- 1Society of Meta-Research and Biomedical Innovation, Cancer Research Working Group, London, United Kingdom
- 2Department of Life Sciences, Faculty of Natural Sciences, Imperial College London, London, United Kingdom
- 3Department of Metabolism, Digestion and Reproduction, Faculty of Medicine, Imperial College London, London, United Kingdom
- 4Department of Obstetrics and Gynaecology, Chelsea and Westminster Hospital National Health Service (NHS) Foundation Trust, London, United Kingdom
- 5Institute of Reproductive and Developmental Biology, Imperial College London, London, United Kingdom
- 6Department of Surgery and Cancer, Faculty of Medicine, Imperial College London, London, United Kingdom
- 7West London Gynaecological Cancer Centre, Imperial College Healthcare National Health Service (NHS) Trust, London, United Kingdom
The investigation of differentially expressed genes (DEGs) and their interactome could provide valuable insights for the development of markers to optimize cervical intraepithelial neoplasia (CIN) screening and treatment. This study investigated patients with cervical disease to identify gene markers whose dysregulated expression and protein interaction interface were linked with CIN and cervical cancer (CC). Literature search of microarray datasets containing cervical epithelial samples was conducted in Gene Expression Omnibus and Pubmed/Medline from inception until March 2021. Retrieved DEGs were used to construct two protein-protein interaction (PPI) networks. Module DEGs that overlapped between CIN and CC samples, were ranked based on 11 topological algorithms. The highest-ranked hub gene was retrieved and its correlation with prognosis, tissue expression and tumor purity in patients with CC, was evaluated. Screening of the literature yielded 9 microarray datasets (GSE7803, GSE27678, GSE63514, GSE6791, GSE9750, GSE29570, GSE39001, GSE63678, GSE67522). Two PPI networks from CIN and CC samples were constructed and consisted of 1704 and 3748 DEGs along 21393 and 79828 interactions, respectively. Two gene clusters were retrieved in the CIN network and three in the CC network. Multi-algorithmic topological analysis revealed PCNA as the highest ranked hub gene between the two networks, both in terms of expression and interactions. Further analysis revealed that while PCNA was overexpressed in CC tissues, it was correlated with favorable prognosis (log-rank P=0.022, HR=0.58) and tumor purity (P=9.86 × 10-4, partial rho=0.197) in CC patients. This study identified that cervical PCNA exhibited multi-algorithmic topological significance among DEGs from CIN and CC samples. Overall, PCNA may serve as a potential gene marker of CIN progression. Experimental validation is necessary to examine its value in patients with cervical disease.
Introduction
Cervical cancer (CC) constitutes one of the most commonly diagnosed gynecological cancers worldwide (1). Progression of CC is characterized by the transition from an initial premalignant state called cervical intraepithelial neoplasia (CIN), that is graded based on the extension of dysplastic abnormalities in the epithelial cells of the cervix. Three stages of pre-malignancy can be defined: CIN-I [low-grade intraepithelial lesion (LSIL)], CIN-II and CIN-III [high-grade intraepithelial lesion (HSIL)] (2, 3). Persistent infection with high-risk human papilloma virus (HPV) is considered necessary for the development of CIN, however the majority of women clear the infection (2, 3). Nevertheless, a fraction of women develop CIN that can progress to CC if not detected and treated (2, 3).
The prolonged period necessary for progression from carcinogenic HPV infection to precancerous CIN to cancer, allows for detection and treatment of these lesions and dramatic reductions in mortality from cancer (4). However, concerns related to the low accuracy of Papanicolaou test and the financial burden pertained to cytological-based screening, have been raised (5). Additionally, overtreatment for CIN remains a matter of considerable discussion (6). Thus, markers for estimating the progression of CIN may be of potential value in the optimization of cervical screening and treatment.
Genetic markers of CIN and CC are poorly defined, but genetic variation likely plays a major role in the different host-viral interactions observed across individuals infected with HPV (7). Cervical carcinogenesis, likely depends on a complex interaction between HPV exposure, genetic predisposition due to inheritance of common risk variants, exposure to further environmental carcinogens and progressive carcinogenic processes such as loss of tumor suppressor genes and apoptotic dysregulation (7, 8). This phenomenon also requires the dynamic interaction of multiple strongly associated genes, as opposed to the dysregulated expression of individual ones. Thus, analysis of differentially expressed genes (DEGs) and their interactome, could be pivotal in the development of markers for the monitoring of CIN progression. Our study focused on examining cervical epithelial gene expression from patients with CIN and CC. The aim was to identify potential gene biomarkers whose dysregulated expression and protein interaction interface were involved in CIN and CC.
Materials and Methods
Our study focused on the identification of genes with a potential role in the progression of pre-cancerous cervical lesions to cervical cancer.
Collection of Microarray Data
The screening of the literature was conducted from inception until March 2021. We initially searched the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) and the search terms included: (cervix OR cervical). Additionally, we performed a search of the National Library of Medicine (NLM) PubMed using the terms: [(differentially expressed genes) AND (cervix OR cervical)]. Two authors (PG and KSK) created the search strategy and conducted the screening of the yielded datasets. Discrepancies in the literature search process were discussed and resolved by MK.
Datasets were restricted based on organism type (Homo sapiens), expression profiling (microarray), sample type (cervical epithelial tissue) and disease state (CIN and CC). No restrictions in terms of language and geographic region were used for dataset retrieval. Datasets lacking expression data for controls were excluded. No exclusion criteria related to the baseline characteristics of patients from which tissue sections were obtained, were applied.
Identification of Differentially Expressed Genes
Cervical epithelial samples from healthy controls were compared to those with CIN or CC and DEGs were identified using ImaGEO (9). Integration of differential gene expression was performed using the random effect model and genes with the strongest average effect across the collected datasets were retrieved. DEGs with P<0.05 corrected by the Benjamini-Hochberg False Discovery Rate were considered as significant. DEGs with Z score>1.96 were regarded as upregulated, while those with Z score<1.96 as downregulated (corresponding to a 5% significance level).
Construction of Protein-Protein Interaction Networks
DEGs from CIN and CC were used separately to construct two networks of encoded proteins using The Search Tool for the Retrieval of Interacting Genes (STRING) (10). The predicted protein-protein interaction (PPI) networks were identified using a medium probabilistic confidence score of >0.4 and mapped with Cytoscape (11). The purpose of applying a reasonably moderate cut-off score was to increase the coverage of all possible protein interactions without overestimating their precision. Non-interacting proteins were excluded from the networks.
