
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol. , 06 December 2022
Sec. Gastrointestinal Cancers: Gastric and Esophageal Cancers
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1058028
This article is part of the Research Topic Reviews in Gastrointestinal Cancers View all 35 articles
Objective: With the prevalence of next-generation sequencing (NGS) technology, a large number of long non-coding RNAs (lncRNAs) have attracted tremendous attention and have been the topic of extensive research on gastric cancer (GC). It was revealed that lncRNAs not only participate in the transduction of various signaling pathways, thus influencing GC genesis and development, but also have the potential for GC diagnosis. Therefore, we aimed to conduct a meta-analysis of previous studies on GC.
Materials and methods: An electronic search was made before August 2021 on databases including PubMed, Embase, and Web of Science. Relevant articles that compare lncRNA expression in GC patients and healthy controls were summarized. We conducted a meta-analysis with the objective of evaluating the ability of lncRNAs in diagnosing GC.
Results: A total of 40 original research studies including 6,772 participants were discussed in this meta-analysis. The overall sensitivity, specificity, and the area under the curve (AUC) were 0.78 (95% CI: 0.75–0.81), 0.79 (95% CI: 0.74–0.83), and 0.85 (95% CI: 0.81–0.87), respectively. The value of pooled diagnostic odds ratios (DORs) was 13.00 (95% CI: 10.00–17.00).
Conclusions: This meta-analysis revealed that serum or plasma lncRNAs have high sensitivity and specificity, which makes lncRNAs clinically feasible in diagnosing GC. The results from this meta-analysis demonstrated that peripheral blood lncRNAs may become novel noninvasive biomarkers in the foreseeable future. At the same time, it should be noted that a greater number of blood samples and more evidence from rigorous multicenter clinical studies are necessary to justify their applicability as cancer biomarkers.
Cancer is the leading cause of death and is a significant obstacle in the pursuit of a higher life expectancy worldwide (1). Unfortunately, the incidence and mortality of cancer are growing rapidly. Gastric cancer (GC) is an important malignant tumor in the digestive tract. According to the latest data, in 2020 alone, there are over 1 million new patients diagnosed with GC and about 769,000 cases die from it (2). It is widely accepted that chronic Helicobacter pylori (H. pylori) infection is the primary cause of GC (3, 4), and the International Agency for Research on Cancer (IARC) cited H. pylori as a group 1 carcinogen (5, 6).
The present treatment strategy for early GC usually depends on endoscopic surgery, while for advanced GC, the treatment methods include surgery, chemotherapy, and immunotherapy (7). Although progress has been achieved in GC treatment, challenges in terms of diagnosis remain. By the time symptoms appear in patients, most of them have already been diagnosed with an advanced stage of cancer (8), which seriously affects their prognosis and 5-year survival rate (9). Currently, gastrointestinal endoscopy operation together with biopsy is the main approach to identifying GC lesions, but detecting small lesions proved to be difficult because of the limited experience of endoscopists (5). In addition, patients find it difficult to undergo endoscopy because it is an invasive procedure and causes discomfort. Consequently, noninvasive biomarkers tend to be a better choice to solve this difficulty. From the traditional point of view, biomarkers in detecting GC can be classified from serum and gastric juice: serum biomarkers included carcinoembryonic antigen (CEA), carbohydrate antigen 199 (CA199), carbohydrate antigen 724 (CA724), and pepsinogen (PG) (10). Gastric juice biomarkers included CA724, CEA, CA199, CA242, and α1-antitrypsin (11, 12). However, the low sensitivity and specificity of these biomarkers in detecting GC limit their further application (13). Therefore, exploring novel biomarkers is of great importance in GC diagnosis.
With the increasing popularity of NGS applications, a large number of studies have been conducted to identify the role of lncRNAs in various tumors over several decades. Long non-coding RNAs, a class of non-coding RNA molecules with a length of more than 200 nt and lacking open reading frames, are closely associated with tumor invasion (14), metastasis, and drug resistance (15) of GC through multiple pathways. Moreover, studies also evaluated the diagnostic value of lncRNAs in distinguishing GC patients from healthy volunteers. These studies have demonstrated that the expression of lncRNAs could be a novel biomarker in screening GC due to their high sensitivity and specificity. Therefore, it is worthwhile to perform a systematic review and summarize the diagnostic values of these lncRNAs.
Some meta-analyses investigated the diagnostic or prognostic value of lncRNAs. However, most of them only focused on one specific lncRNA, such as lncRNA TP73-AS1 (16), lncRNA DLX6-AS1 (17), lncRNA DRAIR (18), and lncRNA HEIH (19). Furthermore, another study used a small number of lncRNAs to determine the diagnostic value of all lncRNAs in GC but ignored the heterogeneity sample differences (20). Considering the weakness of previous studies, a more integrative meta-analysis is necessary to determine GC diagnosis via lncRNAs.
