
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 24 November 2022
Sec. Breast Cancer
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1020121
This article is part of the Research Topic Dissecting Triple Negative Breast Cancer; Advances and Challenges From the Laboratory to the Clinic View all 8 articles
Bacterial inhabitants of the body have the potential to play a role in various stages of cancer initiation, progression, and treatment. These bacteria may be distal to the primary tumour, such as gut microbiota, or local to the tissue, before or after tumour growth. Breast cancer is well studied in this context. Amongst breast cancer types, Triple Negative Breast Cancer (TNBC) is more aggressive, has fewer treatment options than receptor-positive breast cancers, has an overall worse prognosis and higher rates of reoccurrence. Thus, an in-depth understanding of the bacterial influence on TNBC progression and treatment is of high value. In this regard, the Gut Microbiota (GM) can be involved in various stages of tumour progression. It may suppress or promote carcinogenesis through the release of carcinogenic metabolites, sustenance of proinflammatory environments and/or the promotion of epigenetic changes in our genome. It can also mediate metastasis and reoccurrence through interactions with the immune system and has been recently shown to influence chemo-, radio-, and immune-therapies. Furthermore, bacteria have also been found to reside in normal and malignant breast tissue. Several studies have now described the breast and breast tumour microbiome, with the tumour microbiota of TNBC having the least taxonomic diversity among all breast cancer types. Here, specific conditions of the tumour microenvironment (TME) - low O2, leaky vasculature and immune suppression - are supportive of tumour selective bacterial growth. This innate bacterial ability could enable their use as delivery agents for various therapeutics or as diagnostics. This review aims to examine the current knowledge on bacterial relevance to TNBC and potential uses while examining some of the remaining unanswered questions regarding mechanisms underpinning observed effects.
Triple-Negative Breast Cancers (TNBC) characteristically lack, or express at very low levels, human growth factor receptor 2 (HER2), progesterone (PR) and/or estrogen receptors (ER) (1). In 2020, 12% -17% of the 2.3 million new breast cancer (BC) cases and over 685,000 deaths worldwide can be attributed to TNBC (2). TNBC disproportionally affects young premenopausal women with west African ancestry, particularly African American and Ghanaian women (3). Other relevant risk factors include Breast Cancer gene-1/2 (BRCA) mutations, smoking history, and obesity (4, 5). Generally, TNBC originates in the milk duct as ductal carcinoma and less frequently in mammary lobules as lobule carcinoma (6). Based on its genotype profiling and cellular origin, TNBC can be classified into four subtypes: basal subtypes 1/2, mesenchymal subtype and an androgen receptor-expressing luminal subtype (7).
Surgery (lumpectomy or mastectomy) followed by radiation is available for early-stage patients, and immune checkpoint inhibitor therapies are offered on a case-by-case basis (8). For those that have missed the surgical window, the standard treatment for non-metastatic TNBC at the early stage is still nonspecific chemotherapy including platinum, taxane, anthracycline and cyclophosphamide with checkpoint inhibitor immunotherapy, such as atezolizumab or pembrolizumab given as a neoadjuvant where tumors are greater than 2cm in diameter and lymph node-positive (9). Adjuvant capecitabine treatment is standard in the case of residual disease in conjunction with PARP inhibitors olaparib or talazoparib, where BRCA-1/2 mutations are present (10). TNBC is highly invasive and has no standard treatment care options for the metastatic disease stage. Therapeutic schemes for this disease are constrained to conventional cytotoxic chemotherapy with additional immunotherapy targeting programmed death receptor 1 or ligand (PD-1 or PDL-1), as endocrine or receptor based therapies (e.g. HER2, ER and PR) are completely ineffective for TNBC (11). Recently antibody drug conjugates such as sacituzumab govitecan have been approved for metastatic TNBC (12).
Humans are a symbiont of human and microbial cells, with a ratio of 1.3-2.5 bacterial: human cells (13). Although the majority of these microbes reside within our gut, distinctive collections of microbes are also found in most body parts, possibly even including the brain, although present evidence is inconclusive (14). These distinctive microbial signatures are known as the microbiome (15, 16). The term microbiome, as Whipps and co-workers originally postulated, includes not only the community of the microorganisms but also their “theatre of activity”. These ecosystems, created by a multitude of microbes that may include bacteria, archaea, fungi, yeast, and viruses, are site-specific (15). The microbiome of each body part has distinctive characteristics regarding population dynamics and the diversity of microbial species (17). This site-specific diversity and dynamics can be regarded as a health indicator with, high diversity in the gut microbiome generally linked to good health (18). Our microbiomes represent a virtual organ that performs essential body functions that maintain our homeostasis, such as metabolizing nutrients, maintaining the integrity of the mucosal barriers, developing a healthy immune system, modulating a healthy neuronal development (including regulating our moods) and defending us against pathogens (19–21).
Microbes start colonizing our body as early as in the 2nd trimester of fetal development, where low levels of microbial signals can be detected in the fetal gut, skin, placenta, and lung tissue (22). However, the first major colonization event in early life happens at birth, where the mode of delivery determines the neonate microbiome composition to resemble either a vaginal or skin microbiome (23). After this event, our microbiomes are shaped by external factors such as diet, lifestyle, and environmental biodiversity (24). Our microbiome composition varies with age. In early neonatal life, breastfeeding enables the vertical transmission (mother to infant) of bacteria. Thus, neonates exhibit a microbiome composition resembling their mother’s milk. The adult type of GM composition starts appearing at ages 3-5 years (25). At this age, 90% of all species of the adult GM would have already colonized the gut.
Overall, a diverse GM is a healthy and robust GM, well able to perform the multiple tasks that define our health status (see Figure 1) (26). On average, the adult GM is estimated to be composed of 300-500 species (27), comprised of 12 bacterial phyla and one Archaean taxon, with the majority of species belonging to the Bacteroidetes and Firmicutes phyla and a smaller proportion to the Proteo- and Actinobacteria phyla (28, 29). However, GM composition differs between individuals and starting in mid-to-late adulthood (40-50 years of age), it increasingly diverges towards a microbiome that is unique to each individual (28, 30). Compositional uniqueness is more accentuated among the elderly (>65 years old) since, at this life stage, microbiome diversity can decrease significantly. In this age group, uniqueness has been positively associated with a healthy status (24, 30).
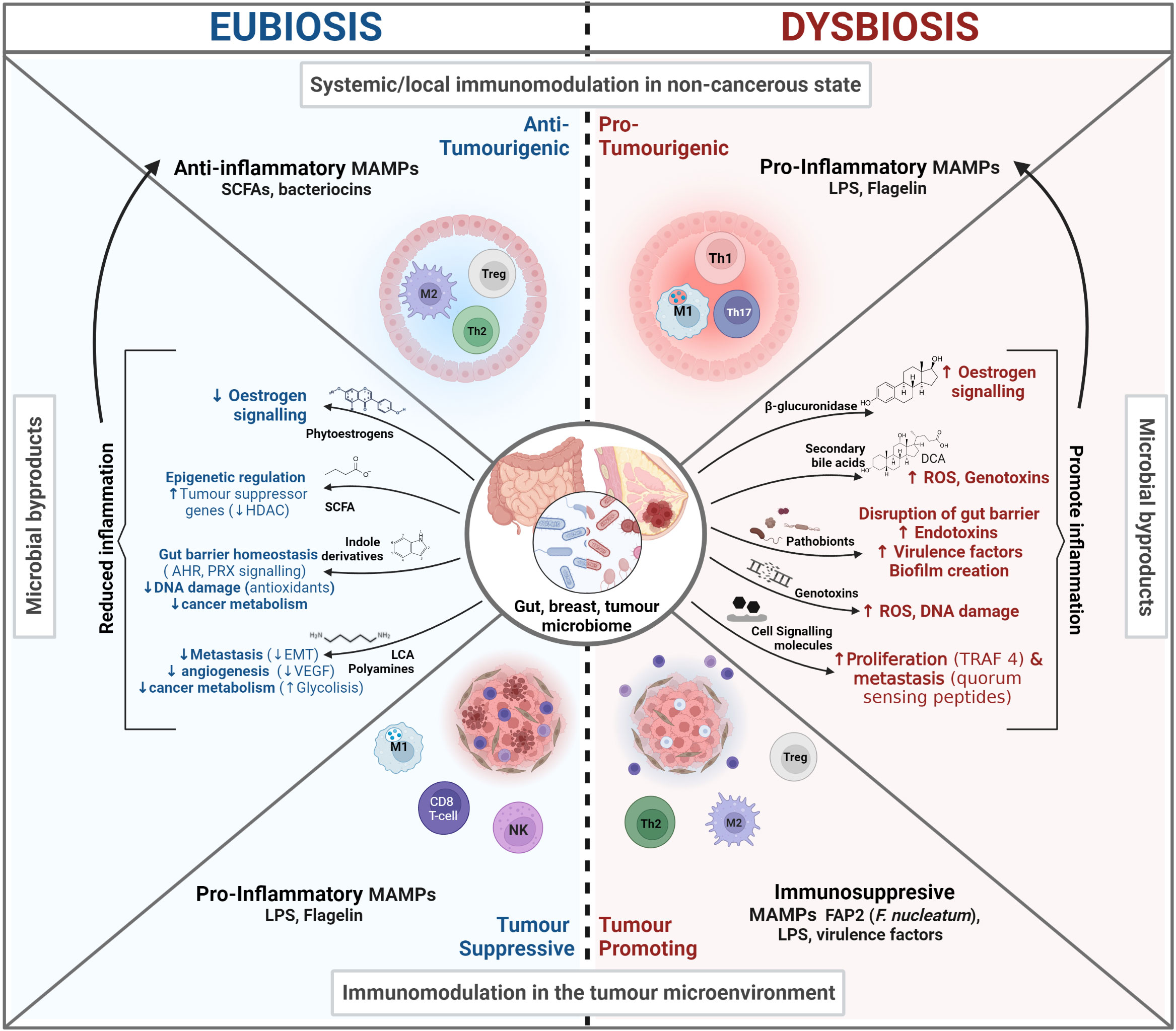
Figure 1 The microbiome influence in Cancer. In eubiosis the microbiome promotes our health status and prevents oncogenesis and tumor progression by influencing the immune system, promoting gut barrier integrity, and influencing cell signaling. Conversely, during dysbiosis there is a loss of barrier integrity that can lead to potentially harmful bacterial translocation, chronic inflammation in distal sites and the production of cancer promoting bacterial by-products.
A healthy microbiome safeguards host-microbiome homeostasis. Here, different microbe populations regulate the abundance of neighboring commensal or pathogenic bacteria by occupying a niche and adjusting the niche environment. Bacteria compete for nutrients, release bacteriocins (peptides which are toxic to and inhibit/regulate the growth of similar or closely related bacterial strains) or bacterial signals (MAMPs, see 1.2) to communicate with the host in order to modulate the release of antimicrobial peptides, mucin and ultimately, immune responses (31). Together, these actions contribute to the formation and maintenance of a healthy GIT mucosal “firewall”, which by segregating the GM from host cells, prevents microbial translocation and adverse immune priming events (32, 33).
Overall, a healthy GM safeguards host-microbiome homeostasis by modulating immune tolerance against gut commensals and eliciting pertinent immune responses (34). These GM-immune cell interactions are essential for the proper development of the gut-associated lymphoid tissues (GALT), which is the largest mass of lymphoid tissue in the body (more in 1.2) (35). The loss of beneficial microbes, expansion of pathobionts (commensals that at higher densities can cause harm), and/or the overall loss of microbial diversity can alter the GM composition in a way in which the abovementioned self-regulation and host-microbiome homeostasis functions are impaired. This altered and impaired GM composition is known as dysbiosis (36). GM Dysbiosis has been found to influence tumorigenesis through multiple mechanisms and interactions, including modulating our immune system, the metabolism of estrogens, and the production of protective or oncogenic metabolites, as described in Figure 1 and sections 2.1.3-2.1.4 (32).
The microbiome promotes the development and maintenance of the GIT mucosa and associated lymphoid tissues by producing microbial motifs (antigens) known as microbe-associated molecular patterns (MAMPs) (e.g., lipopolysaccharide (LPS), short-chain fatty acids (SFCA), and peptidoglycans) (37–39). The more commonly known PAMPs - pathogen-associated molecular patterns - are a subset of MAMPs, which include non-pathogenic microbes. MAMPs are recognized by pattern recognition receptors (PRRs) in Antigen Presenting Cells (APCs)). The GM uses MAMPs to communicate, trains and supports the maturation of the innate immune system, in order to: (i) modulate tolerance by discerning self from non-self (40); (ii) ensure homeostatic levels of innate immune cells (e.g. macrophages and dendritic cells) (41); (iii) bridge the innate and adaptative immune systems, through the production of co-stimulatory signals that induce an adaptive immune system response (42, 43). See Figure 2.
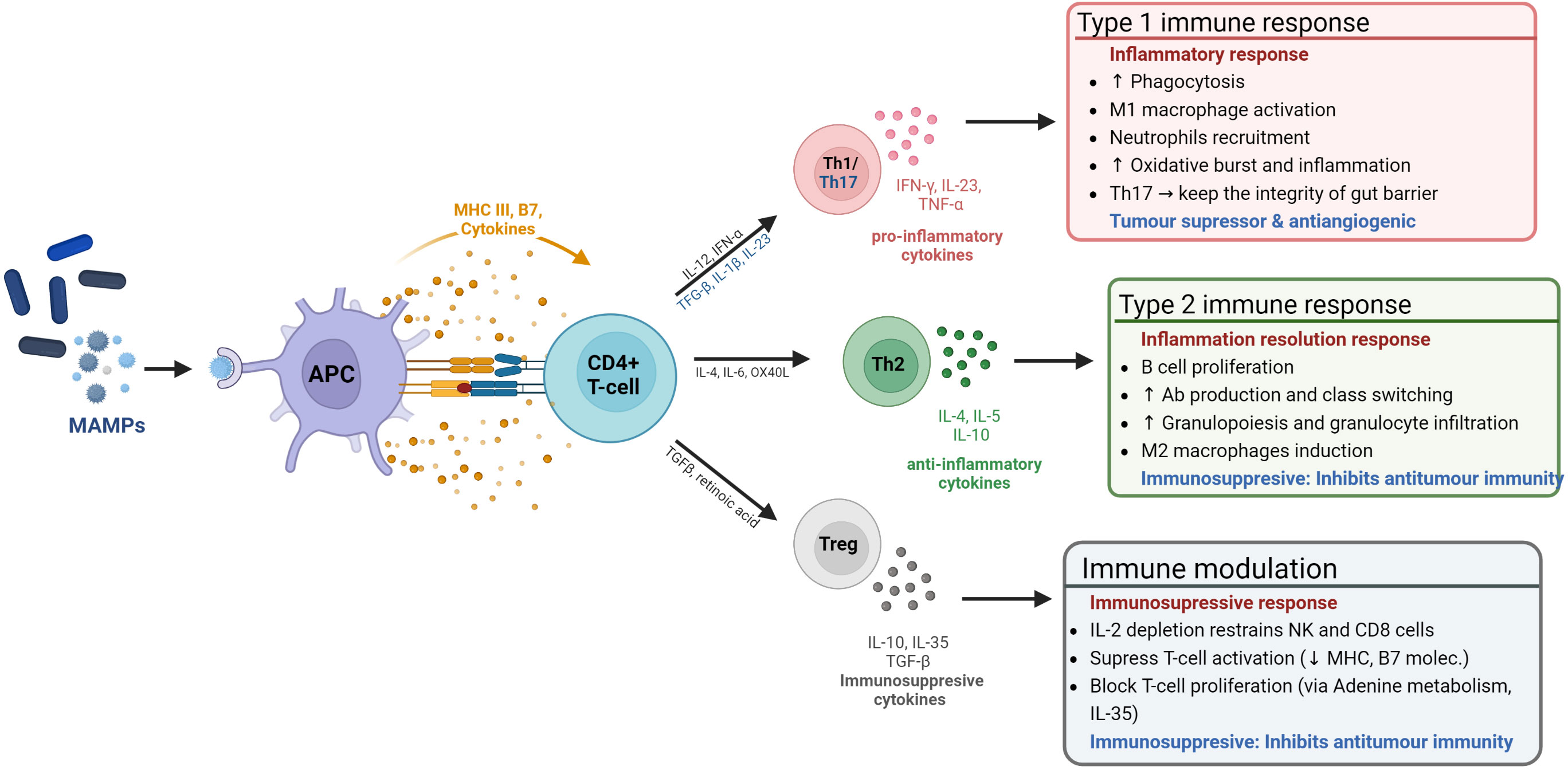
Figure 2 The microbiome influence on the host immune system. The microbiome modulates the immune system through MAMP production. These are sensed by antigen presenting cells (APCs), which process them to be presented to CD4+ naïve T-cells to induce either immune tolerance or an immune response.
