
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 06 January 2022
Sec. Cancer Molecular Targets and Therapeutics
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.780387
This article is part of the Research Topic Molecular Targets and Therapeutic Strategies in Gynecological Cancers View all 7 articles
Yimucao has been used as an herbal medicine to treat gynecological diseases. Common genes of Yimucao active compounds were investigated using network pharmacology. The components and targets of Yimucao were retrieved from the TCMSP database. Cervical cancer targets were collected from GeneCards, TTD, DisGeNET, and KEGG. Cisplatin-related genes were downloaded from GeneWeaver. The protein-protein interaction (PPI) network was created using the STRING database. A drug-bioactive compound-disease-target network was constructed using Cytoscape. GO and KEGG analyses were performed to investigate common targets of quercetin and cisplatin in cervical cancer. We found that quercetin was the highly bioactive compound in Yimucao. The drug-bioactive compound-disease-target network contained 93 nodes and 261 edges. Drug-related key targets were identified, including EGFR, IL6, CASP3, VEGFA, MYC, CCND1, ERBB2, FOS, PPARG, and CASP8. Core targets were primarily related to the response to metal ions, cellular response to xenobiotic stimulus, and transcription factor complex. The KEGG pathway analysis revealed that quercetin and cisplatin may affect cervical cancer through platinum drug resistance and the p53 and HIF-1 pathways. Furthermore, quercetin combined with cisplatin downregulated the expression of EGFR, MYC, CCND1, and ERBB2 proteins and upregulated CASP8 expression in HeLa and SiHa cells. Functionally, quercetin enhanced cisplatin-induced anticancer activity in cervical cancer cells. Our results indicate that quercetin can be used to overcome cisplatin resistance in cervical cancer cells.
More than 500,000 women are diagnosed with cervical cancer each year, which causes more than 300,000 deaths worldwide (1). The standardized incidence rate of cervical cancer is expected to be 13.1 cases per 100,000 women worldwide (2). Advanced or recurrent cervical cancer conveys a poor prognosis with a 10–20% 1-year relative survival rate (3). Current treatment approaches for locally advanced cervical cancer include brachytherapy, external beam radiotherapy, and concurrent chemotherapy based on cisplatin (4). Among them, cisplatin is one of the most widely used chemotherapeutic agents. However, 40% of locally advanced cervical cancer patients still experience disease recurrence (5). An economic cost estimate based on cervical cancer demonstrated that per capita expenditures of cervical cancer treatment ranged from US$368 to 11,400 (6). Cervical cancer places heavy burdens on both families and society. Traditional Chinese medicine (TCM) has been applied to treat cancer, having the advantage of acting on a variety of cancer-related signaling pathways and molecular targets without severe side effects (7).
Yimucao has been used for the uterotonic action, dysmenorrhea, lochia, and other gynecological disorders in China for thousands of years (8). Quercetin is a key compound of Yimucao that exhibits good antitumor activity (9, 10), especially for cervical cancer (11–13). One study used HPLC analysis and reported that Yimucao contents primarily include leonurine hydrochloride, quercetin, rutin, kaempferol, and apigenin (14). Many studies have investigated the effect of quercetin on cervical cancer cell apoptosis. For example, Bishayee and colleagues found that quercetin promotes cervical cancer cell apoptosis and inhibits cell cycle by inducing cytochrome c release and ROS accumulation in G2/M (15). One group confirmed that quercetin nanoparticles play a role in inhibiting cervical cancer progression by inhibiting JAK2, which induces autophagy, apoptosis, and antiproliferative activities (16). Furthermore, previous experiments have shown that quercetin promotes G2 phase arrest and apoptosis by activating the p53 signaling pathway (17). Quercetin might also reduce the MMP2, Ezrin, METTL3, and P-Gp expression in cancer cells, enhancing the antitumor effect of cisplatin by inhibiting the proliferation, migration, and invasion and potentiating apoptosis of cervical cancer cells (18). Most connections between Yimucao and the other therapeutic were obtained from previous experience and long-term clinical observations (8). However, the mechanism for the synergy between Yimucao and cisplatin for the treatment of cervical cancer is largely unexplored. Thus, it is necessary to extend the knowledge regarding the synergistic effects of Yimucao chemical compounds with cisplatin in treating cervical cancer using technological approaches.
Recently, the prediction of effective components and potential targets of Chinese medicine using network pharmacology has become increasingly popular for TCM modernization and internationalization (19–21). Network pharmacology enables a more complete understanding of the mechanism of medicine and disease using multiple authoritative databases (22). Chinese medicine treat many complicated diseases, including cancer, as a disturbance of interlinked complex biological networks and determines the drug mechanism of action in accordance with the network topology (23). For instance, Zhu and colleagues demonstrated that β-elemene inhibits peritoneal effusion production in pancreatic cancer by inhibiting the HIF1α-VEGFA pathway using the network pharmacology (24). They studied the molecular biological mechanism of Kushen compound injection and its anticancer effect using network pharmacology and found that the active ingredients of Kushen induced cancer cell apoptosis through the p53 and PI3K-Akt signaling pathways (25). Network pharmacology was also implemented to predict the mechanism of Fuzhengkangai in EGFR mutation-positive lung adenocarcinomas, and two gene targets, BCL2 and PRKCA, were identified (26). These studies have demonstrated that network pharmacology is a useful predictive tool for exploring the chemical components of Yimucao and its effects on cervical cancer.
In the present study, a network pharmacology method was applied to identify the potential biologically active ingredients in Yimucao and to clarify the mechanism by which the active ingredients of Yimucao, particularly quercetin, enhance the effect of cisplatin on cervical cancer and related key proteins (Figure 1).
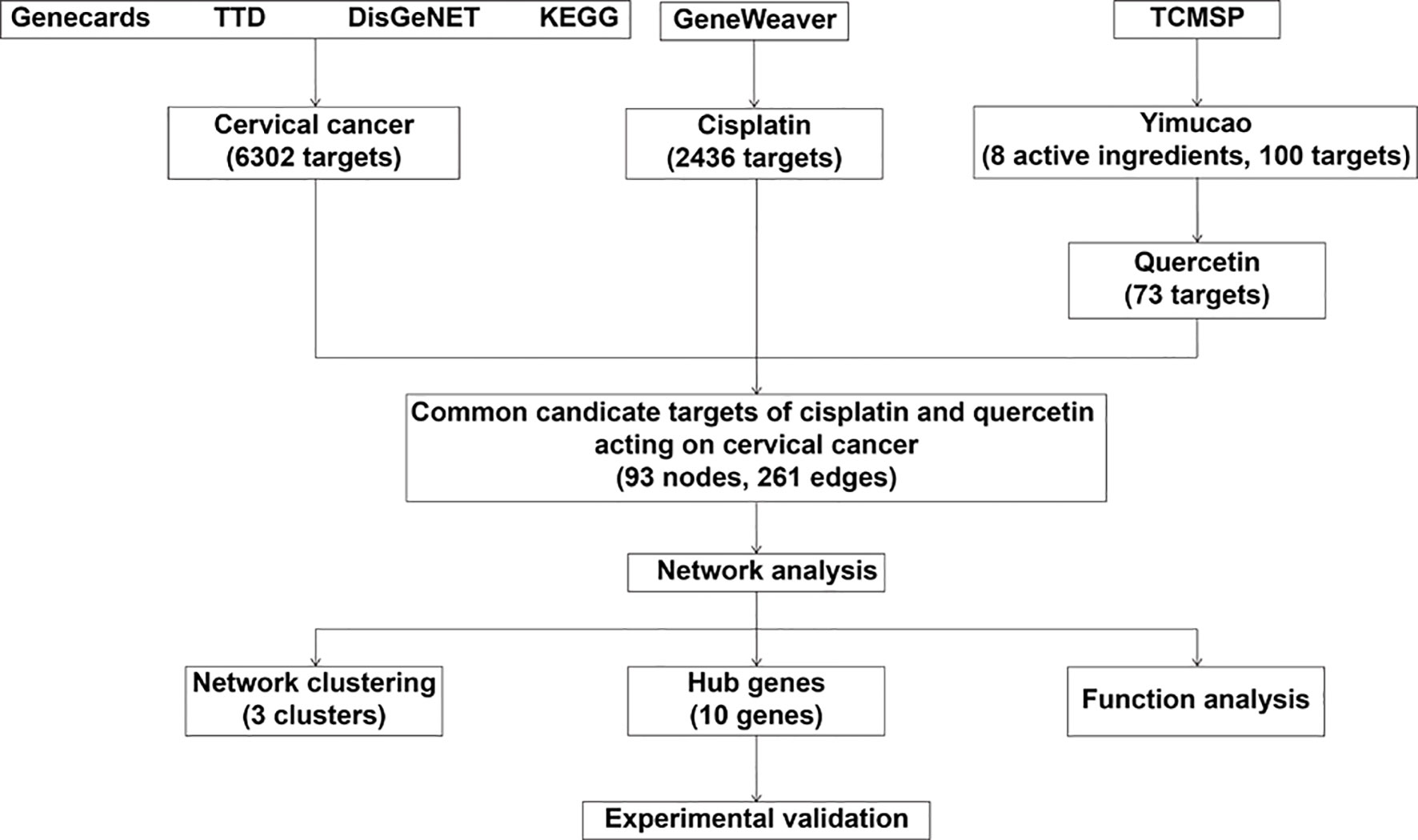
Figure 1 Schematic of the network pharmacological analysis used to identify the potential mechanism by which quercetin enhances cisplatin action in cervical cancer. TTD, Therapeutic Target Database; KEGG, Kyoto Encyclopedia of Genes and Genomes; TCMSP, Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) database.
Information about compounds from Yimucao was identified by searching the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://tcmspw.com/). TCMSP identifies compound–target networks (27). The herbs were input as the keywords to determine all components in TCMSP. Absorption, distribution, metabolism, and excretion (ADME) are strategic processes in drug discovery and development that were used to select the compounds of Yimucao (28). The effect of the compound on the disease is largely determined by the degree of oral bioavailability (OB) (29), and high OB is usually an essential indicator for determining the drug-likeness (DL) index of active substances. Substances with OB ≥ 30% were regarded to have high OB. The first-rank compounds and improved candidate compounds were selected using DL in the initial period of drug development (30). The average DL index in the DrugBank database was 0.18. Substances with a DL index ≥0.18 were regarded to have high druggability. Therefore, compounds with OB ≥ 30% and DL ≥ 0.18 were selected for further analysis in the present study (31).
