- Department of Gastroenterology, The Affiliated Hospital of Qingdao University, Qingdao, China
Cancer is a major threat to human health and longevity. Chemotherapy is an effective approach to inhibit cancer cell proliferation, but a growing number of cancer patients are prone to develop resistance to various chemotherapeutics, including platinum, paclitaxel, adriamycin, and 5-fluorouracil, among others. Significant progress has been made in the research and development of chemotherapeutic drugs over the last few decades, including targeted therapy drugs and immune checkpoint inhibitors; however, drug resistance still severely limits the application and efficacy of these drugs in cancer treatment. Recently, emerging studies have emphasized the role of circular RNAs (circRNAs) in the proliferation, migration, invasion, and especially chemoresistance of cancer cells by regulating the expression of related miRNAs and targeted genes. In this review, we comprehensively summarized the potential roles and mechanisms of circRNAs in cancer drug resistance including the efflux of drugs, apoptosis, intervention with the TME (tumor microenvironment), autophagy, and dysfunction of DNA damage repair, among others. Furthermore, we highlighted the potential value of circRNAs as new therapeutic targets and prognostic biomarkers for cancer.
Introduction
Cancer is a worldwide public health problem and a leading cause of premature death (1). The main reasons for the high mortality of patients and poor cancer prognosis are late diagnosis, tumor invasion and metastasis, and resistance to chemotherapeutic drugs. Resistance to chemotherapy, which involves intrinsic resistance and acquired resistance, has become a serious obstacle towards cancer therapy (2).
Although recent breakthroughs in treatment have contributed to the decline in cancer mortality, such as combined administration of drugs, checkpoint blockade immunotherapies, and targeted therapies for malignant tumors, tumor cells still exhibit a tendency towards drug resistance (1, 3). This becomes problematic as cancer cells become cross-resistant to several drugs with different anti-tumor mechanisms, causing invalid effects of various combination chemotherapies (4). Immune checkpoint inhibitors including programmed cell death 1 (PD1), PD1 ligand 1 (PD- L1), and cytotoxic T lymphocyte antigen 4 (CTLA4) have been successfully used in clinical applications. However, resistance to immune checkpoint blockade appeared simultaneously. For instance, it has been found that STK11/LKB1 mutations act as the main driver of immune escape and intrinsic resistance to PD-1 blockade in KRAS-mutant lung adenocarcinoma (LUAD) (5). Additionally, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) have been extensively studied as key targeted therapies, and within one or two years after treatment with TKIs, patients with activating alterations of the EGFR gene often acquire resistance to TKI therapy (2). Therefore, an insight into the specific molecular mechanisms that mediate drug resistance is crucial for understanding and overcoming drug tolerance in cancers.
Circular RNAs (circRNAs) were first identified in 1976 and belong to a growing list of types of non-coding RNAs, with a circular loop structure (6, 7). Most circRNAs, comprising a single exon or multiple exons, are expressed by known protein-coding genes (8). circRNAs can modulate gene splicing of pre-RNA and transcription, regulate RNA-binding proteins, and act as microRNA (miRNA) “sponges” to play a crucial role in transcriptional regulation (9, 10). Recent studies have revealed that numerous circRNAs are differentially expressed in various cancer patients and are correlated with the tumorigenesis and progression of cancers (11). For example, autophagy-associated circCDYL promotes the malignant progression of breast cancer cells and suppresses the clinical response to chemotherapy in patients with breast cancer (12). Conversely, circ-HuR suppresses the expression of CNBP-facilitated HuR and the progression of gastric cancer, serving as a tumor inhibitor (13). Moreover, accumulating evidence has shown that circRNAs are correlated with tumor chemoresistance and may play a crucial role in the occurrence and regulation of chemoresistance (14). To investigate the emerging role of circRNAs in the drug resistance of cancers, we systematically and comprehensively summarized the major mechanisms of cancer chemoresistance and the molecular mechanisms by which circRNAs enhance or suppress drug resistance in cancers (Figure 1), which indicates that circRNAs may function as potential biomarkers and therapeutic targets for cancer.
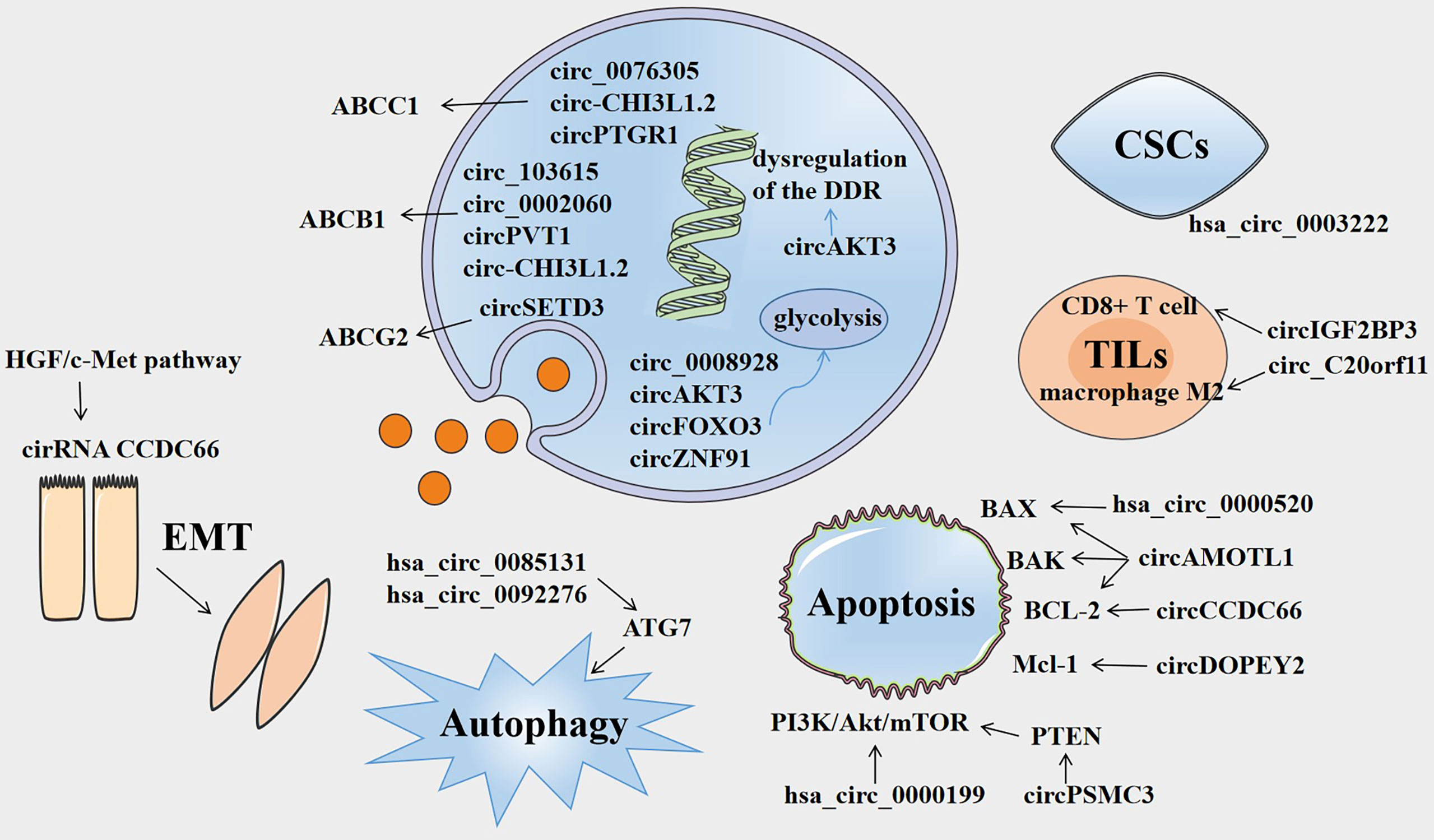
Figure 1 Overview of the involvement of circRNAs in cancer drug resistance. Several circRNAs are involved in drug resistance by influencing ABC efflux transporters, dysregulation of DNA damage response (DDR), glycolysis, epithelial-mesenchymal transition (EMT), autophagy, apoptosis, tumor-infiltrating lymphocytes (TILs), and cancer stem cells (CSCs).
Mechanisms of Drug Resistance
Efflux of Drugs
Drug efflux from cancer cells is a common and important mechanism of resistance or multidrug resistance (MDR), which is inseparable from ATP-binding cassette (ABC) efflux transporters, including MDR-associated protein 2 (MRP2/ABCC2), P-glycoprotein (P-gp/ABCB1), and breast cancer resistance protein (BCRP/ABCG2) (15). ABC efflux transporters, as membrane protein complexes, are often overexpressed in cancer cells and form a defense system against chemotherapeutic drugs as well as a variety of cytotoxic agents, which greatly restricts the effective application of chemotherapy. For example, P-glycoprotein (P-gp) has a wide substrate spectrum that mediates the export of a multitude of drugs, including antibiotics, immunosuppressive agents, and chemotherapeutic drugs (16).
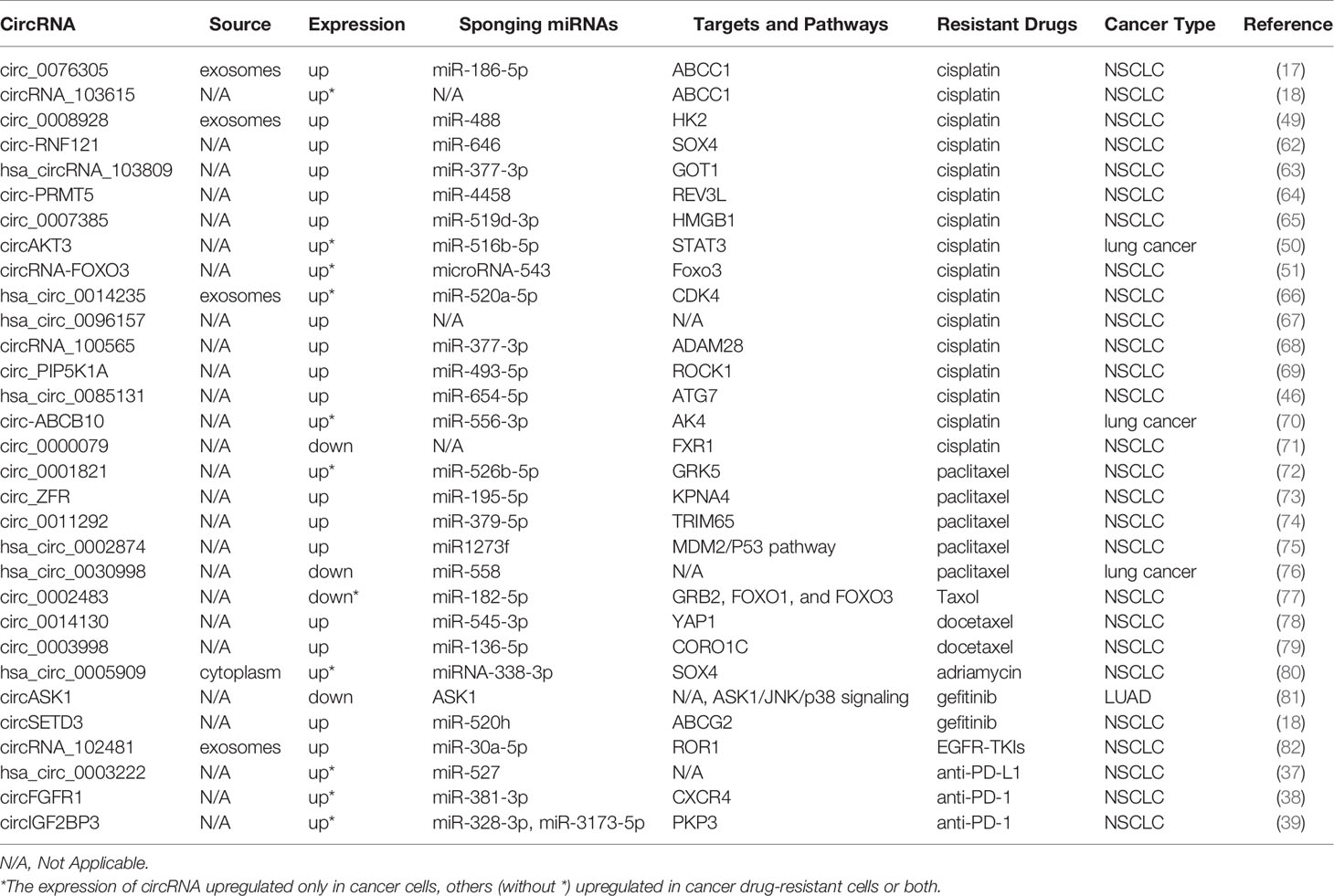
Several researchers have discovered that the function of some circRNAs in drug resistance of cancers is related to ABC efflux transporters. For example, it has been found that circ_0076305 enhances ABCC1 expression by sponging miR-186-5p, thus regulating cisplatin (CDDP) resistance in non-small cell lung cancer (NSCLC) (17). Similarly, ABCB1 overexpression can reverse the effects of circRNA_103615 silencing on CDDP resistance in NSCLC (18). CircSETD3 upregulates the expression of the ABCG2 transporter by binding to miR-520h, mediating gefitinib to be pumped out of NSCLC cells (19). Both circ_0002060 and circPVT1 contribute to drug resistance in osteosarcoma cells by regulating the expression of ABCB1 (20, 21). Additionally, the knockdown of circ-CHI3L1.2 downregulates the expression levels of P-gp, MRP1, and GSTP1 and weakens CDDP resistance in osteosarcoma (22). CircPTGR1 and ABCC1 levels are significantly overexpressed in hepatocellular carcinoma (HCC) cells, and circPTGR1 modulates the 5-FU resistance of HCC cells via the miR-129-5p/ABCC1 axis (23). Therefore, it is crucial to explore the relationships among circRNAs, ABC efflux transporters, and tolerance to anti-cancer drugs in order to find new therapeutic targets for cancer.
