
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 29 September 2021
Sec. Cancer Molecular Targets and Therapeutics
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.757498
This article is part of the Research Topic Molecular Markers and Targeted Therapy for Hepatobiliary Tumors View all 51 articles
Background: Cholangiocarcinoma (CCA), including intrahepatic (iCCA), perihilar (pCCA), and distal (dCCA) CCA, is a highly aggressive malignancy originating from bile duct. The prognosis of CCA is very poor, and the biomarker study is unsatisfactory compared with other common cancers.
Materials and methods: In our study, we investigated the expression of dual-specificity phosphatase 11(DUSP11) in eight pairs of iCCAs, pCCAs, and dCCAs, and their corresponding tumor-adjacent tissues, as well as their tumor-adjacent tissues with qPCR. Moreover, we investigated the expression of DUSP11 in 174 cases of CCAs with immunohistochemistry, including 74 iCCAs, 64 pCCAs, and 36 dCCAs. We classified these patients into subsets with low and high expressions of DUSP11, and evaluated the correlations between the DUSP11 subsets and clinicopathological factors. With univariate and multivariate analyses, we assessed the correlation between DUSP11 and the overall survival (OS) rates in these CCA patients.
Results: In all the CCA subtypes, DUSP11 was elevated in CCAs compared with their paired adjacent tissues. In iCCA, pCCA, and dCCA, the percentages of DUSP11 high expression were 44.59%, 53.85%, and 55.56%, respectively. In iCCA, high DUSP11 expression was significantly associated with an advanced T stage and a poor prognosis. However, the prognostic value of DUSP11 in pCCA and dCCA was not significant. To decrease the statistical error caused by the small sample size of the dCCA cohort, we merged pCCA and dCCA into extracellular CCA (eCCA). In the 101 cases of eCCA, DUSP11 expression was also not significantly associated with the prognosis.
Conclusions: DUSP11 expression was associated with tumor infiltration and the OS rate in iCCA, but not in pCCA and dCCA. DUSP11 was an independent biomarker of iCCA indicating a poor prognosis. Our results suggested that a high expression of DUSP11 was a post-operational risk factor, and detecting DUSP11 could guide the individual treatment for patients with CCA.
Cholangiocarcinoma (CCA) is a highly aggressive malignancy with extremely poor prognoses. Anatomically, CCA is further classified as three subtypes including intrahepatic (iCCA), perihilar (pCCA), and distal (dCCA) CCA based on their origin in the biliary tree (1). pCCA is the most prevalent type of CCA accounting for about 50%–60% of the total CCA cases, while dCCA and iCCA accounted for 20%–30% and 10%, respectively (2). The motility and morbidity of CCA are increasing rapidly in the recent decades, but the treatment options have few progresses (3). China has the highest incidence of CCA worldwide, and whether Chinese CCA patients have special molecular features is still unknown. In general, the rate of radical surgical resection of CCA is quite low, and the treatment options of unresectable or advanced-stage CCA are an urgent need. CCAs usually have poor responses to the classical adjuvant therapies such as chemotherapy or radiotherapy. Till 2020, there emerged the first FDA-approved target drug of CCA, pemigtinib, which was used for CCA with FGFR2 fusion or mutation (4). Overall, the 5-year overall survival rate (OS) of CCA is very unsatisfactory, remaining approximately 30% after radical resection (5). More biomarkers of CCA should be investigated because biomarker discovery is the initiation to find new drug targets and new treatment therapy.
Protein phosphorylation is a key post-translational modification mainly regulated by serine/threonine phosphatase and tyrosine phosphatase. Protein tyrosine phosphatase (PTP) family participates in numerous processes such as signal transduction and cell proliferation, via dephosphorylating the phosphor-tyrosine of substrates (6). In humans, PTPs consist of 107 members and are divided into four main subgroups, which are type-I cysteine-based PTP (including classical PTPs and dual-specific phosphatase), type-II cysteine-based PTP, type-III cysteine-based phosphatases, and aspartic acid-based PTPs (7). These PTPs have different tissue specificities, substrates, and functions. Genetic and epigenetic alterations in the PTP genes can result in aberrant tyrosine phosphorylation, and consequently lead to diverse effects including an uncontrollable cell proliferation and tumorigenesis (8, 9). Interestingly, both the tumor suppressing role and oncogenic functions of PTPs have been showed in cancer, and the putative oncogenic or tumor suppressive functions of PTP are considered to rely on the cellular context.
Dual-specificity phosphatases (DUSPs) have a dephos-phorylating activity to both threonine/serine and tyrosine residues (10). There are 61 DUSPs out of the 107 PTP members, which have heterogeneous forms and functions, and are further classified based on the specific domains and sequence similarity. DUSP11, also known as PIR1 (phosphatase interacting with RNA and ribonucleoprotein 1), is a unique member of atypical DUSPs which could bind directly to RNA and possess RNA 5’-triphosphatase and diphosphatase activities (11, 12). DUSP11 converts the 5’ triphosphate of microRNA precursors to a 5’ monophosphate, and regulates cellular noncoding RNAs levels (12–14). In addition to a catalysis towards RNA, more evidence showed that DUSP11 could also dephosphorylate proteins. For example, DUSP11 could attenuate lipopolysaccharide-induced macrophage activation by targeting TGF-β-activated kinase 1 (15). DUSP11 was considered to participate in cancer progression by several previous studies (16, 17), but its functions and regulation mechanisms in cancer are still unclear to date. The tissue specificity and functions in tumor of DUSP11 have not been well studied.
In our study, we investigated the expression of DUSP11 in 174 cases of CCAs, including 74 iCCAs, 64 pCCAs, and 36 dCCAs. Moreover, we classified the patients into subsets with low and high expressions of DUSP11, and evaluated the clinicopathological factors in these subsets. With univariate and multivariate analyses, we assessed the correlation between DUSP11 and the OS rate in 74 iCCAs, 64 pCCAs, and 36 dCCAs.
A total of 258 patients were diagnosed with CCA in Chinese Academy of Medical Sciences and Peking Union Medical College and National Cancer Center Shenzhen Hospital from 2009 to 2016, which formed the primary cohort. A total of 174 cases of CCAs, including 74 iCCAs, 64 pCCAs, and 36 dCCAs, were selected from the primary cohort into the validation cohort, if they followed the criteria: (1) radical surgery with clear surgical margin was performed; (2) available formalin-fixed tumor tissues for IHC; (3) available follow-ups more than 3 months and complete medical records; and (4) no history of other malignancies. All samples were obtained with a prior consent from patients. The study was approved and supervised by the Ethics Committee of Chinese Academy of Medical Sciences and Peking Union Medical College and National Cancer Center Shenzhen Hospital.
