- 1Department of Oncology, Wuhan Third Hospital, Wuhan, Hubei, China
- 2North Sichuan Medical College, Nanchong, Sichuan, China
- 3Department of Thoracic Surgery, Affiliated Hospital of North Sichuan Medical College, Nanchong, Sichuan, China
- 4Suzhou Kowloon Hospital, Shanghai Jiao Tong University School of Medicine, Suzhou, Jiangsu, China
- 5Department of Oncology, Linfen People’s Hospital, Linfen, Shanxi, China
- 6Tianjin Key Laboratory of Medical Epigenetics, Key Laboratory of Breast Cancer Prevention and Therapy (Ministry of Education), Department of Biochemistry and Molecular Biology, Tianjin Medical University, Tianjin, China
- 7Department of Oncology, Affiliated Hospital of North Sichuan Medical College, Nanchong, Sichuan, China
Recent discoveries have suggested that the F-actin capping protein α1 subunit (CAPZA1) in various human tumors could play a significantly important role in regulating cell proliferation, metastasis, and epithelial–mesenchymal transition. However, the immune-regulating role of CAPZA1 in the initiation and development of lung adenocarcinoma (LUAD) remains unclear. In our research, we first found that CAPZA1 serves as an oncogene in pan-cancers from the TCGA data and higher CAPZA1 expression process unfavorably prognostic value in LUAD based on starBase database, PrognoScan, and LOGpc database. Then, in our analyses, lncRNAs AC026356.1 in LUAD acted as a competitive endogenous RNA (ceRNA) of miR-30d-5p, which might be the possible regulatory miRNA of CAPZA1 based on the starBase database. Finally, we confirmed that CAPZA1 expression had a tightly positive correlation with immune infiltration cells, immune infiltration markers, TMB, MSI, immune score, stromal score, and immune checkpoints, indicating that CAPZA1 was a markedly reliable therapeutic target for immunological antitumor strategies. In conclusion, our investigations revealed that CAPZA1 might function as an immune-associated biomarker in the development and treatment of LUAD, thereby acting as a promising prognostic and therapeutic target against LUAD.
1 Introduction
Lung cancer (LC) has gradually become a leading cause of cancer-related morbidity and mortality worldwide among men and women (1, 2). Lung adenocarcinoma (LUAD) accounts for over 40% of lung cancer, with the most common subtype being lung cancer (3). Despite advances in therapeutic strategies such as surgical techniques, radiotherapy, and chemotherapy, the survival of patients with LUAD is still unfavorable (4). Immunotherapy has been a promising and effective treatment strategy for multiple cancers (5–7), including LUAD (8). However, there are some limitations regarding immune checkpoint inhibitors in this study: Over half of the LUAD patients are not markedly sensitive to PD-1/PD-L1 antitumor immunotherapy, indicating the existence of additional immunotherapeutic biomarkers in the LUAD tumor microenvironment (9). Therefore, we propose identifying immune-associated prognostic biomarkers to conquer LUAD for timely detection, early diagnosis, prognosis, and treatment.
F-actin capping protein α1 subunit (CAPZA1) belongs to the capping protein (CP), which is a family of the actin filament (F-actin) that is a critical component of the cytoskeleton, and the dynamic remodeling of these filaments provides the impetus for cell invasion and migration (10–13). Recent discoveries have suggested that CAPZA1 in various human tumors could regulate cell proliferation, metastasis, and epithelial–mesenchymal transition (14, 15), and elevated CAPZA1 is closely linked to an unfavorable prognosis in gastric cancer (16). For instance, PIM1 phosphorylated CAPZA1 and was responsible for increasing prostate cancer adhesion and migration in vitro (17). Additionally, during the immune process, the actin cytoskeleton plays a pivotal role in controlling B-cell antigen uptake, polarization, presentation, T-lymphocyte activation, and effector functions in the tumor microenvironment (TEM) (18, 19). To the best of our knowledge, the immune-regulating role of CAPZA1 in LUAD development remains incompletely understood.
This current study investigated the expression profiling and prognostic value of CAPZA1 in pan-cancers based on several databases. Then, ncRNAs, containing miRNAs and lncRNAs, participated in the regulation of CAPZA1 in LUAD. High CAPZA1 levels in LUAD were also associated with immune cell infiltration and immune checkpoints, suggesting that ncRNAs-mediated upregulation of CAPZA1 is an immune-related biomarker with a worse impact on clinical outcomes.
2 Material and methods
2.1 Data collection and processing
The mRNA expression data and somatic mutation data of the LUAD in The Cancer Genome Atlas (TCGA) database were acquired using UCSC Xena (https://xena.ucsc.edu/) (20). MSI data was downloaded from the research of Russell Bonneville (21). The flow chart is shown in Supplementary Figure 1.
2.2 StarBase online database
The starBase database (http://starbase.sysu.edu.cn/) (22) was employed to identify differentially expressed CAPZA1 and its prognostic value in many cancers. The expression of CAPZA1 is presented by FPKM (fragments per kilobase of transcript sequence per million base pairs sequenced). Firstly, the starBase database involving PITA, RNA22, miRmap, microT, miRanda, PicTar, and TargetScan was employed to predict potential miRNAs that directly bond to CAPZA1 upstream in LUAD. This was done to speculate about their differential expression and OS. Next, we predicted 16 miRNAs, of which six were negatively correlated with CAPZA1 expression, so we chose the one least correlated, mirRNA-30d-5p. Then, we forecast potential hsa-miR-30d-5p-modulated lncRNAs in LUAD and analyzed the differential expression level and prognosis of the miRNAs screened. There were two lncRNAs that met the requirements for screening eight lncRNAs. Finally, the starBase database was also used to examine the miR-30d-5p-CAPZA1, miR-30d-5p-AC026356.1, and AC026356.1-CAPZA1 interactions. P <0.05 was treated as the significance threshold for all statistical analyses.
2.3 Survival analysis
The CAPZA1 survival analysis of CAPZA1 expression in LUAD was validated through the PrognoScan database (http://www.abren.net/PrognoScan/) (23) and LOGpc (Long-term Outcome and Gene Expression Profiling Database of pan-cancers, https://bioinfo.henu.edu.cn/DatabaseList.jsp). A corrected P-value <0.05 was regarded as a statistically significant difference.
2.4 Immune cell infiltration and immune checkpoint analysis
Additionally, we applied the TIMER online database (https://cistrome.shinyapps.io/timer/) (24), a comprehensive resource for systematic analysis of tumor immune infiltrates, immune infiltrate biomarkers, and immune systems checkpoint across LUAD into CAPZA1-associated the abundance of the immune response. The immunophenoscore (IPS), which can be obtained from the TCIA website (https://tcia.at/home), has been verified to predict patients’ response to immune checkpoint inhibitors (ICIs). A higher IPS has a better outcome with ICI treatment (25).
2.5 Correlation between CAPZA1 and immune characteristics
Recent studies have shown that tumor mutational burden (TMB) predicts the response to the clinical efficacy of anti-PD1/PD-L1 treatment (26). Microsatellite instability (MSI) (27) has also been considered an indispensable biomarker for TME. To further evaluate CAPZA1 in LUAD immunity, we examined the link between CAPZA1 expression and MSI and TMB. Finally, ImmuneScore and StromalScore analyses were carried out via the “estimate” R package.
2.6 Gene set enrichment analysis
GSEA (28) (version 3.0, http://www.broadinstitute.org/gsea/index.jsp) has been utilized for evaluating gene sets or biological pathways, which varied significantly between CAPZA1 in high- and low-expression level groups. In this study, the following settings were used: 500 was the maximum size of the gene set, while 15 was the minimum size of the gene set, with 1,000 as the number of permutations, along with improved gene sets with an FDR (false discovery rate) q-val <0.05 having been measured as significant.
3 Results
3.1 CAPZA1 mRNA expression in pan-cancers
CAPZA1 mRNA levels from 33 cancer types in the TCGA were applied to TIMER. The results indicated that the expression of CAPZA1 was relatively high in 13 cancers, such as BLCA, BRCA, CHOL, ESCA, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, STAD, THCA, and UCEC, while the deletion of CAPZA1 was in KICH and SKCM (Figure 1A). We further utilized the starBase database to verify whether CAPZA1 expression is overexpressed in multiple tumor tissues. Figures 1B–N showed that CAPZA1 mRNA expression in the 13 types were still upregulated.

