- 1Department of Hematology, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 2Institute of Hematology, Zhejiang University, Hangzhou, China
- 3Zhejiang Province Key Laboratory of Hematology Oncology Diagnosis and Treatment, Hangzhou, China
- 4Department of Hematology, The Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China
- 5Acornmed Biotechnology Co., Ltd., Beijing, China
Core Binding Factor (CBF)-AML is one of the most common somatic mutations in acute myeloid leukemia (AML). t(8;21)/AML1-ETO-positive acute myeloid leukemia accounts for 5-10% of all AMLs. In this study, we consecutively included 254 AML1-ETO patients diagnosed and treated at our institute from December 2009 to March 2020, and evaluated molecular mutations by 185-gene NGS platform to explore genetic co-occurrences with clinical outcomes. Our results showed that high KIT VAF(≥15%) correlated with shortened overall survival compared to other cases with no KIT mutation (3-year OS rate 26.6% vs 59.0% vs 69.6%, HR 1.50, 95%CI 0.78-2.89, P=0.0005). However, no difference was found in patients’ OS whether they have KIT mutation in two or three sites. Additionally, we constructed a risk model by combining clinical and molecular factors; this model was validated in other independent cohorts. In summary, our study showed that c-kit other than any other mutations would influence the OS in AML1-ETO patients. A proposed predictor combining both clinical and genetic factors is applicable to prognostic prediction in AML1-ETO patients.
Introduction
The t(8;21)(q22;q22) is the most commonly observed chromosomal translocation in acute myeloid leukemia (AML) patients; it generates the AML1-ETO (AE) fusion protein (1–4). Adult AMLs with AML1-ETO account for approximately 5%–8% (5–7) and are associated morphologically with AML-M2/M4 subtypes. The median age of these patients is considerably lower, and the prognosis is better compared to normal-karyotype AMLs or other chromosome aberrations. This favorable consequence is associated with a higher complete remission (CR) rate and lower relapse incidence (8–10). However, some subtypes of AML1-ETO-postive AML patients were observed with a higher incidence of relapse and poorer outcomes, and the coexisting c-kit activation mutations may be one of the underlying reasons. Adult patients with CBF leukemia had 12.8%–46.1% of c-kit mutations (11–13). Whether or not other genetic mutations will cooperate with AML1-ETO to drive disease progression is still unknown. We do not know, for example, if oncogene mutations such as ASXL2, KMT2A, and TET2 in CN-AMLs have any biological influence in AML1-ETO-postive AML or if the co-occurrences of driver genes with AML1-ETO would contribute to poor clinical outcomes. In fact, it has been reported that mutations in coding driver genes determined by next-generation sequencing (NGS) technology, such as c-kit, cause other molecular disorders that lead to myeloid leukemia. Additionally, the widely established genetic factors can predict clinical outcomes for newly diagnosed AML1-ETO-postive AMLs. Therefore, it is urgently needed to further study the characteristics of the AML1-ETO molecular, particularly in assessing their accompanied genes and their more potential prognostic markers.