A total of eight consecutive iCCAs, pCCAs, and dCCAs, and their corresponding tumor-adjacent tissues were collected for qPCR. Total mRNA was extracted from the frozen tissues using the TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA), and then converted into cDNA using the ReverTra Ace qPCR RT kit (TOYOBO, Japan). Quantitative real-time PCR was performed using the SYBR Green Master (Roche, USA) and Light Cycler Roche 480 PCR instrument. The mRNA level was standardized with the 2-ΔΔCt method by normalization to GAPDH. The primer sequences were as follows:
DUSP11, forward:5’-GGCTGCCGAGTCTTTTCCT-3’,Reverse5’-TTTCCACCTTTCGGGGATGTG-3’; GAPDH, forward:5’-GGAGCGAGATCCCTCCAAAAT-3’, reverse: 5’-GGCTGTTGTCATACTTCTCATGG-3’.
Immunohistochemistry was performed with a streptavidin peroxidase complex method. Briefly, the paraffin-embedded tissues were deparaffinized and rehydrated with xylene and graded alcohol. To inactivate the endogenous peroxidase, 3% hydrogen peroxide was used, and then, the slides were incubated in a citrate buffer (pH = 6.0) for the optimal antigen retrieval. The unspecific binding was blocked by incubation in 1% bovine serum albumin for 30 minutes. Primary antibody of DUSP11 (Santa Cruz Biotechnology, catalog: sc-393220) was used to incubate the tissues at 4°C overnight. Phosphate buffered saline was used to rinse the slides three times, and secondary antibodies labeled with streptavidin-biotin-peroxidase reagent were used to incubate the tissues for 1 hour. After that, slides were treated with the 3,3’-diaminobenzidine solution for 10 minutes for visualization. Slides were counterstained with hematoxylin and mounted at last.
The IHC results were semi-quantified by two independent pathologists who were unaware of the clinical data. The final IHC scores was evaluated as the scores of the percentage of positive-stained cells multiplied by the scores of staining intensity. In brief, the scores of staining intensity were defined as: score 0 for negative staining, score 1 for weak staining, score 2 for moderate staining, and score 3 for strong staining. The scores for positive-stained cells were set as follows: score 1 for <25% of positive cells; score 2 for 25%–50% of positive cells; score 3 for 50%–75% of positive cells; and score 4 for 75%–100% of positive cells. The final IHC score ranged from from score 0 to 12, and was divided into subsets with different DUSP11 expression according to the cut-off, which was defined by the receiver operating characteristic (ROC) curve.
SPSS 25.0 (IBM, Chicago, IL, USA) and GraphPad prism 5.0 software (California Resources Corporation, Los Angeles,CA, USA) were used for statistical analyses. The chi-square test was used to analyze the correlations between DUSP11 and the clinicopathological factors. The univariate analysis was analyzed with the log-rank test, and the survival curves were plotted with the Kaplan-Meier method. The Cox proportional hazards regression model was applied to identify the independent prognostic factors. P-values less than 0.05 in all experiments were considered statistically significant.
The expression of DUSP11 was detected with qPCR in eight pairs of iCCAs, pCCAs, and dCCAs, as well as their corresponding tumor-adjacent tissues (Figure 1A). In these tissues, DUSP11 expressions in iCCAs, pCCAs, and dCCAs were substantially higher than those in their paired adjacent tissues. To better depict the expression of DUSP11 in CCA, the DUSP11 expression was investigated by IHC in 174 cases of CCAs, including 74 iCCAs, 64 pCCAs, and 36 dCCAs. In consistent with the DUSP11 function as a phosphatase toward phosho-RNA, the intracellular localization of DUSP11 was in the cell nucleus in CCA (Figure 1B). In iCCA, pCCA, and dCCA, the percentages of DUSP11 high expression were 44.59%, 53.85%, and 55.56%, respectively (Table 1). The basic information of CCA patients is shown in Table 1, including the sex, age, tumor size, differentiation, and T/N/M/TNM stage of the patients. The results of basic patients characters were consistent with previous studies (5, 18), supporting the validation of our cohort.
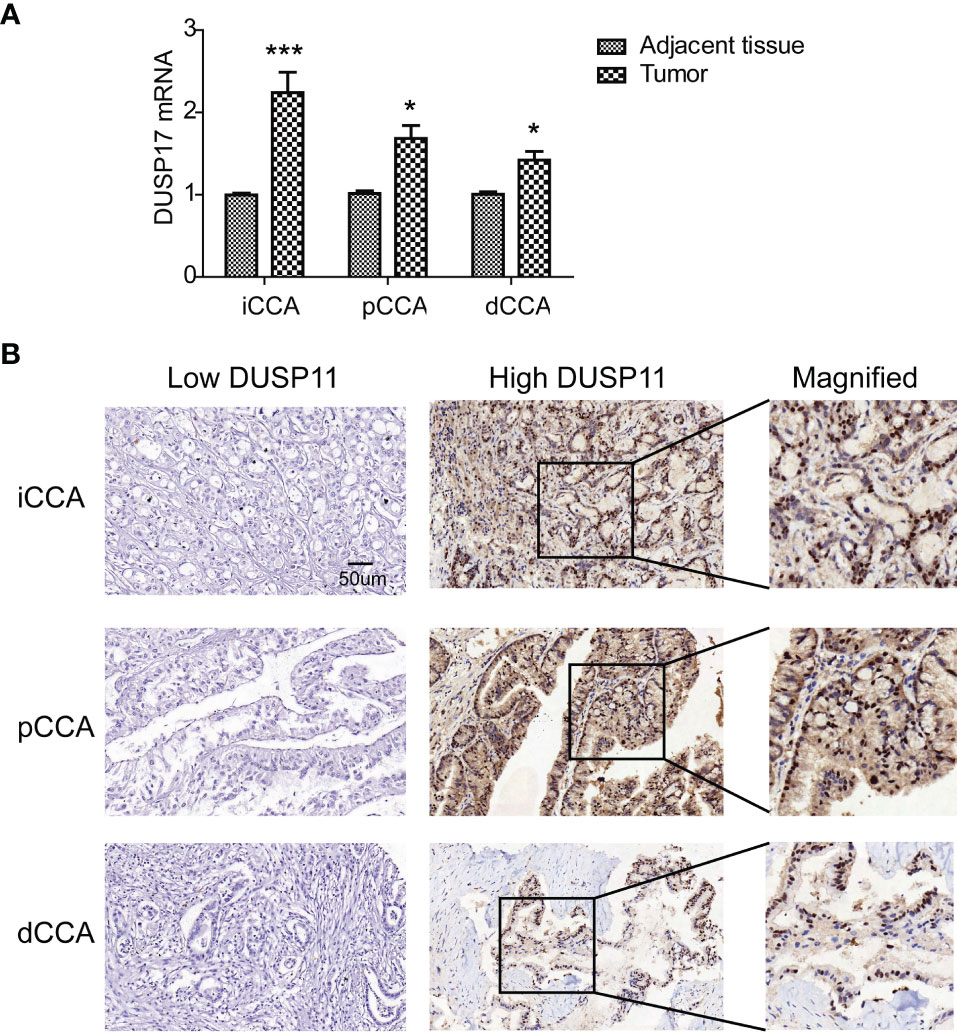
Figure 1 Expression of DUSP11 in CCA tissues. (A) The expression of DUSP11 was detected with qPCR in eight pairs of iCCAs, pCCAs, and dCCAs, as well as their corresponding tumor-adjacent tissues. * represents P < 0.05 and *** represents P < 0.0001, calculated with paired t-test. (B) DUSP11 expression was investigated in 174 cases of CCAs, including 74 iCCAs, 64 pCCAs, and 36 dCCAs, by IHC. Representative images of low and high DUSP11 expressions were displayed.