In order to identify potentially eligible studies that were published before August 2021, two authors (JL and QX) separately conducted an electronic database search, including PubMed, Embase, and Web of science. The following search strategy was used: (Lnc RNA OR long non-coding RNA OR lncR) AND (“stomach neoplasms”[Mesh] OR “gastric cancer” OR “stomach cancer” OR “Gastric Neoplasm” OR “gastric carcinoma” OR “stomach carcinoma” OR “gastric adenocarcinoma” OR “stomach adenocarcinoma”) AND (blood OR serum OR plasma OR circulating) AND (diagnosis OR diagnostic OR diagnose).
For the enrolled articles, the following inclusion criteria must be fulfilled: (1) a comparison was made between GC and healthy controls; (2) the diagnosis of GC was confirmed by a pathologist; (3) the detection technique had to be quantitative real-time PCR and test samples were from serum or plasma; and (4) sufficient data were provided to calculate 2 × 2 tables including TP (true positive), FP (false positive), TN (true negative), and FN (false negative).
The exclusion criteria were as follows: (1) duplicate articles; (2) reviews, meta-analysis, bioinformatics, case reports, and laboratory studies; (3) studies irrelevant to the diagnostic value of lncRNAs or GC; and (4) the full text was not available.
The Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) (21) was applied to evaluate all enrolled articles in the meta-analysis, which mainly depend on the following domains: patient selection, index test, reference standard, and flow and timing. YZ, SB, and YD were responsible for this part of the work.
Two authors (YZ and YD) independently screened the full text of every study and extracted relevant information or data including (1) basic information of the enrolled articles: the first author, publication year, country of origin, ethnicity, specimen type (serum or plasma), lncRNA type, cases, and healthy control group size, mean age, and gender distribution; and (2) sensitivity, specificity, TP, FP, FN, and TN values, which were also extracted from each article.
STATA 16.0 (Stata Corporation, College Station, TX, USA) and Revman 5.4 (The Nordic Cochrane Centre, Copenhagen, Denmark) were used to analyze extracted data. In this diagnostic meta-analysis, forest plots were applied to estimate sensitivity and specificity. The area under the curve (AUC) of the summary receiver operating curve (SROC) was used to calculate the diagnostic efficiency of serum or plasma lncRNAs in GC. According to a previous report, diagnostic efficiency can be divided into low, good, very good, and excellent in terms of AUC values:<0.75, 0.75–0.92, 0.93–0.96, and 0.97 or above (22). Meanwhile, Q test and Higgins I2 statistic were used to estimate the heterogeneity among all included studies. If I2 > 50%, signifying the existence of heterogeneity, then the random-effect model was needed for data consolidation. Otherwise, the fixed-effect model was needed. Finally, the potential bias of publication was estimated by Deeks’ funnel plot. p < 0.05 was considered statistically significant.
