
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 26 January 2022
Sec. Gastrointestinal Cancers: Gastric and Esophageal Cancers
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.822745
This article is part of the Research Topic The Role of non-coding RNAs in Gastrointestinal Cancers View all 18 articles
 Qingfan Yang1,2†
Qingfan Yang1,2† Yu Chen2,3,4†
Yu Chen2,3,4† Rui Guo1†
Rui Guo1† Yalan Dai1,2
Yalan Dai1,2 Liyao Tang3,4
Liyao Tang3,4 Yueshui Zhao2,3,4
Yueshui Zhao2,3,4 Xu Wu2,3,4
Xu Wu2,3,4 Mingxing Li2,3,4
Mingxing Li2,3,4 Fukuan Du2,3,4
Fukuan Du2,3,4 Jing Shen2,3,4
Jing Shen2,3,4 Tao Yi5*
Tao Yi5* Zhangang Xiao2,3,4*
Zhangang Xiao2,3,4* Qinglian Wen1,2*
Qinglian Wen1,2*Gastric cancer has developed as a very common gastrointestinal tumors, with recent effective advancements in the diagnosis and treatment of early gastric cancer. However, the prognosis for gastric cancer remains poor. As a result, there is in sore need of better understanding the mechanisms of gastric cancer development and progression to improve existing diagnostic and treatment options. In recent years, epigenetics has been recognized as an important contributor on tumor progression. Epigenetic changes in cancer include chromatin remodeling, DNA methylation and histone modifications. An increasing number of studies demonstrated that noncoding RNAs (ncRNAs) are associated with epigenetic changes in gastric cancer. Herein, we describe the molecular interactions of histone modifications and ncRNAs in epigenetics. We focus on ncRNA-mediated histone modifications of gene expression associated with tumorigenesis and progression in gastric cancer. This molecular mechanism will contribute to our deeper understanding of gastric carcinogenesis and progression, thus providing innovations in gastric cancer diagnosis and treatment strategies.
Gastric cancer (GC) is ranking the fifth most common cancer in the world, with over one million cases in 2018, nearly two-thirds of which occur in developing countries. GC remains the third leading cause of tumor-related death, just behind lung cancers and colorectal cancers (1–3). GC is a highly heterogeneous disease, and according to Lauren’s classification, it can be classified into two histological subtypes: the intestinal type and the diffuse type. High-throughput technologies have been well developed, and a new molecular classification is proposed by The Cancer Genome Atlas (TCGA), which subdivided GC into chromosomal instability (CIN)、genomic stability (GS)、microsatellite instability (MSI) and Epstein-Barr-Virus positivity (EBV) (4). There are four subtypes according to the WHO classification system, including papillary, tubular, signet ring, and mucinous (5).
GC is a heterogeneous cancer with multifactorial and unique epigenetic and genetic events, but the pathogenesis and molecular mechanisms remain elusive (6, 7). Both genetic and environmental factors are able to participate in the development and progression of GC (8). Genetic mutations have been considered as driving force for cancer development. However, current research data show substantial evidence that epigenetic alterations play an essential role in the development and progression of cancers, including GC (9, 10). Epigenetics refers to alterations in the process of gene expression without changes in DNA sequence, which are heritable, transient and reversible in gene expression (11). Epigenetic changes in cancer include DNA methylation, histone modifications and chromatin remodeling. The occurrence as well as progression of GC can be partially ascribed to environmental factors, including age, high salt intake, a diet low in fruits/vegetables and H. pylori infection (1).. However, environmental effects on GC are controllable, and only by exploring epigenetic regulation in depth can epigenetic alterations be controlled and managed. Histone modification occupies an important place in the regulatory mechanisms of epigenetics, and it is detected mainly in amino acids and carbohydrates, which also plays an essential role in cancer development (12). Recently, a large amount of literature indicates that noncoding RNAs (ncRNAs) are closely related to histone modifications in GC. In this review, the epigenetic alterations of histone post-transcriptional modifications associated with ncRNAs in gastric cancer have been intensively discussed.
Chromatin is a linearly arranged structure composed of millions of nucleosomes consisting mainly of DNA, RNA and histones (13). The fundamental unit of human genetic material is the nucleosome and consists of a histone octamer wrapped by a double-stranded DNA, this octamer contains two copies of histones H2A, H2B, H3 and H4 (14). Post-translational modifications of histones, including multiple covalent modifications such as methylation, acetylation, phosphorylation, ubiquitination, sumoylation, etc. They largely regulate DNA accessibility and gene expression (15). Currently, aberrant regulation of histone modifications has now been investigated in many kinds of cancers, and the role of histone modifications is being extensively studied. In this paper, we briefly describe the various types of histone post-transcriptional modifications and focus on the interaction between ncRNAs and histone acetylation/methylation in GC.
Histone acetylation, a reversible and dynamic process, is inversely regulated by deacetylases (HDACs) and histone acetyltransferases (HATs) (16). Acetylation neutralizes the positively charged lysine side chains to make the DNA structure more open and make it easier for transcription factors to combine with other proteins, which in turn promotes gene expression. In recent years, three major families of HATs have been identified, including MYST family (MOZ, Ybf2, Sas2, TIP60), Gcn5-related N-acetyltransferase family (GNAT) and orphan family (CBP/EP300 and nuclear receptors) (17). HDACs can be classified into four categories: Class I HDACs, consisting of HDACs 1, 2, 3 and 8; Class II HDACs, consisting of HDACs 4, 5, 6, 7, 9 and 10; Class III HDACs, consisting of seven silencing regulatory proteins, including NAD-dependent protein deacetylases and ADP ribosylases, also known as Sirtuins; Class IV HDACs, containing only HDAC11, has sequence similarity compared to class I and II proteins (16). HATs relax chromatin structure and promote transcription through transferring the acetyl group from acetyl coenzyme A to histone amino acid terminus. In contrast, HDACs make the chromatin structure more compact by removing the acetyl group from the lysine terminus, thereby inhibiting transcription. Altered overall levels of histone acetylation have been shown to be connected with many tumor phenotypes (18). In general, hyperacetylation leads to increased gene expression, especially when proto-oncogenes are involved, and gene expression might be activated. Hypoacetylation of oncogenes is usually localized to the promoter simultaneously with DNA methylation, resulting in suppression of gene expression (19).
Histone methylation plays an important role in cell biological fate, such as DNA recombination and damage repair, gene expression and cell differentiation, and so forth (20). Histone methylation, catalyzed by histone methyltransferases (HMTs), occurs mainly on lysine and arginine residues located at histone tails. Arginine can be mono- or dimethylated, while for lysine, apart from mono- and dimethylated, it is able to be trimethylated (21). Histone methylation regulatory genes include protein arginine methyltransferases PRMTs, lysine methyltransferases KMTs (e.g. EZH2 and DOT1L), and histone demethylases HDMs (e.g. LSD1 and KDMs). The function of all KMTs is to bind s-adenosylmethionine (SAM) methyl donors to lysine methyl acceptors and facilitates methyl transfer from SAM to the telomeres (22). Protein arginine methylation reactions are mainly catalyzed by the protein arginine methyltransferase family, including PRMT1-9. These enzymes catalyze the reaction of transferring a methyl group from S-adenosylmethionine (AdoMet) to a guanidino nitrogen of arginine, which results in the formation of methylarginine and S-adenosylhomocysteine (AdoHcy) (23). Lysine methylation occurs mainly on histone H3 and H4, for example, six lysine residues, H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20, are proven to be methylated in histones H3 and H4, respectively. Among them, acetylation can take place in histone H3 for H3K9 and H3K27. Lots of other lysine residues, H3K14, H3K18, H3K23 and H4K5, H4K8, H4K12 and H4K16, can only be acetylated rather than methylated. Methylation/acetylation in H3K9 and H3K27 performs different physiological functions, for instance, acetylation of H3K9 and H3K27 leads to gene transcriptional activation, whereas methylation of H3K9 and H3K27 represses transcription (14).
Histone phosphorylation occurs mainly at serine, tyrosine and threonine residues in the histone tails, which is regulated by various kinases and phosphatases, mediating biological fate such as DNA damage repair, chromatin remodeling, transcriptional activation and apoptosis (24). During mitosis, histone phosphorylation disrupts the balance of interactions between histones and DNA, resulting in an unstable chromatin structure, which ultimately allows the chromatin structure to remodel into homologous chromosomes (21). It is worth noting that histone phosphorylation mediates the transcriptional regulation of genes, particularly those that regulate the cell cycle and cell proliferation. For example, H3Ser10p and H2BSer32p are involved in epidermal growth factor (EGF)-mediated gene transcriptional activation (25, 26). In addition, H3ser10p mediates the transcriptional activation of proto-oncogenes such as c-myc and c-fos (26, 27). Histone H3 threonine 45 (H3T45) is phosphorylated with the participation of protein kinase C1, which promotes acetylation of H3K56 and ultimately regulates apoptosis and DNA replication (28, 29). A growing number of studies have shown that histone phosphorylation is involved in tumorigenesis and progression. For example, histone H4 phosphorylation is closely associated with liver regeneration and hepatocellular carcinoma (30). Increased levels of histone H3S10 phosphorylation is involved in the proliferation of gastric cancer cells and can be an independent prognostic indicator of vascular infiltration in gastric cancer (31, 32).
In post-transcriptional modifications, protein ubiquitination is a common process in cells (33, 34). Ubiquitination is a cascade reaction that relies on Adenosine Triphosphate (ATP) to link ubiquitin to a substrate protein. Ubiquitin activated by ubiquitin activating enzymes E1s is transferred to the ubiquitin binding enzymes E2s and ultimately transferred to the substrate protein by the ubiquitin ligases E3s (35). Among them, the ubiquitin ligases E3s are key enzymes in the ubiquitination process because of their specific recognition of the substrate protein (36). Histone ubiquitination is also involved in DNA damage repair, gene transcriptional regulation and genomic stability (37). Protein deubiquitination is catalyzed by deubiquitinating enzymes (DUB) and the dynamic balance of ubiquitination and deubiquitination plays an important role in cellular homeostasis. Therefore, their aberrant expression has been shown to be involved in the development and progression of many cancers (38–40).
Sumoylation has the same mechanism compared with ubiquitination, allowing for the covalent attachment of small ubiquitin-like modifiers (SUMO) to lysine residues of the substrate protein, which is a reversible post-transcriptional modification (41). Sumoylation acts as a negative regulator involved in the regulation of protein stability, gene transcription and ultimately repression of gene transcription (42, 43). Most of the substrate proteins for sumoylation are nuclear proteins. For example, the sumoylation of transcription factor can recruit chromatin-modifying enzymes to repress gene expression (44). In addition, sumoylation regulates the expression and activity of histone-modifying enzymes, suggesting a close relationship between sumoylation and epigenetic regulation (24). For instance, sumoylation of the transcription factor E2F1 promotes EZH2 transcription by increasing the binding of E2F1 to the EZH2 promoter (45). It has been shown that SUMO-1 is involved in the proliferation and apoptosis of endometrial cancer cells by increasing the level of histone H4 sumoylation (46). Another study shows that the sumoylation of transcription factor ETV1 promotes prostate cancer development (47), suggesting that sumoylation is involved in tumorigenesis and progression.