To screen the potential clinicopathological variables which may be associated with DUSP11 expression, we analyzed the correlation between the clinicopathological variables and DUSP11 with the chi-square test (Table 2). In iCCA, DUSP11 expression was significantly associated with the T stage. High expression of DUSP11 was positively correlated with an advanced T stage (P = 0.008), indicating that DUSP11 may be an attributor to iCCA infiltration. In pCCA and dCCA, no clinicopathological variables exhibited a significant correlation with DUSP11 expression.
We performed univariate analysis to evaluate the prognostic significance of DUSP11 and other clinicopathological factors in CCA. All clinicopathological factors and DUSP11 expression were enrolled into the univariate analysis. The Kaplan-Meier method was used to plot the OS curves, and the log-rank test was used to analyze the statistical difference between the subgroups (Table 3).
In iCCA, a high DUSP11 expression was significantly associated with a low OS rate (P = 0.002). The 5-year OS rates of patients with low and high DUSP11 were 52.6% and 9.4%, respectively (Figure 2A). However, the prognostic significance of DUSP11 in pCCA and dCCA was not remarkable (P = 0.354 and 0.459, respectively), though pCCA and dCCA patients with a high DUSP11 expression seemed to have poorer prognoses compared with those with a low DUSP11 expression (25.2% vs. 47.5% in pCCA, 45.9% vs. 50.0% in dCCA) (Figures 2B, C).
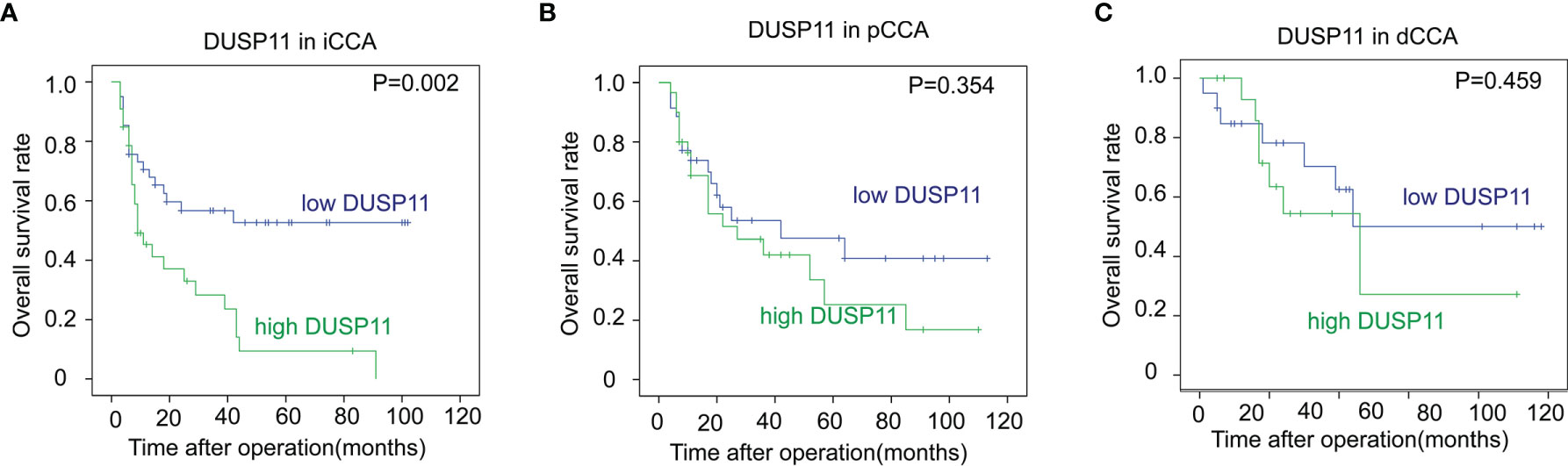
Figure 2 The correlations between DUSP11 expression and the OS rates of CCA. (A–C) The survival curves of low and high expressions of DUSP11 in iCCA (A), pCCA (B), and dCCA (C) were plotted by the Kaplan-Meier method, and the statistical significance was analyzed with the log-rank test.
In iCCA, a large tumor size and advanced N stage and M stage, representing positive lymphatic invasion and distant metastasis, were also indicators of a poor prognosis (P = 0.007, 0.010 and 0.010 respectively) (Figures 3A–C). In addition, an advanced TNM stage was associated with the unfavorable outcome of iCCA as well (P = 0.049) (Figure 3D). In pCCA, poor differentiation was a notable indicator for a poor prognosis (P < 0.001) (Figure 4A). Moreover, advanced T stage, N stage, and M stage were also associated with a poor prognosis of pCCA (P = 0.004, 0.006, and 0.012, respectively) (Figures 4B–D). As expected, patients in an advanced TNM stage had a much poorer outcome than those in an early TNM stage (P = 0.049) (Figure 4E). In dCCA, no factors were defined to be associated with the OS time, which may be attributed to the small number of patients (n = 36).
