
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 26 January 2023
Sec. Cancer Molecular Targets and Therapeutics
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1087989
 Ananna Bhadra Arna1
Ananna Bhadra Arna1 Hardikkumar Patel2
Hardikkumar Patel2 Ravi Shankar Singh1
Ravi Shankar Singh1 Frederick S. Vizeacoumar2
Frederick S. Vizeacoumar2 Anthony Kusalik3
Anthony Kusalik3 Andrew Freywald2
Andrew Freywald2 Franco J. Vizeacoumar4*
Franco J. Vizeacoumar4* Yuliang Wu1*
Yuliang Wu1*DEAD/H-box helicases are implicated in virtually every aspect of RNA metabolism, including transcription, pre-mRNA splicing, ribosomes biogenesis, nuclear export, translation initiation, RNA degradation, and mRNA editing. Most of these helicases are upregulated in various cancers and mutations in some of them are associated with several malignancies. Lately, synthetic lethality (SL) and synthetic dosage lethality (SDL) approaches, where genetic interactions of cancer-related genes are exploited as therapeutic targets, are emerging as a leading area of cancer research. Several DEAD/H-box helicases, including DDX3, DDX9 (Dbp9), DDX10 (Dbp4), DDX11 (ChlR1), and DDX41 (Sacy-1), have been subjected to SL analyses in humans and different model organisms. It remains to be explored whether SDL can be utilized to identity druggable targets in DEAD/H-box helicase overexpressing cancers. In this review, we analyze gene expression data of a subset of DEAD/H-box helicases in multiple cancer types and discuss how their SL/SDL interactions can be used for therapeutic purposes. We also summarize the latest developments in clinical applications, apart from discussing some of the challenges in drug discovery in the context of targeting DEAD/H-box helicases.
Helicases constitute a ubiquitous group of molecular motors that couple the energy from nucleoside triphosphate hydrolysis with the unwinding and/or remodeling of DNA or RNA molecules, and occasionally, with the disruption of protein-nucleic acid interactions. The human genome encodes 95 known helicases; out of them, 64 are RNA helicases and 31 are DNA helicases (1). These enzymes are involved in virtually all aspects of nucleic acid metabolism, including replication, repair, recombination, transcription, splicing, chromosome segregation and telomere maintenance (2–5). To support augmented proliferation and match the requirements of accelerated nucleic acid metabolism, helicases are frequently overexpressed in cancer cells (6). Meantime, naturally occurring loss of function (LOF) mutations in helicases are associated with many diseases, including cancers (7). Therefore, helicases become attractive targets for chemotherapeutic developments. Unfortunately, direct targeting of these molecules may represent a serious challenge, as normal cells are also highly dependent on their cellular functions. Therefore, alternate strategies are required.
Given that many helicases are overexpressed or lost in cancers, identifying their synthetic lethal (SL) and synthetic dosage lethal (SDL) interactions may represent an effective strategy to exploit them for cancer therapeutics. Two genes are said to exhibit SL interaction, if LOF of both these genes affect cellular viability, while neither of them has any effect on their own (8). This concept facilitates the development of targeted therapies that will selectively kill cancer cells, while sparing normal cells. Over the last decade, the most successful clinical application in the field of SL is the development of poly (ADP-ribose) polymerase-1 (PARP-1) inhibitors in BRCA1/2-mutant breast and ovarian cancers (9). In this review, we will highlight the potential of DEAD/H-box helicase in the invention of cancer therapeutics using the SL approach. We will also discuss potential opportunities to implement the SDL approach, where LOF of one gene affects cell viability only when a partner gene is overactivated, to exploit overexpressed DEAD/H-box helicases for cancer therapeutics.
Based on substrate specificity and polarity, helicases are classified as RNA or DNA helicases and as 5ʹ–3ʹ or 3ʹ–5ʹ helicases (10). Based on their conserved motifs, helicases are grouped into six superfamilies (SF1 - SF6) (11). Among them, SF2 is the largest superfamily and is characterized by its 12 “signature” motifs (Q, I, Ia, Ib, Ic, II, III, IV, IVa, V, Va, and VI). SF2 is further classified into several subfamilies, including DEAD/H-box RNA helicases, RecQ-like family, and Snf2-like enzymes based on their sequences, structures, and mechanisms of action (12, 13).
The DEAD/H-box protein family is named after the sequence (Asp-Glu-Ala-Asp/His) in motif II (14). They are all composed of two RecA-like domains, while some are additionally flanked by N- and/or C-terminal accessory domain(s) (Figure 1A). Cooperatively, these domains are involved in RNA binding, ATP binding and hydrolysis, unwinding, strand annealing, and protein-protein interactions (15). There are at least 36 DEAD-box helicases and 14 DEAH-box helicases in humans (16). Despite sharing the conserved helicase core, a substantial difference is observed between DEAD-box and DEAH-box helicases from the biochemical perspective. To disrupt nucleic acid structures, DEAD-box helicases use simple cycles of RNA duplex binding, unwinding, and release, while DEAH-box helicases function only as translocases in the 3’→5’ direction (17). In terms of their nucleic acid-related functions, both DEAD-box and DEAH-box proteins are implicated in virtually every aspect of RNA metabolic processes, including transcription (18–20), ribosomes biogenesis (21, 22), small RNA process (23, 24), pre-mRNA splicing (25, 26), RNA storage and decay (27, 28), nuclear export (29–31), liquid–liquid phase separation (32–34), RNA degradation (35, 36), translation (37–42), and so on (Figure 1B). Some of them are also involved in DNA metabolism, such as DNA repair (43–47). On a biological level, they are involved in innate immunity responses (48), signal transduction (49), cell differentiation and organ development (50, 51), programmed cell death (52), and mitochondrial regulations (53). Dysregulation of the expression or function of these proteins is likely to be one of the reasons behind the development of cancer and various diseases. For instance, mutations in DDX11 (ChlR1) are associated with Warsaw Breakage syndrome (WBS) and Roberts syndrome (54). Germline mutations in DDX3 account for 1%-3% of unexplained intellectual disability cases (55–59). Mutations in DDX6 cause intellectual disability and dysmorphic features (60). Mutations in DDX41 are associated with myelodysplastic syndromes (MDS), acute myeloid leukemia (AML), and myeloid neoplasms (MNs) (61, 62).
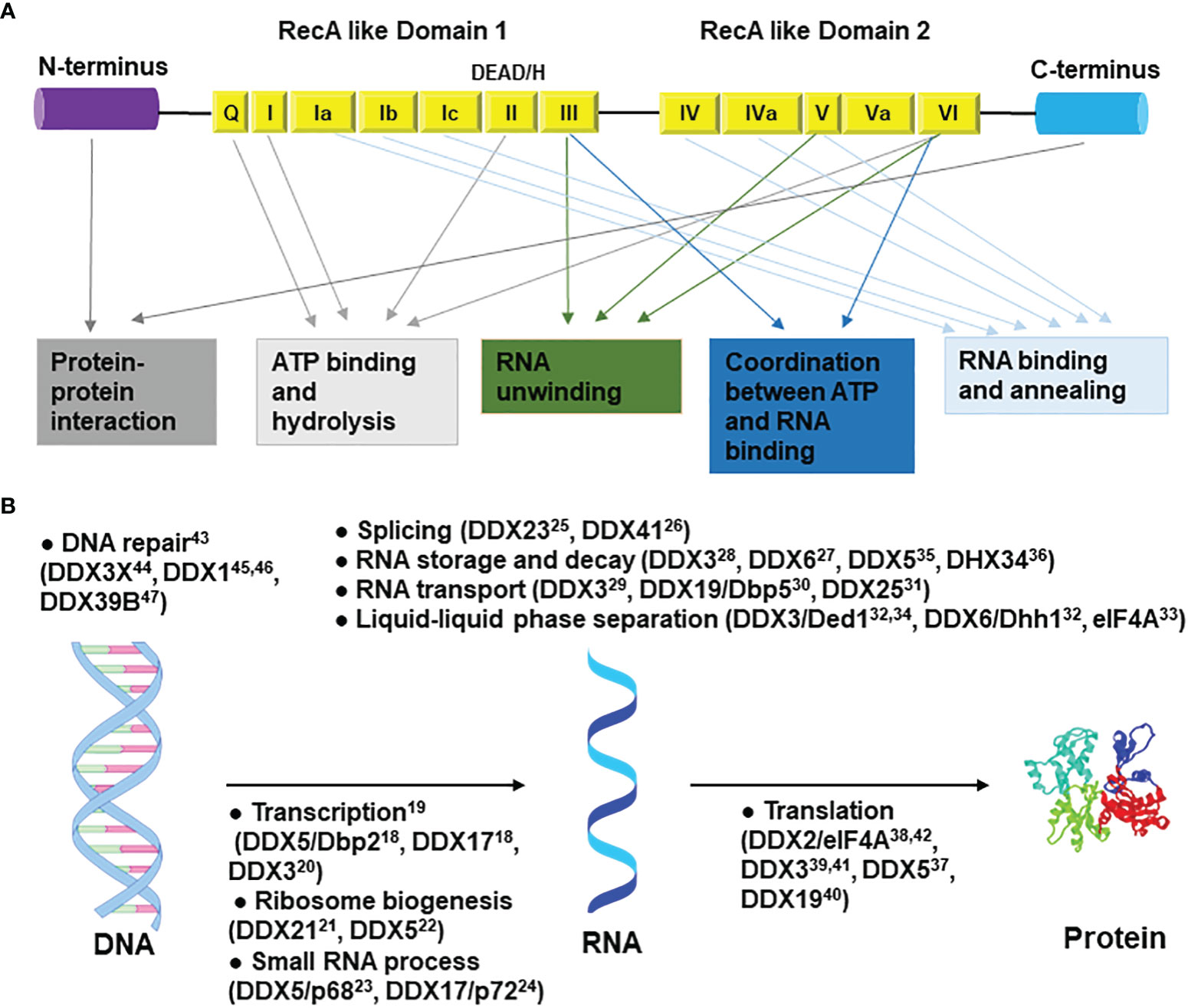
Figure 1 General structure and functions of DEAD/H-box helicases. (A) The N-terminal and C-terminal regions are involved in protein-protein interactions. Motifs Q, I, II, and VI are involved in ATP binding and hydrolysis; motifs III, V, and VI in RNA unwinding; motifs Ia, Ib, Ic, IV, IVa, and V in RNA binding and annealing; motifs III and Va in coordination between ATP and RNA binding. (B) Roles of DEAD/H-box RNA helicases in the central dogma pathway. Only the most studied top two to four helicases are shown in each catalogue. Some roles outside of the central dogma are not shown. Related references are shown as superscripts.
