
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Oncol., 15 December 2022
Sec. Hematologic Malignancies
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1074913
This article is part of the Research TopicCase Reports in Hematological Malignancies : 2022View all 34 articles
 Xue Chen1
Xue Chen1 Fang Wang1
Fang Wang1 Xiaosu Zhou2
Xiaosu Zhou2 Yang Zhang1
Yang Zhang1 Panxiang Cao1
Panxiang Cao1 Xiaoli Ma1
Xiaoli Ma1 Lili Yuan1
Lili Yuan1 Jiancheng Fang1
Jiancheng Fang1 Mingyue Liu1
Mingyue Liu1 Ming Liu1
Ming Liu1 Jiaqi Chen1
Jiaqi Chen1 Qihui Chen3
Qihui Chen3 Ping Wu1
Ping Wu1 Yue Lu4
Yue Lu4 Xiujuan Ma1
Xiujuan Ma1 Hongxing Liu1,2*
Hongxing Liu1,2*In this manuscript, we report torque teno mini virus (TTMV) as a cause of acute promyelocytic leukemia (APL) lacking PML::RARA in a 3-year-old boy. Astolfi et al. firstly identified partial integration of the TTMV genome into RARA intron 2, which resulted in in-frame TTMV::RARA fusion in two APL-like pediatric cases without PML::RARA in November 2021. This fascinating report identified an unexpected exogenous genetic cause of APL and could be of great importance for diagnosing and managing APL. Here we report the third childhood APL-like case caused by TTMV integration and investigate the location and structure of the integrated TTMV sequence. These findings suggest TTMV::RARA is a recurrent cause of APL lacking PML::RARA. Considering the widespread prevalence of TTMV in the population, more TTMV::RARA positive APL-like cases might remain to be identified. Establishing a bioinformatic analysis strategy optimized for the highly variable TTMV genome sequence may facilitate the identification of TTMV::RARA by whole transcript sequencing. An effective PCR protocol to identify TTMV::RARA based on a profound analysis of the conservation of TTMV segments in the fusion transcript is also expected. Also, further investigation is needed to elucidate the oncogenic mechanisms of TTMV integration and the clinical features of TTMV::RARA positive patients.
Astolfi et al. (1, 2) firstly reported torque teno mini virus (TTMV) as a cause of childhood acute promyelocytic leukemia (APL) lacking PML::RARA fusion. They identified partial integration of the TTMV genome into the intron 2 of the human RARA gene, which resulted in in-frame TTMV::RARA fusion transcripts in two APL-like pediatric cases without PML::RARA fusion. This is a fascinating report because it identified an unexpected exogenous genetic cause of APL and could be of great importance for diagnosing and managing APL, especially given the widespread recessive carriage and transmission of TTMV in the population.
To explore whether there were more TTMV::RARA positive APL-like cases, we retrospectively performed a principal component analysis comparing 74 PML::RARA-positive APL cases, 343 oncogenic fusion gene negative AML cases, and 50 healthy controls in our whole-transcriptome sequencing (WTS) cohort (3). The result showed that one AML case was separated from other AML cases while clustered adjacent to the APL cohort (Figure 1A).
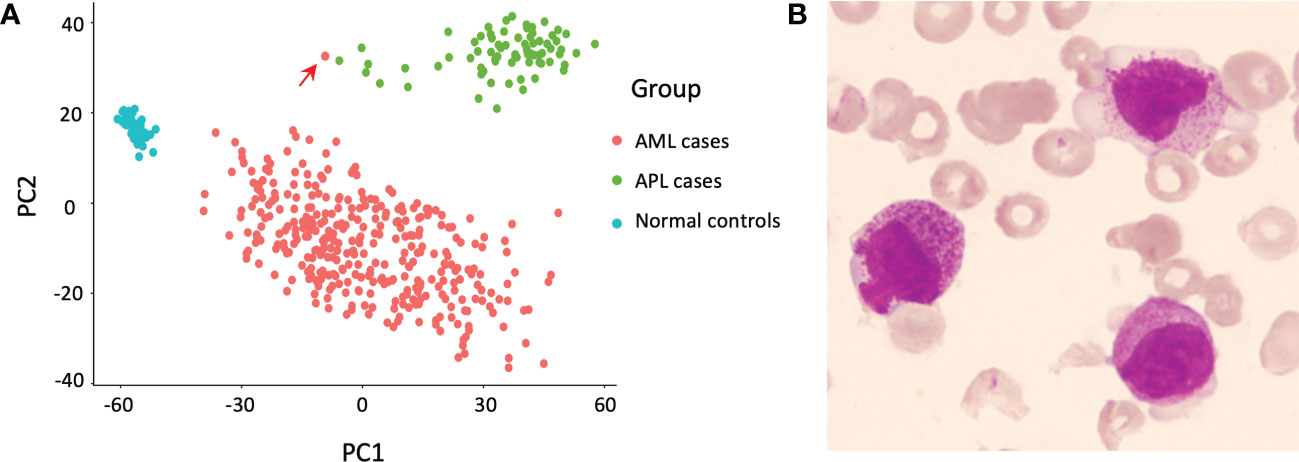
Figure 1 Discovery of the index case. (A) Principal component analysis revealed one relapsed acute myeloid leukemia (AML) case (indicated by the red arrow) separate from other AML cases, but adjacent to the cluster of acute promyelocytic leukemia (APL) cases. (B) Morphology of the bone marrow (BM) smear at relapse, Wright’s stain.