Noncoding RNAs (ncRNAs) are a unique kind of RNA transcripts that are intensively recognized in eukaryotic genomes (48). Although over 75% of the human genome can be transcribed, among them, only ca. 2% of the transcription products are translated into proteins, and the remaining transcripts (which cannot encode proteins) are initially treated as junk transcription products (49). Transformed from junk transcription products to functional molecules that regulate cellular processes, ncRNAs regulate gene expression at different levels during physiological development, including signal transduction, chromatin remodeling, gene transcription and post-transcriptional modifications (50). Through their regulatory networks, ncRNAs can modulate many downstream targets to mediate specific cell biological fate (51). Growing evidence indicates that ncRNAs influence normal cell function and disease progression, especially in cancer development, which are considered as tumor suppressors and oncogenic factors (52, 53). Meanwhile, high-throughput sequencing technologies developed a lot, and increasing studies are showing that a little bit of ncRNAs have small open reading frames (sORFs), which can encode peptides or proteins (48). It has been demonstrated that HOXB-AS3, encoded by lncRNA, is shown to regulate tumor energy metabolism. FBXW7-185aa 18 and PINT-87aa 20, encoded by circRNA, can block tumor cell cycle and inhibit cell expansion. SHPRH-146aa 19, encoded by circRNA, can suppress tumor cell expansion and malignant phenotype. miPEP-200a and miPEP-200b, encoded by miRNAs, regulate epithelial-mesenchymal transition (EMT), leading to tumorigenesis and progression. According to transcription length, ncRNAs are mainly classified as long noncoding RNA (lncRNAs, linear, >200 nucleotides), circular RNA (circRNAs, covalently closed continuous loops), and short ncRNAs (linear, <200 nucleotides) (54).
miRNAs are considered as a class of highly conserved tissue-specific small molecule non-protein-coding RNAs. Mature miRNAs are generated from primary miRNAs (pri-miRNAs) by two consecutive enzymatic reactions and loaded into RNA-induced silencing complexes (RISCs) containing Argonaute 2 (AGO2) (55, 56), which repress messenger RNAs (mRNAs) translation and promote mRNA degradation by directly combining with mRNA, and ultimately inhibit gene expression at the post-transcriptional stage to maintain intracellular homeostasis (57, 58). They are powerful regulatory factors of various biologically important activities such as cell growth and development (59). Regulatory mechanisms of miRNAs action include gene polymorphism, miRNA promoter methylation, interactions with RNA binding proteins (RBPs) or other RNAs, nucleotide modification of miRNA sequences at different stages of maturation, and unsymmetric miRNA strand selection (60) (36). miRNAs can repress the expression and activity of target RNAs at the post-transcriptional level, and the mechanism is the recognition of miRNA response elements (MREs) of target RNAs. MREs are usually located in the 3’ untranslated region (3’UTR) of the RNA, and can also be located in the 5’ untranslated region (5’UTR) and in the coding sequence (CDS). miRNAs recognize and bind sequence-complementary MREs at the 3’UTR of the target transcription product, and their inhibition of protein synthesis mechanism is not fully understood (56). When RNA transcripts have identical MREs on their 3’UTR, they can act as competing endogenous RNAs (ceRNAs), influencing each other’s expression by competing and combining with miRNAs from the same library. Each miRNA can target multiple RNAs, and at the same time, each RNA can be a target of multiple miRNAs. Ultimately, miRNAs can form a complex ceRNET with their associated transcripts (Figure 1A) (61). Herein, we highlight a few example from each category, with Figure 1B providing visual representations of the ceRNA network (62–64). Many physiological and pathological processes, including cardiovascular/metabolic diseases and cancers, are proven to be heavily dependent on miRNAs (65). Therefore, tumor development, such as cell proliferation, motility, apoptosis and angiogenesis, can be affected by aberrant expression of miRNAs (66). Recently, growing studies have demonstrated that deregulation of miRNAs expression is related to gastric cancer by acting as oncogenes and tumor suppressors, which can contribute to the diagnosis and treatment of GC (67–69). What’s more, there is growing evidence suggesting that circulating plasma and serum miRNAs can act as potential biomarkers for non-invasive tumor diagnosis, cancer therapy and prognosis (70–72).
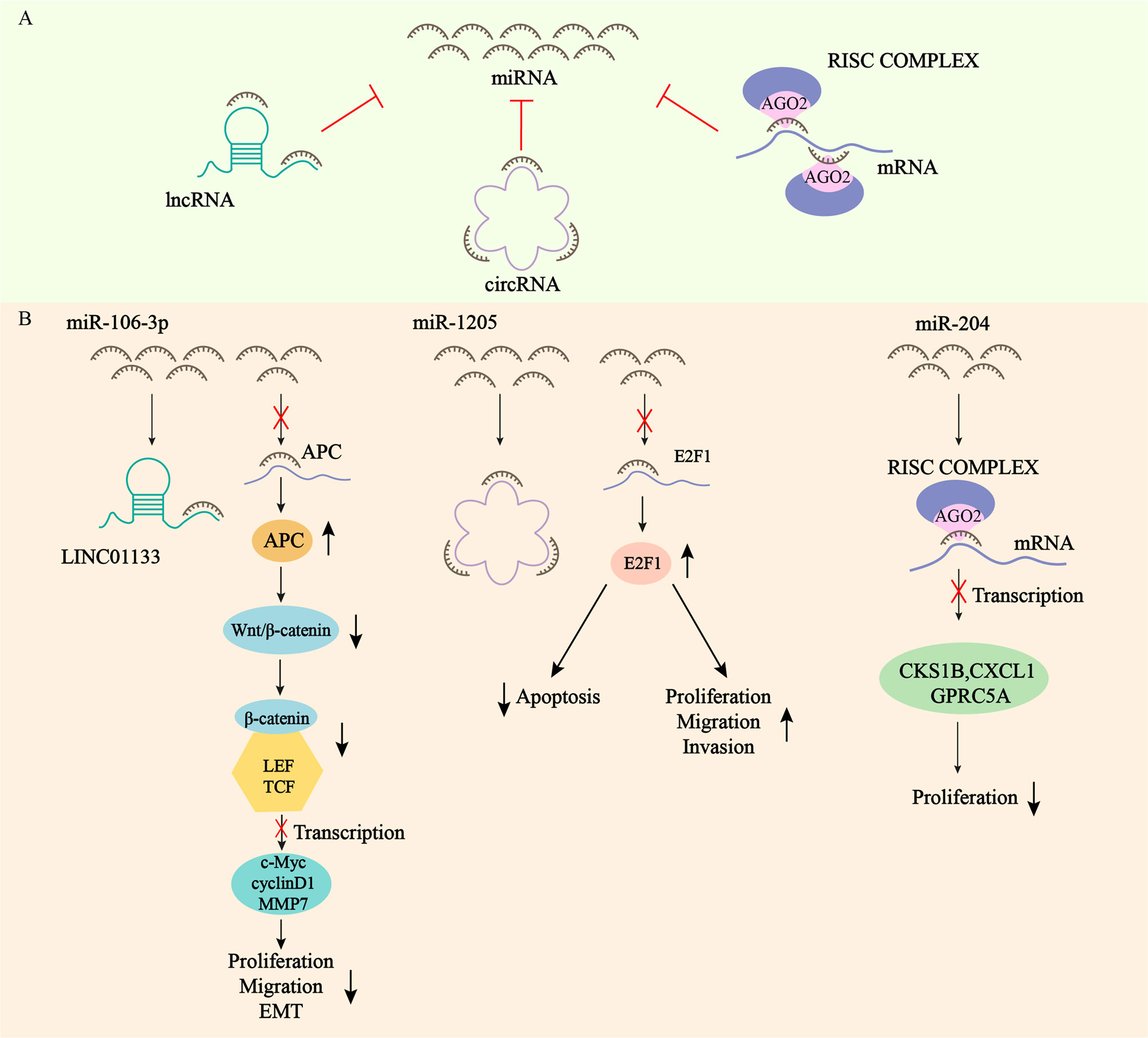
Figure 1 Network of competitive endogenous RNAs. (A) mRNAs, lncRNAs, and circRNAs all contain microRNA (miRNA) response elements (MREs) that can bind to miRNAs and regulate the expression of RNA targets at the post-transcriptional level. (B) LINC01133 exerts oncogenic effect by acting as a sponge for miR-106-3p, which regulates the expression of APC and inactivates the Wnt/β-catenin signaling pathway. circCYFIP2 exerts oncogenic effect by acting as a sponge for miR-1205, which regulates the expression of E2F1. miRNA-204 inhibits the transcription of CKS1B, CXCL1 and GPRC5A mRNAs by binding to AGO2 to form the RISC complex, which ultimately inhibits the proliferation of GC.
lncRNA is defined as a transcript greater than 200 nt in length, and typically lacks the capability of encoding proteins (73). lncRNAs exert a variety of biological functions at the epigenetic level by regulating gene expression. As a result, tumorigenesis and progression are highly dependent on the dysfunctional lncRNAs (74). Recent studies disclose that a lot of lncRNAs are aberrantly regulated in GC, and altered lncRNA expression participates in cancer pathological processes, such as cell proliferation, metastasis, EMT, apoptosis, tumor stemness, drug resistance, etc. Therefore, it has the potential to act as a biomarker and therapeutic target for GC (75–78). Furthermore, it has been shown that the broad expression pattern in cancer, stability in circulating body fluids (plasma and urine) and tumor specificity of lncRNAs imply that subtype/tissue-specific expression of lncRNAs is important for the discovery of novel diagnostic, prognostic and therapeutic targets (79–81).
Circular RNAs (circRNAs) are covalent closed-loop structures formed by post-sniping of precursor mRNAs without 5’ caps or 3’ polyA tails, and they are also a class of endogenous RNAs without function of encoding proteins or peptides (82). Unlike linear RNA, circRNA has a special cyclic covalent bond structure, which makes them evolutionarily conserved and highly stable. Thus, they are inherently resistant to RNA exonucleases as well as RNaseR activity and are widely expressed in complex tissues and cell types at specific stages (83). Literature demonstrates that circRNAs have many biological functions, such as microRNA sponges, transcriptional regulation, regulation of mRNA stability, encoding functional proteins, and binding to RNA-binding proteins (84). Therefore, circRNAs have obvious impacts on many human diseases, such as diabetes, neurological/cardiovascular diseases, and cancers, thus exhibiting great potential as diagnostic and therapeutic targets (85, 86). In the case of GC, the importance of dysregulated circRNAs in gastric carcinogenesis, progression and clinicopathology has been demonstrated, thus showing the value of biomarkers for diagnosis and treatment (87, 88).
Firstly, ncRNAs specify the histone modification pattern of target genes by recruiting tissue modification complexes directed to specific gene loci and might repress cis or trans transcription of target genes (Figure 2); Secondly, ncRNAs regulate target genes by participating in ceRNET as miRNA sponges (Figure 3); Thirdly, they can directly target histone modifiers and affect the alteration of histone or non-histone modifications pattern (Figure 4). Through these mechanisms, they drive the malignant biological behaviors of GC cells and ultimately promote tumor formation and progression (Table 1 and Table 2).
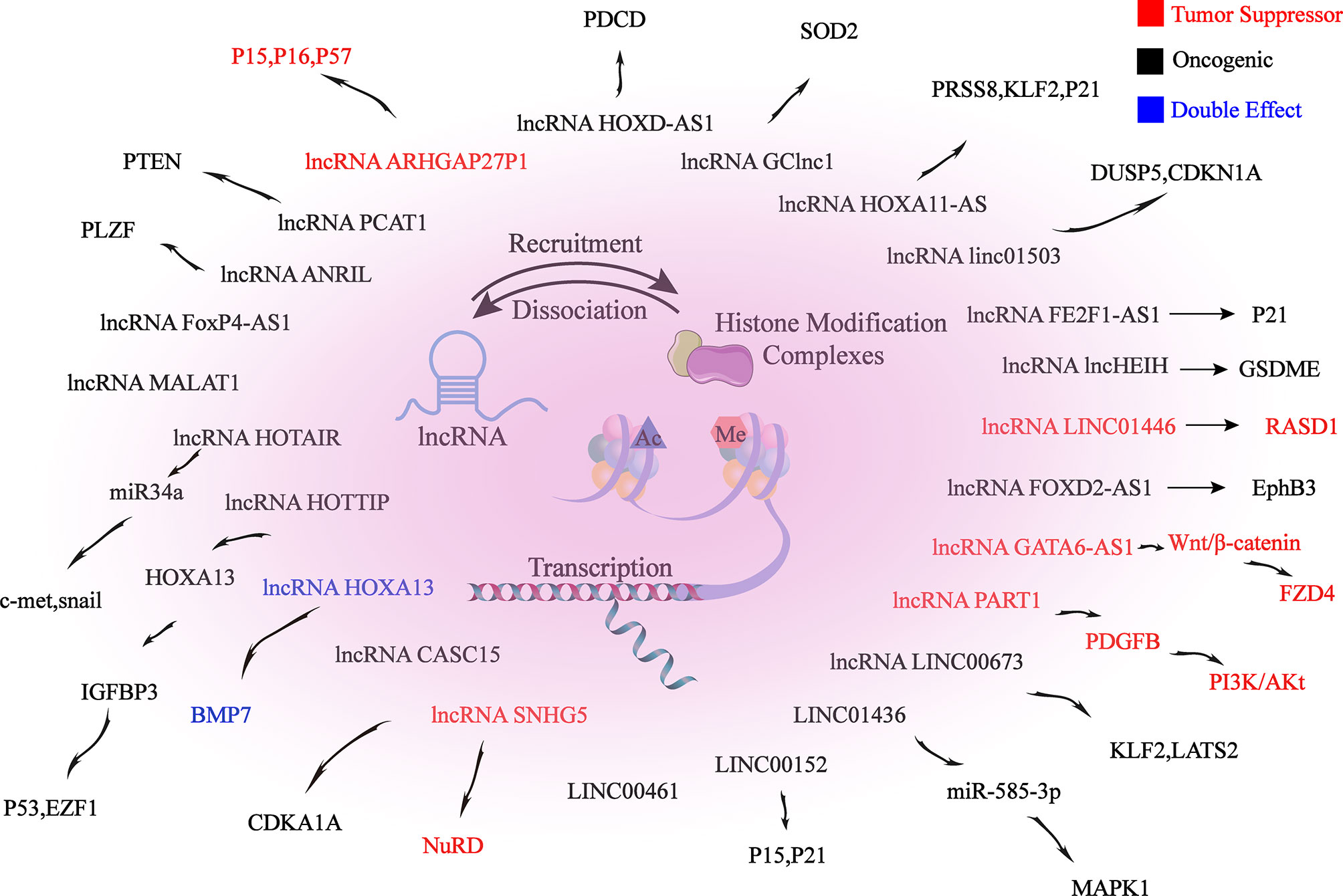
Figure 2 lncRNAs mediate histone modifications of target genes. lncRNAs induce changes in promoter histone modification patterns by recruiting histone modification complexes to target gene promoter-associated binding sites, thereby epigenetically regulating target gene expression, which ultimately alters the biological behaviors of GC.
