- 1School of Clinical Medicine, Weifang Medical University, Weifang, China
- 2Department of Thoracic Surgery, Central Hospital Affiliated to Shandong First Medical University, Jinan, China
- 3Department of Surgery, Khyber Medical University Peshawar, Peshawar, Pakistan
- 4Breast Center, Central Hospital Affiliated to Shandong First Medical University, Jinan, China
According to the latest statistics from the International Agency for Research on Cancer (IARC), lung cancer is one of the most lethal malignancies in the world, accounting for approximately 18% of all cancer-associated deaths. Yet, even with aggressive interventions for advanced lung cancer, the five-year survival rate remains low, at around 15%. The hedgehog signaling pathway is highly conserved during embryonic development and is involved in tissue homeostasis as well as organ development. However, studies have documented an increasing prevalence of aberrant activation of HH signaling in lung cancer patients, promoting malignant lung cancer progression with poor prognostic outcomes. Inhibitors targeting the HH pathway have been widely used in tumor therapy, however, they still cannot avoid the occurrence of drug resistance. Interestingly, natural products, either alone or in combination with chemotherapy, have greatly improved overall survival outcomes for lung cancer patients by acting on the HH signaling pathway because of its unique and excellent pharmacological properties. In this review, we elucidate on the underlying molecular mechanisms through which the HH pathway promotes malignant biological behaviors in lung cancer, as well as the potential of inhibitors or natural compounds in targeting HH signaling for clinical applications in lung cancer therapy.
Introduction
Lung cancer, which is one of the most lethal malignancies worldwide, can be classified into non-small cell lung cancer (NCSLC) or small cell lung cancer (SCLC) based on histopathological type (1–3). Among them, NSCLC accounts for approximately 85% of lung cancer cases (4–6). Regrettably, most lung cancer patients are already in advanced stages at the time of diagnosis, losing the opportunity for surgery. However, with decades of medical development, diverse and individualized treatment strategies for advanced lung cancer, including chemotherapy, radiotherapy, targeted therapy as well as immunotherapy have been developed (7, 8). Among them, molecular targeted therapies targeting driver genes has shown good application prospects, and the corresponding molecular targeted drugs have been developed for common lung cancer driver genes, such as epidermal growth factor receptor (EGFR), mesenchymal lymphoma kinase (ALK) and c-ros oncogene 1 receptor tyrosine kinase (ROS1), significantly prolonging progression-free survival (PFS) and overall survival (OS) outcomes for lung cancer patients (9–11).
The HH signaling pathway is highly evolutionarily conserved, and maintains tissue homeostasis and organ development (12, 13). There is evidence that HH signaling participates in embryonic lung development, regulates epithelial-mesenchymal interactions, differentiation of embryonic neuroendocrine cells, and repair of injured airway tissues (14). Aberrant activation of HH signaling is closely associated with lung cancer development (15). The Kras/YY1/ZNF322/SHH transcriptional axis increases the expression levels of the SHH protein, leading to lung cancer malignant progression by inducing tumor angiogenesis (16).
In lung squamous cell carcinoma (LSCC), HH signaling regulates EMT-associated proteins, such as E-Cadherin and β-catenin to induce the EMT process (17), and Smo gene amplification is one of the mechanisms through which HH signaling is activated to increase the resistance of lung cancer cells to epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) by inducing the EMT process (18). In an EGFR-TKIs-resistant lung cancer mouse model, the HH inhibitor, LDE225 (Sonidegib), in combination with gefitinib significantly inhibited tumor growth than gefitinib alone, completely inhibiting the phosphorylation of PI3K/AKT and MAPK signaling. Moreover, LDE225 enhanced the sensitivity of lung cancer cells to standard chemotherapy (18). In addition, to establish whether HH signaling was activated in lung cancer cells, Gli1 protein levels in advanced NSCLC were assessed. Gli1 protein overexpression was found to be associated with poor prognostic outcomes and immune checkpoint inhibitor resistance (19). Activation of HH signaling is essential for the development of some types of SCLC (14). In an SCLC mouse model, inhibition of HH signaling using cyclopamine significantly reduced tumor growth. HH inhibitors in combination with radiotherapy or chemotherapy regimens provide individualized treatment options for SCLC patients (14).
In this review, we elucidate on the underlying molecular mechanisms of HH signaling in lung cancer development and explore the relevance of HH signaling-related proteins in the prognosis of several types of lung cancer, indicating new directions for research.
The Canonical Pathway of HH Signaling
The hedgehog (HH) gene was first detected in 1980 by Nüsslein-Volhard and Wieschaus while studying the embryonic development of Drosophila (20). As the canonical signaling pathway in vertebrates, the HH-Ptch-Smo-Gli route consists of several essential components, including Sonic hedgehog (SHH), Desert hedgehog (DHH), and Indian hedgehog (IHH), Smoothened (Smo), Patched (Ptch), three glioma-associated oncogene transcription factors (Gli1, Gli2, Gli3), Suppressor of fused (SuFu). Of the three HH homologs, the SHH protein is highly expressed, with the most potent biological effects (21). Smo, a seven-pass transmembrane protein belonging to the G protein-coupled receptor (GPCR) superfamily, can be inhibited by Ptch. Ptch, a twelve-pass transmembrane protein to be the receptor of SHH, can negatively regulate the HH pathway. As downstream effectors of the HH signaling pathway, the Gli zinc-finger transcription factor family plays a crucial role in final activation or inactivation of the HH pathway (22). Gli1 acts as a transcriptional activator because it lacks an N-terminal inhibitory region, while Gli2 and Gli3 both have N-terminal inhibitory regions and C-terminal activation regions that play dual roles of activation and repression (23). The Gli1 protein acts as a key transcription factor, regulating the expressions of downstream target genes of the HH pathway.
In the absence of HH ligands, Ptch receptors constitutively inhibit the activities of Smo proteins (24), leading to the phosphorylation of Gli proteins (Gli2 and Gli3) within the microtubule complex by casein kinase 1a (CK1a), protein kinase A (PKA), and glycogen synthase kinase 3β (GSK3β). The phosphorylated Gli proteins then bind β-transducing repeat-containing protein (β-TrCP) to form the Gli/β-TrCP complex that is sheared into a transcription-repressive form (Gli-R) by the actions of ubiquitin-proteasome (25). Briefly, when the HH signaling pathway is inactivated, Gli-R enters the nucleus, where they bind target gene promoters to repress gene transcriptions.
In the presence of HH ligands, inhibitory effects of Ptch receptors on Smo proteins are alleviated. The Smo protein, activated by CK1, PKA, and GPCR kinase 2 (GRK2)-mediated phosphorylation, translocates to the primary cilium by interacting with β-arrestin (26). As Smo proteins continuously accumulate on the primary cilium, a microtubule complex consisting of Gli and SuFu proteins translocate to the top of the primary cilium (27), where it inhibits Gli proteins C-terminal hydrolysis by interacting with Smo proteins, thereby activating and releasing Gli proteins. The activated Gli protein (Gli-A), which has a transcriptional activation function, enters the nucleus to regulate the expressions of HH pathway target genes (28). Target genes of the HH pathway include Gli1, Ptch1, FoxA2, Bcl-2, Bcl-xl, Myc, and Cyclin family (Figure 1) (29).
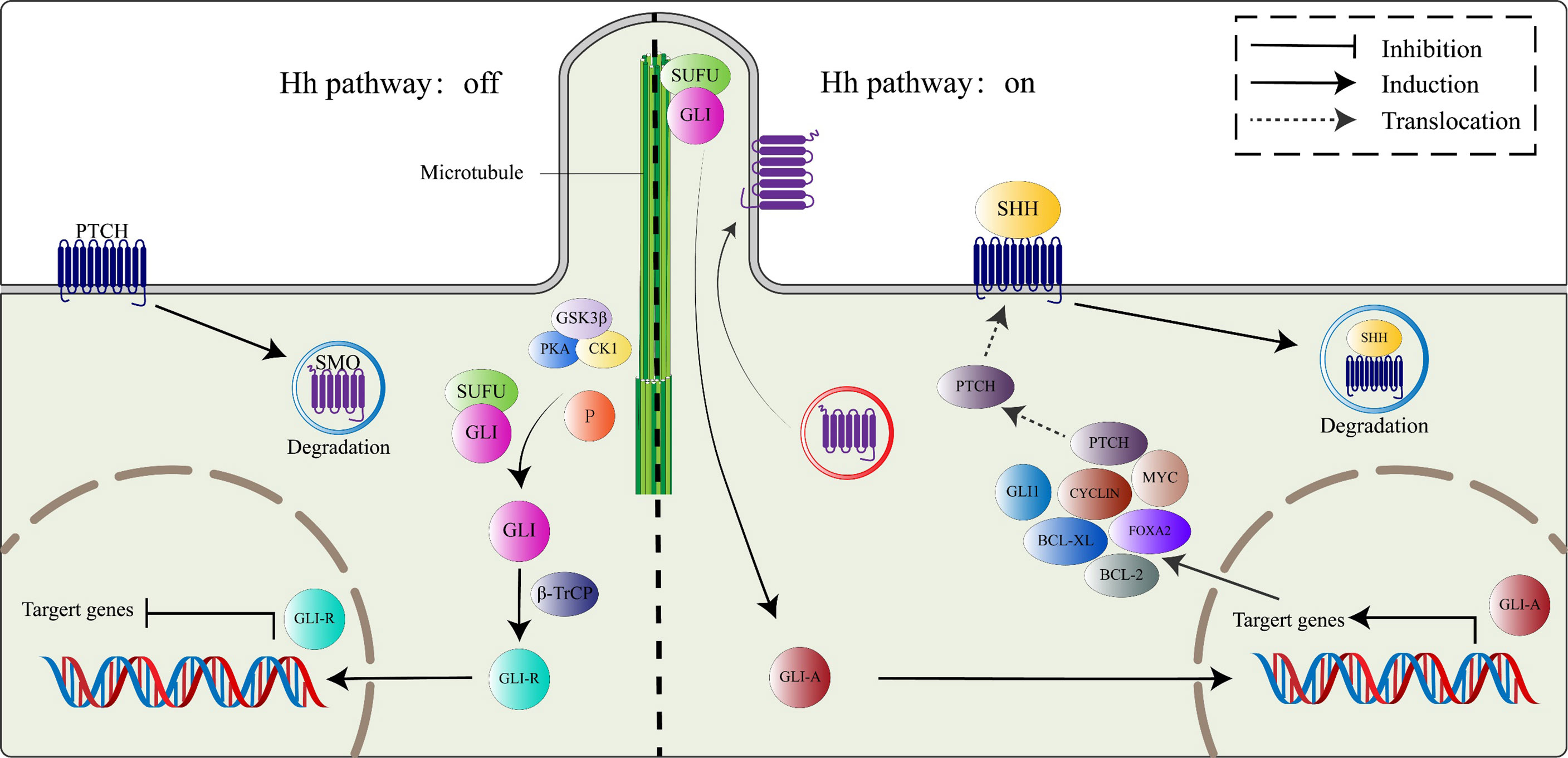
Figure 1 The canonical HH signaling pathway. In the absence of the HH ligand (left), PTCH binds SMO, leading to its degradation. Microtubule complex containing Gli and SUFU is phosphorylated by GSK3β/PKA/CK1 to activate Gli proteins, which subsequently combine with β-TrCP to switch into transcriptional repressor (GLI-R). In the presence of HH ligand (right), SMO restriction by PTCH is relieved, active SMO moves to the primary cilium. Active Smo interacts with the SUFU/GLI complex, localized at the top of the primary cilium, GLI proteins from the complex switch into transcription activators (GLI-A), which then translocate to the nucleus and activate the HH target genes, including PTCH, GLI1, FOXA2, BCL-2, BCL-Xl, MYC, and CYCLIN family among others.
The Noncanonical Pathway of HH Signaling
Noncanonical HH signal transduction is the signaling response to any associated components of the HH pathway, different from the common canonical HH signaling pathway. Based on HH pathway-related components, the noncanonical HH pathway is classified into four types: SHH-mediated, Ptch-mediated, Smo-mediated, and Gli-mediated noncanonical signaling (Figure 2).
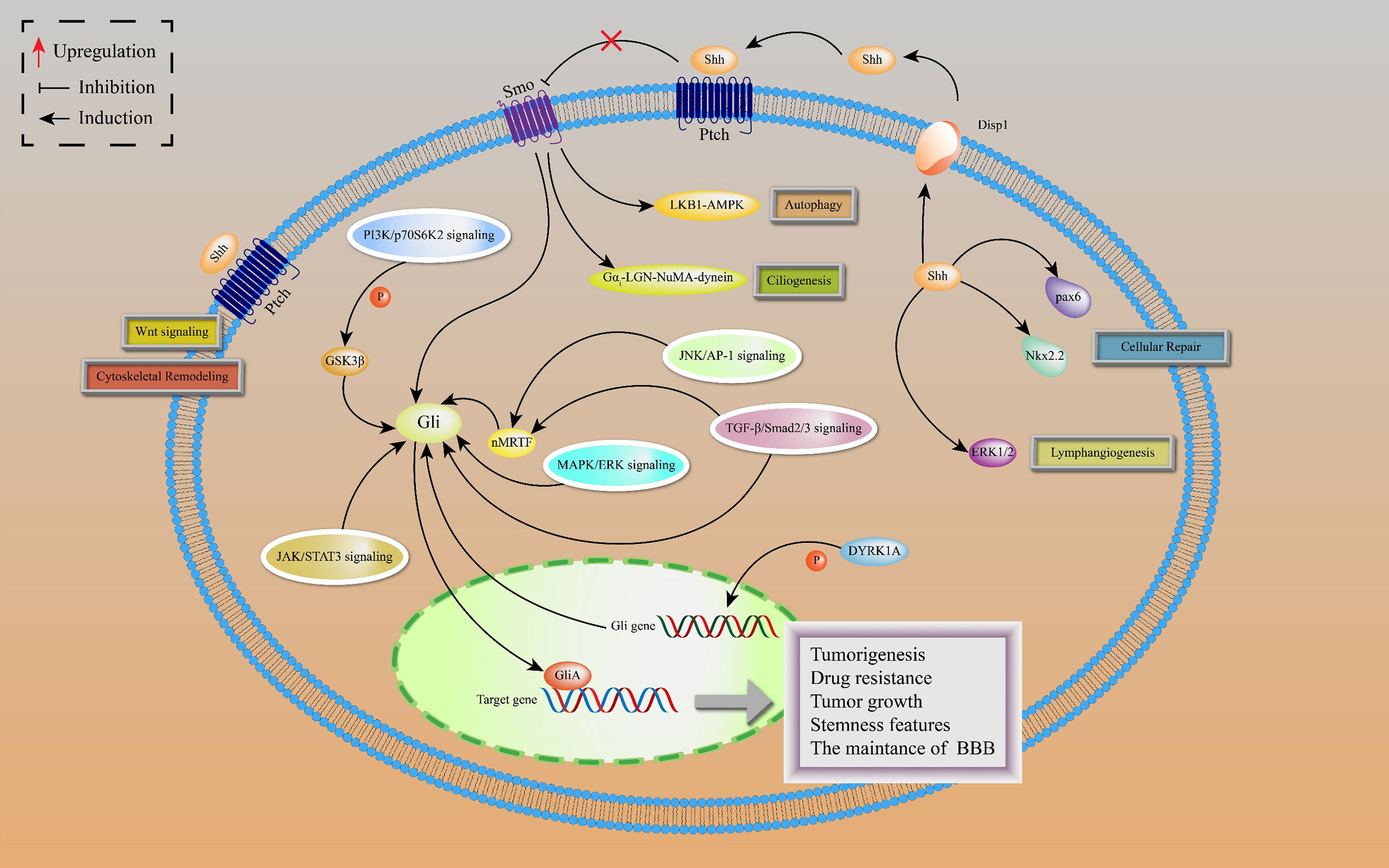
Figure 2 Noncanonical HH signaling transductions are depicted, including SHH-mediated, Ptch-mediated, Smo-mediated and Gli-mediated noncanonical signaling. SHH-mediated noncanonical pathway is associated with cellular repair and lymphangiogenesis. Ptch-mediated but Smo-independent pathway is activated by SHH to induce Wnt signaling and cytoskeletal remodeling. In Smo-mediated but Gli-independent noncanonical pathway, the SHH ligand activates LKB1-AMPK and Gαi-LGN-NuMA-dynein axes in neurons, leading to activation of autophagy and ciliogenesis. There is a crosstalk between Gli activated and Smo-independent pathways as well as with numerous oncogenic pathways, resulting in lung cancer tumorigenesis and development.
