- 1Department of Medical Laboratory Sciences, Faculty of Health Sciences, The Open University of Sri Lanka, Nugegoda, Sri Lanka
- 2Centre for Genomics and Personalised Health, Faculty of Health, School of Biomedical Sciences, Queensland University of Technology, Brisbane, QLD, Australia
- 3Faculty of Engineering and IT, School of Biomedical Engineering, University of Technology Sydney, Sydney, NSW, Australia
- 4Princess Alexandra Hospital, Cancer Care Services, Woolloongabba, QLD, Australia
- 5The University of Queensland Diamantina Institute, The University of Queensland, Brisbane, QLD, Australia
Lung cancer is the leading cause of cancer morbidity and mortality worldwide and early diagnosis is crucial for the management and treatment of this disease. Non-invasive means of determining tumour information is an appealing diagnostic approach for lung cancers as often accessing and removing tumour tissue can be a limiting factor. In recent years, liquid biopsies have been developed to explore potential circulating tumour biomarkers which are considered reliable surrogates for understanding tumour biology in a non-invasive manner. Most common components assessed in liquid biopsy include circulating tumour cells (CTCs), cell-free DNA (cfDNA), circulating tumour DNA (ctDNA), microRNA and exosomes. This review explores the clinical use of circulating tumour biomarkers found in liquid biopsy for screening, early diagnosis and prognostication of lung cancer patients.
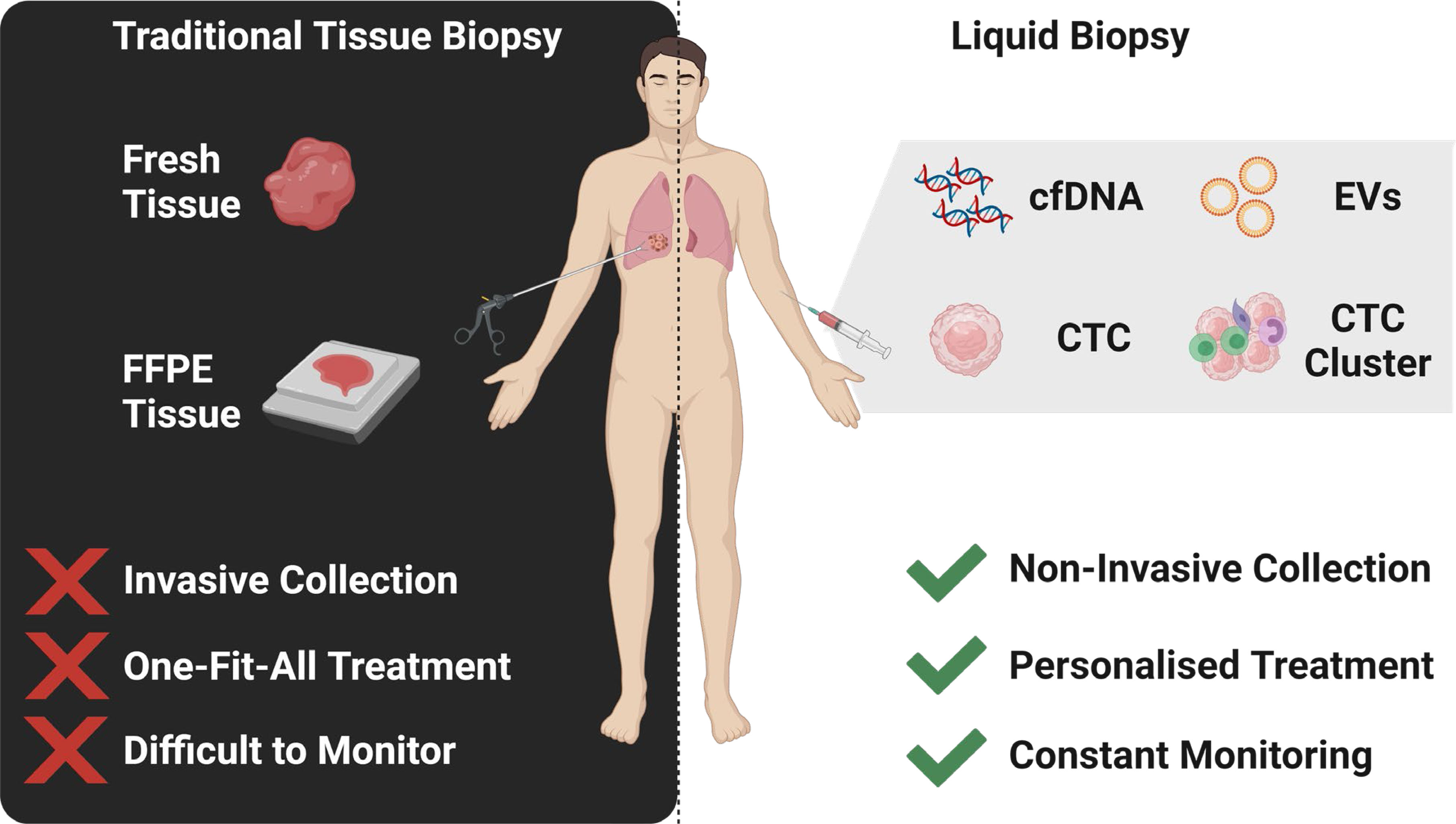
Graphical Abstract The identification of multi-marker analytes in liquid biopsy samples enables a personalised medicine approach to the management of lung cancer.
Introduction
Lung Cancer
Lung cancer is the leading cause of cancer related deaths and accounted for over 2.1 million new cases and 1.8 million deaths in 2018 (1). If the cancer is diagnosed in the early stages (i.e., Stage I – II), the five-year survival rate is estimated to be around 56% (2). However, only 16% of lung cancer cases are diagnosed early, which causes the overall five-year survival rate of lung cancer patients to be less than 20% with adverse clinical outcomes (3, 4). Lung cancer is mainly categorized into two histological groups: non-small cell carcinoma (NSCLC) and small cell lung carcinoma (SCLC) (5). NSCLC is the most prevalent lung cancer type that accounts for 80% to 85% of all lung cancer cases and is further divided into three histological subtypes of adenocarcinoma, squamous cell carcinoma, and large cell (undifferentiated) carcinoma (6). Adenocarcinoma accounts for 40% of all lung cancers and is frequently found in peripheral bronchi (7, 8). Squamous cell carcinoma comprises 25%-30% of all lung cancer cases which arises from the main bronchi and disseminates into the carina. Large cell (undifferentiated) carcinoma represents about 10% and may originate from different part of the lung. SCLC accounts for 10-15% of all lung cancers and it is the most aggressive type with the lowest overall survival (OS) (5, 6).
Liquid Biopsy
Lung cancer patients often suffer from progression of their disease with adverse clinical outcomes, complications and recurrence (9). Therefore, early diagnosis is vital for effective disease management and preventing the advancement of cancer. However, one of the main challenges for managing and treating lung cancer patients is the lack of sensitive early diagnostic methods (10). To date, low-dose spiral computed tomography (LDCT) is the most commonly used approach for lung cancer screening with more than four times higher sensitivity compared to X-ray imaging (3, 11, 12). However, high false-positive results in the early stages of lung cancers and radiation exposure often limits the usage of LDCT (10).
Tumour biopsy is the gold standard for lung cancer diagnosis due to the potential of investigating targeted biomarkers including carcinoembryonic antigen (CEA), fragments of cytokeratin ‐19 (CYFRA21‐1), squamous cell carcinoma antigen (SCC) and neuron‐specific enolase (NSE) (13, 14). However, tumour biopsy is an invasive approach, requiring specialist medical expertise and cannot be routinely performed outside of a hospital setting. Single site biopsy may have a sampling bias by not being representative of the whole tumour (15). Moreover, tumour biopsies represent a single time point snapshot of the tumour, where therapy-induced changes cannot be determined over the course of treatment. Therefore, alternative, more dynamic biomarkers which can be assessed serially over the course of therapy are desirable (14).
Liquid biopsy have emerged as a promising tool to detect tumour biomarkers (Figure 1) in body fluids in a non-invasive manner. They have shown to play a crucial role in lung cancer screening, early diagnosis, monitoring and determining patient prognosis (16). The minimal invasiveness, ease of use and ability to have repeat measurements over time make it a useful companion diagnostic tool. To date, liquid biopsies has been developed as a novel diagnostic approach to explore potential circulating tumour biomarkers and is considered as a reliable way to understand dynamically changing tumour biology under the stressors of treatment. Liquid biopsy biomarkers include cell-free DNA (cfDNA), circulating tumour DNA (ctDNA), microRNA (miRNA), exosomes and circulating tumour cells (CTCs) (3, 12, 17–19). This review provides an overview of these circulating tumour biomarkers and their clinical significance in screening, early diagnosis and prognostication of lung cancer (Figure 2).
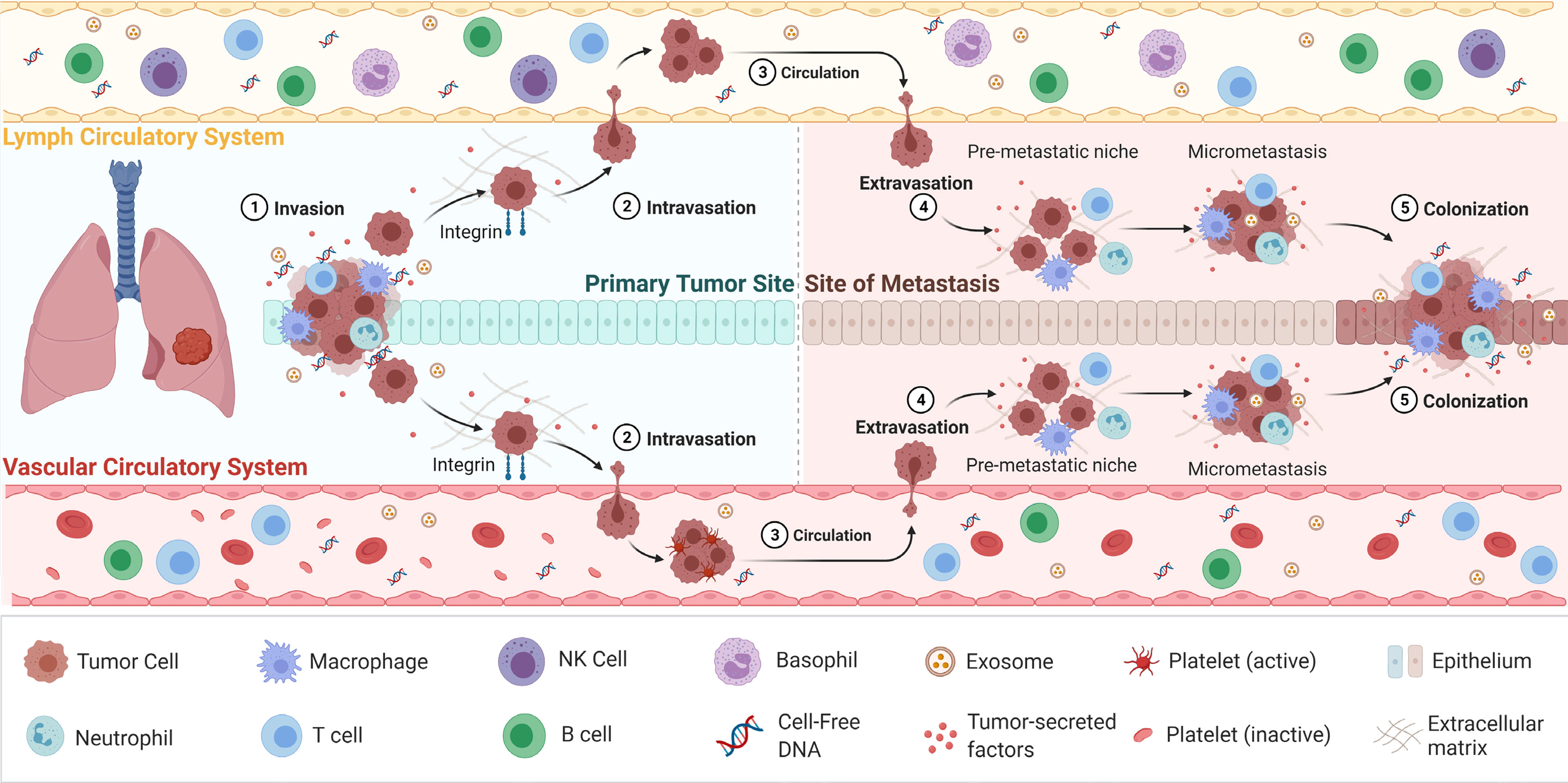
Figure 1 The process of cancer metastasis. Metastasis occurs via the vascular or lymph circulatory systems, where cancer cells from the primary tumour, intravasate into the blood/lymphatics systems and travel through the body, and extravasate at local/distant sites/organs. Adapted from “Overview of Metastatic Cascade”, by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates.