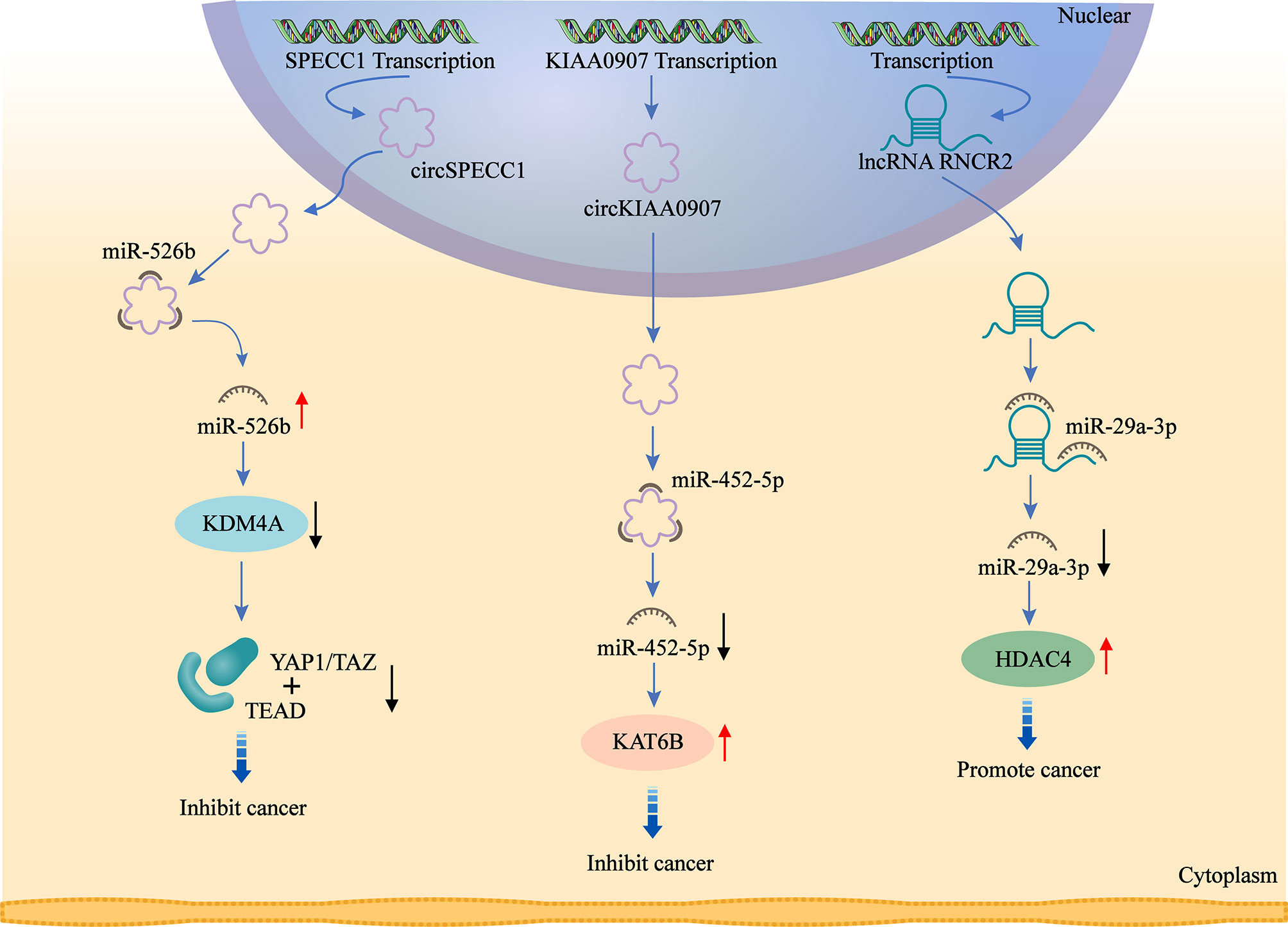
Figure 3 ceRNAs are involved in the regulation of histone modifiers. circ_0000745 acts as a sponge for miR-526b and promotes KDM4A expression. circKIAA0907 acts as a sponge of miR-452-5P and promotes KAT6B expression. lncRNA MIAT acts as a sponge for miR-29a-3p and promotes HDAC4 expression. ceRNAs alter histone and non-histone modifications by regulating the expression of histone modifiers, which affects downstream signaling targets and ultimately exerts their oncogenic or anti-tumor effects.
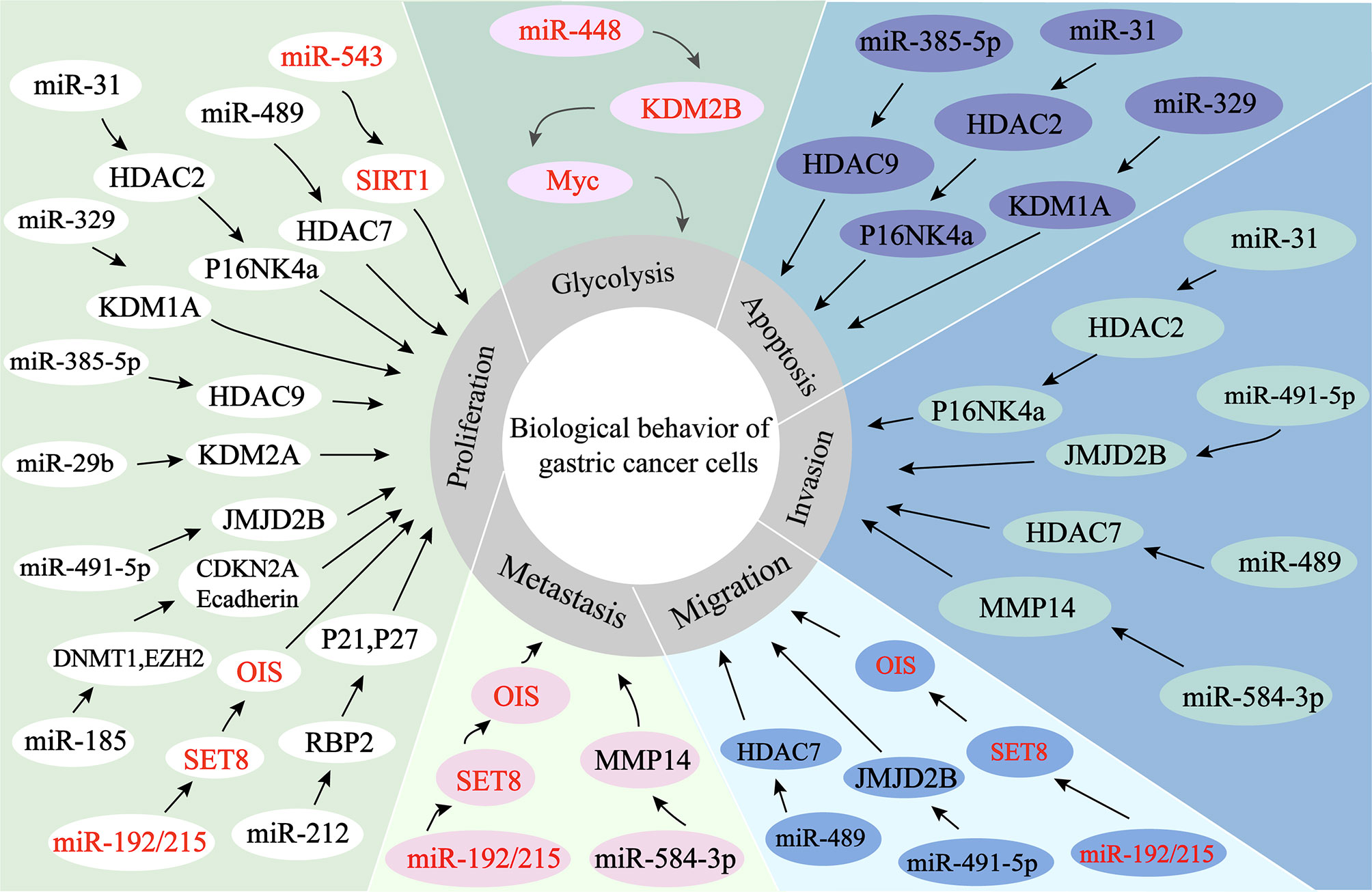
Figure 4 miRNAs regulate histone modification in GC. 1. miR-584-3P recruits EZH2 and EHMT2 to the MMP-14 promoter, thereby inhibiting the expression of MMP-14 and ultimately exerting its oncogenic effects. 2. miRNAs directly target histone modifiers and affect the alteration of histone or non-histone modifications patterns, ultimately leading to changes in the biological behaviors of gastric cancer cells.
The expression of miR-584-3p is decreased in GC. miR-584-3p inhibits MMP-14 transcription by recruiting euchromatic histone lysine methyltransferase 2 (EHMT2) and enhancer of zeste homolog 2 (EZH2) to interact with AGO2, leading to enrichment of repressive epigenetic markers and reduced binding of YY1 to the MMP-14 promoter. YY1 (one member of the GLI-Krüppel protein family) directly targets the matrix metalloproteinase 14 (MMP-14) to promote MMP-14 expression in GC cells and tissues, resulting in tumorigenesis and invasion. miR-584-3p ultimately inhibits GC cell growth, metastasis and angiogenesis by suppressing the expression of MMP14 (90).
In GC cells, miR543 expression is upregulated, promoting cell cycle and proliferation, and thus positively correlates with the clinical phenotype of GC patients. By directly targeting the SIRT1 3 ‘-UTR, miR-543 can inhibit the expression of SIRT1 mRNA, thereby promoting GC cell proliferation and cell cycle progression. SIRT1, a class III histone deacetylase, is downregulated in GC and can inhibit gastric carcinogenesis and progression (90).
miR-489 is significantly downregulated in GC cells and tissues, and the downregulation can promote GC cell proliferation, invasion and migration, which is positively correlated with the prognosis of GC patients. Knockdown of HDAC7, a direct target of miR-489, can inhibit GC development as well as antagonize the effects of miR-489 inhibitors on GC cells. Taken together, by targeting HDAC7 and blocking the PI3K/AKT pathway, miR-489 exerts tumor suppressive effects in GC cell growth (99).
The overexpression of miR-31, which is aberrantly downregulated in GC cell lines, might inhibit proliferation and migration of GC cells, as well as induce apoptosis. miR-31 expression in GC is epigenetically regulated, and down-regulation of miR-31 is related to DNA methylation of promoter. In addition, HDAC2 can act as a direct target of miR-31 through combining with MREs in the 3’-UTR, and its expression is negatively regulated by miR-31. HDAC2 inactivation restores the activity of p16 INK4a, which has anti-tumor effects in gastric cancer (91).
miR-383-5p expression is downregulated in GC cells and tissues. Therefore, miR-383-5p overexpression inhibits proliferation, induces apoptosis, as well as suppresses tumor growth in GC cell lines. HDAC9 is a direct target of miR-383-5p, which can be upregulated in GC cell lines. Therefore, it is closely associated with malignant development of GC. As a result, knockdown of HDAC9 in GC cell lines promotes apoptosis and inhibits proliferation, and microRNA-383-5p can suppress GC development through targeting HDAC9 expression (92).
miRNA-329 expression is reduced in GC. miRNA-329 overexpression in GC cells promotes apoptosis and inhibits the malignant biological behaviors of GC cells. miRNA-329 mimics and siRNAs targeting KDM1A, a downstream target of miRNA-329, reduce the expression of KDM1A and increase the levels of H3K4me1 and H3K4me2. As a result, patient with high expression of miRNA-329 or low expression of KDM1A has a longer overall survival. In summary, microRNA-329 promotes apoptosis and inhibits proliferation, metastasis and growth by negatively regulating KDM1A in GC cells (93).
miR-448 is overexpressed in GC, promotes glycolysis and growth of GC, and is associated with poorer survival. The mechanism is that miR-448 expression is increased in GC, and the increasing expression of miR-448 upregulates the expression of the rate-limiting enzyme of glycolysis, Myc, through directly inhibiting KDM2B, which ultimately stimulates glycolysis (94).
miR-212 expression is aberrantly reduced in human GC cells and tissues. miR-212 is found to inhibit RBP2 expression through directly combining with the 3’UTR site of Retinoblastoma binding protein 2 (RBP2). RBP2, a newly identified histone demethylase, is overexpressed in GC. miR-212 exerts oncogenic effects in gastric cancer by suppressing RBP2 expression and increasing the expression of P21 CIP1 and P27 kip1 to arrest the cell cycle and inhibit cell colony formation (100).
miR-192/215 expression is significantly increased in GC cells and tissues. miR-192/215 can directly combine with the 3’UTR of SET8 to facilitate proliferation and metastasis of GC cells. Histone methyltransferase SET8 (KMT5A), one of the members of SET domain-containing methyltransferase family, is responsible for catalyzing monomethylation of H4K20me. And the expression of SET8 is downregulated in GC cells and tissues. Furthermore, in GC cells, SET8 triggers oncogene-induced senescence (OIS) and induces senescence through p53-dependent DNA damage induced oncogenes. Thus, by inhibiting the senescence signaling pathway, miR-192/215-SET8-p53 axis exacerbates GC development (95).
