
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 05 October 2021
Sec. Gynecological Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.734758
This article is part of the Research Topic Molecular Pathology and Computational Image Analyses in Gynecologic Malignancies View all 10 articles
 Jinfeng Xiong1†
Jinfeng Xiong1† Jing Cheng2†
Jing Cheng2† Hui Shen1
Hui Shen1 Ci Ren1
Ci Ren1 Liming Wang1
Liming Wang1 Chun Gao1
Chun Gao1 Tong Zhu1
Tong Zhu1 Xiaomin Li1
Xiaomin Li1 Wencheng Ding1
Wencheng Ding1 Da Zhu1*
Da Zhu1* Hui Wang1*
Hui Wang1*Human papillomavirus (HPV) integration in the human genome is suggested to be an important cause of cervical cancer. With the development of sequencing technologies, an increasing number of integration “hotspots” have been identified. However, this HPV integration information was derived from analysis of whole cervical cancer tissue, and we know very little about the integration in different cancer cell subgroups or individual cancer cells. This study optimized the preparation of probes and provided a dual-color fluorescence in situ hybridization (FISH) method to detect HPV integration sites in paraffin-embedded cervical cancer samples. We used both HPV probes and site-specific probes: 3p14 (FHIT), 8q24 (MYC), 13q22 (KLF5/KLF12), 3q28 (TP63), and 5p15 (TERT). We detected HPV signals in 75 of the 96 cases of cervical cancer; 62 cases showed punctate signals, and 13 cases showed diffuse punctate signals. We identified 3p14 as a high-frequency HPV integration site in 4 cervical cancer cases. HPV integration at 8p14 occurred in 2 cases of cervical cancer. In the same cervical cancer tissue of sample No.1321, two distinct subgroups of cells were observed based on the HPV probe but showed no difference in cell and nucleus morphology. Our study provides a new method to investigate the frequent HPV integration sites in cervical cancer and reports the heterogeneity within cervical cancer from the perspective of HPV integration.
Cervical cancer remains the leading cause of gynecological tumor-related mortality worldwide and the second most common malignancy in women, with 570,000 women diagnosed with cervical cancer and 311,000 dying from the disease each year (1, 2).
Most HPV infections are cleared by the immune system, but in some cases, the infection persists. Persistent infection with high-risk HPV leads to cervical intraepithelial neoplasia (CIN), which occasionally develops into cervical cancer (3). High-risk HPV types include 16, 18, 31, 33, 58 and other subtypes. HPV16 and 18 infection is common and can be detected in approximately 70% of cervical cancer cases. After infection, the virus can remain in the episome or integrate into the human genome, and the two patterns may coexist (4).
Recent studies have suggested that the integration of HPV in the human genome is an important cause of cervical cancer. Integration can preserve the upstream regulatory region (URR) of the virus. This HPV replication initiation region in the human genome is not stable but activates the cell’s DNA replication and other systems, which is an important “trigger” for cancer (5). The integration of HPV DNA into the human genome causes various genetic changes, such as oncogene amplification, inactivation of tumor suppressor genes, chromosomal rearrangement, and genomic instability. Integration can occur near key genes, leading to increased instability near integration sites in the human cell genome, resulting in local chromatin structural and functional changes and even the formation of new virus-human gene fusion transcripts in response to URRs of the virus (6, 7). In 2015, whole-genome sequencing and high-throughput virus capture sequencing methods identified up to 3,667 HPV integration breakpoints in cervical neoplasms (8). Frequent integration has been reported in genes associated with tumor progression, such as the oncogene MYC. It has been reported that loss of function of the tumor suppressor gene RAD51B after HPV DNA insertion affects DNA repair pathways and genomic instability in tumors (9).
Increasing research on HPV integration has led to the recognition of HPV integration status as a potential biomarker for the prediction of diagnosis, progression, and survival and even as a biomarker for cancer screening (10). With the development of high-throughput sequencing technologies (e.g., whole-genome sequencing, transcriptome sequencing, HIVID, etc.), an increasing number of integration “hotspots” have been identified (8, 9). However, this HPV integration information was derived from analyses of the whole cervical cancer tissue, and we know very little about the integration in different cancer cell subgroups or individual cancer cells. To visually display the more refined HPV integration at the single tumor cell level, this study optimized the preparation of probes and provided a dual-color FISH method to verify the HPV integration sites in paraffin-embedded cervical cancer samples. By preserving the original morphology and spatial structure of tumor tissue, we provide a new method to investigate the relationship between HPV integration and cervical carcinogenesis.
Formalin-fixed and paraffin wax-embedded cervical cancer samples were selected from the Department of Pathology, Tongji Hospital, Wuhan, China. All the samples were reviewed and confirmed independently by two pathologists, and cases with discrepancies were discussed until a consensus was reached. Samples with poor morphology or too many lymphocytes were excluded.
Clinical characteristics were obtained from patient charts. The TNM staging system, which is based on surgical and pathological reports, was used in this study to evaluate cervical cancer patients. We collected a total of 96 samples: 93 squamous cell carcinoma (SCC) samples, 3 adenocarcinoma (AD) samples. The study was approved by the hospital’s ethics committee.