This is particularly relevant for TNBC, wherein the tumor microenvironment is characteristically immunosuppressive, featuring immunosuppressive and pro-metastatic cell types and factors, such as: Type 2 macrophages (M2) and neutrophils (N2), Cancer-Associated Fibroblasts (CAFs), Cancer-Associated Adipocytes (CAAs), and altered extracellular matrix (ECM) (44). In fact, a current TNBC clinical trial (NCT02981303) is attempting to harness these TNBC immunosuppressive features and the immunostimulatory effects of PAMPs, by using a soluble yeast β-1,3/1,6-glucan PAMP (referred to as Imprime), to enhance the effect of immunotherapies after positive pre-clinical trials (45).
Chronic inflammation is a long-term reaction over the course of weeks or even an entire lifetime to inflammatory stimuli with continuous recruitment of monocytes and lymphocytes in addition to local tissue damage caused by the prolonged inflammatory response itself (46). Chronic inflammation contributes to tumorigenesis at all stages of oncogenesis, progression and dissemination, by promoting genomic instability and epigenetic modifications, inducing proliferation, strengthening anti-apoptotic pathways, stimulating angiogenesis and metastasis (47, 48). Roughly 10% of cancers are a result of a non-modifiable factors such as genetic predisposition to tumor development, with the remaining 90% a result of modifiable factors which induce DNA damage through environmental or life-style factors, for example U.V. damage, smoking, diet, alcohol use, obesity and infection status, that directly damage DNA or lead to some form of chronic inflammation (49). Among these, up to 20% of all cancers develop in tissues commonly affected by chronic inflammation (50).
Inflammation is a mechanism mainly orchestrated by the innate immune system to defend the body against pathogens and injuries, which can be perpetuated by exogenous stimuli (e.g. MAMPs) (51, 52). The prolonged/recurrent exposure to stimuli can lead to an uncontrolled infiltration and accumulation of immune cells, which can become polarized towards pro-inflammatory, tumor-promoting cell types (Type 1/17, 2 in Figure 2). It can also lead to the release/production of damage-associated molecular patterns (DAMPs), which signal a status of altered-self and amplify and perpetuate immune reactions through a vicious cycle of inflammation and DAMP production (53, 54).
The most relevant mechanism for bacterial-driven chronic inflammation is gut-barrier failure (33, 55, 56). A healthy barrier prevents the translocation of microbes into compartments where they would elicit a systemic immune response and establish immune memory (57). This anatomical separation is achieved through a multi-level gut-barrier, which ensures (i) segregation: preventing direct contact between the GM and host tissue; and (ii) compartmentalization – ensuring that responses to commensal bacteria are kept locally (56). The disruption of segregation/compartmentalization can lead to leaky-gut syndrome: A change in the permeability of the gut epithelial lining enabling the translocation of microbes or their by-products from the gut-lumen into the bloodstream, where they can be distributed systemically to other body sites and incite inflammatory responses (58). The recurrent injury to the tissue caused by these responses can consequentially prime the immune system (via DAMPs) towards chronic inflammation (59). In fact, a commonality across GM-associated diseases is that they are all a product of barrier disfunction (50, 60). This has been correlated to localized digestive tract diseases, such as in colon (61), liver (62) and pancreatic cancer (63), among others; and systemic inflammatory diseases, such as metabolic (64) autoimmune diseases (65), and cancer in other body sites (32), including the breast (66). A study looking at the inflammatory pathway score using a defined set of inflammatory genes from 3,632 tumors from four BC cohorts came to the conclusion that inflammation was associated with worse outcome overall in the BC cohort, but in TNBC, was associated more positively with tumor clearing immune response and immune cell infiltrations (67).
The GM produces a large array of small molecules during the metabolism of food and xenobiotics (compounds of nonhost origin that enter the gut with the diet, lifestyle or are produced by microbes). These can be in the form of low molecular weight metabolites, peptides, and proteins. In fact, the GM is associated with many biochemical pathways and in the synthesis of specific metabolites that are absorbed into the circulation. In this way, the GM contributes to the host biology a circulating pool of bacterially derived metabolites that can potentially exceed concentrations typically achieved by drugs (10 μM–1 mM) (68). Many of these by-products play critical roles in interbacterial (between different microbial species) and host-GM signaling by engaging with specific host receptors (69). As such, GM-derived metabolites can signal distant organs in the body and facilitate a connection between the host immune and hormone system, brain (the gut-brain axis), and metabolism. The beneficial or detrimental effects of these GM-derived metabolites will depend on the context and the state of the host, considering that the primordial nature of the symbiotic microbiota is to ensure its host health (70). Accordingly, below are outlined some of the pathways, metabolites and/or by-products by which the GM can exert an effect in TNBC.
The risk for developing receptor-positive breast cancers in post-menopausal women is highly associated with the levels of circulating estrogens and the time of exposure (71, 72). Whilst estrogen metabolism is not traditionally considered an essential factor in TNBC, it may be to a degree, as some TNBCs express alternative estrogen receptors. In addition, different jurisdictions have different cut-offs for the expression of either estrogen receptor (ER) and/or progesterone receptor (PR). For example, in the US, TNBC is diagnosed when receptor expression is lower than 1%. On the other hand, in the European Union, TNBC is diagnosed when the expression levels are lower than 10%.
Furthermore, estrogen signaling in TNBC can also be maintained by constitutively active estrogen-related receptors (ERRs) other than the canonical ER-α (73). Among these, estrogen receptors ERβ and the G protein-coupled estrogen receptor 1 (GPER-1) enable some degree of estrogen reactivity in TNBC (74). While the role of GPER in cancer is still inconclusive, new evidence shows that ER-β can have anticancer effects, including for TNBC. For example, a recent study looking into a cohort of 567 TNBC tumors, found that ERβ was expressed in 18% of them. Possible mechanisms for ERβ mediated tumor suppression include the formation of co-repressor complexes that suppress the activity of oncogenic NFκB/RELA (p65) and thus inhibit p65 signaling (75). Additionally, Erβ has been found to downregulate the unfolded protein response (UPR), which enables the survival of cancer cell to endoplasmic reticulum stress induced by poor tumor vascularization (76). Furthermore, ERβ BC cell mitochondrial translocation inhibits TNBC proliferation of vitro and in vivo models via mitoERβ activation (77, 78). Finally, ERβ has been shown to inhibit epithelial to mesenchymal transition (EMT) and the invasiveness of TNBC in vitro (79) and inhibit metastatic TNBC phenotypes by suppressing TGFβ signaling through the regulation of cystatins (80). It is important to highlight that these alternate estrogen receptors can respond to systemic estrogen but, not to current endocrine treatments (81).
Hence, even in TNBC, estrogen can play a role in cancer progression, depending on levels of circulating estrogen, in which bacteria play a role and the cancer’s ability to respond via canonical or non-canonical receptors. In order to excrete estrogen from the body, the liver conjugates estrogen to glucuronic acid which can be then excreted in the bile. The GM has enhanced capacity to increase systemic estrogen levels by increasing enterohepatic circulation (82). GM bacteria can increase levels of systemic estrogen in two ways – first, by blocking the binding of estrogen to glucuronic acid, reducing its inactivation (83). Secondly, estrogens that are marked for excretion through the bile can be deconjugated by bacterial species expressing β-glucuronidases enabling their reuptake (84, 85).
GM bacteria can also metabolize phytoestrogens from dietary polyphenols, which are thought to modulate estrogen metabolism by reducing the systemic levels of circulation estrogen, as product of the inhibition of estrogen synthetase activity and reducing the bioavailability of ERs, for which they compete (86). In this regard. GM species such as Eubacterium limosum activate polyphenols (isoflavones and lignans) by demethylating their hydroxyl groups (87). Enterogenic lignan can then be transformed into bioactive enterolactone by other GM strains, such as members of genus Eggerthella (88). Interestingly, equol, the isoflavone derivative with the greatest estrogenic and antioxidant activity, is only found in one third to one half of humans (thus, only in those harboring equol-producing microbes) (89). The isoflavone daidzein found exclusively in soya beans and other legumes and can be converted to equol by several bacterial species including Slackia, Lactobacillus, Paraeggerthella, Bifidobacterium and Eggerthella sp. among others (89). While the effects of bacterially activated phytoestrogens on receptor positive BC are not yet agreed upon; a recent TNBC clinical trial, comprising 39 patients with invasive TNBC, established that a course of oral S‐equol inhibited proliferation of breast tumor cells, as measured by the cell proliferation marker Ki-67, with a 20% decrease in Ki‐67 expression in almost one third of patients (90).
SCFAs are one of the main metabolites generated by the GM in the large intestine through anaerobic fermentation of indigestible dietary fiber and resistant starch (91). Among these, butyrate is the most important in relation to cancer, which is produced by Firmicutes (92). Cancer-driven histone deacetylase (HDAC) activity can lead to dysregulated epigenetic changes that silence tumor suppressor genes (TSG) facilitating malignant proliferation (93). Butyrate shows the most potent anti-cancer properties, including anti-inflammatory effects, suppression of angiogenesis, histone deacetylase (HDAC) inhibition which can reverse silencing of tumor suppressor genes (TSGs) and apoptosis induction in tumor cells by means of mitochondrial ROS production (94–97). A high fiber diet promotes the maintenance of butyrate-producing bacteria, making it cancer-protective. Conversely, the opposite effect is true; depletion of butyrate-producing bacteria may promote inflammation and tumorigenesis systemically (98). The receptors responsible for detecting SCFAs in TNBC are free the free fatty acid receptors (FFAR) 1 and 2 (99). In vitro TNBC studies have demonstrated that the activation of FFAR2 receptors increases the expression of adhesion proteins (E-cadherin) and inhibits MAPK signaling (via Hippo-Yap pathway inhibition), thus leading to a reduction of actin polymerization and cell invasiveness (100). This is supported by a study showing that the expression of both receptors was reduced in invasive breast carcinoma and metastatic TNBC tumors, relative to normal breast tissue (101).
Lithocholic acid (LCA) is a bile acid metabolite that has been found to exert cancer protective effects. It is produced exclusively in the gut by a few species of anaerobic bacteria in the genus Clostridium from primary bile acids (102). Any bile acids found in breast tissue originate from the gut (103). LCA anticancer properties in relation to BC in general, include reductions in cancer cell proliferation and epithelial to mesenchymal cell transition inhibition acting via the G-protein-coupled bile acid receptor 1 (TGR5) that exerts downstream anti-inflammatory effects (104). It also inhibits angiogenesis by inhibiting vascular endothelial growth factor (VEGF) (105). LCA alters cellular metabolism by inducing glycolysis and increasing mitochondrial oxidative phosphorylation in BC cells that depend on the Warburg effect (104). The Warburg effect is a cancer specific effect, with increased glucose uptake and fermentation to lactate even in the presence of oxygen, proposed to be an evolutionary mechanism to sustain proliferative growth through the generation of essential biomolecules, by-products of glycolysis (94). BC patients have been found to have low levels of the LCA-producing gene 7α/β-hydroxysteroid dehydroxylase (baiH) detected in DNA extracted from stool samples in early cancer cases when compared with healthy controls (104, 106). In addition, microbiota-derived bile acids of GM origin accumulate in breast tumors and correlate with reduced proliferation (107). Other studies found that BC cells treated with LCA in vitro decreased the expression of nuclear factor-2 (NRF2) and increased the expression of Kelch-like ECH associating protein 1 (KEAP1), constitutive androstane receptor (CAR) and inducible nitric oxide synthase (iNOS) (108). All of which has been found to correlate with an improved survival rate of BC patients, except for TNBC (108). In this regard, studies investigating this in the different subtypes of TNBC, in particular, are thus needed.
Polyamines are small polycationic molecules with a wide array of biological functions including gene regulation, stress resistance, cell proliferation and differentiation. These are mainly sourced from the GM, where they derive from bacterial amino-acid metabolism (109). Among these, Cadaverine is a biogenic amine derived from the decarboxylation of lysine and is used by bacteria to buffer the pH of their environment. It is synthesized by lysine decarboxylase (LdcC) and cadaverine A (CadA), enzymes found in species of the bacterial genera Enterococcus, Enterobacter, Escherichia, Proteus, Streptococci, and Shigella among other (110). Cadaverine has been shown to have tumor suppressor roles in breast cancer. Its antitumor effects have been proven in a TNBC murine model (grafted 4T1 tumor cells), with cadaverine found to reduce Epithelial to mesenchymal transition (EMT), an essential process driving tumor progression and metastasis, in which epithelial cells lose their features (cell polarity and cell–cell adhesion) and gain the invasive properties characteristic of mesenchymal stem cells. Consequently, cadaverine inhibited tumor growth, reduced cellular migration and invasion, ultimately reducing metastasis. In addition, cadaverine was also found capable of reducing BC invasion by inhibiting mesenchymal-to-epithelial (MET) transition (reverting mesenchymal tumor cells to a more epithelial like state) via the activation of trace amino acid (TAAR) cell receptors (111). This is supported by results from clinical trials (112)
Indoles are bioactive products of the GM bacterial catabolism of tryptophan. Among these, 3-Indolepropionic acid (IPA), has been the subject of numerous studies due to its anticarcinogenic effects. IPA and other propionic acid species (PAs) derived from phenylalanine and tyrosine are synthesized through aromatic amino-acid transferases and phenyllactate dehydratase found in some species of the bacterial genera Lactobacillus, Akkermansia, Clostridium and Peptostreptococci (68, 113). In general, Indoles are also key interbacterial and GM-Host signaling molecules, act through steroid and xenobiotic receptors (AHRs and PXRs) (114). Indole derivatives prevent carcinogenesis by promoting gut homeostasis (upregulating tight junction, cell turnover, mucin and AMP secretion) (115) and modulate immune tolerance by shifting immune-cell polarization towards anti-inflammatory types. IPA is a free radical scavenger and a potent antioxidant, which prevents DNA damage in non-transformed cells exposed to multiple types of oxidative damage (113, 116). Furthermore, IPA and indoxyl-sulphate (IS) exert cytostatic effects in breast cancer cells (including TNBC) in vitro and in vivo, where they reduce cancer cell stemness, their EMT and proliferation (117). In a recent study, it was found that four new benzo[f]indole-4,9-dione derivatives reduce TNBC cell viability by ROS accumulation in vitro and exert cytotoxic effects on TNBC cells (MDA-MB 231) through the intrinsic apoptosis pathway – activation of the caspase 9 and Bax/Bcl-2 pathway (118).