RUNX3 is a transcription factor that binds directly to the region of miR-29b promoter and cooperates with Smad3, leading to higher activity of miR-29b promoter. Both miR-29b and RUNX3 are decreased in GC tissues, with positively correlated expression. miR-29b directly binds to KDM2A 3′-UTR and negatively regulates the expression of KDM2A, serving as a target of miR-29b. Through this mechanism, the proliferation and migration of GC cells are inhibited (101).
As miR-491-5p is decreased in GC cells, tissues and serum, its high expression can inhibit proliferation and invasion in GC. By targeting JMJD2B (KDM4B), miRNA-491-5p can inhibit malignant biological behaviors in GC cell lines, exhibiting the potential as a gastric cancer-associated marker (96).
The GKN1-miR-185-DNMT1 axis exerts oncogenic effects in gastric cancer through mediating epigenetic alterations and cell cycle. Gastrokine 1 (GKN1) reduces GC cell proliferation, viability, and colony formation, as well as blocks cell cycle. The mechanisms are as follows: 1. GKN1 inhibits DNMT1 and EZH2 expression through upregulation of miR-185 expression. miR-185 negatively regulates the inhibitory actions of GKN1 on tumorigenicity, expression of DNMT1 and EZH2, as well as DNMT1 activity; 2. GKN1-induced miR-185 leads to CDKN2A and E-cadherin re-expression through demethylation of CDKN2A and E-cadherin promoter regions; 3. GKN1 negatively regulates DNMT1 through repressing HDAC1 and inducing Tip60 in a miR-185-free manner (97).
lncRNA SNHG5 is a transcript consisting of six exons of U50HG, which has been reported to be the host gene of snoRNAs U50 and U50’. SNHG5 expression is aberrantly decreased in GC cells and tissues, inhibiting metastasis and proliferation in GC cells. The NuRD complex participates in chromatin remodeling and histone deacetylase with subunits, including MTA1, MTA2, HDAC1 and HDAC2. The mechanism is that the overexpression of SNHG5 affects the formation of the NuRD complex by blocking the transference of MTA2 from the cytoplasm to the nucleus, resulting in significant upregulation of histone H3 and non-histone p53 acetylation levels, which interferes with nucleosome remodeling and the formation of the histone deacetylation complex. As a result, the metastasis and growth of GC cells are suppressed (98).
The expression of lncRNA MALAT1 is aberrantly upregulated in GC cells and tissues. MALAT1 induces EGFL7 protein expression by altering H3 histone acetylation at the region of EGFL7 promoter. Epidermal growth factor-like domain-containing protein 7 (EGFL7), also known as vascular endothelial statin, is encoded by the EGFL7 gene and involved in regulating the formation of vascular ducts and the progression of many cancers. MALAT1 stimulates migration and invasion of GC cell lines through the above mechanism (102).
MIAT expression is significantly increased in GC. Knockdown of lncMIAT inhibits the malignant biological behaviors of GC cells. lncMIAT competitively combines with miR-29a-3p through serving as a sponge for miR-29a-3p, thereby increasing HDAC4 expression and ultimately exerting an oncogenic effect in GC (103).
lncRNA HOXA11-AS is specifically increased in GC, promoting GC cell proliferation, migration and invasion, with apoptosis inhibition. HOXA11-AS is thought to bind directly to LSD1, EZH2, WDR5, STAU1, DNMT1 and AGO2 in tumor cells, acting as a frame for EZH2 and LSD1/DNMT1 to downregulate PRSS8, KLF2 and P21 expression, thus promoting GC. EZH2 can recognize and bind to PRSS8, KLF2 and P21 promoters, which in turn drives H3K27me3 modification. While LSD1 contributes to H3K4 demethylation by binding directly to the PRSS8 promoter. Moreover, HOXA11-AS can also serve as a miR-1297 sponge by binding to miR-1297, releasing repressive effect on EZH2 mRNA. In addition, EZH2 is further recruited by HOXA11-AS to epigenetically inhibit the expression of KLF2 and PRSS8 (104, 109).
lncRNA FEZF1-AS1 is located at chromosome 7, which is a transcript of 2564 bp in length. The transcription factor SP1 specifically recognizes and binds to the FEZF1-AS1 promoter binding site, increasing the expression of FEZF1-AS1 in GC. GC cells are proliferated through the following mechanisms: Firstly, FEZF1-AS1 causes G1-S cell cycle arrest; Secondly, FEZF1-AS1 binds to LSD1, causing p21 promoter H3K4me2 demethylation and inhibiting the expression of the downstream target p21 (110).
In GC, Linc01503 is regulated by the transcription factor 1 (EGR1) and its expression is upregulated. Linc01503 epigenetically represses the expression of cyclin-dependent kinase inhibitor 1A (CDKN1A) and dual-specificity phosphatase 5 (DUSP5) by recruiting LSD1 and EZH2. EZH2 binds to CDKN1A and DUSP5 promoters, driving histone 3 lysine 4 trimethylation (H3K4me3). While LSD1 binds to DUSP5 and CDKN1A promoters, mediating H3K4me2 demethylation. Ultimately, GC cell cycle progression and proliferation can be promoted by Linc01503, with apoptosis inhibited (111).
The expression of HOXD-AS1 is increased in DDP-resistant GC patients. DDP sensitivity of GC cells can be promoted by knockdown of HOXD-AS1, which can promote cisplatin resistance in GC through recruiting EZH2 and upregulating H3K27me3 levels on the region of PDCD4 promoter in GC cells to epigenetically silence PDCD4 (104).
The expression of lncRNA GATA6 antisense RNA 1 (GATA6-AS1) was decreased in GC cells, which promotes the malignant development of GC cells, such as proliferation, metastasis and invasion. Its overexpression can inhibit tumor growth as well as lymph node metastasis (LNM) in GC. GATA6-AS1 can downregulate the expression of FZD4 by specifically recruiting and binding with EZH2 to increase the enrichment of H3K27me3 in the region FZD4 promoter, which in turn inhibits the Wnt/β-catemin downstream pathway, ultimately leading to the blockage of GC progression (112).
lncRNA 01446 (LINC01446), a 3484-bp ncRNA, is located at chromosome 7p12.1. LINC01446 is downregulated in GC, which is correlates with metastasis and poor prognosis in GC patients. This lncRNA can epigenetically repress Ras-related dexamethasone inducible 1 (RASD1) by recruiting LSD1 to the region of RASD1 promoter, ultimately promoting malignant behaviors of tumor cells (105).
lncRNA PART1 is locate at chromosome 5q12.1. PART1 is decreased in tumor cells and tissues, which is closely related to poor prognosis of GC patients. PART1 overexpression inhibits invasion and metastasis of GC cells. Mechanistically, PART1 acts as a role of tumor suppressor by interacting with androgen receptor (AR) and stimulating the PLZF gene expression in GC cells. Then, co-occupancy of EZH2 and PLZF on the PDGFB promoter region enable histone 3 lysine 27 trimethylation (H3K27me3) to epigenetically silence PDGFB and suppress the PI3K/Akt pathway (106).
Rho GTPase-activating protein (ARHGAP) is a family of Rho homologous GTPase activating proteins that exert oncogenic effects by aberrant regulation of Rho/Rac/Cdc42-like GTPases (107). ARHGAP20 gene is confirmed to be located at chromosome 17q24.1, which is decreased in GC cells, tissues and plasma. Low expression of ARHGAP27P1 is related to adverse clinical features, suggesting a poor prognosis for GC patients. Mechanistic investigations showed that ARHGAP27P1 inhibits proliferation and blocks the cell cycle through combining with Jumonji-domain containing 3 (JMJD3), leading to a significant downregulation of H3K27me3 levels, which ultimately epigenetically induces transcription of p15, p16 and p57 genes (126). Highly expressed FOXP4-AS1 can promote proliferation, migration as well as metastasis of GC cells by interacting with EZH2/LSD1 (127).
LINC00461 is upregulated in GC tissues, which is positively correlated to TNM staging and lymphatic metastasis, mediating cell proliferation and apoptosis in GC through interacting with LSD1, thereby aggravating the progression of GC (108).
lncRNA prostate cancer–associated transcript 1 (PCAT‐1) is upregulated in CDDP‐resistant GC cell lines and tissues. Mechanistically, PCAT-1 epigenetically silences PTEN gene by recruiting EZH2 to the region of PTEN promoter, resulting in H3K27 trimethylation in the PTEN gene promoter region, ultimately leading to the progress of cisplatin resistance in GC cells. Trough inhibiting of PCAT-1 and upregulating PTEN expression, cisplatin sensitivity of CDDP‐resistant GC cells can be enhanced (113).
Tumor suppressor PLZF inhibits proliferation, metastasis and invasion of GC cells. The expression of PLZF is downregulated and negatively correlates with the expression level of lncRNA ANRIL in GC. This lncRNA recruits EZH2, a core member of PRC2, in turn catalyzes H3K27 trimethylation, which then drives PLZF gene silencing by increasing DNA methylation of PLZF. In summary, lncRNA ANRIL suppresses EMT as well as cell proliferation through epigenetic regulation of the tumor suppressor PLZF (114).
lncRNA HOTTIP causes hypomethylation of the HoxA13 promoter E1 site and elevates levels of H3K4me3 by decreasing the recruitment of DNA methyltransferases DNMT1 and DNMT3b at the HoxA13 promoter E1 site, increasing the binding of WDR5 and MLL complexes, which ultimately epigenetically activates HoxA13 expression. Meanwhile, IGFBP3 and HOTTIP are shown to be downstream targets of HoxA13 in GC cells. The expression of HoxA13 and HOTTIP shows positive feedback. HoxA13 trans-activated the expression of IGFBP-3 gene by binding to the Hox binding element at the IGFBP-3 promoter region. IGFBP-3 has been reported to inhibit migration, invasion and EMT in GC through inhibiting invasion factors, such as MMP14 and urokinase-type fibrinogen activator (115). However, IGFBP-3 might enhance division and metastasis in GC CS12 cell line, and knockdown of IGFBP-3 inhibits the poor biological behaviors of GC cells (128).
lncRNA HOTAIR is highly upregulated in GC, which promotes metastasis and invasion of GC cells by inducing EMT, and its overexpression predicts poor prognosis. HOTAIR epigenetically represses miR34a and activates the expression of its downstream targets C-Met (HGF/C-Met/Snail pathway) and Snail by recruiting the multiple sparing repressor complex 2 (PRC2, including EZH2, SUZ12, etc.) to bind directly to the region of miR34a promoter and induce H3K27 trimethylation. Thereby GC cell EMT process and tumor metastasis are accelerated (116).
Another study shows that the expression level of the epithelial marker E-cadherin (CDH1) is increased in HOTAIR knockdown GC. By contrast, the expression levels of mesenchymal markers are downregulated in HOTAIR knockdown GC cells, such as Snail, Slug, Twist, N-cadherin and β-catenin. HOTAIR knockdown derepresses the histone marker H3K27me3 for E-cadherin and induces conversion of the E-cadherin promoter to the active marker acetylated H3K27. In addition, the loss of SUZ12 with the involvement of the active acetyltransferase CBP leads to an increase in acetylation of H3K27 (129). In summary, in the region of E-cadherin promoter, HOTAIR can epigenetically repress E-cadherin by turning acetylation of histone H3 lysine 27 into methylation, which ultimately promotes EMT of GC (130).
Reprogramming of cancer cells into induced pluripotent stem cells (iPSCs) might be a novel method to suppress tumorigenesis. Another research reported the successful reprogramming of the human gastric cell lines (CS12s) into induced pluripotent stem cell–like cell lines (CS12iPSLCs) by utilizing Jun dimerization protein 2 (JDP2) and octamer-binding protein 4 (OCT4). The study shows that CS12iPSLCs exhibit reduced tumorigenicity in vivo and reduce colony formation, proliferation, invasion, migration, and drug resistance in vitro. The oncogenic function of Bone morphogenetic protein 7 (BMP7) is switched by lncRNA HOXA13 axis and loses in CS12iPSLCs. H3K4me3 can be increased by recruiting HOXA13, MLL1, WDR5 and HOTTIP at the BMP7 promoter S1 site, thereby activating the expression of BMP7 and promoting the process of GC in CS12. H3K27me3 can be enhanced by recruiting HOXA13, ZEH2, JARID2 and HOTAIR at the BMP7 promoter S2 site, thereby epigenetically suppressing BMP7 expression and inhibiting the progression of GC in CS12iPSLC. In summary, the HOXA13-HOTAIR and HOXA13-HOTTIP axis are recruited to different sites of the BMP7 promoter, leading to different outcomes of GC (117).
lncRNA CASC15 (cancer susceptibility candidate 15) is located at chromosome 6p22.3, which regulates GC cell biological behaviors and is involved in tumorigenesis and progression. The mechanisms are as follows: Firstly, CASC15 regulates GC cell growth partially by serving as a bridge between EZH2 and WDR5 in nucleus, epigenetically silencing CDKN1A. Moreover, in cytoplasm, CASC15 mediates GC cell migration in part by serving as a ceRNA for miR-33a-5p, which competitively binds to ZEB1 (118).
lncRNA LINC00673, an intergenic lncRNA, is located at chromosome 17q25.1. The expression of LINC00673 is aberrantly increased in GC cells. Knockdown of LINC00673 suppresses invasion as well as proliferation, with promoted apoptosis. LINC00673, activated by transcription factor SP1, exerts its oncogenic effects partly through serving as a frame for EZH2 and LSD1 to epigenetically suppress KLF2 and LATS2 genes, promoting the progression of GC (119).
