- 1Department of Gastroenterology, Changhai Hospital, Naval Medical University, Shanghai, China
- 2Department of Emergency, Changhai Hospital, Naval Medical University, Shanghai, China
- 3Shanghai Institute of Pancreatic Diseases, Shanghai, China
Background: As a processing method of RNA precursors, alternative splicing (AS) is critical to normal cellular activities. Aberrant AS events are associated with cancer development and can be promising targets to treat cancer. However, no detailed and unbiased study describes the current state of AS of cancer research. We aim to measure and recognize the current state and trends of AS cancer research in this study.
Methods: The Web of Science Core Collection was used to acquire the articles. Utilizing three bibliometric tools (CiteSpace, VOSviewer, R-bibliometrix), we were able to measure and recognize the influence and collaboration data of individual articles, journals, and co-citations. Analysis of co-occurrence and burst information helped us identify the trending research areas related to AS of cancer.
Results: From 2012 to 2021, the total number of papers on AS of cancer published in 766 academic journals was 3,507, authored by 20,406 researchers in 405 institutions from 80 countries/regions. Research involving AS of cancer genes was primarily conducted in the United States and China; simultaneously, the Chinese Academy of Sciences, Fudan University, and National Cancer Institute were the institutions with strong research capabilities. Scorilas Andreas is the scholar with the most publications, while the most co-citations were generated by Wang, Eric T. Plos One published the most papers on AS of cancer, while J Biol Chem was the most co-cited academic journal in this field. The results of keyword co-occurrence analysis can be divided into three types: molecular (P53, CD44, androgen receptor, srsf3, esrp1), pathological process (apoptosis, EMT, metastasis, angiogenesis, proliferation), and disease (breast cancer, colorectal cancer, prostate cancer, hepatocellular carcinoma, gastric cancer).
Conclusion: Research on AS of cancer has been increasing in intensity over the past decade. Current AS of cancer studies focused on the hallmarks of AS in cancer and AS signatures including diagnostic and therapeutic targets. Among them, the current trends are splicing factors regulating epithelial–mesenchymal transition and other hallmarks, aberrant splicing events in tumors, and further mechanisms. These might give researchers interested in this field a forward-looking perspective and inform further research.
1 Introduction
The spatiotemporal-specific expression of genes determines cellular physiological functions, and abnormal expression of genes is commonly associated with pathological conditions, including carcinogenesis (1). According to the central dogma, RNA is the key molecule for transmitting genetic information, and its stability during transcription and translation ensures successful gene expression and functional protein production (2). The production of mature mRNA from mRNA precursor involves steps such as splicing, adding cap structure, polyadenylation, and nucleobase modification (3). These processing steps regulated by their respective regulatory systems increase the diversity of the transcriptome and proteome.
Alternative splicing (AS) is one of the processing methods of mRNA precursors, including the critical process of removing introns and connecting exons (4). As a result of AS, a single gene is capable of generating multiple transcripts, enhancing the structure and activity of protein domains (5). AS is completed by a synthetic spliceosome protein complex and is regulated by the complicated interaction of cis-elements and trans-factors (6, 7). This precise regulation maintains the balance of different gene transcripts and ensures the accurate process of physiological activities and cell homeostasis (8). Due to abnormal expression and interaction of AS regulatory factors, aberrant splicing leads to the overproduction of tumor-promoting transcripts and the decrease in tumor-suppressing transcripts, both of which play a crucial role in tumor development (9–11).
In recent years, research on the panoramic delineation and regulatory mechanisms of AS events in tumorigenesis has developed rapidly (4, 7, 12). Identifying tumor-promoting transcripts, aberrant splicing factors, and mutated cis-element sequences is capable of providing diagnostic models and guiding the design of small-molecule compounds or oligonucleotide therapeutic drugs in a targeted manner (13–16). These studies have expanded the research direction of new targets and specific drugs for the early diagnosis and treatment of tumors.
Numerous reviews have summarized research on AS in cancer diagnosis and treatment, but to our knowledge, there is no comprehensive picture of AS splicing in cancer. Bibliometrics enables the qualitative and quantitative analyses of data, including contributions and collaborations of authors, institutions, and countries, and also the assessment of research trends (17–19). Thence, this report aimed to use the bibliometric method to assess the overall research trends of AS splicing in tumors and the hot issues over the past decade.
2 Materials and methods
2.1 Data collection
The Web of Science, which is broadly used in bibliometrics, can provide extensive and authoritative global academic data bibliometric software needs (20, 21). We mainly use the Web of Science Core Collection (WoSCC). The data were obtained on 21 July 2022 from the WoSCC database. The search formula was [TS = (alternative splicing)] AND TS = (cancer* OR anticancer* OR tumor* OR oncology OR neoplasm* OR carcinoma* OR lymphoma* OR sarcoma* OR leukemia*). The wildcard character (*) was used to allow variable endings of keywords to capture as much data as possible. There was a limitation on the publication year (2012–2021). The inclusion of English-language literature was limited to original articles and reviews. We downloaded the search results as “Full Record and Cited References” and “Plain Text.” Following this, the files were renamed as “download_*.txt” to be analyzed by the CiteSpace software.
2.2 Data analysis
We used three bibliometric tools, CiteSpace 6.1.R2 Basic (22), R-bibliometrix 4.0.0 (23), and VOSviewer 1.6.18 (24), to conduct bibliometric evaluation and visualization and Microsoft Excel 2021 for statistics and the plotting part of the figure. The first step was to clean our data. For example, “splicing factors” and “splicing factor” were merged as “splicing factor” and “rna splicing,” “pre-mrna splicing,” and “alternative splicing” were unified as “alternative splicing” (25).
CiteSpace is capable of discovering collaboration, keywords, domain research structure, future direction, and evolution as a bibliometric and visual analysis tool in a scientific field (26). Using CiteSpace, we utilize its co-occurrence, timeline, bursts, and dual-map functions to draw a series of figures about countries/regions and institutions, journals, references, citations, and keywords. We use the analysis process and parameter settings recommended by the software developers, where the time span is 2012–2021, the time slice is 1 year, the selection criterion is the g index (k = 25), pruning is none, and the minimum duration of burstness is 2 years. The size of the node in the CiteSpace visualization portrays the number of co-occurrence. Furthermore, linkages show the relationships between the co-occurrences (22). As time passes from 2012 to 2021, the node and line’s colors change from purple to red to symbolize the different years. High betweenness centrality (≥0.10) nodes with purple circles serve as a hub between distinct networks (26–28).
Another bibliometric tool that excels in producing and visualizing knowledge maps is VOSviewer, which displays the kinds of clusters, overlays, or density colors (24). We mainly applied the co-occurrence analysis function of VOSviewer, including authors, journals, references, and their co-citations, as well as keywords. The specific parameters applied are mentioned in the corresponding chapters. In addition, we use the full counting method as a counting method. The meaning of the size of the node is the same as that of CiteSpace. Nodes with the same color belong to the same cluster. Additionally, links show the relationship between co-occurrences, and their degree of thickness is determined by the estimated strength value. The value is related to the number of papers published by the two authors or the frequency of the co-occurrence of the two keywords (24). The co-cited frequency is positively correlated with the word and round sizes as well as the yellow opacity in density maps. The color on the overlay map represents the typical publishing year.
R-bibliometrix is an R package for executing a comprehensive science mapping analysis of scientific literature (23). To be adaptable and make integration with other statistical and graphical R packages easier, R-bibliometrix was programmed in R. Maps of the geographical distribution of the countries/regions were produced using it. The network map displays the current state of research and communication between various countries/regions (23).
Excel 2021 software was used to anatomize the yearly publications and cited frequency. Additionally, we obtained impact factor (IF), journal citation report (JCR), average per item (ACI), journal classification, and author H-index from Web of Science on 1 August 2022.
3 Results
3.1 Annual growth trend
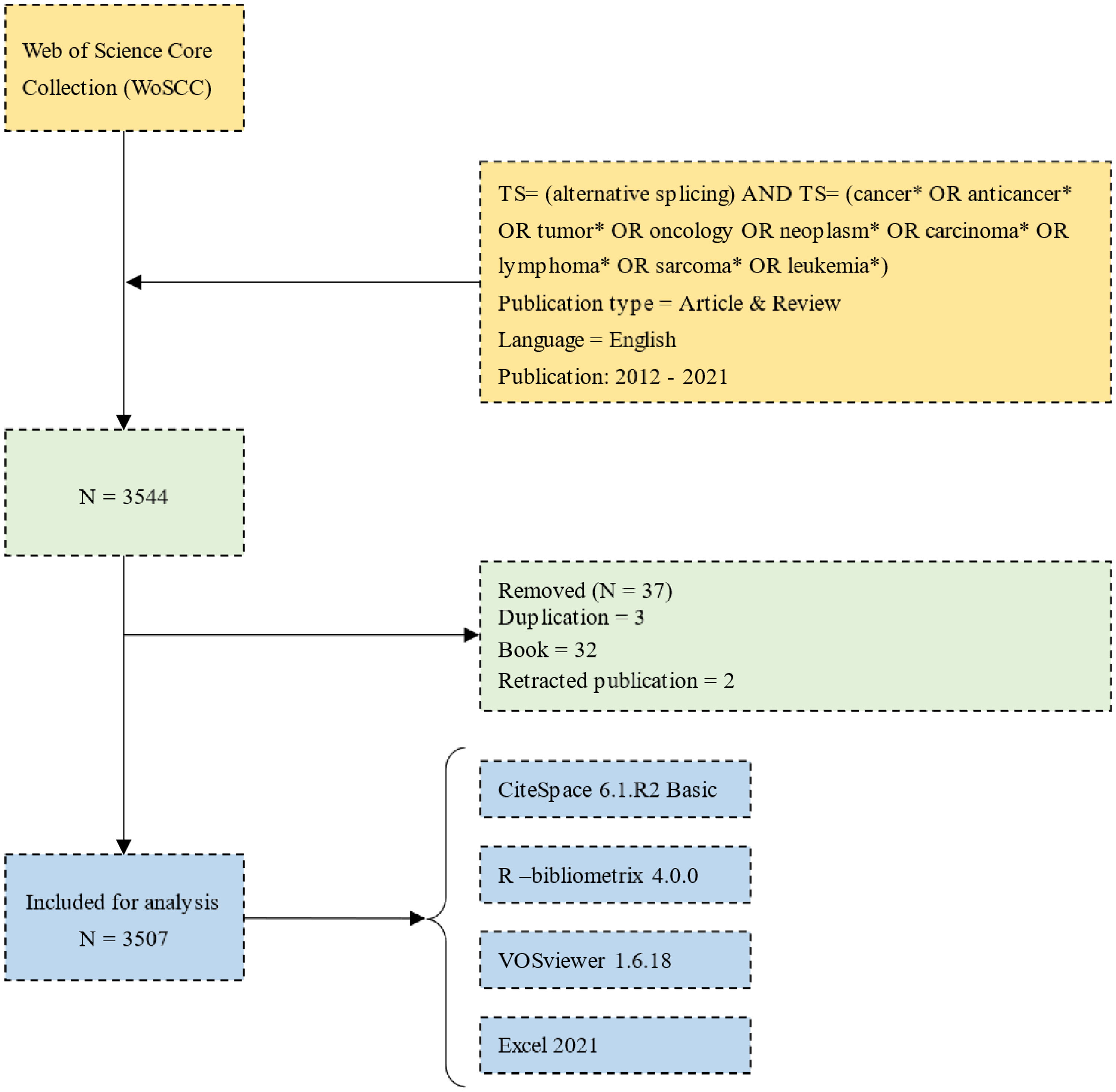
From the WoSCC database, we acquired 3,544 papers, and we ultimately included 3,507 publications that qualified (Figure 1; Supplementary Table 1). The number of publications related to AS of cancer and the frequency of citations have both steadily increased over the past 10 years, as illustrated in Figure 2.