Bacterial artificial chromosome (BAC) plasmids for 8q24 (RP11-1145O20), 3p14 (RP11-191B8), 3q28 (RP11-373I6), 5p15 (RP11-326E20) and 13q22 (RP11-179I20) were purchased from Life Technologies (California, America). The whole-genome plasmid of HPV types 16 and 18 was a gift from Haraud zur Hausen. The probes were labeled by standard nick translation with biotin- or digoxigenin-dUTPs. The biotin-labeled HPV probes and digoxigenin-labeled BAC probes were then coprecipitated with human Cot-1 DNA and salmon sperm DNA. Pelleted probes were then dissolved in a hybridization buffer composed of 50% formamide, 2×SSC, and 10% dextran sulfate. The detailed protocol and parameters of HPV-BAC dual-color probes are provided in the Supplementary Material.
Four-micrometer thick paraffin wax tissue sections were dewaxed, dehydrated with 100% ethanol for 3 min twice before air drying, and pretreated with 3% H2O2 for 10 min at room temperature. The slides were incubated in 1 M NaSCN for 20 min at 80°C, followed by digestion with 4 mg/ml pepsin (1:3000, Sigma) in 0.02 M HCl for 12 min at 37°C. The slides were rinsed 2 times in 2× SSC, postfixed in 4% formaldehyde for 10 min at room temperature, and dehydrated in an ascending ethanol series.
Probes and target DNA were denatured simultaneously for 7 min at 90°C before hybridization overnight at 37°C. After hybridization, the preparations were washed stringently in 50% formamide (3 × 5 min) and 2× SSC at 43°C.
The biotin- and digoxigenin-labeled probes were detected consecutively using the dual-color tyramide signal amplification (TSA) procedure. The biotin-labeled probe was detected by streptavidin-HRP (1:100, Perkin Elmer). Then, the first amplification reaction was carried out under a coverslip by applying 50 μl of Cy3-tyramide (1:50, Perkin Elmer) for 20 min at room temperature. Thereafter, the slides were soaked in blocking reagent (Perkin Elmer) for 15 min at room temperature to block the remaining peroxidase activity. Subsequently, the digoxigenin-labeled probe was detected by anti-digoxigenin-HRP (1:200, Roche), followed by TSA amplification using FITC-tyramide (1:50, Perkin Elmer). Finally, the slides were washed in 4× SSC, dehydrated in an ascending ethanol series, and mounted in Vectashield (Vector Laboratories).
Images were recorded with a fluorescence microscope (Olympus BX53) equipped with FITC, TRITC, and DAPI bandpass filters. A minimum of 100 nuclei in each sample were observed for HPV integration site evaluation. Among the cells containing HPV signals, the ratio of cells with colocalized HPV probe signals and BAC probe signals was calculated. Samples with a ratio greater than 60% were evaluated for HPV integration in a specific site.
HPV and BAC probe hybridization on HPV-positive cell lines (SiHa, HeLa and CaSki) was used as a control for HPV integration site detection. On sample tissue sections, BAC probes were also used as a control for effective hybridization.
HPV16/18 probes were hybridized in SiHa and CaSki cells, two smaller HPV signals were observed in SiHa cells with only two copies of HPV16 (Figure 1A), and 7 to 8 signals of different sizes were observed in dots and clumps in CaSki cells with 500 HPV16/18 copies (Figure 1D). We observed that with the increase in HPV integrated copy number in cells, the fluorescence intensity and area of the HPV signal also increased, but the relationship was not linear. The results of hybridization in SiHa cells prove that our HPV probe has sufficient sensitivity.