Although previous studies have observed poor prognostic roles of the concomitant gene with KIT (9, 10), the impact of the specific mutation sites cooperated with their concomitant genes for the Chinese population, and the most important co-mutations with distinct prognosis in AML1-ETO patients are still controversial. Meanwhile, whether or not some critical co-mutations may translate into treatment decisions in clinical practice still requires investigation. According to European Leukemia Net (ELN), AML1-ETO patients with c-kit mutation tend to have an unfavorable prognosis with higher relapse rates. However, there are various mutation sites, and we do not know which one is the key (14). It is also reported that AML1-ETO patients with c-kit of EV8-11 may suffer from poor outcomes (15). In summary, the aforementioned studies implied that more co-mutations with prognostic significance remain to be discovered.
In recent years, NGS has been used in genomic and epigenetic research with the advantage of high throughput, high sensitivity, and high stability. However, there still lacks uniformed statement about how to precisely identify the risk stratification of patients with AML1-ETO, especially combining with the results of NGS. Herein, we conducted a study of 254 AML1-ETO-positive AML patients to explore the genetic profiling and identify the co-mutated genes with distinct prognosis. In summary, the aim of this study is to construct an easily applicable complex gene model for overall survival (OS) in patients with newly diagnosed AML1-ETO-positive AML.
Methods
Cases
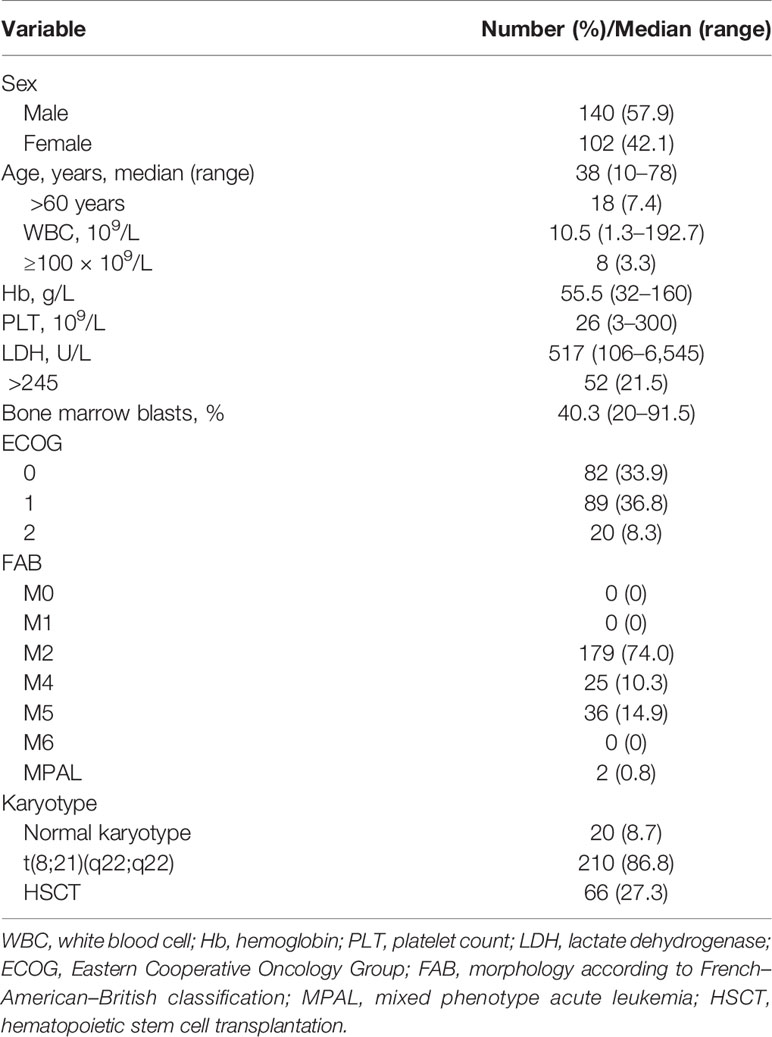
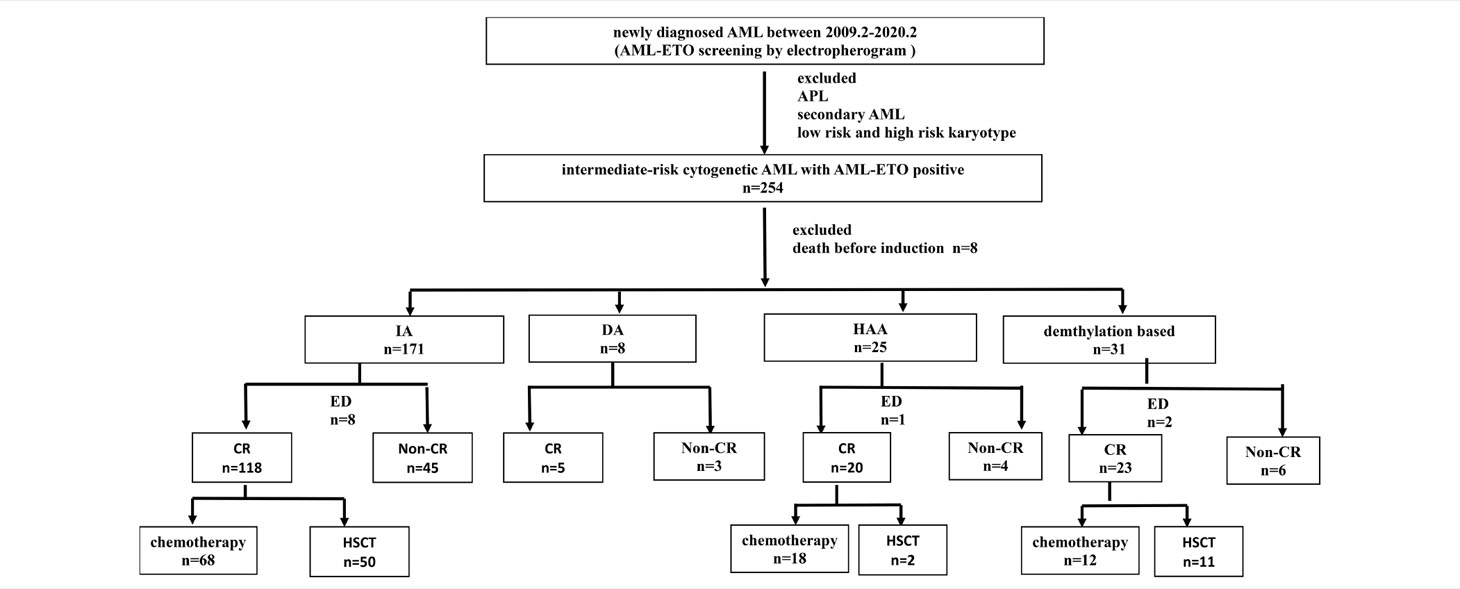
This study included 254 patients with de novo AML with AML1-ETO-positive, who were diagnosed between December 2009 to March 2020 and treated in the First Affiliated Hospital of Zhejiang University (Figure 1). Patients who died before induction or receiving non-standard dose induction therapies were excluded. All patients were informed about the study and provided written consents following the Declaration of Helsinki. The study was approved by the ethics committee of the First Affiliated Hospital of Zhejiang University. Diagnosis of AML was determined in accordance with the 2016 revision of World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia (16). Cytogenetic risks were classified based on the updated risk stratification and management (17). All 254 patients received induction therapy. Among them, 70.9% (n = 180) received IA 3 + 7 regimen [idarubicin 8–10 mg/m2 for 3 days plus cytarabine (Ara-C) 100 mg/m2 for 7 days], 12.2% (n = 31) received demethylation treatment [decitabine 10 mg/m2 for 3 days plus low-dose chemotherapy (Ara-C) 20 mg/m2 for 14 days], and 9.8% (n = 25) received HAA regimen (homoharringtonine 2 mg/m2, Ara-C 100 mg/m2, and aclarubicin 20 mg/day for 7 days). Patients who failed to achieve CR after first induction were treated by reinduction (same regimen if partial remission after the first cycle, crossed regimen if no remission). Patients who relapsed and had suitable donor underwent allogeneic hematopoietic stem cell transplantation (allo-HSCT).
