- 1Department of Oral and Maxillofacial Surgery, The Affiliated Stomatology Hospital of Southwest Medical University, Luzhou, China
- 2Orofacial Reconstruction and Regeneration Laboratory, The Affiliated Stomatology Hospital of Southwest Medical University, Luzhou, China
- 3Department of Oral and Maxillofacial Surgery, The Affiliated Hospital of Southwest Medical University, Luzhou, China
- 4Department of Hematology, The Affiliated Hospital of Southwest Medical University, Luzhou, China
- 5Stem Cell Laboratory, The Affiliated Hospital of Southwest Medical University, Luzhou, China
- 6Department of Pathology, The Affiliated Hospital of Southwest Medical University, Luzhou, China
Purpose: This study aimed to determine the expression profiles of long non-coding RNA (lncRNA), microRNA (miRNA), and mRNA in chemotherapy-resistant B-cell acute lymphoblastic leukemia (B-ALL).
Methods: LncRNA, miRNA, and mRNA profiles were assessed by RNA-seq in diagnostic bone marrow samples from 6 chemotherapy-resistant and 6 chemotherapy-sensitive B-ALL patients. The lncRNA DUXAP8/miR-29a/PIK3CA signaling network was identified as the most dysregulated in chemoresistant patient samples, and its effect on cellular phenotypes, PI3K-AKT-mTOR signaling, and chemosensitivity of doxorubicin (Dox)-resistant Nalm-6 (N6/ADR), and Dox-resistant 697 (697/ADR) cells were assessed. Furthermore, its synergy with inotuzumab ozogamicin treatment was investigated.
Results: 1,338 lncRNAs, 75 miRNAs, and 1620 mRNAs were found to be dysregulated in chemotherapy-resistant B-ALL in comparison to chemotherapy-sensitive B-ALL patient samples. Through bioinformatics analyses and RT-qPCR validation, the lncRNA DUXAP8/miR-29a/PIK3CA network and PI3K-AKT-mTOR signaling were identified as significantly associated with B-ALL chemotherapy resistance. In N6/ADR and 697/ADR cells, LncRNA DUXAP8 overexpression and PIK3CA overexpression induced proliferation and inhibited apoptosis, and their respective knockdowns inhibited proliferation, facilitated apoptosis, and restored Dox chemosensitivity. MiR-29a was shown to affect the lncRNA DUXAP8/PIK3CA network, and luciferase reporter gene assay showed direct binding between lncRNA DUXAP8 and miR-29a, as well as between miR-29a and PIK3CA. Targeting lncRNA DUXAP8/miR-29a/PIK3CA network synergized with inotuzumab ozogamicin’s effect on N6/ADR and 697/ADR cells.
Conclusion: Targeting the lncRNA DUXAP8/miR-29a/PIK3CA network not only induced an apoptotic effect on Dox-resistant B-ALL and restored Dox chemosensitivity via PI3K-AKT-mTOR signaling but also showed synergism with inotuzumab ozogamicin treatment.
Introduction
Acute lymphoblastic leukemia (ALL), as a hematological malignancy with the unrestricted proliferation of abnormal, immature lymphocytes or their progenitors that cause bone marrow element dysregulation, affects 1.7 new cases per 100,000 populations annually and accounts for nearly 2% of all lymphoid neoplasms in the United States (1, 2). ALL mainly occurs in children but less in adults, with the predominant type of B-cell ALL (B-ALL), which accounts for approximately 85% of total ALL cases (1, 2). Along with the improvement of treatment strategies, precision medicine, drug innovations, and so on, the prognosis of ALL is much improved in recent decades (3–6). However, long-term survival benefits are only achieved in the majority of childhood ALL, while only 30%–40% of adult ALL achieves long-term disease-free survival; especially, after relapse or refractoriness, the role of salvage chemotherapy is very limited, with less than 10% of long survival (7, 8). Therefore, the efforts to uncover underlying pathogenesis then discover novel treatment targets for ALL are never ceased.
Long non-coding RNA (lncRNA), a kind of non-coding RNA recognized recently with a length of more than 200 bp, has been reported to regulate a huge amount of biological processes and to be involved in the pathogenesis of most diseases via its interaction with cellular compounds such as miRNA, mRNA, DNA, and proteins (9–13). Besides, lncRNA has been also observed to be deeply involved in the development, progression, and treatment response of hematological malignancies (14–16). In the aspect of B-ALL, a recent comprehensive bioinformatics analysis identifies 1,235 dysregulated lncRNAs engaged in B-ALL subtype classification, and 942 aberrant lncRNAs related to B-ALL relapse (17). In addition, some specific lncRNAs were identified to regulate B-ALL growth and apoptosis, including lncRNA TEX41, lncRNA CRNDE, and lncRNA TCL6 (18–20). However, the function of a large majority of lncRNAs in B-ALL remains obscure, and in particular, seldom studies have investigated the comprehensive lncRNA profile involved in treatment refractoriness, not to mention the detailed molecule mechanism of some key lncRNAs underlying chemotherapy resistance.
Therefore, the current study aimed to investigate lncRNA, miRNA, and mRNA expression profiles related to B-ALL chemotherapy resistance via RNA sequencing, then further to explore the effect and interaction of the lncRNA DUXAP8/miR-29a/PIK3CA network on drug-resistant B-ALL cell proliferation, apoptosis, and chemosensitivity as well as its synergy with inotuzumab ozogamicin.
Methods
Samples
A total of 12 diagnostic bone marrow samples were collected from adult patients with B-ALL treated at our hospital. All bone marrows were sampled before the initiation of induction therapy. Of 12 bone marrow samples, 6 samples were from the adult B-ALL patients who achieved complete remission (CR) following the induction therapy with the VDP (vincristine, doxorubicin, prednisone) regimen, and these patients were considered as chemotherapy-sensitive patients (S-ALL) in the study; then, another 6 samples were from the adult B-ALL patients failing to achieve response at the end of the induction therapy with the VDP regimen, who exhibited a minimal residual disease (MRD) >1 × 10-2 by flow cytometry at the end of the induction therapy, and these patients were considered as chemotherapy-resistant patients (R-ALL) in the study.
The inclusion criteria of patients were as follows: (1) newly diagnosed as B-ALL by ESMO criteria (21); (2) older than 18 years; (3) received VDP regimen induction treatment; (4) had available induction treatment response data; and (5) had accessible bone marrow samples before treatment. The exclusion criteria of patients were as follows: (1) other types of ALL; (2) complicated with or history of other hematological malignancies or solid tumors; and (3) history of chemotherapy. This study was approved by the Ethics Committee of our hospital. The written informed consents were offered by patients.
Workflow of RNA Sequencing Including lncRNA, mRNA, and MicroRNA
Total RNA in R-ALL and S-ALL bone marrow samples was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and quantified with Agilent 2100 (Agilent, Santa Clara, CA, USA). The ribosomal RNA was depleted using the Ribo-Zero rRNA Removal Kit (Illumina, San Diego, CA, USA), and the library of mRNA and lncRNA was constructed using the Illumina TruSeq Stranded Total RNA Library Prep Kit (Illumina, USA) according to the kit’s protocol. For the construction of the miRNA library, a miRNA library kit (Qiagen, Hilden, Germany) was applied. HiSeq 2500 (Illumina, USA) was applied to complete the sequencing of lncRNA, mRNA, and miRNA, respectively. After the acquisition of data, the R project (Version 3.6.3) was applied to complete quantile normalization, data processing, bioinformatics analysis, and graph plotting. Briefly, the Factoextra package and Pheatmap package were acquired to perform principal component analysis (PCA) and heatmap analysis, respectively; the DESeq2 and Limma package was adopted to analyze the mean gene expression, fold change (FC), and differentially expressed lncRNA, mRNA, or miRNA (DElncRNA, DEmRNA, or DEmiRNA). The DElncRNAs, DEmRNAs, and DEmiRNAs were shown with a volcano plot. Gene Ontology (GO) and Kyoko Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted via the DAVID web server. Pearson’s correlation coefficient was used to calculate the correlation between the expression of DElncRNAs and DEmRNAs. The potential target of DEmiRNAs was investigated by miRanda. The competing endogenous RNA (ceRNA) network was displayed using the graph in R packages, which contained the following ceRNAs: (1) top 10 deregulated (5 upregulated and 5 downregulated) DElncRNAs according to log2FC; (2) DEmRNAs, which have a significant correlation with the DElncRNAs mentioned in (1) (Pearson’s correlation coefficient >0.9 or <-0.9); and (3) DEmiRNAs, whose potential targets were DEmRNAs and DElncRNAs mentioned in (1) and (2). The expressions of the selected lncRNA, miRNA, and mRNA were evaluated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
RNA-sequencing raw data in the article are available on the public database “National Omics Data Encyclopedia (NODE) database (https://www.biosino.org/node)” with tracking number OEP003016. The link is https://www.biosino.org/node/project/detail/OEP003016.
Cell Culture
Human B-ALL cells Nalm-6 and 697 were obtained from DSMZ (Braunschweig, Germany). The doxorubicin (Dox)-resistant Nalm-6 (N6/ADR) cells were obtained from ATCC (Rockville, USA). The Dox-resistant 697 (697/ADR) cells were constructed according to a previous study (22). The Nalm-6 and 697 cells were cultured in RPMI-1640 medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (HyClone, USA). The N6/ADR and 697/ADR cells were cultured in 70 nM Dox (Selleck, Houston, UT, USA) containing RPMI-1640 medium supplemented with 10% FBS. The Dox resistance of cells was confirmed by chemosensitivity to Dox assay. The expressions of lncRNA DUXAP8, miR-29a, PIK3CA in Nalm-6, N6/ADR, 697, and 697/ADR cells were evaluated.
LncRNA DUXAP8 Plasmid Transfection
Control lncRNA overexpression plasmid, lncRNA DUXAP8 overexpression plasmid, control lncRNA short hairpin RNA (shRNA) plasmid, and lncRNA DUXAP8 shRNA plasmid were constructed by Shanghai GenePharma Co., Ltd (Shanghai, China), with pEX-2 vector (GenePharma, Shanghai, China) and pGPU6 vector (GenePharma, China), respectively. The plasmids were transfected into N6/ADR or 697/ADR cells in the presence of Lipofectamine™ 2000 (Invitrogen, USA). After transfection, the cells were divided into oeCTL, oeLNC, shCTL, and shLNC groups. The N6/ADR and 697/ADR cells that were not transfected served as blank control.
miR-29a Plasmid Transfection
Control miR shRNA and miR-29a shRNA plasmids were constructed with the pGCMV/miR/inhibit vector (GenePharma) by Shanghai GenePharma Co., Ltd (Shanghai, China). After the construction, the control miR shRNA plasmid, miR-29a shRNA plasmid, control lncRNA shRNA plasmid, and lncRNA DUXAP8 shRNA plasmid were co-transfected into N6/ADR and 697/ADR cells by applying Lipofectamine™ 2000, accordingly. The cells were then divided into 5 groups: (a) blank group, no transfection cells; (b) shCTL group, cells were transfected with control lncRNA shRNA plasmid and control miR shRNA plasmid; (c) shLNC group, cells were transfected with lncRNA DUXAP8 shRNA plasmid and control miR shRNA plasmid; (d) shMIR group, cells were transfected with control lncRNA shRNA plasmid and miR-29a shRNA plasmid; and (e) shLNC + shMIR group, cells were transfected with lncRNA DUXAP8 shRNA plasmid and miR-29a shRNA plasmid.
PIK3CA Plasmid Transfection
The pEX-2 vector and pGCMV/miR vector (GenePharma, China) were applied to construct the control mRNA overexpression, PIK3CA overexpression, control miR overexpression, and miR-29a overexpression plasmids by Shanghai GenePharma Co., Ltd. (Shanghai, China). To perform the co-transfection of control mRNA overexpression plasmid, PIK3CA overexpression plasmid, control miR overexpression plasmid, and miR-29a overexpression plasmid, Lipofectamine™ 2000 was adopted with the manufacturer’s instruction followed. The following groups were generated after transfection: (a) blank group, cells without transfection; (b) oeCTL group, cells were transfected with control mRNA overexpression plasmid and control miR overexpression plasmid; (c) oeMIR group, cells were transfected with miR-29a overexpression plasmid and control mRNA overexpression plasmid; (d) oePIK3CA group, cells were transfected with control miR overexpression plasmid and PIK3CA overexpression plasmid; and (e) oeMIR + oePIK3CA group, cells were transfected with miR-29a overexpression plasmid and PIK3CA overexpression plasmid.