This article has been registered on the International Platform of Registered Systematic Review and Meta-analysis Protocols (INPLASY, https://inplasy.com/); the registration number is INPLASY2022110024.
Through the search strategy described above, there were 476 articles from PubMed, Embase, and Web of Science included. A total of 69 duplicates were removed after a review of titles and abstracts. Next, we carefully read the rest of the articles and found 364 irrelevant publications. In addition, three articles were excluded for inadequate data. Finally, 40 publications including 6,772 participants were involved in this systematic review and meta-analysis. The basic characteristics of the included articles are listed in Table 1, and the flow-process diagram for the literature is presented in Figure 1.
The QUADAS-2 tool embedded in Revman 5.4 was used to assess the quality of each study. As shown in Figures 2A, B, the evaluation criteria mainly focus on patient selection, index test, reference standard, and flow and timing.
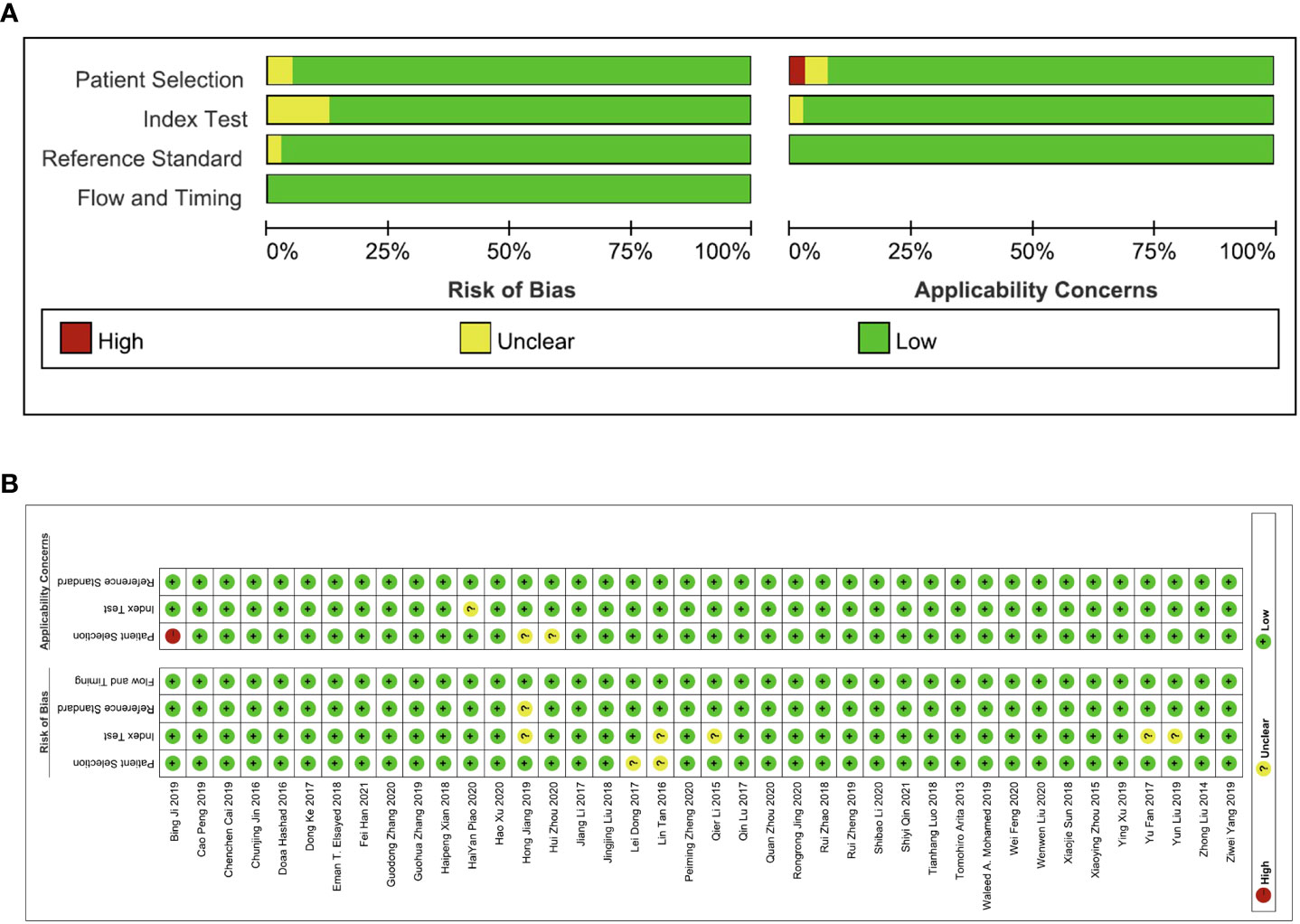
Figure 2 The quality assessment of the included studies via the QUADAS-2 tool. (A) Risk of bias and applicability concerns graph and (B) summary of quality assessment.
We added all included studies to Revman 5.4, and then according to the extracted data, related figures were plotted via STATA 16. There were 52 lncRNAs reported among 40 studies, and their corresponding diagnostic accuracies are shown in Table 2. Overall sensitivity, specificity, and AUC were 0.78 (95% CI: 0.75–0.81), 0.79 (95% CI: 0.74–0.83), and 0.85 (95% CI: 0.81–0.87), respectively, which signifies a great performance for lncRNAs as noninvasive biomarkers to distinguish GC patients. The pooled diagnostic odds ratio (DOR) was 13.00 (95% CI: 10.00–17.00). Meanwhile, the pooled positive likelihood ratio (PLR) and negative likelihood ratio (NLR) were 3.70 (95% CI: 3.00–4.50) and 0.28 (95% CI: 0.24–0.32), respectively.
Deeks’ funnel plot asymmetry test was used to evaluate the publication bias of the enrolled articles. The results demonstrated a low potential for publication bias (p = 0.00).
In clinical practice, there are various noninvasive circulation biomarkers applied when screening GC patients from a healthy population. Of note, invasive diagnostic methods are unable to forecast prognosis and monitor the progress of GC. Meanwhile, the discomfort caused by such invasive tests makes it difficult for patients to accept them, thus limiting their further applications. In addition, traditional biomarkers lack enough specificity and sensitivity to diagnose GC, making their diagnostic efficacies questionable (23). Therefore, developing appropriate noninvasive biomarkers that can be used to diagnose and predict the prognosis of GC patients is of paramount importance. With the prevalence of next-generation sequencing (NGS) technology, a large number of lncRNAs have attracted tremendous attention and have been the topic of extensive research. It was revealed that lncRNAs not only participate in the transduction of various signaling pathways and thus influence cancer development (24), but also have the potential for cancer diagnosis (25, 26).