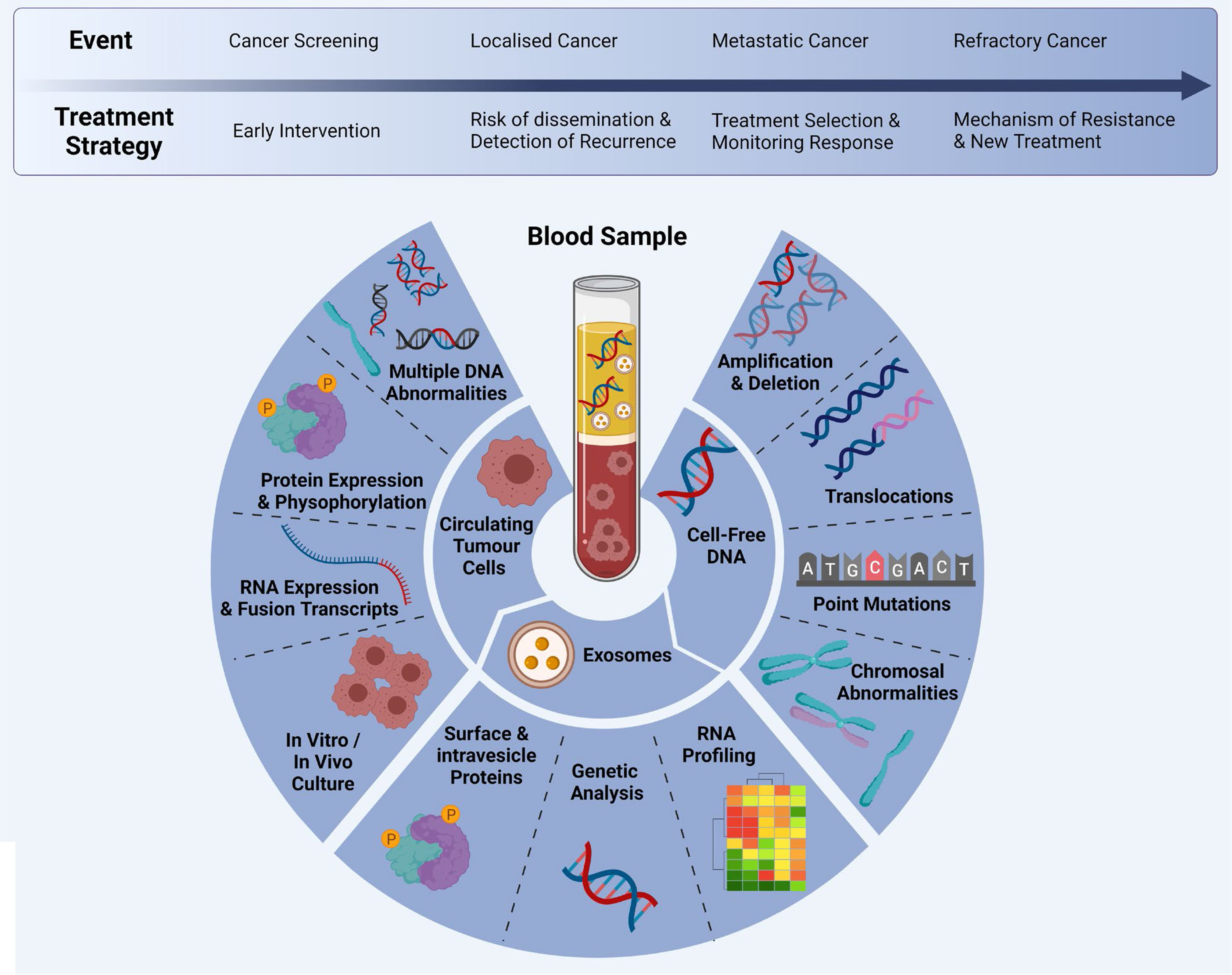
Figure 2 Multi-marker analytes in a liquid biopsy sample: CTCs, cfDNA and exosomes can be analysed at different time points during clinical progression/treatment. Adapted from Ref (20) and created with BioRender.com.
cfDNA and ctDNA
Cell free DNA (cfDNA) is derived from dividing and apoptotic cells as a result of the normal physiological process of tissue remodeling (21–23). In healthy subjects, the amount of cfDNA is quite low and it has been estimated as 5–10 ng/mL in body fluids (24). The half-life of cfDNA is approximately 2 hours (25). The proportion of cell free DNA derived from tumour cells is known as ctDNA (18). The concentration of ctDNA in plasma varies from 0.01% to 90% of total cfDNA (25). The amount of ctDNA present in plasma correlates with the tumour burden, progression free survival (PFS) and OS (10), which is a useful biomarker to monitor NSCLC patients in different stages (26). A study conducted by Newman et al. has been found that a 100% detection rate of ctDNA in stage II–IV NCSLC patients and 50% in early-stage patients (26). Owing to the short half-life of ctDNA, treatment efficacy of patients can be rapidly detected, earlier than radiological changes (25). On the other hand, in order to detect ctDNA in the peripheral circulation, blood samples should be collected within a certain time period to avoid degradation (24). ctDNA can identify specific molecular changes present in the original tumour. These include mutations in oncogenes/tumour suppressor genes and gene amplifications or epigenetic changes (19). Therefore, cfDNA/ctDNA has been used as a prognostic marker for the diagnosis of different types of cancers including lung cancer (17, 27, 28). Furthermore, ctDNA provides a molecular picture of the residual disease which helps in decision making for the commencement of adjuvant chemotherapy after the surgery (29). The concentration of cfDNA is found to be higher in early stage NSCLC patients, recurrent and advanced stage NSCLC patients compared to healthy subjects (26, 29). A study by Pohomaryova et al., reported that the plasma concentration of cfDNA in lung cancer patients is eight times higher than normal healthy adults (30). Several studies have been reported the presence of high cfDNA concentrations has a significant association with the worse clinical outcome (31–34). Furthermore, cfDNA plays an important role for identifying blood tumour mutational burden (bTMB) in NSCLC, indicating the number of somatic mutations in the genome coding regions (35). It has been found that sensitivity and specificity of bTMB assay were 93.9%, 93.9% and 100.0%, compared to tissue TMB (36).
cfDNA mutation analysis is also important to identify specific mutations and molecular targets for personalized therapies (37). This can be beneficial to select patients for immunotherapy and pursue a clinically meaningful improvement in terms of survival (38, 39). The United States Food and Drug Administration (U.S. FDA) approved ctDNA as the first liquid biopsy test for the detection of NSCLC patients with EGFR mutations who were suitable for personalized therapy (Roche Cobas EGFR mutation test v2) (40). This assay can detect multiple mutations in exons 18, 19, 20 and 21 in NSCLC including L858R, T790M, G719X, S7681, and L861Q (41). Using ctDNA, 62.5% of patients were identified with EGFR mutations (exon 19 deletion, exon 20 T790 M insertion and exon 21 L858R mutation) at the baseline, while the rate of EGFR mutation positivity was higher among patients with metastatic disease (42). ctDNA, as a specific tumour marker, has a high specificity of 80-95% for the detection of EGFR mutations, which can inform on the use of tyrosine kinase targeted therapies. However, the sensitivity of this approach is comparatively lower – about 60-85%, which cannot be used to ensure the EGFR mutation does not exist in the patient. In addition to detection of EGFR mutation, ALK rearrangements have been assessed in ctDNA of lung cancer patients (43–45). ALK is a membrane-bound tyrosine kinase receptor encoded by the ALK gene (46, 47) and rearrangement of ALK can be seen in 2-7% of NSCLC patients. Hence, ALK rearrangement has emerged as the second most studied targetable mutation in order to develop a novel treatment approach to lung cancer (48).
In addition to single-gene assays, higher through-put NGS multi-gene readout assays have been developed to identify single-nucleotide variants (SNVs), copy number alterations (CNAs), inserts/deletions and or fusions. These assays include the FoundationOne Liquid CDx, Guardant 360 CDx, MSK-ACCESS, OncoDNA and Archer Reveal. These assays have the advantage of broad genotyping and utility in clinical trials (49). The blood TMB (bTMB) assays have shown concordance with the tumour TMB assays in advanced stage NSCLC, in particular for predicting clinical outcomes to immunotherapy (50, 51).
Usually, at the time of sample collection, tumour-derived DNA is found highly fragmented and mixed with non-tumour DNA/cfDNA. Separation of ctDNA from cfDNA can be pursued using ultrasensitive analytical assays (52). Several testing approaches have been identified; including emulsions, beads, amplification, magnetics (BEAMing), droplet digital PCR (ddPCR) and next generation sequencing (NGS), which can detect down to a few copies of ctDNA (53–55). In contrast, the low amount of ctDNA in plasma has been challenging the clinical applicability of this technique. Therefore, evaluation of ctDNA in other body fluids than plasma may potentially provide a solution to this. The concentration of cfDNA in body fluids is higher near the proximal tumour sites (37). It is worth mentioning that the current guidelines emphasize plasma as a preferred specimen than serum for the detection of ctDNA, as leukocyte lysis takes place during clotting which thereby leads to high contamination rate of germinal DNA than in plasma (56). Furthermore, the specimen should be processed within 6 hours of collection in order to prevent release of DNA from normal blood cell lysis and leukocyte stabilization reagents would be useful in these circumstances (57).