Identification of Clustering Modules and Hub Genes
Highly interconnected clusters or modules in the PPI networks were identified using the Molecular Complex Detection (MCODE) (12). Selection of cut-off was ensued following manual inspection of clusters and a score resulting in the distinct segregation of clusters into groups was considered. Clusters with MCODE score >20 were regarded as significant modules.
The interactions of module DEGs in the PPI networks were analysed using CytoHubba (13). Module DEGs were ranked based on the intersection of 11 established topological algorithms as described by Chin et al., namely: Degree, Closeness, Betweenness, Radiality, Stress, EcCentricity, BottleNeck, Edge Percolated Component (EPC), Maximum Neighborhood Component (MNC), Density of Maximum Neighborhood Component (DMNC) and Maximal Clique Centrality (MCC) (13). The top 10 ranked module DEGs that overlapped in the CIN and CC networks, were considered as hub genes.
Analysis of Prognosis, Expression Level, and Tumor Purity of Hub Genes in CC
The highest ranked hub gene, both in terms of expression and interactions, was retrieved as a potential gene biomarker in CIN progression and was further characterized. Firstly, using publicly available transcriptome data from The Cancer Genome Atlas (TCGA) via the Gene Expression Profiling Interactive Analysis 2 (GEPIA2), its differential expression in CC tissues based on a P<0.05 and |log2 fold change|>2, was assessed (14). Secondly, its prognostic value in patients with CC and controls, which were divided into high and low expression groups, was examined. Correlation with overall survival (OS) and disease-free survival (DFS) were established using a log-rank P<0.05. Thirdly, its association between expression and the tumor microenvironment in terms of purity, was estimated in CC using the partial Spearman’s correlation (partial rho) via the Tumor Immune Estimation Resource 2 (TIMER2) algorithm (15).
Results
Overview of Microarray Datasets
Our search of the GEO database resulted in 38292 datasets, of which 24826 were categorized as human. From these, 214 microarray tissue-containing datasets were retrieved and 116 datasets with either duplicate gene expression sample and series or incompatible platforms, were excluded. Screening of the resulting 98 datasets, based on cancer type and tissue type, revealed one CIN [GSE27678 (16–18)] and five CC datasets [GSE27678, GSE29570 (19), GSE39001 (20), GSE63678 (21), GSE67522 (22, 23)] with cervical epithelial samples (Figure 1A).
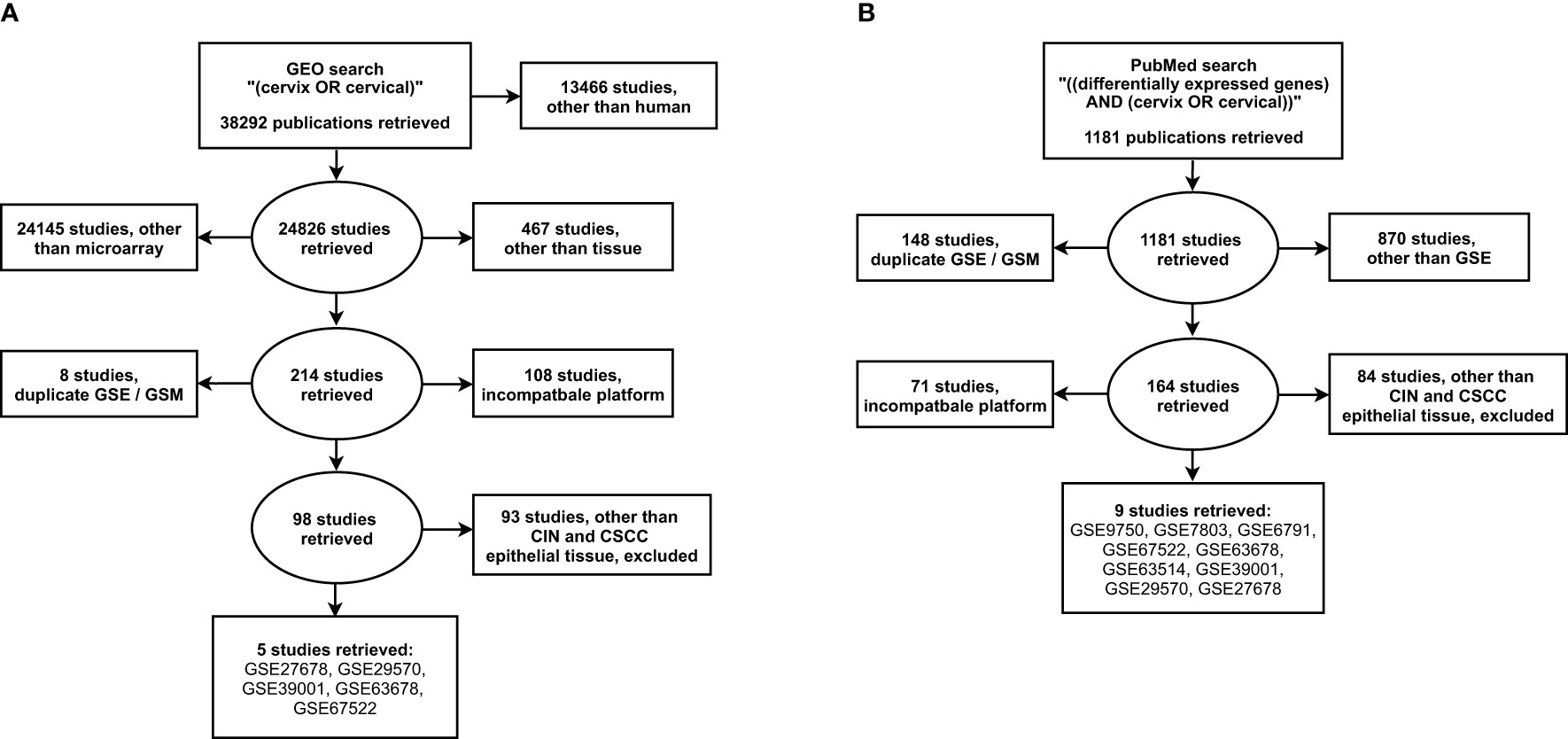
Figure 1 Search strategy for the selection of eligible gene expression studies from the (A) NCBI GEO and the (B) NLM PubMed. GSE, gene expression series; GSM, gene expression sample. NCBI, National Center for Biotechnology Information; GEO, Gene Expression Omnibus; NLM, National Library of Medicine.
Our additional search of the PubMed database, resulted in 1181 datasets, of which 311 corresponded to gene expression studies. From these, 219 microarray datasets with either duplicate gene expression sample and series or incompatible platform were excluded. Screening of the resulting 164 datasets, resulted in three CIN [GSE7803 (24), GSE27678, GSE63514 (25)] and nine CC datasets [GSE6791 (26), GSE7803, GSE9750 (27), GSE27678, GSE29570, GSE39001, GSE63514, GSE63678, GSE67522] with cervical epithelial samples (Figure 1B).
