
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 18 January 2022
Sec. Skin Cancer
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.770325
 Zhanghui Yue1
Zhanghui Yue1 Meng Cao1
Meng Cao1 Anlan Hong1
Anlan Hong1 Qian Zhang1
Qian Zhang1 Guoqiang Zhang2
Guoqiang Zhang2 Zhibin Jin3
Zhibin Jin3 Liang Zhao1
Liang Zhao1 Qiang Wang1
Qiang Wang1 Fang Fang1
Fang Fang1 Yan Wang1*
Yan Wang1* Jianfang Sun1*
Jianfang Sun1*m6A modification is one of the most important post-transcriptional modifications in RNA and plays an important role in promoting translation or decay of RNAs. The role of m6A modifications has been highlighted by increasing evidence in various cancers, which, however, is rarely explored in acral melanoma. Here, we demonstrated that m6A level was highly elevated in acral melanoma tissues, along with the expression of METTL3, one of the most important m6A methyltransferase. Besides, higher expression of METTL3 messenger RNA (mRNA) correlated with a higher stage in primary acral melanoma patients. Knockdown of METTL3 decreased global m6A level in melanoma cells. Furthermore, METTL3 knockdown suppressed the proliferation, migration, and invasion of melanoma cells. In METTL3 knockdown xenograft mouse models, we observed decreased volumes and weights of melanoma tissues. Mechanistically, we found that METTL3 regulates certain m6A-methylated transcripts, thioredoxin domain containing protein 5 (TXNDC5), with the confirmation of RNA-seq, MeRIP-seq, and Western blot. These data suggest that METTL3 may play a key role in the progression of acral melanoma, and targeting the m6A dependent-METTL3 signaling pathway may serve as a promising therapeutic strategy for management of patients of acral melanomas.
Melanoma represents the most invasive skin cancer type and comprises the most skin cancer-related deaths (1). It has distinct clinicopathological subtypes with biological heterogeneity (2). Acral melanoma (AM), defined as melanoma arising from the glabrous skin of palms, soles, and nail beds, is a rare subtype in Caucasians but accounting for more than 70% in Asian population (3). The previous study revealed that the progression of AM has distinct clinical and genetic features, potentially due to diagnosis at a more advanced clinical stage, or due to biological differences favoring tumor aggression, AM usually has poorer prognosis than other types of cutaneous melanoma (4–6). In recent years, targeted therapies and checkpoint immunotherapies have greatly improved the prognosis of advanced melanomas (7, 8). However, more than half of Asian patients are unable to benefit from immunotherapy and targeted therapy (9), which might attribute to lower tumor mutation burden, programmed-death 1 ligand low expression, phenotype plasticity, etc. (10–13). Up to date, most AM patients do not have successful genotype-specific treatment options. Consequently, determining the molecular mechanisms underlying AM progression is critical for improving therapy and diagnosis. The multistep process involves complicated interplays between epigenetics and transcriptomic alterations results in the AM progression. From a clinical perspective, trauma (14), mechanical stress (15, 16), and some other epigenetic stimulus seemed to be associated with the risk of AM develop. Meanwhile, increasing evidence has revealed that AM has greater prevalence of epigenetic alternation than genetic changes (17–19). Hence, targeting epigenetic regulation represents a novel approach for AM treatment.
N6-methyladenosine (m6A), the most prevalent eukaryotic RNA modification, recently emerged as a major post-transcriptional modulator of gene expression (20). It influences the multifaceted process of RNA metabolism, including splicing, stability, and translation efficiency. Dynamic m6A modification participates in diverse important life processes (21). Accumulating evidence in recent years reveals that aberrant m6A RNA modifications levels and dysregulation of m6A regulators underlie various human cancers, including glioblastoma (22), hepatocellular carcinoma (23), cervical cancer (24), leukemia (25), and breast cancer (26). M6A occurs primarily in the consensus RRACH motif (R ¼ GorA; H ¼ A, C, or U) and is dynamically regulated by the “writer,” “eraser,” and “reader” proteins (27, 28). Writers mainly belong to the m6A-METTL-associated complex and include METTL3, METTL14, WTAP, VIRMA, etc. The demethylases FTO and ALKBH5 function as erasers, removing N‐methyl from adenosine (29). The m6A sites are specifically recognized by “Readers,” including YTH family, elF3, and IGF2BP family proteins (30). METTL3 is the core catalytic component and cooperates with other proteins to methylate adenosine. The onset and progression of various cancers are affected by METTL3, either dependent or independent on its m6A RNA methyltransferase activity (31–33). METTL3 principally functions as an oncogene to promote carcinogenesis in various cancers: METTL3 depletion induced cell differentiation and apoptosis to delay acute myeloid leukemia progression by regulating m6A modification on MYC, BCL2, and PTEN mRNAs (25); high expression of METTL3 correlates with poor clinical outcomes in glioblastoma, and the m6A modification on SRSF transcript added by METTL3 leads to human glioblastoma outgrowth and self-renewal (34). In bladder cancer, METTL3 was upregulated in patient samples, and it benefits cancer progression by mediating several m6A modification targets (CDCP1, ITGA6, AFF4, IKBKB, RELA, and MYC), in which YTH family served as the binding protein of the target transcripts (35–37). METTL3 also exhibits tumor suppressor function in some cases: lower expression of METTL3 was detected in renal cell carcinoma patients and predicts a favorable prognosis (38). Nevertheless, not much is known about the biological and pathological role of METTL3-dependent m6A modification in acral melanoma.
In this study, we found that the imbalance of global m6A abundance and dysregulated expressions of m6A regulators were frequent in primary acral melanoma patients. The high expression of METTL3 was positively related to advanced clinical stage in primary acral melanoma. We then confirmed that METTL3 functions as an oncoprotein in primary acral melanoma by both in vitro and in vivo experiments. Further molecular mechanism investigation indicated that TXNDC5 was downregulated by decreasing m6A-levels after METTL3 knockdown and ultimately inhibiting the progression of primary acral melanoma. Thus, our findings extend the understanding of m6A-driven machinery in acral melanoma progression and proposed that the METTL3-m6A-TXNDC5 axis may serve as biomarker and therapeutic strategy in the clinic.
The normal human epidermal melanocyte (HEMa) was extracted from fresh foreskin tissue donated after circumcision in adults and cultured in melanocyte medium with 5% fetal bovine serum (Sciencell, USA). A875, MV3 cell lines were kindly supplied by Prof. Xiulian Xu, who worked in our institute. HMY-1 cell line was a gift from Prof. Yan Kong who worked in Peking University Cancer Hospital and Institute. A375, SK-MEL-28, and A2058 were obtained from the Cell Bank of Type Culture Collection (Guangzhou Cellcook Biotech Co., Ltd., Guangzhou, China). A375 and SK-MEL-28 were derived from cutaneous melanoma, A875 was derived from melanoma brain metastasis tissue, A2058 and MV3 were derived from metastatic lymph node, and HMY-1 was derived from acral melanoma. Except for HMY-1 that originate from Asians, the other cell lines originate from Caucasians. Human melanoma cell lines were cultured in Dulbecco’s modified Eagle’s medium (A375, A875, A2058, MV3, and HMY-1) or minimal essential medium (SK-MEL-28) supplemented with 10% fetal bovine serum (BioInd, Beit HaEmek, Israel) and penicillin-streptomycin solution (Gibco, Grand Island, NY, USA) at 37°C in a 5% CO2 atmosphere. All cell lines were routinely tested for mycoplasma contamination and identified by sequence tandem repeat profiling. Short hairpin RNA (shRNA) sequences were designed by Hanbio Biotechnology Co., Ltd. (Shanghai, China) to target human METTL3. After annealing, double strands of short hairpin RNA (shRNA) were inserted into the lentiviral pHBLV-U6-MCS-CMV-ZsGreen-PGK-PURO vector (Hanbio), named shMETTL3, and the non-targeted control was named NTC. Stable cell lines were established after selection with 1 µg/ml puromycin (Millipore, Billerica, MA, USA). The target gene knockdown efficiency was evaluated via quantitative RT-PCR (qRT-PCR) and Western blot analysis.
This study was approved by the Ethics Committee of the Institute of Dermatology at the Chinese Academy of Medical Sciences and carried out according to the Declaration of Helsinki. Thirty-four pairs of fresh melanoma tumor tissues and adjacent normal specimens were obtained from patients who underwent surgical resection without any preoperative treatment between September 2017 and July 2019. All samples were confirmed by pathological examination according to the American Joint Committee on Cancer (AJCC) Cancer Staging Manual (8th edition). The clinicopathological features of all patients are summarized in Table 1. Written informed consent for the biological studies was obtained from each patient.
