- 1Center of Uterine Cancer Diagnosis & Therapy Research of Zhejiang Province, Department of Obstetrics and Gynecology, the Second Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
- 2Department of Obstetrics and Gynecology, Taizhou Woman and Children’s Hospital of Wenzhou Medical University, Taizhou, China
The abnormally methylated tumor suppressor genes (TSGs) associated with cervical cancer are unclear. DNA methylation data, RNA-seq expression profiles, and overall survival data were downloaded from TCGA CESC database. DMGs and DEGs were obtained through CHAMP and DESeq packages, respectively. TSGs were downloaded from TSGene 2.0. Candidate hypermethylated/down-regulated TSGs were further evaluated and pyrosequencing was used to confirm their difference in methylation levels of selected TSGs in cervical cancer patients. A total of 25946 differentially methylated CpGs corresponding to 2686 hypermethylated genes and 4898 hypomethylated genes between cervical cancer and adjacent normal cervical tissues were found in this study. Besides, 693 DEGs (109 up-regulated and 584 down-regulated) were discovered in cervical cancer tissues. Then, 192 hypermethylated/down-regulated genes were obtained in cervical cancer compared to adjacent tissues. Interestingly, 26 TSGs were found in hypermethylated/down-regulated genes. Among these genes, low expression of MRVI1 and NTRK3 was associated with poor overall survival in cervical cancer. Moreover, GEO data showed that MRVI1 and NTRK3 were significantly decreased in cervical cancer tissues. The expression levels of MRVI1 and NTRK3 were negatively correlated with the methylation levels of their promoter CpG sites. Additionally, elevated methylation levels of MRVI1 and NTRK3 promoter were further verified in cervical cancer tissues by pyrosequencing experiments. Finally, the ROC results showed that the promoter methylation levels of MRVI1 and NTRK3 had the ability to discriminate cervical cancer from healthy samples. The study contributes to our understanding of the roles of MRVI1 and NTRK3 in cervical cancer.
Introduction
Cervical cancer is the fourth most common cancer and the fourth leading cause of cancer death in women (1). The prognosis varies depending on the stage of cervical cancer. Compared with patients with early stage of cervical cancer, the five-year survival period of patients with advanced cervical cancer is much shorter (2). Therefore, the identification predictive biomarkers can help effective targeted therapy and treatment decisions.
Epigenetic processes can be reversed and this principle makes it a potential target for therapeutic intervention (3). Epigenetic variations could change the expression of tumor suppressor genes (TSGs) in cervical cancer (4). DNA methylation is an important part of epigenetics (5, 6) and the regulatory effect of DNA methylation on gene expression has been studied extensively (7, 8). DNA methylation levels could be detected by techniques, including pyrosequencing, methylation-specific polymerase chain reaction, methylation-sensitive high-resolution melting, multiplex ligation-dependent probe amplification (MLPA), and Combined bisulfite restriction analysis (COBRA) and MethyLight (9). Aberrant methylation of TSGs could silence the expression of TSGs to consequently promote tumor formation (10). During recent decades, there have been a massive number of studies about TSGs in cervical cancer (11–13). For example, compared with the control samples, the promoter methylation frequency of TSG (including RARB, CADM1, PAX1, and DAPK1) in patients with invasive cervical cancer is higher (14). The silencing of TSGs is thought to be an early, driving event in the oncogenic process. Even after human papilloma virus (HPV) clearance, the silencing of TSGs by DNA hypermethylation could trigger carcinogenesis of the cervix (15). However, changes in DNA methylation and related abnormal TSGs expression have not been systematically elucidated in cervical cancer.
Gene methylation profiling and gene expression profiling have been utilized to investigate DNA methylation and gene expression in the molecular mechanism, biological process, and biomarker (16–18). Combined analysis of gene expression and DNA methylation data may contribute to identifying potential biomarkers of cervical cancer for treatment. Therefore, in this study, Illumina HumanMethylation450K methylation data and RNA-seq expression profiles from the Cancer Genome Atlas-Cervical Cancer (TCGA-CESC) were integrated for identifying the DMGs and DEGs in cervical cancer. First, TSGs among hypermethylated/down-regulated genes were found. Second, cervical cancer prognosis-related genes were selected and used as candidate cervical cancer-related TSGs. Then, expression levels of these TSGs were subsequently verified in three independent data sets from the Gene Expression Omnibus (GEO) database. Moreover, cervical cancer tissues and paired adjacent normal cervical tissues were collected to verify the methylation levels of these TSGs. Finally, receiver operating characteristic (ROC) curve analysis was used to assess the development of candidate cervical cancer related TSGs. This study aims to find prognostic and diagnostic TSGs related to cervical cancer through data analysis and experimental verification.