In our meta-analysis, we included 40 original research studies including 6,772 participants to evaluate the diagnostic accuracies of lncRNAs for GC. The random-effect model was used in this meta-analysis due to the existence of heterogeneity. According to the AUC value, 5 lncRNAs with one panel of lncRNAs had a high diagnostic value, 30 lncRNAs had a moderate diagnostic value, and 4 lncRNAs had a low value. As shown in the forest plot (Figures 3A, B) and SROC curve (Figure 4), the overall sensitivity, specificity, and AUC were 0.78 (95% CI: 0.75–0.81), 0.79 (95% CI: 0.74–0.83), and 0.85 (95% CI: 0.81–0.87), respectively, which suggest that lncRNAs have a better diagnostic value than traditional tumor markers such as CEA and CA199 (27). Meanwhile, the PLR and NLR in our meta-analysis were 3.70 and 0.28, which implied that circulation lncRNAs had the ability to pick out GC patients from healthy people. As displayed in Figure 5, the results from Deeks’ funnel plot asymmetry test demonstrated a low potential for publication bias (p = 0.00). A meta-analysis enrolled 11 studies reported that circular RNAs had a high sensitivity (0.71) and specificity (0.78) as a tumor marker in the diagnosis of GC (28). Lin et al. conducted another meta-analysis to test the diagnostic potential of circRNAs in GC, and they found that the pooled sensitivity, specificity, and ROC were 0.68, 0.70, and 0.78, respectively (29). As for the microRNAs in diagnosing GC, a meta-analysis from Wei et al. revealed that circulating miRNAs also had the potential to be biomarkers in GC, which have a sensitivity of 0.76, a specificity of 0.81, and an AUC of 0.86 (30). Although the above results suggested that circRNAs and miRNAs had promising applications, we found that lncRNAs were better than them in diagnosing GC. However, the expression level of lncRNAs is a concerning issue in GC diagnosis. Depending on their role in tumor biology, not all lncRNAs are oncogenes. Some of them play a critical role in promoting tumor genesis and regulating tumor cellular properties, while others function as inhibiting factors in the development of tumors. For instance, upregulation of C5orf66-AS1 can decrease cellular activities including proliferation, migration, and invasion (31). By contrast, high expression of CCAT2 facilitates GC cell proliferation and invasion and implies poor prognosis (32). In our meta-analysis, there were 31 lncRNAs that were highly expressed and 9 lncRNAs that were downregulated in GC patients. Hence, choosing which lncRNA for early diagnosis is dependent on the actual situation and different tumors, especially when applying them as biomarkers in a clinical setting. Furthermore, more high-impact and large-scale studies are needed to illuminate the mechanism of abnormal lncRNA expression.
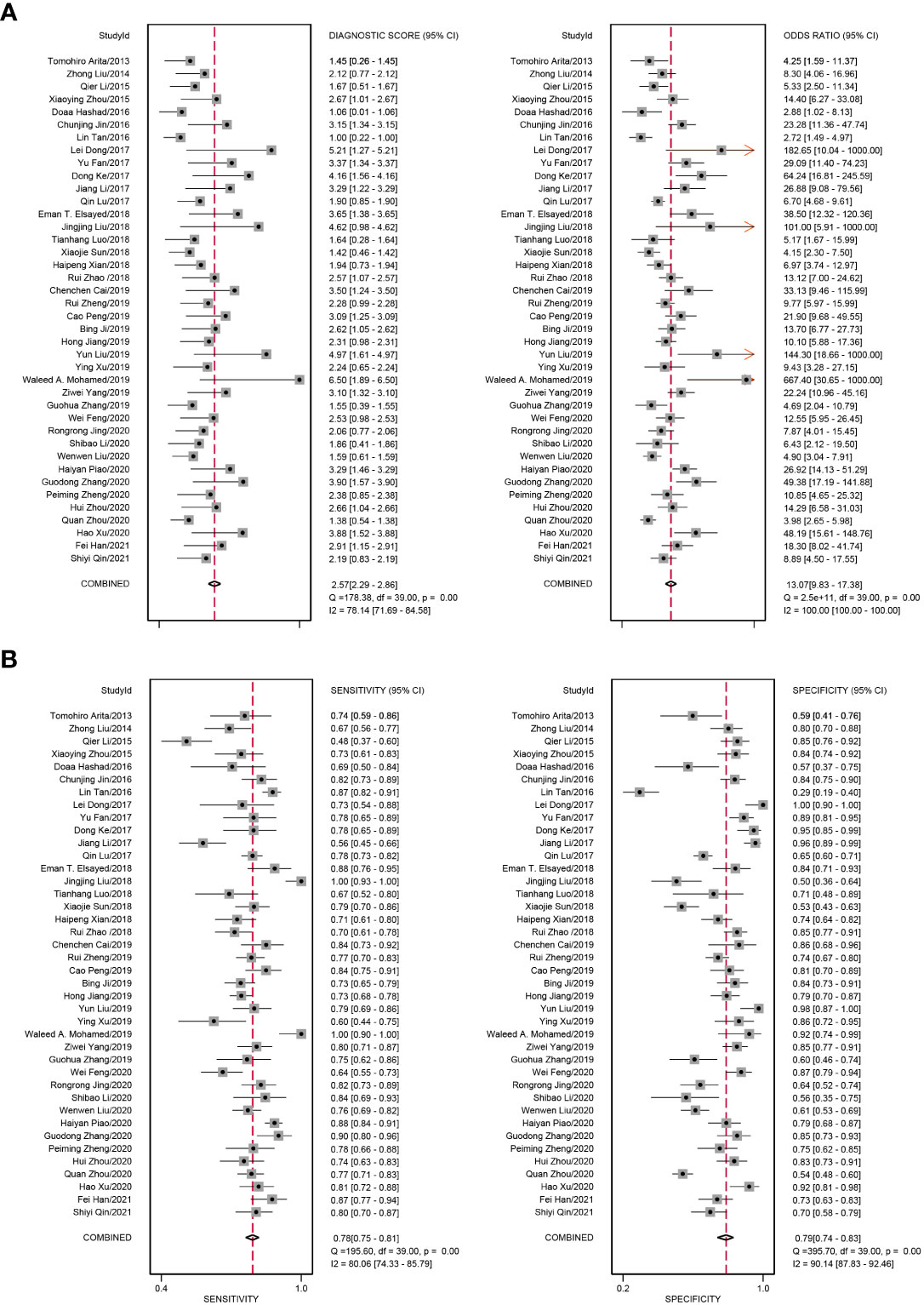
Figure 3 Forest plots of diagnostic accuracy of circulating lncRNAs in GC. (A) The pooled diagnostic score and diagnostic odds ratio (DOR) of circulating lncRNAs in the diagnosis of GC patients. (B) The pooled sensitivity and specificity of circulating lncRNAs in the diagnosis of GC patients.
