- 1Cancer Research Center, School of Medicine, Xiamen University, Xiamen, Fujian, China
- 2Bone & Joint Research Institute, Zhongshan Hospital, School of Medicine, Xiamen University, Xiamen, Fujian, China
- 3School of Pharmaceutical Sciences, Xiamen University, Xiamen, Fujian, China
Bone metastatic cancer-secreted extracellular factors are capable of modifying the bone microenvironment through interacting with bone cells, including osteoblasts. Reticulum ribosome-binding protein 1 (RRBP1) is substantially expressed in certain bone metastatic cancer cells. This study was undertaken to determine whether RRBP1 from bone metastatic cancer cells affects the osteoblastic phenotype expression. Breast and prostate cancer cells, MDA-MB-231 and PC3, were cultured, respectively, followed by collecting conditioned mediums (CMs) and identifying the abundance of RRBP1 in CMs using LC-MS/MS. MC3T3-E1 cells were cultured with a mixed medium (including CMs from shRRBP1-transduced two-type cancer cells) with or without endoplasmic reticulum (ER) stress inhibitor 4-PBA, followed by measuring the levels of osteoblastic phenotype expression and biomarkers of ER stress using western blotting, qPCR, and ARS staining, respectively. Similar experiments were performed in shRrbp1-transduced MC3T3-E1 cells cultured with a mixed medium (including CMs from the two-type cancer cells). Bone formation parameters were measured in the tibia of nude mice injected with shRRBP1-transduced two-type cancer cells using micro-CT analysis. These results showed that RRBP1 is the sole shared high-abundance protein in CMs from the two-type cancer cells, involving osteoblast differentiation. CMs from shRRBP1-transduced two-type cells boosted the osteoblastic phenotype expression partially through increasing ER stress. CMs from the two-type cancer cells partially offset the similar alterations induced by shRrbp1 in MC3T3-E1 cells. Injection with shRRBP1-transduced two-type cells ameliorated the bone lesions in nude mice. Therefore, RRBP1 depletion of bone metastatic cancer enhanced the osteoblastic phenotype expression, suggesting a role of RRBP1 in the bone microenvironment.
Introduction
Bone metastasis is a key element of morbidity and mortality in most patients with advanced cancers, resulting in osteolytic (excess bone resorption)-type lesions, osteoblastic (excess bone formation)-type, and mixed lesions (1). The imbalance between bone resorption and bone formation in the bone microenvironment contributes to bone metastatic lesions (1). Numerous studies have demonstrated that cancer-secreted extracellular factors are capable of modifying the bone microenvironment through interacting with bone cells, including osteoblasts (responsible for bone formation) and osteoclasts (responsible for bone resorption). For example, breast cancer cells metastasize to bone tissue and produce many factors to mediate bone destruction by stimulating osteoclasts (1, 2). Prostate cancer cells supply several osteoblast-stimulating factors to osteoblasts or osteoblast precursor cells to promote bone formation in the bone microenvironment (3–5). Although the role of osteoclasts in the bone microenvironment seems to be more important than that of osteoblasts, increasing evidence report that osteoblasts might play equal, even more, important roles than osteoclasts during bone metastasis of cancer cells. The addition of bisphosphonates, which is aimed at impairing the activity of bone-resorbing osteoclasts, does not extend disease-free survival for patients with osteolytic bone metastatic breast cancer (6). Based on their studies, Dr. Andrea M. Mastro and his colleagues put forward the idea that osteoblasts are more than intermediaries between tumor cancer cells and osteoclasts, as osteoblasts are co-opted into creating a microenvironment that exacerbates bone loss and are prevented from producing matrix proteins for mineralization (7–9). Therefore, the influence of cancer-secreted extracellular factors on osteoblasts is worthy of more attention.
Reticulum ribosome-binding protein 1 (RRBP1), an endoplasmic reticulum (ER) membrane protein, is essential for ribosome binding and the translocation of nascent proteins across the membrane of rough ER (10–12). In addition, RRBP1 is detected in the nucleus and cytoplasm (13), which is involved with the localization of a subset of mRNAs to ER in a ribosome-independent manner to assist in the translation of mRNAs and secretion of various protein products (14). As a potential oncogene, increased RRBP1 expression has been linked to certain bone metastatic cancer cells, including lung, breast, and prostate cancers (15–17). Recently, Zheng et al. reported that RRBP1, together with COL6A1, VCAN, and CREB3L, is annotated as osteoblast differentiation in periodontal ligament stem cells (18). However, whether RRBP1 of bone metastatic cancer cells is responsible for modifying the phenotype expression of osteoblasts in the bone microenvironment remains unknown.
In this study, we identified the abundance of RRBP1 protein in the conditioned mediums (CMs) from human breast cancer cell line MDA-MB-231 and prostate cancer cell line PC3, respectively. We also looked into the effect of CMs from the two-type cancer cells transduced with the shRNA/RRBP1 vector (shRRBP1) on the osteoblastic phenotype expression in MC3T3-E1 preosteoblastic cells, the effect of injection with the two-type cancer cells transduced with shRRBP1 into the tibia medullary cavity of nude mice on bone tissue, and the mechanism of RRBP1 responsible for modifying the osteoblastic phenotype expression associated with ER stress. Our data suggest that depletion of RRBP1 in bone metastatic cancer cells could boost the osteoblastic phenotype expression, partially through the enhancement of ER stress.
Materials and methods
Cell culture
Bone metastatic human breast cancer MDA-MB-231 and prostate cancer PC3, and embryonic kidney HEK293T cell lines were obtained from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). MDA-MB-231 and HEK293T cells were maintained in DMEM with high glucose (Hyclone, Logan, UT, USA), and PC3 cells were maintained in F12 (Hyclone, Logan, UT, USA). The mouse preosteoblastic cell line, MC3T3-E1, purchased from ATCC (Manassas, VA, USA) was cultured in α-MEM (Hyclone, Logan, UT, USA). The mediums were supplemented with 10% fetal bovine serum,100 U/mL penicillin, and 100 μg/mL streptomycin, and the cells were cultured at 37°C in a water-saturated atmosphere of 5% CO2.
Preparation of cancer cell-CMs
MDA-MB-231 or PC3 cells were seeded at a density of 1×107 cells/dish of a 150 mm petri dish in DMEM with high glucose or F12 for 24 h, respectively. After the seeded cells reached 80% confluency, the medium was filtered by 0.44 μm filter and collected as cancer cell-CM from MDA-MB-231 or PC3 cells, respectively. According to subsequent experimental designs, MC3T3-E1 were cultured in a 1:1 mixture of double mineralized medium (α-MEM, containing 10 nM dexamethasone, 2 mM β-GP, and 50 μg/mL vitamin C) and CM from MDA-MB-231 cells (CM-231) or CM from PC3 cells (CM-PC3) as described previously (9, 19).
Mass spectrometry sample preparation
MDA-MB-231 or PC3 cells were seeded at a density of 1 ×107 cells/dish of a 150 mm petri dish in a serum-free medium for 48 h, respectively, which reached 80% confluency. 15 mL CM-231 or CM-PC3 was collected and centrifuged at 3000 x g for 5 min to remove the dead cells and cell debris, respectively. The supernatants were concentrated to 1 mL in the filter unit (Amicon Ultra-4, Millipore, MA, USA) and prepared for LC-MS/MS analysis as described in previous studies (20, 21). Briefly, the supernatants were reduced with 8 M urea (UA), 10 mM DTT, and 50 mM iodoacetamide (IAA), digested with ice-cold 0.01 μg/μL trypsin in 20 mM ammonium bicarbonate buffer (ABC), acidified with 50 μL 50% acetonitrile (ACN)/1% formic acid (FA), and desalted with 0.1% FA and C18. The samples were pooled, dried in a Speedvac (Eppendorf, German), and resuspended with 0.1% FA. The concentration of samples was detected before liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
LC-MS/MS analysis
Orbitrap Fusion Lumos equipped a nanoelectrospray source combined with a nanoscale EASY-nLC 1200 UHPLC system (Thermo Fisher Scientific) was used to perform all MS experiments, as described previously (20, 21). Briefly, CM from MDA-MB-231 cells (CM-231) or CM from PC3 cells (CM-PC3), which was prepared as described above, was separated by a nanoscale RP-HPLC column (75 µm × 25 cm) packed with 2 µm C18 beads. The mobile phase A and B were prepared using 0.1% FA and 80% MS-grade ACN dissolved in double-distilled water. The separation flow rate was set at 350 nL/min and the gradient applied to ranging from 9% to 29% of mobile phase B over 95 min followed by a linear increase to 44% of mobile phase B. A data-dependent manner alternating between full-scan MS and MS2 scans was carried out to acquire raw data. The spray voltage was set at 2.2 kV. The temperature of the ion transfer tube was 300°C. Full scans ranging from 350 to 1800 m/z with 120,000 resolutions were recorded in the Orbitrap analyzer. The Target value for AGC was set to 4 × 105, and the maximal injection time was set to 50 ms. Selected ions were sequentially fragmented in a 3 seconds cycle by HCD with 30% normalized collision energy, specified isolated windows 1.6 m/z, 15,000 resolutions. Unassigned ions or those with a charge of 2+ and >7+ were rejected for MS/MS. AGC of 5 × 104 and 40 ms maximal injection time were used. Dynamic exclusion was set to 40 s. Raw data were processed and annotated using Proteome Discoverer (PD, version 2.2) and reviewed by the SwissProt human proteome database (20259 entries). Only peptides with at least six amino acids in length were considered. The peptide and protein identifications were filtered by PD to control the false discovery rate (FDR) <1%. At least one unique peptide was required for protein identification.