Figure 1 Expression analysis for CAPZA1 in multiple cancers. (A) The expression of CAPZA1 in 33 types of human cancer based on the TCGA cancer and normal data in TIMER. (B–N) CAPZA1 expression in the starBase BLCA. (B), BRCA (C), CHOL (D), ESCA (E), HNSC (F), KIRC (G), KIRP (H), LIHC (I), LUAD (J), LUSC (K), STAD (L), THCA (M), and UCEC (N) tissues compared with corresponding TCGA normal tissues. *p-value < 0.05; ***p-value < 0.001.
3.2 Prognostic potential of CAPZA1 in pan-cancers
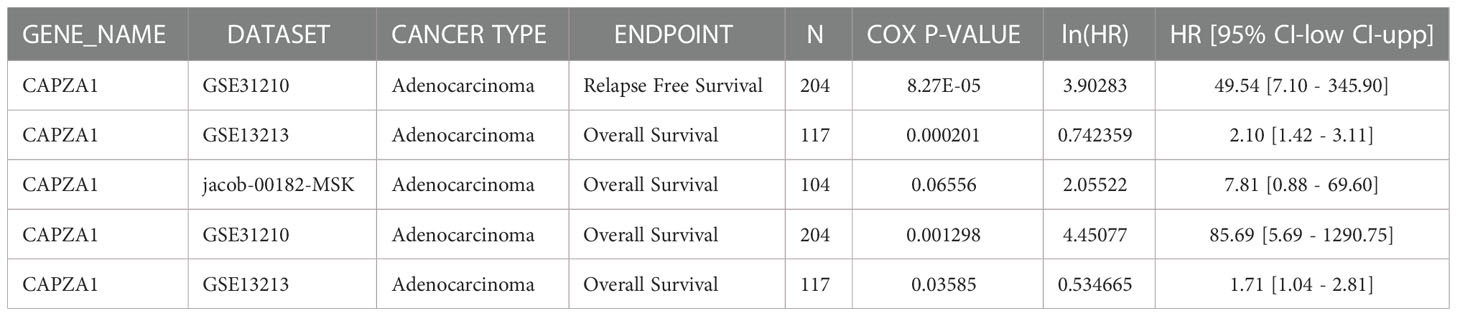
Firstly, the starBase database was employed to explore the prognosis of CAPZA1 in pan-cancer data downloaded from the TCGA. The result indicated that CAPZA1 at high expression afforded worse OS in KIRC, KIRP, LIHC, and LUAD (Figures 2A–M). Then, the fact that patients with high CAPZA1 expression had an unfavorable long-term prognosis in LUAD was also validated in PrognoScan (Table 1) and the LOGpc database (Figures 3A–F). These findings indicated that overexpressed CAPZA1 might be a worse prognostic marker in LUAD.
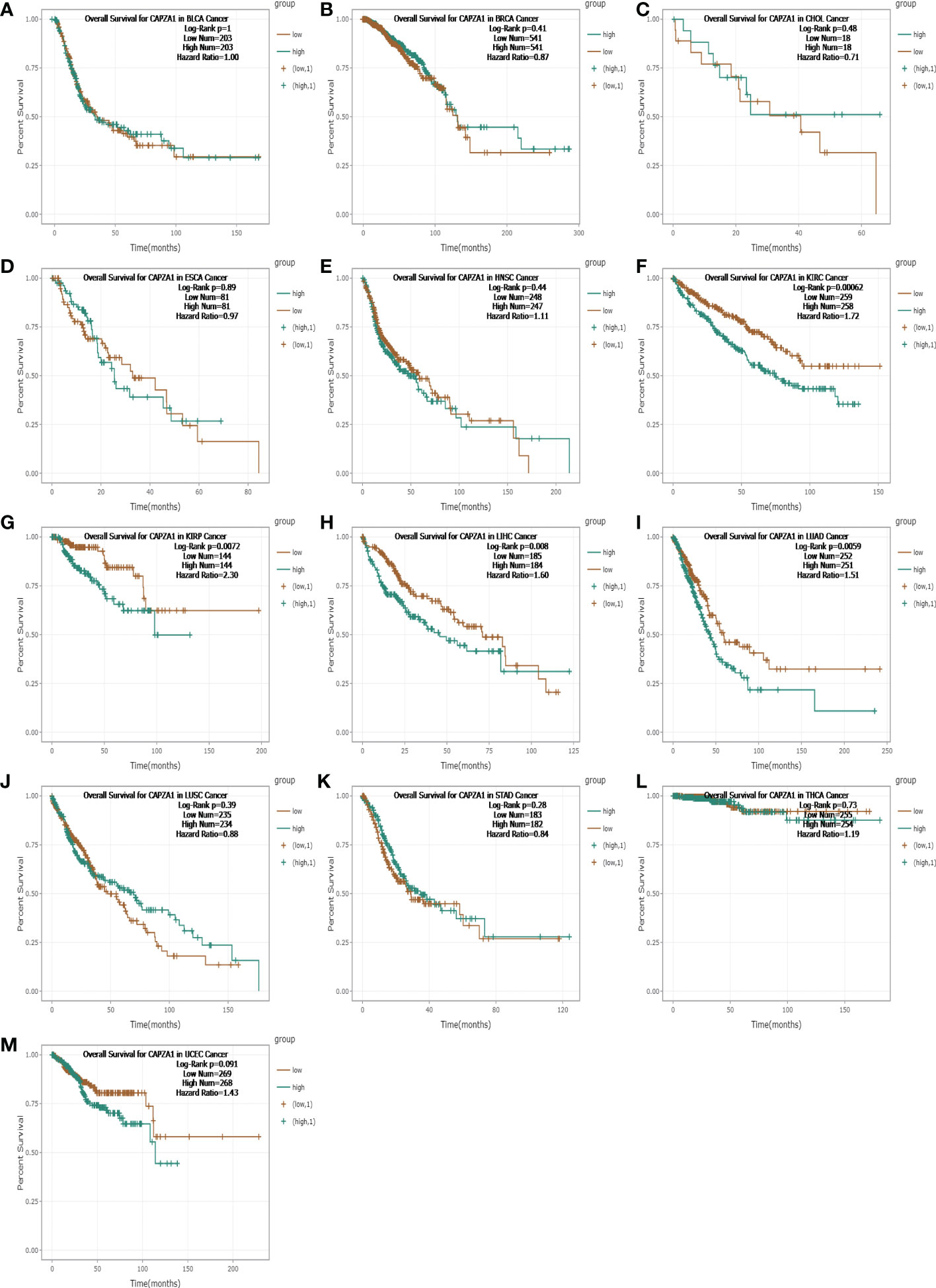
Figure 2 The overall survival (OS) analysis for CAPZA1 in various human cancers determined by starBase. (A–M) The OS plot of CAPZA1 in BLCA. (A), BRCA (B), CHOL (C), ESCA (D), HNSC (E), KIRC (F), KIRP (G), LIHC (H), LUAD (I), LUSC (J), STAD (K), THCA (L), and UCEC (K).