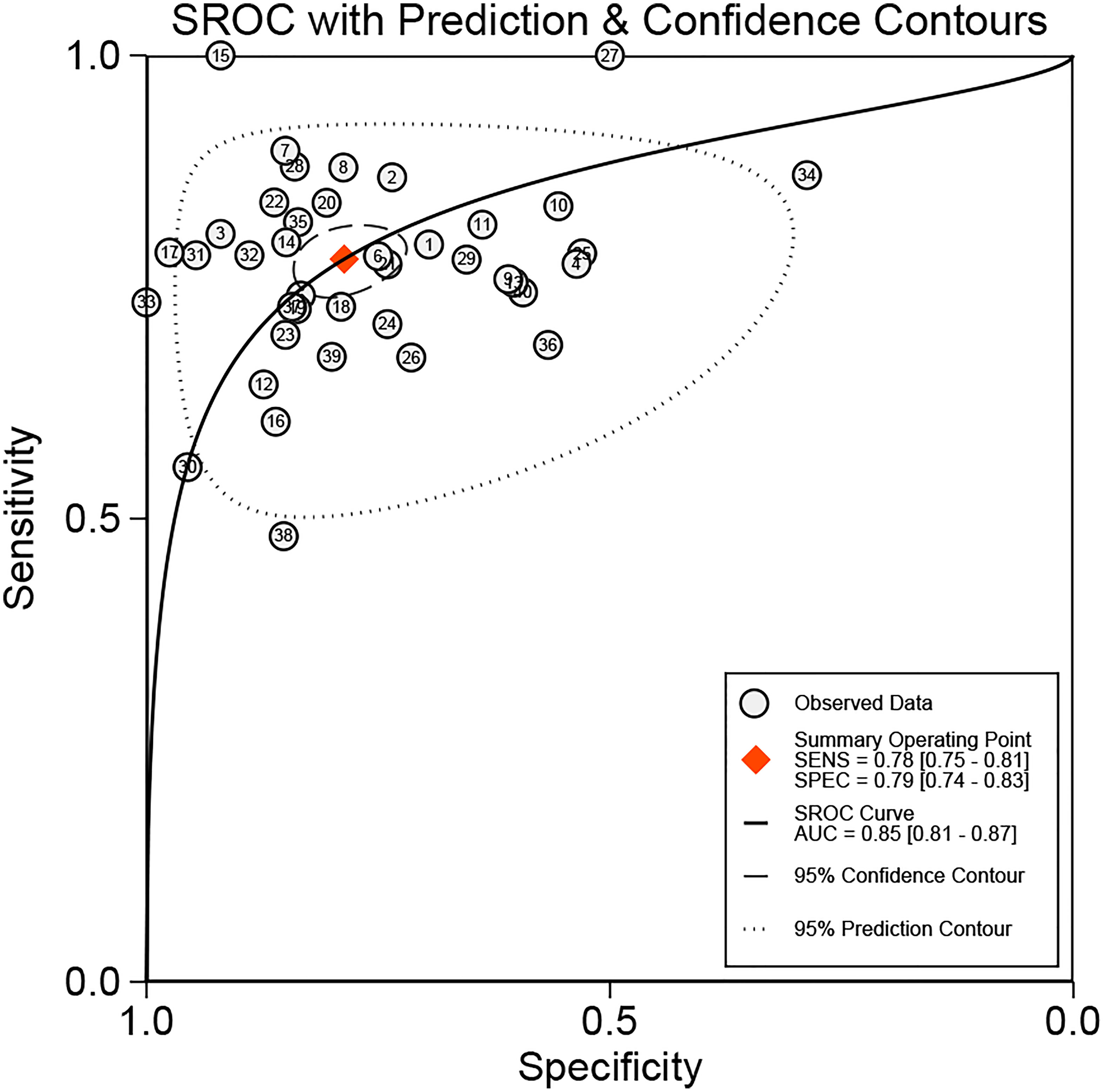
Figure 4 SROC of circulating lncRNAs in the diagnosis of GC patients. SROC, summary receiver operator characteristic curve; AUC, area under the curve.