The index case was a 3-year-old boy initially admitted to a local hospital because of persistent fever and headache. Blood tests showed white blood cell count of 36.76 × 109/L, hemoglobin level of 86 g/L, and platelet count of 117 × 109/L. Prothrombin time and activated partial thromboplastin time were 13.3 s (reference, 9.4 - 12.5 s) and 23.6 s (reference, 25.1 - 38.0 s), respectively. Morphologic examination of bone marrow (BM) smears disclosed infiltration by 73.6% of hyper-granular promyelocytes. Flow cytometry (FCM) revealed 84.1% of myeloblasts (positive for CD13, CD33, CD45, CD117, CD123, CD64, and cMPO; partially positive for CD34; negative for HLA-DR, CD11b, CD16, and other T- or B-lymphoid related markers). The presumptive initial diagnosis of this patient was APL. However, both reverse transcription PCR (RT-PCR) and fluorescence in situ hybridization failed to detect the PML::RARA fusion in the BM, and the karyotype was normal. Next-generation sequencing mutation analysis of 86 genes that are frequently mutated in hematological malignancies revealed FLT3-ITD mutation.
Given the clinical suspicion of APL, the patient was immediately treated with ATRA and hydroxyurea. The white blood cell count increased to 128.52×109/L after four-day treatment, and induction chemotherapy (daunorubicin, cytarabine, and etoposide) was started concomitantly. ATRA was discontinued seven days after ATRA initiation due to the absence of PML::RARA. Follow-up monitoring 21 days after induction chemotherapy showed 13% of myeloblasts and promyelocytes in BM. Then he received consolidation therapy (idarubicin, cytarabine, and etoposide) and achieved complete remission (CR).
However, one month after achieving CR, the patient suffered from recurrent fever and was admitted to our hospital. BM morphology examination showed that 60% of nucleated cells were blast cells with abundant purple granules, irregular nuclei, and cytoplasmatic budding, while negative for Auer rods (Figure 1B). The karyotype was normal, and an in-house established protocol of WTS screening for oncogenic fusion genes reported a negative result (3). The patient was started on induction chemotherapy and achieved remission by day 39. Then he underwent haploidentical hematopoietic stem cell transplantation with his father as the donor but relapsed 50 days later with multiple metastases and gave up treatment.
Following the retrospective cluster analysis, we performed a manual investigation of the RARA transcript sequence in WTS data of this case. Alignment of the WTS data in integrative genomics viewer (IGV) revealed abundant RARA fusion transcripts with a 5’ extension from RARA exon 3, which is not homologous to any human sequence (Figure 2A). Further analysis revealed the exogenous sequence started with 58 nucleotides that aligned to the untranslated region (UTR) of TTMV, followed by 256 nucleotides starting with a start codon (ATG) and revealed a significant alignment to different TTMV isolates (83% coverage, 93% to 94% identity), in-frame with the full RARA exon 3. RT-PCR and Sanger sequencing confirmed the existence of the TTMV::RARA fusion transcript (Figures 2B, C). This full-length TTMV::RARA fusion transcript was predicted to encode a fusion protein containing 488 amino acids (Figure 2D). The putative N terminal 85 polypeptide sequence showed 54% identity and 45% coverage to TTMV open reading frame 2 (ORF2) and displayed the conserved motif WX7HX3CXCX5H, which were shared among all anelloviruses. The RARA part of the fusion protein was consistent with PML::RARA and all other RARA fusions.
