- 1Institute of Translational Medicine, Zhejiang University City College, Hangzhou, China
- 2Medical Genetics Center, Ningbo University School of Medicine, Ningbo, China
- 3Department of Clinical Medicine, Zhejiang University City College School of Medicine, Hangzhou, China
miR-873 is a microRNA located on chromosome 9p21.1. miR-873-5p and miR-873-3p are the two main members of the miR-873 family. Most studies focus on miR-873-5p, and there are a few studies on miR-873-3p. The expression level of miR-873-5p was down-regulated in 14 cancers and up-regulated in 4 cancers. miR-873-5p has many targeted genes, which have unique molecular functions such as catalytic activity, transcription regulation, and binding. miR-873-5p affects cancer development through the PIK3/AKT/mTOR, Wnt/β-Catenin, NF-κβ, and MEK/ERK signaling pathways. In addition, the target genes of miR-873-5p are closely related to the proliferation, apoptosis, migration, invasion, cell cycle, cell stemness, and glycolysis of cancer cells. The target genes of miR-873-5p are also related to the efficacy of several anti-cancer drugs. Currently, in cancer, the expression of miR-873-5p is regulated by a variety of epigenetic factors. This review summarizes the role and mechanism of miR-873-5p in human tumors shows the potential value of miR-873-5p as a molecular marker for cancer diagnosis and prognosis.
Introduction
With the increasing incidence and mortality of cancer worldwide in recent decades, it has become the second leading cause of human death (1). MicroRNA (miRNA) is a set of non-coding RNA (2) less than 25 nucleotides in length. miRNAs can bind to the 3’-untranslated region (3’-UTR) of target mRNA molecules and regulate the expression of target genes, thus playing an important role in cancer (3). The miR-873 family is located on chromosome 9 (chr9:28888878-28888954). Its family includes two main members of the human genome, including hsa-miR-873-5p (miR-873-5p) and hsa-miR-873-3p (miR-873-3p). Their mature sequences are 21 and 22 nucleotides in length, respectively, and are highly conserved (Figure 1). At present, most researches focus on miR-873-5p.
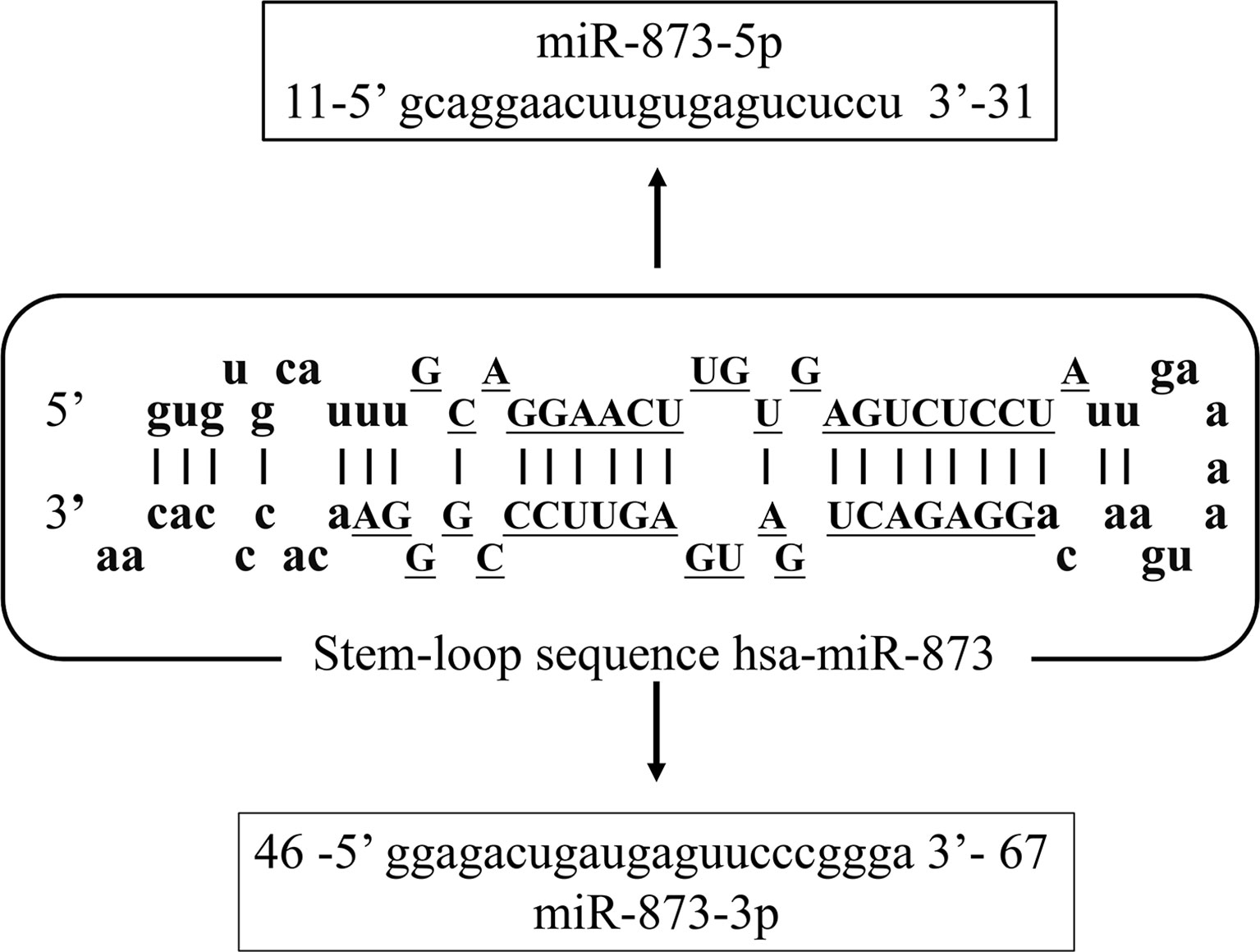
Figure 1 The sequence structure of the miR-873 family. Hsa-mir-873 is located on chromosome 9 (chr9:28888878-28888954). It has two mature sequences, hsa-miR-873-5p (MIMAT0004953, miR-873-5p) and hsa-miR-873-3p (MIMAT0022717, miR-873-3p).
Studies have found that the expression of miR-873-5p is dysregulated in a variety of cancers and plays different roles in different cancers. On the one hand, miR-873-5p is upregulated and carcinogenic in non-small cell lung cancer (NSCLC) (4), and hepatocellular carcinoma (HCC) (5); on the other hand, miR-873-5p is involved in colorectal cancer (CRC) (6) and gastric cancer (GC) (7) are down-regulated and exert a tumor suppressor effect. miR-873-5p can affect cell proliferation (5), apoptosis (6), migration (8), invasion (9), cell stemness (10), and other biological processes by regulating the expression of its target genes. In addition, miR-873-5p also has important clinical significance in drug sensitivity and prognosis of cancer patients (4, 6). miR-873-5p can also be regulated by a variety of epigenetic factors. Among them, the interaction between non-coding RNA (lncRNA or circRNA) and miR-873-5p is mainly researched. This review focuses on studying the biological role of miR-873-5p in tumors, exploring the molecular functional network of its targeted genes, and predicting the potential role of miR-873-5p in the diagnosis and prognosis of human cancer.
The Biological Function of miR-873-5p Target Genes
miR-873-5p can directly bind to the 3’-UTR of target gene mRNA and regulate gene expression after transcription. The target gene of miR-873-5p has unique molecular functions, including catalytic activity, transcription regulation, binding, etc. (Figure 2).
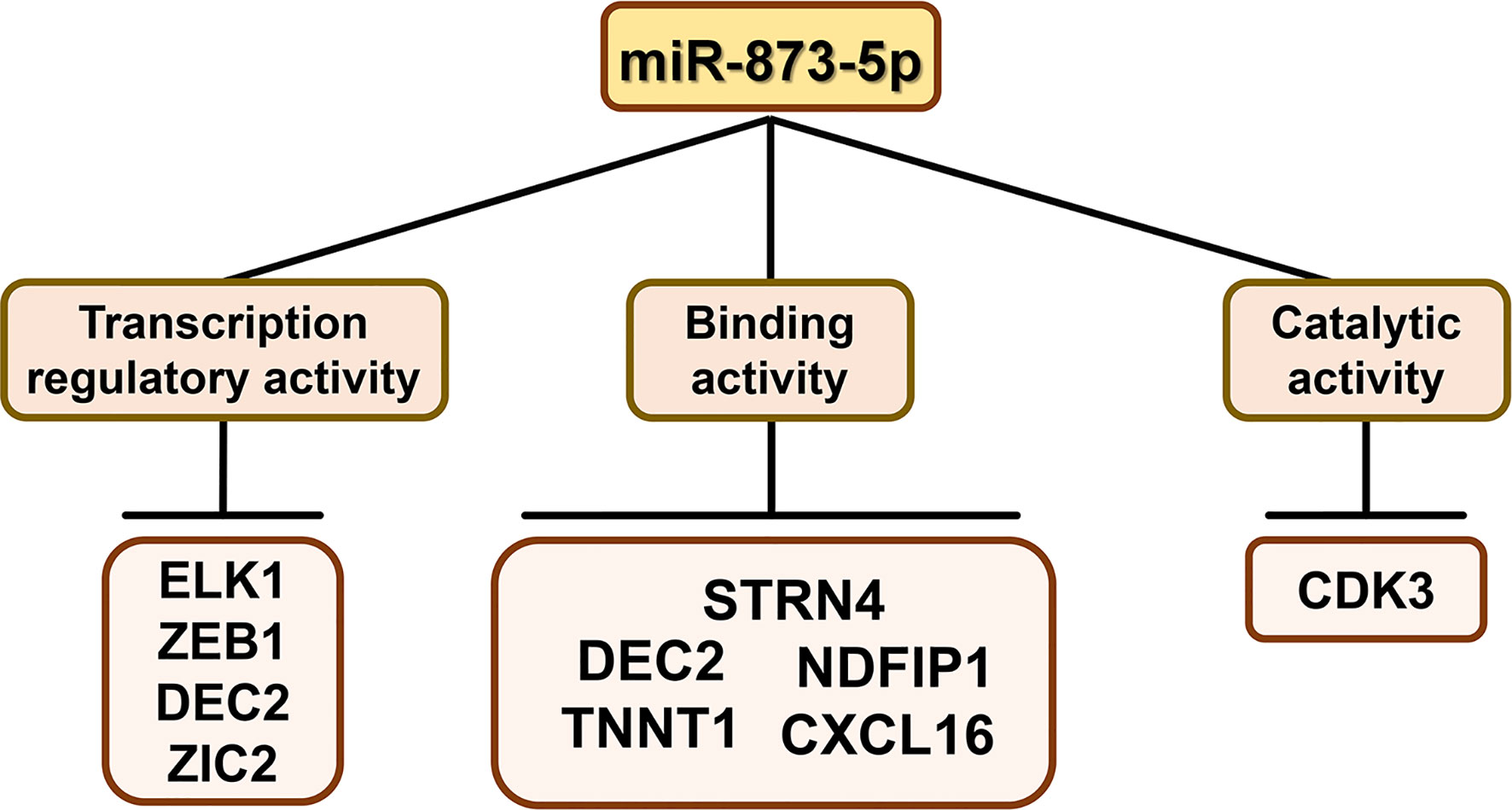
Figure 2 The molecular functions of miR-873-5p target genesThe target genes of miR-873-5p have the molecular functions of binding, catalytic, and transcription regulator activity.
Among the miR-873-5p target genes, CDK3 is a catalytically active gene. CDK3 is a cyclin-dependent kinase, which can phosphorylate the estrogen receptor (ER) and enhance ER activity, thereby promoting the occurrence and development of breast cancer (BC) (11).
Among the miR-873-5p target genes, genes with transcriptional regulatory activity are ELK1, DEC2, ZEB1, and ZIC2. ELK1 is a key transcriptional regulator that mediates the MEK-ERK signal transduction, and it can activate early oncogene expression (12, 13). DEC2 is the basic helix-loop-helix transcription factor of the clock gene. It plays an important role in the circadian rhythm, cell proliferation, and apoptosis, and thus participates in tumor progression (14). ZEB1 is a member of the zinc finger E-box binding protein (ZEB) transcription factor family (15). ZEB1 can bind to the promoter of the liver cancer-derived growth factor (HDGF) and increase the level of HDGF transcription, leading to the pathogenesis of endometrial cancer (EC) (16). ZIC (Cerebellar Zinc Finger Protein) protein has five highly conserved Cys2His2 motifs, which can bind to DNA and thus function as a transcription factor (17). As a member of the ZIC family, ZIC2 can promote tumor growth and metastasis of hepatocellular carcinoma through transcriptional regulation of p21-activated kinase 4 (18). In addition, ZIC2 can bind to the DNA-binding high mobility base box of TCF4, thereby inhibiting the transcriptional activity of β-catenin (19).
The miR-873-5p target genes with binding activity include DEC2, NDFIP1, STRN4, TNNT1, and CXCL16. DEC2 can inhibit its downstream molecules by binding to the E-box (20). NDFIP1 is a membrane protein with small endosomes containing PY motifs, which can transport E3 ligase and its substrate to endosomes (21). STRN4 is a member of the striatin family. It can combine with MINK1 of the germinal center kinase family to form a large complex, which is essential for the process of cytokinesis (22, 23). Troponin T1 (TNNT1) is a subunit of troponin T, which can bind to tropomyosin and anchor the troponin complex at a specific location on striated muscle filaments (24). CXCL16-CXCR6 are chemokines and chemokine receptors, respectively, which can bind to each other (25). The mutual binding of CXCL16 and CXCR6 involves a variety of biological activities, including cell adhesion (26) and anti-tumor immunity (27).
MiR-873-5p Dysregulation in Various Cancers
As shown in Table 1, miR-873-5p is abnormally expressed in 18 types of cancers. Among them, miR-873-5p is up-regulated in 4 types of cancers, including NSCLC (4), lung adenocarcinoma (LUAD) (28, 29), lung cancer (LCA) (5, 8, 30), and Merkel cell carcinoma (MCC) (24627810). miR-873-5p is down-regulated in 14 types of cancers, including nasopharyngeal carcinoma (NPC) (32), lung cancer (LCA) (33), cervical cancer (CC) (34, 35), EC (36), BC (10, 11, 37, 38), pancreatic ductal adenocarcinoma (PDAC) (39), glioblastoma (GM) (9, 40–42), osteosarcoma (OS) (43), papillary thyroid carcinoma(PTC) (44), CRC (6, 45–49), esophageal cancer(ESCA) (50),GC (7, 51, 52), tongue squamous cell carcinoma (TSCC) (53), and pancreatic cancer(PC) (54).
Highly expressed miR-873-5p can inhibit cell proliferation, induce cell apoptosis, inhibit EMT, metastasis, and invasion process, thereby promoting the occurrence and development of cancer. Among the four types of cancers (NSCLC, LUAD, HCC, and MCC), miR-873-5p can promote their progression, indicating that miR-873-5p has tumor suppressor and cancer-promoting effects.