Plasmid construction and transfection
Briefly, PLKO.1 vector was digested with EcoR1(New England BioLabs, Beijing, China) and Age1(New England BioLabs, Beijing, China). The reaction product was recovered with the HiPure Gel Micro Kit (Magen, Guangzhou, China) after Agron gel electrophoresis. The short hairpin RNA sequences for RRBP1 were shown in Table 1. The short hairpin RNA targeting the RRBP1 vector (shRRBP1) was then ligated with T4 DNA ligase. A stable cell line was established under the pressure of puromycin (5 μg/mL, APExBIO, Huston, USA) after lentiviral transfection with shRRBP1 or shRRBP1-2 (22). Before further investigations, the level of RRBP1 was determined by qPCR and western blotting assays. The luciferase reporter gene was expressed by transfecting the pLenti X2 Hygro/pTER shLUC (w607-1) vector into the animal research cells, and stable cell lines were established under the pressure of hygromycin B (0.2 mg/mL, Yeasen, Shanghai, China) and puromycin (5 μg/mL, APExBIO, Huston, USA) (23).
Quantitative real-time PCR assay
Total RNA was extracted by using an RNA extraction kit (Bioflux, Beijing, China) and then subjected to reverse transcription by using Primescript RT Master Mix Kit (Takara, Dalian, China) to synthesize cDNA. Real-time PCR was then performed using a Light Cycler (Roche) with a SYBR Premix Ex Taq II Kit (Takara, Dalian, China). Results were analyzed as described in the previous study (24). The primers used for qPCR was described in Table 2.
Western blotting analysis
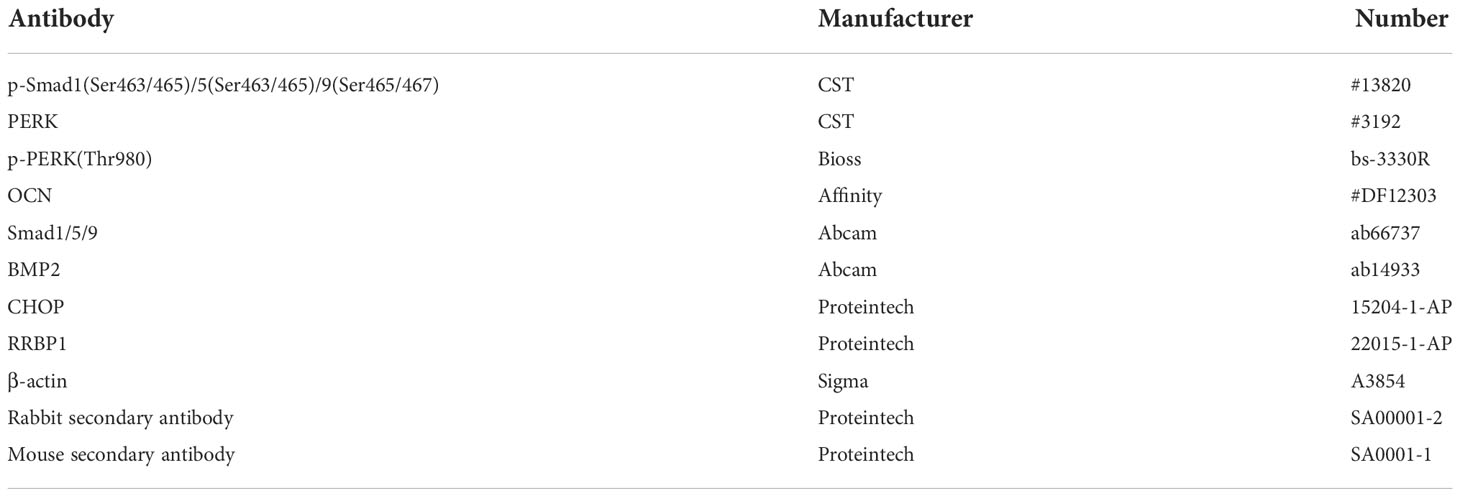
As described previously (22), protein extracts from MDA-MB-231, PC3, and MC3T3-E1 cells were subjected to SDS-PAGE (8-15%) and transferred to a PVDF membrane (GE Healthcare, Hertfordshire, UK), respectively. The membrane was incubated with various primary antibodies (1:1000) as required at 4°C overnight (Table 3), followed by the addition of the corresponding secondary antibodies at room temperature for 1 h. Enhanced chemiluminescence (ECL) detection kit was used to detect antibody reactivity (Pierce, Rockford, IL, USA).
Alizarin red S staining and evaluation of matrix mineralization
According to the manufacturer’s procedure (G1450, Solarbio, Beijing, China), MC3T3-E1 cells subjected to different treatments, including the transduction with shRrbp1 and the addition of CMs from MDA-MB-231 or PC3 cells transduced with or without shRRBP1, were cultured in a 1:1 mixture of double mineralized medium (α-MEM, containing 10 nM dexamethasone, 2 mM β-GP, and 50 μg/mL vitamin C) and CM from MDA-MB-231 cells or CM from PC3 cells for 28 d. The mineralized MC3T3-E1 cells were then washed with PBS three times, fixed with 4% paraformaldehyde for 15 min, washed with deionized water, dyed with 1% ARS solution for 30 min. After being washed with deionized water three times, plates were imaged under an inverted microscope (IX73, Olympus, Japan). Treated with 10% cetylpyridinium chloride destain solution for 1 h, the absorbance of the samples was measured at 562 nm using BIO-RAD 680(Hercules, CA, USA) (25).
In vivo animal model of bone lesions
After being approved by the Committee on the Ethics of Animal Experiments of Xiamen University, all animal studies were conducted according to the regulations of the Institutional Animal Care and Use Committee protocol. Twenty-four 5-week-old female BALB/C nu/nu nude mice were purchased from Shanghai Slac Laboratory Animal Co.Ltd.(Shanghai, China) and randomly assigned to the two groups (12 mice each), such as MDA-MB-231 and PC3 groups. Subsequently, the MDA-MB-231 group was randomly assigned to the two sub-groups (6 mice each), such as 231-shCtrl (shCtrl and pLenti X2 Hygro/pTER shLUC (w607-1)) and 231-shRRBP1 (shRRBP1 and pLenti X2 Hygro/pTER shLUC (w607-1)). Similarly, the PC3 group was randomly assigned to the two sub-groups (6 mice each), such as PC3-shCtrl (shCtrl and pLenti X2 Hygro/pTER shLUC (w607-1)) and PC3-shRRBP1(shRRBP1 and pLenti X2 Hygro/pTER shLUC (w607-1)).100 μL stable MDA-MB-231 or PC3 cells (4×105/mouse) containing the control of shRRBP1 and pLenti X2 Hygro/pTER shLUC (w607-1) vectors or shRRBP1 and pLenti X2 Hygro/pTER shLUC (w607-1) vectors were injected into the tibial medullary cavity of mouse two hind legs with a 1 mL syringe, respectively. Mouse body weight was measured every 5 days. Until 55 days, mice were subjected to measurement of tumor growth with the IVIS Lumin II in vivo imaging (PerkinElmer, Waltham, MA, USA) by using luciferase substrates (Promega, Wisconsin, USA) (23) and then sacrificed for microcomputed tomography (micro-CT) analysis.
Micro-CT analysis
As described previously (26–28), mouse hind legs were fixed with 4% paraformaldehyde for 48 h before micro-CT analysis. The qualitative and semi-quantitative analyses of bone destruction in the two groups were performed using high-resolution micro-CT scanning (Skyscan 1272 In vivo micro-CT imaging system, Bruker, Belgium). Various parameters were measured at thresholding of 85-255. Each group’s representative 3D images and mineralization mapping were carried out using SkyScan CTVox software.
Statistical analysis
All the data are presented as the means ± SEMs. Differences between the groups were examined for statistical significance using t-test (between two groups) and one-way ANOVA following Tukey’s post hoc test (among three groups) with GraphPad Prism 6 software (GraphPad Software, San Diego, CA, USA). Any p values <0.05 were considered statistically significant. Each in vitro experiment was repeated three with similar results.