Exosomes
Exosomes are small extracellular vesicles derived by endocytosis, with 40–100 nm in size (58), which can be found in all the body fluids and capable of transporting DNA, mRNAs, noncoding miRNAs, proteins and lipids (59). It consists of a lipid bilayer that prevents degradation by enzymes such as ribonuclease and high pH conditions. Hence, exosomes play an important role in cell mediated communication in normal healthy cells and disease cells including tumour cells (60). All cell types, including normal, disease or tumour cells can release exosomes into the extracellular space (61). Recent studies suggest a higher number of exosomes in patients with cancer than in healthy adults (62, 63). There is emerging evidence that exosomes have a crucial role in carcinogenesis, cancer progression and metastasis of several tumours, including NSCLC (14).
Exosomes derived from tumour cells can promote carcinogenesis by transferring oncogenic factors and thereby induce malignant transformation (64). Acquiring the oncogenic factors from by non-cancerous cells leads to change of cellular behaviour and share comparable characteristics that the tumour poses (65). Tumour derived exosomes also have the potential of influencing epithelial-mesenchymal transition (EMT) and facilitate metastasis by transferring migratory and metastatic capacity to the non-cancerous cells (66). Moreover, these extracellular vesicles cause the expression of vimentin in normal cells, stimulating EMT and increasing the metastasis ability. This can further facilitate neoangiogenesis by releasing proteins such as fibroblast growth factor (FGF), vascular endothelial growth factor (VEGF), IL-6, and IL-8, and stimulating vascular endothelial cells via miRNAs (67, 68).
The analysis of nucleic acids in exosomes has shown to be a more reliable and sensitive mutation detection approach than using cfDNA/ctDNA. Thakur et al. showed tumour-derived exosomes carry double-stranded DNA which represents the whole genome and the mutation profile of the primary tumour cells (69). Exosomes hold great promise as biomarkers for the early diagnosis and treatment via analysis of nucleic acids and other markers to find clues on primary tumour and metastasis condition of patients. Different exosomal proteins and miRNA have been currently employed for the diagnostic and prognostic utility. Some surface proteins offer promising potential as tumour markers including CD91, CD317 and EGFR (70).
Exosomal microRNA-96 (miRNA-96) fosters lung cancer progression by suppressing the activity of Lim domain 7 (LMO7) protein. LMO7 is a fibrous actin-binding protein functions in the formation and maintenance of actin cytoskeleton. In lung cancers, it functions as a tumour suppressor molecule while a shortage of LMO7 confers a genetic predisposition to lung cancer (67). Studies have been documented that the amount of exosomal miRNA is different between healthy individuals and lung cancer patients at different stages (71, 72). Therefore, exosomal miRNA is imperative as a promising and effective noninvasive candidate biomarker for early diagnosis, tumour profiling and enabling a timely treatment plan in NSCLC (10).
It has been found that EGFR protein can be transferred from tumour cells to non-cancerous cells via exosomes which leading to downregulation of the VEGF pathway (73). Similar to cfDNA, exosomal RNA could be utilized to detect EGFR mutations. It is worth mentioning that exosomal RNA has been found to be more efficient in analyzing EGFR mutations than cfDNA (74).
Although exosomes have provided prospective clinical evidence on cancer, still several constraints limit their clinical utility. One of the major drawbacks is difficulty in purification due to its’ smaller size. Further, few drawbacks are associated with the difficulty to adopt exosome isolation techniques in clinical environment. In this context, several isolation techniques have been employed based on the physical, chemical and biological properties of exosomes (75). Physical methods include ultracentrifugation, ultrafiltration, density gradient separation and size exclusion chromatography (75). All these aforementioned methods are based on the molecular size and density of the exosomes. In the ultracentrifugation, according to the density, blood and other components are separated with different centrifugal speeds and exosomes are separated in the final ultracentrifugation step. Ultrafiltration is another physical method that utilizes the exosome size for separation of these extracellular vesicles (59). Furthermore, Polymeric-based precipitation is one of the promising techniques that used the chemical properties of exosomes by using beads coated with specific antibodies coated.
CTCs
CTCs were first described by an Australian physician, Thomas Ashworth in 1869, where he observed cells similar to the primary tumour were found in the blood of a patient with metastatic disease. Accordingly, metastasis is thought to be facilitated by metastatic precursor cells, known as CTCs, which are tumour cells that detach from a tumour into the vasculature (76). They are extremely rare and approximately 1-10 CTCs can be present per milliliter of whole blood (77). Emerging evidence has demonstrated that the presence of CTCs in the vascular system associate with worse clinical outcomes in terms of OS of cancers (5). It has been shown that patients with NSCLC have ten times higher concentration of CTCs in their blood circulation than in other cancer patients (78–80).
CTCs are considered to play a pivotal role in metastasis of cancer and during which they go through a process called EMT (81, 82). EMT is a complex biological process that results in the gradual lowering of epithelial features of the tumour cells and obtains mesenchymal properties to acquire migratory and invasive metastatic characteristics (83). In this process, tumour cells enter the circulation and reach a distant site where a metastatic deposit is initiated. Here, CTCs regain epithelial characteristics allowing further proliferation and formation of metastases deposits at the distant site (Figure 1) (84). A sensitive and specific isolation method is therefore imperative to provide sufficient and purified CTCs to analyse (85, 86). Since CTCs are extremely rare, their isolation is greatly constrained by technological implications (87, 88). With the advancement of novel technologies for CTC enrichment, in particular that of microfluidics, CTC capture is becoming increasingly efficient. The current CTC enrichment methods are based either on the biological or physical properties of CTCs. The CellSearch (Menarini Silicon Biosystems) platform is the only FDA-approved CTC enumeration method which target epithelial cell adhesion molecule (EpCAM), a transmembrane glycoprotein involved in cell to cell adhesion (89). This platform is approved for breast, colon and prostate cancers. However, this platform has a number of limitations, including that is only captured epithelial CTCs and does not capture mesenchymal shifted CTCs with low or absent EpCAM expression (90, 91). Furthermore, isolation and enrichment of CTCs have been extremely challenging in NSCLC due to the downregulation of the epithelial tumour markers during EMT, which consequently masks the true number of CTCs (10). CTCs often have partial-EMT or hybrid states, which can downregulated markers such as EpCAM (92–94). This is further confounded by the presence of CTC clusters or microemboli which have a higher metastatic capacity compared to single CTCs, including immune evasion strategies by the inclusion of leukocytes (83, 95–97).
To tackle these drawbacks, label-free CTC isolation technologies have been established to isolate CTCs based on size, density and deformability (87, 98). Still, considerable constraints of label-free technologies are inevitable and the usage is limited due to their throughput (99). To overcome this, recently microfluidic technologies have been developed including electrophoresis, hydrodynamic and cross-flow filtration, micropore and micropost trapping, deterministic lateral displacement and inertial focusing systems to capture CTCs (90, 99–103). This has added values through effective cell sorting without need for purification, high system throughputs and ability to analyze functions of CTCs in vitro (90, 99, 104–106).
Isolated CTCs can be analysed through different approaches including molecular and proteomic studies. For instance, captured CTCs can be used for EGFR mutation analysis which will provide a better understanding of the tumour genetic profile similar to ctDNA (10). Studies emphasized that detection of EGFR mutations in CTC could assist in determining prospective therapeutic decisions which ultimately would lead to the advancement of precision medicine and personalized oncology (5, 107). Other than EGFR mutations, programmed death ligand-1 (PD-L1) expression on CTC has also been studied; however, authors have not been established a clear relationship between its expression and cancer progression and prognosis in patients with NSCLC (108, 109).
The isolation of a sufficient amount of CTCs in the blood of NSCLC patients and identify its various biomarkers can aid in early detection of the NSCLC and will also provide real-time monitoring of cancer progression, treatment efficacy and prognosis.
Circulating miRNAs
miRNAs are a type of gene expression regulator that works at the post-translational level with a multi-protein complex known as the RNA-induced silencing complex (miRISC), exerting their function at the 3’-untranslated region (3’UTR) of target complementary messenger RNA (mRNA) sequences (110, 111). miRNAs have been discovered to be single-stranded RNA molecules with a length of 19 to 22 nucleotides (14, 112). It has been well established that each miRNA can regulate and act as a target for multiple mRNAs and that each mRNA, resulting in a cascade of gene regulation (113, 114). Evidence suggests that any dysregulation of miRNAs could have an impact on a variety of diseases, most notably cancer (115). In the context of cancer, miRNA dysregulation has been linked to tumor initiation, growth, and progression, with evidence pointing to miRNAs acting as tumor suppressors and oncogenes (115). In addition, it was found that miRNAs are packed into extracellular vesicles before being released into the extracellular space. These vesicles could be exosome and microvesicles (116–118). Several studies have been conducted to explore the role and application of miRNAs in cancer (75, 119–121). The serum or plasma level of miRNAs can serve as a predictive marker in cancer patients, indicating signs of the disease stage (75, 119). miRNAs are released into the circulation via protein-miRNA complexes, exosomes, tumor-educated platelets (TEP), and apoptotic bodies (122, 123). Accordingly, evidence suggests that cancer therapeutic approaches such as chemotherapy and radiotherapy may affect circulating miRNA levels, implying that miRNAs can serve as biomarkers for response or resistance to therapy (120, 121, 124, 125). In lung cancer, miRNAs have been found to distinguish healthy individuals from cancer patients (126–128). miRNAs may also serve as biomarkers of therapeutic success in NSCLC patients, allowing for better patient management decisions (129). As a result, studies on lung cancer patients revealed that, when compared to LDCT, miRNAs had significantly lower false-positive results when it came to detecting lung cancer patients in their early stages (129). Furthermore, a meta-analysis study from 65 studies (6919 patients with lung cancer and 7064 healthy volunteers), discovered a panel of four miRNAs, including miR-21-5p, miR-126-3p, miR-155-5p, and miR-223-3p, which can be used as a potential biomarker in lung cancer screening (130). Moreover, a different study conducted on early NSCLC patients showed that a number of miRNAs were differentially regulated between short-term survivors and long-term survivors. These miRNAs, which included miR-1, miR-30d, miR-486, and miR-499, were also linked to the OS (127). In line with these findings, Li et al. studied miR-486 and miR-150 on plasma samples from lung cancer patients to explore their early diagnostic value (131). As a result, these miRNAs were discovered to have greater than 80% specificity and sensitivity in discriminating healthy individuals from lung cancer patients (131). Another study on patients with metastatic lung cancer discovered that miR-18a, miR-28-3p, miR-191, miR-145, and miR-328 were associated with 3-year survival (132). In studies on NSCLC tumors, researchers discovered six serum miRNAs from patients that had significantly different expressions when compared to healthy people. Accordingly, miR-15b-5p was found to be overexpressed, while miR-19-3p, miR-92-3p, miR-16-5p, miR-17b-5p, and miR-20a-5p were found to have downregulation (133). Among these miRNAs, miR-16-5p, miR-15b-5p, and miR-20a-5p had the highest sensitivity and specificity values (133). However, despite these promising findings, there are some drawbacks to using miRNAs as diagnostic biomarkers, which are listed in Table 1.