The potential targets of active components in Yimucao were also obtained from the TCMSP database (27). The species of target proteins was limited to Homo sapiens, and their corresponding official symbols were obtained from UniProtKB (http://www.uniprot.org) (32). In addition, cisplatin-related genes were identified using the GeneWeaver database (33), which comprises a number of gene sets from multiple functional genomic data resources, curated publications, and user submissions. A large dataset consisting of 2,436 (GS125959) genes from humans was selected for further analysis.
Cervical cancer-related genes were identified by searching the following databases: GeneCards (https://www.genecards.org/) (34), Therapeutic Target Database (TTD, https://db.idrblab.org/ttd/) (35), DisGeNET (https://www.disgenet.org/) (36), and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) (37), using the keyword “cervical cancer”. In brief, online databases were searched using the disease names to obtain corresponding target genes, and then the targets of cervical cancer were determined by removing duplicate targets.
Cytoscape 3.8.0 was used to construct a component–target network to reflect the complex relationships between hypothetical bioactive ingredients and their targets (38). Cytoscape 3.8.0 is an open-source software platform that is applicable to exhibit complex networks and integrate different types of attribute data. The nodes of the network represent the screened active components and potential targets, while the connection between these nodes represents the biological interactions (39).
Common targets of Yimucao, cisplatin, and cervical cancer were collected as the core targets of Yimucao and cisplatin for cervical cancer. Core genes were identified by constructing a PPI. There are various databases related to PPIs, and the STRING database (https://string-db.org/) covers the greatest number of species and contains more interactive information (40). In this study, the core targets were submitted to STRING, with the species limited to Homo sapiens and confidence score limited to those > 0.4, to obtain PPI data. The value of “degree”, the number of correlations of the node in the whole network, was used as a reference to the importance of the core target.
“Molecular Complex Detection” (MCODE: a plug-in of Cytoscape) was applied to determine the relationships between network clusters to identify hub genes with a high degree of connectivity (41). According to previous literature, advanced options were set with a degree cutoff = 2, a node score cutoff = 0.2, and a K-Core = 2 (42–44).
GO and KEGG pathway enrichment analyses were performed using the R package “ClusterProfifiler” to explore the gene functions (45, 46). The Benjamini Hochberg method was utilized to adjust the P-value. Only functional annotations with an enrichment P-value < 0.05 were chosen for further analyses.
Human cervical cancer cells (HeLa and SiHa) were purchased from the European Collection of Authenticated Cell Cultures. Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) was used to grow cells, at 37°C with 5% CO2 and 95% air.
In our preliminary experiment, the IC30 of quercetin in HeLa cells was 15 μM, and the IC30 of cisplatin was 10 μM, and the IC30 of quercetin and cisplatin in SiHa cells was 25 and 12 μM, respectively. Quercetin (≥95%, HPLC) was obtained from Sigma-Aldrich Company (CAS: 117-39-5, USA). Quercetin was prepared into a 200 nM stock using dimethyl sulfoxide and maintained at −20°C. Then, quercetin was prepared in complete culture medium at 15 and 25 μM to treat cervical cancer cells. Cisplatin (Sigma, USA) was prepared in phosphate-buffered saline (PBS) at a concentration of 10 mM, stored at −20°C, and then used with complete culture medium to prepare 10 and 12 μM to act on cervical cancer cells. Quercetin combined with cisplatin was used to treat cervical cancer Hela and SiHa cells: 15 μM quercetin combined with 10 μM cisplatin treated Hela cells for 24 h, and 25 μM quercetin combined with 12 μM cisplatin affected on SiHa cells for 48 h.
HeLa and SiHa cells were seeded into a six-well plate at a density of 2.5×105 cells/well and divided into four groups (blank control, quercetin, cisplatin, quercetin and cisplatin). Radioimmunoprecipitation assay (RIPA) lysis buffer (60 mM Tris-HCl, pH 6.8, 5% glycerol, 2% SDS) and bicinchoninic acid (BCA) protein determination kits (Thermo, USA) were applied to extract and quantify the total protein in the cells after incubation (24 h for HeLa; 48 h for SiHa cells). Proteins were electrophoresed in polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, USA). Membranes were incubated with primary antibodies at 4°C overnight. Rabbit anti-human EGFR, rabbit anti-human IL-6, rabbit anti-human caspase-3, rabbit anti-human VEGFA, rabbit anti-human c-Myc, rabbit anti-human Cyclin D1, rabbit anti-human HER2/ErbB2, rabbit anti-human c-Fos, rabbit anti-human PPARγ, and mouse anti-human caspase-8 antibodies were purchased from Cell Signaling (Danvers, MA, USA) and used at a 1:1,000 dilution. Rabbit anti-human vinculin antibody (1:3,000; Proteintech, USA) was used as a control. After washing three times with Tris-buffered saline Tween (TBST), PVDF membranes and horseradish peroxidase-conjugated secondary antibody were incubated for 1.5 h at ambient temperature. Vinculin was employed as a loading control. Peroxidase-conjugated goat antirabbit antibody (1:5,000; Biosharp, China) and anti-mouse antibody (1:5,000; Biosharp, China) were used as secondary antibodies. Finally, the bound antibodies were identified using Immobilon Western chemiluminescent HRP substrate (Millipore, USA). Immunoblots were imaged using a Bio-Rad ChemiDoc TM XRS system (Bio-Rad, USA). Image J software version 1.8.0 (National Institutes of Health, USA) was applied to quantify the intensity of the target western blot.
HeLa and SiHa cells were seeded in 96-well plates overnight. HeLa cells were treated with 15 μM quercetin and 15 μM cisplatin for 24 h, and SiHa cells were treated with 25 μM quercetin and 15 μM cisplatin for 48 h. Then, Cell Counting Kit-8 (CCK-8) was used to measure cell viability by a Microplate Reader as described previously (18).
HeLa and SiHa cells were seeded in 6-well plates overnight. HeLa cells were treated with 15 μM quercetin and 10 μM cisplatin for 48 h, and SiHa cells were treated with 30 μM quercetin and 12 μM cisplatin for 48 h. Cell apoptosis was measured by Annexin V-FITC/propidium iodide (PI) method as described previously (18).
Data were entered and analyzed using SPSS 22.0 software (Chicago, USA). The results are expressed as the mean ± standard deviation (SD). Multiple groups were calculated using one-way analysis of variance (ANOVA) followed by Tukey’s posttest. A two-tailed p-value < 0.05 was regarded as statistically significant.
Using TCMSP, a total of 42 active compounds in Yimucao were identified based on the predefined criteria (OB ≥ 30% and DL ≥0.18) (Supplementary Table 1), and 8 active compounds, including quercetin, were identified from the herb. The details of the active compounds and their structures are described in Table 1 and Supplementary Figure 1. Based on the target prediction system in the TCMSP database, 100 putative target genes were identified for Yimucao (Supplementary Table 2). In addition, a total of 2,436 cisplatin-related genes were collected from GeneWeaver. Cervical cancer target genes were retrieved from GeneCards, TTD, DisGeNET, and OMIM. In total, 6,302 potential target genes related to cervical cancer were collected. Eight active compounds and 100 Yimucao target proteins were used to construct a network consisting of 108 nodes and 154 edges (Figure 2). As showed in this compound-target network, nodes with a greater number of edges were more important within the network and are presented as larger in size were those on which to focus. The network with drug-bioactive compounds and disease targets contained 93 nodes (including 1 disease, 1 chemotherapy drug, 1 herb, 8 compounds, and 82 genes) and 261 edges (Figure 3). By intersecting 100 target genes of Yimucao, and 2,436 target genes of cisplatin with the 6,302 potential target genes related to cervical cancer, a total of 53 intersections were obtained (Figure 4A).
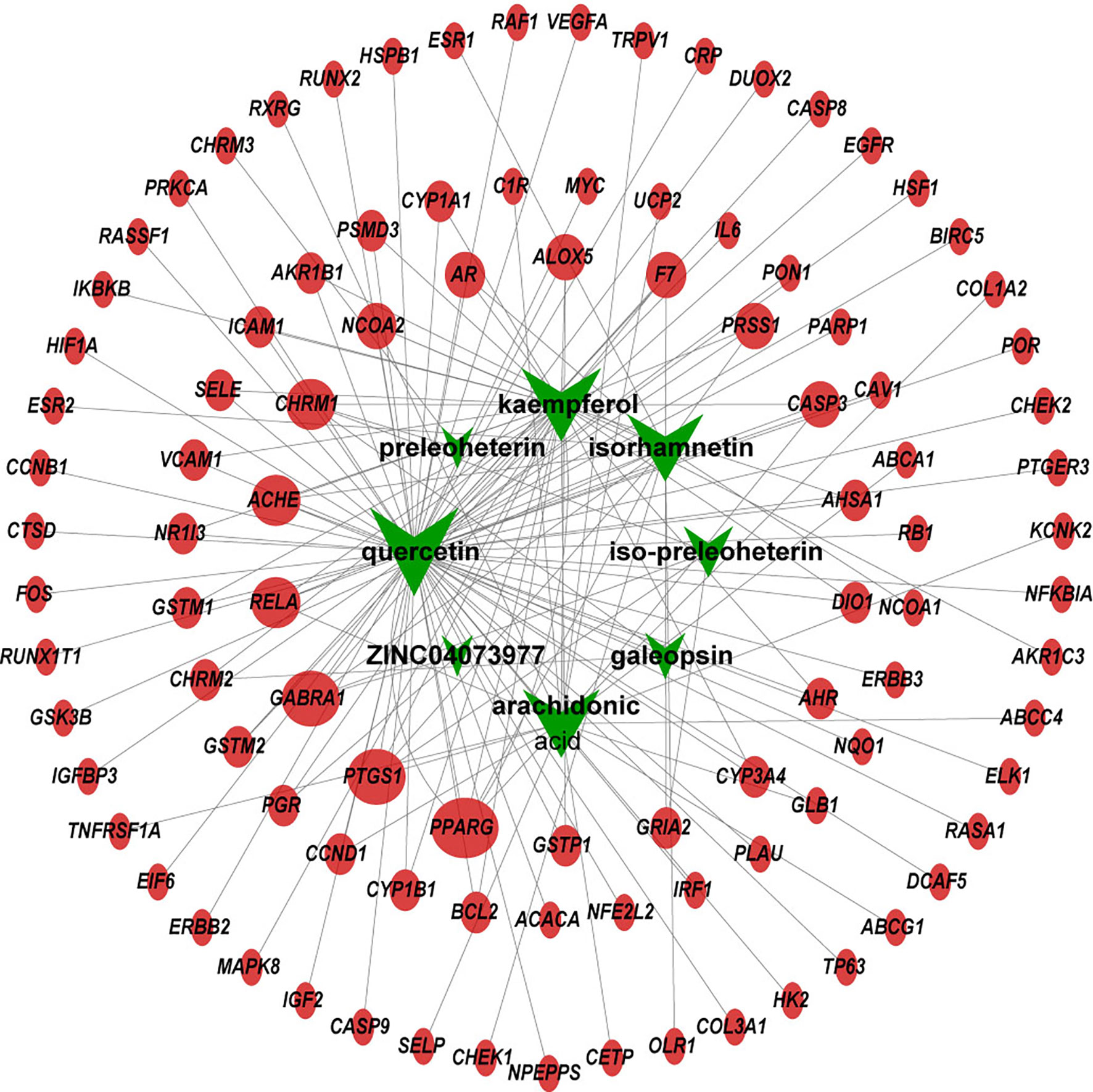
Figure 2 Compound-target network of Yimucao. Green nodes are presented to compounds in Yimucao. Red circle nodes represent for the target genes. The size of the node indicates the size of the degree.