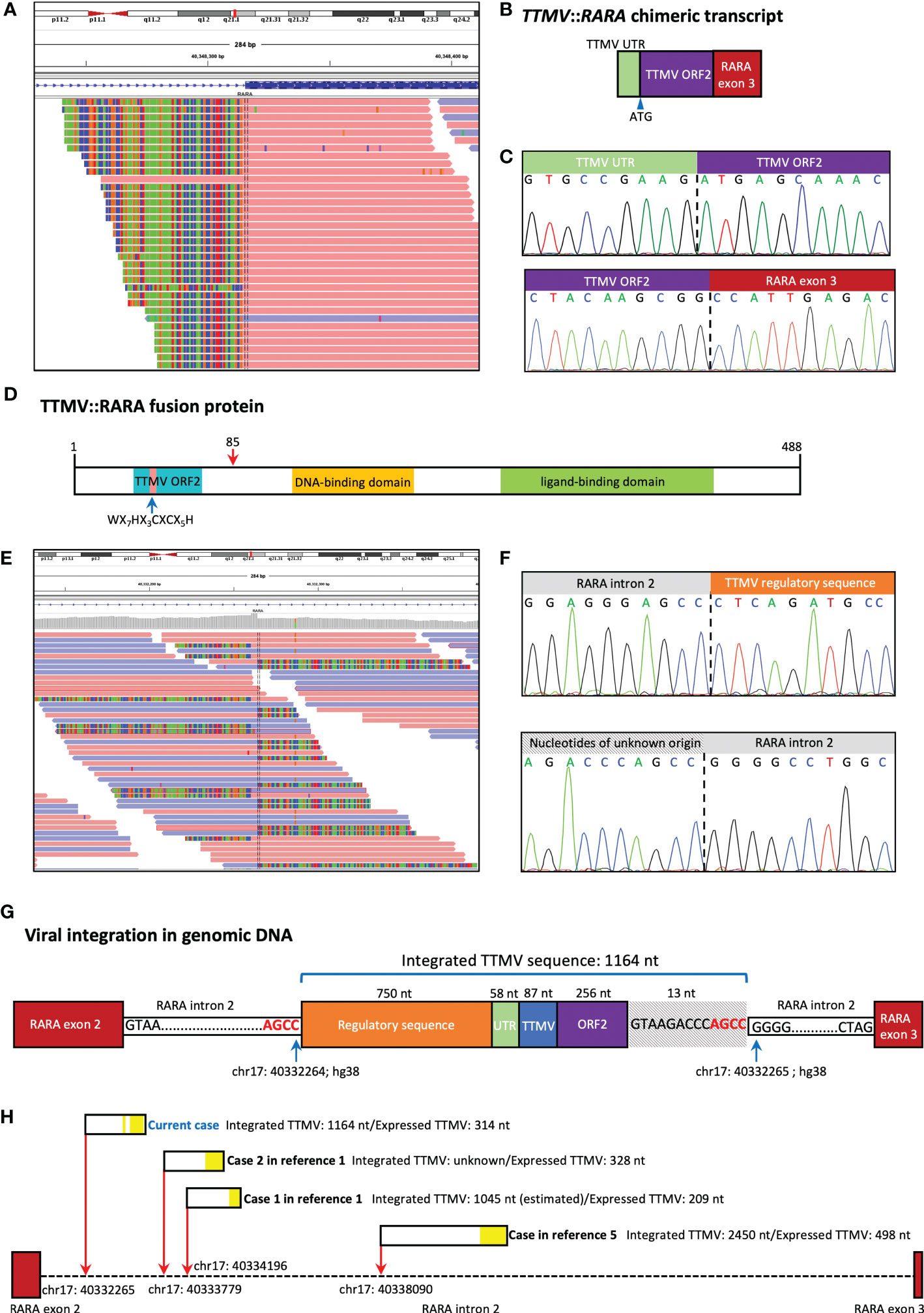
Figure 2 TTMV integration structure of our case. (A) Alignment of the whole transcript sequencing (WTS) data in integrative genomics viewer (IGV) revealed abundant RARA fusion transcripts with a 5’ extension from RARA exon 3, which is not homologous to any human sequence. (B) Schematic representation of the fusion between the TTMV sequence and RARA exon 3. The chimeric transcript includes TTMV open reading frame 2 (ORF2) and an upstream conserved untranslated region (UTR), in-frame with RARA exon 3. (C) Sanger sequencing confirmed the existence of the TTMV::RARA fusion transcript. (D) The predicted structure of the TTMV::RARA fusion protein. The blue arrow indicates the highly conserved TTMV ORF2 motif. The red arrow at position 85 indicates the fusion site. (E) Whole genome sequencing (WGS) revealed the insertion was located at chr17: 40332265 (hg38) in RARA intron 2. (F) Sanger sequencing confirmed the two genomic junction sequences. (G) Schematic representation of the location and structure of the integrated TTMV sequence. (H) TTMV insertion characteristics in the 4 cases reported to date. The red arrows show the insertion positions of TTMV sequence in RARA intron 2 (hg38). The black rectangles show the integrated TTMV sequences, and the yellow blocks within them show the nucleotides expressed in the TTMV::RARA fusion transcript.
