
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 19 October 2021
Sec. Genitourinary Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.753920
This article is part of the Research Topic Precision Medicine and Translational Research in Urological Oncology View all 11 articles
 Xiaoling Lin1,2†
Xiaoling Lin1,2† Yishuo Wu1,2†
Yishuo Wu1,2† Fang Liu1,2
Fang Liu1,2 Rong Na3
Rong Na3 Da Huang3
Da Huang3 Danfeng Xu3
Danfeng Xu3 Jian Gong1,2
Jian Gong1,2 Yao Zhu4
Yao Zhu4 Bo Dai4
Bo Dai4 Dingwei Ye4
Dingwei Ye4 Hongjie Yu5
Hongjie Yu5 Haowen Jiang1,2
Haowen Jiang1,2 Zujun Fang1,2
Zujun Fang1,2 Jie Zheng1,2*
Jie Zheng1,2* Qiang Ding1,2*
Qiang Ding1,2*Introduction: The clinical performance of [–2]proPSA (p2PSA) and its derivatives in predicting the presence and aggressiveness of prostate cancer (PCa) has been well evaluated in prostate biopsy patients. However, no study has been performed to evaluate the common genetic determinants that affect serum level of p2PSA.
Materials and Methods: Here, we performed a two-stage genome-wide association study (GWAS) on the p2PSA level in Chinese men who underwent a transperineal ultrasound-guided prostate biopsy at Huashan Hospital, Shanghai Cancer Center, and Ruijin Hospital in Shanghai, China. Germline variants significantly associated with the p2PSA level in the first stage (n = 886) were replicated in the second stage (n = 1,128). Multivariate linear regression was used to assess the independent contribution of confirmed single nucleotide polymorphisms (SNPs) and known covariates, such as age, to the level of p2PSA.
Results: A novel non-synonymous SNP, rs72725879, in region 8q24.21 of the PRNCR1 gene was significantly associated with the serum level of p2PSA in this two-stage GWAS (p = 2.28 × 10−9). Participants with homozygous “T” alleles at rs72725879 had higher p2PSA levels compared to allele “C” carriers. This variant was also nominally associated with PCa risk (p-combined = 3.44 × 10−18). The association with serum level of p2PSA was still significant after adjusting for PCa risk and age (p = 0.017).
Conclusions: Our study shows that the genetic variants in the 8q24.21 region are associated with the serum level of p2PSA in a large-scale Chinese population. By taking inherited variations between individuals into account, the findings of these genetic variants may help improve the performance of p2PSA in predicting prostate cancer.
Prostate cancer (PCa) is one of the most common tumors in men and one of the leading causes of cancer-related death, conferring 1,111,700 new cases and 307,500 deaths annually worldwide (1). Although the incidence of PCa in Chinese is lower than that in Caucasian and African populations, it has been rising progressively in recent decades, along with increasing mortality (2).
Prostate-specific antigen (PSA) screening is the most widely used biomarker for the early detection and surveillance of PCa. However, PSA can also be affected by prostatitis, benign prostate hyperplasia, age, ethnicity, and genetic factors, which means it is organ-specific rather than cancer-specific. Therefore, its low specificity in clinical applications leads to quite a number of unnecessary biopsies and overdiagnosis of indolent cancers (3). In terms of genetic influence on the serum PSA level, it is estimated that 40% of the variations between individuals can be explained by inherited factors (4). In previous genome-wide association studies (GWAS), multiple inherited variants had been demonstrated to influence the serum levels of PSA in European and Asian populations (5–8).
Regarding the issues of unnecessary biopsies and overdiagnosis caused by PSA screening, new biomarkers with higher specificities and better abilities to discriminate PCa and aggressive PCa are needed. A relatively new biomarker [–2],proPSA (p2PSA), a predominant precursor form of PSA, was found elevated in almost all of the peripheral zone cancers, but was largely undetectable in the transition zone (9). In previous studies, p2PSA and its derivative prostate health index (phi) [(p2PSA/free PSA) × √tPSA] have been proven to have a better discriminating ability in predicting PCa in Caucasians (10–14). Later on, the clinical utilities of p2PSA and phi are also implicated in the Chinese (15–17). However, whether the serum p2PSA level is affected by genetic variance between different individuals remains unknown.
Discovery of novel genetic variants that influence the serum level of p2PSA may improve our understanding of the molecular mechanisms and clinical utility of p2PSA test. Therefore, to identify the genetic variants that influence the serum p2PSA level, we carried out a two-stage GWAS among Chinese men who underwent prostate biopsy.
Our study included two prostate biopsy cohorts from three medical centers, which were genotyped with the same genotyping array platform and denoted as stages 1 and 2.
Stage 1 consisted of 886 subjects from Huashan Hospital, Fudan University, and Shanghai Cancer Center, Fudan University, between 2010 and 2014. Stage 2 included 1,128 subjects from Huashan Hospital, Fudan University, and Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, between 2015 and 2018. All the patients (n = 2,014) underwent initial prostate biopsies at the above-mentioned medical centers from 2010 to 2018.