Results
RRBP1 was the sole shared high-abundance protein involving osteoblast differentiation in CMs from both MDA-MB-231 and PC3 cells
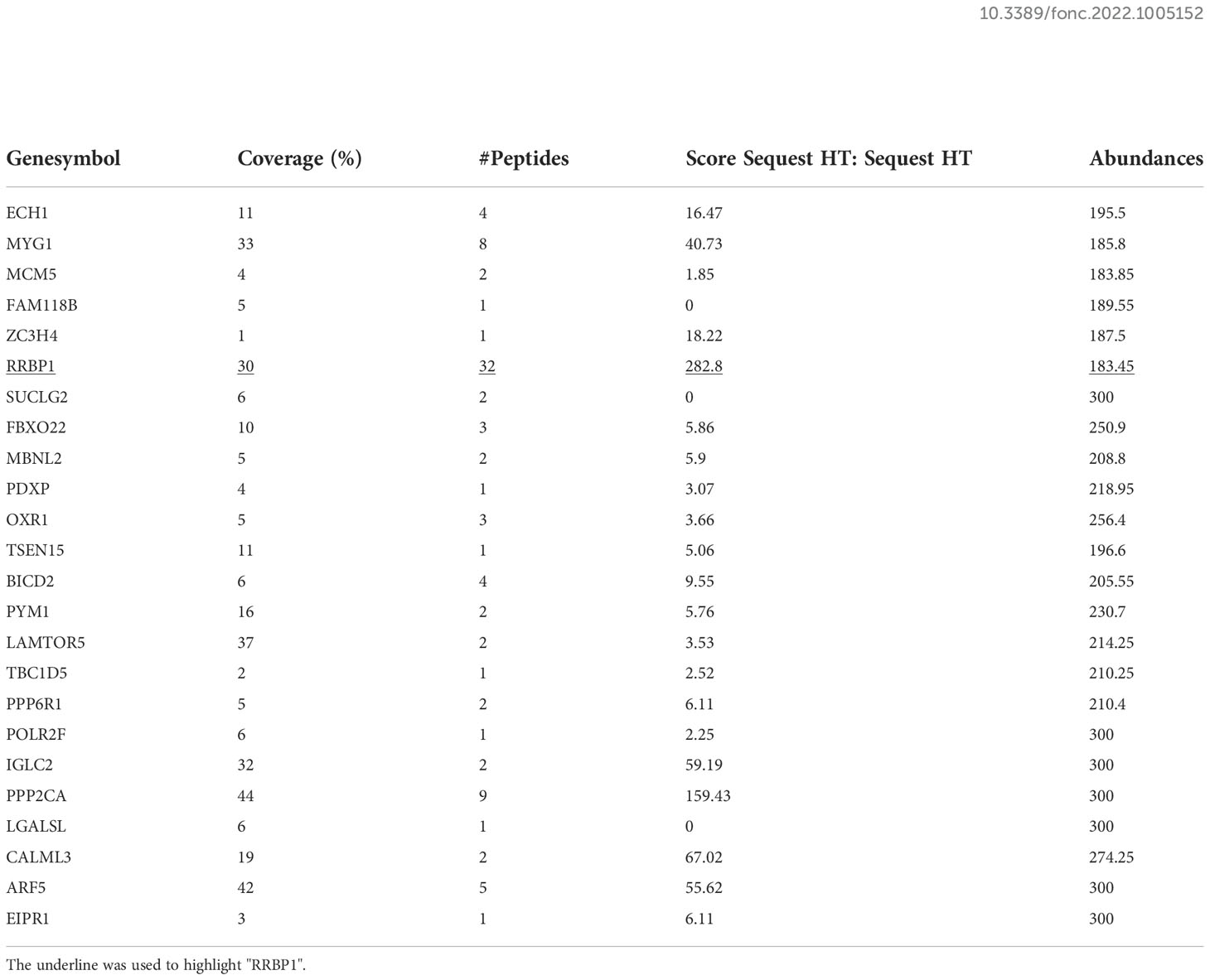
To confirm the crucial elements in the two types of cancer cells to regulate osteoblasts in the bone microenvironment, the soluble proteins in CMs from both MDA-MB-231 and PC3 cells were identified and analyzed using LC-MS/MS method. There were 530 types of proteins in the top 20% of high-abundance proteins from CM of MDA-MB-231 cells (from now CM-231), as well as 573 types of proteins from CM of PC3 cells (from now CM-PC3). 24 types of shared proteins were then identified in 530 types of proteins from CM-231 and 573 types of proteins from CM-PC3 (Supplementary Figure 1). The analysis results of gene ontology (GO) showed that RRBP1 was the sole protein involving osteoblast differentiation in the 24 types of shared proteins (Tables 4, 5).
CMs from both MDA-MB-231 and PC3 cells transduced with shRRBP1 enhanced the osteoblastic phenotype expression in MC3T3-E1 cells
The expression levels of RRBP1 in breast and prostatic adenocarcinoma tumor tissues were analyzed via Gene Expression Profiling Interactive Analysis (GEPIA) database. The results showed a significantly higher expression of RRBP1 in tumor tissues(T), compared with normal tissues(N) (Supplementary Figure 2A). Meanwhile, we observed that the protein expression levels of RRBP1 in the two-type cancer cells were higher than that in MC3T3-E1 cells (Supplementary Figure 2B). Thus, MDA-MB-231 and PC3 cells were then simultaneously transduced with the short hairpin RNA targeting the RRBP1 vector (shRRBP1) for subsequent experiments. As shown in Figures 1A, B, the mRNA and protein levels of RRBP1 were downregulated in MDA-MB-231 or PC3 cells transduced with shRRBP1 or shRRBP1-2, compared with the control vector (shCtrl). MC3T3-E1 cells were then cultured with a mixed medium (1:1 mixture of double mineralized medium(α-MEM) and CMs from two-type cancer cells that were transduced with shRRBP1). The mRNA levels of ALP (alkaline phosphatase, ALP, gene name ALP, an early differentiation marker), BGLAP (osteocalcin, OCN, gene name BGLAP, a later differentiation marker), TNFRSF11B (osteoprotegerin, OPG, gene name TNFRSF11B, a soluble decoy receptor for TNFSF11), and TNFSF11(receptor activator of nuclear factor-κB ligand, RANKL, gene name TNFSF11) were measured using qPCR assay. As shown in Figure 1C, CM from MDA-MB-231 cells transduced with shRRBP1 (CM-231shRRBP1) resulted in significantly increased ALP, BGLAP, TNFRSF11B, and TNFSF11 mRNA levels in MC3T3-E1 cells, compared with CM-231shCtrl. Meanwhile, the protein expression levels of some signal molecules related to osteoblastic phenotype, such as OCN, bone morphogenetic protein 2 (BMP2), and small mother against decapentaplegic (SMAD) 1/5/9 (Smad1/5/9), and matrix mineralization were assessed using western blotting analysis and ARS staining assay, respectively. Compared with CM-231shCtrl, CM-231shRRBP1 increased the levels of OCN, BMP2, and p-Smad1/5/9 and matrix mineralization (Figures 1D, E). Similarly, CM from PC3 cells transduced with shRRBP1 (CM-PC3shRRBP1) resulted in significantly increased ALP, BGLAP, TNFRSF11B, and TNFSF11 mRNA levels in MC3T3-E1 cells, compared with CM-PC3shCtrl (Figure 1F), as well as the levels of OCN, BMP2, p-Smad1/5/9 (Figure 1G) and mineralization (Figure 1H).
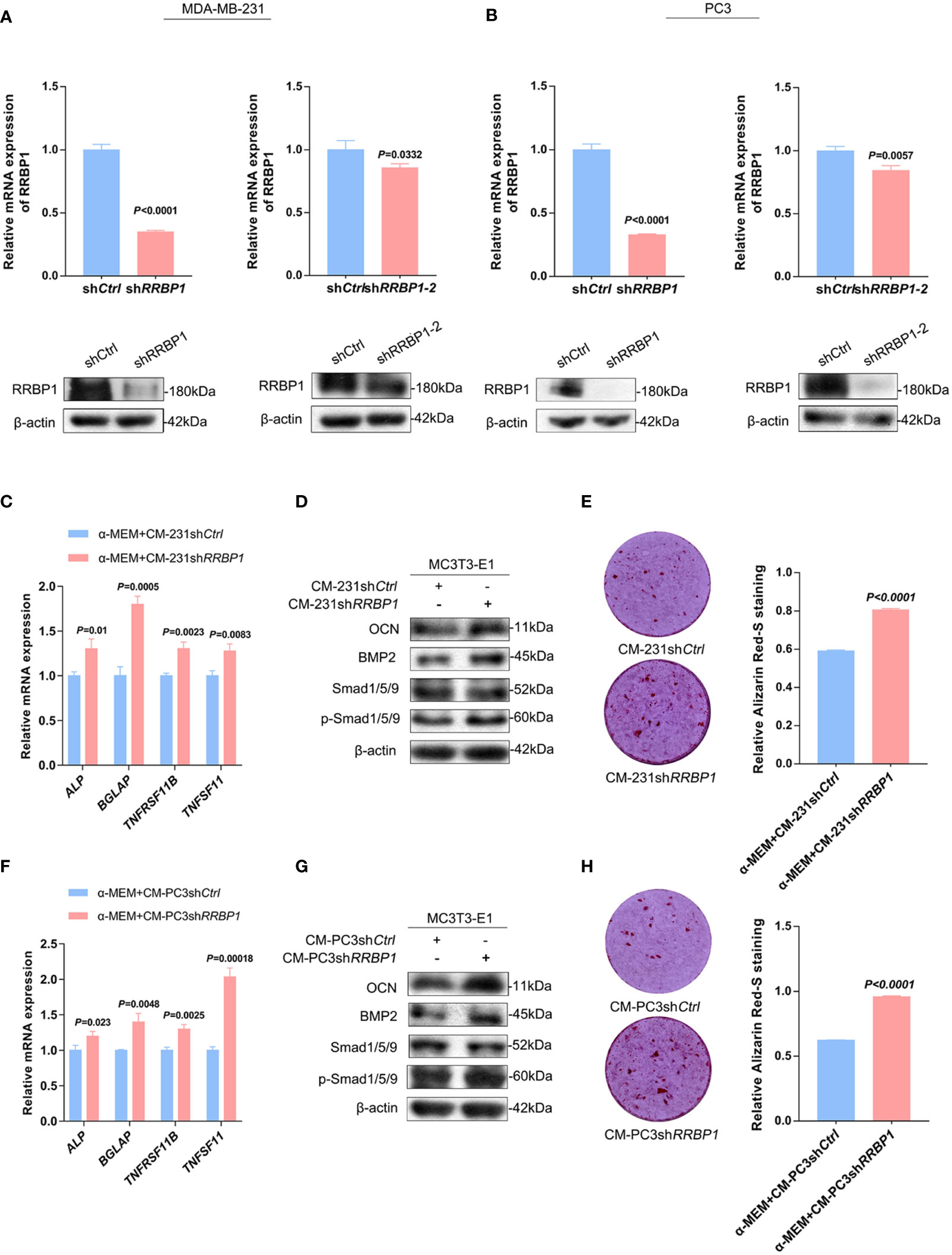
Figure 1 The effect of CM from MDA-MB-231 or PC3 cells transduced with shRRBP1 on the osteoblastic phenotype expression in MC3T3-E1 cells. (A) MDA-MB-231 cells were transduced with shRRBP1 or shRRBP1-2 vectors, respectively. The relative mRNA and protein levels of RRBP1 were measured via qPCR and western blotting assays, respectively. (B) PC3 cells were transduced with shRRBP1 or shRRBP1-2 vectors, respectively. The relative mRNA and protein levels of RRBP1 were measured via qPCR and western blotting assays, respectively. (C, D) MC3T3-E1 cells were cultured in α-MEM+CM-231shCtrl and α-MEM+CM-231shRRBP1 for 7 d, respectively. Relative mRNA levels of ALP, BGLAP, TNFRSF11B, and TNFSF11 were measured via qPCR assay (C). The protein levels of OCN, BMP2, Smad1/5/9, p-Smad1/5/9, and β-actin were measured via western blotting analysis (D). (E) MC3T3-E1 cells were cultured in α-MEM+CM-231 shCtrl and α-MEM+CM-231shRRBP1 for 28 d, respectively, followed by ARS staining and evaluation of mineralization. (F, G) MC3T3-E1 cells were cultured in α-MEM+CM-PC3shCtrl and α-MEM+CM-PC3shRRBP1 for 7 d, respectively. Relative mRNA levels of ALP, BGLAP, TNFRSF11B, and TNFSF11 were measured via qPCR assay (F). The protein levels of OCN, BMP2, Smad1/5/9, p-Smad1/5/9, and β-actin were measured via western blotting analysis (G). (H) MC3T3-E1 cells were cultured in α-MEM+CM-PC3shCtrl and α-MEM+CM-PC3shRRBP1 for 28 d, respectively, followed by ARS staining and evaluation of mineralization. The data are representative of three independent experiments.