Apoptosis
The major aim of cancer chemotherapy is to promote the apoptosis of cancer cells and expose them to anti-cancer drugs. The effector phase of apoptosis involves several pro-apoptotic proteins (e.g., BAX, Bak, Bad) and anti-apoptotic proteins (e.g., BCL-2, BCL-XL, BCL-W) (24). However, the genes in cancer cells commonly demonstrate mutations, including apoptotic genes, which often cause dysfunction. This may result in the occurrence of resistance to chemotherapeutics, as any interference that mediates the activation of anti-apoptotic pathways or suppression of pro-apoptotic signal transduction is a potential mechanism of drug resistance (25).
Certain circRNAs have been shown to regulate the apoptosis of drug-resistant cancer cells by modulating pro- or anti-apoptotic proteins. For example, circAMOTL1 can significantly modulate the expression of the Protein Kinase B (AKT) as well as AKT-related pro-apoptotic (BAX and BAK) factors and anti-apoptotic (BCL-2) proteins, thus mediating the paclitaxel (PTX)-resistant effects in breast cancer (26). CircCCDC66 inhibits apoptosis by targeting miR-618 and resists the release of BCL-2, which is an essential regulator of CDDP resistance in gastric cancer (27). Moreover, hsa_circ_0000520 overexpression increases the expression of the BAX protein and reduces the expression of p-PI3K and p-Akt proteins, ultimately reversing the Herceptin resistance of gastric cancer (28). Re-expression of cDOPEY2 decreases the expression of the anti-apoptotic protein Mcl-1 and substantially strengthens the cell lethality of CDDP by augmenting the apoptotic process in CDDP-resistant esophageal squamous cell carcinoma (ESCC) cells (29). The PI3K/Akt/mTOR signaling pathway plays a crucial role in cell cycle regulation, including cell survival, proliferation, and metabolism, and is intimately related to autophagy and apoptosis (30). Based on the existing literature, PI3K-AKT-mTOR signaling is extensively implicated in chemoresistance and drives the process of malignant tumors (31). Silencing of hsa_circ_0000199 deactivates PI3K/Akt/mTOR signaling to promote apoptosis, enhancing triple-negative breast cancer (TNBC) chemosensitivity (32). PTEN is an important tumor suppressor gene that encodes a phosphatase protein and resists the PI3K/Akt/mTOR anti-apoptotic pathway as an antagonist (33). It has been reported that PTEN expression can be promoted by circPSMC3 by decreasing miR-10a-5p levels, which enhances the esophageal squamous cell carcinoma (ESCC) cells towards gefitinib sensitivity (34).
TME
TME consists of the stromal cells, immune cells, and extracellular matrix and mounting evidence suggests that the TME plays a crucial role in multiple aspects of tumor progression, particularly in therapeutic resistance (35). Acquired resistance induced by the TME primarily acts as an adaptive response by the host towards pharmacological damage (36). Li et al. explored the combined effects of cancer stem cells (CSCs), circRNA (hsa_circ_0003222), and immune checkpoint blockers in NSCLC malignant behavior as well as drug resistance and found that NSCLC resistance to anti-PD-L1-based therapy could be reduced by silencing hsa_circ_0003222 (37). Additionally, circFGFR1 promotes immune evasion of NSCLC cells and enhances tolerance to anti- PD-1- based therapy by interacting with miR-381-3p and upregulating the expression of C-X-C motif chemokine receptor 4 (CXCR4) (38). Tumor-infiltrating lymphocytes (TILs), an important component of the TME, have attracted increasing attention for therapy resistance in recent years. CircIGF2BP3 expression is negatively associated with the infiltration of CD8+ T cells, which induces immune escape from CD8+ T cell-mediated killing. Mechanistically, PKP3 upregulated by circIGF2BP3 combines with FXR1 to stabilize OTUB1 mRNA, which increases PD-L1 abundance by promoting its deubiquitination (39). A study on ovarian cancer has suggested that silencing of circ_C20orf11 suppresses extracellular vesicle (EV)-induced macrophage M2 polarization and enhances sensitivity to CDDP in vivo (40).
In addition, the interactions between cancer cells and TME also play a pivotal role in epithelial-mesenchymal transition (EMT), which has emerged as a significant cancer cell behavior correlated with metastatic potential and chemoresistance (41). The EMT-associated NF-κB/HER2/STAT3 pathway is crucial in radioresistance of breast cancer stem cells (42). However, the relationship between circRNAs and EMT in drug resistance still needs to be investigated. A recent study has found that the HGF/c-Met pathway regulates the expression of circCCDC66 and SAE2 to promote EMT and chemoresistance in lung adenocarcinoma (LADC) cells (43). In order to overcome this resistance, disruption of EMT-related pathways in tumor cells is desirable.
Other Mechanisms
Many other drug resistance mechanisms of cancers, including autophagy, glycolysis, dysfunction of DNA damage repair, altered drug targets, decreased drug influx, and so forth are known (14). However, the relationship between circRNAs and the above mentioned drug resistance mechanisms are still being investigated. Autophagy plays a crucial role in tumorigenesis, progression, and therapeutic intervention in cancers (44). A previous study has found that the knockdown of autophagy-related genes (ATG5 and ATG7) enhances therapeutic cell killing, indicating that autophagy may promote the acquired resistance of cancer cells to chemotherapeutics (45). Hsa_circ_0085131 and hsa_circ_0092276 serve as competitive endogenous RNAs (ceRNAs) to elevate autophagy-associated factor ATG7 expression, thus enhancing CDDP resistance in NSCLC and doxorubicin (DOX) resistance in breast cancer (46, 47). Tumor cells often switch to glycolysis, even under aerobic conditions, and components of the glycolytic pathway, including transporters, enzymes, and metabolites, are involved in inducing drug resistance (48). Some circRNAs mediate chemotherapy resistance by modulating glycolysis in tumor cells. For instance, circ_0008928 silencing enhances CDDP sensitivity in CDDP-resistant NSCLC cells and impedes NSCLC progression and glycolysis by upregulating miR-488 expression and downregulating HK2 expression (49). Similarly, the CDDP-resistance mechanism of circAKT3 and circRNA-FOXO3 is involved in disturbing the glycolysis balance (50, 51). CDDP sensitivity is suppressed partly by circAKT3 by modulating the miR-516b-5p/STAT3 axis in lung cancer cells, whereas CDDP sensitivity is promoted by circRNA-FOXO3 via the miR-543/Foxo3 axis in NSCLC cells. The hypoxia-induced exosomal circZNF91 transferred to normoxic pancreatic cancer cells can interact with miR-23b-3p and upregulate the expression of the deacetylase sirtuin1 (SIRT1), thereby enhancing the deacetylation-dependent stability of HIF-1α, resulting in gemcitabine (GEM) resistance and glycolysis-induced chemoresistance (52). DNA damage is a relatively common and vital cellular event that is implicated in mutations, metabolic dysfunction, cellular or organismic death, and tumorigenesis (53). Upregulation of processes such as DNA damage tolerance (DDT) and DNA damage response (DDR) is advantageous to cancer cells because it allows them to resist damage lesions (54). A large number of processes in cancers activate cellular DDR to remove or repair DNA lesions (54). Dysregulation of DDR is often the route by which tumor cells evade chemotherapy (55). A recent study found that circAKT3 had an impact on DDR in gastric cancer cells and might promote CDDP resistance in gastric cancer through the DDR and PI3K/AKT pathways (56). Alterations in chemotherapeutic targets can have a great influence on drug resistance. Alterations in DNA topoisomerase-II (Topo-II) activity, such as topo-II mutations or downregulation of topo-II protein, can result in resistance to topo-II-targeted drugs, including anthracyclines (57). As for drug influx, the mechanism by which only a small amount of chemotherapeutics enters cells has been elucidated. For instance, methotrexate (MTX), a dihydrofolate reductase (DHFR) inhibitor, enters the cell predominantly via the decreased folate carrier (RFC) (58). Decreased expression and inactivating alterations of the RFC are documented mechanisms of MTX resistance (59). Taken together, diverse mechanisms of drug resistance have been discovered, however more detailed mechanisms of chemoresistance are still largely unknown and require further investigation.
CircRNAs and Drug Resistance
CircRNAs and Lung Cancer Drug Resistance
According to the global cancer statistics of 2020, lung cancer remains a major cause of cancer-related deaths (60). CDDP is one of the most effective anti-cancer drugs and is extensively used in the clinic; however, the development of CDDP resistance seriously hinders the therapeutic effect of cancer (61). The involvement of circRNAs in drug resistance in lung cancer is shown in Table 1. Accumulating evidence has demonstrated that circRNAs are implicated in CDDP resistance in lung cancer. Knockdown or silencing of certain circRNAs suppresses CDDP resistance in NSCLC such as circ-RNF121, circ_0076305, circRNA_103615, circ_0008928, hsa_circRNA_103809, circ-PRMT5, and circ_0007385 (17, 18, 49, 62–65). Additionally, circAKT3 inhibits glycolysis balance and enhances CDDP resistance, whereas circRNA-FOXO3 accelerates glycolysis and promotes CDDP sensitivity (50, 51). Both hsa_circ_0014235 and hsa_circ_0096157 overexpression intensifies the CDDP resistance and facilitates the malignancy of NSCLC cells, including proliferation, migration, and invasion (66, 67). circRNA_100565 is correlated with a poor prognosis of NSCLC, and circRNA_100565 deletion mitigates CDDP resistance, which can be attenuated by miR-377-3p inhibition or ADAM28 overexpression (68). circ_PIP5K1A regulates CDDP sensitivity via the miR-493-5p/ROCK1 axis, and circ_0085131 enhances NSCLC cell drug resistance by targeting miR-654-5p to upregulate ATG7 expression (46, 69). Additionally, the sensitivity of CDDP can be promoted by the knockdown of circ-ABCB10 in lung cancer cells (70). However, circ_0000079 overexpression disturbs the formation of the FXR1/PRCKI complex by regulating FXR1, thereby suppressing cell invasion and CDDP resistance in NSCLC (71).
PTX is also an anti-neoplastic agent widely used to treat several solid tumor types, including lung cancer. Knockdown of circ_0001821 and circ_ZFR reduces the proliferation and metastasis ability but enhances PTX sensibility and apoptosis of NSCLC cells by downregulating the expression of GRK5 and KPNA4 by sponging related miRNAs (72, 73). Similarly, circ_0011292 modulates the miR-379-5p/TRIM65 axis to promote tumorigenesis and enhance PTX resistance in NSCLC (74). It has also been found that hsa_circ_0002874 downregulation can reverse the PTX resistance of NSCLC and induce apoptosis by regulating the miR1273f/MDM2/P53 signaling pathway (75). In contrast to the above circRNAs, circ_0030998 and circ_0002483 overexpression decreases the malignant behavior of lung cancer cells and enhances their sensitivity to PTX by targeting miR-558 and miR-182-5p, respectively (76, 77).
Du et al. explored a special circ_0014130-miR-545-3p-YAP1 pathway in the modulation of drug resistance and malignant behaviors of docetaxel (DTX)-resistant NSCLC cells (78). circ_0003998 knockdown suppresses colony formation and facilitates apoptosis and DTX sensitivity by sponging miR-136-5p to control CORO1C expression in DTX-resistant NSCLC cells (79). It has also been found that circ_0005909 knockdown resists the proliferation, metastasis, and drug resistance of NSCLC cells (80). Moreover, a novel protein encoded by circASK1 activates ASK1-dependent apoptosis, thereby ameliorating gefitinib resistance in LUAD (81). It has been shown that circSETD3 overexpression interferes with gefitinib sensitivity, and circSETD3 interacts with miR-520h and ABCG2 to reduce the intracellular concentration of gefitinib (19).
