
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 03 January 2022
Sec. Neuro-Oncology and Neurosurgical Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.769033
This article is part of the Research TopicMolecular Biology of Biomarkers in Diagnosis and Treatment of Glioblastoma MultiformeView all 25 articles
 Tengfeng Yan1†
Tengfeng Yan1† Daofeng Tian1†
Daofeng Tian1† Junhui Chen1†
Junhui Chen1† Yinqiu Tan1
Yinqiu Tan1 Yue Cheng2
Yue Cheng2 Liguo Ye1
Liguo Ye1 Gang Deng1
Gang Deng1 Baohui Liu1
Baohui Liu1 Fanen Yuan1
Fanen Yuan1 Shenqi Zhang1
Shenqi Zhang1 Linzhi Cai1*
Linzhi Cai1* Qianxue Chen1*
Qianxue Chen1*The Fc Fragment of IgG Binding Protein (FCGBP) has been proven to participate in intestinal tumor immunity. However, the biological role of FCGBP has remained unclear in glioma. The differential expression of FCGBP was explored by Oncomine and GEPIA databases. The effect of FCGBP on prognosis was analyzed via Kaplan–Meier plotter and GEPIA. The Tumor Immune Estimation Resource (TIMER) tool was used to determine the correlations of FCGBP expression with tumor immune infiltration. Firstly, FCGBP was highly expressed in glioma and correlated with a worse prognosis. Gene Ontology (GO) and KEGG pathway enrichment analyses revealed that the differentially expressed genes (DEGs) and co-expression genes of FCGBP were mainly involved in the immune response. Furthermore, FCGBP expression was positively associated with multiple immune cells infiltrates as well as the expression levels of multiple immune markers in glioma. FCGBP co-expression networks mostly participated in the regulation of immune response. Finally, immunohistochemistry (IHC) assays were conducted to explore the expression of FCGBP, PD-L1, CCL2 and CD8 in glioma and correlations between them. We found that PDL1 and FCGBP were synchronously upregulated in glioma tissues. These findings revealed a new mechanism by which FCGBP participates in the immune tolerance of glioma, and implied the potential of FCGBP as a therapeutic target or predictive marker for patients.
Glioma is the most common malignant tumor of the brain and central nervous system (CNS) in adults (1). High-grade glioma is significantly aggressive and highly heterogeneous, and their tumor microenvironment contains immune cells, glioma stem cells, and mesenchymal cells (2–4). With the further clarification of molecular markers, in 2021, the World Health Organization (WHO) updated the classification of CNS tumors, which is conducive to the molecular treatment of glioma (5). Therefore, it is particularly important to explore novel effective molecular markers to better guide the diagnosis and treatment of glioma.
The Fc Fragment of IgG Binding Protein (FCGBP) protein has multiple von Willebrand D (VWD) domains that can bind to gel-forming mucins (6, 7). FCGBP is also a cysteine-rich protein, which has been detected in body fluids (8). However, its molecular function is still unclear and may be related to the body’s innate immunity (9, 10). In cancer, FCGBP has been widely found to be differentially low-expressed (11–13). However, the role of FCGBP is still unknown in glioma.
Tumor microenvironment (TME) is an essential element in tumor development, and consists of numerous components, which together determine the final fate of tumor cells (14). Recently studies have shown that tumor-infiltrating immune cells play a leading role in the TME (15). Immunotherapy mainly focuses on the signal axis of immune checkpoint PD-1 and its ligand PD-L1. They are highly expressed in tumors and can bind to PD-1 on the surface of T cells to limit T cell activation and induce depleted state, leading to tumor immunity escape (16).
In the present study, we aimed to identify the expression of FCGBP and its correlation with prognosis, and possible signaling pathway in glioma. In addition, we used the bioinformatics analysis–based immune cell-specific signatures to examin the relationship between FCGBP expression with tumor-infiltrating immune cells and immune-related molecules, and drew further interest regarding whether FCGBP could act as a novel immune marker for immune therapy of glioma patients.
We used paraffin-embedded glioma tissue microarrays (108 glioma samples). All specimens were taken from inpatients in the Department of Neurosurgery of Wuhan University People’s Hospital from January 2016 to March 2018. Among these patients, none had received any chemotherapy or radiotherapy before surgery. All patients signed an informed consent form. This study was approved by the Ethics Committee of Wuhan University People’s Hospital School of Medicine.
The paraffin-embedded tissue microarray is heated in an oven (60°C) for 90 minutes. Then, put the slides in different concentrations (100%, 95%, and 75%) of xylene (3 × 5 minutes/time) and ethanol for hydration. After washing 3 times with PBS, 3% H2O2 was added to the slide and left at room temperature for 10 minutes. Then completely immerse the slide in 95°C antigen retrieval solution for 10 minutes, and then let it cool naturally. Triton-PBS (100X) was used for 5 minutes, and the slides were blocked with 1% BSA for 30 minutes. After that, add the primary antibody and incubate overnight at 4°C. The next day, the slides were washed with PBS (3×10 minutes/time), and then incubated with HRP goat anti-rabbit/mouse IgG for 1 hour. After that, DAB was dropped on the glass slide, and the color reaction was terminated with tap water. Hematoxylin staining was repeated for 1 min, and the color was separated with 1% hydrochloric acid ethanol solution. Finally, cover the glass slide with neutral balsam and Olympus BX40 microscope (Tokyo, Japan) to acquire images.
The mRNA sequencing data and clinicopathological data of all cases in this study were downloaded the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/) and the Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/tcga/) database. The mRNA expression level of FCGBP gene in glioma was explored on the oncomine platform (www.oncomine.org). GeneMANIA (http://genemania.org/) is a gene network visualization website that was used amount of functional correlation genes to identify a single gene, and predicted gene function and related pathways. A p-value less than 0.05 indicates statistical significance. Identify the genomic changes of FCGBP in TCGA-GBM and TCGA-LGG on the cBioPortal platform (http://www.cbioportal.org/). Tumor Immunity Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) is a TCGA-based website for systematic analysis of the correlation between genes and different immune cell subtypes in different types of cancer and immune molecules. LinkedOmics (http://www.linkedomics.org/login.php) was used to identify differentially expressed genes related to FCGBP (n = 544) in TCGA-GBMLGG. Differentially expressed genes were analyzed using Pearson correlation coefficient. And enriched it in Gene Ontology (GO) and KEGG pathway. TISIDB is a website for gene- and tumor-immune interaction (http://cis.hku.hk/TISIDB/index.php). The detailed clinical characteristics of the included observations are shown in Supplementary Tables 1, 2.
GSEA (http://software.broadinstitute.org/gsea/index.jsp) was used to analyze potential genes related to FCGBP. According to the true pivot genes (top 50% and bottom 50%), they were divided into high and low groups. Then GSEA was used for bio-process enrichment, and used Normalized Enrichment Score (NES) to estimate GSEA enrichment. The significance of enrichment was evaluated at the FDR <0.25 level, p-value <0.05, and FDR <0.25 level.
Spearman correlation analysis was used to test the correlation. According to the 50% cut-off point of gene expression, patients were divided into high group and low group. The difference in survival rate between groups was evaluated by Kaplan-Meier survival analysis and log-rank significance test.
Firstly, we used TIMER data to evaluate the mRNA expression level of FCGBP in different tumors and normal tissues. TIMER data analysis revealed that FCGBP was significantly lower expressed in most tumor tissues compared with normal tissues. However, FCGBP showed a significantly high expression in glioblastoma (GBM) compared with normal tissues (Figure 1A). Through the GEPIA database, we evaluated the transcription level of FCGBP in different tumor tissues and normal tissues (Figure 1B). The results showed that the transcription level of FCGBP in GBM and low-grade glioma (LGG) tissues was significantly higher than normal tissues.
