
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 30 September 2022
Sec. Gastrointestinal Cancers: Hepato Pancreatic Biliary Cancers
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1001807
As a new type of post-translational modification (PTM), lysine 2-hydroxyisobutyrylation (Khib) was firstly identified in histones and functioned as a regulator of transactivation in mammals. However, the role of Khib proteins remains to be investigated. Here, we firstly identified 10,367 Khib sites on 2,325 modified proteins in seven patients with pancreatic cancer by applying liquid chromatography with tandem mass spectrometry (LC-MS/MS) qualitative proteomics techniques. Among them, 27 Khib-modified sites were identified in histones. Bioinformatics analysis revealed that the Khib-modified proteins were mainly distributed in the cytoplasm and enhanced in metabolic pathways, including glycolysis/gluconeogenesis, the tricarboxylic acid cycle (TCA cycle), and fatty acid degradation. In an overlapping comparison of lysine 2-hydroxyisobutyrylation, succinylation, and acetylation in humans, 105 proteins with 80 sites were modified by all three PTMs, suggesting there may be a complex network among the different modified proteins and sites. Furthermore, MG149, which was identified as a Tip60 inhibitor, significantly decreased the total Khib modification level in pancreatic cancer (PC) and strongly suppressed PC’s proliferation, migration, and invasion ability. Overall, our study is the first profiling of lysine 2-hydroxyisobutyrylome and provides a new database for better investigating Khib in PC.
1. This is the first report of Khib in the tumor tissue of patients with pancreatic cancer.
2. The Khib proteins in pancreatic cancer are widely distributed.
3. Inhibiting the total Khib level can suppress pancreatic cancer cells’ proliferation, migration, and invasion ability.
Post-translational modifications (PTM) of proteins contribute to protein function by adding chemical groups to amino acid residues to alter the charge, structure, and molecular weight of the protein (1), thereby affecting protein activity, subcellular location, interaction chaperones, stability, and amplifying/weakening protein function (2). Until now, more than 200 PTMs have been identified, including phosphorylation, ubiquitination, acetylation, succinylation, and 2-hydroxyisobutyrylation (3–5).
It is universally acknowledged that 2-hydroxybutyrate (2-HIBA) is a microbial metabolite detected at micromolar concentrations in human bio-fluids, including blood, saliva, and urine (6). Previous reports demonstrated that elevated 2-HIBA levels were associated with metabolic diseases, including diabetes (7) and obesity (8). 2-hydroxyisobutyrate was a synthetic precursor of 2-hydroxyisobutyryl coenzyme A, which was regulated by carbon sources (9) and identified as the donor of lysine 2-hydroxyisobutyrylation (Khib) (10). Lysine 2-hydroxyisobutyrylation is a novel PTM, which was first detected in histones in 2014 (4) and has been validated by following reports in animals, plants, and microorganisms (11–14). Tip60 was identified as a Khib writer in mammalian cells, which could catalyze Khib modifications in vitro and in vivo. In contrast, histone deacetylase 2 (HDAC2) and histone deacetylase 3 (HDAC3), but not histone deacetylase 1 (HDAC1), acted as erasers to remove Khib modifications in vivo (14). Previous studies have confirmed that Khib is a widespread type of PTM with strict evolutionary conservation, and it could be up-regulated by carbon sources generated by the pentose phosphate pathway and glycolysis/gluconeogenesis (12), which strongly suggestes a close correlation between Khib modification and metabolism. Recently, Khib was reported to be involved in cancer progression. Khib was identified in ENO1 and promoted the ability of glycolysis and proliferation in hepatoma cells (15). Also, Khib in oral squamous cell carcinoma (OSCC) was suggested to promote cancer progression (16).
Pancreatic cancer (PC) is one of the most malignant digestive tumors, with a five-year survival rate of only 9%. In the past decade, the mortality rate of men with PC has increased steadily, rather than decreasing. According to a statistical report, there were up to 420,000 new PC cases and 410,000 deaths worldwide in 2020 (17), and it is predicted that PC may become the second most deadly malignancy in 2030, just after lung cancer (18). Currently, the treatment of PC is mainly surgical resection described as pancreaticoduodenectomy; however, owing to its insidious onset, high malignancy, and rapid progression, the operable rate is far below 20% (19). Post-translational modifications have a vital role in epigenetics and signal transduction, numbers of studies have focused on the contribution of PTMs as diagnostic markers or therapeutic targets for cancer treatment (20). Although there have been many reports on lysine 2-hydroxyisobutyrylation in recent years, the role of Khib in PC development remained unclear.
In the present study, we first reported the Khib sites in human pancreatic cancer via proteomics. Totally, 10,367 lysine 2-hydroxyisobutyrylation modification sites on 2325 proteins were identified, and they mainly enriched in protein synthesis and metabolic pathways. Moreover, MG149, a selective inhibitor of Tip60, was proved to enormously decrease the total Khib level in PC and suppress PC cell proliferation and metastasis abilities. The novel Khib sites and proteins identified in this research not only promote our insights into the functional role of Khib but also provide new perspectives and potential drug targets for future investigations in PC.
To demonstrate the Khib modification stage in PC, the tissues from seven different patients (Table S1) were digested and lysed to extract proteins (Figure S1A), and then Western Blotting with pan- 2-hydroxyisobutyryllysine antibody (pan-anti-Khib) was used to prove the status of Khib in these proteins (Figure 1A). Also, proteins obtained from different pancreatic cell lines (HPDE, SW1990, BXPC, ASPC, Mia PaCa-2, PANC-1) were examined by WB (Figure 1B and Figure S1B). Our analysis showed that the level of Khib modification in different PC tissues or cell lines was slightly different, mainly concentrated at 35—75 kD, indicating Khib was widely distributed and conserved in PC.
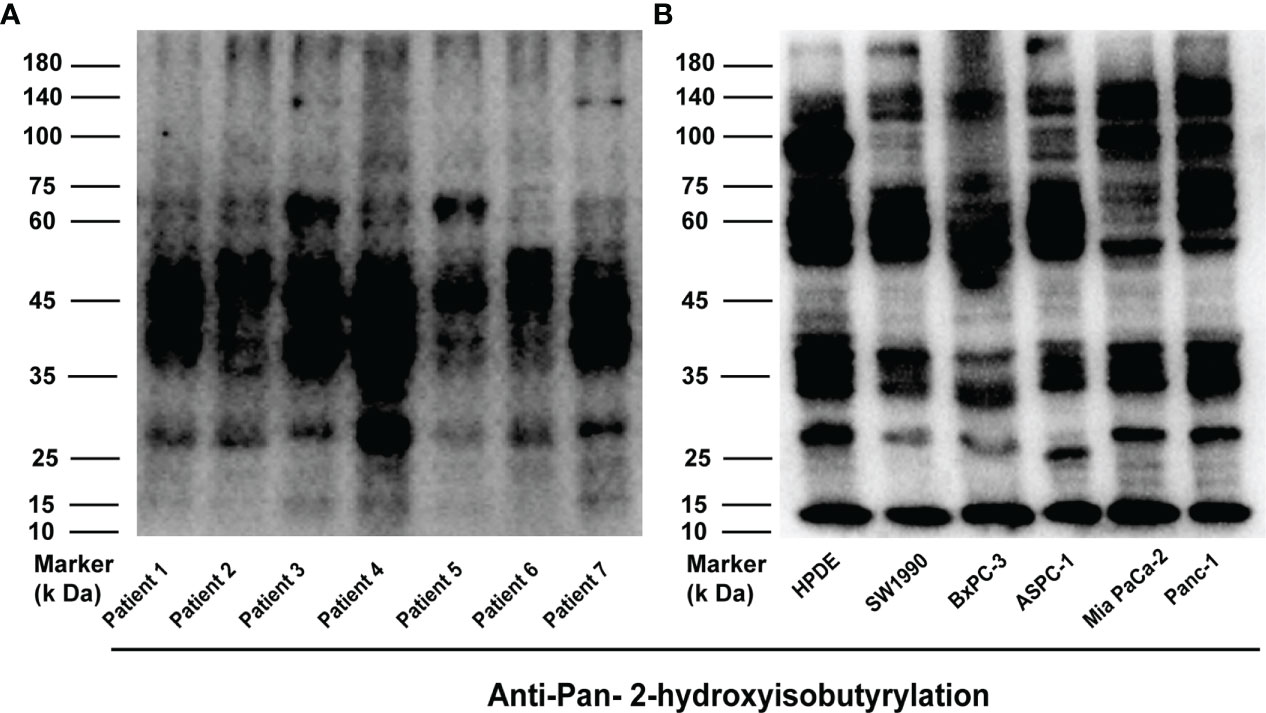
Figure 1 2-hydroxyisobutyrylation (Khib) is a type of widely distributed protein-translational modifications (PTMs). (A, B) Western blot analysis of the Khib proteins in pancreatic cancer (PC) and every cell line.