LINC01436 is highly expressed in gastric cancer tissues and cells. High LINC01436 level is associated with poor patient prognosis. In addition, knockdown of LINC01436 inhibits migration and proliferation, with apoptosis promoted. Mechanistically, LINC01436 mediates trimethylation of H3K27 at the miR-585-3p promoter through enhancing EZH2, epigenetically silencing miR-585-3p expression, which in turn upregulates mitogen-activated protein kinase 1 (MAPK1) expression, ultimately promoting GC progression (120).
lncRNA FOXD2-AS1 expression is significantly increased in GC and positively related to the tumor size, later pathological stage as well as poor prognosis. Overexpression of FOXD2-AS1 can accelerate tumor development and predict poor prognosis in GC patients by EZH2 binding to the region of EphB3 promoter and inducing H3K27me3 modification, or LSD1 combining with the region of EphB3 promoter and inducing demethylation of H3K4, ultimately epigenetically silencing of EphB3 (121).
Overexpression of lncHEIH in GC promotes proliferation, migration, tumorigenesis, and expansion of GC stem cells, leading to tumor malignant transformation. lncHEIH can upregulate EZH2 expression, while recruiting and binding EZH2 in the GSDME promoter region and inducing H3K27me3 modification in this region to epigenetically silence GSDME expression, thereby promoting GC progression (122).
lncRNAs 152 (LINC00152) expression is significantly increased in gastric cancer cells and tissues, which is correlated with lymph node metastasis, depth of tumor invasion, higher TNM stage as well as poor prognosis. LINC00152 epigenetically silences the expression of p15 and p21 through combining with EZH2, ultimately accelerating cell cycle progression and proliferation in GC (123).
lncRNA BC041951, named gastric cancer-associated lncRNA 1 (GClnc1), is upregulated in GC and closely associated with poor prognosis of patients. Mechanistically, lncRNA GClnc1 might serve as a modular scaffold to recruit WD repeat protein 5 (WDR5) and histone acetyltransferase KAT2A to the region of the superoxide dismutase 2 mitochondria (SOD2) gene promoter (one of the target genes of the WDR5/KAT2A complex), increasing the levels of H3K4 trimethylation and H3K9 acetylation in the promoter region of SOD2, thereby epigenetically promoting the expression of SOD2. As a result, GC cell proliferation, invasion, tumor growth can be promoted, in addition, chemotherapy resistance can be controlled (124).
circMRPS35 suppresses GC development through recruiting KAT7 to mediate histone modification. This study shows that circMRPS35 expression is significantly decreased in GC and related to the clinicopathological features and better prognosis of patients. circMRPS35 exerts oncogenic effect by inhibiting invasion and proliferation of GC cells in vitro and in vivo. The mechanism is that circMRPS35 enriches H4K5 acetylation at the regions of FOXO1 and FOXO3a promoters by recruiting the histone acetyltransferase KAT7, which ultimately epigenetically activates the expression of FOXO1 and FOXO3a, and induces subsequent responses in their downstream targets, including upregulation of p21, p27 and E-calmodulin, with downregulation of Twist1 expression, ultimately inhibiting cell proliferation and invasion (125).
Another study showed that circKIAA0907 is downregulated in GC. Upregulation of circKIAA0907 induces cell cycle arrest, proliferation and apoptosis, as well as inhibits autophagy. Mechanistically, circKIAA0907 serves as a sponge of miR-452-5p to increase lysine acetyltransferase 6B (KAT6B) expression in GC. KAT6B is a target gene of miR-452-5p, which has been found to play an regulatory role in many cancers. Shi et al. demonstrates that microRNA-4513 induces EMT and cell proliferation by targeting KAT6B (131). circKIAA0907 inhibits GC development and progression through downregulating miR-452-5p and upregulating KAT6B. This study demonstrates the potential diagnostic and therapeutic value of circKIAA0907, which can serve as a tumor suppressor in GC via the miR-452-5p/KAT6B axis (132).
circ_0000745 (circ_SPECC1), transcribed from the SPECC1 gene, is downregulated in GC tissues and blood. It exerts anti-tumor effects by significantly inhibiting metastasis and proliferation of GC cells, with apoptosis promoted. circ_SPECC1 is shown to bind directly to miR-526b, which in turn affects miR-526b downstream signaling targets. miR-526b is downregulated in GC cells and tissues, inhibiting invasion and proliferation, with apoptosis promoted. Lysine demethylase 4A (KDM4A, also known as JMJD2A), a member of the histone demethylase family, is also an important target of miR-526b, whose expression is shown to be increased in various cancers. Huang et al. (133) demonstrates that KDM4A expression is aberrantly increased in GC tissues and correlated with clinicopathological features. Inhibition of KDM4A expression weakens transformation and growth of GC cells, with apoptosis promoted. In GC, YAP1/TAZ serve as downstream target genes of KDM4A, promoting transcription through binding to the transcription factor TEAD, ultimately leading to tumor formation (134). In summary, circ_SPECC1 can improve the inhibiting effect of miR-526b at the downstream signaling target YAP1/KDM4A, with invasion and growth of GC cells inhibited (135).
Altering the ncRNA histone modification pattern by recruiting histone modifiers into the ncRNA promoter epigenetically regulates ncRNA expression, which in turn changes downstream signaling pathways and target genes. This mechanism ultimately plays an essential role in GC (Table 3).
miR-155 expression is increased in GC and exhibits pro-carcinogenic effects. MRTF-A (MKL1) promotes miR-155 expression by inducing the acetylation of histones and the recruitment of RNA polymerase II in the region of miR-155 promoter through the Wnt-β-catenin pathway. MRTF-A can act as a coactivator of serum response factor (SRF), participating in cell apoptosis, differentiation, proliferation and migration. miR-155 inhibits SOX1 expression through combining with the 3’UTR of SOX1, promoting migration of tumor cells. In summary, MRTF-A/miR-155/SOX1 axis can mediate metastasis of GC (146).
miR-34a and miR-34b/c, derived from miR-34 family, participate in cell cycle arrest, senescence and apoptosis in tumor. Mechanistically, Sirt7 can epigenetically inhibit miR-34a expression by deacetylating H3K18ac, ultimately preventing apoptosis in gastric cancer cells. The expression of Sirt7, a family member of NAD+-dependent protein deacetylases, is significantly upregulated in GC, and its knockdown promotes apoptosis and reduces the growth of tumor (137).
miR-330-3p expression is downregulated in GC. However, the ectopic expression of miR-330-3p decreases migration, proliferation, colony formation, and EMT in tumor cells. Treatment of GC cells with the histone deacetylase inhibitor trichostatin A (TSA) and the DNA methylation inhibitor 5-aza-CdR (AZA) enhances the expression of miR-330-3p. This indicates that the downregulation of miR-330-3p is partially mediated through hypermethylation in the region of the gene promoter. In addition, MSI1 is a target gene of miR-330-3p in GC cells. MSI1, an RNA binding protein (RBP), is upregulated in GC cell lines and tissues through combining with the mRNA 3’UTR sequences, leading to translational repression. Moreover, its expression is negatively related to the expression of miR-330-3p in GC tissues (139).
miR-375 expression is downregulated in GC due to methylation of its promoter and histone deacetylation, which is related to poor prognosis and lymph node metastasis. While miR-375 ectopic expression inhibits cell proliferation and tumor growth. miR-375 specifically and directly inhibits the expression of YAP1, TEAD4 and CTGF through combining with their 3’UTRs. YAP1, a co-activator of transcription, is a crucial downstream signaling molecule of the Hippo pathway. It mainly combines with the transcription factor TEAD and exerts oncogenic effects. The expression of CTGF is mediated by YAP1 and TEAD in GC cells. Overall, miR-375 is downregulated in GC cells due to its promoter methylation, which results in the co-activation of YAP1/TEADs-CTGF in the Hippo signaling pathway to promote gastric carcinogenesis (141).
Histone deacetylase 3 (HDAC3) is significantly upregulated in GC tissues and cells. However, knockdown of HDAC3 inhibits the growth of GC cells. HDAC3 overexpression upregulates miR-454 expression, which is significantly related to malignant clinical features in GC patients, implying that it can act as a biomarker for poor prognosis. Moreover, CHD5 is suggested to be a target of miR-454. The ectopic expression of CHD5 inhibits cell proliferation and tumorigenicity, as well as leads to cellular senescence. CHD5 expression is downregulated in GC and negatively associated with miR-454 levels. In summary, HDAC3 targets CHD5 through miR-454 to regulate the growth of GC cells (142).
Histone modification of the miR-133 family at the promoter region leads to a significant decrease of miR-133 expression in GC. Restoration of miR-133b/a-3p expression can target the anti-apoptotic molecules Bcl-xL and Mcl-1 to inhibit proliferation and induce apoptosis in GC cells, thereby suppressing GC growth. In summary, miR-133b/a-3p is regulated by histone modifications, exerting oncogenic effects in GC through targeting Bcl-xL and Mcl-1 (138).
Histone deacetylase 1 (HDAC1) knockdown inhibits cell metastasis and adhesion in GC cells through upregulating miR-34a. The mechanism is that the HDAC1/miR-34a axis regulates CD44 expression, activation as well as its downstream factors, including LIM domain kinase 1 (LIMK-1), matrix metalloproteinase (MMP)-2, ras homolog family member A (RhoA), and Bcl-2. The former three proteins participate in the organization of the microtubulin and actin cytoskeletons as well as the formation of cellular pseudopods. Thus, the depletion of HDAC1 can decreases the metastatic ability of GC cells through the miR-34a/CD44 axis (143).
LSD1 is thought to be a demethylase of H3K4me1/2, H3K9me1/2 and several non-histone proteins, which is extensively expressed in GC tissues. LSD1 deletion leads to upregulation of the migration inhibitory factor CD9 through decreasing intracellular miR-142-5p expression, which ultimately inhibits GC migration (140).
lncRNA HOXC-AS3, located at chromosome 12q13.13, is defined as an antisense transcript of HOXC10. Gaining of H3K27ac and H3K4me3 activation of the promoter contributes to driving HOXC-AS3 gene in GC cells and tissues. By binding to YBX1, HOXC-AS3 transcriptionally regulates genes, such as MMP7, WNT10B and HDAC5. They are related to the biological behaviors of GC cells, ultimately inducing migration and proliferation in GC. In addition, HDAC5 gene is a member of the histone deacetylase (HDAC) family. Therefore, this study suggests that HOXC-AS3 is regulated by histone modifications, which in turn regulates HDAC5 to promote GC (136).
The lncRNA FENDRR is a gene of length 3099nts and located at Chr3q13.31. The expression of FENDRR is reduced in GC, which is correlated with poor prognosis and clinicopathological characteristics in GC patients. The histone deacetylase 3 (HDAC3) drives metastasis of GC cells through epigenetic silencing of FENDRR, induction of fibronectin1 (FN1) expression, and activation of MMP2/MMP9 (144).
lncRNA HRCEG is decreased in GC cells, which is an HDAC1-regulated RNA affecting GC cell proliferation and EMT transformation. HRCEG expression is negatively regulated by histone deacetylase 1 (HDAC1). The overexpression of HRCEG inhibits GC cells proliferation and EMT process (145).
Recently, growing evidence indicates that epigenetic alteration is a critical factor involved in tumorigenesis and progression. Research progress has been achieved in the use of epigenetic methods to treat cancer. For instance, numerous drugs targeting epigenetic pathways have achieved clinical efficacy, including inhibitors of DNMTs and HDACs. However, the mechanisms of epigenetic alterations in tumorigenesis and progression have not been fully explored. In this review, we attempt to comprehensively describe the important role of ncRNA and histone modification interactions in the pathogenesis of GC, thus providing a solid basis for identifying more specific and effective epigenetic therapeutic agonists/inhibitors. As the field of ncRNAs continues to evolve, there are many technologies that support the feasibility of ncRNA-based therapies. For ncRNAs with down-regulated expression, their expression levels can be enhanced by liposomes/nanoparticles from virus- or plasmid-based expression vectors. For ncRNAs with up-regulated expression, RNAi/shRNAs can be constructed or CRISPR-Cas9 can be used to suppress their expression levels. However, efforts are still needed in ncRNA-based therapy for tumors. Moreover, considering the limited efficacy of monotherapies and the possibility of serious adverse effects, we should achieve more progress in the field of epigenetic drugs combined with other anti-tumor drugs for cancer treatment, which will be a boon for GC patients.