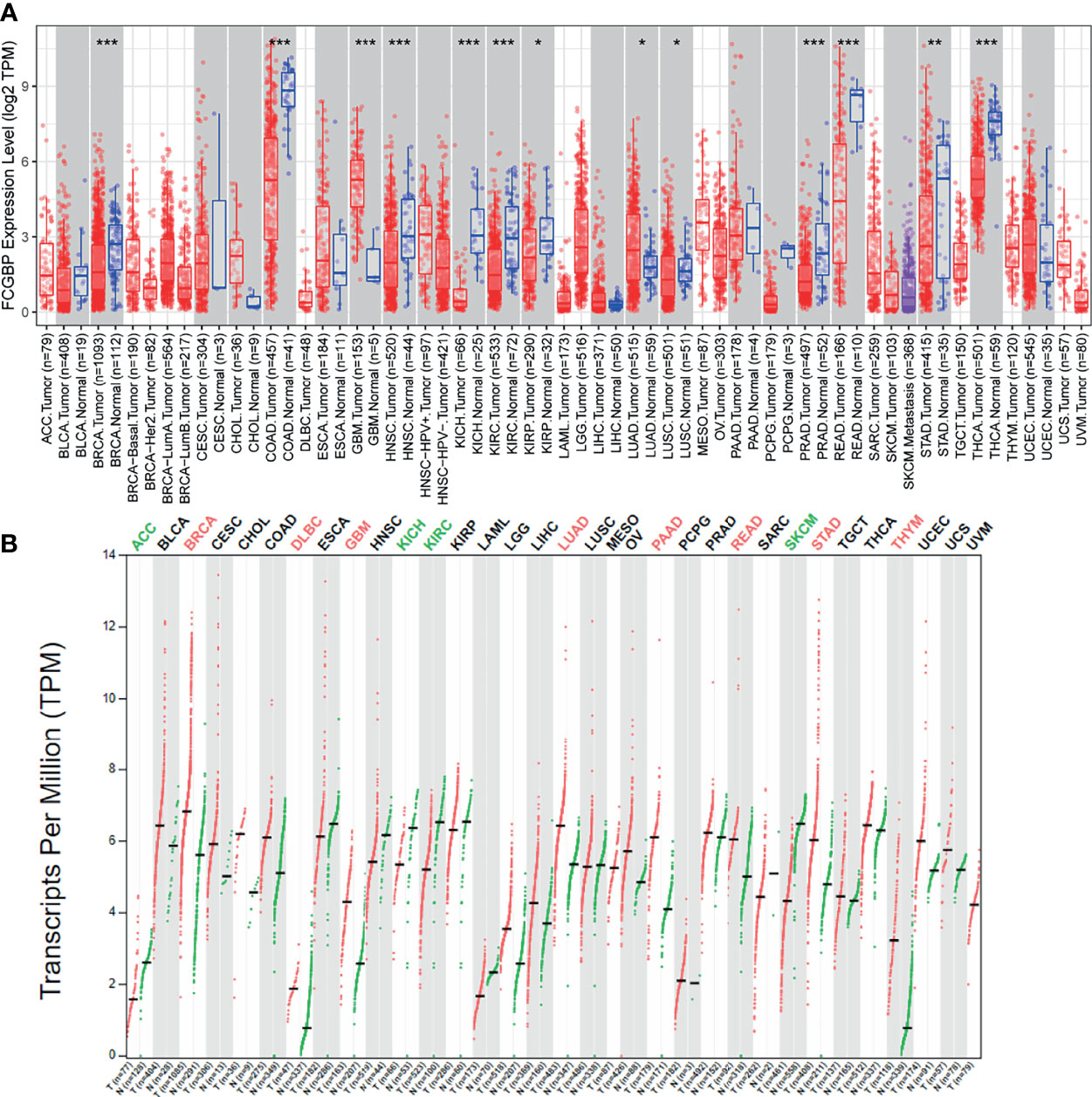
Figure 1 The expression FCGBP in different cancer tissues (A) The mRNA expression levels of FCGBP in different tumor tissues and normal tissues analyzed by TIMER (*P <.05, **P <.01, ***P <.001). (B) Transcription level of FCGBP was determined in different tumor tissues and normal tissues by GEPIA (*P <.05, **P <.01, ***P <.001).
Oncomine online database was utilized to analyze the mRNA expression level of FCGBP in tumor and normal tissues. A total of 38 data sets were obtained, and the expression of FCGBP was up-regulated in 14 data sets. Especially in the central nervous system (CNS), the expression of FCGBP in five glioma datasets were all up-regulated (Figure 2A). Three independent cohorts including 1319 cases were validated in further study [693 cases in Validation cohort 1 (CGGA-693); 301 cases in Validation cohort 2 (CGGA-301); and 325 cases in Validation cohort 3 (CGGA-325)]. Then, all CGGA database analysis found that FCGBP was also significantly up-regulated in glioma, and positively correlated with the grade of glioma (Figure 2B). To verify the upregulation of FCGBP in gliomas, we performed IHC analysis using glioma samples. FCGBP is not detectable in normal brain tissue, but it is expressed in different grades of gliomas, especially high-grade gliomas (Figure 2C).