Based on the antibody enrichment and liquid chromatography with tandem mass spectrometry (LC-MS/MS) methods, proteomic analysis was performed on seven pancreatic cancer tissue samples extracted from patients with PC (Figure 2A). In this study, a total of 10,367 Khib-modified sites in 2,325 proteins were identified with a localization probability > 0.75. The details of all proteins and associated Khib sites identified in this study were listed in Table S2. Our study showed a great depth of detection in Khib proteins compared to previous studies and contributed to a more comprehensive Khib map in PC (Figure 2B). To characterize the extent of Khib coverage in the substrate proteins, we calculated the number of every protein modification site. Most proteins showed 1–3 modification sites, 38%, 17%, and 11%, respectively. In comparison, the percentage of proteins with > 20 sites was 3% (Figure 2C), demonstrating that Khib modifications always have more than one site in most of the proteins, suggesting us to pay attention to the position of modification and possible interrelationships in the subsequent protein modification studies.
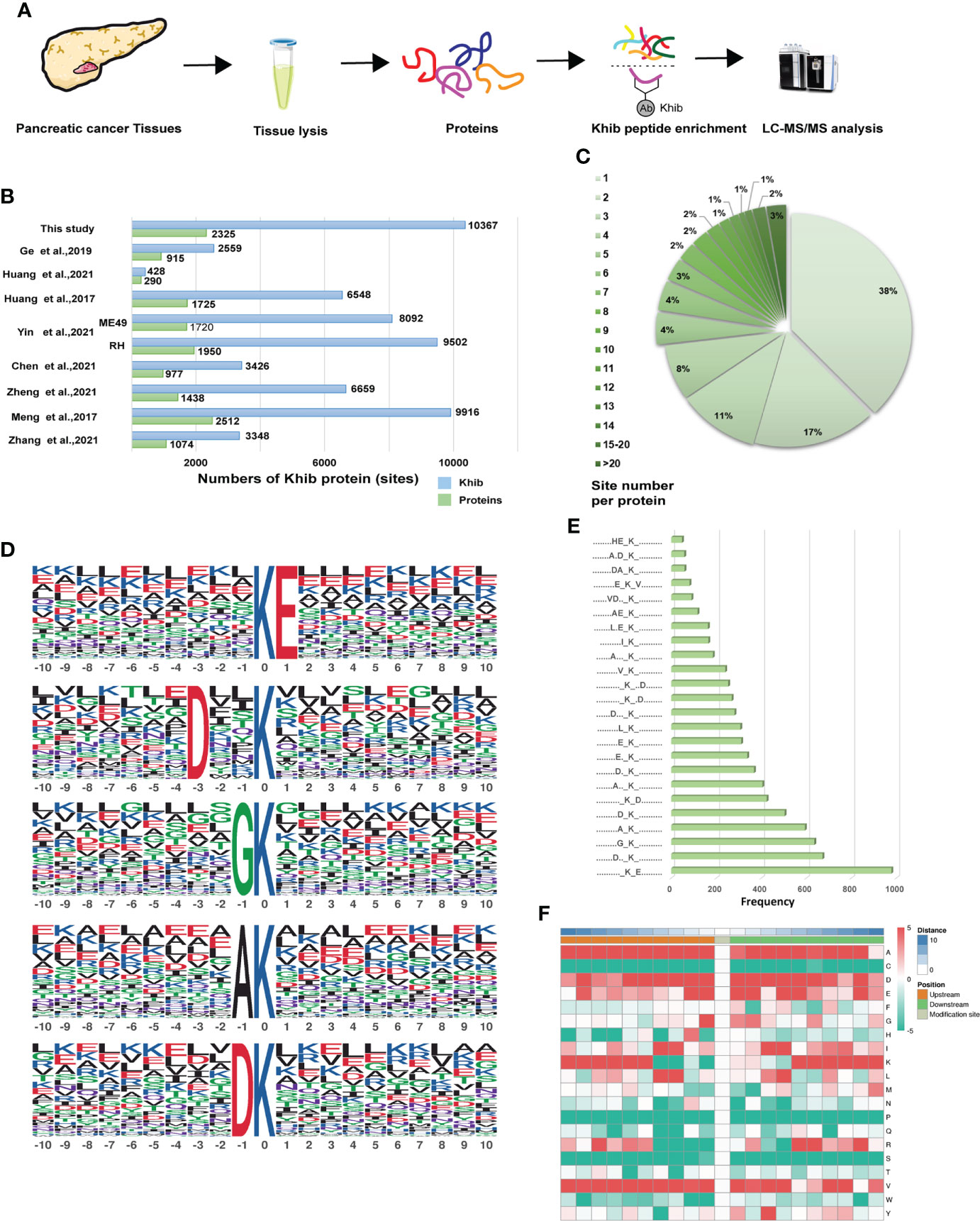
Figure 2 Genome-wide identity and profiling of the Khib proteins and sites in PC. (A) Workflow for the identification of Khib modification in PC. Proteins extracted from the PC tissues were digested into peptides, and then the specific Khib antibody enriched the Khib sites-containing peptides, followed by the liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis. (B) Comparison of the Khib proteins and sites identified in our study and those in previous reports. (C) Numbers and ratios of Khib sites for each protein. (D) The top five Khib motifs. The size of each letter indicates the frequency of that amino acid residue at that position. (E) Frequency of occurrence of each motif among the peptides with Khib modifications. (F) The heatmap of the amino acid profile of the Khib sites, featuring the enrichment (red) and depletion (green) of the amino acids at every position (from -10 to +10) on both sides of the Khib site.
The Motif-X program was performed to detect the sequence motifs in the Khib-modified peptides (Table S3). The five most abundant motifs were [KhibE], [DXXKhib], [GKhib], [AKhib] and [DKhib] (in which X indicates random amino acid residue) with frequency of 978, 672, 636, 594, and 504 respectively (Figures 2D, E). The motif enrichment heatmap showed a significant preference for negatively charged glutamate (E) and aspartate (D) around the modified lysine residue. In contrast to previous studies, alanine (A) and valine (V) showed great preference around the modified lysine residue (Figure 2F), which indicated the complex nature of the potential PTM mechanism in PC.
To systematically describe the Khib modified proteins, Wolfpsort software was applied to analyze the subcellular location of the 2-hydroxyisobutyrylated proteins in PC. There were 37%, 23%, 13%, 13%, and 6% Khib proteins distributed in the cytoplasm, nucleus, mitochondria, extracellular, and plasma membrane, respectively (Figure 3A), suggesting that the Khib modification was widespread in PC. Then, Gene Ontology (GO) analysis, including biological processes, cellular components, and molecular functions, was applied to demonstrate the potential functions and locations of the Khib proteins (Table S4). According to the cellular component data, Khib proteins were enriched in the cytoplasm (27%), organelles (16%), membrane-enclosed lumen (9%), and membrane (9%). In accordance with the molecular function analyses, the Khib proteins were mainly associated with binding (63%) and enzyme activity (21%), revealing that Khib modification may play a key role in protein function regulation, DNA transcription, and metabolic activity. The biological process analyses presented that the Khib proteins were mainly distributed in the metabolism (31%), biological process (13%), stimulation response (12%), and location (7%) (Figure 3B and Figure S3A).
