
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 27 September 2021
Sec. Genitourinary Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.736431
This article is part of the Research Topic Advances in Prostate Cancer: Model Systems, Molecular and Cellular Mechanisms, Early Detection, and Therapies View all 21 articles
Complex heterogeneity is an important characteristic in the development of prostate cancer (PCa), which further leads to the failure of known therapeutic options. PCa research has been hampered by the current in vitro model systems that cannot fully reflect the biological characteristics and clinical diversity of PCa. The tumor organoid model in three-dimensional culture retains the heterogeneity of primary tumor tissues in vitro well and enables high-throughput screening and genome editing. Therefore, the establishment of a PCa organoid model that recapitulates the diverse heterogeneity observed in clinical settings is of great significance for the study of PCa. In this review, we summarize the culture conditions, establishments, and limitations of PCa organoids and further review their application for the study of pathogenesis, drug screening, mechanism of drug resistance, and individualized treatment for PCa. Additionally, we look forward to other potential developmental directions of PCa organoids, such as the interaction between prostate cancer tumor cells and their microenvironment, clinical individualized treatments, heterogeneous transformation model, tumor immunotherapy, and organoid models combined with liquid biopsy. Through this, we provide more effective preclinical experimental schemes using the PCa organoid model.
Prostate cancer (PCa) is one of the most common malignancies among men worldwide (1). In 2020, the incidence of prostate cancer in men was as high as 7.3%, second only to lung cancer. At the same time, the mortality rate reached 3.8% which led PCa to become the fifth major cause of cancer death in men (2). Since most PCa cases are androgen-driven adenocarcinomas, androgen deprivation therapy (ADT) is the main clinical treatment for early-stage PCa (3, 4). Most patients can benefit from treatment at an early stage through a rapid decrease in prostate-specific antigen (PSA) and a reduction in tumor volume. However, after a period of treatment, castration resistance ultimately ensues and the disease may develop into androgen receptor (AR)-castration-resistant prostate cancer (CRPC) or even neuroendocrine prostate cancer (NEPC), an AR-negative small cell neuroendocrine carcinoma (5, 6). The treatment for PCa is challenging due to its complex spatial, morphological, and genetic heterogeneity. Additionally, the oncologic transformation mechanism that results in clinical heterogeneity remains unclear. Therefore, there is an urgent need for prostate cancer preclinical models that can fully reflect the heterogeneity of PCa.
The research models used for the study of PCa mainly include traditional cell lines, conditional reprogramming cells (CRC), organoid models, patient-derived xenografts (PDX), and genetically engineered mouse models (GEMM). Different models have different advantages and disadvantages (Table 1). Traditional PCa cell lines, including LNCaP, VCaP, PC3, 22RV1, DU145, C4-2, and NCI-H660, are widely available and inexpensive. These cell lines show infinity growth, amenability to high-throughput screening and easy genome editing, but a lack of tumor heterogeneity and tumor microenvironment (7–9). In the CRC culture system, the combination of Rho kinase inhibitor Y-27632 and irradiated mouse fibroblast feeder cells enables primary cancer cells to acquire partial stem cell characteristics and the ability to indefinitely proliferate in vitro (10). However, androgen responsiveness of PCa is limited in this system and CRC is susceptible to contamination by feeder cells (11, 12). PDX, an important preclinical model in vivo, recapitulates tumor heterogeneity with high fidelity and correlates highly with patient responses (13). However, PDX is expansive and need a long time to establish (14). Most importantly, PDX is not amenable to high-throughput screening and genome editing (15). GEMM is spontaneous animal model that has been generated to emulate the expected functional consequences of key genomic alterations and has their own complete tumor microenvironment and immune system (16, 17). However, it is not only expensive and time consuming, but also only contains one or two genomic alterations by gene editing and is prone to induce multisystem tumors (18).
Therefore, a suitable model system, which can compensate for the shortcomings of the above-mentioned models, is particularly significant for PCa research. The organoid model might be such a compensatory model. Organoids in three-dimensional (3D) culture are derived from pluripotent stem cells or isolated organ progenitor cells to form organlike structures like the organs in vivo (19). Organoids encapsulate the diverse heterogeneity observed in clinical medicine (13, 14). More importantly, organoids are convenient for genetic manipulation and high-throughput drug screening in vitro (15, 16). Furthermore, they have a high correlation with the drug response of primary tumors in patients. On the other hand, PCa organoids also have some disadvantages, such as low efficiency of establishing organoids (particularly primary PCa), lack of microenvironment and immune system and the contamination of normal cells (17, 18). However, their unique advantages allow PCa organoids to be a great potential in vitro preclinical model, allowing for in-depth analyses of tumor heterogeneity.
In the present review, we review the culture conditions, establishments and limitations of PCa organoids and their applications in pathogenesis, drug screening, mechanism of drug resistance and individualized treatment for PCa. In addition, we further discuss the potential developmental directions of PCa organoids.
The organoid model is a 3D culture of isolated pluripotent stem cells or organ progenitor cells in a matrix such as Matrigel. It provides a similar environment in vitro for cells or tissues to develop into micro-organs. The culture method of PCa has been described. Figure 1 shows the general process of organoid culture for PCa. Sato et al. (26) first developed the universal organoid medium that contains advanced DMEM/F12 medium with epidermal growth factor (EGF), Noggin, and Wnt agonist R-spondin-1. On this basis, Drost et al. (27) continued to add different compounds and growth factors, including anaplastic lymphoma kinase (ALK) 3/4/5 inhibitor A83-01, dihydrotestosterone (DHT), fibroblast growth factor-10 (FGF10), fibroblast growth factor-2 (FGF2), prostaglandin E2 (PGE2), nicotinamide, and p38 inhibitor SB202190, N-acetylcysteine, B27 supplement and Rho kinase inhibitor Y-27632 to culture PCa organoids successfully (Table 2). Among them, growth factors FGF10, FGF2, PGE2, nicotinamide and SB202190 are not required when murine-derived PCa organoids are cultured. These organoids form a glandular structure, have a stable karyotype similar to that of the prostate in vivo, and complete AR signal transduction (28). In this culture system, PCa organoids can be successfully cultured within 2 weeks with an average split ratio of 1:2 every 2 weeks (27). Additionally, it can be applied to the PDX model, maintaining the heterogeneity of PCa (17). Based on these culture conditions, PCa organoids have been successfully established from normal tissues, tumor biopsy samples, circulating tumor cells (CTC), PCa cell lines, PDX models of PCa, human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) (17, 27–32).