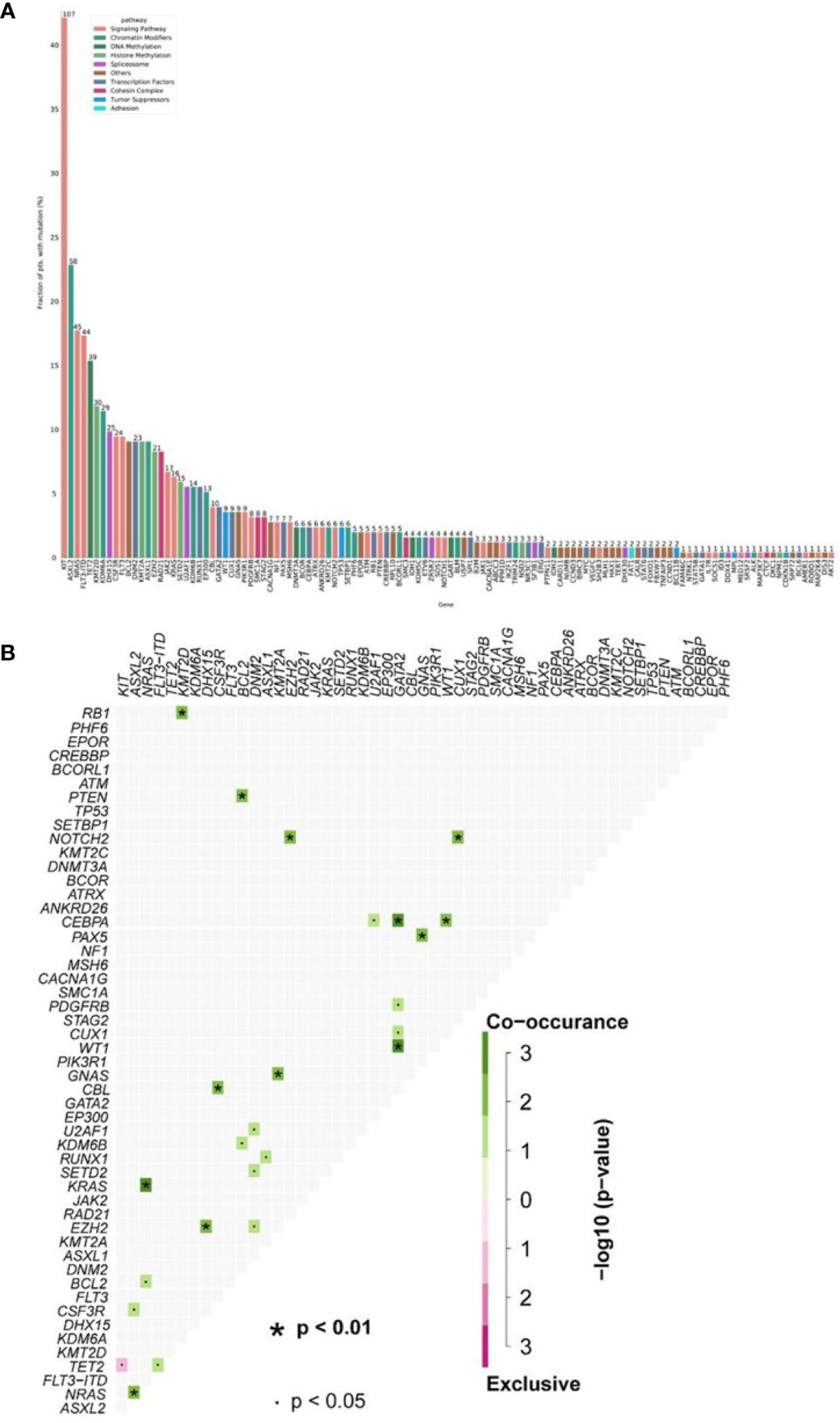
Figure 1 (A) Mutational landscape in 254 AML1-ETO patients. (B) The number of gene mutations per patient. (Each row represents stated gene; each column represents a patient; the right side of the graph annotates the frequency and number of the gene; the upper histogram showed the number of gene mutations per patient; different colors below the graph represent different mutation patterns).
Next-Generation Sequencing Experiment Design and Methods
NGS was performed in all 254 patients using a panel of 185 genes, which covered all the mutation hotspots of acute leukemia (AL), myelodysplastic syndrome (MDS), and myeloproliferative neoplasms (MPNs) (Supplementary Table S1). The gene chip was provided by Acornmed Biotechnology Co., Ltd. Genomic DNAs were extracted from mononuclear cells isolated by Ficoll gradient centrifugation from bone marrow (BM) samples at primary diagnosis by DNeasy 96 Blood & Tissue Kit (Qiagen, Hilden, Germany). Gene library amplification was based on KAPA Library Amplification Kit (KAPA, Boston, MA, USA). Roche NimbleGen kit (Roche, Basel, Switzerland) was used to capture the target region. The hybridized captured samples were sequenced using NovaSeq 6000 PE150.
Preprocessing of raw sequence data and quality control statistics were performed by using an indigenous QC program. Reads were aligned to the hg19 version of the human genome using Burrows–Wheeler Alignment tool (BWA, version 0.7.12). PCR duplicates were marked using the MarkDuplicates tool in Picard. IndelRealigner and BaseRecalibrator on Genome Analysis Toolkit (GATK; version 3.8) were used to conduct realignment and recalibration of the BWA alignment results, respectively. Mutect2 was used for identifying paired-sample variant calling of SNV and INDEL. We obtained candidate variations through background database filtering of normal samples. Pindel (version 0.2.4) (13) was used for detecting CBF. We also compared the quantitative results with electropherogram to ensure the reliability of NGS. All the variants were annotated by ANNOVAR software.
Multiplexed libraries were sequenced using 150-bp paired-end runs on an Illumina Novaseq. To ensure the quality of data, the following criteria were performed to filter raw variant results: Average effective sequencing depth on target per sample ≥1,000x; Allele mutation frequency ≥1% for single-nucleotide variation and insertion or deletion, respectively; all reads were filtered by high Mapping quality (≥30) and Base quality (≥30); the mutant reads need to be supported by positive and negative strands. Furthermore, we excluded synonymous variants, as well as Single Nucleotide Polymorphisms (SNPs) when reported in the 1000 Genomes Project database (October 2014 release) at a population frequency >1% or our in-house SNP databases.
Definition of Clinical End Points
CR was defined as less than 5% bone marrow blasts and showing normal maturation of all cell lineages, no blast in peripheral blood, absolute neutrophil count >1.0 × 10E9/L, platelet count (PLT) >100 × 10E9/L, and no extramedullary leukemia. Early death (ED) referred to all causes of death within 30 days from the first day of induction chemotherapy. Relapse was defined as more than 5% blasts in bone marrow, reappearance of blasts in peripheral blood, or extramedullary leukemia in patients with previously documented CR. OS was defined as the time from diagnosis to death or last follow-up. Relapse-free survival (RFS) was defined as the time from first CR to relapse, censoring at death in CR or last follow-up. The last follow-up was done on December 5, 2020.
Statistical Analysis
Data analysis was performed with SPSS (Version 25) and R (version 3.6.3). Differences in continuous variables were analyzed by Mann–Whitney U-test, and categorical variables were compared by chi-square test or Fisher’s exact test. OS and RFS probabilities were estimated using the Kaplan–Meier survival analysis, and the differences in survival curves were compared by log-rank test. Cox hazard models were used to assess the clinical variables with survival. Factors for univariate analysis included clinical characteristics [sex, age (median 38 years)], laboratory characteristics [white blood cells (WBCs) (median 17.9 × 109/L), hemoglobin (Hb) (median 79 g/l), PLT (median 40 × 109/L), lactate dehydrogenase (median 757 U/L), bone marrow (BM) blasts (median 48%)], and AML1-ETO molecular characteristics [VAF (0.48%), AML1-ETO number (1 or more than 1), length (bps), location (JM/beta1-sheet/other domains)]. A statistical significance level of 0.1 in the univariate analysis was used to select variables in the risk score system. Least Absolute Shrinkage and Selector Operation (LASSO) Cox regression model was used in variable selection and predictive prognostic model construction. A two-tailed p < 0.05 was considered significant.
