
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 17 January 2022
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.801319
This article is part of the Research Topic Non-coding RNAs in Gastrointestinal and Gynecological Cancers: New Insights into the Mechanisms of Cancer Therapeutic Resistance View all 22 articles
 Jinjin Chu1
Jinjin Chu1 Xianzhu Fang2
Xianzhu Fang2 Zhonghou Sun3
Zhonghou Sun3 Linlin Gai1
Linlin Gai1 Wenqing Dai1
Wenqing Dai1 Haibo Li1
Haibo Li1 Xinyi Yan1
Xinyi Yan1 Jinke Du1
Jinke Du1 Lili Zhang1
Lili Zhang1 Lu Zhao1
Lu Zhao1 Donghua Xu1*
Donghua Xu1* Shushan Yan4*
Shushan Yan4*Colorectal cancer (CRC) is the third prevalent cancer worldwide, the morbidity and mortality of which have been increasing in recent years. As molecular targeting agents, anti-epidermal growth factor receptor (EGFR) monoclonal antibodies (McAbs) have significantly increased the progression-free survival (PFS) and overall survival (OS) of metastatic CRC (mCRC) patients. Nevertheless, most patients are eventually resistant to anti-EGFR McAbs. With the intensive study of the mechanism of anti-EGFR drug resistance, a variety of biomarkers and pathways have been found to participate in CRC resistance to anti-EGFR therapy. More and more studies have implicated non-coding RNAs (ncRNAs) primarily including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), are widely involved in tumorigenesis and tumor progression. They function as essential regulators controlling the expression and function of oncogenes. Increasing data have shown ncRNAs affect the resistance of molecular targeted drugs in CRC including anti-EGFR McAbs. In this paper, we have reviewed the advance in mechanisms of ncRNAs in regulating anti-EGFR McAbs therapy resistance in CRC. It provides insight into exploring ncRNAs as new molecular targets and prognostic markers for CRC.
CRC is the third most frequent cancer worldwide. Global cancer statistics in 2020 has shown there are about 1.932 million new cases and 935,000 deaths of CRC worldwide, accounting for 10.0% of the total new cases of cancer and 9.4% of the total cancer-related deaths, respectively (1). Following lung cancer, CRC causes the second highest mortality in cancer patients worldwide (1). The therapeutic strategies for CRC mainly include surgery, chemotherapy, radiotherapy and targeted therapy. Currently, surgery and chemotherapy are still the preferred treatment options for CRC. Nevertheless, patients with metastatic CRC (mCRC) have a poor prognosis (2). The combined chemotherapy and molecular targeted drugs can noticeably increase the progression-free survival (PFS) and overall survival (OS) of mCRC patients (3). As molecular targeted drugs, cetuximab and panitumumab can directly target epidermal growth factor receptor (EGFR). Combined with chemotherapeutic drugs, they are applied to effectively treat mCRC patients carrying wild-type RAS and BRAF (4). Unfortunately, few patients with mCRC are sensitive to anti-EGFR treatment, and most responding patients usually develop resistance to the therapy (5). In recent years, a variety of biomarkers and pathways have been found to participate in regulating the resistance to anti-EGFR therapy, and thus affecting the therapeutic effect and reducing the survival rate of CRC patients (6). Some studies have suggested the potential resistance mechanisms in order to explore strategies for overcoming anti-EGFR resistance (5, 7, 8) (Figure 1).
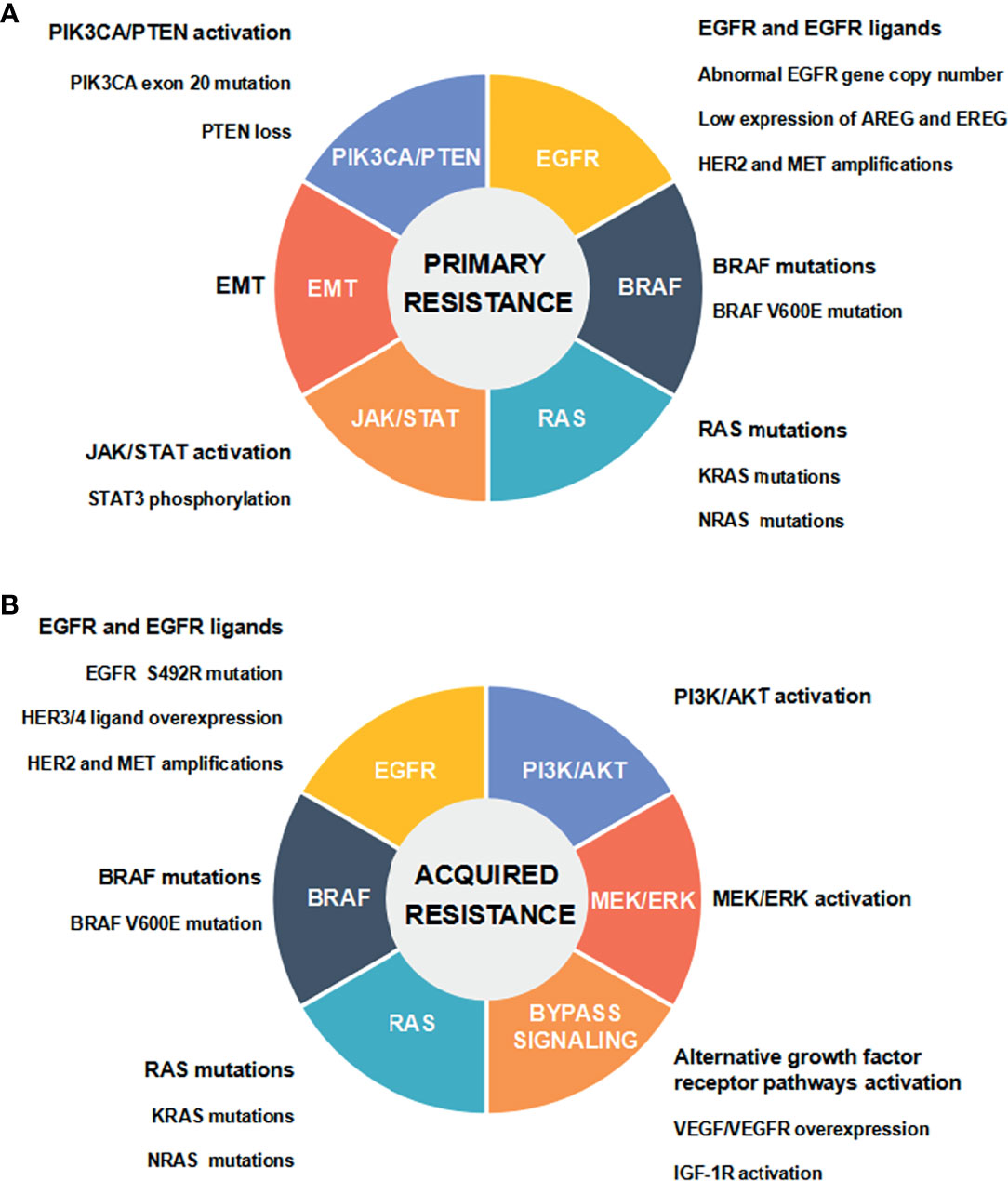
Figure 1 Mechanisms of anti-EGFR drug resistance in CRC. (A) Primary resistance mechanisms. (B) Acquired resistance mechanisms.