The SHH ligands selectively promote lymphatic endothelial cell proliferation to induce lymphangiogenesis after kidney injury by activating extracellular signal-regulated kinase-1 and -2 (ERK1/2) pathway, leading to kidney fibrosis, independently of Smo (30). Moreover, SHH proteins are involved in repairing responses of damaged cerebellum by upregulating the expressions of Nkx2.2 and Pax6 transcription factors in cadmium-exposed rats (31).
The Ptch-mediated noncanonical pathway is a Ptch-mediated Smo-independent pathway. This pathway is accompanied by cytoskeletal remodeling and Wnt signal activation, which provides a possible explanation for the failure of Smo inhibitors in HH-dependent malignant tumors (32). In addition, active Smo induces autophagy and promotes ciliogenesis by acting on LKB1-AMPK and Gαi-LGN-NuMA-dynein axes in some cell lines, including neurons, independently of the Smo-mediated canonical pathway (33).
The Gli-mediated noncanonical pathway is also known as Gli-activated, but Smo-independent pathway. In drug-resistant basal cell carcinoma (BCC), activator protein-1 (AP-1) and transforming growth factor-β (TGFβ) synergistically stimulate a nuclear myocardin-related transcription factor (nMRTF) to enhance transcriptional activities of Gli1 (34). Desmoglein 2 activates signal transducer and activator of transcription 3 (STAT3) to upregulate Gli1 expressions, resulting in BCC occurrence and development (35). Astrocyte-derived TGFβ1/Smad2/3 signaling contributes to blood-brain barrier (BBB) functions by increasing ZO-1 expression via upregulating Gli2 protein levels (36). In addition, dual-specificity tyrosine-regulated kinase 1A (DYRK1A) phosphorylate Gli1 at Ser408 to enhance the transcriptional activities of Gli1, suggesting that DYRK1A regulates noncanonical HH signaling (37).
In lung adenocarcinoma (LAC), KRAS and vascular endothelial growth factor (VEGF) receptor, NRP2, trigger MAPK/ERK/Gli1 signaling cascade to promote tumor progression (38). As a downstream effector of the PI3K pathway, p70S6K2 enhances the transcription activities of Gli1 by modulating the phosphorylation of GSK3β in NCSLC (39). Finally, there are other oncogenic pathways involving noncanonical HH signal transduction, such as AMP-activated protein kinase (AMPK), PI3K-AKT-mTOR signaling, and Protein kinase C (PKC) (40). Figure 3 shows the crosstalk between the HH pathway with various oncogenic pathways.
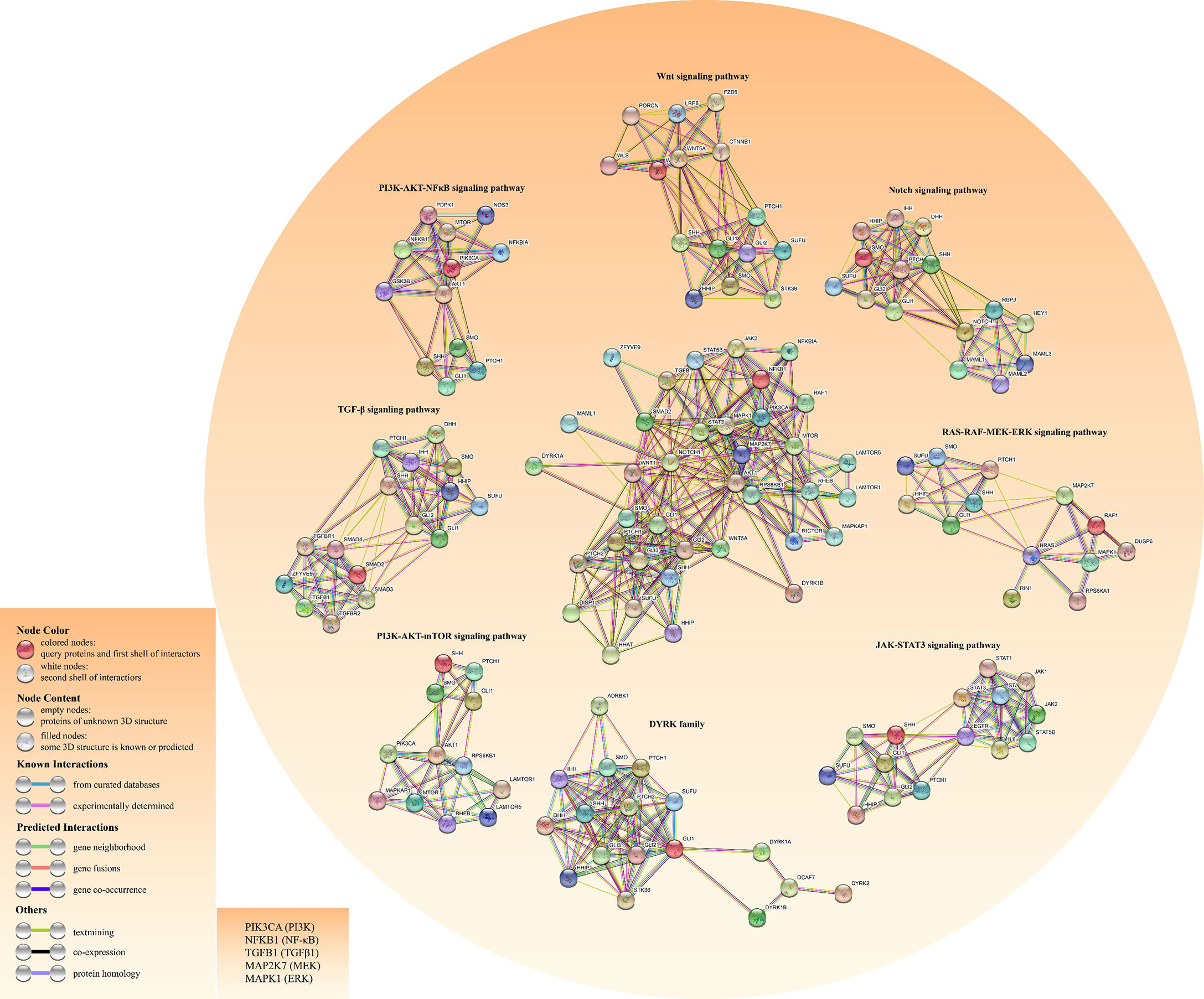
Figure 3 Construction of the crosstalk network for both HH pathway and other cancer-related pathways using the STRING database. The left bottom literal statements are the alternative names of the molecule.
The HH Pathway Promotes the Malignant Progression of Lung Cancer
Abnormal activations of the HH signal are closely associated with lung cancer occurrence and development. They are involved in cell proliferation, invasion, metastasis, drug resistance, stemness characteristics, and tumor microenvironment. Therefore, we summarize the recent findings of HH signaling in lung cancer and discuss the potential for targeting the HH pathways to treat lung cancer.
The HH Pathway Promotes Lung Cancer Cell Proliferation
The HH pathway remarkably induces tumor proliferation (41, 42). HH signaling regulates INSM1 interactions with N-myc to promote SCLC cell growth in a Gli-dependent manner (43). The E3 ligase (HERC4) and liver kinase B1 (LKB1) inhibit cell proliferation in lung cancer by inactivating the HH pathway. Mechanistically, HERC4 has been shown to negatively regulate the HH pathway by destabilizing the Smo protein (44), while the mechanisms through which the HH pathway is inhibited by LKB1 have not been elucidated (45).
HH Pathway Promotes Lung Cancer Cell Invasion and Migration
The HH pathway is involved in cell invasion and migration of multiple tumors (46). SAM- and SH3-domain containing 1 (SASH1) inhibit hepatocellular carcinoma invasion and migration by inactivating the HH and PI3K/AKT pathways in vitro and in vivo (47). In vitro, transmembrane 107 protein (TMEM107) inhibited epithelial-mesenchymal transition (EMT) and invasion by negatively regulating HH signaling in NSCLC (48). Besides, Gli1, the downstream gene of the HH pathway, promotes NSCLC cell invasion and metastasis in vitro and in vivo by inducing the EMT process. The downregulation of Gli1 significantly inhibited tumor growth and enhanced E-cadherin expressions (49). Interestingly, the KRAS mutation activated the Kras/YY1/ZNF322A/SHH axis by triggering the expression of downstream genes, including YY1, ZNF322A, and SHH, which promoted angiogenesis in NSCLC in vitro and in vivo (16). The overexpression of phosphatidylethanolamine‐binding protein 4 (PEBP4) promotes NSCLC cell proliferation and EMT by regulating the HH pathway (50) while fibroblast activation protein α (FAPα) promoted LSCC cell growth, adhesion, and migration in vitro via the PI3K and HH pathways (51).
Hedgehog-interacting proteins (HHIP) negatively regulate the HH pathway by binding SHH proteins (52–54). HHIP serve as tumor suppressors by significantly inhibiting lung cancer cell proliferation, migration, and invasion (53). Similarly, overexpression of signal peptide-CUB-EGF domain-containing protein 2 (SCUBE2) inhibits NSCLC cell proliferation and invasion by modulating the HH pathway. The SCUBE2-mediated inhibition of the HH pathway can be reversed by recombinant SHH proteins (55). GATA-6 impairs the metastatic abilities of LAC cells (56). Moreover, GATA-6 suppresses cell proliferation and migration in LSCC by inhibiting the expressions of SHH at transcriptional levels (57). Additionally, miR‐520b enhances NSCLC cell proliferation and metastasis by activating the SPOP‐Gli2/3 axis (58). Hematopoietic pre-B-cell leukemia transcription factor (PBX)-interacting protein (HPIP) is a nucleo-cytoplasmic shuttling protein (59) that promotes NSCLC cell proliferation, invasion, and migration in vitro and tumor growth in vivo by interfering with the HH pathway (54).
In an A549 cell line, tumor suppressor candidate 3 silencing (TUSC3) inhibited growth, proliferation and induced apoptosis as well as radiation sensitivity (60). In contrast, TUSC3 overexpression in NSCLC promoted cell proliferation, migration, and invasion in vitro and in vivo, possibly by involving HH signaling (61). Leucine zipper transcription factor-like 1 (LZTFL1) inhibits lung cancer tumorigenesis, at least partly by inhibiting HH signaling pathway to maintain epithelial cell differentiation (62). Figure 4 shows HH signaling interaction networks in lung cancer patients.
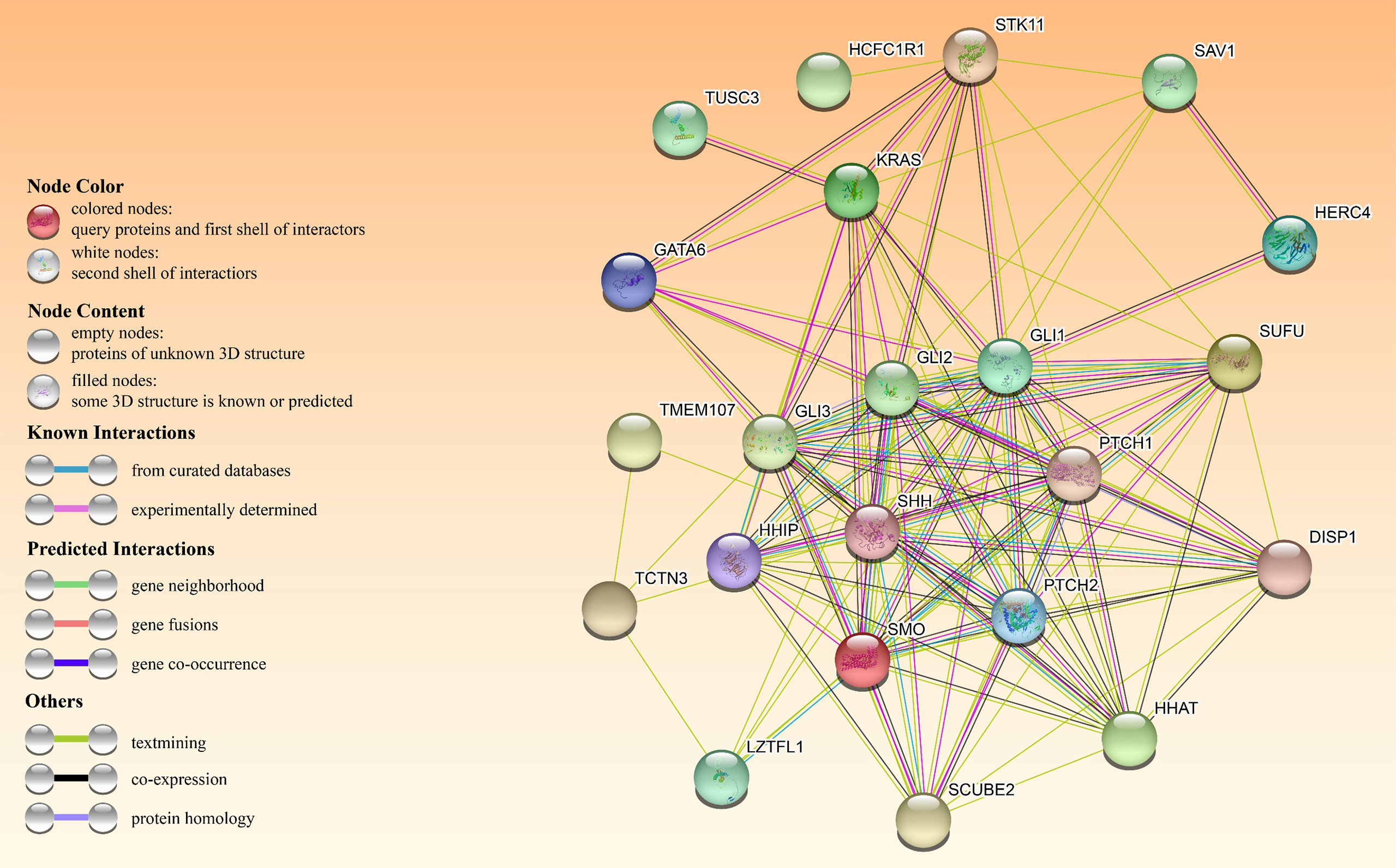
Figure 4 The HH pathway interacts with other oncogenes or tumor suppressors in patients with HH-dependent lung cancer. The interactome in the center was obtained through the STRING database.
HH Pathway and Drug Resistance
Drug resistance is one of the leading causes of tumor treatment failure (63), while hyperactivation of the HH pathway is frequently described in many drugs resistant malignant tumors. The HH pathway promotes cell proliferation as well as invasion in LAC with acquired drug resistance to EGFR-TKIs, partially by acting on HGF and MET signaling (64). Della Corte et al. (65) investigated the mechanisms in acquired resistance to EFGR-TKI during NSCLC treatment. The HH pathway was found to induce drug resistance by regulating the EMT process. Additionally, the active HH pathway was shown to induce EMT and upregulated ABCG2 in EGFR-TKI-resistant NSCLC patients, while the opposite result was observed when the HH pathway was inhibited (66).
MiRNA disorders are prevalent in many malignant tumors, including lung cancer (67, 68). miR-182-5p is the direct target of Gli2, the negative feedback axis of miR-182-5p/Gli2 regulates cell cisplatin resistance in LAC (69).