Results
Differential Methylation and Expression Analysis
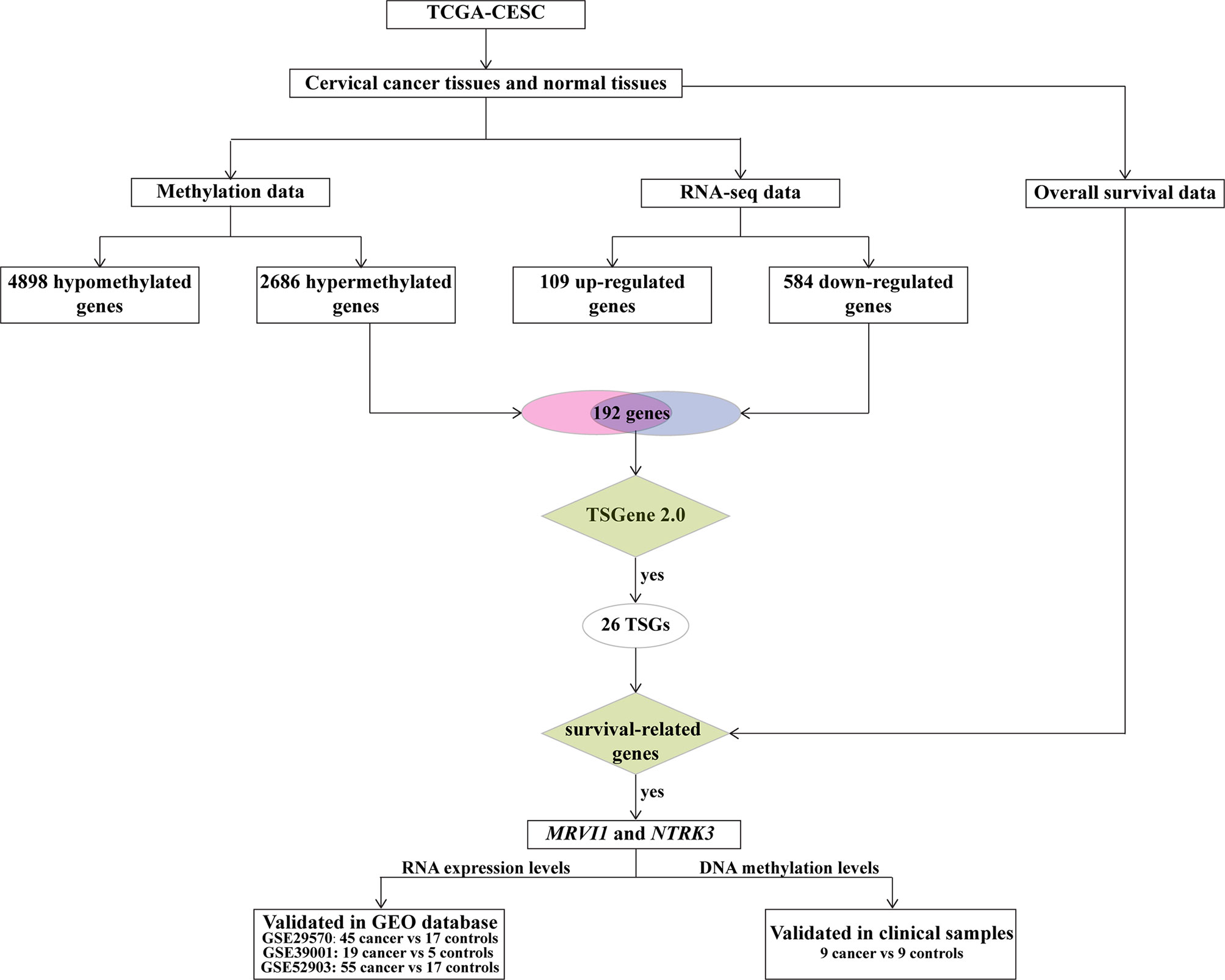
The workflow of our study is displayed in Figure 1. TCGA-CESC was used to identify aberrantly methylation-regulated genes. 12611 hypermethylated CpG sites, which were correspond to 2686 genes, were found in cervical cancer than that in adjacent normal cervical tissues. On the contrary, 13335 hypomethylated CpG sites, which were correspond to 4898 genes, were discovered in cervical cancer compared to adjacent normal cervical tissues (Figures 2A, B). Additionally, a total of 693 DEGs (109 up-regulated and 584 down-regulated) were obtained from TCGA-CESC (Figures 2C, D). Then, 192 hypermethylated/down-regulated genes (Figure 2E) and 60 hypomethylated/up-regulated genes (Figure 2F) were identified. Hypermethylated/down-regulated genes were particularly focused in the current study.

Figure 2 The differentially methylated genes (DMGs) and differentially expression genes (DEGs) from TCGA-CESC database. (A) The volcano plot was constructed using log2fold change and −log10 (padj) values. Red plots represent the up-regulated CpG sites, green plots represent the down-regulated CpG sites, and black plots show those CpG sites that are not differentially expressed. (B) Heatmap of methylation values for top 1000 CpG sites methylated in cervical cancer patients. CpG sites are shown in the vertical columns and the cervical cancer samples in the horizontal rows. High methylation levels are displayed in red and low methylation levels in blue, according to the scale bar in the right of figure. (C) The volcano plot was constructed using log2 fold change and −log10 (padj) values. Red plots represent the up-regulated genes, green plots represent the down-regulated genes, and black plots show those genes that are not differentially expressed. (D) Heatmap of methylation values for DEGs in cervical cancer patients. The genes are displayed in the vertical columns and the cervical cancer samples in the horizontal rows. High expression levels are shown in red and low expression levels in blue, according to the scale bar in the right of the figure. (E) Venn diagrams of the genes relevant to hypermethylated/down-regulated genes. (F) Venn diagrams of the genes relevant to hypormethylated/up-regulated genes.
GO and KEGG Pathway Enrichment of the Down-Regulated DEGs With Hypermethylation
The top 15 significant GO enrichments of biological processes were illustrated in Figure 3A, including extracellular structure organization, multicellular organismal signaling, extracellular matrix organization, and actin filament-based process. There were 13 enrichment pathways, such as vascular smooth muscle contraction, cGMP-PKG signaling pathway, calcium signaling pathway, focal adhesion, ECM-receptor interaction, proteoglycans in cancer, apelin signaling pathway (Figure 3B).

Figure 3 GO and KEGG pathway of the genes relevant to hypermethylation/low expression genes. (A) GO of hypermethylated/down-regulated genes. Names of the top 15 items are indicated by the y-axis. The size of the colored dots is the enriched number of genes in each GO classification. The red dots indicate high enrichment and the blue dots indicate low enrichment. FDR value is expressed by the color order on the right edge. (B) KEGG pathway enrichment of hypermethylated/down-regulated genes. The size of the dot in the KEGG pathway bubble plot shows the enriched genes. High enriched represented by red, otherwise, by blue.
Identification of Candidate TSGs
26 TSGs were discovered in hypermethylated/down-regulated genes (Figure 4A). A total of 2361 cervical cancer survival-related genes were found by Kaplan–Meier analysis using RNA expression data. After integrated TSGs and survival-related genes, 2 overlapping genes (MRVI1 and NTRK3) were discovered and considered as the cervical cancer candidate TSGs (Figure 4B).

Figure 4 Venn diagram of overlapping genes. (A) Venn diagram of overlapping genes in down-regulated genes (blue circle), and hypermethylation genes (red circle), and TSGs (green circle). (B) Venn diagram of overlapping genes in down-regulated genes (blue circle), and hypermethylation genes (red circle), TSGs (green circle), and survival-related genes (yellow circle). TSGs, tumor suppressor genes.
Survival Analysis and Validation of MRVI1 and NTRK3 Expression
The expression levels of MRVI1 and NTRK3 genes were obtained in this study (Figure 5A). As shown in Figures 5B, C, the expression levels of MRVI1 (P = 0.002) and NTRK3 (P = 0.029) were significantly lower in cervical cancer than those in adjacent normal cervical tissues. In addition, patients with low expression of MRVI1 (P = 0.026) and NTRK3 (P = 0.025) had significantly worse survival rates (Figures 5D, E).

Figure 5 Prognostic values of MRVI1 and NTRK3. (A) Hierarchical clustering of MRVI1 and NTRK3; Expression of MRVI1 (B) and NTRK3 (C) in 304 cervical cancer patients and 3 normal cervical tissues. The MRVI1 and NTRK3 expression levels (log2(FPKM+1)) are significantly decreased in cervical cancer patients compared with the normal controls. (D, E) Kaplan–Meier curves show that low expression of MRVI1 and NTRK3 have worse prognosis than that of the high expression of MRVI1 and NTRK3.
