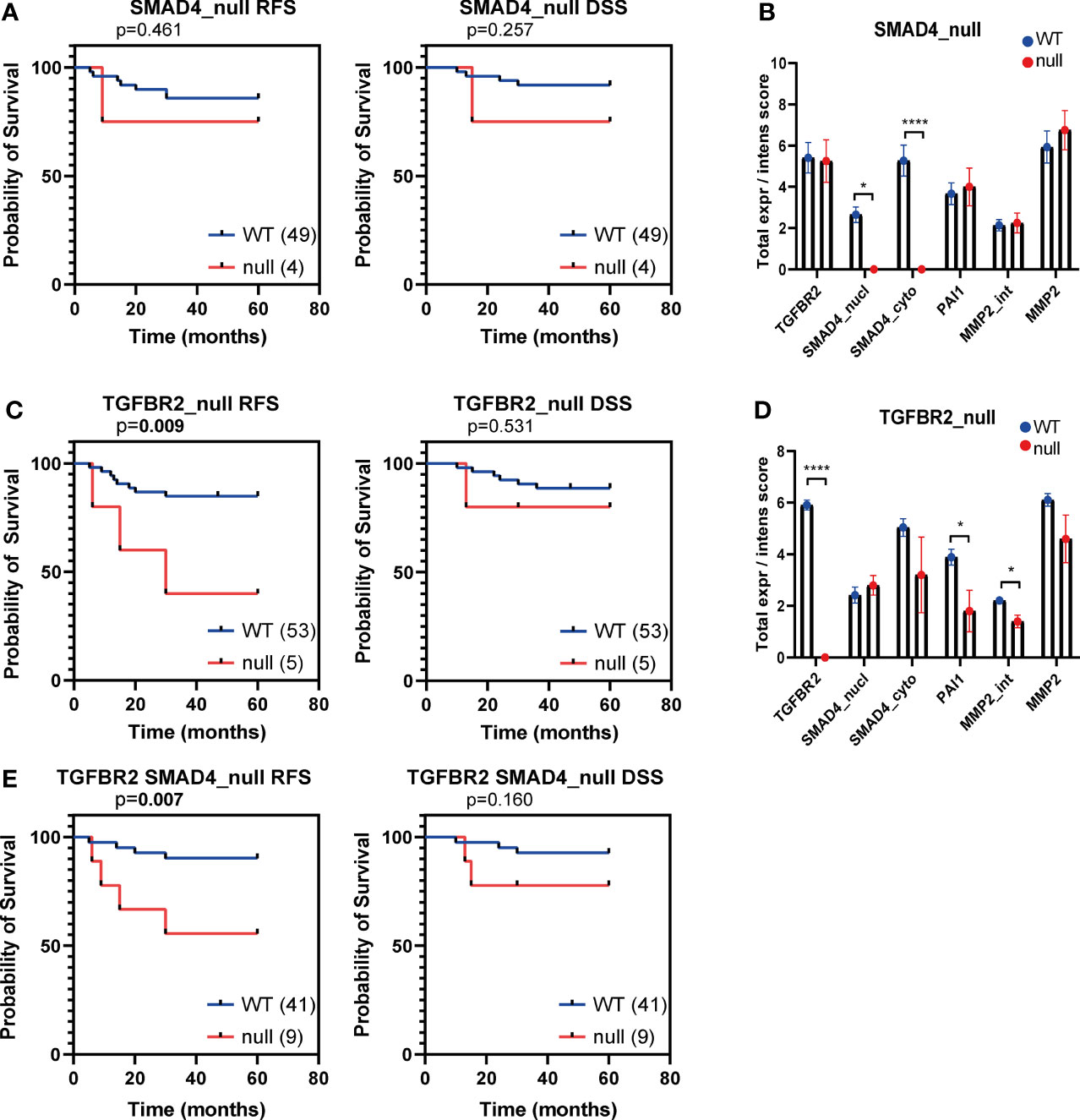
Figure 5 Kaplan Meier curves for recurrence free survival (RFS) and disease specific survival (DSS) for SMAD4null and TβR2 null AC patients samples. Kaplan Meier curves for RFS and DSS of the institutional AC cohort, dichotomized based on SMAD4null (A), TβR2null (C), TβR2null or SMAD4null (E) expression. The number of patient samples included in each group is indicated between brackets in the legend. P values based on log-rank test, bold indicates significance (p ≤ 0.05). Protein expression of TβR2, SMAD4 (nuclear and cytoplasmic), PAI-1 and MMP2 were compared in TβR2 wild type versus null (B) and SMAD4 wild type versus null (D). Significance is indicated with p<0.05 (*) and p<0.0001 (****).