Injection with both MDA-MB-231 and PC3 cells transduced with shRRBP1 ameliorated the bone lesions in nude mice
MDA-MB-231 and PC3 cells transduced with shRRBP1 were injected into the proximal tibial metaphysic in nude mice for 8 w, respectively (Supplementary Figure 3A). As shown in Figures 2A, B, there was no significant difference in tumor size and mouse body weight between the 231-shCtrl and 231-shRRBP1 groups. Micro-CT images were shown in Supplementary Figures 3B, C. The quantitative results of micro-CT analysis showed significantly increased bone mineral density, bone volume fraction (BV/TV), and trabecular number (Tb. N), and no significant alterations in trabecular thickness (Tb. Th) and trabecular space (Tb. Sp) in the 231-shRRBP1 group, compared with the 231-shCtrl group (Figures 2C–G). In the nude mice injected with PC3 cells transduced with shRRBP1 for 8 w, there was a slightly decreased tumor size in the PC3-shRRBP1 group, compared with the PC3-shCtrl group (Figure 3A, p=0.0343), although there was no difference in mouse body weight (Figure 3B). Quantitatively, bone mineral density, BV/TV, Tb. N, and Tb. Th in the PC3-shRRBP1 group were significantly higher than that in the PC3-shCtrl group, while Tb. Sp in the PC3-shRRBP1 group was significantly lower than that in the PC3-shCtrl group (Figures 3C–G).
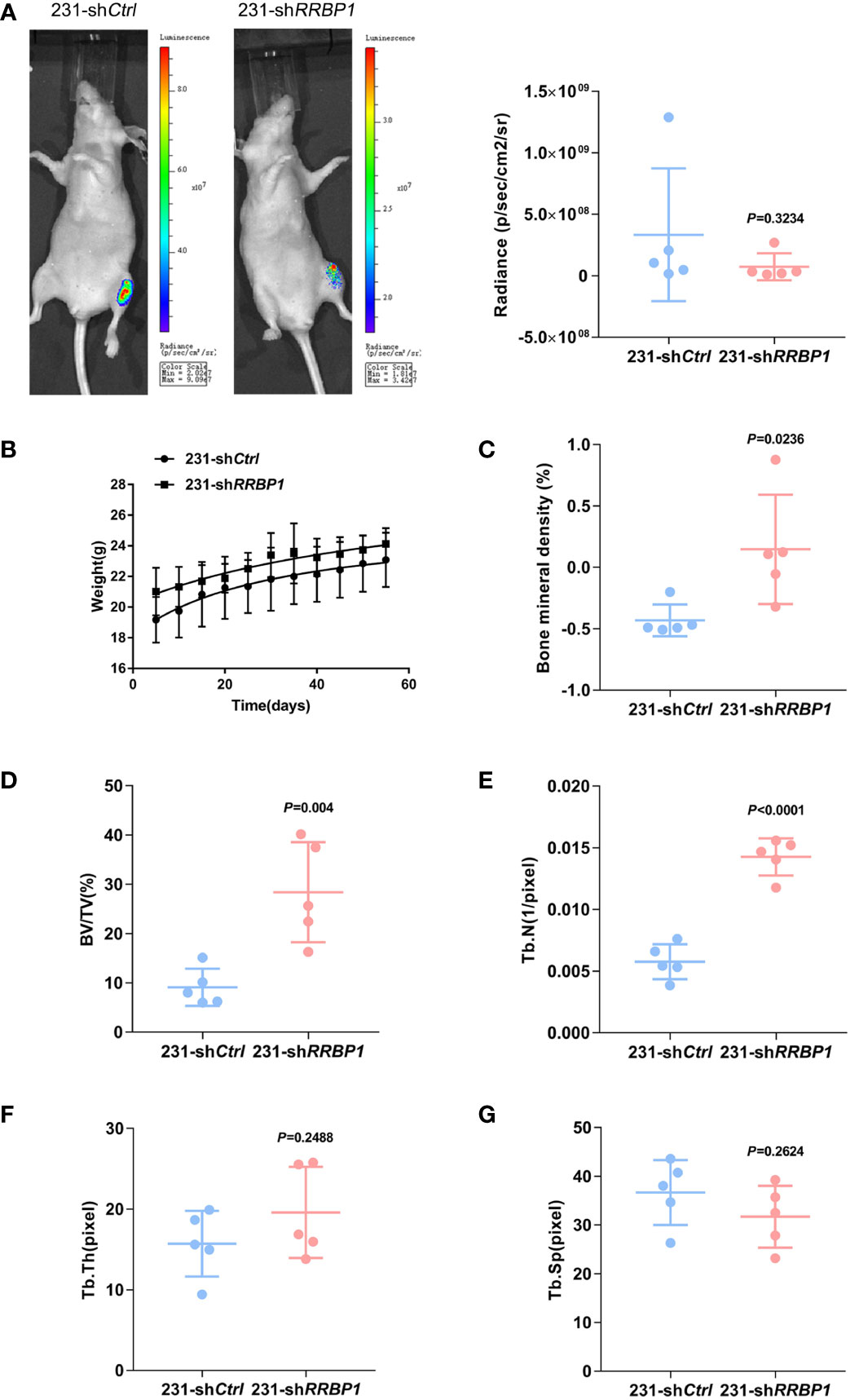
Figure 2 The effect of injection with MDA-MB-231 transduced with shRRBP1 into the proximal metaphysic of the tibia on bone mass in a nude mouse model. MDA-MB-231 cells transduced with shRRBP1 were injected into the proximal metaphysic of the tibia in nude mice for 8 w. (A) The radiance (p/sec/cm2/sr) of tumor mass was measured using the IVIS Lumin II in vivo imagining. (B) Mouse body weight was measured and analyzed using GraphPad Prism 6 software. (C–G) Quantitative analysis of structural parameters of the tibia by high-resolution micro-CT scanning: bone mineral density (C), bone volume/total tissue volume (BV/TV) (D), trabecular number (Tb. N) (E), trabecular thickness (Tb. Th) (H), and Trabecular Space (Tb. Sp) (G).
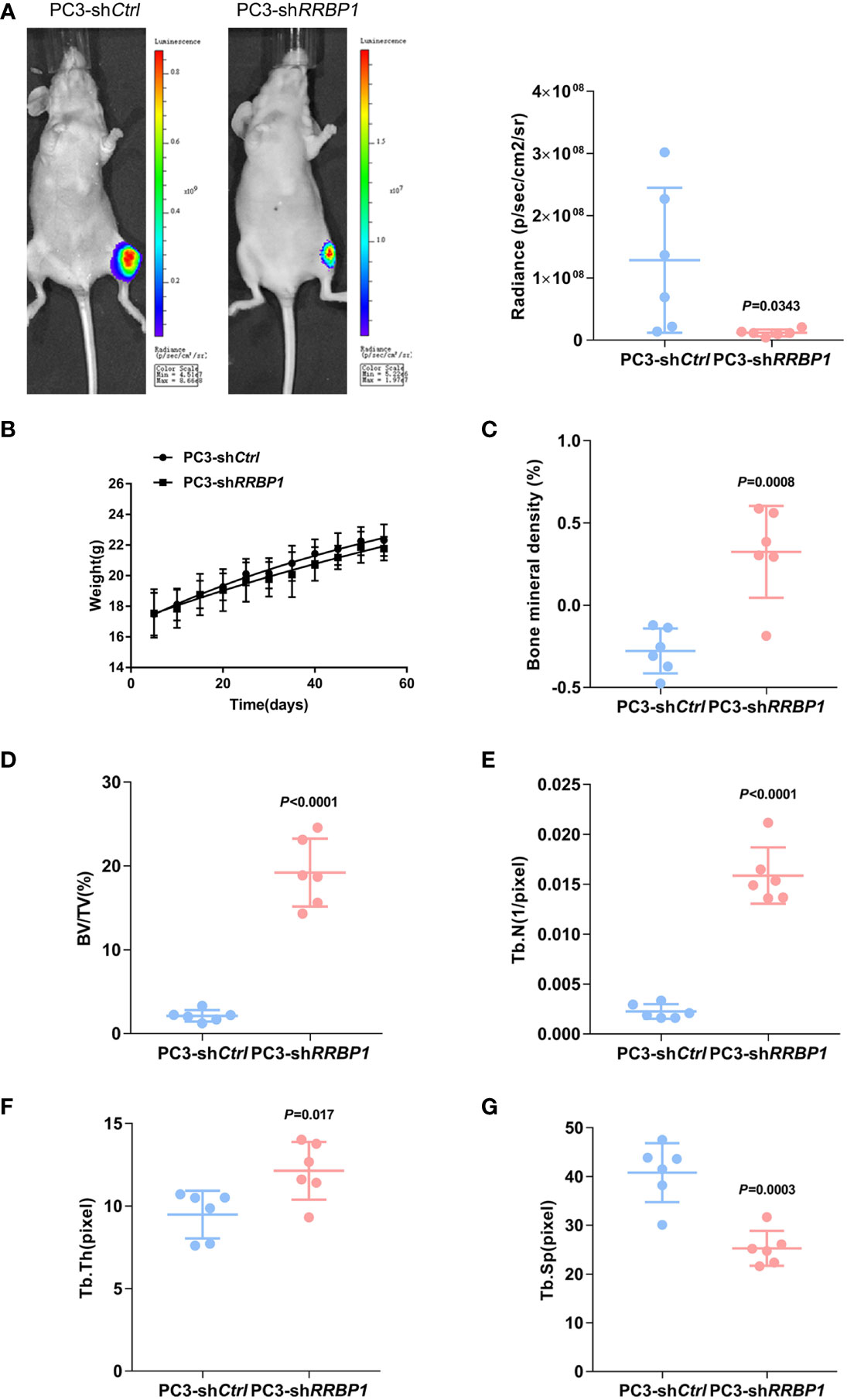
Figure 3 The effect of injection with PC3 transduced with shRRBP1 into the proximal metaphysic of the tibia on bone mass in a nude mouse model. PC3 cells transduced with shRRBP1 were injected into the proximal metaphysic of the tibia in nude mice for 8 w. (A) The radiance (p/sec/cm2/sr) of tumor mass was measured using the IVIS Lumin II in vivo imagining. (B) Mouse body weight was measured and analyzed using GraphPad Prism 6 software. (C–G) Quantitative analysis of structural parameters of the tibia by high-resolution micro-CT scanning: bone mineral density (C), bone volume/total tissue volume (BV/TV) (D), trabecular number (Tb. N) (E), trabecular thickness (Tb. Th) (H), and Trabecular Space (Tb. Sp) (G).