PI3K-AKT-mTOR Pathway Activation
The PIK3CA overexpression plasmid or mTOR activator (MHY1485) (2 μM, MCE, China) was cocultured with lncRNA DUXAP8 knockdown plasmid in N6/ADR and 697/ADR cells, followed by the detection of cell viability via CCK-8 assay, to explore the role of the PI3K-AKT-mTOR pathway in the restored chemosensitivity by lncRNA DUXAP8 knockdown.
RT-qPCR
The TRIzol Reagent was adopted to extract total RNA. The concentration of total RNA was evaluated by NanoDrop 2000 (Thermo, USA). 1 μg total RNA was transcribed into cDNA using the RT Reagent Kit (Takara, Shiga, Japan) and subjected to the following thermal cycling: 37°C for 15 minutes (min) and 85°C for 5 seconds (sec). The qPCR mix (Takara, Japan) was used to complete qPCR. β-Actin was used as an internal reference for lncRNA and mRNA, and U6 served as an internal reference for miRNA. Primers were listed as follows: lncRNA DUXAP8, forward, 5′ ACCAGCCTCACTAGCACTCTC 3′, reverse, 5′ CTTCCAGCCTCAGCCTCCTAA 3′; PIK3CA, forward, 5′ TTCTCAACTGCCAATGGACTGT 3′, reverse, 5′ AGCACGAGGAAGATCAGGAATG 3′; β-actin, forward 5′ GGCACCACACCTTCTACAATGA 3′, reverse, 5′ TCTCCTTAATGTCACGCACGATT 3′; miR-29a, forward, 5′ ACACTCCAGCTGGGTAGCACCATCTGAAAT 3′, reverse, 5′ TGTCGTGGAGTCGGCAATTC 3′; U6, forward, 5′ GCTCGCTTCGGCAGCACATA3′, reverse, 5′ AATATGGAACGCTTCACGAATTTGC 3′.
Western Blot
The cells were lysed with RIPA buffer (Beyotime, Shanghai, China). The BCA protein assay kit (Beyotime, China) was used to determine the protein concentration. After thermal denaturation, 20 μg protein was separated with 4%–20% precast gel (Beyotime, China). The protein was then transferred to a nitrocellulose membrane (PALL, Port Washington, NY, USA). After being blocked, the membrane was incubated with diluted primary antibodies and secondary antibodies, successively. Finally, the protein bands were visualized with the ECL Kit (Beyotime, China). The antibodies’ information was as follows: PIK3CA monoclonal antibody (1: 2,000) (Invitrogen, USA); Bcl-2 monoclonal antibody (1: 500) (Invitrogen, USA); cleaved caspase 3 monoclonal antibody (1: 1,000) (CST, UK); AKT monoclonal antibody (1:1,000) (CST; UK); phospho-AKT (pAKT) monoclonal antibody (1:1,000) (CST, UK); mTOR monoclonal antibody (1:3,000) (Invitrogen, USA); phospho-mTOR (pmTOR) monoclonal antibody (1:1,000) (CST, UK); S6K monoclonal antibody (1:3,000) (CST, UK); phospho-S6K (pS6k) monoclonal antibody (1:1,000) (CST, UK); and anti-rabbit IgG, HRP-linked antibody (1: 10,000) (Invitrogen, USA).
Cell Proliferation and Cell Apoptosis
The Cell Counting Kit-8 (Beyotime, China) was applied to assess cell proliferation. 3 × 103 cells were loaded into a 96-well plate. 10 μl reagent was added and incubated with cells for 2 h after transfection. A microplate reader was used to read optical density (OD) values. The TUNEL Apoptosis Assay Kit (Beyotime, China) was used to assess the cell apoptosis rate according to the instructions, with 2.5 × 104 cells seeded into a 24-well plate.
Chemosensitivity to Dox Assay
For the confirmation of Dox resistance, un-transfected Nalm-6, N6/ADR, 697, and 697/ADR were seeded in a 96-well plate with the number of 1 × 104 and incubated with a range concentration (0, 5, 10, 20, 40, and 80 nM) of Dox for 24 h referring to a previous paper (23). For chemosensitivity detection of transfected cells, the cells were cultured with a range concentration (0, 5, 20, 80 nM) of Dox for 24 h. After the incubation, the cell viability was detected with Cell Counting Kit-8 as described in Cell Proliferation.
Luciferase Reporter Gene Assay
To detect the binding of lncRNA DUXAP8 and miR-29a, the wild type and mutant type of lncRNA DUXAP8 were cloned into the pGL6 vector (Beyotime, China). The wild-type and mutant-type lncRNA DUXAP8 plasmids, miR-29a mimic, and control mimic were co-transfected into 293T cells (ATCC, USA) by Lipofectamine™ 2000. After 48 h of incubation, the cells were lysed and the fluorescence value was detected using Luciferase Reporter Gene Assay Kit (Beyotime, China). To detect the binding of PIK3CA and miR-29a, the PIK3CA wild-type and mutant-type plasmids were constructed with a pGL6 vector. With the application of Lipofectamine™ 2000, the PIK3CA wild-type plasmid, PIK3CA mutant-type plasmid, miR-29a mimic, and control mimic were co-transfected into 293T cells. At 48 h, the fluorescence value was detected as mentioned above.
LncRNA DUXAP8 Modification and Inotuzumab Ozogamicin (CMC-544) Treatment
The control lncRNA overexpression plasmid, lncRNA DUXAP8 overexpression plasmid, control lncRNA shRNA plasmid, and lncRNA DUXAP8 shRNA plasmid were transfected into N6/ADR and 697/ADR cells according to the methods described in the LncRNA DUXAP8 Plasmid Transfection. The N6/ADR and 697/ADR cells that were not transfected served as blank control. Then, CMC-544 (Pfizer Inc., New York, NY, USA) was added into cells with various concentrations of 0, 0.5, 1, 2, 4, and 8 ng/ml (24). After incubation for 48 h, cell viability was evaluated with Cell Counting Kit-8, and the half-maximal inhibitory concentration (IC50) was calculated using Probit regression analysis by SPSS 23.0 (IBM, Armonk, NY, USA). At last, the transfected and blank control cells were incubated with CMC-544 at an amount of 2.670 or 2.794 ng/ml for 48 h, with the apoptosis rate determined by TUNEL Apoptosis Assay Kit. The cell viability and apoptosis detection were carried out as described in Cell Proliferation and Cell Apoptosis.
Statistical Analysis
All data in this study were expressed as mean ± standard deviation. The difference among groups and between two groups was assessed by one-way ANOVA followed by Dunnett’s test or t-test. GraphPad Prism 7.02 (GraphPad Software Inc., La Jolla, CA, USA) was applied to analyze data and make graphs. The statistical significance was defined as p < 0.05. No significance: NS (p>0.05); *p<0.05, **p<0.01, ***p<0.001.
Results
Study Flow
The current study consisted of two main parts: the clinical part and the experimental part (Supplementary Figure 1). In the clinical part, 6 R-ALL patients and S-ALL patients were enrolled to detect the lncRNA, miRNA, and mRNA profiles via RNA-seq and RT-qPCR validation to sort key networks related to chemoresistance. Then, experiments were performed via modifying lncRNA DUXAP8, miR-29a, and PIK3CA to explore their effect (alone or in combination) on Dox-resistant B-ALL cell proliferation, apoptosis, Dox chemosensitivity, etc.
Characteristics
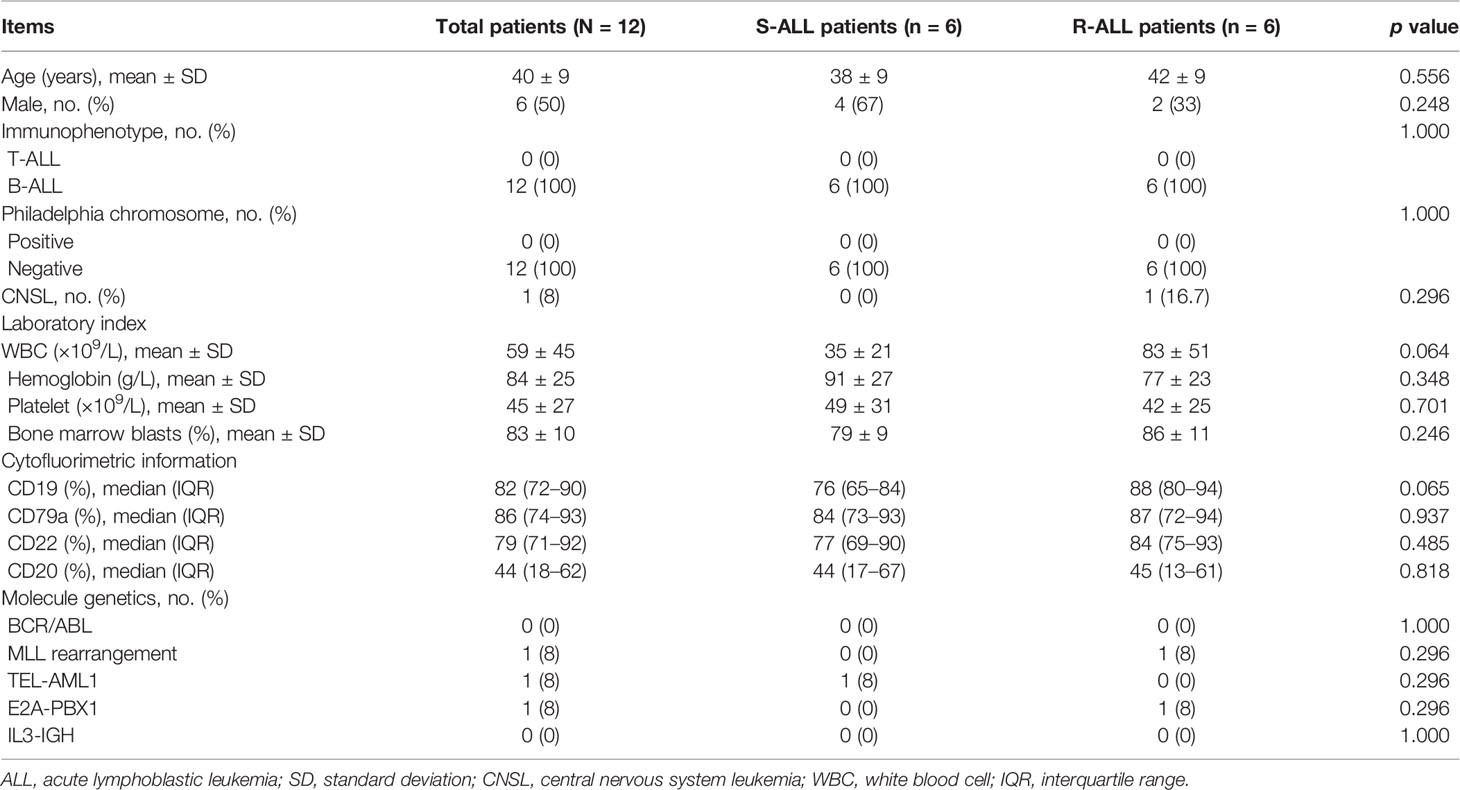
The enrolled B-ALL patients were aged 40 ± 9 years, with 50% males and 50% females. Meanwhile, the mean WBC was 59 ± 45 × 109/l, and the mean bone marrow blast percentage was (83 ± 10)%. The detailed information of their characteristics is shown in Table 1. It was notable that by comparison, S-ALL patients showed a trend of lower WBC (p = 0.064) and CD19 percentage (p = 0.065) compared to R-ALL patients but lacked statistical significance. Furthermore, other characteristics were of no difference between them (Table 1).
LncRNA, miRNA, and mRNA Expression Profiles
The lncRNA expression profile could clearly distinguish R-ALL patients from S-ALL patients via PCA plot analysis (Figure 1A) and showed a good inter-consistent trend in R-ALL patients and S-ALL patients, respectively, by heatmap analysis (Figure 1B). Then, a total of 759 upregulated and 579 downregulated lncRNAs were identified in R-ALL patients compared with S-ALL patients (Figure 1C).