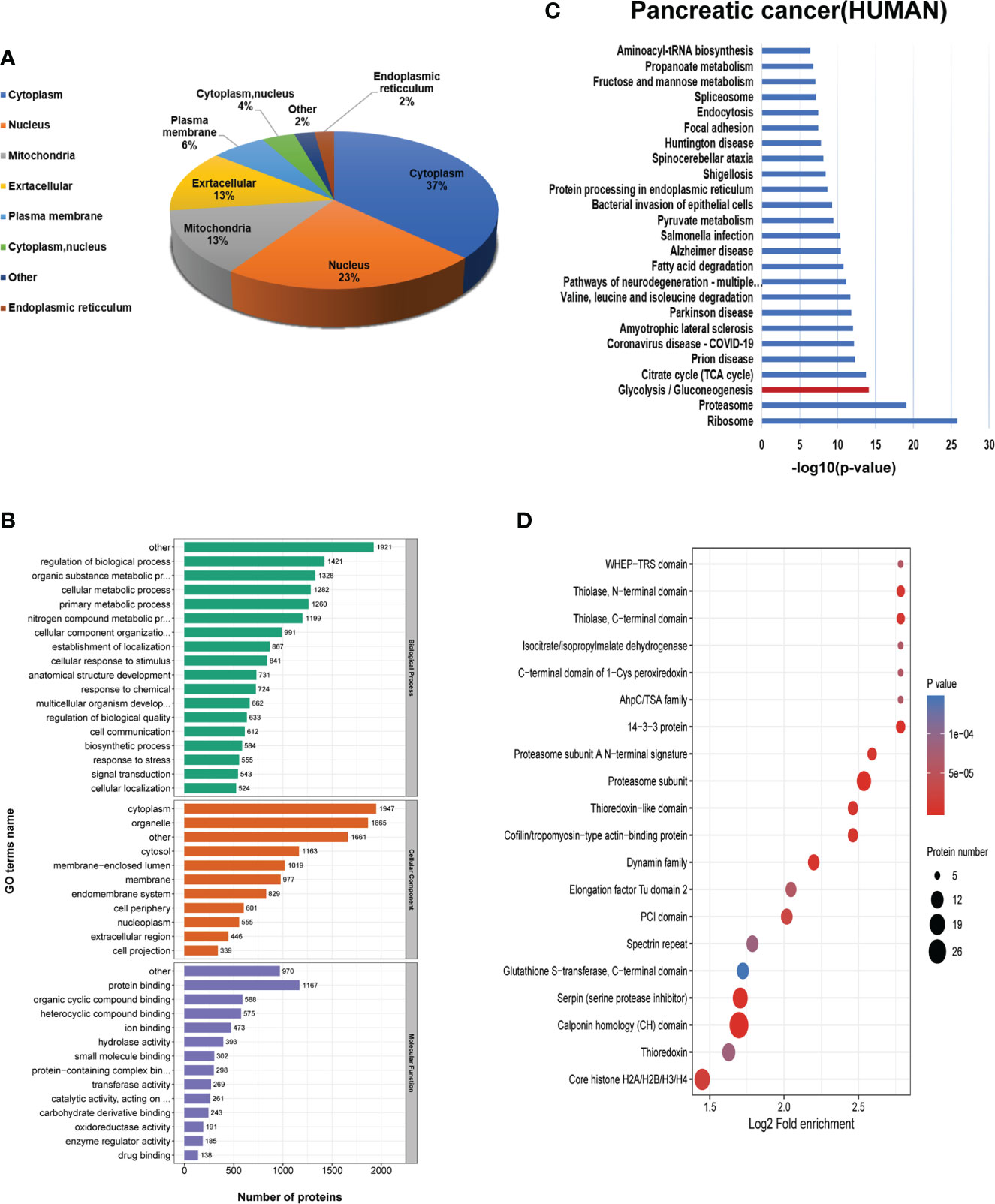
Figure 3 Bioinformatics analysis of the 2-hydroxyisobutyrylation (Khib) proteins. (A) Subcellular localization of the Khib proteins. (B) Gene Ontology (GO) classification analysis of the Khib proteins based on the biological process, molecular function, and cellular component. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the Khib proteins. (D) Protein domain enrichment analysis.
Furthermore, to explore the characteristics and role of the Khib proteins in PC, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed to show that proteins with Khib modification concentrated in metabolic pathways, including glycolysis/gluconeogenesis, TCA cycle, and fatty acid degradation, among others, in accordance with a previous study on Oryza sativa (11) and Ustilaginoidea virens (21) (Figure S3C). Also, ribosomes, proteasomes, and proteins related to multiple diseases were enriched in the KEGG pathways analysis, indicating that the Khib proteins had a close interrelationship with protein synthesis, degradation, bio-metabolism, and disease development (Figures 3C, B). The Khib proteins were observed intensively in the proteasome subunit, 14-3-3 protein, serpin (serine protease inhibitor), cofilin/tropomyosin-type actin-binding protein, thioredoxin-like domain, and Core histone H2A/H2B/H3/H4 in the protein domain enrichment analysis (Figure 3D).
Lysine 2-hydroxyisobutyrylation was first reported in the histones and was critical for regulating chromatin function (4). To investigate the Khib modification status of histones in PC, we listed 27 Khib sites on the histones identified in this study (Table S5), and a comparison was performed with other species, including rice seed (22), mammals (14), and moss (23) (Figure 4A), which revealed multiple overlapping Khib modification sites on the H3 and H4 proteins, and the specific sites H4K13 and H4K78 were identified. Among these, nine sites on H2B were observed, which were mainly distributed between K34-K120 in the globular structure, and it may affect the binding of H2B to DNA by shifting the charge state (4). Overall, different species have their unique Khib sites, suggesting that Khib is not only conserved but is also a unique PTM in histones.
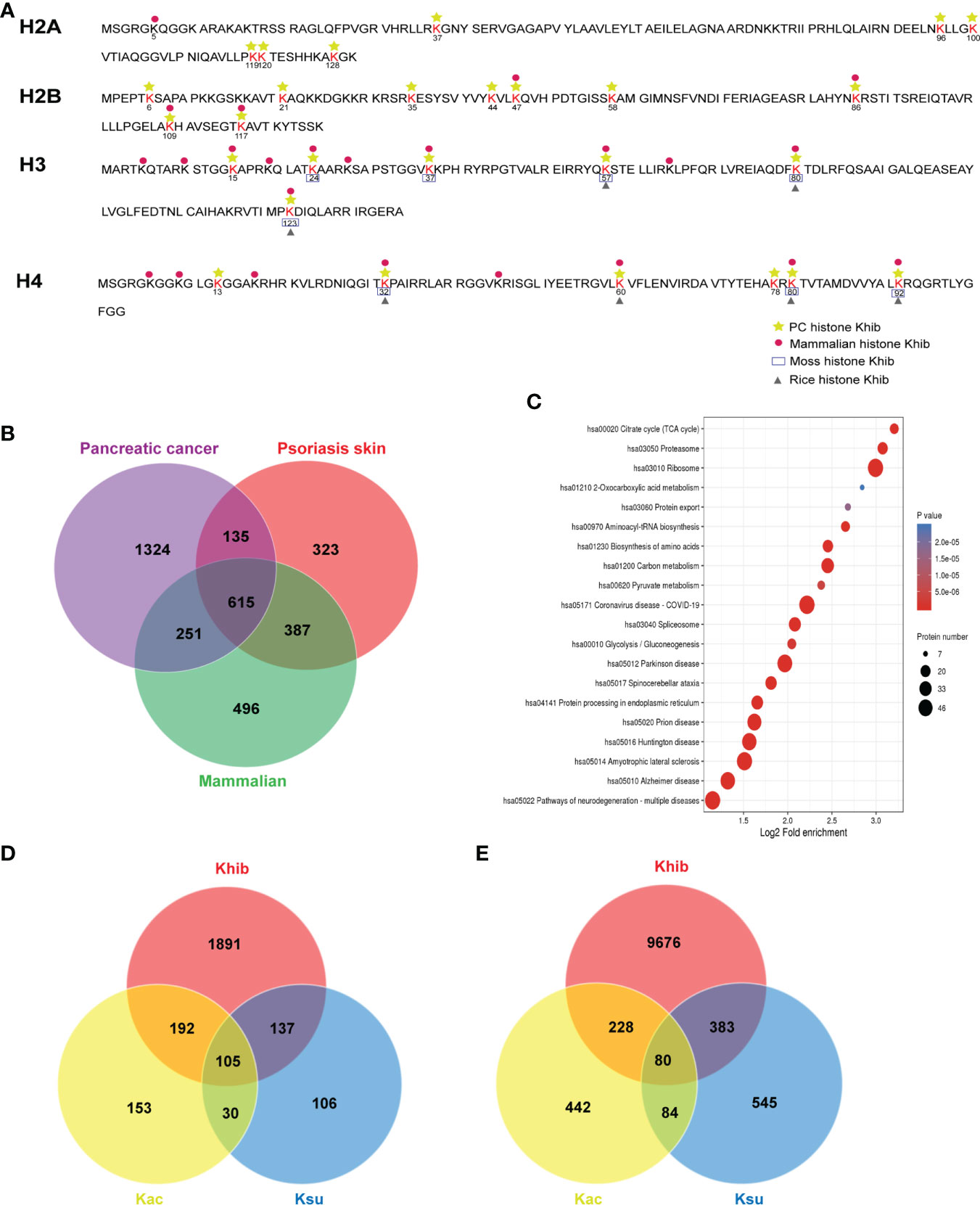
Figure 4 Conservation and uniqueness of the 2-hydroxyisobutyrylation (Khib) proteins in human pancreatic cancer (PC). (A) The comparison of the Khib histones in diverse species. (B) The number of overlapping Khib proteins among PC, psoriasis skin, and mammalian cells. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the common Khib proteins in PC, psoriasis skin, and mammalian cells. (D) The number of overlapping proteins among Khib, lysine succinylation (Ksu), and lysine acetylation (Kac). (E) The number of overlapping modified sites among Khib, Kac, and Ksu.