The overlap between the two literature searches ultimately revealed three CIN (GSE7803, GSE27678, GSE63514) and nine CC datasets (GSE6791, GSE7803, GSE9750, GSE27678, GSE29570, GSE39001, GSE63514, GSE63678, GSE67522). The retrieved datasets included cervical epithelial tissue biopsies from healthy controls (n=130) and patients with either CIN (n=115) or CC (n=218). From the identified CC datasets, four included squamous cell carcicnomas (GSE7803, GSE27678, GSE63514, GSE67522) and three included both squamous cell carcinomas and adenocarcinomas (GSE9750, GSE29570, GSE39001), while in two the histopathology of CC was not specified (GSE6791, GSE63678).
Differentially Expressed Genes in CIN and CC
Integration of differential gene expression across the retrieved datasets revealed 1853 DEGs in CIN patients, when compared to healthy controls. Of these, 1157 DEGs were upregulated, and 696 were downregulated. Further, a total of 3861 DEGs were identified in CC patients when compared to healthy controls. Of which, 1986 were upregulated and 1875 were downregulated. Comparative analysis between these expression profiles revealed 991 overlapping DEGs, 862 unique to CIN and 2870 to CC (Figure 2).
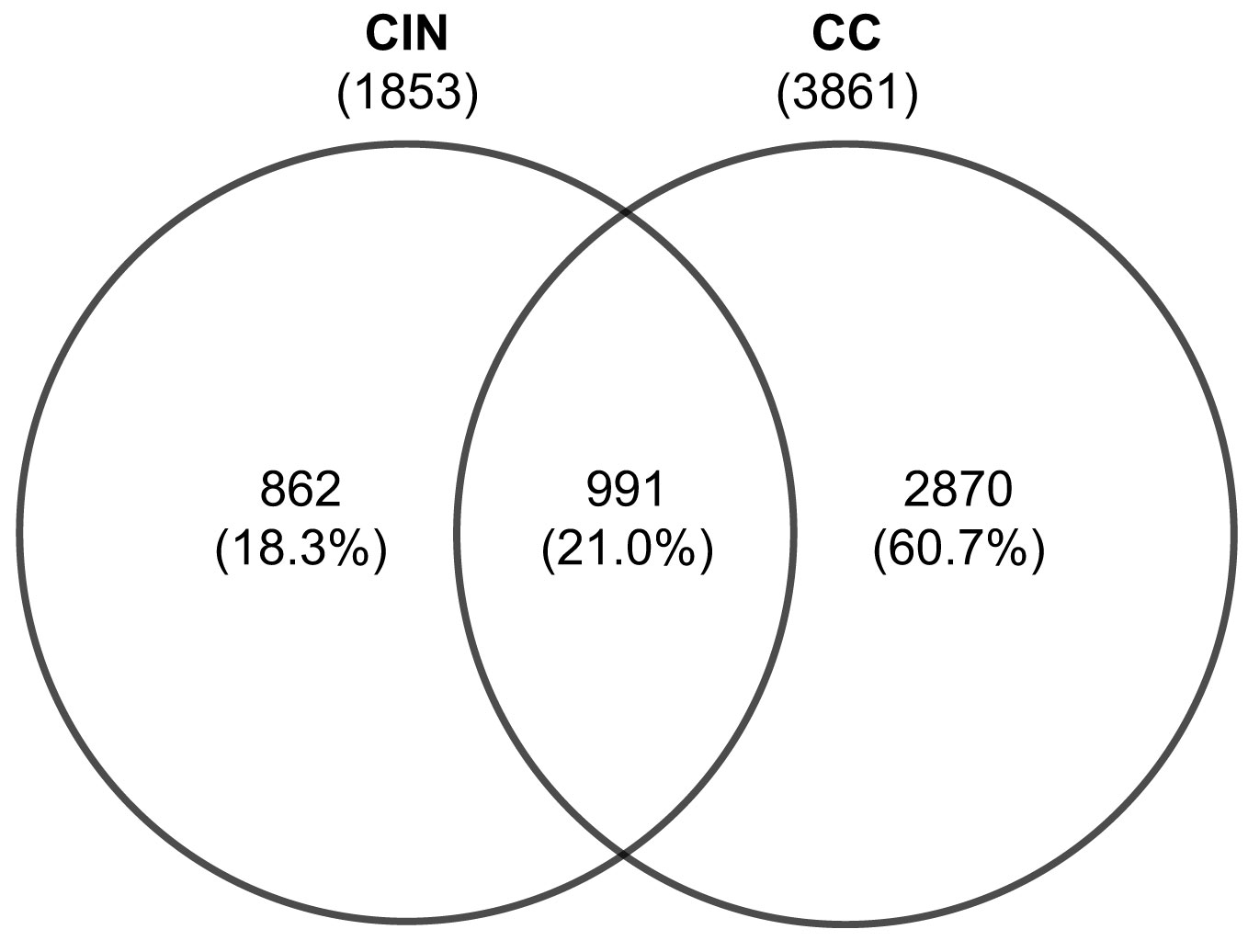
Figure 2 Venn diagram of the differentially expressed genes from cervical epithelial samples in patients with CIN and CC. Values are numbers unless otherwise stated. CC, Cervical cancer; CIN, Cervical intraepithelial neoplasia.
Protein-Protein Interaction Networks and Modules in CIN and CC
Two PPI networks of DEGs from CIN and CC datasets were constructed and consisted a total of 1704 and 3748 DEGs along 21393 and 79828 interactions, respectively (Figures S1A, S2A). Two highly interconnected modules were identified in the CIN network and three in the CC network (Figures S1B, S2B and Tables S1, S2). From the top 10 ranked hub module DEGs in each network, six overlapping genes were revealed: PCNA (proliferating cell nuclear antigen), CDK1 (cyclin dependent kinase 1), MCM4 (minichromosome maintenance complex component 4), BRCA1 (BRCA1 DNA repair associated), MCM5 (minichromosome maintenance complex component 5) and RAD51 (RAD51 recombinase) (Figure 3, Table 1, Tables S3, S4).