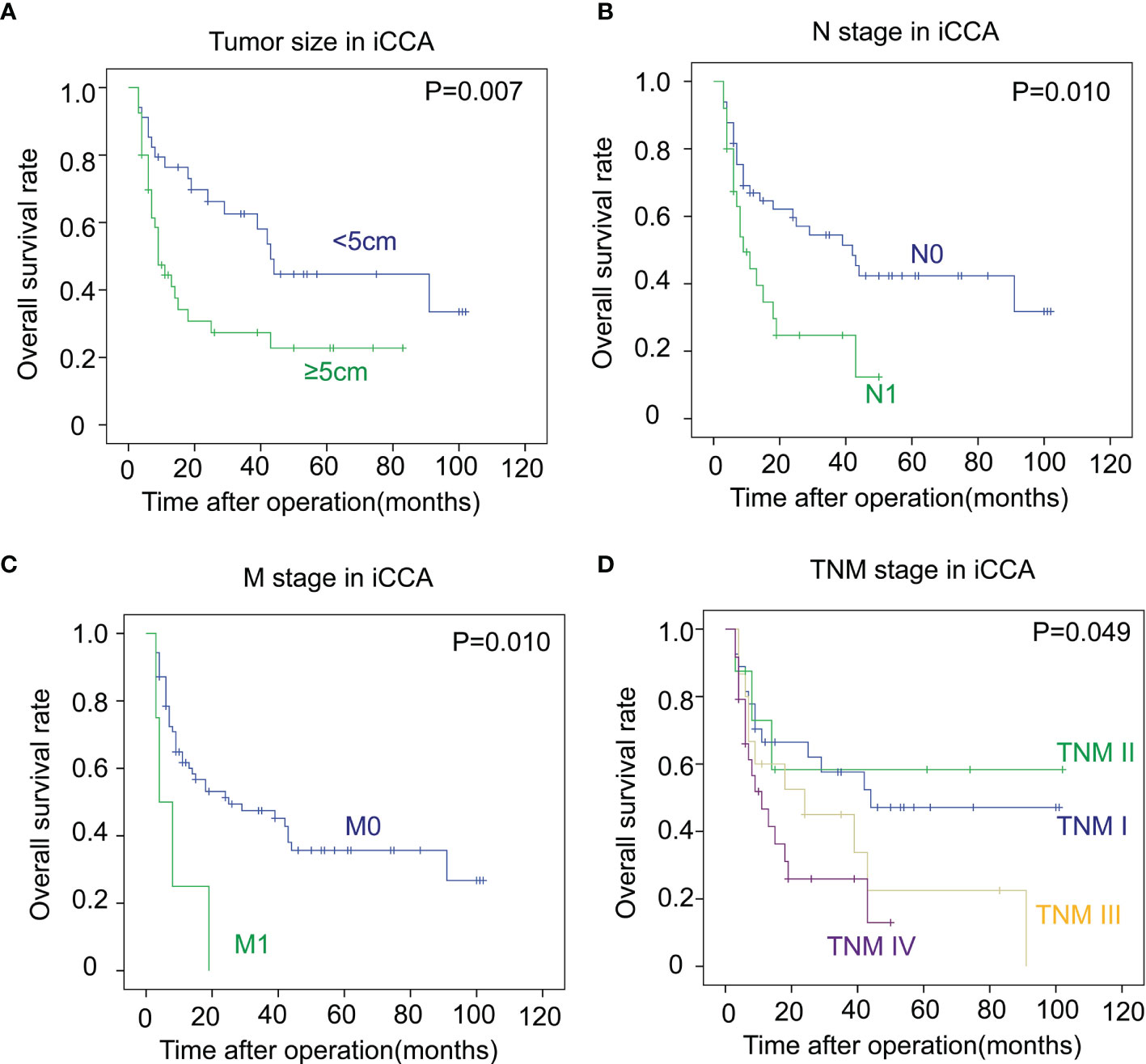
Figure 3 The correlations between the clinicopathological factors and OS rates in iCCA. (A–D) In iCCA, the survival curves of tumor size (A), N stage (B), M stage (C), and TNM stage (D) were plotted by the Kaplan-Meier method, and the statistical significance was analyzed with the log-rank test.
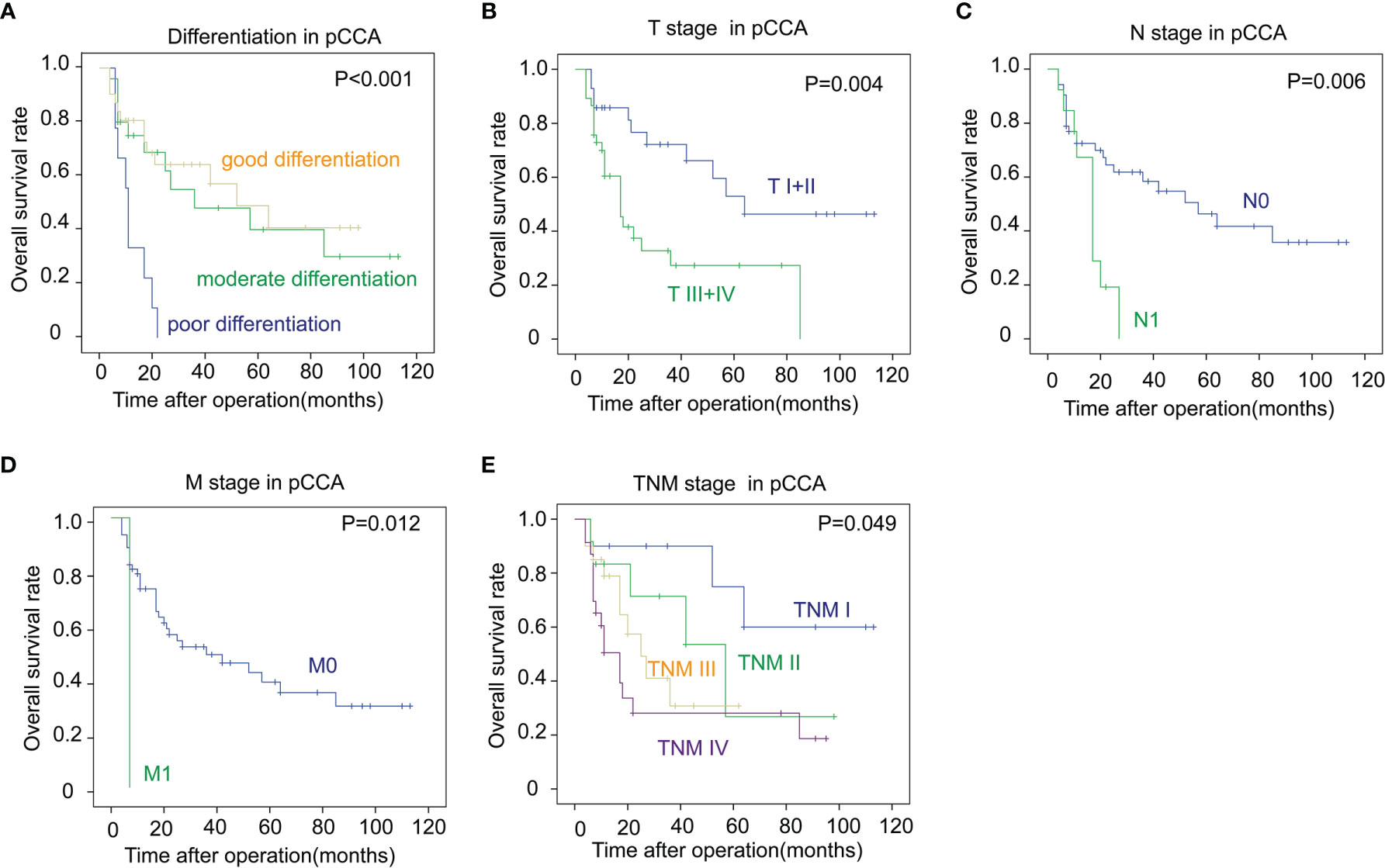
Figure 4 The correlations between the clinicopathological factors and OS rates in pCCA. (A–E) Patients with pCCA were stratified according to the tumor differentiation (A), T stage (B), N stage (C), M stage (D), and TNM stage (E), and the correlations between these factors and the OS rates were analyzed with the log-rank test.