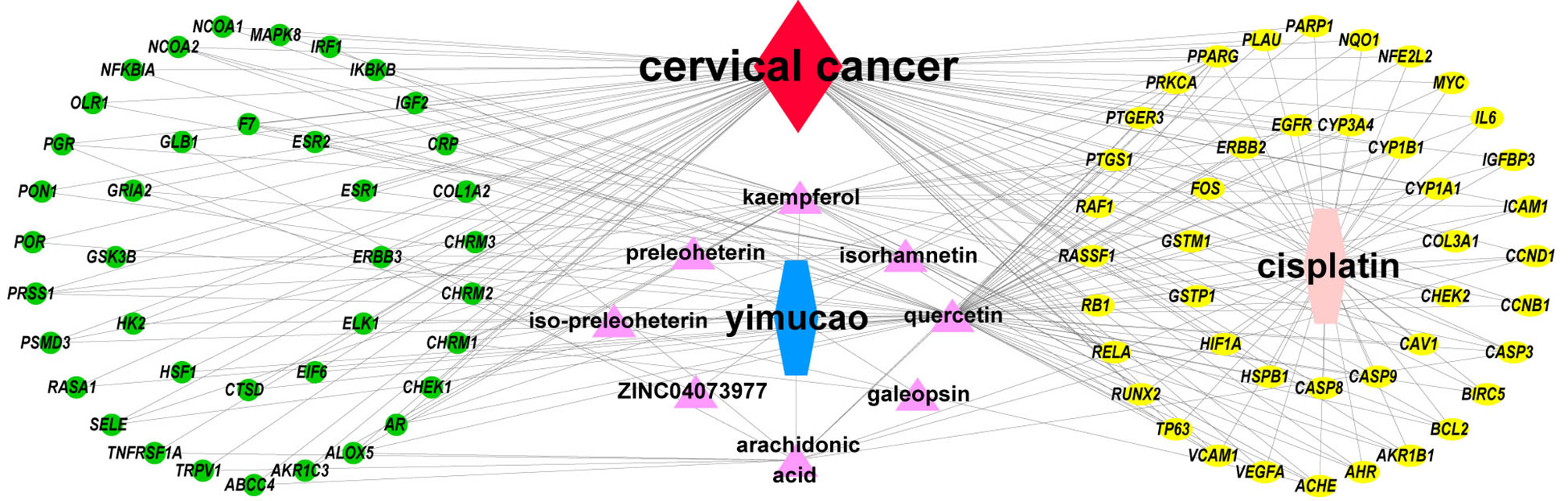
Figure 3 Drug-bioactive compound-disease-target network of combined treatment with Yimucao and cisplatin for cervical cancer. The green circle represents the gene, the yellow circle represents overlapping genes of cisplatin, quercetin, and cervical cancer target genes, and the pink triangle represents compounds of Yimucao. The red diamond indicates cervical cancer. The blue and pink hexagons represent Yimucao and cisplatin, respectively.
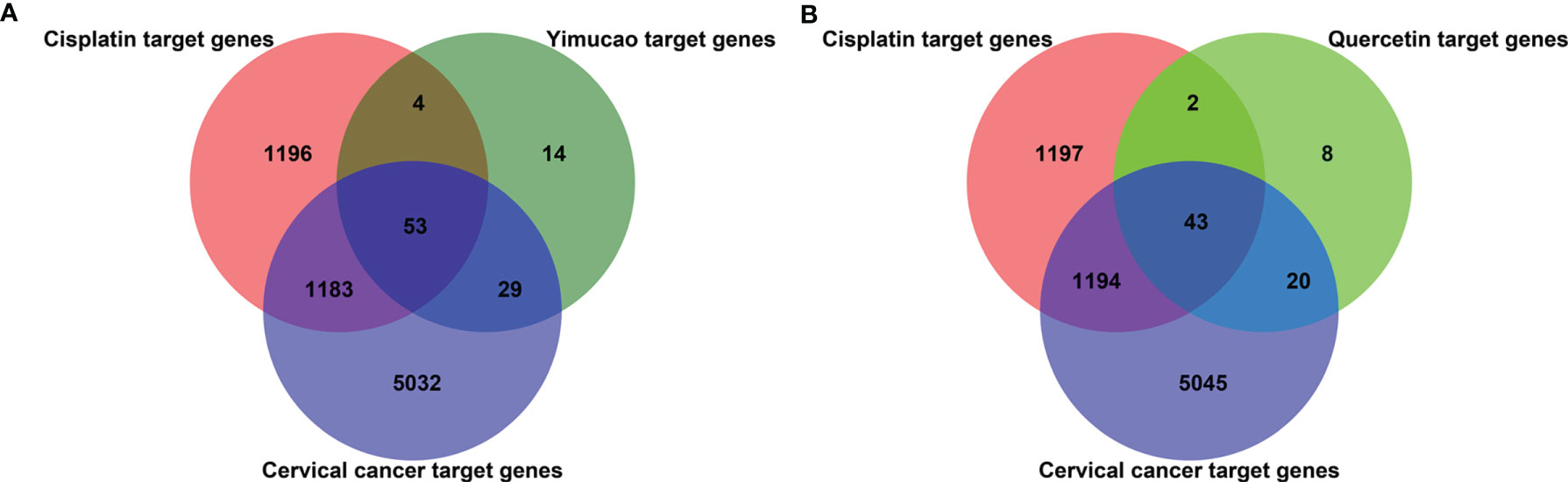
Figure 4 Quercetin and cisplatin target gene prediction for cervical cancer treatment. The overlapping number of cisplatin, Yimucao (A) or quercetin (B), and cervical cancer target genes. The red circle represents cisplatin target genes, the purple circle represents cervical cancer target genes, and the dark green and bright circles represent Yimucao and quercetin target genes, respectively.
Quercetin was the top ingredient node with the greatest number of edges. The intersection of 73 quercetin targets, 2,436 cisplatin targets, and 6,302 potential target genes related to cervical cancer revealed 43 genes. These intersections were considered potential candidate targets of the synergistic effect of quercetin and cisplatin against cervical cancer (Supplementary Table 3 and Figure 4B).
After extracting information regarding the relationships between potential anti-cervical cancer target genes of quercetin and cisplatin from the STRING website, a gene-gene interaction network with 43 nodes and 317 edges was constructed (Figure 5A). In the PPI network, the core genes with higher degree values were EGFR, IL6, CASP3, VEGFA, MYC, CCND1, ERBB2, FOS, PPARG, and CASP8. The number of edges connected to these nodes was relatively high (32, 31, 31, 30, 30, 29, 26, 23, 21, and 20, respectively) (Figure 5B).