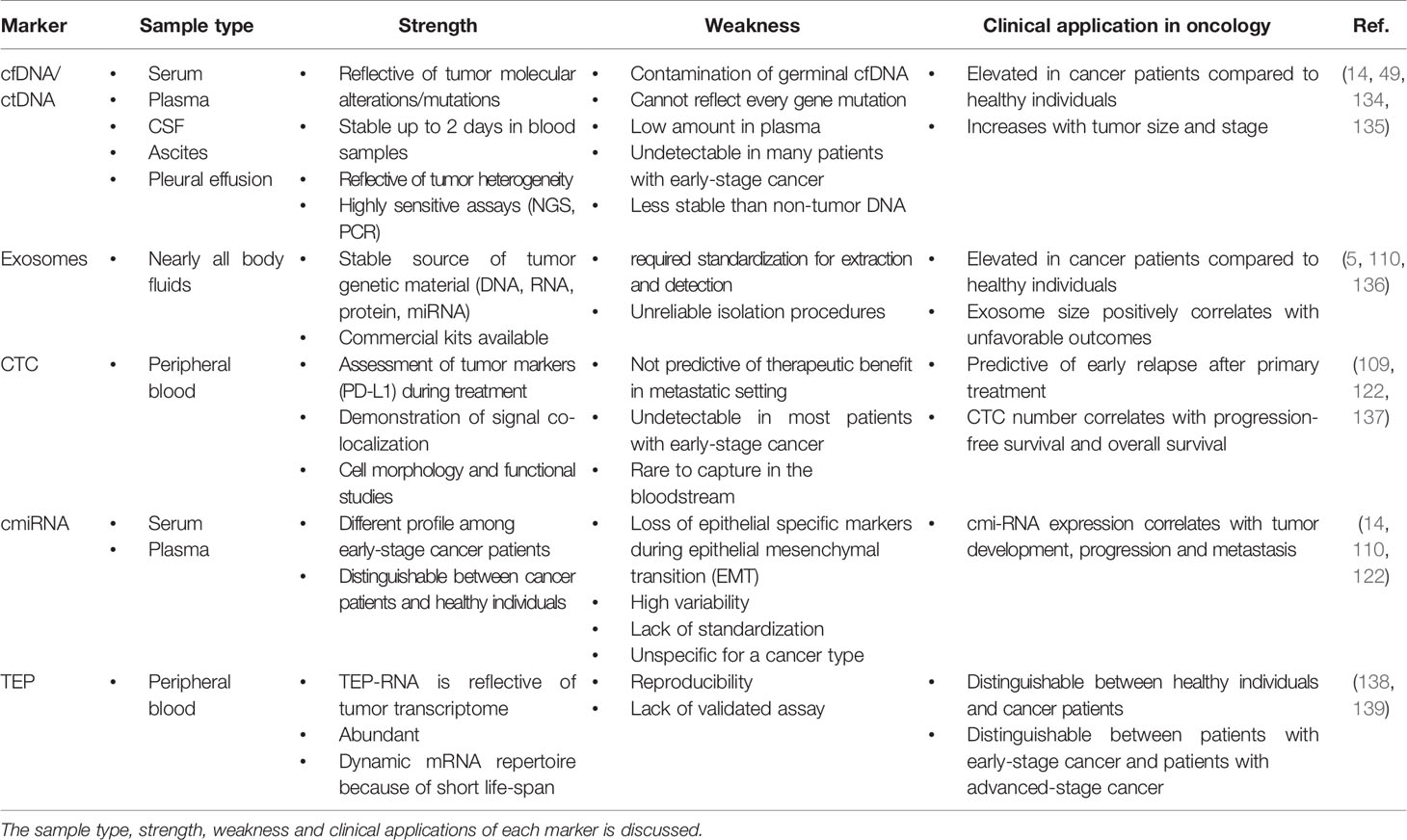
Table 1 Comparison of different liquid biopsy markers including cfDNA/ctDNA, exosome, CTC, cmiRNA and TEP.
Tumor Educated Platelets
Platelets are enucleated cells that participate in coagulation. Platelet activation has also been found in inflammatory diseases like eczema and asthma (140). Evidence suggests that platelets may play an important role in tumorigenesis by helping tumor evasion, angiogenesis, and metastasis (141, 142). Activated platelets secrete α-granules to release transforming growth factor-beta (TGFβ) and adenosine triphosphate (ATP), leading to epithelial mesenchymal transition (EMT) and metastasis (143). On the other hand, tumour cells have been found to induce thrombocytosis by producing growth factors and cytokines such as granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin (IL)-1 and IL-6 (144). Given this information, the interaction between tumour, tumour microenvironment (TME), and platelets account for tumor-educated platelets (TEPs) (145, 146). Accordingly, numerous studies explored the use of TEPs as a liquid biopsy in cancer initiation and progression (147–150). It has been discovered that growth factors released and produced by platelets and tumor cells, such as vascular endothelial growth factor (VEGF), contribute to changes in mRNA expression within platelets, resulting in a specific spliced mRNA signature (151, 152). The mRNA signature could be used as a biomarker to distinguish cancer patients from healthy individuals. According to multiple studies, patients with cancer onset and progression have a highly dynamic mRNA repertoire (138, 153). Therefore, analyzing mRNA profiles may be useful in detecting primary tumors, metastasis, and cancer staging (45, 138). A study that compared patients with localized and metastatic tumors to healthy people discovered that platelet mRNA profiles could distinguish cancer cases from healthy ones with 96% accuracy, 97% sensitivity, and 94% specificity (153). In the context of lung cancer, Geraci and colleagues investigated platelet mRNA profiles in the context of lung cancer using a lung cancer model. As a result, they discovered distinct platelet mRNA gene expression between metastatic and control groups (154). Furthermore, research has revealed that some patients with lung adenocarcinoma have ALK rearrangement, resulting in the EML4–ALK fusion gene product (155, 156). Using reverse transcription-polymerase chain reaction (RT-PCR), Nelson et al. examined blood platelets from 77 NSCLC patients and found 38 cases had an EML4-ALK-rearrangement in platelets with 65% sensitivity and 100% specificity (157). In addition, it was reported that the EML4-ALK rearrangement in platelets had a correlation with PFS and OS in patients who received crizotinib. Accordingly, patients with EML4-ALK+platelets had 3.7 month PFS, whereas patients with EML4-ALK- platelets had 16 month PFS (157). When compared to patients with non-cancerous inflammatory diseases, patients with cancer have a hyperactive state of TEPs, according to functional analysis (158). In addition, using RNA sequencing, the platelet RNAs were investigated in patients with early stage NSCLC and healthy individuals. It was demonstrated that integrin alpha-IIb (ITGA2B) was expressed more in NSCLC patients than healthy individuals (159). As a result of their findings, the researchers concluded that TEP ITGA2B could be a promising marker for detecting patients with early-stage NSCLC (159).
Discussion
Liquid biopsy has shown promise for the early diagnosis and management of lung cancer due to its high sensitivity, specificity, non-invasive sampling and low-risk profile. cfDNA/ctDNA, CTCs, miRNAs and exosomes are considered as potentially actionable biomarkers which are used in liquid biopsy. However, robust characterization of each marker is needed for the comprehensive understanding of their role in NSCLC disease progression, prognostication and determining a tailored treatment regimen. It is clear that certain liquid biopsy analytes would be useful over the course of disease progression. For example, ctDNA and exosomes may be informative early in disease onset, and could be used to identify specific actionable mutations/and early stage disease which cannot be determined using LDCT. Whereas CTCs may be used to identify the risk of developing metastatic disease, and the types of clones which may develop treatment resistance (Figure 3). ctDNA has also shown utility over the course of therapy where the variant allele frequencies can be monitored over time/therapy to determine increases or decreases of tumour specific mutations in response to treatment. ctDNA has been the most studied marker in the field of lung cancer with clinical utility for a number of gene mutations. The detection of tumour specific mutations post therapy may also present a window of therapeutic opportunity where the patient has minimal residual disease (MRD), prior to clinically detectable disease progression with radiological evidence. Whilst larger multi-marker NGS panels have come onto the marker, cross comparisons of blood and tumour studies will be warranted to determine their utility for targeted therapies. Moreover, large prospective clinical trials are needed to provide a better understanding of the clinical utility of liquid biopsy assays.