All the clinicopathological factors were enrolled into the Cox-regression hazard model for multivariate analysis (Table 4). In iCCA, DUSP11 was identified as an independent prognostic biomarker (P = 0.022, 95% confidence interval = 1.13–4.83). The hazard ratio (HR) of a high DUSP11 was 2.33, representing that patients with a high DUSP11 expression had a 2.33-fold time more than patients with a low DUSP11. However, the independent prognostic significance of DUSP11 in pCCA and dCCA was not significant (P = 0.993 and 0.640, respectively).
To eliminate the effects of a small sample size towards the statistical significance, we merged pCCA and dCCA to extrahepatic CCA (eCCA), and performed the univariate analysis. In the 101 cases of eCCA, the prognostic significance of DUSP11 was still not remarkable (P = 0.241), but there existed a trend that a high DUSP11 expression seemed to correlate a low OS rate (Table 5). The 5-year OS rates of low and high DUSP11 were 48.7% and 26.0%, respectively (Figure 5A). Moreover, poor differentiation (P = 0.016), and advanced T stage (P = 0.043), M stage (P = 0.003), and TNM stage (P = 0.012) were all indicators for an unfavorable prognosis of eCCA (Figures 5B–E).
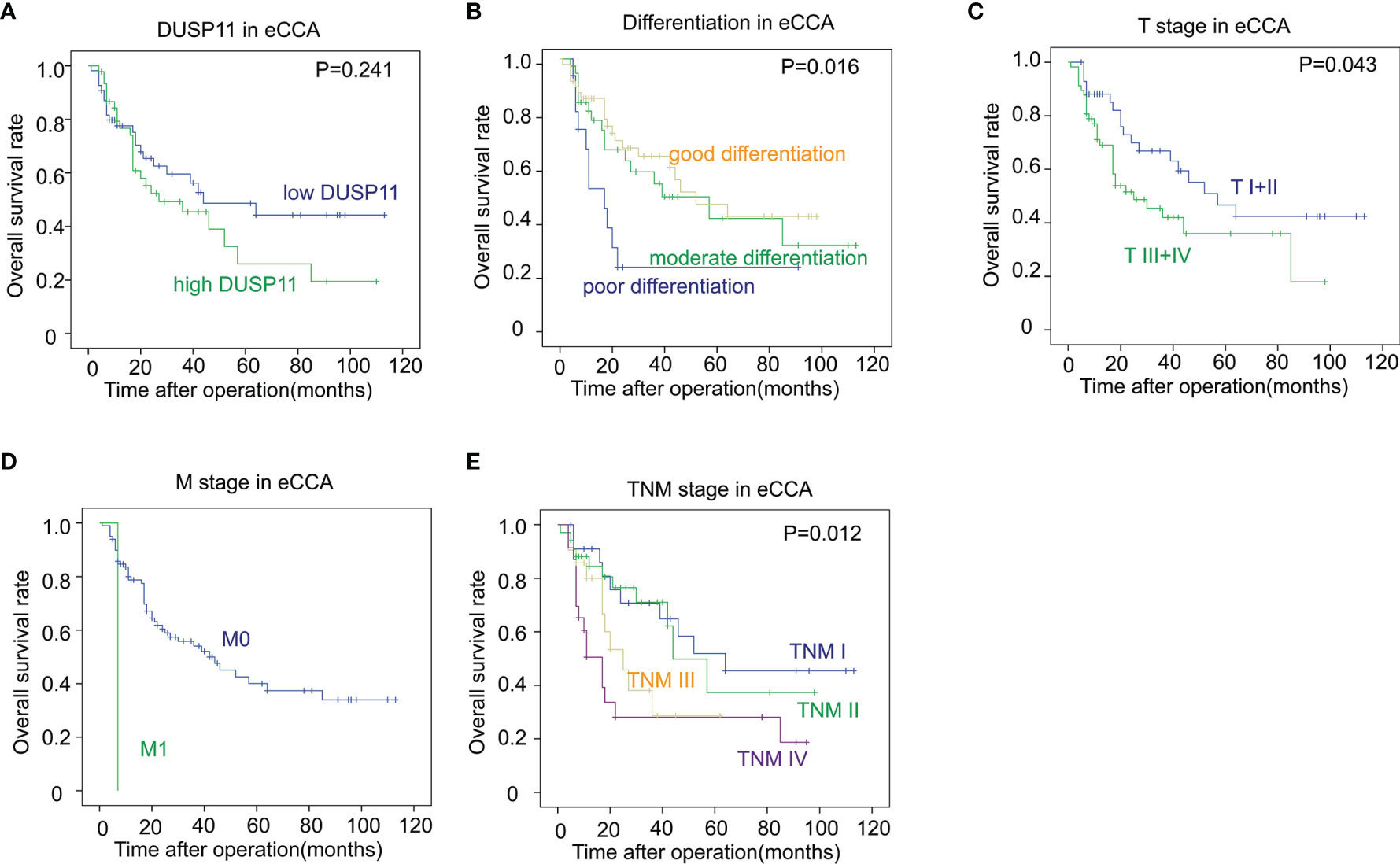
Figure 5 The correlations between DUSP11, clinicopathological factors, and the OS rates in eCCA. (A–E) In the 101 cases of eCCA, the associations between the OS rate and DUSP11 expression (A) or clinicopathological factors including tumor differentiation (B), T stage (C), M stage (D), and TNM stage (E) were analyzed with the log-rank test.
Compared with other common cancer types such as gastric cancer or lung cancer, CCA is characterized by its low rate of radical resection because of the specificity of the hepatic portal (19, 20). The low rate of radical surgery increases the difficulty of specimen obtainment and establishment of a large cohort (20, 21), which is the basement of biomarker identification and new drug target. However, more prognostic biomarkers of CCA are an urgent need to select the high-risk patients and help formulate the precise treatment. Our study collected a total of 174 cases of CCAs, which was a relatively large CCA cohort. We demonstrated for the first time that DUSP11 was an independent prognostic biomarker in CCA, suggesting that a high expression of DUSP11 was a post-operational risk and detecting DUSP11 could guide the individual treatment for patients with CCA.