Exosomes are small extracellular vesicles that play essential roles in immunity, signal transduction, tumor treatment, and drug resistance (83). Yang et al. suggested that tumor-derived exosomal circRNA_102481 participated in EGFR-TKI resistance by sponging microRNA-30a-5p to modulate ROR1 in NSCLC (82). As for immune checkpoint inhibitors, hsa_circ_0003222 and circFGFR1 can promote NSCLC resistance to anti-PD-L1-based and anti-PD-1-based therapies, respectively (37, 38). Furthermore, suppression of circIGF2BP3/PKP3 promotes the therapeutic efficacy of anti-PD-1 in a Lewis LADC mouse model (39).
CircRNAs and Breast Cancer Drug Resistance
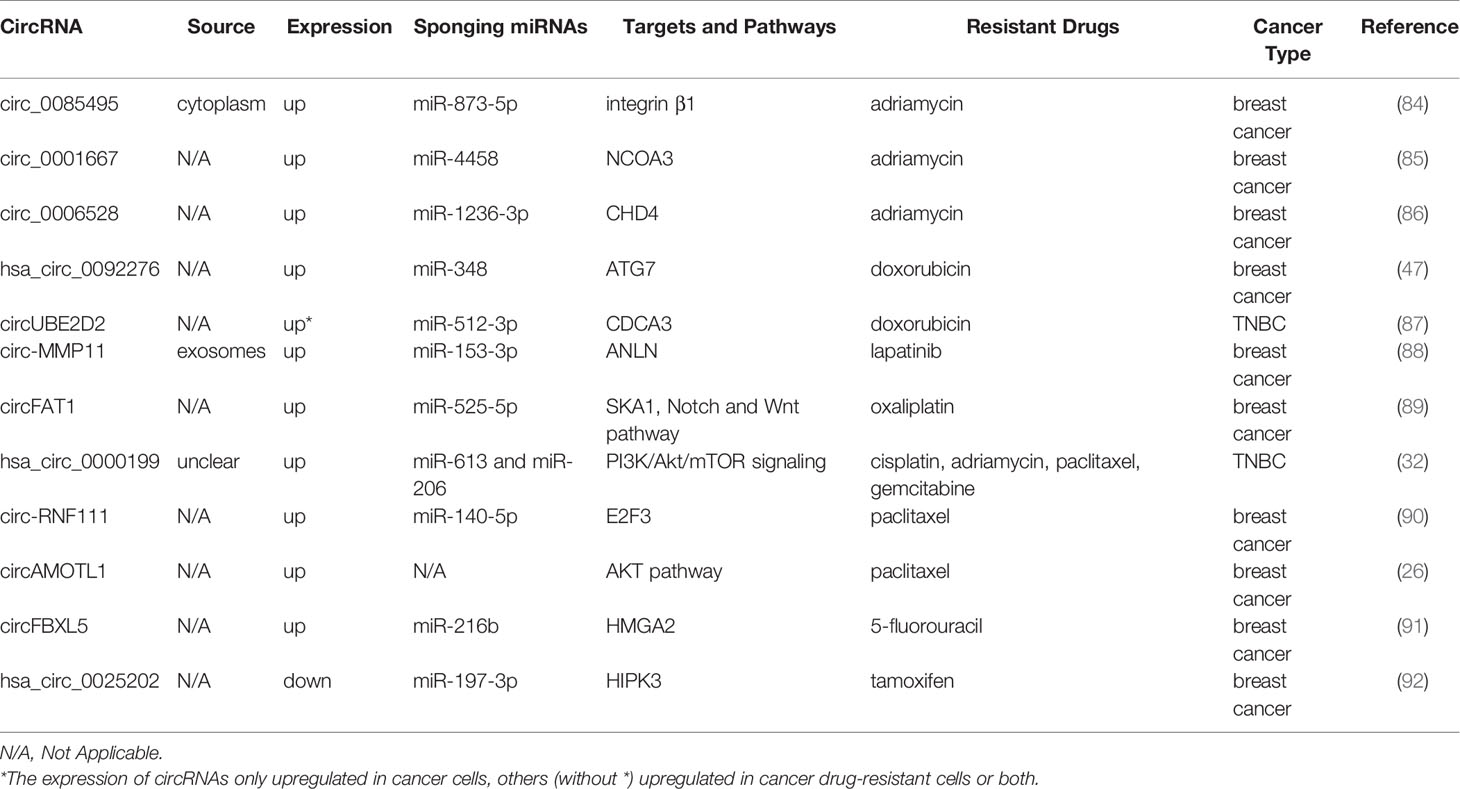
It was reported that there were 2.3 million new cases of breast cancer in female patients, which became the most generally diagnosed cancer, exceeding lung cancer in 2020 (60). Multiple circRNAs are shown to be correlated with breast cancer resistance (Table 2). The resistance to adriamycin (ADM), namely DOX, is closely correlated with therapeutic efficacy in patients with breast cancer and their prognosis (93). The knockdown of circ_0085495, circ_0001667, and circ_0006528 attenuates ADM resistance by related sponging miRNAs, becoming promising therapeutic targets for overcoming ADM resistance in patients with breast cancer (84–86). Similarly, hsa_circ_0092276, which sponges miR-384, regulates ATG7 expression and promotes DOX resistance in breast cancer (47). Additionally, circUBE2D2 depletion induces a tumor-suppressive effect and suppression in DOX resistance, which has been greatly impaired upon miR512-3p downregulation or CDCA3 overexpression (87).
Lapatinib resistance is promoted by circ-MMP11 in breast cancer cells, and mechanically circ-MMP11 regulates ANLN expression by sponging miR-153-3p (88). CircFAT1 facilitates oxaliplatin (OX) resistance in breast cancer by regulating miR-525-5p/SKA1, and the Notch and Wnt pathways can be activated by SKA1, which has been identified by rescue assays, GSEA, and western blotting (89). Li et al. found that silencing hsa_circ_0000199 contributed to TNBC chemosensitivity to multiple drugs (32). In their study, the TNBC cell lines in the si-hsa_circ_0000199 group are prone to become sensitive to chemotherapeutic drugs, including CDDP, adriamycin, paclitaxel, and gemcitabine (GEM). It has also been found that circ-RNF111 increases PTX resistance in breast cancer by elevating E2F3 via sponging miR-140-5p (90). CircAMOTL1 overexpression reduces apoptosis and enhances the invasion of breast cancer cells exposed to PAX (26). Furthermore, 5-FU resistance is promoted by circFBXL5 in breast cancer via the miR-216b/HMGA2 axis (91). Additionally, hsa_circ_0025202 overexpression impedes tumor growth and promotes tamoxifen sensitivity, while miR-197-3p overexpression facilitates cell malignancy and TAM resistance in breast cancer (92).
CircRNA and Gastric Cancer Drug Resistance
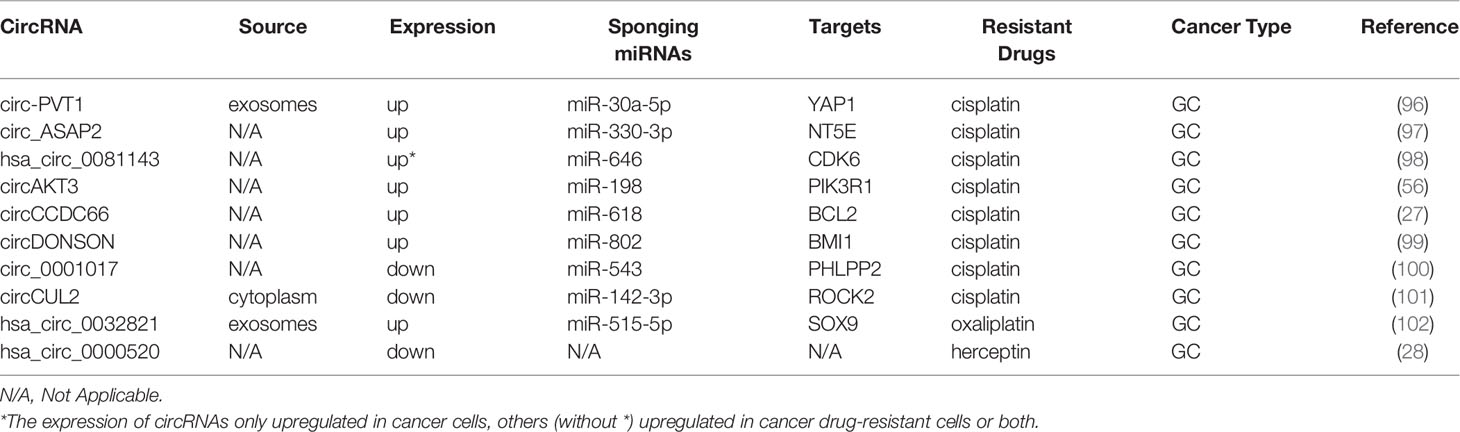
Gastric cancer (GC) is the fifth most common malignant tumor and is the fourth leading cause of cancer-related deaths (60, 94). The 5-year survival rate of advanced GC is approximately 20% (95). Platinum is a basic first-line chemotherapy drug for advanced GC (95). Studies on the mechanisms of circRNAs and resistance to CDDP are presented in Table 3. Exosomal circ-PVT1 regulates invasion, autophagy, and apoptosis and promotes CDDP resistance via the miR-30a-5p/YAP1 axis in GC cells, indicating that exosomal circ-PVT1 may be a valuable therapeutic target in GC (96). Additionally, the sensitivity of GC to CDDP is increased by knockdown of circ_ASAP2 or hsa_circ_0081143, which also represses the progression of GC by acting as ceRNAs (97, 98). It has been found that circAKT3 contributes to CDDP resistance in GC by enhancing DNA damage repair and hindering GC cell apoptosis (56). CircCCDC66 and circDONSON have been shown to induce CDDP resistance in GC by targeting the related miRNA and gene (27, 99). All the above studies demonstrate that certain circRNAs are overexpressed in CDDP-resistant GC and may serve as biomarkers for poor prognosis.
In contrast, Zhang et al. illustrated that upregulated expression of circ_0001017 inhibited malignant biological behaviors of GC and promoted CDDP sensitivity of CDDP-resistant GC cells partially via the miR-543/PHLPP2 axis (100). Similarly, circCUL2, acting as a tumor suppressor, enhances CDDP sensitivity by miR-142-3p/ROCK2-mediated autophagy (101). Exosomal circ_0032821 facilitates OXA resistance in OXA-sensitive GC cells by sponging miR-515-5p to enhance SOX9 expression (102). In addition, Herceptin resistance of GC cells can be reversed by hsa_circ_0000520 overexpression through inhibition of the PI3K-Akt signaling pathway (28).
CircRNAs and Osteosarcoma Drug Resistance
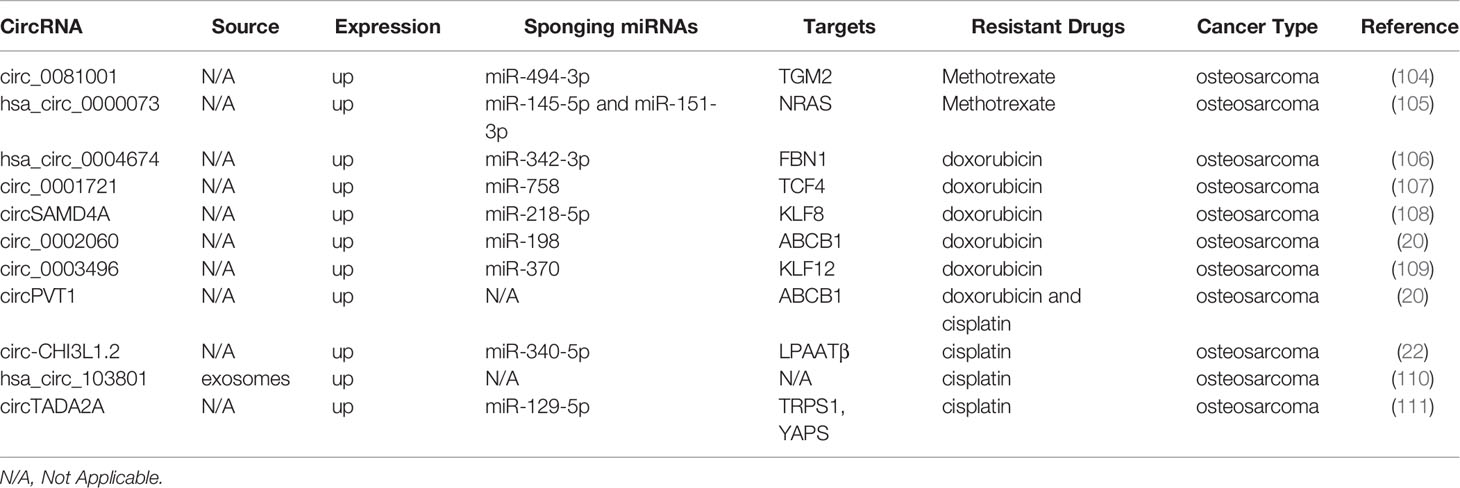
Osteosarcoma is the most widespread primary bone tumor affecting children and adolescents, and effective chemotherapeutic regimens include the combination of high-dose MTX, DOX, and CDDP (103). As shown in Table 4, multiple circRNAs are associated with osteosarcoma drug resistance. Circ_0081001 knockdown promotes MTX sensitivity of osteosarcoma cells through the suppression of miR-494-3p-mediated upregulation of TGM2 (104). Similarly, hsa_circ_0000073 promotes malignant behaviors, including proliferation, invasion, and migration, and facilitates MTX resistance of osteosarcoma cells by sponging miR-145-5p and miR-151-3p to upregulate NRAS (105).