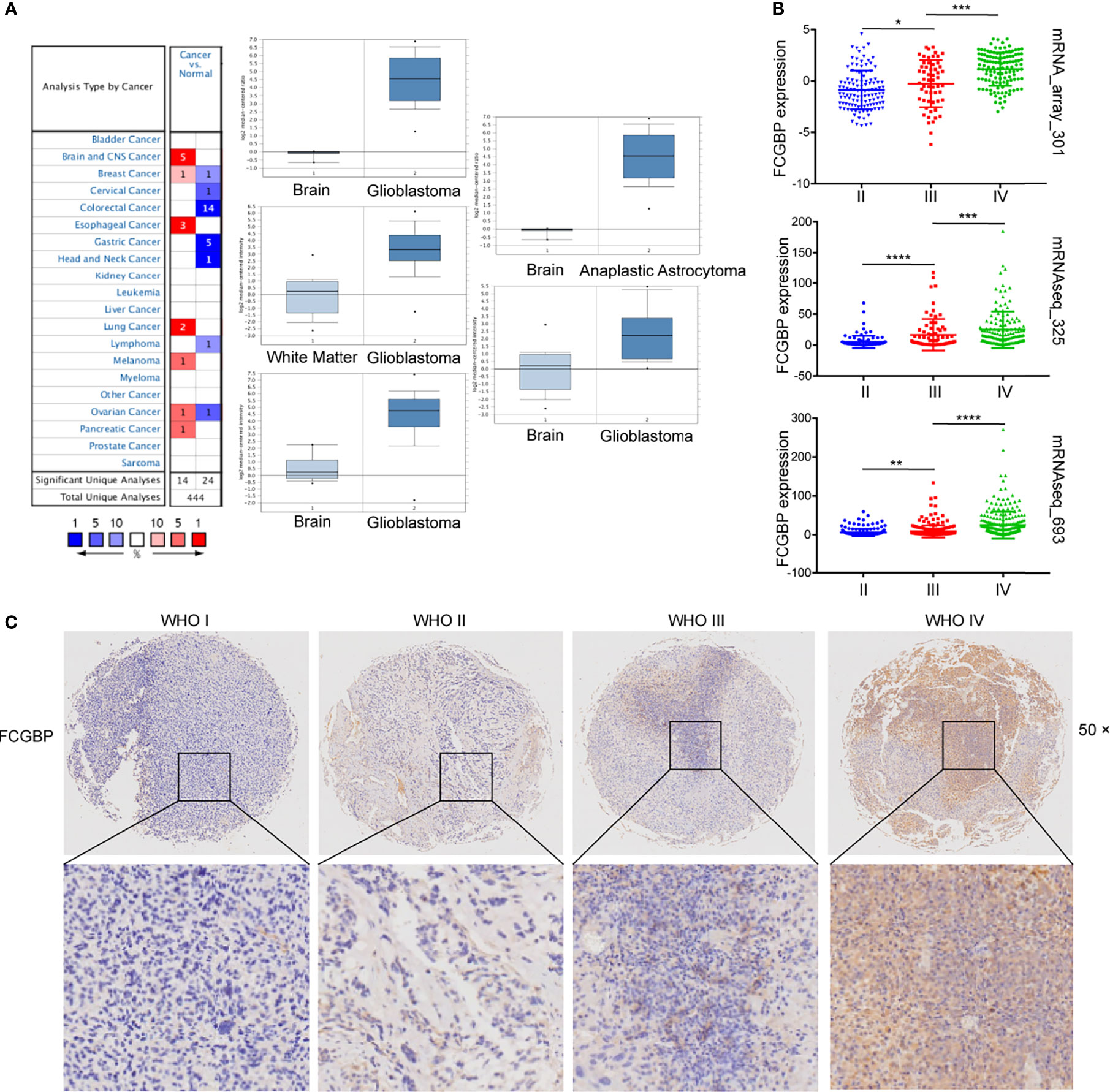
Figure 2 FCGBP is up-regulated in glioma. (A) The mRNA expression level of FCGBP in different tumor tissues and normal tissues analyzed by Oncomine (*P <.05, **P <.01, ***P <.001, ****P <0.0001). (B) The mRNA expression level of FCGBP was determined in glioma tissues by CGGA database (*P <.05, **P <.01, ***P <.001). (C) Typical images of IHC staining of FCGBP in glioma tissues.
Further univariate and multivariate Cox regression analysis showed that FCGBP can be significantly negatively correlated with the prognosis of glioma patients in TCGA and CGGA (Table 1 and 2). Using an online software GEPIA, which can be used to analyze the survival data from TCGA data sets. Survival analysis showed that the expression of FCGBP was significantly correlated with the poor overall survival (OS) and disease-free survival (DFS) of LGG patients, while high FCGBP expression showed no associations with OS or DFS in GBM patients (Figure 3A). Meanwhile, we next scanned the TCGA database, and found that overexpression of FCGBP predicted poor DFS and OS in LGG patients, while high FCGBP expression also showed no associations with OS or DFS in GBM patients (Figure 3B). Survival curve analysis was verified via three independent CGGA cohorts, and revealed that high expression of FCGBP had a significant poor prognosis in glioma (Figure 3C).
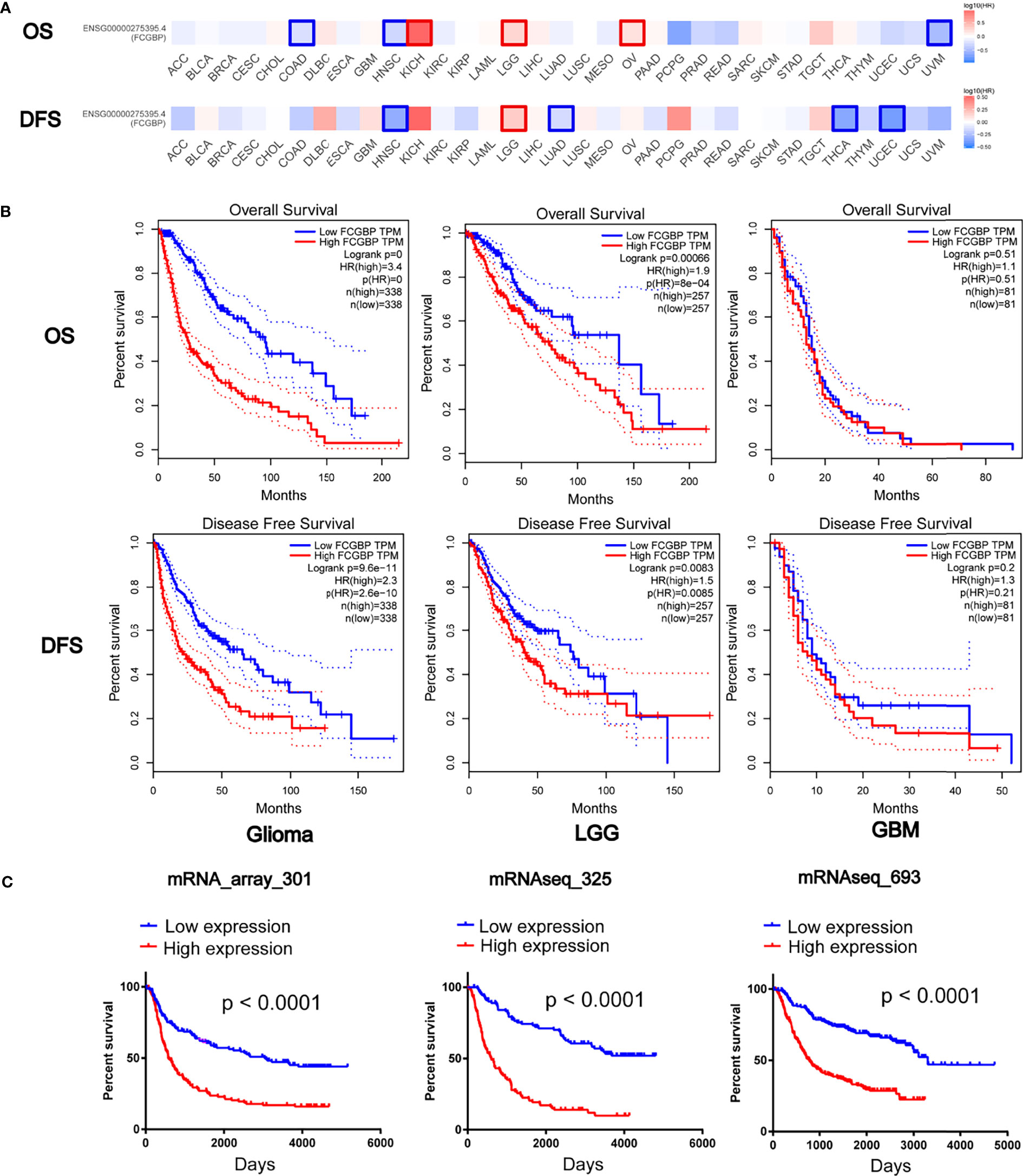
Figure 3 Prognostic potential of FCGBP in glioma. (A) The OS and DFS Map of FCGBP in different cancers by GEPIA. (B) Kaplan‐Meier survival analysis of all glioma, LGG and GBM in TCGA data sets. (C) Kaplan‐Meier survival analysis of all glioma in CGGA data sets.