In the seventh edition of AJCC/UICC in 2007, pCCA and dCCA were separated from eCCA and regarded as distinct subtypes (22). iCCA, pCCA, and dCCA have different morbidities, clinical characteristics, treatment strategies, and prognosis, but whether they have different biological features is still controversial (23). In some occasions, iCCA and eCCA have the same biomarkers such as EGFR and HER2 (24, 25), but several biomarkers exhibited a different prognostic significance in iCCA and eCCA (21, 26). In this study, we demonstrated that DUSP11 expression was correlated with a poor prognosis in iCCA but not eCCA. This result further supported that iCCA and eCCA are two distinct cancer types, which have different biological factors and biomarkers.
In the era of high-throughput sequencing, numerous genetic alterations, such as the mutation, deletion, duplication, or translocation of PTP genes, are reported to be linked with diverse cancer phenotypes (27). The underlying mechanisms of the tumor suppressor or oncogenic role of PTPs in tumorigenesis or tumor progression are not fully understood. The loss or genetic alterations of several PTPs are shown to promote tumorigenesis, proliferation, and metastasis in in vitro and in vivo models, and these PTPs are generally considered to be tumor suppressors, including PTEN in prostate and breast cancer (28–30), SHP1 in leukemia and lymphomas (31, 32), PTPRF in colon, breast, and lung cancer (33, 34), and DUSP4 in breast, pancreas, and thyroid cancer (35–38). On the other hand, several tumor PTPs were identified as tumor suppressors because their genetic variations or loss facilitate tumorigenesis and tumor progression. For example, SHP2 was considered as an oncogene in breast cancer, leukemia, and gliomas (39–43), and PTP4A3 is an oncogene in breast, gastric, and colon cancer (44–46). The studies of DUSP11 in cancer are very few and the role of DUSP11 in tumor progression is nearly in vacancy. This is the first study reporting a definite role of DUSP11 as an oncogene because it is significantly associated with iCCA prognosis. Our results expand the understanding of the role of the DUSP family in cancer, and indicates DUSP11 as a potential drug target of CCA.
Although PTPs are attracting more and more attention as an onco-protein or a tumor suppressor, the improvement of PTP inhibitors as specific inhibitors or target drugs remains challenging. To obtain the specific small-molecule inhibitors are difficult because the catalytic domains of PTPs are very conserved. Till now, only a small proportion of PTPs have specific inhibitors, such as PTP1B, SHP2, and PTPN9 (47–50). However, the interacting proteins, substrates, and molecular catalytic mechanism of DUSP11 are poorly understood, and there is still no available inhibitor of DUSP11. More studies on the expression profile of DUSP11 in cancer would help improve the therapeutic use of its potential inhibitor. We showed the clinical relevance of DUSP11 in CCA and suggested that the inhibitor towards DUSP11 may be a potential therapeutic strategy to CCA.
In conclusion, we, for the first time, investigated the expression of DUSP11 in 174 cases of CCAs, including 74 iCCAs, 64 pCCAs, and 36 dCCAs, and evaluated the clinical significance of DUSP11 by assessing DUSP11 correlation between the clinicopathological factors and prognosis. As a result, we demonstrated that DUSP11 expression was associated with tumor infiltration and the OS rate in iCCA, but not in pCCA or dCCA. DUSP11 was an independent biomarker of iCCA, indicating a poor prognosis. Our results suggested that a high expression of DUSP11 was a post-operational risk factor, and detecting DUSP11 could guide the individual treatment for patients with CCA.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the Ethics Committee of Chinese Academy of Medical Sciences and Peking Union Medical College and National Cancer Center Shenzhen Hospital. The patients/participants provided their written informed consent to participate in this study.
Concept and design: XC. Administrative support: LX and PW. Specimen collection: XC, LX, PW, WZ, WL, and TL. Collection and assembly of data: XC, LX, PW, WZ, WL, and TL. Data analysis and interpretation: LX and PW. All authors contributed to the article and approved the submitted version.
The study is supported by Sanming Project of Medicine in Shenzhen (No. SZSM202011010) and Sanming Project of Medicine in Shenzhen (No. SZSM201911008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Rizvi S, Gores GJ. Emerging Molecular Therapeutic Targets for Cholangiocarcinoma. J Hepatol (2017) 67:632–44. doi: 10.1016/j.jhep.2017.03.026
2. Blechacz B. Cholangiocarcinoma: Current Knowledge and New Developments. Gut liver (2017) 11:13–26. doi: 10.5009/gnl15568
3. Sun R, Liu Z, Qiu B, Chen T, Li Z, Zhang X, et al. Annexin10 Promotes Extrahepatic Cholangiocarcinoma Metastasis by Facilitating EMT via PLA2G4A/PGE2/STAT3 Pathway. EBioMedicine (2019) 47:142–55. doi: 10.1016/j.ebiom.2019.08.062
4. Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, et al. Pemigatinib for Previously Treated, Locally Advanced or Metastatic Cholangiocarcinoma: A Multicentre, Open-Label, Phase 2 Study. Lancet Oncol (2020) 21:671–84. doi: 10.1016/S1470-2045(20)30109-1
5. Xu YF, Yang XQ, Lu XF, Guo S, Liu Y, Iqbal M, et al. Fibroblast Growth Factor Receptor 4 Promotes Progression and Correlates to Poor Prognosis in Cholangiocarcinoma. Biochem Biophys Res Commun (2014) 446:54–60. doi: 10.1016/j.bbrc.2014.02.050
6. Barr AJ, Ugochukwu E, Lee WH, King ON, Filippakopoulos P, Alfano I, et al. Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome. Cell (2009) 136:352–63. doi: 10.1016/j.cell.2008.11.038
7. Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, et al. Protein Tyrosine Phosphatases in the Human Genome. Cell (2004) 117:699–711. doi: 10.1016/j.cell.2004.05.018