Results
Clinical Characteristics of the AML1-ETO-Positive Patients at Diagnosis
Patients’ characteristics and additional molecular and cytogenetic aberrations are shown in Table 1 according to CBF subtype. The median follow-up was 4.5 years.
Of the total 254 patients, the median age was 38 (10–78) years and 41.7% (106/254) were male. AML1-ETO was most common in M2 (68.9%, 175/254), second in M5 (15.0%, 38/254) (Table 1). Consistent with previous findings (7, 14), high WBC count (median 17.9 × 10E9/L, 55.1% over 100 × 10E9/L) and high percentage of BM blasts (median 48.0%, range 3.0%–91.5%) were observed. Among all the patients, 11.8% (30/254) had normal karyotypes, 5.1% (13/254) had -X chromosomes, and 11.0% (28/254) had -Y chromosomes. CRs were achieved in 85.0% (216/254) patients, and 27.6% (70/254) relapsed after CR.
The total number of patients of the HAA group is lower than that of the IA group. The CR rates were different between IA group and HAA group (p = 0.01). Early deaths occurred in 5.1% (13/254) patients. The median follow-up was 53.7 (6–121) months for all patients. Median OS was 11.4 (95% CI 7.8–15.0) months, and median RFS rate was not reached; OS rate at 3 years was 28.4% ± 6.0%, and RFS rate at 3 years was 57.8% ± 8.0%. In addition, 40.8% (20/49) patients underwent HSCT in CR1, which showed significantly superior outcomes than those who did not (OS p = 0.002, RFS p = 0.015, respectively). Details of the patients were summarized in Tables 1 and 2. The OS of HAA plus decitabine regimen was superior than that of IA regimen, but there is no significant difference between HAA regimen and decitabine regimen. Due to the limited number of patients with c-kit gene (3 out of all 31 patients) receiving decitabine regimen (Figure 2), it is not feasible for us to take further evaluation.
Figure 2 Overall survival (OS) in the different regimen patient groups. Red line: HAA regimen, 3-year OS 80.00% ± 12.65%; Green line: HAA/decitabine regimen, 3-year OS 60.12% ± 11.13%; Blue line: IA regimen, 3-year OS 57.21% ± 4.58%.
Mutational Landscape of AML1-ETO-Positive Patients
Thirty-seven different mutant genes were detected in the 254 patients, and the details of mutations were presented in Figure 1. The top five common concomitant mutations were KIT (42%), ASXL1 (23%), NRAS (17.7%), FLT3-ITD (17.3%), and TET2 (15.3%). The median number of co-mutant genes was 3 (0–10), but the number of mutations did not affect the OS and RFS except kit (both p > 0.05). For genes detected in more than 3 patients, we performed gene association analysis and found interesting coexistence and mutual exclusion relationships (Figure 1). For example, kit, FLT3-ITD, and WT 1,IDH2 tend to appear together, while NPM 1-WT1 and NPM 1–RUNX1 appear to be mutually exclusive (p = 0.00008; p = 0.0014, respectively). Here, 132 patients (29.1%) have 2 or more mutations/fusion genes (18). In our study, the frequencies of concomitant genes were WT1 24.4% (62/254) FLT3-ITD 17.3% (44/254). There was a study that showed that mutations of c-kitD835 were associated with poor prognosis and elevated WBCs, especially in patients with inv(16) (19). C-kitD816 was the most common mutation in our study; the frequency was 54% (58/107) in all the c-kit mutation patients. These mutations mostly occur in exon 8 or 17 and are observed in 25% AML1-ETO cases (11). We found 124 KIT mutations in 84 patients, of which 91 mutations were located in exon 17 (73%), mainly SNVs at D816 or N822. In exon 8, we found 24 mutations (19%), mainly in-frame insertions/deletions at positions 416 to 422.
The Numbers of c-kit Concomitant Mutations
C-kit mutations are significant prognostic predictors in CBF-AML patients with t(8;21) and inv(16), which are associated with poor prognosis; however, current findings are inconsistent in this regard (20, 21). Nevertheless, different OS rates have been reported for CBF-AML patients with c-kit mutations compared to others (6) (Figure 3). Current research still has inconsistent conclusions regarding whether c-kit is related to poor prognosis, while our results had clearly shown a relationship.
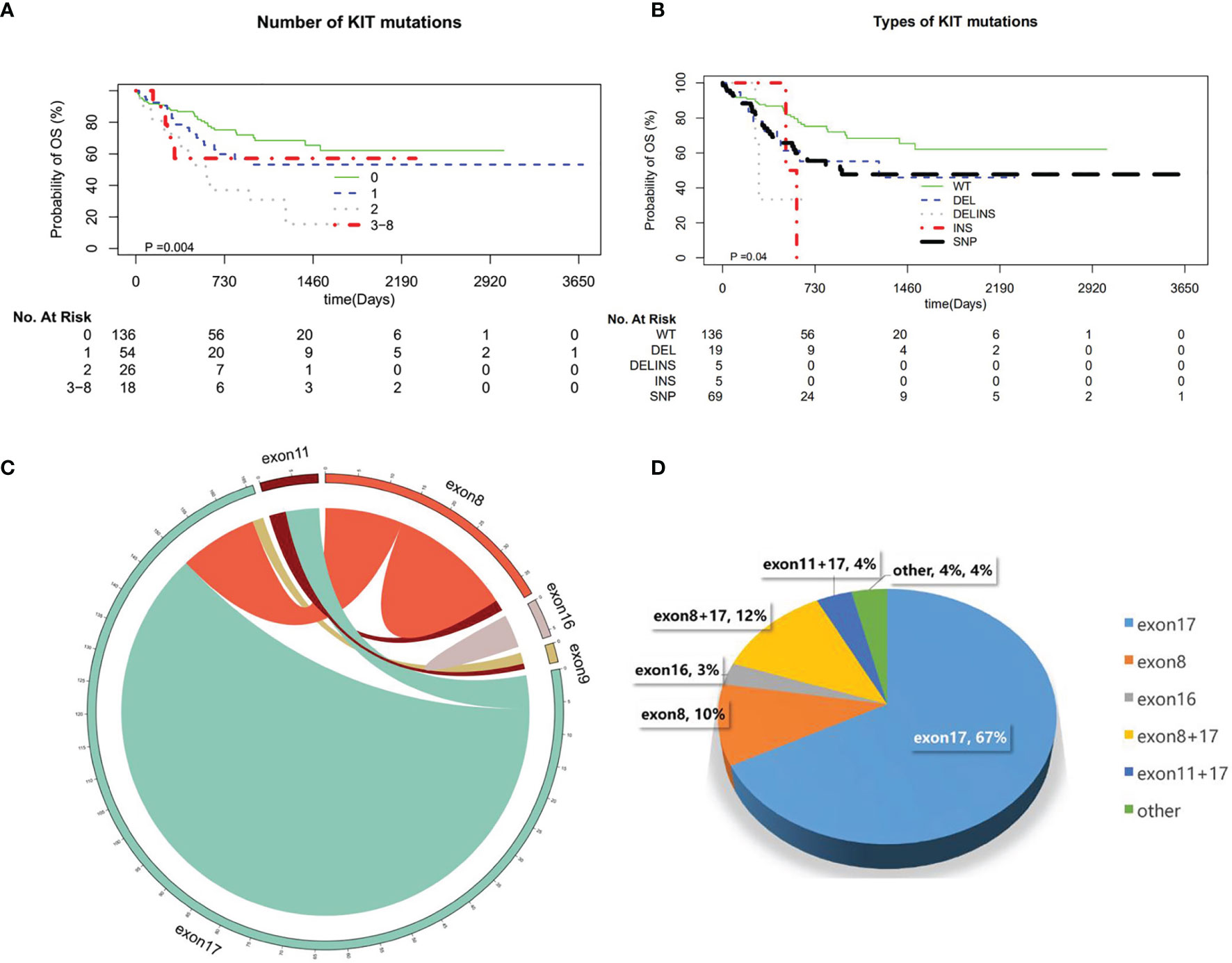
Figure 3 (A) Number of KIT mutations. (B) Types of KIT mutations. (C) The distribution of KIT exons. (D) The scale of KIT exons.
