- 1Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Men’s Health and Reproductive Health Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Skull Base Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 4Department of Pathology, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Circular RNAs (circRNAs) are a group of long non-coding RNAs with enclosed structure generated by back-splicing events. Numerous members of these transcripts have been shown to affect carcinogenesis. Circular RNA itchy E3 ubiquitin protein ligase (circITCH) is a circRNA created from back splicing events in ITCH gene, a protein coding gene on 20q11.22 region. ITCH has a role as a catalyzer for ubiquitination through both proteolytic and non-proteolytic routes. CircITCH is involved in the pathetiology of cancers through regulation of the linear isoform as well as serving as sponge for several microRNAs, namely miR-17, miR-224, miR-214, miR-93-5p, miR-22, miR-7, miR-106a, miR-10a, miR-145, miR-421, miR-224-5p, miR-197 and miR-199a-5p. CircITCH is also involved in the modulation of Wnt/β-catenin and PTEN/PI3K/AKT pathways. Except from a single study in osteosarcoma, circITCH has been found to exert tumor suppressor role in diverse cancers. In the present manuscript, we provided a comprehensive review of investigations that reported function of circITCH in the carcinogenesis.
Introduction
Circular RNAs (circRNAs) are a group of long non-coding RNAs with enclosed structure. This structure is created through establishment of a covalent bond between 5’ and 3’ termini through a back-splicing event in exons of a certain pre-mRNA (1). Several studies have indicated broad expression of circRNAs in mammalian cells in a cell type- or tissue-specific manner (1). CircRNAs have been shown to affect different cellular and biological processes, namely cell proliferation (1), differentiation, pluripotency (2) and epithelial-mesenchymal transition (EMT) (3). Moreover, they can participate in the remodeling of endoplasmic reticulum stress, autophagy and phagocytosis, DNA repair mechanisms as well as drug efflux (4). Different mechanisms have been suggested for circRNAs effects in these processes with the most appreciated one being their function as decoys for microRNAs (miRNAs) or RNA-binding proteins. Through this mechanism, circRNAs can influence expression of genes or translation of proteins with regulatory functions (1). CircRNA have the ability to base pair with other types of RNAs as well (5). Moreover, circRNAs can suppress activity of certain proteins, particularly cell cycle proteins through interacting with them (6). While circRNAs are mainly considered as non-coding RNAs, they might be served as a template for production of proteins under some conditions (5). Cumulatively, circRNAs can influence expression of cellular proteins, interfere with RNA-binding proteins to affect transcription of genes, regulate gene transcription in cis, and modulate splicing events (5). Yet, the competing endogenous function of circRNAs is the chief way through which they exert their biological effects (5). Several studies have emphasized on the role of circRNAs in cancer development and induction of chemo/radioresistance (4, 5).
Circular RNA itchy E3 ubiquitin protein ligase (circITCH) is an example of cancer-related circRNAs which can be used as target for therapeutic interventions. It is created from back splicing events in ITCH gene, a protein coding gene on 20q11.22 region. ITCH has a role as a catalyzer for ubiquitination through both proteolytic and non-proteolytic routes (7). It has been shown to affect tumorigenesis in a context-dependent manner (7). Recent studies have shown involvement of the circRNA from this locus in the carcinogenesis process. In the present manuscript, we provided a comprehensive review of investigations that reported function of circITCH in this process. The evidence regarding the role of circITCH in cancers is classified based on the samples/models used in the original papers to in vitro, in vivo and clinical studies.
Cell Line Studies
Bladder Cancer
CircITCH has been found to be down-regulated in bladder cancer cell lines. Forced over-expression of circITCH could inhibit proliferation, migratory potential, invasive properties and metastatic ability of bladder cancer cells. Functionally, circITCH acts as a sponge for miR-17 and miR-224 to up-regulate expression of their target genes p21 and PTEN. Cumulatively, circITCH functions as a tumor suppressor circRNA in bladder cancer (8).
Breast Cancer
Expression of circITCH has also been shown to be decreased in triple negative breast cancer cell lines. Stable transfection of MDA-MB-231 and BT-549 cells with circITCH-expressing vectors has resulted in inhibition of proliferation, invasiveness and metastatic ability of these cells. Mechanistically, circITCH serves as a molecular sponge for miR-214 and miR-17 leading to enhancement of expression of the linear form of ITCH. This circRNA functionally inactivates Wnt/β-catenin signaling (9).
Cervical Cancer
Expression of circITCH has also been shown to be down-regulated in cervical cancer cell lines. Up-regulation of circITCH in cervical cancer cells has inhibited their proliferation, migration, and invasiveness. Mechanistically, circITCH acts a sponge for miR-93-5p to regulate expression of FOXK2 (10).
Osteosarcoma
Down-regulation of circITCH has also been verified in osteosarcoma cells. Overexpression of circITCH has induced cell apoptosis and decreased cell viability, proliferation, migratory potential and invasive properties of MG63 and Saos-2 osteosarcoma cells. This circRNA could decrease expression of miR-22 in osteosarcoma cells, thus suppressing PTEN/PI3K/AKT and SP-1 signals (11). On the other hand, another study in the hFOB1.19 osteoblast cell line and multiple osteosarcoma cell lines has shown up-regulation of circITCH in neoplastic cells compared with the osteoblast cells. Functionally, circITCH enhanced migration, invasive properties, and growth of these neoplastic cells through sponging miR-7 and increasing expression of EGFR (12).
Figure 1 shows the tumor suppressor role of circITCH in bladder, breast and cervical cancers as well as dual role of this circRNA in osteosarcoma.