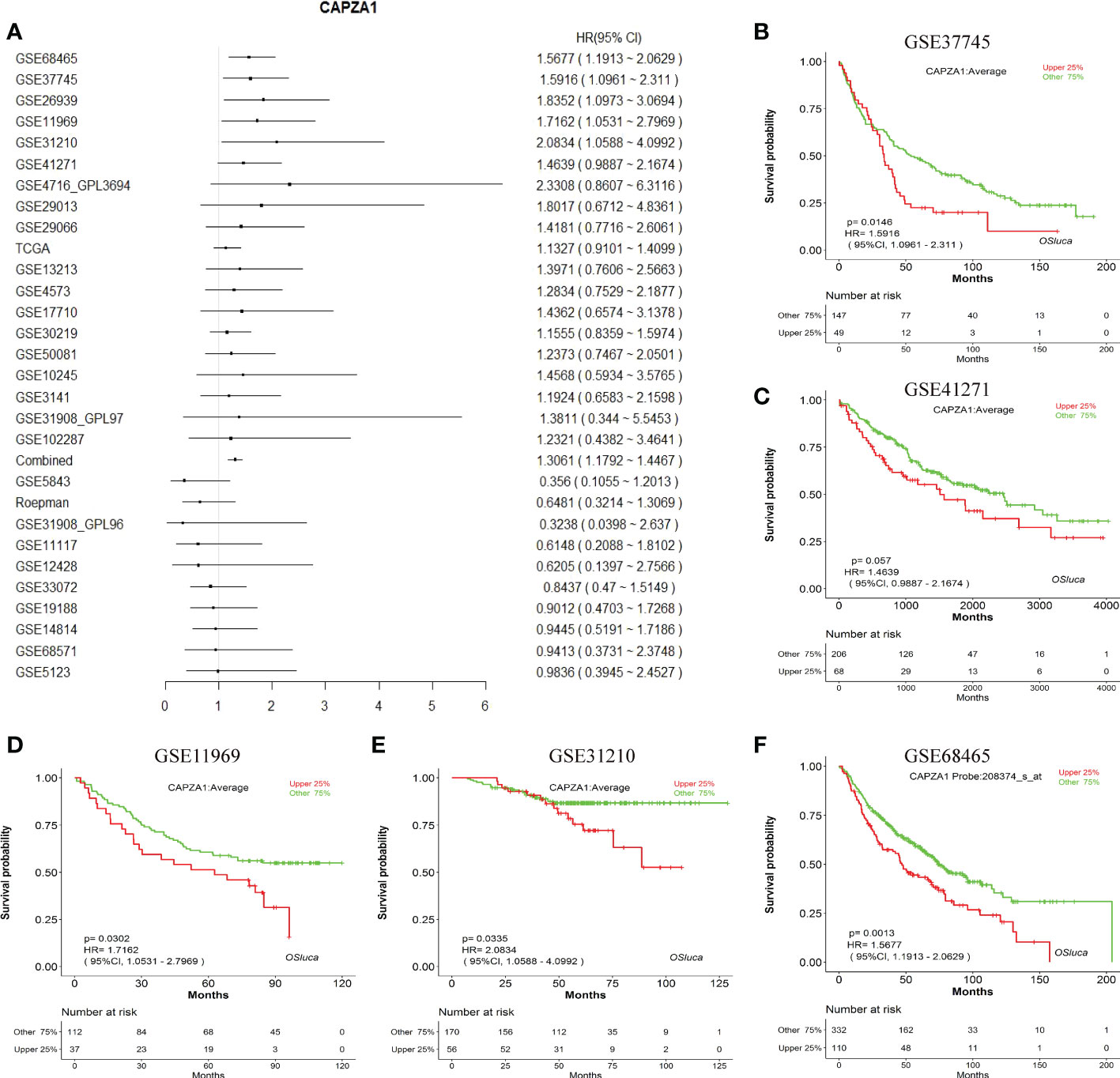
Figure 3 The prognostic value of CAPZA1. (A) Prognostic value of CAPZA1 in multiple data sets. (B–F) The representative survival curves.
3.3 Prediction and analysis of upstream miRNAs of CAPZA1
Noncoding RNAs (ncRNAs) are derived through the genome that fail to translate to proteins but regulate gene expression in cancers, including miRNAs and lncRNAs (29, 30). Thus, we aim to analyze the role of ncRNAs in regulating CAPZA1. Initially, we found 16 potential miRNAs that directly bond to CAPZA1 as its upstream (Figures 4A, B; Supplementary Table 1). As is well known, miRNAs could negatively modulate the expression of target genes by causing their cleavage or translational repression (31). As expected, CAPZA1 reversely correlated with hsa-miR-30a-5p, hsa-miR-30c-5p, hsa-miR-27b-3p, hsa-miR-488-3p, and hsa-miR-30d-5p in LUAD because of correlation analysis (Figure 4B, P <0.05). We then examined the expression and prognosis of the above miRNAs and found that only the hsa-miR-30d-5 in LUAD was at a low level, and its downregulation harbored a better life for patients (Figures 4C–E, P <0.05). These findings reveal that CAPZA1 might be the most potentially targeted mRNA of hsa-miR-30d-5 in LUAD.
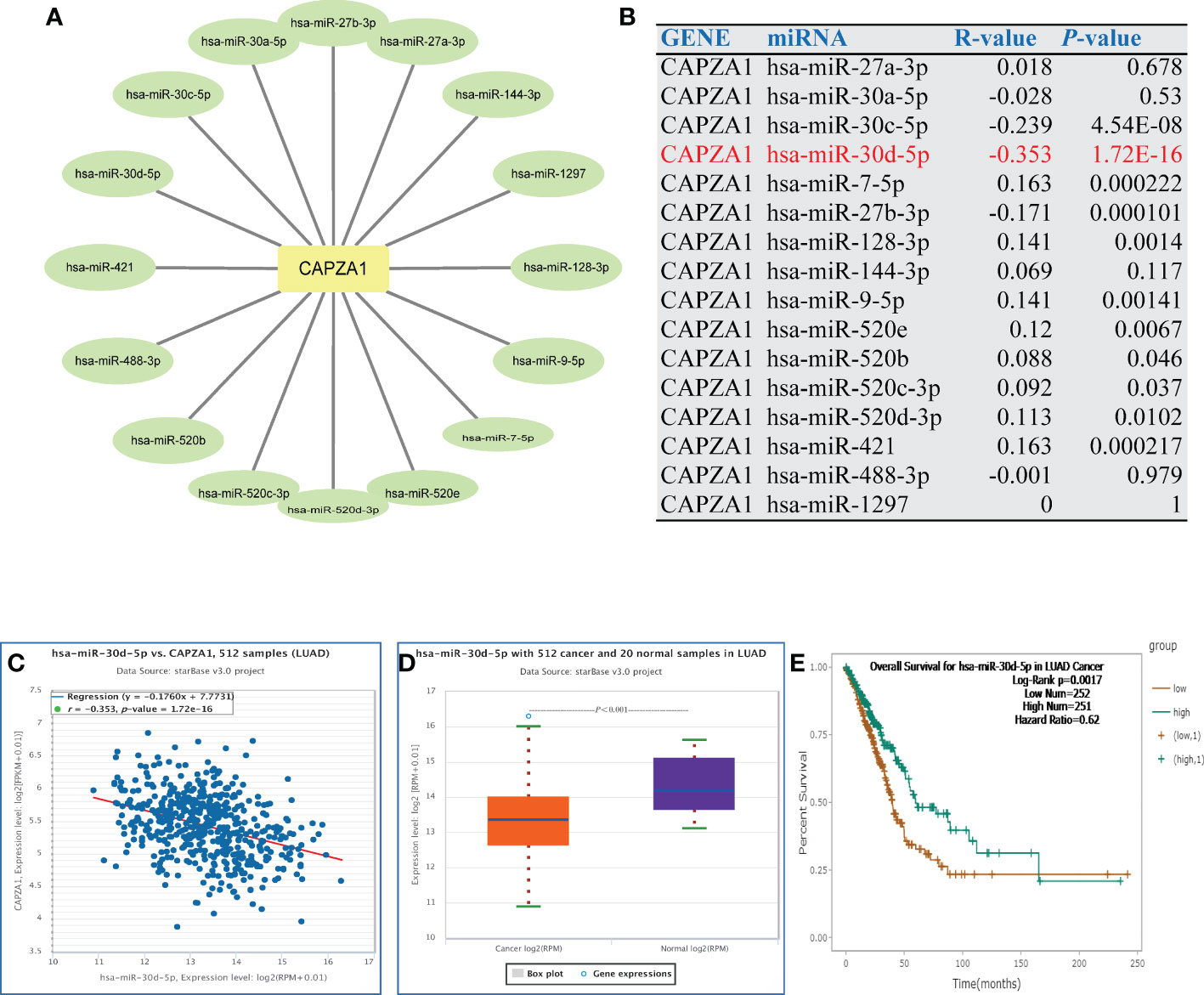
Figure 4 Identification of miR-30d-5p as a potential upstream miRNA of CAPZA1 in LUAD. (A) The miRNA- CAPZA1 regulatory network established by Cytoscape software. (B) The expression correlation between predicted miRNAs and CAPZA1 in LUAD as analyzed by the starBase database. (C) The expression correlation between miR-30d-5p and CAPZA1 in LUAD as analyzed by the starBase database. (D) The expression of miR-30d-5p in LUAD and control normal samples determined by the starBase database. (E) The prognostic value of let-7c-5p in LUAD assessed by the starBase database.