QY and YD were responsible for the review of the literature. QY and YC wrote the manuscript. YC and RG edited the manuscript. QY and YD drew the tables and pictures. TY, QW, and ZX designed the study and made valuable discussions and revisions to the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ZX is thankful to the National Natural Science Foundation of China (Grants 81503093, 81972643, and 81672444) for the financial supports.
1. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric Cancer. Lancet (2020) 396(10251):635–48. doi: 10.1016/S0140-6736(20)31288-5
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
3. Eusebi LH, Telese A, Marasco G, Bazzoli F, Zagari RM. Gastric Cancer Prevention Strategies: A Global Perspective. J Gastroenterol Hepatol (2020) 35(9):1495–502. doi: 10.1111/jgh.15037
4. Link A, Kupcinskas J. MicroRNAs as non-Invasive Diagnostic Biomarkers for Gastric Cancer: Current Insights and Future Perspectives. World J Gastroenterol (2018) 24(30):3313–29. doi: 10.3748/wjg.v24.i30.3313
5. Sexton RE, Al Hallak MN, Diab M, Azmi AS. Gastric Cancer: A Comprehensive Review of Current and Future Treatment Strategies. Cancer Metastasis Rev (2020) 39(4):1179–203. doi: 10.1007/s10555-020-09925-3
6. Sterea AM, Egom EE, El Hiani Y. TRP Channels in Gastric Cancer: New Hopes and Clinical Perspectives. Cell Calcium (2019) 82:102053. doi: 10.1016/j.ceca.2019.06.007
7. Guggenheim DE, Shah MA. Gastric Cancer Epidemiology and Risk Factors. J Surg Oncol (2013) 107(3):230–6. doi: 10.1002/jso.23262
8. Correa P. Gastric Cancer: Overview. Gastroenterol Clin North Am (2013) 42(2):211–7. doi: 10.1016/j.gtc.2013.01.002
9. Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric Cancer: Descriptive Epidemiology, Risk Factors, Screening, and Prevention. Cancer Epidemiol Biomarkers Prev (2014) 23(5):700–13. doi: 10.1158/1055-9965.EPI-13-1057
10. Feng W, Ding Y, Zong W, Ju S. Non-Coding RNAs in Regulating Gastric Cancer Metastasis. Clin Chim Acta (2019) 496:125–33. doi: 10.1016/j.cca.2019.07.003
11. Puneet, Kazmi HR, Kumari S, Tiwari S, Khanna A, Narayan G. Epigenetic Mechanisms and Events in Gastric Cancer-Emerging Novel Biomarkers. Pathol Oncol Res (2018) 24(4):757–70. doi: 10.1007/s12253-018-0410-z
12. Werner RJ, Kelly AD, Issa JJ. Epigenetics and Precision Oncology. Cancer J (2017) 23(5):262–9. doi: 10.1097/PPO.0000000000000281
13. Michalak EM, Burr ML, Bannister AJ, Dawson MA. The Roles of DNA, RNA and Histone Methylation in Ageing and Cancer. Nat Rev Mol Cell Biol (2019) 20(10):573–89. doi: 10.1038/s41580-019-0143-1
14. Song Y, Wu F, Wu J. Targeting Histone Methylation for Cancer Therapy: Enzymes, Inhibitors, Biological Activity and Perspectives. J Hematol Oncol (2016) 9(1):49. doi: 10.1186/s13045-016-0279-9
15. Zhou Z, Lin Z, Pang X, Tariq MA, Ao X, Li P, et al. Epigenetic Regulation of Long non-Coding RNAs in Gastric Cancer. Oncotarget (2018) 9(27):19443–58. doi: 10.18632/oncotarget.23821
16. Li Y, Seto E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb Perspect Med (2016) 6(10):634. doi: 10.1101/cshperspect.a026831
17. Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harb Perspect Biol (2016) 8(4):a019521. doi: 10.1101/cshperspect.a019521
18. Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of Acetylation at Lys16 and Trimethylation at Lys20 of Histone H4 is a Common Hallmark of Human Cancer. Nat Genet (2005) 37(4):391–400. doi: 10.1038/ng1531
19. Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol (2016) 8(9):a019505. doi: 10.1101/cshperspect.a019505
20. Black JC, Van Rechem C, Whetstine JR. Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact. Mol Cell (2012) 48(4):491–507. doi: 10.1016/j.molcel.2012.11.006
21. Qin J, Wen B, Liang Y, Yu W, Li H. Histone Modifications and Their Role in Colorectal Cancer (Review). Pathol Oncol Res (2020) 26(4):2023–33. doi: 10.1007/s12253-019-00663-8
22. McCabe MT, Mohammad HP, Barbash O, Kruger RG. Targeting Histone Methylation in Cancer. Cancer J (2017) 23(5):292–301. doi: 10.1097/PPO.0000000000000283
23. Di Lorenzo A, Bedford MT. Histone Arginine Methylation. FEBS Lett (2011) 585(13):2024–31. doi: 10.1016/j.febslet.2010.11.010
24. Shanmugam MK, Arfuso F, Arumugam S, Chinnathambi A, Jinsong B, Warrier S, et al. Role of Novel Histone Modifications in Cancer. Oncotarget (2018) 9(13):11414–26. doi: 10.18632/oncotarget.23356
25. Lau AT, Lee SY, Xu YM, Zheng D, Cho YY, Zhu F, et al. Phosphorylation of Histone H2B Serine 32 is Linked to Cell Transformation. J Biol Chem (2011) 286(30):26628–37. doi: 10.1074/jbc.M110.215590
26. Choi HS, Choi BY, Cho YY, Mizuno H, Kang BS, Bode AM, et al. Phosphorylation of Histone H3 at Serine 10 is Indispensable for Neoplastic Cell Transformation. Cancer Res (2005) 65(13):5818–27. doi: 10.1158/0008-5472.CAN-05-0197
27. Chadee DN, Hendzel MJ, Tylipski CP, Allis CD, Bazett-Jones DP, Wright JA, et al. Increased Ser-10 Phosphorylation of Histone H3 in Mitogen-Stimulated and Oncogene-Transformed Mouse Fibroblasts. J Biol Chem (1999) 274(35):24914–20. doi: 10.1074/jbc.274.35.24914
28. Darieva Z, Webber A, Warwood S, Sharrocks AD. Protein Kinase C Coordinates Histone H3 Phosphorylation and Acetylation. Elife (2015) 4:e09886. doi: 10.7554/eLife.09886
29. Trevino LS, Wang Q, Walker CL. Phosphorylation of Epigenetic "Readers, Writers and Erasers": Implications for Developmental Reprogramming and the Epigenetic Basis for Health and Disease. Prog Biophys Mol Biol (2015) 118(1-2):8–13. doi: 10.1016/j.pbiomolbio.2015.02.013
30. Besant PG, Attwood PV. Histone H4 Histidine Phosphorylation: Kinases, Phosphatases, Liver Regeneration and Cancer. Biochem Soc Trans (2012) 40(1):290–3. doi: 10.1042/BST20110605
31. Takahashi H, Murai Y, Tsuneyama K, Nomoto K, Okada E, Fujita H, et al. Overexpression of Phosphorylated Histone H3 is an Indicator of Poor Prognosis in Gastric Adenocarcinoma Patients. Appl Immunohistochem Mol Morphol (2006) 14(3):296–302. doi: 10.1097/00129039-200609000-00007
32. Uguen A, Conq G, Doucet L, Talagas M, Costa S, De Braekeleer M, et al. Immunostaining of Phospho-Histone H3 and Ki-67 Improves Reproducibility of Recurrence Risk Assessment of Gastrointestinal Stromal Tumors. Virchows Arch (2015) 467(1):47–54. doi: 10.1007/s00428-015-1763-2
33. Park HB, Kim JW, Baek KH. Regulation of Wnt Signaling Through Ubiquitination and Deubiquitination in Cancers. Int J Mol Sci (2020) 21(11):3904. doi: 10.3390/ijms21113904
34. Antao AM, Tyagi A, Kim KS, Ramakrishna S. Advances in Deubiquitinating Enzyme Inhibition and Applications in Cancer Therapeutics. Cancers (Basel) (2020) 12(6):1579. doi: 10.3390/cancers12061579
35. Sun T, Liu Z, Yang Q. The Role of Ubiquitination and Deubiquitination in Cancer Metabolism. Mol Cancer (2020) 19(1):146. doi: 10.1186/s12943-020-01262-x
36. Tang R, Langdon WY, Zhang J. Regulation of Immune Responses by E3 Ubiquitin Ligase Cbl-B. Cell Immunol (2019) 340:103878. doi: 10.1016/j.cellimm.2018.11.002
37. Thompson LL, Guppy BJ, Sawchuk L, Davie JR, McManus KJ. Regulation of Chromatin Structure via Histone Post-Translational Modification and the Link to Carcinogenesis. Cancer Metastasis Rev (2013) 32(3-4):363–76. doi: 10.1007/s10555-013-9434-8
38. Popovic D, Vucic D, Dikic I. Ubiquitination in Disease Pathogenesis and Treatment. Nat Med (2014) 20(11):1242–53. doi: 10.1038/nm.3739
39. Hahn MA, Dickson KA, Jackson S, Clarkson A, Gill AJ, Marsh DJ. The Tumor Suppressor CDC73 Interacts With the Ring Finger Proteins RNF20 and RNF40 and is Required for the Maintenance of Histone 2B Monoubiquitination. Hum Mol Genet (2012) 21(3):559–68. doi: 10.1093/hmg/ddr490
40. Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T, et al. Estrogen-Dependent Gene Transcription in Human Breast Cancer Cells Relies Upon Proteasome-Dependent Monoubiquitination of Histone H2B. Cancer Res (2011) 71(17):5739–53. doi: 10.1158/0008-5472.CAN-11-1896
41. Hay RT. SUMO: A History of Modification. Mol Cell (2005) 18(1):1–12. doi: 10.1016/j.molcel.2005.03.012
42. Gill G. Post-Translational Modification by the Small Ubiquitin-Related Modifier SUMO has Big Effects on Transcription Factor Activity. Curr Opin Genet Dev (2003) 13(2):108–13. doi: 10.1016/s0959-437x(03)00021-2
43. Shiio Y, Eisenman RN. Histone Sumoylation is Associated With Transcriptional Repression. Proc Natl Acad Sci U S A (2003) 100(23):13225–30. doi: 10.1073/pnas.1735528100
44. Wotton D, Pemberton LF, Merrill-Schools J. SUMO and Chromatin Remodeling. Adv Exp Med Biol (2017) 963:35–50. doi: 10.1007/978-3-319-50044-7_3
45. Du L, Fakih MG, Rosen ST, Chen Y. SUMOylation of E2F1 Regulates Expression of EZH2. Cancer Res (2020) 80(19):4212–23. doi: 10.1158/0008-5472.CAN-20-1259
46. Zheng J, Liu L, Wang S, Huang X. SUMO-1 Promotes Ishikawa Cell Proliferation and Apoptosis in Endometrial Cancer by Increasing Sumoylation of Histone H4. Int J Gynecol Cancer (2015) 25(8):1364–8. doi: 10.1097/IGC.0000000000000501
47. Oh S, Shin S, Janknecht R. Sumoylation of Transcription Factor ETV1 Modulates its Oncogenic Potential in Prostate Cancer. Int J Clin Exp Pathol (2021) 14(7):795–810. doi: 10.1038/s41598-019-44685-3
48. Wang J, Zhu S, Meng N, He Y, Lu R, Yan GR. ncRNA-Encoded Peptides or Proteins and Cancer. Mol Ther (2019) 27(10):1718–25. doi: 10.1016/j.ymthe.2019.09.001