Validation of MRVI1 and NTRK3 Expression Levels by the GEO Database
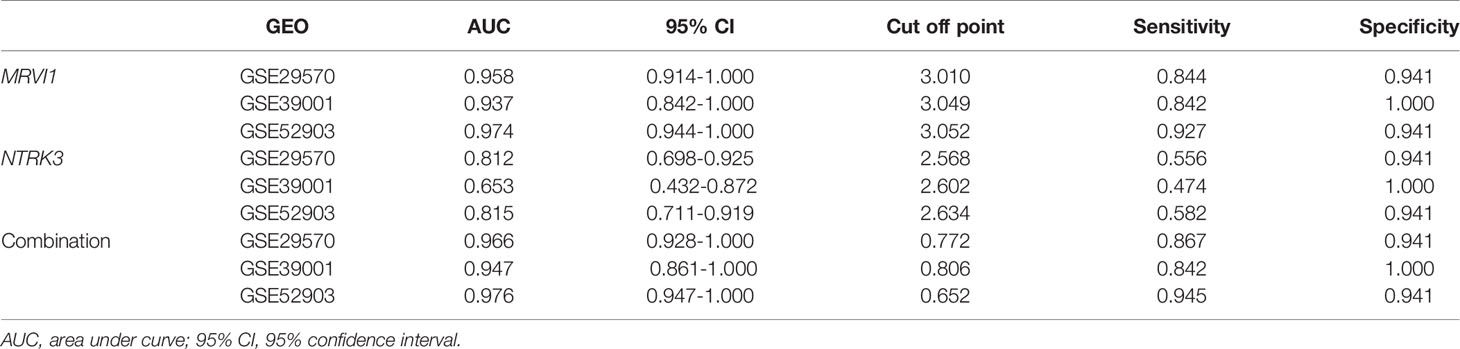
MRVI1 and NTRK3 levels were all significantly lower in cervical cancers compared to normal cervical tissues in three cervical cancer related datasets (p < 0.05, Figures 6A–F). The ROC curves of MRVI1 and NTRK3 gene expression levels to determine cervical cancer were presented in Figures 6G–O. The ROC curve indicated that MRVI1 exhibited high diagnostic efficiency for cervical cancer in normal cervical controls in three datasets (AUC > 0.937). The AUC of the prediction model for NTRK3 was greater than 0.653 in these datasets. The AUC of the combined prediction model of MRVI1 and NTRK3 (AUC > 0.947) was higher than that of the MRVI1 (AUC > 0.937) and NTRK3 (AUC > 0.653) (Figures 6G–O and Table 1). These results suggested that the expression levels of MRVI1 and NTRK3 could distinguish between cervical cancer patients and healthy controls.

Figure 6 Confirmed expression levels and ROC curve analyses for the prediction of cervical cancer. (A–F) Boxplots of gene expression levels in three GEO databases for MRVI1 and NTKR3. The left y-axis shows the mRNA expression levels for MRVI1 or NTRK3. The x-axis represents the two groups (normal tissue and cervical tumor). Each panel represents a different GEO database (GSE29570, GSE39001, and GSE52903). ROC curve of MRVI1 (G–I), NTRK1 (J–L), and combined expression (M–O) for distinguishing between cervical cancer and non-tumor tissues in individual GEO datasets. AUC, the area under the ROC curve; GEO, Gene Expression Omnibus; ROC, receiver operating characteristics.
Correlation Analysis of Promoter Region Methylation Level and Gene Expression Level
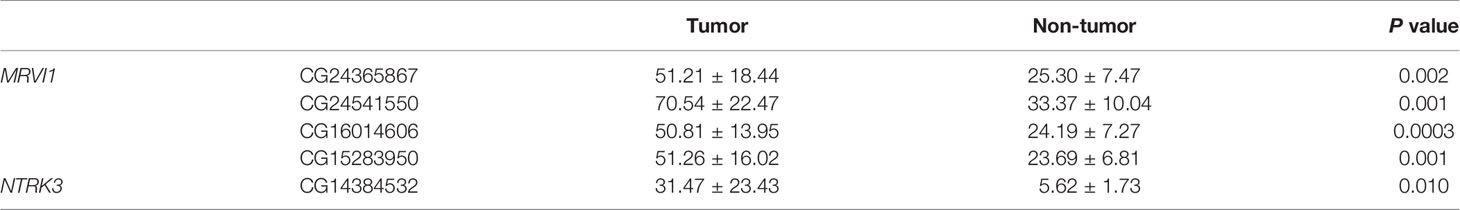
A total of 5 CpG sites were located in the promoter regions of MRVI1 (cg24365867, cg24541550, cg16014606, and cg15283950) and NTRK3 (cg14384532) (Figure 7A). The methylation levels of these CpG sites were up-regulated in cervical tumors compared to controls (p < 0.05, Figures 7B–F). Further correlation analysis showed that the methylation levels of these CpG sites were negatively associated with gene expression for these two genes (p < 0.05, Figure 8).
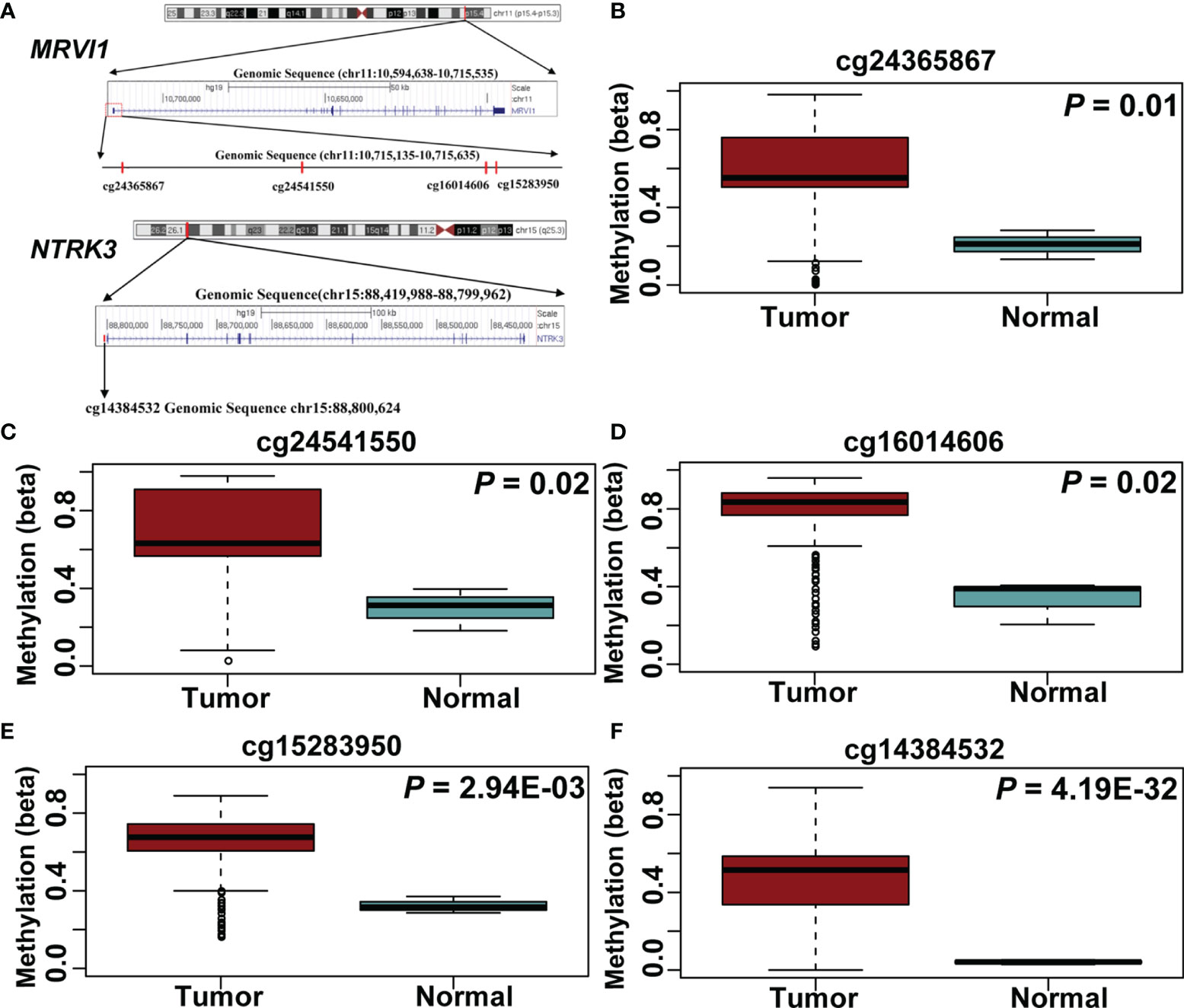
Figure 7 Five CpG sites in MRVI1 and NTRK3 promoter regions. (A) The locations of CpG sites. The level of MRVI1 promoter DNA methylation (B–E) and NTRK3 promoter DNA methylation (F) presented as a box plot in the cervical cancer and normal cervical tissues.