The indications for prostate biopsy were the same across the three centers: 1) PSA >10.0 ng/ml; 2) PSA >4.0 ng/ml with a confirmation within 3 months; 3) PSA level ranging from 4.0 to 10.0 ng/ml, with suspicious %fPSA (free PSA divided by PSA) <0.16; and 4) abnormal findings from digital rectal examination (DRE), ultrasound, or magnetic resonance imaging (MRI) with any level of PSA.
Blood specimens were obtained before biopsies and serum samples were extracted. Serum total PSA (tPSA), free PSA (fPSA), and p2PSA were measured with a Beckman Coulter D×I 800 Immunoassay System (Beckman Coulter, Brea, CA, USA). All assays and quality control (QC) were performed according to the manufacturer’s instructions and standard QC protocols. Specifically, Access Hybritech Calibrators S0, S1–S6, i.e., blank and low to high concentrations, were run as internal known standards before each batch of the measurements. The measurements of the internal known control materials, Access Hybritech QC 1, 2, and 3, were below two standard deviations for each batch.
All epidemiological and clinical pieces of information were collected before biopsy. The patients would be excluded if pathologically diagnosed biopsy outcome was missing or if the tPSA, fPSA, and p2PSA were unable to be tested for bad quality. This study was approved by the institutional review board of each medical center, and written informed consent to participate in the present study was obtained from all participants.
All DNA samples from stages 1 and 2 were extracted from blood samples and genotyped with the same genotyping array platform, Illumina Asian Screening Array (ASA) Beadchip, which included 659,184 single nucleotide polymorphisms (SNPs).
Genotyping QC was conducted together with data from stages 1 and 2. We used the following standard QC procedure to select qualified samples and SNPs for imputation analysis. Samples were excluded if they: i) had a genotyping call rate of <95%; ii) were duplicates or showed familial relationships [identity by state (IBS) >0.99]; and iii) had ambiguous gender. SNPs were removed if they had: i) a genotyping rate of <95% (n = 9,650); ii) a minor allele frequency of <0.01 (n = 152,901); and iii) a p-value <10−3 with the Hardy–Weinberg equilibrium test in patients with negative biopsy results (n = 2,721). After QC analysis, a total of 2,014 (886 in stage 1 and 1,128 in stage 2) samples with 493,912 genotyped SNPs were retained for imputation analysis.
Imputation was performed with the IMPUTE computer program (18) using the 1000 Genomes Project Han Chinese in Beijing (CHB) population as the reference. A total of 17,098,949 SNPs with imputation information score >0.90 were included in the analysis.
GWAS analysis was conducted in stage 1, which included 886 samples with 17,098,949 SNPs. Using the same sample and SNP QC criteria above, 886 samples with 4,552,207 SNPs were left in the association analysis. PCAs were estimated using EIGENSTRAT. Beta values and p-values were estimated using quantitative linear regression for each SNP, adjusting for age and the first two PCAs.
We then performed a replication analysis using data from stage 2. As an independent set, stage 2 included 1,128 samples with 17,098,949 SNPs (including genotyped and imputed). The combined analysis of two-stage data was performed using linear regression, adjusting for age.
The associations between the serum p2PSA level and SNP genotypes were evaluated using a quantitative linear regression model assuming additive effects of the alleles (0, 1, and 2). In the regression models, log-transformed p2PSA levels were used as the dependent variable, each SNP as an independent variable, and age as a covariate. This analysis was performed by the PLINK V.1.90 software package (19). P-values less than 5 × 10−8 and 0.05 were regarded as significant levels in the GWAS and other analyses, respectively.
A principal component approach was used to evaluate population stratification in the first stage with the EIGENSTRAT software (20). The top two eigenvectors were adjusted as covariates in the quantitative linear regression analysis. Quantile–quantile (Q–Q) plots were performed using the R package (http://www.R-project.org). Linkage disequilibrium (LD)-based result clumping analysis was applied to test the independence of the respective SNPs in the 8q24.21 locus using PLINK (19). LocusZoom (21) and haploview (22) were used to create plots of genetic data.
A total of 886 subjects were recruited in the first stage and 1,128 in the second stage. The clinical characteristics of the cohorts in the two stages are described in Table 1. A total of 886 PCa cases were detected in two biopsy cohorts, with an overall positive biopsy rate of 42.9%. No significant difference in the clinical characteristics was observed between the two cohorts (Table 1).
In the first stage, 886 subjects were genotyped with the Illumina Asian Screening Array. After quality control, 4,552,207 SNPs and 877 individuals were eligible for GWAS analysis. We did not observe population structure in our cohort (Supplementary Figure S1). In addition, the Q-Q plots revealed an unadjusted inflation factor of 0.9999 (Supplementary Figure S2), indicating no evidence of systematic bias for the association of logp2PSA phenotype observed in the current study. The Manhattan plot of GWAS for the first stage is shown in Figure 1. We selected all signals associated with the logp2PSA level at p < 1 × 10−5 for replication analysis. A total of 148 SNPs were selected, and 86 of them reached a p-value of <5 × 10−8, which were mainly located in two regions, 8q21.3 and 8q24.21 (Supplementary Table S1).