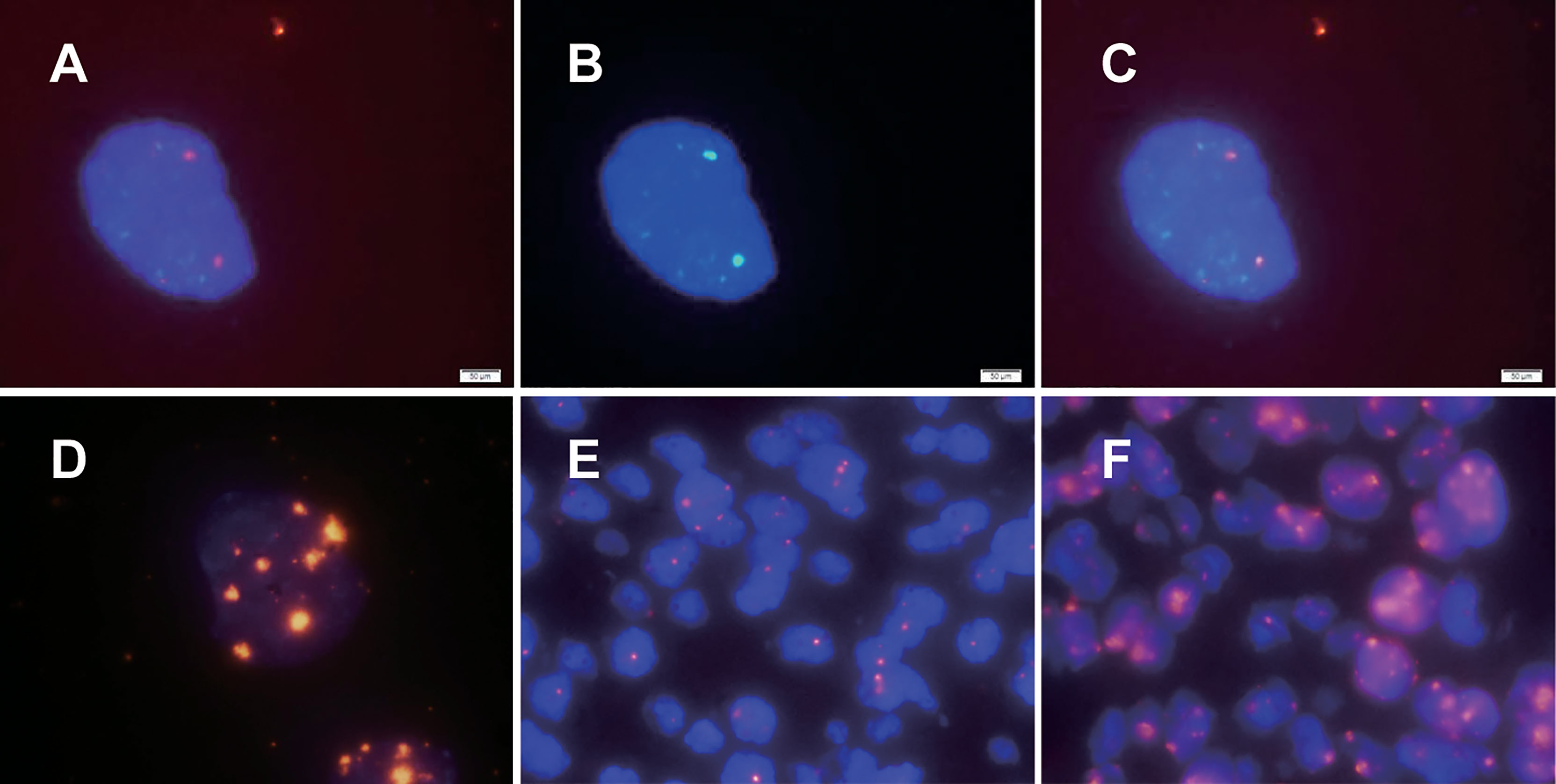
Figure 1 HPV signal patterns in cervical cancer cell lines and cervical cancer paraffin sections detected by dual-color FISH. The fluorescence intensity and area of the HPV signal were observed to increase with increasing HPV integrated copy number in cells, but the relationship was not linear. HPV signals in SiHa and CaSki cells are illustrated in (A, D), respectively. (B) The specific chromosomal site (Bacterial artificial chromosome, BAC) probe targeting the 13q22 in SiHa. As shown in (C), the HPV signal and the specific chromosomal site probe targeting the 13q22 (RP11-315L12) signal were colocalized. There were two main types of HPV signals in paraffin specimens of cervical cancer: punctate signals and diffuse-punctate signals. The punctate signals indicate integrated HPV (E), while diffuse signals indicate the presence of episomal HPV at the same time (F).
In previous studies, only HPV probes were used to study the presence and amount of HPV integration. To further study the integration sites of HPV, we explored the simultaneous hybridization of HPV probes and BAC plasmid probes into cervical cancer cells. Since previous studies suggested that two HPV16 copies of SiHa were integrated near the KLF5 gene at 13q22, our experiment showed that the HPV signal and the BAC plasmid probe targeting the 13q22 (RP11-315L12) signal were colocalized, as expected (Figures 1A–C). Chromosomal site probes can be used to indicate whether HPV is integrated at the site and can be used as a control for whether hybridization is successful. The results from SiHa cells confirmed the credibility of our dual-color probe.
We further explored the application of dual-color probes in paraffin sections of cervical cancer to prepare for possible clinical application. The in situ hybridization methods (including experimental procedures and various parameters) in cultured cells and paraffin sections are completely different. Through a review of the literature and experiments (11–13), we confirmed that both the sodium bisulfite and sodium thiocyanide methods are feasible. We found that the sodium bisulfite method was milder and preserved the nucleus well, but it often failed in aged paraffin sections. The sodium thiocyanide method is not easy to use, but the treatment is more complete, and it also has a higher success rate of hybridization for aged paraffin sections. To study the integration sites of HPV, we used the sodium thiocyanide method in this study. There were two main types of HPV signals in cervical cancer paraffin sections: punctate signals and diffuse-punctate signals. The punctate signal suggests integrated HPV (Figure 1E), and the diffuse signal suggests the presence of episomal HPV at the same time (Figure 1F).
We detected HPV signals in 75 cases of cervical cancer; 62 cases showed punctate signals, and 13 cases showed diffuse punctate signals (Table 1). We did not observe a simple diffuse signal in cervical cancer, which is consistent with the findings that HPV is generally integrated in cervical cancer in previous studies (12, 14).