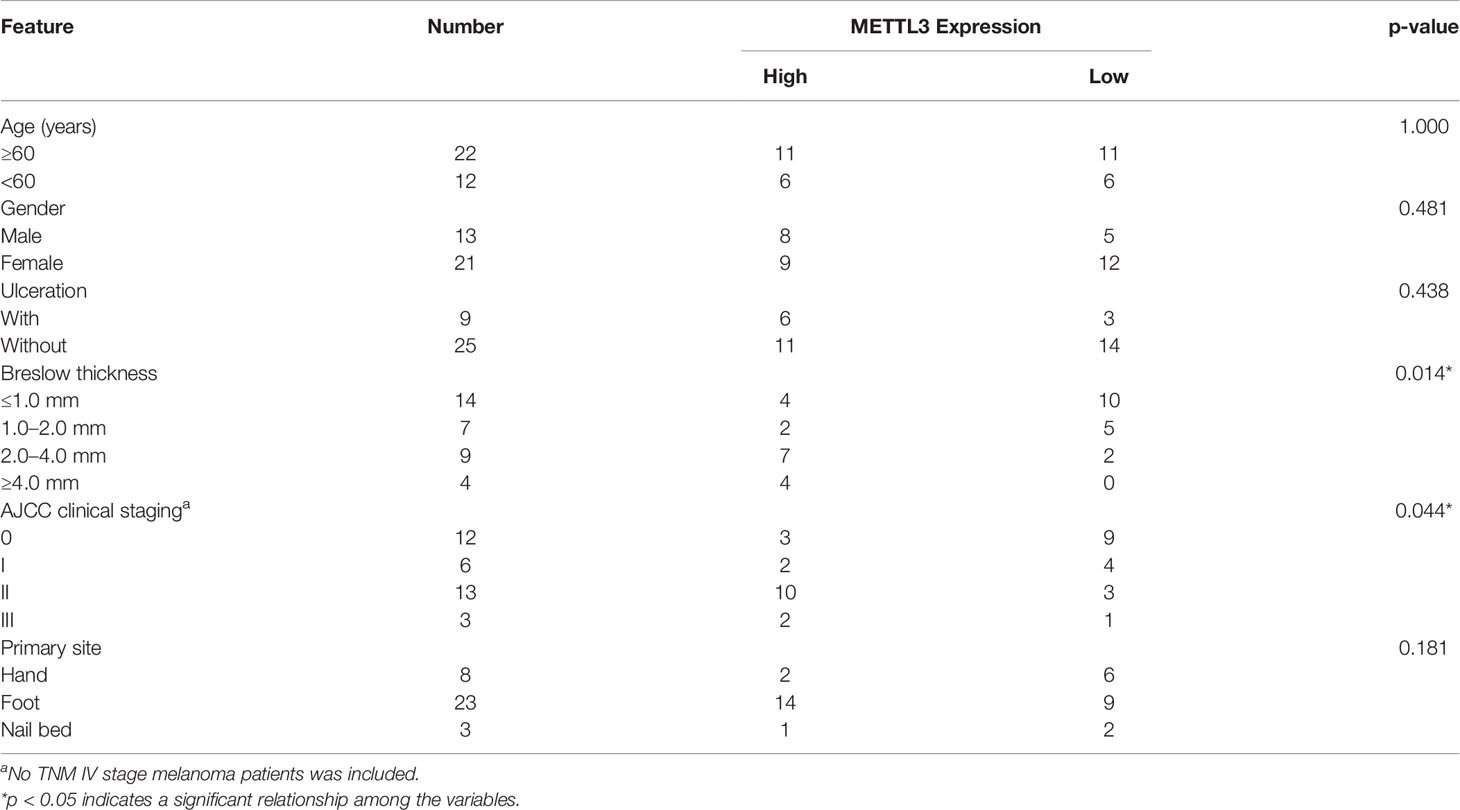
Table 1 Correlation between METTL3 expression and clinicopathologic features of acral melanoma patients (n = 34).
Cells and tissues were lysed using 1× radioimmunoprecipitation assay (RIPA) buffer (9806, Cell Signaling Technology, Danvers, MA, USA) containing protease inhibitors (Complete; Roche, Basel, Switzerland) and phosphatase inhibitors (PhosSTOP; Roche). Equal amounts of proteins were loaded and separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene fluoride membranes (Immobile P; Millipore), and detected by immunoblotting with enhanced chemiluminescence (Chemidoc XRS, Bio-Rad, Hercules, CA, USA). Antibodies used for Western blotting were as follows unless otherwise specified: METTL3 (Abcam, Cambridge, UK; ab195352, 1:1,000) and TXNDC5 (Proteintech, China, 19834-1-AP, 1:10,000); glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Abcam, ab181602, 1:10,000) was used as a loading control.
A total of 2 × 104 cells/well were seeded into a six-well plate. Upon cells reaching 90% confluence, a wound-like gap was made in the cell monolayer using a 200-µl pipette tip. Cells were washed with phosphate-buffered saline three times and incubated for the indicated time. The migration distance of cells at 0, 24, and 48 h was photographed and analyzed using ImageJ software (NIH, Bethesda, MD, USA).
Cell migration and invasion assays were carried out in a 24-well Transwell chamber (Corning, NY, USA) inserts with an 8-μm pore polycarbonate filter. For the migration assays, approximately 5 × 104 cells were suspended in 200 μl serum-free Dulbecco’s modified Eagle’s medium and added to the upper chamber. The lower chamber contained 600 μl complete culture medium with 10% fetal bovine serum. After 24–48 h, the cells were fixed in 4% paraformaldehyde, followed by staining with crystal violet. Cells on the upper surface of the membrane were removed with a cotton swab. Five fields per chamber were selected and photographed randomly using an inverted microscope (Nikon, Tokyo, Japan). For the invasion assay, 100 μl diluted Matrigel (Corning, NY, USA) was dispensed into the upper chamber before seeding the cells, and the remaining process was performed similarly in the migration assay. ImageJ software was used to count the migrated and invaded cells.
We performed qRT-PCR to measure the gene expression levels in AM tissues and cell lines. Total RNA from different samples was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The purity and concentration of RNA were determined using a NanoDrop 2000 spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA samples (500–1,000 ng) were subjected to RT-PCR using the TaKaRa RT-PCR kit (Takara, Shiga, Japan). Expression levels were quantified by qPCR in a LightCycler ® 480 Instrument II device (Roche Applied Science, Mannheim, Germany) using SYBR Master Mix (without ROX Vazyme, Nanjing, China). All primers were provided by General Biotech Co., Ltd. (Shanghai, China). Target gene expression was normalized to the housekeeping gene GAPDH using the ΔCt method, with a relative expression equal to 2−ΔCt. All sequences of the primers used in our study were obtained from primerbank (https://pga.mgh.harvard.edu/primerbank/), and the specificity of the primers were verified by the primer blast function of NCBI. Primers sequences used in this study are listed in Supplementary Table S1.
Tissues were previously fixed in 10% formalin, paraffin embedded, and cut into 4-µm sections. The slides were subjected to dewaxing, rehydration, antigen retrieval, and blocking and then incubated with primary antibody [anti-METTL3 (1:1,000, Abcam) and anti-Ki-67 (1:400, Abcam)] overnight at 4°C. On the following day, after washing with phosphate-buffered saline, the slides were further incubated for 1 h with the secondary antibody at room temperature. Diaminobenzidine and hematoxylin were used to visualize the reaction and counterstain the slide.
For the cell proliferation analysis, 2 × 104 cells were seeded into 12-well plates at 72 h after transfection; green fluorescent protein (GFP)-positive cells were counted on four consecutive days using a countess automated cell counter (Invitrogen). Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) was used to determine the effect of METTL3 on cell viability; the transfected cells were cultured in the 96‐well plate at a density of 4 × 103 cells/well for 4 days. CCK-8 dye solution was added to each well at a certain time point. Following 2 h of incubation at 37°C, the absorbance was measured on a microplate reader (Thermo Fisher Scientific) at 450 nm.
The colony-formation assay was performed using a six-well plate. A total of 600 cells/well transfected cells were incubated for 2 weeks. The colonies were fixed in 4% paraformaldehyde and stained with 1% crystal violet for 30 min.
5-Ethynyl-20-deoxyuridine (EdU) Cell Proliferation Kit with Alexa Fluor 555 (Epizyme, Shanghai, China) was used to detect DNA synthesis and cell proliferation. Cells were incubated at 37°C with Dulbecco’s modified Eagle’s medium (DMEM) containing 10 μM EdU for 2 h. After being fixed with 4% paraformaldehyde, these cells were subsequently stained with Azide 555 Dye Solution and Hoechst 33342. An inverted fluorescent microscope was used to capture three randomly selected fields to visualize the EdU-stained cells.
Total RNA isolation was performed to measure global changes in the m6A modification level. The content of m6A was determined using an EpiQuik m6A RNA Methylation Quantification Kit (cat. P-9005, EpiGentek, Farmingdale, NY, USA) following the manufacturer’s protocol. Briefly, negative and diluted positive control RNA, and 200 ng total RNA, were used to analyze each sample.
The dot blot assay was conducted according to the bio-protocol database (https://en.bio-protocol.org/e2095). Briefly, the indicated amount of total RNA was denatured and spotted onto a Hybond-N+ membrane (GE Healthcare, Little Chalfont, UK) and crosslinked using a UV crosslinker. The membrane was first stained with methylene blue (MB) for 2 h and then washed with RNase-free water for 1 h. Images were acquired using an Epiwhite light as the loading control. After MB was washed with RNase-free water, the membrane was incubated in 5% bovine serum albumin/Tris-buffered saline containing 1% Tween-20 as the blocking buffer and then with an anti-m6A antibody (1:1,000; Abcam) overnight at 4°C. Thereafter, the membrane was washed, incubated with secondary antibody, and washed again. Finally, the membrane was exposed to Hyperfilm ECL, and images were acquired. The m6A signal was quantified by TotalLab software (Amersham Pharmacia Biotech, Amersham, UK) and normalized to MB levels.
Sixteen BALB/c nude mice (male, 6-week-old) were raised under pathogen-free conditions and randomly divided into two groups. A total of 2 × 106 A375 NTC or sh-METTL3#2 cells were subcutaneously inoculated into the right hind flank. Body weight and tumor size were measured every other day. The tumors were harvested at the end of the observation period, and tumor weight and gross images were recorded (11 days after inoculation). The tumors were embedded in RNA later and 10% formalin for further detection. Animals were treated humanely, and all procedures complied with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals or equivalent guidelines and approved by the Institutional Animal Care and Use Committee of our hospital.
The RNA amount and purity of each sample were quantified using a NanoDrop ND-1000 (Wilmington, DE, USA). RNA integrity was assessed with a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) with RNA integrity number >7.0 and confirmed by electrophoresis in a denaturing agarose gel. Approximately 25 μg of total RNA representing a specific adipose type was used to deplete ribosomal RNA according to the manuscript of the Epicentre Ribo-Zero Gold Kit (Illumina, San Diego, CA, USA). Following purification, the ribosomal-depleted RNA was fragmented into small pieces using the Magnesium RNA Fragmentation Module (cat. e6150, New England Biolabs, Ipswich, MA, USA) at 86°C for 7 min. The cleaved RNA fragments were incubated for 2 h at 4°C with an m6A-specific antibody (No. 202003, Synaptic Systems, Goettingen, Germany) in IP buffer (50 mM Tris–HCl, 750 mM NaCl, and 0.5% Igepal CA-630). The immunoprecipitated RNA was reverse-transcribed to prepare the complementary DNA (cDNA) using SuperScript™ II Reverse Transcriptase (cat. 1896649, Invitrogen), which was used to synthesize U-labeled second-stranded DNAs with Escherichia coli DNA polymerase I (cat. m0209, New England Biolabs), RNase H (cat. m0297, New England Biolabs), and dUTP solution (cat. R0133, Thermo Fisher Scientific). An A-base was then added to the blunt ends of each strand to prepare them for ligation to the indexed adapters. Each adapter contained a T-base overhang for ligating the adapter to the A-tailed fragmented DNA. Single- or dual-index adapters were ligated to the fragments, and size selection was performed with AMPureXP beads. After treatment with the heat-labile UDG enzyme (cat. m0280, New England Biolabs) of the U-labeled second-stranded DNAs, the ligated products were amplified with PCR under the following conditions: initial denaturation at 95°C for 3 min; 8 cycles of denaturation at 98°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 30 s; with a final extension at 72°C for 5 min. The average insert size of the final cDNA library was 300 ± 50 bp. Finally, 2 × 150-bp paired-end sequencing (PE150) was performed on an Illumina Novaseq™ 6000 (LC-Bio Technology Co., Ltd., Hangzhou, China) following the vendor’s recommended protocol.