To achieve the hallmark of excessive cell proliferation, DEAD/H-box helicases are often overexpressed in cancer cells (Table 1). For example, DDX3 is highly expressed in breast cancer (124), Ewing sarcoma (68), glioblastoma (125) and gallbladder carcinoma (126). DDX27 is upregulated in gastric tumors (94), colorectal cancer (127), and breast cancer (128). Cancer-specific antigens, DDX43 (helicase antigen gene, HAGE) and DDX53 (cancer-associated gene, CAGE), are overexpressed in almost all cancers (129), while DDX20 (dp103) is highly expressed in breast cancer (130). Nevertheless, controversial findings are reported for some DEAD/H-box helicases. For example, although DDX21 is highly expressed in breast cancer (131), colorectal cancer (89), gastric cancer (132) and neuroblastoma (90), low DDX21 levels are associated with poor clinical outcome of breast cancer patients (91). Moreover, up-regulation of DEAD/H-box helicases correlates with advanced stage and poor prognosis in cancer patients (7). For example, high DDX1 levels are associated with higher pathological grades and poorer prognosis in hepatocellular carcinoma (63) and breast cancer (133). The expression of DDX23 is enhanced in glioma tissues, and this is associated with poor survival of glioma patients (92). In computational analyses of 15 DEAH-box RNA helicases, elevated expression of 12 of them are associated with poor prognosis and worse clinical features in hepatocellular carcinoma (111). Because of their overarching role in RNA metabolism, DEAD/H-box helicases are likely to affect multiple aspects of cell behaviors, including cell proliferation, which may lead to cancer development.
Given that there is only sporadic data available for each of these helicases, we examined the expression of these helicases in 24 different cancer types, using gene data from patient samples available in The Cancer Genome Atlas (TCGA). This revealed that few of the helicases are always lost across the cancers examined, such as DDX3X and DDX6. Some of them are only overexpressed across human cancers, such as DDX27, DDX41 and DDX56, while most of them are both overexpressed and lost in different cancer types (Figure 2). We noticed few inconsistencies between the TCGA data and published findings for some helicases, which might be due to analyzing cell lines instead of patient samples (68, 134, 135). In fact, inconsistencies between data obtained from cancer cell lines and cancer patient tissues are not uncommon and have been reported (136, 137).
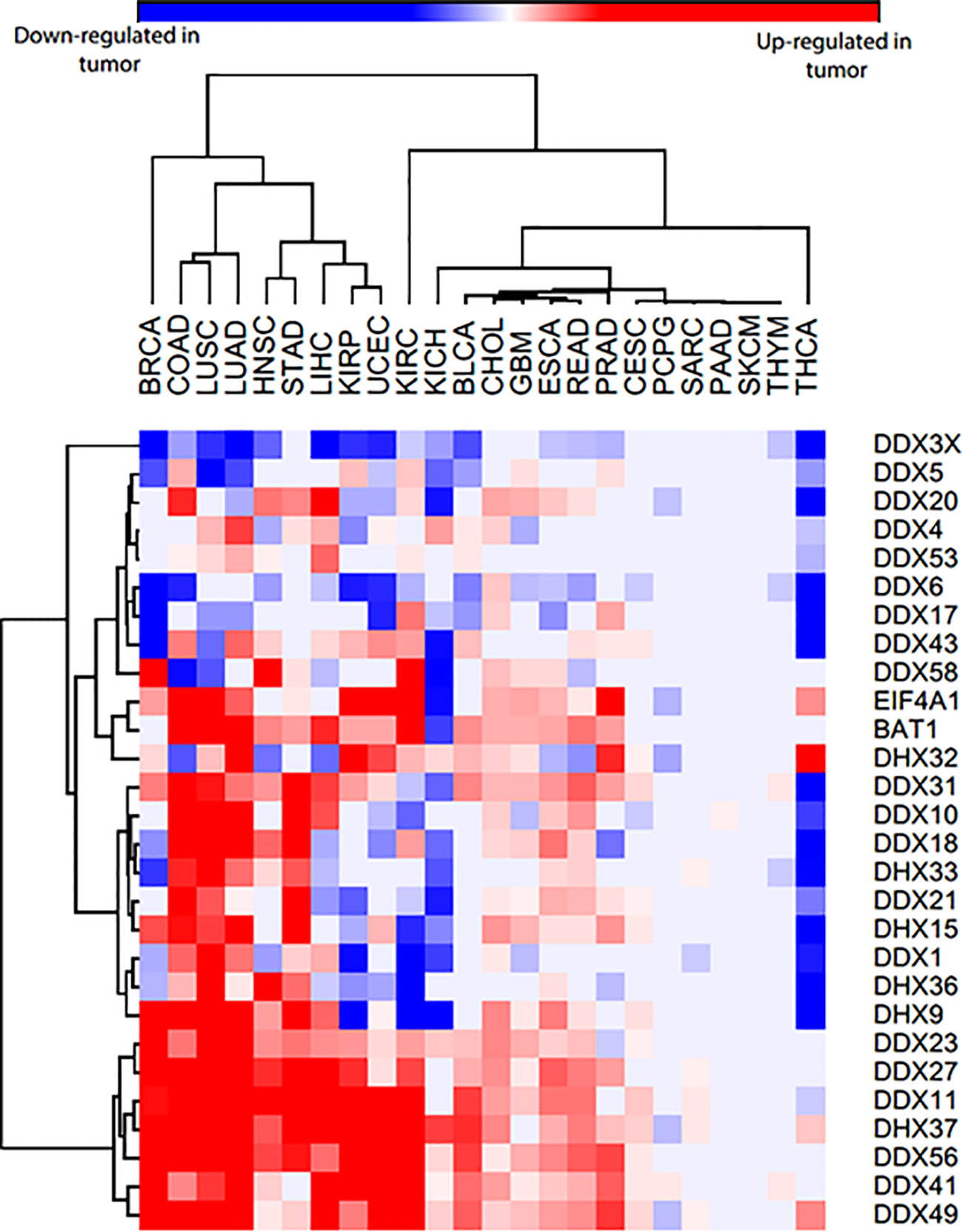
Figure 2 Heatmap of DEAD/H-box helicases expression in 24 different cancers and normal tissues from TCGA patient data. The blue color represents that they are significantly lost in tumor samples compared to normal samples, red color represents that they are significantly overexpressed in tumor samples compared to normal samples, and grey means no significant difference between normal and tumor samples. Cancer type abbreviations used are: BRCA, Breast invasive carcinoma; COAD, Colon adenocarcinoma; LUSC, Lung squamous cell carcinoma; LUAD, Liver hepatocellular carcinoma; HNSC, Head and neck squamous cell carcinoma; STAD, Stomach adenocarcinoma; LIHC, Liver hepatocellular carcinoma; KIRP, Kidney renal papillary cell carcinoma; UCEC, Uterine corpus endometrial carcinoma; KIRC, Kidney renal clear cell carcinoma; KICH, Kidney Chromophobe; BLCA, Bladder urothelial carcinoma; CHOL, Cholangiocarcinoma; GBM, Glioblastoma multiforme; ESCA, Esophageal carcinoma; READ, Rectum adenocarcinoma; PRAD, Prostate adenocarcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; PCPG, Pheochromocytoma and paraganglioma; SARC, Sarcoma; PAAD, Pancreatic adenocarcinoma; SKCM, Skin cutaneous melanoma; THYM, Thymoma; THCA, Thyroid carcinoma. BAT1 is DDX39B and EIF4A1 is DDX2.
Apart from differential expression of these genes, mutations in DEAD/H-box helicases have also been associated with various cancers. For example, somatic mutations in DDX3X are identified in various cancers (138). Mutations in DDX18 (86) or DDX41 (139) are found in AML/MDS patients and germline mutations in RIG-I (DDX58) are found in colon cancer (110). Many missense mutations are enriched in two conserved RecA-like domains (Ref 85–88, TCGA and COSMIC data), and some frameshift mutations completely remove or drastically alter these domains, which impair the ATPase/helicase activity in these proteins. However, variants can also be found in the N- and C-terminal domains; the functional consequences of these mutations remain undetermined. Pharmacological inhibition of these helicases that are overexpressed in cancer may benefit the patients; however, identification of the targeted helicases and development of the compounds are still in their infancy (134, 140–142). Moreover, it is unclear if the inhibition of these molecules may affect normal cell functions. Application of SL or SDL may circumvent these challenges.
SL represents functional relations between pairs of genes whose concomitant alteration-of-function is lethal (8). SL in DNA damage repair pathways is a promising strategy for DNA damage response. BRCA1 and BRCA2 play pivotal roles in homologous recombination (HR) repair that enable precise repair of DNA double-strand breaks (DSBs) using sister chromatids as templates (143). PARP-1 is an abundant nuclear protein in cells and plays a vital role in repairing single-strand breaks (SSBs) (144). PARP inhibitors (PARPis), which repress the catalytic activity of PARP, lead to tumor-specific cell death due to the combined deficiency in the HR and SSB repair pathway evoked by BRCA1/2 mutations and PARP inhibition respectively (145). PARPis are the first and most successful drugs designed to exploit the concept of SL (9), which provides a novel strategy for targeting other genes. In this regard, DEAD/H-box helicases, whose expression is lost in cancer, would provide a ‘tumor-specific context’ in which a second gene (SL partner) becomes a ‘vulnerable target’ that can be used to eliminate cancer cells (Figure 3A).
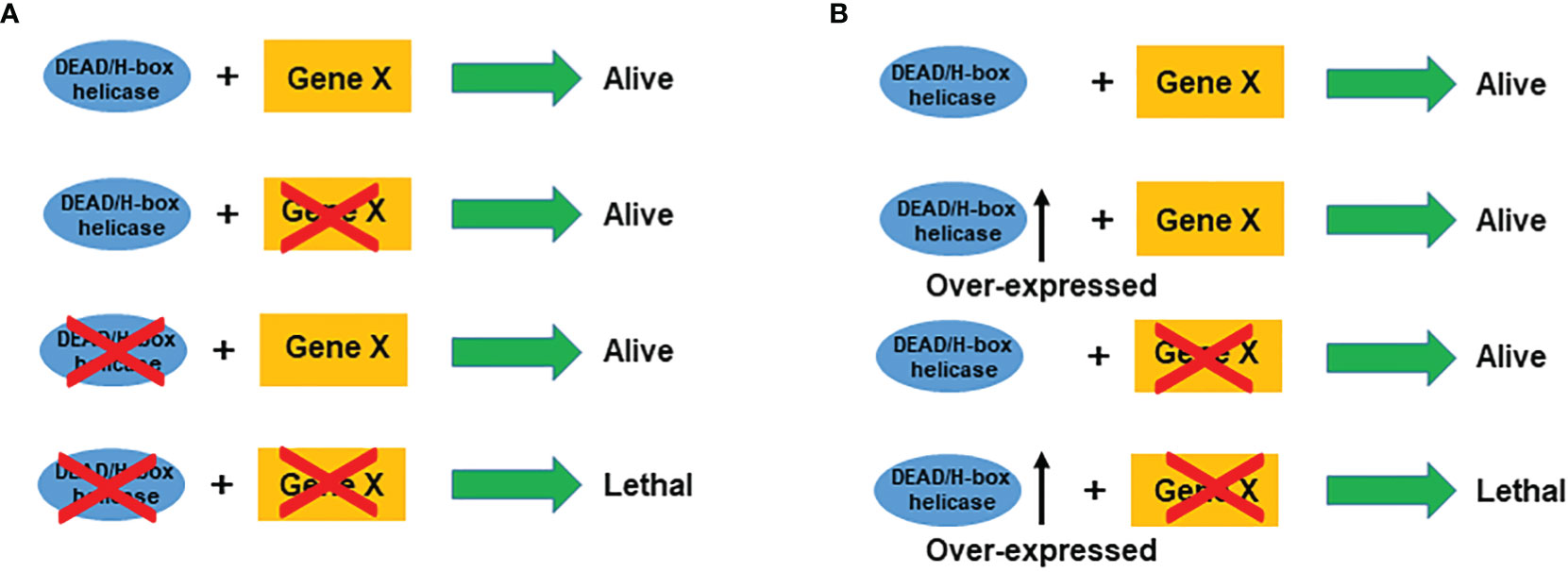
Figure 3 Schematic illustration of synthetic lethality (A) and synthetic dosage lethality (B) for DEAD/H-box helicases. Cross on a gene stands for mutation or inhibition.