3.4 Prediction and analysis of upstream lncRNAs of hsa-miR-30d-5p
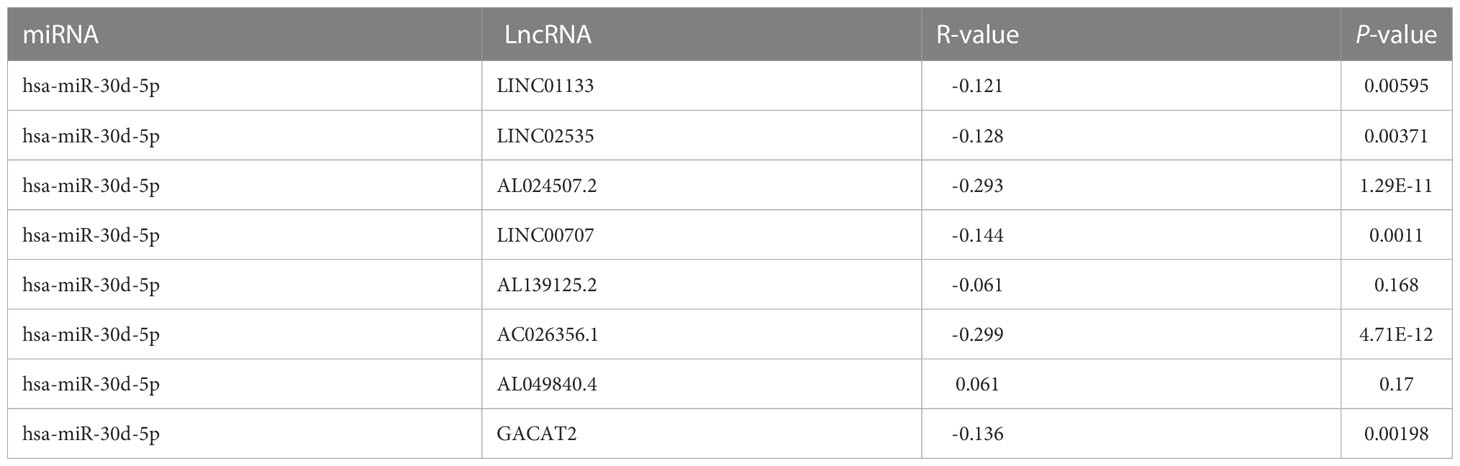
Subsequently, we forecasted potential upstream lncRNAs of hsa-miR-30d-5p in LUAD through the starBase database and observed nine possible lncRNAs (Supplementary Table 2; Supplementary Figure 2). Based on the competitive endogenous RNA (ceRNA) hypothesis, LncRNA sponges various miRNAs to inhibit their expression and reduce the regulatory effect on target mRNA (32). Consequently, only AC026356.1 in LUAD showed higher activity than normal samples, and higher AC026356.1 was reciprocally linked to favorable patients’ OS among all the nine lncRNAs (Figures 5A–P). AC026356.1 lncRNA was positively relevant to CAPZA1 expression whereas negative with hsa-miR-30d-5p using the starBase database (Tables 2, 3). To this end, AC026356.1 in LUAD might be most likely to interact with upstream lncRNAs of the hsa-miR-30d-5p-CAPZA1 axis (Figure 6).
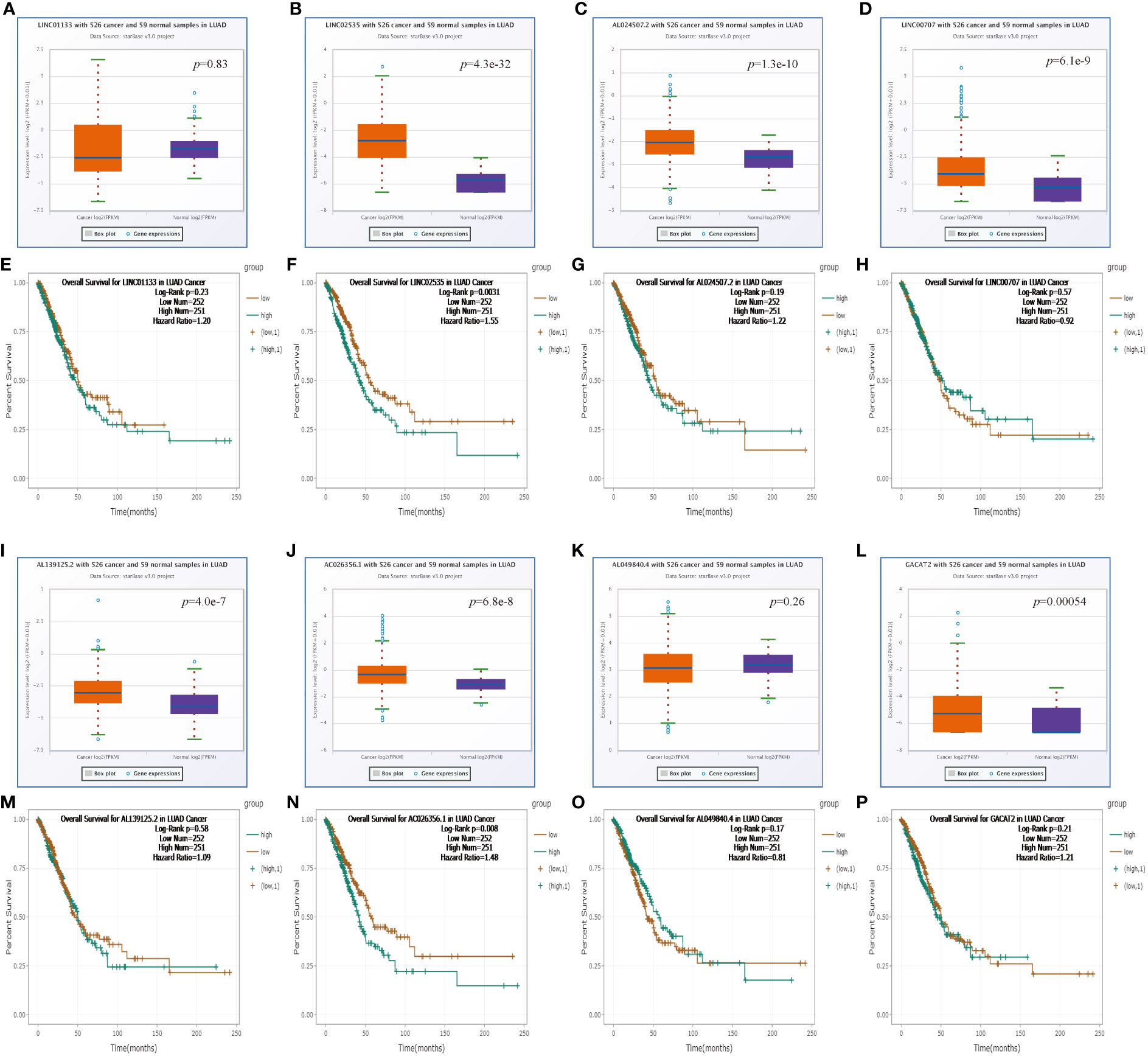
Figure 5 Expression analysis and survival analysis for upstream lncRNAs of miR-30d-5p in LUAD (A–D, I–L). The expression of LINC01133 (A), LINC02535 (B), AL024507.2 (C), LINC00707 (D), AL139125.2 (I), AC026356.1 (J), AL049840.4 (K), and GACAT2 (L) in the starBase database. (E–H, M–P) The OS analysis for LINC01133 (E), LINC02535 (F), AL024507.2 (G), LINC00707 (H), AL139125.2 (M), AC026356.1 (N), AL049840.4 (O), and GACAT2 (P) in LUAD.
3.5 CAPZA1-associated immune infiltration assessment in LUAD
TME harbors comprehensively infiltrating immune cells, which contribute to influencing the development of cancers, so we investigated the relationship between CAPZA1 expression and immune infiltration. As presented in Figure 7A, the CAPZA1 expression level in LUAD was statistically positively related to macrophages, CD4+ T cells, CD8+ T cells, dendritic cells, and neutrophils, whereas the reciprocal association was with B cells and purity. Moreover, there was a significant change in immune cell infiltration under different copy numbers of CAPZA1 in LUAD (Figure 7B).