The cBioPortal online tool was used to analyze the genetic changes of FCGBP in TCGA PanCan Atlas dataset, and we found that FCGBP has a higher mutation frequency in tumors (Figure 4A). In LGG and GBM, the results showed a mutation frequency of 2.5% and 3% (Figure 4B). The gene-gene interaction network of FCGBP was constructed based on the GeneMANIA database. The functional analysis revealed that FCGBP may related to tumor immune response (Figure 4C).
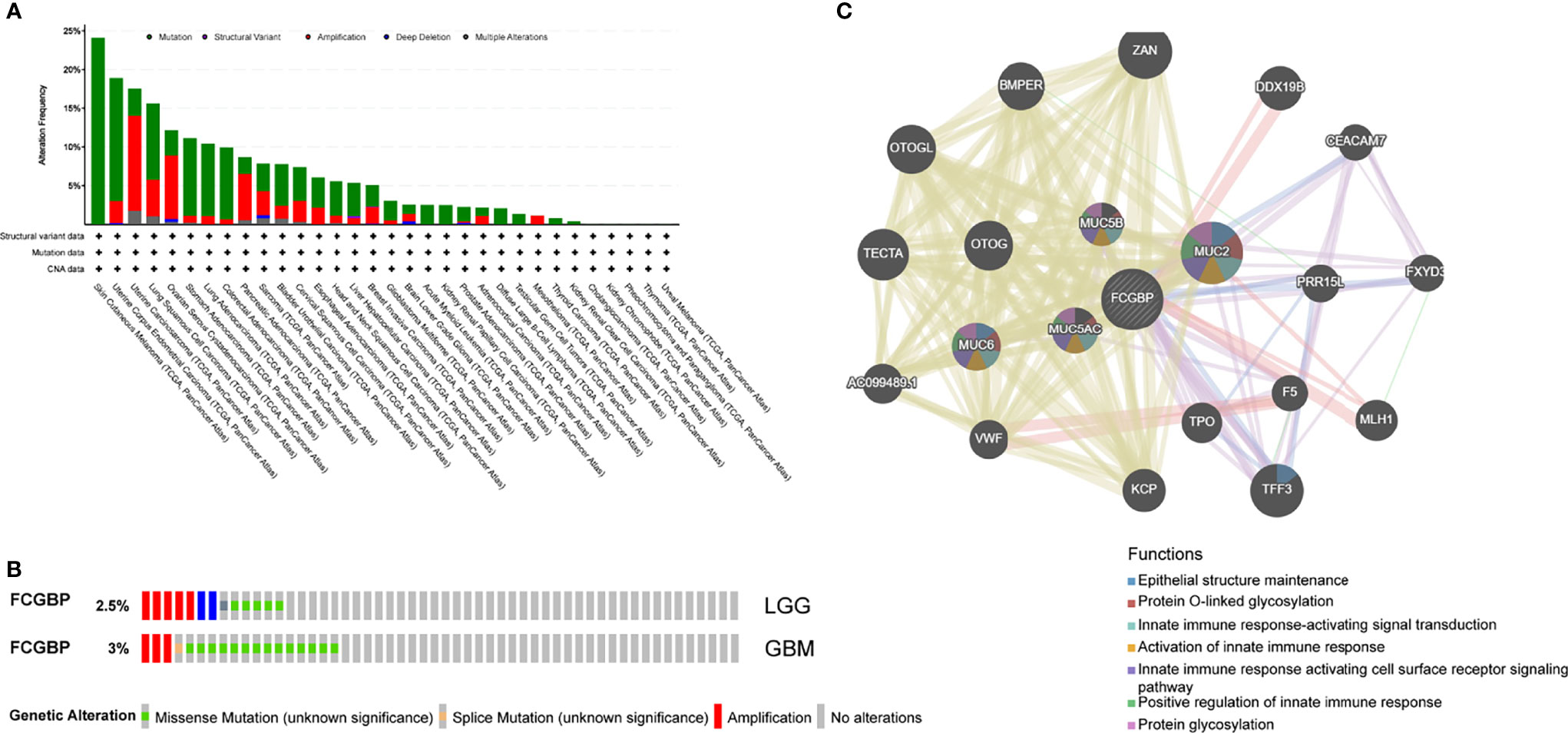
Figure 4 Analysis of genetic changes in glioma, and interacting gene network of FCGBP. (A) Genetic alterations of the FCGBP gene were analyzed by cBioPortal. (B) The mutation frequency of FCGBP in LGG and GBM. (C) FCGBP’s interactive network was generated through the GeneMANIA database.
In order to study the potential function of FCGBP in glioma, we used the LinkedOmics database to explore related molecules of FCGBP. The volcano map shows that red show a significant positive correlation with FCGBP, while green show a significant negative correlation (Figure 5A). And select the top 50% genes with positive or negative correlation and display them in the heatmap (Figure 5B). GO and KEGG pathway analysis of gene set enrichment analysis (GSEA) was showed that FCGBP co-expressed genes are mainly involved in immune response, etc. (Figure 5C). Furthermore, we validated the differentially expressed genes of FCGBP in the CGGA database, and also found that the positively related genes of FCGBP were significantly related to the immune response pathway (Figure S1).
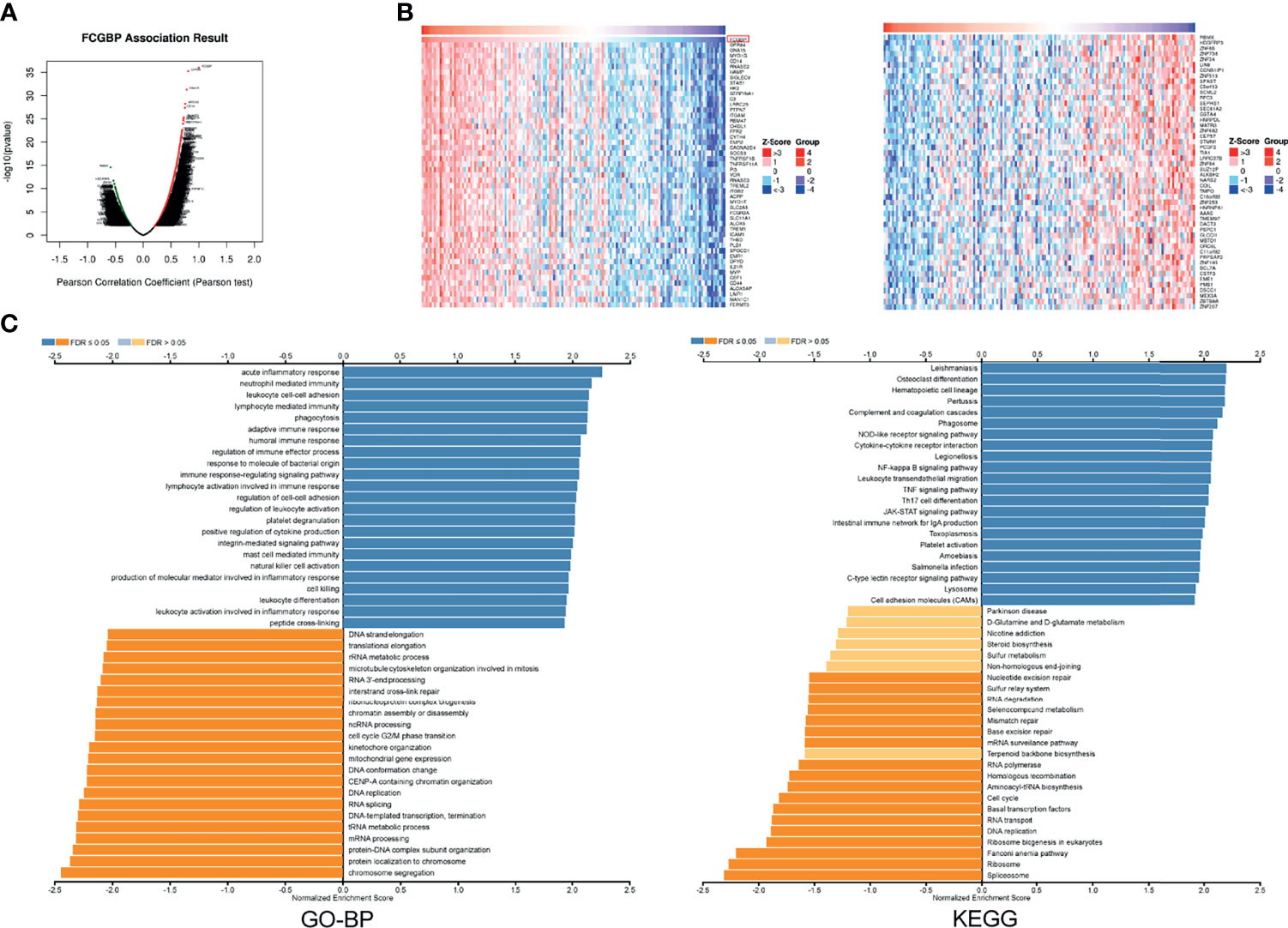
Figure 5 FCGBP co-expression network in glioma (A) The volcano map showed FCGBP-related genes in glioma. (B) The heatmap showed FCGBP-related genes in glioma. (C) Based on the LinkedOmics database, GO annotations and KEGG pathways of genes co-expressed with FCGBP in glioma were significantly enriched.