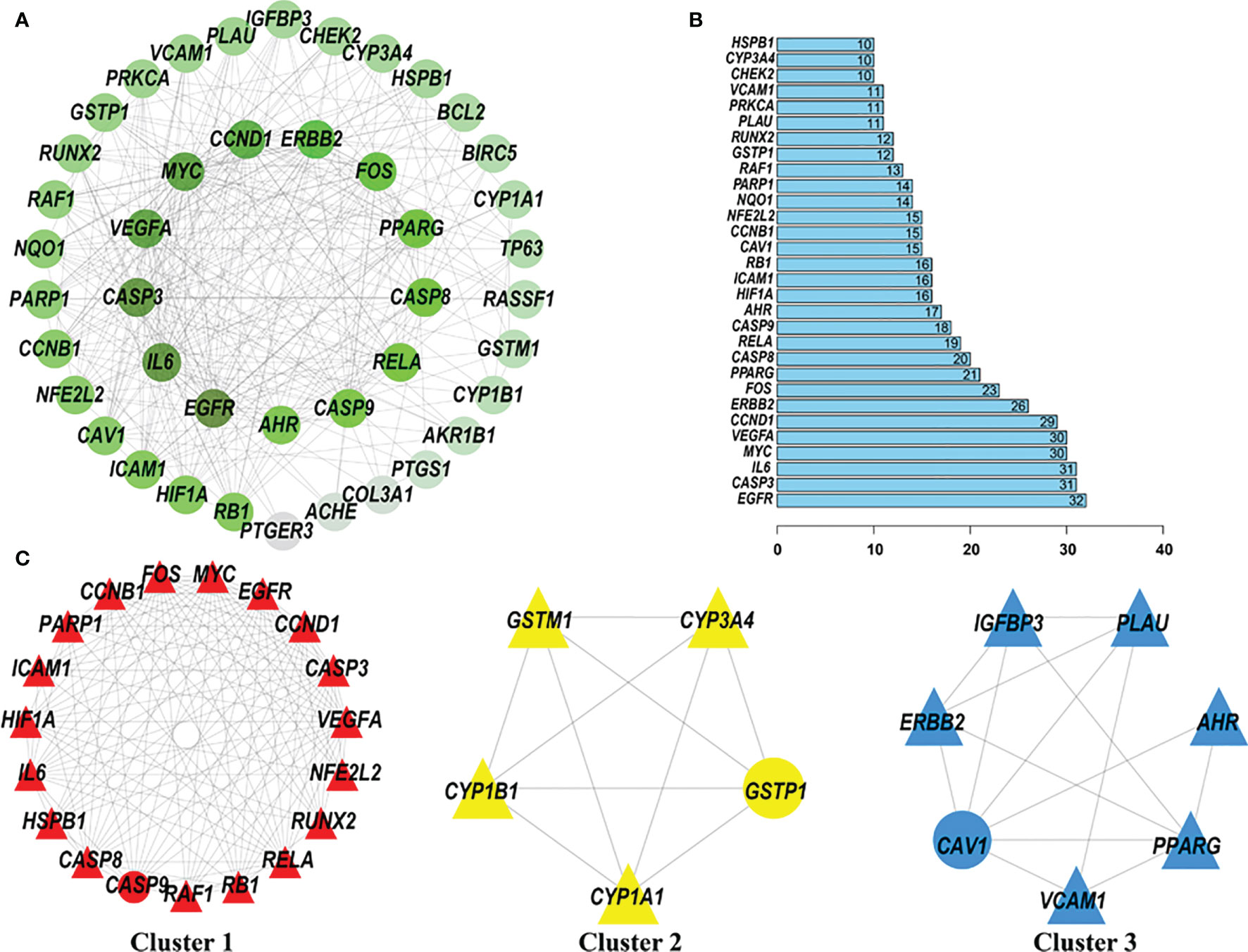
Figure 5 Protein-protein interaction (PPI) network of quercetin and cisplatin anti-cervical cancer genes. The MCODE algorithm was applied to this network to identify neighborhoods where proteins were densely connected. The gradient from dark green to light green represents the change in the number of edges from more to less; three significant modules with a score >4.5 were selected. (A) PPI network of quercetin and cisplatin anti-cervical cancer genes. (B) Histogram of key proteins. The y-axis represents the name of genes, the x-axis represents the number of adjacent genes, and height is the number of gene connections. (C) Module 1, MCODE score = 14.111; Module 2, MCODE score = 5; and Module 3, MCODE score = 4.667.
Then, three subclusters within the PPI network were identified using the MCODE tool (Figure 5C and Table 2). The highest-scoring cluster, cluster 1, contained 19 nodes and 127 edges, including IL6, PARP1, RUNX2, CCND1, RB1, EGFR, CASP9, HIF1A, VEGFA, NFE2L2, CCNB1, MYC, CASP3, CASP8, HSPB1, RAF1, ICAM1, FOS, and RELA. Cluster 2 contained 5 nodes (CYP1B1, GSTM1, CYP1A1, CYP3A4, and GSTP1) and 10 edges. Cluster 3 contained 7 nodes and 14 edges.

Table 2 Cluster of target genes of quercetin and cisplatin anti-cervical cancer protein-protein interaction network.
To further understand the identified candidate target genes, GO enrichment analysis, including biological process (BP), cell composition (CC), and molecular function (MF), was conducted. Enriched GO terms are shown in Figure 6A and Supplementary Table 4. In the BP group, the active targets were essentially connected to “response to metal ion”, “cellular response to xenobiotic stimulus”, “response to radiation”, and “response to oxidative stress”. In the CC group, the genes were primarily enriched in “transcription factor complex”, “cyclin-dependent protein kinase holoenzyme complex”, “nuclear chromatin”, and “RNA polymerase II transcription factor complex”. In the MF group, the majority of genes were enriched in “activating transcription factor binding”, “E-box binding”, “DNA-binding transcription activator activity, RNA polymerase II-specific”, and “repressing transcription factor binding”.
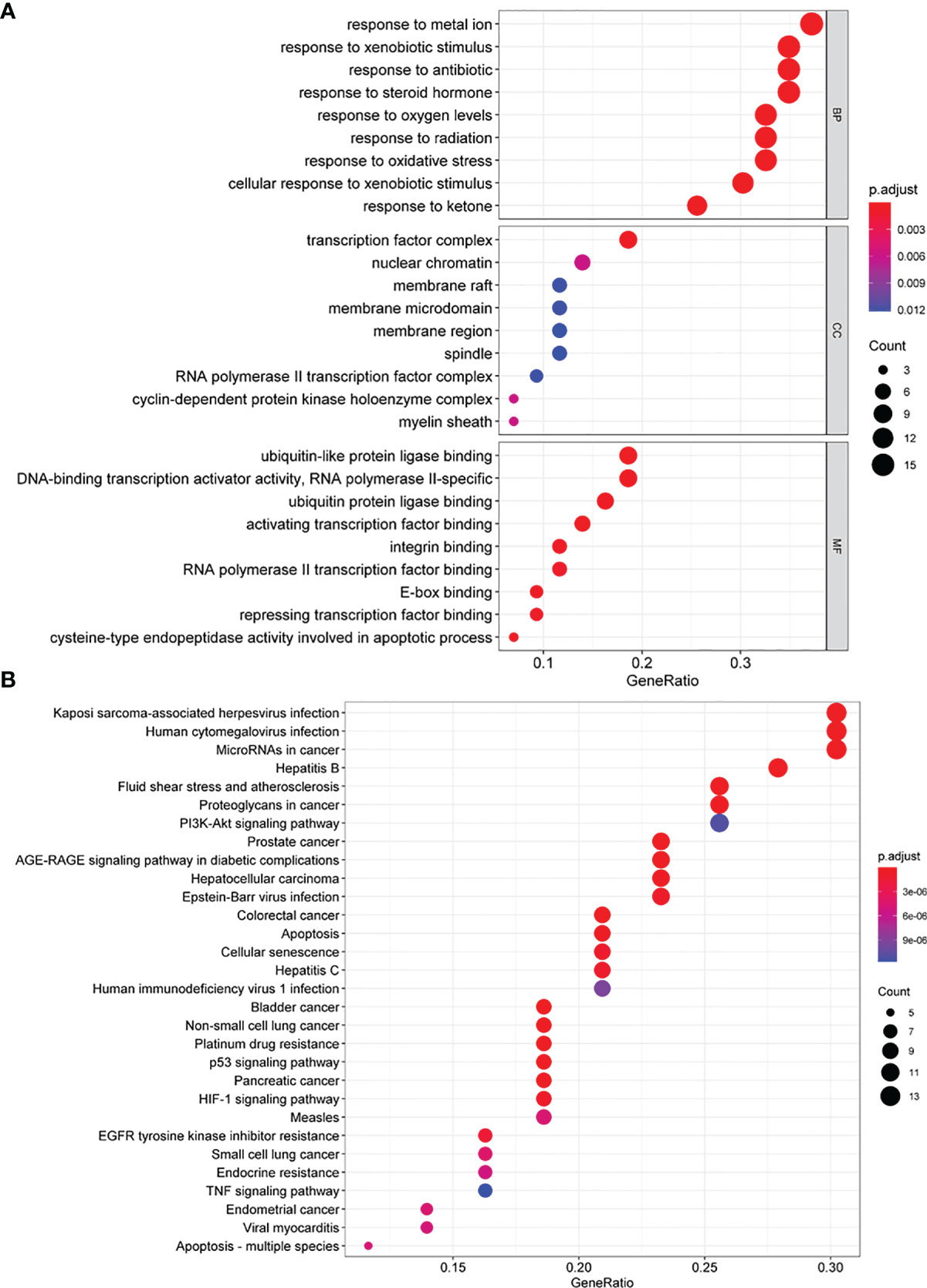
Figure 6 GO and KEGG enrichment analysis of quercetin synergism with cisplatin anti-cervical cancer genes. (A) The GO enrichment analysis of quercetin synergizing with cisplatin anti-cervical cancer genes. Ontology covers three domains: biological process (BP), cellular component (CC), and molecular function (MF). The abscissa GeneRatio represents the proportion of genes of interest in the entry, and the ordinate represents each entry. The size of the dots represents the number of genes annotated in the entry, and the color of the dots represents the corrected P-value of the hypergeometric test. (B) The KEGG enrichment analysis of quercetin synergizing with cisplatin to target anti-cervical cancer genes. The abscissa represents the number of genes annotated in the pathway, the ordinate represents the pathway, and the color of the column represents the corrected P-value. KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology.
KEGG pathway enrichment was used to explore the major effects that associated with the quercetin. A total of 102 signaling pathways (Figure 6B and Supplementary Table 5) were selected (P < 0.05). The principal pathways were related to cancer, including “platinum drug resistance”, “p53 signaling pathway”, “HIF-1 signaling pathway”, “proteoglycans in cancer”, “apoptosis”, “PI3K-Akt signaling pathway”, and “TNF signaling pathway”.
Changes in 10 corresponding key protein levels in cervical cancer cells (HeLa or SiHa cells) in response to different drug combinations were detected by western blot analysis (Figure 7). After treatment with quercetin or cisplatin, the protein expression of EGFR, VEGFA, MYC, and CCND1 in HeLa and SiHa cell lines was significantly decreased, and the expression levels of CASP3, FOS, and CASP8 were significantly increased. In both HeLa and SiHa cell lines, IL6, ERBB2, and PPARG protein levels were significantly decreased after treatment with cisplatin. However, significant downregulation of IL6 and ERBB2 in response to treatment with cisplatin was only observed in SiHa cells, and significant downregulation of PPARG was only found in HeLa cells.