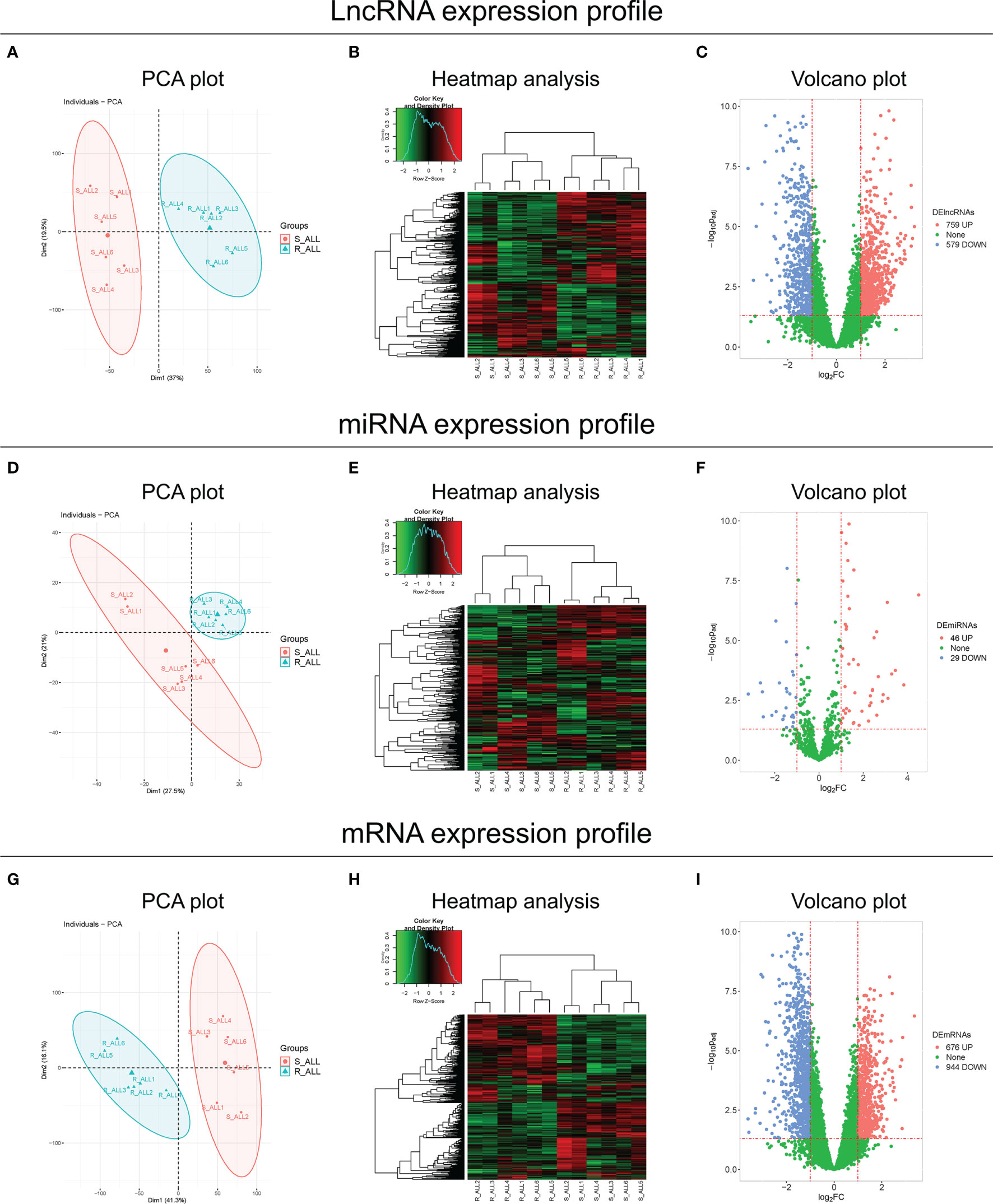
Figure 1 PCA, heatmap, and volcano analyses of RNA sequencing. PCA plot and heatmap analyses of the lncRNA expression profile, and volcano analysis of dysregulated lncRNAs between R-ALL and S-ALL patients (A–C). PCA plot and heatmap analyses of the miRNA expression profile, and volcano analysis of dysregulated miRNAs between R-ALL and S-ALL patients (D–F). PCA plot and heatmap analyses of the mRNA expression profile, and volcano analysis of dysregulated mRNAs between R-ALL and S-ALL patients (G–I). The data were from the comparison of diagnostic B-ALL samples.
Besides, the miRNA expression profile also distinguished R-ALL patients from S-ALL patients via PCA plot analysis (Figure 1D) and exhibited an acceptable inter-consistent trend by heatmap analysis (Figure 1E), with 46 upregulated/29 downregulated miRNAs discovered in R-ALL patients compared to S-ALL patients (Figure 1F). Furthermore, the mRNA expression profile was also able to differentiate R-ALL patients from S-ALL patients (Figure 1G), and disclosed an inter-consistent trend by heatmap analysis (Figure 1H), with 676 upregulated/944 downregulated mRNAs uncovered in R-ALL patients compared to S-ALL patients (Figure 1I). In addition, the detailed information of each dysregulated lncRNA, miRNA, and mRNA is exhibited in Supplementary Tables 1–3, respectively.
Identification of lncRNA DUXAP8/miR-29a/PIK3CA Network and Downstream PI3K-AKT Pathway
KEGG enrichment analyses of dysregulated lncRNA (Figure 2A), miRNA (Figure 2B), and mRNA (Figure 2C) expression profiles were carried out, which observed that they were all closely related to the PI3K-AKT pathway. Inspiringly, a comprehensive ceRNA network was established based on dysregulated lncRNA, miRNA, and mRNA expression profiles, which discovered that the lncRNA DUXAP8/miR-29a/PIK3CA network was a key component (Figure 2D). What is more, PIK3CA is the key component involved in the PI3K-AKT pathway, which is closely engaged in ALL pathogenesis and treatment sensitivity (25–27). Subsequently, RT-qPCR was performed to verify their expressions, which also demonstrated that lncRNA DUXAP8 was upregulated (Figure 2E), miR-29a was downregulated (Figure 2F), and PIK3CA was overexpressed (Figure 2G) in R-ALL patients compared to S-ALL patients (all p < 0.05). Based on the abovementioned information, the lncRNA DUXAP8/miR-29a/PIK3CA network and downstream PI3K-AKT pathway were essential in chemotherapy resistance, then subsequent experiments were performed to explore and validate this issue.
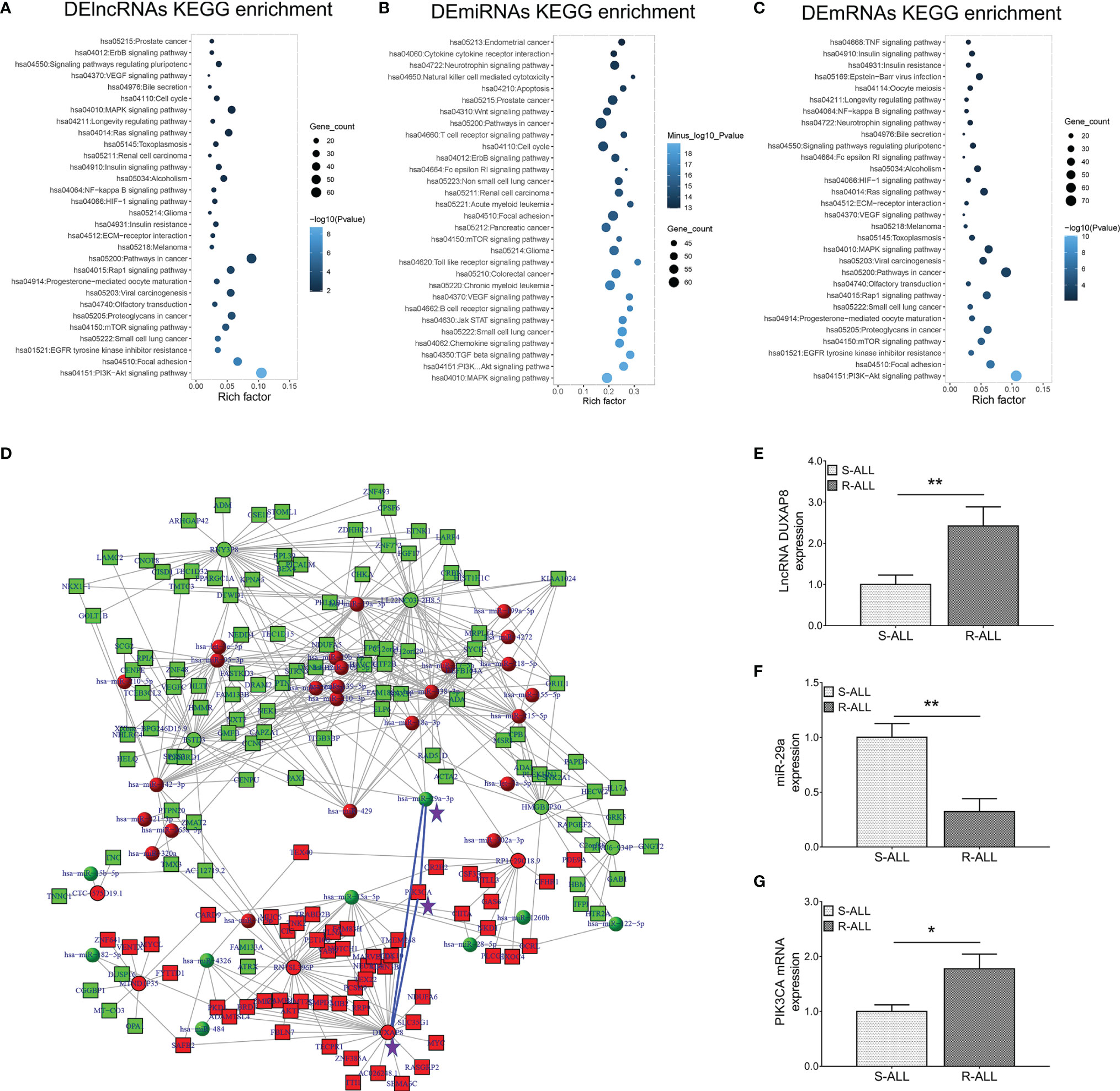
Figure 2 lncRNA–miRNA–mRNA co-expression network and RT-qPCR validation. KEGG (A) enrichment analysis according to DElncRNAs; KEGG (B) enrichment analysis according to DEmiRNAs; KEGG (C) enrichment analysis according to DEmRNAs. LncRNA–miRNA–mRNA co-expression network based on dysregulated genes between R-ALL and S-ALL patients (D). RT-qPCR detection data of lncRNA DUXAP8 (E), miR-29a (F), and PIK3CA (G) expressions between R-ALL and S-ALL patients.
Effect of lncRNA DUXAP8 Modification on Dox-Resistant B-ALL Cells
Firstly, Dox resistance was confirmed in N6/ADR and 697/ADR cells (Supplementary Figures 2A, B), then upregulated lncRNA DUXAP8 and PIK3CA and downregulated miR-29a were observed in N6/ADR and 697/ADR cells (all p < 0.05, Supplementary Figures 2C, D).
After confirmation of transfection efficiency (all p < 0.01, Figures 3A, B), lncRNA DUXAP8 overexpression was observed to promote cell proliferation in N6/ADR (Figure 3C) and 697/ADR (Figure 3D) cells (both p < 0.05). Then, lncRNA DUXAP8 overexpression increased Bcl-2 expression but repressed cleaved caspase-3 expression in N6/ADR (Figures 3E, G) and 697/ADR (Figures 3F, H) cells (all p < 0.05). In addition, lncRNA DUXAP8 overexpression reduced the cell apoptosis rate reflected by TUNEL-positive cell percentage in N6/ADR (Figures 3I, K) and 697/ADR (Figures 3J, L) cells (both p < 0.05). Besides, lncRNA DUXAP8 overexpression decreased Dox chemosensitivity in N6/ADR cells to some extent (Figure 3M) (p < 0.05) but did not affect Dox chemosensitivity in 697/ADR cells (Figure 3N) (p > 0.05). Inversely, lncRNA DUXAP8 knockdown exhibited the opposite effect to its overexpression in regulating the abovementioned cell functions; in particular, lncRNA DUXAP8 knockdown restored Dox chemosensitivity to some extent in N6/ADR (Figure 3M) and 697/ADR (Figure 3N) cells (all p < 0.05).