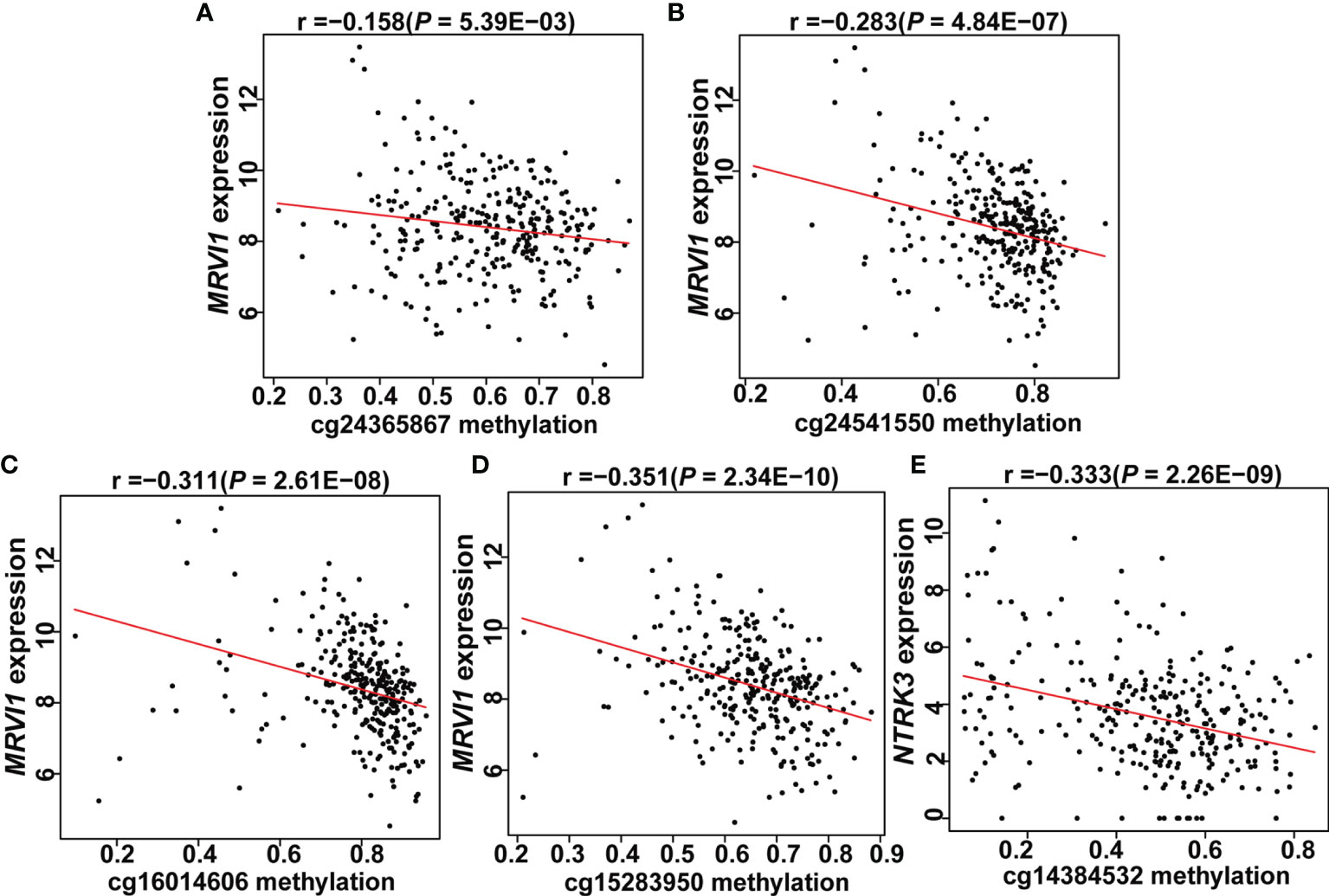
Figure 8 Expression of two genes correlated with promoter CpG site methylation in cervical tissues. (A–D) MRVI1 gene expression was negatively correlated with DNA methylation levels of cg24365867, cg24541550, cg16014606, and cg15283950. (E) NTRK3 gene expression was negatively correlated with DNA methylation level of cg14384532. x-axis: DNA methylation levels; y-axis: relative gene expression levels; red line: linear regression.
Verification of Differences in Promoter Methylation Levels of MRVI1 and NTRK3 by Pyrosequencing Experiments
In order to verify the differential methylation levels of MRVI1 and NTRK3 between cervical cancer and adjacent normal cervical tissues, pyrosequencing experiments were conducted. As shown in Table 2 and Figure 9A, the methylation levels of cg24365867, cg24541550, cg16014606, and cg15283950 of MRVI1 gene in cervical cancer were significantly higher than that in adjacent normal cervical tissues (p < 0.05). Compared with adjacent normal tissues, a significantly elevated methylation level of cg14384532 on NTRK3 was also found in cervical cancer tissues (p < 0.05, Table 2 and Figure 9A). In addition, ROC analysis was performed and AUC was calculated to assess the potential diagnostic value of MRVI1 and NTRK3 using the methylation levels of CpG sites on promoter regions. As shown in Figures 9B–G, five CpG sites had excellent diagnostic performance for discriminating cervical cancer from healthy cervical samples (cg24365867, p = 0.003, AUC = 0.901; cg24541550, p= 0.003, AUC = 0.901; cg16014606, p = 0.002, AUC = 0.914; cg15283950, p = 0.003, AUC = 0.901; cg14384532, p = 0.014, AUC = 0.840; Combined, p = 0.0002, AUC = 1.000).