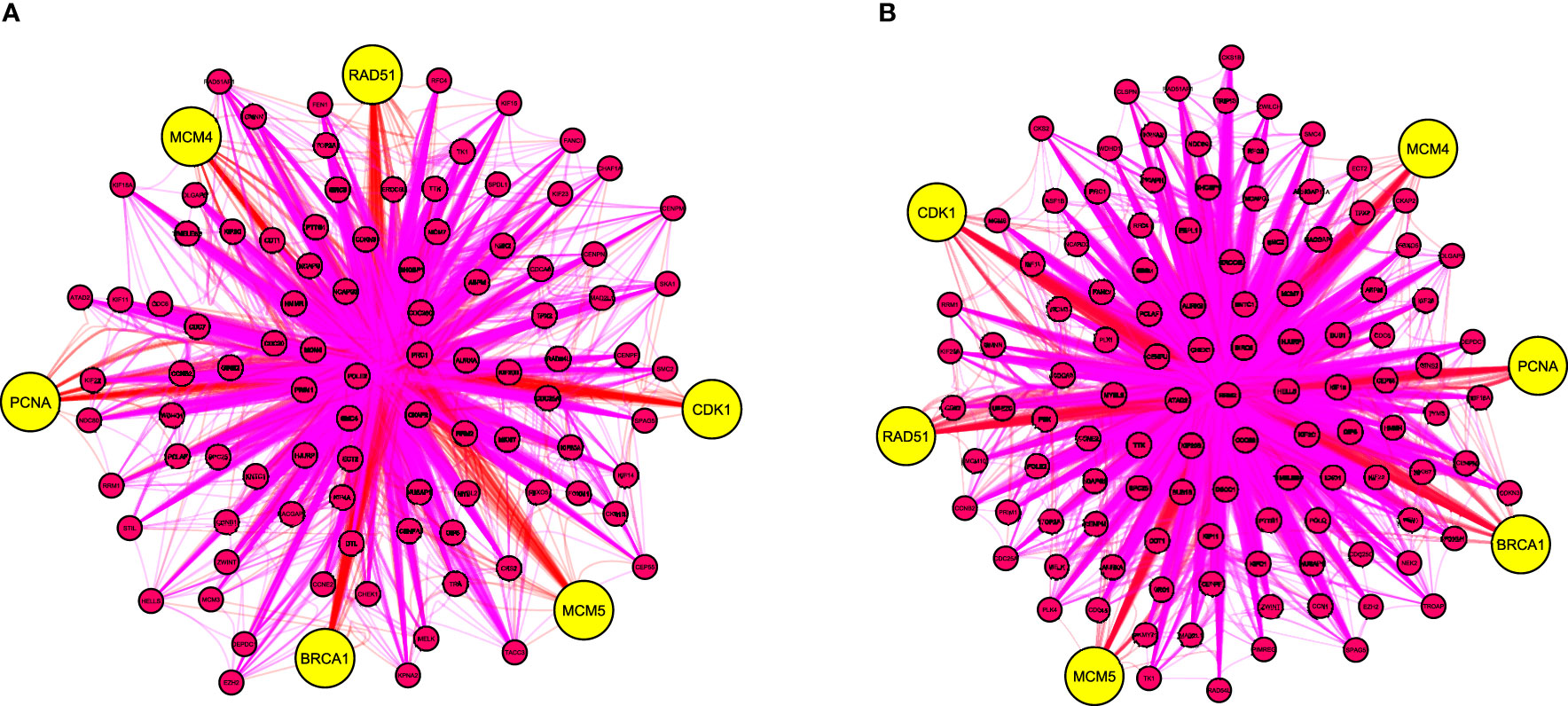
Figure 3 The top 10 overlapping hub genes of clustering modules in the protein-protein interaction network of differentially expressed genes from (A) CIN and (B) CC patients. Yellow nodes indicate hub genes. BRCA1, BRCA1 DNA repair associated; CIN, cervical intraepithelial neoplasia; CC, cervical cancer; CDK1, cyclin dependent kinase 1; MCM4, minichromosome maintenance complex component 4; MCM5, minichromosome maintenance complex component 5; PCNA, proliferating cell nuclear antigen; RAD51, RAD51 recombinase.
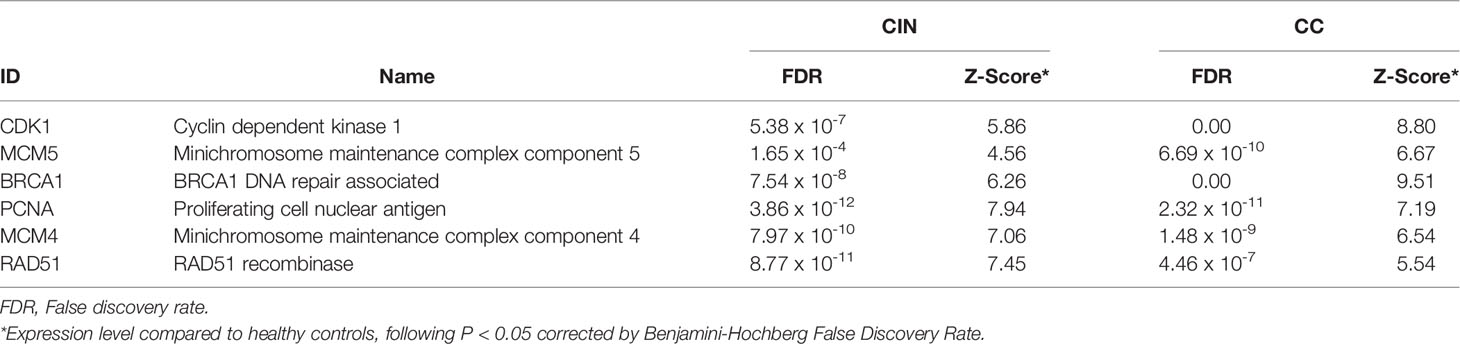
Table 1 The top ranked and overlapping hub genes according to 11 topological algorithms in the protein-protein interaction (PPI) networks of cervical intraepithelial neoplasia (CIN) and cervical cancer (CC) differentially expressed genes.
Prognosis, Expression Level, and Tumor Purity of PCNA in CC
Survival analysis of the TCGA (309 patients) revealed that low expression of PCNA correlated with significantly reduced OS (log-rank P=0.022, HR=0.58) in CC patients, but not with DFS (log-rank P=0.55, HR=1.2) (Figures 4A, B). Expression of PCNA was significantly upregulated in CC tissues when compared to normal (Figure 4C). Pan-cancer analysis demonstrated that dysregulated expression of PCNA was highest in CC among 24 other tumor types (Figure S3). Additionally, the expression level of PCNA was significantly and positively correlated with tumor purity in CC (P=9.86 × 10-4, partial rho=0.197) (Figure 4D).