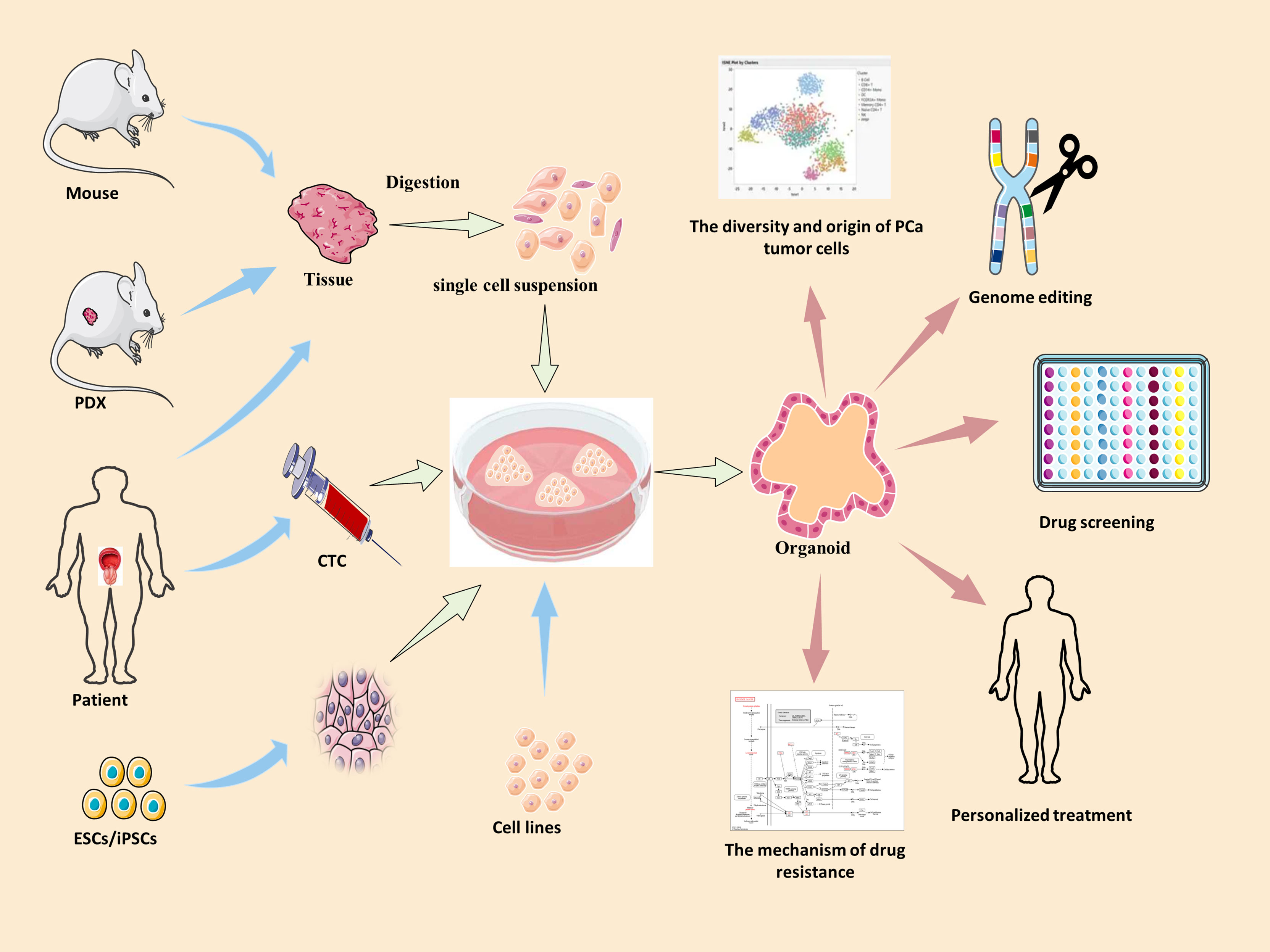
Figure 1 The summary of culture and applications of PCa organoid. Blue arrow →, the specimen source of PCa organoid. Red arrow →, the applications of PCa organoid. PDX, patient-derived xenografts; ESCs, human embryonic stem cells; iPSCs, induced pluripotent stem cells; CTC, circulating tumor cells.
Different growth factors play different roles in the culture of PCa organoids. R-spondin-1 and Noggin enhance the formation and expansion of organoids by promoting Wnt signaling and regulating the bone morphogenetic protein (BMP) signaling pathway, respectively (33, 34). Elimination of R-spondin-1 or Noggin will result in the inhibition of AR expression (28). Due to the high dosage and cost of R-spondin-1 and Noggin, the preparation methods of conditioned medium using modified cells for these factors are sorted in Table 3 (27, 35, 36). EGF has been shown to be essential for prostate epithelial cell line growth derived from organoids, which vastly strengthens cell viability and proliferation (37). The absence of EGF upregulates AR signal transduction at the transcription and translation levels and also increases PSA expression (28). Previous research has suggested that EGF in culture medium could significantly reduce the sensitivity of PCa organoids to androgen resistance (38); thus, it is necessary to eliminate EGF during the pharmacological response of sensitivity to anti-androgen drugs. FGF2, PGE2, nicotinamide, B27 supplement, and transforming growth factor-beta (TGF-β) inhibitor A83-01 promote the proliferation of prostate cells to maintain the long-term growth of organoids (39–41). P38 kinase inhibitor SB202190 inhibits the histological keratinization phenomenon that may be increased by pressure signal transduction (28). DHT and FGF10 improve the efficiency of organoid formation (28). Rho kinase inhibitor Y-27632 can promote the proliferation of epithelial cells (42). It is worth noting that Y-27632 is only added to the medium during establishment of the first generation of organoids, after which there is no need to add it to the subsequent generations, and added when using TrypLE to digest organoids (27). N-acetylcysteine promotes organoid proliferation by inhibiting cell oxidation (43). These growth factors and hormones constitute the basic conditions to support the survival and long-term culture of PCa cells in the 3D culture system. These studies highlight the importance of media modification in culturing organoids. Required additive components differ depending on cell types, gene mutations and phenotypes of PCa. For instance, the addition of 10% FBS helps to promote the proliferation of PCa organoids for patients with bone metastases (44).
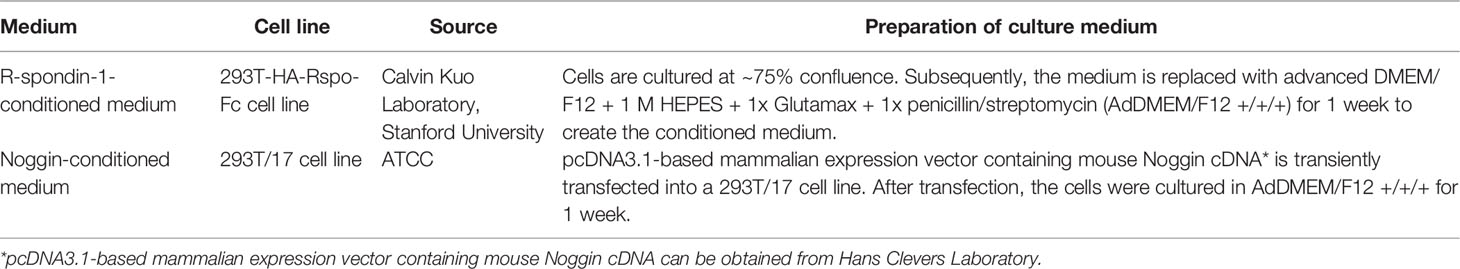
Table 3 Preparation of Noggin-conditioned medium and R-spondin-1-conditioned medium for organoids (27, 35, 36).
The culture of PCa organoids is still under continuous exploration and improvement. It has evolved from a separate organoid culture to a co-culture of organoid and cells related to the tumor microenvironment (45). For example, Richards et al. (46) co-cultured PCa epithelial cells with primary prostate stromal cells to study the direct interaction between epithelial and stromal cells. They found that the addition of stromal cells not only improved the ability to generalize tissue characteristics, but also improved the survival rate and formation efficiency of tumor organs. This indicates that organoid technology can successfully simulate the growth environment, in vivo, for tumor cells through co-culture with the tumor microenvironment. It can also provide a new platform for the study of the interaction of cancer cells with the microenvironment. Unfortunately, only the co-culture of PCa organoids with stromal cells has been studied, and co-culture studies with other tumor microenvironment cells have been reported.