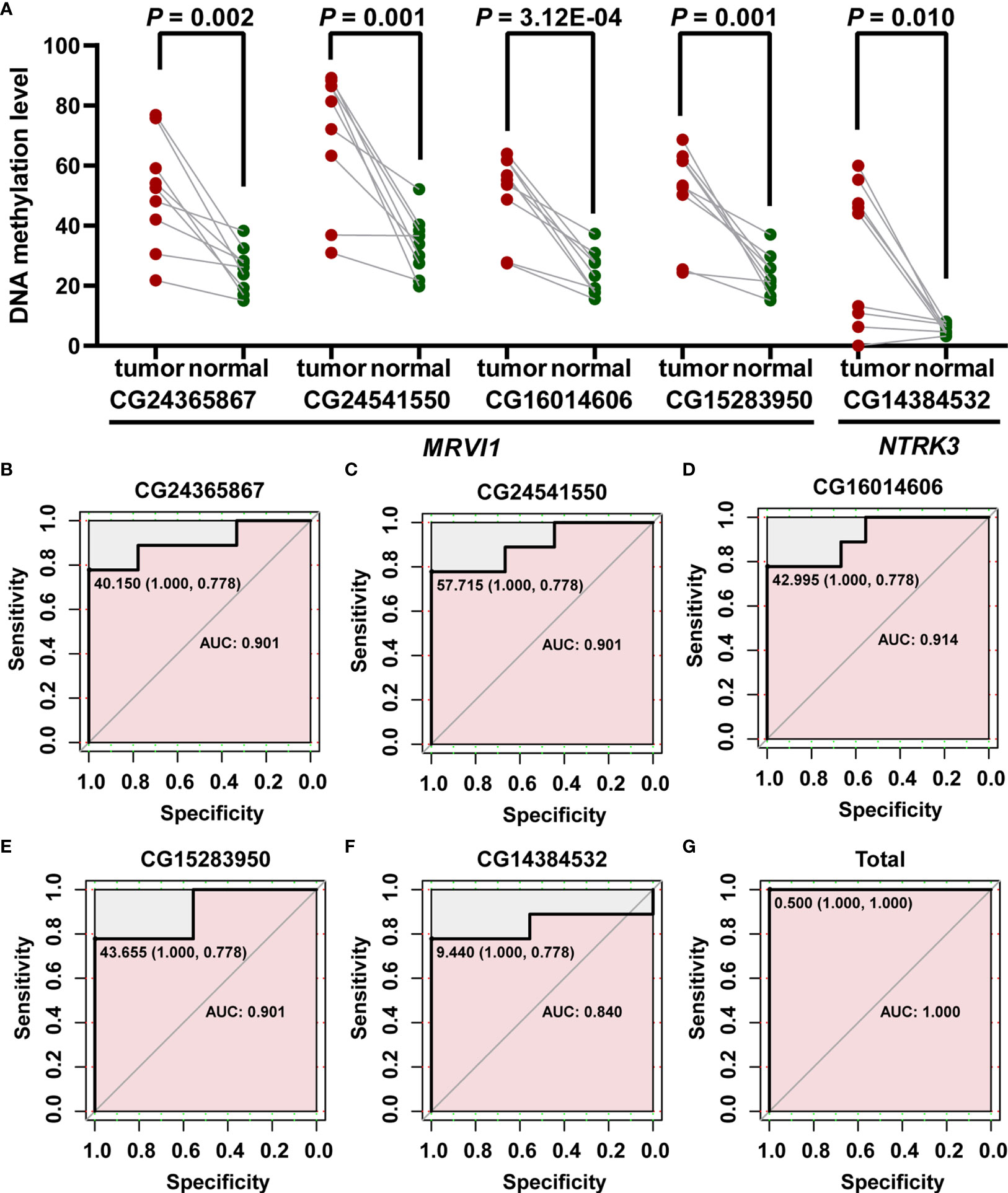
Figure 9 Confirmed methylation levels and ROC curve analyses for the prediction of cervical cancer. (A) The methylation levels of MRVI1 and NTRK3 were significantly higher in cervical cancer tissues than in adjacent normal tissues. ROC curve of methylation levels of MRVI1 (B–E), NTRK1 (F), and combined methylation levels of five CpGs (G) for distinguishing between cervical cancer and non-tumor cervical tissues. AUC, the area under the ROC curve; ROC, receiver operating characteristics.
Discussion
Cervical cancer is one of the most common types of cancer and represents a major global health challenge (1). Since aberrant DNA methylation occurs very early during tumorigenesis (19), it could therefore be used as an early diagnostic biomarker (20). In this study, hypermethylated and significantly lower expressions of TSGs MRVI1 and NTRK3 were discovered in cervical cancers than that in normal cervical tissues using the bioinformatics. The differences of MRVI1 and NTRK3 expressions between cervical cancer specimens and normal cervical tissues were further verified via three GEO datasets. Besides, the low expression of MRVI1 and NTRK3 was negatively associated with high methylation levels of promoter CpG sites. Moreover, promoter hypermethylation levels of MRVI1 and NTRK3 were also found in our clinical cervical cancer samples. ROC curve analyses proved the diagnostic value of MRVI1 and NTRK3 in cervical cancer. Furthermore, low expression of MRVI1 and NTRK3 was associated with poor prognosis of cervical cancer. These results enhanced our understanding of the DNA methylation pattern of TSGs in cervical cancer.
MRVI1 is a protein-coding gene, which has been widely studied in cancer (21, 22). MRVI1 was reported to regulate the cellular release of calcium signal (23), which plays an important role in cancer cell proliferation invasiveness (24). One study discovered that MRVI1 was transcriptionally activated by p53, and p53-induced inhibition of colorectal cancer prognosis was depended on MRVI1 (25). Zhu et al. found that the MRVI1-AS1/ATF3 signalling pathway could increase paclitaxel chemosensitivity by modulating the Hippo-TAZ signalling pathway in nasopharyngeal cancer (21). Another research found that miR-940 could promote proliferation and metastasis of endometrial carcinoma through the regulation of MRVI1 (22). High expression of MRVI1 had a better prognosis than that of the low expression of MRVI1 in endometrial carcinoma (22). Unfortunately, the role of MRVI1 in cervical cancer has not yet been reported. In the current study, the overall survival of cervical cancer patients with low MRVI1 expression was also significantly shorter than those with high MRVI1 expression, which is consistent with previous endometrial carcinoma study.