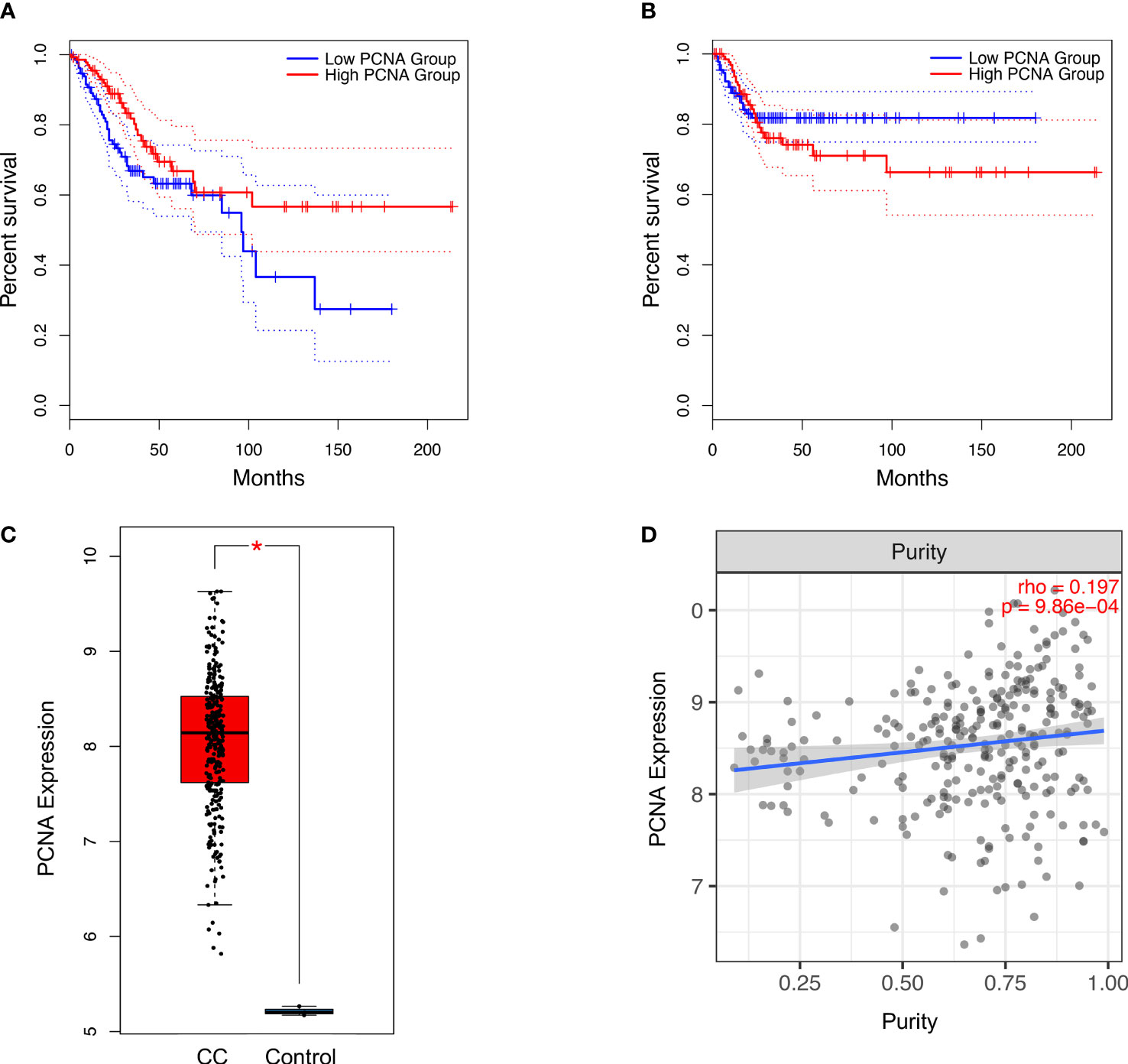
Figure 4 Association of PCNA expression with (A) overall survival (log-rank P=0.022, n(high)=146, n(low)=146) and (B) disease-free survival (log-rank P =0.55, n(high)=146, n(low)=146) in CC patients. Significance was determined using a log-rank P<0.05. Overall expression (C) (transcripts per million) of PCNA based on the TCGA in patients with CC. Significance was determined using a P<0.05 and |log2 fold change|>2. Association between expression level of PCNA and (D) tumour purity in patients with CC. Estimation was determined using the partial Spearman’s correlation (rho). CC, Cervical Cancer; PCNA, Proliferating cell nuclear antigen; TCGA, The Cancer Genome Atlas. *P < 0.05.
Discussion
Analysis of differentially expressed genes from cervical epithelial samples of CIN and CC patients, identified two gene modules in the CIN network and three in the CC network. Multi-algorithmic topological analysis revealed six overlapping hub genes, namely: PCNA, CDK1, MCM4, BRCA1, MCM5 and RAD51. PCNA was the highest ranked hub gene both in terms of expression and interactions, rendering its potential value as a marker of CIN progression.
Findings in the Context of the Literature
PCNA encodes a nuclear protein that is maximally expressed in late G1 and S phases of the cell cycle (28–32). It constitutes a core component of the replication and repair machinery, acting as an auxiliary yet orchestral co-factor of the DNA polymerase δ and ϵ (32–35). By encircling the DNA in a “sliding clamp” formation and recruiting crucial factors to the replication fork, PCNA increases the processivity of the polymerase during replication (32–35). Its role in tumor initiation and progression is linked with perturbed dynamics of DNA synthesis and post-replicative repair, which are all driven from its dysregulated activation (36–38).
A plethora of experimental studies have investigated PCNA expression in CIN and CC (32, 39). Earlier reports have demonstrated significant and positive correlation of PCNA expression with mitotic index and tumor grade (40–44). Additionally, PCNA expression was found to be associated with the presence of oncogenic HPV, possibly due to the suboptimal interaction of the HPV oncoprotein E7 with p21Cip1/Waf1 which physiologically results in PCNA underexpression (45, 46). However, PCNA expression has shown major upregulation upon CIN3 progression and further invasiveness, irrespective of HPV status (47). Therefore, it may be speculated that overexpression of PCNA is primarily associated with CIN progression and to a lesser extent with HPV infection which has a more prominent role in CIN onset.
In spite of being a strong marker of cell proliferation and CIN progression, the prognostic value of PCNA in CC has not been systematically studied and remains a matter of considerable debate (45, 48–50). While PCNA expression did not exhibit prognostic value in two reports by Branca et al. and Costa et al., another study which included 111 CC patients demonstrated association between its overexpression and decreased survival (32, 45, 51). Conversely and in accordance with our findings, a more recent in silico study showed that high expression of PCNA was rather associated with favorable prognosis, although this result was based on the analysis of only 300 CC patients (52). Taken together, the role of PCNA in the prediction of CC prognosis remains inconclusive.