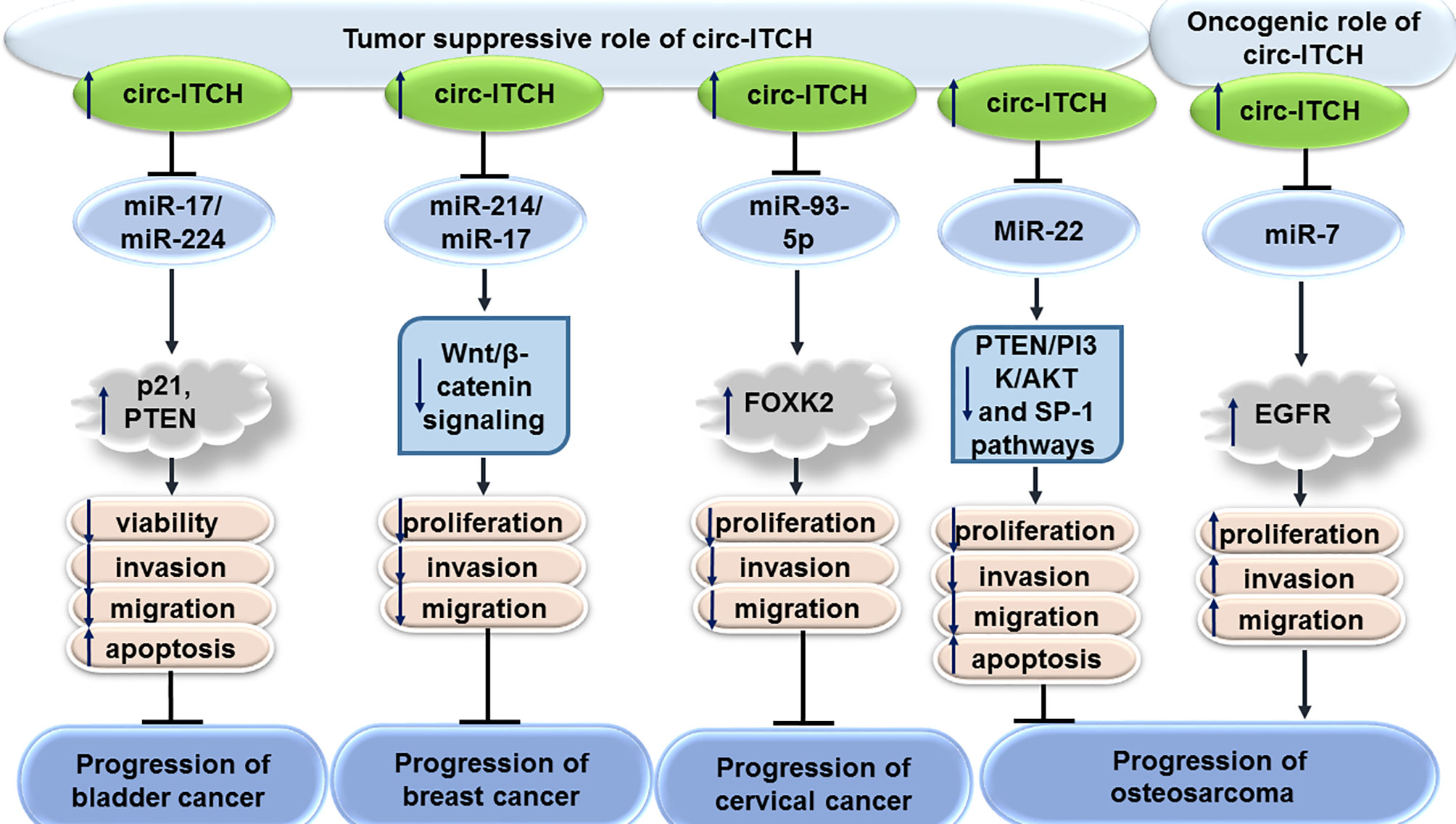
Figure 1 Tumor suppressor role of circITCH in bladder, breast and cervical cancers as well as dual role of this circRNA in osteosarcoma.
Thyroid Cancer
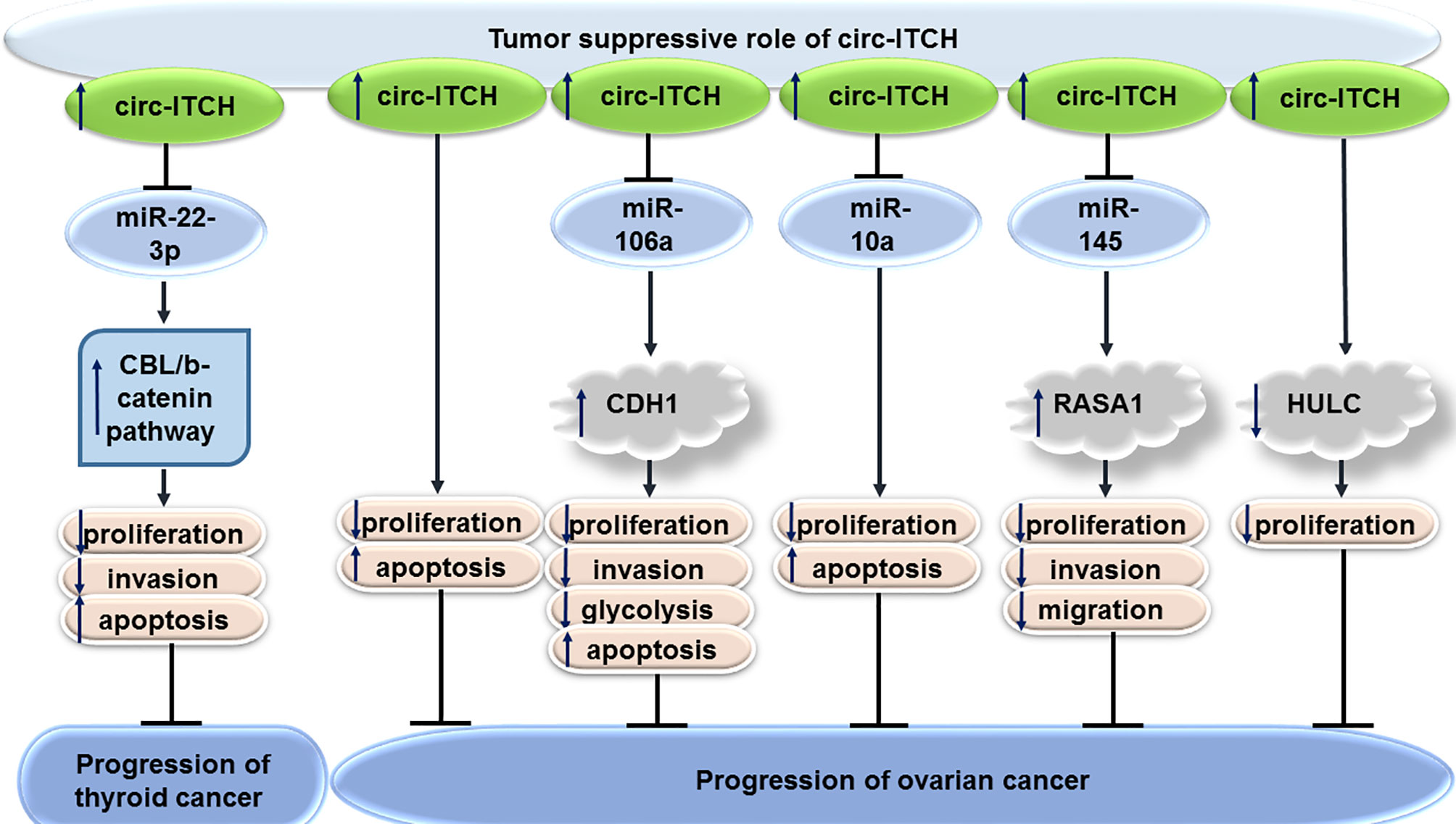
In thyroid cancer cells, forced over-expression of circITCH inhibits cell proliferation and invasive properties, while promoting cell apoptosis. These effects are mediated through sponging miR-22-3p and subsequent up-regulation of levels of CBL, an E3 ligase of nuclear β-catenin. Cumulatively, circITCH affects activity of the Wnt/β-catenin pathway through modulation of CBL levels, therefore suppressing progression of thyroid cancer (13).