Once thought to be sterile, it is now well established that the breast has its own unique microbial signature (119–123). Breast and milk microbiomes are related, which makes the milk microbiome a good predictor for Breast Microbiome (BM) composition in lactating women (124). Milk has been found to accommodate more than 360 species of bacteria, mainly from the Actinobacteria, Bacteroidetes and Firmicutes phyla (125). Due to various exogenous factors (dietary habits, geographic locations, lactating phase, and research methodologies) different studies report different microbial richness and composition at genus/species level. Despite this, Staphylococcus and Streptococcus have been present in 98.7 and 97.7% of the samples analyzed, respectively, and are considered as core genera, followed by lactic-acid specific bacteria (Bifidobacterium sp. and Lactobacillus sp.) (126).
Microbes residing in breast tissue have variable origins, as these can be sourced from different body-parts, through different interactions. In this regard, aerobes and facultative anaerobes from the skin and other epithelial surfaces may gain access to the breast through the nipple-areolar opening. For example, bacteria from the oral mucosa may gain access during breastfeeding and/or sexual contact (127). The growth and persistence of the infiltrated bacteria is sustained by the favourable conditions of breast tissue, featuring (1) nutrient-rich fatty tissue content, (2) diffuse ducts originating from the nipple; and (3) a widespread vasculature and lymphatic systems facilitating their movement (119).
Conversely, the microbial translocation of strictly anaerobic bacteria, which cannot survive in the presence of oxygen (e.g., Bacteroides, and some Lactobacillus and Bifidobacterium species) into breast tissue is far more complex and has been a topic of research and debate for decades. It has been suggested that some bacterial taxa may be translocated from the gut to the mammary tissue via an enteromammary pathway (128). This is supported by evidence from different studies showing that dendritic cells (DCs) can sample and engulf bacteria directly from the lumen (maintaining epithelial barrier integrity) (129) and transport them alive (for up to 60 h) to other lymphoid tissues (57). This was also supported by a seminal study of the origin of human breast milk bacteria. This study revealed that during the perinatal period there is a heightened bacterial translocation to the GALT, which is followed by bacterial colonization of the breast during the immediate postpartum. Here, it was also suggested that bacterial transport was mediated by DCs, which was supported by evidence showing that: (i) the majority of mononuclear cells in the milk originated from the GALT, (ii) staining showing bacteria-DCs co-localization and (iii) culturing of viable bacteria extracted from DCs purified form maternal milk and blood. Other preclinical studies have suggested GIT translocation of certain bacteria, such as bifidobacteria, to distal tumors (130).
Tumor colonization by bacteria is facilitated by the increased permeability of the tumor microenvironment (TME), with leaky vasculature due to rapid angiogenesis. Tumor selective growth of specific facultative and/or anaerobic bacterial strains is supported by the highly hypoxic and nutrient-rich necrotic tumor regions and its characteristic suppressed immune surveillance (131–134). Several recent studies have raised convincing evidence that associates the breast microbiome with cancer; however, its role in tumorigenesis is still a subject of active investigation, as it still unknown whether different microbial signatures are a cause or a product of tumorigenesis driven tissue remodeling (134, 135). Nevertheless, the role of intratumoral bacteria (including in TNBC) in tumor progression is now well-acknowledged and considered an enabling factor of “the hallmarks of cancer” due to their capacity to contribute to genome instability and mutation, and tumor-promoting inflammation (136).
In recent years various investigations have found characteristic microbial signatures associated with the breast cancer tumor microbiota (TM). In general, these studies point out that the microbial composition of breast tissue is dominated by bacteria from Proteobacteria, followed by Firmicutes, Actinobacteria and Bacteroidetes phyla, but BC tumor tissue features an altered composition in terms of abundance. In this regard, BC tumor tissue has an overrepresentation of Proteobacteria (a phylum usually associated with inflammation and disease), with higher abundances of Gammaproteobacteria, especially from the Enterobacteriaceae family and reduced abundance of Actinobacteria and Bacteroidetes. BC TM also has a higher abundance of pathobionts (pro-inflammatory) in the Firmicutes [e.g. Staphylococcus (137), Streptococcus pyogenes (138)) and Actinobacteria (Micrococcus (139), Atopobium (140)] phyla. Furthermore, the BC tumor microbiota showed an increase in abundance of taxa with known carcinogenic effects, such as those belonging to the Fusobacterium genus (121, 122, 141, 142). In fact, taxa from the Enterobacteriaceae, Streptococcaceae, Staphylococcaceae and Micrococcaceae families have been found in pancreatic cancer tumors (143) with Staphylococcaceae species in particular, also present in higher abundance within the tumor microbiota of prostate cancer (144). Likewise, in an ovarian cancer microbiota study, the Gammaproteobacteria Shewanella sp. was detected in 91% of cancer tissue samples (145). On the contrary, bacterial taxa with known anti-carcinogenic effects, such as species from the Bacteroidetes phyla and Lactococcus, and Streptococcus genera, were less abundant in BC TM (146).
For TNBC in particular, the studies described in Table 1, list the microbial signatures found to be enriched in the TNBC TME for each study. The lack of congruity between studies, reflects the lack of defined protocols for the study of breast/tumor microbiomes, wherein, a myriad of variables that can influence the outcomes/results. For example, differences in sample source (fresh-frozen/FFPE, ethnicity/geography), experimental protocols (collection: surgery/biopsy, DNA extraction, hybridization/sequencing), and bioinformatic workflows (146). Nevertheless, these studies offer insights to relevant features that are particular to the TNBC tumor microbiota. First, the TM of TNBC has the least taxonomic diversity among all BC types (148). Second, TNBC has higher abundance of Aggregatibacter and Caulobacter (147, 149) which are biofilm forming bacterial strains that have been previously associated with localised periodontitis and endocarditis (150, 151). Furthermore, these studies found that the microbial profiles found in tumor-adjacent non-cancerous (matched) tissue had a higher abundance of pathobionts and were more similar to the TM than those found in non-matched healthy tissue controls. This indicates that the TME resident tumor microbiota can extend to surrounding tissues and/or that microbial profiles found in the tumor pre-existed tumor formation, which would suggest that these microbes have an active role in tumorigenesis (148). This can be supported by recent evidence from a study involving seven different cancer types and over 1500 FFPE tumor samples, where intratumoral bacteria were found to localize within the tumor and immune cells (152).
All these studies are merely taking a snapshot of the bacteria present at the time of sampling. The bacterial load is of extremely low biomass relative to the TME or the GM for that matter and issues of contamination and sampling techniques are important confounding factors. While our lab and others to improve the relevant methodology (153–156), bacteria at the scene cannot be pinned down to having a causative effect in oncogenesis or are merely bystanders having found a niche where they can survive (135).
In a eubiotic state, the breast microbiota produces metabolites that may confer protection from pathogens, boost immune responses and inhibit tumorigenesis. Across different TNBC-specific studies, a higher abundance of BM commensals (e.g. Streptococcus sp.) that synthesize cadaverine, a known BC tumor suppressor, has been found to correlate with healthy breast tissue (111). A different study highlighted that butyrate-producing strains (e.g. Odoribacter sp.), recognised for their anti-inflammatory effects, were absent in tumor tissues (92, 146). Additionally, several breast tissue resident commensals, such as Staphylococci sp., Lactococcus sp., and Streptococcus sp. produce lantibiotics and other bacteriocins, preventing the potential growth of pathogenic strains that could trigger chronic inflammation. Beyond this, some bacteriocins have also been found to have selective cytotoxicity toward cancer cells (157). For example, Lactobacillus salivarius and L. gasseri, have been shown to clear mastitis infections during lactation when administered as a probiotic (158) and their excreted products (as cell-free supernatant) has been found to inhibit BC cells in vitro (159). Similarly, Bovicin HC5 from Streptococcus bovis HC5 has been shown to induce cell death in BC cell lines in vitro (160).
Furthermore, breast tissue resident bacteria also have the capability to activate the immune system via MAMPs. For example, the MAMP Flagellin (and the principal component of bacterial flagella) activates Toll-receptor 5 (TLR5). This is a PRR recently found to be specifically expressed in the ductal epithelium of normal breast tissues and circulating immature dendritic cells induces the secretion of pro-inflammatory cytokines and chemokines, increases tumor necrosis and neutrophil infiltration, inhibiting cell proliferation and anchorage-independent growth in mouse xenografts of human BC cells (161). In this context, Lactococcus lactis has been shown to recruit and activate natural killer cells (the main innate immunity cytotoxic effector cells toward cancer cells) (122). This has been confirmed in a clinical trial where Lactococcus abundance positively correlated with the number of NK cells recruited (162). Finally, healthy breast tissue is known to harbor bacterial strains, such as species of Lactococcus and Streptococcus, genera known to produce antioxidants and ROS scavengers (e.g. indoles, IPA) that neutralize free radicals and reduce oxidative damage, preventing oncogenesis (119, 163). Supporting all the protective benefits to breast health are extensive clinical studies and meta-analyses correlating the use of antibiotics with a moderately increased risk of BC (164–166).
Some bacterial signatures can induce cellular and immunomodulatory changes that promote oncogenesis, tumor growth and metastasis. Here these are referred to as Pathobionts. Now, while some of these microbes may not be a pathogen in other body parts they behave as such in this specific context, as the capacity of any microbe to act as a pathogen is context dependent, on location and state of immune activation. Some of these effects are listed in Table 2.
Conventionally, breast cancers were deemed to be ‘cold’ in terms of the immune response, in that they generally do not evoke a robust immune response thereby making them less likely to respond to immune checkpoint inhibitor (ICI) therapies. However, TNBC is the most immunogenic out of all the BC types (177). The response to ICI can vary significantly from person to person, with only a modest subset of patients displaying increased survival rates (178). In a phase 1b clinical trial assessing the antitumor activity of an anti-PD-L1 antibody in patients with locally advanced or metastatic breast cancer, the confirmed objective response rate (ORR) for BC was 3.0% and 5.2% for TNBC. Results from this study suggest that the expression of PD-L1 by tumor-infiltrating lymphocytes (TIL) may predict the response to ICI therapy in breast cancer, as patients with TILs expressing PD-L1 showed an increased ORR in the overall population (16.7% vs. 1.6%), an effect that was remarkable in the TNBC subgroup, with an ORR of 22.2% vs. 2.6% (179). This confirmed previous findings in TNBC where a 10% increase in TILs was significantly correlated with a reduced reoccurrence of distant tumors in patients undergoing trastuzumab treatment in a large clinical trial, which is the reason why TILs are used as a biomarker of prognostic outcome (180). TILS can be subdivided into: CD4+ antigen-presenting T-helpers (Th) CD4+ immunosuppressive T-regs, and cytotoxic CD8+ T-cells – involved in direct tumor cell killing (see Figure 2). While high prevalence CD8+ T-cells and Th1 cell correlate with better treatment outcomes, the presence of Tregs and Th2 cells are associated with a poorer prognosis (more in figure 2) (181). In this regard, certain bacterial species known to colonize TNBC tumors (e.g., Fusobacterium nucleatum, and Staphylococcus aureus) have been found to shift TIL populations and induce a tumor suppressing, pro-inflammatory (Type 1) immune response in melanoma. Bacterial peptides (MAMPs) produced by these species are recognised by MHC I and II in APCs and stimulates the production of pro-inflammatory cytokines, which in turn recruit cytotoxic cells and induce pro-inflammatory Th1 cells differentiation (see Figure 2) (182).
Ever-increasing evidence supports the GM’s role in modulating treatment and a toxic response to cancer therapy, with several recent studies precisely demonstrating the response to ICI treatments across numerous cancer types (183–193). Here, different studies list varied species and genera associated with positive treatment outcomes, pointing to the complexity of confounding factors and interactions between the patient, the environment and their microbiome. However, in general, these studies report higher response to therapy and overall survival in patients with diverse microbiome profiles (eubiosis), containing Bifidobacterium, Akkermansia and Enterococcus, among others. On the other hand, patients with a reduced microbiome diversity (dysbiosis) responded poorly to this therapy (186, 189). This was confirmed in GF murine models. Mice receiving FMT from responders improved their response to anti-PD-LI, while those who received an FMT from non-responders developed resistance (185, 194)..
Interestingly, some studies have shown that the therapy can induce detrimental changes to the microbiome that may influence the development of resistance (183). This highlights the relevance of managing the microbiome richness during the course of treatment (193). Immunotherapies can also lead to a wide range of inflammatory side effects, including colitis, thyroid dysfunction, pituitary inflammation, and inflammatory arthritis, among others (195, 196). The combination of low efficacy, high cost and the risk of side effects calls for the development of effective biomarkers, potentially through GM composition. However, to date, there are not sufficient data to support this for TNBC; and thus, more studies are required.
Initial studies showed that cancer radiotherapy (RT) has fewer side effects on germ-free mice (197); further studies confirm that bacteria are responsible for the toxic effects of radiotherapy (198, 199). It was also shown that during macrophage polarization, cell metabolism is altered by shifting the balance between glucose utilization and fatty acid oxidation, influencing the immune response in the TME. Radiotherapy response or resistance is highly dependent on the TME, and this alteration may change the radio sensitivity of cancer cells. M1 macrophages are thought to enhance the radio-sensitivity of BC cells; however, M2 macrophages can activate radio-resistance through IL-4/IL-13-mediated STAT6 phosphorylation and M2 polarization (200). One study found that the vancomycin treatment enhanced the RT efficacy and that butyrate, a metabolite generated by vancomycin-depleted gut bacteria, abolished the vancomycin effect (201). This is another example of context-dependent effects of the microbiome, wherein butyrate, which is regarded as a beneficial metabolite in the context of oncogenesis and cancer progression, can negatively impact the outcome of a cancer therapy. In addition, IPA (see 1.4.4) was found to exert an RT-protective effect on mice (202). Patients who had a toxic reaction to RT were associated with an over-abundance of Clostridium, Rosesporium, and Phascolarctobacterium (203).