EGFR is a kind of HER tyrosine kinase receptor, which is composed of extracellular ligand binding domain, transmembrane hydrophobic domain, and intracellular tyrosine kinase domain. EGFR is selectively activated by binding to epidermal growth factor (EGF) as one of the major ligands. EGFR transmits signals from cytoplasm to nucleus through RAS/RAF/MEK/ERK/MAPK, PI3K/PTEN/AKT/mTOR, and some other intracellular signaling pathways which participate in regulating cancer cell proliferation, invasion, and angiogenesis (9). Abnormal expression and activation of any signal molecules mentioned above may lead to primary (de novo) or acquired (secondary) resistance to anti-EGFR therapy in mCRC (5). Abnormal EGFR gene copy number, protein expression of EGFR ligands, HER2 and MET gene amplifications, and activation of EGFR downstream cascade signaling pathways [including the mutations of RAS/BRAF/PIK3CA, the loss of PTEN, STAT3 phosphorylation, and epithelial-mesenchymal transition (EMT)], have been demonstrated to be associated with the primary resistance to anti-EGFR therapy in CRC (5, 7, 8). It has been well documented that the acquired resistance is attributed to EGFR ectodomain mutation (S492R), genetic alterations in RAS/RAF and other downstream signaling molecules, and the activation of intracellular signaling pathways that are bypassing EGFR and mediated by IGF1R, HER2, MET, and VEGFR (5, 7, 8). Multiple genetic and nongenetic mechanisms drive resistance to anti-EGFR therapy in CRC, with a significant overlap in primary and acquired resistance (8) (Figure 1).
NcRNAs are a type of RNAs which have no protein-coding function. According to the length, they are divided into two classes: small non-coding RNAs (sncRNAs) with a length of 18-200 nt, and long noncoding RNAs (lncRNAs) with a length over 200 nt. NcRNAs are widely involved in cell proliferation, apoptosis, autophagy, EMT, and cell cycle progression (10–14). Accumulated studies have suggested ncRNAs play important roles in tumorigenesis, progression, and anti-EGFR monoclonal antibodies (McAbs) treatment resistance in CRC (15–21). In this review, we have focused on current progress in the underlying molecular mechanisms of ncRNAs in regulating the resistance to anti-EGFR therapy in CRC. We aim to fully explore the potentials of ncRNAs as novel molecular targets and prognostic markers for CRC.
MiRNAs are single-stranded small ncRNAs with a length of 21-25 nt. The synthesis of miRNAs involves multiple biological steps. Firstly, primary miRNAs (pri-miRNAs) are encoded by DNA in the nucleus and transcribed by ribonucleic acid polymerase II. Secondly, long pri-miRNAs are processed by ribonuclease III Drosha, which produces precursor miRNAs (pre-miRNAs) with a length of 60-70 nt. Lastly, pre-miRNAs are cleaved into mature double-stranded miRNAs by ribonuclease III Dicer in the cytoplasm. Then, mature miRNAs participate in forming RNA-induced silencing complex (RISC) (22). MiRNAs induce messenger RNA (mRNA) degradation and translation repression by directly binding to the 3’-untranslated region (3’-UTR) of targeted mRNAs, and act as regulators at the post-transcriptional level during gene expression process (23). They are widely involved in cell proliferation, apoptosis, autophagy, and immune response (10, 11, 18, 21). Accumulated studies have suggested miRNAs participate in the pathogenesis of various diseases including cancers (24–28). MiRNAs act either as oncogenic miRNAs (onco-miRs) or tumor suppressive miRNAs (TS-miRs) with significant tissue- and organ-specificity (29, 30). Many studies have also found that miRNAs participate in regulating the drug resistance in CRC (31, 32). It has been demonstrated miR-31 negatively regulates breast cancer invasion and metastasis (33). However, it negatively regulates the expression of tumor suppressors and thus exerts oncogenic effects in lung cancer (34). In CRC, miR-31 has been documented to promote cancer progression by activating RAS signaling pathway and hypoxia inducible factor 1α (HIF-1α), respectively (35, 36). Taken together, miR-31 is involved in tumor progression and metastasis by serving as a TS-miR or an onco-miR in different malignancies. The diverse roles of miR-31 in cancer may be attributed to different types of cancer cells, specific targets, and other complicated factors. Further research is required to reveal its specific functions in CRC.
MiRNAs, aberrantly expressed in tumor tissues and tumor cells, exert their tumor suppressive- and oncogenic-functions by regulating different targeted genes (Figure 2). When the expression levels of TS-miRs decrease, negative regulation on targeted genes weakens. Besides, increasing expression levels of onco-miRs promotes tumor development, metastasis, and drug resistance through down-regulating tumor suppressive genes.
MiRNAs regulates anti-EGFR drug resistance by directly targeting tumor-related genes involved in EGFR-related signaling pathways in CRC. Abnormal expression of miRNAs is commonly observed in anti-EGFR treatment-resistant CRC cells. Recent studies have shown that miRNAs may predict the prognosis and drug therapeutic efficacy of CRC patients (37–39). The latest studies regarding miRNAs and drug resistance to anti-EGFR therapy in CRC have been described in the following subsections and briefly summarized in Table 1.
EGFR signaling pathway has been confirmed to be aberrantly activated in multiple malignant tumors, which is associated with tumor progression and prognosis. Increasing evidence has implicated miRNAs participate in regulating EGFR signaling pathway and play vital roles in anti-EGFR drug resistance in CRC (Figure 2). Zhou et al. have found miR-133b regulated cell proliferation and invasion in CRC by targeting EGFR (40). Moreover, the combination of miR-133b mimics and cetuximab can effectively suppress the proliferation and invasion of cetuximab-resistant CRC cells (40). Suto et al. have found miR-7 is involved in regulating the EGFR signaling pathway by down-regulating the expression of EGFR and RAF-1, which could inhibit CRC cells proliferation and reverse cetuximab resistance in CRC patients with mutant KRAS (41). Sun and the colleagues have found that miR-302a suppressed CRC metastasis by targeting nuclear factor I B (NFIB) and CD44 and decreasing the activation of NFIB/ITGA6 signaling pathway (42). MiR-302a has also been found to restore the response to cetuximab by inhibiting CD44-induced cancer stem cell (CSC)-like characteristics through EGFR-mediated MAPK and protein kinase B (AKT) signaling pathways (42). These studies have revealed that miRNAs can directly target EGFR (or RAF) in CRC cells, inhibit the activation of its downstream signaling pathways, and thus repress CRC cells growth and invasion. Besides, miR-100 and miR-125b have been found to cooperatively regulate the resistance to cetuximab in CRC through Wnt signaling pathway that has a cross-talk with EGFR pathway (50). MiRNAs are extensively involved in regulating the resistance to cetuximab. Accordingly, miRNAs might serve as markers for predicting anti-EGFR therapy in mCRC patients due to their regulatory effects on EGFR signaling pathway.
KRAS, a member of RAS family, has almost 40% mutation rate in CRC patients. KRAS mutations are predictive biomarkers for the treatment efficacy of anti-EGFR treatment and the outcome of patients with CRC (53). MiRNAs have been widely reported to regulate the therapeutic response and drug sensitivity of CRC patients through KRAS signaling pathway (43–45) (Figure 2).