Ovarian Cancer
Expression of circITCH expression has been found to be down-regulated in numerous epithelial ovarian cancer cell lines versus normal ovarian epithelial cells. This circRNA could inhibit proliferation of SKOV3 and OVCAR-3 cells, while enhancing their apoptosis (14). Another study has shown the role of circITCH in suppression of proliferation, invasiveness, and glycolytic process in ovarian cancer cells through sequestering miR-106a and enhancing expression of CDH1 (15). miR-10a-alpha has also been identified as a target of circITCH in ovarian cancer cells through which circITCH exerts its tumor suppressor effects (16). Moreover, circITCH has been shown to suppress progression of this cancer via influencing miR-145/RASA1 axis (17). Finally, circITCH has been suggested to suppress proliferation of ovarian cells through deceasing expression of HULC (18). Figure 2 shows the tumor suppressor role of circITCH in thyroid and ovarian cancers.
Hepatocellular Carcinoma
CircITCH has also been shown to have tumor suppressor roles in hepatocellular carcinoma. In fact, the effects of lidocaine on inhibition of proliferation of hepatocellular carcinoma cells have been shown to be mediated through restoration of circITCH in these cells. Mechanistically, circITCH modulates expression of CPEB3 through sponging miR-421 (19). CircITCH can also affect progression of hepatocellular carcinoma through sponging miR-224-5p and increasing expression of MafF (20). CircRNAITCH levels have been found to down-regulated in several hepatocellular cancer cell lines compared with normal hepatic L-02 cell line. Up-regulation of circRNAITCH has inhibited proliferation of these cells, suppressed their colony formation aptitude and induced their apoptosis. CircRNAITCH could be used as a sponge for miR-7 and miR-214. Through this route, it regulates Wnt/β-catenin signals and suppresses c-myc and cyclin D1 levels (21).
Glioma
CircITCH has also been shown to inhibit proliferation and invasive potential of glioma cells via sequestering miR-106a-5p and enhancing expression of SASH1 (22). Moreover, it has been reported to serve as a sponge for miR-214 and promote expression of linear ITCH in glioma cells (23).
Oral Squamous Cell Carcinoma
The miR-421/PDCD4 axis has been shown as the downstream axis mediating the role of circITCH in modulation of progression of oral squamous cell carcinoma by regulating (24).
Figure 3 shows the tumor suppressor role of circITCH in hepatocellular carcinoma, glioma and oral squamous cell carcinoma.
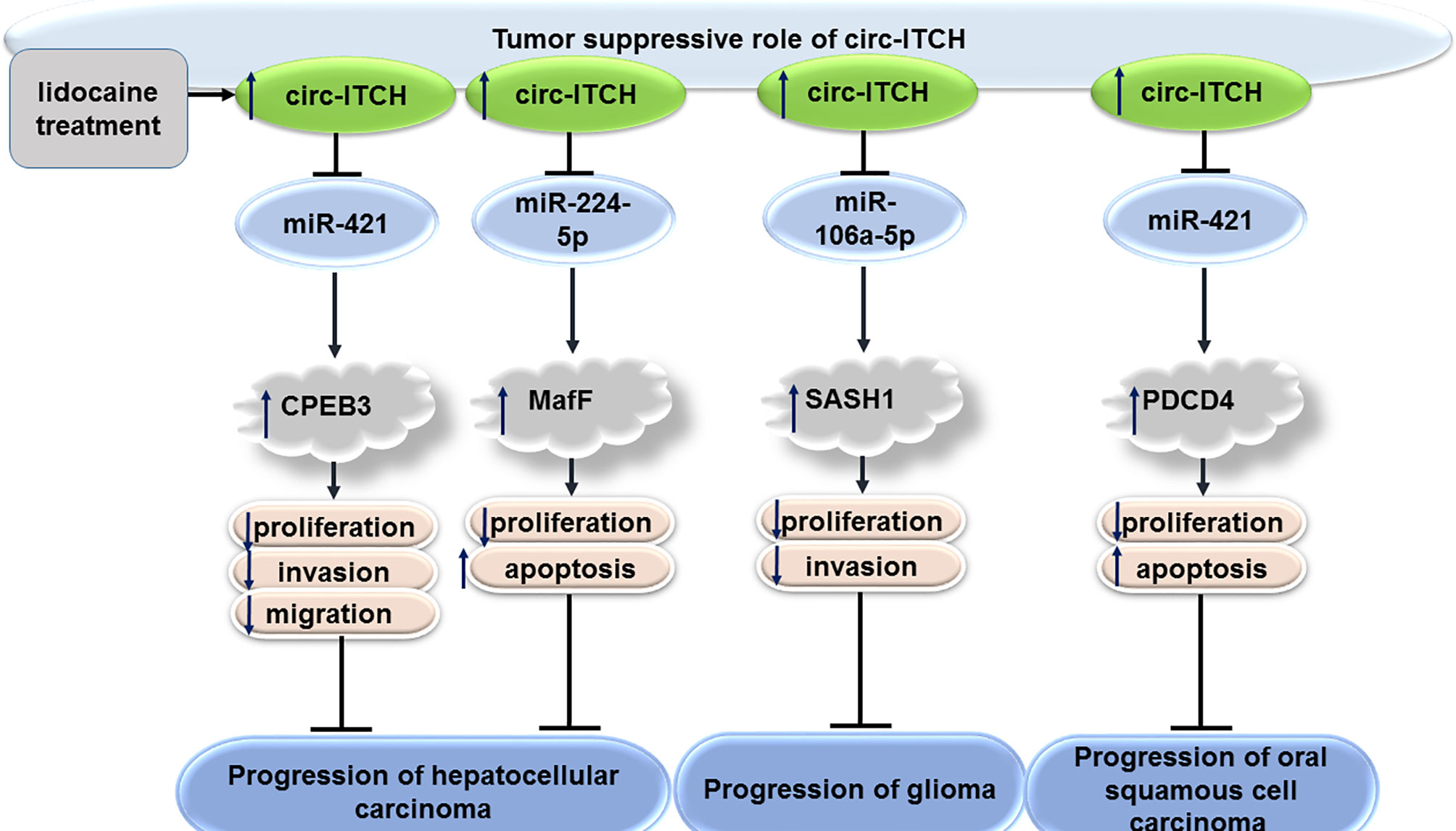
Figure 3 Tumor suppressor role of circITCH in hepatocellular carcinoma, glioma and oral squamous cell carcinoma.