Synthetic dosage lethality (SDL), a variant of SL, decreases cell viability only when a gene is overexpressed and a second gene is inactivated (146). For DEAD/H-box overexpressed cancers, inhibition of its interaction partner will cause lethality in the cancer cells only (Figure 3B). Using the SDL genetic approach, a genome-wide SDL screen identified a deletion in the histone deacetylase, RPD3 gene, as selectively sensitive to the overexpression of yeast TDP1, a tyrosyl-DNA-phosphodiesterase (147). The SDL interaction was conserved in a human rhabdomyosarcoma cell line with innate TDP1 levels, and these cells were sensitive to the treatment with histone deacetylase inhibitors (147). However, so far, no SDL approach has been utilized to target DEAD/H-box helicases.
Although SL/SDL interactions between gene pairs in cancer biology have been viewed from the target perspective, these genetic approaches also have been extensively applied in yeast to discover functional relationships, as they conserve their functions across organisms. Accordingly, we highlight several DEAD/H-box helicases below, that have been studied in model organisms as well as in human cells for synthetic lethal interactions (Table 2).
DDX3 has two paralogs, DDX3X on the X-chromosome and DDX3Y on the Y-chromosome. Sharing more than 90% identity, they are redundant for protein synthesis (155), cell proliferation and survival (156), and temperature-sensitive in hamster cell line (157), but distinct for liquid-liquid phase separation, dissolution, and translation repression (34). Interestingly, the combination of DDX3 inhibitor RK-33 and PARP inhibitor Olaparib causes SL in BRCA1-proficient breast cancer (148). DDX3X is reported to be actively recruited to sites of DNA damage in live cells (44), and it regulates the expression of DNA repair genes (158), indicating DDX3X is essential for DNA repair, such as non-homologous DNA end joining (NHEJ). RK-33 is a small molecule that binds DDX3X and inhibits its helicase activity (134). Mechanistically, RK-33 might inhibit NHEJ repair. Combination of RK-33 and PARPis, which inhibit DDX3X and PARP and block SSB and NHEJ repair pathways respectively, leads to cell death, representing a classical ‘between-pathway’ SL interaction. Whether RK-33 can manipulate any other SL interactions of DDX3X for better outcomes requires further investigation. More recently, analyses of multiple-omics datasets and experimental validation revealed that redundancy exists between DDX3X and DDX3Y (159); therefore, we have to consider the gender of the patient and look for any loss of chromosome Y in male patients before applying RK-33. In addition, it remains to be determined, whether RK-33 binds and inhibits the DDX3Y protein in addition to DDX3X.
It is worth noting that DDX3 has been reported to function as both an oncogene and a tumor suppressor. DDX3 exerts oncogenic roles in glioblastoma (160, 161), meningioma (162), Ewing sarcoma (68), prostate cancer (134), chronic lymphocytic leukemia (163, 164), pancreatic ductal adenocarcinoma (165), and gallbladder cancer (126). In contrast, DDX3 acts as a tumor suppressor in natural killer/T-cell lymphoma (166) and cutaneous squamous cell carcinoma (20). Moreover, the dual role of DDX3 have been reported in same type of cancer, including breast cancer (124, 167–169), hepatocellular carcinoma (170, 171), lung cancer (142, 172), colorectal cancer (70, 173, 174), and head and neck squamous cell carcinoma (69, 175, 176). So far, there are no mechanistic explanations for the complex behavior of DDX3 in these cancers, which appears to be context dependent. For its tumor suppressive function, particularly in cases of LOF mutations, we believe that the SL approach can be used for drug development; for its oncogenic roles, the SDL strategy could be potentially applied.
As mentioned above, SL interactions in yeast reveal functional relationship between gene pairs. DDX9 (Dbp9), encoding an essential nucleolar protein involved in 60S-ribosomal-subunit biogenesis, exhibits an SL relation with DDX6 (Dbp6), a component required for 60S-ribosomal-subunit assembly in yeast (177). Interestingly, dbp6/dbp9 double mutants show synthetic lethality: no viable dbp9/dbp6 double mutants could be recovered, indicating a functional interaction of Dbp9 with Dbp6, and accumulated defects in ribosome biogenesis lead to cell death (149). Our analyses of the TCGA data (Figure 2) show that DDX6 is downregulated in multiple cancers. If the yeast interactions are conserved, inhibition of DDX9 in these cancers may result in a therapeutically relevant SL interaction.
Parkinson’s disease (PD) is characterized by loss of dopaminergic neurons in midbrain and the presence of Lewy inclusion bodies, which are predominantly composed of misfolding and aggregation of the α-synuclein protein. Interestingly, a reciprocal susceptibility was observed in the PD patients towards the occurrence of melanoma (178). Although fundamentally divergent, the common link between the two disorders is the accumulation of α-synuclein into amyloid fibrils. Gerhard Braus’s group expressed human α-synuclein in yeast cells and monitored dosage-dependent toxicity effects on the formation and reduction of inclusions bodies (150). They identified the nucleolar DDX10 (yeast ortholog Dbp4) as a strong enhancer of α-synuclein toxicity. While downregulation of Dbp4 rescued cells from α-synuclein toxicity, overexpression of Dbp4 led to an SL phenotype. These findings provide a novel link between nucleolar processes and α-synuclein-mediated toxicity, with DDX10 emerging as a promising drug target for melanoma.
RNAi-dependent knockdown of DDX11 causes premature sister chromatid separation and a profound delay in mitotic progression of human cells, suggesting that DDX11 is required to establish proper sister chromatid cohesion during the S phase (179). Biallelic DDX11 mutations in humans cause WBS, which is characterized by severe microcephaly, pre- and post-natal growth retardation, and abnormal skin pigmentation (54). Rob Wolthuisb and his team found that DDX11 mutations cause cohesion defects in patient fibroblasts. This discovery prompted them to subject patient’s fibroblasts to a genome-wide siRNA screen to search for genes that are synthetically lethal with mutant DDX11 (151). Screening results revealed several components of the anaphase promoting complex or cyclosome (APC/C) as top hits, and the DDX11 mutant cells proved to be hypersensitive to the inhibition of the APC/C complex. Mechanistically, they found that APC/C inhibition aggravates cohesion defects and causes mitotic death. As the TCGA data indicates that DDX11 is overexpressed in multiple cancers (Figure 2), further analyses of this gene and the effect of its overexpression in sister chromatid cohesion might reveal if APC/C inhibition could still remain a viable therapeutic option. The same group also found that ESCO2 (establishment of cohesion 1 homolog 2) was one of the strongest hits in their siRNA screen, in which they used WBS patient fibroblasts (152). The synthetic lethality between DDX11 and ESCO2 was also observed in yeast orthologues Chl1 (DDX11/ChlR1) and Eco1 (ESCO2) (180), and chicken DDX11 and ESCO2 (153). Thus, some of the conserved interactions observed in yeast may be of high relevance in humans as well.
DDX41 is conserved across species; its orthologs abstrakt in Drosophila (181), Sacy-1 in C. elegans (154), and DrDDX41 in zebrafish (182) have been studied. Both germ line and acquired somatic mutations of DDX41 have been associated with MNs, MDS and AML (62, 139). David Greenstein’s group used a sacy-1 reduction-of-function genetic background in C. elegans, conducted a genome-wide RNAi screen, and identified five clones that produced increased levels of sterility, gamete degeneration, or embryonic lethality (183). The five RNAi clones target the transcripts of three genes: mog-2 (one clone), Y111B2A.25 (one clone), and emb-4 (three clones) (154). Because three phenotypes have been observed in a variety of sacy-1 mutant alleles, individual siRNA in sacy-1 mutant animal confirmed that genes mog-2, Y111B2A.25, and emb-4 are synthetic lethal interactors of sacy-1 (DDX41). Interestingly, similar to DDX41, these three genes encoded proteins that are constitutive components of the spliceosome (154), and it remains to be seen if these interactions are still conserved in humans. As DDX41 is overexpressed in multiple cancers, spliceosome inhibitors should benefit these patients, provided the functional interaction is conserved.
Olaparib, rucaparib, niraparib, talazoparib, and veliparib, classified as PARPis, are, so far, the only SL drugs for cancer patients with BRCA1/2 mutations (9). There is an urgent need for new SL targets in other cancers. Many DEAD/H-box helicases are upregulated in various malignancies, while some of their mutations are also associated with cancers. Therefore, there is a growing interest in DEAD/H-box helicases as plausible targets for anti-cancer therapy. Indeed, identifying and developing SL interactions of DEAD/H-box helicases as therapeutic targets or a direct targeting of these helicases have been attempted (140, 150, 184, 185).
DEAD/H-box helicases have potentials to be targeted for future drug developments due to their unveiled ties with cell toxicity in various cancers (52). The lethal characteristic of helicases becomes more prominent upon their inhibition in overexpressing cancers. Upregulation of DDX5 leads to poor patient outcomes through the promotion of tumorigenesis and tumor recurrence (74). Consequently, depletion of DDX5 leads to the suppression of the mammalian target of rapamycin complex 1 (mTORC1) signaling pathway and induces apoptosis in prostate cancer cells (184). Compounds targeting DDX5, such as Resveratrol and RX-5902 (186), have been developed; however, further studies are required to evaluate anti-cancer effectiveness of these drugs. DHX9 is overexpressed in lung cancer (187) and its suppression is selectively lethal to cancer cells, but is tolerable for normal cells (188). Active screening for DHX9 inhibitors is in progress, as they can potentially be used as therapeutic agents specifically attacking cancer cells (140).
Another exciting prospect is to identify SL and SDL partners that have already been implicated in cell apoptosis in a cancer-specific manner or whose DEAD/H-box helicase partners, when overexpressed, have a putative apoptotic nature. An elevated expression of DDX10 in cancer leads to a poor survival rate in chondrosarcoma patients (78). Additionally, inhibition of α-synuclein displays tumor growth suppression in melanoma cells (189). Since DDX10 is a strong enhancer of α-synuclein-induced toxicity (150), we can employ SDL interaction between DDX10 and α-synuclein and use α-synuclein inhibitors as promising therapeutics for cancer, such as melanoma. Fortunately, various drugs used in PD’s model follow the same principle of α-synuclein inhibition and can be evaluated for cancer therapy (185). However, this may be context dependent as DDX10 is found to be both overexpressed as well as lost in cancers (Figure 2).
Many DEAD/H-box helicases, such as DDX3, DDX5, DDX10, and DDX21, have been reported to act both as oncogenes and tumor suppressors in different contexts. Therefore, in-depth research is essential to understand the intricacy and interplay of their dual antagonistic roles, and to ultimately, to effectively use them as targets for cancer treatment. Generally, the SDL approach might be useful in cancers with over-expressed DEAD/H-box helicases that function as oncogenes, while the SL approach might work with DEAD/H-box helicases that act as tumor suppressors. In both scenarios, the outcome is to promote cancer-specific apoptosis through pharmacological interventions. Identifying and manipulating these interacting partners open new avenues for drug development in cancer-targeted therapy.