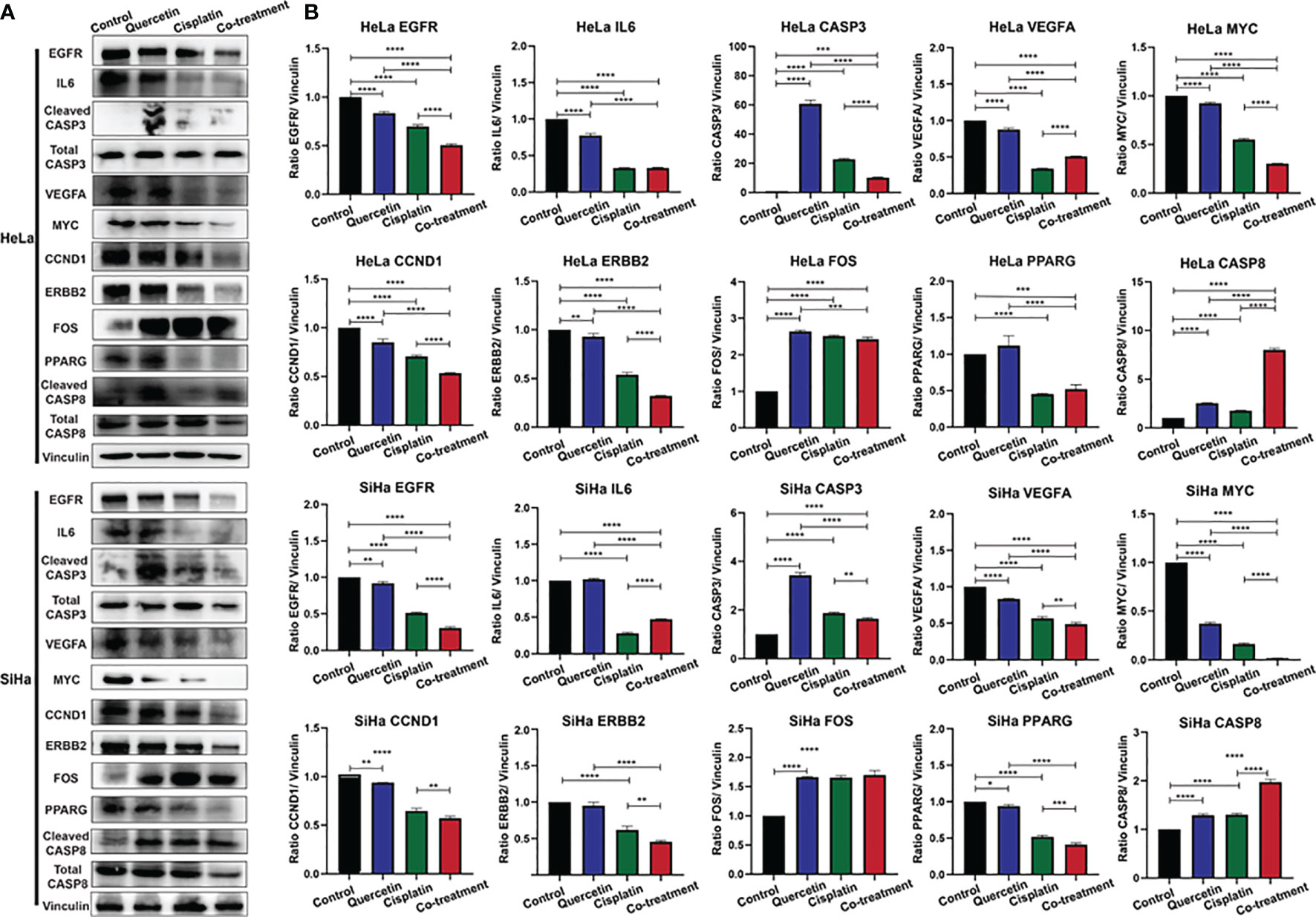
Figure 7 Western blotting was used to detect the relationship between the target protein and cervical cancer. (A) Immunoblot of EGFR, IL6, CASP3, VEGFA, MYC, CCND1, ERBB2, FOS, PPARG, and CASP8 in HeLa cells and SiHa cells. Vinculin was used as a loading control. (B) Densitometry for quantitation of relative differences in band intensity for of EGFR, IL6, CASP3, VEGFA, MYC, CCND1, ERBB2, FOS, PPARG, and CASP8 normalized to vinculin in (A). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To explore the synergistic effect of quercetin on cisplatin in cervical cancer, a combination treatment with quercetin and cisplatin was next explored. Protein levels of EGFR, MYC, CCND1, and ERBB2 in HeLa and SiHa cells were significantly decreased compared to those of HeLa and SiHa cells treated with both drugs. In the combination group, a significant increase in CASP8 protein levels was observed compared to either a single drug in HeLa and SiHa cells. Compared to the control group or quercetin treatment group, significant reductions in the IL6 and PPARG protein levels were observed in the combination group in HeLa and SiHa cells, but increased and decreased levels of IL6 and PPARG in the combination group were only observed in SiHa cells compared to cisplatin treatment alone. In HeLa and SiHa cells, significantly higher CASP3 protein expression was observed in the combination group than in the control group, and decreased levels of CASP3 were observed in the combination group compared to the single drug groups. Compared to the control or quercetin treatment group, VEGFA protein levels were significantly decreased in the combination group in the two cell lines, but increased and decreased levels in the combination group compared to the quercetin treatment group were only found in HeLa cells and SiHa cells, respectively. Compared to the control group, FOS levels were significantly increased in the combination group in the two cell lines, but were significantly decreased in the combination group compared to the quercetin treatment group in HeLa cell.
To determine whether quercetin could promote the antitumor activity of cisplatin in cervical cancer cells, we treated HeLa and SiHa cells with quercetin or cisplatin or combination. The results from CCK-8 assay showed that quercetin inhibited cell viability in both HeLa and SiHa cells. Moreover, quercetin promoted the cisplatin-mediated suppression of cell viability in both HeLa and SiHa cells (Figure 8). Furthermore, we observed that cisplatin induced cell apoptosis in cervical cancer cells, which was further enhanced by quercetin treatment (Figure 9). Taken together, quercetin can enhance cisplatin-mediated antitumor activity in cervical cancer cells via regulating cell viability and apoptosis.
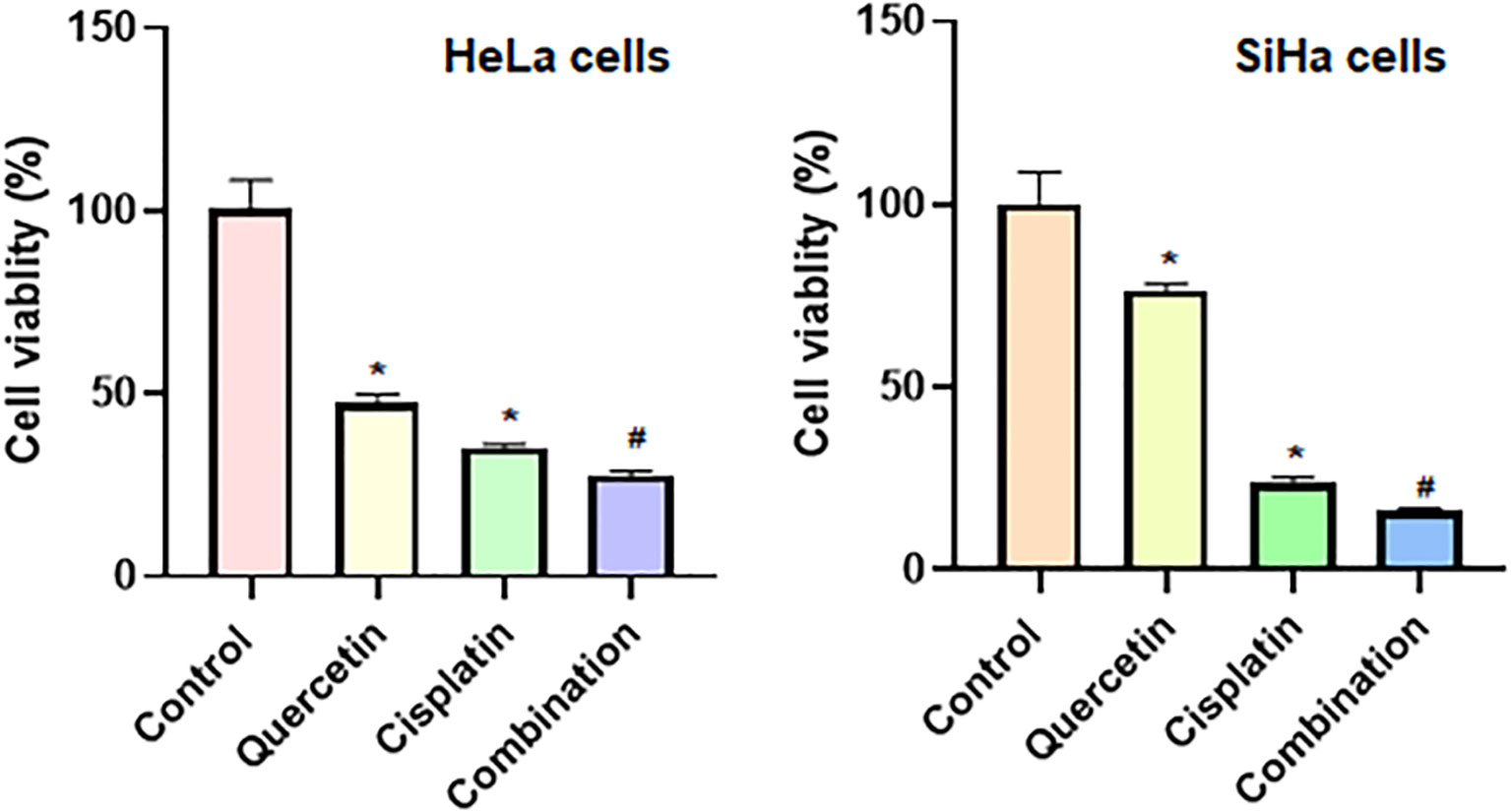
Figure 8 Quercetin enhances cisplatin-induced inhibition of viability of cervical cancer cells. CCK-8 assay was utilized to measure the cell viability in HeLa and SiHa cells after treatments with quercetin or cisplatin or combination. HeLa cells were treated with 15 μM quercetin and 15 μM cisplatin for 24 h, and SiHa cells were treated with 25 μM quercetin and 15 μM cisplatin for 48 h. *P < 0.05 vs control group; #P < 0.05 vs quercetin alone or cisplatin alone.