For patients with TNBC that has spread to the lymph nodes and is more than 1 cm in size, the American Society of Clinical Oncology (ASCO) recommends neoadjuvant chemotherapy. A lack of alternative treatments beyond initial surgeries and radiotherapy to reduce the tumor load means that chemotherapy remains the mainstay of treatment options. Common drugs include platinums, capecitabine doxorubicin, cyclophosphamide, paclitaxel, methotrexate, and fluorouracil (5-FU).
The microbiome affects many of these drugs not only in their efficacy but also in their level of toxic side effects. For platinums to work correctly, they must induce double-stranded DNA breaks, and the microbial production of ROS promote these mechanisms (204). A study in germ-free mice found that their cisplatin antitumor activity could be restored with the introduction of L. acidophilus (205).
A study from our lab examined the effects of bacterial species identified in breast cancer patient tumor samples on thirty standard chemotherapies in vitro. Results demonstrated an increase in the toxicity of six chemotherapeutic drugs, a decrease in nine, including doxorubicin and gemcitabine, and no effect in the remaining 15, with different bacterial species producing different effects (206). This was also verified in an in vivo mouse tumor model, with inhibitory effects on gemcitabine evident in tumors colonized by E. coli. Such findings suggest that response to therapy in BC tumors may be improved by microbiome modulation, and/or that the microbiome profile of a patient should be considered to inform treatment choice. To this end, our lab also demonstrated a proof of concept to utilize the microbiome signature of breast biopsies to infer the malignancy status of the tissue (123). Alternatively, in the context of bacteriotherapy (see later), another study from our lab demonstrated the ability of the natural enzymolome of introduced bacteria to mediate local tumor therapy in a murine model via activation of multiple systemically administered chemotherapeutics (207).
In relation to TNBC, there is only one recent study which looked at the GM of 30 patients with TNBC, identifying Bacteroides and Ruminococcaceae as taxa more abundant in TNBC patients who achieved a complete pathological response (pCR) after treatment with neoadjuvant chemotherapy compared with those who did not achieve a pCR. Patients with a partial response had higher quantities of Bacteroides caccae than those without any response (208). Resistance to the nucleoside analogue gemcitabine was found to be induced by intracellular gammaproteobacteria in a murine model, with treatment efficacy restored through antibiotic ablation of the bacteria (209).
The potentials of our microbiome can be harnessed to treat/prevent TNBC through therapeutic/life-style interventions that modulate our microbiome composition. Additionally, these commensals or their by-products can be used to develop early TNBC detection systems or to build bacterial based therapies. See Figure 3.
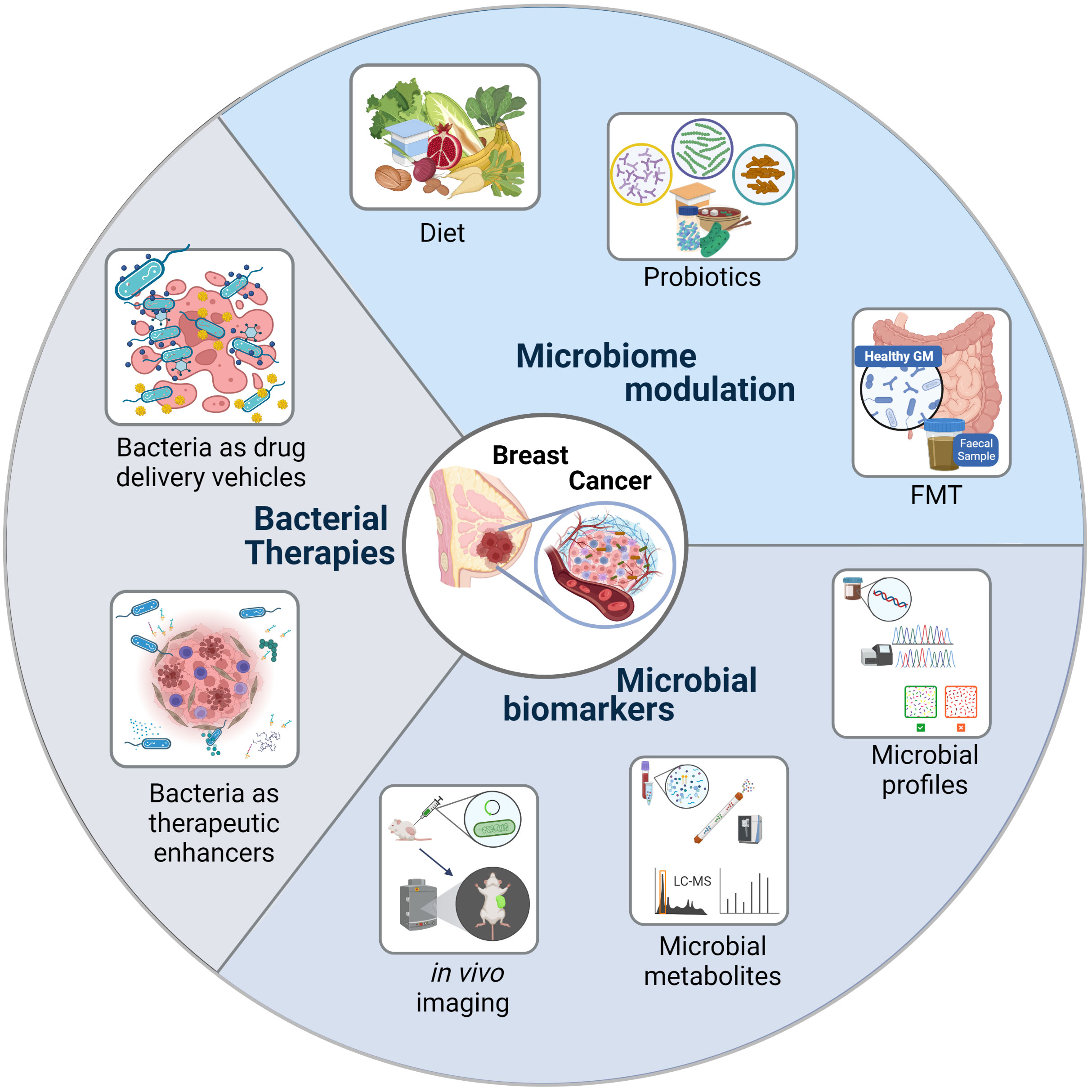
Figure 3 Bacteria as Cancer Theranostics. Harnessing the potentials of the microbiome to treat/prevent triple negative breast cancer.
Prevention is better than cure; therefore, maintaining eubiosis through diet is extremely important. A western diet rich in fat, sugar and low in fiber combined with a sedentary lifestyle has been associated with a higher risk of BC (210). Obesity and gut dysbiosis go hand in hand, with both being associated with chronic inflammation, increased risk of developing BC and failure of cancer immunotherapy (211). In a mouse model of TNBC where obesity was induced via a western diet and compared with lean mice, tumor volumes throughout the experiment were significantly higher in obese animals, as expected, with an association between obesity and enhanced TNBC growth, with significant loss of diversity in the GM, showing a decrease in Bacteroides species, particularly Alistipes (211). Conflicting findings suggest that Alistipes exerts beneficial effects in colitis, has been associated with the increased efficacy of ICI and general activation of innate immunity whereas it has been found to be pathogenic in colorectal cancer (212).
Diet is crucially important in the interplay between the gut microbiome and estrogen metabolism, thereby affecting breast cancer metastasis depending to various degrees on breast cancer type, as a western diet is associated with increased levels of β-glucuronidase feeding estrogens back into the bloodstream. Low fiber has a compounding effect with a reduction in SCFAs such as butyrate which can help protect gut barrier maintenance and reduce inflammation, thereby adding to the pro-tumorigenic effect (213). Increased levels of inflammatory proteins systemically increase insulin resistance and leptin levels, both of which promote carcinogenesis (214). A reduction in adiponectin from adipose tissue contributed to insulin resistance and increased insulin-like growth factor 1 (IGF-1) levels, which can elicit increased cell proliferation (215).
Conversely, a diet high in fiber reduces β-glucuronidase expression, lowers systemic estrogen and increases sex hormone-binding globulin (SGBH) levels with fecal excretion of estrogen (213). A high fiber diet can also drive increased intestinal alkaline phosphatase production, essential for gut barrier integrity (216). Butyrate is also known to inhibit histone deacetylase and tumor progression (217). Retinoic acid derived from vitamin A combined with trichostatin (a synthetic HDACi) has been found to increase the HDAC inhibitory effect in murine xenograft models of BC (218). A diet rich in fiber and polyphenols has been found to enhance the BC survival rate (219, 220).
Probiotics can inhibit pathogenic bacteria colonization, help maintain eubiosis, promote gut barrier maintenance, reduce gut and systemic inflammation, and enhance immune and treatment response to BC (221–223). An in vitro study using the BC MCF-7 cell line found significant inhibition of cell proliferation, increased levels of apoptosis, and cell cycle arrest when using live, heat-killed cells (HKC) or cytoplasmic fractions (CF) of E. fecalis and S. hominis isolated from the breast milk of healthy women (224). Oral supplementation with a probiotic containing Lactobacillus reuteri alone was sufficient to inhibit BC tumorigenesis in murine models genetically predisposed to neoplasia and fed a cancer-promoting western diet through microbially-initiated CD4+/CD25+ lymphocytes (225). Moreover, researchers have found that oral administration of L. acidophilus can modulate immune responses via stimulation of IL-12 and promotion of Th1 production, a potent activator of NK cells, in a murine xenograft model of a breast adenocarcinoma (226). A further in vivo study showed that drinking probiotics containing Lactobacillus helveticus increased IL-10 and decreased IL-6 production in mice, which is vital in BC inhibition (227). A Japanese population-based case-control study involving 306 cases of BC and 662 controls found that regular consumption of probiotics containing Lactobacillus casei was inversely associated with BC incidence (228). Probiotics have also been found as an alternative to antibiotic treatment in cases of mastitis during breast feeding (158, 229, 230).
The most radical yet efficient means to modify the gut microbiome involves fecal microbial transplantation (FMT), a black-box approach, whereby the underlying mechanisms do not need to be fully understood for efficacy to be evaluated. The entire GM from a healthy donor, usually an exceptional responder to the treatment being applied, is transplanted into a recipient undergoing the same treatment. This technique has successfully reversed resistance to ICI treatment in two recent metastatic melanoma trials, associated with an increased abundance of Ruminococcaceae and Bifidobacteriaceae (231, 232). Besides FMT, where the entire donor GM is transplanted, other approaches focus on transplanting coalitions of bacteria (233) or even components of strain-specific bacteria in the case of Enterococcus gallinarum flagellin have been used (234).
Microbial signatures associated with reduced treatment-related toxicity can be taken advantage of using FMT or combinations of specific bacteria in all types of cancer treatments. FMT has been found to alleviate undesirable harmful effects of 5-fluorouracil-based chemotherapy in murine models of colorectal cancer (235). In pre-clinical models, FMT and indole 3-propionic acid have been observed to reduce radiation-associated toxicity (236, 237). Regarding BC in general and, more specifically, TNBC, these studies still need to be conducted.
One recent study found the GM composition at BC diagnosis can serve as a prognostic marker. That α diversity was not predictive of favorable BC prognosis or side effects, and β diversity of the GM was associated with tumor grading but not BC subtype. Furthermore, the relative dominance of C. bolteae, C. asparagiforme, and B. uniformis in stool samples was associated with axillary lymph node invasion (238). It is now evident that the GM can affect all types of cancer treatments. This implies that bacteria associated with positive treatment outcomes can be used as biomarkers of treatment efficacy and toxicity, allowing for the much-touted advent of precise, tailored, personalised treatments (239–241).
Timing is everything or could play a role in chemotherapeutic treatment; one study using metronomic chemotherapy of capecitabine which is lower doses at more frequent intervals, showed promising results with reduced toxic side effects and less drastic changes to GM diversity. They found Blautia obeum to be associated with significantly prolonged PFS and significantly progression-free survival (PFS) with the occurrence of the Slackia genus (242). While chemotherapy can change the bacterial diversity, specific microbiome composition can, in turn, modify the efficacy of chemotherapy. Therefore, it is reasonable that particular probiotic concoctions could be administered adjunctively during chemotherapeutic treatment to augment effectiveness (221).
Oral administration of Bifidobacterium longum RAPO and ICI enhanced anti-PD-1 efficacy in preclinical murine models of TNBC (243). CRISPR-based phage therapy has recently been deployed to selectively wipe out nosocomial Clostridioides difficile infections, which are very difficult to treat without causing massive collateral damage to the patient’s beneficial GM (244). This approach used in TNBC treatment, combined with restoring lost valuable members of the GM, could synergistically rejuvenate patient GM functions and pave the way for establishing the best possible microbial amalgam for positive treatment outcomes. This approach has successfully been applied in murine models of atherosclerosis, with informed modification of the GM using small peptides to remodel the GM from that of one associated with a western diet to that of a low-fat-diet, resulting in reduced atherosclerotic plaques and a lowered pro-inflammatory cytokine profile (245).
In 1813, Vautier observed that cancer patients infected with Clostridium perfringens who developed gas gangrene appeared to be cured of cancer (246). Subsequently, bacterial infections and their effects on cancer were observed over 150 years ago by two German physicians W. Busch in 1868, and F. Fehleisen in 1882 (247, 248), who both independently found improvements in the condition of their patient’s symptoms, after a Streptococcus pyogenes infection. The first attempt to utilize bacteria as a cancer treatment followed shortly after, in 1893, when Dr William Coley combined the lytic compounds of S. pyogenes and Serratia marcescens and injected the mixture named Coley’s toxin into the tumor tissue with partial success in some patients (249).
There is a tricky balance when using bacteria in any therapy to get an effective therapeutic dose without toxic side effects on normal tissues. The advantage of using bacteria to deliver a genetic therapeutic cargo is their natural tumor tropism when systemically administered with high levels of local replication and the ability to persist within the TME. Furthermore, invasive species can invade tumor cells and, upon bacterial lysis, deliver a therapeutic DNA payload in the form of a bacterial plasmid which can be expressed directly in the tumor cell (250). Various payloads can allow for the direct killing of tumor cells or immunomodulation of the host immune system, increasing tumor antigenicity and promoting clearance by the immune system (251). Non-pathogenic Salmonella engineered with a quorum-sensing (QS) switch naturally hone to tumors and only express their therapeutic payload specifically within the TME once they have reached a particular critical density, thereby destroying cancerous tissue only (252). In preclinical models, activation of TLR5 by S. typhimurium flagellin in BC cells activates innate pro-inflammatory response for effective anti-tumor clearing (253). Bacteria can also be engineered to produce intrinsic bacterial biomolecules with known tumoricidal effects, some of which have been proven effective in human breast cancer cell lines (see Supplementary Table 1). These biomolecules include bacterial peptides, bacteriocin compounds, enzymes, or toxins.