The specimen for organoid culture can be derived from any stage of tumor development and only a small piece of tissue or few cells are required to culture in vitro (47). Thus, different phenotypes of PCa organoids have been successfully cultivated. Karthaus et al. (28) used mouse prostate epithelial cells and human prostate specimens to cultivate normal prostate organoids composed of fully differentiated CK5+ basal cells and CK8+ luminal cells. It was found that such organoids retained the expression of AR signaling factors, prostate-specific transcription factor Nkx3.1, basal cell markers p63 and CK5, and luminal cell marker CK8. At the same time, the original tissues of mouse with PTEN knockout and TMPRSS2-ERG fusion gene were cultured in organoids, and the phenotypes were consistent. Additionally, Gao et al. (17) successfully cultivated seven metastatic castration-resistant prostate cancer (mCRPC)-type organoids from biopsy tissue specimens and CTC in patients with metastatic PCa. These organoids capsulate the molecular diversity and clinical heterogeneity of PCa subtypes, including TMPRSS2-ERG fusion, ETS translocation, SPOP mutation, SPINK1 overexpression, FOXA1 and PIK3R1 mutation, CHD1 deletion, PTEN deletion, and AR expression, and retain the genomic characteristics of primary tumors. These organoids reproduce a large repertoire of patient-derived CRPC lines with different genomic alterations to research PCa.
NEPC, a subtype of PCa with a poor prognosis, easily develops therapeutic resistance (48). Currently, there are only a few effective models for the study of neuroendocrine tumors. Puca et al. (49) successfully cultured four neuroendocrine-like PCa organoids from metastatic biopsy specimens, including simple small cell carcinoma and high-grade carcinoma with extensive neuroendocrine differentiation. They reported that the organoid model is a good tool for further understanding the characteristics of NEPC. Currently, androgen-sensitive and hormone-insensitive CRPC organoid models have been established, and CRPC transformation models have also been reported. Additionally, PCa cell lines LNCaP, C4-2B, and single luminal epithelial progenitors can also produce PCa organoids (29, 50). LNCaP produces androgen-dependent models while C4-2B produces a non-androgen-dependent model, each representing different clinical characteristics.
Recently, a novel in vitro modeling system combining the advantages of organoids and PDX has been reported (51). The PDX-organoid model takes less time and costs less than the PDX model and provides an adequate source of tissue for the establishment of organoid models. The LuCap PDX series used surgical specimens from metastatic PCa, successfully producing 21 PDX models (52). Beshiri et al. (30) produced PDX-organoid models with mCRPC characteristics similar to the LuCaP series. These PDX-organoid models retained the genomic heterogeneity of mCRPC and AR-dependent signaling, thus providing a good platform for the study of the pathogenesis, therapeutic options, and individualized treatment for mCRPC. Furthermore, bone metastatic prostate cancer (BMPC) organoid models derived from patients have been successfully established (44).
There are several limitations in the establishment of PCa organoids. Firstly, the overall success rate of PCa organoids is only 15-20% (17), limiting the extensive development of clinically diversified PCa models. A study produced statistics on organoids from 81 PCa specimens with diverse pathological and clinical features (53). The success rate of organoid development from metastatic prostatectomy reached 4/9 while that of organoid development from transurethral resection of the prostate was only 4/14. Another study reported a success rate of 16% (4/25) for organoids cultured from patients with metastatic PCa (49). This indicates that the key to the success of PCa organoids depends on the source and intrinsic characteristics of the sample. Therefore, it is necessary to improve the culture conditions for different origins or phenotypes of PCa specimens to obtain a high success rate. Secondly, the organoids from patients with primary PCa have not been successfully cultured to date. This is probably because tumor cells do not have a selective advantage over normal cells in the current culture medium (27). The optimal combination of culture factors still needs to be explored and verified repeatedly. Thirdly, the lack of availability of clinical samples of mCRPC patients hampers the establishment of a biological resource bank of PCa organoids that includes a wide variety of clinical phenotypes. Fourthly, it is still challenging to maintain the growth of organoids for a long time. Fifthly, PCa organoids contained only epithelial cells and/or stromal cells, and lacked some tumor microenvironment components, such as immune cells and vascular components. PCa organoids with an immune compatible microenvironment have not been developed. Therefore, they cannot be used for immunotherapy research.
As a major technological breakthrough, organoids have been recognized as an important tool for biomedical research. An existing study has shown that PCa organoids can not only recapitulate in situ histology in vitro, but also have a genetic mutational landscape similar to that of prostate cancer (17). This suggests that organoids provide a very representative in vitro model for the study of prostate cancer. Organoids allow research on disease tissue biology, mechanism of disease occurrence and development, drug screening, and personalized treatment in an environment that simulates endogenous cell tissue and organ structure (Figure 1). Therefore, we summarize the applications of prostate cancer organoids with respect to these aspects.
In vitro models provide valuable insights into prostate biology, but current in vitro modeling systems are not representative of the cellular structure of the prostate. The organoid model better mimics prostate epithelial glands by recapitulating epithelial differentiation and cell polarity (50). Single-cell RNA Sequencing (RNA-Seq) analysis of patient-derived prostate epithelial cells revealed that compared to monolayer cultures, organoid cultures contained more distinct cell populations (54, 55). Ten cell types were identified in vitro, including a rare population of putative stem cells marked by high Keratin 13 (KRT13), Lymphocyte Antigen 6D (LY6D) and prostate stem cell antigen (PSCA). These results demonstrated that organoid culture condition has contribute to the survival and proliferation of different cell populations. It has allowed a deeper understanding of the cells present in prostate model systems and the creation of an in-depth atlas of the cellular population.
The origin cell of PCa remains a subject of debate. According to previous studies (56), PCa has two types of origin cells: basal cells and luminal cells. In tissue recombination models, only basal cells reconstitute a complete prostate gland (57, 58). However, it has been shown that luminal cells can generate basal cells through murine lineage-tracing experiments (59). Additionally, in the human prostate, only basal cells have been shown to be effectively transformed by select oncogenes (60). Organoid culture provides a unique model to solve this problem. After the organoid culture of basal and luminal cells of mouse/human prostate tissue were separated by fluorescence-activated cell sorting, both formed prostate-like organoids that retained both basal and luminal epithelial layers and preserved androgen-responsiveness in culture (28). This suggests that both basal and luminal cells have stem-like potential, and organoid culture can maintain stem cell characteristics of PCa. Another study (61) reported that c-MYC/myrAKT1-transduced human prostate basal- and luminal-derived organoids represented histological and molecular features of human PCa. These studies confirm that both human primary basal cells and luminal cells are the origin cells of PCa. Organoids can monitor the early development of PCa in vitro in real-time and directly compare the transformation of basal and luminal cells.
As is well-known, organoids are originally derived from cancer stem cells (CSCs) residing within the tumor bulk of the samples. PCa is a highly heterogeneous tumor harboring multiple cancer cell types. Different from the cell-of-origin that undergoes tumorigenic transformation due to gene mutation, CSCs are a cell population with self-renewal and pluripotent properties driving clonal tumor evolution (62). The existence of CSCs provides theoretical explanations for many molecular characteristics, cancer recurrence, metastasis and treatment resistance of PCa (63). Studies on CSCs have been hampered by the lack of suitable in vitro models. Here, organoids can be used as a good model in vitro for CSCs. Therapies targeting CSCs may lead to more effective cancer treatments for PCa. Therefore, organoid models can help us further understand the composition of normal stem cells and CSCs, which will be important in studying the occurrence and development of prostate cancer.