DOX resistance of osteosarcoma is facilitated by hsa_circ_0004674 through the Wnt/β-catenin pathway via modulation of the miR-342-3p/FBN1 axis (106). Moreover, some circRNAs potentiate DOX resistance and facilitate the progression of osteosarcomas, such as circ_0001721, circSAMD4A, circ_0002060, and circ_0003496 (20, 107–109). Zhu et al. report that resistance to DOX and CDDP can be weakened by suppressing circPVT1 expression in osteosarcoma cells (21). It has been reported that circ-CHI3L1.2 knockdown promotes apoptosis and attenuates resistance of CDDP-resistant osteosarcoma cells (22). Additionally, exosomal hsa_circ_103801 can intensify the facilitating effect of exosomes on the chemoresistance of osteosarcoma cells to CDDP (110). Similarly, circTADA2A can target miR-129-5p, which is competitively bound to TRPS1 and YAP1, thereby regulating osteosarcoma cell proliferation and CDDP resistance (111).
CircRNA and Glioma Drug Resistance
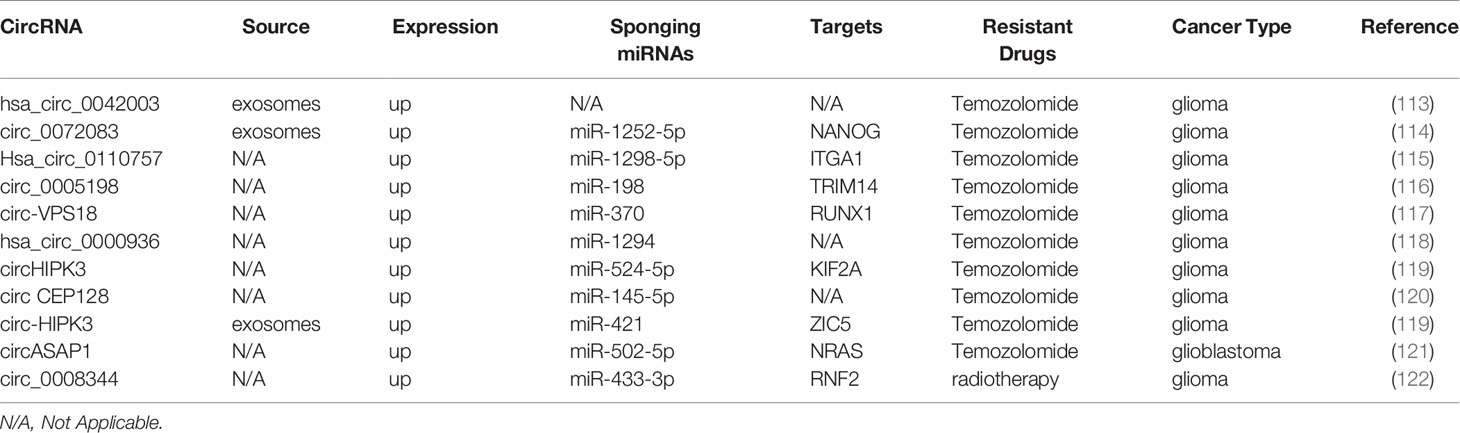
Glioma is the most common form of aggressive intracranial tumors and is characterized by a high rate of mortality, metastasis, and drug resistance (112). Here, we summarized the contribution of circRNAs to chemoresistance in glioma patients (Table 5). It has been shown that exosomal hsa_circ_0042003, mediated by heparanase transfers from temozolomide (TMZ)-resistant glioma cells to drug-sensitive cells, which contributes to TMZ resistance in glioma (113). In glioma, exosomal circ_0072083 modulates NANOG and ALKBH5 by targeting miR-1252-5p and demethylation to control TMZ resistance (114). Hsa_circ_0110757 and circ_0005198 facilitate TMZ resistance and inhibit glioma cell apoptosis through the miR-1298-5p/ITGA1 and miR-198/TRIM14 axis, respectively (115, 116). Consistent with these findings, downregulation of circ-VPS18, hsa_circ_0000936, circHIPK3, and circ CEP128 improve TMZ sensitivity and repress glioma progression (117–120). The highlight is that circHIPK3 downregulation inhibits the PI3K/AKT signaling pathway partly through the miR-524-5p/KIF2A axis (119). Similarly, in TMZ-resistant glioma, exosomal circ-HIPK3 can regulate the miR-421/ZIC5 axis to promote cell progression and TMZ resistance (123). circASAP1 overexpression promotes glioblastoma cell proliferation and TMZ resistance via the circASAP1/miR-502-5p/NRAS regulatory network, indicating that circASAP1 is a potential target for TMZ-resistant glioblastoma therapy (121). circ_0008344 downregulation impedes glioma growth and functions on the miR-433-3p/RNF2 axis to promote radiosensitivity in glioma (122).
CircRNAs and Ovarian Cancer Drug Resistance
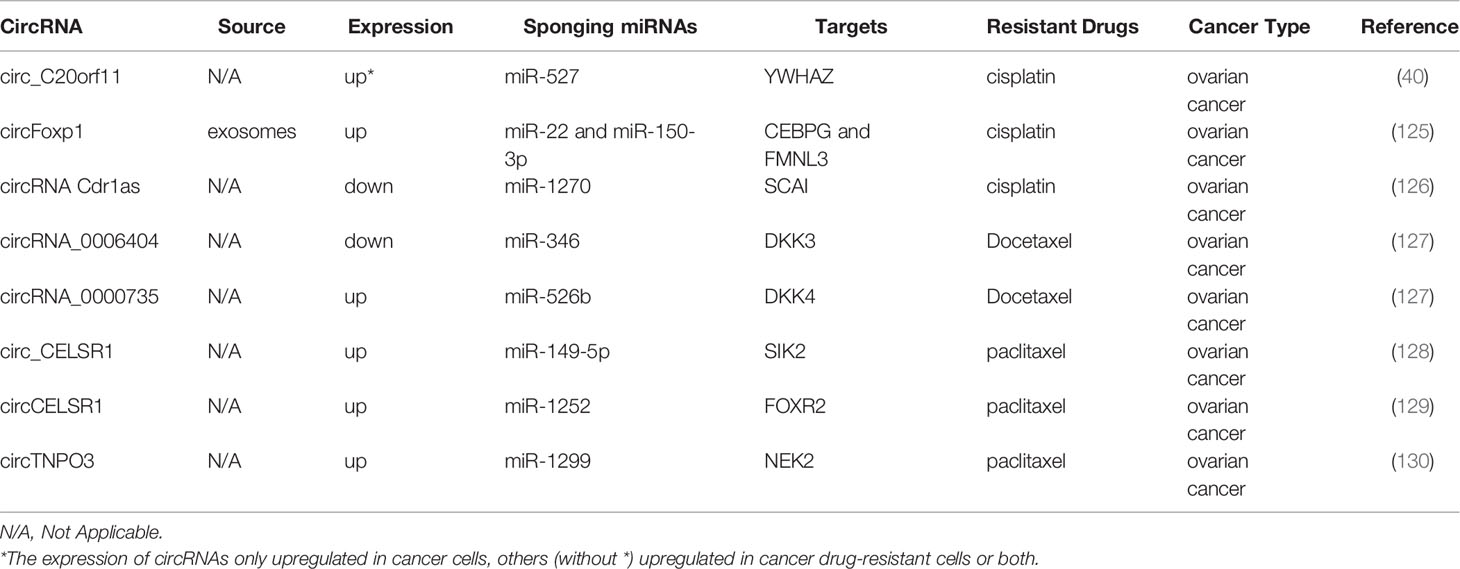
Ovarian cancer is one of the most common gynecologic malignant tumors, and conventional treatment is mainly limited to chemoresistance (124). Table 6 displays the chemoresistance-related circRNAs in ovarian cancer. Both circ_C20orf11 and circulating exosomal circFoxp1 can confer CDDP resistance in ovarian cancer cells (40, 125). In contrast, Cdr1as improves sensitivity to CDDP in ovarian cancer by modulating the miR-1270/SCAI axis (126).
The downregulation of circRNA_0000735 and upregulation of circRNA_0006404 can suppress the expression of p-GP, causing DTX treatment tolerance (127). Two studies demonstrated that circ_CELSR1 was upregulated in PTX-resistant ovarian cancer cells, and circ_CELSR1 silencing impeded PTX resistance in ovarian cancer in vivo (128, 129). Meanwhile, one study revealed that the inhibition of circCELSR1 also resulted in ovarian cancer cell G0/G1 arrest and the promotion of apoptosis (129). Similarly, circTNPO3 upregulates NEK2 expression by sponging miR-1299 to enhance PTX resistance in ovarian cancer cells (130).
CircRNA and Drug Resistance of Other Cancers
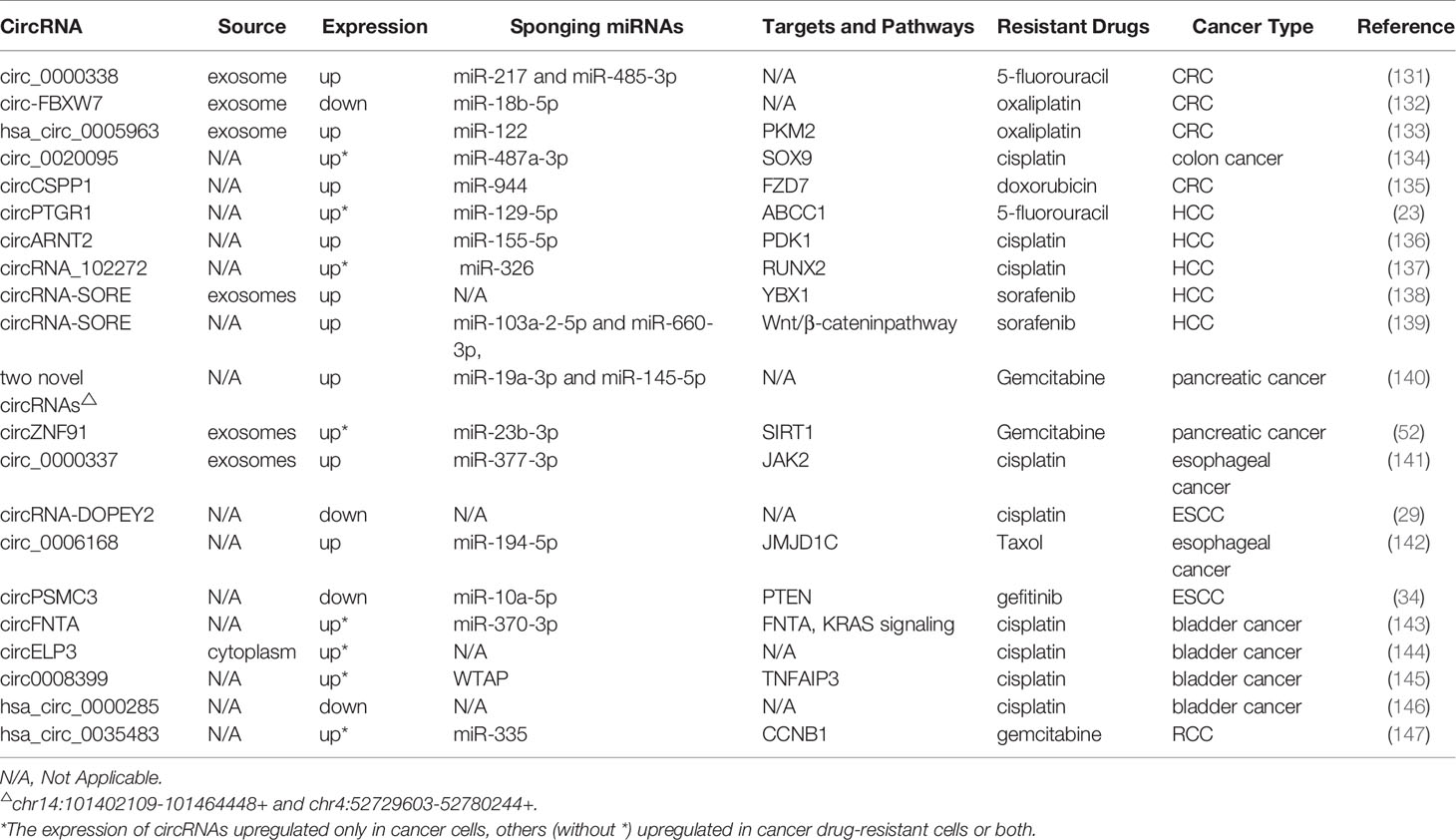
We also summarized a number of studies on circRNAs and drug resistance in other cancers, including hepatocellular carcinoma (HCC), colorectal cancer (CRC), esophageal cancer (EC), pancreatic cancer (PC), and some urinary system tumors (Table 7). As we all know, tolerance to chemotherapeutics is the pivotal cause of recurrence and poor prognosis in colorectal cancer patients. It has been shown that circ_0000338 knockdown sensitizes 5-FU-resistant CRC cells to 5-FU by promoting apoptosis and hindering proliferation (131). Exosomal circ-FBXW7 leads resistant cells sensitive to OX-induced apoptosis, inhibits OX-induced epithelial-mesenchymal transition, and suppresses OX efflux (132). In contrast, hsa_circ_0005963 is transferred by exosomes from OX-resistant CRC cells to OX-sensitive cells, resulting in glycolysis and drug resistance by enhancing PKM2 expression (133). A recent study suggested that circ_0020095 acts as a miR-487a-3p sponge to promote CDDP resistance by increasing the expression of SOX9 (134). In DOX-resistant CRC cells, circCSPP1 knockdown improves DOX sensitivity, attenuates cell malignant behaviors, and induces apoptosis through the miR-944/FZD7 axis (135).