HH Pathway and Lung Cancer CSCs
A small proportion of tumor cells are regarded as CSCs that play a vital role in tumor occurrence, development and recurrence (70, 71). Aberrantly HH signaling is common in CSCs (72, 73). SHH-positive (SHH+) cells in NSCLC, expressing an uncleaved full-length SHH protein, exhibited drug resistance and CSC characteristics. A high abundance of SHH+ cells is a biomarker for worse prognostic outcomes for NSCLC patients (74). Low-folate (LF) is associated with malignant progression of lung cancer. Functionally, the LF microenvironment increases CSCs-like potential, partially through the HH pathway (75). Fibroblast growth factor receptor 1 (FGFR1) amplification in NSCLC is particularly prevalent, especially in LSCC (76, 77), where it stimulates the expressions of Gli2 through ERK pathway activation. The FGFR1/Gli2 axis upregulates the expression levels of CSCs markers (78). Aberrantly activated Notch and HH pathways induce CSCs phenotypes. Notch-Hedgehog positive tumor cells mediate immune evasion by enhancing the functions of regulatory T cells (79).
HH Pathway and the Tumor Microenvironment in Lung Cancer
The tumor microenvironment, a local homeostatic environment composed of tumor cells, stromal cells (including fibroblasts, immune and inflammatory cells) as well as the extracellular matrix, provides the necessary materials for tumor initiation and progression (80). Forkhead box F1(FoxF1) is associated with poor prognostic outcomes in some lung cancer subtypes. FoxF1, a downstream effector of HH signaling, stimulates the secretions of HGF and FGF2 by lung cancer fibroblasts, promoting lung cancer cell growth and migration in vivo (81).
NSCLC cells activate HH signaling in lung fibroblasts in a paracrine manner to stimulate the production of pro-angiogenic and metastatic factors, which induce fibroblast proliferation, invasion and collagen deposition, leading to lung cancer progression (82). Similarly, tumor cell-derived HH ligands stimulate perivascular stromal cells to secrete VEGF-A, promoting tumor angiogenesis in vitro and in vivo (83). CAFs-derived SHH ligands activate HH signaling in NSCLC cells in a paracrine manner, upregulating the expressions of EMT-related genes such as α-SMA and FAP, and enhancing the migration abilities of lung cancer cells (84). CAFs remodel actin cytoskeleton-induced EMT process in an Smo-dependent manner, enhancing the insensitivity of lung cancer cells to EGFR-TKIs (85).
TAMs receiving HH ligands secreted by tumor cells activate KLF4 and STAT3, downstream transcription factors of HH signaling, which drive TAMs M2 polarization, reduce CD8+ T cells, and inhibit their functions, thereby promoting malignant progression in various tumors (86, 87). By co-culturing A549 cells with THP-1-derived macrophages, A549 cells stimulated THP-1-derived macrophages toward TAMs M2 polarization. In contrast, THP-1-derived macrophages upregulate the expressions of stemness-related genes, including Sox2 and NANOG through HH, STAT3 and Notch signaling, increasing the stemness characteristics of lung cancer CSCs (88).
In conclusion, CAFs and TAMs interact with tumor cells to induce a pro-tumorigenic microenvironment, and HH signaling in the tumor microenvironment regulates CAFs, TAMs, and tumor cells in an autocrine and paracrine manner to increase stemness characteristics and promote the EMT process. Targeting HH signaling is a promising strategy to inhibit the formation of the tumor microenvironment.
HH Pathway and Lung Cancer Prognosis
It has not been conclusively determined whether HH pathway-related proteins are associated with lung cancer prognosis. We reviewed recent findings regarding the biological functions of the HH pathway in lung cancer, to establish the link between HH signaling proteins and prognostic outcomes of lung cancer patients (Table 1).
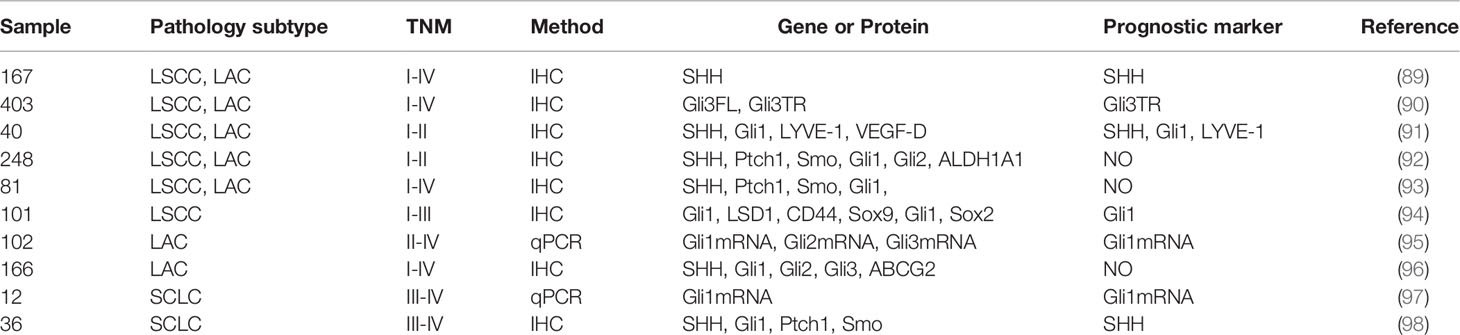
Hwang et al. (91) performed immunohistochemistry (IHC) to assess the expressions of SHH, Gli1, LYVE-1, and VEGF-D in 40 cases of primary NSCLC tissues. HH signaling protein overexpression, especially SHH, represent an independent risk factor for NSCLC. Moreover, SHH protein overexpression is negatively correlated with tumor differentiation and predicts poor prognostic outcomes in LSCC patients (89).
Gli proteins are potential CSCs markers and independent prognostic factors in LSCC. Their overexpression is closely associated with various malignant behaviors of LSCC, including TNM staging, lymph node metastasis, and clinical staging (94). Ishikawa et al. (95) measured the expression levels of Gli1, Gli2, and Gli3 mRNA in surgical samples from stage II-IV LAC patients using q-PCR. Their findings implied that Gli1 mRNA can serve as potential independent biomarker for prognosis in advanced LAC patients. Moreover, Lim et al. (98) evaluated the expression levels of HH signaling-related proteins, including SHH, Ptch, Smo, and Gli1, in extensive-stage SCLC samples. Their data revealed that the other markers have nothing to do with patient prognosis, while SHH proteins are potential markers for PFS and OS. Additionally, it has also been reported that truncated Gli3 (Gli3TR) is essential for LAC initiation and acts as a potential prognostic risk factor (90).
Overexpression of Gli1 tends to imply progressive stages and is associated with unfavorable prognostic outcomes (89, 99). In contrast, Gli1 mRNA overexpression is associated with better survival outcomes for advanced SCLC patients (97). Savani et al. (93) assayed tissue microarray with 81 samples from 42 patients with various NSCLC histologies. Minimum overexpression of HH signaling-related proteins in NSCLC do not correlate with patient outcomes. Additionally, there were no significant correlations between HH signaling-realted proteins, ALDH1A1 and RFS, OS in early-stage NSCLC (92). Kim et al. (96) showed that SHH and Gli1 overexpression imply better OS and PFS in LAC, but they are not independent prognostic factors.
Recent Advances in HH Inhibitors
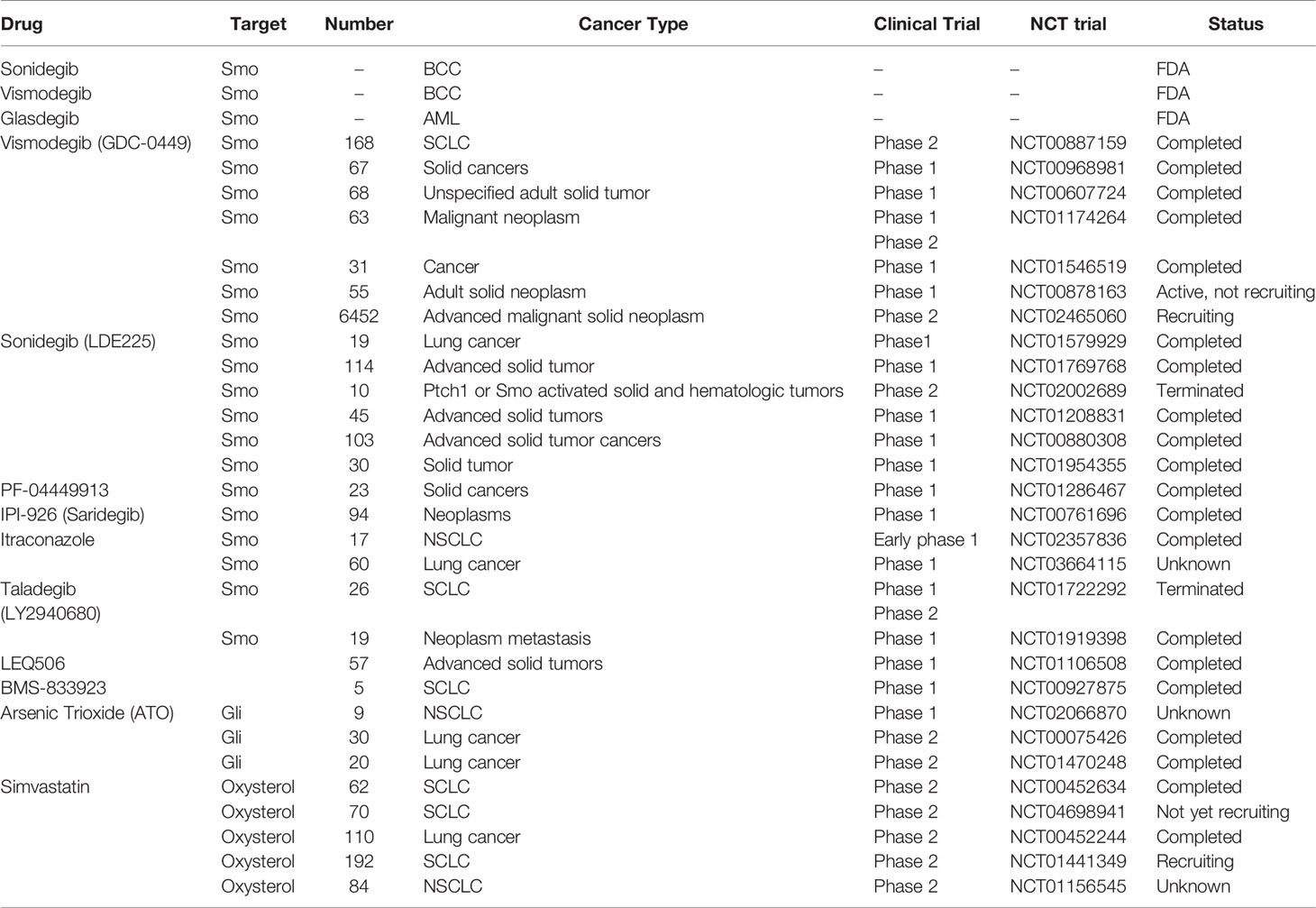
The three currently marketed HH inhibitors, Vismodegib and Sonidegib, are used to treat locally metastatic and advanced BCC (100–104), while Glasdegib is used for the treatment of acute myeloid leukemia (AML) (105, 106). Unfortunately, HH inhibitor resistance, which is attributed to both Smo acquired resistance and activation of noncanonical HH signaling, can never be avoided (107, 108). Therefore, there is an urgent need to develop new HH inhibitors to overcome HH inhibitor resistance and prolong the survival outcomes for patients with advanced HH-dependent tumors. Table 2 shows the HH inhibitors currently in clinical trials for lung cancer.
Itraconazole, a classic antifungal drug, has excellent pharmacological characteristics and safety. It acts on Smo proteins with a site of action distinct from that of Vismodegib, exhibiting potent anti-HH signaling effects (109–111). In a phase 2 trial involving metastatic nonsquamous NSCLC, itraconazole combined with pemetrexed significantly prolonged the overall survival outcomes for patients by up to 32 months, compared to pemetrexed alone (8 months) (112). In a randomized controlled study investigating the effects of itraconazole on clinical outcomes in patients with advanced NSCLC receiving platinum-based chemotherapy, it was confirmed that itraconazole significantly improved 1-year PFS and overall response rates (ORR), but not 1-year OS (113). The rationale for these two studies was based on anti-angiogenic effects of itraconazole. HH signaling is highly associated with tumor angiogenesis, however, a limited number of studies have determined whether itraconazole affects tumor angiogenesis through HH signaling. In advanced SCLC patients, the Smo inhibitor, Sonidegib, combined with standard chemotherapy, improved clinical outcomes were well tolerated. In one case of advanced SCLC with Sox2 amplification, progression free survival after combination therapy was as long as 27 months (114).
In conclusion, HH inhibitors alone or combined with other agents, including chemotherapeutic agents and small molecule targeted agents, have significant therapeutic implications in HH-dependent lung cancer patients. The current clinical trials have some limitations. First, there was no patient stratification before clinical trials; Second, activation mechanisms of canonical and non-canonical HH signaling in lung cancer are poorly understood; Third, there is a lack of large samples, multicenter data for further validation.
HHat Inhibitors
Through autoproteolytic cleavage, full-length SHH proteins generate 25 kDa SHH-C and 19 kDa SHH-N. Then, SHH-N undergoes C-terminal cholesterylation and N-terminal palmitoylation to mature into the SHH-N ligand (22). HHat mediates N-terminal palmitoylation of SHH-N and is essential for SHH-N ligand maturation. HHat inhibitors inhibit SHH-mediated activation of canonical and noncanonical HH pathways by blocking SHH-N ligand palmitoylation. RU-SKI43 (IC50: 0.85 μM), a small molecule inhibitor, exhibited HHat inhibitory effects in vitro and in vivo (115). RU-SKI-201 (IC50: 0.73 μM), optimized based on RU-SKI43, overcame the off-target toxicity of RU-SKI 43 and specifically inhibited HH signaling in multiple tumor cell lines (116). Although both small molecule inhibitors exhibit potent anti-HH signaling activities, their oral availability and pharmacological toxicities should be evaluated at the animal level.
SHH Inhibitors
The inhibition of HH proteins induces all Ptch-induced pro-apoptotic pathways (117). 5E1, a monoclonal antibody to SHH-N, has been widely used in biological experiments. Interestingly, most of the binding sites of 5E1 on SHH-N ligands overlap with HHIP (118). Similar to 5E1, Tolani et al. developed a new therapeutic antibody that targets the full-length SHH protein and SHH-N ligand to inhibit tumor growth (119). Owens et al. identified HL2-m5 (IC50: 230 nM), a macrocyclic peptide inhibitor with a high affinity for SHH ligands, which inhibited HH signaling in vitro without significant pharmacological toxicities (120). This compound should be further validated via animal experiments to assess its oral availability and safety.
Smo Inhibitors
Smo inhibitor resistance is the leading cause of progression in most HH-dependent tumor patients and is mainly attributed to mutations in the drug binding pocket of the Smo protein (121). In a phase I trial, patients with locally metastatic and progressing BCC previously treated with, or not with HH inhibitors exhibited good clinical responses to LY2940680 (Taladegib) and were well tolerated (122). Cyclopamine and its derivatives from natural compounds reverse HH signaling activation by targeting Smo, thereby limiting HH-dependent tumor cell growth (123). As a semisynthetic cyclopamine analogue, IPI-926 (Saridegib) addresses the issues of low potency and poor aqueous solubility of cyclopamine. Significantly, it inhibited tumor progression in HH-dependent medulloblastoma (MB) mice models with no obvious toxicity (124). IPI-926 has certain inhibitory activities against SmoD473H (Smo mutant), which is resistant to GDC-0449 (Vismodegib). When combined with gemcitabine, IPI-926 was shown to enhance anti-tumor activities of gemcitabine by increasing tumor vascular densities, which prolonged the survival outcomes of mice with pancreatic ductal adenocarcinoma (125). In a clinical phase I study involving basal cell carcinoma patients, despite differences in chemical structures between IPI-926 and Vismodegib, both exhibited similar sites of action on the Smo protein and shared same resistance mechanisms (126).
The FDA approved itraconazole for antifungal treatment, and its safety as well as side effects are well understood (111, 127). Compared to Vismodegib, Itraconazole inhibits Smo through different mechanisms. Itraconazole exerts significant anticancer activities in vivo, even targeting Vismodegib-resistant SmoD477G (110). Similar to itraconazole, posaconazole exerts anti-tumor activities by targeting Smo. Interestingly, posaconazole without triazole retains its antagonistic effects on HH signaling and disrupts the inhibitory effect on Cyp3A4 (128). The des-triazole derivatives, based on the posaconazole scaffold, were effective at inhibiting HH signaling and were well tolerated (128).