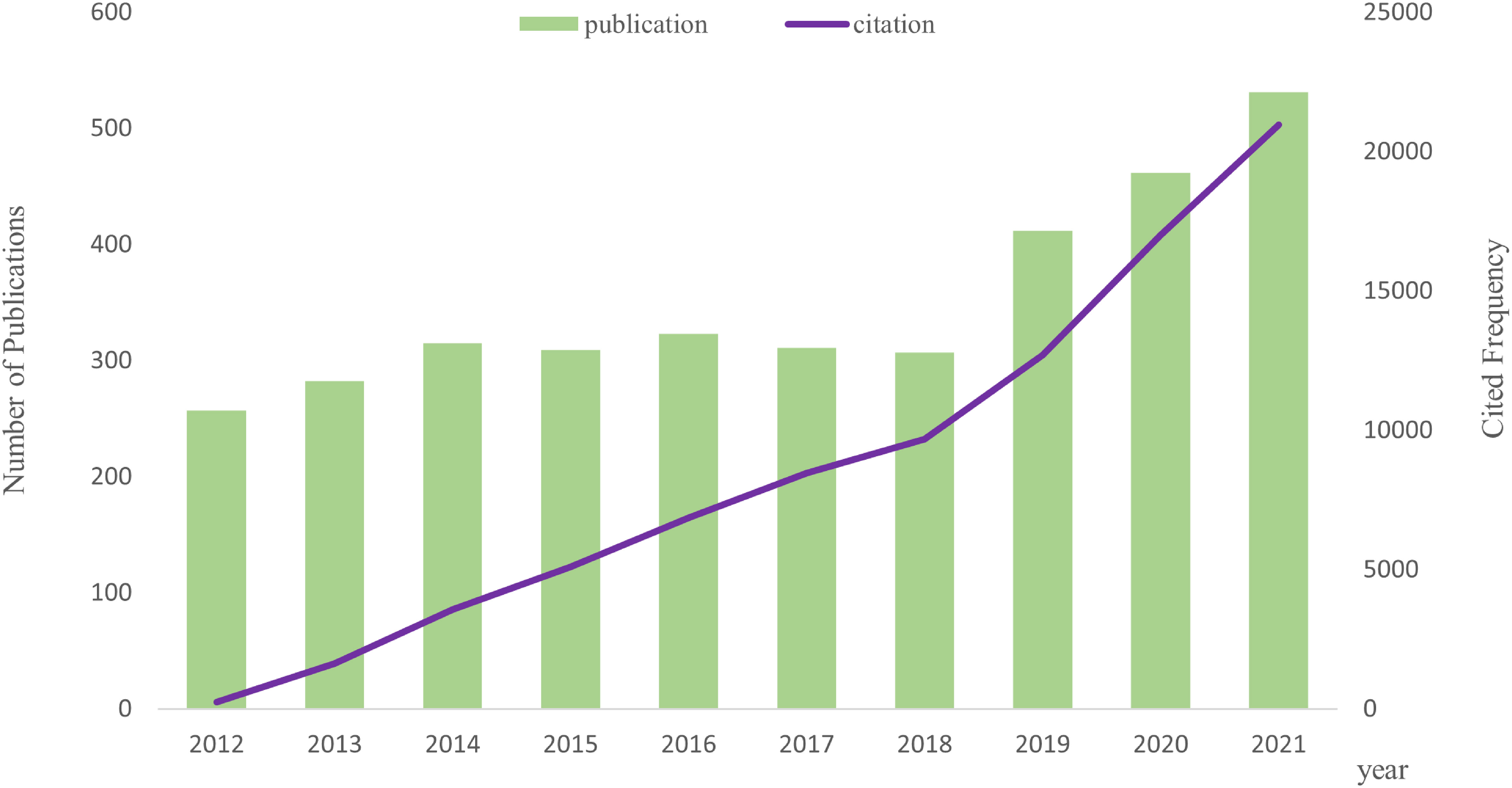
Figure 2 Tendency via bar and line graphs of alternative splicing of cancer publications and cited frequency nearly 10 years.
3.2 Distribution of countries/regions and institutions
There were 3,507 papers in total, representing 80 different countries/regions and 405 institutions. The United States, China, and Germany are the top 3 countries in terms of the number of published articles, with 1,165, 1,013, and 249, respectively (Table 1). China’s centrality, however, was less than 0.10, suggesting that it may not be a “hub” node in the AS of cancer studies (20). In contrast, the United States (centrality = 0.41), England (centrality = 0.25), and Germany (centrality = 0.20) had high centrality, which is depicted in Figure 3A by a purple circle. As shown in Figure 3A, the closeness of the co-occurrence atlas of countries/regions was 0.1864, denoting lively collaboration between them (27). Figure 3C shows the relative proportion of annual publications for the top 10 countries from 2012 to 2021. It was evident that China’s share had gradually increased. A country/region co-authorship network was created by R-bibliometrix (Figure 3D). The network map showed the current state of research and communication activities between these countries/regions. The Chinese Academy of Sciences is the scientific research institution with the largest number of published papers, as shown in Figure 3B; however, its centrality is just 0.07 (n = 76). By contrast, the National Cancer Institute (n = 65, centrality = 0.14), the University of California San Diego (n = 48, centrality = 0.2), and Karolinska Institute (n = 25, centrality = 0.14) had a high centrality. The publication counts, H-index, and ACI of the top 10 most productive institutions are displayed in the bar graph of Figure 3E.
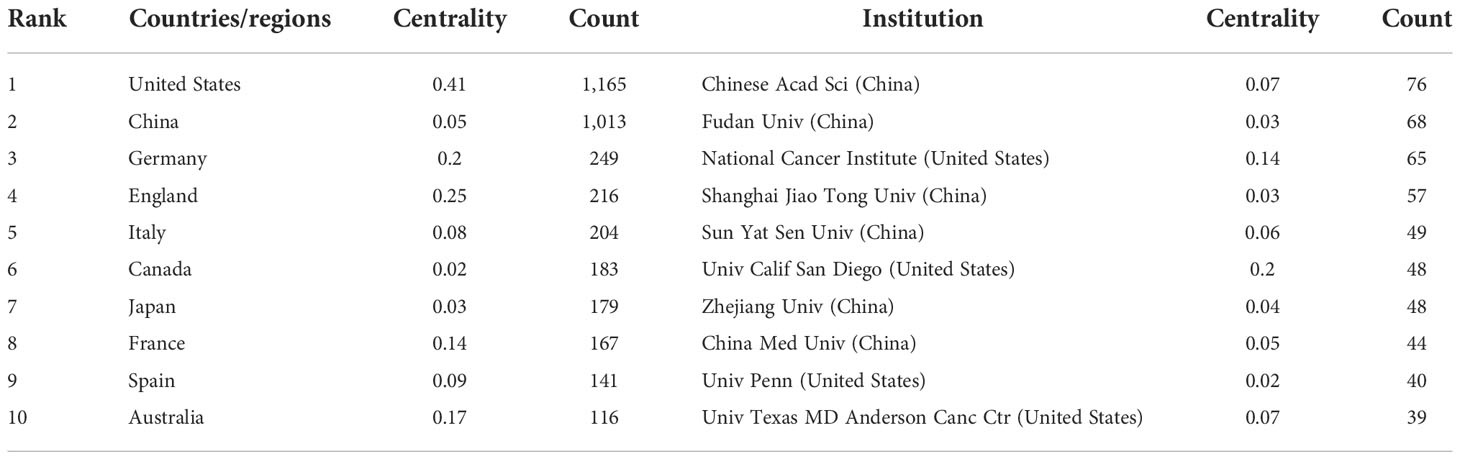
Table 1 Top 10 countries/regions and academic institutions involved in alternative splicing of cancer research.
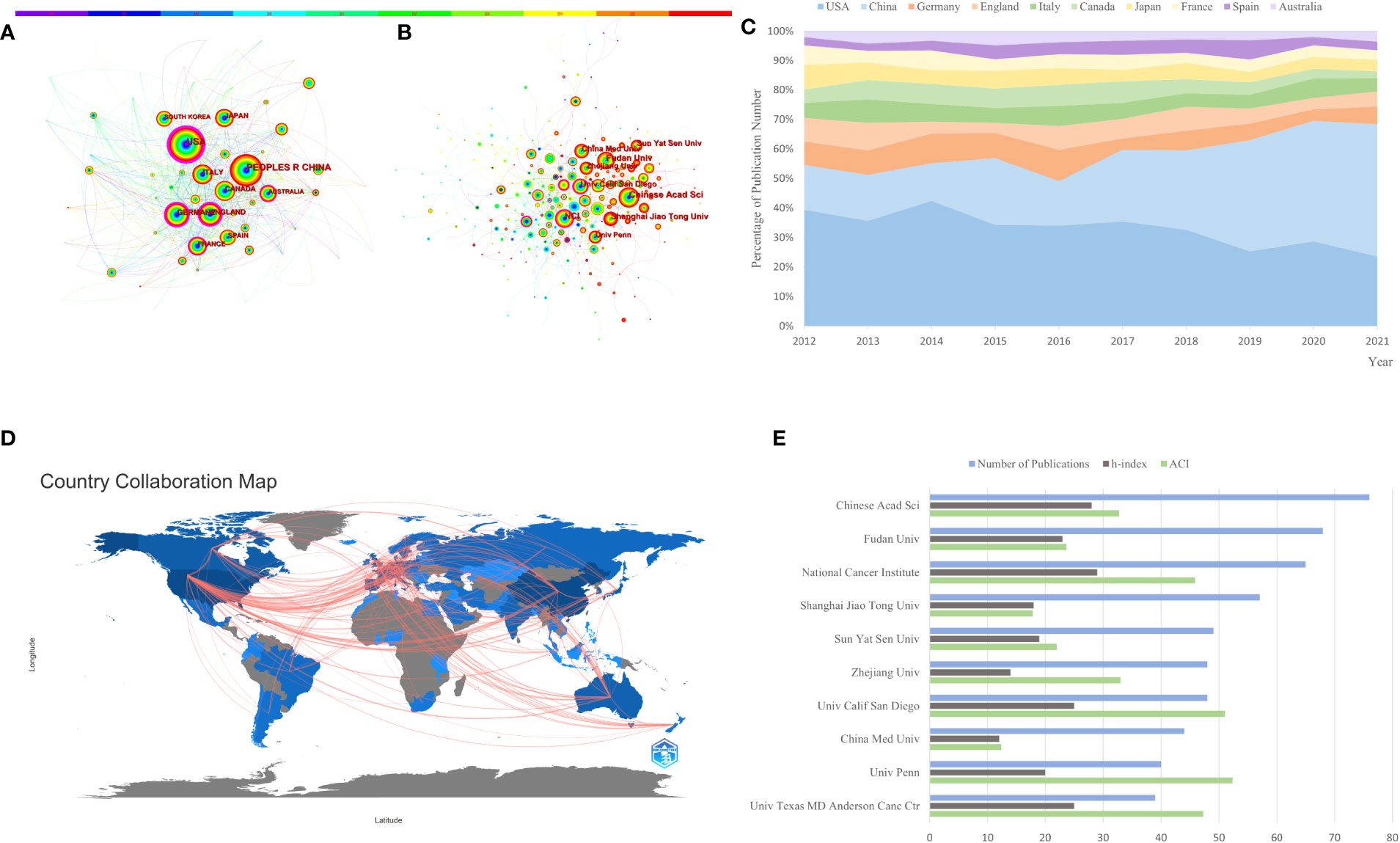
Figure 3 The co-occurrence atlas of (A) countries/regions (n ≥ 100) and (B) academic institutions (n ≥ 40) in alternative splicing of cancer research. As time passes from 2012 to 2021, the color of the node and line changes from purple to red. Purple-round nodes indicate strong betweenness centrality (≥0.1). (C) The relative fraction of annual publications in the top 10 countries from 2012 to 2021. (D) Network diagram of countries/regions (min edges = 2) involved in alternative splicing of cancer research. (E) The top 10 most productive institutions’ publication counts, h-index, and ACI.
