- 1Department of Oncology, Tianjin Medical University General Hospital, Tianjin, China
- 2Department of General Surgery, Tianjin Medical University General Hospital, Tianjin, China
- 3Institute of General Surgery, Tianjin Medical University General Hospital, Tianjin, China
Background: The landscape of intratumor heterogeneity (ITH) is present from the tumor evolution. ITH is a promising clinical indicator, but the association between ITH and prognosis remains controversial. Therefore, a meta-analysis was performed to explore whether ITH can serve as a valuable prognostic indicator in solid tumors.
Methods: All included studies were from PubMed, Embase, Cochrane, and Web of Science databases up to October 10, 2020. Studies based on ITH with available prognostic information were included. Three researchers independently completed study selection and data extraction following PRISMA guidelines. The random-effect model was used for synthesis. Hazard ratio (HR) and 95% confidence intervals (CI) were used with the endpoint defined by overall survival (OS), disease-specific survival (DFS), and progression-free survival (PFS).
Results: A total of 9,804 solid tumor patients from 21 studies were included. Analysis of specific cancers in the TCGA database showed similar results based on different ITH assessment methods, which provided the logical support for data consolidation. Available evidence revealed a negative relationship between ITH and prognosis for a specific cancer (such as lung cancer). However, the OS results from 14 tumor types showed that high ITH associated with shorter survival time [HR 1.65 (95% CI, 1.42–1.91)]. PFS and DFS analyses showed similar results [HR 1.89 (95% CI, 1.41–2.54) and HR 1.87 (95% CI, 1.15–3.04)] in general. The status of tumor metastasis and sampling models were not the confounding factors.
Conclusions: High ITH is associated with worse prognosis in many solid tumors in general although this association was absent for some cancers. ITH is expected to be a promising clinical prognostic factor for the improvement of assessment, treatment, and surveillance strategy.
Introduction
In 1976, Peter C. Nowell (1) laid the foundation for the clonal evolution model of cancer. He hypothesized that tumors arise from a founder cell and that genetic instability gives the tumor the potential to produce multiple “sublines.” In 1977, Fidler et al. (2) inoculated different parts of tumor cells of melanoma cells into mice and produced a variety of lung metastases numbers and proposed that there are distinct “clones” in tumors. In recent years, with the rapid development of next-generation sequencing technology, intratumor heterogeneity (ITH) reflects the subclone information of solid tumors and is used to study tumor metastatic patterns (3–9). In addition, ITH has the potential to be a new clinical prognostic indicator (10, 11). However, the results of previous studies did not provide consistent conclusion regarding the association between ITH and prognosis.
ITH is the genetic diversity reserve of tumors in the face of natural selection, which is largely responsible for the failure of targeted therapies (12). However, a key question is whether this diversity has important impact on a tumor’s natural history or a patient’s outcome (especially those who do not receive targeted therapy). To clarify these issues, we conducted a meta-analysis to explore the relationship between ITH and the prognosis of patients with solid tumors and evaluate the ability of ITH in predicting outcome by stratified confounding factors.
Methods
We performed a systematic review and meta-analysis following PRISMA guidelines (registration numbers CRD42020193878 and INPLASY202060107).
We retrieved studies from PubMed, Embase, Cochrane, and Web of Science databases up to October 10, 2020, using ITH and prognosis as the main keywords. We also screened EMSO, ASCO, and WCLC of recent years to avoid missing updated data. The type of study or language of the study was not limited in the study collection.
Inclusion Criteria
The inclusion criteria were as follows:
1. The study focused on solid tumors and examined ITH.
2. The study adopted a complete system for the assessment of ITH with the following steps: sample acquisition, extraction of genetic information, and evaluation of ITH by a scientific algorithm.
3. The study provided clear prognostic information and retrievable results.
Outcome
All studies included prognostic endpoints including overall survival (OS), disease-specific survival (DFS), and progression-free survival (PFS).
Exclusion Criteria
The exclusion criteria were as follows:
1. The source of the sample was not taken from the tumor region.
2. RNA-ITH was excluded because most data were based on DNA sequencing.
3. Prognostic information was not associated with ITH or could not be extracted.
Screening and Extraction
All studies were reviewed separately by three investigators (ZZ, TY, and XG) according to inclusion and exclusion criteria. In cases in which the results were different, the case was discussed to obtain consensus. The information from the included studies was extracted in accordance with unified standards. When the included cohort was at risk of repeat statistics for the same patients, after considering the number of participants studied, the standardization of genetic information assessment, and the evaluation instruments of ITH, only the most relevant cohort was included in each forest map. Original data were extracted when a clear hazard ratio (HR) and 95% confidence interval (CI) was provided in included studies. If studies just provided the survival curve and the number of patients, the Engauge Tool and HR Calculations Spreadsheet were used to extract the HR and 95% CI (13). We standardized descriptions of the tumor types involved in the study according to the classification of tumor types in the TCGA database. Kaplan–Meier curves or univariate Cox regression data were selected whenever possible.
Study Quality Assessment
We used the Guidelines for Assessing Quality in Prognostic Studies on the Basis of the Framework of Potential Biases in the study of deviation analysis (14). The procedure was evaluated independently by two researchers (ZZ and TY), and disputes were resolved by a third researcher.
Statistical Methods
The characteristics of all included participants were collected. All extracted HR and 95% CI were combined by the R version 4.0.2 with Meta package version 4.13-0. The Random Effects Model was used for all forest maps. Results were obtained through the combined HR and 95% CI. Data heterogeneity is represented by I2, with I2 ≥ 50 considered as high heterogeneity. Sensitivity analysis was performed to explore the effects of high heterogeneity. Subgroup analysis was used to reduce heterogeneity and analyze confounding factors. It was analyzed from the perspectives of OS, PFS, DFS, ITH assessment method, distant metastasis or not, sampling model, and so on. Funnel plots were generated for each forest map to assess publication offset.