Strengths and Limitations
This is the first study that comprehensively examined the protentional role of DEGs and their interactome as gene biomarkers in CIN progression, using 9 publicly available datasets with more than 450 included patients. In doing so, we employed a multi-algorithmic protein-interaction based approach that relied on different levels of filtering.
Our study also had limitations. Cervical epithelial samples from CIN patients were not limited to any specific HPV infection status. Inclusion of a heterogeneous genotypic distribution of HPV prevalence and type among CIN patients, restricts the predictive value of PCNA as a potential marker of CIN progression. However, HPV types share a common genomic organization and thus the number and the different combinations of encoded oncoproteins contributing to the malignant initiation and transformation, are believed to be similar (53). Additionally, it was not possible to control for other potential confounders such as demographic characteristics (e.g. age) and medical comorbidities (e.g. obesity) in included patients, which could have resulted in residual confounding (54).
Included studies employed different expression profiling platforms, a confounding factor known to hinder statistical power and reliability in detection of DEGs - this type of heterogeneity often results in different expression scales that inevitably reduce the number of integrated DEGs, even after normalization (55). We partially addressed unknown cross-study heterogeneity by employing a random effect model for establishing the significance of DEGs between studies (55–59).
Conclusions
The disease burden of CC has significantly decreased in recent years in developed countries, however the financial costs of screening, the limited capacity of cytological-based diagnosis and the competing risk of reproductive consequences following treatment, remain a challenge. Our study identified that cervical PCNA exhibited multi-algorithmic topological significance among DEGs from CIN and CC samples. Overall, PCNA may serve as a potential gene marker of CIN progression. Experimental validation is necessary to examine the screening, diagnostic and prognostic value of PCNA in patients with CIN and CC.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found at the Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) using the following accession numbers: GSE7803, GSE27678, GSE63514, GSE6791, GSE9750, GSE29570, GSE39001, GSE63678, GSE67522.
Author Contributions
The study was conceived and designed by PG and KSK. The data was acquired and collated by PG and KSK, and analyzed by PG. The manuscript was drafted and revised critically for important intellectual content by all authors. All authors gave final approval of the version to be published and have contributed to the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Imperial Open Access Fund for funding the article processing charges for this manuscript. Author PG thanks the “Bodossaki Foundation” and author KSK thanks the “General Michael Arnaoutis Foundation”, for supporting their research activities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.779042/full#supplementary-material
References
1. Sung H, Ferly J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Buckley C, Butler E, Fox H. Cervical Intraepithelial Neoplasia. J Clin Pathol (1982) 35(1):1–13. doi: 10.1136/jcp.35.1.1
3. Tainio K, Athanasiou A, Tikkinen KA, Aaltonen R, Cárdenas J, Glazer-Livson S, et al. Clinical Course of Untreated Cervical Intraepithelial Neoplasia Grade 2 Under Active Surveillance: Systematic Review and Meta-Analysis. BMJ (2018) 360:k499. doi: 10.1136/bmj.k499
4. Catarino R, Petignat P, Dongui G, Vassilakos P. Cervical Cancer Screening in Developing Countries at a Crossroad: Emerging Technologies and Policy Choices. World J Clin Oncol (2015) 6(6):281. doi: 10.5306/wjco.v6.i6.281
5. Mehta V, Vasanth V, Balachandran C. Pap Smear. Indian J Dermatol Venereol Leprol (2009) 75(2):214. doi: 10.4103/0378-6323.48686
6. Sahasrabuddhe VV, Luhn P, Wentzensen N. Human Papillomavirus and Cervical Cancer: Biomarkers for Improved Prevention Efforts. Future Microbiol (2011) 6(9):1083–98. doi: 10.2217/fmb.11.87
7. Bowden SJ, Bodinier B, Kalliala I, Zuber V, Vuckovic D, Doulgeraki T, et al. Genetic Variation in Cervical Preinvasive and Invasive Disease: A Genome-Wide Association Study. Lancet Oncol (2021) 22(4):548–57. doi: 10.1016/S1470-2045(21)00028-0
8. Cancer ICoESoC. Carcinoma of the Cervix and Tobacco Smoking: Collaborative Reanalysis of Individual Data on 13,541 Women With Carcinoma of the Cervix and 23,017 Women Without Carcinoma of the Cervix From 23 Epidemiological Studies. Int J Cancer (2006) 118(6):1481–95. doi: 10.1002/ijc.21493
9. Toro-Domínguez D, Martorell-Marugán J, López-Domínguez R, García-Moreno A, González-Rumayor V, Alarcón-Riquelme ME, et al. ImaGEO: Integrative Gene Expression Meta-Analysis From GEO Database. Bioinformatics (2019) 35(5):880–2. doi: 10.1093/bioinformatics/bty721
10. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: Protein–Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res (2019) 47(D1):D607–13. doi: 10.1093/nar/gky1131
11. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res (2003) 13(11):2498–504. doi: 10.1101/gr.1239303
12. Bader GD, Hogue CW. An Automated Method for Finding Molecular Complexes in Large Protein Interaction Networks. BMC Bioinformatics (2003) 4(1):1–27. doi: 10.1186/1471-2105-4-2
13. Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko M-T, Lin C-Y. Cytohubba: Identifying Hub Objects and Sub-Networks From Complex Interactome. BMC Syst Biol (2014) 8(4):1–7. doi: 10.1186/1752-0509-8-S4-S11
14. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res (2019) 47(W1):W556–60. doi: 10.1093/nar/gkz430
15. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77(21):e108–e10. doi: 10.1158/0008-5472.CAN-17-0307