Our criteria for determining HPV integration were as follows: when the percentage was greater than 60% (cells showing colocalized HPV signals and chromosomal locus probe signals/cells containing HPV signals), we concluded that HPV was integrated at this site (Figures 2A–C); otherwise, there was no integration at this site (Figures 2D–F). Based on a review of the literature on HPV integration sites and genomic alterations in cervical cancer (8, 15), we prepared probes targeting five regions of the human chromosome: 3p14 containing FHIT, 8q24 containing MYC, 13q22 containing KLF5/KLF12, 3q28 containing TP63, and 5p15 containing TERT. We detected 3p14 as a high-frequency HPV integration site in 4 cervical cancers. HPV integration at 8p14 occurred in 2 cases of cervical cancer. We found HPV integration in one patient each at 5p15 and 13q22. We did not detect HPV integration at 3q28, although 3q28 shows the greatest copy number amplification in cervical cancer and is associated with CIN progression (16) (Table 1).
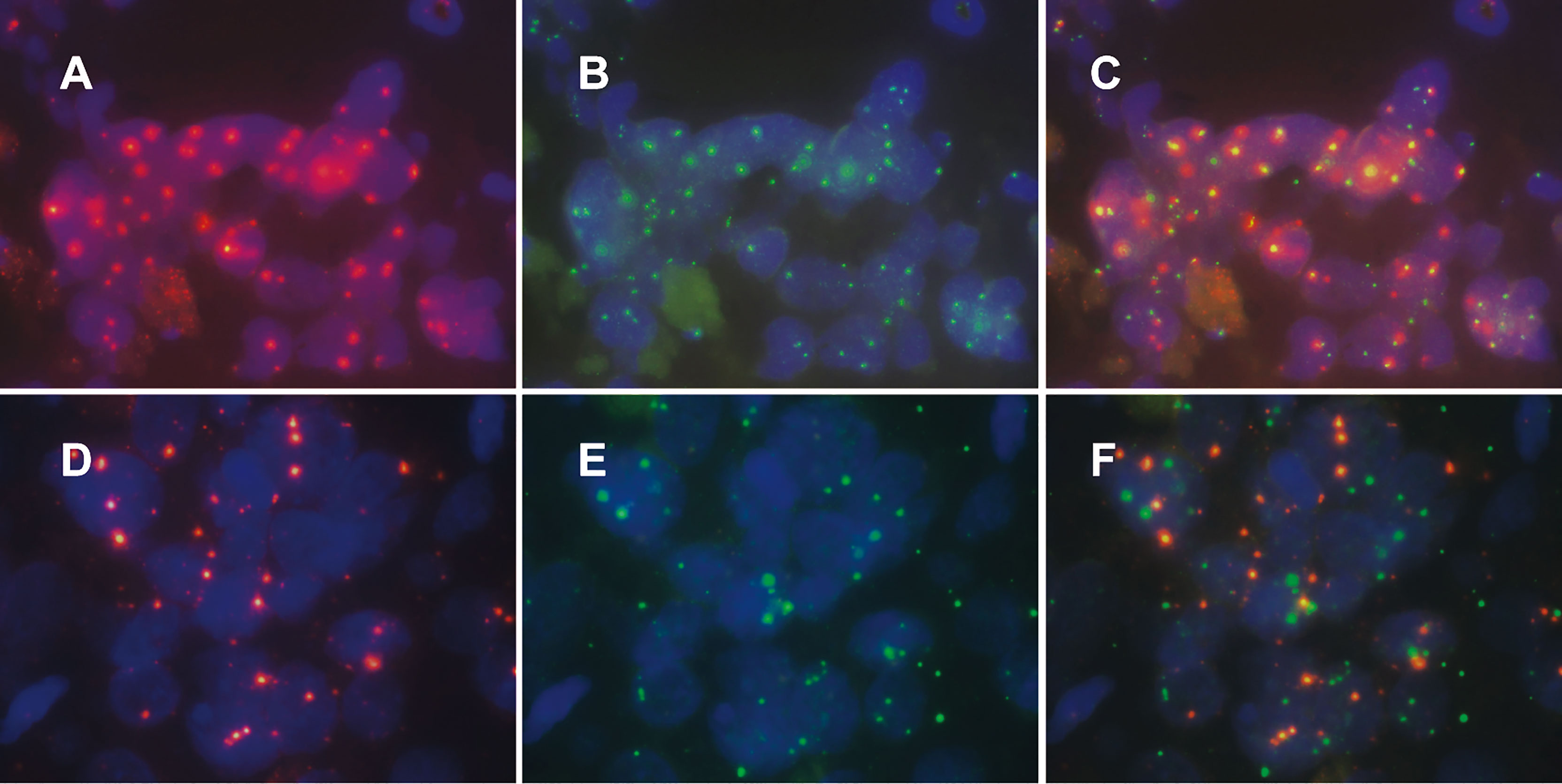
Figure 2 Patterns of human specific chromosomal site probe signals and HPV signals in cervical cancer tissue. Among the cells containing HPV signals, the ratio of cells with colocalized HPV probe signals and specific chromosomal site probe signals was calculated. Samples with a ratio greater than 60% were evaluated for HPV integration in a specific site (A–C); otherwise, we evaluate that HPV was not integrated at this site (D–F).