Simultaneously, the Khib modification in proteins excluding histones showed its features. By comparing the Khib proteins identified in mammalian (HeLa cells) (14), psoriasis skin (24), and PC detected in this study, 615 Khib proteins were observed in all the reports (Figure 4B and Table S6). The KEGG enrichment analysis of these common Khib proteins showed a preferred distribution in the ribosome and metabolism, including glycolysis/gluconeogenesis, carbon metabolism, and TCA cycle (Figure 4C), indicating that Khib modification was conserved among different species. Meanwhile, the Khib proteins, especially those populated in PC, were enriched in the metabolic, phosphatidylinositol 3-kinase (PI3K)-protein kinase B (Akt) signaling, and tight junction pathways (Table S6 and Figure S6), indicating that Khib modification was of great significance in the fields of bio-metabolism in PC
Post-translational modifications played crucial roles in regulating protein functions. It has been reported that various types of PTMs can be observed in one protein or even in the same locus. Compared with the results of previous studies in humans (25, 26), a total of 105 proteins with 80 sites were identified to be modified by Khib, lysine succinylation (Ksu), and lysine acetylation (Kac) (Figure 4D and Table S6). In addition, 242 and 297 Khib proteins presented Ksu and Kac modifications, respectively. In addition, 463 and 308 Khib sites were shared with Ksu and Kac, respectively (Figure 4E and Table S6). Among them, some lysine sites closely related to protein activity showed multiple modifications, suggesting that Khib might be biologically meaningful.
As described in the KEGG analysis, Khib proteins were concentrated in metabolic pathways in PC (Figure 3C). Compared with other species (Figure S3C) (11, 21), glycolysis/gluconeogenesis was observed in the Khib proteins analysis, suggesting that Khib plays a critical role in regulating glucose metabolism. Further, many enzymes in central metabolism, including glycolysis/gluconeogenesis, TCA cycle, and pentose phosphate metabolic pathways, were mapped to study the function of Khib in PC (Figure 5). It is worth noting that some critical enzymes represented multiple modification sites, including PGK, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and lactate dehydrogenase (LDH) (Table S7). Clearly, Khib modifications were prevalent in metabolic enzymes, and the modification status of specific sites affected enzyme activity and served a vital role in various metabolic pathways.
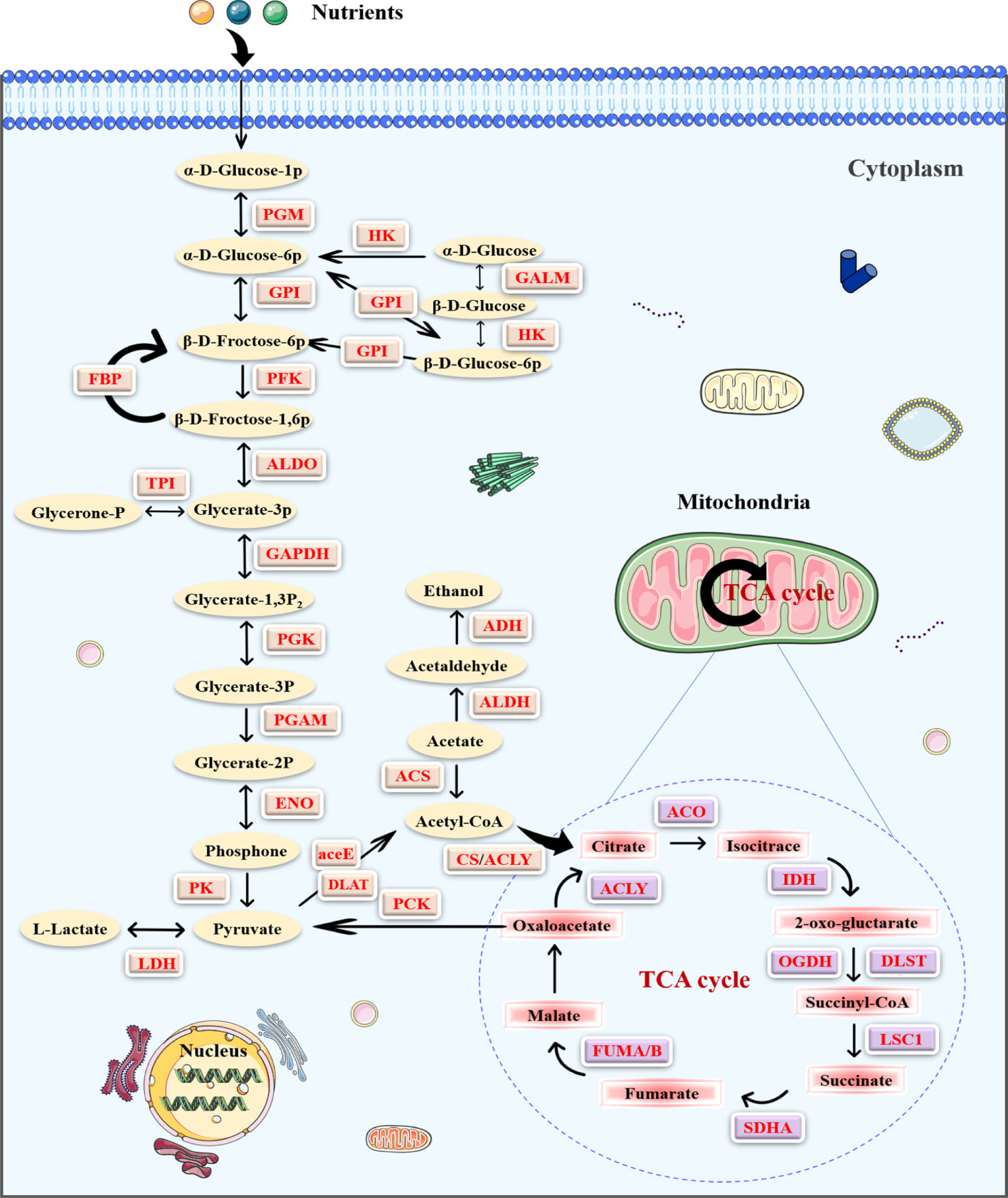
Figure 5 The enrichment of the 2-hydroxyisobutyrylation (Khib) proteins in the central metabolic pathways in pancreatic cancer (PC). The Khib enzymes are marked with red.
MG149 was identified as a selective inhibitor of Tip60 (27), which was known as a “writer” of Khib modification in mammalian cells (14). When treated with MG149, the level of Khib modification in PC cells was significantly decreased (Figures 6A, B and Figure S5). In order to investigate the role of Khib in PC development, the CCK8, colony-forming, wounding, and transwell assays were performed with the SW1990 and ASPC-1 cells. The CCK8 and colony-forming assay results suggested that MG149 contributed to reduced viability in the SW1990 and ASPC-1 cells (Figures 7A, B). Besides, the results of wounding and transwell assay demonstrated that the ability of migration and invasion was significantly suppressed when PC cells were treated with MG149 (Figures 7C, D). Thus, the results above strongly suggested that lysine 2-hydroxyisobutyrylation affected the PC cells’ proliferation, migration, and invasion ability in vitro.