16. Ng G, Winder D, Muralidhar B, Gooding E, Roberts I, Pett M, et al. Gain and Overexpression of the Oncostatin M Receptor Occur Frequently in Cervical Squamous Cell Carcinoma and Are Associated With Adverse Clinical Outcome. J Pathol (2007) 212(3):325–34. doi: 10.1002/path.2184
17. Winder DM, Chattopadhyay A, Muralidhar B, Bauer J, English WR, Zhang X, et al. Overexpression of the Oncostatin M Receptor in Cervical Squamous Cell Carcinoma Cells Is Associated With a Pro-Angiogenic Phenotype and Increased Cell Motility and Invasiveness. J Pathol (2011) 225(3):448–62. doi: 10.1002/path.2968
18. Caffarel MM, Chattopadhyay A, Araujo AM, Bauer J, Scarpini CG, Coleman N. Tissue Transglutaminase Mediates the Pro-Malignant Effects of Oncostatin M Receptor Over-Expression in Cervical Squamous Cell Carcinoma. J Pathol (2013) 231(2):168–79. doi: 10.1002/path.4222
19. Guardado-Estrada M, Medina-Martínez I, Juárez-Torres E, Roman-Bassaure E, Macías L, Alfaro A, et al. The Amerindian mtDNA Haplogroup B2 Enhances the Risk of HPV for Cervical Cancer: De-Regulation of Mitochondrial Genes May Be Involved. J Hum Genet (2012) 57(4):269–76. doi: 10.1038/jhg.2012.17
20. Espinosa AM, Alfaro A, Roman-Basaure E, Guardado-Estrada M, Palma Í, Serralde C, et al. Mitosis Is a Source of Potential Markers for Screening and Survival and Therapeutic Targets in Cervical Cancer. PloS One (2013) 8(2):e55975. doi: 10.1371/journal.pone.0055975
21. Pappa KI, Polyzos A, Jacob-Hirsch J, Amariglio N, Vlachos GD, Loutradis D, et al. Profiling of Discrete Gynecological Cancers Reveals Novel Transcriptional Modules and Common Features Shared by Other Cancer Types and Embryonic Stem Cells. PloS One (2015) 10(11):e0142229. doi: 10.1371/journal.pone.0142229
22. Sharma S, Mandal P, Sadhukhan T, Chowdhury RR, Mondal NR, Chakravarty B, et al. Bridging Links Between Long Noncoding RNA HOTAIR and HPV Oncoprotein E7 in Cervical Cancer Pathogenesis. Sci Rep (2015) 5(1):1–15. doi: 10.1038/srep11724
23. Saha SS, Chowdhury RR, Mondal NR, Roy S, Sengupta S. Expression Signatures of HOX Cluster Genes in Cervical Cancer Pathogenesis: Impact of Human Papillomavirus Type 16 Oncoprotein E7. Oncotarget (2017) 8(22):36591. doi: 10.18632/oncotarget.16619
24. Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ, Trimble CL, et al. Gene Expression Analysis of Preinvasive and Invasive Cervical Squamous Cell Carcinomas Identifies HOXC10 as a Key Mediator of Invasion. Cancer Res (2007) 67(21):10163–72. doi: 10.1158/0008-5472.CAN-07-2056
25. Den Boon JA, Pyeon D, Wang SS, Horswill M, Schiffman M, Sherman M, et al. Molecular Transitions From Papillomavirus Infection to Cervical Precancer and Cancer: Role of Stromal Estrogen Receptor Signaling. Proc Natl Acad Sci (2015) 112(25):E3255–E64. doi: 10.1073/pnas.1509322112
26. Pyeon D, Newton MA, Lambert PF, Den Boon JA, Sengupta S, Marsit CJ, et al. Fundamental Differences in Cell Cycle Deregulation in Human Papillomavirus–Positive and Human Papillomavirus–Negative Head/Neck and Cervical Cancers. Cancer Res (2007) 67(10):4605–19. doi: 10.1158/0008-5472.CAN-06-3619
27. Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, et al. Identification of Copy Number Gain and Overexpressed Genes on Chromosome Arm 20q by an Integrative Genomic Approach in Cervical Cancer: Potential Role in Progression. Genes Chromosomes Cancer (2008) 47(9):755–65. doi: 10.1002/gcc.20577
28. Takasaki Y, Deng J-S, Tan EM. A Nuclear Antigen Associated With Cell Proliferation and Blast Transformation. J Exp Med (1981) 154(6):1899–909. doi: 10.1084/jem.154.6.1899
29. Miyachi K, Fritzler MJ, Tan EM. Autoantibody to a Nuclear Antigen in Proliferating Cells. J Immunol (1978) 121(6):2228–34.
30. Mathews MB, Bernstein RM, Franza BR, Garrels JI. Identity of the Proliferating Cell Nuclear Antigen and Cyclin. Nature (1984) 309(5966):374–6. doi: 10.1038/309374a0
31. Bravo R, Celis JE. A Search for Differential Polypeptide Synthesis Throughout the Cell Cycle of HeLa Cells. J Cell Biol (1980) 84(3):795–802. doi: 10.1083/jcb.84.3.795
32. Branca M, Ciotti M, Giorgi C, Santini D, Di Bonito L, Costa S, et al. Up-Regulation of Proliferating Cell Nuclear Antigen (PCNA) Is Closely Associated With High-Risk Human Papillomavirus (HPV) and Progression of Cervical Intraepithelial Neoplasia (CIN), But Does Not Predict Disease Outcome in Cervical Cancer. Eur J Obstet Gynecol Reprod Biol (2007) 130(2):223–31. doi: 10.1016/j.ejogrb.2006.10.007
33. Kelman Z. PCNA: Structure, Functions and Interactions. Oncogene (1997) 14(6):629–40. doi: 10.1038/sj.onc.1200886
34. Hingorani MM, O’Donnell M. Sliding Clamps: A (Tail) Ored Fit. Curr Biol (2000) 10(1):R25–R9. doi: 10.1016/S0960-9822(99)00252-3
35. Moldovan G-L, Pfander B, Jentsch S. PCNA, the Maestro of the Replication Fork. Cell (2007) 129(4):665–79. doi: 10.1016/j.cell.2007.05.003
36. Shivji MK, Kenny MK, Wood RD. Proliferating Cell Nuclear Antigen Is Required for DNA Excision Repair. Cell (1992) 69(2):367–74. doi: 10.1016/0092-8674(92)90416-A
37. Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, et al. Nuclear Dynamics of PCNA in DNA Replication and Repair. Mol Cell Biol (2005) 25(21):9350–9. doi: 10.1128/MCB.25.21.9350-9359.2005
38. Lehmann AR, Fuchs RP. Gaps and Forks in DNA Replication: Rediscovering Old Models. DNA Repair (2006) 5(12):1495–8. doi: 10.1016/j.dnarep.2006.07.002
39. Astudillo H, Lopez T, Castillo S, Gariglio P, Benitez L. P53, Bcl-2, PCNA Expression, and Apoptotic Rates During Cervical Tumorigenesis. Ann N Y Acad Sci (2003) 1010(1):771–4. doi: 10.1196/annals.1299.138
40. Xue Y, Feng Y, Zhu G, Zhang X. Proliferative Activity in Cervical Intraepithelial Neoplasia and Cervical Carcinoma. Chin Med J (Engl) (1999) 112(4):373–5.
41. Khaled A, Imamura Y, Noriki S, Fukuda M. Early Progression Stage of Malignancy of Uterine Cervical Dysplasia as Revealed by Immunohistochemical Demonstration of Increased DNA-Instability. Eur J Histochem (2000) 44(2):143–56.