3.3 Authors and co-cited authors
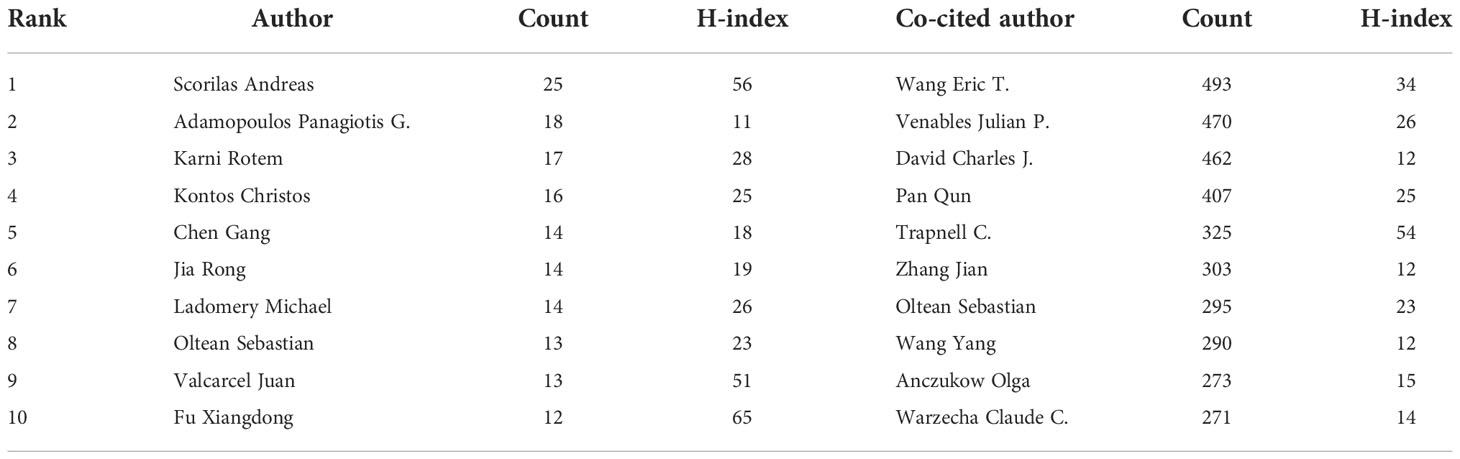
There were 20,406 authors active in AS of cancer research, and 25 of them published 10 or more articles, as shown in Figure 4A and Supplementary Table 2. Scorilas Andreas, a National and Kapodistrian University of Athens scholar, was the most prolific author (n = 25), followed by Adamopoulos Panagiotis G. and Karni Rotem (Table 2). Different colors represent different clusters in Figure 4, a total of 15 (29). Active partnerships, such as Ladomery Michael and Oltean Sebastian, are frequently found in the same cluster. Additionally, there were alliances between two linked authors of different colors, which seemed to be Ghigna Claudia and Fu Xiangdong.
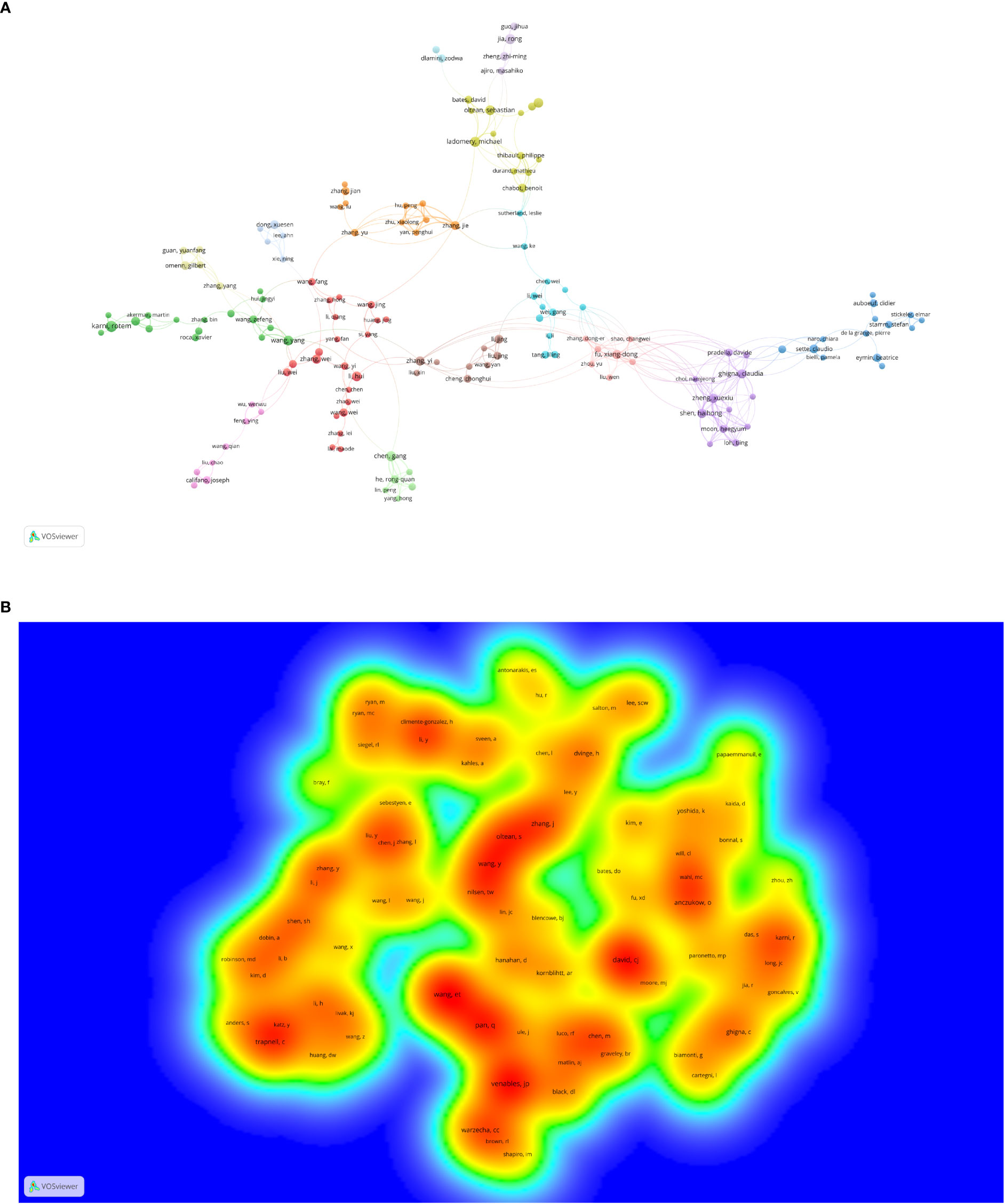
Figure 4 The co-occurrence (A) authors’ (documents ≥5) cluster map and (B) co-cited authors’ (citations ≥100) density map of alternative splicing of cancer research. (A) Nodes with the same color represent that they belong to the same cluster, the size of the node is proportional to the number of articles published by the author, and the thickness of the connection is proportional to the number of articles co-published by two authors. (B) The co-cited frequency is positively correlated with the size and depth of the word and the yellow color, respectively.
Authors who have been cited in one article are known as co-cited authors (30). From Figure 4B and Supplementary Table 2, we can see that 83 authors out of the 85,667 co-cited authors had more than 100 co-citations. They are displayed as a density map in Figure 4B, which made it easy to identify the writers who were co-cited frequently. The hue gets warmer with more citations (31). Wang Eric T., Venables Julian P., and David Charles J. had the most co-citations, as indicated in Table 2 and Figure 4B. The images cannot display all the information due to CiteSpace and VOSviewer visualization’s intrinsic restrictions. As a result, we have supplemented this with detailed information on the graphs in the Supplementary Material.
3.4 Journals and co-cited academic journals
Articles about AS of cancer research have been published in 766 scholarly journals overall. Nine hundred and five papers, or 25.81% of the total publications, were published in the top 15 journals (Table 3). The number of papers published in Plos One is the largest (n = 145, 4.1%), followed by the International Journal of Molecular Sciences (n = 84, 2.4%) and Oncotarget (n = 77, 2.2%). A co-citation map of journals was produced by VOSviewer, as shown in Figure 5A. The required minimum was established at 100 citations, and 292 journals satisfied the requirement.
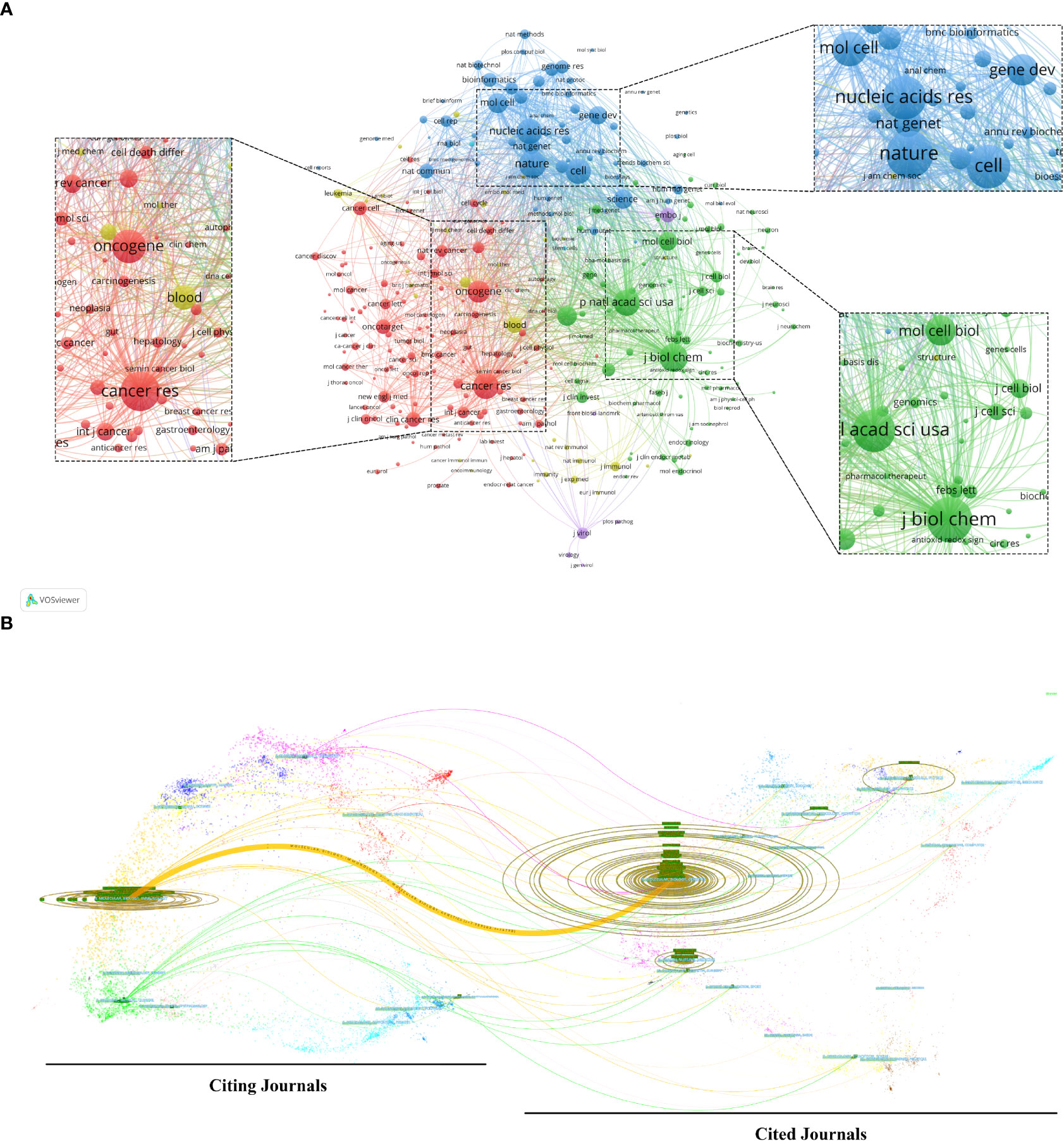
Figure 5 (A) Analysis of journals on alternative splicing of cancer co-citations via VOSviewer. (B) The dual-map overlay of journals on alternative splicing of cancer. The colored channel denotes the citation relationship, with the citing journals on the left and the cited journals on the right.
Of the 6,707 cited journals, 84 have more than 500 citations. Among them, the Journal of Biological Chemistry (n = 7,922), Nature (n = 7,333), and Proceedings of the National Academy of Sciences of the United States of America (PNAS) (n = 6,656) ranked in the top 3 in the number of citations (Table 3). Additionally, 31.16% of all cited sources came from the top 15 co-cited journals.
The dual-map overlay of journals represents the field distribution of citing and cited journals, as seen in Figure 5B (32). Citation relationships are indicated by colored paths, with citing journals on the left and cited journals on the right (22). The principal citation channel indicates that papers published in Molecular/Biological/Immunology journals mainly cite papers published in Molecular/Biological/Genetics journals (Figure 5B).