Results
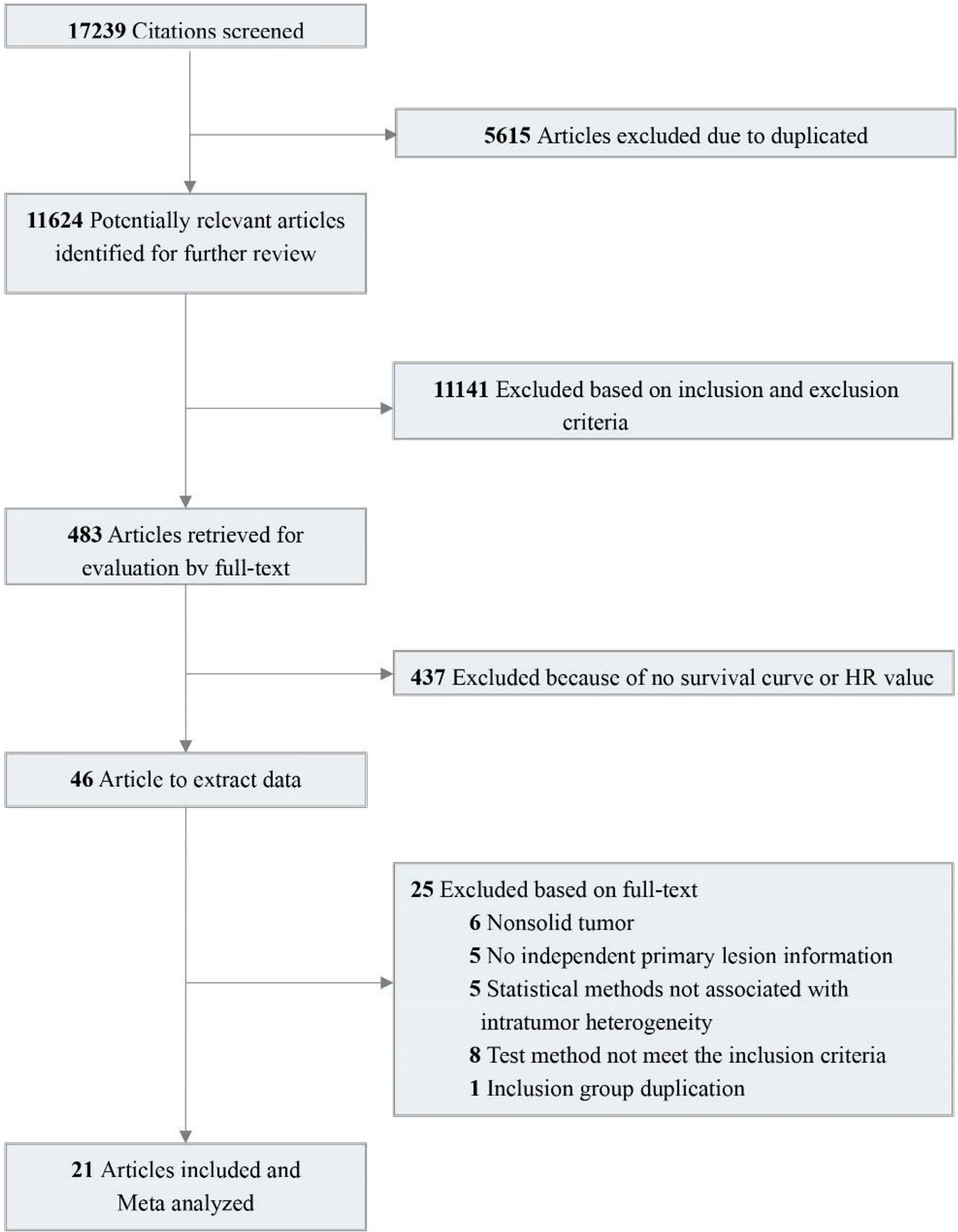
A total of 17,239 relevant studies were retrieved. After removing duplicates, we screened 483 potentially relevant articles by scanning the titles and abstracts. We reviewed the full text and screened the candidate studies according to the inclusion criteria, and 437 studies with no prognostic information were excluded. Of the remaining 46 studies, 25 were excluded using the exclusion criteria. In the end, 21 studies were included (15–33). These studies contain 38 pieces of comparison information that were extracted. Details of the flow chart for study identification are shown in Figure 1.
Quality evaluation: The assessment includes study participation, study attrition, prognostic factor measurement, outcome measurement, confounding measurement and account, and analysis. Sixteen studies were not at risk in the quality assessment. Two studies were considered as high risk in the study participation, and four studies did not describe confounding measurement and account. No study had more than two risks (Supplementary Table S1).
Characteristics of all cohorts: The final study population included 9,804 participants. All studies are retrospective studies published between 2013 and 2020 and involved various tumor types (Supplementary Table S2).