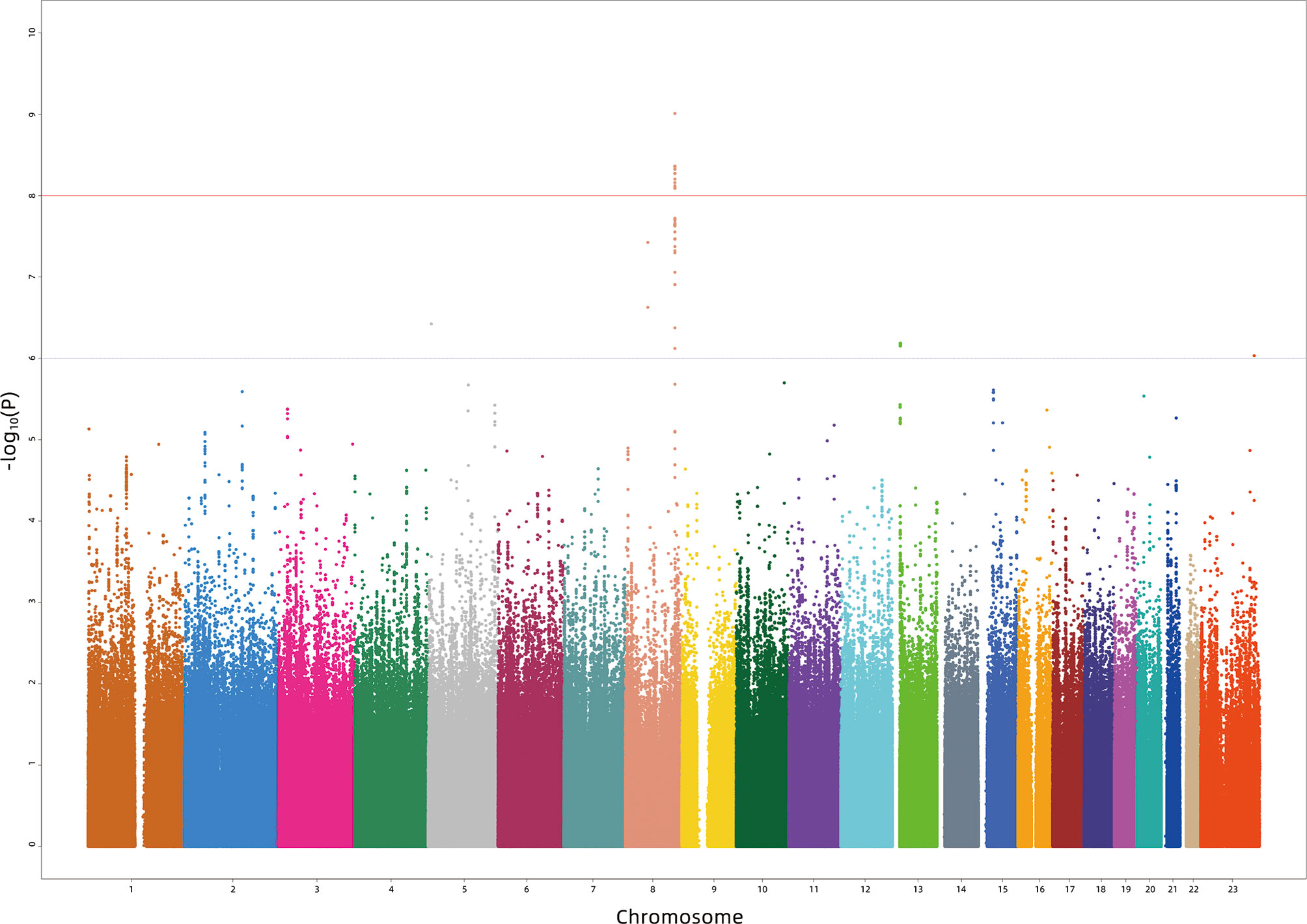
Figure 1 Manhattan plot for the genome-wide association study results for the levels of p2PSA in the Chinese population. The X-axis represents the chromosomal position and the Y-axis represents the –log10 p-value from linear regression. The horizontal dashed line shows the preset threshold of p = 1 × 10−6. The horizontal solid line indicates the preset threshold of p = 1 × 10−8.
In the second stage, 143 out of 148 SNPs remained qualified after QC in 1,128 subjects. Among the 143 candidate SNPs, rs72725879 at the 8q24 locus was confirmed to be significantly associated with the logp2PSA level at a p-value cutoff of 0.0003 (Bonferroni correction of 143 tests). We then combined the data of the two stages and found two SNPs that reached genome-wide significance (p = 2.28 × 10−9 for rs72725879 and p = 5.31 × 10−9 for rs13254738) (Table 2). Therefore, the 8q24.21 region was revealed to be significantly associated with p2PSA levels.