In our study, the frequency of c-kit concomitant genes were 42.0% (107/254), which is the same as in other studies (22). In the patients with c-kit mutations, the one-site mutation was most common. Two sites were rare. There were also 3–8 sites of c-kit mutations. We analyzed the patients with c−KIT mutation into exon 8, 17 and exon 10 groups; the numbers of the mutation had no effect on OS. There is no significant difference in OS whether there are two or three mutation sites. And for the mutation form of c-kit, insertion or deletion has no obvious impacts on OS. Although the mutation mode of INS in our study has a statistical difference in survival, considering the small sample size, it needs a large cohort of patients to further confirm the result.
Molecular Characteristics of the AML1-ETO
The molecular structure characteristics of the AML1-ETO were summarized in Figure 1. The OS and RFS according to different molecular characteristics of AML1-ETO were shown in our results (Figure 4). Except for c-kit mutation, there is no significant difference in the OS of patients regardless of the genetic mutations.

Figure 4 Overall survival (OS) and recurrence-free survival (RFS) in the entire patient group according to different molecular characteristics of AML1-ETO. (A) Mutation with NRAS. (B) Mutation with FLT3-ITD. (C) Mutation with ASXL2S. (D) Mutation with TET2.
The frequency of FLT3-ITD concomitant gene was 17.3% (44/254). The FLT3-ITD is a gene suggested with poor prognosis in AML, especially the FLT3-ITD mutation of higher frequency is an important factor affecting survival. In our study, the proportion of patients with higher frequency of FLT3-ITD mutations was suggested in AML1-ETO patients, while there is no impact on the OS in AML1-ETO patients, regardless of whether they are accompanied by FLT3-ITD mutation. The same results were also shown in AML1-ETO patients with ASXL2, NRAS mutation. That is different from the study of Duployez et al. (15). According to their results, the association of mutations remained associated with the highest hazard of relapse in patients with t(8;21) AML (15). C-kit, ASXL2, NRAS, and FLT3-ITD mutations were most common in our cohort. The OS showed no difference in AML1-ETO patients with these mutations.
A Risk Score Combined Clinical and Molecular Profiles
Patients (n = 75) from the First Affiliated Hospital of Soochow University and Ningbo First Hospital were used as a validation cohort. The median age in the analyzed patients was 42 (6–78) years. The most common recurrent mutations occurred in KIT (n = 84, 50%), ASXL2 (n = 46, 28%), NRAS (n = 37, 22%), FLT3-ITD (n = 35, 21%), and TET2 (n = 30, 18%). We observed that high KIT mutant allele burden predicts poor outcome in t(8:21) AML. High KIT VAF (≥15%) correlated with shortened OS compared to the other KIT mutated cases including low VAF and wild-type KIT [3-year OS 26.6% vs. 59.0% vs. 69.6%, hazard ratio (HR) 1.50, 95% CI 0.78–2.89, p = 0.0005]. In addition, we also identified that some other mutated genes influence the prognosis of patients with t(8;21), such as FLT3-ITD high mutation burden (VAF ≥44% vs. other cases, 3-year OS 30.0% vs. 56.2%, HR 2.94, 95% CI 0.43–20.18, p = 0.056), TET2 high mutation burden (VAF ≥43% vs. other cases, 3-year OS 33.3% vs. 56.5%, HR 2.87, 95% CI 0.66–12.46, p = 0.018), and DHX15 high mutation burden (VAF ≥22% vs. other cases, 3-year OS 15.0% vs. 58.3%, HR 2.65, 95% CI 0.81–8.73, p = 0.011).
In univariate analyses for OS, age >42 years (3-year OS 46.3% vs. 64.4%, HR 1.91, 95% CI 1.14–3.14, p = 0.012), WBC >27.1 × 109/L (3-year OS 34.3% vs. 60.0%, HR 2.59, 95% CI 1.13–5.9, p = 0.001), BM blast >20% (3-year OS 52.2% vs. 92.8%, HR 6.36, 95% CI 2.7–14.97, p = 0.035), LDH >504 U/L (3-year OS 44.1% vs. 67.1%, HR 2.62, 95% CI 1.50–4.59, p = 0.0007), PLT ≤28 × 109/L (3-year OS 47.1% vs. 66.9%, HR 1.89, 95% CI 1.13–3.17, p = 0.019), and Hb ≤87 g/L (3-year OS 49.4% vs. 73.8%, HR 2.20, 95% CI 1.27–3.84, p = 0.019) were significantly associated with poor OS. Six variables were incorporated in our scoring model by LASSO, including age, WBC, PLT, KIT mutation, FLT3-ITD mutation, and TET2 mutation. A risk scoring model was developed by incorporating the weighted coefficients of these variables. The risk score grouped AML1-ETO AML patients into two subgroups: low risk (LR; n = 68) and high risk (HR; n = 86) groups. The 3-year OS rates for LR and HR groups were 72.7% and 43.0% (p < 0.0001; Figure 5). Similar results were also observed in the validation cohort (3-year OS 79.1% vs. 49.5%, p = 0.01; Figure 5). Concordance index (train: 0.708, 95% CI 0.680–0.736; validation: 0.722, 95% CI 0.666–0.778) demonstrated discrimination power well, and calibration plots showed that the nomograms did well compared with an ideal model. Six variables were incorporated in our scoring model by LASSO, including age, WBC, PLT, KIT mutation, FLT3-ITD mutation, and TET2 mutation. A risk scoring model was developed by incorporating the weighted coefficients of these variables. The risk score grouped AML1-ETO AML patients into two subgroups: LR (n = 68) and HR (n = 86) groups. The 3-year OS rates for LR and HR groups were 72.7% and 43.0% (p < 0.0001; Figure 5). Similar results were observed in the validation cohort (3-year OS 79.1% vs. 49.5%, p = 0.01; Figure 5).