Next, the immune cell infiltration was analyzed using the TIMER website, and we found that the expression of FCGBP was positively correlated with the infiltration of B cells, CD4 T cells, Neutrophil and Dendritic cells (Figure 6A). Relationship of FCGBP expression with immune infiltration in tumors was assessed using correlation analysis and TISIDB databases. Furthermore, we found that positive correlation between FCGBP expression and effector memory CD8 + T cells, effector memory CD4 + T cells, Myeloid-derived suppressor cells, macrophages (Figures 6B, C).
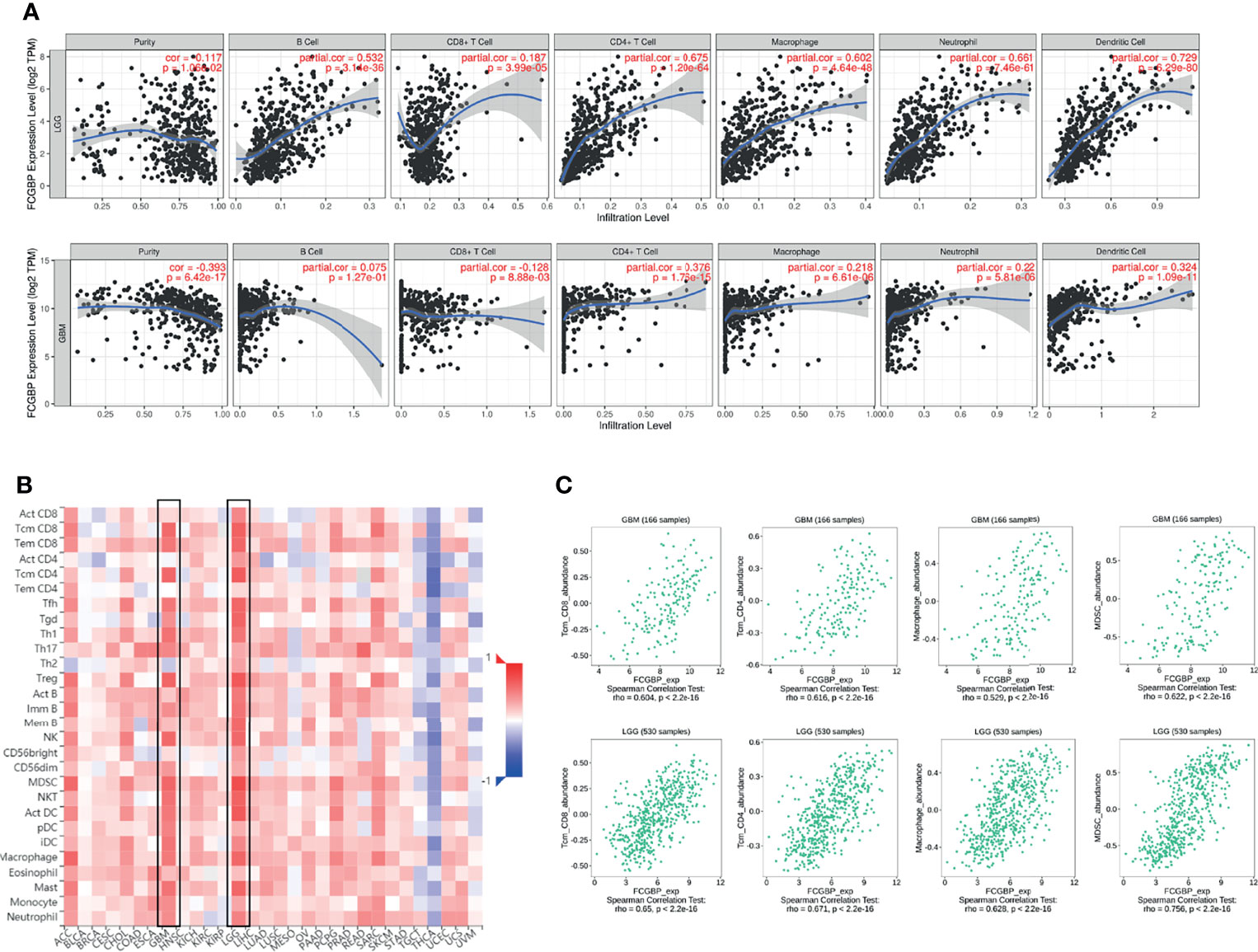
Figure 6 The relationship between the expression of FCGBP and the level of immune infiltration in glioma. (A) The expression of FCGBP is correlated with the level of tumor immune infiltration of B cells, CD8 + T cells, CD4 + T cells, Neutrophil and Dendritic cells in glioma. (B) Correlations between FCGBP expression and immune infiltration levels in TISIDB. (C) The relationship between the expression of FCGBP and CD8 + T cells, effector memory CD4 T cells, Myeloid-derived suppressor cells, macrophages in glioma.
The Spearman’s correlations between FCGBP expression and immunomodulators, and chemokines were analyzed using the TISIDB database. The results showed the correlation between FCGBP expression level and immunosuppressive agents. The top two immunosuppressive agents include HAVCR2 and TGFB1 (Figure 7A). Meanwhile, the correlation between the expression of FCGBP and immunostimulators was analyzed. The top three immunostimulators include CD86, CD40 and TMEM173 (Figure 7B). In addition, we also analyzed the correlation between the expression of FCGBP with chemokines and receptors. The top four significantly positively correlated chemokines include CCL2, CCL5, CXCL10, and CXCL16 (Figure 7C). For the correlation between FCGBP expression and receptors, the top five receptors include CCR5, CCR1, CXCR6, and CX3CR1 (Figure 7D). Therefore, FCGBP could regulate immune molecules.