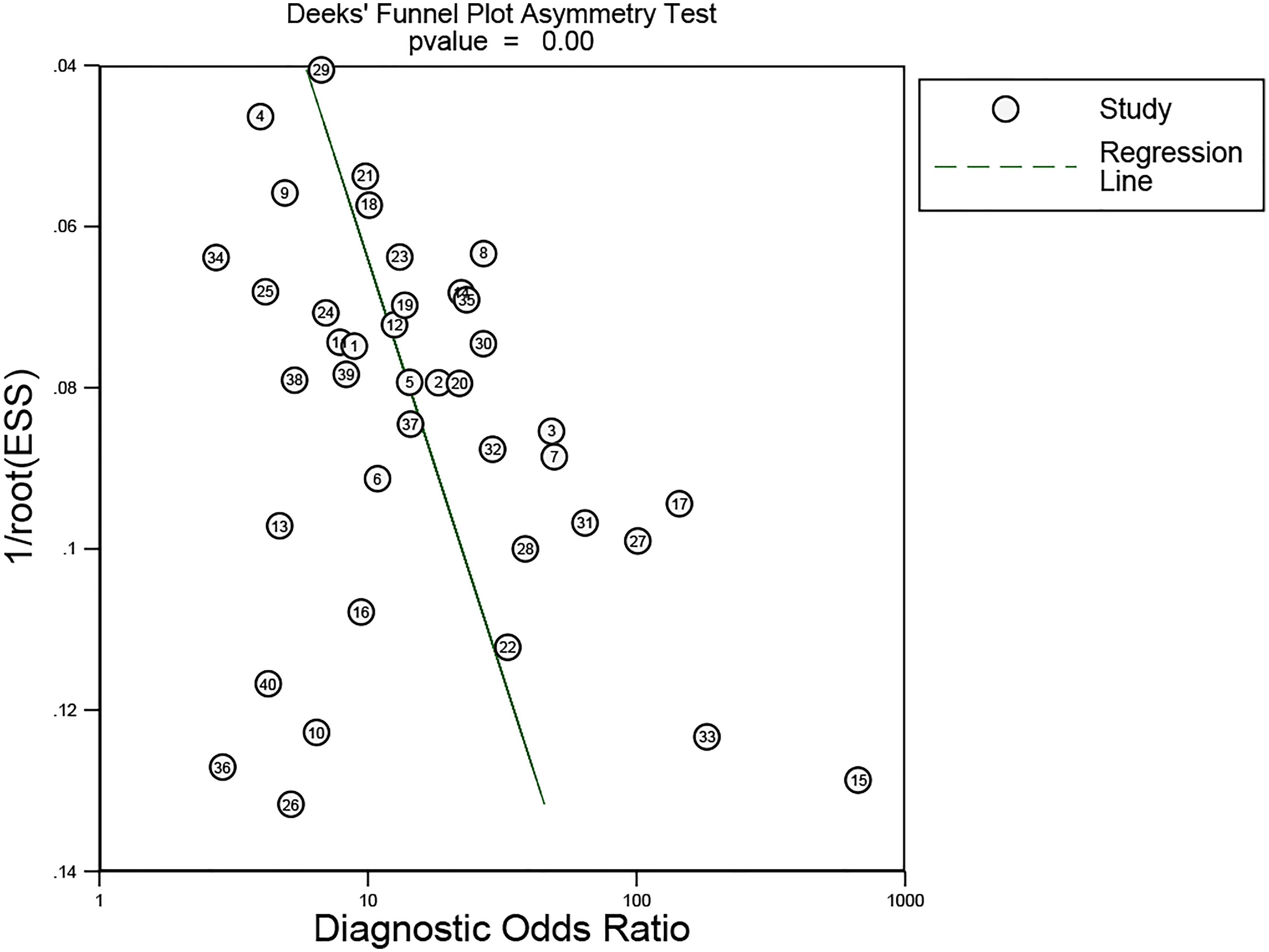
Figure 5 Deek’s funnel plot asymmetry test was used to estimate the publication bias for discrimination of circulating lncRNAs in GC patients.
The research on early GC diagnosis in China began in the 1970s. With the continuous development of medical technology and the efforts of medical workers, the detection rate of early GC in China has improved, but there is still a gap compared with Japan and South Korea, because these countries have the most comprehensive GC prevention and screening programs in the world, and their early GC detection rates have reached 50% and 70% (33), respectively. There are advantages and disadvantages in diagnosing GC with lncRNAs. Traditionally, gastroscopy together with biopsy is the main method in detecting stomach lesions. However, the early diagnosis rate depends on many factors including the endoscopists’ experience and standard operation, patient cooperation during the examination, and visual clarity using endoscopy. LncRNAs are acceptable for patients because of their invasiveness. Moreover, lncRNAs are abundant in the blood. Because of their stable properties (34) and higher sensitivity and specificity than CEA and CA199, they can replace old biomarkers and, thus, can be used as auxiliary biomarkers. This study further examined the diagnostic performance of lncRNAs in GC from the perspective of a noninvasive method, which would assist with the early diagnosis of GC. Compared with previous studies (20, 35), our study had several strengths in terms of study design and data analyses. First, we included more recent eligible articles using a comprehensive and updated search strategy, which improved the precision of the estimated effect size; second, we calculated the diagnostic efficacy in one specific cancer instead of pan-cancer, which could provide more accurate supporting information in GC diagnosis; third, we performed comprehensive analyses to explore the heterogeneity and diagnostic accuracy of circulating lncRNAs in GC. The results of this study indicate that circulating lncRNAs can be used as potential biomarkers for the diagnosis of GC. There are some limitations that should not be overlooked in the present meta-analysis. First, the number of studies included is relatively small, and more studies are needed before a solid conclusion can be drawn. Second, all included studies were case–control studies instead of randomized controlled trials, which may lead to some related biases. In order to acquire high-quality evidence, more randomized controlled trials are needed to avoid biases. Third, most of the included studies were from China and most of the included patients were Asian. This could further affect the generalization of the results, which could be attributed to ethnicity differences.