NTRK3 encodes the TrkC protein, a member of neurotrophic tropomyosin receptor kinase (Trk) family, which autophosphorylates and motivates various signalling pathways such as MAPK and PI3K/AKT pathways (26). Trk aberrations, including gene fusion, gene overexpression, and single nucleotide variation, are involved in the pathogenesis of many cancers, among which NTRK3 gene fusion is extremely confirmed for oncogenic event (27). Unusual activation of NTRK3 and its fusion proteins may balance the epithelial–mesenchymal transition (EMT), oncogenicity, and tumor growth rate via triggering various signalling pathways (28). ETV6-NTRK3 gene fusion acted as a potent oncogene driver and had been presented in the majority of cases of infantile fibrosarcoma (29). Oncogenic fusions in NTRK family receptor tyrosine kinases had been identified in several cancers and could serve as therapeutic targets, for instance in spitz tumors (30), fibrosarcoma (31), gastrointestinal stromal tumors (32), and inflammatory myofibroblastic tumors (33). Conversely, NTRK3 expression was a good prognosis factor in a variety of cancers and more specifically in melanomas (34), neuroblastomas (35), and colorectal cancer (36). NTRK3 expression and activation had been shown to trigger apoptosis in medulloblastoma cells (37). In recent years, SPECC1L-NTRK3 gene fusion was found in cervical sarcoma patients (38). However, the research on NTRK3 gene in cervical cancer is rare. Depending on the present study, NTRK3 expression was significantly lower in cervical cancer specimens than that in normal cervical tissues, and low NTRK3 expression was associated with a poor prognosis. These findings suggested that NTRK3 might likewise serve as a tumor suppressor gene in cervical cancer.
In present study, two TSGs (MRVI1 and NTRK3) were identified via bioinformatics. Nevertheless, a total of 26 hypermethylated/down-regulated TSGs have been discovered, the rest TSGs should be further studied. Although, the hypermethylation levels of MRVI1 and NTRK3 were verified in 9 cervical cancer tissues by pyrosequencing, the large number of clinical samples should be collected in further study. Hypermethylated and down-regulated expression levels of TSGs MRVI1 and NTRK3 have been identified in the current study; however, the detail epigenetic regulatory mechanism under cervical cancer still needs further investigation. In summary, our results revealed that hypermethylation in the promoter regions of MRVI1 and NTRK3 genes might lead to low expression in cervical cancer. Low expression levels of MRVI1 and NTRK3 were associated with poor prognosis of cervical cancer. The methylation levels and expression levels of MRVI1 and NTRK3 had the ability to effectively discriminate cervical cancer from healthy samples. Therefore, MRVI1 and NTRK3 genes may play important roles in the occurrence and prognosis of cervical cancer. It could be further explored and validated as a therapeutic target for cervical cancer. In conclusion, the down-regulation of MRVI1 and NTRK3 may drive cervical cancer through hypermethylation of their promoters. Further studies are needed to draw more attention to the roles of these TSGs in cervical cancer.
Materials and Methods
Data Resources for DNA Methylation, RNA-Seq Data and Clinical Information
Illumina Infinium HumanMethylation450K and RNA-seq expression profiles were downloaded from TCGA-CESC (https://cancergenome.nih.gov/). Methylation data of 307 cervical cancer and 3 normal cervical tissues were collected in the present study. The probes were annotated by using the Bioconductor package with the human genome assembly GRCh37 (hg19). Gene expression profile corresponding to abnormally methylated genes was also download. In addition, the corresponding clinical overall survival data of 291 samples were included. Besides, 3 cervical cancer-related expression datasets (GSE29570, GSE39001, and GSE52903) were obtained from the GEO database. GSE29570 (39) includes the expression data from 17 healthy female exocervix samples and 45 cervical cancer samples, GSE39001 (40) contains data from 5 healthy female cervical samples and 19 cervical cancer samples, and GSE52903 (41) contains expression profiles from a discovery cohort of 17 healthy female cervical samples and 55 cervical cancer samples.
Differential Methylation and Gene Expression Analysis
Between cervical cancer tissues and normal cervical tissues, significant DMGs and DEGs were identified using DESeq package (42) and CHAMP package of R (43), respectively. The false discovery rate (FDR) was adopted to avoid the occurrence of false-positive results. FDR < 0.05 and |Log2 Fold change (Log2FC)| > 1 were used to select significant DMG or DEG.
Gene Ontology (GO) and KEGG Pathway Analysis
GO and KEGG pathway enrichment analysis of hypermethylated/down-regulated genes was performed using the g:Profiler program (44).
Searching for TSGs Associated With Cervical Cancer
Among hypermethylated/down-regulated genes, TSGs were identified based on TSGene 2.0 (45). With the median expression level as the demarcation point, 291 patients with clinical data in TCGA were divided into low-risk group and high-risk group. Kaplan-Meier analysis in the survival package of R (46) was used to compare the difference in overall survival between the two groups. Prognostic-related TSGs were considered as candidate genes for cervical cancer. To solve the problem of a small number of normal tissues in TCGA-CESC, the expression levels of cervical cancer candidate TSGs in three GEO datasets (GSE29570, GSE39001, and GSE52903) were further compared by T test using R.
Pyrosequencing Experiment
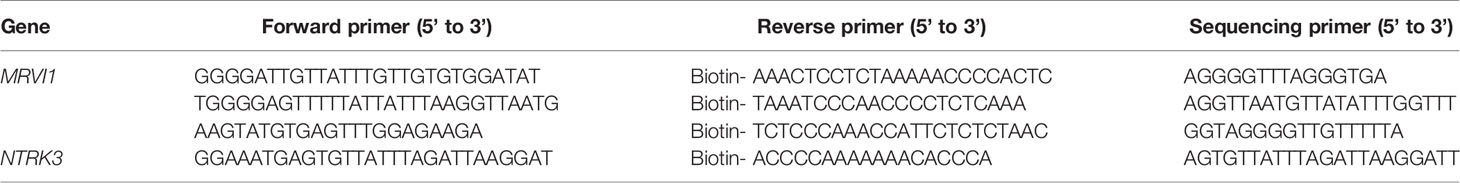
Nine pairs of cervical cancer specimens and adjacent normal cervical tissues were obtained from the Second Affiliated Hospital of Wenzhou Medical University. Cervical cancer patients were diagnosed by experienced pathologists based on the results of surgically removed specimens (Supplementary Table 1). Human genomic DNA was extracted from tissue samples using the Genomic DNA Extraction Kit (Qiagen, Dusseldorf, Germany). DNA concentrations were determined by the Infinite F200 Tecan microplate reader (Tecan, männedorf, Switzerland). Primers were designed using the PyroMark Assay Design Software 2.0 and bisulfite-treated DNA PCR-amplified using the PyroMark PCR kit prior to analysis on a PyroMark Q96 according to manufacturer’s instruction (Qiagen, Dusseldorf, Germany). Sequences of the PCR primers were shown in Table 3. Amplification was carried out as follows: 95°C for 3 min, followed by 40 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min, with a final elongation step at 72°C for 7 min. Raw data were analyzed using PyroMark Q96 software (Qiagen, Dusseldorf, Germany). The research protocol was approved by the Ethics Committee of the Second Affiliated Hospital of Wenzhou Medical University. Written informed consents were obtained from all subjects.