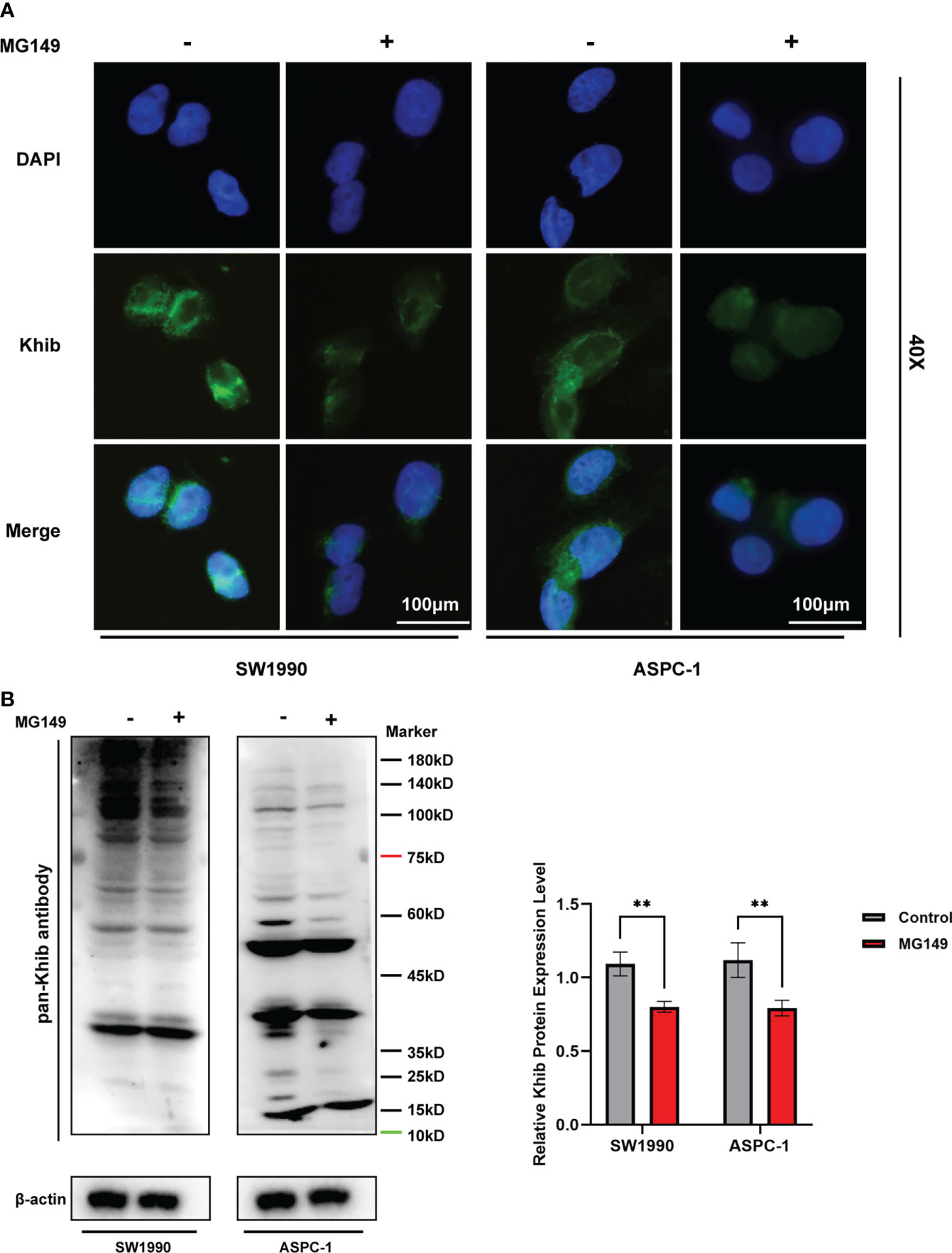
Figure 6 The total 2-hydroxyisobutyrylation (Khib) level in pancreatic cancer (PC) cells were suppressed by MG149. The SW1990 and ASPC-1 cells were treated with MG149 (74 μM), and then the immunofluorescence and Western blot assays were performed with pan-Khib antibody. (A) Immunofluorescence assay for PC cells treated with MG149. Different stains represent specific target genes, Khib (green) and DAPI (blue). Scale bar, 100 μm. (B) Western blotting analysis of the Khib proteins in the SW1990 and ASPC-1 cells treated with MG149. **P < 0.01, determined by independent Student’s t tests. Data are derived from three independent experiments and presented as means ± SDs.
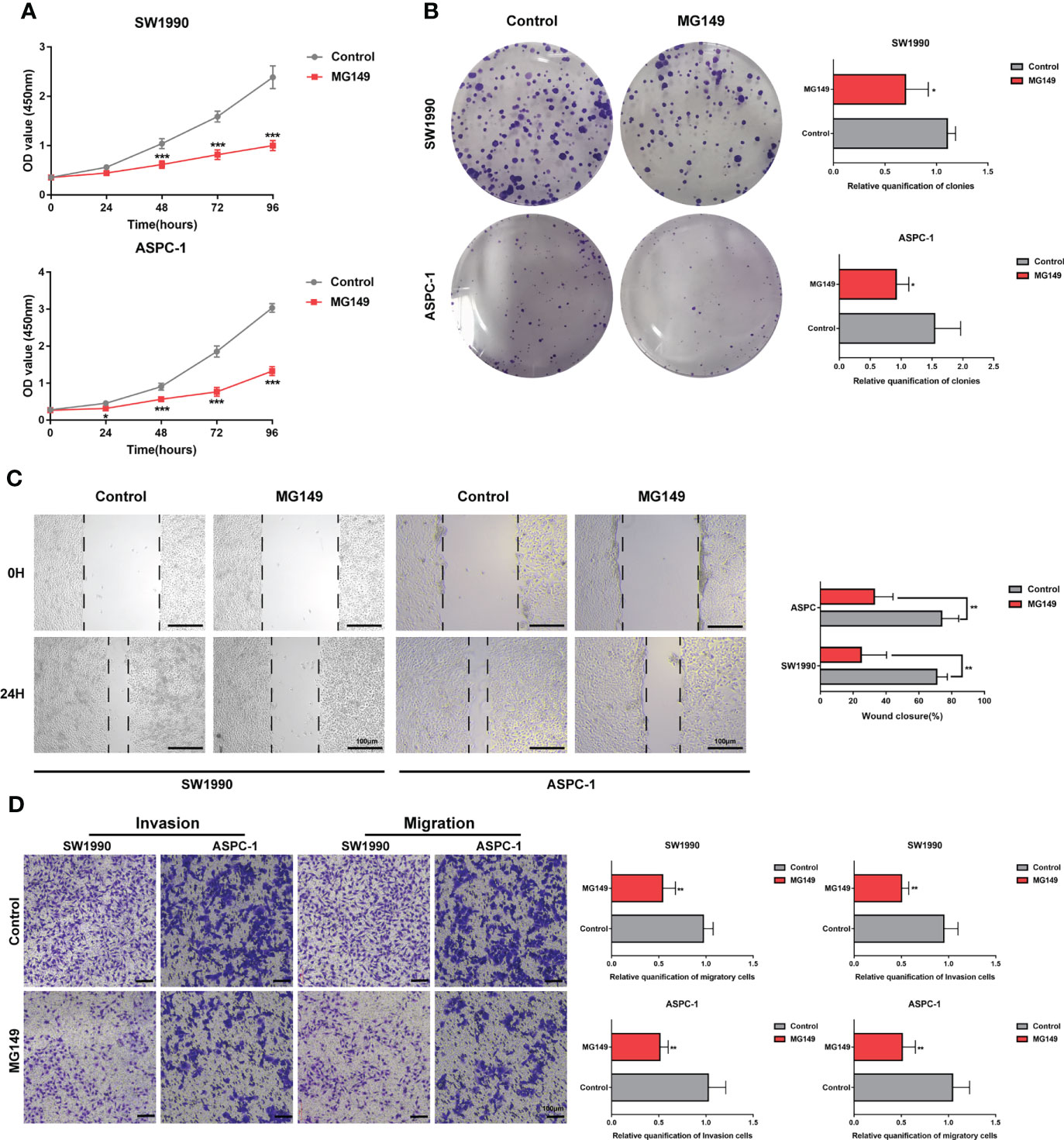
Figure 7 MG149 suppressed PC proliferation, migration, and invasion. (A) CCK8 assay was performed to measure the proliferation ability of PC cells. (B) Representative images of the colony formation assay of the SW1990 and ASPC-1 cells with or without MG149 (74 μM) treatment and culture in a complete culture medium for 14 days. (C) Representative images of the wound healing assays of the SW1990 and ASPC-1 cells with or without MG149 (74 μM) treatment, Scale bar: 100 μm. (D) Transwell assays were performed to examine the effect of MG149 on PC cell migratory and invasion ability. Scale bar: 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
Post-translational modifications of proteins, including phosphorylation, acetylation, and lactylation, alter protein function by adding chemical groups. 2-hydroxyisobutyrylation is a novel type of PTM identified in histones, and it played a crucial role in the regulatory function of chromatin (4) with operational differences from acetylation modifications (28). Khib is prevalent in several species, including plants, microorganisms, and humans. Recently, studies on Khib have been extended to non-histone proteins, and it is suggested to be involved in the development of human diseases. The Khib proteins identified in peripheral blood from patients with immunoglobulin A (IgA) nephropathy aggregate in the interleukin-17 (IL-17) signaling pathway and phagosome class (29), and they were also observed to be enriched in the PI3K-Akt signaling pathway in psoriasis skin lesions, which is associated to psoriasis progression (24). Conversely, Khib has rarely been studied in pancreatic cancer. To investigate the role of Khib in the progression of PC, we identified 10367 Khib sites on 2325 proteins in this study, which were more compared to that in previous studies, and expanded the databases of Khib proteins in PC.