Research concerning TGF-β signalling in cervical AC is scarce. Farley et al., studied TGF-β and TβR protein expression in AC (N=7) and its precursor lesions, and suggested that the neoplastic transformation of the endocervix might be related to deregulated TGF-β, and therewith loss of cell cycle control (39). Fan et al., studied TGF-β protein expression in 66 AC cases (who all received chemo- and immunotherapy prior to surgery), and described a positive expression being an independent predictor for worse survival (40). Previous studies of our group concerning the TGF-β pathway in cervical cancer demonstrated differences in TGF-β pathway activity comparing AC to SCC, however, too few AC samples were included to lead to meaningful conclusions (33). By comparing the expression of a 153-gene TGF-β response signature in AC and SCC patient samples from the TCGA dataset (19), we identified that most AC and SCC cluster separately. This suggests that the TGF-β pathway might indeed play a different role in the tumorigenesis of these cervical carcinoma subtypes. To better characterize cervical AC, we performed a comprehensive IHC analysis of various proteins of the TGF-β canonical pathway and downstream TGF-β pathway targets in a well-defined, consecutive cohort of 62 cervical AC patients. The AC series of our study was carefully classified using an additional mucus staining (14).
First, we assessed the expression of the core TGF-β canonical pathway proteins: TGF-β receptors and SMAD proteins. These proteins, are crucial in the signalling cascade to transfer the extracellular signal from activated TGF-β into transcription of target genes, including PAI-1, αvβ6 integrin, MMP’s and several cell cycle inhibitors (Figure 6). We showed that low expression of the core TGF-β pathway proteins (TβR1, TβR2, pSMAD2 and SMAD4) was associated with a poor prognosis in AC. Interestingly, a similar correlation to disease progression was observed in SCC (29). Previously, our group reported that all cervical SCC tumours show nuclear pSMAD2 expression and cytoplasmic SMAD4 expression (29). In comparison with SCC, here, pSMAD2 and SMAD4 staining was found to be lower in AC (Table 4; Figures 2D, F). Overall, the association of low expression of pSMAD2 and SMAD4 with poor prognosis, suggests that inactivation of the TGF-β pathway in AC might result in hampering cytostatic effects of the pathway.
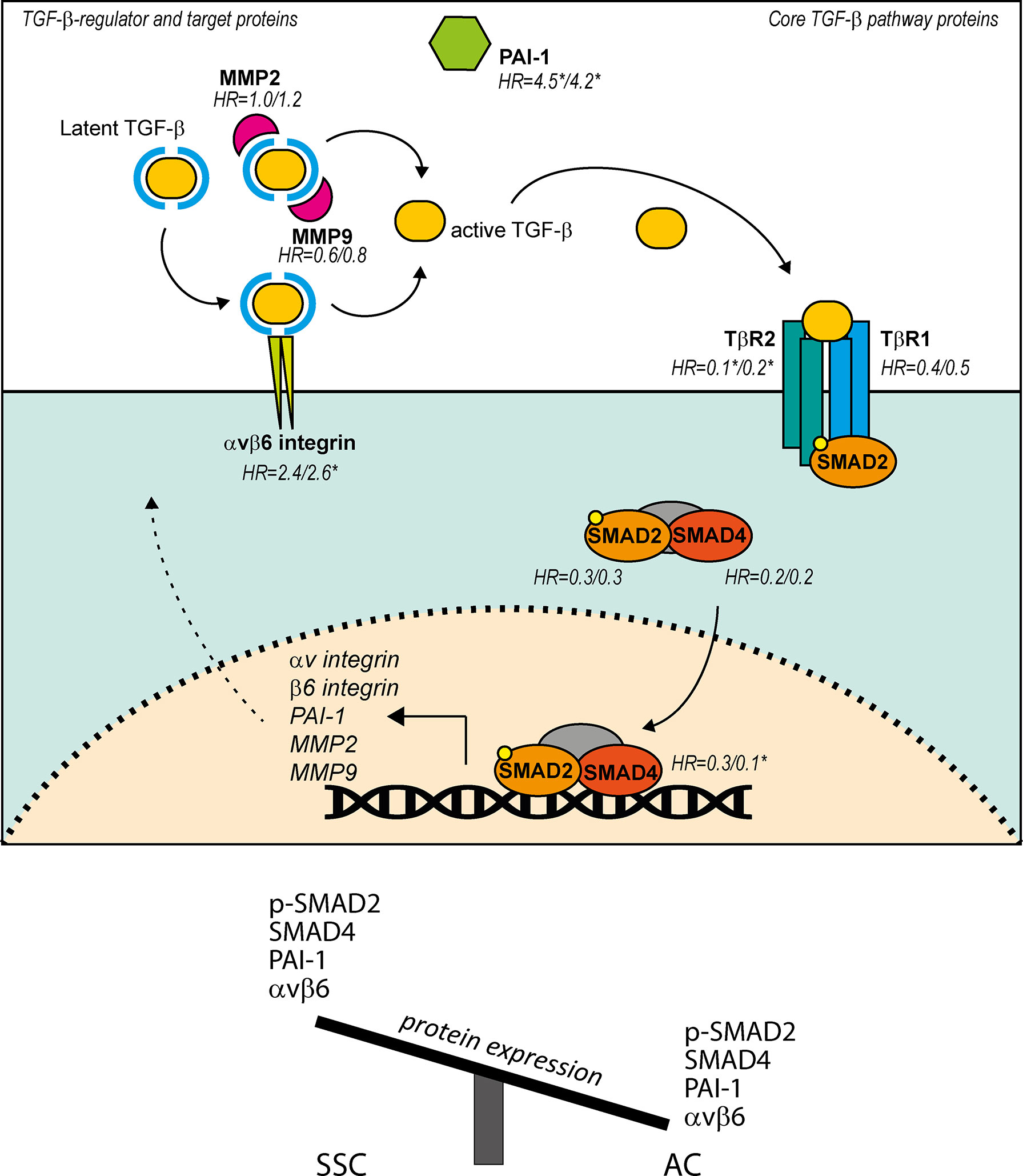
Figure 6 Summary of the presented findings of TGF-β pathway, TGF-β regulator and TGF-β target proteins in AC, and differences in expression between AC and SCC in regard to these proteins. TGF-β pathway starts with binding of TGF-β to TβR2, leading to phosphorylation of TβR1 and subsequent phosphorylation of SMAD2. SMAD2 together with SMAD4 and SMAD3 (not indicated) form a complex and translocate to the nucleus to regulate transcription of TGF-β target genes PAI-1, αv integrin, β6 integrin, MMP2 and MMP9. Avβ6 integrin and MMP2/9 mediate cleavage of latent TGF-β. Hazard ratios (HR) are calculated low over high, and are shown for the proteins investigated in the present study. HRs are shown as RFS/DSS, significance (p ≤ 0.05) is indicated with*.