In contrast to PCR or high-throughput sequencing methods, the FISH method can detect HPV integration without destroying tissue morphology. Using an HPV probe as a guide, we observed major differences (both clonality and heterogeneity) in cancer cells within cervical cancer. For example, in sample No. 1321, two groups of cancer cells in the same cervical cancer tissue showed no difference in cell and nucleus morphology after HE staining and were pathologically identified as the same type of cells (Figures 3A–C). However, two distinct subgroups of cells were observed by the HPV probe. Every cell in the a group showed HPV integration, the HPV signal area was large, and the intensity was high, suggesting intense HPV integration. In the b group, some of the cells showed HPV integration, the HPV signal area was small, and the intensity was low, suggesting low integration (Figures 3D–F).
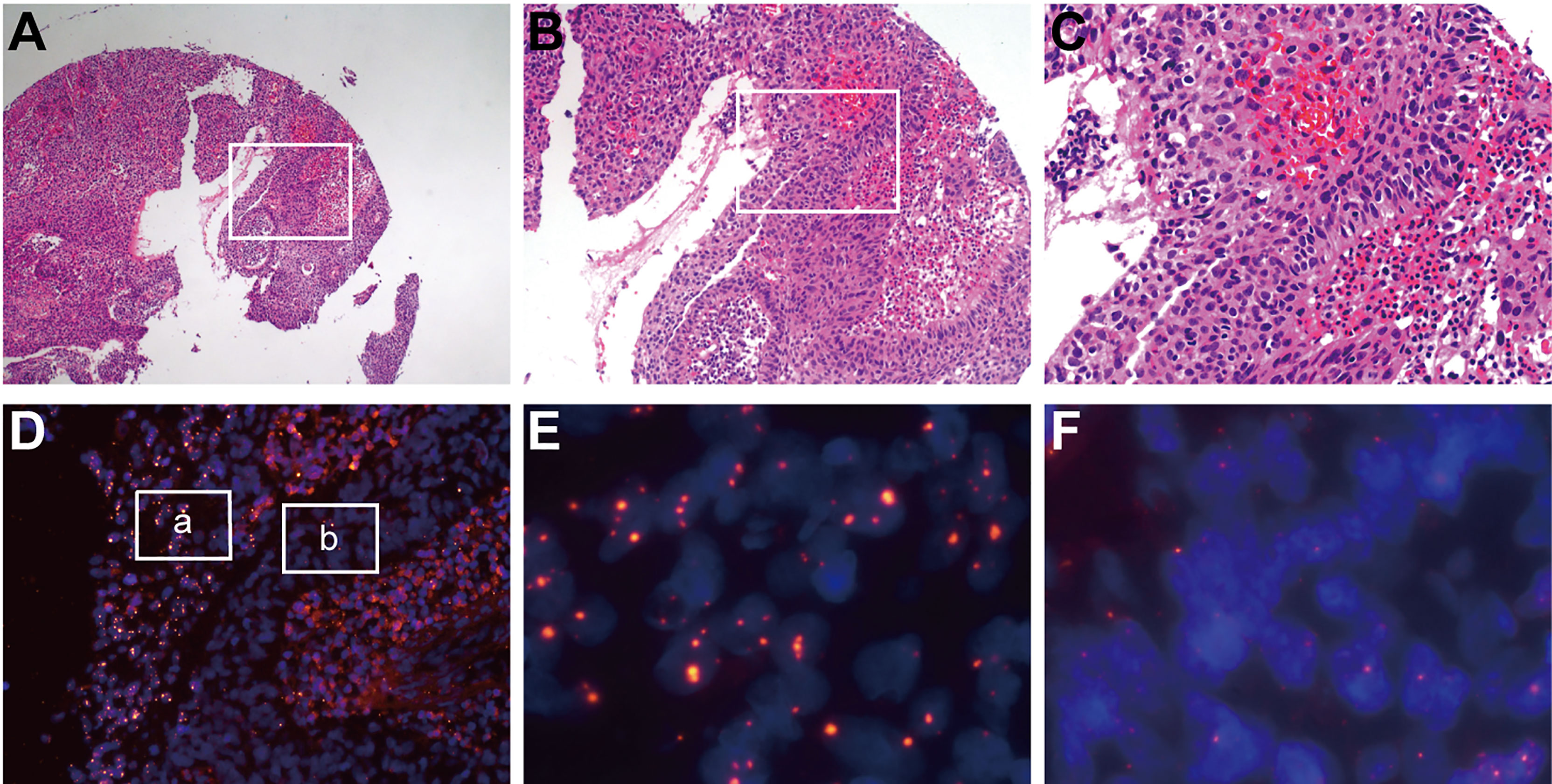
Figure 3 HPV signaling suggests clonality and heterogeneity in cervical cancer tissue. In sample No. 1321, two groups of cancer cells in the same cervical cancer tissue showed no difference in cell and nuclear morphology after HE staining and were pathologically identified as the same type of cells (A–C), which were distinguished significantly by HPV probes (D). Every cell in the “a” group (E) showed HPV integration, the HPV signal area was large, and the intensity was high, suggesting intense HPV integration. In the “b” group (F), some of the cells showed HPV integration, the HPV signal area was small, and the intensity was low, suggesting a low integration.