49. Peschansky VJ, Wahlestedt C. Non-Coding RNAs as Direct and Indirect Modulators of Epigenetic Regulation. Epigenetics (2014) 9(1):3–12. doi: 10.4161/epi.27473
50. Mattick JS, Makunin IV. Non-Coding RNA. Hum Mol Genet (2006) 15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046
51. Anastasiadou E, Jacob LS, Slack FJ. Non-Coding RNA Networks in Cancer. Nat Rev Cancer (2018) 18(1):5–18. doi: 10.1038/nrc.2017.99
52. Zhang P, Wu W, Chen Q, Chen M. Non-Coding RNAs and Their Integrated Networks. J Integr Bioinform (2019) 16(3):20190027. doi: 10.1515/jib-2019-0027
53. Pavet V, Portal MM, Moulin JC, Herbrecht R, Gronemeyer H. Towards Novel Paradigms for Cancer Therapy. Oncogene (2011) 30(1):1–20. doi: 10.1038/onc.2010.460
54. Li D, Zhang J, Li J. Role of miRNA Sponges in Hepatocellular Carcinoma. Clin Chim Acta (2020) 500:10–9. doi: 10.1016/j.cca.2019.09.013
55. Slack FJ, Chinnaiyan AM. The Role of Non-Coding RNAs in Oncology. Cell (2019) 179(5):1033–55. doi: 10.1016/j.cell.2019.10.017
56. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing Endogenous RNA (ceRNA) Cross Talk and Language in ceRNA Regulatory Networks: A New Look at Hallmarks of Breast Cancer. J Cell Physiol (2019) 234(7):10080–100. doi: 10.1002/jcp.27941
57. Mishra S, Yadav T, Rani V. Exploring miRNA Based Approaches in Cancer Diagnostics and Therapeutics. Crit Rev Oncol Hematol (2016) 98:12–23. doi: 10.1016/j.critrevonc.2015.10.003
58. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol (2018) 141(4):1202–7. doi: 10.1016/j.jaci.2017.08.034
59. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An Overview of microRNAs: Biology, Functions, Therapeutics, and Analysis Methods. J Cell Physiol (2019) 234(5):5451–65. doi: 10.1002/jcp.27486
60. Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering Mirnas’ Action Through miRNA Editing. Int J Mol Sci (2019) 20(24):6249. doi: 10.3390/ijms20246249
61. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in Cancer: Possible Functions and Clinical Implications. J Med Genet (2015) 52(10):710–8. doi: 10.1136/jmedgenet-2015-103334
62. Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, et al. LINC01133 as ceRNA Inhibits Gastric Cancer Progression by Sponging miR-106a-3p to Regulate APC Expression and the Wnt/beta-Catenin Pathway. Mol Cancer (2018) 17(1):126. doi: 10.1186/s12943-018-0874-1
63. Lin J, Liao S, Li E, Liu Z, Zheng R, Wu X, et al. Circcyfip2 Acts as a Sponge of miR-1205 and Affects the Expression of Its Target Gene E2F1 to Regulate Gastric Cancer Metastasis. Mol Ther Nucleic Acids (2020) 21:121–32. doi: 10.1016/j.omtn.2020.05.007
64. Shrestha S, Yang CD, Hong HC, Chou CH, Tai CS, Chiew MY, et al. Integrated MicroRNA-mRNA Analysis Reveals miR-204 Inhibits Cell Proliferation in Gastric Cancer by Targeting CKS1B, CXCL1 and GPRC5A. Int J Mol Sci (2017) 19(1):87. doi: 10.3390/ijms19010087
65. Rupaimoole R, Slack FJ. MicroRNA Therapeutics: Towards a New Era for the Management of Cancer and Other Diseases. Nat Rev Drug Discovery (2017) 16(3):203–22. doi: 10.1038/nrd.2016.246
66. Han TS, Hur K, Cho HS, Ban HS. Epigenetic Associations Between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma. Cancers (Basel) (2020) 12(9):2622. doi: 10.3390/cancers12092622
67. Shin VY, Chu KM. MiRNA as Potential Biomarkers and Therapeutic Targets for Gastric Cancer. World J Gastroenterol (2014) 20(30):10432–9. doi: 10.3748/wjg.v20.i30.10432
68. Lin J, Shen J, Yue H, Cao Z. Mirna1835p.1 Promotes the Migration and Invasion of Gastric Cancer AGS Cells by Targeting TPM1. Oncol Rep (2019) 42(6):2371–81. doi: 10.3892/or.2019.7354
69. Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, et al. Exosomal Transfer of Tumor-Associated Macrophage-Derived miR-21 Confers Cisplatin Resistance in Gastric Cancer Cells. J Exp Clin Cancer Res (2017) 36(1):53. doi: 10.1186/s13046-017-0528-y
70. Zhang Z, Dong Y, Hua J, Xue H, Hu J, Jiang T, et al. A five-miRNA Signature Predicts Survival in Gastric Cancer Using Bioinformatics Analysis. Gene (2019) 699:125–34. doi: 10.1016/j.gene.2019.02.058
71. Smirnov A, Fishman V, Yunusova A, Korablev A, Serova I, Skryabin BV, et al. DNA Barcoding Reveals That Injected Transgenes are Predominantly Processed by Homologous Recombination in Mouse Zygote. Nucleic Acids Res (2020) 48(2):719–35. doi: 10.1093/nar/gkz1085
72. Han TS, Hur K, Xu G, Choi B, Okugawa Y, Toiyama Y, et al. MicroRNA-29c Mediates Initiation of Gastric Carcinogenesis by Directly Targeting ITGB1. Gut (2015) 64(2):203–14. doi: 10.1136/gutjnl-2013-306640
73. Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci (2018) 19(5):1310. doi: 10.3390/ijms19051310
74. Chi Y, Wang D, Wang J, Yu W, Yang J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells (2019) 8(9):1015. doi: 10.3390/cells8091015
75. Hu G, Niu F, Humburg BA, Liao K, Bendi S, Callen S, et al. Molecular Mechanisms of Long Noncoding RNAs and Their Role in Disease Pathogenesis. Oncotarget (2018) 9(26):18648–63. doi: 10.18632/oncotarget.24307
76. Wu Q, Ma J, Wei J, Meng W, Wang Y, Shi M. lncRNA SNHG11 Promotes Gastric Cancer Progression by Activating the Wnt/beta-Catenin Pathway and Oncogenic Autophagy. Mol Ther (2021) 29(3):1258–78. doi: 10.1016/j.ymthe.2020.10.011
77. Wei GH, Wang X. lncRNA MEG3 Inhibit Proliferation and Metastasis of Gastric Cancer via P53 Signaling Pathway. Eur Rev Med Pharmacol Sci (2017) 21(17):3850–6.
78. Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D, et al. Impaired Autophagic Degradation of lncRNA ARHGAP5-AS1 Promotes Chemoresistance in Gastric Cancer. Cell Death Dis (2019) 10(6):383. doi: 10.1038/s41419-019-1585-2
79. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res (2017) 77(15):3965–81. doi: 10.1158/0008-5472.CAN-16-2634
80. Sheng Y, Han C, Yang Y, Wang J, Gu Y, Li W, et al. Correlation Between LncRNA-LINC00659 and Clinical Prognosis in Gastric Cancer and Study on its Biological Mechanism. J Cell Mol Med (2020) 24(24):14467–80. doi: 10.1111/jcmm.16069
81. Li X, Du Y, Wang Y. The Value of LncRNA SNHG5 as a Marker for the Diagnosis and Prognosis of Gastric Cancer. Am J Transl Res (2021) 13(5):5420–7.
82. Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA Code: Emerging Implications for Cancer Diagnosis and Treatment. Mol Oncol (2019) 13(4):669–80. doi: 10.1002/1878-0261.12468
83. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and Functional Roles of Circular RNAs in Human Cancer. Mol Cancer (2020) 19(1):30. doi: 10.1186/s12943-020-1135-7
84. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: Functions and Properties of a Novel Potential Biomarker for Cancer. Mol Cancer (2017) 16(1):94. doi: 10.1186/s12943-017-0663-2
85. Yin Y, Long J, He Q, Li Y, Liao Y, He P, et al. Emerging Roles of circRNA in Formation and Progression of Cancer. J Cancer (2019) 10(21):5015–21. doi: 10.7150/jca.30828
86. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat Rev Genet (2019) 20(11):675–91. doi: 10.1038/s41576-019-0158-7
87. Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z, et al. CircRNA_100269 is Downregulated in Gastric Cancer and Suppresses Tumor Cell Growth by Targeting miR-630. Aging (Albany NY) (2017) 9(6):1585–94. doi: 10.18632/aging.101254
88. Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA Hsa_Circ_0000745 may Serve as a Diagnostic Marker for Gastric Cancer. World J Gastroenterol (2017) 23(34):6330–8. doi: 10.3748/wjg.v23.i34.6330
89. Zheng L, Chen Y, Ye L, Jiao W, Song H, Mei H, et al. miRNA-584-3p Inhibits Gastric Cancer Progression by Repressing Yin Yang 1- Facilitated MMP-14 Expression. JSci Rep (2017) 7(1):8967. doi: 10.1038/s41598-017-09271-5
90. Li J, Dong G, Wang B, Gao W, Yang Q. miR-543 Promotes Gastric Cancer Cell Proliferation by Targeting SIRT1. Biochem Biophys Res Commun (2016) 469(1):15–21. doi: 10.1016/j.bbrc.2015.11.062
91. Wei J, Wang Z, Wang Z, Yang Y, Fu C, Zhu J, et al. MicroRNA-31 Function as a Suppressor Was Regulated by Epigenetic Mechanisms in Gastric Cancer. BioMed Res Int (2017) 2017:5348490. doi: 10.1155/2017/5348490
92. Xu G, Li N, Zhang Y, Zhang J, Xu R, Wu Y. MicroRNA-383-5p Inhibits the Progression of Gastric Carcinoma via Targeting HDAC9 Expression. Braz J Med Biol Res (2019) 52(8):e8341. doi: 10.1590/1414-431X20198341
93. Cai L, Chen Q, Fang S, Lian M, Cai M. MicroRNA-329 Inhibits Cell Proliferation and Tumor Growth While Facilitates Apoptosis via Negative Regulation of KDM1A in Gastric Cancer. J Cell Biochem (2018) 119(4):3338–51. doi: 10.1002/jcb.26497
94. Hong X, Xu Y, Qiu X, Zhu Y, Feng X, Ding Z, et al. MiR-448 Promotes Glycolytic Metabolism of Gastric Cancer by Downregulating KDM2B. Oncotarget (2016) 7(16):22092–102. doi: 10.18632/oncotarget.8020
95. Zhang X, Peng Y, Yuan Y, Gao Y, Hu F, Wang J, et al. Histone Methyltransferase SET8 is Regulated by miR-192/215 and Induces Oncogene-Induced Senescence via P53-Dependent DNA Damage in Human Gastric Carcinoma Cells. Cell Death Dis (2020) 11(10):937. doi: 10.1038/s41419-020-03130-4
96. Zhang J, Ren J, Hao S, Ma F, Xin Y, Jia W, et al. MiRNA-491-5p Inhibits Cell Proliferation, Invasion and Migration via Targeting JMJD2B and Serves as a Potential Biomarker in Gastric Cancer. Am J Transl Res (2018) 10(2):525–34.