The presence of an oxysterol binding site in each of the seven transmembrane domains and extracellular cysteine-rich domain (CRD) of the Smo protein is necessary for HH signaling activation (129, 130). Cholesterol molecules can be transferred from the cytoplasm to oxysterol sites on the seven transmembrane regions or extracellular CRD via hydrophobic channels on Smo proteins, mediating Smo activation (129, 130). Therefore, the oxysterol binding site on the Smo protein is a potential HH signaling target. There is a need to determine the mechanisms through which the Smo protein integrates the information conveyed by the two oxysterol binding sites to determine the activities of HH signaling in vivo. As a Bcl-2 inhibitor, ABT-199 is approved by the FDA for treatment of chronic lymphocytic leukemia (ALL). ABT-199 (IC50: 215.9 nM) targets the extracellular CRD of Smo while acting as a potential competitive inhibitor of oxysterol, which inactivates HH signaling (131). In addition, ABT-199 was shown to target the drug binding pocket mutant and/or constitutively activated Smo proteins with excellent pharmacological properties and safety, therefore, its effects in patients with HH inhibitor resistance should be evaluated (131).
Multi-targeted HH inhibitors act on multiple components of HH signaling and/or inhibit multiple noncanonical HH pathways, exhibiting potent inhibitory anti-HH signaling effects. Lospinoso Severini et al. identified compound 22 (IC50: 7.1 μM), which contains an isoflavone scaffold, to be a multi-target HH inhibitor that inhibits MB cell growth by targeting Smo and Gli proteins (132). As a semisynthetic oxysterol analogue, Oxy210 (IC50: 1.83 μM) is a dual inhibitor of TGFβ and HH signaling, with excellent pharmacological properties, but should be further evaluated at the animal level (133). Li et al. and Zhu et al. determined that HH-13, HH-20 (IC50: <0.2 μM), and L-4 (IC50: 2.33 nM) act as potential Smo inhibitors by targeting SmoD473H, respectively (134–136). Moreover, L-4 showed high efficiency, good tolerability, and high oral bioavailability in ICR mice (134).
In conclusion, reliable HH inhibitors from FDA-approved drugs, such as itraconazole and ABT-199 have been screened and their toxic effects as well as side effects evaluated. The Oxysterol site of the Smo protein is essential for activation of HH signaling, and the development of Smo inhibitors, based on the Oxysterol site, can overcome Smo acquired resistance, with great potential for clinical applications.
Gli Inhibitors
Gli inhibitors target Smo downstream genes to overcome Smo acquired resistance and inhibit Gli-dependent noncanonical pathways, thereby achieving better clinical efficacies and improving patient clinical outcomes. GANT58 and GANT61 (IC50: 5 μM) are widely used in biological experiments by targeting the Gli1 transcription factor (137). Notably, GANT61 is more active against HH signaling than GANT58, which is mainly attributed to its ability to interfere with the binding of Gli1 to DNA (137). The Gli-activated but Smo-independent pathway is activated in some types of LSCC, including PI3K/AKT and RAS-MEK signaling. Therefore, GANT61 is more potent than Vismodegib in Gli-dependent LSCC patients (138).
Arsenic trioxide (ATO) (IC50: 2.7 μM), approved by the FDA for acute promyelocytic leukemia (APL), targeted the Gli1 transcription factor and exhibited significant cytotoxicity in mice models of MB and Ewing sarcoma, supporting further pharmacological evaluation in Gli1-dependent malignancies (139). ATO kills SCLC CSCs by downregulating CSCs-related genes, such as Sox2 and c-Myc at least in part by targeting HH signaling (140). The lack of efficacy of ATO in relapsed SCLC patients and SCLC patient-derived xenografts (PDX) mice in phase II clinical trials may be attributed to low proportions of SCLC CSCs and intermittent dosing regimens (141). The therapeutic benefits of ATO in combination with chemotherapeutic agents and daily continuous dosing regimens in SCLC patients should be investigated (141). Due to differences in study methods and different cell lines, it should be determined if the primary target of ATO is Gli1 or Gli2 (142). Pyrvinium (IC50: 10nM) is an FDA-approved casein kinase-1a (CK1a) agonist with high efficacies. Moreover, CK1a can also act as a negative regulator of HH signaling (143). Mechanistically, CK1a downregulates HH target genes by modulating their stability via phosphorylation of Gli transcriptional factors (143).
Studies have found that VDR signaling negatively regulates Gli1 expression through crosstalks with HH signaling (144–148). Vitamin D3-based derivative compounds 16, 21, and 22 modulated Gli1 proteins in vitro by targeting VDR signaling and HH signaling (149). However, the mechanisms through which VDR regulates HH signaling have not been elucidated, which requires further investigations.
Bromodomain (BRD), a class of conserved protein domains that specifically recognize acetylated lysine in histones, promote the enrichment of chromatin remodeling factors and transcription factors to specific gene transcription sites by binding acetylated lysine, altering the activities of RNA polymerase II, and regulating gene expressions (150, 151). Liu et al. developed non-selective BET inhibitors, compounds 25 and 35 (IC50: < 1nM) by further optimization of the BRD4 inhibitor, ABBV-075, which inhibited Gli protein expressions and overcame resistant Smo mutations in MB mice without apparent toxicity (152). Compared to compound 35, compound 25 exhibits stronger efficacies and better safety, however, it should be optimized to improve its pharmacokinetic parameters (152).
In patients with recurrent metastatic BCC, Itraconazole in combination with ATO (ATO-ITRA) reduced Gli1 mRNA levels by 75% from baseline levels (153). Three of the five patients treated with an ATO-ITRA regimen were stable for three months, but did not achieve tumor shrinkage, which was attributed to sequential dosing as well as lower doses. Higher doses and/or daily continuous dosing regimens should be used to evaluate the clinical efficacy of the ATO-ITRA regimen (153).
In conclusion, Gli inhibitors overcome Smo acquired resistance and inhibit Gli activatation via the Smo-independent noncanonical pathway, with robust anti-tumor activities in Gli-dependent tumors. Gli inhibitors (including ATO and GANT61) combined with Smo inhibitors (including Itraconazole and Taladegib) or chemotherapeutic agents represent a promising strategy for patients with HH-dependent tumors (Figure 5).
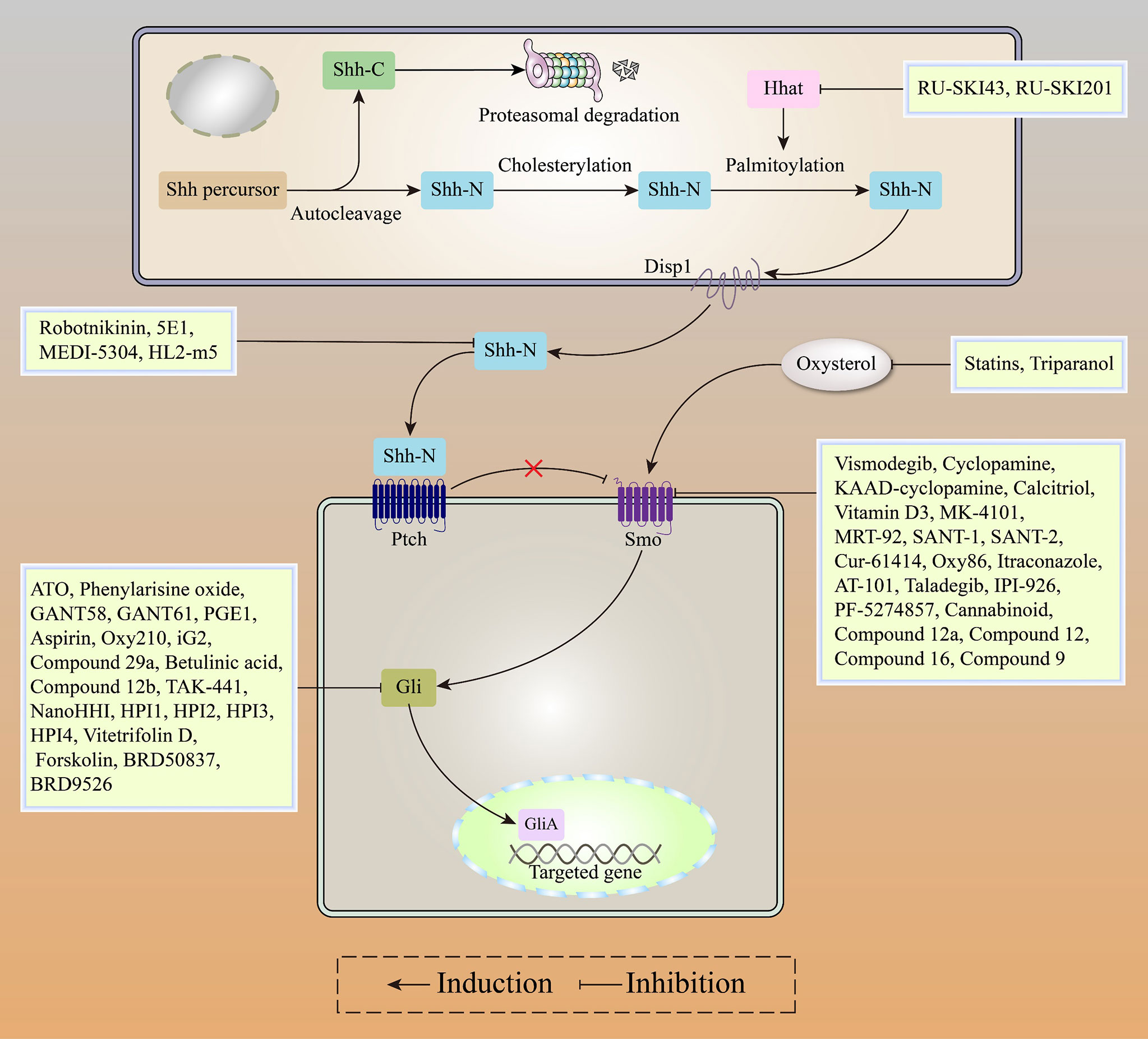
Figure 5 Recent advances in HH inhibitors, including HHat inhibitors, SHH inhibitors, Smo inhibitors, and Gli inhibitors.
The HH Pathway and Natural Products for Lung Cancer Therapy
Apart from being easily accessible, natural products have potent anti-inflammatory, anti-bacterial as well as anti-tumor activities (154, 155). Natural products inhibit cell proliferation, invasion, EMT processes as well as stemness features by targeting HH signaling in various malignancies, including lung cancer [Figure 6 (156, 157)].
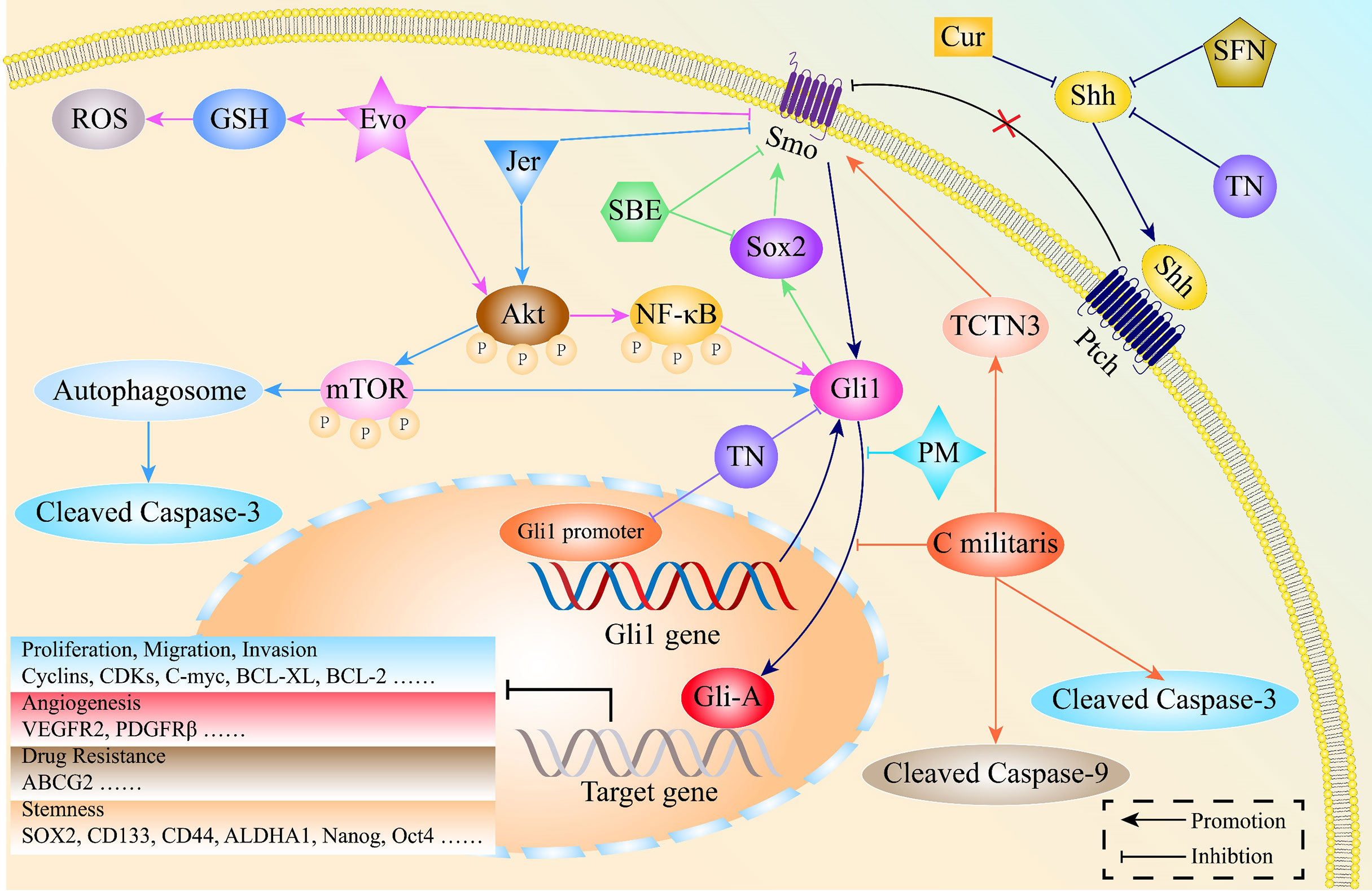
Figure 6 The mechanisms through which natural products exert potent anti-cancer activities by regulating the HH pathway in lung cancer. Jervine (Jer), Evodiamine (Evo), Scutellariabarbata D. Don extraction (SBE), Sulforaphane (SFN), Triptonide (TN), Curcumin (Cur), Pristimerin (PM), Cordyceps militaris Exerts (C militaris), Tectonic protein 3 (TCTN3).
Natural Products Induce Cell Apoptosis in Lung Cancer by Inhibiting the HH Pathway
Autophagy, a process of cellular self-degradation, removes damaged or redundant proteins as well as organelles. It plays an important role in maintaining intracellular homeostasis as well as tumorigenesis (158–161). Autophagy is a double-edged sword that promotes and inhibits tumor development (162–164). Lei et al. reported that Jervine triggers autophagy and increases the expressions of cleaved caspase 3, leading to NSCLC cell apoptosis by acting on AKT/mTOR and HH pathways (165).
The additional natural products that block cell proliferation include Evodiamine, Sulforaphane, and Cordyceps militaris. Evodiamine inhibits cell proliferation by modulation of the AKT/NF-κB and HH pathways in NSCLC (166). Cordyceps militaris induces NSCLC cell apoptosis by blocking Gli1 nuclear translocation via inhibiting the expressions of tectonic protein 3 (TCTN3) (167). In the PC9 cell line, Sulforaphane reversed lung cancer cell resistance to gefitinib by regulating HH signaling (168).
Natural Products Suppress Lung Cancer Cell Invasion and Metastasis by Inhibiting the HH Pathway
As a traditional Chinese medicine, Scutellariabarbata D. Don extraction (SBE)has been used in clinical management for many years because of its reliable and effective anti-cancer activities (169, 170). SBE downregulates the downstream components of HH signaling, leading to tumor cell cycle arrest and increasing lung cancer cell sensitivity to cisplatin (171).