Enhancement of ER stress contributed to the regulation of shRRBP1 from CM-231 or CM-PC3 on the osteoblastic phenotype expression in MC3T3-E1 cells
As a ribosome binding protein, RRBP1 is a member of the ER stress response (16), some ER stress biomarkers, such as the transcription factor C/EBP homologous protein (CHOP) and the double-stranded RNA-dependent protein kinase (PKR)-like ER kinase (PERK), were then assessed in MC3T3-E1 cells cultured by CMs from shRRBP1-transduced two-type cancer cells. The results in Figure 4A showed that CMs from MDA-MB-231 cells transduced with shRRBP1(CM-231shRRBP1) led to increased expression levels of CHOP, p-PERK, and PERK in MC3T3-E1 cells, compared with CM-231shCtrl. Similar results were observed in MC3T3-E1 cells cultured by CMs from PC3 cells transduced with shRRBP1 (CM-PC3shRRBP1) (Figure 4B). To confirm the role of ER stress in regulating osteoblastic phenotype, ER stress inhibitor 4-phenylbutiric acid (4-PBA) was added in the mixed medium prior to MC3T3-E1 cells culture. As shown in Figure 4C, the increased expression levels of OCN, BMP2, and p-Smad1/5/9 induced by CM-231shRRBP1 were down-regulated by the addition of 4-PBA in MC3T3-E1 cells. Similarly, CM-PC3shRRBP1 also increased the levels of OCN, BMP2, and p-Smad1/5/9 and the addition of 4-PBA partially reduced the increased levels of OCN and p-Smad1/5/9, except the level of BMP2 that was up-regulated by 4-PBA (Figure 4D).
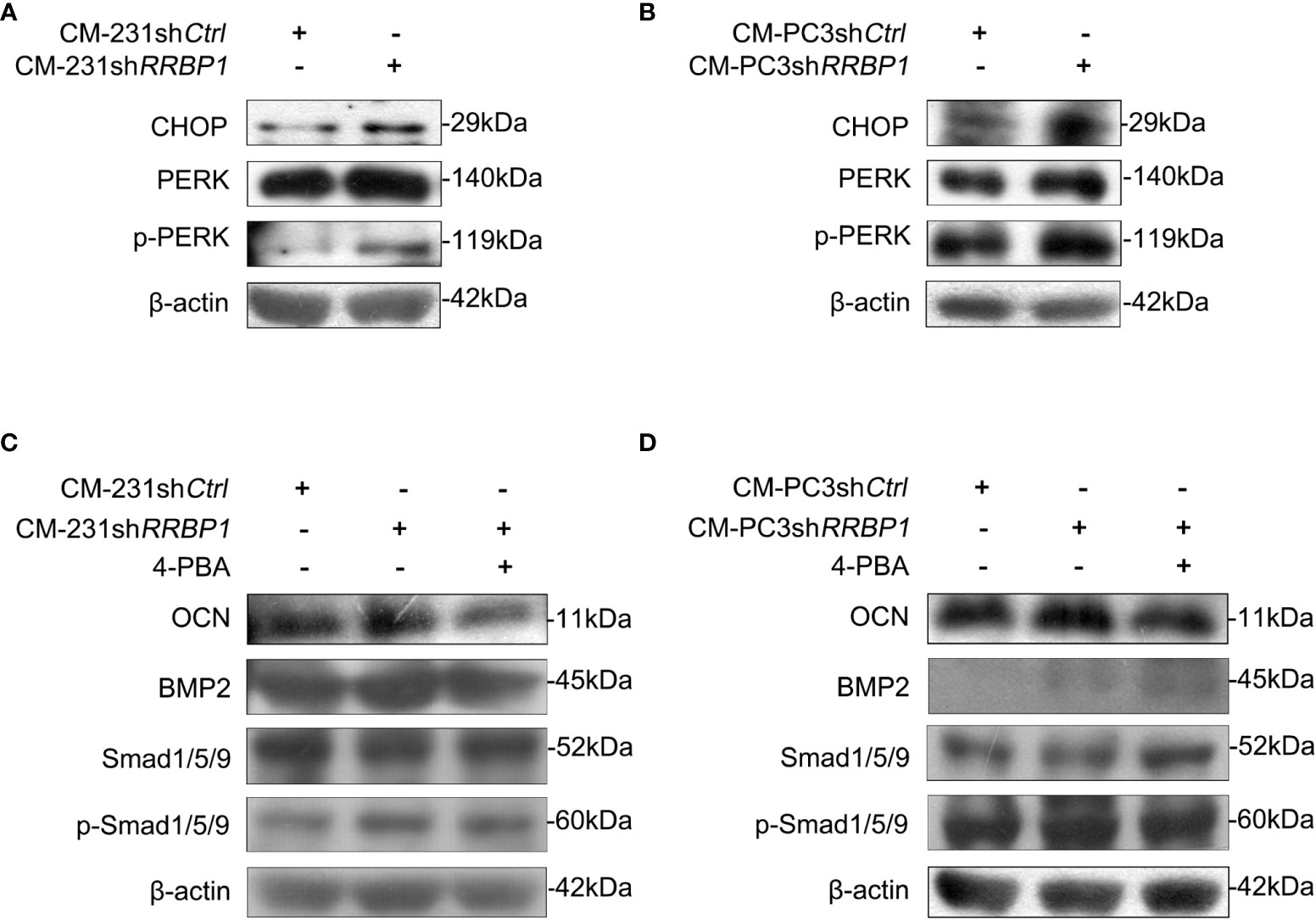
Figure 4 The involvement of ER stress in increased expression of osteoblastic phenotype mediated by CM from MDA-MB-231 or PC3 cells transduced with shRRBP1. (A, B) MC3T3-E1 cells were cultured in α-MEM+CM-231shCtrl and α-MEM+CM-231shRRBP1 or α-MEM+CM-PC3shCtrl and α-MEM+CM-PC3shRRBP1 for 7 d, respectively. The protein levels of CHOP, PERK, p-PERK, and β-actin were measured via western blotting analysis. (C, D) MC3T3-E1 cells were cultured in α-MEM+CM-231shCtrl and α-MEM+CM-231shRRBP1 or α-MEM+CM-PC3shCtrl and α-MEM+CM-PC3shRRBP1 for 5 d, followed by the treatment of 4-PBA (0.5 mM) for 2 d. The protein levels of OCN, BMP2, Smad1/5/9, p-Smad1/5/9, and β-actin were measured via western blotting analysis. The data are representative of three independent experiments.
Effect of RRBP1 depletion with shRrbp1 on the osteoblastic phenotype expression and matrix mineralization in MC3T3-E1 cells were partially offset by CMs from both MDA-MB-231 and PC3 cells
As shown in Figure 5A, RRBP1 depletion with shRrbp1(shRrbp1) suppressed the mRNA and protein expression of RRBP1 in MC3T3-E1 cells. The transduction of shRrbp1 resulted in significantly increased ALP, BGLAP, TNFRSF11B, and TNFSF11 mRNA levels, compared with the shCtrl group (Figure 5B), accompanied with increased levels of OCN, Smad1/5/9, p-Smad1/5/9, and BMP2 (Figure 5C) and matrix mineralization (Figure 5D). To further confirm whether RRBP1 from CM-231 and CM-PC3 affected the osteoblastic phenotype, CM-231 or CM-PC3, as a biological reagent, was added in the medium for culturing MC3T3-E1 cells transduced with shRrbp1. It was observed that compared with the shRrbp1 group, the addition of CM-231 reduced the increased levels of OCN, Smad1/5/9, p-Smad1/5/9, and BMP2 and matrix mineralization induced by shRrbp1 in MC3T3-E1 cells (Figures 6A, C), as well as CM-PC3 (Figures 6B, C). The increased levels of CHOP, PERK, and p-PERK caused by shRrbp1 were attenuated by the addition of either CM-231 or CM-PC3, compared with the shRrbp1 group (Figures 6D, E).