Different ITH Assessment Methods Had Limited Influence on This Study
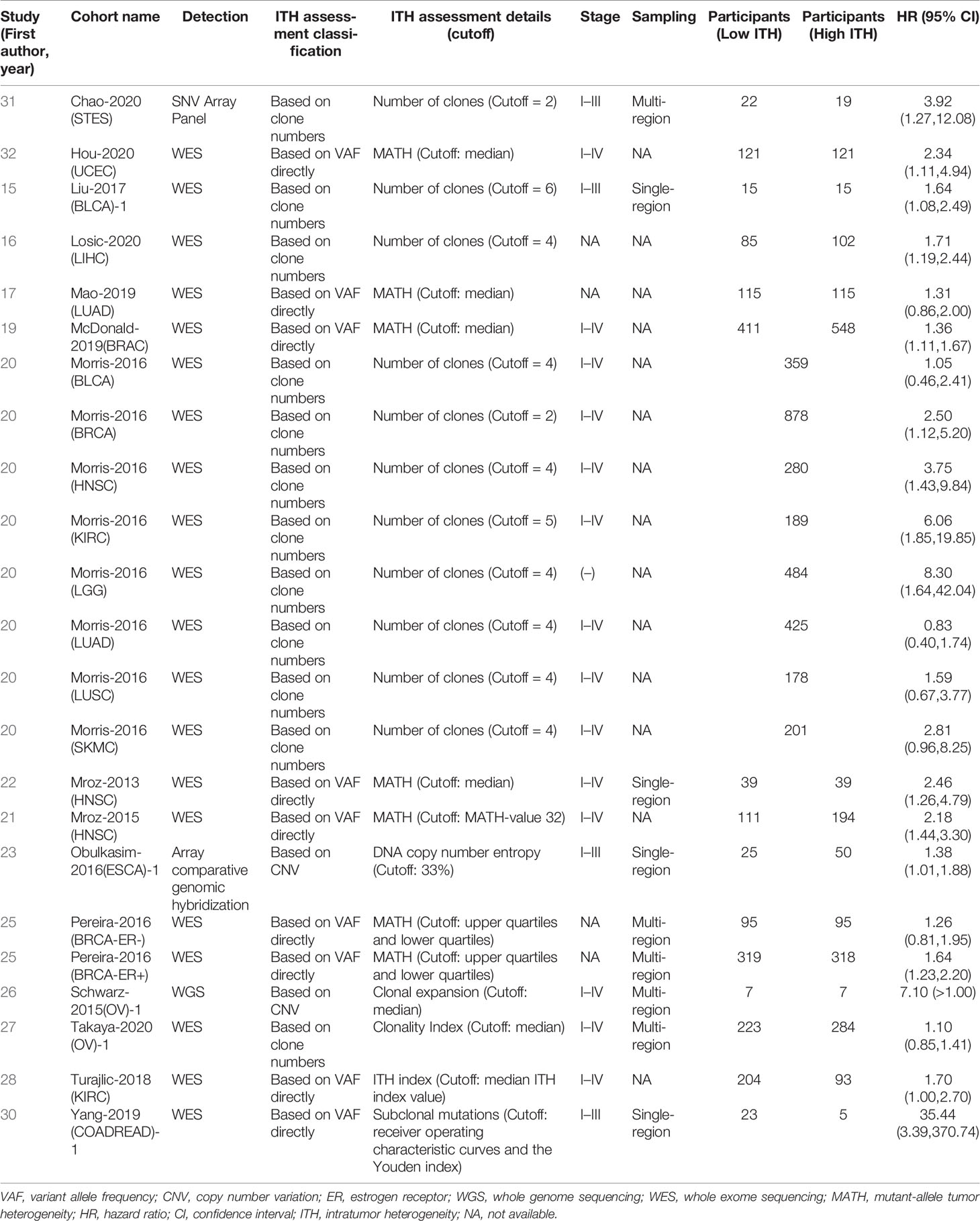
Researchers have used a variety of methods for ITH evaluation and gradually formed some relatively stable platforms. Single-nucleotide variants (SNVs) and small indels are universally investigated DNA variations. The main ITH evaluation method is to calculate variant allele fraction (VAF) based on SNVs and small indel information. Some researchers (18, 19, 23, 30, 34) use VAF to calculate the number of clones by clone detection methods such as PyClone and EXPANDS. Other researchers (20, 22, 24, 25, 28, 31, 33, 35) directly use VAF to directly reflect the information of ITH through methods such as mutant-allele tumor heterogeneity (MATH). Copy number variations (CNVs), as genetic information with high mutation frequency, have also been recently used by a few researchers in the evaluation of ITH (26, 29). Table 1 summarizes the details.
In general, these different ITH evaluation methods are essentially an integrated analysis of SNV, indels, and CNV, and the number of tumor subpopulations was simulated mathematically as the ITH value. Meanwhile, when different ITH assessment methods were used to analyze the same cancer type in TCGA (15, 20, 23, 24), the prognosis results showed consistency. Therefore, we pooled the results to generate an association between ITH and prognosis, even though some studies evaluated ITH differently.
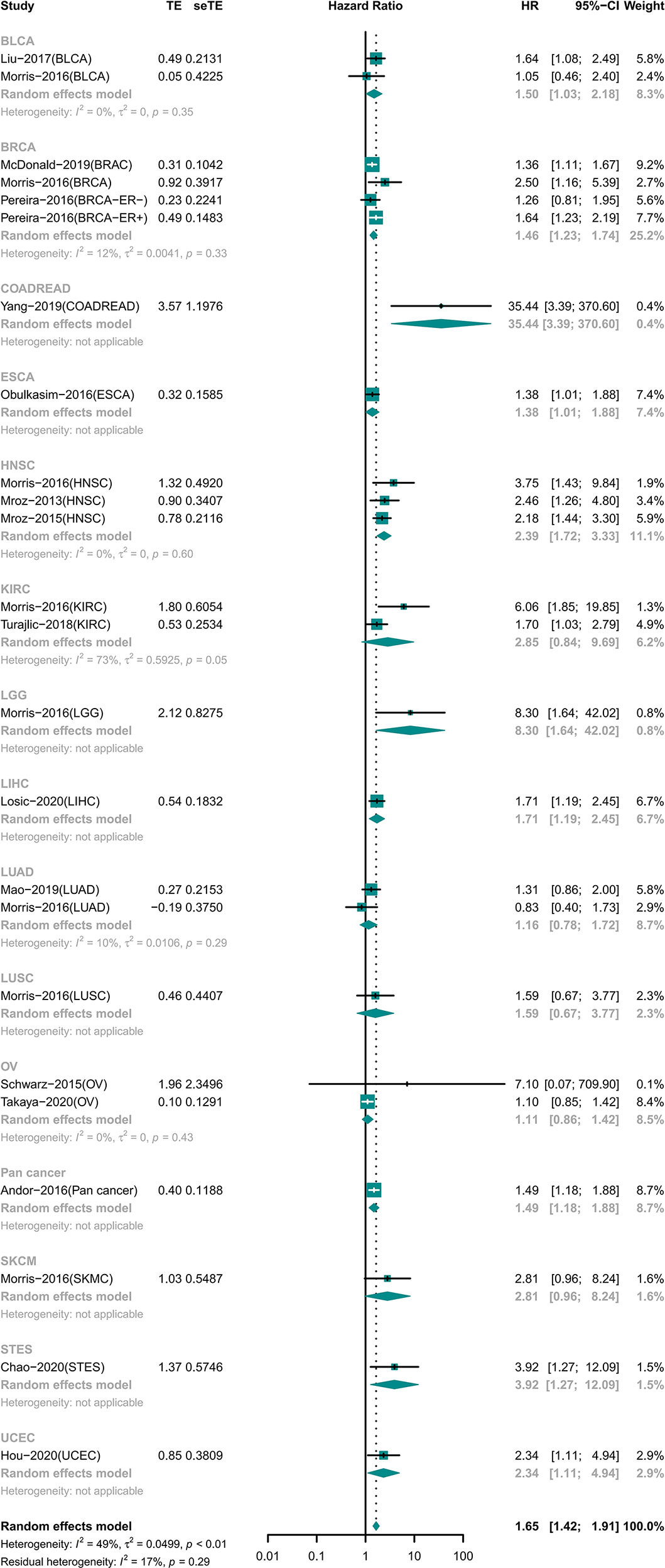
High ITH Was Associated With Worse Prognosis in General
A total of 7,971 participants involving 14 tumor types were examined for OS (15, 18–20, 22–26, 28–31, 33–35). The results showed that high ITH was associated with a high risk of death (HR 1.65 [95% CI, 1.42–1.91]) (Figure 2). However, this correlation was not homogeneous across all tumor types. Subgroup analysis showed that high ITH indicated a worse prognosis in patients with bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon adenocarcinoma or rectum adenocarcinoma (COADREAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), gastroesophageal adenocarcinoma (STET), and uterine corpus endometrial carcinoma (UCEC). Conversely, ITH showed no correlation with prognosis in patients with kidney renal clear cell carcinoma (KIRC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), and skin cutaneous melanoma (SKCM).