As for HCC, the knockdown of circPTGR1 and circARNT2 induces apoptosis of HCC cells and inhibits 5-FU and CDDP resistance, respectively (23, 136). Moreover, circRNA, as a ceRNA, suppresses the activity of miR-326 to upregulate RUNX2 expression and enhance CDDP resistance (137). Two studies elaborate the mechanism of circRNA-SORE in sorafenib resistance in HCC from different perspectives. One study reveals that circRNA-SORE inhibits PRP19-mediated YBX1 degradation, thus affecting the expression of downstream gene targets of YBX1, including ERK, AKT, c-Myc, Raf1, and TGF-β1 (138). Another study demonstrates that circRNA-SORE interacts with miR-103a-2-5p and miR-660-3p by serving as a microRNA sponge, thus activating the Wnt/β-catenin pathway and leading to sorafenib resistance (139). In addition, Huang et al. confirm that overexpression of circMET can promote HCC development and immune tolerance through the miR-30-5p/Snail/DPP4/CXCL10 axis (148).
An increasing number of studies have elucidated the molecular mechanism of circRNAs in the initiation and progression of PC; however, studies on drug resistance mechanisms are in their preliminary exploration stage (149). For example, PC drug-resistant cells (PANC-1-GR) increase sensitivity to GEM after silencing two novel circRNAs (chr14:101402109-101464448+ or chr4:52729603-52780244+) (140). Exosomal circZNF91 transferred to normoxic PC cells can result in GEM and glycolysis chemoresistance (52).
As early as 2016, researchers utilized circRNA microarray to discover that some circRNAs are distinguished in the radioresistant esophageal cell line and the parental cell line, indicating that these dysregulated circRNAs are involved in radiation resistance (150). In esophageal cancer cells, CDDP resistance can be increased by exosomal circ_0000337, which regulates the miR-377-3p/JAK2 axis (141). cDOPEY2 promotes the ubiquitination and degradation of CPEB4 to inhibit CPEB4-mediated Mcl-1 translation, thereby alleviating CDDP resistance (29). Moreover, knockdown of circ_0006168 or JMJD1C plays a crucial role in inhibiting cell proliferation, invasion, and migration and promoting apoptosis, which accelerates the taxol sensitivity of Esophageal squamous cell carcinoma (ESCC) in vitro (142). In contrast, circPSMC3 overexpression inhibits miR-10a-5p expression and increases the sensitivity of ESCC cells to gefitinib (34).
Research on circRNAs in the drug resistance of urinary system tumors is also in its initial stages. It is an AR-mediated ADAR2/circFNTA/miR-370-3p/FNTA pathway that stimulates KRAS to change bladder cancer cell invasion and chemosensitivity to CDDP (143). Hypoxia-elevated circELP3 facilitates CDDP resistance in bladder cancer cells (144). circ0008399 promotes the formation of the WTAP/METTL3/METTL14 m6A methyltransferase complex and increases the expression of TNFAIP3 in an m6A-dependent manner. Additionally, the high expression of circ0008399 and WTAP is correlated with poor prognosis in patients with bladder cancer (145). A study reveals that hsa_circ_0000285 is expressed at significantly lower levels in both bladder cancer tissues and CDDP-resistant patients, but the detailed mechanism of hsa_circ_0000285 in chemosensitivity needs further exploration (146). In addition, hsa_circ_0035483 contributes to GEM-induced autophagy and promotes resistance of renal clear cell carcinoma (RCC) to GEM by regulating the hsa-miR-335/CCNB1 axis (147).
Conclusions and Future Perspectives
Currently, resistance to chemotherapeutic drugs has become an urgent problem impeding the treatment of various cancers. As a novel RNA class, an increasing number of circRNAs have been confirmed to be related to the chemoresistance of cancers. Most circRNAs function as miRNA sponges and form the circRNAs/miRNAs/mRNAs regulatory axis to regulate drug resistance or sensitivity of cancers. The sensitivity of cancer cells to chemotherapeutic drugs can be enhanced by silencing circRNAs that are upregulated in drug-resistant cancer cells or by overexpressing circRNAs that are downregulated in drug-resistant cancer cells.
The underlying mechanisms of chemoresistance-related circRNAs include the efflux of drugs, apoptosis, glycolysis, intervention with the TME, autophagy, and dysfunction of DNA damage repair, among others, which still requires further exploration. In addition, multiple pathways are involved in circRNA-modulated drug resistance in cancers including P53 signaling, KRAS signaling, the Wnt/β-catenin pathway, and the PI3K/Akt/mTOR pathway. New challenges owing to different chemotherapies are inevitable, demanding further elucidation of the involved signaling networks.
Taken together, it is still a challenge to select the key target for the treatment of malignancies from a large number of candidate circRNAs. Most studies on circRNAs and drug resistance are limited to in vitro and in vivo experiments. Therefore, long-term follow-up of patients and analysis of the relationship between circRNAs and drug resistance are warranted in this regard. The drug resistance of cancers may be associated with a systematic and comprehensive regulatory network comprising circRNAs, miRNAs, and target mRNAs. More research is required to elucidate the concrete molecular mechanisms between circRNAs and drug resistance in cancers and to survey the role of circRNAs in clinical practice in the future.
Author Contributions
Xin-YL, QZ, and JG reviewed literature and originally drafted the manuscript. PZ, HL, Z-BT, and C-PZ contributed to editing and embellished the manuscript. Xia-YL approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by the National Natural Science Foundation (No. 81802777), the “Clinical medicine + X” scientific research project of Affiliated Hospital of Qingdao University, and Qingdao Chinese Medicine Technology Project (2021-zyym26).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing. And we thank all the authors for helping with the writing and publication of this article.
References
1. Bray F, Laversanne M, Weiderpass E, Soerjomataram I. The Ever-Increasing Importance of Cancer as a Leading Cause of Premature Death Worldwide. Cancer (2021) 127:3029–30. doi: 10.1002/cncr.33587
2. Kuwano M, Sonoda K, Murakami Y, Watari K, Ono M. Overcoming Drug Resistance to Receptor Tyrosine Kinase Inhibitors: Learning From Lung Cancer. Pharmacol Ther (2016) 161:97–110. doi: 10.1016/j.pharmthera.2016.03.002
3. Vasan N, Baselga J, Hyman DM. A View on Drug Resistance in Cancer. Nature (2019) 575:299–309. doi: 10.1038/s41586-019-1730-1
4. Longley DB, Johnston PG. Molecular Mechanisms of Drug Resistance. J Pathol (2005) 205:275–92. doi: 10.1002/path.1706
5. Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov (2018) 8:822–35. doi: 10.1158/2159-8290.Cd-18-0099
6. Jeck WR, Sharpless NE. Detecting and Characterizing Circular RNAs. Nat Biotechnol (2014) 32:453–61. doi: 10.1038/nbt.2890
7. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-Like Structures. Proc Natl Acad Sci USA (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
8. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded Identification and Characterization of Mammalian Circular RNAs. Genome Biol (2014) 15:409. doi: 10.1186/s13059-014-0409-z
9. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA Sponges and can be Used as a New Class of Biomarker. J Biotechnol (2016) 238:42–51. doi: 10.1016/j.jbiotec.2016.09.011
10. Cheng F, Zheng B, Si S, Wang J, Zhao G, Yao Z, et al. The Roles of CircRNAs in Bladder Cancer: Biomarkers, Tumorigenesis Drivers, and Therapeutic Targets. Front Cell Dev Biol (2021) 9:666863. doi: 10.3389/fcell.2021.666863
11. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: Functions and Properties of a Novel Potential Biomarker for Cancer. Mol Cancer (2017) 16:94. doi: 10.1186/s12943-017-0663-2
12. Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, et al. Autophagy-Associated circRNA circCDYL Augments Autophagy and Promotes Breast Cancer Progression. Mol Cancer (2020) 19:65. doi: 10.1186/s12943-020-01152-2
13. Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y, et al. Circ-HuR Suppresses HuR Expression and Gastric Cancer Progression by Inhibiting CNBP Transactivation. Mol Cancer (2019) 18:158. doi: 10.1186/s12943-019-1094-z
14. Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, et al. Noncoding RNAs in Gastric Cancer: Implications for Drug Resistance. Mol Cancer (2020) 19:62. doi: 10.1186/s12943-020-01185-7
15. Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, et al. Mammalian Drug Efflux Transporters of the ATP Binding Cassette (ABC) Family in Multidrug Resistance: A Review of the Past Decade. Cancer Lett (2016) 370:153–64. doi: 10.1016/j.canlet.2015.10.010
16. Sharom FJ. The P-Glycoprotein Multidrug Transporter. Essays Biochem (2011) 50:161–78. doi: 10.1042/bse0500161
17. Wang X, Wang H, Jiang H, Qiao L, Guo C. Circular RNAcirc_0076305 Promotes Cisplatin (DDP) Resistance of Non-Small Cell Lung Cancer Cells by Regulating ABCC1 Through miR-186-5p. Cancer Biother Radiopharm (2021). doi: 10.1089/cbr.2020.4153