Whole genome sequencing (WGS) and Sanger sequencing confirmed the insertion of 1164 bp exogenous fragment at chr17: 40332265 in RARA intron 2, which aligned to parts of the TTMV genome (Figures 2E, F). There were 750 nucleotides and 87 nucleotides derived from TTMV genomic sequences upstream and downstream of the TTMV UTR region, respectively. Also, there were 13 nucleotides not belonging to any known TTMV isolates downstream of TTMV ORF2, all of which were removed during splicing (Figure 2G). PCR and Sanger sequencing also confirmed the existence of the same genomic fusion sequence in the archived DNA samples at primary diagnosis. The integrated viral sequence possessed the two 15-nt conserved sequences (CGAATGGCTGAGTTT and AGGGGCAATTCGGGC) in the UTR of all TTVs (4). The last four bases (AGCC) of RARA intron 2 sequence 5’ to the insertion site were the same as the four 3’ terminal bases of the inserted sequence, suggesting there might be a microhomologous recombination mechanism that mediated the viral sequence integration (Figure 2G).
We here report the third childhood APL-like case caused by the integration of TTMV fragment into the RARA locus. The first case was a 6-year-old girl diagnosed as hypergranular APL based on the characteristic morphologic features. The patient received CR after treatment combining standard AML induction therapy and ATRA, followed by 3 high-dose cytabine-based courses of consolidation therapy. She relapsed 8 months later and achieved CR after receiving combination of ATRA and ATO. Then she underwent HSCT and was alive at 4 years after HSCT. The second case was a 3-year-old AML child reported in the same literature. TTMV::RARA fusion in this patient was identified by a retrospective in silico analysis in a WTS database of 22 pediatric cytogenetically normal AML cases and the laboratory and clinical data were not detailly described (1, 2). Recently, Sala-Torra et al. (5) reported TTMV::RARA fusion in a 39-year-old man diagnosed as AML with APL characteristics but without PML::RARA. He was initially treated with the standard 7+3 regimen and showed induction failure. He then received mitoxantrone, etoposide, and cytarabine with decitabine and venetoclax with a response. Table 1 provides patient characteristics of all reported cases, including the current case, and the TTMV insertion characteristics in the 4 cases reported to date was summarized in Figure 2H. These findings suggest TTMV::RARA is a recurrent cause of APL lacking PML::RARA fusion.
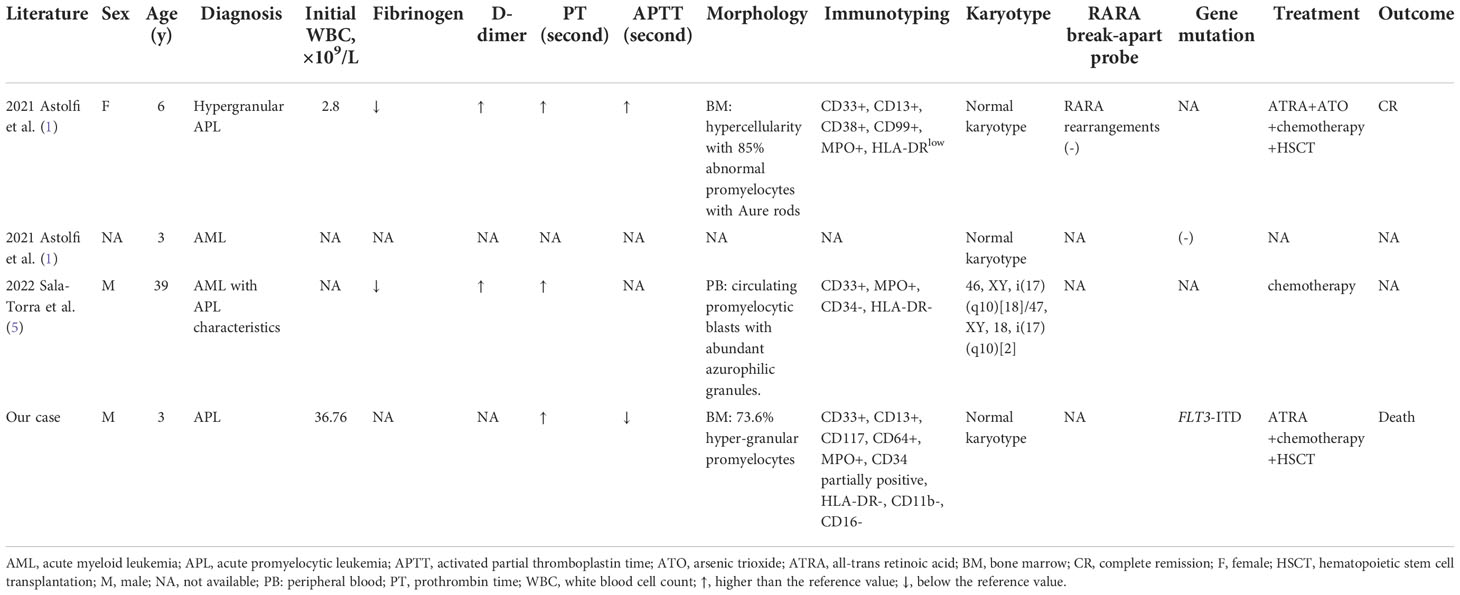
Table 1 Summary of clinical and laboratory characteristics in the 4 reported cases with TTMV::RARA fusion.