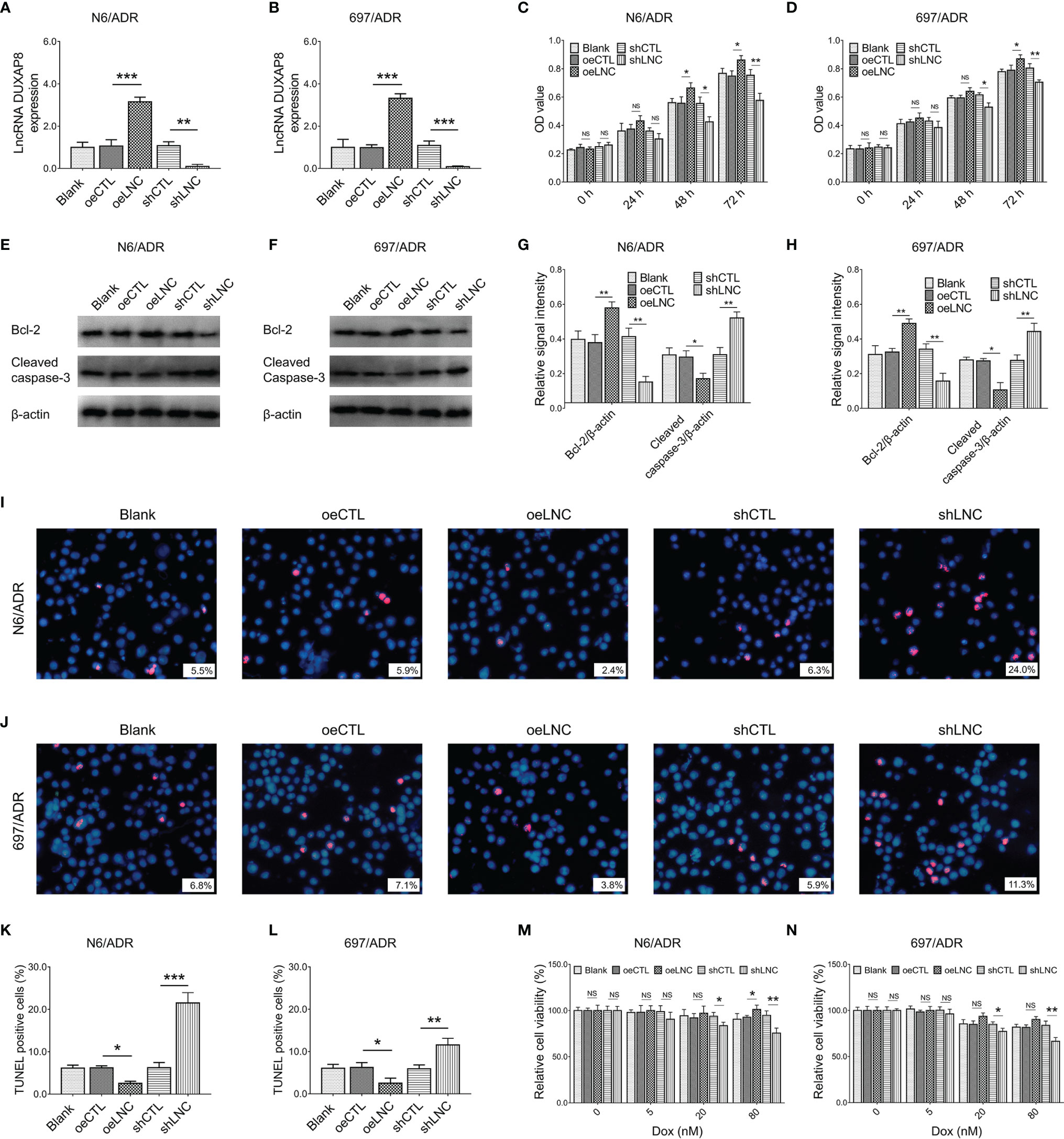
Figure 3 Cell behaviors after lncRNA DUXAP8 modification. LncRNA DUXAP8 (A, B), cell proliferation by CCK-8 (C, D), apoptotic marker expressions (E–H), cell apoptosis rate (I–L), and chemosensitivity to Dox (M, N) among blank, oeCTL, oeLNC, shCTL, and shLNC groups in N6/ADR cells and 697/ADR cells.
Interaction Between lncRNA DUXAP8 and miR-29a
It was then observed that lncRNA DUXAP8 negatively regulated miR-29a expression in both N6/ADR (Figure 4A) and 697/ADR (Figure 4B) cells (all p < 0.05). By the Luciferase reporter gene assay with the designed binding sequence shown in Figure 4C, lncRNA DUXAP8 was found to be directly bound to miR-29a to regulate its expression (p < 0.01, Figure 4D).
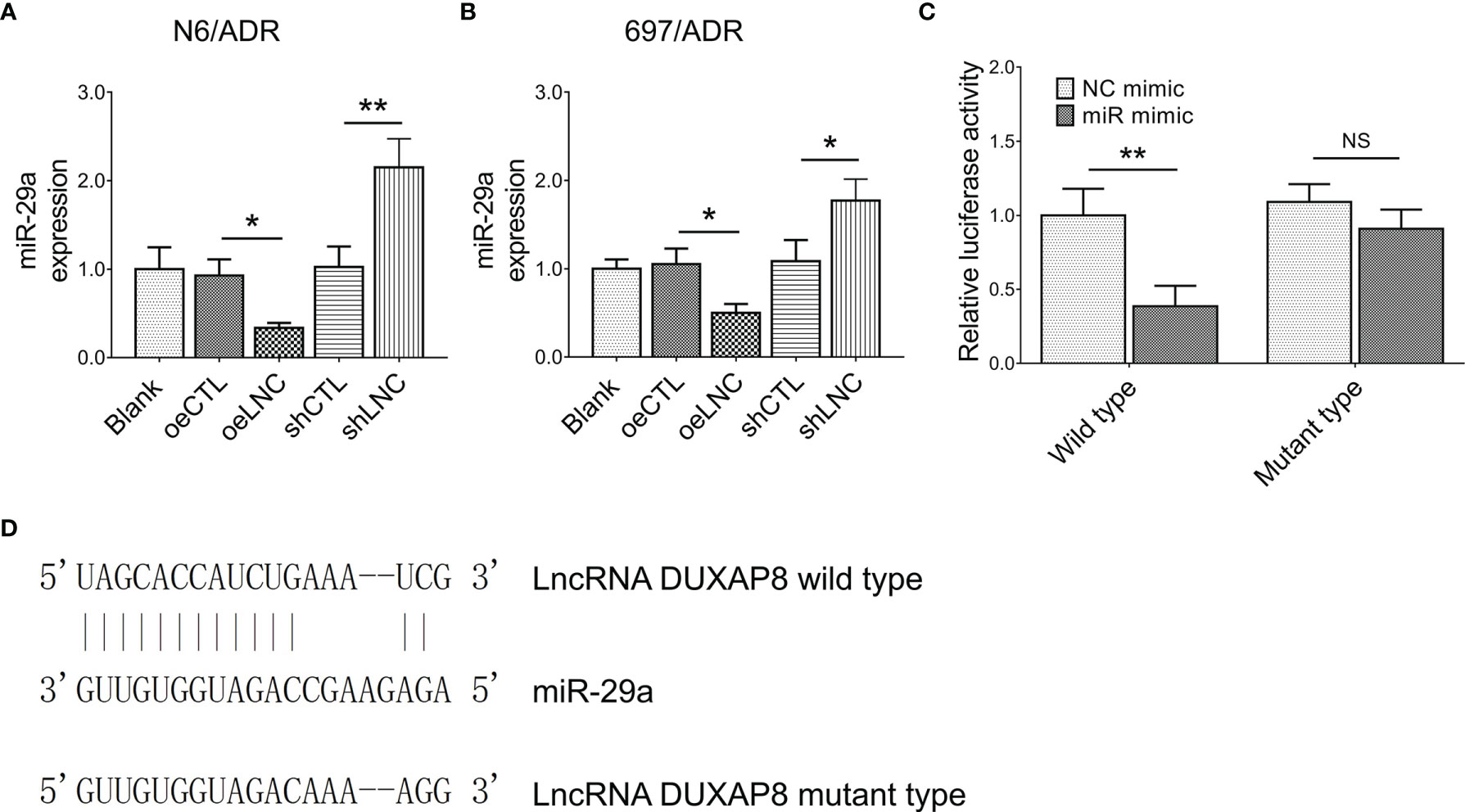
Figure 4 Interaction between lncRNA DUXAP8 and miR-29a. MiR-29a expression among blank, oeCTL, oeLNC, shCTL, and shLNC groups in N6/ADR cells and 697/ADR cells (A, B). Luciferase reporter gene assay results (C) and binding site between lncRNA DUXAP8 and miR-29a (D).
Effect of Modification of lncRNA DUXAP8 and miR-29a on Dox-Resistant B-ALL Cells
MiR-29a knockdown did not affect lncRNA DUXAP8 expression in both N6/ADR (Figure 5A) and 697/ADR (Figure 5B) cells (both p > 0.05), while lncRNA DUXAP8 knockdown increased miR-29a expression in both N6/ADR (Figure 5C) and 697/ADR (Figure 5D) cells (both p < 0.01). Subsequent experiments revealed that miR-29a knockdown enhanced cell proliferation in N6/ADR (Figure 5E) and 697/ADR (Figure 5F) cells (both p < 0.05); it also suppressed the cell apoptosis rate reflected by the TUNEL-positive cell percentage in N6/ADR (Figures 5G, I) and 697/ADR (Figures 5H, J) cells (both p < 0.05); then, it promoted Bcl-2 expression but reduced cleaved caspase-3 expression in N6/ADR (Figures 5K, M) and 697/ADR (Figures 5L, N) cells (all p < 0.05). Besides, miR-29a knockdown decreased Dox chemosensitivity in N6/ADR (Figure 5O) and 697/ADR (Figure 5P) cells (both p < 0.05).