Paralog genes arise from gene duplication, an evolutionary mechanism for creating new genes, which result in two functionally distinct genes, or more frequently, functionally overlapping genes. In fact, 13700 or two-thirds of human protein coding genes are paralogous (190). Paralogs provide both unique opportunities and challenges for the SL approach in developing targeted therapies. If a single gene is depleted, its paralog can compensate by taking over its function (191). Thus, the loss of a single gene is well tolerated by the cell; this phenomenon is called paralog buffering. However, if both paralogs are depleted, there is no mechanism in place to compensate for the lost function and this results in cell lethality (190). Recent CRISPR-based screenings and mining of publicly available data have identified several SL interactions among paralogs (159, 190–194). Paralog dependency is found in CSTF2-CSTF2T, DNAJC15-DNAJC19, FAM50A-FAM50B, and RPP25-RPP25L (159), CCNL1-CCNL2, CDK4-CDK6, MEK1-MEK2, and OXSR1-STK39 (190), CNOT7-CNOT8, COPS7A-COPS7B, CCNE1-CCNE2, and CCNT1-CCNT2 (192), STK38–STK38L and TET1–TET2 gene combinations (194). Paralog redundancy has been identified in CCNL1-CCNL2, OXSR1-STK39, EIF1-EIF1B, G3BP1-G3BP2, GFPT1-GFPT2, and PDS5A-PDS5B (190), MAP2K1-MAP2K2, RAS-RAF, FAM50A-FAM50B (192), sex chromosome genes ZFX-ZFY, DDX3X-DDX3Y, EIF1AX-EIF1AY (159), SAR1A–SAR1B, RAB1A–RAB1B, LDHA–LDHB, RBM26–RBM27 and hnRNPF–hnRNPH3 gene pairs (194) Recently, it was reported that VRK1 is a SL target in VRK2-mutated or silenced cancers (195, 196) and SMARCA2 is a SL target in SMARCA4 mutated cancers (197), suggesting SL is an excellent approach for paralog-related cancer treatment. Although ARID1A and ARID1B are synthetic lethal (198), it was reported that dual ARID1A-ARID1B loss leads to rapid carcinogenesis (199). This finding emphasizes that caution should be executed when developing new paralog-directed SL therapies. Lastly, some genes have more than two paralogs, such as Akt1-Akt2-Akt3 and RAD51-RAD51B-RAD51C-RAD51D-XRCC2-XRCC3, while FRG1 has 23 paralogs (200), which should present challenges to implementing the SL approach.
Multiple paralogues exist in the DEAD/H-box helicase family, including DDX2A-DDX2B, DDX3X-DDX3Y, DDX19A-DDX19B, DDX60-DDX60L and DDX39A-DDX39B. The DDX19A-DDX19B paralog pair engages in the SL interaction, where enhanced DDX19A expression is strictly required as a compensatory response to the low level of DDX19B (191, 193). Besides the DDX3X-DDX3Y and DDX19A-DDX19B pairs, the remaining paralog pairs have not been studied. Thus, computational predictions combined with experimental validation are expected to expand the horizon of uncovering paralog genetic interactions among helicase genes that can be used as potential therapeutic targets.
Although various small molecules have been utilized to target patient-specific molecular alterations for personalized cancer treatment protocols, their efficiency rate is still far from the desired. A drawback we currently face with the personalized treatments is their off-target effects, as these targeted therapeutics fail to effectively differentiate normal cells from the cancerous ones, ultimately leading to cytotoxicity. As we solve the structure-function relationship and interaction network of various DEAD/H-box helicases, it appears that we cannot limit their function as only molecular motors for nucleotides. LOF and overexpression of DEAD/H-box helicases in specific cancers are likely to provide an effective platform for developing more selective treatment approaches by inhibiting SL/SDL partners of these molecules, thus revolutionizing the arena of personalized genotype-based targeted therapeutics. Combination therapy of SL/SDL drugs with known chemotherapeutics might also synergistically improve patient outcomes. These approaches may provide a better window for therapeutic index optimization and minimize undesirable off-target effects associated with drug administration. It can also further optimize patient-specific treatment plans by targeting genetic vulnerabilities associated with their specific mutations in certain cancer subtypes and design solutions to LOF mutations that obviously cannot be targeted by our traditional approaches.
Despite the recent limelight and efforts in SL target discovery, there are only a handful of success stories in SL drug development reaching clinical trials. A molecule targeted by an SL drug may deregulate multiple biological processes, as different pathways share components, leading to more adverse patient outcomes. Thus, more in-depth understanding of the mechanisms is essential to unveil the complexities and heterogeneity of the SL interactome to pinpoint the molecular network and dependencies across various cancer types. The discovery of SL/SDL interacting compounds to enhance drug selectivity and design new effective combination therapies will make this process even more exciting. By investigating DEAD/H-box helicases within the SL/SDL context and implementing the generated knowledge, we can accelerate the process of bringing novel drugs to the bedside and positively impact cancer patient outcomes.
AA: Conceptualization, Data Curation, Writing, Visualization. HP and RS: Data Curation, Revision. FSV: Data Curation (TCGA). AK and AF: Revision. FJV and YW: Conceptualization, Data Curation, Writing and Editing, Visualization, Supervision, Funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Canada Foundation for Innovation (CFI-33364) and Cancer Research Society (CRS-22493) to FJV, and the Natural Sciences and Engineering Research Council of Canada (NSERC) (RGPIN-2019-05487), the Leukemia & Lymphoma Society of Canada (LLSC-356432), the Cancer Research Society (CRS-24139), and the Saskatchewan Health Research Foundation (SHRF-5093) to YW.
The authors apologize to investigators whose contributions could not be appropriately cited because of space limitations.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Tuteja N, Tuteja R. Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery. Eur J Biochem (2004) 271:1835–48. doi: 10.1111/j.1432-1033.2004.04093.x
2. Brosh RM Jr., Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res (2007) 35:7527–44. doi: 10.1093/nar/gkm1008
3. Dillingham MS. Superfamily I helicases as modular components of DNA-processing machines. Biochem Soc Trans (2011) 39:413–23. doi: 10.1042/BST0390413
4. Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet (2010) 44:393–417. doi: 10.1146/annurev-genet-102209-163602
5. Jankowsky E. RNA Helicases at work: binding and rearranging. Trends Biochem Sci (2011) 36:19–29. doi: 10.1016/j.tibs.2010.07.008
6. Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol (2013) 10:121–32. doi: 10.4161/rna.23312
7. Dhar S, Datta A, Brosh RM Jr. DNA Helicases and their roles in cancer. DNA Repair (Amst) (2020) 96:102994. doi: 10.1016/j.dnarep.2020.102994
8. Ashworth A, Lord CJ. Synthetic lethal therapies for cancer: what's next after PARP inhibitors? Nat Rev Clin Oncol (2018) 15:564–76. doi: 10.1038/s41571-018-0055-6
9. Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science (2017) 355:1152–8. doi: 10.1126/science.aam7344
10. Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys (2008) 37:317–36. doi: 10.1146/annurev.biophys.37.032807.125908
11. Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem (2007) 76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300
13. Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol (2010) 20:313–24. doi: 10.1016/j.sbi.2010.03.011
14. Linder P, Fuller-Pace F. Happy birthday: 25 years of DEAD-box proteins. Methods Mol Biol (2015) 1259:17–33. doi: 10.1007/978-1-4939-2214-7_2
15. Putnam AA, Jankowsky E. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim Biophys Acta (2013) 1829:884–93. doi: 10.1016/j.bbagrm.2013.02.002
16. Abdelhaleem M, Maltais L, Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics (2003) 81:618–22. doi: 10.1016/S0888-7543(03)00049-1
17. Gilman B, Tijerina P, Russell R. Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs. Biochem Soc Trans (2017) 45:1313–21. doi: 10.1042/BST20170095
18. Dardenne E, Polay Espinoza M, Fattet L, Germann S, Lambert MP, Neil H, et al. RNA Helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep (2014) 7:1900–13. doi: 10.1016/j.celrep.2014.05.010
19. Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res (2006) 34:4206–15. doi: 10.1093/nar/gkl460
20. Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Lee YH. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res (2006) 66:6579–88. doi: 10.1158/0008-5472.CAN-05-2415
21. Koltowska K, Okuda KS, Gloger M, Rondon-Galeano M, Mason E, Xuan J, et al. The RNA helicase Ddx21 controls vegfc-driven developmental lymphangiogenesis by balancing endothelial cell ribosome biogenesis and p53 function. Nat Cell Biol (2021) 23:1136–47. doi: 10.1038/s41556-021-00784-w
22. Saporita AJ, Chang HC, Winkeler CL, Apicelli AJ, Kladney RD, Wang J, et al. RNA Helicase DDX5 is a p53-independent target of ARF that participates in ribosome biogenesis. Cancer Res (2011) 71:6708–17. doi: 10.1158/0008-5472.CAN-11-1472
23. Li H, Lai P, Jia J, Song Y, Xia Q, Huang K, et al. RNA Helicase DDX5 inhibits reprogramming to pluripotency by miRNA-based repression of RYBP and its PRC1-dependent and -independent functions. Cell Stem Cell (2017) 20:462–477 e6. doi: 10.1016/j.stem.2016.12.002
24. Remenyi J, Bajan S, Fuller-Pace FV, Arthur JS, Hutvagner G. The loop structure and the RNA helicase p72/DDX17 influence the processing efficiency of the mice miR-132. Sci Rep (2016) 6:22848. doi: 10.1038/srep22848
25. Mathew R, Hartmuth K, Mohlmann S, Urlaub H, Ficner R, Luhrmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat Struct Mol Biol (2008) 15:435–43. doi: 10.1038/nsmb.1415
26. Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen B, Hosono N, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell (2015) 27:658–70. doi: 10.1016/j.ccell.2015.03.017
27. Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with p-bodies and stress granules. Mol Biol Cell (2007) 18:1385–96. doi: 10.1091/mbc.e06-12-1120
28. Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J (2012) 441:119–29. doi: 10.1042/BJ20110739
29. Brennan R, Haap-Hoff A, Gu L, Gautier V, Long A, Schroder M. Investigating nucleo-cytoplasmic shuttling of the human DEAD-box helicase DDX3. Eur J Cell Biol (2018) 97:501–11. doi: 10.1016/j.ejcb.2018.08.001
30. Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues JP, Boscheron C, et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J (1999) 18:4332–47. doi: 10.1093/emboj/18.15.4332
31. Sheng Y, Tsai-Morris CH, Gutti R, Maeda Y, Dufau ML. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is a transport protein involved in gene-specific mRNA export and protein translation during spermatogenesis. J Biol Chem (2006) 281:35048–56. doi: 10.1074/jbc.M605086200
32. Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature (2019) 573:144–8. doi: 10.1038/s41586-019-1502-y
33. Tauber D, Tauber G, Khong A, Van Treeck B, Pelletier J, Parker R. Modulation of RNA condensation by the DEAD-box protein eIF4A. Cell (2020) 180:411–426.e16. doi: 10.1016/j.cell.2019.12.031
34. Shen H, Yanas A, Owens MC, Zhang C, Fritsch C, Fare CM, et al. Sexually dimorphic RNA helicases DDX3X and DDX3Y differentially regulate RNA metabolism through phase separation. Mol Cell (2022) 82:2588–603.e9. doi: 10.1016/j.molcel.2022.04.022
35. Geissler V, Altmeyer S, Stein B, Uhlmann-Schiffler H, Stahl H. The RNA helicase Ddx5/p68 binds to hUpf3 and enhances NMD of Ddx17/p72 and Smg5 mRNA. Nucleic Acids Res (2013) 41:7875–88. doi: 10.1093/nar/gkt538
36. Hug N, Aitken S, Longman D, Raab M, Armes H, Mann AR, et al. A dual role for the RNA helicase DHX34 in NMD and pre-mRNA splicing and its function in hematopoietic differentiation. RNA (2022) 28:1224–38. doi: 10.1261/rna.079277.122
37. Sun J, Wu G, Pastor F, Rahman N, Wang WH, Zhang Z, et al. RNA Helicase DDX5 enables STAT1 mRNA translation and interferon signalling in hepatitis b virus replicating hepatocytes. Gut (2021) 0:1–15. doi: 10.1136/gutjnl-2020-323126
38. Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, et al. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol (2011) 12:235–45. doi: 10.1038/nrm3083
39. Ryan CS, Schroder M. The human DEAD-box helicase DDX3X as a regulator of mRNA translation. Front Cell Dev Biol (2022) 10:1033684. doi: 10.3389/fcell.2022.1033684
40. Mikhailova T, Shuvalova E, Ivanov A, Susorov D, Shuvalov A, Kolosov PM, et al. RNA Helicase DDX19 stabilizes ribosomal elongation and termination complexes. Nucleic Acids Res (2017) 45:1307–18. doi: 10.1093/nar/gkw1239
41. Chen HH, Yu HI, Tarn WY. DDX3 modulates neurite development via translationally activating an RNA regulon involved in Rac1 activation. J Neurosci (2016) 36:9792–804. doi: 10.1523/JNEUROSCI.4603-15.2016
42. Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem (1999) 68:913–63. doi: 10.1146/annurev.biochem.68.1.913
43. Cargill M, Venkataraman R, Lee S. DEAD-box RNA helicases and genome stability. Genes (Basel) 12 (2021). doi: 10.3390/genes12101471
44. Cargill MJ, Morales A, Ravishankar S, Warren EH. RNA Helicase, DDX3X, is actively recruited to sites of DNA damage in live cells. DNA Repair (Amst) (2021) 103:103137. doi: 10.1016/j.dnarep.2021.103137
45. Li L, Germain DR, Poon HY, Hildebrandt MR, Monckton EA, McDonald D, et al. DEAD box 1 facilitates removal of RNA and homologous recombination at DNA double-strand breaks. Mol Cell Biol (2016) 36:2794–810. doi: 10.1128/MCB.00415-16
46. Li L, Monckton EA, Godbout R. A role for DEAD box 1 at DNA double-strand breaks. Mol Cell Biol (2008) 28:6413–25. doi: 10.1128/MCB.01053-08
47. Xu Z, Li X, Li H, Nie C, Liu W, Li S, et al. Suppression of DDX39B sensitizes ovarian cancer cells to DNA-damaging chemotherapeutic agents via destabilizing BRCA1 mRNA. Oncogene (2020) 39:7051–62. doi: 10.1038/s41388-020-01482-x
48. Bonaventure B, Goujon C. DExH/D-box helicases at the frontline of intrinsic and innate immunity against viral infections. J Gen Virol (2022) 103:001766. doi: 10.1099/jgv.0.001766
49. Bruns AM, Horvath CM. LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine (2015) 74:198–206. doi: 10.1016/j.cyto.2015.02.010
50. Zhang L, Yang Y, Li B, Scott IC, Lou X. The DEAD-box RNA helicase Ddx39ab is essential for myocyte and lens development in zebrafish. Development (2018) 145. doi: 10.1242/dev.161018
51. Hozumi S, Hirabayashi R, Yoshizawa A, Ogata M, Ishitani T, Tsutsumi M, et al. DEAD-box protein Ddx46 is required for the development of the digestive organs and brain in zebrafish. PLos One (2012) 7:e33675. doi: 10.1371/journal.pone.0033675
52. Samir P, Kanneganti TD. DEAD/H-box helicases in immunity, inflammation, cell differentiation, and cell death and disease. Cells 11 (2022) 11:1608. doi: 10.3390/cells11101608
53. Szczesny RJ, Wojcik MA, Borowski LS, Szewczyk MJ, Skrok MM, Golik P, et al. Yeast and human mitochondrial helicases. Biochim Biophys Acta (2013) 1829:842–53. doi: 10.1016/j.bbagrm.2013.02.009
54. van der Lelij P, Chrzanowska KH, Godthelp BC, Rooimans MA, Oostra AB, Stumm M, et al. Warsaw Breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am J Hum Genet (2010) 86:262–6. doi: 10.1016/j.ajhg.2010.01.008
55. de Castro Fonseca M, de Oliveira JF, Araujo BHS, Canateli C, do Prado PFV, Amorim Neto DP, et al. Molecular and cellular basis of hyperassembly and protein aggregation driven by a rare pathogenic mutation in DDX3X. iScience (2021) 24:102841. doi: 10.1016/j.isci.2021.102841
56. Moresco G, Costanza J, Santaniello C, Rondinone O, Grilli F, Prada E, et al. A novel de novo DDX3X missense variant in a female with brachycephaly and intellectual disability: a case report. Ital J Pediatr (2021) 47:81. doi: 10.1186/s13052-021-01033-4
57. Lennox AL, Hoye ML, Jiang R, Johnson-Kerner BL, Suit LA, Venkataramanan S, et al. Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development. Neuron (2020) 106:404–420.e8. doi: 10.1016/j.neuron.2020.01.042
58. Nicola P, Blackburn PR, Rasmussen KJ, Bertsch NL, Klee EW, Hasadsri L, et al. De novo DDX3X missense variants in males appear viable and contribute to syndromic intellectual disability. Am J Med Genet Part A (2019) 179:570–8. doi: 10.1002/ajmg.a.61061
59. Scala M, Torella A, Severino M, Morana G, Castello R, Accogli A, et al. Three de novo DDX3X variants associated with distinctive brain developmental abnormalities and brain tumor in intellectually disabled females. Eur J Hum Genet (2019) 27:1254–9. doi: 10.1038/s41431-019-0392-7
60. Balak C, Benard M, Schaefer E, Iqbal S, Ramsey K, Ernoult-Lange M, et al. Rare De novo missense variants in RNA helicase DDX6 cause intellectual disability and dysmorphic features and lead to p-body defects and RNA dysregulation. Am J Hum Genet (2019) 105:509–25. doi: 10.1016/j.ajhg.2019.07.010
61. Badar T, Chlon T. Germline and somatic defects in DDX41 and its impact on myeloid neoplasms. Curr Hematol Malig Rep (2022) 17:113–20. doi: 10.1007/s11899-022-00667-3
62. Makishima H, Bowman TV, Godley LA. DDX41-associated susceptibility to myeloid neoplasms. Blood (2022). doi: 10.1182/blood.2022017715
63. Yuan M, Xu J, Cao S, Sun S. DDX1 is a prognostic biomarker and correlates with immune infiltrations in hepatocellular carcinoma. BMC Immunol (2022) 23:59. doi: 10.1186/s12865-022-00533-0
64. Jin Y, Shi J, Wang H, Lu J, Chen C, Yu Y, et al. MYC-associated protein X binding with the variant rs72780850 in RNA helicase DEAD box 1 for susceptibility to neuroblastoma. Sci China Life Sci (2021) 64:991–9. doi: 10.1007/s11427-020-1784-7
65. Lyu S, Lu J, Chen W, Huang W, Huang H, Xi S, et al. High expression of eIF4A2 is associated with a poor prognosis in esophageal squamous cell carcinoma. Oncol Lett (2020) 20:177. doi: 10.3892/ol.2020.12038
66. Gao C, Guo X, Xue A, Ruan Y, Wang H, Gao X. High intratumoral expression of eIF4A1 promotes epithelial-to-mesenchymal transition and predicts unfavorable prognosis in gastric cancer. Acta Biochim Biophys Sin (2020) 52:310–9. doi: 10.1093/abbs/gmz168
67. Shaoyan X, Juanjuan Y, Yalan T, Ping H, Jianzhong L, Qinian W. Downregulation of EIF4A2 in non-small-cell lung cancer associates with poor prognosis. Clin Lung Cancer (2013) 14:658–65. doi: 10.1016/j.cllc.2013.04.011
68. Wilky BA, Kim C, McCarty G, Montgomery EA, Kammers K, DeVine LR, et al. RNA Helicase DDX3: a novel therapeutic target in Ewing sarcoma. Oncogene (2016) 35:2574–83. doi: 10.1038/onc.2015.336
69. Chen HH, Yu HI, Yang MH, Tarn WY. DDX3 activates CBC-eIF3-Mediated translation of uORF-containing oncogenic mRNAs to promote metastasis in HNSCC. Cancer Res (2018) 78:4512–23. doi: 10.1158/0008-5472.CAN-18-0282
70. Su CY, Lin TC, Lin YF, Chen MH, Lee CH, Wang HY, et al. DDX3 as a strongest prognosis marker and its downregulation promotes metastasis in colorectal cancer. Oncotarget (2015) 6:18602–12. doi: 10.18632/oncotarget.4329
71. Chang PC, Chi CW, Chau GY, Li FY, Tsai YH, Wu JC, et al. DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control. Oncogene (2006) 25:1991–2003. doi: 10.1038/sj.onc.1209239
72. D'Oronzo S, Silvestris E, Lovero D, Cafforio P, Duda L, Cormio G, et al. DEAD-box helicase 4 (Ddx4)(+) stem cells sustain tumor progression in non-serous ovarian cancers. Int J Mol Sci (2020) 21:6096. doi: 10.3390/ijms21176096
73. Hashimoto H, Sudo T, Mikami Y, Otani M, Takano M, Tsuda H, et al. Germ cell specific protein VASA is over-expressed in epithelial ovarian cancer and disrupts DNA damage-induced G2 checkpoint. Gynecol Oncol (2008) 111:312–9. doi: 10.1016/j.ygyno.2008.08.014
74. Li K, Zhao G, Yuan H, Zhang J, Li Q, Gong D, et al. Upregulated expression of DDX5 predicts recurrence and poor prognosis in breast cancer. Pathol Res Pract (2022) 229:153736. doi: 10.1016/j.prp.2021.153736
75. Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res (2008) 68:7938–46. doi: 10.1158/0008-5472.CAN-08-0932