Collectively, our meta-analysis revealed that serum or plasma lncRNAs have high sensitivity and specificity, which makes them clinically feasible in diagnosing GC. We believe that peripheral blood lncRNAs may become novel noninvasive biomarkers in the foreseeable future. At the same time, it should be noted that a greater number of blood samples and more evidence from rigorous multicenter clinical studies are necessary to justify their applicability as cancer biomarkers.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Conceptualization: JL and LF. Methodology: JL, QX, YYZ, and YD. Formal analysis: JL and YQZ. Writing—original draft preparation: JL. Writing—review, and editing: JL and SB. Funding acquisition: LF. Resources: XZ. Supervision: LF and XZ. All authors contributed to the article and approved the submitted version.
This study was funded by the National Natural Science Foundation of China (Grant number: 8217100675) and Major Discipline Construction of Minhang District, Shanghai (Grant number: 2020MWDXK03) and Leading Talent Project of Minhang District, Shanghai (Grant number: 2021LJRC03).
All authors gratefully acknowledge the help of their supervisor Professor LF.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Bray F, Laversanne M, Weiderpass E, Soerjomataram I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer (2021) 127:3029–30. doi: 10.1002/cncr.33587
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-a Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
3. Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to helicobacter pylori. Int J Cancer (2015) 136:487–90. doi: 10.1002/ijc.28999
4. Anderson ML. Helicobacter pylori infection. Postgrad Med (1994) 96:40–50. doi: 10.1080/00325481.1994.11945920
5. Yao K, Uedo N, Kamada T, Hirasawa T, Nagahama T, Yoshinaga S, et al. Guidelines for endoscopic diagnosis of early gastric cancer. Dig Endosc (2020) 32:663–98. doi: 10.1111/den.13684
6. Schistosomes, liver flukes and helicobacter pylori. In: IARC working group on the evaluation of carcinogenic risks to humans, vol. 61. Lyon: IARC Monogr Eval Carcinog Risks Hum. (1994) p. 1–241.
7. Song Y, Ye M, Zhou J, Wang Z, Zhu X. Targeting e-cadherin expression with small molecules for digestive cancer treatment. Am J Transl Res (2019) 11:3932–44.
8. Lurje G, Schiesser M, Claudius A, Schneider PM. Circulating tumor cells in gastrointestinal malignancies: Current techniques and clinical implications. J Oncol (2010) 2010:392652. doi: 10.1155/2010/392652
9. Kunz PL, Gubens M, Fisher GA, Ford JM, Lichtensztajn DY, Clarke CA. Long-term survivors of gastric cancer: A California population-based study. J Clin Oncol (2012) 30:3507–15. doi: 10.1200/JCO.2011.35.8028
10. Yang AP, Liu J, Lei HY, Zhang QW, Zhao L, Yang GH. CA72-4 combined with CEA, CA125 and CAl9-9 improves the sensitivity for the early diagnosis of gastric cancer. Clin Chim Acta (2014) 437:183–6. doi: 10.1016/j.cca.2014.07.034
11. Matsuoka T, Yashiro M. Biomarkers of gastric cancer: Current topics and future perspective. World J Gastroenterol (2018) 24:2818–32. doi: 10.3748/wjg.v24.i26.2818
12. Hsu PI, Chen CH, Hsiao M, Wu DC, Lin CY, Lai KH, et al. Diagnosis of gastric malignancy using gastric juice alpha 1-antitrypsin. Cancer Epidemiol Biomarkers Prev (2010) 19:405–11. doi: 10.1158/1055-9965.EPI-09-0609
13. Thomas DS, Fourkala EO, Apostolidou S, Gunu R, Ryan A, Jacobs I, et al. Evaluation of serum CEA, CYFRA21-1 and CA125 for the early detection of colorectal cancer using longitudinal preclinical samples. Br J Cancer (2015) 113:268–74. doi: 10.1038/bjc.2015.202
14. Huang YX, Zhang J, Hou LD, Wang G, Liu H, Zhang R, et al. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway (vol 36, 194, 2017). J Exp Clin Cancer Res (2020) 39:194–208.