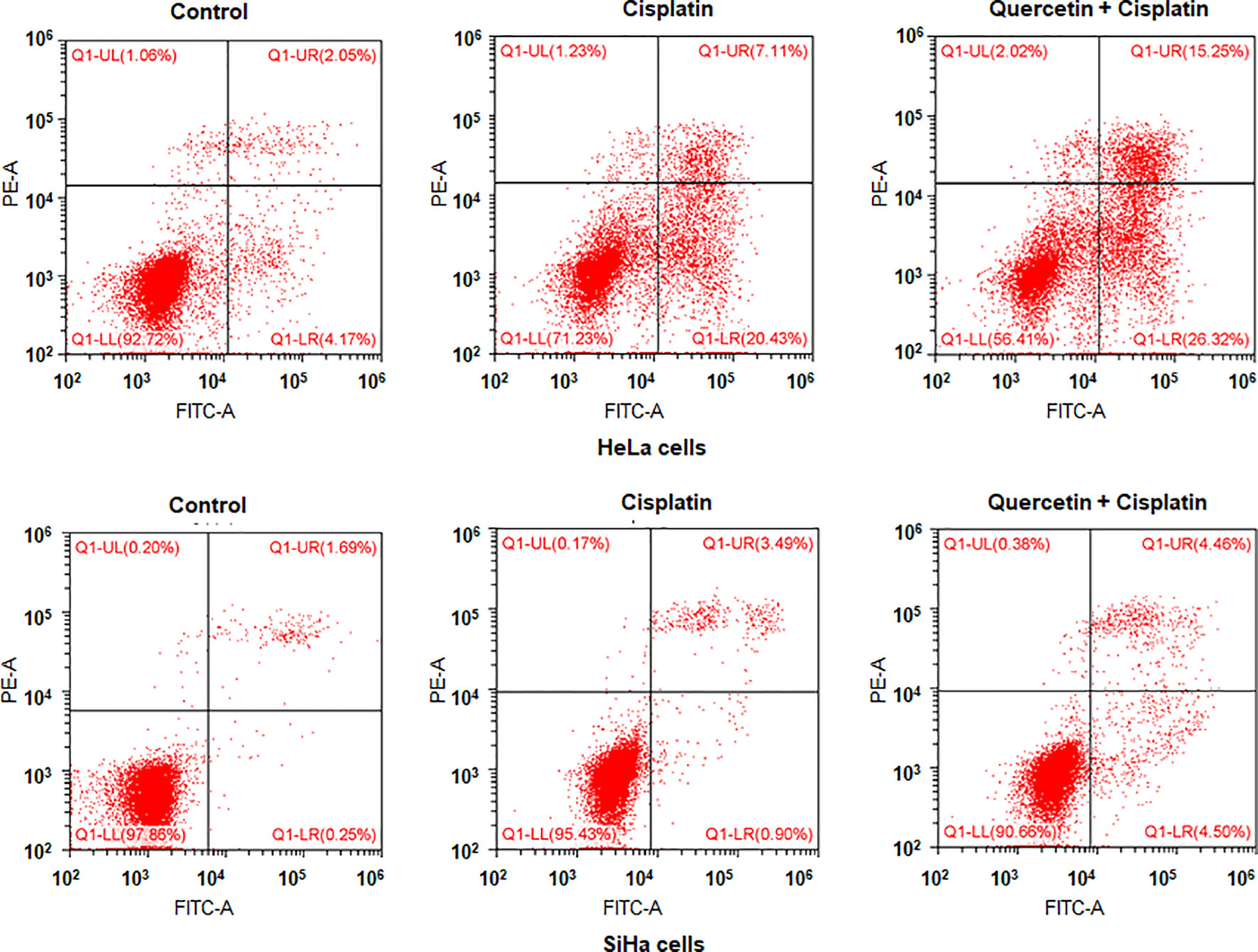
Figure 9 Quercetin enhances cisplatin-induced apoptosis of cervical cancer cells. HeLa cells were treated with 15 μM quercetin and 10 μM cisplatin for 48 h, and SiHa cells were treated with 30 μM quercetin and 12 μM cisplatin for 48 h. Flow cytometry analysis was utilized to measure apoptosis of HeLa and SiHa cells after various treatments.
Our previous study had found that an active ingredient of Yimucao, leonurine, combined with cisplatin exerted synergistic antitumorigenic effects on cervical cancer (47); however, the detailed mechanism of Yimucao’s actions and its active ingredients’ synergism with cisplatin on cervical cancer is currently unknown. In the present study, we attempted to illustrate the synergistic mechanism of Yimucao and cisplatin by applying a network pharmacology approach. We identified 8 active compounds and 100 target proteins of Yimucao. Then, 73 potential targets of quercetin and cisplatin related to cervical cancer were obtained by constructing a biological network of interactions between chemical compounds and protein targets at the molecular systemic level. The predicted results indicated that quercetin and cisplatin interacted with 43 targets related to cervical cancer. The synergistic inhibitory effects of quercetin and cisplatin on cervical cancer cells were observed in our previous study (18). Quercetin and cisplatin modulate the same targets simultaneously, which explains their synergistic mechanisms. The PPI network indicated that EGFR, IL6, CASP3, VEGFA, MYC, CCND1, ERBB2, FOS, PPARG, and CASP8 may play key roles in the effect of quercetin and cisplatin against cervical cancer. GO functional analysis revealed that genes were primarily related to the transcription initiation process, such as cyclin-dependent protein kinase holoenzyme complex, transcription factor complex, activating transcription factor binding, and E-box binding. The KEGG analyses demonstrated that 102 common targets were enriched in signaling pathways, including the platinum drug resistance, p53 signaling pathway, proteoglycans in cancer, apoptosis, PI3K-Akt signaling pathway, TNF signaling pathway, and HIF-1 signaling pathway. In vitro experiments verified that in response to cisplatin or quercetin treatment, expression of EGFR, IL6, VEGFA, MYC, CCND1, ERBB2, and PPARG was significantly reduced, while the expression of CASP3, FOS, and CASP8 was significantly increased. In addition, significant changes in the protein expression of five genes (EGFR, MYC, CCND1, ERBB2, and CASP8) were observed in the cells treated with the combination of both two drugs to cells treated with either drug alone. Our results demonstrated that these five genes were related to the synergistic effect of quercetin and cisplatin.
EGFR belongs to the family of HER tyrosine kinase receptor family and has a regulatory role in cell proliferation, transformation, and survival (48). It was estimated that 90% of cervical cancers overexpress EGFR, and high expression of EGFR is associated with poorer prognosis (49). Propofol has been reported to enhance the antitumor effect of cisplatin through the EGFR/JAK2/STAT3 pathway (50). Meanwhile, it has also been reported that TGF-α and amphiregulin inhibit the Hippo signaling pathway and activate YAP through EGFR to induce the proliferation and migration of cervical cancer cells (51). Two prospective studies revealed that cisplatin combined with anti-EFGR monoclonal antibody followed by surgery in locally advanced cervical cancer patients leads to excellent treatment outcomes (52, 53). ERBB2 is another important therapeutic target of the epidermal growth factor receptor family in the treatment of malignant tumors (54). As a proto-oncogene, ERBB2 is considered to be a therapeutic target for cervical cancer (55). Whole-exome sequencing of cervical carcinoma results confirmed that the ERBB2/PIK3CA/AKT/mTOR pathway was one of the major mechanisms of cervical cancer in tumorigenesis (56). Moreover, ERBB2 activation was reported as one of the key pathways in cisplatin-resistance in ovarian cancer (57, 58). CCND1 belongs to the highly conserved cyclin family and plays a key role in the cell cycle transition from G1 phase to S phase transition (59). MYC is a proto-oncogene that encodes a nuclear phosphoprotein and is involved in cell proliferation, the cell cycle, and apoptosis (60). However, there have been no reports regarding the role of the four genes in quercetin treatment of cervical cancer.
Mutations in the upstream signaling molecules EGFR and ERBB2 have been reported to be associated with increased NF-κB signaling. NF-κB, in turn, promotes transcription of the downstream proliferation-regulating genes CCND1 and MYC, which participate in VEGF-dependent angiogenesis, metastasis, and cellular telomerase immortality (61). Studies have demonstrated that NF-κB is a key factor to confer drug resistance in human cancer (61). The cytoplasmic NF-κB expression is correlated with a trend toward poor outcomes in locally advanced cervical cancer patients after chemoradiation therapy (62). Our results demonstrated that quercetin combined with cisplatin has a synergistic effect on cervical cancer, leading to decreased protein expression of EGFR, MYC, CCND1, and ERBB2. Our network pharmacology research indicated that these four proliferation-related genes are therapeutic targets for quercetin to promote cisplatin-mediated inhibition of proliferation in cervical cancer cells.
Previous studies have shown that cisplatin acts on CASP8 in cervical cancer. Cisplatin in combination with death receptor ligands enhances the expression of CASP8 in the HPV 16-positive cervical cancer cell line SiHa to increase apoptosis (63). In addition, by enhancing exogenous and endogenous apoptotic pathways, cisplatin amplifies CASP8 activation, sensitizing cervical cancer cells to apoptosis induced by rosulumab (64). However, the effect of quercetin on CASP8 in cervical cancer has not been reported. One study showed that loss of CASP8 activation is a potential mechanism for cisplatin resistance in human cancer cells (65). Our results demonstrated that in the treatment of cervical cancer, cisplatin combined with quercetin increases the expression of CASP8, indicating that CASP8 may be the key factor of quercetin in promoting the apoptosis of cervical cancer cells in combination with cisplatin for the treatment of cervical cancer. Indeed, our results demonstrated that quercetin enhanced cisplatin-mediated antitumor activity in cervical cancer cells via regulating cell viability and apoptosis.
This study extends our knowledge regarding the potential role of quercetin in cervical cancer and exhibits a feasible method for revealing potential drugs from Chinese medicinal formulas. However, there are still several limitations in the present work. First, more comprehensive TCM target gene databases are needed to deliver more credible consequences of network pharmacological analysis. Second, animal experiments are needed in order to verify the results of this study. Third, it is necessary to explore the changes in these five genes in response to single-drug therapy or two-drug combination therapy.