Currently, many studies have suggested that HPV integration plays an important role in the development of cervical cancer, but our knowledge of HPV integration is still insufficient. The reasons are as follows: First, HPV integration sites in the human genome are numerous and scattered, and different studies have reported different integration hotspots (6, 8, 17–20). Previous studies have detected high-frequency HPV integration sites and attempted to explain the relationship between HPV integration and the progression of cervical cancer by using high-frequency HPV integration sites. There is no conclusion yet regarding whether HPV integration is the driver of cervical cancer or the consequence of human cell genome instability during cervical cancer development (21). Second, the HPV sequence is not completely integrated into the human cell genome; only fragments are integrated (14). The integrated fragments are different in different samples and different at different sites in the same sample (8). Previous studies suggested that integrated HPV breaks in the E2 region and retains the viral oncogenes E6 and E7 (21), but the results of high-throughput sequencing suggest that this is not always the case (8). It is still unclear whether every integrated HPV fragment has a role. To elucidate the relationship between HPV integration and cervical cancer and the carcinogenic mechanism of HPV integration, we must be able to truly and accurately detect HPV integration.
Previous studies on HPV integration mainly include PCR, high-throughput sequencing, and in situ hybridization, each of which has advantages and disadvantages. PCR technology includes DIPS-PCR, APOT-PCR and others, which use specially designed primers to amplify HPV-human fusion fragments (22, 23). Considering the different specificities of primers, PCR can indeed detect some HPV-human fusion fragments, but some will be missed. High-throughput sequencing technologies, including WGS, RNA-seq, and HIVID, are more efficient and comprehensive in detecting HPV-human fusion fragments (8, 9). High-throughput sequencing technology provides an overall picture of HPV integration in a given tissue. There is a lack of information on differences in HPV integration in the tissue, and validation by other techniques (PCR, FISH) is required.
Previous studies on HPV integration using FISH techniques mainly investigated the association between HPV integration and CIN progression (12, 24). At present, there are few studies on the simultaneous detection of HPV integration and a specific chromosomal locus in cervical cancer paraffin specimens. The main obstacle is the preparation of FISH probes. The specific chromosomal site probe was nick translated from the BAC clones. The BAC was sufficiently long, and even if HPV integration led to the loss of some chromosomal fragments, the chromosomal site-specific probes still showed signals, such as at 3p14. Therefore, regardless of whether the DNA copy number at the integration site increases or decreases, we can effectively detect the HPV integration site with colocalization of two colors.
The advantage of paraffin specimens over primary cultured cancer cells is that paraffin specimens are easier to obtain and retrospectively analyzed, can preserve the morphology of cervical cancer tissue, and can be applied clinically. To simultaneously detect integrated HPV and a specific chromosomal site, our dual-color FISH technique needs to address the following two key points: probe nick translation and hybridization process. The formula of the dual-color probe, time of nick translation, and dose of human COT-1 DNA were optimized through repeated trials in this study. We provide the detailed protocol in the Supplementary Material to help more researchers use this method. In situ hybridization of paraffin sections is an improvement over previous research techniques (11, 12).
No HPV16/18 signal was detected in 21 cervical cancer specimens, the possible reasons for not detecting HPV16/18 signals in some samples are as follows: (1) There was no HPV integration in this sample. (2) As 80% of cervical cancers are infected by HPV16/18, we selected HPV16/18 probe. However, a few samples may not be HPV16/18 integrated but integrated by other subtypes. (3) Considering the intra-tumoral heterogeneity of cervical cancer tissues, some tumor cell clones in the same tissue have HPV integration but some clones do not. The tumor cell population at the paraffin sections we examined may happened to be with no HPV integration.
We used a threshold value for identifying an HPV integration site: in cells containing HPV signals, the ratio of cells with colocalized HPV signals and chromosomal site-specific signals is more than 60%, which is interpreted as HPV integration at a specific site. The application of FISH in fusion gene research could be used as a reference, since fusion gene and HPV integration have similarities. Tomlins used FISH to evaluate the TMPRSS2:ETV1 fusion gene in paraffin sections of prostate cancer and found that, fusion signals were observed in an average of 31% of the 100 cancer cells in the positive case (Supporting Online Material page 3) (25). Therefore, the cut-off value of 60% in this study is a relatively strict standard. Considering that we are the first to evaluate HPV integration using the ratio of cells with colocalization signals, the threshold of this ratio is worthy of further study.
Our selection of five candidate chromosomal sites is based on a review of HPV integration in cervical cancer research (8, 15). FHIT is a tumor suppressor at 3p14, which is significant copy number loss in cervical cancer, as loss of its activity results in replication stress and DNA damage. MYC is an oncogene, and it has been suggested that MYC activation is associated with HPV integration at 8q24. KLF5 is a transcriptional activator at 13q22 and may play a role in cell proliferation. Telomerase expression plays a role in cellular senescence, and deregulation of telomerase expression is found in somatic cells. 3q28 is significant copy number gain in cervical cancer, amplification and overexpression of TP63 at 3q28 is a biomarker of progression from CIN to cervical cancer. In our study, 3p14 and 8q24 were two high-frequency sites of HPV integration. Future studies may be able to reclassify cervical cancer at the molecular level from the perspective of HPV integration.