In a few samples, we observed a complete lack of staining for either TβR2 or SMAD4. These TβR2null and SMAD4null cases were identified in 9% and 8% of our cohort, respectively. In the TCGA data, approximately 12% of AC patients showed a mutation or DNA methylation of SMAD4 (19). This could fit with our findings; the slightly higher percentage might be due to the low sample size in the TCGA dataset (31 AC samples) and of the group deficient in TβR2. In contrast, the TCGA analysis revealed no mutations or DNA methylation for TGFBR2, whereas our results suggest that about 9% of the cases had a complete lack of expression. Again, the small AC sample size in the TCGA cohort, and the small sample size of patients deficient in TβR2, might be the cause. Alternatively, this might be the result of epigenetic or posttranslational silencing. Future research should address these questions.
Next, we investigated TGF-β regulating and target proteins αvβ6 integrin and PAI-1. Avβ6 is a transmembrane cell surface receptor that mediates cell adhesion. It binds and activates latent TGF-β (41). In turn, TGF-β upregulates the expression of αvβ6 integrin on human keratinocytes (42). The serine protease inhibitor PAI-1, regulates the cleavage of plasminogen into plasmin by inhibiting the urokinase- and tissue plasminogen activators (uPA and tPA), which is an important mechanism in the regulation, formation, and degradation of the extracellular matrix (ECM) (33, 43). In normal cervical epithelium, PAI-1 and αvβ6 are not- or weakly expressed, while their expression in cervical intra-epithelial neoplasia is strong (32). Interestingly, in AC we observed predominantly weak PAI-1 and αvβ6 staining. This was in contrast with SCC, where PAI-1 and αvβ6 were strongly expressed (Table 4; Figures 2H, J). Moreover, in AC the characteristic staining pattern of αvβ6 on the tumour-stromal interface was less obvious compared to SCC (32). Despite the overall weak staining, high PAI-1 and αvβ6 expression were associated with a worse outcome. These findings were opposite to the expression of core TGF-β pathway members (TβR and SMAD proteins), suggesting these proteins might be induced by other signalling pathways associated with worse survival outcome (e.g. TNF-α/NFkB) (44). Indeed, neither PAI-1 nor αvβ6 showed correlative (inverse) expression with one of the core TGF-β pathway proteins in AC. This was in contrast to SCC, where TGF-β1 expression inversely correlated with PAI-1 and αvβ6 (31, 32). Thus, although in both AC and SCC high PAI-1 expression correlates with worse outcome, the expression pattern and staining intensity of PAI-1 and of αvβ6 was different between the two subtypes.