3.5 Co-cited references and reference burst
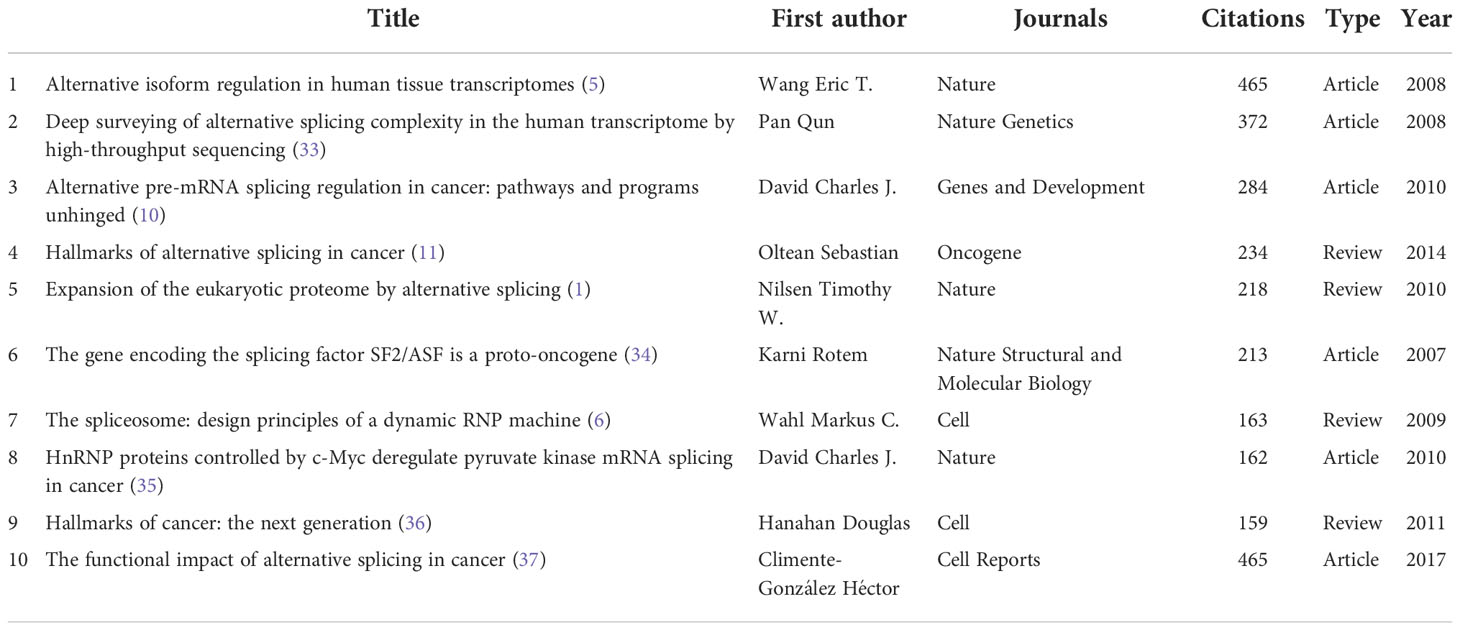
Thirty-seven references out of the 132,804 cited ones were quoted at least 100 times (Supplementary Table 3). The top 10 co-cited references are included in Table 4, with a minimum of 157 co-citations. The article by Wang Eric T. et al. from Nature in 2008 (n = 465) is the one that has received the most co-citations out of all of them. In addition, four of the top 10 were reviews, and six of the top 10 were research articles.
The entire network map might be partitioned into several clusters using the clustering function, and studies inside a cluster might have distinct study themes to studies from other clusters (Figure 6A). Each cluster’s most prevalent terms were designated as cluster labels (38). The references’ timeline view could allow users to see how various research hotspots have changed over time. As shown in Figure 6B, cluster #0 (post-transcriptional regulation), #3 (prognostic alternative), #4 (alternative splicing), #6 (circular RNA), #7 (androgen receptor), and #8 (non-coding RNA malat1) started earlier, while cluster #1 (splicing signature), #2 (aberrant splicing), and #5 (oncogene srsf3) are still ongoing, which could be considered as the frontier.
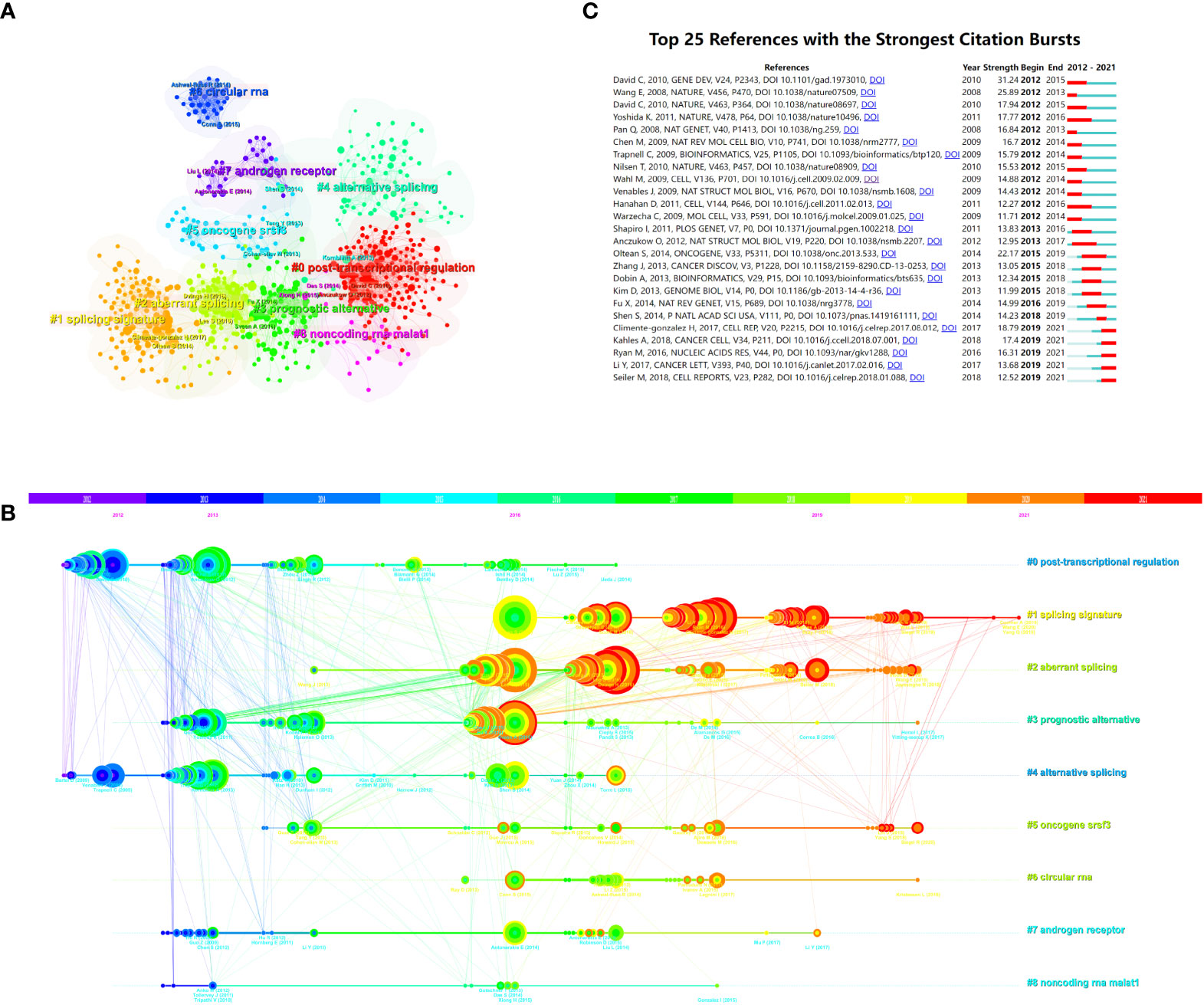
Figure 6 The reference co-citation analysis maps in (A) cluster view and (B) timeline view were produced by CiteSpace. (C) Visualization map of the top 25 references related to alternative splicing of cancer that have received the most citations. (B) Each cluster is represented as a horizontal axis; the larger the number of the cluster label, the smaller the cluster. The linkages show co-cited associations, and the node size represents co-citation frequencies. The node and line colors indicate distinct years. LLR used the title to extract cluster labels. (C) The red bars indicate citation burstness, whereas the blue bars indicate that the reference has been published.
with citation bursts indicate that their citations have grown by leaps and bounds in a particular period (27). Two hundred and fifty references were included in the citation bursts analysis, and the top 25 are listed in Figure 6C. One paper, entitled “Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged,” had the strongest burstness (strength = 31.24) with citation burstness from 2012 to 2015 (10), which was published in Gene Dev by David Charles J. et al. in 2010. It is worth noting that five references (15, 37, 39–41) were still in burstness. Climente-González Héctor et al. (37) explored the functional influence of AS in cancer, Kahles André et al. (15) conducted a comprehensive analysis of AS across tumors, Ryan Michael et al. (39) provided a web-based resource for exploring the AS patterns of TCGA tumors, Li Yuan et al. (40) discovered a series of AS signatures in non-small cell lung cancer, and Seiler Michael et al. (41) analyzed the functional consequences of somatic mutation of splicing factor genes, respectively.
3.6 Keyword analysis of trending research topic
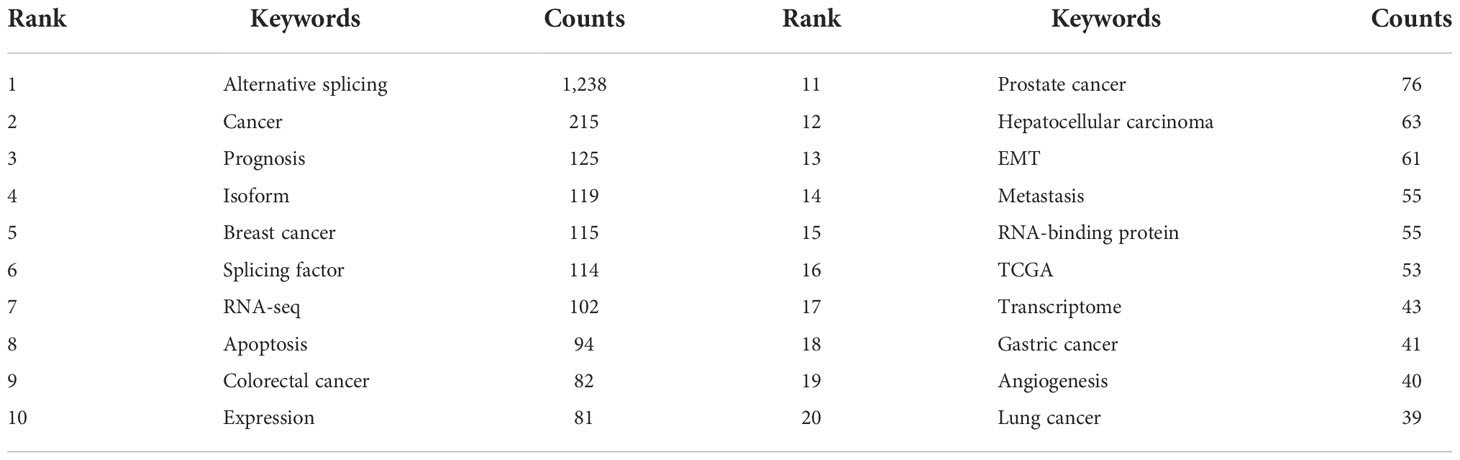
In total, 6,116 keywords were recovered, 121 of which appeared at least 10 times, and there were 31 keywords that appeared at least 30 times. As represented in Table 5, alternative splicing (n = 1,238), cancer (n = 215), and prognosis (n = 125) were the three most popular keywords. We divided the keywords into three categories, namely, molecules, pathological processes, and diseases associated with AS of cancer, and listed the top 15 keywords in Table 6, respectively. Obviously, P53 (n = 39), CD44 (n = 35), androgen receptor (n = 31), srsf3 (n = 24), and esrp1 (n = 16) were several molecular keywords with the highest frequency; apoptosis (n = 94), EMT (n = 61), metastasis (n = 55), angiogenesis (n = 40), proliferation (n = 34), and epigenetics (n = 23) were several pathological process keywords with the highest frequency; and breast cancer (n = 115), colorectal cancer (n = 82), prostate cancer (n = 76), hepatocellular carcinoma (n = 76), and gastric cancer (n = 41) were several disease keywords with the highest frequency in AS of cancer studies.