In the current study, the strongest association effects were observed for two SNPs, rs72725879 and rs13254738, both of which were located in region 8q24.21 of the PRNCR1 (PCa-associated non-coding RNA 1) gene. rs72725879-T and rs13254738-C showed a significant association with increasing serum levels of p2PSA. The two SNPs were correlated with each other (R2 = 0.79). After adjusting the association results for rs72725879 as a covariate, rs13254738 became insignificant (p = 0.054). We then performed an additional univariate analysis for rs72725879 and found that it could explain 2.2% of the total genetic variance for the p2PSA levels.
Variants in the 8q24.21 region have previously been reported to be associated with a risk of PCa (23–30). Due to the potential confounding effects of the p2PSA level and PCa, we also evaluated whether the p2PSA-associated SNPs found in this study were associated with PCa. In the association analysis of the total study population combining the two cohorts, we confirmed the association of rs72725879 with PCa, with a combined odds ratio (OR) of 1.90 and a p-value of 3.44 × 10−18 for the T allele. After adjusting for PCa risk and age as covariates, rs72725879 was still associated with the p2PSA level (p = 0.017) (Supplementary Table S2).
We then grouped the subjects in the two stages into three groups based on their genotypes. The proportions of PCa cases detected in the CC, CT, and TT groups were 24.18%, 33.77%, and 50.76%, respectively. The T allele of rs72725879 showed a significant positive association with the increasing detection rate of PCa in the prostate biopsy cohort (p-trend < 3.92 × 10−21) (Supplementary Figure S3).
Then, we specifically looked into 14 SNPs associated with a risk of PCa within the 8q24.21 region reported by previous GWAS and assessed their effects on the levels of p2PSA (Table 3). The Manhattan plot of 1,236 SNPs within this region is also shown in Figure 2. These 14 SNPs belonged to five LD blocks in this locus according to previously reported results (28). In the association analysis adjusted for age, we revealed that eight SNPs associated with PCa risk also contributed to the serum p2PSA level at a p-value cutoff of 0.0036 (after a Bonferroni correction of 14 tests), although it did not reach a GWAS significance. Among the eight SNPs, rs72725879 in block 2 was found to be in weak LD with rs13252298 (r2 = 0.26) in the Chinese population (Figure 2).
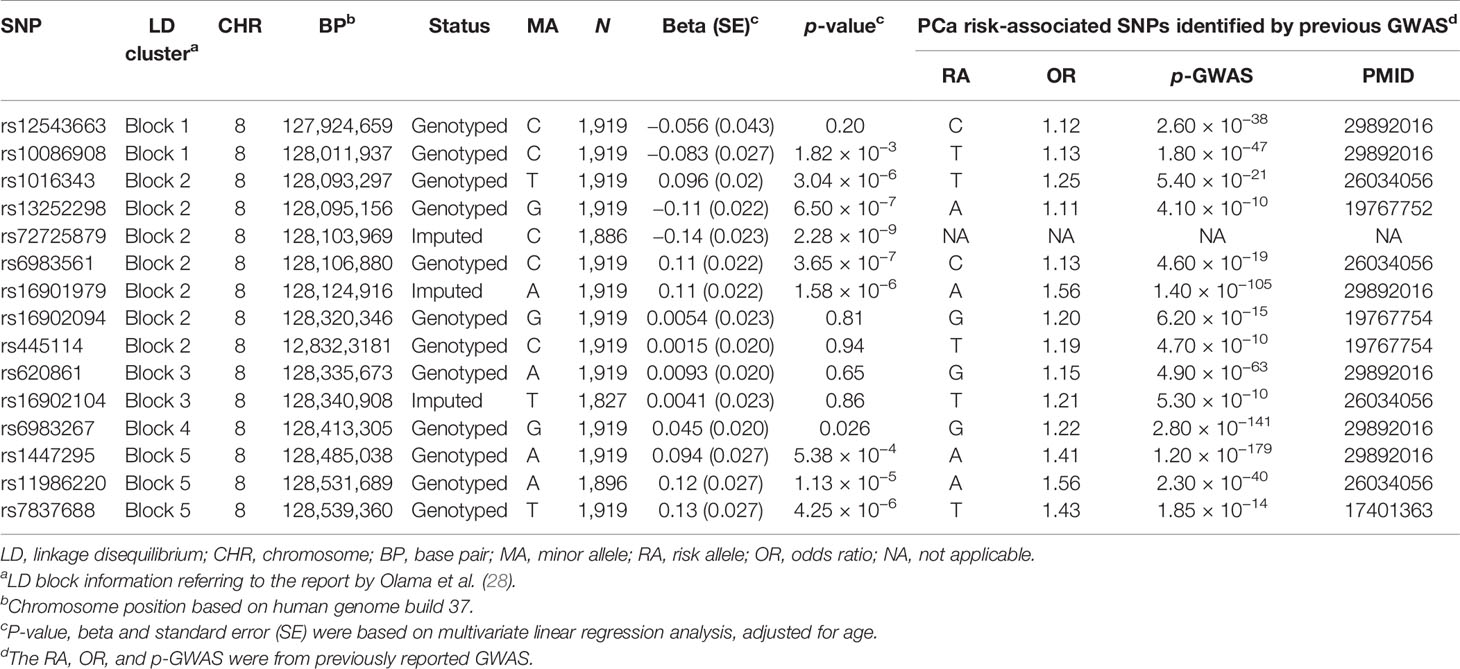
Table 3 Association analysis between logp2PSA and prostate cancer (PCa) risk-associated variants reported by previous genome-wide association study (GWAS) within the region 8q24.21.