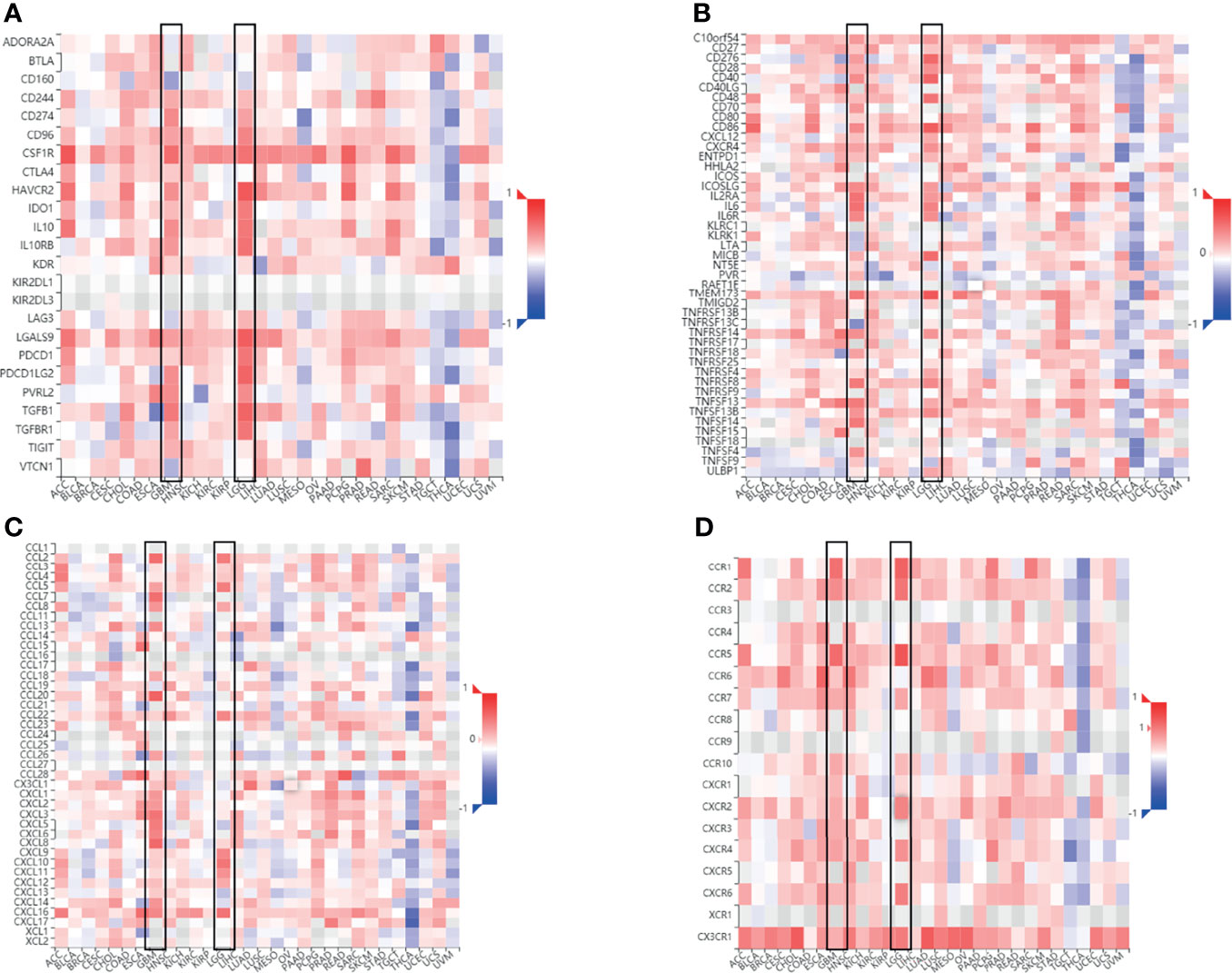
Figure 7 Correlation between FCGBP expression and immune markers. Correlation between FCGBP mRNA expression and immune-related molecules. (A) Correlation between FCGBP and glioma immunosuppressive agents. (B) The correlation between FCGBP and glioma immunostimulants. (C) Correlation between FCGBP and chemokines in gliomas. (D) The correlation between FCGBP and glioma receptors.
Tissue microarrays were constructed as previously described (17). Next, we analyzed the correlation between FCGBP with immune infiltrating molecules and immune checkpoint genes. In addition, the same glioma specimen section was used for CCL2, CD8 and PDL1 IHC staining. The results showed that FCGBP was positively correlated with the expression of CCL2, CD8 and PDL1 (Figure 8). These indicate that FCGBP may have a synergistic effect with immune infiltrating molecules and immune checkpoint members.
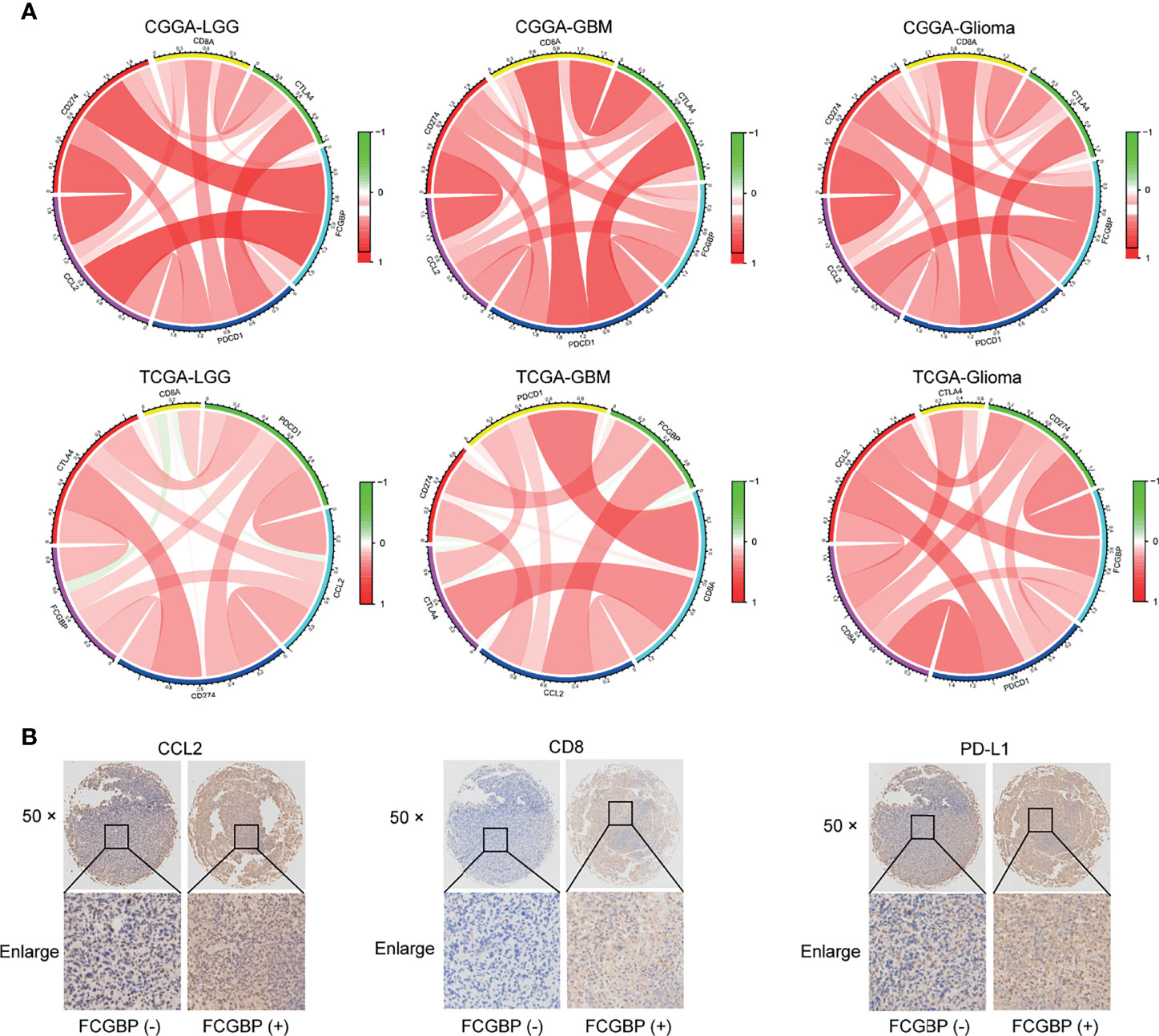
Figure 8 Tissue Microarrays (TMAs) and Immunohistochemistry (IHC). (A) The chord diagram illustrating the correlation of the expression of FCGBP with CD8, CCL2 and immune checkpoints molecules (CTLA4, PD1, PDL1). (B) IHC staining of CCL2, CD8 and PDL1 in glioma tissues.