The Biological Role of miR-873-5p in Human Cancer
MiR-873-5p and Different Signaling Pathways
miR-873-5p can affect the occurrence and development of cancer by participating in the PIK3/AKT/mTOR, Wnt/β-Catenin, NF-κβ, MEK/ERK, and other signaling pathways (Figure 3).
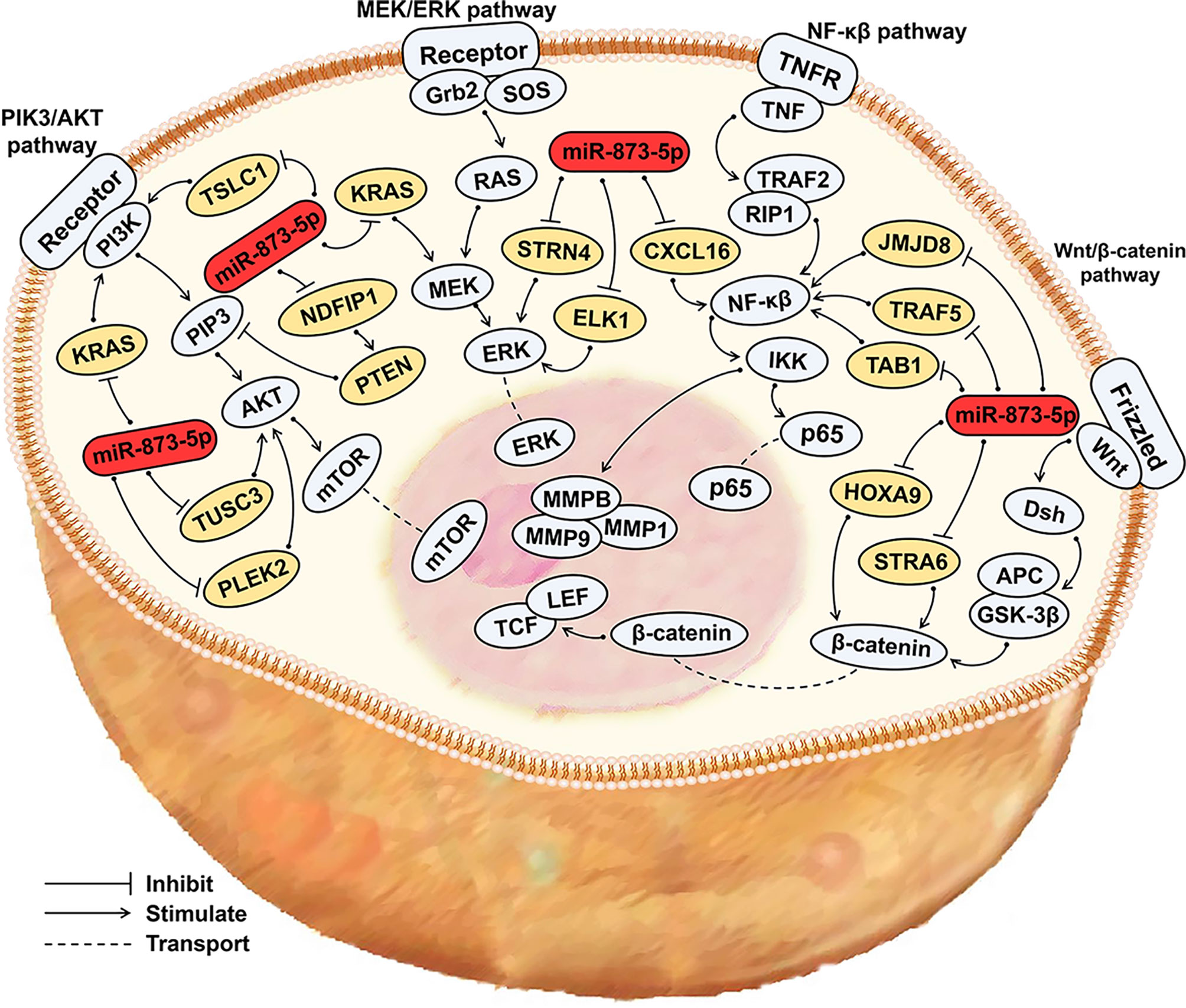
Figure 3 miR-873-5p related signaling pathways in cancer. miR-873-5p can influence cancer development by participating in the PIK3/AKT/mTOR, Wnt/β-Catenin, NF-κβ, MEK/ERK signaling pathways. Blue, signaling pathway; Orange, target gene; Red, miR-873-5p.
The PIK3/AKT signaling pathway is often overactivated in malignant tumors. The PIK3/AKT signaling pathway can participate in cell cycle regulation, promote cell proliferation and metastasis, and inhibit cell apoptosis (55). In HCC, miR-873-5p promotes the development of HCC through the NDFIP1/AKT/mTOR axis (5). miR-873-5p can directly activate PIK3/AKT to promote HCC progression (30). miR-873-5p can down-regulate TUSC3 expression, inhibit the AKT signaling pathway, and thus hinder CRC development (49). In PC, miR-873-5p targets PLEK2 and inhibits the AKT signaling pathway, thereby inhibiting the development of cancer (54).
The Wnt/β-Catenin signaling pathway is important for tumor development, and the dysregulation of the Wnt/β-Catenin signaling pathway may lead to cell proliferation and malignancy (56). miR-873-5p inhibits the expression of HOXA9 and STRA6, and blocks the Wnt/β-Catenin signaling pathway, thereby inhibiting the development of OS and GC (43, 52).
The NF-κβ signaling pathway can inhibit cell apoptosis, and it is closely related to tumor occurrence, growth, and metastasis (57). By inhibiting the expression of JMJD8, TNF receptor-related factor 5 (TRAF5) and TGF-β activated kinase 1 (MAP3K7) binding protein 1 (TAB1), miR-873-5p can inhibit the NF-κβ signaling pathway, thereby hindering the progression of CRC (6, 45). In PTC, miR-873-5p can down-regulate the expression of CXCL16, and can also inhibit the development of PTC through down-regulating the NF-κβ signaling pathway (44).
The MEK/ERK signaling pathway can promote cell proliferation and migration and is involved in the occurrence and development of a variety of cancers (12). In CRC, miR-873-5p targets ELK1 and STRN4, and exerts a tumor suppressor effect through the ERK signaling pathway (47). By down-regulating KRAS expression, miR-873-5p can also inhibit the ERK signaling pathway to suppress the development of PDAC and TNBC (39). In addition, miR-873-5p can also deactivate the PI3K/AKT and ERK signaling pathways to inhibit the development of BC (58).
MiR-873-5p and Cell Cycle
The regulation of the cell cycle is of great significance to the proliferation and apoptosis of cancer cells (Figure 4). Increased expression of miR-873-5p can inhibit the expression of GLI1 and cyclin B, thereby inducing GC cells to arrest the G2/M cell cycle (51). After miR-873-5p targets to inhibit JMJD8, it blocks CRC HCT116 and SW480 cells in the G1-S cell cycle (6). miRNA-873-5p can accelerate the S phase process of HCC cells, thereby promoting cancer cell proliferation (30). Other studies have shown that miR-873-5p can down-regulate STRA6, thereby inducing GC cells to arrest in the G0/G1 cell cycle and increasing cell mortality (30). After miR-873-5p targeted IGF2BP1, GM cells showed significant G0/G1 block and S phase reduction (42).
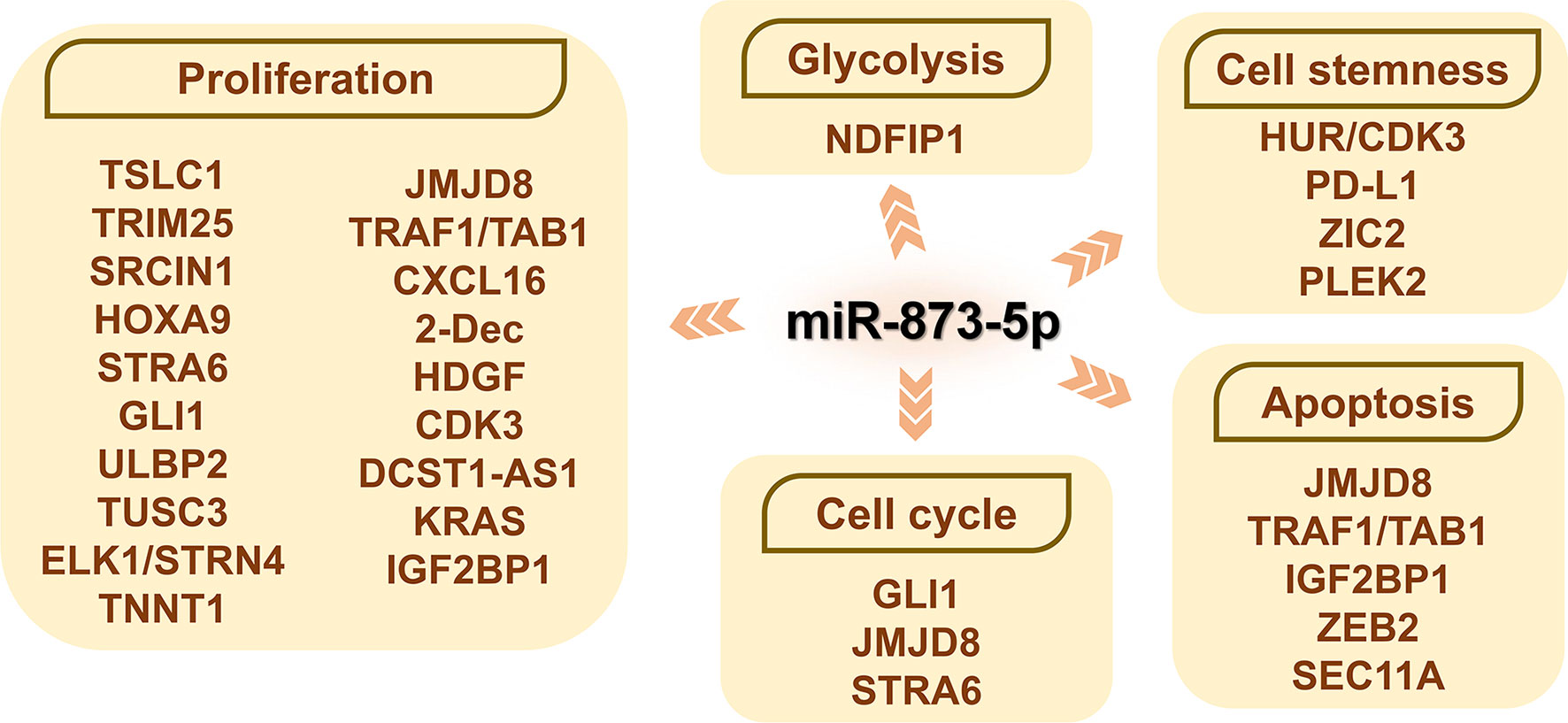
Figure 4 The role of miR-873-5p and its target genes on the cell biology of cancer cells. By promoting or inhibiting cell proliferation and apoptosis, miR-873-5p has both oncogenic or pro-cancer effects in different cancers. miR-873-5p inhibits the aerobic glycolysis of cancer cells by targeting NDFIP1. In addition, miR-873-5p can reduce the stemness of cancer cells by targeting PD-L1 and HUR/CDK3. By targeting GLI1, JMJD8, IGF2BP1, and STRA6, miR-873-5p can inhibit the progression of the cancer cell cycle.
MiR-873-5p and Cell Proliferation and Apoptosis
The targeted genes of miR-873-5p are closely related to the process of tumor cell proliferation and apoptosis (Figure 4).
STRN4 directly acts on protein kinases such as MINK1, TNIK, and MAP4K4. The knockdown of STRN4 inhibits the proliferation of PDAC and CRC cancer cells (59). miR-873-5p can target ELK1 and STRN4 and inhibit the proliferation of CRC LoVo and HCT116 cells through the regulation of the ERK-CyclinD1 signaling pathway (47).
Human cytomegalovirus glycoprotein UL16 binding protein 2 (ULBP2) is an important activation receptor on the surface of natural killer cells. In normal tissues, low levels of ULBP2 can lead to the activation of immune cells (60, 61). In CC C33a cells, miR-873-5p activates immune cells by inhibiting ULBP2 expression, thereby attenuating cell proliferation (34).
Jumonji domain-containing protein 8 (JMJD8) contains a JmjC domain (62) at 74-269 amino acid residues. miR-873-5p can inhibit the NF-κβ signaling pathway by down-regulating the expression of JMJD8 in CRC cells, thereby inhibiting cell proliferation, blocking the G1-S transition, and enhancing the apoptosis of CRC HCT116 and SW480 cells (6). miR-873-5p directly targets the 3’-UTR of TUSC3 to down-regulate its expression and inhibit AKT signaling pathway and CRC cell proliferation (49). TNNT1 expression is closely related to the clinical stage of tumor tissues and can promote the proliferation of cancer cells through metastatic G1/S transition (63). miR-873-5p down-regulates TNNT1 and may inhibit the proliferation of CRC cells (48). Besides, TRAF5 and TAB1 are both key components of the NF-κβ signaling pathway (45). miR-873-5p directly targets TRAF5 and TAB1 to inhibit the NF-κβ signaling pathway, thereby inhibiting the cell proliferation of CRC (45).
KRAS can enhance the AKT and ERK signaling pathways that are related to cell proliferation (64). miR-873-5p inhibits the cell proliferation of PDAC and TNBC tissues (38) by targeting KRAS, thereby inhibiting the ERK and PI3K/AKT signaling pathways (39). miR-873-5p can induce apoptosis of PDAC and TNBC by regulating the Caspase-dependent apoptotic pathway (39). DCST1-AS1 is an oncogenic lncRNA (38). DCST1-AS1 can sponge miR-873-5p and thus reduce the inhibition of miR-873-5p on the expression of IGF2BP1, thereby up-regulating the expression of MYC and promoting the proliferation of TNBC cells (38).
Tripartite motif-containing protein 25 (TRIM25) is a member of TRIM protein, which can target the degradation of MTA-1 (65). MTA-1 is a member of the metastasis-related gene (MTA) family and plays an important role in the proliferation of cancer cells (66). miR-873-5p can inhibit TRIM25 expression, which can promote the proliferation of HCC cells (8). TSLC1 is a new type of tumor suppressor gene, which is related to proliferation, apoptosis, cell cycle, and tumorigenicity of cancer cell (67). The inhibition of TSLC1 by miRNA-873-5p can lead to hyperphosphorylation of PI3K/AKT/mTOR and other signaling pathways to promote HCC cell proliferation (30).