Statistical Analysis
Pearson correlation coefficient was used to correlate promoter methylation levels with candidate TSGs expression levels. ROC curves were used to compare the sensitivity and specificity of the candidate TSGs expression levels and promoter methylation levels in the prediction of cervical cancer. All the data were analyzed using R scripts. A two-tailed p value < 0.05 was considered statistically significant.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
Conceptualization, HJ and XZ. Methodology, KL. Formal analysis, KL, WJ, JL, J-aZ. Investigation and writing, HJ and XZ. Visualization and supervision, XZ. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the grant from the Subject of Integrated Traditional Chinese and Western Medicine in Zhejiang Province (2017-XK-A42). The study sponsors had no involvement in the collection, analysis, and interpretation of data, or in the writing of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.802068/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Sakuragi N. Refining Insight Into Cervical Cancer Progression. Lancet Oncol (2014) 15:371–2. doi: 10.1016/S1470-2045(14)70085-3
3. Mathers JC, Strathdee G, Relton CL. Induction of Epigenetic Alterations by Dietary and Other Environmental Factors. Adv Genet (2010) 71:3–39. doi: 10.1016/B978-0-12-380864-6.00001-8
4. Zhao S. Specific Type Epigenetic Changes in Cervical Cancers. Methods Mol Biol (2015) 1238:733–49. doi: 10.1007/978-1-4939-1804-1_38
5. Teschendorff AE, Relton CL. Statistical and Integrative System-Level Analysis of DNA Methylation Data. Nat Rev Genet (2018) 19:129–47. doi: 10.1038/nrg.2017.86
6. Xie XL, Yu Y, Yuan ZF, Yang J, Ma PP, Li DC, et al. Comparative Analysis on Content and Distribution of CpG Sites in Milk Production Traits and Mastitis-Related Genes in Dairy Cattle. Yi Chuan (2012) 34:437–44. doi: 10.3724/SP.J.1005.2012.00437
7. Husquin LT, Rotival M, Fagny M, Quach H, Zidane N, McEwen LM, et al. Exploring the Genetic Basis of Human Population Differences in DNA Methylation and Their Causal Impact on Immune Gene Regulation. Genome Biol (2018) 19:222. doi: 10.1186/s13059-018-1601-3
8. Long MD, Smiraglia DJ, Campbell MJ. The Genomic Impact of DNA CpG Methylation on Gene Expression; Relationships in Prostate Cancer. Biomolecules (2017) 7. doi: 10.3390/biom7010015
9. Quillien V, Lavenu A, Karayan-Tapon L, Carpentier C, Labussiere M, Lesimple T, et al. Comparative Assessment of 5 Methods (Methylation-Specific Polymerase Chain Reaction, MethyLight, Pyrosequencing, Methylation-Sensitive High-Resolution Melting, and Immunohistochemistry) to Analyze O6-Methylguanine-DNA-Methyltranferase in a Series of 100 Glioblastoma Patients. Cancer (2012) 118:4201–11. doi: 10.1002/cncr.27392
10. Kazanets A, Shorstova T, Hilmi K, Marques M, Witcher M. Epigenetic Silencing of Tumor Suppressor Genes: Paradigms, Puzzles, and Potential. Biochim Biophys Acta (2016) 1865:275–88. doi: 10.1016/j.bbcan.2016.04.001
11. Sin STK, Li Y, Liu M, Ma S, Guan XY. TROP-2 Exhibits Tumor Suppressive Functions in Cervical Cancer by Dual Inhibition of IGF-1R and ALK Signaling. Gynecol Oncol (2019) 152:185–93. doi: 10.1016/j.ygyno.2018.10.039
12. Qin Y, Tang X, Liu M. Tumor-Suppressor Gene NBPF1 Inhibits Invasion and PI3K/mTOR Signaling in Cervical Cancer Cells. Oncol Res (2016) 23:13–20. doi: 10.3727/096504015X14410238486766
13. Guerrero-Setas D, Perez-Janices N, Blanco-Fernandez L, Ojer A, Cambra K, Berdasco M, et al. RASSF2 Hypermethylation is Present and Related to Shorter Survival in Squamous Cervical Cancer. Mod Pathol (2013) 26:1111–22. doi: 10.1038/modpathol.2013.32
14. Tawe L, Grover S, Zetola N, Robertson ES, Gaseitsiwe S, Moyo S, et al. Promoter Hypermethylation Analysis of Host Genes in Cervical Cancer Patients With and Without Human Immunodeficiency Virus in Botswana. Front Oncol (2021) 11:560296. doi: 10.3389/fonc.2021.560296
15. Rogeri CD, Silveira HCS, Causin RL, Villa LL, Stein MD, de Carvalho AC, et al. Methylation of the hsa-miR-124, SOX1, TERT, and LMX1A Genes as Biomarkers for Precursor Lesions in Cervical Cancer. Gynecol Oncol (2018) 150:545–51. doi: 10.1016/j.ygyno.2018.06.014
16. Li Z, Heng J, Yan J, Guo X, Tang L, Chen M, et al. Integrated Analysis of Gene Expression and Methylation Profiles of 48 Candidate Genes in Breast Cancer Patients. Breast Cancer Res Treat (2016) 160:371–83. doi: 10.1007/s10549-016-4004-8
17. van den Dungen MW, Murk AJ, Kok DE, Steegenga WT. Comprehensive DNA Methylation and Gene Expression Profiling in Differentiating Human Adipocytes. J Cell Biochem (2016) 117:2707–18. doi: 10.1002/jcb.25568
18. Zhou R, Man Y. Integrated Analysis of DNA Methylation Profiles and Gene Expression Profiles to Identify Genes Associated With Pilocytic Astrocytomas. Mol Med Rep (2016) 13:3491–7. doi: 10.3892/mmr.2016.4943
19. Hao X, Luo H, Krawczyk M, Wei W, Wang W, Wang J, et al. DNA Methylation Markers for Diagnosis and Prognosis of Common Cancers. Proc Natl Acad Sci USA (2017) 114:7414–9. doi: 10.1073/pnas.1703577114
20. Kim MS, Lee J, Sidransky D. DNA Methylation Markers in Colorectal Cancer. Cancer Metastasis Rev (2010) 29:181–206. doi: 10.1007/s10555-010-9207-6
21. Zhu Y, He D, Bo H, Liu Z, Xiao M, Xiang L, et al. The MRVI1-AS1/ATF3 Signaling Loop Sensitizes Nasopharyngeal Cancer Cells to Paclitaxel by Regulating the Hippo-TAZ Pathway. Oncogene (2019) 38:6065–81. doi: 10.1038/s41388-019-0858-7
22. Zhou Z, Xu YP, Wang LJ, Kong Y. miR-940 Potentially Promotes Proliferation and Metastasis of Endometrial Carcinoma Through Regulation of MRVI1. Biosci Rep (2019) 39. doi: 10.1042/BSR20190077
23. Schlossmann J, Ammendola A, Ashman K, Zong X, Huber A, Neubauer G, et al. Regulation of Intracellular Calcium by a Signalling Complex of IRAG, IP3 Receptor and cGMP Kinase Ibeta. Nature (2000) 404:197–201. doi: 10.1038/35004606