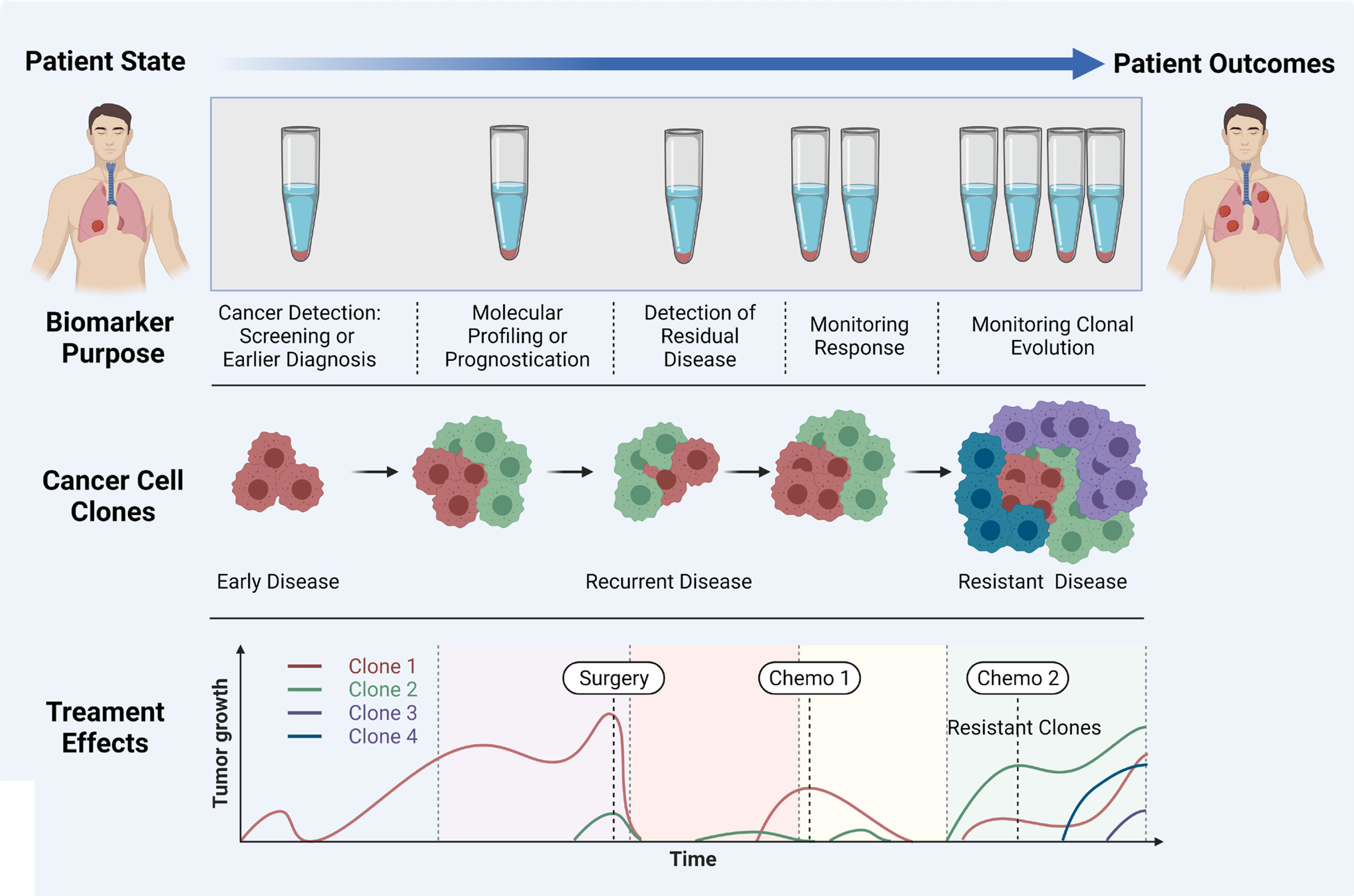
Figure 3 Importance of liquid biopsy at different stages of cancer propagation, indicating the biomarker purpose, evolution of clones, and treatment effects. Adapted from Ref (160) and created with BioRender.com.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
The authors are supported by project grants and fellowships for AK from NHMRC (1157741), Cure Cancer (1182179), and the PA Research Foundation (KOB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Boloker G, Wang C, Zhang J. Updated Statistics of Lung and Bronchus Cancer in United States (2018). J Thorac Dis (2018) 10(3):1158. doi: 10.21037/jtd.2018.03.15
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA: Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
3. Patz EF Jr., Campa MJ, Gottlin EB, Kusmartseva I, Guan XR, Herndon JE. Panel of Serum Biomarkers for the Diagnosis of Lung Cancer. J Clin Oncol (2007) 25(35):5578–83. doi: 10.1200/JCO.2007.13.5392
4. Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, et al. SEER Cancer Statistics Review, 1975-2017. Bethesda, MD: National Cancer Institute (2020) https://seer.cancer.gov/csr/1975_2017/
5. Kapeleris J, Kulasinghe A, Warkiani ME, Vela I, Kenny L, O’Byrne K, et al. The Prognostic Role of Circulating Tumor Cells (Ctcs) in Lung Cancer. Front Oncol (2018) 8:311. doi: 10.3389/fonc.2018.00311
6. Lemjabbar-Alaoui H, Hassan OU, Yang Y-W, Buchanan P. Lung Cancer: Biology and Treatment Options. Biochim Biophys Acta (BBA)-Reviews Cancer (2015) 1856(2):189–210. doi: 10.1016/j.bbcan.2015.08.002
8. Travis WD, Brambilla E, Riely GJ. New Pathologic Classification of Lung Cancer: Relevance for Clinical Practice and Clinical Trials. J Clin Oncol (2013) 31(8):992–1001. doi: 10.1200/JCO.2012.46.9270
9. Polanski J, Jankowska-Polanska B, Rosinczuk J, Chabowski M, Szymanska-Chabowska A. Quality of Life of Patients With Lung Cancer. OncoTargets Ther (2016) 9:1023. doi: 10.2147/OTT.S100685
10. Revelo AE, Martin A, Velasquez R, Kulandaisamy PC, Bustamante J, Keshishyan S, et al. Liquid Biopsy for Lung Cancers: An Update on Recent Developments. Ann Trans Med (2019) 7(15):349. doi: 10.21037/atm.2019.03.28
11. Team NLSTR. Reduced Lung-Cancer Mortality With Low-Dose Computed Tomographic Screening. N Engl J Med (2011) 365(5):395–409. doi: 10.1056/NEJMoa1102873
12. Bauml J, Levy B. Clonal Hematopoiesis: A New Layer in the Liquid Biopsy Story in Lung Cancer. Clin Cancer Res (2018) 24(18):4352–4. doi: 10.1158/1078-0432.CCR-18-0969
13. Chu X-Y, Hou X-B, Song W-A, Xue Z-Q, Wang B, Zhang L-B. Diagnostic Values of SCC, CEA, Cyfra21-1 and NSE for Lung Cancer in Patients With Suspicious Pulmonary Masses: A Single Center Analysis. Cancer Biol Ther (2011) 11(12):995–1000. doi: 10.4161/cbt.11.12.15526
14. Rijavec E, Coco S, Genova C, Rossi G, Longo L, Grossi F. Liquid Biopsy in non-Small Cell Lung Cancer: Highlights and Challenges. Cancers (2020) 12(1):17. doi: 10.3390/cancers12010017
15. Bailey C, Black JR, Reading JL, Litchfield K, Turajlic S, McGranahan N, et al. Tracking Cancer Evolution Through the Disease Course. Cancer Discov (2021) 11(4):916–32. doi: 10.1158/2159-8290.CD-20-1559
16. Lim M, Kim C-J, Sunkara V, Kim M-H, Cho Y-K. Liquid Biopsy in Lung Cancer: Clinical Applications of Circulating Biomarkers (Ctcs and Ctdna). Micromachines (2018) 9(3):100. doi: 10.3390/mi9030100
17. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of Circulating Tumor DNA in Early-and Late-Stage Human Malignancies. Sci Trans Med (2014) 6(224):224ra24–ra24. doi: 10.1158/1538-7445.AM2014-5606
18. Sozzi G, Conte D, Leon M, Cirincione R, Roz L, Ratcliffe C, et al. Quantification of Free Circulating DNA as a Diagnostic Marker in Lung Cancer. J Clin Oncol (2003) 21(21):3902–8. doi: 10.1200/JCO.2003.02.006
19. Santarpia M, Liguori A, D’Aveni A, Karachaliou N, Gonzalez-Cao M, Daffinà MG, et al. Liquid Biopsy for Lung Cancer Early Detection. J Thorac Dis (2018) 10(Suppl 7):S882. doi: 10.21037/jtd.2018.03.81
20. De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol Sci (2019) 40(3):172–86. doi: 10.1016/j.tips.2019.01.006
21. Fernandez-Garcia D, Hills A, Page K, Hastings RK, Toghill B, Goddard KS, et al. Plasma Cell-Free DNA (Cfdna) as a Predictive and Prognostic Marker in Patients With Metastatic Breast Cancer. Breast Cancer Res (2019) 21(1):1–13. doi: 10.1186/s13058-019-1235-8
22. Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Life and Death of Circulating Cell-Free DNA. Cancer Biol Ther (2019) 20(8):1057–67. doi: 10.1080/15384047.2019.1598759
23. Zhu G, Guo YA, Ho D, Poon P, Poh ZW, Wong PM, et al. Tissue-Specific Cell-Free DNA Degradation Quantifies Circulating Tumor DNA Burden. Nat Commun (2021) 12(1):1–11. doi: 10.1038/s41467-021-22463-y
24. Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-Free DNA in Human Blood Plasma: Length Measurements in Patients With Pancreatic Cancer and Healthy Controls. Pancreas (1998) 17(1):89–97. doi: 10.1097/00006676-199807000-00012
25. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating Mutant DNA to Assess Tumor Dynamics. Nat Med (2008) 14(9):985–90. doi: 10.1038/nm.1789
26. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An Ultrasensitive Method for Quantitating Circulating Tumor DNA With Broad Patient Coverage. Nat Med (2014) 20(5):548–54. doi: 10.1038/nm.3519
27. Dawson S-J, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin S-F, et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N Engl J Med (2013) 368(13):1199–209. doi: 10.1056/NEJMoa1213261
28. Spellman PT, Gray JW. Detecting Cancer by Monitoring Circulating Tumor DNA. Nat Med (2014) 20(5):474–5. doi: 10.1038/nm.3564
29. Diaz LA Jr., Bardelli A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J Clin Oncol (2014) 32(6):579. doi: 10.1200/JCO.2012.45.2011
30. Ponomaryova AA, Rykova EY, Cherdyntseva NV, Skvortsova TE, Dobrodeev AY, Zav’yalov AA, et al. Potentialities of Aberrantly Methylated Circulating DNA for Diagnostics and Post-Treatment Follow-Up of Lung Cancer Patients. Lung Cancer (2013) 81(3):397–403. doi: 10.1016/j.lungcan.2013.05.016
31. Hyun MH, Sung JS, Kang EJ, Choi YJ, Park KH, Shin SW, et al. Quantification of Circulating Cell-Free DNA to Predict Patient Survival in non-Small-Cell Lung Cancer. Oncotarget (2017) 8(55):94417. doi: 10.18632/oncotarget.21769
32. Alama A, Coco S, Genova C, Rossi G, Fontana V, Tagliamento M, et al. Prognostic Relevance of Circulating Tumor Cells and Circulating Cell-Free DNA Association in Metastatic Non-Small Cell Lung Cancer Treated With Nivolumab. J Clin Med (2019) 8(7):1011. doi: 10.3390/jcm8071011
33. Nygaard A, Holdgaard P, Spindler KG, Pallisgaard N, Jakobsen A. The Correlation Between Cell-Free DNA and Tumour Burden was Estimated by PET/CT in Patients With Advanced NSCLC. Br J Cancer (2014) 110(2):363–8. doi: 10.1038/bjc.2013.705
34. Lee Y, Park S, Kim WS, Lee JC, Jang SJ, Choi J, et al. Correlation Between Progression-Free Survival, Tumor Burden, and Circulating Tumor DNA in the Initial Diagnosis of Advanced-Stage EGFR-Mutated non-Small Cell Lung Cancer. Thorac Cancer (2018) 9(9):1104–10. doi: 10.1111/1759-7714.12793
35. Ou S-HI, Nagasaka M, Zhu VW. Liquid Biopsy to Identify Actionable Genomic Alterations. Am Soc Clin Oncol Educ Book (2018) 38:978–97. doi: 10.1200/EDBK_199765
36. Fabrizio D, Malboeuf C, Lieber D, Zhong S, He J, White E, et al. Analytic Validation of a Next Generation Sequencing Assay to Identify Tumor Mutational Burden From Blood (Btmb) to Support Investigation of an Anti-PD-L1 Agent, Atezolizumab, in a First Line non-Small Cell Lung Cancer Trial (BFAST). Ann Oncol (2017) 28:v27. doi: 10.1093/annonc/mdx363.018