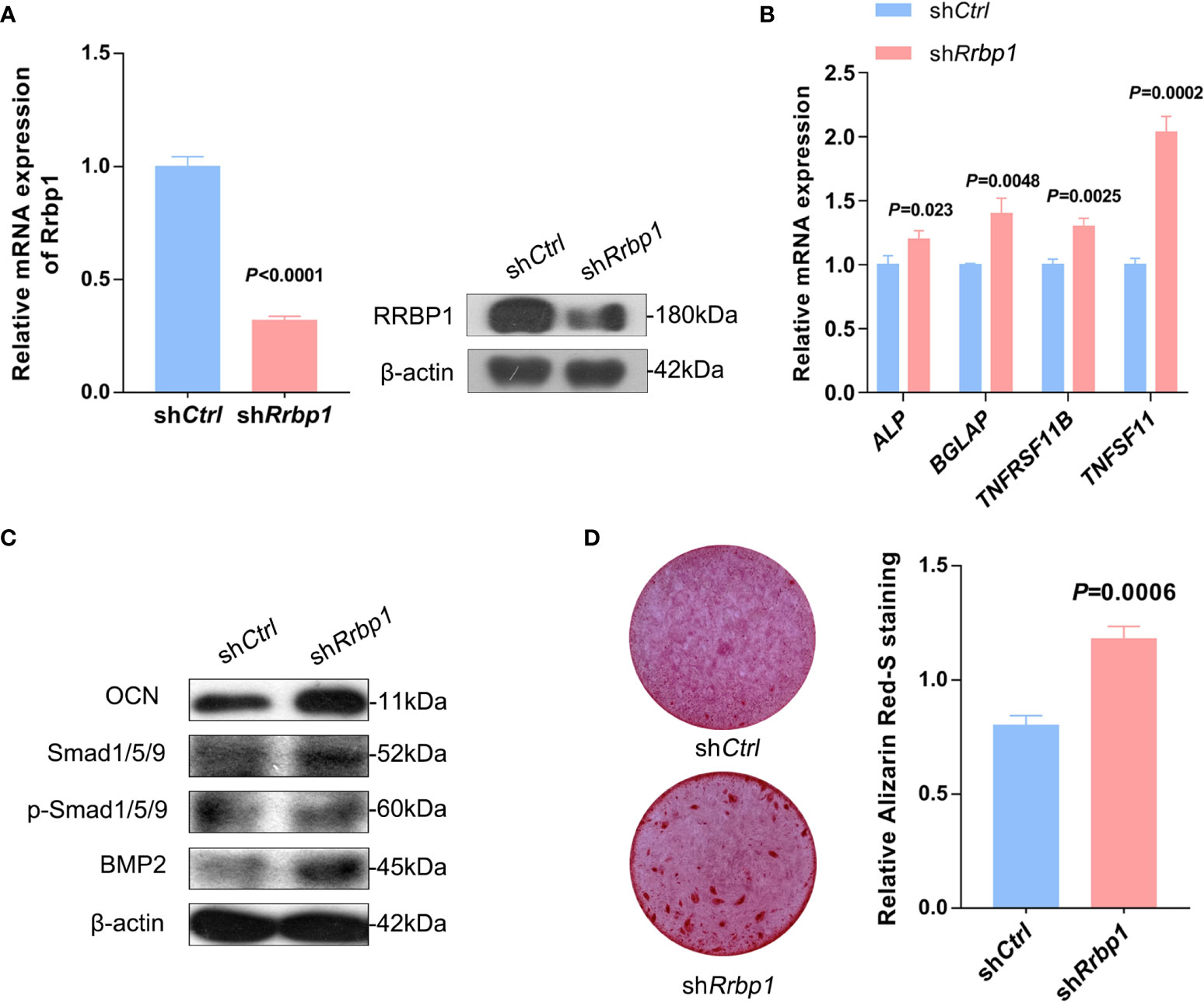
Figure 5 The osteoblastic phenotype expression in MC3T3-E1 cells transduced with shRrbp1. (A–C) MC3T3-E1 cells transduced with shRrbp1 were cultured with a mineralized medium for 7 d. The relative mRNA level of Rrbp1 was measured via qPCR assay and the protein levels of RRBP1 and β-actin were measured via western blotting analysis (A). Relative mRNA levels of ALP, BGLAP, TNFRSF11B, and TNFSF11 were measured via qPCR assay (B). The protein levels of OCN, BMP2, Smad1/5/9, p-Smad1/5/9, and β-actin were measured via western blotting analysis (C). (D) MC3T3-E1 cells transduced with shRrbp1 were cultured with a mineralized medium for 28 d, followed by ARS staining. Left panel: representative images; right panel: absorbance values at 575 nm of different groups. The data are representative of three independent experiments.
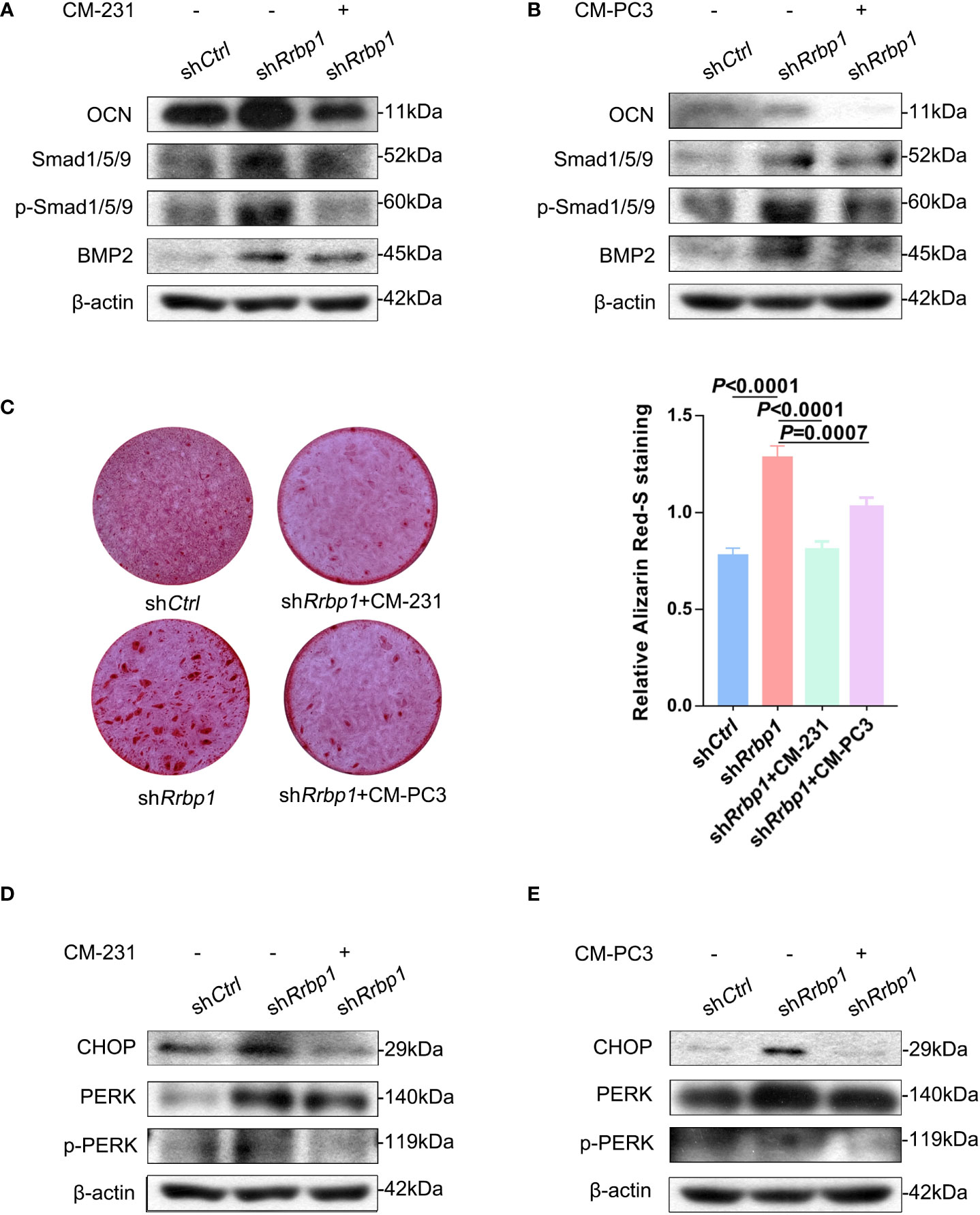
Figure 6 The effect of CM from MDA-MB-231 or PC3 cells on the osteoblastic phenotypic expression in MC3T3-E1 cells transduced with shRrbp1. (A, B) MC3T3-E1 cells transduced with shRrbp1 were cultured in α-MEM+CM-231 or α-MEM+CM-PC3 for 7d, respectively. The protein levels of OCN, BMP2, Smad1/5/9, p-Smad1/5/9, and β-actin were measured via western blotting analysis. (C) MC3T3-E1 cells transduced with shRrbp1 were cultured in α-MEM+ CM-PC3 and α-MEM+CM-PC3 for 28 d, respectively, followed by ARS staining. Left panel: representative images; right panel: absorbance values at 575 nm of different groups. (D, E) MC3T3-E1 cells transduced with shRrbp1 were cultured in α-MEM+CM-231 or α-MEM+CM-PC3 for 7d. The protein levels of CHOP, IREα, p-IREα, PERK, p-PERK, and β-actin were measured via western blotting analysis. The data are representative of three independent experiments.