As an important part of the tumor microenvironment, immune cells have been shown to regulate tumor progression (18). Immunosuppressants began to be gradually used in clinical patients. For example, PD-1 and CTLA-4 antibodies have achieved sustained efficacy in some patients (19). Although new therapies such as immunotherapy have been used, it is still difficult to obtain a good prognosis for GBM patients (20). Tumor heterogeneity may be the main factor limiting GBM patients’ access to effective treatment (21).
FCGBP was previously considered to be one of the components of mucus secreted by goblet cells in the intestine. FCGBP is an important part of the intestinal mucosal immune defense and participates in anti-inflammatory and cell protection (22, 23). Significantly low expression of FCGBP has been observed in intestinal inflammation and tumors, such as ulcerative colitis (24), colorectal adenoma (25), and colorectal cancer (26–28). In addition, FCGBP is also down-regulated in multiple tumors (12, 13, 29, 30), and this imbalance of expression due to different functions that may be involved in different tumors. Through the TCGA and CGGA databases, we first found that FCGBP was highly expressed in glioma tissues and had a significant positive correlation with the grade of glioma. Previous studies have found that ovarian cancer with high FCGBP expression had a worse prognosis, which was consistent with our results (31).
Previous studies have shown that FCGBP may be involved in the immune protection of the intestinal tract (32). Meanwhile, we found that high expression of FCGBP may be a potential poor prognostic factor in this study. Next, KEGG and GO enrichment analysis both show that FCGBP expression is related to immune and inflammation-related biological processes. In the immune microenvironment, tumor-infiltrating immune cells and cancer cells conduct extensive and dynamic crosstalk, which has revealed that certain molecules can participate in this dialogue (33). In this study, we further found that high FCGBP expression is associated with high abundance of immune infiltration, including B cells, CD4+ T cells, neutrophils, macrophages and DCs in LGG. These results indicate at least that FCGBP protein may increase the recruitment of immune cells in LGG, which is not significant in GBM.
The effectiveness of immunotherapy mainly depends on the internal immune microenvironment of the tumor (34). In recent years, immune checkpoint inhibitors have become the most advanced immunotherapy in clinical applications. Among them, death protein 1 (PD-1) and T cell programmed death ligand (PD-L1) have received the most extensive research (35, 36). Through the typical images of immunohistochemistry, we revealed that FCGBP was significantly positively correlated with PD-L1, CD8 and CCL2. This indicated that FCGBP may be related to the up-regulation of immune checkpoints. However, there is no molecular study of FCGBP in glioma cells, further demonstration is needed.
The novelty of this study is that we firstly found that FCGBP has a poor prognostic in glioma, and we also explored the possible mechanism of FCGBP in glioma. We confirmed the correlation between FCGBP and immune infiltration of glioma, and proposed that FCGBP may act a novel immunotherapy biomarker. Therefore, our findings will be helpful to further optimize the immunotherapy of glioma.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
All patients signed an informed consent form. The studies involving human participants were reviewed and approved by the Institutional Ethics Committee of the Faculty of Medicine at Renmin Hospital of Wuhan University.
Conceptualization, TY, DT, and QC. Data curation, TY, JC, and LC. Formal analysis, TY, JC, LC, LY, YC and QC. Funding acquisition, QC. Investigation, BL and QC. Methodology, SZ. Project administration, DT. Software, TY, JC, and YT. Supervision, BL. Visualization, FY. Writing – original draft, GD. Writing – review and editing, TY and LC. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (NO.82072764). Nantong University School-level Research Fund (NO.2019JQ017) and supported by the Fundamental Research Funds for the Central Universities (NO.2042021kf0133).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.769033/full#supplementary-material
1. Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007-2011. Neuro-Oncol 16 Suppl (2014) 4:iv1–63. doi: 10.1093/neuonc/nou223
2. Shi Y, Ping YF, Zhang X, Bian XW. Hostile Takeover: Glioma Stem Cells Recruit TAMs to Support Tumor Progression. Cell Stem Cell (2015) 16:219–20. doi: 10.1016/j.stem.2015.02.008
3. Erreni M, Solinas G, Brescia P, Osti D, Zunino F, Colombo P, et al. Human Glioblastoma Tumours and Neural Cancer Stem Cells Express the Chemokine CX3CL1 and Its Receptor CX3CR1. Eur J Cancer (2010) 46:3383–92. doi: 10.1016/j.ejca.2010.07.022
4. Zhang Q, Xiang W, Yi DY, Xue BZ, Wen WW, Abdelmaksoud A, et al. Current Status and Potential Challenges of Mesenchymal Stem Cell-Based Therapy for Malignant Gliomas. Stem Cell Res Ther (2018) 9:228. doi: 10.1186/s13287-018-0977-z
5. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-oncology (2021) 23:1231–51. doi: 10.1093/neuonc/noab106
6. Lang T, Klasson S, Larsson E, Johansson ME, Hansson GC, Samuelsson T. Searching the Evolutionary Origin of Epithelial Mucus Protein Components-Mucins and FCGBP. Mol Biol Evol (2016) 33:1921–36. doi: 10.1093/molbev/msw066
7. Lang T, Hansson GC, Samuelsson T. An Inventory of Mucin Genes in the Chicken Genome Shows That the Mucin Domain of Muc13 Is Encoded by Multiple Exons and That Ovomucin Is Part of a Locus of Related Gel-Forming Mucins. BMC Genomics (2006) 7:197. doi: 10.1186/1471-2164-7-197
8. Kobayashi K, Ogata H, Morikawa M, Iijima S, Harada N, Yoshida T, et al. Distribution and Partial Characterisation of IgG Fc Binding Protein in Various Mucin Producing Cells and Body Fluids. Gut (2002) 51:169–76. doi: 10.1136/gut.51.2.169