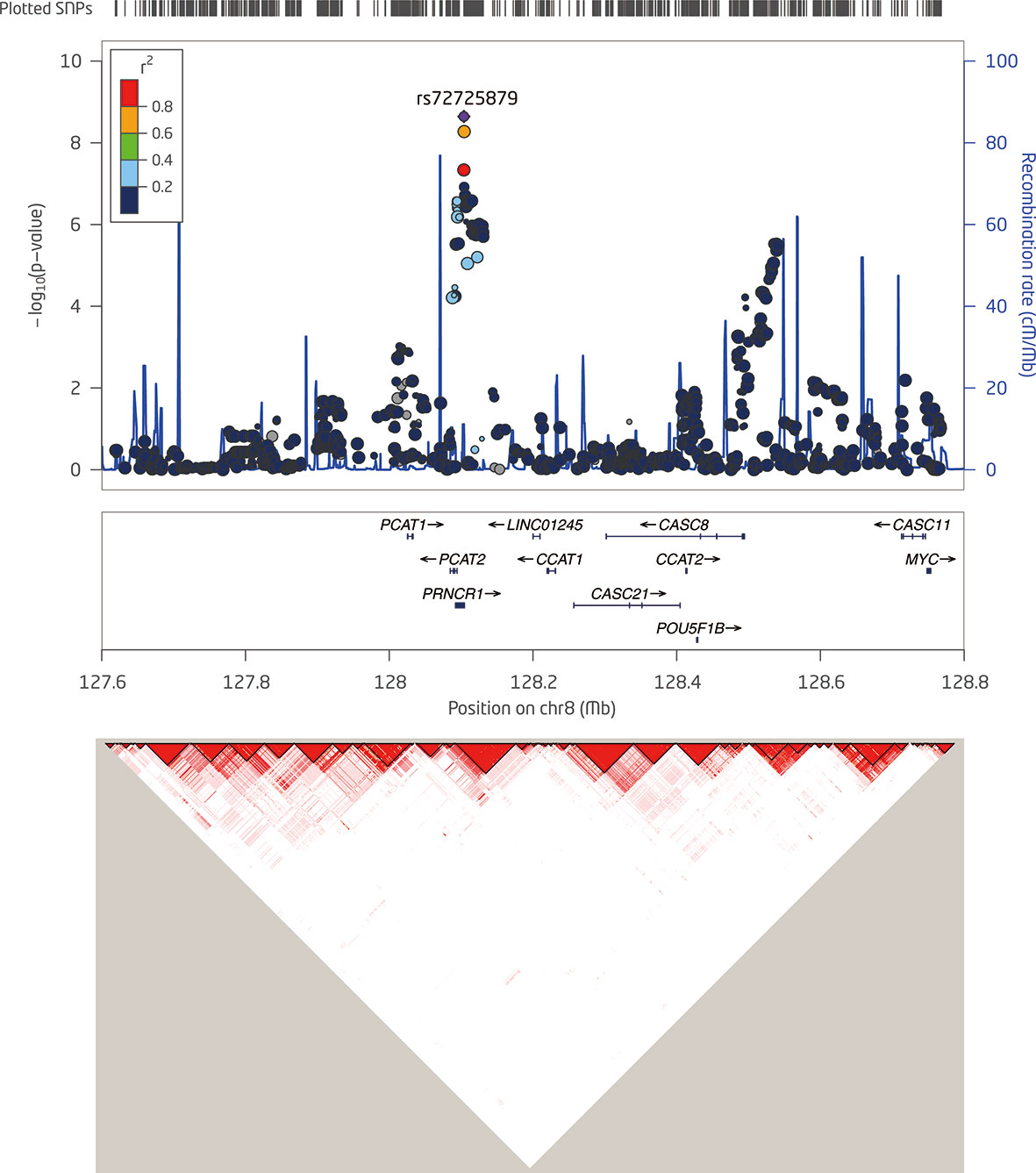
Figure 2 Detailed regional plots of −log10 p-values in the 8q24.21 region shown for logp2PSA. Colors indicate the linkage disequilibrium (LD) strength between rs7272589 and the other single nucleotide polymorphisms (SNPs) assessed. SNPs with red circle are reported to be associated with risk of prostate cancer (PCa) within the 8q24.21 region for the corresponding block. The right Y-axis shows the recombination rate from the 1000 Genomes Project data as reference. LD maps were based on D′ values using data from the two-stage samples.