In agreement with previous studies, 27 Khib sites in histones were identified in this study, of which the modification sites in H2B were mainly concentrated in the spherical structure of K34-K120, which may have affected the binding of H2B to DNA (4). Given that there are fewer studies on Khib in humans, for reasons of homology, we chose Khib data from mammalian cells and psoriasis skin in humans to perform a comparison with Khib proteins in PC. Notably, 615 Khib proteins were found in the above studies, and they were predicted to make great contributions to regulating metabolism pathways, including the TCA cycle, glycolysis, and carbon metabolism. In addition, 105 proteins and 80 sites were found to show three modifications by overlapping analysis with acetylation, succinylation, and 2-hydroxyisobutyrylation. This may result from regulation by the transcriptional co-activator P300, which was suggested to catalyze various modifications of histones, including acetylation, propionylation, butyrylation, and 2-hydroxyisobutyrylation (6, 28, 30, 31). Therefore, it is reasonable to suppose that there is either synergistic or competitive crosstalk among different types of PTMs, suggesting the possibilities of multiple modifications of proteins should be fully considered in studies of proteins, highlighting the importance and necessity of protein modification mapping.
Pancreatic tumor is exposed to severe physical, oxidative, and inflammatory stress. Thus, cancer cells rewire intermediary metabolism to promote unlimited proliferation in a hypoxic-ischemic environment, evade immune surveillance, and develope distant metastases. Previous studies have demonstrated that metabolic reprogramming promotes pancreatic tumorigenesis and metastasis through epigenetic regulation (32, 33). Notably, a total of 66 Khib proteins with 670 sites were observed in central metabolism (Table S7), and KEGG analysis showed an apparent enrichment in metabolic pathways, including glycolysis/gluconeogenesis, TCA cycle, and fatty acid degradation (Figure 3C). These results suggested that Khib may significantly influence PC metabolism and contribute to PC progression. MG149 was identified as a highly selective inhibitor of Tip60, which was known as a writer catalyzing Khib in mammalian cells (14). In order to explore the contribution of Khib to PC development, the SW1990 and ASPC-1 cells were treated with MG149 to decrease the total level of Khib modification. Then, we conducted the CCK8 and colony-forming assay, the results of which showed that MG149 strongly reduced the proliferation viability of the SW1990 and ASPC-1 cells compared to the control group. In addition, the results of the wounding and transwell assay suggested that MG149 suppressed the ability of migration and invasion in the SW1990 and ASPC-1 cells. The results above demonstrated that lysine 2-hydroxyisobutyrylation played an essential role in PC development and reminded us to pay more attention to 2-hydroxyisobutyrylation modification in PC. PTMs impact protein stability and function, and PTM therapies targeting proteins have made considerable advances in cancer (20, 34). It is promising that the inhibitors focusing on Khib proteins may be potential targets for pancreatic cancer therapy.
Since Khib modification is a new type of protein modification, it is still unexplored and lacks mature site-specific antibodies, making further in-depth studies challenging. Indeed, comparing data between cancerous and paraneoplastic tissues should be more convincing; however, given individual patient characteristics, a large number of samples are required. Based on the considerations above, we aimed to explore Khib protein profiles of PC with cancer tissue and provided a new perspective for further studies. In addition, although mass spectrometry has developed rapidly, the information on low-abundance proteins and amino acid residues with low recognition sensitivity was often missed, which made subsequent studies difficult. Bao et al. developed the 2-hydr_Ensemble residue identification algorithm with artificial intelligence approaches, which was more accurate than traditional algorithms in inferring Khib-modified residues (35) and it could be helpful for understanding the functions and further studying the Khib-specific modification sites. In the current study, we first described 10367 Khib-modified sites on 2325 modified proteins in seven persons with pancreatic cancer and then demonstrated that Khib modification played a significant role in PC development. Indeed, the potential mechanisms by which Khib promoted PC progression remained unclear, more samples and studies are needed to elucidate how lysine 2-hydroxyisobutyrylation affects PC development. Our work has undeniably helped to enhance our view of this new lysine PTM. The significant amount of Khib proteins identified in PC is expected to serve as an invaluable database for future pancreatic cancer-related research, providing new insights and potential targets for treating pancreatic cancer.
2-hydroxyisobutyrylation at lysine is a novel type of PTMs, first reported in the histones of HeLa cells. Recently, great studies about Khib modification identified in proteins have been reported in fields of diverse species. This study provides an in-depth insight into the Khib protein in pancreatic carcinoma for the first time and found that the Khib proteins are mainly enriched in bio-metabolic pathways. Further, we have demonstrated that Khib may act as a tumor promoter in PC development and provided a new perspective to explore the multiple functions of 2-hydroxyisobutyrylation and potential treatment targets in PC.
Seven patients with PC who had not been treated with radiotherapy were recruited for our study, and all patients independently signed written informed consent and donated tumor tissues for scientific research. This study was approved by the Tongji Hospital Research Ethics Committee and the Institutional Review Board. The enrolled patients’ demographic characteristics and clinical information are listed in Table S1. After surgery for PC, cancer tissues were rapidly dipped into MACS Tissue Storage Solution (Miltenyi Biotec GmbH, USA) and then stored at -80°C.
Samples were ground to cell powder in liquid nitrogen, and then four volumes of lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail, 3 μM TSA, and 50 mM NAM) were added, followed by sonication three times on ice, after centrifugation (12,000 g at 4°C for 10 min), and collection of the supernatant. Then, the protein concentration was determined by using the BCA kit.
The protein solution was reduced with dithiothreitol (5 mM, 56°C, 30min) and then alkylated with 11 mM iodoacetamide (15 min, room temperature, dark conditions) and diluted with 100 mM TEAB in order to make the sample urea concentration less than 2 M. The first digestion was performed overnight at a trypsin-to-protein ratio of 1:50(trypsin: protein, weight to weight), followed by second digestion of 1:100 for 4 h.
Tryptic peptides dissolved in NETN buffer were incubated overnight with prewashed antibody beads(PTM-802, PTM Bio), followed by washing with NETN buffer and H2O. 0.1% trifluoroacetic acid was used to elute the bound peptides and then combined and vacuum-dried. Finally, all samples were desalted for LC-MS/MS analysis.