Prostate Cancer
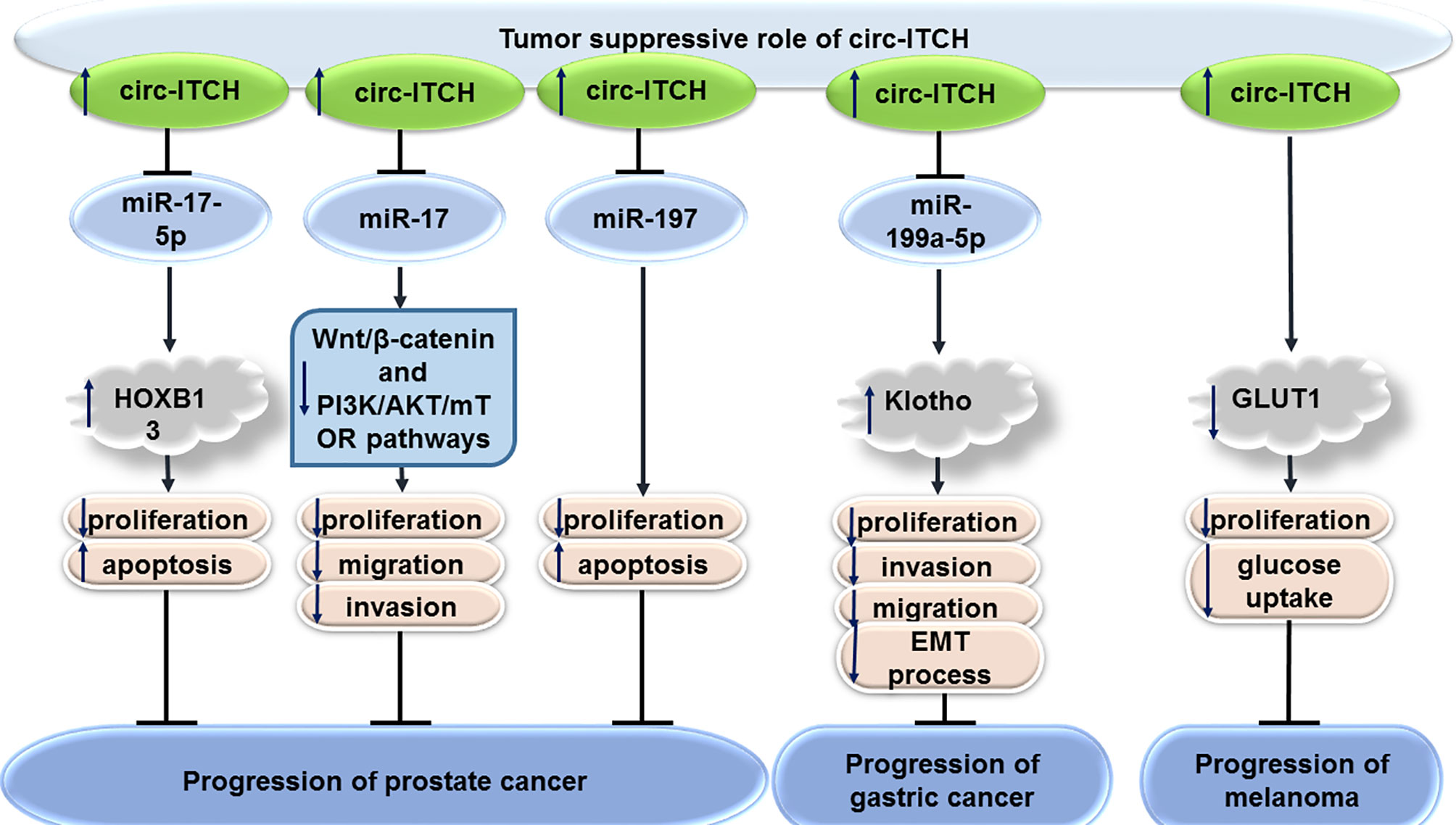
CircITCH exerts tumor suppressor roles in prostate cancer via influencing miR-17-5p/HOXB13 axis (25). Moreover, circITCH can inhibit proliferation, migratory aptitude, and invasiveness of human prostate cancer cells through sequestering miR-17. This circRNA can also down-regulate expression levels of several proteins in the Wnt/β-catenin and PI3K/AKT/mTOR signal transductions in LNCaP and PC-3 cells, as representatives of androgen receptor (AR)-positive and AR-negative cells, respectively (26). miR-197 is another target of circITCH in prostate cancer cells through which it regulates progression of this type of cancer (27).
Gastric Cancer
In addition, circITCH can suppress gastric carcinogenesis through modulation of miR-199a-5p/Klotho axis (28) as well as the Wnt/β-catenin pathway (29).
Melanoma
Finally, circITCH decreases expression of GLUT1 and inhibits uptake of glucose by melanoma cells to suppress their proliferation (30).
Figure 4 shows the tumor suppressor role of circITCH in prostate cancer, gastric cancer and melanoma.