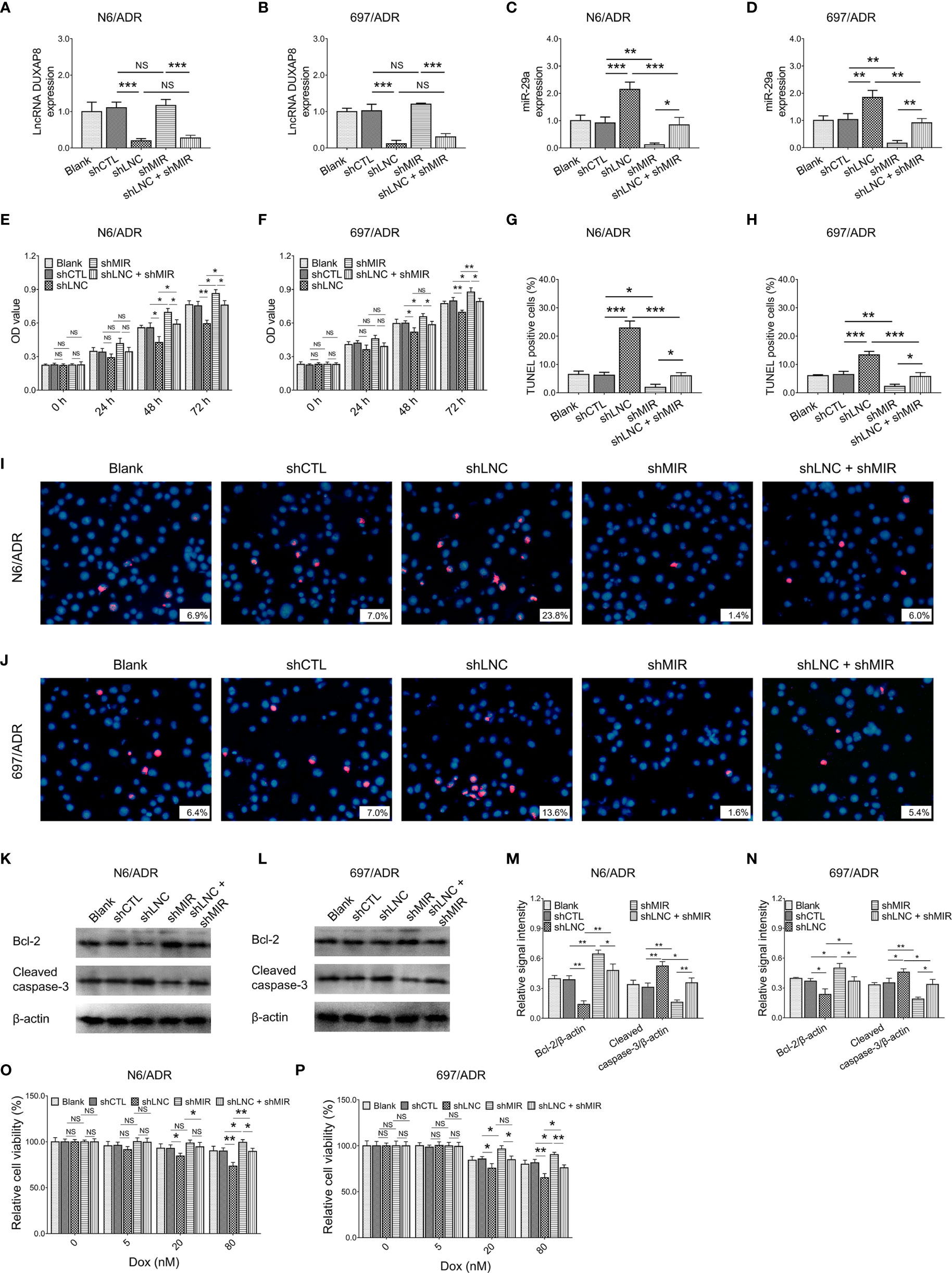
Figure 5 Cell behaviors after lncRNA DUXAP8 and miR-29a modification. LncRNA DUXAP8 (A, B), miR-29a (C, D), cell proliferation by CCK-8 (E, F), cell apoptosis rate (G–J), apoptotic marker expressions (K–N), and chemosensitivity to Dox (O, P) among blank, shCTL, shLNC, shMIR, and shLNC +shMIR groups in N6/ADR cells and 697/ADR cells.
Most notably, miR-29a knockdown compensated for the effects of lncRNA DUXAP8 knockdown on cell proliferation (Figure 5E), cell apoptosis rate reflected by TUNEL-positive cell percentage (Figures 5G, I), apoptotic markers (Figures 5K, M), and Dox chemosensitivity (Figure 5O) in N6/ADR cells (all p < 0.05). Similarly, miR-29a knockdown also compensated for the effects of lncRNA DUXAP8 knockdown on cell proliferation (Figure 5F), cell apoptosis rate reflected by TUNEL-positive cell percentage (Figures 5H, J), apoptotic markers (Figures 5L, N), and Dox chemosensitivity (Figure 5P) in 697/ADR cells (all p < 0.05).
Interaction Among lncRNA DUXAP8, miR-29a, and PIK3CA
LncRNA DUXAP8 knockdown decreased the PIK3CA gene expression in N6/ADR (Figure 6A) and 697/ADR (Figure 6B) cells (both p < 0.05); it also repressed PIK3CA protein expression in N6/ADR (Figures 6C, E) and 697/ADR (Figures 6D, F) cells (both p < 0.05), whereas miR-29a knockdown increased PIK3CA gene expression in N6/ADR (Figure 6A) and 697/ADR (Figure 6B) cells (both p < 0.001), as well as elevated PIK3CA protein expression in N6/ADR (Figures 6C, E) and 697/ADR (Figures 6D, F) cells (both p < 0.01). Subsequent Luciferase reporter gene assay observed that miR-29a was directly bound to PIK3CA to regulate its expression (p < 0.01, Figure 6G), with the designed binding sequence shown in Figure 6H.
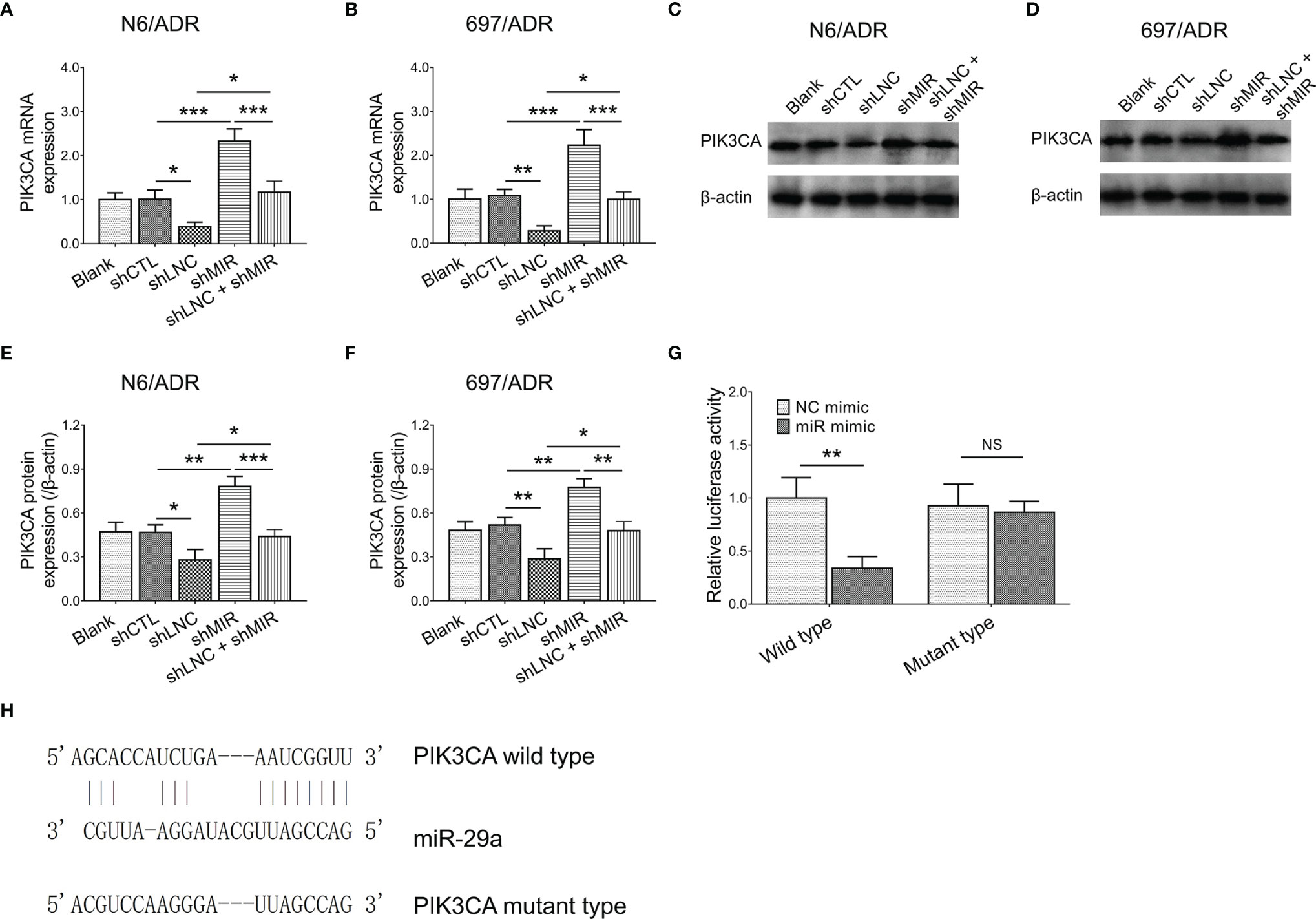
Figure 6 Interaction among lncRNA DUXAP8, miR-29a, and PIK3CA. PIK3CA expression among blank, shCTL, shLNC, shMIR, and shLNC +shMIR groups in N6/ADR cells and 697/ADR cells (A–F). Luciferase reporter gene assay results (G) and binding site between miR-29a and PIK3CA (H).
Effect of Modification of miR-29a and PIK3CA on PI3K-AKT-mTOR Pathway
PIK3CA overexpression did not affect miR-29a expression in N6/ADR (Figure 7A) and 697/ADR (Figure 7B) cells (both p > 0.05), while miR-29a overexpression inhibited PIK3CA expression in N6/ADR (Figure 7C) and 697/ADR (Figure 7D) cells (both p < 0.05). It was then noted that miR-29a overexpression inactivated the PI3K-AKT-mTOR pathway in N6/ADR (Figures 7E, G) and 697/ADR (Figures 7F, H) cells (all p < 0.05), while PIK3CA overexpression activated the PI3K-AKT-mTOR pathway in N6/ADR (Figures 7E, G) and 697/ADR (Figures 7F, H) cells (all p < 0.05).
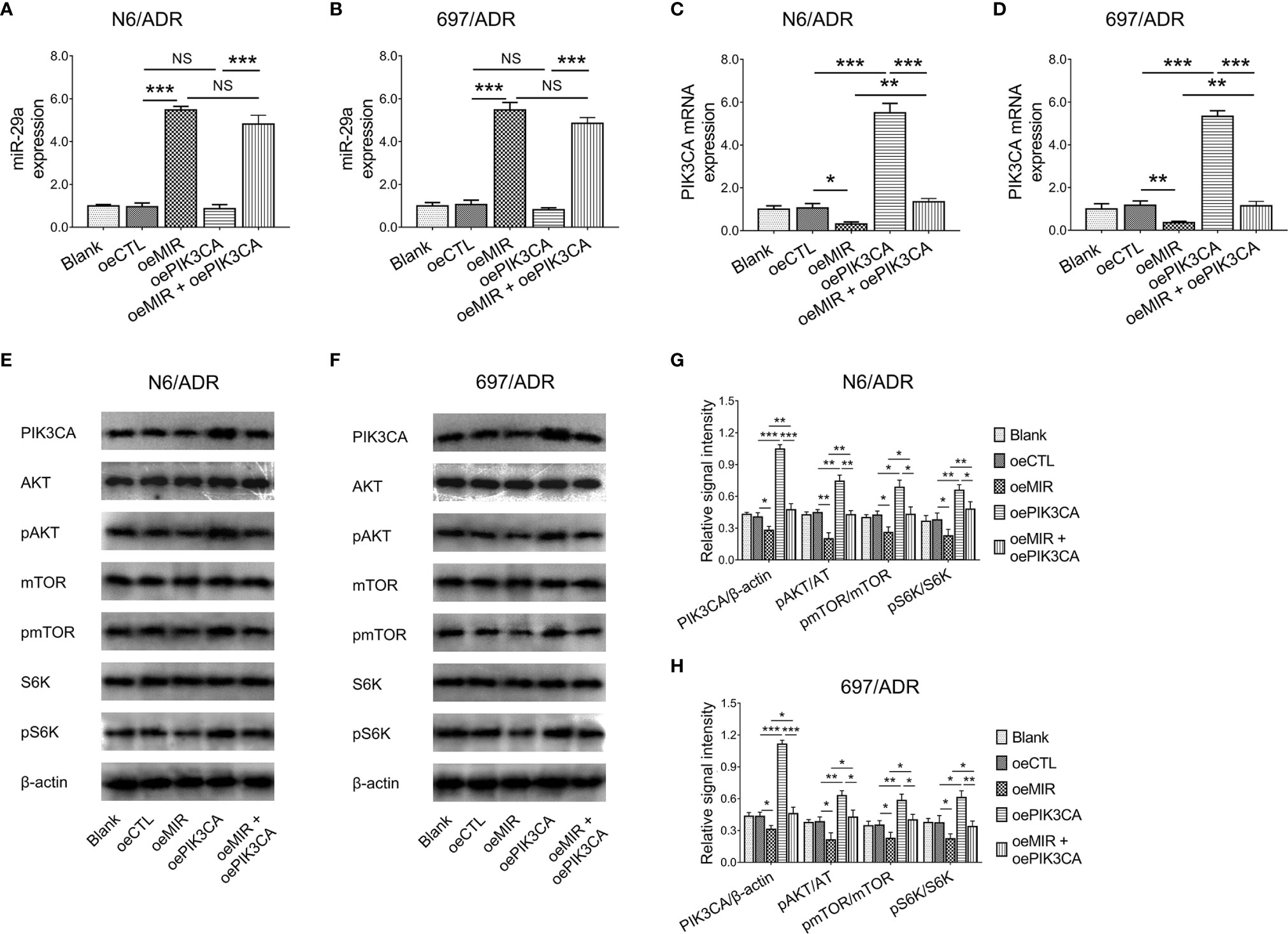
Figure 7 Interaction among miR-29a, PIK3CA, and PI3K-AKT-mTOR signaling. MiR-29a (A, B) and PIK3CA (C, D) expressions among blank, oeCTL, oeMIR, oePIK3CA, and oeMIR + oePIK3CA groups in N6/ADR cells and 697/ADR cells. PI3K-AKT-mTOR signaling among blank, oeCTL, oeMIR, oePIK3CA, and oeMIR + oePIK3CA groups in N6/ADR cells and 697/ADR cells (E–H).