Discussion
Despite it is substantially expressed in bone metastatic cancer cells, the role of RRBP1 in regulating the bone microenvironment was obscure. In this study, our findings showed that RRBP1 was the sole type of shared protein involving osteoblast differentiation in the top 20% of high-abundance proteins from CM-231 and CM-PC3. CMs from both MDA-MB-231 and PC3 cells transduced with shRRBP1 significantly increased the mRNA levels of ALP, BGLAP, TNFRSF11B, and TNFSF11 in MC3T3-E1 cells, as well as matrix mineralization and OCN, BMP2, and p-Smad1/5/9 protein levels. These results indicated that either MDA-MB-231 cell- or PC3 cell-derived RRBP1 suppressed the osteoblastic phenotype expression, at least partially, by interrupting the BMP2/Smad1/5/9 pathway, which is a conserved signaling pathway and plays a key role in osteoblast activation and extracellular matrix calcification (6, 29). Furthermore, the increased mineralization and OCN, p-Smad1/5/9, and BMP2 levels in MC3T3-E1 cells caused by shRrbp1 transduction were offset partially by CMs from MDA-MB-231 and PC3 cells. Additionally, we observed that the injection of both MDA-MB-231 and PC3 cells transduced with shRRBP1 into the proximal metaphysis of the tibia partially significantly increased bone mineral density, BV/TV, and Tb. N in nude mice. As for the inconsistent Tb. Th and Tb. Sp in the tibia of nude mice between the injection of two-type cancer cells transduced with shRRBP1, it might be associated with the differential regulation of other cancer-secreted extracellular factors in the bone environments. Taken together, it is demonstrated that as the sole shared high-abundance protein involving osteoblast differentiation from CMs of MDA-MB-231 and PC3 cells, RRBP1 depletion with shRRBP1 in cancer cells enhanced the osteoblastic phenotype expression and ameliorated the bone lesions induced by bone metastatic cancer cells. Therefore, our findings provided novel key information regarding the role of RRBP1 in bone metastatic cancer. RRBP1 of bone metastatic cancer cells might exacerbate bone lesions by suppressing the osteoblastic phenotype, suggesting that RRBP1 represents an attractive target for the treatment of the bone lesions induced by bone metastatic cancers. Considering the role of osteoclasts in the bone lesions, the other shared high-abundance proteins from CM-231 and CM-PC3 involving osteoclast function might be worthy of attention in our future studies. Combined with recent studies on drug delivery systems (30–32), targeting these high-abundance proteins in bone metastatic cancer cells would be a potential approach for ameliorating bone metastatic lesions.
ER stress induced by various physiological and pathological stimuli could lead to the accumulation of the unfolded or misfold response proteins in the ER lumen, thereby representing a fundamental threat to cell viability (33, 34). In response to ER stress, an adaptive signaling pathway in the ER is triggered to copy with the stress through attenuating protein synthesis, clearing the unfolded/misfolded proteins, and increasing the capacity of ER to fold protein (35). This process is referred to as the unfolded protein response (UPR), which is controlled by three major sensors: IRE1(inositol-requiring 1), ATF6(activating transcription factor 6), and PERK (double-strand RNA-dependent protein kinase (PKR)-like ER kinase). These three ER stress sensors are normally bound by the ER chaperone GRP78/BIP (35–37). When accumulating misfolded proteins in the ER lumen engage GRP78, the three sensors are released to activate their downstream pathways, including PERK/CHOP, thereby relieving ER stress and restoring ER function (35–37). It has been determined that ER stress influences osteoblast differentiation. On the hand, the inducement of ER stress impairs osteoblast differentiation (38, 39). For example, the deficiency of ADP-ribosylation-like factor 6 interacting protein 5 (Arl6ip5) impairs osteoblast differentiation via the inducement of ER stress and enhancement of ER stress-mediated apoptosis (38). Defective autophagy in osteoblasts induces ER stress and causes remarkable bone loss (39). On the other hand, the inducement of ER stress facilities osteoblastic differentiation (34, 40). The treatment of cranial immature osteoblasts with BMP2 induces mild ER stress involving the facilitation of osteogenesis, associated with a high demand for the synthesis and secretion of bone matrix proteins (34). Dr. Chen and his colleagues address that ER stress occurred after tooth extraction and regulating the degree of ER stress can promote bone healing in tooth extraction sockets by increasing the expression of Runx2 and ALP (40). Hence, the indefinite influence of ER stress on osteoblast metabolism may be due to a difference in its degree. Our findings showed that RRBP1 depletion increased CHOP and p-PERK levels in MC3T3-E1 cells cultured with CM from MDA-MB-231 and PC3 cells transduced with shRRBP1 vector, as well as in MC3T3-E1 cells transduced with shRrbp1. These results indicated that RRBP1 depletion induced ER stress via increasing p-PERK and CHOP in MC3T3-E1 cells, similar to a previous study that RRBP1 knockdown by shRNAs causes ER stress in lung cancer cells (16). Furthermore, it was observed that the addition of ER stress inhibitor 4-PBA partially attenuated the promoting effect of CMs from shRRBP1-transduced two-type cancer cells on OCN and p-Smad1/5/9 levels in MC3T3-E1 cells, implicating that RRBP1 depletion-induced ER stress partially aided in the enhancement of osteoblastic phenotype expression in MC3T3-E1 cell. Therefore, it is suggested that RRBP1 depletion might induce mild ER stress, thereby enhancing the osteoblastic phenotype, consistent with the previous studies that mild ER stress facilities osteoblastic differentiation (34, 40). Additionally, increased CHOP, PERK, OCN, p-Smad1/5/9, and BMP2 levels and matrix mineralization induced by shRrbp1 in MC3T3-E1 cells were offset partially by CMs from both MDA-MB-231 and PC3 cells, providing strong evidence for the involvement of RRBP1 from bone metastatic cells in suppressing the osteoblastic phenotype expression associated with decreased ER stress.
Conclusion
In summary, RRBP1 was the sole shared high-abundance protein involving osteoblast differentiation in CMs from both MDA-MB-231 and PC3 cells. CMs from both MDA-MB-231 and PC3 cells transduced with shRRBP1 enhanced the osteoblastic phenotype expression in MC3T3-E1 cells. Meanwhile, the injection of MDA-MB-231 and PC3 cells transduced with shRRBP1 into the tibia medullary cavity partially ameliorated the bone lesions in the tibia of nude mice. Furthermore, RRBP1 depletion-induced ER stress aided in the enhancement of osteoblastic phenotype expression, exhibiting the increased p-PERK and CHOP levels. Therefore, depletion of RRBP1 with shRRBP1 in bone metastatic cancer cells could boost the osteoblastic phenotype expression and ameliorate the bone lesions, partially via the enhancement of ER stress. Bone metastatic cancer-secreted RRBP1 may play an important role in regulating the bone microenvironment, via targeting osteoblasts.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by the Committee on the Ethics of Animal Experiments of Xiamen University.
Author contributions
RC: Data curation, formal analysis, experiment, methodology, writing-original draft of materials and methods. YW: Data curation, software, experiment, methodology. YX: Data curation, experiment. YH: Software, methodology. QL: Resources, funding acquisition. CX: Conceptualization, funding acquisition, writing-original draft of discussion section. BZ: Conceptualization, resources, supervision, writing-original draft of the other sections, writing-review & editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81972091 and 82172412) and the Natural Science Foundation of Fujian Province (No. 2019J01009).
Acknowledgments
We would like to thank MS Jiqian Xia for statistical data analytics support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1005152/full#supplementary-material
Supplementary Figure 1 | Identification of RRBP1 in CM from MDA-MB-231 or PC3 cells using LC-MS/MS analysis. The soluble mediators in CMs from MDA-MB-231 (CM-231) or PC3(CM-PC3) cells were identified and analyzed using LC-MS/MS analysis, respectively. 24 types of shared proteins were then identified in 530 types of proteins from CM-231 and 573 types of proteins from CM-PC3.
Supplementary Figure 2 | The expression of RRBP1 in related tissues and cells. (A) The expression levels of RRBP1 in breast and prostate tumor tissues were analyzed via Gene Expression Profiling Interactive Analysis (GEPIA) database. (B) The protein expression levels of RRBP1 in MDA-MB-231, PC3, and MC3T3-E1 cells were detected via western blotting analysis.
Supplementary Figure 3 | The effect of injection with MDA-MB-231 or PC3 cells transduced with shRRBP1 into the proximal metaphysic of the tibia on the bone lesions in a nude mice model. (A) Schematic diagram illustrating the injection site of MDA-MB-231 cells and PC3 cells transduced with shRRBP1 in a nude mice model. (B, C) Representative micro-CT images of the tibia showing bone mass using high-resolution micro-CT scanning. The effect of injection with MDA-MB-231 cells transduced with shRRBP1 into the proximal metaphysic of the tibia on the bone lesions in a nude mice model (B). The effect of injection with PC3 cells transduced with shRRBP1 into the proximal metaphysic of the tibia on the bone lesions in a nude mice model (C).