Bioinformatic analysis including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment was performed using OmicStudio tools (https://www.omicstudio.cn/tool). Transcript expression levels were dichotomized based on the median expression, and a false discovery rate <0.05 indicated statistical significance. All statistical analyses were performed using SPSS version 22.0 software (SPSS, Inc., Chicago IL, USA). The relationship between METTL3 expression and clinicopathological patient characteristics was analyzed using Pearson χ2 test or Fisher exact test. Individual groups were compared by two-tailed Student’s t-test, and one-way analysis of variance was used for multiple groups. Pearson correlation analysis was used to estimate the relationship between the expression level of METTL3 and TXNDC5. Data are presented as the mean ± SD from at least three biological replicates, with *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0. 0001 indicating the significance levels.
Given that the imbalance of m6A modification exists in various cancers, the m6A level in melanoma remains uncertain. To determine the potential involvement of m6A modifications in melanoma, we first examined m6A levels in 32 pairs of melanoma tumor and adjacent normal tissues. The m6A RNA Methylation Quantification assay showed that m6A modification levels were remarkably elevated in acral melanoma tumor tissues compared with the adjacent normal tissues (Figure 1A, p < 0.05), suggesting that m6A alternations play an indispensable role in the carcinogenesis of melanoma. These results were confirmed in the dot blot assay (Figure 1B).
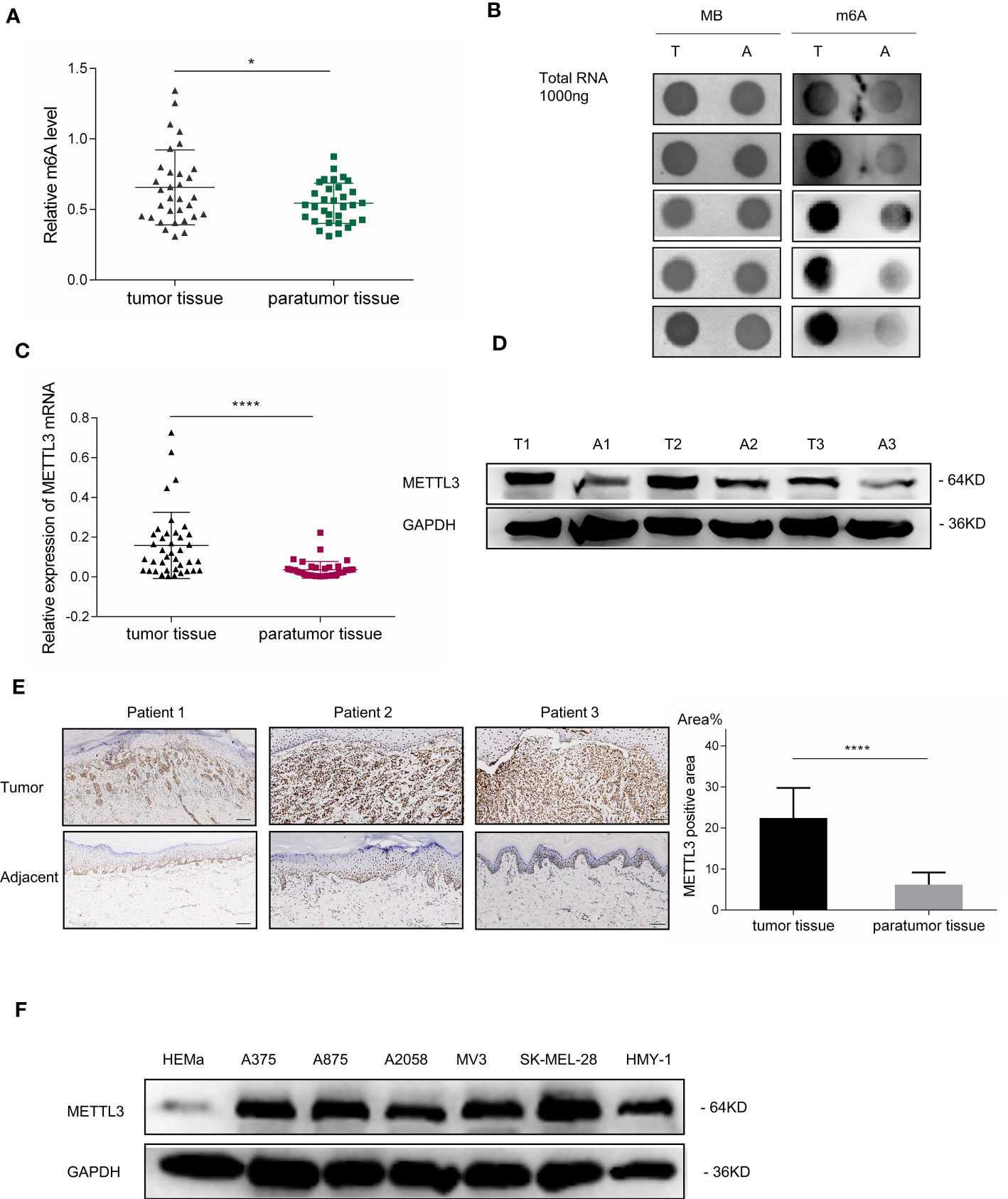
Figure 1 m6A RNA methylation level and METTL3 expression in patients with melanoma. (A) Global m6A quantification of total RNA from 32 melanoma specimens demonstrated the elevation of m6A levels in tumor tissues; scatterplot displays statistical results. Bar charts show the mean ± SD of each group (n = 32). (B) Dot blot of total RNA extract from five pairs of melanoma tumor tissues and adjacent normal tissues stained with an anti-m6A antibody. Methylene blue represents loading control of RNA samples. (C) METTL3 expression in melanoma tumor and adjacent normal tissues determined via qRT- PCR. Bar charts show the mean ± SD of each group (n = 34). (D) Western blotting confirmed METTL3 upregulation in melanoma. GAPDH served as a loading control. (E) IHC revealed masses of METTL3-positive cells in AM tumor tissues. Relatively negative staining was observed in adjacent normal tissues. Bar = 100 μm. Bar chart shows the mean ± SD of each group (n = 12). (F) Western blot analysis indicated that METTL3 protein levels of HEMa were lower in melanoma cells. Data are presented as the mean ± SD from at least three biological replicates, with *p < 0.05, and ****p < 0.0001 indicating the significance levels.
The RNA m6A modification is dynamically modulated by both methyltransferases and demethyltransferases. We hypothesized that the upregulation of m6A modification was caused by the dysregulation of m6A-related genes in melanoma, the expressions of which were analyzed by RT-PCR in 34 pairs of melanoma tumor and adjacent normal tissues. Results showed that the mRNA expression of METTL3, the core m6A methyltransferase, was significantly upregulated (Figure 1C, p < 0.0001), which may be responsible for the upregulation of global m6A levels in melanoma (Table 1).
To further examine METTL3 expression in melanoma, the baseline characteristics of 34 patients with melanoma were considered and are summarized in Table 1, and it was found that the high expression of METTL3 mRNA was related to advanced clinical stages and reflected the clinical significance of METTL3 in melanoma. The increased METTL3 protein levels in tumor tissue were detected by Western blotting (Figure 1D). Immunohistochemistry (IHC) also determined the upregulated protein expression of METTL3 in 12/12 (100%) melanoma tissue samples relative to the corresponding adjacent normal tissues (Figure 1E). In melanoma cell lines, METTL3 was significantly highly expressed at both the mRNA and protein level compared to that in normal human epidermal melanocytes (HEMa) (Supplementary Figure S1A and Figure 1F).
To explore the function of METTL3 in melanoma, we silenced the expression of METTL3 in A375 and A875 cells with shRNA transfection, which reduced METTL3 expression by approximately 90% relative to that in non-targeted control (NTC) cells, as confirmed by qRT-PCR and Western blotting (Figure 2A). Then, global m6A quantification and m6A dot blot assays were performed and demonstrated that m6A modification level was downregulated heavily by METTL3 silencing in A375 and A875 cells (Figures 2B, C).