Other Cancers
miR-7 and miR-214 have been found to be sequestered by circITCH in lung (31) and esophageal cancers (32). In addition to mentioned cancer types, circITCH has tumor suppressor roles in renal cancer (33), multiple myeloma (34) and colorectal cancer (35). Table 1 summarizes expression and function of circITCH in cancer cell lines
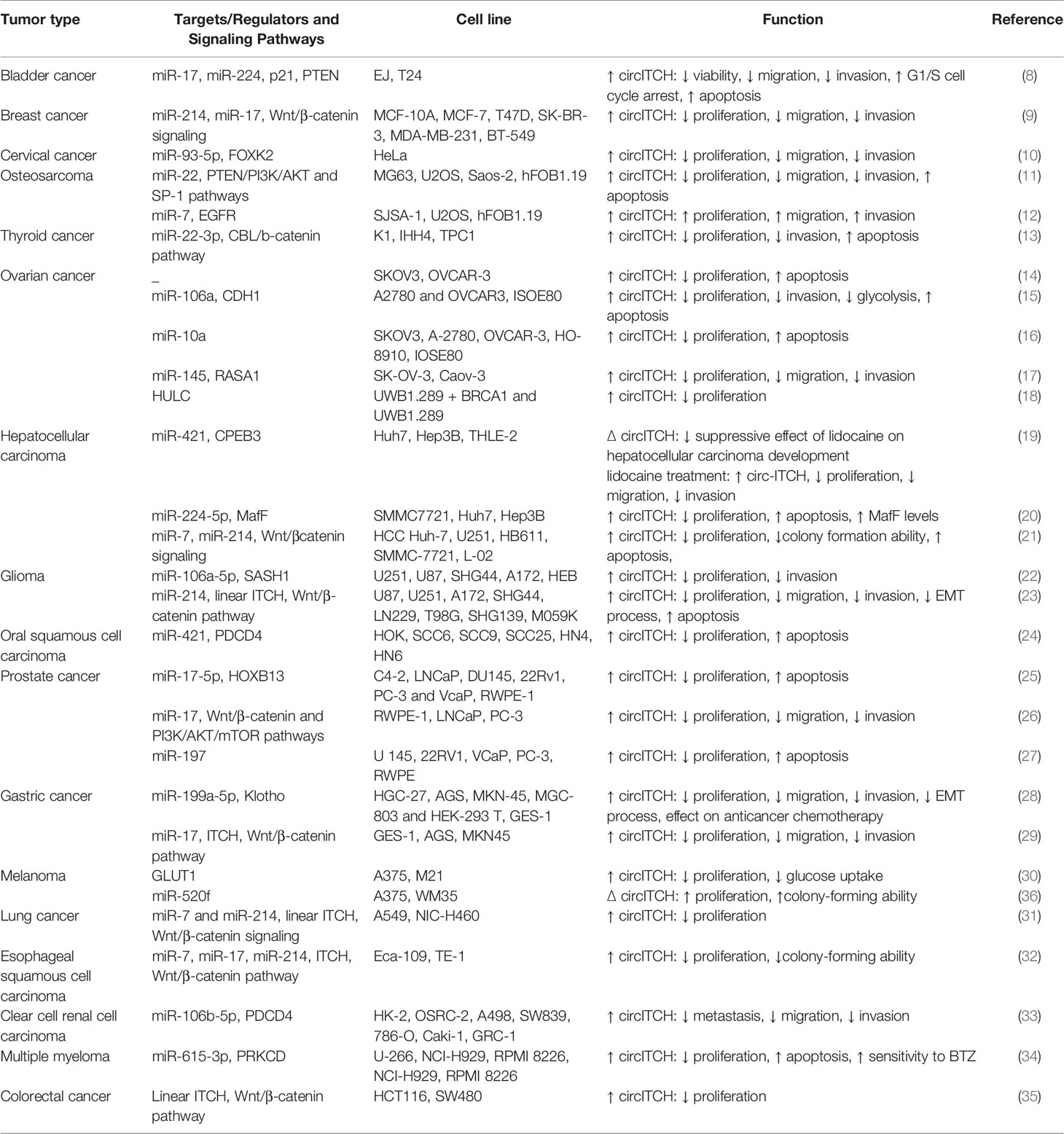
Table 1 Expression and function of circITCH in cancer cell lines (∆: knock-down or deletion, BTZ: Bortezomib).
Animal Studies
Subcutaneous injection of T24 bladder cancer cells transfected with circITCH into the nude mice has shown the impact of this circRNA in reduction of tumor volumes and tumor weight. Notably, expressions of p21 and PTEN have been up-regulated in the tumors originated from circITCH overexpressing cells (8). Other in vivo studies have consistently verified the tumor suppressor roles of circITCH in different animal models (Table 2). Similarly, over-expression of circITCH has increased sensitivity of bortezomib-resistant multiple myeloma cells to this drug in animal models (34).
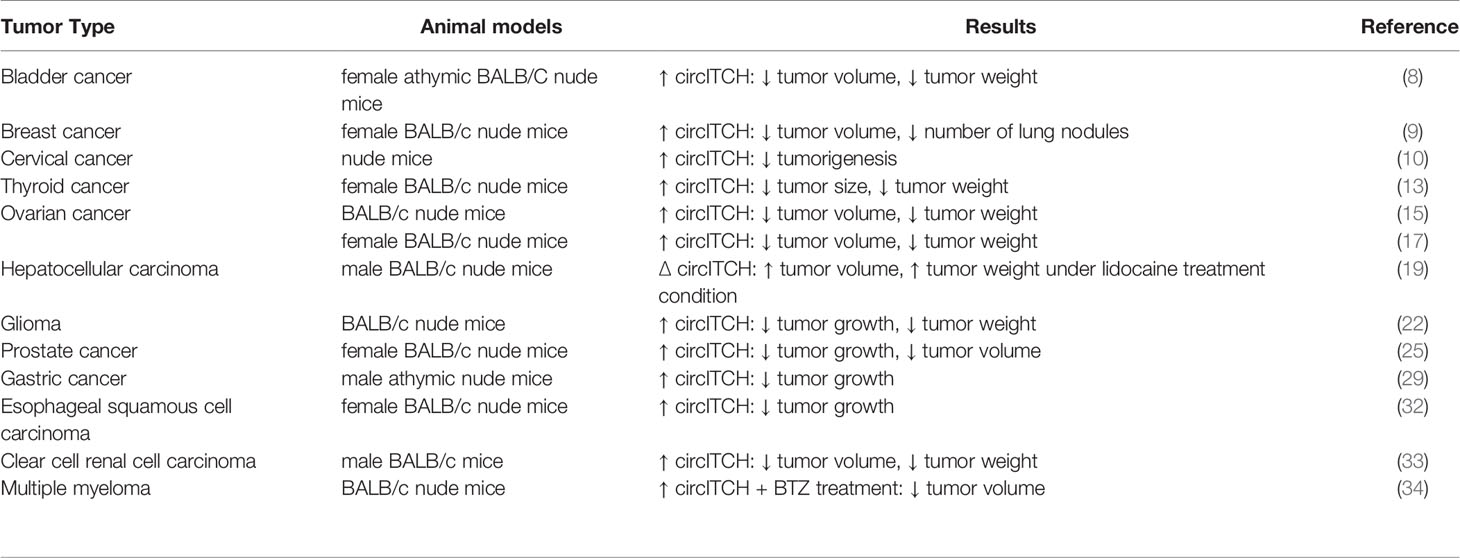
Table 2 Summary of studies which assessed impact of circITCH up-regulation or silencing in animal models (∆: knock-down or deletion, BTZ: Bortezomib).
Clinical Studies
Different studies in samples obtained from patients with diverse types of neoplasms have verified down-regulation of circITCH in neoplastic samples when compared with normal (non-affected) tissues (Table 3). Down-regulation of circITCH in bladder cancer tissues has been correlated with histological grade. In addition, bladder cancer patients who had circITCH down-regulation exhibited poor clinical outcome (8).