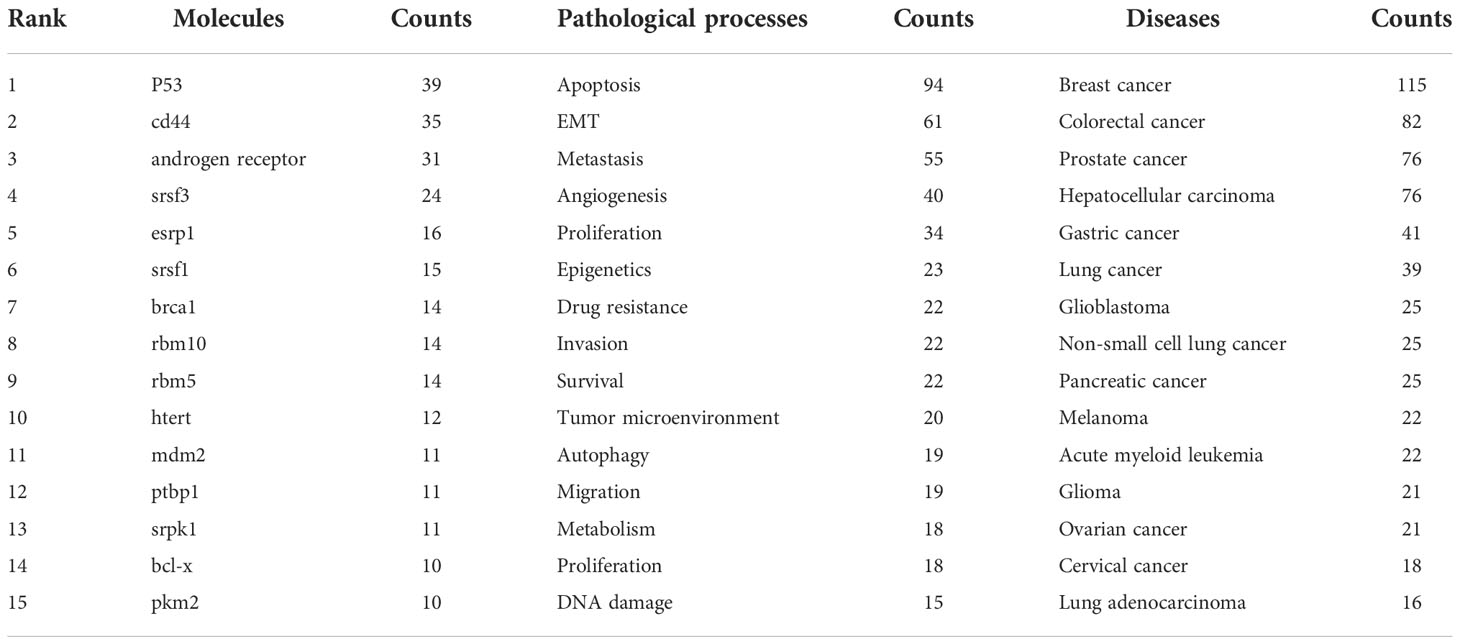
Table 6 Top 15 molecules, pathological processes, and diseases related to alternative splicing of cancer.
Keywords with high co-occurrence counts (n ≥ 10) are displayed as an overlay map in Figure 7A, with the hue denoting the typical year of publication. As we can see, the emerging fields that were given the color yellow include splicing factor, prognosis, immunotherapy, and TCGA (The Cancer Genome Atlas). Each cluster displayed the top 3 keywords over time in the timeline view (Figure 7B). Six of the seven clusters were still active except for cluster #6. Among them, #0 (splicing event) is the biggest cluster, followed by #1 (therapeutic target), #2 (epithelial–mesenchymal transition), and #3 (splicing factor). Supplementary Table 2 provides further details.
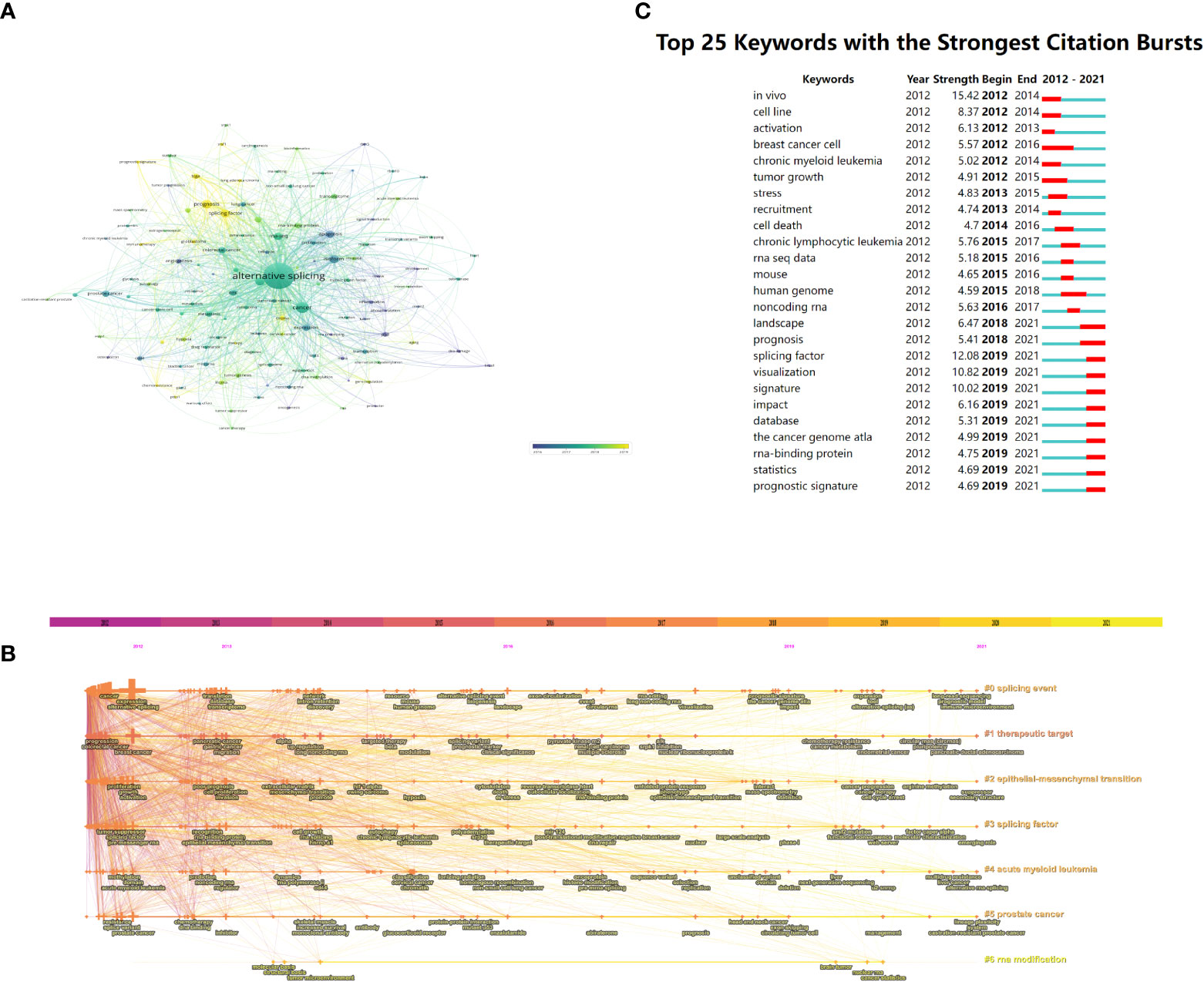
Figure 7 The (A) overlay map (n ≥ 10, max lines = 1,000) and (B) timeline view of keywords associated with alternative splicing of cancer. (C) Greatest citation bursts for the top 25 keywords (sorted by the starting year). (A) The color of the node represents the average publication year, the size of the node is proportional to the counts of co-occurrences of keywords, and the thickness of the link is positively correlated with the number of co-occurrences of the keywords represented by the two nodes. (B) Each cluster is represented as a horizontal axis; the larger the number of the cluster label, the smaller the cluster. LLR used the title to extract cluster labels. (C) The red bars indicate citation burstness.
Bursts of keywords are those that were used a lot during a certain time frame (27). Figure 7C demonstrates that in vivo had the strongest bursts, with a strength of 15.42, followed by a splicing factor at 12.08 and visualization at 10.82. Notably, until 2021, the landscape, prognosis, splicing factor, and other keywords were in burstness. The results of VOSviewer and CiteSpace had a considerable similarity regarding keywords, indicating the reliability of our analysis.
4 Discussion
4.1 General information
Based on the information from the WoSCC database, a total of 3,507 AS of cancer publications by 20,406 writers in 405 institutions from 80 countries/regions were published between 2012 and 2021 in 766 academic journals. Increasing publications indicate that curiosity and attention are growing regarding the AS of cancer. The AS research officially started in 1977, when Roberts R.J. and Sharp P.A. found the phenomenon of “alternative splicing” (42, 43). Since that time, AS research has expanded quickly. Our focus is on the relationship between alternative splicing and cancer. AS research of cancer has steadily increased in steps during the last 10 years, and the relevant published article and cited frequency in 2021 are almost twice and over 80 times those of 2012, respectively.
The volume of published articles is a very important indicator, but the centrality is a crucial indicator for the quality of published articles in country/region analysis, where high centrality nodes (≥0.10) indicate the “hub” influence of particular countries/regions in the worldwide collaboration map (26, 28, 38, 44). The United States and China made the largest contributions to papers on AS of cancer, as shown in Table 1 and Figure 3A. The top 10 institutions by the number of publications were mainly from China and the United States: six were from China and four were from the United States, respectively. China and Chinese institutions, on the other hand, had a centrality of less than 0.1, while the United States had a centrality of 0.41, suggesting that the United States may continue to dominate AS of the cancer field. Furthermore, the United States, England, Germany, Australia, and France had high betweenness centrality, which indicates that they were important hubs in AS of the cancer field’s international collaboration. Moreover, it could be seen that countries/regions and institutions had an active collaboration in terms of network density, respectively.
Scorilas Andreas not only published the most articles related to AS of cancer but is also the top 1 H-index scholar (Table 2), demonstrating his excellent contribution to AS of cancer research. Scorilas is a researcher at the National and Kapodistrian University of Athens, committed to AS, gene transcription, and next-generation sequencing. For the past 10 years, his group has published a series of articles (45–50) that described the process of identification of novel alternative splicing variants using next-generation sequencing methodology and discussed their expression situation and pathophysiological implications. Furthermore, Oltean Sebastian is also a scholar working at the University of Exeter; his team centered on the regulation and form of AS in prostate cancer and therapeutic targets associated with AS in cancer. In 2014, Nature published an Oltean and Bates’s review entitled “Hallmarks of alternative splicing in cancer” (11). They summarized how the numerous phenotypic traits that tumors acquire are influenced by AS and identified a new class of anticancer treatments called alternative splicing inhibitors; This article was co-cited up to 234 times with citation bursts from 2015 to 2019. Significantly, the top 1 co-cited author, Wang Eric T., a professor at the University of Florida, focused on exploring the function of the short and long gamma subunit splice variants of human GABA(A) receptors. He published the top 1 co-cited reference that reviewed the regulation of AS (5), with the second strongest citation burst strength.
According to a journal analysis (Table 3), Plos One was the ninth most referenced journal and published the most AS of cancer studies. The fact that Nucleic Acids Research was among the top 5 academic journals, as well as co-cited journals, shows how important it is to the dissemination of AS of cancer research. The top 10 most-cited papers were published in the journals, which are also generally the most-cited journals. For instance, Nature obtained the second-highest number of co-citations, in part due to three of the top 10 extremely co-cited references (1, 5, 35) (Table 4). Similar to the dual-map study (Figure 5), we found that the majority of the journals were on the subjects of molecular, genetics, and comprehensive fields.
The knowledge network may be represented in part by the co-cited references cited by the papers in the corresponding field (22, 27, 51). The study’s top 10 co-cited articles found that three articles primarily explored the relationship between AS and hallmarks of cancer (11, 36, 37), two were about the discovery of alternative splicing isoforms using high-throughput sequencing (5, 33), two were related to the regulation of AS (6, 10), and two papers focused on several specific molecules, splicing factor SF2/ASF, and heterogeneous nuclear ribonucleoproteins (hnRNPs) controlled by c-Myc, respectively (34, 35). Moreover, one review discussed the mechanisms of proteome expansion by AS (1). As shown in our citation bursts results (Figure 6C), five references in the AS of cancer research are still active: three are related to the function and mechanisms of AS (37), one is a dedicated effort to mine prognostic AS signatures (40), and three present the landscape of AS in TCGA tumors (15, 39, 41).
4.2 The hotspots and trending
4.2.1 Hallmarks of alternative splicing in cancer
A transformation of the AS state occurs concurrently with the acquisition of cancer characteristics during carcinogenesis (11). Erroneous splicing produces tumor-specific isoforms, and the disordered expression of these isoforms propels tumor malignant progression (52). Alterations in isoform-specific splicing patterns of many genes drive tumor cells to acquire sustained proliferative signals, escape growth inhibition, resist cell death, induce angiogenesis, invade and metastasize, and escape immune evasion surveillance (4). As shown in Figure 6C, the results of the reference burst showed that a review by Oltean Sebastian systematically expounded the hallmarks of AS in cancer. Among the keywords of the pathological process (Table 6), it also covered many hot words such as apoptosis, migration, angiogenesis, and proliferation, which were closely related to cancer hallmarks.