PFS was examined in 1,310 participants (17, 27, 29–31). Patients with high ITH were more likely to develop disease progression (HR 1.89 [95% CI, 1.41–2.54]) (Supplementary Figure S1). Subgroup analysis showed that high ITH was associated with poorer PFS for patients with KIRC, COADREAD, and OV.
In examining DFS, a total of 1,413 participants with five tumor types met the inclusion criteria (16, 17, 23, 31, 32). High ITH increased the risk of distant metastasis and recurrence (HR 1.87 [95% CI, 1.15–3.04]) (Supplementary Figure S2). We observed a correlation in patients with prostate adenocarcinoma (PRAD), COADREAD, LGG and glioblastoma multiforme (GBM), and thyroid carcinoma (THCA). Conversely, ITH showed no correlation with prognosis in patients with LUSC and LUAD.
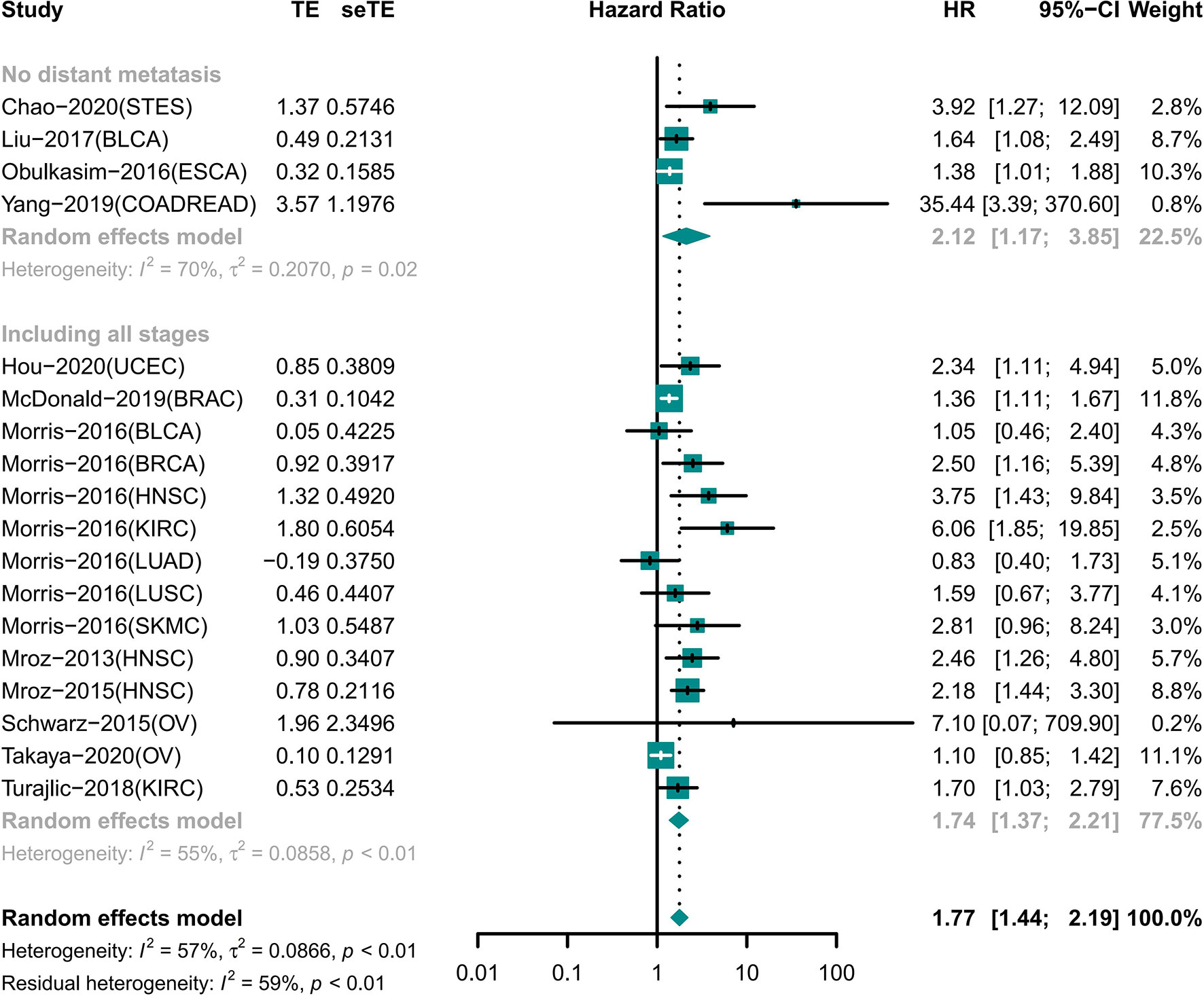
High ITH Indicated Short OS Independent of Distant Metastasis
A total of 18 cohorts involving 12 different tumor types recorded information for different tumor stages (Figure 3). Among the cohorts, 14 cohorts included patients with tumors of any stage involving nine tumor types; the results showed that the OS was shorter in the high ITH group (HR 1.74 [95% CI, 1.37–2.21]) (22–25, 29–31, 35). Four cohorts included information of four tumors with no distant metastasis, and high ITH was still associated with worse OS (HR 2.12 [95% CI, 1.17–3.85]) (18, 26, 33, 34).