In AC, TGF-β expression was found to be higher compared to SCC (33, 45). If TGF-β1 expression is high in AC, but the core pathway proteins and target/regulating proteins are lower in tumour cells, this suggests that TGF-β1 might be acting predominantly on the microenvironment in AC rather than on the tumour cells. TGF-β affects many cell types in the microenvironment, including immune cells, and it can promote tumour progression by evasion of the immune system (24). By activating pro-tumorigenic microenvironment cells like M2 macrophages or by inhibiting CD8 T cell activation, TGF-β1 might result in a tumour promoting effect. Future work should address these points.
Similar to the differences observed in protein expression scores for SCC and AC, our bioinformatical analysis showed that SCC and AC samples fell into separate clusters when looking at the TGF-β response signature. This suggests, that these subsets do show differences in TGF-β signalling. Part of these differences could be explained by the lower staining intensity of pSMAD2, SMAD4, PAI-1 and αvβ6 in SCC. The TGF-β pathway-related protein expression differences observed between AC and SCC are summarized in Figure 6. Interestingly, 6/31 of AC samples clustered in the predominant SCC cluster, indicating that these 6 AC patients might respond to TGF-β similarly to SCC patients, and might potentially have higher TGF-β related protein expression. Further investigation is necessary to identify the differences in TGF-β response between AC and SCC.
In conclusion, in AC, similar to SCC, PAI-1 and αvβ6 integrin are unfavourable prognostic factors although this may not be TGF-β pathway related - given the inverse correlation in protein expression levels. Moreover, we showed that in AC there is a low core TGF-β pathway protein expression compared to SCC, and low expression is associated with poor patient survival and worse prognosis. As such, the use of TGF-β inhibitors to treat AC patients with moderate to strong staining for core TGF-β pathway proteins, would not be recommended, as tumour cell proliferation might increase. Alternatively, for AC patients with high expression of TGF-β target proteins such as PAI-1 and αvβ6 integrin, as well as for SCC patients, TGF-β inhibitors could be of clinical interest. Few clinical trials using these inhibitors are including patients with cervical cancer, and targeting TGF-β remains challenging (46, 47). The role of TGF-β on the immune system is gaining interest, and there is increasing evidence that TGF-β affects immunotherapy response in cervical cancer as well (26, 48–51). Differences between AC and SCC, should be taken into account in these studies, as immune cell recruitment has been found to differ between the two subtypes (51). Future studies including validation in in vitro and in vivo models will be necessary to further elucidate the role of TGF-β in cervical AC and SCC. Our results could also be of relevance for other cancer types with this subclassification, such as lung cancer. Based on the here presented data, characterisation of TGF-β related protein expression in individual AC patients will be important upon patient inclusion in future clinical trials with TGF-β inhibitors, as monotherapy or in combination with other therapies.
Publicly available datasets were analyzed in this study. This data can be found here: TCGA database, discussed in DOI:10.1038/nature21386.
VMS carried out the experiments. RCS and MK performed out the bioinformatical analysis. EMS, ESJ, LR and LM analysed data. Study design was done by VMS, and EJ. VMS and CDdK were responsible for recruitment of patients and collecting samples. VMS and DLM wrote the draft manuscript. ESJ, LR, PtD and CDdK revised the manuscript. All authors had final approval of the submitted and published versions.
This study was supported by Cancer Genomics Netherlands (CGC.NL) to LR and PtD, and the Louise Vehmeijer stichting Amsterdam to ESJ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge Enno Dreef and Natalja ter Haar for their assistance performing the immunological staining’s. We kindly thank Cees Sier for providing the MMP9 antibody. We thank Arko Gorter for his structural comments on the design of this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.797453/full#supplementary-material
Supplementary Table 1 | Overview of the antibodies used in the present study. Monoclonal and polyclonal antibodies used for the immunohistochemistry assays. The antigen retrieval method, the dilution used, and the supplying companies are listed, as well as the references concerning the studies of our group where the antibodies were described previously.