Previous studies have found more than 40 SNPs associated with the serum PSA level. These findings provided important information on the genetic variations in PSA among different individuals and could help improve personalized PSA screening, thereby reducing unnecessary biopsies. Besides PSA, a relatively new biomarker, p2PSA and its derivative phi, have become important biomarkers for PCa diagnosis, especially for men with a PSA level in the range 2.0–10.0 ng/ml. Yet, there is no study on the genetic variants influencing the serum p2PSA level. To our knowledge, this is the first GWAS on the serum p2PSA level in the Chinese population. In this two-stage GWAS in Chinese men who underwent prostate biopsy, we identified one single locus, 8q24.21, associated with the serum p2PSA level at genome-wide significance.
Multiple GWAS and fine-mapping studies had identified that common genetic variations in 8q24 influenced the inherited risk of PCa independently (24–30), while only one SNP (rs17464492) in 8q24 that influences the serum PSA level had been identified in non-Hispanic whites previously (7). In our study, rs72725879 (in region 2) appeared to be the leading SNP that affected the serum p2PSA level in the identified locus, 8q24.21, and it was located in the exon region of a non-coding RNA gene known as PCa-associated non-coding RNA 1 (PRNCR1). This SNP had previously been reported to be associated with PCa in men of African ancestry (30). According to data from The 1000 Genomes Project (1KGP), the risk allele frequency (RAF) of rs72725879 (T) was 0.76 in this study cohort, being higher than that in normal Asian (RAF = 0.66, ASN 1KGP), African (RAF = 0.33, AFR 1KGP), and European (RAF = 0.19, EUR 1KGP) populations. Previous GWAS had also identified that SNPs (rs1016343, rs13252298, rs13254738, and rs6983561) in PRNCR1 were associated with PCa risk (23, 28, 29). In our two-stage combined analysis, SNP rs72725879 and the above three SNPs (rs1016343, rs13252298, and rs6983561) in PRNCR1 were also found associated with PCa risk after adjustment for age (all p < 5.0 × 10−8). PRNCR1, also known as PCAT8, is a long non-coding RNA (lcRNA) that is upregulated in aggressive PCa. This lncRNA could bind to the androgen receptor and enhance the androgen receptor-mediated gene activation programs and proliferation in PCa cells (31). In the current study cohort, the T allele of rs72725879 was also observed to be associated with an increased risk of PCa detected in biopsy. Since a higher PCa risk can have an impact on the serum p2PSA level, it is also possible that the association between rs72725879 and the serum p2PSA level observed in the current study may also reflect some latent or undiagnosed disease.
Distinguishing whether SNPs are associated with p2PSA, PCa, or both is relatively complicated. The levels of p2PSA can be influenced by a number of factors (e.g., age, prostate infection, prostate inflammation, cancerous status, and urological manipulations). Nevertheless, we performed both the association analysis (with p2PSA) after adjusting for age and the association analysis with PCa to address part of these issues. Our results showed that the association between the genetic variant (rs72725879) and the p2PSA level was still significant after adjusting for PCa and age. This indicated that, although the association between PCa and serum p2PSA level was stronger, which is plausible because p2PSA is a diagnostic predictor for PCa, genetic variance also contributed to the baseline p2PSA level among different individuals. It has been reported that PSA-associated SNPs discovered in GWAS could be used to help normalize an individual’s PSA level, and incorporating these genetic factors into the application of PSA screening may increase the ability to classify individuals who should be biopsied (7). Here, we have proven that genetic variants also had an impact on p2PSA, so that personalizing the cutoff value for p2PSA by adjusting for genetic variants that were associated with the p2PSA levels in each individual might enhance the sensitivity and specificity of p2PSA in guiding biopsies.
There were several limitations in our study. Firstly, the study population was relatively small so that some signals might have been missed. Secondly, the overall p2PSA level in the current prostate biopsy cohort would be higher than that in a general population; thus, the findings from our study still need to be further validated in a larger general population. However, approximately 40% of the participants in our study were PCa cases, which enabled us to evaluate the associations between all p2PSA-associated SNPs and PCa.
In the current study, we described the first GWAS in a Chinese prostate biopsy population and identified one single locus at 8q24.21 that was associated with the serum p2PSA level at genome-wide significance. By taking inherited variations between individuals into account, the findings of these genetic variants may help calculate personalized cutoff values for serum p2PSA for patients, thus improving the performance of p2PSA to predict PCa risk.
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Institutional Review Board of Huashan Hospital, Fudan University; the Institutional Review Board of Shanghai Cancer Center, Fudan University; and the Institutional Review Board of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. The patients/participants provided written informed consent to participate in this study.
JZ, QD, and XL directed and designed the study. YW, FL, RN, DH, DX, JG, YZ, BD, DY, and HJ recruited the study subjects and managed the respective projects. XL and HY performed the bioinformatics and statistical analyses. XL and FL performed the genotyping and p2PSA testing. ZF and QD coordinated the project. All authors contributed to the article and approved the submitted version.