Synthetic miR-143 (miR-143#12) inhibits KRAS signaling pathway activation and restores the sensitivity of cetuximab-resistant CRC cells by targeting the KRAS activating protein SOS1 (43). Overexpression of miR-143 or miR-145 can increase the sensitivity to cetuximab by enhancing cetuximab-mediated antibody-dependent cellular cytotoxicity (ADCC) in CRC cells (44). Strippoli et al. have demonstrated miR-31-3p, miR-143 and miR-145 are closely correlated with anti-EGFR treatment resistance in mCRC via regulating RAS-MAPK axis and c-MYC pathway (45). Moreover, miR-143 and miR-145 have been well established to exert tumor-suppressive effects and are beneficial for the efficacy of anti-EGFR treatment in CRC, whereas miR-31-3p comes to the opposite. It has been shown that the overexpression of miR-193a-3p can promote BRAF-mutant CRC cells apoptosis by inhibiting the expression of KRAS and myeloid cell leukemia-1 (Mcl1) through MAPK signal (47). As a tumor suppressor, miR-193a-3p promotes the efficacy of BRAF inhibitor dabrafenib (DAB) and MEK inhibitor trametinib (TRA), and enhances the anti-proliferative effect of combined therapy of DAB, TRA with cetuximab in CRC (47). A recent study has shown that 4-acetyl-antroquinonol B (4-AAQB) inhibits CRC cell proliferation and induces cell apoptosis by up-regulating miR-193a-3p, down-regulating KRAS and inhibiting the activation of KRAS signaling pathway. The combined treatment of 4-AAQB with cetuximab can make KRAS-mutant CRC cells resensitized to cetuximab (46). In addition to KRAS, miR-193a-3p acts on multiple signaling pathways and plays a tumor-suppressive role by regulating the expression of interleukin 17 receptor D (IL17RD) and erb-b2 receptor tyrosine kinase 4 (ERBB4) in CRC (54, 55). And lower expression of miR-193a-3p in CRC tissues predicts poorer PFS independently of the status of BRAF mutation (56). Accordingly, miR-193a-3p may serve as a prognostic biomarker. Its combination with molecular targeted drugs may be a novel therapeutic strategy for BRAF-mutant CRC. Weng et al. have reported that lauric acid can induce miR-378 expression and increase the sensitivity of BRAF- and KRAS-mutant CRC cells to cetuximab by inhibiting KRAS, BRAF, MEK, ERK1/2 protein expressions through the MAPK signaling pathway (48). In addition, they have found that eicosapentaenoic acid ethyl ester (EPA) can also increase the expression of miR-378 in BRAF- and KRAS-mutant CRC cells and resensitize KRAS-mutant CRC cells to cetuximab (49). Taken together, miRNAs play vital roles in regulating the therapeutic response and drug sensitivity of KRAS- or BRAF-mutant CRC through RAS signaling pathway. Potential miRNAs and key molecules in the RAS signaling pathway may serve as promising biomarkers for predicting the efficacy and drug resistance during the targeted therapy in CRC.
The PI3K/AKT signaling pathway is widely involved in carcinogenesis and cancer progression. Aberrant activation of PI3K-AKT can promote CRC invasion and metastasis (57). It has been reported that miRNAs can directly target the PI3K/AKT signaling molecules or signaling pathway regulators, including numerous regulatory proteins (51, 52, 57–60) (Figure 2). MiR-375 and miR-199a-5p promote cetuximab resistance in CRC patients by repressing the expression of PH domain and leucine-rich repeat protein phosphatase 1 (PHLPP1) and positively regulating AKT signaling pathway (51). Nevertheless, some other studies have found miR-375 and miR-199a-5p inhibit CRC cells proliferation and invasion, suggesting their complicated functions in CRC (52, 58–60). It has been well documented that miR-375 suppressed CRC cell proliferation by targeting PIK3CA via the PI3K/AKT pathway (61), while miR-199a-5p inhibited CRC cell survival, proliferation, migration, and invasion by downregulating GCNT2 expression and inhibiting the AKT and ERK signal activation (62). Different roles of miR-375 and miR-199a-5p exerting in CRC, might be attributed to significant tumor heterogeneity among patients. Taken together, miRNAs regulate the progression and drug resistance of CRC by regulating tumor suppressors or oncogenes involved in various signaling pathways including PI3K/AKT signal. However, the precise mechanisms of miR-375 and miR-199a-5p underlying the resistance to anti-EGFR drugs in CRC warrant to be fully elucidated in more future research.
Tumor immune microenvironment is composed of a variety of cells, extracellular matrix and various signaling molecules (63). Imbalance of tumor immune microenvironment is essential for tumor growth, metastasis and prognosis (64). MiRNA-mediated regulation of tumor microenvironment (TME) has been demonstrated to affect cancer growth, angiogenesis, metastasis, and drug resistance exerting either antitumor or tumorigenic effects (65). For instance, a recent study has shown miR-34a promoted the expression of B7-H3 and TNF-α in tumor microenvironment and negatively regulated T cell-mediated immune response, which thus induced immunosuppression and immune escape in CRC (66). MiR-148a-3p and miR-448 respectively down-regulate the expression of calnexin (CANX) and indoleamine 2,3-dioxygenase 1 (IDO1), which enhances CD8+ T cell-mediated immune response in CRC (67, 68).
Cetuximab can induce ADCC by binding to EGFR on cancer cells and CD16 receptor on natural killer (NK) cells and dendritic cells (DCs) (69–71). It stimulates the production of proinflammatory cytokines, such as IFN-γ and TNF-α, and activates cytotoxic T cells in the TME, thereby exerting tumor immunosuppressive effects (69–71). It has been suggested that anti-EGFR therapy and immunotherapy have synergetic and complementary mechanisms. The combination of immune checkpoint inhibitors, chemotherapy with anti-EGFR McAbs in mCRC has shown an encouraging clinical outcome (72). Nevertheless, littler is known about the role of miRNAs in regulating tumor immune microenvironment and thus affecting anti-EGFR drugs resistance in CRC.
LncRNAs are a type of ncRNAs over 200 nt in length. They are mainly formed by RNA polymerase II-catalyzed transcription typically containing a cleavable 3’ poly-A tail (73). According to genomic localization, lncRNAs are grouped into five classes: sense lncRNA, antisense lncRNA, intronic lncRNA, bidirectional lncRNA, and intergenic lncRNA (74). LncRNAs have low sequence conservation and high tissue and organ specificity. As competitive endogenous RNAs (ceRNAs), lncRNAs can directly sponge miRNAs and inhibit their expression. LncRNAs interact with DNA, RNA and protein, acting as regulators of gene expression at multiple levels and play roles in various cell processes, such as genomic imprinting, epigenetic regulation, transcriptional regulation, chromosome conformation, and cell cycle regulation (75). A great deal of data has suggested lncRNAs participate in the pathogenesis of various diseases, including cancer (75–78). Linc00152, SNHG1, SCARNA2, DLEU1 and XIST contribute to colorectal carcinogenesis, metastasis and prognosis of CRC (54, 79–83). In addition, a number of studies have implicated lncRNAs lead to associated with primary or acquired drug resistance in CRC, thereby reducing drug efficacy (84, 85). Nonetheless, the regulatory mechanisms of lncRNAs underlying anti-EGFR therapy resistance in CRC are not clear yet.
Increasing evidence has supported that lncRNAs participate in regulating CRC resistance to anti-EGFR McAbs through multiple signaling pathways (Figure 3). The study by Peng et al. has found that down-regulation of POU5F1P4 in cetuximab-sensitive CRC cells can reduce their sensitivity in mCRC (86). LNC00973 and several other lncRNAs may be involved in cetuximab resistance by regulating glucose metabolism (87). Down-regulation of LNC00973 can improve cetuximab resistance in drug-resistant CRC cells (87). Lu et al. have elaborated that the overexpression of lncRNA MIR100HG-derived miR-100 and miR-125b promotes cetuximab resistance through Wnt/β-catenin pathway in CRC (50) (Tables 1, 2). Recent studies have reported that lncRNA CRART16 is up-regulated in CRC cells with secondary cetuximab-resistance. CRART16 contributes to cetuximab resistance in CRC by up-regulating ERBB3 through miR-371a-5p/MAPK signaling pathway (88). LncRNA HCG18 promotes cell proliferation, migration, and cetuximab resistance in CRC by up-regulating PD-L1 and down-regulating CD8+ T lymphocytes via sponging miR-20b-5p (89). Besides, the study by Yang et al. has shown the evidence that up-regulation of UCA1 in cetuximab-resistant CRC cells and the produced exosomes (90). Moreover, exosomal UCA1 is observed to cause drug resistance in cetuximab-sensitive CRC cells (90). Due to its non-invasive and relatively stable content in serum, exosomal UCA1 is hopefully used as a new biomarker for CRC in the future (Table 2).
Accumulated studies have suggested lncRNAs have been elucidated to serve as ceRNAs by sponging miRNAs, which subsequently regulates miRNAs-mediated anti-EGFR therapy resistance in CRC. In addition, lncRNAs play vital roles in CRC progression, metastasis, and drug resistance. These findings provide therapeutic targets and potential prognostic markers for CRC with regard to lncRNAs. Future studies are warranted to reveal the specific mechanism of lncRNAs involved in CRC progression, metastasis, and drug resistance.