In conclusion, network pharmacology indicated that the primary active ingredients of Yimucao, particularly quercetin, could act on multiple targets in cervical cancer. Quercetin exerted a synergistic effect on cisplatin-treated cervical cancer cells primarily through downregulating the protein levels of EGFR, MYC, CCND1, and ERBB2 and upregulating CASP8. Quercetin promoted cisplatin-induced antitumor activity in cervical cancer cells via regulating cell viability and apoptosis.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Conceptualization, HJ and XZ. Methodology, HJ. Formal analysis, HJ, KL RL, and SX. Investigation, KL, RL, and SX. Writing, HJ and XZ. Visualization and supervision, XZ. All authors contributed to the article and approved the submitted version.
This work was supported by the grant from the Subject of Integrated Traditional Chinese and Western Medicine in Zhejiang Province (2017-XK-A42) and Health Commission of Zhejiang Province (WKJ-ZJ-2036).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.780387/full#supplementary-material
1. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical Cancer. Lancet (2019) 393:169–82. doi: 10.1016/S0140-6736(18)32470-X
2. Arbyn M, Weiderpass E, Bruni L, de Sanjose S, Saraiya M, Ferlay J, et al. Estimates of Incidence and Mortality of Cervical Cancer in 2018: A Worldwide Analysis. Lancet Glob Health (2020) 8:e191–203. doi: 10.1016/S2214-109X(19)30482-6
3. Diaz-Padilla I, Monk BJ, Mackay HJ, Oaknin A. Treatment of Metastatic Cervical Cancer: Future Directions Involving Targeted Agents. Crit Rev Oncol Hematol (2013) 85:303–14. doi: 10.1016/j.critrevonc.2012.07.006
4. Small W Jr., Bacon MA, Bajaj A, Chuang LT, Fisher BJ, Harkenrider MM, et al. Cervical Cancer: A Global Health Crisis. Cancer (2017) 123:2404–12. doi: 10.1002/cncr.30667
5. da Costa SCS, Bonadio RC, Gabrielli FCG, Aranha AS, Dias Genta MLN, Miranda VC, et al. Neoadjuvant Chemotherapy With Cisplatin and Gemcitabine Followed by Chemoradiation Versus Chemoradiation for Locally Advanced Cervical Cancer: A Randomized Phase II Trial. J Clin Oncol (2019) 37:3124–31. doi: 10.1200/JCO.19.00674
6. Van Minh H, My NTT, Jit M. Cervical Cancer Treatment Costs and Cost-Effectiveness Analysis of Human Papillomavirus Vaccination in Vietnam: A PRIME Modeling Study. BMC Health Serv Res (2017) 17:353. doi: 10.1186/s12913-017-2297-x
7. Yan Z, Lai Z, Lin J. Anticancer Properties of Traditional Chinese Medicine. Comb Chem High Throughput Screen (2017) 20:423–9. doi: 10.2174/1386207320666170116141818
8. Zhu YZ, Wu W, Zhu Q, Liu X. Discovery of Leonuri and Therapeutical Applications: From Bench to Bedside. Pharmacol Ther (2018) 188:26–35. doi: 10.1016/j.pharmthera.2018.01.006
9. Kim DH, Khan H, Ullah H, Hassan STS, Smejkal K, Efferth T, et al. MicroRNA Targeting by Quercetin in Cancer Treatment and Chemoprotection. Pharmacol Res (2019) 147:104346. doi: 10.1016/j.phrs.2019.104346
10. Brito AF, Ribeiro M, Abrantes AM, Pires AS, Teixo RJ, Tralhao JG, et al. Quercetin in Cancer Treatment, Alone or in Combination With Conventional Therapeutics? Curr Med Chem (2015) 22:3025–39. doi: 10.2174/0929867322666150812145435
11. Kedhari Sundaram M, Raina R, Afroze N, Bajbouj K, Hamad M, Haque S, et al. Quercetin Modulates Signaling Pathways and Induces Apoptosis in Cervical Cancer Cells. Biosci Rep (2019) 39. doi: 10.1042/BSR20190720
12. Kedhari Sundaram M, Hussain A, Haque S, Raina R, Afroze N. Quercetin Modifies 5’cpg Promoter Methylation and Reactivates Various Tumor Suppressor Genes by Modulating Epigenetic Marks in Human Cervical Cancer Cells. J Cell Biochem (2019) 120:18357–69. doi: 10.1002/jcb.29147
13. Lin TH, Hsu WH, Tsai PH, Huang YT, Lin CW, Chen KC, et al. Dietary Flavonoids, Luteolin and Quercetin, Inhibit Invasion of Cervical Cancer by Reduction of UBE2S Through Epithelial-Mesenchymal Transition Signaling. Food Funct (2017) 8:1558–68. doi: 10.1039/C6FO00551A
14. Li B, Li D, Xue D, Zhao G. The Establishment and Application of Chemical Fingerprint and Multi-Component Quantitative Analysis for Leonuri Herba. J Tianjin Univ Traditional Chin Med (2020) 39:5. doi: 10.11656/j.issn.1673-9043.2020.03.19
15. Bishayee K, Ghosh S, Mukherjee A, Sadhukhan R, Mondal J, Khuda-Bukhsh AR. Quercetin Induces Cytochrome-C Release and ROS Accumulation to Promote Apoptosis and Arrest the Cell Cycle in G2/M, in Cervical Carcinoma: Signal Cascade and Drug-DNA Interaction. Cell Prolif (2013) 46:153–63. doi: 10.1111/cpr.12017
16. Luo CL, Liu YQ, Wang P, Song CH, Wang KJ, Dai LP, et al. The Effect of Quercetin Nanoparticle on Cervical Cancer Progression by Inducing Apoptosis, Autophagy and Anti-Proliferation via JAK2 Suppression. BioMed Pharmacother (2016) 82:595–605. doi: 10.1016/j.biopha.2016.05.029
17. Clemente-Soto AF, Salas-Vidal E, Milan-Pacheco C, Sanchez-Carranza JN, Peralta-Zaragoza O, Gonzalez-Maya L. Quercetin Induces G2 Phase Arrest and Apoptosis With the Activation of P53 in an E6 Expressionindependent Manner in HPVpositive Human Cervical Cancerderived Cells. Mol Med Rep (2019) 19:2097–106. doi: 10.3892/mmr.2019.9850
18. Xu W, Xie S, Chen X, Pan S, Qian H, Zhu X. Effects of Quercetin on the Efficacy of Various Chemotherapeutic Drugs in Cervical Cancer Cells. Drug Des Devel Ther (2021) 15:577–88. doi: 10.2147/DDDT.S291865
19. Casas AI, Hassan AA, Larsen SJ, Gomez-Rangel V, Elbatreek M, Kleikers PWM, et al. From Single Drug Targets to Synergistic Network Pharmacology in Ischemic Stroke. Proc Natl Acad Sci U S A (2019) 116:7129–36. doi: 10.1073/pnas.1820799116
20. Cheng F, Kovacs IA, Barabasi AL. Network-Based Prediction of Drug Combinations. Nat Commun (2019) 10:1197. doi: 10.1038/s41467-019-09186-x
21. Xue J, Shi Y, Li C, Song H. Network Pharmacology-Based Prediction of the Active Ingredients, Potential Targets, and Signaling Pathways in Compound Lian-Ge Granules for Treatment of Diabetes. J Cell Biochem (2019) 120:6431–40. doi: 10.1002/jcb.27933
22. Zhang R, Zhu X, Bai H, Ning K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front Pharmacol (2019) 10:123. doi: 10.3389/fphar.2019.00123
23. Hu JX, Thomas CE, Brunak S. Network Biology Concepts in Complex Disease Comorbidities. Nat Rev Genet (2016) 17:615–29. doi: 10.1038/nrg.2016.87
24. Zhu J, Li B, Ji Y, Zhu L, Zhu Y, Zhao H. Betaelemene Inhibits the Generation of Peritoneum Effusion in Pancreatic Cancer via Suppression of the HIF1AVEGFA Pathway Based on Network Pharmacology. Oncol Rep (2019) 42:2561–71. doi: 10.3892/or.2019.7360
25. He R, Ou S, Chen S, Ding S. Network Pharmacology-Based Study on the Molecular Biological Mechanism of Action for Compound Kushen Injection in Anti-Cancer Effect. Med Sci Monit (2020) 26:e918520. doi: 10.12659/MSM.918520
26. Bing Z, Cheng Z, Shi D, Liu X, Tian J, Yao X, et al. Investigate the Mechanisms of Chinese Medicine Fuzhengkangai Towards EGFR Mutation-Positive Lung Adenocarcinomas by Network Pharmacology. BMC Complement Altern Med (2018) 18:293. doi: 10.1186/s12906-018-2347-x
27. Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: A Database of Systems Pharmacology for Drug Discovery From Herbal Medicines. J Cheminform (2014) 6:13. doi: 10.1186/1758-2946-6-13
28. Su X, Kong L, Lei X, Hu L, Ye M, Zou H. Biological Fingerprinting Analysis of Traditional Chinese Medicines With Targeting ADME/Tox Property for Screening of Bioactive Compounds by Chromatographic and MS Methods. Mini Rev Med Chem (2007) 7:87–98. doi: 10.2174/138955707779317830
29. Chen ML, Shah V, Patnaik R, Adams W, Hussain A, Conner D, et al. Bioavailability and Bioequivalence: An FDA Regulatory Overview. Pharm Res (2001) 18:1645–50. doi: 10.1023/A:1013319408893
30. Tao W, Xu X, Wang X, Li B, Wang Y, Li Y, et al. Network Pharmacology-Based Prediction of the Active Ingredients and Potential Targets of Chinese Herbal Radix Curcumae Formula for Application to Cardiovascular Disease. J Ethnopharmacol (2013) 145:1–10. doi: 10.1016/j.jep.2012.09.051
31. Ning K, Zhao X, Poetsch A, Chen WH, Yang J. Computational Molecular Networks and Network Pharmacology. BioMed Res Int (2017) 2017:7573904. doi: 10.1155/2017/7573904