Effect of Modification of miR-29a and PIK3CA on Dox-Resistant B-ALL Cells
MiR-29a overexpression decreased cell proliferation in N6/ADR (Figure 8A) and 697/ADR (Figure 8B) cells (both p < 0.05), while it increased the cell apoptosis rate reflected by the TUNEL-positive cell percentage in N6/ADR (Figures 8C, E) and 697/ADR (Figures 8D, F) cells (both p < 0.01). MiR-29a overexpression also inhibited Bcl-2 expression but promoted cleaved caspase-3 expression in N6/ADR (Figures 8G, I) and 697/ADR (Figures 8H, J) cells (all p < 0.05). Besides, miR-29a overexpression increased Dox chemosensitivity in N6/ADR (Figure 8K) and 697/ADR (Figure 8L) cells (both p < 0.05). However, PIK3CA overexpression showed the opposite effect as that of miR-29a overexpression in N6/ADR and 697/ADR cells (all p < 0.05).
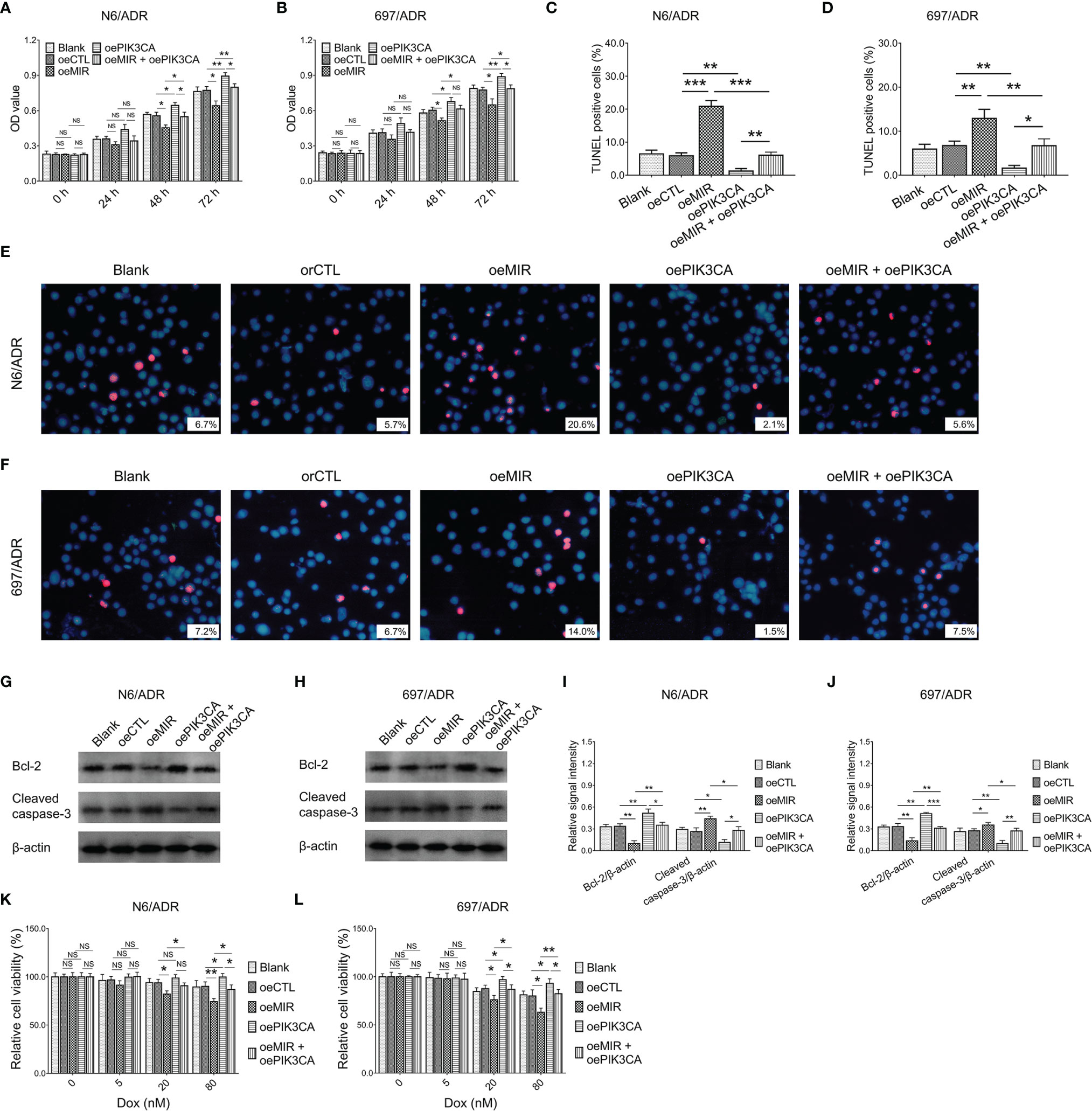
Figure 8 Cell behaviors after miR-29a and PIK3CA modification. Cell proliferation by CCK-8 (A, B), cell apoptosis rate (C–F), apoptotic marker expressions (G–J), and chemosensitivity to Dox (K, L) among blank, oeCTL, oeMIR, oePIK3CA, and oeMIR + oePIK3CA groups in N6/ADR cells and 697/ADR cells.
It was inspiring that PIK3CA overexpression compensated for the effects of miR-29a overexpression on cell proliferation (Figure 8A), cell apoptosis rate reflected by the TUNEL-positive cell percentage (Figures 8C, E), apoptotic markers (Figures 8G, I), and Dox chemosensitivity (Figure 8K) in N6/ADR cells (all p < 0.05). Similarly, PIK3CA overexpression also compensated for the effects of miR-29a overexpression on cell proliferation (Figure 8B), cell apoptosis rate reflected by the TUNEL-positive cell percentage (Figures 8D, F), apoptotic markers (Figures 8H, J), and Dox chemosensitivity (Figure 8L) in 697/ADR cells (all p < 0.05).
In addition, PIK3CA overexpression plasmid or mTOR activator (MHY1485) was cocultured with lncRNA DUXAP8 knockdown plasmid to explore the role of the PI3K-AKT-mTOR pathway in the restored chemosensitivity by lncRNA DUXAP8 knockdown. Then, it was observed that PI3K-AKT-mTOR pathway activation decreased Dox chemosensitivity in N6/ADR (Supplementary Figure 3A) and 697/ADR cells (Supplementary Figure 3B) and also attenuated the effect of lncRNA DUXAP8 knockdown.
LncRNA DUXAP8 Modification and Inotuzumab Ozogamicin on Dox-Resistant B-ALL Cells
Inotuzumab ozogamicin is a novel drug recommended for the treatment of refractory B-ALL, so we further explored whether modification of lncRNA DUXAP8 affected inotuzumab ozogamicin treatment in Dox-resistant B-ALL cells. It was observed that lncRNA DUXAP8 overexpression increased the IC50 of inotuzumab ozogamicin in N6/ADR cells (Figure 9A) (p < 0.05), but not in 697/ADR cells (Figure 9B) (p > 0.05); it also decreased the cell apoptosis rate reflected by the TUNEL-positive cell percentage in inotuzumab ozogamicin-treated N6/ADR cells (Figures 9C, E) (p < 0.05), but not in 697/ADR cells (Figures 9D, F) (p > 0.05). Besides, lncRNA DUXAP8 knockdown decreased the IC50 of inotuzumab ozogamicin in N6/ADR cells (Figure 9A) and 697/ADR cells (Figure 9B) (both p < 0.05), which also enhanced the cell apoptosis rate reflected by the TUNEL-positive cell percentage in inotuzumab ozogamicin-treated N6/ADR cells (Figures 9C, E) and 697/ADR cells (Figures 9D, F) (both p < 0.05).
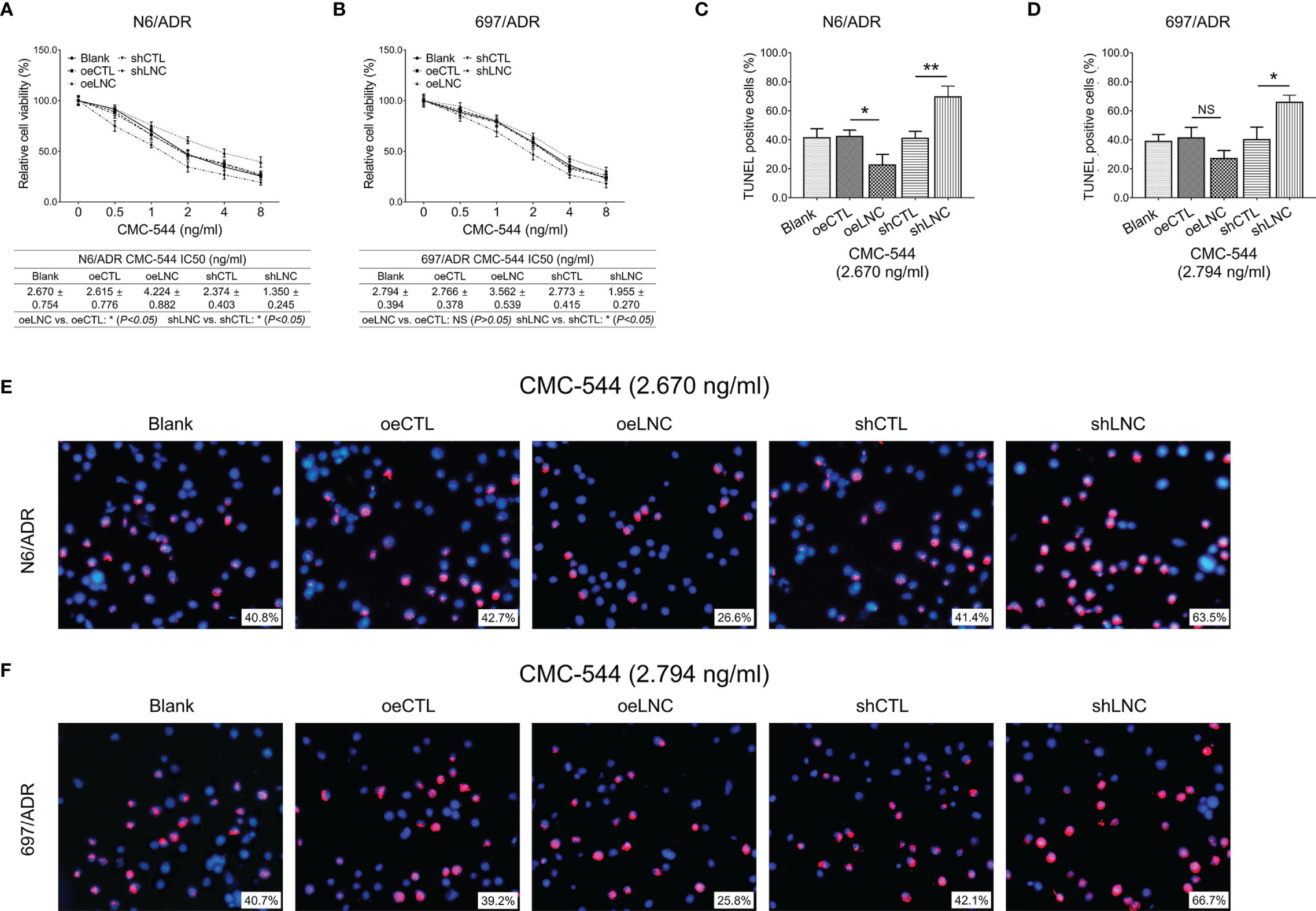
Figure 9 Cell behaviors after lncRNA DUXAP8 modification and inotuzumab ozogamicin treatment. Relative cell viability (A, B) and cell apoptosis rate (C–F) among blank, oeCTL, oeLNC, shCTL, and shLNC groups in inotuzumab ozogamicin treated N6/ADR cells and 697/ADR cells.