Considering the widespread prevalence of TTMV in the population, more TTMV::RARA positive APL-like cases might remain to be identified. Establishing a bioinformatic analysis strategy optimized for the highly variable TTMV genome sequence may facilitate the identification of TTMV::RARA by WTS data analysis. An effective PCR protocol to identify TTMV::RARA is also worth expecting, but this should be based on a profound analysis of the conservation of TTMV segments in the fusion transcript. Also, further investigation is needed to elucidate the oncogenic mechanisms of TTMV integration and the clinical features of TTMV::RARA positive patients. The first reported case was successfully treated with ATRA and ATO and subsequent HSCT. However, as ATRA was coupled with chemotherapy and HSCT, it is almost impossible to assess the contribution of ATRA. The outcomes of the other two reported cases were not available. In addition, our patient in the current case report died after giving up treatment and the efficiency of ATRA was unclear since ATRA was used only for 7 days. Therefore, further studies into the optimal therapeutic approaches for patients with TTMV::RARA are urgently needed.
The datasets presented in this article are not readily available because of ethical/privacy restrictions. Requests to access the datasets should be directed to the corresponding author.
The studies involving human participants were reviewed and approved by The Institutional Review Board and Ethical Committee of the Hebei Yanda Lu Daopei Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
HL designed the research; XC designed molecular studies and wrote the paper; PC, JF, QC, and XiuM performed bioinformatics analysis; FW, YZ, JC, and MingL supervised clinical and experimental findings; XZ, XiaM, LY, and MingyL performed molecular studies; YL was involved in the management of the patient and provided clinical data. PW performed morphological analysis. All authors reviewed the manuscript and contributed to the final draft.
We thank the patient in this study.
Author QC is employed by Beijing Geneprofile Technologies Co,.Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Astolfi A, Masetti R, Indio V, Bertuccio SN, Messelodi D, Rampelli S, et al. Torque teno mini virus as a cause of childhood acute promyelocytic leukemia lacking PML/RARA fusion. Blood (2021) 138(18):1773–7. doi: 10.1182/blood.2021011677
3. Chen X, Wang F, Zhang Y, Ma X, Cao P, Yuan L, et al. Fusion gene map of acute leukemia revealed by transcriptome sequencing of a consecutive cohort of 1000 cases in a single center. Blood Cancer J (2021) 11(6):112. doi: 10.1038/s41408-021-00504-5
4. Okamoto H, Nishizawa T, Tawara A, Peng Y, Takahashi M, Kishimoto J, et al. Species-specific TT viruses in humans and nonhuman primates and their phylogenetic relatedness. Virology (2000) 277(2):368–78. doi: 10.1006/viro.2000.0588
Keywords: acute promyelocytic leukemia, torque teno mini virus, RARA, TTMV::RARA, whole-transcriptome sequencing
Citation: Chen X, Wang F, Zhou X, Zhang Y, Cao P, Ma X, Yuan L, Fang J, Liu M, Liu M, Chen J, Chen Q, Wu P, Lu Y, Ma X and Liu H (2022) Torque teno mini virus driven childhood acute promyelocytic leukemia: The third case report and sequence analysis. Front. Oncol. 12:1074913. doi: 10.3389/fonc.2022.1074913
Received: 20 October 2022; Accepted: 23 November 2022;
Published: 15 December 2022.
Edited by:
Shimin Hu, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Madhu M. Ouseph, Cornell University, United StatesCopyright © 2022 Chen, Wang, Zhou, Zhang, Cao, Ma, Yuan, Fang, Liu, Liu, Chen, Chen, Wu, Lu, Ma and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongxing Liu, c3RhcmxpdUBwa3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.