76. Taniguchi K, Iwatsuki A, Sugito N, Shinohara H, Kuranaga Y, Oshikawa Y, et al. Oncogene RNA helicase DDX6 promotes the process of c-myc expression in gastric cancer cells. Mol Carcinog (2018) 57:579–89. doi: 10.1002/mc.22781
77. Nakagawa Y, Morikawa H, Hirata I, Shiozaki M, Matsumoto A, Maemura K, et al. Overexpression of rck/p54, a DEAD box protein, in human colorectal tumours. Br J Cancer (1999) 80:914–7. doi: 10.1038/sj.bjc.6690441
78. Quan X, Zhao C, Gao Z, Zhang Y, Zhao R, Wang J, et al. DDX10 and BYSL as the potential targets of chondrosarcoma and glioma. Med (Baltimore) (2021) 100:e27669. doi: 10.1097/MD.0000000000027669
79. Zhou X, Liu Z, He T, Zhang C, Jiang M, Jin Y, et al. DDX10 promotes the proliferation and metastasis of colorectal cancer cells via splicing RPL35. Cancer Cell Int (2022) 22:58. doi: 10.1186/s12935-022-02478-1
80. Li J, Liu L, Liu X, Xu P, Hu Q, Yu Y. The role of upregulated DDX11 as a potential prognostic and diagnostic biomarker in lung adenocarcinoma. J Cancer (2019) 10:4208–16. doi: 10.7150/jca.33457
81. Park JS, Lee ME, Jang WS, Rha KH, Lee SH, Lee J, et al. The DEAD/DEAH box helicase, DDX11, is essential for the survival of advanced clear cell renal cell carcinoma and is a determinant of PARP inhibitor sensitivity. Cancers (Basel) (2021) 13:2574. doi: 10.3390/cancers13112574
82. Zhou HZ, Li F, Cheng ST, Xu Y, Deng HJ, Gu DY, et al. DDX17-regulated alternative splicing that produced an oncogenic isoform of PXN-AS1 to promote HCC metastasis. Hepatology (2022) 75:847–65. doi: 10.1002/hep.32195
83. Liu X, Li L, Geng C, Wen S, Zhang C, Deng C, et al. DDX17 promotes the growth and metastasis of lung adenocarcinoma. Cell Death Discovery (2022) 8:425. doi: 10.1038/s41420-022-01215-x
84. Zhang Y, Yu F, Ni B, Li Q, Bae SW, Choi JH, et al. The RNA-binding protein DDX18 promotes gastric cancer by affecting the maturation of MicroRNA-21. Front Oncol (2020) 10:598238. doi: 10.3389/fonc.2020.598238
85. Chen N, Zhang G, Fu J, Wu Q. Identification of key modules and hub genes involved in esophageal squamous cell carcinoma tumorigenesis using WCGNA. Cancer Control (2020) 27:1073274820978817. doi: 10.1177/1073274820978817
86. Payne EM, Bolli N, Rhodes J, Abdel-Wahab OI, Levine R, Hedvat CV, et al. Ddx18 is essential for cell-cycle progression in zebrafish hematopoietic cells and is mutated in human AML. Blood (2011) 118:903–15. doi: 10.1182/blood-2010-11-318022
87. Wang Q, Ye Y, Lin R, Weng S, Cai F, Zou M, et al. Analysis of the expression, function, prognosis and co-expression genes of DDX20 in gastric cancer. Comput Struct Biotechnol J (2020) 18:2453–62. doi: 10.1016/j.csbj.2020.09.002
88. Yang Y, Yang M, Pang H, Qiu Y, Sun T, Wang T, et al. A macrophage differentiation-mediated gene: DDX20 as a molecular biomarker encompassing the tumor microenvironment, disease staging, and prognoses in hepatocellular carcinoma. Oxid Med Cell Longev (2022) 2022:9971776. doi: 10.1155/2022/9971776
89. Tanaka A, Wang JY, Shia J, Zhou Y, Ogawa M, Hendrickson RC, et al. DEAD-box RNA helicase protein DDX21 as a prognosis marker for early stage colorectal cancer with microsatellite instability. Sci Rep (2020) 10:22085. doi: 10.1038/s41598-020-79049-9
90. Putra V, Hulme AJ, Tee AE, Sun JQJ, Atmadibrata B, Ho N, et al. The RNA-helicase DDX21 upregulates CEP55 expression and promotes neuroblastoma. Mol Oncol (2021) 15:1162–79. doi: 10.1002/1878-0261.12906
91. Zhang H, Zhang Y, Chen C, Zhu X, Zhang C, Xia Y, et al. A double-negative feedback loop between DEAD-box protein DDX21 and snail regulates epithelial-mesenchymal transition and metastasis in breast cancer. Cancer Lett (2018) 437:67–78. doi: 10.1016/j.canlet.2018.08.021
92. Yin J, Park G, Lee JE, Choi EY, Park JY, Kim TH, et al. DEAD-box RNA helicase DDX23 modulates glioma malignancy via elevating miR-21 biogenesis. Brain (2015) 138:2553–70. doi: 10.1093/brain/awv167
93. Zhao C, Li Y, Qiu C, Chen J, Wu H, Wang Q, et al. Splicing factor DDX23, transcriptionally activated by E2F1, promotes ovarian cancer progression by regulating FOXM1. Front Oncol (2021) 11:749144. doi: 10.3389/fonc.2021.749144
94. Jin Y, Yang S, Gao X, Chen D, Luo T, Su S, et al. DEAD-box helicase 27 triggers epithelial to mesenchymal transition by regulating alternative splicing of lipoma-preferred partner in gastric cancer metastasis. Front Genet (2022) 13:836199. doi: 10.3389/fgene.2022.836199
95. Xiaoqian W, Bing Z, Yangwei L, Yafei Z, Tingting Z, Yi W, et al. DEAD-box helicase 27 promotes hepatocellular carcinoma progression through ERK signaling. Technol Cancer Res Treat (2021) 20:15330338211055953. doi: 10.1177/15330338211055953
96. Xie Y, Liu Y, Ding J, Li G, Ni B, Pang H, et al. Identification of DDX31 as a potential oncogene of invasive metastasis and proliferation in PDAC. Front Cell Dev Biol (2022) 10:762372. doi: 10.3389/fcell.2022.762372
97. Daizumoto K, Yoshimaru T, Matsushita Y, Fukawa T, Uehara H, Ono M, et al. A DDX31/Mutant-p53/EGFR axis promotes multistep progression of muscle-invasive bladder cancer. Cancer Res (2018) 78:2233–47. doi: 10.1158/0008-5472.CAN-17-2528
98. Zhang H, He C, Guo X, Fang Y, Lai Q, Wang X, et al. DDX39B contributes to the proliferation of colorectal cancer through direct binding to CDK6/CCND1. Cell Death Discovery (2022) 8:30. doi: 10.1038/s41420-022-00827-7
99. Wei J, Lu J, Cao Y, Yao G, Huang Y, Zhao H, et al. DDX39B predicts poor survival and associated with clinical benefit of anti-PD-L1 therapy in ccRCC. Curr Cancer Drug Targets (2021) 21:849–59. doi: 10.2174/1568009621666210811115054
100. Mathieu MG, Linley AJ, Reeder SP, Badoual C, Tartour E, Rees RC, et al. HAGE, a cancer/testis antigen expressed at the protein level in a variety of cancers. Cancer Immun (2010) 10:2.
101. Ma N, Xu HE, Luo Z, Zhou J, Zhou Y, Liu M. Expression and significance of DDX43 in lung adenocarcinoma. Pakistan J Pharm Sci (2017) 30:1491–6.
102. Lian X, Xiang D, Peng C, Chen J, Liao M, Sun G, et al. DDX49 is a novel biomarker and therapeutic target for lung cancer metastases. J Cell Mol Med (2020) 24:1141–5. doi: 10.1111/jcmm.14734
103. Wang X, Gu G, Zhu H, Lu S, Abuduwaili K, Liu C. LncRNA SNHG20 promoted proliferation, invasion and inhibited cell apoptosis of lung adenocarcinoma via sponging miR-342 and upregulating DDX49. Thorac Cancer (2020) 11:3510–20. doi: 10.1111/1759-7714.13693
104. Cho B, Lim Y, Lee DY, Park SY, Lee H, Kim WH, et al. Identification and characterization of a novel cancer/testis antigen gene CAGE. Biochem Biophys Res Commun (2002) 292:715–26. doi: 10.1006/bbrc.2002.6701
105. Xu Y, Wang X, Chu Y, Li J, Wang W, Hu X, et al. Analysis of transcript-wide profile regulated by microsatellite instability of colorectal cancer. Ann Transl Med (2022) 10:169. doi: 10.21037/atm-21-6126
106. Wang J, Wang Y, Wang J, Zhang S, Yu Z, Zheng K, et al. DEAD-box helicase 56 functions as an oncogene promote cell proliferation and invasion in gastric cancer via the FOXO1/p21 Cip1/c-myc signaling pathway. Bioengineered (2022) 13:13970–85. doi: 10.1080/21655979.2022.2084235
107. Zhou H, Du Y, Wei X, Song C, Song J, Xu N, et al. DDX56 transcriptionally activates MIST1 to facilitate tumorigenesis of HCC through PTEN-AKT signaling. Theranostics (2022) 12:6069–87. doi: 10.7150/thno.72471
108. Wolf D, Fiegl H, Zeimet AG, Wieser V, Marth C, Sprung S, et al. High RIG-I expression in ovarian cancer associates with an immune-escape signature and poor clinical outcome. Int J Cancer (2020) 146:2007–18. doi: 10.1002/ijc.32818
109. Beyer S, Muller L, Mitter S, Keilmann L, Meister S, Buschmann C, et al. And EFTUD2 expression predicts poor survival in endometrial cancer. J Cancer Res Clin Oncol (2022). doi: 10.1007/s00432-022-04271-z
110. Song J, Zhao W, Zhang X, Tian W, Zhao X, Ma L, et al. Mutant RIG-I enhances cancer-related inflammation through activation of circRIG-I signaling. Nat Commun (2022) 13:7096. doi: 10.1038/s41467-022-34885-3
111. Chen X, Lin L, Chen G, Yan H, Li Z, Xiao M, et al. High levels of DEAH-box helicases relate to poor prognosis and reduction of DHX9 improves radiosensitivity of hepatocellular carcinoma. Front Oncol (2022) 12:900671. doi: 10.3389/fonc.2022.900671
112. Chellini L, Pieraccioli M, Sette C, Paronetto MP. The DNA/RNA helicase DHX9 contributes to the transcriptional program of the androgen receptor in prostate cancer. J Exp Clin Cancer Res (2022) 41:178. doi: 10.1186/s13046-022-02384-4
113. Chen Y, Chen X, Pan L, Huang Y, Cai Y, Li J, et al. RNA Helicase DHX15 decreases cell apoptosis by NF-kappaB signaling pathway in burkitt lymphoma. Cancer Cell Int (2022) 22:92. doi: 10.1186/s12935-021-02426-5
114. Xie C, Liao H, Zhang C, Zhang S. Overexpression and clinical relevance of the RNA helicase DHX15 in hepatocellular carcinoma. Hum Pathol (2019) 84:213–20. doi: 10.1016/j.humpath.2018.10.006
115. Hu X, Yuan G, Li Q, Huang J, Cheng X, Chen J. DEAH-box polypeptide 32 promotes hepatocellular carcinoma progression via activating the beta-catenin pathway. Ann Med (2021) 53:437–47. doi: 10.1080/07853890.2021.1898674
116. Huang C, Liang X, Huang R, Zhang Z. Up-regulation and clinical relevance of novel helicase homologue DHX32 in colorectal cancer. J Exp Clin Cancer Res (2009) 28:11. doi: 10.1186/1756-9966-28-11
117. Abdelhaleem M. The novel helicase homologue DDX32 is down-regulated in acute lymphoblastic leukemia. Leukemia Res (2002) 26:945–54. doi: 10.1016/S0145-2126(02)00040-1
118. Zhu Y, Du Y, Zhang Y. DHX33 promotes colon cancer development downstream of wnt signaling. Gene (2020) 735:144402. doi: 10.1016/j.gene.2020.144402
119. Tian QH, Zhang MF, Luo RG, Fu J, He C, Hu G, et al. DHX33 expression is increased in hepatocellular carcinoma and indicates poor prognosis. Biochem Biophys Res Commun (2016) 473:1163–9. doi: 10.1016/j.bbrc.2016.04.033
120. Zeng Y, Qin T, Flamini V, Tan C, Zhang X, Cong Y, et al. Identification of DHX36 as a tumour suppressor through modulating the activities of the stress-associated proteins and cyclin-dependent kinases in breast cancer. Am J Cancer Res (2020) 10:4211–33.