Src is a tyrosine kinase that is frequently up-regulated in cancer and is very important for cancer cell proliferation (68, 69). Src Kinase Signaling Inhibitor 1 (SRCIN1) is a tumor suppressor gene that suppresses cancer by inactivating Src in cancer (70). miR-873-5p activates the Src signaling pathway by down-regulating of SRCIN1 expression and promotes the proliferation of LUAD cells (28).
Insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1) is a carcinoembryonic protein that is expressed in various cancers including leukemia (71). IGF2BP1 can stabilize and enhance the expression of c-MYC and MKI67, which are both effective regulators of cell proliferation and apoptosis (72). Overexpression of miR-873-5p in GM cells can significantly down-regulate the expression of IGF2BP1, MKI67, and c-MYC, and lead to cell proliferation inhibition and apoptosis (42). ZEB2 is a transcription factor containing zinc fingers, which is essential in early embryonic development (73). ZEB2 can increase the expression of cyclin A1, cyclin D1, and Bcl-2 in GM cells, thereby promoting the growth of GM cells (74). miR-873-5p down-regulates ZEB2 expression, which can promote GM cell apoptosis (74).
HOXA9 is a member of the mammalian HOX family (75), which is abnormally activated in a variety of cancers such as CRC (76) and GC (77). miR-873-5p directly targets HOXA9 and reduces the expression levels of β-catenin and cyclin D1 through the inactivation of the Wnt/β-catenin signaling pathway, thereby inhibiting OS cell proliferation (43).
Hedgehog (Hh) signaling pathway can participate in the cancer process through mechanisms such as promotion of tumor invasion and metastasis (78, 79). GLI1 is a transcription factor of the Hh signaling pathway and downstream target genes and is usually used as a marker to activate the Hh signaling pathway (80). Studies have found that increased expression of miR-873-5p can inhibit the expression of GLI1 and inhibit the cell proliferation of NSCLC (4), GC (51), and CC (35) through the Hh signaling pathway. STRA6, as a transmembrane protein of RA, is overexpressed in many cancer types (81). Overexpression of STRA6 can upregulate Wnt pathway-related genes, such as β-catenin, MMP-7, and c-myc. miR-873-5p down-regulates the expression of STRA6 in GC and can inhibit GC cell proliferation (52).
The estrogen receptor (ER) is a member of the nuclear receptor superfamily of ligand-activated transcription factors and plays an important role in BC (82). miR-873-5p inhibits ER activity by targeting CDK3, thereby inhibiting the growth of BC cells (11).
As a chemokine, the binding of CXCL16 to its sole receptor CXCR6 can involve biological activities such as cell adhesion (26) and anti-tumor immunity (27). Silencing CXCL16 can inhibit the proliferation and invasion of cancer cells by regulating the NF-κβ signaling pathway (83). Overexpression of miR-873-5p targets CXCL16 and suppresses the NF-κβ signaling pathway in PTC cells, thereby inhibiting PTC cell proliferation (44).
HDGF is a secreted growth factor (84), which can interact with the β-catenin pathway and promote cancer cell proliferation (85). Therefore, miR-873-5p targeted down-regulation of HDGF may inhibit EC cell proliferation through the β-catenin signaling pathway (36). DEC2 plays an important role in circadian rhythm, cell proliferation, and apoptosis, and is also closely related to tumor progression (14). In ESCA, miR-873-5p can inhibit ESCA cell proliferation by targeting the DEC2 gene, thereby affecting the circadian rhythm (14, 50).
MiR-873-5p and Cell Migration, Invasion, and EMT
The migration and invasion of cancer cells are important for the progression of cancer. Epithelial cell-mesenchymal transition (EMT) is a process of epithelial cell changes, which is characterized by weak cell adhesion and enhanced migration ability (86). EMT is an important marker of cancer progression and metastasis of malignant tumors (87) (Figure 5).
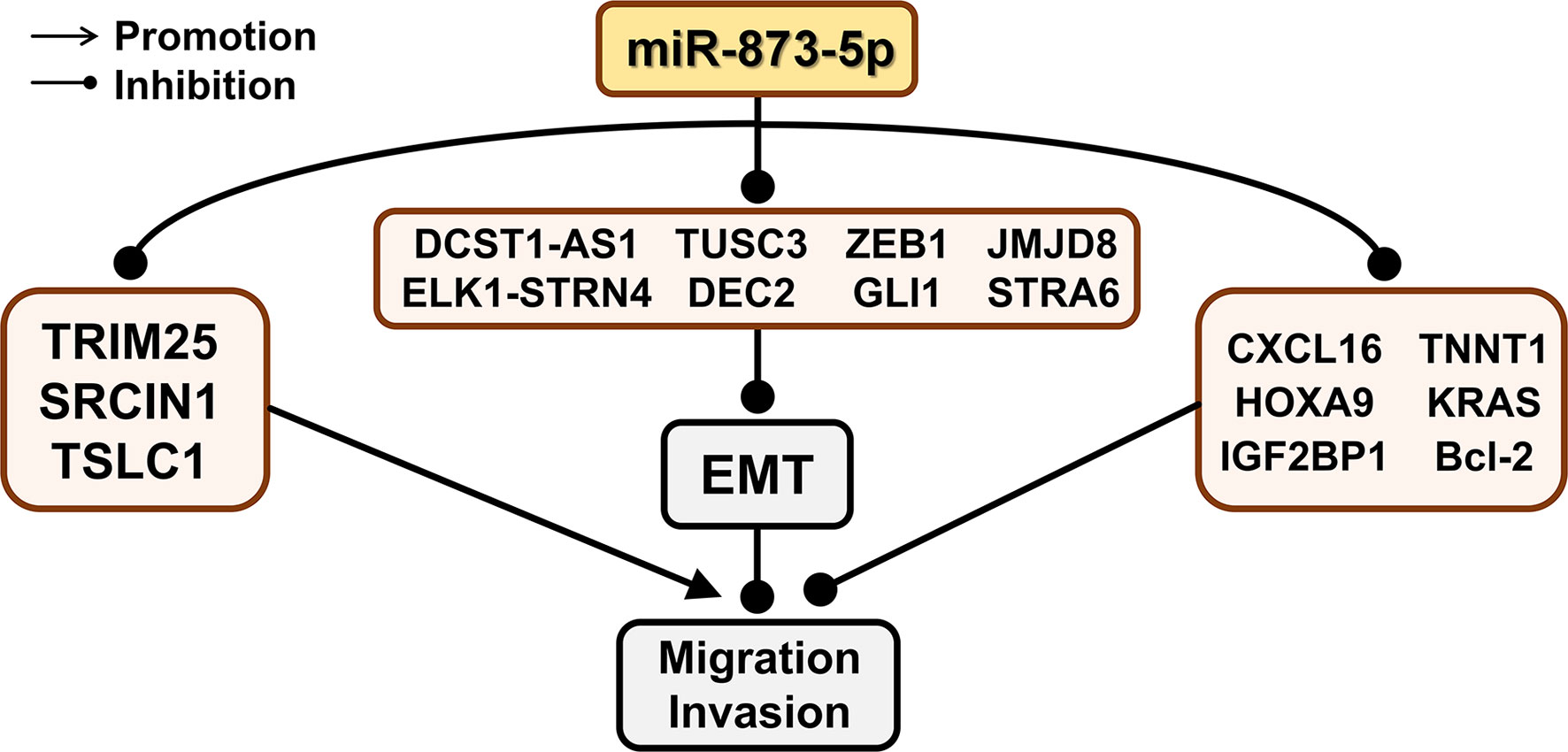
Figure 5 The effect of miR-873-5p target genes on EMT, migration, and invasion of cancer cells. miR-873-5p inhibits the EMT process by inhibiting the expression of ZEB1, TUSC3, DCST1-AS1, JMJD8, ELK1-STRN4, GLI1, STRA6, and DEC2. MiR-873-5p targets TRIM25, SRCIN1, and TSLC1, and promotes cell migration and invasion. In addition, miR-873-5p inhibits the migration and invasion of cancer cells by targeting CXCL16, TNNT1, IGF2BP1, Bcl-2, TNNT1, and HOXA9.
In CRC HCT8 cells, the down-regulation of miR-873-5p corresponds to the up-regulation of ELK1 and STRN4, which leads to the down-regulation of E-cadherin and α-E-catenin and enhances EMT, and ultimately promotes the migration of CRC cells (47). ZEB1 is closely related to migration and EMT (88). In CRC, the up-regulation of miR-873-5p also corresponds to the down-regulation of ZEB1 expression, thereby significantly increasing the levels of E-cadherin, β-catenin, and ZO-1. This leads to a decrease in the levels of N-cadherin and vimentin, which changes the cell phenotype from EMT to MET, thereby inhibiting the EMT process of CRC cells (46). When miR-873-5p targets JMJD8, the expression of E-cadherin and cytokeratin is significantly increased, thereby weakening the EMT effect and inhibiting the migration and invasion of CRC cells (6). TUSC3 may change the EMT of CRC by regulating PI3K/Akt and WNT/β-catenin signaling pathways, thereby changing its metastasis and invasiveness (89). miR-873-5p can negatively regulate the expression of TUSC3, thereby inhibiting the EMT ability of CRC cells (49). TNNT1 is negatively correlated with the expression of E-cadherin in colon adenocarcinoma (90). miR-873-5p can regulate E-cadherin expression by targeting TNNT1, thereby inhibiting CRC cell migration and invasion (48).
When miR-873-5p targets to inhibit GLI1, the expression level of E-cadherin is significantly increased, while the levels of N-cadherin and vimentin are significantly reduced, thereby inhibiting the EMT process of CC cells (35). miR-873-5p can negatively regulate ULBP2 and activate immune cells, thereby reducing the invasion and metastasis of CC cells (34).
In GC cells, miR-873-5p can lead to the downregulation of N-cadherin and vimentin by inhibiting STRA6, thereby inhibiting the EMT process of GC cells, and cell metastasis and invasion (52).
LEF1 is an important transcription factor involved in the activation of the Wnt signaling pathway, which can promote the synthesis of mesenchymal fibronectin and EMT (91). When miR-873-5p binds to DCST1-AS1, the expression of LEF1 is up-regulated, and the EMT of TNBC cells is enhanced to promote cancer cell migration and invasion (38).
In ESCA, miR-873-5p can down-regulate the expression of DEC2, thereby inhibiting the effect of EMT and reducing the migration and invasion of ESCA cells (50, 92).
In PDAC and TNBC, miR-873-5p can target KRAS, thereby inhibiting cell migration and invasion through the ERK/AKT signaling pathway (39). The Wnt/β-catenin signaling pathway is a key mechanism for cell maintenance and development, including cell differentiation, migration, and invasion (93).
miR-873-5p can target HOXA9 and inhibit the migration and invasion of OS cells through suppressing the Wnt/β-catenin signaling pathway (43).
MTA-1 can promote cell metastasis through histone deacetylation and nucleosome remodeling (66). After miR-873-5p inhibits the expression of TRIM25, the function of MTA-1 is enhanced to promote the metastasis and invasion of HCC cells (8). TSLC1 is a specific tumor suppressor involved in cell adhesion and invasion (94). Therefore, in HCC, miR-873-5p can target TSLC1 to increase HCC cell adhesion, thereby promoting HCC cell migration (30).
SRCIN1 is the main regulator of E-cadherin (95), which can regulate the growth and movement of cell (96). miR-873-5p down-regulates the expression of SRCIN1, which can reduce cell adhesion and promote the migration of LUAD cell A549 (28).
IGF2BP1 can enhance the directionality of cell migration in a PTEN-dependent manner. miR-873-5p can down-regulate the expression of PTEN by targeting IGF2BP1, thereby inhibiting the migration ability of GM cells (42). Matrix metalloproteinases (MMP) have been shown to activate and regulate GM cell migration (97). Bcl-2 is an oncogene and it can promote the migration and invasiveness of GM cells by enhancing the activity of MMP (98). miR-873-5p can target Bcl-2 to enhance the activity of MMP and inhibit the migration and invasion of GM cells (9). MMPs are related to the development of cancer, which can promote the degradation of extracellular matrix and cell invasion and metastasis (99, 100). Overexpression of miR-873-5p can inhibit the expression of MMP1, MMP9, and MMP13 by down-regulating CXCL16, thereby inhibiting the migration and invasion of PTC cells (44).
MiR-873-5p and Cell Stemness
Although cancer stem cells (CSCs) only account for a small part of cancer cells, they have the ability to self-renew (101). At present, CSC is considered to be the main factor leading to tumor recurrence and drug resistance (102).
Programmed cell death ligand 1 (PD-L1) is an immune checkpoint molecule and a ligand for PD-1 (103). The expression of PD-L1 is highly correlated with stemness-related genes in BC tissues and is overexpressed in basal BC. Therefore, PD-L1 may promote the stemness of BC cells (104, 105). PD-L1 can activate the PI3K/AKT and ERK signaling pathways in BC (106). And miR-873-5p can target PD-L1 and down-regulate its expression, and then inhibit the stemness of BC cells through the PI3K/Akt and ERK1/2 signaling pathways (10).
HuR is an RNA binding protein that can promote the progression of various tumors (107). HuR can directly bind and up-regulate CDK3 to promote the stemness of LCA (33). miR-873-5p can competitively bind to CDK3 with HuR and reduce CDK3 expression, thereby reducing the stemness of LCA cells (33).
Studies have found that ZIC2 may affect the occurrence and development of tumors through the AKT signaling pathway (108). Up-regulation of miR-873-5p can inhibit the expression of ZIC2 and disrupt the AKT signaling pathway, thereby inhibiting the stemness and tumorigenicity of NPC cells (109). Overexpression of miR-873-5p can silence PLEK2 and inhibit the self-renewal of PC stem cells through the PIK3/AKT signaling pathway, thereby inhibiting the development of PC (54).
MiR-873-5p and Glycolysis
Tumor cells can change their metabolism to adapt to the challenging hypoxic environment (110). Intermediates in glycolysis can be used to meet the biosynthetic needs of rapidly growing tumors (111). AKT/rapamycin (mTOR) activation enables the continued growth and survival of tumor cells that rely on aerobic glycolysis, while the expression of NDFIP1 reduces the AKT/mTOR signaling pathway in cancer cells (5). In HCC, miR-873-5p inhibits the Warburg effect through the NDFIP1/AKT/mTOR axis, thereby inhibiting the aerobic glycolysis of HCC cells (5).
The Role of miR-873-5p in Cancer Treatment
Gefitinib (EGFR-TKI) can reduce viability and proliferation of cancer cells and angiogenesis in NSCLC (Figure 6). However, the resistance of cancer cells to gefitinib has greatly limited its clinical application (4, 112, 113). The enhancement of the GLI1 expression can increase the radiation resistance of NSCLC cells. When GLI1 is silenced, gefitinib can significantly reduce the growth of NSCLC cells (114, 115). The down-regulation of GLI1 by miR-873-5p can reduce the resistance of NSCLC cells to gefitinib, thereby causing NSCLC PC9 cell apoptosis (4).