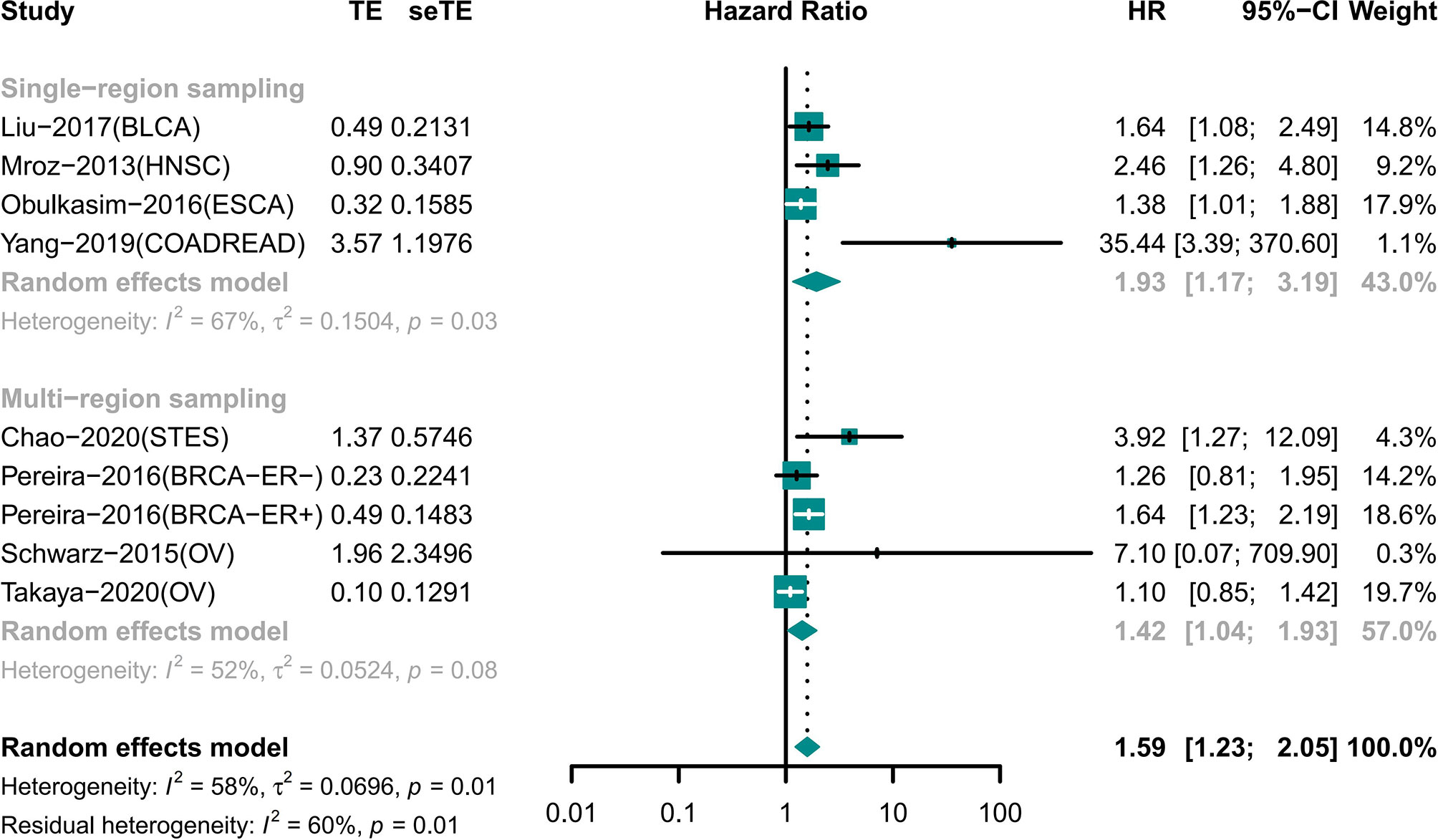
The Results of Single-Region and Multi-Region Sampling Were Similar for the Association of ITH and Prognosis
Night cohorts with precise descriptions of the sampling model were used to determine the impact of the sampling model on the results (Figure 4). These cohorts included patients with BLCA, BRCA, COADREAD, ESCA, HNSC, OV, and STES. Among the cohorts, four cohorts used single-region sampling and the analysis showed that high ITH patients had a worse prognosis (HR 1.93 [95% CI, 1.17–3.19]) (18, 25, 26, 33). Analysis of the other four cohorts using multi-region sampling indicated that patients with a high ITH had a higher risk of death (HR 1.42 [95% CI, 1.04–1.93]) (28–30, 34). The results of the two subgroups were similar.
Publication Bias Analysis
Funnel plots for all forest maps are shown in Supplementary Figure S3.
Discussion
This meta-analysis was an exploratory study probing the relationship between ITH and prognosis. Previous findings that examined the relation of ITH on prognosis have been controversial. The same is true for the prognostic implications of ITH for different tumor types. We demonstrated through a meta-analysis that a high ITH was associated with a poorer prognosis in patients with solid tumors. We stratified results by different solid tumors and found that the relation of ITH with prognosis was different for different tumor types. We also stratified results independently according to various confounding factors and found that various confounding factors did not affect the prognostic relation of ITH. Our findings have deepened the understanding of the field and suggest that ITH is a very promising indicator for clinical applications.