Tryptic peptides were dissolved in 0.1% formic acid (solvent A) and directly loaded onto a reversed-phase analytical column (15-cm length, 75 μm i.d.). As described previously (13), the gradient comprised of solvent B at a constant flow rate of 400 nL/min on an EASY-nLC 1000 UPLC system. Then, the peptides were subjected to the NSI source, followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) coupled online to the UPLC, with the following parameter: 2.0 kV electrospray voltage, 350 to 1800 m/z scan range, and resolution of 70,000 for detection. Selected peptides MS/MS and fragments were detected in the Orbitrap at a resolution of 17,500. Depending on the data, a program of 20 MS/MS scans was carried out alternating after one MS scan with 15.0s dynamic exclusion. Automatic gain control (AGC) was programmed at 5E4. The fixed first mass was set at 100 m/z.
The MS/MS data were manipulated using the MaxQuant search engine (v.1.5.2.8). The detailed steps have been previously described (13).
Gene Ontology annotation was performed using the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). The KEGG database was applied to identify the protein pathway. The Motif-X algorithm was used to analyze the motif characteristics of the modification sites. Wolfpsort software was used to predict subcellular localization.
The human normal pancreatic duct epithelial (HPDE) and human PC cell lines (ASPC-1, BXPC-3, SW1990, and Mia PaCa-2, PANC-1) cell lines used in this study were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The cell line catalog number were shown as below: HPDE (bio-133286), ASPC-1 (bio-72969), BXPC-3 (bio-50655), SW1990 (bio-73389), Mia PaCa-2(bio-74690), PANC-1(bio-73125). HPDE, BXPC, ASPC-1, and SW1990 were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS). Mia PaCa-2 and PANC-1 were cultured in Dulbecco modified Eagle medium (DMEM) with 10% FBS. All cells were cultured in a humidified incubator at 37 °C and 5% CO2.
MG149 was used with a concentration of 74 µM, which was reported as half maximal inhibitory concentration (IC50) of Tip60 (27).
The western blotting assay was performed to detect the expression of the Khib proteins in PC tissues and cell lines. Briefly, cells or tissues were lysed by pre-formulated lysis buffer (RIPA, 1% Protease Inhibitor Cocktail, 3 μM TSA, and 50 mM NAM). The protein concentrations were measured by BCA kit (P0012S; Beyotime Biotechnology, China), and then we analyzed 50μg protein of every sample on 10% SDS-PAGE gels. All blots were visualized using ECL Kit (RM00020P; ABclonal, Wuhan, China) and the intensity of the bands was assessed by Image Lab (Bio-Rad, California, USA). The antibodies used in this study were: pan-lysine 2-hydroxyisobutyrylation antibody (1:1000; PTM-802; PTM Bio, Hangzhou, China), FITC antibody (1:50; AS011; ABclonal, Wuhan, China), β-actin antibody (1:1000; AC026; ABclonal, Wuhan, China). β-actin was used as a loading control and the intensity of the entire bands was assessed with ImageJ2 (National Institute of Mental Health, Bethesda, MD, USA). All experiments were performed three times following the same procedure.
Cells were spread evenly on coverslips and incubated for 24 hours, then fixed in 4% paraformaldehyde at room temperature for 15 min and permeabilized with 0.1% TX-100 for 10 min at room temperature. Then, the cells were blocked with 5% BSA in PBS for 1 hour and incubated overnight at 4°C with pan-Khib antibody (PTM-802; PTM Bio, Hangzhou, China). After washing three times with PBS, cells were incubated with the appropriate secondary antibody for 1 hour at 4°C. Coverslips were stained with DAPI and mounted. Immunofluorescence images were then observed and captured under a fluorescent microscope.
The viability of the SW1990 and ASPC-1 was evaluated with the cell counting kit-8 (Dojindo Laboratories Co. Ltd, Kumamoto, Japan). In brief, both SW1990 and ASPC-1 cells were seeded in 96-well plates at a density of 3 × 103 cells per well. There were five replicates per group (n = 5). At the time points indicated, 10 μL of CCK-8 solution was added to each well and incubated at 37°C and 5% CO2 for 3 hours. Then, the absorbance at 450 nm at different time points with the plate reader (Bio-Tek Elx 800, USA) to assess cell proliferation. All experiments were performed three times following the same procedure.
The SW1990 and ASPC-1 were seeded into 6-well plates with a density of 500 cells per well and cultured in a completed medium at 37°C and 5% CO2 for 14 days. Then, the indicated cells were washed with PBS, fixed with 4% paraformaldehyde, and stained with crystalline violet dye solution. After drying, the wells were photographed with a cell phone. All experiments were repeated three times.
The indicated PC cells were cultured in 6-well plates until the cell density approached 90%; we scratched the wound vertically across the center of the well with the tip of a 200 μL pipette gently. After being washed with PBS three times, the PC cells were cultured with a serum-free medium at 37°C, 5% CO2. The result of the assay was observed and captured with a microscope (Nikon, Japan) at 0 and 24 hours, respectively. All experiments were repeated three times.
To examine cell migration capacity, the PC cells were suspended at a density of 1 × 105/mL in a 24-well transwell plate (8 μm pore size; Corning) containing 200 μL of a serum-free medium in the upper transwell chamber, and 700 μL of a complete medium in the lower chamber. After co-culturing for 24 h, we fixed the PC cells on the sub-membrane surface with 4% paraformaldehyde and stained them with the crystalline violet solution. Counted the number of stained cells with a Nikon light microscope (Nikon, Japan). In the case of the invasion assay, 60 μL of Matrigel matrix gel is placed in the upper transwell chamber (BD Biosciences, America). Other operations performed were the same as cell migration assay described above. All experiments were carried out three times following the same procedure.
One-way analysis of variance (ANOVA) and Duncan’s multiple range test were performed by SPSS v22.0. Quantitative data were analyzed by two-tailed independent Student’s t tests.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material. The mass spectrometry proteomics data in this study were deposited in the ProteomeXchange Consortium via the PRIDE (36) partner repository with the dataset identifier PXD029817 (http://www.ebi.ac.uk/pride).
The studies involving human participants were reviewed and approved by Tongji Hospital Research Ethics Committee and the Institutional Review Board. The patients/participants provided their written informed consent to participate in this study.
YL conceived the idea and designed the study, analyzed data, and wrote the manuscript. XL, performed most of the experiment. KZ, PQ, ZD, provided help for data analysis. JW and WY supervised the entire project.
This study was supported by the National Natural Science Foundation of China (Nos.81874062 and 82072730), and the Youth Program of National Natural Science Foundation of China (81902439).
The study was supported by the National Natural Science Foundation of China (Nos.81874062 and 82072730), and the Youth Program of National Natural Science Foundation of China (81902439). We thank Jingjie PTM BioLab (Hangzhou, China) for technical support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1001807/full#supplementary-material
CCK8, cell counting kit-8; Khib, lysine 2-hydroxyisobutylation; Kac, lysine acetylation; Ksu, lysine succinylation; KEGG, kyoto encyclopedia of genes and genomes; LC-MS/MS, liquid chromatography with tandem mass spectrometry; OSCC, oral squamous cell carcinoma; PTM, post-translational modification; PC, pancreatic cancer; TCA cycle, tricarboxylic acid cycle; 2-HIBA, 2-hydroxybutyrate.