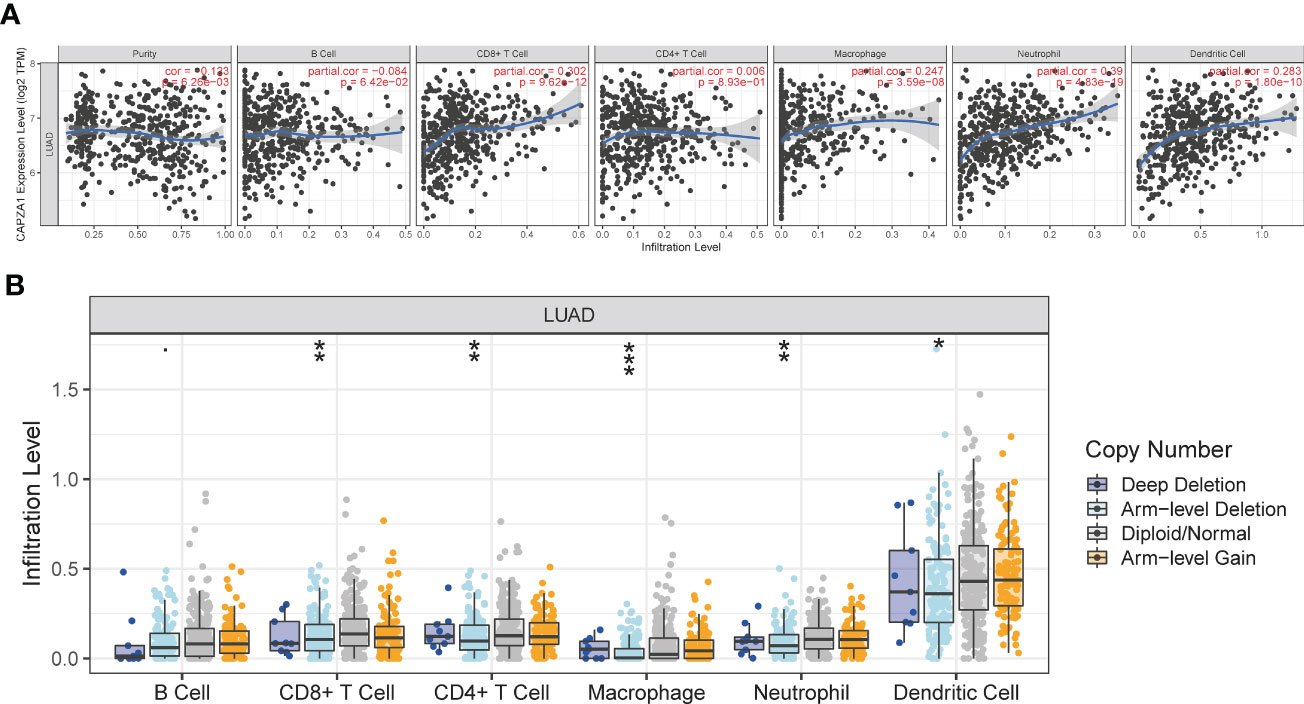
Figure 7 The relationship of immune cell infiltration with CAPZA1 level in LUAD. (A) The correlation of CAPZA1 expression level with B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, or dendritic cell infiltration level in LUAD. (B) The infiltration level of various immune cells under different copy numbers of CAPZA1 in LUAD. *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001.
3.6 The link between CAPZA1 and immune checkpoints, IPS in LUAD
PD1/PD-L1 and CTLA-4 are well-known immune checkpoints that play instrumental roles in cancer treatment modalities due to immune surveillance and escape. To ascertain the part of the immune response of CAPZA1 in LUAD with PD1, PD-L1, or CTLA-4, we utilized TIMER data to analyze the link between CAPZA1 and immune checkpoints. Indeed, high CAPZA1 and AC026356.1 expression was significantly positively correlated with PD1, PD-L1, and CTLA-4 in LUAD, respectively (Figures 8A–F), demonstrating that CAPZA1 can mediate tumorigenesis and progression by regulating tumor immune escape in LUAD. The IPS values (ips_ctla4_neg_pd1_pos, ips_ctla4_pos_pd1_neg, ips_ctla4_pos_pd1_neg, and ips_ctla4_pos_pd1_pos) have been reported in predicting cancer patients’ responses to anti-CTLA-4 treatment. Here, we noted a marked rise in scores in the low-CAPZA1 group (Figures 9A–D), implying that the low-CAPZA1 group is more suitable for anti-CTLA-4 and anti-PD-1/PD-L1 immunotherapy.
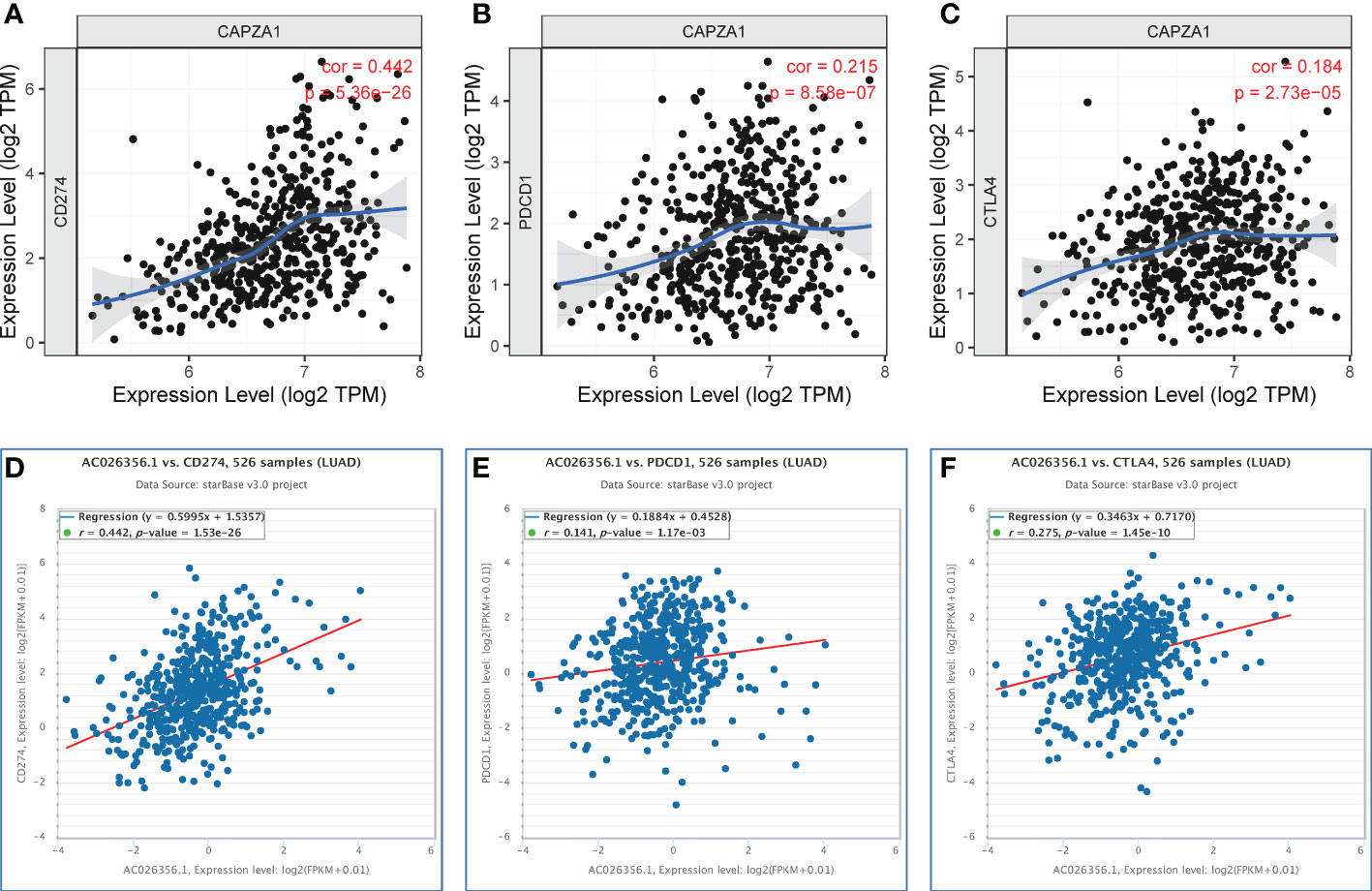
Figure 8 Correlation of CAPZA1 and AC026356.1 expression with PD-1, PD-L1, and CTLA-4 expression in LUAD. (A) Spearman correlation of CAPZA1 with expression of PD-L1 in LUAD using TIMER. (B) Spearman correlation of CAPZA1 with expression of PD-1 in LUAD using TIMER. (C) Spearman correlation of CAPZA1 with expression of CTLA-4 in LUAD using TIMER. (D) The expression correlation of AC026356.1 with PD-L1 in LUAD was determined by the starBase database. (E) The expression correlation of AC026356.1 with PD-1 in LUAD determined by the starBase database. (F) The expression correlation of AC026356.1 with CTLA-4 in LUAD was determined by the starBase database.