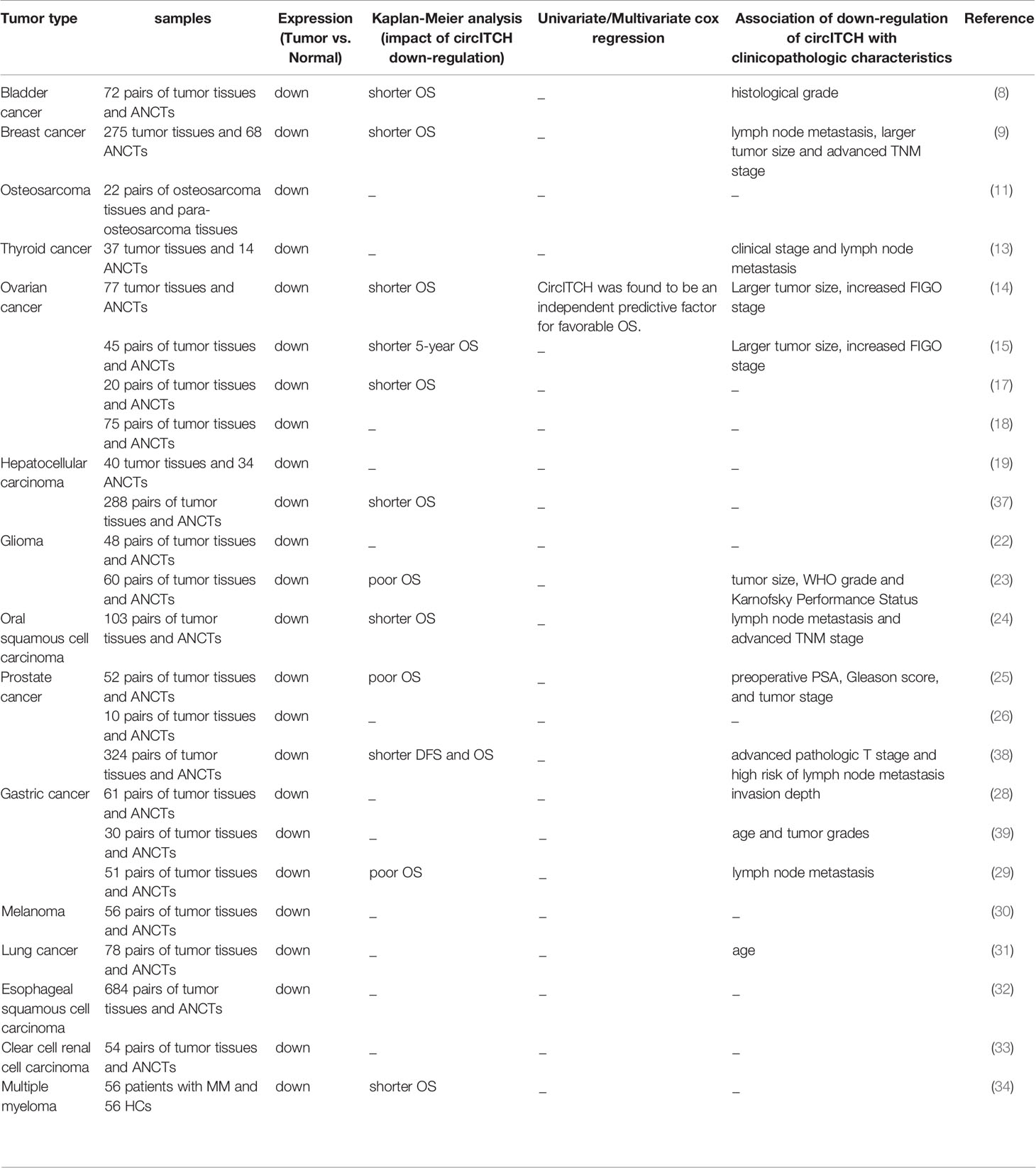
Table 3 Results of studies that reported dysregulation of circITCH in clinical samples (ANCTs, adjacent non-cancerous tissues; OS, Overall survival; FIGO, International Federation of Gynecology and Obstetrics; DFS, disease-free survival; TNM, tumor‐node‐metastasis).
Expression of circITCH has also been reported to be lower in ovarian tumor tissues compared with corresponding non-tumoral tissues. Most notably, expression of circITCH has been inversely correlated with tumor size and FIGO stage in these patients. Based on multivariate Cox analyses, over-expression of circITCH has been identified as an independent predictor of favorable overall survival of patients with ovarian cancer (14).
Cumulatively, decreased levels of circITCH have been correlated with poor outcome in diverse types of cancers, suggesting this circRNA as a prognostic factor in human malignancies.
Discussion
Except from a single study in osteosarcoma, circITCH has been found to exert tumor suppressor role in diverse cancers. This circRNA is involved in the pathetiology of cancers through regulation of the linear isoform as well as serving as sponge for several microRNAs, namely miR-17, miR-224, miR-214, miR-93-5p, miR-22, miR-7, miR-106a, miR-10a, miR-145, miR-421, miR-224-5p, miR-197 and miR-199a-5p. CircITCH also partakes in the modulation of Wnt/β-catenin and PTEN/PI3K/AKT pathways.
A number of miRNAs have been found to interact with circITCH in diverse tissues. For instance, miR-7 has been found to be sponged by circITCH in osteosarcoma, hepatocellular carcinoma, lung cancer and esophageal squamous cell carcinoma. Meanwhile, miR-17 has been detected as a target of this circRNA in bladder, breast, prostate, gastric and esophageal squamous cell cancers. Moreover, circITCH has been shown to sponge miR-214 in breast, lung, hepatocellular carcinoma, glioma and esophageal cancers. Thus, circITCH/miR-7, circITCH/miR-17 and circITCH/miR-214 axes are appropriate therapeutic targets for diverse types of cancers.
The correlation between expression levels of circITCH and clinicopathological data such as tumor size, local invasion, distant metastasis and different staging systems shows the importance of this circRNA in the development or progression of cancers, representing a novel biomarker role for it. Although the impact of circITCH in determination of prognosis of cancer patients is well established, its function as a diagnostic marker is not studied. Since circRNAs are stable transcripts in the circulation, they are expected to reflect cancer course. Thus, future investigations should focus on evaluation of levels of circITCH in plasma of patients with different stages of cancers to find the suitability of this marker for diagnostic purposes as well as patients’ follow-up. The main question in this regard is whether expression level of circITCH is changed after chemo/radiotherapy or tumor excision. If so, it can be used as a marker for early detection of cancer recurrence.
Another question to be answered is the correlation between expression levels of the circular and linear form of ITCH in different types of cancers. The answer to this question can help in better understanding of the mechanism of dysregulation of circITCH in relation to cancer progression.
Since circITCH is mostly considered as a tumor suppressor circRNA, several groups have assessed the impact of forced over-expression of this transcript in cancer cells transplanted into animal models. The results have been mostly promising, yet needing to be approved in clinical settings.
The interactions between circITCH and RNA-binding proteins have not identified in the previous literature. However, the online database circular RNA Interactome (https://circinteractome.nia.nih.gov/) has listed a number of RNA-binding proteins possibly interacting with circRNAs originated from ITCH locus (Table 4).