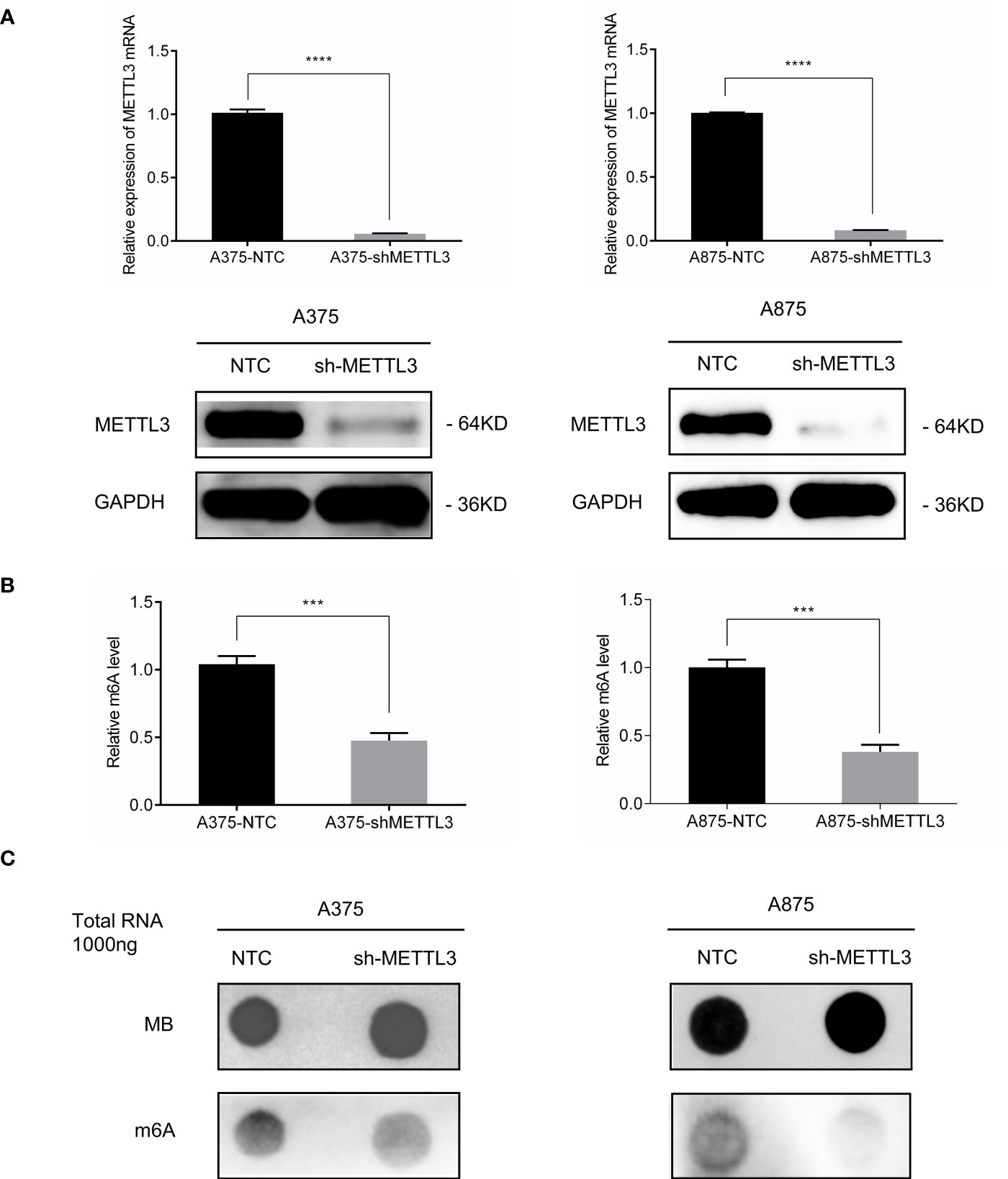
Figure 2 METTL3 silencing decreased global m6A level in melanoma cell lines. (A) METTL3 mRNA and protein levels showing knockdown efficiency in A375 and A875; bar chart showing data from three independent experiments; mean ± SD of each group. (B) m6A modification levels were decreased both in A375-shMETTL3 and A875-shMETTL3 as determined via colorimetric ELISA. (C) Dot blots showing relative total RNA m6A (bottom) and loading control (top). Data are presented as the mean ± SD. ***p < 0.001,****p < 0.0001.
We next assessed the changes in cellular behaviors in A375 and A875 cells caused by METTL3 knockdown. Cell counting, CCK-8 assays were carried out and indicated that the knockdown of METTL3 expression significantly inhibited melanoma cell proliferation (Figures 3A, B and Supplementary Figure S2A). In EdU assays, the number of EdU-positive cells exhibited the decreased tendency in A375/A875 shMETTL3 groups compared to that of NTC groups (Figure 3C), and the ratio of EdU-positive cells is shown in Supplementary Figure S2. Moreover, a similar trend was noted when carrying out in colony formation assays. For A375 and A875 cells, the colony forming rates of NTC cells were 3.9- and 2.7-fold higher, respectively, than those of shMETTL3 cells (Figure 3D), which meant that METTL3 knockdown inhibited melanoma cell proliferation and colony formation ability in vitro. As high expression of METTL3 correlated with advanced stage of melanoma, which was revealed above, we speculated that the upregulation of METTL3 might be involved in migration and invasion of melanoma cells. We therefore compared the migration and invasion abilities of NTC and shMETTL3 cells. Wound healing assays and Transwell analysis indicated that METTL3 knockdown significantly impaired the abilities of migration and invasion in melanoma cells. (Figures 3E–G and Supplementary Figure S2C).
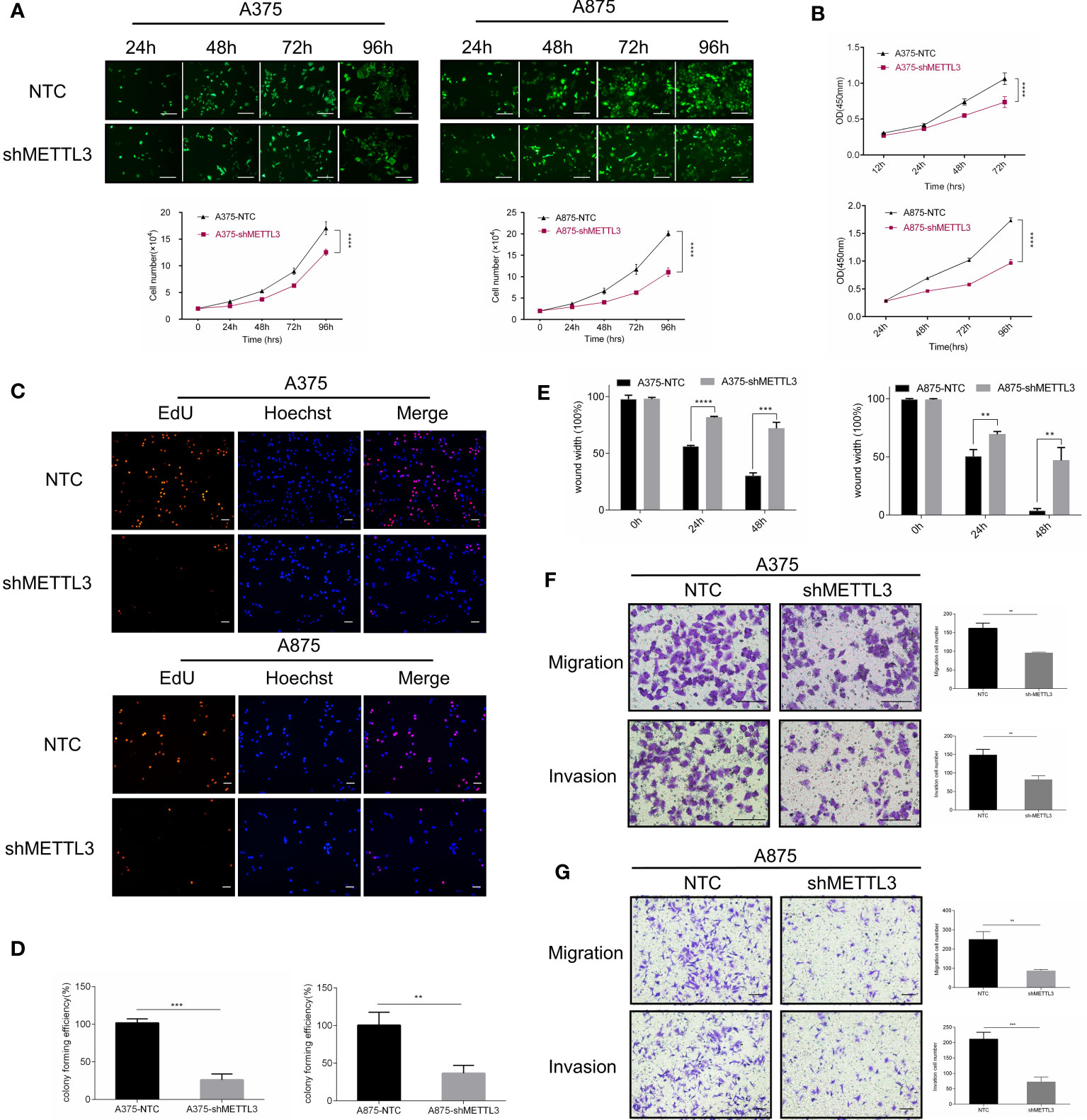
Figure 3 METTL3 knockdown inhibits melanoma cell proliferation, migration, and invasion in vitro. (A) Continuous cell count of A375 and A875 transfected cells with GFP reflected cell proliferation ability. (B) Cell proliferation assay by CCK8 method. (C) EdU cell proliferation, EdU (red), Hoechst (blue), scale bar = 100 μm. (D) Quantification of the results from colony-formation assay. (E) The statistical histogram of wound-healing assays in A375 and A875 transfected cells. Transwell assays with or without Matrigel indicated suppressed migration and invasion of (F) A375 and (G) A875 cells following METTL3 knockdown. Bar = 100 μm. Mean ± SD of each group (n = 3). Significant differences were evaluated using two-way or one-way ANOVA. **P < 0.01, ***P < 0.001, ****P < 0.0001.
To confirm the functional role of METTL3 in vivo, A375 shMETTL3 or NTC cells were subcutaneously inoculated into the right flank of nude mice, respectively (Figure 4A). Eleven days later, the xenograft tumors were collected to measure the tumor volume and weight, which were considerably lower in the shMETTL3 group than the in NTC group, as Figures 4B–D show. IHC was performed subsequently and showed that the expression of Ki-67, the proliferation marker in melanoma, decreased correspondingly in the sh-METTL3 group (Figure 4F). Then, RNA was extracted from xenograft tumors, and global m6A levels were detected, which were significantly lower as well in the sh-METTL3 group than in the NTC group (Figure 4E).