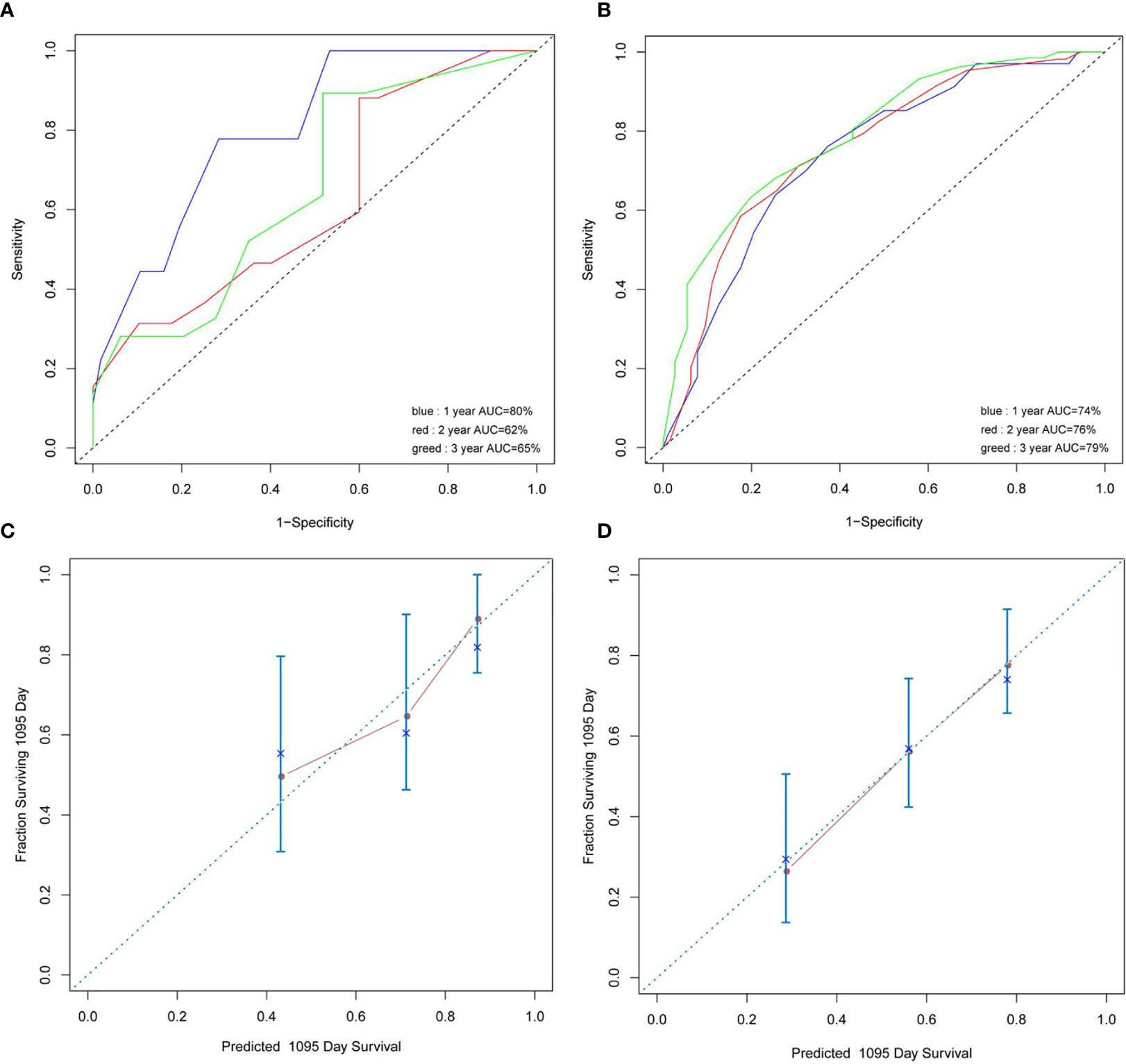
Figure 5 (A, B) Concordance index showed the risk score efficacy. (C, D) The calibration plots showed that the nomograms did well compared with an ideal model.
Discussion
In this study, we analyzed the gene distribution of AML1-ETO patients by NGS platform. It was concluded that Chinese AML1-ETO patients had many coexisting gene mutations, and the interaction was very complicated. Meanwhile, we explored the structural characteristics of AML1-ETO of each patient by NGS platform and found that the number of C-kit had significant effects on prognosis. Since multiple factors could affect the prognosis, we propose, for the first time, gene mutations of a large number of AML1-ETO-positive patients to predict patient outcomes. The databases from this study may be helpful for further investigation of the risk stratification and prognostic prediction regarding patients with AML1-ETO AML.
It is believed that the development of AML is a multistep process, which requires at least two genetic abnormalities for the development of the disease (17). We analyzed the gene expression of the 254 de novo AML1-ETO patients by target sequencing and discovered that most of the co-mutations clustered in methylation-related genes, chromatin-modifying genes, and transcription factor genes. C-kit, ASXL2, NRAS, and FLT3-ITD mutations were most common, while mutations such as Bcl-2 and KMT2A were rare. The distribution according to gene function was similar to reports by Yu et al. and Duployez et al. using NGS, but the contaminant mutations without c-kit did not influence OS in our study (15, 22). Duployez et al. (15) showed that among the 215 patients analyzed, 182 (85%) had at least 1 mutation. In our study, the main mutation genes were c-kit (42%), ASXL2 (23%), NRAS (17.7%), and FLT3-ITD (17.3%). This is consistent with findings in the existing literature. While Duployez et al. (15) showed that diverse cooperating mutations may influence CBF-AML pathophysiology as well as clinical behavior and point to potential unique pathogenesis of t(8;21) vs. inv(16) AML, our study looks at AML-ETO patients in general, with an emphasis on the gene mutational landscape and the risk score combined clinical and molecular profiles, which can be a prognosis suggestion for AML-ETO patients.