Discussion
A previous secondary bioinformatics analysis involving The Cancer Genome Atlas (TCGA) data identifies 469 upregulated and 286 downregulated lncRNAs in ALL patients compared to controls (28). Another study analyzes the GSE67684 dataset from the Gene Expression Omnibus then validates the candidate lncRNAs by RT-qPCR, which observes 21 dysregulated lncRNAs in accordance with ALL patients compared to controls (29). However, no study has explored the aberrant pattern of the lncRNA profile between chemotherapy-resistant B-ALL patients and chemotherapy-sensitive patients. Our current study filled this gap, which observed that 1,338 lncRNAs, 75 miRNAs, and 1,620 mRNAs were dysregulated between chemotherapy-resistant B-ALL patients and chemotherapy-sensitive B-ALL patients. These data would propose new information on the interaction between genetics and chemotherapy resistance in B-ALL.
Since the concept of lncRNA is just initiated in a short time, in-depth studies of its mechanisms underlying ALL are very limited, and only a few studies have reported several specific lncRNAs that are involved in ALL pathogenesis, such as lncRNA MALAT1, lncRNA VPS9D1-AS1, and lncRNA TEX41 (18, 30, 31). However, few studies on lncRNA have been disclosed in the context of B-ALL chemotherapy resistance. LncRNA DUXAP8 is previously reported as an oncogene in several cancers. For instance, lncRNA DUXAP8 promotes neuroblastoma progression by regulating miR-29 mediated NOL4L and downstream Wnt/β-actin pathway (32); it also enhances colorectal cancer proliferation, migration, and invasion by regulating EZH2 and LSD1 (33). In terms of leukemia, only a related paper is available, which observes that lncRNA DUXAP8 regulates acute myeloid leukemia glycolysis and apoptosis through the Wnt/β-actin pathway (34). Furthermore, lncRNA DUXAP8 is also revealed as a regulator of an anticancer agent, such as a PARP inhibitor (35). However, no data in the aspect of lncRNA DUXAP8 in ALL are reported. In our present study, further bioinformatic analyses and RT-qPCR validation observed that lncRNA DUXAP8 was closely related to chemotherapy resistance in B-ALL patients; subsequent experiments also revealed that lncRNA DUXAP8 promotes cell proliferation and inhibits cell apoptosis, then targeting it restored the Dox chemosensitivity in chemoresistant B-ALL cell lines. The possible explanations were as follows: (1) lncRNA DUXAP8 regulated the miR-29a/PIK3CA network and downstream PI3K-AKT pathway, as shown by subsequent molecule mechanism experiments, to modify cell proliferation, apoptosis, and Dox chemosensitivity in B-ALL, and (2) lncRNA DUXAP8 activated several key carcinogenetic pathways such as Wnt/β-actin, AKT/mTOR, and WTAP/Fak, to regulate these cell functions (34, 36, 37).
MiR-29a is a well-known anti-oncogene in both solid tumor and hematological malignancies (38, 39). In leukemias, MiR-29a deficiency activates CD40 signaling/T-cell interaction to engage in chronic lymphocytic leukemia pathogenesis (40); meanwhile, it is greatly dysregulated in acute myeloid leukemia, chronic lymphocytic leukemia, and ALL patients (41–43). In the aspect of PI3K-AKT, it is a documented pathway involved in ALL development, progression, and even treatment sensitivity (25, 44, 45). In our present study, we observed that the lncRNA DUXAP8/miR-29a/PIK3CA network and downstream PI3K-AKT pathway were closely related to chemotherapy resistance in B-ALL patients. Then, comprehensive experiments were performed to demonstrate that the lncRNA DUXAP8/miR-29a/PIK3CA network regulates cell proliferation and apoptosis and targeting the network could restore the Dox chemosensitivity in chemoresistant B-ALL cell lines. The most possible explanation was further excavated, which observed that the effect of the lncRNA DUXAP8/miR-29a/PIK3CA network in chemoresistant B-ALL resulted from activation of downstream PI3K-AKT-mTOR signaling. This finding provided a novel target network for chemoresistant B-ALL, which was exciting.
For chemotherapy-refractory or relapsed B-ALL patients, salvage treatment options are lacking and urgently need investigation. Currently, inotuzumab ozogamicin, an anti-CD22 antibody–drug conjugate, achieves encouraging efficacy and acceptable tolerance as a salvage treatment regimen in refractory or relapsed B-ALL patients (46–50). In the current study, it was further discovered that targeting lncRNA DUXAP8 and inotuzumab ozogamicin had synergistic effects in killing chemoresistant B-ALL cells. These would provide a new train of thought regarding refractory B-ALL treatment.
Despite the interesting findings initiated in our present study, some limitations existed as well: firstly, the validation of expression of the lncRNA DUXAP8/miR-29a/PIK3CA network could be further conducted in a larger sample size of B-ALL patients. Secondly, targeting lncRNA DUXAP8 restored the Dox chemosensitivity in Dox-resistant B-ALL cell lines via miR-29a/PIK3CA and subsequent PI3K-AKT-mTOR signaling, while whether other potential mechanisms beyond this could be further investigated. Thirdly, overall changes induced by modification in lncRNA DUXAP8 were relatively minor, then the synergistic effect of modifying lnc DUXAP8 on inotuzumab ozogamicin was relatively minor and observed only in one tested cell line, so further validation experiments were needed in the future.
In conclusion, targeting the lncRNA DUXAP8/miR-29a/PIK3CA network not only exhibits a good killing effect on Dox-resistant B-ALL and restores Dox chemosensitivity via PI3K-AKT-mTOR signaling but also synergizes with inotuzumab ozogamicin. These imply the effectiveness of this network as a treatment target for chemotherapy-refractory B-ALL.
Data Availability Statement
The original contributions presented in the study are publicly available. These data can be accessed using the following link/accession number: https://www.biosino.org/node/search; OEP003016.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of The Affiliated Hospital of Southwest Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LZ and JT conceived and designed the study. LZ, SZ, XL, and JT collected and analyzed the data. LZ, TZ, and JT prepared the figures. LZ, SZ, and XL wrote the manuscript. TZ and JT edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Sichuan Science and Technology Program (No. 2019YFS0301), Doctor Research Foundation of The Affiliated Hospital of Southwest Medical University (No. 19032; No. 19079), The Key Research Project from Health and Family Planning Commission of Sichuan Province (No. 18ZD014), and the Applied Basic Research Project of Luzhou Science and Technology Bureau (2019LZXNYDJ54;2020LZXNYDJ48 and 2020-JYJ-50).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.773601/full#supplementary-material
Supplementary Figure 1 | Study flow chart.
Supplementary Figure 2 | Expression of lncRNA DUXAP8, miR-29a and PIK3CA in N6/ADR and 697/ADR cells. Confirmation of the Dox resistance status of N6/ADR (A) and 697/ADR cells (B). Expression of lncRNA DUXAP8, miR-29a and PIK3CA in N6/ADR and 697/ADR cells compared to normal NALM6 and 697 cells, respectively (C, D).
Supplementary Figure 3 | Role of PI3K-AKT-mTOR pathway in this restored chemosensitivity. Relative cell viability among Blank, CTL, shLNC, oePIK3CA, shLNC+oePIK3CA, MHY1485, shLNC+MHY1485 groups in N6/ADR cells (A) and 697/ADR cells (B) under 0, 5, 20, 80 nM Dox treatment.
References
2. Baljevic M, Jabbour E, O’Brien S, Kantarjian HM. Acute Lymphoblastic Leukemia. In: Kantarjian HM, Wolff RA, editors. The MD Anderson Manual of Medical Oncology (3 ed.). New York: McGraw-Hill Education (2016).
3. Sasaki K, Jabbour E, Short NJ, Jain N, Ravandi F, Pui CH, et al. Acute Lymphoblastic Leukemia: A Population-Based Study of Outcome in the United States Based on the Surveillance, Epidemiology, and End Results (SEER) Database, 1980-2017. Am J Hematol (2021) 96(6):650–8. doi: 10.1002/ajh.26156
4. Pui CH. Precision Medicine in Acute Lymphoblastic Leukemia. Front Med (2020) 14(6):689–700. doi: 10.1007/s11684-020-0759-8
5. Papadantonakis N, Advani AS. Recent Advances and Novel Treatment Paradigms in Acute Lymphocytic Leukemia. Ther Adv Hematol (2016) 7(5):252–69. doi: 10.1177/2040620716652289
6. Man LM, Morris AL, Keng M. New Therapeutic Strategies in Acute Lymphocytic Leukemia. Curr Hematol Malig Rep (2017) 12(3):197–206. doi: 10.1007/s11899-017-0380-3
7. Thomas X, Le Jeune C. Treating Adults With Acute Lymphocytic Leukemia: New Pharmacotherapy Options. Expert Opin Pharmacother (2016) 17(17):2319–30. doi: 10.1080/14656566.2016.1250884
8. Gavralidis A, Brunner AM. Novel Therapies in the Treatment of Adult Acute Lymphoblastic Leukemia. Curr Hematol Malig Rep (2020) 15(4):294–304. doi: 10.1007/s11899-020-00591-4
9. Kazimierczyk M, Wrzesinski J. Long Non-Coding RNA Epigenetics. Int J Mol Sci (2021) 22(11):6166. doi: 10.3390/ijms22116166
10. Aliperti V, Skonieczna J, Cerase A, Long Non-Coding RNA. (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders. Noncoding RNA (2021) 7(2):36. doi: 10.3390/ncrna7020036
11. Khan S, Masood M, Gaur H, Ahmad S, Syed MA. Long Non-Coding RNA: An Immune Cells Perspective. Life Sci (2021) 271:119152. doi: 10.1016/j.lfs.2021.119152
12. Fu D, Shi Y, Liu JB, Wu TM, Jia CY, Yang HQ, et al. Targeting Long Non-Coding RNA to Therapeutically Regulate Gene Expression in Cancer. Mol Ther Nucleic Acids (2020) 21:712–24. doi: 10.1016/j.omtn.2020.07.005
13. Ghafouri-Fard S, Gholipour M, Taheri M. The Emerging Role of Long Non-Coding RNAs and Circular RNAs in Coronary Artery Disease. Front Cardiovasc Med (2021) 8:632393. doi: 10.3389/fcvm.2021.632393
14. Li J, Zou J, Wan X, Sun C, Peng F, Chu Z, et al. The Role of Noncoding RNAs in B-Cell Lymphoma. Front Oncol (2020) 10(577890). doi: 10.3389/fonc.2020.577890