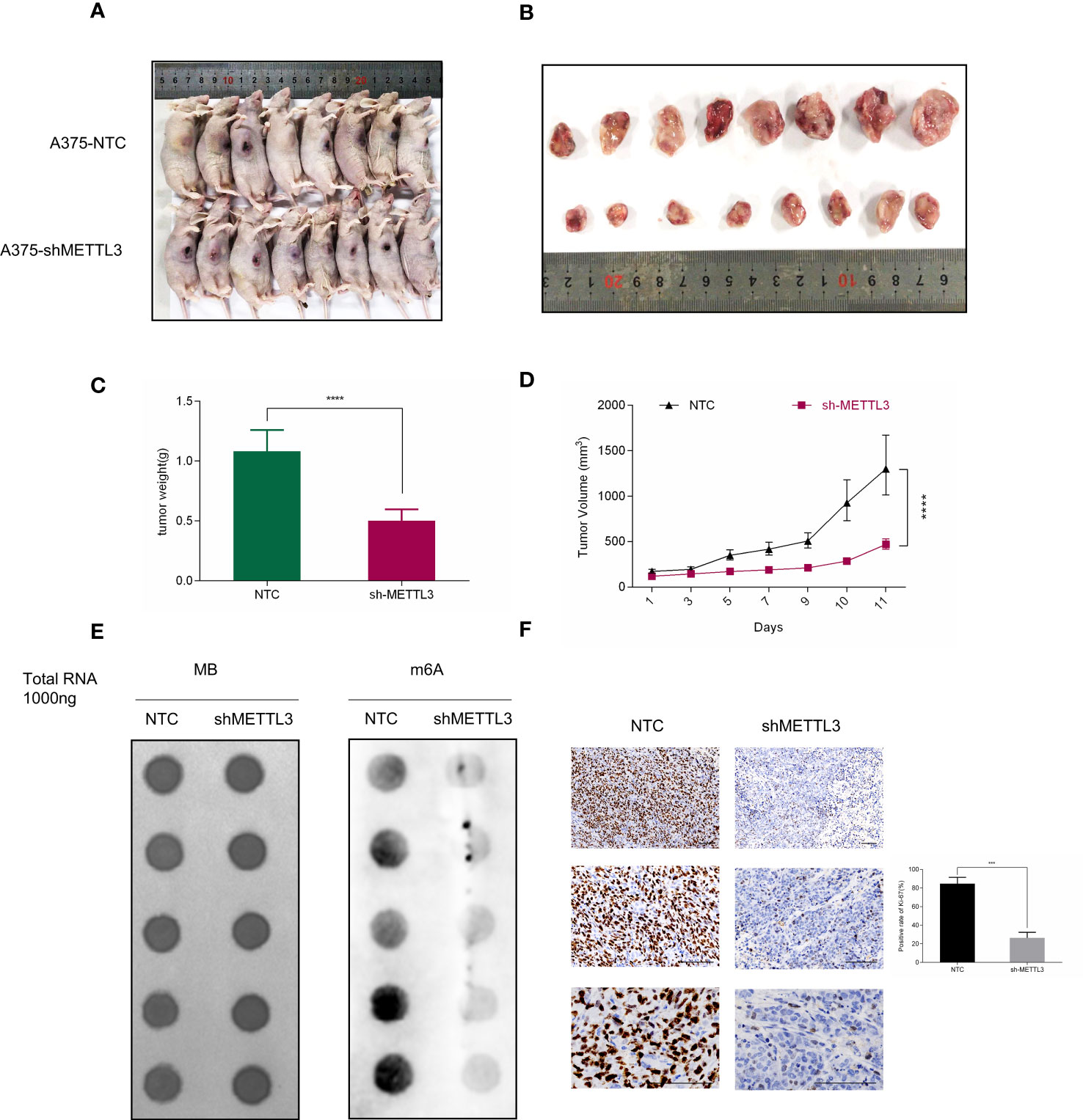
Figure 4 METTL3 depletion delays tumor growth in nude mice. (A, B) Representative photographs and tumor size are shown at 11 days after inoculation. (C) Xenografts derived from A375-shMETTL3 cells demonstrated (C) tumor weight and (D) volume that were much lower than their A375 NTC cell-derived counterparts. (E) m6A level of METLL3 knockdown group xenografts was much lower than in control tumors. (F) Representative Ki67 staining of xenograft tumor section and quantification of Ki67-positive cells are shown. Bar = 100 μm. Data are presented as the mean ± SD (n = 5). Significant differences were evaluated using Student t-test. ***P < 0.001, ****P < 0.0001.
To determine the regulatory role of METTL3 in gene expression, we performed RNA-seq in A375 shMETTL3 and NTC cells. A total of 33,013 genes transcripts were identified, and the expression of these transcripts was quantified. Upon comparison with NTC cells, 232 significantly differentially expressed genes (DEGs) were identified in METTL3-depleted A375 cells (sh) (Figure 5A). Nearly 72% (167 of 232) of these genes were downregulated regardless of m6A modifications (fold-change ≥2 and p < 0.05) (Figure 5B). Furthermore, Gene Ontology (GO) analysis revealed that DEGs involved in the immune response, cell adhesion, and inflammatory response were significantly upregulated after METTL3 depletion (Figure 5C). Genes downregulated in sh-METTL3 cells were related to GO terms associated with cell cycle and negative regulation of apoptotic process (Figure 5E). KEGG pathway enrichment associated upregulated genes with the cytokine receptor interaction, NOD-like receptor, P53, and nuclear factor kappa B (NF-κB) signaling pathway, which were closely related to the tumor micro-environment, cell cycle, and growth. Pathways enriched for downregulated genes included the cGMP-PKG signaling pathway, insulin pathway, natural killer cell-mediated cytotoxicity, and HSV-1 infection (Figures 5D, F). Taken together, METTL3 exhibited cancer-promoting effects based on transcriptional regulation in melanoma.
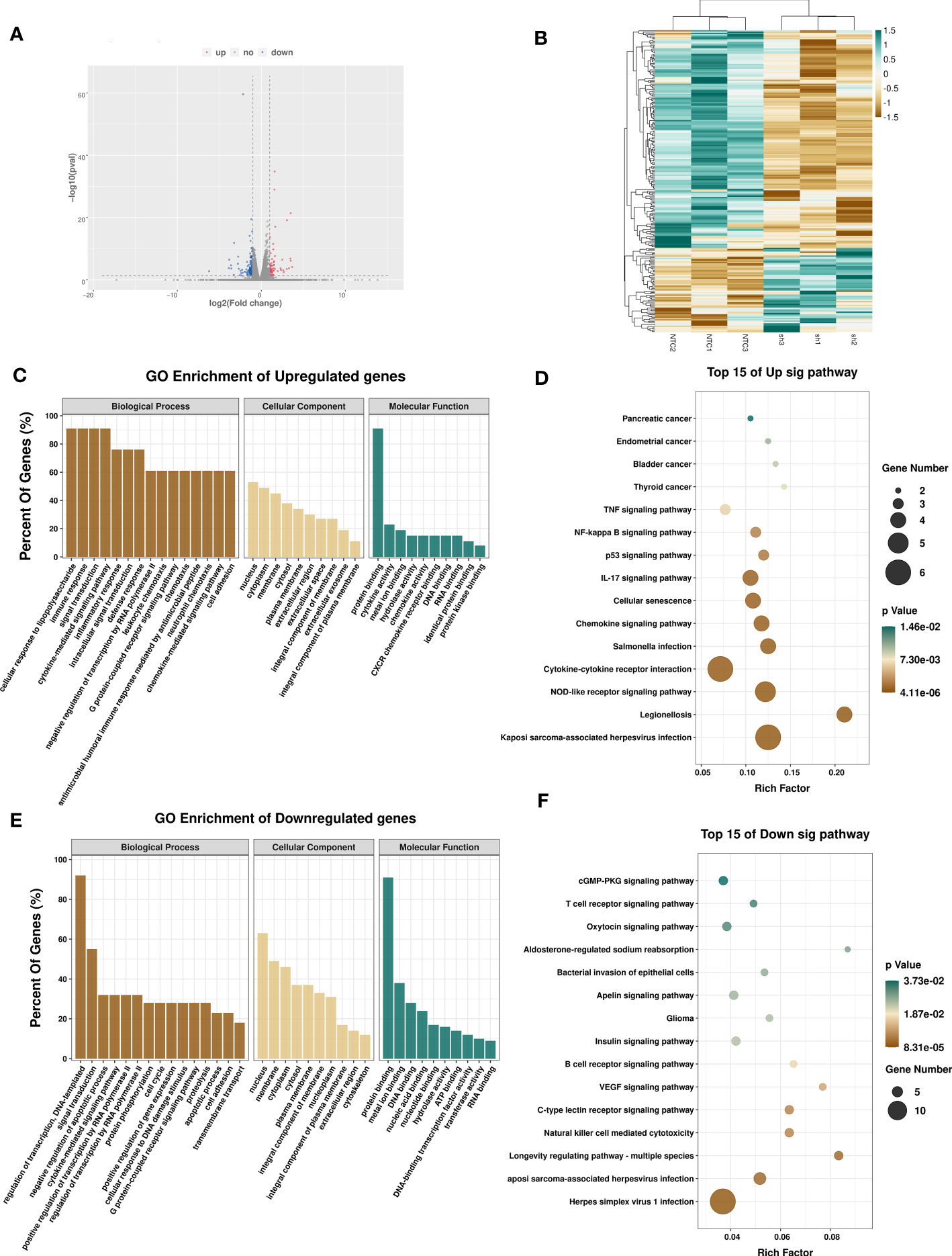
Figure 5 Effects of METTL3 on transcription profile of melanoma cells. (A) Volcano plot showing significantly upregulated (red) and downregulated (blue) genes between A375-NTC (NTC) and A375-shMETTL3(sh) groups. (B) Heatmap showing differentially expressed transcripts between NTC and sh groups. GO and KEGG analyses of significantly upregulated (C, D) and downregulated (E, F) genes identified via RNA-seq suggesting the involvement of DEGs in signal transduction.
As the core methyltransferase, METTL3 regulates m6A modification, as suggested by the decrease in m6A-modified RNA under METTL3 depletion. To clarify this observation, we performed methylated RNA immunoprecipitation sequencing (MeRIP-seq) to map the m6A modification in sh-METTL3 and NTC cells (Figure 6A). Raw sequencing reads were processed to discard adaptor sequences and low-quality bases using the Trimmomatic v0.36 tool. The m6A consensus RRACH motif exhibited high enrichment in sh-METTL3 and NTC cells (Figure 6B). Importantly, in shMETTL3 (sh) cells, nearly 82% (65 of 80) of differentially m6A-modified transcripts were downregulated (fold-change ≥2 and p < 0.05) (Figure 6C). Read distribution analysis showed that the NTC and METTL3 knockdown groups has a similar pattern for the total m6A distribution, and reads from m6A-IP samples were highly accumulated around the noncoding area, coding sequence, and within 3′-untranslated regions (3′-UTRs) under all experimental conditions (Figure 6D). By integrating the m6A input RNA-seq dataset and 232 DEGs previously obtained, 31 genes were found to be affected by the methylation of m6A catalyzed by METTL3, with 23 genes downregulated and 9 genes upregulated, indicating their potential as METTL3 targets (Figure 6E).