CircRNAs are novel covalently closed circular single-stranded ncRNAs discovered in recent years, mainly formed by exon reverse splicing of pre-mRNA. According to the sequence origin, circRNAs are grouped into exonic circRNAs, circular intronic RNAs, and exon-intron circRNAs (91). They exist stably in plasma, serum, saliva, and other body fluids, and are widely expressed in various types of cells with cell- and tissue-specificity (92, 93). Acting as ceRNAs, circRNAs can competitively bind with miRNAs and regulate gene expression via interacting with miRNAs or RNA-binding proteins (RBPs). They exert essential effects on the progression of multiple diseases including cancer (17, 94–99).
Increasing evidence has supported that circRNAs participate in regulating tumorigenesis and drug resistance of CRC (100, 101). Chen et al. have found that circ-PRKDC acted as a miR-375 sponge and targeted FOXM1, and enhanced CRC cells resistance to 5-fluorouracil (5-FU) through the Wnt/β-catenin signaling pathway (102) (Table 3, Figure 4). CircRNAs of circ_0007031, circ_0007006, and circ_0000504 have been found to modulate 5-FU resistance of CRC cells by regulating AKT3 via the AKT signaling pathway, while circ_0048234 can sponge miR-671-5p in 5-FU-resistant CRC cells via the EGFR signaling pathway (103) (Table 3, Figure 4). ATP-binding cassette (ABC) transporters, such as ABCB1, ABCC1, and ABCG2, have been reported to play crucial roles in CRC drug resistance by increasing drug efflux out of cancer cells (105). Inhibition expression of ABC transporters is an effective approach to reverse drug resistance in cancer cells (105, 106). A number of ncRNAs have been demonstrated to be involved in regulating ABC transporters in drug-resistant cancer cells by regulating EGFR and its downstream signaling pathways (107, 108). Circ_0007031 has been documented to induce 5-FU resistance by modulating the expression of ABC transporter ABCC5 through miR-133b/ABCC5 axis in CRC (100). MiR-7 functions as a regulator of anti-EGFR therapy resistance in CRC. It has been shown that ciRS-7 regulated CRC cell growth and invasion by sponging miR-7 and upregulating EGFR and IGF-1R expression (109). Similarly, CiRS-7 can function as ceRNA for miR-7 to activate EGFR/RAF1/MAPK pathway in CRC (110). The study by Zeng et al. has reported circHIPK3 sponged miR-7 to upregulate the expression of several oncogenes, such as FAK, IGF1R, EGFR, and YY1, through the PI3K/AKT and MEK/ERK signaling pathways that contributing to drug resistance in CRC (104). Additionally, inhibition of circHIPK3 can reverse the resistance to cetuximab by targeting miR-7 in CRC cells (104) (Figure 4). All these findings have provided novel insights into the understanding of drug resistance mechanisms regarding circRNAs. Nevertheless, more studies are warranted to estimating the involvement and mechanism of circRNAs in regulating the resistance to anti-EGFR therapy in CRC.
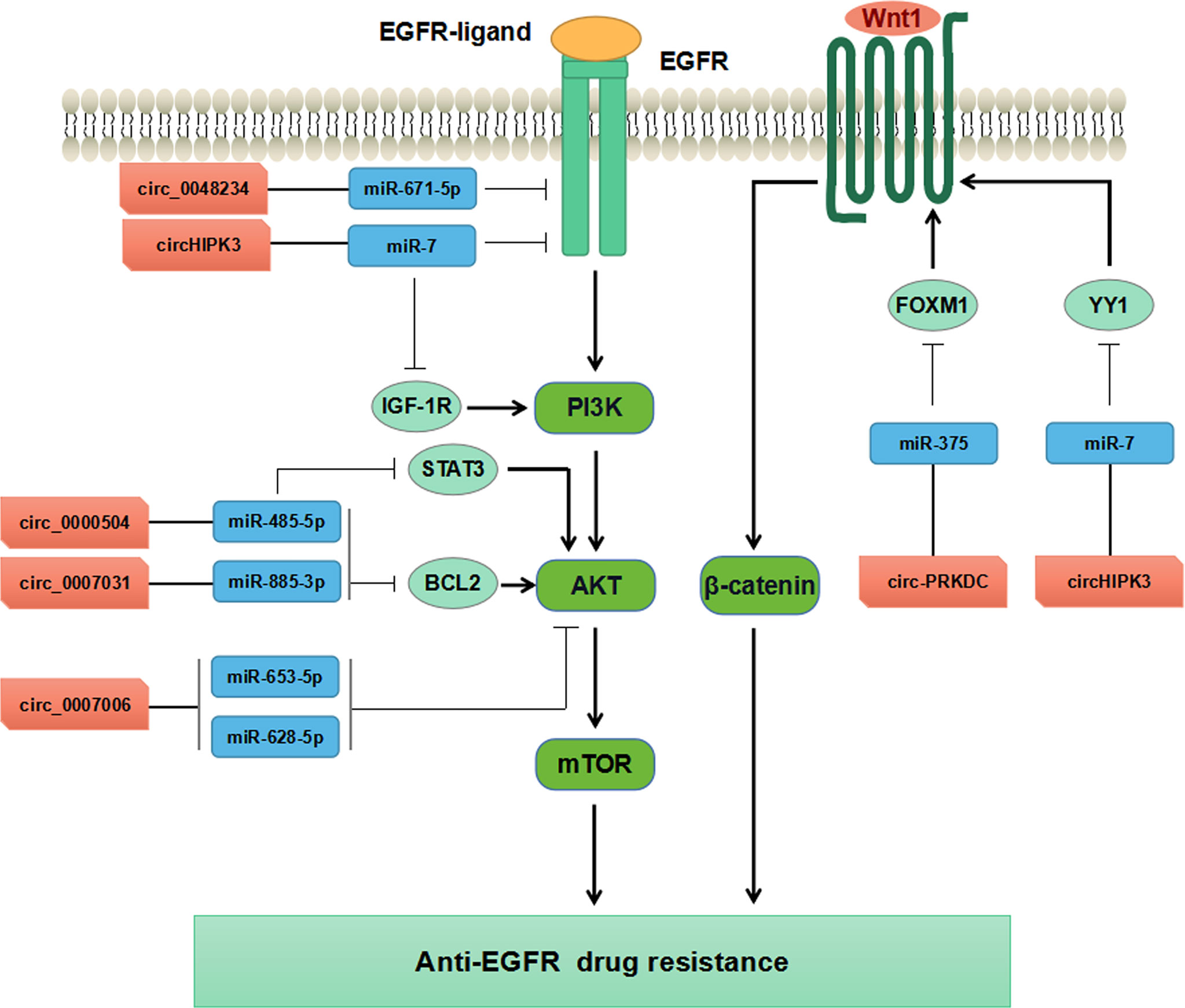
Figure 4 Mechanisms of circRNAs regulating CRC drug resistance by modulating AKT, EGFR, and WNT pathways.
Drug resistance remains a major challenge for CRC treatment. The mechanisms underlying CRC resistance to anti-EGFR therapy are complicated. Increasing studies have shown that ncRNAs play crucial roles in regulating the resistance to anti-EGFR therapy in CRC, primarily including miRNAs, lncRNAs and circRNAs, which have been identified as either oncogenes or tumor suppressors (111). Currently available studies have supported ncRNAs participate in modulating anti-EGFR drug resistance based on miRNAs-mRNAs, lncRNAs-miRNAs-mRNAs, or circRNAs-miRNAs-mRNAs regulatory networks through the EGFR signaling pathway, RAS signaling pathway, and PI3K/AKT signaling pathway. Accordingly, ncRNAs may function as novel biomarkers in predicting the efficacy and resistance of anti-EGFR therapy in CRC. Nevertheless, the molecular mechanisms of ncRNAs involved in anti-EGFR therapy resistance still warrant to be further elucidated in CRC. Further studies need to focus on investigating new therapeutic strategies based on ncRNAs regulatory networks combing with anti-EGFR targeted therapy in CRC.
SY, DX, and JC wrote the draft and revised it. JC, XF, ZS, LG, WD, HL, XY, JD, LLZ, and LZ collected the data and designed the tables and figures. All authors read and approved the submitted version.