Tumor angiogenesis is closely associated with multiple malignant behaviors of lung cancer, such as proliferation, invasion, and metastasis. Inhibition of tumor angiogenesis is of great clinical significance. However, angiogenesis inhibitors that target VEGF/VEGFR2 did not significantly improve the five-year survival rate for cancer patients (172, 173). Therefore, there is a need to develop novel angiogenesis inhibitors that lack toxic effects. Pristimerin, a compound isolated from Celastrus aculeatus Merr, exhibits multiple pharmacological activities, including anti-inflammatory, anti-microbial, anti-tumor, and anti-peroxidative effects (174, 175). In NSCLC, Pristimerin affects the maturation of tumor angiogenesis by regulating the translocation of its downstream gene (Gli1) to the nucleus through HH signaling (176).
Natural Products Eliminate Lung Cancer CSCs by Inhibiting the HH Pathway
Triptonide, a natural compound separated from Tripterygium wilfordii Hook, exhibits potent anti-inflammatory (177) and anti-tumor efficacies (178, 179). It has been shown to inhibit the proliferation, invasion, as well as migration of lung cancer cells and eliminates stem-like signatures by reducing the expression levels of Gli1 at both transcriptional and translational levels via inhibiting Gli1 promoter activities (180).
Curcumin is a potent compound isolated from the rootstalk of Curcuma longa (181, 182). It was shown to effectively eliminate various cancer stem cells and reduce their CSCs-like characteristics (183–185). Curcumin has powerful inhibitory effects on lung cancer CSCs in vitro. Mechanistically, Curcumin downregulates the expression levels of CSCs markers, including CD133, CD44, ALDHA1, Nanog and Oct4, by inhibiting the HH and Wnt/β-catenin pathways (186). In addition, SBE suppresses stemness-like phenotypes in NSCLC by targeting the Sox2/Smo/Gli1 positive feedback loop (187).
Conclusions and Future Directions
The HH pathway plays a pivotal role in lung cancer oncogenesis and development. The HH pathway is involved in drug resistance and CSCs. In resistant-EGFR-TKIs lung cancer cells, aberrant activations of the HH pathway are frequently observed. Therefore, the combination of HH inhibitors and EGFR-TKIs provide a novel treatment strategy for advanced lung cancer patients.
The noncanonical HH pathway, especially the Gli-activated but Smo-independent signaling, promotes the malignant progression of lung cancer, including cell proliferation, invasion, migration, metastasis, EMT and stemness features. Thus, it is necessary to block the noncanonical HH signaling in patients with HH-dependent tumors.
Additionally, we summarized recent findings regarding the molecular mechanisms of the HH pathway in promoting lung cancer progression and discussed the correlation between HH pathway-related proteins and prognostic outcomes for lung cancer patients. HH pathway-related proteins have been shown to predict prognostic outcomes in advanced NSCLC patients, and overexpressions of SHH and Gli1 are strongly associated with poor prognostic outcomes. Therefore, studies should focus on prognostic values of HH pathway-related proteins in patients with late-stage lung cancer.
Studies are aimed at developing novel Smo and Gli inhibitors. However, although Vismodegib and Sonidegib have shown good efficacies in clinical treatment, they remain at risk of drug resistance, which is mainly attributed to mutations in the drug binding pocket of the Smo protein. Itraconazole exerts potent anti-tumor activities in Vismodegib-resistant tumors because it has a different active site on Smo than Vismodegib. Furthermore, the safety, pharmacokinetics and toxicity of itraconazole as a traditional antifungal drug are well understood, resulting in significant cost saving in drug development. The clinical efficacy and toxicity of Itraconazole in treatment of lung cancer has been assessed. A Phase II study conducted by Rudin et al. investigated the implications of itraconazole in combination with pemetrexed in lung cancer therapy. The results showed that itraconazole significantly prolonged the overall survival outcomes of patients by up to 32 months, compared to pemetrexed alone (8 months), with mild adverse effects. Notably, the preclinical trial was designed based on the principle that itraconazole inhibits tumor angiogenesis. However, it was not determined whether itraconazole affects malignant progression by inhibiting lung cancer angiogenesis through the HH pathway. By targeting the downstream transcription factors of HH signaling, Gli inhibitors overcome Smo mutations and block Gli-activated but Smo-independent noncanonical signaling, with promising clinical applications in HH-dependent tumors. ATO, an FDA-approved drug for APL treatment, exerts potent antitumor activities by targeting Gli1/2 proteins and has excellent pharmacological properties. Itraconazole, in combination with ATO, significantly downregulated Gli1 mRNA expression levels and prolonged PFS in BCC patients (3 months). However, due to sequential dosing and lower doses, the ATO-ITRA regimen did not downsize the tumor. Studies on ATO-ITRA may allow us to test higher doses or continuous dosing regimens to assess clinical efficacies in HH-dependent tumor patients.
In addition, some natural compounds have been used in oncology treatment for many years, and are characterized by wide sources, low-toxicity and broad-spectrum anti-tumor properties. A phase II trial of whether curcumin in combination with Lovaza (made with fish oils) reduces the sizes of lung nodules will be conducted soon. Interestingly, despite the general disadvantages of poor oral bioavailability and low potency of some natural compounds such as curcumin, researchers have developed a number of nano-delivery systems for curcumin, which have greatly improved its aqueous solubility and anti-tumor potency. In addition, various natural compounds can act on multiple targets and are involved in extensive oncogenic signaling networks, which should be investigated further. In future, more natural extracts will be developed, but further optimization and modification for these compounds to enhance their anti-tumor activities, oral bioavailability and reduce drug toxicity will be a great importance.
Author Contributions
Z-GS and NZ designed the work. CM wrote the manuscript. KH and IU prepared the figures and tables. Q-KZ drafted and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Shandong Provincial Natural Science Foundation (grant no. ZR2020MH204), the 19th batch of science and technology innovation development plan of Jinan in 2020 (Clinical medicine science and technology innovation plan, grant no.202019032), and the second group of science and technology projects of Jinan Health Committee (grant no. 2020-3-15).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Ashrafizadeh M, Najafi M, Makvandi P, Zarrabi A, Farkhondeh T, Samarghandian S. Versatile Role of Curcumin and its Derivatives in Lung Cancer Therapy. J Cell Physiol (2020) 235(12):9241–68. doi: 10.1002/jcp.29819
3. Ashrafizadeh M, Mirzaei S, Hushmandi K, Rahmanian V, Zabolian A, Raei M, et al. Therapeutic Potential of AMPK Signaling Targeting in Lung Cancer: Advances, Challenges and Future Prospects. Life Sci (2021) 278:119649. doi: 10.1016/j.lfs.2021.119649
4. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2016. CA Cancer J Clin (2016) 66(1):7–30. doi: 10.3322/caac.21332
5. Abadi AJ, Zarrabi A, Gholami MH, Mirzaei S, Hashemi F, Zabolian A, et al. Small in Size, But Large in Action: microRNAs as Potential Modulators of PTEN in Breast and Lung Cancers. Biomolecules (2021) 11(2):304. doi: 10.3390/biom11020304
6. Ashrafizadeh M, Zarrabi A, Hushmandi K, Hashemi F, Moghadam ER, Owrang M, et al. Lung Cancer Cells and Their Sensitivity/Resistance to Cisplatin Chemotherapy: Role of microRNAs and Upstream Mediators. Cell Signal (2021) 78:109871. doi: 10.1016/j.cellsig.2020.109871
7. Herbst RS, Morgensztern D, Boshoff C. The Biology and Management of Non-Small Cell Lung Cancer. Nature (2018) 553(7689):446–54. doi: 10.1038/nature25183
8. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, et al. Lung Cancer: Current Therapies and New Targeted Treatments. Lancet (2017) 389(10066):299–311. doi: 10.1016/s0140-6736(16)30958-8
9. Chen R, Manochakian R, James L, Azzouqa AG, Shi H, Zhang Y, et al. Emerging Therapeutic Agents for Advanced Non-Small Cell Lung Cancer. J Hematol Oncol (2020) 13(1):58. doi: 10.1186/s13045-020-00881-7
10. Yang S, Zhang Z, Wang Q. Emerging Therapies for Small Cell Lung Cancer. J Hematol Oncol (2019) 12(1):47. doi: 10.1186/s13045-019-0736-3
11. Custodio A, Méndez M, Provencio M. Targeted Therapies for Advanced Non-Small-Cell Lung Cancer: Current Status and Future Implications. Cancer Treat Rev (2012) 38(1):36–53. doi: 10.1016/j.ctrv.2011.04.001
12. Chi S, Huang S, Li C, Zhang X, He N, Bhutani MS, et al. Activation of the Hedgehog Pathway in a Subset of Lung Cancers. Cancer Lett (2006) 244(1):53–60. doi: 10.1016/j.canlet.2005.11.036
13. Beachy PA, Karhadkar SS, Berman DM. Tissue Repair and Stem Cell Renewal in Carcinogenesis. Nature (2004) 432(7015):324–31. doi: 10.1038/nature03100
14. Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog Signalling Within Airway Epithelial Progenitors and in Small-Cell Lung Cancer. Nature (2003) 422(6929):313–7. doi: 10.1038/nature01493
15. Huang S, Yang L, An Y, Ma X, Zhang C, Xie G, et al. Expression of Hedgehog Signaling Molecules in Lung Cancer. Acta Histochem (2011) 113(5):564–9. doi: 10.1016/j.acthis.2010.06.003
16. Lin C-C, Kuo IY, Wu L-T, Kuan W-H, Liao S-Y, Jen J, et al. Dysregulated Kras/YY1/ZNF322A/SHH Transcriptional Axis Enhances Neo-Angiogenesis to Promote Lung Cancer Progression. Theranostics (2020) 10(22):10001–15. doi: 10.7150/thno.47491
17. Yue D, Li H, Che J, Zhang Y, Tseng H-HK, Jin JQ, et al. Hedgehog/Gli Promotes Epithelial-Mesenchymal Transition in Lung Squamous Cell Carcinomas. J Exp Clin Cancer Res: CR (2014) 33(1):34. doi: 10.1186/1756-9966-33-34
18. Della Corte CM, Bellevicine C, Vicidomini G, Vitagliano D, Malapelle U, Accardo M, et al. SMO Gene Amplification and Activation of the Hedgehog Pathway as Novel Mechanisms of Resistance to Anti-Epidermal Growth Factor Receptor Drugs in Human Lung Cancer. Clin Cancer Res: an Off J Am Assoc Cancer Res (2015) 21(20):4686–97. doi: 10.1158/1078-0432.CCR-14-3319
19. Mehlman C, Takam Kamga P, Costantini A, Julié C, Dumenil C, Dumoulin J, et al. Baseline Hedgehog Pathway Activation and Increase of Plasma Wnt1 Protein Are Associated With Resistance to Immune Checkpoint Inhibitors in Advanced Non-Small-Cell Lung Cancer. Cancers (2021) 13(5):1107. doi: 10.3390/cancers13051107
20. Nüsslein-Volhard C, Wieschaus E. Mutations Affecting Segment Number and Polarity in Drosophila. Nature (1980) 287(5785):795–801. doi: 10.1038/287795a0
21. Varjosalo M, Taipale J. Hedgehog: Functions and Mechanisms. Genes Dev (2008) 22(18):2454–72. doi: 10.1101/gad.1693608
22. Briscoe J, Thérond PP. The Mechanisms of Hedgehog Signalling and its Roles in Development and Disease. Nat Rev Mol Cell Biol (2013) 14(7):416–29. doi: 10.1038/nrm3598
23. Ruiz i Altaba A. Catching a Gli-Mpse of Hedgehog. Cell (1997) 90(2):193–6. doi: 10.1016/s0092-8674(00)80325-6
24. Taipale J, Cooper MK, Maiti T, Beachy PA. Patched Acts Catalytically to Suppress the Activity of Smoothened. Nature (2002) 418(6900):892–7. doi: 10.1038/nature00989
25. Zhang Q, Shi Q, Chen Y, Yue T, Li S, Wang B, et al. Multiple Ser/Thr-Rich Degrons Mediate the Degradation of Ci/Gli by the Cul3-HIB/SPOP E3 Ubiquitin Ligase. Proc Natl Acad Sci USA (2009) 106(50):21191–6. doi: 10.1073/pnas.0912008106
26. Kovacs JJ, Whalen EJ, Liu R, Xiao K, Kim J, Chen M, et al. Beta-Arrestin-Mediated Localization of Smoothened to the Primary Cilium. Science (2008) 320(5884):1777–81. doi: 10.1126/science.1157983
27. Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The Output of Hedgehog Signaling is Controlled by the Dynamic Association Between Suppressor of Fused and the Gli Proteins. Genes Dev (2010) 24(7):670–82. doi: 10.1101/gad.1902910
28. Liu A, Wang B, Niswander LA. Mouse Intraflagellar Transport Proteins Regulate Both the Activator and Repressor Functions of Gli Transcription Factors. Dev (Cambridge Engl) (2005) 132(13):3103–11. doi: 10.1242/dev.01894
29. Vokes SA, Ji H, Wong WH, McMahon AP. A Genome-Scale Analysis of the Cis-Regulatory Circuitry Underlying Sonic Hedgehog-Mediated Patterning of the Mammalian Limb. Genes Dev (2008) 22(19):2651–63. doi: 10.1101/gad.1693008
30. Zhuo H, Zhou D, Wang Y, Mo H, Yu Y, Liu Y. Sonic Hedgehog Selectively Promotes Lymphangiogenesis After Kidney Injury Through Noncanonical Pathway. Am J Physiol Renal Physiol (2019) 317(4):F1022–F33. doi: 10.1152/ajprenal.00077.2019