The mechanisms that drive the pathogenesis of PCa and the series of clinical transformations are not well understood. Gene editing at the organoid level can simulate the effects of different gene mutations on the occurrence, development, and heterogeneous transformation of PCa in vitro. The easy handling of PCa organoid cultures in vitro also facilitates the editing of specific genes related to human diseases using lentivirus transfection, plasmid transformation, or the CRISPR/Cas9 gene-editing system (64, 65). Chua et al. (50) demonstrated that combined PTEN deletion and KrasG12D activation in organoids derived from CARNs (castration-resistant Nkx3.1-expressing cells) produced similar phenotypes to donor tumors of PCa in vivo. This proved that the deletion of PTEN and the routine mutation of KRAS are important in the induction of prostate tumors. The tumor origin cells and the chronological sequence of oncogenic events play an important role in defining the disease status. Using PCa organoids, Pietrzak et al. (66) proved that loss of the TIP5 transcription factor could trigger PTEN-loss mediated oncogenic transformation in prostate luminal cells, but becomes dispensable once the transformation is established. This suggests that TIP5-mediated chromatin states can control key developmental pathways and tumor suppressor genes, which could drive the development of cancer. In summary, transforming normal prostate organoids into cancerous organoids in vitro provides the optimal tool to identify the molecular subtypes of PCa in the genomic analysis of primary tumors.
Considering the heterogeneity of PCa, it is difficult to identify the true driver mutations. However, organoid technology displays a unique advantage. Because normal tissue-derived and tumor tissue-derived organoids from the same patient can be established simultaneously, the discrepancy in gene expression between the two can be compared to identify possible driver mutations. Furthermore, organoid modeling and gene editing can verify the screened mutant genes to study their influence on PCa. Normal tissue-derived organoid has relatively stable genetic information and can be used as a control model for studying tumor mutations.
Since PCa organoids can maintain genetic stability and heterogeneity, coupled with its easy high-throughput screening, it has become an ideal model for drug toxicity and efficacy evaluation (67). To explore whether CRPC-derived organoids were suitable for drug testing, they were used to determine the sensitivity to enzalutamide, everolimus, and BKM-120 (17). It was found that AR-amplified MSK-PCa2 organoid was extremely sensitive to enzalutamide and resistant to other drugs. Moreover, MSK-PCa2 organoid with PTEN loss and PIK3R1 mutation was sensitive to everolimus and BKM-120, consistent with the results in vivo. This result demonstrates the utility of CRPC-derived organoids in assessing drug sensitivity, similar to findings in clinical trials. Further, these results suggest that organoids can be used for drug screening and modeling individualized treatment. Four CRPC-neuroendocrine (CRPC-NE) organoids and two CRPC-adenocarcinoma (CRPC-Aden) organoids were established for high-throughput sequencing of 129 chemotherapeutics and targeted drugs (49). The results demonstrated that the AR antagonist, enzalutamide and taxane chemotherapies, cabazitaxel and docetaxel were effective for CRPC-Aden organoids. A small number of drugs, such as pozotinib and vandetanib, were found to have significant activity in CRPC-NE organoids.
Organoid models are effective in vitro drug testing platforms for identifying potential pharmacological treatments and screening inhibitors targeting different phenotypes of PCa. For instance, Jansson et al. (68) screened 110 drugs from CRPC LuCaP PDX-derived organoids and showed that HSP90 inhibitors had a significant inhibitory effect on CRPC. Further studies have demonstrated that the HSP90 inhibitor, ganetespib, decreased tumor growth by inhibiting multiple targets including the factors involved in AR and PI3K pathways. Using established PTEN/TP53 null LuCaP 136 tumors, they found that compared with any single therapy, the combination of ganetespib and castration significantly inhibited tumor growth and led to a delay in castration resistance.
Gene mutation, chromosome amplification, and chromosome rearrangement are the main reasons for the development of drug resistance in tumors (69). Patient-derived organoids (PDO) of drug resistance can be established according to the different mutations in patients to explore targets for improving the prognosis of patients with PCa, which is incomparable with existing PCa cell lines. It has been confirmed that there are at least three general mechanisms for the development of resistance in CRPC (38). First, genetic mutations, such as AR, ETS, TP53, and PTEN gene mutations, lead to the activation of the AR signaling pathway (70). Second, the activation of bypass signals, such as the glucocorticoid receptor pathway, compensates for the loss of the AR signal (71). Third, during treatment, tumor cells acquire resistance by switching lineages from a cell type, dependent on the drug target, to another, which is not. This may be represented by cases of PCa that are AR-negative or neuroendocrine specific (72, 73). The corresponding heterogeneous organoid models will represent their respective resistance mechanisms. Moreover, to clarify the molecular mechanism of PCa resistance, some groups have used PCa organoids as a platform for rapid detection of the effects of different mutations on anti-tumor drugs using CRISPR/Cas9 or lentivirus transfection technology for gene editing. For instance, Pappas et al. (38) used the PCa organoid model to demonstrate that the loss of p53 did not induce resistance to androgenic molecules but Pten deficiency increased resistance to androgenic drugs. However, the dual loss of p53 and Pten resulted in complete resistance to the second generation of anti-androgen drugs. Dai et al. (74) found that PCa-associated SPOP mutations conferred resistance to bromodomain and extra-terminal (BET) inhibitors by stabilizing bromodomain-containing protein 4 (BRD4). The development of resistance and relapse in CRPC tumor cells is a major problem in the field of PCa research. Since 3D organoid cultures simulate the emergence of treatment-resistant residual tumors, it provides a more effective platform for study. Dhimolea et al. (75) reported that treatment-resistant residual tumor cells in organoids, xenografts, and cancer patients entered a dormant diapause-like adaptation to reduce apoptosis priming by suppressing MYC activity or inhibiting MYC transcriptional co-activator BRD4, which could weaken drug cytotoxicity and induce the tumor to be resistant to treatment. Therefore, organoid culture can be useful in constructing a heterogeneous PCa model in vitro to study the influence of specific genes on PCa resistance with the help of gene-editing technology. It is beneficial for developing first-line therapeutic drugs or screening new therapeutic targets.
Organoids, as preclinical cancer models, can be used to achieve precision medicine through in vitro therapeutic screening of individual patient samples. Several studies have demonstrated that drug response to organoids may predict clinical outcomes (76, 77). Although many clinical trials related to PDO models are carried out, there are limited reports about PCa. Beltran H’s team cultured CRPC-NE samples from patients into organoids for high-throughput drug screening. One CRPC-NE organoid, OWCM155, showed significant sensitivity to aurora kinase inhibitor alisertib (49), a result concordant with what was being observed from the clinical response of corresponding patients in the phase II trial of alisertib for CRPC-NE (NCT01482962) (78). On the other hand, the CRPC-NE organoid, OWCM154, did not react to alisertib in vitro, nor did the patient in phase II clinical trial. In additional, Beshiri et al. (30) found that LuCaP-derived organoids with BRCA2 deficiency were sensitive to olaparib, which was consistent with the response observed in clinical setting (79). These data indicate that organoids may be useful tools as patient ‘avatars’ for clinical trials and applied to develop new strategies for precision medicine in cancer. However, not all drug responses of organoids are consistent with clinical responses. Karkampouna at al. (80). cultured PDOs for drug screening. The organoids from an advanced PCa patient (case P82) as well as from a primary PCa patient (case P134) were both resistant to enzalutamide in vitro, while cases P82 and P134, were sensitive and resistant, respectively, to enzalutamide. The correlation between drug responses of organoids and clinical responses may depend on the size or site of the patient sample. Larger sample sizes would be needed to increase the confidence level in the data. The high heterogeneity of PCa and the drug profiles correlated with disease stage, sometimes leads to incompatibility between the results of the drug response in organoids and the clinical response of the patient.