Despite the large potential of bacteria-based mediated cancer treatments, the risks of adverse unmanageable side effects have tempered their use. Currently, the BCG vaccine is the standard for treating patients suffering from the Non-muscle invasive bladder cancer (254). 50 to 70% of patients have a positive outcome, with approximately 5% suffering adverse effects, including sepsis (255). Clinical trials with positive results include intra-tumoral administration with spores of the attenuated strain of Clostridium novyi (C. novyi-NT) to treat one patient suffering from advanced leiomyosarcoma, with promising results (256). Attenuated L. monocytogenes have also been found to be safe and effective in treating patients with advanced mesothelioma, lung, pancreatic, and ovarian cancers (257, 258). Pre-clinical studies using Bifidobacterium expressing cytosine deaminase (CD), which converted prodrug 5-fluorocytosine (5-FC) into chemotherapeutic agent (5-FU), was found to be effective in treating breast cancer when administered systemically in animal models (259–261).
The associations between TNBC and the microbiota are intricate, not yet fully understood, but undeniable. It is still not apparent whether changes in the GM and BM microbial composition are drivers of carcinogenesis or a response to tumor development. The effects are bidirectional, making it much more challenging to tease out these complex relationships. Furthermore, another issue is that these studies are just a snapshot in time. Bacteria have a short lifespan and quick generation time, changing over the course of the day with the body’s own circadian rhythms. Guilty microbes may have been and gone, whilst their effects rage on, especially in the case of DNA damage or epigenetic changes to the hosts’ genome, enabling cancer progression. Context is also critical concerning space and time; a bacterium in the gut may be beneficial but, when found in the TME, may exert the opposite effect. This also applies to distinct types of cancer. Bacterial metabolites can function as hormones, are highly pleiotropic and context-dependent, and can be produced distal to the tumor site while having systemic immune-modulatory effects.
Despite associations having been found in various microbiome-wide association studies, very little is known regarding the actual biochemical mechanisms. In the context of TNBC, even less is known, but lessons learned from studies involving other BC types are undoubtedly valuable. FMT also shows that even without the underlying mechanisms being fully understood, bacteria can still be used safely to improve treatment efficacy and reduce the toxic side effects of all types of cancer treatments currently used in TNBC.
More extensive clinical studies explicitly concerning TNBC are required to further elucidate the microbiome’s role and see past all of the confounding factors. Indeed, dietary changes should be taken on board to prevent cancer first, with probiotics (symbiont microbes) playing a role in returning the body to a state of normobiosis. Known associations can now be used as therapeutic and prognostic biomarkers, with enormous potential. The promise of completely personalized theranostics, however, is still a way off. Bacteriotherapy beyond FMT is further away, with plenty of evidence in in vitro and murine studies but lacking any clinical trials because of safety concerns. Genetic manipulation to reduce potential risks continues to make progress which, when matched to bacteria’s ability to selectively replicate within the TME, may in time overcome treatment hesitancy.
Indeed, monitoring of TNBC patients’ microbiomes may become standard practice where feasible, with modulation to a more beneficial state where possible. Altering the microbiome has potential to be the most unintrusive and safe means to change the body’s metabolism, improve treatment efficacy and reduce toxic side effects. Large-scale profiling of the GM and the associated metabolome of TNBC patients relative to that of healthy individuals will allow the development of theranostic biomarkers and treatments to improve the clinical prognosis and quality of life for TNBC patients.
CD and YFB wrote the manuscript, CD prepared the tables and YFB the figures. MT revised the manuscript. All authors contributed to the article and approved the submitted version.
The authors wish to acknowledge support relevant to this manuscript from Science Foundation Ireland (18/SP/3522) and Breakthrough Cancer Research, as part of the Precision Oncology Ireland consortium.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1020121/full#supplementary-material
1. National Cancer Institute (NCI). Surveillance epidemiology, end results (SEER) program: SEER*Stat database: Incidence - SEER research data, Nov 2018 Sub (1975-2016) <Katrina/Rita population adjustment> - linked to county attributes - total U.S., 1969-2017 counties, national cancer institute, DCCPS, surveillance research program. November 2018;based on the November 2018 submission. Bethesda, MD: NCI (2018).
2. Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast. (2022) 66:15–23. doi: 10.1016/j.breast.2022.08.010
3. Siddharth S, Sharma D. Racial disparity and triple-negative breast cancer in African-American women: A multifaceted affair between obesity, biology, and socioeconomic determinants. Cancers (Basel) (2018) 10(12):5. doi: 10.3390/cancers10120514
4. Kohler BA, Sherman RL, Howlader N, Jemal A, Ryerson AB, Henry KA, et al. Annual report to the nation on the status of cancer, 1975-2011, featuring incidence of breast cancer subtypes by race/ethnicity, poverty, and state. J Natl Cancer Instit (2015) 107(6):djv048. doi: 10.1093/jnci/djv048
5. Nwagu GC, Bhattarai S, Swahn M, Ahmed S, Aneja R. Prevalence and mortality of triple-negative breast cancer in West Africa: Biologic and sociocultural factors. JCO Global Oncol (2021) 2021(7):1129–40. doi: 10.1200/GO.21.00082
6. Derakhshan F, Reis-Filho JS. Pathogenesis of triple-negative breast cancer. Annu Rev Pathol (2022) 17:181–204. doi: 10.1146/annurev-pathol-042420-093238
7. Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PloS One (2016) 11(6):e0157368. doi: 10.1371/journal.pone.0157368
8. MacDonald I, Nixon NA, Khan OF. Triple-negative breast cancer: A review of current curative intent therapies. Curr Oncol (2022) 29(7):4768–78. doi: 10.3390/curroncol29070378
9. Li Y, Zhang H, Merkher Y, Chen L, Liu N, Leonov S, et al. Recent advances in therapeutic strategies for triple-negative breast cancer. J Hematol Oncol (2022) 15(1):121. doi: 10.1186/s13045-022-01341-0
10. Liu X, Wu K, Zheng D, Luo C, Fan Y, Zhong X, et al. Efficacy and safety of PARP inhibitors in advanced or metastatic triple-negative breast cancer: A systematic review and meta-analysis. Front Oncol (2021) 11. doi: 10.3389/fonc.2021.742139
11. Almansour NM. Triple-negative breast cancer: A brief review about epidemiology, risk factors, signaling pathways, treatment and role of artificial intelligence. Front Mol Biosci (2022) 9. doi: 10.3389/fmolb.2022.836417
12. Koster KL, Huober J, Joerger M. New antibody-drug conjugates (ADCs) in breast cancer-an overview of ADCs recently approved and in later stages of development. Explor Target Anti Ther (2022) 3(1):27–36. doi: 10.37349/etat.2022.00069
13. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PloS Biol (2016) 14(8):e1002533. doi: 10.1371/journal.pbio.1002533
14. Link CD. Is there a brain microbiome? Neurosci Insights (2021) 16:26331055211018709. doi: 10.1177/26331055211018709
15. Whipps JM, Lewis K, Cooke R. Mycoparasitism and plant disease control. Fungi Biol control Syst (1988) 1988:161–87.
16. Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev (2012) 70(suppl_1):S38–44. doi: 10.1111/j.1753-4887.2012.00493.x
17. Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded human microbiome project. Nature. (2017) 550(7674):61–6. doi: 10.1038/nature23889
18. Amato KR, Arrieta M-C, Azad MB, Bailey MT, Broussard JL, Bruggeling CE, et al. The human gut microbiome and health inequities. Proc Natl Acad Sci (2021) 118(25):e2017947118. doi: 10.1073/pnas.2017947118
19. Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol (2015) 21(29):8787–803. doi: 10.3748/wjg.v21.i29.8787
20. Appleton J. The gut-brain axis: Influence of microbiota on mood and mental health. Integr Med (Encinitas) (2018) 17(4):28–32.
21. Dash S, Syed YA, Khan MR. Understanding the role of the gut microbiome in brain development and its association with neurodevelopmental psychiatric disorders. Front Cell Dev Biol (2022) 10:880544. doi: 10.3389/fcell.2022.880544
22. Mishra A, Lai GC, Yao LJ, Aung TT, Shental N, Rotter-Maskowitz A, et al. Microbial exposure during early human development primes fetal immune cells. Cell. (2021) 184(13):3394–409.e20. doi: 10.1016/j.cell.2021.04.039
23. Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends Mol Med (2015) 21(2):109–17. doi: 10.1016/j.molmed.2014.12.002
24. Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. (2012) 488(7410):178–84. doi: 10.1038/nature11319
25. Rodríguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis (2015) 26:26050–. doi: 10.3402/mehd.v26.26050
26. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. (2012) 489(7415):220–30. doi: 10.1038/nature11550
27. Eisenstein M. The hunt for a healthy microbiome. Nature. (2020) 577(7792):S6–8. doi: 10.1038/d41586-020-00193-3
28. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J (2017) 474(11):1823–36. doi: 10.1042/BCJ20160510
30. Wilmanski T, Diener C, Rappaport N, Patwardhan S, Wiedrick J, Lapidus J, et al. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat Metab (2021) 3(2):274–86. doi: 10.1038/s42255-021-00348-0
31. Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. Modulation of the gut microbiota to improve innate resistance. Curr Opin Immunol (2018) 54:137–44. doi: 10.1016/j.coi.2018.08.003
32. Ge Y, Wang X, Guo Y, Yan J, Abuduwaili A, Aximujiang K, et al. Gut microbiota influence tumor development and alter interactions with the human immune system. J Exp Clin Cancer Res (2021) 40(1):42. doi: 10.1186/s13046-021-01845-6
33. Mezouar S, Chantran Y, Michel J, Fabre A, Dubus J-C, Leone M, et al. Microbiome and the immune system: From a healthy steady-state to allergy associated disruption. Hum Microbio J (2018) 10:11–20. doi: 10.1016/j.humic.2018.10.001
34. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. (2014) 157(1):121–41. doi: 10.1016/j.cell.2014.03.011
35. Claud EC, Walker WA. Chapter 5 - the intestinal microbiota and the microbiome. In: Polin RA, Neu J, editors. Gastroenterology and nutrition: Neonatology questions and controversies. Philadelphia, U.S.A.: W.B. Saunders, Elsevier (2008). p. 73–92.
36. Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol (2014) 16(7):1024–33. doi: 10.1111/cmi.12308
37. Toor D, Wsson MK, Kumar P, Karthikeyan G, Kaushik NK, Goel C, et al. Dysbiosis disrupts gut immune homeostasis and promotes gastric diseases. Int J Mol Sci (2019) 20(10):6–9. doi: 10.3390/ijms20102432
38. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol (2011) 30(1):16–34. doi: 10.3109/08830185.2010.529976
39. Janeway CA Jr.Approaching the asymptote? evolution and revolution in immunology Cold Spring Harbor Symp quantitative Biol (1989) 54 Pt 1:1–13 doi: 10.1101/SQB.1989.054.01.003
40. Medzhitov R, Janeway CA. Innate immunity: impact on the adaptive immune response. Curr Opin Immunol (1997) 9(1):4–9. doi: 10.1016/S0952-7915(97)80152-5
41. Gorjifard S, Goldszmid RS. Microbiota–myeloid cell crosstalk beyond the gut. J Leukocyte Biol (2016) 100(5):865–79. doi: 10.1189/jlb.3RI0516-222R
42. Li D, Wu M. Pattern recognition receptors in health and diseases. Signal Transduct Target Ther (2021) 6(1):291. doi: 10.1038/s41392-021-00687-0
43. Amarante-Mendes GP, Adjemian S, Branco LM, Zanetti LC, Weinlich R, Bortoluci KR. Pattern recognition receptors and the host cell death molecular machinery. Front Immunol (2018) 9. doi: 10.3389/fimmu.2018.02379
44. Fan Y, He S. The characteristics of tumor microenvironment in triple negative breast cancer. Cancer Manag Res (2022) 14:1–17. doi: 10.2147/CMAR.S316700
45. Chan ASH, Kangas TO, Qiu X, Uhlik MT, Fulton RB, Ottoson NR, et al. Imprime PGG enhances anti-tumor effects of tumor-targeting, anti-angiogenic, and immune checkpoint inhibitor antibodies. Front Oncol (2022) 12. doi: 10.3389/fonc.2022.869078
47. Mantovani A. Inflammation by remote control. Nature. (2005) 435(7043):752–3. doi: 10.1038/435752a
48. Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev (2018) 32(19-20):1267–84. doi: 10.1101/gad.314617.118
49. Singh N, Baby D, Rajguru JP, Patil PB, Thakkannavar SS, Pujari VB. Inflammation and cancer. Ann Afr Med (2019) 18(3):121–6. doi: 10.4103/aam.aam_56_18
50. Francescone R, Hou V, Grivennikov SI. Microbiome, inflammation, and cancer. Cancer J (2014) 20(3):181–9. doi: 10.1097/PPO.0000000000000048
51. Serhan CN, Ward PA, Gilroy DW, Ayoub SS. Fundamentals of inflammation. New York, United States: Cambridge University Press (2010).
52. Abudukelimu A, Barberis M, Redegeld FA, Sahin N, Westerhoff HV. Predictable irreversible switching between acute and chronic inflammation. Front Immunol (2018) 9:1596. doi: 10.3389/fimmu.2018.01596
53. Roh JS, Sohn DH. Damage-associated molecular patterns in inflammatory diseases. Immune Netw (2018) 18(4):e27. doi: 10.4110/in.2018.18.e27
54. Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukocyte Biol (2007) 81(1):1–5. doi: 10.1189/jlb.0306164
55. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer (2013) 13(11):800–12. doi: 10.1038/nrc3610
56. Thoo L, Noti M, Krebs P. Keep calm: the intestinal barrier at the interface of peace and war. Cell Death Disease (2019) 10(11):849. doi: 10.1038/s41419-019-2086-z
57. Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. (2004) 303(5664):1662–5. doi: 10.1126/science.1091334
58. Paray BA, Albeshr MF, Jan AT, Rather IA. Leaky gut and autoimmunity: An intricate balance in individuals health and the diseased state. Int J Mol Sci (2020) 21(24):4. doi: 10.3390/ijms21249770
59. Murch S. 5 - gastrointestinal mucosal immunology and mechanisms of inflammation. In: Wyllie R, Hyams JS, Kay M, editors. Pediatric gastrointestinal and liver disease (Sixth edition). Philadelphia: Elsevier (2021). p. 40–52.e3.