121. Cui Y, Li Z, Cao J, Lane J, Birkin E, Dong X, et al. The G4 resolvase DHX36 possesses a prognosis significance and exerts tumour suppressing function through multiple causal regulations in non-small cell lung cancer. Front Oncol (2021) 11:655757. doi: 10.3389/fonc.2021.655757
122. Liu Z, Ye Y, Liu Y, Liu Y, Chen H, Shen M, et al. RNA Helicase DHX37 facilitates liver cancer progression by cooperating with PLRG1 to drive superenhancer-mediated transcription of cyclin D1. Cancer Res (2022) 82:1937–52. doi: 10.1158/0008-5472.CAN-21-3038
123. Huang K, Pang T, Tong C, Chen H, Nie Y, Wu J, et al. Integrative expression and prognosis analysis of DHX37 in human cancers by data mining. BioMed Res Int (2021) 2021:6576210. doi: 10.1155/2021/6576210
124. Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P Jr., et al. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene (2008) 27:3912–22. doi: 10.1038/onc.2008.33
125. Brai A, Riva V, Clementi L, Falsitta L, Zamperini C, Sinigiani V, et al. Targeting DDX3X helicase activity with BA103 shows promising therapeutic effects in preclinical glioblastoma models. Cancers (Basel) (2021) 13:5569. doi: 10.3390/cancers13215569
126. Miao X, Yang ZL, Xiong L, Zou Q, Yuan Y, Li J, et al. Nectin-2 and DDX3 are biomarkers for metastasis and poor prognosis of squamous cell/adenosquamous carcinomas and adenocarcinoma of gallbladder. Int J Clin Exp Pathol (2013) 6:179–90.
127. Yang C, Li D, Bai Y, Song S, Yan P, Wu R, et al. DEAD-box helicase 27 plays a tumor-promoter role by regulating the stem cell-like activity of human colorectal cancer cells. Onco Targets Ther (2019) 12:233–41. doi: 10.2147/OTT.S190814
128. Li S, Ma J, Zheng A, Song X, Chen S, Jin F. DEAD-box helicase 27 enhances stem cell-like properties with poor prognosis in breast cancer. J Transl Med (2021) 19:334. doi: 10.1186/s12967-021-03011-0
129. Martelange V, De SC, De PE, Lurquin C, Boon T. Identification on a human sarcoma of two new genes with tumor-specific expression. Cancer Res (2000) 60:3848–55.
130. Shin EM, Hay HS, Lee MH, Goh JN, Tan TZ, Sen YP, et al. DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J Clin Invest (2014) 124:3807–24. doi: 10.1172/JCI73451
131. Zhang Y, Baysac KC, Yee LF, Saporita AJ, Weber JD. Elevated DDX21 regulates c-jun activity and rRNA processing in human breast cancers. Breast Cancer Res (2014) 16:449. doi: 10.1186/s13058-014-0449-z
132. Cao J, Wu N, Han Y, Hou Q, Zhao Y, Pan Y, et al. DDX21 promotes gastric cancer proliferation by regulating cell cycle. Biochem Biophys Res Commun (2018) 505:1189–94. doi: 10.1016/j.bbrc.2018.10.060
133. Germain DR, Graham K, Glubrecht DD, Hugh JC, Mackey JR, Godbout R. DEAD box 1: a novel and independent prognostic marker for early recurrence in breast cancer. Breast Cancer Res Treat (2011) 127:53–63. doi: 10.1007/s10549-010-0943-7
134. Xie M, Vesuna F, Tantravedi S, Bol GM, Heerma van Voss MR, Nugent K, et al. RK-33 radiosensitizes prostate cancer cells by blocking the RNA helicase DDX3. Cancer Res (2016) 76:6340–50. doi: 10.1158/0008-5472.CAN-16-0440
135. Hashimoto K, Nakagawa Y, Morikawa H, Niki M, Egashira Y, Hirata I, et al. Co-Overexpression of DEAD box protein rck/p54 and c-myc protein in human colorectal adenomas and the relevance of their expression in cultured cell lines. Carcinogenesis (2001) 22:1965–70. doi: 10.1093/carcin/22.12.1965
136. Webber JT, Kaushik S, Bandyopadhyay S. Integration of tumor genomic data with cell lines using multi-dimensional network modules improves cancer pharmacogenomics. Cell Syst (2018) 7:526–536 e6. doi: 10.1016/j.cels.2018.10.001
137. Warren A, Chen Y, Jones A, Shibue T, Hahn WC, Boehm JS, et al. Global computational alignment of tumor and cell line transcriptional profiles. Nat Commun (2021) 12:22. doi: 10.1038/s41467-020-20294-x
138. Bol GM, Xie M, Raman V. DDX3, a potential target for cancer treatment. Mol Cancer (2015) 14:188. doi: 10.1186/s12943-015-0461-7
139. Churpek JE, Smith-Simmer K. DDX41-associated familial myelodysplastic syndrome and acute myeloid leukemia. Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, Amemiya A, editors. Seattle (WA: GeneReviews((R (2021).
140. Cencic R, Senechal P, Pelletier J. Establishment of a primary screening assay for the DHX9 helicase. Comb Chem High Throughput Screen (2015) 18:855–61. doi: 10.2174/1386207318666151019093433
141. Yoneyama-Hirozane M, Kondo M, Matsumoto SI, Morikawa-Oki A, Morishita D, Nakanishi A, et al. High-throughput screening to identify inhibitors of DEAD box helicase DDX41. SLAS Discovery advancing Life Sci R D (2017) 22:1084–92. doi: 10.1177/2472555217705952
142. Bol GM, Vesuna F, Xie M, Zeng J, Aziz K, Gandhi N, et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol Med (2015) 7:648–69. doi: 10.15252/emmm.201404368
143. Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol (2015) 7:a016600. doi: 10.1101/cshperspect.a016600
144. Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr (2014) 24:15–28. doi: 10.1615/CritRevEukaryotGeneExpr.2013006875
145. Kaur SD, Chellappan DK, Aljabali AA, Tambuwala M, Dua K, Kapoor DN. Recent advances in cancer therapy using PARP inhibitors. Med Oncol (2022) 39:241. doi: 10.1007/s12032-022-01840-7
146. Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics (1996) 143:95–102. doi: 10.1093/genetics/143.1.95
147. Duffy S, Fam HK, Wang YK, Styles EB, Kim JH, Ang JS, et al. Overexpression screens identify conserved dosage chromosome instability genes in yeast and human cancer. Proc Natl Acad Sci U.S.A. (2016) 113:9967–76. doi: 10.1073/pnas.1611839113
148. Heerma van Voss MR, Brilliant JD, Vesuna F, Bol GM, van der Wall E, van Diest PJ, et al. Combination treatment using DDX3 and PARP inhibitors induces synthetic lethality in BRCA1-proficient breast cancer. Med Oncol (2017) 34:33. doi: 10.1007/s12032-017-0889-2
149. Daugeron MC, Kressler D, Linder P. Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p. RNA (2001) 7:1317–34. doi: 10.1017/S1355838201010640
150. Popova B, Wang D, Patz C, Akkermann D, Lazaro DF, Galka D, et al. DEAD-box RNA helicase Dbp4/DDX10 is an enhancer of alpha-synuclein toxicity and oligomerization. PLos Genet (2021) 17:e1009407. doi: 10.1371/journal.pgen.1009407
151. de Lange J, Faramarz A, Oostra AB, de Menezes RX, van der Meulen IH, Rooimans MA, et al. Defective sister chromatid cohesion is synthetically lethal with impaired APC/C function. Nat Commun (2015) 6:8399. doi: 10.1038/ncomms9399
152. Faramarz A, Balk JA, van Schie JJM, Oostra AB, Ghandour CA, Rooimans MA, et al. Non-redundant roles in sister chromatid cohesion of the DNA helicase DDX11 and the SMC3 acetyl transferases ESCO1 and ESCO2. PLos One (2020) 15:e0220348. doi: 10.1371/journal.pone.0220348
153. Abe T, Kawasumi R, Arakawa H, Hori T, Shirahige K, Losada A, et al. Chromatin determinants of the inner-centromere rely on replication factors with functions that impart cohesion. Oncotarget (2016) 7:67934–47. doi: 10.18632/oncotarget.11982
154. Tsukamoto T, Gearhart MD, Kim S, Mekonnen G, Spike CA, Greenstein D. Insights into the involvement of spliceosomal mutations in myelodysplastic disorders from analysis of SACY-1/DDX41 in caenorhabditis elegans. Genetics (2020) 214:869–93. doi: 10.1534/genetics.119.302973
155. Venkataramanan S, Gadek M, Calviello L, Wilkins K, Floor SN. DDX3X and DDX3Y are redundant in protein synthesis. RNA (2021) 27:1577–88. doi: 10.1261/rna.078926.121
156. Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. Identification and characterization of essential genes in the human genome. Science (2015) 350:1096–101. doi: 10.1126/science.aac7041
157. Sekiguchi T, Iida H, Fukumura J, Nishimoto T. Human DDX3Y, the y-encoded isoform of RNA helicase DDX3, rescues a hamster temperature-sensitive ET24 mutant cell line with a DDX3X mutation. Exp Cell Res (2004) 300:213–22. doi: 10.1016/j.yexcr.2004.07.005
158. Chan CH, Chen CM, Lee YW, You LR. DNA Damage, liver injury, and tumorigenesis: Consequences of DDX3X loss. Mol Cancer Res MCR (2019) 17:555–66. doi: 10.1158/1541-7786.MCR-18-0551
159. Koferle A, Schlattl A, Hormann A, Thatikonda V, Popa A, Spreitzer F, et al. Interrogation of cancer gene dependencies reveals paralog interactions of autosome and sex chromosome-encoded genes. Cell Rep (2022) 39:110636. doi: 10.1016/j.celrep.2022.110636
160. Sun M, Song L, Zhou T, Gillespie GY, Jope RS. The role of DDX3 in regulating snail. Biochim Biophys Acta (2011) 1813:438–47. doi: 10.1016/j.bbamcr.2011.01.003
161. Hueng DY, Tsai WC, Chiou HY, Feng SW, Lin C, Li YF, et al. DDX3X biomarker correlates with poor survival in human gliomas. Int J Mol Sci (2015) 16:15578–91. doi: 10.3390/ijms160715578
162. Tsai WC, Hueng DY, Lin CR, Yang TCK, Nieh S, Gao HW. Applying DDX3X biomarker to discriminate atypical from benign meningiomas in tissue microarray. Appl Immunohistochem Mol Morphol (2018) 26:263–7. doi: 10.1097/PAI.0000000000000422
163. Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. New Engl J Med (2011) 365:2497–506. doi: 10.1056/NEJMoa1109016
164. Ojha J, Secreto CR, Rabe KG, Van Dyke DL, Kortum KM, Slager SL, et al. Identification of recurrent truncated DDX3X mutations in chronic lymphocytic leukaemia. Br J Haematol (2015) 169:445–8. doi: 10.1111/bjh.13211
165. Liang S, Yang Z, Li D, Miao X, Yang L, Zou Q, et al. The clinical and pathological significance of nectin-2 and DDX3 expression in pancreatic ductal adenocarcinomas. Dis Markers (2015) 2015:379568. doi: 10.1155/2015/379568
166. Jiang L, Gu ZH, Yan ZX, Zhao X, Xie YY, Zhang ZG, et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet (2015) 47:1061–6. doi: 10.1038/ng.3358
167. Bol GM, Raman V, van der Groep P, Vermeulen JF, Patel AH, van der Wall E, et al. Expression of the RNA helicase DDX3 and the hypoxia response in breast cancer. PLos One (2013) 8:e63548. doi: 10.1371/journal.pone.0063548
168. Cannizzaro E, Bannister AJ, Han N, Alendar A, Kouzarides T. DDX3X RNA helicase affects breast cancer cell cycle progression by regulating expression of KLF4. FEBS Lett (2018) 592:2308–22. doi: 10.1002/1873-3468.13106
169. van der Pol CC, Moelans CB, Manson QF, Batenburg MCT, van der Wall E, Borel Rinkes I, et al. Cytoplasmic DDX3 as prognosticator in male breast cancer. Virchows Arch (2021) 479:647–55. doi: 10.1007/s00428-021-03107-4
170. Li HK, Mai RT, Huang HD, Chou CH, Chang YA, Chang YW, et al. DDX3 represses stemness by epigenetically modulating tumor-suppressive miRNAs in hepatocellular carcinoma. Sci Rep (2016) 6:28637. doi: 10.1038/srep28637
171. Huang JS, Chao CC, Su TL, Yeh SH, Chen DS, Chen CT, et al. Diverse cellular transformation capability of overexpressed genes in human hepatocellular carcinoma. Biochem Biophys Res Commun (2004) 315:950–8. doi: 10.1016/j.bbrc.2004.01.151
172. Wu DW, Lee MC, Wang J, Chen CY, Cheng YW, Lee H. DDX3 loss by p53 inactivation promotes tumor malignancy via the MDM2/Slug/E-cadherin pathway and poor patient outcome in non-small-cell lung cancer. Oncogene (2014) 33:1515–26. doi: 10.1038/onc.2013.107
173. Heerma van Voss MR, Vesuna F, Trumpi K, Brilliant J, Berlinicke C, de Leng W, et al. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget (2015) 6:28312–26. doi: 10.18632/oncotarget.4873
174. Fu R, Yang P, Li Z, Liu W, Amin S, Li Z. Avenanthramide a triggers potent ROS-mediated anti-tumor effects in colorectal cancer by directly targeting DDX3. Cell Death Dis (2019) 10:593. doi: 10.1038/s41419-019-1825-5
175. Heerma van Voss MR, van Kempen PM, Noorlag R, van Diest PJ, Willems SM, Raman V. DDX3 has divergent roles in head and neck squamous cell carcinomas in smoking versus non-smoking patients. Oral Dis (2015) 21:270–1. doi: 10.1111/odi.12299
176. Lee CH, Lin SH, Yang SF, Yang SM, Chen MK, Lee H, et al. Low/negative expression of DDX3 might predict poor prognosis in non-smoker patients with oral cancer. Oral Dis (2014) 20:76–83. doi: 10.1111/odi.12076
177. Sailer C, Jansen J, Sekulski K, Cruz VE, Erzberger JP, Stengel F. A comprehensive landscape of 60S ribosome biogenesis factors. Cell Rep (2022) 38:110353. doi: 10.1016/j.celrep.2022.110353
178. Dean DN, Lee JC. Linking parkinson's disease and melanoma: Interplay between alpha-synuclein and Pmel17 amyloid formation. Mov Disord (2021) 36:1489–98. doi: 10.1002/mds.28655
179. Parish JL, Rosa J, Wang X, Lahti JM, Doxsey SJ, Androphy EJ. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J Cell Sci (2006) 119:4857–65. doi: 10.1242/jcs.03262
180. Borges V, Smith DJ, Whitehouse I, Uhlmann F. An Eco1-independent sister chromatid cohesion establishment pathway in s. cerevisiae. Chromosoma (2013) 122:121–34. doi: 10.1007/s00412-013-0396-y
181. Irion U, Leptin M. Developmental and cell biological functions of the drosophila DEAD-box protein abstrakt. Curr Biol (1999) 9:1373–81. doi: 10.1016/S0960-9822(00)80082-2
182. Ma JX, Li JY, Fan DD, Feng W, Lin AF, Xiang LX, et al. Identification of DEAD-box RNA helicase DDX41 as a trafficking protein that involves in multiple innate immune signaling pathways in a zebrafish model. Front Immunol (2018) 9:1327. doi: 10.3389/fimmu.2018.01327
183. Kim S, Govindan JA, Tu ZJ, Greenstein D. SACY-1 DEAD-box helicase links the somatic control of oocyte meiotic maturation to the sperm-to-oocyte switch and gamete maintenance in caenorhabditis elegans. Genetics (2012) 192:905–28. doi: 10.1534/genetics.112.143271
184. Taniguchi T, Iizumi Y, Watanabe M, Masuda M, Morita M, Aono Y, et al. Resveratrol directly targets DDX5 resulting in suppression of the mTORC1 pathway in prostate cancer. Cell Death Dis (2016) 7:e2211. doi: 10.1038/cddis.2016.114
185. Ueda J, Uemura N, Sawamura M, Taguchi T, Ikuno M, Kaji S, et al. Perampanel inhibits alpha-synuclein transmission in parkinson's disease models. Mov Disord (2021) 36:1554–64. doi: 10.1002/mds.28558
186. Kost GC, Yang MY, Li L, Zhang Y, Liu CY, Kim DJ, et al. 1-(3,5-Dimethoxyphenyl)-4-[(6-Fluoro-2-Methoxyquinoxalin-3-yl)Aminocarbonyl] piperazine (RX-5902), interferes with beta-catenin function through Y593 phospho-p68 RNA helicase. J Cell Biochem (2015) 116:1595–601. doi: 10.1002/jcb.25113
187. Cao S, Sun R, Wang W, Meng X, Zhang Y, Zhang N, et al. RNA Helicase DHX9 may be a therapeutic target in lung cancer and inhibited by enoxacin. Am J Transl Res (2017) 9:674–82.
188. Lee T, Pelletier J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget (2016) 7:42716–39. doi: 10.18632/oncotarget.8446
189. Shekoohi S, Rajasekaran S, Patel D, Yang S, Liu W, Huang S, et al. Knocking out alpha-synuclein in melanoma cells dysregulates cellular iron metabolism and suppresses tumor growth. Sci Rep (2021) 11:5267. doi: 10.1038/s41598-021-84443-y
190. Parrish PCR, Thomas JD, Gabel AM, Kamlapurkar S, Bradley RK, Berger AH. Discovery of synthetic lethal and tumor suppressor paralog pairs in the human genome. Cell Rep (2021) 36:109597. doi: 10.1016/j.celrep.2021.109597
191. De Kegel B, Ryan CJ. Paralog buffering contributes to the variable essentiality of genes in cancer cell lines. PLos Genet (2019) 15:e1008466. doi: 10.1371/journal.pgen.1008466
192. Dede M, McLaughlin M, Kim E, Hart T. Multiplex enCas12a screens detect functional buffering among paralogs otherwise masked in monogenic Cas9 knockout screens. Genome Biol (2020) 21:262. doi: 10.1186/s13059-020-02173-2
193. Feng X, Tang M, Dede M, Su D, Pei G, Jiang D, et al. Genome-wide CRISPR screens using isogenic cells reveal vulnerabilities conferred by loss of tumor suppressors. Sci Adv (2022) 8:eabm6638. doi: 10.1126/sciadv.abm6638
194. Gonatopoulos-Pournatzis T, Aregger M, Brown KR, Farhangmehr S, Braunschweig U, Ward HN, et al. Genetic interaction mapping and exon-resolution functional genomics with a hybrid Cas9-Cas12a platform. Nat Biotechnol (2020) 38:638–48. doi: 10.1038/s41587-020-0437-z
195. Shields JA, Meier SR, Bandi M, Mulkearns-Hubert EE, Hajdari N, Ferdinez MD, et al. VRK1 is a synthetic-lethal target in VRK2-deficient glioblastoma. Cancer Res (2022) 82:4044–57. doi: 10.1158/0008-5472.CAN-21-4443
196. So J, Mabe NW, Englinger B, Chow KH, Moyer SM, Yerrum S, et al. VRK1 as a synthetic lethal target in VRK2 promoter-methylated cancers of the nervous system. JCI Insight (2022) 7:e158755. doi: 10.1172/jci.insight.158755
197. Cantley J, Ye X, Rousseau E, Januario T, Hamman BD, Rose CM, et al. Selective PROTAC-mediated degradation of SMARCA2 is efficacious in SMARCA4 mutant cancers. Nat Commun (2022) 13:6814. doi: 10.1038/s41467-022-34562-5
198. McDonald ER, Weck A, Schlabach MR, Billy E, Mavrakis KJ, et al. Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by Large-scale, deep RNAi screening. Cell (2017) 170:577–592.e10. doi: 10.1016/j.cell.2017.07.005
199. Wang Z, Chen K, Jia Y, Chuang JC, Sun X, Lin YH, et al. Dual ARID1A/ARID1B loss leads to rapid carcinogenesis and disruptive redistribution of BAF complexes. Nat Cancer (2020) 1:909–22. doi: 10.1038/s43018-020-00109-0
Keywords: DEAD/H-box helicase, synthetic lethality, synthetic dosage lethality, therapeutic target, drug development, cancer
Citation: Arna AB, Patel H, Singh RS, Vizeacoumar FS, Kusalik A, Freywald A, Vizeacoumar FJ and Wu Y (2023) Synthetic lethal interactions of DEAD/H-box helicases as targets for cancer therapy. Front. Oncol. 12:1087989. doi: 10.3389/fonc.2022.1087989
Received: 02 November 2022; Accepted: 28 December 2022;
Published: 26 January 2023.
Edited by:
Dalia Barsyte-Lovejoy, University of Toronto, CanadaReviewed by:
Dominic Owens, University of Toronto, CanadaCopyright © 2023 Arna, Patel, Singh, Vizeacoumar, Kusalik, Freywald, Vizeacoumar and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuliang Wu, eXVsaWFuZy53dUB1c2Fzay5jYQ==; Franco J. Vizeacoumar, ZnJhbmNvLnZpemVhY291bWFyQHVzYXNrLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.