18. Liang H, Lin Z, Lin H, Zhao L, Huang W. circRNA_103615 Contributes to Tumor Progression and Cisplatin Resistance in NSCLC by Regulating ABCB1. Exp Ther Med (2021) 22:934. doi: 10.3892/etm.2021.10366
19. Huang Y, Dai Y, Wen C, He S, Shi J, Zhao D, et al. Circsetd3 Contributes to Acquired Resistance to Gefitinib in Non-Small-Cell Lung Cancer by Targeting the miR-520h/ABCG2 Pathway. Mol Ther Nucleic Acids (2020) 21:885–99. doi: 10.1016/j.omtn.2020.07.027
20. Ji Y, Liu J, Zhu W, Ji J. Circ_0002060 Enhances Doxorubicin Resistance in Osteosarcoma by Regulating the miR-198/ABCB1 Axis. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2020.4240
21. Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed Circpvt1, a Potential New Circular RNA Biomarker, Contributes to Doxorubicin and Cisplatin Resistance of Osteosarcoma Cells by Regulating ABCB1. Int J Biol Sci (2018) 14:321–30. doi: 10.7150/ijbs.24360
22. Zhang Z, Zhou Q, Luo F, Zhou R, Xu J, Xiao J, et al. Circular RNA Circ-CHI3L1.2 Modulates Cisplatin Resistance of Osteosarcoma Cells via the miR-340-5p/Lpaatβ Axis. Hum Cell (2021) 34:1558–68. doi: 10.1007/s13577-021-00564-6
23. Li X, Zhang T. Circular RNA PTGR1 Regulates 5-FU Resistance and Development of Hepatocellular Carcinoma Cells by Modulating miR-129-5p/ABCC1 Axis. Cell Biol Int (2021). doi: 10.1002/cbin.11635
24. Lomonosova E, Chinnadurai G. BH3-Only Proteins in Apoptosis and Beyond: An Overview. Oncogene (2008) 27 Suppl 1:S2–19. doi: 10.1038/onc.2009.39
25. Kryczka J, Kryczka J, Czarnecka-Chrebelska KH, Brzeziańska-Lasota E. Molecular Mechanisms of Chemoresistance Induced by Cisplatin in NSCLC Cancer Therapy. Int J Mol Sci (2021) 22(16):8885. doi: 10.3390/ijms22168885
26. Ma J, Fang L, Yang Q, Hibberd S, Du WW, Wu N, et al. Posttranscriptional Regulation of AKT by Circular RNA Angiomotin- Like 1 Mediates Chemoresistance Against Paclitaxel in Breast Cancer Cells. Aging (Albany NY) (2019) 11:11369–81. doi: 10.18632/aging.102535
27. Zhang Q, Miao Y, Fu Q, Hu H, Chen H, Zeng A, et al. CircRNACCDC66 Regulates Cisplatin Resistance in Gastric Cancer via the miR-618/BCL2 Axis. Biochem Biophys Res Commun (2020) 526:713–20. doi: 10.1016/j.bbrc.2020.03.156
28. Lv X, Li P, Wang J, Gao H, Hei Y, Zhang J, et al. Hsa_Circ_0000520 Influences Herceptin Resistance in Gastric Cancer Cells Through PI3K-Akt Signaling Pathway. J Clin Lab Anal (2020) 34:e23449. doi: 10.1002/jcla.23449
29. Liu Z, Gu S, Wu K, Li L, Dong C, Wang W, et al. CircRNA-DOPEY2 Enhances the Chemosensitivity of Esophageal Cancer Cells by Inhibiting CPEB4-Mediated Mcl-1 Translation. J Exp Clin Cancer Res (2021) 40:361. doi: 10.1186/s13046-021-02149-5
30. Tewari D, Patni P, Bishayee A, Sah AN, Bishayee A. Natural Products Targeting the PI3K-Akt-mTOR Signaling Pathway in Cancer: A Novel Therapeutic Strategy. Semin Cancer Biol (2019). doi: 10.1016/j.semcancer.2019.12.008
31. Burris HA. 3rd. Overcoming Acquired Resistance to Anticancer Therapy: Focus on the PI3K/AKT/mTOR Pathway. Cancer Chemother Pharmacol (2013) 71:829–42. doi: 10.1007/s00280-012-2043-3
32. Li H, Xu W, Xia Z, Liu W, Pan G, Ding J, et al. Hsa_circ_0000199 Facilitates Chemo-Tolerance of Triple-Negative Breast Cancer by Interfering With miR-206/613-Led PI3K/Akt/mTOR Signaling. Aging (Albany NY) (2021) 13:4522–51. doi: 10.18632/aging.202415
33. Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J. miR-21 Confers Cisplatin Resistance in Gastric Cancer Cells by Regulating PTEN. Toxicology (2013) 306:162–8. doi: 10.1016/j.tox.2013.02.014
34. Zhu H, Du F, Cao C. Restoration of Circpsmc3 Sensitizes Gefitinib-Resistant Esophageal Squamous Cell Carcinoma Cells to Gefitinib by Regulating miR-10a-5p/PTEN Axis. Cell Biol Int (2021) 45:107–16. doi: 10.1002/cbin.11473
35. Pitt JM, Vetizou M, Daillere R, Roberti MP, Yamazaki T, Routy B, et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity (2016) 44:1255–69. doi: 10.1016/j.immuni.2016.06.001
36. Sun Y. Tumor Microenvironment and Cancer Therapy Resistance. Cancer Lett (2016) 380:205–15. doi: 10.1016/j.canlet.2015.07.044
37. Li C, Zhang J, Yang X, Hu C, Chu T, Zhong R, et al. Hsa_Circ_0003222 Accelerates Stemness and Progression of non-Small Cell Lung Cancer by Sponging miR-527. Cell Death Dis (2021) 12:807. doi: 10.1038/s41419-021-04095-8
38. Zhang PF, Pei X, Li KS, Jin LN, Wang F, Wu J, et al. Circular RNA Circfgfr1 Promotes Progression and Anti-PD-1 Resistance by Sponging miR-381-3p in non-Small Cell Lung Cancer Cells. Mol Cancer (2019) 18:179. doi: 10.1186/s12943-019-1111-2
39. Liu Z, Wang T, She Y, Wu K, Gu S, Li L, et al. N(6)-Methyladenosine-Modified Circigf2bp3 Inhibits CD8(+) T-Cell Responses to Facilitate Tumor Immune Evasion by Promoting the Deubiquitination of PD-L1 in non-Small Cell Lung Cancer. Mol Cancer (2021) 20:105. doi: 10.1186/s12943-021-01398-4
40. Yin J, Huang HY, Long Y, Ma Y, Kamalibaike M, Dawuti R, et al. Circ_C20orf11 Enhances DDP Resistance by Inhibiting miR-527/YWHAZ Through the Promotion of Extracellular Vesicle-Mediated Macrophage M2 Polarization in Ovarian Cancer. Cancer Biol Ther (2021) 22(7–9):440–54. doi: 10.1080/15384047.2021.1959792
41. Erin N, Grahovac J, Brozovic A, Efferth T. Tumor Microenvironment and Epithelial Mesenchymal Transition as Targets to Overcome Tumor Multidrug Resistance. Drug Resist Update (2020) 53:100715. doi: 10.1016/j.drup.2020.100715
42. Nantajit D, Lin D, Li JJ. The Network of Epithelial-Mesenchymal Transition: Potential New Targets for Tumor Resistance. J Cancer Res Clin Oncol (2015) 141:1697–713. doi: 10.1007/s00432-014-1840-y
43. Joseph NA, Chiou SH, Lung Z, Yang CL, Lin TY, Chang HW, et al. The Role of HGF-MET Pathway and CCDC66 cirRNA Expression in EGFR Resistance and Epithelial-to-Mesenchymal Transition of Lung Adenocarcinoma Cells. J Hematol Oncol (2018) 11:74. doi: 10.1186/s13045-018-0557-9
44. Choi AM, Ryter SW, Levine B. Autophagy in Human Health and Disease. N Engl J Med (2013) 368:651–62. doi: 10.1056/NEJMra1205406
45. White E. Deconvoluting the Context-Dependent Role for Autophagy in Cancer. Nat Rev Cancer (2012) 12:401–10. doi: 10.1038/nrc3262
46. Kong R. Circular RNA Hsa_Circ_0085131 is Involved in Cisplatin-Resistance of non-Small-Cell Lung Cancer Cells by Regulating Autophagy. Cell Biol Int (2020) 44:1945–56. doi: 10.1002/cbin.11401
47. Wang Q, Liang D, Shen P, Yu Y, Yan Y, You W. Hsa_circ_0092276 Promotes Doxorubicin Resistance in Breast Cancer Cells by Regulating Autophagy via miR-348/ATG7 Axis. Transl Oncol (2021) 14:101045. doi: 10.1016/j.tranon.2021.101045
48. Marcucci F, Rumio C. Glycolysis-Induced Drug Resistance in Tumors-A Response to Danger Signals? Neoplasia (2021) 23:234–45. doi: 10.1016/j.neo.2020.12.009
49. Shi Q, Ji T, Ma Z, Tan Q, Liang J. Serum Exosomes-Based Biomarker Circ_0008928 Regulates Cisplatin Sensitivity, Tumor Progression, and Glycolysis Metabolism by miR-488/HK2 Axis in Cisplatin-Resistant Nonsmall Cell Lung Carcinoma. Cancer Biother Radiopharm (2021). doi: 10.1089/cbr.2020.4490
50. Xu Y, Jiang T, Wu C, Zhang Y. CircAKT3 Inhibits Glycolysis Balance in Lung Cancer Cells by Regulating miR-516b-5p/STAT3 to Inhibit Cisplatin Sensitivity. Biotechnol Lett (2020) 42:1123–35. doi: 10.1007/s10529-020-02846-9
51. Zhang Y, Ge P, Zhou D, Xing R, Bai L. Circular RNA FOXO3 Accelerates Glycolysis and Improves Cisplatin Sensitivity in Lung Cancer Cells. Via miR-543/Foxo3 Axis Oncol Lett (2021) 22:839. doi: 10.3892/ol.2021.13100
52. Zeng Z, Zhao Y, Chen Q, Zhu S, Niu Y, Ye Z, et al. Hypoxic Exosomal HIF-1α-Stabilizing Circznf91 Promotes Chemoresistance of Normoxic Pancreatic Cancer Cells via Enhancing Glycolysis. Oncogene (2021). doi: 10.1038/s41388-021-01960-w
53. Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular Mechanisms of Mammalian DNA Repair and the DNA Damage Checkpoints. Annu Rev Biochem (2004) 73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723
54. Haynes B, Saadat N, Myung B, Shekhar MP. Crosstalk Between Translesion Synthesis, Fanconi Anemia Network, and Homologous Recombination Repair Pathways in Interstrand DNA Crosslink Repair and Development of Chemoresistance. Mutat Res Rev Mutat Res (2015) 763:258–66. doi: 10.1016/j.mrrev.2014.11.005
55. Jeggo PA, Pearl LH, Carr AM. DNA Repair, Genome Stability and Cancer: A Historical Perspective. Nat Rev Cancer (2016) 16:35–42. doi: 10.1038/nrc.2015.4
56. Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, et al. Circular RNA AKT3 Upregulates PIK3R1 to Enhance Cisplatin Resistance in Gastric Cancer. Via miR-198 Suppression Mol Cancer (2019) 18:71. doi: 10.1186/s12943-019-0969-3
57. Friche E, Danks MK, Schmidt CA, Beck WT. Decreased DNA Topoisomerase II in Daunorubicin-Resistant Ehrlich Ascites Tumor Cells. Cancer Res (1991) 51:4213–8.
58. Bertino JR. Karnofsky Memorial Lecture. Ode to Methotrexate. J Clin Oncol (1993) 11:5–14. doi: 10.1200/jco.1993.11.1.5
59. Guo W, Healey JH, Meyers PA, Ladanyi M, Huvos AG, Bertino JR, et al. Mechanisms of Methotrexate Resistance in Osteosarcoma. Clin Cancer Res (1999) 5:621–7.
60. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
61. Shen DW, Pouliot LM, Hall MD, Gottesman MM. Cisplatin Resistance: A Cellular Self-Defense Mechanism Resulting From Multiple Epigenetic and Genetic Changes. Pharmacol Rev (2012) 64:706–21. doi: 10.1124/pr.111.005637
62. Liu Y, Zhai R, Hu S, Liu J. Circular RNA Circ-RNF121 Contributes to Cisplatin (DDP) Resistance of non-Small-Cell Lung Cancer Cells by Regulating the miR-646/SOX4 Axis. Anticancer Drugs (2022) 33(1):e186–97. doi: 10.1097/cad.0000000000001184
63. Zhu X, Han J, Lan H, Lin Q, Wang Y, Sun X. A Novel Circular RNA Hsa_circRNA_103809/miR-377-3p/GOT1 Pathway Regulates Cisplatin-Resistance in non-Small Cell Lung Cancer (NSCLC). BMC Cancer (2020) 20:1190. doi: 10.1186/s12885-020-07680-w
64. Pang J, Ye L, Zhao D, Zhao D, Chen Q. Circular RNA PRMT5 Confers Cisplatin-Resistance via miR-4458/REV3L Axis in non-Small-Cell Lung Cancer. Cell Biol Int (2020) 44:2416–26. doi: 10.1002/cbin.11449
65. Ye Y, Zhao L, Li Q, Xi C, Li Y, Li Z. Circ_0007385 Served as Competing Endogenous RNA for miR-519d-3p to Suppress Malignant Behaviors and Cisplatin Resistance of non-Small Cell Lung Cancer Cells. Thorac Cancer (2020) 11:2196–208. doi: 10.1111/1759-7714.13527
66. Xu X, Tao R, Sun L, Ji X. Exosome-Transferred Hsa_Circ_0014235 Promotes DDP Chemoresistance and Deteriorates the Development of non-Small Cell Lung Cancer by Mediating the miR-520a-5p/CDK4 Pathway. Cancer Cell Int (2020) 20:552. doi: 10.1186/s12935-020-01642-9
67. Lu H, Xie X, Wang K, Chen Q, Cai S, Liu D, et al. Circular RNA Hsa_Circ_0096157 Contributes to Cisplatin Resistance by Proliferation, Cell Cycle Progression, and Suppressing Apoptosis of non-Small-Cell Lung Carcinoma Cells. Mol Cell Biochem (2020) 475:63–77. doi: 10.1007/s11010-020-03860-1
68. Zhong Y, Lin H, Li Q, Liu C, Shen J. CircRNA_100565 Contributes to Cisplatin Resistance of NSCLC Cells by Regulating Proliferation, Apoptosis and Autophagy via miR-337-3p/ADAM28 Axis. Cancer biomark (2021) 30:261–73. doi: 10.3233/cbm-201705
69. Feng N, Guo Z, Wu X, Tian Y, Li Y, Geng Y, et al. Circ_PIP5K1A Regulates Cisplatin Resistance and Malignant Progression in non-Small Cell Lung Cancer Cells and Xenograft Murine Model via Depending on miR-493-5p/ROCK1 Axis. Respir Res (2021) 22:248. doi: 10.1186/s12931-021-01840-7
70. Wu Z, Gong Q, Yu Y, Zhu J, Li W. Knockdown of Circ-ABCB10 Promotes Sensitivity of Lung Cancer Cells to Cisplatin via miR-556-3p/AK4 Axis. BMC Pulm Med (2020) 20:10. doi: 10.1186/s12890-019-1035-z
71. Chen C, Zhang M, Zhang Y. Circ_0000079 Decoys the RNA-Binding Protein FXR1 to Interrupt Formation of the FXR1/PRCKI Complex and Decline Their Mediated Cell Invasion and Drug Resistance in NSCLC. Cell Transplant (2020) 29:963689720961070. doi: 10.1177/0963689720961070
72. Liu Y, Li C, Liu H, Wang J. Circ_0001821 Knockdown Suppresses Growth, Metastasis, and TAX Resistance of non-Small-Cell Lung Cancer Cells by Regulating the miR-526b-5p/GRK5 Axis. Pharmacol Res Perspect (2021) 9:e00812. doi: 10.1002/prp2.812
73. Li J, Fan R, Xiao H. Circ_ZFR Contributes to the Paclitaxel Resistance and Progression of non-Small Cell Lung Cancer by Upregulating KPNA4 Through Sponging miR-195-5p. Cancer Cell Int (2021) 21:15. doi: 10.1186/s12935-020-01702-0
74. Guo C, Wang H, Jiang H, Qiao L, Wang X. Circ_0011292 Enhances Paclitaxel Resistance in Non-Small Cell Lung Cancer by Regulating miR-379-5p/TRIM65 Axis. Cancer Biother Radiopharm (2020) 33(1):e166–77. doi: 10.1089/cbr.2019.3546
75. Xu J, Ni L, Zhao F, Dai X, Tao J, Pan J, et al. Overexpression of Hsa_Circ_0002874 Promotes Resistance of non-Small Cell Lung Cancer to Paclitaxel by Modulating miR-1273f/MDM2/p53 Pathway. Aging (Albany NY) (2021) 13:5986–6009. doi: 10.18632/aging.202521
76. Li X, Feng Y, Yang B, Xiao T, Ren H, Yu X, et al. A Novel Circular RNA, Hsa_Circ_0030998 Suppresses Lung Cancer Tumorigenesis and Taxol Resistance by Sponging miR-558. Mol Oncol (2021) 15:2235–48. doi: 10.1002/1878-0261.12852
77. Li X, Yang B, Ren H, Xiao T, Zhang L, Li L, et al. Hsa_circ_0002483 Inhibited the Progression and Enhanced the Taxol Sensitivity of non-Small Cell Lung Cancer by Targeting miR-182-5p. Cell Death Dis (2019) 10:953. doi: 10.1038/s41419-019-2180-2
78. Du D, Cao X, Duan X, Zhang X. Blocking Circ_0014130 Suppressed Drug Resistance and Malignant Behaviors of Docetaxel Resistance-Acquired NSCLC Cells via Regulating miR-545-3p-YAP1 Axis. Cytotechnology (2021) 73:571–84. doi: 10.1007/s10616-021-00478-z
79. Zhang W, Song C, Ren X. Circ_0003998 Regulates the Progression and Docetaxel Sensitivity of DTX-Resistant Non-Small Cell Lung Cancer Cells by the miR-136-5p/CORO1C Axis. Technol Cancer Res Treat (2021) 20:1533033821990040. doi: 10.1177/1533033821990040
80. Song HM, Meng D, Wang JP, Zhang XY. circRNA Hsa_Circ_0005909 Predicts Poor Prognosis and Promotes the Growth, Metastasis, and Drug Resistance of Non-Small-Cell Lung Cancer via the miRNA-338-3p/SOX4 Pathway. Dis Markers (2021) 2021:8388512. doi: 10.1155/2021/8388512
81. Wang T, Liu Z, She Y, Deng J, Zhong Y, Zhao M, et al. A Novel Protein Encoded by Circask1 Ameliorates Gefitinib Resistance in Lung Adenocarcinoma by Competitively Activating ASK1-Dependent Apoptosis. Cancer Lett (2021) 520:321–31. doi: 10.1016/j.canlet.2021.08.007
82. Yang B, Teng F, Chang L, Wang J, Liu DL, Cui YS, et al. Tumor-Derived Exosomal circRNA_102481 Contributes to EGFR-TKIs Resistance via the miR-30a-5p/ROR1 Axis in non-Small Cell Lung Cancer. Aging (Albany NY) (2021) 13:13264–86. doi: 10.18632/aging.203011
83. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem (2019) 88:487–514. doi: 10.1146/annurev-biochem-013118-111902
84. Xie H, Zheng R. Circ_0085495 Knockdown Reduces Adriamycin Resistance in Breast Cancer Through miR-873-5p/Integrin β1 Axis. Anticancer Drugs (2021) 33(1):e166–77. doi: 10.1097/cad.0000000000001174
85. Cui Y, Fan J, Shi W, Zhou Z. Circ_0001667 Knockdown Blocks Cancer Progression and Attenuates Adriamycin Resistance by Depleting NCOA3 via Releasing miR-4458 in Breast Cancer. Drug Dev Res (2021). doi: 10.1002/ddr.21845
86. Hao J, Du X, Lv F, Shi Q. Knockdown of Circ_0006528 Suppresses Cell Proliferation, Migration, Invasion, and Adriamycin Chemoresistance via Regulating the miR-1236-3p/CHD4 Axis in Breast Cancer. J Surg Res (2021) 260:104–15. doi: 10.1016/j.jss.2020.10.031
87. Dou D, Ren X, Han M, Xu X, Ge X, Gu Y, et al. CircUBE2D2 (Hsa_Circ_0005728) Promotes Cell Proliferation, Metastasis and Chemoresistance in Triple-Negative Breast Cancer by Regulating miR-512-3p/CDCA3 Axis. Cancer Cell Int (2020) 20:454. doi: 10.1186/s12935-020-01547-7
88. Wu X, Ren Y, Yao R, Zhou L, Fan R. Circular RNA Circ-MMP11 Contributes to Lapatinib Resistance of Breast Cancer Cells by Regulating the miR-153-3p/ANLN Axis. Front Oncol (2021) 11:639961. doi: 10.3389/fonc.2021.639961
89. Yao Y, Li X, Cheng L, Wu X, Wu B. Circular RNA FAT Atypical Cadherin 1 (Circfat1)/microRNA-525-5p/Spindle and Kinetochore-Associated Complex Subunit 1 (SKA1) Axis Regulates Oxaliplatin Resistance in Breast Cancer by Activating the Notch and Wnt Signaling Pathway. Bioengineered (2021) 12:4032–43. doi: 10.1080/21655979.2021.1951929
90. Zang H, Li Y, Zhang X, Huang G. Circ-RNF111 Contributes to Paclitaxel Resistance in Breast Cancer by Elevating E2F3 Expression. Via miR-140-5p Thorac Cancer (2020) 11:1891–903. doi: 10.1111/1759-7714.13475
91. Zhu M, Wang Y, Wang F, Li L, Qiu X. CircFBXL5 Promotes the 5-FU Resistance of Breast Cancer via Modulating miR-216b/HMGA2 Axis. Cancer Cell Int (2021) 21:384. doi: 10.1186/s12935-021-02088-3
92. Li H, Li Q, He S. Hsa_circ_0025202 Suppresses Cell Tumorigenesis and Tamoxifen Resistance via miR-197-3p/HIPK3 Axis in Breast Cancer. World J Surg Oncol (2021) 19:39. doi: 10.1186/s12957-021-02149-x
93. Zeng H, Wang L, Wang J, Chen T, Li H, Zhang K, et al. microRNA-129-5p Suppresses Adriamycin Resistance in Breast Cancer by Targeting SOX2. Arch Biochem Biophys (2018) 651:52–60. doi: 10.1016/j.abb.2018.05.018
94. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2020. CA Cancer J Clin (2020) 70:7–30. doi: 10.3322/caac.21590
95. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric Cancer. Lancet (2020) 396:635–48. doi: 10.1016/s0140-6736(20)31288-5
96. Yao W, Guo P, Mu Q, Wang Y. Exosome-Derived Circ-PVT1 Contributes to Cisplatin Resistance by Regulating Autophagy, Invasion, and Apoptosis Via miR-30a-5p/YAP1 Axis in Gastric Cancer Cells. Cancer Biother Radiopharm (2021) 36:347–59. doi: 10.1089/cbr.2020.3578
97. Sun Y, Ma J, Lin J, Sun D, Song P, Shi L, et al. Circular RNA Circ_ASAP2 Regulates Drug Sensitivity and Functional Behaviors of Cisplatin-Resistant Gastric Cancer Cells by the miR-330-3p/NT5E Axis. Anticancer Drugs (2021) 32(9):950–61. doi: 10.1097/cad.0000000000001087
98. Xue M, Li G, Fang X, Wang L, Jin Y, Zhou Q. Hsa_Circ_0081143 Promotes Cisplatin Resistance in Gastric Cancer by Targeting miR-646/CDK6 Pathway. Cancer Cell Int (2019) 19:25. doi: 10.1186/s12935-019-0737-x
99. Liu Y, Xu J, Jiang M, Ni L, Ling Y. CircRNA DONSON Contributes to Cisplatin Resistance in Gastric Cancer Cells by Regulating miR-802/BMI1 Axis. Cancer Cell Int (2020) 20:261. doi: 10.1186/s12935-020-01358-w
100. Zhang J, Zha W, Qian C, Ding A, Mao Z. Circular RNA Circ_0001017 Sensitizes Cisplatin-Resistant Gastric Cancer Cells to Chemotherapy by the miR-543/PHLPP2 Axis. Biochem Genet (2021). doi: 10.1007/s10528-021-10110-6
101. Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, et al. Circcul2 Regulates Gastric Cancer Malignant Transformation and Cisplatin Resistance by Modulating Autophagy Activation via miR-142-3p/ROCK2. Mol Cancer (2020) 19:156. doi: 10.1186/s12943-020-01270-x
102. Zhong Y, Wang D, Ding Y, Tian G, Jiang B. Circular RNA Circ_0032821 Contributes to Oxaliplatin (OXA) Resistance of Gastric Cancer Cells by Regulating SOX9 via miR-515-5p. Biotechnol Lett (2021) 43:339–51. doi: 10.1007/s10529-020-03036-3
103. Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. J Clin Oncol (2015) 33:3029–35. doi: 10.1200/jco.2014.59.4895
104. Wei W, Ji L, Duan W, Zhu J. Circular RNA Circ_0081001 Knockdown Enhances Methotrexate Sensitivity in Osteosarcoma Cells by Regulating miR-494-3p/TGM2 Axis. J Orthop Surg Res (2021) 16:50. doi: 10.1186/s13018-020-02169-5
105. Li X, Liu Y, Zhang X, Shen J, Xu R, Liu Y, et al. Circular RNA Hsa_Circ_0000073 Contributes to Osteosarcoma Cell Proliferation, Migration, Invasion and Methotrexate Resistance by Sponging miR-145-5p and miR-151-3p and Upregulating NRAS. Aging (Albany NY) (2020) 12:14157–73. doi: 10.18632/aging.103423
106. Bai Y, Li Y, Bai J, Zhang Y. Hsa_circ_0004674 Promotes Osteosarcoma Doxorubicin Resistance by Regulating the miR-342-3p/FBN1 Axis. J Orthop Surg Res (2021) 16:510. doi: 10.1186/s13018-021-02631-y