37. Villatoro S, Mayo-de-las-Casas C, Jordana-Ariza N, Viteri-Ramírez S, Garzón-Ibañez M, Moya-Horno I, et al. Prospective Detection of Mutations in Cerebrospinal Fluid, Pleural Effusion, and Ascites of Advanced Cancer Patients to Guide Treatment Decisions. Mol Oncol (2019) 13(12):2633–45. doi: 10.1002/1878-0261.12574
38. Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With non–Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol (2019) 5(5):696–702. doi: 10.1001/jamaoncol.2018.7098
39. Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood-Based Tumor Mutational Burden as a Predictor of Clinical Benefit in non-Small-Cell Lung Cancer Patients Treated With Atezolizumab. Nat Med (2018) 24(9):1441–8. doi: 10.1038/s41591-018-0134-3
40. Keppens C, Palma JF, Das PM, Scudder S, Wen W, Normanno N, et al. Detection of EGFR Variants in Plasma: A Multilaboratory Comparison of a Real-Time PCR EGFR Mutation Test in Europe. J Mol Diagn (2018) 20(4):483–94. doi: 10.1016/j.jmoldx.2018.03.006
41. Yang J, Hui Y, Zhang Y, Zhang M, Ji B, Tian G, et al. Application of Circulating Tumor DNA as a Biomarker for non-Small Cell Lung Cancer. Front. Oncol (2021) 11:725938. doi: 10.3389/fonc.2021.725938
42. Akamatsu H, Koh Y, Okamoto I, Fujimoto D, Bessho A, Azuma K, et al. Clinical Significance of Monitoring EGFR Mutation in Plasma Using Multiplexed Digital PCR in EGFR Mutated Patients Treated With Afatinib (West Japan Oncology Group 8114LTR Study). Lung Cancer (2019) 131:128–33. doi: 10.1016/j.lungcan.2019.03.021
43. Cui S, Zhang W, Xiong L, Pan F, Niu Y, Chu T, et al. Use of Capture-Based Next-Generation Sequencing to Detect ALK Fusion in Plasma Cell-Free DNA of Patients With Non-Small-Cell Lung Cancer. Oncotarget (2017) 8(2):2771. doi: 10.18632/oncotarget.13741
44. Bordi P, Tiseo M, Rofi E, Petrini I, Restante G, Danesi R, et al. Detection of ALK and KRAS Mutations in Circulating Tumor DNA of Patients With Advanced ALK-Positive NSCLC With Disease Progression During Crizotinib Treatment. Clin Lung Cancer (2017) 18(6):692–7. doi: 10.1016/j.cllc.2017.04.013
45. McCoach CE, Blakely CM, Banks KC, Levy B, Chue BM, Raymond VM, et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non–Small Cell Lung Cancer. Clin Cancer Res (2018) 24(12):2758–70. doi: 10.1158/1078-0432.CCR-17-2588
46. Ivan D, Prieto VG. Use of Immunohistochemistry in the Diagnosis of Melanocytic Lesions: Applications and Pitfalls. Future Oncol (2010) 6(7):1163–75. doi: 10.2217/fon.10.81
47. Huang H. Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor With Many Faces. Int J Mol Sci (2018) 19(11):3448. doi: 10.3390/ijms19113448
48. Rosas G, Ruiz R, Araujo JM, Pinto JA, Mas L. ALK Rearrangements: Biology, Detection and Opportunities of Therapy in non-Small Cell Lung Cancer. Crit Rev Oncol/Hematol (2019) 136:48–55. doi: 10.1016/j.critrevonc.2019.02.006
49. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid Biopsy Enters the Clinic — Implementation Issues and Future Challenges. Nat Rev Clin Oncol (2021) 18(5):297–312. doi: 10.1038/s41571-020-00457-x
50. Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol (2019) 5(5):696–702. doi: 10.1001/jamaoncol.2018.7098
51. Jia Q, Chiu L, Wu S, Bai J, Peng L, Zheng L, et al. Tracking Neoantigens by Personalized Circulating Tumor DNA Sequencing During Checkpoint Blockade Immunotherapy in Non-Small Cell Lung Cancer. Adv Sci (Weinheim Baden-Wurttemberg Germany) (2020) 7(9):1903410. doi: 10.1002/advs.201903410
52. Eslami-S Z, Cortés-Hernández LE, Alix-Panabières C. The Metastatic Cascade as the Basis for Liquid Biopsy Development. Front Oncol (2020) 10:1055. doi: 10.3389/fonc.2020.01055
53. Esposito Abate R, Pasquale R, Fenizia F, Rachiglio AM, Roma C, Bergantino F, et al. The Role of Circulating Free DNA in the Management of NSCLC. Expert Rev Anticancer Ther (2019) 19(1):19–28. doi: 10.1080/14737140.2019.1548938
54. Vanni I, Coco S, Truini A, Rusmini M, Dal Bello MG, Alama A, et al. Next-Generation Sequencing Workflow for NSCLC Critical Samples Using a Targeted Sequencing Approach by Ion Torrent PGM™ Platform. Int J Mol Sci (2015) 16(12):28765–82. doi: 10.3390/ijms161226129
55. Dono M, De Luca G, Lastraioli S, Anselmi G, Dal Bello MG, Coco S, et al. Tag-Based Next Generation Sequencing: A Feasible and Reliable Assay for EGFR T790M Mutation Detection in Circulating Tumor DNA of non Small Cell Lung Cancer Patients. Mol Med (2019) 25(1):1–13. doi: 10.1186/s10020-019-0082-5
56. Normanno N, Denis MG, Thress KS, Ratcliffe M, Reck M. Guide to Detecting Epidermal Growth Factor Receptor (EGFR) Mutations in Ctdna of Patients With Advanced non-Small-Cell Lung Cancer. Oncotarget (2017) 8(7):12501. doi: 10.18632/oncotarget.13915
57. Kang Q, Henry NL, Paoletti C, Jiang H, Vats P, Chinnaiyan AM, et al. Comparative Analysis of Circulating Tumor DNA Stability in K3EDTA, Streck, and Cellsave Blood Collection Tubes. Clin Biochem (2016) 49(18):1354–60. doi: 10.1016/j.clinbiochem.2016.03.012
58. Zheng H, Zhan Y, Liu S, Lu J, Luo J, Feng J, et al. The Roles of Tumor-Derived Exosomes in non-Small Cell Lung Cancer and Their Clinical Implications. J Exp Clin Cancer Res (2018) 37(1):1–11. doi: 10.1186/s13046-018-0901-5
59. Vanni I, Alama A, Grossi F, Dal Bello MG, Coco S. Exosomes: A New Horizon in Lung Cancer. Drug Discov Today (2017) 22(6):927–36. doi: 10.1016/j.drudis.2017.03.004
60. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-Mediated Transfer of Mrnas and Micrornas is a Novel Mechanism of Genetic Exchange Between Cells. Nat Cell Biol (2007) 9(6):654–9. doi: 10.1038/ncb1596
61. Masaoutis C, Mihailidou C, Tsourouflis G, Theocharis S. Exosomes in Lung Cancer Diagnosis and Treatment. From translating Res into Future Clin Practice Biochimie (2018) 151:27–36. doi: 10.1016/j.biochi.2018.05.014
62. Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal Microrna: A Diagnostic Marker for Lung Cancer. Clin Lung Cancer (2009) 10(1):42–6. doi: 10.3816/CLC.2009.n.006
63. Logozzi M, Mizzoni D, Di Raimo R, Fais S. Exosomes: A Source for New and Old Biomarkers in Cancer. Cancers (2020) 12(9):2566. doi: 10.3390/cancers12092566
64. Kharaziha P, Ceder S, Li Q, Panaretakis T. Tumor Cell-Derived Exosomes: A Message in a Bottle. Biochim Biophys Acta (BBA)-Reviews Cancer (2012) 1826(1):103–11. doi: 10.1016/j.bbcan.2012.03.006
65. Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, et al. In Vivo Imaging Reveals Extracellular Vesicle-Mediated Phenocopying of Metastatic Behavior. Cell (2015) 161(5):1046–57. doi: 10.1016/j.cell.2015.04.042
66. Kahlert C, Kalluri R. Exosomes in Tumor Microenvironment Influence Cancer Progression and Metastasis. J Mol Med (2013) 91(4):431–7. doi: 10.1007/s00109-013-1020-6
67. Wu H, Zhou J, Mei S, Wu D, Mu Z, Chen B, et al. Circulating Exosomal Microrna-96 Promotes Cell Proliferation, Migration and Drug Resistance by Targeting LMO7. J Cell Mol Med (2017) 21(6):1228–36. doi: 10.1111/jcmm.13056
68. Kholia S, Ranghino A, Garnieri P, Lopatina T, Deregibus MC, Rispoli P, et al. Extracellular Vesicles as New Players in Angiogenesis. Vasc Pharmacol (2016) 86:64–70. doi: 10.1016/j.vph.2016.03.005
69. Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-Stranded DNA in Exosomes: A Novel Biomarker in Cancer Detection. Cell Res (2014) 24(6):766–9. doi: 10.1038/cr.2014.44
70. Yamashita T, Kamada H, Kanasaki S, Maeda Y, Nagano K, Abe Y, et al. Epidermal Growth Factor Receptor Localized to Exosome Membranes as a Possible Biomarker for Lung Cancer Diagnosis. Die Pharmazie-An Int J Pharm Sci (2013) 68(12):969–73.