60. Stolfi C, Maresca C, Monteleone G, Laudisi F. Implication of intestinal barrier dysfunction in gut dysbiosis and diseases. Biomedicines. (2022) 10(2):289. doi: 10.3390/biomedicines10020289
61. Genua F, Raghunathan V, Jenab M, Gallagher WM, Hughes DJ. The role of gut barrier dysfunction and microbiome dysbiosis in colorectal cancer development. Front Oncol (2021) 11:626349. doi: 10.3389/fonc.2021.626349
62. Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol (2017) 14(9):527–39. doi: 10.1038/nrgastro.2017.72
63. Akshintala VS, Talukdar R, Singh VK, Goggins M. The gut microbiome in pancreatic disease. Clin Gastroenterol Hepatol (2019) 17(2):290–5. doi: 10.1016/j.cgh.2018.08.045
64. Tilg H, Zmora N, Adolph TE, Elinav E. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol (2020) 20(1):40–54. doi: 10.1038/s41577-019-0198-4
65. Kinashi Y, Hase K. Partners in leaky gut syndrome: Intestinal dysbiosis and autoimmunity. Front Immunol (2021) 12. doi: 10.3389/fimmu.2021.673708
66. Ruo SW, Alkayyali T, Win M, Tara A, Joseph C, Kannan A, et al. Role of gut microbiota dysbiosis in breast cancer and novel approaches in prevention, diagnosis, and treatment. Cureus. (2021) 13(8):e17472. doi: 10.7759/cureus.17472
67. Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, et al. Inflammation is associated with worse outcome in the whole cohort but with better outcome in triple-negative subtype of breast cancer patients. J Immunol Res (2020) 2020:5618786. doi: 10.1155/2020/5618786
68. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. (2017) 551(7682):648–52. doi: 10.1038/nature24661
69. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. (2012) 336(6086):1262–7. doi: 10.1126/science.1223813
70. Agus A, Clément K, Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. (2021) 70(6):1174–82. doi: 10.1136/gutjnl-2020-323071
71. Russo J, Russo IH. The role of estrogen in the initiation of breast cancer. J Steroid Biochem Mol Biol (2006) 102(1-5):89–96. doi: 10.1016/j.jsbmb.2006.09.004
72. Fuhrman BJ, Feigelson HS, Flores R, Gail MH, Xu X, Ravel J, et al. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab (2014) 99(12):4632–40. doi: 10.1210/jc.2014-2222
73. Treeck O, Schüler-Toprak S, Ortmann O. Estrogen actions in triple-negative breast cancer. Cells. (2020) 9(11):3–8. doi: 10.3390/cells9112358
74. Hsu LH, Chu NM, Lin YF, Kao SH. G-Protein coupled estrogen receptor in breast cancer. Int J Mol Sci (2019) 20(2):3–5. doi: 10.3390/ijms20020306
75. Aspros KGM, Carter JM, Hoskin TL, Suman VJ, Subramaniam M, Emch MJ, et al. Estrogen receptor beta repurposes EZH2 to suppress oncogenic NFκB/p65 signaling in triple negative breast cancer. NPJ Breast Cancer (2022) 8(1):20. doi: 10.1038/s41523-022-00387-0
76. Rajapaksa G, Nikolos F, Bado I, Clarke R, Gustafsson J-Å, Thomas C. ERβ decreases breast cancer cell survival by regulating the IRE1/XBP-1 pathway. Oncogene. (2015) 34(31):4130–41. doi: 10.1038/onc.2014.343
77. Yang SH, Liu R, Perez EJ, Wen Y, Stevens SM Jr., Valencia T, et al. Mitochondrial localization of estrogen receptor beta. Proc Natl Acad Sci United States America (2004) 101(12):4130–5. doi: 10.1073/pnas.0306948101
78. Song I-S, Jeong YJ, Jeong SH, Kim JE, Han J, Kim T-H, et al. Modulation of mitochondrial ERβ expression inhibits triple-negative breast cancer tumor progression by activating mitochondrial function. Cell Physiol Biochem (2019) 52:468–85. doi: 10.33594/000000034
79. Bado I, Nikolos F, Rajapaksa G, Gustafsson J, Thomas C. ERβ decreases the invasiveness of triple-negative breast cancer cells by regulating mutant p53 oncogenic function. Oncotarget. (2016) 7(12):13599–611. doi: 10.18632/oncotarget.7300
80. Reese JM, Bruinsma ES, Nelson AW, Chernukhin I, Carroll JS, Li Y, et al. ERβ-mediated induction of cystatins results in suppression of TGFβ signaling and inhibition of triple-negative breast cancer metastasis. Proc Natl Acad Sci (2018) 115(41):E9580–E9. doi: 10.1073/pnas.1807751115
81. Li K, Zong D, Sun J, Chen D, Ma M, Jia L. Rewiring of the endocrine network in triple-negative breast cancer. Front Oncol (2022) 12:830894. doi: 10.3389/fonc.2022.830894
82. Pellock SJ, Redinbo MR. Glucuronides in the gut: Sugar-driven symbioses between microbe and host. J Biol Chem (2017) 292(21):8569–76. doi: 10.1074/jbc.R116.767434
83. Shen R-L, Dang X-Y, Dong J-L, Hu X-Z. Effects of oat β-glucan and barley β-glucan on fecal characteristics, intestinal microflora, and intestinal bacterial metabolites in rats. J Agric Food Chem (2012) 60(45):11301–8. doi: 10.1021/jf302824h
84. McIntosh FM, Maison N, Holtrop G, Young P, Stevens VJ, Ince J, et al. Phylogenetic distribution of genes encoding β-glucuronidase activity in human colonic bacteria and the impact of diet on fecal glycosidase activities. Environ Microbiol (2012) 14(8):1876–87. doi: 10.1111/j.1462-2920.2012.02711.x
85. Gloux K, Berteau O, El Oumami H, Béguet F, Leclerc M, Doré J. A metagenomic β-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc Natl Acad Sci (2011) 108(supplement_1):4539–46. doi: 10.1073/pnas.1000066107
86. Parida S, Sharma D. The microbiome-estrogen connection and breast cancer risk. Cells. (2019) 8(12):8. doi: 10.3390/cells8121642
87. Possemiers S, Heyerick A, Robbens V, De Keukeleire D, Verstraete W. Activation of prestrogens from hops (Humulus lupulus l.) by intestinal microbiota; conversion of isoxanthohumol into 8-prenylnaringenin. J Agric Food Chem (2005) 53(16):6281–8. doi: 10.1021/jf0509714
88. Maruo T, Sakamoto M, Ito C, Toda T, Benno Y. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus eggerthella. Int J Syst Evol Microbiol (2008) 58(Pt 5):1221–7. doi: 10.1099/ijs.0.65404-0
89. Mayo B, Vázquez L, Flórez AB. Equol: A bacterial metabolite from the daidzein isoflavone and its presumed beneficial health effects. Nutrients. (2019) 11(9):1–4. doi: 10.3390/nu11092231
90. Lathrop KI, Kaklamani VG, Brenner AJ, Li R, Nazarullah A, Hackman S, et al. Novel estrogen receptor beta agonist s-equol decreases tumor proliferation in patients with triple negative breast cancer (TNBC). J Clin Oncol (2020) 38(15_suppl):560. doi: 10.1200/JCO.2020.38.15_suppl.560
91. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res (2013) 54(9):2325–40. doi: 10.1194/jlr.R036012
92. Levy M, Thaiss CA, Elinav E. Metabolites: messengers between the microbiota and the immune system. Genes Dev (2016) 30(14):1589–97. doi: 10.1101/gad.284091.116
93. Glozak M, Seto E. Histone deacetylases and cancer. Oncogene. (2007) 26(37):5420–32. doi: 10.1038/sj.onc.1210610
94. Liberti MV, Locasale JW. The warburg effect: How does it benefit cancer cells? Trends Biochem Sci (2016) 41(3):211–8. doi: 10.1016/j.tibs.2015.12.001
95. Salimi V, Shahsavari Z, Safizadeh B, Hosseini A, Khademian N, Tavakoli-Yaraki M. Sodium butyrate promotes apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment. Lipids Health Disease (2017) 16(1):208. doi: 10.1186/s12944-017-0593-4
96. Nakkarach A, Foo HL, Song AA-L, Mutalib NEA, Nitisinprasert S, Withayagiat U. Anti-cancer and anti-inflammatory effects elicited by short chain fatty acids produced by escherichia coli isolated from healthy human gut microbiota. Microb Cell Factor (2021) 20(1):36. doi: 10.1186/s12934-020-01477-z
97. Wu X, Wu Y, He L, Wu L, Wang X, Liu Z. Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. J Cancer (2018) 9(14):2510–7. doi: 10.7150/jca.25324
98. Rea D, Coppola G, Palma G, Barbieri A, Luciano A, Del Prete P, et al. Microbiota effects on cancer: from risks to therapies. Oncotarget. (2018) 9(25):17915. doi: 10.18632/oncotarget.24681
99. Le Poul E, Loison C, Struyf S, Springael J-Y, Lannoy V, Decobecq M-E, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem (2003) 278(28):25481–9. doi: 10.1074/jbc.M301403200
100. Thirunavukkarasan M, Wang C, Rao A, Hind T, Teo YR, Siddiquee AA, et al. Short-chain fatty acid receptors inhibit invasive phenotypes in breast cancer cells. PloS One (2017) 12(10):e0186334. doi: 10.1371/journal.pone.0186334
101. Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumors reveals novel subgroups. Nature. (2012) 486(7403):346–52. doi: 10.1038/nature10983
102. Ridlon JM, Bajaj JS. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm Sin B (2015) 5(2):99–105. doi: 10.1016/j.apsb.2015.01.006
103. Javitt NB, Budai K, Raju U, Levitz M, Miller D, Cahan A. Breast-gut connection: origin of chenodeoxycholic acid in breast cyst fluid. Lancet (1994) 343(8898):633–5. doi: 10.1016/S0140-6736(94)92635-2
104. Mikó E, Vida A, Kovács T, Ujlaki G, Trencsényi G, Márton J, et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim Biophys Acta (BBA)-Bioenerg (2018) 1859(9):958–74. doi: 10.1016/j.bbabio.2018.04.002
105. Sampsell K, Hao D, Reimer RA. The gut microbiota: A potential gateway to improved health outcomes in breast cancer treatment and survivorship. Int J Mol Sci (2020) 21(23):9239. doi: 10.3390/ijms21239239
106. Long SL, Gahan CG, Joyce SA. Interactions between gut bacteria and bile in health and disease. Mol aspects Med (2017) 56:54–65. doi: 10.1016/j.mam.2017.06.002
107. Wu R, Yu I, Tokumaru Y, Asaoka M, Oshi M, Ishikawa T, et al. Decreased bile acid metabolism and association with prognosis reflecting microbiome in tumor microenvironment involved in cancer cell proliferation in breast cancer. J Clin Oncol (2022) 40(16_suppl):e12539–e. doi: 10.1200/JCO.2022.40.16_suppl.e12539
108. Kovács P, Csonka T, Kovács T, Sári Z, Ujlaki G, Sipos A, et al. Lithocholic acid, a metabolite of the microbiome, increases oxidative stress in breast cancer. Cancers (Basel) (2019) 11(9):16–24. doi: 10.3390/cancers11091255
109. Tofalo R, Cocchi S, Suzzi G. Polyamines and gut microbiota. Front Nutr (2019) 6. doi: 10.3389/fnut.2019.00016
110. Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M. Remaining mysteries of molecular biology: the role of polyamines in the cell. J Mol Biol (2015) 427(21):3389–406. doi: 10.1016/j.jmb.2015.06.020
111. Kovács T, Mikó E, Vida A, Sebő É, Toth J, Csonka T, et al. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci Rep (2019) 9(1):1–14. doi: 10.1038/s41598-018-37664-7
112. Vattai A, Akyol E, Kuhn C, Hofmann S, Heidegger H, von Koch F, et al. Increased trace amine-associated receptor 1 (TAAR1) expression is associated with a positive survival rate in patients with breast cancer. J Cancer Res Clin Oncol (2017) 143(9):1637–47. doi: 10.1007/s00432-017-2420-8
113. Konopelski P, Mogilnicka I. Biological effects of indole-3-Propionic acid, a gut microbiota-derived metabolite, and its precursor tryptophan in mammals’ health and disease. Int J Mol Sci (2022) 23(3):7. doi: 10.3390/ijms23031222
114. Rothhammer V, Quintana FJ. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol (2019) 19(3):184–97. doi: 10.1038/s41577-019-0125-8
115. Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe (2018) 23(6):716–24. doi: 10.1016/j.chom.2018.05.003
116. Karbownik M, Stasiak M, Zasada K, Zygmunt A, Lewinski A. Comparison of potential protective effects of melatonin, indole-3-propionic acid, and propylthiouracil against lipid peroxidation caused by potassium bromate in the thyroid gland. J Cell Biochem (2005) 95(1):131–8. doi: 10.1002/jcb.20404
117. Kovács T, Mikó E, Ujlaki G, Yousef H, Csontos V, Uray K, et al. The involvement of oncobiosis and bacterial metabolite signaling in metastasis formation in breast cancer. Cancer Metastasis Rev (2021) 40(4):1223–49. doi: 10.1007/s10555-021-10013-3
118. Guerra FS, Dias FRF, Cunha AC, Fernandes PD. Benzo[f]indole-4,9-dione derivatives effectively inhibit the growth of triple-negative breast cancer. Molecules. (2021) 26(15):2–7. doi: 10.3390/molecules26154414
119. Urbaniak C, Cummins J, Brackstone M, Macklaim JM, Gloor GB, Baban CK, et al. Microbiota of human breast tissue. Appl Environ Microbiol (2014) 80(10):3007–14. doi: 10.1128/AEM.00242-14
120. Wang H, Altemus J, Niazi F, Green H, Calhoun BC, Sturgis C, et al. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget. (2017) 8(50):88122–38. doi: 10.18632/oncotarget.21490
121. Xuan C, Shamonki JM, Chung A, DiNome ML, Chung M, Sieling PA, et al. Microbial dysbiosis is associated with human breast cancer. PloS One (2014) 9(1):e83744. doi: 10.1371/journal.pone.0083744
122. Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The microbiota of breast tissue and its association with breast cancer. Appl Environ Microbiol (2016) 82(16):5039–48. doi: 10.1128/AEM.01235-16
123. Hogan G, Eckenberger J, Narayanen N, Walker SP, Claesson MJ, Corrigan M, et al. Biopsy bacterial signature can predict patient tissue malignancy. Sci Rep (2021) 11(1):18535. doi: 10.1038/s41598-021-98089-3
124. Eslami-S Z, Majidzadeh-A K, Halvaei S, Babapirali F, Esmaeili R. Microbiome and breast cancer: New role for an ancient population. Front Oncol (2020) 10(120). doi: 10.3389/fonc.2020.00120
125. Hunt KM, Foster JA, Forney LJ, Schütte UM, Beck DL, Abdo Z, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PloS One (2011) 6(6):e21313. doi: 10.1371/journal.pone.0021313
126. Lackey KA, Williams JE, Meehan CL, Zachek JA, Benda ED, Price WJ, et al. What’s normal? microbiomes in human milk and infant feces are related to each other but vary geographically: the INSPIRE study. Front Nutr (2019) 6:45. doi: 10.3389/fnut.2019.00045
127. OConnor H, MacSharry J, Bueso YF, Lindsay S, Kavanagh EL, Tangney M, et al. Resident bacteria in breast cancer tissue: pathogenic agents or harmless commensals? Discovery Med (2018) 26(142):93–102.