This work is supported by funds from the National Natural Science Foundation, China (82003042, 81870237, 82171790, 32000075) and Natural Science Foundation, Shandong Province (ZR2020KC001, ZR2019QH012, ZR2020QC012).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Wang H. MicroRNAs and Apoptosis in Colorectal Cancer. Int J Mol Sci (2020) 21:5353. doi: 10.3390/ijms21155353
3. Pei X, Liu Y, Sun L, Zhang J, Fang Y, Liao X, et al. Outcome of Molecular Targeted Agents Plus Chemotherapy for Second-Line Therapy of Metastatic Colorectal Cancer: A Meta-Analysis of Randomized Trials. Clin Colorectal Cancer (2016) 15:e149–e56. doi: 10.1016/j.clcc.2016.03.005
4. Fakih MG. Metastatic Colorectal Cancer: Current State and Future Directions. J Clin Oncol (2015) 33:1809–24. doi: 10.1200/JCO.2014.59.7633
5. Zhao B, Wang L, Qiu H, Zhang M, Sun L, Peng P, et al. Mechanisms of Resistance to Anti-EGFR Therapy in Colorectal Cancer. Oncotarget (2017) 8:3980–4000. doi: 10.18632/oncotarget.14012
6. Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-Type KRAS is Required for Panitumumab Efficacy in Patients With Metastatic Colorectal Cancer. J Clin Oncol (2008) 26:1626–34. doi: 10.1200/JCO.2007.14.7116
7. Schirripa M, Lenz HJ. Colorectal Cancer: Overcoming Resistance to Anti-EGFR Therapy - Where do We Stand? Nat Rev Gastroenterol Hepatol (2016) 13:258–9. doi: 10.1038/nrgastro.2016.52
8. Dienstmann R, Salazar R, Tabernero J. Overcoming Resistance to Anti-EGFR Therapy in Colorectal Cancer. Am Soc Clin Oncol Educ Book (2015) 35:e149–56. doi: 10.14694/EdBook_AM.2015.35.e149
9. Bianco R, Gelardi T, Damiano V, Ciardiello F, Tortora G. Rational Bases for the Development of EGFR Inhibitors for Cancer Treatment. Int J Biochem Cell Biol (2007) 39:1416–31. doi: 10.1016/j.biocel.2007.05.008
10. Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, et al. Autophagy Suppresses Tumorigenesis of Hepatitis B Virus-Associated Hepatocellular Carcinoma Through Degradation of microRNA-224. Hepatol (2014) 59:505–17. doi: 10.1002/hep.26659
11. Zhou K, Liu M, Cao Y. New Insight Into microRNA Functions in Cancer: Oncogene-microRNA-Tumor Suppressor Gene Network. Front Mol Biosci (2017) 4:46. doi: 10.3389/fmolb.2017.00046
12. Yue QY, Zhang Y. Effects of Linc00460 on Cell Migration and Invasion Through Regulating Epithelial-Mesenchymal Transition (EMT) in non-Small Cell Lung Cancer. Eur Rev Med Pharmacol Sci (2018) 22:1003–10.
13. Song TF, Huang LW, Yuan Y, Wang HQ, He HP, Ma WJ, et al. LncRNA MALAT1 Regulates Smooth Muscle Cell Phenotype Switch via Activation of Autophagy. Oncotarget (2018) 9:4411–26. doi: 10.18632/oncotarget.23230
14. Li YF, Pei FL, Cao MZ. CircRNA_101951 Promotes Migration and Invasion of Colorectal Cancer Cells by Regulating the KIF3A-Mediated EMT Pathway. Exp Ther Med (2020) 19:3355–61. doi: 10.3892/etm.2020.8600
15. Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX, Yang L. Circular RNA circVAPA is Up-Regulated and Exerts Oncogenic Properties by Sponging miR-101 in Colorectal Cancer. BioMed Pharmacother (2019) 112:108611. doi: 10.1016/j.biopha.2019.108611
16. Shabaninejad Z, Vafadar A, Movahedpour A, Ghasemi Y, Namdar A, Fathizadeh H, et al. Circular RNAs in Cancer: New Insights Into Functions and Implications in Ovarian Cancer. J Ovarian Res (2019) 12:84. doi: 10.1186/s13048-019-0558-5
17. Zeng K, Wang S. Circular RNAs: The Crucial Regulatory Molecules in Colorectal Cancer. Pathol Res Pract (2020) 216:152861. doi: 10.1016/j.prp.2020.152861
18. Slattery ML, Mullany LE, Sakoda LC, Wolff RK, Samowitz WS, Herrick JS. Dysregulated Genes and miRNAs in the Apoptosis Pathway in Colorectal Cancer Patients. Apoptosis (2018) 23:237–50. doi: 10.1007/s10495-018-1451-1
19. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
20. Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong H, et al. Long non-Coding RNAs in Colorectal Cancer. Oncotarget (2016) 7:5226–39. doi: 10.18632/oncotarget.6446
21. Chen W, Cai G, Liao Z, Lin K, Li G, Li Y. miRNA-766 Induces Apoptosis of Human Colon Cancer Cells Through the P53/Bax Signaling Pathway by MDM4. Exp Ther Med (2019) 17:4100–8. doi: 10.3892/etm.2019.7436
22. O'brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) (2018) 9:402. doi: 10.3389/fendo.2018.00402
23. Ha M, Kim VN. Regulation of microRNA Biogenesis. Nat Rev Mol Cell Biol (2014) 15:509–24. doi: 10.1038/nrm3838
24. Rizzuti M, Filosa G, Melzi V, Calandriello L, Dioni L, Bollati V, et al. MicroRNA Expression Analysis Identifies a Subset of Downregulated miRNAs in ALS Motor Neuron Progenitors. Sci Rep (2018) 8:10105. doi: 10.1038/s41598-018-28366-1
25. Rupaimoole R, Slack FJ. MicroRNA Therapeutics: Towards a New Era for the Management of Cancer and Other Diseases. Nat Rev Drug Discov (2017) 16:203–22. doi: 10.1038/nrd.2016.246
26. Liu SQ, Jiang S, Li C, Zhang B, Li QJ. miR-17-92 Cluster Targets Phosphatase and Tensin Homology and Ikaros Family Zinc Finger 4 to Promote TH17-Mediated Inflammation. J Biol Chem (2014) 289:12446–56. doi: 10.1074/jbc.M114.550723
27. Dragomir MP, Kopetz S, Ajani JA, Calin GA. Non-Coding RNAs in GI Cancers: From Cancer Hallmarks to Clinical Utility. Gut (2020) 69:748–63. doi: 10.1136/gutjnl-2019-318279
28. Goh SY, Chao YX, Dheen ST, Tan E-K, Tay SS-W. Role of MicroRNAs in Parkinson’s Disease. Int J Mol Sci (2019) 20:5649. doi: 10.3390/ijms20225649
29. Mayr C, Beyreis M, Wagner A, Pichler M, Neureiter D, Kiesslich T. Deregulated MicroRNAs in Biliary Tract Cancer: Functional Targets and Potential Biomarkers. BioMed Res Int (2016) 2016:4805270. doi: 10.1155/2016/4805270
30. Wen D, Danquah M, Chaudhary AK, Mahato RI. Small Molecules Targeting microRNA for Cancer Therapy: Promises and Obstacles. J Control Release (2015) 219:237–47. doi: 10.1016/j.jconrel.2015.08.011
31. Fanini F, Fabbri M. MicroRNAs and Cancer Resistance: A New Molecular Plot. Clin Pharmacol Ther (2016) 99:485–93. doi: 10.1002/cpt.353
32. Maleki M, Golchin A, Javadi S, Khelghati N, Morovat P, Asemi Z, et al. Role of Exosomal miRNA in Chemotherapy Resistance of Colorectal Cancer: A Systematic Review. Chem Biol Drug Des (2021). doi: 10.1111/cbdd.13947
33. Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, et al. A Pleiotropically Acting microRNA, miR-31, Inhibits Breast Cancer Metastasis. Cell (2009) 137:1032–46. doi: 10.1016/j.cell.2009.03.047
34. Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, et al. MicroRNA-31 Functions as an Oncogenic microRNA in Mouse and Human Lung Cancer Cells by Repressing Specific Tumor Suppressors. J Clin Invest (2010) 120:1298–309. doi: 10.1172/JCI39566
35. Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, et al. MicroRNA-31 Activates the RAS Pathway and Functions as an Oncogenic MicroRNA in Human Colorectal Cancer by Repressing RAS P21 GTPase Activating Protein 1 (RASA1). J Biol Chem (2013) 288:9508–18. doi: 10.1074/jbc.M112.367763
36. Chen T, Yao LQ, Shi Q, Ren Z, Ye LC, Xu JM, et al. MicroRNA-31 Contributes to Colorectal Cancer Development by Targeting Factor Inhibiting HIF-1alpha (FIH-1). Cancer Biol Ther (2014) 15:516–23. doi: 10.4161/cbt.28017
37. Sur D, Cainap C, Burz C, Havasi A, Chis IC, Vlad C, et al. The Role of miRNA -31-3p and miR-31-5p in the Anti-EGFR Treatment Efficacy of Wild-Type K-RAS Metastatic Colorectal Cancer. Is it Really the Next Best Thing in miRNAs? J BUON (2019) 24:1739–46.