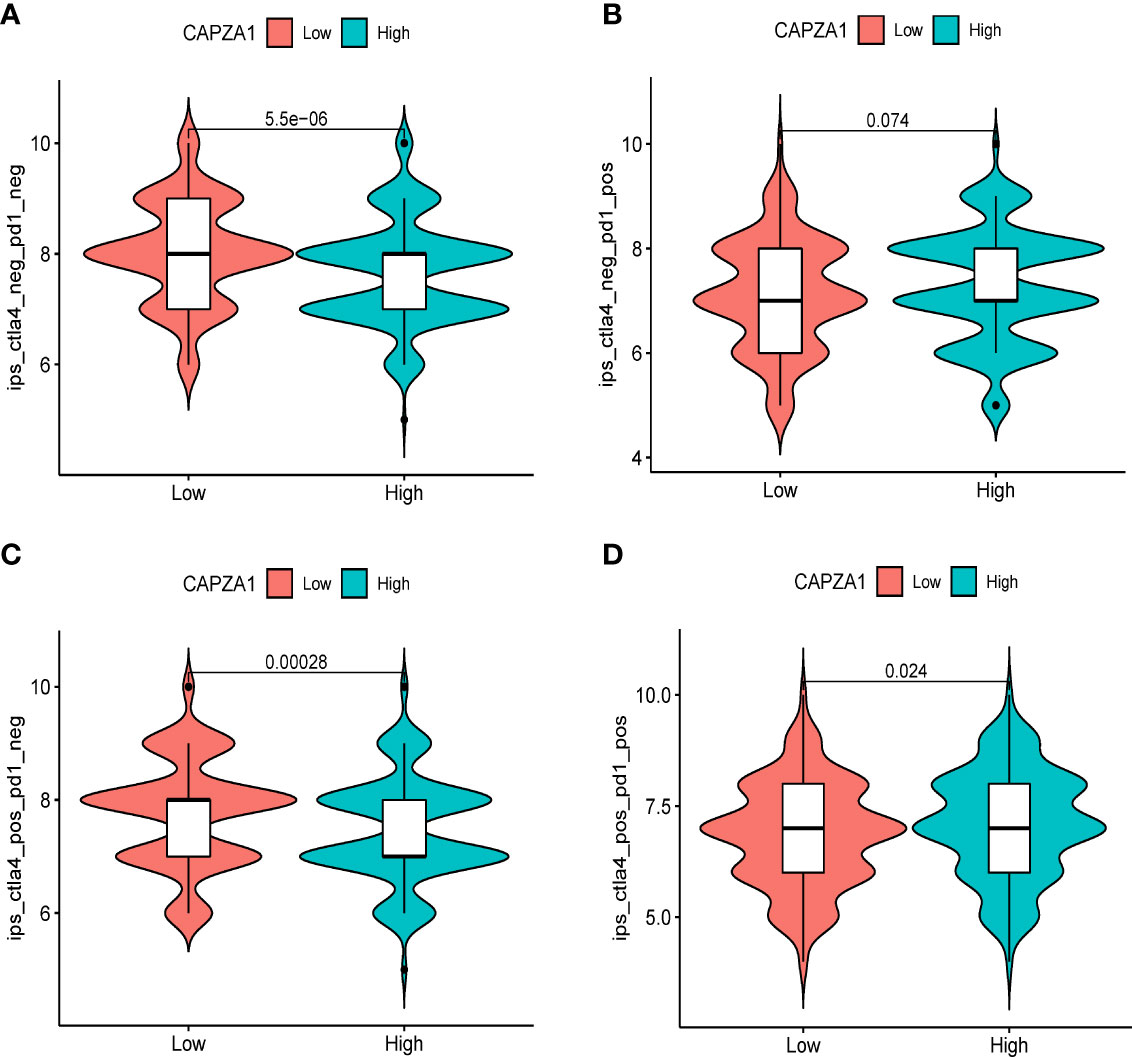
Figure 9 Correlation of CAPZA1 expression with the immunophenoscore (IPS) in HCC. (A) ips_ctla4_neg_pd1_neg, (B) ips_ctla4_neg_pd1_pos, (C) ips_ctla4_pos_pd1_neg, and (D) ips_ctla4_pos_pd1_pos.
3.7 The correlation of CAPZA1 with immunotherapy and potential mechanisms
We further confirmed that CAPZA1 in LUAD was significantly positively correlated with the TMB, MSI, immune score, stromal score, and estimateScores (Figure 10A–E). GSEA revealed that CAPZA1 might be involved in cancer-related critical pathways such as cell cycle, P53 signal pathway, mismatch repair, Nod-like receptor signal pathway, and Toll-like receptor signal pathway (Figure 10F).
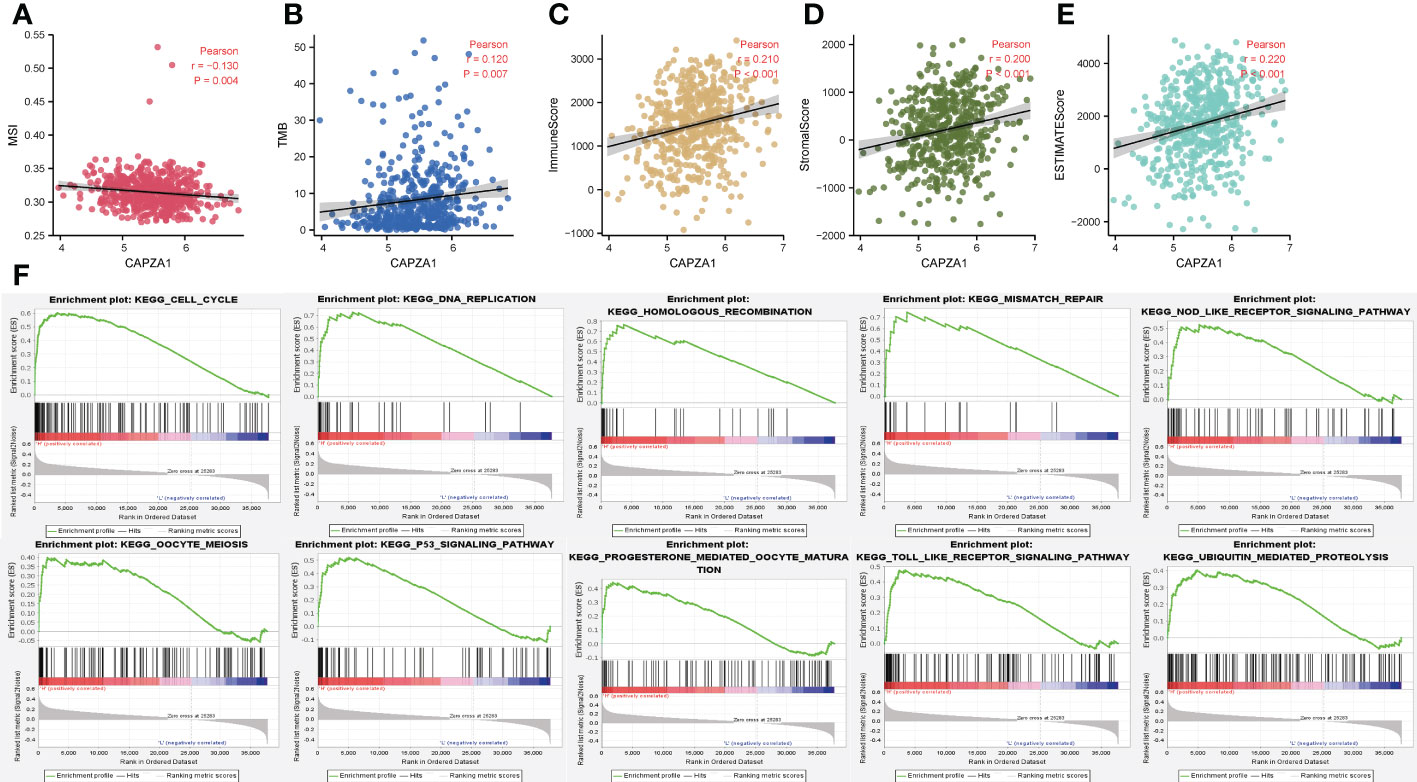
Figure 10 The correlation of CAPZA1 with immunotherapy and potential mechanisms. (A) Pearson correlation of CAPZA1 with MSI in LUAD. (B) Pearson correlation of CAPZA1 with TMB in LUAD. (C) Pearson correlation of CAPZA1 with ImmuneScore in LUAD. (D) Pearson correlation of CAPZA1 with StromalScore in LUAD. (E) Pearson correlation of CAPZA1 with estimateScores in LUAD. (F) Potential signaling pathway.