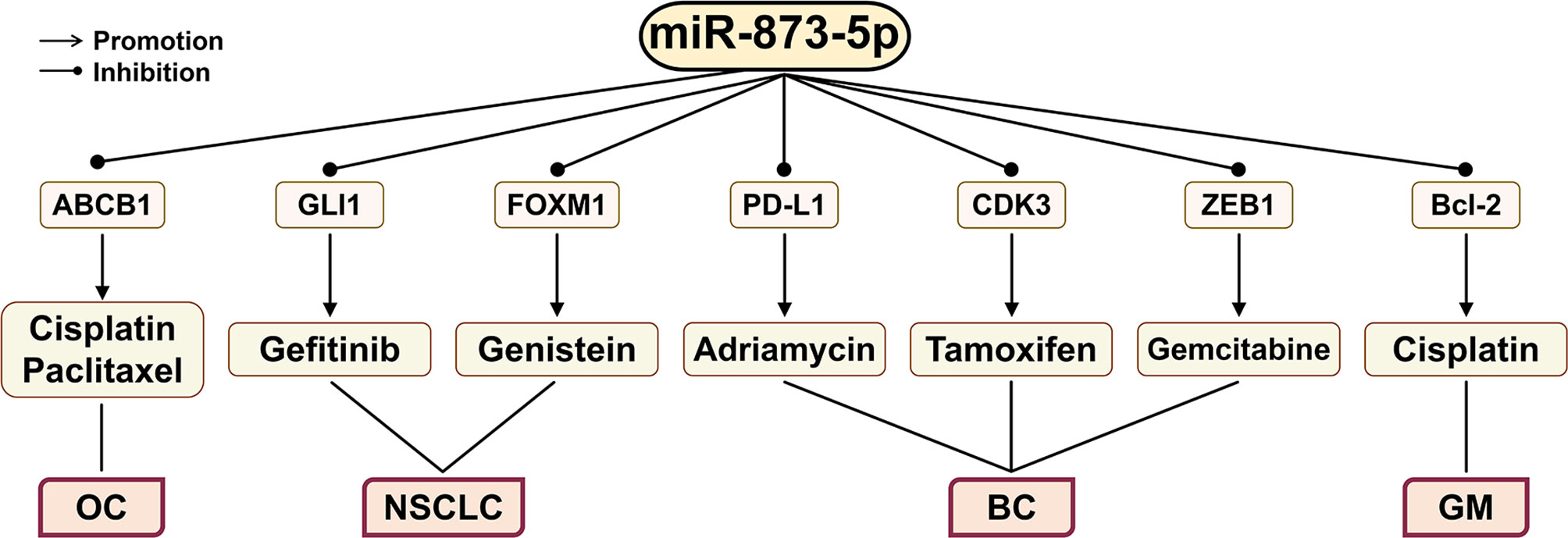
Figure 6 The effect of miR-873-5p on the efficacy of different cancer chemotherapy drugs through its target genes. miR-873-5p inhibits the expression of ABCB1, Bcl-2, GLI1, FOXM1, PD-L1, CDK3, and ZEB1, thereby improving the inhibitory effects of various anticancer drugs on cancer cells. OC, ovarian cancer; GM, glioblastomas; LC, lung cancer; BC, breast cancer.
The main treatments for BC include surgery, targeted therapy, radiotherapy, and chemotherapy. For TNBC, chemotherapy is the only treatment (10). CSCs may contribute to the chemoresistance of cancer (116). By activating the PI3K/Akt and ERK1/2 signaling pathways, the PD-1/PD-L1 axis can promote the stemness and drug resistance of BC cells. miR-873-5p targeted inhibition of PD-L1 expression can attenuate the resistance of BC cells to Adriamycin (10). In addition, miR-873-5p may also inhibit ERa phosphorylation by targeting CDK3, thereby restoring the sensitivity of BC drug-resistant cells to tamoxifen (11).
Norbiliin (NCTD) is a dimethyl analog of phthalazine, which can inhibit the biological functions of cell proliferation and angiogenesis in a variety of cancers (117–119). NCTD can overcome tamoxifen resistance by targeting the miR-873-5p/CDK3 axis in BC cells (120).
Gemcitabine is a chemotherapy drug that is derived from deoxycytidine and is commonly used to treat BC patients (121). ZEB1 plays a key role in promoting the development of CSCs, and its overexpression is related to cancer chemoresistance (15). miR-873-5p can bind to the 3’-UTR of ZEB1 to directly inhibit its expression, thereby enhancing the cell growth inhibition induced by gemcitabine treatment (37).
Ovarian cancer (OC) is mostly treated with cisplatin and paclitaxel, but OC cancer cells often develop resistance to these drugs (122). The ABC superfamily transporter and P-glycoprotein (MDR1) play a key role in the multidrug resistance (MDR) of cancer. They can mediate the outflow of various chemical drugs, such as anticancer drugs (123–125). Overexpression of miR-873-5p increases the sensitivity of OC cells to cisplatin and paclitaxel by targeting ABCB1 to down-regulate the expression of MDR1 (126).
GM is the most common primary brain tumor in adults, and cisplatin is currently a chemical drug widely used to treat GM (127, 128). A study has found that inhibiting the expression of Bcl-2 can enhance the sensitivity of GM to cisplatin (129). miR-873-5p can enhance the sensitivity of GM cells to cisplatin by targeting Bcl-2 (9).
In addition, genistein is a soy-derived isoflavone that can play a beneficial role in cancer treatment (130). Genistein can inhibit the progression of NSCLC by regulating the circ_0031250/miR-873-5p/FOXM1 axis (131).
The Regulation of miR-873-5p in Human Cancer
Current studies have found that methyltransferase, circRNA, and lncRNA are involved in the regulation of miR-873-5p in human cancer (Figure 7).
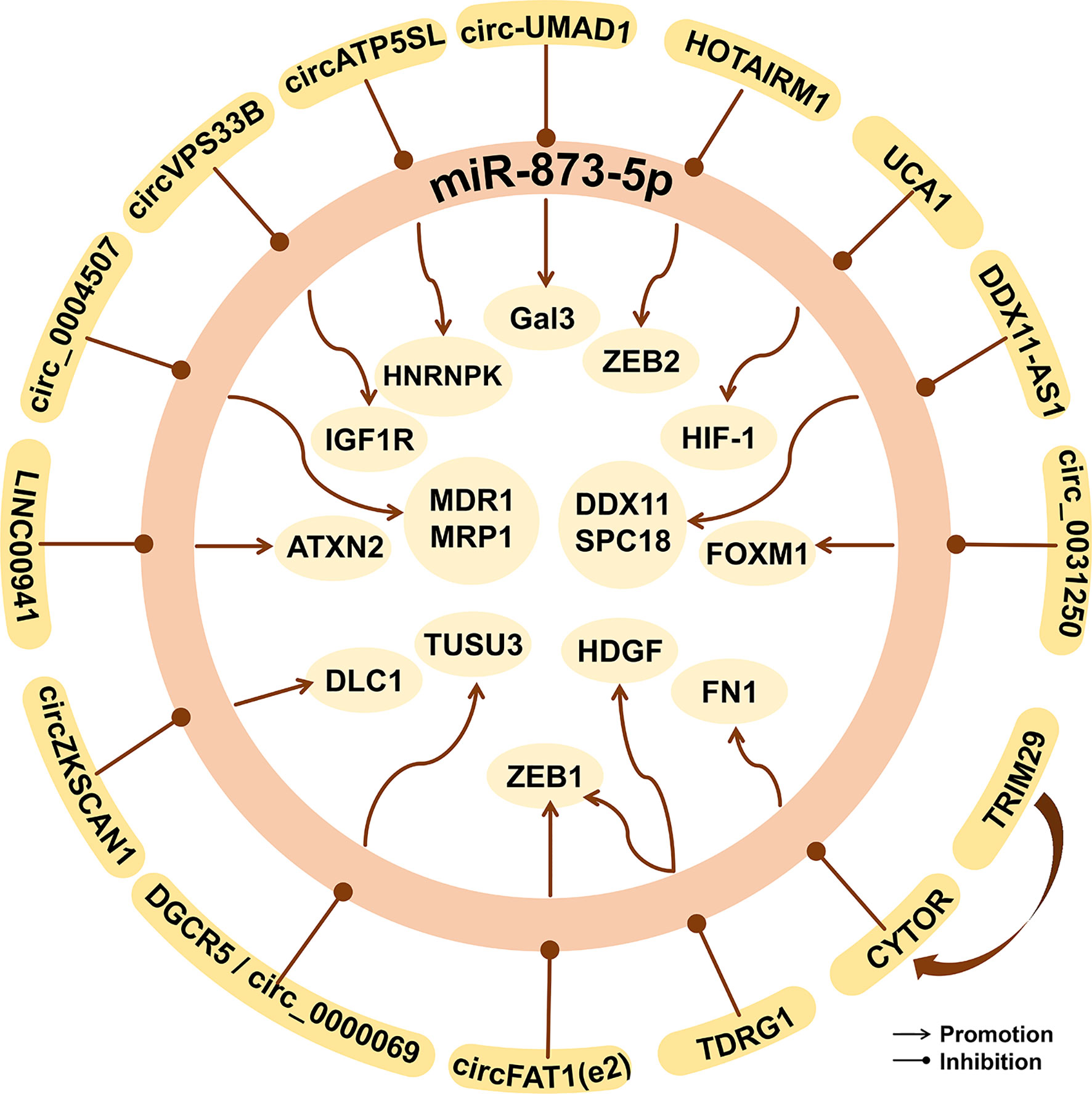
Figure 7 The epigenetic factors of miR-873-5p in human cancer. miR-873-5p can be targeted and regulated by lncRNAs, circRNAs, and other proteins, thus affecting downstream gene expression and playing an important role in cancer.
CircRNA is a new type of non-coding RNA that can bind miRNAs to stop their regulation of target genes (132). Hsa_circ_0000069 can sponge miR-873-5p, which can promote the expression of TUSC3, thereby promoting the proliferation, migration, and invasion of CC cells (133). In Neuroblastoma, circDGKB can sponge miR-873-5p to increase the expression of ZEB1 and GLI1, and promote the occurrence and development of cancer (134). circ-UMAD1 can sponge miR-873-5p, thereby up-regulating the expression of Galectin-3 and inducing lymphatic metastasis of PTC (135). circFAT1(e2) can promote the proliferation, metastasis, and invasion of PTC cells by inhibiting the miR-873-5p/ZEB1 axis, thereby exerting a carcinogenic effect (136). Knockout of circ_0004507 can up-regulate the expression of miR-873-5p and inhibit the progression of laryngeal cancer (137). circ_0031250 can promote the proliferation, migration, and invasion of NSCLC cells by inhibiting the miR-873-5p/FOXM1 axis (131). circZKSCAN1 can inhibit the progression, proliferation, migration, and invasion of HCC by down-regulating the miR-873-5p/DLC1 axis, thereby hindering the occurrence and development of HCC (138). Infant hemangioma (IH) is one of the most benign endothelial tumors in infants and young children. circATP5SL can eliminate the inhibition of IGF1R by sponging miR-873-5p, thereby promoting IH cell invasion, proliferation, and migration (139). circVPS33B accelerates tumor cells’ proliferation, migration, and growth by down-regulating the miR-873-5p/HNRNPK axis in invasive GC (140).
LncRNA MCF2L-AS1 can promote CSC-like characteristics of NSCLC cells by down-regulating the expression level of miR-873-5p, thereby exerting carcinogenic effects (141). YY1 is a member of the YY family. It is a zinc finger protein and is overexpressed in a variety of cancers (142). YY1 can down-regulate the level of miR-873-5p, thereby activating the PI3K/AKT and ERK signaling pathways, thereby promoting the stemness of cancer cells (58). LncRNA CYTOR can regulate the expression of genes in the nucleus, thereby participating in the occurrence and development of cancers such as CRC (143). By up-regulating lncRNA CYTOR, TRIM29 inhibits pre-mir-873-5p to produce miR-873-5p, thereby up-regulating FN1 and promoting the migration and invasion of PTC cells (144).
The expression of lncRNA DGCR5 is significantly reduced in LC. DGCR5 shares the same binding site of miR-873-5p with TUSC3 (145). Ki-67 and MMP-3, MMP-9 are the markers of cell proliferation, cell migration, and invasion (100, 146). The binding of DGCR5 to miR-873-5p reduces the expression of TUSC3, Ki-67, MMP-3, and MMP-9, and thus decreases the proliferation and migration ability of LC cells (145). LncRNA TDRG1 is a proto-oncogene for CC (147) and endometrial cancer (148). The expression of lncRNA TDRG1 is up-regulated in human GC tissues and is related to the clinical prognosis of GC patients (149). As an important regulator of cancer, HDGF can be down-regulated through the EMT signaling pathway and the MMP-2 and MMP-9 signaling pathways (150). TDRG1 can target the miR-873-5p/HDGF axis, thereby promoting the tumor phenotype of GC cells (149). In addition, TDRG1 up-regulates the expression of ZEB1 by targeting miR-873-5p, thereby promoting tumorigenesis and the development of NSCLC cell lines (151). LncRNA HOTAIRM1 inhibits the miR-873-5p expression and promotes the expression of ZEB2 in GM, thereby inhibiting tumor cell apoptosis (74).
Competitive endogenous RNA (ceRNA) is considered to be a mechanism in post-transcriptional regulation and is related to tumor progression (152, 153). In OS, miR-873-5p targets to inhibit the expression of DDX11, and thus reduces the expression of MMP2, MMP9, N-cadherin, but increases the expression of E-cadherin, thereby inhibiting the migration and EMT process of OS cell lines (154). LncRNA DDX11-AS1 is up-regulated in GC tissues and cell lines, and its expression increases with the development of TNM stages and lymph node metastasis (155). LncRNA DDX11-AS1 as a ceRNA can bind to miR-873-5p and up-regulate the expression of DDX11 in OS and SPC18 in GC, thereby promoting the occurrence and development of OS (154) and GC (155).
Single nucleotide polymorphisms (SNPs) can change the secondary structure of lncRNA, thereby affecting the interaction between lncRNA and its interacting miRNA, and ultimately increasing the risk of cancer (156). The rs12982687 site of lncRNA UCA1 can affect the binding of miR-873-5p, thereby increasing the function of HIF-1 signal transduction, promoting the proliferation and migration of CRC cells (157).