31. P. M MM, Shahi MH, Tayyab M, Farheen S, Khanam N, Tabassum S, et al. Cadmium-Induced Neurodegeneration and Activation of Noncanonical Sonic Hedgehog Pathway in Rat Cerebellum. J Biochem Mol Toxicol (2019) 33(4):e22274. doi: 10.1002/jbt.22274
32. Faria A, Akyala AI, Parikh K, Brüggemann LW, Spek CA, Cao W, et al. Smoothened-Dependent and -Independent Pathways in Mammalian Noncanonical Hedgehog Signaling. J Biol Chem (2019) 294(25):9787–98. doi: 10.1074/jbc.RA119.007956
33. Akhshi T, Trimble WS. A Non-Canonical Hedgehog Pathway Initiates Ciliogenesis and Autophagy. J Cell Biol (2021) 220(1):e202004179. doi: 10.1083/jcb.202004179
34. Yao CD, Haensel D, Gaddam S, Patel T, Atwood SX, Sarin KY, et al. AP-1 and TGFß Cooperativity Drives Non-Canonical Hedgehog Signaling in Resistant Basal Cell Carcinoma. Nat Commun (2020) 11(1):5079. doi: 10.1038/s41467-020-18762-5
35. Jin R, Wang X, Zang R, Liu C, Zheng S, Li H, et al. Desmoglein-2 Modulates Tumor Progression and Osimertinib Drug Resistance Through the EGFR/Src/PAK1 Pathway in Lung Adenocarcinoma. Cancer Lett (2020) 483:46–58. doi: 10.1016/j.canlet.2020.04.001
36. Fu J, Li L, Huo D, Zhi S, Yang R, Yang B, et al. Astrocyte-Derived TGFβ1 Facilitates Blood-Brain Barrier Function via Non-Canonical Hedgehog Signaling in Brain Microvascular Endothelial Cells. Brain Sci (2021) 11(1):77. doi: 10.3390/brainsci11010077
37. Ehe BK, Lamson DR, Tarpley M, Onyenwoke RU, Graves LM, Williams KP. Identification of a DYRK1A-Mediated Phosphorylation Site Within the Nuclear Localization Sequence of the Hedgehog Transcription Factor GLI1. Biochem Biophys Res Commun (2017) 491(3):767–72. doi: 10.1016/j.bbrc.2017.07.107
38. Po A, Silvano M, Miele E, Capalbo C, Eramo A, Salvati V, et al. Noncanonical GLI1 Signaling Promotes Stemness Features and In Vivo Growth in Lung Adenocarcinoma. Oncogene (2017) 36(32):4641–52. doi: 10.1038/onc.2017.91
39. Mizuarai S, Kawagishi A, Kotani H. Inhibition of p70s6K2 Down-Regulates Hedgehog/GLI Pathway in Non-Small Cell Lung Cancer Cell Lines. Mol Cancer (2009) 8:44. doi: 10.1186/1476-4598-8-44
40. Pietrobono S, Gagliardi S, Stecca B. Non-Canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Transcription Factors Beyond Smoothened. Front Genet (2019) 10:556. doi: 10.3389/fgene.2019.00556
41. Huang S, He J, Zhang X, Bian Y, Yang L, Xie G, et al. Activation of the Hedgehog Pathway in Human Hepatocellular Carcinomas. Carcinogenesis (2006) 27(7):1334–40. doi: 10.1093/carcin/bgi378
42. Ma X, Chen K, Huang S, Zhang X, Adegboyega PA, Evers BM, et al. Frequent Activation of the Hedgehog Pathway in Advanced Gastric Adenocarcinomas. Carcinogenesis (2005) 26(10):1698–705. doi: 10.1093/carcin/bgi130
43. Chen C, Breslin MB, Lan MS. Sonic Hedgehog Signaling Pathway Promotes INSM1 Transcription Factor in Neuroendocrine Lung Cancer. Cell Signal (2018) 46:83–91. doi: 10.1016/j.cellsig.2018.02.014
44. Sun X, Sun B, Cui M, Zhou Z. HERC4 Exerts an Anti-Tumor Role Through Destabilizing the Oncoprotein Smo. Biochem Biophys Res Commun (2019) 513(4):1013–8. doi: 10.1016/j.bbrc.2019.04.113
45. Song K, Zheng G, Zhao Y. Liver Kinase B1 Suppresses Growth of Lung Cancer Cells Through Sonic Hedgehog Signaling Pathway. Cell Biol Int (2018) 42(8):994–1005. doi: 10.1002/cbin.10965
46. Li B, Huang W, Cao N, Lou G. Forkhead-Box R2 Promotes Metastasis and Growth by Stimulating Angiogenesis and Activating Hedgehog Signaling Pathway in Ovarian Cancer. J Cell Biochem (2018) 119(9):7780–9. doi: 10.1002/jcb.27148
47. Sun C, Zhang Z, He P, Zhou Y, Xie X. Involvement of PI3K/Akt Pathway in the Inhibition of Hepatocarcinoma Cell Invasion and Metastasis Induced by SASH1 Through Downregulating SHH-Gli1 Signaling. Int J Biochem Cell Biol (2017) 89:95–100. doi: 10.1016/j.biocel.2017.06.006
48. Xu H, Dun S, Gao Y, Ming J, Hui L, Qiu X. TMEM107 Inhibits EMT and Invasion of NSCLC Through Regulating the Hedgehog Pathway. Thorac Cancer (2020) 12(1):79–89. doi: 10.1111/1759-7714.13715
49. Li H, Yue D, Jin JQ, Woodard GA, Tolani B, Luh TM, et al. Gli Promotes Epithelial-Mesenchymal Transition in Human Lung Adenocarcinomas. Oncotarget (2016) 7(49):80415–25. doi: 10.18632/oncotarget.11246
50. Jian W, Bai Y, Li X, Kang J, Lei Y, Xue Y. Phosphatidylethanolamine-Binding Protein 4 Promotes the Epithelial-to-Mesenchymal Transition in non-Small Cell Lung Cancer Cells by Activating the Sonic Hedgehog Signaling Pathway. J Cell Biochem (2019) 120(4):5386–95. doi: 10.1002/jcb.27817
51. Jia J, Martin TA, Ye L, Meng L, Xia N, Jiang WG, et al. Fibroblast Activation Protein-α Promotes the Growth and Migration of Lung Cancer Cells via the PI3K and Sonic Hedgehog Pathways. Int J Mol Med (2018) 41(1):275–83. doi: 10.3892/ijmm.2017.3224
52. Queiroz KCS, Spek CA, Peppelenbosch MP. Targeting Hedgehog Signaling and Understanding Refractory Response to Treatment With Hedgehog Pathway Inhibitors. Drug Resist Updates: Rev Commentaries Antimicrob Anticancer Chemother (2012) 15(4):211–22. doi: 10.1016/j.drup.2012.05.002
53. Zhao J-G, Wang J-F, Feng J-F, Jin X-Y, Ye W-L. HHIP Overexpression Inhibits the Proliferation, Migration and Invasion of Non-Small Cell Lung Cancer. PloS One (2019) 14(11):e0225755. doi: 10.1371/journal.pone.0225755
54. Pan J, Qin Y, Zhang M. HPIP Promotes non-Small Cell Lung Cancer Cell Proliferation, Migration and Invasion Through Regulation of the Sonic Hedgehog Signaling Pathway. Biomed Pharmacother = Biomed Pharmacother (2016) 77:176–81. doi: 10.1016/j.biopha.2015.12.012
55. Yang B, Miao S, Li Y. SCUBE2 Inhibits the Proliferation, Migration and Invasion of Human Non-Small Cell Lung Cancer Cells Through Regulation of the Sonic Hedgehog Signaling Pathway. Gene (2018) 672:143–9. doi: 10.1016/j.gene.2018.06.012
56. Cheung WKC, Zhao M, Liu Z, Stevens LE, Cao PD, Fang JE, et al. Control of Alveolar Differentiation by the Lineage Transcription Factors GATA6 and HOPX Inhibits Lung Adenocarcinoma Metastasis. Cancer Cell (2013) 23(6):725–38. doi: 10.1016/j.ccr.2013.04.009
57. Xu L, Deng S, Xiong H, Shi W, Luo S, Chen L. GATA-6 Transcriptionally Inhibits SHH to Repress Cell Proliferation and Migration in Lung Squamous Cell Carcinoma. Int J Biochem Cell Biol (2019) 115:105591. doi: 10.1016/j.biocel.2019.105591
58. Liu X, Liu J, Zhang X, Tong Y, Gan X. MiR-520b Promotes the Progression of Non-Small Cell Lung Cancer Through Activating Hedgehog Pathway. J Cell Mol Med (2019) 23(1):205–15. doi: 10.1111/jcmm.13909
59. Abramovich C, Chavez EA, Lansdorp PM, Humphries RK. Functional Characterization of Multiple Domains Involved in the Subcellular Localization of the Hematopoietic Pbx Interacting Protein (HPIP). Oncogene (2002) 21(44):6766–71. doi: 10.1038/sj.onc.1205784
60. Li Y-G, Liang N-X, Qin Y-Z, Ma D-J, Huang C-J, Liu L, et al. Effects of RNAi-Mediated TUSC3 Silencing on Radiation-Induced Autophagy and Radiation Sensitivity of Human Lung Adenocarcinoma Cell Line A549 Under Hypoxic Condition. Tumour Biol: J Int Soc Oncodev Biol Med (2016). doi: 10.1007/s13277-016-5458-3
61. Gu Y, Pei X, Ren Y, Cai K, Guo K, Chen J, et al. Oncogenic Function of TUSC3 in non-Small Cell Lung Cancer Is Associated With Hedgehog Signalling Pathway. Biochim Et Biophys Acta Mol Basis Dis (2017) 1863(7):1749–60. doi: 10.1016/j.bbadis.2017.05.005
62. Wei Q, Chen Z-H, Wang L, Zhang T, Duan L, Behrens C, et al. LZTFL1 Suppresses Lung Tumorigenesis by Maintaining Differentiation of Lung Epithelial Cells. Oncogene (2016) 35(20):2655–63. doi: 10.1038/onc.2015.328
63. Feizabadi MS. Modeling Multi-Mutation and Drug Resistance: Analysis of Some Case Studies. Theor Biol Med Model (2017) 14(1):6. doi: 10.1186/s12976-017-0052-y
64. Lin E-H, Kao Y-R, Lin C-A, Kuo T-Y, Yang S-P, Hsu C-F, et al. Hedgehog Pathway Maintains Cell Survival Under Stress Conditions, and Drives Drug Resistance in Lung Adenocarcinoma. Oncotarget (2016) 7(17):24179–93. doi: 10.18632/oncotarget.8253
65. Della Corte CM, Malapelle U, Vigliar E, Pepe F, Troncone G, Ciaramella V, et al. Efficacy of Continuous EGFR-Inhibition and Role of Hedgehog in EGFR Acquired Resistance in Human Lung Cancer Cells With Activating Mutation of EGFR. Oncotarget (2017) 8(14):23020–32. doi: 10.18632/oncotarget.15479
66. Bai X-Y, Zhang X-C, Yang S-Q, An S-J, Chen Z-H, Su J, et al. Blockade of Hedgehog Signaling Synergistically Increases Sensitivity to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancer Cell Lines. PloS One (2016) 11(3):e0149370. doi: 10.1371/journal.pone.0149370
67. Wang H, Yan C, Shi X, Zheng J, Deng L, Yang L, et al. MicroRNA-575 Targets BLID to Promote Growth and Invasion of Non-Small Cell Lung Cancer Cells. FEBS Lett (2015) 589(7):805–11. doi: 10.1016/j.febslet.2015.02.013
68. Yu G, Zhong N, Chen G, Huang B, Wu S. Downregulation of PEBP4, a Target of miR-34a, Sensitizes Drug-Resistant Lung Cancer Cells. Tumour Biol: J Int Soc Oncodev Biol Med (2014) 35(10):10341–9. doi: 10.1007/s13277-014-2284-3
69. Seidl C, Panzitt K, Bertsch A, Brcic L, Schein S, Mack M, et al. MicroRNA-182-5p Regulates Hedgehog Signaling Pathway and Chemosensitivity of Cisplatin-Resistant Lung Adenocarcinoma Cells via Targeting GLI2. Cancer Lett (2020) 469:266–76. doi: 10.1016/j.canlet.2019.10.044
70. Oren O, Smith BD. Eliminating Cancer Stem Cells by Targeting Embryonic Signaling Pathways. Stem Cell Rev Rep (2017) 13(1):17–23. doi: 10.1007/s12015-016-9691-3
71. Koury J, Zhong L, Hao J. Targeting Signaling Pathways in Cancer Stem Cells for Cancer Treatment. Stem Cells Int (2017) 2017:2925869. doi: 10.1155/2017/2925869
72. Xu M, Gong A, Yang H, George SK, Jiao Z, Huang H, et al. Sonic Hedgehog-Glioma Associated Oncogene Homolog 1 Signaling Enhances Drug Resistance in CD44(+)/Musashi-1(+) Gastric Cancer Stem Cells. Cancer Lett (2015) 369(1):124–33. doi: 10.1016/j.canlet.2015.08.005
73. Li C, Du Y, Yang Z, He L, Wang Y, Hao L, et al. GALNT1-Mediated Glycosylation and Activation of Sonic Hedgehog Signaling Maintains the Self-Renewal and Tumor-Initiating Capacity of Bladder Cancer Stem Cells. Cancer Res (2016) 76(5):1273–83. doi: 10.1158/0008-5472.CAN-15-2309
74. Giroux Leprieur E, Tolani B, Li H, Leguay F, Hoang NT, Acevedo LA, et al. Membrane-Bound Full-Length Sonic Hedgehog Identifies Cancer Stem Cells in Human non-Small Cell Lung Cancer. Oncotarget (2017) 8(61):103744–57. doi: 10.18632/oncotarget.21781
75. Chen W-J, Huang R-FS. Low-Folate Stress Reprograms Cancer Stem Cell-Like Potentials and Bioenergetics Metabolism Through Activation of mTOR Signaling Pathway to Promote In Vitro Invasion and In Vivo Tumorigenicity of Lung Cancers. J Nutr Biochem (2018) 53:28–38. doi: 10.1016/j.jnutbio.2017.10.001
76. Kim HR, Kim DJ, Kang DR, Lee JG, Lim SM, Lee CY, et al. Fibroblast Growth Factor Receptor 1 Gene Amplification Is Associated With Poor Survival and Cigarette Smoking Dosage in Patients With Resected Squamous Cell Lung Cancer. J Clin Oncol: Off J Am Soc Clin Oncol (2013) 31(6):731–7. doi: 10.1200/JCO.2012.43.8622
77. Seo AN, Jin Y, Lee HJ, Sun P-L, Kim H, Jheon S, et al. FGFR1 Amplification is Associated With Poor Prognosis and Smoking in Non-Small-Cell Lung Cancer. Virchows Archiv: Int J Pathol (2014) 465(5):547–58. doi: 10.1007/s00428-014-1634-2
78. Ji W, Yu Y, Li Z, Wang G, Li F, Xia W, et al. FGFR1 Promotes the Stem Cell-Like Phenotype of FGFR1-Amplified Non-Small Cell Lung Cancer Cells Through the Hedgehog Pathway. Oncotarget (2016) 7(12):15118–34. doi: 10.18632/oncotarget.7701
79. Chang WH, Lai AG. Aberrations in Notch-Hedgehog Signalling Reveal Cancer Stem Cells Harbouring Conserved Oncogenic Properties Associated With Hypoxia and Immunoevasion. Br J Cancer (2019) 121(8):666–78. doi: 10.1038/s41416-019-0572-9
80. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
81. Saito R-A, Micke P, Paulsson J, Augsten M, Peña C, Jönsson P, et al. Forkhead Box F1 Regulates Tumor-Promoting Properties of Cancer-Associated Fibroblasts in Lung Cancer. Cancer Res (2010) 70(7):2644–54. doi: 10.1158/0008-5472.CAN-09-3644
82. Bermudez O, Hennen E, Koch I, Lindner M, Eickelberg O. Gli1 Mediates Lung Cancer Cell Proliferation and Sonic Hedgehog-Dependent Mesenchymal Cell Activation. PloS One (2013) 8(5):e63226. doi: 10.1371/journal.pone.0063226
83. Chen W, Tang T, Eastham-Anderson J, Dunlap D, Alicke B, Nannini M, et al. Canonical Hedgehog Signaling Augments Tumor Angiogenesis by Induction of VEGF-A in Stromal Perivascular Cells. Proc Natl Acad Sci USA (2011) 108(23):9589–94. doi: 10.1073/pnas.1017945108
84. Choe C, Shin YS, Kim SH, Jeon MJ, Choi SJ, Lee J, et al. Tumor-Stromal Interactions With Direct Cell Contacts Enhance Motility of Non-Small Cell Lung Cancer Cells Through the Hedgehog Signaling Pathway. Anticancer Res (2013) 33(9):3715–23.