Overall, our analysis shows that OS, PFS, and DFS results indicate that the relationship between high ITH and worse prognosis is forceful. Seventeen solid tumors were included in these analyses. ITH was correlated with prognosis in 12 solid tumors. ITH may be the effect of time accumulation (36). Taking COADREAD as an example, the gradual progression from adenoma to adenocarcinoma contributes to the accumulation of ITH (37, 38), and the relation of ITH on prognosis may be more easily reflected in this cancer type. However, ITH was not associated with prognosis in patients with SKCM, KIRC, LUAD, LUSC, and OV in OS. Some tumors metastasize in the early stages (39, 40), and highly malignant tumors may have a shorter evolution time (41). ITH may not have enough time to develop to detect when tumor is diagnosed (41). Besides, because of the limitations of sequencing depth and purity of tumors, a large number of subclones may not be detected (42). In addition, various solid tumors can be further classified by pathological types. Although more research is needed to clarify the heterogeneity between tumor types, the available results are encouraging.
The challenge in ITH research is that there is no unified standard to calculate ITH, which greatly limits the clinical application. However, the vast majority of studies showed that there was no significant difference in the relationship between different ITH assessment methods and prognosis, regardless of single-region sampling or multi-region sampling. For example, in LUAD, different ITH evaluation methods [clone numbers (15), MATH (20), and subclonal populations (23)] show that ITH is not associated with prognosis. This may be because the principle of calculating the ITH is similar, which is based on SNV and CNV, and reflects the numbers of subpopulation in tumors. Of course, efforts have been made to develop more reasonable ITH algorithms, and a promising approach to evaluating ITH has been recently reported (43), Stefan et al. made a contribution to characterize ITH across cancer types based on the PCAWG dataset, including SNVs, indels, SVs, and CNVs, as well as subclonal drivers, subclonal selection, and mutation signatures. The assessment of prognosis based on the new method should be expected.
In the included studies, regardless of whether participants had distant metastasis, most of the samples used for sequencing were taken from the primary lesion. The correlation between ITH and prognosis was confirmed in tumors without distant metastasis. The increased number of subclones is associated with a greater possibility of distant metastasis, recurrence, and drug resistance, thus affecting the prognosis (44). However, for tumors in which distant metastases progress and more mutations appear, the results from primary tumor may not be sufficient. For patients with metastasis at the time of diagnosis, there may be differences in subclone composition between the metastatic lesion and primary lesion (45). Kim et al. proposed that the genetic distance between the metastatic and primary lesion may affect the prognosis (46). However, Reiter et al. (47) analyzed the ITH of distant metastases and found that CRC distant metastases are derived from subclones of the primary tumor. Anyway, so far, the results show that that detection in the primary lesion is sufficient to evaluate the role of ITH in prognosis.
Our results showed that single-site sampling and multi-region sampling were similar in terms of prognosis assessment. There was no evidence to suggest which sampling type was better because the subgroups did not come from the same cohorts. Huang et al. found that deep targeted sequencing of a single tumor specimen seems to be sufficient to evaluate ITH (48). However, tumor subclones have different spatial distribution (49). Zhang et al. reported that single-region sampling only reflects about 76% of the total mutation in multi-region sampling (50). Therefore, at present, when evaluating the ITH, sampling a single region may be sufficient in terms of validity or cost. Of course, if conditions permit, evaluating a diversified ITH provides more complete information.
It should be noted that these condition factors may not be perfectly matched. We analyzed the influence of different staging and sampling methods on the results based on all the information available in the original study, although this may somewhat damage the confidence level. The results presented by the available data are already suggestive; of course, more evidence is needed to verify our conclusions on staging and sampling methods in the future.
Limitations
(1) Although the number of included cohorts and participants was large, the results did not cover all solid tumor types (2). Multiple cancer types and participant differences in the number of participants between TCGA cohort and small cohort studies lead to heterogeneity and publication bias risk in this study (3). For some tumor types, few cohorts were included, which was not enough to draw a positive or negative conclusion (4). The duration of follow-up varied from study to study; however, a minimum follow-up of 4 years is acceptable (5). Evaluation of ITH still requires a more uniform approach that could be better used in the clinic (6). Multiple confounders were analyzed in our meta-analysis. However, more strictly matching data are needed in the future to verify our conclusions (7). Some cancers (e.g., lung cancer) have shown that ITH is not prognostic and should be given more attention (8). For the moment, widespread clinical application could be a challenge. However, as tumor-based sequencing technologies become more widely available and ITH-related modeling algorithms continue to be developed, this problem is expected to be solved in the future.