8. Julien SG, Dube N, Hardy S, Tremblay ML. Inside the Human Cancer Tyrosine Phosphatome. Nat Rev Cancer (2011) 11:35–49. doi: 10.1038/nrc2980
9. Wang HM, Xu YF, Ning SL, Yang DX, Li Y, Du YJ, et al. The Catalytic Region and PEST Domain of PTPN18 Distinctly Regulate the HER2 Phosphorylation and Ubiquitination Barcodes. Cell Res (2014) 24:1067–90. doi: 10.1038/cr.2014.99
10. Patterson KI, Brummer T, O’Brien PM, Daly RJ. Dual-Specificity Phosphatases: Critical Regulators With Diverse Cellular Targets. Biochem J (2009) 418:475–89. doi: 10.1042/BJ20082234
11. Deshpande T, Takagi T, Hao L, Buratowski S, Charbonneau H. Human PIR1 of the Protein-Tyrosine Phosphatase Superfamily has RNA 5’-Triphosphatase and Diphosphatase Activities. J Biol Chem (1999) 274:16590–4. doi: 10.1074/jbc.274.23.16590
12. Yuan Y, Li DM, Sun H. PIR1, a Novel Phosphatase That Exhibits High Affinity to RNA . Ribonucleoprotein Complexes. J Biol Chem (1998) 273:20347–53. doi: 10.1074/jbc.273.32.20347
13. Burke JM, Kincaid RP, Nottingham RM, Lambowitz AM, Sullivan CS. DUSP11 Activity on Triphosphorylated Transcripts Promotes Argonaute Association With Noncanonical Viral microRNAs and Regulates Steady-State Levels of Cellular Noncoding RNAs. Genes Dev (2016) 30:2076–92. doi: 10.1101/gad.282616.116
14. Burke JM, Sullivan CS. DUSP11 - An RNA Phosphatase That Regulates Host and Viral Non-Coding RNAs in Mammalian Cells. RNA Biol (2017) 14:1457–65. doi: 10.1080/15476286.2017.1306169
15. Yang CY, Chuang HC, Tsai CY, Xiao YZ, Yang JY, Huang RH, et al. DUSP11 Attenuates Lipopolysaccharide-Induced Macrophage Activation by Targeting Tak1. J Immunol (2020) 205:1644–52. doi: 10.4049/jimmunol.2000334
16. Cai Y, Wu Q, Liu Y, Wang J. AZIN1-AS1, A Novel Oncogenic LncRNA, Promotes the Progression of Non-Small Cell Lung Cancer by Regulating MiR-513b-5p and DUSP11. OncoTargets Ther (2020) 13:9667–78. doi: 10.2147/OTT.S261497
17. Caprara G, Zamponi R, Melixetian M, Helin K. Isolation and Characterization of DUSP11, a Novel P53 Target Gene. J Cell Mol Med (2009) 13:2158–70. doi: 10.1111/j.1582-4934.2008.00616.x
18. Chen T, Li K, Liu Z, Liu J, Wang Y, Sun R, et al. WDR5 Facilitates EMT and Metastasis of CCA by Increasing HIF-1alpha Accumulation in Myc-Dependent and Independent Pathways. Mol Ther J Am Soc Gene Ther (2021) 29:2134–50. doi: 10.1016/j.ymthe.2021.02.017
19. DeOliveira ML, Cunningham SC, Cameron JL, Kamangar F, Winter JM, Lillemoe KD, et al. Cholangiocarcinoma: Thirty-One-Year Experience With 564 Patients at a Single Institution. Ann Surg (2007) 245:755–62. doi: 10.1097/01.sla.0000251366.62632.d3
20. Liu Z, Sun R, Zhang X, Qiu B, Chen T, Li Z, et al. Transcription Factor 7 Promotes the Progression of Perihilar Cholangiocarcinoma by Inducing the Transcription of C-Myc and FOS-Like Antigen 1. EBioMedicine (2019) 45:181–91. doi: 10.1016/j.ebiom.2019.06.023
21. Li Z, Liu J, Chen T, Sun R, Liu Z, Qiu B, et al. HMGA1-TRIP13 Axis Promotes Stemness and Epithelial Mesenchymal Transition of Perihilar Cholangiocarcinoma in a Positive Feedback Loop Dependent on C-Myc. J Exp Clin Cancer Res CR (2021) 40:86. doi: 10.1186/s13046-021-01890-1
22. Farges O, Fuks D, Le Treut YP, Azoulay D, Laurent A, Bachellier P, et al. AJCC 7th Edition of TNM Staging Accurately Discriminates Outcomes of Patients With Resectable Intrahepatic Cholangiocarcinoma: By the AFC-IHCC-2009 Study Group. Cancer (2011) 117:2170–7. doi: 10.1002/cncr.25712
23. Xu YF, Liu ZL, Pan C, Yang XQ, Ning SL, Liu HD, et al. HMGB1 Correlates With Angiogenesis and Poor Prognosis of Perihilar Cholangiocarcinoma via Elevating VEGFR2 of Vessel Endothelium. Oncogene (2019) 38:868–80. doi: 10.1038/s41388-018-0485-8
24. Claperon A, Mergey M, Nguyen Ho-Bouldoires TH, Vignjevic D, Wendum D, Chretien Y, et al. EGF/EGFR Axis Contributes to the Progression of Cholangiocarcinoma Through the Induction of an Epithelial-Mesenchymal Transition. J Hepatol (2014) 61:325–32. doi: 10.1016/j.jhep.2014.03.033
25. Pellat A, Vaquero J, Fouassier L. Role of ErbB/HER Family of Receptor Tyrosine Kinases in Cholangiocyte Biology. Hepatology (2018) 67:762–73. doi: 10.1002/hep.29350
26. Qiu B, Chen T, Sun R, Liu Z, Zhang X, Li Z, et al. Sprouty4 Correlates With Favorable Prognosis in Perihilar Cholangiocarcinoma by Blocking the FGFR-ERK Signaling Pathway and Arresting the Cell Cycle. EBioMedicine (2019) 50:166–77. doi: 10.1016/j.ebiom.2019.11.021
27. Andersen JN, Jansen PG, Echwald SM, Mortensen OH, Fukada T, Del Vecchio R, et al. A Genomic Perspective on Protein Tyrosine Phosphatases: Gene Structure, Pseudogenes, and Genetic Disease Linkage. FASEB J Off Publ Fed Am Soc Exp Biol (2004) 18:8–30. doi: 10.1096/fj.02-1212rev
28. Hollander MC, Blumenthal GM, Dennis PA. PTEN Loss in the Continuum of Common Cancers, Rare Syndromes and Mouse Models. Nat Rev Cancer (2011) 11:289–301. doi: 10.1038/nrc3037
29. Kinross KM, Montgomery KG, Kleinschmidt M, Waring P, Ivetac I, Tikoo A, et al. An Activating Pik3ca Mutation Coupled With Pten Loss Is Sufficient to Initiate Ovarian Tumorigenesis in Mice. J Clin Invest (2012) 122:553–7. doi: 10.1172/JCI59309
30. Petrocelli T, Slingerland JM. PTEN Deficiency: A Role in Mammary Carcinogenesis. Breast Cancer Res BCR (2001) 3:356–60. doi: 10.1186/bcr322
31. Cheng J, Zhang D, Zhou C, Marasco WA. Down-Regulation of SHP1 and Up-Regulation of Negative Regulators of JAK/STAT Signaling in HTLV-1 Transformed Cell Lines and Freshly Transformed Human Peripheral Blood CD4+ T-Cells. Leukemia Res (2004) 28:71–82. doi: 10.1016/S0145-2126(03)00158-9
32. Oka T, Ouchida M, Koyama M, Ogama Y, Takada S, Nakatani Y, et al. Gene Silencing of the Tyrosine Phosphatase SHP1 Gene by Aberrant Methylation in Leukemias/Lymphomas. Cancer Res (2002) 62:6390–4.