85. Choe C, Shin Y-S, Kim C, Choi S-J, Lee J, Kim SY, et al. Crosstalk With Cancer-Associated Fibroblasts Induces Resistance of non-Small Cell Lung Cancer Cells to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibition. OncoTargets Ther (2015) 8:3665–78. doi: 10.2147/OTT.S89659
86. Petty AJ, Li A, Wang X, Dai R, Heyman B, Hsu D, et al. Hedgehog Signaling Promotes Tumor-Associated Macrophage Polarization to Suppress Intratumoral CD8+ T Cell Recruitment. J Clin Invest (2019) 129(12):5151–62. doi: 10.1172/JCI128644
87. Petty AJ, Dai R, Lapalombella R, Baiocchi RA, Benson DM, Li Z, et al. Hedgehog-Induced PD-L1 on Tumor-Associated Macrophages Is Critical for Suppression of Tumor-Infiltrating CD8+ T Cell Function. JCI Insight (2021) 6(6):e146707. doi: 10.1172/jci.insight.146707
88. Zhang X, Zhu M, Hong Z, Chen C. Co-Culturing Polarized M2 Thp-1-Derived Macrophages Enhance Stemness of Lung Adenocarcinoma A549 Cells. Ann Trans Med (2021) 9(8):709. doi: 10.21037/atm-21-1256
89. Xu L, Xiong H, Shi W, Zhou F, Zhang M, Hu G, et al. Differential Expression of Sonic Hedgehog in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. Neoplasma (2019) 66(5):839–46. doi: 10.4149/neo_2018_181228N1002
90. Bai X-Y, Lin J-Y, Zhang X-C, Xie Z, Yan H-H, Chen Z-H, et al. High Expression of Truncated GLI3 is Associated With Poor Overall Survival in Patients With non-Small Cell Lung Cancer. Cancer Biomark: Section A Dis Markers (2013) 13(1):37–47. doi: 10.3233/CBM-130312
91. Hwang J, Kang MH, Yoo YA, Quan YH, Kim HK, Oh SC, et al. The Effects of Sonic Hedgehog Signaling Pathway Components on Non-Small-Cell Lung Cancer Progression and Clinical Outcome. World J Surg Oncol (2014) 12:268. doi: 10.1186/1477-7819-12-268
92. Raz G, Allen KE, Kingsley C, Cherni I, Arora S, Watanabe A, et al. Hedgehog Signaling Pathway Molecules and ALDH1A1 Expression in Early-Stage non-Small Cell Lung Cancer. Lung Cancer (Amsterdam Netherlands) (2012) 76(2):191–6. doi: 10.1016/j.lungcan.2011.10.015
93. Savani M, Guo Y, Carbone DP, Csiki I. Sonic Hedgehog Pathway Expression in non-Small Cell Lung Cancer. Ther Adv Med Oncol (2012) 4(5):225–33. doi: 10.1177/1758834012450362
94. Cui Y, Cui C-A, Yang Z-T, Ni W-D, Jin Y, Xuan Y-H. Gli1 Expression in Cancer Stem-Like Cells Predicts Poor Prognosis in Patients With Lung Squamous Cell Carcinoma. Exp Mol Pathol (2017) 102(2):347–53. doi: 10.1016/j.yexmp.2017.03.004
95. Ishikawa M, Sonobe M, Imamura N, Sowa T, Shikuma K, Date H. Expression of the GLI Family Genes Is Associated With Tumor Progression in Advanced Lung Adenocarcinoma. World J Surg Oncol (2014) 12:253. doi: 10.1186/1477-7819-12-253
96. Kim JE, Kim H, Choe J-Y, Sun P, Jheon S, Chung J-H. High Expression of Sonic Hedgehog Signaling Proteins Is Related to the Favorable Outcome, EGFR Mutation, and Lepidic Predominant Subtype in Primary Lung Adenocarcinoma. Ann Surg Oncol (2013) 20 Suppl 3:S570–6. doi: 10.1245/s10434-013-3022-6
97. Kozirovskis V, Zandberga E, Magone M, Purkalne G, Linē A, Vikmanis U. High Expression of GLI1 Is Associated With Better Survival in Advanced SCLC. Exp Oncol (2020) 42(1):75–7. doi: 10.32471/exp-oncology.2312-8852.vol-42-no-1.14266
98. Lim S, Lim SM, Kim MJ, Park SY, Kim JH. Sonic Hedgehog Pathway as the Prognostic Marker in Patients With Extensive Stage Small Cell Lung Cancer. Yonsei Med J (2019) 60(10):898–904. doi: 10.3349/ymj.2019.60.10.898
99. Jian-Hui C, Er-Tao Z, Si-Le C, Hui W, Kai-Ming W, Xin-Hua Z, et al. CD44, Sonic Hedgehog, and Gli1 Expression Are Prognostic Biomarkers in Gastric Cancer Patients After Radical Resection. Gastroenterol Res Pract (2016) 2016:1013045. doi: 10.1155/2016/1013045
100. Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, et al. Efficacy and Safety of Vismodegib in Advanced Basal-Cell Carcinoma. N Engl J Med (2012) 366(23):2171–9. doi: 10.1056/NEJMoa1113713
101. Frampton JE, Basset-Séguin N. Vismodegib: A Review in Advanced Basal Cell Carcinoma. Drugs (2018) 78(11):1145–56. doi: 10.1007/s40265-018-0948-9
102. Migden MR, Guminski A, Gutzmer R, Dirix L, Lewis KD, Combemale P, et al. Treatment With Two Different Doses of Sonidegib in Patients With Locally Advanced or Metastatic Basal Cell Carcinoma (BOLT): A Multicentre, Randomised, Double-Blind Phase 2 Trial. Lancet Oncol (2015) 16(6):716–28. doi: 10.1016/s1470-2045(15)70100-2
103. Migden MR, Chang ALS, Dirix L, Stratigos AJ, Lear JT. Emerging Trends in the Treatment of Advanced Basal Cell Carcinoma. Cancer Treat Rev (2018) 64:1–10. doi: 10.1016/j.ctrv.2017.12.009
104. Brancaccio G, Pea F, Moscarella E, Argenziano G. Sonidegib for the Treatment of Advanced Basal Cell Carcinoma. Front Oncol (2020) 10:582866. doi: 10.3389/fonc.2020.582866
105. Hoy SM. Glasdegib: First Global Approval. Drugs (2019) 79(2):207–13. doi: 10.1007/s40265-018-1047-7
106. Norsworthy KJ, By K, Subramaniam S, Zhuang L, Del Valle PL, Przepiorka D, et al. FDA Approval Summary: Glasdegib for Newly Diagnosed Acute Myeloid Leukemia. Clin Cancer Res (2019) 25(20):6021–5. doi: 10.1158/1078-0432.Ccr-19-0365
107. Brinkhuizen T, Reinders MG, van Geel M, Hendriksen AJ, Paulussen AD, Winnepenninckx VJ, et al. Acquired Resistance to the Hedgehog Pathway Inhibitor Vismodegib Due to Smoothened Mutations in Treatment of Locally Advanced Basal Cell Carcinoma. J Am Acad Dermatol (2014) 71(5):1005–8. doi: 10.1016/j.jaad.2014.08.001
108. Katoh M. Genomic Testing, Tumor Microenvironment and Targeted Therapy of Hedgehog-Related Human Cancers. Clin Sci (Lond) (2019) 133(8):953–70. doi: 10.1042/cs20180845
109. Kim J, Tang JY, Gong R, Kim J, Lee JJ, Clemons KV, et al. Itraconazole, a Commonly Used Antifungal That Inhibits Hedgehog Pathway Activity and Cancer Growth. Cancer Cell (2010) 17(4):388–99. doi: 10.1016/j.ccr.2010.02.027
110. Kim J, Aftab BT, Tang JY, Kim D, Lee AH, Rezaee M, et al. Itraconazole and Arsenic Trioxide Inhibit Hedgehog Pathway Activation and Tumor Growth Associated With Acquired Resistance to Smoothened Antagonists. Cancer Cell (2013) 23(1):23–34. doi: 10.1016/j.ccr.2012.11.017
111. Pace JR, Teske KA, Chau LQ, Dash RC, Zaino AM, Wechsler-Reya RJ, et al. Structure-Activity Relationships for Itraconazole-Based Triazolone Analogues as Hedgehog Pathway Inhibitors. J Med Chem (2019) 62(8):3873–85. doi: 10.1021/acs.jmedchem.8b01283
112. Rudin CM, Brahmer JR, Juergens RA, Hann CL, Ettinger DS, Sebree R, et al. Phase 2 Study of Pemetrexed and Itraconazole as Second-Line Therapy for Metastatic Nonsquamous Non–Small-Cell Lung Cancer. J Thorac Oncol (2013) 8(5):619–23. doi: 10.1097/JTO.0b013e31828c3950
113. Mohamed AW, Elbassiouny M, Elkhodary DA, Shawki MA, Saad AS. The Effect of Itraconazole on the Clinical Outcomes of Patients With Advanced non-Small Cell Lung Cancer Receiving Platinum-Based Chemotherapy: A Randomized Controlled Study. Med Oncol (2021) 38(3):23. doi: 10.1007/s12032-021-01475-0
114. Pietanza MC, Litvak AM, Varghese AM, Krug LM, Fleisher M, Teitcher JB, et al. A Phase I Trial of the Hedgehog Inhibitor, Sonidegib (LDE225), in Combination With Etoposide and Cisplatin for the Initial Treatment of Extensive Stage Small Cell Lung Cancer. Lung Cancer (2016) 99:23–30. doi: 10.1016/j.lungcan.2016.04.014
115. Petrova E, Rios-Esteves J, Ouerfelli O, Glickman JF, Resh MD. Inhibitors of Hedgehog Acyltransferase Block Sonic Hedgehog Signaling. Nat Chem Biol (2013) 9(4):247–9. doi: 10.1038/nchembio.1184
116. Rodgers UR, Lanyon-Hogg T, Masumoto N, Ritzefeld M, Burke R, Blagg J, et al. Characterization of Hedgehog Acyltransferase Inhibitors Identifies a Small Molecule Probe for Hedgehog Signaling by Cancer Cells. ACS Chem Biol (2016) 11(12):3256–62. doi: 10.1021/acschembio.6b00896
117. Bissey P-A, Mathot P, Guix C, Jasmin M, Goddard I, Costechareyre C, et al. Blocking SHH/Patched Interaction Triggers Tumor Growth Inhibition Through Patched-Induced Apoptosis. Cancer Res (2020) 80(10):1970–80. doi: 10.1158/0008-5472.CAN-19-1340
118. Maun HR, Wen X, Lingel A, de Sauvage FJ, Lazarus RA, Scales SJ, et al. Hedgehog Pathway Antagonist 5E1 Binds Hedgehog at the Pseudo-Active Site. J Biol Chem (2010) 285(34):26570–80. doi: 10.1074/jbc.M110.112284
119. Tolani B, Hoang NT, Acevedo LA, Giroux Leprieur E, Li H, He B, et al. Preclinical Characterization of Therapeutic Antibodies Targeted at the Carboxy-Terminus of Sonic Hedgehog. Oncotarget (2018) 9(18):14311–23. doi: 10.18632/oncotarget.24510
120. Owens AE, de Paola I, Hansen WA, Liu Y-W, Khare SD, Fasan R. Design and Evolution of a Macrocyclic Peptide Inhibitor of the Sonic Hedgehog/Patched Interaction. J Am Chem Soc (2017) 139(36):12559–68. doi: 10.1021/jacs.7b06087
121. Atwood SX, Sarin KY, Whitson RJ, Li JR, Kim G, Rezaee M, et al. Smoothened Variants Explain the Majority of Drug Resistance in Basal Cell Carcinoma. Cancer Cell (2015) 27(3):342–53. doi: 10.1016/j.ccell.2015.02.002
122. Bendell J, Andre V, Ho A, Kudchadkar R, Migden M, Infante J, et al. Phase I Study of LY2940680, a Smo Antagonist, in Patients With Advanced Cancer Including Treatment-Naïve and Previously Treated Basal Cell Carcinoma. Clin Cancer Res (2018) 24(9):2082–91. doi: 10.1158/1078-0432.CCR-17-0723
123. Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, et al. Effects of Oncogenic Mutations in Smoothened and Patched can be Reversed by Cyclopamine. Nature (2000) 406(6799):1005–9. doi: 10.1038/35023008
124. Tremblay MR, Lescarbeau A, Grogan MJ, Tan E, Lin G, Austad BC, et al. Discovery of a Potent and Orally Active Hedgehog Pathway Antagonist (IPI-926). J Med Chem (2009) 52(14):4400–18. doi: 10.1021/jm900305z
125. Catenacci DVT, Junttila MR, Karrison T, Bahary N, Horiba MN, Nattam SR, et al. Randomized Phase Ib/II Study of Gemcitabine Plus Placebo or Vismodegib, a Hedgehog Pathway Inhibitor, in Patients With Metastatic Pancreatic Cancer. J Clin Oncol: Off J Am Soc Clin Oncol (2015) 33(36):4284–92. doi: 10.1200/JCO.2015.62.8719
126. Jimeno A, Weiss GJ, Miller WH, Gettinger S, Eigl BJC, Chang ALS, et al. Phase I Study of the Hedgehog Pathway Inhibitor IPI-926 in Adult Patients With Solid Tumors. Clin Cancer Res: Off J Am Assoc Cancer Res (2013) 19(10):2766–74. doi: 10.1158/1078-0432.CCR-12-3654
127. Shi W, Nacev BA, Aftab BT, Head S, Rudin CM, Liu JO. Itraconazole Side Chain Analogues: Structure-Activity Relationship Studies for Inhibition of Endothelial Cell Proliferation, Vascular Endothelial Growth Factor Receptor 2 (VEGFR2) Glycosylation, and Hedgehog Signaling. J Med Chem (2011) 54(20):7363–74. doi: 10.1021/jm200944b
128. Teske KA, Dash RC, Morel SR, Chau LQ, Wechsler-Reya RJ, Hadden MK. Development of Posaconazole-Based Analogues as Hedgehog Signaling Pathway Inhibitors. Eur J Med Chem (2019) 163:320–32. doi: 10.1016/j.ejmech.2018.11.056
129. Nedelcu D, Liu J, Xu Y, Jao C, Salic A. Oxysterol Binding to the Extracellular Domain of Smoothened in Hedgehog Signaling. Nat Chem Biol (2013) 9(9):557–64. doi: 10.1038/nchembio.1290
130. Deshpande I, Liang J, Hedeen D, Roberts KJ, Zhang Y, Ha B, et al. Smoothened Stimulation by Membrane Sterols Drives Hedgehog Pathway Activity. Nature (2019) 571(7764):284–8. doi: 10.1038/s41586-019-1355-4
131. Wang J, Zhang Y, Huang W-J, Yang J, Tang W-G, Huang T-M, et al. ABT-199 Inhibits Hedgehog Pathway by Acting as a Competitive Inhibitor of Oxysterol, Rather as a BH3 Mimetic. Acta Pharmacol Sin (2021) 42(6):1005–13. doi: 10.1038/s41401-020-00504-4
132. Lospinoso Severini L, Quaglio D, Basili I, Ghirga F, Bufalieri F, Caimano M, et al. A Smo/Gli Multitarget Hedgehog Pathway Inhibitor Impairs Tumor Growth. Cancers (2019) 11(10):1518. doi: 10.3390/cancers11101518
133. Stappenbeck F, Wang F, Tang L-Y, Zhang YE, Parhami F. Inhibition of Non-Small Cell Lung Cancer Cells by Oxy210, an Oxysterol-Derivative That Antagonizes TGFβ and Hedgehog Signaling. Cells (2019) 8(10):E1297. doi: 10.3390/cells8101297
134. Zhu M, Wang H, Wang C, Fang Y, Zhu T, Zhao W, et al. L-4, a Well-Tolerated and Orally Active Inhibitor of Hedgehog Pathway, Exhibited Potent Anti-Tumor Effects Against Medulloblastoma In Vitro and In Vivo. Front Pharmacol (2019) 10:89. doi: 10.3389/fphar.2019.00089
135. Li Q-R, Zhao H, Zhang X-S, Lang H, Yu K. Novel-Smoothened Inhibitors for Therapeutic Targeting of Naïve and Drug-Resistant Hedgehog Pathway-Driven Cancers. Acta Pharmacol Sin (2019) 40(2):257–67. doi: 10.1038/s41401-018-0019-5
136. Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, et al. Small-Molecule Inhibitors Reveal Multiple Strategies for Hedgehog Pathway Blockade. Proc Natl Acad Sci USA (2009) 106(33):14132–7. doi: 10.1073/pnas.0907134106