In our study, the keyword co-occurrence analysis by VOSviewer (Figure 7A) and cluster label (#2) from CiteSpace (Figure 7B) both highlighted epithelial–mesenchymal transition (EMT), during which relatively quiescent, tightly connected epithelial cells acquire highly motile and invasive mesenchymal properties (53, 54). As epithelial cancers progress, cancer cells develop aggressive and migratory characteristics that allow them to invade nearby tissues and disseminate to distant organs (55), and 90% of cancer deaths are caused by this metastasis development process (56). EMT is linked to the reprogramming of multiple genes’ expression. E-cadherin, claudins, and occludins are examples of epithelial-specific genes that are inhibited by the SNAIL proteins (SNAIL1 and SNAIL2) (57, 58). N-cadherin, fibronectin, and matrix metalloproteases are examples of mesenchymal-specific genes that can be stimulated by the bHLH transcription factors (TWIST1 and TWIST2) and ZEB proteins (ZEB1 and ZEB2) (59–61). There is a ton of evidence to support the idea that AS events cause mesenchymal and epithelial cells to differ proteomically (62). According to reports, the modulation of a number of splicing factors is crucial to the EMT process (63). Numerous pre-mRNA targets can be regulated by a single AS factor. As a result, variations in their expression levels may have an impact on multiple aspects of the development of EMT (53).
The fifth keyword of Molecules in our study is esrp1 (Table 6). Depending on where their binding sites (UGG-rich motifs) are located in their RNA targets, ESRP proteins have a positional effect and either promote or repress exon inclusion (64, 65). esrp1 and esrp2 are two epithelial-restricted splicing regulators (66). During the activation of EMT programs, ESRPs regulate a network of epithelial regulators, and AS has a significant impact on the physical connections between isoforms (67). The isomers may have completely different effects. By boosting P120’s affinity for E-cadherin, P120 isoforms 3 and 4 can help epithelial cells adhere to one another (68). In contrast, p120 isoform 1 promotes RAC1 activity and stimulates cell migration and invasiveness by blocking the RHOA–ROCK signaling pathway (69).
Another molecule in Table 6 closely related to EMT is CD44. Various extracellular matrix elements are bound by the cell surface glycoprotein that CD44 encodes for (70). Mesenchymal CD44 splicing isoforms can be produced more readily when esrp1 is inhibited by ZEB1 (71). Notably, the change from the epithelial isoform (CD44v) to CD44s reveals a crucial function in EMT (72).
Other hallmarks of cancers are also closely related to AS. Epidermal growth factor receptor (EGFR) is an important molecule that affects cell proliferation and motility. EGFR lacking the fourth exon after AS can consistently activate the proliferation of cancer cells (73). Loss of p53 function causes tumors to escape growth suppressors, and a splice variant of p53 without tumor-suppressor function even competes with wild-type p53 (74). Bcl-x has two splice isoforms, including the pro-apoptotic Bcl-xS and the anti-apoptotic Bcl-xL, which is a common aberrant AS event in several types of cancer (75). Similarly, molecules of the vascular endothelial growth factor (VEGF) family commonly have multiple splice forms, which are closely associated with abnormal tumor angiogenesis (76). Pyruvate kinase (PKM) is an important molecule in cellular metabolism, and its splice isomers, PKM1 and PKM2, are expressed in the adult and embryonic stages, respectively; PKM2 is aberrantly expressed in a variety of cancers (35). The same strategy is used by tumors to evade immune destruction, as HLA and MHC-I molecules have a variety of aberrant splice isomers that assist tumors to escape immune recognition (77, 78).
Alternative splicing occurs in the vast majority of genes, and many of the spliced isoforms are closely associated with cancer hallmarks (11). An understanding of the relationship between abnormal AS regulatory mechanisms and cancer hallmarks will facilitate the development of specific targeted drugs to inhibit the progression of cancer phenotypes (79–81).
4.2.2 Alternative splicing signature of cancer
In our research, signature-related words appeared many times. For example, one of the cluster labels of the reference is splicing signature (#1, Figure 6B), therapeutic target (#1, Figure 7B) is in the keywords cluster labels, and signature and prognostic signature are in the keywords burst (Figure 7C). In the last 10 years, transcriptome sequencing at the genome-wide level has demonstrated that AS is closely modulated in a tissue- and developmental stage-specific way, and it has also been shown that AS is frequently dysregulated in a variety of human cancer types (63, 82–85). As a result, the study of AS events as tumor indicators and therapeutic targets has gained a lot of attention. Additionally, the range of tumor biomarkers and therapeutic targets has been considerably broadened by AS (86).
Currently, it is understood that the principal causes of tumorigenesis are splicing abnormalities, which include genetic changes in the spliced gene and changed expression of either or both of the key regulators or core components of the precursor messenger RNA (pre-mRNA) splicing machinery (8, 87). This also provides a theoretical basis for AS to become a tumor marker and a therapeutic target.
The core issues in oncology continue to be the early detection and diagnosis of cancer as well as the selection of the best-customized treatment for each patient. It is now possible to identify genome-wide AS thanks to the advancement of high-throughput sequencing methods, particularly RNA sequencing (88–93). Numerous potential benefits of RNA-seq include its capacity to estimate the great amount of both acknowledged and original alternative transcripts, as well as its ability to offer a finer resolution, deeper coverage, and greater accuracy (93). Due to advancements in sequencing and bioinformatics technology, a number of cancer-specific AS events with potential prognostic and predictive significance in clinical situations have been found thus far (40, 94–98). For instance, hormone-directed therapy is less successful in castration-resistant prostate cancer patients who carry the alternatively spliced androgen receptor variation 7 (99). It was proposed that the tumors with increased background expression of PKM2 show a more aggressive phenotype and poor response to chemotherapy in pancreatic ductal adenocarcinoma patients undergoing radical surgery and adjuvant chemotherapy (100). A prospective therapeutic target in colorectal cancer is CD44 variation 6, an independent negative prognostic factor (101–103). Similar to our research, these molecules appear in the molecular keywords (Table 6), indicating that they have potential clinical translation value and are research hotspots.
Numerous effective therapeutic approaches have been developed as a result of the discovery of cancer-specific AS mutations. First, reversing faulty RNA splicing has been made possible by inhibiting post-translational modifications of splicing factors or RNA-binding proteins, particularly with the help of small-molecule inhibitors that target protein kinases (104, 105). The dual-specificity Cdc2-like kinases and SR-rich protein-specific kinases are these agents’ two primary targets (106). Second, a critical treatment focus is the adjustment of signaling pathways that control AS events (107). For instance, the PI3K/AKT/mTOR pathway inhibitors MK2206 and BEZ235 can alter splicing results (108, 109). Third, antisense oligonucleotides can hinder the splicing machinery’s ability to reach the regulatory regions in the pre-mRNA for therapeutic purposes and encourage the purge of the targeted mRNA by endogenous cellular nucleases (110, 111). Bcl-x is a critical gene, ranked 14th in our molecular keywords (Table 6). Bcl-x antisense oligonucleotides were created to encourage a splicing transition that favors the generation of pro-apoptotic Bcl-xS rather than anti-apoptotic Bcl-xL (112). Fourth, AS isoform proteins unique to tumors have always been prospective therapeutic targets. Certain tactics have been devised to use immunotherapies to target cancer-specific isoforms (86). The EGFR isoforms de4 and vIII are among the most extensively researched therapeutic targets (113).
In the realm of tumor research, the identification and therapy of cancers are enduring focus topics. More significant biomarkers may be represented by AS signature spectra or characteristics. These AS signatures can be used as biomarkers for tumor diagnosis or to develop more effective drug candidates.
4.2.3 Mechanism of alternative splicing
As shown in Table 6, it can be seen that splicing-related proteins such as srsf3, esrp1, srsf1, ptbp1, rbm10, and rbm5 have constantly been research hotspots. Among them, srsf3 (#5, Figure 6B) ranks sixth in the reference cluster label, and the splicing factor (#3, Figure 7B) is also one of the keyword cluster labels. The keyword splicing factor is also reflected in the keywords burst (strength = 12.08, Figure 7C), with burstness until 2021. This part mainly discusses the important role of splicing factors in AS. Splicing factors are auxiliary proteins that take part in the splicing of pre-mRNA.
Trans-acting splicing factors, which bind to sequence motifs connected to the stimulation (enhancers) or inhibition (silencers) of splicing, usually control AS. These motifs can be found in exons and introns, and they frequently have the greatest impact near splice sites (114, 115).
Our research indicates that splicing factors from the families of hnRNPs and serine/arginine-rich proteins (SR proteins) have been a focus of this field’s research. Many splicing factors ranked high in the molecular keywords (Table 6) belong to the SR proteins, RNA-binding motif (RBM) proteins, etc. An article with research on the mechanism of hnRNPs is the eighth cited reference (Table 4) and also ranks fifth in the strength of the reference burst (Figure 6C).
In addition to a carboxy-terminal arginine/serine-rich domain that contributes to protein–protein interactions, SR proteins have one or two copies of an RNA recognition motif domain at the amino terminus that offers RNA-binding specificity (116, 117). The majority of SR proteins function as splicing activators, helping the spliceosome to recognize exons and enable exon inclusion by binding to pre-mRNA at exonic splicing enhancers. SR proteins frequently face competition from splicing repressors such as hnRNPs. By binding to exonic or intronic splicing silencers, hnRNPs obstruct spliceosome elements’ access and suppress splice site choice. RNA-binding domains and somewhat unstructured domains, which are likely involved in protein–protein interactions, are both present in hnRNPs in a comparable manner. Exon skipping is prevented by SR proteins’ concentration-dependent inhibition of hnRNPs’ activity (115, 118, 119).
In conclusion, many RNA-binding proteins, such as SR proteins and hnRNPs, bind splicing enhancers and silencers (120). Furthermore, some members of RBM proteins also play important physiological roles as splicing factors. Similar to our study, among them, there are more reports about RBM4 (121), RBM5 (122), and RBM10 (123). Similar to SR protein and hnRNPs protein, RBMs can also regulate the occurrence of splicing events alone or cooperate with other splicing factors to regulate the splicing process (124).
4.3 Strengths and limitations
Overall, as far as we know, this article may be the first to apply bibliometric methods to comprehensively examine papers associated with AS of cancer research published in the last 10 years. The bibliometric method offers a fresh and unbiased perspective on the changing research hotspots and fashion in contrast to conventional reviews (21). Simultaneously, we conducted an investigation using various bibliometric tools, which could produce more prosperous outcomes across numerous dimensions (21, 125). This study will educate the public about the significance of AS of cancer, offer scholars a complete image of AS of cancer study, and additionally provide a detailed and impartial direction for the field’s upcoming growth. This study unavoidably has certain shortcomings. First, we only obtained the WoSCC database’s English-language articles, leaving out non-English or non-WoSCC items. However, WoSCC’s English articles are the most often utilized data source in bibliometrics; thus, to some extent, they may represent the majority of the field (21, 126). Secondly, some studies report that bibliometric methods are inevitably biased because they are based on natural language processing (17, 20). Our findings, however, are comparable to recent conventional reviews while offering more comprehensive and unbiased data (4, 11, 86).