107. Guan H, Xu H, Chen J, Wu W, Chen D, Chen Y, et al. Circ_0001721 Enhances Doxorubicin Resistance and Promotes Tumorigenesis in Osteosarcoma Through miR-758/TCF4 Axis. Cancer Cell Int (2021) 21:336. doi: 10.1186/s12935-021-02016-5
108. Wei W, Ji L, Duan W, Zhu J. CircSAMD4A Contributes to Cell Doxorubicin Resistance in Osteosarcoma by Regulating the miR-218-5p/KLF8 Axis. Open Life Sci (2020) 15:848–59. doi: 10.1515/biol-2020-0079
109. Xie C, Liang G, Xu Y, Lin E. Circular RNA Hsa_Circ_0003496 Contributes to Tumorigenesis and Chemoresistance in Osteosarcoma Through Targeting (microRNA) miR-370/Krüppel-Like Factor 12 Axis. Cancer Manag Res (2020) 12:8229–40. doi: 10.2147/cmar.S253969
110. Pan Y, Lin Y, Mi C. Cisplatin-Resistant Osteosarcoma Cell-Derived Exosomes Confer Cisplatin Resistance to Recipient Cells in an Exosomal Circ_103801-Dependent Manner. Cell Biol Int (2021) 45:858–68. doi: 10.1002/cbin.11532
111. Zhang J, Ma X, Zhou R, Zhou Y. TRPS1 and YAP1 Regulate Cell Proliferation and Drug Resistance of Osteosarcoma via Competitively Binding to the Target of Circtada2a - miR-129-5p. Onco Targets Ther (2020) 13:12397–407. doi: 10.2147/ott.S276953
112. Gusyatiner O, Hegi ME. Glioma Epigenetics: From Subclassification to Novel Treatment Options. Semin Cancer Biol (2018) 51:50–8. doi: 10.1016/j.semcancer.2017.11.010
113. Si J, Li W, Li X, Cao L, Chen Z, Jiang Z. Heparanase Confers Temozolomide Resistance by Regulation of Exosome Secretion and Circular RNA Composition in Glioma. Cancer Sci (2021) 112(9):3491–06. doi: 10.1111/cas.14984
114. Ding C, Yi X, Chen X, Wu Z, You H, Chen X, et al. Warburg Effect-Promoted Exosomal Circ_0072083 Releasing Up-Regulates NANGO Expression Through Multiple Pathways and Enhances Temozolomide Resistance in Glioma. J Exp Clin Cancer Res (2021) 40:164. doi: 10.1186/s13046-021-01942-6
115. Li H, Liu Q, Chen Z, Wu M, Zhang C, Su J, et al. Hsa_circ_0110757 Upregulates ITGA1 to Facilitate Temozolomide Resistance in Glioma by Suppressing hsa-miR-1298-5p. Cell Death Dis (2021) 12:252. doi: 10.1038/s41419-021-03533-x
116. Deng Y, Zhu H, Xiao L, Liu C, Meng X. Circ_0005198 Enhances Temozolomide Resistance of Glioma Cells Through miR-198/TRIM14 Axis. Aging (Albany NY) (2020) 13:2198–211. doi: 10.18632/aging.202234
117. Li W, Ma Q, Liu Q, Yan P, Wang X, Jia X. Circ-VPS18 Knockdown Enhances TMZ Sensitivity and Inhibits Glioma Progression by MiR-370/RUNX1 Axis. J Mol Neurosci (2021) 71:1234–44. doi: 10.1007/s12031-020-01749-8
118. Hua L, Huang L, Zhang X, Feng H. Downregulation of Hsa_Circ_0000936 Sensitizes Resistant Glioma Cells to Temozolomide by Sponging miR-1294. J Biosci (2020) 45:101. doi: 10.1007/s12038-020-00072-z
119. Yin H, Cui X. Knockdown of Circhipk3 Facilitates Temozolomide Sensitivity in Glioma by Regulating Cellular Behaviors Through miR-524-5p/KIF2A-Mediated PI3K/AKT Pathway. Cancer Biother Radiopharm (2020) 36(7):556–67. doi: 10.1089/cbr.2020.3575
120. Hua L, Huang L, Zhang X, Feng H, Shen B. Knockdown of Circular RNA CEP128 Suppresses Proliferation and Improves Cytotoxic Efficacy of Temozolomide in Glioma Cells by Regulating miR-145-5p. Neuroreport (2019) 30:1231–8. doi: 10.1097/wnr.0000000000001326
121. Wei Y, Lu C, Zhou P, Zhao L, Lyu X, Yin J, et al. EIF4A3-Induced Circular RNA ASAP1 Promotes Tumorigenesis and Temozolomide Resistance of Glioblastoma via NRAS/MEK1/ERK1-2 Signaling. Neuro Oncol (2021) 23:611–24. doi: 10.1093/neuonc/noaa214
122. Di L, Zhao X, Ding J. Knockdown of Circ_0008344 Contributes to Radiosensitization in Glioma via miR-433-3p/RNF2 Axis. J Biosci (2021) 46:101. doi: 10.1007/s12038-021-00198-8
123. Han C, Wang S, Wang H, Zhang J. Exosomal Circ-HIPK3 Facilitates Tumor Progression and Temozolomide Resistance by Regulating miR-421/ZIC5 Axis in Glioma. Cancer Biother Radiopharm (2020) 36(7):537–48. doi: 10.1089/cbr.2019.3492
124. Wang JY, Lu AQ, Chen LJ. LncRNAs in Ovarian Cancer. Clin Chim Acta (2019) 490:17–27. doi: 10.1016/j.cca.2018.12.013
125. Luo Y, Gui R. Circulating Exosomal Circfoxp1 Confers Cisplatin Resistance in Epithelial Ovarian Cancer Cells. J Gynecol Oncol (2020) 31:e75. doi: 10.3802/jgo.2020.31.e75
126. Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as Upregulates SCAI to Suppress Cisplatin Resistance in Ovarian Cancer via miR-1270 Suppression. Mol Ther Nucleic Acids (2019) 18:24–33. doi: 10.1016/j.omtn.2019.07.012
127. Chen YY, Tai YC. Hsa_circ_0006404 and Hsa_Circ_0000735 Regulated Ovarian Cancer Response to Docetaxel Treatment via Regulating P-GP Expression. Biochem Genet (2021). doi: 10.1007/s10528-021-10080-9
128. Wei S, Qi L, Wang L. Overexpression of Circ_CELSR1 Facilitates Paclitaxel Resistance of Ovarian Cancer by Regulating miR-149-5p/SIK2 Axis. Anticancer Drugs (2021) 32:496–507. doi: 10.1097/cad.0000000000001058
129. Zhang S, Cheng J, Quan C, Wen H, Feng Z, Hu Q, et al. Circcelsr1 (Hsa_Circ_0063809) Contributes to Paclitaxel Resistance of Ovarian Cancer Cells by Regulating FOXR2 Expression. Via miR-1252 Mol Ther Nucleic Acids (2020) 19:718–30. doi: 10.1016/j.omtn.2019.12.005
130. Xia B, Zhao Z, Wu Y, Wang Y, Zhao Y, Wang J. Circular RNA Circtnpo3 Regulates Paclitaxel Resistance of Ovarian Cancer Cells by miR-1299/NEK2 Signaling Pathway. Mol Ther Nucleic Acids (2020) 21:780–91. doi: 10.1016/j.omtn.2020.06.002
131. Zhao K, Cheng X, Ye Z, Li Y, Peng W, Wu Y, et al. Exosome-Mediated Transfer of Circ_0000338 Enhances 5-Fluorouracil Resistance in Colorectal Cancer Through Regulating MicroRNA 217 (miR-217) and miR-485-3p. Mol Cell Biol (2021) 41(5):e00517–20. doi: 10.1128/mcb.00517-20
132. Xu Y, Qiu A, Peng F, Tan X, Wang J, Gong X. Exosomal Transfer of Circular RNA FBXW7 Ameliorates the Chemoresistance to Oxaliplatin in Colorectal Cancer by Sponging miR-18b-5p. Neoplasma (2021) 68:108–18. doi: 10.4149/neo_2020_200417N414
133. Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, et al. Exosome-Delivered circRNA Promotes Glycolysis to Induce Chemoresistance Through the miR-122-PKM2 Axis in Colorectal Cancer. Mol Oncol (2020) 14:539–55. doi: 10.1002/1878-0261.12629
134. Sun Y, Cao Z, Shan J, Gao Y, Liu X, Ma D, et al. Hsa_circ_0020095 Promotes Oncogenesis and Cisplatin Resistance in Colon Cancer by Sponging miR-487a-3p and Modulating Sox9. Front Cell Dev Biol (2020) 8:604869. doi: 10.3389/fcell.2020.604869
135. Xi L, Liu Q, Zhang W, Luo L, Song J, Liu R, et al. Circular RNA Circcspp1 Knockdown Attenuates Doxorubicin Resistance and Suppresses Tumor Progression of Colorectal Cancer via miR-944/FZD7 Axis. Cancer Cell Int (2021) 21:153. doi: 10.1186/s12935-021-01855-6
136. Li Y, Zhang Y, Zhang S, Huang D, Li B, Liang G, et al. circRNA Circarnt2 Suppressed the Sensitivity of Hepatocellular Carcinoma Cells to Cisplatin by Targeting the miR-155-5p/PDK1 Axis. Mol Ther Nucleic Acids (2021) 23:244–54. doi: 10.1016/j.omtn.2020.08.037
137. Guan Y, Zhang Y, Hao L, Nie Z. CircRNA_102272 Promotes Cisplatin-Resistance in Hepatocellular Carcinoma by Decreasing MiR-326 Targeting of RUNX2. Cancer Manag Res (2020) 12:12527–34. doi: 10.2147/cmar.S258230
138. Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, et al. CircRNA-SORE Mediates Sorafenib Resistance in Hepatocellular Carcinoma by Stabilizing YBX1. Signal Transduct Target Ther (2020) 5:298. doi: 10.1038/s41392-020-00375-5
139. Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N(6)-Methyladenosine-Modified CircRNA-SORE Sustains Sorafenib Resistance in Hepatocellular Carcinoma by Regulating Beta-Catenin Signaling. Mol Cancer (2020) 19:163. doi: 10.1186/s12943-020-01281-8
140. Shao F, Huang M, Meng F, Huang Q. Circular RNA Signature Predicts Gemcitabine Resistance of Pancreatic Ductal Adenocarcinoma. Front Pharmacol (2018) 9:584. doi: 10.3389/fphar.2018.00584
141. Zang R, Qiu X, Song Y, Wang Y. Exosomes Mediated Transfer of Circ_0000337 Contributes to Cisplatin (CDDP) Resistance of Esophageal Cancer by Regulating JAK2 via miR-377-3p. Front Cell Dev Biol (2021) 9:673237. doi: 10.3389/fcell.2021.673237
142. Qu F, Wang L, Wang C, Yu L, Zhao K, Zhong H. Circular RNA Circ_0006168 Enhances Taxol Resistance in Esophageal Squamous Cell Carcinoma by Regulating miR-194-5p/JMJD1C Axis. Cancer Cell Int (2021) 21:273. doi: 10.1186/s12935-021-01984-y
143. Chen J, Sun Y, Ou Z, Yeh S, Huang CP, You B, et al. Androgen Receptor-Regulated circFNTA Activates KRAS Signaling to Promote Bladder Cancer Invasion. EMBO Rep (2020) 21:e48467. doi: 10.15252/embr.201948467
144. Su Y, Yang W, Jiang N, Shi J, Chen L, Zhong G, et al. Hypoxia-Elevated Circelp3 Contributes to Bladder Cancer Progression and Cisplatin Resistance. Int J Biol Sci (2019) 15:441–52. doi: 10.7150/ijbs.26826
145. Wei W, Sun J, Zhang H, Xiao X, Huang C, Wang L, et al. Circ0008399 Interaction With WTAP Promotes Assembly and Activity of the M6a Methyltransferase Complex and Promotes Cisplatin Resistance in Bladder Cancer. Cancer Res (2021) 81(24):6142–56. doi: 10.1158/0008-5472.Can-21-1518
146. Chi BJ, Zhao DM, Liu L, Yin XZ, Wang FF, Bi S, et al. Downregulation of Hsa_Circ_0000285 Serves as a Prognostic Biomarker for Bladder Cancer and is Involved in Cisplatin Resistance. Neoplasma (2019) 66:197–202. doi: 10.4149/neo_2018_180318N185
147. Yan L, Liu G, Cao H, Zhang H, Shao F. Hsa_circ_0035483 Sponges hsa-miR-335 to Promote the Gemcitabine-Resistance of Human Renal Cancer Cells by Autophagy Regulation. Biochem Biophys Res Commun (2019) 519:172–8. doi: 10.1016/j.bbrc.2019.08.093
148. Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, et al. Circular RNA circMET Drives Immunosuppression and Anti-PD1 Therapy Resistance in Hepatocellular Carcinoma via the miR-30-5p/Snail/DPP4 Axis. Mol Cancer (2020) 19:92. doi: 10.1186/s12943-020-01213-6
149. Yao X, Mao Y, Wu D, Zhu Y, Lu J, Huang Y, et al. Exosomal Circ_0030167 Derived From BM-MSCs Inhibits the Invasion, Migration, Proliferation and Stemness of Pancreatic Cancer Cells by Sponging miR-338-5p and Targeting the Wif1/Wnt8/β-Catenin Axis. Cancer Lett (2021) 512:38–50. doi: 10.1016/j.canlet.2021.04.030
Keywords: circular RNAs, drug resistance, cancer, chemotherapy, therapeutic targets
Citation: Liu X-Y, Zhang Q, Guo J, Zhang P, Liu H, Tian Z-B, Zhang C-P and Li X-Y (2022) The Role of Circular RNAs in the Drug Resistance of Cancers. Front. Oncol. 11:790589. doi: 10.3389/fonc.2021.790589
Received: 13 October 2021; Accepted: 13 December 2021;
Published: 05 January 2022.
Edited by:
Qingbin Cui, University of Toledo, United StatesReviewed by:
Xiu-Yan Huang, Shanghai Jiao Tong University, ChinaChanglong Xu, Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, China
Copyright © 2022 Liu, Zhang, Guo, Zhang, Liu, Tian, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Yu Li, bGl4aWFveXUwNUAxNjMuY29t
 Xin-Yuan Liu
Xin-Yuan Liu Qi Zhang
Qi Zhang Xiao-Yu Li
Xiao-Yu Li