Conclusions
High ITH is associated with worse prognosis in many solid tumors in general, although this association was absent for some cancers. ITH is expected to be a promising clinical prognostic factor for the improvement of assessment, treatment, and surveillance strategy.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
TY: conceptualization and writing—original draft. XG: conceptualization and visualization, and writing—review and editing. SZ, ZZ, XZ, and CL: data curation. GL: conceptualization, project administration, supervision, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Gabrielle White Wolf, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.744064/full#supplementary-material
References
1. Nowell PC. The Clonal Evolution of Tumor Cell Populations. Science (1976) 194(4260):23–8. doi: 10.1126/science.959840
2. Fidler IJ, Kripke ML. Metastasis Results From Preexisting Variant Cells Within a Malignant Tumor. Science (1977) 197(4306):893–5. doi: 10.1126/science.887927
3. Swanton C. Intratumor Heterogeneity: Evolution Through Space and Time. Cancer Res (2012) 72(19):4875–82. doi: 10.1158/0008-5472.CAN-12-2217
4. Schwartz R, Schaffer AA. The Evolution of Tumour Phylogenetics: Principles and Practice. Nat Rev Genet (2017) 18(4):213–29. doi: 10.1038/nrg.2016.170
5. McBride M, Rida PC, Aneja R. Turning the Headlights on Novel Cancer Biomarkers: Inspection of Mechanics Underlying Intratumor Heterogeneity. Mol Aspects Med (2015) 45:3–13. doi: 10.1016/j.mam.2015.05.001
6. Hu Z, Ding J, Ma Z, Sun R, Seoane J, Shaffer J, et al. Quantitative Evidence for Early Metastatic Seeding in Colorectal Cancer. Nat Genet (2019) 51(7):1113–22. doi: 10.1038/s41588-019-0423-x
7. Sottoriva A, Kang H, Ma Z, Graham T, Salomon M, Zhao J, et al. A Big Bang Model of Human Colorectal Tumor Growth. Nat Genet (2015) 47(3):209–16. doi: 10.1038/ng.3214
8. Navin NE, Hicks J. Tracing the Tumor Lineage. Mol Oncol (2010) 4(3):267–83. doi: 10.1016/j.molonc.2010.04.010
9. Davis A, Gao R, Navin N. Tumor Evolution: Linear, Branching, Neutral or Punctuated? Biochim Biophys Acta Rev Cancer (2017) 1867(2):151–61. doi: 10.1016/j.bbcan.2017.01.003
10. Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of Tumor Mutation Burden as an Immunotherapy Biomarker: Utility for the Oncology Clinic. Ann Oncol (2019) 30(1):44–56. doi: 10.1093/annonc/mdy495
11. Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-Cell-Autonomous Driving of Tumour Growth Supports Sub-Clonal Heterogeneity. Nature (2014) 514(7520):54–8. doi: 10.1038/nature13556
12. Hobor S, Van Emburgh BO, Crowley E, Misale S, Di Nicolantonio F, Bardelli A. TGFalpha and Amphiregulin Paracrine Network Promotes Resistance to EGFR Blockade in Colorectal Cancer Cells. Clin Cancer Res (2014) 20(24):6429–38. doi: 10.1158/1078-0432.CCR-14-0774
13. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical Methods for Incorporating Summary Time-to-Event Data Into Meta-Analysis. Trials (2007) 8:16. doi: 10.1186/1745-6215-8-16
14. Hayden JA, Cote P, Bombardier C. Evaluation of the Quality of Prognosis Studies in Systematic Reviews. Ann Intern Med (2006) 144(6):427–37. doi: 10.7326/0003-4819-144-6-200603210-00010
15. Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, et al. Pan-Cancer Analysis of the Extent and Consequences of Intratumor Heterogeneity. Nat Med (2016) 22(1):105–13. doi: 10.1038/nm.3984
16. Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med (2017) 376(22):2109–21. doi: 10.1056/NEJMoa1616288
17. Joung JG, Oh BY, Hong HK, Al-Khalidi H, Al-Alem F, Lee HO, et al. Tumor Heterogeneity Predicts Metastatic Potential in Colorectal Cancer. Clin Cancer Res (2017) 23(23):7209–16. doi: 10.1158/1078-0432.CCR-17-0306
18. Liu D, Abbosh P, Keliher D, Reardon B, Miao D, Mouw K, et al. Mutational Patterns in Chemotherapy Resistant Muscle-Invasive Bladder Cancer. Nat Commun (2017) 8(1):2193. doi: 10.1038/s41467-017-02320-7
19. Losic B, Craig AJ, Villacorta-Martin C, Martins-Filho SN, Akers N, Chen X, et al. Intratumoral Heterogeneity and Clonal Evolution in Liver Cancer. Nat Commun (2020) 11(1):291. doi: 10.1038/s41467-019-14050-z
20. Mao H. Clinical Relevance of Mutant-Allele Tumor Heterogeneity and Lung Adenocarcinoma. Ann Transl Med (2019) 7(18):432. doi: 10.21037/atm.2019.08.112
21. Masoodi T, Siraj AK, Siraj S, Azam S, Qadri Z, Parvathareddy SK, et al. Evolution and Impact of Subclonal Mutations in Papillary Thyroid Cancer. Am J Hum Genet (2019) 105(5):959–73. doi: 10.1016/j.ajhg.2019.09.026
22. McDonald KA, Kawaguchi T, Qi Q, Peng X, Asaoka M, Young J, et al. Tumor Heterogeneity Correlates With Less Immune Response and Worse Survival in Breast Cancer Patients. Ann Surg Oncol (2019) 26(7):2191–9. doi: 10.1245/s10434-019-07338-3
23. Morris LG, Riaz N, Desrichard A, Şenbabaoğlu Y, Hakimi AA, Makarov V, et al. Pan-Cancer Analysis of Intratumor Heterogeneity as a Prognostic Determinant of Survival. Oncotarget (2016) 7(9):10051–63. doi: 10.18632/oncotarget.7067
24. Mroz EA, Tward AD, Hammon RJ, Ren Y, Rocco JW. Intra-Tumor Genetic Heterogeneity and Mortality in Head and Neck Cancer: Analysis of Data From the Cancer Genome Atlas. PloS Med (2015) 12(2):e1001786. doi: 10.1371/journal.pmed.1001786
25. Mroz EA, Tward AD, Pickering CR, Myers JN, Ferris RL, Rocco JW. High Intratumor Genetic Heterogeneity is Related to Worse Outcome in Patients With Head and Neck Squamous Cell Carcinoma. Cancer (2013) 119(16):3034–42. doi: 10.1002/cncr.28150
26. Obulkasim A, Ylstra B, van Essen HF, Benner C, Stenning S, Langley R, et al. Reduced Genomic Tumor Heterogeneity After Neoadjuvant Chemotherapy is Related to Favorable Outcome in Patients With Esophageal Adenocarcinoma. Oncotarget (2016) 7(28):44084–95. doi: 10.18632/oncotarget.9857