9. Hoffmann W. Trefoil Factor Family (TFF) Peptides and their Different Roles in the Mucosal Innate Immune Defense and More: An Update. Curr Med Chem. 2021;28(36):7387–7399. doi: 10.2174/0929867328666210215114140
10. Lencer WI, Blumberg RS. A Passionate Kiss, Then Run: Exocytosis and Recycling of IgG by FcRn. Trends Cell Biol (2005) 15:5–9. doi: 10.1016/j.tcb.2004.11.004
11. Xiong L, Wen Y, Miao X, Yang Z. NT5E and FcGBP as Key Regulators of TGF-1-Induced Epithelial-Mesenchymal Transition (EMT) Are Associated With Tumor Progression and Survival of Patients With Gallbladder Cancer. Cell Tissue Res (2014) 355:365–74. doi: 10.1007/s00441-013-1752-1
12. Gazi MH, He M, Cheville JC, Young CY. Downregulation of IgG Fc Binding Protein (Fc gammaBP) in Prostate Cancer. Cancer Biol Ther (2008) 7:70–5. doi: 10.4161/cbt.7.1.5131
13. O’Donovan N, Fischer A, Abdo EM, Simon F, Peter HJ, Gerber H, et al. Differential Expression of IgG Fc Binding Protein (FcgammaBP) in Human Normal Thyroid Tissue, Thyroid Adenomas and Thyroid Carcinomas. J Endocrinol (2002) 174:517–24. doi: 10.1677/joe.0.1740517
14. Quail DF, Joyce JA. The Microenvironmental Landscape of Brain Tumors. Cancer Cell (2017) 31:326–41. doi: 10.1016/j.ccell.2017.02.009
15. Giraldo NA, Sanchez-Salas R, Peske JD, Vano Y, Becht E, Petitprez F, et al. The Clinical Role of the TME in Solid Cancer. Br J Cancer (2019) 120:45–53. doi: 10.1038/s41416-018-0327-z
16. Lingel H, Brunner-Weinzierl MC. CTLA-4 (CD152): A Versatile Receptor for Immune-Based Therapy. Semin Immunol (2019) 42:101298. doi: 10.1016/j.smim.2019.101298
17. Han S, Xia J, Qin X, Han S, Wu A. Phosphorylated SATB1 Is Associated With the Progression and Prognosis of Glioma. Cell Death Dis (2013) 4:e901. doi: 10.1038/cddis.2013.433
18. Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, et al. Immune Cells Within the Tumor Microenvironment: Biological Functions and Roles in Cancer Immunotherapy. Cancer Lett (2020) 470:126–33. doi: 10.1016/j.canlet.2019.11.009
19. Tian Y, Zhai X, Yan W, Zhu H, Yu J. Clinical Outcomes of Immune Checkpoint Blockades and the Underlying Immune Escape Mechanisms in Squamous and Adenocarcinoma NSCLC. Cancer Med (2021) 10:3–14. doi: 10.1002/cam4.3590
20. Jain KK. A Critical Overview of Targeted Therapies for Glioblastoma. Front Oncol (2018) 8:419. doi: 10.3389/fonc.2018.00419
21. Friedmann-Morvinski D. Glioblastoma Heterogeneity and Cancer Cell Plasticity. Crit Rev Oncog (2014) 19:327–36. doi: 10.1615/CritRevOncog.2014011777
22. Kobayashi K, Blaser MJ, Brown WR. Identification of a Unique IgG Fc Binding Site in Human Intestinal Epithelium. J Immunol (1989) 143:2567–74.
23. Selbach M, Mann M. Protein Interaction Screening by Quantitative Immunoprecipitation Combined With Knockdown (QUICK). Nat Methods (2006) 3:981–3. doi: 10.1038/nmeth972
24. Kim M, Lee S, Yang SK, Song K, Lee I. Differential Expression in Histologically Normal Crypts of Ulcerative Colitis Suggests Primary Crypt Disorder. Oncol Rep (2006) 16:663–70. doi: 10.3892/or.16.4.663
25. Lee S, Bang S, Song K, Lee I. Differential Expression in Normal-Adenoma-Carcinoma Sequence Suggests Complex Molecular Carcinogenesis in Colon. Oncol Rep (2006) 16:747–54. doi: 10.3892/or.16.4.747
26. Zhu H, Wu TC, Chen WQ, Zhou LJ, Wu Y, Zeng L, et al. Screening for Differentially Expressed Genes Between Left- and Right-Sided Colon Carcinoma by Microarray Analysis. Oncol Lett (2013) 6:353–8. doi: 10.3892/ol.2013.1414
27. Yang W, Shi J, Zhou Y, Liu T, Zhan F, Zhang K, et al. Integrating Proteomics and Transcriptomics for the Identification of Potential Targets in Early Colorectal Cancer. Int J Oncol (2019) 55:439–50. doi: 10.3892/ijo.2019.4833
28. Zhang GL, Pan LL, Huang T, Wang JH. The Transcriptome Difference Between Colorectal Tumor and Normal Tissues Revealed by Single-Cell Sequencing. J Cancer (2019) 10:5883–90. doi: 10.7150/jca.32267
29. Zhuang Q, Shen A, Liu L, Wu M, Shen Z, Liu H, et al. Prognostic and Immunological Roles of Fc Fragment of IgG Binding Protein in Colorectal Cancer. Oncol Lett (2021) 22:526. doi: 10.3892/ol.2021.12787
30. Onstenk W, Sieuwerts AM, Mostert B, Lalmahomed Z, Bolt-de Vries JB, van Galen A, et al. Molecular Characteristics of Circulating Tumor Cells Resemble the Liver Metastasis More Closely Than the Primary Tumor in Metastatic Colorectal Cancer. Oncotarget (2016) 7:59058–69. doi: 10.18632/oncotarget.10175
31. Wang K, Guan C, Shang X, Ying X, Mei S, Zhu H, et al. A Bioinformatic Analysis: The Overexpression and Clinical Significance of FCGBP in Ovarian Cancer. Aging (2021) 13:7416–29. doi: 10.18632/aging.202601
32. Risques RA, Lai LA, Himmetoglu C, Ebaee A, Li L, Feng Z, et al. Ulcerative Colitis-Associated Colorectal Cancer Arises in a Field of Short Telomeres, Senescence, and Inflammation. Cancer Res (2011) 71:1669–79. doi: 10.1158/0008-5472.CAN-10-1966
33. Grivennikov SI, Greten FR, Karin M. Immunity, Inflammation, and Cancer. Cell (2010) 140:883–99. doi: 10.1016/j.cell.2010.01.025
34. Wu T, Dai Y. Tumor Microenvironment and Therapeutic Response. Cancer Lett (2017) 387:61–8. doi: 10.1016/j.canlet.2016.01.043
35. Qi Y, Liu B, Sun Q, Xiong X, Chen Q. Immune Checkpoint Targeted Therapy in Glioma: Status and Hopes. Front Immunol (2020) 11:578877. doi: 10.3389/fimmu.2020.578877
Keywords: FCGBP, glioma, immune infiltration, prognostic biomarker, PD-L1
Citation: Yan T, Tian D, Chen J, Tan Y, Cheng Y, Ye L, Deng G, Liu B, Yuan F, Zhang S, Cai L and Chen Q (2022) FCGBP Is a Prognostic Biomarker and Associated With Immune Infiltration in Glioma. Front. Oncol. 11:769033. doi: 10.3389/fonc.2021.769033
Received: 01 September 2021; Accepted: 07 December 2021;
Published: 03 January 2022.
Edited by:
Lei Jin, The University of Newcastle, AustraliaReviewed by:
Yiming Tu, Nanjing Medical University, ChinaCopyright © 2022 Yan, Tian, Chen, Tan, Cheng, Ye, Deng, Liu, Yuan, Zhang, Cai and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qianxue Chen, Y2hlbnF4NjY2QHdodS5lZHUuY24=; Linzhi Cai, Y2FpbGluemhpQHlhaG9vLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.