MiR-873-5p and the Prognosis of Cancer Patients
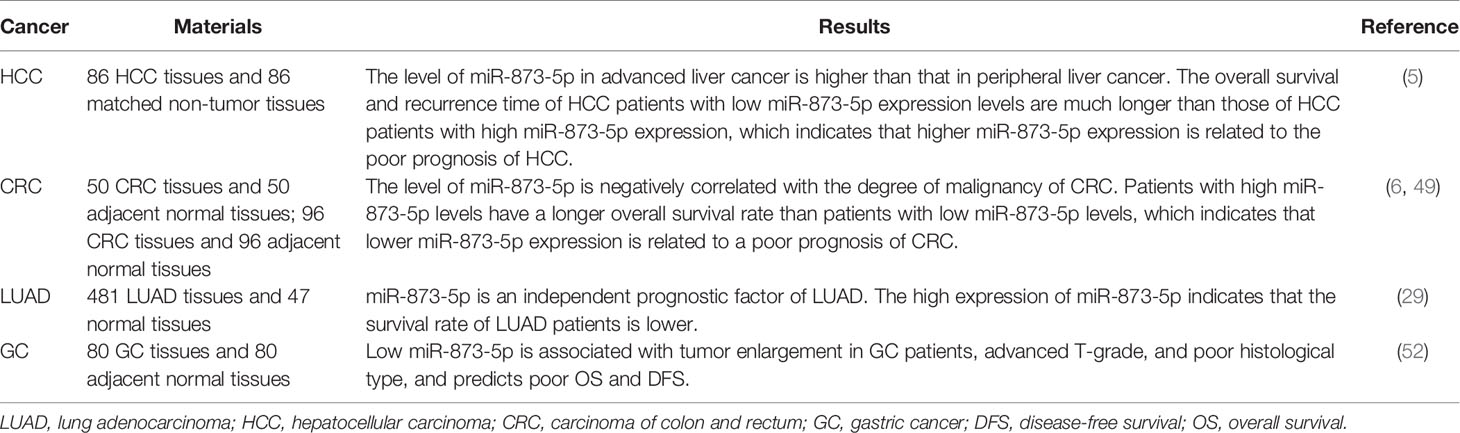
At present, many studies have found that miR-873-5p is significantly related to the prognosis of cancer patients (Table 2). Compared with normal tissues, the expression level of miR-873-5p is increased not only in HCC tissues but also in advanced HCC. Increased expression of miR-873-5p in HCC is positively correlated with lymph node metastasis and metastasis stage, but negatively correlated with tumor differentiation, indicating that miR-873-5p may be related to the aggressiveness and poor prognosis of HCC (5). In addition, low expression of miR-873-5p is associated with poor prognosis of LUAD (29).
Decreased expression of miR-873-5p is an indicator of poor prognosis in CRC patients (49). In addition, the level of miR-873-5p is negatively correlated with the degree of malignancy of CRC, and high levels of miR-873-5p are significantly correlated with a longer overall survival rate of patients (6). In GC, low expression of miR-873-5p is associated with large tumors, advanced T grade, poor histological type, poor overall survival, and short recurrence-free survival (52). In CC, the overall survival rate of patients with low miR-873-5p expression is lower than that of patients with high miR-873-5p expression (34).
The Role of miR-873-3p in Cancer
Proliferation-specific fork head box m1 (FOXM1) has been identified as an important cell cycle regulator, which can control the transition of cells from G1 to S phase and cell progression to M phase (158). miR-873-3p can significantly reduce the mRNA and protein levels of FOXM1. Therefore, miR-873-3p targets FOXM1 to inhibit LCA cell proliferation through its cell cycle regulation function (159). In BC, circTP63 binds to miR-873-3p and prevents its targeted inhibition of FOXM1, thereby inducing the progression and growth of estrogen receptor-positive BC (160). LINC00941 up-regulates the expression of ATXN2 by competitively binding miR-873-3p, stimulates the proliferation and metastasis of pancreatic adenocarcinoma, and promotes its occurrence and development (161) (Figure 8).
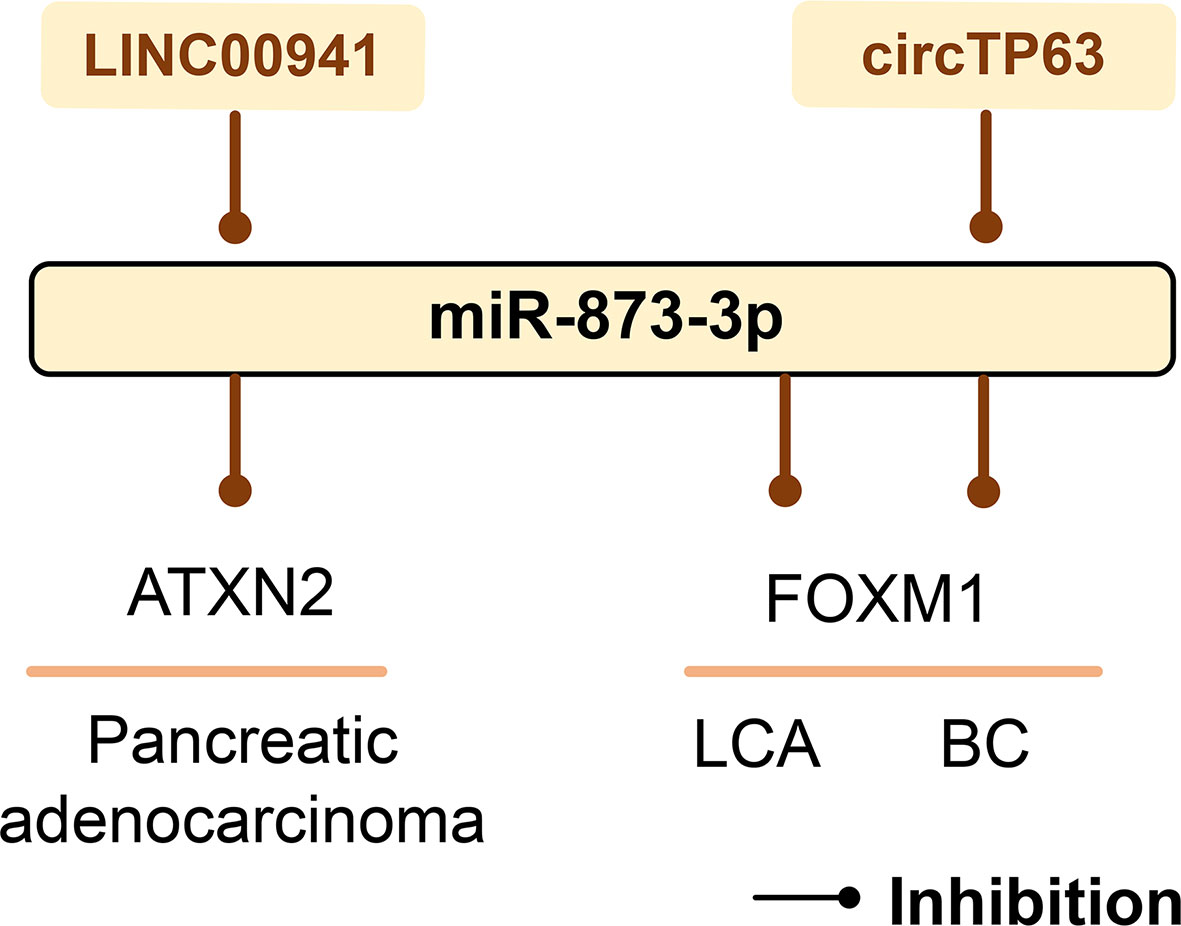
Figure 8 The role of miR-873-3p in human cancer. mir-873-3p plays an important role in pancreatic adenocarcinoma, LCA, and BC by regulating target genes. LINC00941 and circTP63 can sponge miR-873-3p and thus affect cancer development. LCA, lung cancer; BC, breast cancer.
Conclusions and Perspectives
miR-873-5p is widely involved in the progression of cancer, its expression is dysregulated in most cancer tissues and cell lines. Besides, the target gene of miR-873-5p has a series of molecular regulation functions, such as catalytic activity, transcription regulation, and binding. In cancer, miR-873-5p affects cancer development through the PIK3/AKT/mTOR, Wnt/β-Catenin, NF-κβ, MEK/ERK signaling pathways. miR-873-5p involves a variety of biological processes through the regulation of target genes, such as cell proliferation and apoptosis, EMT, cell migration and invasion, cell cycle, and cell stemness. miR-873-5p can also inhibit or promote the effects of cancer drugs by regulating its target genes. miR-873-5p can also be used as a specific diagnostic and prognostic indicator for various cancers. Finally, this review also summarizes epigenetic regulatory factors of miR-873-5p, including lncRNA, circRNA, methyltransferase, etc., which are also involved in the occurrence and development of various cancers.
However, there are still many deficiencies in the research on miR-873-5p. First of all, current studies have shown that miR-873-5p is dysregulated in 18 kinds of cancers, and it can cause cancer or suppress cancer. However, existing studies have not proven that miR-873-5p is cancer-specific. This will limit the application of miR-873-5p for cancer diagnosis, and it needs to be further explored. Second, the specific mechanism of miR-873-5p in some cancers has not been studied. Besides, more preclinical studies and clinical trials are needed to explore the effects of miR-873-5p on the efficacy of anticancer drugs. Finally, most studies are involved with miR-873-5p, and the research on miR-873-3p is very lacking.
Here we show that miR-873-5p plays a significant role in the initiation and progression of key biological and pathological processes in human cancers. Therefore, miR-873-5p can be the main research focus in the fight against human cancers. This review mainly summarizes the research progress of miR-873-5p in human cancers, which will expand our understanding of the molecular and cellular biological mechanisms of miR-873-5p.
Author Contributions
SD, ZH, and YZ conceived the review. CZ and YZ collated and analyzed the literature. SD and YZ helped complete diagrams and writing papers. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Kumari S, Sharma N, Sahi SV. Advances in Cancer Therapeutics: Conventional Thermal Therapy to Nanotechnology-Based Photothermal Therapy. Pharmaceutics (2021) 13(8). doi: 10.3390/pharmaceutics13081174
2. Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: Synthesis, Mechanism, Function, and Recent Clinical Trials. Biochim Biophys Acta (2010) 1803(11):1231–43. doi: 10.1016/j.bbamcr.2010.06.013
3. Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell (2009) 136(2):215–33. doi: 10.1016/j.cell.2009.01.002
4. Jin S, He J, Li J, Guo R, Shu Y, Liu P. MiR-873 Inhibition Enhances Gefitinib Resistance in non-Small Cell Lung Cancer Cells by Targeting Glioma-Associated Oncogene Homolog 1. Thorac Cancer (2018) 9(10):1262–70. doi: 10.1111/1759-7714.12830
5. Zhang Y, Zhang C, Zhao Q, Wei W, Dong Z, Shao L, et al. The miR-873/NDFIP1 Axis Promotes Hepatocellular Carcinoma Growth and Metastasis Through the AKT/mTOR-Mediated Warburg Effect. Am J Cancer Res (2019) 9(5):927–44.
6. Wang L, Jiang F, Ma F, Zhang B. MiR-873-5p Suppresses Cell Proliferation and Epithelial-Mesenchymal Transition via Directly Targeting Jumonji Domain-Containing Protein 8 Through the NF-kappaB Pathway in Colorectal Cancer. J Cell Commun Signaling (2019) 13(4):549–60. doi: 10.1007/s12079-019-00522-w
7. Pereira A, Moreira F, Vinasco-Sandoval T, Cunha A, Vidal A, Ribeiro-Dos-Santos AM, et al. Mirnome Reveals New Insights Into the Molecular Biology of Field Cancerization in Gastric Cancer. Front Genet (2019) 10:592. doi: 10.3389/fgene.2019.00592
8. Li YH, Zhong M, Zang HL, Tian XF. The E3 Ligase for Metastasis Associated 1 Protein, TRIM25, Is Targeted by microRNA-873 in Hepatocellular Carcinoma. Exp Cell Res (2018) 368(1):37–41. doi: 10.1016/j.yexcr.2018.04.010
9. Chen X, Zhang Y, Shi Y, Lian H, Tu H, Han S, et al. MiR-873 Acts as a Novel Sensitizer of Glioma Cells to Cisplatin by Targeting Bcl-2. Int J Oncol (2015) 47(4):1603–11. doi: 10.3892/ijo.2015.3143
10. Gao L, Guo Q, Li X, Yang X, Ni H, Wang T, et al. MiR-873/PD-L1 Axis Regulates the Stemness of Breast Cancer Cells. EBioMedicine (2019) 41:395–407. doi: 10.1016/j.ebiom.2019.02.034
11. Cui J, Yang Y, Li H, Leng Y, Qian K, Huang Q, et al. MiR-873 Regulates ERalpha Transcriptional Activity and Tamoxifen Resistance via Targeting CDK3 in Breast Cancer Cells. Oncogene (2015) 34(30):3895–907. doi: 10.1038/onc.2014.430
12. Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, et al. DNA-Independent PARP-1 Activation by Phosphorylated ERK2 Increases Elk1 Activity: A Link to Histone Acetylation. Mol Cell (2007) 25(2):297–308. doi: 10.1016/j.molcel.2006.12.012
13. Chai Y, Chipitsyna G, Cui J, Liao B, Liu S, Aysola K, et al. C-Fos Oncogene Regulator Elk-1 Interacts With BRCA1 Splice Variants BRCA1a/1b and Enhances BRCA1a/1b-Mediated Growth Suppression in Breast Cancer Cells. Oncogene (2001) 20(11):1357–67. doi: 10.1038/sj.onc.1204256
14. Sato F, Bhawal UK, Yoshimura T, Muragaki Y. DEC1 and DEC2 Crosstalk Between Circadian Rhythm and Tumor Progression. J Cancer (2016) 7(2):153–9. doi: 10.7150/jca.13748
15. Zhang P, Sun Y, Ma L. ZEB1: At the Crossroads of Epithelial-Mesenchymal Transition, Metastasis and Therapy Resistance. Cell Cycle (2015) 14(4):481–7. doi: 10.1080/15384101.2015.1006048
16. Xiao YY, Lin L, Li YH, Jiang HP, Zhu LT, Deng YR, et al. ZEB1 Promotes Invasion and Metastasis of Endometrial Cancer by Interacting With HDGF and Inducing Its Transcription. Am J Cancer Res (2019) 9(11):2314–30.