32. Zhang M, Yuan Y, Zhou W, Qin Y, Xu K, Men J, et al. Network Pharmacology Analysis of Chaihu Lizhong Tang Treating non-Alcoholic Fatty Liver Disease. Comput Biol Chem (2020) 86:107248. doi: 10.1016/j.compbiolchem.2020.107248
33. Baker E, Bubier JA, Reynolds T, Langston MA, Chesler EJ. GeneWeaver: Data Driven Alignment of Cross-Species Genomics in Biology and Disease. Nucleic Acids Res (2016) 44:D555–9. doi: 10.1093/nar/gkv1329
34. Safran M, Solomon I, Shmueli O, Lapidot M, Shen-Orr S, Adato A, et al. GeneCards 2002: Towards a Complete, Object-Oriented, Human Gene Compendium. Bioinformatics (2002) 18:1542–3. doi: 10.1093/bioinformatics/18.11.1542
35. Li YH, Yu CY, Li XX, Zhang P, Tang J, Yang Q, et al. Therapeutic Target Database Update 2018: Enriched Resource for Facilitating Bench-to-Clinic Research of Targeted Therapeutics. Nucleic Acids Res (2018) 46:D1121–7. doi: 10.1093/nar/gkx1076
36. Pinero J, Bravo A, Queralt-Rosinach N, Gutierrez-Sacristan A, Deu-Pons J, Centeno E, et al. DisGeNET: A Comprehensive Platform Integrating Information on Human Disease-Associated Genes and Variants. Nucleic Acids Res (2017) 45:D833–9. doi: 10.1093/nar/gkw943
37. Wixon J, Kell D. The Kyoto Encyclopedia of Genes and Genomes–KEGG. Yeast (2000) 17:48–55. doi: 10.1002/(SICI)1097-0061(200004)17:1<48::AID-YEA2>3.0.CO;2-H
38. Qin T, Wu L, Hua Q, Song Z, Pan Y, Liu T. Prediction of the Mechanisms of Action of Shenkang in Chronic Kidney Disease: A Network Pharmacology Study and Experimental Validation. J Ethnopharmacol (2020) 246:112128. doi: 10.1016/j.jep.2019.112128
39. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res (2003) 13:2498–504. doi: 10.1101/gr.1239303
40. C. The UniProt. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res (2017) 45:D158–69. doi: 10.1093/nar/gkw1099
41. Bader GD, Hogue CW. An Automated Method for Finding Molecular Complexes in Large Protein Interaction Networks. BMC Bioinf (2003) 4:2. doi: 10.1186/1471-2105-4-2
42. Kulshrestha A, Suman S, Ranjan R. Network Analysis Reveals Potential Markers for Pediatric Adrenocortical Carcinoma. Onco Targets Ther (2016) 9:4569–81. doi: 10.2147/OTT.S108485
43. Cheng R, Qi L, Kong X, Wang Z, Fang Y, Wang J. Identification of the Significant Genes Regulated by Estrogen Receptor in Estrogen Receptor-Positive Breast Cancer and Their Expression Pattern Changes When Tamoxifen or Fulvestrant Resistance Occurs. Front Genet (2020) 11:538734. doi: 10.3389/fgene.2020.538734
44. Zeng ML, Zhu XJ, Liu J, Shi PC, Kang YL, Lin Z, et al. An Integrated Bioinformatic Analysis of the S100 Gene Family for the Prognosis of Colorectal Cancer. BioMed Res Int (2020) 2020:4746929. doi: 10.1155/2020/4746929
45. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res (2017) 45:D353–61. doi: 10.1093/nar/gkw1092
46. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat Genet (2000) 25:25–9. doi: 10.1038/75556
47. Lin M, Pan C, Xu W, Li J, Zhu X. Leonurine Promotes Cisplatin Sensitivity in Human Cervical Cancer Cells Through Increasing Apoptosis and Inhibiting Drug-Resistant Proteins. Drug Des Devel Ther (2020) 14:1885–95. doi: 10.2147/DDDT.S252112
48. Citri A, Yarden Y. EGF-ERBB Signalling: Towards the Systems Level. Nat Rev Mol Cell Biol (2006) 7:505–16. doi: 10.1038/nrm1962
49. Cerciello F, Riesterer O, Sherweif M, Odermatt B, Ciernik IF. Is EGFR a Moving Target During Radiotherapy of Carcinoma of the Uterine Cervix? Gynecol Oncol (2007) 106:394–9. doi: 10.1016/j.ygyno.2007.04.019
50. Li H, Lu Y, Pang Y, Li M, Cheng X, Chen J. Propofol Enhances the Cisplatin-Induced Apoptosis on Cervical Cancer Cells via EGFR/JAK2/STAT3 Pathway. BioMed Pharmacother (2017) 86:324–33. doi: 10.1016/j.biopha.2016.12.036
51. He C, Mao D, Hua G, Lv X, Chen X, Angeletti PC, et al. The Hippo/YAP Pathway Interacts With EGFR Signaling and HPV Oncoproteins to Regulate Cervical Cancer Progression. EMBO Mol Med (2015) 7:1426–49. doi: 10.15252/emmm.201404976
52. Qing D, Wu Y, Liu X, Jiang H, Zhu C, Liu P, et al. Long-Term Results of Concurrent Chemoradiotherapy Combined With Anti-EGFR Monoclonal Antibody Prior to Surgery in Locally Advanced Cervical Cancer: A Single-Institute Prospective Study. Cancer Manag Res (2020) 12:12309–17. doi: 10.2147/CMAR.S282372
53. Lu H, Wu Y, Liu X, Jiang H, Pang Q, Peng L, et al. A Prospective Study on Neoadjuvant Chemoradiotherapy Plus Anti-EGFR Monoclonal Antibody Followed by Surgery for Locally Advanced Cervical Cancer. Onco Targets Ther (2018) 11:3785–92. doi: 10.2147/OTT.S164071
54. Conesa-Zamora P, Torres-Moreno D, Isaac MA, Perez-Guillermo M. Gene Amplification and Immunohistochemical Expression of ERBB2 and EGFR in Cervical Carcinogenesis. Correlation With Cell-Cycle Markers and HPV Presence. Exp Mol Pathol (2013) 95:151–5. doi: 10.1016/j.yexmp.2013.06.011
55. Xiang S, Sun Z, He Q, Yan F, Wang Y, Zhang J. Aspirin Inhibits ErbB2 to Induce Apoptosis in Cervical Cancer Cells. Med Oncol (2010) 27:379–87. doi: 10.1007/s12032-009-9221-0
56. Zammataro L, Lopez S, Bellone S, Pettinella F, Bonazzoli E, Perrone E, et al. Whole-Exome Sequencing of Cervical Carcinomas Identifies Activating ERBB2 and PIK3CA Mutations as Targets for Combination Therapy. Proc Natl Acad Sci USA (2019) 116:22730–6. doi: 10.1073/pnas.1911385116
57. Yang L, Zhao H, Yin X, Liang H, Zheng Z, Shen Q, et al. Exploring Cisplatin Resistance in Ovarian Cancer Through Integrated Bioinformatics Approach and Overcoming Chemoresistance With Sanguinarine. Am J Transl Res (2020) 12:923–39.
58. Inoue M, Koga F, Yoshida S, Tamura T, Fujii Y, Ito E, et al. Significance of ERBB2 Overexpression in Therapeutic Resistance and Cancer-Specific Survival in Muscle-Invasive Bladder Cancer Patients Treated With Chemoradiation-Based Selective Bladder-Sparing Approach. Int J Radiat Oncol Biol Phys (2014) 90:303–11. doi: 10.1016/j.ijrobp.2014.05.043
59. Yue Y, Zhou K, Li J, Jiang S, Li C, Men H. MSX1 Induces G0/G1 Arrest and Apoptosis by Suppressing Notch Signaling and is Frequently Methylated in Cervical Cancer. Onco Targets Ther (2018) 11:4769–80. doi: 10.2147/OTT.S165144
60. Ji W, Lou W, Hong Z, Qiu L, Di W. Genomic Amplification of HPV, H-TERC and C-MYC in Liquid-Based Cytological Specimens for Screening of Cervical Intraepithelial Neoplasia and Cancer. Oncol Lett (2019) 17:2099–106. doi: 10.3892/ol.2018.9825
61. Tilborghs S, Corthouts J, Verhoeven Y, Arias D, Rolfo C, Trinh XB, et al. The Role of Nuclear Factor-Kappa B Signaling in Human Cervical Cancer. Crit Rev Oncol Hematol (2017) 120:141–50. doi: 10.1016/j.critrevonc.2017.11.001
62. Altoos B, Small C, Dalal P, Attieh D, El Achy S, Parimi V, et al. Prognostic Significance of Nuclear Factor Kappa B Expression in Locally Advanced Cervical Cancer Patients Treated Definitively With Concurrent Chemoradiation. Am J Clin Oncol (2020) 43:47–51. doi: 10.1097/COC.0000000000000626
63. Tan S, Hougardy BM, Meersma GJ, Schaap B, de Vries EG, van der Zee AG, et al. Human Papilloma Virus 16 E6 RNA Interference Enhances Cisplatin and Death Receptor-Mediated Apoptosis in Human Cervical Carcinoma Cells. Mol Pharmacol (2012) 81:701–9. doi: 10.1124/mol.111.076539
64. Wu XX, Kakehi Y. Enhancement of Lexatumumab-Induced Apoptosis in Human Solid Cancer Cells by Cisplatin in Caspase-Dependent Manner. Clin Cancer Res (2009) 15:2039–47. doi: 10.1158/1078-0432.CCR-08-2667
Keywords: quercetin, cisplatin, cervical cancer, network pharmacology, treatment
Citation: Ji H, Li K, Xu W, Li R, Xie S and Zhu X (2022) Prediction of the Mechanisms by Which Quercetin Enhances Cisplatin Action in Cervical Cancer: A Network Pharmacology Study and Experimental Validation. Front. Oncol. 11:780387. doi: 10.3389/fonc.2021.780387
Received: 21 September 2021; Accepted: 08 December 2021;
Published: 06 January 2022.
Edited by:
Lingying Wu, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaReviewed by:
Vaishali Aggarwal, University of Pittsburgh, United StatesCopyright © 2022 Ji, Li, Xu, Li, Xie and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueqiong Zhu, d3p6eHFAd3poZWFsdGguY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.