Our results are similar with those in the Hong Kong center. More importantly, our study included the largest number of patients. Some studies showed that the most common mutation types combined with AML1-ETO patients are C-KIT, NRAS, and FLT3. Among all combined mutations, C-KIT has a poor prognosis and NRAS is meaningless. Our research is consistent with this result. Our study also found that FLT3-ITD in all patients did not affect the survival of AML1-ETO patients if the mutation load is not considered. When we proposed a threshold value, we found that 44% can be used as the value for the mutation load. The patients above this value correlated with poorer OS. This result can be used as the FLT3-ITD with AML1-ETO mutation threshold for Chinese people.
CBF-AMLs are commonly associated with favorable prognosis; however, this prognosis can be changed. With some kinds of gene mutations, correspondingly, only 50% of CBF-AML patients are able to preserve long-term remission without any relapse. Our results showed that the 3-year OS of HAA group was 80.00% ± 12.65%, while that of the IA group was 57.21% ± 4.58%. We proposed that the HAA regimen had more survival benefits than IA regimen, which was consistent with the result of Cao et al. (23). Seyhan et al. showed that RAS genes and identified their association with doxorubicin and etoposide sensitivity. They also found that GF2R, CTSA, and ATP6AP2 were gene biomarkers, which can subgroup AML patients into distinct good and bad prognostic groups (24). We try to find more drug-sensitive biomarker genes for AML-ETO patients and improve AML patient efficiency.
Previous studies have shown that AML1-ETO combined with FLT3-ITD would affect the survival of patients; however, our results suggested that the combination of FLT3-ITD did not affect the OS in this kind of patients. Our study included 254 AML1-ETO-positive AML patients. Although our study is a retrospective one, the follow-up time is long. It is also a study with a larger number of cases included.
The results of several studies showed the gene file of AML1-ETO and some prognostic effects of molecular traits of AML1-ETO. In our study, we included 254 AML1-ETO AML patients, and we found controversial results. This is probably due to the heterogeneity of the patients, difference in treatment approaches, and the effect of co-mutations.
Up to now, little information is available for contaminant mutation and predicting the prognosis of so many AML1-ETO patients in China. In this study, only c-kit mutation was able to distinguish patients with good and poor prognosis among many mutation genes concomitant with AML1-ETO. The numbers of c-kit or the presence of FLT3-ITD could not influence the OS of AML1-ETO patients. And the age, WBC count, and blast cell count are related with the OS of AML1-ETO patients. This new model may help doctors to interfere in advance to improve the prognosis and reduce the mortality of patients.
There are still some limitations in the current study. First, due to its retrospective nature, induction and consolidation regimens cannot be completely unified. Furthermore, some included patients had a high loss ratio of survival data. A larger sample size of prospective study is required to further verify the results. In this study, multi-gene sequencing and comprehensive prognostic analysis were performed for AML1-ETO-positive Chinese patients. These findings not only provided important information of the molecular structure characteristics of AML1-ETO, but they also revealed the important contaminant mutated genes with distinct clinical outcomes.
Data Availability Statement
The sequencing data presented in our study are publicly available in the NCBI Sequence Read Archive (SRA) repository, with accession number PRJNA784000.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of the First Affiliated Hospital of Zhejiang University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JJ designed this study. MY, YZ, BZ, HM, WY, HT, JH and HZ collected and integrated the clinical materials. FL, SC, LL and JQ provided support for NGS. JW and CW displayed the statistical analysis of data. MY, YZ, CH, and JJ contributed to the final draft, and all authors reviewed and approved the final draft.
Funding
This work was supported by the Zhejiang Province Natural Science Foundation of China grants (LY16H160009).
Conflict of Interest
LL, JQ, FL, SC, and CW are employed by Acornmed Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Xiaoxia Hu and Xianxi Liu for their help in the initial phase of this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.783114/full#supplementary-material
Supplementary Table S1 | A panel of 185 genes, which covered all the mutation hotspots of acute leukemia (AL), myelodysplastic syndrome (MDS), and myeloproliferative neoplasms (MPNs).
References
1. Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, et al. Identification of Breakpoints in T(8;21) Acute Myelogenous Leukemia and Isolation of a Fusion Transcript, AML1/ETO, With Similarity to Drosophila Segmentation Gene, Runt. Blood (1992) 80(7):1825–31. doi: 10.1182/blood.V80.7.1825.bloodjournal8071825
2. Andrieu V, Radford-Weiss I, Troussard X, Chane C, Valensi F, Guesnu F, et al. Molecular Detection of T(8;21)/AML1-ETO in AML M1/M2:correlation With Cytogenetics, Morphology and Immunophenotype. Br J.Haematol (1996) 92:855–65. doi: 10.1046/j.1365-2141.1996.415954.x
3. Moroi K, Sato T. Comparison Between Procaine and Isocarboxazid Metabolism In Vitro by a Liver Microsomal Amidase-Esterase. Biochem.Pharmacol (1975) 24:1517–21. doi: 10.1016/0006-2952(75)90029-5
4. Rowley JD. Recurring Chromosome Abnormalities in Leukemia Andlymphoma. Semin Hematol (1990) 27:122–36.
5. Pollyea DA, Kohrt HE, Medeiros BC. Acute Myeloid Leukemia in the Elderly: A Review. Br J Hematol (2010) 152:524–42. doi: 10.1111/j.1365-2141.2010.08470.x
6. Riera L, Marmont F, Toppino D, Frairia C, Sismondi F, Audisio E, et al. Core Binding Factor Acute Myeloid Leukemia and C-KIT Mutations. Oncol Rep (2013) 29:1867–72. doi: 10.3892/or.2013.2328
7. Cairoli R, Beghini A, Grillo G, Nadali G, Elice F, Ripamonti CB, et al. Prognostic Impact of C-KIT Mutations in Core Bindingfactor Leukemias: An Italian Retrospective Study. Blood (2006) 107:3463–8. doi: 10.1182/blood-2005-09-3640
8. Bloomfield CD, Lawrence D, Byrd JC, Carroll A, Pettenati MJ, Tantravahi R, et al. Frequency of Prolonged Remission Duration After High-Dose Cytarabine Intensification in Acute Myeloid Leukemia Varies by Cytogenetic Subtype. Cancer Res (1998) 58:4173–9.