Moreover, the integrated HPV signal can be used as a guide for the discovery of cervical cancer cell subgroups with different integrated viral loads. These subgroups may reveal the origin, metastasis, and recurrence of cervical cancer. This finding suggests that previous PCR and high-throughput sequencing methods for detecting HPV integration sites in entire cervical cancer tissue cannot accurately reveal the characteristics of different subgroups of cancer cells. This limitation may be why previous studies often report many integration sites, but they cannot determine whether these integration sites are the initiating factors before carcinogenesis or the consequence of genome instability after carcinogenesis.
In clinical applications, high-throughput sequencing technologies such as WGS and HIVID can be combined with FISH to detect HPV integration after cervical cancer tissues are obtained by surgery. On the one hand, FISH can be used to verify sequencing results. Since the determination of HPV integration sites by sequencing is based on the similarity of base sequences, sequencing reads with low specificity or containing repetitive sequences will lead to sequence alignment errors, which can be verified by FISH. On the other hand, FISH can be used to detect whether the cancer cells containing a specific HPV integration site are the majority or the minority, which can be indirectly shown by the abundance of the human-virus fusion sequence in the sequencing results, while it can be intuitively observed by FISH. If cancer cells containing a specific HPV integration site are the majority in the tissue, HPV integration at this site may be the key factor promoting cervical cancer. If they are the minority, HPV integration at this site may be a consequence of genome instability in cancer cells.
In conclusion, our study provides a method for the detection of HPV integration sites in paraffin-embedded cervical cancer samples using dual-color FISH and reports the heterogeneity within cervical cancer from the perspective of HPV integration. Our study provides new methods and ideas for research on HPV integration in cervical carcinogenesis.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by Ethics Committee of Tongji Hospital. The patients/participants provided their written informed consent to participate in this study.
HW and DZ designed and supervised the research together with JX. DZ and HS provided technical support. DZ, JX, CR, LW, and WD performed the experiments. CG, TZ, and XL obtained specimens. JC collected clinical data. HW, DZ, JX, and HS provided analysis and interpretation of data. The manuscript was drafted by DZ and JX. All authors contributed to the article and approved the submitted version.
This work was supported by funds from National Natural Science Foundation of China (81830074, 81974412, 81772786, 81502253, 81902667 and 82002763).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.734758/full#supplementary-material.
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int J Cancer (2015) 136:E359–86. doi: 10.1002/ijc.29210
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
3. Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human Papillomavirus and Cervical Cancer. Lancet (2013) 382:889–99. doi: 10.1016/S0140-6736(13)60022-7
4. Oyervides-Munoz MA, Perez-Maya AA, Rodriguez-Gutierrez HF, Gomez-Macias GS, Fajardo-Ramirez OR, Trevino V, et al. Understanding the HPV Integration and its Progression to Cervical Cancer. Infect Genet Evol (2018) 61:134–44. doi: 10.1016/j.meegid.2018.03.003
5. Kadaja M, Isok-Paas H, Laos T, Ustav E, Ustav M. Mechanism of Genomic Instability in Cells Infected With the High-Risk Human Papillomaviruses. PloS Pathog (2009) 5:e1000397. doi: 10.1371/journal.ppat.1000397
6. Kraus I, Driesch C, Vinokurova S, Hovig E, Schneider A, von Knebel Doeberitz M, et al. The Majority of Viral-Cellular Fusion Transcripts in Cervical Carcinomas Cotranscribe Cellular Sequences of Known or Predicted Genes. Cancer Res (2008) 68:2514–22. doi: 10.1158/0008-5472.CAN-07-2776
7. Lu X, Lin Q, Lin M, Duan P, Ye L, Chen J, et al. Multiple-Integrations of HPV16 Genome and Altered Transcription of Viral Oncogenes and Cellular Genes Are Associated With the Development of Cervical Cancer. PloS One (2014) 9:e97588. doi: 10.1371/journal.pone.0097588
8. Hu Z, Zhu D, Wang W, Li W, Jia W, Zeng X, et al. Genome-Wide Profiling of HPV Integration in Cervical Cancer Identifies Clustered Genomic Hot Spots and a Potential Microhomology-Mediated Integration Mechanism. Nat Genet (2015) 47:158–63. doi: 10.1038/ng.3178
9. Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, et al. Landscape of Genomic Alterations in Cervical Carcinomas. Nature (2014) 506:371–5. doi: 10.1038/nature12881