42. Steinbeck RG, Heselmeyer KM, Moberger HB, Auer GU. The Relationship Between Proliferating Cell Nuclear Antigen (PCNA), Nuclear DNA Content and Mutant P53 During Genesis of Cervical Carcinoma. Acta Oncol (1995) 34(2):171–6. doi: 10.3109/02841869509093952
43. Park JS, Rhyu KS, Kim CJ, Kim HS, Han KT, Ahn HK, et al. Presence of Oncogenic HPV DNAs in Cervical Carcinoma Tissues and Pelvic Lymph Nodes Associating With Proliferating Cell Nuclear Antigen Expression. Gynecol Oncol (1996) 60(3):418–23. doi: 10.1006/gyno.1996.0066
44. Karakitsos P, Kyroudes A, Apostolaki C, Paizi P, Voulgaris Z, Alekou G, et al. The Evaluation of PCNA/Cyclin Expression in Cervical Intraepithelial Lesions. Gynecol Oncol (1994) 55(1):101–7. doi: 10.1006/gyno.1994.1256
45. Tjalma WA, Weyler JJ, Bogers JJ, Pollefliet C, Baay M, Goovaerts GC, et al. The Importance of Biological Factors (Bcl-2, Bax, P53, PCNA, MI, HPV and Angiogenesis) in Invasive Cervical Cancer. Eur J Obstet Gynecol Reprod Biol (2001) 97(2):223–30. doi: 10.1016/S0301-2115(00)00541-8
46. Funk JO, Waga S, Harry JB, Espling E, Stillman B, Galloway DA. Inhibition of CDK Activity and PCNA-Dependent DNA Replication by P21 Is Blocked by Interaction With the HPV-16 E7 Oncoprotein. Genes Dev (1997) 11(16):2090–100. doi: 10.1101/gad.11.16.2090
47. Wang J-L, Zheng B-Y, Li X-D, Ångström T, Lindström MS, Wallin K-L. Predictive Significance of the Alterations of P16ink4a, P14arf, P53, and Proliferating Cell Nuclear Antigen Expression in the Progression of Cervical Cancer. Clin Cancer Res (2004) 10(7):2407–14. doi: 10.1158/1078-0432.CCR-03-0242
48. Tjalma W, Weyler J, Pollefliet C, Bogers J, Van Marck E, van Dam P, et al. The Evaluation of Proliferative Activity in CIN III and Microinvasive Cervical Cancer and Its Role in Recurrence. Eur J Obstet Gynecol Reprod Biol (2001) 94(2):270–5. doi: 10.1016/S0301-2115(00)00333-X
49. Maeda M, Simoes M, Wakamatsu A, Longatto Filho A, Oyafuso M, De Mello E, et al. Relevance of the Rates of PCNA, Ki-67 and P53 Expression According to the Epithelial Compartment in Cervical Lesions. Pathologica (2001) 93(3):189–95.
50. Oka K, Hoshi T, Arai T. Prognostic Significance of the PC10 Index as a Prospective Assay for Cervical Cancer Treated With Radiation Therapy Alone. Cancer (1992) 70(6):1545–50. doi: 10.1002/1097-0142(19920915)70:6<1545::AID-CNCR2820700617>3.0.CO;2-S
51. Costa S, Terzano P, Santini D, Ceccarelli C, Martoni A, Angelelli B, et al. Neoadjuvant Chemotherapy in Cervical Carcinoma: Regulators of Cell Cycle, Apoptosis, and Proliferation as Determinants of Response to Therapy and Disease Outcome. Am J Clin Pathol (2001) 116(5):729–37. doi: 10.1309/8B4E-57PR-T50F-VRQT
52. Li X, Tian R, Gao H, Yan F, Ying L, Yang Y, et al. Identification of Significant Gene Signatures and Prognostic Biomarkers for Patients With Cervical Cancer by Integrated Bioinformatic Methods. Technol Cancer Res Treat (2018) 17:1533033818767455. doi: 10.1177/1533033818767455
53. Garnett T, Duerksen-Hughes P. Modulation of Apoptosis by Human Papillomavirus (HPV) Oncoproteins. Arch Virol (2006) 151(12):2321–35. doi: 10.1007/s00705-006-0821-0
54. Wee CC, Phillips RS, McCarthy EP. BMI and Cervical Cancer Screening Among White, African-American, and Hispanic Women in the United States. Obes Res (2005) 13(7):1275–80. doi: 10.1038/oby.2005.152
55. Lyu Y, Li Q. A Semi-Parametric Statistical Model for Integrating Gene Expression Profiles Across Different Platforms. BMC Bioinformatics (2016). BioMed Central. 17(S1):5 doi: 10.1186/s12859-015-0847-y
56. Johnson WE, Li C, Rabinovic A. Adjusting Batch Effects in Microarray Expression Data Using Empirical Bayes Methods. Biostatistics (2007) 8(1):118–27. doi: 10.1093/biostatistics/kxj037
57. Dillies M-A, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, et al. A Comprehensive Evaluation of Normalization Methods for Illumina High-Throughput RNA Sequencing Data Analysis. Brief Bioinform (2013) 14(6):671–83. doi: 10.1093/bib/bbs046
58. Hansen KD, Irizarry RA, Wu Z. Removing Technical Variability in RNA-Seq Data Using Conditional Quantile Normalization. Biostatistics (2012) 13(2):204–16. doi: 10.1093/biostatistics/kxr054
Keywords: cervical intraepithelial neoplasia, CIN, cervical cancer, gene biomarkers, cervical disease
Citation: Giannos P, Kechagias KS, Bowden S, Tabassum N, Paraskevaidi M and Kyrgiou M (2021) PCNA in Cervical Intraepithelial Neoplasia and Cervical Cancer: An Interaction Network Analysis of Differentially Expressed Genes. Front. Oncol. 11:779042. doi: 10.3389/fonc.2021.779042
Received: 17 September 2021; Accepted: 28 October 2021;
Published: 26 November 2021.
Edited by:
Sarah M. Temkin, Anne Arundel Medical Center, United StatesReviewed by:
Komsun Suwannarurk, Thammasat University, ThailandClarissa Polen-De, Mayo Clinic, United States
Copyright © 2021 Giannos, Kechagias, Bowden, Tabassum, Paraskevaidi and Kyrgiou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Konstantinos S. Kechagias, a29uc3RhbnRpbm9zLmtlY2hhZ2lhczE4QGltcGVyaWFsLmFjLnVr
†These authors have contributed equally to this work and share first authorship
 Panagiotis Giannos
Panagiotis Giannos Konstantinos S. Kechagias
Konstantinos S. Kechagias Sarah Bowden3,5
Sarah Bowden3,5 Maria Kyrgiou
Maria Kyrgiou