9. Marcucci G, Caligiuri MA, Bloomfield CD. Molecular and Clinical Advances in Core Binding Factor Primary Acute Myeloid Leukemia: A Paradigm for Translational Research in Malignant Hematology. Cancer Invest (2000) 18:768–80. doi: 10.3109/07357900009012209
10. Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment Cytogenetic Abnormalities are Predictive of Induction Success, Cumulative Incidence of Relapse, and Overall Survival in Adult Patients With De Novo Acute Myeloid Leukemia: Results From Cancer and Leukemia Group B (CALGB 8461). Blood (2002) 100:4325–36. doi: 10.1182/blood-2002-03-0772
11. Malaise M, Steinbach D, Corbacioglu S. Clinical Implications of C-Kit Mutations in Acute Myelogenous Leukemia. Curr Hematol Malig Rep (2009) 4:77–82. doi: 10.1007/s11899-009-0011-8
12. Park SH, Chi HS, Min SK, Park BG, Jang S, Park CJ. Prognostic Impact of C-KIT Mutations in Core Binding Factor Acute Myeloid Leukemia. Leuk Res (2011) 35:1376–83. doi: 10.1016/j.leukres.2011.06.003
13. Paschka P, Marcucci G, Ruppert AS, Mro´zek K, Chen H, Kittles RA, et al. Adverse Prognostic Significance of KIT Mutations in Adult Acute Myeloid Leukemia With Inv(16) and T(8;21): A Cancer and Leukemia Group B Study. J Clin Oncol (2006) 24:3904–11. doi: 10.1200/JCO.2006.06.9500
14. Nguyen S, Leblanc T, Fenaux P, Witz F, Blaise D, Pigneux A, et al. A White Blood Cell Index as the Main Prognostic Factor in T(8;21) Acute Myeloid Leukemia (AML): A Survey of 161 Cases From the French AML Intergroup. Blood (2002) 99:3517–23. doi: 10.1182/blood.V99.10.3517
15. Duployez N, Marceau-Renaut A, Boissel N, Petit A, Bucci M, Geffroy S, et al. Comprehensive Mutational Profiling of Core Binding Factor Acute Myeloid Leukemia. Blood (2016) 127(20):2451–9. doi: 10.1182/blood-2015-12-688705
16. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood (2016) 127(20):2391–405. doi: 10.1182/blood-2016-03-643544
17. Estey EH. Acute Myeloid Leukemia: 2021 Update on Risk-Stratification and Management. Am J Hematol (2020) 95(11):1368–98. doi: 10.1002/ajh.25975
18. Zhao CX, Wang JM, Li JM, Zou SH, Chen FY, Liang AB, et al. [Using Next Generation Sequencing Technology to Analyze Gene Mutations in Patients with Acute Myeloid Leukemia and the Impact on Prognosis]. Zhonghua Yi Xue Za Zhi (2019) 99(40):3145–51. doi: 10.3760/cma.j.issn.0376-2491.2019.40.005
19. Ayatollahi H, Shajiei A, Sadeghian MH, Sheikhi M, Yazdandoust E, Ghazanfarpour M, et al. Hematol Oncol Stem Cell Ther. Hematol Oncol Stem Cell Ther (2017) 10(1):1–7. doi: 10.1016/j.hemonc.2016.08.005
20. Huh J, Kim HJ, Jung CW, Kim HJ, Kim SH, Kim YK, et al. A Genome-Wide Single-Nucleotide Polymorphism-Array can Improve the Prognostic Stratification of the Core Binding Factor Acute Myeloid Leukemia. Am J Hematol (2012) 87:961–8. doi: 10.1002/ajh.23281
21. Park SH, Lee HJ, Kim IS, Kang JE, Lee EY, Kim HJ, et al. Incidences and Prognostic Impact of C-KIT, WT1, CEBPA, and CBL Mutations, and Mutations Associated With Epigenetic Modification in Core Binding Factor Acute Myeloid Leukemia:A Multicenter Study in a Korean Population. Ann Lab Med (2015) 35:288–97. doi: 10.3343/alm.2015.35.3.288
22. Yu G, Yin C, Wu F, Jiang L, Zheng Z, Xu D, et al. Gene Mutation Profile and Risk Stratification in AML1−ETO−positive Acute Myeloid Leukemia Based on Next−Generation Sequencing. Oncol Rep (2019) 42:2333–44. doi: 10.3892/or.2019.7375
23. Cao J, Feng H, Ding NN, Wu QY, Chen C, Niu MS, et al. Homoharringtonine Combined With Aclarubicin and Cytarabine Synergistically Induces Apoptosis in T(8;21) Leukemia Cells and Triggers Caspase-3-Mediated Cleavage of the AML1-ETO Oncoprotein. Cancer Med (2016) 5(11):3205–13. doi: 10.1002/cam4.913
Keywords: acute myeloid leukemia, AML-ETO, next-generation sequencing, prognosis, LASSO Cox regression
Citation: Yang M, Zhao B, Wang J, Zhang Y, Hu C, Liu L, Qin J, Lou F, Cao S, Wang C, Yu W, Tong H, Meng H, Huang J, Zhu H and Jin J (2022) A Predictor Combining Clinical and Genetic Factors for AML1-ETO Leukemia Patients. Front. Oncol. 11:783114. doi: 10.3389/fonc.2021.783114
Received: 25 September 2021; Accepted: 07 December 2021;
Published: 14 January 2022.
Edited by:
Anna Maria Testi, Sapienza University of Rome, ItalyReviewed by:
Matteo Molica, S. Eugenio Hospital, ItalyIbrahim C. Haznedaroglu, Hacettepe University Hospital, Turkey
Felicetto Ferrara, Hospital Antonio Cardarelli, Italy
Copyright © 2022 Yang, Zhao, Wang, Zhang, Hu, Liu, Qin, Lou, Cao, Wang, Yu, Tong, Meng, Huang, Zhu and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Jin, amllajA1MDNAemp1LmVkdS5jbg==
 Min Yang
Min Yang Bide Zhao4
Bide Zhao4 Yi Zhang
Yi Zhang Chao Hu
Chao Hu Lixia Liu
Lixia Liu Jiayue Qin
Jiayue Qin Chengcheng Wang
Chengcheng Wang Hongyan Tong
Hongyan Tong Jian Huang
Jian Huang Honghu Zhu
Honghu Zhu Jie Jin
Jie Jin