128. Perez PF, Doré J, Leclerc M, Levenez F, Benyacoub J, Serrant P, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics (2007) 119(3):e724–32. doi: 10.1542/peds.2006-1649
129. Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol (2001) 2(4):361–7. doi: 10.1038/86373
130. Cronin M, Morrissey D, Rajendran S, El Mashad SM, van Sinderen D, O’Sullivan GC, et al. Orally administered bifidobacteria as vehicles for delivery of agents to systemic tumors. Mol Ther (2010) 18(7):1397–407. doi: 10.1038/mt.2010.59
131. Sznol M, Lin SL, Bermudes D, Zheng LM, King I. Use of preferentially replicating bacteria for the treatment of cancer. J Clin Invest (2000) 105(8):1027–30. doi: 10.1172/JCI9818
132. Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer (2010) 10(11):785–94. doi: 10.1038/nrc2934
133. Morrissey D, O’Sullivan GC, Tangney M. Tumour targeting with systemically administered bacteria. Curr Gene Ther (2010) 10(1):3–14. doi: 10.2174/156652310790945575
134. Cummins J, Tangney M. Bacteria and tumours: causative agents or opportunistic inhabitants? Infect Agents Cancer (2013) 8(1):1–8. doi: 10.1186/1750-9378-8-11
135. Cummins J, Tangney M. Bacteria and tumours: causative agents or opportunistic inhabitants? Infect Agent Cancer (2013) 8(1):11. doi: 10.1186/1750-9378-8-11
136. Hanahan D. Hallmarks of cancer: New dimensions. Cancer Discover (2022) 12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059
137. Xie W, Huang Y, Xie W, Guo A, Wu W. Bacteria peptidoglycan promoted breast cancer cell invasiveness and adhesiveness by targeting toll-like receptor 2 in the cancer cells. PloS One (2010) 5(5):e10850. doi: 10.1371/journal.pone.0010850
138. Stevens DL, Bryant AE. Severe group a streptococcal infections. In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes: Basic biology to clinical manifestations. Oklahoma City (OK: University of Oklahoma Health Sciences Center © The University of Oklahoma Health Sciences Center (2016).
139. Lu W, He F, Lin Z, Liu S, Tang L, Huang Y, et al. Dysbiosis of the endometrial microbiota and its association with inflammatory cytokines in endometrial cancer. Int J Cancer (2021) 148(7):1708–16. doi: 10.1002/ijc.33428
140. Caselli E, Soffritti I, D’Accolti M, Piva I, Greco P, Bonaccorsi G. Atopobium vaginae and porphyromonas somerae induce proinflammatory cytokines expression in endometrial cells: A possible implication for endometrial cancer? Cancer Manag Res (2019) 11:8571–5. doi: 10.2147/CMAR.S217362
141. Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci Rep (2016) 6(1):30751. doi: 10.1038/srep30751
142. Thompson KJ, Ingle JN, Tang X, Chia N, Jeraldo PR, Walther-Antonio MR, et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PloS One (2017) 12(11):e0188873. doi: 10.1371/journal.pone.0188873
143. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discovery (2018) 8(4):403–16. doi: 10.1158/2159-8290.CD-17-1134
144. Cavarretta I, Ferrarese R, Cazzaniga W, Saita D, Lucianò R, Ceresola ER, et al. The microbiome of the prostate tumor microenvironment. Eur Urol (2017) 72(4):625–31. doi: 10.1016/j.eururo.2017.03.029
145. Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Alwine JC, et al. The ovarian cancer oncobiome. Oncotarget. (2017) 8(22):36225–45. doi: 10.18632/oncotarget.16717
146. Tzeng A, Sangwan N, Jia M, Liu C-C, Keslar KS, Downs-Kelly E, et al. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med (2021) 13(1):60–. doi: 10.1186/s13073-021-00874-2
147. Banerjee S, Wei Z, Tan F, Peck KN, Shih N, Feldman M, et al. Distinct microbiological signatures associated with triple negative breast cancer. Sci Rep (2015) 5:15162–. doi: 10.1038/srep15162
148. Banerjee S, Wei Z, Tian T, Bose D, Shih NNC, Feldman MD, et al. Prognostic correlations with the microbiome of breast cancer subtypes. Cell Death Disease (2021) 12(9):831. doi: 10.1038/s41419-021-04092-x
149. Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Peck KN, et al. Distinct microbial signatures associated with different breast cancer types. Front Microbiol (2018) 9:951. doi: 10.3389/fmicb.2018.00951
150. Fine DH, Kaplan JB, Kachlany SC, Schreiner HC. How we got attached to actinobacillus actinomycetemcomitans: A model for infectious diseases. Periodontol 2000 (2006) 42(1):114–57. doi: 10.1111/j.1600-0757.2006.00189.x
151. Goldberg MH, Katz J. Infective endocarditis caused by fastidious oro-pharyngeal HACEK micro-organisms. J Oral Maxillofac surg (2006) 64(6):969–71. doi: 10.1016/j.joms.2006.02.027
152. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science. (2020) 368(6494):973–80. doi: 10.1126/science.aay9189
153. Walker SP, Tangney M, Claesson MJ. Sequence-based characterization of intratumoral bacteria-a guide to best practice. Front Oncol (2020) 10:179. doi: 10.3389/fonc.2020.00179
154. Walker SP, Barrett M, Hogan G, Flores Bueso Y, Claesson MJ, Tangney M. Non-specific amplification of human DNA is a major challenge for 16S rRNA gene sequence analysis. Sci Rep (2020) 10(1):16356. doi: 10.1038/s41598-020-73403-7
155. Flores Bueso Y, Walker SP, Hogan G, Claesson MJ, Tangney M. Protoblock - a biological standard for formalin fixed samples. Microbiome. (2020) 8(1):122. doi: 10.1186/s40168-020-00901-1
156. Flores Bueso Y, Walker SP, Tangney M. Characterization of FFPE-induced bacterial DNA damage and development of a repair method. Biol Methods Protoc (2020) 5(1):bpaa015. doi: 10.1093/biomethods/bpaa015
157. Kaur S, Kaur S. Bacteriocins as potential anticancer agents. Front Pharmacol (2015) 6:272. doi: 10.3389/fphar.2015.00272
158. Jiménez E, Fernández L, Maldonado A, Martín R, Olivares M, Xaus J, et al. Oral administration of lactobacillus strains isolated from breast milk as an alternative for the treatment of infectious mastitis during lactation. Appl Environ Microbiol (2008) 74(15):4650–5. doi: 10.1128/AEM.02599-07
159. Srikham K, Daengprok W, Niamsup P, Thirabunyanon M. Characterization of streptococcus salivarius as new probiotics derived from human breast milk and their potential on proliferative inhibition of liver and breast cancer cells and antioxidant activity. Front Microbiol (2021) 12. doi: 10.3389/fmicb.2021.797445
160. Yaghoubi A, Khazaei M, Hasanian SM, Avan A, Cho WC, Soleimanpour S. Bacteriotherapy in breast cancer. Int J Mol Sci (2019) 20(23):5880. doi: 10.3390/ijms20235880
161. Cai Z, Sanchez A, Shi Z, Zhang T, Liu M, Zhang D. Activation of toll-like receptor 5 on breast cancer cells by flagellin suppresses cell proliferation and tumor growth. Cancer Res (2011) 71(7):2466–75. doi: 10.1158/0008-5472.CAN-10-1993
162. Li H, Du X, Yan L, Tang Z, Zhang L, Zheng Q, et al. Low abundance of lactococcus lactis in human colorectal cancer is associated with decreased natural killer cells. Nutr Cancer (2022) 74(3):938–46. doi: 10.1080/01635581.2021.1944649
163. Koller VJ, Marian B, Stidl R, Nersesyan A, Winter H, Simić T, et al. Impact of lactic acid bacteria on oxidative DNA damage in human derived colon cells. Food Chem Toxicol (2008) 46(4):1221–9. doi: 10.1016/j.fct.2007.09.005
164. Friedman GD, Oestreicher N, Chan J, Quesenberry CP Jr., Udaltsova N, Habel LA. Antibiotics and risk of breast cancer: Up to 9 years of follow-up of 2.1 million women. cancer epidemiol biomarkers prev. Cancer Epidemiology, Biomarkers & Prevention (2006) 15(11):2102–6. doi: 10.1158/1055-9965.EPI-06-0401
165. Velicer CM, Heckbert SR, Lampe JW, Potter JD, Robertson CA, Taplin SH. Antibiotic use in relation to the risk of breast cancer. Jama (2004) 291(7):827–35. doi: 10.1001/jama.291.7.827
166. Simin J, Tamimi RM, Engstrand L, Callens S, Brusselaers N. Antibiotic use and the risk of breast cancer: A systematic review and dose-response meta-analysis. Pharmacol Res (2020) 160:105072. doi: 10.1016/j.phrs.2020.105072
167. Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis. (2009) 30(7):1073–81. doi: 10.1093/carcin/bgp127
168. Wee ZN, Yatim SM, Kohlbauer VK, Feng M, Goh JY, Bao Y, et al. IRAK1 is a therapeutic target that drives breast cancer metastasis and resistance to paclitaxel. Nat Commun (2015) 6:8746. doi: 10.1038/ncomms9746
169. Wu K, Zhang H, Fu Y, Zhu Y, Kong L, Chen L, et al. TLR4/MyD88 signaling determines the metastatic potential of breast cancer cells. Mol Med Rep (2018) 18(3):3411–20. doi: 10.3892/mmr.2018.9326
170. Vitiello GA, Cohen DJ, Miller G. Harnessing the microbiome for pancreatic cancer immunotherapy. Trends Cancer (2019) 5(11):670–6. doi: 10.1016/j.trecan.2019.10.005
171. Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and mincle-induced immune suppression. Nature. (2016) 532(7598):245–9. doi: 10.1038/nature17403
172. Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, et al. Breast cancer colonization by fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun (2020) 11(1):3259. doi: 10.1038/s41467-020-16967-2
173. Rousseau A, Rio M-C, Alpy F. TRAF4, at the crossroad between morphogenesis and cancer. Cancers (Basel) (2011) 3(2):2734–49. doi: 10.3390/cancers3022734
174. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of fusobacterium persistence and antibiotic response in colorectal cancer. Science. (2017) 358(6369):1443–8. doi: 10.1126/science.aal5240
175. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
176. Park J, Wysocki RW, Amoozgar Z, Maiorino L, Fein MR, Jorns J, et al. Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci Trans Med (2016) 8(361):361ra138–361ra138. doi: 10.1126/scitranslmed.aag1711
177. Hendrickx W, Simeone I, Anjum S, Mokrab Y, Bertucci F, Finetti P, et al. Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. OncoImmunology. (2017) 6(2):e1253654. doi: 10.1080/2162402X.2016.1253654
178. García-Aranda M, Redondo M. Immunotherapy: a challenge of breast cancer treatment. Cancers (Basel) (2019) 11(12):1822. doi: 10.3390/cancers11121822
179. Dirix LY, Takacs I, Jerusalem G, Nikolinakos P, Arkenau HT, Forero-Torres A, et al. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN solid tumor study. Breast Cancer Res Treat (2018) 167(3):671–86. doi: 10.1007/s10549-017-4537-5
180. Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann oncol: Off J Eur Soc Med Oncol (2014) 25(8):1544–50. doi: 10.1093/annonc/mdu112
181. Qureshi S, Chan N, George M, Ganesan S, Toppmeyer D, Omene C. Immune checkpoint inhibitors in triple negative breast cancer: The search for the optimal biomarker. biomark Insights (2022) 17:11772719221078774. doi: 10.1177/11772719221078774
182. Kalaora S, Nagler A, Nejman D, Alon M, Barbolin C, Barnea E, et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature. (2021) 592(7852):138–43. doi: 10.1038/s41586-021-03368-8
183. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
184. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
185. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. (2018) 359(6371):104–8. doi: 10.1126/science.aao3290
186. Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia (2017) 19(10):848–55. doi: 10.1016/j.neo.2017.08.004
187. Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol (2017) 28(6):1368–79. doi: 10.1093/annonc/mdx108
188. Routy B, Le Chatelier E, Derosa L, Duong CP, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science. (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
189. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
190. Li W, Deng Y, Chu Q, Zhang P. Gut microbiome and cancer immunotherapy. Cancer Letters (2019) 447:41–7. doi: 10.1016/j.canlet.2019.01.015
191. Temraz S, Nassar F, Nasr R, Charafeddine M, Mukherji D, Shamseddine A. Gut microbiome: A promising biomarker for immunotherapy in colorectal cancer. Int J Mol Sci (2019) 20(17):4155. doi: 10.3390/ijms20174155
192. Fessler J, Matson V, Gajewski TF. Exploring the emerging role of the microbiome in cancer immunotherapy. J ImmunoTher Cancer (2019) 7(1):108. doi: 10.1186/s40425-019-0574-4
193. Shui L, Yang X, Li J, Yi C, Sun Q, Zhu H. Gut microbiome as a potential factor for modulating resistance to cancer immunotherapy. Front Immunol (2020) 10(2989). doi: 10.3389/fimmu.2019.02989
194. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
195. Naidoo J, Cappelli LC, Forde PM, Marrone KA, Lipson EJ, Hammers HJ, et al. Inflammatory arthritis: a newly recognized adverse event of immune checkpoint blockade. oncol (2017) 22(6):627–30. doi: 10.1634/theoncologist.2016-0390
196. Martins F, Sofiya L, Sykiotis GP, Lamine F, Maillard M, Fraga M, et al. Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat Rev Clin Oncol (2019) 16(9):563–80. doi: 10.1038/s41571-019-0218-0
197. McLaughlin MM, Dacquisto MP, Jacobus DP, Horowitz RE. Effects of the germfree state on responses of mice to whole-body irradiation. Radiat Res (1964) 23(3):333–49. doi: 10.2307/3571614
198. Touchefeu Y, Montassier E, Nieman K, Gastinne T, Potel G, Bruley des Varannes S, et al. Systematic review: the role of the gut microbiota in chemotherapy-or radiation-induced gastrointestinal mucositis–current evidence and potential clinical applications. Alimentary Pharmacol Ther (2014) 40(5):409–21. doi: 10.1111/apt.12878
199. Wang A, Ling Z, Yang Z, Kiela PR, Wang T, Wang C, et al. Gut microbial dysbiosis may predict diarrhea and fatigue in patients undergoing pelvic cancer radiotherapy: a pilot study. PloS One (2015) 10(5):e0126312. doi: 10.1371/journal.pone.0126312
200. Rahal OM, Wolfe AR, Mandal PK, Larson R, Tin S, Jimenez C, et al. Blocking interleukin (IL) 4-and IL13-mediated phosphorylation of STAT6 (Tyr641) decreases M2 polarization of macrophages and protects against macrophage-mediated radioresistance of inflammatory breast cancer. Int J Radiat Oncol Biol Physics (2018) 100(4):1034–43. doi: 10.1016/j.ijrobp.2017.11.043
201. Uribe-Herranz M, Rafail S, Beghi S, Gil-de-Gómez L, Verginadis I, Bittinger K, et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J Clin Invest (2020) 130(1):466–79. doi: 10.1172/JCI124332
202. Xiao H-W, Cui M, Li Y, Dong J-L, Zhang S-Q, Zhu C-C, et al. Gut microbiota-derived indole 3-propionic acid protects against radiation toxicity via retaining acyl-CoA-binding protein. Microbiome. (2020) 8(1):69. doi: 10.1186/s40168-020-00845-6
203. Reis Ferreira M, Andreyev HJN, Mohammed K, Truelove L, Gowan SM, Li J, et al. Microbiota- and radiotherapy-induced gastrointestinal side-effects (MARS) study: A Large pilot study of the microbiome in acute and late-radiation enteropathy. Clin Cancer research: an Off J Am Assoc Cancer Res (2019) 25(21):6487–500. doi: 10.1158/1078-0432.CCR-19-0960
204. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. (2013) 342(6161):967–70. doi: 10.1126/science.1240527
205. Armstrong H, Bording-Jorgensen M, Dijk S, Wine E. The complex interplay between chronic inflammation, the microbiome, and cancer: understanding disease progression and what we can do to prevent it. Cancers (Basel) (2018) 10(3):83. doi: 10.3390/cancers10030083
206. Lehouritis P, Cummins J, Stanton M, Murphy CT, McCarthy FO, Reid G, et al. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci Rep (2015) 5(1):1–12. doi: 10.1038/srep14554
207. Lehouritis P, Stanton M, McCarthy FO, Jeavons M, Tangney M. Activation of multiple chemotherapeutic prodrugs by the natural enzymolome of tumour-localised probiotic bacteria. J Control Release (2016) 222:9–17. doi: 10.1016/j.jconrel.2015.11.030
208. Swoboda AT, Sharma A, Olopade OI, Nanda R, Gilbert J. Characterizing the gut microbiome of patients with triple-negative breast cancer. J Clin Oncol (2019) 37(15_suppl):e14186–e. doi: 10.1200/JCO.2019.37.15_suppl.e14186
209. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. (2017) 357(6356):1156–60. doi: 10.1126/science.aah5043
210. Bodai BI, Nakata TE. Breast cancer: Lifestyle, the human gut Microbiota/Microbiome, and survivorship. Perm J (2020) 24:19.129. doi: 10.7812/TPP/19.129
211. Hossain F, Majumder S, David J, Bunnell BA, Miele L. Obesity modulates the gut microbiome in triple-negative breast cancer. Nutrients. (2021) 13(10):12. doi: 10.3390/nu13103656
212. Parker BJ, Wearsch PA, Veloo AC, Rodriguez-Palacios A. The genus alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol (2020) 11:906. doi: 10.3389/fimmu.2020.00906
213. Mirzaei R, Afaghi A, Babakhani S, Sohrabi MR, Hosseini-Fard SR, Babolhavaeji K, et al. Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed Pharmacother (2021) 139:111619. doi: 10.1016/j.biopha.2021.111619
214. Sánchez-Jiménez F, Pérez-Pérez A, de la Cruz-Merino L, Sánchez-Margalet V. Obesity and breast cancer: Role of leptin. Front Oncol (2019) 9. doi: 10.3389/fonc.2019.00596
215. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med (2013) 19(5):576–85. doi: 10.1038/nm.3145
216. Singh SB, Lin HC. Role of intestinal alkaline phosphatase in innate immunity. Biomolecules. (2021) 11(12):1784. doi: 10.3390/biom11121784
217. Delage B, Dashwood RH. Dietary manipulation of histone structure and function. Annu Rev Nutr (2008) 28:347. doi: 10.1146/annurev.nutr.28.061807.155354
218. Sirchia SM, Ren M, Pili R, Sironi E, Somenzi G, Ghidoni R, et al. Endogenous reactivation of the RARβ2 tumor suppressor gene epigenetically silenced in breast Cancer1. Cancer Res (2002) 62(9):2455–61.