Future Perspectives
CircITCH, as a tumor suppressor circRNA can be utilized in therapeutic regimens for cancers. Delivery methods include nanoparticles and exosome-based methods (40). Artificial circRNAs have been successfully used as miRNA sponges in recent years (41). Thus, synthetic circITCH molecules with the potential of sponging oncogenic miRNAs can be used for attenuation of carcinogenic process. Yet, this method should be appraised in cell lines and animal models. Finally, the interactions between circITCH and RNA-binding proteins should be assessed in future investigations.
Author Contributions
SG-F wrote the draft and revised it. MT designed and supervised the study. TK and EJ collected the data and designed the tables and figures. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cao H, Liu Z, Huang P, Yue Y, Xi J. LncRNA-RMRP Promotes Proliferation, Migration and Invasion of Bladder Cancer via MiR-206. Eur Rev Med Pharmacol Sci (2019) 23(3):1012–21. doi: 10.26355/eurrev_201902_16988
2. Yu C-Y, Li T-C, Wu Y-Y, Yeh C-H, Chiang W, Chuang C-Y, et al. The Circular RNA Circbirc6 Participates in the Molecular Circuitry Controlling Human Pluripotency. Nat Commun (2017) 8(1):1–15. doi: 10.1038/s41467-017-01216-w
3. Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J, et al. Circirak3 Sponges MiR-3607 to Facilitate Breast Cancer Metastasis. Cancer Lett (2018) 430:179–92. doi: 10.1016/j.canlet.2018.05.033
4. Cui C, Yang J, Li X, Liu D, Fu L, Wang X. Functions and Mechanisms of Circular Rnas in Cancer Radiotherapy and Chemotherapy Resistance. Mol Cancer (2020) 19(1):1–16. doi: 10.1186/s12943-020-01180-y
5. Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs Function as CeRNAs to Regulate and Control Human Cancer Progression. Mol Cancer (2018) 17(1):1–11. doi: 10.1186/s12943-018-0827-8
6. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 Circular RNA Retards Cell Cycle Progression via Forming Ternary Complexes With P21 and CDK2. Nucleic Acids Res (2016) 44(6):2846–58. doi: 10.1093/nar/gkw027
7. Yin Q, Wyatt CJ, Han T, Smalley KS, Wan L. ITCH as a Potential Therapeutic Target in Human Cancers. In: Seminars in Cancer Biology. Academic Press, Elsevier (2020).
8. Yang C, Yuan W, Yang X, Li P, Wang J, Han J, et al. Circular RNA Circ-ITCH Inhibits Bladder Cancer Progression by Sponging Mir-17/MiR-224 and Regulating P21, PTEN Expression. Mol Cancer (2018) 17(1):1–12. doi: 10.1186/s12943-018-0771-7
9. Tang L, Wang Y, Wang H, Xu B, Ji H, Xu G, et al. Long Noncoding-RNA Component of Mitochondrial RNA Processing Endoribonuclease Is Involved in the Progression of Cholangiocarcinoma by Regulating MicroRNA-217. Cancer Sci (2019) 110(7):2166. doi: 10.1111/cas.14074
10. Li J, Guo R, Liu Q, Sun J, Wang H. Circular RNA Circ-ITCH Inhibits the Malignant Behaviors of Cervical Cancer by MicroRNA-93-5p/FOXK2 Axis. Reprod Sci (2020) 27(3):860–8. doi: 10.1007/s43032-020-00140-7
11. Ren C, Liu J, Zheng B, Yan P, Sun Y, Yue B. The Circular RNA Circ-ITCH Acts as a Tumour Suppressor in Osteosarcoma via Regulating Mir-22. Artif Cells nanomedicine Biotechnol (2019) 47(1):3359–67. doi: 10.1080/21691401.2019.1649273
12. Li H, Lan M, Liao X, Tang Z, Yang C. Circular RNA Cir-ITCH Promotes Osteosarcoma Migration and Invasion Through Cir-ITCH/miR-7/EGFR Pathway. Technol Cancer Res Treat (2020) 19:1533033819898728. doi: 10.1177/1533033819898728
13. Wang M, Chen B, Ru Z, Cong L. Circrna Circ-ITCH Suppresses Papillary Thyroid Cancer Progression Through miR-22-3p/CBL/β-Catenin Pathway. Biochem Biophys Res Commun (2018) 504(1):283–8. doi: 10.1016/j.bbrc.2018.08.175
14. Luo L, Gao Y, Sun X. Circ-ITCH Correlates With Small Tumor Size, Decreased FIGO Stage and Prolonged Overall Survival, and It Inhibits Cells Proliferation While Promotes Cells Apoptosis in Epithelial Ovarian Cancer. Cancer Biomarkers (2018) 23(4):505–13. doi: 10.3233/CBM-181609
15. Lin C, Xu X, Yang Q, Liang L, Qiao S. Circular RNA ITCH Suppresses Proliferation, Invasion, and Glycolysis of Ovarian Cancer Cells by Up-Regulating CDH1 via Sponging Mir-106a. Cancer Cell Int (2020) 20(1):1–13. doi: 10.1186/s12935-020-01420-7
16. Luo L, Gao Y, Sun X. Circular RNA ITCH Suppresses Proliferation and Promotes Apoptosis in Human Epithelial Ovarian Cancer Cells by Sponging Mir-10a-Alpha. Eur Rev Med Pharmacol Sci (2018) 22(23):8119–26. doi: 10.26355/eurrev_201812_16503
17. Hu J, Wang L, Chen J, Gao H, Zhao W, Huang Y, et al. The Circular RNA Circ-ITCH Suppresses Ovarian Carcinoma Progression Through Targeting miR-145/RASA1 Signaling. Biochem Biophys Res Commun (2018) 505(1):222–8. doi: 10.1016/j.bbrc.2018.09.060
18. Yan H, Xiang H, Sun B, Feng F, Chen P. Circular RNA-ITCH Inhibits the Proliferation of Ovarian Carcinoma by Downregulating LncRNA HULC. Reprod Sci (Thousand Oaks Calif). (2020) 27(1):375–9. doi: 10.1007/s43032-019-00049-w
19. Zhao L, Ma N, Liu G, Mao N, Chen F, Li J. Lidocaine Inhibits Hepatocellular Carcinoma Development by Modulating Circ_ITCH/Mir-421/CPEB3 Axis. Digestive Dis Sci (2021) 12:1–14. doi: 10.1007/s10620-020-06787-1
20. Wu M, Deng X, Zhong Y, Hu L, Zhang X, Liang Y, et al. Maff Is Regulated via the Circ-ITCH/MiR-224-5p Axis and Acts as a Tumor Suppressor in Hepatocellular Carcinoma. Oncol Res (2020) 28(3):299. doi: 10.3727/096504020X15796890809840
21. Yang B, Zhao J, Huo T, Zhang M, Wu X. Effects of CircRNA-ITCH on Proliferation and Apoptosis of Hepatocellular Carcinoma Cells Through Inhibiting Wnt/β-Catenin Signaling Pathway. J BUON. (2020) 25(3):1368–74.