Supplementary Table 2 | The 153-gene TGF-β response signature as defined by Padua et al. (38). The gene names are indicated, as well as their up- or downregulated status in the original signature.
AC, cervical adenocarcinoma; ASC, cervical adenosquamous carcinoma; avβ6, alpha-v beta-6 integrin; DSS, Disease specific survival; FIGO, International Federation of Gynaecology and Obstetrics; HPV, Human Papillomavirus; MMP2, matrix metalloproteinase 2; MMP9, matrix metalloproteinase 9; PAI-1, plasminogen activator inhibitor 1; (p-)SMAD, (phosphorylated-) mothers against decapentaplegic homolog family members; RFS, Recurrence free survival; SCC, cervical squamous cell carcinoma; TCGA, The Cancer Genome Atlas; TGF-β, Transforming Growth Factor–beta; TβR1, Transforming growth factor-beta type I receptor; TβR2, Transforming growth factor-beta type 2 receptor.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Hausen Hz. Papillomaviruses in the Causation of Human Cancers - a Brief Historical Account. Virology (2009) 384:260–5. doi: 10.1016/j.virol.2008.11.046
3. Tindle RW. Immune Evasion in Human Papillomavirus-Associated Cervical Cancer. Nat Rev Cancer (2002) 2:59–65. doi: 10.1038/nrc700
4. Wells M NJ, Ostor AG, Crum CP, Franceschi S, Tommasino M. Tumours of the Uterine Cervix. In: Young R, Kurman RJ, Carcangiu ML, Herrington CS, editors. WHO Classification of Tumours of Female Reproductive Organs, 4th ed. Lyon, France: IARC (2014). p. 169–207.
5. Watson M, Saraiya M, Benard V, Coughlin SS, Flowers L, Cokkinides V, et al. Burden of Cervical Cancer in the United States, 1998-2003. Cancer (2008) 113:2855–64. doi: 10.1002/cncr.23756
6. Bulk S, Visser O, Rozendaal L, Verheijen RHM, Meijer CJLM. Incidence and Survival Rate of Women With Cervical Cancer in the Greater Amsterdam Area. Br J Cancer (2003) 89:834–9. doi: 10.1038/sj.bjc.6601157
7. Sasieni P, Castanon A, Cuzick J. Screening and Adenocarcinoma of the Cervix. Int J Cancer (2009) 125:525–9. doi: 10.1002/ijc.24410
8. Sasieni P, Adams J. Changing Rates of Adenocarcinoma and Adenosquamous Carcinoma of the Cervix in England. Lancet (2001) 357:1490–3. doi: 10.1016/S0140-6736(00)04646-8
9. Vizcaino AP, Moreno V, Bosch FX, Muñoz N, Barros-Dios XM, Borras J, et al. International Trends in Incidence of Cervical Cancer: II. Squamous-Cell Carcinoma. Int J Cancer (2000) 89:429–35. doi: 10.1002/(sici)1097-0215(20000501)86:3<429::aid-ijc20>3.0.co;2-d
10. Smith HO, Tiffany MF, Qualls CR, Key CR. The Rising Incidence of Adenocarcinoma Relative to Squamous Cell Carcinoma of the Uterine Cervix in the United States - A 24-Year Population-Based Study. Gynecol Oncol (2000) 78:97–105. doi: 10.1006/gyno.2000.5826
11. Williams NL, Werner TL, Jarboe EA, Gaffney DK. Adenocarcinoma of the Cervix: Should We Treat It Differently? Curr Oncol Rep (2015) 17. doi: 10.1007/s11912-015-0440-6
12. van der Horst J, Siebers AG, Bulten J, Massuger LF, de Kok IMCM. Increasing Incidence of Invasive and in Situ Cervical Adenocarcinoma in the Netherlands During 2004–2013. Cancer Med (2017) 6:416–23. doi: 10.1002/cam4.971
13. Galic V, Herzog TJ, Lewin SN, Neugut AI, Burke WM, Lu YS, et al. Prognostic Significance of Adenocarcinoma Histology in Women With Cervical Cancer. Gynecol Oncol (2012) 125:287–91. doi: 10.1016/j.ygyno.2012.01.012