15. Carrasco-Leon A, Amundarain A, Gomez-Echarte N, Prosper F, Agirre X. The Role of lncRNAs in the Pathobiology and Clinical Behavior of Multiple Myeloma. Cancers (Basel) (2021) 13(8):1976. doi: 10.3390/cancers13081976
16. Wurm AA, Pina C. Long Non-Coding RNAs as Functional and Structural Chromatin Modulators in Acute Myeloid Leukemia. Front Oncol (2019) 9:899. doi: 10.3389/fonc.2019.00899
17. James AR, Schroeder MP, Neumann M, Bastian L, Eckert C, Gokbuget N, et al. Long Non-Coding RNAs Defining Major Subtypes of B Cell Precursor Acute Lymphoblastic Leukemia. J Hematol Oncol (2019) 12(1):8. doi: 10.1186/s13045-018-0692-3
18. Orlandella FM, Smaldone G, Salvatore G, Vitagliano L, Cianflone A, Parasole R, et al. The lncRNA TEX41 Is Upregulated in Pediatric B-Cells Acute Lymphoblastic Leukemia and It Is Necessary for Leukemic Cell Growth. biomark Res (2021) 9(1):54. doi: 10.1186/s40364-021-00307-7
19. Wang W, Wu F, Ma P, Gan S, Li X, Chen L, et al. LncRNA CRNDE Promotes the Progression of B-Cell Precursor Acute Lymphoblastic Leukemia by Targeting the miR-345-5p/CREB Axis. Mol Cells (2020) 43(8):718–27. doi: 10.14348/molcells.2020.0065
20. Cuadros M, Andrades A, Coira IF, Balinas C, Rodriguez MI, Alvarez-Perez JC, et al. Expression of the Long Non-Coding RNA TCL6 Is Associated With Clinical Outcome in Pediatric B-Cell Acute Lymphoblastic Leukemia. Blood Cancer J (2019) 9(12):93. doi: 10.1038/s41408-019-0258-9
21. Hoelzer D, Bassan R, Dombret H, Fielding A, Ribera JM, Buske C, et al. Acute Lymphoblastic Leukaemia in Adult Patients: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann Oncol (2016) 27(suppl 5):v69–82. doi: 10.1093/annonc/mdw025
22. Treichel RS, Olken S. The Relationship Between Multi-Drug Resistance and Resistance to Natural-Killer-Cell and Lymphokine-Activated Killer-Cell Lysis in Human Leukemic Cell Lines. Int J Cancer (1992) 50(2):305–10. doi: 10.1002/ijc.2910500223
23. Song C, Ge Z, Ding Y, Tan BH, Desai D, Gowda K, et al. IKAROS and CK2 Regulate Expression of BCL-XL and Chemosensitivity in High-Risk B-Cell Acute Lymphoblastic Leukemia. Blood (2020) 136(13):1520–34. doi: 10.1182/blood.2019002655
24. de Vries JF, Zwaan CM, De Bie M, Voerman JS, den Boer ML, van Dongen JJ, et al. The Novel Calicheamicin-Conjugated CD22 Antibody Inotuzumab Ozogamicin (CMC-544) Effectively Kills Primary Pediatric Acute Lymphoblastic Leukemia Cells. Leukemia (2012) 26(2):255–64. doi: 10.1038/leu.2011.206
25. Sanchez VE, Nichols C, Kim HN, Gang EJ, Kim YM. Targeting PI3K Signaling in Acute Lymphoblastic Leukemia. Int J Mol Sci (2019) 20(2):412. doi: 10.3390/ijms20020412
26. Shi YF, Huang ZY, Huang YS, Dong RJ, Xing CY, Yu K, et al. Interaction Between CD9 and PI3Kp85 Activates the PI3K/AKT Signaling Pathway in Blineage Acute Lymphoblastic Leukemia. Oncol Rep (2021) 46(1):140. doi: 10.3892/or.2021.8091
27. Neri LM, Cani A, Martelli AM, Simioni C, Junghanss C, Tabellini G, et al. Targeting the PI3K/Akt/mTOR Signaling Pathway in B-Precursor Acute Lymphoblastic Leukemia and Its Therapeutic Potential. Leukemia (2014) 28(4):739–48. doi: 10.1038/leu.2013.226
28. Wang W, Lyu C, Wang F, Wang C, Wu F, Li X, et al. Identification of Potential Signatures and Their Functions for Acute Lymphoblastic Leukemia: A Study Based on the Cancer Genome Atlas. Front Genet (2021) 12:656042. doi: 10.3389/fgene.2021.656042
29. Li S, Bian H, Cao Y, Juan C, Cao Q, Zhou G, et al. Identification of Novel lncRNAs Involved in the Pathogenesis of Childhood Acute Lymphoblastic Leukemia. Oncol Lett (2019) 17(2):2081–90. doi: 10.3892/ol.2018.9832
30. Song Y, Guo NH, Zheng JF. LncRNA-MALAT1 Regulates Proliferation and Apoptosis of Acute Lymphoblastic Leukemia Cells via miR-205-PTK7 Pathway. Pathol Int (2020) 70(10):724–32. doi: 10.1111/pin.12993
31. Xiao S, Xu N, Ding Q, Huang S, Zha Y, Zhu H. LncRNA VPS9D1-AS1 Promotes Cell Proliferation in Acute Lymphoblastic Leukemia Through Modulating GPX1 Expression by miR-491-5p and miR-214-3p Evasion. Biosci Rep (2020) 40(10):BSR20193461. doi: 10.1042/BSR20193461
32. Nie L, Li C, Zhao T, Wang Y, Liu J. LncRNA Double Homeobox A Pseudogene 8 (DUXAP8) Facilitates the Progression of Neuroblastoma and Activates Wnt/beta-Catenin Pathway via microRNA-29/Nucleolar Protein 4 Like (NOL4L) Axis. Brain Res (2020) 1746:146947. doi: 10.1016/j.brainres.2020.146947
33. Gong A, Huang Z, Ge H, Cai Y, Yang C. The Carcinogenic Complex lncRNA DUXAP8/EZH2/LSD1 Accelerates the Proliferation, Migration and Invasion of Colorectal Cancer. J BUON (2019) 24(5):1830–6.
34. Zhai H, Zhao J, Pu J, Zhao P, Wei J. LncRNA-DUXAP8 Regulation of the Wnt/beta-Catenin Signaling Pathway to Inhibit Glycolysis and Induced Apoptosis in Acute Myeloid Leukemia. Turk J Haematol (2021) 38(4):264–72. doi: 10.4274/tjh.galenos.2021.2020.0769
35. Hu Y, Zhang X, Zai HY, Jiang W, Xiao L, Zhu Q. lncRNA DUXAP8 Facilitates Multiple Malignant Phenotypes and Resistance to PARP Inhibitor in HCC via Upregulating Foxm1. Mol Ther Oncolytics (2020) 19:308–22. doi: 10.1016/j.omto.2020.10.010
36. Ji X, Tao R, Sun LY, Xu XL, Ling W. Down-Regulation of Long Non-Coding RNA DUXAP8 Suppresses Proliferation, Metastasis and EMT by Modulating miR-498 Through TRIM44-Mediated AKT/mTOR Pathway in Non-Small-Cell Lung Cancer. Eur Rev Med Pharmacol Sci (2020) 24(6):3152–65. doi: 10.26355/eurrev_202003_20682
37. Li JR, Liu L, Luo H, Chen ZG, Wang JH, Li NF. Long Noncoding RNA DUXAP8 Promotes Pancreatic Carcinoma Cell Migration and Invasion Via Pathway by miR-448/WTAP/Fak Signaling Axis. Pancreas (2021) 50(3):317–26. doi: 10.1097/MPA.0000000000001751
38. Dey S, Kwon JJ, Liu S, Hodge GA, Taleb S, Zimmers TA, et al. miR-29a Is Repressed by MYC in Pancreatic Cancer and Its Restoration Drives Tumor-Suppressive Effects via Downregulation of LOXL2. Mol Cancer Res (2020) 18(2):311–23. doi: 10.1158/1541-7786.MCR-19-0594
39. Wang XS, Gong JN, Yu J, Wang F, Zhang XH, Yin XL, et al. MicroRNA-29a and microRNA-142-3p Are Regulators of Myeloid Differentiation and Acute Myeloid Leukemia. Blood (2012) 119(21):4992–5004. doi: 10.1182/blood-2011-10-385716
40. Sharma S, Pavlasova GM, Seda V, Cerna KA, Vojackova E, Filip D, et al. miR-29 Modulates CD40 Signaling in Chronic Lymphocytic Leukemia by Targeting TRAF4: An Axis Affected by BCR Inhibitors. Blood (2021) 137(18):2481–94. doi: 10.1182/blood.2020005627
41. Wang F, Wang XS, Yang GH, Zhai PF, Xiao Z, Xia LY, et al. miR-29a and miR-142-3p Downregulation and Diagnostic Implication in Human Acute Myeloid Leukemia. Mol Biol Rep (2012) 39(3):2713–22. doi: 10.1007/s11033-011-1026-5
42. Raeisi F, Mahmoudi E, Dehghani-Samani M, Hosseini SSE, Ghahfarrokhi AM, Arshi A, et al. Differential Expression Profile of miR-27b, miR-29a, and miR-155 in Chronic Lymphocytic Leukemia and Breast Cancer Patients. Mol Ther Oncolytics (2020) 16:230–7. doi: 10.1016/j.omto.2020.01.004
43. Zanette DL, Rivadavia F, Molfetta GA, Barbuzano FG, Proto-Siqueira R, Silva WA Jr., et al. miRNA Expression Profiles in Chronic Lymphocytic and Acute Lymphocytic Leukemia. Braz J Med Biol Res (2007) 40(11):1435–40. doi: 10.1590/S0100-879X2007001100003
44. Evangelisti C, Chiarini F, Cappellini A, Paganelli F, Fini M, Santi S, et al. Targeting Wnt/beta-Catenin and PI3K/Akt/mTOR Pathways in T-Cell Acute Lymphoblastic Leukemia. J Cell Physiol (2020) 235(6):5413–28. doi: 10.1002/jcp.29429
45. Hales EC, Taub JW, Matherly LH. New Insights Into Notch1 Regulation of the PI3K-AKT-Mtor1 Signaling Axis: Targeted Therapy of Gamma-Secretase Inhibitor Resistant T-Cell Acute Lymphoblastic Leukemia. Cell Signal (2014) 26(1):149–61. doi: 10.1016/j.cellsig.2013.09.021
46. Sigmund AM, Sahasrabudhe KD, Bhatnagar B. Evaluating Blinatumomab for the Treatment of Relapsed/Refractory ALL: Design, Development, and Place in Therapy. Blood Lymphat Cancer (2020) 10:7–20. doi: 10.2147/BLCTT.S223894
47. DeAngelo DJ, Advani AS, Marks DI, Stelljes M, Liedtke M, Stock W, et al. Inotuzumab Ozogamicin for Relapsed/Refractory Acute Lymphoblastic Leukemia: Outcomes by Disease Burden. Blood Cancer J (2020) 10(8):81. doi: 10.1038/s41408-020-00345-8
48. Jabbour E, Gokbuget N, Advani A, Stelljes M, Stock W, Liedtke M, et al. Impact of Minimal Residual Disease Status in Patients With Relapsed/Refractory Acute Lymphoblastic Leukemia Treated With Inotuzumab Ozogamicin in the Phase III INO-VATE Trial. Leuk Res (2020) 88:106283. doi: 10.1016/j.leukres.2019.106283
49. Kantarjian HM, DeAngelo DJ, Stelljes M, Liedtke M, Stock W, Gokbuget N, et al. Inotuzumab Ozogamicin Versus Standard of Care in Relapsed or Refractory Acute Lymphoblastic Leukemia: Final Report and Long-Term Survival Follow-Up From the Randomized, Phase 3 INO-VATE Study. Cancer (2019) 125(14):2474–87. doi: 10.1002/cncr.32116
Keywords: B-cell acute lymphoblastic leukemia, lncRNA expression profile, chemotherapy resistance, lncRNA DUXAP8/miR-29a/PIK3CA network, PI3K-AKT-mTOR signaling
Citation: Zhang L, Zhou S, Zhou T, Li X and Tang J (2022) Targeting the lncRNA DUXAP8/miR-29a/PIK3CA Network Restores Doxorubicin Chemosensitivity via PI3K-AKT-mTOR Signaling and Synergizes With Inotuzumab Ozogamicin in Chemotherapy-Resistant B-Cell Acute Lymphoblastic Leukemia. Front. Oncol. 12:773601. doi: 10.3389/fonc.2022.773601
Received: 13 September 2021; Accepted: 28 January 2022;
Published: 02 March 2022.
Edited by:
Patrycja Dubielecka, Brown University, United StatesReviewed by:
Nashwa El-Khazragy, Ain Shams University, EgyptLuca Lo Nigro, Azienda Ospedaliero Universitaria Policlinico - San Marco, Italy
Copyright © 2022 Zhang, Zhou, Zhou, Li and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junling Tang, UG9seWNsb25lUkVTX1RKTEBzd211LmVkdS5jbg==
 Li Zhang1,2,3
Li Zhang1,2,3 Junling Tang
Junling Tang