17. Houtmeyers R, Souopgui J, Tejpar S, Arkell R. The ZIC Gene Family Encodes Multi-Functional Proteins Essential for Patterning and Morphogenesis. Cell Mol Life Sci: CMLS (2013) 70(20):3791–811. doi: 10.1007/s00018-013-1285-5
18. Lu SX, Zhang CZ, Luo RZ, Wang CH, Liu LL, Fu J, et al. Zic2 Promotes Tumor Growth and Metastasis via PAK4 in Hepatocellular Carcinoma. Cancer Lett (2017) 402:71–80. doi: 10.1016/j.canlet.2017.05.018
19. Pourebrahim R, Houtmeyers R, Ghogomu S, Janssens S, Thelie A, Tran HT, et al. Transcription Factor Zic2 Inhibits Wnt/beta-Catenin Protein Signaling. J Biol Chem (2011) 286(43):37732–40. doi: 10.1074/jbc.M111.242826
20. Azmi S, Ozog A, Taneja R. Sharp-1/DEC2 Inhibits Skeletal Muscle Differentiation Through Repression of Myogenic Transcription Factors. J Biol Chem (2004) 279(50):52643–52. doi: 10.1074/jbc.M409188200
21. Mund T, Pelham HR. Regulation of PTEN/Akt and MAP Kinase Signaling Pathways by the Ubiquitin Ligase Activators Ndfip1 and Ndfip2. Proc Natl Acad Sci USA (2010) 107(25):11429–34. doi: 10.1073/pnas.0911714107
22. Hyodo T, Ito S, Hasegawa H, Asano E, Maeda M, Urano T, et al. Misshapen-Like Kinase 1 (MINK1) Is a Novel Component of Striatin-Interacting Phosphatase and Kinase (STRIPAK) and is Required for the Completion of Cytokinesis. J Biol Chem (2012) 287(30):25019–29. doi: 10.1074/jbc.M112.372342
23. Laine A, Martin B, Luka M, Mir L, Auffray C, Lucas B, et al. Foxo1 Is a T Cell-Intrinsic Inhibitor of the RORgammat-Th17 Program. J Immunol (2015) 195(4):1791–803. doi: 10.4049/jimmunol.1500849
24. Kuroda T, Yasuda S, Nakashima H, Takada N, Matsuyama S, Kusakawa S, et al. Identification of a Gene Encoding Slow Skeletal Muscle Troponin T as a Novel Marker for Immortalization of Retinal Pigment Epithelial Cells. Sci Rep (2017) 7(1):8163. doi: 10.1038/s41598-017-08014-w
25. Hu W, Liu Y, Zhou W, Si L, Ren L. CXCL16 and CXCR6 are Coexpressed in Human Lung Cancer In Vivo and Mediate the Invasion of Lung Cancer Cell Lines In Vitro. PloS One (2014) 9(6):e99056. doi: 10.1371/journal.pone.0099056
26. Hara T, Katakai T, Lee JH, Nambu Y, Nakajima-Nagata N, Gonda H, et al. A Transmembrane Chemokine, CXC Chemokine Ligand 16, Expressed by Lymph Node Fibroblastic Reticular Cells has the Potential to Regulate T Cell Migration and Adhesion. Int Immunol (2006) 18(2):301–11. doi: 10.1093/intimm/dxh369
27. Hojo S, Koizumi K, Tsuneyama K, Arita Y, Cui Z, Shinohara K, et al. High-Level Expression of Chemokine CXCL16 by Tumor Cells Correlates With a Good Prognosis and Increased Tumor-Infiltrating Lymphocytes in Colorectal Cancer. Cancer Res (2007) 67(10):4725–31. doi: 10.1158/0008-5472.CAN-06-3424
28. Gao Y, Xue Q, Wang D, Du M, Zhang Y, Gao S. miR-873 Induces Lung Adenocarcinoma Cell Proliferation and Migration by Targeting SRCIN1. Am J Trans Res (2015) 7(11):2519–26.
29. Zheng R, Mao W, Du Z, Zhang J, Wang M, Hu M. Three Differential Expression Profiles of miRNAs as Potential Biomarkers for Lung Adenocarcinoma. Biochem Biophys Res Commun (2018) 507(1-4):377–82. doi: 10.1016/j.bbrc.2018.11.046
30. Han G, Zhang L, Ni X, Chen Z, Pan X, Zhu Q, et al. MicroRNA-873 Promotes Cell Proliferation, Migration, and Invasion by Directly Targeting TSLC1 in Hepatocellular Carcinoma. Cell Physiol Biochem: Int J Exp Cell Physiol Biochem Pharmacol (2018) 46(6):2261–70. doi: 10.1159/000489594
31. Ning MS, Kim AS, Prasad N, Levy SE, Zhang H, Andl T. Characterization of the Merkel Cell Carcinoma Mirnome. J Skin Cancer (2014) 2014:289548. doi: 10.1155/2014/289548
32. Lv B, Li F, Liu X, Lin L. The Tumor-Suppressive Role of microRNA-873 in Nasopharyngeal Carcinoma Correlates With Downregulation of ZIC2 and Inhibition of AKT Signaling Pathway. Cancer Gene Ther (2021) 28(1-2):74–88. doi: 10.1038/s41417-020-0185-8
33. Zhang Y, Yang L, Ling C, Heng W. HuR Facilitates Cancer Stemness of Lung Cancer Cells via Regulating miR-873/CDK3 and miR-125a-3p/CDK3 Axis. Biotechnol Lett (2018) 40(4):623–31. doi: 10.1007/s10529-018-2512-9
34. Liang HX, Li YH. MiR-873, as a Suppressor in Cervical Cancer, Inhibits Cells Proliferation, Invasion and Migration via Negatively Regulating ULBP2. Genes Genomics (2020) 42(4):371–82. doi: 10.1007/s13258-019-00905-8
35. Feng J, Wang T. MicroRNA-873 Serves a Critical Role in Human Cervical Cancer Proliferation and Metastasis via Regulating Glioma-Associated Oncogene Homolog 1. Exp Ther Med (2020) 19(2):1243–50. doi: 10.3892/etm.2019.8348
36. Wang Q, Zhu W. MicroRNA-873 Inhibits the Proliferation and Invasion of Endometrial Cancer Cells by Directly Targeting Hepatoma-Derived Growth Factor. Exp Ther Med (2019) 18(2):1291–8. doi: 10.3892/etm.2019.7713
37. Wang G, Dong Y, Liu H, Ji N, Cao J, Liu A, et al. Loss of miR-873 Contributes to Gemcitabine Resistance in Triple-Negative Breast Cancer via Targeting ZEB1. Oncol Lett (2019) 18(4):3837–44. doi: 10.3892/ol.2019.10697
38. Tang L, Chen Y, Tang X, Wei D, Xu X, Yan F. Long Noncoding RNA DCST1-AS1 Promotes Cell Proliferation and Metastasis in Triple-Negative Breast Cancer by Forming a Positive Regulatory Loop With miR-873-5p and MYC. J Cancer (2020) 11(2):311–23. doi: 10.7150/jca.33982
39. Mokhlis HA, Bayraktar R, Kabil NN, Caner A, Kahraman N, Rodriguez-Aguayo C, et al. The Modulatory Role of MicroRNA-873 in the Progression of KRAS-Driven Cancers. Mol Ther Nucleic Acids (2019) 14:301–17. doi: 10.1016/j.omtn.2018.11.019
40. Skalsky RL, Cullen BR. Reduced Expression of Brain-Enriched microRNAs in Glioblastomas Permits Targeted Regulation of a Cell Death Gene. PloS One (2011) 6(9):e24248. doi: 10.1371/journal.pone.0024248
41. Li K, Zhang Q, Niu D, Xing H. Mining Mirnas’ Expressions in Glioma Based on GEO Database and Their Effects on Biological Functions. BioMed Res Int (2020) 2020:5637864. doi: 10.1155/2020/5637864
42. Wang RJ, Li JW, Bao BH, Wu HC, Du ZH, Su JL, et al. MicroRNA-873 (miRNA-873) Inhibits Glioblastoma Tumorigenesis and Metastasis by Suppressing the Expression of IGF2BP1. J Biol Chem (2015) 290(14):8938–48. doi: 10.1074/jbc.M114.624700
43. Liu Y, Wang Y, Yang H, Zhao L, Song R, Tan H, et al. MicroRNA873 Targets HOXA9 to Inhibit the Aggressive Phenotype of Osteosarcoma by Deactivating the Wnt/betacatenin Pathway. Int J Oncol (2019) 54(5):1809–20. doi: 10.3892/ijo.2019.4735
44. Wang Z, Liu W, Wang C, Ai Z. miR-873-5p Inhibits Cell Migration and Invasion of Papillary Thyroid Cancer via Regulation of CXCL16. OncoTargets Ther (2020) 13:1037–46. doi: 10.2147/OTT.S213168
45. Gong H, Fang L, Li Y, Du J, Zhou B, Wang X, et al. Mir873 Inhibits Colorectal Cancer Cell Proliferation by Targeting TRAF5 and TAB1. Oncol Rep (2018) 39(3):1090–8. doi: 10.3892/or.2018.6199
46. Li G, Xu Y, Wang S, Yan W, Zhao Q, Guo J. MiR-873-5p Inhibits Cell Migration, Invasion and Epithelial-Mesenchymal Transition in Colorectal Cancer via Targeting ZEB1. Pathol Res Pract (2019) 215(1):34–9. doi: 10.1016/j.prp.2018.10.008
47. Fan C, Lin B, Huang Z, Cui D, Zhu M, Ma Z, et al. MicroRNA-873 Inhibits Colorectal Cancer Metastasis by Targeting ELK1 and STRN4. Oncotarget (2019) 10(41):4192–204. doi: 10.18632/oncotarget.24115
48. Chen Y, Wang J, Wang D, Kang T, Du J, Yan Z, et al. TNNT1, Negatively Regulated by miR-873, Promotes the Progression of Colorectal Cancer. J Gene Med (2020) 22(2):e3152. doi: 10.1002/jgm.3152
49. Zhu Y, Zhang X, Qi M, Zhang Y, Ding F. miR-873-5p Inhibits the Progression of Colon Cancer via Repression of Tumor Suppressor Candidate 3/AKT Signaling. J Gastroenterol Hepatol (2019) 34(12):2126–34. doi: 10.1111/jgh.14697
50. Liang Y, Zhang P, Li S, Li H, Song S, Lu B. MicroRNA-873 Acts as a Tumor Suppressor in Esophageal Cancer by Inhibiting Differentiated Embryonic Chondrocyte Expressed Gene 2. Biomed Pharmacother Biomed Pharmacother (2018) 105:582–9. doi: 10.1016/j.biopha.2018.05.152
51. Cao D, Yu T, Ou X. MiR-873-5P Controls Gastric Cancer Progression by Targeting Hedgehog-GLI Signaling. Die Pharmazie (2016) 71(10):603–6. doi: 10.1691/ph.2016.6618
52. Lin L, Xiao J, Shi L, Chen W, Ge Y, Jiang M, et al. STRA6 Exerts Oncogenic Role in Gastric Tumorigenesis by Acting as a Crucial Target of miR-873. J Exp Clin Cancer Res: CR (2019) 38(1):452. doi: 10.1186/s13046-019-1450-2
53. Yao Y, Liu XQ, Yang FY, Mu JW. MiR-873-5p Modulates Progression of Tongue Squamous Cell Carcinoma via Targeting SEC11A. Oral Dis (2021). doi: 10.1111/odi.13830
54. Yang XL, Ma YS, Liu YS, Jiang XH, Ding H, Shi Y, et al. microRNA-873 Inhibits Self-Renewal and Proliferation of Pancreatic Cancer Stem Cells Through Pleckstrin-2-Dependent PI3K/AKT Pathway. Cell Signalling (2021) 84:110025. doi: 10.1016/j.cellsig.2021.110025
55. Xu J, Gong L, Qian Z, Song G, Liu J. ERBB4 Promotes the Proliferation of Gastric Cancer Cells via the PI3K/Akt Signaling Pathway. Oncol Rep (2018) 39(6):2892–8. doi: 10.3892/or.2018.6343
56. Chen J, Rajasekaran M, Hui KM. Atypical Regulators of Wnt/beta-Catenin Signaling as Potential Therapeutic Targets in Hepatocellular Carcinoma. Exp Biol Med (2017) 242(11):1142–9. doi: 10.1177/1535370217705865
57. Pahl HL. Activators and Target Genes of Rel/NF-kappaB Transcription Factors. Oncogene (1999) 18(49):6853–66. doi: 10.1038/sj.onc.1203239
58. Guo Q, Wang T, Yang Y, Gao L, Zhao Q, Zhang W, et al. Transcriptional Factor Yin Yang 1 Promotes the Stemness of Breast Cancer Cells by Suppressing miR-873-5p Transcriptional Activity. Mol Ther Nucleic Acids (2020) 21:527–41. doi: 10.1016/j.omtn.2020.06.018
59. Wong M, Hyodo T, Asano E, Funasaka K, Miyahara R, Hirooka Y, et al. Silencing of STRN4 Suppresses the Malignant Characteristics of Cancer Cells. Cancer Sci (2014) 105(12):1526–32. doi: 10.1111/cas.12541
60. Champsaur M, Lanier LL. Effect of NKG2D Ligand Expression on Host Immune Responses. Immunol Rev (2010) 235(1):267–85. doi: 10.1111/j.0105-2896.2010.00893.x
61. Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, et al. ULBPs, Novel MHC Class I-Related Molecules, Bind to CMV Glycoprotein UL16 and Stimulate NK Cytotoxicity Through the NKG2D Receptor. Immunity (2001) 14(2):123–33. doi: 10.1016/S1074-7613(01)00095-4
62. Yeo KS, Tan MC, Wong WY, Loh SW, Lam YL, Tan CL, et al. JMJD8 is a Positive Regulator of TNF-Induced NF-kappaB Signaling. Sci Rep (2016) 6:34125. doi: 10.1038/srep34125
63. Shi Y, Zhao Y, Zhang Y, AiErken N, Shao N, Ye R, et al. TNNT1 Facilitates Proliferation of Breast Cancer Cells by Promoting G1/S Phase Transition. Life Sci (2018) 208:161–6. doi: 10.1016/j.lfs.2018.07.034
64. Hubbard PA, Moody CL, Murali R. Allosteric Modulation of Ras and the PI3K/AKT/mTOR Pathway: Emerging Therapeutic Opportunities. Front Physiol (2014) 5:478. doi: 10.3389/fphys.2014.00478
65. Zang HL, Ren SN, Cao H, Tian XF. The Ubiquitin Ligase TRIM25 Inhibits Hepatocellular Carcinoma Progression by Targeting Metastasis Associated 1 Protein. IUBMB Life (2017) 69(10):795–801. doi: 10.1002/iub.1661
66. Nicolson GL, Nawa A, Toh Y, Taniguchi S, Nishimori K, Moustafa A. Tumor Metastasis-Associated Human MTA1 Gene and its MTA1 Protein Product: Role in Epithelial Cancer Cell Invasion, Proliferation and Nuclear Regulation. Clin Exp Metastasis (2003) 20(1):19–24. doi: 10.1023/A:1022534217769