14. Spaans VM, Trietsch MD, Peters AAW, Osse M, ter Haar N, Fleuren GJ, et al. Precise Classification of Cervical Carcinomas Combined With Somatic Mutation Profiling Contributes to Predicting Disease Outcome. PLoS One (2015) 10. doi: 10.1371/journal.pone.0133670
15. Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, et al. Landscape of Genomic Alterations in Cervical Carcinomas. Nature (2014) 506:371–5. doi: 10.1038/nature12881
16. Samuels S, Spaans VM, Osse M, Peters LAW, Kenter GG, Fleuren GJ, et al. Human Leukocyte Antigen-DR Expression is Significantly Related to an Increased Disease-Free and Disease-Specific Survival in Patients With Cervical Adenocarcinoma. Int J Gynecol Cancer (2016) 26:1503–9. doi: 10.1097/IGC.0000000000000783
17. Punt S, van Vliet ME, Spaans VM, de Kroon CD, Fleuren GJ, Gorter A, et al. FoxP3+ and IL-17+ Cells are Correlated With Improved Prognosis in Cervical Adenocarcinoma. Cancer Immunol Immunother (2015) 64:745–53. doi: 10.1007/s00262-015-1678-4
18. Spaans VM, Peters AAW, Fleuren GJ, Jordanova ES. HLA-E Expression in Cervical Adenocarcinomas: Association With Improved Long-Term Survival. J Transl Med (2012) 10. doi: 10.1186/1479-5876-10-184
19. Burk RD, Chen Z, Saller C, Tarvin K, Carvalho AL, Scapulatempo-Neto C, et al. Integrated Genomic and Molecular Characterization of Cervical Cancer. Nature (2017) 543:378–84. doi: 10.1038/nature21386
20. Khelil M, Griffin H, Bleeker MCG, Steenbergen RDM, Zheng K, Saunders-Wood T, et al. Delta-Like Ligand-Notch1 Signalling is Selectively Modulated by HPV16 E6 to Promote Squamous Cell Proliferation and Correlates With Cervical Cancer Prognosis. Cancer Res (2021) 8. doi: 10.1158/0008-5472.can-20-1996
21. Wright AA, Howitt BE, Myers AP, Dahlberg SE, Palescandolo E, Van Hummelen P, et al. Oncogenic Mutations in Cervical Cancer: Genomic Differences Between Adenocarcinomas and Squamous Cell Carcinomas of the Cervix. Cancer (2013) 119:3776–83. doi: 10.1002/cncr.28288
22. Batlle E, Massagué J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity (2019) 50:924–40. doi: 10.1016/j.immuni.2019.03.024
23. Martinez J, Zhang XH-F. BMP/Coco Antagonism as a Deterministic Factor of Metastasis Dormancy in Lung. Breast Cancer Res (2013) 15:302. doi: 10.1186/bcr3350
24. Meulmeester E, ten Dijke P. The Dynamic Roles of TGF-β in Cancer. J Pathol (2011) 223:206–19. doi: 10.1002/path.2785
25. van den Bulk J, de Miranda NFCC, ten Dijke P. Therapeutic Targeting of TGF-β in Cancer: Hacking a Master Switch of Immune Suppression. Clin Sci (Lond). (2021) 135:35–52. doi: 10.1042/CS20201236
26. Birrer MJ, Fujiwara K, Oaknin A, Randall L, Ojalvo LS, Valencia C, et al. The Changing Landscape of Systemic Treatment for Cervical Cancer: Rationale for Inhibition of the TGF-β and PD-L1 Pathways. Front Oncol (2022) 12:814169. doi: 10.3389/FONC.2022.814169
27. Zhang L, Zhou F, ten Dijke P. Signaling Interplay Between Transforming Growth Factor-β Receptor and PI3K/AKT Pathways in Cancer. Trends Biochem Sci (2013) 38:612–20. doi: 10.1016/j.tibs.2013.10.001
28. Massagué J. Tgfβ Signalling in Context. Nat Rev Mol Cell Biol (2012) 13:616–30. doi: 10.1038/nrm3434
29. Kloth JN, Kenter GG, Spijker HS, Uljee S, Corver WE, Jordanova ES, et al. Expression of Smad2 and Smad4 in Cervical Cancer: Absent Nuclear Smad4 Expression Correlates With Poor Survival. Mod Pathol (2008) 21:866–75. doi: 10.1038/modpathol.2008.62