4 Discussion
LUAD is one of the significant invasive diseases limiting the life span of humans (33–35). Identifying the molecular mechanisms underlying LUAD carcinogenesis is becoming increasingly instrumental in searching for novel therapeutic targets and biomarkers. Previous studies have confirmed that CAPZA1 elicits pivotal roles in regulating the onset and development of human cancers (36, 37). However, the role of CAPZA1 in LUAD remains incompletely known and needs further study.
Our study first examined the finding that CAPZA1 was significantly overexpressed at mRNA levels in 13 tumors compared with normal tissues, which was validated in the starBase database. Moreover, survival and prognosis analysis displayed that higher CAPZA1 correlated with worse survival in KIRC, KIRP, LUAD, and LIHC using the starBase database. In addition, only the prognosis of CAPZA1 in LUAD had been validated in the PrognoScan and LOGpc databases, which might shed light on novel targets for LUAD individualized treatment.
Mechanisms related to ceRNA are modulated by ncRNAs with direct or indirect effects, including gene expression regulation (38). Our study used the starBase database to find possible miRNAs interacting with CAPZA1 RNA binding proteins, thereby modulating CAPZA1 mRNA stability. The result revealed that 16 miRNAs were finally found. Further analysis shows that only hsa-miR-30d-5p in LUAD was screened since it was low and its downregulation favored a better life for patients. Most of the research related to this miRNA has proved to be tumor-suppressive in many cancers, such as prostate cancer (39), renal cell carcinoma (RCC) (40), non-small cell lung cancer (NSCLC) (41), esophageal squamous cell carcinoma (ESCC) (42), and gallbladder (43). For instance, downregulated miR-30d-5p has been linked to favorable clinical outcomes in patients with RCC, and overexpression of miR-30d-5p attenuates RCC cell line proliferation and cell-cycle G1/S transition and facilitates apoptosis, which is denoted as a tumor-suppressor (40). CAPZA1 might be the most potentially targeted mRNA of hsa-miR-30d-5 in LUAD.
Recently, lncRNAs have been considered miRNA sponges that contribute to the regulation of cancers (44). Based on this, nine possible lncRNAs in LUAD function upstream of the hsa-miR-30d-5-CAPZA1 axis. Following survival analysis and correlation analysis, only AC026356.1 showed higher activity than normal samples, and higher AC026356.1 was reciprocally linked to favorable patients’ OS among all these lncRNAs. More importantly, AC026356.1 lncRNA was positively relevant to CAPZA1 expression whereas negative with hsa-miR-30d-5p, uncovering that AC026356.1 is most likely upstream lncRNAs of the hsa-miR-30d-5p-CAPZA1 axis.
Extensive evidence has proved that the tumor-infiltrating immune cells exert a crucial function on patients’ prognosis and immunotherapy (45). For example, activating tumor infiltrating CD8 T cells were able to contribute to antitumor responses (44). In our analysis, CAPZA1 in LUAD was positively relevant to the infiltration of most immune cells, especially macrophages, CD4+ T cells, CD8+ T cells, dendritic cells, and neutrophils, which is also validated based on some immune infiltration markers. Also, immunotherapy, mainly represented by immune checkpoint inhibitors (ICIs), has shown promising efficacy in treating many cancers (46). Thus, the linked immune checkpoints, including PD1, PD-L1, or CTLA-4 with CAPZA1, were assessed in LUAD, which has been successfully validated by IPS analysis, indicating that it might exert a pivotal role in restoring immune cytotoxic activity. Similarly, this correlation had the same trend, indicating that CAPZA1 might serve as a new tumor immunotherapy response predictor in LUAD. We further confirmed that CAPZA1 in LUAD was significantly positively correlated with the TMB, MSI, immune score, and stromal score, indicating that CAPZA1 could regulate immune infiltration function in LUAD. Moreover, GSEA revealed the tumor-associated and even tumor-promoting roles of CAPZA1, encompassing cancer-related critical pathways such as cell cycle, P53 signal pathway, mismatch repair, Nod-like receptor signal pathway, and Toll-like receptor signal pathway. It also displayed that CAPZA1 played a crucial role in tumor progression.
Nevertheless, some restrictions are supposed to be highlighted in our investigations. Firstly, considering tissue specificity, we fail to enable our conclusions to be reapplied to other tumors. Next, we need to validate the roles of CAPZA1 in LUAD on antitumor immunotherapy and tumor immunity based on more basic experiments. Finally, it is imperative to gather more robust data or samples for LUAD due to the lack of available public datasets.
For the first time, collectively, we comprehensively determine that CAPZA1 serves as an oncogene in pan-cancers and has a high CAPZA1 expression process that has an unfavorably high prognostic value in LUAD based on the starBase and PrognoScan databases. Our findings also uncovered that CAPZA1 might play an essential role in tumor immunity during the development of LUAD, such as immune infiltration cells, immune infiltration markers, TMB, MSI, immune score, stromal score, and immune checkpoints, and maybe a reliable therapeutic target for immunological antitumor strategies.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: 1. https://xena.ucsc.edu/, 2. http://starbase.sysu.edu.cn/, 3. http://www.abren.net/PrognoScan/, 4. https://cistrome.shinyapps.io/timer/, 5. https://bioinfo.henu.edu.cn/DatabaseList.jsp.
Author contributions
The project was designed and conceived by HPZ. HPZ analyzed the data. TQ and HYZ wrote the manuscript. WX and YM participated in the revision process, added analysis content, completed the picture layout, and answered relevant questions. ZY directed the revision process of the article and the revision of the article’s language. HPZ guided the execution of the study and revised the paper. All authors have read and approved the final version of this manuscript.
Funding
This study was supported by the grants from the CBY22-QDA19 of North Sichuan Medical College.