137. Lauth M, Bergström A, Shimokawa T, Toftgård R. Inhibition of GLI-Mediated Transcription and Tumor Cell Growth by Small-Molecule Antagonists. Proc Natl Acad Sci USA (2007) 104(20):8455–60. doi: 10.1073/pnas.0609699104
138. Huang L, Walter V, Hayes DN, Onaitis M. Hedgehog-GLI Signaling Inhibition Suppresses Tumor Growth in Squamous Lung Cancer. Clin Cancer Res: Off J Am Assoc Cancer Res (2014) 20(6):1566–75. doi: 10.1158/1078-0432.CCR-13-2195
139. Beauchamp EM, Ringer L, Bulut G, Sajwan KP, Hall MD, Lee Y-C, et al. Arsenic Trioxide Inhibits Human Cancer Cell Growth and Tumor Development in Mice by Blocking Hedgehog/GLI Pathway. J Clin Invest (2011) 121(1):148–60. doi: 10.1172/JCI42874
140. Chang K-J, Yin J-Z, Huang H, Li B, Yang M-H. Arsenic Trioxide Inhibits the Growth of Cancer Stem Cells Derived From Small Cell Lung Cancer by Downregulating Stem Cell-Maintenance Factors and Inducing Apoptosis via the Hedgehog Signaling Blockade. Trans Lung Cancer Res (2020) 9(4):1379–96. doi: 10.21037/tlcr-20-467
141. Owonikoko TK, Zhang G, Kim HS, Stinson RM, Bechara R, Zhang C, et al. Patient-Derived Xenografts Faithfully Replicated Clinical Outcome in a Phase II Co-Clinical Trial of Arsenic Trioxide in Relapsed Small Cell Lung Cancer. J Transl Med (2016) 14(1):111. doi: 10.1186/s12967-016-0861-5
142. Raju GP. Arsenic: A Potentially Useful Poison for Hedgehog-Driven Cancers. J Clin Invest (2011) 121(1):14–6. doi: 10.1172/JCI45692
143. Li B, Fei DL, Flaveny CA, Dahmane N, Baubet V, Wang Z, et al. Pyrvinium Attenuates Hedgehog Signaling Downstream of Smoothened. Cancer Res (2014) 74(17):4811–21. doi: 10.1158/0008-5472.CAN-14-0317
144. Dormoy V, Béraud C, Lindner V, Coquard C, Barthelmebs M, Brasse D, et al. Vitamin D3 Triggers Antitumor Activity Through Targeting Hedgehog Signaling in Human Renal Cell Carcinoma. Carcinogenesis (2012) 33(11):2084–93. doi: 10.1093/carcin/bgs255
145. Maschinot CA, Hadden MK. Synthesis and Evaluation of Vitamin D3 Analogues With C-11 Modifications as Inhibitors of Hedgehog Signaling. Bioorg Med Chem Lett (2017) 27(17):4011–4. doi: 10.1016/j.bmcl.2017.07.060
146. Hadden MK. Hedgehog and Vitamin D Signaling Pathways in Development and Disease. Vitam Horm (2016) 100:231–53. doi: 10.1016/bs.vh.2015.10.006
147. Banerjee U, Ghosh M, Kyle Hadden M. Evaluation of Vitamin D3 A-Ring Analogues as Hedgehog Pathway Inhibitors. Bioorg Med Chem Lett (2012) 22(3):1330–4. doi: 10.1016/j.bmcl.2011.12.081
148. Banerjee U, DeBerardinis AM, Hadden MK. Design, Synthesis, and Evaluation of Hybrid Vitamin D3 Side Chain Analogues as Hedgehog Pathway Inhibitors. Bioorg Med Chem (2015) 23(3):548–55. doi: 10.1016/j.bmc.2014.12.005
149. Maschinot CA, Chau LQ, Wechsler-Reya RJ, Hadden MK. Synthesis and Evaluation of Third Generation Vitamin D3 Analogues as Inhibitors of Hedgehog Signaling. Eur J Med Chem (2019) 162:495–506. doi: 10.1016/j.ejmech.2018.11.028
150. Wu SY, Chiang CM. The Double Bromodomain-Containing Chromatin Adaptor Brd4 and Transcriptional Regulation. J Biol Chem (2007) 282(18):13141–5. doi: 10.1074/jbc.R700001200
151. Ferri E, Petosa C, McKenna CE. Bromodomains: Structure, Function and Pharmacology of Inhibition. Biochem Pharmacol (2016) 106:1–18. doi: 10.1016/j.bcp.2015.12.005
152. Liu X, Zhang Y, Li Y, Wang J, Ding H, Huang W, et al. Development of Hedgehog Pathway Inhibitors by Epigenetically Targeting GLI Through BET Bromodomain for the Treatment of Medulloblastoma. Acta Pharm Sin B (2021) 11(2):488–504. doi: 10.1016/j.apsb.2020.07.007
153. Ally MS, Ransohoff K, Sarin K, Atwood SX, Rezaee M, Bailey-Healy I, et al. Effects of Combined Treatment With Arsenic Trioxide and Itraconazole in Patients With Refractory Metastatic Basal Cell Carcinoma. JAMA Dermatol (2016) 152(4):452–6. doi: 10.1001/jamadermatol.2015.5473
154. Ashrafizadeh M, Javanmardi S, Moradi-Ozarlou M, Mohammadinejad R, Farkhondeh T, Samarghandian S, et al. Natural Products and Phytochemical Nanoformulations Targeting Mitochondria in Oncotherapy: An Updated Review on Resveratrol. Biosci Rep (2020) 40(4):BSR20200257. doi: 10.1042/bsr20200257
155. Wang K, Chen Q, Shao Y, Yin S, Liu C, Liu Y, et al. Anticancer Activities of TCM and Their Active Components Against Tumor Metastasis. Biomed Pharmacother = Biomed Pharmacother (2021) 133:111044. doi: 10.1016/j.biopha.2020.111044
156. Yang Y, Nguyen TT, Pereira I, Hur J-S, Kim H. Lichen Secondary Metabolite Physciosporin Decreases the Stemness Potential of Colorectal Cancer Cells. Biomolecules (2019) 9(12):797. doi: 10.3390/biom9120797
157. Song L, Liu D, Zhao Y, He J, Kang H, Dai Z, et al. Sinomenine Reduces Growth and Metastasis of Breast Cancer Cells and Improves the Survival of Tumor-Bearing Mice Through Suppressing the SHH Pathway. Biomed Pharmacother = Biomed Pharmacother (2018) 98:687–93. doi: 10.1016/j.biopha.2017.12.065
158. Ashrafizadeh M, Ahmadi Z, Farkhondeh T, Samarghandian S. Autophagy as a Molecular Target of Quercetin Underlying its Protective Effects in Human Diseases. Arch Physiol Biochem (2019) 128(1):1–9. doi: 10.1080/13813455.2019.1671458
159. Ashrafizadeh M, Mohammadinejad R, Tavakol S, Ahmadi Z, Roomiani S, Katebi M. Autophagy, Anoikis, Ferroptosis, Necroptosis, and Endoplasmic Reticulum Stress: Potential Applications in Melanoma Therapy. J Cell Physiol (2019) 234(11):19471–9. doi: 10.1002/jcp.28740
160. Ashrafizadeh M, Ahmadi Z, Farkhondeh T, Samarghandian S. Autophagy Regulation Using Luteolin: New Insight Into its Anti-Tumor Activity. Cancer Cell Int (2020) 20(1):537. doi: 10.1186/s12935-020-01634-9
161. Ashrafizadeh M, Ahmadi Z, Farkhondeh T, Samarghandian S. Modulatory Effects of Statins on the Autophagy: A Therapeutic Perspective. J Cell Physiol (2020) 235(4):3157–68. doi: 10.1002/jcp.29227
162. Kim KH, Lee MS. Autophagy–A Key Player in Cellular and Body Metabolism. Nat Rev Endocrinol (2014) 10(6):322–37. doi: 10.1038/nrendo.2014.35
163. Chang H, Zou Z. Targeting Autophagy to Overcome Drug Resistance: Further Developments. J Hematol Oncol (2020) 13(1):159. doi: 10.1186/s13045-020-01000-2
164. Mele L, Del Vecchio V, Liccardo D, Prisco C, Schwerdtfeger M, Robinson N, et al. The Role of Autophagy in Resistance to Targeted Therapies. Cancer Treat Rev (2020) 88:102043. doi: 10.1016/j.ctrv.2020.102043
165. Lei W, Huo Z. Jervine Inhibits Non-Small Cell Lung Cancer (NSCLC) Progression by Suppressing Hedgehog and AKT Signaling via Triggering Autophagy-Regulated Apoptosis. Biochem Biophys Res Commun (2020) 533(3):397–403. doi: 10.1016/j.bbrc.2020.08.023
166. Lin L, Ren L, Wen L, Wang Y, Qi J. Effect of Evodiamine on the Proliferation and Apoptosis of A549 Human Lung Cancer Cells. Mol Med Rep (2016) 14(3):2832–8. doi: 10.3892/mmr.2016.5575
167. Jo E, Jang H-J, Shen L, Yang KE, Jang MS, Huh YH, et al. Cordyceps Militaris Exerts Anticancer Effect on Non-Small Cell Lung Cancer by Inhibiting Hedgehog Signaling via Suppression of TCTN3. Integr Cancer Ther (2020) 19:1534735420923756. doi: 10.1177/1534735420923756
168. Wang F, Wang W, Li J, Zhang J, Wang X, Wang M. Sulforaphane Reverses Gefitinib Tolerance in Human Lung Cancer Cells via Modulation of Sonic Hedgehog Signaling. Oncol Lett (2018) 15(1):109–14. doi: 10.3892/ol.2017.7293
169. Jiang Q, Li Q, Chen H, Shen A, Cai Q, Lin J, et al. Scutellaria Barbata D. Don Inhibits Growth and Induces Apoptosis by Suppressing IL-6-Inducible STAT3 Pathway Activation in Human Colorectal Cancer Cells. Exp Ther Med (2015) 10(4):1602–8. doi: 10.3892/etm.2015.2692
170. Marconett CN, Morgenstern TJ, San Roman AK, Sundar SN, Singhal AK, Firestone GL. BZL101, a Phytochemical Extract From the Scutellaria Barbata Plant, Disrupts Proliferation of Human Breast and Prostate Cancer Cells Through Distinct Mechanisms Dependent on the Cancer Cell Phenotype. Cancer Biol Ther (2010) 10(4):397–405. doi: 10.4161/cbt.10.4.12424
171. Du J, Chen W, Yang L, Dai J, Guo J, Wu Y, et al. Disruption of SHH Signaling Cascade by SBE Attenuates Lung Cancer Progression and Sensitizes DDP Treatment. Sci Rep (2017) 7(1):1899. doi: 10.1038/s41598-017-02063-x
172. Lupo G, Caporarello N, Olivieri M, Cristaldi M, Motta C, Bramanti V, et al. Anti-Angiogenic Therapy in Cancer: Downsides and New Pivots for Precision Medicine. Front Pharmacol (2016) 7:519. doi: 10.3389/fphar.2016.00519
173. Rahbari NN, Kedrin D, Incio J, Liu H, Ho WW, Nia HT, et al. Anti-VEGF Therapy Induces ECM Remodeling and Mechanical Barriers to Therapy in Colorectal Cancer Liver Metastases. Sci Transl Med (2016) 8(360):360ra135. doi: 10.1126/scitranslmed.aaf5219
174. Lee S-O, Kim J-S, Lee M-S, Lee H-J. Anti-Cancer Effect of Pristimerin by Inhibition of HIF-1α Involves the SPHK-1 Pathway in Hypoxic Prostate Cancer Cells. BMC Cancer (2016) 16:701. doi: 10.1186/s12885-016-2730-2
175. Zhao H, Wang C, Lu B, Zhou Z, Jin Y, Wang Z, et al. Pristimerin Triggers AIF-Dependent Programmed Necrosis in Glioma Cells via Activation of JNK. Cancer Lett (2016) 374(1):136–48. doi: 10.1016/j.canlet.2016.01.055
176. Lei X, Zhong Y, Huang L, Li S, Fu J, Zhang L, et al. Identification of a Novel Tumor Angiogenesis Inhibitor Targeting SHH/Gli1 Signaling Pathway in Non-Small Cell Lung Cancer. Cell Death Dis (2020) 11(4):232. doi: 10.1038/s41419-020-2425-0
177. He L, Liang Z, Zhao F, Peng L, Chen Z. Modulation of IL-37 Expression by Triptolide and Triptonide in THP-1 Cells. Cell Mol Immunol (2015) 12(4):515–8. doi: 10.1038/cmi.2014.92
178. Wang Z, Ma D, Wang C, Zhu Z, Yang Y, Zeng F, et al. Triptonide Inhibits the Pathological Functions of Gastric Cancer-Associated Fibroblasts. Biomed Pharmacother (2017) 96:757–67. doi: 10.1016/j.biopha.2017.10.046
179. Han H, Du L, Cao Z, Zhang B, Zhou Q. Triptonide Potently Suppresses Pancreatic Cancer Cell-Mediated Vasculogenic Mimicry by Inhibiting Expression of VE-Cadherin and Chemokine Ligand 2 Genes. Eur J Pharmacol (2018) 818:593–603. doi: 10.1016/j.ejphar.2017.11.019
180. Zhang M, Tan S, Yu D, Zhao Z, Zhang B, Zhang P, et al. Triptonide Inhibits Lung Cancer Cell Tumorigenicity by Selectively Attenuating the SHH-Gli1 Signaling Pathway. Toxicol Appl Pharmacol (2019) 365:1–8. doi: 10.1016/j.taap.2019.01.002
181. Ashrafizadeh M, Zarrabi A, Hashemipour M, Vosough M, Najafi M, Shahinozzaman M, et al. Sensing the Scent of Death: Modulation of microRNAs by Curcumin in Gastrointestinal Cancers. Pharmacol Res (2020) 160:105199. doi: 10.1016/j.phrs.2020.105199
182. Ashrafizadeh M, Zarrabi A, Hashemi F, Zabolian A, Saleki H, Bagherian M, et al. Polychemotherapy With Curcumin and Doxorubicin via Biological Nanoplatforms: Enhancing Antitumor Activity. Pharmaceutics (2020) 12(11):1084. doi: 10.3390/pharmaceutics12111084
183. Tsai C-F, Hsieh T-H, Lee J-N, Hsu C-Y, Wang Y-C, Kuo K-K, et al. Curcumin Suppresses Phthalate-Induced Metastasis and the Proportion of Cancer Stem Cell (CSC)-Like Cells via the Inhibition of AhR/ERK/SK1 Signaling in Hepatocellular Carcinoma. J Agric Food Chem (2015) 63(48):10388–98. doi: 10.1021/acs.jafc.5b04415
184. Almanaa TN, Geusz ME, Jamasbi RJ. Effects of Curcumin on Stem-Like Cells in Human Esophageal Squamous Carcinoma Cell Lines. BMC Complement Altern Med (2012) 12:195. doi: 10.1186/1472-6882-12-195
185. Abadi AJ, Mirzaei S, Mahabady MK, Hashemi F, Zabolian A, Hashemi F, et al. Curcumin and its Derivatives in Cancer Therapy: Potentiating Antitumor Activity of Cisplatin and Reducing Side Effects. Phytother Res (2021) 36(1):189–213. doi: 10.1002/ptr.7305
186. Zhu J-Y, Yang X, Chen Y, Jiang Y, Wang S-J, Li Y, et al. Curcumin Suppresses Lung Cancer Stem Cells via Inhibiting Wnt/β-Catenin and Sonic Hedgehog Pathways. Phytother Res: PTR (2017) 31(4):680–8. doi: 10.1002/ptr.5791
Keywords: lung cancer, Hh pathway, proliferation, invasion, metastasis, natural product
Citation: Ma C, Hu K, Ullah I, Zheng Q-K, Zhang N and Sun Z-G (2022) Molecular Mechanisms Involving the Sonic Hedgehog Pathway in Lung Cancer Therapy: Recent Advances. Front. Oncol. 12:729088. doi: 10.3389/fonc.2022.729088
Received: 22 June 2021; Accepted: 03 March 2022;
Published: 01 April 2022.
Edited by:
Carminia Maria Della Corte, University of Campania Luigi Vanvitelli, ItalyReviewed by:
Savvas Petanidis, Aristotle University of Thessaloniki, GreeceMilad Ashrafizadeh, Sabancı University, Turkey
Eswari Dodagatta-Marri, University of California, San Francisco, United States
Ali Zarrabi, University of Istinye, Turkey
Copyright © 2022 Ma, Hu, Ullah, Zheng, Zhang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Gang Sun, c3Vuc3pnQDEyNi5jb20=; Nan Zhang, emxrem4yMDE2QDEyNi5jb20=
 Chao Ma
Chao Ma Kang Hu
Kang Hu Irfan Ullah
Irfan Ullah Qing-Kang Zheng
Qing-Kang Zheng Nan Zhang
Nan Zhang Zhi-Gang Sun
Zhi-Gang Sun