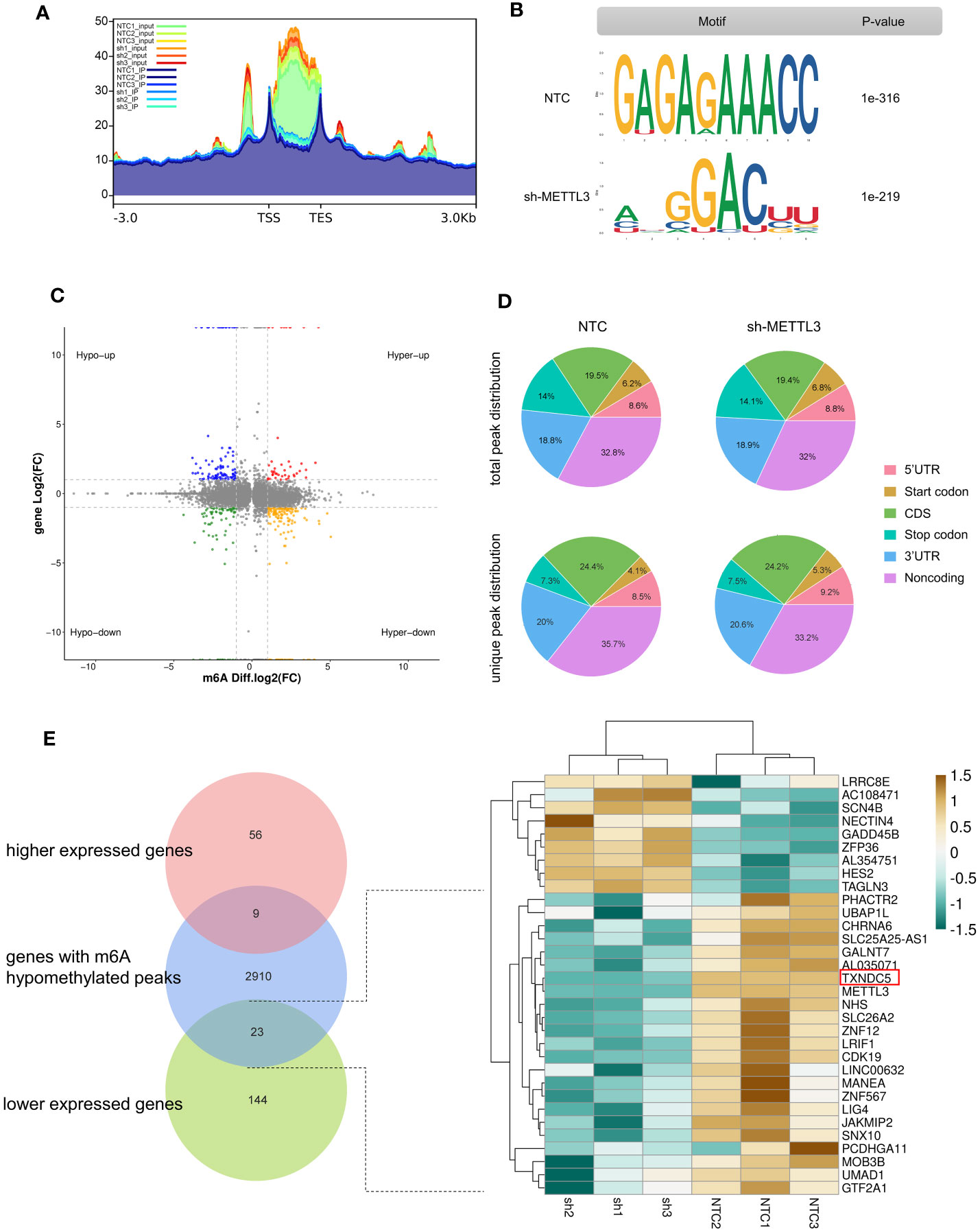
Figure 6 Effects of METTL3 are mediated via m6A methylation. (A) Metagene analysis showing different m6A peak distribution between NTC and METTL3 knockdown groups. (B) “RRACH” as the consensus motif identified by HOMER with m6A peaks in A375 cells with or without METTL3 knockdown. (C) Distribution of genes showing significant changes in both m6A level (log2 fold-change) and gene expression level (log2 fold-change, p < 0.05) in NTCs and sh-METTL3 A375 cells. (D) Proportion of m6A peak distribution in 5′-UTR, start codon, noncoding, coding sequence, stop codon, and 3′-UTR regions in the entire set of RNA transcripts; the total and unique m6A peaks in both groups are shown. (E) Venn diagram and heatmap illustrating genes as potential targets of METTL3 identified by MeRIP-seq and RNA-seq.
To further screening the target genes of METTL3, Western blotting was performed to preliminary verify the potential targets from MeRIP-seq and RNA-seq. Intriguingly, consistent with the gene expression data, knockdown of METTL3 downregulated TXNDC5 at both the mRNA and protein levels (Figure 7A). According to The Cancer Genome Atlas (TCGA) melanoma dataset, intratumoral TXNDC5 expression was positively correlated with METTL3 expression (Figure 7B). Moreover, in A375 NTC/shMETTL3 cells, peak calling analysis distinguished an m6A peak enrichment in the 5′-UTR of TXNDC5 mRNA that was diminished upon METTL3 knockdown (Figure 7C). Meanwhile, we validated TXNDC5 higher expression in melanoma cell lines and melanoma tissues compared with HEMa and adjacent normal tissues, respectively (Figures 7D, E). Taken together, TXNDC5 may be the downstream target of METTL3-m6A in melanoma and may play an important role in melanoma carcinogenesis.
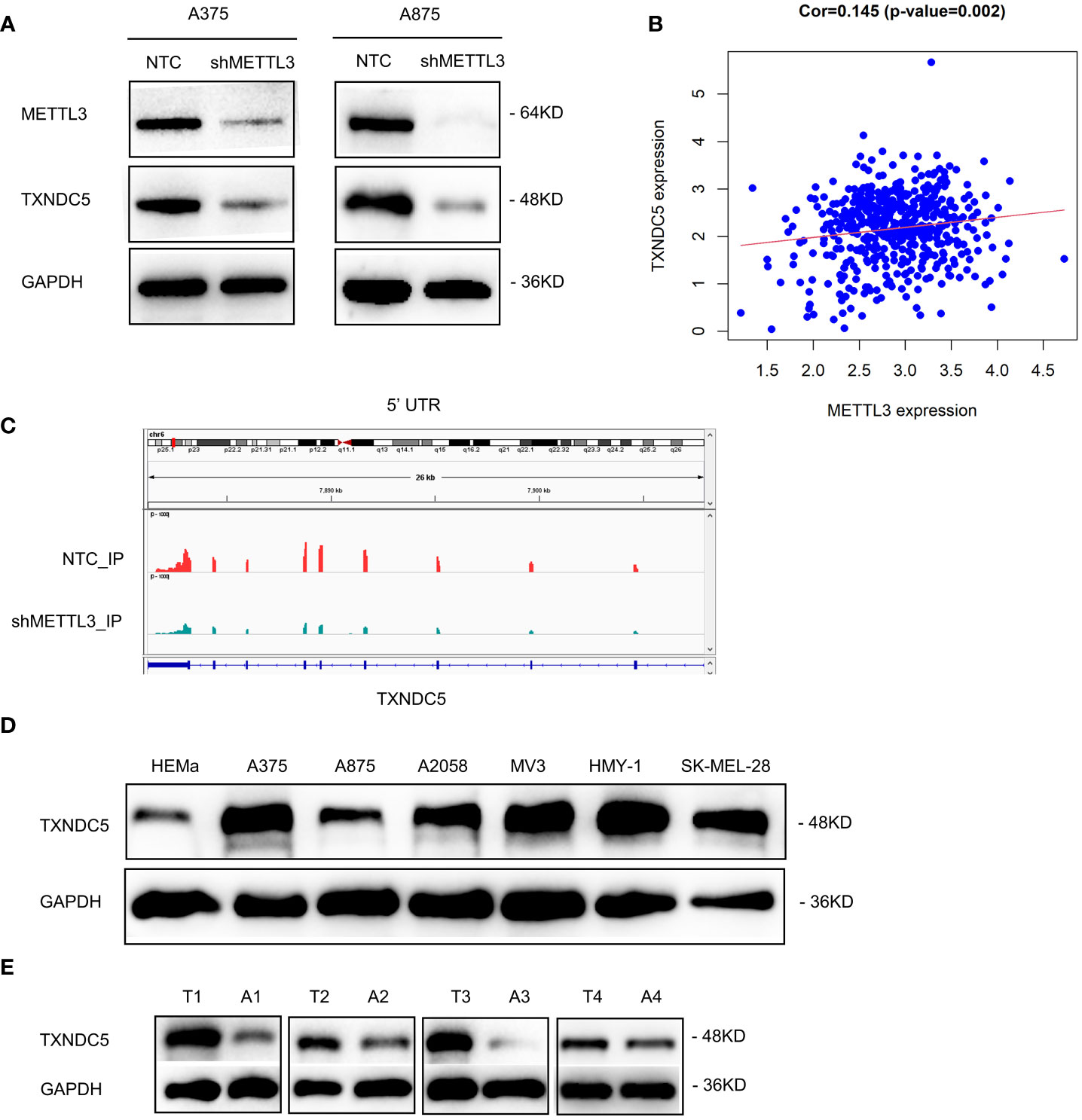
Figure 7 METTL3 targeted TXNDC5 in melanoma regulation. (A) METTL3 knockdown downregulated the protein level of TXNDC5. (B) METTL3 expression was positively correlated with TXNDC5 expression in melanoma in TCGA. (C) database.m6A abundance on TXNDC5 mRNA was plotted by the IGV. METTL3 knockdown diminishes m6A modification of TXNDC5 mRNA in A375 cells. (D, E) TXNDC5 was significantly upregulated in melanoma cell lines and samples.
Here, we observed increased m6A modification levels in acral melanoma tumor tissues. To our knowledge, this is the first report of m6A dysregulation in primary acral melanoma tissues. Abnormal expression of the core methyltransferase METTL3 was suggested as an underlying cause of this phenomenon. Moreover, high METTL3 mRNA expression was significantly associated with clinical staging in melanoma. No melanoma cases in this study received any drug treatment. Thus, identification in their specimens from primary sites suggested specific changes in m6A modification and methyltransferase METTL3 levels occurring in the early stages of acral melanoma pathogenesis. As such, our findings support the use of METTL3 as a biomarker of acral melanoma initiation. Previously, the critical role of METTL3 mainly reflected in the great impact on cell proliferation, growth, survival, invasion, and angiogenesis (39–42). However, the regulation mechanism of METTL3 through m6A modification is diverse and elusive. In our study, METTL3 was found to promote melanoma development by enhancing cell growth and invasion in vivo and in vitro. RNA-seq analysis under METTL3 knockdown revealed DEGs involved in the cell cycle, apoptosis process, immune response, cell adhesion, and defense response, supporting the tumor-promoting effect of METTL3, and reflected that the transcription regulation underlies the tumor-promotor role METTL3.