24. Bong AHL, Monteith GR. Calcium Signaling and the Therapeutic Targeting of Cancer Cells. Biochim Biophys Acta Mol Cell Res (2018) 1865:1786–94. doi: 10.1016/j.bbamcr.2018.05.015
25. Ma L, Wang H, Sun Y, Yang D, Pu L, Zhang X. P53-Induced MRVI1 Mediates Carcinogenesis of Colorectal Cancer. Scand J Gastroenterol (2020) 55:824–33. doi: 10.1080/00365521.2020.1782465
26. Okamura R, Boichard A, Kato S, Sicklick JK, Bazhenova L, Kurzrock R. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis Oncol (2018) 2018. doi: 10.1200/PO.18.00183
27. Khotskaya YB, Holla VR, Farago AF, Mills Shaw KR, Meric-Bernstam F, Hong DS. Targeting TRK Family Proteins in Cancer. Pharmacol Ther (2017) 173:58–66. doi: 10.1016/j.pharmthera.2017.02.006
28. Jin W. Roles of TrkC Signaling in the Regulation of Tumorigenicity and Metastasis of Cancer. Cancers (Basel) (2020) 12. doi: 10.3390/cancers12010147
29. Bielack SS, Cox MC, Nathrath M, Apel K, Blattmann C, Holl T, et al. Rapid, Complete and Sustained Tumour Response to the TRK Inhibitor Larotrectinib in an Infant With Recurrent, Chemotherapy-Refractory Infantile Fibrosarcoma Carrying the Characteristic ETV6-NTRK3 Gene Fusion. Ann Oncol (2019) 30:viii31–5. doi: 10.1093/annonc/mdz382
30. Yeh I, Tee MK, Botton T, Shain AH, Sparatta AJ, Gagnon A, et al. NTRK3 Kinase Fusions in Spitz Tumours. J Pathol (2016) 240:282–90. doi: 10.1002/path.4775
31. Church AJ, Calicchio ML, Nardi V, Skalova A, Pinto A, Dillon DA, et al. Recurrent EML4-NTRK3 Fusions in Infantile Fibrosarcoma and Congenital Mesoblastic Nephroma Suggest a Revised Testing Strategy. Mod Pathol (2018) 31:463–73. doi: 10.1038/modpathol.2017.127
32. Shi E, Chmielecki J, Tang CM, Wang K, Heinrich MC, Kang G, et al. FGFR1 and NTRK3 Actionable Alterations in "Wild-Type" Gastrointestinal Stromal Tumors. J Transl Med (2016) 14:339. doi: 10.1186/s12967-016-1075-6
33. Yamamoto H, Yoshida A, Taguchi K, Kohashi K, Hatanaka Y, Yamashita A, et al. ALK, ROS1 and NTRK3 Gene Rearrangements in Inflammatory Myofibroblastic Tumours. Histopathology (2016) 69:72–83. doi: 10.1111/his.12910
34. Xu X, Tahan SR, Pasha TL, Zhang PJ. Expression of Neurotrophin Receptor Trk-C in Nevi and Melanomas. J Cutan Pathol (2003) 30:318–22. doi: 10.1034/j.1600-0560.2003.00068.x
35. Bouzas-Rodriguez J, Cabrera JR, Delloye-Bourgeois C, Ichim G, Delcros JG, Raquin MA, et al. Neurotrophin-3 Production Promotes Human Neuroblastoma Cell Survival by Inhibiting TrkC-Induced Apoptosis. J Clin Invest (2010) 120:850–8. doi: 10.1172/JCI41013
36. Luo Y, Kaz AM, Kanngurn S, Welsch P, Morris SM, Wang J, et al. NTRK3 is a Potential Tumor Suppressor Gene Commonly Inactivated by Epigenetic Mechanisms in Colorectal Cancer. PloS Genet (2013) 9:e1003552. doi: 10.1371/journal.pgen.1003552
37. Kim JY, Sutton ME, Lu DJ, Cho TA, Goumnerova LC, Goritchenko L, et al. Activation of Neurotrophin-3 Receptor TrkC Induces Apoptosis in Medulloblastomas. Cancer Res (1999) 59:711–9.
38. Hodgson A, Pun C, Djordjevic B, Turashvili G. NTRK-Rearranged Cervical Sarcoma: Expanding the Clinicopathologic Spectrum. Int J Gynecol Pathol (2021) 40:73–7. doi: 10.1097/PGP.0000000000000669
39. Guardado-Estrada M, Medina-Martinez I, Juarez-Torres E, Roman-Bassaure E, Macias L, Alfaro A, et al. The Amerindian mtDNA Haplogroup B2 Enhances the Risk of HPV for Cervical Cancer: De-Regulation of Mitochondrial Genes may be Involved. J Hum Genet (2012) 57:269–76. doi: 10.1038/jhg.2012.17
40. Espinosa AM, Alfaro A, Roman-Basaure E, Guardado-Estrada M, Palma I, Serralde C, et al. Mitosis is a Source of Potential Markers for Screening and Survival and Therapeutic Targets in Cervical Cancer. PloS One (2013) 8:e55975. doi: 10.1371/journal.pone.0055975
41. Medina-Martinez I, Barron V, Roman-Bassaure E, Juarez-Torres E, Guardado-Estrada M, Espinosa AM, et al. Impact of Gene Dosage on Gene Expression, Biological Processes and Survival in Cervical Cancer: A Genome-Wide Follow-Up Study. PloS One (2014) 9:e97842. doi: 10.1371/journal.pone.0097842
42. Anders S, Huber W. Differential Expression Analysis for Sequence Count Data. Genome Biol (2010) 11:R106. doi: 10.1186/gb-2010-11-10-r106
43. Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, et al. ChAMP: Updated Methylation Analysis Pipeline for Illumina BeadChips. Bioinformatics (2017) 33:3982–4. doi: 10.1093/bioinformatics/btx513
44. Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, et al. G:Profiler-A Web Server for Functional Interpretation of Gene Lists (2016 Update). Nucleic Acids Res (2016) 44:W83–9. doi: 10.1093/nar/gkw199
45. Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. TSGene 2.0: An Updated Literature-Based Knowledgebase for Tumor Suppressor Genes. Nucleic Acids Res (2016) 44:D1023–31. doi: 10.1093/nar/gkv1268
Keywords: MRVI1, NTRK3, cervical cancer, TSGs, DNA methylation
Citation: Ji H, Li K, Jiang W, Li J, Zhang J-a and Zhu X (2022) MRVI1 and NTRK3 Are Potential Tumor Suppressor Genes Commonly Inactivated by DNA Methylation in Cervical Cancer. Front. Oncol. 11:802068. doi: 10.3389/fonc.2021.802068
Received: 26 October 2021; Accepted: 24 December 2021;
Published: 24 January 2022.
Edited by:
Ram N. Ganapathi, Carolinas Healthcare System, United StatesReviewed by:
Chandraditya Chakraborty, Dana-Farber Cancer Institute and Harvard Medical School, United StatesStephanie M. McGregor, University of Wisconsin-Madison, United States
Copyright © 2022 Ji, Li, Jiang, Li, Zhang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueqiong Zhu, d3p6eHFAd3poZWFsdGguY29t
†These authors have contributed equally to this work
 Huihui Ji
Huihui Ji Kehan Li
Kehan Li Wenxiao Jiang
Wenxiao Jiang Jingwei Li
Jingwei Li Jian-an Zhang
Jian-an Zhang Xueqiong Zhu
Xueqiong Zhu