As a novel in vitro preclinical disease model, the PCa organoid has potential for broad applications. This model is not only convenient for in vitro applications, but also demonstrates genetic stability and heterogeneity. Not only can it be applied for multi-model verification of PCa pathogenesis and drug screening combined with the PDX animal model, but also for observing the growth and development of tumor cells or tissues in vitro and studying the effects of different mutations on the occurrence and development of diseases. This model provides effective guidance for studying the heterogeneous transformation mechanism of PCa, especially to understand the molecular mechanisms underlying androgen resistance and to screen potential therapeutic targets. From the perspective of drug screening and individualized treatment, different primary or metastatic PCa organoids can be established using PCa samples from different sources. Combined with high-throughput screening technology, anti-PCa drugs can be assessed in batches. The response of patients with PCa to different drugs can be predicted effectively, which will accelerate the development of therapeutic regimens for heterogeneous characteristics of PCa. In PCa organoids, CRISPR/Cas9 and shRNA techniques have been used to study the mechanism of drug resistance. This will allow the study of the functions of drug resistance genes, and will ultimately enable the improvement of first-line therapeutic drugs and the development of novel individualized drugs. Inevitably, PCa organoids still have some challenges including low efficiency of establishing organoids (particularly primary PCa) from human samples, the optimal combination of medium factors that need to be added and the maintenance of culturing those organoids for a long time. Additionally, there are new application directions for PCa organoids.
Exploring the interaction between the tumor and its microenvironment is an important aspect of oncology research. Organoid culture of the vascular system in vitro simulates the interaction between tumor cells and the vascular system while that of the nervous system revealed the interaction between cells and the nervous system. Immunocytes are co-cultured with tumor organoids to reveal the relationship between tumor cells and immune infiltrating cells (81). It is necessary to further study the role of fibroblasts, immune cells, and endothelial cells in the tumor microenvironment so that the structure of PCa organoids can better represent the composition of PCa in vivo.
The lack of in vitro tumor models maintaining the characteristics of tumor cells in vivo has become a bottleneck in the realization of personalized therapy and precision therapy for patients with cancer. Organoids are an ideal model for drug testing and screening because of their ability to be cultured in vitro for a long time to maximize the characterization of tumor cells in vivo. Therefore, to better reflect the clinical heterogeneity of PCa, it is necessary to improve the culture conditions in the future to better support the growth of different types of PCa cells. In 2014, world’s first prostate tumor organ bank was established (18). These organoids were used to test a variety of experimental drugs and cancer drugs, finding that tumor organs with different genetic backgrounds had different sensitivities to these drugs.
Patients with CRPC that have high treatment selection pressure can lead to a significant increase in PCa heterogeneity (82), such as changes in AR and PSA expression levels, AR mutations, and the occurrence of the NEPC phenotype. Therefore, more CRPC models are needed to represent different resistance mechanisms, especially dynamic models to reflect their transformation characteristics. The existing organoid models only reflect the static characteristics of a patient at a certain stage and are insufficient in simulating the diversity and disease progression of PCa. Therefore, it is inevitable to develop a model based on the same specimen that can be induced to reflect the whole process of PCa transformation, including the development of CRPC, the transformation from adenocarcinoma to NEPC, and the occurrence of metastasis. This aids tremendously in investigating progression from pre-neoplastic to neoplastic to metastatic states. Additionally, a paired comparison model can be generated through organoid culture to compare genotypes and phenotypes and explore the mechanism of their heterogeneity.
The application of tumor organoids in tumor immunology offers researchers another attractive preclinical option. Studies (83–85) have found that tumor organoids can be co-cultured with immune cells isolated from autologous tumor tissues or peripheral blood of healthy donors. Further studies on different immune cells should be performed, including amplification of tumor organoids in vitro to explore the interaction between the immune system and tumors. In the model of tumor immunotherapy, compared with immortalized tumor cell lines and humanized immune-oncology models (86, 87), the co-culture of tumor organoids and immune cells from tumor tissues better represents the response of patients to immunotherapy. The co-culture system of tumor organoids and immune cells has a short experimental cycle and does not have the problem of human-mouse immune compatibility, which is expected for an ideal model for tumor immunotherapy.
The patient derived PDX model of PCa was established using a specimen from surgical resection or autopsy, highlighting the need for more specimens. At the same time, the heterogeneity within the tumor is one of the challenges of traditional tissue biopsy. Liquid biopsy is expected to overcome these issues. CTC can be used as an alternative to invasive biopsies to study the heterogeneity of tumors (88). However, liquid biopsy has disadvantages in the detection and enrichment of CTC since only a few CTC can usually be obtained. Tumor organoids can be grown from only limited amounts of specimens to solve this problem. Gao et al. (17) confirmed the feasibility of using a small amount of CTC from peripheral blood of patients with PCa to culture tumor organoids. Exome sequencing showed that PCa organoids derived from CTC retained molecular diversity consistent with primary tumors, including TMPRSS2-ERG fusion, SPOP mutation, SPINK1 overexpression, and CHD1 loss. This is the only successful case of derivation of organoids from blood samples reported thus far. Liquid biopsy combined with tumor organoids may create new opportunities for minimally invasive studies and may facilitate the inclusion of PDO in personalized medical procedures for cancers.
In conclusion, it is necessary to continuously optimize the culture conditions of PCa organoids, constructing a tumor microenvironment more suitable for the different mutations in PCa. Further improving the success rate of PCa organoids is beneficial for obtaining a wider range of phenotypes and genotypes. This will allow the construction of models for hormone naïve PCa, CRPC, and mCRPC to fully demonstrate the clinical heterogeneity of PCa. This will also allow the formation of a PCa organoid bank to maximize its utility in mirroring clinical characteristics and become the preferred preclinical disease model of PCa.
LZ, YZ, and CS: conceptualization. LZ, CZ, and CS: writing. LZ, CZ, and CS: revising. All authors contributed to the article and approved the submitted version.
This study was funded by the National Natural Science Foundation Program of China (No. 31772546 and 32070532) and Laboratory Animal Foundation Program No. SYDW[2017]-02.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Editage (www.editage.cn) for English language editing.
PCa, prostate cancer; ADT, androgen deprivation therapy; PSA, prostate-specific antigen; AR, androgen receptor; CRPC, castration-resistant prostate cancer; NEPC, neuroendocrine prostate cancer; CRC, conditional reprogramming cells; PDX, patient-derived xenografts; GEMM, genetically engineered mouse models; 3D, three-dimensional; EGF, epidermal growth factor; ALK, anaplastic lymphoma kinase; DHT, dihydrotestosterone; FGF10, fibroblast growth factor-10; FGF2, fibroblast growth factor-2; PGE2, prostaglandin E2; CTC, circulating tumor cells; ESCs, embryonic stem cells; iPSCs, induced pluripotent stem cells; BMP, bone morphogenetic protein; TGF-β, transforming growth factor-beta; mCRPC, metastatic castration-resistant prostate cancer; BMPC, bone metastatic prostate cancer; RNA-Seq, RNA Sequencing; KRT13, Keratin 13; LY6D, Lymphocyte Antigen 6D; PSCA, prostate stem cell antigen; CSCs, cancer stem cells; CARNs, castration-resistant Nkx3.1-expressing cells; CRPC-NE, CRPC-neuroendocrine; CRPC-Aden, CRPC-adenocarcinoma; PDO, patient-derived organoids; BRD4, bromodomain-containing protein 4.