38. Mekenkamp LJ, Tol J, Dijkstra JR, De Krijger I, Vink-Borger ME, Van Vliet S, et al. Beyond KRAS Mutation Status: Influence of KRAS Copy Number Status and microRNAs on Clinical Outcome to Cetuximab in Metastatic Colorectal Cancer Patients. BMC Cancer (2012) 12:292. doi: 10.1186/1471-2407-12-292
39. Igarashi H, Kurihara H, Mitsuhashi K, Ito M, Okuda H, Kanno S, et al. Association of MicroRNA-31-5p With Clinical Efficacy of Anti-EGFR Therapy in Patients With Metastatic Colorectal Cancer. Ann Surg Oncol (2015) 22:2640–8. doi: 10.1245/s10434-014-4264-7
40. Zhou J, Lv L, Lin C, Hu G, Guo Y, Wu M, et al. Combinational Treatment With microRNA−133b and Cetuximab has Increased Inhibitory Effects on the Growth and Invasion of Colorectal Cancer Cells by Regulating EGFR. Mol Med Rep (2015) 12:5407–14. doi: 10.3892/mmr.2015.4046
41. Suto T, Yokobori T, Yajima R, Morita H, Fujii T, Yamaguchi S, et al. MicroRNA-7 Expression in Colorectal Cancer is Associated With Poor Prognosis and Regulates Cetuximab Sensitivity via EGFR Regulation. Carcinogenesis (2015) 36:338–45. doi: 10.1093/carcin/bgu242
42. Sun L, Fang Y, Wang X, Han Y, Du F, Li C, et al. miR-302a Inhibits Metastasis and Cetuximab Resistance in Colorectal Cancer by Targeting NFIB and CD44. Theranostics (2019) 9:8409–25. doi: 10.7150/thno.36605
43. Akao Y, Kumazaki M, Shinohara H, Sugito N, Kuranaga Y, Tsujino T, et al. Impairment of K-Ras Signaling Networks and Increased Efficacy of Epidermal Growth Factor Receptor Inhibitors by a Novel Synthetic miR-143. Cancer Sci (2018) 109:1455–67. doi: 10.1111/cas.13559
44. Gomes SE, Simoes AE, Pereira DM, Castro RE, Rodrigues CM, Borralho PM. miR-143 or miR-145 Overexpression Increases Cetuximab-Mediated Antibody-Dependent Cellular Cytotoxicity in Human Colon Cancer Cells. Oncotarget (2016) 7:9368–87. doi: 10.18632/oncotarget.7010
45. Strippoli A, Cocomazzi A, Basso M, Cenci T, Ricci R, Pierconti F, et al. C-MYC Expression Is a Possible Keystone in the Colorectal Cancer Resistance to EGFR Inhibitors. Cancers (Basel) (2020) 12:638. doi: 10.3390/cancers12030638
46. Chu YC, Tsai TY, Yadav VK, Deng L, Huang CC, Tzeng YM, et al. 4-Acetyl-Antroquinonol B Improves the Sensitization of Cetuximab on Both Kras Mutant and Wild Type Colorectal Cancer by Modulating the Expression of Ras/Raf/miR-193a-3p Signaling Axis. Int J Mol Sci (2021) 22:7508. doi: 10.3390/ijms22147508
47. Hiraide S, Takahashi M, Yoshida Y, Yamada H, Komine K, Ishioka C. Tumor Suppressor miR-193a-3p Enhances Efficacy of BRAF/MEK Inhibitors in BRAF-Mutated Colorectal Cancer. Cancer Sci (2021) 112:3856–70. doi: 10.1111/cas.15075
48. Weng WH, Leung WH, Pang YJ, Hsu HH. Lauric Acid can Improve the Sensitization of Cetuximab in KRAS/BRAF Mutated Colorectal Cancer Cells by Retrievable microRNA-378 Expression. Oncol Rep (2016) 35:107–16. doi: 10.3892/or.2015.4336
49. Weng WH, Leung WH, Pang YJ, Kuo LW, Hsu HH. EPA Significantly Improves Anti-EGFR Targeted Therapy by Regulating miR-378 Expression in Colorectal Cancer. Oncol Lett (2018) 16:6188–94. doi: 10.3892/ol.2018.9408
50. Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, et al. lncRNA MIR100HG-Derived miR-100 and miR-125b Mediate Cetuximab Resistance via Wnt/beta-Catenin Signaling. Nat Med (2017) 23:1331–41. doi: 10.1038/nm.4424
51. Mussnich P, Rosa R, Bianco R, Fusco A, D'angelo D. MiR-199a-5p and miR-375 Affect Colon Cancer Cell Sensitivity to Cetuximab by Targeting PHLPP1. Expert Opin Ther Targets (2015) 19:1017–26. doi: 10.1517/14728222.2015.1057569
52. Alam KJ, Mo JS, Han SH, Park WC, Kim HS, Yun KJ, et al. MicroRNA 375 Regulates Proliferation and Migration of Colon Cancer Cells by Suppressing the CTGF-EGFR Signaling Pathway. Int J Cancer (2017) 141:1614–29. doi: 10.1002/ijc.30861
53. Angius A, Pira G, Scanu AM, Uva P, Sotgiu G, Saderi L, et al. MicroRNA-425-5p Expression Affects BRAF/RAS/MAPK Pathways In Colorectal Cancers. Int J Med Sci (2019) 16:1480–91. doi: 10.7150/ijms.35269
54. Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 Functions as a Competing Endogenous RNA to Confer Oxaliplatin Resistance and Holds Prognostic Values in Colon Cancer. Mol Ther (2016) 24:2064–77. doi: 10.1038/mt.2016.180
55. Pekow J, Meckel K, Dougherty U, Huang Y, Chen X, Almoghrabi A, et al. miR-193a-3p is a Key Tumor Suppressor in Ulcerative Colitis-Associated Colon Cancer and Promotes Carcinogenesis Through Upregulation of IL17RD. Clin Cancer Res (2017) 23:5281–91. doi: 10.1158/1078-0432.CCR-17-0171
56. Takahashi H, Takahashi M, Ohnuma S, Unno M, Yoshino Y, Ouchi K, et al. microRNA-193a-3p is Specifically Down-Regulated and Acts as a Tumor Suppressor in BRAF-Mutated Colorectal Cancer. BMC Cancer (2017) 17:723. doi: 10.1186/s12885-017-3739-x
57. Liang L, Gao C, Li Y, Sun M, Xu J, Li H, et al. miR-125a-3p/FUT5-FUT6 Axis Mediates Colorectal Cancer Cell Proliferation, Migration, Invasion and Pathological Angiogenesis via PI3K-Akt Pathway. Cell Death Dis (2017) 8:e2968. doi: 10.1038/cddis.2017.352
58. Mao Q, Quan T, Luo B, Guo X, Liu L, Zheng Q. MiR-375 Targets KLF4 and Impacts the Proliferation of Colorectal Carcinoma. Tumour Biol (2016) 37:463–71. doi: 10.1007/s13277-015-3809-0
59. Tian L, Chen M, He Q, Yan Q, Zhai C. MicroRNA199a5p Suppresses Cell Proliferation, Migration and Invasion by Targeting ITGA3 in Colorectal Cancer. Mol Med Rep (2020) 22:2307–17. doi: 10.3892/mmr.2020.11323
60. Xu L, Wen T, Liu Z, Xu F, Yang L, Liu J, et al. MicroRNA-375 Suppresses Human Colorectal Cancer Metastasis by Targeting Frizzled 8. Oncotarget (2016) 7:40644–56. doi: 10.18632/oncotarget.9811
61. Wang Y, Tang Q, Li M, Jiang S, Wang X. MicroRNA-375 Inhibits Colorectal Cancer Growth by Targeting PIK3CA. Biochem Biophys Res Commun (2014) 444:199–204. doi: 10.1016/j.bbrc.2014.01.028