67. Liu D, Feng X, Wu X, Li Z, Wang W, Tao Y, et al. Tumor Suppressor in Lung Cancer 1 (TSLC1), a Novel Tumor Suppressor Gene, is Implicated in the Regulation of Proliferation, Invasion, Cell Cycle, Apoptosis, and Tumorigenicity in Cutaneous Squamous Cell Carcinoma. Tumour Biol: J Int Soc Oncodevelopmental Biol Med (2013) 34(6):3773–83. doi: 10.1007/s13277-013-0961-2
68. Liu W, Yue F, Zheng M, Merlot A, Bae DH, Huang M, et al. The Proto-Oncogene C-Src and its Downstream Signaling Pathways Are Inhibited by the Metastasis Suppressor, NDRG1. Oncotarget (2015) 6(11):8851–74. doi: 10.18632/oncotarget.3316
69. Le XF, Bast RC Jr. Src Family Kinases and Paclitaxel Sensitivity. Cancer Biol Ther (2011) 12(4):260–9. doi: 10.4161/cbt.12.4.16430
70. Kennedy S, Clynes M, Doolan P, Mehta JP, Rani S, Crown J, et al. SNIP/p140Cap mRNA Expression is an Unfavourable Prognostic Factor in Breast Cancer and is Not Expressed in Normal Breast Tissue. Br J Cancer (2008) 98(10):1641–5. doi: 10.1038/sj.bjc.6604365
71. Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong M, Singh V, et al. RNA-Binding Protein IGF2BP1 Maintains Leukemia Stem Cell Properties by Regulating HOXB4, MYB, and ALDH1A1. Leukemia (2020) 34(5):1354–63. doi: 10.1038/s41375-019-0656-9
72. Gutschner T, Hammerle M, Pazaitis N, Bley N, Fiskin E, Uckelmann H, et al. Insulin-Like Growth Factor 2 mRNA-Binding Protein 1 (IGF2BP1) is an Important Protumorigenic Factor in Hepatocellular Carcinoma. Hepatology (2014) 59(5):1900–11. doi: 10.1002/hep.26997
73. Goossens S, Janzen V, Bartunkova S, Yokomizo T, Drogat B, Crisan M, et al. The EMT Regulator Zeb2/Sip1 is Essential for Murine Embryonic Hematopoietic Stem/Progenitor Cell Differentiation and Mobilization. Blood (2011) 117(21):5620–30. doi: 10.1182/blood-2010-08-300236
74. Lin YH, Guo L, Yan F, Dou ZQ, Yu Q, Chen G. Long non-Coding RNA HOTAIRM1 Promotes Proliferation and Inhibits Apoptosis of Glioma Cells by Regulating the miR-873-5p/ZEB2 Axis. Chin Med J (2020) 133(2):174–82. doi: 10.1097/CM9.0000000000000615
75. Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, et al. Loss of Expression of the Hoxa-9 Homeobox Gene Impairs the Proliferation and Repopulating Ability of Hematopoietic Stem Cells. Blood (2005) 106(12):3988–94. doi: 10.1182/blood-2005-05-2003
76. Wang X, Bu J, Liu X, Wang W, Mai W, Lv B, et al. miR-133b Suppresses Metastasis by Targeting HOXA9 in Human Colorectal Cancer. Oncotarget (2017) 8(38):63935–48. doi: 10.18632/oncotarget.19212
77. Ma YY, Zhang Y, Mou XZ, Liu ZC, Ru GQ, Li E. High Level of Homeobox A9 and PBX Homeobox 3 Expression in Gastric Cancer Correlates With Poor Prognosis. Oncol Lett (2017) 14(5):5883–9. doi: 10.3892/ol.2017.6937
78. Armas-Lopez L, Zuniga J, Arrieta O, Avila-Moreno F. The Hedgehog-GLI Pathway in Embryonic Development and Cancer: Implications for Pulmonary Oncology Therapy. Oncotarget (2017) 8(36):60684–703. doi: 10.18632/oncotarget.19527
79. Lemjabbar-Alaoui H, Dasari V, Sidhu SS, Mengistab A, Finkbeiner W, Gallup M, et al. Wnt and Hedgehog are Critical Mediators of Cigarette Smoke-Induced Lung Cancer. PloS One (2006) 1:e93. doi: 10.1371/journal.pone.0000093
80. Pietrobono S, Santini R, Gagliardi S, Dapporto F, Colecchia D, Chiariello M, et al. Targeted Inhibition of Hedgehog-GLI Signaling by Novel Acylguanidine Derivatives Inhibits Melanoma Cell Growth by Inducing Replication Stress and Mitotic Catastrophe. Cell Death Dis (2018) 9(2):142. doi: 10.1038/s41419-017-0142-0
81. Karunanithi S, Levi L, DeVecchio J, Karagkounis G, Reizes O, Lathia JD, et al. RBP4-STRA6 Pathway Drives Cancer Stem Cell Maintenance and Mediates High-Fat Diet-Induced Colon Carcinogenesis. Stem Cell Rep (2017) 9(2):438–50. doi: 10.1016/j.stemcr.2017.06.002
82. Yager JD, Davidson NE. Estrogen Carcinogenesis in Breast Cancer. New Engl J Med (2006) 354(3):270–82. doi: 10.1056/NEJMra050776
83. Liang K, Liu Y, Eer D, Liu J, Yang F, Hu K. High CXC Chemokine Ligand 16 (CXCL16) Expression Promotes Proliferation and Metastasis of Lung Cancer via Regulating the NF-kappaB Pathway. Med Sci Monitor: Int Med J Exp Clin Res (2018) 24:405–11. doi: 10.12659/MSM.906230
84. Kishima Y, Yoshida K, Enomoto H, Yamamoto M, Kuroda T, Okuda Y, et al. Antisense Oligonucleotides of Hepatoma-Derived Growth Factor (HDGF) Suppress the Proliferation of Hepatoma Cells. Hepato-Gastroenterology (2002) 49(48):1639–44.
85. Tang J, Shi H, Li H, Zhen T, Dong Y, Zhang F, et al. The Interaction of Hepatoma-Derived Growth Factor and Beta-Catenin Promotes Tumorigenesis of Synovial Sarcoma. Tumour Biol: J Int Soc Oncodevelopmental Biol Med (2016) 37(8):10287–301. doi: 10.1007/s13277-016-4905-5
86. De Craene B, Berx G. Regulatory Networks Defining EMT During Cancer Initiation and Progression. Nat Rev Cancer (2013) 13(2):97–110. doi: 10.1038/nrc3447
87. Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, et al. A Transient, EMT-Linked Loss of Basement Membranes Indicates Metastasis and Poor Survival in Colorectal Cancer. Gastroenterology (2006) 131(3):830–40. doi: 10.1053/j.gastro.2006.06.016
88. Yao X, Wang Y, Duan Y, Zhang Q, Li P, Jin R, et al. IGFBP2 Promotes Salivary Adenoid Cystic Carcinoma Metastasis by Activating the NF-Kappab/ZEB1 Signaling Pathway. Cancer Lett (2018) 432:38–46. doi: 10.1016/j.canlet.2018.06.008
89. Gu Y, Wang Q, Guo K, Qin W, Liao W, Wang S, et al. TUSC3 Promotes Colorectal Cancer Progression and Epithelial-Mesenchymal Transition (EMT) Through WNT/beta-Catenin and MAPK Signalling. J Pathol (2016) 239(1):60–71. doi: 10.1002/path.4697
90. Hao YH, Yu SY, Tu RS, Cai YQ. TNNT1, a Prognostic Indicator in Colon Adenocarcinoma, Regulates Cell Behaviors and Mediates EMT Process. Biosci Biotechnol Biochem (2020) 84(1):111–7. doi: 10.1080/09168451.2019.1664891
91. Ye X, Brabletz T, Kang Y, Longmore GD, Nieto MA, Stanger BZ, et al. Upholding a Role for EMT in Breast Cancer Metastasis. Nature (2017) 547(7661):E1–3. doi: 10.1038/nature22816
92. Liu Q, Wu Y, Seino H, Haga T, Yoshizawa T, Morohashi S, et al. Correlation Between DEC1/DEC2 and Epithelialmesenchymal Transition in Human Prostate Cancer PC3 Cells. Mol Med Rep (2018) 18(4):3859–65. doi: 10.3892/mmr.2018.9367
93. Serman L, Nikuseva Martic T, Serman A, Vranic S. Epigenetic Alterations of the Wnt Signaling Pathway in Cancer: A Mini Review. Bosnian J Basic Med Sci (2014) 14(4):191–4. doi: 10.17305/bjbms.2014.4.205
94. Chen K, Wang G, Peng L, Liu S, Fu X, Zhou Y, et al. CADM1/TSLC1 Inactivation by Promoter Hypermethylation Is a Frequent Event in Colorectal Carcinogenesis and Correlates With Late Stages of the Disease. Int J Cancer (2011) 128(2):266–73. doi: 10.1002/ijc.25356
95. Di Stefano P, Leal MP, Tornillo G, Bisaro B, Repetto D, Pincini A, et al. The Adaptor Proteins P140cap and P130cas as Molecular Hubs in Cell Migration and Invasion of Cancer Cells. Am J Cancer Res (2011) 1(5):663–73.
96. Cabodi S, del Pilar Camacho-Leal M, Di Stefano P, Defilippi P. Integrin Signalling Adaptors: Not Only Figurants in the Cancer Story. Nat Rev Cancer (2010) 10(12):858–70. doi: 10.1038/nrc2967
97. Deryugina EI, Bourdon MA, Luo GX, Reisfeld RA, Strongin A. Matrix Metalloproteinase-2 Activation Modulates Glioma Cell Migration. J Cell Sci (1997) 110(Pt 19):2473–82. doi: 10.1242/jcs.110.19.2473
98. Wick W, Wagner S, Kerkau S, Dichgans J, Tonn JC, Weller M. BCL-2 Promotes Migration and Invasiveness of Human Glioma Cells. FEBS Lett (1998) 440(3):419–24. doi: 10.1016/S0014-5793(98)01494-X
99. Kessenbrock K, Wang CY, Werb Z. Matrix Metalloproteinases in Stem Cell Regulation and Cancer. Matrix Biol: J Int Soc Matrix Biol (2015) 44–46:184–90. doi: 10.1016/j.matbio.2015.01.022
100. Shay G, Lynch CC, Fingleton B. Moving Targets: Emerging Roles for MMPs in Cancer Progression and Metastasis. Matrix Biol: J Int Soc Matrix Biol (2015) 44-46:200–6. doi: 10.1016/j.matbio.2015.01.019
101. Won HY, Lee JY, Shin DH, Park JH, Nam JS, Kim HC, et al. Loss of Mel-18 Enhances Breast Cancer Stem Cell Activity and Tumorigenicity Through Activating Notch Signaling Mediated by the Wnt/TCF Pathway. FASEB J: Off Publ Fed Am Societies Exp Biol (2012) 26(12):5002–13. doi: 10.1096/fj.12-209247
102. Butler SJ, Richardson L, Farias N, Morrison J, Coomber BL. Characterization of Cancer Stem Cell Drug Resistance in the Human Colorectal Cancer Cell Lines HCT116 and SW480. Biochem Biophys Res Commun (2017) 490(1):29–35. doi: 10.1016/j.bbrc.2017.05.176
103. Ishibashi M, Tamura H, Sunakawa M, Kondo-Onodera A, Okuyama N, Hamada Y, et al. Myeloma Drug Resistance Induced by Binding of Myeloma B7-H1 (PD-L1) to PD-1. Cancer Immunol Res (2016) 4(9):779–88. doi: 10.1158/2326-6066.CIR-15-0296
104. Almozyan S, Colak D, Mansour F, Alaiya A, Al-Harazi O, Qattan A, et al. PD-L1 Promotes OCT4 and Nanog Expression in Breast Cancer Stem Cells by Sustaining PI3K/AKT Pathway Activation. Int J Cancer (2017) 141(7):1402–12. doi: 10.1002/ijc.30834
105. Soliman H, Khalil F, Antonia S. PD-L1 Expression Is Increased in a Subset of Basal Type Breast Cancer Cells. PloS One (2014) 9(2):e88557. doi: 10.1371/journal.pone.0088557
106. Liu S, Chen S, Yuan W, Wang H, Chen K, Li D, et al. PD-1/PD-L1 Interaction Up-Regulates MDR1/P-Gp Expression in Breast Cancer Cells via PI3K/AKT and MAPK/ERK Pathways. Oncotarget (2017) 8(59):99901–12. doi: 10.18632/oncotarget.21914
107. Zhang W, Vreeland AC, Noy N. RNA-Binding Protein HuR Regulates Nuclear Import of Protein. J Cell Sci (2016) 129(21):4025–33. doi: 10.1242/jcs.192096
108. Wang J, Ma W, Liu Y. Long non-Coding RNA HULC Promotes Bladder Cancer Cells Proliferation But Inhibits Apoptosis via Regulation of ZIC2 and PI3K/AKT Signaling Pathway. Cancer Biomarkers: Section A Dis Markers (2017) 20(4):425–34. doi: 10.3233/CBM-170188
109. Lv B, Li F, Liu X, Lin L. The Tumor-Suppressive Role of microRNA-873 in Nasopharyngeal Carcinoma Correlates With Downregulation of ZIC2 and Inhibition of AKT Signaling Pathway. Cancer Gene Ther (2020). doi: 10.1038/s41417-020-0185-8
110. Qu C, He, Lu X, Dong L, Zhu Y, Zhao Q, et al. Salt-Inducible Kinase (SIK1) Regulates HCC Progression and WNT/beta-Catenin Activation. J Hepatol (2016) 64(5):1076–89. doi: 10.1016/j.jhep.2016.01.005
111. Song H, Pu J, Wang L, Wu L, Xiao J, Liu Q, et al. ATG16L1 Phosphorylation is Oppositely Regulated by CSNK2/casein Kinase 2 and PPP1/protein Phosphatase 1 Which Determines the Fate of Cardiomyocytes During Hypoxia/Reoxygenation. Autophagy (2015) 11(8):1308–25. doi: 10.1080/15548627.2015.1060386
112. Lee DH. Treatments for EGFR-Mutant Non-Small Cell Lung Cancer (NSCLC): The Road to a Success, Paved With Failures. Pharmacol Ther (2017) 174:1–21. doi: 10.1016/j.pharmthera.2017.02.001
113. Duan X, Shi J. Advance in microRNAs and EGFR-TKIs Secondary Resistance Research in non-Small Cell Lung Cancer. Zhongguo Fei Ai Za Zhi Chin J Lung Cancer (2014) 17(12):860–4. doi: 10.3779/j.issn.1009-3419.2014.12.07
114. Xie SY, Li G, Han C, Yu YY, Li N. RKIP Reduction Enhances Radioresistance by Activating the Shh Signaling Pathway in Non-Small-Cell Lung Cancer. OncoTargets Ther (2017) 10:5605–19. doi: 10.2147/OTT.S149200
115. Bora-Singhal N, Perumal D, Nguyen J, Chellappan S. Gli1-Mediated Regulation of Sox2 Facilitates Self-Renewal of Stem-Like Cells and Confers Resistance to EGFR Inhibitors in Non-Small Cell Lung Cancer. Neoplasia (2015) 17(7):538–51. doi: 10.1016/j.neo.2015.07.001
116. Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. Let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells. Cell (2007) 131(6):1109–23. doi: 10.1016/j.cell.2007.10.054
117. Jin D, Wu Y, Shao C, Gao Y, Wang D, Guo J. Norcantharidin Reverses Cisplatin Resistance and Inhibits the Epithelial Mesenchymal Transition of Human Nonsmall Lung Cancer Cells by Regulating the YAP Pathway. Oncol Rep (2018) 40(2):609–20. doi: 10.3892/or.2020.7830
118. Mo L, Zhang X, Shi X, Wei L, Zheng D, Li H, et al. Norcantharidin Enhances Antitumor Immunity of GM-CSF Prostate Cancer Cells Vaccine by Inducing Apoptosis of Regulatory T Cells. Cancer Sci (2018) 109(7):2109–18. doi: 10.1111/cas.13639
119. Zhang JT, Sun W, Zhang WZ, Ge CY, Liu ZY, Zhao ZM, et al. Norcantharidin Inhibits Tumor Growth and Vasculogenic Mimicry of Human Gallbladder Carcinomas by Suppression of the PI3-K/MMPs/Ln-5gamma2 Signaling Pathway. BMC Cancer (2014) 14:193. doi: 10.1186/1471-2407-14-193
120. Zhang X, Zhang B, Zhang P, Lian L, Li L, Qiu Z, et al. Norcantharidin Regulates ERalpha Signaling and Tamoxifen Resistance via Targeting miR-873/CDK3 in Breast Cancer Cells. PloS One (2019) 14(5):e0217181. doi: 10.1371/journal.pone.0217181
121. Papa AL, Sidiqui A, Balasubramanian SU, Sarangi S, Luchette M, Sengupta S, et al. PEGylated Liposomal Gemcitabine: Insights Into a Potential Breast Cancer Therapeutic. Cell Oncol (2013) 36(6):449–57. doi: 10.1007/s13402-013-0146-4
122. Kumar S, Kumar A, Shah PP, Rai SN, Panguluri SK, Kakar SS. MicroRNA Signature of Cis-Platin Resistant vs. Cis-Platin Sensitive Ovarian Cancer Cell Lines. J Ovarian Res (2011) 4(1):17. doi: 10.1186/1757-2215-4-17
123. Qiu JG, Zhang YJ, Li Y, Zhao JM, Zhang WJ, Jiang QW, et al. Trametinib Modulates Cancer Multidrug Resistance by Targeting ABCB1 Transporter. Oncotarget (2015) 6(17):15494–509. doi: 10.18632/oncotarget.3820
124. Hung TH, Hsu SC, Cheng CY, Choo KB, Tseng CP, Chen TC, et al. Wnt5A Regulates ABCB1 Expression in Multidrug-Resistant Cancer Cells Through Activation of the Non-Canonical PKA/beta-Catenin Pathway. Oncotarget (2014) 5(23):12273–90. doi: 10.18632/oncotarget.2631
125. Gisel A, Valvano M, El Idrissi IG, Nardulli P, Azzariti A, Carrieri A, et al. miRNAs for the Detection of Multidrug Resistance: Overview and Perspectives. Molecules (2014) 19(5):5611–23. doi: 10.3390/molecules19055611
126. Wu DD, Li XS, Meng XN, Yan J, Zong ZH. MicroRNA-873 Mediates Multidrug Resistance in Ovarian Cancer Cells by Targeting ABCB1. Tumour Biol: J Int Soc Oncodevelopmental Biol Med (2016) 37(8):10499–506. doi: 10.1007/s13277-016-4944-y
127. Rocha CR, Garcia CC, Vieira DB, Quinet A, de Andrade-Lima LC, Munford V, et al. Glutathione Depletion Sensitizes Cisplatin- and Temozolomide-Resistant Glioma Cells In Vitro and In Vivo. Cell Death Dis (2015) 6:e1727. doi: 10.1038/cddis.2015.101
128. Baraniskin A, Kuhnhenn J, Schlegel U, Maghnouj A, Zollner H, Schmiegel W, et al. Identification of microRNAs in the Cerebrospinal Fluid as Biomarker for the Diagnosis of Glioma. Neuro-Oncology (2012) 14(1):29–33. doi: 10.1093/neuonc/nor169
129. Zhu CJ, Li YB, Wong MC. Expression of Antisense Bcl-2 cDNA Abolishes Tumorigenicity and Enhances Chemosensitivity of Human Malignant Glioma Cells. J Neurosci Res (2003) 74(1):60–6. doi: 10.1002/jnr.10722
130. Komeil IA, El-Refaie WM, Gowayed MA, El-Ganainy SO, El Achy SN, Huttunen KM, et al. Oral Genistein-Loaded Phytosomes With Enhanced Hepatic Uptake, Residence and Improved Therapeutic Efficacy Against Hepatocellular Carcinoma. Int J Pharmaceut (2021) 601:120564. doi: 10.1016/j.ijpharm.2021.120564
131. Yu Y, Xing Y, Zhang Q, Zhang Q, Huang S, Li X, et al. Soy Isoflavone Genistein Inhibits Hsa_Circ_0031250/miR-873-5p/FOXM1 Axis to Suppress Non-Small-Cell Lung Cancer Progression. IUBMB Life (2021) 73(1):92–107. doi: 10.1002/iub.2404
132. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
133. Zhang S, Chen Z, Sun J, An N, Xi Q. CircRNA Hsa_circRNA_0000069 Promotes the Proliferation, Migration and Invasion of Cervical Cancer Through miR-873-5p/TUSC3 Axis. Cancer Cell Int (2020) 20:287. doi: 10.1186/s12935-020-01387-5
134. Yang J, Yu L, Yan J, Xiao Y, Li W, Xiao J, et al. Circular RNA DGKB Promotes the Progression of Neuroblastoma by Targeting miR-873/GLI1 Axis. Front Oncol (2020) 10:1104. doi: 10.3389/fonc.2020.01104
135. Yu W, Ma B, Zhao W, Liu J, Yu H, Tian Z, et al. The Combination of circRNA-UMAD1 and Galectin-3 in Peripheral Circulation Is a Co-Biomarker for Predicting Lymph Node Metastasis of Thyroid Carcinoma. Am J Trans Res (2020) 12(9):5399–415.
136. Liu J, Li H, Wei C, Ding J, Lu J, Pan G, et al. Circfat1(E2) Promotes Papillary Thyroid Cancer Proliferation, Migration, and Invasion via the miRNA-873/ZEB1 Axis. Comput Math Methods Med (2020) 2020:1459368. doi: 10.1155/2020/1459368
137. Yi X, Chen W, Li C, Chen X, Lin Q, Lin S, et al. Circular RNA Circ_0004507 Contributes to Laryngeal Cancer Progression and Cisplatin Resistance by Sponging miR-873 to Upregulate Multidrug Resistance 1 and Multidrug Resistance Protein 1. Head Neck (2021) 43(3):928–41. doi: 10.1002/hed.26549
138. Li J, Bao S, Wang L, Wang R. CircZKSCAN1 Suppresses Hepatocellular Carcinoma Tumorigenesis by Regulating miR-873-5p/Downregulation of Deleted in Liver Cancer 1. Digestive Dis Sci (2021). doi: 10.1007/s10620-020-06789-z
139. Wei Z, Yuan X, Ding Q, Xu Y, Hong L, Wang J. CircATP5SL Promotes Infantile Haemangiomas Progression via IGF1R Regulation by Targeting miR-873-5p. Am J Trans Res (2021) 13(3):1322–36.
140. Lu Y, Cheng J, Cai W, Zhuo H, Wu G, Cai J. Inhibition of circRNA Circvps33b Reduces Warburg Effect and Tumor Growth Through Regulating the miR-873-5p/HNRNPK Axis in Infiltrative Gastric Cancer. OncoTargets Ther (2021) 14:3095–108. doi: 10.2147/OTT.S292575
141. Li S, Lin L. Long Noncoding RNA MCF2L-AS1 Promotes the Cancer Stem Cell-Like Traits in Non-Small Cell Lung Cancer Cells Through Regulating miR-873-5p Level. Environ Toxicol (2021) 36(7):1457–65. doi: 10.1002/tox.23142
142. Shi Y, Seto E, Chang LS, Shenk T. Transcriptional Repression by YY1, a Human GLI-Kruppel-Related Protein, and Relief of Repression by Adenovirus E1A Protein. Cell (1991) 67(2):377–88. doi: 10.1016/0092-8674(91)90189-6
143. Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 Functions as a Competing Endogenous RNA to Confer Oxaliplatin Resistance and Holds Prognostic Values in Colon Cancer. Mol Ther: J Am Soc Gene Ther (2016) 24(12):2064–77. doi: 10.1038/mt.2016.180
144. Wu T, Zhang DL, Wang JM, Jiang JY, Du X, Zeng XY, et al. TRIM29 Inhibits miR-873-5P Biogenesis via CYTOR to Upregulate Fibronectin 1 and Promotes Invasion of Papillary Thyroid Cancer Cells. Cell Death Dis (2020) 11(9):813. doi: 10.1038/s41419-020-03018-3
145. Luo J, Zhu H, Jiang H, Cui Y, Wang M, Ni X, et al. The Effects of Aberrant Expression of LncRNA DGCR5/miR-873-5p/TUSC3 in Lung Cancer Cell Progression. Cancer Med (2018). doi: 10.1002/cam4.1566
146. Scholzen T, Gerdes J. The Ki-67 Protein: From the Known and the Unknown. J Cell Physiol (2000) 182(3):311–22. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9
147. Jiang H, Liang M, Jiang Y, Zhang T, Mo K, Su S, et al. The lncRNA TDRG1 Promotes Cell Proliferation, Migration and Invasion by Targeting miR-326 to Regulate MAPK1 Expression in Cervical Cancer. Cancer Cell Int (2019) 19:152. doi: 10.1186/s12935-019-0872-4
148. Chen S, Wang LL, Sun KX, Liu Y, Guan X, Zong ZH, et al. LncRNA TDRG1 Enhances Tumorigenicity in Endometrial Carcinoma by Binding and Targeting VEGF-A Protein. Biochim Biophys Acta Mol Basis Dis (2018) 1864(9 Pt B):3013–21. doi: 10.1016/j.bbadis.2018.06.013
149. Ma Y, Xu XL, Huang HG, Li YF, Li ZG. LncRNA TDRG1 Promotes the Aggressiveness of Gastric Carcinoma Through Regulating miR-873-5p/HDGF Axis. Biomed Pharmacother Biomed Pharmacother (2020) 121:109425. doi: 10.1016/j.biopha.2019.109425
150. Li M, Shen J, Wu X, Zhang B, Zhang R, Weng H, et al. Downregulated Expression of Hepatoma-Derived Growth Factor (HDGF) Reduces Gallbladder Cancer Cell Proliferation and Invasion. Med Oncol (2013) 30(2):587. doi: 10.1007/s12032-013-0587-7
151. Hu X, Mu Y, Wang J, Zhao Y. LncRNA TDRG1 Promotes the Metastasis of NSCLC Cell Through Regulating miR-873-5p/ZEB1 Axis. J Cell Biochem (2019). doi: 10.1002/jcb.29559
152. Lian Y, Xiong F, Yang L, Bo H, Gong Z, Wang Y, et al. Long Noncoding RNA AFAP1-AS1 Acts as a Competing Endogenous RNA of miR-423-5p to Facilitate Nasopharyngeal Carcinoma Metastasis Through Regulating the Rho/Rac Pathway. J Exp Clin Cancer Res: CR (2018) 37(1):253. doi: 10.1186/s13046-018-0918-9
153. Feng K, Liu Y, Xu LJ, Zhao LF, Jia CW, Xu MY. Long Noncoding RNA PVT1 Enhances the Viability and Invasion of Papillary Thyroid Carcinoma Cells by Functioning as ceRNA of microRNA-30a Through Mediating Expression of Insulin Like Growth Factor 1 Receptor. Biomed Pharmacother Biomed Pharmacother (2018) 104:686–98. doi: 10.1016/j.biopha.2018.05.078
154. Zhang H, Lin J, Chen J, Gu W, Mao Y, Wang H, et al. DDX11-AS1 Contributes to Osteosarcoma Progression via Stabilizing DDX11. Life Sci (2020) 254:117392. doi: 10.1016/j.lfs.2020.117392
155. Ren Z, Liu X, Si Y, Yang D. Long non-Coding RNA DDX11-AS1 Facilitates Gastric Cancer Progression by Regulating miR-873-5p/SPC18 Axis. Artif Cells Nanomed Biotechnol (2020) 48(1):572–83. doi: 10.1080/21691401.2020.1726937
156. Gong J, Liu W, Zhang J, Miao X, Guo AY. lncRNASNP: A Database of SNPs in lncRNAs and Their Potential Functions in Human and Mouse. Nucleic Acids Res (2015) 43(Database issue):D181–6. doi: 10.1093/nar/gku1000
157. Fu Y, Zhang Y, Cui J, Yang G, Peng S, Mi W, et al. SNP Rs12982687 Affects Binding Capacity of lncRNA UCA1 With miR-873-5p: Involvement in Smoking-Triggered Colorectal Cancer Progression. Cell Commun Signaling: CCS (2020) 18(1):37. doi: 10.1186/s12964-020-0518-0
158. Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, et al. The Forkhead Box M1 Transcription Factor Stimulates the Proliferation of Tumor Cells During Development of Lung Cancer. Cancer Res (2006) 66(4):2153–61. doi: 10.1158/0008-5472.CAN-05-3003
159. Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, et al. Circtp63 Functions as a ceRNA to Promote Lung Squamous Cell Carcinoma Progression by Upregulating FOXM1. Nat Commun (2019) 10(1):3200. doi: 10.1038/s41467-019-11162-4
160. Deng Y, Xia J, Xu YE. Circular RNA Circtp63 Enhances Estrogen Receptor-Positive Breast Cancer Progression and Malignant Behaviors Through the miR-873-3p/FOXM1 Axis. Anti-Cancer Drugs (2021) 32(1):44–52. doi: 10.1097/CAD.0000000000001010
Keywords: miR-873-5p, cancer, cell, prognosis, signaling pathway
Citation: Zou Y, Zhong C, Hu Z and Duan S (2021) MiR-873-5p: A Potential Molecular Marker for Cancer Diagnosis and Prognosis. Front. Oncol. 11:743701. doi: 10.3389/fonc.2021.743701
Received: 19 July 2021; Accepted: 20 September 2021;
Published: 05 October 2021.
Edited by:
Zong Sheng Guo, Roswell Park Comprehensive Cancer Center, United StatesReviewed by:
Sara Sergio, University of Salento, ItalyWei Zhang, Northwestern University, United States
Copyright © 2021 Zou, Zhong, Hu and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiwei Duan, ZHVhbnN3QHp1Y2MuZWR1LmNu
 Yuhao Zou1,2
Yuhao Zou1,2 Shiwei Duan
Shiwei Duan