This work was funded by grants from the Clinical Science and Technology Innovation Project of Shanghai Shen Kang Hospital Development Center to Qiang Ding (no. SHDC12015105), and Clinical Research Project of Shanghai Health Commission to YW (No.20214Y0511).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.753920/full#supplementary-material
Supplementary Figure 1 | The principal component analysis was conducted in 886 samples in the first stage and 1,978 from 1000 Genomes data by the first two principal components.
Supplementary Figure 2 | The quantile-quantile (Q-Q) plot of the expected and observed P values using imputed SNPs data in stage 1.
Supplementary Figure 3 | Plot for PCa detection rate by the genotypes of rs72725879.
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global Cancer Statistics, 2012. CA Cancer J Clin (2015) 65:87–108. doi: 10.3322/caac.21262
2. Liu X, Yu C, Bi Y, Zhang ZJ. Trends and Age-Period-Cohort Effect on Incidence and Mortality of Prostate Cancer From 1990 to 2017 in China. Public Health (2019) 172:70–80. doi: 10.1016/j.puhe.2019.04.016
3. Loeb S, Bjurlin MA, Nicholson J, Tammela TL, Penson DF, Carter HB, et al. Overdiagnosis and Overtreatment of Prostate Cancer. Eur Urol (2014) 65:1046–55. doi: 10.1016/j.eururo.2013.12.062
4. Pilia G, Chen WM, Scuteri A, Orru M, Albai G, Dei M, et al. Heritability of Cardiovascular and Personality Traits in 6,148 Sardinians. PloS Genet (2006) 2:e132. doi: 10.1371/journal.pgen.0020132
5. Sun J, Tao S, Gao Y, Peng T, Tan A, Zhang H, et al. Genome-Wide Association Study Identified Novel Genetic Variant on SLC45A3 Gene Associated With Serum Levels Prostate-Specific Antigen (PSA) in a Chinese Population. Hum Genet (2013) 132:423–9. doi: 10.1007/s00439-012-1254-3
6. Terao C, Terada N, Matsuo K, Kawaguchi T, Yoshimura K, Hayashi N, et al. A Genome-Wide Association Study of Serum Levels of Prostate-Specific Antigen in the Japanese Population. J Med Genet (2014) 51:530–6. doi: 10.1136/jmedgenet-2014-102423
7. Hoffmann TJ, Passarelli MN, Graff RE, Emami NC, Sakoda LC, Jorgenson E, et al. Genome-Wide Association Study of Prostate-Specific Antigen Levels Identifies Novel Loci Independent of Prostate Cancer. Nat Commun (2017) 8:14248. doi: 10.1038/ncomms14248
8. Li W, Bicak M, Sjoberg DD, Vertosick E, Dahlin A, Melander O, et al. Genome-Wide Association Study Identifies Novel Single Nucleotide Polymorphisms Having Age-Specific Effect on Prostate-Specific Antigen Levels. Prostate (2020) 80:1405–12. doi: 10.1002/pros.24070
9. Mikolajczyk SD ML, Wang TJ, Rittenhouse HG, Marks LS, Song W, Wheeler TM, et al. A Precursor Form of Prostate-Specific Antigen Is More Highly Elevated in Prostate Cancer Compared With Benign Transition Zone Prostate Tissue. Cancer Res (2000) 60:756–9.
10. Guazzoni G, Nava L, Lazzeri M, Scattoni V, Lughezzani G, Maccagnano C, et al. Prostate-Specific Antigen (PSA) Isoform P2psa Significantly Improves the Prediction of Prostate Cancer at Initial Extended Prostate Biopsies in Patients With Total PSA Between 2.0 and 10 Ng/Ml: Results of a Prospective Study in a Clinical Setting. Eur Urol (2011) 60:214–22. doi: 10.1016/j.eururo.2011.03.052
11. Catalona WJ, Partin AW, Sanda MG, Wei JT, Klee GG, Bangma CH, et al. A Multicenter Study of [-2]Pro-Prostate Specific Antigen Combined With Prostate Specific Antigen and Free Prostate Specific Antigen for Prostate Cancer Detection in the 2.0 to 10.0 Ng/Ml Prostate Specific Antigen Range. J Urol (2011) 185:1650–5. doi: 10.1016/j.juro.2010.12.032
12. Guazzoni G, Lazzeri M, Nava L, Lughezzani G, Larcher A, Scattoni V, et al. Preoperative Prostate-Specific Antigen Isoform P2psa and its Derivatives, %P2psa and Prostate Health Index, Predict Pathologic Outcomes in Patients Undergoing Radical Prostatectomy for Prostate Cancer. Eur Urol (2012) 61:455–66. doi: 10.1016/j.eururo.2011.10.038
13. Lazzeri M, Briganti A, Scattoni V, Lughezzani G, Larcher A, Gadda GM, et al. Serum Index Test %[-2]proPSA and Prostate Health Index Are More Accurate Than Prostate Specific Antigen and %fPSA in Predicting a Positive Repeat Prostate Biopsy. J Urol (2012) 188:1137–43. doi: 10.1016/j.juro.2012.06.017