1. Huang KY, Su MG, Kao HJ, Hsieh YC, Jhong JH, Cheng KH, et al. dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins. Nucleic Acids Res (2016) 44(D1):D435–46. doi: 10.1093/nar/gkv1240
2. Lin H, Su X, He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem Biol (2012) 7(6):947–60. doi: 10.1021/cb3001793
3. Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. (2007) 6(5):812–9. doi: 10.1074/mcp.M700021-MCP200
4. Dai L, Peng C, Montellier E, Lu Z, Chen Y, Ishii H, et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat Chem Biol (2014) 10(5):365–70. doi: 10.1038/nchembio.1497
5. Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol (2011) 7(1):58–63. doi: 10.1038/nchembio.495
6. Huang H, Tang S, Ji M, Tang Z, Shimada M, Liu X, et al. p300-mediated lysine 2-hydroxyisobutyrylation regulates glycolysis. Mol Cell (2018) 70(4):663–78 e6. doi: 10.1016/j.molcel.2018.04.011
7. Vangipurapu J, Fernandes Silva L, Kuulasmaa T, Smith U, Laakso M. Microbiota-related metabolites and the risk of type 2 diabetes. Diabetes Care (2020) 43(6):1319–25. doi: 10.2337/dc19-2533
8. Calvani R, Miccheli A, Capuani G, Tomassini Miccheli A, Puccetti C, Delfini M, et al. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes (Lond). (2010) 34(6):1095–8. doi: 10.1038/ijo.2010.44
9. Rohwerder T, Muller RH. Biosynthesis of 2-hydroxyisobutyric acid (2-HIBA) from renewable carbon. Microb Cell Fact. (2010) 9:13. doi: 10.1186/1475-2859-9-13
10. Chen XF, Chen X, Tang X. Short-chain fatty acid, acylation and cardiovascular diseases. Clin Sci (Lond). (2020) 134(6):657–76. doi: 10.1042/CS20200128
11. Xue C, Qiao Z, Chen X, Cao P, Liu K, Liu S, et al. Proteome-wide analyses reveal the diverse functions of lysine 2-hydroxyisobutyrylation in oryza sativa. Rice (N Y). (2020) 13(1):34. doi: 10.1186/s12284-020-00389-1
12. Dong H, Guo Z, Feng W, Zhang T, Zhai G, Palusiak A, et al. Systematic identification of lysine 2-hydroxyisobutyrylated proteins in Proteus mirabilis. Mol Cell Proteomics. (2018) 17(3):482–94. doi: 10.1074/mcp.RA117.000430
13. Zhang N, Zhang L, Li L, Geng J, Zhao L, Ren Y, et al. Global profiling of 2-hydroxyisobutyrylome in common wheat. Genomics Proteomics Bioinf (2021) S1672-0229(21):00007-3. doi: 10.1016/j.gpb.2020.06.008
14. Huang H, Luo Z, Qi S, Huang J, Xu P, Wang X, et al. Landscape of the regulatory elements for lysine 2-hydroxyisobutyrylation pathway. Cell Res (2018) 28(1):111–25. doi: 10.1038/cr.2017.149
15. Yuan Y, Yuan HF, Geng Y, Zhao LN, Yun HL, Wang YF, et al. Aspirin modulates 2-hydroxyisobutyrylation of ENO1K281 to attenuate the glycolysis and proliferation of hepatoma cells. Biochem Biophys Res Commun (2021) 560:172–8. doi: 10.1016/j.bbrc.2021.04.083
16. Zhang Z, Xie H, Zuo W, Tang J, Zeng Z, Cai W, et al. Lysine 2-hydroxyisobutyrylation proteomics reveals protein modification alteration in the actin cytoskeleton pathway of oral squamous cell carcinoma. J Proteomics. (2021) 249:104371. doi: 10.1016/j.jprot.2021.104371
17. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin (2020) 70(1):7–30. doi: 10.3322/caac.21590
18. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res (2014) 74(11):2913–21. doi: 10.1158/0008-5472.CAN-14-0155
19. Spanknebel K, Conlon KC. Advances in the surgical management of pancreatic cancer. Cancer J (2001) 7(4):312–23.
20. Hsu JM, Li CW, Lai YJ, Hung MC. Posttranslational modifications of PD-L1 and their applications in cancer therapy. Cancer Res (2018) 78(22):6349–53. doi: 10.1158/0008-5472.CAN-18-1892
21. Chen X, Xu Q, Duan Y, Liu H, Chen X, Huang J, et al. Ustilaginoidea virens modulates lysine 2-hydroxyisobutyrylation in rice flowers during infection. J Integr Plant Biol (2021) 63(10):1801–14. doi: 10.1111/jipb.13149
22. Meng X, Xing S, Perez LM, Peng X, Zhao Q, Redona ED, et al. Proteome-wide analysis of lysine 2-hydroxyisobutyrylation in developing rice (Oryza sativa) seeds. Sci Rep (2017) 7(1):17486. doi: 10.1038/s41598-017-17756-6
23. Yu Z, Ni J, Sheng W, Wang Z, Wu Y. Proteome-wide identification of lysine 2-hydroxyisobutyrylation reveals conserved and novel histone modifications in physcomitrella patens. Sci Rep (2017) 7(1):15553. doi: 10.1038/s41598-017-15854-z
24. Ge H, Li B, Chen W, Xu Q, Chen S, Zhang H, et al. Differential occurrence of lysine 2-hydroxyisobutyrylation in psoriasis skin lesions. J Proteomics. (2019) 205:103420. doi: 10.1016/j.jprot.2019.103420
25. Guo Z, Pan F, Peng L, Tian S, Jiao J, Liao L, et al. Systematic proteome and lysine succinylome analysis reveals the enhanced cell migration by hyposuccinylation in esophageal squamous cell cancer. Mol Cell Proteomics (2021) 20:100053. doi: 10.1074/mcp.RA120.002150
26. Kozuka-Hata H, Kitamura A, Hiroki T, Aizawa A, Tsumoto K, Inoue JI, et al. System-wide analysis of protein acetylation and ubiquitination reveals a diversified regulation in human cancer cells. Biomolecules (2020) 10(3):411. doi: 10.3390/biom10030411
27. Ghizzoni M, Wu J, Gao T, Haisma HJ, Dekker FJ, George Zheng Y. 6-alkylsalicylates are selective Tip60 inhibitors and target the acetyl-CoA binding site. Eur J Med Chem (2012) 47(1):337–44. doi: 10.1016/j.ejmech.2011.11.001
28. Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol (2017) 18(2):90–101. doi: 10.1038/nrm.2016.140
29. Huang S, Zheng F, Lin H, Zhou X, Xu H, Zhang C, et al. Quantitative proteomics analysis of lysine 2-hydroxyisobutyrylation in IgA nephropathy. Clin Proteomics. (2021) 18(1):7. doi: 10.1186/s12014-021-09314-0
30. Xu X, Liu T, Yang J, Chen L, Liu B, Wei C, et al. The first succinylome profile of trichophyton rubrum reveals lysine succinylation on proteins involved in various key cellular processes. BMC Genomics (2017) 18(1):577. doi: 10.1186/s12864-017-3977-y
31. Zheng H, Song N, Zhou X, Mei H, Li D, Li X, et al. Proteome-wide analysis of lysine 2-hydroxyisobutyrylation in candida albicans. mSystems (2021) 6(1):e01129-20. doi: 10.1128/mSystems.01129-20
32. McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet (2017) 49(3):367–76. doi: 10.1038/ng.3753
33. Carrer A, Trefely S, Zhao S, Campbell SL, Norgard RJ, Schultz KC, et al. Acetyl-CoA metabolism supports multistep pancreatic tumorigenesis. Cancer Discovery (2019) 9(3):416–35. doi: 10.1158/2159-8290.CD-18-0567
34. Chen L, Liu S, Tao Y. Regulating tumor suppressor genes: Post-translational modifications. Signal Transduct Target Ther (2020) 5(1):90. doi: 10.1038/s41392-020-0196-9
35. Bao WZ, Yang B, Chen BT. 2-hydr_Ensemble: Lysine 2-hydroxyisobutyrylation identification with ensemble method. Chemometr Intell Lab (2021) 215(4):104351. doi: 10.1016/j.chemolab.2021.104351
Keywords: PTMs, post-translational modifications, Khib, lysine 2-hydroxyisobutyrylation, pancreatic cancer, proteomics
Citation: Lu Y, Li X, Zhao K, Qiu P, Deng Z, Yao W and Wang J (2022) Global landscape of 2-hydroxyisobutyrylation in human pancreatic cancer. Front. Oncol. 12:1001807. doi: 10.3389/fonc.2022.1001807
Received: 24 July 2022; Accepted: 12 September 2022;
Published: 30 September 2022.
Edited by:
Rui Vitorino, University of Aveiro, PortugalReviewed by:
Simone Sidoli, Albert Einstein College of Medicine, United StatesCopyright © 2022 Lu, Li, Zhao, Qiu, Deng, Yao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Yao, eXcxMzU1N0AxNjMuY29t; Jianming Wang, d2ptMThqZ21AYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.