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71:7–33. doi: 10.3322/caac.21654
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
3. Mottet N, van den Bergh RCN, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment With Curative Intent. Eur Urol (2021) 79:243–62. doi: 10.1016/j.eururo.2020.09.042
4. Cornford P, van den Bergh RCN, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer. Part II-2020 Update: Treatment of Relapsing and Metastatic Prostate Cancer. Eur Urol (2021) 79:263–82. doi: 10.1016/j.eururo.2020.09.046
5. Karantanos T, Corn PG, Thompson TC. Prostate Cancer Progression After Androgen Deprivation Therapy: Mechanisms of Castrate Resistance and Novel Therapeutic Approaches. Oncogene (2013) 32:5501–11. doi: 10.1038/onc.2013.206
6. Ma T, Bai S, Qi Y, Zhan Y, Ungerleider N, Zhang DY, et al. Increased Transcription and High Translation Efficiency Lead to Accumulation of Androgen Receptor Splice Variant After Androgen Deprivation Therapy. Cancer Lett (2021) 504:37–48. doi: 10.1016/j.canlet.2020.12.037
7. Sobel RE, Sadar MD. Cell Lines Used in Prostate Cancer Research: A Compendium of Old and New Lines–Part 1. J Urol (2005) 173:342–59. doi: 10.1097/01.ju.0000141580.30910.57
8. van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, et al. Molecular Characterization of Human Prostate Carcinoma Cell Lines. Prostate (2003) 57:205–25. doi: 10.1002/pros.10290
9. Namekawa T, Ikeda K, Horie-Inoue K, Inoue S. Application of Prostate Cancer Models for Preclinical Study: Advantages and Limitations of Cell Lines, Patient-Derived Xenografts, and Three-Dimensional Culture of Patient-Derived Cells. Cells (2019) 8:74. doi: 10.3390/cells8010074
10. Palechor-Ceron N, Krawczyk E, Dakic A, Simic V, Yuan H, Blancato J, et al. Conditional Reprogramming for Patient-Derived Cancer Models and Next-Generation Living Biobanks. Cells (2019) 8:1327. doi: 10.3390/cells8111327
11. Timofeeva OA, Palechor-Ceron N, Li G, Yuan H, Krawczyk E, Zhong X, et al. Conditionally Reprogrammed Normal and Primary Tumor Prostate Epithelial Cells: A Novel Patient-Derived Cell Model for Studies of Human Prostate Cancer. Oncotarget (2017) 8:22741–58. doi: 10.18632/oncotarget.13937
12. Wu X, Wang S, Li M, Li J, Shen J, Zhao Y, et al. Conditional Reprogramming: Next Generation Cell Culture. Acta Pharm Sin B (2020) 10:1360–81. doi: 10.1016/j.apsb.2020.01.011
13. Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-Tumoral Heterogeneity. Cell (2020) 180:188–204.e22. doi: 10.1016/j.cell.2019.11.036
14. Kopper O, de Witte CJ, Lõhmussaar K, Valle-Inclan JE, Hami N, Kester L, et al. An Organoid Platform for Ovarian Cancer Captures Intra- and Interpatient Heterogeneity. Nat Med (2019) 25:838–49. doi: 10.1038/s41591-019-0422-6
15. Kondo J, Inoue M. Application of Cancer Organoid Model for Drug Screening and Personalized Therapy. Cells (2019) 8:470. doi: 10.3390/cells8050470
16. Mazzocchi A, Soker S, Skardal A. 3D Bioprinting for High-Throughput Screening: Drug Screening, Disease Modeling, and Precision Medicine Applications. Appl Phys Rev (2019) 6:011302. doi: 10.1063/1.5056188
17. Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid Cultures Derived From Patients With Advanced Prostate Cancer. Cell (2014) 159:176–87. doi: 10.1016/j.cell.2014.08.016
18. Bahmad HF, Cheaito K, Chalhoub RM, Hadadeh O, Monzer A, Ballout F, et al. Sphere-Formation Assay: Three-Dimensional In Vitro Culturing of Prostate Cancer Stem/Progenitor Sphere-Forming Cells. Front Oncol (2018) 8:347. doi: 10.3389/fonc.2018.00347
19. Vela I, Chen Y. Prostate Cancer Organoids: A Potential New Tool for Testing Drug Sensitivity. Expert Rev Anticancer Ther (2015) 15:261–3. doi: 10.1586/14737140.2015.1003046
20. Okada S, Vaeteewoottacharn K, Kariya R. Application of Highly Immunocompromised Mice for the Establishment of Patient-Derived Xenograft (PDX) Models. Cells (2019) 8:889. doi: 10.3390/cells8080889
21. Arriaga JM, Abate-Shen C. Genetically Engineered Mouse Models of Prostate Cancer in the Postgenomic Era. Cold Spring Harb Perspect Med (2019) 9:a030528. doi: 10.1101/cshperspect.a030528
22. Walrath JC, Hawes JJ, Van Dyke T, Reilly KM. Genetically Engineered Mouse Models in Cancer Research. Adv Cancer Res (2010) 106:113–64. doi: 10.1016/S0065-230X(10)06004-5
23. Parisotto M, Metzger D. Genetically Engineered Mouse Models of Prostate Cancer. Mol Oncol (2013) 7:190–205. doi: 10.1016/j.molonc.2013.02.005
24. Lancaster MA, Knoblich JA. Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science (2014) 345:1247125. doi: 10.1126/science.1247125
25. Lin D, Wyatt AW, Xue H, Wang Y, Dong X, Haegert A, et al. High Fidelity Patient-Derived Xenografts for Accelerating Prostate Cancer Discovery and Drug Development. Cancer Res (2014) 74:1272–83. doi: 10.1158/0008-5472.CAN-13-2921-T
26. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures In Vitro Without a Mesenchymal Niche. Nature (2009) 459:262–5. doi: 10.1038/nature07935
27. Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, et al. Organoid Culture Systems for Prostate Epithelial and Cancer Tissue. Nat Protoc (2016) 11:347–58. doi: 10.1038/nprot.2016.006
28. Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, et al. Identification of Multipotent Luminal Progenitor Cells in Human Prostate Organoid Cultures. Cell (2014) 159:163–75. doi: 10.1016/j.cell.2014.08.017
29. Ma L, Li J, Nie Q, Zhang Q, Liu S, Ge D, et al. Organoid Culture of Human Prostate Cancer Cell Lines Lncap and C4-2B. Am J Clin Exp Urol (2017) 5:25–33.