Recent findings regarding the function of m6A in cancer progression are controversial. Ma et al. reported that the N6-adenosine methyltransferase METTL14 was significantly downregulated in hepatocellular carcinoma, in parallel with an overall decrease in m6A-modified RNA, which in turn affected the tumor suppressor miR-126, ultimately promoting the metastasis and invasion of hepatocellular carcinoma (23). In contrast, Chen et al. found that overexpression of the methyltransferase METTL3 was related to the poor prognosis of hepatocellular carcinoma, promoting its proliferation, migration, and lung metastasis through the m6A-dependent YTHDF2-SOCS2 pathway (43). Previous studies of m6A levels and METTL3 in melanoma have also yielded inconsistent results. For example, m6A levels were increased in addition to METTL3, and demethylase ALKBH5 was found to be significantly down- and upregulated, respectively, in ocular melanoma tumor tissues and cell lines (44). In contrast, a study of uveal melanoma showed the opposite results, reporting upregulation of both m6A modifications and METTL3 and highlighting their role as cancer-promoting factors (45). Yang et al. found that FTO expression was elevated in cutaneous melanoma tissues, with FTO knockdown inhibiting cell proliferation, migration, and metastasis. Knockdown of METTL3/METTL14 in Mel624 cells had the opposite effects (46). The discrepancies between these previous findings underscore the complexity of m6A modification and its regulatory enzymes in human cancers. This may be because of the involvement different reader proteins that facilitate m6A modification-associated signaling, different cellular functions regulated by the target genes, and distinct mRNA regions of m6A distribution. Further studies are required to evaluate these controversial results. This also indicates the great significance to explore the downstream targets of METTL3.
TXNDC5 is a copious enzyme in fibroblast-enriched endoplasmic reticulum (47). As a critical member of the protein disulfide isomerase (PDI) family, the aberrant overexpression of TXNDC5 protein has been investigated in multiple human tumors, including esophageal squamous cell carcinoma (48), cervical cancer (49), lung cancer (50), and hepatocellular carcinoma (51). In a previous study, multiple mechanisms have been described whereby TXNDC5 contributes to cancer development, which is mainly evident in promoting tumor survival and growth in hypoxia environment, altering tumor microenvironment to assists tumor cell metastasis and invasion, and interacting with plasma membrane receptors to control oncogenic cellular response (52). Our study conducted MeRIP-seq to explore the potential mRNAs regulated by METTL3 and found a series of genes whose transcripts were subjected to METTL3-mediated m6A modification. We identified that TXNDC5 was decreased in RNA level and m6A level after METTL3 knockdown. Similarly, the enhanced protein expression of TXNDC5 was also confirmed in melanoma tissues and cell lines. Analysis of the underlying mechanism of METTL3 suggested that it affected melanoma cell growth and invasion by regulating TXNDC5. These results suggest that METTL3 may affect the tumorigenesis of melanoma by modulating transcripts of TXNDC5 in an m6A-dependent manner.
Few studies have explored the functions of METTL3 inside melanoma cells. Wu et al. found that UCK2 overexpression was regulated by m6A-METTL3 axis in melanoma metastasis (53); Chang et al. proposed that miR-302a-3p targets and suppresses the expression of METTL3 to inhibit melanoma cell progression (54). In accordance with this, elevated METTL3 expression was detected in human melanoma cell lines, which led to increased m6A activity, colony formation, and invasion of melanoma cells through MMP2 and N-cadherin accumulation (55). In consistence with their studies, we also identified the critical role of METTL3 in melanoma cell growth, migration, and invasion. However, our research obtained first insight into the dysregulation of m6A modification and METTL3 in Chinese acral melanoma tissues and revealed the clinic significance of METTL3. We also found that TXNDC5 served as a novel target of METTL3-m6A axis in acral melanoma.
Regrettably, due to the limited AM population, the risk prediction role of METTL3 in the progression of AM may not be fully revealed in the current study; more advanced patients will be incorporate in our future studies. In conclusion, we revealed the critical role of METTL3-mediated m6A modification in acral melanoma progression; increased m6A level was confirmed in acral melanoma samples. METTL3 was essential for acral melanoma carcinogenesis and progression, as featured by promoting cancer cell proliferation, migration, and invasion. Importantly, we uncovered that METTL3 affects melanoma progression by targeting m6A-TXNDC5 mRNA, and it is hoped that these findings will contribute to a potential therapeutic strategy in acral melanoma.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE183967
The studies involving human participants were reviewed and approved by Ethics Committee of the Institute of Dermatology at the Chinese Academy of Medical Sciences. The patients/participants provided their written informed consent to participate in this study.
Conceptualization: JS and YW. Data curation: ZY, MC, and AH. Formal analysis: ZY, QZ, and MC. Funding acquisition: JS and YW. Investigation: ZY, QZ, and MC. Methodology: QZ and ZJ. Project administration: JS and YW. Resources: JS, YW, ZJ, GZ, QW, LZ, and FF. Software: QZ and AH. Supervision: JS, YW, and QW. Validation: MC, AH, and QZ. Visualization: ZY and MC. Writing—original draft preparation: ZY and AH. Writing—review and editing: YW and JS.
This work was supported by the National Natural Science Foundation of China (grant number 81872216), PUMC Youth Fund (grant number 3332020105), and Nanjing Incubation Program for National Clinical Research Center (grant number 2019060001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank Professor Yan Kong of Peking University Cancer Hospital & Institute and Professor Xiulian Xu of Hospital of Dermatology, Chinese Academy of Medical Sciences for providing us human melanoma cell lines. We also wish to acknowledge Professor Jennifer Yunyan Zhang of Duke University Medical Center for helpful comments and suggestions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.770325/full#supplementary-material
AM, acral melanoma; HEMa, human epidermal melanocytes; m6A, N6-methyladenosine; METTL3, methyltransferase like 3; TXNDC5, thioredoxin domain containing protein 5; qRT-PCR, quantitative real-time reverse transcriptase PCR; NTC, non-targeted control; IHC, immunohistochemistry; DEG, differentially expressed gene; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MeRIP-Seq, methylated RNA immunoprecipitation sequencing; RNA-seq, RNA sequencing; CDS, coding sequence; 3′-UTR, 3 primer untranslated region; 5′-UTR, 5 primer untranslated region.
1. Houghton AN, Polsky D. Focus on Melanoma. Cancer Cell (2002) 2:275–8. doi: 10.1016/s1535-6108(02)00161-7
2. Schadendorf D, van Akkooi ACJ, Berking C, Griewank KG, Gutzmer R, Hauschild A, et al. Melanoma. Lancet (2018) 392:971–84. doi: 10.1016/S0140-6736(18)31559-9
3. Chi Z, Li S, Sheng X, Si L, Cui C, Han M, et al. Clinical Presentation, Histology, and Prognoses of Malignant Melanoma in Ethnic Chinese: A Study of 522 Consecutive Cases. BMC Cancer (2011) 11:85. doi: 10.1186/1471-2407-11-85
4. Kuchelmeister C, Schaumburg-Lever G, Garbe C. Acral Cutaneous Melanoma in Caucasians: Clinical Features, Histopathology and Prognosis in 112 Patients. Br J Dermatol (2000) 143:275–80. doi: 10.1046/j.1365-2133.2000.03651.x
5. Borkowska AM, Szumera-Cieckiewicz A, Spalek MJ, Teterycz P, Czarnecka AM, Rutkowski PL. Clinicopathological Features and Prognostic Factors of Primary Acral Melanomas in Caucasians. J Clin Med (2020) 9(9):2996. doi: 10.3390/jcm9092996
6. Sondermann W, Zimmer L, Schadendorf D, Roesch A, Klode J, Dissemond J. Initial Misdiagnosis of Melanoma Located on the Foot is Associated With Poorer Prognosis. Med (Baltimore) (2016) 95:e4332. doi: 10.1097/MD.0000000000004332
7. Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of Mutated, Activated BRAF in Metastatic Melanoma. N Engl J Med (2010) 363:809–19. doi: 10.1056/NEJMoa1002011
8. Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med (2015) 373:23–34. doi: 10.1056/NEJMoa1504030
9. Kong Y, Sheng X, Wu X, Yan J, Ma M, Yu J, et al. Frequent Genetic Aberrations in the CDK4 Pathway in Acral Melanoma Indicate the Potential for CDK4/6 Inhibitors in Targeted Therapy. Clin Cancer Res (2017) 23:6946–57. doi: 10.1158/1078-0432.CCR-17-0070
10. Nakamura Y, Ishitsuka Y, Tanaka R, Okiyama N, Saito A, Watanabe R, et al. Acral Lentiginous Melanoma and Mucosal Melanoma Expressed Less Programmed-Death 1 Ligand Than Cutaneous Melanoma: A Retrospective Study of 73 Japanese Melanoma Patients. J Eur Acad Dermatol Venereol (2019) 33:e424–e6. doi: 10.1111/jdv.15742
11. Sheen YS, Tan KT, Tse KP, Liao YH, Lin MH, Chen JS, et al. Genetic Alterations in Primary Melanoma in Taiwan. Br J Dermatol (2020) 182:1205–13. doi: 10.1111/bjd.18425
12. Yeh I, Jorgenson E, Shen L, Xu M, North JP, Shain AH, et al. Targeted Genomic Profiling of Acral Melanoma. J Natl Cancer Inst (2019) 111:1068–77. doi: 10.1093/jnci/djz005
13. Arozarena I, Wellbrock C. Phenotype Plasticity as Enabler of Melanoma Progression and Therapy Resistance. Nat Rev Cancer (2019) 19:377–91. doi: 10.1038/s41568-019-0154-4