5 Conclusion
In conclusion, research on AS of cancer has steadily advanced step by step with active cooperation over the past 10 years, with the possibility that the United States will continue to hold the lead in this field. Scorilas Andreas and Wang Eric T. were the authors with the most publications and co-citations in AS of the cancer field, respectively. Currently, AS of cancer research is predominantly centered on hallmarks of AS in cancer, AS signatures, and therapeutic targets, as well as further mechanisms underlying AS. Among them, splicing factors that regulate EMT or other hallmarks, aberrant AS signatures and therapeutic targets in cancer research, and the mechanism of AS may become popular and fruitful directions. These could offer directions and fresh perspectives for future studies in the AS of cancer.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
L-WW, Y-NP, BT, YB, and Z-SL designed this study. BT, YB, DJ-B, and YG collected the data and performed the analysis. XZ, S-WZ, and Y-HZ provided support in the data curation, validation, and visualization. BT, YB, and D-JB wrote the original draft. L-WW and Z-SL reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82103268), Shanghai Sailing Program (No. 21YF1459100), China Postdoctoral Science Foundation (No. 48067), the start-up funds for postdoctoral in Second Military Medical University and the start-up Scientific Research Fund of Young Teachers in Changhai Hospital (No. 2019QNB01), Shanghai Science and Technology Innovation Action Program (No. 21Y31900100) and 234 clinical research fund of Changhai hospital (No. 2019YXK006).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1068805/full#supplementary-material
References
1. Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature (2010) 463:457–63. doi: 10.1038/nature08909
3. Bonnal SC, López-Oreja I, Valcárcel J. Roles and mechanisms of alternative splicing in cancer — implications for care. Nat Rev Clin Oncol (2020) 17:457–74. doi: 10.1038/s41571-020-0350-x
4. Zhang Y, Qian J, Gu C, Yang Y. Alternative splicing and cancer: a systematic review. Signal Transduct Target Ther (2021) 6:78. doi: 10.1038/s41392-021-00486-7
5. Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature (2008) 456:470–6. doi: 10.1038/nature07509
6. Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell (2009) 136:701–18. doi: 10.1016/j.cell.2009.02.009
7. Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, et al. An RNA map predicting Nova-dependent splicing regulation. Nature (2006) 444:580–6. doi: 10.1038/nature05304
8. Urbanski LM, Leclair N, Anczuków O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA (2018) 9:e1476. doi: 10.1002/wrna.1476
9. Qi F, Li Y, Yang X, Wu Y-P, Lin L-J, Liu X-M. Significance of alternative splicing in cancer cells. Chin Med J (Engl) (2020) 133:221–8. doi: 10.1097/CM9.0000000000000542
10. David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev (2010) 24:2343–64. doi: 10.1101/gad.1973010
11. Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene (2014) 33:5311–8. doi: 10.1038/onc.2013.533
12. Zhang YJ, Yan LB, Zeng J, Zhou H, Liu HR, Yu G, et al. Pan-cancer analysis of clinical relevance of alternative splicing events in 31 human cancers. Oncogene (2019) 38:6678–95. doi: 10.1038/s41388-019-0910-7
13. Frankiw L, Baltimore D, Li G. Alternative mRNA splicing in cancer immunotherapy. Nat Rev Immunol (2019) 19:675–87. doi: 10.1038/s41577-019-0195-7
14. Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer (2005) 5:845–56. doi: 10.1038/nrc1739
15. Kahles A, Lehmann K-V, Toussaint NC, Hüser M, Stark SG, Sachsenberg T, et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell (2018) 34:211–224.e6. doi: 10.1016/j.ccell.2018.07.001
16. Sciarrillo R, Wojtuszkiewicz A, Assaraf YG, Jansen G, Kaspers GJL, Giovannetti E, et al. The role of alternative splicing in cancer: From oncogenesis to drug resistance. Drug Resist Update Rev Comment Antimicrob Anticancer Chemother (2020) 53:100728. doi: 10.1016/j.drup.2020.100728
17. Yan W-T, Lu S, Yang Y-D, Ning W-Y, Cai Y, Hu X-M, et al. Research trends, hot spots and prospects for necroptosis in the field of neuroscience. Neural Regener Res (2021) 16:1628–37. doi: 10.4103/1673-5374.303032
18. Dong Q, Liang Q, Chen Y, Li J, Lu L, Huang X, et al. Bibliometric and visual analysis of vascular calcification research. Front Pharmacol (2021) 12:690392. doi: 10.3389/fphar.2021.690392
19. Chen C, Song M. Visualizing a field of research: A methodology of systematic scientometric reviews. PLos One (2019) 14:e0223994. doi: 10.1371/journal.pone.0223994
20. Zhang J, Song L, Jia J, Tian W, Lai R, Zhang Z, et al. Knowledge mapping of necroptosis from 2012 to 2021: A bibliometric analysis. Front Immunol (2022) 13:917155. doi: 10.3389/fimmu.2022.917155
21. Zhang J, Song L, Xu L, Fan Y, Wang T, Tian W, et al. Knowledge domain and emerging trends in ferroptosis research: A bibliometric and knowledge-map analysis. Front Oncol (2021) 11:686726. doi: 10.3389/fonc.2021.686726
22. Chen C. CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. J Am Soc Inf Sci Technol (2006) 57:359–77. doi: 10.1002/asi.20317
23. Aria M, Cuccurullo C. Bibliometrix: An R-tool for comprehensive science mapping analysis. J Informetr (2017) 11:959–75. doi: 10.1016/j.joi.2017.08.007
24. van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics (2010) 84:523–38. doi: 10.1007/s11192-009-0146-3
25. Paunkov A, Chartoumpekis DV, Ziros PG, Sykiotis GP. A bibliometric review of the Keap1/Nrf2 pathway and its related antioxidant compounds. Antioxid Basel Switz (2019) 8:E353. doi: 10.3390/antiox8090353
26. Chen C. Searching for intellectual turning points: Progressive knowledge domain visualization. Proc Natl Acad Sci (2004) 101:5303–10. doi: 10.1073/pnas.0307513100
27. Chen C. Science mapping: A systematic review of the literature. J Data Inf Sci (2017) 2:1–40. doi: 10.1515/jdis-2017-0006
28. Chen C, Hu Z, Liu S, Tseng H. Emerging trends in regenerative medicine: a scientometric analysis in CiteSpace. Expert Opin Biol Ther (2012) 12:593–608. doi: 10.1517/14712598.2012.674507
29. Cheng K, Guo Q, Shen Z, Yang W, Wang Y, Sun Z, et al. Bibliometric analysis of global research on cancer photodynamic therapy: Focus on nano-related research. Front Pharmacol (2022) 13:927219. doi: 10.3389/fphar.2022.927219
30. Li C, Wu K, Wu J. A bibliometric analysis of research on haze during 2000-2016. Environ Sci pollut Res Int (2017) 24:24733–42. doi: 10.1007/s11356-017-0440-1
31. Daim TU, Rueda G, Martin H, Gerdsri P. Forecasting emerging technologies: Use of bibliometrics and patent analysis. Technol Forecast Soc Change (2006) 73:981–1012. doi: 10.1016/j.techfore.2006.04.004
32. Chen C, Leydesdorff L. Patterns of connections and movements in dual-map overlays: A new method of publication portfolio analysis. J Assoc Inf Sci Technol (2014) 65:334–51. doi: 10.1002/asi.22968
33. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet (2008) 40:1413–5. doi: 10.1038/ng.259
34. Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol (2007) 14:185–93. doi: 10.1038/nsmb1209
35. David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-myc deregulate pyruvate kinase mRNA splicing in cancer. Nature (2010) 463:364–8. doi: 10.1038/nature08697
36. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
37. Climente-González H, Porta-Pardo E, Godzik A, Eyras E. The functional impact of alternative splicing in cancer. Cell Rep (2017) 20:2215–26. doi: 10.1016/j.celrep.2017.08.012
38. Ma L, Ma J, Teng M, Li Y. Visual analysis of colorectal cancer immunotherapy: A bibliometric analysis from 2012 to 2021. Front Immunol (2022) 13:843106. doi: 10.3389/fimmu.2022.843106
39. Ryan M, Wong WC, Brown R, Akbani R, Su X, Broom B, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res (2016) 44:D1018–1022. doi: 10.1093/nar/gkv1288
40. Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z, et al. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett (2017) 393:40–51. doi: 10.1016/j.canlet.2017.02.016
41. Seiler M, Peng S, Agrawal AA, Palacino J, Teng T, Zhu P, et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep (2018) 23:282–296.e4. doi: 10.1016/j.celrep.2018.01.088
42. Berget SM, Moore C, Sharp PA. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA*. Proc Natl Acad Sci (1977) 74:3171–5. doi: 10.1073/pnas.74.8.3171
43. Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5’ ends of adenovirus 2 messenger RNA. Cell (1977) 12:1–8. doi: 10.1016/0092-8674(77)90180-5
44. Liu S, Xia K, Liu X, Duan Y, Hu M, Xia H, et al. Bibliometric analysis of birt-Hogg-Dubé syndrome from 2001 to 2021. Front Med (2022) 9:857127. doi: 10.3389/fmed.2022.857127
45. Adamopoulos PG, Theodoropoulou MC, Scorilas A. Alternative splicing detection tool-a novel PERL algorithm for sensitive detection of splicing events, based on next-generation sequencing data analysis. Ann Transl Med (2018) 6:244. doi: 10.21037/atm.2018.06.32
46. Adamopoulos PG, Kontos CK, Scorilas A. Identification and molecular cloning of novel transcripts of the human kallikrein-related peptidase 10 (KLK10) gene using next-generation sequencing. Biochem Biophys Res Commun (2017) 487:776–81. doi: 10.1016/j.bbrc.2017.04.078
47. Adamopoulos PG, Kontos CK, Tsiakanikas P, Scorilas A. Identification of novel alternative splice variants of the BCL2L12 gene in human cancer cells using next-generation sequencing methodology. Cancer Lett (2016) 373:119–29. doi: 10.1016/j.canlet.2016.01.019
48. Adamopoulos PG, Kontos CK, Scorilas A, Sideris DC. Identification of novel alternative transcripts of the human ribonuclease kappa (RNASEK) gene using 3 ‘ RACE and high-throughput sequencing approaches. Genomics (2020) 112:943–51. doi: 10.1016/j.ygeno.2019.06.010
49. Adamopoulos PG, Mavrogiannis AV, Kontos CK, Scorilas A. Novel alternative splice variants of the human protein arginine methyltransferase 1 (PRMT1) gene, discovered using next-generation sequencing. Gene (2019) 699:135–44. doi: 10.1016/j.gene.2019.02.072
50. Adamopoulos PG, Tsiakanikas P, Adam EE, Scorilas A. Unraveling novel survivin mRNA transcripts in cancer cells using an in-house developed targeted high-throughput sequencing approach. Genomics (2021) 113:573–81. doi: 10.1016/j.ygeno.2020.09.053
51. Lu C, Liu M, Shang W, Yuan Y, Li M, Deng X, et al. Knowledge mapping of angelica sinensis (Oliv.) diels (Danggui) research: A scientometric study. Front Pharmacol (2020) 11:294. doi: 10.3389/fphar.2020.00294
52. Biamonti G, Catillo M, Pignataro D, Montecucco A, Ghigna C. The alternative splicing side of cancer. Semin Cell Dev Biol (2014) 32:30–6. doi: 10.1016/j.semcdb.2014.03.016
53. Pradella D, Naro C, Sette C, Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer (2017) 16:8. doi: 10.1186/s12943-016-0579-2
54. Piera-Velazquez S, Jimenez SA. Endothelial to mesenchymal transition: Role in physiology and in the pathogenesis of human diseases. Physiol Rev (2019) 99:1281–324. doi: 10.1152/physrev.00021.2018
55. Nieto MA, Huang RY-J, Jackson RA, Thiery JP. EMT: 2016. Cell (2016) 166:21–45. doi: 10.1016/j.cell.2016.06.028
56. Mehlen P, Puisieux A. Metastasis: A question of life or death. Nat Rev Cancer (2006) 6:449–58. doi: 10.1038/nrc1886
57. Ohkubo T, Ozawa M. The transcription factor snail downregulates the tight junction components independently of e-cadherin downregulation. J Cell Sci (2004) 117:1675–85. doi: 10.1242/jcs.01004
58. Wang Y, Shi J, Chai K, Ying X, Zhou BP. The role of snail in EMT and tumorigenesis. Curr Cancer Drug Targets (2013) 13:963–72. doi: 10.2174/15680096113136660102
59. Kang Y, Massagué J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell (2004) 118:277–9. doi: 10.1016/j.cell.2004.07.011
60. Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, et al. The two-handed e box binding zinc finger protein SIP1 downregulates e-cadherin and induces invasion. Mol Cell (2001) 7:1267–78. doi: 10.1016/s1097-2765(01)00260-x
61. Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, et al. DeltaEF1 is a transcriptional repressor of e-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene (2005) 24:2375–85. doi: 10.1038/sj.onc.1208429
62. Mathias RA, Simpson RJ. Towards understanding epithelial-mesenchymal transition: a proteomics perspective. Biochim Biophys Acta (2009) 1794:1325–31. doi: 10.1016/j.bbapap.2009.05.001
63. Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLos Genet (2011) 7:e1002218. doi: 10.1371/journal.pgen.1002218
64. Warzecha CC, Shen S, Xing Y, Carstens RP. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol (2009) 6:546–62. doi: 10.4161/rna.6.5.9606
65. Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J (2010) 29:3286–300. doi: 10.1038/emboj.2010.195
66. Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell (2009) 33:591–601. doi: 10.1016/j.molcel.2009.01.025
67. Yang Y, Park JW, Bebee TW, Warzecha CC, Guo Y, Shang X, et al. Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-Mesenchymal transition. Mol Cell Biol (2016) 36:1704–19. doi: 10.1128/MCB.00019-16
68. Ishiyama N, Lee S-H, Liu S, Li G-Y, Smith MJ, Reichardt LF, et al. Dynamic and static interactions between p120 catenin and e-cadherin regulate the stability of cell-cell adhesion. Cell (2010) 141:117–28. doi: 10.1016/j.cell.2010.01.017
69. Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, et al. A p120 catenin isoform switch affects rho activity, induces tumor cell invasion, and predicts metastatic disease. J Biol Chem (2008) 283:18344–54. doi: 10.1074/jbc.M801192200
70. Naor D, Wallach-Dayan SB, Zahalka MA, Sionov RV. Involvement of CD44, a molecule with a thousand faces, in cancer dissemination. Semin Cancer Biol (2008) 18:260–7. doi: 10.1016/j.semcancer.2008.03.015
71. Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest (2016) 126:3219–35. doi: 10.1172/JCI76725
72. Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest (2011) 121:1064–74. doi: 10.1172/JCI44540
73. Wang H, Zhou M, Shi B, Zhang Q, Jiang H, Sun Y, et al. Identification of an exon 4-deletion variant of epidermal growth factor receptor with increased metastasis-promoting capacity. Neoplasia N Y N (2011) 13:461–71. doi: 10.1593/neo.101744
74. Okumura N, Yoshida H, Kitagishi Y, Nishimura Y, Matsuda S. Alternative splicings on p53, BRCA1 and PTEN genes involved in breast cancer. Biochem Biophys Res Commun (2011) 413:395–9. doi: 10.1016/j.bbrc.2011.08.098
75. Cloutier P, Toutant J, Shkreta L, Goekjian S, Revil T, Chabot B. Antagonistic effects of the SRp30c protein and cryptic 5’ splice sites on the alternative splicing of the apoptotic regulator bcl-x. J Biol Chem (2008) 283:21315–24. doi: 10.1074/jbc.M800353200
76. Joukov V, Pajusola K, Kaipainen A, Chilov D, Lahtinen I, Kukk E, et al. A novel vascular endothelial growth factor, VEGF-c, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. EMBO J (1996) 15:290–8. doi: 10.1002/j.1460-2075.1996.tb00521.x
77. Rouas-Freiss N, Bruel S, Menier C, Marcou C, Moreau P, Carosella ED. Switch of HLA-G alternative splicing in a melanoma cell line causes loss of HLA-G1 expression and sensitivity to NK lysis. Int J Cancer (2005) 117:114–22. doi: 10.1002/ijc.21151
78. Rodríguez-Cruz TG, Liu S, Khalili JS, Whittington M, Zhang M, Overwijk W, et al. Natural splice variant of MHC class I cytoplasmic tail enhances dendritic cell-induced CD8+ T-cell responses and boosts anti-tumor immunity. PLos One (2011) 6:e22939. doi: 10.1371/journal.pone.0022939
79. Kole R, Krainer AR, Altman S. RNA Therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discovery (2012) 11:125–40. doi: 10.1038/nrd3625
80. Rigo F, Seth PP, Bennett CF. Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv Exp Med Biol (2014) 825:303–52. doi: 10.1007/978-1-4939-1221-6_9
81. McClorey G, Wood MJ. An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr Opin Pharmacol (2015) 24:52–8. doi: 10.1016/j.coph.2015.07.005
82. Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol (2009) 16:670–6. doi: 10.1038/nsmb.1608
83. Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics (2006) 7:325. doi: 10.1186/1471-2164-7-325
84. Misquitta-Ali CM, Cheng E, O’Hanlon D, Liu N, McGlade CJ, Tsao MS, et al. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol Cell Biol (2011) 31:138–50. doi: 10.1128/MCB.00709-10
85. Lapuk A, Marr H, Jakkula L, Pedro H, Bhattacharya S, Purdom E, et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res MCR (2010) 8:961–74. doi: 10.1158/1541-7786.MCR-09-0528
86. Bessa C, Matos P, Jordan P, Gonçalves V. Alternative splicing: Expanding the landscape of cancer biomarkers and therapeutics. Int J Mol Sci (2020) 21(23):9032. doi: 10.3390/ijms21239032
87. Rahman MA, Nasrin F, Bhattacharjee S, Nandi S. Hallmarks of splicing defects in cancer: Clinical applications in the era of personalized medicine. Cancers (2020) 12:E1381. doi: 10.3390/cancers12061381
88. Zhao S, Fung-Leung W-P, Bittner A, Ngo K, Liu X. Comparison of RNA-seq and microarray in transcriptome profiling of activated T cells. PLos One (2014) 9:e78644. doi: 10.1371/journal.pone.0078644
89. Zhang C, Dower K, Zhang B, Martinez RV, Lin L-L, Zhao S. Computational identification and validation of alternative splicing in ZSF1 rat RNA-seq data, a preclinical model for type 2 diabetic nephropathy. Sci Rep (2018) 8:7624. doi: 10.1038/s41598-018-26035-x
90. Zhang C, Zhang B, Lin L-L, Zhao S. Evaluation and comparison of computational tools for RNA-seq isoform quantification. BMC Genomics (2017) 18:583. doi: 10.1186/s12864-017-4002-1
91. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet (2009) 10:57–63. doi: 10.1038/nrg2484
92. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods (2008) 5:621–8. doi: 10.1038/nmeth.1226
93. Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet (2016) 17:257–71. doi: 10.1038/nrg.2016.10
94. Hu Y-X. Systematic profiling of alternative splicing signature reveals prognostic predictor for cervical cancer. J Transl Med (2019) 17(1):379. doi: 10.1186/s12967-019-02140-x
95. Zhu J, Chen Z, Yong L. Systematic profiling of alternative splicing signature reveals prognostic predictor for ovarian cancer. Gynecol Oncol (2018) 148:368–74. doi: 10.1016/j.ygyno.2017.11.028
96. Yang C, Wu Q, Huang K, Wang X, Yu T, Liao X, et al. Genome-wide profiling reveals the landscape of prognostic alternative splicing signatures in pancreatic ductal adenocarcinoma. Front Oncol (2019) 9:511. doi: 10.3389/fonc.2019.00511
97. Zhang D, Hu Q, Liu X, Ji Y, Chao H-P, Liu Y, et al. Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat Commun (2020) 11:2089. doi: 10.1038/s41467-020-15815-7
98. Xiong Y, Deng Y, Wang K, Zhou H, Zheng X, Si L, et al. Profiles of alternative splicing in colorectal cancer and their clinical significance: A study based on large-scale sequencing data. EBioMedicine (2018) 36:183–95. doi: 10.1016/j.ebiom.2018.09.021
99. Qu Y, Dai B, Ye D, Kong Y, Chang K, Jia Z, et al. Constitutively active AR-V7 plays an essential role in the development and progression of castration-resistant prostate cancer. Sci Rep (2015) 5:7654. doi: 10.1038/srep07654
100. Calabretta S, Bielli P, Passacantilli I, Pilozzi E, Fendrich V, Capurso G, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene (2016) 35:2031–9. doi: 10.1038/onc.2015.270
101. Fan C-W, Wen L, Qiang Z-D, Chen T, Zhou Z-G, Mo X-M, et al. Prognostic significance of relevant markers of cancer stem cells in colorectal cancer - a meta analysis. Hepatogastroenterology (2012) 59:1421–7. doi: 10.5754/hge10727
102. Saito S, Okabe H, Watanabe M, Ishimoto T, Iwatsuki M, Baba Y, et al. CD44v6 expression is related to mesenchymal phenotype and poor prognosis in patients with colorectal cancer. Oncol Rep (2013) 29:1570–8. doi: 10.3892/or.2013.2273
103. Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell (2014) 14:342–56. doi: 10.1016/j.stem.2014.01.009
104. Lee SC-W, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med (2016) 22:976–86. doi: 10.1038/nm.4165
105. Antonopoulou E, Ladomery M. Targeting splicing in prostate cancer. Int J Mol Sci (2018) 19:E1287. doi: 10.3390/ijms19051287
106. Aubol BE, Wu G, Keshwani MM, Movassat M, Fattet L, Hertel KJ, et al. Release of SR proteins from CLK1 by SRPK1: A symbiotic kinase system for phosphorylation control of pre-mRNA splicing. Mol Cell (2016) 63:218–28. doi: 10.1016/j.molcel.2016.05.034
107. Black AJ, Gamarra JR, Giudice J. More than a messenger: Alternative splicing as a therapeutic target. Biochim Biophys Acta Gene Regul Mech (2019) 1862:194395. doi: 10.1016/j.bbagrm.2019.06.006
108. Sanidas I, Polytarchou C, Hatziapostolou M, Ezell SA, Kottakis F, Hu L, et al. Phosphoproteomics screen reveals akt isoform-specific signals linking RNA processing to lung cancer. Mol Cell (2014) 53:577–90. doi: 10.1016/j.molcel.2013.12.018
109. Passacantilli I, Frisone P, De Paola E, Fidaleo M, Paronetto MP. hnRNPM guides an alternative splicing program in response to inhibition of the PI3K/AKT/mTOR pathway in Ewing sarcoma cells. Nucleic Acids Res (2017) 45:12270–84. doi: 10.1093/nar/gkx831
110. Levin AA. Treating disease at the RNA level with oligonucleotides. N Engl J Med (2019) 380:57–70. doi: 10.1056/NEJMra1705346
111. Havens MA, Hastings ML. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res (2016) 44:6549–63. doi: 10.1093/nar/gkw533
112. Mercatante DR, Bortner CD, Cidlowski JA, Kole R. Modification of alternative splicing of bcl-x pre-mRNA in prostate and breast cancer cells. analysis of apoptosis and cell death. J Biol Chem (2001) 276:16411–7. doi: 10.1074/jbc.M009256200
113. Moscatello DK, Holgado-Madruga M, Godwin AK, Ramirez G, Gunn G, Zoltick PW, et al. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res (1995) 55:5536–9.
114. Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA N Y N (2008) 14:802–13. doi: 10.1261/rna.876308
115. Fu X-D, Ares M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet (2014) 15:689–701. doi: 10.1038/nrg3778
116. Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev (1992) 6:837–47. doi: 10.1101/gad.6.5.837
117. Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, et al. Protein-protein interactions and 5’-splice-site recognition in mammalian mRNA precursors. Nature (1994) 368:119–24. doi: 10.1038/368119a0
118. Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA (2012) 3:1–12. doi: 10.1002/wrna.100
119. Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J (2009) 417:15–27. doi: 10.1042/BJ20081501
120. Singh R, Valcárcel J. Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol (2005) 12:645–53. doi: 10.1038/nsmb961
121. Wang Y, Chen D, Qian H, Tsai YS, Shao S, Liu Q, et al. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell (2014) 26:374–89. doi: 10.1016/j.ccr.2014.07.010
122. Sutherland LC, Wang K, Robinson AG. RBM5 as a putative tumor suppressor gene for lung cancer. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer (2010) 5:294–8. doi: 10.1097/JTO.0b013e3181c6e330
123. Bechara EG, Sebestyén E, Bernardis I, Eyras E, Valcárcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell (2013) 52:720–33. doi: 10.1016/j.molcel.2013.11.010
124. Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA Splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer (2016) 16:413–30. doi: 10.1038/nrc.2016.51
125. Shen J, Shen H, Ke L, Chen J, Dang X, Liu B, et al. Knowledge mapping of immunotherapy for hepatocellular carcinoma: A bibliometric study. Front Immunol (2022) 13:815575. doi: 10.3389/fimmu.2022.815575
Keywords: bibliometric, alternative splicing, cancer, CiteSpace, VOSviewer, R-bibliometrix
Citation: Tian B, Bian Y, Bian D-J, Gao Y, Zhang X, Zhou S-W, Zhang Y-H, Pang Y-N, Li Z-S and Wang L-W (2022) Knowledge mapping of alternative splicing of cancer from 2012 to 2021: A bibliometric analysis. Front. Oncol. 12:1068805. doi: 10.3389/fonc.2022.1068805
Received: 13 October 2022; Accepted: 24 November 2022;
Published: 14 December 2022.
Edited by:
Zhiqiang Liu, Tianjin Medical University, ChinaReviewed by:
Rong Lin, Huazhong University of Science and Technology, ChinaRong Wan, Shanghai Jiao Tong University School of Medicine, China
Copyright © 2022 Tian, Bian, Bian, Gao, Zhang, Zhou, Zhang, Pang, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ya-Nan Pang, MTU3MjEyNTk0NzJAMTYzLmNvbQ==; Zhao-Shen Li, emhzbEB2aXAuMTYzLmNvbQ==; Luo-Wei Wang, d2FuZ2x1b3dlaW1kQDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
 Bo Tian
Bo Tian Yan Bian
Yan Bian De-Jian Bian
De-Jian Bian Ye Gao
Ye Gao Xun Zhang1
Xun Zhang1 Ya-Nan Pang
Ya-Nan Pang Zhao-Shen Li
Zhao-Shen Li