Acknowledgments
We appreciatively thank the UCSC, TCGA, StarBase, TIMER, PrognoScan, and LOGpc databases for the availability of the data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1025192/full#supplementary-material
Supplementary Figure 1 | Flow chart
Supplementary Figure 2 | The lncRNAs of the miR-30d-5p regulatory network are established by cytoscape software.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin (2020) 70(1):7–30. doi: 10.3322/caac.21590
3. Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis (2018) 9(2):117. doi: 10.1038/s41419-017-0063-y
4. Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WE, et al. The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (Eighth) edition of the TNM classification for lung cancer. J Thorac Oncol (2016) 11(1):39–51. doi: 10.1016/j.jtho.2015.09.009
5. Aikins ME, Xu C, Moon JJ. Engineered nanoparticles for cancer vaccination and immunotherapy. Acc Chem Res (2020) 53(10):2094–105. doi: 10.1021/acs.accounts.0c00456
6. Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol (2020) 20(11):651–68. doi: 10.1038/s41577-020-0306-5
7. Liu SV, Giaccone G. Lung cancer: First-line immunotherapy in lung cancer - taking the first step. Nat Rev Clin Oncol (2016) 13(10):595–6. doi: 10.1038/nrclinonc.2016.148
8. Passaro A, Spitaleri G, Gyawali B, de Marinis F. Immunotherapy in non-Small-Cell lung cancer patients with performance status 2: Clinical decision making with scant evidence. J Clin Oncol (2019) 37(22):1863–7. doi: 10.1200/jco.18.02118
9. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell (2017) 168(4):707–23. doi: 10.1016/j.cell.2017.01.017
10. Svitkina T. The actin cytoskeleton and actin-based motility. Cold Spring Harb Perspect Biol (2018) 10(1):a018267. doi: 10.1101/cshperspect.a018267
11. Cooper JA, Sept D. New insights into mechanism and regulation of actin capping protein. Int Rev Cell Mol Biol (2008) 267:183–206. doi: 10.1016/s1937-6448(08)00604-7
12. Edwards M, Zwolak A, Schafer DA, Sept D, Dominguez R, Cooper JA. Capping protein regulators fine-tune actin assembly dynamics. Nat Rev Mol Cell Biol (2014) 15(10):677–89. doi: 10.1038/nrm3869
13. Waddle JA, Cooper JA, Waterston RH. The alpha and beta subunits of nematode actin capping protein function in yeast. Mol Biol Cell (1993) 4(9):907–17. doi: 10.1091/mbc.4.9.907
14. Huang D, Cao L, Zheng S. CAPZA1 modulates EMT by regulating actin cytoskeleton remodelling in hepatocellular carcinoma. J Exp Clin Cancer Res (2017) 36(1):13. doi: 10.1186/s13046-016-0474-0
15. Huang D, Cao L, Xiao L, Song JX, Zhang YJ, Zheng P, et al. Hypoxia induces actin cytoskeleton remodeling by regulating the binding of CAPZA1 to f-actin via PIP2 to drive EMT in hepatocellular carcinoma. Cancer Lett (2019) 448:117–27. doi: 10.1016/j.canlet.2019.01.042
16. Lee YJ, Jeong SH, Hong SC, Cho BI, Ha WS, Park ST, et al. Prognostic value of CAPZA1 overexpression in gastric cancer. Int J Oncol (2013) 42(5):1569–77. doi: 10.3892/ijo.2013.1867
17. Santio NM, Vainio V, Hoikkala T, Mung KL, Lång M, Vahakoski R, et al. PIM1 accelerates prostate cancer cell motility by phosphorylating actin capping proteins. Cell Commun Signal (2020) 18(1):121. doi: 10.1186/s12964-020-00618-6
18. Burbage M, Keppler SJ. Shaping the humoral immune response: Actin regulators modulate antigen presentation and influence b-T interactions. Mol Immunol (2018) 101:370–6. doi: 10.1016/j.molimm.2018.07.026
19. Pérez-Martínez M, Gordón-Alonso M, Cabrero JR, Barrero-Villar M, Rey M, Mittelbrunn M, et al. F-actin-binding protein drebrin regulates CXCR4 recruitment to the immune synapse. J Cell Sci (2010) 123(Pt 7):1160–70. doi: 10.1242/jcs.064238
20. Lee CM, Barber GP, Casper J, Clawson H, Diekhans M, Gonzalez JN, et al. UCSC genome browser enters 20th year. Nucleic Acids Res (2020) 48(D1):D756–d761. doi: 10.1093/nar/gkz1012
21. Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen H-Z, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol (2017) 1:1–15. doi: 10.1200/PO.17.00073
22. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Res (2014) 42(Database issue):D92–7. doi: 10.1093/nar/gkt1248
23. Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med Genomics (2009) 2:18. doi: 10.1186/1755-8794-2-18
24. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res (2017) 77(21):e108–10. doi: 10.1158/0008-5472.Can-17-0307
25. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep (2017) 18(1):248–62. doi: 10.1016/j.celrep.2016.12.019
26. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med (2017) 9(1):34. doi: 10.1186/s13073-017-0424-2
27. Li K, Luo H, Huang L, Luo H, Zhu X. Microsatellite instability: a review of what the oncologist should know. Cancer Cell Int (2020) 20:16. doi: 10.1186/s12935-019-1091-8
28. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA (2005) 102(43):15545–50. doi: 10.1073/pnas.0506580102
29. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov (2021) 20(8):629–51. doi: 10.1038/s41573-021-00219-z
30. Goodall GJ, Wickramasinghe VO. RNA In cancer. Nat Rev Cancer (2021) 21(1):22–36. doi: 10.1038/s41568-020-00306-0
31. Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer (2011) 11(12):849–64. doi: 10.1038/nrc3166
32. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell (2011) 146(3):353–8. doi: 10.1016/j.cell.2011.07.014
33. Borczuk AC. Prognostic considerations of the new world health organization classification of lung adenocarcinoma. Eur Respir Rev (2016) 25(142):364–71. doi: 10.1183/16000617.0089-2016
34. Liu D, Vokes NI, Van Allen EM. Toward molecularly driven precision medicine in lung adenocarcinoma. Cancer Discov (2017) 7(6):555–7. doi: 10.1158/2159-8290.Cd-17-0355
35. Gillette MA, Satpathy S, Cao S, Dhanasekaran SM, Vasaikar SV, Krug K, et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell (2020) 182(1):200–225.e35. doi: 10.1016/j.cell.2020.06.013
36. Tsugawa H, Mori H, Matsuzaki J, Sato A, Saito Y, Imoto M, et al. CAPZA1 determines the risk of gastric carcinogenesis by inhibiting helicobacter pylori CagA-degraded autophagy. Autophagy (2019) 15(2):242–58. doi: 10.1080/15548627.2018.1515530
37. Li J, Zhang W, Gao J, Du M, Li H, Li M, et al. E3 ubiquitin ligase UBR5 promotes the metastasis of pancreatic cancer via destabilizing f-actin capping protein CAPZA1. Front Oncol (2021) 11:634167. doi: 10.3389/fonc.2021.634167
38. Sun YM, Chen YQ. Principles and innovative technologies for decrypting noncoding RNAs: from discovery and functional prediction to clinical application. J Hematol Oncol (2020) 13(1):109. doi: 10.1186/s13045-020-00945-8
39. Song Y, Song C, Yang S. Tumor-suppressive function of miR-30d-5p in prostate cancer cell proliferation and migration by targeting NT5E. Cancer Biother Radiopharm (2018) 33(5):203–11. doi: 10.1089/cbr.2018.2457
40. Liang L, Yang Z, Deng Q, Jiang Y, Cheng Y, Sun Y, et al. miR-30d-5p suppresses proliferation and autophagy by targeting ATG5 in renal cell carcinoma. FEBS Open Bio (2021) 11(2):529–40. doi: 10.1002/2211-5463.13025
41. Hosseini SM, Soltani BM, Tavallaei M, Mowla SJ, Tafsiri E, Bagheri A, et al. Clinically significant dysregulation of hsa-miR-30d-5p and hsa-let-7b expression in patients with surgically resected non-small cell lung cancer. Avicenna J Med Biotechnol (2018) 10(2):98–104.
42. Zhu Y, Liu J, Fan L, Wang F, Bu L, Ma T, et al. Serum expression and significance of MicroRNA-30d-5p in esophageal squamous cell carcinoma. Int J Clin Exp Pathol (2017) 10(8):8677–85.
43. Liu K, Xu Q. LncRNA PVT1 regulates gallbladder cancer progression through miR-30d-5p. J Biol Regul Homeost Agents (2020) 34(3):875–83. doi: 10.23812/20-180-a-32
44. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature (2014) 505(7483):344–52. doi: 10.1038/nature12986
45. Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer (2018) 18(3):139–47. doi: 10.1038/nrc.2017.117
Keywords: LUAD, CAPZA1, ceRNA, prognosis, immunotherapy
Citation: Qin T, Xiang W, Mao Y, Zhai H, Yang Z and Zhang H (2023) NcRNA-regulated CAPZA1 associated with prognostic and immunological effects across lung adenocarcinoma. Front. Oncol. 12:1025192. doi: 10.3389/fonc.2022.1025192
Received: 22 August 2022; Accepted: 05 December 2022;
Published: 04 January 2023.
Edited by:
Feng Xu, Shantou University, ChinaReviewed by:
Yuan Zhuhui, Peking University Third Hospital, ChinaTao Yang, Guizhou University of Traditional Chinese Medicine, China
Copyright © 2023 Qin, Xiang, Mao, Zhai, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihao Yang, eWFuZ2R1bm1pbkBmb3htYWlsLmNvbQ==; Hongpan Zhang, aG9uZ3BhbnpoYW5nc2NjbkAxNjMuY29t
†These authors have contributed equally to this work
 Tingting Qin
Tingting Qin Wanping Xiang2,3†
Wanping Xiang2,3† Hongpan Zhang
Hongpan Zhang