15. Wei ZJ, Chen L, Meng L, Han WX, Huang L, Xu AM. LncRNA HOTAIR promotes the growth and metastasis of gastric cancer by sponging miR-1277-5p and upregulating COL5A1. Gastric Cancer (2020) 23:1018–32. doi: 10.1007/s10120-020-01091-3
16. Zhong Y, Zhao M, Yu Y, Li Q, Wang F, Wu P, et al. Prognostic value and therapeutic potential of the long noncoding RNA TP73-AS1 in cancers: A systematic review and meta-analysis. Sci Rep (2020) 10:9053. doi: 10.1038/s41598-020-65726-2
17. Tian SB, Liu JL, Kong S, Peng LP. LncRNA DLX6-AS1 as a potential molecular biomarker in the clinicopathology and prognosis of various cancers: A meta-analysis. Bioscience Rep (2020) 40:1–9. doi: 10.1042/BSR20193532
18. Jin T. LncRNA DRAIR is a novel prognostic and diagnostic biomarker for gastric cancer. Mamm Genome (2021) 32:503–7. doi: 10.1007/s00335-021-09911-2
19. Chen X, Sun X, Li X, Xu L, Yu WY. LncRNA-HEIH is a novel diagnostic and predictive biomarker in gastric cancer. Genet Testing Mol Biomarkers (2021) 25:284–92. doi: 10.1089/gtmb.2020.0270
20. Cao F, Hu Y, Chen Z, Han W, Lu W, Xu J, et al. Circulating long noncoding RNAs as potential biomarkers for stomach cancer: A systematic review and meta-analysis. World J Surg Oncol (2021) 19:89. doi: 10.1186/s12957-021-02194-6
21. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med (2011) 155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009
22. Jones CM, Athanasiou T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. Ann Thorac Surg (2005) 79:16–20. doi: 10.1016/j.athoracsur.2004.09.040
23. Ye DM, Xu G, Ma W, Li Y, Luo W, Xiao Y, et al. Significant function and research progress of biomarkers in gastric cancer. Oncol Lett (2020) 19:17–29.
24. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene (2017) 36:5661–7. doi: 10.1038/onc.2017.184
25. Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: A potential novel class of cancer biomarkers. Front Genet (2015) 6:145. doi: 10.3389/fgene.2015.00145
26. Maass PG, Luft FC, Bahring S. Long non-coding RNA in health and disease. J Mol Med (Berl) (2014) 92:337–46. doi: 10.1007/s00109-014-1131-8
27. Feng F, Tian YZ, Xu GH, Liu Z, Liu SS, Zheng GZ, et al. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer (2017) 17:737–42. doi: 10.1186/s12885-017-3738-y
28. Jiang F, Hong FL, Shah MW, Shen XB. Circular RNAs as diagnostic biomarkers in gastric cancer: A meta-analysis review. Pathol Res Practice (2019) 215:152419–26. doi: 10.1016/j.prp.2019.04.011
29. Lin YF, Luo XL, Cui ZL, Chen Y. Diagnostic potential for circular RNAs in gastric carcinoma: A meta-analysis. Clin Laboratory (2019) 65:343–51. doi: 10.7754/Clin.Lab.2018.180810
30. Wei H, Pu K, Liu XG, Li BX, Zhang HS, Wang H, et al. The diagnostic value of circulating microRNAs as a biomarker for gastric cancer: A meta-analysis. Oncol Rep (2019) 41:87–102.
31. Zhou Q, Li H, Jing J, Yuan Y, Sun L. Evaluation of C5orf66-AS1 as a potential biomarker for predicting early gastric cancer and its role in gastric carcinogenesis. Oncotargets Ther (2020) 13:2795–805. doi: 10.2147/OTT.S239965
32. Wu SW, Hao YP, Qiu JH, Zhang DB, Yu CG, Li WH. High expression of long non-coding RNA CCAT2 indicates poor prognosis of gastric cancer and promotes cell proliferation and invasion. Minerva Med (2017) 108:317–23. doi: 10.23736/S0026-4806.17.04703-6
33. Ren W, Yu J, Zhang ZM, Song YK, Li YH, Wang L. Missed diagnosis of early gastric cancer or high-grade intraepithelial neoplasia. World J Gastroenterol (2013) 19:2092–6. doi: 10.3748/wjg.v19.i13.2092
34. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res (2017) 77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634
Keywords: gastric cancer, lncRNA, diagnosis, systematic review, meta - analysis
Citation: Li J, Zhang Y, Xu Q, Zhang Y, Bei S, Ding Y, Zhang X and Feng L (2022) Diagnostic value of circulating lncRNAs for gastric cancer: A systematic review and meta-analysis. Front. Oncol. 12:1058028. doi: 10.3389/fonc.2022.1058028
Received: 30 September 2022; Accepted: 08 November 2022;
Published: 06 December 2022.
Edited by:
Emilio Francesco Giunta, Università degli Studi della Campania Luigi Vanvitelli, ItalyReviewed by:
Xiaoqi Yang, Huazhong University of Science and Technology, ChinaCopyright © 2022 Li, Zhang, Xu, Zhang, Bei, Ding, Zhang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohong Zhang, NzYwNDU0ODA2QHFxLmNvbQ==; Li Feng, ZmVuZ2xpX21oQDE2My5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.