33. Shimozato O, Waraya M, Nakashima K, Souda H, Takiguchi N, Yamamoto H, et al. Receptor-Type Protein Tyrosine Phosphatase Kappa Directly Dephosphorylates CD133 and Regulates Downstream AKT Activation. Oncogene (2015) 34:1949–60. doi: 10.1038/onc.2014.141
34. Stevenson WS, Best OG, Przybylla A, Chen Q, Singh N, Koleth M, et al. DNA Methylation of Membrane-Bound Tyrosine Phosphatase Genes in Acute Lymphoblastic Leukaemia. Leukemia (2014) 28:787–93. doi: 10.1038/leu.2013.270
35. Balko JM, Cook RS, Vaught DB, Kuba MG, Miller TW, Bhola NE, et al. Profiling of Residual Breast Cancers After Neoadjuvant Chemotherapy Identifies DUSP4 Deficiency as a Mechanism of Drug Resistance. Nat Med (2012) 18:1052–9. doi: 10.1038/nm.2795
36. Balko JM, Schwarz LJ, Bhola NE, Kurupi R, Owens P, Miller TW, et al. Activation of MAPK Pathways Due to DUSP4 Loss Promotes Cancer Stem Cell-Like Phenotypes in Basal-Like Breast Cancer. Cancer Res (2013) 73:6346–58. doi: 10.1158/0008-5472.CAN-13-1385
37. Hijiya N, Tsukamoto Y, Nakada C, Tung Nguyen L, Kai T, Matsuura K, et al. Genomic Loss of DUSP4 Contributes to the Progression of Intraepithelial Neoplasm of Pancreas to Invasive Carcinoma. Cancer Res (2016) 76:2612–25. doi: 10.1158/0008-5472.CAN-15-1846
38. Mazumdar A, Poage GM, Shepherd J, Tsimelzon A, Hartman ZC, Den Hollander P, et al. Analysis of Phosphatases in ER-Negative Breast Cancers Identifies DUSP4 as a Critical Regulator of Growth and Invasion. Breast Cancer Res Treat (2016) 158:441–54. doi: 10.1007/s10549-016-3892-y
39. Bunda S, Burrell K, Heir P, Zeng L, Alamsahebpour A, Kano Y, et al. Inhibition of SHP2-Mediated Dephosphorylation of Ras Suppresses Oncogenesis. Nat Commun (2015) 6:8859. doi: 10.1038/ncomms9859
40. Dance M, Montagner A, Salles JP, Yart A, Raynal P. The Molecular Functions of Shp2 in the Ras/Mitogen-Activated Protein Kinase (ERK1/2) Pathway. Cell Signalling (2008) 20:453–9. doi: 10.1016/j.cellsig.2007.10.002
41. Xu R, Yu Y, Zheng S, Zhao X, Dong Q, He Z, et al. Overexpression of Shp2 Tyrosine Phosphatase Is Implicated in Leukemogenesis in Adult Human Leukemia. Blood (2005) 106:3142–9. doi: 10.1182/blood-2004-10-4057
42. Zhou X, Coad J, Ducatman B, Agazie YM. SHP2 Is Up-Regulated in Breast Cancer Cells and in Infiltrating Ductal Carcinoma of the Breast, Implying Its Involvement in Breast Oncogenesis. Histopathology (2008) 53:389–402. doi: 10.1111/j.1365-2559.2008.03103.x
43. Zhou XD, Agazie YM. Inhibition of SHP2 Leads to Mesenchymal to Epithelial Transition in Breast Cancer Cells. Cell Death Differ (2008) 15:988–96. doi: 10.1038/cdd.2008.54
44. Cramer JM, Zimmerman MW, Thompson T, Homanics GE, Lazo JS, Lagasse E. Deletion of Ptp4a3 Reduces Clonogenicity and Tumor-Initiation Ability of Colitis-Associated Cancer Cells in Mice. Stem Cell Res (2014) 13:164–71. doi: 10.1016/j.scr.2014.05.004
45. Fiordalisi JJ, Dewar BJ, Graves LM, Madigan JP, Cox AD. Src-Mediated Phosphorylation of the Tyrosine Phosphatase PRL-3 Is Required for PRL-3 Promotion of Rho Activation, Motility and Invasion. PloS One (2013) 8:e64309. doi: 10.1371/journal.pone.0064309
46. Walls CD, Iliuk A, Bai Y, Wang M, Tao WA, Zhang ZY. Phosphatase of Regenerating Liver 3 (PRL3) Provokes a Tyrosine Phosphoproteome to Drive Prometastatic Signal Transduction. Mol Cell Proteomics MCP (2013) 12:3759–77. doi: 10.1074/mcp.M113.028886
47. Erbe DV, Wang S, Zhang YL, Harding K, Kung L, Tam M, et al. Ertiprotafib Improves Glycemic Control and Lowers Lipids via Multiple Mechanisms. Mol Pharmacol (2005) 67:69–77. doi: 10.1124/mol.104.005553
48. Lantz KA, Hart SG, Planey SL, Roitman MF, Ruiz-White IA, Wolfe HR, et al. Inhibition of PTP1B by Trodusquemine (MSI-1436) Causes Fat-Specific Weight Loss in Diet-Induced Obese Mice. Obes (Silver Spring) (2010) 18:1516–23. doi: 10.1038/oby.2009.444
49. Xu YF, Chen X, Yang Z, Xiao P, Liu CH, Li KS, et al. PTP-MEG2 Regulates Quantal Size and Fusion Pore Opening Through Two Distinct Structural Bases and Substrates. EMBO Rep (2021) 22:e52141. doi: 10.15252/embr.202052141
Keywords: DUSP11, prognosis, biomarker, intrahepatic cholangiocarcinoma, extrahepatic cholangiocarcinoma
Citation: Xu L, Wang P, Zhang W, Li W, Liu T and Che X (2021) Dual-Specificity Phosphatase 11 Is a Prognostic Biomarker of Intrahepatic Cholangiocarcinoma. Front. Oncol. 11:757498. doi: 10.3389/fonc.2021.757498
Received: 12 August 2021; Accepted: 31 August 2021;
Published: 29 September 2021.
Edited by:
Yunfei Xu, Shandong University, ChinaReviewed by:
Lanping Wang, Shandong First Medical University, ChinaCopyright © 2021 Xu, Wang, Zhang, Li, Liu and Che. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Che, Y2hleHUwMTkyQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.