62. Chao CC, Wu PH, Huang HC, Chung HY, Chou YC, Cai BH, et al. Downregulation of miR-199a/B-5p is Associated With GCNT2 Induction Upon Epithelial-Mesenchymal Transition in Colon Cancer. FEBS Lett (2017) 591:1902–17. doi: 10.1002/1873-3468.12685
63. Junttila MR, De Sauvage FJ. Influence of Tumour Micro-Environment Heterogeneity on Therapeutic Response. Nat (2013) 501:346–54. doi: 10.1038/nature12626
64. Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, et al. The Lung Microenvironment: An Important Regulator of Tumour Growth and Metastasis. Nat Rev Cancer (2019) 19:9–31. doi: 10.1038/s41568-018-0081-9
65. Lee SS, Cheah YK. The Interplay Between MicroRNAs and Cellular Components of Tumour Microenvironment (TME) on Non-Small-Cell Lung Cancer (NSCLC) Progression. J Immunol Res (2019) 2019:3046379. doi: 10.1155/2019/3046379
66. Meng F, Yang M, Chen Y, Chen W, Wang W. miR-34a Induces Immunosuppression in Colorectal Carcinoma Through Modulating a SIRT1/NF-Kappab/B7-H3/TNF-Alpha Axis. Cancer Immunol Immunother (2021) 70:2247–59. doi: 10.1007/s00262-021-02862-2
67. Zheng J, Yang T, Gao S, Cheng M, Shao Y, Xi Y, et al. miR-148a-3p Silences the CANX/MHC-I Pathway and Impairs CD8(+) T Cell-Mediated Immune Attack in Colorectal Cancer. FASEB J (2021) 35:e21776. doi: 10.1096/fj.202100235R
68. Lou Q, Liu R, Yang X, Li W, Huang L, Wei L, et al. miR-448 Targets IDO1 and Regulates CD8(+) T Cell Response in Human Colon Cancer. J Immunother Cancer (2019) 7:210. doi: 10.1186/s40425-019-0691-0
69. Trivedi S, Srivastava RM, Concha-Benavente F, Ferrone S, Garcia-Bates TM, Li J, et al. Anti-EGFR Targeted Monoclonal Antibody Isotype Influences Antitumor Cellular Immunity in Head and Neck Cancer Patients. Clin Cancer Res (2016) 22:5229–37. doi: 10.1158/1078-0432.CCR-15-2971
70. Trotta AM, Ottaiano A, Romano C, Nasti G, Nappi A, De Divitiis C, et al. Prospective Evaluation of Cetuximab-Mediated Antibody-Dependent Cell Cytotoxicity in Metastatic Colorectal Cancer Patients Predicts Treatment Efficacy. Cancer Immunol Res (2016) 4:366–74. doi: 10.1158/2326-6066.CIR-15-0184
71. Pecci F, Cantini L, Bittoni A, Lenci E, Lupi A, Crocetti S, et al. Beyond Microsatellite Instability: Evolving Strategies Integrating Immunotherapy for Microsatellite Stable Colorectal Cancer. Curr Treat Options Oncol (2021) 22:69. doi: 10.1007/s11864-021-00870-z
72. Bourhis J, Stein A, Paul De Boer J, Van Den Eynde M, Gold KA, Stintzing S, et al. Avelumab and Cetuximab as a Therapeutic Combination: An Overview of Scientific Rationale and Current Clinical Trials in Cancer. Cancer Treat Rev (2021) 97:102172. doi: 10.1016/j.ctrv.2021.102172
73. Hombach S, Kretz M. Non-Coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol (2016) 937:3–17. doi: 10.1007/978-3-319-42059-2_1
74. Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell (2009) 136:629–41. doi: 10.1016/j.cell.2009.02.006
75. Saha P, Verma S, Pathak RU, Mishra RK. Long Noncoding RNAs in Mammalian Development and Diseases. Adv Exp Med Biol (2017) 1008:155–98. doi: 10.1007/978-981-10-5203-3_6
76. Luo J, Qu J, Wu DK, Lu ZL, Sun YS, Qu Q. Long non-Coding RNAs: A Rising Biotarget in Colorectal Cancer. Oncotarget (2017) 8:22187–202. doi: 10.18632/oncotarget.14728
77. Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, et al. Genetic Variants at the 9p21 Locus Contribute to Atherosclerosis Through Modulation of ANRIL and CDKN2A/B. Atherosclerosis (2012) 220:449–55. doi: 10.1016/j.atherosclerosis.2011.11.017
78. Johnson R. Long non-Coding RNAs in Huntington's Disease Neurodegeneration. Neurobiol Dis (2012) 46:245–54. doi: 10.1016/j.nbd.2011.12.006
79. Zhu Y, Li B, Liu Z, Jiang L, Wang G, Lv M, et al. Up-Regulation of lncRNA SNHG1 Indicates Poor Prognosis and Promotes Cell Proliferation and Metastasis of Colorectal Cancer by Activation of the Wnt/beta-Catenin Signaling Pathway. Oncotarget (2017) 8:111715–27. doi: 10.18632/oncotarget.22903
80. Liu T, Han Z, Li H, Zhu Y, Sun Z, Zhu A. LncRNA DLEU1 Contributes to Colorectal Cancer Progression via Activation of KPNA3. Mol Cancer (2018) 17:118. doi: 10.1186/s12943-018-0873-2
81. Li M, Wang Q, Xue F, Wu Y. lncRNA-CYTOR Works as an Oncogene Through the CYTOR/miR-3679-5p/MACC1 Axis in Colorectal Cancer. DNA Cell Biol (2019) 38:572–82. doi: 10.1089/dna.2018.4548
82. Zhang XT, Pan SX, Wang AH, Kong QY, Jiang KT, Yu ZB. Long Non-Coding RNA (lncRNA) X-Inactive Specific Transcript (XIST) Plays a Critical Role in Predicting Clinical Prognosis and Progression of Colorectal Cancer. Med Sci Monit (2019) 25:6429–35. doi: 10.12659/MSM.915329
83. Qian J, Garg A, Li F, Shen Q, Xiao K. lncRNA LUNAR1 Accelerates Colorectal Cancer Progression by Targeting the Mir4953p/MYCBP Axis. Int J Oncol (2020) 57:1157–68. doi: 10.3892/ijo.2020.5128
84. Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, et al. Long non-Coding RNA CCAL Regulates Colorectal Cancer Progression by Activating Wnt/beta-Catenin Signalling Pathway via Suppression of Activator Protein 2alpha. Gut (2016) 65:1494–504. doi: 10.1136/gutjnl-2014-308392
85. Wang H, Guan Z, He K, Qian J, Cao J, Teng L. LncRNA UCA1 in Anti-Cancer Drug Resistance. Oncotarget (2017) 8:64638–50. doi: 10.18632/oncotarget.18344
86. Peng K, Liu R, Yu Y, Liang L, Yu S, Xu X, et al. Identification and Validation of Cetuximab Resistance Associated Long Noncoding RNA Biomarkers in Metastatic Colorectal Cancer. BioMed Pharmacother (2018) 97:1138–46. doi: 10.1016/j.biopha.2017.11.031
87. Jing C, Ma R, Cao H, Wang Z, Liu S, Chen D, et al. Long Noncoding RNA and mRNA Profiling in Cetuximab-Resistant Colorectal Cancer Cells by RNA Sequencing Analysis. Cancer Med (2019) 8:1641–51. doi: 10.1002/cam4.2004