22. Chen W, Wu M, Cui S-T, Zheng Y, Liu Z, Luo L-S. CircRNA Circ-ITCH Inhibits the Proliferation and Invasion of Glioma Cells Through Targeting the MiR-106a-5p/SASH1 Axis. Cell Transplant (2021) 30:0963689720983785. doi: 10.1177/0963689720983785
23. Li F, Ma K, Sun M, Shi S. Identification of the Tumor-Suppressive Function of Circular RNA ITCH in Glioma Cells Through Sponging miR-214 and Promoting Linear ITCH Expression. Am J Transl Res (2018) 10(5):1373–86.
24. Hao C, Wangzhou K, Liang Z, Liu C, Wang L, Gong L, et al. Circular RNA ITCH Suppresses Cell Proliferation But Induces Apoptosis in Oral Squamous Cell Carcinoma by Regulating MiR-421/PDCD4 Axis. Cancer Manage Res (2020) 12:5651. doi: 10.2147/CMAR.S258887
25. Wang X, Wang R, Wu Z, Bai P. Circular RNA ITCH Suppressed Prostate Cancer Progression by Increasing HOXB13 Expression via Spongy MiR-17-5p. Cancer Cell Int (2019) 19:328. doi: 10.1186/s12935-019-0994-8
26. Li S, Yu C, Zhang Y, Liu J, Jia Y, Sun F, et al. Circular RNA Cir-ITCH Is a Potential Therapeutic Target for the Treatment of Castration-Resistant Prostate Cancer. BioMed Res Int (2020) 2020:7586521. doi: 10.1155/2020/7586521
27. Yuan Y, Chen X, Huang E. Upregulation of Circular RNA Itchy E3 Ubiquitin Protein Ligase Inhibits Cell Proliferation and Promotes Cell Apoptosis Through Targeting miR-197 in Prostate Cancer. Technol Cancer Res Treat (2019) 18:1533033819886867. doi: 10.1177/1533033819886867
28. Wang Y, Wang H, Zheng R, Wu P, Sun Z, Chen J, et al. Circular RNA ITCH Suppresses Metastasis of Gastric Cancer via Regulating MiR-199a-5p/Klotho Axis. Cell Cycle (2021) 20(5-6):522–36. doi: 10.1080/15384101.2021.1878327
29. Peng Y, Wang HH. Cir-ITCH Inhibits Gastric Cancer Migration, Invasion and Proliferation by Regulating the Wnt/β-Catenin Pathway. Sci Rep (2020) 10(1):1–13. doi: 10.1038/s41598-020-74452-8
30. Lin Q, Jiang H, Lin D. Circular RNA ITCH Downregulates GLUT1 and Suppresses Glucose Uptake in Melanoma to Inhibit Cancer Cell Proliferation. J Dermatol Treat (2021) 32(2):231–5. doi: 10.1080/09546634.2019.1654069
31. Wan L, Zhang L, Fan K, Cheng Z-X, Sun Q-C, Wang J-J. Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/β-Catenin Pathway. BioMed Res Int (2016) 2016. doi: 10.1155/2016/1579490
32. Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has Inhibitory Effect on ESCC by Suppressing the Wnt/β-Catenin Pathway. Oncotarget (2015) 6(8):6001. doi: 10.18632/oncotarget.3469
33. Gao P, Huang Y, Hou Y, Li Q, Wang H. Circular RNA ITCH Is a Tumor Suppressor in Clear Cell Renal Cell Carcinoma Metastasis Through miR-106b-5p/PDCD4 Axis. J Immunol Res (2021) 2021. doi: 10.1155/2021/5524344
34. Liu J, Du F, Chen C, Li D, Chen Y, Xiao X, et al. CircRNA ITCH Increases Bortezomib Sensitivity Through Regulating the Mir-615-3p/PRKCD Axis in Multiple Myeloma. Life Sci (2020) 262:118506. doi: 10.1016/j.lfs.2020.118506
35. Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. Cir-ITCH Plays an Inhibitory Role in Colorectal Cancer by Regulating the Wnt/β-Catenin Pathway. PloS One (2015) 10(6):e0131225. doi: 10.1371/journal.pone.0131225
36. Sun M-x, An Q, Chen L-m, Guo L. MiR-520f Regulated Itch Expression and Promoted Cell Proliferation in Human Melanoma Cells. Dose-Response (2020) 18(2):1559325820918450. doi: 10.1177/1559325820918450
37. Guo W, Zhang J, Zhang D, Cao S, Li G, Zhang S, et al. Polymorphisms and Expression Pattern of Circular RNA Circ-ITCH Contributes to the Carcinogenesis of Hepatocellular Carcinoma. Oncotarget (2017) 8(29):48169. doi: 10.18632/oncotarget.18327
38. Huang E, Chen X, Yuan Y. Downregulated Circular RNA Itchy E3 Ubiquitin Protein Ligase Correlates With Advanced Pathologic T Stage, High Lymph Node Metastasis Risk and Poor Survivals in Prostate Cancer Patients. Cancer Biomarkers (2019) 26(1):41–50. doi: 10.3233/CBM-182111
39. Ghasemi S, Emadi-Baygi M, Nikpour P. Down-Regulation of Circular RNAITCH and Circhipk3 in Gastric Cancer Tissues. Turkish J Med Sci (2019) 49(2):687–95. doi: 10.3906/sag-1806-50
40. He AT, Liu J, Li F, Yang BB. Targeting Circular Rnas as a Therapeutic Approach: Current Strategies and Challenges. Signal Transduction Targeted Ther (2021) 6(1):1–14. doi: 10.1038/s41392-021-00569-5
Keywords: circular RNA, circITCH, cancer, expression, ncRNAs
Citation: Ghafouri-Fard S, Khoshbakht T, Taheri M and Jamali E (2021) CircITCH: A Circular RNA With Eminent Roles in the Carcinogenesis. Front. Oncol. 11:774979. doi: 10.3389/fonc.2021.774979
Received: 13 September 2021; Accepted: 30 September 2021;
Published: 15 October 2021.
Edited by:
Peixin Dong, Hokkaido University, JapanReviewed by:
Youliang Wang, Beijing Institute of Technology, ChinaFukang Sun, Shanghai Jiao Tong University, China
Copyright © 2021 Ghafouri-Fard, Khoshbakht, Taheri and Jamali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Elena Jamali, ZWxlbmEuamFtYWxpQHlhaG9vLmNvbQ==
 Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Tayyebeh Khoshbakht
Tayyebeh Khoshbakht Mohammad Taheri
Mohammad Taheri Elena Jamali4*
Elena Jamali4*