10. Liu L, Ying C, Zhao Z, Sui L, Zhang X, Qian C, et al. Identification of Reliable Biomarkers of Human Papillomavirus 16 Methylation in Cervical Lesions Based on Integration Status Using High-Resolution Melting Analysis. Clin Epigenet (2018) 10:10. doi: 10.1186/s13148-018-0445-8
11. Hopman AH, Kamps MA, Smedts F, Speel EJ, Herrington CS, Ramaekers FC. HPV In Situ Hybridization: Impact of Different Protocols on the Detection of Integrated HPV. Int J Cancer (2005) 115:419–28. doi: 10.1002/ijc.20862
12. Hopman AHN, Smedts F, Dignef W, Ummelen M, Sonke G, Mravunac M, et al. Transition of High-Grade Cervical Intraepithelial Neoplasia to Micro-Invasive Carcinoma Is Characterized by Integration of HPV 16/18 and Numerical Chromosome Abnormalities. J Pathol (2004) 202:23–33. doi: 10.1002/path.1490
13. Algeciras-Schimnich A, Policht F, Sitailo S, Song M, Morrison L, Sokolova I. Evaluation of Quantity and Staining Pattern of Human Papillomavirus (HPV)-Infected Epithelial Cells in Thin-Layer Cervical Specimens Using Optimized HPV-CARD Assay. Cancer Cytopathol (2007) 111:330–8. doi: 10.1002/cncr.22946
14. Pett M, Coleman N. Integration of High-Risk Human Papillomavirus: A Key Event in Cervical Carcinogenesis? J Pathol (2007) 212:356–67. doi: 10.1002/path.2192
15. Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic Review of Genomic Integration Sites of Human Papillomavirus Genomes in Epithelial Dysplasia and Invasive Cancer of the Female Lower Genital Tract. Cancer Res (2004) 64:3878–84. doi: 10.1158/0008-5472.CAN-04-0009
16. Zhu D, Jiang XH, Jiang YH, Ding WC, Zhang CL, Shen H, et al. Amplification and Overexpression of TP63 and MYC as Biomarkers for Transition of Cervical Intraepithelial Neoplasia to Cervical Cancer. Int J Gynecol Cancer (2014) 24:643–8. doi: 10.1097/IGC.0000000000000122
17. Melsheimer P, Vinokurova S, Wentzensen N, Bastert G, von Knebel Doeberitz M. DNA Aneuploidy and Integration of Human Papillomavirus Type 16 E6/E7 Oncogenes in Intraepithelial Neoplasia and Invasive Squamous Cell Carcinoma of the Cervix Uteri. Clin Cancer Res (2004) 10:3059–63. doi: 10.1158/1078-0432.CCR-03-0565
18. Schmitz M, Driesch C, Beer-Grondke K, Jansen L, Runnebaum IB, Durst M. Loss of Gene Function as a Consequence of Human Papillomavirus DNA Integration. Int J Cancer (2012) 131(5):E593–602. doi: 10.1002/ijc.27433
19. Schmitz M, Driesch C, Jansen L, Runnebaum IB, Durst M. Non-Random Integration of the HPV Genome in Cervical Cancer. PloS One (2012) 7:e39632. doi: 10.1371/journal.pone.0039632
20. Das P, Thomas A, Mahantshetty U, Shrivastava SK, Deodhar K, Mulherkar R. HPV Genotyping and Site of Viral Integration in Cervical Cancers in Indian Women. PloS One (2012) 7:e41012. doi: 10.1371/journal.pone.0041012
21. Woodman CB, Collins SI, Young LS. The Natural History of Cervical HPV Infection: Unresolved Issues. Nat Rev Cancer (2007) 7:11–22. doi: 10.1038/nrc2050
22. Luft F, Klaes R, Nees M, Durst M, Heilmann V, Melsheimer P, et al. Detection of Integrated Papillomavirus Sequences by Ligation-Mediated PCR (DIPS-PCR) and Molecular Characterization in Cervical Cancer Cells. Int J Cancer (2001) 92:9–17. doi: 10.1002/1097-0215(200102)9999:9999<::AID-IJC1144>3.0.CO;2-L
23. Klaes R, Woerner SM, Ridder R, Wentzensen N, Duerst M, Schneider A, et al. Detection of High-Risk Cervical Intraepithelial Neoplasia and Cervical Cancer by Amplification of Transcripts Derived From Integrated Papillomavirus Oncogenes. Cancer Res (1999) 59:6132–6.
24. Van Tine BA, Kappes JC, Banerjee NS, Knops J, Lai L, Steenbergen RD, et al. Clonal Selection for Transcriptionally Active Viral Oncogenes During Progression to Cancer. J Virol (2004) 78:11172–86. doi: 10.1128/JVI.78.20.11172-11186.2004
Keywords: human papillomavirus, HPV integration, FISH, cervical cancer, site-specific
Citation: Xiong J, Cheng J, Shen H, Ren C, Wang L, Gao C, Zhu T, Li X, Ding W, Zhu D and Wang H (2021) Detection of HPV and Human Chromosome Sites by Dual-Color Fluorescence In Situ Hybridization Reveals Recurrent HPV Integration Sites and Heterogeneity in Cervical Cancer. Front. Oncol. 11:734758. doi: 10.3389/fonc.2021.734758
Received: 01 July 2021; Accepted: 08 September 2021;
Published: 05 October 2021.
Edited by:
Umberto Malapelle, University of Naples Federico II, ItalyReviewed by:
Congrong Liu, Peking University Health Science Center, ChinaCopyright © 2021 Xiong, Cheng, Shen, Ren, Wang, Gao, Zhu, Li, Ding, Zhu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Da Zhu, MTUzOTM4ODAxQHFxLmNvbQ==; Hui Wang, aHVpdDcxQHNvaHUuY29t
†These authors contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.