97. Yoon JH, Choi YJ, Choi WS, Ashktorab H, Smoot DT, Nam SW, et al. GKN1-miR-185-DNMT1 Axis Suppresses Gastric Carcinogenesis Through Regulation of Epigenetic Alteration and Cell Cycle. Clin Cancer Res (2013) 19(17):4599–610. doi: 10.1158/1078-0432.CCR-12-3675
98. Zhao L, Guo H, Zhou B, Feng J, Li Y, Han T, et al. Long non-Coding RNA SNHG5 Suppresses Gastric Cancer Progression by Trapping MTA2 in the Cytosol. Oncogene (2016) 35(44):5770–80. doi: 10.1038/onc.2016.110
99. Zhang H, Li L, Yuan C, Wang C, Gao T, Zheng Z. MiR-489 Inhibited the Development of Gastric Cancer via Regulating HDAC7 and PI3K/AKT Pathway. World J Surg Oncol (2020) 18(1):73. doi: 10.1186/s12957-020-01846-3
100. Jiping Z, Ming F, Lixiang W, Xiuming L, Yuqun S, Han Y, et al. MicroRNA-212 Inhibits Proliferation of Gastric Cancer by Directly Repressing Retinoblastoma Binding Protein 2. J Cell Biochem (2013) 114(12):2666–72. doi: 10.1002/jcb.24613
101. Kong Y, Zou S, Yang F, Xu X, Bu W, Jia J, et al. RUNX3-Mediated Up-Regulation of miR-29b Suppresses the Proliferation and Migration of Gastric Cancer Cells by Targeting KDM2A. Cancer Lett (2016) 381(1):138–48. doi: 10.1016/j.canlet.2016.07.038
102. Deng QJ, Xie LQ, Li H. Overexpressed MALAT1 Promotes Invasion and Metastasis of Gastric Cancer Cells via Increasing EGFL7 Expression. Life Sci (2016) 157:38–44. doi: 10.1016/j.lfs.2016.05.041
103. Li Y, Wang K, Wei Y, Yao Q, Zhang Q, Qu H, et al. lncRNA-MIAT Regulates Cell Biological Behaviors in Gastric Cancer Through a Mechanism Involving the miR-29a-3p/HDAC4 Axis. Oncol Rep (2017) 38(6):3465–72. doi: 10.3892/or.2017.6020
104. Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res (2016) 76(21):6299–310. doi: 10.1158/0008-5472.CAN-16-0356
105. Lian Y, Yan C, Lian Y, Yang R, Chen Q, Ma D, et al. Long Intergenic non-Protein-Coding RNA 01446 Facilitates the Proliferation and Metastasis of Gastric Cancer Cells Through Interacting With the Histone Lysine-Specific Demethylase LSD1. Cell Death Dis (2020) 11(7):522. doi: 10.1038/s41419-020-2729-0
106. Han H, Wang S, Meng J, Lyu G, Ding G, Hu Y, et al. Long Noncoding RNA PART1 Restrains Aggressive Gastric Cancer Through the Epigenetic Silencing of PDGFB via the PLZF-Mediated Recruitment of EZH2. Oncogene (2020) 39(42):6513–28. doi: 10.1038/s41388-020-01442-5
107. Nakamura F. FilGAP and its Close Relatives: A Mediator of Rho-Rac Antagonism That Regulates Cell Morphology and Migration. Biochem J (2013) 453(1):17–25. doi: 10.1042/BJ20130290
108. Shi X, You X, Zeng WC, Deng YJ, Hong HL, Huang OX, et al. Knockdown of LINC00461 Inhibits Cell Proliferation and Induces Apoptosis in Gastric Cancer by Targeting LSD1. Eur Rev Med Pharmacol Sci (2019) 23(24):10769–75. doi: 10.26355/eurrev_201912_19779
109. Liu Z, Chen Z, Fan R, Jiang B, Chen X, Chen Q, et al. Over-Expressed Long Noncoding RNA HOXA11-AS Promotes Cell Cycle Progression and Metastasis in Gastric Cancer. Mol Cancer (2017) 16(1):82. doi: 10.1186/s12943-017-0651-6
110. Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M, et al. LincRNAFEZF1-AS1 Represses P21 Expression to Promote Gastric Cancer Proliferation Through LSD1-Mediated H3K4me2 Demethylation. Mol Cancer (2017) 16(1):39. doi: 10.1186/s12943-017-0588-9
111. Ma Z, Gao X, Shuai Y, Wu X, Yan Y, Xing X, et al. EGR1-Mediated Linc01503 Promotes Cell Cycle Progression and Tumorigenesis in Gastric Cancer. Cell Prolif (2021) 54(1):e12922. doi: 10.1111/cpr.12922
112. Li ZT, Zhang X, Wang DW, Xu J, Kou KJ, Wang ZW, et al. Overexpressed lncRNA GATA6-AS1 Inhibits LNM and EMT via FZD4 Through the Wnt/beta-Catenin Signaling Pathway in GC. Mol Ther Nucleic Acids (2020) 19:827–40. doi: 10.1016/j.omtn.2019.09.034
113. Li H, Ma X, Yang D, Suo Z, Dai R, Liu C. PCAT-1 Contributes to Cisplatin Resistance in Gastric Cancer Through Epigenetically Silencing PTEN via Recruiting EZH2. J Cell Biochem (2020) 121(2):1353–61. doi: 10.1002/jcb.29370
114. Wang JB, Jin Y, Wu P, Liu Y, Zhao WJ, Chen JF, et al. Tumor Suppressor PLZF Regulated by lncRNA ANRIL Suppresses Proliferation and Epithelial Mesenchymal Transformation of Gastric Cancer Cells. Oncol Rep (2019) 41(2):1007–18. doi: 10.3892/or.2018.6866
115. Roy R, Yang J, Moses MA. Matrix Metalloproteinases as Novel Biomarkers and Potential Therapeutic Targets in Human Cancer. J Clin Oncol (2009) 27(31):5287–97. doi: 10.1200/JCO.2009.23.5556
116. Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH, et al. LincHOTAIR Epigenetically Silences Mir34a by Binding to PRC2 to Promote the Epithelial-to-Mesenchymal Transition in Human Gastric Cancer. Cell Death Dis (2015) 6:e1802. doi: 10.1038/cddis.2015.150
117. Wu DC, Wang SSW, Liu CJ, Wuputra K, Kato K, Lee YL, et al. Reprogramming Antagonizes the Oncogenicity of HOXA13-Long Noncoding RNA HOTTIP Axis in Gastric Cancer Cells. Stem Cells (2017) 35(10):2115–28. doi: 10.1002/stem.2674
118. Wu Q, Xiang S, Ma J, Hui P, Wang T, Meng W, et al. Long non-Coding RNA CASC15 Regulates Gastric Cancer Cell Proliferation, Migration and Epithelial Mesenchymal Transition by Targeting CDKN1A and ZEB1. Mol Oncol (2018) 12(6):799–813. doi: 10.1002/1878-0261.12187
119. Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F, et al. Long Noncoding RNA LINC00673 Is Activated by SP1 and Exerts Oncogenic Properties by Interacting With LSD1 and EZH2 in Gastric Cancer. Mol Ther (2017) 25(4):1014–26. doi: 10.1016/j.ymthe.2017.01.017
120. Xu Y, Dong M, Wang J, Zhao W, Jiao M. LINC01436 Inhibited miR-585-3p Expression and Upregulated MAPK1 Expression to Promote Gastric Cancer Progression. Dig Dis Sci (2021) 66(6):1885–94. doi: 10.1007/s10620-020-06487-w
121. Xu TP, Wang WY, Ma P, Shuai Y, Zhao K, Wang YF, et al. Upregulation of the Long Noncoding RNA FOXD2-AS1 Promotes Carcinogenesis by Epigenetically Silencing EphB3 Through EZH2 and LSD1, and Predicts Poor Prognosis in Gastric Cancer. Oncogene (2018) 37(36):5020–36. doi: 10.1038/s41388-018-0308-y
122. Lu Y, Hou K, Li M, Wu X, Yuan S. Exosome-Delivered LncHEIH Promotes Gastric Cancer Progression by Upregulating EZH2 and Stimulating Methylation of the GSDME Promoter. Front Cell Dev Biol (2020) 8:571297. doi: 10.3389/fcell.2020.571297
123. Chen WM, Huang MD, Sun DP, Kong R, Xu TP, Xia R, et al. Long Intergenic non-Coding RNA 00152 Promotes Tumor Cell Cycle Progression by Binding to EZH2 and Repressing P15 and P21 in Gastric Cancer. Oncotarget (2016) 7(9):9773–87. doi: 10.18632/oncotarget.6949
124. Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discovery (2016) 6(7):784–801. doi: 10.1158/2159-8290.CD-15-0921
125. Jie M, Wu Y, Gao M, Li X, Liu C, Ouyang Q, et al. CircMRPS35 Suppresses Gastric Cancer Progression via Recruiting KAT7 to Govern Histone Modification. Mol Cancer (2020) 19(1):56. doi: 10.1186/s12943-020-01160-2
126. Zhang G, Xu Y, Zou C, Tang Y, Lu J, Gong Z, et al. Long Noncoding RNA ARHGAP27P1 Inhibits Gastric Cancer Cell Proliferation and Cell Cycle Progression Through Epigenetically Regulating P15 and P16. Aging (Albany NY) (2019) 11(20):9090–110. doi: 10.18632/aging.102377
127. Chen RY, Ju Q, Feng LM, Yuan Q, Zhang L. The Carcinogenic Complex lncRNA FOXP4-AS1/EZH2/LSD1 Accelerates Proliferation, Migration and Invasion of Gastric Cancer. Eur Rev Med Pharmacol Sci (2019) 23(19):8371–6. doi: 10.26355/eurrev_201910_19148
128. Wang SS, Wuputra K, Liu CJ, Lin YC, Chen YT, Chai CY, et al. Oncogenic Function of the Homeobox A13-Long Noncoding RNA HOTTIP-Insulin Growth Factor-Binding Protein 3 Axis in Human Gastric Cancer. Oncotarget (2016) 7(24):36049–64. doi: 10.18632/oncotarget.9102
129. Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, et al. CBP-Mediated Acetylation of Histone H3 Lysine 27 Antagonizes Drosophila Polycomb Silencing. Development (2009) 136(18):3131–41. doi: 10.1242/dev.037127
130. Song Y, Wang R, Li LW, Liu X, Wang YF, Wang QX, et al. Long non-Coding RNA HOTAIR Mediates the Switching of Histone H3 Lysine 27 Acetylation to Methylation to Promote Epithelial-to-Mesenchymal Transition in Gastric Cancer. Int J Oncol (2019) 54(1):77–86. doi: 10.3892/ijo.2018.4625
131. Ding H, Shi Y, Liu X, Qiu A. MicroRNA-4513 Promotes Gastric Cancer Cell Proliferation and Epithelial-Mesenchymal Transition Through Targeting KAT6B. Hum Gene Ther Clin Dev (2019) 30(3):142–8. doi: 10.1089/humc.2019.094
132. Zhu L, Wang C, Lin S, Zong L. CircKIAA0907 Retards Cell Growth, Cell Cycle, and Autophagy of Gastric Cancer In Vitro and Inhibits Tumorigenesis In Vivo via the miR-452-5p/KAT6B Axis. Med Sci Monit (2020) 26:e924160. doi: 10.12659/MSM.924160
133. Hu CE, Liu YC, Zhang HD, Huang GJ. JMJD2A Predicts Prognosis and Regulates Cell Growth in Human Gastric Cancer. Biochem Biophys Res Commun (2014) 449(1):1–7. doi: 10.1016/j.bbrc.2014.04.126
134. Zhang J, Zhou Y, Tang PMK, Cheng ASL, Yu J, To KF, et al. Mechanotransduction and Cytoskeleton Remodeling Shaping YAP1 in Gastric Tumorigenesis. Int J Mol Sci (2019) 20(7):1576. doi: 10.3390/ijms20071576
135. Chen LH, Wang LP, Ma XQ. Circ_SPECC1 Enhances the Inhibition of miR-526b on Downstream KDM4A/YAP1 Pathway to Regulate the Growth and Invasion of Gastric Cancer Cells. Biochem Biophys Res Commun (2019) 517(2):253–9. doi: 10.1016/j.bbrc.2019.07.065
136. Zhang E, He X, Zhang C, Su J, Lu X, Si X, et al. A Novel Long Noncoding RNA HOXC-AS3 Mediates Tumorigenesis of Gastric Cancer by Binding to YBX1. Genome Biol (2018) 19(1):154. doi: 10.1186/s13059-018-1523-0
137. Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu S, et al. Sirt7 Promotes Gastric Cancer Growth and Inhibits Apoptosis by Epigenetically Inhibiting miR-34a. Sci Rep (2015) 5:9787. doi: 10.1038/srep09787
138. Liu Y, Zhang X, Zhang Y, Hu Z, Yang D, Wang C, et al. Identification of Mirnomes in Human Stomach and Gastric Carcinoma Reveals miR-133b/a-3p as Therapeutic Target for Gastric Cancer. Cancer Lett (2015) 369(1):58–66. doi: 10.1016/j.canlet.2015.06.028
139. Guan A, Wang H, Li X, Xie H, Wang R, Zhu Y, et al. MiR-330-3p Inhibits Gastric Cancer Progression Through Targeting MSI1. Am J Transl Res (2016) 8(11):4802–11.
140. Zhao LJ, Fan QQ, Li YY, Ren HM, Zhang T, Liu S, et al. LSD1 Deletion Represses Gastric Cancer Migration by Upregulating a Novel miR-142-5p Target Protein CD9. Pharmacol Res (2020) 159:104991. doi: 10.1016/j.phrs.2020.104991
141. Kang W, Huang T, Zhou Y, Zhang J, Lung RWM, Tong JHM, et al. miR-375 is Involved in Hippo Pathway by Targeting YAP1/TEAD4-CTGF Axis in Gastric Carcinogenesis. Cell Death Dis (2018) 9(2):92. doi: 10.1038/s41419-017-0134-0
142. Xu G, Zhu H, Zhang M, Xu J. Histone Deacetylase 3 is Associated With Gastric Cancer Cell Growth via the miR-454-Mediated Targeting of CHD5. Int J Mol Med (2018) 41(1):155–63. doi: 10.3892/ijmm.2017.3225
143. Lin L, Jiang H, Huang M, Hou X, Sun X, Jiang X, et al. Depletion of Histone Deacetylase 1 Inhibits Metastatic Abilities of Gastric Cancer Cells by Regulating the miR-34a/CD44 Pathway. Oncol Rep (2015) 34(2):663–72. doi: 10.3892/or.2015.4010
144. Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, et al. Decreased Expression of the Long non-Coding RNA FENDRR is Associated With Poor Prognosis in Gastric Cancer and FENDRR Regulates Gastric Cancer Cell Metastasis by Affecting Fibronectin1 Expression. J Hematol Oncol (2014) 7:63. doi: 10.1186/s13045-014-0063-7
145. Wu S, Wu E, Wang D, Niu Y, Yue H, Zhang D, et al. LncRNA HRCEG, Regulated by HDAC1, Inhibits Cells Proliferation and Epithelial-Mesenchymal-Transition in Gastric Cancer. Cancer Genet (2020) 241:25–33. doi: 10.1016/j.cancergen.2019.12.007
Keywords: ncRNA, histone modification, gastric cancer, epigenetic modifications, mechanisms
Citation: Yang Q, Chen Y, Guo R, Dai Y, Tang L, Zhao Y, Wu X, Li M, Du F, Shen J, Yi T, Xiao Z and Wen Q (2022) Interaction of ncRNA and Epigenetic Modifications in Gastric Cancer: Focus on Histone Modification. Front. Oncol. 11:822745. doi: 10.3389/fonc.2021.822745
Received: 26 November 2021; Accepted: 28 December 2021;
Published: 26 January 2022.
Edited by:
Muzafar Ahmad Macha, Islamic University of Science and Technology, IndiaReviewed by:
Qi Shengcai, Shanghai Stomatology Prevention Hospital, ChinaCopyright © 2022 Yang, Chen, Guo, Dai, Tang, Zhao, Wu, Li, Du, Shen, Yi, Xiao and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Yi, eWl0YW9AaGtidS5lZHUuaGs=; Zhangang Xiao, emhhbmdhbmd4aWFvQHN3bXUuZWR1LmNu; Qinglian Wen, d3FsNzMxMTVAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.