References
1. Guise TA, Mohammad KS, Clines G, Stebbins EG, Wong DH, Higgins LS, et al. Basic mechanisms responsible for osteolytic and osteoblastic bone metastases. Clin Cancer Res (2006) 12(20 Pt 2):6213s–6s. doi: 10.1158/1078-0432.CCR-06-1007
2. Chen YC, Sosnoski DM, Mastro AM. Breast cancer metastasis to the bone: mechanisms of bone loss. Breast Cancer Res (2010) 12(6):215. doi: 10.1186/bcr2781
3. Yin JJ, Mohammad KS, Käkönen SM, Harris S, Wu-Wong JR, Wessale JL, et al. A causal role for endothelin-1 in the pathogenesis of osteoblastic bone metastases. Proc Natl Acad Sci USA (2003) 100(19):10954–9. doi: 10.1073/pnas.1830978100
4. Dai J, Kitagawa Y, Zhang J, Yao Z, Mizokami A, Cheng S, et al. Vascular endothelial growth factor contributes to the prostate cancer-induced osteoblast differentiation mediated by bone morphogenetic protein. Cancer Res (2004) 64(3):994–9. doi: 10.1158/0008-5472.can-03-1382
5. Dolloff NG, Shulby SS, Nelson AV, Stearns ME, Johannes GJ, Thomas JD, et al. Bone-metastatic potential of human prostate cancer cells correlates with Akt/PKB activation by alpha platelet-derived growth factor receptor. Oncogene (2005) 24(45):6848–54. doi: 10.1038/sj.onc.1208815
6. Jongwattanapisan P, Terajima M, Miguez PA, Querido W, Nagaoka H, Sumida N, et al. Identification of the effector domain of biglycan that facilitates BMP-2 osteogenic function. Sci Rep (2018) 8(1):7022. doi: 10.1038/s41598-018-25279-x
7. Mercer RR, Miyasaka C, Mastro AM. Metastatic breast cancer cells suppress osteoblast adhesion and differentiation. Clin Exp Metastasis (2004) 21(5):427–35. doi: 10.1007/s10585-004-1867-6
8. Mercer RR. Mastro AM cytokines secreted by bone-metastatic breast cancer cells alter the expression pattern of f-actin and reduce focal adhesion plaques in osteoblasts through PI3K. Exp Cell Res (2005) 310(2):270–81. doi: 10.1016/j.yexcr.2005.07.029
9. Kinder M, Chislock E, Bussard KM, Shuman L, Mastro AM. Metastatic breast cancer induces an osteoblast inflammatory response. Exp Cell Res (2008) 314(1):173–83. doi: 10.1016/j.yexcr.2007.09.021
10. Savitz AJ, Meyer DI. 180-kD ribosome receptor is essential for both ribosome binding and protein translocation. J Cell Biol (1993) 120(4):853–63. doi: 10.1083/jcb.120.4.853
11. Benyamini P, Webster P, Meyer DI. Knockdown of p180 eliminates the terminal differentiation of a secretory cell line. Mol Biol Cell (2009) 20(2):732–44. doi: 10.1091/mbc.e08-07-0682
12. Ueno T, Tanaka K, Kaneko K, Taga Y, Sata T, Irie S, et al. Enhancement of procollagen biosynthesis by p180 through augmented ribosome association on the endoplasmic reticulum in response to stimulated secretion. J Biol Chem (2010) 285(39):29941–50. doi: 10.1074/jbc.M109.094607
13. Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell (2006) 127(3):635–48. doi: 10.2147/CMAR.S186632
14. Cui XA, Zhang H, Palazzo AF. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PloS Biol (2012) 10(5):e1001336. doi: 10.1371/journal.pbio.1001336
15. Telikicherla D, Marimuthu A, Kashyap MK, Ramachandra YL, Mohan S, Roa JC, et al. Overexpression of ribosome binding protein 1 (RRBP1) in breast cancer. Clin Proteomics (2012) 9(1):7. doi: 10.1186/1559-0275-9-7
16. Tsai HY, Yang YF, Wu AT, Yang CJ, Liu YP, Jan YH, et al. Endoplasmic reticulum ribosome-binding protein 1 (RRBP1) overexpression is frequently found in lung cancer patients and alleviates intracellular stress-induced apoptosis through the enhancement of GRP78. Oncogene (2013) 32(41):4921–31. doi: 10.1038/onc.2012.514
17. Li T, Wang Q, Hong X, Li H, Yang K, Li J, et al. RRBP1 is highly expressed in prostate cancer and correlates with prognosis. Cancer Manage Res (2019) 11:3021–7. doi: 10.2147/CMAR.S186632
18. Zheng Y, Li X, Huang Y, Jia L, Li W. Time series clustering of mRNA and lncRNA expression during osteogenic differentiation of periodontal ligament stem cells. PeerJ (2018) 6:e5214. doi: 10.7717/peerj.5214
19. Yuen HF, Chiu YT, Chan KK, Chan YP, Chua CW, McCrudden CM, et al. Prostate cancer cells modulate osteoblast mineralisation and osteoclast differentiation through id-1. Br J Cancer (2010) 102(2):332–41. doi: 10.1038/sj.bjc.6605480
20. Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods (2009) 6(5):359–62. doi: 10.1038/nmeth.1322
21. Peng BL, Li WJ, Ding JC, He YH, Ran T, Xie BL, et al. A hypermethylation strategy utilized by enhancer-bound CARM1 to promote estrogen receptor α-dependent transcriptional activation and breast carcinogenesis. Theranostics (2020) 10(8):3451–73. doi: 10.7150/thno.39241
22. Zhang B, Wang F, Dai L, Cai H, Zhan Y, Gang S, et al. Lentivirus-mediated PLCγ1 gene short-hairpin RNA suppresses tumor growth and metastasis of human gastric adenocarcinoma. Oncotarget (2016) 7(7):8043–54. doi: 10.18632/oncotarget.6976
23. Lau CP, Wong KC, Huang L, Li G, Tsui SK, Kumta SM. A mouse model of luciferase-transfected stromal cells of giant cell tumor of bone. Connect Tissue Res (2015) 56(6):493–503. doi: 10.3109/03008207.2015.1075519
24. Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, et al. Protein kinase c α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell (2013) 24(3):347–64. doi: 10.1016/j.ccr.2013.08.005
25. Gregory CA, Gunn WG, Peister A, Prockop DJ. An alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal Biochem (2004) 329(1):77–84. doi: 10.1016/j.ab.2004.02.002
26. Chen X, Wang Y, Chen R, Qu N, Zhang B, Xia C. Suppressing PLCγ1 enhances osteogenic and chondrogenic potential of BMSCs. Biochem Biophys Res Commun (2020) 532(2):292–9. doi: 10.1016/j.bbrc.2020.08.049
27. Radhakrishnan J, Manigandan A, Chinnaswamy P, Subramanian A, Sethuraman S. Gradient nano-engineered in situ forming composite hydrogel for osteochondral regeneration. Biomaterials (2018) 162:82–98. doi: 10.1016/j.biomaterials
28. Samadi N, Thapa D, Salimi M, Parkhimchyk A, Tabatabaei N. Low-cost active thermography using cellphone infrared cameras: from early detection of dental caries to quantification of THC in oral fluid. Sci Rep (2020) 10(1):7857. doi: 10.1038/s41598-020-64796-6
29. Byon CH, Javed A, Dai Q, Kappes JC, Clemens TL, Darley-Usmar VM, et al. Oxidative stress induces vascular calcification through modulation of the osteogenic transcription factor Runx2 by AKT signaling. J Biol Chem (2008) 283(22):15319–27. doi: 10.1074/jbc.M800021200
30. Singh RK, Patel KD, Mahapatra C, Kang MS, Kim HW. C-dot generated bioactive organosilica nanospheres in theranostics: Multicolor luminescent and photothermal properties combined with drug delivery capacity. ACS Appl Mater Interfaces (2016) 8(37):24433–44. doi: 10.1021/acsami.6b07494
31. Patel KD, Kim TH, Mandakhbayar N, Singh RK, Jang JH, Lee JH, et al. Coating biopolymer nanofibers with carbon nanotubes accelerates tissue healing and bone regeneration through orchestrated cell- and tissue-regulatory responses. Acta Biomater (2020) 108:97–110. doi: 10.1016/j.actbio.2020.03.012
32. Singh RK, Yoon DS, Mandakhbayar N, Li C, Kurian AG, Lee NH, et al. Diabetic bone regeneration with nanoceria-tailored scaffolds by recapitulating cellular microenvironment: Activating integrin/TGF-β co-signaling of MSCs while relieving oxidative stress. Biomaterials (2022) 288:121732. doi: 10.1016/j.biomaterials.2022.121732
33. Giorgi C, De Stefani D, Bononi A, Rizzuto R, Pinton P. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. Int J Biochem Cell Biol (2009) 41(10):1817–27. doi: 10.1016/j.biocel.2009.04.010
34. Saito A, Ochiai K, Kondo S, Tsumagari K, Murakami T, Cavener DR, et al. Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem (2011) 286(6):4809–18. doi: 10.1074/jbc.M110.152900
35. Almanza A, Carlesso A, Chintha C, Creedican S, Doultsinos D, Leuzzi B, et al. Endoplasmic reticulum stress signalling - from basic mechanisms to clinical applications. FEBS J (2019) 286(2):241–78. doi: 10.1111/febs.14608
36. Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol (2000) 2(6):326–32. doi: 10.1038/35014014
37. Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of golgi localization signals. Dev Cell (2002) 3(1):99–111. doi: 10.1016/s1534-5807(02)00203-4
38. Wu Y, Yang M, Fan J, Peng Y, Deng L, Ding Y, et al. Deficiency of osteoblastic Arl6ip5 impaired osteoblast differentiation and enhanced osteoclastogenesis via disturbance of ER calcium homeostasis and induction of ER stress-mediated apoptosis. Cell Death Dis (2014) 5(10):e1464. doi: 10.1038/cddis.2014.427
39. Li H, Li D, Ma Z, Qian Z, Kang X, Jin X, et al. Defective autophagy in osteoblasts induces endoplasmic reticulum stress and causes remarkable bone loss. Autophagy (2018) 14(10):1726–41. doi: 10.1080/15548627.2018.1483807
Keywords: RRBP1, conditioned mediums (CMs), bone metastatic cancer cells, the osteoblastic phenotype expression, MC3T3-E1 cells, endoplasmic reticulum (ER) stress
Citation: Chen R, Wang Y, Xu Y, He Y, Li Q, Xia C and Zhang B (2022) RRBP1 depletion of bone metastatic cancer cells contributes to enhanced expression of the osteoblastic phenotype. Front. Oncol. 12:1005152. doi: 10.3389/fonc.2022.1005152
Received: 28 July 2022; Accepted: 24 November 2022;
Published: 09 December 2022.
Edited by:
Rentian Feng, University of Pittsburgh, United StatesReviewed by:
Xuefang Hao, Inner Mongolia University for Nationalities, ChinaRajendra Kumar Singh, Institute of Tissue Regeneration Engineering (ITREN), South Korea
Jianying Zhang, University of Pittsburgh, United States
Copyright © 2022 Chen, Wang, Xu, He, Li, Xia and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Li, c3VubnltYXlscUB4bXUuZWR1LmNu; Chun Xia, Y2h1bnhpYUB4bXUuZWR1LmNu; Bing Zhang, Y3Jpc3RhbDY2QHhtdS5lZHUuY24=
†These authors have contributed equally to this work
 Rui Chen
Rui Chen Yue Wang2†
Yue Wang2† Yaohui He
Yaohui He Chun Xia
Chun Xia Bing Zhang
Bing Zhang