30. Beshiri ML, Tice CM, Tran C, Nguyen HM, Sowalsky AG, Agarwal S, et al. A PDX/Organoid Biobank of Advanced Prostate Cancers Captures Genomic and Phenotypic Heterogeneity for Disease Modeling and Therapeutic Screening. Clin Cancer Res (2018) 24:4332–45. doi: 10.1158/1078-0432.CCR-18-0409
31. Calderon-Gierszal EL, Prins GS. Directed Differentiation of Human Embryonic Stem Cells Into Prostate Organoids In Vitro and Its Perturbation by Low-Dose Bisphenol a Exposure. PloS One (2015) 10:e0133238. doi: 10.1371/journal.pone.0133238
32. Hepburn AC, Curry EL, Moad M, Steele RE, Franco OE, Wilson L, et al. Propagation of Human Prostate Tissue From Induced Pluripotent Stem Cells. Stem Cells Transl Med (2020) 9:734–45. doi: 10.1002/sctm.19-0286
33. Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-Spondins Function as Ligands of the Orphan Receptors LGR4 and LGR5 to Regulate Wnt/Beta-Catenin Signaling. Proc Natl Acad Sci USA (2011) 108:11452–7. doi: 10.1073/pnas.1106083108
34. Cook C, Vezina CM, Allgeier SH, Shaw A, Yu M, Peterson RE, et al. Noggin Is Required for Normal Lobe Patterning and Ductal Budding in the Mouse Prostate. Dev Biol (2007) 312:217–30. doi: 10.1016/j.ydbio.2007.09.038
35. Broutier L, Andersson-Rolf A, Hindley CJ, Boj SF, Clevers H, Koo BK, et al. Culture and Establishment of Self-Renewing Human and Mouse Adult Liver and Pancreas 3D Organoids and Their Genetic Manipulation. Nat Protoc (2016) 11:1724–43. doi: 10.1038/nprot.2016.097
36. Yan HHN, Siu HC, Law S, Ho SL, Yue SSK, Tsui WY, et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell (2018) 23:882–897.e11. doi: 10.1016/j.stem.2018.09.016
37. Cheaito K, Bahmad HF, Jalloul H, Hadadeh O, Msheik H, El-Hajj A, et al. Epidermal Growth Factor Is Essential for the Maintenance of Novel Prostate Epithelial Cells Isolated From Patient-Derived Organoids. Front Cell Dev Biol (2020) 8:571677. doi: 10.3389/fcell.2020.571677
38. Pappas KJ, Choi D, Sawyers CL, Karthaus WR. Prostate Organoid Cultures as Tools to Translate Genotypes and Mutational Profiles to Pharmacological Responses. J Vis Exp (2019) 152:e60346. doi: 10.3791/60346
39. Kato M, Ishii K, Iwamoto Y, Sasaki T, Kanda H, Yamada Y, et al. Activation of FGF2-FGFR Signaling in the Castrated Mouse Prostate Stimulates the Proliferation of Basal Epithelial Cells. Biol Reprod (2013) 89:81. doi: 10.1095/biolreprod.112.107516
40. Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-Term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology (2011) 141:1762–72. doi: 10.1053/j.gastro.2011.07.050
41. Vo BT, Morton D Jr, Komaragiri S, Millena AC, Leath C, Khan SA. Tgf-β Effects on Prostate Cancer Cell Migration and Invasion Are Mediated by PGE2 Through Activation of PI3K/AKT/Mtor Pathway. Endocrinology (2013) 154:1768–79. doi: 10.1210/en.2012-2074
42. Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK Inhibitor and Feeder Cells Induce the Conditional Reprogramming of Epithelial Cells. Am J Pathol (2012) 180:599–607. doi: 10.1016/j.ajpath.2011.10.036
43. Khanna S, Mitra S, Lakhera PC, Khandelwal S. N-Acetylcysteine Effectively Mitigates Cadmium-Induced Oxidative Damage and Cell Death in Leydig Cells In Vitro. Drug Chem Toxicol (2016) 39:74–80. doi: 10.3109/01480545.2015.1028068
44. Lee S, Burner DN, Mendoza TR, Muldong MT, Arreola C, Wu CN, et al. Establishment and Analysis of Three-Dimensional (3D) Organoids Derived From Patient Prostate Cancer Bone Metastasis Specimens and Their Xenografts. J Vis Exp (2020) 156:e60367. doi: 10.3791/60367
45. Eder T, Eder IE. 3D Hanging Drop Culture to Establish Prostate Cancer Organoids. Methods Mol Biol (2017) 1612:167–75. doi: 10.1007/978-1-4939-7021-6_12
46. Richards Z, McCray T, Marsili J, Zenner ML, Manlucu JT, Garcia J, et al. Prostate Stroma Increases the Viability and Maintains the Branching Phenotype of Human Prostate Organoids. iScience (2019) 12:304–17. doi: 10.1016/j.isci.2019.01.028
47. Xia X, Li F, He J, Aji R, Gao D. Organoid Technology in Cancer Precision Medicine. Cancer Lett (2019) 457:20–7. doi: 10.1016/j.canlet.2019.04.039
48. Wang HT, Yao YH, Li BG, Tang Y, Chang JW, Zhang J. Neuroendocrine Prostate Cancer (NEPC) Progressing From Conventional Prostatic Adenocarcinoma: Factors Associated With Time to Development of NEPC and Survival From NEPC Diagnosis-a Systematic Review and Pooled Analysis. J Clin Oncol (2014) 32:3383–90. doi: 10.1200/JCO.2013.54.3553
49. Puca L, Bareja R, Prandi D, Shaw R, Benelli M, Karthaus WR, et al. Patient Derived Organoids to Model Rare Prostate Cancer Phenotypes. Nat Commun (2018) 9:2404. doi: 10.1038/s41467-018-04495-z
50. Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, et al. Single Luminal Epithelial Progenitors can Generate Prostate Organoids in Culture. Nat Cell Biol (2014) 16:951–61. doi: 10.1038/ncb3047
51. Huang L, Bockorny B, Paul I, Akshinthala D, Frappart PO, Gandarilla O, et al. PDX-Derived Organoids Model In Vivo Drug Response and Secrete Biomarkers. JCI Insight (2020) 5:e135544. doi: 10.1172/jci.insight.135544
52. Nguyen HM, Vessella RL, Morrissey C, Brown LG, Coleman IM, Higano CS, et al. Lucap Prostate Cancer Patient-Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease and Serve as Models for Evaluating Cancer Therapeutics. Prostate (2017) 77:654–71. doi: 10.1002/pros.23313
53. Servant R, Garioni M, Vlajnic T, Blind M, Pueschel H, Müller DC, et al. Prostate Cancer Patient-Derived Organoids: Detailed Outcome From a Prospective Cohort of 81 Clinical Specimens. J Pathol (2021) 254:543–55. doi: 10.1002/path.5698
54. McCray T, Moline D, Baumann B, Vander Griend DJ, Nonn L. Single-Cell RNA-Seq Analysis Identifies a Putative Epithelial Stem Cell Population in Human Primary Prostate Cells in Monolayer and Organoid Culture Conditions. Am J Clin Exp Urol (2019) 7:123–38.