30. Kloth JN, Fleuren GJ, Oosting J, de Menezes RX, Eilers PHC, Kenter GG. Substantial Changes in Gene Expression of Wnt, MAPK and Tnfα Pathways Induced by TGF-β1 in Cervical Cancer Cell Lines. Carcinogenesis (2005) 26:1493–502. doi: 10.1093/carcin/bgi110
31. Hazelbag S, Gorter A, Kenter GG, Van Den Broek L, Fleuren G. Transforming Growth Factor-β1 Induces Tumor Stroma and Reduces Tumor Infiltrate in Cervical Cancer. Hum Pathol (2002) 33:1193–9. doi: 10.1053/hupa.2002.130109
32. Hazelbag S, Kenter GG, Gorter A, Dreef EJ, Koopman LA, Violette SM, et al. Overexpression of the αvβ6 Integrin in Cervical Squamous Cell Carcinoma is a Prognostic Factor for Decreased Survival. J Pathol (2007) 212:316–24. doi: 10.1002/path.2168
33. Hazelbag S, Kenter GG, Gorter A, Fleuren GJ. Prognostic Relevance of TGF-β1 and PAI-1 in Cervical Cancer. Int J Cancer (2004) 112:1020–8. doi: 10.1002/ijc.20512
34. Noordhuis MG, Fehrmann RSN, Wisman GBA, Nijhuis ER, Van Zanden JJ, Moerland PD, et al. Involvement of the TGF-β and β-Catenin Pathways in Pelvic Lymph Node Metastasis in Early-Stage Cervical Cancer. Clin Cancer Res (2011) 17:1317–30. doi: 10.1158/1078-0432.CCR-10-2320
35. Do Ki K, Tong SY, Huh CY, Lee JM, Lee SK, Chi SG. Expression and Mutational Analysis of TGF-β/Smads Signaling in Human Cervical Cancers. J Gynecol Oncol (2009) 20:117–21. doi: 10.3802/jgo.2009.20.2.117
36. Ruiter DJ, Ferrier CM, van Muijen GNP, Henzen-Logmans SC, Kennedy S, Kramer MD, et al. Quality Control of Immunohistochemical Evaluation of Tumour-Associated Plasminogen Activators and Related Components. Eur J Cancer (1998) 34:1334–40. doi: 10.1016/S0959-8049(98)00151-8
37. Sier CFM, Zuidwijk K, Zijlmans HJMAA, Hanemaaijer R, Mulder-Stapel AA, Prins FA, et al. EMMPRIN-Induced MMP-2 Activation Cascade in Human Cervical Squamous Cell Carcinoma. Int J Cancer (2006) 118:2991–8. doi: 10.1002/ijc.21778
38. Padua D, Zhang XHF, Wang Q, Nadal C, Gerald WL, Gomis RR, et al. Tgfβ Primes Breast Tumors for Lung Metastasis Seeding Through Angiopoietin-Like 4. Cell (2008) 133:66–77. doi: 10.1016/j.cell.2008.01.046
39. Farley J, Gray K, Nycum L, Prentice M, Birrer MJ, Jakowlew SB. Endocervical Cancer Is Associated With an Increase in the Ligands and Receptors for Transforming Growth Factor- and a Contrasting Decrease in p27Kip1. Gynecol Oncol (2000) 78:113–22. doi: 10.1006/gyno.2000.5879
40. Fan DM, Tian XY, Wang RF, Yu JJ. The Prognosis Significance of TGF-β1 and ER Protein in Cervical Adenocarcinoma Patients With Stage Ib ~ IIa. Tumor Biol (2014) 35:11237–42. doi: 10.1007/s13277-014-2110-y
41. Munger JS, Huang X, Kawakatsu H, Griffiths MJD, Dalton SL, Wu J, et al. The Integrin αvβ6 Binds and Activates Latent Tgfβ1: A Mechanism for Regulating Pulmonary Inflammation and Fibrosis. Cell (1999) 96:319–28. doi: 10.1016/S0092-8674(00)80545-0
42. Zambruno G, Marchisio PC, Marconi A, Vaschieri C, Melchiori A, Giannetti A, et al. Transforming Growth Factor-β1 Modulates β1 and β5 Integrin Receptors and Induces the De Novo Expression of the Avβ6 Heterodimer in Normal Human Keratinocytes: Implications for Wound Healing. J Cell Biol (1995) 129:853–65. doi: 10.1083/jcb.129.3.853