14. Zhang N, Wang L, Zhu GN, Sun DJ, He H, Luan Q, et al. The Association Between Trauma and Melanoma in the Chinese Population: A Retrospective Study. J Eur Acad Dermatol Venereol (2014) 28:597–603. doi: 10.1111/jdv.12141
15. Costello CM, Pittelkow MR, Mangold AR. Acral Melanoma and Mechanical Stress on the Plantar Surface of the Foot. N Engl J Med (2017) 377:395–6. doi: 10.1056/NEJMc1706162
16. Gong HZ, Zhang S, Zheng HY, Qu T, Li J. The Role of Mechanical Stress in the Formation of Plantar Melanoma: A Retrospective Analysis of 72 Chinese Patients With Plantar Melanomas and a Meta-Analysis. J Eur Acad Dermatol Venereol (2020) 34:90–6. doi: 10.1111/jdv.15933
17. Pradhan D, Jour G, Milton D, Vasudevaraja V, Tetzlaff MT, Nagarajan P, et al. Aberrant DNA Methylation Predicts Melanoma-Specific Survival in Patients With Acral Melanoma. Cancers (Basel) (2019) 11(12):2031. doi: 10.3390/cancers11122031
18. Fazio M, van Rooijen E, Dang M, van de Hoek G, Ablain J, Mito JK, et al. SATB2 Induction of a Neural Crest Mesenchyme-Like Program Drives Melanoma Invasion and Drug Resistance. Elife (2021) 10:e64370. doi: 10.7554/eLife.64370
19. Scheeren FA, van Doorn R. Inhibition of the Epigenetic Reader BRD4 Reduces SIRPalpha-Mediated Phagocytosis and Melanoma Invasion. J Invest Dermatol (2021) 141:252–4. doi: 10.1016/j.jid.2020.07.023
20. Shi H, Wei J, He C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol Cell (2019) 74:640–50. doi: 10.1016/j.molcel.2019.04.025
21. Liu J, Harada BT, He C. Regulation of Gene Expression by N(6)-Methyladenosine in Cancer. Trends Cell Biol (2019) 29:487–99. doi: 10.1016/j.tcb.2019.02.008
22. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. M(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-Like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell (2017) 31:591–606 e6. doi: 10.1016/j.ccell.2017.02.013
23. Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 Suppresses the Metastatic Potential of Hepatocellular Carcinoma by Modulating N(6) -Methyladenosine-Dependent Primary MicroRNA Processing. Hepatology (2017) 65:529–43. doi: 10.1002/hep.28885
24. Wang Q, Guo X, Li L, Gao Z, Su X, Ji M, et al. N(6)-Methyladenosine METTL3 Promotes Cervical Cancer Tumorigenesis and Warburg Effect Through YTHDF1/HK2 Modification. Cell Death Dis (2020) 11:911. doi: 10.1038/s41419-020-03071-y
25. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-Methyladenosine (M(6)A)-Forming Enzyme METTL3 Controls Myeloid Differentiation of Normal Hematopoietic and Leukemia Cells. Nat Med (2017) 23:1369–76. doi: 10.1038/nm.4416
26. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. YTHDF3 Induces the Translation of M(6)A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell (2020) 38:857–71 e7. doi: 10.1016/j.ccell.2020.10.004
27. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3' UTRs and Near Stop Codons. Cell (2012) 149:1635–46. doi: 10.1016/j.cell.2012.05.003
28. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the Human and Mouse M6a RNA Methylomes Revealed by M6a-Seq. Nature (2012) 485:201–6. doi: 10.1038/nature11112
29. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in Nuclear RNA is a Major Substrate of the Obesity-Associated FTO. Nat Chem Biol (2011) 7:885–7. doi: 10.1038/nchembio.687
30. Zhao Y, Shi Y, Shen H, Xie W. M(6)A-Binding Proteins: The Emerging Crucial Performers in Epigenetics. J Hematol Oncol (2020) 13:35. doi: 10.1186/s13045-020-00872-8
31. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA Circularization by METTL3-Eif3h Enhances Translation and Promotes Oncogenesis. Nature (2018) 561:556–60. doi: 10.1038/s41586-018-0538-8
32. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The M(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell (2016) 62:335–45. doi: 10.1016/j.molcel.2016.03.021
33. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-Mediated M(6)A Modification of HDGF mRNA Promotes Gastric Cancer Progression and has Prognostic Significance. Gut (2020) 69:1193–205. doi: 10.1136/gutjnl-2019-319639
34. Li F, Yi Y, Miao Y, Long W, Long T, Chen S, et al. N(6)-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res (2019) 79:5785–98. doi: 10.1158/0008-5472.CAN-18-2868
35. Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H, Wu M, et al. The M(6)A Methyltransferase METTL3 Promotes Bladder Cancer Progression via AFF4/NF-Kappab/MYC Signaling Network. Oncogene (2019) 38:3667–80. doi: 10.1038/s41388-019-0683-z
36. Yang F, Jin H, Que B, Chao Y, Zhang H, Ying X, et al. Dynamic M(6)A mRNA Methylation Reveals the Role of METTL3-M(6)A-CDCP1 Signaling Axis in Chemical Carcinogenesis. Oncogene (2019) 38:4755–72. doi: 10.1038/s41388-019-0755-0
37. Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, et al. N(6)-Methyladenosine Modification of ITGA6 mRNA Promotes the Development and Progression of Bladder Cancer. EBioMedicine (2019) 47:195–207. doi: 10.1016/j.ebiom.2019.07.068
38. Li X, Tang J, Huang W, Wang F, Li P, Qin C, et al. The M6A Methyltransferase METTL3: Acting as a Tumor Suppressor in Renal Cell Carcinoma. Oncotarget (2017) 8:96103–16. doi: 10.18632/oncotarget.21726
39. Chamorro-Jorganes A, Sweaad WK, Katare R, Besnier M, Anwar M, Beazley-Long N, et al. METTL3 Regulates Angiogenesis by Modulating Let-7e-5p and miRNA-18a-5p Expression in Endothelial Cells. Arterioscler Thromb Vasc Biol (2021) 41:e325–e37. doi: 10.1161/ATVBAHA.121.316180
40. Yao MD, Jiang Q, Ma Y, Liu C, Zhu CY, Sun YN, et al. Role of METTL3-Dependent N(6)-Methyladenosine mRNA Modification in the Promotion of Angiogenesis. Mol Ther (2020) 28:2191–202. doi: 10.1016/j.ymthe.2020.07.022
41. Chen Y, Pan C, Wang X, Xu D, Ma Y, Hu J, et al. Silencing of METTL3 Effectively Hinders Invasion and Metastasis of Prostate Cancer Cells. Theranostics (2021) 11:7640–57. doi: 10.7150/thno.61178
42. Wang W, Shao F, Yang X, Wang J, Zhu R, Yang Y, et al. METTL3 Promotes Tumour Development by Decreasing APC Expression Mediated by APC mRNA N(6)-Methyladenosine-Dependent YTHDF Binding. Nat Commun (2021) 12:3803. doi: 10.1038/s41467-021-23501-5
43. Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-Methyladenosine Methyltransferase-Like 3 Promotes Liver Cancer Progression Through YTHDF2-Dependent Posttranscriptional Silencing of SOCS2. Hepatology (2018) 67:2254–70. doi: 10.1002/hep.29683
44. Jia R, Chai P, Wang S, Sun B, Xu Y, Yang Y, et al. M(6)A Modification Suppresses Ocular Melanoma Through Modulating HINT2 mRNA Translation. Mol Cancer (2019) 18:161. doi: 10.1186/s12943-019-1088-x
45. Luo G, Xu W, Zhao Y, Jin S, Wang S, Liu Q, et al. RNA M(6) A Methylation Regulates Uveal Melanoma Cell Proliferation, Migration, and Invasion by Targeting C-Met. J Cell Physiol (2020) 235:7107–19. doi: 10.1002/jcp.29608
46. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. M(6)A mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 Blockade. Nat Commun (2019) 10:2782. doi: 10.1038/s41467-019-10669-0
47. Shih YC, Chen CL, Zhang Y, Mellor RL, Kanter EM, Fang Y, et al. Endoplasmic Reticulum Protein TXNDC5 Augments Myocardial Fibrosis by Facilitating Extracellular Matrix Protein Folding and Redox-Sensitive Cardiac Fibroblast Activation. Circ Res (2018) 122:1052–68. doi: 10.1161/CIRCRESAHA.117.312130
48. Wang H, Yang X, Guo Y, Shui L, Li S, Bai Y, et al. HERG1 Promotes Esophageal Squamous Cell Carcinoma Growth and Metastasis Through TXNDC5 by Activating the PI3K/AKT Pathway. J Exp Clin Cancer Res (2019) 38:324. doi: 10.1186/s13046-019-1284-y
49. Xu B, Li J, Liu X, Li C, Chang X. TXNDC5 is a Cervical Tumor Susceptibility Gene That Stimulates Cell Migration, Vasculogenic Mimicry and Angiogenesis by Down-Regulating SERPINF1 and TRAF1 Expression. Oncotarget (2017) 8:91009–24. doi: 10.18632/oncotarget.18857
50. Chawsheen HA, Jiang H, Ying Q, Ding N, Thapa P, Wei Q. The Redox Regulator Sulfiredoxin Forms a Complex With Thioredoxin Domain-Containing 5 Protein in Response to ER Stress in Lung Cancer Cells. J Biol Chem (2019) 294:8991–9006. doi: 10.1074/jbc.RA118.005804
51. Park MS, Kim SK, Shin HP, Lee SM, Chung JH. TXNDC5 Gene Polymorphism Contributes to Increased Risk of Hepatocellular Carcinoma in the Korean Male Population. Anticancer Res (2013) 33:3983–7.
52. Chawsheen HA, Ying Q, Jiang H, Wei Q. A Critical Role of the Thioredoxin Domain Containing Protein 5 (TXNDC5) in Redox Homeostasis and Cancer Development. Genes Dis (2018) 5:312–22. doi: 10.1016/j.gendis.2018.09.003
53. Wu H, Xu H, Jia D, Li T, Xia L. METTL3-Induced UCK2 M(6)A Hypermethylation Promotes Melanoma Cancer Cell Metastasis via the WNT/beta-Catenin Pathway. Ann Transl Med (2021) 9:1155. doi: 10.21037/atm-21-2906
54. Chang X, Lin YY, Bai LN, Zhu W. miR-302a-3p Suppresses Melanoma Cell Progression via Targeting METTL3. J Chemother (2021) 21:1–12. doi: 10.1080/1120009X.2021.1953886
Keywords: Mettl3 (methyltransferase like 3), melanoma, TXNDC5, m6A (N6-methyladenose), progression
Citation: Yue Z, Cao M, Hong A, Zhang Q, Zhang G, Jin Z, Zhao L, Wang Q, Fang F, Wang Y and Sun J (2022) m6A Methyltransferase METTL3 Promotes the Progression of Primary Acral Melanoma via Mediating TXNDC5 Methylation. Front. Oncol. 11:770325. doi: 10.3389/fonc.2021.770325
Received: 03 September 2021; Accepted: 15 December 2021;
Published: 18 January 2022.
Edited by:
Xunwei Wu, Shandong University, ChinaReviewed by:
Jianming Zhang, Shanghai Jiao Tong University, ChinaCopyright © 2022 Yue, Cao, Hong, Zhang, Zhang, Jin, Zhao, Wang, Fang, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Wang, ZHJ3YW5neWFuQDE2My5jb20=; Jianfang Sun, ZmFuZ21pbjU3NThAYWxpeXVuLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.