55. Henry GH, Malewska A, Joseph DB, Malladi VS, Lee J, Torrealba J, et al. A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra. Cell Rep (2018) 25:3530–3542.e5. doi: 10.1016/j.celrep.2018.11.086
56. Packer JR, Maitland NJ. The Molecular and Cellular Origin of Human Prostate Cancer. Biochim Biophys Acta (2016) 1863:1238–60. doi: 10.1016/j.bbamcr.2016.02.016
57. Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP, Witte ON. Trop2 Identifies a Subpopulation of Murine and Human Prostate Basal Cells With Stem Cell Characteristics. Proc Natl Acad Sci USA (2008) 105:20882–7. doi: 10.1073/pnas.0811411106
58. Garraway IP, Sun W, Tran CP, Perner S, Zhang B, Goldstein AS, et al. Human Prostate Sphere-Forming Cells Represent a Subset of Basal Epithelial Cells Capable of Glandular Regeneration In Vivo. Prostate (2010) 70:491–501. doi: 10.1002/pros.21083
59. Wang ZA, Mitrofanova A, Bergren SK, Abate-Shen C, Cardiff RD, Califano A, et al. Lineage Analysis of Basal Epithelial Cells Reveals Their Unexpected Plasticity and Supports a Cell-of-Origin Model for Prostate Cancer Heterogeneity. Nat Cell Biol (2013) 15:274–83. doi: 10.1038/ncb2697
60. Stoyanova T, Cooper AR, Drake JM, Liu X, Armstrong AJ, Pienta KJ, et al. Prostate Cancer Originating in Basal Cells Progresses to Adenocarcinoma Propagated by Luminal-Like Cells. Proc Natl Acad Sci USA (2013) 110:20111–6. doi: 10.1073/pnas.1320565110
61. Park JW, Lee JK, Phillips JW, Huang P, Cheng D, Huang J, et al. Prostate Epithelial Cell of Origin Determines Cancer Differentiation State in an Organoid Transformation Assay. Proc Natl Acad Sci USA (2016) 113:4482–7. doi: 10.1073/pnas.1603645113
62. Rycaj K, Tang DG. Cell-of-Origin of Cancer Versus Cancer Stem Cells: Assays and Interpretations. Cancer Res (2015) 75:4003–11. doi: 10.1158/0008-5472.CAN-15-0798
63. Skvortsov S, Skvortsova II, Tang DG, Dubrovska A. Concise Review: Prostate Cancer Stem Cells: Current Understanding. Stem Cells (2018) 36:1457–74. doi: 10.1002/stem.2859
64. Koo BK, Stange DE, Sato T, Karthaus W, Farin HF, Huch M, et al. Controlled Gene Expression in Primary Lgr5 Organoid Cultures. Nat Methods (2011) 9:81–3. doi: 10.1038/nmeth.1802
65. Nie J, Hashino E. Organoid Technologies Meet Genome Engineering. EMBO Rep (2017) 18:367–76. doi: 10.15252/embr.201643732
66. Pietrzak K, Kuzyakiv R, Simon R, Bolis M, Bär D, Aprigliano R, et al. TIP5 Primes Prostate Luminal Cells for the Oncogenic Transformation Mediated by PTEN-Loss. Proc Natl Acad Sci USA (2020) 117:3637–47. doi: 10.1073/pnas.1911673117
67. Weeber F, Ooft SN, Dijkstra KK, Voest EE. Tumor Organoids as a Pre-Clinical Cancer Model for Drug Discovery. Cell Chem Biol (2017) 24:1092–100. doi: 10.1016/j.chembiol.2017.06.012
68. Jansson KH, Tucker JB, Stahl LE, Simmons JK, Fuller C, Beshiri ML, et al. High-Throughput Screens Identify HSP90 Inhibitors as Potent Therapeutics That Target Inter-Related Growth and Survival Pathways in Advanced Prostate Cancer. Sci Rep (2018) 8:17239. doi: 10.1038/s41598-018-35417-0
69. Fojo T. Multiple Paths to a Drug Resistance Phenotype: Mutations, Translocations, Deletions and Amplification of Coding Genes or Promoter Regions, Epigenetic Changes and Micrornas. Drug Resist Update (2007) 10:59–67. doi: 10.1016/j.drup.2007.02.002
70. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell (2015) 161:1215–28. doi: 10.1016/j.cell.2015.06.053
71. Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid Receptor Confers Resistance to Antiandrogens by Bypassing Androgen Receptor Blockade. Cell (2013) 155:1309–22. doi: 10.1016/j.cell.2013.11.012
72. Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 Cooperate to Suppress Prostate Cancer Lineage Plasticity, Metastasis, and Antiandrogen Resistance. Science (2017) 355:78–83. doi: 10.1126/science.aah4199
73. Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, et al. SOX2 Promotes Lineage Plasticity and Antiandrogen Resistance in TP53- and RB1-Deficient Prostate Cancer. Science (2017) 355:84–8. doi: 10.1126/science.aah4307
74. Dai X, Gan W, Li X, Wang S, Zhang W, Huang L, et al. Prostate Cancer-Associated SPOP Mutations Confer Resistance to BET Inhibitors Through Stabilization of BRD4. Nat Med (2017) 23:1063–71. doi: 10.1038/nm.4378
75. Dhimolea E, de Matos Simoes R, Kansara D, Al’Khafaji A, Bouyssou J, Weng X, et al. An Embryonic Diapause-Like Adaptation With Suppressed Myc Activity Enables Tumor Treatment Persistence. Cancer Cell (2021) 39:240–256.e11. doi: 10.1016/j.ccell.2020.12.002
76. Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discovery (2017) 7:462–77. doi: 10.1158/2159-8290.CD-16-1154
77. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-Derived Organoids Model Treatment Response of Metastatic Gastrointestinal Cancers. Science (2018) 359:920–6. doi: 10.1126/science.aao2774
78. Beltran H, Oromendia C, Danila DC, Montgomery B, Hoimes C, Szmulewitz RZ, et al. A Phase II Trial of the Aurora Kinase a Inhibitor Alisertib for Patients With Castration-Resistant and Neuroendocrine Prostate Cancer: Efficacy and Biomarkers. Clin Cancer Res (2019) 25:43–51. doi: 10.1158/1078-0432.CCR-18-1912
79. Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med (2015) 373:1697–708. doi: 10.1056/NEJMoa1506859
80. Karkampouna S, La Manna F, Benjak A, Kiener M, De Menna M, Zoni E, et al. Patient-Derived Xenografts and Organoids Model Therapy Response in Prostate Cancer. Nat Commun (2021) 12:1117. doi: 10.1038/s41467-021-21300-6
81. Xu R, Zhou X, Wang S, Trinkle C. Tumor Organoid Models in Precision Medicine and Investigating Cancer-Stromal Interactions. Pharmacol Ther (2021) 218:107668. doi: 10.1016/j.pharmthera.2020.107668
82. Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The Mutational Landscape of Lethal Castration-Resistant Prostate Cancer. Nature (2012) 487:239–43. doi: 10.1038/nature11125
83. Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of Tumor-Reactive T Cells by Co-Culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell (2018) 174:1586–1598.e12. doi: 10.1016/j.cell.2018.07.009
84. Cattaneo CM, Dijkstra KK, Fanchi LF, Kelderman S, Kaing S, van Rooij N, et al. Tumor Organoid-T-Cell Coculture Systems. Nat Protoc (2020) 15:15–39. doi: 10.1038/s41596-019-0232-9
85. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell (2018) 175:1972–1988.e16. doi: 10.1016/j.cell.2018.11.021
86. Yuki K, Cheng N, Nakano M, Kuo CJ. Organoid Models of Tumor Immunology. Trends Immunol (2020) 41:652–64. doi: 10.1016/j.it.2020.06.010
87. Guo W, Zhang C, Qiao T, Zhao J, Shi C. Strategies for the Construction of Mouse Models With Humanized Immune System and Evaluation of Tumor Immune Checkpoint Inhibitor Therapy. Front Oncol (2021) 11:673199. doi: 10.3389/fonc.2021.673199
Keywords: prostate cancer (PCa), organoid model, heterogeneity, pathogenesis, drug screening, individualized treatment
Citation: Zhou L, Zhang C, Zhang Y and Shi C (2021) Application of Organoid Models in Prostate Cancer Research. Front. Oncol. 11:736431. doi: 10.3389/fonc.2021.736431
Received: 05 July 2021; Accepted: 09 September 2021;
Published: 27 September 2021.
Edited by:
Andrew Goldstein, University of California, Los Angeles, United StatesReviewed by:
Larisa Nonn, University of Illinois at Chicago, United StatesCopyright © 2021 Zhou, Zhang, Zhang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changhong Shi, Y2hhbmdob25nQGZtbXUuZWR1LmNu; Yongbin Zhang, eW9uZ2JpbnpoYW5nQGd6dWNtLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.