27. Oh BY, Shin HT, Yun JW, Kim KT, Kim J, Bae JS, et al. Intratumor Heterogeneity Inferred From Targeted Deep Sequencing as a Prognostic Indicator. Sci Rep (2019) 9(1):4542. doi: 10.1038/s41598-019-41098-0
28. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The Somatic Mutation Profiles of 2,433 Breast Cancers Refines Their Genomic and Transcriptomic Landscapes. Nat Commun (2016) 7:11479. doi: 10.1038/ncomms11479
29. Schwarz RF, Ng CK, Cooke SL, Newman S, Temple J, Piskorz AM, et al. Spatial and Temporal Heterogeneity in High-Grade Serous Ovarian Cancer: A Phylogenetic Analysis. PloS Med (2015) 12(2):e1001789. doi: 10.1371/journal.pmed.1001789
30. Takaya H, Nakai H, Sakai K, Nishio d, Murakami K, Mandai M, et al. Intratumor Heterogeneity and Homologous Recombination Deficiency of High-Grade Serous Ovarian Cancer are Associated With Prognosis and Molecular Subtype and Change in Treatment Course. Gynecol Oncol (2020) 156(2):415–22. doi: 10.1016/j.ygyno.2019.11.013
31. Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T, et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell (2018) 173(3):595–610 e511. doi: 10.1016/j.cell.2018.03.043
32. Wu P, Yang W, Ma J, Zhang J, Liao M, Xu L, et al. Mutant-Allele Tumor Heterogeneity in Malignant Glioma Effectively Predicts Neoplastic Recurrence. Oncol Lett (2019) 18(6):6108–16. doi: 10.3892/ol.2019.10978
33. Yang J, Lin Y, Huang Y, Jin J, Zou S, Zhang X, et al. Genome Landscapes of Rectal Cancer Before and After Preoperative Chemoradiotherapy. Theranostics (2019) 9(23):6856–66. doi: 10.7150/thno.37794
34. Chao J, Bedell V, Lee J, Li MS, Chu P, Yuan YC, et al. Association Between Spatial Heterogeneity Within Nonmetastatic Gastroesophageal Adenocarcinomas and Survival. JAMA Netw Open (2020) 3(4):e203652. doi: 10.1001/jamanetworkopen.2020.3652
35. Hou Y, Li T, Gan W, Lv S, Zeng Z, Yan Z, et al. Prognostic Significance of Mutant-Allele Tumor Heterogeneity in Uterine Corpus Endometrial Carcinoma. Ann Transl Med (2020) 8(6):339. doi: 10.21037/atm.2020.02.136
36. Goto T, Hirotsu Y, Amemiya K, Mochizuki H, Omata M. Understanding Intratumor Heterogeneity and Evolution in NSCLC and Potential New Therapeutic Approach. Cancers (Basel) (2018) 10(7). doi: 10.3390/cancers10070212
37. Brenner H, Kloor M, Pox CP. Colorectal Cancer. Lancet (London England) (2014) 383(9927):1490–502. doi: 10.1016/S0140-6736(13)61649-9
38. Zheng Z, Yu T, Zhao X, Gao X, Zhao Y, Liu G. Intratumor Heterogeneity: A New Perspective on Colorectal Cancer Research. Cancer Med (2020) 9(20)7637–45. doi: 10.1002/cam4.3323
39. Turajlic S, Swanton C. Metastasis as an Evolutionary Process. Science (2016) 352(6282):169–75. doi: 10.1126/science.aaf2784
40. Zhao ZM, Zhao B, Bai Y, Iamarino A, Gaffney SG, Schlessinger J, et al. Early and Multiple Origins of Metastatic Lineages Within Primary Tumors. Proc Natl Acad Sci U.S.A. (2016) 113(8):2140–5. doi: 10.1073/pnas.1525677113
41. Naxerova K, Reiter JG, Brachtel E, Lennerz JK, van de Wetering M, Rowan A, et al. Origins of Lymphatic and Distant Metastases in Human Colorectal Cancer. Science (2017) 357(6346):55–60. doi: 10.1126/science.aai8515
42. Shi W, Ng CKY, Lim RS, Jiang T, Kumar S, Li X, et al. Reliability of Whole-Exome Sequencing for Assessing Intratumor Genetic Heterogeneity. Cell Rep (2018) 25(6):1446–57. doi: 10.1016/j.celrep.2018.10.046
43. Dentro SC, Leshchiner I, Haase K, Tarabichi M, Wintersinger J, Deshwar AG, et al. Characterizing Genetic Intra-Tumor Heterogeneity Across 2,658 Human Cancer Genomes. Cell (2021) 184(8):2239–54.e39. doi: 10.1016/j.cell.2021.03.009
44. McGranahan N, Swanton C. Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer Cell (2015) 27(1):15–26. doi: 10.1016/j.ccell.2014.12.001
45. Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, et al. The Evolutionary History of Lethal Metastatic Prostate Cancer. Nature (2015) 520(7547):353–7. doi: 10.1038/nature14347
46. Kim TM, Jung SH, An CH, Lee SH, Baek IP, Kim MS, et al. Subclonal Genomic Architectures of Primary and Metastatic Colorectal Cancer Based on Intratumoral Genetic Heterogeneity. Clin Cancer Res (2015) 21(19):4461–72. doi: 10.1158/1078-0432.CCR-14-2413
47. Reiter JG, Hung WT, Lee IH, Nagpal S, Giunta P, Degner S, et al. Lymph Node Metastases Develop Through a Wider Evolutionary Bottleneck Than Distant Metastases. Nat Genet (2020) 52(7):692–700. doi: 10.1038/s41588-020-0633-2
48. Huang A, Zhao X, Yang XR, Li FQ, Zhou XL, Wu K, et al. Circumventing Intratumoral Heterogeneity to Identify Potential Therapeutic Targets in Hepatocellular Carcinoma. J Hepatol (2017) 67(2):293–301. doi: 10.1016/j.jhep.2017.03.005
49. Mamlouk S, Childs LH, Aust D, Heim D, Melching F, Oliveira C, et al. DNA Copy Number Changes Define Spatial Patterns of Heterogeneity in Colorectal Cancer. Nat Commun (2017) 8:14093. doi: 10.1038/ncomms14093
Keywords: intratumor heterogeneity, solid tumors, prognosis, clinical prognosis index, meta-analysis
Citation: Yu T, Gao X, Zheng Z, Zhao X, Zhang S, Li C and Liu G (2021) Intratumor Heterogeneity as a Prognostic Factor in Solid Tumors: A Systematic Review and Meta-Analysis. Front. Oncol. 11:744064. doi: 10.3389/fonc.2021.744064
Received: 21 July 2021; Accepted: 17 September 2021;
Published: 15 October 2021.
Edited by:
Davide Zella, University of Maryland, Baltimore, United StatesReviewed by:
Tope Oyelade, University College London, United KingdomWei Zhao, National Cancer Institute (NCI), United States
Copyright © 2021 Yu, Gao, Zheng, Zhao, Zhang, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Liu, bGcxMDU5QHRtdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Tao Yu1†
Tao Yu1† Gang Liu
Gang Liu