219. Schultz PN, Beck ML, Stava C, Vassilopoulou-Sellin R. Health profiles in 5836 long-term cancer survivors. Int J Cancer (2003) 104(4):488–95. doi: 10.1002/ijc.10981
220. Reich M, Lesur A, Perdrizet-Chevallier C. Depression, quality of life and breast cancer: a review of the literature. Breast Cancer Res Treat (2008) 110(1):9–17. doi: 10.1007/s10549-007-9706-5
221. Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA: Cancer J Clin (2017) 67(4):326–44. doi: 10.3322/caac.21398
222. Taherian M, Mahin Samadi P, Rastegar H, Faramarzi MA, Rostami-Nejad M, Yazdi MH, et al. An overview on probiotics as an alternative strategy for prevention and treatment of human diseases. Iranian J Pharm research: IJPR (2019) 18(Suppl1):31–50. doi: 10.22037/ijpr.2020.112232.13620
223. Mendoza L. Potential effect of probiotics in the treatment of breast cancer. Oncol Rev (2019) 13(2):136. doi: 10.4081/oncol.2019.422
224. Hassan Z, Mustafa S, Rahim RA, Isa NM. Anti-breast cancer effects of live, heat-killed and cytoplasmic fractions of enterococcus fecalis and staphylococcus hominis isolated from human breast milk. In Vitro Cell Dev Biol-Animal (2016) 52(3):337–48. doi: 10.1007/s11626-015-9978-8
225. Lakritz JR, Poutahidis T, Levkovich T, Varian BJ, Ibrahim YM, Chatzigiagkos A, et al. Beneficial bacteria stimulate host immune cells to counteract dietary and genetic predisposition to mammary cancer in mice. Int J cancer (2014) 135(3):529–40. doi: 10.1002/ijc.28702
226. Yazdi MH, Dallal MMS, Hassan ZM, Holakuyee M, Amiri SA, Abolhassani M, et al. Oral administration of lactobacillus acidophilus induces IL-12 production in spleen cell culture of BALB/c mice bearing transplanted breast tumour. Br J Nutr (2010) 104(2):227–32. doi: 10.1017/S0007114510000516
227. De Leblanc ADM, Matar C, Thériault C, Perdigón G. Effects of milk fermented by lactobacillus helveticus R389 on immune cells associated to mammary glands in normal and a breast cancer model. Immunobiology. (2005) 210(5):349–58. doi: 10.1016/j.imbio.2005.05.024
228. Toi M, Hirota S, Tomotaki A, Sato N, Hozumi Y, Anan K, et al. Probiotic beverage with soy isoflavone consumption for breast cancer prevention: a case-control study. Curr Nutr Food Science (2013) 9(3):194–200. doi: 10.2174/15734013113099990001
229. Arroyo R, Martín V, Maldonado A, Jiménez E, Fernández L, Rodríguez JM. Treatment of infectious mastitis during lactation: antibiotics versus oral administration of lactobacilli isolated from breast milk. Clin Infect Diseases (2010) 50(12):1551–8. doi: 10.1086/652763
230. Fernández L, Cárdenas N, Arroyo R, Manzano S, Jiménez E, Martín V, et al. Prevention of infectious mastitis by oral administration of lactobacillus salivarius PS2 during late pregnancy. Clin Infect Dis (2016) 62(5):568–73. doi: 10.1093/cid/civ974
231. Davar D, Dzutsev Amiran K, McCulloch John A, Rodrigues Richard R, Chauvin J-M, Morrison Robert M, et al. Fecal microbiota transplant overcomes resistance to anti–PD-1 therapy in melanoma patients. Science. (2021) 371(6529):595–602. doi: 10.1126/science.abf3363
232. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. (2021) 371(6529):602–9. doi: 10.1126/science.abb5920
233. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. (2019) 565(7741):600–5. doi: 10.1038/s41586-019-0878-z
234. Lauté-Caly DL, Raftis EJ, Cowie P, Hennessy E, Holt A, Panzica DA, et al. The flagellin of candidate live biotherapeutic enterococcus gallinarum MRx0518 is a potent immunostimulant. Sci Rep (2019) 9(1):1–14. doi: 10.1038/s41598-018-36926-8
235. Chang C-W, Lee H-C, Li L-H, Chiang Chiau J-S, Wang T-E, Chuang W-H, et al. Fecal microbiota transplantation prevents intestinal injury, upregulation of toll-like receptors, and 5-fluorouracil/oxaliplatin-induced toxicity in colorectal cancer. Int J Mol Sci (2020) 21(2):386. doi: 10.3390/ijms21020386
236. Xiao H-W, Cui M, Li Y, Dong J-L, Zhang S-Q, Zhu C-C, et al. Gut microbiota-derived indole 3-propionic acid protects against radiation toxicity via retaining acyl-CoA-binding protein. Microbiome. (2020) 8(1):1–17. doi: 10.1186/s40168-020-00845-6
237. Cui M, Xiao H, Li Y, Zhou L, Zhao S, Luo D, et al. Fecal microbiota transplantation protects against radiation-induced toxicity. EMBO Mol Med (2017) 9(4):448–61. doi: 10.15252/emmm.201606932
238. Terrisse S, Derosa L, Iebba V, Ghiringhelli F, Vaz-Luis I, Kroemer G, et al. Intestinal microbiota influences clinical outcome and side effects of early breast cancer treatment. Cell Death Differentiation (2021) 28(9):2778–96. doi: 10.1038/s41418-021-00784-1
239. Panebianco C, Andriulli A, Pazienza V. Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome. (2018) 6(1):92. doi: 10.1186/s40168-018-0483-7
240. Gately S. Human microbiota and personalized cancer treatments: role of commensal microbes in treatment outcomes for cancer patients. Precis Med Cancer Therapy: Springer (2019) p:253–64. doi: 10.1007/978-3-030-16391-4_10
241. Wilkinson EM, Ilhan ZE, Herbst-Kralovetz MM. Microbiota–drug interactions: Impact on metabolism and efficacy of therapeutics. Maturitas. (2018) 112:53–63. doi: 10.1016/j.maturitas.2018.03.012
242. Guan X, Ma F, Sun X, Li C, Li L, Liang F, et al. Gut microbiota profiling in patients with HER2-negative metastatic breast cancer receiving metronomic chemotherapy of capecitabine compared to those under conventional dosage. Front Oncol (2020) 10. doi: 10.3389/fonc.2020.00902
243. Kim H, Oh R, Park S, Ji GE, Park MS, Kim S-E. Combination therapy of bifidobacterium longum RAPO with anti-PD-1 treatment enhances anti-tumor immune response in association with gut microbiota modulation. Curr Dev Nutr (2021) 5(Suppl 2):1131–. doi: 10.1093/cdn/nzab061_015
244. Selle K, Fletcher JR, Tuson H, Schmitt DS, McMillan L, Vridhambal GS, et al. In vivo targeting of clostridioides difficile using phage-delivered CRISPR-Cas3 antimicrobials. mBio. (2020) 11(2):e00019–20. doi: 10.1128/mBio.00019-20
245. Chen PB, Black AS, Sobel AL, Zhao Y, Mukherjee P, Molparia B, et al. Directed remodeling of the mouse gut microbiome inhibits the development of atherosclerosis. Nat Biotechnol (2020) 38(11):1288–97. doi: 10.1038/s41587-020-0549-5
246. Barbé S, Van Mellaert L, Anné J. The use of clostridial spores for cancer treatment. J Appl Microbiol (2006) 101(3):571–8. doi: 10.1111/j.1365-2672.2006.02886.x
247. Fehleisen F. Ueber die züchtung der erysipelkokken auf künstlichem nährboden und ihre Übertragbarkeit auf den menschen. Dtsch Med Wochenschr (1882) 8(31):553–4.
248. Busch W. Aus der sitzung der medicinischen section vom 13 November 1867. Berl Klin Wochenschr (1868) 5(5):137.
249. Coley W. Disappearance of a recurrent carcinoma after injections of mixed toxins. Ann Surg (1912) 55:897–8.
250. Baban CK, Cronin M, O’Hanlon D, O’Sullivan GC, Tangney M. Bacteria as vectors for gene therapy of cancer. Bioengineered bugs (2010) 1(6):385–94. doi: 10.4161/bbug.1.6.13146
251. Amer MH. Gene therapy for cancer: present status and future perspective. Mol Cell therapies (2014) 2(1):1–19. doi: 10.1186/2052-8426-2-27
252. Swofford Charles A, Van Dessel N, Forbes Neil S. Quorum-sensing salmonella selectively trigger protein expression within tumors. Proc Natl Acad Sci (2015) 112(11):3457–62. doi: 10.1073/pnas.141455811
253. Cai Z, Sanchez A, Shi Z, Zhang T, Liu M, Zhang D. Activation of toll-like receptor 5 on breast cancer cells by flagellin suppresses cell proliferation and tumor GrowthAntitumor activity of TLR5 signaling in breast cancer cells. Cancer Res (2011) 71(7):2466–75. doi: 10.1158/0008-5472.CAN-10-1993
254. Sieow BF-L, Wun KS, Yong WP, Hwang IY, Chang MW. Tweak to treat: Reprograming bacteria for cancer treatment. Trends Cancer (2021) 7(5):447–64. doi: 10.1016/j.trecan.2020.11.004
255. Alexandroff AB, Jackson AM, O’Donnell MA, James K. BCG Immunotherapy of bladder cancer: 20 years on. Lancet (1999) 353(9165):1689–94. doi: 10.1016/S0140-6736(98)07422-4
256. Roberts NJ, Zhang L, Janku F, Collins A, Bai R-Y, Staedtke V, et al. Intratumoral injection of clostridium novyi-NT spores induces antitumor responses. Sci Trans Med (2014) 6(249):249ra111–249ra111. doi: 10.1126/scitranslmed.3008982
257. Maciag PC, Radulovic S, Rothman J. The first clinical use of a live-attenuated listeria monocytogenes vaccine: a phase I safety study of lm-LLO-E7 in patients with advanced carcinoma of the cervix. Vaccine. (2009) 27(30):3975–83. doi: 10.1016/j.vaccine.2009.04.041
258. Le DT, Brockstedt DG, Nir-Paz R, Hampl J, Mathur S, Nemunaitis J, et al. A live-attenuated listeria vaccine (ANZ-100) and a live-attenuated listeria vaccine expressing mesothelin (CRS-207) for advanced cancers: Phase I studies of safety and immune InductionLive-attenuated listeria vaccines for advanced cancer. Clin Cancer Res (2012) 18(3):858–68. doi: 10.1158/1078-0432.CCR-11-2121
259. Fujimori M. Anaerobic bacteria as a gene delivery system for breast cancer therapy. Nihon rinsho Japanese J Clin Med (2008) 66(6):1211–8.
260. Fujimori M. Genetically engineered bifidobacterium as a drug delivery system for systemic therapy of metastatic breast cancer patients. Breast cancer (2006) 13(1):27–31. doi: 10.2325/jbcs.13.27
Keywords: microbiome, triple-negative-breast-cancer, microbiome & dysbiosis, bacterial theranostics, microbiome modulation
Citation: Devoy C, Flores Bueso Y and Tangney M (2022) Understanding and harnessing triple-negative breast cancer-related microbiota in oncology. Front. Oncol. 12:1020121. doi: 10.3389/fonc.2022.1020121
Received: 15 August 2022; Accepted: 31 October 2022;
Published: 24 November 2022.
Edited by:
Siun Walsh, Mater Misericordiae University Hospital, IrelandReviewed by:
Francesca Bianchi, University of Milan, ItalyCopyright © 2022 Devoy, Flores Bueso and Tangney. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark Tangney, bS50YW5nbmV5QHVjYy5pZQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.