71. Fortunato O, Gasparini P, Boeri M, Sozzi G. Exo-Mirnas as a New Tool for Liquid Biopsy in Lung Cancer. Cancers (2019) 11(6):888. doi: 10.3390/cancers11060888
72. Wu J, Shen Z. Exosomal Mirnas as Biomarkers for Diagnostic and Prognostic in Lung Cancer. Cancer Med (2020) 9(19):6909–22. doi: 10.1002/cam4.3379
73. Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial Expression of Autocrine VEGF Upon the Uptake of Tumor-Derived Microvesicles Containing Oncogenic EGFR. Proc Natl Acad Sci (2009) 106(10):3794–9. doi: 10.1073/pnas.0804543106
74. Fujita Y, Suda K, Kimura H, Matsumoto K, Arao T, Nagai T, et al. Highly Sensitive Detection of EGFR T790M Mutation Using Colony Hybridization Predicts Favorable Prognosis of Patients With Lung Cancer Harboring Activating EGFR Mutation. J Thorac Oncol (2012) 7(11):1640–4. doi: 10.1097/JTO.0b013e3182653d7f
75. Zhang Y, Roth JA, Yu H, Ye Y, Xie K, Zhao H, et al. A 5-Microrna Signature Identified From Serum Microrna Profiling Predicts Survival in Patients With Advanced Stage non-Small Cell Lung Cancer. Carcinogenesis (2019) 40(5):643–50. doi: 10.1093/carcin/bgy132
76. Jin H, Varner J. Integrins: Roles in Cancer Development and as Treatment Targets. Br J Cancer (2004) 90(3):561–5. doi: 10.1038/sj.bjc.6601576
77. Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of Rare Circulating Tumour Cells in Cancer Patients by Microchip Technology. Nature (2007) 450(7173):1235–9. doi: 10.1038/nature06385
78. Tanaka F, Yoneda K, Hasegawa S. Circulating Tumor Cells (Ctcs) in Lung Cancer: Current Status and Future Perspectives. Lung Cancer: Targets Ther (2010) 1:77. doi: 10.2147/LCTT.S6828
79. Krebs MG, Sloane R, Priest L, Lancashire L, Hou J-M, Greystoke A, et al. Evaluation and Prognostic Significance of Circulating Tumor Cells in Patients With non–Small-Cell Lung Cancer. J Clin Oncol (2011) 29(12):1556–63. doi: 10.1200/JCO.2010.28.7045
80. Alix-Panabières C, Pantel K. Characterization of Single Circulating Tumor Cells. FEBS Lett (2017) 591(15):2241–50. doi: 10.1002/1873-3468.12662
81. Lee JM, Dedhar S, Kalluri R, Thompson EW. The Epithelial–Mesenchymal Transition: New Insights in Signaling, Development, and Disease. J Cell Biol (2006) 172(7):973–81. doi: 10.1083/jcb.200601018
82. Zheng Q-M, Chen X-Y, Bao Q-F, Yu J, Chen L-H. ILK Enhances Migration and Invasion Abilities of Human Endometrial Stromal Cells by Facilitating the Epithelial–Mesenchymal Transition. Gynecol Endocrinol (2018) 34(12):1091–6. doi: 10.1080/09513590.2018.1498477
83. Herath S, Razavi Bazaz S, Monkman J, Ebrahimi Warkiani M, Richard D, O’Byrne K, et al. Circulating Tumor Cell Clusters: Insights Into Tumour Dissemination and Metastasis. Expert Rev Mol Diagn (2020) 20(11):1139–47. doi: 10.1080/14737159.2020.1846523
84. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell (2009) 139(5):871–90. doi: 10.1016/j.cell.2009.11.007
85. Alix-Panabières C, Pantel K. Circulating Tumor Cells: Liquid Biopsy of Cancer. Clin Chem (2013) 59(1):110–8. doi: 10.1373/clinchem.2012.194258
86. Alix-Panabières C, Pantel K. Challenges in Circulating Tumour Cell Research. Nat Rev Cancer (2014) 14(9):623–31. doi: 10.1038/nrc3820
87. Alix-Panabières C. EPISPOT Assay: Detection of Viable Dtcs/Ctcs in Solid Tumor Patients. Minimal Residual Dis Circulating Tumor Cells Breast Cancer (2012) 195:69–76. doi: 10.1007/978-3-642-28160-0_6
88. Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating Tumor Cells: Approaches to Isolation and Characterization. J Cell Biol (2011) 192(3):373–82. doi: 10.1083/jcb.201010021
89. Königsberg R, Obermayr E, Bises G, Pfeiler G, Gneist M, Wrba F, et al. Detection of Epcam Positive and Negative Circulating Tumor Cells in Metastatic Breast Cancer Patients. Acta Oncol (2011) 50(5):700–10. doi: 10.3109/0284186X.2010.549151
90. Kulasinghe A, Zhou J, Kenny L, Papautsky I, Punyadeera C. Capture of Circulating Tumour Cell Clusters Using Straight Microfluidic Chips. Cancers (2019) 11(1):89. doi: 10.3390/cancers11010089
91. Pantel K, Alix-Panabières C. Circulating Tumour Cells in Cancer Patients: Challenges and Perspectives. Trends Mol Med (2010) 16(9):398–406. doi: 10.1016/j.molmed.2010.07.001
92. Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben-Jacob E, et al. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front. Oncol (2015) 5:155. doi: 10.3389/fonc.2015.00155
93. Semaan A, Bernard V, Kim DU, Lee JJ, Huang J, Kamyabi N, et al. Characterisation of Circulating Tumour Cell Phenotypes Identifies a Partial-EMT Sub-Population for Clinical Stratification of Pancreatic Cancer. Br J Cancer (2021) 124(12):1970–7. doi: 10.1038/s41416-021-01350-9
94. Kulasinghe A, Perry C, Jovanovic L, Nelson C, Punyadeera C. Circulating Tumour Cells in Metastatic Head and Neck Cancers. Int J Cancer (2015) 136(11):2515–23. doi: 10.1002/ijc.29108
95. Au SH, Storey BD, Moore JC, Tang Q, Chen YL, Javaid S, et al. Clusters of Circulating Tumor Cells Traverse Capillary-Sized Vessels. Proc Natl Acad Sci U S A (2016) 113(18):4947–52. doi: 10.1073/pnas.1524448113
96. Kulasinghe A, Kapeleris J, Kimberley R, Mattarollo SR, Thompson EW, Thiery JP, et al. The Prognostic Significance of Circulating Tumor Cells in Head and Neck and Non-Small-Cell Lung Cancer. Cancer Med (2018) 7(12):5910–9. doi: 10.1002/cam4.1832
97. Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, et al. Neutrophils Escort Circulating Tumour Cells to Enable Cell Cycle Progression. Nature (2019) 566(7745):553–7. doi: 10.1038/s41586-019-0915-y
98. Ding L, Radfar P, Rezaei M, Warkiani ME. An Easy-to-Operate Method for Single-Cell Isolation and Retrieval Using a Microfluidic Static Droplet Array. Microchimica Acta (2021) 188(8):242. doi: 10.1007/s00604-021-04897-9
99. Zeinali M, Lee M, Nadhan A, Mathur A, Hedman C, Lin E, et al. High-Throughput Label-Free Isolation of Heterogeneous Circulating Tumor Cells and CTC Clusters From Non-Small-Cell Lung Cancer Patients. Cancers (2020) 12(1):127. doi: 10.3390/cancers12010127
100. Rostami P, Kashaninejad N, Moshksayan K, Saidi MS, Firoozabadi B, Nguyen N-T. Novel Approaches in Cancer Management With Circulating Tumor Cell Clusters. J Sci: Adv Mater Dev (2019) 4(1):1–18. doi: 10.1016/j.jsamd.2019.01.006
101. Alshareef M, Metrakos N, Juarez Perez E, Azer F, Yang F, Yang X, et al. Separation of Tumor Cells With Dielectrophoresis-Based Microfluidic Chip. Biomicrofluidics (2013) 7(1):011803. doi: 10.1063/1.4774312
102. Tang Y, Shi J, Li S, Wang L, Cayre YE, Chen Y. Microfluidic Device With Integrated Microfilter of Conical-Shaped Holes for High Efficiency and High Purity Capture of Circulating Tumor Cells. Sci Rep (2014) 4(1):1–7. doi: 10.1038/srep06052
103. Wan S, Kim TH, Smith KJ, Delaney R, Park G-S, Guo H, et al. New Labyrinth Microfluidic Device Detects Circulating Tumor Cells Expressing Cancer Stem Cell Marker and Circulating Tumor Microemboli in Hepatocellular Carcinoma. Sci Rep (2019) 9(1):1–11. doi: 10.1038/s41598-019-54960-y
104. Khoo BL, Warkiani ME, Tan DS-W, Bhagat AAS, Irwin D, Lau DP, et al. Clinical Validation of an Ultra High-Throughput Spiral Microfluidics for the Detection and Enrichment of Viable Circulating Tumor Cells. PLoS One (2014) 9(7):e99409. doi: 10.1371/journal.pone.0099409
105. Fachin F, Spuhler P, Martel-Foley JM, Edd JF, Barber TA, Walsh J, et al. Monolithic Chip for High-Throughput Blood Cell Depletion to Sort Rare Circulating Tumor Cells. Sci Rep (2017) 7(1):1–11. doi: 10.1038/s41598-017-11119-x
106. Zhou J, Kulasinghe A, Bogseth A, O’Byrne K, Punyadeera C, Papautsky I. Isolation of Circulating Tumor Cells in non-Small-Cell-Lung-Cancer Patients Using a Multi-Flow Microfluidic Channel. Microsystems Nanoeng (2019) 5(1):1–12. doi: 10.1038/s41378-019-0045-6
107. Wang C, Mu Z, Chervoneva I, Austin L, Ye Z, Rossi G, et al. Longitudinally Collected Ctcs and CTC-Clusters and Clinical Outcomes of Metastatic Breast Cancer. Breast Cancer Res Treat (2017) 161(1):83–94. doi: 10.1007/s10549-016-4026-2
108. Kulasinghe A, Perry C, Kenny L, Warkiani ME, Nelson C, Punyadeera C. PD-L1 Expressing Circulating Tumour Cells in Head and Neck Cancers. BMC Cancer (2017) 17(1):1–6. doi: 10.1186/s12885-017-3316-3
109. Mazel M, Jacot W, Pantel K, Bartkowiak K, Topart D, Cayrefourcq L, et al. Frequent Expression of PD-L1 on Circulating Breast Cancer Cells. Mol Oncol (2015) 9(9):1773–82. doi: 10.1016/j.molonc.2015.05.009
110. Freitas C, Sousa C, Machado F, Serino M, Santos V, Cruz-Martins N, et al. The Role of Liquid Biopsy in Early Diagnosis of Lung Cancer. Front Oncol (2021) 11:1130. doi: 10.3389/fonc.2021.634316
111. Esquela K, Slack J. Oncomirs Mirnas With a Role in Cancer. Nat Rev Cancer (2006) 6:259–569. doi: 10.1038/nrc1840
112. Ambros V. The Functions of Animal Micrornas. Nature (2004) 431(7006):350–5. doi: 10.1038/nature02871
113. Lim L, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray Analysis Shows That Some Micrornas Downregulate Large Numbers of Target Mrnas. Nature (2005) 433:769–73. doi: 10.1038/nature03315
114. Krek A, Grü n D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial Microrna Target Predictions. Nat Genet (2005) 37:495–500. doi: 10.1038/ng1536
115. Zhang B, Pan X, Cobb GP, Anderson TA. Micrornas as Oncogenes and Tumor Suppressors. Dev Biol (2007) 302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028
116. Turchinovich A, Weiz L, Burwinkel B. Extracellular Mirnas: The Mystery of Their Origin and Function. Trends Biochem Sci (2012) 37(11):460–5. doi: 10.1016/j.tibs.2012.08.003
117. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 Complexes Carry a Population of Circulating Micrornas Independent of Vesicles in Human Plasma. Proc Natl Acad Sci (2011) 108(12):5003–8. doi: 10.1073/pnas.1019055108
118. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. Micrornas are Transported in Plasma and Delivered to Recipient Cells by High-Density Lipoproteins. Nat Cell Biol (2011) 13(4):423–33. doi: 10.1038/ncb2210
119. Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, et al. Serum Microrna Signatures Identified in a Genome-Wide Serum Microrna Expression Profiling Predict Survival of non-Small-Cell Lung Cancer. J Clin Oncol (2010) 28(10):1721–6. doi: 10.1200/JCO.2009.24.9342
120. Pigati L, Yaddanapudi SC, Iyengar R, Kim D-J, Hearn SA, Danforth D, et al. Selective Release of Microrna Species From Normal and Malignant Mammary Epithelial Cells. PLoS One (2010) 5(10):e13515. doi: 10.1371/journal.pone.0013515
121. Okada H, Kohanbash G, Lotze MT. Micrornas in Immune Regulation—Opportunities for Cancer Immunotherapy. Int J Biochem Cell Biol (2010) 42(8):1256–61. doi: 10.1016/j.biocel.2010.02.002
122. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating Liquid Biopsies Into the Management of Cancer. Nat Rev Clin Oncol (2017) 14(9):531–48. doi: 10.1038/nrclinonc.2017.14
123. Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical Relevance of Circulating Cell-Free Micrornas in Cancer. Nat Rev Clin Oncol (2014) 11(3):145–56. doi: 10.1038/nrclinonc.2014.5
124. Nagasaka M, Uddin MH, Al-Hallak MN, Rahman S, Balasubramanian S, Sukari A, et al. Liquid Biopsy for Therapy Monitoring in Early-Stage non-Small Cell Lung Cancer. Mol Cancer (2021) 20(1):1–16. doi: 10.1186/s12943-021-01371-1
125. Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R, et al. Circulating Micrornas in Cancer: Potential and Challenge. Front Genet (2019) 10:626. doi: 10.3389/fgene.2019.00626
126. Wang P, Yang D, Zhang H, Wei X, Ma T, Cheng Z, et al. Early Detection of Lung Cancer in Serum by a Panel of Microrna Biomarkers. Clin Lung Cancer (2015) 16(4):313–9.e1. doi: 10.1016/j.cllc.2014.12.006
127. Montani F, Marzi MJ, Dezi F, Dama E, Carletti RM, Bonizzi G, et al. Mir-Test: A Blood Test for Lung Cancer Early Detection. JNCI: J Natl Cancer Institute (2015) 107(6):djv063. doi: 10.1093/jnci/djv063
128. Wozniak MB, Scelo G, Muller DC, Mukeria A, Zaridze D, Brennan P. Circulating Micrornas as Non-Invasive Biomarkers for Early Detection of Non-Small-Cell Lung Cancer. PLoS One (2015) 10(5):e0125026. doi: 10.1371/journal.pone.0125026
129. Umu SU, Langseth H, Bucher-Johannessen C, Fromm B, Keller A, Meese E, et al. A Comprehensive Profile of Circulating Rnas in Human Serum. RNA Biol (2018) 15(2):242–50. doi: 10.1080/15476286.2017.1403003
130. Yang Y, Hu Z, Zhou Y, Zhao G, Lei Y, Li G, et al. The Clinical Use of Circulating Micrornas as non-Invasive Diagnostic Biomarkers for Lung Cancers. Oncotarget (2017) 8(52):90197. doi: 10.18632/oncotarget.21644
131. Li W, Wang Y, Zhang Q, Tang L, Liu X, Dai Y, et al. Microrna-486 as a Biomarker for Early Diagnosis and Recurrence of Non-Small Cell Lung Cancer. PLoS One (2015) 10(8):e0134220. doi: 10.1371/journal.pone.0134220
132. Wang Y, Gu J, Roth JA, Hildebrandt MA, Lippman SM, Ye Y, et al. Pathway-Based Serum Microrna Profiling and Survival in Patients With Advanced Stage Non–Small Cell Lung Cancer. Cancer Res (2013) 73(15):4801–9. doi: 10.1158/0008-5472.CAN-12-3273
133. Fan L, Qi H, Teng J, Su B, Chen H, Wang C, et al. Identification of Serum Mirnas by Nano-Quantum Dots Microarray as Diagnostic Biomarkers for Early Detection of Non-Small Cell Lung Cancer. Tumor Biol (2016) 37(6):7777–84. doi: 10.1007/s13277-015-4608-3
134. Marques JF, Junqueira-Neto S, Pinheiro J, Machado JC, Costa JL. Induction of Apoptosis Increases Sensitivity to Detect Cancer Mutations in Plasma. Eur J Cancer (2020) 127:130–8. doi: 10.1016/j.ejca.2019.12.023
135. Abbosh C, Birkbak NJ, Swanton C. Early Stage NSCLC—Challenges to Implementing Ctdna-Based Screening and MRD Detection. Nat Rev Clin Oncol (2018) 15(9):577–86. doi: 10.1038/s41571-018-0058-3
136. Chemi F, Rothwell DG, McGranahan N, Gulati S, Abbosh C, Pearce SP, et al. Pulmonary Venous Circulating Tumor Cell Dissemination Before Tumor Resection and Disease Relapse. Nat Med (2019) 25(10):1534–9. doi: 10.1038/s41591-019-0593-1
137. Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, et al. The Identification and Characterization of Breast Cancer Ctcs Competent for Brain Metastasis. Sci Trans Med (2013) 5(180):180ra48–ra48. doi: 10.1126/scitranslmed.3005109
138. Lomnytska M, Pinto R, Becker S, Engström U, Gustafsson S, Björklund C, et al. Platelet Protein Biomarker Panel for Ovarian Cancer Diagnosis. Biomark Res (2018) 6(1):1–14. doi: 10.1186/s40364-018-0118-y
139. Chen R, Xu X, Qian Z, Zhang C, Niu Y, Wang Z, et al. The Biological Functions and Clinical Applications of Exosomes in Lung Cancer. Cell Mol Life Sci (2019) 76(23):4613–33. doi: 10.1007/s00018-019-03233-y
140. Page C, Pitchford S. Platelets and Allergic Inflammation. Clin Exp Allergy (2014) 44(7):901–13. doi: 10.1111/cea.12322
141. Wojtukiewicz MZ, Sierko E, Hempel D, Tucker SC, Honn KV. Platelets and Cancer Angiogenesis Nexus. Cancer Metastasis Rev (2017) 36(2):249–62. doi: 10.1007/s10555-017-9673-1
142. Heeke S, Mograbi B, Alix-Panabières C, Hofman P. Never Travel Alone: The Crosstalk of Circulating Tumor Cells and the Blood Microenvironment. Cells (2019) 8(7):714. doi: 10.3390/cells8070714
143. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical Features of Patients Infected With 2019 Novel Coronavirus in Wuhan, China. Lancet (2020) 395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5
144. Suzuki A, Takahashi T, Nakamura K, Tsuyuoka R, Okuno Y, Enomoto T, et al. Thrombocytosis in Patients With Tumors Producing Colony-Stimulating Factor. Blood (1992) 80(8):2052–9. doi: 10.1182/blood.V80.8.2052.bloodjournal8082052
145. Sadeghi Rad H, Monkman J, Warkiani ME, Ladwa R, O’Byrne K, Rezaei N, et al. Understanding the Tumor Microenvironment for Effective Immunotherapy. Med Res Rev (2021) 41(3):1474–98. doi: 10.1002/med.21765
146. Kalita-de Croft P, Sadeghi Rad H, Gasper H, O’Byrne K, Lakhani SR, Kulasinghe A. Spatial Profiling Technologies and Applications for Brain Cancers. Expert Rev Mol Diagn (2021) 21(3):323–32. doi: 10.1080/14737159.2021.1900735
147. Li D, Yang W, Zhang Y, Yang JY, Guan R, Xu D, et al. Genomic Analyses Based on Pulmonary Adenocarcinoma in Situ Reveal Early Lung Cancer Signature. BMC Med Genomics (2018) 11(5):89–97. doi: 10.1186/s12920-018-0413-3
148. Liefaard M, Lips E, Best M, Sol N, Rookus M, Sonke G, et al. RNA Signatures From Tumor-Educated Platelets (TEP) Enable Detection of Early-Stage Breast Cancer. Ann Oncol (2019) 30:iii13. doi: 10.1093/annonc/mdz095.036
149. Xue L, Xie L, Song X, Song X. Identification of Potential Tumor-Educated Platelets RNA Biomarkers in non-Small-Cell Lung Cancer by Integrated Bioinformatical Analysis. J Clin Lab Anal (2018) 32(7):e22450. doi: 10.1002/jcla.22450
150. Sheng M, Dong Z, Xie Y. Identification of Tumor-Educated Platelet Biomarkers of non-Small-Cell Lung Cancer. OncoTargets Ther (2018) 11:8143. doi: 10.2147/OTT.S177384
151. Sabrkhany S, Kuijpers MJ, van Kuijk SM, Sanders L, Pineda S, Damink SWO, et al. A Combination of Platelet Features Allows Detection of Early-Stage Cancer. Eur J Cancer (2017) 80:5–13. doi: 10.1016/j.ejca.2017.04.010
152. Plantureux L, Mège D, Crescence L, Dignat-George F, Dubois C, Panicot-Dubois L. Impacts of Cancer on Platelet Production, Activation and Education and Mechanisms of Cancer-Associated Thrombosis. Cancers (2018) 10(11):441. doi: 10.3390/cancers10110441
153. Best M, Sol N, Kooi I, Tannous B, Wesseling P. RNA-Seq of Tumor-Educated Platelets Enables Article RNA-Seq of Tumor-Educated Platelets Enables. Cancer Cell (2015) 28:666–76. doi: 10.1016/j.ccell.2015.09.018
154. Calverley DC, Phang TL, Choudhury QG, Gao B, Oton AB, Weyant MJ, et al. Significant Downregulation of Platelet Gene Expression in Metastatic Lung Cancer. Clin Trans Sci (2010) 3(5):227–32. doi: 10.1111/j.1752-8062.2010.00226.x
155. Rodig SJ, Mino-Kenudson M, Dacic S, Yeap BY, Shaw A, Barletta JA, et al. Unique Clinicopathologic Features Characterize ALK-Rearranged Lung Adenocarcinoma in the Western Population. Clin Cancer Res (2009) 15(16):5216–23. doi: 10.1158/1078-0432.CCR-09-0802
156. Dacic S. Molecular Genetic Testing for Lung Adenocarcinomas: A Practical Approach to Clinically Relevant Mutations and Translocations. J Clin Pathol (2013) 66(10):870–4. doi: 10.1136/jclinpath-2012-201336
157. Nilsson RJA, Karachaliou N, Berenguer J, Gimenez-Capitan A, Schellen P, Teixido C, et al. Rearranged EML4-ALK Fusion Transcripts Sequester in Circulating Blood Platelets and Enable Blood-Based Crizotinib Response Monitoring in non-Small-Cell Lung Cancer. Oncotarget (2016) 7(1):1066. doi: 10.18632/oncotarget.6279
158. Sol N, Wurdinger T. Platelet RNA Signatures for the Detection of Cancer. Cancer Metastasis Rev (2017) 36(2):263–72. doi: 10.1007/s10555-017-9674-0
159. Xing S, Zeng T, Xue N, He Y, Y-z L, Li H-l, et al. Development and Validation of Tumor-Educated Blood Platelets Integrin Alpha 2b (ITGA2B) RNA for Diagnosis and Prognosis of Non-Small-Cell Lung Cancer Through RNA-Seq. Int J Biol Sci (2019) 15(9):1977. doi: 10.7150/ijbs.36284
Keywords: Circulating tumour cells, cell-free DNA, circulating tumour DNA, microRNA and exosomes, lung cancer, liquid biopsy
Citation: Herath S, Sadeghi Rad H, Radfar P, Ladwa R, Warkiani M, O’Byrne K and Kulasinghe A (2022) The Role of Circulating Biomarkers in Lung Cancer. Front. Oncol. 11:801269. doi: 10.3389/fonc.2021.801269
Received: 25 October 2021; Accepted: 21 December 2021;
Published: 21 January 2022.
Edited by:
Pasquale Pisapia, University of Naples Federico II, ItalyReviewed by:
Simon Chang-Hao Tsao, The University of Melbourne, AustraliaSandra V. Fernandez, Fox Chase Cancer Center, United States
Copyright © 2022 Herath, Sadeghi Rad, Radfar, Ladwa, Warkiani, O’Byrne and Kulasinghe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arutha Kulasinghe, YXJ1dGhhLmt1bGFzaW5naGVAdXEuZWR1LmF1
 Sayuri Herath1
Sayuri Herath1 Habib Sadeghi Rad
Habib Sadeghi Rad Majid Warkiani
Majid Warkiani Ken O’Byrne
Ken O’Byrne Arutha Kulasinghe
Arutha Kulasinghe