88. Zhang X, Wen L, Chen S, Zhang J, Ma Y, Hu J, et al. The Novel Long Noncoding RNA CRART16 Confers Cetuximab Resistance in Colorectal Cancer Cells by Enhancing ERBB3 Expression via miR-371a-5p. Cancer Cell Int (2020) 20:68. doi: 10.1186/s12935-020-1155-9
89. Xu YJ, Zhao JM, Ni XF, Wang W, Hu WW, Wu CP. LncRNA HCG18 Suppresses CD8(+) T Cells to Confer Resistance to Cetuximab in Colorectal Cancer via miR-20b-5p/PD-L1 Axis. Epigenomics (2021) 13:1281–97. doi: 10.2217/epi-2021-0130
90. Yang YN, Zhang R, Du JW, Yuan HH, Li YJ, Wei XL, et al. Predictive Role of UCA1-Containing Exosomes in Cetuximab-Resistant Colorectal Cancer. Cancer Cell Int (2018) 18:164. doi: 10.1186/s12935-018-0660-6
91. Chen LL, Yang L. Regulation of circRNA Biogenesis. RNA Biol (2015) 12:381–8. doi: 10.1080/15476286.2015.1020271
92. Barrett SP, Salzman J. Circular RNAs: Analysis, Expression and Potential Functions. Development (2016) 143:1838–47. doi: 10.1242/dev.128074
93. Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y, et al. Circular RNAs in Cancer: Emerging Functions in Hallmarks, Stemness, Resistance and Roles as Potential Biomarkers. Mol Cancer (2019) 18:90. doi: 10.1186/s12943-019-1002-6
94. Lukiw WJ. Circular RNA (circRNA) in Alzheimer's Disease (AD). Front Genet (2013) 4:307. doi: 10.3389/fgene.2013.00307
95. Liu Q, Shuai M, Xia Y. Knockdown of EBV-Encoded circRNA Circrpms1 Suppresses Nasopharyngeal Carcinoma Cell Proliferation and Metastasis Through Sponging Multiple miRNAs. Cancer Manag Res (2019) 11:8023–31. doi: 10.2147/CMAR.S218967
96. Li O, Kang J, Zhang JJ, Wang J, Hu LW, Li L, et al. Circle RNA FOXP1 Promotes Cell Proliferation in Lung Cancer by Regulating miR-185-5p/Wnt1 Signaling Pathway. Eur Rev Med Pharmacol Sci (2020) 24:6767–78. doi: 10.26355/eurrev_202006_21665
97. Zhan W, Liao X, Chen Z, Li L, Tian T, Yu L, et al. Circular RNA Hsa_circRNA_103809 Promoted Hepatocellular Carcinoma Development by Regulating miR-377-3p/FGFR1/ERK Axis. J Cell Physiol (2020) 235:1733–45. doi: 10.1002/jcp.29092
98. Zhang S, Wang W, Wu X, Zhou X. Regulatory Roles of Circular RNAs in Coronary Artery Disease. Mol Ther Nucleic Acids (2020) 21:172–9. doi: 10.1016/j.omtn.2020.05.024
99. Zhao Y, Zhong R, Deng C, Zhou Z. Circle RNA Circabcb10 Modulates PFN2 to Promote Breast Cancer Progression, as Well as Aggravate Radioresistance Through Facilitating Glycolytic Metabolism Via miR-223-3p. Cancer Biother Radiopharm (2021) 36:477–90. doi: 10.1089/cbr.2019.3389
100. He X, Ma J, Zhang M, Cui J, Yang H. Circ_0007031 Enhances Tumor Progression and Promotes 5-Fluorouracil Resistance in Colorectal Cancer Through Regulating miR-133b/ABCC5 Axis. Cancer biomark (2020) 29:531–42. doi: 10.3233/cbm-200023
101. Liu N, Jiang F. Chen Z. A Preliminary Study on the Pathogenesis of Colorectal Cancer by Constructing a Hsa-circRNA-0067835-miRNA-mRNA Regulatory Network. Onco Targets Ther (2021) 14:4645–58. doi: 10.2147/OTT.S319300
102. Chen H, Pei L, Xie P, Guo G. Circ-PRKDC Contributes to 5-Fluorouracil Resistance of Colorectal Cancer Cells by Regulating miR-375/FOXM1 Axis and Wnt/β-Catenin Pathway. Onco Targets Ther (2020) 13:5939–53. doi: 10.2147/ott.S253468
103. Xiong W, Ai YQ, Li YF, Ye Q, Chen ZT, Qin JY, et al. Microarray Analysis of Circular RNA Expression Profile Associated With 5-Fluorouracil-Based Chemoradiation Resistance in Colorectal Cancer Cells. BioMed Res Int (2017) 2017:8421614. doi: 10.1155/2017/8421614
104. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 Promotes Colorectal Cancer Growth and Metastasis by Sponging miR-7. Cell Death Dis (2018) 9:417. doi: 10.1038/s41419-018-0454-8
105. Hu T, Li Z, Gao CY, Cho CH. Mechanisms of Drug Resistance in Colon Cancer and its Therapeutic Strategies. World J Gastroenterol (2016) 22:6876–89. doi: 10.3748/wjg.v22.i30.6876
106. Gao Q, Li XX, Xu YM, Zhang JZ, Rong SD, Qin YQ, et al. IRE1alpha-Targeting Downregulates ABC Transporters and Overcomes Drug Resistance of Colon Cancer Cells. Cancer Lett (2020) 476:67–74. doi: 10.1016/j.canlet.2020.02.007
107. Jiang L, Wang P, Sun YJ, Wu YJ. Ivermectin Reverses the Drug Resistance in Cancer Cells Through EGFR/ERK/Akt/NF-kappaB Pathway. J Exp Clin Cancer Res (2019) 38:265. doi: 10.1186/s13046-019-1251-7
108. Wang Y, Wang Y, Qin Z, Cai S, Yu L, Hu H, et al. The Role of non-Coding RNAs in ABC Transporters Regulation and Their Clinical Implications of Multidrug Resistance in Cancer. Expert Opin Drug Metab Toxicol (2021) 17:291–306. doi: 10.1080/17425255.2021.1887139
109. Tang W, Ji M, He G, Yang L, Niu Z, Jian M, et al. Silencing CDR1as Inhibits Colorectal Cancer Progression Through Regulating microRNA-7. Onco Targets Ther (2017) 10:2045–56. doi: 10.2147/OTT.S131597
110. Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res (2017) 23:3918–28. doi: 10.1158/1078-0432.CCR-16-2541
Keywords: miRNA, lncRNA, circRNA, CRC, EGFR, drug resistance
Citation: Chu J, Fang X, Sun Z, Gai L, Dai W, Li H, Yan X, Du J, Zhang L, Zhao L, Xu D and Yan S (2022) Non-Coding RNAs Regulate the Resistance to Anti-EGFR Therapy in Colorectal Cancer. Front. Oncol. 11:801319. doi: 10.3389/fonc.2021.801319
Received: 25 October 2021; Accepted: 20 December 2021;
Published: 17 January 2022.
Edited by:
Peixin Dong, Hokkaido University, JapanReviewed by:
Zhe-Sheng Chen, St. John’s University, United StatesCopyright © 2022 Chu, Fang, Sun, Gai, Dai, Li, Yan, Du, Zhang, Zhao, Xu and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shushan Yan, Znl5YW5zc0B3Zm1jLmVkdS5jbg==; Donghua Xu, eHVkaEB3Zm1jLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.