14. Fossati N, Buffi NM, Haese A, Stephan C, Larcher A, McNicholas T, et al. Preoperative Prostate-Specific Antigen Isoform P2psa and Its Derivatives, %P2psa and Prostate Health Index, Predict Pathologic Outcomes in Patients Undergoing Radical Prostatectomy for Prostate Cancer: Results From a Multicentric European Prospective Study. Eur Urol (2015) 68:132–8. doi: 10.1016/j.eururo.2014.07.034
15. Na R, Ye D, Liu F, Chen H, Qi J, Wu Y, et al. Performance of Serum Prostate-Specific Antigen Isoform [-2]proPSA (P2psa) and the Prostate Health Index (PHI) in a Chinese Hospital-Based Biopsy Population. Prostate (2014) 74:1569–75. doi: 10.1002/pros.22876
16. Chiu PK, Roobol MJ, Teoh JY, Lee WM, Yip SY, Hou SM, et al. Prostate Health Index (PHI) and Prostate-Specific Antigen (PSA) Predictive Models for Prostate Cancer in the Chinese Population and the Role of Digital Rectal Examination-Estimated Prostate Volume. Int Urol Nephrol (2016) 48:1631–7. doi: 10.1007/s11255-016-1350-8
17. Na R, Ye D, Qi J, Liu F, Helfand BT, Brendler CB, et al. Prostate Health Index Significantly Reduced Unnecessary Prostate Biopsies in Patients With PSA 2-10 Ng/mL and PSA >10 Ng/Ml: Results From a Multicenter Study in China. Prostate (2017) 77:1221–9. doi: 10.1002/pros.23382
18. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A New Multipoint Method for Genome-Wide Association Studies by Imputation of Genotypes. Nat Genet (2007) 39:906–13. doi: 10.1038/ng2088
19. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet (2007) 81:559–75. doi: 10.1086/519795
20. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal Components Analysis Corrects for Stratification in Genome-Wide Association Studies. Nat Genet (2006) 38:904–9. doi: 10.1038/ng1847
21. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: Regional Visualization of Genome-Wide Association Scan Results. Bioinformatics (2010) 26:2336–7. doi: 10.1093/bioinformatics/btq419
22. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics (2005) 21:263–5. doi: 10.1093/bioinformatics/bth457
23. Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome-Wide Association Study Identifies a Second Prostate Cancer Susceptibility Variant at 8q24. Nat Genet (2007) 39:631–7. doi: 10.1038/ng1999
24. Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-Wide Association Study of Prostate Cancer Identifies a Second Risk Locus at 8q24. Nat Genet (2007) 39:645–9. doi: 10.1038/ng2022
25. Zheng SL, Sun J, Cheng Y, Li G, Hsu FC, Zhu Y, et al. Association Between Two Unlinked Loci at 8q24 and Prostate Cancer Risk Among European Americans. J Natl Cancer Inst (2007) 99:1525–33. doi: 10.1093/jnci/djm169
26. Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, et al. Multiple Regions Within 8q24 Independently Affect Risk for Prostate Cancer. Nat Genet (2007) 39:638–44. doi: 10.1038/ng2015
27. Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple Newly Identified Loci Associated With Prostate Cancer Susceptibility. Nat Genet (2008) 40:316–21. doi: 10.1038/ng.90
28. Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, et al. Multiple Loci on 8q24 Associated With Prostate Cancer Susceptibility. Nat Genet (2009) 41:1058–60. doi: 10.1038/ng.452
29. Hoffmann TJ, Van Den Eeden SK, Sakoda LC, Jorgenson E, Habel LA, Graff RE, et al. A Large Multiethnic Genome-Wide Association Study of Prostate Cancer Identifies Novel Risk Variants and Substantial Ethnic Differences. Cancer Discovery (2015) 5:878–91. doi: 10.1158/2159-8290.CD-15-0315
30. Han Y, Rand KA, Hazelett DJ, Ingles SA, Kittles RA, Strom SS, et al. Prostate Cancer Susceptibility in Men of African Ancestry at 8q24. J Natl Cancer Inst (2016) 108. doi: 10.1093/jnci/djv431
Keywords: genome-wide association study, p2PSA, polymorphism, prostate cancer, Chinese
Citation: Lin X, Wu Y, Liu F, Na R, Huang D, Xu D, Gong J, Zhu Y, Dai B, Ye D, Yu H, Jiang H, Fang Z, Zheng J and Ding Q (2021) A Germline Variant at 8q24 Contributes to the Serum p2PSA Level in a Chinese Prostate Biopsy Cohort. Front. Oncol. 11:753920. doi: 10.3389/fonc.2021.753920
Received: 05 August 2021; Accepted: 27 September 2021;
Published: 19 October 2021.
Edited by:
Benyi Li, University of Kansas Medical Center, United StatesCopyright © 2021 Lin, Wu, Liu, Na, Huang, Xu, Gong, Zhu, Dai, Ye, Yu, Jiang, Fang, Zheng and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Ding, cWlhbmdkX3Vyb2xvZ3lAMTI2LmNvbQ==; Jie Zheng, YXRsYW50aWNhQDE2My5jb20=
†These authors have contributed equally to this study
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.