43. Higgins PJ, Czekay RP, Wilkins-Port CE, Higgins SP, Freytag J, Overstreet JM, et al. PAI-1: An Integrator of Cell Signaling and Migration. Int J Cell Biol (2011) 2011. doi: 10.1155/2011/562481
44. Garg AK, Jhingran A, Klop AH, Aggarwal BB, Kunnumakkara AB, Broadus RR, et al. Expression of Nuclear Transcription Factor Kappa B in Locally Advanced Human Cervical Cancer Treated With Definitive Chemoradiation. Int J Radiat Oncol Biol Phys (2010) 78:1331–6. doi: 10.1016/j.ijrobp.2009.09.044
45. Santin AD, Hermonat PL, Hiserodt JC, Fruehauf J, Schranz V, Barclay D, et al. Differential Transforming Growth Factor-β Secretion in Adenocarcinoma and Squamous Cell Carcinoma of the Uterine Cervix. Gynecol Oncol (1997) 64:477–80. doi: 10.1006/gyno.1996.4579
46. Birrer MJ, Mileshkin LR, Fujiwara K, Ray-Coquard I, Alexandre J, Okamoto A, et al. 879tip Phase II Study of Bintrafusp Alfa, a Bifunctional Fusion Protein Targeting TGF-β and PD-L1, in Platinum-Experienced Advanced Cervical Cancer. Ann Oncol (2020) 31:S644–5. doi: 10.1016/J.ANNONC.2020.08.1018
47. Liu S, Ren J, ten Dijke P. Targeting Tgfβ Signal Transduction for Cancer Therapy. Signal Transduction Targeting Ther (2021) 6. doi: 10.1038/S41392-020-00436-9
48. Ni Y, Soliman A, Joehlin-Price A, Rose PG, Vlad A, Edwards RP, et al. High TGF-β Signature Predicts Immunotherapy Resistance in Gynecologic Cancer Patients Treated With Immune Checkpoint Inhibition. NPJ Precis Oncol (2021) 5:1–11. doi: 10.1038/s41698-021-00242-8
49. García-Rocha R, Moreno-Lafont M, Mora-García ML, Weiss-Steider B, Montesinos JJ, Piña-Sánchez P, et al. Mesenchymal Stromal Cells Derived From Cervical Cancer Tumors Induce TGF-β1 Expression and IL-10 Expression and Secretion in the Cervical Cancer Cells, Resulting in Protection From Cytotoxic T Cell Activity. Cytokine (2015) 76:382–90. doi: 10.1016/J.CYTO.2015.09.001
50. Strauss J, Braiteh FS, Calvo E, de Miguel M, Cervantes A, Edenfield WJ, et al. Evaluation of Bintrafusp Alfa, a Bifunctional Fusion Protein Targeting TGF-β and PD-L1, in Cervical Cancer: Data From Phase 1 and Phase 2 Studies. J Clin Oncol ASCO Annual Meeting I (2021) 39:5509–9. doi: 10.1200/JCO.2021.39.15_suppl.5509
51. Rotman J, Heeren AM, Gassama AA, Lougheed SM, Pocorni N, Stam AGM, et al. Adenocarcinoma of the Uterine Cervix Shows Impaired Recruitment of Cdc1 and CD8 + T Cells and Elevated β-Catenin Activation Compared With Squamous Cell Carcinoma. Clin Cancer Res (2020) 26:3791–802. doi: 10.1158/1078-0432.CCR-19-3826
Keywords: cervical cancer, adenocarcinoma, squamous cell carcinoma, TGFBR2, SMAD4, transforming growth factor-β (TGF-β)
Citation: Marvin DL, Spaans VM, de Kroon CD, Slieker RC, Khelil M, ten Dijke P, Ritsma L and Jordanova ES (2022) Low Transforming Growth Factor-β Pathway Activity in Cervical Adenocarcinomas. Front. Oncol. 12:797453. doi: 10.3389/fonc.2022.797453
Received: 18 October 2021; Accepted: 12 May 2022;
Published: 08 June 2022.
Edited by:
Giancarlo Castellano, Institut de Recerca Biomèdica August Pi i Sunyer (IDIBAPS), SpainReviewed by:
Oscar Gee Wan Wong, The University of Hong Kong, Hong Kong SAR, ChinaCopyright © 2022 Marvin, Spaans, de Kroon, Slieker, Khelil, ten Dijke, Ritsma and Jordanova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ekaterina S. Jordanova, ZS5qb3JkYW5vdmFAYW1zdGVyZGFtdW1jLm5s
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.